Abstract
Background
Eriocaulon is a wetland plant genus with important ecological value, and one of the famous taxonomically challenging groups among angiosperms, mainly due to the high intraspecific diversity and low interspecific variation in the morphological characters of species within this genus. In this study, 22 samples representing 15 Eriocaulon species from China, were sequenced and combined with published samples of Eriocaulon to test the phylogenetic resolution using the complete chloroplast genome. Furthermore, comparative analyses of the chloroplast genomes were performed to investigate the chloroplast genome evolution of Eriocaulon.
Results
The 22 Eriocaulon chloroplast genomes and the nine published samples were proved highly similar in genome size, gene content, and order. The Eriocaulon chloroplast genomes exhibited typical quadripartite structures with lengths from 150,222 bp to 151,584 bp. Comparative analyses revealed that four mutation hotspot regions (psbK-trnS, trnE-trnT, ndhF-rpl32, and ycf1) could serve as effective molecular markers for further phylogenetic analyses and species identification of Eriocaulon species. Phylogenetic results supported Eriocaulon as a monophyletic group. The identified relationships supported the taxonomic treatment of section Heterochiton and Leucantherae, and the section Heterochiton was the first divergent group. Phylogenetic tree supported Eriocaulon was divided into five clades. The divergence times indicated that all the sections diverged in the later Miocene and most of the extant Eriocaulon species diverged in the Quaternary. The phylogeny and divergence times supported rapid radiation occurred in the evolution history of Eriocaulon.
Conclusion
Our study mostly supported the taxonomic treatment at the section level for Eriocaulon species in China and demonstrated the power of phylogenetic resolution using whole chloroplast genome sequences. Comparative analyses of the Eriocaulon chloroplast genome developed molecular markers that can help us better identify and understand the evolutionary history of Eriocaulon species in the future.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-023-04034-z.
Keywords: Eriocaulon, Phylogenetic relationships, Chloroplast genome, Divergence time
Background
The family Eriocaulaceae includes 11 genera and about 1,400 species that occur primarily in the neotropics [1, 2]. Molecular phylogenetic studies showed that Eriocaulaceae is divided into two subfamilies (Eriocauloideae and Paepalanthoideae) [1]. The Eriocauloideae includes two genera, Eriocaulon L. and Mesanthemum Körn. Mesanthemum is only distributed in Africa, and most Eriocaulon species are confined to tropical and subtropical regions. There are three centers of species diversity, namely Africa (contains 111 species), the Americas (contains 122 species), and Asia (contains 220 species) [3, 4].
Eriocaulon, which includes about 470 species, is characterized by diplostemonous flowers with twice as many stamens as petals, nectar glands on the apices of the petals, and staminate and pistillate flowers with free petals [1, 5, 6]. These nectaries produce fluid that attracts insects, indicating insect pollination [5]. The species of the genus are mainly perennial herbs that grow in moist habitats or shallow wetlands. As a species-rich and widely distribution genus of wetland plants, Eriocaulon plays a significant role in the ecosystem [7].
The taxonomy of Eriocaulon is very difficult due to the high intraspecific diversity and low interspecific variation in the morphological characters within the genus [2, 8–11]. Hooker referred to the Eriocaulon as “the most difficult of classification, presenting no good sectional characters.” Several systematic studies have focused on Eriocaulon in Australia [9, 10] and India [12] using the molecular and morphological evidence. In the last ten years, many new Eriocaulon species have been described in India [13–17], Southeast Asia [18–21], and Brazil [22–24].
According to the Flora of China, there are 35 species in China [8]. Two subgenera (Trimeranthus Nakai and Eriocaulon Nakai) were classified of the Chinese species [25] Subgen. Trimeranthus was further divided into three sections: sect. Spathopeplus Nakai, sect. Leucocephala Nakai, and sect. Macrocaulon Ruhl.. Based on the morphological characteristics of the seeds and flowers, we established an infrageneric system [3]. The infrageneric classification recognized two subgenera and 10 sections. Phylogenetic analyses of the Eriocaulaceae strongly supported the monophyly of Eriocaulon [1, 2]. Only a few molecular studies sought to resolve phylogenetic relationships at the species level within this widespread genus. Davies et al. [9] resolved the taxonomy of the E. carsonii complex in Australia with amplified fragment length polymorphism (AFLP) genetic markers. Recently, Larridon et al. [4] used five markers, including four chloroplast markers and one nuclear marker, to create the first molecular phylogenetic study for the genus. Darshetkar et al. [12] focused on the Indian Eriocaulon species that 552 accessions from 66 Eriocaulon species were analyzed. This phylogenetic study used ITS and trnL–F, yielding three major clades of Indian Eriocaulon species. However, the phylogenetic relationships of Chinese Eriocaulon species are poorly understood. The markers used in the Eriocaulon phylogeny offer less information and the phylogenetic relationships are poorly resolved. More genetic markers are needed to access the phylogenetic relationships of Eriocaulon species in China.
The chloroplast genome is smaller than the plant mitochondrial and nuclear genomes, and the chloroplasts play a crucial role in photosynthesis [26, 27]. The chloroplast genome exhibits a conserved quadripartite structure of a large single-copy (LSC), a small single-copy (SSC), and two inverted repeat regions (IRs). Most angiosperms exhibit maternal inheritance [28, 29], and the chloroplast genomes are structurally stable during evolution, with mutation rates that are between those shown in the mitochondrial and nuclear genomes [30]. Therefore, the chloroplast genome provides an ideal model for genomic evolution and molecular markers for resolving phylogenetic relationships [31–33]. The chloroplast sequences were the first to be used in molecular evolution [34], and considerable attention has been paid to the evolutionary rate variations among genes or lineages in the chloroplast genome [31].
Recently, more and more studies have shown that variation in chloroplast genomes provides effective information that can be used to resolve phylogenetic relationships at multiple taxonomy levels, especially in taxonomically complex groups [35, 36]. For example, chloroplast genome data had resolved the systematic positions of enigmatic taxa in Saxifragales [37] and shed lights on the intergeneric relationships and spatio-temporal evolutionary history of Melocanninae (Poaceae) [36]. Moreover, the chloroplast genome sequences showed variations at the intraspecies levels, and revealed the genetic difference and diversity of endangered species [38, 39] and cultivated species [40, 41].
In this study, we assembled the whole chloroplast genomes of 22 Eriocaulon samples and combined them with nine published samples in GenBank. These samples included half of the species in China, and the taxonomic status of some species were unresolved. Furthermore, we analyzed most of the chloroplast gene sequences in GenBank. Our specific goals were as follows: (a) to compare the chloroplast genome structures within the genus Eriocaulon; (b) to identify the mutation hotspot regions as potential chloroplast markers for species identification and phylogeny; (c) to infer and test the phylogenetic relationships and divergence time among the Eriocaulon species in China using the whole chloroplast genome; (d) to include the chloroplast gene sequences from GenBank to infer the deep relationships of Eriocaulon species in the world.
Results
General features of Eriocaulon chloroplast genomes
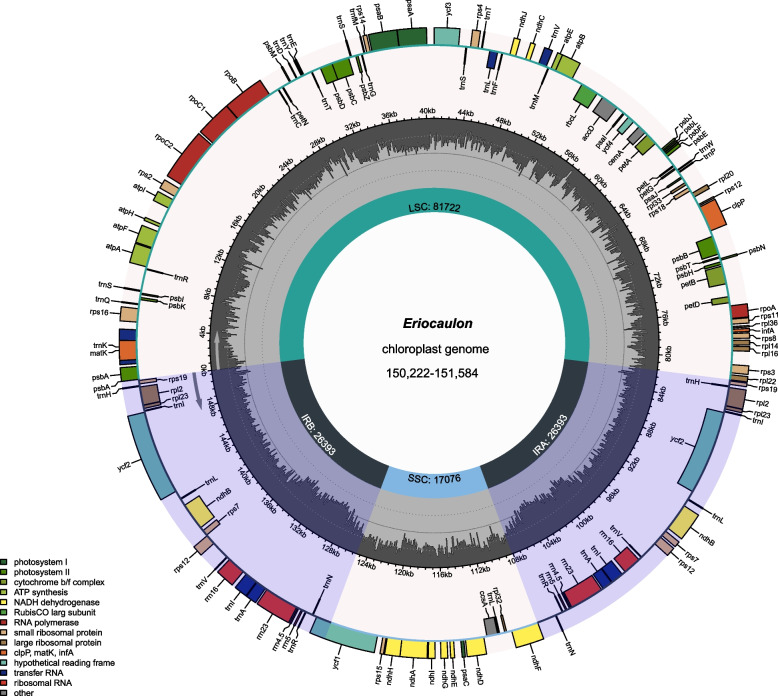
The length of the 31 chloroplast genomes varied from 150,222 bp (E. sp. 02) to 151,584 bp (E. australe 01) (Table 1 and Table S1). The Eriocaulon chloroplast genome exhibited typical quadripartite structures (Fig. 1). The IR regions (ranging from 25,950 bp (E. schochianum) to 26,532 bp (E. decemflorum 01)) were separated by an LSC region ranging from 80,367 bp (E. oryzetorum) to 81,722 bp (E. australe 01) and an SSC region ranging from 16,890 (E. decemflorum 02) to 17,104 bp (E. australe 03). The GC content in the Eriocaulon chloroplast genomes was 35.7–35.9% (Table 1). There were 113 unique genes in the chloroplast genome of Eriocaulon species, including 79 protein-coding genes, 30 tRNA genes, and four rRNA genes. Among the protein-coding genes, 44 genes were associated with photosynthesis, and 25 were related to self-replication.
Table 1.
Summary of the chloroplast genomes for 31 Eriocaulon samples
| Section | Species | Total length (bp) | LSC (bp) | IR (bp) | SSC (bp) | Total GC content (%) | Total genes | Protein coding genes | rRNA genes | tRNA genes |
|---|---|---|---|---|---|---|---|---|---|---|
| Apoda | E. alpestre 01 | 151,372 | 81,354 | 26,513 | 16,992 | 35.8 | 114 | 80 | 4 | 30 |
| Apoda | E. alpestre 02 | 151,398 | 81,340 | 26,514 | 17,030 | 35.8 | 114 | 80 | 4 | 30 |
| Apoda | E. alpestre 03 | 151,343 | 81,361 | 26,483 | 17,016 | 35.8 | 114 | 80 | 4 | 30 |
| Heterochiton | E. australe 01 | 151,584 | 81,722 | 26,393 | 17,076 | 35.7 | 114 | 80 | 4 | 30 |
| Heterochiton | E. australe 02 | 150,770 | 81,403 | 26,132 | 17,103 | 35.7 | 114 | 80 | 4 | 30 |
| Heterochiton | E. australe 03 | 151,498 | 81,592 | 26,401 | 17,104 | 35.8 | 114 | 80 | 4 | 30 |
| Simplices | E. brownianum | 151,432 | 81,533 | 26,457 | 16,985 | 35.8 | 114 | 80 | 4 | 30 |
| Apoda | E. buergerianum 01 | 151,383 | 81,331 | 26,507 | 17,038 | 35.8 | 114 | 80 | 4 | 30 |
| Apoda | E. buergerianum 02 | 151,366 | 81,334 | 26,507 | 17,018 | 35.8 | 114 | 80 | 4 | 30 |
| Apoda | E. buergerianum 03 | 151,389 | 81,347 | 26,507 | 17,028 | 35.8 | 114 | 80 | 4 | 30 |
| Leucantherae | E. cinereum 01 | 150,725 | 80,741 | 26,523 | 16,938 | 35.8 | 114 | 80 | 4 | 30 |
| Leucantherae | E. cinereum 02 | 150,708 | 80,733 | 26,523 | 16,929 | 35.9 | 114 | 80 | 4 | 30 |
| Nasmythia | E. decemflorum 01 | 151,408 | 81,424 | 26,532 | 16,920 | 35.9 | 114 | 80 | 4 | 30 |
| Nasmythia | E. decemflorum 02 | 151,032 | 81,152 | 26,495 | 16,890 | 35.9 | 114 | 80 | 4 | 30 |
| Apoda | E. faberi 01 | 151,361 | 81,329 | 26,507 | 17,018 | 35.8 | 114 | 80 | 4 | 30 |
| Apoda | E. faberi 02 | 151,361 | 81,329 | 26,507 | 17,018 | 35.8 | 114 | 80 | 4 | 30 |
| - | E. fistulosum | 151,415 | 81,493 | 26,460 | 17,002 | 35.9 | 114 | 80 | 4 | 30 |
| Anisopetalae | E. henryanum 01 | 151,051 | 81,171 | 26,451 | 16,978 | 35.8 | 114 | 80 | 4 | 30 |
| Anisopetalae | E. henryanum 02 | 151,010 | 81,119 | 26,456 | 16,979 | 35.8 | 114 | 80 | 4 | 30 |
| Apoda | E. miquelianum | 151,344 | 81,304 | 26,507 | 17,026 | 35.8 | 114 | 80 | 4 | 30 |
| Simplices | E. nantoense | 151,389 | 81,509 | 26,442 | 16,996 | 35.8 | 114 | 80 | 4 | 30 |
| Simplices | E. nepalense 01 | 150,947 | 81,064 | 26,451 | 16,981 | 35.8 | 114 | 80 | 4 | 30 |
| Simplices | E. nepalense 02 | 150,973 | 81,090 | 26,451 | 16,981 | 35.9 | 114 | 80 | 4 | 30 |
| Simplices | E. oryzetorum | 150,255 | 80,367 | 26,441 | 17,006 | 35.8 | 114 | 80 | 4 | 30 |
| Simplices | E. schochianum | 150,450 | 81,570 | 25,950 | 16,980 | 35.8 | 114 | 80 | 4 | 30 |
| Simplices | E. sp. 01 | 151,462 | 81,579 | 26,449 | 16,985 | 35.8 | 114 | 80 | 4 | 30 |
| Leucantherae | E. sp. 02 | 150,222 | 80,611 | 26,349 | 16,913 | 35.9 | 114 | 80 | 4 | 30 |
| Leucantherae | E. tokinense 01 | 150,951 | 81,185 | 26,411 | 16,944 | 35.7 | 114 | 80 | 4 | 30 |
| Leucantherae | E. tokinense 02 | 150,900 | 81,146 | 26,411 | 16,932 | 35.7 | 114 | 80 | 4 | 30 |
| Disepala | E. truncatum 01 | 151,052 | 81,117 | 26,452 | 17,031 | 35.9 | 114 | 80 | 4 | 30 |
| Disepala | E. truncatum 02 | 151,057 | 81,122 | 26,452 | 17,031 | 35.9 | 114 | 80 | 4 | 30 |
Fig. 1.
Structural and gene map of the Eriocaulon chloroplast genome
The boundaries between IR and SC regions were compared in the 18 Eriocaulon species (Fig. 1). The Eriocaulon SC/IR junctions were highly conserved. the LSC/IRb junction was located in rpl22, while the IRb/SSC junction was located in the ndhF, and IRb expanded progressively from the IR regions to ndhF. The IRa/SSC junction was found within the ycf1 and the IRa/LSC border was adjacent to the psbA.
For all Eriocaulon species, 64 types of codons encoding 20 amino acids were detected (Figure S1). The total number of codons was 22,336–22,571. AUU was the most-used codon (982–1,000 instances), whereas CGG was the least (65–71 instances). The RSCU values are shown in Figure S1, and the values for all codons ranged from 0.26 to 2.27 in the Eriocaulon chloroplast genome. The RSCU values of 30 codons were greater than 1.00 in all Eriocaulon chloroplast genomes and all of them ended with A/U, except for UUG.
SSR polymorphisms and long repeat structure
We total identified 777 SSRs in the 18 Eriocaulon chloroplast genomes (Table 2). The number of SSRs in Eriocaulon ranged from 33 to 58, with an average of 43. Dinucleotide repeats were the most common (37.07%), followed by mononucleotide repeats (22.13%), tetranucleotide repeats (21.75%), and trinucleotide repeats (14.41%); pentanucleotide and hexanucleotide repeats were the least common (2.32%). Most of the SSRs were located in the intergenic region of the LSC.
Table 2.
Numbers and types of SSRs in the 18 Eriocaulon chloroplast genomes
| Species | mono- | di- | tri- | tetra- | penta- | hexa- | Total |
|---|---|---|---|---|---|---|---|
| E. alpestre | 11 | 18 | 3 | 12 | 0 | 2 | 46 |
| E. australe | 16 | 21 | 10 | 10 | 1 | 0 | 58 |
| E. brownianum | 8 | 18 | 8 | 10 | 2 | 1 | 47 |
| E. buergerianum | 12 | 16 | 4 | 12 | 0 | 2 | 46 |
| E. cinereum | 9 | 15 | 4 | 7 | 0 | 0 | 35 |
| E. decemflorum | 10 | 14 | 5 | 8 | 1 | 1 | 39 |
| E. faberi | 13 | 17 | 4 | 12 | 0 | 2 | 48 |
| E. fistulosum | 8 | 10 | 5 | 8 | 2 | 0 | 33 |
| E. henryanum | 8 | 18 | 7 | 10 | 0 | 1 | 44 |
| E. miquelianum | 12 | 15 | 6 | 11 | 0 | 2 | 46 |
| E. nantoense | 8 | 19 | 7 | 8 | 1 | 1 | 44 |
| E. nepalense | 8 | 17 | 7 | 10 | 2 | 44 | |
| E. oryzetorum | 8 | 14 | 4 | 4 | 2 | 1 | 33 |
| E. schochianum | 7 | 18 | 8 | 10 | 3 | 1 | 47 |
| E. sp. 01 | 7 | 18 | 8 | 11 | 2 | 2 | 48 |
| E. sp. 02 | 9 | 16 | 5 | 7 | 0 | 0 | 37 |
| E. tokinense | 7 | 12 | 10 | 14 | 0 | 1 | 44 |
| E. truncatum | 11 | 12 | 7 | 5 | 2 | 1 | 38 |
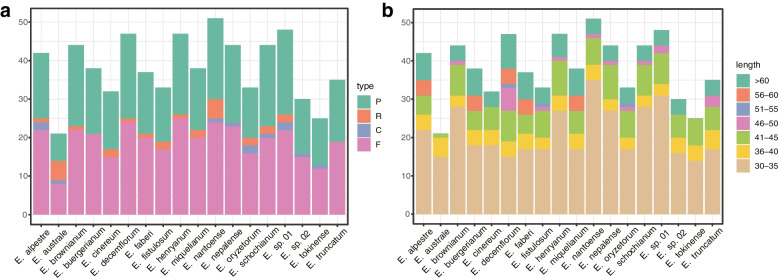
Four categories of long repeats—forward, reverse, complement, and palindromic—were detected (Fig. 2). There were 8–25 forward repeats, 0–2 reverse repeats, 0–5 complement repeats, and 7—22 palindromic repeats. E. australe had the lowest (21) and E. nantoense had the highest (51) number of repeats. The repeat sizes ranged from 30 to 86 bp. More than half of the repeats were 30–35 bp long, while only three repeats were 51–55 bp long.
Fig. 2.
Long-repeat sequences in the Eriocaulon chloroplast genomes. a Total number of the four repeat types. b Number of repeats by length
Eriocaulon chloroplast genome variation
The mVISTA results showed the Eriocaulon chloroplast genome had collineation, no rearrangement, and high sequence similarity (Figure S2). The Eriocaulon chloroplast genomes aligned with a length of 159,226 bp, including 16,502 variable sites (10.36%), and 14,365 parsimony-informative sites (9.02%). The overall nucleotide diversity (π) was 0.02448 (Table 3). The SSC regions had the highest variation and the IR had the lowest sequence divergence. The mean interspecies and intraspecies genetic distances were 0.0279 and 0.0012, respectively. Eriocaulon sp. 01 and E. brownianum had the lowest genetic distance value (0.003) and E. australe and E. decemflorum had the highest (0.0431). Eriocaulon australe had the highest intraspecies genetic distance (0.0048) among the three samples.
Table 3.
Sequence divergence of the Eriocaulon chloroplast genomes
| Regions | Aligned length (bp) | Variable sites | Information sites | Nucleotide Diversity | Number of Haplotypes | ||
|---|---|---|---|---|---|---|---|
| Numbers | % | Numbers | % | ||||
| LSC | 87,335 | 11,501 | 13.17% | 10,006 | 11.46% | 0.03131 | 30 |
| SSC | 17,756 | 2946 | 16.59% | 2580 | 14.53% | 0.04268 | 30 |
| IR | 27,053 | 1030 | 3.81% | 892 | 3.30% | 0.00848 | 28 |
| Whole chloroplast genome | 159,226 | 16,502 | 10.36% | 14,365 | 9.02% | 0.02448 | 30 |
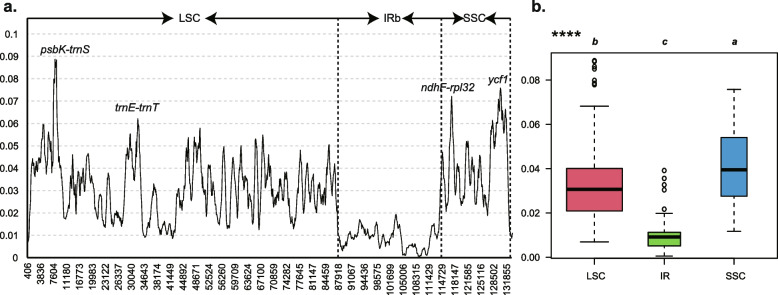
Mutation hotspots in the Eriocaulon chloroplast genome was identified using the slide window method, and the results are presented in Fig. 3a. The π values ranged from 0 to 0.08872 within an 800-bp window. The π values > 0.06 was defined the mutation hotspots regions. Four peaks were identified, including three noncoding regions (psbK-trnS, trnE-trnT, and ndhF-rpl32) and one coding region (ycf1). Two regions (psbK-trnS and trnE-trnT) were located in the LSC region and the other two (ndhF-rpl32 and ycf1) in the SSC region. The psbK-trnS region exhibited the highest π value. This result also showed that the SSC regions had the highest variation and the IR had the lowest sequence divergence (Fig. 3b).
Fig. 3.
The nucleotide diversity (π) values in the Eriocaulon chloroplast genome. Window size: 800 bp, step size: 100 bp. a The π values of the windows. b Boxplots of π-value differences among the LSC, IR, and SSC regions
Molecular evolution of the Eriocaulon chloroplast genomes
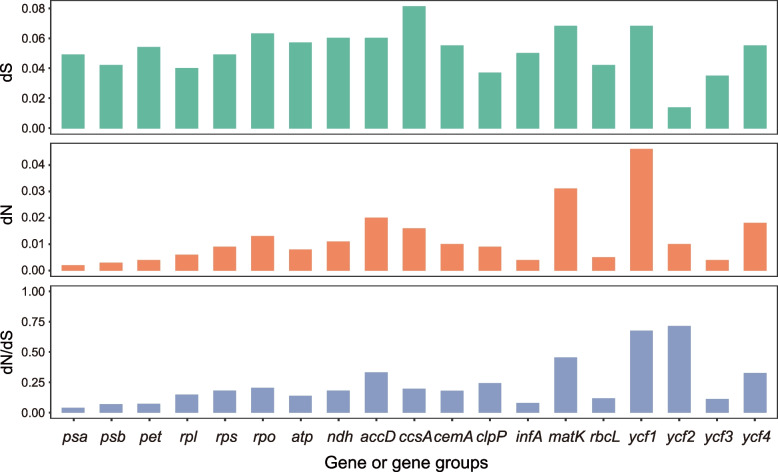
The dS, dN, and ω values for the 79 protein-coding genes are shown in Supplemental Table S2. The highest dN value was 0.046 in the ycf1 gene, and the highest dS value was 0.105 in the rps15 gene. All the ω values were less than 0.5, indicating the genes were under purifying selection. The t test showed the values of dS, dN, and ω in the genes had significant differences, indicating variable molecular evolution rate among the genes. Among the gene groups, the rps group had the highest ω values and the psa group had the lowest (Fig. 4). The t test supported the difference of mutation rates among the gene groups.
Fig. 4.
The evolutionary values of dN, dS and ratio (ω) in Eriocaulon
Phylogenetic relationships of Eriocaulon
The whole chloroplast genome dataset contained 31 Eriocaulon chloroplast genome samples and one outgroup of Paepalanthus alpinus, among which, 164,361 bp were aligned nucleotide sites, including 27,016 variable sites. The 83-gene dataset contained 73,559 nucleotide sites, including 9,654 variable sites and 5,133 parsimony-informative sites. The phylogenetic relationships of Eriocaulon based on the two datasets showed similar topologies (Figure S3). All Eriocaulon species formed a monophyletic group (BS = 100/PP = 1) and all relationships among the major clades were strongly supported. All samples of the same species also formed a clade.
The section Heterochiton was the first divergent group of Eriocaulon and was sister to the remaining species. The section Leucantherae, including three species (E. cinereum, E. sp. 02, and E. tokinense), was the second divergent group and was strongly supported. The section Simplices (including E. henryanum, which belongs to section Anisopetalae) was a sister to Disepala. The section Apoda formed a monophyletic group with high support values (BS = 100/PP = 1) and was a sister to the section Nasmythia. The phylogenetic position of E. fistulosum, from Australia, was uncertain due to the lower support values (BS = 42/PP = 0.8 in the whole chloroplast genome dataset and BS = 72/PP = 0.99 in the 83-gene dataset). The branch lengths of sections Apoda and Simplices were very short, indicating that these groups may have undergone rapid radiation.
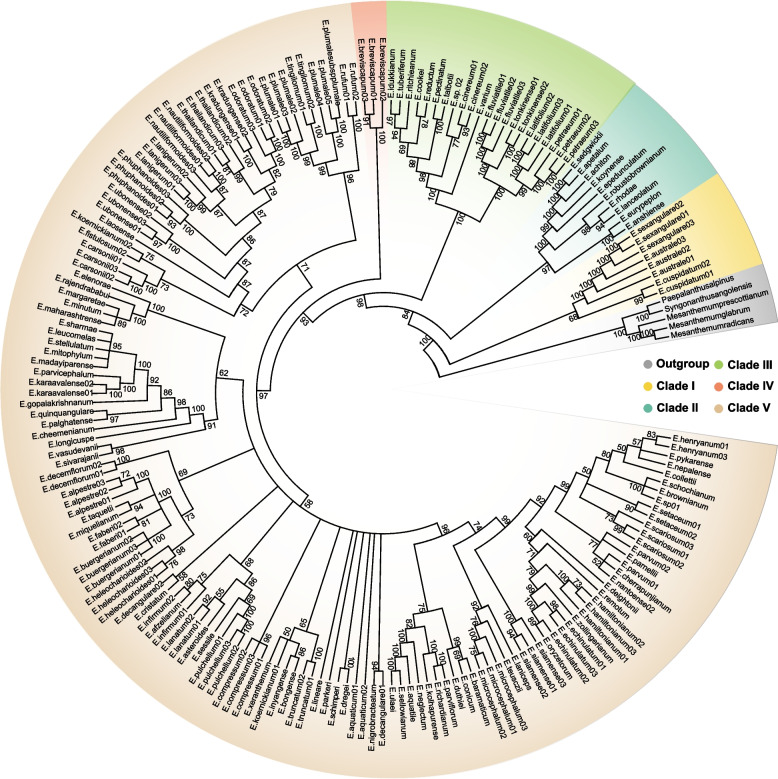
The chloroplast gene dataset contained 197 samples and 121 species of Eriocaulon (Table S3). The dataset of five genes included 5,322 aligned sites of which 917 were variable sites. Five clades were supported by the ML tree in Eriocaulon (Fig. 5). The Clade I contained the species from the section Heterochiton, which was the first divergent group and was sister to the remaining clades. Clade II included ten species which were mainly distributed in India. Clade III consisted of the species of section Leucantherae. Clade IV was the singleton, containing E. breviscapum. The major Eriocaulon species were in the clade V and the subclades in this clade were less well supported.
Fig. 5.
Phylogenetic trees of Eriocaulon using the five chloroplast genes
Divergence time estimate
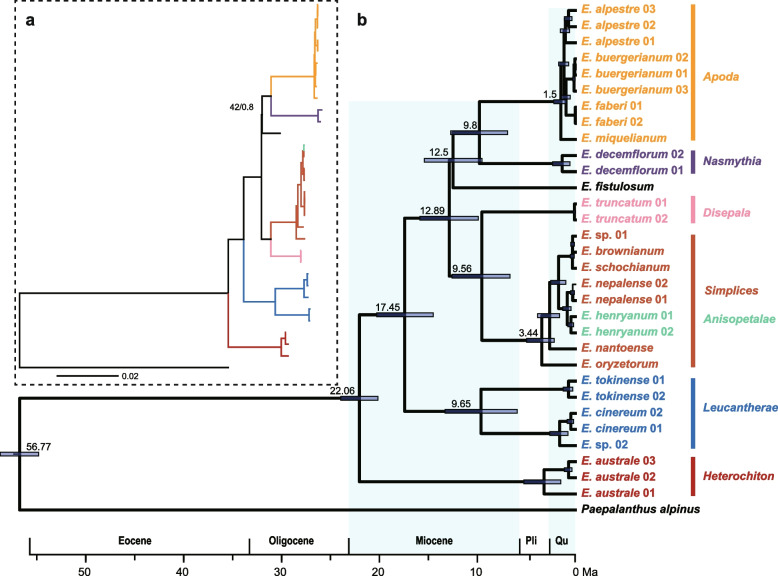
Using the 83-gene datasets, the divergence time suggests that the stem and crown ages of Eriocaulon were 56.77 Ma (95% highest posterior densities (HPD): 55.88–62.91 Ma) in the early Eocene and 22.06 Ma (95% HPD) during the later Oligocene (Fig. 6). The stem and crown ages of section Leucantherae were 17.45 Ma and 9.65 Ma. The split between the section Anisopetalae and section Disepala occurred at 9.56 Ma, during the later Miocene. The split between the section Apoda and Nasmythia occurred at 9.8 Ma.
Fig. 6.
Phylogeny and divergence times of Eriocaulon. a the tree topology of Eriocaulon using the whole chloroplast genome dataset. The number above the lines indicates the ML bootstrap values (BS) and BI posterior probability (PP). BS = 100 and PP = 1.0 are not shown. b divergence times of Eriocaulon. The blue bars correspond to the 95% highest posterior density (HPD)
Using the five chloroplast gene dataset, the crown age of Eriocaulon was 22.3 Ma, the five clades were divergent from 17.01 Ma to 21.24 Ma showing rapid radiation. Most of species was divergent less than 10 M, starting at the later Miocene (Figure S4). These results indicated that all of the sections or clades had diverged in the later Miocene and most of the extant Eriocaulon species diverged in the Quaternary.
Discussion
Chloroplast genome evolution of Eriocaulon
This study is the first to attempt a comparative analysis of Eriocaulon chloroplast genomes. The 31 Eriocaulon chloroplast genomes were very similar in overall structure, gene numbers, content and order. However, the length of the chloroplast genome showed noticeable differences compared with other lineages within the genus [32, 42, 43]. The Eriocaulon chloroplast genome size ranged from 150,222 bp to 151,584 bp, while the LSC region ranged from 80,367 bp to 81,722 bp (Table 1). The length differences occurred mainly in the LSC regions, while the coding region showed less variation. This suggested that the chloroplast genome size variation of Eriocaulon species mainly occurred in the non-coding regions within the LSC region.
Sequences with higher GC content are more stable and have lower mutation rates. Among angiosperms, the overall GC content typically accounts for 30–40% of the chloroplast genome, and the IR region exhibits higher GC content than the LSC and SSC regions [40, 44, 45]. The overall GC content in the Eriocaulon chloroplast genomes was 35.7–35.9% and the rRNA genes in the IR regions had a high level of GC content (55.2%), which contributed to the high GC content in the IR region overall (43.2%) compared with that of the LSC region (32.7%) and SSC region (27.8%).
Long sequence repeats in the genomes contribute to genome rearrangement [46–48]. In the Eriocaulon chloroplast genomes, 21 (E. australe) to 51 (E. nantoense) repeats were found in each species. Four types of sequence repeat occur; in previous studies, forward repeats were the most abundant in the chloroplast genome. However, we found almost as many palindromic repeats as forward repeats in the studied species (Fig. 2). SSRs are very abundant in the chloroplast genome and most of them are universal at the interspecies level within the genus or even the family [49, 50]. In the Eriocaulon chloroplast genomes, we found 33 to 58 SSR loci. Other studies have shown that the most abundant SSRs were A/T-rich mononucleotide repeats, which was consistent with the chloroplast genome’s common polyA or polyT repeats and rare G or C repeats [35, 51–53]. Dinucleotide repeats were the most common type in the Eriocaulon chloroplast genomes (Table 2) and had high AT content.
Chloroplast markers for Eriocaulon
As a famously difficult taxonomic group, effective molecular markers are necessary to rapidly assess genetic divergence and identify species. However, universal or common molecular markers are ineffective for this group [4, 12]. The mutation events are not random and are concentrated in hotspot regions in the chloroplast genome sequences, so variable markers or species barcodes can be identified in the chloroplast genome [32, 54]. Based on the nucleotide diversity analyses, we proposed four regions with high π values with high potential as markers to resolve taxonomic issues in Eriocaulon and function as DNA barcodes for species identification.
The intergenic region psbK-trnS possess the highest π values (Fig. 3), however, this marker is little used in plant phylogeny. The intergenic region trnE-trnT is about 800 bp long and is used in Camassia (Agavaceae) [55], Chamaecrista sect. Xerocalyx [56], and the family Solanaceae [57]. However, this space often contains large A/T-rich regions that may lead to low sequence quality in some groups [58]. In the Eriocaulon, we detected an SSR structure (repeat type: AT) within some species. The ndhF-rpl32, located in the SSC region with an alignment length of 1,496 bp, has a long history of use in specie identification and plant phylogenetic studies [59]. This region has previously displayed a high level of genetic divergence and is probably the most variable marker at low taxonomic level. The two regions in the coding gene ycf1 (ycf1a and ycf1b) are the most variable markers in several plant lineages and are more variable than matK and rbcL combination [32, 60]. Recently, ycf1 has been used as the core DNA barcode in the study of plant phylogeny [61–63]. Based on our study, these four divergent markers may be helpful for further phylogenetic and species identification of Eriocaulon species.
Phylogenetics and divergence time of Eriocaulon
The relationships derived by using two chloroplast genome datasets were consistent. The phylogenetic resolution of Eriocaulon species has been greatly improved in comparison with recently published results [4, 12], with most nodes having 100% support values (Figure S3). However, the five chloroplast genes had the lower resolution and supports Eriocaulon species was divided into five clades (Fig. 5). Molecular phylogeny partly supported the taxonomic classification at the section level for the Chinese species in our previous study based on their morphological characteristics (Fig. 5) [3], such as the seed surfaces and calyces of female flowers.
Ma [25] classified the 28 Chinese species of Eriocaulon into the two subgenera Trimeranthus and Eriocaulon sensu (monotypic: E. decemflorum Maxim.), according to their flower numbers. We recognized two subgenera of East Asian species [3]. The subgenus Spathopeplus Koern, which included seven sections (Macrocaulon, Simplices, Anisopetalae, Heterochiton, Disepala, Leucantherae, and Nasmythia), has the sepals of the female flowers fused to some extent into a spathe. The subgenus Trimeranthus Nakai, which included three sections (Macropoda, Apoda, and Nudicuspa), has free female sepals. Molecular phylogenetic relationships did not support both taxonomic treatments of the subgenera (Fig. 6 and Figure S3) and not all of the subgenera were monophyletic groups. The section Heterochiton included three species in East Asia (clade I in Fig. 5), large herbs that grow 20–60 cm high. This section was the first divergent group in Eriocaulon (Fig. 5) [4]. The sections Simplices and Anisopetalae formed a clade that was supported by their morphological characteristics (Fig. 6 and Figure S3), such as three female sepals with a reduction of the median sepal. There are many more species in section Simplices and it is difficult to distinguish them using morphology, as in the E. nepalense complex (comprising E. nepalense, E. huzulaefolium, and E. nantoense). Eriocaulon decemflorum (section Nasmythia) was retrieved as a single-species lineage (Figure S3). This result supports its position as the only member of section Nasmythia based on its reduced, dimerous flowers and seed ornamentation structure. The subclades of the Clade V were poorly resolved using the five chloroplast genes (Fig. 5). Larridon et al. [4] divided the Clade V into approximately seven branches, however, owing to the lower supported values, these results were not solid and adding more molecular data is essential for phylogeny of this famous taxonomically challenging group.
Phylogenetic and divergence time analysis indicated that the Eriocaulon species may have undergone rapid radiation. The divergence time analysis results indicated that Eriocaulon originated in the early Eocene (Fig. 6). There were two significant periods of rapid diversification of Eriocaulon. The first was in the early Miocene, which led to the major lineages of the extant Eriocaulon species. During this period, due to the higher temperatures [64–67], suitable habitats for Eriocaulon were fragmented through aridification, which led to the first rapid radiation. The second period was in the Quaternary, which led to most of the extant Eriocaulon species. After 5 Ma, the global temperature decreased sharply after a short period of global warming [68], providing a diverse range of habitats and further increasing the species diversity of Eriocaulon.
Conclusions
In this work, we sequenced and assembled the complete chloroplast genome sequences of 22 samples representing 15 Eriocaulon species. By adding published samples of Eriocaulon, comparative genomics indicated that the Eriocaulon chloroplast genomes were relatively conserved and four mutation hotspot regions emerged as potential variable molecular markers for inferring phylogenetic relationships and species identification. Phylogenetic analysis based on the chloroplast genome supported part of the results of our previous taxonomic treatment study at the section level using morphological characteristics. The world Eriocaulon species were divided into five clades and underwent the rapid radiation. Divergence time analysis revealed that Eriocaulon originated in the early Eocene and diversified in the later Miocene. Overall, this study demonstrated that the whole chloroplast genome sequences displayed variable information to resolve phylogenetic relationships in this difficult-to-characterize genus.
Methods
Sample collection and sequencing
We collected 22 samples representing 15 species in China. The sample details are shown in Table S1 and the voucher specimens were deposited at the Museum of Beijing Forestry University. Zhixiang Zhang identified all samples. We also downloaded all of the published complete chloroplast genomes of Eriocaulon from GenBank. In total, we obtained 31 samples representing 18 Eriocaulon species (Table S1).
Fresh leaves dried in silica gel for DNA extraction. The total genomic DNA was extracted with the mCTAB method [69]. NanoDrop 2000 microspectrophotometer was used to quantify the DNA concentration and quality. Genomic DNA was fragmented randomly into 350 bp segments with an ultrasonicator. A paired-end library was constructed with an insert size of 350 bp and sequenced with the Illumina Hiseq Xten sequencing system at Novegene Co. Ltd. in Tianjin. Approximately 5.0 Gb of raw data were generated for each sample.
Chloroplast genome assembly and annotation
To obtain high-quality clean reads, Trimmomatic v0.36 [70] was run to cut and remove the adaptors and low-quality reads. GetOrganelle [71] was used to assemble the chloroplast genome and the k-mer length was set to 95. Clean reads were mapped to the assembled chloroplast genome using Geneious Prime (Biomatters Ltd., Auckland, New Zealand) to validate the sequence errors. The complete chloroplast genome was annotated using the perl script Plann [72] with the Eriocaulon henryanum (OK539718) as the reference. The errors in the start and stop codon positions of the protein genes were manually checked and adjusted using Geneious Prime [73].
Chloroplot [74] was employed to draw the chloroplast genome structure of Eriocaulon. All of the new sequenced and annotated complete chloroplast genomes were deposited in GenBank and the accession numbers were shown in Table S1. Geneious Prime was used to extract the protein-coding genes of Eriocaulon chloroplast genomes. Relative synonymous codon usage (RSCU) indicated the ratio of the observed frequency of a particular codon to the expected frequency of that codon. The codon frequency and RSCU were calculated using MEGA X and codon frequency distribution was illustrated using TBtools [75] with the form of a heatmap.
Chloroplast genome sequence divergence analysis
To visualize the sequence divergence among the Eriocaulon species, the mVISTA program was used to compare the 18 Eriocaulon species’ chloroplast genomes. The annotation of Eriocaulon alpestre (OK539714) was used as a reference. To identify the mutation hotspot regions and quantize the sequence divergence, we aligned the 20 chloroplast genomes with MAFFT v7.0. Variable and parsimony-informative sites, and nucleotide diversity (π) in the aligned sequences were used to evaluate sequence divergence. Variable and parsimony-informative sites were calculated with MEGA X [76]. The π value was calculated with the software DnaSP v6 [77] using the sliding window method. The window length was set to 800 bp with a 100-bp step size.
Simple sequence repeats and repeat structure analysis
Four types of repeat sequences, forward, palindromic, reverse, and complement repeats, were identified by the REPuter online program [78] with the parameters of a repeat size of ≥ 30 bp and a Hamming distance of 3. SSRs were identified using the PERL script microsatellite identification (MISA) software [79], with the threshold number of repeats set as ≥ 10 repeat units for mononucleotides, ≥ 5 for dinucleotides, ≥ 4 for trinucleotides, and ≥ 3 for tetranucleotides, pentanucleotides, and hexanucleotides.
Molecular evolution of the chloroplast genome of Eriocaulon. We used the ratio (ω) of non-synonymous (dN) to synonymous (dS) substitutions to analyze the role of natural selection in driving the molecular evolution of the Eriocaulon chloroplast genome. The ω value is an indicator of natural selection of the protein-coding genes. The values ω > 1, ω = 1, and ω < 1 indicate positive, neutral, and negative selection, respectively. All the protein-coding genes were aligned with the MAFFT and deleted the stop codon. The dN, dS and ω values were calculated using the MEGA X [76]. We analyzed all the 79 protein-coding genes and the gene groups with some function, such as atp, psa, pet, and rpo.
Phylogenetic analyses
Both maximum likelihood (ML) and Bayesian inference (BI) methods were performed to infer the phylogeny relationships of Eriocaulon. We used two datasets to infer phylogenic relationships: the complete chloroplast genome sequences and the 83-genes (including 79 protein-coding genes and four rRNA genes) of the 32 samples, with Paepalanthus alpinus as the outgroup. The nucleotide sequences of the 79 common protein-coding genes were extracted from each chloroplast genome, aligned, and concatenated.
Best-fitting models of nucleotide substitution were selected using ModelFinder [80]. ML analyses were performed in RAxML-NG [81] with 500 bootstrap replicates (BS). The BI analysis was performed in Mrbayes v3.2 [82] with two independent Markov chain Monte Carlo chains. Each chain began with a random tree with 2,000,000 generations. The first 25% of the sampled trees were discarded as burn-in, and the Bayesian posterior probabilities (PP) were calculated using the remaining trees.
Phylogenetic analyses using the chloroplast gene sequences from GenBank
Five chloroplast genes of rbcL, rpoB, matK, rpoC1, and trnL-F of Eriocaulon were downloaded from GenBank database. All the genes were aligned using MAFFT, and concatenated by the information of specimen voucher in order to ensure these sequences from the same individual using PhyloSuite v1.2.2 [83]. The ML tree was reconstructed using the IQ-TREE v2 and the supported values were assessed used the ultrafast bootstrap approximation (UFBoot) methods [84].
Fossil priors and divergence time estimate
Divergence time was estimated using BEAST v2.5.1 [85] with two priors based on the concatenated 83-gene dataset and the five chloroplast gene dataset (keep one sample of each species). Following Larridon et al. [4], two priors were used: (i) the crown age of Eriocaulaceae was 56 Ma (the root of the tree); (ii) the crown age of Eriocaulon was 21.66 Ma.
Uncorrelated log-normal distribution relaxed molecular clock models were selected to account for rate variability among clades. The nucleotide substitution model and the prior tree model were set to GTR and Yule models, respectively. Both priors were set under the normal distribution. The MCMC run had a chain length of 500,000,000 generations with sampling every 10,000 generations. Tracer 1.6 [86] was used to evaluate convergence and ensure a sufficient and effective sample size for all parameters surpassing 200. The maximum clade credibility tree was produced using TreeAnnotator v2.4 after discarding the first 10% of the generations.
Supplementary Information
Additional file 1: Figure S1. The RSCU values of the coding genes in the Eriocaulon chloroplast genome.
Additional file 2: Figure S2. mVISTA-based sequence identity plot of 18 Eriocaulon species, using E. alpestre as a reference.
Additional file 3: Figure S3. Phylogenetic trees of Eriocaulon. a. The whole chloroplast genome dataset. b. The 83-genes dataset. The number above the lines indicates the ML bootstrap values (BS) and BI posterior probability (PP). BS=100 and PP=1.0 are not shown.
Additional file 4: Figure S4. Divergence times of Eriocaulon using the five chloroplast genes.
Additional file 5: Table S1. Sampling information for the Eriocaulon samples in this study.
Additional file 6: Table S2. The dN, dS, and ω values of 79 protein-coding genes.
Additional file 7: Table S3. The GenBank information of the chloroplast genes used in inferring the Eriocaulon phylogeny.
Acknowledgements
Not applicable.
Authors’ contributions
WD and ZZ conceived and designed the experiments; RD, ZZ, KL and XL contributed sample material; EL, WD, YG, KL, RD and XL performed the experiments and analyzed the data; WD, EL and ZZ wrote and reviewed the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by Science and Technology Basic Resources Investigation Program of China (Grant No. 2021FY100200), and the Second Tibetan Plateau Scientific Expedition and Research (STEP) program (Grant No. 2019QZKK050202).
Availability of data and materials
New sequenced and other published chloroplast genome sequences can be found in GenBank (https://www.ncbi.nlm.nih.gov/genbank/), and the accession numbers showed in Table S1.
Declarations
Ethics approval and consent to participate
The collecting of all samples in this study followed the Regulations on the Protection of Wild Plants of China, the IUCN Policy Statement on Research Involving Species at Risk of Extinction and the Convention on the Trade in Endangered Species of Wild Fauna and Flora. All methods were carried out in accordance with relevant guidelines and regulations.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Wenpan Dong, Email: wpdong@bjfu.edu.cn.
Zhixiang Zhang, Email: zxzhang@bjfu.edu.cn.
References
- 1.de Andrade MJG, Giulietti AM, Rapini A, de Queiroz LP, Conceição AdS, de Almeida PRM, van den Berg C. A comprehensive phylogenetic analysis of Eriocaulaceae: evidence from nuclear (ITS) and plastid (psbA-trnH and trnL-F) DNA sequences. Taxon. 2010;59(2):379–88.
- 2.Giulietti AM, Andrade MJG, Scatena VL, Trovó M, Coan AI, Sano PT, Santos FA, Borges RL, van den Berg C. Molecular phylogeny, morphology and their implications for the taxonomy of Eriocaulaceae. Rodriguésia. 2012;63:001–019. doi: 10.1590/S2175-78602012000100001. [DOI] [Google Scholar]
- 3.Zhang Z. Monographie der Gattung Eriocaulon in Ostasien: Dissertationes Botanicae. 1999. [Google Scholar]
- 4.Larridon I, Tanaka N, Liang Y, Phillips SM, Barfod AS, Cho S-H, Gale SW, Jobson RW, Kim Y-D, Li J, et al. First molecular phylogenetic insights into the evolution of Eriocaulon (Eriocaulaceae, Poales) J Plant Res. 2019;132(5):589–600. doi: 10.1007/s10265-019-01129-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Judd WS, Campbell CS, Kellogg EA, Stevens PF, Donoghue MJ. Plant systematics: a phylogenetic approach. Ecología mediterránea. 1999;25(2):215. [Google Scholar]
- 6.Ashwini MD, Mandar ND, Rao GR, Shubhada T, Konickal MP, Ritesh Kumar C. Eriocaulon karaavalense Eriocaulaceae), a new species from india based on morphological and molecular evidence. Ann Bot Fenn. 2019;56(4–6):305–316. [Google Scholar]
- 7.Horiuchi Y, Kamijo T, Tanaka N. Biological and ecological constraints to the reintroduction of Eriocaulon heleocharioides (Eriocaulaceae): a species extinct in the wild. J Nat Conserv. 2020;56:125866. doi: 10.1016/j.jnc.2020.125866. [DOI] [Google Scholar]
- 8.Ma W, Zhang Z, Thomas S. Eriocaulaceae. Flora of China. 2000;24:7–17. [Google Scholar]
- 9.Davies RJP, Craigie AI, Mackay DA, Whalen MA, Cheong JPE, Leach GJ. Resolution of the taxonomy of Eriocaulon (Eriocaulaceae) taxa endemic to Australian mound springs, using morphometrics and AFLP markers. Aust Syst Bot. 2007;20(5):428–447. doi: 10.1071/SB07019. [DOI] [Google Scholar]
- 10.Leach GJ. A revision of Australian Eriocaulon (Eriocaulaceae) Telopea. 2017;20:205–259. [Google Scholar]
- 11.Leach GJ. Synopsis of the genus Eriocaulon (Eriocaulaceae) for New Guinea. Aust Syst Bot. 2018;31(6):420–432. [Google Scholar]
- 12.Darshetkar AM, Datar MN, Prabhukumar KM, Kim SY, Tamhankar S, Choudhary RK. Systematic analysis of the genus Eriocaulon L. in India based on molecular and morphological evidence. System Biodivers. 2021;19(7):693–723. doi: 10.1080/14772000.2021.1914764. [DOI] [Google Scholar]
- 13.Sunil CN, Kumar VVN. A new species of Eriocaulon (Eriocaulaceae) from Western Ghats. India Webbia. 2015;70(2):211–215. doi: 10.1080/00837792.2015.1050175. [DOI] [Google Scholar]
- 14.Sunil CN, Ratheesh Narayanan MK, Sivadasan M, Alfarhan AH, Abdul Jaleel V. Eriocaulon vandaanamense sp. nov. (Eriocaulaceae) from Kerala India. Nord J Bot. 2015;33(2):155–158. doi: 10.1111/njb.00658. [DOI] [Google Scholar]
- 15.Khanna K, Kumar A. Three new species of Eriocaulon L.(Eriocaulaceae) from India. Biol Forum Int J. 2019;11:21–26. [Google Scholar]
- 16.Nampy S, Akhil MK. Eriocaulon sanjappae (Eriocaulaceae), a new species from the southern Western Ghats, India. Nord J Bot. 2021;39(9).
- 17.Harishma KH, Mohan V, Nampy S. Eriocaulon pandeyana (Eriocaulaceae), a new species from southern Western Ghats. India Phytotaxa. 2022;539(3):273–279. doi: 10.11646/phytotaxa.539.3.6. [DOI] [Google Scholar]
- 18.Souladeth P, Prajaksood A, Parnell JAN, Newman MF. Typification of names in Eriocaulon in the flora of Thailand and flora of Cambodia, Laos and Vietnam. Edinb J Bot. 2017;74(1):5–13. doi: 10.1017/S0960428616000238. [DOI] [Google Scholar]
- 19.Souladeth P, Tagane S, Newman MF, Prajaksood A. Two new species of Eriocaulon (Eriocaulaceae) from Laos. Kew Bull. 2020;75(4):56. doi: 10.1007/s12225-020-09909-0. [DOI] [Google Scholar]
- 20.Khorngton S, Souladeth P, Prajaksood A. Eriocaulon longibracteatum (Eriocaulaceae), a new species from Thailand and Cambodia. Kew Bull. 2020;75(1):20. doi: 10.1007/s12225-020-9879-1. [DOI] [Google Scholar]
- 21.Souladeth P, Newman MF, Prajaksood A. Two new species of Eriocaulon (Eriocaulaceae) from Cambodia. Kew Bull. 2022;77(1):127–137. doi: 10.1007/s12225-022-10005-8. [DOI] [Google Scholar]
- 22.Oliveira ALRD, Bove CP. Eriocaulon albosetaceum: a new species of Eriocaulaceae from the Brazilian Cerrado. Webbia. 2019;74(1):15–21. doi: 10.1080/00837792.2019.1572964. [DOI] [Google Scholar]
- 23.de Oliveira ALR, Bove CP. Two new species of Eriocaulon from the Tocantins-Araguaia river basin. Brazil Syst Bot. 2011;36(3):605–609. doi: 10.1600/036364411X583592. [DOI] [Google Scholar]
- 24.Oliveira ALRD, Bove CP. Eriocaulon L. from Brazil: An annotated checklist and taxonomic novelties. Acta Botanica Brasilica. 2015;29:175–189. doi: 10.1590/0102-33062014abb3668. [DOI] [Google Scholar]
- 25.Ma W. New materials of Eriocaulon L. from China. J Syst Evol. 1991;29(4):289. [Google Scholar]
- 26.Savolainen V, Goudet J. Rate of gene sequence evolution and species diversification in flowering plants: a re-evaluation. Proc R Soc Lond B. 1998;265(1396):603–607. doi: 10.1098/rspb.1998.0337. [DOI] [Google Scholar]
- 27.Zapata JM, Guera A, Esteban-Carrasco A, Martin M, Sabater B. Chloroplasts regulate leaf senescence: delayed senescence in transgenic ndhF-defective tobacco. Cell Death Differ. 2005;12(10):1277–1284. doi: 10.1038/sj.cdd.4401657. [DOI] [PubMed] [Google Scholar]
- 28.Corriveau JL, Coleman AW. Rapid screening method to detect potential biparental inheritance of plastid DNA and results for over 200 angiosperm species. Am J Bot. 1988;75(10):1443–1458. doi: 10.1002/j.1537-2197.1988.tb11219.x. [DOI] [Google Scholar]
- 29.Zhang Q, Liu Y. Sodmergen: Examination of the cytoplasmic DNA in male reproductive cells to determine the potential for cytoplasmic inheritance in 295 angiosperm species. Plant Cell Physiol. 2003;44(9):941–951. doi: 10.1093/pcp/pcg121. [DOI] [PubMed] [Google Scholar]
- 30.Wolfe KH, Perry AS. Nucleotide substitution rates in legume chloroplast DNA depend on the presence of the inverted repeat. J Mol Evol. 2002;55(5):501–508. doi: 10.1007/s00239-002-2333-y. [DOI] [PubMed] [Google Scholar]
- 31.Dong W, Xu C, Wen J, Zhou S. Evolutionary directions of single nucleotide substitutions and structural mutations in the chloroplast genomes of the family Calycanthaceae. BMC Evol Biol. 2020;20(1):96. doi: 10.1186/s12862-020-01661-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong WP, Sun JH, Liu YL, Xu C, Wang YH, Suo Z, Zhou SL, Zhang ZX, Wen J. Phylogenomic relationships and species identification of the olive genus Olea (Oleaceae) J Syst Evol. 2022;60(6):1263–1280. doi: 10.1111/jse.12802. [DOI] [Google Scholar]
- 33.Dong W, Li E, Liu Y, Xu C, Wang Y, Liu K, Cui X, Sun J, Suo Z, Zhang Z, et al. Phylogenomic approaches untangle early divergences and complex diversifications of the olive plant family. BMC Biol. 2022;20(1):92. doi: 10.1186/s12915-022-01297-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clegg MT, Gaut BS, Learn GH, Morton BR. Rates and patterns of chloroplast DNA evolution. Proc Nat Acad Sci USA. 1994;91(15):6795–6801. doi: 10.1073/pnas.91.15.6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li L, Hu Y, He M, Zhang B, Wu W, Cai P, Huo D, Hong Y. Comparative chloroplast genomes: insights into the evolution of the chloroplast genome of Camellia sinensis and the phylogeny of Camellia. BMC Genomics. 2021;22(1):138. doi: 10.1186/s12864-021-07427-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou MY, Liu JX, Ma PF, Yang JB, Li DZ. Plastid phylogenomics shed light on intergeneric relationships and spatiotemporal evolutionary history of Melocanninae (Poaceae: Bambusoideae) J Syst Evol. 2022;60(3):640–652. doi: 10.1111/jse.12843. [DOI] [Google Scholar]
- 37.Dong W, Xu C, Wu P, Cheng T, Yu J, Zhou S, Hong D-Y. Resolving the systematic positions of enigmatic taxa: Manipulating the chloroplast genome data of Saxifragales. Mol Phylogenet Evol. 2018;126:321–330. doi: 10.1016/j.ympev.2018.04.033. [DOI] [PubMed] [Google Scholar]
- 38.Shang C, Li E, Yu Z, Lian M, Chen Z, Liu K, Xu L, Tong Z, Wang M, Dong W. Chloroplast Genomic Resources and Genetic Divergence of Endangered Species Bretschneidera sinensis (Bretschneideraceae). Front Ecol Evol. 2022;10:873100.
- 39.Torre S, Sebastiani F, Burbui G, Pecori F, Pepori AL, Passeri I, Ghelardini L, Selvaggi A, Santini A. Novel Insights Into Refugia at the Southern Margin of the Distribution Range of the Endangered Species Ulmus laevis. Front Plant Sci. 2022;13:826158. doi: 10.3389/fpls.2022.826158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao S, Xu P, Deng Y, Dai X, Zhao L, Heider B, Zhang A, Zhou Z, Cao Q. Comparative analysis of chloroplast genomes of cultivars and wild species of sweetpotato (Ipomoea batatas [L.] Lam) BMC Genomics. 2021;22(1):262. doi: 10.1186/s12864-021-07544-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu H, Zhao W, Hua W, Liu J. A large-scale population based organelle pan-genomes construction and phylogeny analysis reveal the genetic diversity and the evolutionary origins of chloroplast and mitochondrion in Brassica napus L. BMC Genomics. 2022;23(1):339. doi: 10.1186/s12864-022-08573-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dong W, Liu Y, Xu C, Gao Y, Yuan Q, Suo Z, Zhang Z, Sun J. Chloroplast phylogenomic insights into the evolution of Distylium (Hamamelidaceae) BMC Genomics. 2021;22(1):293. doi: 10.1186/s12864-021-07590-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun J, Wang S, Wang Y, Wang R, Liu K, Li E, Qiao P, Shi L, Dong W, Huang L, et al. Phylogenomics and Genetic Diversity of Arnebiae Radix and Its Allies (Arnebia, Boraginaceae) in China. Front Plant Sc. 2022;13:920826. doi: 10.3389/fpls.2022.920826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li D-M, Li J, Wang D-R, Xu Y-C, Zhu G-F. Molecular evolution of chloroplast genomes in subfamily Zingiberoideae (Zingiberaceae) BMC Plant Biol. 2021;21(1):558. doi: 10.1186/s12870-021-03315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li B, Liu T, Ali A, Xiao Y, Shan N, Sun J, Huang Y, Zhou Q, Zhu Q. Complete chloroplast genome sequences of three aroideae species (Araceae): lights into selective pressure, marker development and phylogenetic relationships. BMC Genomics. 2022;23(1):218. doi: 10.1186/s12864-022-08400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Do HDK, Kim JH. A dynamic tandem repeat in monocotyledons inferred from a comparative analysis of chloroplast genomes in Melanthiaceae. Front Plant Sci. 2017;8:693. doi: 10.3389/fpls.2017.00693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mehmood F, Shahzadi I, Ali Z, Islam M, Naeem M, Mirza B, Lockhart PJ, Ahmed I, Waheed MT. Correlations among oligonucleotide repeats, nucleotide substitutions, and insertion–deletion mutations in chloroplast genomes of plant family Malvaceae. J Syst Evol. 2020;59(2):388–402. [Google Scholar]
- 48.Wang M, Wang X, Sun J, Wang Y, Ge Y, Dong W, Yuan Q, Huang L. Phylogenomic and evolutionary dynamics of inverted repeats across Angelica plastomes. BMC Plant Biol. 2021;21(1):26. doi: 10.1186/s12870-020-02801-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li B, Lin F, Huang P, Guo W, Zheng Y. Development of nuclear SSR and chloroplast genome markers in diverse Liriodendron chinense germplasm based on low-coverage whole genome sequencing. Biol Res. 2020;53(1):21. doi: 10.1186/s40659-020-00289-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu D, Abe J, Gai J, Shimamoto Y. Diversity of chloroplast DNA SSRs in wild and cultivated soybeans: evidence for multiple origins of cultivated soybean. Theor Appl Genet. 2002;105(5):645–653. doi: 10.1007/s00122-002-0972-7. [DOI] [PubMed] [Google Scholar]
- 51.Jung J, Kim C, Kim JH. Insights into phylogenetic relationships and genome evolution of subfamily Commelinoideae (Commelinaceae Mirb.) inferred from complete chloroplast genomes. BMC Genomics. 2021;22(1):231. doi: 10.1186/s12864-021-07541-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu K, Lin C, Lee SY, Mao L, Meng K. Comparative analysis of complete Ilex (Aquifoliaceae) chloroplast genomes: insights into evolutionary dynamics and phylogenetic relationships. BMC Genomics. 2022;23(1):203. doi: 10.1186/s12864-022-08397-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu S, Wang Z, Su Y, Wang T. Comparative genomic analysis of Polypodiaceae chloroplasts reveals fine structural features and dynamic insertion sequences. BMC Plant Biol. 2021;21(1):31. doi: 10.1186/s12870-020-02800-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dong W, Xu C, Liu Y, Shi J, Li W, Suo Z. Chloroplast phylogenomics and divergence times of Lagerstroemia (Lythraceae) BMC Genomics. 2021;22:434. doi: 10.1186/s12864-021-07769-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fishbein M, Kephart SR, Wilder M, Halpin KM, Datwyler SL. Phylogeny of Camassia (Agavaceae) inferred from plastid rpl16 intron and trnD–trnY–trnE–trnT intergenic spacer DNA sequences: implications for species delimitation. Syst Bot. 2010;35(1):77–85. doi: 10.1600/036364410790862588. [DOI] [Google Scholar]
- 56.Torres DC, Lima JPMS, Fernandes AG, Nunes EP, Grangeiro TB. Phylogenetic relationships within Chamaecrista sect. Xerocalyx (Leguminosae, Caesalpinioideae) inferred from the cpDNA trnE-trnT intergenic spacer and nrDNA ITS sequences. Genet Mol Biol. 2011;34:244–51. [DOI] [PMC free article] [PubMed]
- 57.Melotto-Passarin DM, Berger IJ, Dressano K, De Martin VdF, Oliveira GCX, Bock R, Carrer H. Phylogenetic relationships in Solanaceae and related species based on cpDNA sequence from plastid trnE-trnT region. Crop Breed Appl Biotech. 2008;8(1):85–95.
- 58.Deguilloux MF, Dumolin-Lapègue S, Gielly L, Grivet D, Petit RJ. A set of primers for the amplification of chloroplast microsatellites in Quercus. Mol Ecol Notes. 2003;3(1):24–27. doi: 10.1046/j.1471-8286.2003.00339.x. [DOI] [Google Scholar]
- 59.Dong W, Liu J, Yu J, Wang L, Zhou S. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS ONE. 2012;7(4):e35071. doi: 10.1371/journal.pone.0035071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dong W, Xu C, Li C, Sun J, Zuo Y, Shi S, Cheng T, Guo J, Zhou S. ycf1, the most promising plastid DNA barcode of land plants. Sci Rep. 2015;5:8348. doi: 10.1038/srep08348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Neubig KM, Abbott JR. Primer development for the plastid region ycf1 in Annonaceae and Other Magnoliids. Am J Bot. 2010;97(6):E52–E55. doi: 10.3732/ajb.1000128. [DOI] [PubMed] [Google Scholar]
- 62.Handy SM, Parks MB, Deeds JR, Liston A, de Jager LS, Luccioli S, Kwegyir-Afful E, Fardin-Kia AR, Begley TH, Rader JI, et al. Use of the chloroplast gene ycf1 for the genetic differentiation of Pine Nuts obtained from consumers experiencing dysgeusia. J Agric Food Chem. 2011;59(20):10995–11002. doi: 10.1021/jf203215v. [DOI] [PubMed] [Google Scholar]
- 63.Neubig K, Whitten W, Carlsward B, Blanco M, Endara L, Williams N, Moore M. Phylogenetic utility of ycf1 in orchids: a plastid gene more variable than matK. Plant Syst Evol. 2009;277(1):75–84. doi: 10.1007/s00606-008-0105-0. [DOI] [Google Scholar]
- 64.Zachos J. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science. 2001;292(5517):686–693. doi: 10.1126/science.1059412. [DOI] [PubMed] [Google Scholar]
- 65.Wolfe JA. A Paleobotanical interpretation of tertiary climates in the Northern Hemisphere: data from fossil plants make it possible to reconstruct Tertiary climatic changes, which may be correlated with changes in the inclination of the earth's rotational axis. Am Sci. 1978;66(6):694–703. [Google Scholar]
- 66.Miller KG, Fairbanks RG. Evidence for Oligocene-middle Miocene abyssal circulation changes in the western North Atlantic. Nature. 1983;306(5940):250–253. doi: 10.1038/306250a0. [DOI] [Google Scholar]
- 67.Keller G, Barron JA. Paleoceanographic implications of Miocene deep-sea hiatuses. GSA Bull. 1983;94(5):590–613. doi: 10.1130/0016-7606(1983)94<590:PIOMDH>2.0.CO;2. [DOI] [Google Scholar]
- 68.Buchardt B. Oxygen isotope palaeotemperatures from the tertiary period in the North Sea area. Nature. 1978;275(5676):121–123. doi: 10.1038/275121a0. [DOI] [Google Scholar]
- 69.Li J, Wang S, Jing Y, Wang L, Zhou S. A modified CTAB protocol for plant DNA extraction. Chin Bull Bot. 2013;48(1):72–78. doi: 10.3724/SP.J.1259.2013.00072. [DOI] [Google Scholar]
- 70.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020;21(1):241. doi: 10.1186/s13059-020-02154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang DI, Cronk QCB. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 2015;3(8):1500026. doi: 10.3732/apps.1500026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):167–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zheng S, Poczai P, Hyvönen J, Tang J, Amiryousefi A. Chloroplot: an online program for the versatile plotting of organelle genomes. Front Genet. 2020;11:576124. doi: 10.3389/fgene.2020.576124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13(8):1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 76.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rozas J, Ferrer-Mata A, Sanchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sanchez-Gracia A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol. 2017;34(12):3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- 78.Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29(22):4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Beier S, Thiel T, Münch T, Scholz U, Mascher M. MISA-web: a web server for microsatellite prediction. Bioinformatics. 2017;33(16):2583–2585. doi: 10.1093/bioinformatics/btx198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14(6):587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 2019;35(21):4453–4455. doi: 10.1093/bioinformatics/btz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang D, Gao F, Jakovlic I, Zou H, Zhang J, Li WX, Wang GT. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 2020;20(1):348–355. doi: 10.1111/1755-0998.13096. [DOI] [PubMed] [Google Scholar]
- 84.Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37(5):1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bouckaert R, Heled J, Kuhnert D, Vaughan T, Wu CH, Xie D, Suchard MA, Rambaut A, Drummond AJ. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comp Biol. 2014;10(4):e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rambaut A, Suchard M, Xie D, Drummond A. Tracer v1. 6. 2014. Available from http://beast.bio.ed.ac.uk/Tracer.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. The RSCU values of the coding genes in the Eriocaulon chloroplast genome.
Additional file 2: Figure S2. mVISTA-based sequence identity plot of 18 Eriocaulon species, using E. alpestre as a reference.
Additional file 3: Figure S3. Phylogenetic trees of Eriocaulon. a. The whole chloroplast genome dataset. b. The 83-genes dataset. The number above the lines indicates the ML bootstrap values (BS) and BI posterior probability (PP). BS=100 and PP=1.0 are not shown.
Additional file 4: Figure S4. Divergence times of Eriocaulon using the five chloroplast genes.
Additional file 5: Table S1. Sampling information for the Eriocaulon samples in this study.
Additional file 6: Table S2. The dN, dS, and ω values of 79 protein-coding genes.
Additional file 7: Table S3. The GenBank information of the chloroplast genes used in inferring the Eriocaulon phylogeny.
Data Availability Statement
New sequenced and other published chloroplast genome sequences can be found in GenBank (https://www.ncbi.nlm.nih.gov/genbank/), and the accession numbers showed in Table S1.