Abstract
It has been reported that Arabidopsis chloroplast accD transcripts undergo RNA editing and that loss of accD-C794 RNA editing does not affect plant growth under normal conditions. To date, the exact biological role of accD-C794 editing has remained elusive. Here, we reveal an unexpected role for accD-C794 editing in response to heat stress. Loss of accD-C794 editing results in a yellow and dwarf phenotype with decreased chloroplast gene expression under heat stress, and artificial improvement of C794-edited accD gene expression enhances heat tolerance in Arabidopsis. These data suggest that accD-C794 editing confers heat tolerance in planta. We also found that treatment with the product of acetyl coenzyme A carboxylase (ACCase) could allay mutant phenotypic characteristics and showed that a mutation in the CAC3 gene for the α-subunit of ACCase was associated with dwarfism under heat stress. These observations indicate that defective accD-C794 editing may be intrinsic to reduced ACCase activity, thereby contributing to heat sensitivity. ACCase catalyzes the committed step of de novo fatty acid (FA) biosynthesis. FA content analysis revealed that unsaturated oleic (C18:1) and linoleic acids (C18:2) were low in the accD-C794 editing-defective mutant but high in the C794-edited accD-overexpressing plants compared with the wild type. Supplying exogenous C18:1 and C18:2 could rescue the mutant phenotype, suggesting that these FAs play an essential role in tolerance to heat stress. Transmission electron microscopy observations showed that heat stress seriously affected the membrane architecture in accD editing-defective mutants but not in accD-overexpressing plants. These results provide the first evidence that accD-C794 editing regulates FA biosynthesis for maintenance of membrane structural homeostasis under heat stress.
Key words: chloroplast RNA editing, RARE1, accD, fatty acid, heat stress
Arabidopsis chloroplast accD transcripts undergo RNA editing. Editing efficiency of the accD-C794 site is positively correlated with plant heat tolerance. Editing of accD-C794 confers heat tolerance by regulating ACCase activity and fatty acid content for the maintenance of membrane structural homeostasis.
Introduction
RNA editing involves post-transcriptional modifications, including nucleotide insertion, deletion, transitions, and transversions (Brennicke et al., 1999). In plants, one significant editing process within organellar mRNAs involves cytidine (C) to uridine (U) transition (C→U) (Chateigner-Boutin and Small, 2010; Yan et al., 2018; Small et al., 2020), with 619 and 43 editing sites identified in the mitochondrion and chloroplast of Arabidopsis, respectively (Ruwe et al., 2013; Leu et al., 2016). C→U editing events were originally proposed to involve an editosome complex that generally comprises pentatricopeptide repeat (PPR) proteins and non-PPR protein editing factors (Small et al., 2020; Sun et al., 2016; Yan et al., 2018). According to statistical analysis based on the plant editosome database (https://ngdc.cncb.ac.cn/ped/home), a total of 106 PPRs are involved in Arabidopsis organellar RNA editing, among which 47 PPRs participate in chloroplast RNA editing and the remainder are associated with mitochondria. In addition to PPRs, most RNA-editing sites require one or more non-PPR proteins, for example, multiple organellar RNA-editing factors (MORFs) (Takenaka et al., 2012; Bayer-Csaszar et al., 2017), organelle RNA recognition motif (ORRM) proteins (Shi et al., 2016), organelle zinc-finger (OZ) proteins (Sun et al., 2015), protoporphyrinogen IX oxidase 1 (PPO1) (Zhang et al., 2014), porphobilinogen deaminase HEMC (Huang et al., 2017), and chloroplast ribonucleoprotein 31A (CP31A) (Tillich et al., 2009). During RNA editing, transcriptome activation is considered to involve specific PPRs that interact at RNA sequences proximal to C targets aided by non-PPR recruitment of auxiliary cofactors either as editing activators or as regulators of editing efficiency (Small et al., 2020).
Recent evidence suggests that RNA editing is an essential response to abiotic stresses (Liu et al., 2018; Cui et al., 2019; Chu and Wei, 2020; Zhang et al., 2020). Interestingly, overall editing levels of several gene transcripts, such as matK and ndhB, decreased with increasing temperature in chloroplasts of Vitis vinifera (Zhang et al., 2020). Furthermore, transcriptome differences between heat-shocked and cold-stressed Arabidopsis showed that C→U editing rates were significantly affected by heat or cold stress (Chu and Wei, 2020), and editing of chloroplast rps8, rpl2, and atpA in Oryza sativa seems to correlate with cold-sensitive phenotypes (Cui et al., 2019; Liu et al., 2018). Nonetheless, the importance of RNA editing in plant growth is not apparent in genetic studies, and the molecular mechanisms that underlie the essential roles of RNA editing during temperature stress remain largely unknown.
In higher vascular plants, the ubiquitous unsaturated fatty acids (UFAs) C18:1, C18:2, and linolenic acid (C18:3) (Harwood, 1988; He et al., 2020) exert a biologically significant effect on organellar membrane structure, fluidity, and function under abiotic stress (He et al., 2018; Li et al., 2020). FA biosynthesis is initiated in chloroplasts and is tightly regulated by the accD gene (Thelen and Ohlrogge, 2002; Hölzl and Dörmann, 2019). This gene encodes the β-carboxyl transferase (β-CT) subunit of acetyl coenzyme A carboxylase (ACCase) (Sasaki et al., 1997), which is required for the conversion of acetyl-CoA into malonyl-CoA during the initial rate-limiting step of FA synthesis (Sasaki and Nagano, 2004). Two isoforms of ACCase are apparent within plant cells. The first is a cytosolic ACCase monomer, and the second is a heterotetrameric ACCase found in plastids (Sasaki et al., 1993; Konishi and Sasaki, 1994). Apart from the β-CT subunit, the heterotetrameric ACCase contains three additional subunits, including the biotin carboxyl carrier protein 1, biotin carboxylase, and the α-subunit of carboxyltransferase (α-CT), which are encoded by the nuclear genes CAC1, CAC2, and CAC3, respectively (Sasaki and Nagano, 2004; Yu et al., 2017). A previous study in Nicotiana sp. revealed that elevated accD expression was accompanied by increased ACCase content in plastids and significantly elevated levels of FAs in leaves (Madoka et al., 2002). Normal expression of accD requires RNA editing at the C794 and C1568 sites, which are located in the exon and the 3′ UTR, respectively, of the accD transcript (Chateigner-Boutin and Small, 2007). Editing at the former site is known to require the presence of a specific PPR protein, designated RARE1 (Robbins et al., 2009). However, the relationship between accD-C794 editing, ACCase, FA biosynthesis, and stress resistance remains largely unknown.
In the present study, we obtained an Arabidopsis rare1 mutant with abolished accD-C794 editing and discovered that it exhibits a yellow and dwarf phenotype compared with the wild type under heat stress. Further investigation showed that heat tolerance in plants was strongly associated with the extent of accD-C794 editing. Moreover, exogenous malonyl-CoA application effectively rescued the rare1 phenotype under heat stress, and mutation of the CAC3 gene significantly contributed to heat-sensitive dwarfism in Arabidopsis plants. Thus, deficiency in accD-C794 editing dramatically influences plastid heteromeric ACCase activity. Further consequences of altered accD-C794 editing were observed in FA biosynthesis and the structural integrity of biological membranes, revealing that unsaturated C18:1 and C18:2 FAs play a vital role in membrane homeostasis under heat-stressed conditions. Our findings thus reveal an unexpected and novel role for C794 editing in the regulation of ACCase, FA synthesis, and heat-stress responses.
Results
RARE1 loss of function causes a heat-sensitive growth defect
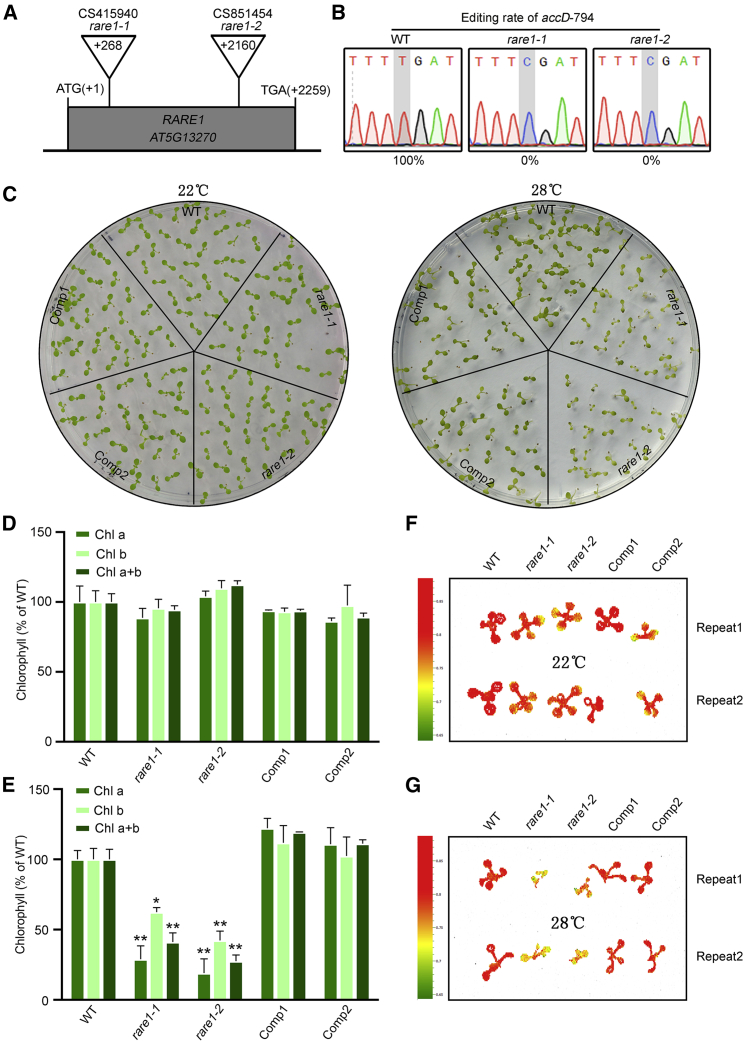
The PPR protein RARE1 is responsible for editing of the accD-C794 site (Robbins et al., 2009). To define the function of RARE1 in plants, we engineered two different mutants, each carrying a T-DNA insertion within RARE1, namely rare1-1 (CS415940) and rare1-2 (CS851454) (Figure 1A and Supplemental Figure 1). Subsequent DNA sequencing analysis showed that C794 editing of chloroplast accD transcripts was abolished in each case (Figure 1B). Both rare1 lines displayed a phenotype not dissimilar to that of wild-type plants under normal growth conditions (Figure 1C), as noted elsewhere (Robbins et al., 2009). Strikingly, at an elevated temperature of 28°C, all rare1 lines displayed yellow foliage and a slowed growth rate, whereas the more resilient wild-type plants retained typical green foliage. This mutant phenotype could be rescued by pRARE1::RARE1-c-Myc complementation (Figure 1C). Biochemical analysis revealed that there were no significant differences in chlorophyll content among the wild type, both rare1 lines, and complementary transgenic plants at 22°C (Figure 1D). However, at an elevated temperature of 28°C, the contents of chlorophyll a, chlorophyll b, and total chlorophyll were significantly reduced in the rare1 lines compared with wild-type plants and the complemented lines (Figure 1E). Measurement of the Fv/Fm value further validated the above results in vitro (Figure 1F and 1G). Therefore, heat stress is severely detrimental to chlorophyll synthesis in rare1 lines, which develop a yellow leaf phenotype.
Figure 1.
Arabidopsis rare1 mutants with abolished accD-C794 editing displayed a heat-stress-sensitive phenotype.
(A) A schematic map of the RARE1 locus, illustrating the T-DNA insertion sites within the rare1 mutants.
(B) The editing rate of chloroplast accD transcripts at the C794 site in WT and rare1 mutants.
(C) Seedling phenotypes of wild-type (WT), rare1, and pRARE1:RARE1-c-Myc transgenic plants (complemented) grown at 22°C or 28°C.
(D and E) Chlorophyll content in plants grown at 22°C (D) or 28°C (E).
(F and G) Fv/Fm values of WT, rare1, and complemented lines grown at 22°C (F) or 28°C (G). Error bars indicate SD from three independent repeats. Bars marked with an asterisk (Student’s t-test; ∗p ≤ 0.05, ∗∗p ≤ 0.01) differ significantly from the WT sample.
RARE1 gene deficiency influences chloroplast gene expression
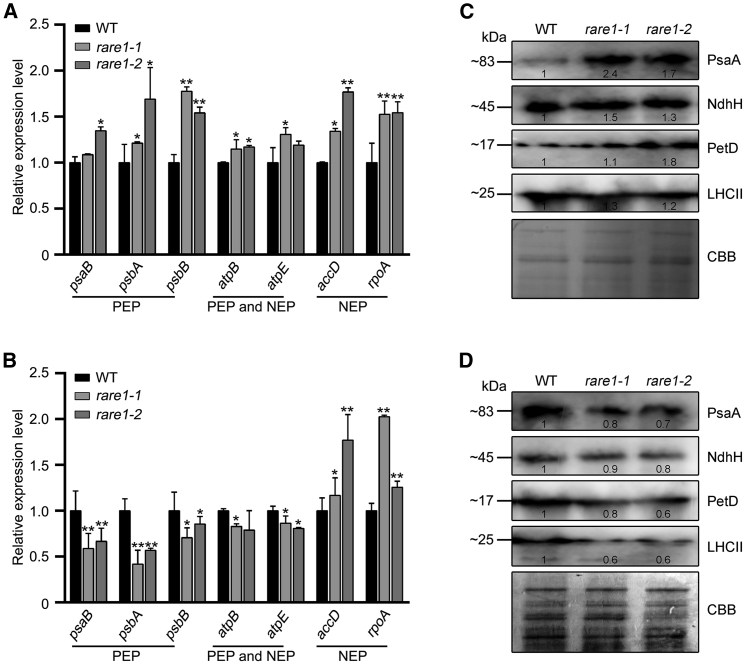
In our initial experiments, we assessed differences in expression among plastid-encoded RNA polymerase (PEP)-dependent genes (psaB, psbA, and psbB), nuclear-encoded RNA polymerase (NEP)-dependent genes (accD and rpoA), and genes that are both PEP- and NEP-dependent (atpB and atpE) in wild-type and rare1 mutant Arabidopsis plants. Under normal growth conditions, almost all transcript levels of chloroplast genes were significantly higher in the rare1 mutants than in the wild-type plants (Figure 2A). When expression levels of these genes were analyzed in each line cultivated at 28°C, PEP-related genes (psaB, psbA, psbB, atpB, and atpE) were repressed, whereas expression levels of accD and rpoA NEP-dependent genes were elevated (Figure 2B). Expression patterns of chloroplast transcripts in the rare1 mutants grown at 28°C were similar to those in PEP-defective mutants described elsewhere (Gao et al., 2011; Yagi et al., 2012). In a series of immunoblotting experiments, we detected the effect of abnormal chloroplast gene transcription on accumulation of photosynthesis-related proteins in the rare1 mutants, including the core subunit of photosystem I (PsaA), subunit H of NAD(P)H dehydrogenase (NdhH), subunit IV of the cytochrome b6f complex (PetD), and light-harvesting chlorophyll a/b complex II. In these experiments, PsaA, NdhH, and PetD levels were drastically increased in the rare1 mutants compared with wild-type plants under normal growth conditions (Figure 2C). By contrast, the accumulation of these photosynthesis-related proteins was reduced in mutant plants under heat-stress conditions (Figure 2D). These results suggest that the RARE1 mutation leads to significant defects in chloroplast development under heat stress and markedly increases the accumulation of chloroplast functional components under normal conditions.
Figure 2.
Effects of RARE1 mutation on chloroplast gene expression.
(A) qRT–PCR analysis of chloroplast genes in WT and rare1 plants cultured at 22°C. Representative genes, including plastid-encoded polymerase (PEP)-dependent, nuclear-encoded polymerase (NEP)-dependent, and PEP- and NEP-dependent genes encoded in chloroplasts, were used to analyze the impact of the RARE1 mutation.
(B) Changes in transcript levels of chloroplast genes in WT and rare1 plants grown at 28°C. Data represent the mean of three replicates ±SD.
(C and D) Detection of chloroplast protein accumulation in WT and rare1 mutants grown at 22°C (C) or 28°C (D). Bars denote standard errors.
Asterisks (∗) in (A) and (B) indicate a significant difference from the WT (∗p ≤ 0.05; ∗∗p ≤ 0.01).
Overexpression of the C794-edited accD gene enhances plant heat tolerance
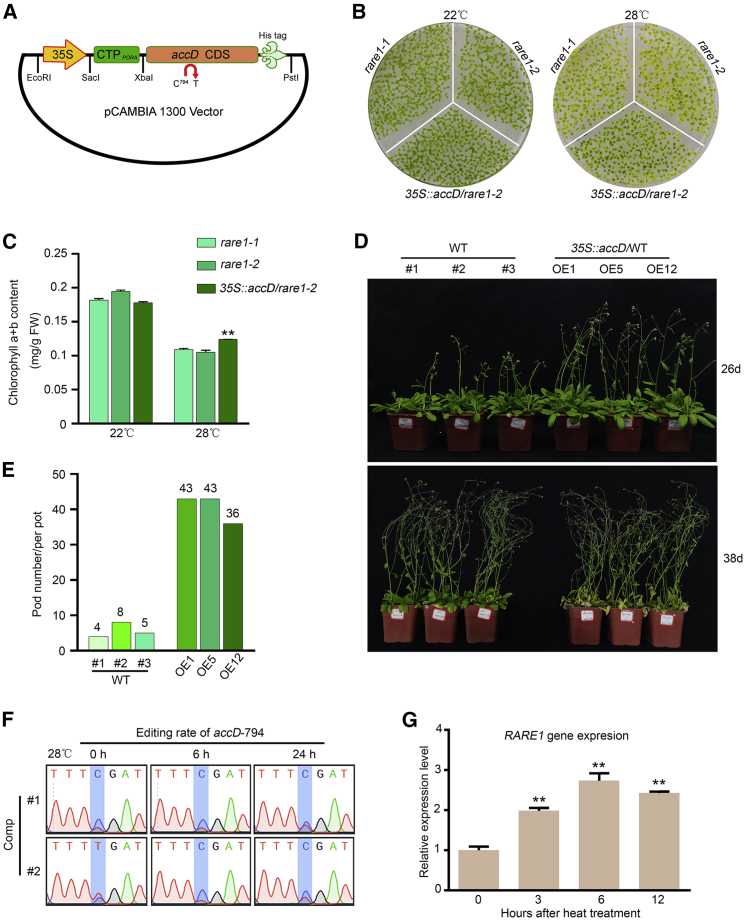
To determine whether this accD-C794 editing deficiency confers heat-sensitive defects upon rare1 mutants, we prepared a 35S:accD-His construct in which the coding sequence of C794-edited accD was N-terminally fused with the chloroplast transit peptide from the protochlorophyllide oxidoreductase A (PORA) gene under the control of the CaMV 35S promoter (Figure 3A). The resultant transgenic plants overexpressing the C794-edited accD gene in the rare1 mutant or wild-type background displayed a heat-tolerant phenotype compared with the original plants (Figure 3B and Supplemental Figure 2). Phenotype investigation of plants overexpressing C794-unedited accD under 28°C further demonstrated that the effect was not due to overexpression (Supplemental Figure 2). Furthermore, analysis of chlorophyll content showed that the transgenic plants with edited accD retained more chlorophyll than the rare1 mutants under heat-stress conditions (Figure 3C). These results indicate that accD-C794 RNA editing is associated with heat tolerance in Arabidopsis. In addition, we found that three independent overexpressing lines of C794-edited accD (accD overexpressed [OE]) in the wild-type background exhibited significantly higher heat tolerance (Figure 3D and Supplemental Figure 3). Under heat stress, accD-OE plants grew more rapidly and better than wild-type plants (Figure 3D). We also compared heat-stress responses between wild-type and accD-OE plants at the reproductive stage and observed a greater abundance of siliques in the transgenic plants compared with the controls (Figure 3E). We also analyzed the RNA-editing rate at the accD-C794 site and the expression of RARE1 after heat treatment. The accD-C794 editing rate and RARE1 expression increased as the treatment time increased (Figure 3F and 3G). These findings suggest that the expression level of the edited accD gene contributes to heat tolerance in Arabidopsis.
Figure 3.
Overexpression of the C794-edited accD gene conferred heat tolerance.
(A) Schematic illustration of the accD-fusion construct. Expression of the edited accD gene fused with the chloroplast transit peptide of RPOA and a His-tag is driven by the 35S promoter.
(B) Phenotype of C794-edited accD-overexpressing transgenic plants in the rare1 background.
(C) Chlorophyll contents in transgenic seedlings grown at 22°C or 28°C. Data represent the mean of three replicates ±SD. Asterisks (∗) indicate a statistical difference (∗∗p ≤ 0.01).
(D) Heat-tolerance assay of three edited accD transgenic lines in the WT background.
(E) The number of fertile siliques in the WT and transgenic plants under heat stress.
(F) Effect of heat treatment on editing rate at the accD-C794 site in two independent RARE1-complemented plants with incomplete accD editing.
(G) Effect of heat treatment on RARE1 gene expression.
The heat-sensitive phenotype of the rare1 mutant is associated with a defect in ACCase activity
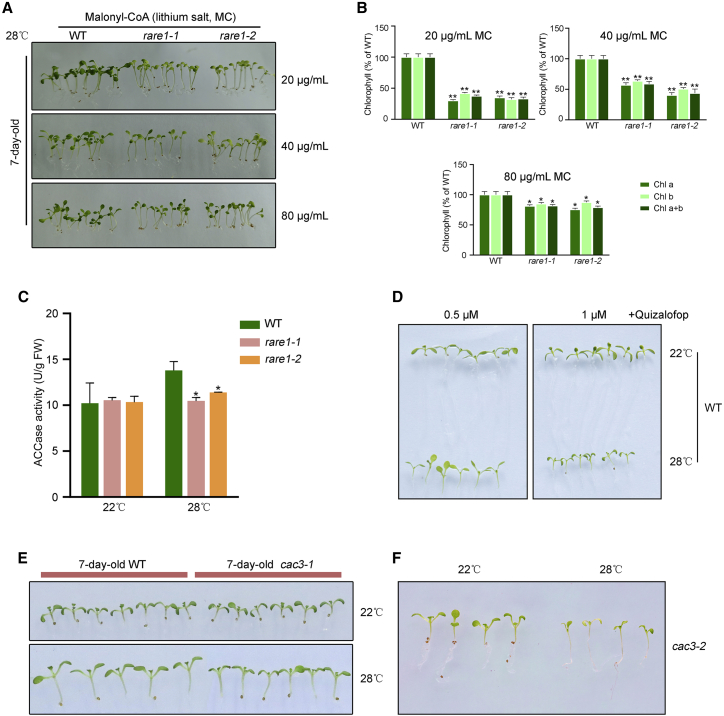
It is well documented that accD encodes a core subunit of ACCase required for FA biosynthesis (Sasaki et al., 1997). To examine the possibility that abolished RNA editing of the accD transcript results in ACCase inactivation under heat stress, we assessed the effects of malonyl-CoA supplements on the rare1 phenotype. We observed that exogenous supplementation with malonyl-CoA alleviated the heat-induced defects of rare1, as indicated by a slight recovery from the yellow, dwarf phenotype on medium with a low concentration of malonyl-CoA lithium salt to near-wild-type characteristics at 28°C supplemented with 80 μg/ml malonyl-CoA lithium salt (Figure 4A). Levels of chlorophyll in rare1 plants were highly correlated with the concentration of malonyl-CoA lithium salt under the described conditions (Figure 4B). An enzyme activity assay demonstrated that ACCase activity was lower in the rare1 mutant compared with the wild type under heat-stress conditions (Figure 4C). To further investigate the association between the heat sensitivity of rare1 and ACCase dysfunction, we analyzed the growth phenotype of wild-type seedlings treated with an inhibitor of plastid ACCase. Inhibition of ACCase caused a dwarf phenotype at 28°C but not at 22°C (Figure 4D). We also identified and characterized a T-DNA insertion line of the CAC3 gene encoding the α-CT subunit of ACCase, which we named cac3-1. Sequencing analysis revealed that the T-DNA insertion in the non-coding 3′ UTR of CAC3 significantly reduced expression from neighboring sequences but not the coding region (Supplemental Figure 4). The cac3-1 mutants displayed near-wild-type phenotypes at 22°C but adopted a dwarf habit at 28°C (Figure 4E). A novel allelic mutant of CAC3 (cac3-2) obtained by CRISPR-Cas9 genome editing also exhibited a severe dwarf phenotype under heat-stress conditions (Figure 4F and Supplemental Figure 5). These findings suggest that loss of accD-C794 editing directly impairs the activity of ACCase, which directly manifests as the heat-sensitive phenotype of rare1 mutants.
Figure 4.
ACCase activity is tightly correlated with heat tolerance.
(A) Phenotypic analysis of rare1 mutants grown on Murashige and Skoog medium supplemented with varying concentrations of malonyl-CoA lithium salt.
(B) Chlorophyll content of rare1 plants grown with malonyl-CoA supplementation. Asterisks (∗) indicate a significant difference between the WT and the rare1 mutants (∗p ≤ 0.05; ∗∗p ≤ 0.01).
(C) ACCase activity in the WT and the rare1 mutants at 22°C or 28°C.
(D) Effect of a plastid ACCase inhibitor on the phenotype of the WT under 22°C or 28°C.
(E and F) Growth phenotypes of cac3 mutants at 22°C and 28°C.
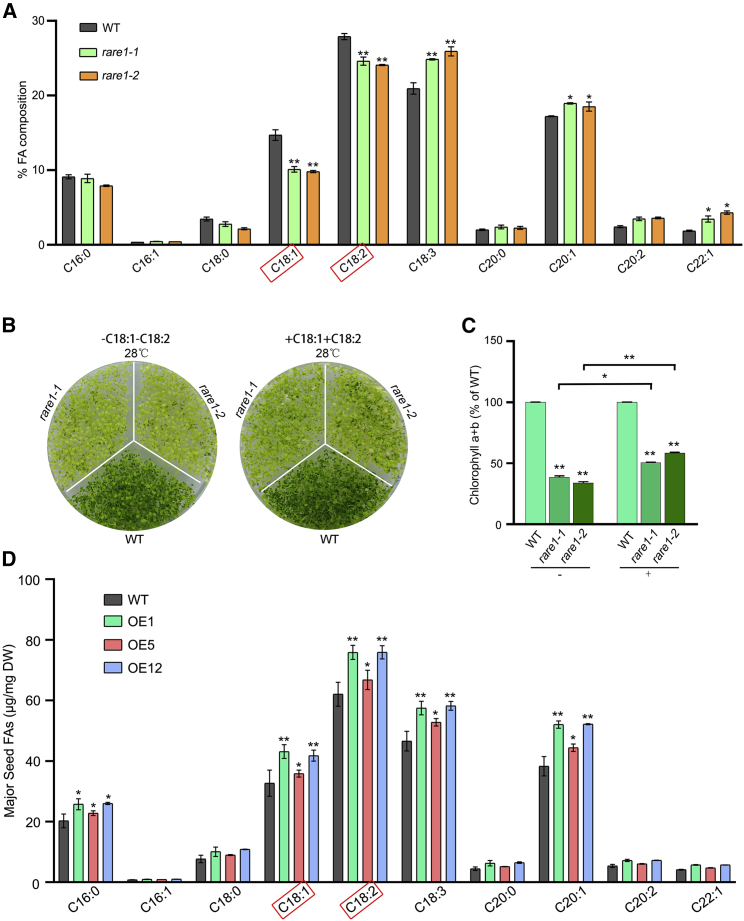
Levels of the UFAs C18:1 and C18:2 correlate with heat tolerance
UFAs play an essential role in plant adaptation to environmental stresses and presumably contribute to the heat sensitivity of rare1 plants. We therefore made gas chromatography-mass spectrometry comparisons of UFA levels between wild-type and rare1 plants, and the abundances of C18:1, C18:2, and C18:3 UFAs differed significantly between the genotypes. Levels of C18:1 and C18:2 were lower in the rare1 mutants, whereas the C18:3 content was higher (cf. wild type; Figure 5A). Furthermore, when exogenous C18:1 and C18:2 were applied to rare1 plants at a concentration of 1 μg/ml under heat stress, the development of the rare1 yellow phenotype was less pronounced (Figure 5B and Supplemental Figure 6B). By contrast, no phenotype differences were observed between the wild type and rare1 under normal conditions (Supplemental Figure 6A). We also measured chlorophyll content in rare1 mutants cultured at 28°C in the presence of C18:1 and C18:2 supplements and found that levels of chlorophyll in the rare1 mutants increased compared with the control plants (Figure 5C). Transgenic plants overexpressing the C794-edited accD gene in the wild-type background exhibited enhanced heat tolerance (Figure 3D). Therefore, we also measured FA levels in the seeds of three independent transgenic lines and found that their total FA content was 11.7%–28.3% higher than that of wild-type seeds (Supplemental Figure 7). Further analysis of the FA composition revealed that C18:1 and C18:2 levels were significantly higher in seeds of the transgenic lines (Figure 5D). These results suggest that C18:1 and C18:2 UFAs are responsible for RNA-editing-related heat tolerance in Arabidopsis.
Figure 5.
Analysis of fatty acid composition in rare1 mutants and accD-overexpressing lines.
(A) Comparison of fatty acid composition between the WT and the rare1 mutants.
(B) Phenotypic observation of the WT and the rare1 mutants grown with C18:1 and C18:2 supplementation at 28°C.
(C) Changes in chlorophyll content of the WT and the rare1 mutants grown with or without C18:1 and C18:2. The chlorophyll content of WT seedlings is set to 100%.
(D) Comparison of fatty acid composition between the WT and the accD-overexpressing (OE) plants in the WT background. Asterisks (∗) indicate significantly higher or lower levels relative to the WT (∗p ≤ 0.05; ∗∗p ≤ 0.01).
Heat stress causes membrane damage in rare1 mutant plants
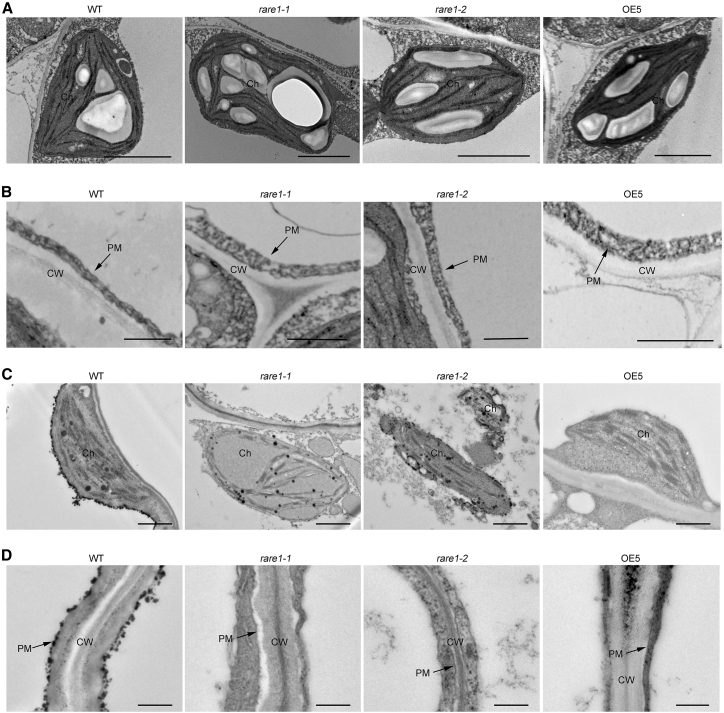
UFAs are important components of phospholipids in biomembranes and help to maintain membrane fluidity. We examined the structures of organelles and plasma membranes in wild-type, rare1, and C794-edited accD-OE leaf cells using transmission electron microscopy (TEM) (Figure 6). There were no significant differences in chloroplast and plasma membrane structure among wild-type, rare1, and C794-edited accD-OE plants under normal conditions (Figure 6A and 6B). Interestingly, chloroplasts in wild-type and OE cells appeared as predominantly lunate entities with abundant thylakoid membranes. However, corresponding plastids in rare1 mutants appeared as partially lysed, morphologically irregular organelles with a lower abundance of thylakoid membranes (Figure 6C). We also examined plasmalemma structures in cells from the same plants at 28°C and noted that whereas cellular surfaces in wild-type and accD-OE plants stained deep black, staining of the corresponding rare1 membranes was significantly less pronounced (Figure 6D). These results suggest that biological membrane integrity is maintained under heat stress in wild-type and OE plants, but not in rare1 plants.
Figure 6.
Transmission electron micrographs of biological membranes in the WT, the rare1 mutants, and the edited accD-OE transgenic plants.
(A and C) Chloroplast ultrastructure of the WT, the rare1 mutants, and the OE plants grown at 22°C (A) or 28°C (C). Ch, chloroplast. OE, the C794-edited accD-OE transgenic line.
(B and D) Plasma membrane structure of the WT, the rare1 mutants, and the accD-OE plants under 22°C (B) or 28°C (D). PM, plasma membrane; CW, cell wall.
Scale bars: 2 μm in (A) and (C) and 500 nm in (B) and (D).
Discussion
The chloroplast accD gene encodes the β-CT subunit of heteromeric ACCase, which catalyzes the first committed step in FA biosynthesis. After transcription, accD transcripts undergo RNA editing at two sites, C794 and C1568. In fact, almost all accD transcripts in Arabidopsis exist in the C794-edited form. C794-edited accD seems to be more beneficial to plant growth. Nevertheless, Robbins et al. (2009) reported that an Arabidopsis mutant lacking accD-C794 editing exhibits a wild-type phenotype under growth-room conditions. To date, the function and importance of accD-C794 editing in plants remain unclear. We have engineered two alleles of RARE1 that are mutated in accD-C794 editing capabilities, and the corresponding plants display a yellow, dwarf phenotype when cultivated at 28°C, suggesting that editing is necessary for normal responses to heat stress. At the same time, it is possible that other RARE1-mediated processes may be involved in the heat-induced phenotype of the rare1 mutants. To address this issue, we overexpressed C794-edited accD in both rare1 mutants and the wild type and observed enhanced heat tolerance compared with the control plants (Figure 3). This evidence provides support for the notion that accD-C794 editing confers tolerance to heat stress in Arabidopsis.
Editing at the C794 site underlies the S265→L substitution in the AccD protein, but it is still considered to be non-essential for full heteromeric ACCase enzyme activity because the abolition of C794 RNA editing does not affect plant growth, ACCase activity, and total seed FA content under normal conditions (Figures 1C and 4C and Supplemental Figure 8). It is possible that levels of cytoplasmic homomeric ACCase are sufficient to satisfy the FA demand for plant growth under normal conditions. Here, we also demonstrated that Arabidopsis plants carrying a mutation in the α-CT subunit of ACCase encoded by the CAC3 gene display a phenotype similar to that of wild-type plants under normal conditions (Figure 4E). Despite this result, previous studies have shown that mutations in the ACC1 and ACC2 genes, which encode two isoforms of homomeric ACCase, produce a lethal embryonic phenotype (Baud et al., 2003). Thus, we speculate that homomeric ACCase has greater biological significance than its heterotetrameric counterpart for plant growth under normal conditions.
The plastid heteromeric ACCase has primary sequence similarity to the prokaryotic Escherichia coli isomer and is considered to be of cyanobacterial evolutionary origin (Nikolau et al., 2003). Previous reports have shown that a mutation in the BCCP protein subunit of ACCase in E. coli confers a temperature-sensitive phenotype (Cronan, 2001). Coupled with observations that Arabidopsis cac3 mutants lacking the ACCase α-CT subunit are also phenotypically heat sensitive (Figure 4E and 4F), these reports lead us to surmise that the plastid heterotetrameric ACCase plays a central role in normal plant growth under heat stress. This notion is further supported by the finding that the rare1 mutants showed significantly lower lipid contents under heat stress (Supplemental Figure 9). It is noteworthy that Arabidopsis plants carrying the CAC3 mutation and accD-C794 editing deficiency also showed phenotypic similarities during heat stress and that exogenous supplementation with malonyl-CoA, the product of ACCase, partially rescued the phenotype of C794-editing-deficient plants (Figure 4A). These results suggest that accD-C794 editing deficiency reduces the plastid ACCase activity. In addition, the editing rate of accD-C794 appears to be directly proportional to the heat tolerance of plants (Figure 3F). These observations suggest that heteromeric ACCase confers heat tolerance and that accD-C794 editing is critical for the function of heteromeric ACCase under heat stress.
C18:1, C18:2, and C18:3 are the predominant UFAs in most plants (Harwood, 1988) and are associated with both abiotic and biotic plant stresses (He et al., 2020). We observed a decrease in C18:1 and C18:2 UFA content in the accD-C794-editing-deficient plants, whereas levels of these UFAs increased in the C794-edited accD-OE plants (Figure 5A and 5D). In addition, the accD-C794-editing-deficient mutants are phenotypically heat sensitive, whereas edited accD-OE plants exhibit enhanced heat tolerance (Figures 1C and 3). These findings suggest that the heat-stress phenotype is related to altered C18:1 and C18:2 contents in these plants. This notion is further supported by C18:1 and C18:2 supplementation studies of the accD-C794-editing-deficient mutants under heat stress (Figure 5B and Supplemental Figure 6B).
Membrane lipids contain high levels of UFAs, which are essential for homeostatic maintenance of membrane structure. Previous reports have indicated that the plasmalemmas surrounding thermo-tolerant yeast cells are enriched in C18:1 content, with proportionately lower levels of C18:2 and C18:3, in that order (Swan and Watson, 1999). Moreover, C18:1 is reported to be crucial for the maintenance of cell membrane integrity and fluidity, thus contributing to improved survival rates of Lactobacillus plantarum after freeze drying (Wang et al., 2020). In this study, accD-C794-editing-deficient mutants with low proportions of C18:1 and C18:2 showed a heat-induced yellow, dwarf phenotype. In addition, TEM observations of heat-stress-treated plant tissues revealed morphologically compromised chloroplast membrane integrity and partial lipid staining of cellular plasma membranes in these plants (Figure 6), highlighting the importance of C18:1 and C18:2 for biological membrane system integrity under heat stress.
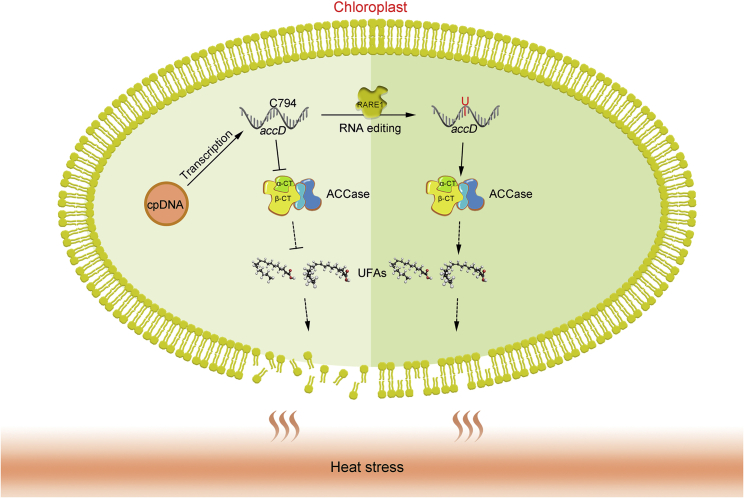
In summary, the data presented here suggest that accD-C794 editing positively affects ACCase activity; this activity is essential for FA biosynthesis, which in turn correlates directly with the proportions of C18:1 and C18:2 UFAs that are integral to the biological membrane homeostasis required for correct physiological functioning and heat tolerance (Figure 7).
Figure 7.
A working model showing how accD-C794 editing influences the regulation of ACCase activity and fatty acid biosynthesis.
Chloroplast accD transcripts undergo C→U RNA editing at the C794 site, which depends on the PPR protein RARE1. Loss of RARE1 function causes defects in C794 editing of accD transcripts. The extent of editing of accD, which encodes the α-CT of heterotetrametric plastid ACCase, is positively correlated with ACCase activity, thereby influencing UFA biosynthesis and ultimately regulating plastid membrane integrity under heat stress.
Methods
Plant materials and growth conditions
The rare1 (CS415940 and CS851454) mutants and cac3 mutant (SALK_207356) used in this study were obtained from the Arabidopsis Biological Resource Center. Individual mutant plants were genotyped by PCR from genomic DNA with oligonucleotide primers designed in locations flanking both ends of the T-DNA insertion, along with a third primer located on the left border of the T-DNA insertion. For phenotypic characterization, chlorophyll detection, and immunoblot analyses, Arabidopsis seeds were surface sterilized, plated on Murashige and Skoog medium (Sigma) containing 1% sucrose, incubated at 4°C for 2 days, and then grown under long-day conditions (16 h light and 8 h darkness) at 22°C or 28°C for 7 days. For exogenous application experiments, seedlings were grown on 1% sucrose-containing Murashige and Skoog medium with malonyl-CoA lithium salt (Sigma) or UFAs (Aladdin) under the above-mentioned growth conditions.
Chloroplast RNA-editing analysis
Total RNA was purified using the TRIzol reagent (Thermo Fisher Scientific), and first-strand cDNA was synthesized using a HiScript III 1st Strand cDNA Synthesis Kit (+gDNA wiper) (Vazyme). Specific cDNA fragments containing editing sites were amplified and sequenced directly. RNA-editing efficiency was estimated from the relative height of nucleotide peaks. The primer sequences are listed in Supplemental Table 1.
Quantitative RT-PCR
Quantitative RT-PCR was carried out as described previously (Huang et al., 2020). The AtActin2 gene was used as an internal control. The sequence information used for quantitative RT-PCR is presented in Supplemental Table 1.
Immunoblot analyses
Total protein was extracted from the wild type, rare1 mutants, and accD-OE transgenic plants for immunological detection as described previously (Huang et al., 2020). The amount of total protein was measured by a Bradford assay, and 40 μg total protein was loaded for each sample. The polyclonal antibodies for PsaA, NdhH, PetD, and light-harvesting chlorophyll a/b complex II were obtained as a gift from Dr. Hai-Bo Xiong (The Institute of Botany, Chinese Academy of Sciences). The anti-His antibody was obtained from Abcam.
Lipid analysis
Total FAs were extracted and converted to FA methyl esters in methanol solution containing 5% sulfuric acid. Samples were treated with ultrasound for 30 min at 42°C and then incubated at 65°C for 3 h. After that, FA methyl esters were extracted into hexane and 0.9% (w/v) NaCl and analyzed on an Agilent Technologies GCMS-TQ8050 system with a 100 m × 0.25 mm ID SH-Rt-2560 column (Shimadzu).
TEM
Leaves from 7-day-old wild-type, rare1, and accD-OE plants were fixed with 2.5% glutaraldehyde in phosphate buffer (pH 7.4) followed by osmium tetroxide and then dehydrated in an ethanol series. Ultrathin sections were obtained using a diamond knife and mounted on copper grids. The grids were then stained with uranyl acetate and alkaline lead citrate and examined using a JEM-1400 (JEOL) transmission electron microscope.
ACCase activity analysis
Fresh tissue (about 0.1 g) of whole seedlings from 7-day-old wild-type and rare1 mutants was ground thoroughly using a TissueLyser Ⅱ (Qiagen). The ACCase activity assay was carried out using an ACC assay kit (Zcibio) according to the manufacturer’s descriptions. Absorbance at 660 nm was measured with a Multimode Microplate Reader (Tecan).
Funding
This work was supported by the National Natural Science Foundation of China (91317312 and 31900387) and the Natural Science Foundation of Hunan Province (2020JJ4037 and 2021JJ40243).
Author contributions
C.H. and L.-T.X. conceived the project and designed the research. C.H., D.L., Z.-A.L., Z.-F.L., and H.-O.L. performed experiments. C.H., Y.S., Q.L., and R.-Z.W. analyzed data. C.H., D.P.M., and L.-T.X. wrote the paper, which all authors edited and approved.
Acknowledgments
We thank Dr. Hai-Bo Xiong (The Institute of Botany, Chinese Academy of Sciences) for kindly providing PsaA, NdhH, PetD, and light-harvesting chlorophyll a/b complex II antibodies. No conflict of interest is declared.
Published: October 10, 2022
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Supplemental information
References
- Baud S., Guyon V., Kronenberger J., Wuillème S., Miquel M., Caboche M., Lepiniec L., Rochat C. Multifunctional acetyl-CoA carboxylase 1 is essential for very long chain fatty acid elongation and embryo development in Arabidopsis. Plant J. 2003;33:75–86. doi: 10.1046/j.1365-313x.2003.016010.x. [DOI] [PubMed] [Google Scholar]
- Bayer-Császár E., Haag S., Jörg A., Glass F., Härtel B., Obata T., Meyer E.H., Brennicke A., Takenaka M. The conserved domain in MORF proteins has distinct affinities to the PPR and E elements in PPR RNA editing factors. Biochim. Biophys. Acta. Gene Regul. Mech. 2017;1860:813–828. doi: 10.1016/j.bbagrm.2017.05.004. [DOI] [PubMed] [Google Scholar]
- Brennicke A., Marchfelder A., Binder S. RNA editing. FEMS Microbiol. Rev. 1999;23:297–316. doi: 10.1111/j.1574-6976.1999.tb00401.x. [DOI] [PubMed] [Google Scholar]
- Chateigner-Boutin A.L., Small I. A rapid high-throughput method for the detection and quantification of RNA editing based on high-resolution melting of amplicons. Nucleic Acids Res. 2007;35:e114. doi: 10.1093/nar/gkm640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chateigner-Boutin A.L., Small I. Plant RNA editing. RNA Biol. 2010;7:213–219. doi: 10.4161/rna.7.2.11343. [DOI] [PubMed] [Google Scholar]
- Chu D., Wei L. Reduced C-to-U RNA editing rates might play a regulatory role in stress response of Arabidopsis. J. Plant Physiol. 2020;244:153081. doi: 10.1016/j.jplph.2019.153081. [DOI] [PubMed] [Google Scholar]
- Cronan J.E., Jr. The biotinyl domain of Escherichia coli acetyl-CoA carboxylase. Evidence that the "thumb" structure id essential and that the domain functions as a dimer. J. Biol. Chem. 2001;276:37355–37364. doi: 10.1074/jbc.M106353200. [DOI] [PubMed] [Google Scholar]
- Cui X., Wang Y., Wu J., Han X., Gu X., Lu T., Zhang Z. The RNA editing factor DUA1 is crucial to chloroplast development at low temperature in rice. New Phytol. 2019;221:834–849. doi: 10.1111/nph.15448. [DOI] [PubMed] [Google Scholar]
- Gao Z.P., Yu Q.B., Zhao T.T., Ma Q., Chen G.X., Yang Z.N. A functional component of the transcriptionally active chromosome complex, Arabidopsis pTAC14, interacts with pTAC12/HEMERA and regulates plastid gene expression. Plant Physiol. 2011;157:1733–1745. doi: 10.1104/pp.111.184762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hölzl G., Dörmann P. Chloroplast lipids and their biosynthesis. Annu. Rev. Plant Biol. 2019;70:51–81. doi: 10.1146/annurev-arplant-050718-100202. [DOI] [PubMed] [Google Scholar]
- Harwood J.L. Fatty acid metabolism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1988;39:101–138. doi: 10.1146/annurev.pp.39.060188.000533. [DOI] [Google Scholar]
- He M., He C.Q., Ding N.Z. Abiotic stresses: general defenses of land plants and chances for engineering multistress tolerance. Front. Plant Sci. 2018;9:1771. doi: 10.3389/fpls.2018.01771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M., Qin C.X., Wang X., Ding N.Z. Plant unsaturated fatty acids: biosynthesis and regulation. Front. Plant Sci. 2020;11:390. doi: 10.3389/fpls.2020.00390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., David M.P., Cao Z.-L., Xiao L.-T. Mutation of chloroplast CHLM contributes to down-regulation of multiple stress response genes in Arabidopsis. Plant Growth Regul. 2020;91:209–219. doi: 10.1007/s10725-020-00600-9. [DOI] [Google Scholar]
- Huang C., Yu Q.-B., Li Z.-R., Ye L.-S., Xu L., Yang Z.-N. Porphobilinogen deaminase HEMC interacts with the PPR-protein AtECB2 for chloroplast RNA editing. Plant J. 2017;92:546–556. doi: 10.1111/tpj.13672. [DOI] [PubMed] [Google Scholar]
- Konishi T., Sasaki Y. Compartmentalization of two forms of acetyl-CoA carboxylase in plants and the origin of their tolerance toward herbicides. Proc. Natl. Acad. Sci. USA. 1994;91:3598–3601. doi: 10.1073/pnas.91.9.3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu K.-C., Hsieh M.-H., Wang H.-J., Hsieh H.-L., Jauh G.-Y. Distinct role of Arabidopsis mitochondrial P-type pentatricopeptide repeat protein-modulating editing protein, PPME, in nad1 RNA editing. RNA Biol. 2016;13:593–604. doi: 10.1080/15476286.2016.1184384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Liu L.N., Meng Q., Fan H., Sui N. The roles of chloroplast membrane lipids in abiotic stress responses. Plant Signal. Behav. 2020;15:1807152. doi: 10.1080/15592324.2020.1807152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Lan J., Huang Y., Cao P., Zhou C., Ren Y., He N., Liu S., Tian Y., Nguyen T., et al. WSL5, a pentatricopeptide repeat protein, is essential for chloroplast biogenesis in rice under cold stress. J. Exp. Bot. 2018;69:3949–3961. doi: 10.1093/jxb/ery214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madoka Y., Tomizawa K.I., Mizoi J., Nishida I., Nagano Y., Sasaki Y. Chloroplast transformation with modified accD operon increases acetyl-CoA carboxylase and causes extension of leaf longevity and increase in seed yield in tobacco. Plant Cell Physiol. 2002;43:1518–1525. doi: 10.1093/pcp/pcf172. [DOI] [PubMed] [Google Scholar]
- Nikolau B.J., Ohlrogge J.B., Wurtele E.S. Plant biotin-containing carboxylases. Arch. Biochem. Biophys. 2003;414:211–222. doi: 10.1016/s0003-9861(03)00156-5. [DOI] [PubMed] [Google Scholar]
- Robbins J.C., Heller W.P., Hanson M.R. A comparative genomics approach identifies a PPR-DYW protein that is essential for C-to-U editing of the Arabidopsis chloroplast accD transcript. RNA. 2009;15:1142–1153. doi: 10.1261/rna.1533909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruwe H., Castandet B., Schmitz-Linneweber C., Stern D.B. Arabidopsis chloroplast quantitative editotype. FEBS Lett. 2013;587:1429–1433. doi: 10.1016/j.febslet.2013.03.022. [DOI] [PubMed] [Google Scholar]
- Sasaki Y., Nagano Y. Plant acetyl-CoA carboxylase: structure, biosynthesis, regulation, and gene manipulation for plant breeding. Biosci. Biotechnol. Biochem. 2004;68:1175–1184. doi: 10.1271/bbb.68.1175. [DOI] [PubMed] [Google Scholar]
- Sasaki Y., Kozaki A., Hatano M. Link between light and fatty acid synthesis: thioredoxin-linked reductive activation of plastidic acetyl-CoA carboxylase. Proc. Natl. Acad. Sci. USA. 1997;94:11096–11101. doi: 10.1073/pnas.94.20.11096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y., Hakamada K., Suama Y., Nagano Y., Furusawa I., Matsuno R. Chloroplast-encoded protein as a subunit of acetyl-CoA carboxylase in pea plant. J. Biol. Chem. 1993;268:25118–25123. doi: 10.1016/s0021-9258(19)74577-3. [DOI] [PubMed] [Google Scholar]
- Shi X., Bentolila S., Hanson M.R. Organelle RNA recognition motif-containing (ORRM) proteins are plastid and mitochondrial editing factors in Arabidopsis. Plant Signal. Behav. 2016;11:e1167299. doi: 10.1080/15592324.2016.1167299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small I.D., Schallenberg-Rüdinger M., Takenaka M., Mireau H., Ostersetzer-Biran O. Plant organellar RNA editing: what 30 years of research has revealed. Plant J. 2020;101:1040–1056. doi: 10.1111/tpj.14578. [DOI] [PubMed] [Google Scholar]
- Sun T., Bentolila S., Hanson M.R. The unexpected diversity of plant organelle RNA editosomes. Trends Plant Sci. 2016;21:962–973. doi: 10.1016/j.tplants.2016.07.005. [DOI] [PubMed] [Google Scholar]
- Sun T., Shi X., Friso G., Van Wijk K., Bentolila S., Hanson M.R. A zinc finger motif-containing protein is essential for chloroplast RNA editing. PLoS Genet. 2015;11:e1005028. doi: 10.1371/journal.pgen.1005028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan T.M., Watson K. Stress tolerance in a yeast lipid mutant: membrane lipids influence tolerance to heat and ethanol independently of heat shock proteins and trehalose. Can. J. Microbiol. 1999;45:472–479. doi: 10.1139/w99-033. [DOI] [PubMed] [Google Scholar]
- Takenaka M., Zehrmann A., Verbitskiy D., Kugelmann M., Härtel B., Brennicke A. Multiple organellar RNA editing factor (MORF) family proteins are required for RNA editing in mitochondria and plastids of plants. Proc. Natl. Acad. Sci. USA. 2012;109:5104–5109. doi: 10.1073/pnas.1202452109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelen J.J., Ohlrogge J.B. Metabolic engineering of fatty acid biosynthesis in plants. Metab. Eng. 2002;4:12–21. doi: 10.1006/mben.2001.0204. [DOI] [PubMed] [Google Scholar]
- Tillich M., Hardel S.L., Kupsch C., Armbruster U., Delannoy E., Gualberto J.M., Lehwark P., Leister D., Small I.D., Schmitz-Linneweber C. Chloroplast ribonucleoprotein CP31A is required for editing and stability of specific chloroplast mRNAs. Proc. Natl. Acad. Sci. USA. 2009;106:6002–6007. doi: 10.1073/pnas.0808529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Chen P., Yu X., Xia Y., Yan L.T., Ai L. C18:1 improves the freeze-drying resistance of Lactobacillus plantarum by maintaining the cell membrane. ACS Appl. Bio Mater. 2020;3:4933–4940. doi: 10.1021/acsabm.0c00444. [DOI] [PubMed] [Google Scholar]
- Yagi Y., Ishizaki Y., Nakahira Y., Tozawa Y., Shiina T. Eukaryotic-type plastid nucleoid protein pTAC3 is essential for transcription by the bacterial-type plastid RNA polymerase. Proc. Natl. Acad. Sci. USA. 2012;109:7541–7546. doi: 10.1073/pnas.1119403109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Zhang Q., Yin P. RNA editing machinery in plant organelles. Sci. China Life Sci. 2018;61:162–169. doi: 10.1007/s11427-017-9170-3. [DOI] [PubMed] [Google Scholar]
- Yu Q., Lutz K.A., Maliga P. Efficient plastid transformation in Arabidopsis. Plant Physiol. 2017;175:186–193. doi: 10.1104/pp.17.00857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A., Jiang X., Zhang F., Wang T., Zhang X. Dynamic response of RNA editing to temperature in grape by RNA deep sequencing. Funct. Integr. Genomics. 2020;20:421–432. doi: 10.1007/s10142-019-00727-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Tang W., Hedtke B., Zhong L., Liu L., Peng L., Lu C., Grimm B., Lin R. Tetrapyrrole biosynthetic enzyme protoporphyrinogen IX oxidase 1 is required for plastid RNA editing. Proc. Natl. Acad. Sci. USA. 2014;111:2023–2028. doi: 10.1073/pnas.1316183111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.