Abstract
Fecal microbiota transplantation (FMT) of human fecal samples into germ-free (GF) mice is useful for establishing causal relationships between the gut microbiota and human phenotypes. However, due to the intrinsic differences between human and mouse intestines and the different diets of the two organisms, it may not be possible to replicate human phenotypes in mice through FMT; similarly, treatments that are effective in mouse models may not be effective in humans. In this study, we aimed to identify human gut microbes that undergo significant and consistent changes (i.e., in relative abundances) after transplantation into GF mice in multiple experimental settings. We collected 16S rDNA-seq data from four published studies and analyzed the gut microbiota profiles from 1713 human–mouse pairs. Strikingly, on average, we found that only 47% of the human gut microbes could be re-established in mice at the species level, among which more than 1/3 underwent significant changes (referred to as “variable taxa”). Most of the human gut microbes that underwent significant changes were consistent across multiple human–mouse pairs and experimental settings. Consequently, about 1/3 of human samples changed their enterotypes, i.e., significant changes in their leading species after FMT. Mice fed with a controlled diet showed a lower enterotype change rate (23.5%) than those fed with a noncontrolled diet (49.0%), suggesting a possible solution for rescue. Most of the variable taxa have been reported to be implicated in human diseases, with some recognized as the causative species. Our results highlight the challenges of using a mouse model to replicate human gut microbiota-associated phenotypes, provide useful information for researchers using mice in gut microbiota studies, and call for additional validations after FMT. An online database named FMT-DB is publicly available at http://fmt2mice.humangut.info/#/.
Keywords: Germ-free mice, Fecal microbiota transplantation, Gut microbe, Enterotype, 16S rDNA
Introduction
In recent years, it has been well established that alterations in the gut microbiota are linked to many aspects of human health and diseases [1], [2], [3], [4], [5], with many alterations playing causative roles. For example, microbiota may play fundamental roles in the induction, training, and function of the host immune system, and several studies have shown that gut microbes are associated with the occurrence and development of diseases [6], [7], [8]. However, establishing causal relationships between gut microbiota alterations and human phenotypes has proven to be difficult, despite advances in computational and experimental techniques [9].
Fecal microbiota transplantation (FMT) of human fecal samples (fresh or frozen) into germ-free (GF) mice is one of the few available methods to establish causal relationships between human phenotypes and altered gut microbiota [10], [11]. These humanized mice could be used to replicate human phenotypes at both physiological and molecular levels, to study the relative contribution of the respective microbiota to host dysfunctions or disease phenotypes, and to test the effects of perturbations of certain species (often by the addition of lab-cultured species into mice) on the phenotypes of interest [12], [13]. These humanized mice are thus also extremely valuable for finding possible intervention methods for human diseases. Numerous successful applications of such methods have been reported in the past few years [14], [15], [16], [17]. For example, recent studies have shown that the addition of Bifidobacterium longum, Collinsella aerofaciens, and/or Enterococcus faecium to GF mice receiving FMTs from nonresponding patients can greatly increase the efficacy of anti-programmed death-ligand 1 (anti-PD-L1) therapy [18]. Similarly, age-associated differences in IgA responses can be replicated in young GF mice, and those that receive FMTs from infants can be used to demonstrate the influence of genetic factors on the development of the gut microbiota and mucosal IgA responses [19].
However, engraftment of fecal microbial communities from human feces into GF mice results in only a partial resemblance to the donor microbiota [14], favoring phylotypes that get adapted to the recipient species, due to the genetic, behavioral, physiological, and anatomical differences between the guts of mice and humans [20], [21]. For example, mouse intestinal villi are taller than their human counterparts, and the intestinal pH of mice is lower than that of humans [22]. Additionally, mice have a large cecum, which is an important site for fermentation, while the human cecum is relatively small [20], [21]. Consequently, native gut microbes in mice and humans are vastly different. For example, a recent study has shown that only 4% of human and mouse gut microbes shared 95% identity and a coverage of 90% [10], [11], [20], [23]. These results are consistent with the fact that most pathogens are able to infect multiple hosts, but some are highly adapted to a single host [24].
Having become aware of the differences between human and mouse guts and their important implications, researchers have taken measures to reduce the impacts of these differences on FMT by using pilot experimental methods to select the most suitable model animals for studying certain bacteria and/or feeding mice with human food [25]. The use of other models has also been considered recently [26]. Many studies have shown that gut microbes can be (at least partially) influenced by diet [27]. For example, an increased abundance of Prevotella is associated with increased dietary fiber in humans [28]. These efforts are, unfortunately, not pain-free and call for a systematic analysis of the alterations in human fecal samples after transplantation into mice.
To address these issues, we conducted a systematic meta-analysis using published human-to-mouse FMT datasets. We collected 16S rDNA-seq data from 1713 human–mouse pairs from four published studies (Table 1) [19], [27], [29], [30]. These data contained fecal microbiota sequencing information of both the human (before FMT) and the corresponding recipient mouse (after FMT). We compared the changes in the relative abundances of the same species/genus in these human–mouse pairs and identified consistently changed species. These species had relative abundances higher than 0.1% and showed significant changes (with |Log2 median FC| > 1; FC, fold change) after FMT, which were abundantly present in a significant proportion of human–mouse pairs. In addition, we also identified variable species, i.e., those with significantly changed relative abundances after FMT. Our results would be informative to researchers who use (or plan to use) GF mice in their gut microbiota studies. In addition, our analysis also calls for additional validations of the species of interest after FMT, e.g., to check whether the significantly changed species in different phenotype groups are still significantly changed after FMT. To date, this type of validation has been mostly ignored.
Table 1.
List of studies matching our search criteria
| NCBI BioProject accession No. | No. of Human–mouse pairs | Phenotype of donors | Amplified target region | Sequencing platform | Read length (bp) | Published date | Ref. |
|---|---|---|---|---|---|---|---|
| PRJNA314018 | 420 | Obese and healthy | V3-16S rRNA | Ion Torrent | 200 | 2-Mar-2016 | [29] |
| PRJEB7604 | 161 | Obese and healthy | V4-16S rRNA | Illumina | 250 | 28-Oct-2014 | [30] |
| PRJEB11697 | 184 | Healthy | V4-16S rRNA | Illumina | 250 × 2 | 15-Mar-2016 | [19] |
| PRJEB15481 | 948 | Healthy | V4-16S rRNA | Illumina | 250 × 2 | 21-Dec-2016 | [27] |
Results
Less than 50% of human gut microbes at the species level could be re-established in mice after FMT
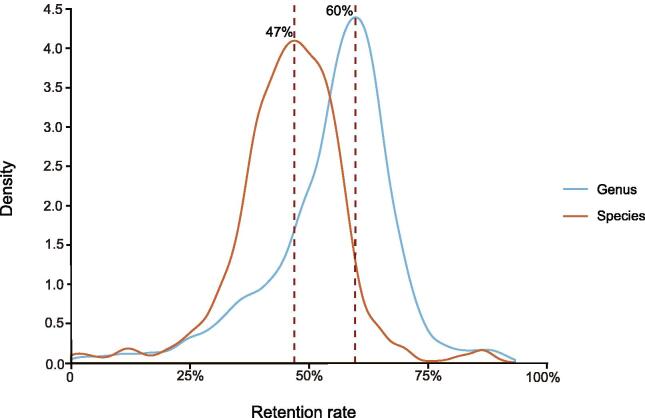
We collected data from a total of 1713 human-to-mouse FMT experiments and analyzed the relative microbe abundances in fecal samples from the human donors and the corresponding GF mouse recipients (see the Materials and methods for details). For each human–mouse pair, we computed the relative abundances before and after FMT. We first checked how many gut microbes could be re-established in mice after FMT. We limited our analysis to taxa that were supported by at least five sequencing reads in both samples of the human–mouse pairs. As shown in Figure 1, on average, only 47% of human gut microbes could be re-established in the mouse gut at the species level after FMT; at the genus level, 60% could be re-established. These numbers are consistent with some previous results [29] but significantly differ from others. For example, using 64 human–mouse pairs, Turnbaugh et al. [31] found that 50%–90% of human gut microbes at the genus level could be re-established in mice after FMT. However, these re-establishment rates were all based on a much smaller number of FMT experiments.
Figure 1.
47%(at the species level)and 60%(at the genus level)of human gut microbes could be re-established in the mouse gut after FMT
We limited our analysis to taxa that were supported by at least five sequencing reads in both human and mouse samples in each of the human–mouse pairs. FMT, fecal microbiota transplantation.
Over one-third of the re-established gut microbes are significantly and consistently changed in relative abundances after FMT into GF mice
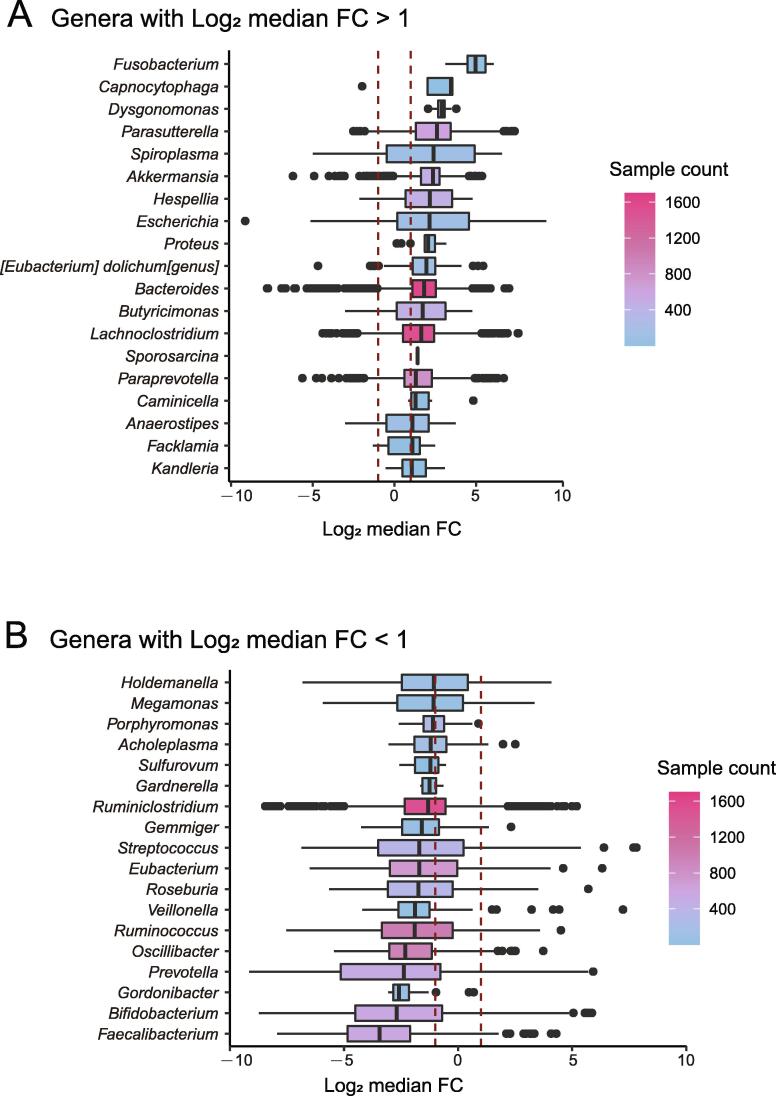
To capture consistent changes in gut microbes, we also required a species or genus to be present in at least five FMT pairs and under two experimental conditions with a minimal relative abundance of 0.1% in each sample (before and after FMT). We estimated that over 1/3 of the re-established gut microbes were significantly and consistently changed in relative abundances after FMT at both the genus and species levels. At the genus level, 38.9% (37/95) of the genera identified in our study were significantly changed after FMT (i.e., the median abundance changes were at least two-fold) (Figure 2). At the species level, 38.5% (84/218) of the species were significantly changed after FMT (Figure S1). We refer to these significantly changed taxonomic groups as “variable genera” and “variable species”, respectively, and refer to them together as “variable taxa”; conversely, we refer to others as “stable taxa”. As shown in Figure 3 and Figure S2, the variable taxa showed consistent changes across the experimental conditions.
Figure 2.
Variable genera show significant changes in relative abundances after FMT
A. Box plot showing variable genera with significantly increased changes in relative abundances after FMT. Log2 median FC > 1. B. Box plot showing variable genera with significantly decreased changes in relative abundances after FMT. Log2 median FC < −1. Log2 median FC, log2-transformed median fold change.
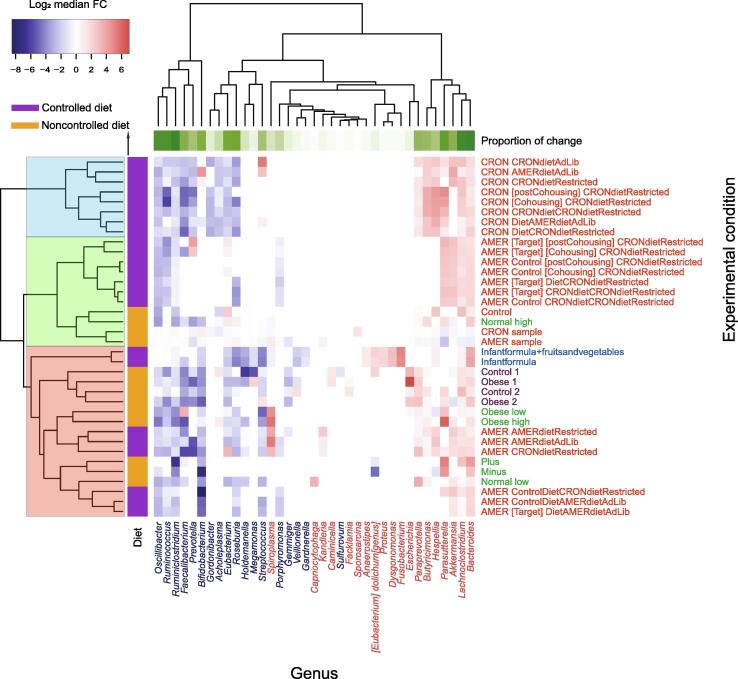
Figure 3.
Variable genera show consistent changes across the experimental conditions
A heatmap was plotted based on the Log2 median FC values of each variable genus under 33 experimental conditions (excluding 3 control groups) obtained from experimental condition groups of four NCBI BioProjects (Table 1). Experimental conditions from the same study are marked with the same color (labeled on the right). Genera with significantly increased and decreased abundances are marked in red and blue, respectively (labeled at the bottom). Row-side colors show whether controlled diets were used in the experiments, while column-side colors show the proportion of the experimental conditions in which the genus significantly changed.
Changes in enterotypes are relatively stable over time but are significantly affected by diet
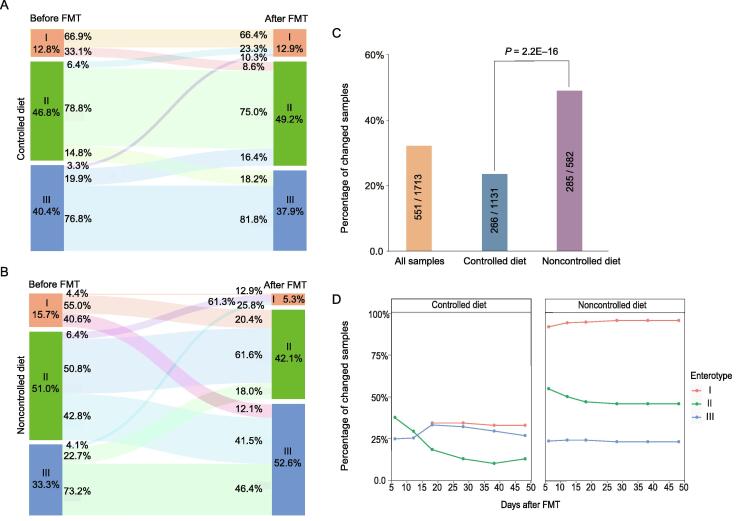
We also checked the changes in enterotypes after FMT. Human gut microbes can be classified into three enterotypes, each with a distinct leading species [32]. Our results showed that the mixed data from humans and mice could also be classified into three enterotypes, as shown in Figure S3. Although the concept of enterotypes has recently been hotly debated, they are nevertheless a useful approach to better understand complex biological problems [33]. Changing from one enterotype to another often indicates significant alterations in the overall gut microbiota profile [32]. We calculated the enterotypes for all the fecal samples before and after FMT using a method previously described (http://enterotype.embl.de/enterotypes.html). The overall classification of all samples is shown in Figure S3. Strikingly, we found that the enterotypes of 32.2% (551/1713) of the human samples changed after FMT. These results are consistent with the results that 1/3 of the re-established species and/or genera were significantly altered in relative abundances after FMT. As shown in Figure S4, 57.2% of the type I samples changed their enterotypes after FMT, followed by type II (31.2%) and type III (24.3%).
We next examined the factors that contribute to the enterotype changes. A previous study has shown that diet has a strong influence on intestinal microbes [34]. After dividing the human–mouse pairs into two groups according to whether mice were fed with a controlled diet (i.e., human food; see Materials and methods for details), we found that mice fed with a controlled diet showed a significantly lower enterotype change rate (23.5%) than those fed with a noncontrolled diet (49.0%) (Figure 4A–C). As shown in Figure 4B and C, in the noncontrolled diet group, about 95.6%, 49.2%, and 26.8% of the human samples of enterotypes I, II, and III changed their enterotypes after FMT into GF mice, respectively. These numbers decreased to 33.1%, 21.2%, and 23.2%, respectively, in the controlled diet group (Figure 4A), largely due to the decreased changes in the leading taxa of the respective enterotypes (Table S1). These results are consistent with the previous results that diet has a significant impact on the gut microbiota [34], [35], and may provide useful hints for the better retention of the human gut microbiota in humanized mice. However, we found that a controlled diet may induce additional variable taxa at both the species and genus levels (Figure S5), indicating the complex reactions of the mouse gut microbiota to human diets and the limited capacity of controlled diets for recipient mice.
Figure 4.
Changes in enterotypes after FMT to mice and the effects of diets
A. Enterotype changes in the “controlled diet” group. The recipient mice were fed with human food. B. Enterotype changes in the “noncontrolled diet” group. The recipient mice were fed with mouse food. C. Enterotype change rates of all samples, samples in the “controlled diet” group, and samples in the “noncontrolled diet” group. D. Enterotype change rates of samples in the “controlled diet” group (left) and “noncontrolled diet” group (right) at different days after FMT, respectively.
We also determined whether the gut microbiota profiles could be different at different time points after FMT into GF mice. Surprisingly, dividing the human–mouse pairs into subgroups according to the days after FMT, we found that the enterotype change rate was relatively stable over time, with the exception of human samples with initial enterotype II (the green lines in Figure 4D), which had the highest rate of enterotype change during the first 10–20 days. We found similar trends in GF mice fed with controlled and noncontrolled diets (Figure 4D). These results suggest a possible “shock” period immediately after transplantation of the fecal microbiota, followed by adaptation and a stable state [19], [36].
Species in different genera have different change trends
We next determined whether species in the same genus have similar change trends after FMT. We identified four typical patterns (patterns I–IV) among a total of 27 genera that contained multiple species. Pattern I consists of nine genera; in this group, most species and, frequently, the corresponding genera are “stable taxa” (Figure 5A, Figure S6A). Pattern II also consists of five stable genera (Figure 5B, Figure S6B). However, these genera include increased or both increased and decreased species and, sometimes, stable species; the overall abundances of the genera were not changed after FMT. Pattern III (Figure 5C, Figure S6C) and Pattern IV (Figure 5D, Figure S6D) consist of five increased and eight decreased “variable genera”, respectively; the included species are either increased/decreased variable species or stable species. These genera account for 33.33%, 18.52%, 18.52%, and 29.63% of the multi-species genera, respectively. These results indicate that there are distinct species preferences for genera between humans and mice.
Figure 5.
Species in the same genus may undergo distinct changes after FMT
Four typical patterns of species abundance change trends in a certain genus. A. Pattern I. B. Pattern II. C. Pattern III. D. Pattern IV.
Most variable taxa are linked to human health and diseases
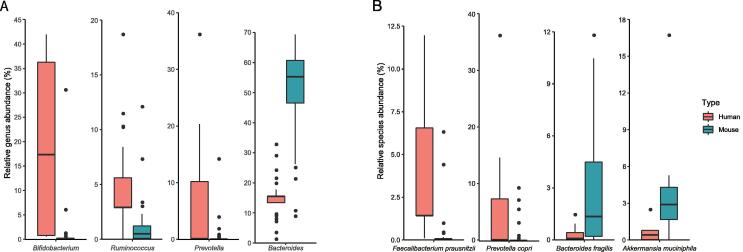
Strikingly, we found that most of the identified variable taxa have been reported to be implicated in various human diseases and/or can be used in disease intervention and treatment (Table 2, Table S1). Figure 6 shows eight selected variable taxa (including four genera and four species) that have been relatively well studied in human diseases [7], [16], [17], [18], [28], [37], [38], [39], [40], [41], [42], [43], [44], [45], [46], [47], [48], [49], [50], [51], [52], [53], [54], [55], [56], [57], [58], [59], [60], [61], [62], [63], [64], [65], [66], [67], [68], [69], [70], [71], [72], [73], [74], [75], [76], [77], [78]. Figures S7 and S8 shows all the variable genera and species that showed significant changes after FMT. Bifidobacterium has been reported to be implicated in obesity and can protect humans from enteropathogenic infection through the production of acetate [41]. In addition, Bifidobacterium, Akkermansia muciniphila, and other species have been shown to promote antitumor immunity and facilitate anti-PD-L1 efficacy [42]. Ruminococcus obeum has been shown to be able to competitively inhibit and control Vibrio cholerae infection [51]. Prevotella copri can induce insulin resistance, aggravate glucose intolerance, and augment circulating levels of branched-chain amino acids [43]. Faecalibacterium prausnitzii has anti-inflammatory effects on murine models, partly because its secreted metabolites are able to block NF-κB activation and IL-8 production [71].
Table 2.
Selected variable taxa and their responses to diet changes and associations with human diseases
| Variable taxon | Log2 median FC (all samples) | Log2 median FC (controlled diet group) | Associated disease | Refs. |
|---|---|---|---|---|
| Variable genus | ||||
| Bifidobacterium | −2.68 | −1.85 | Obese | [37], [38], [39] |
| Behcet’s disease | [37], [38], [40] | |||
| Autism spectrum disorders | [7], [37], [38] | |||
| Enteropathogenic infection | [37], [38], [41] | |||
| Cancer | [18], [37], [38], [42] | |||
| Prevotella | −2.37 | −2.12 | Type 2 diabetes | [28], [43] |
| Obese | [28], [44] | |||
| Rheumatoid arthritis | [45] | |||
| Autoinflammatory disease | [46] | |||
| Bechet’s disease | [40] | |||
| Chronic kidney disease | [47] | |||
| Ruminococcus | −1.9 | −1.86 | Obese | [48], [49], [50] |
| Inflammatory bowel disease | [48], [51], [52], [53] | |||
| Psoriatic arthritis | [53] | |||
| Autism spectrum disorders | [7] | |||
| Vibrio Cholerae infection | [51] | |||
| Cancer | [54] | |||
| Streptococcus | −1.7 | −1.87 | Pediatric asthma | [55] |
| Allergic diseases | [55] | |||
| Colorectal cancer | [56] | |||
| Autism spectrum disorders | [7] | |||
| Bacteroides | 1.83 | 1.83 | Abscess formation | [57] |
| Bacteremia | [57] | |||
| Autism spectrum disorders | [7] | |||
| Type2 diabetes | [43] | |||
| Cancer | [58] | |||
| Variable species | ||||
| Bacteroides fragilis | 3.00 | 3.31 | Cancer | [59], [60], [61], [62] |
| Type 2 diabetes | [62] | |||
| Collinsella aerofaciens | −1.52 | −1.52 | Inflammatory bowel disease | [63] |
| Rheumatic diseases | [51] | |||
| Obese | [64] | |||
| Prevotella copri | −2.67 | −2.33 | Type 2 diabetes | [43], [65] |
| Metabolic disorder | [43], [66], [67] | |||
| Rheumatoid arthritis | [67], [68], [69] | |||
| Cancer | [65] | |||
| Faecalibacterium prausnitzii | −3.44 | −2.9 | Type 2 diabetes | [17] |
| Metabolic disorder | [16], [70] | |||
| Gut dysbiosis | [15], [71] | |||
| Inflammatory bowel disease | [16] | |||
| Cancer | [72] | |||
| Akkermansia muciniphila | 2.36 | 2.41 | Metabolic disorder | [73], [74] |
| Obese | [74], [75] | |||
| Cancer | [76] | |||
| Type 2 diabetes | [17], [77] | |||
| Inflammatory bowel disease | [74] | |||
| Rheumatoid arthritis | [78] | |||
Note: FC, fold change.
Figure 6.
Representative variable genera and species showing significant changes after FMT
A. and B. Box plots show the relative abundances of representative variable genera (A) and species (B) in humans (before FMT) and mice (after FMT), respectively. Please see Table 2 and Table S1 for further details of their associations with human diseases.
These results highlight the challenges of using mouse models in gut microbiota studies: it is difficult not only to reproduce human phenotypes (at both the philological and molecular levels) in recipient mice if the suspected causal species is significantly decreased in abundance after FMT but also to transfer the treatment methods back to humans if the interventional species adapts much better to mice than to humans (e.g., Akkermansia muciniphila).
Discussion
Mice have been widely used in human disease studies; so far, FMT from humans to mice was one of the few available experimental methods for establishing causal relationships between altered microbial abundances in the human gut and diseases. However, due to the intrinsic differences between mouse and human guts, transplantation of fecal microbial communities from human feces into GF mice can only re-establish a portion of the donor microbiota, most of which have adapted to the recipient species [14]. Despite the widespread awareness of the differences between humans and mice and a handful of differential species identified in small-scale studies, a systematic analysis of the alterations of human gut microbiota after FMT into mice has yet to be conducted.
In this study, we collected and analyzed 1713 FMT experiments and identified relative microbial abundances before and after transplantation (i.e., human–mouse pairs). We focused on species that were abundantly present in more than 400 human–mouse pairs with a relative abundance higher than 0.1% and showing a significant change after FMT (with |Log2 median FC| > 1). By meeting these criteria, the changes are more likely to be consistent across experimental conditions. Strikingly, we found that over one-third of human gut microbes were significantly and consistently changed after FMT at both the species and genus levels, including leading taxa in enterotype analysis. Human feces transplanted into recipient mice fed with human food (the “controlled diet” group) showed a lower decrease in enterotype changes, suggesting a possible method for reducing such differences. However, a controlled diet may induce additional variable taxa at both the species and genus levels, indicating that the complex reactions of the mouse gut microbiota to human diets and a limited capacity of controlled diets for recipient mice.
Conclusion
Our results highlight the challenges of selecting mice as animal models in gut microbiota studies, and imply that it would be difficult to transfer findings in mice directly to humans due to the preferential adaptations of the variable taxa to mouse and human guts.
Strikingly, most of the variable taxa were implicated in human diseases. Our results, thus, are informative to researchers that use (or plan to use) GF mice in their gut microbiota and disease-association studies. In addition, our results also call for additional validations of the species of interest after FMT. For example, researchers should check whether significantly changed species in different human phenotype groups are still significantly changed after FMT. In other words, fecal samples from both human patients and healthy controls should be transplanted into GF mice; researchers should then be concerned not only whether the phenotypes of interest are replicated in mice but also whether the differentially abundant taxa in GF mice receiving patient and healthy feces are the same as those found in human samples. To date, this type of validation has been mostly ignored.
Materials and methods
Data
We performed an extensive search in the PubMed and NCBI Sequence Read Archive (SRA) databases to search for publications and/or deposited metagenomic sequencing data of human-to-mouse FMT experiments, which were published from January 1, 2014 to April 24, 2018. Because the number of published data of the relevant experiments was relatively small, we used two keywords, “microbiota transplanted into mice” and “human fecal microbiome mice”, to expand the range of our search results. We obtained a total of 58 studies, among which only 13 were human-to-mouse FMT experiments (Figure S9). We excluded experiments in which recipient mice were genetically modified, not GF, or treated with antibiotics; we also excluded experiments using RNA-seq or non-16S rDNA-seq and studies contained less than ten samples (Figure S9). Finally, we identified four studies that met our search criteria (Table 1), all of which used 16S rDNA-seq data to survey gut microbiota before and after FMT. These studies included a total of 1713 human–mouse pairs. All of the runIDs from the four NCBI BioProjects are provided in Table S2 and can be downloaded from NCBI. Mice were fed with 36 different types of diets. According to the feeding conditions, these diet groups were divided into two groups: the controlled diet group, which contained mice fed with human food, and the noncontrolled diet group, which contained other human–mouse pairs. The number of samples in each of the 36 diet groups ranged from 1 to 107, with median values of 20 and 49 for the controlled diet and noncontrolled diet groups, respectively (see Figure S10 for a density plot of the distribution of the sample sizes). We downloaded 16S rDNA-seq data from the NCBI SRA database using the command-line tool fastq-dump of the SRA tools (https://github.com/ncbi/sra-tools, accessed in July 2018). We obtained the related meta-data, including the human–mouse pairs, experimental conditions, and dates of sampling after FMT, from the corresponding publications and/or the NCBI SRA database.
Data processing and taxonomic assignment
We used FastQC (ver. 0.11.8; downloaded from http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) to evaluate the overall quality of the downloaded data, followed by Trimmomatic (ver. 0.35) [79] to remove vector sequences and low-quality bases. We directly used single-ended sequencing reads for subsequent analyses and merged the pair-ended reads using Casper (ver. 0.8.2) [80]. We then used Qiime (qiime2-2018.6) [81] to search for and remove possible chimeras.
We used MAPseq (ver. 1.2; July 16, 2017) [82] to analyze the obtained clean data and to assign taxonomic classification information to the reads, and used the full-length rRNA sequences downloaded from the official website of MAPseq (https://www.meringlab.org/software/mapseq/) as a reference dataset. It has previously been shown that at the genus level, MAPseq has a higher accuracy than other popular tools, such as Qiime [81] and Mothur [83]. MAPseq is also advantageous in our study compared with de novo clustering methods. The clean reads often have uneven ends after the removal of low-quality and/or sequencing-primer sequences. 16S rDNA sequences belonging to the same species/genus cannot be reliably clustered together; consequently, the retention rate, i.e., the proportion of human gut microbes that can be found in the recipient mice after FMT, will be significantly underestimated. This underestimation is also part of the reason why a few popular metagenomic databases, including EBI Metagenomics [84] and GMrepo [85], have adopted MAPseq as the main tool for taxonomy assignment. We removed reads with a cutoff value at the genus level of less than 0.4 (the combined score) as recommended by the authors of MAPseq [82]. We then calculated the relative abundances at both the genus and species levels for each sample, with total abundance values of 100%.
The workflow of our analysis pipeline is shown in Figure S11. All the human–mouse pairs used in this study along with their NCBI runIDs in the NCBI SRA database, enterotypes, experimental conditions, and diet types are listed in Table S2.
Statistical analysis
We uploaded all the processed data into R (ver. 3.5.1; downloaded from https://www.r-project.org; accessed in July 2018) and further analyzed them using the following packages: xlsx (ver. 0.6.1), dplyr (ver. 0.8.0.1), reshape2 (ver. 0.8.0), foreach (ver. 1.4.4), and doParallel (ver. 1.0.14); we used packages ggplot2 (ver. 3.2.1), netwokD3 (ver. 0.4), and heatmap3 (ver. 1.1.6) to visualize the results. We performed chisq.test and other statistical analyses using built-in functions. To focus more on abundant species/genus and avoid dramatic FCs in the calculation due to low-abundance species, we removed genera and species with relative abundances of less than 0.1% from subsequent analyses. To make the results more accurate, we also removed genera and species that were supported by less than 5 reads.
We used a web-based tool (http://enterotype.embl.de/enterotypes.html) to determine the enterotype for each sample by using the relative abundances of that sample as the input.
Code availability
All the processed data and R scripts are available at https://github.com/whchenlab/2019-liyz-human-to-mouse-FMT.
Data availability
The online database FMT-DB is publicly available at http://fmt2mice.humangut.info/#/.
CRediT author statement
Yanze Li: Formal analysis, Investigation, Writing - original draft, Writing - review & editing. Wenming Cao: Visualization, Investigation. Na L Gao: Methodology. Xing-Ming Zhao: Writing - original draft. Wei-Hua Chen: Conceptualization, Project administration, Writing - original draft, Writing - review & editing. All authors have read and approved the final manuscript.
Competing interests
The authors declare no conflict of financial interests.
Acknowledgments
We are very grateful to Qianhui Zhu (Institute of Microbiology, Chinese Academy of Sciences, China) and Chengwei Zhang (Institute of Microbiology, Chinese Academy of Sciences, China) for their help in downloading 16S rDNA-seq data. We thank Balakrishnan Subramanian (Huazhong University of Science and Technology, China) for providing technical support on building the online database. This work was partially supported by the National Key R&D Program of China (Grant Nos. 2018YFC0910502 and 2018YFC0910500 to WHC), the National Natural Science Foundation of China (Grant Nos. 61932008, 61772368, and 61572363), the Natural Science Foundation of Shanghai, China (Grant No. 17ZR1445600), and the Shanghai Municipal Science and Technology Major Project, China (Grant No. 2018SHZDZX01).
Handled by Yigang Tong
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation and Genetics Society of China.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gpb.2020.06.024.
Contributor Information
Xing-Ming Zhao, Email: xmzhao@fudan.edu.cn.
Wei-Hua Chen, Email: weihuachen@hust.edu.cn.
Supplementary data
The following are the Supplementary data to this article:
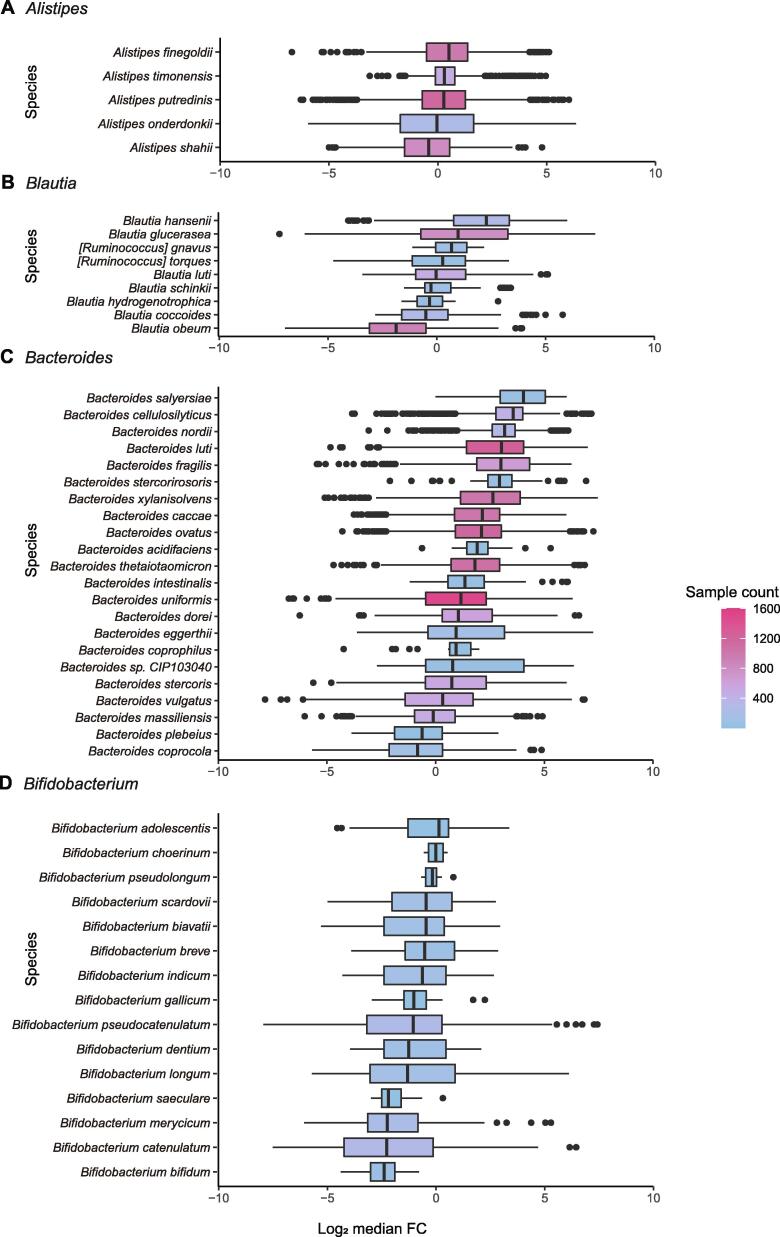
Variable species show significant changes in relative abundance after FMT A. Box plot showing variable species with significantly increased changes in relative abundances after FMT. Log2 median FC > 1. B. Box plot showing variable species with significantly decreased changes in relative abundances after FMT. Log2 median FC < −1.
Variable species show consistent changes across the experimental conditions
Enterotypes of all the human fecal samples sequenced before and after FMT A. Enterotype model showing with principal coordinates analysis (PCoA). B. Enterotype model showing with between-class analysis (BCA). Cluster labels 1, 2, and 3 correspond to enterotype I, enterotype II, and enterotype III, respectively.
Enterotype changes in all samples after FMT
Complex interactions of the mouse gut microbiota with human diets
Four types of multi-species genera with distinct change patterns at the species levels
Variable genera and their relative abundances in humans (before FMT) and mice (after FMT)
Variable species and their relative abundances in humans (before FMT) and mice (after FMT)
A graphical workflow showing our searching and excluding criteria
Density plot of the distributions of the sample sizes
Workflow of our analysis pipeline Boxes and circles indicate raw or processed data; arrows connecting these objects indicate the direction of the analyses, with the text labels corresponding to the tools used to perform such analyses. Detailed usage of these tools and their versions can be found in the “Materials and Methods” section.
A complete list of variable taxa and their responses to diet changes and associations with human diseases
A complete list of human–mouse pairs and their experimental conditions, enterotypes, and diet types
References
- 1.Clemente J.C., Ursell L.K., Parfrey L.W., Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmidt T.S.B., Raes J., Bork P. The human gut microbiome: from association to modulation. Cell. 2018;172:1198–1215. doi: 10.1016/j.cell.2018.02.044. [DOI] [PubMed] [Google Scholar]
- 3.Lee Y.K., Mazmanian S.K. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science. 2010;330:1768–1773. doi: 10.1126/science.1195568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu H., Esteve E., Tremaroli V., Khan M.T., Caesar R., Manneras-Holm L., et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med. 2017;23:850–858. doi: 10.1038/nm.4345. [DOI] [PubMed] [Google Scholar]
- 5.Wang J., Thingholm L.B., Skieceviciene J., Rausch P., Kummen M., Hov J.R., et al. Genome-wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat Genet. 2016;48:1396–1406. doi: 10.1038/ng.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belkaid Y., Hand T. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strati F., Cavalieri D., Albanese D., De Felice C., Donati C., Hayek J., et al. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome. 2017;5:24. doi: 10.1186/s40168-017-0242-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung H., Pamp S.J., Hill J.A., Surana N.K., Edelman S.M., Troy E.B., et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang D., Leung R.K., Guan W., Au W.W. Involvement of gut microbiome in human health and disease: brief overview, knowledge gaps and research opportunities. Gut Pathog. 2018;10:3. doi: 10.1186/s13099-018-0230-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wrzosek L., Ciocan D., Borentain P., Spatz M., Puchois V., Hugot C., et al. Transplantation of human microbiota into conventional mice durably reshapes the gut microbiota. Sci Rep. 2018;8:6854. doi: 10.1038/s41598-018-25300-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seekatz A.M., Aas J., Gessert C.E., Rubin T.A., Saman D.M., Bakken J.S., et al. Recovery of the gut microbiome following fecal microbiota transplantation. mBio. 2014;5:e00893-14. doi: 10.1128/mBio.00893-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hormannsperger G., Schaubeck M., Haller D. Intestinal microbiota in animal models of inflammatory diseases. ILAR J. 2015;56:179–191. doi: 10.1093/ilar/ilv019. [DOI] [PubMed] [Google Scholar]
- 13.Turnbaugh P.J., Hamady M., Yatsunenko T., Cantarel B.L., Duncan A., Ley R.E., et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arrieta M.C., Walter J., Finlay B.B. Human microbiota-associated mice: a model with challenges. Cell Host Microbe. 2016;19:575–578. doi: 10.1016/j.chom.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 15.Le Bastard Q., Ward T., Sidiropoulos D., Hillmann B.M., Chun C.L., Sadowsky M.J., et al. Fecal microbiota transplantation reverses antibiotic and chemotherapy-induced gut dysbiosis in mice. Sci Rep. 2018;8:619. doi: 10.1038/s41598-018-24342-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miquel S., Martín R., Lashermes A., Gillet M., Meleine M., Gelot A., et al. Anti-nociceptive effect of Faecalibacterium prausnitzii in non-inflammatory IBS-like models. Sci Rep. 2016;6:19399. doi: 10.1038/srep19399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin J., Li Y., Cai Z., Li S., Zhu J., Zhang F., et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 18.Matson V., Fessler J., Bao R., Chongsuwat T., Zha Y., Alegre M.L., et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359:104–108. doi: 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Planer J.D., Peng Y., Kau A.L., Blanton L.V., Ndao I.M., Tarr P.I., et al. Development of the gut microbiota and mucosal IgA responses in twins and gnotobiotic mice. Nature. 2016;534:263–266. doi: 10.1038/nature17940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hugenholtz F., de Vos W.M. Mouse models for human intestinal microbiota research: a critical evaluation. Cell Mol Life Sci. 2018;75:149–160. doi: 10.1007/s00018-017-2693-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen T.L., Vieira-Silva S., Liston A., Raes J. How informative is the mouse for human gut microbiota research? Dis Model Mech. 2015;8:1–16. doi: 10.1242/dmm.017400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McConnell E.L., Basit A.W., Murdan S. Measurements of rat and mouse gastrointestinal pH, fluid and lymphoid tissue, and implications for in-vivo experiments. J Pharm Pharmacol. 2008;60:63–70. doi: 10.1211/jpp.60.1.0008. [DOI] [PubMed] [Google Scholar]
- 23.Wang J., Kalyan S., Steck N., Turner L.M., Harr B., Künzel S., et al. Analysis of intestinal microbiota in hybrid house mice reveals evolutionary divergence in a vertebrate hologenome. Nat Commun. 2015;6:6440. doi: 10.1038/ncomms7440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baumler A., Fang F.C. Host specificity of bacterial pathogens. Cold Spring Harb Perspect Med. 2013;3:a010041. doi: 10.1101/cshperspect.a010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar R., Herold J.L., Taylor J., Xu J., Xu Y. Variations among Streptococcus gallolyticus subsp. gallolyticus strains in connection with colorectal cancer. Sci Rep. 2018;8:1514. doi: 10.1038/s41598-018-19941-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coelho L.P., Kultima J.R., Costea P.I., Fournier C., Pan Y., Czarnecki-Maulden G., et al. Similarity of the dog and human gut microbiomes in gene content and response to diet. Microbiome. 2018;6:72. doi: 10.1186/s40168-018-0450-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Griffin N.W., Ahern P.P., Cheng J., Heath A.C., Ilkayeva O., Newgard C.B., et al. Prior dietary practices and connections to a human gut microbial metacommunity alter responses to diet interventions. Cell Host Microbe. 2017;21:84–96. doi: 10.1016/j.chom.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kovatcheva-Datchary P., Nilsson A., Akrami R., Lee Y., De Vadder F., Arora T., et al. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of Prevotella. Cell Metab. 2015;22:971–982. doi: 10.1016/j.cmet.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 29.Zhang L., Bahl M.I., Roager H.M., Fonvig C.E., Hellgren L.I., Frandsen H.L., et al. Environmental spread of microbes impacts the development of metabolic phenotypes in mice transplanted with microbial communities from humans. ISME J. 2017;11:676–690. doi: 10.1038/ismej.2016.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goodrich J.K., Waters J.L., Poole A.C., Sutter J.L., Koren O., Blekhman R., et al. Human genetics shape the gut microbiome. Cell. 2014;159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turnbaugh P.J., Ridaura V.K., Faith J.J., Rey F.E., Knight R., Gordon J.I. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arumugam M., Raes J., Pelletier E., Le Paslier D., Yamada T., Mende D.R., et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Costea P.I., Hildebrand F., Arumugam M., Backhed F., Blaser M.J., Bushman F.D., et al. Enterotypes in the landscape of gut microbial community composition. Nat Microbiol. 2018;3:8–16. doi: 10.1038/s41564-017-0072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.David L.A., Maurice C.F., Carmody R.N., Gootenberg D.B., Button J.E., Wolfe B.E., et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu G.D., Chen J., Hoffmann C., Bittinger K., Chen Y.Y., Keilbaugh S.A., et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Staley C., Kaiser T., Beura L.K., Hamilton M.J., Weingarden A.R., Bobr A., et al. Stable engraftment of human microbiota into mice with a single oral gavage following antibiotic conditioning. Microbiome. 2017;5:87. doi: 10.1186/s40168-017-0306-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Esaiassen E., Hjerde E., Cavanagh J.P., Simonsen G.S., Klingenberg C., Ledeboer N.A. Bifidobacterium bacteremia: clinical characteristics and a genomic approach to assess pathogenicity. J Clin Microbiol. 2017;55:2234–2248. doi: 10.1128/JCM.00150-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Callaghan A., van Sinderen D. Bifidobacteria and their role as members of the human gut microbiota. Front Microbiol. 2016;7:925. doi: 10.3389/fmicb.2016.00925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salazar N., Dewulf E.M., Neyrinck A.M., Bindels L.B., Cani P.D., Mahillon J., et al. Inulin-type fructans modulate intestinal Bifidobacterium species populations and decrease fecal short-chain fatty acids in obese women. Clin Nutr. 2015;34:501–507. doi: 10.1016/j.clnu.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 40.Shimizu J., Kubota T., Takada E., Takai K., Fujiwara N., Arimitsu N., et al. Bifidobacteria abundance-featured gut microbiota compositional change in patients with Behcet’s disease. PLoS One. 2016;11:e0153746. doi: 10.1371/journal.pone.0153746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fukuda S., Toh H., Hase K., Oshima K., Nakanishi Y., Yoshimura K., et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 42.Sivan A., Corrales L., Hubert N., Williams J.B., Aquino-Michaels K., Earley Z.M., et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pedersen H.K., Gudmundsdottir V., Nielsen H.B., Hyotylainen T., Nielsen T., Jensen B.A., et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016;535:376–381. doi: 10.1038/nature18646. [DOI] [PubMed] [Google Scholar]
- 44.Hjorth M.F., Roager H.M., Larsen T.M., Poulsen S.K., Licht T.R., Bahl M.I., et al. Pre-treatment microbial Prevotella-to-Bacteroides ratio, determines body fat loss success during a 6-month randomized controlled diet intervention. Int J Obes (Lond) 2018;42:284. doi: 10.1038/ijo.2018.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pianta A., Arvikar S., Strle K., Drouin E.E., Wang Q., Costello C.E., et al. Evidence of the immune relevance of Prevotella copri, a gut microbe, in patients with rheumatoid arthritis. Arthritis Rheumatol. 2017;69:964–975. doi: 10.1002/art.40003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lukens J.R., Gurung P., Vogel P., Johnson G.R., Carter R.A., McGoldrick D.J., et al. Dietary modulation of the microbiome affects autoinflammatory disease. Nature. 2014;516:246–249. doi: 10.1038/nature13788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lun H., Yang W., Zhao S., Jiang M., Xu M., Liu F., et al. Altered gut microbiota and microbial biomarkers associated with chronic kidney disease. Microbiologyopen. 2019;8:e00678. doi: 10.1002/mbo3.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.La Reau A.J., Suen G. The Ruminococci: key symbionts of the gut ecosystem. J Microbiol. 2018;56:199–208. doi: 10.1007/s12275-018-8024-4. [DOI] [PubMed] [Google Scholar]
- 49.Le Chatelier E., Nielsen T., Qin J., Prifti E., Hildebrand F., Falony G., et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 50.Cann I., Bernardi R.C., Mackie R.I. Cellulose degradation in the human gut: Ruminococcus champanellensis expands the cellulosome paradigm. Environ Microbiol. 2016;18:307–310. doi: 10.1111/1462-2920.13152. [DOI] [PubMed] [Google Scholar]
- 51.Hsiao A., Ahmed A.M., Subramanian S., Griffin N.W., Drewry L.L., Petri W.A., et al. Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature. 2014;515:423–426. doi: 10.1038/nature13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu T.C., Stappenbeck T.S. Genetics and pathogenesis of inflammatory bowel disease. Annu Rev Pathol. 2016;11:127–148. doi: 10.1146/annurev-pathol-012615-044152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scher J.U., Ubeda C., Artacho A., Attur M., Isaac S., Reddy S.M., et al. Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis Rheumatol. 2015;67:128–139. doi: 10.1002/art.38892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gopalakrishnan V., Spencer C.N., Nezi L., Reuben A., Andrews M.C., Karpinets T.V., et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Johnson C.C., Ownby D.R. The infant gut bacterial microbiota and risk of pediatric asthma and allergic diseases. Transl Res. 2017;179:60–70. doi: 10.1016/j.trsl.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boleij A., van Gelder M.M., Swinkels D.W., Tjalsma H. Clinical importance of Streptococcus gallolyticus infection among colorectal cancer patients: systematic review and meta-analysis. Clin Infect Dis. 2011;53:870–878. doi: 10.1093/cid/cir609. [DOI] [PubMed] [Google Scholar]
- 57.Wexler H.M. Bacteroides: the good, the bad, and the nitty-gritty. Clin Microbiol Rev. 2007;20:593–621. doi: 10.1128/CMR.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vetizou M., Pitt J.M., Daillere R., Lepage P., Waldschmitt N., Flament C., et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Choi V.M., Herrou J., Hecht A.L., Teoh W.P., Turner J.R., Crosson S., et al. Activation of Bacteroides fragilis toxin by a novel bacterial protease contributes to anaerobic sepsis in mice. Nat Med. 2016;22:563–567. doi: 10.1038/nm.4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deng H., Li Z., Tan Y., Guo Z., Liu Y., Wang Y.e., et al. A novel strain of Bacteroides fragilis enhances phagocytosis and polarises M1 macrophages. Sci Rep. 2016;6:29401. doi: 10.1038/srep29401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gagnaire A., Nadel B., Raoult D., Neefjes J., Gorvel J.P. Collateral damage: insights into bacterial mechanisms that predispose host cells to cancer. Nat Rev Microbiol. 2017;15:109–128. doi: 10.1038/nrmicro.2016.171. [DOI] [PubMed] [Google Scholar]
- 62.Zakharzhevskaya N.B., Vanyushkina A.A., Altukhov I.A., Shavarda A.L., Butenko I.O., Rakitina D.V., et al. Outer membrane vesicles secreted by pathogenic and nonpathogenic Bacteroides fragilis represent different metabolic activities. Sci Rep. 2017;7:5008. doi: 10.1038/s41598-017-05264-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Owens B. Gut bacteria link to immunotherapy sparks interest. Nat Biotechnol. 2018;36:121–123. doi: 10.1038/nbt0218-121. [DOI] [PubMed] [Google Scholar]
- 64.de la Cuesta-Zuluaga J., Corrales-Agudelo V., Carmona J.A., Abad J.M., Escobar J.S. Body size phenotypes comprehensively assess cardiometabolic risk and refine the association between obesity and gut microbiota. Int J Obes (Lond) 2018;42:424–432. doi: 10.1038/ijo.2017.281. [DOI] [PubMed] [Google Scholar]
- 65.Thaiss C.A., Zmora N., Levy M., Elinav E. The microbiome and innate immunity. Nature. 2016;535:65–74. doi: 10.1038/nature18847. [DOI] [PubMed] [Google Scholar]
- 66.Honda K., Littman D.R. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535:75–84. doi: 10.1038/nature18848. [DOI] [PubMed] [Google Scholar]
- 67.Lloyd-Price J., Mahurkar A., Rahnavard G., Crabtree J., Orvis J., Hall A.B., et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature. 2017;550:61–66. doi: 10.1038/nature23889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bernard N.J. Prevotella copri associated with new-onset untreated RA. Nat Rev Rheumatol. 2014;10:2. doi: 10.1038/nrrheum.2013.187. [DOI] [PubMed] [Google Scholar]
- 69.Gut microbes linked to arthritis. Nature 2013;503:169.
- 70.Tremaroli V., Backhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 71.Lopez-Siles M., Duncan S.H., Garcia-Gil L.J., Martinez-Medina M. Faecalibacterium prausnitzii: from microbiology to diagnostics and prognostics. ISME J. 2017;11:841–852. doi: 10.1038/ismej.2016.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rossi O., van Berkel L.A., Chain F., Tanweer Khan M., Taverne N., Sokol H., et al. Faecalibacterium prausnitzii A2–165 has a high capacity to induce IL-10 in human and murine dendritic cells and modulates T cell responses. Sci Rep. 2016;6:18507. doi: 10.1038/srep18507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chelakkot C., Choi Y., Kim D.K., Park H.T., Ghim J., Kwon Y., et al. Akkermansia muciniphila-derived extracellular vesicles influence gut permeability through the regulation of tight junctions. Exp Mol Med. 2018;50:e450. doi: 10.1038/emm.2017.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dao M.C., Everard A., Aron-Wisnewsky J., Sokolovska N., Prifti E., Verger E.O., et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology. Gut. 2016;65:426–436. doi: 10.1136/gutjnl-2014-308778. [DOI] [PubMed] [Google Scholar]
- 75.Schneeberger M., Everard A., Gómez-Valadés A.G., Matamoros S., Ramírez S., Delzenne N.M., et al. Akkermansia muciniphila inversely correlates with the onset of inflammation, altered adipose tissue metabolism and metabolic disorders during obesity in mice. Sci Rep. 2015;5:16643. doi: 10.1038/srep16643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cani P.D., de Vos W.M. Next-generation beneficial microbes: the case of Akkermansia muciniphila. Front Microbiol. 2017;8:1765. doi: 10.3389/fmicb.2017.01765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Plovier H., Everard A., Druart C., Depommier C., Van Hul M., Geurts L., et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med. 2017;23:107–113. doi: 10.1038/nm.4236. [DOI] [PubMed] [Google Scholar]
- 78.Stoll M.L., Pierce M.K., Watkins J.A., Zhang M., Weiss P.F., Weiss J.E., et al. Akkermansia muciniphila is permissive to arthritis in the K/BxN mouse model of arthritis. Genes Immun. 2019;20:158–166. doi: 10.1038/s41435-018-0024-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kwon S., Lee B., Yoon S. CASPER: context-aware scheme for paired-end reads from high-throughput amplicon sequencing. BMC Bioinformatics. 2014;15:S10. doi: 10.1186/1471-2105-15-S9-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Matias Rodrigues J.F., Schmidt T.S.B., Tackmann J., von Mering C., Birol I. MAPseq: highly efficient k-mer search with confidence estimates, for rRNA sequence analysis. Bioinformatics. 2017;33:3808–3810. doi: 10.1093/bioinformatics/btx517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schloss P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B., et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mitchell A.L., Scheremetjew M., Denise H., Potter S., Tarkowska A., Qureshi M., et al. EBI Metagenomics in 2017: enriching the analysis of microbial communities, from sequence reads to assemblies. Nucleic Acids Res. 2018;46:D726–D735. doi: 10.1093/nar/gkx967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wu S., Sun C., Li Y., Wang T., Jia L., Lai S., et al. GMrepo: a database of curated and consistently annotated human gut metagenomes. Nucleic Acids Res. 2020;48:D545–D553. doi: 10.1093/nar/gkz764. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Variable species show significant changes in relative abundance after FMT A. Box plot showing variable species with significantly increased changes in relative abundances after FMT. Log2 median FC > 1. B. Box plot showing variable species with significantly decreased changes in relative abundances after FMT. Log2 median FC < −1.
Variable species show consistent changes across the experimental conditions
Enterotypes of all the human fecal samples sequenced before and after FMT A. Enterotype model showing with principal coordinates analysis (PCoA). B. Enterotype model showing with between-class analysis (BCA). Cluster labels 1, 2, and 3 correspond to enterotype I, enterotype II, and enterotype III, respectively.
Enterotype changes in all samples after FMT
Complex interactions of the mouse gut microbiota with human diets
Four types of multi-species genera with distinct change patterns at the species levels
Variable genera and their relative abundances in humans (before FMT) and mice (after FMT)
Variable species and their relative abundances in humans (before FMT) and mice (after FMT)
A graphical workflow showing our searching and excluding criteria
Density plot of the distributions of the sample sizes
Workflow of our analysis pipeline Boxes and circles indicate raw or processed data; arrows connecting these objects indicate the direction of the analyses, with the text labels corresponding to the tools used to perform such analyses. Detailed usage of these tools and their versions can be found in the “Materials and Methods” section.
A complete list of variable taxa and their responses to diet changes and associations with human diseases
A complete list of human–mouse pairs and their experimental conditions, enterotypes, and diet types
Data Availability Statement
The online database FMT-DB is publicly available at http://fmt2mice.humangut.info/#/.