Abstract
Motivation
Single-cell/nuclei RNA-sequencing (scRNA-seq) technologies can simultaneously quantify gene expression in thousands of cells across the genome. However, the majority of the noncoding RNAs, such as microRNAs (miRNAs), cannot currently be profiled at the same scale. MiRNAs are a class of small noncoding RNAs and play an important role in gene regulation. MiRNAs originate from the processing of primary transcripts, known as primary-microRNAs (pri-miRNAs). The pri-miRNA transcripts, independent of their cognate miRNAs, can also function as long noncoding RNAs, code for micropeptides or even interact with DNA, acting like enhancers. Therefore, it is apparent that the significance of scRNA-seq pri-miRNA profiling expands beyond using pri-miRNA as proxies of mature miRNAs. However, there are no computational methods that allow profiling and quantification of pri-miRNAs at the single-cell-type resolution.
Results
We have developed a simple yet effective computational framework to profile pri-MiRNAs from single-cell RNA-sequencing datasets (PPMS). Based on user input, PPMS can profile pri-miRNAs at cell-type resolution. PPMS can be applied to both newly produced and publicly available datasets obtained via single cell or single-nuclei RNA-seq. It allows users to (i) investigate the distribution of pri-miRNAs across cell types and cell states and (ii) establish a relationship between the number of cells/reads sequenced and the detection of pri-miRNAs. Here, to demonstrate its efficacy, we have applied PPMS to publicly available scRNA-seq data generated from (i) individual chambers (ventricles and atria) of the human heart, (ii) human pluripotent stem cells during their differentiation into cardiomyocytes (the heart beating cells) and (iii) hiPSCs-derived cardiomyocytes infected with severe acute respiratory syndrome coronavirus 2.
Introduction
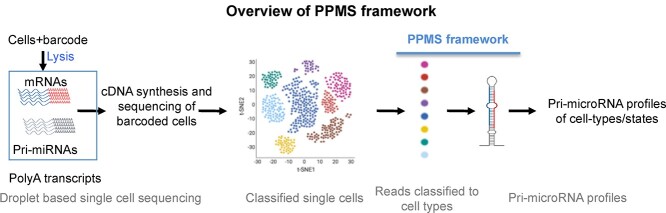
MicroRNAs (miRNAs) are a class of short noncoding RNAs that repress the translation of target genes via semi complimentary base-pairing [1]. Cells use miRNAs to control the production of proteins and regulate critical biological pathways in both parental and non-parental cells alike to mediate cell-to-cell communications [1, 2]. Canonically, the biogenesis of miRNAs begins with the transcription of primary miRNA (pri-miRNA) from the intronic regions of protein-coding genes. However, a small minority of miRNAs originate from the exons of protein-coding genes and long noncoding RNAs [3]. While still in the nucleus, the pri-miRNA transcript (>1000 nucleotides) is processed by the Microprocessor complex, containing DROSHA, DGCR8 and other proteins, into precursor miRNAs (pre-miRNAs), which can be exported to the cytoplasm and further processed by DICER1 into mature single-stranded (−3p and –5p) mature miRNAs (17–24 nucleotides; [4]). Mature miRNAs can silence messenger RNAs (mRNAs) via base-pairing with semi complementary sequences [5]. miRNAs also play an important role in cell–cell communications and can be shuttled from their parent cells to recipient cells, typically but not exclusively via extracellular vesicles [6–8]. Both miRNA biogenesis and cell-to-cell communication are altered by intra- and extracellular environmental perturbations, contributing to disease development [9]. Beyond their capacity to act as miRNA precursors, pri-miRNAs can (i) act as long noncoding RNAs [10], (ii) encode small peptides [11] and (iii) have enhancer-like DNA interactions [10]. In bulk RNA-sequencing datasets, pri-miRNAs are less abundant than mRNAs and are often filtered out from the data analysis due to the lack of necessary deep-sequencing coverage [12]. We conceptualize that dissecting pri-miRNA expression across different cell types and states would expand the mechanistic understanding of diseases, supporting the development of both cell-type-personalized RNA therapeutics and clinical biomarkers. We have developed a computational framework to profile pri-miRNAs using single-cell/nuclei RNA-sequencing (scRNA-seq) datasets (PPMS, Figure 1). PPMS enables the user to detect, quantify and analyze pri-miRNAs at a single-cell-type resolution.
Figure 1.
Panel (A) provides an overview of the framework. Prior to the sequencing, the RNAs in the single cells are barcoded to represent individual cells. Single-cell transcriptomic data can be used to classify cells into respective cell types. Next, we use this information to collate cells to represent cell types and then map reads to the miRNA transcripts in the genome to profile pri-miRNAs for respective cell types.
Approach
ScRNA-seq technologies have revolutionized molecular studies of heterogeneous tissues by providing tools to identify biological pathways and new cell types that underpin key developmental processes [13]. The application of scRNA-seq technologies has evolved from qualitative to quantitative (differential expression) analyses, broadening their scope to facilitate investigation of various scientific questions. Gene expression profiling of tissues at single-cell (or nuclei) resolution requires isolation of single-cells/nuclei, nucleic acid amplification and sequencing of individual cells/nuclei. For example, fluorescence-activated cell sorting can sort single cells/nuclei and isolate them in microtiter plates to profile hundreds of cells. These numbers are however insufficient to evaluate cellular heterogeneity and reliable gene expression profiles of respective cell types. These limitations are therefore solved by microfluidic droplet technologies that can amplify single cells/nuclei in a droplet and capture thousands of cells. Therefore, we demonstrated the efficacy of the PPMS framework on scRNA-seq data obtained from microfluidic droplet technologies.
To develop PPMS, we took advantage of the following: (i) the majority of pri-miRNAs are polyadenylated [3, 14], (ii) high-throughput droplet-based scRNA-seq techniques profile the −3′ end of RNA transcripts, which carry the poly-A tail [15] and (iii) in a scRNA-seq dataset, transcriptional signatures help to classify an individual cell/nucleus into their respective cell types or cellular states [16].
Even if scRNA-seq data are sparse and expression of pri-miRNAs is at low levels [17], our approach attained sufficient read coverage to profile pri-miRNAs by aggregating classified cells into the respective cell types to reduce sparsity. As a prerequisite, PPMS requires the user to map and process scRNA-seq datasets to annotate cells into respective cell types or cellular states. PPMS extracts pri-miRNAs in two steps: (i) it classifies reads to respective cell types or states, and (ii) cell types or states specific reads are then assembled to pri-miRNA transcripts using StringTie [18].
To show the practical usefulness of our approach, here, we have applied PPMS to scRNA-seq datasets (see methods section) generated by high-throughput droplet-microfluidics based methods. More generally, PPMS can be applied to any single-cell/nuclei/cell-type RNA-sequencing method, for example, the low-throughput SMART-seq2 method, capable of profiling full-length RNA-sequencing at single-cell resolution [19].
Methods
PPMS framework
To extract pri-miRNAs, PPMS requires to input: (i) mapped BAM (Binary Alignment Map, i.e. the comprehensive raw data of sequencing) as a standard alignment output file format and (ii) cell-type annotation represented by individual cell barcodes. As an output, PPMS provides: (i) cell-type classified BAM files, (ii) normalized (counts per million, CPM) and raw read counts per cell type, (iii) normalized (CPM) and raw pri-miRNAs transcript counts per cell type estimated via UMI (unique molecular identifiers) and (iv) differentially expressed pri-miRNAs (Supplementary Figures 1 and 2, see Supplementary Data available online). For further details, see Supplementary Data available online. The output can be easily exported as text files to enable additional analyses and data mining of single-cell level pri-miRNAs.
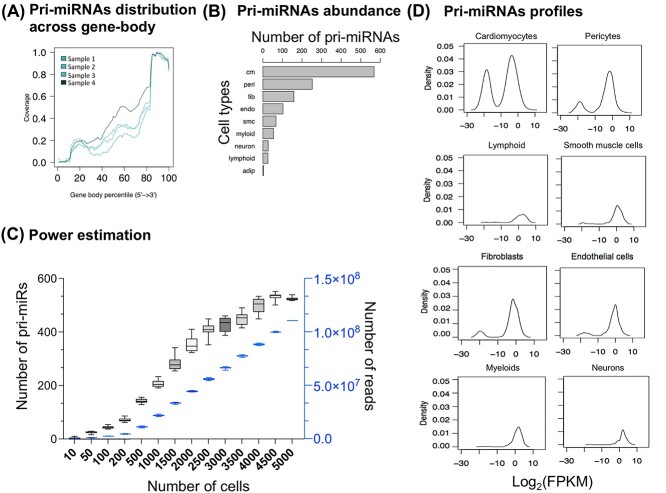
Figure 2.
Panels (A–D) summarize various features of the profiled pri-miRNAs (see text for details).
Implementation of PPMS framework
PPMS framework has been implemented in R and is available on GitHub (https://github.com/SrivastavaLab-ICL/PPMS). PPMS can run in two stand-alone versions: (i) graphical version (Supplementary Figures 1 and 2, see Supplementary Data available online), implemented in R using R shiny package, and run as a GUI in a web browser and (ii) Command-line version implemented using Python and shell scripting (Supplementary Figures 1 and 2, see Supplementary Data available online).
(a) Graphical version: the web browser interface option is for users who are not well-versed in programming skills and want to analyze single-cell RNA-seq datasets (Supplementary Figure 1, see Supplementary Data available online). The graphical version of PPMS can profile pri-miRNAs either in a single sample mode (Supplementary Figure 2A, see Supplementary Data available online) or multi-sample mode (Supplementary Figure 2B, see Supplementary Data available online). Both modes require the user to input respective BAM (binary alignment map) and cell annotation files. In single sample mode, the user can select the experiment type, i.e. single-cell or single-cell-type and simply press the ‘Run’ button. On the other hand, the multi-sample run mode would require cell-type input to generate a normalized/raw read/UMI count matrix for all the input sample files. The PPMS framework will run in the background to generate cell-type classified BAM files and normalized (counts per million, CPM)/raw read/UMI counts per pri-miRNAs. Once the run is complete, the user can save the results on the local computer for further analysis. Next, the user can employ our interface to perform differential gene expression analysis or use it later with their favorite tools.
(b) Command-line version: in addition to the graphical user interface version, we have also implemented a command-line version for users who are more versed in computer programming and would like to embed PPMS in their analysis workflow. The command-line version is essentially a wrapper shell script, PPMS.sh. PPMS.sh shell script requires (1) full path of the BAM file, (2) cell annotation file, (3) number of processors to use and (4) out file prefix. An example of the command:
sh PPMS.sh <BAM file> <annotation file> <number of processors> <prefix for the out files> All the results will be saved in the same directory, and PPMS will delete all its generated temporary files.
Application of PPMS on publicly available scRNA-seq datasets
We implemented PPMS onto the following scRNA-seq datasets: (i) Atlas of cells in the adult human heart [20], (ii) subsequent stages of human cardiomyocyte maturation from differentiating pluripotent stem cells profiled at a single-cell resolution (array express accession number: E-MTAB-6268) [21] and (iii) hiPSCs-derived cardiomyocytes infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV2) virus (gene expression omnibus accession number: GSE156754; [22]). See Supplementary Data available online for the methods used to analyze scRNA-seq and bulk RNA-seq (Supplementary text methods section).
Application of PPMS to the cells in the adult human heart
We aimed to demonstrate that PPMS can identify and characterize pri-miRNAs at single-cell type level and detect their differential expression. We first applied PPMS on the publicly available Human Cell Atlas (HCA) scRNA-seq dataset (array express accession number: ERP123138) generated by sequencing single-nucleus transcriptomes from the four chambers (left and right atria and left and right ventricles) of the human heart [20]. PPMS identified 329, 278, 477 and 444 pri-miRNAs in left and right atria and left and right ventricles, respectively, with pri-miRNA expression ranging from 1 to 8.5 counts (CPM).
Characteristics of pri-miRNA expression and power considerations
We first explored the pri-miRNA profiles obtained from the human heart to check if the pri-miRNA signal originates from an independent transcript. We profiled the pri-miRNA signal across the gene body (Figure 2A) and observed a higher signal from the 3′ end of the transcript. This observation is consistent with the single-cell workflow, which profiles signals from the transcript’s polyadenylated (3′) end. We also investigated how pri-miRNA expression varied across cardiac cell types and observed that the number of pri-miRNAs was directly proportional to the number of cells profiled (Figure 2B). This observation led us to explore the relationship between the number of cells and the pri-miRNAs detected. We down-sampled (10 bootstrap samples) the most abundant cell-type (cardiomyocytes) and observed that to obtain a stable pri-miRNA profile, we need data from >3000 cells sequenced at an average coverage of 66 million reads (Figure 2C, Supplementary text methods section). Next, we plotted log2(FPKM) and observed a bimodal distribution that shifts towards the right (higher expression) as low abundant cells such as myeloid and neuronal cells are profiled (Figure 2D). Noteworthy, we report a significant concordance of pri-miRNAs expression between (i) single-cells and single-nuclei datasets from the same hearts (Supplementary Figure 3A, see Supplementary Data available online) and between (ii) bulk and single-nuclei datasets from the left ventricles (Supplementary Figure 3B, see Supplementary Data available online).
Differential transcription of pri-miRNAs across the human heart chambers
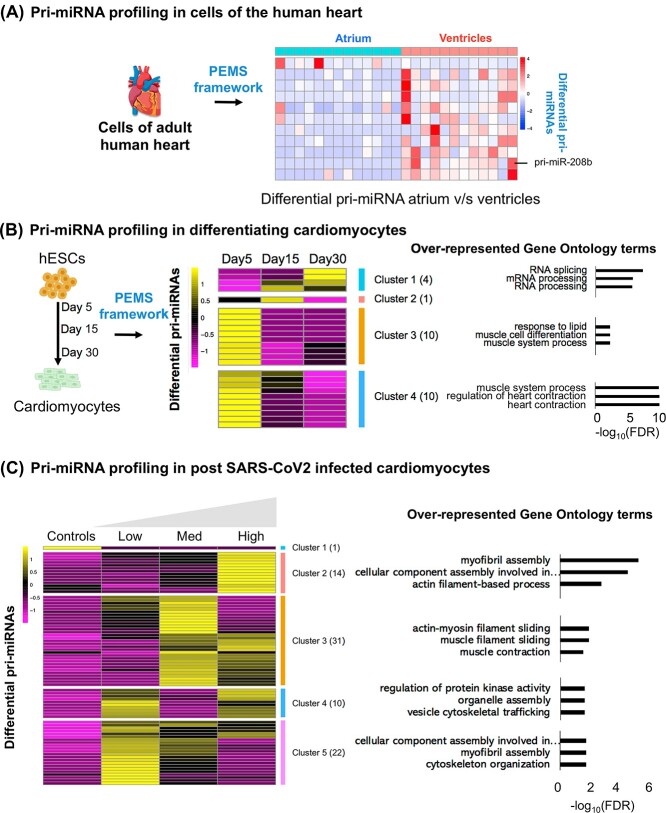
Differential expression (DE) analysis of pri-miRNAs between the cardiomyocytes obtained from the ventricles and atria was possible. It resulted in 11 differential pri-miRNAs (false discovery rate (FDR) < 0.05, Supplementary Table 1, Supplementary Figure 4A, B, see Supplementary Data available online and Figure 3A). Consistent with a previous report where bulk RNA-seq analyzed miRNA expression, PPMS identified higher expression of pri-miR-208b in the cardiomyocytes resident in the ventricle versus atria [23].
Figure 3.
(A) Heatmap summarizes pri-miRNAs differentially expressed between atria and ventricles in the human heart. (B) This panel summarizes the pri-miRNAs that are differentially expressed across various stages of differentiating cardiomyocytes. (C) The heatmap summarizes differential pri-miRNA in post-SARS-CoV2 infected cardiomyocytes. The right panel shows overrepresented gene ontology terms of miRNA target genes. The functional enrichment analysis was performed using WebGestalt R package [31].
To evaluate the cardiac relevance of the pri-miRNAs emerging from the analyses of the human heart, we predicted the targets of differentially expressed pri-miRNAs (see methods section of supplementary text for detail on target prediction). The targets of miRNAs are overrepresented for gene ontology terms related to ion transport, regulation of heart development and cell adhesion (Supplementary Figure 4C and Supplementary Tables 2 and 3, see Supplementary Data available online).
Expression of pri-miRNAs during cardiomyocyte differentiation
As an additional application of PPMS, we analyzed scRNA-seq data from human induced pluripotent stem cells (iPSC)-derived cardiomyocytes (iPSC-CM) at different cellular states of differentiation (Days 5, 15 and 30; [21]). See Supplementary Data available online for details on the processing and identification of cardiomyocytes. PPMS identified 293, 193 and 228 pri-miRNAs on Days 5, 15 and 30 of cardiomyocyte differentiation, respectively, with pri-miRNA expression ranging from 1 to 81 CPM. DE analysis in cardiomyocytes on Days 15 and 30 with respect to cardiomyocytes on Day 5 resulted in 12 differential pri-miRNAs. The DE pri-miRNAs were relevant to cardiomyocyte differentiation (FDR < 0.05, Supplementary Table 4, Figure 3B, Supplementary Figure 5A and B, see Supplementary Data available online). For example, miR-133a is highly expressed in cardiomyocytes, where it regulates hypertrophic response, whereas the miRNAs in the Let-7 family are essential for maturation and adult-like metabolism in stem cell-derived cardiomyocytes [24, 25]. The targets of DE pri-miRNAs were overrepresented for pathways linked to cardiovascular terms such as cardiac arrhythmias and hypertrophy (Supplementary Figure 5A and B, see Supplementary Data available online). To explore this further, we observed that the targets of DE pri-miRNAs demonstrated different expression patterns across the three-time points (Figure 3B). We clustered the DE pri-miRNAs in four broad groups (Figure 3B). The target genes of miRNAs in all other clusters are overrepresented for terms linked to cardiac biology (Figure 3B). For example, the target genes of miRNAs in clusters 3 and 4 were overrepresented for muscle cell differentiation and heart contraction, respectively (Figure 3B, Supplementary Tables 5 and 6, see Supplementary Data available online).
PPMS also provided an opportunity to compare pri-miRNAs during cardiomyocyte differentiation with fetal and adult human hearts (Supplementary Figure 5C, see Supplementary Data available online for detailed cell-type annotation of fetal hearts and analysis). We finally observed that pri-miRNAs of iPSC-CM were highly correlated (Spearman’s correlation > 0.6) with adult and fetal human hearts (Supplementary Figure 5C and D, see Supplementary Data available online).
Expression of pri-miRNAs in hiPSCs-derived cardiomyocytes infected with SARS-CoV2 virus
To demonstrate the efficacy of the PPMS framework in a disease setting, we explored the impact of the SARS-CoV2 virus on cultured cardiomyocytes. Heart injury is common in patients hospitalized for coronavirus disease of 2019 (COVID-19), but the underlining role played by microRNAs is primarily unknown [26]. To this aim, we analyzed human iPSC-CM infected with three concentrations of SARS-CoV2 [22]. By mapping sequencing data to the SARS-CoV2 genome, the virus was detected in the SARS-CoV2-infected cardiomyocytes only (Supplementary Figure 6, see Supplementary Data available online). PPMS identified 151 pri-miRNAs differentially expressed between infected versus noninfected cells (Figure 3C, Supplementary Table 7 and text for details). Similar to the iPSC-CMs maturation dataset, the DE pri-miRNAs were clustered into five groups. The targets of miRNAs in respective groups were overrepresented for gene ontology terms linked to cardiac function, such as myofibril assembly and muscle contraction (FDR < 0.05, Figure 3C, Supplementary Tables 8 and 9, see Supplementary Data available online). Next, we checked if the DE pri-miRNAs could act as cardiometabolic biomarkers that rise with COVID-19 severity. Therefore, we overlapped our DE pri-miRNAs with circulating miRNAs published in Gutmann et al., 2022 [27]. Interestingly, four of our DE pri-miRNAs (MIR199B, MIR1290, MIR125A and MIR32) overlap with higher levels of miRNAs in the plasma of patients infected with SARS-CoV2 virus.
Limitations
In addition to intrinsic data limitations such as the low sequencing coverage, PPMS cannot profile pri-miRNAs that are not polyadenylated (including 16 long noncoding RNAs hosting miRNAs) [3]. The PPMS framework can detect pri-miRNAs forming single transcriptional units (Supplementary Figure 7A and B, see Supplementary Data available online). However, in the cases where individual pri-miRNAs are co-transcribed from a cluster like the miR-17-92 cluster [3], PPMS might not be able to differentiate the individual pri-miRNAs (Supplementary Figure 7C, see Supplementary Data available online).
Key Points
We developed a framework to profile pri-miRNAs in single cells to enable researchers to explore the role of microRNA at single-cell types/cell state levels.
Our framework, PPMS, has a user-friendly graphical interface for researchers who are not well versed in programming.
PPMS provides users with the power and experimental design considerations by establishing a relationship between the number of cells/reads sequenced and the detection of pri-miRNAs.
The application of PPMS to the human hearts profiled at single-cell resolution led to the identification of the cardiac chamber and cell-type-specific regulation of microRNAs.
Profiling of pri-miRNAs identified dysregulation of cardiac protective microRNAs in cardiomyocytes infected with the SARS-CoV2 virus.
Supplementary Material
Acknowledgments
We are grateful to Dr Michael Lee and Dr Michela Noseda (NHLI, Imperial College London, UK) for their support in procuring the single-cell dataset for the adult human hearts from the Human Cell Atlas. We thank Mr Jeremy Hill for the English proofreading of the manuscript.
Author Biographies
Jiahui Ji is a PhD student at National Heart and Lung Institute (NHLI), Imperial College London. Her research mainly focuses on single-cell multi-omics analysis, cell-cell communication, diabetes and vascular diseases, as well as non-coding RNAs.
Maryam Anwar is a Bioinformatician at National Heart and Lung Institute at Imperial College London. Her research focuses on developing and implementing data analyses and integration pipelines for various omics data.
Enrico Petretto is Director of the Centre for Computational Biology (CCB) and leads the Systems Genetics Group at Duke-NUS Medical School in Singapore, which was established as a landmark collaboration between two world-ranking institutions of higher education: Duke University (USA) and the National University of Singapore (NUS). He is also Theme Leader for Bioinformatics and Systems Genetics at the Institute of Molecular and Translational Cardiology (IMTC) for the study of Brugada Syndrome and Sudden Death, Policlinico San Donato University and Research Hospital in Milan (Italy), Adjunct Professor at the Institute of Big Data and Artificial Intelligence, China Pharmaceutical University (CPU) in Nanjing (China) and Senior Lecturer at Imperial College London. His research focuses on Systems Genetics to study the genetic regulation of cellular pathways and gene networks that drive human disease. His lab uses Systems Biology and integrative genomics approaches to identify new drug targets for various complex disease, including cardiometabolic (e.g., cardiomyopathies, Brugada Syndrome) and fibrotic disease, diabetic nephropathy, cancer and important brain disorders such as epilepsy and Alzheimer’s disease.
Costanza Emanueli is a chair in Cardiovascular Science at NHLI, Imperial College London. Her research focus on Cardiovascular Disease and Regenerative Medicine, and particularly on ischemic disease, diabetes, noncoding RNAs, and cell-to-cell communication.
Prashant Kumar Srivastava is a lecturer in cardiovascular bioinformatics and medical statistics at National Heart and Lung Institute, Imperial College London. His primary interest is in using computational approaches to the analysis of complex genetic/epigenetic modifications to model their impact on gene expression and develop strategies for therapeutic interventions in complex cardiac and brain disorders.
Contributor Information
Jiahui Ji, National Heart and Lung Institute, Imperial College London, UK.
Maryam Anwar, National Heart and Lung Institute, Imperial College London, UK.
Enrico Petretto, London Institute of Medical Sciences, MRC, UK; Duke-NUS Medical School, Singapore; Institute of Big Data and Artificial Intelligence, China Pharmaceutical University (CPU), 211198 Nanjing, China.
Costanza Emanueli, National Heart and Lung Institute, Imperial College London, UK.
Prashant Kumar Srivastava, National Heart and Lung Institute, Imperial College London, UK.
Funding
This study was funded through the following grants: (i) British Heart Foundation Programme Grant (RG/15/5/31446) and Personal Chair Awards (CH/15/1/31199) to CE. (ii) The European Union’s Horizon 2020 research and innovation programme under grant agreement no. 101016072 was awarded to CE, EP and PKS. (iii) Diabetes UK early-career grants (20/0006187) were awarded to PKS and (iv) The Royal Society (RGS\R2\202413) Research Grant awarded to PKS.
References
- 1. Gomes CPDC, Schroen B, Kuster GM, et al. . Regulatory RNAs in heart failure. Circulation 2020;141:313–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boon RA, Vickers KC. Intercellular transport of microRNAs. Arterioscler Thromb Vasc Biol 2013;33:186–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dhir A, Dhir S, Proudfoot NJ, et al. . Microprocessor mediates transcriptional termination in long noncoding microRNA genes. Nat Struct Mol Biol 2015;22:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lee Y, Ahn C, Han J, et al. . The nuclear RNase III Drosha initiates microRNA processing. Nature 2003;425:415–9. [DOI] [PubMed] [Google Scholar]
- 5. Ambros V. The functions of animal microRNAs. Nature 2004 431:70062004;431:350–5. [DOI] [PubMed] [Google Scholar]
- 6. Turchinovich A, Samatov TR, Tonevitsky AG, et al. . Circulating miRNAs: cell-cell communication function? Front Genet 2013;4:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kang W, Fromm B, Houben AJ, et al. . MapToCleave: high-throughput profiling of microRNA biogenesis in living cells. Cell Rep 2021;37(7):37. [DOI] [PubMed] [Google Scholar]
- 8. Roden C, Gaillard J, Kanoria S, et al. . Novel determinants of mammalian primary microRNA processing revealed by systematic evaluation of hairpin-containing transcripts and human genetic variation. Genome Res 2017;27:374–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schwarzenbach H, Gahan PB. MicroRNA shuttle from cell-to-cell by exosomes and its impact in cancer. Non-Coding RNA 2019;5(1):28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. He D, Wu D, Muller S, et al. . miRNA-independent function of long noncoding pri-miRNA loci. Proc Natl Acad Sci USA 2021;118:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Prel A, Dozier C, Combier JP, et al. . Evidence that regulation of pri-mirna/mirna expression is not a general rule of mipeps function in humans. Int J Mol Sci 2021;22(7):3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chang TC, Pertea M, Lee S, et al. . Genome-wide annotation of microRNA primary transcript structures reveals novel regulatory mechanisms. Genome Res 2015;25:1401–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Villani AC, Satija R, Reynolds G, et al. . Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 2017;356(6335):356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 2004;10:1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zilionis R, Nainys J, Veres A, et al. . Single-cell barcoding and sequencing using droplet microfluidics. Nat Protoc 2016 12:12016;12:44–73. [DOI] [PubMed] [Google Scholar]
- 16. Trapnell C. Defining cell types and states with single-cell genomics. Genome Res 2015;25:1491–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lähnemann D, Köster J, Szczurek E, et al. . Eleven grand challenges in single-cell data science. Genome Biol 2020 21:12020;21:1–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pertea M, Pertea GM, Antonescu CM, et al. . StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 2015 33:32015;33:290–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Picelli S, Faridani OR, Björklund ÅK, et al. . Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc 2013 9:12014;9:171–81. [DOI] [PubMed] [Google Scholar]
- 20. Litviňuková M, Talavera-López C, Maatz H, et al. . Cells of the adult human heart. Nature 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Friedman CE, Nguyen Q, Lukowski SW, et al. . Single-cell transcriptomic analysis of cardiac differentiation from human PSCs reveals HOPX-dependent cardiomyocyte maturation. Cell Stem Cell 2018;23:586–598.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bermejo JAP, Kang S, Rockwood SJ, et al. . SARS-CoV-2 infection of human iPSC-derived cardiac cells reflects cytopathic features in hearts of patients with COVID-19. Sci Transl Med 2021;13(590):eabf7872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kakimoto Y, Tanaka M, Kamiguchi H, et al. . MicroRNA deep sequencing reveals chamber-specific miR-208 family expression patterns in the human heart. Int J Cardiol 2016;211:43–8. [DOI] [PubMed] [Google Scholar]
- 24. Kuppusamy KT, Jones DC, Sperber H, et al. . Let-7 family of microRNA is required for maturation and adult-like metabolism in stem cell-derived cardiomyocytes. Proc Natl Acad Sci USA 2015; 112: E2785–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Carè A, Catalucci D, Felicetti F, et al. . MicroRNA-133 controls cardiac hypertrophy. Nat Med 2007;13:613–8. [DOI] [PubMed] [Google Scholar]
- 26. Nishiga M, Wang DW, Han Y, et al. . COVID-19 and cardiovascular disease: from basic mechanisms to clinical perspectives. Nat Rev Cardiol 2020 17:92020;17:543–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gutmann C, Khamina K, Theofilatos K, et al. . Association of cardiometabolic microRNAs with COVID-19 severity and mortality. Cardiovasc Res 2022;118:461–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li AL, Lv JB, Gao L. MiR-181a mediates Ang II-induced myocardial hypertrophy by mediating autophagy. Eur Rev Med Pharmacol Sci 2017;21:5462–70. [DOI] [PubMed] [Google Scholar]
- 29. Hu F, Zhang S, Chen X, et al. . MiR-219a-2 relieves myocardial ischemia-reperfusion injury by reducing calcium overload and cell apoptosis through HIF1α/ NMDAR pathway. Exp Cell Res 2020;395(1):395. [DOI] [PubMed] [Google Scholar]
- 30. Lai X, Vera J. MicroRNA clusters. Encycl Syst Biol 2013;1310–4. [Google Scholar]
- 31. Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res 2005;33(Web Server issue):W741–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.