Abstract
Computational approaches for drug discovery such as quantitative structure–activity relationship (QSAR) rely on structural similarities of small molecules to infer biological activity, but are often limited to identifying new drug candidates in the chemical spaces close to known ligands. Here we report a biological activity-based modeling (BABM) approach, in which compound activity profiles established across multiple assays are used as signatures to predict compound activity in other assays or against a new target. This approach was validated by identifying candidate antivirals for Zika and Ebola based on high throughput screening data. BABM models were then applied to predict 311 compounds with potential anti-SARS-CoV-2 activity. 32% of the predicted compounds had antiviral activity in a cell culture live virus assay, the most potent compounds showing an IC50 in the nanomolar range. Most of the confirmed anti-SARS-CoV-2 compounds were found to be viral entry inhibitors and/or autophagy modulators. The confirmed compounds have the potential to be further developed into anti-SARS-CoV-2 therapies.
Editorial summary:
Activity profiles generated from quantitative high-throughput screening improve drug candidate prediction
Introduction
The early-stage drug discovery process relies on target identification, assay development, and high throughput screening (HTS) to identify lead compounds for chemical optimization and further preclinical development. Traditional HTS campaigns are often limited to 1-2 million compounds due to the high costs and operational bottle necks that limit the chance for lead identification.1,2 However, recent advances in computational technologies have made it possible to virtually screen millions of compounds for potential biological activity.2 Existing virtual screening (VS) methods can be grouped into two broad categories: ligand-based VS and target structure-based VS. Both methods depend on chemical structure information to make predictions while the target-based approach in addition requires the availability of detailed target protein information. These severe dependencies have tended to limit the applicability of such methods to querying only in the close structural vicinity of already known ligand structures and drug targets.
A critical advance that enabled the development of activity rather than structural paradigm described here was the large-scale application of quantitative HTS (qHTS)3, where every compound is tested in a broad concentration response format. The high-quality data from qHTS are thus substantially richer for use in computational modeling to predict activities of large compound libraries against new assays or new drug targets. In the past 15 years, our in-house collections of over half a million compounds have been screened in a wide spectrum of biological assays in qHTS format,4 resulting in compound activity profiles that enabled the development of a biological activity-based modeling (BABM) approach complementary to traditional structure-based approaches. Among our in-house libraries, the NCATS Pharmaceutical Collection (NPC)5 and the Library of Pharmacologically Active Compounds (LOPAC) have been screened in nearly every one of our ~2,000 assays providing the most comprehensive set of activity profiles that comprise an ideal training dataset for machine learning models.
Unlike traditional QSAR approaches (part of the ligand-based VS category),6,7 where similarity in chemical structure is used to infer biological activity, BABM builds on the hypothesis that compounds that show similar activity patterns tend to share similar targets or mechanisms of action.8,9 In this approach, each assay is treated as an independent descriptor. Analogous to structure descriptors, where the presence and absence of certain structure features or properties are used to represent a compound, the presence and absence of activities against a panel of assays form the activity profile or signature of a compound. If extracted from across multiple screening campaigns each at massive scale, such activity signatures can then be applied to infer compound activity in a completely new assay or against a completely new target.
A fundamental difference compared to traditional QSAR modeling is thus that BABM does not require any chemical structure information to make predictions, such that its application domain is not limited to small molecules with well-defined structures. In fact, BABM can be applied to any substances with available biological profiling, including macromolecules and mixtures (e.g. natural products). Of particular note is that compounds showing similar activities do not necessarily share similar structures.10 Thus the BABM approach has no intrinsic limitations in discovering new chemical scaffolds.11 These new scaffolds can then serve as starting points for lead identification efforts and be used to construct new QSAR models for lead optimization.
The global pandemic of the highly contagious coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2),12 presented an urgent need for new methods that can quickly and systematically screen large compound libraries for new drug candidates. In this context, we first applied BABM to generate prediction models for two infectious diseases, Zika13 and Ebola,14 to test the robustness of BABM and its applicability to different assay and data types, and to benchmark against traditional QSAR methods. The BABM model identified actives that were experimentally verified with high confirmation rates (~50-80%). The approach was then applied to build prediction models for SARS-CoV-2. To build prediction models for these disease targets, we selected training data that included both qHTS assay data (SARS-CoV-2 and Zika virus (ZIKV) non-structural protein NS115) and data collected from published literature (SARS-CoV-2 and Ebola virus (EBOV)16). These models, mostly trained on the qHTS activity profiles of the NPC and LOPAC library compounds, were applied to predict the activity of all ~0.5 million compounds in our in-house library. Models were constructed using BABM and the performances were compared with those of traditional QSAR models as well as a combination of both activity and structural features. A little over 300 compounds identified by the BABM models as potential anti-SARS-CoV-2 leads were then tested in a live virus assay with ~100 confirmed (>30%), validating the utility and accuracy of the BABM approach. The confirmed anti-SARS-CoV-2 compounds were further investigated for their potential antiviral mechanisms in terms of viral entry inhibition,17 SARS-CoV-2 main protease inhibition,18 and autophagy modulation.19 Some of the experimentally confirmed lead compounds may have the potential to be further developed into new antiviral therapies.
Results
Model performance and validation
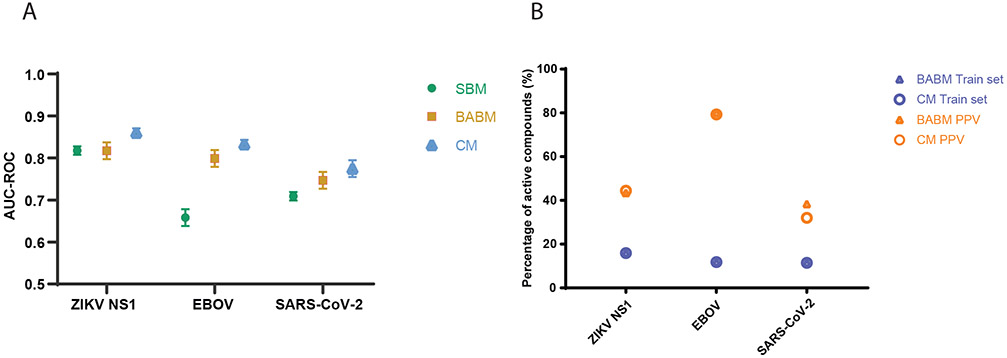
Table 1 provides an overview of the three viral targets (SARS-CoV-2, ZIKV, EBOV) used for modeling. The entire model training, testing and validation process is illustrated in Figure 1. Model performance was measured by the area under the ROC curve (AUC-ROC; see Online Methods section for details). The majority of the models performed well on their corresponding test sets with mean AUC-ROC values >0.8 (Figure 2A and Supplementary Table 1). The structure-activity combined models (CM) showed the best performances compared to the models built on activity (BABM) or structure (SBM) alone with mean AUC-ROC values >0.83. Of the BABM models using data from different assay panels and compound libraries, the BABM-S and BABM-M models all showed good performances with mean AUC-ROC values of 0.79 and 0.84, respectively (Supplementary Table 1). The BABM-G models with the smallest assay panel for training showed the lowest AUC-ROC values averaging 0.75. The structure-based models generally showed lower performances than the CM and BABM models with a mean AUC-ROC of 0.72. Supplementary Figure 1 shows example ROC curves from each of the three types of models.
Table 1.
Overview of viral targets used for modeling and summary statistics of model identified active compounds.
| Model | Data Source | Data Type | Active Enrichment (fold)* |
Potency Range** |
Structure Diversity*** |
|---|---|---|---|---|---|
| SARS-CoV-2 | In house/Literature | HTS/Literature | 2.8-3.3 | 0.5 μM - 32 μM | 0.07-0.23 |
| ZIKV NS1 a | In house | qHTS | 2.7-27.5 | 1 nM - 63 μM | 0.04-0.26 |
| EBOV | Literature | HTS | 6.5-6.8 | 0.4 μM - 25 μM | 0.07-0.21 |
The NS1 protein is a nonstructural protein that is not present in the virus itself but only appears in the host cells when virus starts replication. The NS1 assay detects compounds that block NS1 protein production, which may inhibit virus entry to cells, and virus RNA and virus protein productions in cells.
Fold = hit rate of model predicted actives/hit rate of assay used for model training
Potency range of experimentally confirmed model predicted actives
Tanimoto similarity was calculated between each model predicted active and active compounds in the training set. The values shown here are the range of the average Tanimoto scores for the model predicted actives.
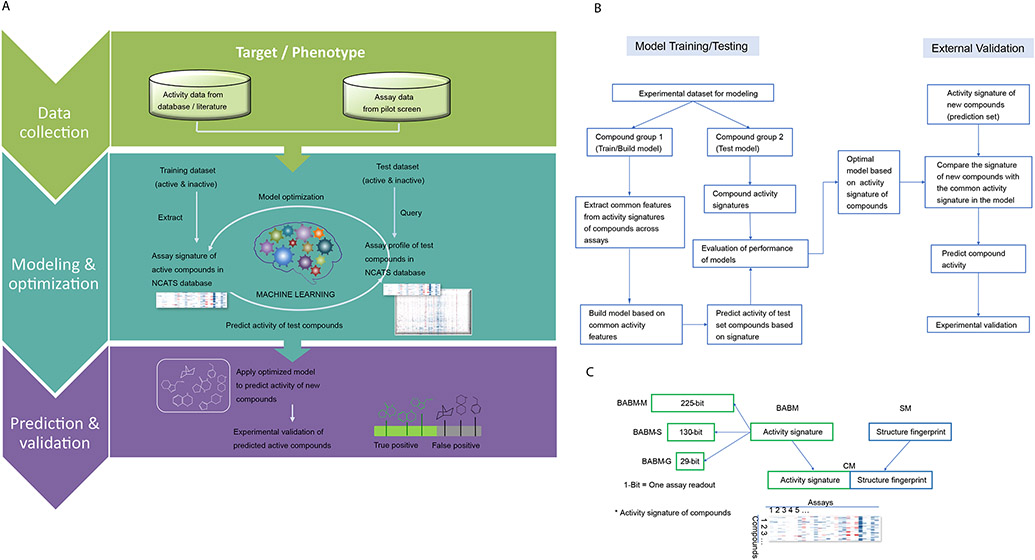
Figure 1.
A. Biological activity-based modeling (BABM) process overview. For any biological target of interest, T (e.g., SARS-CoV-2, ZIKV NS1, EBOV), the model identifies the activity pattern of active vs. inactive compounds based on the training data, which are activity profiles of a set of compounds across a diverse panel of assays including T. The active signature is then matched against the activity profiles of a new set of compounds across the same assay panel. The ability of the model to use this signature to correctly identify actives from the new compound set is first tested using part of the data with known T activity (the test set). An AUC-ROC value is calculated using the test set to evaluate the model performance. The model is then applied to a set of compounds with unknown T activity (prediction set; e.g., Sytravon, MLS, Genesis). Predictions are made on the new compounds based on their activity profile similarity to that of the active signature for T. The predicted T actives are further validated experimentally for their activity against T. Comparing experimental results with model predictions, true positives (TP) and false positives (FP) are counted to determine the performance of the model. In the heat maps, each row represents a compound, each column is an assay, and the heat map is colored by the compound activity. B. Detailed flowchart of the modeling process. C. Types of signatures and fingerprints used in different models.
Figure 2.
Model performance and experimental validation. A. Model performances on the test sets measured by AUC-ROC values. Mean AUC-ROC values from ten randomly generated test sets are plotted with the error bars indicating the SD values. B. Model performances measured by external experimental validation PPV (colored in different shades of brown) in comparison to training set active rates (e.g., original assay hit rate; colored in different shades of blue) (Supplementary Table 2). Model selected compounds are significantly enriched with true actives. Model type: SBM = Structure based model; BABM = Activity-based model (Sytravon); CM = Combined model (SBM+BABM).
To further validate the models and identify new compounds with antiviral activity, a subset of model predicted actives was selected for each viral target for experimental validation (see Online Methods for details). For ZIKV, 1,676 selected actives predicted by the model were tested in the original NS1 assay15 that generated the data to train the models. To validate the EBOV models, the EBOV-eGFP infection assay15,20 was applied to test 96 selected model predicted EBOV actives. All 96 compounds were first inspected at 30 μM for potential cytotoxicity, resulting in 62 compounds with <50% cell killing, which were further tested for EBOV infection inhibition. The EBOV inhibition activity of these 62 compounds were used to evaluate model performance. The positive predictive value [PPV = TP/(TP+FP)], i.e., the fraction of model predicted actives that are experimentally confirmed, was calculated for each model (Figure 2B and Supplementary Table 1). The model PPVs ranged from 30% (SBM for NS1) to 89% (CM-G for EBOV). The EBOV models showed higher PPVs (~80%) than the NS1 models (~40%). Compared to the active rates in their corresponding training datasets (i.e. original assay hit rate), all model predicted active sets were significantly enriched with true active compounds (two-tailed Fisher’s exact test: p<10−10) (Supplementary Table 1). For example, the active rate of the EBOV BABM-S model training set was 11.8% and the corresponding model PPV (i.e. experimental validation set active rate) was 80%. Thus the enrichment of actives by the EBOV BABM-S model was 6.8-fold (80/11.8). The enrichment of actives for all models (Table 1) ranged from 2.7-fold (BABM-S for NS1; p<10−20) to 27.5-fold (SBM for NS1; p<10−20).
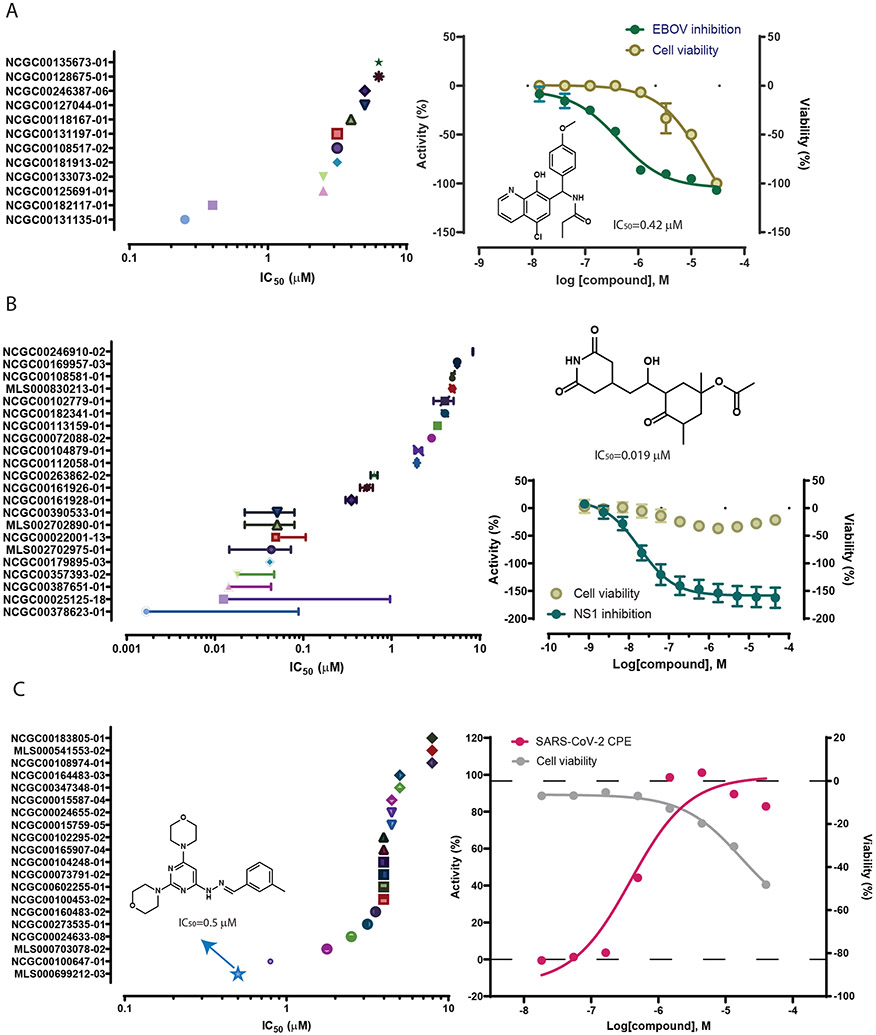
Most models showed enrichments between 5- and 10-fold when compared to the active rates in the training set. The potency ranges of the experimentally confirmed actives are summarized in Table 1 and Figure 3. The models identified potent compounds for all three disease targets with IC50s in the nanomolar range (Figure 3). Experimental validation data for all models are provided as Supplementary Data 1.
Figure 3.
Experimental validation results from the sencondary confirmation of model predicted actives. A. Potencies and examples of compounds confirmed in the EBOV inhibition assay with minimal cytotoxicity. Replicate data are presented as mean±SD. B. Potencies and examples of compounds confirmed in the ZIKV NS1 inhibition assay with minimal cytotoxicity. Replicate data (n = 3) are presented as mean±SD. C. Potencies and examples of compounds confirmed in the anti-SARS-CoV-2 CPE assay.
Identification of anti-EBOV and anti-ZIKV compounds
Of the 50 compounds with anti-EBOV activity confirmed at 30 μM, we selected 27 that showed >90% inhibition of EBOV infection with minimal cytotoxicity (>80% cell viability) to test in concentration-response format (0.17 nM to 30 μM; 1:3 fold dilution; triplicate) to determine their EBOV inhibition potency. All 27 compounds showed concentration dependent inhibition of EBOV infection with IC50s ranging from 25 nM to 25 μM (Figure 3A; Supplementary Data 2). Seven of these compounds were potent with IC50 ≤5 μM and were not apparently cytotoxic or at least six times more potent in the EBOV inhibition assay compared to the cell viability counter assay. Two of the seven compounds, umifenovir and difeterol, are known drugs (see Supplemental Information for details). The other five compounds have no previously reported anti-EBOV activity.
A subset (170) of the experimentally confirmed NS1-assay active compounds with relatively potent NS1 signal inhibition activity (IC50 <10 μM) and no apparent cytotoxicity were selected for secondary confirmation with compounds tested at 11 concentrations in triplicate (Figure 3B; Supplementary Data 2). Ten of the 170 compounds did not show activity in the secondary confirmation assay yielding a confirmation rate of 94% for the NS1 assay. 29 compounds showed potent inhibition with IC50 <1 μM, 17 of which were not apparently cytotoxic or at least three times more potent in the NS1 assay. A number of these potent compounds are known drugs or bioactive compounds (see Supplemental Information for details). The other eight potent compounds can potentially be developed into new antiviral therapies.
Identification of anti-SARS-CoV-2 compounds
The activity of 311 compounds predicted by the SARS-CoV-2 BABM models were tested in the live virus CPE assay, 99 of which were confirmed as active, yielding a hit rate of 32% (Figure 2B and Supplementary Table 1). The model PPVs ranged from 32% (CM-S) to 38% (BABM-S). Compared to the active rates in their corresponding training datasets, all model predicted active sets were significantly enriched with true active compounds (two-tailed Fisher’s exact test: p<10−3) (Figure 3C and Supplementary Table 1). Compared to the hit rate of the original NPC screen (11%), the models were able to improve the hit rate by 2.8- to 3.3-fold (Table 1). The SBM was not used for compound selection because its performance (average AUC-ROC = 0.71) during model training and testing did not meet the 0.75 cutoff. Nonetheless, the SBM predictions made on the 311 compounds were used to assess the performance of the SBM on the experiment validation set in comparison with the BABM models (Figure 2B and Supplementary Table 1). The PPV of the SBM was 31.6%, lowest of all SARS-CoV-2 models. The potency ranges of the experimentally confirmed actives are summarized in Table 1 and Figure 3. Experimental validation data for all 311 compounds are provided in Supplementary Data 1 and 3. The structures and WFS scores of ~5,000 compounds predicted as active by at least one of the SARS-CoV-2 BABM models are provided as Supplementary Data 4.
The experimentally confirmed SARS-CoV-2 active compounds were further tested at 8 concentrations (instead of 3 concentrations in the primary screen) to get more accurate potency measures (Supplementary Data 2 and 5). Nine of the 94 compounds became inactive in the secondary confirmation assay yielding a confirmation rate of 90% for the SARS-CoV-2 CPE assay. The most potent compound (MLS000699212-03; Benzaldehyde, 3-methyl-, 2-(2,6-di-4-morpholinyl-4-pyrimidinyl) hydrazone) had an IC50 of 500 nM. This compound showed slight cytotoxicity inhibiting 55% cell viability with an IC50 of 14 μM, indicating a large therapeutic window (selectivity index or SI = 28). This compound has only one published study, which is a patent on a compound series described as autophagy modulators for treating neurodegenerative diseases.21 Autophagy has been implicated in the entry of coronavirus into host cells, including SARS-CoV, MERS-CoV and SARS-CoV-222,23. Another potent compound with IC50 <1 μM (800 nM) is a synthetic compound with no previous literature report (NCGC00100647-01; N2,N4-bis(3-methylphenyl)-6-(4-morpholinyl)-1,3,5-Triazine-2,4-diamine). In addition, 13 compounds had IC50 <5 μM, 8 of which are known drugs or bioactives (see Supplemental Information for details), and the other 5 are compounds without any well annotated biological activity. Some of the known anti-SARS-CoV-2 compounds reported in the literature, especially those currently in clinical trials for COVID-19, were also screened in our CPE assay with varying potencies,24 for example, remdesivir (10 μM), chloroquine (6.5 μM), lopinavir (12.6 μM), azithromycin (48 μM), apilimod (23 nM), and emetine (46 nM). In comparison, the potencies of the anti-SARS-CoV-2 compounds identified by our models fall within the range of the known anti-SARS-CoV-2 compounds.
Antiviral mechanism of anti-SARS-CoV-2 compounds
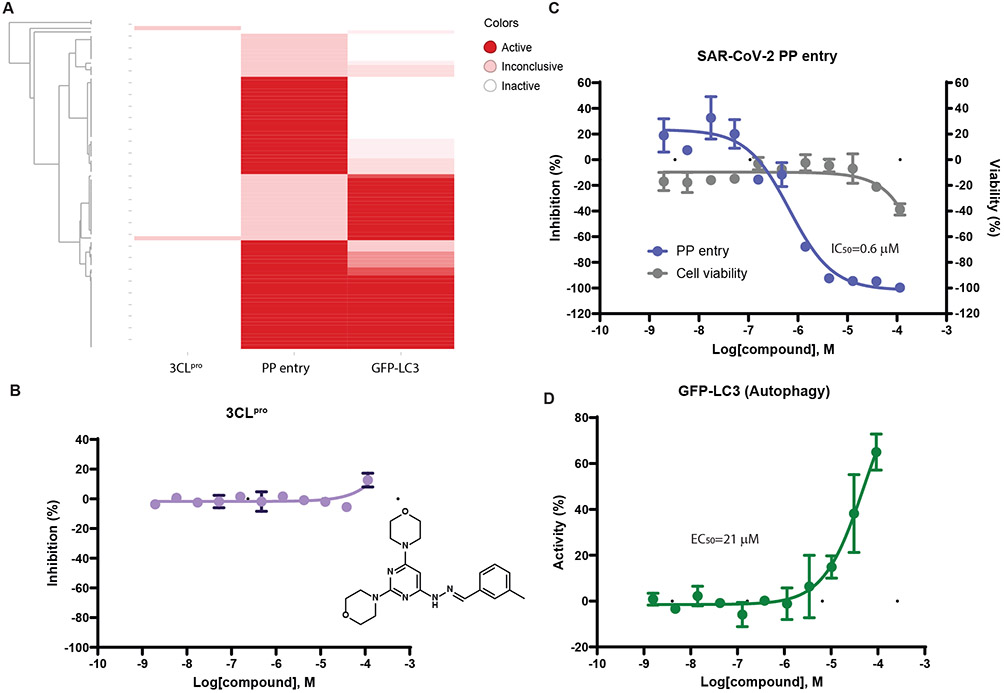
There are multiple targets for therapeutics intervention against SARS-CoV-2 infection including viral entry into host cells, proteolysis of viral polypeptide by the 3C-like protease to release the non-structural proteins, and autophagy pathway in host cells.25 We further investigated the potential antiviral mechanism of the 85 experimentally confirmed anti-SARS-CoV-2 compounds using three assays, the SARS-CoV-2 pseudotyped particle (PP) entry assay,26-28 the SARS-CoV-2 3C-like protease (3CLpro) assay,18 and the GFP-LC3 assay for autophagy modulators (see Supplemental Information for details).29 Out of the 85 anti-SARS-CoV-2 compounds, 53 were viral entry inhibitors determined by the PP entry assay, 35 were identified as autophagy modulators in the GFP-LC3 assay by all three parameters, and 52 were active in at least one autophagy parameter (Figure 4A). Two compounds showed marginal activity in the 3CLpro assay. The results from all three assays are summarized in Supplementary Data 2. These results suggest that autophagy plays a major role in the antiviral activity of the model identified anti-SARS-CoV-2 compounds. Most of these compounds are viral entry inhibitors, and 3CLpro inhibition (related to viral replication) is not a major antiviral mechanism of these compounds. The most potent anti-SARS-CoV-2 compound, MLS000699212, showed potent inhibition (IC50 = 592 nM) of viral cell entry and was active in all three parameters of the autophagy assay, indicating a dual mechanism of action (Figure 4).
Figure 4.
Mechanistic study of experimentally connfirmed anti-SARS-CoV-2 compounds. A. Activity distribution of compounds in the SARS-CoV-2 PP entry, 3CLpro, and GFP-LC3 autophagy assays. Concentration response curves of the most potent compound, MLS000699212, in the SARS-CoV-2 3CLpro (B),PP entry (C), and GFP-LC3 autophagy (D) assays.
Discussion
Traditional QSAR models rely on chemical structure similarity to infer biological activity and thus are limited in their power to discover new chemical scaffolds. Consequently biological activity predictions made on chemicals with structure types not included in the training set are often not reliable – this is commonly referred to as the “applicability domain” (AD) issue.30 QSAR models are thus fundamentally restricted by their ADs, namely by the chemical spaces within which the models were originally trained. Incorporating biological response patterns into the models helps to alleviate this issue by expanding the model AD to cover structurally dissimilar chemicals that share similar activity profiles. Activity-based modeling is a relatively new concept, especially when applied to drug discovery. The prerequisite of activity-based modeling is the availability of sets of compounds tested consistently across multiple biological assays with the results serving as compound descriptors or fingerprints. This is enabled by the recent advances in HTS technologies that have produced a tremendous amount of biological activity data in a relatively short amount of time. As a center specialized in HTS, NCATS has a data repository that hosts biological response data on over half a million compounds tested against thousands of assays mostly in qHTS format, which form a rich set of activity profiles at unprecedented scale (over 130 million wells screened over the last 4 years) .3,31 We show here that a subset of these data could be used to build activity-based models to identify antiviral compounds for Zika, Ebola and COVID-19.
Compared to traditional QSAR models built with chemical structure data alone, the BABM identified compounds that are structurally distinct from the training set and the compounds identified by the SBM (see Supplemental Information for details), demonstrating the advantage of the BABM in discovering new chemical types. Combining traditional structure-based models with BABM can maximize the chance of identifying the best lead compounds as new candidates for any therapeutic target of interest. Both the BABM and CM models used activity data in other assays as descriptors for training while the CM used structure features in addition. The model predictions were further validated experimentally. Using the ZIKV NS1 models for example, even though the BABM identified a larger portion of the experimentally confirmed actives (i.e., was more sensitive), the CM had a lower false positive rate (i.e., was more specific). Adding structure information helped the CM to achieve a slightly improved PPV. For all three viral targets modeled in this study, the CM models achieved the best overall performance compared to the SBM and BABM models. More intriguingly, the sizes of the training sets for all the models were much smaller than the prediction sets on which the models were applied, 30- to 100-fold for the BABM and CM models, and up to 300-fold for the SBM (Supplementary Table 2). That models built on a small training set performed well on predicting a much larger and more diverse set of compounds with accuracies on par with or better than most in silico screening approaches further demonstrated that the models were robust enough to be applicable to large and diverse compound collections to identify new leads.1,32
The SARS-CoV-2 BABM models identified ~100 compounds that were experimentally verified to show antiviral activity in a live virus assay. The results from further mechanism of action studies showed that most of these compounds inhibited SARS-CoV-2 cell entry, and/or modulated the autophagy process in host cells. Models built for Zika and Ebola also identified new lead compounds. In addition, we provided the prediction results of ~5,000 compounds that were predicted as active by the SARS-CoV-2 BABM models as a resource to the scientific community to develop new anti-COVID-19 therapies. The activity-based approach was demonstrated here to be able to be rapidly applied to identify lead compounds for new targets or disease phenotypes.
As a complement to structure-based approaches, either ligand or target structure-based, the additional information provided by activity data is shown here to significantly improve the predictive power of VS models. Furthermore, the assays, as part of the activity signature, that contributed the most to the predictive power of the BABM models could provide clues to the underlying targets or mechanisms of the disease for which the models were built, such as COVID-19.33 The chemical scaffolds identified by BABM from an existing screening library can also be incorporated into QSAR models to screen other chemical libraries more efficiently with no bioactivity profiles available. Of note is that, in addition to HTS libraries, the general concept of BABM can be extended to any type of biological data, such as genomics and proteomics data,34 data generated on mixtures or antibodies, and clinical data, where clearly defined structure information is not available. As such the BABM approach shows the promise of broad applications in different areas of biology.
Online Methods
SARS-CoV-2 cytopathic effect (CPE) assay
Vero-E6 cells (ATCC® VeroE6 CRL-1586) previously selected for high ACE2 expression 35 (grown in EMEM, 10% FBS, and 1% Penicillin/Streptomycin) were cultured in T175 flasks and passaged at 95% confluency. Cells were washed once with PBS and dissociated from the flask using TrypLE. Cells were counted prior to seeding. A CPE assay previously used to measure antiviral effects against SARS-CoV 36 was adapted for performance in 384 well plates to measure CPE of SARS CoV-2 with the following modifications. Cells, harvested and suspended at 160,000 cells/ml in MEM/1% PSG/1% HEPES supplemented 2% HI FBS, were batch inoculated with SARS CoV-2 (USA_WA1/2020) at M.O.I. of approximately 0.002 which resulted in approximately 5% cell viability 72 h post infection. Compound solutions in DMSO were acoustically dispensed into assay ready plates (ARPs) as 3 point 1:5 titrations (or 8 point 1:3 titrations for confirmation screen). ARPs were stored at −20°C and shipped to BSL3 facility (Southern Research Institute, Birmingham, AL) for CPE assay. ARPs were brought to room temperature and 5μl of assay media was dispensed to all wells. The plates were transported into the BSL-3 facility were a 25 μL aliquot of virus inoculated cells (4000 Vero E6 cells/well) was added to each well in columns 3-24. The wells in columns 23-24 contained virus infected cells only (no compound treatment). A 25 μL aliquot of uninfected cells was added to columns 1-2 of each plate for the cell only (no virus) controls. After incubating plates at 37°C with 5% CO2 and 90% humidity for 72 h, 30 μL of Cell Titer-Glo (Promega, Madison, WI) was added to each well. Following incubation at room temperature for 10 minutes the plates were sealed with a clear cover, surface decontaminated, and luminescence was read using a Perkin Elmer Envision (Waltham, MA) plate reader to measure cell viability.
NS1 TR-FRET assay
HEK293 cells were maintained in EMEM medium with 10% fetal bovine serum, 1% pen/strep (Gibco, Cat. # 15140–122). Cells were seeded at 1000 cells/3 μL/well in the white 1536-well plate and incubated at 37 °C with 5% CO2 overnight. Compounds in dilution were added to cells at 23 nL/well and incubated for one hour followed by addition of 2 μL/well of the prototypic ZIKV strain, MR766 solution to cells (MOI = 0.5). After an incubation at 37 °C for 24 h, 2.5 μL/well of detection reagent mixture of two labeled anti-ZIKV NS1 antibodies was added to assay plates. TR-FRET signals were measured using an Envision plate reader (PerkinElmer). Compounds were tested as 7 point 1:5 titrations in the primary screen and 11 point 1:3 titrations in triplicate in the confirmation screen. Data were normalized by using the control wells (without addition of ZIKV) as a negative control (0% NS1) and positive wells (with ZIKV) as 100% NS1 level.
ATP content assay for cell viability and compound cytotoxicity
Cells were seeded in the 1536- well assay plates and incubated for 16 hours at 37°C with 5% CO2. Test compounds dissolved in DMSO were added to assay plates at a volume of 23 nl/well by an automated pintool workstation (Wako Automation, San Diego, CA). Compounds were incubated with cells for 48 hours at 37°C with 5% CO2. ATPlite, the ATP monitoring reagent (PerkinElmer), was then transferred to the assay plates and incubated for 15 minutes at RT. The resulting luminescence was measured using the PHERAstar FSX plate reader (BMG Labtech, Cary, NC, USA). Data was normalized using the wells without cells as a control for 100% cell killing, and cell-containing wells with DMSO control were used as full cell viability (0% cell killing).
EBOV-eGFP infection assay
As described previously, 15,20 vero E6 cells were maintained in Dulbecco's modified Eagle medium (DMEM) (HyClone) supplemented with 10% fetal bovine serum (FBS) (Sigma-Aldrich). The following Ebola virus was used: Ebola virus NML/H.sapiens-lab/COD/1976/Mayinga-eGFP-p3 (EBOV/May-eGFP) (derived from an Ebola virus, family Filoviridae, genus Ebolavirus, species Zaire ebolavirus, GenBank accession No NC_002549). All work with infectious virus was performed in the biosafety level 4 (BSL-4) facility at the National Microbiology Laboratory (NML) of the Public Health Agency of Canada (PHAC) in the Canadian Science Centre for Human and Animal Health (CSCHAH), Winnipeg, Canada. All procedures were conducted in accordance with international protocols appropriate for this level of biosafety. The toxicity of compounds was evaluated in Vero E6 cells by using the PrestoBlue cell viability reagent, which is a resazurin dye-based assay (Life Technologies, Canada). Cells were plated, allowed to adhere overnight, and then treated with various compound concentrations for 2 h. Control cells received an equivalent volume of 10% dimethyl sulfoxide (DMSO) only. PrestoBlue cell viability reagent was added according to the manufacturer's protocol. Viability was determined by comparing fluorescence readings of treated cells to those of untreated controls.
3CLpro enzyme assay and counter screen18
SARS-CoV-2 3CLpro, sensitive internally quenched fluorogenic substrate, and assay buffer were obtained from BPS Bioscience (San Diego, CA, USA). The enzyme was expressed in E. coli expression system with a molecular weight of 34 kDa. The peptide substrate contains 14 amino sequence (KTSAVLQSGFRKME) with Dabcyl and Edans attached on its N- and C-termini, respectively. The reaction buffer is composed of 20 mM Tris-HCl (pH 7.3), 100 mM NaCl, 1 mM EDTA, 0.01 % BSA (bovine serum albumin), and 1 mM 1,4-dithio-D, L-threitol (DTT). The 3CLpro enzyme assay was carried out in 1536-well black, medium binding microplates (Greiner BioOne, Monroe, NC, USA) with a total volume of 4 μL that includes 2 μL 2X enzyme (50 nM) in reaction buffer and 2 μL 2X substrate (20 μM). The experiment was conducted at room temperature (RT). In brief, 2 μL/well enzyme was firstly added into 1536-well plate. Compounds in DMSO were then transferred as 23 nL/well with an automated pintool workstation (WAKO Scientific Solutions, San Diego, CA). The compounds and enzyme were incubated for 30 min at RT. Afterwards, 2 μL/well substrate was dispensed into assay plate, followed by 1 hr incubation for the enzyme reaction. The fluorescent intensity was measured on a PHERAstar FSX plate reader (BMG Labtech, Cary, NC, USA) with Ex=340 nm/Em=460 nm. A counter-screen assay to eliminate the fluorescence quenching compounds was carried out by dispensing 4 μL of substrate containing fluorescent Edans fragment, SGFRKME-Edans, into 1536-well assay plates in the absence of enzyme. Compounds were pin transferred as 23 nL/well and the fluorescence signal was read. Compounds were tested as 11 point 1:3 titrations in duplicate for both enzyme assay and counter screen.
Pseudotyped particle (PP) entry assay in 1536-well format
Cell line and cell culture:
HEK293 cell line with stable expression of human ACE2 (HEK293-ACE2) was generated by Codex BioSolutions (Gaithersburg, MD).37 In short, Expi293F cells (ThermoFisher) were seeded into cells a 6-well plate with 70-80% confluency. For each well, the cells were transfected with 2.5 ug pCMV_ACE2_IRES_Puromycin plasmid (Codex BioSolutions) using Lipofectamine 3000 (ThermoFisher). Twenty-four hours later, the cells were disassociated with trypsin and transferred into 100-mm dishes. The cells were selected with 1 ug/ml Puromycin for 2-3 weeks. Single colonies were picked into 24-well plates containing 1 ml of DMEM 10% FBS supplemented with 1 ug/ml Puromycin. Western blot was performed to screen the ACE2 expression clones with an ACE2 specific antibody. The positive clones were further confirmed with SARS-CoV2-S PP entry assay.
Pseudotyped particle (PP) generation:
Pseudotyped particles (PPs), SARS-CoV2-S PP, VSV-G PP and delEnv (bald) PP were custom produced by Codex Biosolutions (Gaithersburg, MD) using previously reported methods using a murine leukemia virus (MLV) pseudotyping system.26,27 The SARS-CoV2-S construct with Wuhan-Hu-1 sequence (BEI #NR-52420) was C-terminally truncated by 19 amino acids to reduce ER retention28 for pseudotyping.
PP entry assay:
HEK293-ACE2 cells were seeded in white, solid bottom 1536-well microplates (Greiner BioOne) at 2000 cells/well in 2 μL/well medium, and incubated at 37 °C with 5% CO2 overnight (~16 h). Compounds were titrated 1:3 in DMSO and dispensed via pintool at 23 nL/well to assay plates. Cells were incubated with test articles for 1 h at 37 °C with 5% CO2, before 2 μL/well of PP was added. The plates were then spinoculated by centrifugation at 1500 rpm (453 xg) for 45 min, and incubated for 48 h at 37 °C 5% CO2 to allow cell entry of PP and expression of luciferase reporter. After the incubation, the supernatant was removed with gentle centrifugation using a Blue Washer (BlueCat Bio). Then 4 μL/well of Bright-Glo Luciferase detection reagent (Promega) was added to assay plates and incubated for 5 min at room temperature. The luminescence signal was measured using a PHERAStar plate reader (BMG Labtech). Compounds were tested as 11 point 1:3 titrations in duplicate. Data was normalized with wells containing PPs as 100%, and wells containing control delEnv PP (no spike protein) as 0%.
GFP-LC3 high-content assay
As previously described,29 GFP-LC3 MEF (mouse embryonic fibroblasts) cells were dispensed at 800 cells/5 μl/well in 1536-well tissue culture-treated black/clear bottom, collagen coated plates (Corning, Acton, MA) using a Flying Reagent Dispenser (FRD, Aurora Discovery, Carlsbad, CA). The assay plates with cells were incubated at 37°C with 5% CO2 for 5 h, followed by addition of 23 nL of compound or control, chloroquine diphosphate (CQ), into the assay wells using a Wako Pintool station (Wako Automation, San Diego, CA). After 18-h incubation at 37°C with 5% CO2, the cells were fixed with 4% (v/v) paraformaldehyde (EMS, Hatfield, PA) and nuclei were stained with Hoechst 33342 (Invitrogen, Madison, WI) for 30 min at room temperature. After washing twice with phosphate buffered saline (PBS) using Blue Washer (Blue Cat Bio, Concord, MA), the assay plates were imaged for GFP-LC3 puncta formation using an Operatta CLS (Perkin Elmer) through 20x objective in confocal format. EGFP channel (Excitation 460-490nm/Emission 500-550nm) and DAPI (Excitation 355-385nm/Emission 430-500nm) were used to measure the fluorescence intensities. Images were acquired from each well for one center field (around 25% of a single well area in a 1536-well plate) and analyzed with software of Operetta Harmony 4.6. The compartment analysis algorithm was used to identify the nuclei, apply a cytoplasmic mask and quantitate GFP spots in the GFP channel. A nuclear mask was generated from DAPI stained nuclei. Autophagosomal membrane-associated GFP-LC3 (puncta) was detected as GFP-fluorescent vesicular objects that exceeded a threshold defined by untreated cells and that were located exclusively in the cytoplasmic area. Data was expressed as three output parameters i.e. “% of positive cells”, “Total Spot Area - Mean per Well” and “Relative Spot Intensity - Mean per Well”. Compounds were tested as 11 point 1:3 titrations in triplicate.
In vitro assay and structure data
qHTS data generated on the NPC from the CPE assay (https://opendata.ncats.nih.gov/covid19/index.html) as well as compounds reported as active from recent anti-SARS-CoV-2 repurposing screens38-40 and drugs proposed by the scientific community as potential COVID-19 therapies41-44 were used to train the SARS-CoV-2 models. The detailed qHTS data analysis process including data normalization, correction, classification of concentration response curves, and activity assignment was described previously 45. Briefly, concentration response curves were fit to a four-parameter Hill equation yielding concentrations of half-maximal inhibition (IC50) and maximal response (efficacy) values 3,46. From the CPE assay, compounds that showed concentration dependent response with >30% efficacy were considered active. Other compounds were considered inactive. Literature reported anti-SARS-CoV-2 compounds were considered active.
qHTS data generated in-house at NCATS were used to train the models for ZIKV NS1. NS1 activity data15 were generated in qHTS format on three bioactive collections: the Library of Pharmacologically Active Compounds (LOPAC, 1,280 compounds), the NCATS Pharmaceutical Collection (NPC, 2,816 approved and investigational drugs) 5, and the Mechanism Interrogation PlatE (MIPE, 1,866 cancer drugs with known mechanism of action) 47. Compounds that showed inhibition in both the ratio and 615 nm readouts were considered active. Compounds that were inactive in the ratio readout were considered inactive. Other compounds were considered inconclusive and excluded from modeling. A NCATS in-house collection, the Genesis library, of ~90K diverse compounds was also screened for NS1 activity at a single concentration (14 μM). From these results, compounds that showed >30% inhibition in both the ratio and 615 nm readouts were considered active and other compounds were considered inactive.
The activity data on ~2,600 drugs screened in an EBOV assay from a literature report were used to train the EBOV activity models.16 These compounds were mapped to 2,065 unique compounds in the NCATS compound library. The anti-EBOV activities (active or inactive) of these compounds were assigned according to the literature report.16 All compounds and their assay activities (1 = active, 0 = inactive) used to train the SARS-CoV-2, ZIKV NS1 and EBOV models are provided as Supplementary Data 6.
A subset of the compounds in the bioactive collections, NPC and LOPAC in particular, were screened in nearly all the assays available at NCATS. Two NCATS in-house diverse compound libraries, Sytravon, which contains ~44,000 compounds, and Genesis, which contains ~90,000 compounds, and a subset (~100,000 compounds) of the other NCATS bioactive libraries and a large diverse compound library (MLS), were also screened in subpanels of the NCATS assay portfolio. The bioactive compound activity profiles in the assays that also screened the Sytravon (130 readouts), Genesis library (39 readouts), or MLS (225 readouts) were used to train and test the activity-based models (BABM-S or BABM-G). Structure fingerprints were generated for all compounds using the ChemoTyper48 for the structure-based models (SBM). Structure data on all the compounds with target activity data available were used to train and test the SBM. The compositions of these datasets are summarized in Supplementary Table 2 and the different types of models based on these datasets are summarized in Table 2 and illustrated in Figure 1C. The assay activity-based models (BABM-S, BABM-G, BABM-M) and the activity-structure combined models (CM-S, CM-G, CM-M) were applied to predict the target activity of the compounds with activity profiles available from the Sytravon/Genesis/MLS assays (Figure 1). In the combined models, the activity profile and the structure fingerprint were concatenated to form a new fingerprint for each compound. The SBM was applied to predict the target activity of all ~600K compounds in the NCATS compound library. For activity-based models, only compounds that showed activity in at least 10% of the Sytravon, Genesis or MLS assay panel were kept for analyses. Here, the definition of “active” is not as strict as what would normally be considered as a “hit” for lead identification. Any type of concentration dependent activity observed, regardless of potency or efficacy, was labeled as “active”. As such, compounds that showed activities in multiple assays are not compounds that deemed “promiscuous” in the traditional sense.
Table 2.
Models built on different training datasets
| Assay Data Source/Model Type |
Chemical Structure | Assay Activity | Activity and Structure Combined |
|---|---|---|---|
| MLS | SBM | BABM-M | CM-M |
| Sytravon | SBM | BABM-S | CM-S |
| Genesis | SBM | BABM-G | CM-G |
SBM = Structure based model
BABM-M = Activity-based model (MLS)
BABM-S = Activity-based model (Sytravon)
BABM-G = Activity-based model (Genesis)
CM-M = Combined model (SBM+BABM-M)
CM-S = Combined model (SBM+BABM-S)
CM-G = Combined model (SBM+BABM-G)
Modeling
The Weighted Feature Significance (WFS) method previously developed at NCATS49 was applied to construct the models. Briefly, WFS is a two-step scoring algorithm. In the first step, a two-tailed Fisher’s exact test is used to determine the significance of enrichment for each feature in the active compounds compared to inactive compounds, and a p-value is calculated for all the features present in the data set. For structure data, the feature value was set to 1 for compounds containing that structural feature and 0 for compounds that do not have that feature. For assay activity data, each assay readout was treated as a feature and the feature value was set to 1 for “active” compounds and 0 for inactive compounds. If a feature is less frequent in the active compound set than the inactive compound set, then its p-value is set to 1. These p-values form what we call a “comprehensive” feature fingerprint, which is then used to score each compound for its active potential according to Equation (1), where pi is the p-value for feature i; C is the set of all features present in a compound; M is the set of features encoded in the “comprehensive” feature fingerprint (i.e., features present in at least one active compound); N is the number of features; and α is the weighting factor, which is set to 1 in all the models described here so that all assay features and structure features are treated equally. A high WFS score indicates a strong potential to be active.
| (1) |
For each model, compounds were randomly split into two groups of approximately equal sizes, one used for training and the other for testing. The randomization was conducted 10 times to generate 10 different training and test sets to evaluate the robustness of the models. Model performance was assessed by calculating the area under the receiver operating characteristic (ROC) curve (AUC-ROC), which is a plot of sensitivity [TP/(TP+FN)] versus (1-specificity [TN/(TN+FP)]) 50. A perfect model would have an AUC-ROC of 1 whereas an AUC-ROC of 0.5 indicates a random classifier. The random data split and model training and testing were repeated ten times, and the average AUC-ROC values were calculated for each model. For external experimental validation of models, model performance was measured by the positive predictive value (PPV = TP/(TP+FP)). Statistical significance was determined by the two-tailed Fisher’s exact test comparing model PPV with the active rate in the training dataset for the corresponding target being modeled.
Selection of model predicted actives
Models with AUC-ROC >0.75 were considered for compound selection. WFS score cutoff values for model predicted actives were determined using the ROC curves where both sensitivity and specificity were optimized. Only compounds that scored higher than the cutoff values were considered candidates for follow up selection. Due to the limitations of different assays and resources, for each target we selected compounds with the largest possible structure diversity that could fit into one 1,536-well plate for experimental validation. When the candidate pool was much larger than the target number of compounds, the candidates were narrowed down based on structure type. For this purpose, the entire NCATS in-house compound library was clustered based on structure similarity (729-bit ChemoTyper48 fingerprints) using the self-organizing map (SOM) algorithm51. From the clusters that contain model predicted actives, a fraction of the active compounds was selected from each cluster based on the WFS score and the number of models that predicted the compound as active. Because the EBOV assay could only test ~100 compounds, the anti-EBOV candidates were manually inspected and narrowed down further based on literature reports, structure novelty and ADME properties. In most cases the selection was driven by availability of physical samples. All compounds that met the WFS score cutoff from a model were selected when less than 1,408 compounds had physical samples available for cherry picking. The SARS-CoV-2 CPE assay (live virus) could only be run in 384-well format. Limited by the testing space available and physical sample availability, only 311 model predicted compounds were selected for experimental confirmation in the SARS-CoV-2 live virus assay.
Statistical analysis and illustrations
Principal component analysis (PCA) was performed within R package version 3.4.3. The first three principal components (PCs), PC1, PC2 and PC3 were calculated based on the 729 ChemoTyper fingerprints. 3D PCA plots were generated using the first three PCs in TIBCO® Spotfire® version 7.11.1 (Somerville, MA). Concentration response curve plots were generated using Prism GraphPad 8 (San Diego, CA) with IC50 values calculated using a three-parameter logistic regression.
Supplementary Material
Acknowledgements
This work was supported by the Intramural Research Programs of the National Center for Advancing Translational Sciences, National Institutes of Health. The authors would like to thank H. Guo, X. Hu, and M. Shen for assistance with CPE assay data processing, and R. Eastman, Z. Itkin, and P. Shinn for compound management and plating.
Footnotes
Competing financial interests
The authors declare no competing financial interests.
Code availability
Source codes used to generate the modeling results are included in supplementary material.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The datasets generated during and/or analysed during the current study are included in this published article (and its supplementary information files) and available in the NCATS Open Science Data Portal of COVID-19 (https://opendata.ncats.nih.gov/covid19/index.html) and PubChem (https://pubchem.ncbi.nlm.nih.gov/#query=ncats&tab=bioassay).
References
- 1.Lavecchia A & Di Giovanni C Virtual screening strategies in drug discovery: a critical review. Curr Med Chem 20, 2839–2860 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Gloriam DE Bigger is better in virtual drug screens. Nature 566, 193–194, doi: 10.1038/d41586-019-00145-6 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Inglese J et al. Quantitative high-throughput screening: a titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc Natl Acad Sci U S A 103, 11473–11478, doi:0604348103 [pii] 10.1073/pnas.0604348103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.PubChem. NCATS qHTS assay data, <https://www.ncbi.nlm.nih.gov/pcassay?term=NCGC%5Bsourcename%5D&cmd=search> (2020).
- 5.Huang R et al. The NCGC pharmaceutical collection: a comprehensive resource of clinically approved drugs enabling repurposing and chemical genomics. Sci Transl Med 3, 80ps16, doi:3/80/80ps16 [pii] 10.1126/scitranslmed.3001862 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hansch C Quantitative approach to biochemical structure-activity relationships. Accounts Chem. Res 2, 232–239, doi: 10.1021/ar50020a002 (1969). [DOI] [Google Scholar]
- 7.Cherkasov A et al. QSAR modeling: where have you been? Where are you going to? J Med Chem 57, 4977–5010, doi: 10.1021/jm4004285 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho MH et al. A bioluminescent cytotoxicity assay for assessment of membrane integrity using a proteolytic biomarker. Toxicol In Vitro 22, 1099–1106, doi:S0887-2333(08)00048-9 [pii] 10.1016/j.tiv.2008.02.013 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang R et al. Modelling the Tox21 10 K chemical profiles for in vivo toxicity prediction and mechanism characterization. Nat Commun 7, 10425, doi: 10.1038/ncomms10425 ncomms10425 [pii] (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petrone PM et al. Rethinking molecular similarity: comparing compounds on the basis of biological activity. ACS Chem Biol 7, 1399–1409, doi: 10.1021/cb3001028 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Riniker S, Wang Y, Jenkins JL & Landrum GA Using information from historical high-throughput screens to predict active compounds. J Chem Inf Model 54, 1880–1891, doi: 10.1021/ci500190p (2014). [DOI] [PubMed] [Google Scholar]
- 12.Chen N et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 395, 507–513, doi: 10.1016/S0140-6736(20)30211-7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu M et al. Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat Med 22, 1101–1107, doi: 10.1038/nm.4184 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kouznetsova J et al. Identification of 53 compounds that block Ebola virus-like particle entry via a repurposing screen of approved drugs. Emerg Microbes Infect 3, e84, doi: 10.1038/emi.2014.88 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang S et al. Emetine inhibits Zika and Ebola virus infections through two molecular mechanisms: inhibiting viral replication and decreasing viral entry. Cell Discov 4, 31, doi: 10.1038/s41421-018-0034-1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johansen LM et al. A screen of approved drugs and molecular probes identifies therapeutics with anti-Ebola virus activity. Sci Transl Med 7, 290ra289, doi: 10.1126/scitranslmed.aaa5597 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Chen CZ et al. Identifying SARS-CoV-2 entry inhibitors through drug repurposing screens of SARS-S and MERS-S pseudotyped particles. bioRxiv, 2020.2007.2010.197988, doi: 10.1101/2020.07.10.197988 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu W et al. Identification of SARS-CoV-2 3CL Protease Inhibitors by a Quantitative High-Throughput Screening. ACS Pharmacology & Translational Science 3, 1008–1016, doi: 10.1021/acsptsci.0c00108 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorshkov K et al. The SARS-CoV-2 cytopathic effect is blocked with autophagy modulators. bioRxiv, 2020.2005.2016.091520, doi: 10.1101/2020.05.16.091520 (2020). [DOI] [Google Scholar]
- 20.Qiu X et al. Prophylactic Efficacy of Quercetin 3-beta-O-d-Glucoside against Ebola Virus Infection. Antimicrob Agents Chemother 60, 5182–5188, doi: 10.1128/AAC.00307-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Depamphilis ML & Parsons LN Autophagy modulators for treating neurodegenerative diseases. WO2016204988A1 (2016). [Google Scholar]
- 22.Gorshkov K et al. The SARS-CoV-2 cytopathic effect is blocked with autophagy modulators bioRxiv, doi: 10.1101/2020.05.16.091520 (2020). [DOI] [Google Scholar]
- 23.Yang N & Shen HM Targeting the Endocytic Pathway and Autophagy Process as a Novel Therapeutic Strategy in COVID-19. Int J Biol Sci 16, 1724–1731, doi: 10.7150/ijbs.45498 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen CZ et al. Drug Repurposing Screen for Compounds Inhibiting the Cytopathic Effect of SARS-CoV-2. bioRxiv, 2020.2008.2018.255877, doi: 10.1101/2020.08.18.255877 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shyr ZA, Gorshkov K, Chen CZ & Zheng W Drug discovery strategies for SARS-CoV-2. Journal of Pharmacology and Experimental Therapeutics, JPET-MR-2020-000123, doi: 10.1124/jpet.120.000123 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Millet JK et al. Production of Pseudotyped Particles to Study Highly Pathogenic Coronaviruses in a Biosafety Level 2 Setting. J Vis Exp, doi: 10.3791/59010 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Millet JK & Whittaker GR Murine Leukemia Virus (MLV)-based Coronavirus Spike-pseudotyped Particle Production and Infection. Bio-protocol 6, e2035, doi: 10.21769/BioProtoc.2035 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ou X et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun 11, 1620, doi: 10.1038/s41467-020-15562-9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y et al. A cell-based quantitative high-throughput image screening identified novel autophagy modulators. Pharmacol Res 110, 35–49, doi: 10.1016/j.phrs.2016.05.004 S1043-6618(16)30046-9 [pii] (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Domenico G, Giuseppe Felice M, Marco C, Angelo C & Orazio N Applicability Domain for QSAR Models: Where Theory Meets Reality. International Journal of Quantitative Structure-Property Relationships (IJQSPR) 1, 45–63, doi: 10.4018/IJQSPR.2016010102 (2016). [DOI] [Google Scholar]
- 31.Thomas CJ et al. The pilot phase of the NIH Chemical Genomics Center. Curr Top Med Chem 9, 1181–1193, doi:CTMC-Abs-014-9-13 [pii] (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McInnes C Virtual screening strategies in drug discovery. Curr Opin Chem Biol 11, 494–502, doi: 10.1016/j.cbpa.2007.08.033 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Zhu H et al. Mining of high throughput screening database reveals AP-1 and autophagy pathways as potential targets for COVID-19 therapeutics. arXiv, 2007.12242[q-bio.QM] (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Konig R et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell 135, 49–60, doi: 10.1016/j.cell.2008.07.032 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li W et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426, 450–454, doi: 10.1038/nature02145 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Severson WE et al. Development and validation of a high-throughput screen for inhibitors of SARS CoV and its application in screening of a 100,000-compound library. Journal of biomolecular screening 12, 33–40, doi: 10.1177/1087057106296688 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao T et al. A trimeric human angiotensin-converting enzyme 2 as an anti-SARS-CoV-2 agent in vitro. bioRxiv, 2020.2009.2018.301952, doi: 10.1101/2020.09.18.301952 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Touret F et al. In vitro screening of a FDA approved chemical library reveals potential inhibitors of SARS-CoV-2 replication. bioRxiv, 2020.2004.2003.023846, doi: 10.1101/2020.04.03.023846 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weston S, Haupt R, Logue J, Matthews K & Frieman MB FDA approved drugs with broad anti-coronaviral activity inhibit SARS-CoV-2. bioRxiv, 2020.2003.2025.008482, doi: 10.1101/2020.03.25.008482 (2020). [DOI] [Google Scholar]
- 40.Riva L et al. A Large-scale Drug Repositioning Survey for SARS-CoV-2 Antivirals. bioRxiv, 2020.2004.2016.044016, doi: 10.1101/2020.04.16.044016 (2020). [DOI] [Google Scholar]
- 41.Caly L, Druce JD, Catton MG, Jans DA & Wagstaff KM The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res 178, 104787, doi: 10.1016/j.antiviral.2020.104787 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y et al. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet 395, 1569–1578, doi: 10.1016/S0140-6736(20)31022-9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geleris J et al. Observational Study of Hydroxychloroquine in Hospitalized Patients with Covid-19. N Engl J Med, doi: 10.1056/NEJMoa2012410 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bleasel MD & Peterson GM Emetine, Ipecac, Ipecac Alkaloids and Analogues as Potential Antiviral Agents for Coronaviruses. Pharmaceuticals (Basel) 13, doi: 10.3390/ph13030051 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang R in High-Throughput Screening Assays in Toxicology Vol. 1473 Methods in Molecular Biology (eds Zhu Hao & Xia Menghang) Ch. 12, (Humana Press, 2016). [Google Scholar]
- 46.Wang Y, Jadhav A, Southal N, Huang R & Nguyen DT A grid algorithm for high throughput fitting of dose-response curve data. Curr Chem Genomics 4, 57–66, doi: 10.2174/1875397301004010057 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mathews Griner LA et al. High-throughput combinatorial screening identifies drugs that cooperate with ibrutinib to kill activated B-cell-like diffuse large B-cell lymphoma cells. Proc Natl Acad Sci U S A 111, 2349–2354, doi: 10.1073/pnas.1311846111 1311846111 [pii] (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang C et al. New publicly available chemical query language, CSRML, to support chemotype representations for application to data mining and modeling. J Chem Inf Model 55, 510–528, doi: 10.1021/ci500667v (2015). [DOI] [PubMed] [Google Scholar]
- 49.Huang R et al. Weighted feature significance: a simple, interpretable model of compound toxicity based on the statistical enrichment of structural features. Toxicol Sci 112, 385–393, doi:kfp231 [pii] 10.1093/toxsci/kfp231 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zweig MH & Campbell G Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clin Chem 39, 561–577 (1993). [PubMed] [Google Scholar]
- 51.Kohonen T Self-organizing neural projections. Neural networks : the official journal of the International Neural Network Society 19, 723–733 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are included in this published article (and its supplementary information files) and available in the NCATS Open Science Data Portal of COVID-19 (https://opendata.ncats.nih.gov/covid19/index.html) and PubChem (https://pubchem.ncbi.nlm.nih.gov/#query=ncats&tab=bioassay).