Abstract
In male mice, the transcription factors STRA8 and MEISON initiate meiosis I. We report that STRA8/MEISON activates the transcription factors A-MYB and TCFL5, which together reprogram gene expression after spermatogonia enter into meiosis. TCFL5 promotes transcription of genes required for meiosis, mRNA turnover, miR-34/449 production, meiotic exit, and spermiogenesis. This transcriptional architecture is conserved in rhesus macaque, suggesting TCFL5 plays a central role in meiosis and spermiogenesis in placental mammals. Tcfl5em1/em1 mutants are sterile, and spermatogenesis arrests at the mid- or late-pachytene stage of meiosis. Moreover, Tcfl5+/em1 mutants produce fewer motile sperm.
Keywords: TCFL5, spermatogenesis, meiosis, A-MYB, transcription factors network, miR-34/449
In brief
The testis-specific transcription factor, TCFL5, expressed in pachytene spermatocytes regulates the meiotic gene expression program in collaboration with the transcription factor A-MYB.
Introduction
Gametogenesis converts diploid progenitor germ cells into haploid gametes specialized for sexual reproduction. Male gametogenesis encompasses a stepwise developmental pathway of meiotic cell divisions and differentiation to generate mature sperm from self-renewing spermatogenic stem cells (Handel and Schimenti, 2010). Because haploid sperm are mostly transcriptionally inert (Grunewald et al., 2005; Kierszenbaum and Tres, 1975; Ostermeier et al., 2002), meiotic cells must express not only gene products required for the orderly progression and completion of meiosis, but also produce the transcripts encoding proteins required to build mature sperm (Bolcun-Filas et al., 2011; Handel and Schimenti, 2010).
A-MYB binds promoters, super enhancers, and endogenous retroviral enhancers that promote transcription of meiotic genes (Maezawa et al., 2020; Sakashita et al., 2020). Although A-Myb-mutant germ cells fail to express meiotic genes and arrest early in meiosis I (Bolcun-Filas et al., 2011), A-MYB directly binds the promoters of less than one-quarter of meiotic genes (Bolcun-Filas et al., 2011; Li et al., 2013), suggesting that one or more additional transcription factors act downstream or in parallel to A-MYB to regulate meiotic gene transcription.
Meiotic distal super enhancers often contain the DNA-binding motif of the testis-specific transcription factor TCFL5 (Maezawa et al., 2020), and Tcfl5 homozygous and heterozygous mutant mice are male sterile (Galán-Martínez et al., 2022; Xu et al., 2022). Here, we report that, in response to retinoic acid (RA), STRA8/MEIOSIN initiate the transcription of A-Myb and Tcfl5. TCFL5 promotes the transcription of genes required for meiosis, meiotic exit, as well as genes related to spermiogenesis. Moreover, the A-MYB/TCFL5 regulatory architecture establishes a coherent feedforward loop to ensure transcription of miR-34/449—a family of miRNAs with essential roles in spermatogenesis (Comazzetto et al., 2014)—during male meiosis I. Without TCFL5, male mouse germ cells arrest at the mid- or late-pachynema. This transcriptional architecture is evolutionarily conserved in the old-world monkey rhesus macaque, suggesting it predates the divergence of rodents and primates. Our findings identify a transcriptional circuit that regulates meiotic gene expression.
Materials and methods
Mice
Mice were maintained and used according to the guidelines of the Institutional Animal Care and Use Committee of the University of Massachusetts Medical School (A201900331). The committee specifically approved this study. C57BL/6J mice (RRID: IMSR_JAX:000664) were used as wild-type controls. A-Mybrepro9 (MGI3512907) were a gift from John Schimenti, and were maintained in a C57BL/6 background.
Tcfl5em1/em1 mutant mice was generated and their genotypes were screened as described in Yu et al. (2022). Briefly, two sgRNAs (20 ng/μl each) targeting sequences in Tcfl5 intron 1 (5′-GCA GUC UGG GUA CUA GAUA G-3′) and intron 3 (5′-AUU CAC UCA AAC AAC AAG AG-3′) and Cas9 mRNA (50 ng/μl, TriLink Biotechnologies, L-7206) were injected into the pronucleus and cytoplasm of fertilized eggs (Transgenic Animal Modeling Core, University of Massachusetts Medical School), and 15–25 blastocysts were transferred into uterus of pseudo-pregnant ICR females at day E2.5. Tcfl5+/FLAG was generated as described in Yu et al. (2022).
Isolation of mouse germ cells by FACS
Germ cell sorting was as described (Gainetdinov et al., 2018; Yu et al., 2022). Briefly, we de-capsulated the testis specimens and incubated with 1× Gey′s Balanced Salt Solution (GBSS, Sigma, G9779) containing 0.4 mg/ml collagenase type 4 (Worthington; LS004188) at 33°C for 15 min. Thereafter, we washed the seminiferous tubules twice with 1× GBSS, and incubated them with 1× GBSS containing 0.5 mg/ml trypsin and 1 μg/ml DNase I at 33°C for 15 min. Next, the seminiferous tubules were gently pipetted up and down for 3 min through a Pasteur pipette to homogenize at 4°C on. Trypsin was inactivated with fetal bovine serum (FBS; f.c. 7.5% [v/v]), and the cell suspension was passed through a pre-wetted 70 μm cell strainer. Cells were recovered by centrifugation at 300 × g at 4°C for 10 min, resuspended in 1× GBSS containing 5% (v/v) FBS, 1 μg/ml DNase I, and 5 μg/ml Hoechst 33342 (Thermo Fisher, 62249), and incubated at 33°C for 45 min rotating at 150 rpm. Finally, we added propidium iodide (0.2 μg/ml, f.c.; Thermo Fisher, P3566) directly to the cells which were further filtered through a pre-wetted 40 μm cell strainer. Cell sorting (FACS Core, University of Massachusetts Medical School) was performed as described (Bastos et al., 2005; Yu et al., 2022).
Testis histology
Wild-type and Tcfl5em1/em1 mutant testis tissue was fixed with Bouin’s solution, and paraffin embedded tissue sectioned at 5 μm thickness, and then stained with hematoxylin and eosin (H&E) solutions (Morphology Core Facility, University of Massachusetts Medical School).
Sperm motility
Cauda epidydimal sperm were incubated in warm EmbryoMax HTF media at 37°C, 5% CO2. A drop of sperm was removed from the suspension and pipetted into a sperm counting glass chamber, then assayed by CASA or video acquisition. CASA was conducted using an IVOS II instrument (Hamilton Thorne, Beverly, MA) with the following settings: 100 frames acquired at 60 Hz; minimal contrast = 50; 4-pixel minimal cell size; minimal static contrast = 5; 0%straightness (STR) threshold; 10 μm/s VAP Cutoff; prog. min VAP, 20 μm/s; 10 μm/s VSL Cutoff; 5-pixel cell size; cell intensity = 90; static head size = 0.30–2.69; static head intensity = 0.10–1.75; static elongation = 10–94; slow cells motile = yes; 0.68 magnification; LED illumination intensity = 3000; IDENT illumination intensity = 3603; 37°C. The raw data files (i.e., .dbt files for motile sperm and .dbx files for static sperm) containing tracks ≥ 45 frames were used for sperm motility analysis. CASAnova was utilized to determine fractions of progressive and hyperactivated sperm from .dbt files of motile sperm, as previously described (Goodson et al., 2011)
Immunofluorescence
Immunofluorescent (IF) staining of FACS-purified germ cells was as described (Gainetdinov et al., 2018). Briefly, freshly sorted germ cells were incubated in 25 mM sucrose solution at room temperature for 20 min and fixed with 1% (w/v) formaldehyde solution containing 0.15% (v/v) Triton X-100 at room temperature for 2 h. After washing the slides with (i) 1 × PBS containing 0.4% (v/v) Photo-Flo 200 (Kodak, 1464510), (ii) 1× PBS containing 0.1% (v/v) Triton X-100, and (iii) 1× PBS containing 0.3% (w/v) BSA, 1% (v/v) donkey serum (Sigma, D9663), and 0.05% (v/v) Triton X-100 (10 min each wash), slides were incubated with Rabbit polyclonal anti-SYCP3 (Abcam, ab15093; 1:1,000 dilution) and mouse monoclonal anti-γH2AX (Millipore, 05–636; 1:1,000 dilution) primary antibodies in 1 × PBS containing 3% (w/v) BSA, 10% (v/v) donkey serum, and 0.5% (v/v) Triton X-100 in a humidifying chamber at room temperature overnight. Next, slides were washed sequentially as (i–iii, above), then incubated with donkey anti-rabbit IgG (H+L) Alexa Flour 488 (Thermo Fisher Scientific, A-21206; 1:2,000) and anti-mouse IgG (H+L) Alexa Flour 594 (Thermo Fisher Scientific, A-21203; 1:2,000) secondary antibodies at room temperature for 1 h. The slides were washed three times with 1 × PBS containing 0.4% (v/v) Photo-Flo 200 (10 min each wash) and once with 0.4% (v/v) Photo-Flo 200 for 10 min. Immunofluorescent images were captured using Leica SP8 with 63×, 1.4 NA oil immersion objective with post-acquisition deconvolution.
For IF staining of cryopreserved testis sections, slides were fixed with 1 × PBS containing 0.1% (w/v) sodium periodate (NaIO4), 3% (w/v) l-lysine and 3% (v/v) paraformaldehyde at room temperature for 20 min. Slides were blocked with 2.5% goat serum. Next, slides were incubated with rabbit polyclonal anti-SYCP3 (Abcam, ab15093; 1:300 dilution) and mouse monoclonal anti-γH2AX (Millipore, 05–636; 1:300 dilution) antibodies at 4°C overnight. After primary antibody incubation, slides were washed twice with 1 × PBS containing 0.05% (v/v) Tween-20 and incubated with goat anti-rabbit IgG (H+L) Alexa Flour 568 (Thermo Fisher Scientific, A-11011; 1:500 dilution) and goat anti-mouse IgG (H+L) Alexa Flour 488 (Thermo Fisher Scientific, A-32723; 1:500 dilution) secondary antibodies at room temperature for 2 h. Next, nuclei were counterstained with 1 mg/ml DAPI (Thermo Fisher Scientific, 62248), the slides were sealed using gold antifade mountant solution (Fisher scientific, P36930) and covered with a #1 coverslip. Immunofluorescent images were captured using a Leica DMi8 Leica microscope equipped with a 63×, 1.4 NA oil immersion objective.
TUNEL histochemical staining
Click-iT TUNEL Colorimetric IHC Detection kit (Thermo Fisher, C10625) was used to detect DNA breaks according to the manufacturer’s protocol. In brief, Bouin solution-fixed, paraffin-embedded slides were de-paraffinized in three changes of xylene for 5 min each, gradually re-hydrated in 100% (v/v), 95% (v/v), and 70% (v/v) ethanol for 5 min each, and then washed in 1× PBS for 5 min. After pre-treating the slides with 20 μg/ml Proteinase K at room temperature for 15 min, slides were washed with water twice (2 min each). Positive control slides were treated with 0.5 U Turbo DNase (Thermo Fisher, AM2238) at room temperature for 30 min. Slides were then incubated with TdT reaction buffer (component J) containing terminal deoxynucleotidyl transferase (component L) in a humidified chamber at 37°C for 1 h. The reaction was quenched with 2x SSC for 15 min, then washed twice in PBS. Peroxidase activity was quenched in 3% (v/v) H2O2 at room temperature for 5 min. Slides were incubated with biotin azide and copper sulfate in a humidified chamber at 37°C for 30 min, then stained with peroxidase substrate at room temperature for 10 min. Nuclei were counterstained with methyl green, and the slides sealed with EcoMount (Biocare Medical, EM897L). Images were captured using Leica DMi8 microscope equipped with a 20× objective.
Western blotting
Frozen testis tissues were homogenized in a Dounce homogenizer using 20 strokes of pestle B in RIPA lysis buffer (25 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1% (v/v) NP-40, 1% sodium deoxycholate, and 0.1% (w/v) SDS) containing 1× homemade protease inhibitor cocktail (1 mM 4-(2-Aminoethyl)benzenesulfonyl fluoride hydrochloride (Sigma; A8456), 0.3 μm Aprotinin, 40 μm Betanin hydrochloride, 10 μm. E-64 (Sigma; E3132), 10 μm Leupeptin hemisulfate). Lysed tissue was then sonicated (Branson Digital Sonifier; 450 Cell Disruptor) to break nuclei. After sonication, the samples were centrifuged at 20,000 × g at 4°C for 30 min. The supernatant was transferred to a new tube, and protein concentration measured using the Pierce BCA Protein Assay Kit (ThermoFisher; 23225). Total protein (50 μg) from each sample was mixed with 1/4 volume of loading dye (106 mM Tris-HCl, pH 6.8, 141 mM Tris base, 2% SDS, 10% v/v glycerol, 0.51 mM EDTA, 0.22 mM SERVA Blue G and 0.175 mM Phenol Red) containing 0.2 M dithiothreitol and heated at 95°C for 6 min to denature proteins. Samples were resolved by electrophoresis through a 4–20% polyacrylamide gradient SDS gel (Thermo Fisher, XP04205BOX), and the gel transferred to PVDF membrane (Millipore, IPVH00010). The membrane was blocked with Blocking Buffer (Rockland Immunochemicals, MB-070) at room temperature for 1.5 h and incubated with rabbit polyclonal anti-A-MYB (Sigma, HPA008791; 1:1,000 dilution), rabbit polyclonal anti-MILI (Abcam, ab36764; 1:1,000 dilution), or mouse monoclonal anti-FLAG (Sigma, F3165; 1:1,000) antibody at 4°C overnight. Next, the membrane was washed three times (30 min each) with 1× PBS-T (0.1% (v/v) Tween-20 in 1× PBS) at room temperature; incubated with secondary donkey anti-rabbit IRDye 680RD antibody (LI-COR, 926-68073; 1:15,000 dilution) at room temperature for 30 min; washed three times (30 min each) with 1× PBS-T at room temperature, and signal detected using the Odyssey Infrared Imaging System (LI-COR). As a loading control, membrane was incubated with mouse anti-ACTIN antibody (Santa Cruz Biotechnology, sc-47778; 1:10,000 dilution) at room temperature for 2 h, followed by goat anti-mouse IRDye 800CW secondary antibody (LI-COR, 926-32210; 1:15,000 dilution) as described above.
Global-run-on sequencing
Nuclei isolation
One and a half million FACS-purified primary spermatocytes were pelleted by centrifuging at 400g for 10 min at 4°C and gently resuspended in 1 ml swelling buffer (10 mM Tris-HCl pH 7.5 (w/v), 2 mM MgCl2 (w/v), 3 mM CaCl2 (w/v)). We added additional 9 ml swelling buffer dropwise to the resuspended cells and swirled gently to mix further and incubated on ice for five minutes. Cells were subsequently centrifuged at 400g for 10 min at 4°C, the supernatant was removed, and the cell pellet was gently resuspended in 500 μl prechilled lysis buffer 1 (9 mM Tris-HCl pH 7.5 (w/v), 1.8 mM MgCl2 (w/v), 2.7 mM CaCl2 (w/v), 10% Glycerol (v/v), 1x protease inhibitor cocktail (PIC), 0.4 U/μl RNAsIn Promega N2615). 500 μl prechilled lysis buffer 2 (9 mM Tris-HCl pH 7.5 (w/v), 1.8 mM MgCl2 (w/v), 2.7 mM CaCl2 (w/v), 10% Glycerol (v/v), 10% Igepal CA-360 (v/v), 1x PIC, 0.4 U/μl RNAsIn Promega N2615) were added dropwise and the mix was incubated for 5 min on ice. 9 ml prechilled lysis buffer 3 (9 mM Tris-HCl pH 7.5 (w/v), 1.8 mM MgCl2 (w/v), 2.7 mM CaCl2 (w/v), 10% Glycerol (v/v), 0.005% Igepal CA-360 (v/v), 1x PIC, 0.4 U/μl RNAsIn Promega N2615) was added, dropwise, to the lysate and the tube was gently swirled to mix. The mixed lysate was centrifuged at 600g for 5 min at 4°C and the supernatant was removed. The nuclei-containing pellet was resuspended in 1 ml prechilled Lysis Buffer 3 and additional 9 ml prechilled lysis Buffer 3 were added to the mixture dropwise. The nuclei were centrifuged at 600g for 5 min at 4°C, the supernatant was removed and the pellet was resuspended in 1 ml of freezing buffer (50 mM Tris-HCl pH 8.0 (w/v), 5mM MgCl2 (w/v), 0.1 mM EDTA pH 8.0 (w/v), 40% Glycerol (v/v), 1x PIC, 0.4 U/μl RNAsIn Promega N2615) and transferred to a fresh 1.7 ml Eppendorf tube. The isolated nuclei were centrifuged at 900g for 6 min at 4°C, the supernatant was removed and the pellet was resuspended in 100 μl freezing buffer. Isolated nuclei were flash frozen in liquid nitrogen and stored at −80°C until the Run-On was performed.
Nuclear Run-On
100 μl previously frozen isolated nuclei were thawed for 5 min on ice, mixed with 100 μl 2x Nuclear Run-On (NRO) reaction buffer (10 mM Tris-HCl pH 8.0, 5 mM MgCl2, 1 mM DTT, 300 mM KCl, 0.5 mM ATP, 0.5 mM GTP, 0.5 mM CTP, 0.5 mM Br-UTP, 1% Lauroylsarcosine sodium salt solution (Sigma; L7414), 1U/μl RNAsIn (Promega; N2615), 1x PIC) by pipetting gently and the mixture was immediately incubated at 30°C for 30 min. 24 μl 10x TURBO DNase buffer and 10 μl TURBO DNase (Thermo Fisher Scientific; AM2238) were mixed with the NRO reaction by pipetting and the mixture was incubated at 37°C for 20 min. RNA was extracted with 1 ml Trizol reagent (Life Technologies; 15596026), following the manufacturer’s instructions, precipitated with isopropanol and washed with 80% Ethanol before resuspending in 30 μl water. RNA was quantified using the Qubit RNA HS assay kit (Thermo Fisher Scientific; Q32855).
Br-UTP Immunoprecipitation.
50 μl Dynabeads MyOne Streptavidin T1 beads were washed by incubating with 1 ml IP buffer (150 mM NaCl, 50 mM Tris-HCl pH 8.0, 0.05% Tween-20, 1 mM EDTA, 1 mM DTT, 1x PIC, 1U/μl RNAsIn Promega N2615) at room temperature for 5 min using end-over-end tumbler. The wash solution was removed, and the beads were blocked by incubating with 300 μl blocking buffer (150 mM NaCl, 50 mM Tris-HCl pH 8.0, 0.05% Tween-20, 1 mM EDTA, 1 mM DTT, 0.1% Polyvinylpyrrolidone) for 1 h using end-over-end tumbler. Subsequently the blocking solution was removed, and the beads were kept at 4°C until required for purification.
30 μl RNA previously precipitated from the NRO reaction was incubated at 65°C for 5 min and immediately placed on ice for 3 min. Additional RNAsIn was added to 250 μl of IP buffer (final concentration 1U/μl) containing 0.2ug/μl of anti-BrdU biotin-conjugated antibody (Sigma; MAB3262B). The RNA was coupled to the antibody by rotating in the solution for 1 h at 4°C and subsequently added to the previously blocked beads. Coupling of blocked beads with RNA-antibody complex was performed by incubating at 4°C for 30 min using end-over-end tumbler. The antibody and RNA-coupled beads were washed with 500 μl IP buffer 5 times for 5 min each. Following the removal of last wash, RNA was extracted using Trizol reagent (Life Technologies; 15596026) precipitated in 10 μl water. RNA library preparation was performed as in “Long RNA library construction” below.
Long RNA library construction and analysis
We extracted total RNA from frozen testis samples and from sorted germ cells using mirVana miRNA isolation kit (Thermo Fisher, AM1560). To quantify absolute molecule of transcripts, we added 1 μl of 1:100 dilution of ERCC spike-in mix 1 (Thermo Fisher, 4456740, LOT00418382) to 0.5–1μg total RNA, before RNA library construction from FACS-purified germ cells. Library construction was performed as described (Yu et al., 2022; Özata et al., 2020) and sequenced as 79 + 79 nt paired-end reads using a NextSeq500 (Illumina).
RNA-seq analysis is performed as described in ref. Yu et al. (2022). Transcript abundance was reported as reads per million uniquely mapped reads per thousand nucleotides (RPKM) calculated using homemade Bash scripts.
Transcript abundance in molecules per cell was performed as described (Gainetdinov et al., 2018). Because each sample contained ~623,291,645 molecules of ERCC spike-in mix, the abundance of each gene = (number of mapped reads × 623291645)/(number of cells used to prepare the library × the number of reads mapping to the ERCC spike-in sequences). RNA sequencing statistics are provided in Table S1. All analyses used Ensembl-v86 gene annotations.
Chromatin immunoprecipitation and sequencing
Chromatin Immunoprecipitation (ChIP) was performed as described (Li et al., 2013; Yu et al., 2022; Özata et al., 2020). Briefly, we minced the frozen testis tissue on dry ice and fixed them in 1.7 ml tubes in ice-cold PBS containing 2% (w/v) formaldehyde at room temperature for 30 min using tumbling end-over-end. Fixed tissue was crushed in ChIP lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl, pH 8.1) with a Dounce homogenizer using 40 strokes pestle B (Kimble-Chase, Vineland, USA). We sonicated the samples to shear chromatin to 150–200 bp (Covaris, E220). Sonicated lysate was then diluted 1:10 with ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8.1, 167 mM NaCl) and immunoprecipitated using 5 μg anti-TCFL5 (Sigma, HPA055223, lot # 000000907) or 5.5 μg anti-A-MYB (Sigma, HPA008791, lot # C105987) antibody. After immunoprecipitation, DNA was extracted with phenol:chloroform:isoamyl alcohol (25:24:1; pH 8), and ChIP-seq libraries generated as described (Yu et al., 2022; Özata et al., 2020). Libraries were sequenced as 79 + 79 nt paired-end reads using a NextSeq500 (Illumina).
ChIP-seq analysis using MACS 1.1.2 (ref. Zhang et al. (2008)) with parameters -keep-dup all -q 0.001 was performed as described in refs. (Yu et al., 2021; Yu et al., 2022; Özata et al., 2020). ChIP sequencing statistics are provided in Table S1.
The distance (bp) of transcription start site to the nearest TCFL5 or A-MYB peak summit for each gene was calculated. Genes that retain TCFL5 or A-MYB peak within the range of 500 bp around their transcription start site were considered as TCFL5- or A-MYB-regulated genes.
CUT&RUN Sequencing
Cleavage Under Targets and Release Using Nuclease (CUT&RUN) sequencing from FACS-purified germ cells was performed exactly as described in ref. Yu et al. (2022). CUT&RUN fragments were sequenced using 79 + 79 nt paired-end reads on a NextSeq500 (Illumina). After mapping raw reads to the mouse genome (mm10) using Bowtie 2.2.5 (ref. Langmead and Salzberg (2012)) with parameters —very-sensitive —no-unal —no-mixed —no-discordant -|10, A-MYB and TCFL5 peaks were identified using Sparse Enrichment Analysis for CUT&RUN (SEARC Meers et al. (2019)) in “norm relaxed” mode. Without antibody was used as the background control.
To be considered bound by TCFL5, a gene was required (1) to have a SEARC-identified peak within 500 bp of its transcription start site in both CUT&RUN replicates; or (2) to have a SEARC-identified peak within 500 bp of its transcription start site in one of the two CUT&RUN replicates and a MACS2-identified peak within 500 bp of its transcription start site in two of three ChIP-seq replicates. To be considered bound by A-MYB, a gene was required (1) to have a SEARC-identified peak within 500 bp of its transcription start site in both CUT&RUN replicates; or (2) to have a SEARC-identified peak within 500 bp of its transcription start site in one of the two CUT&RUN replicates and a MACS2-identified peak within 500 bp of its transcription start site in ChIP-seq. CUT&RUN sequencing statistics are provided in Table S1.
Results
TCFL5, a testis-specific transcription factor first expressed at the pachytene stage of meiosis I
To determine the expression profile of Tcfl5 mRNA, we used publicly available RNA sequencing data from various mouse tissues (Merkin et al., 2012) (Fig. 1A and 1B); to measure TCFL5 protein abundance, we used Tcfl5+/FLAG knock-in mice, in which one copy of TCFL5 protein was tagged with an amino terminal 3XFLAG sequence (“FLAG-TCFL5”). FLAG-TCFL5 was detected in testis but not in other tissues we examined (Fig. 1C; Fig. S1A).
Fig. 1. TCFL5 is specifically expressed in primary spermatocytes of testis.
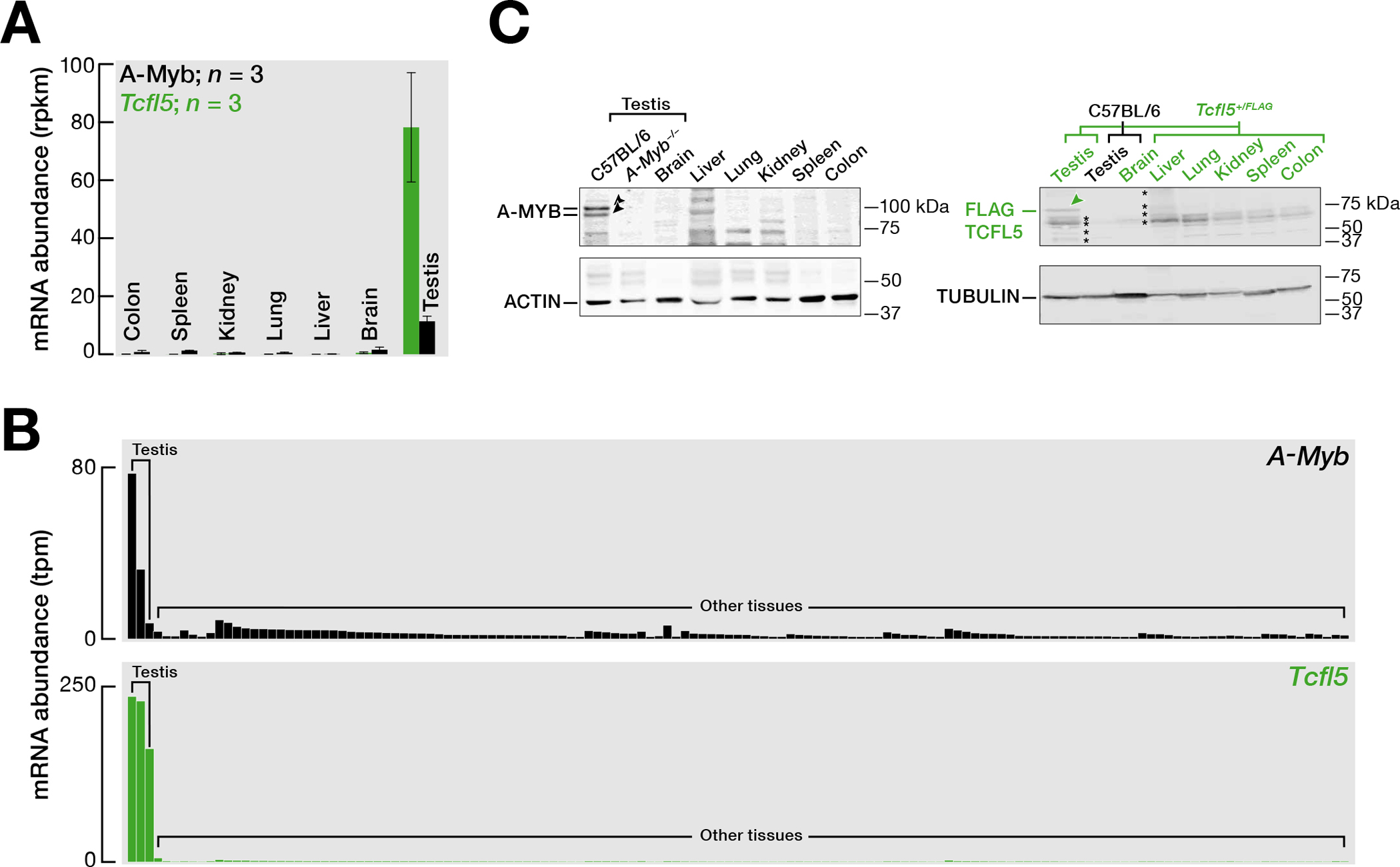
(A) Steady-state mRNA abundance of A-Myb and Tcfl5 measured by RNA sequencing of various mouse tissues (Merkin et al., 2012). The bar represents the mean mRNA abundance of A-Myb and Tcfl5 from three independent trials. Whiskers show standard error.
(B) mRNA abundance of A-Myb and Tcfl5 measured by RNA sequencing of various tissues. Data is from the mouse ENCODE project.
(C) Abundance of A-MYB and FLAG–TCFL5 proteins in various mouse tissues. FLAG-TCFL5 protein was detected in various tissues from Tcfl5+/FLAG mouse. ACTIN serves as a loading control. Each lane contained 50 μg testis protein. See Fig. S1A for uncropped western blot images. Asterisks represent background from the secondary antibody: See Fig. S1B.
Consistent with the testis-specific expression of human TCFL5 (Maruyama et al., 1998) and the primary spermatocyte-specific expression of mouse Tcfl5 (Yu et al., 2022), TCFL5 was not detected in purified spermatogonia, was abundant in primary spermatocytes (comprising pachytene and diplotene spermatocytes), and present at a lower level in secondary spermatocytes (Fig. S1C). MILI, which is present in both spermatogonia and spermatocytes, served as a control (Aravin et al., 2006; Aravin et al., 2007; Aravin et al., 2008; Gainetdinov et al., 2018; Kuramochi-Miyagawa et al., 2008; Li et al., 2013; Ozata et al., 2019; Özata et al., 2020).
STRA8/MEIOSIN initiates a coherent feedforward loop to ensure A-MYB and TCFL5 expression
A-MYB promotes its own transcription and directly initiates transcription of Tcfl5; TCFL5 responds by reinforcing its own transcription and by increasing the steady-state abundance of A-MYB via positive transcriptional feedback circuit (Li et al., 2013; Yu et al., 2022; Özata et al., 2020). Since the retinoic acid-responsive transcription factor STRA8 and its interacting partner MEIOSIN trigger the entrance of spermatogonia into meiosis I (Ishiguro et al., 2020; Kojima et al., 2019). Our re-analysis of publicly available STRA8 and MEIOSIN ChIP-seq data (Kojima et al., 2019) identified a prominent STRA8 peak within the promoter region of A-Myb and prominent MEIOSIN peaks within the promoter regions of both A-Myb and Tcfl5 (Fig. 2). Because A-Myb expression precedes that of Tcfl5 (Yu et al., 2022) and the concentration of retinoic acid drops immediately after entry into meiosis I and remains low until late meiosis I (Endo et al., 2017), we propose that in response to the short burst of retinoic acid, the STRA8/MEIOSIN complex initiates A-MYB expression, whereas TCFL5 production is delayed until A-MYB reaches a concentration threshold. Once the A-MYB concentration reaches its steady-state level, high Tcfl5 expression persists even after retinoic acid levels decline. This behavior likely reflects the organization of STRA8/MEIOSIN, A-MYB, and TCFL5 into a coherent feed feedforward loop (Alon, 2007). Finally, binding of MEISOIN to the Tcfl5 promoter suggests that MEIOSIN itself may play a STRA8-independent role in the progression of meiosis, consistent with observations that the pre-leptotene block is more severe in Meiosin−/− than in Stra8−/− mice (Ishiguro et al., 2020).
Fig. 2. STRA8/MEIOSIN axis initiates the transcription of A-Myb and Tcfl5.
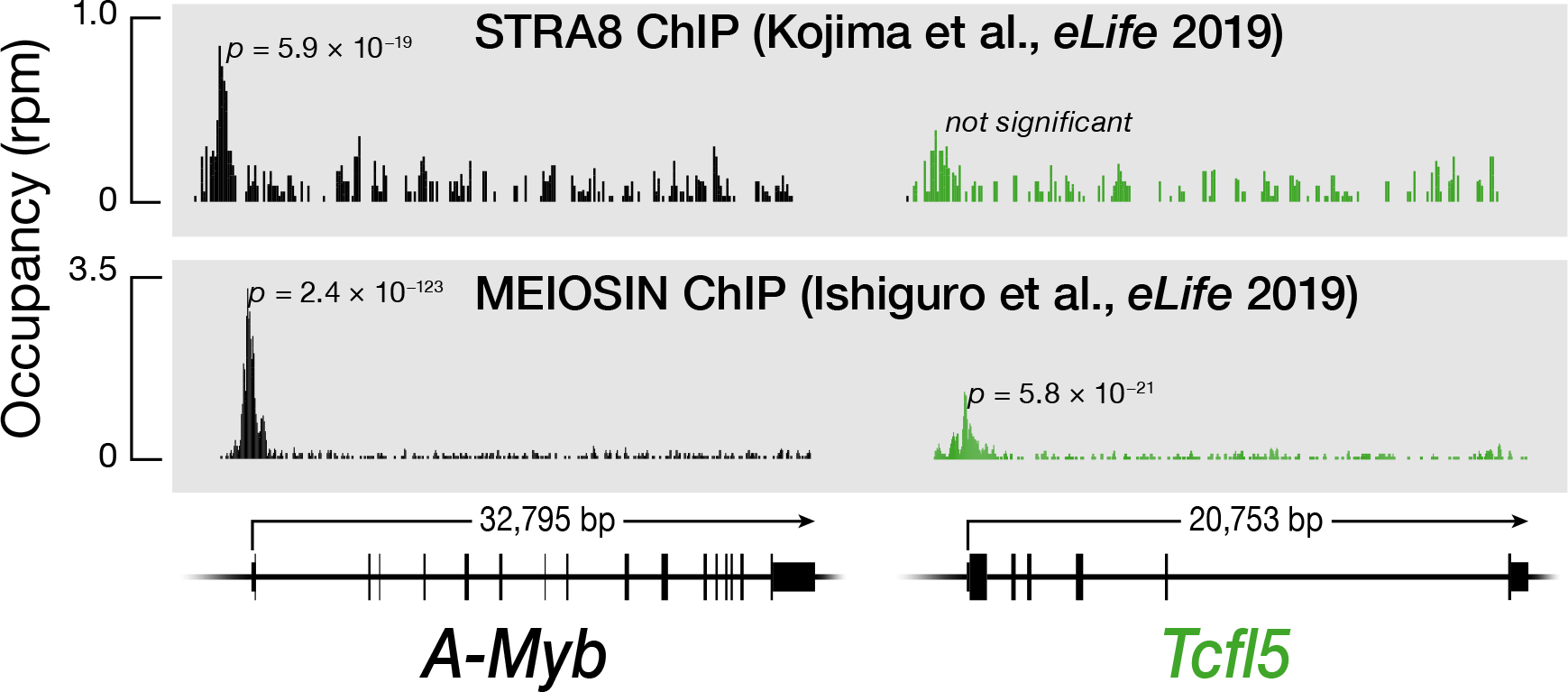
STRA8, MEIOSIN, A-MYB, and TCFL5 ChIP-seq peaks at the promoters of the A-Myb and Tcfl5 genes.
TCFL5-deficient germ cells arrest later than A-Myb mutants
Tcfl5em1/em1 mutant testes lack epididymal sperm (Fig. 3A, B). Consistent with the testis-specific expression of Tcfl5, we observed no abnormalities in Tcfl5em1/em1 females. Tcfl5em1/em1 mutant testes arrest spermatogenesis at meiosis I and contain no spermatids or spermatozoa (Fig. 3B). Strikingly, the developing pachytene spermatocytes in Tcfl5em1/em1 mutants formed abnormal multinucleated symplasts (Fig. 3B). These giant cells contain multiple nuclei within a common cytoplasm, suggesting incomplete cytokinesis. TUNEL staining detected no increase in apoptotic germ cells compared to wild-type (Fig. S2A).
Fig. 3. Tcfl5em1/em1 mutant germ cells arrest at the mid-pachytene stage of meiosis I, and Tcfl5+/em1 mutant mice fail to produce functional sperm.
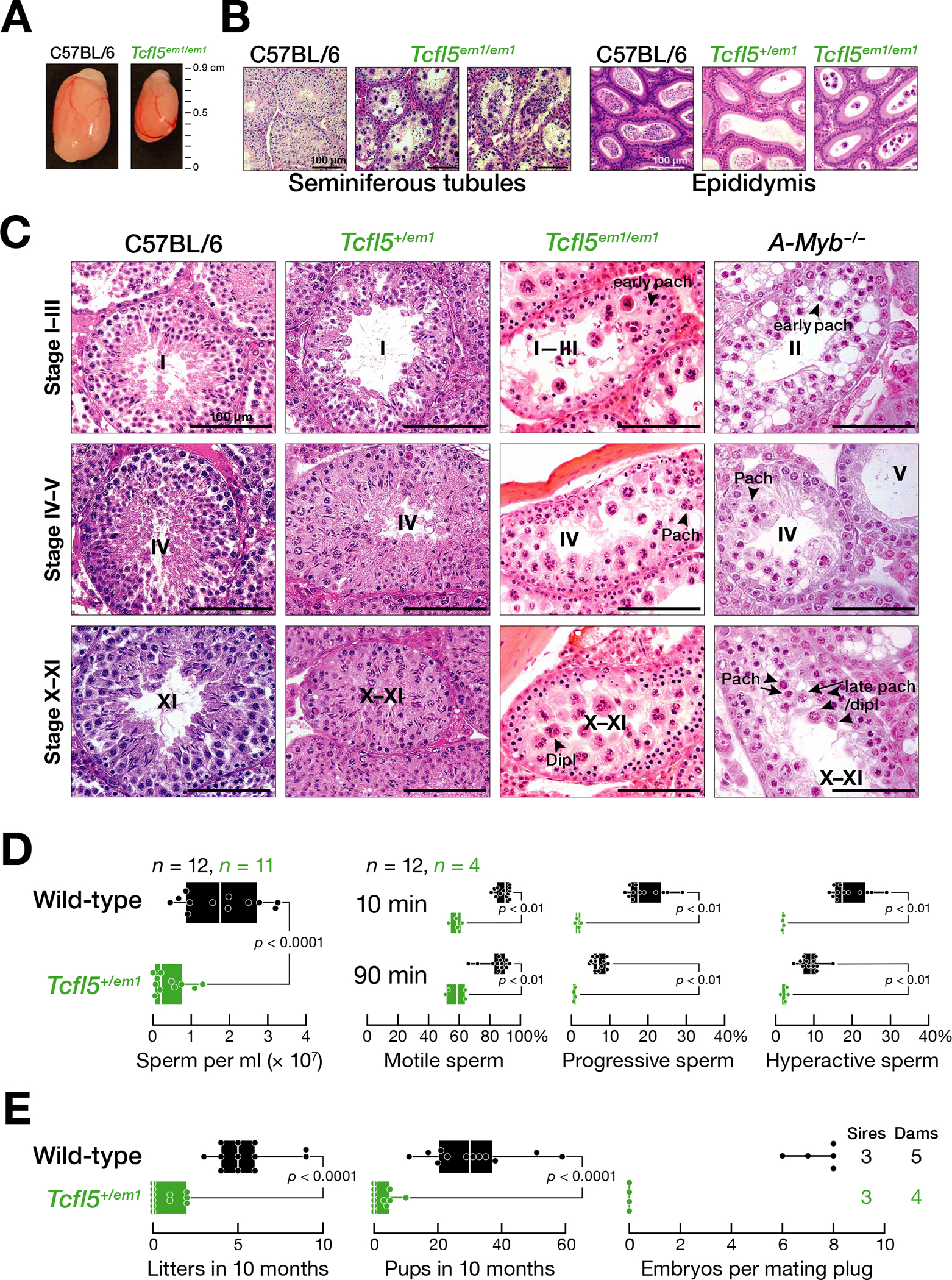
(A) Comparison of testes from four-month-old C57BL/6 and Tcfl5em1/em1 mice.
(B) Hematoxylin and eosin staining of testis and epididymis sections from C57BL/6, Tcfl5em1/em1, and Tcfl5+/em1 mice.
(C) Seminiferous tubules at different epithelial stages from C57BL/6, Tcfl5em1/em1, Tcfl5+/em1, and A-Myb−/− mice.
(D) Characterization of sperm from Tcfl5em1/em1 and C57BL/6 caudal epididymis.
(E) Viable litters and total pups sired in 10 months by two-to-eight-month-old male mice, and the number of embryos present at 14.5 days post-coitus in C57BL/6 females mated to Tcfl5+/em1 or C57BL/6 males.
In (D) and (E), vertical lines denote median, and whiskers mark the minimum and maximum values. Each dot represents an individual male. Significance was measured using a two-tailed unpaired Mann-Whitney U test.
Immunostaining to detect the synaptonemal complex protein SYCP3 showed that the chromosomes of the Tcfl5em1/em1 multinucleated pachytene spermatocytes are of normal length and appearance (Fig. S2B, C). In contrast, γH2AX, a marker for double-strand DNA breaks, was inappropriately present on the autosomal chromosomes of ~40% of pachytene spermatocytes of Tcfl5em1/em1 mutant testis (Fig. S2B–D). Similarly, 40% of A-Myb−/− pachytene spermatocytes featured cloud-like γH2AX staining over the autosomes (Fig. S2C, D), consistent with unrepaired double-stranded DNA breaks persisting in unsynapsed chromatin (Bolcun-Filas et al., 2011). Given that TCFL5 participates in a positive feedback loop that reinforces transcription of A-Myb, the presence of γH2AX staining on autosomal chromosomes in Tcfl5em1/em1 likely reflects the reduced abundance of A-MYB in the absence of TCFL5 (Yu et al., 2022).
A-Myb−/− mutant testis had a loose, defective epithelium by stage II (evidenced by spermatogonial density and size), and some pachytene spermatocytes underwent apoptosis at stages IV and X-XI (Fig. 3C). Compared to stage II tubules, stage IV and stage V tubules contained fewer early pachytene cells. By stage X–XI, few late pachytene or diplotene spermatocytes—judged by their increased size compared to early pachytene cells—were present. In contrast, the epithelium of Tcfl5em1/em1 testes was intact at stage II, and loosening of the attachment of pachytene spermatocytes to the epithelium was not observed before stage IV. The stage XI tubules in Tcfl5em1/em1 mutants contained clusters of three or four diplotene spermatocytes, some of which showed cytoplasm eosin staining, suggesting the onset of apoptosis (Fig. 3C).
TCFL5 is haploinsufficient
Heterozygous Tcfl5+/em1 testes are not normal. Tcfl5+/em1 have ~5-fold fewer caudal epididymal sperm (two-sided unpaired Mann-Whitney-Wilcoxon U test, p < 0.0001) (Fig 3B, D) and fewer motile and progressive sperm. Few sperm were hyperactivated when incubated in capacitating conditions (Fig. 3D). When mated with C57BL/6 females, adult Tcfl5+/em1 males sired fewer viable litters and fewer pups than C57BL/6 males (two-sided unpaired Mann-Whitney-Wilcoxon U test, both p < 0.0001) (Fig. 3E). Moreover, the embryos sired by Tcfl5+/em1 males often failed to develop. For example, three Tcfl5+/em1 males were each housed with two C57BL/6 females, and embryos isolated 14.5 days after the appearance of a mating plug. No embryos were detected among the C57BL/6 females paired with Tcfl5+/em1 males (n = 4 dams and 3 sires); females paired with C57BL/6 males all carried healthy embryos (7.4 ± 0.9, n = 5 dams and 3 sires) (Fig. 3E).
To better understand the underlying molecular defects in Tcfl5+/em1 heterozygotes, we measured steady-state RNA abundance in primary spermatocytes purified from adult testes (Fig. 4A). The mRNA abundance of 4,653 genes in primary spermatocytes were significantly lower in Tcfl5+/em1 compared with C57BL/6 (mutant ÷ control < 0.5 and FDR < 0.05) (Fig. 4A; Table S2A). Notably, the abundance of Tcfl5 mRNA itself was >5-fold lower in Tcfl5+/em1 primary spermatocytes than in wild-type (mean change = 0.17 ± 0.06). CUT&RUN analysis of TCFL5 DNA binding in wild-type revealed that TCFL5 bound near the transcription start site for 2,173 of 4,653 genes whose transcripts were reduced in Tcfl5+/em1 primary spermatocytes (Fig. S3A).
Fig. 4. TCFL5 regulates male meiosis I and spermiogenesis.
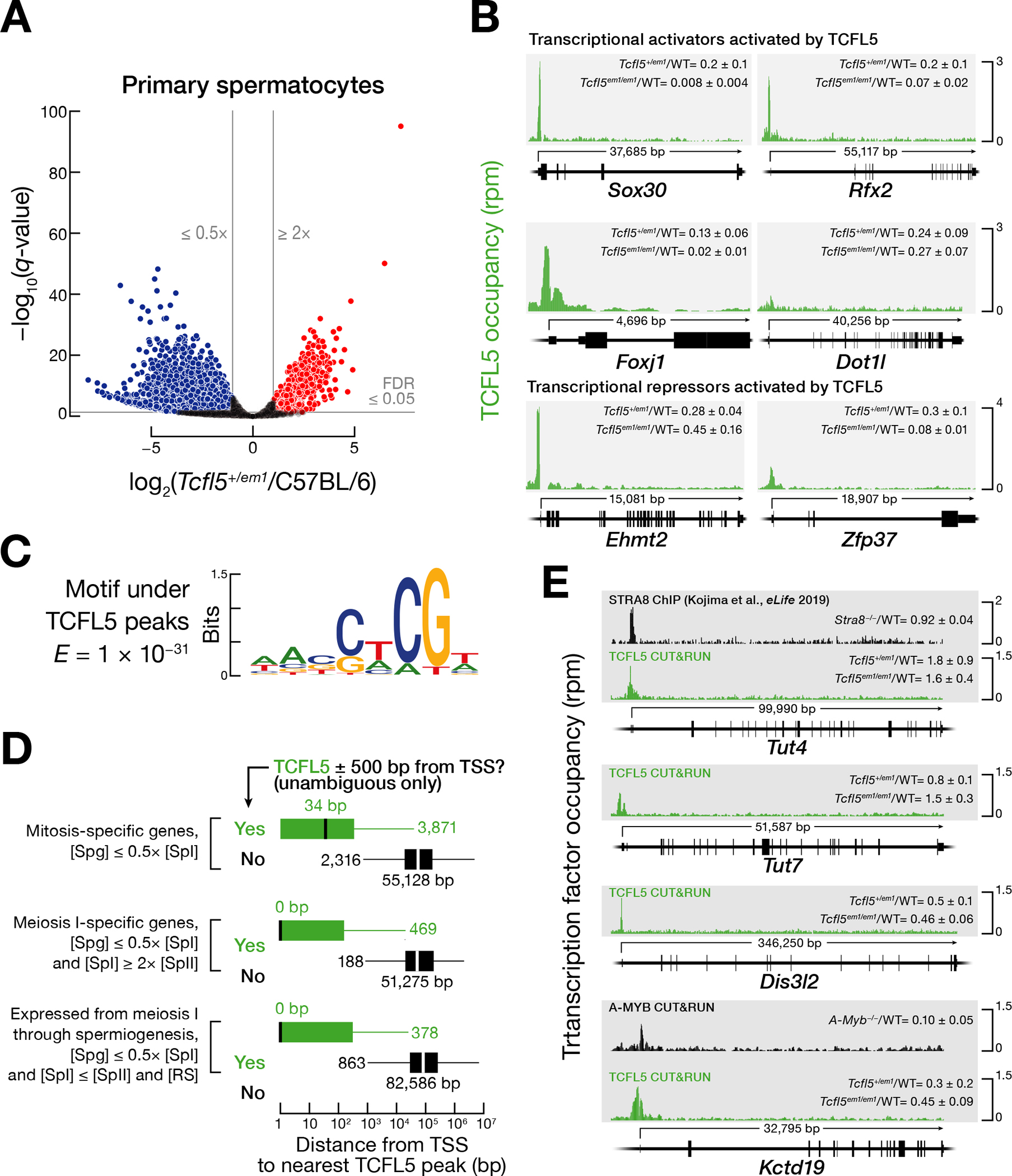
(A) The volcano plot shows the genes whose mRNA abundance increased or decreased significantly in FACS-purified primary spermatocytes from Tcfl5+/em1 heterozygotes (n = 3), compared to C57BL/6 (n = 3).
(B) TCFL5 CUT&RUN peaks at the promoters of Sox30, Rfx2, Foxj1, Dot1l, Ehmt2, and Zfp37. WT denotes C57BL/6.
(C) MEME-reported canonical TCFL5 binding motif underlying the TCFL5 CUT&RUN peaks.
(D) Distance (mean of two trials) from the nearest TCFL5 CUT&RUN peak to the transcription start site (TSS) for three categories of genes: mitosis-specific, meiosis I-specific, and genes whose expression starts at meiosis and persists through spermiogenesis. Genes were classified as not bound by TCFL5 if they had no significant TCFL5 peak ± 500 bp from the transcription start site in any of the CUT&RUN or ChIP-seq datasets. Vertical black lines: median; whiskers: maximum and minimum values, excluding outliers.
(E) STRA8 ChIP-seq and TCFL5 and A-MYB CUT&RUN peaks at the promoters of Tut4/7, Dis3l2, and Kctd19. WT denotes C57BL/6.
Tcfl5 haploinsufficiency made obtaining Tcfl5em1/em1 males daunting: Ten heterozygous breeding pairs produced only 28 pups over five years, of which only four were male homozygotes. Moreover, adult Tcfl5em1/em1 whole testes contain a higher fraction of primary spermatocytes than wild-type, so comparisons of transcript abundance between Tcfl5em1/em1 and wild-type testes underestimate decreases in TCFL5-dependent gene expression.
To further test the idea that TCFL5 is a transcriptional activator, we measured the change in the transcription rate of genes in the absence of one Tcfl5 allele by performing global-run-on sequencing of FACS-purified primary spermatocytes from Tcfl5+1/em1 heterozygous mutants and C57BL/6 controls. The nascent transcript abundance of 227 genes in primary spermatocytes were significantly lower in Tcfl5+/em1 compared with C57BL/6 (mutant ÷ control < 0.5 and FDR < 0.05) (Fig. S3B; Table S2B). In primary spermatocytes from C57BL/6 controls, the median rate of transcription of genes bound by TCFL5, A-MYB, or both was higher than for genes bound by neither protein (Wilcoxon signed-rank test p-value < 2.2 × 10−16) (Fig. S3C). CUT&RUN detected TCFL5 at the promoters of 77 of 227 genes whose nascent transcript abundance was reduced in Tcfl5+/em1 primary spermatocytes (Fig. S3D; Table S2B).
TCFL5 activates transcription of transcription factor genes
Of the 1,304 genes whose steady-state transcript abundance was reduced more than twofold in Tcfl5em1/em1 mutant mice whole testes compared to wild-type (median change = 0.2 ± 0.1) and whose promoters were bound by TCFL5, the transcript abundance of 1,058 genes was also reduced in Tcfl5+/em1 primary spermatocytes (median change = 0.26 ± 0.09). Of these genes, the promoters of only ~42% (443 of 1,058) were occupied by A-MYB, suggesting that >600 genes are transcriptionally activated by TCFL5 rather than A-MYB.
Beyond those genes directly activated by TCFL5, many others are likely regulated by transcription factors that are themselves under TCFL5 control. In fact, 73 chromatin modifying enzymes or sequence-specific, DNA-binding transcription factors whose promoters are bound by TCFL5 showed significantly reduced mRNA abundance in both Tcfl5+/em1 primary spermatocytes and Tcfl5em1/em1 mutant testes compared to wild-type, strong evidence that they require TCFL5 for their activation (Fig. S4A; Table S2A). These include Sox30, Rfx2, and Foxj1, which encode transcription factors likely to participate in spermiogenesis, as well as Dot1l, which encodes a conserved methyltransferase that methylates lysine 79 in histone H3 at a subset of enhancer elements and regulates homologous recombination during meiosis in C. elegans (Feng et al., 2002; Godfrey et al., 2019; Lascarez-Lagunas et al., 2020) (Fig. 4B). SOX30 is required for male fertility, and both SOX30 and RFX2 activate the expression of genes that participate in spermiogenesis (Feng et al., 2017; Zhang et al., 2018; Wu et al., 2016). The forkhead family transcription factor FOXJ1 activates transcription of ciliary genes (Stubbs et al., 2008; Yu et al., 2008) and regulates sperm motility in frog testes (Beckers et al., 2018). In addition to these transcriptional activators, the 73 TCFL5-regulated transcription factor genes include two repressors, EHMT2 and ZFP37 (Tachibana et al., 2007; Payen et al., 1998) (Fig. 4B). Dimethylation of lysine 9 of histone H3 by EHMT2 has been proposed to ensure that recombination occurs at the correct genomic loci (Tachibana et al., 2007).
Mapping TCFL5 binding across the genome
We identified genes with high TCFL5 binding occupancy using Sparse Enrichment Analysis for CUT&RUN (SEARC Meers et al. (2019)) to examine CUT&RUN data from FACS-purified primary spermatocytes. SEARC identified 9,290 genes, including 7,664 protein-coding and 332 non-coding genes, with a TCFL5 peak within 500 bp of their transcription start site (Table S3). The sequence motif associated with the CUT&RUN peaks defined the TCFL5 consensus binding site as WANSWCGW (W = A or T; S = G or C; expected value by chance, E = 1 × 10−31) (Fig. 4C).
To better understand the role of TCFL5 in regulating meiotic gene expression, we classified genes according to their transcript concentration: i.e., molecules per cell corrected for cell volume (Fig. S4B; Table S4A, B). Mitosis-specific genes were defined as those whose transcript concentration in spermatogonia was more than twice that in primary spermatocytes (8,209 genes). Meiosis I-specific genes were required to have a transcript concentration in primary spermatocytes more than twice that in either spermatogonia or secondary spermatocytes (792 genes). mRNAs required during spermiogenesis are mostly transcribed during meiosis (Grunewald et al., 2005; Kierszenbaum and Tres, 1975; Ostermeier et al., 2002). A third category comprised genes turned on at meiosis I and whose mRNAs persisted through spermiogenesis; these were defined as having a transcript concentration in primary spermatocytes that was more than twice that in spermatogonia but was essentially unchanged in secondary spermatocytes and round spermatids (1,466 genes).
The promoters of ~47% of mitosis-specific genes were bound by TCFL5 (3,871 of 8,209 genes; median distance from transcription start site to the nearest TCFL5 peak = 34 bp), suggesting TCFL5 sustains their expression after the onset of meiosis I (Fig. 4D; Table S3). Approximately 59% of the promoters of the meiosis I-specific genes were bound by TCFL5 (469 of 792 genes; median distance from transcription start site to the nearest TCFL5 peak = 0 bp); these genes are likely turned on or up by TCFL5 early in meiosis I. Among the 1,466 genes expressed at meiosis I whose expression persisted through spermiogenesis, TCFL5 bound to the promoters of ~26% (378 of 1,466 genes; median distance from transcription start site to the nearest TCFL5 peak = 0 bp) (Fig. 4D). ChIP-seq of TCFL5 using whole adult testis corroborated the CUT&RUN data from sorted cells (Fig. S4C; Table S3).
Notably, TCFL5 binds the promoters of Tut4 (Zcchc11), Tut7 (Zcchc6), and Dis3l2 (Fig. 4E). TUT4 and TUT7 are terminal nucleotide transferases that uridylate zygotene-expressed mRNAs whose 3′ untranslated regions contain AU-rich elements (AUUUA; AREs), marking them for destruction by the 3′-to-5′ exoribonuclease DIS3L2 as cells transit from the zygotene to the pachytene stage (Morgan et al., 2019). The mRNA abundance of Tut7 and Dis3l2 increased >5-fold in primary spermatocytes, compared to spermatogonia, whereas Tut4 increased only modestly (~1.6-fold). The expression pattern of Tut7 and Dis3l2 suggests that these genes are activated by TCFL5 bound to their promoters. Consistent with this idea, Dis3l2 mRNA decreased twofold in Tcfl5em1/em1, relative to C57BL/6 (mean decrease = 0.46 ± 0.06 in whole testes and 0.5 ± 0.1 primary spermatocytes) (Fig. 4E). Unexpectedly, Tut4 mRNA abundance increased ~50% in both Tcfl5em1/em1 whole testes and Tcfl5+/em1 primary spermatocytes (mean increase = 1.6 ± 0.4 in whole testes and 1.8 ± 0.9 in primary spermatocytes). Although we cannot exclude the possibility that TCFL5 represses expression of Tut4 but activates Dis3l2, we propose an alternative explanation. TCFL5 may activate the expression of all three, but the expected decrease of Tut4 and Tut7 mRNA in Tcfl5 mutants is obscured by an increase in their mRNA stability because Tut4 and Tut7 mRNAs are themselves ARE-dependent targets of TUT4/TUT7-catalyzed uridylation and DIS3L2 degradation. In this view, the decrease in Dis3l2 expression in Tcfl5 mutants causes Tut4 and Tut7 mRNAs to become more stable. Supporting this view, the Tut4 3′ UTR contains three canonical AREs, and the 1030-nt long Tut7 3′ UTR both contains one canonical ARE and is longer than typical for testis (median = 639 nt), a general feature of TUT4/7 targets (median = 1,703 nt) (Morgan et al., 2019).
Finally, the set of promoters bound by TCFL5 includes Kctd19 (Fig. 4E), a gene whose protein product is essential for meiotic exit (Horisawa-Takada et al., 2021; Oura et al., 2021). The Kctd19 promoter is also bound by A-MYB. The abundance of Kctd19 mRNA increases >200-fold when spermatogonia differentiate into spermatocytes (Table S4A). Kctd19 mRNA abundance declined >3-fold in Tcfl5+1/em1 and 10-fold in A-Myb−/− primary spermatocytes.
A-MYB/TCFL5 regulatory architecture establishes a coherent feedforward loop to amplify miR-34/449 production
The miR-34/449 miRNA family comprises two highly conserved clusters, miR-34a/b/c and miR-449a/b/c, whose miRNAs share a single seed sequence, the miRNA region that determines target specificity. The miR-34/449 family is essential for spermatogenesis and for ciliogenesis of efferent ductules epithelium (Bao et al., 2012; Comazzetto et al., 2014; Marcet et al., 2011; Song et al., 2014; Wu et al., 2014). While miR-34a (chromosome 4) is detectable at low levels in spermatogonia and through later stages of spermatogenesis(Bao et al., 2012), the expression of miR-34b/c and miR-449a/b/c increases >10-fold at the onset of meiosis I (Comazzetto et al., 2014). Our data identified prominent TCFL5 peaks on the promoters of the miR-34b and miR-34c genes; A-MYB bound to the promoter of the miR-34b locus only (Fig. 5A). The miR-449a/b/c cluster resides in the second intron of Cdc20b gene, and both A-MYB and TCFL5 bound to the promoter of Cdc20b (Fig. 5A). By contrast, we detect no A-MYB nor TCFL5 binding around the promoter of miR-34a. Finally, we detected TCFL5 peaks near the transcription start sites of the promoters of five essential miRNA biogenesis genes: Drosha, Dgcr8, Dicer, Xpo5, and Ago2, significantly more than expected by chance by comparison to protein-coding genes generally (Chi-square test p = < 2.0 × 10−3) (Fig. 5B). A-MYB bound to the promoters of Xpo5 and Tarbp2, a Dicer partner protein (Fig. 5C). In summary, the A-MYB/TCFL5 regulatory architecture activates the transcription of miRNA genes essential for spermatogenesis and genes encoding miRNA biogenesis proteins, thereby generating a feedforward loop predicted to ensure accumulation of miR-34/miR-449 family miRNAs during meiosis I.
Fig. 5. A-MYB/TCFL5 regulatory architecture establishes coherent feedforward loop to burst the expression of miR34/miR449 family, and a model for the transcriptional architecture of mouse male meiotic cells.
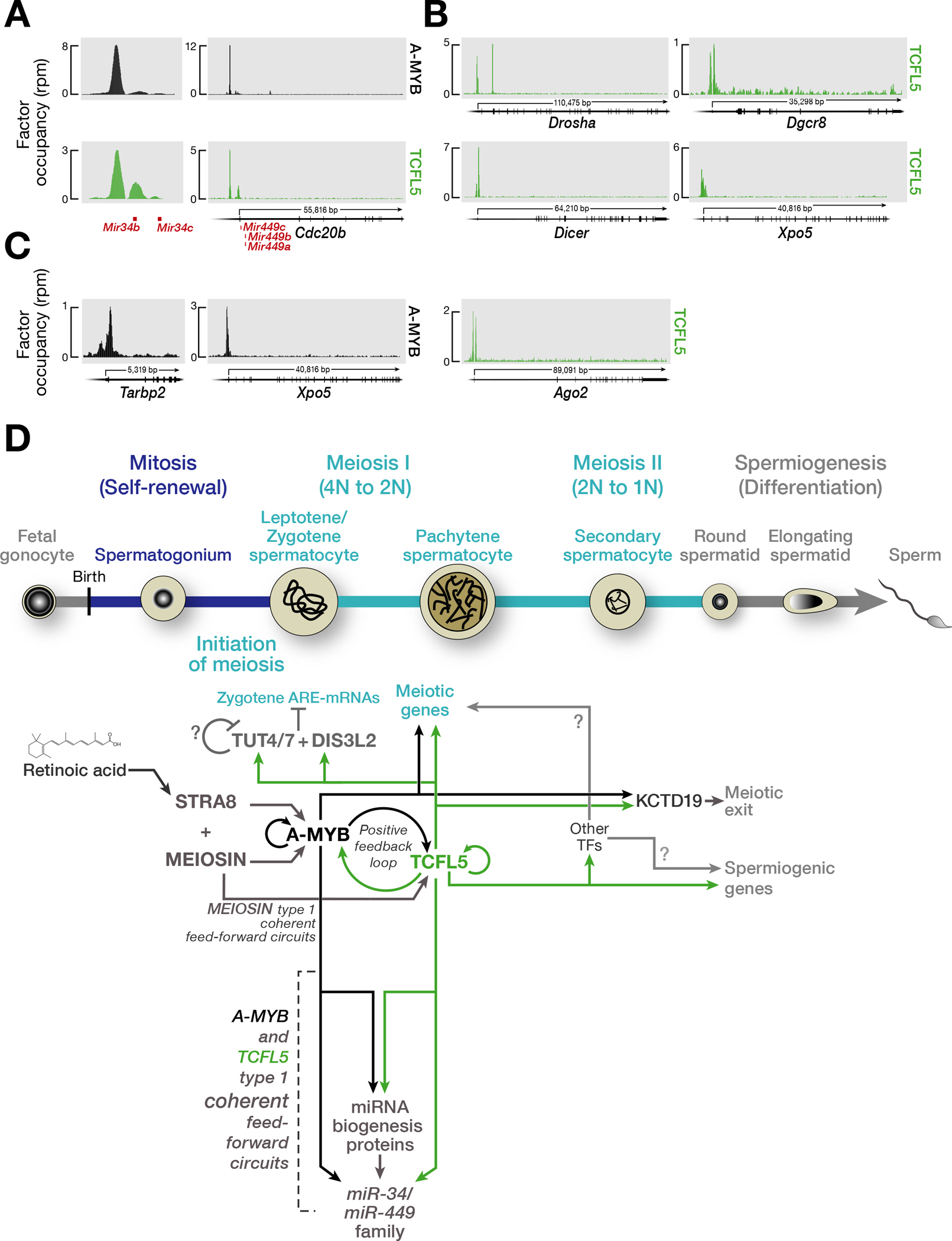
(A) A-MYB and TCFL5 CUT&RUN peaks at the promoters of miR-34b/c and miR-449a/b/c genes.
(B,C) A-MYB and TCFL5 occupancies around the promoters of genes encoding miRNA maturation proteins: TCFL5 occupancy (B) A-MYB occupancy (C).
(D) The model incorporates hypotheses from refs. (Bolcun-Filas et al., 2011; Galán-Martínez et al., 2022; Horisawa-Takada et al., 2021; Ishiguro et al., 2020; Kojima et al., 2019; Oura et al., 2021; Xu et al., 2022; Yu et al., 2022) and this study. The developmental progression of spermatogenesis is aligned with the time of expression of various proteins. The figure highlights the proposed sequential roles of STRA8, MEIOSIN, A-MYB, and TCFL5, calling attention to the underlying architecture of the transcriptional circuits regulating male meiosis in mouse.
The regulatory architecture that drives miR-34/449 production in rodents is conserved in rhesus macaque
We recently demonstrated that TCFL5 orthologs are present in the genomes of amniotes (Yu et al., 2022). We then tested whether TCFL5 and A-MYB similarly organize a coherent feedforward loop that ensures the robust production of MIR34/449 in rhesus macaque. A-MYB and TCFL5 ChIP-seq identified significant peaks at the promoters of MIR34B/C and CDC20B, whose second intron hosts MIR449A/B (Fig. S5A). Furthermore, the promoters of five of the six key miRNA biogenesis genes featured a TCFL5 or an A-MYB peak near their transcription start sites (DROSHA, DGCR8, DICER, TARBP2, and XPO5; Chi-square test p = < 2.0 × 10−3) (Fig. S5B). Together, these data suggest that the regulation miR-34/449 miRNA family by A-MYB/TCFL5 transcriptional architecture was found in the last common ancestor of Glires and primates.
DISCUSSION
Our data, together with those from previous studies (Bolcun-Filas et al., 2011; Galán-Martínez et al., 2022; Horisawa-Takada et al., 2021; Ishiguro et al., 2020; Kojima et al., 2019; Oura et al., 2021; Xu et al., 2022; Yu et al., 2022), suggest a model for the evolutionarily conserved transcriptional architecture by which A-MYB and TCFL5 collaborate to reprogram gene expression at the onset of male meiosis I (Fig. 5D). Immediately before meiosis begins, a burst of retinoic acid induces transcription of Stra8 and Meiosin, whose protein products play essential roles in the initiation of meiosis I. STRA8/MEIOSIN, in turn, activates A-Myb transcription during the leptotene or zygotene phases of meiotic prophase I, a period characterized by general transcriptional quiescence. By the onset of the pachynema, A-MYB initiates transcription of Tcfl5, and A-MYB and TCFL5 mutually reinforce their own transcription via interlocking positive feedback loops (Yu et al., 2022). Together A-MYB and TCFL5 directly promote transcription of hundreds of genes encoding proteins required for meiosis and spermiogenesis. These include components of the miRNA biogenesis machinery and the miR-34/449 miRNA family, promoting the accumulation of miR-34/449 family miRNAs.
Although STRA8 is expressed in early and intermediate spermatogonia, it does not suffice to initiate meiosis in these cells (Endo et al., 2015; Zhou et al., 2008). STRA8 requires its partner protein MEIOSIN to drive the transcription of genes required for meiotic initiation and progression—including A-Myb (Ishiguro et al., 2020) (Fig. 2). Our re-analysis of publicly available STRA8 (Kojima et al., 2019) and MEIOSIN ChIP-seq data (Ishiguro et al., 2020) revealed that only MEIOSIN binds the Tclfl5 promoter, suggesting that MEIOSIN can both collaborate with STRA8 and function independently.
What mechanisms underlie the reduced fertility phenotype observed in the absence of a single allele of Tcfl5? TCFL5 is a basic Helix-Loop-Helix (bHLH) transcription factor (Siep et al., 2004). bHLH transcription factors often regulate gene expression in a tissue-specific manner (Bhattacharya and Baker, 2011) and require interacting partners to function (Bhattacharya and Baker, 2011; Shively et al., 2019). Could then TCFL5 require a co-factor? Does reduced intracellular concentration of TCFL5 compromise the function of its co-factor in Tcfl5+/em1 mutant mice? However, the tissue-specific activity of bHLH transcription factors can be conferred by the aid of broadly expressed co-factors, whose expression threshold can be the rate limiting factor for activity of bHLH transcription factors. Along the parallel lines, expression of possible co-factor might be directly regulated by TCFL5. Halving the intracellular concentration of TCFL5 in Tcfl5+/em1 mutant mice may therefore result in amplified negative effect on TCFL5 function itself.
Supplementary Material
Statistical analyses.
All statistics were calculated using R console (https://www.rstudio.com); graphs were generated using R console or Prism 8.4.3 (GraphPad Software, LLC.). For box plots, boxes represent the first and third quartiles, except in Figure 3, where whiskers show the maximum and minimum values excluding outliers. Outliers are defined as data points with values > third quartile + 1.5 × IQR or lower than the first quartile − 1.5 × IQR, where IQR (interquartile range) is the maximum third quartile minus the first quartile minimum. Two-sided Mann-Whitney-Wilcoxon U test was used to calculate p-values. q-values in Figure 4A correspond to Benjamini-Hochberg FDR-corrected p-values
Acknowledgements
We thank S. Pechold, T. Giehl, B. Gosselin, Y. Gu and T. Krumpoch at UMMS FACS Core for their expert mouse germ cell sorting; K. Orwig for providing rhesus macaque testis specimens; C. Baer for her expert help with Leica SP8 Lightning confocal microscopy; Tipping and C. Andersson for technical assistance; and members of the Zamore and Ozata laboratories for discussions and critical comments on the manuscript.
Funding
This work was supported in part by NIGMS grants R37 GM062862 and R35 GM136275 (P.D.Z.), NICHHD grant P01 HD078253 (Z.W. and P.D.Z.), and Swedish Research Council grant 2020-03818 (D.M.O), Carltryggersstiftelse CTS 21:1158 (D.M.O). The UMMS FACS Core is supported in part by NIH grant S10OD028576.
Footnotes
Declaration of interest
The authors declare no competing interests.
Accession numbers
Sequencing data are available from the National Center for Biotechnology Information Sequence Read Archive using Gene Expression Omnibus accession number GSE166104.
References
- Alon U (2007). Network motifs: theory and experimental approaches. Nat Rev Genet 8, 450–461. [DOI] [PubMed] [Google Scholar]
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, Chien M, Russo JJ, Ju J, Sheridan R, Sander C, Zavolan M, and Tuschl T (2006). A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442, 203–207. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth KF, Bestor T, and Hannon GJ (2008). A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 31, 785–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, and Hannon GJ (2007). Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316, 744–747. [DOI] [PubMed] [Google Scholar]
- Bao J, Li D, Wang L, Wu J, Hu Y, Wang Z, Chen Y, Cao X, Jiang C, Yan W, and Xu C (2012). MicroRNA-449 and microRNA-34b/c function redundantly in murine testes by targeting E2F transcription factor-retinoblastoma protein (E2F-pRb) pathway. J Biol Chem 287, 21686–21698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos H, Lassalle B, Chicheportiche A, Riou L, Testart J, Allemand I, and Fouchet P (2005). Flow cytometric characterization of viable meiotic and postmeiotic cells by Hoechst 33342 in mouse spermatogenesis. Cytometry A 65, 40–49. [DOI] [PubMed] [Google Scholar]
- Beckers A, Ott T, Schuster-Gossler K, Boldt K, Alten L, Ueffing M, Blum M, and Gossler A (2018). The evolutionary conserved FOXJ1 target gene Fam183b is essential for motile cilia in Xenopus but dispensable for ciliary function in mice. Sci Rep 8, 14678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, and Baker NE (2011). A network of broadly expressed HLH genes regulates tissue-specific cell fates. Cell 147, 881–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolcun-Filas E, Bannister LA, Barash A, Schimenti KJ, Hartford SA, Eppig JJ, Handel MA, Shen L, and Schimenti JC (2011). A-MYB (MYBL1) transcription factor is a master regulator of male meiosis. Development 138, 3319–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comazzetto S, Di Giacomo M, Rasmussen KD, Much C, Azzi C, Perlas E, Morgan M, and O’Carroll D (2014). Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 Loci. PLoS Genet 10, e1004597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T, Freinkman E, de Rooij DG, and Page DC (2017). Periodic production of retinoic acid by meiotic and somatic cells coordinates four transitions in mouse spermatogenesis. Proc Natl Acad Sci U S A 114, E10132–E10141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T, Romer KA, Anderson EL, Baltus AE, de Rooij DG, and Page DC (2015). Periodic retinoic acid-STRA8 signaling intersects with periodic germ-cell competencies to regulate spermatogenesis. Proc Natl Acad Sci U S A 112, E2347–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng CA, Spiller C, Merriner DJ, O’Bryan MK, Bowles J, and Koopman P (2017). SOX30 is required for male fertility in mice. Sci Rep 7, 17619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, and Zhang Y (2002). Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol 12, 1052–1058. [DOI] [PubMed] [Google Scholar]
- Gainetdinov I, Colpan C, Arif A, Cecchini K, and Zamore PD (2018). A Single Mechanism of Biogenesis, Initiated and Directed by PIWI Proteins, Explains piRNA Production in Most Animals. Mol Cell 71, 775–790.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galán-Martínez J, Berenguer I, Del Carmen Maza M, Stamatakis K, Gironès N, and Fresno M (2022). TCFL5 deficiency impairs the pachytene to diplotene transition during spermatogenesis in the mouse. Sci Rep 12, 10956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey L, Crump NT, Thorne R, Lau IJ, Repapi E, Dimou D, Smith AL, Harman JR, Telenius JM, Oudelaar AM, Downes DJ, Vyas P, Hughes JR, and Milne TA (2019). DOT1L inhibition reveals a distinct subset of enhancers dependent on H3K79 methylation. Nat Commun 10, 2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodson SG, Zhang Z, Tsuruta JK, Wang W, and O’Brien DA (2011). Classification of mouse sperm motility patterns using an automated multiclass support vector machines model. Biol Reprod 84, 1207–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald S, Paasch U, Glander HJ, and Anderegg U (2005). Mature human spermatozoa do not transcribe novel RNA. Andrologia 37, 69–71. [DOI] [PubMed] [Google Scholar]
- Handel MA, and Schimenti JC (2010). Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat Rev Genet 11, 124–136. [DOI] [PubMed] [Google Scholar]
- Horisawa-Takada Y, Kodera C, Takemoto K, Sakashita A, Horisawa K, Maeda R, Shimada R, Usuki S, Fujimura S, Tani N, Matsuura K, Akiyama T, Suzuki A, Niwa H, Tachibana M, Ohba T, Katabuchi H, Namekawa SH, Araki K, and Ishiguro KI (2021). Meiosis-specific ZFP541 repressor complex promotes developmental progression of meiotic prophase towards completion during mouse spermatogenesis. Nat Commun 12, 3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro KI, Matsuura K, Tani N, Takeda N, Usuki S, Yamane M, Sugimoto M, Fujimura S, Hosokawa M, Chuma S, Ko MSH, Araki K, and Niwa H (2020). MEIOSIN Directs the Switch from Mitosis to Meiosis in Mammalian Germ Cells. Dev Cell 52, 429–445.e10. [DOI] [PubMed] [Google Scholar]
- Kierszenbaum AL, and Tres LL (1975). Structural and transcriptional features of the mouse spermatid genome. J Cell Biol 65, 258–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima ML, de Rooij DG, and Page DC (2019). Amplification of a broad transcriptional program by a common factor triggers the meiotic cell cycle in mice. eLife 8, e43738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, Hata K, Li E, Matsuda Y, Kimura T, Okabe M, Sakaki Y, Sasaki H, and Nakano T (2008). DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev 22, 908–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lascarez-Lagunas LI, Herruzo E, Grishok A, San-Segundo PA, and Colaiácovo MP (2020). DOT-1.1-dependent H3K79 methylation promotes normal meiotic progression and meiotic checkpoint function in C. elegans. PLoS Genet 16, e1009171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XZ, Roy CK, Dong X, Bolcun-Filas E, Wang J, Han BW, Xu J, Moore MJ, Schimenti JC, Weng Z, and Zamore PD (2013). An Ancient Transcription Factor Initiates the Burst of piRNA Production during Early Meiosis in Mouse Testes. Mol Cell 50, 67–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maezawa S, Sakashita A, Yukawa M, Chen X, Takahashi K, Alavattam KG, Nakata I, Weirauch MT, Barski A, and Namekawa SH (2020). Super-enhancer switching drives a burst in gene expression at the mitosis-to-meiosis transition. Nat Struct Mol Biol 27, 978–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcet B, Chevalier B, Luxardi G, Coraux C, Zaragosi LE, Cibois M, Robbe-Sermesant K, Jolly T, Cardinaud B, Moreilhon C, Giovannini-Chami L, Nawrocki-Raby B, Birembaut P, Waldmann R, Kodjabachian L, and Barbry P (2011). Control of vertebrate multiciliogenesis by miR-449 through direct repression of the Delta/Notch pathway. Nat Cell Biol 13, 693–699. [DOI] [PubMed] [Google Scholar]
- Maruyama O, Nishimori H, Katagiri T, Miki Y, Ueno A, and Nakamura Y (1998). Cloning of TCFL5 encoding a novel human basic helix-loop-helix motif protein that is specifically expressed in primary spermatocytes at the pachytene stage. Cytogenet Cell Genet 82, 41–45. [DOI] [PubMed] [Google Scholar]
- Meers MP, Tenenbaum D, and Henikoff S (2019). Peak calling by Sparse Enrichment Analysis for CUT&RUN chromatin profiling. Epigenetics Chromatin 12, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkin J, Russell C, Chen P, and Burge CB (2012). Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science 338, 1593–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan M, Kabayama Y, Much C, Ivanova I, Di Giacomo M, Auchynnikava T, Monahan JM, Vitsios DM, Vasiliauskaitė L, Comazzetto S, Rappsilber J, Allshire RC, Porse BT, Enright AJ, and O’Carroll D (2019). A programmed wave of uridylation-primed mRNA degradation is essential for meiotic progression and mammalian spermatogenesis. Cell Res 29, 221–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostermeier GC, Dix DJ, Miller D, Khatri P, and Krawetz SA (2002). Spermatozoal RNA profiles of normal fertile men. Lancet 360, 772–777. [DOI] [PubMed] [Google Scholar]
- Oura S, Koyano T, Kodera C, Horisawa-Takada Y, Matsuyama M, Ishiguro KI, and Ikawa M (2021). KCTD19 and its associated protein ZFP541 are independently essential for meiosis in male mice. PLoS Genet 17, e1009412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozata DM, Gainetdinov I, Zoch A, O’Carroll D, and Zamore PD (2019). PIWI-interacting RNAs: small RNAs with big functions. Nat Rev Genet 20, 89–108. [DOI] [PubMed] [Google Scholar]
- Özata DM, Yu T, Mou H, Gainetdinov I, Colpan C, Cecchini K, Kaymaz Y, Wu PH, Fan K, Kucukural A, Weng Z, and Zamore PD (2020). Evolutionarily conserved pachytene piRNA loci are highly divergent among modern humans. Nat Ecol Evol 4, 156–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payen E, Verkerk T, Michalovich D, Dreyer SD, Winterpacht A, Lee B, De Zeeuw CI, Grosveld F, and Galjart N (1998). The centromeric/nucleolar chromatin protein ZFP-37 may function to specify neuronal nuclear domains. J Biol Chem 273, 9099–9109. [DOI] [PubMed] [Google Scholar]
- Sakashita A, Maezawa S, Takahashi K, Alavattam KG, Yukawa M, Hu YC, Kojima S, Parrish NF, Barski A, Pavlicev M, and Namekawa SH (2020). Endogenous retroviruses drive species-specific germline transcriptomes in mammals. Nat Struct Mol Biol 27, 967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shively CA, Liu J, Chen X, Loell K, and Mitra RD (2019). Homotypic cooperativity and collective binding are determinants of bHLH specificity and function. Proc Natl Acad Sci U S A 116, 16143–16152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siep M, Sleddens-Linkels E, Mulders S, van Eenennaam H, Wassenaar E, Van Cappellen WA, Hoogerbrugge J, Grootegoed JA, and Baarends WM (2004). Basic helix-loop-helix transcription factor Tcfl5 interacts with the Calmegin gene promoter in mouse spermatogenesis. Nucleic Acids Res 32, 6425–6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song R, Walentek P, Sponer N, Klimke A, Lee JS, Dixon G, Harland R, Wan Y, Lishko P, Lize M, Kessel M, and He L (2014). miR-34/449 miRNAs are required for motile ciliogenesis by repressing cp110. Nature 510, 115–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs JL, Oishi I, Izpisúa Belmonte JC, and Kintner C (2008). The forkhead protein Foxj1 specifies node-like cilia in Xenopus and zebrafish embryos. Nat Genet 40, 1454–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Nozaki M, Takeda N, and Shinkai Y (2007). Functional dynamics of H3K9 methylation during meiotic prophase progression. EMBO J 26, 3346–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Bao J, Kim M, Yuan S, Tang C, Zheng H, Mastick GS, Xu C, and Yan W (2014). Two miRNA clusters, miR-34b/c and miR-449, are essential for normal brain development, motile ciliogenesis, and spermatogenesis. Proc Natl Acad Sci U S A 111, E2851–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Hu X, Li Z, Wang M, Li S, Wang X, Lin X, Liao S, Zhang Z, Feng X, Wang S, Cui X, Wang Y, Gao F, Hess RA, and Han C (2016). Transcription Factor RFX2 Is a Key Regulator of Mouse Spermiogenesis. Sci Rep 6, 20435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Zhang Y, Qin D, Gui Y, Wang S, Du G, Yang F, Li L, Yuan S, Wang M, and Wu X (2022). Transcription factor-like 5 is a potential DNA- and RNA-binding protein essential for maintaining male fertility in mice. J Cell Sci 135, jcs259036. [DOI] [PubMed] [Google Scholar]
- Yu T, Biasini A, Cecchini K, Saflund M, Mou H, Arif A, Eghbali A, de Rooij D, Weng Z, Zamore PD, and Ozata DM (2022). A-MYB/TCFL5 regulatory architecture ensures the production of pachytene piRNAs in placental mammals. RNA rna.079472.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Fan K, Özata DM, Zhang G, Fu Y, Theurkauf WE, Zamore PD, and Weng Z (2021). Long first exons and epigenetic marks distinguish conserved pachytene piRNA clusters from other mammalian genes. Nat Commun 12, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Ng CP, Habacher H, and Roy S (2008). Foxj1 transcription factors are master regulators of the motile ciliogenic program. Nat Genet 40, 1445–1453. [DOI] [PubMed] [Google Scholar]
- Zhang D, Xie D, Lin X, Ma L, Chen J, Zhang D, Wang Y, Duo S, Feng Y, Zheng C, Jiang B, Ning Y, and Han C (2018). The transcription factor SOX30 is a key regulator of mouse spermiogenesis. Development 145, dev164723. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, and Liu XS (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Li Y, Nie R, Friel P, Mitchell D, Evanoff RM, Pouchnik D, Banasik B, McCarrey JR, Small C, and Griswold MD (2008). Expression of stimulated by retinoic acid gene 8 (Stra8) and maturation of murine gonocytes and spermatogonia induced by retinoic acid in vitro. Biol Reprod 78, 537–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


