Abstract
Microbial resistance to antibiotics is an ancient and dynamic issue that has brought a situation reminiscent of the pre-antibiotic era to the limelight. Currently, antibiotic resistance and the associated infections are widespread and pose significant global health and economic burden. Thus, the misuse of antibiotics, which has increased resistance, has necessitated the search for alternative therapeutic agents for combating resistant pathogens. Antimicrobial peptides (AMPs) hold promise as a viable therapeutic approach against drug-resistant pathogens. AMPs are oligopeptides with low molecular weight. They have broad-spectrum antimicrobial activities against pathogenic microorganisms. AMPs are nonspecific and target components of microbes that facilitate immune response by acting as the first-line defense mechanisms against invading pathogenic microbes. The diversity and potency of AMPs make them good candidates for alternative use. They could be used alone or in combination with several other biomaterials for improved therapeutic activity. They can also be employed in vaccine production targeting drug-resistant pathogens. This review covers the opportunities and advances in AMP discovery and development targeting antimicrobial resistance (AMR) bacteria. Briefly, it presents an overview of the global burden of the antimicrobial resistance crisis, portraying the global magnitude, challenges, and consequences. After that, it critically and comprehensively evaluates the potential roles of AMPs in addressing the AMR crisis, highlighting the major potentials and prospects.
Keywords: Antimicrobial peptides, Bacteria, antimicrobial resistance (AMR)
Introduction
Bacterial resistance to antibiotics and the rapid increase in infectious diseases due to antimicrobial resistant (AMR) bacteria have brought to the international attention a situation reminiscent of the pre-antibiotic era. Currently, antibiotic resistance and the associated infections are widespread and pose severe health and economic burden globally, necessitating a massive demand for alternative treatment approaches [1]. Additionally, many emerging alternative treatment options are at different stages of preclinical and clinical trials [2-5]. Interestingly, antimicrobial peptides (AMPs) hold promises as valuable therapeutic options against drug-resistant pathogens [6]. AMPs are low molecular-weight oligopeptides that are found in plants, animals, and humans [7]. They play a crucial role in the host’s innate immune response and have broad-spectrum antimicrobial activity [8]. AMPs have an amino acid length ranging from 15 to 150 [9]. They are cationic, having a net charge of +3 and an average hydrophilic composition of 42% [10]. AMPs, also called the host defense peptides or the innate defense regulatory peptides, are nonspecific and target components of microbes that facilitate immunological response [11], acting as the first-line defense mechanism against invading pathogens [12].
Furthermore, AMPs are amphipathic and evolutionarily conserved peptides. Their antibacterial activity involves electrostatic interactions with intracellular bacterial components and cell membranes. This is accomplished by forming ionic pores or transient gaps, leading to bacterial membrane permeability alterations. Apart from their broad-spectrum activity, peptides are thermally stable and water-soluble [9,13] and are involved in intracellular angiogenesis (blood vessel formation), inflammation responses, and cell signaling in the host cell [14].
Although there are established disadvantages in the application of AMPs, such as resistance [15], toxicity [16], immunogenicity, and hemolytic activity to host cells [16-18], susceptibility to proteolysis [19,20], poor pharmacokinetics [21], undesirable or nonspecific interactions with host cells [21], stability, and selectivity [22]; these issues make the AMPs inefficient to reach the target and exert their action [23]. However, AMP diversity and potency make them good candidates for antibiotic alternatives [9]. Also, the pronounced selectivity of AMP for bacterial cells over host cells [24] makes them an indispensable option in tackling the bacterial AMR crisis. Currently, scientists are recording success in an effort to mitigate all the challenges associated with their use. The current review presents the opportunities and advances in AMPs discovery and development targeting AMR bacteria. First, we reflect on the global burden of the AMR crisis, portraying the global magnitude, challenges, and consequences. Next, we critically and comprehensively highlight the potential roles of AMPs in addressing the AMR crisis.
The Global Burden of Antimicrobial Resistance (AMR) Crisis
Antibiotics used during the 1930s were effective in managing infectious diseases and substantially resulted in a decrease in the rates of morbidity and mortality. However, microbial resistance to antibiotics was soon recorded immediately after penicillin’s discovery. Despite the emergence of resistance, there was little concern because of the continuous production of new drug derivatives to manage the issue. This caused the myth that infectious diseases caused by bacteria would be overcome in no distant time [25,26]. Although newer antibiotics were developed after that, efforts made by scientists to develop newer and more efficient antibiotics were not enough to mitigate the impending AMR crisis, which Alexander Fleming earlier predicted during his 1945 Nobel Prize lecture [27]. Thus, multi-drug resistant infectious diseases became unavoidable, and the already-available antibiotics lost efficacy and effectiveness [28].
Sadly, antibiotic resistance in healthcare-associated infections is currently predominant and quite alarming in many countries. The emergence of multi-drug resistant pathogens harboring different resistant genes has been a major concern. Several recent studies show the global magnitude of the issue. For example, the emergence of extended-spectrum beta-lactam (ESBL) among Klebsiella pneumoniae isolates from Iraq [29], plasmid-mediated quinolone resistance genes among Escherichia coli and Pseudomonas aeruginosa isolates from Iran, where about 85% of all E. coli isolates were reported to have at least one plasmid-mediated quinolone gene [30]. Additionally, ESBL-harboring Salmonella typhi have been isolated from patients in Nigeria infected with typhoid fever [31].
Several factors promote the emergence of antimicrobial-resistant organisms. Both medical and non-medical use of antibiotics has been seen as key players in this regard. According to the Centers for Disease Control and Prevention (CDC) [32], antibiotics given during treatment duration are wrong 30%-50% of the time. Luyt et al. [33] also reported that in the intensive care unit, about 30%-60% of drugs prescribed are highly unnecessary. However, the non-medical use of antibiotics remains a major concern [34]. In addition, the quest to provide a safe and adequate food supply for the ever-increasing world population at different quarters has raised the demand for antibiotics in the agricultural sector [35]. Currently, many critically important antibiotics widely used in human medicine, such as penicillin, tetracyclines, aminoglycosides, sulphonamides, macrolides, and quinolones, are used in agricultural and aquaculture practices [36]. It has been estimated that in the United States alone, about 80% of the available antibiotics are not used clinically; instead, they are used in animal agriculture as either a growth supplement or for infection control [37,38].
Many countries use antibiotics in livestock production (eg, poultry) [39,40]. This use has been a welcome development for livestock farmers due to improved poultry performance and subsequent economic gains. In fact, the past decade has seen rapid growth in global livestock production with an increased inclination towards intensive systems, where antimicrobial use is elemental to production processes. Little wonder, two-thirds of the future increase in antimicrobial use is projected for animal production [41,42]. In several countries, antibiotics are consumed indirectly by livestock. In 2010, the estimation was about 63,151 tons [41], with each kilogram of meat harvested from chicken, cattle, or pigs containing 148mg, 45mg, and 172mg of antibiotics, respectively. Consequently, this is projected to increase by 67% by the year 2030 [38,41,43]. Moreover, highly populated and rapidly developing countries risk a double increase [41]. In livestock production, poultry records the highest antimicrobial use, followed by pigs and dairy cattle [44]. Currently, China stands as the largest producer and consumer of antibiotics globally, with more than 50% going into poultry and other livestock [39,45]. Making up 45% of global veterinary antimicrobial consumers in 2017, China preceded Brazil, the US, Thailand, and India. Together with Iran, Mexico, Spain, and Argentina, these countries accounted for 75% of the antimicrobials used in livestock production [46].
Common antibiotics used in intensive poultry production, especially in North America, include bacitracin, tetracycline, virginiamycin, tylosin, salinomycin, and bambermycin [47]. Tetracyclines are commonplace among US livestock producers, accounting for more than two-thirds of antimicrobials used in poultry production [48]. The increasing use of antimicrobials in livestock and aquaculture is of great concern with the imminent danger of AMR [49,50], especially as most of these antimicrobials are used in human medicine [51]. Statistically, about 95% of antibiotics ingested by livestock are released in unchanged form when the livestock excrete waste. The waste enters the natural environment and eventually causes disease and facilitates the rapid increase in antimicrobial-resistant zoonotic pathogens in the environment [52,53].
Primarily, overuse of antibiotics is the primary cause of resistance evolution. Although the overuse of antibiotics is strongly discouraged, the problem of over-prescription across the globe still exists [54]. Across countries, antibiotic prescriptions differ. For low-and middle-income countries, the majority of the populace acquires antibiotics over the counter without a prescription or proper diagnosis [55]. This practice also leads to a high consumption rate of antibiotics. Research shows that antibiotics prescribed to humans are used at home and finally end up in the sewage. The presence of antibiotics in sewage and wastewater makes urban wastewater treatment plants (WWTPs) and sewage a significant source of antibiotic-resistant bacteria (ARB) and antibiotic-resistant genes (ARG) [56]. It is worth noting that although antimicrobial-resistant organisms occur naturally in the environment, human activities and the presence of antibiotics in sewage and wastewater contribute to the distribution of drug-resistant bacterial genes to the environment [56,57]. Some purification processes in the WWTPs are inefficient as they cannot eliminate the resistant genes of concern. Most WWTPs are hotspots for the spread of antibiotic resistance determinants in the environment, which is a great threat as final effluent is usually introduced into surface waters. WWTPs also constitute a crucial interface between humans and the environment. WWTPs harbor a large microbial community, enhancing the exchange of ARGs by horizontal gene transfer (HGT) [58,59].
Also, the use of antibiotics in agriculture allows the permeation of antibiotics into soil and water sources from crop treatments. Currently, the aquatic environment has become a reservoir for ARB and ARG and serves as a channel that sustains and enhances the transformation and persistence of antibiotic residues and ARB and their genes. The aquatic environment harbors genetic elements such as plasmids, integrons, transposons, and other mobile genetic elements that enable bacteria to acquire resistance capacity and quick adaptation in the presence of antimicrobials to which they are supposed to have been susceptible [60]. Integrated farming, drug residue accumulation, and consumption of contaminated food and food products are other major factors that lead to ABR and ARGs dissemination [61]. The dissemination of both the ARB and ARGs to humans can occur directly through the exposure of humans to infected animals and biological substances and indirectly through the consumption of contaminated food products (eg, meat, dairy products, and eggs).
All these described issues show that antimicrobial drug resistance and the associated infections will pose a more severe health and economic burden if adequate plans are not put in to overcome the crisis. Notably, the reality is that bacteria are very versatile and adaptive. So, AMR seems likely to increase in the near future unless all hands are on deck towards renewing research efforts, unraveling more virulence and resistant markers, and designing more effective alternative treatment approaches. So far, the use of antimicrobial peptides, together with other emerging biomaterials, is offering hope in reducing the AMR crisis. These AMPs have good activity against bacterial pathogens, and they are promising alternatives to antibiotics.
Classification of Antimicrobial Peptides
Gramicidin was the first described AMP. It was discovered in 1939 from Bacillus brevis isolated from the soil and was shown to be effective against bacterial pathogens [62-65]. Currently, there are several catalogued AMP [66], and different criteria are used to classify antimicrobial peptides. They can be classified based on source (mammalian, amphibians, insects, microorganisms) [66-68], the activity (antiviral, antiparasitic, antibacterial, antifungal, antihuman immunodeficiency virus (HIV), antitumor, anticancer) [69,70], structural characteristics [70], and amino acid constituents [71]. Interestingly, bacteria can produce some antibacterial peptides. The most researched and notable of these peptides are the bacteriocins. Several studies have shown the antibacterial potential of bacteriocins. Although the antimicrobial potentials of bacteriocins are beyond the scope of this review, readers are encouraged to read the following papers [72,73].
Based on their cell target activity, AMPs are categorized into two groups: the extracellular targeting group (damage outer cell membrane) and the intracellular targeting groups [11]. The membrane-disrupting peptides exert their effects in multiple mechanisms [10]. Some peptides can also act both on the intracellular and extracellular target and can also switch from one mechanism to another depending on the peptide concentration, membrane makeup of the target pathogen, and the existing growth phase [11]. So far, peptide classification has been limited by diverse information and nomenclature. Different methods for classifying peptides from plants, animals, and bacteria are also utilized. However, a recent report by Wang [74] describes unifying the classification of AMP in the AMP database. The criteria for this unified classification include the source of the peptide, method of biosynthesis, biological potentials, sequence of amino acid, and mechanism of action, in addition to their three-dimensional structure.
Mechanisms of Action of Peptides
Peptides exert their effect either by compromising the integrity of the bacterial cell wall or through interferences with intracellular metabolic processes [10,75]. Also, peptides’ exact mechanisms of action against all pathogens are not the same. While the initiation of interaction depends solely on electrostatic forces [76,77], the effects AMPs exert on the membrane are dependent on the properties of the peptides, such as the structure, size, net charge, amphipathicity, and composition of the membrane itself [76].
Generally, two major mechanisms are considered in the activity of peptides: direct cell killing through cell membrane disruption and immunomodulatory action. About three different models, such as the Barrel-stave, Carpet, and Toroidal-pore have been described over the years to define the mechanism of cell membrane disruption by AMPs [23,78,79] (Figure 1). In bacteria species, AMPs initiate cell disruption in three major steps; first, the attraction phase involves the interaction of the cationic peptides with the bacterial surface. The cationic peptide binds to the bacterial outer membrane surfaces using lipopolysaccharide as a receptor. Since the AMPs are positively charged, attachment makes it displace cationic ions such as Ca2+ and Mg2+, causing a disruption of the outer membrane, allowing the AMPs to reach the intracellular target (negatively charged phospholipids) [23,80]. The second is the attachment, which involves the penetration of the polysaccharides and their attachment and interaction with cellular targets such as the protective capsular coats (lipopolysaccharides in gram-negative bacteria or the teichoic and lipoteichoic acids in gram-positive bacteria) [23]. Once successful in crossing the cell membrane, AMPs interact with the lipids in the cytoplasmic membrane, such as phosphatidylglycerol and cardiolipin, destabilizing the bonds of the phospholipids, resulting in disintegration or permeability [79]. Furthermore, there is the insertion stage, where the peptides position themselves within the membrane bilayer, leading to membrane thinning, curvature, and disruption of the membrane barrier [79].
Figure 1.
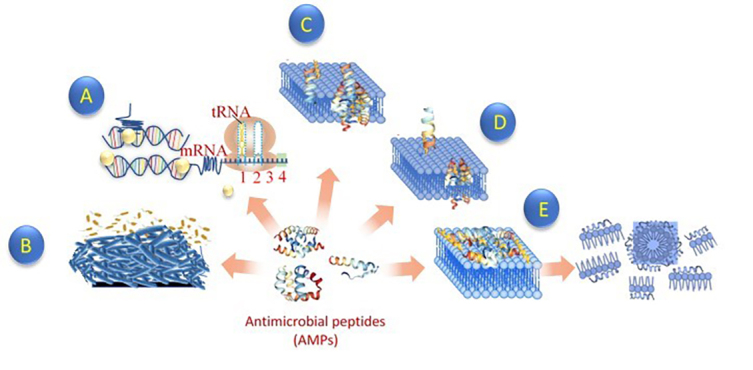
Mechanism of action of the antimicrobial peptide against bacteria. AMP interacts with bacterial membrane via electrostatic interactions. This approach makes it difficult for bacteria to develop resistance to it. Some AMP acts on membrane, while others do not. Membrane peptides have cationic peptides that disrupt the bacterial membrane when interacting with it. On the other hand, non-membrane peptides translocate across the membrane without causing any damage. There is also some AMP that creates trans-membrane pores on the membrane. Others AMP does not disrupt cell functions as they translocate across the membrane. Protein synthesis, several enzymatic activities, cell signaling activities, and other critical intercellular functions are also disrupted by AMPs (A). Some AMP also disrupts biofilm formation (B). While interacting with bacterial membrane, some AMP usually undergoes some conformational changes. Currently, three different models are used to define AMP mechanisms of action across the bacterial membrane. (C) Barrel stave model: aggregation of AMP with each other. This aggregate is inserted into the lipid bilayer of the cell and then arranged parallel to the phospholipids leading to the formation of a channel (D). Toroidal pore model: AMPs embedded in the cell membrane. Accumulation of these AMPs leads to the formation of the ring hole (E). Carpet model: AMP accumulates on the cell surface leading to damage to the membrane in the form of a detergent [161,162].
In addition to permeabilizing the lipid bilayers, recent studies show AMPs with the non-lipid target inside the cell [81]. Moreover, inside the cytoplasm, peptides can further exert complex effects like inhibition of protein, nucleic acid biosynthesis, enzymatic activities, cell wall synthesis, protease activity, and inhibition of DNA and RNA [71,77,78,82]. The non-membrane targeting group of peptides usually employs this mechanism. This group of AMPs can penetrate freely into bacterial membrane without causing any damage [83]. This is because they have non-membrane permeabilizing (non-lytic) activity [84,85]. For this group of peptides, bacterial cell membrane permeabilization and subsequent cell killing occur rapidly and simultaneously [86]. However, non-lytic peptides often have a lag phase period between membrane permeabilization and cell killing [87]. Although the mechanism of non-lytic peptide interaction is still not completely clear, it is known that this group of peptides enters the cell via spontaneous translocation [88,89]. This translocation event leads to the formation of pores in the membrane through which the peptides pass. The route is only transiently breached, and the smooth movement of the peptide across helps to sustain membrane integrity.
Another mechanism that has been reported in proline-rich AMP is membrane translocation facilitated by proteins secreted by the bacteria [90,91]. While some AMP acts on bacterial membranes and others target intracellular components, some peptides have multiple modes of action. For example, a recent study reported a peptide NP-6 with dual mechanisms of action against Staphylococcus aureus [92]. In addition to compromising the bacterial membrane integrity with a resulting increase in membrane permeability, the peptide also binds to the DNA and RNA and disrupts intracellular β-galactosidase activity in S. aureus.
In immunomodulation, peptides act through the recruitment and activation of leukocytes, autoinflammation, degradation of protective coats such as lipopolysaccharides, and phagocytosis [93]. AMPs can cause a reduction in the expression of inflammatory cytokines, while modulation of the expression of chemokines has also been reported [77] (Figure 2). Allomyrinasin obtained from Dynastid beetle was shown to induce anti-inflammatory activities [94]. In a study by Nagaoka et al. [95], LL-37 was reported to trigger the production of pro-inflammatory cytokines (IL-1B) by suppressing LPS/ATP-induced macrophages pyroptosis. This AMP was also reported to trigger neutrophils to produce ectosomes (antimicrobial microvesicles), demonstrating the potential therapeutic potential of cathelicidin LL-37. AWRK6, a synthetic cationic peptide from AMP, inhibits LPS-induced inflammatory response [96]. In a more recent investigation, gcIFN-20 peptide suppresses bacterial load and mortalities significantly. Importantly, using both in vivo and in vitro approaches, LPS-induced pro-inflammatory cytokines were also inhibited [97]. This study by Xiao et al. demonstrates that AMP can interact with LPS, consequently leading to LPS aggregation and neutralization and thereby showing the therapeutic potential of AMPs. In a study by Wei et al. [98], the cathelicidin-PY peptide not only damaged bacterial membrane but also inhibited the production of nitric oxide (NO). There was also inhibition of TNF-α, IL-6, and monocyte chemoattractant proteins-1 (MCP-1). The peptide was also shown to bind to LPS, triggering the activation of Toll-like receptors (TLR4) inflammatory response pathways.
Figure 2.
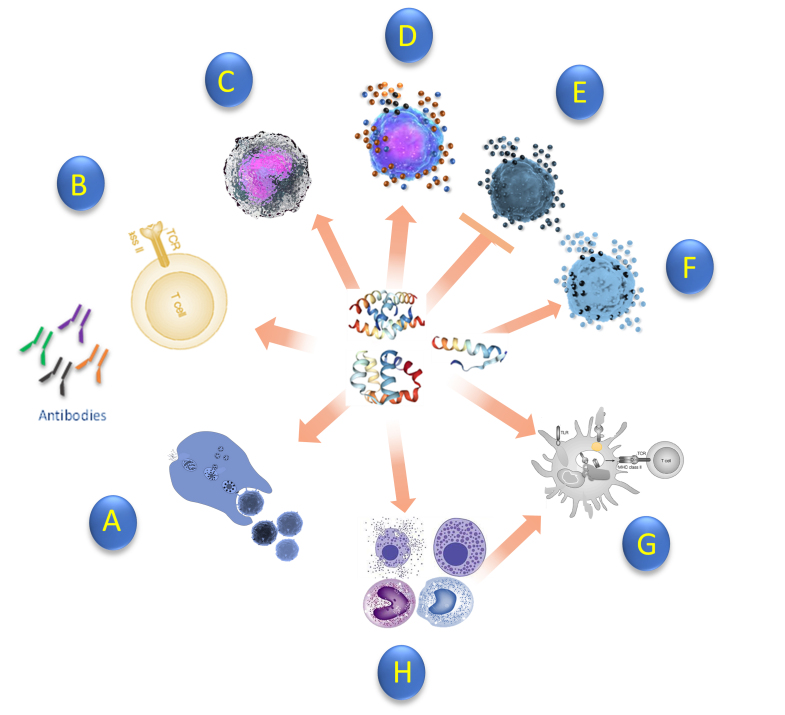
Immunological response of antimicrobial peptide against bacteria. (A) Enhance phagocytosis (B) Naïve T cell (C) Induce chemokines (D) Induce pro-inflammatory cytokines (E) Suppress pro-inflammatory cytokines (F) Induce anti-inflammatory cytokines (G) Promote dendritic cell differentiation (H) Recruitment of monocytes, mast cells, and Macrophages which promotes cell differentiation. AMP exhibits antibacterial and immunomodulatory activities against pathogens. Peptides can also enhance bacterial phagocytosis of IgG-opsonized bacteria and suppress neutrophil apoptosis [163,164].
A novel peptide sparanegtin was also reported to exhibit antimicrobial activity against both gram-positive and gram-negative bacteria. Interestingly, the peptide was also reported to display an immunoprotective role in addition to exerting immunomodulatory effects in an in vivo infection model experiment [99]. Other studies reporting host antimicrobial immune responses when treated with peptides are also available [100,101]. Therefore, more studies detailing the mechanisms of action of membrane and non-membrane targeting peptides are needed to design better peptides targeting extensively drug-resistant pathogens. Understanding the exact mechanisms of the actions of peptides is the major avenue or route to easily using peptides in treating AMR infections.
Application of Peptides Against Drug-Resistant Bacteria
Multi-drug resistant pathogens can be made to be susceptible to antibiotics when treated with AMP. The cell-membrane permeabilization activity of AMP makes it possible for a peptide to sensitize bacteria to antibiotics. The peptide basically acts like an enhancer, enhancing the activity of antibiotics originally not effective against resistant pathogens [102]. Rázquin-Olazarán et al. [103] demonstrated the potentiating activity of peptides. Three different commercially prepared peptides, namely P4-8, P4-9, and P4-18, were used against multi-drug resistant P. aeruginosa (ps4). This pathogen was found to employ different antibiotic resistance mechanisms, including the acquisition of cephalosporinase AmpC and the efflux pump MexAB-OprM. Exposure to P4-9 triggered rapid changes in the bacterial cell membrane. Interestingly, the descendants of the cells that survived the exposure remarkedly showed permeability to certain hydrophobic compounds up to 20 h, even when grown in the absence of the peptide. The peptide P4-9 did potentiate the effect of antibiotic novobiocin in a process the authors referred to as “Post-Antibiotic Effect-associated Permeabilization.” Similarly, administration of other antibiotics such as macrolide, cephalosporins, ciprofloxacin, rifampin, and fosfomycin after the withdrawal of P4-9 also showed “Post-Antibiotic Effect-associated Permeabilization” at a varying magnitude. This study demonstrates the potential of AMP in overcoming drug-resistant strategies employed by pathogens. It explicitly demonstrates the therapeutic outcomes of a combined AMP-antibiotic treatment. Therefore, AMP can induce a post-antibiotics effect. This effect could be enough to influence the susceptibility of the bacteria to antibiotics. More importantly, a peptide might not even be needed to enhance antibiotics activity, provided the target bacteria sustains its sensitization to the antibiotics previously induced by a peptide.
In another encouraging investigation demonstrating the therapeutic potentials of AMP, D-enantiomeric protease-resistant peptides (DJK-6) were used to both prevent and eradicate preformed biofilms in the K. pneumonia-resistant strain [104]. The peptide potentiated the activity with β-lactam antibiotics meropenem, imipenem, and cefepime, inhibiting K. pneumonia. The ability of the peptides to prevent and/or disrupt biofilms formed by this pathogen demonstrates the therapeutic potential of AMP. A similar study using D-enantiomeric peptides showed that the peptides eradicated wild-type and multi-drug resistant biofilm and protected against infection due to P. aeruginosa [105]. A recent study by Denardi et al. [106] reported the antibacterial activity of several AMPs (MSI-78, h-Lf1-11, LL-37, magainin-2, and fengycin 2B). From the results, the AMPs showed bactericidal activity against all the tested clinical bacterial strains (E. coli, MRSA, P. aeruginosa, Acinetobacter baumannii, and carbapenem-resistant K. pneumoniae).
Furthermore, a brevinin-1 peptide derived from the skin secretion of frog and its analog was tested against diverse pathogens. Properties such as the antibiofilm, antimicrobial, and hemolytic activity of the peptides and their analogs were evaluated. The peptide was designed by adjusting the conformational structure, hydrophobicity, and net charge. This approach led to the production of a peptide with enhanced therapeutic potential. The peptide and its derivatives had a broad-spectrum activity against the pathogens, inhibited biofilm formation, and eradicated mature biofilms of Methicillin-resistant S. aureus (MRSA) and Enterococcus faecalis. These peptides were also proven to permeate E. coli outer membrane dose-dependently. In the in vitro analysis, the analog of the peptide also reduced mortality in the Galleria mellonella larva infected with MRSA [107].
On the other hand, AMP can also be modified for improved therapeutic potential. Specifically, chemical modifications and the use of non-coded amino acids could be viable approaches to overcome the limitations associated with the use of peptides. For example, a study by Li et al. [108] using a modified N6NH2 peptide showed promising antimicrobial activity. This peptide was modified to increase its stability and antibacterial activity. Analog (N6PNH2 and V112N6NH2) were generated by substituting some amino acids in formulating the AMP. At a higher concentration of 2xMIC, the parent peptides and their analog, with the exception of V112N6NH2, showed bacteriolytic activity against Aeromonas veronii, with DN6NH2 having the strongest activity. In addition, biofilm formation was significantly decreased. The analog Guo-N6NH2 displayed low toxicity in the mice and protected the infected mice from bacterial killing. For the mechanisms of action of the peptide, it was reported that the analogs exhibited different antimicrobial activity through cell permeabilization, interaction with DNA, and the formation of outer membrane vesicles (OMVs) [108]. An interesting finding of the study was that the formulated peptide had a significantly higher outer membrane penetration ability. Also, the peptide prevents bacterial translocation, inhibits proinflammatory cytokines, and increases the expression of anti-inflammatory cytokines, which all help to alleviate multiple-organ damage. This study provided a novel method for peptide design targeting multi-drug resistant pathogens. A more recent study by Shang et al. [109] showed that tryptophan-containing peptides could attenuate the quorum sensing in P. aeruginosa MRPA0108 by downregulating the expression of some genes (lasA, lasB, rhlA, and rhlB) in a dose-dependent manner. The peptides disrupted LasB elastase expression and LasA protease enzymes by 24% and 44%, respectively. It also prevented the rhl gene from producing pyocyanin and rhamnolipids by 73% and 44%, respectively, and Psl (a crucial biofilm matrix polysaccharide). Overall, the study demonstrated the roles of AMP in changing bacterial tolerance to antibiotics.
In more studies demonstrating the antibiofilm potential of AMP, a recent study by Elsalem et al. [110] reported an engineered cationic AMP, WLBU2, having a broad-spectrum activity against several multidrug-resistant pathogens with significant inhibition of biofilm formation. The AMP showed limited host cytotoxicity and can be synergistically combined with antibiotics for improved therapeutic activity. A previous study by Elsalem et al. [111] also reported the therapeutic potential of the WIBU2 peptide. Ordinarily, treating infection due to biofilm using conventional antibiotics usually poses some challenges. Most examples in this review demonstrate the dual antibacterial activity of AMP, a feature not commonly seen in conventional antibiotics.
A recent study by Degasperi et al. [112] also showed that an elastase-activated D-BMAP18 peptide exhibits good antimicrobial and anti-inflammatory potentials with low cell toxicity. Another recent study by Koch and his colleague showed that deep mutational scanning of DNA-encoded AMPs is a viable approach for optimizing AMPs. This was demonstrated using AMP, Bac7. The Bac7 in the study exhibited strong activity against drug-resistant E. coli. Interestingly, the activity was less dependent on SbmA (an essential inner membrane protein transporter commonly used by gram-negative bacteria to move AMPs rich in a proline to the cytosol, where they inhibit translation). The peptide also showed strong inhibition of ribosomes with low cytotoxicity, making it a desirable therapeutic candidate [113]. Another study by Lu et al. [114] showed that the Nigrocin-PN with low hemolytic and cytotoxic activity has a broad-spectrum activity against multi-drug resistant pathogens. The AMP in the study did not only show efficient antimicrobial activity but also substantially ameliorated pulmonary inflammation triggered by K. pneumonia in vivo. Nigrocin-PN also showed synergistic effects with antibiotics (ampicillin) by delaying resistance acquisition by S. aureus. The mechanism of action of this AMP was via bacterial membrane disruption, thus, demonstrating the mechanisms of action of AMP in combination with antibiotics. Also, the “Rana box” function was studied, and interestingly, it played a crucial role in significantly lowering the toxicity of the AMP without any influence on the antibacterial activity of the peptide.
At this point, it is interesting to emphasize that using a computational modeling approach can allow for efficient AMP design. In addition to predicting AMP function, this in silico approach is particularly useful because in vitro testing can be expensive and time-consuming. Combining in silico prediction tools with in vitro testing can be crucial in identifying AMP with good antimicrobial activity. This approach is fundamental in engineering an effective AMP and optimizing existing AMP. Several researchers have used several computational approaches with promising therapeutic results [115-117]. Recently, Bobde et al. [118] used Ab initio computational approach to design a novel narrow-spectrum PHNX AMP against drug-resistant E. coli and S. aureus. The designed biomaterial showed significant activity against E. coli than S. aureus but displayed some hemolysis in the cytotoxicity experiment involving human red blood cells (RBCs). Also, Tram et al. [119] in their study designed de novo synthetic peptides (BTT2-4 and BTT6). The peptide (BTT1) showed broad-spectrum activity against colistin-resistant Enterobacterales and MRSA. The interaction of the peptide with LPS was sufficient to induce inner membrane lysis of the bacterial cell. Interestingly, in addition to being non-cytotoxic, the peptide activity was more profound in the presence of saline and trypsin, which are proteolytic molecules that usually inhibit the activity of peptides.
So far, few synthetic AMP has advanced to clinical therapeutic application despite the promises. In order to bridge this gap, more studies are needed at all levels. More in vivo studies involving other gram-negative and gram-positive pathogens are needed to profile the full range of AMP activity completely. Moreover, substituting the appropriate AMP residue could help to improve their therapeutic index. However, despite the potential of AMP, bacterial resistance to AMP has been reported in several studies [7,120] (Figure 3). In addition, most of the mechanism’s bacteria use to evade antibiotics attack are shared with AMP. However, despite these resistance issues, AMP offers better potential, especially when combined or conjugated with other biomaterials.
Figure 3.
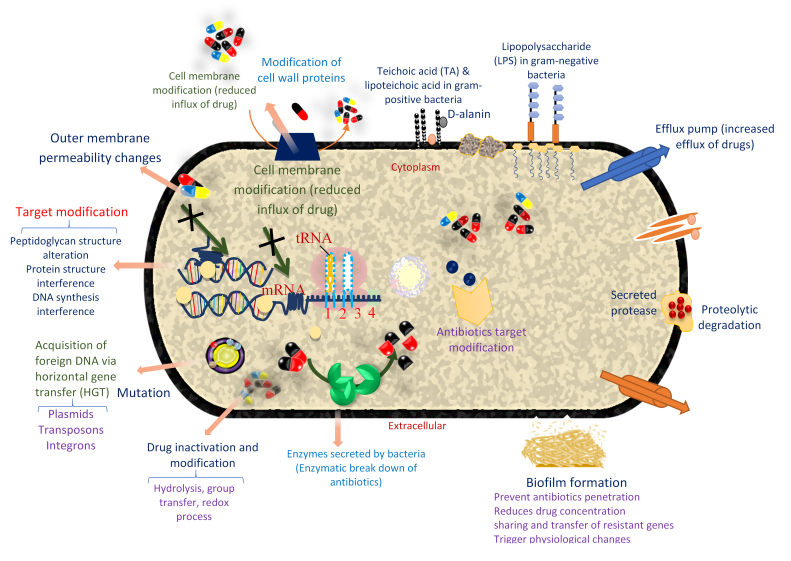
Mechanism of bacteria resistance to antibiotics and AMP. Resistance to antibiotics is generally due to cell membrane modification, target modification, enzymatic breakdown of antibiotics, drug inactivation and modification, mutation, and acquisition of foreign DNA via HGT and biofilm formation. Some of these resistance mechanisms in antibiotics are shared by AMP. Specifically, in AMP, resistance is due to extracellular protease-mediated degradation, altered cell surface changes, repulsion of AMP through changes in the cell wall and membrane surface changes, biofilm formation, modification of host cellular processes, and LPS modification. There is also AMP sequestration/inactivation and AMP-induced gene induction/downregulation. Inactivation of some genes can also lead to loss of LPS production and reduction in the binding of AMP. The anionic feature of bacterial cell membrane makes them a good binding site for cationic AMP. Therefore, teichoic acid modification reduces the negative charge in the bacterial cell membrane. Resistance to AMP is also due to the active efflux of AMPs. D-alanine alteration of teichoic and lipoteichoic acids in gram-positive bacteria is another AMP resistance mechanism in some bacteria. Moreover, some pathogens, especially the gram-negatives known for the presence of diverse polysaccharides such as K. pneumonia, resist peptides by forming capsular polysaccharides [23,165].
Finally, the fact that AMP can act against diverse pathogens with very little concern of toxicity and ease of manipulation to reduce undesirable toxic effects, as we are demonstrating, makes them particularly more promising than other biomaterials for tackling AMR pathogens. For instance, in nanomaterials, toxicity to host cells and their negative effects on human microbiota is a critical concern [121]. Moreover, the ability of the biological system to clear nanomaterials which often leads to long-term accumulation resulting in some detrimental effects is also a major concern [122]. In bacteriophages, the narrow host range is a critical factor in their use as an antimicrobial agent because most infections are polymicrobial. Moreover, phages alone are usually not enough to induce an immunological reaction [123]. Sometimes, virulent genes can even be transferred to the target bacteria, making them even more pathogenic [124]. Although scientists are exploring diverse means to mitigate these limitations, AMP also offers several good advantages when compared to other biomaterials. In AMP, there is slower emergence of resistance. Moreover, they have broad-spectrum activity, can easily modulate immunological reactions, and have lower synthetic costs than other biomaterials.
The immunomodulatory activity of AMP is currently being harnessed in vaccine formulation targeting AMR as AMP has the potential for use in vaccine design. Some emerging technologies use AMP as a vaccine or as adjuvant to induce immunological responses against microbes. Usually, the first consideration in using AMP for a vaccine is to identify the immunodominant region of the epitope or peptide capable of triggering adequate immune responses [125]. For selecting candidate peptides, in silico bioinformatics approach has been helpful [126]. After identifying, selecting, and constructing candidate epitopes or antigens, there is the synthesis of the antigenic peptides. This is followed by conjugation to adjuvants or carrier molecules for bioactivities improvement. Finally, there is immuno-profiling of the constructs and other preclinical and clinical trial studies. For peptide vaccines, adjuvants and matching delivery systems are usually needed to improve the therapeutic value [127]. So far, several peptide vaccines have shown immunological effects against diverse drug-resistant pathogens. Few examples of these have been seen in LL-37 used against S. aureus, and E. coli infection [128,129], and hLF1-11 used against E. coli and P. aeruginosa [130]. Therefore, using AMP in a vaccine is a promising approach to fully harnessing AMP’s potential in alleviating the AMR crisis.
Combination of Antimicrobial Peptides with Other Biomaterials and Other Advances in AMP’s Design
Combination therapy is the simultaneous use of two or more antibiotics or adjuvant(s) [131]. The combination of antimicrobials is vital as it reduces dosages, attenuates negative side effects, and enhances the selectivity of compounds [75]. Due to the rapid killing activity of AMP against diverse pathogens, they have been considered a potential alternative for combating multi-drug resistance when used in a single or combined with other biomaterials [8,132]. In addition, antimicrobial peptides can be used alongside an antibiotic during treatment. This method usually leads to an increase in the lifetime of antibiotics. It also reduces the dosage of peptides needed for significant activity and potentiates the effect of antibiotics [133]. Many studies have reported the antimicrobial effect of peptides and their combination with common antibiotics against clinical infections [107,134] (Table 1).
Table 1. Synergistic Combination of AMPs with Other Biomaterials (eg, Antibiotics).
| AMP | Synergistic molecules | Target organisms | References |
| Nisin | Colistin | Pseudomonas biofilms | [157] |
| 4-hexylresorcinol | Polymyxin | K. pneumonia KPM9, A. baumannii, P. aeruginosa | [134] |
| Dermaseptin | Dermaseptin | E. coli, P. aeruginosa, S. aureus | [166] |
| Brevinin-1 | MRSA, E. coli, Enterofecalis | [107] | |
| N6NH2 + Analog | Rifampicin+ Streptomycin+sulfate +doxycycline | A. veronii, ACCC61732 | [108] |
| Tridecaptin B | rifamcipin | A. baumannii | [138] |
| Tridecaptin M | Rifampicin, vancomycin, and ceftazidime | Extremely drug-resistant A. baumannii | [167] |
| Ranalexin | Endopeptidase lysostaphin | S. aureus (MRSA) | [138] |
| Cholestryramine | daptomycin | E. faecium BL00239-1, PR00708-14 | [168] |
| Melimine (Mel4) | Melimine +ciprofloxacin, Mel 4+ciprofloxacin | P. aeruginosa 27853 | [169] |
| Dermaseptin | Dermaseptin | E. coli, P. aeruginosa, S. aureus | [170] |
| Lactoferricin | Ciprofloxacin, ceftazidime | P. aeruginosa | [157] |
| P10 | Doripenem/Ceftazidim | P. aeruginosa, A. baumannii | [157] |
| Gad-1 | Kanamycin, ciprofloxacin | P. aeruginosa | [171] |
In combination with an antibiotic such as streptomycin sulfate, rifampicin, kanamycin sulfate, and doxycycline hyclate, N6NH2 showed synergistic activity against A. veronii and an additive effect with all the antibiotics as previously reported. The therapy showed no antagonistic activity with any antibiotics [108]. In addition, a multidrug-resistant Staphylococcus epidermidis (MRSE) strain was subjected to treatment with two peptides I1WL5W and I4WL5W. The two peptides were synergistic with erythromycin, ampicillin, penicillin, and tetracycline and had an additive effect with ceftazidime. In combination with penicillin, both peptides displayed high synergistic activity against biofilm formation and minimal activity with the other antibiotics. Notably, the peptides increased the permeability activity of penicillin. An in vivo experiment in the study using the infection model of a multi-drug resistant strain showed a positive effect as there was a very high healing potency on the lesion inflicted on the model [133]. Tumor necrosis factor (TNF-α) and IL-6 in the serum of the model showed that MRSE induced a systemic proinflammatory cytokines response with a rise in the levels of the proinflammatory cytokines TNF-α and IL-6. After treatment with a combination of penicillin and 11WL5W and penicillin and 14WL5W, there was an inhibition of the expression of these cytokines by 96.8% and 95.6%, respectively [133].
Another study by Kampshoff et al. [135] evaluated the synergistic effect of three AMPs with different antibiotics. The result showed a significant synergistic effect against drug-resistant P. aeruginosa when melamine and ciprofloxacin were combined. Tridecaptins is a polypeptide used in the treatment of gram-negative bacteria and has previously been reported to have a very low intrinsic activity against A. baumannii and cannot efficiently permeabilize the outer membrane [136,137]. Further study was conducted by the author using tridecaptin M peptide in combination with rifampicin against the same strain, A. baumannii ATCC 19606, to ascertain the efficacy of the combinatorial therapy. The efficacy of the combination was tested against the pathogen using a blood infection model. Interestingly, tridecaptin was able to permeabilize the pathogen’s membrane but was insufficient to cause cell death. By using the checkerboard assay, the peptide significantly reduces the dosage of rifampicin. In the rabbit blood, the combination of rifampicin and tridecaptin eradicated the pathogen by ~1.5 logs CFU within 4 hours of infection [138]. Antimicrobials such as those containing polypeptides families (colistin B, polymyxin B1, polymyxin B2, bacitracin, and colistin A) have also been used in combination therapy with AMP for the treatment of drug-resistant pathogens. They have also been reported to have synergistic activity against K. pneumonia, P. aeruginosa, and A. baumannii, which were all multi-drug resistant [139].
Finally, encapsulation of AMP in carbon nanotubes could help to improve their interaction with biomolecules and enhance their therapeutic potential [140-142]. Furthermore, conjugating AMP with biomaterials such as hydrogels, nanoparticles, and electrospun fibers could help to overcome the limitations associated with several AMPs [143]. AMPs have also been successfully decorated with nanoparticles and polymers with enhanced antimicrobial activity [144]. Several other recent advances in recombinant tactics and molecular engineering offer the potential for improved AMP activity against AMR pathogens [145]. Reports are also available on the chemical modification of AMPs for improved therapeutic potentials [146,147]. However, for an up-to-date understanding of the advances in the design of AMPs conjugates targeting AMR pathogens, readers are highly encouraged to read Silva et al. [148].
Conclusion and Future Perspectives
Despite the potential of AMPs in helping to solve global AMR issues, several challenges remain to be addressed. Limited stability is a major issue in the full translation of AMPs for clinical use. AMPs are often susceptible to degradation by protease [16,149]. Thus, their use for tropical applications is often limited. This issue can be overcome by adjusting or modifying their sequences. The high extraction costs [77], lack of specificity and cytotoxicity [16], poor bioavailability, and short half-lives [150] are just some of the major issues. Moreover, like antibiotics, organisms have several mechanisms through which they evade the action of antimicrobial peptides (Figure 3). So far, not much has been learned about the mechanisms of bacterial resistance to AMPs. This issue is a major setback to the full translation of most AMP studies into clinical use. As recently reviewed by Vimberg et al. [151], bacterial membrane plays a crucial role in resistance to AMP, and this area warrants further investigation. Considering that AMPs have been proposed as potential alternatives to antibiotics, deep insight and understanding into their possible mechanism of resistance by bacteria will be of great significance in properly engineering and modifying AMPs for use against extensive drug-resistant pathogens.
Interestingly, AMPs have pharmacodynamic features that make them reduce resistant evolution in target pathogens. Also, they can effectively be used synergistically with one another and with other biomaterials (antibiotics, nanomaterials, phages, etc.) for improved therapeutic value. Currently, several AMPs for use against bacterial infectious agents are at different stages of clinical trials (Table 2). Although most of these AMPs with broad-spectrum activity against AMR bacterial infections have been reported, only a few have reached the market. Unfavorable pharmacokinetics, cytotoxicity, and several other issues hinder successful clinical trials. Encapsulating AMPs with other biomaterials could help prevent degradation by proteases and enhance their stability [152]. For example, AMPs could be encapsulated with nanomaterials [153], as we have previously mentioned in this paper. AMPs can also be implanted into drug delivery vehicles, thus, enhancing specific target binding [154].
Table 2. Selected AMPs in Different Stages of Clinical Trials.
| AMP | Clinical applications | Administration | Mechanisms of action | Stage of development | Company |
| LTX-109 (lytixar) | Skin infection caused by gram-negative bacteria and gram-positive bacteria such as methicillin resistant and sensitive S. aureus nasal carriage | Topical | Disrupt bacteria via permeabilization of membrane | Phase 2 | Lytix Biopharma |
| Omiganan (MB1-226, MX-594AN) | Anti-inflammatory | Topical | Catheter-associated infections | Phase 3 (completed) | Migenix |
| XF-73 | Infections due to Staphylococcus during surgeries, nasal carriage | Topical | Disrupt bacteria via permeabilization of membrane | Phase 2 (recruiting) | Destiny Pharma |
| Surotomycin | Diarrhea due to C. difficile | Oral | Depolaries bacterial membrane | Phase 3 (completed) | Cubist Pharmaceuticals, Merck and Co |
| P2TA | Infections caused by necrotizing soft tissues | Intravenous | Immunomodulation | Phase 3 | Atox Bio |
| Opebacan | Wound, burn, meningococcal infections | Intravenous | Disrupt bacterial membrane via permeabilization | Phase 2 | Xoma |
| DPK 060 | Infections due to eczematous lesions | Tropical | - | Phase 2 | DermaGen AB |
| Murepavadin (POL 7080) | Lower respiratory infections due to P. aeruginosa and ventilator-associated Pneumonia | Intravenous | Disrupt LPS and transport protein D | Phase 3 | Polyphor |
| PMX-30063 (brilacidin) | Bacterial skin infections | Intravenous or tropical | Disrupt bacterial membrane via permeabilization | Phase 2 | Innovation pharmaceuticals |
| Histatin-1 and -3, P-113 (histamin derivatives) | Chronic infections due to P. aeruginosa, gingivitis, and periodontal diseases | Tropical | Generation of ROS | Phase 1,2,3 | Demgen |
| Human Lactoferrin derived peptide (hLF1-11) | Infectious diseases due to gram-negative bacteria | Intravenous | Disrupt cells by binding to DNA | Phase 1 (completed) | AM-Pharma |
| C15G2, Novispirin analogue | Dental caries infections due to Streptococcus mutans | Oral | Selective disruption of membrane and intracellular targets | Phase 2 | |
| Brilacidin (PMX-30063), defensin mimetic | Broad spectrum antibacterial therapy | Oral | Disruption of bacterial membrane | Phase 2 | |
| Opebacan (Rbp121, neuprex) | LPS/endotoxin of gram-negative bacteria | Intravenous | Damage to bacterial membrane | Phase 2 | |
| XOMA-629 (XMP-629) | LPS/endotoxins of gram-negative bacteria | Tropical | Damage to bacterial membrane | Phase 2 | |
| Novarifyn (NP432) | Bacterial infection | Tropical | Disruption of bacterial membrane | Phase 1 | |
| AMP PL-18 | Bacterial vaginosis and mixed vaginitis | Tropical | - | Phase 1 | Protelight Pharmaceuticals Australia PTY LTD |
Furthermore, the synergistic combination of AMP with other biomaterials has shown promise in treating biofilm infection caused by S. aureus [155] and P. aeruginosa [156]. Also, combining AMP with amphiphilic hydrogels and other nanomaterials offers an attractive approach. For example, this approach has been studied by Atefyekta et al. [150]; in their study, AMP was covalently bonded to mesoporous hydrogels and tested in vivo in S. epidermidis, S. aureus, P. aeruginosa, MDR E. coli, and MRSA. The AMP-modified mesoporous hydrogel showed a promising bacterial killing effect and also increased stability as its antimicrobial activity was sustained up to 48h in human blood serum. In a recent exciting investigation by Jahangiri et al. [157], the therapeutic potential of AMP in combination with conventional antibiotics was also demonstrated. It was shown that the combined effect of AMP (Nisin and P10) with five common antibiotics (ciprofloxacin, colistin, tobramycin, ceftazidime, and doripenem) was highly synergistic as the combination showed strong inhibitory activity against extensively drug-resistant A. baumannii and colistin-resistant P. aeruginosa [157]. All these promising preclinical results involving AMP and other biomaterials are enough shreds of evidence that the advances in research and technology could offer a solid basis for engineering innovative AMP-based antimicrobials. The combination of AMP with other biomaterials will not only eliminate drug-resistant bacteria but will prevent resistant development and enhance the individual biomaterials’ therapeutic effects.
The scientific community is in the age of synthetic biology. Currently, biological materials are engineered or manipulated to exhibit some features they ordinarily do not possess. Therefore, genetic engineering incorporating different biomaterials should be embraced as an integral approach to producing high-quality and highly effective AMP and other molecules needed to overcome the drug-resistant crisis. AMPs should be synergistically combined with other emerging biomaterials (nanomaterials, phages, antibodies, etc.) while adapting structural modification of AMPs in addition to in silico prediction of their activity which will help in better utilization of AMPs. Importantly, a strategy to overcome some limitations of AMP could be to incorporate unnatural amino acids, as successfully investigated in several studies [158]. Natural amino acid rearrangement could also help in overcoming the challenges. For example, Wang et al. [159] work using a systematic amino acid rearrangement generated AMP with high proteolytic resistance with a membrane-disruptive mechanism in E. coli.
In summary, although AMPs are promising agents with good antimicrobial activity against AMR pathogens, research should be intensified to overcome the challenges limiting their full implementation and translation into clinics. Specifically, further study on the design and formulation of AMP incorporating nanomaterial is needed. AMP-conjugated nanoparticles are a desirable approach because of the possibility of fine-tuning the combination. Moreover, exploiting the emerging genomic options and bioinformatic tools could help to predict the activity of AMPs even before synthesizing them; this could help to prevent unnecessary synthetic efforts and wastage of funds. Efforts should also be fortified to improve the AMPs database. For the latest updates on the evolution of the AMP database, readers are encouraged to read the recent paper by Wang et al. [160].
Therefore, reducing the burden and impact of AMR to the barest minimum could be possible, and AMPs are one of the sharpest tools with the potential to help achieve this feat. However, as previously mentioned, other emerging approaches to tackling AMR bacterial infections should also be adequately given attention. Generally, there is no single weapon and tactic to overcome the AMR crisis. Thus, these emerging approaches should complement AMPs in the ongoing efforts to find a solution to the rapidly increasing AMR infectious agents.
Glossary
- AMP
Antimicrobial peptide
- AMR
Antimicrobial resistance
- ESBL
Extended spectrum β-lactamases
- WWTPs
Wastewater treatment plants
- HGT
Horizontal gene transfer
- LPS
Lipopolysaccharide
- MRSA
Methicillin-resistant Staphylococcus aureus
- MRSE
Multidrug-resistant Staphylococcus epidermidis
- NO
Nitric oxide
- TNF-α
Tumor necrosis factor
- IL
Interleukin
Author Contributions
IEM (ORCID: https://www.orcid.org/0000-0003-4715-8019) wrote the first draft of the paper. EIN (ORCID: https://www.orcid.org/0000-0003-4432-0885) critically reviewed it to improve content/quality. All authors read and approved the final version of the manuscript.
Conflict of interest
The authors report no conflict of interest.
Funding
Authors received no funding for this study.
References
- Mba IE, Okeke OP, Sharndama HC, Osondu-Chuka GO, Ukomadu J, Ugwu C. Antimicrobial resistance: revisiting the mechanisms of resistance. Access Microbiol. 2022;4(5):po0577. 10.1099/acmi.ac2021.po0053 [DOI] [Google Scholar]
- Wang CH, Hsieh YH, Powers ZM, Kao CY. Defeating Antibiotic-Resistant Bacteria: Exploring Alternative Therapies for a Post-Antibiotic Era. Int J Mol Sci. 2020. Feb;21(3):1061. 10.3390/ijms21031061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mba IE, Nweze EI. Nanoparticles as therapeutic options for treating multidrug-resistant bacteria: research progress, challenges, and prospects. World J Microbiol Biotechnol. 2021. May;37(6):108. 10.1007/s11274-021-03070-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mba IE, Nweze EI. Application of nanotechnology in the treatment of infectious diseases: an overview. In: Hameed S, Rehman S. Nanotechnology for infectious diseases. Singapore: Springer; 2022. 10.1007/978-981-16-9190-4_2 [DOI] [Google Scholar]
- Anyaegbunam NJ, Anekpo CC, Anyaegbunam ZK, Doowuese Y, Chinaka CB, Odo OJ, et al. The resurgence of phage-based therapy in the era of increasing antibiotic resistance: from research progress to challenges and prospects. Microbiol Res. 2022. Nov;264:127155. 10.1016/j.micres.2022.127155 [DOI] [PubMed] [Google Scholar]
- Magana M, Pushpanathan M, Santos AL, Leanse L, Fernandez M, Ioannidis A, et al. The value of antimicrobial peptides in the age of resistance. Lancet Infect Dis. 2020. Sep;20(9):e216–30. 10.1016/S1473-3099(20)30327-3 [DOI] [PubMed] [Google Scholar]
- Moravej H, Moravej Z, Yazdanparast M, Heiat M, Mirhosseini A, Moosazadeh Moghaddam M, et al. Antimicrobial Peptides: Features, Action, and Their Resistance Mechanisms in Bacteria. Microb Drug Resist. 2018. Jul/Aug;24(6):747–67. 10.1089/mdr.2017.0392 [DOI] [PubMed] [Google Scholar]
- Rima M, Rima M, Fajloun Z, Sabatier JM, Bechinger B, Naas T. Antimicrobial Peptides: A Potent Alternative to Antibiotics. Antibiotics (Basel). 2021. Sep;10(9):1095. 10.3390/antibiotics10091095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro BP, Zasloff M, Rolff J. Antimicrobial peptides: application informed by evolution. Science. 2020. May;368(6490):eaau5480. 10.1126/science.aau5480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra B, Reiling S, Zarena D, Wang G. Host defense antimicrobial peptides as antibiotics: design and application strategies. Curr Opin Chem Biol. 2017. Jun;38:87–96. 10.1016/j.cbpa.2017.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rončević T, Puizina J, Tossi A. Antimicrobial peptides as anti-infective agents in pre-post-antibiotic era? Int J Mol Sci. 2019. Nov;20(22):5713. 10.3390/ijms20225713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell M, Wenc AK, Donaghy CM, Wasche DV, Abissi I, Naing MD, et al. Exploring synergy and its role in antimicrobial peptide biology. Methods Enzymol. 2022;663:99–130. 10.1016/bs.mie.2021.09.017 [DOI] [PubMed] [Google Scholar]
- Wang C, Hong T, Cui P, Wang J, Xia J. Antimicrobial peptides towards clinical application: delivery and formulation. Adv Drug Deliv Rev. 2021. Aug;175:113818. 10.1016/j.addr.2021.05.028 [DOI] [PubMed] [Google Scholar]
- Annunziato G, Costantino G. Antimicrobial peptides (AMPs): a patent review (2015-2020). Expert Opin Ther Pat. 2020. Dec;30(12):931–47. [DOI] [PubMed] [Google Scholar]
- Abdi M, Mirkalantari S, Amirmozafari N. Bacterial resistance to antimicrobial peptides. J Pept Sci. 2019. Nov;25(11):e3210. 10.1002/psc.3210 [DOI] [PubMed] [Google Scholar]
- Greco I, Molchanova N, Holmedal E, Jenssen H, Hummel BD, Watts JL, et al. Correlation between hemolytic activity, cytotoxicity and systemic in vivo toxicity of synthetic antimicrobial peptides. Sci Rep. 2020. Aug;10(1):13206. 10.1038/s41598-020-69995-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinakumar R, Walkenhorst WF, Wimley WC. Broad-spectrum antimicrobial peptides by rational combinatorial design and high-throughput screening: the importance of interfacial activity. J Am Chem Soc. 2009. Jun;131(22):7609–17. 10.1021/ja8093247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbensgaard A, Mordhorst H, Overgaard MT, Aarestrup FM, Hansen EB. Dissection of the antimicrobial and hemolytic activity of Cap18: generation of Cap18 derivatives with enhanced specificity. PLoS One. 2018. May;13(5):e0197742. 10.1371/journal.pone.0197742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr CG, Wimley WC. Antimicrobial peptides are degraded by the cytosolic proteases of human erythrocytes. Biochim Biophys Acta Biomembr. 2017. Dec;1859(12):2319–26. 10.1016/j.bbamem.2017.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer DK, Torres MD, Duay SS, Lovie E, Simpson L, von Köckritz-Blickwede M, et al. Antimicrobial Susceptibility Testing of Antimicrobial Peptides to Better Predict Efficacy. Front Cell Infect Microbiol. 2020. Jul;10:326. 10.3389/fcimb.2020.00326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretta A, Scieuzo C, Petrone AM, Salvia R, Manniello MD, Franco A, et al. Antimicrobial Peptides: A New Hope in Biomedical and Pharmaceutical Fields. Front Cell Infect Microbiol. 2021. Jun;11:668632. 10.3389/fcimb.2021.668632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Sahoo N, Bhunia A. Antimicrobial Peptides and their Pore/Ion Channel Properties in Neutralization of Pathogenic Microbes. Curr Top Med Chem. 2016;16(1):46–53. 10.2174/1568026615666150703115454 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S, Bharath Prasad AS, Mehta CH, Nayak UY. Antimicrobial peptide polymers: no escape to ESKAPE pathogens-a review. World J Microbiol Biotechnol. 2020. Aug;36(9):131. 10.1007/s11274-020-02907-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimley WC. Describing the mechanism of antimicrobial peptide action with the interfacial activity model. ACS Chem Biol. 2010. Oct;5(10):905–17. 10.1021/cb1001558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison L, Zembower TR. Antimicrobial Resistance. Gastrointest Endosc Clin N Am. 2020. Oct;30(4):619–35. 10.1016/j.giec.2020.06.004 [DOI] [PubMed] [Google Scholar]
- Aslam B, Khurshid M, Arshad MI, Muzammil S, Rasool M, Yasmeen N, et al. Antibiotic Resistance: One Health One World Outlook. Front Cell Infect Microbiol. 2021. Nov;11:771510. 10.3389/fcimb.2021.771510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming A. Penicillin [Nobel Prize Lecture, December 11, 1945]. Nobel lectures in physiology or medicine, 1942-1962. World Scientific Publishing Company, Singapore, 1999. [Google Scholar]
- Zaman SB, Hussain MA, Nye R, Mehta V, Mamun KT, Hossain N. A Review on Antibiotic Resistance: Alarm Bells are Ringing. Cureus. 2017. Jun;9(6):e1403. 10.7759/cureus.1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raouf FE, Benyagoub E, Alkhudhairy MK, Akrami S, Saki M. Extended-spectrum beta-lactamases among Klebsiella pneumoniae from Iraqi patients with community-acquired pneumonia. Rev Assoc Med Bras. 2022. Jun;68(6):833–7. 10.1590/1806-9282.20220222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saki M, Farajzadeh Sheikh A, Seyed-Mohammadi S, Asareh Zadegan Dezfuli A, Shahin M, Tabasi M, et al. Occurrence of plasmid-mediated quinolone resistance genes in Pseudomonas aeruginosa strains isolated from clinical specimens in southwest Iran: a multicentral study. Sci Rep. 2022. Feb;12(1):2296. 10.1038/s41598-022-06128-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akinyemi KO, Al-Khafaji N, Al-Alaq FT, Fakorede CO, Al-Dahmoshi H, Iwalokun BA, Akpabio I, Alkafaas SS, Saki M. Extended-spectrum Beta-lactamases Encoding Genes among Salmonella Enterica serovar Typhi Isolates in Patients with Typhoid Fever from four Academic Medical Centers Lagos, Nigeria. Revista de investigacion clinica; organo del Hospital de Enfermedades de la Nutricion. 2022;74(3):165–171. [DOI] [PubMed] [Google Scholar]
- CDC. Office of Infectious Disease Antibiotic Resistance Threats in the United States. Atlanta, GA. Retrieved from: https://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf. 2013.
- Luyt CE, Bréchot N, Trouillet JL, Chastre J. Antibiotic stewardship in the intensive care unit. Crit Care. 2014. Aug;18(5):480. 10.1186/s13054-014-0480-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellberg B, Gilbert DN. The future of antibiotics and resistance: a tribute to a career of leadership by John Bartlett. Clin Infect Dis. 2014. Sep;59(2 Suppl 2):S71–5. 10.1093/cid/ciu392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manyi-Loh C, Mamphweli S, Meyer E, Okoh A. Antibiotic Use in Agriculture and Its Consequential Resistance in Environmental Sources: Potential Public Health Implications. Molecules. 2018. Mar;23(4):795. 10.3390/molecules23040795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. Critically Important Antimicrobials for Human Medicine; 2017. (5th revision 2016). http://apps.who.int/iris/bitstream/10665/255027/1/9789241512220-eng.pdf?ua=1
- Aslam B, Wang W, Arshad MI, Khurshid M, Muzammil S, Rasool MH, et al. Antibiotic resistance: a rundown of a global crisis. Infect Drug Resist. 2018. Oct;11:1645–58. 10.2147/IDR.S173867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallinga D, Smit LA, Davis MF, Casey JA, Nachman KE. A Review of the Effectiveness of Current US Policies on Antimicrobial Use in Meat and Poultry Production. Curr Environ Health Rep. 2022. Jun;9(2):339–54. 10.1007/s40572-022-00351-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Sangthong R, McNeil E, Tang R, Chongsuvivatwong V. Antibiotic use in chicken farms in northwestern China. Antimicrob Resist Infect Control. 2020. Jan;9(1):10. 10.1186/s13756-019-0672-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosain MZ, Kabir SM, Kamal MM. Antimicrobial uses for livestock production in developing countries. Vet World. 2021. Jan;14(1):210–21. 10.14202/vetworld.2021.210-221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Boeckel TP, Brower C, Gilbert M, Grenfell BT, Levin SA, Robinson TP, et al. Global trends in antimicrobial use in food animals. Proc Natl Acad Sci USA. 2015. May;112(18):5649–54. 10.1073/pnas.1503141112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayalakshmi K, Veeraselvam M, Ramkumar PK, et al. A review on antimicrobial resistance, diagnosis and an alternative approach. J Entomol Zool Stud. 2021;9(1):1058–71. [Google Scholar]
- Samreen A, Ahmad I, Malak HA, Abulreesh HH. Environmental antimicrobial resistance and its drivers: a potential threat to public health. J Glob Antimicrob Resist. 2021. Dec;27:101–11. 10.1016/j.jgar.2021.08.001 [DOI] [PubMed] [Google Scholar]
- Cuong NV, Padungtod P, Thwaites G, Carrique-Mas JJ. Antimicrobial usage in animal production: A review of the literature with a focus on low-and middle-income countries. Antibiotics (Basel). 2018. Aug;7(3):75. 10.3390/antibiotics7030075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Founou LL, Founou RC, Essack SY. Antibiotic Resistance in the Food Chain: A Developing Country-Perspective. Front Microbiol. 2016. Nov;7:1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiseo K, Huber L, Gilbert M, Robinson TP, Van Boeckel TP. Global Trends in Antimicrobial Use in Food Animals from 2017 to 2030. Antibiotics (Basel). 2020. Dec;9(12):918. 10.3390/antibiotics9120918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehdi Y, Létourneau-Montminy MP, Gaucher ML, Chorfi Y, Suresh G, Rouissi T, Brar SK, Côté C, Ramirez AA, Godbout S. Use of antibiotics in broiler production: Global impacts and alternatives. Animal nutrition (Zhongguo xu mu shou yi xue hui). 2018;4(2):170–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquillo MG, Carlos J, Hernandez A, et al. Antibiotic and synthetic growth promoters in animal diets: review of impact and analytical methods. Food Control. 2017;72:255–67. 10.1016/j.foodcont.2016.03.001 [DOI] [Google Scholar]
- Van Boeckel TP, Glennon EE, Chen D, Gilbert M, Robinson TP, Grenfell BT, et al. Reducing antimicrobial use in food animals. Science. 2017. Sep;357(6358):1350–2. 10.1126/science.aao1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Huang L, Wang Q, Zeng H, Xu J, Chen Z. Antibiotics in aquaculture ponds from Guilin, South of China: Occurrence, distribution, and health risk assessment. Environ Res. 2022. Mar;204(Pt B):112084. [DOI] [PubMed] [Google Scholar]
- Al Amin M, Hoque MN, Siddiki AZ, Saha S, Kamal MM. Antimicrobial resistance situation in animal health of Bangladesh. Vet World. 2020. Dec;13(12):2713–27. 10.14202/vetworld.2020.2713-2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabello FC, Godfrey HP, Buschmann AH, Dölz HJ. Aquaculture as yet another environmental gateway to the development and globalisation of antimicrobial resistance. Lancet Infect Dis. 2016. Jul;16(7):e127–33. 10.1016/S1473-3099(16)00100-6 [DOI] [PubMed] [Google Scholar]
- Khmelevtsova LE, Sazykin IS, Azhogina TN, Sazykina MA. The dissemination of antibiotic resistance in various environmental objects (Russia). Environ Sci Pollut Res Int. 2020. Dec;27(35):43569–81. 10.1007/s11356-020-10231-2 [DOI] [PubMed] [Google Scholar]
- Ventola CL. The antibiotic resistance crisis: part 1: causes and threats. P&T. 2015. Apr;40(4):277–83. [PMC free article] [PubMed] [Google Scholar]
- Sartelli M, C Hardcastle T, Catena F, Chichom-Mefire A, Coccolini F, Dhingra S, et al. Antibiotic Use in Low and Middle-Income Countries and the Challenges of Antimicrobial Resistance in Surgery. Antibiotics (Basel). 2020. Aug;9(8):497. 10.3390/antibiotics9080497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triggiano F, Calia C, Diella G, Montagna MT, De Giglio O, Caggiano G. The Role of Urban Wastewater in the Environmental Transmission of Antimicrobial Resistance: The Current Situation in Italy (2010-2019). Microorganisms. 2020. Oct;8(10):1567. 10.3390/microorganisms8101567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liguori K, Keenum I, Davis BC, Calarco J, Milligan E, Harwood VJ, et al. Antimicrobial Resistance Monitoring of Water Environments: A Framework for Standardized Methods and Quality Control. Environ Sci Technol. 2022. Jul;56(13):9149–60. 10.1021/acs.est.1c08918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell MM, Truelstrup Hansen L, Jamieson RC, Neudorf KD, Yost CK, Tong A. Removal of antibiotic resistance genes in two tertiary level municipal wastewater treatment plants. Sci Total Environ. 2018. Dec;643:292–300. 10.1016/j.scitotenv.2018.06.212 [DOI] [PubMed] [Google Scholar]
- Manoharan RK, Ishaque F, Ahn YH. Fate of antibiotic resistant genes in wastewater environments and treatment strategies - A review. Chemosphere. 2022. Jul;298:134671. 10.1016/j.chemosphere.2022.134671 [DOI] [PubMed] [Google Scholar]
- González-Plaza JJ, Blau K, Milaković M, Jurina T, Smalla K, Udiković-Kolić N. Antibiotic-manufacturing sites are hot-spots for the release and spread of antibiotic resistance genes and mobile genetic elements in receiving aquatic environments. Environ Int. 2019. Sep;130:104735. 10.1016/j.envint.2019.04.007 [DOI] [PubMed] [Google Scholar]
- Fletcher S. Understanding the contribution of environmental factors in the spread of antimicrobial resistance. Environ Health Prev Med. 2015. Jul;20(4):243–52. 10.1007/s12199-015-0468-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos RJ. Studies on a bactericidal agent extracted from a soil bacillus: I. Preparation of the agent. Its activity in vitro. J Exp Med. 1939. Jun;70(1):1–10. 10.1084/jem.70.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos RJ. Studies on a bactericidal agent extracted from a soil bacillus: II. Protective effect of the bactericidal agent against experimental pieuococcus infections in mice. J Exp Med. 1939. Jun;70(1):11–7. 10.1084/jem.70.1.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarges R, Witkop B, Gramicidin AV. The Structure of Valine- and Isoleucine-Gramicidin A. J Am Chem Soc. 1965. a May;87(9):2011–20. 10.1021/ja01087a027 [DOI] [PubMed] [Google Scholar]
- Sarges R, Witkop B. Gramicidin. Vii. The Structure of Valine- and Isoleucine-Gramicidin B. J Am Chem Soc. 1965. b May;87(9):2027–30. 10.1021/ja01087a029 [DOI] [PubMed] [Google Scholar]
- Bin Hafeez A, Jiang X, Bergen PJ, Zhu Y. Antimicrobial Peptides: An Update on Classifications and Databases. Int J Mol Sci. 2021. Oct;22(21):11691. 10.3390/ijms222111691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone K, Camarda K, Spencer P, Tamerler C. Antimicrobial peptide similarity and classification through rough set theory using physicochemical boundaries. BMC Bioinformatics. 2018. Dec;19(1):469. 10.1186/s12859-018-2514-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti M. Cathelicidins, multifunctional peptides of the innate immunity. J Leukoc Biol. 2004. Jan;75(1):39–48. 10.1189/jlb.0403147 [DOI] [PubMed] [Google Scholar]
- Wang G. Improved methods for classification, prediction, and design of antimicrobial peptides. Methods Mol Biol. 2015;1268:43–66. 10.1007/978-1-4939-2285-7_3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haney EF, Straus SK, Hancock RE. Reassessing the host defense peptide landscape. Front Chem. 2019. Feb;7:43. 10.3389/fchem.2019.00043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huan Y, Kong Q, Mou H, Yi H. Antimicrobial peptides: classification, design, application and research progress in multiple fields. Front Microbiol. 2020. Oct;11:582779. 10.3389/fmicb.2020.582779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikindas ML, Weeks R, Drider D, Chistyakov VA, Dicks LM. Functions and emerging applications of bacteriocins. Curr Opin Biotechnol. 2018. Feb;49:23–8. 10.1016/j.copbio.2017.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younas S, Mazhar B, Liaqat I, Ali S, Tahir HM, Ali NM. Bacteriocin Production by Lactobacilli and Their Role as Antibacterial Tool against Common Pathogens. J Oleo Sci. 2022. Apr;71(4):541–50. 10.5650/jos.ess21424 [DOI] [PubMed] [Google Scholar]
- Wang G. Unifying the classification of antimicrobial peptides in the antimicrobial peptide database. Methods Enzymol. 2022;663:1–18. 10.1016/bs.mie.2021.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zharkova MS, Orlov DS, Golubeva OY, Chakchir OB, Eliseev IE, Grinchuk TM, et al. Application of Antimicrobial Peptides of the Innate Immune System in Combination With Conventional Antibiotics-A Novel Way to Combat Antibiotic Resistance? Front Cell Infect Microbiol. 2019. Apr;9:128. 10.3389/fcimb.2019.00128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenssen H, Hamill P, Hancock RE. Peptide antimicrobial agents. Clin Microbiol Rev. 2006. Jul;19(3):491–511. 10.1128/CMR.00056-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalzgraff A, Brandenburg K, Weindl G. Antimicrobial peptides and their therapeutic potential for bacterial skin infections and wounds. Front Pharmacol. 2018. Mar;9:281. 10.3389/fphar.2018.00281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Somma A, Moretta A, Canè C, Cirillo A, Duilio A. Antimicrobial and antibiofilm peptides. Biomolecules. 2020. Apr;10(4):652. 10.3390/biom10040652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raheem N, Straus SK. Mechanisms of Action for Antimicrobial Peptides With Antibacterial and Antibiofilm Functions. Front Microbiol. 2019. Dec;10:2866. 10.3389/fmicb.2019.02866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Yang N, Mao R, Hao Y, Teng D, Wang J. In Vitro Pharmacodynamics and Bactericidal Mechanism of Fungal Defensin-Derived Peptides NZX and P2 against Streptococcus agalactiae. Microorganisms. 2022. Apr;10(5):881. 10.3390/microorganisms10050881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari S, Booth V. Antimicrobial Peptide Mechanisms Studied by Whole-Cell Deuterium NMR. Int J Mol Sci. 2022. Mar;23(5):2740. 10.3390/ijms23052740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inui Kishi RN, Stach-Machado D, Singulani JL, Dos Santos CT, Fusco-Almeida AM, Cilli EM, et al. Evaluation of cytotoxicity features of antimicrobial peptides with potential to control bacterial diseases of citrus. PLoS One. 2018. Sep;13(9):e0203451. 10.1371/journal.pone.0203451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scocchi M, Mardirossian M, Runti G, Benincasa M. Non-Membrane Permeabilizing Modes of Action of Antimicrobial Peptides on Bacteria. Curr Top Med Chem. 2016;16(1):76–88. 10.2174/1568026615666150703121009 [DOI] [PubMed] [Google Scholar]
- Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002. Jan;415(6870):389–95. 10.1038/415389a [DOI] [PubMed] [Google Scholar]
- Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005. Mar;3(3):238–50. 10.1038/nrmicro1098 [DOI] [PubMed] [Google Scholar]
- Yount NY, Bayer AS, Xiong YQ, Yeaman MR. Advances in antimicrobial peptide immunobiology. Biopolymers. 2006;84(5):435–58. 10.1002/bip.20543 [DOI] [PubMed] [Google Scholar]
- Schneider T, Kruse T, Wimmer R, Wiedemann I, Sass V, Pag U, et al. Plectasin, a fungal defensin, targets the bacterial cell wall precursor Lipid II. Science. 2010. May;328(5982):1168–72. 10.1126/science.1185723 [DOI] [PubMed] [Google Scholar]
- Matsuzaki K. Magainins as paradigm for the mode of action of pore forming polypeptides. Biochim Biophys Acta. 1998. Nov;1376(3):391–400. 10.1016/S0304-4157(98)00014-8 [DOI] [PubMed] [Google Scholar]
- Cardoso MH, Meneguetti BT, Costa BO, Buccini DF, Oshiro KG, Preza SL, et al. Non-Lytic Antibacterial Peptides That Translocate Through Bacterial Membranes to Act on Intracellular Targets. Int J Mol Sci. 2019. Oct;20(19):4877. 10.3390/ijms20194877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otvos L Jr. The short proline-rich antibacterial peptide family. Cell Mol Life Sci. 2002. Jul;59(7):1138–50. 10.1007/s00018-002-8493-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scocchi M, Tossi A, Gennaro R. Proline-rich antimicrobial peptides: converging to a non-lytic mechanism of action. Cell Mol Life Sci. 2011. Jul;68(13):2317–30. 10.1007/s00018-011-0721-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Li J, Tang H, Li Q, Shen G, Li S, et al. Antibacterial Peptide NP-6 Affects Staphylococcus aureus by Multiple Modes of Action. Int J Mol Sci. 2022. Jul;23(14):7812. 10.3390/ijms23147812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk A, van Eldik M, Veldhuizen EJ, Tjeerdsma-van Bokhoven HL, de Zoete MR, Bikker FJ, et al. Immunomodulatory and anti-inflammatory activities of chicken cathelicidin-2 derived peptides. PLoS One. 2016. Feb;11(2):e0147919. 10.1371/journal.pone.0147919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Seo M, Lee HJ, Baek M, Kim IW, Kim SY, et al. Anti-Inflammatory Activity of Antimicrobial Peptide Allomyrinasin Derived from the Dynastid Beetle, Allomyrina dichotoma. J Microbiol Biotechnol. 2019. May;29(5):687–95. 10.4014/jmb.1809.09031 [DOI] [PubMed] [Google Scholar]
- Nagaoka I, Tamura H, Reich J. Therapeutic Potential of Cathelicidin Peptide LL-37, an Antimicrobial Agent, in a Murine Sepsis Model. Int J Mol Sci. 2020. Aug;21(17):5973. 10.3390/ijms21175973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Jin L, Wang H, Tai S, Liu H, Zhang D. AWRK6, A Synthetic Cationic Peptide Derived from Antimicrobial Peptide Dybowskin-2CDYa, Inhibits Lipopolysaccharide-Induced Inflammatory Response. Int J Mol Sci. 2018. Feb;19(2):600. 10.3390/ijms19020600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X, Lu H, Zhu W, Zhang Y, Huo X, Yang C, et al. A Novel Antimicrobial Peptide Derived from Bony Fish IFN1 Exerts Potent Antimicrobial and Anti-Inflammatory Activity in Mammals. Microbiol Spectr. 2022. Apr;10(2):e0201321. 10.1128/spectrum.02013-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Yang J, He X, Mo G, Hong J, Yan X, et al. Structure and function of a potent lipopolysaccharide-binding antimicrobial and anti-inflammatory peptide. J Med Chem. 2013. May;56(9):3546–56. 10.1021/jm4004158 [DOI] [PubMed] [Google Scholar]
- Zhu X, Chen F, Li S, Peng H, Wang KJ. A Novel Antimicrobial Peptide Sparanegtin Identified in Scylla paramamosain Showing Antimicrobial Activity and Immunoprotective Role In Vitro and Vivo. Int J Mol Sci. 2021. Dec;23(1):15. 10.3390/ijms23010015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Cai S, Qiao X, Wu M, Guo Z, Wang R, et al. As-CATH1-6, novel cathelicidins with potent antimicrobial and immunomodulatory properties from Alligator sinensis, play pivotal roles in host antimicrobial immune responses. Biochem J. 2017. Aug;474(16):2861–85. 10.1042/BCJ20170334 [DOI] [PubMed] [Google Scholar]
- Granslo HN, Aarag Fredheim EG, Esaiassen E, Christophersen L, Jensen PØ, Mollnes TE, Moser C, Flaegstad T, Klingenberg C, Cavanagh JP. The synthetic antimicrobial peptide LTX21 induces inflammatory responses in a human whole blood model and a murine peritoneum model. APMIS: acta pathologica, microbiologica, et immunologica Scandinavica. 2019;127(6):475–483. [DOI] [PubMed] [Google Scholar]
- Ferrer-Espada R, Shahrour H, Pitts B, Stewart PS, Sánchez-Gómez S, Martínez-de-Tejada G. A permeability-increasing drug synergizes with bacterial efflux pump inhibitors and restores susceptibility to antibiotics in multi-drug resistant Pseudomonas aeruginosa strains. Sci Rep. 2019. Mar;9(1):3452. 10.1038/s41598-019-39659-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rázquin-Olazarán I, Shahrour H, Martínez-de-Tejada G. A synthetic peptide sensitizes multi-drug resistant Pseudomonas aeruginosa to antibiotics for more than two hours and permeabilizes its envelope for twenty hours. J Biomed Sci. 2020. Aug;27(1):85. 10.1186/s12929-020-00678-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro SM, de la Fuente-Núñez C, Baquir B, Faria-Junior C, Franco OL, Hancock RE. Antibiofilm peptides increase the susceptibility of carbapenemase-producing Klebsiella pneumoniae clinical isolates to β-lactam antibiotics. Antimicrob Agents Chemother. 2015. Jul;59(7):3906–12. 10.1128/AAC.00092-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente-Núñez C, Reffuveille F, Mansour SC, Reckseidler-Zenteno SL, Hernández D, Brackman G, et al. D-enantiomeric peptides that eradicate wild-type and multidrug-resistant biofilms and protect against lethal Pseudomonas aeruginosa infections [published correction appears in Chem Biol. 2015 17;22(9):1280-2]. Chem Biol. 2015. Feb;22(2):196–205. 10.1016/j.chembiol.2015.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denardi LB, de Arruda Trindade P, Weiblen C, Ianiski LB, Stibbe PC, Pinto SC, et al. In vitro activity of the antimicrobial peptides h-Lf1-11, MSI-78, LL-37, fengycin 2B, and magainin-2 against clinically important bacteria. Braz J Microbiol. 2022. Mar;53(1):171–7. 10.1007/s42770-021-00645-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Liu Y, Gao Y, Wang Y, Xia Q, Zhong R, et al. Enhanced antimicrobial activity of N-terminal derivatives of a novel Brevinin-1 peptide from the skin secretion of Odorrana schmackeri. Toxins (Basel). 2020. Jul;12(8):484. 10.3390/toxins12080484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Wang Z, Han H, Teng D, Mao R, Hao Y, et al. Dual antibacterial activities and biofilm eradication of a marine peptide-N6NH2 and its analogs against multidrug-resistant aeromonas veronii. Int J Mol Sci. 2020. Dec;21(24):9637. 10.3390/ijms21249637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang D, Han X, Du W, Kou Z, Jiang F. Trp-containing antibacterial peptides impair quorum sensing and biofilm development in multidrug-resistant Pseudomonas aeruginosa and exhibit synergistic effects with antibiotics. Front Microbiol. 2021. Feb;12:611009. 10.3389/fmicb.2021.611009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsalem L, Khasawneh A, Al Sheboul S. WLBU2 Antimicrobial Peptide as a Potential Therapeutic for Treatment of Resistant Bacterial. Turk J Pharm Sci. 2022. Feb;19(1):110–6. 10.4274/tjps.galenos.2020.43078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsalem L, Al Sheboul S, Khasawneh A. Synergism between WLBU2 peptide and antibiotics against methicillin-resistant Staphylococcus aureus and extended-spectrum beta-lactamase-producing Enterobacter cloacae. J Appl Biomed. 2021. Mar;19(1):14–25. 10.32725/jab.2021.001 [DOI] [PubMed] [Google Scholar]
- Degasperi M, Sgarra R, Mardirossian M, Pacor S, Maschio M, Scocchi M. Elastase-Activated Antimicrobial Peptide for a Safer Pulmonary Treatment of Cystic Fibrosis Infections. Antibiotics (Basel). 2022. Feb;11(3):319. 10.3390/antibiotics11030319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch P, Schmitt S, Heynisch A, Gumpinger A, Wüthrich I, Gysin M, et al. Optimization of the antimicrobial peptide Bac7 by deep mutational scanning. BMC Biol. 2022. May;20(1):114. 10.1186/s12915-022-01304-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Liu L, Ma C, Di L, Chen T. A novel antimicrobial peptide found in Pelophylax nigromaculatus. J Genet Eng Biotechnol. 2022. May;20(1):76. 10.1186/s43141-022-00366-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okella H, Georrge JJ, Ochwo S, Ndekezi C, Koffi KT, Aber J, et al. New putative antimicrobial candidates: in silico design of fish-derived antibacterial peptide-motifs. Front Bioeng Biotechnol. 2020. Dec;8:604041. 10.3389/fbioe.2020.604041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, Starr CG, Troendle E, Wiedman G, Wimley WC, Ulmschneider JP, et al. Simulation-guided rational de novo design of a small pore-forming antimicrobial peptide. J Am Chem Soc. 2019. Mar;141(12):4839–48. 10.1021/jacs.8b11939 [DOI] [PubMed] [Google Scholar]
- Cardoso MH, Orozco RQ, Rezende SB, Rodrigues G, Oshiro KG, Cândido ES, et al. Computer-aided design of antimicrobial peptides: are we generating effective drug candidates? Front Microbiol. 2020. Jan;10:3097. 10.3389/fmicb.2019.03097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobde SS, Alsaab FM, Wang G, Van Hoek ML. Ab initio Designed Antimicrobial Peptides Against Gram-Negative Bacteria. Front Microbiol. 2021. Nov;12:715246. 10.3389/fmicb.2021.715246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tram ND, Selvarajan V, Boags A, Mukherjee D, Marzinek JK, Cheng B, et al. Manipulating turn residues on de novo designed β-hairpin peptides for selectivity against drug-resistant bacteria. Acta Biomater. 2021. Nov;135:214–24. 10.1016/j.actbio.2021.09.004 [DOI] [PubMed] [Google Scholar]
- Jangir PK, Ogunlana L, MacLean RC. Evolutionary constraints on the acquisition of antimicrobial peptide resistance in bacterial pathogens. Trends Microbiol. 2021. Dec;29(12):1058–61. 10.1016/j.tim.2021.03.007 [DOI] [PubMed] [Google Scholar]
- Khan ST, Musarrat J, Al-Khedhairy AA. Countering drug resistance, infectious diseases, and sepsis using metal and metal oxides nanoparticles: current status. Colloids Surf B Biointerfaces. 2016. Oct;146:70–83. 10.1016/j.colsurfb.2016.05.046 [DOI] [PubMed] [Google Scholar]
- Lin Z, Monteiro-Riviere NA, Riviere JE. Pharmacokinetics of metallic nanoparticles. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2015. Mar-Apr;7(2):189–217. 10.1002/wnan.1304 [DOI] [PubMed] [Google Scholar]
- Torres-Barceló C, Hochberg ME. Evolutionary rationale for phages as complements of antibiotics. Trends Microbiol. 2016. Apr;24(4):249–56. 10.1016/j.tim.2015.12.011 [DOI] [PubMed] [Google Scholar]
- Krylov V, Shaburova O, Krylov S, Pleteneva E. A genetic approach to the development of new therapeutic phages to fight pseudomonas aeruginosa in wound infections. Viruses. 2012. Dec;5(1):15–53. 10.3390/v5010015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demento SL, Siefert AL, Bandyopadhyay A, Sharp FA, Fahmy TM. Pathogen-associated molecular patterns on biomaterials: a paradigm for engineering new vaccines. Trends Biotechnol. 2011. Jun;29(6):294–306. 10.1016/j.tibtech.2011.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baindara P, Mandal SM. Antimicrobial Peptides and Vaccine Development to Control Multi-drug Resistant Bacteria. Protein Pept Lett. 2019;26(5):324–31. 10.2174/0929866526666190228162751 [DOI] [PubMed] [Google Scholar]
- Guryanova SV, Ovchinnikova TV. Immunomodulatory and Allergenic Properties of Antimicrobial Peptides. Int J Mol Sci. 2022. Feb;23(5):2499. 10.3390/ijms23052499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirioni O, Giacometti A, Ghiselli R, Bergnach C, Orlando F, Silvestri C, et al. LL-37 protects rats against lethal sepsis caused by gram-negative bacteria. Antimicrob Agents Chemother. 2006. May;50(5):1672–9. 10.1128/AAC.50.5.1672-1679.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinstraesser L, Hirsch T, Schulte M, Kueckelhaus M, Jacobsen F, Mersch EA, et al. Innate defense regulator peptide 1018 in wound healing and wound infection. PLoS One. 2012;7(8):e39373. 10.1371/journal.pone.0039373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papareddy P, Kalle M, Sørensen OE, Malmsten M, Mörgelin M, Schmidtchen A. The TFPI-2 derived peptide EDC34 improves outcome of gram-negative sepsis. PLoS Pathog. 2013;9(12):e1003803. 10.1371/journal.ppat.1003803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domalaon R, De Silva PM, Kumar A, Zhanel GG, Schweizer F. The anthelmintic drug niclosamide synergizes with colistin and reverses colistin resistance in Gram-negative bacilli. Antimicrob Agents Chemother. 2019. Mar;63(4):e02574–18. 10.1128/AAC.02574-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- León-Buitimea A, Garza-Cárdenas CR, Garza-Cervantes JA, Lerma-Escalera JA, Morones-Ramírez JR. The Demand for New Antibiotics: Antimicrobial Peptides, Nanoparticles, and Combinatorial Therapies as Future Strategies in Antibacterial Agent Design. Front Microbiol. 2020. Jul;11:1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang D, Liu Y, Jiang F, Ji F, Wang H, Han X. Synergistic antibacterial activity of designed Trp-containing antibacterial peptides in combination with antibiotics against multidrug-resistant Staphylococcus epidermidis. Front Microbiol. 2019. Nov;10:2719. 10.3389/fmicb.2019.02719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev YA, Tutel’yan AV, Loiko NG, Buck J, Sidorenko SV, Lazareva I, et al. The use of 4-Hexylresorcinol as antibiotic adjuvant. PLoS One. 2020. Sep;15(9):e0239147. 10.1371/journal.pone.0239147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampshoff F, Willcox MD, Dutta D. A pilot study of the synergy between two antimicrobial peptides and two common antibiotics. Antibiotics (Basel). 2019. May;8(2):60. 10.3390/antibiotics8020060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangra M, Kaur M, Podia M, Tambat R, Singh V, Chandal N, et al. Purification and biological activity of natural variants synthesized by tridecaptin M gene cluster and in vitro drug-kinetics of this antibiotic class. Sci Rep. 2019. a Dec;9(1):18870. 10.1038/s41598-019-54716-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangra M, Kaur M, Tambat R, Rana R, Maurya SK, Khatri N, et al. Tridecaptin M, a new variant discovered in mud bacterium, shows activity against colistin-and extremely drug-resistant Enterobacteriaceae. Antimicrob Agents Chemother. 2019. b May;63(6):e00338–19. 10.1128/AAC.00338-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangra M, Raka V, Nandanwar H. In Vitro Evaluation of Antimicrobial Peptide Tridecaptin M in Combination with Other Antibiotics against Multidrug Resistant Acinetobacter baumannii. Molecules. 2020. Jul;25(14):3255. 10.3390/molecules25143255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco L, Ambroa A, Trastoy R, Bleriot I, Moscoso M, Fernández-Garcia L, et al. In vitro and in vivo efficacy of combinations of colistin and different endolysins against clinical strains of multi-drug resistant pathogens. Sci Rep. 2020. Apr;10(1):7163. 10.1038/s41598-020-64145-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehaghani MZ, Bagheri B, Yousefi F, et al. Boron nitride nanotube as an antimicrobial peptide carrier: a theoretical insight. Int J Nanomedicine. 2021. b;16:1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarghami Dehaghani M, Yousefi F, Bagheri B, Seidi F, Hamed Mashhadzadeh A, Rabiee N, et al. α-helical antimicrobial peptide encapsulation and release from boron nitride nanotubes: a computational study. Int J Nanomedicine. 2021. a Jun;16:4277–88. 10.2147/IJN.S313855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarghami Dehaghani M, Yousefi F, Seidi F, Sajadi SM, Rabiee N, Habibzadeh S, et al. Dynamics of Antimicrobial Peptide Encapsulation in Carbon Nanotubes: The Role of Hydroxylation. Int J Nanomedicine. 2022. Jan;17:125–36. 10.2147/IJN.S335380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai A, Ferrão R, Palma P, Patricio T, Parreira P, Anes E, et al. Antimicrobial peptide-based materials: opportunities and challenges. J Mater Chem B Mater Biol Med. 2022. Apr;10(14):2384–429. 10.1039/D1TB02617H [DOI] [PubMed] [Google Scholar]
- Maleki Dizaj S, Salatin S, Khezri K, Lee JY, Lotfipour F. Targeting Multidrug Resistance With Antimicrobial Peptide-Decorated Nanoparticles and Polymers. Front Microbiol. 2022. Mar;13:831655. 10.3389/fmicb.2022.831655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deo S, Turton KL, Kainth T, Kumar A, Wieden HJ. Strategies for improving antimicrobial peptide production. Biotechnol Adv. 2022. Oct;59:107968. 10.1016/j.biotechadv.2022.107968 [DOI] [PubMed] [Google Scholar]
- Rezende SB, Oshiro KG, Júnior NG, Franco OL, Cardoso MH. Advances on chemically modified antimicrobial peptides for generating peptide antibiotics. Chem Commun (Camb). 2021. Nov;57(88):11578–90. 10.1039/D1CC03793E [DOI] [PubMed] [Google Scholar]
- Han Y, Zhang M, Lai R, Zhang Z. Chemical modifications to increase the therapeutic potential of antimicrobial peptides. Peptides. 2021. Dec;146:170666. 10.1016/j.peptides.2021.170666 [DOI] [PubMed] [Google Scholar]
- Silva AR, Guimarães MS, Rabelo J, Belén LH, Perecin CJ, Farías JG, et al. Recent advances in the design of antimicrobial peptide conjugates. J Mater Chem B Mater Biol Med. 2022. May;10(19):3587–600. 10.1039/D1TB02757C [DOI] [PubMed] [Google Scholar]
- Svendsen JS, Grant TM, Rennison D, Brimble MA, Svenson J. Very Short and Stable Lactoferricin-Derived Antimicrobial Peptides: Design Principles and Potential Uses. Acc Chem Res. 2019. Mar;52(3):749–59. 10.1021/acs.accounts.8b00624 [DOI] [PubMed] [Google Scholar]
- Atefyekta S, Blomstrand E, Rajasekharan AK, Svensson S, Trobos M, Hong J, et al. Antimicrobial Peptide-Functionalized Mesoporous Hydrogels. ACS Biomater Sci Eng. 2021. Apr;7(4):1693–702. 10.1021/acsbiomaterials.1c00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vimberg V, Buriánková K, Mazumdar A, Branny P, Novotná GB. Role of membrane proteins in bacterial resistance to antimicrobial peptides. Med Res Rev. 2022. May;42(3):1023–36. 10.1002/med.21869 [DOI] [PubMed] [Google Scholar]
- Obuobi S, Tay HK, Tram ND, Selvarajan V, Khara JS, Wang Y, et al. Facile and efficient encapsulation of antimicrobial peptides via crosslinked DNA nanostructures and their application in wound therapy. J Control Release. 2019. Nov;313:120–30. 10.1016/j.jconrel.2019.10.013 [DOI] [PubMed] [Google Scholar]
- Moorcroft SC, Roach L, Jayne DG, Ong ZY, Evans SD. Nanoparticle-Loaded Hydrogel for the Light-Activated Release and Photothermal Enhancement of Antimicrobial Peptides. ACS Appl Mater Interfaces. 2020. Jun;12(22):24544–54. 10.1021/acsami.9b22587 [DOI] [PubMed] [Google Scholar]
- Sarma P, Mahendiratta S, Prakash A, Medhi B. Specifically targeted antimicrobial peptides: A new and promising avenue in selective antimicrobial therapy. Indian J Pharmacol. 2018. Jan-Feb;50(1):1–3. 10.4103/ijp.IJP_218_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez J, Martin-Serrano Á, Gómez-Casanova N, Falanga A, Galdiero S, Javier de la Mata F, et al. Effect of the Combination of Levofloxacin with Cationic Carbosilane Dendron and Peptide in the Prevention and Treatment of Staphylococcus aureus Biofilms. Polymers (Basel). 2021. Jun;13(13):2127. 10.3390/polym13132127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosler S, Karaaslan E. Inhibition and destruction of Pseudomonas aeruginosa biofilms by antibiotics and antimicrobial peptides. Peptides. 2014. Dec;62:32–7. 10.1016/j.peptides.2014.09.021 [DOI] [PubMed] [Google Scholar]
- Jahangiri A, Neshani A, Mirhosseini SA, Ghazvini K, Zare H, Sedighian H. Synergistic effect of two antimicrobial peptides, Nisin and P10 with conventional antibiotics against extensively drug-resistant Acinetobacter baumannii and colistin-resistant Pseudomonas aeruginosa isolates. Microb Pathog. 2021. Jan;150:104700. 10.1016/j.micpath.2020.104700 [DOI] [PubMed] [Google Scholar]
- Svenson J, Karstad R, Flaten GE, Brandsdal BO, Brandl M, Svendsen JS. Altered activity and physicochemical properties of short cationic antimicrobial peptides by incorporation of arginine analogues. Mol Pharm. 2009. May-Jun;6(3):996–1005. 10.1021/mp900057k [DOI] [PubMed] [Google Scholar]
- Wang J, Song J, Yang Z, He S, Yang Y, Feng X, et al. Antimicrobial Peptides with High Proteolytic Resistance for Combating Gram-Negative Bacteria. J Med Chem. 2019. Mar;62(5):2286–304. 10.1021/acs.jmedchem.8b01348 [DOI] [PubMed] [Google Scholar]
- Wang G, Zietz CM, Mudgapalli A, Wang S, Wang Z. The evolution of the antimicrobial peptide database over 18 years: milestones and new features. Protein Sci. 2022. Jan;31(1):92–106. 10.1002/pro.4185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phoenix DA, Dennison SR, Harris F. Bacterial resistance to host defence peptides. In: Epand RM. Host Defense Peptides and Their Potential as Therapeutic Agents. Switzerland: Springer; 2016. pp. 161–204. 10.1007/978-3-319-32949-9_7 [DOI] [Google Scholar]
- Boswell MT, Cockeran R. Effect of antimicrobial peptides on planktonic growth, biofilm formation and biofilm-derived bacterial viability of Streptococcus pneumoniae. S Afr J Infect Dis. 2021. Jan;36(1):226. 10.4102/sajid.v36i1.226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan M, van der Does AM, Tang X, Lindbom L, Agerberth B, Haeggström JZ. Antimicrobial peptide LL-37 promotes bacterial phagocytosis by human macrophages. J Leukoc Biol. 2014. Jun;95(6):971–81. 10.1189/jlb.0513304 [DOI] [PubMed] [Google Scholar]
- Li J, Chen J, Yang G, Tao L. Sublancin protects against methicillin-resistant Staphylococcus aureus infection by the combined modulation of innate immune response and microbiota. Peptides. 2021. Jul;141:170533. 10.1016/j.peptides.2021.170533 [DOI] [PubMed] [Google Scholar]
- Pandit G, Sarkar T, S R V, Debnath S, Satpati P, Chatterjee S. Delineating the Mechanism of Action of a Protease Resistant and Salt Tolerant Synthetic Antimicrobial Peptide against Pseudomonas aeruginosa. ACS Omega. 2022. Apr;7(18):15951–68. 10.1021/acsomega.2c01089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Heeney D, Srisengfa Y, Golomb B, Griffey S, Marco M. Bacteriocin biosynthesis contributes to the anti-inflammatory capacities of probiotic Lactobacillus plantarum. Benef Microbes. 2018. Feb;9(2):333–44. 10.3920/BM2017.0096 [DOI] [PubMed] [Google Scholar]
- Zaïri A, Ferrières L, Latour-Lambert P, Beloin C, Tangy F, Ghigo JM, et al. In vitro activities of dermaseptins K4S4 and K4K20S4 against Escherichia coli, Staphylococcus aureus, and Pseudomonas aeruginosa planktonic growth and biofilm formation. Antimicrob Agents Chemother. 2014;58(4):2221–8. 10.1128/AAC.02142-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley VJ, Kinnear CL, Sim DG, Olson SN, Jackson LM, Hansen E, et al. An adjunctive therapy administered with an antibiotic prevents enrichment of antibiotic-resistant clones of a colonizing opportunistic pathogen. eLife. 2020. Dec;9:e58147. 10.7554/eLife.58147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasir M, Dutta D, Willcox MD. Activity of Antimicrobial Peptides and Ciprofloxacin against Pseudomonas aeruginosa Biofilms. Molecules. 2020. Aug;25(17):3843. 10.3390/molecules25173843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechinger B, Juhl DW, Glattard E, Aisenbrey C. Revealing the Mechanisms of Synergistic Action of Two Magainin Antimicrobial Peptides. Front Med Technol. 2020. Dec;2:615494. 10.3389/fmedt.2020.615494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portelinha J, Angeles-Boza AM. The Antimicrobial Peptide Gad-1 Clears Pseudomonas aeruginosa Biofilms under Cystic Fibrosis Conditions. ChemBioChem. 2021. May;22(9):1646–55. 10.1002/cbic.202000816 [DOI] [PubMed] [Google Scholar]