Abstract
Globally, colorectal cancer (CRC) is one of the most typical lethal cancers. One of the main factors for better outcomes in CRC management is the early detection of the disease. As an integral component of human metabolism and homeostasis, gut microbiome has recently been a subject of extensive research for its role in the pathogenesis, diagnosis, and treatment of CRC.
Microbial dysbiosis (the decrease in beneficial gut flora and the increase of detrimental populations) leads to chronic inflammation and genetic alteration in the host cells, triggering and promoting CRC carcinogenesis. Identifying these microbial changes in depth would potentially isolate the pathogenic microbiota species and establish biomarker models for early detection of CRC. On the other hand, modifying these microbial changes would help formulate preventative and therapeutic strategies for CRC, developing a more precise CRC management plan according to each patient's microbial print. This essay explains gut microbiome composition, microbial changes (dysbiosis) in CRC carcinogenesis, the probability of creating microbiome-based CRC biomarkers, and potential microbiome-targeted treatment options.
Keywords: colorectal cancer, target therapy, diagnosis, biomarker, crc, cancer, colorectal, microbiome
Introduction and background
Colorectal cancer (CRC) is the fourth most common cancer in the UK and accounts for 12% of all new cancer cases [1]. Considered a "silent disease", early diagnosis of CRC is a chief factor that influences survival; therefore, establishing reliable non-invasive CRC biomarkers is required [2].
Currently available CRC screening options include faecal occult blood tests (FOBTs), faecal immunochemical tests (FITs), and subsequent colonoscopy if FOBT or FIT are positive [3]. FOBT's sensitivity is still limited because CRCs do not always bleed or only bleed intermittently [4]. While colonoscopy represents the standard gold method for diagnosing CRC, it is challenging to use it as a screening test due to its high cost and invasiveness. Thus, there is a need to find other feasible CRC screening methods [5].
The intestinal microbiome, gut flora, had primarily been hidden in the blind spot of the medical research community until the last 10 years [6]. There is a rapid proliferation of interest in studying this "forgotten organ" and correlating its function to several human pathological changes, such as cancers, especially after the advances in DNA/RNA sequencing [7]. Current evidence support that specific intestinal microbes drive CRC development and progression, yet their pathogenic mechanisms are still unclear. Microbial analysis can also identify some faecal microbial markers of CRC that could help in the early diagnosis [8].
This study aims to explore some information about the microbial community and how microbiota changes could trigger CRC pathogenesis, the different techniques for microbial analysis, and the potential use of microbiome as a biomarker to detect CRC and as an adjunct component of its treatment.
Review
Gut microbiota composition
Symbiotic Microbial Composition
Gut microbiota includes the microorganisms, such as bacteria, viruses, archaea, and eukaryotic organisms, that inhabit the human gastrointestinal tract and manipulates several physiological functions [9]. Meanwhile, microbiome refers to the gut microbiota's collective genes, genomes, and metabolic products in the host environment [10]. Holobiont refers to the biological entity involving a host and its inherited symbiont microbiota, while hologenome describes the collective genomes of the host genome and associated microbial genomes [11]. A healthy gut microbiota composition (symbiosis or eubiosis) is essential for maintaining normal gut nutrition, metabolism, cell proliferation, immune system development, and protection against pathogenesis [12].
The microbial structure is affected by various factors, such as dietary carcinogens, smoking, alcohol, and other environmental factors. Chief among factors affecting gut microbiome composition is the modern western lifestyle (associated with increased fast food and stress levels) that leads to reduced beneficial bacteria and enriched pathogenic species [13]. The high calorific content of the western diet (high fat and carbohydrates) causes microbial structure changes (dysbiosis) and increases the risk of developing obesity [14] and carcinogenesis [15]. On the other hand, fasting can increase the diversity of bacteria (symbiosis) in your gut, which is essential for your immune and overall health [16]. Cignarella et al. have reported that intermittent fasting, ideally 16 hours of fasting and 8 hours of diet, resulted in increased enrichment of the Lactobacillaceae bacteria families (probiotic) that have beneficial effects on health, including the exclusion of pathogens, immunomodulation, and the production of a healthy bacterial molecule [17,18]. Intermittent fasting also has potent immunomodulatory effects that are at least partially mediated by the gut microbiome [19].
Microbial Dysbiosis
Microbial dysbiosis refers to pathological alterations of gut microbiota compositions, resulting in several disease states [15]. Dysbiosis has been associated with a wide range of diseases, including type 1 diabetes mellitus, inflammatory bowel disease (IBD), allergic disorders, metabolic syndrome, non-alcoholic fatty liver disease, obesity, and CRC in both human and animal models [20-22]. One of the necessary consequences of gut microbial dysregulation is cancer development due to an increased percentage of harmful bacterial microbiota that produces pro-carcinogenic substances and destroys the gut barrier [23]. Microbiota-induced inflammation and genotoxicity eventually induce carcinogenesis and the development of CRC [24].
Microbiome and CRC interplay
Correlation between gut microbial changes and CRC formation has been established in several studies recently [25-28] because of up-to-date gene sequencing techniques [26]. Microbiota within tumour tissue has a specific bacterial composition compared to normal healthy areas. Bacterial species such as Bacteroides fragilis, Escherichia coli, and Fusobacterium nucleatum are more abundant in the CRC microenvironment [29-31], and this potentially could drive using microbiota as CRC biomarkers [32].
Mechanism of CRC Development by Microbial Dysbiosis
Many recent studies have highlighted the link between CRC and gut microbiome alteration [29], with several hypotheses as to the causal role of microbes in CRC development [33]. Yet, the exact pathogenic mechanisms are still unclear [28]. Gut microbiota could influence colorectal carcinogenesis through various mechanisms that are described below.
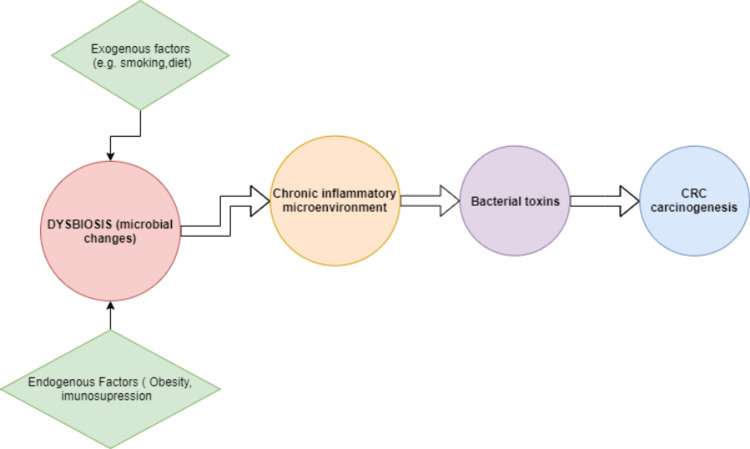
Microbial-induced inflammation: Some evidence suggests that gut microbiota induce chronic inflammation, which might trigger carcinogenesis [34]. Zhang et al. reported that CRC-associated microbial changes and subsequent gradual inflammation might gradually form a microenvironment that triggers CRC [35]. F. nucleatum, a common oral bacterium, has also been associated with CRC progression, metastasis, and chemoresistance [36]. As a pro-carcinogenic bacterium, F. nucleatum codes a virulence factor, FadA, that can activate the β-catenin pathway, which drives the initiation and progression of CRC. E. coli has also presented abundance in CRC with a significant role in promoting neoplasia [37]. Figure 1 explains how the microbiome could produce inflammatory changes that initiate CRC development.
Figure 1. Dysbiosis-induced CRC development.
CRC: colorectal cancer.
Nevertheless, there is not enough evidence to say that inflammation or the presence of bacteria or bacterial metabolites alone is enough to promote tumour growth [38]. However, it is still an active area of inquiry to determine which microbial species are most responsible for carcinogenesis.
Driver-passenger model: Two classes of the microbial community are involved in CRC pathogenesis, namely, the "driver" community and the "passenger" community [39]. The "driver" community (pro-carcinogenic indigenous bacteria) cause microenvironmental alterations that potentially initiate carcinogenesis by producing carcinogenic toxins that damage DNA in colonic epithelial cells [17]. Enterococcus faecalis represents an example of a driver microbiome as it releases extracellular superoxide that causes intracellular DNA carcinogenic alterations within the colonic mucosa [17]. Subsequently, the "passenger" community (opportunistic colonising bacteria) outcompetes the driver microbes and augments tumour progression and growth. The commonest passenger bacteria to colonise in CRC tissue is Fusobacterium spp. [39].
Host-microbial genetic interaction: Traditional theories of cancer aetiology focus on the mechanism of altering mammalian genetics by external risk factors such as smoking [9]. However, with advanced computerised diagnostics, internal host-microbial genetic interactions have been of study interest [40]. There is evidence that host-to-microorganism interactions activate procarcinogenic signalling pathways that trigger molecular alterations (genomic and epigenetic changes), stimulation of adenoma-carcinoma sequence, and then CRC development and progression [41].
Recently, host-microbe interactions in CRC tumours have been studied to prove an association between specific tumour mutations and distinct microbial, metabolic, and interaction profiles [42]. Burns et al. also found statistically significant associations between loss-of-function mutations in tumour genes and shifts in the abundances of specific bacterial taxa, suggesting a potential functional genetic interaction between bacteria and tumour profiles [28].
These mechanistic components have the potential to be modulated for therapeutic or prophylactic purposes in the context of CRC. Nevertheless, these studies only show correlations and cannot directly cause effects. Thus, it is unclear whether the microbiome is altered before or after the appearance of specific mutations.
Microbial analysis techniques
Most human microbial bacteria are uncultivable, so microbiome studies on CRC patients have relied on molecular-based methods [39]. Targeted genotyping (e.g. 16S ribosomal RNA (rRNA) based) and metagenomics are the most widely utilised methods for microbial analysis.
Gene Amplicon Sequencing
Over the past few decades, gene amplicon sequencing has been the primary technique for studying microbial-specific marker/target genes that function in evolutionary transitions and changes [43]. 16S rRNA sequencing is the gold standard targeted gene sequencing approach for identifying microbial community composition and assessing genetic diversity, especially for unculturable organisms [44]. Moreover, 16S rRNA gene sequencing analysis has assisted in correlating between alterations in microbial community (dysbiosis) functions and certain diseases, including Crohn's disease, ulcerative colitis, diabetes, and gastrointestinal cancers [45-47]. However, the 16S rRNA gene sequences approach does not target the whole genome realistically. It is limited in assessing molecular host-microbiota and microbiome-microbiome interactions that reflect the biological microbial community [48].
Metagenomics
Metagenomics comprehensively catalogues all microorganisms present (unculturable and culturable, known and unknown) in complex environmental samples [49]. The metagenomic analysis provides a functional analysis of microbial communities, such as polygenetic analysis and taxonomic classification. As a result, metagenomics outperforms 16S rRNA target gene sequencing in defining microbiota ecosystems, which gives a unimodal single-gene analysis [50].
Metagenomics is also reliable for studying the microbial community's genomic linkages between function and phylogeny (evolutionary history and relationships among or within groups of organisms) [50]. Metagenomics, complementing metatranscriptomic or metaproteomic techniques, could describe more expressed microbial activities [51].
Current studies focus mainly on the identification and profiling of microbial composition. Still, the microbial community is more complex and requires including the molecular interactions with the host and in-between microbiota to reflect the actual biological microenvironment [52].
Limitations of microbial analysis
Methodological Limitations
Current microbial analysis designs have some challenges. Amongst them is the absence of a gold-standard unified methodology of studying. The sample size is usually tiny and variable from one centre to another, which produces the most non-reliable and non-representative outcomes. Samples are mainly collected from faecal content without mucosal biopsies, which partially reflect the gut microbiome community [53]. Other challenges of getting an effective sample include handling (e.g. type (faecal or mucosal), collection, contamination, transportation, storage, and time to analysis), and nucleic acid analysis (e.g. methods for DNA extraction, selecting regions and depths of sequencing, varying polymerase chain reaction (PCR) primers for 16S microbial analysis, and variable methods for assignment of taxonomy) [54].
Gut microorganisms are considered difficult to culture [55]. Non-bacterial microbial components (i.e. virome, mycobiome, and protozoans) are less studied in current research due to a lack of facilities, despite having an established role in CRC stage progression [54]. Faecal samples are a non-invasive approach for screening tests; however, mucosal samples are more potent for identifying specific species of bacteria associated with CRC initiation and growth [56].
The lack of unified metadata and high-processing computer equipment, for high-volume data and statistical analysis, also affects the results' accuracy, reproducibility, and interpretability. Advanced computational tools for functional analysis that consider host-microbial molecular interactions are required to get more reliable results [57].
Exogenous and Endogenous Variability
Another challenge is that case-control studies are affected by the host (age, sex, and genotype) and environmental (e.g. diet and lifestyle) factors that produce biased results [58]. The existing database also focuses only on the western population that does not consider the heterogenicity of these variables. Therefore, it is crucial to include these factors, such as diet, lifestyle, and smoking, when examining microbiota's role in CRC and other diseases [59]. Therefore, some studies suggested a new strategy for microbial analysis in CRC by using tumour and normal tissue samples from the same CRC patient [60,61]. Burns et al. [62], for example, have found different microbial communities with specific functional pathways in CRC tissue compared to nearby healthy tissue. Therefore, it is essential to include these exogenous and endogenous differences with personalised study patterns to set reliable human microbial datasets (Table 1) [57].
Table 1. Challenges and required developments for better microbial outcomes in CRC.
CRC: colorectal cancer; PCR: polymerase chain reaction.
| Methodology | Limitation | Development |
| Population sample | Heterogenicity (i.e. geography and lifestyle); most current studies include western populations [59]. | Comprehensive and diverse studies for a better microbial database [57]. |
| Study design | Case-control: Controls are affected by host and environmental factors (i.e. diet and genetics) [60]. | Individualised approach (i.e. paired diseased-healthy tissue samples, diet, and metabolic analysis) for a personalised diagnosis and personalised therapy (i.e. pre/probiotics) [63]. |
| Sample collection | Faecal samples (partially reflect gut microbiome) [54]. | Tissue (mucosal) samples for a better understanding of environmental processes and biological interactions [64]. |
| Microbial sample | Non-bacterial microbial components (i.e. virome, mycobiome, and protozoans). Non-bacterial microbial dataset (limited): gut virome dysbiosis (differ in CRC stages, patients - control) [54]. | Taxonomy-based analysis: CRC-associated bacterial taxa + less covered taxonomic groups (i.e. fungi and viruses) [23]. Distinct sequencing techniques (i.e. regions, depth, and PCR) avoid heterogenicity bias [57]. |
| Data analysis | Independent taxons analysis: Without considering ecological correlation -> decretive host-microbial interaction [65]. | Functional-based analysis: Combined omics (i.e. metagenomic, metatranscriptomics, metaproteomics, and metabolomics) approaches of a mechanistic host-microbial interaction for direct causal effect [23]. |
Future Challenges
To get more specific bioinformatics about CRC-related microbiota, studies should involve the environmental functions and interactions within the microbial micro-environment along with the anatomical compositions [62]. In recent research, omics datasets (genomics, transcriptomics, proteomics, metabolomics, metagenomics, phenomics, etc.) have been included to describe microbial biological processes more accurately. Metatranscriptomics assess microbiota communities' taxonomic signature and function; meanwhile, metaproteomics analyses the microbiome-associated protein profiles to reflect the bodily functions under different environmental conditions [66]. Multi-omics data analysis requires highly advanced computational and technological resources with complex algorithms and software to process a high volume of data and correlate multiple variables and interactions [67]. These data would explain specific biological and environmental interactions that describe microbiomes' role in CRC development and create better therapeutic and biotechnological applications.
Clinical applications of microbiota in CRC
Microbial Biomarkers
Several biomarkers (genetic, blood, molecular, and imaging biomarkers ) are crucial in early detection, prognostication, and risk stratification in CRC [68]. Getting clinically reliable predictive biomarkers could improve the accuracy of predicting clinical outcomes such as survival, tumour recurrence, and metastasis. Personalised CRC treatment algorithms would be applied in clinical practice to get the best results for each individual's disease characteristics [69]. Studying microbial changes, therefore, could provide a diagnostic and predictive CRC biomarker soon [70].
Recent studies suggest that gut microbiome analysis of stool samples significantly differentiated between healthy individuals and patients with adenoma vs. carcinoma samples by identifying either the enrichment or depletion of specific bacterial populations within faecal samples [71]. Subsequently, the gut microbiome could serve as a screening tool for CRC detection [72]. By combining microbial biomarker tests with the FITs, sensitivity for CRC detection and accuracy of treatment outcomes prediction might increase [73].
The faecal metabolome is the metabolites, such as short-chain fatty acids, produced from microbial interactions to maintain the homeostasis of host metabolism [74]. Metabolomic analysis of faecal samples might also be potential clinical CRC biomarkers [36]. Xinhao et al. concluded that an increased abundance of specific gut microbiota (e.g. B. fragilis) was significantly associated with increased levels of particular metabolites (called metabolome), such as adrenic acid and decanoic acid, in CRC patients [75]. Faecal metabolomic analysis has produced some identified biomarkers for CRC diagnosis and therapeutic evaluation; however, more research studies are necessary to get more data on metabolomes [76].
Local volatile organic compound (VOC) is another potential CRC biomarker that reflects gut microbial compositions [77]. It is the gaseous molecules produced during bacterial fermentation in the gut and then emitted from urine and faeces that have shown specific signatures that reflect gut microbial compositions and functions [78]. With gas chromatography-mass spectrometry (GC-MS), faecal or urinary VOC analysis could also serve as a novel screening tool for CRC detection [79-81]. Vernia et al., in a multicentric study, recruited CRC patients screened with colonoscopy to analyse VOC in their breath. They reported that VOC models could detect patients with CRC with an area under the curve (AUC) of 0.84 with 95% sensitivity and 64% specificity [82]. VOC analysis, therefore, represents a promising non-invasive tool for CRC screening [82].
Gut Microbiome and CRC Precision Therapy
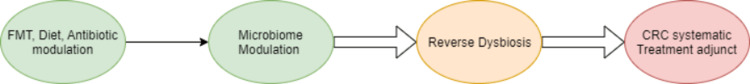
Modern microbial analytical techniques have improved our understanding of the mechanism of CRC formation linked to dysbiosis [83]. It is hypothesised that using antibiotics may also result in microbial dysbiosis and immune system issues, which speed up CRC progression [84]. Therefore, reversing these dysbiotic changes may aid in preventing and treating them [85]. Potential therapeutic strategies include dietary changes, pre/probiotics, faecal microbiota transplantation (FMT), and antibiotics [86].
Some studies suggest that dietary modifications, including ingesting more fibre, may reduce the incidence of colon cancer and work in conjunction with traditional treatments [87-89]. Probiotics, live beneficial flora, may treat CRC by combating CRC-driver microbiota, increasing gut microbial diversity [90], improving immunity homoeostasis, reducing chronic inflammation, and decreasing carcinogenic metabolites [87]. Through gut microbiome reconstruction, FMT can improve bile acid metabolism and immunotherapy efficacy and subsequently serve as a natural remedy for CRC [91]. A healthy environment, good food, exercise, weight control, and avoiding or alleviating stress with relaxation techniques are also essential preventative strategies against developing CRC (Figure 2) [85]. Obesity, diabetes, irritable bowel syndrome, IBD, depression, and cardiovascular disease are also studied for microbiome-based therapy [92-97].
Figure 2. Microbiome modulation and CRC treatment.
FMT: faecal microbiota transplantation; CRC: colorectal cancer.
Future studies will need to improve ways for modulating the microbiome and provide major unsolved questions (e.g. CRC-microbiota causal link) that will be addressed in existing and future research [98]. Combining endogenous host variables (e.g. host genetics and the microbiome) with exogenous environmental factors (e.g. nutrition and smoking) can significantly impact the treatment response of CRC [9].
Conclusions
A substantial body of research has established a strong relationship between microbial alterations (dysbiosis) and CRC carcinogenesis. However, the precise microbial-host interactions in CRC development remain elusive and influenced by various cofactors (exogenous and endogenous).
Some studies have expressed that microbiome alterations can be modified to treat CRC. However, more sophisticated molecular-based analysis and prospective interventional studies may yield more specific CRC microbial biomarkers and personalized therapeutic techniques for CRC management.
The content published in Cureus is the result of clinical experience and/or research by independent individuals or organizations. Cureus is not responsible for the scientific accuracy or reliability of data or conclusions published herein. All content published within Cureus is intended only for educational, research and reference purposes. Additionally, articles published within Cureus should not be deemed a suitable substitute for the advice of a qualified health care professional. Do not disregard or avoid professional medical advice due to content published within Cureus.
Footnotes
The authors have declared that no competing interests exist.
References
- 1.Cancer Research UK. Bowel cancer statistics. [ Oct; 2022 ]. 2022. https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/bowel-cancer https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/bowel-cancer
- 2.Colorectal cancer. Kuipers EJ, Grady WM, Lieberman D, et al. Nat Rev Dis Primers. 2015;1:15065. doi: 10.1038/nrdp.2015.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Advances in fecal tests for colorectal cancer screening. Schreuders EH, Grobbee EJ, Spaander MC, Kuipers EJ. Curr Treat Options Gastroenterol. 2016;14:152–162. doi: 10.1007/s11938-016-0076-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fecal occult blood tests in colorectal cancer screening: systematic review and meta-analysis of traditional and new-generation fecal immunochemical tests. Meklin J, SyrjÄnen K, Eskelinen M. Anticancer Res. 2020;40:3591–3604. doi: 10.21873/anticanres.14349. [DOI] [PubMed] [Google Scholar]
- 5.Colorectal cancer screening: tests, strategies, and perspectives. Stracci F, Zorzi M, Grazzini G. Front Public Health. 2014;2:210. doi: 10.3389/fpubh.2014.00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The gut flora as a forgotten organ. O'Hara AM, Shanahan F. EMBO Rep. 2006;7:688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Effects of probiotics on gut microbiota: mechanisms of intestinal immunomodulation and neuromodulation. Hemarajata P, Versalovic J. Therap Adv Gastroenterol. 2013;6:39–51. doi: 10.1177/1756283X12459294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The intestinal microbiota and colorectal cancer. Cheng Y, Ling Z, Li L. Front Immunol. 2020;11:615056. doi: 10.3389/fimmu.2020.615056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colorectal carcinogenesis: an archetype of gut microbiota-host interaction. Alexander JL, Scott AJ, Pouncey AL, Marchesi J, Kinross J, Teare J. Ecancermedicalscience. 2018;12:865. doi: 10.3332/ecancer.2018.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Revised estimates for the number of human and bacteria cells in the body. Sender R, Fuchs S, Milo R. PLoS Biol. 2016;14:0. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Host-microbiota interactions: from holobiont theory to analysis. Simon JC, Marchesi JR, Mougel C, Selosse MA. Microbiome. 2019;7:5. doi: 10.1186/s40168-019-0619-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Role of the microbiota in immunity and inflammation. Belkaid Y, Hand TW. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Impact of human aging and modern lifestyle on gut microbiota. Valle Gottlieb MG, Closs VE, Junges VM, Schwanke CH. Crit Rev Food Sci Nutr. 2018;58:1557–1564. doi: 10.1080/10408398.2016.1269054. [DOI] [PubMed] [Google Scholar]
- 14.Gut microbiota and lifestyle interventions in NAFLD. Houghton D, Stewart CJ, Day CP, Trenell M. Int J Mol Sci. 2016;17:447. doi: 10.3390/ijms17040447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bile acid dysregulation, gut dysbiosis, and gastrointestinal cancer. Tsuei J, Chau T, Mills D, Wan YJ. Exp Biol Med (Maywood) 2014;239:1489–1504. doi: 10.1177/1535370214538743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Impacts of gut bacteria on human health and diseases. Zhang YJ, Li S, Gan RY, Zhou T, Xu DP, Li HB. Int J Mol Sci. 2015;16:7493–7519. doi: 10.3390/ijms16047493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enterococcus faecalis induces aneuploidy and tetraploidy in colonic epithelial cells through a bystander effect. Wang X, Allen TD, May RJ, Lightfoot S, Houchen CW, Huycke MM. Cancer Res. 2008;68:9909–9917. doi: 10.1158/0008-5472.CAN-08-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lactobacillaceae and cell adhesion: genomic and functional screening. Turpin W, Humblot C, Noordine ML, Thomas M, Guyot JP. PLoS One. 2012;7:0. doi: 10.1371/journal.pone.0038034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Intermittent fasting confers protection in CNS autoimmunity by altering the gut microbiota. Cignarella F, Cantoni C, Ghezzi L, et al. Cell Metab. 2018;27:1222–1235. doi: 10.1016/j.cmet.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Current understanding of dysbiosis in disease in human and animal models. DeGruttola AK, Low D, Mizoguchi A, Mizoguchi E. Inflamm Bowel Dis. 2016;22:1137–1150. doi: 10.1097/MIB.0000000000000750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gut microbiome and gastrointestinal cancer: les liaisons dangereuses. Tözün N, Vardareli E. J Clin Gastroenterol. 2016;50:0–6. doi: 10.1097/MCG.0000000000000714. [DOI] [PubMed] [Google Scholar]
- 22.Microbiota dysbiosis and barrier dysfunction in inflammatory bowel disease and colorectal cancers: exploring a common ground hypothesis. Yu LC. J Biomed Sci. 2018;25:79. doi: 10.1186/s12929-018-0483-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colorectal cancer mutational profiles correlate with defined microbial communities in the tumor microenvironment. Burns MB, Montassier E, Abrahante J, et al. PLoS Genet. 2018;14:0. doi: 10.1371/journal.pgen.1007376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Microbiome, inflammation and colorectal cancer. Chen J, Pitmon E, Wang K. Semin Immunol. 2017;32:43–53. doi: 10.1016/j.smim.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 25.The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Gopalakrishnan V, Helmink BA, Spencer CN, Reuben A, Wargo JA. Cancer Cell. 2018;33:570–580. doi: 10.1016/j.ccell.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Wong SH, Yu J. Nat Rev Gastroenterol Hepatol. 2019;16:690–704. doi: 10.1038/s41575-019-0209-8. [DOI] [PubMed] [Google Scholar]
- 27.Gut microbiome and its role in colorectal cancer. Rebersek M. BMC Cancer. 2021;21:1325. doi: 10.1186/s12885-021-09054-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gut microbiota dysbiosis drives the development of colorectal cancer. Fan X, Jin Y, Chen G, Ma X, Zhang L. Digestion. 2021;102:508–515. doi: 10.1159/000508328. [DOI] [PubMed] [Google Scholar]
- 29.Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Mima K, Nishihara R, Qian ZR, et al. Gut. 2016;65:1973–1980. doi: 10.1136/gutjnl-2015-310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Castellarin M, Warren RL, Freeman JD, et al. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.The association between fecal enterotoxigenic B. fragilis with colorectal cancer. Haghi F, Goli E, Mirzaei B, Zeighami H. BMC Cancer. 2019;19:879. doi: 10.1186/s12885-019-6115-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quantitative profiling of colorectal cancer-associated bacteria reveals associations between fusobacterium spp., enterotoxigenic Bacteroides fragilis (ETBF) and clinicopathological features of colorectal cancer. Viljoen KS, Dakshinamurthy A, Goldberg P, Blackburn JM. PLoS One. 2015;10:0. doi: 10.1371/journal.pone.0119462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adenomatous polyps are driven by microbe-instigated focal inflammation and are controlled by IL-10-producing T cells. Dennis KL, Wang Y, Blatner NR, et al. Cancer Res. 2013;73:5905–5913. doi: 10.1158/0008-5472.CAN-13-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Intestinal inflammation targets cancer-inducing activity of the microbiota. Arthur JC, Perez-Chanona E, Mühlbauer M, et al. Science. 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Changes in gut microbiota and plasma inflammatory factors across the stages of colorectal tumorigenesis: a case-control study. Zhang Y, Yu X, Yu E, et al. BMC Microbiol. 2018;18:92. doi: 10.1186/s12866-018-1232-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fusobacterium nucleatum acts as a pro-carcinogenic bacterium in colorectal cancer: from association to causality. Wang S, Liu Y, Li J, et al. Front Cell Dev Biol. 2021;9:710165. doi: 10.3389/fcell.2021.710165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Transcriptional regulation of Wnt/β-catenin pathway in colorectal cancer. Bian J, Dannappel M, Wan C, Firestein R. Cells. 2020;9:2125. doi: 10.3390/cells9092125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.The interrelationships of the gut microbiome and inflammation in colorectal carcinogenesis. Cho M, Carter J, Harari S, Pei Z. Clin Lab Med. 2014;34:699–710. doi: 10.1016/j.cll.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Tjalsma H, Boleij A, Marchesi JR, Dutilh BE. Nat Rev Microbiol. 2012;10:575–582. doi: 10.1038/nrmicro2819. [DOI] [PubMed] [Google Scholar]
- 40.Genetic determinants of the gut microbiome in UK twins. Goodrich JK, Davenport ER, Beaumont M, et al. Cell Host Microbe. 2016;19:731–743. doi: 10.1016/j.chom.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Distinct microbes, metabolites, and ecologies define the microbiome in deficient and proficient mismatch repair colorectal cancers. Hale VL, Jeraldo P, Chen J, et al. Genome Med. 2018;10:78. doi: 10.1186/s13073-018-0586-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Distinct gut microbiome patterns associate with consensus molecular subtypes of colorectal cancer. Purcell RV, Visnovska M, Biggs PJ, Schmeier S, Frizelle FA. Sci Rep. 2017;7:11590. doi: 10.1038/s41598-017-11237-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Comparative analysis of amplicon and metagenomic sequencing methods reveals key features in the evolution of animal metaorganisms. Rausch P, Rühlemann M, Hermes BM, et al. Microbiome. 2019;7:133. doi: 10.1186/s40168-019-0743-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rapid and highly-specific generation of targeted DNA sequencing libraries enabled by linking capture probes with universal primers. Pel J, Leung A, Choi WW, et al. PLoS One. 2018;13:0. doi: 10.1371/journal.pone.0208283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.A metagenome-wide association study of gut microbiota in type 2 diabetes. Qin J, Li Y, Cai Z, et al. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 46.Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Ferreira RM, Pereira-Marques J, Pinto-Ribeiro I, Costa JL, Carneiro F, Machado JC, Figueiredo C. Gut. 2018;67:226–236. doi: 10.1136/gutjnl-2017-314205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Defective ATG16L1-mediated removal of IRE1α drives Crohn's disease-like ileitis. Tschurtschenthaler M, Adolph TE, Ashcroft JW, et al. J Exp Med. 2017;214:401–422. doi: 10.1084/jem.20160791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Colorectal cancer and the human gut microbiome: reproducibility with whole-genome shotgun sequencing. Vogtmann E, Hua X, Zeller G, et al. PLoS One. 2016;11:0. doi: 10.1371/journal.pone.0155362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Current challenges and best-practice protocols for microbiome analysis. Bharti R, Grimm DG. Brief Bioinform. 2021;22:178–193. doi: 10.1093/bib/bbz155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Metagenomics - a guide from sampling to data analysis. Thomas T, Gilbert J, Meyer F. Microb Inform Exp. 2012;2:3. doi: 10.1186/2042-5783-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Metaproteomics: studying functional gene expression in microbial ecosystems. Wilmes P, Bond PL. Trends Microbiol. 2006;14:92–97. doi: 10.1016/j.tim.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 52.Gut microbiota-produced tryptamine activates an epithelial G-protein-coupled receptor to increase colonic secretion. Bhattarai Y, Williams BB, Battaglioli EJ, et al. Cell Host Microbe. 2018;23:775–785. doi: 10.1016/j.chom.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Flemer B, Lynch DB, Brown JM, et al. Gut. 2017;66:633–643. doi: 10.1136/gutjnl-2015-309595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Quantitation of faecal Fusobacterium improves faecal immunochemical test in detecting advanced colorectal neoplasia. Wong SH, Kwong TN, Chow TC, et al. Gut. 2017;66:1441–1448. doi: 10.1136/gutjnl-2016-312766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.A high-throughput organoid microinjection platform to study gastrointestinal microbiota and luminal physiology. Williamson IA, Arnold JW, Samsa LA, et al. Cell Mol Gastroenterol Hepatol. 2018;6:301–319. doi: 10.1016/j.jcmgh.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.The gut microflora assay in patients with colorectal cancer: in feces or tissue samples? Rezasoltani S, Dabiri H, Asadzadeh-Aghdaei H, Sepahi AA, Modarressi MH, Nazemalhosseini-Mojarad E. https://pubmed.ncbi.nlm.nih.gov/30996824/ Iran J Microbiol. 2019;11:1–6. [PMC free article] [PubMed] [Google Scholar]
- 57.Systematic review: gut microbiota in fecal samples and detection of colorectal neoplasms. Amitay EL, Krilaviciute A, Brenner H. Gut Microbes. 2018;9:293–307. doi: 10.1080/19490976.2018.1445957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Conducting a microbiome study. Goodrich JK, Di Rienzi SC, Poole AC, et al. Cell. 2014;158:250–262. doi: 10.1016/j.cell.2014.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meta-analysis of the human gut microbiome from urbanized and pre-agricultural populations. Mancabelli L, Milani C, Lugli GA, Turroni F, Ferrario C, van Sinderen D, Ventura M. Environ Microbiol. 2017;19:1379–1390. doi: 10.1111/1462-2920.13692. [DOI] [PubMed] [Google Scholar]
- 60.Microbiota organization is a distinct feature of proximal colorectal cancers. Dejea CM, Wick EC, Hechenbleikner EM, et al. Proc Natl Acad Sci U S A. 2014;111:18321–18326. doi: 10.1073/pnas.1406199111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Towards the human colorectal cancer microbiome. Marchesi JR, Dutilh BE, Hall N, Peters WH, Roelofs R, Boleij A, Tjalsma H. PLoS One. 2011;6:0. doi: 10.1371/journal.pone.0020447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Virulence genes are a signature of the microbiome in the colorectal tumor microenvironment. Burns MB, Lynch J, Starr TK, Knights D, Blekhman R. Genome Med. 2015;7:55. doi: 10.1186/s13073-015-0177-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Microbiota and phage therapy: future challenges in medicine. Paule A, Frezza D, Edeas M. Med Sci (Basel) 2018;6:86. doi: 10.3390/medsci6040086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rectal swabs for analysis of the intestinal microbiota. Budding AE, Grasman ME, Eck A, Bogaards JA, Vandenbroucke-Grauls CM, van Bodegraven AA, Savelkoul PH. PLoS One. 2014;9:0. doi: 10.1371/journal.pone.0101344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Microbiome and colorectal cancer: roles in carcinogenesis and clinical potential. Saus E, Iraola-Guzmán S, Willis JR, Brunet-Vega A, Gabaldón T. Mol Aspects Med. 2019;69:93–106. doi: 10.1016/j.mam.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Integrated omics: tools, advances and future approaches. Misra BB, Langefeld CD, Olivier M, Cox LA. J Mol Endocrinol. 2018;68:0–45. doi: 10.1530/JME-18-0055. [DOI] [PubMed] [Google Scholar]
- 67.Challenges, strategies, and perspectives for reference-independent longitudinal multi-omic microbiome studies. Martínez Arbas S, Busi SB, Queirós P, et al. Front Genet. 2021;12:666244. doi: 10.3389/fgene.2021.666244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Biomarkers in colorectal cancer: current research and future prospects. Ogunwobi OO, Mahmood F, Akingboye A. Int J Mol Sci. 2020;21:5311. doi: 10.3390/ijms21155311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Colorectal cancer tumour markers and biomarkers: recent therapeutic advances. Lech G, Słotwiński R, Słodkowski M, Krasnodębski IW. World J Gastroenterol. 2016;22:1745–1755. doi: 10.3748/wjg.v22.i5.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Faecal microbial biomarkers in early diagnosis of colorectal cancer. Olovo CV, Huang X, Zheng X, Xu M. J Cell Mol Med. 2021;25:10783–10797. doi: 10.1111/jcmm.17010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.First steps towards combining faecal immunochemical testing with the gut microbiome in colorectal cancer screening. Grobbee EJ, Lam SY, Fuhler GM, et al. United European Gastroenterol J. 2020;8:293–302. doi: 10.1177/2050640619890732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.The human gut microbiome as a screening tool for colorectal cancer. Zackular JP, Rogers MA, Ruffin MT 4th, Schloss PD. Cancer Prev Res (Phila) 2014;7:1112–1121. doi: 10.1158/1940-6207.CAPR-14-0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Using fecal immunochemical tubes for the analysis of the gut microbiome has the potential to improve colorectal cancer screening. Krigul KL, Aasmets O, Lüll K, Org T, Org E. Sci Rep. 2021;11:19603. doi: 10.1038/s41598-021-99046-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.The fecal metabolome as a functional readout of the gut microbiome. Zierer J, Jackson MA, Kastenmüller G, et al. Nat Genet. 2018;50:790–795. doi: 10.1038/s41588-018-0135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alterations of the gut microbiome and fecal metabolome in colorectal cancer: implication of intestinal metabolism for tumorigenesis. Du X, Li Q, Tang Z, et al. Front Physiol. 2022;13:854545. doi: 10.3389/fphys.2022.854545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.A comprehensive metabolomics analysis of fecal samples from advanced adenoma and colorectal cancer patients. Telleria O, Alboniga OE, Clos-Garcia M, Nafría-Jimenez B, Cubiella J, Bujanda L, Falcón-Pérez JM. Metabolites. 2022;12:550. doi: 10.3390/metabo12060550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Breathomics in lung disease. van der Schee MP, Paff T, Brinkman P, van Aalderen WM, Haarman EG, Sterk PJ. Chest. 2015;147:224–231. doi: 10.1378/chest.14-0781. [DOI] [PubMed] [Google Scholar]
- 78.Functional genomic analyses of the gut microbiota for CRC screening. Konstantinov SR, Kuipers EJ, Peppelenbosch MP. Nat Rev Gastroenterol Hepatol. 2013;10:741–745. doi: 10.1038/nrgastro.2013.178. [DOI] [PubMed] [Google Scholar]
- 79.Volatile organic compounds emitted from faeces as a biomarker for colorectal cancer. Bond A, Greenwood R, Lewis S, et al. Aliment Pharmacol Ther. 2019;49:1005–1012. doi: 10.1111/apt.15140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Detection of colorectal cancer (CRC) by urinary volatile organic compound analysis. Arasaradnam RP, McFarlane MJ, Ryan-Fisher C, et al. PLoS One. 2014;9:0. doi: 10.1371/journal.pone.0108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Colorectal cancer and adenoma screening using urinary volatile organic compound (VOC) detection: early results from a single-centre bowel screening population (UK BCSP) Mozdiak E, Wicaksono AN, Covington JA, Arasaradnam RP. Tech Coloproctol. 2019;23:343–351. doi: 10.1007/s10151-019-01963-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Are volatile organic compounds accurate markers in the assessment of colorectal cancer and inflammatory bowel diseases? A review. Vernia F, Valvano M, Fabiani S, Stefanelli G, Longo S, Viscido A, Latella G. Cancers (Basel) 2021;13:2361. doi: 10.3390/cancers13102361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Extensive impact of non-antibiotic drugs on human gut bacteria. Maier L, Pruteanu M, Kuhn M, et al. Nature. 2018;555:623–628. doi: 10.1038/nature25979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Antibiotic use and colorectal cancer: a causal association? Ma W, Chan AT. Gut. 2020;69:1913–1914. doi: 10.1136/gutjnl-2019-319792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gut microbiota modulation: a novel strategy for prevention and treatment of colorectal cancer. Fong W, Li Q, Yu J. Oncogene. 2020;39:4925–4943. doi: 10.1038/s41388-020-1341-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Intestinal microbiota: a novel perspective in colorectal cancer biotherapeutics. Ding C, Tang W, Fan X, Wu G. Onco Targets Ther. 2018;11:4797–4810. doi: 10.2147/OTT.S170626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.The role of the gut microbiome in colorectal cancer development and therapy response. Sánchez-Alcoholado L, Ramos-Molina B, Otero A, et al. Cancers (Basel) 2020;12:1406. doi: 10.3390/cancers12061406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Role of diet in colorectal cancer incidence: umbrella review of meta-analyses of prospective observational studies. Veettil SK, Wong TY, Loo YS, Playdon MC, Lai NM, Giovannucci EL, Chaiyakunapruk N. JAMA Netw Open. 2021;4:0. doi: 10.1001/jamanetworkopen.2020.37341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dietary fibre protective against colorectal cancer patients in Asia: a meta-analysis. Masrul M, Nindrea RD. Open Access Maced J Med Sci. 2019;7:1723–1727. doi: 10.3889/oamjms.2019.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Probiotics and colon cancer. Drago L. Microorganisms. 2019;7:66. doi: 10.3390/microorganisms7030066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fecal microbiota transplantation in cancer management: current status and perspectives. Chen D, Wu J, Jin D, Wang B, Cao H. Int J Cancer. 2019;145:2021–2031. doi: 10.1002/ijc.32003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fecal microbiota transplantation in metabolic syndrome: history, present and future. de Groot PF, Frissen MN, de Clercq NC, Nieuwdorp M. Gut Microbes. 2017;8:253–267. doi: 10.1080/19490976.2017.1293224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.The impact of low-FODMAPs, gluten-free, and ketogenic diets on gut microbiota modulation in pathological conditions. Reddel S, Putignani L, Del Chierico F. Nutrients. 2019;11:373. doi: 10.3390/nu11020373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.The ketogenic diet as a treatment paradigm for diverse neurological disorders. Stafstrom CE, Rho JM. Front Pharmacol. 2012;3:59. doi: 10.3389/fphar.2012.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.The gut-brain axis and the microbiome: clues to pathophysiology and opportunities for novel management strategies in irritable bowel syndrome (IBS) Quigley EM. J Clin Med. 2018;7:6. doi: 10.3390/jcm7010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Colorectal cancer in inflammatory bowel disease: the risk, pathogenesis, prevention and diagnosis. Kim ER, Chang DK. World J Gastroenterol. 2014;20:9872–9881. doi: 10.3748/wjg.v20.i29.9872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Role of gut microbiota-generated short-chain fatty acids in metabolic and cardiovascular health. Chambers ES, Preston T, Frost G, Morrison DJ. Curr Nutr Rep. 2018;7:198–206. doi: 10.1007/s13668-018-0248-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Modulating the microbiome to improve therapeutic response in cancer. McQuade JL, Daniel CR, Helmink BA, Wargo JA. Lancet Oncol. 2019;20:0–91. doi: 10.1016/S1470-2045(18)30952-5. [DOI] [PubMed] [Google Scholar]