Summary
Metazoan tissue specification is associated with integration of macrophage lineage cells in sub-tissular niches to promote tissue development and homeostasis. Oncogenic transformation, most prevalently of epithelial cell lineages, results in maladaptation of resident tissue macrophage differentiation pathways to generate parenchymal and interstitial tumor-associated macrophages that largely foster cancer progression. In addition to growth factors, nutrients that can be consumed, stored, recycled or converted to signaling molecules, have emerged as crucial regulators of macrophage responses in tumor. Here we review how nutrient acquisition through plasma membrane transporters and engulfment pathways control tumor-associated macrophage differentiation and function. We also discuss how nutrient metabolism regulates tumor-associated macrophages, and how these processes may be targeted for cancer therapy.
eTOC Blurb
Oncogenic transformation drives maladaptation of resident tissue macrophage differentiation pathways to generate tumor-associated macrophages (TAMs) that largely promote cancer progression. Zhang, Ji, and Li review how metabolism controls TAM development and function. They discuss how nutrients acquired through plasma membrane transporters and engulfment pathways are metabolized to control TAMs, and how these processes may be targeted for cancer therapy.
Introduction
Metazoan is characterized by cell differentiation and organization as tissues. The tissue mass is composed mostly of parenchymal cells that execute tissue-specific functions, such as epithelial cells that make up all body surfaces and many internal glands to mediate filtration, absorption, excretion, secretion and barrier protective functions. The interstitial part of tissue consists of endothelial cells, fibroblasts, nerves and acellular extracellular matrix (ECM) with infrastructural functions that are largely tissue-agnostic. Cells of the hematopoietic lineage including macrophages are further recruited, with resident tissue macrophages (RTMs) adapted to sub-tissular parenchymal and interstitial niches, promoting tissue development and homeostasis aside from the classical roles of macrophages in host defense against infections1–9. Parenchymal cells, particularly those of the epithelium lineages, have a high turn-over rate, and are susceptible to cell transformation10, accounting for 80–90% human malignancies (https://training.seer.cancer.gov/disease/categories/classification.html). The macrophage compartment in the tumor tissue can as well undergo dynamic remodeling with tumor-associated macrophages (TAMs) making up to 50% of the tumor mass11–14.
Tumors are fast growing and metabolically demanding tissues and rewiring of metabolic pathways in genetically altered cancer cells has been well documented15. Acquisition of nutrients delivered systematically and generated locally by highly adaptable TAMs constitutes another major facet of the metabolic network in the tumor microenvironment16–21. Of note, in addition to transporter-mediated nutrient uptake, TAMs are highly capable of scavenging nutrients through engulfment that can be further associated with their detoxification function, befitting the professional phagocyte identity of macrophages. In this review, we will discuss TAM responses from the perspective of cancer as a tissue-level disease with a focus on how nutrient uptake and metabolism regulate TAM differentiation and function.
TAMs as maladapted RTMs
RTMs and TAMs in healthy and tumorous epithelial tissues have been extensively profiled showing cross-tissue transcriptome similarities of macrophage subsets associated with parenchymal and interstitial localizations22–26, suggesting that there are unique features related to tissue architecture to regulate the differentiation and function of parenchymal RTMs or TAMs (pRTMs, pTAMs), and interstitial RTMs or TAMs (iRTMs, iTAMs) (Figure 1A).
Figure 1. TAM subsets and metabolic crosstalk in the tumor microenvironment.
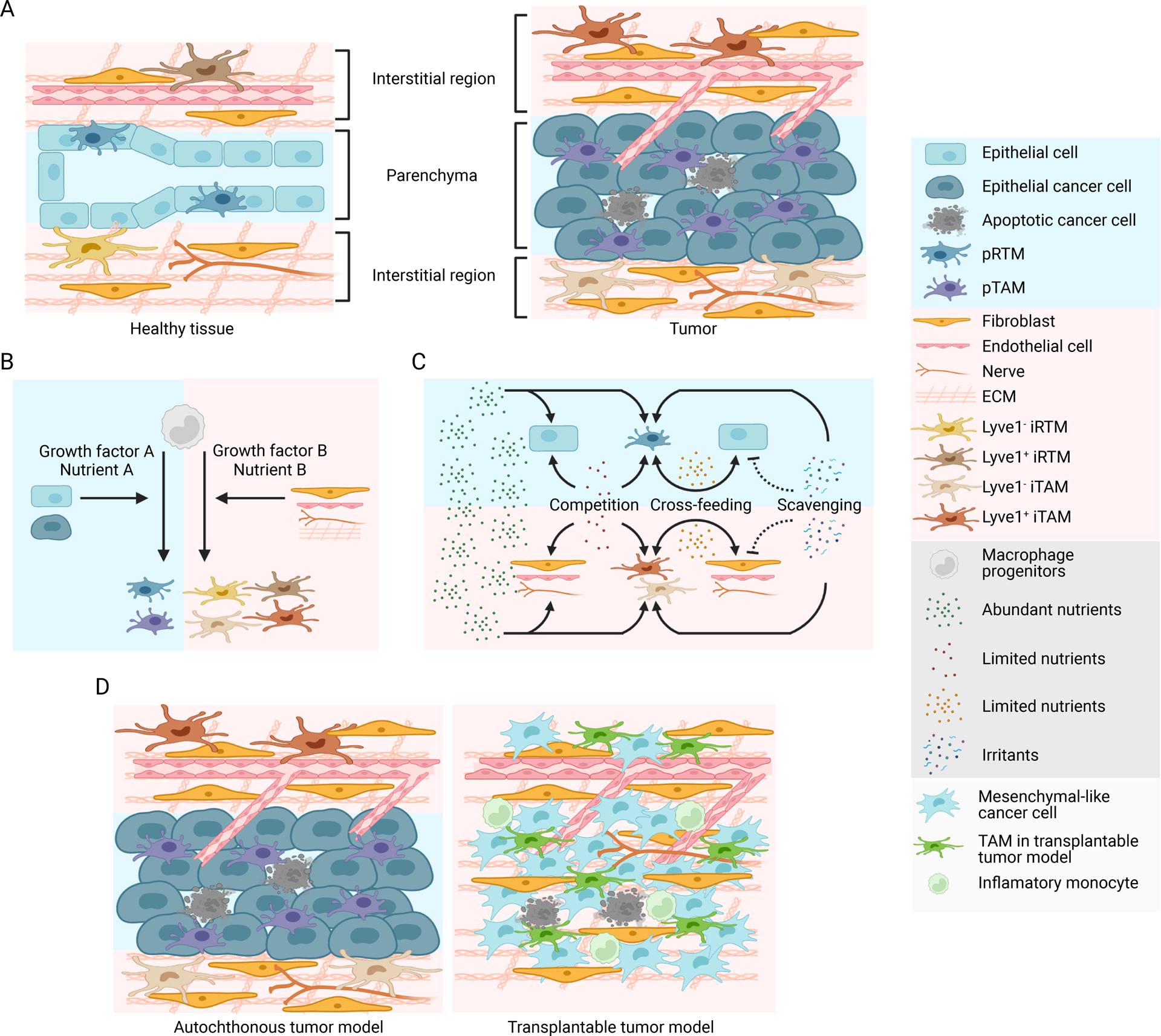
A) A simplified schematic depicting healthy and tumorous epithelial tissues with phenotypically distinct resident tissue macrophages (RTMs) and tumor-associated macrophages (TAMs) localized in sub-tissular interstitial and parenchymal niches. Epithelial cell transformation is associated with expansion and phenotypic adaptation of cancer cell-associated parenchymal TAMs (pTAMs), while interstitial TAMs (iTAMs) with or without expression of the scavenger receptor Lyve1 are also adapted in interstitial regions composed predominantly of fibroblasts, endothelial cells, nerves, and acellular extracellular matrix (ECM).
B) RTM and TAM differentiation from macrophage progenitors is driven by tissue niche factors, including growth factors and nutrients as discussed in1 for RTMs. The parenchymal and interstitial niche factors regulate differentiation of pRTMs/pTAMs and iRTMs/iTAMs, respectively.
C) Modes of metabolic crosstalk between TAMs, cancer cells and other cell types in the tumor microenvironment. Abundant nutrients are taken up by all cell types with no restriction, while limited nutrients can be cross-fed or competed between TAMs, cancer cells, and other cell types in the tumor stroma, promoting or inhibiting tumor growth, respectively. TAMs can also scavenge irritants that otherwise impair tumor tissue fitness and suppress tumor development.
D) A number of animal models have been used to study TAM responses in epithelial cancers. Autochthonous murine tumor models involve transformation of endogenous epithelial lineage cells, and preserve the parenchymal and interstitial tissue architecture with pTAMs and iTAMs differentiated in distinct sub-tissular niches. Transplantable murine tumor models involve inoculation of cancer cell lines propagated in vitro into target tissues, which fails to recapitulate the tumor tissue architecture. In addition, many cancer cell lines derived from epithelial tumors acquire mesenchymal phenotype during in vitro propagation. TAM responses in these models are often associated with acute influx of a large number of inflammatory monocytes with TAM differentiation poorly resembling that induced in human tumor.
The sub-tissular dichotomous differentiation phenotypes of RTMs and TAMs are best demonstrated in mouse mammary gland, where macrophages are dynamically regulated in response to tissue remodeling during development, reproduction cycle as well as sporadic tumor growth driven by oncogenic cell transformation. In healthy mammary gland, iRTMs numerically dominate, while pRTMs are locally enriched alongside epithelial mammary gland branches27. iRTMs are initially derived from fetal liver monocytes and are largely maintained by self-renewal throughout the postnatal development28. The interstitial region is highly enriched for ECM compared to the mammary gland ductal structure formed by layers of epithelial cells. Depletion of macrophages by administration of an inhibitor against colony stimulating factor 1 receptor (CSF1R) causes increased level of interstitial ECM proteins, including collagen and Hyaluronic acid (HA)29. The respective scavenger receptors for collagen and HA are mannose receptor C-type 1 (Mrc1) and lymphatic vessel endothelial hyaluronan receptor 1 (Lyve1)30,31, markers for iRTMs22,23. Although Mrc1 and Lyve1 expression largely overlaps, Mrc1 is more broadly expressed in iRTMs. Of note, Mrc1+Lyve1− iRTMs are enriched in interstitial regions close to the mammary gland epithelium29, where iRTMs may interact with the Notch receptor ligand delta-like 1 (Dll1) expressed by epithelial stem cells32 to suppress Lyve1 expression33. These findings imply further heterogeneity of interstitial niches governing the differentiation of iRTM subsets and suggest prominent functions of iRTMs in ECM remodeling.
Localized between luminal and basal layers of ductal epithelium, the embryonic monocyte-derived pRTMs are a rare population in virgin mammary gland27. However, during pregnancy and lactation, pRTMs are differentiated from circulating monocytes, and undergo massive expansion to accommodate the expanded epithelium27, which is likely because local proliferation of pRTMs is outpaced by the expanding parenchyma niche, causing the de novo pRTM differentiation from monocytes. Short term depletion of pRTMs results in the accumulation of apoptotic alveolar cells and enlargement of mammary gland lumen during post-lactation involution, indicating a critical scavenger function of pRTMs and its role in tissue remodeling. Together, these studies reveal that iRTMs and pRTMs are differentiated in distinct sub-tissular niches of the mammary gland to fulfill specialized functions during development and the reproductive cycle.
Before the characterization of mammary tissue iRTMs and pRTMs, the dichotomous differentiation of macrophages had been revealed in a transgenic model of mammary tumors driven by the polyoma middle T (PyMT) oncoprotein34. Unlike in healthy mammary gland of young mice, circulating monocytes contribute to not only the highly expanded vascular cell adhesion molecule 1 (Vcam1)+ TAMs but also Mrc1+ mammary tissue macrophages in PyMT mammary tumors34, which are localized in the intratumor parenchymal and peritumor interstitial regions, respectively35, and herein renamed as pTAMs and iTAMs. These findings suggest that the interstitial niche for macrophages undergoes remodeling during tumor progression, which is in line with the observation that iTAMs at various stages of tumor progression are transcriptionally deviated from iRTMs in mammary tissue22. Phenotypically similar iTAMs that express the ECM scavenge receptors MRC1 and LYVE1 as well as the folate receptor beta (FOLR2) are also present in human breast tumors22, which are distinct from pTAMs that express high levels of the lipid endocytosis receptor triggering receptor expressed on myeloid cells 2 (TREM2)22. Of note, Trem2 is highly induced in pTAMs from PyMT tumors, and is only minimally expressed in pRTMs from healthy lactating mammary glands (our unpublished observation), suggesting phenotypical adaptation of pTAMs in the tumor microenvironment. Interestingly, Trem2 is also induced in macrophages associated with neuronal and metabolic disorders36–40. As damage occurs in diseased tissues, it is conceivable that Trem2 is induced in macrophages to facilitate the clearance of tissue damage-associated lipids. Thus, pTAM differentiation is not only specified by ‘hard-wired’ signals associated with an expanding parenchymal epithelial niche similar to that of pRTM, but also regulated by ‘on-demand’ signals such as damage-associated molecules present in tumorous tissues.
Tissue-level specification of macrophage differentiation underscores the importance of using autochthonous tumor models such as transgenic cancer models to study TAM responses (Figure 1D). Yet, transplantation tumor models have been widely used in the field. Although these models could recapitulate some aspects of TAM responses including Trem2 expression41, they fall short in reproducing critical aspects of human cancer patient TAM biology. For instance, transplanted cancer cells, even through the orthotopic route, often fail to be integrated to the endogenous tissue that provides critical signals for TAM differentiation42. In addition, most of the commonly used murine cancer cell lines of epithelial origin show a mesenchymal phenotype43, and will not provide the same niche signal for pTAM differentiation. It is also important to note that human cancers are genetically heterogeneous, driving distinct immune responses in tumor44. In some cases, oncogenic events may disrupt or alter the tissue niche specifying the differentiation of TAMs as maladapted RTMs, causing TAM depletion or acquisition of a distinct differentiation pathway, and will also be better defined in autochthonous tumor models. Therefore, in order to best recapitulate human TAM biology, it is crucial to use autochthonous tumor models to preserve tissue-level regulation of macrophage responses in cancer.
Macronutrient uptake and metabolism in control of TAM responses
In addition to growth factor signals that drive tissue niche-associated TAM differentiation, nutrients delivered systemically through circulation and generated locally in the tumor tissue affect the metabolic and functional states of TAMs (Figure 1B and 1C). Nutrient control of TAM responses is affected by several factors including the tumor tissue origin, cancer cell oncogenomic profiles and stages of tumor progression, while tumor model choice is another confounding factor. In this section, we will discuss how TAM differentiation and function are regulated by macronutrients acquired through plasma membrane transporters (Figure 2).
Figure 2. Macronutrient uptake and metabolism control of TAM responses.
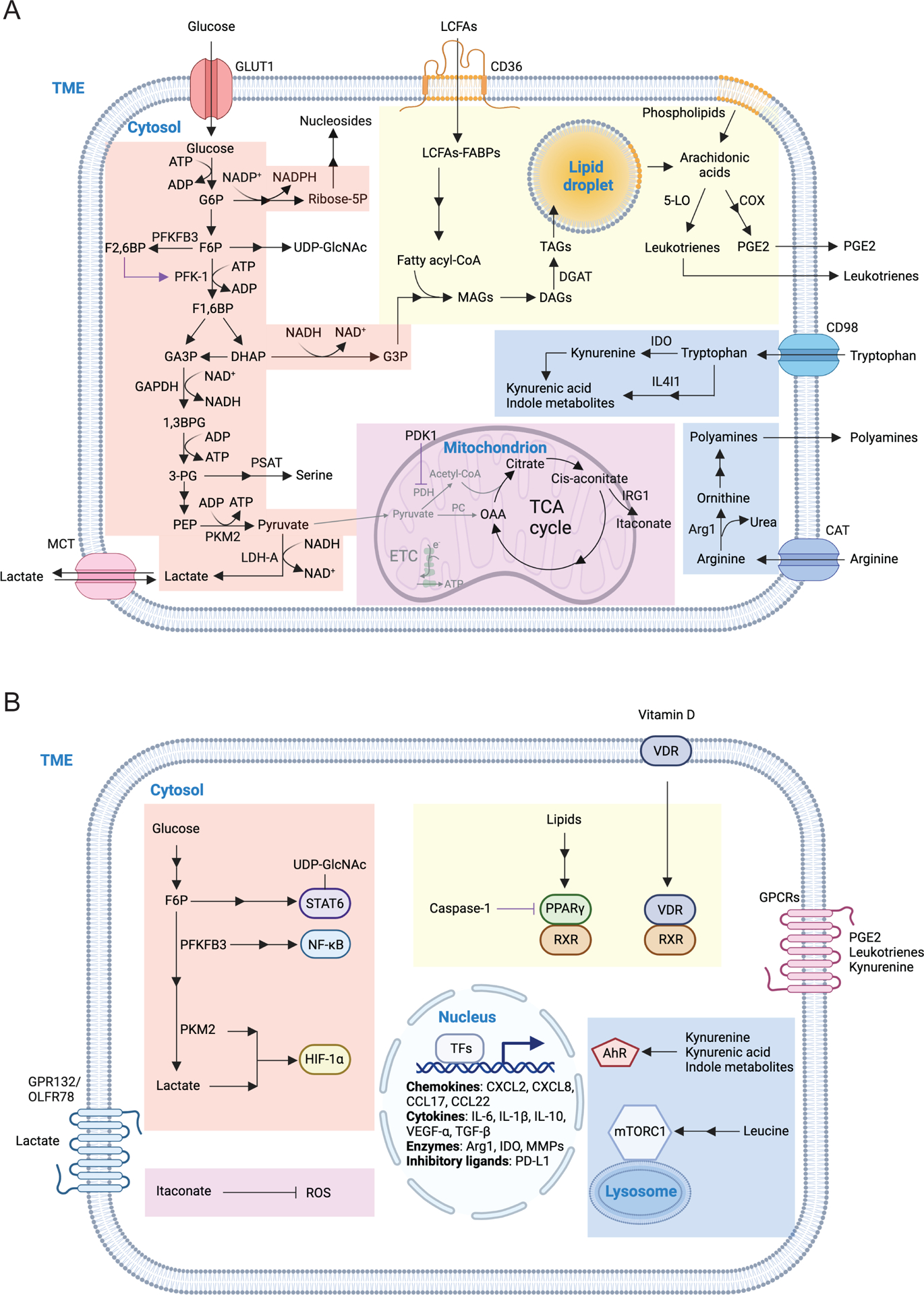
A) Macronutrients including glucose, lipids and amino acids are taken up by tumor-associated macrophages (TAMs) in the tumor microenvironment (TME) and are catabolized or converted to biosynthetic intermediates or signaling metabolites to regulate TAM responses. Glucose acquired through glucose transporter 1 (GLUT1) undergoes glycolysis to produce adenosine triphosphate (ATP) and generates metabolic intermediates to support several biosynthetic pathways. Pyruvate is the end-product of glycolysis, and is mainly reduced to lactate in the cytosol, rather than enter the mitochondrion to complete the tricarboxylic acid (TCA) cycle as a likely consequence of low oxygen level in the TME. Instead, itaconate can be converted from the TCA intermediate cis-aconitate with tumor-promoting functions. Long-chain fatty acids (LCFAs) are acquired via CD36-mediated lipid uptake and contribute to lipogenesis and lipid droplet formation in TAMs, while prostaglandin E2 (PGE2) and leukotrienes are converted from phospholipids, and act as bioactive signaling lipids. Amino acids arginine and tryptophan acquired from plasma membrane transporters can also be converted to bioactive metabolites to regulate tumor progression.
B) Signaling functions of metabolites in TAMs. Metabolites of the glycolytic pathway can promote activation of transcription factors including signal transducer and activator of transcription 6 (STAT6), nuclear factor kappa B (NF-κB), and hypoxia-inducible factor 1-alpha (HIF-1α). In addition, lactate can be sensed by the G protein-coupled receptors (GPCRs) GPR132 and OLFR78, while the TCA cycle metabolite itaconate suppresses the generation of reactive oxygen species (ROS). Lipid-sensing peroxisome proliferator-activated receptor-γ (PPAR-γ) is subject to caspase-1-mediated cleavage to prevent fatty acid oxidation, while lipid-derived metabolites including vitamin D, PGE2, and leukotrienes can bind to vitamin D receptor (VDR) and GPCR family members to induce cellular signaling. Moreover, cytosolic amino acids such as leucine promotes activation of the metabolic regulator mammalian target of rapamycin complex 1 (mTORC1), while the tryptophan metabolites kynurenine, kynurenic acid, and indole metabolites are sensed by aryl hydrocarbon receptor (AhR) and GPCR family member to regulate TAM responses.
1,3BPG, 1,3-bisphosphoglycerate; 3-PG, 3-phosphoglycerate; 5-LO, 5-lipoxygenase; α-KG, alpha-ketoglutarate; ADP, adenosine diphosphate; Arg1, arginase 1; CAT, cationic amino acid transporter; CCL, chemokine (C-C motif) ligand; COX, cyclooxygenase; CXCL, chemokine (C-X-C motif) ligand; DAGs, diacylglycerols; DGAT, diglyceride acyltransferase; DHAP, dihydroxyacetone phosphate; ETC, electron transport chain; F1,6BP, fructose-1,6-bisphosphate; F2,6BP, fructose-2,6-bisphosphate; F6P, fructose 6-phosphate; FABPs, fatty acid-binding proteins; G3P, glycerol 3-phosphate; G6P, glucose 6-phosphate; GA3P, glyceraldehyde 3-phosphate; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; IL, interleukin; IL4I1, interleukin-4 induced 1; IRG1, immune-responsive gene 1; LDH-A, lactate dehydrogenase-A; MAGs, monoacylglycerols; MCT, monocarboxylate transporter; MGLL, monoacylglycerol lipase; MMPs, matrix metalloproteinase; NAD, nicotinamide adenine dinucleotide; NADP, nicotinamide adenine dinucleotide phosphate; OAA, oxaloacetate; PC, pyruvate carboxylase; PDH, pyruvate dehydrogenase; PDK1, pyruvate dehydrogenase kinase 1; PD-L1, programmed death-ligand 1; PEP, phosphoenolpyruvate; PFK-1, 6-phosphofructokinase-1; PFKFB3, 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3; PKM2, pyruvate kinase M2; PSAT, phosphoserine aminotransferase; Ribose-5P, ribose 5-phosphate; RXR, retinoid-X receptor; TAGs, triacylglycerols; TFs, transcription factors; TGF-β, transforming growth factor-beta; UDP-GlcNAc, uridine diphosphate-N-acetylglucosamine; VEGF-α, vascular endothelial growth factor-alpha.
Carbohydrate serves as a major energy and carbon source. Glucose as the most abundant monosaccharide undergoes glycolysis to generate adenosine triphosphate (ATP) and carbon intermediates to support TAM metabolism45. In vivo glucose uptake assays showed that CD11b+ myeloid cells including F4/80+ TAMs have the highest capacity to take up glucose in transplantation tumor models46. Consistent with this observation, histological analysis showed that glucose transporter 1 (GLUT1) is highly expressed in TAMs localized in hypoxic regions of both transplanted and autochthonous murine breast tumor tissues47. In human hepatocellular carcinoma (HCC) samples, high GLUT1 expression is also observed in TAMs compared to macrophages in nontumor regions48. In an orthotopic murine pancreatic ductal adenocarcinoma (PDAC) transplantation tumor model, lysozyme M-cre (LysMcre)-mediated GLUT1 depletion in TAMs suppresses tumor development through natural killer (NK) cell- and CD8+ T cell-dependent mechanisms49. Although this study cannot directly prove that TAM glucose uptake promote cancer progression as LysMcre can also target neutrophils, it suggests that the immunosuppressive function of TAMs is dependent on GLUT1.
Glucose metabolism initiates with a multi-step process of glycolysis (Figure 2A). Glycolytic activities and mRNA expressions of glycolytic enzymes including glucose 6-phosphate isomerase (GPI), phosphofructokinase-B1 (PFKB1), aldolase-A (ALDOA), phosphoglycerate kinase (PGK), and pyruvate kinase-M2 (PKM2) are increased in peritoneal macrophages following transplantation of PDAC tumors49. In addition, TAMs from PyMT tumors express high levels of hexokinase-2 (HK2) and PFKL50. Furthermore, high glycolytic activities are observed in CD14+ monocytes/macrophages from both tumor parenchymal and peritumoral regions of human HCC samples51,52. Enhanced glycolysis in TAMs may be induced by cancer cell-derived factors, as both human and murine macrophages cultured with cancer cells or cancer cell-derived supernatant display enhanced glycolytic activities in several settings50,53,54. More mechanistical studies should be performed to further clarify how TAM glycolysis is induced in the afore-described tumor models, and whether glycolysis is differently regulated in pTAMs and iTAMs. It is also important to note that lower level of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) activity is observed in TAMs from human colon tumors than colonic RTMs55. Although lack of analysis of the whole glycolytic pathway, this study suggests that TAM glucose metabolism varies in different tumor models.
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (PFKFB3) catalyzes the conversion of fructose-6-phosphate (F6P) to fructose-2,6-bisphosphate (F2,6BP) that functions as a potent allosteric activator of the glycolytic enzyme 6-phosphofructokinase-1 (PFK-1). Inhibition of PFKFB3 impairs the production of C-X-C motif chemokine ligand 2 (CXCL2) and CXCL851 as well as expression of programmed death-ligand 1 (PD-L1)52 in HCC supernatant-treated human monocytes in vitro, which has been associated with activation of nuclear factor κ B (NF-κB) signaling51,52. Glycolysis-dependent PD-L1 expression in human HCC TAMs has also been shown to be dependent on PKM256, which may be mediated by the stabilization of hypoxia-inducible factor-1α (HIF)-1α57. These observations suggest that TAM glycolysis may promote the immunosuppressive function of TAMs through the induction of chemokine and PD-L1 expression by modulating TAM signaling (Figure 2B). The signaling regulation function of glycolysis in T cells has recently been shown to be rooted in glycolytic ATP production58,59. Whether such mechanisms operate in TAMs is also open for investigation.
The end-product of glycolysis, pyruvate, has distinct metabolic fates under different conditions. The tumor parenchyma has often low oxygen levels. In this case, pyruvate in pTAMs is predicted to be mainly reduced by lactate dehydrogenase (LDH) to generate lactate in the cytosol, rather than enter mitochondrion to fuel tricarboxylic acid (TCA) cycle or replenish TCA cycle metabolites mediated by pyruvate dehydrogenase (PDH) or pyruvate carboxylase (PC), respectively. Under hypoxic conditions, HIF-1α-induced PDH kinase 1 (PDK1) phosphorylates and inactivates PDH, and thus promotes shunting of pyruvate to the lactate pathway. Indeed, increased PDK1 expression was observed in TAMs from human HCC samples51,52. In addition, PC expression is diminished in TAMs from both mouse and human melanomas, and pharmaceutic activation of PC in TAMs suppresses mouse melanoma progression60. These studies imply that mitochondrial pyruvate metabolism is attenuated in TAMs, and lactate generation may be the dominant metabolic fate of pyruvate. Indeed, increased LDH-A expression was observed in TAMs from both human and murine cancers49,51,52. In addition, primary human macrophage co-cultured with MCF-7 cancer cells upregulated LDH-A61, suggesting that cancer cell-derived factors could also promote pyruvate to lactate conversion. Depletion of LDH-A prevents lactate production in TAMs, decreases expression of PD-L1 and the proangiogenic vascular endothelial growth factor-α (VEGF)-α, and inhibits tumor progression in a K-Ras-mediated lung cancer model62.
Lactate can be exported or imported via monocarboxylate transporters (MCTs) belonging to the SLC16A family63. MCT1 and MCT2 can transport molecules with one carboxylate such as lactate, pyruvate and ketone bodies bidirectionally dependent on the concentration gradient of substrates, while MCT3 and MCT4 are efficient lactate exporters63. Histological analysis showed that MCT1 and MCT4 expression is positively associated with CD163-expressing TAMs in tumors from human breast cancer64 and oral squamous cell carcinoma65, respectively, but their function remains to be determined. Aside from its metabolic function, lactate can act as a signaling molecule sensed by membrane receptors expressed on TAMs. G protein-coupled receptor 132 (GPR132) is a pH-sensing GPCR, and lactate activation of GPR132 triggers TAM expression of a number of chemokines including C-C motif ligand 17 (CCL17) and CCL22 to promote breast cancer metastasis66. In addition, the odorant receptor Olfr78 in TAMs can work together with GPR132 to sense lactate and promote tumor growth and metastasis in a lung transplantation tumor model67. Tumor-derived lactate has also been shown to promote VEGF-α expression in TAMs via HIF-1α68. Furthermore, lactate can modify histone and regulate gene expression through lactylation69. Those findings suggest that lactate may directly regulate TAM signaling and gene expression to promote tumor development.
In addition to glucose catabolism, the metabolic intermediates of glycolysis can be shunted towards a number of anabolic pathways (Figure 2A). Of note, G6P can go through the pentose phosphate pathway (PPP) to generate nicotinamide adenine dinucleotide phosphate (NADPH) as well as ribose 5-phosphate, which is the precursor for de novo purine and pyrimidine nucleotide biosynthesis. Single-cell RNA-sequencing (scRNA-seq) studies revealed that the terminal differentiated Trem2+ TAMs exhibit high purine metabolism than other macrophages in an MC38 liver metastasis model, and high purine metabolism in TAMs is associated with poor clinical outcomes70, but whether the PPP pathway supports the tumor-promoting function of TAMs remains to be determined.
F6P together with glutamine can be shunted to the hexosamine biosynthesis pathway (HBP) and form uridine diphosphate-N-acetylglucosamine (UDP-GlcNAc), which acts as glycosyl donors in glycosylation reactions including O-linked-N-acetylglucosaminylation (O-GlcNAcylation) mediated by O-GlcNAc transferase (OGT). In a B16 transplantation tumor model, MHC-IIlow TAMs have high OGT expression. O-GlcNAcylation of the protease capthepsin B maintains its high expression and promotes lung cancer metastasis and chemoresistance71. TAMs under hyperglycemia conditions promote tumor progression in an MC38 transplantation tumor model, which is also likely dependent on O-GlcNAcylation as glutamine antagonist or the HBP inhibitor 6-Diazo-5-oxo-L-norleucine (DON) can rescue the tumor phenotype72. Of note, in a transplantation mammary tumor model, the cellular UDP-GlcNAc level is associated with the immunosuppressive function of TAMs, which may be dependent on glycosylation of the transcription factor signal transducer and activator of transcription 6 (STAT6)73, implying a signaling function of glycosylation in control of TAM responses (Figure 2B).
Another glycolytic intermediate dihydroxyacetone phosphate (DHAP) can be converted to glycerol 3-phosphate (G3P) by glycerol-3-phosphate dehydrogenase (GPD1). G3P can be further catalyzed by G3P acyltransferases (GPATs) to form phosphatidic acid, which is essential for triacylglycerol (TAG) biogenesis, and the TAG metabolic pathway has an important function in TAMs (see below).
The glycolytic intermediate 3-phosphoglycerate (3-PG) can be catalyzed by phosphoglycerate dehydrogenase (PHGDH) and phosphoserine aminotransferase (PSAT) to form serine and participate in the one-carbon metabolism pathway. Compared to RTMs, TAMs from human lung carcinoma have elevated activities of the serine/glycine one-carbon metabolism pathway74. In a B16-F10 transplantation tumor model, LysMcre-mediated PSAT1 depletion diminishes the TAM population, and suppresses tumor growth74, suggesting that the de novo serine synthesis pathway is critical to support TAM responses.
The mitochondrial TCA cycle serves as a hub to connect several metabolic pathways including glucose metabolism (Figure 2A). Completion of the TCA cycle produces NADH and flavin adenine dinucleotide (FADH2) that are mostly oxidized through the electron transport chain (ETC) with the built-up proton gradient driving mitochondrial ATP production through the process of oxidative phosphorylation (OXPHOS)75. As the tumor microenvironment is relatively hypoxic, the oxygen level may not be high enough to support a robust ETC flow in TAMs76. Yet, in an MC38 transplantation tumor model, the abundance of TCA cycle metabolites increases in TAMs during tumor progression77, suggesting that high TCA flow in TAMs may provide metabolite intermediates to support metabolic events other than OXPHOS. Itaconate is derived from cis-aconitate by aconitate decarboxylase 1 (ACOD1), best known as immune-responsive gene 1 protein (IRG1). IRG1 expression and itaconate production in peritoneal macrophages are elevated following peritoneal tumor inoculation78, which is in line with the observation that IRG1 expression in monocytes from human peritoneal tumor ascites is increased78. Knockdown of IRG1 in peritoneal macrophages reduces the tumor burden78, supporting a pro-tumor function of TAM-produced itaconate. Moreover, elevated IRG1 expression is observed in TAMs isolated from GL261 glioma-bearing mice, and IRG1-deficient TAMs from late-stage glioblastoma express high levels of transcripts that encode proteins involved in antigen presentation and inflammatory responses79. As itaconate can function as an anti-inflammatory metabolite by inhibiting succinate dehydrogenase (SDH) activity to prevent mitochondrial reactive oxygen species (mROS) production80, future studies will reveal whether similar mechanisms operate in TAMs to promote tumor development (Figure 2B). Succinate is another metabolite with important immunomodulatory functions. Cancer cell-derived succinate activates succinate receptor SUCNR1 to trigger tumorigenic TAM signaling via the PI3K-HIF-1α axis81. In addition, intracellular succinate promotes IL-1β production by stabilizing HIF-1α82, and enhances mROS production by fueling TCA cycle and ETC through SDH83 in macrophages, but its function in TAMs remains to be clarified. Thus, TAMs exhibit enhanced glucose consumption to support several metabolic pathways and regulate cell signaling, which appears to promote the immunosuppressive function of TAMs.
Lipid droplet formation in TAMs occurs in several human and murine malignancies84–89, suggesting that TAMs actively acquire and/or synthesize lipid, but have low lipid degradation activity. This is likely because mitochondrion-driven fatty acid oxidation (FAO) is not robust in the low-oxygen tumor microenvironment. CD36, also known as fatty acid translocase, is a member of class B scavenger receptor family and can directly import multiple substrates including long-chain fatty acids (LCFAs)90. CD36-mediated lipid uptake promotes TAM differentiation and supports tumor growth84. In a murine liver metastasis model, macrophages in the liver can also take up tumor microvesicles via CD36, which may contribute to the establishment of a premetastatic niche91. Cytosolic citrate can be converted to acetyl-CoA by ATP citrate synthase (ACLY), which is critical for de novo fatty acid synthesis92. In MC38 and 3LLR transplantation tumor models, LysMcre-mediated ACLY depletion does not affect tumor growth, and only slightly affects TAM phenotypes93. Those observations suggest that fatty acid uptake but not citrate-mediated de novo fatty acid synthesis supports the tumor-promoting function of TAMs (Figure 2A).
The imported cytoplasmic LCFAs bind to fatty acid-binding proteins (FABPs) that facilitate LCFA transportation to subcellular compartments94. Epidermal FABP (E-FABP, or FABP5) and adipocyte/macrophage FABP (A-FABP, or FABP4) are highly expressed in macrophages, but they appear to have distinct functions in control of murine breast cancer growth and metastasis95,96. FABP5 suppresses E0771 breast cancer growth and metastasis through mechanisms that are dependent on TAM expression of interferon-β (IFN-β), and the frequency of FABP5-expressing TAMs is negatively associated with human breast cancer progression95. On the contrary, FABP4 promotes E0771 and PyMT breast cancer progression and metastasis through mechanisms that are dependent on TAM expression of interleukin-6 (IL-6)96. Whether the opposing functions of FABP4 and FABP5 are caused by their distinct roles in different subsets of pTAMs and iTAMs is unknown. The underlying mechanisms by which FABPs regulate cytokine expression are also open for future investigation.
Peroxisome proliferator-activated receptors (PPARs) including PPAR-α, PPAR-β/δ and PPAR-γ are a group of nuclear receptor proteins that bind to cytosolic lipid ligands (Figure 2B). Once activated, PPARs enter the nucleus and form heterodimers with retinoid-X receptor (RXR) to induce expression of target genes including those involved in lipid catabolism. Caspase 1 in TAMs could cleave PPAR-γ to inhibit FAO, and caspase 1 deficiency decreases lipid accumulation in TAMs resulting in diminished PyMT mammary tumor growth87. As caspase 1 is produced as a latent enzyme, and can be activated by the inflammasome pathway, whether and how inflammasome is induced in TAMs to regulate lipid metabolism remain to be determined.
As a major component of lipid droplet, TAG can be broken down into diacylglycerols (DAGs) by adipose triglyceride lipase (ATGL), hydrolyzed into monoacylglycerols (MAGs) by hormone sensitive lipase (HSL), and further catalyzed into free fatty acids and glycerol by monoacylglycerol lipase (MGLL). Low MGLL expression is detected in TAMs, and overexpression of MGLL prevents lipid accumulation in TAMs, causing CD8+ T cell-dependent tumor suppression in an MC38 transplantation tumor model85. MGLL promotes the degradation of 2-arachidonoylglycerol (2-AG), a ligand for cannabinoid receptor 2 (CB2), and thereby reverses the CB2-mediated immunosuppression in TAMs85. MAGs can be converted to DAG and TAG by acyltransferases with diglyceride acyltransferase (DGAT) catalyzing the formation of TAG from DAG (Figure 2A). DGAT1-mediated TAG synthesis enhances the production of proinflammatory mediators including prostaglandins E2 (PGE2) and IL-1β in macrophages97. As PGE2 inhibits anti-tumor immune responses (see below), the DGAT pathway in TAMs may have pro-tumor functions. Indeed, liposome-mediated delivery of a DGAT inhibitor to phagocytes suppresses MCA205 fibrosarcoma tumor development, which is associated with reduced lipid droplet formation and increased CD8+ T cell proliferation in tumor86. Together, these findings demonstrate an important function for TAG biosynthesis in promoting the immunosuppressive function of TAMs.
A number of lipids and lipid derivatives function as important signaling molecules (Figure 2). Membrane phospholipids can release arachidonic acids to generate PGE2 catalyzed by cyclooxygenase (COX) enzymes COX1 or COX2 and PGE synthase (PGES). PGE2 binds to PGE2 receptors, and thereby activates the downstream cAMP and Ca2+-mediated signaling pathways. Expression of genes in the arachidonic acid-PGE2 pathway is positively associated with TREM2+ pTAMs in human esophageal squamous cell carcinoma (ESCC)98. In addition, COX2 is highly induced in myeloid cells infiltrating the transplanted SW780 bladder tumors99, and in CD68+ macrophages infiltrating human melanoma100. Administration of microsomal PGES-1 (mPGES-1) and COX2 inhibitors or overexpression of the PGE2-degrading enzyme 15-hydroxyprostaglandin dehydrogenase (15-PGDH) diminishes PGE2 production and inhibits PD-L1 expression in myeloid lineage cells in vitro101. In a macrophage T cell co-culture system, PGE2 produced by macrophages attenuates CD4+ T cell proliferation102, but the in vivo function of TAM-produced PGE2 remains to be determined. Furthermore, mPGES-1 is barely detectable in TAMs from human neuroblastoma tumors103, suggesting that the COX/mPGES-1/PGE2 pathway may only be important in some tumor types. In addition to PGE2, arachidonic acids can be catalyzed by lipoxygenase such as 5-lipoxygenase (5-LO) to generate leukotrienes. 5-LO expression in TAMs from mouse and human primary breast tumors is lower than that in monocyte-derived macrophages generated in vitro, and 5-LO downregulation is dependent on apoptotic cell engulfment, which may inhibit T cell recruitment and thus exert an immunosuppressive function104. However, high expression of 5-LO and production of leukotriene B4 (LTB4) in alveolar macrophages promote HCC metastasis in lung105. Thus, macrophage production of leukotrienes may have opposing functions in control of tumor development. Aside from phospholipid-derived signaling molecules, cholesterol can be processed to generate vitamin D. Vitamin D binds to vitamin D receptor (VDR) and forms a heterodimer with RXR to control target gene expression. Inhibition of vitamin D-VDR binding in TAMs suppresses tumor progression in a transplantation breast cancer model106, but the underlying mechanisms remain to be determined.
Ketone bodies including acetone, acetoacetate and β-hydroxybutyrate (BHB) are derived from fatty acids and can be converted to acetyl-CoA and fuel the TCA cycle. Ketogenic diets containing BHB target intestinal epithelial cells to suppress colorectal cancer (CRC) development107. BHB has also been shown to inhibit inflammatory responses triggered by the NLRP3 inflammasome in macrophage108, while hepatocyte-produced acetoacetate can be oxidized in macrophages to inhibit the high-fat diet (HFD)-induced liver fibrosis109. Of note, BHB can bind to GPR109a and function as a signaling metabolite in macrophages110, which promotes tissue repair in injury models including ischemic strokes111, alcohol-induced liver injury112, and DSS-induced colitis113. Nonetheless, the functions of ketone bodies in control of TAM responses have yet to be revealed.
Amino acids are a special class of macronutrients that are used for protein biosynthesis and are converted to other metabolite for cellular regulation. Some cytosolic amino acids can also function as signaling molecules, notably, participating in the activation of the metabolic regulator mammalian target of rapamycin complex 1 (mTORC1) through a lysosomal Rag GTPase-mediated nutrient-sensing pathway114 (Figure 2B). Branched-chain amino acids (valine, leucine, isoleucine) and aromatic amino acids are taken up by the heterodimer amino acid transporter CD98 composed of the heavy chain SLC3A2 and the light chain SLC7A5115. As leucine is one of the most critical amino acids that activates mTORC1116, pharmaceutical inhibition of SLC7A5 diminishes mTORC1-mediated glycolysis and inflammatory cytokine production in activated macrophages in vitro117. Nonetheless, a role for CD98-mediated leucine uptake in control of mTORC1 signaling and metabolism in TAMs remains to be determined.
Tryptophan can also be imported through CD98, and further catabolized to kynurenine, with indoleamine 2,3-dioxygenase (IDO) being the rate-limiting enzyme (Figure 2). Depletion of tryptophan by IDO1-expressing human monocyte-derived macrophages suppresses T cell proliferation and activation in vitro118. Macrophage expression of IDO1 has also been shown to promote immune tolerance to apoptotic cells119. Nonetheless, whether the IDO1-meidated tryptophan catabolism non-redundantly contributes to the immunosuppressive function of TAMs remains to be determined. Cancer cells can also express IDO and produce kynurenine to activate aryl hydrocarbon receptor (AhR) in TAMs120. Kynurenine activation of AhR induces Kruppel-like factor 4 (KLF4) expression, but suppresses NF-κB activation, and AhR-deficient TAMs are poorly recruited to tumor, resulting in impaired growth of GL261 glioma cells120. Another study revealed that IDO-overexpressing B16 melanoma display an immunosuppressive phenotype, which is in part dependent on the kynurenine-AhR-mediated regulatory T (Treg) cell-TAM interplay121. Furthermore, tryptophan can be degraded by microbiota and generate indole-containing metabolites to activate AhR in TAMs, which promotes PDAC tumor progression by suppressing intra-tumoral CD8+ T cell function122. Interleukin-4-induced-1 (IL4I1), an L-amino-acid oxidase, has recently been identified as a potent activator of the AhR pathway by promoting tryptophan catabolism to indole metabolites and kynurenic acid (Figure 2)123. scRNA-seq analyses revealed that an enriched IL4I1+IDO1+PD-L1+ TAM subset is associated with T cell dysfunction in a number of human tumors24. Although the in vivo function of IL4I1 in TAMs has yet to be determined, those findings reveal an alternative metabolic pathway that may account for the immunosuppressive function of tryptophan catabolism. In addition to AhR, kynurenine can bind to the cell surface receptor GPR35124. LysMcre-mediated GPR35 depletion suppresses tumor development in both genetic and carcinogen-induced CRC models in part mediated by attenuated tumor angiogenesis125. These studies suggest that tryptophan metabolites may be sensed by both intracellular and plasma membrane-localized receptors in TAMs to promote tumor development.
Positively charged amino acids including arginine are taken up by cationic amino acid transporters (CATs) SLC7A1–4126. Arginases including arginase 1 (Arg1) convert arginine to urea and ornithine as part of the urea cycle for nitrogen excretion. Ornithine can also be catalyzed by ornithine decarboxylase (ODC) to form putrescine, which is the precursor for polyamine biosynthesis (Figure 2A). Polyamines including spermidine and spermine promote cell proliferation and maintain tissue homeostasis127. Arg1 is highly expressed in TAMs128, which is in part dependent on HIF-1α signaling, lactate, and granulocyte macrophage colony-stimulating factor (GM-CSF)129 68,130,131. Uptake of the metabolite creatine through the creatine transporter Slc6a8 has also been shown to sustain Arg1 expression in macrophages132, but its role in TAMs is undefined. Arg1-expressing myeloid cells from transplanted Lewis lung carcinoma (LLC) tumors inhibit T cell proliferation in an in vitro culture system133,134. SLC7A2-mediated arginine uptake in tumor myeloid cells also suppresses T cell proliferation ex vivo135. Those observations suggest that the Arg1-mediated arginine consumption in TAMs may have an immunosuppressive function. In a number of transplantation tumor models, pharmaceutical inhibition of arginase by CB-1158 enhances arginine concentration and cytotoxic immune cell infiltration and inhibits tumor development136. Moreover, LysMcre-mediated Arg1 deletion suppresses murine PDAC progression, which is associated with increased cytotoxic CD8+ T cell infiltration and activation137. The TAM regulation function of Arg1 may also act through polyamine. For instance, the arginine-polyamine pathway is induced in tumor-infiltrating myeloid cells to promote their survival, and depletion of polyamine prolongs mouse survival in transplantation models of brain tumor138,139. These findings suggest a tumor-promoting role of myeloid Arg1, but its specific function in TAMs remains to be determined.
Glutamine is an important non-essential amino aid that can be converted to glutamate by glutaminase (GLS). Conversely, glutamate can generate glutamine via glutamine synthetase (GS). Pharmaceutical inhibition of GLS by JHU083 suppresses transplanted 4T1 tumor progression and reprograms TAMs to a pro-inflammatory state associated with high expression of tumor necrosis factor-α (TNF-α) and co-stimulatory molecules140, but it was unknown whether such reprograming was due to the blockade of GLS in TAMs. Of note, LysMcre-mediated GLS depletion attenuates macrophage engulfment of apoptotic cells141, and the impaired efferocytosis may account for the inflammatory phenotype of TAMs, as efferocytosis is largely anti-inflammatory (see below). CSF1Rcre-mediated depletion of GS does not affect primary LLC tumor growth, but impairs cancer cell metastasis, which was associated with high abundance of glutamate and succinate under the condition of GS inhibition in macrophages with succinate mainly derived from glucose, but not glutamine142. These observations suggest that glutamine/glutamate metabolism may interact with glucose metabolism to regulate macrophage responses, but the exact functions of such metabolic crosstalk in TAMs remain to be investigated.
Engulfment-mediated nutrient acquisition in control of TAM responses
Macrophages can manifest robust engulfment activity to support their scavenger function and provide an alternative route of nutrient acquisition. The transcription factor PU.1 specifies a core macrophage gene expression program including those involved in macrophage phagocytosis such as tyrosine-protein kinase Mer (MerTK) for efferocytosis and CD64 (FCγR1A) for antibody-dependent phagocytosis143,144. In addition, specialized scavenger receptor gene expression programs support discrete engulfment activity of macrophage subsets, including ECM scavenger receptors Mrc1 and Lyve1 for iTAMs, and the lipid scavenger receptor Trem2 for pTAMs. While interactions between scavenger receptors and their ligands trigger intracellular receptor signaling to promote phagocytosis and regulate inflammatory responses, the engulfed cargo generates nutrients that can be further metabolized and engaged in cell signaling (Figure 3).
Figure 3. Engulfment-mediated nutrient acquisition control of TAM responses.
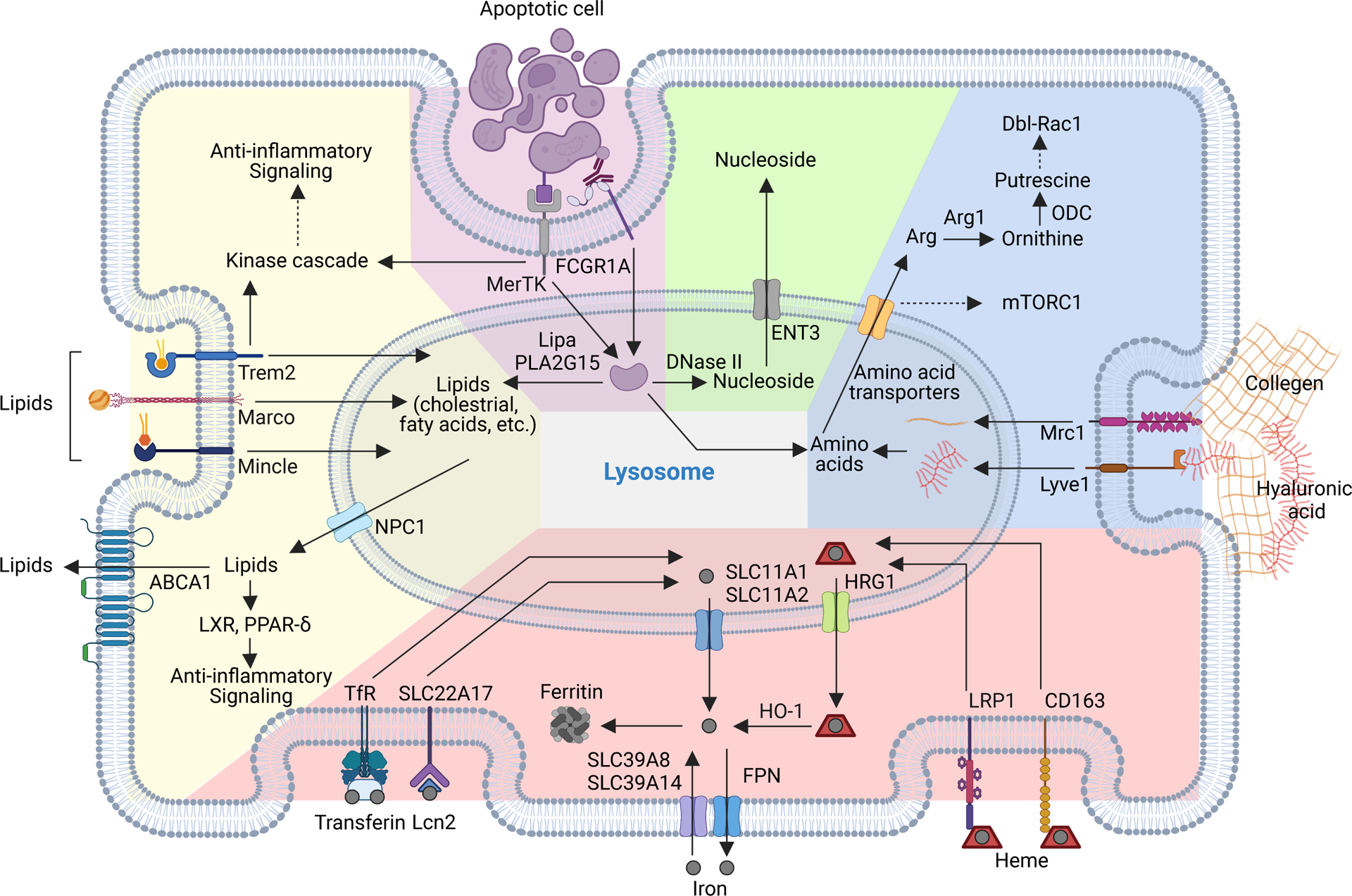
Apoptotic cell as well as lipids, extracellular matrix (ECM) proteins and iron complexes can be phagocytosed or endocytosed in tumor-associated macrophages (TAMs), and nutrients are further generated in the lysosome and exported to the cytosol to support metabolism and signaling responses. Upon ligand binding, efferocytosis receptors such as MER proto-oncogene, tyrosine kinase (MerTK) or lipid-scavenging receptors such as triggering receptor expressed on myeloid cells 2 (Trem2) can activate intracellular kinase signaling cascades and induce expression of anti-inflammatory mediators. The recycled lipids can also induce anti-inflammatory signaling or exported outside of TAMs to promote cross-feeding. Degradation of apoptotic bodies and ECM proteins generate amino acids in the lysosome and may activate the metabolic regulator mammalian target of rapamycin complex 1 (mTORC1). Arginine is a major amino acid generated from ECM degradation and can activate Rac1 through its downstream metabolites. Engulfed apoptotic cells also contain nucleic acids, proper degradation of which is critical to prevent activation of nucleic acid-innate immune sensing pathways. TAMs can either uptake or release iron in both free and complex forms and regulate iron metabolism through competition or crossfeeding in tumor.
ABCA1, ATP Binding cassette subfamily A member 1; CD163, CD163 molecule; DNase II, deoxyribonuclease 2, lysosomal; ENT3, equilibrative nucleoside transporter 3; FPN1, ferroportin 1; SLC39A8, solute carrier family 39 member 8; HO-1, heme oxygenase 1; HRG1, heme-responsive gene 1 protein homolog; FCGR1A, Fc gamma receptor Ia; Lcn2, lipocalin 2; LIPA, lipase A, lysosomal acid type A; LRP1, LDL receptor related protein 1; Lyve1, lymphatic vessel endothelial hyaluronan receptor 1; Marco, macrophage receptor with collagenous structure; Mertk, MER proto-Oncogene, tyrosine kinase; Mincle, macrophage-inducible C-type lectin; Mrc1, mannose receptor C-type 1; NPC1, NPC intracellular cholesterol transporter 1; PLA2G15, phospholipase A2 group XV; SLC11A1, solute carrier family 11 member 1; SLC11A2, solute carrier family 11 member 2; SLC22A17, solute carrier family 22 member 17; SLC39A14, solute carrier family 39 member 14; TfR, transferrin receptor; Trem2, triggering receptor expressed on myeloid cells 2.
A major scavenger function of RTM is apoptotic cell clearance, or efferocytosis. A number of receptors can mediate efferocytosis, including Tyro3, Axl and MerTK that are receptor tyrosine kinases sharing similar structures with two immunoglobulin-like repeats and two fibronectin type III repeats in the extracellular domain145. Upon ligand binding and receptor dimerization, autophosphorylation of intracellular kinase domain activates signaling and gene transcription programs that promote membrane and cytoskeleton remodeling in support of efferocytosis146. MerTK is indispensable for apoptotic cell clearance in mice147, while Tyro3, Axl and MerTK collectively promote efferocytosis and suppress autoimmunity148. In a PyMT transgenic mammary tumor model, impaired clearance of apoptotic cells in mice on a MerTK-deficient background is associated with defective expression of anti-inflammatory cytokines and diminished cancer cell metastasis to lung149. Whether the tumor phenotype can be attributed to the depletion of MerTK in TAMs remains to be determined, as epithelial (cancer) cells can express MerTK and engulf apoptotic cells150. Nonetheless, in a CT26 transplantation tumor model, radiation-triggered tumor therapy is potentiated in MerTK-deficient recipient mice151, and MerTK deficiency inhibits the development of Starry-sky B-cell lymphoma, where MerTK expression is highly restricted in TAMs152. These findings support a critical function for MerTK-mediated TAM efferocytosis in fostering cancer progression.
The T-cell immunoglobulin and mucin domain-containing protein (TIM) family receptors can also mediate clearance of apoptotic cells with TIM-4 being the major family member expressed on antigen presentation cells (APCs) including RTMs in liver, heart, intestine, adipose tissue and the peritoneal cavity153–157. In patients with metastatic non-small cell lung cancer (NSCLC) to serous body cavities, the frequency of TIM-4-expressing cavity-resident macrophages is inversely associated with the frequency of CD8+ T cells that express the ectonucleotide triphosphate diphosphohydrolase CD39158. Importantly, anti-TIM-4 synergizes with anti-PD-1 to revive T cell responses and suppress peritoneal cancer cell metastasis in a murine model158. Loss of peritoneal TIM-4+ TAMs is also associated with elevated T cell immunity and tumor inhibition for ID8 ovarian cancer cell line injected into mouse peritoneal cavity159. These observations further support an important function of TAM efferocytosis in promoting tumor development.
The metabolic outcome of macrophage efferocytosis has started to be revealed. Engulfment of apoptotic cells and their processing in the lysosome drastically increases the intracellular lipid level (Figure 3). High amounts of free intracellular cholesterol activate the transcription factor liver X receptor (LXR) that enhances expression of ATP-binding cassette transporter A1 (ABCA1) to promote cholesterol efflux, and MerTK to support continuous efferocytosis160. Activation of PPAR-δ, another lipid-sensing transcription factor, enables cellular adaptation to efferocytosis-derived lipids by promoting fatty acid metabolism161,162. Mice deficient in LXR or PPAR-δ accumulate dead cells in many tissues due to failure of efferocytosis, revealing a critical role of lipid signaling and metabolism in sustaining efferocytosis163. In addition, LXR and PPAR-δ induce expression of anti-inflammatory cytokines including transforming growth factor-β1 (TGF-β1) and IL-10162, promoting the resolution of inflammation. In support of a critical role of apoptotic cell-derived lipids in metabolic reprogramming of macrophage, depletion of lysosomal lipidase lipidase A (LIPA) and phospholipase A2 group 15 (PLA2G15) that hydrolyze cholesteryl esters and phospholipids, respectively, impairs activation of LXR and PPAR-δ164,165. Inhibition of Niemann-Pick disease type C1 (NPC1)-mediated sterol transport from lysosome to cytosol as well prevents activation of LXR during efferocytosis166. Diminished expression of proinflammatory cytokines IL-1β and IL-6 during efferocytosis has also been shown to be mediated by the Rac GTPase-dependent import of polyamine through pinocytosis139, but the molecular mechanisms and functions of this non-selective liquid phase engulfment pathway need to be further characterized in macrophages.
Lysosomal clearance of apoptotic cell DNA proceeds with DNA degradation and nucleoside export via equilibrative nucleoside transporter 3 (ENT3) (Figure 3), which is critical for the maintenance of an anti-inflammatory state of macrophage, as depletion of lysosomal DNase II triggers expression of the proinflammatory cytokine TNF-α in part through unmethylated CpG DNA-induced activation of toll-like receptor 9 (TLR9)167,168. Furthermore, efferocytosis generates short peptides and free amino acids that are exported from lysosome to cytosol169. Among all amino acids, arginine is the most upregulated amino acid following engulfment of apoptotic cells170. Efferocytosis-derived arginine and ornithine can further boost efferocytosis through the stabilization of mRNA encoding the GTP-exchange factor (GEF) Dbl and activation of the small GTPase Rac1, following their conversion to putrescine by Arg1 and ODC170 (Figure 3). Thus, apoptotic cell-derived lipids and amino acids can be sensed and metabolized to sustain efferocytosis.
The afore-discussed metabolic outcomes of macrophage efferocytosis have yet to be directly evaluated in TAMs. It also remains to be determined whether efferocytosis is differentially regulated in iTAMs and pTAMs, and how cancer cell transformation mechanisms and tumor microenvironment signals affect TAM efferocytosis. Furthermore, the functional role of apoptotic cell clearance by TAMs can be modulated by phagolysosome cargo sorting with the LC3-associated phagocytosis promoting tumor immune tolerance171. How different apoptotic cell scavenge receptors engage different phagolysosome pathways to impact TAM function warrants further investigation.
The endocytosis receptor Mrc1 marks iRTMs and iTAMs34,172, and mediates internalization of the ECM protein collagen173–175 (Figure 3). In a lung tumor model, Mrc1+ iTAMs display a matrix catabolism transcriptome signature175, suggesting that collagens can be effectively engulfed and proteolyzed by iTAMs. Indeed, Mrc1+ iTAMs are localized at the peripheral collagen-rich tumor region131,176, and function to remodel the collagen matrix, which promotes cancer cell invasion and metastasis176,177. iTAMs can also express Lyve1, an endocytosis receptor for the ECM molecule HA30 (Figure 3). Notably, Lyve1 expression marks a subset of iRTMs localized in close proximity to the blood vasculature across tissues178. In a PyMT transgenic mammary tumor model, depletion of Lyve1+ iTAMs inhibits tumor development in association with disruption of a proangiogenic niche179.
The metabolic outcome of Mrc1- and Lyve1-mediated ECM scavenging is poorly understood. Engulfment and proteolysis of ECM proteins in the lysosome generate high amounts of amino acids that may activate the metabolic regulator mTORC1 (Figure 3). In support of the hypothesis, iTAMs are metabolically more active with a larger cell size than pTAMs22. Interestingly, LysMcre-mediated Raptor knockout promotes the accumulation of immune suppressive lung interstitial macrophages and Lewis lung cancer metastasis180. As mentioned above, cytosolic amino acids acquired through plasma membrane amino acid transporters can be sensed by Rag GTPases to promote mTORC1 signaling. However, mTORC1 activation by the lysosome-derived amino acids appears to be Rag GTPase-independent181,182. Future studies will reveal such mechanisms and define how the lysosome-derived amino acids from scavenged ECMs regulate iTAM differentiation and function.
Trem2 is a scavenger receptor of the immunoglobulin superfamily and can recognize endogenous phospholipids as its ligands183–185 (Figure 3). Trem2+ macrophages are present in damaged tissues39, and accumulate high levels of lipids in association with enrichment of a lipid metabolism gene expression signature186. In a murine model of diet-induced non-alcoholic steatohepatitis (NASH), the interstitial region-localized Kupffer cells and monocyte-derived macrophages upregulate Trem2187, suggesting that Trem2 is induced in diverse populations of macrophages to clear damage-associated lipids. In contrast to NASH, Trem2 expression is limited to pTAMs in tumor, implying that damage-associated lipids are predominantly released by cancer cells in the tumor parenchyma. In transplantation tumor models, ablation of Trem2 or treatment with a Trem2 blocking antibody alters the TAM phenotype and synergizes with anti-PD-1 to revive anti-tumor T cell responses41,188,189. In a lung cancer model, Trem2 deficiency triggers NK cell-mediated suppression of tumor growth190. Collectively, these studies demonstrate a critical role for Trem2 in mediating the immunosuppressive function of TAMs.
The metabolic outcome of Trem2-mediated lipid scavenging in macrophages has started to be revealed. Trem2-deficient microglial fail to upregulate lipid metabolism, which can be rescued by an agonist for the lipid-sensing transcription factor LXR40, suggesting that lipids scavenged by Trem2 are important signaling molecules. However, Trem2 ligands in the tumor microenvironment need be further characterized to understand their function in nutrient metabolism and signaling in TAMs. In addition to Trem2, a number of other lipid scavenger receptors including macrophage receptor with collagenous structure (Marco) and macrophage-inducible C-type lectin (Mincle) have been reported to control TAM responses (Figure 3). Marco-dependent lipid uptake in TAMs is associated with induction of an LXR gene expression program that tracks with short disease-free survival in prostate cancer patients with Marco targetable for cancer therapy in preclinical models191. Furthermore, Mincle-mediated lipid uptake supports the pro-tumor function of TAMs through the induction of an X-box binding protein 1 (XBP1)-mediated endoplasmic reticulum (ER) stress response192. Thus, blockade of the lipid scavenger function of TAMs may provide novel therapeutic strategies to inhibit their pro-tumor functions.
Iron is a trace element nutrient that primarily utilized by erythrocytes, but equally critical in all other cell types193. In addition to SLC39A8 and SLC39A14 transporter-mediated uptake of free iron, iron can be acquired through efferocytosis or endocytosis via specific receptors for ion in complex forms including that bound to transferrin, lipocalin, and heme194,195 (Figure 3). Iron can be used as a metabolic cofactor, stored in macrophages in the form of a ferritin complex, or released through the transporter ferroportin (FPN)196. In models of microbial infection and tissue damage, iron-sequestering and iron-donating macrophage phenotypes have been observed at inflammatory and resolving phases of immune responses, respectively197,198. Iron metabolism in TAMs has also important functions in control of cancer progression. High level of iron accumulation in TAMs at the tumor edge, likely iTAMs, is associated with a pro-inflammatory phenotype and predicts favorable outcomes in multiple cancer types199–201. Yet, an iron-secreting and anti-inflammatory phenotype was observed in TAMs at the tumor core, likely pTAMs202, and loading TAMs with iron repolarizes them to a pro-inflammatory phenotype and diminishes tumor growth203. Furthermore, LysMcre-mediated knockout of low-density lipoprotein (LDL) receptor related protein 1 (LRP1 or CD91), the endocytic receptor for heme, promotes TAM infiltration to the tumor parenchyma and angiogenesis in an implanted mouse pancreatic adenocarcinoma model204.
The metabolic outcome of engulfment-mediated ion uptake in TAMs has also started to be elucidated. TAMs at the tumor margin express high level of heme oxygenase 1 (HO-1) that degrades intracellular heme exported out of the lysosome via heme-responsive gene 1 protein homolog (HRG1)205. LysMcre-mediated depletion of HO-1 triggers an immunostimulatory phenotype, suppresses angiogenesis in a transplantation model of sarcoma205, inhibits cancer cell dissemination in a model of melanoma lung metastasis205 and enhances anti-tumor vaccine efficacy in a subcutaneous thymoma model206. Thus, the TAM regulation phenotype of iron is dependent on iron metabolic pathways. While iron level modulates TAM polarization, iron metabolism crosstalk between TAMs and cancer cells may also be critical to regulate tumor growth. In a prostate tumor model, the anti-tumor effect of an iron-chelating reagent is negatively associated with infiltration of iron-laden TAMs207, suggesting that TAMs maintain local iron homeostasis. In breast tumor models, TAMs at the tumor periphery express high levels of the iron carrier lipocalin 2 (Lcn2), that promotes iron transfer to cancer cells in an in vitro coculture system and cancer progression in vivo208,209. Leptomeningeal metastasis cancer cells also secret Lcn2 and express its receptor SLC22A17 to compete with macrophages for iron acquisition in the nutrient sparse subarachnoid space210. Therefore, iron is a limiting micronutrient in the tumor microenvironment, and TAMs can recycle iron to support cancer cell growth.
Concluding remarks
TAMs are maladapted RTMs with sub-tissular niche factors including the dynamically fluctuating nutrient source being a critical regulator of TAM differentiation and function. While the plasma membrane transporter-mediated uptake of glucose appears to majorly support the bioenergetic and biosynthetic needs of TAMs, lipids acquired via transporters or scavenger receptors are mostly stored in lipid droplets as a likely means to detoxify inflammatory lipids produced in the tumor tissue. A number of metabolites generated in glucose and lipid metabolism pathways as well as those converted from amino acids can also act as signaling molecules to promote scavenger and anti-inflammation functions of TAMs. Aside from nutrient consumption, storage and conversion to signaling molecules in TAMs, nutrients acquired through engulfment can be further exchanged with neighboring cells including cancer cells as a likely means to support their metabolic needs. Collectively, the nutrient acquisition and metabolism pathways appear to enable the tumor-promoting activities of TAMs as maladaptation of the tissue maintenance programs of RTMs. Nonetheless, the definitive functions of many nutrient acquisition and metabolism pathways in TAMs, including their differential regulation in pTAM and iTAM subsets, remain to be investigated, as autochthonous tumor models that recapitulate the tissue architecture associated with cell transformation have yet to be used in most studies. Of note, in a transgenic model of breast cancer, the immunosuppressive function of pTAMs is associated with their ability to present tumor-associated antigens to CD8+ T cells and induce T cell exhaustion211. Whether such a tolerogenic function of pTAMs is metabolically regulated is open for investigation.
The pro-tumor activities of TAMs hamper patient responses to conventional chemotherapy and radiotherapy as well as immunotherapy. Notably, blockade of efferocytosis by anti-Mertk enhances TAM uptake of the endogenously produced immune stimulant 2’3-cyclic GMP-AMP (cGAMP) that activates stimulator of interferon genes (STING) signaling and synergizes with immune checkpoint inhibitors to suppress tumor growth212. Depletion of the potassium channel Kir2.1 in TAMs also diminishes efferocytosis, which promotes the accumulation of intratumoral cGAMP and the induction of type I IFN production leading to enhanced anti-tumor CD8+ T cell responses213. Administration of the immune stimulant unmethylated CpG oligonucleotides also reprograms lipid metabolism in TAMs and facilitates phagocytic clearance of cancer cells both in vitro and in vivo regardless of their expression of the ‘don’t eat me’ signal CD47214. Furthermore, TAMs may inhibit tumor growth by competing with cancer cells or other crucial stromal cells such as the endothelium for essential nutrients47,210. Reprograming TAMs by disengaging the tissue-supporting role and promoting the nutrient-competing function may as well represent a new cancer therapy approach. Considering the diverse cell transformation mechanisms and heterogenous immune profiles in tumors of different tissue origin, the functions of TAMs can thus be contextual. A deep understanding of nutrient acquisition and metabolism pathways in TAMs and their metabolic interaction with other components in the tumor microenvironment will help guide the development of mechanism-based cancer therapies by targeting the highly adaptable innate immune cell lineage.
Acknowledgements
We apologize to the authors whose work we could not cite owing to the limited space. We thank members of the M.O.L. laboratory for helpful discussions. This publication is supported by National Institute of Health (R01 CA198289-01 to M.O.L.), Howard Hughes Medical Institute Faculty Scholar Award (M.O.L.), CLIP grant from Cancer Research Institute (M.O.L.), the Geoffrey Beene Cancer Research Center, the Alan and Sandra Gerry Metastasis and Tumor Ecosystems Center (M.O.L.), the Memorial Sloan Kettering Cancer Center (MSKCC) Support Grant/Core Grant (P30 CA08748) and the Cancer Research Institute Irvington Postdoctoral Fellowship (X.Z. and L.J.). The figures were created with BioRender.com
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare no competing interests.
Reference
- 1.Okabe Y, and Medzhitov R (2016). Tissue biology perspective on macrophages. Nat Immunol 17, 9–17. 10.1038/ni.3320. [DOI] [PubMed] [Google Scholar]
- 2.Guilliams M, Thierry GR, Bonnardel J, and Bajenoff M (2020). Establishment and Maintenance of the Macrophage Niche. Immunity 52, 434–451. 10.1016/j.immuni.2020.02.015. [DOI] [PubMed] [Google Scholar]
- 3.Blériot C, Chakarov S, and Ginhoux F (2020). Determinants of Resident Tissue Macrophage Identity and Function. Immunity 52, 957–970. 10.1016/j.immuni.2020.05.014. [DOI] [PubMed] [Google Scholar]
- 4.Lavin Y, Mortha A, Rahman A, and Merad M (2015). Regulation of macrophage development and function in peripheral tissues. Nat Rev Immunol 15, 731–744. 10.1038/nri3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jenkins SJ, and Allen JE (2021). The expanding world of tissue-resident macrophages. Eur J Immunol 51, 1882–1896. 10.1002/eji.202048881. [DOI] [PubMed] [Google Scholar]
- 6.Buechler MB, Fu W, and Turley SJ (2021). Fibroblast-macrophage reciprocal interactions in health, fibrosis, and cancer. Immunity 54, 903–915. 10.1016/j.immuni.2021.04.021. [DOI] [PubMed] [Google Scholar]
- 7.Nobs SP, and Kopf M (2021). Tissue-resident macrophages: guardians of organ homeostasis. Trends Immunol 42, 495–507. 10.1016/j.it.2021.04.007. [DOI] [PubMed] [Google Scholar]
- 8.Kierdorf K, Prinz M, Geissmann F, and Perdiguero EG (2015). Development and function of tissue resident macrophages in mice. Semin Immunol 27, 369–378. 10.1016/j.smim.2016.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haldar M, and Murphy KM (2014). Origin, development, and homeostasis of tissue-resident macrophages. Immunol Rev 262, 25–35. 10.1111/imr.12215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Birtwell D, Luebeck G, and Maley CC (2020). The evolution of metapopulation dynamics and the number of stem cells in intestinal crypts and other tissue structures in multicellular bodies. Evol Appl 13, 1771–1783. 10.1111/eva.13069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelly P, Davison R, Bliss E, and McGee J (1988). Macrophages in human breast disease: a quantitative immunohistochemical study. Brit J Cancer 57, 174–177. 10.1038/bjc.1988.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noy R, and Pollard JW (2014). Tumor-Associated Macrophages: From Mechanisms to Therapy. Immunity 41, 49–61. 10.1016/j.immuni.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeNardo DG, and Ruffell B (2019). Macrophages as regulators of tumour immunity and immunotherapy. Nat Rev Immunol 19, 369–382. 10.1038/s41577-019-0127-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franklin RA, and Li MO (2016). Ontogeny of Tumor-Associated Macrophages and Its Implication in Cancer Regulation. Trends Cancer 2, 20–34. 10.1016/j.trecan.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pavlova NN, Zhu J, and Thompson CB (2022). The hallmarks of cancer metabolism: Still emerging. Cell Metab 34, 355–377. 10.1016/j.cmet.2022.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vitale I, Manic G, Coussens LM, Kroemer G, and Galluzzi L (2019). Macrophages and Metabolism in the Tumor Microenvironment. Cell Metab 30, 36–50. 10.1016/j.cmet.2019.06.001. [DOI] [PubMed] [Google Scholar]
- 17.Wang S, Liu R, Yu Q, Dong L, Bi Y, and Liu G (2019). Metabolic reprogramming of macrophages during infections and cancer. Cancer Lett 452, 14–22. 10.1016/j.canlet.2019.03.015. [DOI] [PubMed] [Google Scholar]
- 18.Mehla K, and Singh PK (2019). Metabolic Regulation of Macrophage Polarization in Cancer. Trends Cancer 5, 822–834. 10.1016/j.trecan.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Netea-Maier RT, Smit JWA, and Netea MG (2018). Metabolic changes in tumor cells and tumor-associated macrophages: A mutual relationship. Cancer Lett 413, 102–109. 10.1016/j.canlet.2017.10.037. [DOI] [PubMed] [Google Scholar]
- 20.Rabold K, Netea MG, Adema GJ, and Netea-Maier RT (2017). Cellular metabolism of tumor-associated macrophages – functional impact and consequences. Febs Lett 591, 3022–3041. 10.1002/1873-3468.12771. [DOI] [PubMed] [Google Scholar]
- 21.Chen D, Zhang X, Li Z, and Zhu B (2021). Metabolic regulatory crosstalk between tumor microenvironment and tumor-associated macrophages. Theranostics 11, 1016–1030. 10.7150/thno.51777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramos RN, Missolo-Koussou Y, Gerber-Ferder Y, Bromley CP, Bugatti M, Núñez NG, Boari JT, Richer W, Menger L, Denizeau J, et al. (2022). Tissue-resident FOLR2+ macrophages associate with CD8+ T cell infiltration in human breast cancer. Cell. 10.1016/j.cell.2022.02.021. [DOI] [PubMed] [Google Scholar]
- 23.Eraslan G, Drokhlyansky E, Anand S, Fiskin E, Subramanian A, Slyper M, Wang J, Wittenberghe NV, Rouhana JM, Waldman J, et al. (2022). Single-nucleus cross-tissue molecular reference maps toward understanding disease gene function. Science 376, eabl4290. 10.1126/science.abl4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mulder K, Patel AA, Kong WT, Piot C, Halitzki E, Dunsmore G, Khalilnezhad S, Irac SE, Dubuisson A, Chevrier M, et al. (2021). Cross-tissue single-cell landscape of human monocytes and macrophages in health and disease. Immunity 54, 1883–1900 e5. 10.1016/j.immuni.2021.07.007. [DOI] [PubMed] [Google Scholar]
- 25.Zhang L, Li Z, Skrzypczynska KM, Fang Q, Zhang W, O’Brien SA, He Y, Wang L, Zhang Q, Kim A, et al. (2020). Single-Cell Analyses Inform Mechanisms of Myeloid-Targeted Therapies in Colon Cancer. Cell 181, 442–459.e29. 10.1016/j.cell.2020.03.048. [DOI] [PubMed] [Google Scholar]
- 26.Cheng S, Li Z, Gao R, Xing B, Gao Y, Yang Y, Qin S, Zhang L, Ouyang H, Du P, et al. (2021). A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell 184, 792–809.e23. 10.1016/j.cell.2021.01.010. [DOI] [PubMed] [Google Scholar]
- 27.Dawson CA, Pal B, Vaillant F, Gandolfo LC, Liu Z, Bleriot C, Ginhoux F, Smyth GK, Lindeman GJ, Mueller SN, et al. (2020). Tissue-resident ductal macrophages survey the mammary epithelium and facilitate tissue remodelling. Nat Cell Biol 22, 546–558. 10.1038/s41556-020-0505-0. [DOI] [PubMed] [Google Scholar]
- 28.Jäppinen N, Félix I, Lokka E, Tyystjärvi S, Pynttäri A, Lahtela T, Gerke H, Elima K, Rantakari P, and Salmi M (2019). Fetal-derived macrophages dominate in adult mammary glands. Nat Commun 10, 281. 10.1038/s41467-018-08065-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Chaffee TS, LaRue RS, Huggins DN, Witschen PM, Ibrahim AM, Nelson AC, Machado HL, and Schwertfeger KL (2020). Tissue-resident macrophages promote extracellular matrix homeostasis in the mammary gland stroma of nulliparous mice. Elife 9, e57438. 10.7554/elife.57438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prevo R, Banerji S, Ferguson DJP, Clasper S, and Jackson DG (2001). Mouse LYVE-1 Is an Endocytic Receptor for Hyaluronan in Lymphatic Endothelium*. J Biol Chem 276, 19420–19430. 10.1074/jbc.m011004200. [DOI] [PubMed] [Google Scholar]
- 31.Napper CE, Drickamer K, and Taylor ME (2006). Collagen binding by the mannose receptor mediated through the fibronectin type II domain. Biochem J 395, 579–586. 10.1042/bj20052027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chakrabarti R, Celià-Terrassa T, Kumar S, Hang X, Wei Y, Choudhury A, Hwang J, Peng J, Nixon B, Grady JJ, et al. (2018). Notch ligand Dll1 mediates cross-talk between mammary stem cells and the macrophageal niche. Science 360. 10.1126/science.aan4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murtomaki A, Uh MK, Choi YK, Kitajewski C, Borisenko V, Kitajewski J, and Shawber CJ (2013). Notch1 functions as a negative regulator of lymphatic endothelial cell differentiation in the venous endothelium. Development 140, 2365–2376. 10.1242/dev.083865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Franklin RA, Liao W, Sarkar A, Kim MV, Bivona MR, Liu K, Pamer EG, and Li MO (2014). The cellular and molecular origin of tumor-associated macrophages. Science 344, 921–925. 10.1126/science.1252510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laviron M, Petit M, Weber-Delacroix E, Combes AJ, Arkal AR, Barthélémy S, Courau T, Hume DA, Combadière C, Krummel MF, et al. (2022). Tumor-associated macrophage heterogeneity is driven by tissue territories in breast cancer. Cell Reports 39, 110865. 10.1016/j.celrep.2022.110865. [DOI] [PubMed] [Google Scholar]
- 36.Zhou Y, Song WM, Andhey PS, Swain A, Levy T, Miller KR, Poliani PL, Cominelli M, Grover S, Gilfillan S, et al. (2020). Human and mouse single-nucleus transcriptomics reveal TREM2-dependent and TREM2-independent cellular responses in Alzheimer’s disease. Nat Med 26, 131–142. 10.1038/s41591-019-0695-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jaitin DA, Adlung L, Thaiss CA, Weiner A, Li B, Descamps H, Lundgren P, Bleriot C, Liu Z, Deczkowska A, et al. (2019). Lipid-Associated Macrophages Control Metabolic Homeostasis in a Trem2-Dependent Manner. Cell 178, 686–698.e14. 10.1016/j.cell.2019.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Do TH, Ma F, Andrade PR, Teles R, Silva B.J. de A., Hu C, Espinoza A, Hsu J-E, Cho C-S, Kim M, et al. (2022). TREM2 macrophages induced by human lipids drive inflammation in acne lesions. Sci Immunol 7. 10.1126/sciimmunol.abo2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ulland TK, Song WM, Huang SC-C, Ulrich JD, Sergushichev A, Beatty WL, Loboda AA, Zhou Y, Cairns NJ, Kambal A, et al. (2017). TREM2 Maintains Microglial Metabolic Fitness in Alzheimer’s Disease. Cell 170, 649–663.e13. 10.1016/j.cell.2017.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nugent AA, Lin K, Lengerich B. van, Lianoglou S, Przybyla L, Davis SS, Llapashtica C, Wang J, Kim DJ, Xia D, et al. (2020). TREM2 Regulates Microglial Cholesterol Metabolism upon Chronic Phagocytic Challenge. Neuron 105, 837–854.e9. 10.1016/j.neuron.2019.12.007. [DOI] [PubMed] [Google Scholar]
- 41.Molgora M, Esaulova E, Vermi W, Hou J, Chen Y, Luo J, Brioschi S, Bugatti M, Omodei AS, Ricci B, et al. (2020). TREM2 Modulation Remodels the Tumor Myeloid Landscape Enhancing Anti-PD-1 Immunotherapy. Cell 182, 886–900.e17. 10.1016/j.cell.2020.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guerin MV, Finisguerra V, Eynde B.J.V. den, Bercovici N, and Trautmann A (2020). Preclinical murine tumor models: A structural and functional perspective. Elife 9, e50740. 10.7554/elife.50740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhong W, Myers JS, Wang F, Wang K, Lucas J, Rosfjord E, Lucas J, Hooper AT, Yang S, Lemon LA, et al. (2020). Comparison of the molecular and cellular phenotypes of common mouse syngeneic models with human tumors. Bmc Genomics 21, 2. 10.1186/s12864-019-6344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wellenstein MD, and Visser K.E. de (2018). Cancer-Cell-Intrinsic Mechanisms Shaping the Tumor Immune Landscape. Immunity 48, 399–416. 10.1016/j.immuni.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Q, Wang J, Yadav DK, Bai X, and Liang T (2021). Glucose Metabolism: The Metabolic Signature of Tumor Associated Macrophage. Front Immunol 12, 702580. 10.3389/fimmu.2021.702580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reinfeld BI, Madden MZ, Wolf MM, Chytil A, Bader JE, Patterson AR, Sugiura A, Cohen AS, Ali A, Do BT, et al. (2021). Cell-programmed nutrient partitioning in the tumour microenvironment. Nature 593, 282–288. 10.1038/s41586-021-03442-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wenes M, Shang M, Matteo MD, Goveia J, Martin-Perez R, Serneels J, Prenen H, Ghesquiere B, Carmeliet P, and Mazzone M (2016). Macrophage Metabolism Controls Tumor Blood Vessel Morphogenesis and Metastasis. Cell Metab 24, 701–715. 10.1016/j.cmet.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 48.Ning WR, Jiang D, Liu XC, Huang YF, Peng ZP, Jiang ZZ, Kang T, Zhuang SM, Wu Y, and Zheng L (2022). Carbonic anhydrase XII mediates the survival and prometastatic functions of macrophages in human hepatocellular carcinoma. J Clin Invest 132, e153110. 10.1172/jci153110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Penny HL, Sieow JL, Gun SY, Lau MC, Lee B, Tan J, Phua C, Toh F, Nga Y, Yeap WH, et al. (2021). Targeting Glycolysis in Macrophages Confers Protection Against Pancreatic Ductal Adenocarcinoma. Int J Mol Sci 22, 6350. 10.3390/ijms22126350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu D, Chang C, Lu N, Wang X, Lu Q, Ren X, Ren P, Zhao D, Wang L, Zhu Y, et al. (2017). Comprehensive Proteomics Analysis Reveals Metabolic Reprogramming of Tumor-Associated Macrophages Stimulated by the Tumor Microenvironment. J Proteome Res 16, 288–297. 10.1021/acs.jproteome.6b00604. [DOI] [PubMed] [Google Scholar]
- 51.Peng ZP, Jiang ZZ, Guo HF, Zhou MM, Huang YF, Ning WR, Huang JH, Zheng L, and Wu Y (2020). Glycolytic activation of monocytes regulates the accumulation and function of neutrophils in human hepatocellular carcinoma. J Hepatol 73, 906–917. 10.1016/j.jhep.2020.05.004. [DOI] [PubMed] [Google Scholar]
- 52.Chen DP, Ning WR, Jiang ZZ, Peng ZP, Zhu LY, Zhuang SM, Kuang DM, Zheng L, and Wu Y (2019). Glycolytic activation of peritumoral monocytes fosters immune privilege via the PFKFB3-PD-L1 axis in human hepatocellular carcinoma. J Hepatol 71, 333–343. 10.1016/j.jhep.2019.04.007. [DOI] [PubMed] [Google Scholar]
- 53.Penny HL, Sieow JL, Adriani G, Yeap WH, Ee PSC, Luis BS, Lee B, Lee T, Mak SY, Ho YS, et al. (2016). Warburg metabolism in tumor-conditioned macrophages promotes metastasis in human pancreatic ductal adenocarcinoma. Oncoimmunology 5, e1191731. 10.1080/2162402x.2016.1191731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arts RJ, Plantinga TS, Tuit S, Ulas T, Heinhuis B, Tesselaar M, Sloot Y, Adema GJ, Joosten LA, Smit JW, et al. (2016). Transcriptional and metabolic reprogramming induce an inflammatory phenotype in non-medullary thyroid carcinoma-induced macrophages. Oncoimmunology 5, e1229725. 10.1080/2162402x.2016.1229725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miller A, Nagy C, Knapp B, Laengle J, Ponweiser E, Groeger M, Starkl P, Bergmann M, Wagner O, and Haschemi A (2017). Exploring Metabolic Configurations of Single Cells within Complex Tissue Microenvironments. Cell Metab 26, 788–800 e6. 10.1016/j.cmet.2017.08.014. [DOI] [PubMed] [Google Scholar]
- 56.Lu LG, Zhou ZL, Wang XY, Liu BY, Lu JY, Liu S, Zhang GB, Zhan MX, and Chen Y (2022). PD-L1 blockade liberates intrinsic antitumourigenic properties of glycolytic macrophages in hepatocellular carcinoma. Gut, gutjnl-2021–326350. 10.1136/gutjnl-2021-326350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Palsson-McDermott EM, Curtis AM, Goel G, Lauterbach MAR, Sheedy FJ, Gleeson LE, Bosch M.W.M. van den, Quinn SR, Domingo-Fernandez R, Johnston DGW, et al. (2015). Pyruvate Kinase M2 Regulates Hif-1alpha Activity and IL-1beta Induction and Is a Critical Determinant of the Warburg Effect in LPS-Activated Macrophages. Cell Metab 21, 347. 10.1016/j.cmet.2015.01.017. [DOI] [PubMed] [Google Scholar]
- 58.Xu K, Yin N, Peng M, Stamatiades EG, Chhangawala S, Shyu A, Li P, Zhang X, Do MH, Capistrano KJ, et al. (2021). Glycolytic ATP fuels phosphoinositide 3-kinase signaling to support effector T helper 17 cell responses. Immunity 54, 976–987 e7. 10.1016/j.immuni.2021.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu K, Yin N, Peng M, Stamatiades EG, Shyu A, Li P, Zhang X, Do MH, Wang Z, Capistrano KJ, et al. (2021). Glycolysis fuels phosphoinositide 3-kinase signaling to bolster T cell immunity. Science 371, 405–410. 10.1126/science.abb2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shu Y, Yang N, Cheng N, Zou Z, Zhang W, Bei Y, Shi Q, Qin M, Zhu WG, and Shen P (2022). Intervening pyruvate carboxylase stunts tumor growth by strengthening anti-tumor actions of tumor-associated macrophages. Signal Transduct Target Ther 7, 34. 10.1038/s41392-021-00807-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Frank AC, Raue R, Fuhrmann DC, Sirait-Fischer E, Reuse C, Weigert A, Lutjohann D, Hiller K, Syed SN, and Brune B (2021). Lactate dehydrogenase B regulates macrophage metabolism in the tumor microenvironment. Theranostics 11, 7570–7588. 10.7150/thno.58380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seth P, Csizmadia E, Hedblom A, Vuerich M, Xie H, Li M, Longhi MS, and Wegiel B (2017). Deletion of Lactate Dehydrogenase-A in Myeloid Cells Triggers Antitumor Immunity. Cancer Res 77, 3632–3643. 10.1158/0008-5472.can-16-2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Manoharan I, Prasad PD, Thangaraju M, and Manicassamy S (2021). Lactate-Dependent Regulation of Immune Responses by Dendritic Cells and Macrophages. Front Immunol 12, 691134. 10.3389/fimmu.2021.691134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li B, Yang Q, Li Z, Xu Z, Sun S, Wu Q, and Sun S (2020). Expression of Monocarboxylate Transporter 1 in Immunosuppressive Macrophages Is Associated With the Poor Prognosis in Breast Cancer. Frontiers Oncol 10, 574787. 10.3389/fonc.2020.574787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bisetto S, Whitaker-Menezes D, Wilski NA, Tuluc M, Curry J, Zhan T, Snyder CM, Martinez-Outschoorn UE, and Philp NJ (2018). Monocarboxylate Transporter 4 (MCT4) Knockout Mice Have Attenuated 4NQO Induced Carcinogenesis; A Role for MCT4 in Driving Oral Squamous Cell Cancer. Frontiers Oncol 8, 324. 10.3389/fonc.2018.00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen P, Zuo H, Xiong H, Kolar MJ, Chu Q, Saghatelian A, Siegwart DJ, and Wan Y (2017). Gpr132 sensing of lactate mediates tumor-macrophage interplay to promote breast cancer metastasis. Proc National Acad Sci 114, 580–585. 10.1073/pnas.1614035114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vadevoo SMP, Gunassekaran GR, Lee C, Lee N, Lee J, Chae S, Park JY, Koo J, and Lee B (2021). The macrophage odorant receptor Olfr78 mediates the lactate-induced M2 phenotype of tumor-associated macrophages. Proc National Acad Sci 118, e2102434118. 10.1073/pnas.2102434118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Colegio OR, Chu NQ, Szabo AL, Chu T, Rhebergen AM, Jairam V, Cyrus N, Brokowski CE, Eisenbarth SC, Phillips GM, et al. (2014). Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature 513, 559–563. 10.1038/nature13490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang D, Tang Z, Huang H, Zhou G, Cui C, Weng Y, Liu W, Kim S, Lee S, Perez-Neut M, et al. (2019). Metabolic regulation of gene expression by histone lactylation. Nature 574, 575–580. 10.1038/s41586-019-1678-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li S, Yu J, Huber A, Kryczek I, Wang Z, Jiang L, Li X, Du W, Li G, Wei S, et al. (2022). Metabolism drives macrophage heterogeneity in the tumor microenvironment. Cell Reports 39, 110609. 10.1016/j.celrep.2022.110609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shi Q, Shen Q, Liu Y, Shi Y, Huang W, Wang X, Li Z, Chai Y, Wang H, Hu X, et al. (2022). Increased glucose metabolism in TAMs fuels O-GlcNAcylation of lysosomal Cathepsin B to promote cancer metastasis and chemoresistance. Cancer Cell 40, 1207–1222.e10. 10.1016/j.ccell.2022.08.012. [DOI] [PubMed] [Google Scholar]
- 72.Mantuano NR, Stanczak MA, Oliveira IA, Kirchhammer N, Filardy AA, Monaco G, Santos RC, Fonseca AC, Fontes M, S. B,C Jr., et al. (2020). Hyperglycemia Enhances Cancer Immune Evasion by Inducing Alternative Macrophage Polarization through Increased O-GlcNAcylation. Cancer Immunol Res 8, 1262–1272. 10.1158/2326-6066.cir-19-0904. [DOI] [PubMed] [Google Scholar]
- 73.Hinshaw DC, Hanna A, Lama-Sherpa T, Metge B, Kammerud SC, Benavides GA, Kumar A, Alsheikh HA, Mota M, Chen D, et al. (2021). Hedgehog Signaling Regulates Metabolism and Polarization of Mammary Tumor-Associated Macrophages. Cancer Res 81, 5425–5437. 10.1158/0008-5472.can-20-1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Raines LN, Zhao H, Wang Y, Chen HY, Gallart-Ayala H, Hsueh PC, Cao W, Koh Y, Alamonte-Loya A, Liu PS, et al. (2022). PERK is a critical metabolic hub for immunosuppressive function in macrophages. Nat Immunol 23, 431–445. 10.1038/s41590-022-01145-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ryan DG, and O’Neill LAJ (2020). Krebs Cycle Reborn in Macrophage Immunometabolism. Annu Rev Immunol 38, 289–313. 10.1146/annurev-immunol-081619-104850. [DOI] [PubMed] [Google Scholar]
- 76.Henze A-T, and Mazzone M (2016). The impact of hypoxia on tumor-associated macrophages. J Clin Invest 126, 3672–3679. 10.1172/jci84427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Umemura N, Sugimoto M, Kitoh Y, Saio M, and Sakagami H (2020). Metabolomic profiling of tumor-infiltrating macrophages during tumor growth. Cancer Immunol Immunother 69, 2357–2369. 10.1007/s00262-020-02622-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weiss JM, Davies LC, Karwan M, Ileva L, Ozaki MK, Cheng RY, Ridnour LA, Annunziata CM, Wink DA, and McVicar DW (2018). Itaconic acid mediates crosstalk between macrophage metabolism and peritoneal tumors. J Clin Invest 128, 3794–3805. 10.1172/jci99169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pires-Afonso Y, Muller A, Grzyb K, Oudin A, Yabo YA, Sousa C, Scafidi A, Poli A, Cosma A, Halder R, et al. (2022). Elucidating tumour-associated microglia/macrophage diversity along glioblastoma progression and under ACOD1 deficiency. Mol Oncol. 10.1002/1878-0261.13287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.O’Neill LAJ, and Artyomov MN (2019). Itaconate: the poster child of metabolic reprogramming in macrophage function. Nat Rev Immunol 19, 273–281. 10.1038/s41577-019-0128-5. [DOI] [PubMed] [Google Scholar]
- 81.Wu JY, Huang TW, Hsieh YT, Wang YF, Yen CC, Lee GL, Yeh CC, Peng YJ, Kuo YY, Wen HT, et al. (2020). Cancer-Derived Succinate Promotes Macrophage Polarization and Cancer Metastasis via Succinate Receptor. Mol Cell 77, 213–227 e5. 10.1016/j.molcel.2019.10.023. [DOI] [PubMed] [Google Scholar]
- 82.Tannahill GM, Curtis AM, Adamik J, Palsson-McDermott EM, McGettrick AF, Goel G, Frezza C, Bernard NJ, Kelly B, Foley NH, et al. (2013). Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature 496, 238–242. 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mills EL, Kelly B, Logan A, Costa ASH, Varma M, Bryant CE, Tourlomousis P, Dabritz JHM, Gottlieb E, Latorre I, et al. (2016). Succinate Dehydrogenase Supports Metabolic Repurposing of Mitochondria to Drive Inflammatory Macrophages. Cell 167, 457–470 e13. 10.1016/j.cell.2016.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Su P, Wang Q, Bi E, Ma X, Liu L, Yang M, Qian J, and Yi Q (2020). Enhanced Lipid Accumulation and Metabolism Are Required for the Differentiation and Activation of Tumor-Associated Macrophages. Cancer Res 80, 1438–1450. 10.1158/0008-5472.can-19-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xiang W, Shi R, Kang X, Zhang X, Chen P, Zhang L, Hou A, Wang R, Zhao Y, Zhao K, et al. (2018). Monoacylglycerol lipase regulates cannabinoid receptor 2-dependent macrophage activation and cancer progression. Nat Commun 9, 2574. 10.1038/s41467-018-04999-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wu H, Han Y, Sillke YR, Deng H, Siddiqui S, Treese C, Schmidt F, Friedrich M, Keye J, Wan J, et al. (2019). Lipid droplet-dependent fatty acid metabolism controls the immune suppressive phenotype of tumor-associated macrophages. Embo Mol Med 11, e10698. 10.15252/emmm.201910698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Niu Z, Shi Q, Zhang W, Shu Y, Yang N, Chen B, Wang Q, Zhao X, Chen J, Cheng N, et al. (2017). Caspase-1 cleaves PPARgamma for potentiating the pro-tumor action of TAMs. Nat Commun 8, 766. 10.1038/s41467-017-00523-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schumann T, Adhikary T, Wortmann A, Finkernagel F, Lieber S, Schnitzer E, Legrand N, Schober Y, Nockher WA, Toth PM, et al. (2015). Deregulation of PPARbeta/delta target genes in tumor-associated macrophages by fatty acid ligands in the ovarian cancer microenvironment. Oncotarget 6, 13416–13433. 10.18632/oncotarget.3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.He Y, Dong Y, Zhang X, Ding Z, Song Y, Huang X, Chen S, Wang Z, Ni Y, and Ding L (2022). Lipid Droplet-Related PLIN2 in CD68(+) Tumor-Associated Macrophage of Oral Squamous Cell Carcinoma: Implications for Cancer Prognosis and Immunotherapy. Frontiers Oncol 12, 824235. 10.3389/fonc.2022.824235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Silverstein RL, and Febbraio M (2009). CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci Signal 2, re3. 10.1126/scisignal.272re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pfeiler S, Thakur M, Grunauer P, Megens RTA, Joshi U, Coletti R, Samara V, Muller-Stoy G, Ishikawa-Ankerhold H, Stark K, et al. (2019). CD36-triggered cell invasion and persistent tissue colonization by tumor microvesicles during metastasis. Faseb J 33, 1860–1872. 10.1096/fj.201800985r. [DOI] [PubMed] [Google Scholar]
- 92.Yan J, and Horng T (2020). Lipid Metabolism in Regulation of Macrophage Functions. Trends Cell Biol 30, 979–989. 10.1016/j.tcb.2020.09.006. [DOI] [PubMed] [Google Scholar]
- 93.Goede K.E. de, Verberk SGS, Baardman J, Harber KJ, Kooyk Y. van, Winther M.P.J. de, Schetters STT, and Bossche J.V. den (2021). Myeloid-Specific Acly Deletion Alters Macrophage Phenotype In Vitro and In Vivo without Affecting Tumor Growth. Cancers 13, 3054. 10.3390/cancers13123054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Furuhashi M, and Hotamisligil GS (2008). Fatty acid-binding proteins: role in metabolic diseases and potential as drug targets. Nat Rev Drug Discov 7, 489–503. 10.1038/nrd2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang Y, Sun Y, Rao E, Yan F, Li Q, Zhang Y, Silverstein KA, Liu S, Sauter E, Cleary MP, et al. (2014). Fatty acid-binding protein E-FABP restricts tumor growth by promoting IFN-beta responses in tumor-associated macrophages. Cancer Res 74, 2986–2998. 10.1158/0008-5472.can-13-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hao J, Yan F, Zhang Y, Triplett A, Zhang Y, Schultz DA, Sun Y, Zeng J, Silverstein KAT, Zheng Q, et al. (2018). Expression of Adipocyte/Macrophage Fatty Acid-Binding Protein in Tumor-Associated Macrophages Promotes Breast Cancer Progression. Cancer Res 78, 2343–2355. 10.1158/0008-5472.can-17-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Castoldi A, Monteiro LB, Bakker N. van T., Sanin DE, Rana N, Corrado M, Cameron AM, Hässler F, Matsushita M, Caputa G, et al. (2020). Triacylglycerol synthesis enhances macrophage inflammatory function. Nat Commun 11, 4107. 10.1038/s41467-020-17881-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xu M, Wang X, Li Y, Geng X, Jia X, Zhang L, and Yang H (2021). Arachidonic Acid Metabolism Controls Macrophage Alternative Activation Through Regulating Oxidative Phosphorylation in PPARgamma Dependent Manner. Front Immunol 12, 618501. 10.3389/fimmu.2021.618501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Eruslanov E, Daurkin I, Vieweg J, Daaka Y, and Kusmartsev S (2011). Aberrant PGE(2) metabolism in bladder tumor microenvironment promotes immunosuppressive phenotype of tumor-infiltrating myeloid cells. Int Immunopharmacol 11, 848–855. 10.1016/j.intimp.2011.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kale S, Raja R, Thorat D, Soundararajan G, Patil TV, and Kundu GC (2014). Osteopontin signaling upregulates cyclooxygenase-2 expression in tumor-associated macrophages leading to enhanced angiogenesis and melanoma growth via alpha9beta1 integrin. Oncogene 33, 2295–2306. 10.1038/onc.2013.184. [DOI] [PubMed] [Google Scholar]
- 101.Prima V, Kaliberova LN, Kaliberov S, Curiel DT, and Kusmartsev S (2017). COX2/mPGES1/PGE2 pathway regulates PD-L1 expression in tumor-associated macrophages and myeloid-derived suppressor cells. Proc National Acad Sci 114, 1117–1122. 10.1073/pnas.1612920114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hsiao YW, Li CF, Chi JY, Tseng JT, Chang Y, Hsu LJ, Lee CH, Chang TH, Wang SM, Wang DD, et al. (2013). CCAAT/enhancer binding protein delta in macrophages contributes to immunosuppression and inhibits phagocytosis in nasopharyngeal carcinoma. Sci Signal 6, ra59. 10.1126/scisignal.2003648. [DOI] [PubMed] [Google Scholar]
- 103.Larsson K, Kock A, Idborg H, Henriksson MA, Martinsson T, Johnsen JI, Korotkova M, Kogner P, and Jakobsson P-J (2015). COX/mPGES-1/PGE2 pathway depicts an inflammatory-dependent high-risk neuroblastoma subset. Proc National Acad Sci 112, 8070–8075. 10.1073/pnas.1424355112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ringleb J, Strack E, Angioni C, Geisslinger G, Steinhilber D, Weigert A, and Brune B (2018). Apoptotic Cancer Cells Suppress 5-Lipoxygenase in Tumor-Associated Macrophages. J Immunol 200, 857–868. 10.4049/jimmunol.1700609. [DOI] [PubMed] [Google Scholar]
- 105.Nosaka T, Baba T, Tanabe Y, Sasaki S, Nishimura T, Imamura Y, Yurino H, Hashimoto S, Arita M, Nakamoto Y, et al. (2018). Alveolar Macrophages Drive Hepatocellular Carcinoma Lung Metastasis by Generating Leukotriene B4. J Immunol 200, 1839–1852. 10.4049/jimmunol.1700544. [DOI] [PubMed] [Google Scholar]
- 106.Staquicini FI, Hajitou A, Driessen WH, Proneth B, Cardo-Vila M, Staquicini DI, Markosian C, Hoh M, Cortez M, Hooda-Nehra A, et al. (2021). Targeting a cell surface vitamin D receptor on tumor-associated macrophages in triple-negative breast cancer. Elife 10, e65145. 10.7554/elife.65145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dmitrieva-Posocco O, Wong AC, Lundgren P, Golos AM, Descamps HC, Dohnalová L, Cramer Z, Tian Y, Yueh B, Eskiocak O, et al. (2022). β-Hydroxybutyrate suppresses colorectal cancer. Nature 605, 160–165. 10.1038/s41586-022-04649-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Youm Y-H, Nguyen KY, Grant RW, Goldberg EL, Bodogai M, Kim D, D’Agostino D, Planavsky N, Lupfer C, Kanneganti TD, et al. (2015). The ketone metabolite β-hydroxybutyrate blocks NLRP3 inflammasome–mediated inflammatory disease. Nat Med 21, 263–269. 10.1038/nm.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Puchalska P, Martin SE, Huang X, Lengfeld JE, Daniel B, Graham MJ, Han X, Nagy L, Patti GJ, and Crawford PA (2019). Hepatocyte-Macrophage Acetoacetate Shuttle Protects against Tissue Fibrosis. Cell Metab 29, 383–398 e7. 10.1016/j.cmet.2018.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, Thangaraju M, Prasad PD, Manicassamy S, Munn DH, et al. (2014). Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 40, 128–139. 10.1016/j.immuni.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rahman M, Muhammad S, Khan MA, Chen H, Ridder DA, Muller-Fielitz H, Pokorna B, Vollbrandt T, Stolting I, Nadrowitz R, et al. (2014). The beta-hydroxybutyrate receptor HCA2 activates a neuroprotective subset of macrophages. Nat Commun 5, 3944. 10.1038/ncomms4944. [DOI] [PubMed] [Google Scholar]
- 112.Chen Y, Ouyang X, Hoque R, Garcia-Martinez I, Yousaf MN, Tonack S, Offermanns S, Dubuquoy L, Louvet A, Mathurin P, et al. (2018). beta-Hydroxybutyrate protects from alcohol-induced liver injury via a Hcar2-cAMP dependent pathway. J Hepatol 69, 687–696. 10.1016/j.jhep.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Huang C, Wang J, Liu H, Huang R, Yan X, Song M, Tan G, and Zhi F (2022). Ketone body beta-hydroxybutyrate ameliorates colitis by promoting M2 macrophage polarization through the STAT6-dependent signaling pathway. Bmc Med 20, 148. 10.1186/s12916-022-02352-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liu GY, and Sabatini DM (2020). mTOR at the nexus of nutrition, growth, ageing and disease. Nat Rev Mol Cell Bio 21, 183–203. 10.1038/s41580-019-0199-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cantor JM, and Ginsberg MH (2012). CD98 at the crossroads of adaptive immunity and cancer. J Cell Sci 125, 1373–1382. 10.1242/jcs.096040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dodd KM, and Tee AR (2012). Leucine and mTORC1: a complex relationship. Am J Physiol-endoc M 302, E1329–42. 10.1152/ajpendo.00525.2011. [DOI] [PubMed] [Google Scholar]
- 117.Yoon BR, Oh YJ, Kang SW, Lee EB, and Lee WW (2018). Role of SLC7A5 in Metabolic Reprogramming of Human Monocyte/Macrophage Immune Responses. Front Immunol 9, 53. 10.3389/fimmu.2018.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Munn DH, Shafizadeh E, Attwood JT, Bondarev I, Pashine A, and Mellor AL (1999). Inhibition of T cell proliferation by macrophage tryptophan catabolism. J Exp Medicine 189, 1363–1372. 10.1084/jem.189.9.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ravishankar B, Liu H, Shinde R, Chandler P, Baban B, Tanaka M, Munn DH, Mellor AL, Karlsson MC, and McGaha TL (2012). Tolerance to apoptotic cells is regulated by indoleamine 2,3-dioxygenase. Proc National Acad Sci 109, 3909–3914. 10.1073/pnas.1117736109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Takenaka MC, Gabriely G, Rothhammer V, Mascanfroni ID, Wheeler MA, Chao CC, Gutierrez-Vazquez C, Kenison J, Tjon EC, Barroso A, et al. (2019). Control of tumor-associated macrophages and T cells in glioblastoma via AHR and CD39. Nat Neurosci 22, 729–740. 10.1038/s41593-019-0370-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Campesato LF, Budhu S, Tchaicha J, Weng CH, Gigoux M, Cohen IJ, Redmond D, Mangarin L, Pourpe S, Liu C, et al. (2020). Blockade of the AHR restricts a Treg-macrophage suppressive axis induced by L-Kynurenine. Nat Commun 11, 4011. 10.1038/s41467-020-17750-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hezaveh K, Shinde RS, Klotgen A, Halaby MJ, Lamorte S, Ciudad MT, Quevedo R, Neufeld L, Liu ZQ, Jin R, et al. (2022). Tryptophan-derived microbial metabolites activate the aryl hydrocarbon receptor in tumor-associated macrophages to suppress anti-tumor immunity. Immunity 55, 324–340 e8. 10.1016/j.immuni.2022.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sadik A, Patterson LFS, Ozturk S, Mohapatra SR, Panitz V, Secker PF, Pfander P, Loth S, Salem H, Prentzell MT, et al. (2020). IL4I1 Is a Metabolic Immune Checkpoint that Activates the AHR and Promotes Tumor Progression. Cell 182, 1252–1270 e34. 10.1016/j.cell.2020.07.038. [DOI] [PubMed] [Google Scholar]
- 124.Wang J, Simonavicius N, Wu X, Swaminath G, Reagan J, Tian H, and Ling L (2006). Kynurenic acid as a ligand for orphan G protein-coupled receptor GPR35. J Biol Chem 281, 22021–22028. 10.1074/jbc.m603503200. [DOI] [PubMed] [Google Scholar]
- 125.Pagano E, Elias JE, Schneditz G, Saveljeva S, Holland LM, Borrelli F, Karlsen TH, Kaser A, and Kaneider NC (2022). Activation of the GPR35 pathway drives angiogenesis in the tumour microenvironment. Gut 71, 509–520. 10.1136/gutjnl-2020-323363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Halaby MJ, and McGaha TL (2021). Amino Acid Transport and Metabolism in Myeloid Function. Front Immunol 12, 695238. 10.3389/fimmu.2021.695238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.A. C,R Jr., Stewart TM, and Pegg AE (2018). Polyamine metabolism and cancer: treatments, challenges and opportunities. Nat Rev Cancer 18, 681–695. 10.1038/s41568-018-0050-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Grzywa TM, Sosnowska A, Matryba P, Rydzynska Z, Jasinski M, Nowis D, and Golab J (2020). Myeloid Cell-Derived Arginase in Cancer Immune Response. Front Immunol 11, 938. 10.3389/fimmu.2020.00938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Doedens AL, Stockmann C, Rubinstein MP, Liao D, Zhang N, DeNardo DG, Coussens LM, Karin M, Goldrath AW, and Johnson RS (2010). Macrophage expression of hypoxia-inducible factor-1 alpha suppresses T-cell function and promotes tumor progression. Cancer Res 70, 7465–7475. 10.1158/0008-5472.can-10-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Su X, Xu Y, Fox GC, Xiang J, Kwakwa KA, Davis JL, Belle JI, Lee WC, Wong WH, Fontana F, et al. (2021). Breast cancer-derived GM-CSF regulates arginase 1 in myeloid cells to promote an immunosuppressive microenvironment. J Clin Invest 131. 10.1172/jci145296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Carmona-Fontaine C, Deforet M, Akkari L, Thompson CB, Joyce JA, and Xavier JB (2017). Metabolic origins of spatial organization in the tumor microenvironment. P Natl Acad Sci Usa 114, 2934–2939. 10.1073/pnas.1700600114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ji L, Zhao X, Zhang B, Kang L, Song W, Zhao B, Xie W, Chen L, and Hu X (2019). Slc6a8-Mediated Creatine Uptake and Accumulation Reprogram Macrophage Polarization via Regulating Cytokine Responses. Immunity 51, 272–284 e7. 10.1016/j.immuni.2019.06.007. [DOI] [PubMed] [Google Scholar]
- 133.Rodriguez PC, Zea AH, DeSalvo J, Culotta KS, Zabaleta J, Quiceno DG, Ochoa JB, and Ochoa AC (2003). L-arginine consumption by macrophages modulates the expression of CD3 zeta chain in T lymphocytes. J Immunol 171, 1232–1239. 10.4049/jimmunol.171.3.1232. [DOI] [PubMed] [Google Scholar]
- 134.Rodriguez PC, Quiceno DG, Zabaleta J, Ortiz B, Zea AH, Piazuelo MB, Delgado A, Correa P, Brayer J, Sotomayor EM, et al. (2004). Arginase I production in the tumor microenvironment by mature myeloid cells inhibits T-cell receptor expression and antigen-specific T-cell responses. Cancer Res 64, 5839–5849. 10.1158/0008-5472.can-04-0465. [DOI] [PubMed] [Google Scholar]
- 135.Bozkus CC, Elzey BD, Crist SA, Ellies LG, and Ratliff TL (2015). Expression of Cationic Amino Acid Transporter 2 Is Required for Myeloid-Derived Suppressor Cell-Mediated Control of T Cell Immunity. J Immunol 195, 5237–5250. 10.4049/jimmunol.1500959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Steggerda SM, Bennett MK, Chen J, Emberley E, Huang T, Janes JR, Li W, MacKinnon AL, Makkouk A, Marguier G, et al. (2017). Inhibition of arginase by CB-1158 blocks myeloid cell-mediated immune suppression in the tumor microenvironment. J Immunother Cancer 5, 101. 10.1186/s40425-017-0308-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Menjivar RE, Nwosu ZC, Du W, Donahue KL, Espinoza C, Brown K, Velez-Delgado A, Yan W, Lima F, Bischoff A, et al. (2022). Arginase 1 is a key driver of immune suppression in pancreatic cancer. Biorxiv, 2022.06.21.497084. 10.1101/2022.06.21.497084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Miska J, Rashidi A, Lee-Chang C, Gao P, Lopez-Rosas A, Zhang P, Burga R, Castro B, Xiao T, Han Y, et al. (2021). Polyamines drive myeloid cell survival by buffering intracellular pH to promote immunosuppression in glioblastoma. Sci Adv 7, eabc8929. 10.1126/sciadv.abc8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.McCubbrey AL, McManus SA, McClendon JD, Thomas SM, Chatwin HB, Reisz JA, D’Alessandro A, Mould KJ, Bratton DL, Henson PM, et al. (2022). Polyamine import and accumulation causes immunomodulation in macrophages engulfing apoptotic cells. Cell Reports 38, 110222. 10.1016/j.celrep.2021.110222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Oh MH, Sun IH, Zhao L, Leone RD, Sun IM, Xu W, Collins SL, Tam AJ, Blosser RL, Patel CH, et al. (2020). Targeting glutamine metabolism enhances tumor-specific immunity by modulating suppressive myeloid cells. J Clin Invest 130, 3865–3884. 10.1172/jci131859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Merlin J, Ivanov S, Dumont A, Sergushichev A, Gall J, Stunault M, Ayrault M, Vaillant N, Castiglione A, Swain A, et al. (2021). Non-canonical glutamine transamination sustains efferocytosis by coupling redox buffering to oxidative phosphorylation. Nat Metabolism 3, 1313–1326. 10.1038/s42255-021-00471-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Palmieri EM, Menga A, Martin-Perez R, Quinto A, Riera-Domingo C, Tullio GD, Hooper DC, Lamers WH, Ghesquiere B, McVicar DW, et al. (2017). Pharmacologic or Genetic Targeting of Glutamine Synthetase Skews Macrophages toward an M1-like Phenotype and Inhibits Tumor Metastasis. Cell Reports 20, 1654–1666. 10.1016/j.celrep.2017.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Eichbaum QG, Iyer R, Raveh DP, Mathieu C, and Ezekowitz RA (1994). Restriction of interferon gamma responsiveness and basal expression of the myeloid human Fc gamma R1b gene is mediated by a functional PU.1 site and a transcription initiator consensus. J Exp Medicine 179, 1985–1996. 10.1084/jem.179.6.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Walsh AD, Johnson LJ, Harvey AJ, Kilpatrick TJ, and Binder MD (2020). Identification and Characterisation of cis-Regulatory Elements Upstream of the Human Receptor Tyrosine Kinase Gene MERTK. Adv Neurol 7, 3–16. 10.3233/bpl-200102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lemke G, and Rothlin CV (2008). Immunobiology of the TAM receptors. Nat Rev Immunol 8, 327–336. 10.1038/nri2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Myers KV, Amend SR, and Pienta KJ (2019). Targeting Tyro3, Axl and MerTK (TAM receptors): implications for macrophages in the tumor microenvironment. Mol Cancer 18, 94. 10.1186/s12943-019-1022-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Seitz HM, Camenisch TD, Lemke G, Earp HS, and Matsushima GK (2007). Macrophages and Dendritic Cells Use Different Axl/Mertk/Tyro3 Receptors in Clearance of Apoptotic Cells. J Immunol 178, 5635–5642. 10.4049/jimmunol.178.9.5635. [DOI] [PubMed] [Google Scholar]
- 148.Lu Q, Gore M, Zhang Q, Camenisch T, Boast S, Casagranda F, Lai C, Skinner MK, Klein R, Matsushima GK, et al. (1999). Tyro-3 family receptors are essential regulators of mammalian spermatogenesis. Nature 398, 723–728. 10.1038/19554. [DOI] [PubMed] [Google Scholar]
- 149.Stanford JC, Young C, Hicks D, Owens P, Williams A, Vaught DB, Morrison MM, Lim J, Williams M, Brantley-Sieders DM, et al. (2014). Efferocytosis produces a prometastatic landscape during postpartum mammary gland involution. J Clin Invest 124, 4737–4752. 10.1172/jci76375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sandahl M, Hunter DM, Strunk KE, Earp HS, and Cook RS (2010). Epithelial cell-directed efferocytosis in the post-partum mammary gland is necessary for tissue homeostasis and future lactation. Bmc Dev Biol 10, 122–122. 10.1186/1471-213x-10-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Crittenden MR, Baird J, Friedman D, Savage T, Uhde L, Alice A, Cottam B, Young K, Newell P, Nguyen C, et al. (2016). Mertk on tumor macrophages is a therapeutic target to prevent tumor recurrence following radiation therapy. Oncotarget 7, 78653–78666. 10.18632/oncotarget.11823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Farnworth-McHugh S, Barth N, Melville L, Paterson M, Lynch C, Holland P, Dransfield I, and Gregory C (2020). Potential Oncogenic Effect of the MERTK-Dependent Apoptotic-Cell Clearance Pathway in Starry-Sky B-Cell Lymphoma. Front Immunol 11, 1759. 10.3389/fimmu.2020.01759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dick SA, Macklin JA, Nejat S, Momen A, Clemente-Casares X, Althagafi MG, Chen J, Kantores C, Hosseinzadeh S, Aronoff L, et al. (2019). Self-renewing resident cardiac macrophages limit adverse remodeling following myocardial infarction. Nat Immunol 20, 29–39. 10.1038/s41590-018-0272-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Shaw TN, Houston SA, Wemyss K, Bridgeman HM, Barbera TA, Zangerle-Murray T, Strangward P, Ridley AJL, Wang P, Tamoutounour S, et al. (2018). Tissue-resident macrophages in the intestine are long lived and defined by Tim-4 and CD4 expression. J Exp Medicine 215, 1507–1518. 10.1084/jem.20180019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Chen Q, and Ruedl C (2020). Obesity retunes turnover kinetics of tissue-resident macrophages in fat. J Leukocyte Biol 107, 773–782. 10.1002/jlb.1ma1219-275r. [DOI] [PubMed] [Google Scholar]
- 156.Wong K, Valdez PA, Tan C, Yeh S, Hongo J-A, and Ouyang W (2010). Phosphatidylserine receptor Tim-4 is essential for the maintenance of the homeostatic state of resident peritoneal macrophages. Proc National Acad Sci 107, 8712–8717. 10.1073/pnas.0910929107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ni M, Zhang J, Sosa R, Zhang H, Wang H, Jin D, Crowley K, Naini B, Reed FE, Busuttil RW, et al. (2021). T-Cell Immunoglobulin and Mucin Domain-Containing Protein-4 Is Critical for Kupffer Cell Homeostatic Function in the Activation and Resolution of Liver Ischemia Reperfusion Injury. Hepatology 74, 2118–2132. 10.1002/hep.31906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Chow A, Schad S, Green MD, Hellmann MD, Allaj V, Ceglia N, Zago G, Shah NS, Sharma SK, Mattar M, et al. (2021). Tim-4+ cavity-resident macrophages impair anti-tumor CD8+ T cell immunity. Cancer Cell 39, 973–988.e9. 10.1016/j.ccell.2021.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Xia H, Li S, Li X, Wang W, Bian Y, Wei S, Grove S, Wang W, Vatan L, Liu JR, et al. (2020). Autophagic adaptation to oxidative stress alters peritoneal residential macrophage survival and ovarian cancer metastasis. Jci Insight 5, e141115. 10.1172/jci.insight.141115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.A-Gonzalez N, Bensinger SJ, Hong C, Beceiro S, Bradley MN, Zelcer N, Deniz J, Ramirez C, Díaz M, Gallardo G, et al. (2009). Apoptotic Cells Promote Their Own Clearance and Immune Tolerance through Activation of the Nuclear Receptor LXR. Immunity 31, 245–258. 10.1016/j.immuni.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Mukundan L, Odegaard JI, Morel CR, Heredia JE, Mwangi JW, Ricardo-Gonzalez RR, Goh YPS, Eagle AR, Dunn SE, Awakuni JUH, et al. (2009). PPAR-δ senses and orchestrates clearance of apoptotic cells to promote tolerance. Nat Med 15, 1266–1272. 10.1038/nm.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Rőszer T, Menéndez-Gutiérrez MP, Lefterova MI, Alameda D, Núñez V, Lazar MA, Fischer T, and Ricote M (2011). Autoimmune Kidney Disease and Impaired Engulfment of Apoptotic Cells in Mice with Macrophage Peroxisome Proliferator-Activated Receptor γ or Retinoid X Receptor α Deficiency. J Immunol 186, 621–631. 10.4049/jimmunol.1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Schneider C, King RM, and Philipson L (1988). Genes specifically expressed at growth arrest of mammalian cells. Cell 54, 787–793. 10.1016/s0092-8674(88)91065-3. [DOI] [PubMed] [Google Scholar]
- 164.Viaud M, Ivanov S, Vujic N, Duta-Mare M, Aira L-E, Barouillet T, Garcia E, Orange F, Dugail I, Hainault I, et al. (2018). Lysosomal Cholesterol Hydrolysis Couples Efferocytosis to Anti-Inflammatory Oxysterol Production. Circ Res 122, 1369–1384. 10.1161/circresaha.117.312333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mota AC, Dominguez M, Weigert A, Snodgrass RG, Namgaladze D, and Brüne B (2021). Lysosome-Dependent LXR and PPARδ Activation Upon Efferocytosis in Human Macrophages. Front Immunol 12, 637778. 10.3389/fimmu.2021.637778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Snodgrass RG, Benatzy Y, Schmid T, Namgaladze D, Mainka M, Schebb NH, Lütjohann D, and Brüne B (2021). Efferocytosis potentiates the expression of arachidonate 15-lipoxygenase (ALOX15) in alternatively activated human macrophages through LXR activation. Cell Death Differ 28, 1301–1316. 10.1038/s41418-020-00652-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kawane K, Ohtani M, Miwa K, Kizawa T, Kanbara Y, Yoshioka Y, Yoshikawa H, and Nagata S (2006). Chronic polyarthritis caused by mammalian DNA that escapes from degradation in macrophages. Nature 443, 998–1002. 10.1038/nature05245. [DOI] [PubMed] [Google Scholar]
- 168.Saito Y, Hikita H, Nozaki Y, Kai Y, Makino Y, Nakabori T, Tanaka S, Yamada R, Shigekawa M, Kodama T, et al. (2019). DNase II activated by the mitochondrial apoptotic pathway regulates RIP1-dependent non-apoptotic hepatocyte death via the TLR9/IFN-β signaling pathway. Cell Death Differ 26, 470–486. 10.1038/s41418-018-0131-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Rudnik S, and Damme M (2021). The lysosomal membrane—export of metabolites and beyond. Febs J 288, 4168–4182. 10.1111/febs.15602. [DOI] [PubMed] [Google Scholar]
- 170.Yurdagul A, Subramanian M, Wang X, Crown SB, Ilkayeva OR, Darville L, Kolluru GK, Rymond CC, Gerlach BD, Zheng Z, et al. (2020). Macrophage Metabolism of Apoptotic Cell-Derived Arginine Promotes Continual Efferocytosis and Resolution of Injury. Cell Metab 31, 518–533.e10. 10.1016/j.cmet.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Cunha LD, Yang M, Carter R, Guy C, Harris L, Crawford JC, Quarato G, Boada-Romero E, Kalkavan H, Johnson MDL, et al. (2018). LC3-Associated Phagocytosis in Myeloid Cells Promotes Tumor Immune Tolerance. Cell 175, 429–441.e16. 10.1016/j.cell.2018.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.A-Gonzalez N, Quintana JA, García-Silva S, Mazariegos M, Aleja A.G. de la, Nicolás-Ávila JA, Walter W, Adrover JM, Crainiciuc G, Kuchroo VK, et al. (2017). Phagocytosis imprints heterogeneity in tissue-resident macrophages. J Exp Med 214, 1281–1296. 10.1084/jem.20161375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Martinez-Pomares L, Wienke D, Stillion R, McKenzie EJ, Arnold JN, Harris J, McGreal E, Sim RB, Isacke CM, and Gordon S (2006). Carbohydrate-independent recognition of collagens by the macrophage mannose receptor. Eur J Immunol 36, 1074–1082. 10.1002/eji.200535685. [DOI] [PubMed] [Google Scholar]
- 174.Madsen DH, Ingvarsen S, Jürgensen HJ, Melander MC, Kjøller L, Moyer A, Honoré C, Madsen CA, Garred P, Burgdorf S, et al. (2011). The Non-phagocytic Route of Collagen Uptake A DISTINCT DEGRADATION PATHWAY*. J Biol Chem 286, 26996–27010. 10.1074/jbc.m110.208033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Madsen DH, Jürgensen HJ, Siersbæk MS, Kuczek DE, Cloud LG, Liu S, Behrendt N, Grøntved L, Weigert R, and Bugge TH (2017). Tumor-Associated Macrophages Derived from Circulating Inflammatory Monocytes Degrade Collagen through Cellular Uptake. Cell Reports 21, 3662–3671. 10.1016/j.celrep.2017.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Haque ASMR, Moriyama M, Kubota K, Ishiguro N, Sakamoto M, Chinju A, Mochizuki K, Sakamoto T, Kaneko N, Munemura R, et al. (2019). CD206+ tumor-associated macrophages promote proliferation and invasion in oral squamous cell carcinoma via EGF production. Sci Rep-uk 9, 14611. 10.1038/s41598-019-51149-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Linde N, Casanova-Acebes M, Sosa MS, Mortha A, Rahman A, Farias E, Harper K, Tardio E, Torres IR, Jones J, et al. (2018). Macrophages orchestrate breast cancer early dissemination and metastasis. Nat Commun 9, 21. 10.1038/s41467-017-02481-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Chakarov S, Lim HY, Tan L, Lim SY, See P, Lum J, Zhang X-M, Foo S, Nakamizo S, Duan K, et al. (2019). Two distinct interstitial macrophage populations coexist across tissues in specific subtissular niches. Science 363. 10.1126/science.aau0964. [DOI] [PubMed] [Google Scholar]
- 179.Opzoomer JW, Anstee JE, Dean I, Hill EJ, Bouybayoune I, Caron J, Muliaditan T, Gordon P, Sosnowska D, Nuamah R, et al. (2021). Macrophages orchestrate the expansion of a proangiogenic perivascular niche during cancer progression. Sci Adv 7, eabg9518. 10.1126/sciadv.abg9518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ding C, Sun X, Wu C, Hu X, Zhang H-G, and Yan J (2018). Tumor Microenvironment Modulates Immunological Outcomes of Myeloid Cells with mTORC1 Disruption. J Immunol Baltim Md 1950 202, 1623–1634. 10.4049/jimmunol.1801112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Hesketh GG, Papazotos F, Pawling J, Rajendran D, Knight JDR, Martinez S, Taipale M, Schramek D, Dennis JW, and Gingras A-C (2020). The GATOR–Rag GTPase pathway inhibits mTORC1 activation by lysosome-derived amino acids. Science 370, 351–356. 10.1126/science.aaz0863. [DOI] [PubMed] [Google Scholar]
- 182.Meng D, Yang Q, Jeong M-H, Curukovic A, Tiwary S, Melick CH, Lama-Sherpa TD, Wang H, Huerta-Rosario M, Urquhart G, et al. (2022). SNAT7 regulates mTORC1 via macropinocytosis. P Natl Acad Sci Usa 119, e2123261119. 10.1073/pnas.2123261119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Atagi Y, Liu C-C, Painter MM, Chen X-F, Verbeeck C, Zheng H, Li X, Rademakers R, Kang SS, Xu H, et al. (2015). Apolipoprotein E Is a Ligand for Triggering Receptor Expressed on Myeloid Cells 2 (TREM2)*. J Biol Chem 290, 26043–26050. 10.1074/jbc.m115.679043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Yeh FL, Wang Y, Tom I, Gonzalez LC, and Sheng M (2016). TREM2 Binds to Apolipoproteins, Including APOE and CLU/APOJ, and Thereby Facilitates Uptake of Amyloid-Beta by Microglia. Neuron 91, 328–340. 10.1016/j.neuron.2016.06.015. [DOI] [PubMed] [Google Scholar]
- 185.Bailey CC, DeVaux LB, and Farzan M (2015). The Triggering Receptor Expressed on Myeloid Cells 2 Binds Apolipoprotein E*. J Biol Chem 290, 26033–26042. 10.1074/jbc.m115.677286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Liu Z, Gao Z, Li B, Li J, Ou Y, Yu X, Zhang Z, Liu S, Fu X, Jin H, et al. (2022). Lipid-associated macrophages in the tumor-adipose microenvironment facilitate breast cancer progression. Oncoimmunology 11, 2085432. 10.1080/2162402x.2022.2085432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Seidman JS, Troutman TD, Sakai M, Gola A, Spann NJ, Bennett H, Bruni CM, Ouyang Z, Li RZ, Sun X, et al. (2020). Niche-Specific Reprogramming of Epigenetic Landscapes Drives Myeloid Cell Diversity in Nonalcoholic Steatohepatitis. Immunity 52, 1057–1074.e7. 10.1016/j.immuni.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Katzenelenbogen Y, Sheban F, Yalin A, Yofe I, Svetlichnyy D, Jaitin DA, Bornstein C, Moshe A, Keren-Shaul H, Cohen M, et al. (2020). Coupled scRNA-Seq and Intracellular Protein Activity Reveal an Immunosuppressive Role of TREM2 in Cancer. Cell 182, 872–885.e19. 10.1016/j.cell.2020.06.032. [DOI] [PubMed] [Google Scholar]
- 189.Binnewies M, Pollack JL, Rudolph J, Dash S, Abushawish M, Lee T, Jahchan NS, Canaday P, Lu E, Norng M, et al. (2021). Targeting TREM2 on tumor-associated macrophages enhances immunotherapy. Cell Reports 37, 109844. 10.1016/j.celrep.2021.109844. [DOI] [PubMed] [Google Scholar]
- 190.Reyes-Torres I, Park M, Magen A, Hamon P, Sanchez-Paulete AR, Humblin E, Grout J, Nair A, Troncoso L, Hamel S, et al. (2021). TREM2 Sensing of Tumor Cell Efferocytosis Promotes a Macrophage Molecular State that Limits NK Cell Antitumor Immunity. Ssrn Electron J. 10.2139/ssrn.3900125. [DOI] [Google Scholar]
- 191.Masetti M, Carriero R, Portale F, Marelli G, Morina N, Pandini M, Iovino M, Partini B, Erreni M, Ponzetta A, et al. (2021). Lipid-loaded tumor-associated macrophages sustain tumor growth and invasiveness in prostate cancer. J Exp Medicine 219, e20210564. 10.1084/jem.20210564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Conza GD, Tsai C-H, Gallart-Ayala H, Yu Y-R, Franco F, Zaffalon L, Xie X, Li X, Xiao Z, Raines LN, et al. (2021). Tumor-induced reshuffling of lipid composition on the endoplasmic reticulum membrane sustains macrophage survival and pro-tumorigenic activity. Nat Immunol 22, 1403–1415. 10.1038/s41590-021-01047-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Collins JF, Prohaska JR, and Knutson MD (2010). Metabolic crossroads of iron and copper. Nutr Rev 68, 133–147. 10.1111/j.1753-4887.2010.00271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Sukhbaatar N, and Weichhart T (2018). Iron Regulation: Macrophages in Control. Pharm 11, 137. 10.3390/ph11040137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Wisgrill L, Wessely I, Spittler A, Förster-Waldl E, Berger A, and Sadeghi K (2018). Human lactoferrin attenuates the proinflammatory response of neonatal monocyte-derived macrophages. Clin Exp Immunol 192, 315–324. 10.1111/cei.13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Soares MP, and Hamza I (2016). Macrophages and Iron Metabolism. Immunity 44, 492–504. 10.1016/j.immuni.2016.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Winn NC, Volk KM, and Hasty AH (2020). Regulation of tissue iron homeostasis: the macrophage “ferrostat.” Jci Insight 5, e132964. 10.1172/jci.insight.132964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Philippot Q, Deslée G, Adair-Kirk TL, Woods JC, Byers D, Conradi S, Dury S, Perotin JM, Lebargy F, Cassan C, et al. (2014). Increased Iron Sequestration in Alveolar Macrophages in Chronic Obtructive Pulmonary Disease. Plos One 9, e96285. 10.1371/journal.pone.0096285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Leftin A, Ben-Chetrit N, Klemm F, Joyce JA, and Koutcher JA (2017). Iron imaging reveals tumor and metastasis macrophage hemosiderin deposits in breast cancer. Plos One 12, e0184765. 10.1371/journal.pone.0184765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Leftin A, Ben-Chetrit N, Joyce JA, and Koutcher JA (2019). Imaging endogenous macrophage iron deposits reveals a metabolic biomarker of polarized tumor macrophage infiltration and response to CSF1R breast cancer immunotherapy. Sci Rep-uk 9, 857. 10.1038/s41598-018-37408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Thielmann CM, Silva M.C. da, Muley T, Meister M, Herpel E, and Muckenthaler MU (2019). Iron accumulation in tumor-associated macrophages marks an improved overall survival in patients with lung adenocarcinoma. Sci Rep-uk 9, 11326. 10.1038/s41598-019-47833-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Marques O, Porto G, Rêma A, Faria F, Paula AC, Gomez-Lazaro M, Silva P, Silva B.M. da, and Lopes C (2016). Local iron homeostasis in the breast ductal carcinoma microenvironment. Bmc Cancer 16, 187. 10.1186/s12885-016-2228-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Silva M.C. da, Breckwoldt MO, Vinchi F, Correia MP, Stojanovic A, Thielmann CM, Meister M, Muley T, Warth A, Platten M, et al. (2017). Iron Induces Anti-tumor Activity in Tumor-Associated Macrophages. Front Immunol 8, 1479. 10.3389/fimmu.2017.01479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Staudt ND, Jo M, Hu J, Bristow JM, Pizzo DP, Gaultier A, VandenBerg SR, and Gonias SL (2013). Myeloid Cell Receptor LRP1/CD91 Regulates Monocyte Recruitment and Angiogenesis in Tumors. Cancer Res 73, 3902–3912. 10.1158/0008-5472.can-12-4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Consonni FM, Bleve A, Totaro MG, Storto M, Kunderfranco P, Termanini A, Pasqualini F, Ali C, Pandolfo C, Sgambelluri F, et al. (2021). Heme catabolism by tumor-associated macrophages controls metastasis formation. Nat Immunol 22, 595–606. 10.1038/s41590-021-00921-5. [DOI] [PubMed] [Google Scholar]
- 206.Alaluf E, Vokaer B, Detavernier A, Azouz A, Splittgerber M, Carrette A, Boon L, Libert F, Soares MP, Moine AL, et al. (2020). Heme oxygenase-1 orchestrates the immunosuppressive program of tumor-associated macrophages. Jci Insight 5. 10.1172/jci.insight.133929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Leftin A, Zhao H, Turkekul M, Stanchina E. de, Manova K, and Koutcher JA (2017). Iron deposition is associated with differential macrophage infiltration and therapeutic response to iron chelation in prostate cancer. Sci Rep-uk 7, 11632. 10.1038/s41598-017-11899-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Duan X, He K, Li J, Cheng M, Song H, Liu J, and Liu P (2018). Tumor associated macrophages deliver iron to tumor cells via Lcn2. Int J Physiology Pathophysiol Pharmacol 10, 105–114. [PMC free article] [PubMed] [Google Scholar]
- 209.Mertens C, Schnetz M, Rehwald C, Grein S, Elwakeel E, Weigert A, Brüne B, and Jung M (2021). Iron-Bound Lipocalin-2 from Tumor-Associated Macrophages Drives Breast Cancer Progression Independent of Ferroportin. Metabolites 11, 180. 10.3390/metabo11030180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Chi Y, Remsik J, Kiseliovas V, Derderian C, Sener U, Alghader M, Saadeh F, Nikishina K, Bale T, Iacobuzio-Donahue C, et al. (2020). Cancer cells deploy lipocalin-2 to collect limiting iron in leptomeningeal metastasis. Science 369, 276–282. 10.1126/science.aaz2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Nixon BG, Kuo F, Ji L, Liu M, Capistrano K, Do M, Franklin RA, Wu X, Kansler ER, Srivastava RM, et al. (2022). Tumor-associated macrophages expressing the transcription factor IRF8 promote T cell exhaustion in cancer. Immunity 55, 2044–2058.e5. 10.1016/j.immuni.2022.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Zhou Y, Fei M, Zhang G, Liang W-C, Lin W, Wu Y, Piskol R, Ridgway J, McNamara E, Huang H, et al. (2020). Blockade of the Phagocytic Receptor MerTK on Tumor-Associated Macrophages Enhances P2X7R-Dependent STING Activation by Tumor-Derived cGAMP. Immunity 52, 357–373.e9. 10.1016/j.immuni.2020.01.014. [DOI] [PubMed] [Google Scholar]
- 213.Chen S, Cui W, Chi Z, Xiao Q, Hu T, Ye Q, Zhu K, Yu W, Wang Z, Yu C, et al. (2022). Tumor-associated macrophages are shaped by intratumoral high potassium via Kir2.1. Cell Metab. 10.1016/j.cmet.2022.08.016. [DOI] [PubMed] [Google Scholar]
- 214.Liu M, O’Connor RS, Trefely S, Graham K, Snyder NW, and Beatty GL (2019). Metabolic rewiring of macrophages by CpG potentiates clearance of cancer cells and overcomes tumor-expressed CD47-mediated ‘don’t eat me signal’. Nat Immunol 20, 265–275. 10.1038/s41590-018-0292-y. [DOI] [PMC free article] [PubMed] [Google Scholar]


