Summary
Single-cell RNA sequencing (scRNA-seq) and spatially resolved transcriptomics (SRT) have experienced rapid development in recent years. The findings of spaceflight-based scRNA-seq and SRT investigations are likely to improve our understanding of life in space and our comprehension of gene expression in various cell systems and tissue dynamics. However, compared to their Earth-based counterparts, gene expression experiments conducted in spaceflight have not experienced the same pace of development. Out of the hundreds of spaceflight gene expression datasets available, only a few used scRNA-seq and SRT. In this perspective piece, we explore the growing importance of scRNA-seq and SRT in space biology and discuss the challenges and considerations relevant to robust experimental design to enable growth of these methods in the field.
Keywords: spaceflight, RNA-sequencing, transcriptomics, single-cell, spatially resolved transcriptomics, spatial
In this perspective, Overbey et al. summarize the current datasets available that have profiled mammalian spaceflight samples using single-cell RNA sequencing and spatially resolved transcriptomics. They also discuss special considerations needed for tissue preservation and data analysis for these sample types.
Introduction
For decades, the health risks associated with space exploration have been increasingly documented and characterized. For some of these risks, countermeasures have been sufficiently developed to sustain human health in low-Earth orbit, the orbital distance from Earth that includes the International Space Station (ISS). However, as humankind plans for a permanent presence on the moon and seeks to advance to Mars, the limits to our countermeasures and the research gaps that have insofar remained unaddressed will create health challenges that rival the engineering challenges of long-duration space missions.
To gain a comprehensive understanding of how spaceflight affects the human body, the molecular responses of the body to spaceflight factors must be robustly profiled. Recent advances in omics technologies have already revolutionized our approach to understanding disease1,2,3 and have created new pathways for tackling health challenges on Earth.4,5,6 However, the spaceflight environment presents a unique set of factors where the genomic impact is largely unknown. Furthermore, the microgravity and radiation environment of space remains difficult to model on Earth. Therefore, given that the opportunities to study the influence of spaceflight on the human genome have historically been, and still are, limited, it is of paramount importance to perform all omics experiments in spaceflight to the highest standards.
The ISS is currently the only research laboratory available off-world and the source of the majority of spaceflight omics experiments performed to date. Most mammalian omics studies aboard the ISS have used Mus musculus, the standard laboratory rodent, as a model organism.7 To scale the number of mammalian studies performed in space, NASA launched their rodent research (RR) program in 2014, with the first omics experiment published in the NASA funded GeneLab database in 2015.8,9 Since the program began, the RR program has launched over a dozen missions generating a steady stream of new omics data on a variety of murine tissues and cell systems. These missions have proven essential for understanding the molecular response of terrestrial life to the environmental stressors of space by providing gene expression data for a subset of mouse tissues. These studies have helped illuminate changes within biological systems including the mitochondria,10 musculoskeletal system,11 circadian rhythm,12 and microRNA (miRNA) profiles.13
The majority of experiments from rodent tissue obtained via the RR missions have been subject to bulk RNA sequencing (RNA-seq), where the whole tissue is homogenized and RNA is extracted. Bulk transcriptomic assays are currently the most common omics assay performed in space biology, with more than 270 transcriptomic datasets available from spaceflight and spaceflight-analogue experiments in the NASA GeneLab database.7 Of these studies, microarray-based experiments have been used to obtain approximately half of the gene expression data stored in GeneLab. As the cost of next-generation sequencing (NGS) decreased, RNA-seq started to replace microarrays as the predominant method for spaceflight transcriptomic studies. However, given recent advances in the field of genomics, bulk RNA-seq is being displaced by higher-resolution methods rapidly on Earth. State-of-the-art tools can now resolve gene expression at the single-cell level via single-cell RNA-seq (scRNA-seq)14 or maintain 2D topography via spatially resolved transcriptomics (SRT).15 While these two types of approaches are becoming standard practice for gene expression research conducted on Earth, the collection protocols for cells and tissues obtained from experiments in spaceflight have not yet been optimized for the application of these methods.
In this perspective piece, we present the current status of scRNA-seq and SRT in spaceflight experiments, the optimal tissue preservation protocols that should be implemented to ensure the success of these experiments, and the downstream computational tools that are used to obtain high-resolution gene expression insights.
scRNA-seq: Importance to spaceflight and prior ISS study designs
In less than a decade, the field of scRNA-seq has matured and produced multiple robust protocols for examining the gene expression of individual cells. This has resulted in a paradigm shift in the field of transcriptomics that has enabled the identification of new cell types, uncovered rare cell populations, revealed cellular heterogeneity, charted cell-lineage trajectories during development and disease, and profiled the perturbation of cellular states.16,17,18,19,20,21
scRNA-seq also has great potential when applied to cell systems and tissues exposed to spaceflight. While gene expression changes have been observed in cell systems and tissues, the exact cell populations driving these expression differences have not yet been characterized. For example, large changes in gene expression have been observed in the murine retina,22 but the differences attributable to specific retinal subpopulations, such as rod photoreceptors, cone photoreceptors, retinal ganglion cells, bipolar cells, amacrine cells, and horizontal cells, have yet to be pinpointed. Similarly, differences in immune function have been observed during spaceflight in the past,23,24,25 where the application of scRNA-seq would have enabled further dissection of specific gene expression differences in the cell populations of, for example, peripheral blood mononuclear cells (PBMCs), thymus tissue, and bone marrow. Overall, the possibility of identifying exact cell populations that are dysregulated during spaceflight enables the development of more precise therapeutics and countermeasures.
While there is a spectrum of approaches for scRNA-seq that have been applied across ground-based experiments, only two platforms (Chromium by 10X Genomics and Rhapsody Single-Cell Analysis System by BD Biosciences) have been applied to samples flown in spaceflight.26,27 These were applied to two experiments: the second rodent research reference mission (RRRM-2) and the NASA Twins Study. In both, single-cell suspensions were obtained, cells were physically isolated from one another, and polyadenylated mRNA was captured using oligo(dT)s that were uniquely labeled per cell to differentiate mRNA of different cells in the experiment.
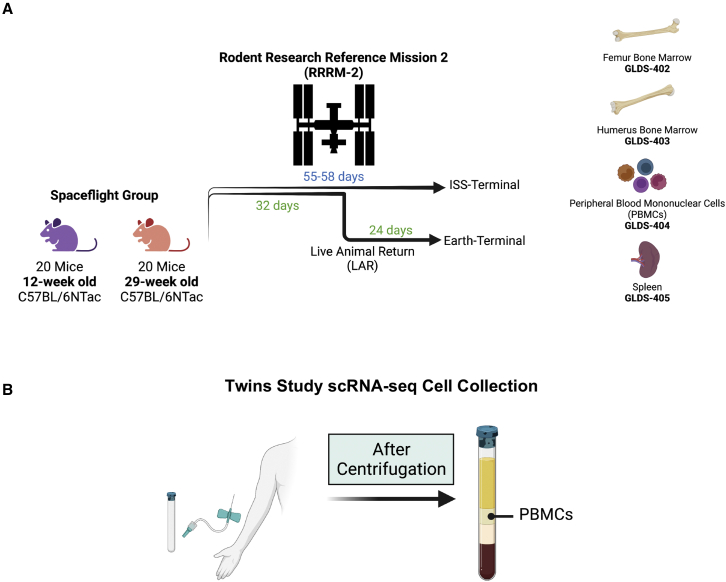
In the RRRM-2 experiment, 40 mice were flown to the ISS and were part of four distinct groups distinguished by age, duration spent on the ISS, and whether the animal was ISS-terminal or Earth-acclimatized after live animal return (LAR) (Figure 1A). A matching set of 40 mice were also housed on Earth to serve as a ground control. Out of the 80 mice between the ground control and spaceflight groups, 16 animals that were part of the LAR group were selected for single-cell sequencing. Single-cell sequencing of spleen tissue, PBMCs, humerus bone marrow, and femur bone marrow was performed using the 10X Genomics Chromium platform. Data for these experiments are publicly available in GeneLab28,29,30,31 (Table 1) and analysis of these datasets is underway in NASA’s Analysis Working Groups (https://genelab.nasa.gov/awg/charter).
Figure 1.
scRNA-seq datasets from spaceflight samples
(A) Design for the rodent research reference mission 2 (RRRM-2) experiment. In RRRM-2, two age groups of mice were launched to the ISS. After 32 days, half of the mice from each age group returned to Earth, where they lived 24 additional days before dissection. This was time-matched with the remaining groups of mice in orbit, which were euthanized and frozen in-flight. Sixteen mice total were selected for scRNA-seq using the 10X Genomics platform. Four cell/tissue types were selected for scRNA-seq, and they each have an associated GeneLab Dataset (GLDS) publicly available: femur bone marrow (GLDS-402), humerus bone marrow (GLDS-403), PBMCs (GLDS-404), and spleen tissue (GLDS-405).
(B) PBMCs from human blood were obtained before and after spaceflight on the ISS as part of the NASA Twins Study. Whole blood was obtained using venipuncture into a sodium citrate cell processing tube (CPT) and PBMCs were obtained from the buffy coat after centrifugation of the CPT.
Table 1.
scRNA-seq datasets from spaceflight
| ID | Link | Mission | Tissue | Organism |
|---|---|---|---|---|
| GLDS-402 | https://genelab-data.ndc.nasa.gov/genelab/accession/GLDS-402/ | RRRM-2 | femur bone marrow | C57BL/6NTac mice |
| GLDS-403 | https://genelab-data.ndc.nasa.gov/genelab/accession/GLDS-403/ | RRRM-2 | humerus bone marrow | C57BL/6NTac mice |
| GLDS-404 | https://genelab-data.ndc.nasa.gov/genelab/accession/GLDS-404/ | RRRM-2 | PBMCs | C57BL/6NTac mice |
| GLDS-405 | https://genelab-data.ndc.nasa.gov/genelab/accession/GLDS-405/ | RRRM-2 | Spleen | C57BL/6NTac mice |
| LSDA | available upon request to the NASA LSDA | NASA Twin Study | PBMCs | human |
A complete list of scRNA-seq datasets from samples exposed to spaceflight. Four datasets are from the RRRM-2 mission and are available on GeneLab. One dataset is from the NASA Twin Study and is part of the NASA Life Sciences Data Archive (LSDA). Due to privacy concerns around human genomic data, the Twin Study data is not publicly available but can be requested. The LSDA has a public-facing portal where data requests can be initiated (https://nlsp.nasa.gov/explore/entryform/lsda_data_requests/lsda_data_requests). The LSDA team provides the appropriate processes, tools, and secure infrastructure for archival of experimental data and dissemination while complying with applicable rules, regulations, policies, and procedures governing the management and archival of sensitive data and information.
The only other published experiment that has used scRNA-seq from a spaceflight study is the NASA Twins Study.32 In this experiment, a set of identical twins participated in a set of omics assays. One twin served as a ground control and remained on Earth while the other twin spent 340 days aboard the ISS. For scRNA-seq, whole blood was collected via venipuncture into cell processing tubes (CPTs) containing sodium citrate as a preservative (Figure 1B). The CPTs were returned immediately to the lab and centrifuged to collect PBMCs. The BD Biosciences Single-Cell Analysis System was used to collect single-cell expression data (Table 1). One important caveat to this study is that there was no in-flight time point, and blood was collected for scRNA-seq pre-launch and post-landing.13,33 The time it takes to return cargo, such as CPTs, to Earth is one of the factors limiting the ease of single-cell experiments. CPTs can be spun-down and frozen before returning to Earth, but this direct freezing process will lyse cells. With cell lysis, it is still possible to generate a sample-wide RNA-seq profile but not at single-cell resolution. To generate single-cell profiles from spaceflight samples, cells will need to be either returned to Earth immediately or cells will need to be preserved using a process that prevents lysis.
Spatially resolved transcriptomics: Leveraging legacy samples and steps for proper experimental design prior to spaceflight
While scRNA-seq has greatly improved gene expression experiments, it does have one major limitation: the loss of locational information due to breakdown of tissues into single-cell suspensions. This makes it difficult to identify gene expression changes that may correlate with the topology of the tissue and limits our ability to decipher cell-to-cell interactions and microenvironment-level changes. In order to retain locational information, a variety of SRT methods have been developed in recent years (see Table 2 for a list of recent SRT methods)15,34,35 and applied to a range of sample and tissue types.36,37,38,39
Table 2.
SRT methods
| SRT Technique | Sample type | Preservation technique | Resolution | References |
|---|---|---|---|---|
| ISS (padlock probe & RCA) |
cells and tissue | FF or FFPE | single cell (transcript level) | Ke et al., 201340 |
| FISH | cells and tissue | FF | single cell (transcript level) | Lubeck et al., 201441 |
| Tomo-seq | tissue slice | FF | multiple cells | Junker et al., 201442 |
| TIVA | intact live cells | none | single cell | Lovatt et al., 201443 |
| FISSEQ | cells and tissue | FF or FFPE | single cell | (Lee et al., 2015)44 |
| MERFISH/MER SCOPE | cells and tissue | FF or FFPE | single cell (transcript level/100 nm) | Moffitt et al., 201645 |
| LCM-seq | tissue | FF or FFPE | multiple cells | Nichterwitz et al., 201646 |
| 10X Genomics Visium | cells and tissue | FF or FFPE | multiple cells (55 μm) | Ståhl et al., 201647 |
| Geo-seq | cells and tissue | FF or FFPE | multiple cells | Chen et al., 201748 |
| STARmap | tissue | FF | single cell | Wang et al., 201849 |
| HDST | tissue | FF | near single cell (2 μm) | Vickovic et al., 201950 |
| DBiT-Seq | cells and tissue | FF or FFPE | near single cell (10 μm) | Liu et al., 202051 |
| NanoString GeoMx | tissue | FF or FFPE | multiple cells | Zollinger et al., 202052 |
| SlideSeq2 | tissue | FF | near single cell (10 μm) | Stickels et al., 202153 |
| BGI scStereo-seq | tissue | FF | single cell (transcript level/500 nm) | Xia et al., 202154 |
| Resolve Biosciences Molecular Cartography | tissue | FF | single cell (transcript level/200 nm) | Groiss et al., 202155 |
| NanoString CosMx | tissue | FF or FFPE | single cell (transcript level/50 nm) | He et al., 202156 |
An evolving list of SRT methods. Acronyms used: ISS, in situ sequencing; LCM-seq, laser capture microdissection; TIVA, transcriptome in vivo analysis; HDST, high definition spatial transcriptomics; FF, flash frozen; FFPE, formalin-fixed paraffin-embedded. Note that this list is not extensive, and there are several more SRT approaches available. Preference was given to newly developed or unique methods.
Due to various complexities in data collection and analysis, SRT methods are less mature than scRNA-seq methods. Additionally, because of the requirement that tissues remain intact during the SRT workflow, these methods are also more sensitive to the tissue types and structures selected for study. This poses an extra challenge when SRT methods are to be applied to legacy space-flown samples that were collected and stored without planning SRT studies from the start of the missions and their morphology is not fully preserved. For these reasons, we believe improvements can be made in the tissue collection process to better preserve tissue morphology. Specifically, when designing future studies to be performed in spaceflight, where specimens will undergo SRT analysis, experiments should first be optimized on Earth before being translated to experiments performed in space. The overall testing cycle for SRT techniques can be divided into three phases: (1) tissue evaluation, (2) tissue preparation, and (3) 2D RNA analysis.
Tissue evaluation will require selection of the proper area of interest of the tissue, which may vary in length, width, and thickness depending on the SRT method of interest. Tissue preparation will require testing to determine the optimal duration of exposure to the fixative agent, which may vary depending on tissue type and method of fixation (e.g., flash frozen [FF], formalin-fixed paraffin-embedded [FFPE] tissue, or a different method). It should be noted that most SRT methods to date have been validated with FF and FFPE tissues (Table 2). The third phase, 2D RNA analysis, will analyze the RNA spatial distribution in the tissue under study by applying one of the SRT methods available at the time of the experiment (Table 2). Due to the scarce availability of tissues flown in spaceflight, it is critical that SRT researchers employ a testing cycle before proceeding to their spaceflight samples.
Tissue preservation: Challenges in spaceflight and recommendations for improvement
Good sample preservation is instrumental to the application of scRNA-seq and SRT on tissues from spaceflight. This stands in contrast to bulk RNA-seq, where RNA is collected and pooled from hundreds of thousands to millions of cells. The sheer volume of RNA collected from these many cells means that partial tissue degradation can occur and still yield enough RNA for sequencing. The tolerance threshold for RNA degradation is much lower for scRNA-seq and SRT due to the high-resolution nature of these methods, compared to bulk RNA-seq. For scRNA-seq and SRT, sufficient RNA for sequencing must be obtained from a smaller number of cells.
RNA is known to degrade quickly and can undergo rapid profile changes and degradation in relatively short periods of time resulting in measurable differences within 5 to 10 min of remaining at room temperature postmortem.57 Additionally, RNA that is not preserved rapidly is subject to degradation that can create distinct RNA-seq profiles, and different RNA transcripts can degrade at different rates.58,59 To account for this, samples intended for scRNA-seq must be preserved rapidly after euthanasia. If the experiment requires single-cell sequencing, the cells should be immediately placed in cryopreservation media and slowly brought to freezing temperature to ensure cells do not lyse during the freezing process. Additionally, tissues should be dissociated prior to freezing. Dissociation after freezing is likely to cause damage to cell membranes and increase the likelihood of RNA leakage due to the stresses of the freezing process.
An alternative to immediate dissociation is the application of single-nucleus RNA-seq (snRNA-seq) where isolation of nuclei can be performed on frozen material, and the nuclei are subsequently used as input for single-cell sequencing.60 In this case, tissue samples can forgo cryopreservation media and should be brought down to freezing temperatures as quickly as possible, ideally snap-freezing the samples in liquid nitrogen if available. Comparisons of snRNA-seq and scRNA-seq in mouse lungs have shown that they have similar gene detection rates, and dissociation bias is reduced in snRNA-seq.61 Likewise, comparisons in kidneys showed similar gene detection rates.62 The recently completed Fly Cell Atlas63 utilized snRNA-seq instead of scRNA-seq because it can be applied consistently even in problematic tissue type (e.g., antennae, wings, fat bodies) and multinucleated cells (e.g., muscle) and because 70%–90% of transcriptomic information is retained with snRNA-seq.63 Moreover, nuclei can be applied as input for single-cell multiomics studies where both the RNA and chromatin accessibility information is captured from the same nucleus simultaneously. Thus, this approach can be extended in spaceflight experiments in the future.
Similarly, samples intended for SRT should be frozen immediately after dissection, ideally in a protective embedding medium. This approach preserves both RNA quality and morphology of the tissue. Most of legacy tissues obtained from the ISS did not undergo immediate embedding and freezing for morphology preservation. In the majority of cases, individual organs were not dissected in-flight, but instead were frozen as a whole carcass on the ISS. These samples were returned to Earth where they were thawed, dissected, and re-frozen until ready for downstream sequencing approaches, mostly in bulk. Due to the necessity of organ dissection back in the laboratory on Earth, many rodent samples exposed to the space environment have undergone this double-freezing procedure.64,65,66,67,68 However, this process might lead to partial RNA degradation and deformation of cell and tissue anatomical shape if specific strategies for tissue morphology preservation were not taken into account at the time of freezing. Any deformation can make application of spatial methods very challenging and, in some cases, unfit for SRT methods.
There is also evidence that whole-carcass preservation leads to different gene expression profiles than tissues dissected in-flight. One study has demonstrated that whole-carcass preservation in-flight leads to greater variability in RNA integrity numbers (RINs) than tissue dissected in-flight.69 Another study compared bulk RNA-seq profiles between mouse liver tissue dissected and frozen directly on the ISS compared to mice preserved as frozen whole carcasses with their liver tissue dissected back on Earth.70 This study notes that whole-carcass preservation in RNAlater can better preserve the tissues; however, there are a few caveats to this approach. The first caveat is that RNAlater has only had limited success in preserving cells for scRNA-seq on Earth71 and will require further optimization before its application can be recommended for such experiments in spaceflight. The second consideration is that RNAlater can induce a unique gene expression response due to the sulfates present in the solution.72 These gene expression responses may be uneven in SRT methods due to the diffusion gradient generated as RNAlater disperses across a tissue. Third, RNAlater is not a fixative agent and has not been tested to our knowledge at the time of writing for compatibility with fixative reagents used in various SRT methods.
Despite the uncertainties in using RNAlater in scRNA-seq and SRT experiments, there is precedent for preserving nucleic acids with RNAlater on the ISS for other sequencing experiments (Table 3). Other reagents with a success in preserving cells and/or nucleic acids on the ISS include using NOTOXhisto, 10% neutral buffer formation (NBF), 4% paraformaldehyde (PFA), and the sequential use of trypsin followed by RNAprotect (Table 3). However, these preservatives are not necessarily suitable for scRNA-seq and SRT due to the lack of testing with scRNA-seq and SRT platforms or due to cell lysis that occurs during the preservation process. The current options with a track record on the ISS are the slow and fast freeze methods to store the tissue at a temperature that will preserve RNA and spatial tissue structure. −80°C freezers available on the ISS can be used to bring tissues down to freezing temperatures, but without a snap-freezing step, this process occurs slowly. To solve this problem, a rapid-freeze instrument was added to the ISS to rapidly bring sample temperatures down to −185°C, to compete with snap-freezing methods that make use of liquid nitrogen that is easily available and safely applied on Earth but not a safe option in a space-based lab73,74,75,76 (Table 3).
Table 3.
Preservation strategies aboard the ISS
| Tissue Preservation on ISS | Properties | Reference |
|---|---|---|
| RNAlater | (1) non-toxic (2) stabilizes cellular RNA (3) eliminates the requirement of immediate freezing of samples |
Choi et al., 2016; Gupta et al., 2015; Herranz et al., 2019; Manzano et al., 2020; Paul et al., 2012, 2005; Schultz et al., 2013; Vandenbrink et al., 2019; Villacampa et al., 2021; Wnorowski et al., 201977,78,79,80,81,82,83,84,85,86 |
| NOTOXhisto | (1) cell-fixative (2) substitute for formalin |
Balsamo et al., 2014; Cockell et al., 202087,88 |
| 10% neutral buffer formation (NBF) | (1) less toxic (2) low cost (3) penetrates tissue relatively fast (4) random cross linking of proteins (5) chemical modification of nucleic acids |
Zamarioli et al., 202189 |
| 4% paraformaldehyde (PFA) | (1) preservation of morphology (2) limited use for RNA extraction (3) formaldehyde is toxic |
Huang et al., 202090 |
| Trypsin, RNAprotect, and −95°C storage |
(1) stabilizes cellular RNA (2) lyses cells |
Huang et al., 202090 |
| Slow freeze (−80°C freezers or colder) |
(1) good for cellular preservation (2) requires specialized equipment (3) formation of ice crystal artifacts |
Choi et al., 2020; Hong et al., 202169,91 |
| Rapid freeze to −185°C | (1) rapid freezing through conduction (2) good for cellular preservation (3) requires specialized equipment (4) high Cost (5) avoids ice crystal formation |
Andersen et al., 2005; Saravia-Butler et al., 2020b, 2020c, 2020a73,74,75,76 |
A list of strategies used for cell, tissue, and nucleic acids preservations that have been previously used in experiments in spaceflight.
There are two options that can help address these challenges moving forward: in-flight tissue dissection and LAR. In-flight tissue dissection will allow tissues of interest to be rapidly frozen, as smaller pieces of tissue freeze faster than a whole carcass, and will avoid the double freeze-thaw that tissues undergo if they are dissected from a frozen carcass back on Earth. The drawback to this approach is that tissue dissection is time-intensive and will require specialized crew training to teach dissection competency. Despite the time commitment required for in-flight dissection, it has been performed in select cases, such as hindlimb dissection during the RR-6 mission,92 though the data from this in-flight dissection are not publicly available. The second approach, LAR, returns organisms to Earth while still alive where they are euthanized after returning to a ground-based lab.93,94 This allows tissues to be preserved according to the highest standards but presents a different set of limitations. Using LAR, organisms have their circadian rhythms disrupted and experience the stressful conditions of atmospheric re-entry. Moreover, depending on the study design of the mission, LAR organisms have the opportunity to begin reacclimating to Earth’s gravity and radiation landscape. Therefore, all three aspects can impact their transcriptomic profiles.
Overall, in spite of the associated limitations, using one of these two options will help circumvent some of the challenges related to the applicability of scRNA-seq and SRT methods on spaceflight samples.
Advancing off-world capabilities: Considerations for performing scRNA-seq and SRT in spaceflight
After the challenge of achieving more rigorous preservation standards for scRNA-seq and SRT methods is met, the next goal should be to increase the capabilities of molecular biology experiments in space. Ideally, RNA extractions, PCR, and even scRNA-seq and SRT itself should be performed during spaceflight. Achieving this will require the development of workflows compatible with microgravity and more sophisticated workstations for performing molecular biology experiments.
The capabilities for performing molecular biology experiments have increased dramatically over the past decade. In 2016, the first biomolecule sequencer, the Oxford Nanopore MinION, was demonstrated to work in microgravity aboard the ISS.95 Since then, the repertoire of molecular biology experiments performed in-flight has expanded to include environmental microbial sampling, plasmid transformation, DNA extraction, PCR amplification, and library prep.96,97,98 While the original biomolecular sequencer study sequenced Mus musculus DNA libraries (prepared on Earth),95 others have been primarily defined in scope of microbial organisms and not to multicellular model organisms, such as C. elegans, Drosophila melanogaster, and Mus musculus, or to human samples. In order to build a spaceflight lab capable of performing scRNA-seq and SRT (methods that are most frequently applied to multicellular organisms) the molecular biology pipeline for handling tissues post-dissection throughout the sequencing process must be more well defined.
Additionally, due to the sensitive nature of RNA, additional laboratory decontamination protocols may need translation to spaceflight. RNase inhibitors will need to be available to wipe down workstations, pipettes, gloves, and any other surfaces the sample may come in contact with. A microbially decontaminated environment would also enhance the quality of RNA obtained, particularly the use of 70% ethanol, isopropanol, or another disinfectant, given the microbe-rich surfaces aboard the ISS.96,97,99 However, the use of ethanol and isopropanol on the ISS is an issue because aerosolized alcohol interferes with the environmental control and life support systems (ECLSSs) in the closed-loop space station environment (J. Perry et al., 2016, 46th International Conference on Environmental Systems, conference).
Moreover, the workstation used for handling samples should have minimal airflow that would pass dust or other airborne particulates across the workstation. Some of these considerations may be met via use of the Microgravity Science Glovebox100 aboard the ISS to control airflow and by using benzalkonium chloride (BZK) wipes in place of alcohol-based disinfectants. Establishing and following general guidelines, regularly applied when preparing samples for scRNA-seq and SRT, will help create consistency in sample retrieval, prevent RNA degradation, and obtain high-quality data from spaceflight samples in which RNA quantity and quality are not compromised. One caveat to performing scRNA-seq and SRT methods in spaceflight is that it will require additional crew time to complete experiments that are typically done on Earth after sample return. Robotic automation can help overcome these challenges by reducing the amount of time that crew will need to spend executing experiments (Figure 2A). For example, automated imaging has already been performed aboard the ISS.101 Additionally, automation of library preparation steps has been performed on Earth from a spatially barcoded chip array similar to the 10X Genomics Visium format102 as well as from 10X Genomics Visium arrays.103 However, due to strict volume and mass limitations in-flight, compact solutions may need to be investigated, as would specific testing to accommodate microgravity-driven changes in fluidic behavior. Automating these processes and translating them to spaceflight-compatible hardware will alleviate potential time-costs placed on the crew by performing these experiments in-flight in their entirety.
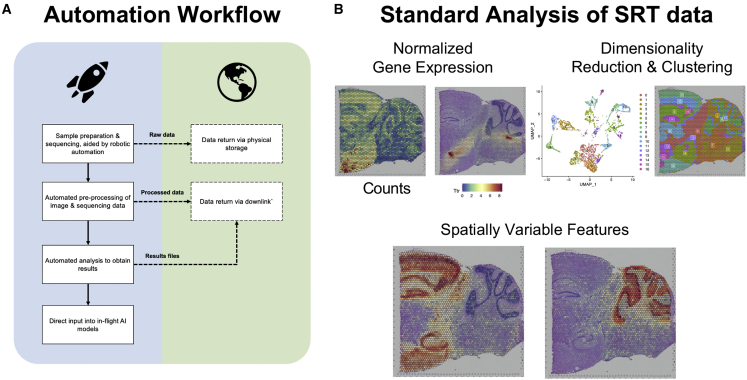
Figure 2.
Computational analysis of spatial transcriptomics data
(A) Shows the flow of data that could be unlocked with increased computational automation. The right column shows sequential steps of proposed computational automation onboard spacecraft, with the dotted lines to boxes on the left showing opportunities to transfer data back to Earth.
(B) Example of data analysis workflow from the 10X Genomics Visium on the “Mouse Brain Serial Section 2 (Sagittal-Posterior)” dataset available through the 10X Genomics Dataset portal (https://www.10xgenomics.com/resources/datasets). Dataset analyzed with Seurat.
Another challenge involves the abundance of data generated by sequencing and imaging experiments, which would all need to be stored in-flight and downlinked to Earth. This is particularly relevant to SRT data, which involves either the generation of large fastq files or multiple high-resolution images. To mitigate data downlink challenges, automated software pipelines could be executed in-flight and could convert data into formats with decreased file size. This processed data could then be downlinked to Earth instead of larger, raw files. Thus, onboard data processing could decrease delays in data analysis caused by the transfer of large files. Full transcriptomics analysis pipelines could even be conducted in space, where results, such as differentially expressed genes (DEGs) and enriched pathways, could be automatically extracted from transcriptomic data in-flight (Figure 2A). Additionally, these results could also be used as feedback to optimize experimental parameters in-flight and increase the quality of future transcriptomic experiments performed in space.
Bioinformatic pipelines and considerations for SRT in spaceflight experiments
Transcriptomic analyses for bulk RNA-seq and scRNA-seq are relatively well defined compared to SRT. For bulk RNA-seq, NASA GeneLab has developed an RNA-seq consensus pipeline (RCP) to standardize processing of short-read RNA-seq data,104 which builds upon standard recommendations in the field.105 The RCP is accessible at the NASA GeneLab_Data_Processing Github repository. However, a standardized pipeline for scRNA-seq is not yet available from NASA GeneLab or international space omics consortia, such as ISSOP (International Standards for Space Omics Processing) or the ESA-funded Space Omics Topical Team. Data analysis for scRNA-seq presents many unique challenges related to inferring cell trajectories, clustering cell types, identifying and filtering doublets (when two cells are mistakenly considered as a single cell), handling of sparsity in gene expression count tables, and defining flexible statistical frameworks for discovering complex differential patterns in gene expression.106 Despite these challenges, standardized methods, analysis frameworks, and tutorials are available,107,108,109 which can be extended to analysis of data from space flight experiments.
In comparison, the establishment of best practices for the analysis of SRT data is a work in progress. The advent of SRT allows for solving the spatial distribution of transcripts, as well as locating cell types and cell states in complex tissues, organs, and organoids. Standardizing analysis across a wide variety of tissue configurations and the large number of emerging technologies is a difficult and complex task. Knowledge of the types of software tools available for each method can help inform experimental selection decisions about which SRT method best matches the specific scientific questions under investigation.
Widely, there are two types of SRT data: single-cell resolution and multi-cell pixel resolution. SRT single-cell resolution data enable the researchers to quantify absolute transcript counts per cell and produce cell gene count tables comparable to scRNA-seq experiments. However, the data are often imaging-based and thus limited in the amount of genes detected in one experiment. On the other hand, the SRT multi-cell pixel resolution data present multiple cells per region of interest and use cell type deconvolution algorithms and label transfer/projection from additional reference scRNA-seq datasets to resolve the cell type composition of the whole-transcriptome region of interest. Regardless of the resolution level of the SRT technique selected, available software packages, for example Seurat110 and Squidpy,111 offer compatibility with several SRT technologies because the structure of the data is similar, meaning that the majority of information comes from transcript counts. Spatial coordinates and cell-level information can be stored as metadata portions of the data object. Both tools include methods for QC (quality control) and preprocessing, normalization, dimensionality reduction, clustering, and visualization in spatial coordinates and finding spatially variable and spatial marker genes (Figure 2B). However, these do not support data processing and analysis for all SRT methods.
Specialized tools have also been developed for cluster analysis,112,113 fusion transcript detection,114 visualization,115 deconvolution of cell-type mixtures,116 inference of extracellular interacting genes,117 identification of sub-tissue architecture/anatomization, finding spatially variable genes,118,119,120 quantifying spatial heterogeneity,121 imputation of missing mRNA expression,122 or improvement of gene expression quantification using spatial, temporal and experimental coordinates. Moreover, comprehensive tools such as STutility builds on the Seurat framework to facilitate analyzing and visualizing multiple samples from the 10X Genomics Visium platform.123 Integration across data types also gives these tools increased investigative potential. For example, integration of single-cell and spatial transcriptomics data, achieved with tools such as cell2location124 and stereoscope125 to name a few, can enhance spatial transcriptomics resolution to an unprecedented level.126 Moreover, integration of gene expression data with histology information and spatial location using machine learning (ML) is also under active research.127 For instance, the framework Squidpy is interconnected with Scanpy and the ML Python ecosystem.111
Despite these advances, challenges associated with metadata standards and data processing exist, including reliable cell segmentation, cell-type identification, integration of expression patterns to anatomical features, finding complex patterns, integration of data from different specimens, time points or perturbations, development of standardized metrics and benchmarks, as well as integration across multiple technologies.128 Other analytical challenges remain for the analysis of SRT data, including those related to sample mapping and image registration and segmentation, data representation, imputation of sparse spatial data, integration of multi-modal data, and automatic inference of knowledge for the combination of tissue structure and molecular data.111
While the above challenges exist for both terrestrial and spaceflight SRT data, some are more specific to spaceflight studies due to the problems related to sample collection and processing, such as the formation of ice crystals,129 which can cause morphology changes and tissue damage leading to altered spatial information. In data derived from spaceflight experiments, true morphological and transcriptional variability that results from exposure to microgravity and radiation should not be confused with artifacts created by sample collection, preservation, and transportation. Therefore, if artifacts due to confounding factors cannot be sufficiently minimized via sample processing protocols or specific experimental design, software tools aiming to remove confounding factors (for example, cells affected by ice crystals), improve transcript quantification, and reconstruct morphology and architecture of damaged tissue using image processing techniques will be essential. Specifically, reconstruction of deformed images using image registration or image deconvolution/restoration techniques and the development of computational tools to elucidate space-driven modification of tissue organization will warrant investigation.
Future prospects for omics technology in spaceflight
The aging ISS, destined to reach its end of life by 2030, has catalyzed an era of “commercial space,” which promises rapid development of the final frontier. To avoid a gap in continuous human presence in space, four private companies are aiming to build and operate commercial space stations in low-Earth orbit. Axiom Space will launch and attach the first segment of its modular space station to the ISS in 2024 and build the remainder of the station while attached to the ISS. The completed Axiom Station will detach to become a free flier by 2030 (https://techcrunch.com/2021/11/30/nasa-details-intent-to-replace-the-international-space-station-with-a-commercial-space-station-by-2030/). Blue Origin has partnered with Sierra Space to build Orbital Reef, while StarLab will be born out of a partnership between Nanoracks, Voyager Space, and Lockheed Martin. The fourth, yet to be named, space station development effort is being led by Northrup Grumman (https://news.northropgrumman.com/news/releases/northrop-grumman-signs-agreement-with-nasa-to-design-space-station-for-low-earth-orbit). Furthermore, advances in rocket technology, including reusable engines, have lowered launch costs significantly and increased the cadence of spaceflights to and from the ISS. For example, SpaceX Crew-4 launched to the ISS less than 2 days after Ax-1, Axiom’s private astronaut mission to the ISS, returned to Earth.
Research in space has been fundamental to our understanding of how to survive and thrive in the weightless environment. Our knowledge and sophistication of scientific investigations in space is continuously strengthened by advances in hardware technology and operations. Numerous hardware options for biological investigations that require cell culture capabilities and maintenance of live specimen in spaceflight, for instance, are already available. A few examples of include Biopack,130 SABL from BioServe Space Technologies (T. Niederwieser, 2015, 45th International Conference on Environmental Systems, conference), Kubik,131 NanoRacks Frame-3, STaARS-1 EF132, and the CubeLab from Space Tango.133 A more current system called the ScienceTaxi (https://www.yurigravity.com/our-service) developed by Yuri Gravity can be integrated into the ISS and into the newer stations coming online; an autonomous version of the ScienceTaxi is in development. This small sampling of hardware options is indicative of the demand for laboratory equipment that is operational in a weightless space environment and the importance of biological experiments conducted in space.
Now, in this new era of commercial space with new space stations in development, there is an unprecedented opportunity to leverage past research experiences in space to re-think and re-design future technologies and laboratories for spaceflight investigations that are on-par with Earth technologies, yet not limited by gravity-based thinking. For example, the speed of technology and methodology innovation for scRNA-seq and SRT is advancing the field rapidly for terrestrial investigations. Future scRNA-seq and SRT methodologies will enable simultaneous multi-omic measurements from individual cells and spatial regions. The potential of these developments has already been commercially realized for single-cell methodologies by 10X Genomics, whose Single Cell Multiome ATAC + Gene Expression kits offer simultaneous measurement of scRNA-seq and chromatin accessibility (ATAC-seq) data from the same group of cells.134,135 Similar developments are in motion for SRT where co-mapping of mRNAs and proteins in a formaldehyde-fixed tissue section via NGS has already been achieved.51 Moreover, spatially resolved ATAC-seq has been developed,136,137 but does not yet allow for simultaneous measurement of RNA. Given the trajectory of the single-cell and spatial fields toward simultaneous measurements of multiple genomic features, it is anticipated that the need for improved cell and tissue preservation protocols in spaceflight will persist.
In addition to these creative workarounds, there is an opportunity to design and invent new paradigms for conducting omics and other biological research in space. Space scientists must define the requirements for on-orbit research protocols and outline the specific types of research investigations important to conduct in space, the tools and capabilities necessary for those space investigations, and the criteria for acceptable high-quality science on orbit. These requirements will serve as a guide for commercial space station developers in defining their own orbital lab designs, including hardware capabilities and services. Armed with an understanding of the types of equipment required to conduct the research, commercial space station developers would be better equipped to consider partnerships with terrestrial technology developers that are leading the rapid advances in omics technology and encourage them to adapt their technologies for a weightless space environment by developing future iterations of their technology with space constraints in mind.
Although launch costs are reducing and launch frequency is increasing, these costs based on mass and volume are still substantial and prohibitive. Offering end-to-end omics capabilities on orbit that are grounded in the concept of “cloud labs” will be a paradigm shift for space research and science. Cloud labs are essentially a virtual lab bench that allow the scientist to control the wet-lab experiment virtually and execute it remotely via an orchestra of automated robotic systems.138 Presently on the ISS and likely in the foreseeable future, several key operational constraints limit rapid, repeatable research in orbit: (1) limited human-tended time for executing experimental protocols, (2) variability in the on-orbit operator’s scientific and technical experience and expertise, (3) a large distance between the principal investigator (PI) and the scientific payload, and (4) inability to rapidly iterate on experimental protocols in real-time. A cloud lab in space will enable the PI to (1) control the experiment via the virtual lab bench, (2) conduct high-quality science by eliminating a variety of confounding variables that are introduced by returning samples to the ground for processing, and (3) iterate on the scientific protocol based on real-time observations of data. Furthermore, on-orbit edge processing and cloud-computing capabilities will accelerate data analysis whereby processed results can be returned to the PI via a secure downlink from the space station. Addition of such capabilities will eliminate the need for large data downloads—a practice that is slow, costly, and requires extremely high bandwidths.
While an upfront investment of capital is required for a state-of-the-art orbital lab, a high return on investment can be expected. Improved on-orbit capabilities that are easier to access and operate will generate greater interest in space-based research investigations, which in turn will increase the demand to leverage spaceflight as an extreme environment for research—for example, the general adaptability of the human body, which may result in the discovery of new molecular mechanisms. It is expected that the reduction in costs to execute orbital science will translate into lower prices for the researcher, thereby generating even greater interest in space. Taken together, these developments will drive a feedforward loop toward greater insights for extending humanity’s footprint in space and improving life on the ground.
Even with rapid advances in genomics and spaceflight technology, experimentation in space will remain costly and time-consuming in the foreseeable future. Thus, careful design of experiments is crucial. One factor that may alleviate the risk of these experiments is in silico modeling. Perturbations on transcription and gene expression can be modeled using ML algorithms to test hypotheses prior to launching an experiment into space.139 Adapting these models to be more amenable to the environmental factors of spaceflight can help use limited spaceflight resources more efficiently. Additionally, to aid with experimental design, a specific SRT method should be selected before experiment launch to implement its optimized tissue preservation protocol, and the development status of available computational tools should be rigorously assessed to understand the expected analysis types and data resolutions available. Given the rapid advancements in SRT analysis, ideal tools and capabilities will grow quickly over time. This sector will continue to push for more state-of-the-art knowledge and state-of-the-art technology never attempted before to develop innovative approaches for improving life sciences research in space. It is also a high-risk, high-reward sector, where projects tend to be expensive to fund and maintain but have the potential to unlock discoveries essential for humans to become a multi-planetary species. Through joint collaborations and partnerships and by building on previous milestones, resources, and tools that have been established for spaceflight, we can continue to rapidly advance our in-flight genomics capabilities.
Results that emerge from scRNA-seq and SRT experiments conducted in spaceflight are expected to increase our understanding of life in space, genetic control of fundamental processes that are altered by spaceflight, and genomic control mechanisms in cellular biology. Research groups and scientists in the field of space biosciences will continue to generate robust multi-omic profiles of both in vitro and in vivo biospecimens available from previous missions and biospecimen samples returned from future missions. The quality of these data will increase as better preservation protocols and innovative orbital lab solutions are implemented. The results will allow us to pinpoint cell-type-specific and locational disturbances in gene expression and will facilitate the development of countermeasures against spaceflight-associated dysregulation and disease. This goal is also in line with current international collaborative efforts to expand our presence in LEO (low Earth orbit), return to the moon, and progress forward to Mars.
Acknowledgments
H.C., P.M., D.B., R.H., N.J.S., J.B., and S.G. are members of the ESA Space Omics Topical Team, funded by the ESA grant/contract 4000131202/20/NL/PG/pt “Space Omics: Towards an integrated ESA/NASA – omics database for spaceflight and ground facilities experiments” awarded to R.H., which was the main funding source for this work. H.C. is also supported by the Horizon Centre for Doctoral Training at the University of Nottingham (UKRI grant no. EP/S023305/1). S.G. is supported by the Swedish Research Council VR grant 2020-04864. E.G.O. is supported through NASA Postdoctoral Fellowship 80NSSC21K0316. We thank Lindsay A. Rutter, Masafumi Muratani, the ISSOP, and the ESA Space Omics Topical Team for fruitful discussions. We would also like to thank Sarah Castro-Wallace for the personal communication on current ISS research.
Author contributions
Conceptualization, E.G.O., S.D., H.C., P.M., R.K., R.T.S., J.B., and S.G.; investigation, E.G.O., S.D., H.C., P.M., Z.A., S.F., R.K., D.B., R.T.S., J.P., D.C., J.M.G., S.V.C., C.E.M., J.B., and S.G.; writing – original draft, E.G.O., S.D., H.C., P.M., Z.A., S.F., R.K., R.T.S., J.B., and S.G.; writing – review & editing, all authors; visualization, E.G.O., H.C., and P.M.; supervision, S.G.; funding acquisition, J.M.G., S.V.C., C.E.M., R.H., N.J.S., and S.G.
Declarations of interests
C.E.M. is a co-Founder of Biotia and Onegevity Health. D.B. is the CSO at yuri and a co-founder at Poppy Health. S.G. is a scientific advisor to 10X Genomics Inc., which holds IP rights to the ST technology and holds stock options.
References
- 1.Hasin Y., Seldin M., Lusis A. Multi-omics approaches to disease. Genome Biol. 2017;18:83. doi: 10.1186/s13059-017-1215-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manzoni C., Kia D.A., Vandrovcova J., Hardy J., Wood N.W., Lewis P.A., Ferrari R. Genome, transcriptome and proteome: the rise of omics data and their integration in biomedical sciences. Brief. Bioinform. 2018;19:286–302. doi: 10.1093/bib/bbw114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mathé E., Hays J., Stover D., Chen J. The omics revolution continues: the Maturation of high-throughput biological data sources. Yearb. Med. Inform. 2018;27:211–222. doi: 10.1055/s-0038-1667085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu M., Zhan X. The crucial role of multiomic approach in cancer research and clinically relevant outcomes. EPMA J. 2018;9:77–102. doi: 10.1007/s13167-018-0128-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vlaanderen J., Moore L.E., Smith M.T., Lan Q., Zhang L., Skibola C.F., Rothman N., Vermeulen R. Application of OMICS technologies in occupational and environmental health research; current status and projections. Occup. Environ. Med. 2010;67:136–143. doi: 10.1136/oem.2008.042788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang J., Yan Y., Zhong W. Application of omics technology to combat the COVID-19 pandemic. MedComm. 2020;2:381–401. doi: 10.1002/mco2.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berrios D.C., Galazka J., Grigorev K., Gebre S., Costes S. NASA GeneLab: interfaces for the exploration of space omics data. Nucleic Acids Res. 2021;49:D1515–D1522. doi: 10.1093/nar/gkaa887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Globus R., Cadena S., Galazka J. Rodent Research-1 (RR1) National Lab Validation Flight: Mouse Liver Transcriptomic, Proteomic, and Epigenomic Data. NASA GeneLab. 2015 doi: 10.26030/K5C1-JD05. [DOI] [Google Scholar]
- 9.Globus R., Galazka J.M., Marcu O., Gebre S.G., Polo S.-H.L., Saravia-Butler A.M., Fogle H.W., Bense H., Chakravarty K., Chen R.B., et al. Rodent Research-1 (RR1) NASA Validation Flight: Mouse Liver Transcriptomic, Proteomic, and Epigenomic Data. NASA GeneLab. 2015 doi: 10.26030/JQ04-0N51. [DOI] [Google Scholar]
- 10.da Silveira W.A., Fazelinia H., Rosenthal S.B., Laiakis E.C., Kim M.S., Meydan C., Kidane Y., Rathi K.S., Smith S.M., Stear B., et al. Comprehensive multi-omics analysis reveals mitochondrial stress as a central biological Hub for spaceflight impact. Cell. 2020;183:1185–1201.e20. doi: 10.1016/j.cell.2020.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cahill T., Cope H., Bass J.J., Overbey E.G., Gilbert R., da Silveira W.A., Paul A.M., Mishra T., Herranz R., Reinsch S.S., et al. Mammalian and Invertebrate models as Complementary tools for gaining Mechanistic insight on muscle responses to spaceflight. Int. J. Mol. Sci. 2021;22:9470. doi: 10.3390/ijms22179470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujita S.-I., Rutter L., Ong Q., Muratani M. Integrated RNA-seq analysis Indicates Asynchrony in Clock genes between tissues under spaceflight. Life. 2020;10:196. doi: 10.3390/life10090196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malkani S., Chin C.R., Cekanaviciute E., Mortreux M., Okinula H., Tarbier M., Schreurs A.S., Shirazi-Fard Y., Tahimic C.G., Rodriguez D.N., et al. Circulating miRNA spaceflight Signature reveals targets for countermeasure development. Cell Rep. 2020;33 doi: 10.1016/j.celrep.2020.108448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen G., Ning B., Shi T. Single-cell RNA-seq technologies and related computational data analysis. Front. Genet. 2019;10:317. doi: 10.3389/fgene.2019.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Asp M., Bergenstråhle J., Lundeberg J. Spatially resolved transcriptomes-next generation tools for tissue exploration. Bioessays. 2020;42 doi: 10.1002/bies.201900221. [DOI] [PubMed] [Google Scholar]
- 16.Aizarani N., Saviano A., Sagar, Mailly L., Durand S., Herman J.S., Pessaux P., Baumert T.F., Grun D. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature. 2019;572:199–204. doi: 10.1038/s41586-019-1373-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montoro D.T., Haber A.L., Biton M., Vinarsky V., Lin B., Birket S.E., Yuan F., Chen S., Leung H.M., Villoria J., et al. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature. 2018;560:319–324. doi: 10.1038/s41586-018-0393-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parikh K., Antanaviciute A., Fawkner-Corbett D., Jagielowicz M., Aulicino A., Lagerholm C., Davis S., Kinchen J., Chen H.H., Alham N.K., et al. Colonic epithelial cell diversity in health and inflammatory bowel disease. Nature. 2019;567:49–55. doi: 10.1038/s41586-019-0992-y. [DOI] [PubMed] [Google Scholar]
- 19.Park J., Shrestha R., Qiu C., Kondo A., Huang S., Werth M., Li M., Barasch J., Susztak K. Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science. 2018;360:758–763. doi: 10.1126/science.aar2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trapnell C. Defining cell types and states with single-cell genomics. Genome Res. 2015;25:1491–1498. doi: 10.1101/gr.190595.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilbrey-Clark A., Roberts K., Teichmann S.A. Cell Atlas technologies and insights into tissue architecture. Biochem. J. 2020;477:1427–1442. doi: 10.1042/bcj20190341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Overbey E.G., da Silveira W.A., Stanbouly S., Nishiyama N.C., Roque-Torres G.D., Pecaut M.J., Zawieja D.C., Wang C., Willey J.S., Delp M.D., et al. Spaceflight influences gene expression, photoreceptor integrity, and oxidative stress-related damage in the murine retina. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-49453-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akiyama T., Horie K., Hinoi E., Hiraiwa M., Kato A., Maekawa Y., Takahashi A., Furukawa S. How does spaceflight affect the acquired immune system? NPJ Microgravity. 2020;6:14. doi: 10.1038/s41526-020-0104-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baqai F.P., Gridley D.S., Slater J.M., Luo-Owen X., Stodieck L.S., Ferguson V., Chapes S.K., Pecaut M.J. Effects of spaceflight on innate immune function and antioxidant gene expression. J. Appl. Physiol. 2009;106:1935–1942. doi: 10.1152/japplphysiol.91361.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crucian B.E., Chouker A., Simpson R.J., Mehta S., Marshall G., Smith S.M., Zwart S.R., Heer M., Ponomarev S., Whitmire A., et al. Immune system dysregulation during spaceflight: potential countermeasures for Deep space exploration missions. Front. Immunol. 2018;9:1437. doi: 10.3389/fimmu.2018.01437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shum E.Y., Walczak E.M., Chang C., Christina Fan H. Quantitation of mRNA transcripts and proteins using the BD Rhapsody™ single-cell analysis system. Adv. Exp. Med. Biol. 2019;1129:63–79. doi: 10.1007/978-981-13-6037-4_5. [DOI] [PubMed] [Google Scholar]
- 27.Zheng G.X.Y., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R., Ziraldo S.B., Wheeler T.D., McDermott G.P., Zhu J., et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017;8 doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galazka J.M., Juran C., Cekanaviciute E., Polo S.-H.L., Saravia-Butler A.M., Boyko V., Stodieck L., Roberts M., Costes S.V., Gebre S.G., Ferguson V. Single Cell Transcriptional Profiling of Spleens from Mice Flown on Rodent Research Reference Mission-2 (RRRM-2) NASA GeneLab. 2021 doi: 10.26030/7P34-NF10. [DOI] [Google Scholar]
- 29.Galazka J.M., et al. Single Cell Transcriptional Profiling of Peripheral Blood Mononuclear Cells (PBMCs) from Mice Flown on Rodent Research Reference Mission-2 (RRRM-2) NASA GeneLab. 2021 doi: 10.26030/0R91-9V15. [DOI] [Google Scholar]
- 30.Galazka J.M., Juran C., Cekanaviciute E., Polo S.-H.L., Saravia-Butler A.M., Boyko V., Stodieck L., Roberts M., Costes S.V., Gebre S.G., Ferguson V. Single Cell Transcriptional Profiling of Humerus Bone Marrow from Mice Flown on Rodent Research Reference Mission-2 (RRRM-2) NASA GeneLab. 2021 doi: 10.26030/KKF4-P733. [DOI] [Google Scholar]
- 31.Galazka J.M., Juran C., Cekanaviciute E., Polo S.-H.L., Saravia-Butler A.M., Boyko V., Stodieck L., Roberts M., Costes S.V., Gebre S.G., Ferguson V. Single Cell Transcriptional Profiling of Femur Bone Marrow from Mice Flown on Rodent Research Reference Mission-2 (RRRM-2) NASA GeneLab. 2021 doi: 10.26030/R127-MC55. [DOI] [Google Scholar]
- 32.Garrett-Bakelman F.E., Darshi M., Green S.J., Gur R.C., Lin L., Macias B.R., McKenna M.J., Meydan C., Mishra T., Nasrini J., et al. The NASA Twins Study: A multidimensional analysis of a year-long human spaceflight. Science. 2019;364:eaau8650. doi: 10.1126/science.aau8650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gertz M.L., Chin C.R., Tomoiaga D., MacKay M., Chang C., Butler D., Afshinnekoo E., Bezdan D., Schmidt M.A., Mozsary C., et al. Multi-omic, single-cell, and Biochemical profiles of astronauts guide Pharmacological strategies for returning to gravity. Cell Rep. 2020;33 doi: 10.1016/j.celrep.2020.108429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liao J., Lu X., Shao X., Zhu L., Fan X. Uncovering an Organ’s molecular architecture at single-cell resolution by spatially resolved transcriptomics. Trends Biotechnol. 2021;39:43–58. doi: 10.1016/j.tibtech.2020.05.006. [DOI] [PubMed] [Google Scholar]
- 35.Maniatis S., Petrescu J., Phatnani H. Spatially resolved transcriptomics and its applications in cancer. Curr. Opin. Genet. Dev. 2021;66:70–77. doi: 10.1016/j.gde.2020.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berglund E., Saarenpaa S., Jemt A., Gruselius J., Larsson L., Bergenstrahle L., Lundeberg J., Giacomello S. Automation of Spatial Transcriptomics library preparation to enable rapid and robust insights into spatial organization of tissues. BMC Genom. 2020;21:298. doi: 10.1186/s12864-020-6631-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Butler D., Mozsary C., Meydan C., Foox J., Rosiene J., Shaiber A., Danko D., Afshinnekoo E., MacKay M., Sedlazeck F.J., et al. Shotgun transcriptome, spatial omics, and isothermal profiling of SARS-CoV-2 infection reveals unique host responses, viral diversification, and drug interactions. Nat. Commun. 2021;12:1660. doi: 10.1038/s41467-021-21361-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giacomello S. A new era for plant science: spatial single-cell transcriptomics. Curr. Opin. Plant Biol. 2021;60 doi: 10.1016/j.pbi.2021.102041. [DOI] [PubMed] [Google Scholar]
- 39.Park J., Foox J., Hether T., Danko D.C., Warren S., Kim Y., Reeves J., Butler D.J., Mozsary C., Rosiene J., et al. System-wide transcriptome damage and tissue identity loss in COVID-19 patients. Cell Rep Med. 2022;3 doi: 10.1016/j.xcrm.2022.100522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ke R., Mignardi M., Pacureanu A., Svedlund J., Botling J., Wahlby C., Nilsson M. In situ sequencing for RNA analysis in preserved tissue and cells. Nat. Methods. 2013;10:857–860. doi: 10.1038/nmeth.2563. [DOI] [PubMed] [Google Scholar]
- 41.Lubeck E., Coskun A.F., Zhiyentayev T., Ahmad M., Cai L. Single-cell in situ RNA profiling by sequential hybridization. Nat. Methods. 2014;11:360–361. doi: 10.1038/nmeth.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Junker J., Noel E., Guryev V., Peterson K., Shah G., Huisken J., McMahon A., Berezikov E., Bakkers J., van Oudenaarden A. Genome-wide RNA Tomography in the zebrafish embryo. Cell. 2014;159:662–675. doi: 10.1016/j.cell.2014.09.038. [DOI] [PubMed] [Google Scholar]
- 43.Lovatt D., Ruble B.K., Lee J., Dueck H., Kim T.K., Fisher S., Francis C., Spaethling J.M., Wolf J.A., Grady M.S., et al. Transcriptome in vivo analysis (TIVA) of spatially defined single cells in live tissue. Nat. Methods. 2014;11:190–196. doi: 10.1038/nmeth.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee J.H., Daugharthy E.R., Scheiman J., Kalhor R., Ferrante T.C., Terry R., Turczyk B.M., Yang J.L., Lee H.S., Aach J., et al. Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nat. Protoc. 2015;10:442–458. doi: 10.1038/nprot.2014.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moffitt J.R., Hao J., Wang G., Chen K.H., Babcock H.P., Zhuang X. High-throughput single-cell gene-expression profiling with multiplexed error-robust fluorescence in situ hybridization. Proc. Natl. Acad. Sci. USA. 2016;113:11046–11051. doi: 10.1073/pnas.1612826113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nichterwitz S., Chen G., Aguila Benitez J., Yilmaz M., Storvall H., Cao M., Sandberg R., Deng Q., Hedlund E. Laser capture microscopy coupled with Smart-seq2 for precise spatial transcriptomic profiling. Nat. Commun. 2016;7 doi: 10.1038/ncomms12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ståhl P.L., Salmen F., Vickovic S., Lundmark A., Navarro J.F., Magnusson J., Giacomello S., Asp M., Westholm J.O., Huss M., et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353:78–82. doi: 10.1126/science.aaf2403. [DOI] [PubMed] [Google Scholar]
- 48.Chen J., Suo S., Tam P.P., Han J.D.J., Peng G., Jing N. Spatial transcriptomic analysis of cryosectioned tissue samples with Geo-seq. Nat. Protoc. 2017;12:566–580. doi: 10.1038/nprot.2017.003. [DOI] [PubMed] [Google Scholar]
- 49.Wang X., Allen W.E., Wright M.A., Sylwestrak E.L., Samusik N., Vesuna S., Evans K., Liu C., Ramakrishnan C., Liu J., et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science. 2018;361:eaat5691. doi: 10.1126/science.aat5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vickovic S., Eraslan G., Salmen F., Klughammer J., Stenbeck L., Schapiro D., Aijo T., Bonneau R., Bergenstrahle L., Navarro J.F., et al. High-definition spatial transcriptomics for in situ tissue profiling. Nat. Methods. 2019;16:987–990. doi: 10.1038/s41592-019-0548-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Y., Yang M., Deng Y., Su G., Enninful A., Guo C.C., Tebaldi T., Zhang D., Kim D., Bai Z., et al. High-spatial-resolution multi-omics sequencing via Deterministic barcoding in tissue. Cell. 2020;183:1665–1681.e18. doi: 10.1016/j.cell.2020.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zollinger D.R., Lingle S.E., Sorg K., Beechem J.M., Merritt C.R. GeoMx™ RNA assay: high Multiplex, digital, spatial analysis of RNA in FFPE tissue. Methods Mol. Biol. 2020;2148:331–345. doi: 10.1007/978-1-0716-0623-0_21. [DOI] [PubMed] [Google Scholar]
- 53.Stickels R.R., Murray E., Kumar P., Li J., Marshall J.L., Di Bella D.J., Arlotta P., Macosko E.Z., Chen F. Highly sensitive spatial transcriptomics at near-cellular resolution with Slide-seqV2. Nat. Biotechnol. 2021;39:313–319. doi: 10.1038/s41587-020-0739-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xia K., Sun H.-X., Li J., Li J., Zhao Y., Chen R., Liu G., Chen Z., Yin R., Hao S., et al. Single-cell Stereo-seq enables cell type-specific spatial transcriptome characterization in Arabidopsis leaves. bioRxiv. 2021 doi: 10.1101/2021.10.20.465066. Preprint at. [DOI] [Google Scholar]
- 55.Groiss S., Pabst D., Faber C., Meier A., Bogdoll A., Unger C., Nilges B., Strauss S., Foderl-Hobenreich E., Hardt M., et al. Highly resolved spatial transcriptomics for detection of rare events in cells. bioRxiv. 2021 doi: 10.1101/2021.10.11.463936. Preprint at. [DOI] [Google Scholar]
- 56.He S., Bhatt R., Brown C., Brown E.A., Buhr D.L., Chantranuvatana K., Danaher P., Dunaway D., Garrison R.G., Geiss G., et al. High-plex multiomic analysis in FFPE at Subcellular level by spatial molecular imaging. bioRxiv. 2021 doi: 10.1101/2021.11.03.467020. Preprint at. [DOI] [Google Scholar]
- 57.Sidova M., Tomankova S., Abaffy P., Kubista M., Sindelka R. Effects of post-mortem and physical degradation on RNA integrity and quality. Biomol Detect Quantif. 2015;5:3–9. doi: 10.1016/j.bdq.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garneau N.L., Wilusz J., Wilusz C.J. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 59.Yang E., van Nimwegen E., Zavolan M., Rajewsky N., Schroeder M., Magnasco M., Darnell J.E. Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes. Genome Res. 2003;13:1863–1872. doi: 10.1101/gr.1272403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eraslan G., Drokhlyansky E., Anand S., Fiskin E., Subramanian A., Slyper M., Wang J., Van Wittenberghe N., Rouhana J.M., Waldman J., et al. Single-nucleus cross-tissue molecular reference maps toward understanding disease gene function. Science. 2022;376 doi: 10.1126/science.abl4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koenitzer J.R., Wu H., Atkinson J.J., Brody S.L., Humphreys B.D. Single-nucleus RNA-sequencing profiling of mouse lung. Reduced dissociation bias and improved rare cell-type detection compared with single-cell RNA sequencing. Am. J. Respir. Cell Mol. Biol. 2020;63:739–747. doi: 10.1165/rcmb.2020-0095ma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu H., Kirita Y., Donnelly E.L., Humphreys B.D. Advantages of single-nucleus over single-cell RNA sequencing of adult kidney: rare cell types and novel cell states revealed in Fibrosis. J. Am. Soc. Nephrol. 2019;30:23–32. doi: 10.1681/asn.2018090912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li H., Janssens J., De Waegeneer M., Kolluru S.S., Davie K., Gardeux V., Saelens W., David F.P.A., Brbic M., Spanier K., et al. Fly Cell Atlas: a single-nucleus transcriptomic atlas of the adult fruit fly. Science. 2022;375:eabk2432. doi: 10.1126/science.abk2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Galazka J.M., Polo S.-H.L., Saravia-Butler A.M., Charles H., Boyko V., Degoricija L., Narang S., Roberts M., Stodieck L., Gebre S.G., Costes S.V. Transcriptional Profiling of Livers from Mice Flown on Rodent Research Reference Mission-1 (RRRM-1) NASA GeneLab. 2021 doi: 10.26030/k766-s627. [DOI] [Google Scholar]
- 65.Smith R., Cramer M., Globus R., Galazka J. Rodent Research-3-CASIS: Mouse Liver Transcriptomic, Proteomic, and Epigenomic Data. NASA GeneLab. 2017 doi: 10.26030/9k6w-4c28. [DOI] [Google Scholar]
- 66.Smith R., Cramer M., Globus R., Galazka J. Rodent Research-3-CASIS: Mouse Eye Transcriptomic and Proteomic Data. NASA GeneLab. 2018 doi: 10.26030/pys4-6t29. [DOI] [Google Scholar]
- 67.Smith R., Cramer M., Globus R., Galazka J. Rodent Research-3-CASIS: Mouse Kidney Transcriptomic, Proteomic, and Epigenomic Data. NASA GeneLab. 2018 doi: 10.26030/q8vt-7p92. [DOI] [Google Scholar]
- 68.Smith R., Cramer M., Globus R., Galazka J. Rodent Research-3-CASIS: Mouse Retina Transcriptomic Data. NASA GeneLab. 2018 doi: 10.26030/pev7-5695. [DOI] [Google Scholar]
- 69.Choi S.Y., Saravia-Butler A., Shirazi-Fard Y., Leveson-Gower D., Stodieck L.S., Cadena S.M., Beegle J., Solis S., Ronca A., Globus R.K. Validation of a new rodent experimental system to investigate Consequences of long duration space Habitation. Sci. Rep. 2020;10:2336. doi: 10.1038/s41598-020-58898-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lai Polo S.-H., Saravia-Butler A.M., Boyko V., Dinh M.T., Chen Y.C., Fogle H., Reinsch S.S., Ray S., Chakravarty K., Marcu O., et al. RNAseq analysis of rodent spaceflight experiments is confounded by sample collection techniques. iScience. 2020;23 doi: 10.1016/j.isci.2020.101733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pimpalwar N., Czuba T., Smith M.L., Nilsson J., Gidlof O., Smith J.G. Methods for isolation and transcriptional profiling of individual cells from the human heart. Heliyon. 2020;6 doi: 10.1016/j.heliyon.2020.e05810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kruse C.P.S., Basu P., Luesse D.R., Wyatt S.E. Transcriptome and proteome responses in RNAlater preserved tissue of Arabidopsis thaliana. PLoS One. 2017;12 doi: 10.1371/journal.pone.0175943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Andersen T., Aubry C., Benschop T., Crippa G., De Parolis M.N., Quemerais S., Renz M., Ryckebosch O., Schawer J., Seidel A., et al. A −180° C Cryogenic Freezer for the International Space Station. SAE. 2005 doi: 10.4271/2005-01-2903. [DOI] [Google Scholar]
- 74.Saravia-Butler A.M., Galazka J., Torres M.L., Chen Y.-C., Choi S., Klotz R., Perreau S., Gower D.L., Shirazi Y., Polo S.-H.L., et al. Transcriptional analysis of soleus from mice preserved with the Rapid Freeze hardware. NASA GeneLab. 2020 doi: 10.26030/e1ea-gx05. [DOI] [Google Scholar]
- 75.Saravia-Butler A.M., Galazka J., Torres M.L., Chen Y.-C., Choi S., Klotz R., Perreau S., Gower D.L., Shirazi Y., Polo S.-H.L., et al. Transcriptional analysis of livers from mice preserved with the Rapid Freeze hardware. NASA GeneLab. 2020 doi: 10.26030/ya8v-7043. [DOI] [Google Scholar]
- 76.Saravia-Butler A.M., Galazka J., Torres M.L., Chen Y.-C., Choi S., Klotz R., Perreau S., Gower D.L., Shirazi Y., Polo S.-H.L., et al. Transcriptional analysis of spleens from mice preserved with the Rapid Freeze hardware. NASA GeneLab. 2020 doi: 10.26030/dty4-1b66. [DOI] [Google Scholar]
- 77.Choi S., Ray H.E., Lai S.-H., Alwood J.S., Globus R.K. Preservation of multiple mammalian tissues to Maximize science return from ground based and spaceflight experiments. PLoS One. 2016;11 doi: 10.1371/journal.pone.0167391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gupta V., Holets-Bondar L., Roby K.F., Enders G., Tash J.S. A tissue retrieval and postharvest processing regimen for rodent reproductive tissues compatible with long-term storage on the international space station and postflight biospecimen sharing program. BioMed Res. Int. 2015;2015:1–12. doi: 10.1155/2015/475935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Herranz R., Vandenbrink J.P., Villacampa A., Manzano A., Poehlman W.L., Feltus F.A., Kiss J.Z., Medina F.J. RNAseq analysis of the response of Arabidopsis thaliana to fractional gravity under blue-light stimulation during spaceflight. Front. Plant Sci. 2019;10:1529. doi: 10.3389/fpls.2019.01529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Manzano A., Villacampa A., Saez-Vasquez J., Kiss J.Z., Medina F.J., Herranz R. The importance of Earth reference controls in spaceflight -omics research: characterization of nucleolin mutants from the Seedling Growth experiments. iScience. 2020;23 doi: 10.1016/j.isci.2020.101686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Paul A.-L., Levine H.G., McLamb W., Norwood K.L., Reed D., Stutte G.W., William Wells H., Ferl R.J. Plant molecular biology in the space station era: utilization of KSC fixation tubes with RNAlater. Acta Astronaut. 2005;56:623–628. doi: 10.1016/j.actaastro.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 82.Paul A.-L., Zupanska A.K., Ostrow D.T., Zhang Y., Sun Y., Li J.L., Shanker S., Farmerie W.G., Amalfitano C.E., Ferl R.J. Spaceflight transcriptomes: unique responses to a novel environment. Astrobiology. 2012;12:40–56. doi: 10.1089/ast.2011.0696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schultz E.R., Kelley K.L., Paul A., Ferl R.J. A method for preparing spaceflight RNAlater-fixed Arabidopsis thaliana (Brassicaceae) tissue for scanning electron microscopy. Appl. Plant Sci. 2013;1 doi: 10.3732/apps.1300034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vandenbrink J.P., Herranz R., Poehlman W.L., Alex Feltus F., Villacampa A., Ciska M., Javier Medina F., Kiss J.Z. RNA-seq analyses of Arabidopsis thaliana seedlings after exposure to blue-light phototropic stimuli in microgravity. Am. J. Bot. 2019;106:1466–1476. doi: 10.1002/ajb2.1384. [DOI] [PubMed] [Google Scholar]
- 85.Villacampa A., Ciska M., Manzano A., Vandenbrink J.P., Kiss J.Z., Herranz R., Medina F.J. From spaceflight to Mars g-levels: Adaptive response of A. thaliana seedlings in a reduced gravity environment is enhanced by red-light photostimulation. Int. J. Mol. Sci. 2021;22:899. doi: 10.3390/ijms22020899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wnorowski A., Sharma A., Chen H., Wu H., Shao N.Y., Sayed N., Liu C., Countryman S., Stodieck L.S., Rubins K.H., et al. Effects of spaceflight on human induced Pluripotent stem cell-derived Cardiomyocyte structure and function. Stem Cell Rep. 2019;13:960–969. doi: 10.1016/j.stemcr.2019.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Balsamo M., Barravecchia I., Mariotti S., Merenda A., De Cesari C., Vukich M., Angeloni D. Molecular and cellular characterization of space flight effects on Microvascular Endothelial cell function – PreparatoryWork for the SFEF project. Microgravity Sci. Technol. 2014;26:351–363. doi: 10.1007/s12217-014-9399-4. [DOI] [Google Scholar]
- 88.Cockell C.S., Santomartino R., Finster K., Waajen A.C., Eades L.J., Moeller R., Rettberg P., Fuchs F.M., Van Houdt R., Leys N., et al. Space station biomining experiment demonstrates rare earth element extraction in microgravity and Mars gravity. Nat. Commun. 2020;11:5523. doi: 10.1038/s41467-020-19276-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zamarioli A., Campbell Z.R., Maupin K.A., Childress P.J., Ximenez J.P.B., Adam G., Chakraborty N., Gautam A., Hammamieh R., Kacena M.A. Analysis of the effects of spaceflight and local administration of thrombopoietin to a femoral defect injury on distal skeletal sites. NPJ Microgravity. 2021;7:12. doi: 10.1038/s41526-021-00140-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huang P., Russell A.L., Lefavor R., Durand N.C., James E., Harvey L., Zhang C., Countryman S., Stodieck L., Zubair A.C. Feasibility, potency, and safety of growing human mesenchymal stem cells in space for clinical application. NPJ Microgravity. 2020;6:16. doi: 10.1038/s41526-020-0106-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hong X., Ratri A., Choi S.Y., Tash J.S., Ronca A.E., Alwood J.S., Christenson L.K. Effects of spaceflight aboard the International Space Station on mouse estrous cycle and ovarian gene expression. NPJ Microgravity. 2021;7:11. doi: 10.1038/s41526-021-00139-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Galazka J.M., Polo S.-H.L., Saravia-Butler A.M., Fogle H.W., Bense N., Boyko V., Dinh M.T., Costes S.V., Gebre S.G. Transcriptional Analysis of Lung from Mice Flown on the RR-6 Mission. NASA GeneLab. 2019 doi: 10.26030/d56m-7e45. [DOI] [Google Scholar]
- 93.Galazka J.M., Polo S.-H.L., Saravia-Butler A.M., Fogle H.W., Bense N., Chen Y.-C., Narang S., Dinh M.T., Costes S.V., Gebre S.G., et al. Effect of spaceflight on liver from mice flown on the ISS for 33 days: Transcriptional analysis. NASA GeneLab. 2019 doi: 10.26030/fmkc-8h31. [DOI] [Google Scholar]
- 94.Galazka J.M., Degoricija L., Polo S.-H.L., Saravia-Butler A.M., Boyko V., Dinh M.T., Houseman C., Costes S.V., Gebre S.G. Transcriptional profiling of thymus from mice flown on the RR-9 Mission. NASA GeneLab. 2021 doi: 10.26030/5mv4-fe66. [DOI] [Google Scholar]
- 95.Castro-Wallace S.L., Chiu C.Y., John K.K., Stahl S.E., Rubins K.H., McIntyre A.B.R., Dworkin J.P., Lupisella M.L., Smith D.J., Botkin D.J., et al. Nanopore DNA sequencing and genome assembly on the International Space Station. Sci. Rep. 2017;7:18022. doi: 10.1038/s41598-017-18364-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Burton A.S., Stahl S.E., John K.K., Jain M., Juul S., Turner D.J., Harrington E.D., Stoddart D., Paten B., Akeson M., Castro-Wallace S.L. Off earth identification of bacterial populations using 16S rDNA nanopore sequencing. Genes. 2020;11:76. doi: 10.3390/genes11010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stahl-Rommel S., Jain M., Nguyen H.N., Arnold R.R., Aunon-Chancellor S.M., Sharp G.M., Castro C.L., John K.K., Juul S., Turner D.J., et al. Real-time culture-Independent microbial profiling onboard the international space station using Nanopore sequencing. Genes. 2021;12:106. doi: 10.3390/genes12010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stahl-Rommel S., Li D., Sung M., Li R., Vijayakumar A., Atabay K.D., Bushkin G.G., Castro C.L., Foley K.D., Copeland D.S., et al. A CRISPR-based assay for the study of eukaryotic DNA repair onboard the International Space Station. PLoS One. 2021;16 doi: 10.1371/journal.pone.0253403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Checinska Sielaff A., Urbaniak C., Mohan G.B.M., Stepanov V.G., Tran Q., Wood J.M., Minich J., McDonald D., Mayer T., Knight R., et al. Characterization of the total and viable bacterial and fungal communities associated with the International Space Station surfaces. Microbiome. 2019;7:50. doi: 10.1186/s40168-019-0666-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Spivey R., Jeter L., Vonk C. 45th AIAA Aerospace Sciences Meeting and Exhibit. American Institute of Aeronautics and Astronautics; 2007. The microgravity science glovebox (MSG), a resource for gravity-dependent phenomena research on the international space station (ISS) [DOI] [Google Scholar]
- 101.Thiel C.S., Tauber S., Seebacher C., Schropp M., Uhl R., Lauber B., Polzer J., Neelam S., Zhang Y., Ullrich O. Real-time 3D high-resolution microscopy of human cells on the international space station. Int. J. Mol. Sci. 2019;20:2033. doi: 10.3390/ijms20082033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jemt A., Salmen F., Lundmark A., Mollbrink A., Fernandez Navarro J., Stahl P.L., Yucel-Lindberg T., Lundeberg J. An automated approach to prepare tissue-derived spatially barcoded RNA-sequencing libraries. Sci. Rep. 2016;6 doi: 10.1038/srep37137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Stenbeck L., Taborsak-Lines F., Giacomello S. Enabling automated and reproducible spatially resolved transcriptomics at scale. Heliyon. 2022;8 doi: 10.1016/j.heliyon.2022.e09651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Overbey E.G., Saravia-Butler A.M., Zhang Z., Rathi K.S., Fogle H., da Silveira W.A., Barker R.J., Bass J.J., Beheshti A., Berrios D.C., et al. NASA GeneLab RNA-seq consensus pipeline: standardized processing of short-read RNA-seq data. iScience. 2021;24 doi: 10.1016/j.isci.2021.102361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Conesa A., Madrigal P., Tarazona S., Gomez-Cabrero D., Cervera A., McPherson A., Szczesniak M.W., Gaffney D.J., Elo L.L., Zhang X., Mortazavi A. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016;17:13. doi: 10.1186/s13059-016-0881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lähnemann D., Koster J., Szczurek E., McCarthy D.J., Hicks S.C., Robinson M.D., Vallejos C.A., Campbell K.R., Beerenwinkel N., Mahfouz A., et al. Eleven grand challenges in single-cell data science. Genome Biol. 2020;21:31. doi: 10.1186/s13059-020-1926-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Andrews T.S., Kiselev V.Y., McCarthy D., Hemberg M. Tutorial: guidelines for the computational analysis of single-cell RNA sequencing data. Nat. Protoc. 2021;16:1–9. doi: 10.1038/s41596-020-00409-w. [DOI] [PubMed] [Google Scholar]
- 108.Luecken M.D., Theis F.J. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol. Syst. Biol. 2019;15 doi: 10.15252/msb.20188746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Stuart T., Satija R. Integrative single-cell analysis. Nat. Rev. Genet. 2019;20:257–272. doi: 10.1038/s41576-019-0093-7. [DOI] [PubMed] [Google Scholar]
- 110.Hao Y., Hao S., Andersen-Nissen E., Mauck W.M., Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Palla G., Spitzer H., Klein M., Fischer D., Schaar A.C., Kuemmerle L.B., Rybakov S., Ibarra I.L., Holmberg O., Virshup I., et al. Squidpy: a scalable framework for spatial omics analysis. Nat. Methods. 2022;19:171–178. doi: 10.1038/s41592-021-01358-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bergenstråhle J., Bergenstråhle L., Lundeberg J. SpatialCPie: an R/Bioconductor package for spatial transcriptomics cluster evaluation. BMC Bioinf. 2020;21:161. doi: 10.1186/s12859-020-3489-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhao E., Stone M.R., Ren X., Guenthoer J., Smythe K.S., Pulliam T., Williams S.R., Uytingco C.R., Taylor S.E.B., Nghiem P., et al. Spatial transcriptomics at subspot resolution with BayesSpace. Nat. Biotechnol. 2021;39:1375–1384. doi: 10.1038/s41587-021-00935-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Friedrich S., Sonnhammer E.L.L. Fusion transcript detection using spatial transcriptomics. BMC Med. Genomics. 2020;13:110. doi: 10.1186/s12920-020-00738-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fernández Navarro J., Lundeberg J., Ståhl P.L. ST viewer: a tool for analysis and visualization of spatial transcriptomics datasets. Bioinformatics. 2019;35:1058–1060. doi: 10.1093/bioinformatics/bty714. [DOI] [PubMed] [Google Scholar]
- 116.Cable D.M., Murray E., Zou L.S., Goeva A., Macosko E.Z., Chen F., Irizarry R.A. Robust decomposition of cell type mixtures in spatial transcriptomics. Nat. Biotechnol. 2021;40:517–526. doi: 10.1038/s41587-021-00830-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yuan Y., Bar-Joseph Z. GCNG: graph convolutional networks for inferring gene interaction from spatial transcriptomics data. Genome Biol. 2020;21:300. doi: 10.1186/s13059-020-02214-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Andersson A., Lundeberg J. sepal: identifying transcript profiles with spatial patterns by diffusion-based modeling. Bioinformatics. 2021;37:2644–2650. doi: 10.1093/bioinformatics/btab164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Svensson V., Teichmann S.A., Stegle O. SpatialDE: identification of spatially variable genes. Nat. Methods. 2018;15:343–346. doi: 10.1038/nmeth.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Xu Y., McCord R.P. CoSTA: unsupervised convolutional neural network learning for spatial transcriptomics analysis. BMC Bioinf. 2021;22:397. doi: 10.1186/s12859-021-04314-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Levy-Jurgenson A., Tekpli X., Yakhini Z. Assessing heterogeneity in spatial data using the HTA index with applications to spatial transcriptomics and imaging. Bioinformatics. 2021;37:3796–3804. doi: 10.1093/bioinformatics/btab569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Li Z., Song T., Yong J., Kuang R. Imputation of spatially-resolved transcriptomes by graph-regularized tensor completion. PLoS Comput. Biol. 2021;17 doi: 10.1371/journal.pcbi.1008218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bergenstråhle J., Larsson L., Lundeberg J. Seamless integration of image and molecular analysis for spatial transcriptomics workflows. BMC Genom. 2020;21:482. doi: 10.1186/s12864-020-06832-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kleshchevnikov V., Shmatko A., Dann E., Aivazidis A., King H.W., Li T., Elmentaite R., Lomakin A., Kedlian V., Gayoso A., et al. Cell2location maps fine-grained cell types in spatial transcriptomics. Nat. Biotechnol. 2022;40:661–671. doi: 10.1038/s41587-021-01139-4. [DOI] [PubMed] [Google Scholar]
- 125.Andersson A., Bergenstrahle J., Asp M., Bergenstrahle L., Jurek A., Fernandez Navarro J., Lundeberg J. Single-cell and spatial transcriptomics enables probabilistic inference of cell type topography. Commun Biol. 2020;3:565. doi: 10.1038/s42003-020-01247-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lohoff T., Ghazanfar S., Missarova A., Koulena N., Pierson N., Griffiths J.A., Bardot E.S., Eng C.H.L., Tyser R.C.V., Argelaguet R., et al. Integration of spatial and single-cell transcriptomic data elucidates mouse organogenesis. Nat. Biotechnol. 2022;40:74–85. doi: 10.1038/s41587-021-01006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hu J., Schroeder A., Coleman K., Chen C., Auerbach B.J., Li M. Statistical and machine learning methods for spatially resolved transcriptomics with histology. Comput. Struct. Biotechnol. J. 2021;19:3829–3841. doi: 10.1016/j.csbj.2021.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Atta L., Fan J. Computational challenges and opportunities in spatially resolved transcriptomic data analysis. Nat. Commun. 2021;12:5283. doi: 10.1038/s41467-021-25557-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chakraborty N., Schmitt C.W., Honnold C.L., Moyler C., Butler S., Nachabe H., Gautam A., Hammamieh R. Protocol improvement for RNA extraction from Compromised frozen specimens generated in Austere conditions: a Path forward to transcriptomics-Pathology systems integration. Front. Mol. Biosci. 2020;7:142. doi: 10.3389/fmolb.2020.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.van Loon J.J.W.A., BIOPACK J.J.W.A. BIOPACK: the ground controlled late access biological research facility. J. Gravit. Physiol. 2004;11:57–65. [PubMed] [Google Scholar]
- 131.Bizet F., Pereda-Loth V., Chauvet H., Gerard J., Eche B., Girousse C., Courtade M., Perbal G., Legue V. Both gravistimulation onset and removal trigger an increase of cytoplasmic free calcium in statocytes of roots grown in microgravity. Sci. Rep. 2018;8:11442. doi: 10.1038/s41598-018-29788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Shaka S., Carpo N., Tran V., Cepeda C., Espinosa-Jeffrey A. Microgravity significantly influences neural stem cells size and numbers: Implications for long-term space missions. Stem Cells Res. Dev. Ther. 2021;7:1–9. doi: 10.24966/srdt-2060/100088. [DOI] [Google Scholar]
- 133.Alonzo M., El Khoury R., Nagiah N., Thakur V., Chattopadhyay M., Joddar B. 3D biofabrication of a cardiac tissue construct for sustained longevity and function. ACS Appl. Mater. Interfaces. 2022;14:21800–21813. doi: 10.1021/acsami.1c23883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Trevino A.E., Muller F., Andersen J., Sundaram L., Kathiria A., Shcherbina A., Farh K., Chang H.Y., Pașca A.M., Kundaje A., et al. Chromatin and gene-regulatory dynamics of the developing human cerebral cortex at single-cell resolution. Cell. 2021;184:5053–5069.e23. doi: 10.1016/j.cell.2021.07.039. [DOI] [PubMed] [Google Scholar]
- 135.Wang K., Xiao Z., Yan Y., Ye R., Hu M., Bai S., Sei E., Qiao Y., Chen H., Lim B., et al. Simple oligonucleotide-based multiplexing of single-cell chromatin accessibility. Mol. Cell. 2021;81:4319–4332.e10. doi: 10.1016/j.molcel.2021.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fan R., Deng Y., Bartosovic M., Ma S., Zhang D., Liu Y., Qin X., Su G., Xu M., Halene S., et al. Spatial-ATAC-seq: spatially resolved chromatin accessibility profiling of tissues at genome scale and cellular level. Research Square. 2021 doi: 10.21203/rs.3.rs-593462/v1. [DOI] [Google Scholar]
- 137.Thornton C.A., Mulqueen R.M., Torkenczy K.A., Nishida A., Lowenstein E.G., Fields A.J., Steemers F.J., Zhang W., McConnell H.L., Woltjer R.L., et al. Spatially mapped single-cell chromatin accessibility. Nat. Commun. 2021;12:1274. doi: 10.1038/s41467-021-21515-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Arnold C. Cloud labs: where robots do the research. Nature. 2022;606:612–613. doi: 10.1038/d41586-022-01618-x. [DOI] [PubMed] [Google Scholar]
- 139.Ji Y., Lotfollahi M., Wolf F.A., Theis F.J. Machine learning for perturbational single-cell omics. Cell Syst. 2021;12:522–537. doi: 10.1016/j.cels.2021.05.016. [DOI] [PubMed] [Google Scholar]