SUMMARY
Pathogen-mediated damage to the intestinal epithelium activates compensatory growth and differentiation repair programs in progenitor cells. Accelerated progenitor growth replenishes damaged tissue and maintains barrier integrity. Despite the importance of epithelial renewal to intestinal homeostasis, we know little about the effects of pathogen-commensal interactions on progenitor growth. We found that the enteric pathogen, Vibrio cholerae, blocks critical growth and differentiation pathways in Drosophila progenitors, despite extensive damage to epithelial tissue. We showed that inhibition of epithelial repair requires interactions between the Vibrio cholerae type six secretion system and a community of symbiotic bacteria, and that elimination of the gut microbiome is sufficient to restore homeostatic growth in infected intestines. Together, this work highlights the importance of pathogen-symbiont interactions for intestinal immune responses and outlines a previously undescribed impact of the type six secretion system on pathogenesis.
INTRODUCTION
The digestive tract is inhabited by a dense polymicrobial community that is important for many aspects of host biology. For instance, these communities induce the differentiation of immune cells, aid in the development of lymphoid tissues, and activate microbe-specific transcriptional responses along the gut (Bouskra et al., 2008; Ivanov et al., 2008; Sommer et al., 2015). Although our understanding of the effects of the microbiome have advanced steadily, comparatively little is known about how interactions among bacteria influence host responses to pathogenesis. Because of the gut’s physiological similarity to mammals, and a comparatively simple microbiome, the intestine of Drosophila melanogaster is widely used to study host-microbe interactions. (Broderick and Lemaitre, 2012; Miguel-Aliaga et al., 2018). As the fly microbiome is cultivable there are simple protocols for the generation of gnotobiotic flies that contain a defined consortium of bacteria (Douglas, 2018; Koyle et al., 2016; Ma et al., 2015). Therefore, it is possible to measure how interactions between bacterial species, including complex high-order interactions of more than two species, impact the host (Gould et al., 2018).
To regulate the intestinal microbiota, mammals and insects integrate physical, chemical, and immune defenses with homeostatic epithelial renewal, effectively restricting the growth and dissemination of intestinal microbes. In Drosophila, enteric bacteria promote bactericidal reactive oxygen species generation, and production of antimicrobial peptides that prevent overgrowth of gut bacterial populations (Ha et al., 2005; Ryu et al., 2006; Tzou et al., 2000; Zaidman-Rémy et al., 2006). Damage to the epithelium by intestinal pathogens, or reactive oxygen species, engages reparative growth programs in intestinal progenitor cells (IPCs) (Amcheslavsky et al., 2009; Buchon et al., 2009a; Jiang et al., 2009). Typically, infection stimulates IPC proliferation via the activation of the Epidermal Growth Factor (EGF) and Janus Kinase/Signal Transducer and Activator of Transcription (JAK/STAT) pathways (Buchon et al., 2009b, 2010; Cronin et al., 2009; Jiang et al., 2009, 2011). Immune effectors and regenerative proliferation are essential immune responses to pathogenic microbes (Miguel-Aliaga et al., 2018). However, it is important to consider the influence of symbiotic bacteria on host defenses. For example, Clostridium difficile infection is associated with shifts in symbiotic bacteria diversity (Samarkos et al., 2018), and a decrease in the abundance of Firmicutes and Bacteroidetes alongside an expansion of Enterobacteriaceae (Peterfreund et al., 2012).
Approximately, 25 percent of sequenced Gram-negative bacteria encode a type six secretion system (T6SS) that injects toxic effectors into susceptible prey (Bingle et al., 2008; Das and Chaudhuri, 2003; Mougous et al., 2006; Pukatzki et al., 2006). T6SS-encoded effectors cover a range of biological functions that include phospholipid hydrolysis, actin-crosslinking, pore-formation, and peptidoglycan degradation (Miyata et al., 2011; Pukatzki et al., 2007; Russell et al., 2011, 2013). Together, these effectors permit T6SS-mediated attacks on eukaryotic and prokaryotic targets in a range of environments and hosts (Schwarz et al., 2010). Interactions between the T6SS and neighboring cells contribute to disease caused by several pathogenic bacteria. For example, the T6SS of Campylobacter jejuni is thought to interact with eukaryotic cells to support in vivo colonization (Lertpiriyapong et al., 2012). In the guts of larval zebrafish, the T6SS of Vibrio cholerae modifies intestinal movements via a eukaryotic effector to displace symbiotic Aeromonas veronii (Logan et al., 2018). Alternatively, Salmonella enterica Serovar Typhimurium uses a T6SS to outcompete Gram-negative commensals and enhance colonization of the mouse intestine (Sana et al., 2016). In Galleria mellonella, the T6SS of Acinetobacter baumannii interacts with the microbiome to diminish host viability (Repizo et al., 2015). Thus, antagonistic interbacterial interactions mediated by the T6SS have measurable impacts on the virulence of intestinal pathogens. However, it remains unclear how such interbacterial interactions influence host responses to bacterial challenge.
Recently, the T6SS was demonstrated to contribute to pathogenesis of Vibrio cholerae (V. cholerae) via interactions with the intestinal microbiome. In the infant mouse model, the T6SS of V. cholerae enhances the development of diarrheal symptoms through interactions with symbiotic E.coli (Zhao et al., 2018). Likewise, the T6SS of V. cholerae acts on Gram-negative symbionts in the Drosophila model of to accelerate host death (Fast et al., 2018a). Drosophila is an established model for the characterization of V. cholerae pathogenesis (Blow et al., 2005). As in humans, adult flies are naturally susceptible to infection with V. cholerae and develop diarrhea-like symptoms upon infection (Blow et al., 2005). Here, we used the Drosophila – Vibrio model to test how interactions between intestinal symbionts and V. cholerae influence host responses to intestinal challenge.
We found that the T6SS of V. cholerae disrupted intestinal homeostasis by blocking the regeneration of the gut epithelium. As part of a normal intestinal immune response, the gut epithelium is renewed via the proliferation of IPCs in response to infection (Bonfini et al., 2016; Buchon et al., 2009a, 2009b, 2010; Jiang et al., 2011). However, despite significant intestinal damage and extensive epithelial shedding, we did not detect an increase in IPC proliferation in guts infected with V. cholerae with a T6SS. Instead, we found that the T6SS impairs growth and differentiation signals required for epithelial renewal. Strikingly, T6SS-dependent arrest of epithelial repair was the result of interactions between the microbiome and the T6SS, as ablation of the microbiome restored epithelial regeneration in response to V. cholerae. Furthermore, this inhibition of renewal was not the result of a bilateral interaction between V. cholerae and a single symbiotic species, but instead required interactions between V. cholerae and a multi-species consortium of intestinal symbionts. In particular, we found that interactions between V. cholerae and a community of three common fly symbionts are sufficient to inhibit epithelial repair, demonstrating that complex symbiont-pathogen interactions have measurable impacts on defences against pathogenic bacteria. Together, the work presented here identifies an arrest of IPC proliferation that requires interactions between the T6SS of V. cholerae and the intestinal microbiome.
RESULTS
The T6SS promotes epithelial shedding.
In Drosophila, enteric infection results in the delamination and expulsion of damaged epithelial cells (Buchon et al., 2010; Zhai et al., 2018). To test the effect of the T6SS on epithelial delamination, we measured epithelial shedding in the guts of adult CB>mCD8::GFP flies infected with wildtype V. cholerae (C6706) or an isogenic C6706ΔvasK mutant, that carries an in-frame deletion in the essential T6SS gene that encodes the VasK protein (Pukatzki et al., 2006). In this fly line, delaminating cells are marked with the induction of GFP (Zhai et al., 2018). Previously, we found that the T6SS contributes to the intestinal pathogenesis of V. cholerae (Fast et al., 2018a). Based on this work, we examined epithelial cell shedding in the guts of flies infected with V. cholerae for 24 hours as flies have been robustly colonized by V. cholerae at this time point, develop disease symptoms, but remain viable. In mock- infected, control flies, we observed few delaminating cells in the posterior midgut (Fig. 1Aa–c). In these flies, we mostly detected instances of one or two delaminating cells per gut with 90% of guts containing ten or fewer shedding cells (Fig. 1B). Infection with C6706ΔvasK promoted a modest increase in shedding. Specifically, we observed clusters of GFP-positive cells that typically contained fewer than ten cells per cluster, with 40% of guts containing more than ten shedding cells (Fig 1Ad–f, Fig. 1B). Infection with C6706 caused a more severe delamination phenotype that was readily visible throughout the posterior midgut (Fig. 1Ag–i). In this challenge, infected guts had multiple patches of large numbers of delaminating cells. For example, whereas 5% of samples infected with C6706ΔvasK had greater than 20 shedding cells in the posterior midgut, 45% of all samples infected with C6706 contained 20 or more shedding cells (Fig. 1B). Furthermore, challenge with C6706 caused greater than 40 shedding cells per posterior midgut in 10% of infected samples, a phenotype that was absent from intestines infected with C6706ΔvasK (Fig. 1B). Comparisons between treatment groups confirmed that infection with C6706 not only greatly increased the number of shedding cells per area relative to unchallenged guts (P = 4.0×10−6), but also increased the number of shedding cells compared to C6706ΔvasK (P=0.007, Fig. 1C). Together, these data demonstrate that the V. cholerae T6SS significantly enhances epithelial shedding in infected Drosophila hosts.
Figure 1. The T6SS promotes epithelial shedding.
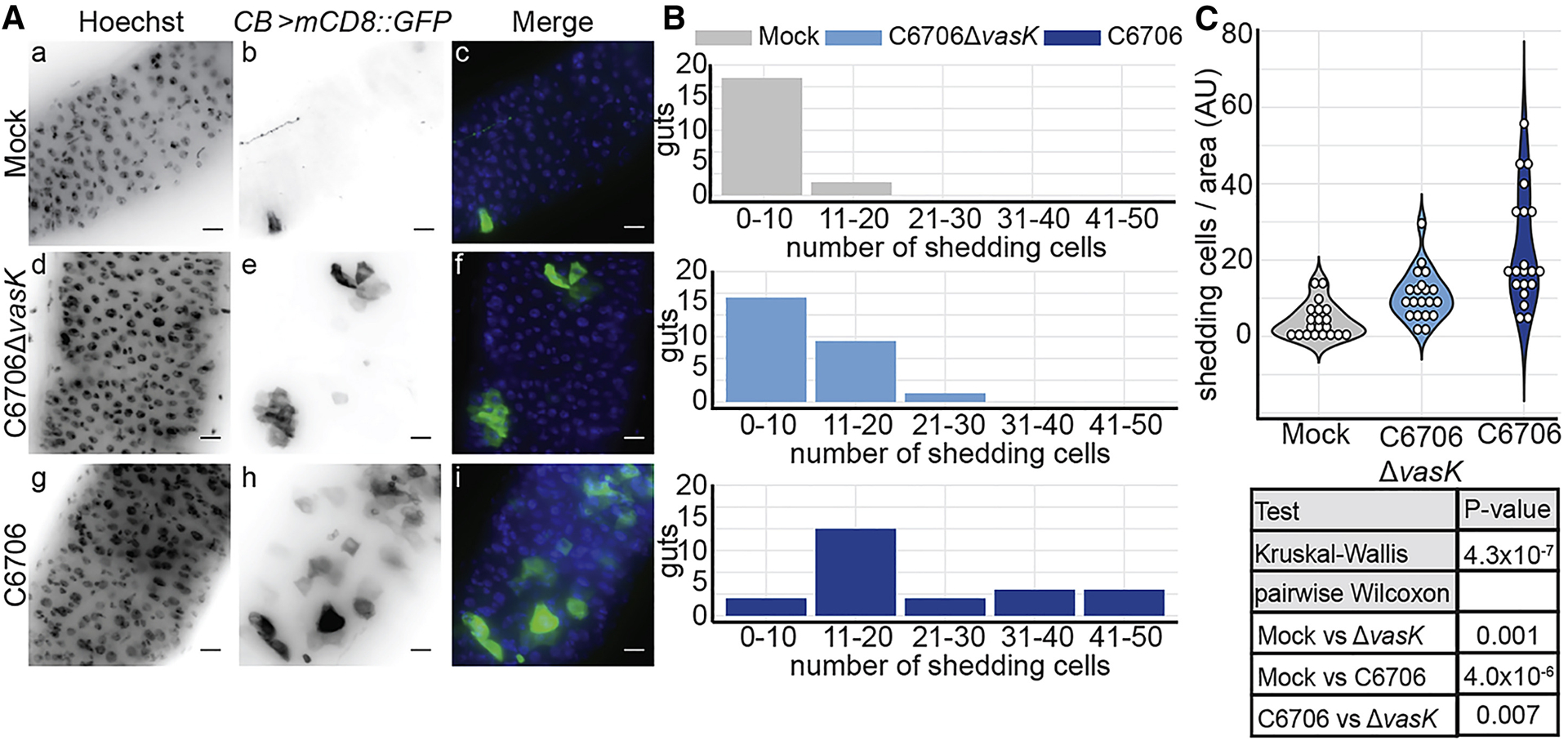
(A) Immunofluorescence images of the posterior midguts of CB>mCD8::GFP flies mock infected or infected with C6706ΔvasK, or C6706. Hoechst marks DNA (blue) and GFP marks shedding intestinal cells (green). Scale bars are 10μm. (B) Histogram of the number of shedding cells in the posterior midguts from (A). (C) Quantification of shedding cells per unit surface area from (A). Each dot represents a measurement from a single fly gut.
Disrupted intestinal homeostasis in response to the T6SS.
In Drosophila melanogaster, intestinal damage and epithelial shedding promotes compensatory growth of IPCs to maintain the epithelial barrier (Bonfini et al., 2016). As there was extensive T6SS-dependent sloughing of epithelial cells, we tested if the T6SS promotes homeostatic growth of IPCs. To address this, we used the esgts>GFP fly line to visualize GFP-positive IPCs in sagittal sections prepared from the posterior midguts of flies infected with C6706 or C6706ΔvasK. The midguts of control flies had a clear intestinal lumen surrounded by an intact epithelium (Fig. 2Aa–d). Consistent with Fig. 1, infection with C6706ΔvasK stimulated a modest shedding of cellular material (asterisks) into the intestinal lumen without an apparent loss of barrier integrity (Fig. 2Ae–h). Challenge with C6706 once again promoted extensive shedding of epithelial cells and cellular debris into the lumen (Fig. 2Ai–l), as well as the appearance of numerous breaks along the basement membrane (arrowheads), suggesting pathogen-dependent damage to the epithelial barrier.
Figure 2. Disrupted intestinal homeostasis in response to the T6SS.
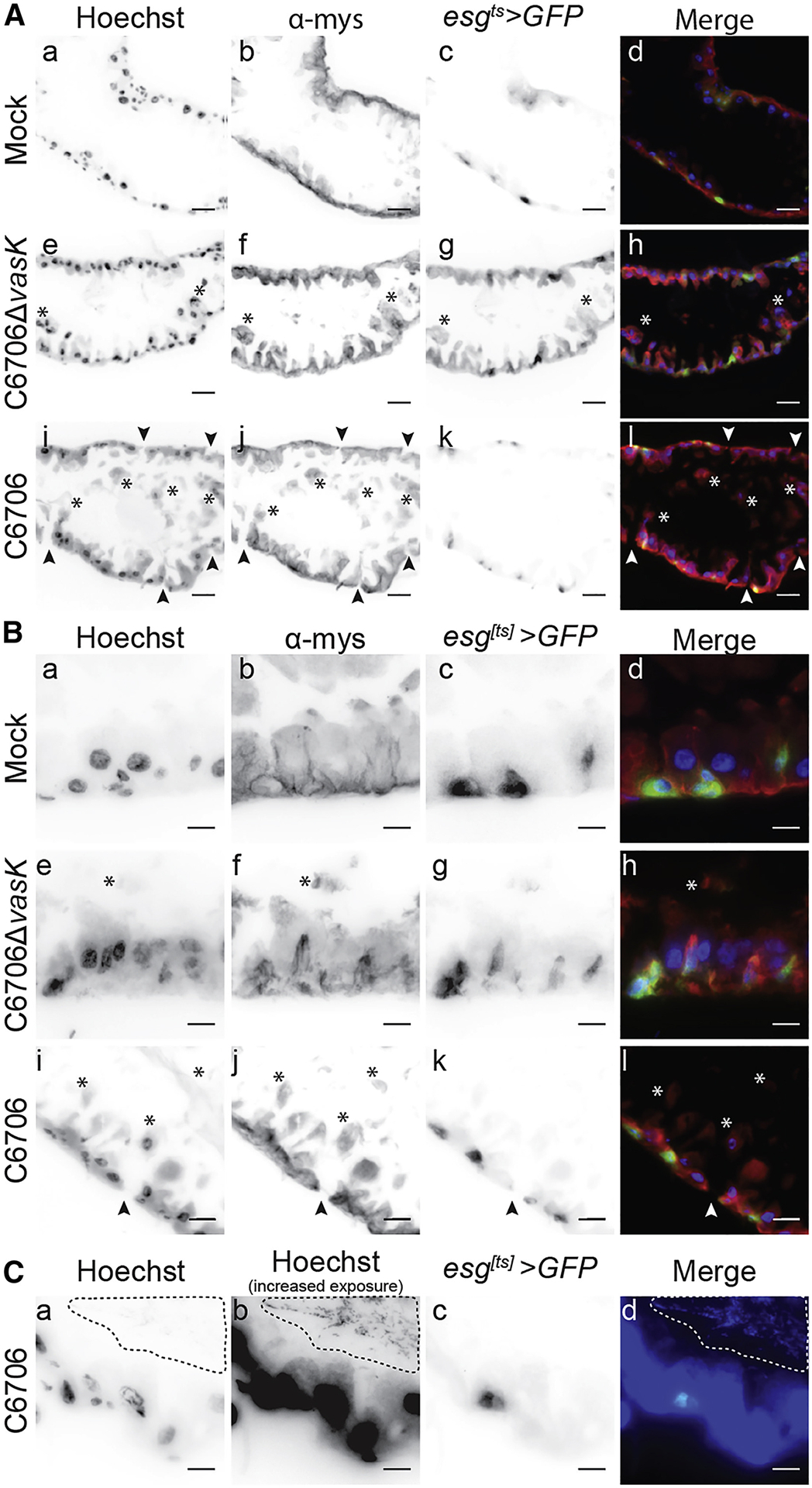
(A-C) Immunofluorescence of sagittal sections prepared from the posterior midgut of esgts>GFP flies mock infected or infected with C6706ΔvasK, or C6706. Hoechst marks DNA (blue), GFP marks IPCs (green), and α-mys marks the β-integrin, myospheroid (mys, red). Arrowheads indicate damage to the intestinal epithelium and asterisks denote cellular matter in the lumen. (C) Visualization of intestinal bacteria via increased exposure of Hoechst stain. The dotted line circles bacteria in the lumen. Scale bars are (A) 25μm and (B & C) 10μm.
As we observed epithelial damage and shedding cells in V. cholerae-infected intestines, we determined if V. cholerae promoted compensatory growth of IPCs. In mock-infected flies, we observed the regular distribution of small GFP-positive IPCs along the basement membrane of the midgut (Fig 2Ba–d). Infection with C6706ΔvasK caused an accumulation of GFP-positive IPCs, consistent with enhanced epithelial renewal in response to infection (Fig. 2B e–h). In contrast, despite extensive shedding of cellular material (Fig 1) and obvious epithelial damage (Fig. 2A), guts challenged with C6706 did not appear to have elevated numbers of IPCs (Fig. 2B i–l). Instead, these cells were similar to the basal GFP-positive cells of mock infected flies (Fig 2Ca–d), despite an immediate proximity of lumenal bacteria to the epithelium (dotted outline). Together, these results demonstrate that C6706ΔvasK provokes shedding of intestinal cells along with the accumulation of esg positive IPCs in a manner consistent with a conventional intestinal immune response to pathogenic bacteria In contrast, we did not observe signs of epithelial renewal in flies infected with C6706, despite widespread intestinal damage, raising the possibility that the V. cholerae T6SS uncouples epithelial shedding from intestinal regeneration.
The T6SS modifies IPC transcriptional responses to V. cholerae.
Given the apparent absence of IPC growth in C6706-infected flies, we used RNA sequencing (RNA-seq) analysis to identify the intestinal response to infection with C6706 (Sup Fig. 1). We found that the host response to C6706 is characterized by the activation of antibacterial defenses, re-programming of metabolic pathways, and the expression of a large cohort of genes required for the generation and assembly of mature ribosomes. Many of these responses match our understanding of the fly transcriptional response to pathogenic bacteria (Sup Fig. 1, 4 (Buchon et al., 2009a; Dutta et al., 2015; Troha et al., 2018). However, and in contrast to classical responses to enteric challenge, we did not detect changes in mRNA levels characteristic of JAK-STAT or EGF responses, two pathways that are intimately linked with homeostatic renewal of a damaged epithelium.
The apparent absence of homeostatic growth signals in C6706-infected intestines prompted us to directly determine the transcriptional response of IPCs to V. cholerae infection. For this experiment, we performed RNA-seq on IPCs purified from the guts of adult esg[ts]/+ flies that we challenged with C6706 or C6706ΔvasK (Fig. 3A). As a control, we sequenced the transcriptome of purified IPCs from uninfected esg[ts]/+ flies. Principle component analysis showed that samples from uninfected flies and those from flies infected with C6706ΔvasK grouped relatively closely. In contrast, samples from C6706-infected flies grouped away from both uninfected and C6706ΔvasK-infected flies (Fig. 3B). Differential gene expression analysis confirmed minimal overlaps between the transcriptomes of C6706 and C6706ΔvasK-infected flies relative to uninfected controls (Fig. 3C). From there, we examined Gene Ontology (GO) term enrichment among the differentially upregulated and downregulated genes. Here, we also compared C6706ΔvasK to C6706 to identify changes in IPC transcriptional responses specific to the T6SS (Fig. 3F). Of note, comparison of the transcription profile of C6706-challenged IPCs relative to uninfected IPCs revealed a downregulation of biological processes involved in growth and mitosis. This included a significant downregulation of processes such as cell proliferation and nuclear division (Fig. 3G). In contrast, this downregulation of growth processes was absent when we compared the transcriptional profile of C6706ΔvasK-infected IPCs to that of uninfected IPCs (Fig. 3H). Instead, we detected a significant enrichment of mitotic processes in flies infected with C6706ΔvasK relative to flies challenged with C6706 (Fig. 3I). Together, these data suggest that IPCs have distinct transcriptional response to wildtype and T6SS-deficient V. cholerae. In particular, we found that the T6SS inhibits the expression of genes required for growth and renewal of the epithelium.
Figure 3. The T6SS modifies IPC transcriptional responses to V. cholerae.
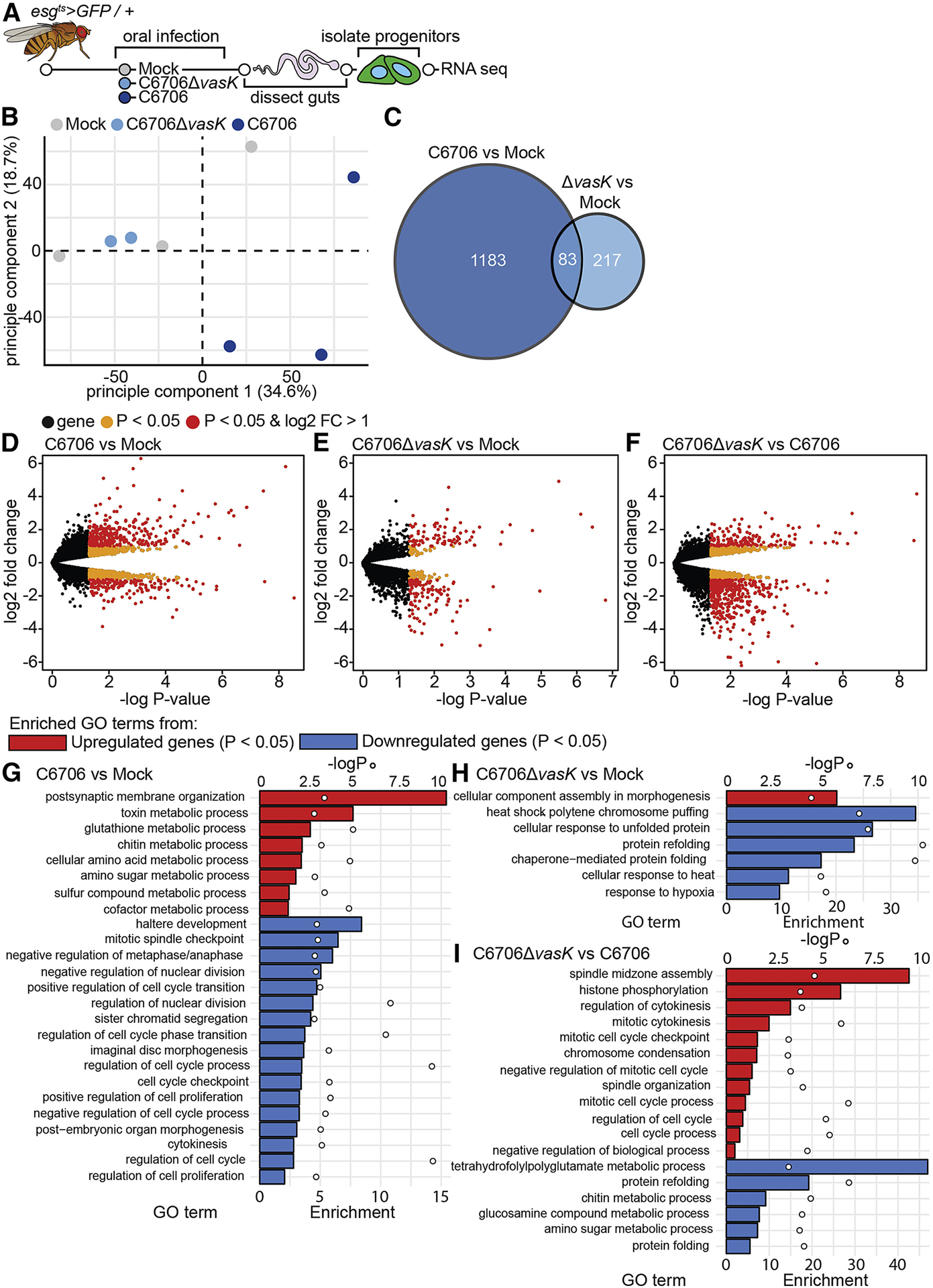
(A) Schematic representation of the RNA-sequencing of IPCs isolated from V. cholerae infected guts. (B) Principle component analysis from the counts per million obtained from RNA-sequencing of IPCs isolated from guts mock infected or infected with C6706 or C6706ΔvasK. (C) Venn diagram of differentially expressed genes (P<0.05) from comparisons of C6706 to Mock and C6706ΔvasK to Mock. (D-F) Volcano plots of differentially expressed genes from comparisons of (D) C6706 to Mock, (E) C6706ΔvasK to Mock, and (F) C6706ΔvasK to C6706. Each dot represents a single gene. Yellow indicates a P<0.05, red indicates P<0.05 and log2 fold change >1 or <−1. (G-I) Gene Ontology analysis from up or down regulated differently expressed genes (P<0.05) from comparisons of (G) C6706 to Mock, (H) C6706ΔvasK to Mock, and (I) C6706ΔvasK to C6706. (G,H,I) Bars (bottom X-axis) represent enrichment scores and circles (top X-axis) represent −logP values for each enriched GO term.
To further characterize T6SS-dependent impacts on epithelial renewal, we determined the transcriptional profile of the whole intestinal response to infection with C6706ΔvasK (Sup Fig. 2A). In general terms, we noticed substantial overlaps between host responses to C6706 and C6706ΔvasK (Sup Fig. 2B). For example, similar to C6706, C6706ΔvasK caused differential expression of genes required for the control of intestinal immunity, metabolism, and the generation of mature ribosomes (Sup Fig. 2C). However, we also observed T6SS-dependent effects on the expression of genes required for epithelial growth and renewal, including decapentaplegic pathway elements, and core components of the cell cycle progression machinery (Sup Fig. 3) (Guo et al., 2013; Tian and Jiang, 2014; Zhou et al., 2015). Specifically, infection with C6706 resulted in a downregulation of critical cell cycle activators relative to challenge with C6706ΔvasK. Combined, these data implicate the T6SS in the regulation of host epithelial renewal.
IPCs fail to facilitate epithelial repair upon intestinal challenge with V. cholerae.
Epithelial damage activates the JAK/STAT and the EGFR pathways to stimulate epithelial repair. We observed increased levels of mRNA of several genes indicative of JAK/STAT and EGF pathway activation in IPCs from C6706ΔvasK-infected flies compared to those from C6706-infected counterparts (Fig. 4A). Furthermore, infection with C6706ΔvasK led to an increase in the expression of cell cycle activators in the IPC population (Figure 4A). These data suggest enhanced IPC growth in progenitors of flies challenged with C6706ΔvasK relative to C6706. Indeed, we observed the transcription signature of diminished EGF and JAK/STAT activity in IPCs purified from flies infected with C6706 relative to IPCs purified from uninfected controls. Specifically, we noted diminished expression of the EGF pathway transcription factor pointed (pnt) and the EGF receptor (EGFR) itself in IPCs from flies infected with C6706 compared to IPCs from uninfected controls (Fig. 4A). Similarly, we noted a reduction in the relative proportions of mRNAs that encode central components of the JAK/STAT pathway. In the JAK/STAT pathway, binding of interleukin-like ligands to the receptor Domeless (dome) induces signalling through the kinase Hopscotch (hop), and results in the transcription of Socs36E (Zeidler and Bausek, 2013). We observed diminished mRNA levels of all three of these signaling components in IPCs from C6706-challenged flies relative to uninfected controls. Furthermore, we detected significant drops in mRNA that encode prominent cell cycle genes, such as the CDC25 ortholog, stg, the S-phase cyclin dependent kinase 2 (Cdk2), and the essential M phase cyclin CyclinB3 (CycB3). In summary, we detected T6SS-dependent decreases in mRNA of genes in pathways responsible for epithelial renewal alongside diminished levels of cell cycle genes, consistent with a failure of intestinal renewal in flies infected with wild-type V. cholerae.
Figure 4. IPCs fail to facilitate epithelial repair upon intestinal challenge with V. cholerae.
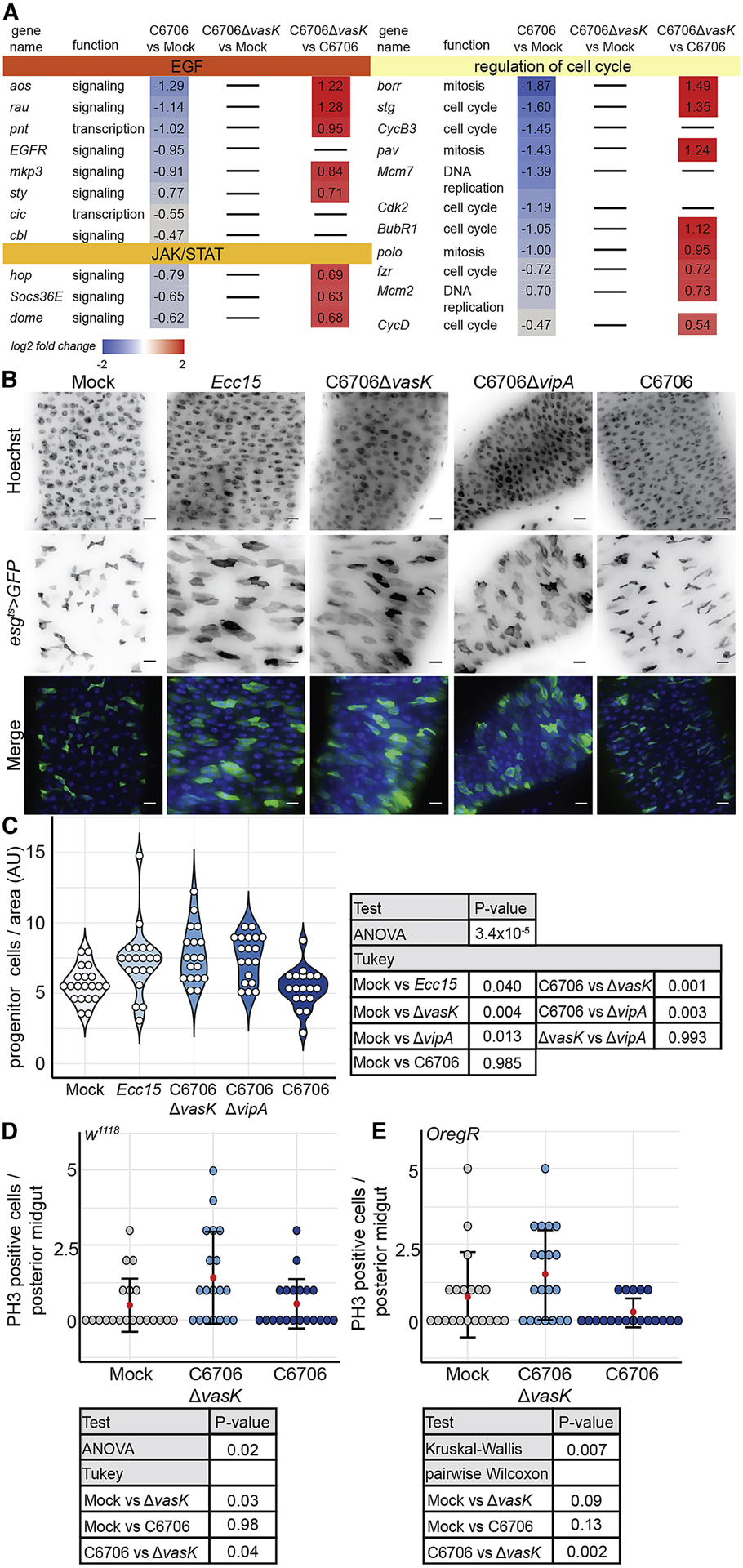
(A) Genes that regulate IPC growth and cell cycle from RNA-seq of IPCs of flies mock infected or infected with C6706 or C6706ΔvasK. (B) Immunofluorescence of the posterior midguts of esgts>GFP flies mock infected or infected with Ecc15, C6706ΔvasK, C6706ΔvipA, or C6706. Hoechst marks DNA (blue) and GFP marks esg positive IPCs (green). Scale bars are 10μm. (C) Quantification of the number of IPCs per unit surface area from (B). Each dot represents a measurement from a single fly gut. (D-E) Quantification of the number of PH3 positive cells in the posterior midguts of (D) w1118 or (E) OregR flies that were mock infected or infected with C6706ΔvasK, or C6706.
To directly test this hypothesis, we examined IPC growth in guts infected with C6706, or with C6706ΔvasK, in two different functional assays. First, we quantified the number of IPCs per area in guts of infected flies as a measure of IPC expansion. As a control, we quantified the number of IPCs in guts of flies infected with the Gram-negative fly pathogen Erwinia carotovora carotovora 15 (Ecc15), a known activator of IPC growth (Buchon et al., 2009b). In agreement with previous reports, infection with Ecc15 promoted a significant increase in the number of IPCs per area (P=0.04, Fig. 4B, C). Similarly, guts infected with C6706ΔvasK had greater numbers of IPCs per area than uninfected controls (P=0.004, Fig. 4B, C). This phenotype was not specific to the vasK T6SS mutation, as we observed a near-identical expansion of IPCs in intestines challenged with V. cholerae with a null mutation in the vipA gene, an essential component of the T6SS outer sheath (P=0.013, Fig. 4B, C) (Zheng et al., 2011). In contrast, guts infected with C6706 had significantly fewer IPCs per area than guts infected with either C6706ΔvasK or C6706ΔvipA (P<0.001 and P<0.003 respectively, Fig. 4B, C). Furthermore, there was no difference in the number of IPCs per area between uninfected flies and those infected with C6706 (P=0.985, Fig. 4B, C), indicating a failure of renewal that requires the T6SS. Next, we quantified mitotic PH3 positive cells in the posterior midguts of two different wildtype fly strains, w1118, and Oregon R, that we infected with C6706ΔvasK or C6706. In both fly backgrounds, infection with C6706ΔvasK prompted an increase in the number of mitotic cells in the posterior midgut. In contrast, both wildtype fly strains had significantly fewer mitotic cells in C6706-infected guts compared to C6706Δvask-challenged counterparts (P=0.04 and P=0.002, Fig. 4D, E).
Collectively, these data demonstrate that the transcriptional response of IPCs to V. cholerae is significantly altered by the presence of a functional T6SS. This difference in response to the T6SS is highlighted by a significant downregulation of pathways critical for intestinal renewal, diminished IPC proliferation, and failed epithelial renewal.
Impaired IPC differentiation in response to the T6SS
IPC proliferation is accompanied by signals through the Notch-Delta axis that direct the generation and differentiation of transitory enteroblasts (Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006, 2007). Our analysis of the RNA-seq data suggested T6SS-dependent effects on Notch pathway activity. For example, we detected an increase in the levels of mRNA of the Notch-response gene, Enhancer of split (E(spl)), as well as Delta (Dl) itself in IPCs from C6706ΔvasK-infected guts relative to C6706-infected guts (Fig. 5A). Furthermore, we noticed a suppression of E(spl) genes and Dl in IPCs from flies infected with C6706 compared to uninfected controls (Fig. 5A). As genes in the E(spl) complex are primary transcriptional targets of the Notch pathway, these data suggest a potential impairment of IPC differentiation programs by the T6SS (Bailey and Posakony, 1995).
Figure 5. Impaired IPC differentiation in response to the T6SS.
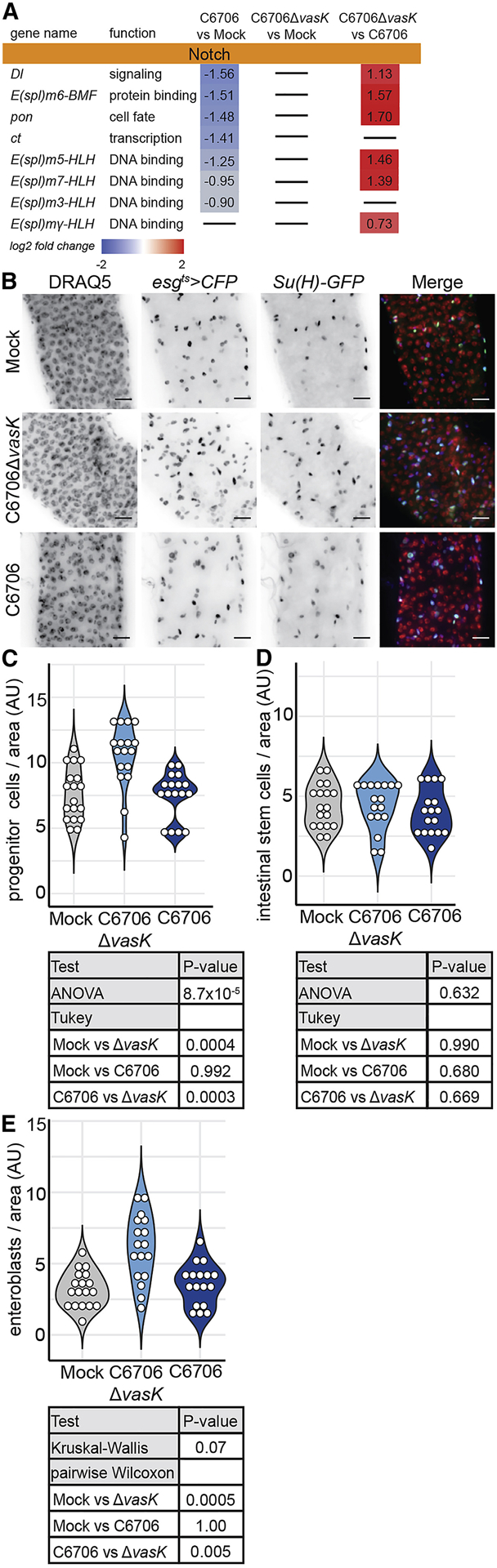
(A) Differentially regulated genes in the Notch signaling pathway, from RNA-sequencing of IPCs from flies mock infected or infected with C6706ΔvasK or C6706 (B) Immunofluorescence of the posterior midguts of esgts>CFP, Su(H)-GFP flies mock infected or infected with C6706ΔvasK, or C6706. DRAQ5 marks DNA (red), CFP marks esg positive IPCs (blue), and GFP marks Su(H) positive enteroblasts. Scale bars are 10μm. (C) Quantification of the number of IPCs (CFP-positive), per unit surface area from (B). Each dot represents a measurement from a single fly gut. (D) Quantification of the number of intestinal stem cells (CFP-positive, GFP-negative) per unit surface area from (B). (E) Quantification of the number of enteroblasts (CFP-positive, GFP-positive) per unit surface area from (B).
To test if IPC differentiation responds differently to the presence of a T6SS, we quantified the number of enteroblasts and stem cells in the posterior midguts of esgGAL4, UAS-CFP; Su(H)-GFP flies that we infected with C6706 or C6706ΔvasK. In the absence of infection, we detected approximately equal numbers of intestinal stem cells (CFP-positive, GFP-negative) and enteroblasts (EB) (CFP-positive, GFP-positive) in the posterior midgut (Fig. 5B, D, E). Consistent with Figure 4, infection with C6706ΔvasK stimulated an expansion of IPCs (Fig. 5B, C). This expansion of IPCs was the result of an increased population of enteroblasts (P = 0.0004, Fig. 5E), not stem cells (Fig. 5D), consistent with the generation of undifferentiated enteroblasts required to renew the intestinal epithelium. In contrast, guts infected with C6706 contained significantly fewer IPCs per area than their C6706ΔvasK-infected counterparts (P = 0.0003, Fig. 5B, C). There was no difference in the number of intestinal stem cells between C6706 or C6706ΔvasK infected guts (Fig. 5D). Instead, there was a significant drop in the number of enteroblasts per unit area in guts challenged with C6706 relative to those infected with C6706ΔvasK (P = 0.005, Fig. 5B, E), indicating that the T6SS likely prevents the generation of enteroblasts.
Together, the data presented here uncover a T6SS-dependent failure of epithelial renewal in V. cholerae-infected flies. We find that flies activate conventional growth and differentiation programs in response to C6706ΔvasK. This response is absent from intestines challenged with pathogenic V. cholerae with a functional T6SS. Instead, we find that despite extensive damage and increased epithelial shedding, IPCs fail to induce genes required for IPC proliferation. This failure of gene expression was accompanied by an lack of ISC proliferation along with an absence of enteroblast differentiation, culminating in impaired epithelial regeneration.
T6SS-dependent failure in epithelial renewal requires intestinal symbionts.
T6SS effectors are toxic to eukaryotic and prokaryotic cells (Joshi et al., 2017). For example, interactions between the V. cholerae T6SS and eukaryotic cells have been implicated in intestinal inflammation, and interactions between the T6SS and the endogenous microbiome are linked to the virulence of V. cholerae (Fast et al., 2018a; Ma and Mekalanos, 2010; Zhao et al., 2018). This prompted us to ask if the IPC response to the T6SS is a function of direct interactions between the T6SS and host cells, or instead requires interactions between the T6SS and the intestinal microbiota.
To test this, we measured epithelial renewal in the guts of germ-free (GF) flies that were infected with C6706 or C6706ΔvasK. Flies were considered GF if commensal load was eliminated below the limit of detection such that no microbial colonies were visible upon plating whole fly homogenates on agar permissive for the growth of Drosophila symbiotic species. Similar to conventionally reared (CR) flies, which host a community of symbiotic microbes, infection of GF flies with C6706ΔvasK stimulated an expansion of IPCs relative to uninfected controls (P = 0.00004, Fig. 6A, B). Enteric infection of GF flies with C6706 resulted in an expansion of IPCs in a manner nearly identical to that of C6706ΔvasK-infected intestines. Indeed, we found no significant difference in the number of IPCs per area between C6706 and C6706ΔvasK-infected GF flies (P = 0.658, Fig. 6A, B). To test if interactions between the T6SS and the gut microbiota prevent infection-dependent induction of epithelial renewal, we generated germ-free flies by two different methods and measured epithelial regeneration in guts infected with C6706. Specifically, we measured the number of IPCs per area in adult germ-free flies that we generated either by administration of antibiotics to adult flies, or by hypochlorite dechorionation and sterilization of embryos. Here, we found that infection with C6706 promoted a significant expansion of IPCs, regardless of the method used to generate germ-free flies (P=0.0004, P=0.001, Fig. 6C), and there was no significant difference in the number of IPCs per area between antibiotic-treated or axenic flies infected with C6706 (P = 0.950, Fig. 6C). Together these results indicate that interactions between the T6SS of V. cholerae, and the endogenous microbiome of Drosophila, prevent the activation of conventional epithelial repair pathways.
Figure 6. IPC suppression of growth in response to the T6SS requires intestinal symbionts.
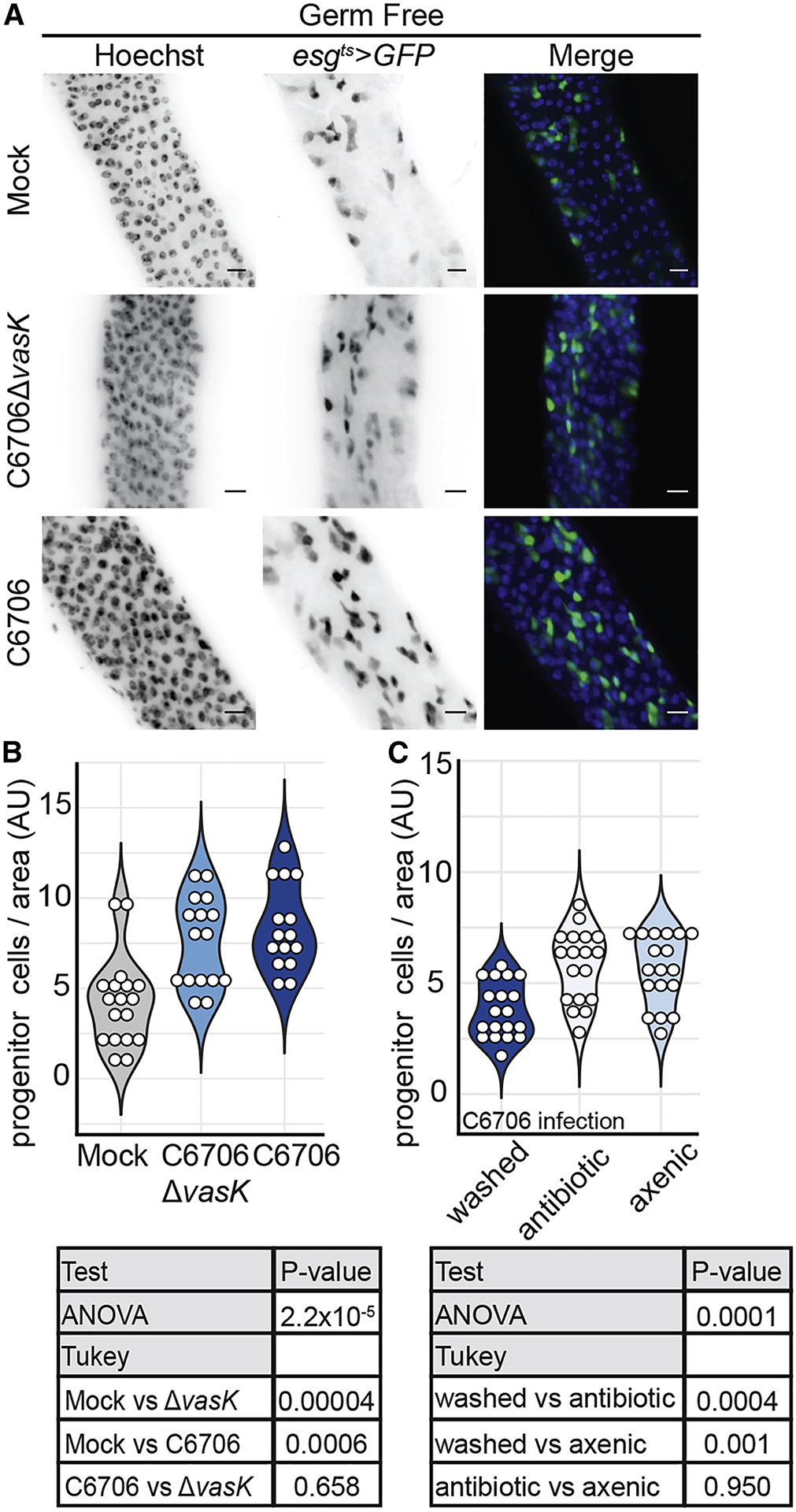
(A) Immunofluorescence of the posterior midguts of germ free esgts>GFP flies mock infected or infected with C6706ΔvasK, or C6706. Hoechst marks DNA (blue) and GFP marks esg positive IPCs (green). Scale bars are 10μm. (B) Quantification of the number of IPCs per unit surface area from (A). Each dot represents a measurement from a single fly gut. (C) Quantification of the number of IPCs per unit surface area in esgts >GFP flies infected with C6706. Flies were made germ-free either by the administration of antibiotics to adults (antibiotic) or by bleaching of embryos (axenic).
T6SS suppression of epithelial renewal requires higher-order microbiome interactions.
As the failure of epithelial renewal in response to the T6SS requires gut microbes, we asked if interactions with specific members of the Drosophila microbiome were responsible for T6SS-mediated loss of epithelial regeneration. We previously showed that the T6SS of V. cholerae targets the Gram-negative fly symbiont Acetobacter pasteurianus (Ap) for destruction, while the Gram-positive symbiont Lactobacillus brevis (Lb) is refractory to T6SS-mediated elimination (Fast et al., 2018a). As Lb is insensitive to the T6SS, we hypothesized that interactions between C6706 and Lb would fail to prevent epithelial repair. To test this hypothesis, we measured the number of IPCs in the guts of infected adult flies that we associated exclusively with Lb. For each bacterial association, we performed a parallel control infection of CR flies with the same cultures of C6706 and C6706ΔvasK. In each control infection, C6706ΔvasK promoted a regenerative response that significantly increased the number of IPCs. In contrast, challenge with C6706 consistently impaired IPC proliferation (Fig. 7A,C,D,F, G, I). We observed similar amounts of epithelial renewal in the intestines of Lb mono-associated flies infected with C6706 or C6706ΔvasK (Fig. 7B, C P=0.999), indicating that interactions between V. cholerae and Lb alone do not affect epithelial renewal. We then tested the ability of Ap to modify renewal. Given the sensitivity of Ap to T6SS-dependent killing, we expected diminished epithelial regeneration in Ap-associated flies challenged with C6706. However, contrary to our prediction, we did not detect a difference in the number of IPCs between Ap-associated guts infected with C6706 or C6706ΔvasK (P=0.996, Fig. 7 E. F). Instead, we found that C6706 promoted IPC proliferation when confronted with an intestine populated exclusively by Ap, indicating that T6SS-Ap alone does not impact epithelial renewal.
Figure 7. T6SS suppression of epithelial renewal requires higher-order microbiome interactions.
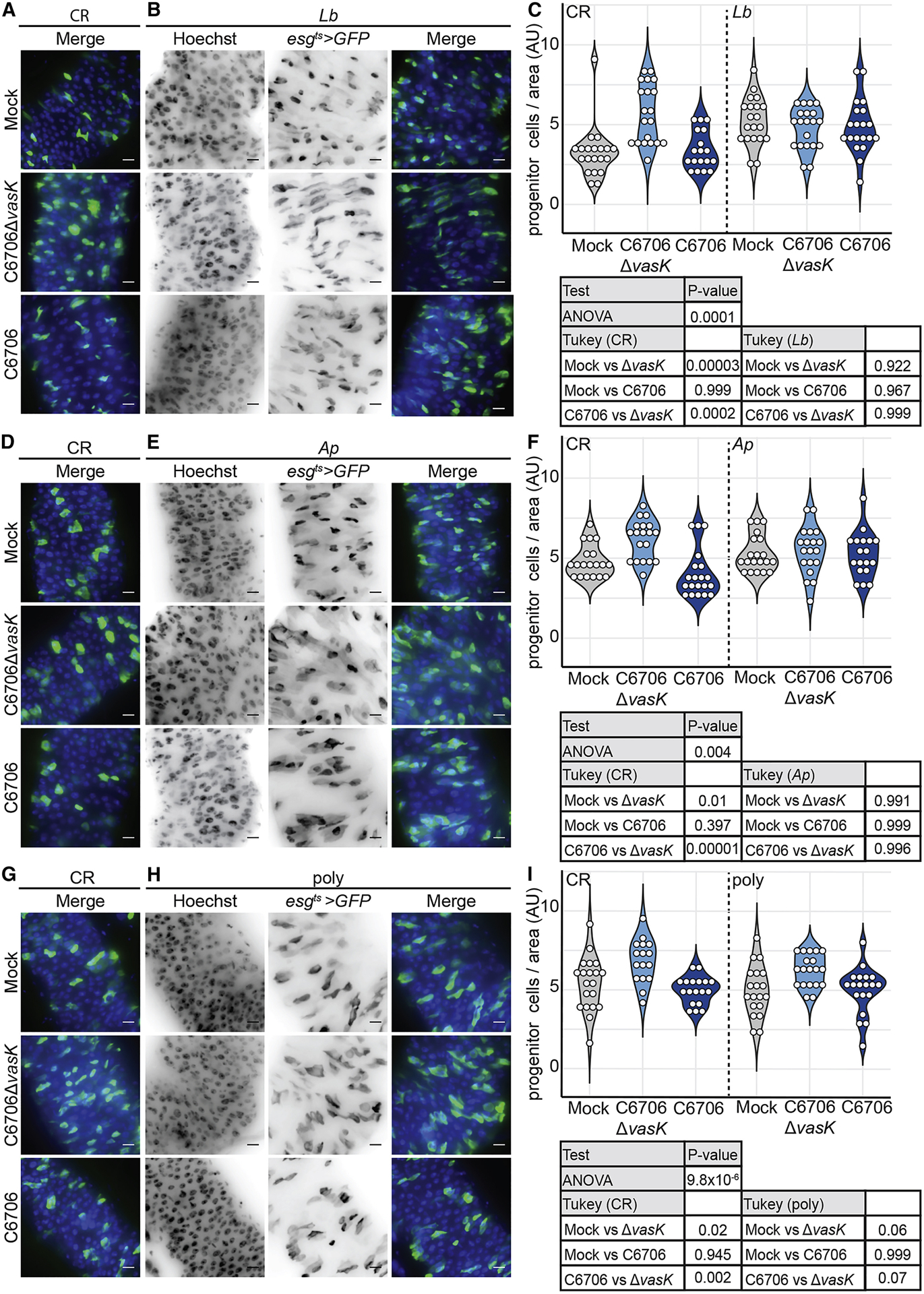
Immunofluorescence of posterior midguts of (A,D,G) CR, (B) Lb mono-associated, (E) Ab mono-associated, or (H) poly-associated esgts >GFP flies mock infected or infected with C6706ΔvasK, or C6706. Hoechst marks DNA (blue) and GFP marks esg positive IPCs (green). Scale bars are 10μm. Quantification of the number of IPCs per unit surface area in the guts of (C,F,I) CR, (C) Lb mono-associated, (F) Ap mono-associated, or (I) poly-associated flies. 2–3 day old virgin female flies were raised on antibiotics 5 day at 25°C to eliminate the microbiome. Germ free flies were then associated with microbial populations as indicated.
Recently, higher-order interactions among polymicrobial communities have been demonstrated to significantly influence host phenotypes in response to bacteria (Gould et al., 2018). This led us to ask if T6SS-dependent interruption of renewal requires a more complex community of symbiotic bacteria. To test this, we associated adult Drosophila with a 1:1:1 mixture of three common fly symbionts, Ap, Lb, and Lactobacillus plantarum (Lp), and quantified IPC numbers in the guts of flies that we infected with C6706 or C6706ΔvasK. Similar to what we observed in CR flies, guts infected with C6706Δvask had increased numbers of IPCs per area, indicating that poly-association with Ap, Lb, and Lp, is sufficient to reproduce physiologically relevant intestinal growth phenotypes in response to infection. In contrast, we did not see a difference in the number of IPCs between guts infected with C6706 and uninfected controls in poly-associated flies (Fig. 7H,I). Furthermore, we found an appreciable, although not statistically significant, difference in the number of IPCs between poly-associated guts infected with C6706 and C6706ΔvasK. These data suggest that interactions between the T6SS and individual symbiotic species are not sufficient to modify IPC repair responses to V. cholerae. Instead, a failure of epithelial renewal in response to the T6SS is a function of interactions between the T6SS and a consortium of intestinal symbionts. These results uncover negative effect of the T6SS on epithelial regeneration programs, mediated by complex interactions between the T6SS and the intestinal microbiome.
DISCUSSION
Enteric infections initiate host responses that halt the expansion and dissemination of pathogenic bacteria. Renewal of the epithelium is achieved by the coordinated expulsion of damaged cells, and the accelerated proliferation of IPCs (Miguel-Aliaga et al., 2018). However, it is unclear how interactions among gut-resident bacteria influence this response. Here, we determined how interactions between the T6SS of an enteric pathogen and intestinal symbionts influence gut transcriptional responses, epithelial shedding, and IPC proliferation. We found that a T6SS-deficient V. cholerae activates classical defense and repair responses in the host. Specifically, C6706ΔvasK promoted transcription of antimicrobial peptides (Sup Fig. 4), shedding of epithelial cells, and IPC proliferation and differentiation. In contrast, infection with C6706, which encodes a fully operational T6SS, significantly altered host responses to infection, indicating a previously unknown effect of the T6SS on host intestinal immunity.
While infection with C6706 promoted antimicrobial peptide transcription (Sup Fig. 4), epithelial repair responses were phenotypically distinct to the outcomes of infection with a T6SS null mutant. In particular, C6706 caused extensive epithelial shedding, but failed to activate regeneration pathways critical for intestinal repair. Strikingly, we found that interactions between the T6SS and the microbiome were responsible for the lack of intestinal regeneration. Specifically, the absence of epithelial growth was the result of complex interactions that required a consortium of symbiotic bacteria. Previously, we showed that interactions between the symbiotic A pasteurianus and the T6SS of V. cholerae reduce host viability (Fast et al., 2018a). However, interactions between Ap and V. cholerae alone do not prevent induction of intestinal regeneration. The change of an effect of a single symbiotic species by the presence of other bacteria is consistent with a recent report that diversity significantly impacts the effects of individual symbiotic species on host physiology (Gould et al., 2018), and suggest that host killing, and the impairment of IPC proliferation in response to the T6SS are independent consequences of infection with V. cholerae.
Since the establishment of Drosophila as a model for oral infection with V. cholerae (Blow et al., 2005) researchers have identified a rich network of immune, metabolic, and growth-regulatory events that influence disease progression. For example, V. cholerae activates the antibacterial Immune Deficiency (IMD) pathway in infected flies (Berkey et al., 2009), a signal transduction cassette that induces expression of antimicrobial peptides in response to bacterial peptidoglycan (Bosco-Drayon et al., 2012; Tzou et al., 2000). IMD pathway mutants have extended viability after infection with V. cholerae, implicating host immune activity in pathogenesis of the bacteria (Berkey et al., 2009). At the same time, infections with V. cholerae impact intestinal levels of acetate (Hang et al., 2014), succinate (Kamareddine et al., 2018), and methionine sulfoxide (Vanhove et al., 2017) with consequences for host insulin signaling, lipid homeostasis, and epithelial renewal in the host. Interestingly, the ability of V. cholerae to suppress epithelial renewal is reverted by mutational inactivation of IMD (Wang et al., 2013), suggesting functional links between immune activity and IPC growth in infected flies. In contrast to our studies, several strains of C6706 cause limited disease in flies and fail to block epithelial renewal (Kamareddine et al., 2018). We speculate that this is a function of differences in quorum sensing between the C6706 strains used in the respective studies, as the strain of C6706 used in this work has low expression of the quorum-sensing mater regulator, HapR (Fast et al., 2018a; Stutzmann and Blokesch, 2016), and HapR mutations convert non-pathogenic strains to lethal strains that have the ability to block IPC growth (Kamareddine et al., 2018). We believe that the fly will serve as an excellent model to identify the extent to which T6SS activity modifies the disease phenotypes described above.
Oral infection of Drosophila with large doses of Pseudomonas entomophila induce a translational blockade that diminishes repair in the gut (Bonfini et al., 2016; Chakrabarti et al., 2012). However, in contrast to Pseudomonas entomophila, V. cholerae-mediated inhibition of epithelial renewal requires interactions between the T6SS and the gut microbiota. We do not know how interactions between C6706, and the microbiome inhibit IPC-mediated repair, although we consider several possible explanations for this effect. For example, the gut is sensitive to growth cues received or generated through host-microbe interactions (Broderick et al., 2014; Buchon et al., 2009b; Jones et al., 2013; Shin et al., 2011), raising the possibility that V. cholerae prevents IPC proliferation by modifying microbiota-derived pro-growth cues. Consistent with this hypothesis, other studies have documented the effects of V. cholerae on the availability of microbial metabolites with downstream effects on epithelial renewal (Kamareddine et al., 2018; Vanhove et al., 2017). In the future, it will be interesting to determine if the T6SS affects the bioavailability of microbiota-derived metabolites.
Interactions between symbionts and the pathogen may also support anti-eukaryotic function of the T6SS. In this scenario, V. cholerae may be required for microbiota-dependent shedding of differentiated epithelial cells, exposing underlying IPCs to intoxication by T6SS effectors such as the actin crosslinker, VgrG-1 (Pukatzki et al., 2006, 2007). Putative links between shedding and IPC access are consistent with a role for the IMD pathway in the shedding of damaged epithelial cells (Zhai et al., 2018). Flies with null mutations in the IMD pathway outlive wild-type flies when infected with V. cholerae (Berkey et al., 2009). Thus, it is possible that null mutations in the IMD pathway prevent excess epithelial shedding, and thereby maintain a barrier that protects IPCs from exposure to V. cholerae. This is supported by our recent data that inhibition of IMD pathway activity exclusively in enterocytes extended the viability of flies infected with C6706 (Shin et al., 2019). Future studies should consider examining the role of eukaryotic toxins as downstream mediators of T6SS-dependent killing.
In summary, the work presented here demonstrates that complex interactions between intestinal symbionts and enteric invaders combine to influence critical components of the intestinal immune response. While the effects of pathogenic bacteria on epithelial repair have been described previously, our work takes in to consideration how interactions between bacterial species within a complex community structure affects this process and uncover a previously unknown effect of the T6SS. Given the diversity of intestinal microbial communities, we believe these findings represent a valuable contribution to the understanding of the effects of the microbiome on host immunity.
STAR METHODS
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Bacterial strains and culture conditions
All Drosophila symbiotic bacterial strains were isolated from wild type lab flies in the Foley lab at the University of Alberta. Lactobacillus plantarum KP (DDBJ/EMBL/GenBank chromosome 1 accession CP013749 and plasmids 1–3 for accession numbers CP013750, CP013751, and CP013752, respectively), Lactobacillus brevis EF (DDBJ/EMBL/GeneBank accession LPXV00000000), and Acetobacter pasteurianus AD (DDBJ/EMBL/GeneBank accession LPWU00000000). Lactobacillus plantarum KP, Lactobacillus brevis EF, and Acetobacter pasteurianus AD have previously been described (Fast et al., 2018b; Petkau et al., 2016). Lactobacillus plantarum was grown in MRS broth (Sigma Lot: BCBS2861V) at 29°C for 24hours. Lactobacillus brevis was grown in MRS broth at 29°C for 48hours. Acetobacter pasteurianus was grown in MRS broth at 29°C with shaking for 48hours. Vibrio cholerae C6706, C6706ΔvasK, and C6706ΔvipA have previously been described (Pukatzki et al., 2006; Zheng et al., 2011). Vibrio strains were grown in Lysogeny Broth (LB) (1% tryptone, 0.5% yeast extract, 0.5% NaCl) at 37°C with shaking in the presence of 100 μg/ml streptomycin. Erwinia carotovora carotovora15 (Basset et al., 2000) was grown in LB (Difco Luria Broth Base, Miller. BD, DF0414-07-3) medium at 29°C with shaking for 24hours. Specific details and procedures are indicated below.
Drosophila stocks and rearing
All fly stocks were maintained at either 18°C or 25°C on standard Bloomington cornmeal medium (Lakovaara, 1969). Standard cornmeal medium: 225g agar, 2850g yellow cornmeal, 675g yeast, 390g soy flour, 3L light corn syrup, 39L water, and 188ml propionic acid. Fresh food was prepared weekly. All experimental flies were adult virgin females. Fly lines used in this study were w; upd2_CB-GAL4, UAS-mCD8:: GFP; (Zhai et al., 2018), w; esg-Gal4, tub-Gal80TS, UAS-GFP; (referred to as esgts, (Micchelli and Perrimon, 2006), w1118, Oregon-R (Bloomington 25211), and w; esg-Gal4, tub-Gal80TS, UAS-CFP, Su(H)-GFP;.
To make germ free flies by antibiotic treatment, freshly eclosed adult flies were raised on autoclaved standard medium that was supplemented with an antibiotic solution just prior pouring food into vials (100 g/ml ampicillin (Sigma BCBK5679V), 100 g/ml metronidazole (Sigma SLBG3633V), 50 g/ml vancomycin (Sigma 057M4022V) dissolved in 50% ethanol, and 100 g/ml neomycin (Sigma 071M0117V) dissolved in water) (Ryu et al., 2008). Conventionally reared counterparts were raised on autoclaved standard cornmeal medium.
To generate axenic flies, embryos were laid on apple juice plates over a 16-h period and collected. The following steps were performed in a sterile tissue culture hood. Embryos were rinsed from the plate with sterile phosphate-buffered saline (PBS). Embryos were placed in a in a 10% solution of 7.4% sodium hypochlorite (Clorox 02408961) for 2.5 minutes, then placed into fresh 10% sodium hypochlorite solution for 2.5 minutes, and then washed with 70% ethanol for 1 minute. Embryos were then rinsed 3 times with sterile water, placed onto sterile food, and maintained at 25°C in a sterilized incubator (Koyle et al., 2016). Prior to infection or symbiont association, microbial elimination from adult flies was confirmed for every vial of axenic or germ-free flies by plating whole-fly homogenates on agar plates permissive for the growth of Lactobacillus and Acetobacter.
METHOD DETAILS
Generation of gnotobiotic Drosophila
Virgin females were raised on antibiotic-supplemented fly food for 5 days at 25°C with 12/12 hour dark/light cycles. On day 5 of antibiotic treatment, a fly from each vial was homogenized in MRS broth and plated on MRS and GYC agar plates to ensure eradication of the microbiome. Flies were starved in sterile empty vials for 2 h prior to bacterial association. For mono-associations, the optical density at 600 nm (OD600) of bacterial liquid cultures was measured and then the culture was spun down and resuspended in 5% sucrose in PBS to a final OD600 of 50. For poly-associations, bacterial cultures of A. pasteurianus, L. brevis, and L. plantarum were prepared to an OD600 of 50 in 5% sucrose in PBS as described above. The bacterial cultures were then mixed at a 1:1:1 ratio. For all bacterial associations, 12 flies/vial were associated with 1 ml of bacterial suspension on autoclaved cotton plugs (Fisher Scientific Canada, 14127106) in sterile fly vials. Flies were fed the bacteria-sucrose mixture for 16 h at 25°C and then flipped onto autoclaved food and raised for 5 days at 29°C. Conventionally reared control flies were given mock associations of 1 ml of 5% sucrose in sterile PBS for 16 h at 25°C. To ensure bacterial association, a sample fly from every vial was homogenized in MRS broth and plated on MRS 1 day prior to infection.
Immunofluorescence
Flies were washed with 95% ethanol and dissected in PBS to isolate adult intestines. Guts were fixed for 1hour at room temperature in 8% formaldehyde in PBS. Guts were rinsed in PBS for 20 minutes at room temperature and blocked overnight in PBT + 3% bovine serum albumin (BSA) (Sigma-Aldrich A3059–10G) (PBS, 0.2% Triton-X) at 4°C. Guts were stained overnight at 4°C in PBT + 3% BSA with appropriate primary antibodies, washed with PBT and stained for 1 hour at room temperature with appropriate secondary antibodies. Guts were rinsed with PBT and then stained with DNA dye for 10 minutes at room temperature. Guts were then rinsed in PBT and a final wash in PBS. Guts were mounted on slides in Fluoromount (Sigma-Aldrich F4680), and R4/R5 region of the posterior midgut was visualized. For sagittal sections, the posterior midgut was excised from dissected whole guts and imbedded in clear frozen section compound (VWR, 95057–838). Guts were cryosectioned in 10μm sections at the Alberta Diabetes Institute Histocore at the University of Alberta. Sectioned guts were fixed in 4% formaldehyde for 20 minutes at room temperature, rinsed with PBS, and then blocked overnight at 4°C in 5% normal goat serum, 1% BSA, and 0.1% tween-20. Sections were rinsed in 1% BSA and 0.1% tween-20, and then stained for 1 hour at room temperature with primary antibodies in blocking buffer. Samples were rinsed and then stained for 1 hour at room temperature with appropriate secondary antibodies and nuclear stain, followed by a final rinse in blocking buffer. All guts were visualized with a spinning disk confocal microscope (Quorum WaveFX; Quorum Technologies Inc.). The primary antibodies used in this study were as follows: anti-PH3 (1:1000, Millipore Sigma (Ser10), 06–570), anti-GFP (1:1000, ThermoFisher, G10362), anti-myospheroid (1:100, CF.6G11 was deposited to the DSHB by Brower, D. DSHB Hybridoma Product CF.6G11). The secondary antibodies used in this study were goat anti-rabbit Alexa Fluor 488 (1:1000, ThermoFisher, A-11008) and goat anti-mouse Alexa Fluor 568 (1:1000, ThermoFisher, A-11004). DNA stains used in this study were Hoechst 33258 (1:500, Molecular Probes Life Technologies, 02C1–2) and DRAQ5 (1:400, Invitrogen, 508DR0200G).
Oral infection
All infections in this study were administered orally. Virgin female flies were separated from male flies after eclosion and placed on autoclaved standard Bloomington food for 5 days at 29°C without flipping. Flies were starved 2 hours prior to infection. For Vibrio infections, V. cholerae was grown on LB plates (1% tryptone, 0.5% yeast extract, 0.5% NaCl, 1.5% agar) at 37°C in the presence of 100 μg/ml streptomycin (Sigma SLBK5521V). Colonies were suspended in LB broth and diluted to a final OD600 of 0.125. For each infection group, groups twelve flies were placed in four vials containing one third of a cotton plug soaked with 3ml of sterile LB (Mock) or with LB containing V. cholerae. For infection with Erwinia, Ecc15 was grown in medium at 29°C with shaking for 24hours and gathered by centrifugation. The pellet was then re-suspended in the residual LB, and 1ml of the suspension was pipetted onto a thin slice of a cotton plug at the bottom of a sterile fly vial. For all infections in this study all flies were kept on their respective infections for 24hours.
Progenitor cell isolation and RNA extraction
IPC isolation by fluorescence activated cell sorting (FACS) was adapted from (Dutta et al., 2013). In brief, three biological replicates consisting of 100 fly guts per replicate with the malpighian tubules and crop removed were dissected into diethyl pyrocarbonate (DEPC) PBS and placed on ice. Guts were dissociated with 1mg/ml of elastase at 27°C with gentle shaking and periodic pipetting for 1hour. IPCs were sorted based on GFP fluorescence and size with a BD FACSAria IIIu. All small GFP positive cells were collected into a tube containing DEPC PBS. Cells were pelleted at 500G for 20 minutes and then resuspended in 500μl of Trizol (ThermoFisher 155596026). Samples were stored at −80°C until all samples from each group were collected. RNA was isolated via a standard Trizol chloroform extraction. Purified RNA was sent on dry ice to the Lunenfeld-Tanenbaum Research Institute (Toronto, Canada) for library construction and sequencing. The sample quality was evaluated using Agilent Bioanalyzer 2100. TaKaRa SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing was used to prepare full length cDNA. The quality and quantity of the purified cDNA was measure with Bioanalyzer and Qubit 2.0. Libraries were sequenced on the Illumina HiSeq3000 platform. For RNA-sequencing of whole guts, RNA was extracted in biological triplicate consisting of 10 dissected whole guts per replicate. RNA was purified by standard TRIZOL chloroform protocol. Purified RNA was sent on dry ice to Novogene (California, USA) for poly-A pulling, library construction and sequencing with Illumina Platform PE150 (NOVAseq 600). The sample quality was evaluated before and after library construction using an Agilent Bioanalyzer 2100.
Quantification of cells per gut area.
Mounted whole guts were loaded on a spinning disk confocal microscope (Quorum WaveFX; Quorum Technologies Inc.) for visualization. The R4-R5 region of posterior midgut of each sample was located by identifying the midgut hindgut transition and moving 1–2 frames anterior from the attachment site of the malpighian tubules. The top and bottom of the intestine were located and marked. Guts were then imaged as z-slices through the depth of the entire tissue. Images were acquired using Velocity Software (Quorum Technologies). All intestines damaged by the dissection process were excluded from quantification. The collected z-slices were split into individual fluorescent channels and compressed into single images with Fiji software (Schindelin et al., 2012). To quantify cells, compressed images spanning the width of the intestine were inverted and cells were counted manually by marking each cell with a colored dot. In cases were multiple cells were in close proximity to one another, the nuclear channel was used to identify the number of nuclei present within the cluster. Area of the gut was measured by tracing the intestinal outline in the nuclear channel. The outlined area was then measured with the measure analyzation tool in Fiji, as outlined previously (Petkau et al., 2017). Quantification of cells from figures 1 and 5 were reanalyzed in a double-blinded study to confirm the findings.
Read processing, alignment, differential expression, and GO analysis
For RNAseq studies, we obtained on average 30 million reads per biological replicate. We used FASTQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/, version 0.11.3) to evaluate the quality of raw, paired-end reads, and trimmed adaptors and reads of less than 36 base pairs in length from the raw reads using Trimmomatic (version 0.36) (Bolger et al., 2014). HISAT2 ((version 2.1.0) (Kim et al., 2015) was used to align reads to the Drosophila transcriptome- bdgp6 (https://ccb.jhu.edu/software/hisat2/index.shtml), and converted the resulting BAM files to SAM flies using Samtools (version 1.8) (Li et al., 2009). Converted files were counted with Rsubread (version 1.24.2) (Liao et al., 2013) and loaded into EdgeR (McCarthy et al., 2012; Robinson et al., 2010). In EdgeR, genes with counts less than 1 count per million were filtered and libraries normalized for size. Normalized libraries were used to call genes that were differentially expressed among treatments. For IPC RNA-seq, genes with P-value < 0.05 were defined as differentially expressed genes. For whole gut RNA-seq, Genes with P-value < 0.01 and FDR < 0.01 were defined as differentially expressed genes Principle component analysis was performed on normalized libraries using Factoextra (version 1.0.5) (Alboukadel and Mundt, 2017), and Gene Ontology enRIchment anaLysis and visuaLizAtion tool (GOrilla) was used to determine Gene Ontology (GO) term enrichment (Eden et al., 2009). Specifically, differentially expressed genes were compared in a two-list unraked comparison to all genes output from edgeR as a background set. Redundant GO terms were removed.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis and data visualization
All graphs, plots, Venn diagrams, and GO-term lists were constructed using R (version 3.5.1) via R-studio (version 1.1.463) with ggplot2 (version 3.1.1). All figures were assembled using Adobe Illustrator. All statistical analysis was completed with R. Normality of data was determined by Bartlett test for equal variances. For normal data, one-way Analysis of Variance (ANOVA) was used to determine overall statistical difference and a Tukey’s test for Honest Significant Differences was used for multiple comparisons. For non-normal data, a Kruskal-Wallis test was used to determine overall statistical difference and pairwise Willcoxon tests with a Bonferroni correction for multiple comparisons was used for multiple comparisons. Details of the specific test used for each data panel can be found in the table below each panel. Statistical significance was set at p ≤ 0.05.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful for CB>mCD8:: GFP flies provided by Dr. Bruno Lemaitre, esg[ts]>GFP flies provided by Dr. Bruce Edgar, and esg[ts]>CFP, Su(H)-GFP flies provided by Dr. Lucy O’Brien. Vibrio cholera C6706 was provided by Dr. John Mekalanos and Ecc15 was provided by Dr. Nicolas Buchon. We acknowledge microscopy support from Dr. Stephen Ogg and Greg Plummer at the Cell Imaging Centre at the University of Alberta. We acknowledge the FACS support provided by Dr. Aja Rieger at the Flow Cytometry Facility at University of Alberta. We acknowledge Kin Chan at the Network Biology Collaborative Centre (nbcc.lunenfeld.ca) for the IPC RNA-Seq service. We are grateful to Dr. Andrew Simmonds for critical reading of the manuscript. This research was funded by grants from the Canadian Institutes of Health Research to EF (MOP77746) and to SP (MOP 137106). D.F, A.G, and B.K were supported by Nation Science and Engineering Research Council scholarships. M.F was supported by Alberta Innovates Technology Futures scholarship.
Footnotes
DECLARATION OF INTEREST
The authors declare no competing interests
Data availability
Gene expression data have been submitted to the NCBI GEO database (GSE136069).
REFERENCES
- Alboukadel K, and Mundt F (2017). factoextra: Extract and Visualize the Results of Multivariate Data Analyses
- Amcheslavsky A, Jiang J, and Ip YT (2009). Tissue damage-induced Intestinal stem cell division in Drosophila. Cell Stem Cell 4, 49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey AM, and Posakony JW (1995). Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev 9, 2609–2622. [DOI] [PubMed] [Google Scholar]
- Basset A, Khush RS, Braun A, Gardan L, Boccard F, Hoffmann JA, and Lemaitre B (2000). The phytopathogenic bacteria Erwinia carotovora infects Drosophila and activates an immune response. Proc. Natl. Acad. Sci 97, 3376–3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkey CD, Blow N, and Watnick PI (2009). Genetic analysis of Drosophila melanogaster susceptibility to intestinal Vibrio cholerae infection. Cell. Microbiol 11, 461–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingle LE, Bailey CM, and Pallen MJ (2008). Type VI secretion: a beginner’s guide. Curr. Opin. Microbiol 11, 3–8. [DOI] [PubMed] [Google Scholar]
- Blow NS, Salomon RN, Garrity K, Reveillaud I, Kopin A, Jackson FR, and Watnick PI (2005). Vibrio cholerae infection of Drosophila melanogaster mimics the human disease cholera. PLoS Pathog 1, 0092–0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfini A, Liu X, and Buchon N (2016). From pathogens to microbiota: How Drosophila intestinal stem cells react to gut microbes. Dev. Comp. Immunol 64, 22–38. [DOI] [PubMed] [Google Scholar]
- Bosco-Drayon V, Poidevin M, Boneca IG, Narbonne-Reveau K, Royet J, and Charroux B (2012). Peptidoglycan sensing by the receptor PGRP-LE in the Drosophila gut induces immune responses to infectious bacteria and tolerance to microbiota. Cell Host Microbe 12, 153–165. [DOI] [PubMed] [Google Scholar]
- Bouskra D, Brézillon C, Bérard M, Werts C, Varona R, Boneca IG, and Eberl G (2008). Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature 456, 507–510. [DOI] [PubMed] [Google Scholar]
- Broderick NA, and Lemaitre B (2012). Gut-associated microbes of Drosophila melanogaster. Gut Microbes 3, 307–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick NA, Buchon N, and Lemaitre B (2014). Microbiota-induced changes in Drosophila melanogaster host gene expression and gut morphology. MBio 5, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchon N, Broderick NA, Poidevin M, Pradervand S, and Lemaitre B (2009a). Drosophila intestinal response to bacterial infection: activation of host defense and stem cell proliferation. Cell Host Microbe 5, 200–211. [DOI] [PubMed] [Google Scholar]
- Buchon N, Broderick N, Chakrabarti S, and Lemaitre B (2009b). Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes Dev 23, 2333–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchon N, Broderick NA, Kuraishi T, and Lemaitre B (2010). Drosophila EGFR pathway coordinates stem cell proliferation and gut remodeling following infection. BMC Biol 8, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S, Liehl P, Buchon N, and Lemaitre B (2012). Infection-induced host translational blockage Inhibits immune responses and epithelial renewal in the Drosophila gut. Cell Host Microbe 12, 60–70. [DOI] [PubMed] [Google Scholar]
- Cronin SJF, Nehme NT, Limmer S, Liegeois S, Pospisilik JA, Schramek D, Leibbrandt A, Simoes R. de M., Gruber S, Puc U, et al. (2009). Genome-wide RNAi screen identifies genes involved in intestinal pathogenic bacterial infection. Science 325, 340–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, and Chaudhuri K (2003). Identification of a unique IAHP (IcmF associated homologous proteins) cluster in Vibrio cholerae and other proteobacteria through in silico analysis . Silico Biol 3 287–300. [PubMed] [Google Scholar]
- Douglas AE (2018). The Drosophila model for microbiome research. Lab Anim. (NY) 47, 157–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta D, Xiang J, Edgar BA, Dutta D, Xiang J, and Edgar BA (2013). RNA Expression Profiling from FACS-Isolated Cells of the Drosophila Intestine. In Current Protocols in Stem Cell Biology, (Hoboken, NJ, USA: John Wiley & Sons, Inc.), pp. 2F.2.1–2F.2.12. [DOI] [PubMed] [Google Scholar]
- Dutta D, Dobson AJ, Houtz PL, Gläßer C, Revah J, Korzelius J, Patel PH, Edgar BA, and Buchon N (2015). Regional cell-specific transcriptome mapping reveals regulatory complexity in the adult Drosophila midgut. Cell Rep 12, 346–358. [DOI] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, and Yakhini Z (2009). GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fast D, Kostiuk B, Foley E, and Pukatzki S (2018a). Commensal pathogen competition impacts host viability. Proc. Natl. Acad. Sci. U. S. A 115, 7099–7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fast D, Duggal A, and Foley E (2018b). Monoassociation with Lactobacillus plantarum disrupts intestinal homeostasis in adult Drosophila melanogaster. MBio 9, e01114–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould AL, Zhang V, Lamberti L, Jones EW, Obadia B, Korasidis N, Gavryushkin A, Carlson JM, Beerenwinkel N, and Ludington WB (2018). Microbiome interactions shape host fitness. Proc. Natl. Acad. Sci 115, E11951–E11960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Driver I, and Ohlstein B (2013). Injury-induced BMP signaling negatively regulates Drosophila midgut homeostasis. J. Cell Biol 201, 945–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha E-M, Oh C-T, Bae YS, and Lee W-J (2005). A direct role for dual oxidase in Drosophila gut immunity. Science (80-.) 310, 847–850. [DOI] [PubMed] [Google Scholar]
- Hang S, Purdy AE, Robins WP, Wang Z, Mandal M, Chang S, Mekalanos JJ, and Watnick PI (2014). The acetate switch of an intestinal pathogen disrupts host insulin signaling and lipid metabolism. Cell Host Microbe 16, 592–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Frutos R. de L., Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, and Littman DR (2008). Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4, 337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Patel PH, Kohlmaier A, Grenley MO, McEwen DG, and Edgar BA (2009). Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell 137, 1343–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Grenley MO, Bravo M-J, Blumhagen RZ, and Edgar BA (2011). EGFR/Ras/MAPK signaling mediates adult midgut epithelial homeostasis and regeneration in Drosophila. Cell Stem Cell 8, 84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RM, Luo L, Ardita CS, Richardson AN, Kwon YM, Mercante JW, Alam A, Gates CL, Wu H, Swanson PA, et al. (2013). Symbiotic Lactobacilli stimulate gut epithelial proliferation via Nox-mediated generation of reactive oxygen species. EMBO J 32, 3017–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A, Kostiuk B, Rogers A, Teschler J, Pukatzki S, and Yildiz FH (2017). Rules of engagement: the type VI secretion system in Vibrio cholerae. Trends Microbiol 25, 267–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamareddine L, Wong ACN, Vanhove AS, Hang S, Purdy AE, Kierek-Pearson K, Asara JM, Ali A, Morris JG Jr, and Watnick PI (2018). Activation of Vibrio cholerae quorum sensing promotes survival of an arthropod host. Nat. Microbiol 3, 243–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, and Salzberg SL (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyle ML, Veloz M, Judd AM, Wong AC-N, Newell PD, Douglas AE, and Chaston JM (2016). Rearing the fruit fly Drosophila melanogaster under axenic and gnotobiotic conditions. J. Vis. Exp 113, e54219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakovaara S (1969). Malt as a culture medium for Drosophila species. Drosoph. Inf. Serv 44, 128. [Google Scholar]
- Lertpiriyapong K, Gamazon ER, Feng Y, Park DS, Pang J, Botka G, Graffam ME, Ge Z, and Fox JG (2012). Campylobacter jejuni type VI secretion system: roles in adaptation to deoxycholic acid, host cell adherence, invasion, and in vivo colonization. PLoS One 7, e42842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2013). The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res 41, e108–e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan SL, Thomas J, Yan J, Baker RP, Shields DS, Xavier JB, Hammer BK, and Parthasarathy R (2018). The Vibrio cholerae type VI secretion system can modulate host intestinal mechanics to displace gut bacterial symbionts. Proc. Natl. Acad. Sci 115, E3779–E3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma AT, and Mekalanos JJ (2010). In vivo actin cross-linking induced by Vibrio cholerae type VI secretion system is associated with intestinal inflammation. Proc. Natl. Acad. Sci. U. S. A 107, 4365–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Storelli G, Mitchell M, and Leulier F (2015). Studying host–microbiota hutualism in Drosophila: Harnessing the power of gnotobiotic flies. Biomed. J 38, 285–293. [DOI] [PubMed] [Google Scholar]
- McCarthy DJ, Chen Y, and Smyth GK (2012). Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40, 4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micchelli CA, and Perrimon N (2006). Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature 439, 475–479. [DOI] [PubMed] [Google Scholar]
- Miguel-Aliaga I, Jasper H, and Lemaitre B (2018). Anatomy and physiology of the digestive tract of Drosophila melanogaster. Genetics 210, 357–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata ST, Kitaoka M, Brooks TM, McAuley SB, and Pukatzki S (2011). Vibrio cholerae requires the type VI secretion system virulence factor VasX to kill Dictyostelium discoideum. Infect. Immun 79, 2941–2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordoñez CL, Lory S, et al. (2006). A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312, 1526–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein B, and Spradling A (2006). The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature 439, 470–474. [DOI] [PubMed] [Google Scholar]
- Ohlstein B, and Spradling A (2007). Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential Notch Signaling. Science (80-.) 315, 988–992. [DOI] [PubMed] [Google Scholar]
- Peterfreund GL, Vandivier LE, Sinha R, Marozsan AJ, Olson WC, Zhu J, and Petkau K, Fast D, Duggal A, and Foley E (2016). Comparative evaluation of the genomes of three common Drosophila-associated bacteria. Biol. Open 5, 1305–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkau K, Ferguson M, Guntermann S, and Foley E (2017). Constitutive immune cctivity promotes tumorigenesis in Drosophila intestinal progenitor cells. Cell Rep 20, 1784–1793. [DOI] [PubMed] [Google Scholar]
- Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, and Mekalanos JJ (2006). Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U. S. A 103, 1528–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukatzki S, Ma AT, Revel AT, Sturtevant D, and Mekalanos JJ (2007). Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl. Acad. Sci. U. S. A 104, 15508–15513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repizo GD, Gagné S, Foucault-Grunenwald M-L, Borges V, Charpentier X, Limansky AS, Gomes JP, Viale AM, and Salcedo SP (2015). Differential role of the T6SS in Acinetobacter baumannii virulence. PLoS One 10, e0138265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, and Mougous JD (2011). Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475, 343–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AB, LeRoux M, Hathazi K, Agnello DM, Ishikawa T, Wiggins PA, Wai SN, and Mougous JD (2013). Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature 496, 508–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu J-H, Ha E-M, Oh C-T, Seol J-H, Brey PT, Jin I, Lee DG, Kim J, Lee D, and Lee W-J (2006). An essential complementary role of NF-κB pathway to microbicidal oxidants in Drosophila gut immunity. EMBO J 25, 3693–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu J-H, Kim S-H, Lee H-Y, Bai JY, Nam Y-D, Bae J-W, Lee DG, Shin SC, Ha E-M, and Lee W-J (2008). Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science 319, 777–782. [DOI] [PubMed] [Google Scholar]
- Samarkos M, Mastrogianni E, and Kampouropoulou O (2018). The role of gut microbiota in Clostridium difficile infection. Eur. J. Intern. Med 50, 28–32. [DOI] [PubMed] [Google Scholar]
- Sana TG, Flaugnatti N, Lugo KA, Lam LH, Jacobson A, Baylot V, Durand E, Journet L, Cascales E, and Monack DM (2016). Salmonella Typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proc. Natl. Acad. Sci. U. S. A 113, E5044–E5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz S, West TE, Boyer F, Chiang W-C, Carl MA, Hood RD, Rohmer L, Tolker-Nielsen T, Skerrett SJ, and Mougous JD (2010). Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog 6, e1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin M, Jones LO, Panteluk A, and Foley E (2019). Cell-specific regulation of intestinal immunity in Drosophila. BioRxiv [Google Scholar]
- Shin SC, Kim S-H, You H, Kim B, Kim AC, Lee K-A, Yoon J-H, Ryu J-H, and Lee W-J (2011). Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science (80-.) 334, 670–674. [DOI] [PubMed] [Google Scholar]
- Sommer F, Nookaew I, Sommer N, Fogelstrand P, and Bäckhed F (2015). Site-specific programming of the host epithelial transcriptome by the gut microbiota. Genome Biol 16, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutzmann S, and Blokesch M (2016). Circulation of a quorum-sensing-impaired variant of Vibrio cholerae strain C6706 masks important phenotypes. MSphere 1, e00098–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian A, and Jiang J (2014). Intestinal epithelium-derived BMP controls stem cell self-renewal in Drosophila adult midgut. Elife 3, e01857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troha K, Im JH, Revah J, Lazzaro BP, and Buchon N (2018). Comparative transcriptomics reveals CrebA as a novel regulator of infection tolerance in D. melanogaster. PLOS Pathog 14, e1006847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzou P, Ohresser S, Ferrandon D, Capovilla M, Reichhart J-M, Lemaitre B, Hoffmann JA, and Imler J-L (2000). Tissue-specific inducible expression of antimicrobial peptide genes in Drosophila surface epithelia. Immunity 13, 737–748. [DOI] [PubMed] [Google Scholar]
- Vanhove AS, Hang S, Vijayakumar V, Wong AC, Asara JM, and Watnick PI (2017). Vibrio cholerae ensures function of host proteins required for virulence through consumption of luminal methionine sulfoxide. PLOS Pathog 13, e1006428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Hang S, Purdy AE, and Watnick PI (2013). Mutations in the IMD pathway and mustard counter Vibrio cholerae suppression of intestinal stem cell division in Drosophila. MBio 4, e00337–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidman-Rémy A, Hervé M, Poidevin M, Pili-Floury S, Kim M-S, Blanot D, Oh B-H, Ueda R, Mengin-Lecreulx D, and Lemaitre B (2006). The Drosophila amidase PGRP-LB modulates the immune response to bacterial infection. Immunity 24, 463–473. [DOI] [PubMed] [Google Scholar]
- Zeidler MP, and Bausek N (2013). The Drosophila JAK-STAT pathway. JAK-STAT 2, e25353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Z, Boquete J-P, and Lemaitre B (2018). Cell-specific Imd-NF-κB responses enable simultaneous antibacterial immunity and intestinal epithelial cell shedding upon bacterial infection. Immunity 48, 897–910. [DOI] [PubMed] [Google Scholar]
- Zhao W, Caro F, Robins W, and Mekalanos JJ (2018). Antagonism toward the intestinal microbiota and its effect on Vibrio cholerae virulence. Science 359, 210–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Ho B, and Mekalanos JJ (2011). Genetic analysis of anti-amoebae and anti-bacterial activities of the type VI secretion system in Vibrio cholerae. PLoS One 6, e23876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Florescu S, Boettcher A-L, Luo L, Dutta D, Kerr G, Cai Y, Edgar BA, and Boutros M (2015). Dpp/Gbb signaling is required for normal intestinal regeneration during infection. Dev. Biol 399, 189–203. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Gene expression data have been submitted to the NCBI GEO database (GSE136069).


