Abstract
The water-soluble inositol phosphates (InsPs) represent a functionally diverse group of small-molecule messengers involved in a myriad of cellular processes. Despite their centrality, our understanding of human InsP metabolism is incomplete because the available analytical toolset to characterize and quantify InsPs in complex samples is limited. Here, we have synthesized and applied symmetrically and unsymmetrically 13C-labeled myo-inositol and inositol phosphates. These probes were utilized in combination with nuclear magnetic resonance spectroscopy (NMR) and capillary electrophoresis mass spectrometry (CE-MS) to investigate InsP metabolism in human cells. The labeling strategy provided detailed structural information via NMR—down to individual enantiomers—which overcomes a crucial blind spot in the analysis of InsPs. We uncovered a novel branch of InsP dephosphorylation in human cells which is dependent on MINPP1, a phytase-like enzyme contributing to cellular homeostasis. Detailed characterization of MINPP1 activity in vitro and in cells showcased the unique reactivity of this phosphatase. Our results demonstrate that metabolic labeling with stable isotopomers in conjunction with NMR spectroscopy and CE-MS constitutes a powerful tool to annotate InsP networks in a variety of biological contexts.
Short abstract
Using chemoenzymatically synthesized myo-inositol (phosphate) isotopomers, unprecedented insight into these central eukaryotic messengers and their metabolism in human cells was obtained.
Introduction
Myo-inositol polyphosphates (InsPs) are ubiquitous, water-soluble small molecules found in all eukaryotes. InsPs are involved in a wide spectrum of biological functions as they are key to fundamental physiological processes. A well-characterized example is inositol-1,4,5-trisphosphate (Ins(1,4,5)P3) as a Ca2+ release factor. More recently, InsPs were shown to regulate the activity of class I histone deacetylases as well as Bruton’s tyrosine kinase (Btk), which implies a wider role for InsPs in transcriptional regulation and in governing intracellular signal transduction.1−3
The InsPs vary greatly with respect to their phosphorylation patterns, and over 20 different InsPs are currently thought to be part of mammalian InsP metabolism.4−7 The most abundant InsPs in mammalian cells are inositol-1,3,4,5,6-pentakisphosphate (InsP5[2OH]) and inositol hexakisphosphate (also called phytic acid, InsP6), with cellular concentrations ranging from the lower micromolar range to >100 μM in human cells and even in the sub-millimolar range in slime molds.8,9 InsP5[2OH] and InsP6 are precursors for the biosynthesis of inositol pyrophosphates (PP-InsPs), which have recently drawn increasing attention due to their dense phosphorylation patterns and their involvement in central signaling processes.10 InsP6 is also found in a growing number of proteins and protein complexes as a structural cofactor or as a “molecular glue” for protein–protein interactions.11−14
While the kinase-mediated pathways of InsP biosynthesis are fairly well studied, there is limited information on dephosphorylation of InsPs in mammalian cells,15 especially with respect to the higher phosphorylated members. To date, MINPP1 (Multiple Inositol Polyphosphate Phosphatase 1) is the only recognized enzyme in the human genome capable of dephosphorylating InsP6.16,17 MINPP1 is related to phytases, a highly conserved group of enzymes in many other organisms that can dephosphorylate various InsPs.18 MINPP1 has been shown to play a role in apoptosis, ER-related stress, and bone and cartilage tissue formation.17,19 Recently, MINPP1 was connected to a genetic disorder: patients with loss-of-function mutations in MINPP1 exhibit pontocerebellar hypoplasia (PCH), a neurodegenerative disease severely impacting cognitive functions and life expectancy.20,21 Therefore, it is important to understand the molecular mechanisms of MINPP1-governed functions in healthy and diseased states.
Although MINPP1 is annotated as a 3-phosphatase, i.e., it predominantly removes the phosphoryl group at the 3-position of InsP6,22 MINPP1 is also able to dephosphorylate several InsPs at different positions with varying affinities and kinetics.23,24 The current assumption is that MINPP1 dephosphorylates InsP6 to hitherto only sparsely annotated InsP4/3 species (Figure 1a).6,7,25−27 However, this activity has only been demonstrated in vitro and in intact cells overexpressing a cytosolic variant of MINPP1. Whether this activity is relevant in vivo and which InsP intermediates are exactly involved is still not clear.25 Furthermore, there is no consensus how MINPP1 accesses its InsP substrates. While early studies suggest MINPP1 to be localized to the ER,28,29 others have also shown alternative localizations into the Golgi, in lysosomes, or even secreted in exosomes.30,31
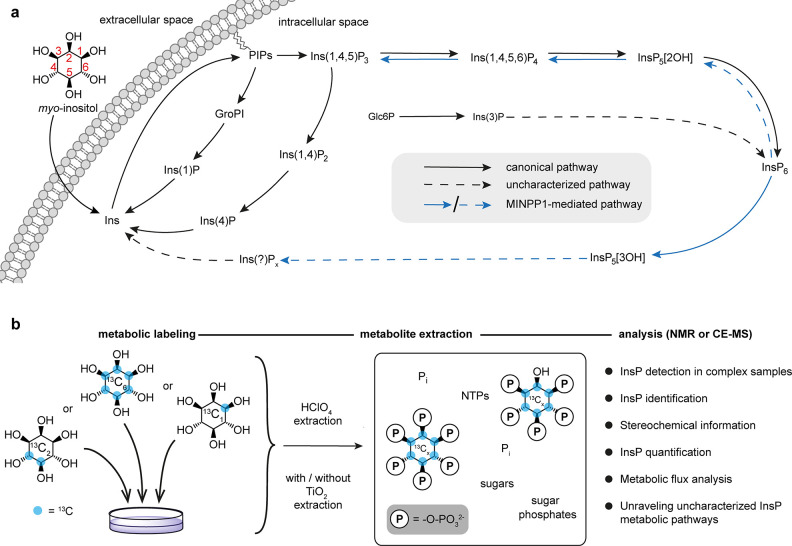
Figure 1.
Probing InsP metabolism with myo-inositol isotopomers. (a) Simplified overview of InsP metabolism with MINPP1-mediated processes highlighted. It is assumed that MINPP1 dephosphorylates InsP6 and various other InsPs down to sparsely annotated InsP3 isomers. PIPs, phosphatidylinositol phosphates; GroPI, glycerophosphoinositol; Glc6P, glucose-6-phosphate; MINPP1, multiple inositol polyphosphate phosphatase1; Ins(X,Y)Pz, myo-inositol with z phosphoryl groups at positions X,Y; InsP5[XOH], inositol pentakisphosphate with a hydroxyl group at position X. IUPAC numbering convention of the positions on the inositol scaffold is shown in red. (b) Workflow for the analysis of cellular InsP pools through metabolic labeling: human cells are grown in medium devoid of nonlabeled myo-inositol but supplemented with an isotopomer of myo-inositol ([13C6]Ins, 4,5[13C2]myo-inositol, 1[13C1]myo-inositol, or 3[13C1]myo-inositol) which are incorporated into the cellular InsP pool. Metabolites are then extracted, resulting in a complex sample containing all water-soluble biomolecules, such as nucleotide triphosphates (NTPs), inorganic phosphate (Pi), and the labeled InsPs. This mixture can be analyzed via NMR or CE-MS exploiting NMR activity and mass difference of the 13C label.
Probing and quantifying InsP metabolites and their interconversion is still a challenging task due to the limitations of current analytical tools. Many established methods for the detection and analysis of InsPs rely on some form of physicochemical separation of different InsPs from a complex mixture. The most common methods are strong-anion exchange chromatography (SAX-HPLC)-based fractionation in combination with radiolabeling and scintillation counting, high-density polyacrylamide electrophoresis with cationic staining, or, more recently, capillary electrophoresis coupled to mass spectrometry (CE-MS).26,32−37 Most of these methods are sensitive and powerful for the analysis of highly phosphorylated InsPs, but the separation and detection of lower InsPs (i.e., InsP1, InsP2, and InsP3 species) in a mixture with isobaric sugar phosphates remain difficult. Our group recently established a metabolic labeling strategy using isotopically labeled [13C6]myo-inositol. Analysis of the extracted metabolites by 2D nuclear magnetic resonance (NMR) spectroscopy enabled the quantification of higher phosphorylated InsPs (InsP6, InsP5[2OH], and PP-InsPs; Figure 1b) without the need for analytical separation.38 In addition, 2D-NMR measurements provide important information on the InsP phosphorylation patterns and should be able to detect the whole range of InsP metabolites, including the lower phosphorylated species.
Here, we combined fully 13C-labeled and asymmetrically 13C-labeled isotopomers of myo-inositol and InsPs in both biochemical and cellular metabolic labeling experiments. Making use of their inherent properties (position-specific NMR activity and different molecular masses), we uncovered an uncharacterized branch of human InsP metabolism. Ins(2,3)P2 and Ins(2)P were identified as major InsPs species in human cells, and their levels are dependent on MINPP1 activity toward InsP6 in vitro and in cellula. Through in vitro characterization, computational kinetic modeling, and metabolic flux via CE-MS analysis, we dissect the complex reactivity of MINPP1. We envision that this combined application of myo-inositol isotopomers in NMR and CE-MS experiments will help unravel complex InsP networks in different biological contexts in the future.
Results
InsP Phosphorylation Patterns Are Well Resolved by BIRD-{1H,13C}HMQC NMR Spectra
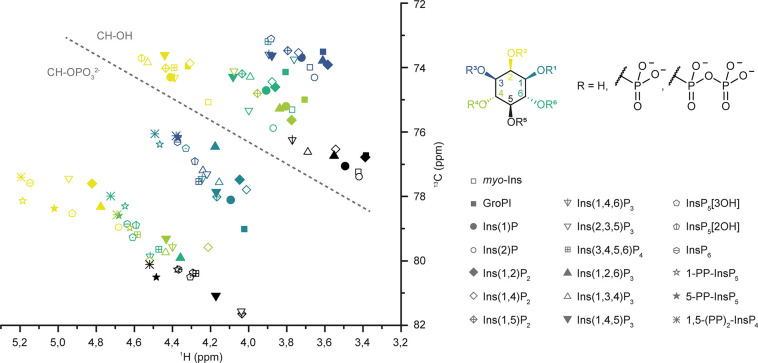
The analysis of complex mixtures of InsP metabolites still constitutes a significant analytical challenge. To identify inositol-derived signals in biological samples via NMR in a methodical way, BIRD-{1H,13C}HMQC-NMR spectra of 19 different InsPs and PP-InsPs (commercially available or synthesized) were recorded and assigned. The collective data of these spectra illustrate that the NMR signals of InsPs cluster in a systematic manner (Figures 2 and S1). NMR signals corresponding to methine groups adjacent to a nonphosphorylated hydroxyl substituent (CH–OH) are separated from methine group signals with a phosphate substituent (CH–O–PO32–), which are collectively shifted downfield in both 1H and 13C dimensions. Within these two groups, clusters for the different positions on the myo-inositol ring are apparent. The 2- and 5-positions form clusters of their own, while positions 1 and 3 as well as positions 4 and 6 are intertwined due to the symmetry plane of the myo-inositol ring. These combined spectra illustrate that a complete set of NMR signals of an InsP can be used to determine the phosphorylation pattern, and thus the identity, of a given InsP. In the case of chiral InsPs, their NMR spectra cannot be used for a definitive assignment but can narrow the identity down to a pair of enantiomers. For distinguishing two InsP enantiomers, a desymmetrization strategy has to be employed, such as unsymmetrical isotopic labeling of the myo-inositol ring with 13C, as will be discussed below.
Figure 2.
HMQC signals of InsPs with different phosphorylation patterns cluster systematically. Collection of BIRD-{1H,13C}HMQC NMR data of various InsP standards in metabolic extract buffer conditions (saturated KClO4 in D2O, pH* 6.0). HMQC signals of different InsPs are represented with symbols, while the position on the inositol ring is color-coded. HMQC signals cluster together depending on phosphorylation status (dotted line) and position on the inositol ring. CH groups bearing the pyrophosphate moiety of PP-InsPs or the 1-glyceryl phosphate group of GroPI cluster with the phosphorylated CH groups and were treated accordingly for creating bagplots (Figure S1).
Ins(2,3)P2 and Ins(2)P Are Major Mammalian Metabolites
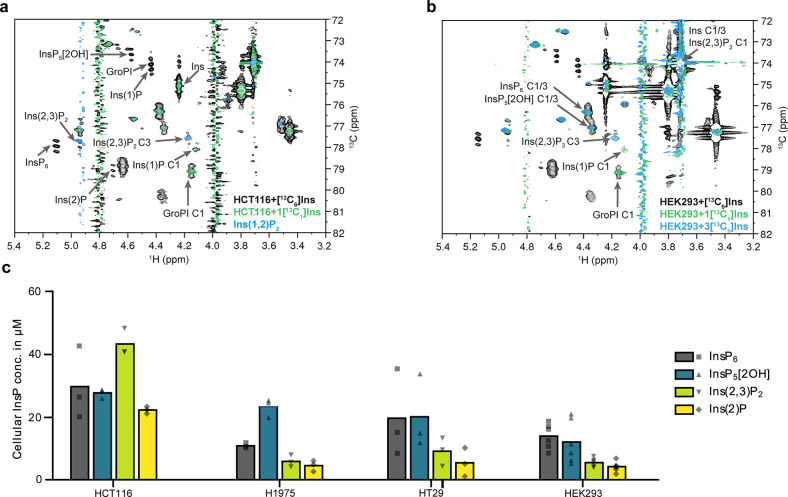
We next performed metabolic labeling of human cell lines (HEK293, HCT116, HT29, H1Hela, H1975) with [13C6]myo-inositol (Figure 1b).38 In brief, cells were grown in a custom medium based on DMEM which contains no natural [12C]myo-inositol but is instead supplemented with [13C6]myo-inositol or an isotopomer of choice (see below). After the cells incorporated the 13C label into their InsP pool to equilibrium (over 2 passages), cells were harvested and their water-soluble metabolites extracted and analyzed by BIRD-{1H,13C}HMQC-NMR. This NMR experiment detects 13CH groups selectively over nonlabeled CH groups, making it particularly suitable for measuring the 13C-labeled InsP pool within a complex background. The information from Figure 2 allowed us to annotate all detectable 13C-labeled species from such extracts. Quantification was performed through relative integration of the signal corresponding to the 2-position against an internal standard and back-calculated to packed cell volumes. The annotation of the different InsPs in an HCT116 metabolic extract is shown exemplarily in Figure 3a (for full annotation see Figure S2). The same set of InsP species was observed in all other cell lines as well (Figures 3b and S3): the major labeled species include InsP6, InsP5[2OH], 1/3-glycerophospho-myo-inositol (1/3-GroPI), inositol 1- or 3-monophosphate (Ins(1/3)P), inositol 1,2- or 2,3-bisphosphate (Ins(1/3,2)P2), inositol 2-monophosphate (Ins(2)P), and myo-inositol. All of these metabolite assignments were validated through spike-in experiments with commercially available InsP standards into 13C-labeled metabolic extracts (Figure S4). Interestingly, labeling of Schizosaccharomyces pombe (Figure S5) revealed a somewhat different metabolite composition.
Figure 3.
Identification and quantification of major InsPs in human cells. (a) Overlay of BIRD-{1H,13C}HMQC-NMR spectra of metabolic extracts from HCT116 cells which were labeled with either [13C6]myo-inositol (black spectrum) or 1[13C1]myo-inositol (green) and a reference spectrum of Ins(1,2)P2 (blue). Annotation of identified InsPs was limited to the most important signals for clarity. Complete annotation is provided in Figure S2. (b) Overlay of BIRD-{1H,13C}HMQC-NMR spectra of metabolic extracts from HEK293 cells which were labeled with [13C6]myo-inositol (black), 1[13C1]myo-inositol (green), or 3[13C1]myo-inositol (blue). Annotation was limited to C1/3 positions for clarity. 1[13C1]myo-inositol-labeled 1 positions of GroPI and Ins(1)P confirm their enantiomeric identity. In contrast, the phosphorylated 3 position of Ins(2,3)P2 is confirmed by labeling with 3[13C1]myo-inositol. (c) Scatter dot plot of quantified InsPs from metabolic extracts of various cells (HCT116, n = 3; H1975, n = 3; HT29, n = 3; HEK293, n = 6, biological replicates) with bars representing the means.
In order to differentiate the possible enantiomers in the mammalian InsP pool, we synthesized asymmetrically 13C-labeled myo-inositols following our previously published protocol.38 Using the singly labeled isotopomer 1[13C1]myo-inositol and doubly labeled 4,5[13C2]Ins, respectively, we repeated the metabolic labeling in HEK293 and HCT116 cells. Focusing on the 1[13C1]myo-inositol labeling, the resulting spectra (Figures 3a, 3b, and S6a) show that the signals that correspond to the phosphorylated 1/3-positions of 1/3-GroPI and Ins(1/3)P are labeled, i.e., the enantiomers present in mammalian cells are 1-GroPI and Ins(1)P. The phosphorylated 1/3-position of Ins(1/3,2)P2 is not labeled, which identifies Ins(2,3)P2 as the prevalent enantiomer, an observation that was reproducible in both cell lines. To confirm this conclusion, HEK293 cells were also labeled with 3[13C1]myo-inositol. Now, the phosphorylated position of the putative InsP2 remains labeled, unambiguously identifying Ins(2,3)P2 as the main InsP2 enantiomer present in human cell lines (Figure 3b).
GroPI and Ins(1)P are established products of cellular phosphatidylinositide turnover;7,39 their detection was therefore anticipated. The presence of Ins(2,3)P2 and Ins(2)P in the micromolar range (especially in HCT116 cells, see Figure 3c) was an unexpected observation. Ins(2,3)P2 and Ins(2)P have not been associated with any established InsP-related pathway so far. Although Ins(2)P and Ins(1/3,2)P2 were detected in 1995 by Mitchell and colleagues,7,40 these metabolites received little attention and have been neglected since then.
Overall, the structural information contained in the HMQC-NMR spectra could be used to assign all detectable 13C-labeled species in mammalian cells, and in combination with the asymmetrical inositol isotopomers, enantiomers could be resolved spectroscopically. This analysis uncovered high amounts of previously poorly characterized lower InsPs, which were not easily accessible with other analytical methods.
Formation of Ins(2,3)P2 and Ins(2)P Is Dependent on MINPP1
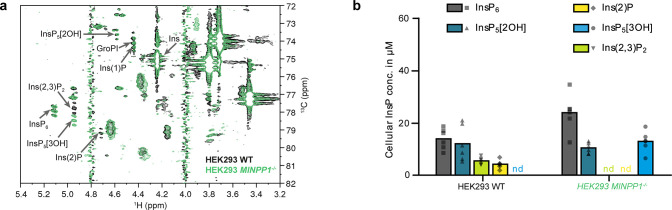
In the biosynthetic pathway toward InsP6 there are no InsP intermediates that are phosphorylated at the 2-position. The 2-phosphoryl group of InsP6 is installed only in the last step, in which IPPK (inositol pentakisphosphate 2-kinase) converts InsP5[2OH] to InsP6. Ins(2,3)P2 and Ins(2)P may therefore be generated downstream of InsP6. A central InsP phosphatase is the mammalian phytase-like enzyme MINPP1, the only recognized InsP6 phosphatase. To investigate possible relationships between Ins(2,3)P2, Ins(2)P, and MINPP1, we turned our attention to cells lacking MINPP1. MINPP1–/– HEK293 cells were labeled with [13C6]myo-inositol, and the metabolites were analyzed by NMR (Figure 4). The MINPP1–/– cells exhibited slightly elevated InsP6 levels and accumulated one new InsP species, which was assigned as InsP5[3OH] or its enantiomer InsP5[1OH] (Figure S4e). InsP5[1/3OH] was not present in any investigated WT cell line. Labeling of MINPP1–/– HEK293 cells with the asymmetric isotopomers 1[13C1]myo-inositol, 3[13C1] myo-inositol, and 4,5[13C2]myo-inositol unambiguously identified the InsP5 in question as InsP5[3OH] (Figure S6b, S6d, and S6e).
Figure 4.
Identification of InsPs in HEK293 and MINPP1–/– HEK293 cells. (a) Overlay of [13C6]myo-inositol-labeled HEK293 (black) and MINPP1–/– HEK293 cells (green). Ins(2,3)P2 and Ins(2)P are not observable in MINPP1–/– cells; instead, InsP5[3OH] accumulates. (b) Scatter dot plot of quantified InsPs from these cell lines (WT, n = 6 same data as in Figure 3c for illustrative purposes; MINPP1–/–, n = 6, biological replicates). Bars represent the means, nd = not detected. Enantiomer-specific identification of InsP5[3OH] is shown in Figure S6.
Strikingly, another change observed in the MINPP1–/– cell extracts was the complete absence of Ins(2,3)P2 and Ins(2)P, establishing a connection between MINPP1 and these lower phosphorylated InsPs. The lack of an undefined InsP2 species was also noted in a previous analysis of the same cell line using a radiolabeling approach.21 Taking into consideration the only sparsely annotated intermediates and products of MINPP1-mediated dephosphorylation of InsP6, it seemed possible that MINPP1 could generate Ins(2,3)P2 and Ins(2)P directly from InsP6.
MINPP1 Dephosphorylates InsP5[2OH] and InsP6 via Fully Distinct Pathways
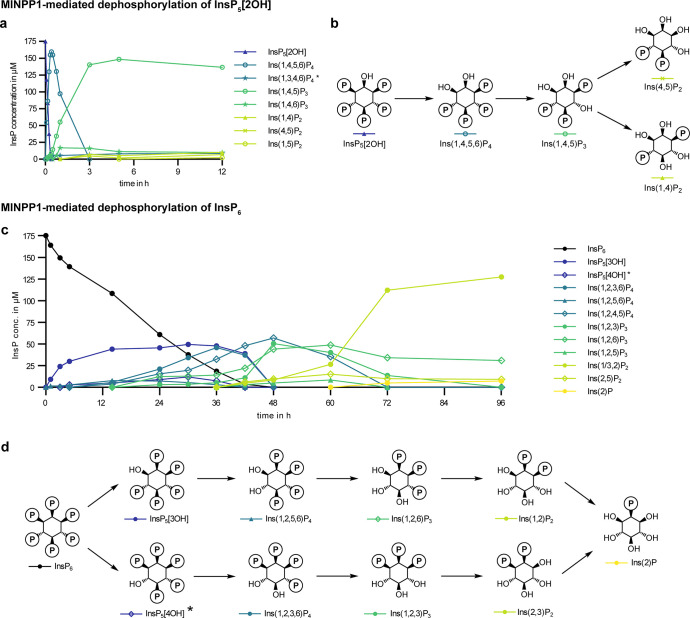
To validate this hypothesis, we next sought to investigate the in vitro activity of MINPP1 against different InsPs. The expression and purification of recombinant MINPP1 in E. coli was optimized to isolate protein yields compatible with biochemical reactions on an NMR scale (Figures S7 and S8). Next, MINPP1 was incubated with fully 13C6-labeled InsP5[2OH], and the reaction was monitored using 2D NMR measurements. In the first experiments we chose a substrate concentration of 50 μM, which is in the middle to upper range of physiological concentrations (Figure S9).7,34,41 To enable the detection and assignment of all intermediates, we subsequently increased the substrate concentration to 175 μM, which did not alter the overall outcome (Figure 5a). The structures of the intermediates were identified using the information from Figure 2 and additional cross-correlation NMR and spike-in experiments where necessary. In agreement with the annotation of MINPP1 as a 3-phosphatase, the first major intermediates for InsP5[2OH] dephosphorylation are Ins(1,4,5,6)P4 and subsequently Ins(1,4,5)P3. MINPP1 therefore directly reverses the phosphorylation reactions catalyzed by IPMK (inositol phosphate multikinase).24,42,43 Ins(1,4,5)P3 is subsequently converted slowly to a mixture of different InsP1/2s (Figure 5b; a full scheme with all minor intermediates is shown in Figure S10).
Figure 5.
Dephosphorylation of InsP5[2OH] and InsP6 by MINPP1 in vitro. (a) Progress curves of MINPP1 reaction with 175 μM [13C6]InsP5[2OH] showing the first 12 h of the reaction (for full scope of progress curves and progress curves at 50 μM substrate concentration see SI). Progress curves shown here are representative of two replicates. (b) Simplified reaction scheme of the MINPP1-mediated dephosphorylation of InsP5[2OH]. Complete reaction scheme that includes all minor intermediates is in Figure S10. (c) Progress curves of MINPP1 reaction with 175 μM [13C6]InsP6 with simplified reaction scheme depicting the two main reaction paths. Progress curves shown here are representative of two replicates. Corresponding progress curve for 50 μM substrate concentration in Figure S13. (d) Simplified reaction scheme for the dephosphorylation of InsP6. Complete reaction scheme that includes all intermediates is in Figure S11. Note that the two enantiomers Ins(1,2)P2 and Ins(2,3)P2 are quantified together. (*) Structure of these InsPs could not be assigned with certainty due to low abundance and interference of more abundant signals.
We then proceeded to probe MINPP1-mediated dephosphorylation of InsP6. In contrast to InsP5[2OH] as a substrate, we observed a complex mixture of intermediates (Figure 5c). In addition, the overall conversion of InsP6 was visibly slower. The two major reaction paths are depicted in Figure 5d (complete scheme in Figure S11): One dephosphorylation sequence proceeds via InsP5[3OH] and Ins(1,2,6)P3 as intermediates, and a second pathway generates Ins(1,2,3)P3 as an intermediate via an unidentified (due to low abundance) InsP5 isomer. Importantly, Ins(1/3,2)P2 and Ins(2)P were observed as the final products of the dephosphorylation of InsP6, validating that MINPP1 is capable of generating these InsPs directly from InsP6.
To assess which enantiomers were formed during MINPP1-mediated dephosphorylation of InsP6, we synthesized 1[13C1]InsP6.38 The InsP5, InsP4, and InsP3 intermediates which are produced by MINPP1 from 1[13C1]InsP6 are enantiopure, as no dephosphorylation of the 1-position was observed (detailed explanation is given in Figures S12a and S12b). Surprisingly though, a mixture of Ins(1,2)P2 and Ins(2,3)P2 was formed during the later stages of the reaction (Figures S12c and S12d). The rather high ratio of Ins(2,3)P2 to Ins(1,2)P2 suggests that Ins(1,2)P2 is formed exclusively via Ins(1,2,6)P3, and Ins(1,2,3)P3 is selectively converted to Ins(2,3)P2. Both InsP2s are, in turn, dephosphorylated to Ins(2)P. Our in vitro assessment of MINPP1 activity thus confirms the notion that MINPP1 can directly generate the novel cellular InsP species from InsP6.
Another interesting observation, which runs counter to assumptions on MINPP1 activity,6,25,44−46 is that the dephosphorylation sequences for InsP6 and InsP5[2OH] do not share any overlap (compare Figures S10, S11, S20, S26, and S27) because MINPP1 seems to be incapable of removing the phosphoryl group at the 2-position. Likely, the charged phosphoryl group on the only axial position of the myo-inositol scaffold plays a role in positioning the InsPs inside MINPP1’s catalytic pocket.47
MINPP1 Exhibits Different Kinetic Properties toward InsP5[2OH] and InsP6
To characterize the kinetic properties of MINPP1, we next numerically determined the reaction rates of the dephosphorylation steps from the respective experimental data based on a time-independent rate model. We formulated the kinetics of the reaction network as a Master equation and approximated the corresponding rate matrix with a least-squares method that iteratively optimized the rates with respect to the scaled experimental data.48,49 The reaction rates for the MINPP1 reaction starting with InsP5[2OH] as a substrate are shown in Figure 6b and were calculated from the experimental data (Figure 5a) and the corresponding network (Figure S10). The calculated rates predict progress curves (Figures 6a and S25) that are in good agreement with the experimental data, which supports the assumption of time-independent rates and thus the absence of inhibition processes. The highest reaction rate (k_20 equaling 330 nmol/(min mg enzyme)) also corresponds to the canonical MINPP1 activity toward InsP5[2OH] in the literature.23
Figure 6.
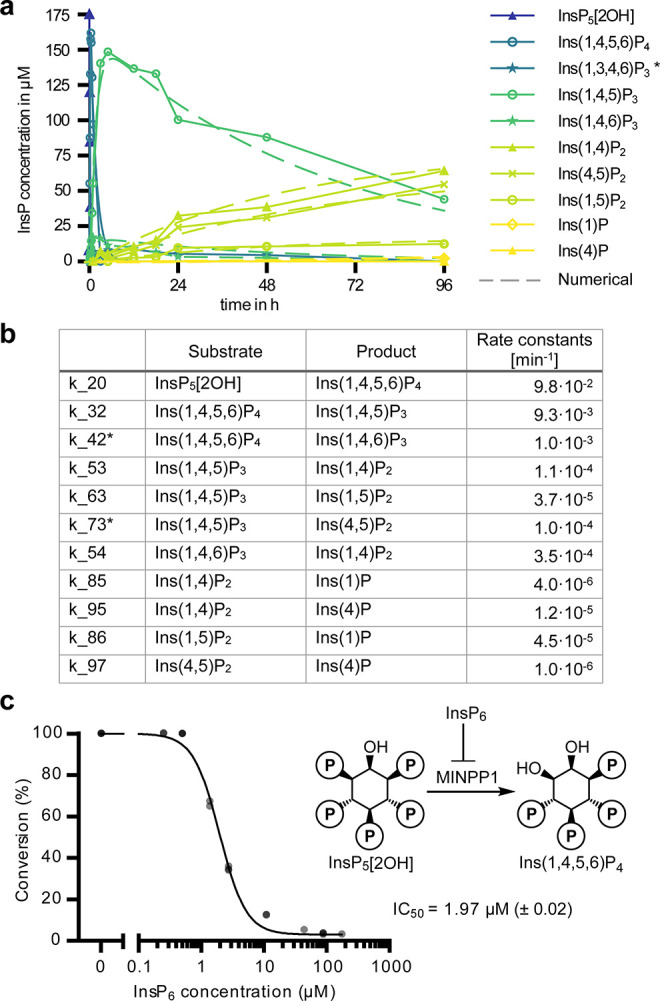
Numerical assessment of MINPP1 reaction rates. (a) Experimental and numerically approximated progress curves of MINPP1 dephosphorylation reactions with 175 μM InsP5[2OH]. Solid lines represent the experimental data (same data as in Figure 5a). Dashed lines represent the progress curves predicted by the numerically determined reaction rates. (b) Numerically determined reaction rates representative of two replicates. Reaction rates marked with an asterisk (*) are subject to constraints. SI also includes attempted numerical approximation of the MINPP1 reaction with InsP6. (c) Demonstration that InsP6 can inhibit dephosphorylation of InsP5[2OH] (175 μM) by MINPP1 (0.5 μM) with high potency. IC50 value is reported with standard error of log10 IC50 in brackets.
However, in the case of InsP6, the computational analysis of the experimental data (Figure 5c) with the network assumption depicted in Figure S11 yielded poor results; only the consumption of InsP6 could be numerically analyzed with a rate of 9.3 × 10–4 min–1 (see SI). The poor fits indicate that the rates in the InsP6 dephosphorylation network might not be time independent but are instead affected by inhibition processes that implicitly introduce a time dependence. Because of its relative stability and slow dephosphorylation, it seemed possible that InsP6 could act as an inhibitor for the dephosphorylation of the MINPP1-generated intermediates.23 This notion is further reinforced by the fact that the conversion of the intermediates progressed notably faster with lower InsP6 starting concentrations (Figure S13). To test this, [13C6]InsP5[2OH] was incubated with MINPP1 in the presence of different amounts of [13C6]InsP6. Indeed, a clear inhibitory effect of InsP6 on the dephosphorylation of InsP5[2OH] by MINPP1 was observed with an apparent IC50 value of 2 μM (Figure 6c). With changing substrate concentrations, the IC50 value also changed as predicted by the Cheng–Prusoff equation, indicating that this inhibition is likely competitive (Figure S14).50
Ins(2,3)P2 and InsP5[3OH] Are Biosynthetically Derived from InsP6 In Cells
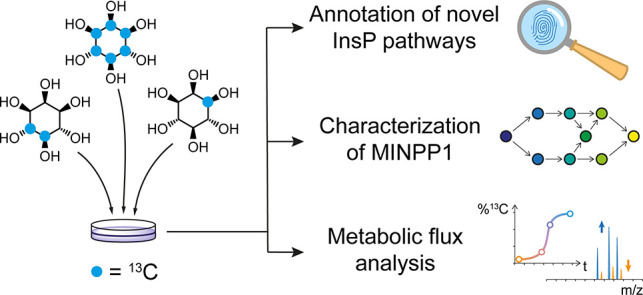
With the biochemical confirmation that MINPP1 can generate InsP5[3OH], Ins(2,3)P2, and Ins(2)P in vitro, we sought to perform metabolic flux analysis to confirm this reaction sequence in living cells. HEK293 or MINPP1–/– HEK293 cells were labeled with [13C6]myo-inositol to equilibrium and subsequently exposed to medium containing 4,5[13C2]myo-inositol for various periods of time before harvesting (Figure 7a). These two isotopomers were chosen to enable analysis by CE-MS: A mass difference of at least 2 Da allows the distinction of the differently labeled InsPs but also the differentiation of Ins(2,3)P2 from other highly abundant, nonlabeled sugar bisphosphates. Following cell lysis, InsP mixtures were extracted with TiO2 beads and analyzed via CE-MS to monitor the incorporation of the 13C2-isotopomers and the decrease of the 13C6-isotopomers simultaneously.
Figure 7.
Metabolic flux analysis via time-dependent isotopic exchange of InsPs in HEK293 and MINPP1–/– HEK293 cells. (a) General workflow of the metabolic flux analysis. (b and c) Ratios of 6-fold 13C-labeled and doubly 13C-labeled InsPs HEK293 (b) and MINPP1–/– HEK293 (c) cells in TiO2-extracted cell lysates. Data of two biological replicates are plotted individually, and means are connected with lines. All extracts contained a constant ∼3% of nonlabeled InsP species, likely stemming from de novo inositol synthesis (Figure S16). Ins(2,3)P2 in MINPP1–/– cells and InsP5[3OH] in WT cells were below the limit of detection. (d) Updated overview of MINPP1-mediated InsP metabolism in human cells. As shown in this work, MINPP1 can dephosphorylate both InsP6 (blue arrows) and InsP5[2OH] (light blue arrows) via through two distinct, nonoverlapping metabolic pathways. Question mark hints toward unidentified phosphatase activities, which might explain the accumulation of InsP5[3OH] observed in MINPP1–/– cells or how Ins(2,3)P2 accumulates selectively in cells while both enantiomers are generated in vitro. Asterisks indicate that we cannot rule out the existence of additional phosphatases that might assist MINPP1-mediated dephosphorylation of InsP6.
CE-MS analysis readily detected the expected [13C6]- and [13C2]InsP species. In addition, all samples contained around 3% of nonlabeled InsPs (12C6), which presumably stems from inositol neogenesis from glucose-6-phosphate.51 The metabolic flux analysis (Figure 7b) indicates that exogenous myo-inositol is incorporated first into the pool of InsP5[2OH], then into InsP6, and last into Ins(2,3)P2 (whose chemical identity was also confirmed with standards in CE-MS measurements, Figure S15). This incorporation sequence supports the hypothesis that Ins(2,3)P2 is indeed derived from InsP6 in human cells and is not an intermediate in the biosynthesis of InsP5[2OH] or InsP6 (Figure 7d).
In MINPP1–/– HEK293 cells, no Ins(2,3)P2 was observed above the limit of detection, although the sensitivity of CE-MS is superior to NMR. Thus, CE-MS analysis confirms that generation of Ins(2,3)P2 is dependent on MINPP1. Similarly, in the biosynthetic sequence, InsP5[3OH] is generated after InsP6 (Figure 7c and 7d), hinting at an unidentified 3-phosphatase activity acting on InsP6, which has been suggested in the past.16 Nevertheless, InsP5[3OH] was not detectable in HEK293 WT cells.
Discussion
We have expanded the detection and identification of complex InsP mixtures using different isotopomers of myo-inositol, InsP5[2OH], and InsP6 in both cellular and biochemical settings. Detection via NMR spectroscopy provided important structural information, enabling the assignment of previously poorly characterized InsPs. Application of asymmetrically labeled 1[13C1]myo-inositol, 3[13C1]myo-inositol, and 4,5[13C2]myo-inositol readily facilitated the distinction of enantiomers in a complex sample, which has remained an analytical challenge to this day. InsP isotopomers with different masses also proved to be useful tools when used in combination with CE-MS analysis, as the higher sensitivity of this technique allows for detailed metabolic flux analyses.
Taking advantage of our labeled myo-inositol isotopomers and InsPs, we uncovered a branch of human InsP metabolism mediated by MINPP1, which was confirmed through in-depth characterization of MINPP1’s reactivity in vitro and in cellula. The in vitro data illustrated that InsP5[2OH] is the preferred substrate for MINPP1, compared to InsP6. Under identical reaction conditions, InsP5[2OH] was depleted with an apparent reaction rate that is 2 orders of magnitude higher than the rate for InsP6 (9.8 × 10–2 versus 9.3 × 10–4 min–1 or ∼330 versus ∼3 nmol/(min mg enzyme), respectively). These activities are in line with previous kinetic analyses of mammalian MINPP1 (211 and 12 nmol/(min mg enzyme), respectively).23 The subsequent slow dephosphorylation of Ins(1,4,5)P3 in vitro (rate constant of 10–4 min–1 or 0.3 nmol/(min mg)) is likely not biologically significant as there are several other Ins(1,4,5)P3 dephosphorylating enzymes with 4–5 magnitudes higher activity (5300–25 000 nmol/(min mg)).54 Interestingly, depletion of cellular MINPP1 did not significantly alter InsP5[2OH] levels, suggesting that other enzymes are able to dephosphorylate InsP5[2OH] in a cellular setting.52,53
In contrast to the straightforward reaction paths for InsP5[2OH] dephosphorylation by MINPP1, the dephosphorylation of InsP6 occurs via an intricate network of intermediates. The first observable intermediates can be attributed to the 3-phosphatase activity of MINPP1; however, a significant portion of InsP6 must initially be dephosphorylated at a different position because the symmetrical Ins(1,2,3)P3 accumulates as an intermediate. Despite this complicated dephosphorylation network, the InsP6 dephosphorylation sequence converges to two final compounds, Ins(1/3,2)P2 and Ins(2)P in vitro. In all human cells we tested, Ins(2,3)P2 and Ins(2)P were present at notable concentrations and constitute a hitherto uncharacterized part of mammalian InsP metabolism. It was somewhat surprising that Ins(2,3)P2 is the predominant InsP2 species within cells, given that MINPP1 is annotated as a 3-phosphatase. Our in vitro data demonstrate that MINPP1 is capable of producing both enantiomers, Ins(1,2)P2 and Ins(2,3)P2, via the aforementioned dephosphorylation pathways from InsP6. It thus seems feasible that Ins(1,2)P2 can also be generated by MINPP1 in cells but may be depleted faster to Ins(2)P by either MINPP1 (which could be modified in its activity through post-translational modifications or different isoforms31) or a separate phosphatase altogether.
Remarkably, the many different dephosphorylation products of InsP6 do not overlap with any intermediates of InsP5[2OH] dephosphorylation, because MINPP1 appears incapable of removing the phosphoryl group at the 2-position of the inositol ring (Figure 7d). While MINPP1 converts InsP5[2OH] to its biosynthetic precursors Ins(1,3,4,5)P4 and Ins(1,4,5)P3 in vitro, InsP6 on the other hand is exclusively dephosphorylated to metabolites, which keep the phosphoryl group at the 2-position. This data is in stark contrast to the common assumption that MINPP1 would convert InsP6 to InsP5[2OH], as is often depicted in overview schemes on InsP metabolism.6,25,44−46 It was shown in the past that the phosphoryl group at the 2-position of the myo-inositol ring (the only axial position) can play an important role for proper recognition of InsPs by protein binding partners.55,56 Our data further corroborates the importance of the phosphorylation status of the 2-position (and thus IPPK activity) because it appears that InsPs may be “sorted” into the known and reversible InsP network (when InsPs contain a free hydroxyl group at the 2-position) or InsPs enter the slower, and potentially irreversible, MINPP1-mediated circuit where they remain phosphorylated at the 2-position.
While this sorting could be accomplished solely by the preferred dephosphorylation by MINPP1, the accessibility to the two different substrates InsP5[2OH] and InsP6 likely also plays a role. We found that the dephosphorylation of InsP5[2OH] was strongly inhibited by low concentrations of InsP6 in vitro (Figures 6c and S15). In the cellular context, this potent inhibitory effect of the abundant InsP6 metabolite raises the question if, and how, MINPP1 can dephosphorylate InsP5[2OH] at all. MINPP1 would need to access localized pools of said InsPs that are tightly regulated to either avoid or make use of the inhibitory effect. Interestingly, MINPP1 is thought to predominantly localize to the ER,28 so how it accesses cytosolic (and presumably nuclear) InsPs is a question that has yet to be answered. While some studies have shown that MINPP1 (isoforms) might also be localized in cellular compartments other than the ER (Figure S17),30 or could be even secreted,31 tools to measure intracellular concentrations of different InsPs with spatial resolution are currently not available. An intriguing avenue for regulation could be that MINPP1 remains localized to intracellular organelles (ER or lysosomes) into which InsPs are controllably translocated and then dephosphorylated. This dephosphorylation could potentially proceed all the way to myo-inositol—with the aid of additional phosphatases—which could then be released through inositol transporters like SLC2A13 (HMIT). HMIT is known to be localized in intracellular membranes due to its ER-retention sequence and internalization sequence.30,31,57−59
Using asymmetrically isotope-labeled myo-inositol, it was possible to assign the uncharacterized InsP5 isomer that accumulates in MINPP1–/– cells as InsP5[3OH]. This accumulation appears counterintuitive, since MINPP1 is currently the only known enzyme in the human genome capable of generating InsP5[3OH]. Nevertheless, Chi et al. also observed a residual 3-phosphatase activity in MINPP1–/– mice.16 An analogous activity in human cells could be responsible for producing InsP5[3OH] from InsP6, as illustrated by our CE-MS-based metabolic flux analysis. Elucidating the identity of this 3-phosphatase will be of interest in the future as it constitutes an additional point of regulation within the InsP network. Furthermore, two recently reported cell lines with elevated intracellular phosphate levels were shown to contain a nonannotated InsP5 isomer (which we assume is also InsP5[1/3OH] based on the SAX-HPLC elution profiles).51,60 Once the absolute configuration of these InsP5 isomers has been determined, and ideally the enzymatic activities responsible for generating these isomers, the impact of cellular phosphate homeostasis on InsP signaling could be further explored.
The physiological role of the herein described dephosphorylation pathway for InsP6 and its intermediates has yet to be explored. The InsPs produced by MINPP1 could be part of a recycling system converting InsP6 back to Ins(2)P, which might be converted to myo-inositol by an inositol monophosphatase (although the lithium-sensitive human enzymes IMPA1/2 are not known to act on Ins(2)P4,61). As MINPP1 is a homologue of phytases, which take part in inositol recycling/scavenging, this possibility does not seem far fetched.18 We cannot exclude the existence of other unknown phosphatases that contribute to this dephosphorylation pathway; however, the accumulation of InsP5[3OH] in MINPP1–/– cells suggests that MINPP1 is obligatory for the dephosphorylation of InsP5[3OH]. Furthermore, the complete absence of Ins(2,3)P2 in MINPP1–/– cells indicates that MINPP1 must carry out the key dephosphorylation of InsP6 on the path toward Ins(2,3)P2. In addition, it remains to be investigated which enzymes can utilize the herein identified Ins(1/3,2)P2 as substrates. Whether any of the InsP6-derived MINPP1 products have signaling functions themselves is also an open question. It is possible that some MINPP1-generated InsPs (or the lack thereof) could be important contributing factors in MINPP1-regulated processes, i.e., ER stress, endochondral ossification, and neuronal function.17,19,21 For example, it would be interesting to investigate if the hyperaccumulation of InsP5[3OH] or the absence of Ins(2,3)P2 and Ins(2)P is partially responsible for causing PCH in patients with MINPP1 loss-of-function mutations.20,21 Ucuncu et al. proposed that hyperaccumulation of InsP6 in neuronal cells of PCH patients might be a mechanistic cause of this disease by chelating iron ions.21 In fact, all InsP species which possess the 1,2,3-phosphorylated motif might be capable of binding iron ions.62 In contrast to their reported 3–4-fold increase of [3H]InsP6 levels (normalized against total tritiated PIPs) in MINPP1–/– HEK293 cells compared to WT cells, we only observed a slight increase using the same cell line but normalizing against packed cell volume. This discrepancy points toward several interesting possibilities: (a) PIP levels, or the incorporation of exogenous myo-inositol, could be (indirectly) influenced by MINPP1 activity, (b) radioactivity-induced cell stress could have an effect on MINPP1 expression,17 or (c) knockout of MINPP1 changes the cell shape/volume. To differentiate between these possibilities, different quantification methods (e.g., normalization against total protein or DNA concentration) should be compared in the future, and the composition of PIP isotopomers during metabolic labeling experiments could be probed with mass spectrometry-based methods.63−65
As a next step, the combination of inositol isotopomers, NMR and CE-MS which we used in this study could be useful to probe InsP metabolism in a variety of biological contexts. For example, it could be investigated how the InsP pool changes during ER-related stress, during which MINPP1 is upregulated, and how this might correlate with the onset of apoptosis.17 Another interesting application would be to determine the fate of inositol (phosphates) in pathogenic parasites such as T. cruzi, in which InsP metabolism is essential for the developmental cycle.66 The question if or how InsP metabolism of the host cell and the parasite influences each other might lead to new therapeutic avenues for these parasitoses. The dissection of InsP degradation in an extracellular context, namely, how InsPs contained in food are converted by digestive processes or the gut microbiome and if the resulting metabolites might have beneficial or detrimental effects on health, is also a fascinating question.47,67,68 With the tools and methods reported here, these topics now become addressable.
Methods
All experimental and computational methods are described in the Supporting Information.
Acknowledgments
M.N.T. and S.K. were funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy (EXC 2008-390540038) UniSysCat. S.K. was partially funded by Deutsche Forschungsgemeinschaft (DFG) through grant CRC 1114 “Scaling Cascades in Complex Systems”, Project Number 235221301, Project B05 “Origin of scaling cascades in protein dynamics”. We thank Peter Schmieder for providing valuable guidance with NMR topics, Lena von Oertzen and Kathrin Motzny for their assistance in cell culture matters, Robert Puschmann for initial synthesis of myo-inositols, and all members of the Fiedler lab for proofreading.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscentsci.2c01032.
Author Contributions
M.N.T. performed most biochemical and in vitro experiments, data analyses, and organic syntheses. S.K. performed all kinetic modeling analyses under the supervision of B.K. Z.F. contributed to initial testing of MINPP1 expression and reactivity optimization. D.Q. and G.L. measured all CE-MS samples under the supervision of H.J. N.M. performed the cellular fractions and Western blots. D.F. and M.N.T. conceived the project with input from A.S. D.F. and M.N.T. supervised the experimental work by Z.F. M.N.T. and D.F. prepared the initial draft of the paper with S.K. contributing the kinetic modeling part, and all authors contributed to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Watson P. J.; Fairall L.; Santos G. M.; Schwabe J. W. R. Structure of HDAC3 Bound to Co-Repressor and Inositol Tetraphosphate. Nature 2012, 481 (7381), 335–340. 10.1038/nature10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.; Vogan E. M.; Nocka L. M.; Rosen C. E.; Zorn J. A.; Harrison S. C.; Kuriyan J. Autoinhibition of Bruton’s Tyrosine Kinase (Btk) and Activation by Soluble Inositol Hexakisphosphate. Elife 2015, 4, e06074. 10.7554/eLife.06074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blind R. D. Structural Analyses of Inositol Phosphate Second Messengers Bound to Signaling Effector Proteins. Adv. Biol. Regul. 2020, 75, 100667. 10.1016/j.jbior.2019.100667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. Toward Understanding the Origin and Evolution of Cellular Organisms. Protein Sci. 2019, 28 (11), 1947–1951. 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine R. F.; Schell M. J. Back in the Water: The Return of the Inositol Phosphates. Nat. Rev. Mol. Cell Biol. 2001, 2 (5), 327–338. 10.1038/35073015. [DOI] [PubMed] [Google Scholar]
- Chatree S.; Thongmaen N.; Tantivejkul K.; Sitticharoon C.; Vucenik I. Role of Inositols and Inositol Phosphates in Energy Metabolism. Molecules 2020, 25 (21), 5079. 10.3390/molecules25215079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker C. J.; Wright J.; Hughes P. J.; Kirk C. J.; Michell R. H. Complex Changes in Cellular Inositol Phosphate Complement Accompany Transit through the Cell Cycle. Biochem. J. 2004, 380 (2), 465–473. 10.1042/bj20031872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D.; Wilson M. S.; Eisenbeis V. B.; Harmel R. K.; Riemer E.; Haas T. M.; Wittwer C.; Jork N.; Gu C.; Shears S. B.; Schaaf G.; Kammerer B.; Fiedler D.; Saiardi A.; Jessen H. J. Analysis of Inositol Phosphate Metabolism by Capillary Electrophoresis Electrospray Ionization Mass Spectrometry. Nat. Commun. 2020, 11, 1–12. 10.1038/s41467-020-19928-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letcher A. J.; Schell M. J.; Irvine R. F. Do Mammals Make All Their Own Inositol Hexakisphosphate?. Biochem. J. 2008, 416 (2), 263–270. 10.1042/BJ20081417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen Trung M.; Furkert D.; Fiedler D. Versatile Signaling Mechanisms of Inositol Pyrophosphates. Curr. Opin. Chem. Biol. 2022, 70, 102177. 10.1016/j.cbpa.2022.102177. [DOI] [PubMed] [Google Scholar]
- Dick R. A.; Zadrozny K. K.; Xu C.; Schur F. K. M.; Lyddon T. D.; Ricana C. L.; Wagner J. M.; Perilla J. R.; Ganser-Pornillos B. K.; Johnson M. C.; Pornillos O.; Vogt V. M. Inositol Phosphates Are Assembly Co-Factors for HIV-1. Nature 2018, 560 (7719), 509–512. 10.1038/s41586-018-0396-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson P. J.; Millard C. J.; Riley A. M.; Robertson N. S.; Wright L. C.; Godage H. Y.; Cowley S. M.; Jamieson A. G.; Potter B. V. L.; Schwabe J. W. R. Insights into the Activation Mechanism of Class I HDAC Complexes by Inositol Phosphates. Nat. Commun. 2016 71 2016, 7 (1), 1–13. 10.1038/ncomms11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H.; Yan Y.; Luo Y.; So W. Y.; Wei X.; Zhang X.; Yang X.; Zhang J.; Su Y.; Yang X.; Zhang B.; Zhang K.; Jiang N.; Chow B. K. C.; Han W.; Wang F.; Rao F. IP6-Assisted CSN-COP1 Competition Regulates a CRL4-ETV5 Proteolytic Checkpoint to Safeguard Glucose-Induced Insulin Secretion. Nat. Commun. 2021 121 2021, 12 (1), 1–13. 10.1038/s41467-021-22941-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H.; Zhang X.; Liu L.; Fu Q.; Zang C.; Ding Y.; Su Y.; Xu Z.; He S.; Yang X.; Wei X.; Mao H.; Cui Y.; Wei Y.; Zhou C.; Du L.; Huang N.; Zheng N.; Wang T.; Rao F. Basis for Metabolite-Dependent Cullin-RING Ligase Deneddylation by the COP9 Signalosome. Proc. Natl. Acad. Sci. U. S. A. 2020, 117 (8), 4117–4124. 10.1073/pnas.1911998117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhn M. Turn and Face the Strange: A New View on Phosphatases. ACS Cent. Sci. 2020, 6 (4), 467–477. 10.1021/acscentsci.9b00909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi H.; Yang X.; Kingsley P. D.; O’Keefe R. J.; Puzas J. E.; Rosier R. N.; Shears S. B.; Reynolds P. R. Targeted Deletion of Minpp1 Provides New Insight into the Activity of Multiple Inositol Polyphosphate Phosphatase In Vivo. Mol. Cell. Biol. 2000, 20 (17), 6496–6507. 10.1128/MCB.20.17.6496-6507.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilaparty S. P.; Agarwal R.; Singh P.; Kannan K.; Ali N. Endoplasmic Reticulum Stress-Induced Apoptosis Accompanies Enhanced Expression of Multiple Inositol Polyphosphate Phosphatase 1 (Minpp1): A Possible Role for Minpp1 in Cellular Stress Response. Cell Stress Chaperones 2016, 21 (4), 593–608. 10.1007/s12192-016-0684-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilaparty S. P.; Singh A.; Baltosser W. H.; Ali N. Computational Analysis Reveals a Successive Adaptation of Multiple Inositol Polyphosphate Phosphatase 1 in Higher Organisms through Evolution. Evol. Bioinform. Online 2014, 10, 239–250. 10.4137/EBO.S18948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffrey J. J.; Hidaka K.; Matsuda M.; Hirata M.; Shears S. B. The Human and Rat Forms of Multiple Inositol Polyphosphate Phosphatase: Functional Homology with a Histidine Acid Phosphatase up-Regulated during Endochondral Ossification. FEBS Lett. 1999, 442 (1), 99–104. 10.1016/S0014-5793(98)01636-6. [DOI] [PubMed] [Google Scholar]
- Appelhof B.; Wagner M.; Hoefele J.; Heinze A.; Roser T.; Koch-Hogrebe M.; Roosendaal S. D.; Dehghani M.; Mehrjardi M. Y. V.; Torti E.; Houlden H.; Maroofian R.; Rajabi F.; Sticht H.; Baas F.; Wieczorek D.; Jamra R. A. Pontocerebellar Hypoplasia Due to Bi-Allelic Variants in MINPP1. Eur. J. Hum. Genet. 2021, 29, 411. 10.1038/s41431-020-00749-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ucuncu E.; Rajamani K.; Wilson M. S. C.; Medina-Cano D.; Altin N.; David P.; Barcia G.; Lefort N.; Banal C.; Vasilache-Dangles M. T.; Pitelet G.; Lorino E.; Rabasse N.; Bieth E.; Zaki M. S.; Topcu M.; Sonmez F. M.; Musaev D.; Stanley V.; Bole-Feysot C.; Nitschké P.; Munnich A.; Bahi-Buisson N.; Fossoud C.; Giuliano F.; Colleaux L.; Burglen L.; Gleeson J. G.; Boddaert N.; Saiardi A.; Cantagrel V. MINPP1 Prevents Intracellular Accumulation of the Chelator Inositol Hexakisphosphate and Is Mutated in Pontocerebellar Hypoplasia. Nat. Commun. 2020, 11 (1), 6087. 10.1038/s41467-020-19919-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene ID: 9562, Homo sapiens, MINPP1 multiple inositol-polyphosphate phosphatase 1. Gene [Internet]. National Library of Medicine (US), National Center for Biotechnology Information: Bethesda, MD; https://www.ncbi.nlm.nih.gov/gene/9562 (accessed 2022. –08–26).
- Nogimori K.; Hughes P. J.; Glennon M. C.; Hodgson M. E.; Putney J. W.; Shears S. B. Purification of an Inositol (1,3,4,5)-Tetrakisphosphate 3-Phosphatase Activity from Rat Liver and the Evaluation of Its Substrate Specificity. J. Biol. Chem. 1991, 266 (25), 16499–16506. 10.1016/S0021-9258(18)55328-X. [DOI] [PubMed] [Google Scholar]
- Craxton A.; Caffrey J. J.; Burkhart W.; Safrany T. S.; Shears B. S. Molecular Cloning and Expression of a Rat Hepatic Multiple Inositol Polyphosphate Phosphatase. Biochem. J. 1997, 328 (1), 75–81. 10.1042/bj3280075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.; Leibiger B.; Yang S. N.; Caffery J. J.; Shears S. B.; Leibiger I. B.; Barker C. J.; Berggren P. O. Cytosolic Multiple Inositol Polyphosphate Phosphatase in the Regulation of Cytoplasmic Free Ca2+ Concentration. J. Biol. Chem. 2003, 278 (47), 46210–46218. 10.1074/jbc.M303743200. [DOI] [PubMed] [Google Scholar]
- Barker C. J.; Illies C.; Berggren P.-O.. HPLC Separation of Inositol Polyphosphates. In Inositol Phosphates and Lipids; Barker C. J., Ed.; Humana Press: New York, 2010; pp 21–46 10.1007/978-1-60327-175-2_2. [DOI] [PubMed] [Google Scholar]
- Shears S. B.A Short Historical Perspective of Methods in Inositol Phosphate Research. In Inositol Phosphates; Miller G., Ed.; Springer US: New York, 2020; pp 1–28 10.1007/978-1-0716-0167-9_1. [DOI] [PubMed] [Google Scholar]
- Ali N.; Craxton A.; Shears S. B. Hepatic Ins(1,3,4,5)P4 3-Phosphatase Is Compartmentalized inside Endoplasmic Reticulum. J. Biol. Chem. 1993, 268 (9), 6161–6167. 10.1016/S0021-9258(18)53233-6. [DOI] [PubMed] [Google Scholar]
- Craxton A.; Ali N.; Shears S. B. Comparison of the Activities of a Multiple Inositol Polyphosphate Phosphatase Obtained from Several Sources: A Search for Heterogeneity in This Enzyme. Biochem. J. 1995, 305 (2), 491–498. 10.1042/bj3050491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windhorst S.; Lin H.; Blechner C.; Fanick W.; Brandt L.; Brehm M. A.; Mayr G. W. Tumour Cells Can Employ Extracellular Ins(1,2,3,4,5,6)P6 and Multiple Inositol-Polyphosphate Phosphatase 1 (MINPP1) Dephosphorylation to Improve Their Proliferation. Biochem. J. 2013, 450 (1), 115–125. 10.1042/BJ20121524. [DOI] [PubMed] [Google Scholar]
- Zubair M.; Hamzah R.; Griffin R.; Ali N. Identification and Functional Characterization of Multiple Inositol Polyphosphate Phosphatase1 (Minpp1) Isoform-2 in Exosomes with Potential to Modulate Tumor Microenvironment. PLoS One 2022, 17, e0264451. 10.1371/journal.pone.0264451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N. W.; Marmelstein A. M.; Fiedler D. Chemical Tools for Interrogating Inositol Pyrophosphate Structure and Function. Chem. Soc. Rev. 2016, 45, 6311–6326. 10.1039/C6CS00193A. [DOI] [PubMed] [Google Scholar]
- Wilson M. S. C.; Bulley S. J.; Pisani F.; Irvine R. F.; Saiardi A. A Novel Method for the Purification of Inositol Phosphates from Biological Samples Reveals That No Phytate Is Present in Human Plasma or Urine. Open Biol. 2015, 5 (3), 150014. 10.1098/rsob.150014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D.; Eisenbeis V. B.; Saiardi A.; Jessen H. J. Absolute Quantitation of Inositol Pyrophosphates by Capillary Electrophoresis Electrospray Ionization Mass Spectrometry. J. Vis. Exp. 2021, 2021 (174), 1–13. 10.3791/62847. [DOI] [PubMed] [Google Scholar]
- Losito O.; Szijgyarto Z.; Resnick A. C.; Saiardi A. Inositol Pyrophosphates and Their Unique Metabolic Complexity: Analysis by Gel Electrophoresis. PLoS One 2009, 4 (5), e5580. 10.1371/journal.pone.0005580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M. S. C.; Saiardi A. Importance of Radioactive Labelling to Elucidate Inositol Polyphosphate Signalling. Top. Curr. Chem. 2017, 375 (1), 1–21. 10.1007/s41061-016-0099-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr G. W. A Novel Metal-Dye Detection System Permits Picomolar-Range h.p.l.c. Analysis of Inositol Polyphosphates from Non-Radioactively Labelled Cell or Tissue Specimens. Biochem. J. 1988, 254, 585–591. 10.1042/bj2540585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmel R. K.; Puschmann R.; Nguyen Trung M.; Saiardi A.; Schmieder P.; Fiedler D. Harnessing 13C-Labeled Myo-Inositol to Interrogate Inositol Phosphate Messengers by NMR. Chem. Sci. 2019, 10 (20), 5267–5274. 10.1039/C9SC00151D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corda D.; Zizza P.; Varone A.; Filippi B. M.; Mariggiò S. The Glycerophosphoinositols: Cellular Metabolism and Biological Functions. Cell. Mol. Life Sci. 2009, 66 (21), 3449–3467. 10.1007/s00018-009-0113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker C. J.; French P. J.; Moore A. J.; Nilsson T.; Berggren P. O.; Bunce C. M.; Kirk C. J.; Michell R. H. Inositol 1,2,3-Trisphosphate and Inositol 1,2- and/or 2,3-Bisphosphate Are Normal Constituents of Mammalian Cells. Biochem. J. 1995, 306 (2), 557–564. 10.1042/bj3060557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountford J. C.; Bunce C. M.; French P. J.; Michell R. H.; Brown G. Intracellular Concentrations of Inositol, Glycerophosphoinositol and Inositol Pentakisphosphate Increase during Haemopoietic Cell Differentiation. BBA - Mol. Cell Res. 1994, 1222 (1), 101–108. 10.1016/0167-4889(94)90030-2. [DOI] [PubMed] [Google Scholar]
- Chang S. C.; Miller A. L.; Feng Y.; Wente S. R.; Majerus P. W. The Human Homolog of the Rat Inositol Phosphate Multikinase Is an Inositol 1,3,4,6-Tetrakisphosphate 5-Kinase. J. Biol. Chem. 2002, 277 (46), 43836–43843. 10.1074/jbc.M206134200. [DOI] [PubMed] [Google Scholar]
- Saiardi A.; Nagata E.; Luo H. R.; Sawa A.; Luo X.; Snowman A. M.; Snyder S. H. Mammalian Inositol Polyphosphate Multikinase Synthesizes Inositol 1,4,5-Trisphosphate and an Inositol Pyrophosphate. Proc. Natl. Acad. Sci. U. S. A. 2001, 98 (5), 2306–2311. 10.1073/pnas.041614598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu C.; Liu J.; Liu X.; Zhang H.; Luo J.; Wang H.; Locasale J. W.; Shears S. B. Metabolic Supervision by PPIP5K, an Inositol Pyrophosphate Kinase/Phosphatase, Controls Proliferation of the HCT116 Tumor Cell Line. Proc. Natl. Acad. Sci. U. S. A. 2021, 118 (10), 1–8. 10.1073/pnas.2020187118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M. P.; Potter B. V. L. The Enzymes of Human Diphosphoinositol Polyphosphate Metabolism. FEBS J. 2014, 281 (1), 14–33. 10.1111/febs.12575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.; Shortreed M.; Wenger C.; Frey B.; Schaffer L.; Scalf M.; Smith L. Global Post-Translational Modification Discovery. J. Proteome Res. 2017, 16 (4), 1383–1390. 10.1021/acs.jproteome.6b00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acquistapace I. M.; Thompson E. J.; Kühn I.; Bedford M. R.; Brearley C. A.; Hemmings A. M. Insights to the Structural Basis for the Stereospecificity of the Escherichia Coli Phytase, AppA. Int. J. Mol. Sci. 2022, 23 (11), 6346. 10.3390/ijms23116346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner C. W.Handbook of Stochastic Methods for Physics, Chemistry, and the Natural Sciences, 2nd ed.; Springer Berlin Heidelberg, 1985. [Google Scholar]
- Van Kampen N. G.Stochastic Processes, 1st ed.; North-Holland Publishing Co.: Amsterdam, New York, Oxford, 1981. [Google Scholar]
- Yung-Chi C.; Prusoff W. H. Relationship between the Inhibition Constant (KI) and the Concentration of Inhibitor Which Causes 50 per Cent Inhibition (I50) of an Enzymatic Reaction. Biochem. Pharmacol. 1973, 22 (23), 3099–3108. 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- Desfougères Y.; Wilson M. S. C.; Laha D.; Miller G. J.; Saiardi A. ITPK1Mediates the Lipid-Independent Synthesis of Inositol Phosphates Controlled by Metabolism. Proc. Natl. Acad. Sci. U. S. A. 2019, 116 (49), 24551–24561. 10.1073/pnas.1911431116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain P. P.; Qian X.; Stiles A. R.; Cho J.; Jones D. H.; Lesley S. A.; Grabau E. A.; Shears S. B.; Spraggon G. Integration of Inositol Phosphate Signaling Pathways via Human ITPK1. J. Biol. Chem. 2007, 282 (38), 28117–28125. 10.1074/jbc.M703121200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffrey J. J.; Darden T.; Wenk M. R.; Shears S. B. Expanding Coincident Signaling by PTEN through Its Inositol 1,3,4,5,6-Pentakisphosphate 3-Phosphatase Activity. FEBS Lett. 2001, 499 (1–2), 6–10. 10.1016/S0014-5793(01)02500-5. [DOI] [PubMed] [Google Scholar]
- Zhang X.; Jefferson A. B.; Auethavekiat V.; Majerus P. W. The Protein Deficient in Lowe Syndrome Is a Phosphatidylinositol-4,5-Bisphosphate 5-Phosphatase. Proc. Natl. Acad. Sci. U. S. A. 1995, 92 (11), 4853–4856. 10.1073/pnas.92.11.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furkert D.; Hostachy S.; Nadler-Holly M.; Fiedler D. Triplexed Affinity Reagents to Sample the Mammalian Inositol Pyrophosphate Interactome. Cell Chem. Biol. 2020, 27 (8), 1097–1108. 10.1016/j.chembiol.2020.07.017. [DOI] [PubMed] [Google Scholar]
- Wild R.; Gerasimaite R.; Jung J.-Y.; Truffault V.; Pavlovic I.; Schmidt A.; Saiardi A.; Jessen H. J.; Poirier Y.; Hothorn M.; Mayer A. Control of Eukaryotic Phosphate Homeostasis by Inositol Polyphosphate Sensor Domains. Science 2016, 352 (6288), 986–990. 10.1126/science.aad9858. [DOI] [PubMed] [Google Scholar]
- Su X. B.; Ko A.-L. A.; Saiardi A. Regulations of Myo-Inositol Homeostasis: Mechanisms, Implications, and Perspectives. Adv. Biol. Regul. 2022, 100921. 10.1016/j.jbior.2022.100921. [DOI] [PubMed] [Google Scholar]
- Uldry M.; Steiner P.; Zurich M.-G.; Béguin P.; Hirling H.; Dolci W.; Thorens B. Regulated Exocytosis of an H+/Myo-Inositol Symporter at Synapses and Growth Cones. EMBO J. 2004, 23 (3), 531–540. 10.1038/sj.emboj.7600072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel E. Di; Kew J. N.; Maycox P. R. Investigation of the H+-Myo-Inositol Transporter (HMIT) as a Neuronal Regulator of Phosphoinositide Signalling. Biochem. Soc. Trans. 2009, 37 (5), 1139–1143. 10.1042/BST0371139. [DOI] [PubMed] [Google Scholar]
- López-Sánchez U.; Tury S.; Nicolas G.; Wilson M. S.; Jurici S.; Ayrignac X.; Courgnaud V.; Saiardi A.; Sitbon M.; Battini J. L. Interplay between Primary Familial Brain Calcification-Associated SLC20A2 and XPR1 Phosphate Transporters Requires Inositol Polyphosphates for Control of Cellular Phosphate Homeostasis. J. Biol. Chem. 2020, 295 (28), 9366–9378. 10.1074/jbc.RA119.011376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENZYME: 3.1.3.25. KEGG [Internet]; https://www.genome.jp/entry/3.1.3.25 (accessed 2022. –08–26).
- Veiga N.; Torres J.; Mansell D.; Freeman S.; Domínguez S.; Barker C. J.; Díaz A.; Kremer C. Chelatable Iron Pool”: Inositol 1,2,3-Trisphosphate Fulfils the Conditions Required to Be a Safe Cellular Iron Ligand. J. Biol. Inorg. Chem. 2009, 14 (1), 51–59. 10.1007/s00775-008-0423-2. [DOI] [PubMed] [Google Scholar]
- Kim Y.; Shanta S. R.; Zhou L.-H.; Kim K. P. Mass Spectrometry Based Cellular Phosphoinositides Profiling and Phospholipid Analysis: A Brief Review. Exp. Mol. Med. 2010, 42 (1), 1. 10.3858/emm.2010.42.1.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettitt T. R.; Dove S. K.; Lubben A.; Calaminus S. D. J.; Wakelam M. J. O. Analysis of Intact Phosphoinositides in Biological Samples. J. Lipid Res. 2006, 47 (7), 1588–1596. 10.1194/jlr.D600004-JLR200. [DOI] [PubMed] [Google Scholar]
- Milne S. B.; Ivanova P. T.; DeCamp D.; Hsueh R. C.; Brown H. A. A Targeted Mass Spectrometric Analysis of Phosphatidylinositol Phosphate Species. J. Lipid Res. 2005, 46 (8), 1796–1802. 10.1194/jlr.D500010-JLR200. [DOI] [PubMed] [Google Scholar]
- Mantilla B. S.; Amaral L. D. D.; Jessen H. J.; Docampo R. The Inositol Pyrophosphate Biosynthetic Pathway of Trypanosoma Cruzi. ACS Chem. Biol. 2021, 16 (2), 283–292. 10.1021/acschembio.0c00759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K.; Vucenik I.; Shamsuddin A. M. [3H]Phytic Acid (Inositol Hexaphosphate) Is Absorbed and Distributed to Various Tissues in Rats. J. Nutr. 1993, 123 (4), 713–720. 10.1093/jn/123.4.713. [DOI] [PubMed] [Google Scholar]
- Bui T. P. N.; Mannerås-Holm L.; Puschmann R.; Wu H.; Troise A. D.; Nijsse B.; Boeren S.; Bäckhed F.; Fiedler D.; deVos W. M. Conversion of Dietary Inositol into Propionate and Acetate by Commensal Anaerostipes Associates with Host Health. Nat. Commun. 2021, 12 (1), 1–16. 10.1038/s41467-021-25081-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.