Abstract
Prolonged periods of drought triggered by climate change hamper plant growth and cause substantial agricultural yield losses every year. In addition to drought, salinity is one of the major abiotic stresses that severely affect crop health and agricultural production. Plant responses to drought and salinity involve multiple processes that operate in a spatiotemporal manner, such as stress sensing, perception, epigenetic modifications, transcription, post-transcriptional processing, translation, and post-translational changes. Consequently, drought and salinity stress tolerance are polygenic traits influenced by genome-environment interactions. One of the ideal solutions to these challenges is the development of high-yielding crop varieties with enhanced stress tolerance, together with improved agricultural practices. Recently, genome-editing technologies, especially clustered regularly interspaced short palindromic repeats (CRISPR) tools, have been effectively applied to elucidate how plants deal with drought and saline environments. In this work, we aim to portray that the combined use of CRISPR-based genome engineering tools and modern genomic-assisted breeding approaches are gaining momentum in identifying genetic determinants of complex traits for crop improvement. This review provides a synopsis of plant responses to drought and salinity stresses at the morphological, physiological, and molecular levels. We also highlight recent advances in CRISPR-based tools and their use in understanding the multi-level nature of plant adaptations to drought and salinity stress. Integrating CRISPR tools with modern breeding approaches is ideal for identifying genetic factors that regulate plant stress-response pathways and for the introgression of beneficial traits to develop stress-resilient crops.
Key words: CRISPR/Cas, drought tolerance, genome editing, polygenic traits, salt tolerance, trait introgression
This review provides a comprehensive analysis of the latest studies and discoveries on plant responses unique to significant abiotic stresses; i.e., drought and salinity. The authors further describe novel insights into the combined use of CRISPR-based tools and modern breeding techniques that will expedite the identification of the genetic factors regulating plant stress-response pathways for crop improvement.
Introduction
Agricultural scientists are expected to face several challenges in the coming years. Some examples include the design of climate-resilient crop varieties, understanding the effects of plant-environment interactions on genetic adaptations, improving crop yields, linking plant genotype to phenotype, and developing efficient high-throughput plant phenotyping methods (Varshney et al., 2021a). Increasing human populations and climate change are the primary obstacles to meeting global food and energy demands proposed as sustainable development goals by the United Nations. Climate change triggers dramatic shifts in water availability and distribution via aberrations in seasonal precipitation, evaporation, and weather patterns; such factors cause imbalances in the salinity and nutrient availability of cultivated soils at global and regional levels (Cai et al., 2014; Konapala et al., 2020). The accretion of these factors gradually transforms once-fertile lands into problematic to barren lands. Ultimately, such events impede crop productivity and commercial agriculture around the world. In addition to climate change, several inanimate factors contribute to abiotic stress in plants, such as lack of water availability over a period of time, sudden or prolonged exposure to high or low temperatures, and higher salt and heavy metal content in the soil (Zafar et al., 2020). Changing rainy seasons will also alter the magnitude and recurrence of drought-like situations, significantly affecting agricultural productivity (Cai et al., 2014). Farmers lose more crop yields annually from drought alone than from all pathogens (Gupta et al., 2020).
Salinity stress ranks second to drought in terms of global constraints that limit crop productivity and land use for agriculture (Gong et al., 2020). About one-fifth of the irrigated land around the globe is estimated to be affected by higher salt accumulation (Morton et al., 2019). In a broad sense, water scarcity and soil salinization are inter-linked processes (Zhu, 2002). Salinization of naturally occurring non-saline soils increases because of irrigation practices and climate change-triggered rising sea levels or higher evaporation rates (Van Zelm et al., 2020). Moreover, the excessive use of chemicals and fertilizers has led to deterioration in soil health by increasing salt accumulation. Such problematic soils are increasing, with a minimum of 10% annual growth; to exacerbate this situation, about half of presently cultivated soils may become salinized by 2050 (Jamil et al., 2011). Overall, drought and salinity are major abiotic factors that severely affect agricultural yields. Therefore, to meet burgeoning food and energy demands, engineering of drought- and salt-tolerant crop varieties is essential under the impeding global warming and climate change conditions.
Plant scientists strive to develop crop varieties with stress-adaptive traits that thrive under changing climates (Henry, 2020). Such climate-smart crops will ensure a steady supply of food and energy products. The stacking of beneficial alleles in elite varieties was achieved through breeding in the past. Hybridization, mutation breeding, and improved agronomic practices increased global food production during the 1960s, leading to the “green revolution.” These traditional approaches are still helpful for developing varieties resilient to changing environmental conditions (Shelake et al., 2019a). However, conventional breeding methods are cumbersome, time consuming, and often fail to produce the desired outcomes, specifically with regard to abiotic stress tolerance, owing to complex genetic inheritance, pleiotropic effects, and unpredictable gene-environment interactions (Varshney et al., 2021b). In the 1990s, transgenic technology and RNA interference were used to produce genetically modified (GM) varieties with enhanced traits such as insect pest resistance, herbicide tolerance, abiotic stress or disease resistance, and biofortification (Singh et al., 2009; Kumar et al., 2020). However, GM varieties are subject to regulatory hurdles and have limited applications (Ahmad et al., 2021).
Recent advances have enabled the generation of targeted mutations at desired loci in plant genomes (Shelake et al., 2019a). For instance, modern breeding techniques include genome editing (GE) tools that enable fast-track introgression of novel beneficial traits into crop varieties; these approaches are commonly referred to as new plant breeding technologies (NPBTs). Modern breeding techniques can be complemented by use of the latest NPBT approaches. Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein (CRISPR/Cas) is the newest addition to the GE arsenal (Jinek et al., 2012). The engineered CRISPR/Cas system can enable editing of a desired locus in a complex genome. CRISPR-based tools have been used effectively for genome engineering in model plants and crop species in a range of fundamental to applied research (Pramanik et al., 2021). Technical breakthroughs in CRISPR-based approaches have made it possible to create transgene-free GE plants. Newer regulations are being formulated to handle the commercial release of GE products, including crop varieties deemed non-GM crops, by consideration on a case-by-case basis (Menz et al., 2020). Recently, genomic-assisted breeding (GAB) and CRISPR-based GE studies have shown promising outcomes in the exploration of plant stress-response pathways.
Moreover, CRISPR tools contribute to discovering the inherent molecular aspects of drought/salinity stress tolerance in plants (Joshi et al., 2020a; Zafar et al., 2020; Ahmad et al., 2021). This review provides an overview of plant responses to drought and salinity stresses at the molecular, physiological, and morphological levels. We discuss CRISPR-based techniques, CRISPR applications for basic and applied research on drought and salinity, and crucial aspects of developing stress-adaptive transgene-free GE varieties by combining genomics-assisted breeding approaches.
Plant responses to drought stress
Drought and salinity stress tolerance are polygenic traits. Plants have evolved several ways to tackle stressful events, which involve the activation of overlapping pathways at morphological, physiological, and molecular levels in response to different abiotic stresses. In addition, various abiotic stresses share some common characteristics of stress perception and effects on plant growth or development. For example, abiotic stresses like drought or salinity usually trigger osmotic stress in plant cells. Several components are often associated with more than one type of abiotic-stress-related pathway; these include sensors, receptors, phytohormones, transcription factors (TFs), kinases, phosphatases, and microRNAs (miRNAs) (Gong et al., 2020; Hussain et al., 2021). Drought stress is instigated by low atmospheric and soil humidity under higher ambient temperatures following water deficit for normal plant growth (Lipiec et al., 2013). Plants adapt their water balance dynamically under drought conditions to reduce water loss or improve water uptake (Mickelbart et al., 2015; Lamaoui et al., 2018; Joshi et al., 2020a; Varshney et al., 2021b).
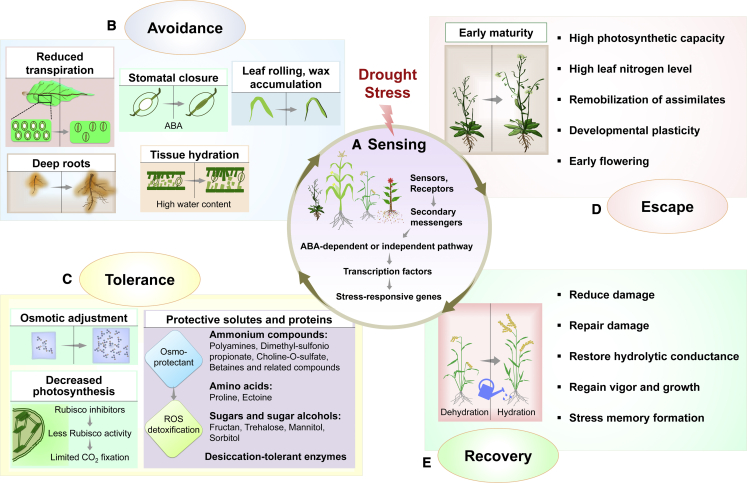
Drought stress perception and tolerance are complex quantitative phenomena governed by multiple genes and biochemical pathways at various stages of plant life progression (Bhat et al., 2020). The multi-level processes include sensing, signaling, and molecular changes to various layers of the central dogma, such as epigenetic modification, transcription, translation, and protein post-translational modifications (Zhang et al., 2021). Once a plant senses drought stress, plant responses can be grouped into four main categories: avoidance, tolerance, escape, and recovery (Figure 1).
Figure 1.
Plant responses to drought stress.
(A–E) Four critical processes involved in plant drought-stress tolerance after sensing the stress (A) are depicted, along with major changes used by plants in each of the processes, which include avoidance (B), tolerance (C), escape (D), and recovery (E). Specific plant species may respond to drought stress differently, and plants use more than one type of process for growth and survival.
An encounter with any stress activates cellular response pathways by altering the physical or chemical state of biomolecules in the cell. Although assigning the role of stress sensor to a particular molecule is complex, some are characterized using indirect approaches. Different primary drought-sensing modes have been suggested in the past (Zhang et al., 2021). For example, a mutant library of putative sensors is expected to alter the production of secondary messengers like Ca2+, reactive oxygen species (ROS), nitric oxide, and phospholipids, enabling screening of the resultant phenotypes. Such an approach has been applied in Arabidopsis to find the first putative osmotic stress sensor, OSCA1, a plasma membrane (PM) protein that forms Ca2+-permeable channels under hyperosmotic conditions (Yuan et al., 2014). Subsequently, several members of the OSCA family have been identified in different plants, including 15 in Arabidopsis (Yuan et al., 2014), 11 in rice (Li et al., 2015), 12 in maize (Ding et al., 2019), 13 in mung bean (Yin et al., 2021), and 42 in wheat (Tong et al., 2022). The OSCA family members may be functionally redundant, and further characterization would be the starting point to exploring the role of OSCA homologs in different stress-response pathways. Drought avoidance is the water-saving strategy that plants adopt under water-deficit conditions (Ilyas et al., 2021). Drought avoidance is achieved through morpho-physiological changes that involve stomatal regulation, limited vegetative growth via a reduced leaf number or leaf area, wax accumulation, an enhanced root system to take up available water, and avoidance of dehydration by lower water use.
Drought tolerance is the ability of a plant to thrive under severe water-deficit conditions through the modulation of physiological processes at different developmental stages (Fang and Xiong, 2015). One or more combinatorial drought tolerance strategies used by plants include low stomatal density, activation of stress-responsive genes, and alteration of signal transduction and metabolic pathways. Osmoprotectants such as amino acids, ammonium compounds, and sugar molecules help with ROS detoxification and maintenance of osmotic pressure, leading to a drought tolerance phenotype (Singh et al., 2015a). Water-deficit conditions also decrease photosynthetic rate and limit assimilate partitioning, presumably owing to enhanced acid invertase activity (Kim et al., 2000). Phytohormones like abscisic acid (ABA) and gibberellin (GA) mediate the signaling cascades that stimulate stress-related gene expression triggered by TFs, rapid stomatal closure, and changes in plant development under drought, salt, and other abiotic stress conditions (Cutler et al., 2010; Colebrook et al., 2014; Roca Paixão et al., 2019; Illouz-Eliaz et al., 2020; Wu et al., 2020).
The mechanism of stomatal opening is regulated via ABA and ABRE, involving a network of TFs. TFs from diverse families, for example, dehydration responsive element binding (DREB), bZIP, WHY, AP2/ERF, HD-Zip, bHLH, AREB/ABF, MYB, NAC, and WDR TFs, play multiple roles in drought stress tolerance and represent a crucial avenue for the development of drought-tolerant plant varieties (Joshi et al., 2016; Goel et al., 2019). Drought escape refers to plant adaptation to water deficit by shortening the life cycle or growth period. Flowering time is an ideal adaptive trait for drought-escape conditions. A transcriptome and phenotypic selection approach identified OsMADS18 (a MADS-box TF) as a drought-escape gene, highlighting the importance of early flowering in drought escape (Groen et al., 2020). Farmers prefer plant varieties with short lifespans that can escape climatic drought conditions.
Some plant genotypes survive the drought period and resume their vigor and growth; this ability is called drought recovery. Plants develop drought stress memory during repeated stresses, allowing them to respond quickly and provide enhanced protection (Jacques et al., 2021). The development of stress memory involves epigenetic changes and physiological adaptations that alter photosynthesis, cellular protective functions, osmotic adjustment, energy mechanisms, maintenance of water status, and plant interactions with soil microbes.
Plant responses to salinity stress
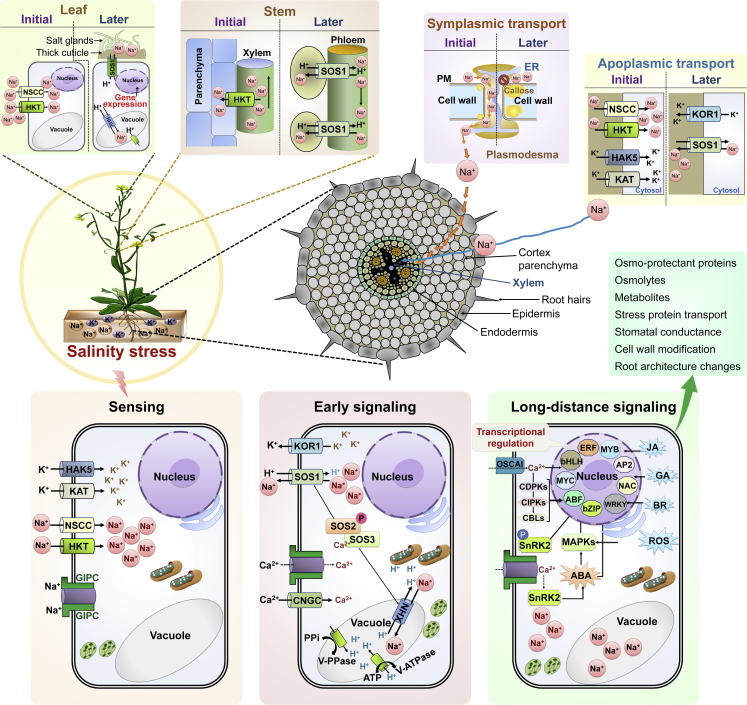
Excessive salt accumulation in plant cells causes salinity stress, hindering growth and development (Singh et al., 2021). During salt stress, an imbalance between cytosolic free sodium (Na+) and potassium (K+) ions is the main factor linked with ion toxicity. Higher salt ion concentrations in the soil limit water and nutrient uptake owing to hyperosmotic stress (Lamers et al., 2020; Van Zelm et al., 2020). Consequently, plant productivity is hampered by reduced photosynthesis, leading to agricultural losses. To survive in highly saline soils, plants have evolved mechanisms that reduce ion uptake in roots, compartmentalize excess ions inside the cell, and adjust osmotic pressure (Mickelbart et al., 2015). Critical aspects of plant responses to salinity stress are emphasized in Figure 2.
Figure 2.
Overview of salinity stress sensing, signaling, and response in plants.
Salinity stress is mainly caused by toxic sodium (Na+), potassium (K+), and chloride (Cl−) ions from sodium chloride (NaCl) and potassium chloride (KCl). In the current model, plant sensing, signaling, and response in the context of Na+ and K+ cytotoxicity are illustrated. Descriptions or full forms of the TF, gene, and family abbreviations are provided in the main text and supplemental information.
Similar to drought, plant responses to surplus salt involve adaptations at multiple levels, such as morphological (lateral shoot growth inhibition, early flowering, and root modifications), physiological (ion sequestration, osmotic adjustment, stomatal conductance, Na+/K+ discrimination), and biochemical (accumulation of polyamines, sugars, and proline, changes in hormone levels, induction of antioxidant machinery and metabolic pathways for detoxification of cytotoxic compounds) adaptations. Key aspects of plant responses to salt stress involve sensing excessive salt, early Ca2+ and ROS signaling cascades that activate various stress-responsive genes to avoid cell damage, and loading the resulting signaling molecules for long-distance transport via apoplastic and symplastic pathways across different tissues (Figure 2). The perception of salinity stress, i.e., sensing higher salt (Na+/K+) concentrations, is arguably the least understood phenomenon despite recent advances in plant salt-stress biology (Van Zelm et al., 2020). Recently, the MOCA1 gene encoding a glucuronosyltransferase for glycosyl inositol phosphorylceramide (GIPC) sphingolipids, PM-localized proteins, was revealed to be a vital plant-specific salt sensor (Jiang et al., 2019).
Initially, Na+ uptake is facilitated by transporters and channels such as nonselective cation channels (NSCCs), cyclic nucleotide-gated channels (CNGCs), glutamate receptors (GLRs), and high-affinity K+ transporters (HKTs) (Tester and Davenport, 2003; Hanin et al., 2016; Qiu et al., 2020a, 2020b). Similarly, K+ influx from the apoplast into the cytoplasm is carried out by channels like the Arabidopsis K+ transporter (AKT1) and high-affinity K+ uptake transporter (HAK). Plant cells recruit Ca2+ into the cytoplasm by CNGC and GIPC-interacting channels to deal with hyperaccumulated Na+/K+ ions (Hanin et al., 2016; Zhang et al., 2021). Along with Ca2+, ROS and 3′,5′-cyclic guanosine monophosphate (cGMP) constitute the three major salt-signaling molecules that activate salt-response pathways (Van Zelm et al., 2020).
After perceiving salt stress, early signaling responses comprise activation of Ca2+ signaling, ion transport, ROS-mediated signaling cascades, phospholipid modifications, and higher protein kinase activities (Chourasia et al., 2021). The process of Ca2+ signaling involves changes in Ca2+ spikes at three levels: instant at the cellular level (as early as 10 s after salt application) and fast- and late-response Ca2+ waves (Choi et al., 2014; Van Zelm et al., 2020). As depicted in Figure 2, Ca2+ binding to calcineurin B-like proteins (CBLs) instigates protein phosphorylation via a signaling cascade mediated by Ca2+-dependent protein kinases (CDPKs) and CBL-interacting protein kinases (CIPKs). Different CBL-CIPKs have been found to coordinate various cellular pathways once activated by Ca2+ and ROS signaling (Manishankar et al., 2018). The salt overly sensitive (SOS) pathway is the best-characterized CBL-CIPK pathway involved in plant salt tolerance (Qiu et al., 2020a, 2020b; Van Zelm et al., 2020; Chen et al., 2021b). In brief, after perception of salt stress, a spike of Ca2+ is generated in the cytoplasm of root cells and activates the SOS signaling pathway. SOS3 (CBL4) becomes activated after binding with Ca2+ and interacts with SOS2 (CIPK24). The SOS3/SOS2 complex then stimulates SOS1 (NHX7) through phosphorylation in the presence of Ca2+. SOS1 is a membrane-localized Na+/H+ antiporter in shoot and root cells (Halfter et al., 2000). Stele-localized SOS1 exports Na+ out of the cell in exchange for H+ at the root–soil interface, providing the first line of salt resistance (Shi et al., 2002). Elevated Na+ is transported to the vacuole by NHX antiporters for storage. Later, the excess Na+ and K+ ions are pumped out of the cytoplasm back into the apoplast by SOS1 and K+ outward rectifying channel (KOR) transporters, respectively.
Long-distance signaling with different molecular players and pathways leads to salinity tolerance by producing osmo-protectant proteins, metabolites, osmolytes, and transporters, regulating stomatal conductance, modifying the cell wall, and reorganizing root architecture. Plants use two pathways to reduce ion toxicity based on either restricting entry into plant cells via roots or sequestering inter- and intracellular ions into compartmentalized bodies such as vacuoles (Lou et al., 2018). Na+ ions enter the root system by either the symplastic pathway or apoplastic transport. Na+/K+ homeostasis is a crucial factor for plant survival that is facilitated by transporters, antiporters, and channels under salt-stress conditions. Recently, several components of salt-homeostasis pathways across plant cells, tissues, and organs have been characterized. These components include protein kinases (CIPK, CDPK), TFs (NAC, WRKY, SERF, bHLH, bZIP, MYB, YABBY, MADS-box, HD-Zip), symporters, SOS antiporters, NHX exchangers, HKT transporters and carrier proteins, and cellular detoxification pathways (glyoxalase pathway) (Mustafiz et al., 2011; Gupta et al., 2018; Van Zelm et al., 2020; Hussain et al., 2021; Sun et al., 2021). Components of the response to salinity and other stresses have been reviewed in recent literature, including summaries of protein kinase–mediated signal transduction (Chen et al., 2021b), interactions of TFs with ABA-dependent and -independent pathways during salt stress (Hussain et al., 2021), and the role of ion transporters in plant growth and stress responses (Tan et al., 2022).
Ca2+ is a leading player in long-distance signaling. Ca2+ enters the cytoplasm via OSCA1-like transporters or GIPC-interacting channels, thereby directly or indirectly regulating the activity of TFs involved in salinity tolerance via a cascade of CBLs, CIPKS, and CDPKs. ABA is one of the critical phytohormones involved in salt-stress defense; it integrates and coordinates signaling during salinity stress and activates whole-plant-level changes involving plant metabolic pathways, ion transport, and developmental processes (Yu et al., 2020). ABA signaling directly influences the activities of TFs or acts via the MAPK signaling cascade that regulates the expression of genes involved in salt-stress mechanisms. Salinity stress is also accompanied by elevated production of ROS (singlet oxygen, superoxide, hydroxyl radicals, and hydrogen peroxide), which modulates the transcription of salt-responsive genes. Elevated levels of plant hormones like jasmonic acid (JA), gibberellic acid (GA), and brassinosteroids (BR) have been found to control TF activities directly or indirectly during long-distance signaling.
The SNF1-related protein kinases (SnRKs) are plant-specific enzymes that play a crucial role in modulating ABA-dependent and ABA-independent signaling pathways. Ca2+-dependent activation of SnRK2 by osmotic and salt stress regulates ABA signaling (Fàbregas et al., 2020; Chen et al., 2021b). ABA-nonactivated kinases (SnRK2.4 and SnRK2.10) are activated in response to salt stress and affect the plant morphology of Arabidopsis (Mazur et al., 2021). Recent studies have shown that cytokinins are also associated with tolerance to abiotic stresses, including salinity (Wang et al., 2019). When plants are subjected to abiotic stresses, genes related to cytokinin signaling are spatiotemporally expressed, possibly connecting local signals to systemic plant responses (Zwack and Rashotte, 2015; Joshi et al., 2018). Overall, the role of phytohormones in abiotic stresses is not yet fully understood, owing to the complex network of associated pathways. In the later phase of the salt-response mechanism, salt-induced signaling pathways cause adaptive changes. Produced solutes mediate osmotic balance and modification of growth and developmental processes at tissue, organ, and whole-plant levels (Singh et al., 2021).
Salt transport occurs through both symplastic and apoplastic routes in root and shoot systems. Symplastic transport occurs through the plasmodesmata (Picard, 2003), and apoplastic transport is mediated by ion-specific transporter proteins (Hall et al., 2006; Munns et al., 2012). Plasmodesmata are symplastic nanochannels that connect adjacent cells and allow the passive or active movement of several molecules, including Na+ ions (Iswanto et al., 2021). After activation of stress signaling, callose accumulation closes these nanochannels and regulates ion movement. On the other hand, Na+ and K+ transporters load ions into the xylem from the root cells for further apoplastic translocation towards the stem and leaves. The process of Na+ loading/unloading in the xylem is a critical aspect of the salt-stress response mechanism. With the progression of salinity stress and subsequent signaling, ions are pumped out of cells through SOS1 and KOR1 in different tissues. Na+ ions are unloaded into the xylem parenchyma cells by HKTs and other transporters. Antiporters like SOS1 flush them out into the phloem while unloading. In leaves, NSCC and HKT family members participate in Na+ movement into the cell. Plants have evolved unique epidermal structures like salt glands and a thick cuticle layer to maintain ion homeostasis (Litalien and Zeeb, 2020). Excess Na+ ions are secreted into the salt glands for storage and exclusion in some species, like halophytes. Overall, targeting plant genes that transduce early signals to activate downstream genes involved in stress signaling would enable the engineering of stress-resilient crop varieties.
CRISPR-based genome engineering tools
Basics of the CRISPR/Cas system
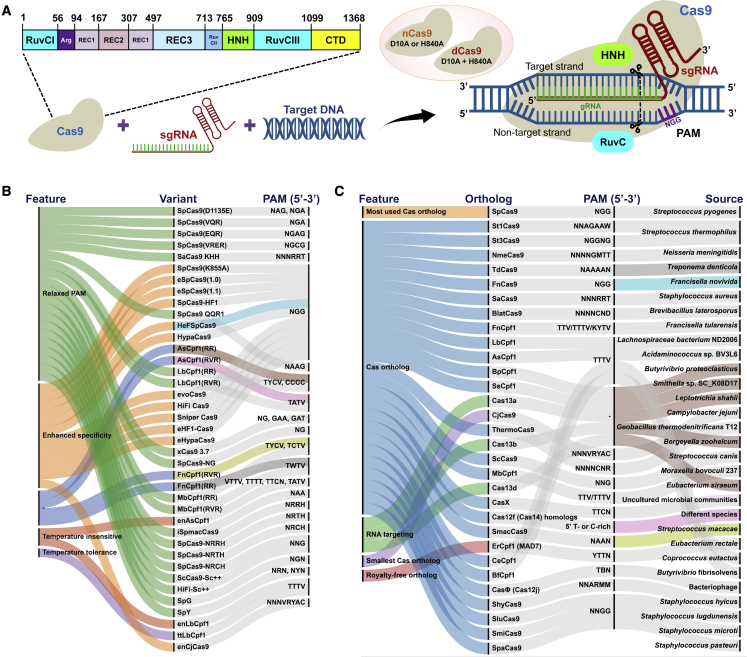
Prokaryotes, including bacteria and archaea, contain a CRISPR array acquired as part of their immune system against viruses. The components of native CRISPR/Cas machinery have been repurposed for targeting desired genomic locations and precision editing (Cong et al., 2013; Mali et al., 2013). The engineered CRISPR/Cas9 system consists of two main components: endonuclease Cas9 enzyme and RNA (Figure 3A).
Figure 3.
Basics of the CRISPR/Cas9 system, Cas variants, and orthologs.
(A) Mode of action of the CRISPR/Cas9 system is explained using SpCas9 as a model. The domain organization of the Cas9 enzyme consists of the recognition lobe (REC1, REC2, REC3), bridge helix (Arg), two nuclease domains (HNH, RuvC), and C-terminal protospacer adjacent motif (PAM) interacting domain (CTD). A ribonucleoprotein complex of the Cas9 enzyme (fully functional or impaired form, nickase or dead) with a single guide RNA (sgRNA) searches for the complementary target DNA region. Recognition of target DNA containing the PAM leads to structural rearrangements in Cas9, forming an R-loop in the Cas9-sgRNA-Target DNA complex. Two functional nuclease domains of wild-type Cas9 generate double-stranded breaks (DSBs) in the target DNA. The error-prone repair of DSBs produces insertions or deletions (indels), causing knockout of the encoded genes.
(B) Cas variants developed for CRISPR-based tool development.
(C) Cas orthologs used for CRISPR-based tool development. N is any nucleotide, R is A/G, M is A/C, D is A/G/T, B is G/T/C, W is A/T, V is G/C/A, R is A/G, and Y is C/T.
The optimized RNA component, also called single guide RNA (sgRNA), is a combination of CRISPR RNA (crRNA) and a trans-activating RNA (tracrRNA). The crRNA includes an RNA fragment of about 20 bp, a complementary sequence of the targeted site, and is usually cloned at the 5′ end of the sgRNA. Recognition of the sgRNA-binding region and protospacer adjacent motif (PAM; typically 3–5 bp) in the genome is followed by multimolecular structural reorganization between the protein-RNA-DNA complex; i.e., Cas-sgRNA-target DNA (Nishimasu et al., 2014). Two endonuclease domains of the Cas enzyme cleave the DNA and generate double-stranded breaks (DSBs) in the targeted region, leading to programmable editing. The ultimate advantages of CRISPR-based GE technology are delivery of CRISPR components at the desired locus in the complex genomic architecture and highly efficient precision editing. Multiplex editing (simultaneously targeting multiple loci) is another feature of CRISPR and other GE tools that enables the engineering of polygenic or multiple traits (Clasen et al., 2016; McCarty et al., 2020).
Cas9 variants and orthologs with novel properties
The most commonly used Cas orthologs are Cas9 and Cpf1 (Cas12a). Cas9 from Streptococcus pyogenes (SpCas9) recognizes NGG as a PAM and produces blunt-end DSBs; it is the most commonly used Cas for animal and plant genome engineering (Figure 3A). Cas12a requires only a crRNA that recognizes the T-rich PAM site, and cleavage generates staggered-end DSBs (Zetsche et al., 2015). Point mutations in SpCas9 and Cas12a have created several variants with higher precision or relaxed PAM requirements (Figure 3B). Apart from SpCas9 and Cas12a, several Cas orthologs from the microbial community have been optimized for GE, recognizing distinct PAMs, displaying different editing specificities, and targeting RNA or DNA (Figure 3C). An RNA-guided RNA-targeting enzyme such as Cas13 enables programmable RNA editing (Abudayyeh et al., 2016). Overall, the growing CRISPR toolbox, including reagents with distinct features, has broadened the scope of crop genome engineering at different layers of the central dogma responsible for various plant traits or phenotypes.
CRISPR-based tools for gene, chromosome, and genome engineering
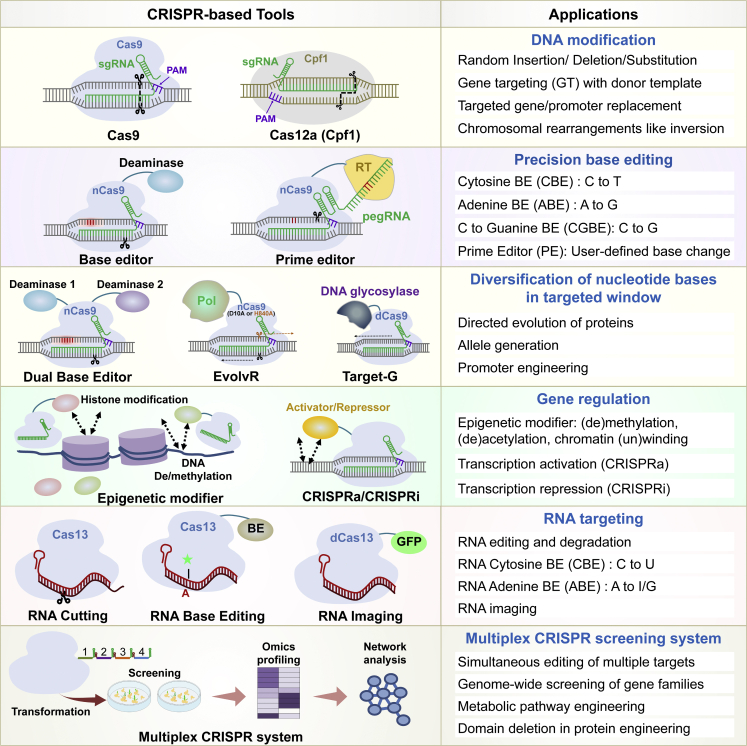
Several CRISPR-based tools have been developed in the last 10 years, enabling desired editing in the targeted genomic loci and beyond (Pramanik et al., 2021). Some examples include DNA base editors (BEs), epigenetic modifiers, prime editors (PEs), transcription regulators (CRISPRa and CRISPRi), and targeted random mutation tools (Dual BEs, Target-G, EvolvR) (Figure 4). EvolvR and Target-G are efficient for the diversification of the target locus in microbes, but their use in plants remains to be demonstrated.
Figure 4.
CRISPR-based tools and their applications for plant GE.
CRISPR-based tools and their applications for DNA modification (indel generation), precise nucleotide substitution, random mutagenesis of a targeted region for novel allele generation, regulation of gene expression, RNA targeting, and CRISPR-based mutant library generation are summarized. Relevant information and references for Cas variants and orthologs are provided in the supplemental information.
Fusion of various effector molecules to partially impaired (Cas9 nickase, nCas9) or nuclease-deficient (dead Cas9, dCas9) Cas9 has been employed as a shuttle system to deliver the CRISPR-fused-cargo at the genomic site of interest. RNA-targeting Cas proteins enable several applications beyond simple RNA editing, such as nucleic acid detection, RNA degradation, RNA base editing, pathogen detection, and live RNA imaging, as summarized in recently reviewed literature (Shelake et al., 2019a, 2019b; Pramanik et al., 2021). Plants cope with stress by several mechanisms that are governed by coding and non-coding regions in the plant genome and epigenetic marks as molecular signatures hidden in DNA packaging. The above-mentioned CRISPR-based tools can exploit various molecular mechanisms mediated by these genomic elements.
One of the significant applications of the CRISPR technique in plant molecular breeding is its capacity to produce chromosome rearrangements, including inversion, translocation, duplication, or deletion, which enable the alteration of genetic linkages, generation, or disruption of open reading frames (ORFs) that govern critical agronomic traits (Huang and Puchta, 2021). CRISPR tools can induce different chromosomal rearrangements and produce stable GE lines in a single generation (Schmidt et al., 2020; Lu et al., 2021). For instance, programmed inversion of a 911-kb region on chromosome 1 and duplication of a 338-kb region on chromosome 2 facilitated a promoter swap between two genes and the creation of a novel gene cassette, conferring novel traits in rice (Lu et al., 2021). Such precise editing at the chromosome level was considered an unrealistic task in the past. In this way, CRISPR tools are challenging several traditional notions, greatly expanding the potential of CRISPR tools for crop improvement.
Application of CRISPR tools for plant stress studies
CRISPR-mediated plant studies related to abiotic stresses can be grouped into three clusters based on the type of target DNA in the plant genome: structural genes, regulatory genes, and cis-regulatory elements (CREs) of structural or regulatory genes (Zafar et al., 2020). Structural genes play a pivotal role in enhancing plant stress tolerance. Regulatory genes encode proteins that contribute indirectly to stress tolerance, such as TFs, kinases, phosphatases, ion transporters, and miRNAs. CRISPR-mediated GE experiments may either stress susceptible or tolerant phenotypes, signifying the function of the target gene as a positive or negative regulator of a specific stress, respectively. In the following sections, we summarize recent CRISPR-based plant GE studies that have helped to reveal the underlying mechanisms involved in drought or salinity (Table 1) and their potential use in the design of stress-resilient crop varieties.
Table 1.
CRISPR-based drought and salinity stress-related GE studies in plants.
| Plant | Target | Type | Protein/Function | Editing outcome | Stress response | Reference |
|---|---|---|---|---|---|---|
| Drought | ||||||
| Arabidopsis | OST2 | SG | PM proton ATPase | new alleles (insertion, deletion) | tolerance | (Osakabe et al., 2016) |
| Arabidopsis | miR169a | RG, miR | involved in adaptation to multiple stresses | KO (934 bp deletion) | tolerance | (Zhao et al., 2016) |
| Arabidopsis | AVP1 | RG, phosphatase | vacuolar H+-pyrophosphatase | CRISPRa (transcription activation) | tolerance | (Park et al., 2017) |
| Arabidopsis | AREB1 | RG, TF | ABA-responsive TF | epigenetic modification (chromatin unwinding) | tolerance | (Roca Paixão et al., 2019) |
| Arabidopsis | TRE1 | RG | trehalase hydrolyzes trehalose into two molecules of D-glucose | KO (insertion, deletion) | tolerance | (Nuñez-Muñoz et al., 2021) |
| Arabidopsis | STL1 | SG | dirigent protein localized to the Casparian strip | KO (deletion) | tolerance | (Wang et al., 2022) |
| Maize | ARGOS8 | RG, promoter | negative regulator of ethylene response | swapping native promoter with GOS2 promoter | tolerance | (Shi et al., 2017) |
| Maize | abh2 | RG | abscisic acid 8′-hydroxylase mediates stomatal opening | KO (insertion, deletion) | tolerance | (Liu et al., 2020c) |
| Populus | NF-YB21 | RG, TF | positively regulates root growth | KO (insertion, deletion) | sensitive | (Zhou et al., 2020) |
| Populus | GNC | RG, TF | mediates stomatal closure | KO (insertion, deletion) | sensitive | (Shen et al., 2021) |
| Potato | FLORE | RG, lncRNA | counterpart of StCDF1 (cycling DOF factor) | promoter knockdown (deletion of 770 and 952 bp) | sensitive | (Ramírez Gonzales et al., 2021) |
| Rapeseed | A6.RGA | RG | DELLA protein, negative regulator of GA signaling | KO (insertion, deletion) | tolerance | (Wu et al., 2020) |
| Rice | DERF1, PMS3, MSH1, MYB5, SPP | RG | genes related to amino acid synthesis and drought tolerance | KO (insertion, deletion) | tolerance | (Zhang et al., 2014) |
| Rice | SAPK2 | RG, kinase | osmotic stress/ABA-activated protein kinase | KO (deletion) | sensitive | (Lou et al., 2017) |
| Rice | NAC14 | RG, TF | involved in growth and developmental changes | KO (deletion) | sensitive | (Shim et al., 2018) |
| Rice | SRL1, SRL2 | SG | regulate leaf morphology | KO (deletion) | tolerance | (Liao et al., 2019) |
| Rice | ERA1 | RG | regulates ABA signaling and the dehydration response | KO (insertion) | tolerance | (Ogata et al., 2020) |
| Rice | ERF83 | RG, TF | regulates drought response genes | KO (insertion) | sensitive | (Jung et al., 2021) |
| Rice | GPX1 | RG | redox sensor and transducer | KO (deletion) | sensitive | (Zhou et al., 2021) |
| Rice | bZIP68 | RG, TF | involved in ABA-independent osmotic stress response | KO (deletion) | sensitive | (Zhou et al., 2021) |
| Rice | SAP | SG | senescence-associated protein (SAP) | KO (insertion, deletion) | sensitive | (Park et al., 2022) |
| Tomato | MAPK3 | RG, Kinase | signaling molecule in stress response | KO (insertion, deletion, substitution) | sensitive | (Wang et al., 2017) |
| Tomato | NPR1 | RG, TF | receptor of salicylic acid | KO (insertion, deletion, substitution) | sensitive | (Li et al., 2019) |
| Tomato | GID1a | RG | GA receptor | KO (insertion) | tolerance | (Illouz-Eliaz et al., 2020) |
| Tomato | LBD40 | RG | involved in JA-mediated stress response | KO (insertion) | tolerance | (Liu et al., 2020a) |
| Wheat | DREB2, DREB3, ERF3 | RG, TF | DREBs, ethylene responsive factor (ERF) | KO (insertion, deletion) | sensitive | (Kim et al., 2018) |
| Wheat | NAC071-A | RG, TF | involved in growth and developmental changes | promoter KO (disruption of MYB recognition sites CAGTTA, TAACTG) | sensitive | (Mao et al., 2022) |
| Wheat | MYBL1 | RG, TF | involved in growth and developmental changes | KO (deletion) | sensitive | (Mao et al., 2022) |
| Salinity | ||||||
| Arabidopsis | C/VIF1 | RG | regulates cell wall and vacuolar invertase activities | KO (insertion, deletion) | tolerance | (Yang et al., 2020) |
| Arabidopsis | SAUR41 | RG | auxin response gene, modulates cell expansion, ion homeostasis | KO (insertion, deletion) | sensitive | (Qiu et al., 2020a, 2020b) |
| Arabidopsis | ACQOS | SG | encodes four nucleotide-binding leucine-rich repeat proteins involved in osmo-tolerance | KO (insertion, deletion) | sensitive | (Kim et al., 2021) |
| Barley | ITPK1 | RG, kinase | involved in phosphate storage and abiotic-stress signaling | KO (insertion, deletion) | sensitive | (Vlčko and Ohnoutková, 2020) |
| Barley | HVP10 | SG | involved in Na+ sequestration in the tonoplast | KO (insertion) | sensitive | (Fu et al., 2021) |
| Cotton | AITR genes | RG, TF | DPA4 is involved in plant development, SOD7 regulates seed size | KO (deletion) | tolerance | (Wang et al., 2021a) |
| Maize | CLCg | SG, IT | involved in chloride transport | KO (insertion, deletion) | sensitive | (Luo et al., 2021) |
| Maize | HKT1 | SG, IT | controls root-shoot Na+ delivery | KO (deletion) | sensitive | (Zhang et al., 2018a) |
| Maize | STL1 | SG | dirigent protein localized to the Casparian strip | KO (insertion, deletion) | tolerance | (Wang et al., 2022) |
| Pumpkin | RBOHD | SG | NADPH oxidase | KO (deletion) | sensitive | (Huang et al., 2019) |
| Rice | RAV2 | RG, TF | functions in the regulation of developmental processes | promoter element deletion | tolerance | (Duan et al., 2016) |
| Rice | MIR528 | RG, miR | regulates salinity stress through downregulation of salinity-related genes | KO (insertion, deletion, substitution) | sensitive | (Zhou et al., 2017) |
| Rice | SAPK1, SAPK2 | RG, kinase | osmotic stress/ABA-activated protein kinases | KO (insertion, deletion) | sensitive | (Lou et al., 2018) |
| Rice | BBS1 | RG, kinase | receptor-like cytoplasmic kinase | KO (deletion of 581 bp) | sensitive | (Zeng et al., 2018) |
| Rice | RR9, RR10 | RG | negatively regulate cytokinin signaling | KO (deletion) | tolerance | (Wang et al., 2019) |
| Rice | DOF15 | RG, TF | regulates primary root elongation | KO (insertion, deletion) | sensitive | (Qin et al., 2019) |
| Rice | SPL10 | RG, TF | regulates trichome development | KO (insertion, deletion) | tolerance | (Lan et al., 2019) |
| Rice | RR22 | RG, TF | involved in cytokinin-dependent gene expression | KO (insertion, deletion, substitution) | tolerance | (Zhang et al., 2019; Han et al., 2022) |
| Rice | NAC041 | RG, TF | involved in salt stress | KO (insertion, deletion) | sensitive | (Bo et al., 2019) |
| Rice | NCA1a, NCA1b | SG | chaperone protein modulates catalase activity | KO (insertion, deletion) | sensitive | (Liu et al., 2019) |
| Rice | GTγ-2 | RG, TF | binds to GT-1 promoter element and regulates growth and development | KO (insertion) | sensitive | (Liu et al., 2020d) |
| Rice | PIL14 | RG, TF | directly binds to the promoter of cell elongation-related genes | KO (deletion) | sensitive | (Mo et al., 2020) |
| Rice | PQT3 | RG | E3 ubiquitin ligase | KO (insertion, deletion) | tolerance | (Alfatih et al., 2020) |
| Rice | BG3 | RG | purine permease, cytokinin transport | KO (insertion, deletion) | sensitive | (Yin et al., 2020) |
| Rice | FLN2 | RG, kinase | involved in sugar partitioning | KO (insertion) | sensitive | (Chen et al., 2020) |
| Rice | SOS1 | SG, IT | Na+/H+ antiporter mediating Na+ transport | insertion of 60-bp translational enhancer | tolerance | (Lu et al., 2020) |
| Rice | GI | RG | circadian clock component | KO (deletion) | tolerance | (Wang et al., 2021b) |
| Rice | three ELF4 homologs | RG | circadian clock components | KO (deletion) | sensitive | (Wang et al., 2021b) |
| Rice | bHLH024 | RG, TF | basic helix–loop–helix TF involved in growth and stress responses | KO (deletion) | tolerance | (Alam et al., 2022) |
| Soybean | NHX5 | SG, IT | Na+/H+ exchanger (NHX) transmembrane protein | KO (insertion, deletion) | sensitive | (Sun et al., 2021) |
| Tomato | HAK20 | SG, IT | involved in K+ and Na+ homeostasis | KO (insertion, deletion) | sensitive | (Wang et al., 2020a) |
| Tomato | HyPRP1 | RG | transmembrane protein involved in multiple stresses | domain deletion | tolerance | (Tran et al., 2021) |
| Tomato | SOS1 | SG, IT | Na+/H+ antiporter mediating Na+ transport | KO (insertion, deletion) | sensitive | (Wang et al., 2021c) |
| Tomato | ABIG1 | RG, TF | homeodomain-leucine zipper (HD-ZIP) TF | KO (deletion) | tolerance | (Ding et al., 2022) |
| Wheat | Two HAG homologs | RG | modulate ROS production and signaling | KO (insertion, deletion) | sensitive | (Zheng et al., 2021) |
| Multiple stresses (including drought, salinity, or both stresses simultaneously) | ||||||
| Arabidopsis | UGT79B2, UGT79B | SG | UDP-glycosyltransferases | KO (insertion) | sensitive to drought, salinity, cold | (Li et al., 2017) |
| Arabidopsis | DPA4, SOD7, AITR2, AITR5, AITR6 | RG, TF | DPA4 is involved in plant development, SOD7 regulates seed size | KO (insertion, deletion) | increased seed size and drought tolerance | (Chen et al., 2019) |
| Arabidopsis | ArathEULS3 | SG | Euonymus europaeus-related lectin | KO (insertion, deletion) | tolerance to osmotic stress, salinity | (Dubiel et al., 2020) |
| Arabidopsis | AIW1, AIW2 | RG, TF | regulate multiple aspects of growth and development | KO (insertion, deletion) | sensitive to ABA but no difference in salt stress | (Wang et al., 2020b) |
| Arabidopsis | AITR family | RG, TF | ABA-induced transcriptional repressors | KO (insertion, deletion) | tolerance to drought, salinity | (Chen et al., 2021a) |
| Arabidopsis | WRKY3, WRKY4 | RG, TF | TFs involved in stress responses | KO (deletion) | sensitive to drought, Me-JA stress | (Li et al., 2021) |
| Rice | DST | RG, TF | zinc-finger TF | KO (insertion, deletion) | tolerance to drought, salinity | (Santosh Kumar et al., 2020) |
| Rice | PQT3 | SG | E3 ubiquitin ligase | KO (insertion, deletion) | tolerance to salinity, oxidative stress | (Alfatih et al., 2020) |
| Rice | NAC006 | RG, TF | involved in growth and developmental changes | KO (insertion, deletion) | sensitive to drought, heat | (Wang et al., 2020c) |
| Rice | miR535 | RG, miR | involved in plant growth and development | KO (deletion) | tolerance to drought, salinity | (Yue et al., 2020) |
| Soybean | MYB118 | RG, TF | MYB TF | KO (substitution) | sensitive to drought, salinity | (Du et al., 2018) |
| Tomato | ARF4 | RG, TF | auxin response factor | KO (deletion) | tolerance to osmotic stress, salinity | (Bouzroud et al., 2020) |
| Tomato | GRXS14, GRXS15, GRXS16, GRXS17 | SG | components of ROS scavenging network | KO (insertion, deletion) | sensitive to heat, chilling, drought, heavy metal toxicity, nutrient deficiency | (Kakeshpour et al., 2021) |
SG, structural gene; RG, regulatory gene; TF, transcription factor; KO, knockout; CRISPRa, CRISPR activation system; miR, microRNA; lncRNA, long non-coding RNA; DOF, DNA-binding with one-finger TF family; IT, ion transporter.
Exploring drought stress mechanisms using CRISPR tools
Drought directly restricts usable water supply for plants and thus has the most drastic growth-limiting effects. Owing to the polygenic nature of drought tolerance mechanisms, various genetic factors have been studied to understand their roles in drought response or to obtain drought-tolerant phenotypes. A range of strategies have been explored, including drought avoidance (stomatal closure, leaf rolling), TF editing (single or multiple copies in diploid or polyploid genomes), altering the expression of various genes (by epigenetic modification, promoter engineering, or miRNA editing), and modifying phytohormone signaling or catabolism pathways. Recent CRISPR-mediated drought studies that describe one of these strategies and their outcomes are summarized in the following paragraphs.
The early responses of plants to drought include ROS production and ABA-induced stomatal closure; these responses elicit long-distance signaling and help plants to cope with water loss and reduce water demand, respectively. PM-localized H+-ATPases generate proton gradients, energizing secondary transporters to bring ions and metabolites inside the cell (Palmgren, 2001). CRISPR/SpCas9 was successfully used to create novel alleles of OST2 (previously AHA1) in Arabidopsis, and the resulting GE lines displayed enhanced response by altered stomatal closure under drought stress (Osakabe et al., 2016). Promotion of adaxial leaf rolling by increasing bulliform cell number is a critical morphological trait for drought avoidance. A leaf-rolling phenotype was obtained by knockout (KO) of SRL1 and SRL2, which encode putative glycosylphosphatidylinositol (GPI)-anchored proteins (Liao et al., 2019). Homozygous GE lines exhibited higher production of antioxidant enzymes that reduced ROS activity, as well as better yield performance than the wild type.
TFs have been crucial candidates for understanding plant stress tolerance mechanisms. In particular, TFs from the previously mentioned families stimulate or suppress the transcription of many genes, imparting stress tolerance by modulating developmental and biochemical pathways (Nawkar et al., 2018; Gong et al., 2020). Thus, the CREs that determine the functions of TFs can reveal the fundamental mechanisms of quantitative traits and may assist in employing them for crop improvement. For instance, NPR1, a master regulator of plant defense response in tomatoes, was successfully inactivated by the CRISPR tool, and the resulting plants showed significant changes in drought responses (Li et al., 2019). The slnpr1 mutants displayed greater stomatal opening, more oxidative stress, and reduced antioxidant potential. Furthermore, the expression of several drought-responsive genes (GST, DREB) was severely affected, implying that SlNPR1 has a positive role in tomato drought tolerance. In Populus, CRISPR-based studies of PdNF-YB21 (Zhou et al., 2020) and GNC (Shen et al., 2021) were performed to understand the functions of these TFs in drought-related processes. Similarly, targeting of drought-responsive TFs such as NAC14 from the NAC family (Liao et al., 2019) and ERF83 from the AP2/ERF family (Jung et al., 2021) in rice using CRISPR tools produced drought-sensitive phenotypes, suggesting that both genes are positive regulators of drought-stress tolerance. Another CRISPR study revealed the pivotal roles of the antioxidant enzyme glutathione peroxidase and bZIP68 (GPX1-bZIP68) in redox sensing that activated osmotic stress-triggered signaling in rice (Zhou et al., 2021). The GPX1-bZIP68 module was found to confer higher osmotic or drought tolerance via an ABA-independent stress-response pathway.
Genetic engineering of polyploid plant genomes is complicated because of their complex genomic organization. In this regard, CRISPR tools could provide an ideal platform for polyploid crop GE because of their unique capabilities, like higher precision and multiplex editing. Kim and coworkers examined the effectiveness of CRISPR-based mutagenesis of two drought-related TFs (DREB2 and ERF3) in protoplasts of polyploid bread wheat (Kim et al., 2018). Editing both of the targeted genes demonstrated the vast potential of CRISPR tools for the manipulation of polypoid plant genomes. A recent study characterized a new member of the NAC family, NAC071-A (Mao et al., 2022). Using the dual-sgRNA approach, deletion of two MYB promoter CREs (CAGTTA and TAACTG) confirmed NAC071-A as a positive regulator of drought stress tolerance in wheat. In addition, CRISPR-generated KO of MYBL1 validated its role as an MYB transcriptional activator of NAC071-A expression, improving drought tolerance.
Epigenetic regulatory mechanisms, including chromatin remodeling, DNA/RNA methylation, and histone modification, play essential roles in response to environmental changes (Chang et al., 2020). Until recently, the molecular details of how epigenetic modifications coordinate stress-response mechanisms were elusive. AREB1 is a well-known positive regulator in the ABA signaling pathway and stress response in Arabidopsis, rice, and soybean (Oh et al., 2005; Yoshida et al., 2010; Barbosa et al., 2013). The histone acetyltransferase (HAT) enzyme relaxes chromatin folding and promotes enhanced gene expression. CRISPR activation (CRISPRa) and epigenetic modification approaches were combined to design a dCas9-HAT fusion system that upregulated AREB1 expression, thereby boosting drought tolerance (Roca Paixão et al., 2019). GE lines exhibited stunted growth but higher chlorophyll content and rapid stomatal dynamics under drought stress. This is a classic example of epigenetic modification for improved drought tolerance. The enhanced expression of AVP1 is another example of CRISPRa potential for the development of drought-tolerant plants (Park et al., 2017). AVP1 has been implicated in auxin transport and proton pumping across the tonoplast, which indirectly enhances the activity of PM-localized H+-ATPases. Up to a five-fold increase in AVP1 expression was reported, leading to improved performance under drought conditions. In the future, ABA pathway-related genes can be targeted using CRISPRa tools that provide novel avenues for the design and development of drought-response strategies.
Regulatory elements including intronic and 5′ or 3′ untranslated regions (UTRs) regulate desired gene expression levels. Promoter and CRE editing are attractive strategies for reprograming transcription. For example, in maize, the native ARGOS8 promoter was replaced with the constitutive GOS2 promoter in the 5′ UTR of ARGOS8 using dual sgRNAs (Shi et al., 2017). ARGOS8 is involved in the negative regulation of ethylene responses and interacts with ethylene response factors. CRISPR-generated GOS promoter-ARGOS8 GE lines exhibited improved drought tolerance and increased crop yield.
Small and long non-coding RNAs (lncRNAs) have emerged as crucial GE targets for crop improvement. miRNAs have been found to control the expression of target mRNAs by providing instructions to repress post-transcriptional expression or direct degradation (Zhang, 2015). In Arabidopsis, simultaneous deletion and gene replacement using a novel combinatory dual-sgRNA/SpCas9 vector generated mir169a mutants that displayed enhanced drought tolerance phenotypes (Zhao et al., 2016). The CDF TF family members control water homeostasis and drought stress conditions (Corrales et al., 2017). A recent study in potato investigated the combined role of CDF1 and its lncRNA counterpart gene FLORE by CRISPR targeting of promoter motifs in the FLORE locus (Ramírez Gonzales et al., 2021). Exposure of GE lines to drought stress demonstrated that CDF1 regulates FLORE expression by binding to its promoter motifs, whereas FLORE acts as a natural antisense transcript that regulates CDF1 transcription. Therefore, miRNAs or lncRNAs can be targeted by CRISPR tools to understand their roles in stress tolerance.
ABA signaling components such as ERA1, which encodes the β-subunit of farnesyltransferase, regulate the dehydration response in Arabidopsis and rice. CRISPR-edited ERA1 mutant lines displayed enhanced tolerance to drought stress (Ogata et al., 2020). In tomato, CRISPR-mediated study of the GA receptor GID1a showed that GID1a contributes to improved water-holding capacity and stomatal conductance (Illouz-Eliaz et al., 2020). The CRISPR-generated gid1 mutants displayed normal growth with decreased transpiration rates and improved recovery from transient water stress without compromising crop yield. The rapeseed genome contains four RGA genes involved in synthesizing DELLA proteins, which inhibit GA signaling. The KO mutant of A6.RGA showed enhanced drought tolerance, confirming this gene as a negative regulator of drought stress-related GA signaling (Wu et al., 2020). CRISPR-derived mutant lines of LBD40 shed light on previously unknown functional aspects of the LBD family (Liu et al., 2020a). The SlLBD40 protein participates in JA signaling and confers improved drought tolerance. In microbes and plants, non-reducing trehalose functions as an osmoprotectant against osmotic stress. Through genetic engineering, enhanced trehalose biosynthesis in rice resulted in tolerance to drought, salinity, and soil sodicity (Joshi et al., 2020b). Therefore, modifying the catabolic pathway of trehalose by targeting trehalase (the enzyme that hydrolyzes trehalose) is an attractive way to develop drought-tolerant GE lines. Arabidopsis GE mutants for TRE1, which encodes the trehalase enzyme, exhibited enhanced drought tolerance (Nuñez-Muñoz et al., 2021).
Exploring plant responses to salinity stress using CRISPR tools
Higher salt concentrations pose a considerable threat to plant growth and development by impeding physiological processes through osmotic stress, nutrient imbalance, ionic toxicity, and oxidative stress (Shrivastava and Kumar, 2015). A combination of biochemical, physiological, and molecular processes regulates available salt concentrations by adjusting ionic tolerance, tissue-specific tolerance, and osmotic tolerance. CRISPR targeting of TFs is a general approach for gaining molecular insights into their regulatory roles in salt-response pathways. KO of salt-stress-related proteins, including TFs, causes either improved stress tolerance or higher susceptibility, depending on their effects on downstream regulatory pathways. In this section, CRISPR-mediated salt-related studies are discussed by considering the targeted genes as positive (sensitive) or negative (tolerance) regulators, depending on the KO effect on plant phenotype.
Genetic factors that contribute to improved stress tolerance are considered to be positive regulators. For instance, RAV2 encodes a TF from the AP2/ERF family in rice. CRISPR-mediated deletion of GT-1 (GAAAAA), a promoter element in the RAV2 locus, confirmed its involvement in salt response (Duan et al., 2016). The GT-1 element was identified as a regulatory region, given that its deletion eliminated salt-responsive RAV2 expression. Several TFs act as positive salt-stress-responsive factors, and salt-sensitive phenotypes were observed in GE lines of DOF15 (Qin et al., 2019), NAC041 (Bo et al., 2019), GTγ-2 (Liu et al., 2020d), and PIL14 (Mo et al., 2020). Examples of CRISPR-mediated targeting of kinase and phosphatase genes include FLN2 in rice (Chen et al., 2020), BBS1 in rice (Zeng et al., 2018), and ITPK1 in barley (Vlčko and Ohnoutková, 2020); the edited plants showed salt-responsive phenotypes with reduced tolerance. Apart from TFs or kinases, other salt-stress-responsive factors have been studied using CRISPR tools; examples include MIR528 in rice (Zhou et al., 2017), RBOHD in pumpkin (Huang et al., 2019), NCA1a/OsNCA1b in rice (Liu et al., 2019), SAUR41 in Arabidopsis (Qiu et al., 2020a, 2020b), the ACQOS gene cluster in Arabidopsis (Kim et al., 2021), BG3 in rice (Yin et al., 2020), three ELF4 homologs in rice (Wang et al., 2021b), HAG1 in hexaploid wheat (Zheng et al., 2021), and HVP10 in barley (Fu et al., 2021).
Some genes function as negative regulators of plant response to salt and other abiotic stresses. RR22, which encodes a type-B response regulator involved in cytokinin signaling, was knocked down using CRISPR/Cas9, thereby improving rice salt tolerance (Zhang et al., 2019). Transgene-free homozygous GE lines were obtained by T1 segregation, which is likely to offer the possibility of trait introgression in related varieties within a short time. The CRISPR-derived SPL10 mutant, a member of the SPL family, displayed a salt-tolerant phenotype, suggesting that OsSPL10 is a negative regulator of rice response to salt stress (Lan et al., 2019). In addition, SPL10 was found to positively regulate trichome formation. Another study reported improved agronomic performance under drought and salt stress by novel base-insertion mutants of DST, which encodes a zinc-finger TF, in indica rice cv. MTU1010 (Santosh Kumar et al., 2020). Additional examples of negative regulators studied with CRISPR tools include C/VIF1 in rice, encoding a proteinaceous inhibitor of fructosidases involved in ABA-mediated responses (Yang et al., 2020); HyPRP1 in tomato, encoding a hybrid proline-rich cell wall-embedded structural protein (Tran et al., 2021); PQT3 in rice, encoding an E3 ubiquitin ligase involved in oxidative and salt stress (Alfatih et al., 2020); RR9 and RR10 in rice, encoding type-A RR proteins implicated in cytokinin signaling (Wang et al., 2019); and a rice homolog of GI, an evening-phased circadian clock component (Wang et al., 2021b).
Transporter proteins facilitate the trafficking of ions and other molecules over short (cell to cell) or long distances (across tissues and organs) in the plant body (Deshmukh et al., 2021). Plants maintain appropriate Na+/K+ balance via ion transporters to endure salt stress (Figure 3). In this regard, plant ion transporters are critical to understanding plant salt-stress response mechanisms. Na+ transporter families include ENA, NHX, HKT, ATPases, and cation exchangers, and K+ transporter families consist of voltage-gated channels/shakers, the KT/HAK/KUP family, VGICs, and KCO channels (Tan et al., 2022). Using CRISPR-mediated mutagenesis, frameshift (5-bp deletion) and truncation (34-bp deletion) mutants of HKT1 were obtained (Zhang et al., 2018a) to characterize the function of the HKT1 locus in the polypoid maize genome. Reduced salt tolerance in GE lines confirmed HKT1 as a positive regulator that promotes Na+ removal from the leaf by transporting Na+ from xylem sap (root to shoot). Similarly, functional aspects of the ion transporters NHX5 (Sun et al., 2021) and CLCg (Luo et al., 2021) were examined using CRISPR tools in the salt responses of polyploid soybean and maize species. CRISPR screening of mutants for HAK20 (Wang et al., 2020a) and SOS1 (Wang et al., 2021c) confirmed that genetic variations in these loci also mediate phenotypic changes associated with salinity tolerance in tomato crops. Notably, disruption of the CRT/DRE promoter element (CCGAC) recognized by the CBF/DREB family of TFs produced a susceptible phenotype under salt stress.
Exploring multiple stress responses with CRISPR tools
CRISPR-based multi-stress plant studies, including those on drought or salinity, are summarized in this section (Table 1). Phytohormones such as ABA, GA, and cytokinins play a significant role in tolerance to multiple biotic and abiotic stresses. Thus, genes related to phytohormone biosynthesis and signaling usually influence plant responses to various stresses. For instance, SAPK1 and SAPK2 enzymes from the SnRK2 family mediate ABA signaling in rice. Loss-of-function sapk2 mutants produced by CRISPR/Cas9 were insensitive to ABA (Lou et al., 2017). The sapk2 mutants displayed high sensitivity to dehydration and ROS, highlighting the role of SAPK2 in drought stress. Later, double-mutant (sapk1/sapk2) lines were exposed to osmotic and salt stresses and exhibited sensitive phenotypes (Lou et al., 2018). These observations indicate that SAPK1 and SAPK2 together act as positive regulators of tolerance to osmotic and salt stress.
Kinases from the MAPK family play crucial roles in plant responses to biotic and abiotic stresses. Protein kinases add phosphate groups to substrate proteins, thereby regulating their functions during various cellular processes (Wang et al., 2020d). A CRISPR-based approach was used to reveal functional aspects of the MAPK3 enzyme in tomato. KO of MAPK3 led to a drought-sensitive phenotype (Wang et al., 2017) and also decreased resistance to the necrotrophic pathogen Botrytis cinerea (Zhang et al., 2018b), suggesting that MAPK3 is a positive regulator of drought tolerance and pathogen resistance. By contrast, MAPK3 mutants showed enhanced thermotolerance compared with WT plants, indicating that MAPK3 is a negative regulator of heat stress tolerance (Yu et al., 2019). It is therefore crucial to consider the trade-off between distinct stress responses and cellular functions while targeting genes involved in multiple cellular processes.
Gene expression and analysis in KO lines of MYB118, which encodes a nuclear-localized TF, revealed its involvement in plant responses to drought, salt, and high temperature. GE plants exhibited reduced tolerance to these stresses, indicating a positive role for MYB118 in the maintenance of osmotic homeostasis and cellular oxidation status (Du et al., 2018). Some more examples of multi-functional TFs targeted by CRISPR include ARF4, improving tolerance to salinity and osmotic stress in tomato (Bouzroud et al., 2020); NAC006 in rice, reducing drought and heat tolerance (Wang et al., 2020c); AIW1 and AIW2 in Arabidopsis, conferring hypersensitivity to ABA but having no effect on salt-stress tolerance (Wang et al., 2020b); six AITR (1, 2, 3, 4, 5, and 6) genes in Arabidopsis, resulting in enhanced drought and salt tolerance (Chen et al., 2021a); and WRKY3 and WRKY4 in Arabidopsis, reducing tolerance to salt stress and methyl jasmonate (Me-JA) treatment (Li et al., 2021). The OsmiR535 gene from the highly conserved miRNA family is a well-known regulator of plant thermotolerance. CRISPR-derived miR535 loss-of-function mutants performed better under drought, salt, high ABA, and osmotic stresses (Yue et al., 2020). Arabidopsis quintuple-mutant lines (AITR2, AITR5, AITR6, DPA4, and SOD7) were generated by multiplex GE and showed increased seed size and drought tolerance (Chen et al., 2019).
Regulation of ROS moieties is an alternative strategy employed by plants to overcome abiotic stresses. Glutaredoxins (GRXs) from the thioredoxin superfamily function as ROS scavengers. Multiplex editing of four GRX genes (GRXS14, GRXS15, GRXS16, and GRXS17) in tomato produced a combination of double, triple, and quadruple mutants that exhibited hypersensitive phenotypes under abiotic stresses like drought, heat, heavy metal toxicity, nutrient deficiency, chilling, and light (Kakeshpour et al., 2021). Another study demonstrating the effective use of the multiplex editing approach included double-KO lines (ugt79b2/ugt79b) of UGT homologs in Arabidopsis (Li et al., 2017). These genes encode glycosyltransferase enzymes involved in the anthocyanin biosynthesis pathway and abiotic-stress responses. Double-mutant lines were hypersensitive to various abiotic stresses, demonstrating the crucial functions of UGT79B2 and UGT79B in anthocyanin production and subsequent stress tolerance. Negative (instead of positive) regulators of plant stress responses would be ideal CRISPR targets for future development of multi-stress-tolerant crops.
Integrating modern breeding approaches with CRISPR tools
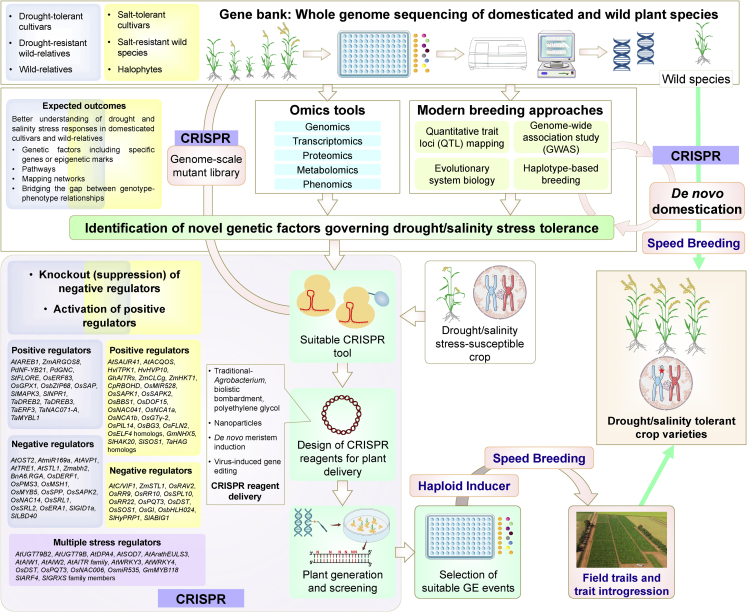
The main goal of researchers in exploring plant stress pathways is to develop stress-resilient varieties by modulating the relevant genomic aspects of the desired crop species. Despite recent progress in understanding genetic factors linked to plant stress, there is still a need to gain more insights into the basis of genetic variations responsible for contrasting phenotypes under drought or salinity stress in related varieties of the same crop species. Alternatively, orthologs of the same gene may govern different traits in related plant species, thus requiring molecular characterization. This section evaluates the applications of CRISPR technology and GAB approaches for the development of plant drought/salinity stress tolerance (Figure 5).
Figure 5.
Overview of the integration of genomic-assisted breeding and other approaches with CRISPR technology to develop drought/salinity-tolerant crops.
Known positive and negative regulators of drought (blue box) and salinity (yellow box) stress tolerance are ideal targets for crop improvement. Descriptions of different aspects depicted here are provided in the main text.
Finding regulatory genetic factors of drought/salinity stress in plants is a major challenge. Several domesticated plant species have lost the genetic variations responsible for quantitative traits during selective breeding, although these traits still reside in wild relatives (Wang et al., 2020a). Alternatively, domesticated species acquire a set of new heritable variations linked to improved fitness under stress conditions (Groen et al., 2022). With advances in genome sequencing methods, high-quality whole-genome sequences of domesticated species and their wild relatives are available in the public domain (Pazhamala et al., 2021). Genomic comparisons between cultivated and wild species may facilitate the identification of these genetic factors that can be introduced into commercial cultivars using CRISPR tools. Through CRISPR engineering, wild relatives that are naturally tolerant to drought/salinity stress can be domesticated by inserting mutations found in the gene pool of the cultivated species, a method described as de novo domestication. This approach enables the preservation of genetic diversity, as shown by recent studies in tomato (Zsögön et al., 2018) and rice (Yu et al., 2021a, 2021b).
The availability of whole-genome sequences for important crops has also facilitated the identification of large gene families, members of which are known to play important roles in plant stress tolerance (Kushwaha et al., 2009; Arya et al., 2014; Singh et al., 2015b; Bhuria et al., 2019). However, determining the exact function of each family member is a challenging task. To this end, precision GE tools have been used to develop large KO mutant libraries that enable easier identification of causal genes with greater specificity (Lu et al., 2017; Liu et al., 2020b). The function of LRR-XII family genes was investigated by developing a CRISPR library in tomato (Jacobs et al., 2017). In a single transformation with three sgRNAs per gene, 54 genes were targeted, resulting in 31 GE plants.
Similarly, targeted mutagenesis of an entire gene family may be achieved in order to functionally characterize all of its members. Moreover, a high-throughput approach for genome-scale mutagenesis of rice genes was developed using an sgRNA library, and more than 91 000 targeted loss-of-function mutants were generated (Lu et al., 2017). This pooled CRISPR library approach for genome-wide mutagenesis also enables the development of mutant libraries in non-model plant species. Recently, quantitative trait loci (QTL)-based mapping, evolutionary systems biology, genome-wide association studies (GWAS), and haplotype-based breeding have proven helpful in the discovery of polygenic adaptations that distinguish stress-prone and resistant cultivars. Among the significant genetic targets associated with plant stress are the CREs, which can drive dose-dependent gene expression, wild species domestication, and breeding innovation (Swinnen et al., 2016; Springer et al., 2019). Integration of CRISPR tools with some novel approaches shows excellent potential for expediting the rate of crop improvement; for instance, through speed breeding (Watson et al., 2018), de novo meristem induction (Maher et al., 2020), and nanomaterial-mediated delivery (Demirer et al., 2021). In particular, speed breeding (with controlled light and temperature conditions) can accelerate the introgression of improved drought/salinity tolerance traits into elite cultivars by either GAB approaches or rapid screening of CRISPR-generated GE lines.
Introgression of drought or salinity tolerance traits into elite cultivars by crossing with haploid inducer (HI) lines is highly desirable. HI lines generate viable plants that contain a half set of chromosomes, and chromosome doubling produces homozygous double HI plants. CRISPR-generated in vivo HI lines ideally serve this purpose, enabling one-step trait introgression into elite crop varieties without the loss of commercial traits during genetic segregation (Kelliher et al., 2019; Khanday et al., 2019).
Perspectives and challenges
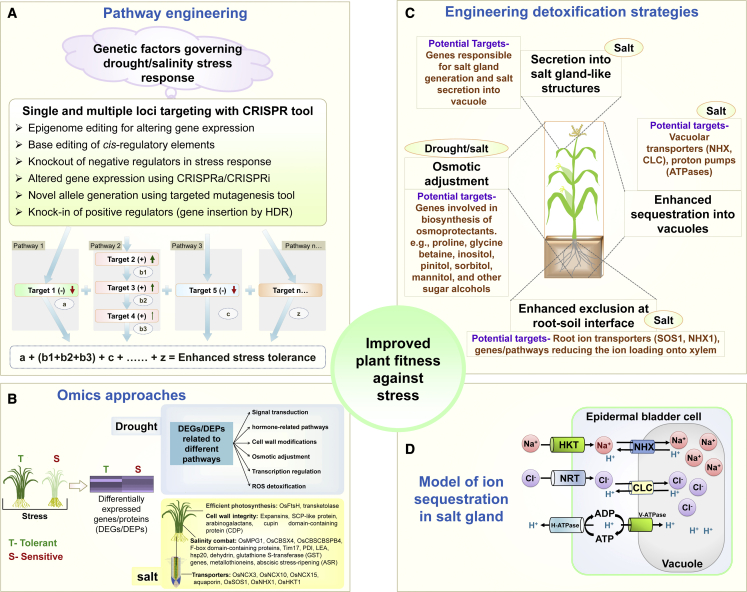
Molecular information on the functions of multiple genes and the interactions among members of different protein families during plant stress is limited, posing challenges for the selection of suitable GE targets. Hence, functional validation of stress-governing genes and protein families in different crops is required. In the following section, we discuss various aspects of drought/salinity stress that would enable the engineering of stress-tolerant crops in the future. These aspects include exploration of genetic factors related to drought/salinity stress, control of gene expression, engineering of multiple pathways, manipulation of natural stress-tolerant genetic materials, and strategies to exclude toxic molecules from the cytosol. Some of these approaches are depicted in Figure 6. Finally, we briefly discuss potential areas for improvement of CRISPR applications in plants.
Figure 6.
Strategies proposed for the engineering of plant tolerance to drought and salinity stress using CRISPR-based tools.
(A) Single or multiple genetic elements can be targeted using desired CRISPR tools. Genes from different pathways responsible for drought or salinity tolerance in plants can be either knocked out (negative element: target 1 or 5, red arrow) or knocked in (positive element: target 2, green arrow). In hypothetical pathway 2, the thickness of the green arrows depicts the major or minor role of the edited target gene in the stress condition. Epigenetic modifiers and CRISPR activation/interference (CRISPRa/CRISPRi) tools provide another means of regulating gene expression without introducing any mutations. In addition, base substitution tools such as base editors enable editing of cis-regulatory elements or generation of novel alleles. Targeting a single gene from a specific pathway may not be sufficient for developing stress tolerance in the case of polygenic traits like drought and salinity. Therefore, targeting of multiple genes from functionally related pathways would be ideal for achieving improved drought and salinity stress tolerance.
(B) Differentially expressed genes found in comparative omics analyses of stress-tolerant and sensitive cultivars are ideal candidates for GE to enable further characterization and exploitation in modern breeding. Details are provided in the supplemental information.
(C) Detoxification strategies are described in the main text.
(D) In the salt gland model, HKT1 transports Na+ and NRT transports Cl− to the gland cell; i.e., the epidermal bladder cell. NHX1 and CLC sequester excess Na+ and Cl− in the vacuole. H-ATPase and V-ATPase generate proton gradients to drive ion transport.
Exploring the basics of stress-responsive genetic factors
Fragmented information on stress sensors, early signaling mechanisms, and regulation of gene expression are significant challenges for understanding the intricacies of abiotic-stress response and tolerance mechanisms. Our understanding of the connecting links between identified or unknown sensors, early signaling processes, and crosstalk among different pathways is still incomplete. Another challenging aspect being dealt with by researchers is the fact that various protein family members exhibit redundant functions in stress-response pathways (Van Zelm et al., 2020). Moreover, plant scientists aim to understand how members of various gene families, like protein kinases, TFs, and ion transporters, influence each other’s functions during encounters with single or multiple stresses. Thus, further characterization of different gene family members in commercial crops would be the starting point for exploring the roles of stress-responsive factors in plant life and for their subsequent use in the design of stress-resilient varieties for the future. CRISPR-mediated targeted mutagenesis of genes with unknown or redundant functions may help to characterize their roles in plant stress biology, as evidenced by the newly identified AITR family (Chen et al., 2019, 2021a; Wang et al., 2021a).
Epigenetic regulation and related mechanisms are central to the fate of gene expression and priming or memory development (Agarwal et al., 2020). Chromatin structure dictates the underlying gene expression, presumed to be active, dead, or repressed in various situations. Information about epigenetic changes and their transfer from one generation to the next is essential for realizing the whole scenario of stress memory development. Therefore, memory or priming is a critical aspect governed by epigenetic modifications, imparting an enhanced capacity to combat future stresses (Varotto et al., 2020). In this regard, CRISPR tools like epigenetic modifiers possess immense potential for enabling controlled gene expression by writing, erasing, or reading the epigenetic marks involved in stress-response pathways. Epigenetic modifiers are attractive tools for shedding light on how epigenetic changes may contribute to the development of multi-stress-tolerant crop varieties.
Multiple pathway engineering
Although some CRISPR-based studies have shown that stress-tolerant phenotypes can be generated by targeting single genes, several studies have implied that tolerance to drought and salinity stress may require simultaneous targeting of multiple genes, owing to the polygenic nature of tolerance. Thus, in many cases, single-gene targeting may not be a “silver-bullet” approach to the development of abiotic-stress-resilient crop varieties. Multiple biochemical pathways contribute to plant stress tolerance; therefore, editing a specific gene may affect the functions of proteins in downstream pathways. For example, targeting Na+ sequestration in vacuoles is of paramount importance to the development of plant salinity tolerance. Targeting vacuolar NHX exchanger genes may not be sufficient to achieve salt-stress tolerance. Therefore, the NHX exchangers must be fueled by a concurrent increase in H+-ATPase operation. Second, Na+ back-leak from the vacuole into the cytosol must be prevented to avoid a futile cycle, and this can be achieved by closely regulating slow and fast vacuolar channels. Thus, simultaneous targeting of multiple genes may enable plant tolerance to salinity. Editing multiple genes from the same or inter-linked pathways would avoid the risk of negative impacts on growth or yield by engineering the plant’s tolerance to drought and salinity stresses (Figure 6A). As shown in Figure 6, an additive effect of targeted genes from multiple pathways would contribute to enhanced stress tolerance. Furthermore, sequential editing of several genes involved in a biochemical or signaling pathway can overcome the barrier of rate-limiting steps (as summarized in hypothetical pathway 2 in Figure 6A).
Harnessing natural genetic resources to develop drought/salinity tolerance
CRISPR-based de novo domestication of wild species could offer a faster means of crop improvement, as demonstrated in wild tomato (Zsögön et al., 2018) and rice (Yu et al., 2021a, 2021b). Furthermore, crop wild relatives are valuable genetic resources for crop improvement. Comparing susceptible and stress-tolerant cultivars or wild relatives could explain how multi-level adaptations help tolerant species to survive under stressful conditions. Omics-based comparisons between genotypes with different stress responses is a reliable approach for uncovering uniquely upregulated or downregulated genes/proteins (Figure 6B). Recently, several studies have evaluated drought stress-related parameters in different crops and their wild relatives, such as rice (Neelam et al., 2018), wheat (Liu et al., 2015), sorghum (Cowan et al., 2020), cotton (Yu et al., 2021a, 2021b), tomato (Bolger et al., 2014; Rigano et al., 2016; Egea et al., 2018), eggplant (Kouassi et al., 2021; Plazas et al., 2022), sweet potato (Nhanala and Yencho, 2021), chickpea (Harish et al., 2020), and Vigna sp. (Iseki et al., 2018). Drought-tolerant wild sorghum showed higher root dhurrin content than cultivated species (Cowan et al., 2020), and gene expression differences in hormone signaling and amino acid metabolism in wild tomato (Solanum pennellii) were crucial for reducing water loss (Egea et al., 2018). Recently, differentially expressed genes or proteins (DEGs/DEPs) were investigated in omics-based drought studies of rice, leading to the identification of genes related to ROS detoxification, signal transduction, cell wall modifications, hormone signaling, transcription regulation, and osmotic adjustment (Baldoni, 2022). Several studies have indicated that the engineering of antioxidative systems may be crucial for drought acclimatization (Laxa et al., 2019). For salt stress in rice, transcriptome and proteome studies have revealed DEGs/DEPs that play a role in ROS detoxification, maintenance of efficient photosynthesis, cell wall integrity, and transport of salt ions across membranes within or outside the cell (Cotsaftis et al., 2011; Li et al., 2018; Farooq et al., 2021). Specific DEGs/DEPs found in rice are depicted in Figure 6B and further elaborated in Supplemental Figure 1. Thus, omics-based identification of stress-responsive genes could provide ideal targets for GE to characterize and further modulate related pathways.
Based on their salt tolerance levels, plants can be categorized as glycophytes (salt sensitive, tolerant of no more than 200 mM NaCl) and halophytes (tolerant of >200 mM NaCl); most cultivated species are glycophytes. Currently, halophyte species are being explored for biotechnology applications that offer several benefits apart from salt tolerance, including pollutant removal, reclamation of degraded lands, and use as high-nutrition fodder or medicinal supplements (Munir et al., 2022). Although glycophytes and halophytes share salt-tolerance mechanisms, the latter have evolved unique features to mitigate higher salt concentrations. For example, halophytes maintain higher osmolyte levels (sugar alcohols, proline) and provide osmoprotection (Gong et al., 2005). Glycophytes are more equipped for higher accumulation of Na+ in shoots rather than roots while maintaining an optimal K+/Na+ ratio (Orsini et al., 2010). The multiple mechanisms of salt tolerance found in Lycium humile (tolerant of up to 750 mM NaCl) from the Solanaceae family are attributed to ABA accumulation, higher proline content and antioxidant capacity, and replacement of spongy parenchyma cells with larger parenchyma (water-storage) cells that enable Na+ storage (Palchetti et al., 2021). In addition, omics approaches have recently been used to investigate the basis of salt tolerance in halophytes (Jiao et al., 2018; Guo et al., 2021). Different omics studies have identified new members or entirely novel gene families implicated in metabolic pathways that help halophytes to cope with a saline environment; they present ideal targets for GE applications. Although comparative omics approaches provide information about DEGs/DEPs in response to abiotic stresses, further screening of the obtained DEGs/DEPs for GE is a significant challenge. Therefore, an improved understanding of these genes and pathways is critical for informed GE-based crop improvement and can help design and interpret studies of DEGs/DEPs that provide a rich resource for stress-resilient crop development.
Recently, natural allelic variants have been identified in various cultivars through QTL and GWAS approaches. Beneficial allelic variations found in wild relatives can be introduced into elite cultivars using introgressive hybridization. However, this process can be time and labor intensive (Shelake et al., 2019a). Also, there is always a risk of losing some beneficial traits during the breeding process. In this regard, GE tools provide a better means of incorporating beneficial alleles with greater accuracy than introgression breeding, as demonstrated in the case of HAK20 (Wang et al., 2020a) and SOS1 (Wang et al., 2021c) in tomato species.
Engineering detoxification strategies
Stress induces the production of ROS and reactive nitrogen species in different organelles and cell types; these species also function as signaling molecules to activate gene expression and related pathways. Upregulation of antioxidant systems is critical for redox homeostasis during drought stress. For instance, detoxifying enzymes such as ascorbate peroxidase and catalase help to reduce ROS toxicity (Laxa et al., 2019). However, at the moment, differences in the antioxidant systems of sensitive and tolerant genotypes during drought stress are not fully understood, thus limiting GE applications. ROS and related antioxidant systems also play an essential role in plant growth and development, and the spatiotemporal expression patterns of chosen targets for GE must therefore be considered.
Different salt detoxification strategies include salt exclusion at the root-soil interface, maintenance of osmotic balance within cells, sequestration of excess ions in the vacuole, and secretion into specialized salt-gland-like structures (Figure 6C) (Litalien and Zeeb, 2020). Among these, engineering salt-gland-like structures could be an attractive strategy (Figure 6D). Understanding the evolution and function of salt glands may ultimately help to design salt-tolerant crops. Salt glands are unicellular, bicellular, or multicellular structures reported in over 50 halophytes across 14 families (Dassanayake and Larkin, 2017). They originate from the epidermis and structurally resemble plant trichomes. Salt glands in halophytes can be classified into two types on the basis of functional aspects. The first type directly secretes salt ions onto the leaf surface, and the second type stores salt ions in an evolved epidermal bladder cell (EBC) by collecting them into the vacuoles (Figure 6D). The key transporters involved in the EBC have recently been characterized in the halophyte Chenopodium quinoa (Bohm et al., 2018). Engineering trichomes into functional salt-gland cells in glycophytes has recently been proposed and discussed in the literature, along with potential candidates for sequestering excess ions in the EBC (Shabala et al., 2014; Dassanayake and Larkin, 2017; Zhao et al., 2020). EBC engineering alone has many limitations and must therefore be combined with other detoxification strategies to avoid adverse effects on growth and development.
Several recent studies have also explored the potential use of the plant and soil microbiome for enhanced plant tolerance to abiotic stresses, including drought and salinity (Leontidou et al., 2020; An et al., 2022; Koshila Ravi et al., 2022). In the future, combining GE-based approaches with microbial use may constitute an ecofriendly and sustainable approach for higher agricultural crop productivity.
Fine-tuning CRISPR-based tools
Some aspects of CRISPR technology warrant further fine-tuning and upgrades for the application, efficacy, and precision of CRISPR tools in plants; these include selection and in-depth understanding of suitable targets, choice and availability of Cas enzymes, functional sgRNAs without off-target effects in the genome, delivery method of CRISPR reagents, and genotyping of GE lines. Optimization of newer CRISPR tools for different plant species is another challenging endeavor. At first, newer CRISPR-based tools are generally invented for bacterial and mammalian GE, then later adopted for plant use (Shelake et al., 2022). Therefore, further optimizations are required to enable the use of newly developed CRISPR technologies in separate plant cultivars or species.
The editing outcome is mainly dependent on endogenous DNA repair machinery and target locus accessibility in the host genomic architecture. For instance, prime editing is a promising technique for rewriting short DNA lengths, but it is still challenging to use in plants because of lower efficiency (Jiang et al., 2020). Therefore, additional efforts are needed to further improve the PE tool for plant use. Also, current base editing tools cannot install all types of base substitutions at targeted genomic regions. Tools for precisely targeted insertion are another aspect of CRISPR application that requires further improvements, enabling CRE insertions to either activate or suppress the expression of downstream genes (Lu et al., 2020).
Target selection and structural aspects of the sgRNA sequence are also crucial factors in successful GE studies. Chromatin context, DNA repair mechanisms recruited at the targeted sites, expression levels of CRISPR machinery, or a combination of these factors determine the GE outcomes (Weiss et al., 2022). The sgRNA can be designed based on prior knowledge of the genome sequence to avoid off-targets or failure due to several potential factors. In addition, off-targeting can be avoided by backcrossing and segregation (Kadam et al., 2018). In summary, bridging the knowledge gap in drought/salinity-related signaling pathways will help to design GE crops with multiple-stress tolerance and no trade-offs among beneficial traits.
Concluding remarks
We have provided an outlook on plant responses to drought and salinity stresses and described the potential use of CRISPR tools along with GAB approaches for the development of stress-resilient crop varieties. Integrated understanding of omics approaches, such as genomics, transcriptomics, proteomics, phenomics, and metabolomics, provides a unique opportunity to map the flow of genetic information across central dogma processes during stress biology. In this regard, CRISPR technology is a boon for elucidating gene functions and fine-tuning plant response pathways for tolerance to drought, salinity, and other stresses in agriculture. The expanding CRISPR toolbox enables the targeting of any process that regulates plant traits by precise and efficient genome engineering. In the near future, CRISPR tools will be a valuable resource helping plant biologists to engineer stress-resistant crop varieties for changing climatic conditions.
Funding
This work was supported by the National Research Foundation of Korea (grants NRF 2021R1I1A3057067, 2021R1A5A8029490, 2020M3A9I4038352, and 2020R1A6A1A03044344). A.K.S. thanks the Indian Council of Agricultural Research, Ministry of Agriculture and Farmers Welfare, Government of India for funding in the laboratory.
Author contributions
R.M.S. and J.-Y.K. conceived the idea. R.M.S. researched and designed the manuscript structure. R.M.S. wrote the initial draft. U.S.K., R.K., D.P., and A.K.S. contributed to writing and editing. All authors made direct intellectual contributions to the editing before final submission and approved the manuscript.
Acknowledgments
No conflict of interest is declared.
Published: August 3, 2022
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Contributor Information
Rahul Mahadev Shelake, Email: rahultnau@gmail.com.
Jae-Yean Kim, Email: kimjy@gnu.ac.kr.
Supplemental information
References
- Abudayyeh O.O., Gootenberg J.S., Konermann S., Joung J., Slaymaker I.M., Cox D.B.T., Shmakov S., Makarova K.S., Semenova E., Minakhin L., et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:aaf5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal G., Kudapa H., Ramalingam A., Choudhary D., Sinha P., Garg V., Singh V.K., Patil G.B., Pandey M.K., Nguyen H.T., et al. Epigenetics and epigenomics: underlying mechanisms, relevance, and implications in crop improvement. Funct. Integr. Genomics. 2020;20:739–761. doi: 10.1007/s10142-020-00756-7. [DOI] [PubMed] [Google Scholar]
- Ahmad S., Tang L., Shahzad R., Mawia A.M., Rao G.S., Jamil S., Wei C., Sheng Z., Shao G., Wei X., et al. CRISPR-based crop improvements: a way forward to achieve zero hunger. J. Agric. Food Chem. 2021;69:8307–8323. doi: 10.1021/acs.jafc.1c02653. [DOI] [PubMed] [Google Scholar]
- Alam M.S., Kong J., Tao R., Ahmed T., Alamin M., Alotaibi S.S., Abdelsalam N.R., Xu J.H. CRISPR/Cas9 mediated knockout of the OsbHLH024 transcription factor improves salt stress resistance in rice (Oryza sativa L.) Plants. 2022;11:1184. doi: 10.3390/plants11091184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfatih A., Wu J., Jan S.U., Zhang Z.S., Xia J.Q., Xiang C.B. Loss of rice PARAQUAT TOLERANCE 3 confers enhanced resistance to abiotic stresses and increases grain yield in field. Plant Cell Environ. 2020;43:2743–2754. doi: 10.1111/pce.13856. [DOI] [PubMed] [Google Scholar]
- An X., Wang Z., Teng X., Zhou R., Wang X., Xu M., Lian B. Rhizosphere bacterial diversity and environmental function prediction of wild salt-tolerant plants in coastal silt soil. Ecol. Indic. 2022;134:108503. [Google Scholar]
- Arya P., Kumar G., Acharya V., Singh A.K. Genome-wide identification and expression analysis of NBS-encoding genes in Malus x domestica and expansion of NBS genes family in Rosaceae. PLoS One. 2014;9:e107987. doi: 10.1371/journal.pone.0107987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldoni E. Improving drought tolerance: can comparative transcriptomics support strategic rice breeding? Plant Stress. 2022;3:100058. [Google Scholar]
- Barbosa E.G.G., Leite J.P., Marin S.R.R., Marinho J.P., de Fátima Corrêa Carvalho J., Fuganti-Pagliarini R., Farias J.R.B., Neumaier N., Marcelino-Guimarães F.C., de Oliveira M.C.N., et al. Overexpression of the ABA-Dependent AREB1 Transcription Factor from Arabidopsis thaliana improves soybean tolerance to water deficit. Plant Mol. Biol. Report. 2013;31:719–730. [Google Scholar]
- Bhat J.A., Deshmukh R., Zhao T., Patil G., Deokar A., Shinde S., Chaudhary J. Harnessing high-throughput phenotyping and genotyping for enhanced drought tolerance in crop plants. J. Biotechnol. 2020;324:248–260. doi: 10.1016/j.jbiotec.2020.11.010. [DOI] [PubMed] [Google Scholar]
- Bhuria M., Goel P., Kumar S., Singh A.K. Genome-wide identification and expression profiling of genes encoding universal stress proteins (USP) identify multi-stress responsive USP genes in Arabidopsis thaliana. Plant Physiol. Rep. 2019;24:434–445. [Google Scholar]
- Bo W., Zhaohui Z., Huanhuan Z., Xia W., Binglin L., Lijia Y., Xiangyan H., Deshui Y., Xuelian Z., Chunguo W., et al. Targeted mutagenesis of NAC transcription factor gene, OsNAC041, leading to salt sensitivity in rice. Rice Sci. 2019;26:98–108. [Google Scholar]
- Böhm J., Messerer M., Müller H.M., Scholz-Starke J., Gradogna A., Scherzer S., Maierhofer T., Bazihizina N., Zhang H., Stigloher C., et al. Understanding the molecular basis of salt sequestration in epidermal bladder cells of Chenopodium quinoa. Curr. Biol. 2018;28:3075–3085.e7. doi: 10.1016/j.cub.2018.08.004. [DOI] [PubMed] [Google Scholar]
- Bolger A., Scossa F., Bolger M.E., Lanz C., Maumus F., Tohge T., Quesneville H., Alseekh S., Sørensen I., Lichtenstein G., et al. The genome of the stress-tolerant wild tomato species Solanum pennellii. Nat. Genet. 2014;46:1034–1038. doi: 10.1038/ng.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzroud S., Gasparini K., Hu G., Barbosa M.A.M., Rosa B.L., Fahr M., Bendaou N., Bouzayen M., Zsögön A., Smouni A., et al. Down regulation and loss of auxin response factor 4 function using CRISPR/Cas9 alters plant growth, stomatal function and improves tomato tolerance to salinity and osmotic stress. Genes. 2020;11 doi: 10.3390/genes11030272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W., Borlace S., Lengaigne M., Van Rensch P., Collins M., Vecchi G., Timmermann A., Santoso A., Mcphaden M.J., Wu L., et al. Increasing frequency of extreme El Niño events due to greenhouse warming. Nat. Clim. Chang. 2014;4:111–116. [Google Scholar]
- Chang Y.N., Zhu C., Jiang J., Zhang H., Zhu J.K., Duan C.G. Epigenetic regulation in plant abiotic stress responses. J. Integr. Plant Biol. 2020;62:563–580. doi: 10.1111/jipb.12901. [DOI] [PubMed] [Google Scholar]
- Chen G., Hu J., Dong L., Zeng D., Guo L., Zhang G., Zhu L., Qian Q. The tolerance of salinity in rice requires the presence of a functional copy of FLN2. Biomolecules. 2019;10:17. doi: 10.3390/biom10010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Zhang N., Zhang Q., et al. Genome editing to integrate seed size and abiotic stress tolerance traits in Arabidopsis reveals a role for DPA4 and SOD7 in the regulation of inflorescence architecture. Int. J. Mol. Sci. 2019;20:26955. doi: 10.3390/ijms20112695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Zhang N., Zhou G., Hussain S., Ahmed S., Tian H., Wang S. Knockout of the entire family of AITR genes in Arabidopsis leads to enhanced drought and salinity tolerance without fitness costs. BMC Plant Biol. 2021;21:137. doi: 10.1186/s12870-021-02907-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Ding Y., Yang Y., Song C., Wang B., Yang S., Guo Y., Gong Z. Protein kinases in plant responses to drought, salt, and cold stress. J. Integr. Plant Biol. 2021;63:53–78. doi: 10.1111/jipb.13061. [DOI] [PubMed] [Google Scholar]
- Choi W.G., Toyota M., Kim S.H., Hilleary R., Gilroy S. Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proc. Natl. Acad. Sci. USA. 2014;111:6497–6502. doi: 10.1073/pnas.1319955111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chourasia K.N., Lal M.K., Tiwari R.K., Dev D., Kardile H.B., Patil V.U., Kumar A., Vanishree G., Kumar D., Bhardwaj V., et al. Salinity stress in potato: understanding physiological, biochemical and molecular responses. Life. 2021;11:545. doi: 10.3390/life11060545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clasen B.M., Stoddard T.J., Luo S., Demorest Z.L., Li J., Cedrone F., Tibebu R., Davison S., Ray E.E., Daulhac A., et al. Improving cold storage and processing traits in potato through targeted gene knockout. Plant Biotechnol. J. 2016;14:169–176. doi: 10.1111/pbi.12370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colebrook E.H., Thomas S.G., Phillips A.L., Hedden P. The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 2014;217:67–75. doi: 10.1242/jeb.089938. [DOI] [PubMed] [Google Scholar]
- Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrales A.-R., Carrillo L., Lasierra P., Nebauer S.G., Dominguez-Figueroa J., Renau-Morata B., Pollmann S., Granell A., Molina R.-V., Vicente-Carbajosa J., et al. Multifaceted role of cycling DOF factor 3 (CDF3) in the regulation of flowering time and abiotic stress responses in Arabidopsis. Plant Cell Environ. 2017;40:748–764. doi: 10.1111/pce.12894. [DOI] [PubMed] [Google Scholar]
- Cotsaftis O., Plett D., Johnson A.A.T., Walia H., Wilson C., Ismail A.M., Close T.J., Tester M., Baumann U. Root-specific transcript profiling of contrasting rice genotypes in response to salinity stress. Mol. Plant. 2011;4:25–41. doi: 10.1093/mp/ssq056. [DOI] [PubMed] [Google Scholar]
- Cowan M.F., Blomstedt C.K., Norton S.L., Henry R.J., Møller B.L., Gleadow R. Crop wild relatives as a genetic resource for generating low-cyanide, drought-tolerant Sorghum. Environ. Exp. Bot. 2020;169:103884. [Google Scholar]
- Cutler S.R., Rodriguez P.L., Finkelstein R.R., Abrams S.R. Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- Dassanayake M., Larkin J.C. Making plants break a sweat: the structure, function, and evolution of plant salt glands. Front. Plant Sci. 2017;8:724. doi: 10.3389/fpls.2017.00406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirer G.S., Silva T.N., Jackson C.T., Thomas J.B., W Ehrhardt D., Rhee S.Y., Mortimer J.C., Landry M.P., Landry M.P. Nanotechnology to advance CRISPR–Cas genetic engineering of plants. Nat. Nanotechnol. 2021;16:243–250. doi: 10.1038/s41565-021-00854-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh R., Rana N., Liu Y., Zeng S., Agarwal G., Sonah H., Varshney R., Joshi T., Patil G.B., Nguyen H.T. Soybean transporter database: a comprehensive database for identification and exploration of natural variants in soybean transporter genes. Physiol. Plant. 2021;171:756–770. doi: 10.1111/ppl.13287. [DOI] [PubMed] [Google Scholar]
- Ding F., Qiang X., Jia Z., Li L., Hu J., Yin M., Xia S., Chen B., Qi J., Li Q., et al. Knockout of a novel salt responsive gene SlABIG1 enhance salinity tolerance in tomato. Environ. Exp. Bot. 2022;200:104903. [Google Scholar]
- Ding S., Feng X., Du H., Wang H. Genome-wide analysis of maize OSCA family members and their involvement in drought stress. PeerJ. 2019;7:e6765. doi: 10.7717/peerj.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y.T., Zhao M.J., Wang C.T., et al. Identification and characterization of GmMYB118 responses to drought and salt stress. BMC Plant Biol. 2018;18:320. doi: 10.1186/s12870-018-1551-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubiel M., De Coninck T., Osterne V.J.S., et al. The ArathEULS3 lectin ends up in stress granules and can follow an unconventional route for secretion. Int. J. Mol. Sci. 2020;21:1659. doi: 10.3390/ijms21051659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y.-B., Li J., Qin R.-Y., Xu R.-F., Li H., Yang Y.-C., Ma H., Li L., Wei P.-C., Yang J.-B. Identification of a regulatory element responsible for salt induction of rice OsRAV2 through ex situ and in situ promoter analysis. Plant Mol. Biol. 2016;90:49–62. doi: 10.1007/s11103-015-0393-z. [DOI] [PubMed] [Google Scholar]
- Egea I., Albaladejo I., Meco V., et al. The drought-tolerant Solanum pennellii regulates leaf water loss and induces genes involved in amino acid and ethylene/jasmonate metabolism under dehydration. Sci. Rep. 2018;8:2791. doi: 10.1038/s41598-018-21187-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fàbregas N., Yoshida T., Fernie A.R. Role of Raf-like kinases in SnRK2 activation and osmotic stress response in plants. Nat. Commun. 2020;11:6184. doi: 10.1038/s41467-020-19977-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Xiong L. General mechanisms of drought response and their application in drought resistance improvement in plants. Cell. Mol. Life Sci. 2015;72:673–689. doi: 10.1007/s00018-014-1767-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooq M., Park J.-R., Jang Y.-H., Kim E.-G., Kim K.-M. Rice cultivars under salt stress Show differential expression of genes related to the regulation of Na+/K+ balance. Front. Plant Sci. 2021;12:680131. doi: 10.3389/fpls.2021.680131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L., Wu D., Zhang X., Xu Y., Kuang L., Cai S., Zhang G., Shen Q. Vacuolar H+-pyrophosphatase HVP10 enhances salt tolerance via promoting Na+ translocation into root vacuoles. Plant Physiol. 2022;188:1248–1263. doi: 10.1093/plphys/kiab538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel P., Bhuria M., Sinha R., Sharma T.R., Singh A.K. Molecular Approaches in Plant Biology and Environmental Challenges. Energy, Environment, and Sustainability. Springer; 2019. Promising transcription factors for salt and drought tolerance in plants; pp. 7–50. [Google Scholar]
- Gong Q., Li P., Ma S., Indu Rupassara S., Bohnert H.J. Salinity stress adaptation competence in the extremophile Thellungiella halophila in comparison with its relative Arabidopsis thaliana. Plant J. 2005;44:826–839. doi: 10.1111/j.1365-313X.2005.02587.x. [DOI] [PubMed] [Google Scholar]
- Gong Z., Xiong L., Shi H., Yang S., Herrera-Estrella L.R., Xu G., Chao D.Y., Li J., Wang P.Y., Qin F., et al. Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 2020;63:635–674. doi: 10.1007/s11427-020-1683-x. [DOI] [PubMed] [Google Scholar]
- Groen S.C., Ćalić I., Joly-Lopez Z., Platts A.E., Choi J.Y., Natividad M., Dorph K., Mauck W.M., Bracken B., Cabral C.L.U., et al. The strength and pattern of natural selection on gene expression in rice. Nature. 2020;578:572–576. doi: 10.1038/s41586-020-1997-2. [DOI] [PubMed] [Google Scholar]
- Groen S.C., Joly-Lopez Z., Platts A.E., Natividad M., Fresquez Z., Mauck W.M., Quintana M.R., Cabral C.L.U., Torres R.O., Satija R., et al. Evolutionary systems biology reveals patterns of rice adaptation to drought-prone agro-ecosystems. Plant Cell. 2022;34:759–783. doi: 10.1093/plcell/koab275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo R., Zhao L., Zhang K., Lu H., Bhanbhro N., Yang C. Comparative genomics and transcriptomics of the extreme halophyte puccinellia tenuiflora provides insights into salinity tolerance differentiation between halophytes and glycophytes. Front. Plant Sci. 2021;12:649001. doi: 10.3389/fpls.2021.649001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A., Rico-Medina A., Caño-Delgado A.I. The physiology of plant responses to drought. Science. 2020;368:266–269. doi: 10.1126/science.aaz7614. [DOI] [PubMed] [Google Scholar]
- Gupta B.K., Sahoo K.K., Ghosh A., Tripathi A.K., Anwar K., Das P., Singh A.K., Pareek A., Sopory S.K., Singla-Pareek S.L. Manipulation of glyoxalase pathway confers tolerance to multiple stresses in rice. Plant Cell Environ. 2018;41:1186–1200. doi: 10.1111/pce.12968. [DOI] [PubMed] [Google Scholar]
- Halfter U., Ishitani M., Zhu J.K. The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. USA. 2000;97:3735–3740. doi: 10.1073/pnas.040577697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall D., Evans A.R., Newbury H.J., Pritchard J. Functional analysis of CHX21: a putative sodium transporter in Arabidopsis. J. Exp. Bot. 2006;57:1201–1210. doi: 10.1093/jxb/erj092. [DOI] [PubMed] [Google Scholar]
- Han X., Chen Z., Li P., Xu H., Liu K., Zha W., Li S., Chen J., Yang G., Huang J., et al. Development of novel rice germplasm for salt-tolerance at seedling stage using CRISPR-Cas9. Sustainability. 2022;14:2621. [Google Scholar]
- Hanin M., Ebel C., Ngom M., Laplaze L., Masmoudi K. New insights on plant salt tolerance mechanisms and their potential use for breeding. Front. Plant Sci. 2016;7:1787. doi: 10.3389/fpls.2016.01787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harish D., Bharadwaj C., Kumar T., Patil B.S., Pal M., Hegde V.S., Sarker A. Identification of stable drought tolerant landraces of chickpea (Cicer arietinum) under multiple environments. Indian J. Agric. Sci. 2020;90:1575–1581. [Google Scholar]
- Henry R.J. Innovations in plant genetics adapting agriculture to climate change. Curr. Opin. Plant Biol. 2020;56:168–173. doi: 10.1016/j.pbi.2019.11.004. [DOI] [PubMed] [Google Scholar]
- Huang T.K., Puchta H. Novel CRISPR/Cas applications in plants: from prime editing to chromosome engineering. Transgenic Res. 2021;30:529–549. doi: 10.1007/s11248-021-00238-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Cao H., Yang L., Chen C., Shabala L., Xiong M., Niu M., Liu J., Zheng Z., Zhou L., et al. Tissue-specific respiratory burst oxidase homolog-dependent H2O2 signaling to the plasma membrane H+-ATPase confers potassium uptake and salinity tolerance in Cucurbitaceae. J. Exp. Bot. 2019;70:5879–5893. doi: 10.1093/jxb/erz328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain Q., Asim M., Zhang R., Khan R., Farooq S., Wu J. Transcription factors interact with ABA through gene expression and signaling pathways to mitigate drought and salinity stress. Biomolecules. 2021;11:1159. doi: 10.3390/biom11081159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illouz-Eliaz N., Nissan I., Nir I., Ramon U., Shohat H., Weiss D. Mutations in the tomato gibberellin receptors suppress xylem proliferation and reduce water loss under water-deficit conditions. J. Exp. Bot. 2020;71:3603–3612. doi: 10.1093/jxb/eraa137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyas M., Nisar M., Khan N., Hazrat A., Khan A.H., Hayat K., Fahad S., Khan A., Ullah A. Drought tolerance strategies in plants: a mechanistic approach. J. Plant Growth Regul. 2021;40:926–944. [Google Scholar]
- Iseki K., Takahashi Y., Muto C., Naito K., Tomooka N. Diversity of drought tolerance in the genus vigna. Front. Plant Sci. 2018;9:729. doi: 10.3389/fpls.2018.00729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iswanto A.B.B., Shelake R.M., Vu M.H., Kim J.Y., Kim S.H. Genome editing for plasmodesmal biology. Front. Plant Sci. 2021;12:679140. doi: 10.3389/fpls.2021.679140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs T.B., Zhang N., Patel D., Martin G.B. Generation of a collection of mutant tomato lines using pooled CRISPR libraries. Plant Physiol. 2017;174:2023–2037. doi: 10.1104/pp.17.00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques C., Salon C., Barnard R.L., Vernoud V., Prudent M. Drought stress memory at the plant cycle level: a review. Plants (Basel) 2021;10:1873. doi: 10.3390/plants10091873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamil A., Riaz S., Ashraf M., Foolad M.R. Gene expression profiling of plants under salt stress. CRC Crit. Rev. Plant Sci. 2011;30:435–458. [Google Scholar]
- Jiang Y.Y., Chai Y.P., Lu M.H., et al. Prime editing efficiently generates W542L and S621I double mutations in two ALS genes in maize. Genome Biol. 2020;21:257. doi: 10.1186/s13059-020-02170-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z., Zhou X., Tao M., Yuan F., Liu L., Wu F., Wu X., Xiang Y., Niu Y., Liu F., et al. Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx. Nature. 2019;572:341–346. doi: 10.1038/s41586-019-1449-z. [DOI] [PubMed] [Google Scholar]
- Jiao Y., Bai Z., Xu J., Zhao M., Khan Y., Hu Y., Shi L. Metabolomics and its physiological regulation process reveal the salt-tolerant mechanism in Glycine soja seedling roots. Plant Physiol. Biochem. 2018;126:187–196. doi: 10.1016/j.plaphy.2018.03.002. [DOI] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi R.K., Bharat S.S., Mishra R. Engineering drought tolerance in plants through CRISPR/Cas genome editing. 3 Biotech. 2020;10:400–414. doi: 10.1007/s13205-020-02390-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi R., Sahoo K.K., Singh A.K., Anwar K., Pundir P., Gautam R.K., Krishnamurthy S.L., Sopory S.K., Pareek A., Singla-Pareek S.L. Enhancing trehalose biosynthesis improves yield potential in marker-free transgenic rice under drought, saline, and sodic conditions. J. Exp. Bot. 2020;71:653–668. doi: 10.1093/jxb/erz462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi R., Sahoo K.K., Tripathi A.K., Kumar R., Gupta B.K., Pareek A., Singla-Pareek S.L. Knockdown of an inflorescence meristem-specific cytokinin oxidase – OsCKX2 in rice reduces yield penalty under salinity stress condition. Plant Cell Environ. 2018;41:936–946. doi: 10.1111/pce.12947. [DOI] [PubMed] [Google Scholar]
- Joshi R., Wani S.H., Singh B., Bohra A., Dar Z.A., Lone A.A., Pareek A., Singla-Pareek S.L. Transcription factors and plants response to drought stress: current understanding and future Directions. Front. Plant Sci. 2016;7:1029. doi: 10.3389/fpls.2016.01029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S.E., Bang S.W., Kim S.H., Seo J.S., Yoon H.B., Kim Y.S., Kim J.K. Overexpression of OsERF83, a vascular tissue-specific transcription factor gene, confers drought tolerance in rice. Int. J. Mol. Sci. 2021;22:7656. doi: 10.3390/ijms22147656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam U.S., Shelake R.M., Chavhan R.L., Suprasanna P. Concerns regarding “off-target” activity of genome editing endonucleases. Plant Physiol. Biochem. 2018;131:22–30. doi: 10.1016/j.plaphy.2018.03.027. [DOI] [PubMed] [Google Scholar]
- Kakeshpour T., Tamang T.M., Motolai G., Fleming Z.W., Park J.E., Wu Q., Park S. CGFS-type glutaredoxin mutations reduce tolerance to multiple abiotic stresses in tomato. Physiol. Plant. 2021;173:1263–1279. doi: 10.1111/ppl.13522. [DOI] [PubMed] [Google Scholar]
- Kelliher T., Starr D., Su X., Tang G., Chen Z., Carter J., Wittich P.E., Dong S., Green J., Burch E., et al. One-step genome editing of elite crop germplasm during haploid induction. Nat. Biotechnol. 2019;37:287–292. doi: 10.1038/s41587-019-0038-x. [DOI] [PubMed] [Google Scholar]
- Khanday I., Skinner D., Yang B., Mercier R., Sundaresan V. A male-expressed rice embryogenic trigger redirected for asexual propagation through seeds. Nature. 2019;565:91–95. doi: 10.1038/s41586-018-0785-8. [DOI] [PubMed] [Google Scholar]
- Kim D., Alptekin B., Budak H. CRISPR/Cas9 genome editing in wheat. Funct. Integr. Genomics. 2018;18:31–41. doi: 10.1007/s10142-017-0572-x. [DOI] [PubMed] [Google Scholar]
- Kim J.Y., Mahé A., Brangeon J., Prioul J.L. A maize vacuolar invertase, IVR2, is induced by water stress. Organ/tissue specificity and diurnal modulation of expression. Plant Physiol. 2000;124:71–84. doi: 10.1104/pp.124.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.T., Choi M., Bae S.J., Kim J.S. The functional association of ACQOS/VICTR with salt stress resistance in arabidopsis thaliana was confirmed by CRISPR-mediated mutagenesis. Int. J. Mol. Sci. 2021;22:11389. doi: 10.3390/ijms222111389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konapala G., Mishra A.K., Wada Y., Mann M.E. Climate change will affect global water availability through compounding changes in seasonal precipitation and evaporation. Nat. Commun. 2020;11:3044. doi: 10.1038/s41467-020-16757-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshila Ravi R., Prema Sundara Valli P., Muthukumar T. Physiological characterization of root endophytic Fusarium haematococcum for hydrolytic enzyme production, nutrient solubilization and salinity tolerance. Biocatal. Agric. Biotechnol. 2022;43:102392. [Google Scholar]
- Kouassi A.B., Kouassi K.B.A., Sylla Z., Plazas M., Fonseka R.M., Kouassi A., Fonseka H., N'guetta A.S., Prohens J. Genetic parameters of drought tolerance for agromorphological traits in eggplant, wild relatives, and interspecific hybrids. Crop Sci. 2021;61:55–68. [Google Scholar]
- Kumar K., Gambhir G., Dass A., Tripathi A.K., Singh A., Jha A.K., Yadava P., Choudhary M., Rakshit S. Genetically modified crops: current status and future prospects. Planta. 2020;251:91. doi: 10.1007/s00425-020-03372-8. [DOI] [PubMed] [Google Scholar]
- Kushwaha H.R., Singh A.K., Sopory S.K., Singla-Pareek S.L., Pareek A. Genome wide expression analysis of CBS domain containing proteins in Arabidopsis thaliana (L.) Heynh and Oryza sativa L. reveals their developmental and stress regulation. BMC Genom. 2009;10:200. doi: 10.1186/1471-2164-10-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamaoui M., Jemo M., Datla R., Bekkaoui F. Heat and drought stresses in crops and approaches for their mitigation. Front. Chem. 2018;6:26. doi: 10.3389/fchem.2018.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers J., van der Meer T., Testerink C. How plants sense and respond to stressful environments. Plant Physiol. 2020;182:1624–1635. doi: 10.1104/pp.19.01464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan T., Zheng Y., Su Z., Yu S., Song H., Zheng X., Lin G., Wu W. OsSPL10, a SBP-Box gene, plays a dual role in salt tolerance and trichome formation in rice (Oryza sativa L.) G3. 2019;9:4107–4114. doi: 10.1534/g3.119.400700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxa M., Liebthal M., Telman W., Chibani K., Dietz K.J. The role of the plant antioxidant system in drought tolerance. Antioxidants. 2019;8:E94. doi: 10.3390/antiox8040094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontidou K., Genitsaris S., Papadopoulou A., et al. Plant growth promoting rhizobacteria isolated from halophytes and drought-tolerant plants: genomic characterisation and exploration of phyto-beneficial traits. Sci. Rep. 2020;10:14857. doi: 10.1038/s41598-020-71652-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Li X., Jiang M. CRISPR/Cas9-mediated mutagenesis of WRKY3 and WRKY4 function decreases salt and Me-JA stress tolerance in Arabidopsis thaliana. Mol. Biol. Rep. 2021;48:5821–5832. doi: 10.1007/s11033-021-06541-4. [DOI] [PubMed] [Google Scholar]
- Li P., Li Y.J., Zhang F.J., Zhang G.Z., Jiang X.Y., Yu H.M., Hou B.K. The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. Plant J. 2017;89:85–103. doi: 10.1111/tpj.13324. [DOI] [PubMed] [Google Scholar]
- Li R., Liu C., Zhao R., Wang L., Chen L., Yu W., Zhang S., Sheng J., Shen L. CRISPR/Cas9-mediated SlNPR1 mutagenesis reduces tomato plant drought tolerance. BMC Plant Biol. 2019;19:38. doi: 10.1186/s12870-018-1627-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Yuan F., Wen Z., et al. Genome-wide survey and expression analysis of the OSCA gene family in rice. BMC Plant Biol. 2015;15:261. doi: 10.1186/s12870-015-0653-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.-F., Zheng Y., Vemireddy L.R., Panda S.K., Jose S., Ranjan A., Panda P., Govindan G., Cui J., Wei K., et al. Comparative transcriptome and translatome analysis in contrasting rice genotypes reveals differential mRNA translation in salt-tolerant Pokkali under salt stress. BMC Genom. 2018;19:935. doi: 10.1186/s12864-018-5279-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S., Qin X., Luo L., Han Y., Wang X., Usman B., Nawaz G., Zhao N., Liu Y., Li R. CRISPR/Cas9-induced mutagenesis of semi-rolled leaf1, 2 confers curled leaf phenotype and drought tolerance by influencing protein expression patterns and ROS scavenging in rice (Oryza sativa L.) Agronomy. 2019;9:728. [Google Scholar]
- Lipiec J., Doussan C., Nosalewicz A., Kondracka K. Effect of drought and heat stresses on plant growth and yield: a review. Int. Agrophys. 2013;27:463–477. [Google Scholar]
- Litalien A., Zeeb B. Curing the earth: a review of anthropogenic soil salinization and plant-based strategies for sustainable mitigation. Sci. Total Environ. 2020;698:134235. doi: 10.1016/j.scitotenv.2019.134235. [DOI] [PubMed] [Google Scholar]
- Liu H., Sultan M.A.R.F., Liu X.L., Zhang J., Yu F., Zhao H.X. Physiological and comparative proteomic analysis reveals different drought responses in roots and leaves of drought-tolerant wild wheat (Triticum boeoticum) PLoS One. 2015;10 doi: 10.1371/journal.pone.0121852. e0121852–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.-J., Jian L., Xu J., Zhang Q., Zhang M., Jin M., Peng Y., Yan J., Han B., Liu J., et al. High-throughput CRISPR/Cas9 mutagenesis streamlines trait gene identification in maize. Plant Cell. 2020;32:1397–1413. doi: 10.1105/tpc.19.00934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Cui L., Xie Z., Zhang Z., Liu E., Peng X. Two NCA1 isoforms interact with catalase in a mutually exclusive manner to redundantly regulate its activity in rice. BMC Plant Biol. 2019;19:105–110. doi: 10.1186/s12870-019-1707-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Zhang J., Xu J., Li Y., Guo L., Wang Z., Zhang X., Zhao B., Guo Y.D., Zhang N. CRISPR/Cas9 targeted mutagenesis of SlLBD40, a lateral organ boundaries domain transcription factor, enhances drought tolerance in tomato. Plant Sci. 2020;301:110683. doi: 10.1016/j.plantsci.2020.110683. [DOI] [PubMed] [Google Scholar]
- Liu S., Li C., Wang H., Wang S., Yang S., Liu X., Yan J., Li B., Beatty M., Zastrow-Hayes G., et al. Mapping regulatory variants controlling gene expression in drought response and tolerance in maize. Genome Biol. 2020;21:163. doi: 10.1186/s13059-020-02069-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Wu D., Shan T., Xu S., Qin R., Li H., Negm M., Wu D., Li J. The trihelix transcription factor OsGTγ-2 is involved adaption to salt stress in rice. Plant Mol. Biol. 2020;103:545–560. doi: 10.1007/s11103-020-01010-1. [DOI] [PubMed] [Google Scholar]
- Lou D., Wang H., Liang G., Yu D. OsSAPK2 confers abscisic acid sensitivity and tolerance to drought stress in rice. Front. Plant Sci. 2017;8:993. doi: 10.3389/fpls.2017.00993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou D., Wang H., Yu D. The sucrose non-fermenting-1-related protein kinases SAPK1 and SAPK2 function collaboratively as positive regulators of salt stress tolerance in rice. BMC Plant Biol. 2018;18:203–217. doi: 10.1186/s12870-018-1408-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Tian Y., Shen R., Yao Q., Wang M., Chen M., Dong J., Zhang T., Li F., Lei M., et al. Targeted, efficient sequence insertion and replacement in rice. Nat. Biotechnol. 2020;38:1402–1407. doi: 10.1038/s41587-020-0581-5. [DOI] [PubMed] [Google Scholar]
- Lu Y., Wang J., Chen B., Mo S., Lian L., Luo Y., Ding D., Ding Y., Cao Q., Li Y., et al. A donor-DNA-free CRISPR/Cas-based approach to gene knock-up in rice. Nat. plants. 2021;7:1445–1452. doi: 10.1038/s41477-021-01019-4. [DOI] [PubMed] [Google Scholar]
- Lu Y., Ye X., Guo R., Huang J., Wang W., Tang J., Tan L., Zhu J.-K., Chu C., Qian Y. Genome-wide targeted mutagenesis in rice using the CRISPR/Cas9 system. Mol. Plant. 2017;10:1242–1245. doi: 10.1016/j.molp.2017.06.007. [DOI] [PubMed] [Google Scholar]
- Luo M., Zhang Y., Li J., Zhang P., Chen K., Song W., Wang X., Yang J., Lu X., Lu B., et al. Molecular dissection of maize seedling salt tolerance using a genome-wide association analysis method. Plant Biotechnol. J. 2021;19:1937–1951. doi: 10.1111/pbi.13607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher M.F., Nasti R.A., Vollbrecht M., Starker C.G., Clark M.D., Voytas D.F. Plant gene editing through de novo induction of meristems. Nat. Biotechnol. 2020;38:84–89. doi: 10.1038/s41587-019-0337-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manishankar P., Wang N., Köster P., Alatar A.A., Kudla J. Calcium signaling during salt stress and in the regulation of ion homeostasis. J. Exp. Bot. 2018;69:4215–4226. doi: 10.1093/jxb/ery201. [DOI] [PubMed] [Google Scholar]
- Mao H., Li S., Chen B., Jian C., Mei F., Zhang Y., Li F., Chen N., Li T., Du L., et al. Variation in cis-regulation of a NAC transcription factor contributes to drought tolerance in wheat. Mol. Plant. 2022;15:276–292. doi: 10.1016/j.molp.2021.11.007. [DOI] [PubMed] [Google Scholar]
- Mazur R., Maszkowska J., Anielska-Mazur A., Garstka M., Polkowska-Kowalczyk L., Czajkowska A., Zmienko A., Dobrowolska G., Kulik A. The SnRK2.10 kinase mitigates the adverse effects of salinity by protecting photosynthetic machinery. Plant Physiol. 2021;187:2785–2802. doi: 10.1093/plphys/kiab438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty N.S., Graham A.E., Studená L., Ledesma-Amaro R. Multiplexed CRISPR technologies for gene editing and transcriptional regulation. Nat. Commun. 2020;11:1281. doi: 10.1038/s41467-020-15053-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menz J., Modrzejewski D., Hartung F., Wilhelm R., Sprink T. Genome edited crops touch the market: a view on the global development and regulatory environment. Front. Plant Sci. 2020;11:1–17. doi: 10.3389/fpls.2020.586027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickelbart M.V., Hasegawa P.M., Bailey-Serres J. Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat. Rev. Genet. 2015;16:237–251. doi: 10.1038/nrg3901. [DOI] [PubMed] [Google Scholar]
- Mo W., Tang W., Du Y., Jing Y., Bu Q., Lin R. Phytochrome-interacting factor-like14 and Slender rice1 interaction controls seedling growth under salt stress. Plant Physiol. 2020;184:506–517. doi: 10.1104/pp.20.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton M.J.L., Awlia M., Al-Tamimi N., Saade S., Pailles Y., Negrão S., Tester M. Salt stress under the scalpel-dissecting the genetics of salt tolerance. Plant J. 2019;97:148–163. doi: 10.1111/tpj.14189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munir N., Hasnain M., Roessner U., Abideen Z. Strategies in improving plant salinity resistance and use of salinity resistant plants for economic sustainability. Crit. Rev. Environ. Sci. Technol. 2022;52:2150–2196. [Google Scholar]
- Munns R., James R.A., Xu B., Athman A., Conn S.J., Jordans C., Byrt C.S., Hare R.A., Tyerman S.D., Tester M., et al. Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nat. Biotechnol. 2012;30:360–364. doi: 10.1038/nbt.2120. [DOI] [PubMed] [Google Scholar]
- Mustafiz A., Singh A.K., Pareek A., Sopory S.K., Singla-Pareek S.L. Genome-wide analysis of rice and Arabidopsis identifies two glyoxalase genes that are highly expressed in abiotic stresses. Funct. Integr. Genomics. 2011;11:293–305. doi: 10.1007/s10142-010-0203-2. [DOI] [PubMed] [Google Scholar]
- Nawkar G.M., Lee E.S., Shelake R.M., Park J.H., Ryu S.W., Kang C.H., Lee S.Y. Activation of the transducers of unfolded protein response in plants. Front. Plant Sci. 2018;9:214. doi: 10.3389/fpls.2018.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelam K., Sahi G.K., Kumar K., Singh K. Identification of drought stress tolerance in wild species germplasm of rice based on leaf and root morphology. Plant Genet. Resour. 2018;16:289–295. [Google Scholar]
- Nhanala S.E.C., Yencho G.C. Assessment of the potential of wild Ipomoea spp. for the improvement of drought tolerance in cultivated sweetpotato Ipomoea batatas (L.) Lam. Crop Sci. 2021;61:234–249. [Google Scholar]
- Nishimasu H., Ran F.A., Hsu P.D., Konermann S., Shehata S.I., Dohmae N., Ishitani R., Zhang F., Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuñez-Muñoz L., Vargas-Hernández B., Hinojosa-Moya J., Ruiz-Medrano R., Xoconostle-Cázares B. Plant drought tolerance provided through genome editing of the trehalase gene. Plant Signal. Behav. 2021;16:1877005. doi: 10.1080/15592324.2021.1877005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata T., Ishizaki T., Fujita M., Fujita Y. CRISPR/Cas9-targeted mutagenesis of OsERA1 confers enhanced responses to abscisic acid and drought stress and increased primary root growth under nonstressed conditions in rice. PLoS One. 2020;15 doi: 10.1371/journal.pone.0243376. e0243376–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S.-J., Song S.I., Kim Y.S., Jang H.-J., Kim S.Y., Kim M., Kim Y.-K., Nahm B.H., Kim J.-K. Arabidopsis CBF3/DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiol. 2005;138:341–351. doi: 10.1104/pp.104.059147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsini F., D’Urzo M.P., Inan G., Serra S., Oh D.-H., Mickelbart M.V., Consiglio F., Li X., Jeong J.C., Yun D.-J., et al. A comparative study of salt tolerance parameters in 11 wild relatives of Arabidopsis thaliana. J. Exp. Bot. 2010;61:3787–3798. doi: 10.1093/jxb/erq188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe Y., Watanabe T., Sugano S.S., et al. Optimization of CRISPR/Cas9 genome editing to modify abiotic stress responses in plants. Sci. Rep. 2016;6:26685. doi: 10.1038/srep26685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palchetti M.V., Reginato M., Llanes A., Hornbacher J., Papenbrock J., Barboza G.E., Luna V., Cantero J.J. New insights into the salt tolerance of the extreme halophytic species Lycium humile (Lycieae, Solanaceae) Plant Physiol. Biochem. 2021;163:166–177. doi: 10.1016/j.plaphy.2021.03.054. [DOI] [PubMed] [Google Scholar]
- Palmgren M.G. Plant plasma membrane H+-ATPases: powerhouses for nutrient uptake. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001;52:817–845. doi: 10.1146/annurev.arplant.52.1.817. [DOI] [PubMed] [Google Scholar]
- Park J.J., Dempewolf E., Zhang W., Wang Z.Y. RNA-guided transcriptional activation via CRISPR/dCas9 mimics overexpression phenotypes in Arabidopsis. PLoS One. 2017;12 doi: 10.1371/journal.pone.0179410. e0179410–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.R., Kim E.G., Jang Y.H., Jan R., Farooq M., Ubaidillah M., Kim K.M. Applications of CRISPR/Cas9 as new strategies for short breeding to drought gene in rice. Front. Plant Sci. 2022;13:850441. doi: 10.3389/fpls.2022.850441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazhamala L.T., Kudapa H., Weckwerth W., Millar A.H., Varshney R.K. Systems biology for crop improvement. Plant Genome. 2021;14:1–23. doi: 10.1002/tpg2.20098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard W.F. The role of cytoplasmic streaming in symplastic transport. Plant Cell Environ. 2003;26:1–15. [Google Scholar]
- Plazas M., González-Orenga S., Nguyen H.T., Morar I.M., Fita A., Boscaiu M., Prohens J., Vicente O. Growth and antioxidant responses triggered by water stress in wild relatives of eggplant. Sci. Hortic. (Amsterdam) 2022;293:110685. [Google Scholar]
- Pramanik D., Shelake R.M., Kim M.J., Kim J.Y. CRISPR-mediated engineering across the central dogma in plant biology for basic research and crop improvement. Mol. Plant. 2021;14:127–150. doi: 10.1016/j.molp.2020.11.002. [DOI] [PubMed] [Google Scholar]
- Qin H., Wang J., Chen X., Wang F., Peng P., Zhou Y., Miao Y., Zhang Y., Gao Y., Qi Y., et al. Rice OsDOF15 contributes to ethylene-inhibited primary root elongation under salt stress. New Phytol. 2019;223:798–813. doi: 10.1111/nph.15824. [DOI] [PubMed] [Google Scholar]
- Qiu T., Qi M., Ding X., Zheng Y., Zhou T., Chen Y., Han N., Zhu M., Bian H., Wang J. The SAUR41 subfamily of SMALL AUXIN UP RNA genes is abscisic acid inducible to modulate cell expansion and salt tolerance in Arabidopsis thaliana seedlings. Ann. Bot. 2020;125:805–819. doi: 10.1093/aob/mcz160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X.M., Sun Y.Y., Ye X.Y., Li Z.G. Signaling role of glutamate in plants. Front. Plant Sci. 2020;10:1743. doi: 10.3389/fpls.2019.01743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez Gonzales L., Shi L., Bergonzi S.B., Oortwijn M., Franco-Zorrilla J.M., Solano-Tavira R., Visser R.G.F., Abelenda J.A., Bachem C.W.B. Potato CYCLING DOF FACTOR 1 and its lncRNA counterpart StFLORE link tuber development and drought response. Plant J. 2021;105:855–869. doi: 10.1111/tpj.15093. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Rigano M.M., Arena C., Di Matteo A., Sellitto S., Frusciante L., Barone A. Eco-physiological response to water stress of drought-tolerant and drought-sensitive tomato genotypes. Plant Biosyst. 2016;150:682–691. [Google Scholar]
- Roca Paixão J.F., Gillet F.X., Ribeiro T.P., Bournaud C., Lourenço-Tessutti I.T., Noriega D.D., Melo B.P., de Almeida-Engler J., Grossi-de-Sa M.F. Improved drought stress tolerance in Arabidopsis by CRISPR/dCas9 fusion with a histone acetyltransferase. Sci. Rep. 2019;9:1–9. doi: 10.1038/s41598-019-44571-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santosh Kumar V.V., Verma R.K., Yadav S.K., Yadav P., Watts A., Rao M.V., Chinnusamy V. CRISPR-Cas9 mediated genome editing of drought and salt tolerance (OsDST) gene in indica mega rice cultivar MTU1010. Physiol. Mol. Biol. Plants. 2020;26:1099–1110. doi: 10.1007/s12298-020-00819-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt C., Fransz P., Rönspies M., Dreissig S., Fuchs J., Heckmann S., Houben A., Puchta H. Changing local recombination patterns in Arabidopsis by CRISPR/Cas mediated chromosome engineering. Nat. Commun. 2020;11:4418. doi: 10.1038/s41467-020-18277-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabala S., Bose J., Hedrich R. Salt bladders: do they matter? Trends Plant Sci. 2014;19:687–691. doi: 10.1016/j.tplants.2014.09.001. [DOI] [PubMed] [Google Scholar]
- Shelake R.M., Pramanik D., Kim J.Y. Evolution of plant mutagenesis tools: a shifting paradigm from random to targeted genome editing. Plant Biotechnol. Rep. 2019;13:423–445. [Google Scholar]
- Shelake R.M., Pramanik D., Kim J.-Y. Exploration of plant-microbe interactions for sustainable agriculture in CRISPR era. Microorganisms. 2019;7:269. doi: 10.3390/microorganisms7080269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelake R.M., Pramanik D., Kim J.-Y. In Vivo rapid investigation of crispr-based base editing components in Escherichia coli (IRI-CCE): a platform for evaluating base editing tools and their components. Int. J. Mol. Sci. 2022;23:1145. doi: 10.3390/ijms23031145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C., Zhang Y., Li Q., Liu S., He F., An Y., Zhou Y., Liu C., Yin W., Xia X. PdGNC confers drought tolerance by mediating stomatal closure resulting from NO and H2O2 production via the direct regulation of PdHXK1 expression in Populus. New Phytol. 2021;230:1868–1882. doi: 10.1111/nph.17301. [DOI] [PubMed] [Google Scholar]
- Shi H., Quintero F.J., Pardo J.M., Zhu J.-K. The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell. 2002;14:465–477. doi: 10.1105/tpc.010371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Gao H., Wang H., Lafitte H.R., Archibald R.L., Yang M., Hakimi S.M., Mo H., Habben J.E. ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 2017;15:207–216. doi: 10.1111/pbi.12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim J.S., Oh N., Chung P.J., Kim Y.S., Choi Y.D., Kim J.K. Overexpression of OsNAC14 improves drought tolerance in rice. Front. Plant Sci. 2018;9:310. doi: 10.3389/fpls.2018.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastava P., Kumar R. Soil salinity: a serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J. Biol. Sci. 2015;22:123–131. doi: 10.1016/j.sjbs.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A.K., Kumar R., Tripathi A.K., Gupta B.K., Pareek A., Singla-Pareek S.L. Genome-wide investigation and expression analysis of Sodium/Calcium exchanger gene family in rice and Arabidopsis. Rice. 2015;8:54. doi: 10.1186/s12284-015-0054-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A.K., Sopory S.K., Wu R., Singla-Pareek S.L. Abiotic Stress Adaptation in Plants. Springer Netherlands; Dordrecht: 2009. Transgenic approaches; pp. 417–450. [Google Scholar]
- Singh M., Kumar J., Singh S., Singh V.P., Prasad S.M. Roles of osmoprotectants in improving salinity and drought tolerance in plants: a review. Rev. Environ. Sci. Biotechnol. 2015;14:407–426. [Google Scholar]
- Singh M., Nara U., Kumar A., Choudhary A., Singh H., Thapa S. Salinity tolerance mechanisms and their breeding implications. J. Genet. Eng. Biotechnol. 2021;19:173. doi: 10.1186/s43141-021-00274-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer N., de León N., Grotewold E. Challenges of translating gene regulatory information into agronomic improvements. Trends Plant Sci. 2019;24:1075–1082. doi: 10.1016/j.tplants.2019.07.004. [DOI] [PubMed] [Google Scholar]
- Sun T., Ma N., Wang C., Fan H., Wang M., Zhang J., Cao J., Wang D. A golgi-localized sodium/hydrogen exchanger positively regulates salt tolerance by maintaining higher K+/Na+ ratio in soybean. Front. Plant Sci. 2021;12:1–15. doi: 10.3389/fpls.2021.638340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinnen G., Goossens A., Pauwels L. Lessons from domestication: targeting cis-regulatory elements for crop improvement. Trends Plant Sci. 2016;21:506–515. doi: 10.1016/j.tplants.2016.01.014. [DOI] [PubMed] [Google Scholar]
- Tan P., Du X., Shang Y., Zhu K., Joshi S., Kaur K., Khare T., Kumar V. Ion transporters and their exploration for conferring abiotic stress tolerance in plants. Plant Growth Regul. 2022;96:1–23. [Google Scholar]
- Tester M., Davenport R. Na+ tolerance and Na+ transport in higher plants. Ann. Bot. 2003;91:503–527. doi: 10.1093/aob/mcg058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong K., Wu X., He L., Qiu S., Liu S., Cai L., Rao S., Chen J. Genome-wide identification and expression profile of OSCA gene family members in Triticum aestivum L. Int. J. Mol. Sci. 2021;23:469. doi: 10.3390/ijms23010469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran M.T., Doan D.T.H., Kim J., Song Y.J., Sung Y.W., Das S., Kim E.-J., Son G.H., Kim S.H., Van Vu T., et al. CRISPR/Cas9-based precise excision of SlHyPRP1 domain(s) to obtain salt stress-tolerant tomato. Plant Cell Rep. 2021;40:999–1011. doi: 10.1007/s00299-020-02622-z. [DOI] [PubMed] [Google Scholar]
- Van Zelm E., Zhang Y., Testerink C. Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 2020;71:403–433. doi: 10.1146/annurev-arplant-050718-100005. [DOI] [PubMed] [Google Scholar]
- Varotto S., Tani E., Abraham E., Krugman T., Kapazoglou A., Melzer R., Radanović A., Miladinović D. Epigenetics: possible applications in climate-smart crop breeding. J. Exp. Bot. 2020;71:5223–5236. doi: 10.1093/jxb/eraa188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney R.K., Barmukh R., Roorkiwal M., Qi Y., Kholova J., Tuberosa R., Reynolds M.P., Tardieu F., Siddique K.H.M. Breeding custom-designed crops for improved drought adaptation. Adv. Genet. 2021;2:e202100017. doi: 10.1002/ggn2.202100017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney R.K., Bohra A., Yu J., Graner A., Zhang Q., Sorrells M.E. Designing future crops: genomics-assisted breeding comes of age. Trends Plant Sci. 2021;26:631–649. doi: 10.1016/j.tplants.2021.03.010. [DOI] [PubMed] [Google Scholar]
- Vlčko T., Ohnoutková L. Allelic variants of CRISPR/Cas9 induced mutation in an inositol trisphosphate 5/6 kinase gene manifest different phenotypes in barley. Plants. 2020;9:195. doi: 10.3390/plants9020195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Zhong Z., Wang X., Han X., Yu D., Wang C., Song W., Zheng X., Chen C., Zhang Y. Knockout of the OsNAC006 transcription factor causes drought and heat sensitivity in rice. Int. J. Mol. Sci. 2020;21:22888. doi: 10.3390/ijms21072288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Chen L., Li R., Zhao R., Yang M., Sheng J., Shen L. Reduced drought tolerance by CRISPR/Cas9-mediated SlMAPK3 mutagenesis in tomato plants. J. Agric. Food Chem. 2017;65:8674–8682. doi: 10.1021/acs.jafc.7b02745. [DOI] [PubMed] [Google Scholar]
- Wang P., Hsu C.C., Du Y., Zhu P., Zhao C., Fu X., Zhang C., Paez J.S., Macho A.P., Tao W.A., et al. Mapping proteome-wide targets of protein kinases in plant stress responses. Proc. Natl. Acad. Sci. USA. 2020;117:3270–3280. doi: 10.1073/pnas.1919901117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Xun H., Wang W., Ding X., Tian H., Hussain S., Dong Q., Li Y., Cheng Y., Wang C., et al. Mutation of GmAITR genes by CRISPR/Cas9 genome editing results in enhanced salinity stress tolerance in soybean. Front. Plant Sci. 2021;12 doi: 10.3389/fpls.2021.779598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.C., Lin T.C., Kieber J., Tsai Y.C. Response Regulators 9 and 10 negatively regulate salinity tolerance in rice. Plant Cell Physiol. 2019;60:2549–2563. doi: 10.1093/pcp/pcz149. [DOI] [PubMed] [Google Scholar]
- Wang X., He Y., Wei H., Wang L. A clock regulatory module is required for salt tolerance and control of heading date in rice. Plant Cell Environ. 2021;44:3283–3301. doi: 10.1111/pce.14167. [DOI] [PubMed] [Google Scholar]
- Wang X., Wang W., Wang Y., Zhou G., Liu S., Li D., Adnan, Hussain S., Hussain S., Ahmed S., Zhang C., et al. AIW1 and AIW2, two ABA-induced WD40 repeat-containing transcription repressors function redundantly to regulate ABA and salt responses in Arabidopsis. J. Plant Interact. 2020;15:196–206. [Google Scholar]
- Wang Y., Cao Y., Liang X., et al. A dirigent family protein confers variation of Casparian strip thickness and salt tolerance in maize. Nat. Commun. 2022;13:2222. doi: 10.1038/s41467-022-29809-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Hong Y., Li Y., Shi H., Yao J., Liu X., Wang F., Huang S., Zhu G., Zhu J. Natural variations in SlSOS1 contribute to the loss of salt tolerance during tomato domestication. Plant Biotechnol. J. 2021;19:20–22. doi: 10.1111/pbi.13443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Hong Y., Zhu G., Li Y., Niu Q., Yao J., Hua K., Bai J., Zhu Y., Shi H., et al. Loss of salt tolerance during tomato domestication conferred by variation in a Na+/K+ transporter. EMBO J. 2020;39:1–14. doi: 10.15252/embj.2019103256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson A., Ghosh S., Williams M.J., Cuddy W.S., Simmonds J., Rey M.-D., Asyraf Md Hatta M., Hinchliffe A., Steed A., Reynolds D., et al. Speed breeding is a powerful tool to accelerate crop research and breeding. Nat. plants. 2018;4:23–29. doi: 10.1038/s41477-017-0083-8. [DOI] [PubMed] [Google Scholar]
- Weiss T., Crisp P.A., Rai K.M., Song M., Springer N.M., Zhang F. Epigenetic Features Drastically Impact CRISPR-Cas9 Efficacy in Plants. Plant Physiol. 2022 doi: 10.1093/plphys/kiac285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Yan G., Duan Z., Wang Z., Kang C., Guo L., Liu K., Tu J., Shen J., Yi B., et al. Roles of the Brassica napus DELLA Protein BnaA6.RGA, in modulating drought tolerance by interacting with the ABA signaling component BnaA10. ABF2. Front. Plant Sci. 2020;11:577. doi: 10.3389/fpls.2020.00577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Chen S., Cheng Y., Zhang N., Ma Y., Wang W., Tian H., Li Y., Hussain S., Wang S. Cell wall/vacuolar inhibitor of fructosidase 1 regulates ABA response and salt tolerance in Arabidopsis. Plant Signal. Behav. 2020;15:1744293. doi: 10.1080/15592324.2020.1744293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L., Zhang M., Wu R., Chen X., Liu F., Xing B. Genome-wide analysis of OSCA gene family members in Vigna radiata and their involvement in the osmotic response. BMC Plant Biol. 2021;21:408–412. doi: 10.1186/s12870-021-03184-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W., Xiao Y., Niu M., Meng W., Li L., Zhang X., Liu D., Zhang G., Qian Y., Sun Z., et al. ARGONAUTE2 enhances grain length and salt tolerance by activating BIG GRAIN3 to modulate cytokinin distribution in rice. Plant Cell. 2020;32:2292–2306. doi: 10.1105/tpc.19.00542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T., Fujita Y., Sayama H., Kidokoro S., Maruyama K., Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010;61:672–685. doi: 10.1111/j.1365-313X.2009.04092.x. [DOI] [PubMed] [Google Scholar]
- Yu D., Ke L., Zhang D., Wu Y., Sun Y., Mei J., Sun J., Sun Y. Multi-omics assisted identification of the key and species-specific regulatory components of drought-tolerant mechanisms in Gossypium stocksii. Plant Biotechnol. J. 2021;19:1690–1692. doi: 10.1111/pbi.13655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Lin T., Meng X., Du H., Zhang J., Liu G., Chen M., Jing Y., Kou L., Li X., et al. A route to de novo domestication of wild allotetraploid rice. Cell. 2021;184:1156–1170.e14. doi: 10.1016/j.cell.2021.01.013. [DOI] [PubMed] [Google Scholar]
- Yu W., Wang L., Zhao R., et al. Knockout of SlMAPK3 enhances tolerance to heat stress involving ROS homeostasis in tomato plants. BMC Plant Biol. 2019;19:354. doi: 10.1186/s12870-019-1939-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Duan X., Luo L., Dai S., Ding Z., Xia G. How plant hormones mediate salt stress responses. Trends Plant Sci. 2020;25:1117–1130. doi: 10.1016/j.tplants.2020.06.008. [DOI] [PubMed] [Google Scholar]
- Yuan F., Yang H., Xue Y., Kong D., Ye R., Li C., Zhang J., Theprungsirikul L., Shrift T., Krichilsky B., et al. OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature. 2014;514:367–371. doi: 10.1038/nature13593. [DOI] [PubMed] [Google Scholar]
- Yue E., Cao H., Liu B. Osmir535, a potential genetic editing target for drought and salinity stress tolerance in Oryza sativa. Plants. 2020;9:13377. doi: 10.3390/plants9101337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafar S.A., Zaidi S.S.E.A., Gaba Y., Singla-Pareek S.L., Dhankher O.P., Li X., Mansoor S., Pareek A. Engineering abiotic stress tolerance via CRISPR/Cas-mediated genome editing. J. Exp. Bot. 2020;71:470–479. doi: 10.1093/jxb/erz476. [DOI] [PubMed] [Google Scholar]
- Zeng D.D., Yang C.C., Qin R., Alamin M., Yue E.K., Jin X.L., Shi C.H. A guanine insert in OsBBS1 leads to early leaf senescence and salt stress sensitivity in rice (Oryza sativa L.) Plant Cell Rep. 2018;37:933–946. doi: 10.1007/s00299-018-2280-y. [DOI] [PubMed] [Google Scholar]
- Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., Van Der Oost J., Regev A., et al. Cpf1 is a single rna-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zhang J., Wei P., et al. The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol J. 2014;12:797–807. doi: 10.1111/pbi.12200. [DOI] [PubMed] [Google Scholar]
- Zhang A., Liu Y., Wang F., Li T., Chen Z., Kong D., Bi J., Zhang F., Luo X., Wang J., et al. Enhanced rice salinity tolerance via CRISPR/Cas9-targeted mutagenesis of the OsRR22 gene. Mol. Breed. 2019;39:47. doi: 10.1007/s11032-019-0954-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B. MicroRNA: a new target for improving plant tolerance to abiotic stress. J. Exp. Bot. 2015;66:1749–1761. doi: 10.1093/jxb/erv013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zhu J., Gong Z., Zhu J.K. Abiotic stress responses in plants. Nat. Rev. Genet. 2021;23:104–119. doi: 10.1038/s41576-021-00413-0. [DOI] [PubMed] [Google Scholar]
- Zhang M., Cao Y., Wang Z., Wang Z.Q., Shi J., Liang X., Song W., Chen Q., Lai J., Jiang C. A retrotransposon in an HKT1 family sodium transporter causes variation of leaf Na+ exclusion and salt tolerance in maize. New Phytol. 2018;217:1161–1176. doi: 10.1111/nph.14882. [DOI] [PubMed] [Google Scholar]
- Zhang S., Wang L., Zhao R., Yu W., Li R., Li Y., Sheng J., Shen L. Knockout of SlMAPK3 reduced disease resistance to Botrytis cinerea in tomato plants. J. Agric. Food Chem. 2018;66:8949–8956. doi: 10.1021/acs.jafc.8b02191. [DOI] [PubMed] [Google Scholar]
- Zhao C., Zhang H., Song C., Zhu J.-K., Shabala S. Mechanisms of plant responses and adaptation to soil salinity. Innovation. 2020;1:100017. doi: 10.1016/j.xinn.2020.100017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Zhang C., Liu W., et al. An alternative strategy for targeted gene replacement in plants using a dual-sgRNA/Cas9 design. Sci. Rep. 2016;6:23890. doi: 10.1038/srep23890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Lin J., Liu X., Chu W., Li J., Gao Y., An K., Song W., Xin M., Yao Y., et al. Histone acetyltransferase TaHAG1 acts as a crucial regulator to strengthen salt tolerance of hexaploid wheat. Plant Physiol. 2021;186:1951–1969. doi: 10.1093/plphys/kiab187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Zhang F., Zhai F., Su Y., Zhou Y., Ge Z., Tilak P., Eirich J., Finkemeier I., Fu L., et al. Rice GLUTATHIONE PEROXIDASE1-mediated oxidation of bZIP68 positively regulates ABA-independent osmotic stress signaling. Mol. Plant. 2022;15:651–670. doi: 10.1016/j.molp.2021.11.006. [DOI] [PubMed] [Google Scholar]
- Zhou J., Deng K., Cheng Y., et al. CRISPR-Cas9 based genome editing reveals new insights into microRNA function and regulation in rice. Front. Plant Sci. 2017;8:1598. doi: 10.3389/fpls.2017.01598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Zhang Y., Wang X., Han X., An Y., Lin S., Shen C., Wen J., Liu C., Yin W., et al. Root-specific NF-Y family transcription factor, PdNF-YB21, positively regulates root growth and drought resistance by abscisic acid-mediated indoylacetic acid transport in Populus. New Phytol. 2020;227:407–426. doi: 10.1111/nph.16524. [DOI] [PubMed] [Google Scholar]
- Zhu J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zsögön A., Čermák T., Naves E.R., Notini M.M., Edel K.H., Weinl S., Freschi L., Voytas D.F., Kudla J., Peres L.E.P. De novo domestication of wild tomato using genome editing. Nat. Biotechnol. 2018;36:1211–1216. doi: 10.1038/nbt.4272. [DOI] [PubMed] [Google Scholar]
- Zwack P.J., Rashotte A.M. Interactions between cytokinin signalling and abiotic stress responses. J. Exp. Bot. 2015;66:4863–4871. doi: 10.1093/jxb/erv172. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.