Abstract
Infant growth trajectory may influence later-life obesity. Human milk provides a wide range of nutritional and bioactive components that are vital for infant growth. Compared to formula-fed infants, breastfed infants are less likely to develop later-onset obesity, highlighting the potential role of bioactive components present in human milk. Components of particular interest are the human milk microbiota, human milk oligosaccharides (HMOs), short-chain fatty acids (SCFAs), and antimicrobial proteins, each of which influence the infant gut microbiome, which in turn has been associated with infant body composition. SCFAs and antimicrobial proteins from human milk may also systemically influence infant metabolism. Although inconsistent, multiple studies have reported associations between HMOs and infant growth, while studies on other bioactive components in relation to infant growth are sparse. Moreover, these microbiome-related components may interact with each other within the mammary gland. Here, we review the evidence around the impact of human milk microbes, HMOs, SCFAs, and antimicrobial proteins on infant growth. Breastfeeding is a unique window of opportunity to promote optimal infant growth, with aberrant growth trajectories potentially creating short- and long-term public health burdens. Therefore, it is important to understand how bioactive components of human milk influence infant growth.
Keywords: infant growth, human milk, human milk microbiome, infant gut microbiome, human milk oligosaccharides, short chain fatty acids, lactoferrin, lysozyme
1. Introduction
Infant growth trajectory impacts both short- and long-term health outcomes. Excessive weight gain during infancy has been associated with increased obesity risk, while stunted growth has also been associated with obesity, along with delayed cognitive and motor skill development and increased mortality [1,2,3,4,5]. Infant body mass index (BMI) trajectories as early as 1 year can predict childhood obesity at 5 years of age [6]. Half of obese children and 70–80% of obese adolescents remain obese into adulthood [7]; therefore, early higher infant BMI can be a risk factor for later-life obesity. In addition, infants who have catch-up growth during infancy are also more likely to develop obesity [5], suggesting that enduring biological changes may occur during disturbances of infant growth trajectories.
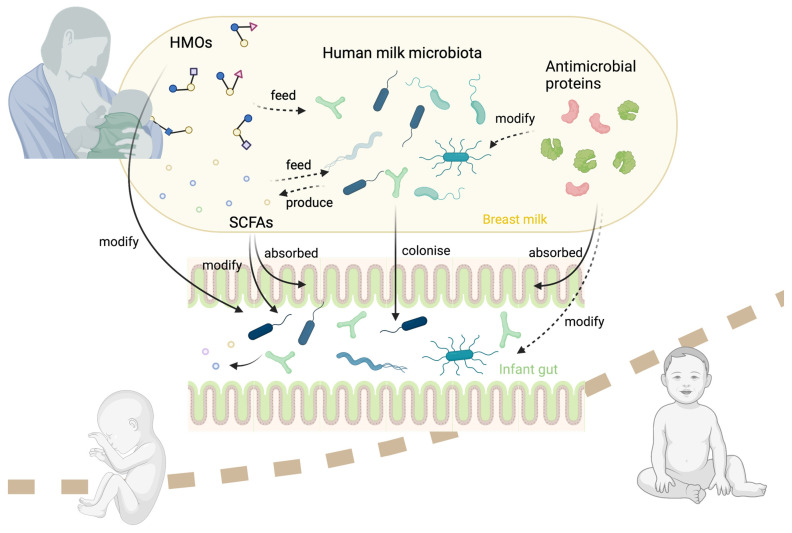
As the optimal source of nutrients for infants, human milk is recommended by the World Health Organisation as the exclusive food source for the first 6 months of life. In addition to its protective effect against a wide array of transmissible and noncommunicable diseases, breastfeeding also promotes healthy infant growth through its macro- and micronutrient content (including carbohydrates, fat, protein, fatty acids, vitamins, and minerals) and bioactive content (including hormones, growth factors, cytokines, microbes, metabolites, and oligosaccharides) [8,9,10]. Given increasing evidence demonstrating a link between the early-life microbiome and later-life body composition, this review will discuss the influence of the milk microbiota and microbiome-modulating factors (human milk oligosaccharides (HMOs), milk short-chain fatty acids (SCFAs), and antimicrobial proteins) on infant growth (Figure 1). In particular, we will focus on the potential of these components of human milk to modulate the infant gut microbiome, and the role of the gut microbiome in infant growth. By synergising data on various microbiome-related human milk components, this narrative review will provide novel insights into the impact of human milk on infant growth. Importantly, given that these microbiome and microbial-related components may interact within the lactating breast, their influence on the infant gut microbiome and growth should be considered holistically.
Figure 1.
Potential interactions between human milk oligosaccharides (HMOs), microbiota, antimicrobial proteins, and short-chain fatty acids (SCFAs), within the mammary gland, and their influence on infant growth via the infant gut microbiota.
2. Review Methodology
A search was performed in PubMed (English) with the following keywords: human/breast milk microbiome, human/breast milk oligosaccharides, human/breast milk short-chain fatty acids, short-chain fatty acids, lactoferrin, lysozyme, breastfeeding, infant growth, infant body composition, infant anthropometrics, infant weight, and infant gut microbiome. The search covers the period prior to November 2022.
3. The Infant Gut Microbiome Can Influence Infant Growth
The gut microbiota has been demonstrated to affect body composition via mechanisms including energy harvesting and metabolic signalling [11,12]. Interestingly, animal studies have revealed that antibiotic treatment in infancy can alter the gut microbiota and lead to adiposity, and that adiposity persists to adulthood after the gut microbiota has recovered [13,14]. Similar results are also reported in human studies [15,16]. Therefore, it is likely that the early gut microbiota can influence infant growth and exert lifelong impacts on host metabolism.
Multiple large cohort studies have reported an association between the infant gut microbiota and growth [17,18,19,20,21]. Higher levels of Bacteroides spp., especially Bacteroides fragilis, and lower levels of Staphylococcus have been consistently reported to be associated with higher BMI in infancy, and to be predictive of childhood obesity [17,18,20,21]. One study of 49 children observed lower numbers of faecal Bifidobacterium and higher numbers of faecal Staphylococcus aureus at 6 and 12 months in infants who later developed overweight and obesity at 7 years of age [22]. In the CHILD cohort, increased diversity (richness) of the infant gut microbiota at 3–4 months of age was associated with higher risk of overweight at 1 year of age [23]. Conversely, another study of the same cohort later found that lower gut microbial diversity (Shannon index) at 1 year of age was associated with a rapid increase in BMI over the first 5 years of age [24]. The disparate results from these two studies may be due to the different time points at which faecal samples were analysed. Indeed, in the same cohort, diversity at 3 months was negatively associated with diversity at 1 year, perhaps suggesting that high diversity in early life may hamper the colonisation of new bacteria later in infancy [24]. While the relationship between the infant gut microbiome and growth remains complex, it may be that the cumulative change in the gut microbiome, rather than the state of the microbiome at a particular time point, influences infant growth [24]. Indeed, the primary influence may be the development of the gut microbiome as an ecological system and the sequential order of colonisation and succession within the infant gut. From this perspective, factors that influence the fitness of the microbiome, such as substrates for and products of bacterial metabolism and antibacterial compounds, should be considered when analysing the relationship between the infant gut microbiome and growth. An early window of opportunity exists for the development of the gut microbiota in relation to body composition, and perturbation during this period can lay the foundations for differing growth trajectories for life.
4. Development of Infant Gut Microbiota and Breastfeeding
Although the “sterile womb” theory is widely accepted, recent findings suggested that infants may be exposed to a low titre of microbes prior to delivery, with viable bacteria and bacterial DNA identified in placentas and amniotic fluid from healthy-term pregnancies [25,26]. However, this theory remains controversial, with opposing results also published [27,28]. Regardless of whether microbial exposure begins in utero, neonates harbour a low-biomass and low-diversity gut microbiome at birth, which develops in terms of diversity, complexity, and microbial load over time until a relatively stable stage is reached [29,30]. However, due to the short-term study design of many studies of the early-life gut microbiome (typically 1–3 years), it is difficult to say when the gut microbiome “matures” to an adult-like state. Recent evidence suggests that the gut microbiome of school-age children (5–12 years), differs significantly to that of adults in terms of both composition and function [31,32,33]. Gut microbiome development is therefore likely to be a gradual process that continues throughout childhood. Nonetheless, the first 2–3 years of life appear to be a particularly dynamic and unstable time within the gut microbiome [31,32,33,34]. From birth, the richness and complexity of the infant gut microbiota gradually increases and eventually reaches a stage that is relatively ecologically stable. During this time, the composition of infant gut microbiome can be influenced by multiple factors, including mode of delivery at birth, gestational age, feeding mode, maternal diet, and antibiotic usage [35,36,37,38,39]. Of all these factors, breastfeeding has been reported to be the most impactful factor, followed by birth mode [32]. Similarly, in another large cohort study of 903 children, intake of human milk was the primary factor associated with the composition and function of the infant gut microbiome [34]. Additionally, consumption of human milk was associated with lower diversity and higher levels of Bifidobacterium spp. in the infant gut [34]. While higher gut microbiome diversity is generally considered optimal for adults, low gut diversity is considered optimal during infancy, especially during breastfeeding. Infants who are breastfed exhibit lower gut microbiome diversity than those who receive formula [23,40], with a strong dominance of Bifidobacterium species [41,42,43,44]. Therefore, early maturation, predicted by higher diversity, may potentially impact the overall development of the gut microbiota negatively. Another study showed that the cessation of breastfeeding, rather than the introduction of solid food, is pivotal in infant gut microbiome diversification, highlighting the role of the bioactive components in human milk [45]. These bioactive components include HMOs, microbes, short-chain fatty acids, and antimicrobial proteins, which together influence the assembly of the infant gut microbiome and infant growth.
5. The Human Milk Microbiome
The human milk microbiome comprises all kingdoms of microorganisms, including bacteria, archaea, microeukaryotes, and viruses, with the bacterial community being the most abundant and well characterised [45]. These microbes may originate from maternal body sites (gut, skin, or the secretions from the pregnant mammary gland) [45,46,47] or from the infant oral cavity via retrograde flow during milk ejections that occur during breastfeeding [48]. In human milk, the most prevalent and abundant bacterial genera are Streptococcus and Staphylococcus, which together typically make up over half of the total profile [49]. Other taxa vary between populations and geographical locations, likely due to maternal and environmental factors, and only constitute a small portion of the milk microbiota [50].
5.1. Human Milk Microbes Colonise the Infant Gut
The infant gut microbiota are acquired both vertically from mother to infant, and horizontally from shared environments and social contacts [51]. One important function of human milk microbiota is the contribution via vertical transmission of microbes from mother to infant, as a small number of shared bacterial strains have been repetitively identified in mother–infant pairs, particularly bifidobacterial strains [52,53,54,55]. Milani et al. reported vertical transmission of Bifidobacterium breve and Bifidobacterium longum subsp. longum strains from milk to the infant gut, which was confirmed by strain isolation and whole-genome sequencing [55]. While B. longum was present in both maternal stool and milk samples, B. breve was only found in milk but not in the maternal gut, although it may have been undetectable in the maternal gut due to low abundance [55]. B. breve has been reported to only contribute to 0.07% of the maternal gut microbiota, but 28.44% and 67.7% of the milk and infant gut microbiota, respectively [56]. However, it should be noted that the mechanism by which these microbes are transferred from the maternal gut to the mammary gland remains unclear. Interestingly, these two species, B. longum and B. breve, together dominate the bifidobacterial genus of exclusively breastfed infants and persist in the infant gut at 6 months of age [57]. This implies a role of milk microbes in the development of the infant gut microbiome, although strain-level studies repeatedly identify only a very small number of shared taxa [46,52,55,58].
5.2. The Potential Role of the Human Milk Microbiota in Infant Growth
The human milk microbiome may influence infant growth by shaping the infant gut microbiota, as indicated by albeit weak evidence to date. Currently, only one study has assessed the relationship between the human milk microbiota and infant growth. Cheema et al. reported significant associations between the milk microbiome and infant body composition at 3 months of age (n = 60) [59]. In this study, the relative abundances of Staphylococcus epidermidis, Streptococcus parasanguis, and Streptococcus lactarius were positively associated with infant anthropometry, adiposity, and fat-free mass, and S. epidermidis was negatively associated with infant length. Associations between the milk microbiome and infant body composition also varied by maternal HMO secretor status. In infants of non-secretor mothers (those which lack the Se gene for the production of fucosylated HMOs), Streptococcus mitis was negatively associated with anthropometry and S. parasanguis was positively associated with BMI-for-age z-score. Interestingly, they also reported associations between maternal anthropometry and body composition with specific milk microbes. Maternal weight, fat mass, fat-free mass, and fat mass index were negatively associated with the relative abundance of S. epidermidis, and fat-free mass was positively associated with Veillonella nakazawae. These results indicate a possible cross-generation transmission of body composition via milk microbiota. Future research should include matching human milk and infant faecal samples with whole-genome sequencing analysis, along with human milk HMO composition, to increase our evidence on associations between the human milk microbiome and composition as a mediator of infant growth.
5.3. The Milk Microbiome as a Potential Contributor to the Intergenerational Transmission of Body Composition
Altered gut bacterial patterns observed in overweight mothers are echoed in their milk microbiome and in infants who later become overweight. Collado et al. reported that the total counts of Bacteroides and Staphylococcus were significantly higher in the gut of overweight women, as measured by fluorescent in situ hybridisation (FISH) and quantitative polymerase chain reaction (qPCR) [60]. During pregnancy, women who experienced higher weight gain had higher counts of Bacteroides and lower counts of Bifidobacterium in their gut. In the same overweight population, higher counts of Staphylococcus and Lactobacillus and lower counts of Bifidobacteria were found in milk samples collected at 1 month and 6 months postnatally [61]. Similarly, lower counts of Bifidobacteria and higher counts of Staphylococcus aureus findings were reported in the stool samples of a group of infants who later became overweight in childhood [22]. These findings suggest that overweight mothers harbor a distinct gut microbiota profile that is reflected in their milk microbiome. Such differences in the maternal microbiome may have consequences for infant gut microbiome colonisation and body composition. However, it remains unclear whether this similarity in the gut microbiome of overweight mothers and their infants who later become overweight is transmitted via the milk microbiome or influenced by other factors such as a shared environment.
Current knowledge implies a plausible route for the transmission of body composition from mother to offspring: overweight and obesity status may change the maternal gut microbiome during pregnancy, which may influence the infant gut microbiome via the milk microbiota during breastfeeding, and ultimately impact infant growth. However, while studies support the notion that gut bacteria influence infant growth [17,19], and that bacteria are vertically transferred from mother to infant via milk [55], the effects of horizontal transmission and host–gene interactions on the development of infant gut microbiota are also likely important [51].
6. Human Milk Oligosaccharides (HMOs)
Beyond the milk microbiome, other components in milk, such as HMOs, have additional effects on the infant gut microbiome and potentially infant growth. HMOs are a group of structurally distinct glycans in human milk [62]. They are the third most abundant solid in human milk after lactose and lipids, and are more abundant than protein [63]. More than 200 different HMOs have been identified, though a group of 19 make up more than 90% of the HMO profile [64]. The composition and concentration of HMOs varies between each mother and across lactation [65,66]. Genetically, the HMO profile can be classified into four groups according to the expression of the genes Se and Le, which are responsible for the expression of two enzymes involved in the synthesis of fucosylated HMOs, α1-2-fucosyltransferase (FUT2) (encoded by the Se gene) and α1-3/4-fucosyltransferase (FUT3) (encoded by the Le gene) [62]. Although the nongenetic factors that influence HMO composition remain largely unknown, some studies have suggested that HMO composition is associated with stage of lactation [67,68,69,70,71], maternal diet [43,70,72,73,74,75,76], and maternal BMI [59,70,73,77,78,79].
Despite the high abundance of HMOs in human milk, they are largely indigestible by the infant. Instead, they serve as an energy source for the microbiota in the infant gut, mainly Bifidobacterium, which degrade HMOs to create metabolites, including short-chain fatty acids (SCFAs) [64]. A recent in vitro study cultivated infant gut microbiota together with three groups of HMO(s), 2′FL only, 2′FL + LNnT, and a mixture of six HMOs (2′FL, LNnT, LNT, diFL, 3′SL, 6′SL) [80]. They found that although all HMOs increased SCFA levels, only the latter two promoted the growth of Bifidobacterium, while the predominant SCFA producer, Ruminococcus, was particularly boosted by the group of six HMOs. These results suggest that HMOs may function synergistically to have a greater influence on colonisation of the infant gut microbiome.
To degrade HMOs into monosaccharides, multiple enzymes from microbes are required for the breakdown of different linkages. In the gut, certain bacterial species express the whole set of enzymes for the digestion of all HMOs, such as Bifidobacterium infantis, Bacteroides fragilis, and Bacteroides vulgatus, while some are only capable of metabolising a subset of the HMOs, such as Bifidobacterium breve and Bifidobacterium longum [81]. The breakdown of HMOs usually occurs in an ordered manner, with modifications removed before the core structures can be degraded [82]. Therefore, some bacteria, such as B. breve, can only function in the presence of bacteria that perform the preceding breakdown steps [83]. This cross-feeding behaviour, especially in the bifidobacterial genus, results in a synergistic thriving of beneficial bacteria. During breastfeeding, these HMO-consuming bacteria thrive, and as a result, suppress the growth of other bacteria, including potential pathogens. Therefore, each mother’s characteristic HMO profile may shape the infant gut microbiome in an individualised way.
HMOs Are Associated with Infant Growth
Multiple studies, although inconsistent, have identified various associations between individual HMOs and/or HMO diversity and infant growth [77,83,84,85,86,87,88,89,90,91]. Most of the previous studies have measured HMOs using high-performance liquid chromatography–mass spectrometry (HPLC-MS) in conjunction with fluorescent derivatisation [59,77,78,84,85,86,87,88,90,91], and two have used nano-LC-chip/time-of-flight (TOF)-MS [83,89]. However, differences in study populations and design, including sample time point(s) and reporting styles for infant growth data, make it difficult to identify any consensus between studies, with contradictory associations reported for some HMOs (Table 1).
Table 1.
Summary of associations between HMOs and infant growth outcomes. Weight includes growth outcomes of weight z score (weight compared to a standard of the same sex), change in weight z score, weight for age z score (weight compared to a standard of the same age and sex), and change in weight for age z score; length includes length z score, change in length z score, length for age z score, and change in length for age z score. Weight for length includes both weight for length z score (weight compared to a standard of the same length and sex) and change in weight for length z score. Head circumference includes head circumference z score, change in head circumference z score, head circumference for age z score, and change in head circumference for age z score. Body mass index includes growth outcomes of body mass index and body mass index for age z score. Fat mass includes fat mass, percentage of fat mass (weight of body fat), and fat mass index (calculated as (fat mass)/height2). Fat-free mass includes fat-free mass (weight of body par that do not contain fat), percentage of fat-free mass, and fat-free mass index (calculated as (fat-free mass)/height2).
| Anthropometrics | |
|---|---|
| Weight |
Positive 3′SL * [59,78,87,89], LNnT [84], 2′FL * [59,77,87], LNFP II [78], 3FL * [59,78,87], DSLNT [84], DFLac * [59,77,87], LSTb ** [78], DFLNH * [59], DSLNH ** [78], DFLNT [59], 6′GL [91] |
|
Negative LNnT [77,87], 6′SL ** [59], LNFP II [84], LNT [83], LNFP I [85], LSTb [87], LSTc [89], DFLNH [77], MFLNH III [91] | |
| Length |
Positive 3′SL * [59,86], LNnT [86], 2′FL [87,90], 3FL ** [59], DFLNH * [59], A-tetra [90], DFLNT [59], LNnDFH [91] |
|
Negative 3′SL [88], LNnT [59,86,87,88], LNT [86], 3FL [91], LNFP I [86], LSTb [87], MFLNH III [91], LNFP V [86], FLNH [86] | |
| Weight for length |
Positive 3′SL [88], LNFP II [78], LNT [78], 3FL ** [78], LSTb ** [78], LSTc [88], LDFT [83], IFLNH 1 [83] |
|
Negative LNFP II [83], LNT [83], LSTa [83], DFLNHc [83] | |
| Head circumference |
Positive 3′SL [77,83], 6′SL [90], LNFP III [88], DFLac [77], MFLNH III [88], LDFT [91], A-tetra [88], LNDFH I [91], LNnDFH [91] |
|
Negative 6′SL [83], LNFP III [89], LNT [83], LNFP I [89], DFLNH [77], MFLNH III [91], LNnFP [86], DFLNHa [89], LNFP V [86] | |
| Body mass index |
Positive DFLac ** [59], LSTb ** [59], DFLNT ** [59] |
|
Negative LNnT [87], 2′FL [86], 6′SL [77], LNT [86], LNnFP [86], LNFP V [86] | |
| Body composition | |
| Fat mass |
Positive 3′SL ** [78], 2′FL * [59], 6′SL ** [78], LNFP III ** [78], LNFP II * [78,85], DSLNT * [78,85], LSTb ** [78], FDSLNH [85], DSLNH ** [78], DFLNT [59] |
|
Negative LNnT [77,85], 6′SL ** [59], LNFP III ** [59] LNFP I [85], DFLNH [77] | |
| Fat-free mass |
Positive 3′SL * [59,90], 3FL ** [59], DFLac ** [59], DFLNH * [59], DFLNT ** [59] |
|
Negative LNT [90], LNFP I [85], LSTc [90] | |
| Fat mass/fat-free mass ratio |
Negative LNFP III ** [59] |
Abbreviations: 3′SL: 3′-Sialyllactose, LNnT: Lacto-n-neotatraose, 2’FL: 2′-Fucosyllactose, 6’SL: 6′-Sialyllactose, LNFP III: Lacto-N-fucopentaose III, LNFP II: Lacto-N-fucopentaose II, LNT: Lacto-N-Tetraose, 3FL: 3-Fucosyllactose, DSLNT: disialyllacto-N-tetraose, DFLac: difucosyllactose, LNFP I: Lacto-N-fucopentaose I, LSTb: sialyl-lacto-N-tetraose, LSTc: sialyl-lacto-N-tetraose, DFLNH: Difucosyllacto-N-hexaose, MFLNH III: Monofucosyllacto-N-hexaose III, LDFT: lactodifucotetraose, A-tetra: A-tetrasaccharide, IFLNH I: fucosyl-paralacto-N-hexaose I, FDSLNH: Fucosyl-disialyllacto-N-hexose, DSLNH: disialyllacto-N-hexaose, DFLNT: difucosyllacto-N-tetrose, LNDFH I: Lacto-N-difucohexaose I, LNnDFH: lactoN-neodifucohexaose, 6′GL: 6′galactosyllactose, LNnFP: lacto-N-neofucopentaose, DFLNHa: difucosyllacto-N-hexaose (a), LSTa: LS-Tetrasaccharide a, DFLNHc: difucosyllacto-N-hexaose c, LNFP V: lacto-N-fucopentaose-V, FLNH: fucosyllacto-N-hexaose. * Associations with both concentration and infant intake. ** Associations with infant intake only.
Associations between infant anthropometrics and the most abundant HMO, 3′SL (3′ sialyllactose), are widely reported [59,77,78,83,86,87,89,90]. Concentrations of 3′SL have been positively associated with infant weight [89], length [77], head circumference [77,83], weight for length [88], fat mass [78], and fat-free mass [59,90], except in one study that reported a negative association with infant length [88]. This discrepancy may be due to the time at which infant anthropometrics were measured (birth to 4 months in one study, and 5 and 9 months in the other) [77,88]. Further research is needed to determine if the relationship between HMOs and infant body composition changes over the course of lactation.
Further, relationships are likely to vary depending on whether HMO concentration or intake is measured [45,59]. Intake of 3′SL at 2 months of age has been positively associated with infant weight, length, and fat-free mass, which is consistent with studies measuring 3′SL concentrations [59,78,90]. However, results in relation to intake are not always consistent with that of the concentration. While 3′SL intake was also associated with infant fat mass [78], such association has not been reported in concentration studies. Cheema et al. measured both HMO concentrations and intakes and found that except for 3′SL, none of the associations between HMO intakes and infant growth held true for concentrations, suggesting that concentration itself is not adequate for assessing associations between HMO and infant growth [59]. However, results also differ between studies in which intakes were measured. While both Saben et al. and Cheema et al. identified positive associations between 3′SL and 3FL (3 fucosyllactose) and infant growth, 6′SL (6′ sialyllactose) was positively associated with infant fat mass in one study and negatively in the other [59,78]. Similarly, many associations were identified in one study but not the other. This discrepancy is likely due to differences in the measurement of milk intake [59,78]. Saben et al. estimated the intake by measuring body weight before and after one feed and the feeding frequency, while Cheema et al. measured the total intake over 24 h. Nevertheless, the collective evidence demonstrates that infant intake of HMOs appears to be one of the factors that can regulate infant growth.
Maternal secretor status influences HMO composition, particularly 2′FL (2′ fucosyllactose) and LNnT (lacto-N-neotetraose), both of which have been added to commercial formulas [92]. 2′FL concentration has been positively associated with infant weight [77,87], length [89,90], and fat mass [77] in both healthy and malnourished populations. 2′FL intake also has been positively associated with weight and fat mass [59]. Another typical secretor, HMO LNnT, which is negatively correlated with 2′FL, is negatively associated with infant weight [77,87], length [86,87], BMI [77], fat mass [77], and fat mass percentage [85]. An opposing result with infant weight gain has also been reported, but only in non-secretor mothers [84]. Although some studies have reported different outcomes between individual HMOs and infant growth depending on maternal secretor status, secretor status by itself does not predict infant growth trajectory. Therefore, the role of HMOs in infant growth may be independent of secretion status, or potentially these results have been impaired by measuring concentrations instead of total infant intakes.
7. Short-Chain Fatty Acids
Short-chain fatty acids are the microbial metabolites of fibre fermentation that are produced in the colon. Some SCFAs, particularly butyrate, are absorbed locally by colonocytes, while the rest are transported to the portal vein and metabolised by the liver or distributed systemically around the body [93]. Although only a small concentration of SCFAs enter the peripheral circulation, they participate in a wide range of metabolic processes by regulating gene expression and binding to G-protein-coupled receptors (GPRs) [94]. In adults, SCFAs have been proposed to affect appetite control, energy harvesting, energy expenditure, and glucose homeostasis [95,96]. In vitro, acetate, butyrate, and propionate have been shown to stimulate the production of the satiety hormone peptide YY and glucagon-like peptide 1 [97,98]. Rodent studies have indicated that acetate can cross the blood–brain barrier and reach the hypothalamus, where it suppresses appetite [99]. Moreover, greater levels of energy expenditure after SCFA administration have been observed in both mouse studies and human studies [100,101,102] indicating potential for interventions designed to optimise the development of body composition early in life. This evidence supports the direct influence of SCFAs on host metabolism through multiple mechanisms.
In addition to direct influence, SCFAs may also indirectly influence host metabolism by impacting the gut microbiome. SCFAs are not only products of but also substrates for microbial metabolisms. Unlike acetate, which can be produced by a wide range of bacteria, pathways for propionate and butyrate production are relatively conserved in a few bacterial genera [103,104]. During the production of SCFAs, intermediate products such as succinate and lactate will be further utilised by a subset of bacteria. Additionally, acetate can be directly utilised by the butyrate-producing bacteria through the acetyl-CoA pathway [105]. This cross-feeding behaviour during the production of SCFA has been shown to promote the growth of certain bacteria and the diversity of microbiota in the gut, which in turn may influence host metabolism [106].
Human Milk SCFAs and Infant Growth
The SCFAs acetate, butyrate, and formate have been identified in human milk [107,108,109]. Given that SCFAs participate in host metabolism, it is likely that SCFAs in human milk may influence infant growth. Currently, only one study has assessed the associations between milk SCFA levels and infant growth outcomes. Prentice et al. analysed SCFA levels in milk samples taken 4–8 weeks postpartum (N = 619) [107]. Child weight, length, and skinfold thicknesses (triceps, subscapular, flank, quadriceps) were measured at 3, 12, and 24 months of age. Human milk butyrate was negatively associated with infant BMI and skinfold thicknesses at 12 months of age, formate was negatively associated with infant BMI at all time points, and acetate was negatively associated with infant skinfold thickness at 3 and 24 months of age. These results highlighted the potential of SCFAs in human milk to influence infant growth, even beyond the period of exclusive breast feeding.
However, due to the dearth of studies on human milk SCFAs, their functions and mechanisms of action remain largely unclear. SCFAs may be transported to the milk from the maternal gut via the circulatory system, or they may be produced locally since milk contains both microbes and HMOs. Compared to the SCFAs identified in faeces and blood, the major SCFA propionate has not been identified in human milk [107,108,109]. Current studies focus on the function of SCFAs produced in the colon, but whether they can survive when consumed by infants in milk and reach the gut intact remains unknown. Therefore, more mechanistic studies are needed to identify both the source and function of human milk SCFAs.
8. Antimicrobial Proteins—Lactoferrin and Lysozyme
Lactoferrin and lysozyme are two of the most studied antimicrobial proteins in human milk [110]. Lactoferrin functions as carrier of iron in human milk [111,112]. It has been shown to inhibit infections of pathogenic bacteria [113,114,115], viruses [116,117,118], fungi [119,120,121], and parasites [122,123,124]. Conversely, lactoferrin peptides also have a strong bifidogenic effect and promote some species of lactobacilli, both of which are thought of as beneficial infant gut taxa [125,126,127]. As one of the major enzymes of human milk, lysozyme lyses Gram-positive bacteria, and when in corporation with lactoferrin, some Gram-negative bacteria as well [128,129]. A piglet model has shown that consumption of lysozyme-rich milk results in an increase in Bifidobacteriaceae and Lactobacillaceae and a decrease in pathogens in the gut [130]. The influence of lysozyme on the gut microbiome was also identified in a drosophila model [131]. Therefore, these antimicrobial proteins may influence infant growth indirectly through the modification of the infant gut microbiome. Apart from their antimicrobial properties, lactoferrin also stimulates intestinal and bone cell proliferation [132,133,134], which may directly influence infant growth.
Evidence for lactoferrin and lysozyme influencing infant growth is sparse. Bovine lactoferrin supplementation in formula (N = 10–12) has been associated with increased infant weight (6 months) and length (4 and 6 months) [135]. Similar results (increased length/height) were reported in a group of older children (12–36 months; N = 26) who received direct bovine lactoferrin supplementation [136]. However, it is difficult to make any conclusions as human milk lactoferrin is not comparable to bovine lactoferrin. Lysozyme supplementation of donor milk (N = 64) and higher concentrations in human milk (N = 42) are both associated with increased infant weight [137,138]. Gridneva et al. measured the total intake of lactoferrin and lysozyme (N = 20; 2–12 months) and found that the intake of lactoferrin was negatively associated with fat-free mass and lysozyme positively with fat mass [139]. Although weak evidence supports the associations between lactoferrin and lysozyme and infant growth, the mechanisms remain to be unveiled. Further studies are needed to elucidate the association between antimicrobial proteins in human milk and infant growth.
9. Interactions of Microbiome and Microbiome-Related Components within the Lactating Mammary Gland
As both HMOs and SCFAs can influence the infant gut microbiota, they may also modify the human milk microbiota through the same mechanisms. HMOs may feed a certain group of bacteria in human milk, creating SCFAs as a by-product (Figure 1). SCFAs may also be metabolised by bacteria within the lactating mammary gland. In addition, as known for their antimicrobial property, lactoferrin and lysozyme may also participate in this interaction.
Although it is currently unclear whether digestion of HMOs occurs within the mammary gland, several studies have suggested that the composition of HMOs is associated with the milk microbiota composition. Aakko et al. observed a positive correlation between total HMO concentration and counts of Bifidobacterium spp. in colostrum in a small study (N = 11) [140]. This association was more pronounced when HMOs were grouped by structure. Positive associations were identified between B. breve and sialylated HMOs and between B. longum and non-fucosylated/non-sialylated HMOs. Similarly, another larger cohort study has demonstrated an association between milk microbiota and individual HMOs (N = 393) [141]. Specifically, the relative abundance of Bifidobacterium was negatively associated with the concentration of DLNH, and in non-secretors only the relative abundance of Staphylococcus was positively associated with concentration of sialylated HMOs. These findings together imply a potential role of HMO in modifying the milk microbiota before reaching the infant gut.
Unlike studies in HMOs, to date no study has examined the associations between microbiota and SCFAs in human milk. If SCFAs are produced from bacterial HMO fermentation within the mammary gland, they may be associated with both the HMO and microbiota content of human milk. Milk SCFAs may also act as a substrate for milk bacterial metabolism independently of HMOs.
Although currently no studies have been carried out to assess the influence of lactoferrin and lysozyme on the human milk microbiome, they also have the potential to influence microbiome composition for their antimicrobial properties. They may interact with the human milk microbiota in a similar manner as in the gut microbiome, as shown in animal and human studies [130,131]. Like other microbiome-related products in human milk, antimicrobial proteins are likely to participate in the regulation of microbiota in human milk and infant gut.
Further studies are needed to elucidate the association between milk SCFAs, microbiota, HMOs, and antimicrobial proteins. If they interact with each other within the mammary gland, they may influence infant growth in an integrated manner. This highlights the importance of viewing lactation as a biological system, with human milk as a whole, promoting healthy infant growth. This may also partially explain why associations between HMOs and infant growth/body composition are inconsistent, as other human milk components may be involved.
10. Summary and Future Directions
In this review, we summarised the current evidence regarding the potential function of the human milk microbiome and microbiome-related products on infant growth, potentially via the modification of the infant gut microbiome and other mechanisms. As these components may interact with one another, they may synergistically influence the human milk microbiome, the infant gut microbiome, and infant growth. While numerous studies have assessed the influence of HMOs on infant growth, evidence for other components is limited. Given the fact that the infant gut microbiome is dynamic in early life, longitudinal studies of larger sample sizes are needed to assess associations between these milk components, the infant microbiome, and infant growth. Importantly, given the potential interactions between these components, an integrative approach is required to holistically assess the impact of human milk on infant growth. This review also highlighted the importance of analysing intakes rather than concentrations of human milk components. Future studies should measure daily intake when possible, to more accurately assess the impact of human milk components on infant growth. Integrated longitudinal studies are required to ascertain the influence of the human milk microbiota and microbiome-related products on growth throughout infancy and beyond, with a focus on their interactions with the infant gut microbiome.
Author Contributions
Writing—original draft preparation, J.M.; writing—review and editing, L.S., D.G. and D.J.P.; supervision, L.S., D.G., D.J.P. and C.T.L. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
J.M., L.S., C.T.L. and D.G. are supported by an unrestricted research grant from Medela AG, administered by The University of Western Australia. This funding body had no input into manuscript design, data interpretation, or the decision to publish.
Funding Statement
J.M., L.S., C.T.L. and D.G. are supported by an unrestricted research grant from Medela AG, administered by The University of Western Australia. J.M. was supported by an additional SIRF (Scholarships for International Research Fees) scholarship from The University of Western Australia and Ad Hoc Postgraduate Scholarship from Medela AG. D.J.P. was supported by the Telethon Kids Institute Ascend Fellowship.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cheung Y., Yip P., Karlberg J. Fetal Growth, Early Postnatal Growth and Motor Development in Pakistani Infants. Int. J. Epidemiol. 2001;30:66–72. doi: 10.1093/ije/30.1.66. [DOI] [PubMed] [Google Scholar]
- 2.Belfort M.B., Rifas-Shiman S.L., Rich-Edwards J.W., Kleinman K.P., Oken E., Gillman M.W. Infant Growth and Child Cognition at 3 Years of Age. Pediatrics. 2008;122:e689–e695. doi: 10.1542/peds.2008-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ong K., Loos R. Rapid Infancy Weight Gain and Subsequent Obesity: Systematic Reviews and Hopeful Suggestions. Acta Paediatr. 2006;95:904–908. doi: 10.1080/08035250600719754. [DOI] [PubMed] [Google Scholar]
- 4.Belfort M.B., Rifas-Shiman S.L., Sullivan T., Collins C.T., McPhee A.J., Ryan P., Kleinman K.P., Gillman M.W., Gibson R.A., Makrides M. Infant Growth Before and After Term: Effects on Neurodevelopment in Preterm Infants. Pediatrics. 2011;128:e899–e906. doi: 10.1542/peds.2011-0282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffman D.J., Sawaya A.L., Verreschi I., Tucker K.L., Roberts S.B. Why Are Nutritionally Stunted Children at Increased Risk of Obesity? Studies of Metabolic Rate and Fat Oxidation in Shantytown Children from São Paulo, Brazil. Am. J. Clin. Nutr. 2000;72:702–707. doi: 10.1093/ajcn/72.3.702. [DOI] [PubMed] [Google Scholar]
- 6.Gittner L.S., Ludington-Hoe S.M., Haller H.S. Utilising Infant Growth to Predict Obesity Status at 5 Years: Infant Obesity. J. Paediatr. Child Health. 2013;49:564–574. doi: 10.1111/jpc.12283. [DOI] [PubMed] [Google Scholar]
- 7.Dietz W.H. Childhood Weight Affects Adult Morbidity and Mortality. J. Nutr. 1998;128:411S–414S. doi: 10.1093/jn/128.2.411S. [DOI] [PubMed] [Google Scholar]
- 8.Gillman M.W. Risk of Overweight Among Adolescents Who Were Breastfed as Infants. JAMA. 2001;285:2461. doi: 10.1001/jama.285.19.2461. [DOI] [PubMed] [Google Scholar]
- 9.Dietz W.H. Breastfeeding May Help Prevent Childhood Overweight. JAMA. 2001;285:2506. doi: 10.1001/jama.285.19.2506. [DOI] [PubMed] [Google Scholar]
- 10.Dewey K.G. Is Breastfeeding Protective Against Child Obesity? J. Hum. Lact. 2003;19:9–18. doi: 10.1177/0890334402239730. [DOI] [PubMed] [Google Scholar]
- 11.Turnbaugh P.J., Ley R.E., Mahowald M.A., Magrini V., Mardis E.R., Gordon J.I. An Obesity-Associated Gut Microbiome with Increased Capacity for Energy Harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 12.Le Chatelier E., Nielsen T., Qin J., Prifti E., Hildebrand F., Falony G., Almeida M., Arumugam M., Batto J.-M., Kennedy S., et al. Richness of Human Gut Microbiome Correlates with Metabolic Markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 13.Cox L.M., Yamanishi S., Sohn J., Alekseyenko A.V., Leung J.M., Cho I., Kim S.G., Li H., Gao Z., Mahana D., et al. Altering the Intestinal Microbiota during a Critical Developmental Window Has Lasting Metabolic Consequences. Cell. 2014;158:705–721. doi: 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho I., Yamanishi S., Cox L., Methé B.A., Zavadil J., Li K., Gao Z., Mahana D., Raju K., Teitler I., et al. Antibiotics in Early Life Alter the Murine Colonic Microbiome and Adiposity. Nature. 2012;488:621–626. doi: 10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang M., Differding M.K., Benjamin-Neelon S.E., Østbye T., Hoyo C., Mueller N.T. Association of Prenatal Antibiotics with Measures of Infant Adiposity and the Gut Microbiome. Ann. Clin. Microbiol. Antimicrob. 2019;18:18. doi: 10.1186/s12941-019-0318-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Azad M.B., Bridgman S.L., Becker A.B., Kozyrskyj A.L. Infant Antibiotic Exposure and the Development of Childhood Overweight and Central Adiposity. Int. J. Obes. 2014;38:1290–1298. doi: 10.1038/ijo.2014.119. [DOI] [PubMed] [Google Scholar]
- 17.Scheepers L.E.J.M., Penders J., Mbakwa C.A., Thijs C., Mommers M., Arts I.C.W. The Intestinal Microbiota Composition and Weight Development in Children: The KOALA Birth Cohort Study. Int. J. Obes. 2015;39:16–25. doi: 10.1038/ijo.2014.178. [DOI] [PubMed] [Google Scholar]
- 18.Vael C., Verhulst S.L., Nelen V., Goossens H., Desager K.N. Intestinal Microflora and Body Mass Index during the First Three Years of Life: An Observational Study. Gut. Pathog. 2011;3:8. doi: 10.1186/1757-4749-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stanislawski M.A., Dabelea D., Wagner B.D., Iszatt N., Dahl C., Sontag M.K., Knight R., Lozupone C.A., Eggesbø M. Gut Microbiota in the First 2 Years of Life and the Association with Body Mass Index at Age 12 in a Norwegian Birth Cohort. mBio. 2018;9:e01751-18. doi: 10.1128/mBio.01751-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korpela K., Renko M., Vänni P., Paalanne N., Salo J., Tejesvi M.V., Koivusaari P., Ojaniemi M., Pokka T., Kaukola T., et al. Microbiome of the First Stool and Overweight at Age 3 Years: A Prospective Cohort Study. Pediatr. Obes. 2020;15:e12680. doi: 10.1111/ijpo.12680. [DOI] [PubMed] [Google Scholar]
- 21.White R.A., Bjørnholt J.V., Baird D.D., Midtvedt T., Harris J.R., Pagano M., Hide W., Rudi K., Moen B., Iszatt N., et al. Novel Developmental Analyses Identify Longitudinal Patterns of Early Gut Microbiota That Affect Infant Growth. PLoS Comput. Biol. 2013;9:e1003042. doi: 10.1371/journal.pcbi.1003042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalliomäki M., Carmen Collado M., Salminen S., Isolauri E. Early Differences in Fecal Microbiota Composition in Children May Predict Overweight. Am. J. Clin. Nutr. 2008;87:534–538. doi: 10.1093/ajcn/87.3.534. [DOI] [PubMed] [Google Scholar]
- 23.Forbes J.D., Azad M.B., Vehling L., Tun H.M., Konya T.B., Guttman D.S., Field C.J., Lefebvre D., Sears M.R., Becker A.B., et al. Association of Exposure to Formula in the Hospital and Subsequent Infant Feeding Practices With Gut Microbiota and Risk of Overweight in the First Year of Life. JAMA Pediatr. 2018;172:e181161. doi: 10.1001/jamapediatrics.2018.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reyna M.E., Petersen C., Dai D.L.Y., Dai R., Becker A.B., Azad M.B., Miliku K., Lefebvre D.L., Moraes T.J., Mandhane P.J., et al. Longitudinal Body Mass Index Trajectories at Preschool Age: Children with Rapid Growth Have Differential Composition of the Gut Microbiota in the First Year of Life. Int. J. Obes. 2022;46:1351–1358. doi: 10.1038/s41366-022-01117-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiménez E., Fernández L., Marín M.L., Martín R., Odriozola J.M., Nueno-Palop C., Narbad A., Olivares M., Xaus J., Rodríguez J.M. Isolation of Commensal Bacteria from Umbilical Cord Blood of Healthy Neonates Born by Cesarean Section. Curr. Microbiol. 2005;51:270–274. doi: 10.1007/s00284-005-0020-3. [DOI] [PubMed] [Google Scholar]
- 26.Aagaard K., Ma J., Antony K.M., Ganu R., Petrosino J., Versalovic J. The Placenta Harbors a Unique Microbiome. Sci. Transl. Med. 2014;6:237ra65. doi: 10.1126/scitranslmed.3008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kennedy K.M., Gerlach M.J., Adam T., Heimesaat M.M., Rossi L., Surette M.G., Sloboda D.M., Braun T. Fetal Meconium Does Not Have a Detectable Microbiota before Birth. Nat. Microbiol. 2021;6:865–873. doi: 10.1038/s41564-021-00904-0. [DOI] [PubMed] [Google Scholar]
- 28.Kuperman A., Zimmerman A., Hamadia S., Ziv O., Gurevich V., Fichtman B., Gavert N., Straussman R., Rechnitzer H., Barzilay M., et al. Deep Microbial Analysis of Multiple Placentas Shows No Evidence for a Placental Microbiome. BJOG Int. J. Obstet. Gynaecol. 2020;127:159–169. doi: 10.1111/1471-0528.15896. [DOI] [PubMed] [Google Scholar]
- 29.Bergström A., Skov T.H., Bahl M.I., Roager H.M., Christensen L.B., Ejlerskov K.T., Mølgaard C., Michaelsen K.F., Licht T.R. Establishment of Intestinal Microbiota during Early Life: A Longitudinal, Explorative Study of a Large Cohort of Danish Infants. Appl. Env. Microbiol. 2014;80:2889–2900. doi: 10.1128/AEM.00342-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagpal R., Tsuji H., Takahashi T., Nomoto K., Kawashima K., Nagata S., Yamashiro Y. Ontogenesis of the Gut Microbiota Composition in Healthy, Full-Term, Vaginally Born and Breast-Fed Infants over the First 3 Years of Life: A Quantitative Bird’s-Eye View. Front. Microbiol. 2017;8:1388. doi: 10.3389/fmicb.2017.01388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hollister E.B., Riehle K., Luna R.A., Weidler E.M., Rubio-Gonzales M., Mistretta T.-A., Raza S., Doddapaneni H.V., Metcalf G.A., Muzny D.M., et al. Structure and Function of the Healthy Pre-Adolescent Pediatric Gut Microbiome. Microbiome. 2015;3:36. doi: 10.1186/s40168-015-0101-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ou Y., Belzer C., Smidt H., de Weerth C. Development of the Gut Microbiota in Healthy Children in the First Ten Years of Life: Associations with Internalizing and Externalizing Behavior. Gut. Microbes. 2022;14:2038853. doi: 10.1080/19490976.2022.2038853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roswall J., Olsson L.M., Kovatcheva-Datchary P., Nilsson S., Tremaroli V., Simon M.-C., Kiilerich P., Akrami R., Krämer M., Uhlén M., et al. Developmental Trajectory of the Healthy Human Gut Microbiota during the First 5 Years of Life. Cell Host Microbe. 2021;29:765–776.e3. doi: 10.1016/j.chom.2021.02.021. [DOI] [PubMed] [Google Scholar]
- 34.Stewart C.J., Ajami N.J., O’Brien J.L., Hutchinson D.S., Smith D.P., Wong M.C., Ross M.C., Lloyd R.E., Doddapaneni H., Metcalf G.A., et al. Temporal Development of the Gut Microbiome in Early Childhood from the TEDDY Study. Nature. 2018;562:583–588. doi: 10.1038/s41586-018-0617-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chernikova D.A., Madan J.C., Housman M.L., Zain-ul-abideen M., Lundgren S.N., Morrison H.G., Sogin M.L., Williams S.M., Moore J.H., Karagas M.R., et al. The Premature Infant Gut Microbiome during the First 6 Weeks of Life Differs Based on Gestational Maturity at Birth. Pediatr. Res. 2018;84:71–79. doi: 10.1038/s41390-018-0022-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lundgren S.N., Madan J.C., Emond J.A., Morrison H.G., Christensen B.C., Karagas M.R., Hoen A.G. Maternal Diet during Pregnancy Is Related with the Infant Stool Microbiome in a Delivery Mode-Dependent Manner. Microbiome. 2018;6:109. doi: 10.1186/s40168-018-0490-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitchell C.M., Mazzoni C., Hogstrom L., Bryant A., Bergerat A., Cher A., Pochan S., Herman P., Carrigan M., Sharp K., et al. Delivery Mode Affects Stability of Early Infant Gut Microbiota. Cell Rep. Med. 2020;1:100156. doi: 10.1016/j.xcrm.2020.100156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gasparrini A.J., Crofts T.S., Gibson M.K., Tarr P.I., Warner B.B., Dantas G. Antibiotic Perturbation of the Preterm Infant Gut Microbiome and Resistome. Gut. Microbes. 2016;7:443–449. doi: 10.1080/19490976.2016.1218584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cong X., Judge M., Xu W., Diallo A., Janton S., Brownell E.A., Maas K., Graf J. Influence of Feeding Type on Gut Microbiome Development in Hospitalized Preterm Infants. Nurs. Res. 2017;66:123–133. doi: 10.1097/NNR.0000000000000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pannaraj P.S., Li F., Cerini C., Bender J.M., Yang S., Rollie A., Adisetiyo H., Zabih S., Lincez P.J., Bittinger K., et al. Association Between Breast Milk Bacterial Communities and Establishment and Development of the Infant Gut Microbiome. JAMA Pediatr. 2017;171:647. doi: 10.1001/jamapediatrics.2017.0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshioka H., Iseki K., Fujita K. Development and Differences of Intestinal Flora in the Neonatal Period in Breast-Fed and Bottle-Fed Infants. Pediatrics. 1983;72:317–321. doi: 10.1542/peds.72.3.317. [DOI] [PubMed] [Google Scholar]
- 42.Zhao J., Yi W., Liu B., Dai Y., Jiang T., Chen S., Wang J., Feng B., Qiao W., Liu Y., et al. MFGM Components Promote Gut Bifidobacterium Growth in Infant and in Vitro. Eur. J. Nutr. 2022;61:277–288. doi: 10.1007/s00394-021-02638-5. [DOI] [PubMed] [Google Scholar]
- 43.Ho N.T., Li F., Lee-Sarwar K.A., Tun H.M., Brown B.P., Pannaraj P.S., Bender J.M., Azad M.B., Thompson A.L., Weiss S.T., et al. Meta-Analysis of Effects of Exclusive Breastfeeding on Infant Gut Microbiota across Populations. Nat. Commun. 2018;9:4169. doi: 10.1038/s41467-018-06473-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma J., Li Z., Zhang W., Zhang C., Zhang Y., Mei H., Zhuo N., Wang H., Wang L., Wu D. Comparison of Gut Microbiota in Exclusively Breast-Fed and Formula-Fed Babies: A Study of 91 Term Infants. Sci. Rep. 2020;10:15792. doi: 10.1038/s41598-020-72635-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stinson L.F., Sindi A.S.M., Cheema A.S., Lai C.T., Mühlhäusler B.S., Wlodek M.E., Payne M.S., Geddes D.T. The Human Milk Microbiome: Who, What, When, Where, Why, and How? Nutr. Rev. 2021;79:529–543. doi: 10.1093/nutrit/nuaa029. [DOI] [PubMed] [Google Scholar]
- 46.Jost T., Lacroix C., Braegger C.P., Rochat F., Chassard C. Vertical Mother-Neonate Transfer of Maternal Gut Bacteria via Breastfeeding: Mother-Neonate Bacterial Transfer. Env. Microbiol. 2014;16:2891–2904. doi: 10.1111/1462-2920.12238. [DOI] [PubMed] [Google Scholar]
- 47.Fernández L., Langa S., Martín V., Maldonado A., Jiménez E., Martín R., Rodríguez J.M. The Human Milk Microbiota: Origin and Potential Roles in Health and Disease. Pharmacol. Res. 2013;69:1–10. doi: 10.1016/j.phrs.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 48.McGuire M.K., McGuire M.A. Got Bacteria? The Astounding, yet Not-so-Surprising, Microbiome of Human Milk. Curr. Opin. Biotechnol. 2017;44:63–68. doi: 10.1016/j.copbio.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 49.Fitzstevens J.L., Smith K.C., Hagadorn J.I., Caimano M.J., Matson A.P., Brownell E.A. Systematic Review of the Human Milk Microbiota. Nutr. Clin. Pr. 2017;32:354–364. doi: 10.1177/0884533616670150. [DOI] [PubMed] [Google Scholar]
- 50.Gomez-Gallego C., Garcia-Mantrana I., Salminen S., Collado M.C. The Human Milk Microbiome and Factors Influencing Its Composition and Activity. Semin. Fetal Neonatal Med. 2016;21:400–405. doi: 10.1016/j.siny.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 51.Moeller A.H., Suzuki T.A., Phifer-Rixey M., Nachman M.W. Transmission Modes of the Mammalian Gut Microbiota. Science. 2018;362:453–457. doi: 10.1126/science.aat7164. [DOI] [PubMed] [Google Scholar]
- 52.Asnicar F., Manara S., Zolfo M., Truong D.T., Scholz M., Armanini F., Ferretti P., Gorfer V., Pedrotti A., Tett A., et al. Studying Vertical Microbiome Transmission from Mothers to Infants by Strain-Level Metagenomic Profiling. mSystems. 2017;2:e00164-16. doi: 10.1128/mSystems.00164-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Solís G., de los Reyes-Gavilan C.G., Fernández N., Margolles A., Gueimonde M. Establishment and Development of Lactic Acid Bacteria and Bifidobacteria Microbiota in Breast-Milk and the Infant Gut. Anaerobe. 2010;16:307–310. doi: 10.1016/j.anaerobe.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 54.Murphy K., Curley D., O’Callaghan T.F., O’Shea C.-A., Dempsey E.M., O’Toole P.W., Ross R.P., Ryan C.A., Stanton C. The Composition of Human Milk and Infant Faecal Microbiota Over the First Three Months of Life: A Pilot Study. Sci. Rep. 2017;7:40597. doi: 10.1038/srep40597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Milani C., Mancabelli L., Lugli G.A., Duranti S., Turroni F., Ferrario C., Mangifesta M., Viappiani A., Ferretti P., Gorfer V., et al. Exploring Vertical Transmission of Bifidobacteria from Mother to Child. Appl. Env. Microbiol. 2015;81:7078–7087. doi: 10.1128/AEM.02037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kordy K., Gaufin T., Mwangi M., Li F., Cerini C., Lee D.J., Adisetiyo H., Woodward C., Pannaraj P.S., Tobin N.H., et al. Contributions to Human Breast Milk Microbiome and Enteromammary Transfer of Bifidobacterium Breve. PLoS ONE. 2020;15:e0219633. doi: 10.1371/journal.pone.0219633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lou Y.C., Olm M.R., Diamond S., Crits-Christoph A., Firek B.A., Baker R., Morowitz M.J., Banfield J.F. Infant Gut Strain Persistence Is Associated with Maternal Origin, Phylogeny, and Traits Including Surface Adhesion and Iron Acquisition. Cell Rep. Med. 2021;2:100393. doi: 10.1016/j.xcrm.2021.100393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Duranti S., Lugli G.A., Mancabelli L., Armanini F., Turroni F., James K., Ferretti P., Gorfer V., Ferrario C., Milani C., et al. Maternal Inheritance of Bifidobacterial Communities and Bifidophages in Infants through Vertical Transmission. Microbiome. 2017;5:66. doi: 10.1186/s40168-017-0282-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheema A.S., Gridneva Z., Furst A.J., Roman A.S., Trevenen M.L., Turlach B.A., Lai C.T., Stinson L.F., Bode L., Payne M.S., et al. Human Milk Oligosaccharides and Bacterial Profile Modulate Infant Body Composition during Exclusive Breastfeeding. IJMS. 2022;23:2865. doi: 10.3390/ijms23052865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Collado M.C., Isolauri E., Laitinen K., Salminen S. Distinct Composition of Gut Microbiota during Pregnancy in Overweight and Normal-Weight Women. Am. J. Clin. Nutr. 2008;88:894–899. doi: 10.1093/ajcn/88.4.894. [DOI] [PubMed] [Google Scholar]
- 61.Collado M.C., Laitinen K., Salminen S., Isolauri E. Maternal Weight and Excessive Weight Gain during Pregnancy Modify the Immunomodulatory Potential of Breast Milk. Pediatr. Res. 2012;72:77–85. doi: 10.1038/pr.2012.42. [DOI] [PubMed] [Google Scholar]
- 62.Bode L. Human Milk Oligosaccharides: Every Baby Needs a Sugar Mama. Glycobiology. 2012;22:1147–1162. doi: 10.1093/glycob/cws074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bode L. The Functional Biology of Human Milk Oligosaccharides. Early Hum. Dev. 2015;91:619–622. doi: 10.1016/j.earlhumdev.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 64.Zivkovic A.M., German J.B., Lebrilla C.B., Mills D.A. Human Milk Glycobiome and Its Impact on the Infant Gastrointestinal Microbiota. Proc. Natl. Acad. Sci. USA. 2011;108:4653–4658. doi: 10.1073/pnas.1000083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Soyyılmaz B., Mikš M.H., Röhrig C.H., Matwiejuk M., Meszaros-Matwiejuk A., Vigsnæs L.K. The Mean of Milk: A Review of Human Milk Oligosaccharide Concentrations throughout Lactation. Nutrients. 2021;13:2737. doi: 10.3390/nu13082737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thum C., Wall C.R., Weiss G.A., Wang W., Szeto I.M.-Y., Day L. Changes in HMO Concentrations throughout Lactation: Influencing Factors, Health Effects and Opportunities. Nutrients. 2021;13:2272. doi: 10.3390/nu13072272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thurl S., Munzert M., Boehm G., Matthews C., Stahl B. Systematic Review of the Concentrations of Oligosaccharides in Human Milk. Nutr. Rev. 2017;75:920–933. doi: 10.1093/nutrit/nux044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chaturvedi P., Warren C.D., Altaye M., Morrow A.L., Ruiz-Palacios G., Pickering L.K., Newburg D.S. Fucosylated Human Milk Oligosaccharides Vary between Individuals and over the Course of Lactation. Glycobiology. 2001;11:365–372. doi: 10.1093/glycob/11.5.365. [DOI] [PubMed] [Google Scholar]
- 69.Coppa G., Pierani P., Zampini L., Carloni I., Carlucci A., Gabrielli O. Oligosaccharides in Human Milk during Different Phases of Lactation. Acta Paediatr. 2007;88:89–94. doi: 10.1111/j.1651-2227.1999.tb01307.x. [DOI] [PubMed] [Google Scholar]
- 70.Han S.M., Derraik J.G.B., Binia A., Sprenger N., Vickers M.H., Cutfield W.S. Maternal and Infant Factors Influencing Human Milk Oligosaccharide Composition: Beyond Maternal Genetics. J. Nutr. 2021;151:1383–1393. doi: 10.1093/jn/nxab028. [DOI] [PubMed] [Google Scholar]
- 71.Zhou Y., Sun H., Li K., Zheng C., Ju M., Lyu Y., Zhao R., Wang W., Zhang W., Xu Y., et al. Dynamic Changes in Human Milk Oligosaccharides in Chinese Population: A Systematic Review and Meta-Analysis. Nutrients. 2021;13:2912. doi: 10.3390/nu13092912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Seferovic M.D., Mohammad M., Pace R.M., Engevik M., Versalovic J., Bode L., Haymond M., Aagaard K.M. Maternal Diet Alters Human Milk Oligosaccharide Composition with Implications for the Milk Metagenome. Sci Rep. 2020;10:22092. doi: 10.1038/s41598-020-79022-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Azad M.B., Robertson B., Atakora F., Becker A.B., Subbarao P., Moraes T.J., Mandhane P.J., Turvey S.E., Lefebvre D.L., Sears M.R., et al. Human Milk Oligosaccharide Concentrations Are Associated with Multiple Fixed and Modifiable Maternal Characteristics, Environmental Factors, and Feeding Practices. J. Nutr. 2018;148:1733–1742. doi: 10.1093/jn/nxy175. [DOI] [PubMed] [Google Scholar]
- 74.Quin C., Vicaretti S.D., Mohtarudin N.A., Garner A.M., Vollman D.M., Gibson D.L., Zandberg W.F. Influence of Sulfonated and Diet-Derived Human Milk Oligosaccharides on the Infant Microbiome and Immune Markers. J. Biol. Chem. 2020;295:4035–4048. doi: 10.1074/jbc.RA119.011351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Meyer K.M., Mohammad M., Bode L., Chu D.M., Ma J., Haymond M., Aagaard K. 20: Maternal Diet Structures the Breast Milk Microbiome in Association with Human Milk Oligosaccharides and Gut-Associated Bacteria. Am. J. Obstet. Gynecol. 2017;216:S15. doi: 10.1016/j.ajog.2016.11.911. [DOI] [Google Scholar]
- 76.Selma-Royo M., González S., Gueimonde M., Chang M., Fürst A., Martínez-Costa C., Bode L., Collado M.C. Maternal Diet Is Associated with Human Milk Oligosaccharide Profile. Mol. Nutr. Food Res. 2022;66:2200058. doi: 10.1002/mnfr.202200058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Larsson M.W., Lind M.V., Laursen R.P., Yonemitsu C., Larnkjær A., Mølgaard C., Michaelsen K.F., Bode L. Human Milk Oligosaccharide Composition Is Associated With Excessive Weight Gain During Exclusive Breastfeeding—An Explorative Study. Front. Pediatr. 2019;7:297. doi: 10.3389/fped.2019.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saben J.L., Sims C.R., Abraham A., Bode L., Andres A. Human Milk Oligosaccharide Concentrations and Infant Intakes Are Associated with Maternal Overweight and Obesity and Predict Infant Growth. Nutrients. 2021;13:446. doi: 10.3390/nu13020446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Samuel T.M., Binia A., de Castro C.A., Thakkar S.K., Billeaud C., Agosti M., Al-Jashi I., Costeira M.J., Marchini G., Martínez-Costa C., et al. Impact of Maternal Characteristics on Human Milk Oligosaccharide Composition over the First 4 Months of Lactation in a Cohort of Healthy European Mothers. Sci. Rep. 2019;9:11767. doi: 10.1038/s41598-019-48337-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Natividad J.M., Marsaux B., Rodenas C.L.G., Rytz A., Vandevijver G., Marzorati M., Van den Abbeele P., Calatayud M., Rochat F. Human Milk Oligosaccharides and Lactose Differentially Affect Infant Gut Microbiota and Intestinal Barrier In Vitro. Nutrients. 2022;14:2546. doi: 10.3390/nu14122546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Asakuma S., Hatakeyama E., Urashima T., Yoshida E., Katayama T., Yamamoto K., Kumagai H., Ashida H., Hirose J., Kitaoka M. Physiology of Consumption of Human Milk Oligosaccharides by Infant Gut-Associated Bifidobacteria. J. Biol. Chem. 2011;286:34583–34592. doi: 10.1074/jbc.M111.248138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Masi A.C., Stewart C.J. Untangling Human Milk Oligosaccharides and Infant Gut Microbiome. iScience. 2022;25:103542. doi: 10.1016/j.isci.2021.103542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Egan M., O’Connell Motherway M., Kilcoyne M., Kane M., Joshi L., Ventura M., van Sinderen D. Cross-feeding by Bifidobacterium breve UCC2003 during co-cultivation with Bifidobacterium bifidum PRL2010 in a mucin-based medium. BMC Microbiol. 2014;14:282. doi: 10.1186/s12866-014-0282-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Berger P.K., Plows J.F., Jones R.B., Alderete T.L., Yonemitsu C., Ryoo J.H., Bode L., Goran M.I. Human Milk Oligosaccharides and Hispanic Infant Weight Gain in the First 6 Months. Obesity. 2020;28:1519–1525. doi: 10.1002/oby.22884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Alderete T.L., Autran C., Brekke B.E., Knight R., Bode L., Goran M.I., Fields D.A. Associations between Human Milk Oligosaccharides and Infant Body Composition in the First 6 Mo of Life. Am. J. Clin. Nutr. 2015;102:1381–1388. doi: 10.3945/ajcn.115.115451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Menzel P., Vogel M., Austin S., Sprenger N., Grafe N., Hilbert C., Jurkutat A., Kiess W., Binia A. Concentrations of Oligosaccharides in Human Milk and Child Growth. BMC Pediatr. 2021;21:481. doi: 10.1186/s12887-021-02953-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lagström H., Rautava S., Ollila H., Kaljonen A., Turta O., Mäkelä J., Yonemitsu C., Gupta J., Bode L. Associations between Human Milk Oligosaccharides and Growth in Infancy and Early Childhood. Am. J. Clin. Nutr. 2020;111:769–778. doi: 10.1093/ajcn/nqaa010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Binia A., Lavalle L., Chen C., Austin S., Agosti M., Al-Jashi I., Pereira A.B., Costeira M.J., Silva M.G., Marchini G., et al. Human Milk Oligosaccharides, Infant Growth, and Adiposity over the First 4 Months of Lactation. Pediatr. Res. 2021;90:684–693. doi: 10.1038/s41390-020-01328-y. [DOI] [PubMed] [Google Scholar]
- 89.Davis J.C.C., Lewis Z.T., Krishnan S., Bernstein R.M., Moore S.E., Prentice A.M., Mills D.A., Lebrilla C.B., Zivkovic A.M. Growth and Morbidity of Gambian Infants Are Influenced by Maternal Milk Oligosaccharides and Infant Gut Microbiota. Sci. Rep. 2017;7:40466. doi: 10.1038/srep40466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rozé J.-C., Hartweg M., Simon L., Billard H., Chen Y., Austin S., Boscher C., Moyon T., Darmaun D., Garcia Rodenas C.L., et al. Human Milk Oligosaccharides in Breast Milk and 2-Year Outcome in Preterm Infants: An Exploratory Analysis. Clin. Nutr. 2022;41:1896–1905. doi: 10.1016/j.clnu.2022.07.024. [DOI] [PubMed] [Google Scholar]
- 91.Samuel T.M., Hartweg M., Lebumfacil J.D., Buluran K.B., Lawenko R.B., Estorninos E.M., Binia A., Sprenger N. Dynamics of Human Milk Oligosaccharides in Early Lactation and Relation with Growth and Appetitive Traits of Filipino Breastfed Infants. Sci. Rep. 2022;12:17304. doi: 10.1038/s41598-022-22244-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vandenplas Y., Berger B., Carnielli V., Ksiazyk J., Lagström H., Sanchez Luna M., Migacheva N., Mosselmans J.-M., Picaud J.-C., Possner M., et al. Human Milk Oligosaccharides: 2′-Fucosyllactose (2′-FL) and Lacto-N-Neotetraose (LNnT) in Infant Formula. Nutrients. 2018;10:1161. doi: 10.3390/nu10091161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Koh A., De Vadder F., Kovatcheva-Datchary P., Bäckhed F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell. 2016;165:1332–1345. doi: 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 94.Tan J., McKenzie C., Potamitis M., Thorburn A.N., Mackay C.R., Macia L. Advances in Immunology. Volume 121. Elsevier; Amsterdam, The Netherlands: 2014. The Role of Short-Chain Fatty Acids in Health and Disease; pp. 91–119. [DOI] [PubMed] [Google Scholar]
- 95.Canfora E.E., Jocken J.W., Blaak E.E. Short-Chain Fatty Acids in Control of Body Weight and Insulin Sensitivity. Nat. Rev. Endocrinol. 2015;11:577–591. doi: 10.1038/nrendo.2015.128. [DOI] [PubMed] [Google Scholar]
- 96.Canfora E.E., Meex R.C.R., Venema K., Blaak E.E. Gut Microbial Metabolites in Obesity, NAFLD and T2DM. Nat. Rev. Endocrinol. 2019;15:261–273. doi: 10.1038/s41574-019-0156-z. [DOI] [PubMed] [Google Scholar]
- 97.Larraufie P., Martin-Gallausiaux C., Lapaque N., Dore J., Gribble F.M., Reimann F., Blottiere H.M. SCFAs Strongly Stimulate PYY Production in Human Enteroendocrine Cells. Sci. Rep. 2018;8:74. doi: 10.1038/s41598-017-18259-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chambers E.S., Viardot A., Psichas A., Morrison D.J., Murphy K.G., Zac-Varghese S.E.K., MacDougall K., Preston T., Tedford C., Finlayson G.S., et al. Effects of Targeted Delivery of Propionate to the Human Colon on Appetite Regulation, Body Weight Maintenance and Adiposity in Overweight Adults. Gut. 2015;64:1744–1754. doi: 10.1136/gutjnl-2014-307913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Frost G., Sleeth M.L., Sahuri-Arisoylu M., Lizarbe B., Cerdan S., Brody L., Anastasovska J., Ghourab S., Hankir M., Zhang S., et al. The Short-Chain Fatty Acid Acetate Reduces Appetite via a Central Homeostatic Mechanism. Nat. Commun. 2014;5:3611. doi: 10.1038/ncomms4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chambers E.S., Byrne C.S., Aspey K., Chen Y., Khan S., Morrison D.J., Frost G. Acute Oral Sodium Propionate Supplementation Raises Resting Energy Expenditure and Lipid Oxidation in Fasted Humans. Diabetes Obes. Metab. 2018;20:1034–1039. doi: 10.1111/dom.13159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Canfora E.E., van der Beek C.M., Jocken J.W.E., Goossens G.H., Holst J.J., Olde Damink S.W.M., Lenaerts K., Dejong C.H.C., Blaak E.E. Colonic Infusions of Short-Chain Fatty Acid Mixtures Promote Energy Metabolism in Overweight/Obese Men: A Randomized Crossover Trial. Sci. Rep. 2017;7:2360. doi: 10.1038/s41598-017-02546-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gao Z., Yin J., Zhang J., Ward R.E., Martin R.J., Lefevre M., Cefalu W.T., Ye J. Butyrate Improves Insulin Sensitivity and Increases Energy Expenditure in Mice. Diabetes. 2009;58:1509–1517. doi: 10.2337/db08-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Louis P., Flint H.J. Diversity, Metabolism and Microbial Ecology of Butyrate-Producing Bacteria from the Human Large Intestine. FEMS Microbiol. Lett. 2009;294:1–8. doi: 10.1111/j.1574-6968.2009.01514.x. [DOI] [PubMed] [Google Scholar]
- 104.Reichardt N., Duncan S.H., Young P., Belenguer A., McWilliam Leitch C., Scott K.P., Flint H.J., Louis P. Phylogenetic Distribution of Three Pathways for Propionate Production within the Human Gut Microbiota. ISME J. 2014;8:1323–1335. doi: 10.1038/ismej.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Duncan S.H., Holtrop G., Lobley G.E., Calder A.G., Stewart C.S., Flint H.J. Contribution of Acetate to Butyrate Formation by Human Faecal Bacteria. Br. J. Nutr. 2004;91:915–923. doi: 10.1079/BJN20041150. [DOI] [PubMed] [Google Scholar]
- 106.Henson M.A., Phalak P. Suboptimal Community Growth Mediated through Metabolite Crossfeeding Promotes Species Diversity in the Gut Microbiota. PLoS Comput. Biol. 2018;14:e1006558. doi: 10.1371/journal.pcbi.1006558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Prentice P.M., Schoemaker M.H., Vervoort J., Hettinga K., Lambers T.T., van Tol E.A.F., Acerini C.L., Olga L., Petry C.J., Hughes I.A., et al. Human Milk Short-Chain Fatty Acid Composition Is Associated with Adiposity Outcomes in Infants. J. Nutr. 2019;149:716–722. doi: 10.1093/jn/nxy320. [DOI] [PubMed] [Google Scholar]
- 108.Paparo L., Nocerino R., Ciaglia E., Di Scala C., De Caro C., Russo R., Trinchese G., Aitoro R., Amoroso A., Bruno C., et al. Butyrate as a Bioactive Human Milk Protective Component against Food Allergy. Allergy. 2021;76:1398–1415. doi: 10.1111/all.14625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Stinson L.F., Gay M.C.L., Koleva P.T., Eggesbø M., Johnson C.C., Wegienka G., du Toit E., Shimojo N., Munblit D., Campbell D.E., et al. Human Milk From Atopic Mothers Has Lower Levels of Short Chain Fatty Acids. Front. Immunol. 2020;11:1427. doi: 10.3389/fimmu.2020.01427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lönnerdal B. Nutritional and Physiologic Significance of Human Milk Proteins. Am. J. Clin. Nutr. 2003;77:1537S–1543S. doi: 10.1093/ajcn/77.6.1537S. [DOI] [PubMed] [Google Scholar]
- 111.Kawakami H., Lonnerdal B. Isolation and Function of a Receptor for Human Lactoferrin in Human Fetal Intestinal Brush-Border Membranes. Am. J. Physiol.—Gastrointest. Liver Physiol. 1991;261:G841–G846. doi: 10.1152/ajpgi.1991.261.5.G841. [DOI] [PubMed] [Google Scholar]
- 112.Lönnerdal B., Iyer S. Lactoferrin: Molecular Structure and Biological Function. Annu. Rev. Nutr. 1995;15:93–110. doi: 10.1146/annurev.nu.15.070195.000521. [DOI] [PubMed] [Google Scholar]
- 113.Jahani S., Shakiba A., Jahani L. The Antimicrobial Effect of Lactoferrin on Gram-Negative and Gram-Positive Bacteria. Int. J. Infect. 2015;2:e27594. doi: 10.17795/iji27594. [DOI] [Google Scholar]
- 114.Farnaud S., Evans R.W. Lactoferrin—A Multifunctional Protein with Antimicrobial Properties. Mol. Immunol. 2003;40:395–405. doi: 10.1016/S0161-5890(03)00152-4. [DOI] [PubMed] [Google Scholar]
- 115.Arnold R.R., Brewer M., Gauthier J.J. Bactericidal Activity of Human Lactoferrin: Sensitivity of a Variety of Microorganisms. Infect. Immun. 1980;28:893–898. doi: 10.1128/iai.28.3.893-898.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Berlutti F., Pantanella F., Natalizi T., Frioni A., Paesano R., Polimeni A., Valenti P. Antiviral Properties of Lactoferrin—A Natural Immunity Molecule. Molecules. 2011;16:6992–7018. doi: 10.3390/molecules16086992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jenssen H. Anti Herpes Simplex Virus Activity of Lactoferrin/Lactoferricin—An Example of Antiviral Activity of Antimicrobial Protein/Peptide. Cell. Mol. Life Sci. 2005;62:3002–3013. doi: 10.1007/s00018-005-5228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ikeda M., Nozaki A., Sugiyama K., Tanaka T., Naganuma A., Tanaka K., Sekihara H., Shimotohno K., Saito M., Kato N. Characterization of Antiviral Activity of Lactoferrin against Hepatitis C Virus Infection in Human Cultured Cells. Virus Res. 2000;66:51–63. doi: 10.1016/S0168-1702(99)00121-5. [DOI] [PubMed] [Google Scholar]
- 119.Fernandes K.E., Carter D.A. The Antifungal Activity of Lactoferrin and Its Derived Peptides: Mechanisms of Action and Synergy with Drugs against Fungal Pathogens. Front. Microbiol. 2017;8:2. doi: 10.3389/fmicb.2017.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Soukka T., Tenovuo J., Lenander-Lumikari M. Fungicidal Effect of Human Lactoferrin against Candida Albicans. FEMS Microbiol. Lett. 1992;90:223–228. doi: 10.1111/j.1574-6968.1992.tb05156.x. [DOI] [PubMed] [Google Scholar]
- 121.Kirkpatrick C.H., Green I., Rich R.R., Schade A.L. Inhibition of Growth of Candida Albicans by Iron-Unsaturated Lactoferrin: Relation to Host-Defense Mechanisms in Chronic Mucocutaneous Candidiasis. J. Infect. Dis. 1971;124:539–544. doi: 10.1093/infdis/124.6.539. [DOI] [PubMed] [Google Scholar]
- 122.Aguilar-Diaz H., Canizalez-Roman A., Nepomuceno-Mejia T., Gallardo-Vera F., Hornelas-Orozco Y., Nazmi K., Bolscher J.G.M., Carrero J.C., Leon-Sicairos C., Leon-Sicairos N. Parasiticidal Effect of Synthetic Bovine Lactoferrin Peptides on the Enteric Parasite Giardia Intestinalis. Biochem. Cell Biol. 2017;95:82–90. doi: 10.1139/bcb-2016-0079. [DOI] [PubMed] [Google Scholar]
- 123.López-Soto F., León-Sicairos N., Nazmi K., Bolscher J.G., de la Garza M. Microbicidal Effect of the Lactoferrin Peptides Lactoferricin17–30, Lactoferrampin265–284, and Lactoferrin Chimera on the Parasite Entamoeba Histolytica. Biometals. 2010;23:563–568. doi: 10.1007/s10534-010-9295-3. [DOI] [PubMed] [Google Scholar]
- 124.Weinberg G.A. Iron Chelators as Therapeutic Agents against Pneumocystis Carinii. Antimicrob. Agents Chemother. 1994;38:997–1003. doi: 10.1128/AAC.38.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Liepke C., Adermann K., Raida M., Mägert H.-J., Forssmann W.-G., Zucht H.-D. Human Milk Provides Peptides Highly Stimulating the Growth of Bifidobacteria: Human Milk Peptides as Bifidus Factors. Eur. J. Biochem. 2002;269:712–718. doi: 10.1046/j.0014-2956.2001.02712.x. [DOI] [PubMed] [Google Scholar]
- 126.Kim W.-S., Ohashi M., Tanaka T., Kumura H., Kim G.-Y., Kwon I.-K., Goh J.-S., Shimazaki K. Growth-Promoting Effects of Lactoferrin on L. Acidophilus and Bifidobacterium Spp. Biometals. 2004;17:279–283. doi: 10.1023/B:BIOM.0000027705.57430.f1. [DOI] [PubMed] [Google Scholar]
- 127.Oda H., Wakabayashi H., Yamauchi K., Abe F. Lactoferrin and Bifidobacteria. Biometals. 2014;27:915–922. doi: 10.1007/s10534-014-9741-8. [DOI] [PubMed] [Google Scholar]
- 128.Ellison R.T., Giehl T.J. Killing of Gram-Negative Bacteria by Lactoferrin and Lysozyme. J. Clin. Invest. 1991;88:1080–1091. doi: 10.1172/JCI115407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chipman D.M., Sharon N. Mechanism of Lysozyme Action: Lysozyme Is the First Enzyme for Which the Relation between Structure and Function Has Become Clear. Science. 1969;165:454–465. doi: 10.1126/science.165.3892.454. [DOI] [PubMed] [Google Scholar]
- 130.Maga E.A., Desai P.T., Weimer B.C., Dao N., Kültz D., Murray J.D. Consumption of Lysozyme-Rich Milk Can Alter Microbial Fecal Populations. Appl. Env. Microbiol. 2012;78:6153–6160. doi: 10.1128/AEM.00956-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Marra A., Hanson M.A., Kondo S., Erkosar B., Lemaitre B. Drosophila Antimicrobial Peptides and Lysozymes Regulate Gut Microbiota Composition and Abundance. mBio. 2021;12:e00824-21. doi: 10.1128/mBio.00824-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Blais A., Fan C., Voisin T., Aattouri N., Dubarry M., Blachier F., Tomé D. Effects of Lactoferrin on Intestinal Epithelial Cell Growth and Differentiation: An in Vivo and in Vitro Study. Biometals. 2014;27:857–874. doi: 10.1007/s10534-014-9779-7. [DOI] [PubMed] [Google Scholar]
- 133.Naot D., Grey A., Reid I.R., Cornish J. Lactoferrin—A Novel Bone Growth Factor. Clin. Med. Res. 2005;3:93–101. doi: 10.3121/cmr.3.2.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nichols B.L., Mckee K.S., Henry J.F., Putman M. Human Lactoferrin Stimulates Thymidine Incorporation into DNA of Rat Crypt Cells. Pediatr. Res. 1987;21:563–567. doi: 10.1203/00006450-198706000-00011. [DOI] [PubMed] [Google Scholar]
- 135.Hernell O., Lönnerdal B. Iron Status of Infants Fed Low-Iron Formula: No Effect of Added Bovine Lactoferrin or Nucleotides. Am. J. Clin. Nutr. 2002;76:858–864. doi: 10.1093/ajcn/76.4.858. [DOI] [PubMed] [Google Scholar]
- 136.Ochoa T.J., Chea-Woo E., Campos M., Pecho I., Prada A., McMahon R.J., Cleary T.G. Impact of Lactoferrin Supplementation on Growth and Prevalence of Giardia Colonization in Children. Clin. Infect. Dis. 2008;46:1881–1883. doi: 10.1086/588476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bol’shakova A.M., Shcherbakova E.G., Ivanova S.D., Medvedeva M.M., Zhuravleva T.P. Lysozyme in the feeding of premature infants with mixed pathology. Antibiotiki. 1984;29:784–790. [PubMed] [Google Scholar]
- 138.Braun O.H., Sandkühler H. Relationships Between Lysozyme Concentration of Human Milk, Bacteriologic Content, and Weight Gain of Premature Infants. J. Pediatric Gastroenterol. Nutr. 1985;4:583–586. doi: 10.1097/00005176-198508000-00015. [DOI] [PubMed] [Google Scholar]
- 139.Gridneva Z., Lai C.T., Rea A., Tie W.J., Ward L.C., Murray K., Hartmann P.E., Geddes D.T. Human Milk Immunomodulatory Proteins Are Related to Development of Infant Body Composition during the First Year of Lactation. Pediatr. Res. 2021;89:911–921. doi: 10.1038/s41390-020-0961-z. [DOI] [PubMed] [Google Scholar]
- 140.Aakko J., Kumar H., Rautava S., Wise A., Autran C., Bode L., Isolauri E., Salminen S. Human Milk Oligosaccharide Categories Define the Microbiota Composition in Human Colostrum. Benef. Microbes. 2017;8:563–567. doi: 10.3920/BM2016.0185. [DOI] [PubMed] [Google Scholar]
- 141.Moossavi S., Atakora F., Miliku K., Sepehri S., Robertson B., Duan Q.L., Becker A.B., Mandhane P.J., Turvey S.E., Moraes T.J., et al. Integrated Analysis of Human Milk Microbiota With Oligosaccharides and Fatty Acids in the CHILD Cohort. Front. Nutr. 2019;6:58. doi: 10.3389/fnut.2019.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.