Abstract
Objective
Insulin is a principal metabolic hormone. It regulates a plethora of metabolic pathways in peripheral tissues. The highly homologous insulin-like growth factor 1 (IGF-1), on the other hand, is important for development and growth. Recent studies have shown that insulin and IGF-1 signaling plays fundamental roles in the brain. Loss of insulin or IGF-1 receptors in astrocytes leads to altered glucose handling, mitochondrial metabolism, neurovascular coupling, and behavioral abnormalities in mice. Here, we aim to investigate molecular mechanisms by which insulin and IGF-1 signaling regulates astrocyte functions.
Methods
IR-flox and IRKO primary astrocytes were treated with 100 nM insulin or IGF-1 for 6 h, and their transcriptomes were analyzed. Astrocytes with either IR deletion, IGF1R deletion or both were used to examine receptor-dependent transcriptional regulations using qPCR. Additional immunoblotting and confocal imaging studies were performed to functionally validate pathways involved in protein homeostasis.
Results
Using next-generation RNA sequencing, we show that insulin significantly regulates the expression of over 1,200 genes involved in multiple functional processes in primary astrocytes. Insulin-like growth factor 1 (IGF-1) triggers a similar robust transcriptional regulation in astrocytes. Thus, over 50% of the differentially expressed genes are regulated by both ligands. As expected, these commonly regulated genes are highly enriched in pathways involved in lipid and cholesterol biosynthesis. Additionally, insulin and IGF-1 induce the expression of genes involved in ribosomal biogenesis, while suppressing the expression of genes involved in autophagy, indicating a common role of insulin and IGF-1 on protein homeostasis in astrocytes. Insulin-dependent suppression of autophagy genes, including p62, Ulk1/2, and several Atg genes, is blunted only when both IR and IGF1R are deleted.
Conclusions
In summary, insulin and IGF-1 potently suppress autophagy in astrocytes through transcriptional regulation. Both IR and IGF1R can elicit ligand-dependent transcriptional suppression of autophagy. These results demonstrate an important role of astrocytic insulin/IGF-1 signaling on proteostasis. Impairment of this regulation in insulin resistance and diabetes may contribute to neurological complications related to diabetes.
Keywords: Insulin, IGF-1, Astrocytes, Transcription, Autophagy, Proteostasis
Graphical abstract
Highlights
-
•
Insulin and IGF-1 elicit common and differential regulation on gene transcription in astrocytes.
-
•
Both insulin and IGF-1 transcriptionally regulate lipid and protein homeostasis, and many other pathways in astrocytes.
-
•
Insulin and IGF-1 potently suppress autophagy through transcriptional regulation in astrocytes.
-
•
Insulin receptors and IGF-1 receptors in astrocytes can potently suppress autophagy pathway.
1. Introduction
Insulin is essential for maintaining glucose and energy homeostasis throughout the body, whereas IGF-1 is mainly responsible for development and growth [1,2]. Insulin and IGF-1 primarily bind to their cognate receptors under physiological conditions, however, both ligands can also act on each other's receptors with a lower binding affinity. Upon ligand stimulations, insulin receptors (IRs) and IGF-1 receptors (IGF1Rs) elicit an array of intracellular signaling cascades, including the common IRS-1-PI3K-Akt and Shc-Ras-MAPK cascades [3]. The activation of these signaling cascades rapidly regulate protein activities relevant to various cellular functions. They can also trigger transcriptional regulation of many genes to elicit long term effects in cells [4,5]. Functionally, impaired insulin signaling or decreased insulin sensitivity leads to dysregulation of cellular processes such as cellular metabolism, homeostasis of subcellular organelles and cell–cell communications, ultimately contributing to organ malfunction and complications related to diabetes [1]. Impaired IGF-1 signaling, on the other hand, has been associated with developmental defects and growth retardation [6].
A growing body of evidence demonstrates the critical roles of insulin and IGF-1 in the brain. Known as a circulating hormone, insulin can cross the blood brain barrier, and may also be produced locally by choroid plexus [[7], [8], [9], [10]]. In the brain, insulin acts on both neurons and glial cells alike as both contain insulin receptors. Insulin signaling regulates phosphorylation of AMPA and NMDA receptors in excitatory neurons to maintain proper synaptic plasticity, an essential feature of learning and cognition at the cellular level [11,12]. Insulin is also critical in regulating the Akt-GSK3β signaling cascade. Loss of insulin receptors in the brain leads to dephosphorylation and hyperactivation of GSK3β, and an increased level of hyperphosphorylated Tau [13], which is prone to form neurofibrillary tangles in neurons [14]. Studies have also demonstrated the specific roles of insulin action in different types of neurons and neural circuits. Thus, loss of IR in different hypothalamic neurons affects appetite, and metabolism of liver and adipose tissues [[15], [16], [17]], whereas loss of IR in the hippocampus has been shown to impair cognitive function in rodents [18,19].
IGF-1 can be produced in the brain or from circulation as well. It is a well-known neuroprotective agent. The IGF-1-IGF1R system is involved in neuronal differentiation, brain development, and protection against brain injuries [20]. Loss of brain IGF1R leads to severe microcephaly and growth retardation [21]. Conversely, elevated IGF-1 signaling in the brain increases brain weight during development [22], and protects neurons against traumatic brain injuries in adult mice by facilitating neurogenesis and neuronal survival [23,24].
As the most abundant glial cells within the brain, astrocytes possess a plethora of crucial functions including neurotransmitter synthesis and release, brain blood flow regulation, mitochondrial homeostasis, and metabolism [[25], [26], [27]]. Many of these functions are dynamically regulated by systemic and local signals [[28], [29], [30]]. Dysregulation of astrocyte functions may play crucial roles in the development of many neurological conditions [31,32]. Our previous studies have shown that insulin regulates exocytosis of ATP in astrocytes to modulate dopaminergic neurons and anxiety and depressive-like behavior in mice [33]. In the hypothalamus, insulin action in astrocytes has been shown to regulate brain glucose sensing, neurovascular coupling, food intake, thermogenesis, and fertility [[34], [35], [36], [37], [38]]. Recent studies have also demonstrated crucial roles of astrocytic IGF-1 signaling in neurovascular function, amyloid-β uptake and cognition in mice [39,40]. Additionally, insulin and IGF-1 synergistically regulate GLUT-1 membrane translocation and glucose uptake in astrocytes [36]. Hence, astrocytic insulin and IGF-1 signaling appears to share common cellular functions, while preserving unique features. A detailed comparison of these two systems in astrocytes has yet to be undertaken.
Here, we used RNA sequencing to systemically compare transcriptional regulations by insulin and IGF-1 in primary astrocytes. We identified several ligand-specific regulations, though most of the gene regulations were common to both insulin and IGF-1. The commonly regulated genes were enriched in pathways involved in cholesterol biosynthesis, autophagy, ribosomal biogenesis, gap junctions, among many others. Upon ligand stimulation, the activation of IRs and IGF1Rs may elicit different transcriptional regulations. In the present study, we focused on the insulin-IR axis and used genetic tools to specifically delete endogenous IR in astrocytes. By comparing the transcriptomes between IR-flox and IRKO astrocytes, we identified insulin-induced regulations that were required by IR. Most of the regulations, however, can be mediated through endogenous IGF1R. We performed functional validations on autophagy, a pathway that is potently suppressed by both insulin and IGF-1. We found that the dramatic suppression of autophagy by insulin and IGF-1 can only be blunted in IR/IGF1R double knockout astrocytes, indicating crosstalk between IR and IGF1R signaling. Together, these results demonstrate that insulin and IGF-1 regulate a broad network of transcriptional programs in astrocytes, highlighted by the pathways involved in protein homeostasis.
2. Methods
2.1. Animals
All animal studies were conducted in compliance with the regulations and ethics guidelines of the NIH and were approved by the IACUC of the New York Institute of Technology College of Osteopathic Medicine. IR-flox and IGF1R-flox mice on a C57BL6/J background were kindly provided by Dr. C. Ronald Kahn's lab. Homozygous IR/IGF1R double floxed mice were generated by crossing IR-flox with IGF1R-flox mice.
2.2. Reagents and materials
Adenovirus encoding CMV promoter-driven GFP:Cre or GFP alone were purchased from Vector Biolabs. Adenovirus encoding mRFP-GFP tandem fluorescent-tagged LC3 (tfLC3) reporter was kindly provided by Dr. Qiangrong Liang (23). Recombinant human insulin solution was purchased from Sigma, whereas recombinant human IGF-1 was from PeproTech. Bafilomycin A1 was from Cell Signaling Technology. Rabbit anti-p62 (#23214), rabbit anti-LC3A/B (#12741), rabbit anti-Ulk1 (#8054), rabbit anti-IGF1Rβ (#9750), rabbit anti-GAPDH (#5174) were purchased from Cell Signaling Technology. Rabbit anti-IRβ (#sc-711) was from Santa Cruz. Mouse anti-GFAP (#MAB306) and mouse anti-NeuN (#MAB377B) were purchased from Millipore. Rabbit anti-GFAP (#ab7260) was purchased from Abcam. Rabbit anti-Iba-1 (#019-19741) was purchased from Fujifilm-Wako. HRP-conjugated goat-anti-rabbit IgG secondary antibodies were purchased from GE Healthcare.
2.3. Primary astrocyte cultures
Primary cortical astrocyte cultures were prepared from IR-flox, IGF1R-flox or IR/IGF1R double floxed newborn pups as previously described (19). Briefly, cortices were surgically dissected from newborn pups, and pooled for the following digestion in Hibernate-A media supplemented with 4 mg/ml papain (Sigma) and 33 U/ml DNase I (Sigma) on a shaker at 37 °C for 30 min. Dissociated cells were plated on a T75 flask and cultured in DMEM/F12 (Gibco) plus 10% FBS and 1× pen/strep (Gibco). Contaminating non-astrocytes were depleted by vigorously shaking the following day. Culture media was changed every 3 days. Upon confluency, astrocytes were trypsinized and replated onto 6-well plates (Corning) or 8-chamber slides (ibidi). To induce flox allele recombination, IR-flox, IGF1R-flox, and double floxed astrocytes were infected with adenovirus encoding Cre:GFP (1 × 109 GC/ml) overnight, and cultured for an additional 5 days before experiments. To generate the control cells, the same floxed astrocytes were infected with adenovirus encoding GFP alone. In studies assessing autophagy flux by mRFP-GFP tandem fluorescent-tagged LC3 (tfLC3) reporter, adenovirus encoding Cre or Luciferase were used to generate KO and control cells.
2.4. Insulin and IGF-1 treatments on cultured astrocytes
To assess transcriptional regulation, primary IRKO, IGF1RKO and double knockout (DKO) astrocytes, along with the appropriated single or double IR/IGF1R floxed control astrocytes were serum starved overnight with DMEM/F12 medium containing 0.1% BSA, followed by 100 nM insulin, 100 nM IGF-1 or vehicle treatment for 6 h. To examine the protein expression after insulin and IGF-1 treatment, astrocytes were serum starved for 4 h, followed by 100 nM insulin, 100 nM IGF-1 or vehicle treatment overnight to allow sufficient time for cells to reach equilibrium on protein levels. To prevent rapid degradation of proteins through autophagy, cells were co-treated with 100 nM Bafilomycin A1.
2.5. RNA isolation and sequencing
Total RNA was extracted from primary cultured IR-flox and IRKO astrocytes treated with 100 nM insulin, IGF-1 or vehicle for 6 h using QIAzol reagent (Qiagen). RNA quality and quantification were verified using a 2100 Agilent Bioanalyzer. Total RNA samples (2 μg, RNA integrity score > 7) were submitted for library preparation and next-generation sequencing by Novogen. Briefly, mRNAs were purified from total RNAs using poly-T oligo-conjugated magnetic beads. Non-stranded cDNA libraries were prepared and subjected to sequencing (paired-end 150 bp).
2.6. Bioinformatic analysis
The RNA-seq data analyses were performed as previously described [41,42]. Reads in FASTQ files were aligned to mouse reference genome (mm10) using STAR read aligner (STAR 2.7.7a) [43]. The numbers of reads aligned to the exon regions of each gene were quantified by a Perl script. The normalization and comparison of RNA-seq were performed using edgeR [44]. Genes with average count-per-million <0.5 were filtered out. The absolute value of log2 fold change (log2FC) greater than 1.5 and false discovery rate (FDR) less than 0.05 were set as the thresholds to determine significantly differentially expressed genes (DEGs). Data plotting was carried out using the R version 4.0.4. The DESeq2 [45] was used to perform the principal component analysis (PCA). The colorRampPalette in mixOmics was used to compute the distances between variables to plot the clustering image map (CIMs, heatmap). Pathway enrichment analysis was performed using g:Profiler (version_e106_eg53_p16 _65fcd97) and was carried out to detect the most enriched Gene Ontology (GO), including Biological Process, Cellular Component, Molecular Function, KEGG, and Reactome. The custom background used in g:profiler comprised genes expressed with the Benjamini–Hochberg FDR algorithm was used with a significance threshold of 0.05. The gene ratio was computed by the intersection size divided by the query size. The gene ratio and intersection size were used as input to generate a dot plot for enriched GO terms. The autophagy pathway analysis of the DEGs was carried out using the KEGG database (KEGG:04140; Autophagy: animal) to generate heat maps.
2.7. RT-PCR and quantitative real-time PCR
500 ng RNA was reverse transcribed using a High-Capacity cDNA Reverse Transcription kit (Applied Biosystems). Quantitative real time PCR was performed using the SYBR Green PCR master mix (CoWin Biosciences), and analyzed in a QuantStudio 3 (Applied Biosystems). All the primer sequences used for this study were listed in Supplemental Table 1.
2.8. Preparation of total cell lysates and immunoblotting
Cells were washed immediately with ice-cold 1× PBS once after treatment. Then, cells were lysed in 1× RIPA lysis buffer complemented with 1× protease inhibitor cocktail (Bimake). Protein concentrations were determined using a BCA kit (ThermoFisher). Lysates (10–20 μg) were resolved on SDS-PAGE gels, transferred to PVDF membranes. PVDF membranes were blocked in 1× PBS supplemented with 0.1% Tween-20 and 1% casein (PCT) at room temperature for 1 h, and incubated with the indicated primary antibodies in PCT overnight at 4 °C. Following the primary antibody incubation, membranes were washed three times with 1× PBS supplemented with 0.1% Tween-20 (1× PBST), incubated with HRP-conjugated secondary antibody (anti-rabbit IgG, NA934; 1:20,000, GE Healthcare) in 1× PBST for 1 h. The chemiluminescent signals were detected using Immobilon Western Chemiluminescent HRP Substrate (Millipore) in an Amersham Imager 680 (GE Healthcare). Densitometric analyses were performed by ImageJ on three independent experiments. Due to the signal variance among different replicates, densitometric signals of each condition were normalized to the intensity of the starvation + Baf A1 lane of the same replicate.
2.9. Autophagy flux by confocal microscopy
IR/IGF1R-flox and DKO astrocytes cultured in 8-chamber slides were infected with adenovirus encoding tfLC3 reporter (1 × 108 GC/ml) overnight and cultured for an additional 24 h. Then astrocytes were kept in normal culture conditions or serum starved for 5 h. Following serum depletion, one third of the wells were treated with 100 nM insulin overnight; one third of the cells were treated with 100 nM IGF-1 overnight; and the rest of the cells were kept in serum-free medium overnight. To induce the maximal autophagy flux, the IR/IGF1R-flox and DKO astrocytes under different treatment conditions were incubated in 1× HBSS in the absence of amino acids for 4 h before live cell imaging. The green and red fluorescence of these cells were captured by Zeiss LSM 980 with Airyscan 2 confocal microscope. GFP+ and RFP+ puncta in the astrocytes were identified and segmented by Ilastik [46]. The resulting segmentation files were analyzed and quantified in ImageJ using the “analyze particles” function. The total area of each cell analyzed was outlined and measured after contrast/brightness adjustment in ImageJ. The GFP+ and RFP+ puncta of each astrocyte were normalized to total cell area.
2.10. Statistical analysis
Data are presented as means ± SEM. For two population comparison, 2-tailed Student's t tests were performed. When comparing multiple treatments, one-way ANOVA followed by Tukey's post hoc test was performed. In the analyses comparing both genotype and treatment, two-way ANOVA followed by Tukey's post hoc test was performed. Significance level was set at P < 0.05.
2.11. Data availability
RNA-seq data of IR-flox and IRKO astrocytes with or without 100 nM insulin or IGF-1 treatment is deposited in Gene Expression Omnibus under accession number GSE213062 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE213062). All the computational scripts used in this study are available at GitHub repository (https://github.com/Jerry-Zhao/2022CaiLabInsIgf1).
3. Results
3.1. Insulin and IGF-1 elicit robust regulations on gene transcription in astrocytes
To explore the transcriptional regulation of insulin and IGF-1 on primary cultured astrocytes, we serum-starved astrocytes overnight and stimulated these cells with insulin, IGF-1 or vehicle for 6 h. To maximize the transcriptional response, we chose to stimulate astrocytes with 100 nM insulin or IGF-1, which would effectively activate both endogenous IR and IGF1R by either ligand. Total cellular RNAs from these astrocytes were isolated, enriched for poly-A tailed RNAs, and subjected to RNA Seq analysis. The normalized sequence counts demonstrated the dramatic enrichment of astrocyte-specific markers, including Gfap, Slc1a3, Aqp4, Apoe and Aldh1l1 (Supplemental Figure S1A) [47]. Meanwhile, the transcripts of the markers for other major cell types in the brain, such as microglia, oligodendrocytes, neurons, and endothelial cells, remained very low or undetectable (Supplemental Figure S1A) [[47], [48], [49]]. Immunostaining showed that over 97% of the cultured cells express Gfap. 1.34% of the cells were Iba-1+, representing a low level of microglia in the culture (Supplemental Figures S1B–D). Additionally, 0.92% of the cultured cells were negative for both Gfap and Iba-1 (Supplemental Figures S1B–D). Together, the vast majority of the cultured cells were astrocytes.
While astrocytes express abundant insulin receptors, IGF-1 receptors, and their major substrates Irs1 and Irs2, the expression levels of Igf1r and Irs2 were about 3-fold higher than Insr and Irs1 (Supplemental Figures S1E–H). Importantly, all were transcriptionally suppressed following insulin and IGF-1 stimulation (Supplemental Figures S1E–H), suggesting a ligand-induced desensitization.
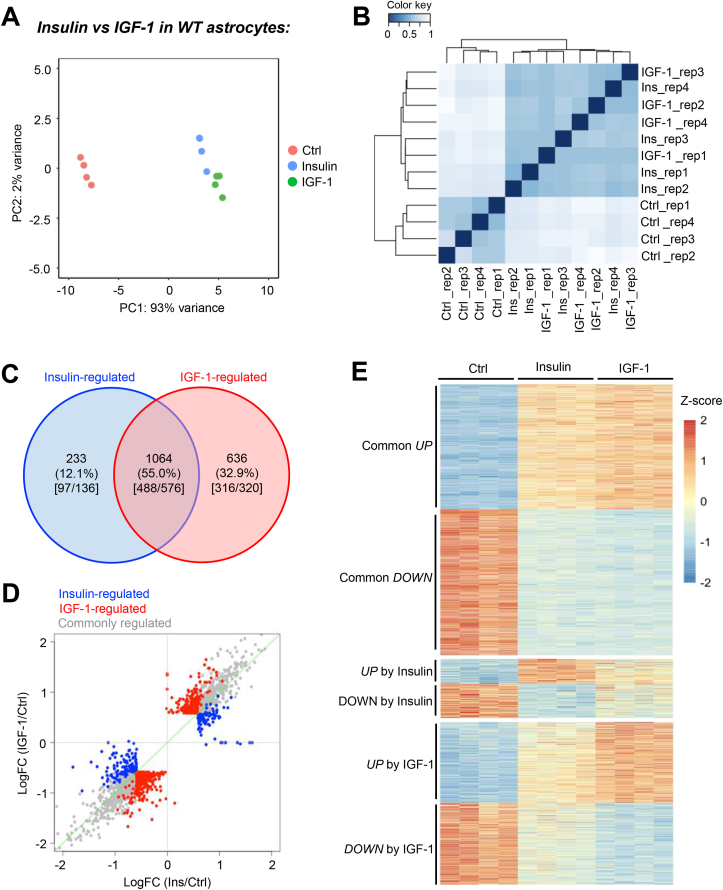
The unbiased principal component analysis showed that the transcriptomes of ligand-stimulated astrocytes clearly differentiated from that of the vehicle-treated group, indicating a robust transcriptional regulation in response to insulin and IGF-1 (Figure 1A). Insulin- and IGF-1-treated samples were slightly separated, indicating ligand-specific regulations (Figure 1A). Pearson correlation analysis confirmed a predominant effect by ligand stimulation (Figure 1B). Of the 1,933 differentially expressed genes among the groups (FDR < 0.05, |FC| > 1.5), 1064 (55%) were regulated by both insulin and IGF-1 (Figure 1C, D, and Supplemental Table 2). 488 of these genes were upregulated following stimulation, while 576 were downregulated (Figure 1C, D). Insulin specifically regulated the expression of 233 genes (12.1%), whereas IGF-1 uniquely regulated the expression of another 636 genes (32.9%) (Figure 1C, D). An approximately equal number of genes were either up- or down-regulated following ligand stimulation. Figure 1E demonstrates the relative expression of genes either commonly, or uniquely regulated by insulin and IGF-1. Together, the bioinformatic analyses clearly demonstrate the intertwined nature of insulin and IGF-1 signaling in astrocytes, while also highlighting the individual transcriptional effects each ligand alone possesses.
Figure 1.
Insulin and IGF-1 potently regulate transcription in astrocytes. A. Principal component analysis (PCA) of transcriptomes in primary cultured astrocytes at baseline or following 100 nM insulin or IGF-1 stimulation for 6 h. B. Pearson correlation showing transcriptional groupings for control, insulin-stimulated and IGF-1-stimulated astrocytes. C. VENN diagram showing unique and common genes regulated by insulin or IGF-1 (FDR < 0.05, |FC| > 1.5). Numbers in the square bracket indicate the up-regulated genes versus down-regulated genes. D. Scattered plot showing ligand stimulated transcriptional changes by insulin and IGF-1. Gray dots represent commonly regulated genes. Blue dots represent insulin-specific regulations, whereas red dots represent IGF-1-specific regulations. E. Heatmap of all significantly regulated genes (FDR < 0.05, |FC| > 1.5) and clustered by their responses to insulin and IGF-1.
3.2. Insulin and IGF-1 show common and ligand-specific regulations on gene transcription in astrocytes
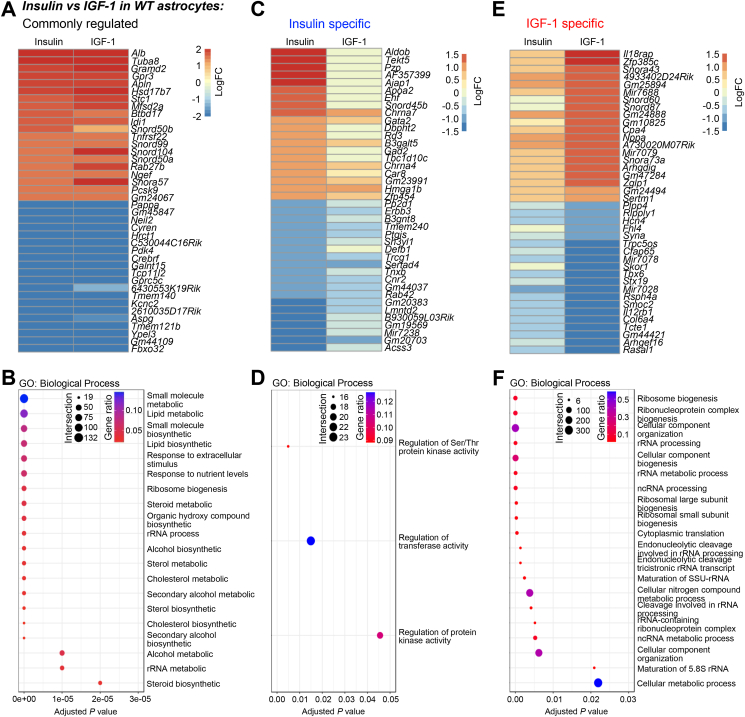
Among the top commonly regulated genes by insulin and IGF-1, many were known to be involved in various critical cellular processes (Figure 2A). Thus, both insulin and IGF-1 upregulated secreted proteins/peptides, albumin (Alb) and apelin (Apln); microtubule subunit, tubulin alpha subunit isoform 8 (Tuba8); cholesterol biosynthesis enzyme, hydroxysteroid 17-beta dehydrogenase 7 (Hsd18b7); and G protein-coupled receptor 3 (Gpr3). The top down-regulated genes by both ligands were exemplified by metalloproteinase pappalysin 1 (Pappa), which has been shown to cleave IGFBPs [50]; pyruvate dehydrogenase kinase 4 (Pdk4); G protein-coupled receptor Gprc5c; and voltage-gated potassium channel Kcnc2. Notably, the most suppressed gene by insulin and IGF-1 was F-box protein 32 (Fbxo32), also known as Atrogin1. Fbxo32 is an E3 ubiquitin ligase and has been shown to be one of the most insulin-suppressed proteins in skeletal muscle [51]. In astrocytes, the expression of Fbxo32 was decreased by ∼90% in response to insulin and IGF-1, suggesting a conserved insulin-Fbxo32 axis to regulate protein ubiquitination and degradation in different cell types in the body. In addition, many non-coding RNAs, including long non-coding RNAs (Gm44109 and 2610035D17Rik), and snoRNAs (Snord50b and Snord99), were among the top regulated genes, revealing previously unrecognized regulation of non-coding RNAs by insulin and IGF-1 in astrocytes.
Figure 2.
Pathways transcriptionally regulated by insulin and IGF-1. A. Heatmap showing the Log2 (fold change) of the top regulated genes by both 100 nM insulin and IGF-1 stimulation. B. The most enriched GO: biological processes commonly regulated by insulin and IGF-1. C. Heatmap showing the Log2 (fold change) of the top regulated genes only by 100 nM insulin stimulation. D. The most enriched GO: biological processes only regulated by insulin. E. Heatmap showing the Log2 (fold change) of the top regulated genes only by 100 nM IGF-1 stimulation. F. The most enriched GO: biological processes only regulated by IGF-1. The size of the dots in the GO plots indicates the number of regulated genes within the pathway, while the color of the dots represents the proportion of the regulated genes over total genes in the pathway.
To further examine the functional impact of these transcriptional regulations in astrocytes, we performed unbiased pathway enrichment analysis. We found that genes commonly regulated by insulin and IGF-1 were enriched in many biosynthetic and metabolic pathways. These included lipid and cholesterol biosynthesis, ribosome biogenesis, and alcohol metabolic pathways (Figure 2B, and Supplemental Table 3). Besides these typical metabolic pathways, insulin- and IGF-1-regulated genes were enriched in autophagy, gap junctions and their related intracellular trafficking, pathways of neurodegeneration, Alzheimer's disease, among others (Supplemental Figures S2A and B). Notably, insulin and IGF-1 significantly suppressed the majority of the genes involved in apoptosis (Supplemental Figures S2C and D, and Supplemental Table 4).
To identify potential transcriptional factors responsible for insulin/IGF-1-mediated gene regulation, we performed TF enrichment analysis. We found that the commonly regulated genes were enriched for motifs of many transcriptional factors like ZF5, E2F-1, FOXN4 (Supplemental Figure S3A, and Supplemental Table 5). As expected, motifs for FoxO1 and Elk1 were also significantly enriched in genes commonly regulated by insulin and IGF-1 (Supplemental Figure S3B).
The comprehensive RNA Seq analysis also revealed ligand-specific gene regulations. Among the most induced genes following insulin stimulation alone was aldolase b (Aldob) which is crucial for fructose modification for feeding into glycolysis (Figure 2C). Interestingly, increases in Aldob mRNA levels in pancreatic islet cells have been associated with impaired insulin release [52], perhaps showing negative feedback in this signaling cascade. Another strong increase was seen in glutamate decarboxylase 2 (Gad2) which is essential for GABA synthesis. One of the most downregulated genes was Erbb3 encoding for a receptor tyrosine kinase in the epidermal growth factor family (Figure 2C). On the pathway level, genes specific to insulin stimulation were enriched in pathways involved in regulations of protein kinases, transferases and serine/threonine protein kinases (Figure 2D, and Supplemental Table 3). This is in accordance with our knowledge of insulin's primary mechanism for signal transduction and activation of tyrosine kinases [53].
The expression of Il18rap, an accessory subunit of interleukin-18 receptor, was significantly increased only by IGF-1 treatment (Figure 2E). Genes exclusively down-regulated by IGF-1 were exemplified by Skor1 which has ties to restless leg syndrome and neurodevelopment (Figure 2E) [54]. All the genes exclusively regulated by IGF-1 were primarily enriched in pathways related to ribosomal RNA processing and ribosome biogenesis (Figure 2F, and Supplemental Table 3), which aligns with the crucial role of IGF-1 signaling in growth and development [6].
3.3. Insulin-induced transcriptional regulation in IR knockout astrocytes
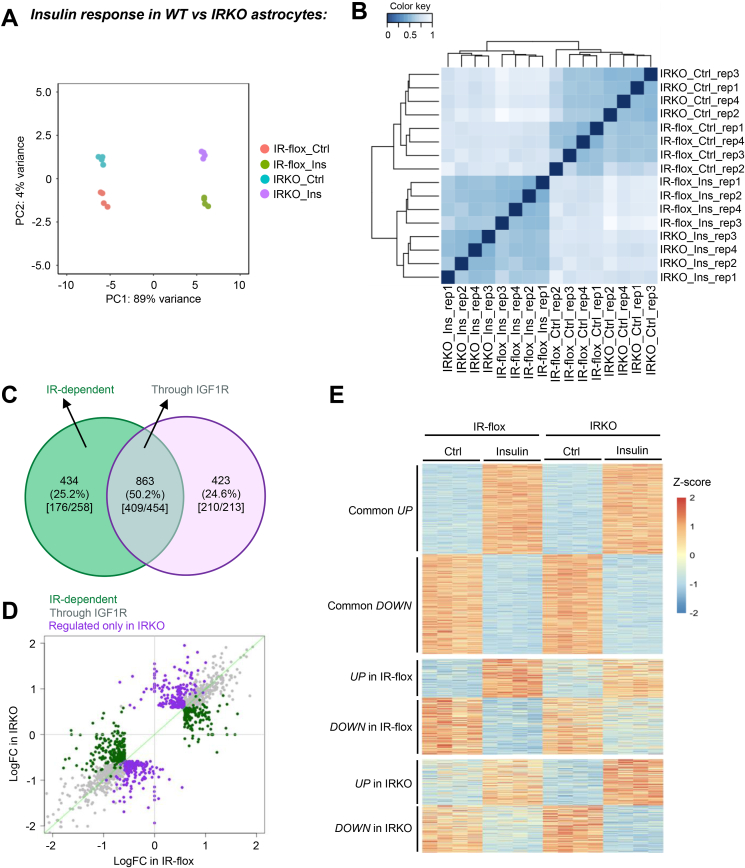
Insulin and IGF-1 preferentially bind to their cognate receptors, but also each other's receptors at higher concentrations, such as the 100 nM used in the present study. Given the co-expression of IR and IGF1R in astrocytes, we next sought to examine the receptor-dependent effects and the relative importance of ligand-receptor cross activation. We focused on the insulin-IR system. Primary astrocytes from IR-flox pups were cultured and infected with adenovirus encoding Cre fused with GFP to trigger flox allele recombination and IR deletion [33]. In parallel, IR-flox astrocytes infected with adenovirus encoding GFP alone were used as controls. Loss of IR in astrocytes led to global gene transcription alterations at basal condition, as the transcriptomes of IR-flox and IRKO cells were separated on the principal component 2 (PC2) in the principal component analysis (Figure 3A). Following 6 h of 100 nM insulin treatment, both IR-flox and IRKO astrocytes showed robust transcriptomic changes, indicated by a rightward shift on PC1 axis (Figure 3A). Pearson correlation analysis showed a predominant effect by insulin stimulation in both IR-flox and IRKO astrocytes (Figure 3B).
Figure 3.
IR-dependent gene regulations. A. Principal component analysis (PCA) of transcriptomes in IR-flox and insulin receptor knockout (IRKO) astrocytes with or without 100 nM insulin stimulation for 6 h. B. Pearson correlation showing the clusters of the four distinct conditions. C. VENN diagram showing significantly up- or down-regulated genes by insulin in the presence and absence of IR (FDR < 0.05, |FC| > 1.5). Numbers in the square bracket indicate the up-regulated genes versus down-regulated genes. D. Scattered plot showing insulin-stimulated transcriptional changes in IR-flox and IRKO astrocytes. Gray dots represent genes regulated by insulin in both IR-flox and IRKO astrocytes. Green dots represent genes regulated by insulin specifically in IR-flox astrocytes, while purple dots represent genes regulated only in IRKO astrocytes. E. Heatmap of all significantly regulated genes (FDR < 0.05, |FC| > 1.5) and clustered by the responses in different cell types.
Of the 1,720 genes significantly regulated by insulin in either IR-flox and IRKO astrocytes (FDR < 0.05, |FC| > 1.5), 434 genes were only significantly regulated by insulin when the IR was present (25.2%), demonstrating the necessity of IR for the regulation of this set of genes in astrocytes (Figure 3C, D, and Supplemental Table 6). On the other hand, 863 genes (50.2%) were still strongly regulated by insulin even in the absence of IR (Figure 3C, D). These data suggest that 100 nM insulin can elicit robust transcriptional regulations in astrocytes through endogenous IGF-1 receptors. Interestingly, the remaining 423 genes were only significantly regulated by insulin in the absence of IR (Figure 3C, D). The heat map in Figure 3E demonstrates the gene regulations by insulin in IR-flox and IRKO astrocytes.
Functionally, genes significantly regulated by insulin in both IR-flox and IRKO astrocytes were enriched in pathways involved in sterol and cholesterol biosynthesis, lipid biosynthesis, autophagy, intracellular trafficking of gap junctions (Supplemental Figure S4, and Supplemental Table 7). The preserved regulation of these pathways by insulin in the absence of endogenous IR strongly suggests the involvement of IGF1R. Interestingly, these same pathways were also highly regulated by IGF-1 in wild-type astrocytes. Together, these data indicate the important and common roles of insulin-IR and IGF-1-IGF1R signaling for lipid and cholesterol metabolism, autophagy and gap junctions in astrocytes.
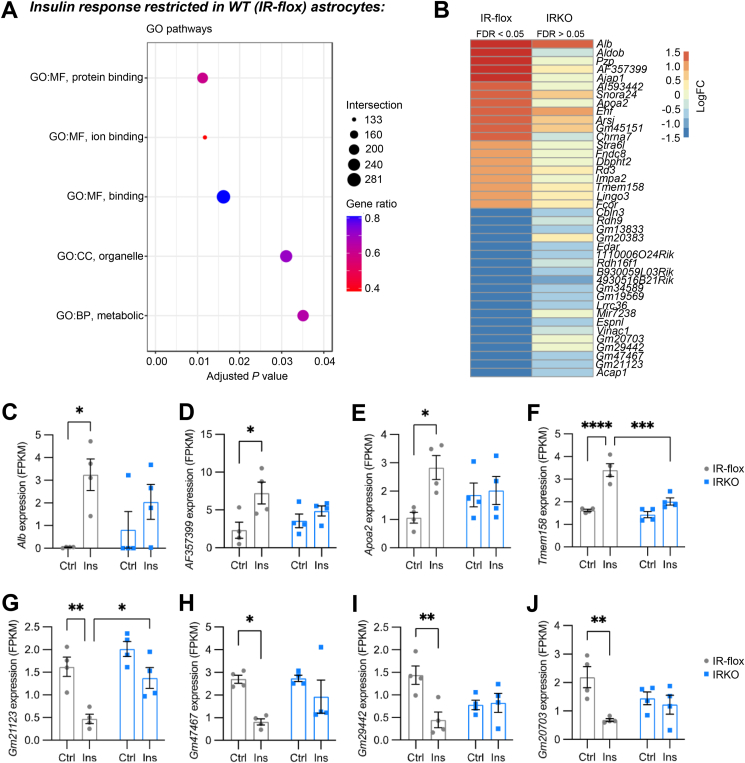
The genes regulated by insulin in IR-flox but not in IRKO astrocytes were enriched in a few very general pathways such as protein and ion binding, as well as metabolic pathways (Figure 4A). On the individual gene level, the top 40 up- or down-regulated genes by insulin in IR-flox astrocytes, but not in IRKO astrocytes, included 25 protein-coding genes and 15 non-coding RNAs. Albumin (Alb), which is usually considered a liver-specific gene, was not expressed in IR-flox astrocytes under basal conditions. However, upon insulin stimulation, Alb was the most up-regulated transcripts and expressed at a meaningful level in astrocytes (Figure 4C). Importantly, the strong induction of Alb expression by insulin was not preserved in IRKO astrocytes. Similarly, IR was required for the insulin-induced expression of AF357399, a protein coding gene with unknown function (Figure 4D); Apoa2, an apolipoprotein responsible for cholesterol balance (Figure 4E); and transmembrane protein Tmem158 (Figure 4F). Interestingly, of the 15 non-coding RNAs that were only regulated in IR-flox astrocytes, 11 were specifically down-regulated, and were classified as long non-coding RNAs (lncRNAs), indicating specific coupling of these lncRNAs to IR signaling in astrocytes (Figure 4G–J).
Figure 4.
Gene regulation required by IR. A. GO pathways significantly regulated by insulin only in IR-flox astrocytes. The size of the dots indicates the number of regulated genes within the pathway, while the color of the dots represents the proportion of the regulated genes over total genes in the pathway. B. Heatmap showing the Log2 (fold change) of the top genes only regulated by insulin in IR-flox astrocytes. C–F. mRNA abundance of representative genes induced by insulin only in IR-flox astrocytes, including Alb (C), AF357399 (D), Apoa2 (E) and Tmem158 (F). G-J. mRNA abundance of representative genes suppressed by insulin only in IR-flox astrocytes, including Gm21123 (G), Gm47467 (H), Gm29442 (I) and Gm20703 (J). Data are shown as mean ± SEM. Two-way ANOVA followed by Tukey's multiple comparison test. ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001; ∗∗∗∗, P < 0.0001. N = 4.
Of note, genes that were uniquely regulated by insulin in IRKO astrocytes were enriched in the Ras and Rap1 signaling pathways (Supplemental Figure S5A), indicating specific regulation on small GTPase signaling cascades by insulin likely through IGF1R. The top 40 genes only up- or down-regulated in IRKO astrocytes included both protein coding genes and a significant amount of non-coding RNAs (Supplemental Figure S5B). Thus, among the top 40 insulin-regulated transcripts in IRKO astrocytes included 15 non-coding RNAs, 13 of which were significantly up-regulated. Strikingly, these up-regulated non-coding RNAs included 9 snoRNAs, indicating a unique regulation of ribosomal RNA processing and ribosomal biogenesis by insulin through IGF1R. The normalized transcript counts of representative genes exclusively regulated by insulin in IRKO astrocytes were illustrated in Supplemental Figures S5C–J.
3.4. Insulin and IGF-1 transcriptionally regulate pathways involved in proteostasis in astrocytes
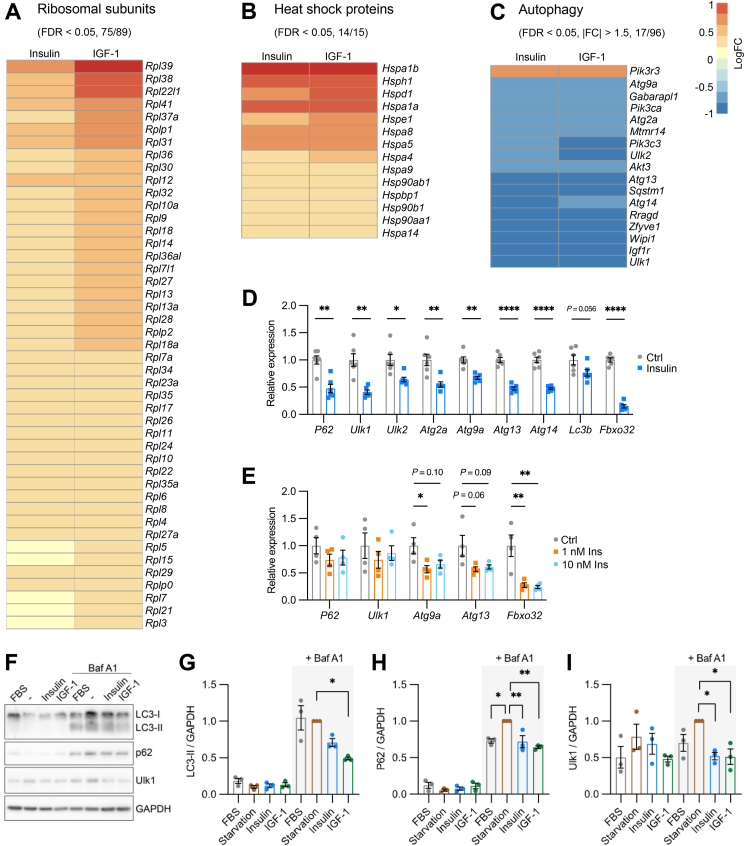
The unbiased bioinformatic approach above showed that both insulin and IGF-1 elicited strong transcriptional regulation on pathways involved in protein homeostasis. Thus, the top regulated pathways by both ligands included ribosomal biogenesis, rRNA processing, as well as autophagy, representing both the synthesis and degradation of proteins. We next looked into the detailed expression levels of individual genes within these pathways. Of the 89 ribosomal subunits detectable in our RNA Seq dataset, insulin and IGF-1 significantly regulated 75 subunits (FDR < 0.05), all of which were up-regulated (Figure 5A). In general, IGF-1 induced a larger fold-change on ribosomal subunits than insulin. Similarly, mitochondrial-specific ribosomal subunits were predominantly up-regulated by insulin and IGF-1 (Supplementary Figure S6A). In addition to ribosomal biogenesis, the majority of the heat shock proteins were dramatically up-regulated (Figure 5B). This up-regulation was, at least in part, due to an increase in Hsf1 expression following insulin or IGF-1 treatment (Supplemental Figure S6B). Heat shock proteins play a diverse range of critical functions in cells, including protein folding, stress response, and providing scaffolding for signaling complexes. The up-regulation of heat shock proteins may help facilitate the correct folding of newly synthesized polypeptides or misfolded proteins, thus contributing to protein homeostasis.
Figure 5.
Insulin and IGF-1 regulate pathways involved in proteostasis. A. Heatmap showing the Log2 (fold change) of the significantly regulated ribosomal subunits by 100 nM insulin or IGF-1 treatment for 6 h (FDR < 0.05). Numbers in the parenthesis indicate significantly regulated genes over total genes detectable in the RNA Seq. B. Heatmap showing the Log2 (fold change) of the significantly regulated heat shock proteins by 100 nM insulin or IGF-1 treatment (FDR < 0.05). Numbers in the parenthesis indicate significantly regulated genes over total genes detectable in the RNA Seq. C. Heatmap showing the Log2 (fold change) of the autophagy genes significantly regulated by at least 50% by 100 nM insulin or IGF-1 treatment (FDR < 0.05, |FC| > 1.5). Numbers in the parenthesis indicate genes significantly regulated by at least 50% over total genes detectable in the RNA Seq. D. Relative mRNA expression of genes involved in autophagy and ubiquitination in astrocytes following 100 nM insulin or vehicle treatment for 6 h. Tbp was used as internal control. Data are shown as mean ± SEM. Two-tailed Student's t-test. ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗∗, P < 0.0001. N = 6. E. Relative mRNA expression of p62, Ulk1, Atg9a, Atg13, and Fbxo32 in astrocytes following 0, 1, and 10 nM insulin treatment for 6 h. Tbp was used as internal control. Data are shown as mean ± SEM. One-way ANOVA followed by Tukey's multiple comparison test. ∗, P < 0.05; ∗∗, P < 0.01. N = 4. F, Immunoblotting showing the protein expression of LC3-I/II, p62 and Ulk1 in astrocytes subjected to overnight serum starvation, 100 nM insulin or IGF-1 stimulation, with or without 100 nM Bafilomycin A1 co-treatment. G–I. Densitometric analysis of LC3-II (G), p62 (H) and Ulk1 (I). Data are shown as mean ± SEM. One-way ANOVA followed by Tukey's multiple comparison test for the conditions with Baf A1 co-treatment. ∗, P < 0.05; ∗∗, P < 0.01. N = 3.
When assessing catabolism, insulin and IGF-1 showed remarkable regulations on genes involved in autophagy. Among the 96 genes included in the KEGG autophagy – animal pathway, 51 were significantly suppressed by insulin and IGF-1, whereas 25 were induced (Supplementary Figure S6C). Of the 17 genes whose expressions were changed by at least 50%, all except one, Pik3r3, were dramatically suppressed (Figure 5C). These included key upstream regulators and core enzymes to form autophagosome, such as Ulk1, Ulk2, Sqstm1, and many Atg genes. Notably, sequestosome 1 (commonly known as p62) provides a docking site for ubiquitinated proteins, therefore routing ubiquitinated proteins to autophagosomes for degradation. Together, these transcriptomic results demonstrate an overall shift toward an anabolic state of protein homeostasis in astrocytes following insulin or IGF-1stimulation.
3.5. Insulin and IGF-1 signaling potently suppress autophagy through transcriptional regulation
To validate the RNA sequencing data on autophagy, we investigated the transcriptional responses of astrocytes to insulin using qPCR. 100 nM insulin stimulation triggered a dramatic downregulation of a diverse assortment of autophagy genes by ∼50% (Figure 5D). These included p62, and Ulk1/2 critical for transducing signals from upstream regulators such as mTOR or AMPK, and multiple Atg genes which have a diverse range of actions including membrane tethering and autophagosome fusion (Figure 5D). Meanwhile, the expression of a well-known autophagy marker gene, Lc3b, showed a trend toward reduction following insulin stimulation (Figure 5D). When stimulated by 100 nM IGF-1, the same group of autophagy genes were all significantly down-regulated (Supplemental Figures 7D–G). Consistent with the RNA Seq data, the mRNA level of Fbxo32 dramatically decreased by almost 95% in insulin treated astrocytes compared with that in astrocytes without stimulation (Figure 5D). The consistent down-regulation of p62 and Fbxo32 further indicated the role of insulin to suppress autophagy selective for proteins in astrocytes. Insulin's suppression of autophagy mediators was potent. Thus, many of the same autophagy genes, as well as Fbxo32, can be efficiently suppressed by insulin at 1 nM, a concentration close to the postprandial plasma insulin levels in humans (Figure 5E) [55,56].
We next examined if these changes encompassed protein levels and autophagy flux, or if they were limited to genomic transcription modulation. LC3 is initially synthesized as an inactive cytosolic protein (LC3-I). During the activation of autophagy process, LC3-I is subjected to multi-step enzymatic reactions by Atg 3, Atg4, and Atg7 to form lipid-conjugated LC3-II, which is specifically inserted into the newly generated autophagosome [57]. Thus, the abundance of LC3-II is commonly used to gauge autophagy flux. To prevent rapid degradation and allow accumulation of autophagy proteins, we co-treated astrocytes with 100 nM bafilomycin A1 (Baf A1) to block autophagosome – lysosome fusion. As expected, astrocytes without Baf A1 treatment showed low proteins levels of LC3-II and p62 (Figure 5F). Following Baf A1 treatment, however, the expression of LC3-II and p62 were dramatically increased in total protein extracts of astrocytes cultured in normal culture conditions containing 10% FBS (Figure 5F). Importantly, while serum starvation moderately increased the protein levels of p62 and Ulk1, both insulin and IGF-1 stimulation significantly decreased the protein contents of p62 and Ulk1 by ∼30% and 50%, respectively, in comparison with serum starved group (Figure 5F, H and I). The LC3-II levels were also decreased by 29% (P = 0.16) in insulin-treated astrocytes and by 51% (P = 0.01) in IGF-1-treated astrocytes (Figure 5F, G). Together, these results demonstrate that both insulin and IGF-1 regulate autophagy flux by suppressing the expression of many upstream kinases and enzymes of autophagy.
3.6. Both insulin and IGF-1 receptors contribute to the regulation of autophagy genes
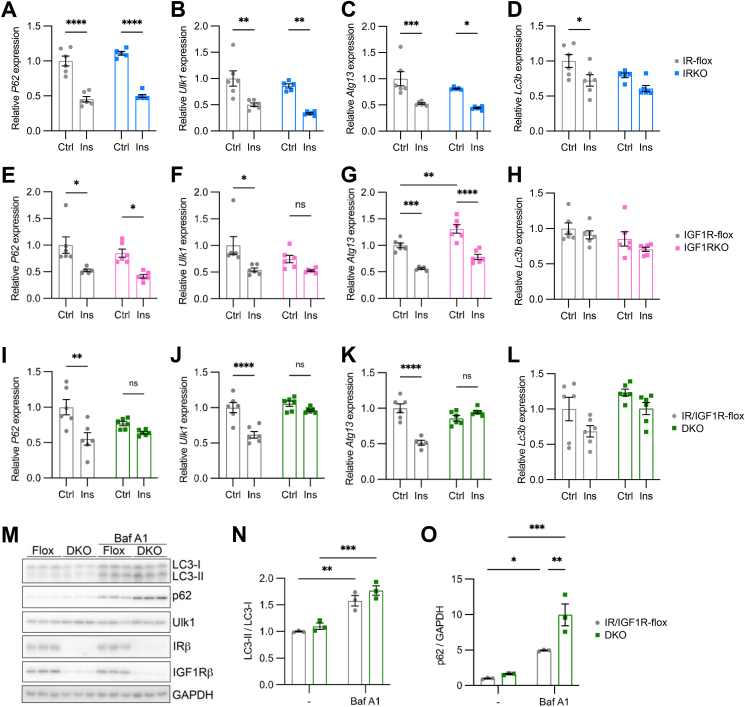
To further determine the relative importance of both IR and IGF1R on autophagy, we applied the same Cre-dependent flox allele recombination to specifically delete IR, IGF-1R or both in primary cultured astrocytes. IR-flox astrocytes infected with adenovirus encoding Cre:GFP showed an almost complete loss of transcripts containing floxed exon 4 of IR compared with control IR-flox astrocytes infected with adenovirus encoding GFP alone (Supplementary Figure S7A). Accompanied with IR loss in astrocytes, the mRNA levels of IGF1R showed a trend toward an increase. Consistent with our RNA Seq analysis comparing IR-flox and IRKO astrocytes, quantitative real-time PCR showed that insulin suppressed representative autophagy genes, such as p62, Ulk1, and Atg13 by around 50% in both IR-flox and IRKO astrocytes (Figure 6A–C), suggesting that endogenous IGF1R was sufficient to mediate the insulin-dependent suppression of autophagy in astrocytes. The expression of Lc3b was modestly decreased in both IR-flox and IRKO astrocytes treated with insulin (Figure 6D).
Figure 6.
Both IR and IGFR contribute to autophagy gene regulation. A–D. Relative mRNA expression of p62 (A), Ulk1 (B), Atg13 (C) and Lc3b (D) in IR-flox and IRKO astrocytes treated with 100 nM insulin for 6 h. Tbp was used as internal control. Data are shown as mean ± SEM. Two-way ANOVA followed by Tukey's multiple comparison test. ∗, P < 0.05; ∗∗, P < 0.01. ∗∗∗, P < 0.001; ∗∗∗∗, P < 0.0001. N = 5–6. E–H. Relative mRNA expression of p62 (E), Ulk1 (F), Atg13 (G) and Lc3b (H) in IGF1R-flox and IGF1RKO astrocytes treated with 100 nM insulin for 6 h. Tbp was used as internal control. Data are shown as mean ± SEM. Two-way ANOVA followed by Tukey's multiple comparison test. ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001; ∗∗∗∗, P < 0.0001. N = 5–6. I-L. Relative mRNA expression of p62 (I), Ulk1 (J), Atg13 (K) and Lc3b (L) in IR/IGF1R-flox and DKO astrocytes treated with 100 nM insulin for 6 h. Tbp was used as internal control. Data are shown as mean ± SEM. Two-way ANOVA followed by Tukey's multiple comparison test. ∗∗, P < 0.01; ∗∗∗∗, P < 0.0001. N = 5–6. M. Immunoblotting showing the protein expression of LC3-I/II, p62, Ulk1, IRβ and IGF1Rβ in astrocytes with or without 100 nM Bafilomycin A1 treatment overnight. N–O. Densitometric analysis of LC3-II/LC3-I ratio (N) and p62 (O). Data are shown as mean ± SEM. Two-way ANOVA followed by Tukey's multiple comparison test. ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001. N = 3.
When IGF1R-flox astrocytes were infected with adenovirus encoding Cre:GFP, the expression of transcripts containing floxed exon 3 of IGF1R decreased by more than 95%, whereas the mRNA levels of IR was slightly increased in IGF1RKO astrocytes (Supplementary Figure S7B). In general, IGF-1-mediated suppression of autophagy genes was preserved in IGF1RKO astrocytes, but with a slightly lower percentage reduction when compared with IGF1R-flox astrocytes (Supplemental Figures 7D–G). These data suggest that despite the lower expression of IR in astrocytes, IGF-1 can signal through IR to suppress autophagy.
Since the expression of IGF1R is 3 times higher than IR in astrocytes, we sought to examine whether insulin-induced suppression of autophagy is primarily mediated by IGF1R. The expression of p62 was decreased by around 50% in both IGF1R-flox and IGF1RKO astrocytes (Figure 6E), indicating endogenous IR was sufficient to fully suppress autophagy by insulin. The expression of Ulk1 was significantly suppressed by insulin in IGF1R-flox astrocytes, whereas its expression only showed a non-significant reduction by ∼30% in IGF1RKO astrocytes (Figure 6F), suggesting a slightly stronger suppression of Ulk1 by IGF1R. Loss of IGF1R in astrocytes up-regulated the expression of Atg13 by 30% in astrocytes at basal condition (Figure 6G). In response to insulin stimulation, however, both IGF1R-flox and IGF1RKO astrocytes showed ∼40% reduction in Atg13 expression compared to baseline (Figure 6G). Thus, IGF1R contributed to the basal expression of Atg13 in astrocytes, whereas IR alone was sufficient to suppress Atg13 expression in response to insulin. Meanwhile, Lc3b expression was not significantly changed in IGF1RKO astrocytes at both basal and insulin-stimulated states (Figure 6H). Together, these data suggest that endogenous insulin receptors are sufficient to suppress autophagy in astrocytes despite having a lower level of expression than IGF1R.
In stark contrast, loss of both endogenous IR and IGF1R (Supplementary Figure S7C) completely blunted insulin-induced suppression of p62, Ulk1, and Atg13 in astrocytes (Figure 6I–K), whereas the expression of Lc3b was not altered by insulin in both IR/IGF1R-flox and IR/IGF1R double knockout astrocytes (Figure 6L). In the normal culture media containing 10% FBS, bafilomycin A1 dramatically increased the protein expression of LC3-I and II in astrocytes, as well as the LC3-II to LC3-I ratio as expected (Figure 6M, N). Their protein levels, however, were not significantly altered in the absence of IR and IGF1R (Figure 6M, N). Hence, loss of both IR and IGF1R does not impact the formation and maturation of autophagosomes in normal culture media supplemented with 10% FBS. Notably, DKO astrocytes showed a dramatic accumulation of p62 in the presence of Baf A1 (Figure 6M, O), strongly suggesting a robust suppression of autophagic flux selective for ubiquitinated proteins by IR/IGF1R signaling.
3.7. Both IR and IGF1R signaling contribute to autophagic flux in astrocytes
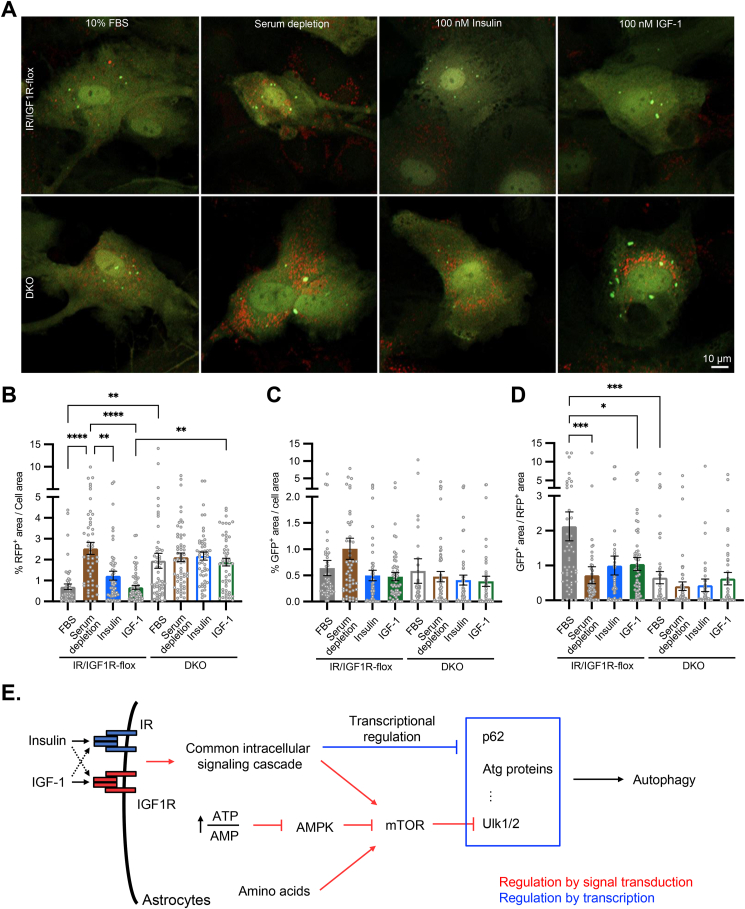
We next sought to validate the functional impact of insulin and IGF-1 signaling on autophagic flux in astrocytes using mRFP-GFP tandem fluorescent-tagged LC3 (tfLC3) reporter [58]. The tfLC3 emits both green and red fluorescence when embedded into an autophagosome with neutral pH. However, once an autophagosome fused with a lysosome to form autophagolysosome, the acidic environment quenches the GFP signal, leading to only red fluorescence from tfLC3 reporter. After both IR/IGF1R-flox and DKO astrocytes were infected with adenovirus encoding tfLC3, astrocytes were placed in normal culture media with 10% FBS, or subjected to serum depletion, 100 nM insulin or IGF-1 treatment overnight. Following the serum depletion or insulin/IGF-1 stimulation, all cells were incubated in Hank's balanced salt solution (HBSS) lacking amino acids for 4 h to induce autophagy flux.
In IR/IGF1R-flox astrocytes, serum depletion increased the RFP+ puncta by ∼2.5-fold compared with normal culture condition (Figure 7A, B), whereas the GFP+ puncta showed a modest increase in serum starved astrocytes (Figure 7A, C). As a result, the GFP+/RFP+ ratio was dramatically decreased by serum depletion in IR/IGF1R-flox astrocytes (Figure 7D), demonstrating rapid autophagosome-lysosome fusion in serum starved astrocytes. Both insulin and IGF-1 stimulations dramatically decreased intracellular RFP+ puncta, to a degree similar to that of the cells under normal culture condition (Figure 7B). The less accumulation of RFP+ LC3 in astrocytes treated with insulin or IGF-1 indicated decreased autophagic flux, consistent with the decreased expression of autophagy genes like p62, Ulk1, Atg13, among others. In agreement with these data, the astrocytes without endogenous IR and IGF1R showed significantly higher intracellular RFP+ puncta in all different treatment groups (Figure 7A, B), due to the loss of IR/IGF1R-mediated suppression on the expression of autophagy genes.
Figure 7.
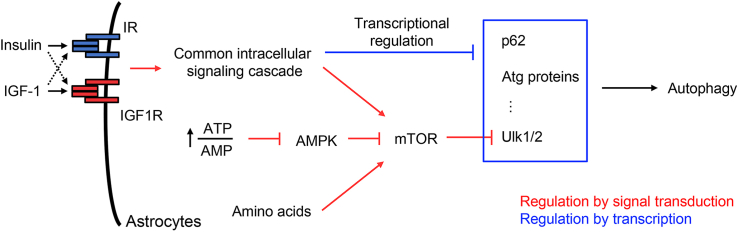
Both IR and IGFR contribute to autophagic flux in astrocytes. A. Representative confocal images showing red and green puncta of IR/IGF1R-flox and DKO astrocytes expressing tfLC3 reporter in normal culture condition (10% FBS), overnight serum depletion, and overnight treatment of 100 nM insulin or IGF-1 following 5 h serum depletion. To induce maximal autophagy flux, all cells were incubated with 1× HBSS for 4 h before confocal live cell imaging. Scale bar: 10 μm. B–D. Quantification of RFP+ puncta over total cell area (B), GFP+ puncta over total cell area (C), and GFP+ to RFP+ ratio (D) in IR/IGF1R-flox and DKO astrocytes under different treatments indicated. Data are shown as mean ± SEM. One-way ANOVA followed by Tukey's multiple comparison test. ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001; ∗∗∗∗, P < 0.0001. N = 50–61 individual cells. E. Illustration showing that while multiple upstream signaling cascades converge on mTOR kinase activity to regulate autophagy flux, insulin and IGF-1 trigger strong transcriptional suppression on many genes directly involved in autophagy in astrocytes. Collectively, autophagy in astrocytes is tightly regulated by metabolic states and nutrient levels.
Taken together, insulin and IGF-1 induce robust transcriptional regulation in astrocytes to regulate multiple cellular processes, including the suppression of autophagy. Both IR and IGF1R mediate the suppression of key autophagy genes, including p62, Ulk1, Ulk2, and Atg13. Thus, in addition to the acute regulation of mTOR kinase activity by insulin/IGF-1, ATP/AMP ratio, and the abundance of intracellular amino acid pool, insulin and IGF-1 can trigger transcriptional regulation to elicit prolonged suppression on autophagy in astrocytes (Figure 7E).
4. Discussion
Insulin is critical for the balance of anabolic and catabolic states throughout peripheral tissues, in addition to modulating blood glucose levels [1]. The discovery that insulin is capable of both crossing the blood brain barrier while remaining biologically active and that the central nervous system contains insulin receptors and closely related IGF-1 receptors opened the door for investigation into insulin's less well characterized actions in the central nervous system [59]. IGF-1, on the other hand, is known to function as a neurotrophic factor and play crucial roles in neural differentiation, survival, and development [20]. In the present study, we examined insulin- and IGF-1-stimulated transcriptomic changes in primary cultured IR-flox or IR knockout astrocytes using RNA sequencing. This unbiased approach allowed us to assess the functional consequences of insulin and IGF-1 signaling in astrocytes. We noted changes in genomic transcription, protein levels, and confocal targets following insulin or IGF-1 stimulation in astrocytes that had varying levels of insulin and IGF-1 receptors. The variety of conditions and stimuli allowed us to better understand a crucial, currently poorly defined signaling cascade in astrocytes.
Our present data demonstrate that insulin and IGF-1 signaling controls a broad transcriptional network related to cellular metabolism in astrocytes. In particular, insulin predominantly enhances the expression of the majority of the cholesterol biosynthetic enzymes. This is in agreement with our recent study demonstrating a potent and robust transcriptional up-regulation of the same pathway in the brains of the mice subjected to hyperinsulinemic-euglycemic clamps [60]. The brain almost exclusively relies on local cholesterol production. Astrocytes are considered one of the major reservoirs of cholesterol in the brain. Loss of cholesterol synthesis in astrocytes impairs normal brain development while also inducing cognitive dysfunction in mice [61]. On the other hand, excess cholesterol may also be detrimental, since cholesterol-enriched plasma membrane microdomains in neurons have been shown to facilitate the production of amyloid-β, and contribute to the pathogenesis of Alzheimer's disease [62]. The regulation of cholesterol biosynthesis by insulin and IGF-1 in astrocytes may be critical for normal brain development, synaptic plasticity, and cognitive function. The impairment of astrocytic insulin signaling in diabetes and insulin resistant states may lead to the disruption of brain cholesterol homeostasis and increase the risks to develop neurodegenerative disease like Alzheimer's disease.
Autophagy is a “self-eating” process that allows for selective recycling of cellular components [63]. It is known to be tightly controlled by multiple signaling cascades. For instance, branched-chain amino acid levels, ATP/AMP ratio, and hormones like insulin and IGF-1 have been reported to converge on mTOR activity to regulate autophagy [64,65]. Downstream of mTOR, Ulk1/2 initiates phosphorylation events on key Atg family proteins, which aid in membrane tethering and autophagosome fusion [66,67]. Our data demonstrate that in addition to spatial and temporal phosphorylation events, autophagy in astrocytes is subjected to potent transcriptional regulation by insulin and IGF-1 signaling.
P62 specifically recruits ubiquitinated proteins to autophagosomes [68]. The potent suppression of p62 expression strongly indicates that insulin and IGF-1 regulate selective autophagy for the degradation of ubiquitinated proteins. In agreement with this, Fbxo32, an E3-ubiquitin ligase, is the most suppressed gene by insulin and IGF-1 in astrocytes. The suppression of autophagy and protein degradation by insulin/IGF-1 signaling has also been demonstrated in skeletal muscles. Loss of IR and IGF1R in skeletal muscle leads to uncontrolled autophagy and proteolysis, contributing to severe muscle atrophy in these mice [69]. While the functional significance of insulin/IGF-1-dependent suppression of selective autophagy in astrocytes requires further investigation, it is plausible to speculate that this regulation may be important for protein and neurotransmitter homeostasis in the brain. Dysregulation of selective autophagy pathways in astrocytes under an insulin resistant state may be a contributing factor for many diabetes-related neurological complications.
In addition to the potent effect on autophagy, insulin and IGF-1 signaling show strong transcriptional regulation on ribosomal biogenesis. Ribosomal biogenesis and rRNA processing are among the top regulated biological processes by both insulin and IGF-1. 85% of the ribosomal subunits are significantly up-regulated by insulin and IGF-1, whereas none are suppressed. Further supporting the elevation of ribosomal biogenesis, the top up-regulated genes by insulin and IGF-1 include many small nucleolar RNAs (snoRNAs), which are known to facilitate the modification and folding of the ribosomal RNAs. Together, the predominant up-regulation of ribosomal biogenesis strongly indicate that insulin/IGF-1 signaling promotes protein synthesis [70]. Notably, IGF-1 appears to have a stronger induction on genes involved in ribosomal biogenesis, which is consistent with the conventional understanding that IGF-1 is a more potent inducer of protein synthesis, cell growth and proliferation [69,71].
Collectively, the induction of ribosomal biogenesis and the suppression of selective autophagy suggests the important role of insulin and IGF-1 in protein homeostasis by shifting the balance toward an anabolic preference in astrocytes. This is also supported by the up-regulation of the majority of the heat shock proteins in our RNA Seq dataset. Thus, in conjunction with increased protein translation, insulin and IGF-1 tune up the chaperones to facilitate proper folding of newly synthesized polypeptides. Alternatively, increased expression of heat shock proteins may also increase the cellular capacity to re-fold the damaged or misfolded proteins, which would also promote protein homeostasis in astrocytes [72].
Our data also strongly suggest astrocytic insulin and IGF-1 signaling are critical for maintaining mitochondrial homeostasis; with their role being analogous to the one seen in proteostasis. Both insulin and IGF-1 predominantly up-regulate the expression of mitochondrial specific ribosomal subunits. Among the highly regulated heat shock proteins, Hspd1 (Hsp60) and Hspe1 (Hsp10) are exclusively localized in mitochondria and important for mitochondrial stress response [73,74]. Astrocytic insulin and IGF-1 signaling may also regulate mitophagy through p62 and ubiquitination of mitochondrial-residing Parkin [75]. Together, our data suggest that a major role of astrocytic insulin and IGF-1 signaling is to balance the anabolic and catabolic states of protein and mitochondria. Additional investigations are needed to distinguish protein and mitochondrial homeostasis using specific models.
Gap junctions between astrocytes and hemichannels in astrocytes have been shown to release a variety of small molecules and neurotransmitters. They play crucial roles in the normal brain development, synaptic plasticity, and pathogenesis of neurological diseases [76,77]. Here, we show that insulin and IGF-1 transcriptionally regulate gap junction related pathways in astrocytes, suggesting a potentially novel mechanism by which insulin signaling contributes to cell–cell communications in the brain. Notably, our previous studies have shown that insulin induces ATP release through exocytosis by mediating Munc18c tyrosine phosphorylation and SNARE complex assembly [33]. Collectively, these data suggest that astrocytic insulin signaling regulates astrocyte-derived signals through multiple mechanisms to modulate neuronal plasticity and neural circuits.
Considering the largely overlapping transcriptional regulation shared by insulin and IGF-1, it makes the plethora of ligand-specific responses in astrocytes all the more interesting. Genes specifically regulated by IGF-1 are enriched in pathways related to ribosomal biogenesis and rRNA processing, indicating a stronger effect of IGF-1 on protein synthesis and cell growth. In contrast, many of the top regulated genes specific to insulin include those involved in cellular metabolism. For instance, Aldob, which encodes for aldolase B and lipid binding protein, Apoa2, are among the most induced genes by insulin, but not by IGF-1. Therefore, insulin appears to have a stronger regulatory role for metabolism in astrocytes. This is in agreement with many previous studies in peripheral tissues and cell cultures [71,78,79]. Multiple mechanisms may contribute to these ligand-specific differences. In astrocytes, the expressions of Igf1r and Irs2 are ∼3 fold higher than Insr and Irs1, indicating a higher signaling capacity of IGF1R-IRS2 cascade. Although both IR and IGF1R can be activated by 100 nM of ligands, the binding affinity and kinetics of insulin and IGF-1 on IR, IGF1R and hybrid receptors are different, which can likely lead to distinct downstream transcriptional regulations. Additionally, IR and IGF1R themselves harbor distinct signaling properties that could explain the unique transcriptional regulations. Supporting this, preadipocytes only expressing IR show higher Ser/Thr phosphorylation on proteins related to mTOR and Akt cascades following ligand stimulation, while cells only expressing IGF1R display higher phosphorylation on proteins involved in cell cycle and mitosis after activation [78]. Notably, even unoccupied IRs and IGF1Rs during the prolonged serum starvation can regulate distinct basal phosphorylation of numerous proteins, which may prime the cells for different transcriptional responses upon insulin-IR activation versus IGF-1-IGF1R activation [78].
By comparing the global transcriptomes of IR-flox and IRKO astrocytes, we have found that a large portion of the transcriptional regulation induced by insulin can go through endogenous IGF1R, providing evidence of ligand-receptor cross activation in astrocytes. Indeed, only when both receptors were knocked out did we see a complete loss of insulin response, such as the suppression of autophagic genes. Additionally, a number of genes require IR for insulin-induced regulation. The most regulated genes in this class include many metabolic enzymes, such as Aldob and Apoa2. Notably, Alb, the gene encoding for albumin, is the most insulin-induced gene in IR-flox astrocytes, whereas this regulation was lost in IRKO astrocytes. While the crucial roles of circulating albumin in fluid distribution, protein, ion and lipid balance, and drug binding in adult mammals have been well established [80], whether insulin-induced Alb expression has any functional significance in the brain remains to be investigated. It is worth noting that fetal brains and choroid plexus have been reported to produce albumin [81]. While our present data support this observation and indicate astrocytes may also produce albumin under the regulation of insulin signaling in neonatal brains, whether such insulin-dependent regulation is also preserved in adult brains awaits further investigations. Somewhat unexpectedly, a third group of genes show transcriptional changes by insulin only when the insulin receptor is absent. These genes could be oppositely regulated by IR and IGF1R. Alternatively, loss of IR may disrupt the baseline signaling and expression, such that removing the insulin receptor essentially lifts any basal regulation of transcription. Differential signaling properties of an IR/IGF1R hybrid receptor may also be a possible mechanism. While the present study was primarily focused on the effect of IR loss in astrocytes, similar receptor-dependent regulations are expected for IGF1R. All of these warrant further investigations.
The present study is one of the first attempts to systemically interrogate insulin and IGF-1 action in astrocytes. The transcriptomic analyses reveal potentially important functional pathways regulated by insulin and IGF-1, many of which await further investigation using in vivo models. A growing body of evidence support sexual dimorphism in central insulin action [33,82,83]. The potential sex-dependent insulin/IGF-1 responses in astrocytes remain unexplored in the present study. Given the functional heterogeneity of astrocytes in different brain regions and at the different developmental stages, future studies are needed to investigate the spatiotemporal effects of insulin and IGF-1 on astrocytes.
In summary, insulin and IGF-1 signaling regulate a broad transcriptional network in astrocytes, including pathways involved in lipid and cholesterol metabolism, protein homeostasis, and gap junctions. Further, both IR and IGF1R can elicit many of these transcriptional regulations, exemplified by the suppression of autophagy. These data demonstrate a common and converging role of IR and IGF1R signaling in regulating lipid and protein homeostasis in astrocytes. Together, these findings expand our understanding of how astrocytes can react to local and systemic signals and contribute to brain metabolism and function. They also provide new insights into ways that insulin resistance in the brain may contribute to neurological disorders, especially those related to diabetes.
Author contributions
S.J.G. designed research, performed experiments, analyzed the data, and wrote the paper. S.M., C.O.S., S.T., H.L., K.L., C.W., Y.H., Q.H., and Y.Z. helped with experiments and data analysis. W.C. designed research, wrote the paper, and supervised the project.
Acknowledgments
This work was supported by NIH grants R01 MH125903 and K01 DK120740 (to W.C.). We thank NYIT Imaging Center for assistance with confocal imaging. Dr. Weikang Cai is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2022.101647.
Conflict of interest
The authors declare no conflict of interests.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
I have shared the link to the raw sequencing data and codes in "Data Availability" Section.
References
- 1.Petersen M.C., Shulman G.I. Mechanisms of insulin action and insulin resistance. Physiol Rev. 2018;98(4):2133–2223. doi: 10.1152/physrev.00063.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu J.P., Baker J., Perkins A.S., Robertson E.J., Efstratiadis A. Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r) Cell. 1993;75(1):59–72. [PubMed] [Google Scholar]
- 3.Taniguchi C.M., Emanuelli B., Kahn C.R. Critical nodes in signalling pathways: insights into insulin action. Nat Rev Mol Cell Biol. 2006;7(2):85–96. doi: 10.1038/nrm1837. [DOI] [PubMed] [Google Scholar]
- 4.Haeusler R.A., McGraw T.E., Accili D. Biochemical and cellular properties of insulin receptor signalling. Nat Rev Mol Cell Biol. 2018;19(1):31–44. doi: 10.1038/nrm.2017.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Batista T.M., Garcia-Martin R., Cai W., Konishi M., O'Neill B.T., Sakaguchi M., et al. Multi-dimensional transcriptional remodeling by physiological insulin in vivo. Cell Rep. 2019;26(12):3429–3443. doi: 10.1016/j.celrep.2019.02.081. e3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abuzzahab M.J., Schneider A., Goddard A., Grigorescu F., Lautier C., Keller E., et al. IGF-I receptor mutations resulting in intrauterine and postnatal growth retardation. N Engl J Med. 2003;349(23):2211–2222. doi: 10.1056/NEJMoa010107. [DOI] [PubMed] [Google Scholar]
- 7.Gray S.M., Aylor K.W., Barrett E.J. Unravelling the regulation of insulin transport across the brain endothelial cell. Diabetologia. 2017;60(8):1512–1521. doi: 10.1007/s00125-017-4285-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Konishi M., Sakaguchi M., Lockhart S.M., Cai W., Li M.E., Homan E.P., et al. Endothelial insulin receptors differentially control insulin signaling kinetics in peripheral tissues and brain of mice. Proc Natl Acad Sci U S A. 2017;114(40):E8478–E8487. doi: 10.1073/pnas.1710625114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kleinridders A., Ferris H.A., Cai W., Kahn C.R. Insulin action in brain regulates systemic metabolism and brain function. Diabetes. 2014;63(7):2232–2243. doi: 10.2337/db14-0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mazucanti C.H., Liu Q.R., Lang D., Huang N., O'Connell J.F., Camandola S., et al. Release of insulin produced by the choroid plexis is regulated by serotonergic signaling. JCI Insight. 2019;4(23) doi: 10.1172/jci.insight.131682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christie J.M., Wenthold R.J., Monaghan D.T. Insulin causes a transient tyrosine phosphorylation of NR2A and NR2B NMDA receptor subunits in rat hippocampus. J Neurochem. 1999;72(4):1523–1528. doi: 10.1046/j.1471-4159.1999.721523.x. [DOI] [PubMed] [Google Scholar]
- 12.Ahmadian G., Ju W., Liu L., Wyszynski M., Lee S.H., Dunah A.W., et al. Tyrosine phosphorylation of GluR2 is required for insulin-stimulated AMPA receptor endocytosis and LTD. EMBO J. 2004;23(5):1040–1050. doi: 10.1038/sj.emboj.7600126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schubert M., Gautam D., Surjo D., Ueki K., Baudler S., Schubert D., et al. Role for neuronal insulin resistance in neurodegenerative diseases. Proc Natl Acad Sci U S A. 2004;101(9):3100–3105. doi: 10.1073/pnas.0308724101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mandelkow E.M., Mandelkow E. Biochemistry and cell biology of tau protein in neurofibrillary degeneration. Cold Spring Harb Perspect Med. 2012;2(7):a006247. doi: 10.1101/cshperspect.a006247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Konner A.C., Janoschek R., Plum L., Jordan S.D., Rother E., Ma X., et al. Insulin action in AgRP-expressing neurons is required for suppression of hepatic glucose production. Cell Metab. 2007;5(6):438–449. doi: 10.1016/j.cmet.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 16.Shin A.C., Filatova N., Lindtner C., Chi T., Degann S., Oberlin D., et al. Insulin receptor signaling in POMC, but not AgRP, neurons controls adipose tissue insulin action. Diabetes. 2017;66(6):1560–1571. doi: 10.2337/db16-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loh K., Zhang L., Brandon A., Wang Q., Begg D., Qi Y., et al. Insulin controls food intake and energy balance via NPY neurons. Mol Metab. 2017;6(6):574–584. doi: 10.1016/j.molmet.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soto M., Cai W., Konishi M., Kahn C.R. Insulin signaling in the hippocampus and amygdala regulates metabolism and neurobehavior. Proc Natl Acad Sci U S A. 2019;116(13):6379–6384. doi: 10.1073/pnas.1817391116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grillo C.A., Piroli G.G., Lawrence R.C., Wrighten S.A., Green A.J., Wilson S.P., et al. Hippocampal insulin resistance impairs spatial learning and synaptic plasticity. Diabetes. 2015;64(11):3927–3936. doi: 10.2337/db15-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Torres-Aleman I. Toward a comprehensive neurobiology of IGF-I. Dev Neurobiol. 2010;70(5):384–396. doi: 10.1002/dneu.20778. [DOI] [PubMed] [Google Scholar]
- 21.Kappeler L., Magalhaes Filho C.M., Dupont J., Leneuve P., Cervera P., Perin L., et al. Brain IGF-1 receptors control mammalian growth and lifespan through a neuroendocrine mechanism. PLoS Biol. 2008;6(10):e254. doi: 10.1371/journal.pbio.0060254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Popken G.J., Hodge R.D., Ye P., Zhang J., Ng W., O'Kusky J.R., et al. In vivo effects of insulin-like growth factor-I (IGF-I) on prenatal and early postnatal development of the central nervous system. Eur J Neurosci. 2004;19(8):2056–2068. doi: 10.1111/j.0953-816X.2004.03320.x. [DOI] [PubMed] [Google Scholar]
- 23.Madathil S.K., Carlson S.W., Brelsfoard J.M., Ye P., D’Ercole A.J., Saatman K.E. Astrocyte-specific overexpression of insulin-like growth factor-1 protects hippocampal neurons and reduces behavioral deficits following traumatic brain injury in mice. PLOS One. 2013;8(6):e67204. doi: 10.1371/journal.pone.0067204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carlson S.W., Madathil S.K., Sama D.M., Gao X., Chen J., Saatman K.E. Conditional overexpression of insulin-like growth factor-1 enhances hippocampal neurogenesis and restores immature neuron dendritic processes after traumatic brain injury. J Neuropathol Exp Neurol. 2014;73(8):734–746. doi: 10.1097/NEN.0000000000000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sofroniew M.V. Astrocyte reactivity: subtypes, states, and functions in CNS innate immunity. Trends Immunol. 2020;41(9):758–770. doi: 10.1016/j.it.2020.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haydon P.G., Carmignoto G. Astrocyte control of synaptic transmission and neurovascular coupling. Physiol Rev. 2006;86(3):1009–1031. doi: 10.1152/physrev.00049.2005. [DOI] [PubMed] [Google Scholar]
- 27.Nagai J., Yu X., Papouin T., Cheong E., Freeman M.R., Monk K.R., et al. Behaviorally consequential astrocytic regulation of neural circuits. Neuron. 2021;109(4):576–596. doi: 10.1016/j.neuron.2020.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diaz-Castro B., Bernstein A.M., Coppola G., Sofroniew M.V., Khakh B.S. Molecular and functional properties of cortical astrocytes during peripherally induced neuroinflammation. Cell Rep. 2021;36(6) doi: 10.1016/j.celrep.2021.109508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lutomska L.M., Miok V., Krahmer N., Gonzalez Garcia I., Gruber T., Le Thuc O., et al. Diet triggers specific responses of hypothalamic astrocytes in time and region dependent manner. Glia. 2022;70(11):2062–2078. doi: 10.1002/glia.24237. [DOI] [PubMed] [Google Scholar]
- 30.Boisvert M.M., Erikson G.A., Shokhirev M.N., Allen N.J. The aging astrocyte transcriptome from multiple regions of the mouse brain. Cell Rep. 2018;22(1):269–285. doi: 10.1016/j.celrep.2017.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sadick J.S., O'Dea M.R., Hasel P., Dykstra T., Faustin A., Liddelow S.A. Astrocytes and oligodendrocytes undergo subtype-specific transcriptional changes in Alzheimer's disease. Neuron. 2022;110(11):1788–1805 e1710. doi: 10.1016/j.neuron.2022.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee H.G., Wheeler M.A., Quintana F.J. Function and therapeutic value of astrocytes in neurological diseases. Nat Rev Drug Discov. 2022;21(5):339–358. doi: 10.1038/s41573-022-00390-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cai W., Xue C., Sakaguchi M., Konishi M., Shirazian A., Ferris H.A., et al. Insulin regulates astrocyte gliotransmission and modulates behavior. J Clin Investig. 2018;128(7):2914–2926. doi: 10.1172/JCI99366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia-Caceres C., Quarta C., Varela L., Gao Y., Gruber T., Legutko B., et al. Astrocytic insulin signaling couples brain glucose uptake with nutrient availability. Cell. 2016;166(4):867–880. doi: 10.1016/j.cell.2016.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manaserh I.H., Chikkamenahalli L., Ravi S., Dube P.R., Park J.J., Hill J.W. Ablating astrocyte insulin receptors leads to delayed puberty and hypogonadism in mice. PLoS Biol. 2019;17(3) doi: 10.1371/journal.pbio.3000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fernandez A.M., Hernandez-Garzon E., Perez-Domper P., Perez-Alvarez A., Mederos S., Matsui T., et al. Insulin regulates astrocytic glucose handling through cooperation with IGF-I. Diabetes. 2017;66(1):64–74. doi: 10.2337/db16-0861. [DOI] [PubMed] [Google Scholar]
- 37.Manaserh I.H., Maly E., Jahromi M., Chikkamenahalli L., Park J., Hill J. Insulin sensing by astrocytes is critical for normal thermogenesis and body temperature regulation. J Endocrinol. 2020;247(1):39–52. doi: 10.1530/JOE-20-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fernandez A.M., Martinez-Rachadell L., Navarrete M., Pose-Utrilla J., Davila J.C., Pignatelli J., et al. Insulin regulates neurovascular coupling through astrocytes. Proc Natl Acad Sci U S A. 2022;119(29) doi: 10.1073/pnas.2204527119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Logan S., Pharaoh G.A., Marlin M.C., Masser D.R., Matsuzaki S., Wronowski B., et al. Insulin-like growth factor receptor signaling regulates working memory, mitochondrial metabolism, and amyloid-beta uptake in astrocytes. Mol Metab. 2018;9:141–155. doi: 10.1016/j.molmet.2018.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tarantini S., Balasubramanian P., Yabluchanskiy A., Ashpole N.M., Logan S., Kiss T., et al. IGF1R signaling regulates astrocyte-mediated neurovascular coupling in mice: implications for brain aging. Geroscience. 2021;43(2):901–911. doi: 10.1007/s11357-021-00350-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson B.S., Zhao Y.T., Fasolino M., Lamonica J.M., Kim Y.J., Georgakilas G., et al. Biotin tagging of MeCP2 in mice reveals contextual insights into the Rett syndrome transcriptome. Nat Med. 2017;23(10):1203–1214. doi: 10.1038/nm.4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moon S., Zhao Y.T. Spatial, temporal and cell-type-specific expression profiles of genes encoding heparan sulfate biosynthesis enzymes and proteoglycan core proteins. Glycobiology. 2021;31(10):1308–1318. doi: 10.1093/glycob/cwab054. [DOI] [PubMed] [Google Scholar]
- 43.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kulkarni A., Dong A., Kulkarni V.V., Chen J., Laxton O., Anand A., et al. Differential regulation of autophagy during metabolic stress in astrocytes and neurons. Autophagy. 2020;16(9):1651–1667. doi: 10.1080/15548627.2019.1703354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cahoy J.D., Emery B., Kaushal A., Foo L.C., Zamanian J.L., Christopherson K.S., et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28(1):264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gerrits E., Heng Y., Boddeke E., Eggen B.J.L. Transcriptional profiling of microglia; current state of the art and future perspectives. Glia. 2020;68(4):740–755. doi: 10.1002/glia.23767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song H.W., Foreman K.L., Gastfriend B.D., Kuo J.S., Palecek S.P., Shusta E.V. Transcriptomic comparison of human and mouse brain microvessels. Sci Rep. 2020;10(1) doi: 10.1038/s41598-020-69096-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lawrence J.B., Oxvig C., Overgaard M.T., Sottrup-Jensen L., Gleich G.J., Hays L.G., et al. The insulin-like growth factor (IGF)-dependent IGF binding protein-4 protease secreted by human fibroblasts is pregnancy-associated plasma protein-A. Proc Natl Acad Sci U S A. 1999;96(6):3149–3153. doi: 10.1073/pnas.96.6.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O'Neill B.T., Bhardwaj G., Penniman C.M., Krumpoch M.T., Suarez Beltran P.A., Klaus K., et al. FoxO transcription factors are critical regulators of diabetes-related muscle atrophy. Diabetes. 2019;68(3):556–570. doi: 10.2337/db18-0416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gerst F., Jaghutriz B.A., Staiger H., Schulte A.M., Lorza-Gil E., Kaiser G., et al. The expression of aldolase B in islets is negatively associated with insulin secretion in humans. J Clin Endocrinol Metab. 2018;103(12):4373–4383. doi: 10.1210/jc.2018-00791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hubbard S.R. The insulin receptor: both a prototypical and atypical receptor tyrosine kinase. Cold Spring Harb Perspect Biol. 2013;5(3):a008946. doi: 10.1101/cshperspect.a008946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sarayloo F., Spiegelman D., Rochefort D., Akcimen F., De Barros Oliveira R., Dion P.A., et al. SKOR1 has a transcriptional regulatory role on genes involved in pathways related to restless legs syndrome. Eur J Hum Genet. 2020;28(11):1520–1528. doi: 10.1038/s41431-020-0670-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bouche C., Lopez X., Fleischman A., Cypess A.M., O'Shea S., Stefanovski D., et al. Insulin enhances glucose-stimulated insulin secretion in healthy humans. Proc Natl Acad Sci U S A. 2010;107(10):4770–4775. doi: 10.1073/pnas.1000002107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsukuda K., Kikuchi M., Irie S., Eto T., Yamada A., Matsuguma K., et al. Evaluation of the 24-hour profiles of physiological insulin, glucose, and C-peptide in healthy Japanese volunteers. Diabetes Technol Ther. 2009;11(8):499–508. doi: 10.1089/dia.2009.0029. [DOI] [PubMed] [Google Scholar]
- 57.Runwal G., Stamatakou E., Siddiqi F.H., Puri C., Zhu Y., Rubinsztein D.C. LC3-positive structures are prominent in autophagy-deficient cells. Sci Rep. 2019;9(1) doi: 10.1038/s41598-019-46657-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kobayashi S., Xu X., Chen K., Liang Q. Suppression of autophagy is protective in high glucose-induced cardiomyocyte injury. Autophagy. 2012;8(4):577–592. doi: 10.4161/auto.18980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Woods S.C., Seeley R.J., Baskin D.G., Schwartz M.W. Insulin and the blood-brain barrier. Curr Pharm Des. 2003;9(10):795–800. doi: 10.2174/1381612033455323. [DOI] [PubMed] [Google Scholar]
- 60.Cai W., Zhang X., Batista T.M., Garcia-Martin R., Softic S., Wang G., et al. Peripheral insulin regulates a broad network of gene expression in hypothalamus, hippocampus, and nucleus accumbens. Diabetes. 2021;70(8):1857–1873. doi: 10.2337/db20-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ferris H.A., Perry R.J., Moreira G.V., Shulman G.I., Horton J.D., Kahn C.R. Loss of astrocyte cholesterol synthesis disrupts neuronal function and alters whole-body metabolism. Proc Natl Acad Sci U S A. 2017;114(5):1189–1194. doi: 10.1073/pnas.1620506114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang H., Kulas J.A., Wang C., Holtzman D.M., Ferris H.A., Hansen S.B. Regulation of beta-amyloid production in neurons by astrocyte-derived cholesterol. Proc Natl Acad Sci U S A. 2021;118(33) doi: 10.1073/pnas.2102191118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Glick D., Barth S., Macleod K.F. Autophagy: cellular and molecular mechanisms. J Pathol. 2010;221(1):3–12. doi: 10.1002/path.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Russell R.C., Yuan H.X., Guan K.L. Autophagy regulation by nutrient signaling. Cell Res. 2014;24(1):42–57. doi: 10.1038/cr.2013.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.He C., Klionsky D.J. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zachari M., Ganley I.G. The mammalian ULK1 complex and autophagy initiation. Essays Biochem. 2017;61(6):585–596. doi: 10.1042/EBC20170021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wesselborg S., Stork B. Autophagy signal transduction by ATG proteins: from hierarchies to networks. Cell Mol Life Sci. 2015;72(24):4721–4757. doi: 10.1007/s00018-015-2034-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu W.J., Ye L., Huang W.F., Guo L.J., Xu Z.G., Wu H.L., et al. p62 links the autophagy pathway and the ubiqutin-proteasome system upon ubiquitinated protein degradation. Cell Mol Biol Lett. 2016;21:29. doi: 10.1186/s11658-016-0031-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.O'Neill B.T., Lauritzen H.P., Hirshman M.F., Smyth G., Goodyear L.J., Kahn C.R. Differential role of insulin/IGF-1 receptor signaling in muscle growth and glucose homeostasis. Cell Rep. 2015;11(8):1220–1235. doi: 10.1016/j.celrep.2015.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kressler D., Hurt E., Bassler J. A puzzle of life: crafting ribosomal subunits. Trends Biochem Sci. 2017;42(8):640–654. doi: 10.1016/j.tibs.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 71.Cai W., Sakaguchi M., Kleinridders A., Gonzalez-Del Pino G., Dreyfuss J.M., O'Neill B.T., et al. Domain-dependent effects of insulin and IGF-1 receptors on signalling and gene expression. Nat Commun. 2017;8 doi: 10.1038/ncomms14892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vabulas R.M., Raychaudhuri S., Hayer-Hartl M., Hartl F.U. Protein folding in the cytoplasm and the heat shock response. Cold Spring Harb Perspect Biol. 2010;2(12) doi: 10.1101/cshperspect.a004390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kleinridders A., Lauritzen H.P., Ussar S., Christensen J.H., Mori M.A., Bross P., et al. Leptin regulation of Hsp60 impacts hypothalamic insulin signaling. J Clin Investig. 2013;123(11):4667–4680. doi: 10.1172/JCI67615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wardelmann K., Rath M., Castro J.P., Blumel S., Schell M., Hauffe R., et al. Central acting Hsp10 regulates mitochondrial function, fatty acid metabolism, and insulin sensitivity in the hypothalamus. Antioxidants. 2021;10(5) doi: 10.3390/antiox10050711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Geisler S., Holmstrom K.M., Skujat D., Fiesel F.C., Rothfuss O.C., Kahle P.J., et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12(2):119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 76.Giaume C., Naus C.C., Saez J.C., Leybaert L. Glial connexins and pannexins in the healthy and diseased brain. Physiol Rev. 2021;101(1):93–145. doi: 10.1152/physrev.00043.2018. [DOI] [PubMed] [Google Scholar]
- 77.Bennett M.V., Contreras J.E., Bukauskas F.F., Saez J.C. New roles for astrocytes: gap junction hemichannels have something to communicate. Trends Neurosci. 2003;26(11):610–617. doi: 10.1016/j.tins.2003.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nagao H., Cai W., Wewer Albrechtsen N.J., Steger M., Batista T.M., Pan H., et al. Distinct signaling by insulin and IGF-1 receptors and their extra- and intracellular domains. Proc Natl Acad Sci U S A. 2021;118(17) doi: 10.1073/pnas.2019474118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li Q., Fu J., Xia Y., Qi W., Ishikado A., Park K., et al. Homozygous receptors for insulin and not IGF-1 accelerate intimal hyperplasia in insulin resistance and diabetes. Nat Commun. 2019;10(1):4427. doi: 10.1038/s41467-019-12368-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zoanni B., Brioschi M., Mallia A., Gianazza E., Eligini S., Carini M., et al. Novel insights about albumin in cardiovascular diseases: focus on heart failure. Mass Spectrom Rev. 2021:e21743. doi: 10.1002/mas.21743. [DOI] [PubMed] [Google Scholar]
- 81.Dziegielewska K.M., Evans C.A., New H., Reynolds M.L., Saunders N.R. Synthesis of plasma proteins by rat fetal brain and choroid plexus. Int J Dev Neurosci. 1984;2(3):215–222. doi: 10.1016/0736-5748(84)90015-7. [DOI] [PubMed] [Google Scholar]
- 82.Bruning J.C., Gautam D., Burks D.J., Gillette J., Schubert M., Orban P.C., et al. Role of brain insulin receptor in control of body weight and reproduction. Science. 2000;289(5487):2122–2125. doi: 10.1126/science.289.5487.2122. [DOI] [PubMed] [Google Scholar]
- 83.Kleinridders A., Cai W., Cappellucci L., Ghazarian A., Collins W.R., Vienberg S.G., et al. Insulin resistance in brain alters dopamine turnover and causes behavioral disorders. Proc Natl Acad Sci U S A. 2015;112(11):3463–3468. doi: 10.1073/pnas.1500877112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data of IR-flox and IRKO astrocytes with or without 100 nM insulin or IGF-1 treatment is deposited in Gene Expression Omnibus under accession number GSE213062 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE213062). All the computational scripts used in this study are available at GitHub repository (https://github.com/Jerry-Zhao/2022CaiLabInsIgf1).
I have shared the link to the raw sequencing data and codes in "Data Availability" Section.