Abstract
Background:
Psychiatric disorders have been associated with advanced epigenetic age in DNA methylation, yet this relationship has not been studied in the brain transcriptome. We examined transcriptomic age using an RNA-based algorithm recently developed by Ren and Kuan (“RNAAgeCalc”) and the associations between posttraumatic stress disorder (PTSD), major depressive disorder (MDD), and alcohol use disorder (AUD) with age-adjusted RNA age (“RNA age residuals”) in three brain regions: dorsolateral prefrontal cortex, ventromedial prefrontal cortex (vmPFC), and motor cortex.
Methods:
RNA sequencing was used to measure gene expression in postmortem brain tissue from the VA National PTSD Brain Bank (n = 94; 59% male).
Results:
Linear models revealed that diagnoses of PTSD and/or MDD were positively associated with RNA age residuals in vmPFC only (p-adj = .012). Three genes in the RNAAgeCalc algorithm (KCNJ16, HYAL2, and CEBPB) were also differentially expressed in association with PTSD/MDD in vmPFC (p-adj = 6.45E-05 to .02). Enrichment analysis revealed that inflammatory and immune-related pathways were overrepresented (p-adj < .05) among the 43 genes in RNAAgeCalc that were also at least nominally associated with PTSD/MDD in vmPFC relative to the 448 RNAAgeCalc genes. Endothelial and mural cells were negatively associated with RNA age residuals in vmPFC (both p-adj = .028) and with PTSD/MDD (both p-adj = .017).
Conclusions:
Results highlight the importance of inflammation and immune system dysregulation in the link between psychopathology and accelerated cellular aging and raise the possibility that blood-brain barrier degradation may play an important role in stress-related accelerated brain aging.
Keywords: RNA, accelerated aging, transcriptomic age, PTSD, major depression, inflammation
Introduction
A substantial body of research suggests that psychiatric and traumatic stress are associated with advanced biological age, defined as metrics of cellular aging that outpace chronological aging. This includes work suggesting that trauma exposure and psychiatric disorders are associated with shortened telomeres (Darrow et al., 2016), advanced epigenetic age as estimated from DNA methylation (DNAm) data (Wolf et al., 2016, 2018; Wolf, Morrison, et al., 2019), and greater scores on a composite biological age metric defined by a variety of metabolic, inflammatory, and physiological measurements (Belsky et al., 2017). Posttraumatic stress disorder (PTSD; Wolf et al., 2016), major depressive disorder (MDD; Protsenko et al., 2021), and alcohol-use disorders (AUD; Bøstrand et al., 2022; Wolf, Logue, et al., 2019) have all been associated with advanced DNAm age in blood in cross-sectional studies. A longitudinal study found that PTSD symptoms and AUD were also associated with an increased pace of epigenetic aging over time (Wolf, Logue, et al., 2019). Recent work has extended this to brain tissue and, using data from the same cohort that is the focus of this study, found associations between advanced DNAm age and AUD (Wolf et al., 2020), and PTSD (in those at risk by virtue of klotho genotype and older age; Wolf et al., 2020) in motor cortex; associations between MDD and advanced DNAm age in prefrontal tissue have also been reported in a distinct cohort (Han et al., 2018). This work is important as advanced biological age is associated with metabolic and immune dysregulation, increased inflammation (Morrison, Logue, et al., 2019; Quach et al., 2017), and reduced neural integrity (Levine et al., 2015; Wolf et al., 2016), which may play a critical role in linking psychiatric stress to early onset metabolic and cardiovascular diseases, neuropathology, and premature mortality.
Recently, Ren and Kuan (2020) developed a gene expression age calculator, which they called “RNAAgeCalc,” from transcriptome-wide RNA sequence data available in the Genotype-Tissue Expression (GTEx) database (https://gtexportal.org/home/). They optimized the prediction of chronological age across 26 tissue types and developed both all-tissue and tissue-specific RNA age algorithms. The all-tissue algorithm was based on 1,616 genes. Pathway analyses indicated that the genes included in the RNAAgeCalc algorithm were enriched for processes relating to interferon activity and membrane adhesion molecules (among upregulated genes) and for RNA transport, mitochondrial function, and ribosome biogenesis functions (among downregulated genes). The estimates generated by the tissue-specific algorithms tended to show stronger associations with chronological age relative to those derived from the all-tissue model (Ren & Kuan, 2020), with estimates from the model developed in brain tissue (based on 472 genes) evidencing a Pearson correlation of r = .82 with chronological age. Advanced transcriptomic age (i.e., estimates that exceed chronological age) in brain tissue was associated with increased somatic mutations in cancer samples and cancer-related mortality (Ren & Kuan, 2020). Prior to the development of the Ren and Kuan sequence-based algorithms, the only published RNA age algorithm was based on microarray expression data (Peters et al., 2015), which showed weaker correlations with chronological age (Ren & Kuan, 2020).
Two studies have examined associations between psychopathology and transcriptomic age in blood using RNA sequence data. Kuan et al. (2021) reported significantly higher age-adjusted RNA age estimates (using the Ren & Kuan, 2020 algorithm) in current PTSD cases compared to individuals without a current or prior PTSD diagnosis among 324 World Trade Center responders. Similarly, Cole et al. (2021) found that individuals with MDD were more likely to have over-estimated transcriptomic age relative to chronological age as compared to healthy controls; transcriptomic age was measured using a novel index derived from gene expression values obtained in peripheral blood mononuclear cells. In addition, one study calculated RNA age using microarray expression (not sequence) data from whole blood samples and found that depression during early adulthood was associated with advanced RNA age in African Americans (Carter et al., 2019). However, no study has examined these associations in post-mortem brain tissue or evaluated the potential genetic and biological pathways that may link psychopathology to accelerated transcriptomic aging. This is important because a better understanding of the specific biological pathways associated with psychiatric stress (e.g., in the methylome, transcriptome, inflammasome, metabolome) can help to identify the types of biological dysregulation involved. This can also help to identify risk for disease, track responses to interventions, and suggest new treatment avenues based on the pathophysiology of biological aging. The primary aim of this study was to examine associations between PTSD, MDD, and AUD and age-adjusted transcriptomic age in three brain regions using data from the VA National PTSD Brain Bank (Friedman et al., 2017). We have previously studied this same cohort to test associations between psychopathology and advanced DNAm age (Wolf et al., 2020) and to evaluate the gene expression correlates of advanced DNAm age (Wolf et al., 2021). A second aim was to identify the genes and related biological processes that drive associations between PTSD, MDD, and/or AUD and age-adjusted transcriptomic age and to evaluate associations between age-adjusted transcriptomic age and estimates of specific neural cell type counts. We also compared RNA age with Horvath-defined DNAm age (Horvath, 2013) and with two brain specific DNAm age indices (Choi et al., 2019; Shireby et al., 2020) to better understand the relationship between cellular aging in the methylome and transcriptome. We hypothesized that psychiatric stress would be associated with advanced transcriptomic age relative to age-at-death and that immune and inflammation-related biological processes would contribute to advanced transcriptomic age.
Materials and Methods
Participants and Procedures
Postmortem left-hemisphere brain tissue samples from N = 117 donors were acquired from the VA National PTSD Brain Bank (Friedman et al., 2017); they were originally collected by the Lieber Institute for Brain Development at Johns Hopkins University (Mighdoll et al., 2018). We examined expression in three brain regions: dorsolateral prefrontal cortex (dlPFC; Brodmann Area 9/46, taken at the level of the genu of the corpus callosum), ventromedial prefrontal cortex (vmPFC; Brodmann Area 12/32, taken at the level of the genu), and motor cortex (Brodmann Area 4, taken at the level of the superior central sulcus).
A standardized protocol was followed to determine cause and manner of death per Maryland state medical examinations. Neuropathological exams were performed by board-certified neuropathologists and included review of medical records and interviews with next-of-kin to evaluate psychiatric and medical diagnoses. Measures administered to next-of-kin to determine decedent diagnoses included the MINI International Neuropsychiatric Interview 6.0, a version of the PTSD Checklist for DSM-5 for post-mortem assessments, and the Lieber Psychological Autopsy Interview, which involves interviews with mental health clinicians (Mighdoll et al., 2018). Psychiatric diagnostic determinations, including PTSD, MDD, and AUD, were additionally evaluated by a minimum of two independent board-certified psychiatrists, who made confidence ratings in the PTSD diagnoses along a 1–5 scale. Only PTSD diagnoses based on confidence rating scores of at least 3 were included as positive PTSD cases. Toxicology evaluations were also performed by board-certified neuropathologists. Decedents with neurodegenerative disease, history or evidence of severe traumatic brain injury, or neuritic pathology were excluded from this brain bank (Mighdoll et al., 2018).
Among the 117 decedents, 94 had RNA data that passed all quality control (QC) checks in one or more region and were included in these analyses. Demographic characteristics of this sample are shown in Table 1. Analyses were focused primarily on cases with PTSD and/or MDD (n = 70 or 74.5%) versus healthy controls (n = 24 or 25.5%) because of substantial overlap across these two diagnostic groups. Only 4.3% (n = 4) of the sample had PTSD without MDD and 34.0% (n = 32) had MDD without PTSD. Comorbid PTSD and MDD was evident among 36.2% (n = 34) of the sample.
Table 1.
Sample Demographic Characteristics (n = 94)
| Variable | N (%) or Mean (SD) |
|---|---|
| Sex (male) | 55 (58.5) |
| Age (years) | 42.49 (11.12) |
| Race: White | 73 (77.7) |
| Race: Black | 21 (22.3) |
| Cigarette Use | 57 (60.6) |
| AUD | 29 (30.9) |
| PTSD or MDD | 70 (74.5) |
| Healthy Control | 24 (25.5) |
| PTSD no MDD | 4 (4.3) |
| MDD no PTSD | 32 (34.0) |
| PTSD + MDD | 34 (36.2) |
Note. Race determined by next-of-kin interview and medical records (as opposed to genotype). AUD = alcohol-use disorder; PTSD = posttraumatic stress disorder; MDD = major depressive disorder.
Genotyping and DNAm Methods and Statistical Procedures
Genotypes were obtained from motor cortex DNA samples using the Illumina HumanOmni2.5–8 array. DNAm from each brain region was assayed on the Illumina Infinium MethylationEPIC array. DNAm age estimates were calculated using the Horvath algorithm (Horvath, 2013) for all three regions. We also estimated DNAm age using two recently developed tissue-specific algorithms that were developed for human brain (Choi et al., 2019) and human cortical tissue (Shireby et al., 2020). Details of DNA extraction, QC procedures, genotype and DNAm data cleaning and imputation, ancestry assignment and ancestral principal component (PC) estimation, DNAm cell type estimates, and DNAm age (Horvath DNAm age, Choi brain-specific DNAm age, and Shireby cortical DNAm age) calculation are reviewed in the Supplementary Materials and were previously described (Logue et al., 2021; Morrison, Miller, et al., 2019; Wolf et al., 2020, 2021).
RNA Methods and Statistical Procedures
As described by Logue et al. (2021), RNA was extracted from 25mg of frozen blocks of dlPFC, vmPFC, and motor cortex tissue for library preparation. A Hiseq 2500 was used to sequence the libraries and produced paired-end 75-bp reads (see Supplementary Materials). Raw reads were first trimmed using Trimmomatic (Bolger et al., 2014) and then mapped to the hg38 reference genome (Schneider et al., 2017) by STAR (Dobin et al., 2013), followed by transcript-level quantification with Kallisto (Bray et al., 2016). Gene-level quantification was achieved by collapsing transcript abundance estimates using the Bioconductor package tximport (Soneson et al., 2016). Log-transformed counts were generated using the regularized log transformation (rLog) method introduced in DEseq2 (Love et al., 2014) as a source to detect outliers, estimate RNA quality, and calculate cell types balance. RNA integrity number (RIN) values were not available for all brain bank samples, thus quality surrogate variables (qSVs) were generated using Bioconductor package sva in R to assess RNA degradation (Leek et al., 2012); these were previously shown to be correlated with RIN values in a subset with these data (Logue et al., 2021). Scores for seven cell types (astrocytes, endothelial cells, microglia, mural cells, neurons, oligodendrocytes, and red blood cells) for each sample in each brain region were estimated using BrainInABlender (Hagenauer et al., 2018). These estimates reflect the expression of cell-type marker genes, and indicate the proportion of a given cell type in a sample (Hagenauer et al., 2018). Samples were excluded if they had less than 50% uniquely mapped reads or were considered outliers (see Supplementary Materials). Bipolar disorder cases were further removed from the samples that passed QC such that analyses were based on n = 94 in dlPFC, n = 87 in vmPFC, and n = 90 in motor cortex. Additional QC procedures are described in the Supplementary Materials.
Bioconductor package RNAAgeCalc (Ren & Kuan, 2020) was used to generate brain-specific RNA age estimates. Of the 472 genes included in RNAAgeCalc, raw counts of expression from 457 genes in dlPFC, vmPFC, and motor cortex were used as the input and 15 additional genes that were not measured were imputed by the algorithm. RNA age for each brain region was estimated using the RNAAgeCacl “predict_age” function in R (see Supplementary Materials). Five (1.06%) of the 472 genes in RNAAgeCalc overlapped genes with probes in the Horvath DNAm age algorithm.
Data Analyses
RNA age residuals for each brain region were calculated by regressing RNA age estimates on age-at-death and saving the residuals. Positive residuals indicate advanced transcriptomic age compared to age-at-death while negative residuals index slowed transcriptomic age. We followed the same basic procedure to calculate DNAm age residuals in each region per three algorithms (Horvath, Choi, and Shireby) and compared their associations with RNA age residuals via correlation. We used multiple regression to examine PTSD/MDD and AUD in association with RNA age residuals, controlling for the first three qSVs, sex, and cigarette use. For effects of interest (PTSD/MDD and AUD) with at least nominal significance (p < .05), we also report a false-discovery rate (FDR; Benjamini & Hochberg, 1995) corrected p-value, incorporating a correction across the three brain regions examined. Follow-up analyses added additional covariates to the model to examine potential confounds of significant associations, including ancestry (top 3 PCs), post-mortem interval (PMI), body mass index (BMI), cell type proportions, manner of death (suicide or substance-related), anti-depressant use at time-of-death, and the total number of different types of traumatic experiences.
Psychiatric diagnoses that were significantly associated with RNA age residuals were further examined in association with expression of the genes included in the RNAAgeCalc algorithm in order to better understand the specific genes that might contribute to the effect. This was achieved by conducting a transcriptome-wide analysis of expression (n = 34,205 genes in vmPFC) versus the psychiatric variable of interest and extracting results for the genes in RNAAgeCalc with sufficient expression levels (of 472 RNAAgeCalc genes, 448 of them met the criteria defined by having more than 1 read count in at least 30 subjects). This analysis controlled for age at the time of death, sex, the first three ancestry PCs, PMI, sequencing run ID, the first three qSVs, and seven cell type estimates. Multiple testing correction was achieved via FDR for the extracted candidates. Transcriptome-wide analysis was performed using the Bioconductor package DESeq2 (Love et al., 2014) in R. We also conducted a biological enrichment analysis of the RNAAgeCalc genes that were at least nominally (p < .05) associated with the psychiatric variables of interest relative to the 448 RNAAgeCalc genes (as the background list) in order to test for enrichment of particular biological pathways and functions within the algorithm genes. Enrichment analysis was performed using the Bioconductor GOSeq package (Young et al., 2010) in R, and the 448 RNAAgeCalc genes were supplied as background genes.
Next, we conducted multiple regression analyses to examine psychiatric diagnoses that were significantly associated with RNA age residuals as predictors of each cell type score (calculated via BrainInABlender), controlling for sex, sequencing run ID, and first three qSVs. We use the same analytic approach to examine associations between RNA age residuals and each cell type score. We corrected p-values across the number of cell types examined using FDR. In order to test whether certain genes in the RNAAgeCalc algorithm were driving the associations between advanced transcriptomic age and cell type composition, transcriptome-wide analysis was performed to model the relationships between gene expression and cell type scores. Sex, sequencing run ID, and the first three qSVs were included as covariates. Results for genes in the RNAAgeCalc algorithm were extracted from the transcriptome-wide analysis and p-values were corrected across the number of candidates and cell types (448 × 7) via FDR.
Results
Associations between RNA Age, DNAm Age, and Chronological Age
The raw RNA age estimates for each brain region correlated strongly with age-at-death (rs = .69 to .76, ps < .001), raw Horvath DNAm age estimates (rs = .63 to .69, ps < .001), and raw Choi and Shireby DNAm age estimates (rs = .59 to .72, ps < .001; Supplementary Figure S1). Examination of the RNA age residuals (i.e., with the influence of age-at-death removed from the variance) showed that these indices of slowed to advanced transcriptomic age were distinct from all three measures of DNAm age residuals (rs = −.03 to .17, smallest p = .09; Supplementary Figure S1). RNA age residuals from different brain regions were weakly correlated with each other (rs = .24 to .35; all p < .05; Supplementary Figure S1).
Associations between Psychopathology and RNA Age Residuals
There was a significant association between PTSD/MDD (but not AUD) and RNA age residuals in vmPFC that withstood correction for multiple testing (β = .39, p = .004, p-adj = .012; Table 2; Figure 1). The effect was not significant in dlPFC or motor cortex. In a series of six additional models, we included sets of additional covariates. The PTSD/MDD effect was still significant in vmPFC when additionally controlling for BMI, PMI, the top three ancestry PCs, seven cell type scores, manner of death, anti-depressant use at time-of-death, and total trauma exposure; the standardized coefficient for the PTSD/MDD term changed minimally (mean absolute value Δβ = .02) with these other covariates included in the model (see Supplementary Materials). Comparison of different comorbid psychiatric groups revealed that the strongest effect was observed among those with PTSD + MDD (β = .50, p = .003; see Supplementary Materials). Based on these results, all subsequent analyses were carried out in vmPFC only in order to limit the number of statistical tests that were conducted.
Table 2.
Results of Regression Analyses Predicting RNA Age Residuals in Three Brain Regions
| Variable | dlPFC | vmPFC | Motor | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B | SE | β | P | P-adj | B | SE | β | P | P-adj | B | SE | β | P | P-adj | |
| qSV1 | .03 | .04 | .08 | .46 | .02 | .03 | .06 | .61 | .03 | .03 | .11 | .32 | |||
| qSV2 | −.04 | .06 | −.07 | .56 | −.04 | .06 | −.08 | .49 | .04 | .06 | .09 | .45 | |||
| qSV3 | −.14 | .11 | −.15 | .21 | .004 | .10 | .01 | .97 | −.04 | .10 | −.04 | .71 | |||
| Sex | .81 | 1.25 | .08 | .52 | −1.56 | 1.16 | −.15 | .18 | −.91 | 1.08 | −.10 | .40 | |||
| Cigarette-use | .37 | 1.33 | .03 | .78 | −1.05 | 1.27 | −.10 | .41 | −.10 | 1.15 | −.01 | .93 | |||
| AUD | −1.50 | 1.35 | −.13 | .27 | −1.03 | 1.25 | −.10 | .41 | .12 | 1.16 | .01 | .92 | |||
| PTSD/MDD | .38 | 1.62 | .03 | .82 | .82 | 4.66 | 1.58 | .39 | .004 | .012 | 1.49 | 1.39 | .14 | .29 | .435 |
Note. qSV = quality surrogate variable; AUD = alcohol-use disorder; PTSD/MDD = posttraumatic stress disorder or major depressive disorder; dlPFC = dorsolateral prefrontal cortex; vmPFC = ventromedial prefrontal cortex; B = unstandardized coefficient; SE = standard error; β = standardized coefficient; p-adj= FDR adjusted p-value across three brain regions.
Figure 1.
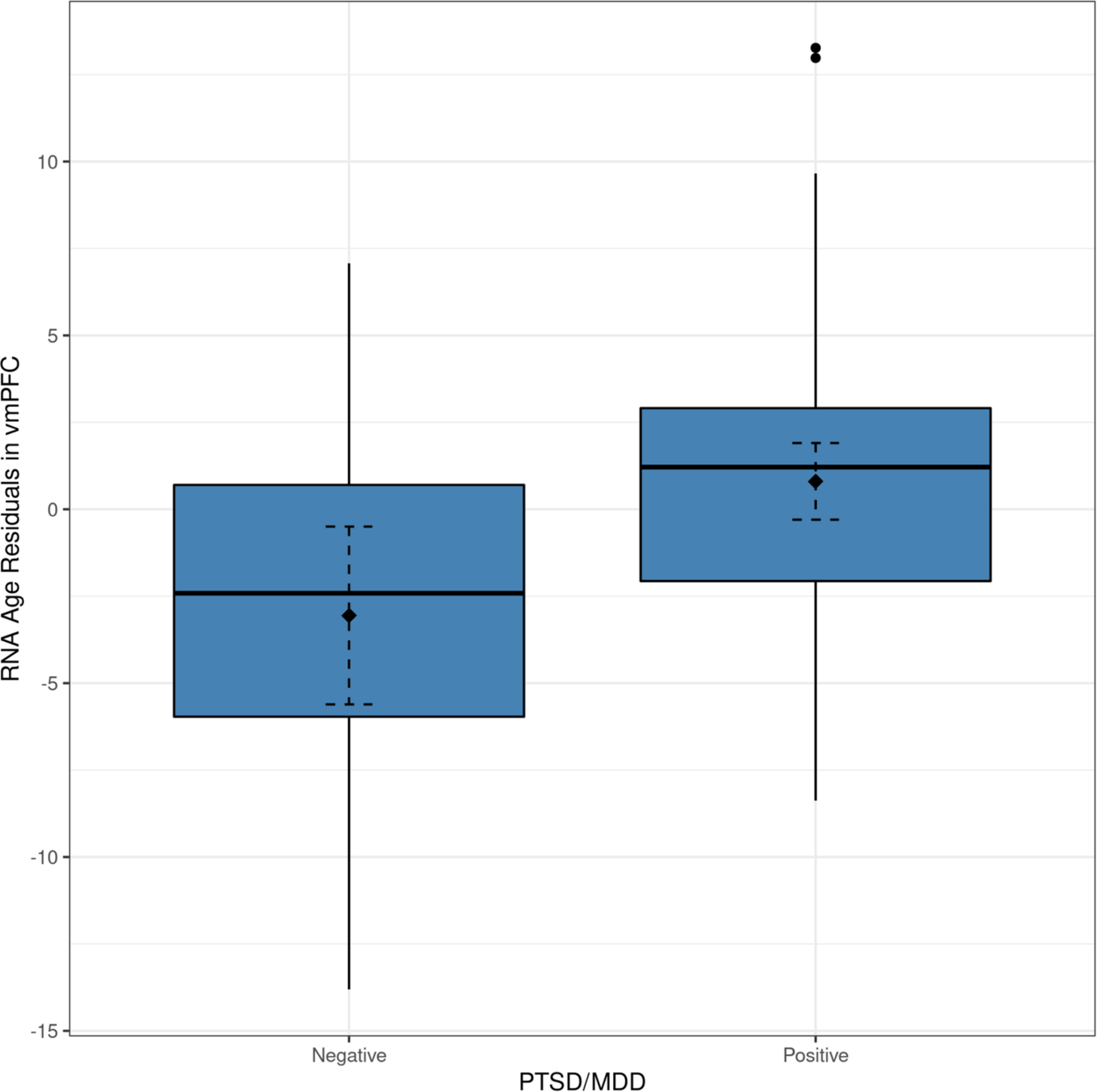
The figure shows a box-plot demonstrating the association between PTSD/MDD diagnoses and RNA age residuals in vmPFC. The filled diamond and the dashed error bars represent the mean value of the RNA age residuals and the 95% confidence interval of the mean. PTSD/MDD = posttraumatic stress disorder or major depressive disorder.
Associations Among PTSD/MDD and Expression of Genes in the RNA age Algorithm in vmPFC
We next examined associations between PTSD/MDD and vmPFC expression of the 448 genes included in the RNAAgeCalc algorithm that passed our QC criteria. Three corrected significant genes emerged: KCNJ16, which was down-regulated in PTSD/MDD cases (p = 1.439E-07, p-adj = 6.449E-05), and CEBPB (p = .0001, p-adj = .017) and HYAL2 (p = 9.792E-05, p-adj = .017), which were upregulated (Table 3; Table S1). We submitted the genes that were at least nominally associated with PTSD/MDD (n = 43) to a biological enrichment analysis using GoSeq with the 448 RNAAgeCalc genes as the background list. We found enrichment of 13 biological processes (including immune function and inflammation, cell death and apoptosis, and cell proliferation) and one molecular function (transcription factor binding) that withstood correction for multiple testing (ps = 6.844E-05 - .001; all p-adj < .05; Tables S2 and S3).
Table 3.
Genes in the RNAAgeCalc Algorithm with Corrected Significant Associations with PTSD/MDD in vmPFC
| Gene | Log2FoldChange | lfcSE | P | P-adj | RNA age coefficient |
|---|---|---|---|---|---|
| KCNJ16 | −.839 | .159 | 1.439E-07 | 6.449E-05 | .369 |
| HYAL2 | .606 | .155 | 9.792E-05 | .017 | .410 |
| CEBPB | .591 | .153 | 1.122E-04 | .017 | .212 |
Note. Log2FoldChange = log2 fold change between the groups of PTSD/MDDs and controls; lfcSE = standard error of the log2FoldChange estimate; p-adj = FDR adjusted p-value across number of RNAAgeCalc genes included in the transcriptome-wide analysis (n = 448); KCNJ16 = potassium inwardly rectifying channel subfamily J member 16; HYAL2 = hyaluronidase 2; CEBPB = CCAAT enhancer binding protein beta.
Associations with Sample Cell Type Composition
Analysis of PTSD/MDD in association with vmPFC cell type scores revealed that PTSD/MDD was associated with endothelial (B = −.294, p = .004, p-adj = .017) and mural cell scores (B = −.234, p = .005, p-adj = .017; Table S4 and Supplementary Materials). RNA age residuals were also significantly associated with endothelial and mural cell scores in vmPFC (both p-adj = .028; Table 4 and 1 Supplementary Materials1). We evaluated if the cell type associations in vmPFC might be a simple reflection of the genes included in RNAAgeCalc and the association between these cell types and RNA age. Overrepresentation analysis of genes in the RNAAgeCalc algorithm revealed significant enrichment of endothelial cell markers (n = 8, p-adj = .005; Table S5), however there was no association between raw RNA age estimates and endothelial (p = .144), or mural cell scores (p = .201; Table S6) in these samples. RNA age estimates were nominally associated with oligodendrocyte cell scores (p = .013, p-adj = .092; Table S6 and Supplementary Materials). There was no association between age-at-death and any cell type scores (Table S7). We further evaluated the 448 genes in RNAAgeCalc in vmPFC and found that expression of 334 genes evidenced corrected significant associations with at least one of the seven cell type scores (Table S8), suggesting that cell type composition made a substantial contribution to the genes in the RNAAgeCalc algorithm in this brain region.
Table 4.
Association between RNA age Residuals and Cell Type Estimates in vmPFC
| Cell type | B | SE | P | P-adj |
|---|---|---|---|---|
| Astrocytes | −.014 | .008 | .089 | .146 |
| Endothelial | −.022 | .008 | .008 | .028 |
| Microglia | −.022 | .013 | .089 | .146 |
| Mural | −.019 | .007 | .005 | .028 |
| Neurons | .012 | .009 | .195 | .227 |
| Oligodendrocytes | .023 | .014 | .104 | .146 |
| RBC | −.001 | .009 | .880 | .880 |
Note. Regressions controlled for sex, sequencing run IDs, and first three qSVs. Cell type content scores were estimated via BraininaBlender. Significant effects are shown in bold font. B = unstandardized coefficient; SE = standard error; P-adj = FDR adjusted p-value across number of cell types; Endothelial = endothelial cells; Mural = mural cells; RBC = red blood cells.
Discussion
Broadening our understanding of biological aging and its association with psychiatric disorders is critical for understanding the physiology of accelerated biological aging and developing treatments to reduce the cumulative burden of accelerated biological aging on disease and premature mortality. This study is the first to investigate psychiatric correlates of transcriptomic age in postmortem brain tissue. We found that brain transcriptomic age was highly correlated with age-at-death across dlPFC, vmPFC, and motor cortex. Further, deviations in transcriptomic age relative to age-at-death were associated with psychopathology: the presence of PTSD and/or MDD (though not AUD) was associated with advanced transcriptomic age in vmPFC. Additionally, we found evidence for immune and inflammation system involvement in the link between PTSD/MDD and advanced epigenetic aging in the transcriptome. Results also revealed reductions in endothelial and mural cells in association with both PTSD/MDD and advanced transcriptomic age. The PTSD/MDD cellular aging effects were observed in vmPFC and not in dlPFC or motor cortex. The vmPFC, which is critical for emotion processing and has been associated with depression and PTSD (Myers-Schulz & Koenigs, 2012), also shows signs of differential functioning in the earliest stages of Alzheimer’s disease (Dillen et al., 2017), and appears to be a central hub linking peripheral measures of inflammation to altered functional connectivity across numerous brain regions (Felger et al., 2016; Mehta et al., 2018; Yin et al., 2019). Collectively, this raises the possibility that vmPFC is particularly vulnerable to stress-related accelerated aging and associated alterations in inflammatory processes.
Psychopathology and Accelerated Aging
These results are consistent with prior studies showing associations between multiple psychiatric phenotypes, including PTSD, AUD, MDD, and anxiety, and advanced DNAm age in blood and brain tissue (Bøstrand et al., 2022; Han et al., 2018; Marečková et al., 2020; Protsenko et al., 2021; Wolf et al., 2018, 2020) and with research suggesting associations between PTSD and MDD and transcriptomic age in blood (Kuan et al., 2021; Cole et al., 2021; Carter et al., 2019). This study extends this association to the brain transcriptome, demonstrating the robustness of these associations across the epigenome and tissue type. Collectively, these studies suggest that associations between psychopathology and advanced epigenetic age are unlikely to be disorder-specific. Rather, psychiatric stress may be a common risk factor for accelerated aging, with increasing risk as a function of disease comorbidity and severity.
Inflammation, Immune Function, and Accelerated Aging
We examined expression among genes included in the transcriptomic age algorithm in the vmPFC to determine their individual associations with PTSD/MDD. We found three genes that evidenced significant associations with PTSD/MDD and thus may have contributed to the link between PTSD/MDD and age-adjusted RNA age estimates: CEBPB, KCNJ16, and HYAL2. CEBPB, which contributed positively to RNA age, was also significantly upregulated among those with PTSD/MDD. CEBPB is the beta isoform of the CCAAT Enhancer Binding Protein (CEBP) family of transcription factors. A study using RNA sequenced data from postmortem brain tissue suggested that CEBPB mRNA was significantly upregulated in cases with mild neurocognitive disorder compared to controls, and that CEBPB protein levels were elevated in the astroglia of postmortem brains of patients with human immunodeficiency virus-associated neurocognitive disorders (Canchi et al., 2020). In addition, the gene targets of the CEBPB transcription factor were significantly enriched for immune, inflammation, metabolic, and autophagy pathways in astrocytes, which strongly suggests its role in regulating neuroinflammation, metabolism, and the immune system (Canchi et al., 2020). Consistent with this, we found that CEBPB contributed to the significant enrichment of inflammatory and immune-related pathways among genes that were included in the RNAAgeCalc algorithm. CEBPB may accelerate brain transcriptomic aging via regulation of immune and inflammatory pathways, potentially contributing to neurocognitive decline. CEBPB may be a therapeutic target to reduce neuroinflammation and slow the pace of aging.
The two other genes in the algorithm that were associated with PTSD/MDD are less well studied. HYAL2 (hyaluronidase 2) is related to inflammatory responses and cardiometabolic pathology. Expression of HYAL2 has been linked to acute coronary syndromes (Vinci et al., 2021) and inflammatory cytokines, chemokines, and angiogenic factors in bladder cancer tissue (Dominguez-Gutierrez et al., 2021). KCNJ16 (potassium inwardly rectifying channel subfamily J member 16) is important for potassium transport into kidney cells and has been associated with metabolic acidosis (Webb et al., 2021) as well as hypotension, hypokalemia, and associated mortality in rats fed a high salt diet (Palygin et al., 2017). KCNJ16 is also expressed in the brain; rodent studies suggest that it is downregulated in hippocampal tissue following chronic restraint stress (Ren et al., 2021), consistent with our results suggesting that this gene was downregulated in association with PTSD/MDD. More broadly, potassium channel pathways are altered in the face of chronic stress (Ren et al., 2021) and treatment with the anti-seizure medication retigabine, which operates as a voltage-gated potassium channel opener, has recently been shown to reduce depressive symptoms in rodent models (Ren et al., 2021) and in initial human trials (Costi et al., 2021). Thus, HYAL2 may be part of the inflammation pathway linking PTSD and MDD to accelerated transcriptomic aging while KCNJ16 may reflect an independent pathway related to the impact of chronic stress and depression on potassium channel gating, with downstream effects on neural integrity and brain aging.
PTSD/MDD, Transcriptomic Age, and Neural Cell Types in vmPFC
The proportion of endothelial and mural cells in vmPFC was reduced among PTSD/MDD cases versus controls and these cell types were also negatively associated with RNA age residuals in vmPFC (the latter association replicated in dlPFC, but not motor cortex). The association between RNA age residuals and endothelial and mural cell proportions did not appear to be due to underlying associations between RNA age and these cell types in vmPFC. Specifically, although the individual genes in the RNA age algorithm evidenced associations with cell type composition in vmPFC, the RNA age scores, which reflect a weighted average of the contribution of each gene, did not show associations with endothelial or mural cell proportions in vmPFC. PTSD/MDD-related accelerated aging in vmPFC may involve degradation of endothelial and mural cells and this may hold particular relevance to understanding the pathology of accelerated aging given associations between these cell types and blood-brain barrier (BBB) integrity (Daneman & Prat, 2015).
Prior work suggests that brain endothelial cells (EC), linked by complex tight junctions, are a key element of the BBB, and mural cells are closely attached to the endothelial layer of blood vessels (Daneman & Prat, 2015). An intact BBB serves to protect the brain from toxins circulating in the periphery via selective permeability (Zlokovic, 2008). Specifically, the restrictive permeability achieved by functioning ECs and mural cells under normal conditions limits the migration of plasma components, red blood cells, leukocytes, and immune cells from the blood vessels into the brain, preventing the generation of neurotoxic products and neuroinflammation (Zlokovic, 2008). Reductions in these cell types in association with advanced transcriptomic aging could suggest a role for BBB permeability in accelerated aging, and presumably, in risk for neuroinflammation and neurodegeneration. Consistent with this, aging is associated with increased senescent brain ECs in mice (Kiss et al., 2020). Similarly, mural cells are decreased in post-mortem human brain tissue samples from Alzheimer’s disease (AD) cases compared to neurologically healthy controls and are strongly correlated with BBB breakdown (Sengillo et al., 2013). Related to this, loss of pericytes, which are a type of vascular mural cell found in the smallest of blood vessels (e.g., capillaries; (Sweeney et al., 2016)) has been shown to be associated with neuronal degeneration and neuroinflammation in mice (Bell et al., 2010). Finally, vmPFC-specific downregulation of a BBB integrity gene (claudin-5) and associated blood vessel damage consistent with BBB permeability was recently reported in the postmortem female depressed brain with similar results evident in the prefrontal cortex of stressed mice (Dion-Albert et al., 2022). Together with the results of our study showing associations between PTSD/MDD and decreased endothelial and mural cells in vmPFC, this raises the possibility that psychiatric stress may be associated with altered homeostasis across the peripheral and central nervous systems (PNS and CNS, respectively) via decreased integrity of the BBB and associated accelerated aging in the transcriptome. The vmPFC may be particularly sensitive to the effects of stress-related cellular aging and associated BBB damage. This BBB breakdown could allow for immune deficiencies, infiltration of inflammatory molecules across the PNS and CNS, decreased cell proliferation, and increased apoptosis.
Limitations
Our study has several limitations. The sample size was small, and we did not have an appropriate replication sample, given the challenges associated with brain tissue acquisition. We also could not separate the effects of PTSD from those of MDD due to substantial comorbidity across the diagnoses. That said, the literature does not suggest that accelerated aging is specific to one psychiatric disorder; future research in larger samples can help to address this question. Sample size concerns also prevented us from including all potential confounds in the model at the same time. This was a cross-sectional study and we are unable to determine causality or the temporal direction of effects, even in the biomarker to biomarker associations (e.g., cell type composition and RNA age residuals). We cannot determine if advanced transcriptomic age is part of the mechanistic process of accelerated aging or simply a biomarker for it. We did not have peripheral samples on the same cohort to test if the results were consistent across the PNS and CNS. We did not have access to additional brain regions that might be important for evaluation of psychiatric conditions and accelerated aging in the transcriptome and it is unclear if the associations we observed are specific to vmPFC (beyond dlPFC and motor cortex). We also did not have single cell sequence data to examine expression more accurately in specific cell types (e.g., endothelial cells). Finally, it is possible that the biomarker to biomarker associations are specific to the PTSD/MDD cohort rather than reflecting a general brain aging process.
Conclusions
This is the first study to evaluate the contribution of psychiatric stress to transcriptomic aging in postmortem brain tissue, extending prior research which suggested associations between psychopathology and advanced cellular aging in DNAm and in gene expression derived from blood samples. We found that PTSD/MDD (and especially their comorbidity) was associated with advanced transcriptomic aging in vmPFC and, in this same region, advanced transcriptomic age and PTSD/MDD were associated with decreased endothelial and mural cells, two cell types that contribute to the integrity of the BBB. We also observed that the genes that contribute to vmPFC transcriptomic aging and that were also associated with PTSD/MDD were enriched for processes relating to immune function, inflammation, cell death and apoptosis, and cellular proliferation. This further suggests that the search for treatments for cellular aging might be best focused on anti-inflammatory therapeutics and those that normalize disturbances to cell repair and growth. Advanced DNAm age was unrelated to advanced transcriptomic age in this study, however, research suggests that both biomarkers are associated with psychiatric disorders and thus may be manifestations of a broader underlying biological aging process that contributes to neuroinflammation, neuropathology, and premature death.
Supplementary Material
Acknowledgements
The Traumatic Stress Brain Research Group are: Matthew J. Friedman, MD. PhD (Chair); Victor E. Alvarez, MD; David Benedek, MD; Christopher Brady, PhD; Alicia Che, PhD; Dianne A. Cruz, MS; David A. Davis, PhD; Matthew J. Girgenti, PhD; Ellen Hoffman, MD, PhD; Paul E. Holtzheimer, MD; Bertrand R. Huber, MD, PhD; Alfred Kaye, MD, PhD; John H. Krystal, MD; Adam T. Labadorf, PhD; Terence M. Keane, PhD; Neil Kowall, MD; Mark W. Logue, PhD; Ann McKee, MD; Brian Marx, PhD; Deborah Mash, MD; Mark W. Miller, PhD; Crystal Noller, PhD; Janitza Montalvo-Ortiz, PhD; William K. Scott, PhD; Thor Stein, MD, PhD; Robert Ursano, MD; Douglas E. Williamson, PhD; Erika J. Wolf, PhD; Keith A. Young, PhD
Funding
This work was supported by the National Institute on Aging grant number 1R21AG061367 to EJW and VA BLR&D Merit Award grant number 1I01BX003477 to MWL. Sage Hawn and Zoe Neale are supported by National Institute of Mental Health award 5T32MH019836. This work was also supported by the National Center for PTSD. Genotype and methylation data were generated with the support of resources at the Pharmacogenomics Analysis Laboratory (Research Service, Central Arkansas Veterans Healthcare System, Little Rock, Arkansas), a core research laboratory funded by the Cooperative Studies Program, Research and Development, Department of Veterans Affairs. The contents of this manuscript do not represent the views of the U.S. Department of Veterans Affairs, the National Institutes of Health, or the United States Government.
Footnotes
Conflicts of Interest
Dr. Wolf owns stock in Illumina, Inc. All other named authors report no financial or other conflicts of interest in relationship to the contents of this article.
We also examined associations between RNA age residuals and cell type scores in dlPFC and motor cortex as a follow-up (Table S9). RNA age residuals were negatively associated with endothelial, mural, microglia, and astrocyte cell scores and positively associated with oligodendrocytes in dlPFC (Table S9). No significant associations emerged in motor cortex.
References
- Bell RD, Winkler EA, Sagare AP, Singh I, LaRue B, Deane R, & Zlokovic B. v. (2010). Pericytes Control Key Neurovascular Functions and Neuronal Phenotype in the Adult Brain and during Brain Aging. Neuron, 68(3). 10.1016/j.neuron.2010.09.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsky DW, Caspi A, Cohen HJ, Kraus WE, Ramrakha S, Poulton R, & Moffitt TE (2017). Impact of early personal-history characteristics on the Pace of Aging: implications for clinical trials of therapies to slow aging and extend healthspan. Aging Cell, 16(4). 10.1111/acel.12591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, & Hochberg Y (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society: Series B (Methodological), 57(1). 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- Bolger AM, Lohse M, & Usadel B (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics, 30(15). 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bøstrand SMK, Vaher K, de Nooij L, Harris MA, Cole JH, Cox SR, Marioni RE, McCartney DL, Walker RM, McIntosh AM, Evans KL, Whalley HC, Wootton RE, & Clarke TK (2022). Associations between alcohol use and accelerated biological ageing. Addiction Biology, 27(1). 10.1111/adb.13100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, & Pachter L (2016). Near-optimal probabilistic RNA-seq quantification. Nature Biotechnology, 34(5). 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- Canchi S, Swinton MK, Rissman RA, & Fields JA (2020). Transcriptomic analysis of brain tissues identifies a role for CCAAT enhancer binding protein β in HIV-associated neurocognitive disorder. Journal of Neuroinflammation, 17(1). 10.1186/s12974-020-01781-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter SE, Ong ML, Simons RL, Gibbons FX, Lei MK, & Beach SRH (2019). The Effect of Early Discrimination on Accelerated Aging Among African Americans. Health Psychology. 10.1037/hea0000788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Joe S, & Nam H (2019). Development of tissue-specific age predictors using DNA methylation data. Genes, 10(11). 10.3390/genes10110888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JJ, McColl A, Shaw R, Lynall ME, Cowen PJ, de Boer P, Drevets WC, Harrison N, Pariante C, Pointon L, Goodyear C, Bullmore E, & Cavanagh J (2021). No evidence for differential gene expression in major depressive disorder PBMCs, but robust evidence of elevated biological ageing. Translational Psychiatry, 11(1). 10.1038/s41398-021-01506-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costi S, Morris LS, Kirkwood KA, Hoch M, Corniquel M, Vo-Le B, Iqbal T, Chadha N, Pizzagalli DA, Whitton A, Bevilacqua L, Jha MK, Ursu S, Swann AC, Collins KA, Salas R, Bagiella E, Parides MK, Stern ER, … Murrough JW (2021). Impact of the KCNQ2/3 channel opener ezogabine on reward circuit activity and clinical symptoms in depression: Results from a randomized controlled trial. American Journal of Psychiatry, 178(5). 10.1176/appi.ajp.2020.20050653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneman R, & Prat A (2015). The blood–brain barrier. Cold Spring Harbor Perspectives in Biology, 7(1). 10.1101/cshperspect.a020412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrow SM, Verhoeven JE, Révész D, Lindqvist D, Penninx BWJH, Delucchi KL, Wolkowitz OM, & Mathews CA (2016). The Association between Psychiatric Disorders and Telomere Length: A Meta-Analysis Involving 14,827 Persons. In Psychosomatic Medicine (Vol. 78, Issue 7). 10.1097/PSY.0000000000000356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillen KNH, Jacobs HIL, Kukolja J, Richter N, von Reutern B, Onur ÖA, Langen KJ, & Fink GR (2017). Functional Disintegration of the Default Mode Network in Prodromal Alzheimer’s Disease. Journal of Alzheimer’s Disease, 59(1). 10.3233/JAD-161120 [DOI] [PubMed] [Google Scholar]
- Dion-Albert L, Cadoret A, Doney E, Kaufmann FN, Dudek KA, Daigle B, Parise LF, Cathomas F, Samba N, Hudson N, Lebel M, Consortium S, Campbell M, Turecki G, Mechawar N, & Menard C (2022). Vascular and blood-brain barrier-related changes underlie stress responses and resilience in female mice and depression in human tissue. Nature Communications, 13(1), 164. 10.1038/s41467-021-27604-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, & Gingeras TR (2013). STAR: Ultrafast universal RNA-seq aligner. Bioinformatics, 29(1). 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Gutierrez PR, Kwenda EP, Donelan W, O’Malley P, Crispen PL, & Kusmartsev S (2021). Hyal2 Expression in Tumor-Associated Myeloid Cells Mediates Cancer-Related Inflammation in Bladder Cancer. Cancer Research, 81(3). 10.1158/0008-5472.CAN-20-1144 [DOI] [PubMed] [Google Scholar]
- Felger JC, Li Z, Haroon E, Woolwine BJ, Jung MY, Hu X, & Miller AH (2016). Inflammation is associated with decreased functional connectivity within corticostriatal reward circuitry in depression. Molecular Psychiatry, 21(10). 10.1038/mp.2015.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman MJ, Huber BR, Brady CB, Ursano RJ, Benedek DM, Kowall NW, & McKee AC (2017). VA’s National PTSD Brain Bank: a National Resource for Research. In Current Psychiatry Reports (Vol. 19, Issue 10). 10.1007/s11920-017-0822-6 [DOI] [PubMed] [Google Scholar]
- Hagenauer MH, Schulmann A, Li JZ, Vawter MP, Walsh DM, Thompson RC, Turner CA, Bunney WE, Myers RM, Barchas JD, Schatzberg AF, Watson SJ, & Akil H (2018). Inference of cell type content from human brain transcriptomic datasets illuminates the effects of age, manner of death, dissection, and psychiatric diagnosis. PLoS ONE, 13(7). 10.1371/journal.pone.0200003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han LKM, Aghajani M, Clark SL, Chan RF, Hattab MW, Shabalin AA, Zhao M, Kumar G, Xie LY, Jansen R, Milaneschi Y, Dean B, Aberg KA, van den Oord EJCG, & Penninx BWJH (2018). Epigenetic aging in major depressive disorder. American Journal of Psychiatry, 175(8). 10.1176/appi.ajp.2018.17060595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S (2013). DNA methylation age of human tissues and cell types. Genome Biology, 14(10). 10.1186/gb-2013-14-10-r115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T, Nyúl-Tóth Á, Balasubramanian P, Tarantini S, Ahire C, DelFavero J, Yabluchanskiy A, Csipo T, Farkas E, Wiley G, Garman L, Csiszar A, & Ungvari Z (2020). Single-cell RNA sequencing identifies senescent cerebromicrovascular endothelial cells in the aged mouse brain. GeroScience, 42(2). 10.1007/s11357-020-00177-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuan PF, Ren X, Clouston S, Yang X, Jonas K, Kotov R, Bromet E, & Luft BJ (2021). PTSD is associated with accelerated transcriptional aging in World Trade Center responders. Translational Psychiatry, 11(1). 10.1038/s41398-021-01437-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT, Johnson WE, Parker HS, Jaffe AE, & Storey JD (2012). The SVA package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics, 28(6). 10.1093/bioinformatics/bts034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine ME, Lu AT, Bennett DA, & Horvath S (2015). Epigenetic age of the pre-frontal cortex is associated with neuritic plaques, amyloid load, and Alzheimer’s disease related cognitive functioning. Aging, 7(12). 10.18632/aging.100864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logue MW, Zhou Z, Morrison FG, Wolf EJ, Daskalakis NP, Chatzinakos C, Georgiadis F, Labadorf AT, Girgenti MJ, Young KA, Williamson DE, Zhao X, Grenier JG, Huber BR, & Miller MW (2021). Gene expression in the dorsolateral and ventromedial prefrontal cortices implicates immune-related gene networks in PTSD. Neurobiology of Stress, 15. 10.1016/j.ynstr.2021.100398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, & Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology, 15(12). 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marečková K, Pačínková A, Klasnja A, Shin J, Andrýsková L, Stano-Kozubík K, Pausová Z, Brázdil M, & Paus T (2020). Epigenetic clock as a correlate of anxiety. NeuroImage: Clinical, 28. 10.1016/j.nicl.2020.102458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta ND, Haroon E, Xu X, Woolwine BJ, Li Z, & Felger JC (2018). Inflammation negatively correlates with amygdala-ventromedial prefrontal functional connectivity in association with anxiety in patients with depression: Preliminary results. Brain, Behavior, and Immunity, 73. 10.1016/j.bbi.2018.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mighdoll MI, Deep-Soboslay A, Bharadwaj RA, Cotoia JA, Benedek DM, Hyde TM, & Kleinman JE (2018). Implementation and clinical characteristics of a posttraumatic stress disorder brain collection. In Journal of Neuroscience Research (Vol. 96, Issue 1). 10.1002/jnr.24093 [DOI] [PubMed] [Google Scholar]
- Morrison FG, Logue MW, Guetta R, Maniates H, Stone A, Schichman SA, McGlinchey RE, Milberg WP, Miller MW, & Wolf EJ (2019). Investigation of bidirectional longitudinal associations between advanced epigenetic age and peripheral biomarkers of inflammation and metabolic syndrome. Aging, 11(11). 10.18632/aging.101992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison FG, Miller MW, Wolf EJ, Logue MW, Maniates H, Kwasnik D, Cherry JD, Svirsky S, Restaino A, Hildebrandt A, Aytan N, Stein TD, Alvarez VE, McKee AC, & Huber BR (2019). Reduced interleukin 1A gene expression in the dorsolateral prefrontal cortex of individuals with PTSD and depression. Neuroscience Letters, 692. 10.1016/j.neulet.2018.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers-Schulz B, & Koenigs M (2012). Functional anatomy of ventromedial prefrontal cortex: Implications for mood and anxiety disorders. In Molecular Psychiatry (Vol. 17, Issue 2). 10.1038/mp.2011.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palygin O, Levchenko V, Ilatovskaya D. v., Pavlov TS, Pochynyuk OM, Jacob HJ, Geurts AM, Hodges MR, & Staruschenko A (2017). Essential role of Kir5.1 channels in renal salt handling and blood pressure control. JCI Insight, 2(18). 10.1172/JCI.INSIGHT.92331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters MJ, Joehanes R, Pilling LC, Schurmann C, Conneely KN, Powell J, Reinmaa E, Sutphin GL, Zhernakova A, Schramm K, Wilson YA, Kobes S, Tukiainen T, Ramos YF, Göring HHH, Fornage M, Liu Y, Gharib SA, Stranger BE, … Singleton AB (2015). The transcriptional landscape of age in human peripheral blood. Nature Communications, 6. 10.1038/ncomms9570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protsenko E, Yang R, Nier B, Reus V, Hammamieh R, Rampersaud R, Wu GWY, Hough CM, Epel E, Prather AA, Jett M, Gautam A, Mellon SH, & Wolkowitz OM (2021). “GrimAge,” an epigenetic predictor of mortality, is accelerated in major depressive disorder. Translational Psychiatry, 11(1). 10.1038/s41398-021-01302-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quach A, Levine ME, Tanaka T, Lu AT, Chen BH, Ferrucci L, Ritz B, Bandinelli S, Neuhouser ML, Beasley JM, Snetselaar L, Wallace RB, Tsao PS, Absher D, Assimes TL, Stewart JD, Li Y, Hou L, Baccarelli AA, … Horvath S (2017). Epigenetic clock analysis of diet, exercise, education, and lifestyle factors. Aging, 9(2). 10.18632/aging.101168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Guo J, Zhu S, Wang Q, Gao R, Zhao C, Feng C, Qin C, He Z, Qin C, Wang Z, & Zang L (2021). The role of potassium channels in chronic stress-induced brain injury. Biological and Pharmaceutical Bulletin, 44(2). 10.1248/bpb.b20-00504 [DOI] [PubMed] [Google Scholar]
- Ren X, & Kuan PF (2020). RNAAgeCalc: A multi-tissue transcriptional age calculator. PLoS ONE, 15(8 August). 10.1371/journal.pone.0237006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider VA, Graves-Lindsay T, Howe K, Bouk N, Chen HC, Kitts PA, Murphy TD, Pruitt KD, Thibaud-Nissen F, Albracht D, Fulton RS, Kremitzki M, Magrini V, Markovic C, McGrath S, Steinberg KM, Auger K, Chow W, Collins J, … Church DM (2017). Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Research, 27(5). 10.1101/gr.213611.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengillo JD, Winkler EA, Walker CT, Sullivan JS, Johnson M, & Zlokovic B. v. (2013). Deficiency in mural vascular cells coincides with blood-brain barrier disruption in alzheimer’s disease. Brain Pathology, 23(3). 10.1111/bpa.12004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shireby GL, Davies JP, Francis PT, Burrage J, Walker EM, Neilson GWA, Dahir A, Thomas AJ, Love S, Smith RG, Lunnon K, Kumari M, Schalkwyk LC, Morgan K, Brookes K, Hannon E, & Mill J (2020). Recalibrating the epigenetic clock: Implications for assessing biological age in the human cortex. Brain, 143(12). 10.1093/brain/awaa334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneson C, Love MI, & Robinson MD (2016). Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Research, 4. 10.12688/F1000RESEARCH.7563.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney MD, Ayyadurai S, & Zlokovic B. v. (2016). Pericytes of the neurovascular unit: Key functions and signaling pathways. In Nature Neuroscience (Vol. 19, Issue 6). 10.1038/nn.4288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinci R, Pedicino D, D’Aiello A, Ciampi P, Ponzo M, Bonanni A, Russo G, Montone RA, Massetti M, Crea F, & Liuzzo G (2021). Platelet hyaluronidase 2 enrichment in acute coronary syndromes: a conceivable role in monocyte-platelet aggregate formation. Journal of Enzyme Inhibition and Medicinal Chemistry, 36(1). 10.1080/14756366.2021.1900159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb BD, Hotchkiss H, Prasun P, Gelb BD, & Satlin L (2021). Biallelic loss-of-function variants in KCNJ16 presenting with hypokalemic metabolic acidosis. European Journal of Human Genetics, 29(10). 10.1038/s41431-021-00883-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf EJ, Chen C. di, Zhao X, Zhou Z, Morrison FG, Daskalakis NP, Stone A, Schichman S, Grenier JG, Fein-Schaffer D, Huber BR, Abraham CR, Miller MW, & Logue MW (2020). Klotho, PTSD, and advanced epigenetic age in cortical tissue. Neuropsychopharmacology. 10.1038/s41386-020-00884-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf EJ, Logue MW, Hayes JP, Sadeh N, Schichman SA, Stone A, Salat DH, Milberg W, McGlinchey R, & Miller MW (2016). Accelerated DNA methylation age: Associations with PTSD and neural integrity. Psychoneuroendocrinology, 63. 10.1016/j.psyneuen.2015.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf EJ, Logue MW, Morrison FG, Wilcox ES, Stone A, Schichman SA, McGlinchey RE, Milberg WP, & Miller MW (2019). Posttraumatic psychopathology and the pace of the epigenetic clock: A longitudinal investigation. Psychological Medicine, 49(5). 10.1017/S0033291718001411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf EJ, Maniates H, Nugent N, Maihofer AX, Armstrong D, Ratanatharathorn A, Ashley-Koch AE, Garrett M, Kimbrel NA, Lori A, VA Mid-Atlantic MIRECC Workgroup, Aiello AE, Baker DG, Beckham JC, Boks MP, Galea S, Geuze E, Hauser MA, Kessler RC, … Logue MW (2018). Traumatic stress and accelerated DNA methylation age: A meta-analysis. Psychoneuroendocrinology, 92. 10.1016/j.psyneuen.2017.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf EJ, Morrison FG, Sullivan DR, Logue MW, Guetta RE, Stone A, Schichman SA, McGlinchey RE, Milberg WP, & Miller MW (2019). The goddess who spins the thread of life: Klotho, psychiatric stress, and accelerated aging. Brain, Behavior, and Immunity, 80. 10.1016/j.bbi.2019.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf EJ, Zhao X, Hawn SE, Morrison FG, Zhou Z, Fein-Schaffer D, Huber B, Miller MW, & Logue MW (2021). Gene expression correlates of advanced epigenetic age and psychopathology in postmortem cortical tissue. Neurobiology of Stress, 15. 10.1016/j.ynstr.2021.100371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Xu X, Chen G, Mehta ND, Haroon E, Miller AH, Luo Y, Li Z, & Felger JC (2019). Inflammation and decreased functional connectivity in a widely-distributed network in depression: Centralized effects in the ventral medial prefrontal cortex. Brain, Behavior, and Immunity, 80. 10.1016/j.bbi.2019.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, & Oshlack A (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biology, 11(2). 10.1186/gb-2010-11-2-r14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlokovic B. v. (2008). The Blood-Brain Barrier in Health and Chronic Neurodegenerative Disorders. In Neuron (Vol. 57, Issue 2). 10.1016/j.neuron.2008.01.003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


