Abstract
In cases of severe wildlife population decline, a key question is whether recovery efforts will be impeded by genetic factors such as inbreeding depression. Decades of excess mortality from gillnet fishing have driven Mexico's vaquita porpoise (Phocoena sinus) to ~10 remaining individuals. We analyzed whole genome sequences from 20 vaquitas and integrated genomic and demographic information into stochastic, individual-based simulations to quantify the species' recovery potential. Our analysis suggests the vaquita's historical rarity has resulted in a low burden of segregating deleterious variation, reducing the risk of inbreeding depression. Similarly, genome-informed simulations suggest the vaquita can recover if bycatch mortality is immediately halted. This study provides hope for vaquitas and other naturally rare endangered species and highlights the utility of genomics in predicting extinction risk.
One-sentence summary:
Whole genome sequencing and genomics-based population viability analyses suggest the vaquita is not doomed to extinction.
A central question for populations that have undergone severe declines is whether recovery is possible, or if it may be hindered by deleterious genetic factors (1). Perhaps the most immediate genetic threat in populations of very small size (<25 individuals) is the deterioration of fitness due to inbreeding depression (2, 3). Thus, predicting the threat of inbreeding depression under various genetic and demographic conditions is essential for the conservation of endangered species.
The critically endangered vaquita porpoise (Phocoena sinus), found only in the northernmost Gulf of California, Mexico, has declined from ~600 individuals in 1997 to around 10 individuals at present (4). This precipitous decline has been driven by incidental mortality in fishing gillnets (bycatch) ((4, 5); Fig. 1A). Efforts to reduce the intensity of illegal gillnet fishing and implement stronger protections for vaquitas have not been successful, and vaquitas are now considered the most endangered marine mammal (4). A recent viability analysis found that the vaquita population could theoretically rebound if bycatch mortality is eliminated (6). However, the degree to which genetic factors may prevent a robust recovery is unknown, leading some to argue that the species is doomed to extinction from genetic threats (see discussion in (1, 7, 8)).
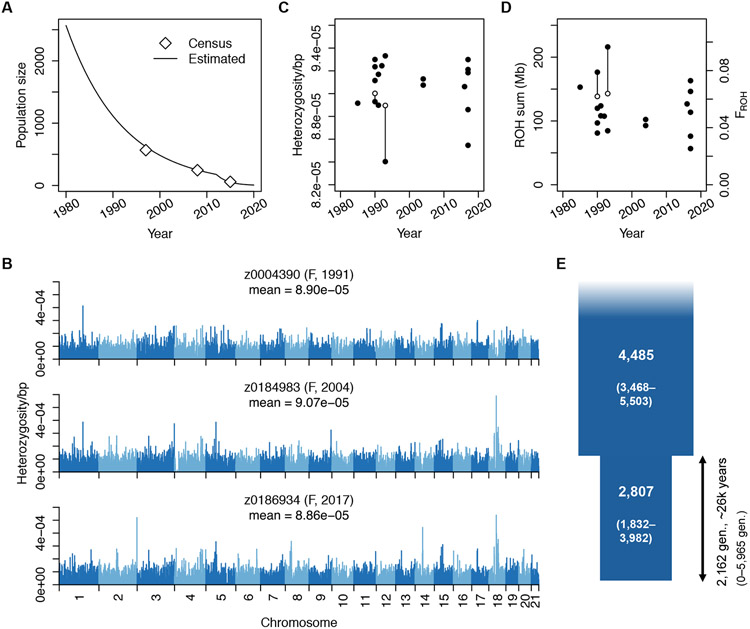
Fig. 1. Vaquita genome-wide diversity and demographic history.
(A) Model of vaquita census population size based on previous surveys (5) shows a dramatic recent decline. (B) Bar plots of per-site heterozygosity in 1-Mb genomic windows in three individuals (one from each sampling period; see fig. S1 for all) show little variability within or between individuals. (C, D) Genome-wide heterozygosity and ROH burden are consistent between sampling periods. Lines connect mother-fetus pairs; open symbols indicate offspring. (E) Two-epoch demographic model inferred with ∂a∂i. Parameter 95% confidence intervals indicated in parentheses.
Population viability analysis (PVA) has long been an important tool for modelling extinction risk (9). However, it is often challenging to parameterize PVA models for highly endangered species where information on the potential impact of inbreeding depression is limited. Genomic data offer a potential solution, as they can be used to estimate the fundamental genetic and demographic parameters underlying inbreeding depression. Although the potential applications of genomics in conservation have been widely discussed (10, 11), genomics remain underutilized in forecasts of population viability and extinction risk.
To investigate the impact of the vaquita’s recent decline and to quantify the species’ recovery potential, we sequenced genomic DNA of 19 archival tissue samples to high depth (total n = 20 including genome from (12), mean coverage = 60X; table S1). Samples were obtained across three time periods: 1985-1993, 2004, and 2016-2017, spanning ~3 vaquita generations (assuming a generation time of 11.9 years; (13)) and an estimated ~99% decline in population size (Fig. 1A, (5)). All 20 vaquita genomes contain uniformly low heterozygosity (mean = 9.04x10−5, standard deviation (S.D.) = 2.44x10−6 heterozygotes/site; Fig. 1B and fig. S1), consistent with a previous estimate from a single individual (12). Additionally, genome-wide diversity appears stable over the sampling period (Fig. 1B, C), as expected given the short duration of the decline.
We also investigated whether vaquita genomes show signs of recent inbreeding. We found that the mean cumulative fraction of vaquita genomes in long (≥1 Mb) runs of homozygosity (ROH) is 5.42% (S.D. = 1.7%), implying a low average inbreeding coefficient of FROH = 0.05 (Fig. 1D and fig. S2). Furthermore, ROH in our sample are relatively short (mean length 1.59-3.18 Mb), suggesting that they trace to a common ancestor from roughly 15-31 generations ago (178-369 years; (5)). This result indicates that these ROH are a consequence of the vaquita’s historically limited population size rather than recent inbreeding. Finally, we found limited evidence for close relatives in our dataset, aside from two known mother-fetus pairs (fig. S3).
To better characterize the vaquita’s long-term demographic history, we used the distribution of allele frequencies to perform model-based demographic inference. Overall, we found good fit for a two-epoch model in which the vaquita effective population size (Ne) declined from 4,485 to 2,807 individuals ~2,162 generations ago (~25.7 KYA; (5); Fig. 1E, figs. S4 and S5, tables S2 to S4). Thus, vaquitas have persisted at relatively small population sizes for at least tens of thousands of years, resulting in uniformly low genome-wide diversity that is among the lowest documented in any species to date (12). Here, we use ‘long-term small population size’ to mean Ne on the order of a few thousand individuals over thousands of generations, as opposed to ‘small population size’ meaning Ne ≤100, as in some other contexts (e.g., (14, 15))).
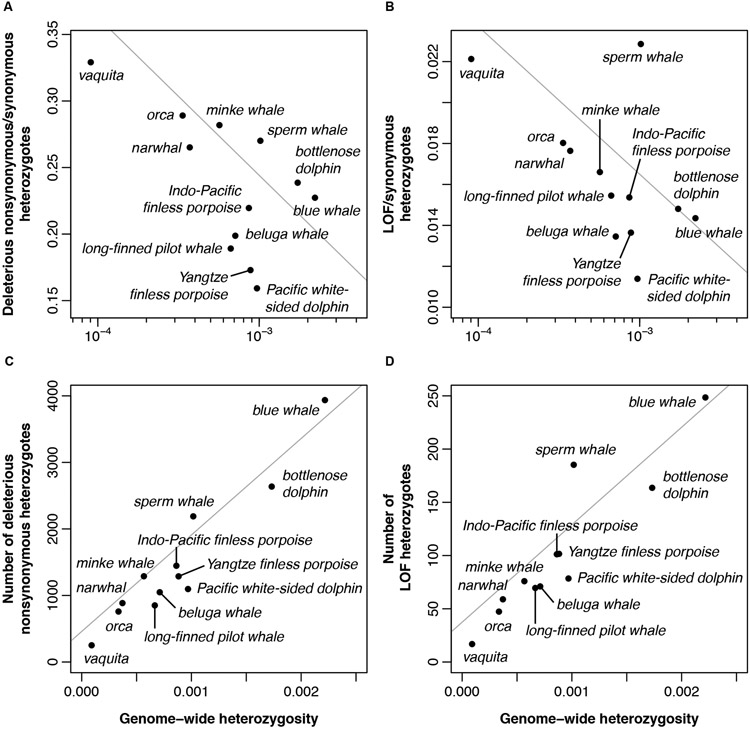
A predicted consequence of long-term small population size is the reduced efficacy of purifying selection against weakly deleterious alleles with selection coefficients <<1/(2*Ne) (14, 15). Such alleles can drift to high frequencies and become fixed, potentially contributing to reduced fitness. To investigate this, we compared the burden of putatively deleterious protein-coding variants in vaquitas with 11 other cetacean species (table S5, fig. S6). Specifically, we focused on nonsynonymous mutations at sites under strong evolutionary constraint (16), and loss-of-function (LOF) mutations that are predicted to disrupt gene function. We used the ratio of deleterious to synonymous variants as a proxy for the efficacy of purifying selection (5) and used genome-wide heterozygosity as a proxy for Ne (Fig. 2A, B and fig. S7). The ratio of deleterious variants is significantly negatively correlated with Ne (phylogenetic generalized least squares (PGLS) regression, pdel. = 1.32x10−2, pLOF = 7.88x10−3), consistent with expectation. Among all species in our study, vaquitas have the highest proportional burden of deleterious alleles. Compared to the species with the next lowest diversity (orca, Orcinus orca), ratios for deleterious and LOF mutations in vaquitas are 1.14x and 1.23x higher, respectively. Furthermore, we demonstrate using simulations that this elevated ratio is minimally impacted by the vaquita’s recent population decline, and is instead attributable to its historical population size (fig. S9; (5)). Similar trends exist for homozygous deleterious mutations, which includes variants that may be fixed in the species (fig. S8). Thus, elevated ratios of deleterious to neutral variation among polymorphisms (heterozygotes) and substitutions (homozygotes) in vaquitas are consistent with an accumulation of weakly deleterious alleles under long-term small population size. The remaining vaquita individuals appear healthy and are actively reproducing (17, 18), suggesting the species’ fitness has not been severely compromised by its longstanding elevated burden of weakly deleterious alleles.
Fig. 2. Deleterious variation in vaquitas and other cetaceans.
Ratios of deleterious nonsynonymous (A) and LOF (B) heterozygotes to synonymous heterozygotes are significantly negatively correlated with genome-wide heterozygosity (per bp, log-scaled). Total numbers of deleterious nonsynonymous (C) and LOF (D) heterozygotes per genome are significantly positively correlated with genome-wide heterozygosity (per bp). Grey lines show phylogeny-corrected regressions (excluding the Indo-Pacific finless porpoise (5)).
A larger concern for vaquita recovery is future fitness declines due to inbreeding depression, given the inevitability of inbreeding in any recovery scenario. However, the risk of inbreeding depression (or “inbreeding load”) is predicted to be reduced in species with long-term small population size because 1) increased homozygosity exposes recessive strongly deleterious alleles to selection more frequently, and 2) drift decreases the absolute number of segregating recessive deleterious variants (19, 20). To assess the potential for future inbreeding depression in vaquitas relative to other cetaceans, we quantified the total number of heterozygous deleterious alleles per genome, which reflect alleles that could contribute to inbreeding depression when made homozygous through inbreeding. We found that the total number of heterozygous putatively deleterious alleles per genome is positively correlated with genome-wide diversity (PGLS pdel. = 5.57x10−6, pLOF = 1.91x10−5) (Fig. 2C, D). Among all cetaceans in our study, vaquitas harbor the fewest deleterious heterozygotes per genome. Compared to the orca, vaquitas have 0.33x and 0.36x the number of deleterious and LOF heterozygotes, respectively. Similar trends are evident in all mutation classes, including conserved noncoding regions (fig. S10). Thus, although vaquitas have an elevated proportion of deleterious relative to neutral variants (Fig. 2A, B, fig. S8), they nevertheless have a low absolute number of segregating deleterious variants (Fig. 2C, D), implying a low inbreeding load.
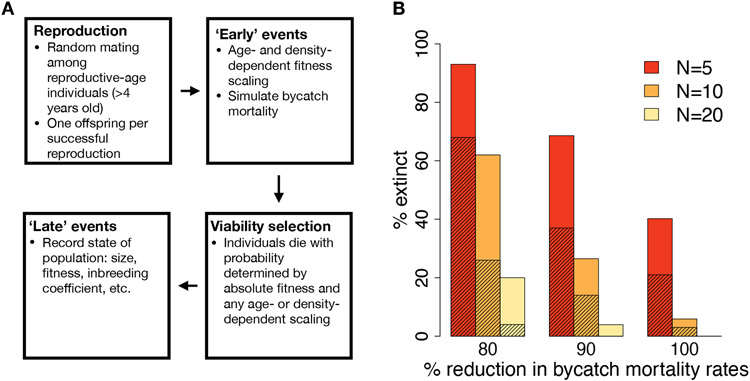
To model potential recovery scenarios for the vaquita, we combined our genomic results with information about vaquita life history to parameterize stochastic, individual-based simulations using SLiM3 ((5, 21); Fig. 3A, fig. S11). These simulations were designed to model vaquita protein-coding regions, incorporating both neutral mutations and recessive deleterious mutations, the latter of which are thought to underlie inbreeding depression (3, 22). We used our genomic dataset to estimate a vaquita mutation rate (fig. S12) as well as a distribution of selection coefficients for new mutations (fig. S13), and assumed an inverse relationship between dominance and selection coefficients (5). Importantly, our model allows for deleterious mutations to drift to fixation and impact fitness (figs. S14 to S16; (5)). We used our demographic model (Fig. 1E) to simulate the historical vaquita population (figs. S17 and S18), then initiated a bottleneck by introducing stochastic bycatch mortality at a rate calibrated to the empirical rate of recent decline as of 2018 (Fig. 1A and fig. S19; (5)). Finally, we allowed for recovery by reducing the bycatch mortality rate after the population reached a ‘threshold population size’ of 10 or fewer individuals, based on the current estimated population size.
Fig. 3. Model schematic and extinction rates under various simulation parameters.
(A) Diagram of events that occur during one year in our SLiM simulation model. (B) Percent of replicates going extinct over the next 50 years under varying recovery parameters. Shading indicates extinction rates when only neutral mutations are simulated, and “N” represents the threshold population size.
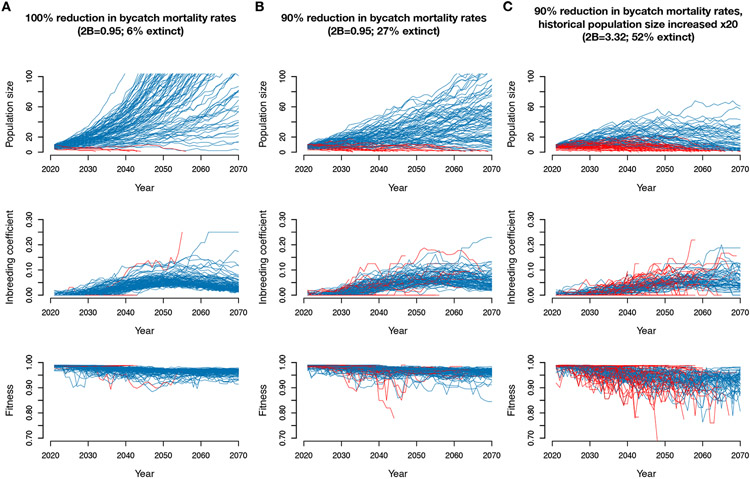
We first used this model to examine the impact of varying levels of bycatch mortality on extinction risk over the next 50 years. We estimate a high probability of recovery if bycatch mortality ceases entirely, with only 6% of simulation replicates going extinct (Figs. 3B, 4A). In addition, simulated populations that persist exhibit substantial growth, with a mean population size in 2070 of 298.7 individuals (S.D. = 218.2; Fig. 4A). However, if bycatch mortality rates are decreased by just 90%, extinction rates increase to 27% (Figs. 3B and 4B), with more limited recovery in population sizes (mean of 49.2 individuals in 2070, S.D. = 34.4; Fig. 4B). Finally, if bycatch mortality rates are decreased by just 80%, extinction occurs in 62% of simulation replicates. Thus, recovery potential critically depends on reducing bycatch mortality rates, with even moderate levels of bycatch resulting in a high likelihood of extinction.
Fig. 4. Simulation trajectories under various recovery scenarios.
(A) Simulation trajectories under empirically-inferred historical demographic parameters assuming a reduction in bycatch mortality of 100%. (B) Simulation trajectories with bycatch mortality rate decreased by only 90%. (C) Simulation trajectories with historical population size increased x20 and assuming a decrease in bycatch mortality of 90%. For all simulations, we assumed a population size threshold of 10 individuals. Replicates that went extinct are colored red and replicates that persisted are colored blue.
Next, we examined the importance of the threshold population size, given uncertainty in the 2018 estimate of 10 individuals (4). As expected, extinction rates decrease when assuming a threshold population size of 20 and increase when assuming a threshold population size of 5 (Fig. 3B). These results emphasize that the number of remaining vaquita individuals is also a critical factor underlying extinction risk.
To quantify the inbreeding load in our model, we estimated the ‘number of diploid lethal equivalents’ (or 2B), which characterizes the rate at which fitness is lost with increasing levels of inbreeding (2, 23). Typically, inbreeding load is quantified by comparing estimates of individual fitness and inbreeding in natural populations (2, 24); however, such data do not exist for most species, including the vaquita. Under our simulation parameters, we estimate an inbreeding load of 2B = 0.95 in vaquitas (table S6), significantly lower than the median empirical estimate for mammals of 6.2 (24), likely due to the vaquita’s relatively small historical Ne. Nevertheless, simulations that exclude deleterious mutations result in a significantly lower extinction rate (Fig. 3B), confirming that inbreeding depression impacts recovery potential in our model.
To further explore how the inbreeding load in our model depends on historical demography, we ran simulations with the historical Ne increased x20. We found an increased extinction rate of 52%, compared to 27% with our empirical population size parameters, with minimal recovery for replicates that persisted (mean of 16.2 individuals in 2070, S.D. = 14.5, Fig. 4C). Additionally, with this larger historical Ne, we observe a greatly increased inbreeding load of 2B = 3.32 (fig. S20 and table S6). These findings further demonstrate the importance of the vaquita’s natural rarity as a factor underlying their low inbreeding load and increased potential for recovery.
Given the uncertainty in many of our model parameters, we conducted sensitivity analyses varying the calving interval, mutation rate, distribution of dominance and selection coefficients, and target size for deleterious mutations (5). Although these factors influence extinction probabilities, recovery remains the likely outcome (>50% probability) in nearly all cases when assuming a threshold population size of 10 and a 90% reduction of bycatch mortality (fig. S21 and table S6). Two notable exceptions to this are for models with a higher mutation rate, where we observed a 55% extinction rate compared to 27% in our ‘base’ model, and for models with decreased calving interval, where we also observed a 55% extinction rate (fig. S21 and table S6). Thus, although uncertainty exists in our projections, the overall conclusion that recovery is possible if bycatch is greatly reduced remains robust to our model assumptions. Finally, we note that our simulations do not consider factors such as reduced adaptive potential or increased susceptibility to disease caused by low genetic variability, which may impact future persistence. Vaquitas have survived with low diversity for tens of thousands of years and have endured environmental changes in the past (12), suggesting that these factors alone do not doom the species to extinction. Conceivably, low diversity in the vaquita may limit the species’ capacity to adapt to increasing global change over the long term, but this risk is challenging to quantify and should not preclude recovery efforts in the short term.
In conclusion, our results suggest there is a high potential for vaquita recovery in the absence of gillnet mortality, refuting the view that the species is doomed to extinction by genetic factors. Our approach leverages genomic data and methodology to forecast population viability and extinction risk, enabling a more nuanced assessment of the threat of genetic factors to persistence. The key aspect of the vaquita that our analysis reveals is that its historical population size was large enough to prevent the fixation of all but weakly deleterious alleles, and small enough to reduce the inbreeding load from recessive strongly deleterious mutations. Numerous other examples of species rebounding from bottlenecks of similar magnitude to that of the vaquita have been documented (reviewed in (1)). For example, many parallels exist between the vaquita and Channel Island foxes, which similarly have exceptionally low genetic diversity, yet were able to rebound from severe recent bottlenecks without apparent signs of inbreeding depression (25). Together, these examples challenge the assumption that populations that have experienced catastrophic declines are genetically doomed and provide hope for the recovery of endangered species that are naturally rare. Finally, our analysis demonstrates the potential for genomics-informed population viability modelling, which may have widespread applications given the increasing feasibility of genomic sequencing for non-model species amid a worsening extinction crisis (26).
Supplementary Material
Acknowledgments
We thank the CanSeq150 project for use of the long-finned pilot whale and Pacific white-sided dolphin genomes. Y. Bukhman generously provided early access to the blue whale genome. We thank J. Mah, P. Nuñez, and M. Lin for providing scripts, and B. Haller for assistance with simulations. We thank the Southwest Fisheries Science Center’s Marine Mammal and Sea Turtle Research Collection for use of archival vaquita tissue samples. All samples were imported to the US under appropriate CITES and US Marine Mammal Protection Act permits.
Funding:
We thank Frances Gulland, The Marine Mammal Center, and NOAA Fisheries for funding genome resequencing. C.C.K. and K.E.L. were supported by National Institutes of Health (NIH) grant R35GM119856 (to K.E.L.). A.C.B. was supported by the Biological Mechanisms of Healthy Aging Training Program NIH T32AG066574. S.N.M. was supported by the Mexican National Council for Science and Technology (CONACYT) Postdoctoral Fellowship 724094 and the Mexican Secretariat of Agriculture and Rural Development Postdoctoral Fellowship.
Footnotes
Competing interests: The authors declare no competing interests.
Data and materials availability: Vaquita raw sequence reads have been deposited in the Sequence Read Archive (SRA) under BioProject PRJNA751981 (see table S1 for details). Accession information for publicly available cetacean genomes is provided in table S5. Scripts used for sequence data processing and analysis are available at doi:10.5281/zenodo.6303135. Scripts for simulations are available at doi:10.5281/zenodo.6308771.
References and Notes
- 1.Wiedenfeld DA et al. , Conserv. Biol 35, 1388–1395 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keller L, Waller DM, Trends Ecol. Evol 17, 19–23 (2002). [Google Scholar]
- 3.Charlesworth D, Willis JH, Nat. Rev. Genet 10, 783–796 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Jaramillo-Legorreta AM et al. , R. Soc. Open Sci 6 (2019), doi: 10.1098/rsos.190598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Materials and methods and supplementary text are available as supplementary materials.
- 6.Cisneros-Mata MA, Delgado JA, Rodríguez-Félix D, Rev. Biol. Trop 69, 588–600 (2021). [Google Scholar]
- 7.Taylor BL, Rojas-Bracho L, Mar. Mammal Sci 15, 1004–1028 (1999). [Google Scholar]
- 8.Sonne C, Diaz-Jaimes P, Adams DH, Science (80-. ) 373, 863–864 (2021). [DOI] [PubMed] [Google Scholar]
- 9.Brook BW et al. , Nature. 404, 385–387 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Allendorf FW, Hohenlohe PA, Luikart G, Nat. Rev. Genet 11, 697–709 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Lewin HA et al. , Proc. Natl. Acad. Sci. U. S. A 115, 4325–4333 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morin PA et al. , Mol. Ecol. Resour 21, 1008–1020 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor BL, Chivers SJ, Larese J, Perrin WF, “Generation length and percent mature estimates for IUCN assessments of cetaceans” (2007). [Google Scholar]
- 14.Lynch M, Conery IJ, Burger R, Am. Nat 146, 489–518 (1995). [Google Scholar]
- 15.Kimura M, Maruyama T, Crow JF, Genetics, 1303–1312 (1963). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ng PC, Henikoff S, Genome Res. 11, 863–874 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor BL et al. , Mar. Mammal Sci 35, 1603–1612 (2019). [Google Scholar]
- 18.Gulland F et al. , Vet. Rec 187, 1–4 (2020).33188043 [Google Scholar]
- 19.Kyriazis CC, Wayne RK, Lohmueller KE, Evol. Lett 5, 33–47 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glémin S, Evolution (N. Y) 57, 2678–2687 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Haller BC, Messer PW, Mol. Biol. Evol 36, 632–637 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCune AR et al. , Science (80-. ) 296, 2398–2401 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Morton NE, Crow JF, Muller HJ, Proc. Natl. Acad. Sci 42, 855–863 (1956). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ralls K, Ballou JD, Templeton A, Ralls K, Ballou JD, Soc. Conserv. Biol 2, 185–193 (1988). [Google Scholar]
- 25.Robinson JA, Brown C, Kim BY, Lohmueller KE, Wayne RK, Curr. Biol 28, 3487–3494.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ceballos G, Ehrlich PR, Raven PH, Proc. Natl. Acad. Sci. U. S. A 117, 13596–13602 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scripts for sequence data processing, (available at doi: 10.5281/zenodo.6303135). [DOI] [Google Scholar]
- 28.Scripts for simulations, (available at doi: 10.5281/zenodo.6303135). [DOI] [Google Scholar]
- 29.Miller SA, Dykes DD, Polesky HF, Nucleic Acids Res. 16, 1215 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyer M, Kircher M, Cold Spring Harb. Protoc 5 (2010), doi: 10.1101/pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- 31.Kircher M, Sawyer S, Meyer M, Nucleic Acids Res. 40, 1–8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van der Auwera GA et al. , Curr. Protoc. Bioinforma, 1–33 (2013). [Google Scholar]
- 33.Li H, arXiv:1303.3997v2 00, 1–3 (2013). [Google Scholar]
- 34.Li H, Bioinformatics. 27, 2987–2993 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smit A, Hubley R, Green P, RepeatMasker Open-4.0 2013-2015, (available at http://www.repeatmasker.org). [Google Scholar]
- 36.Benson G, Nucleic Acids Res. 27, 573–580 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jaramillo-Legorreta AM, Rojas-Bracho L, Gerrodette T, Mar. Mammal Sci 15, 957–973 (1999). [Google Scholar]
- 38.Gerrodette T et al. , Mar. Mammal Sci 27, 79–100 (2011). [Google Scholar]
- 39.Taylor BL et al. , Conserv. Lett 10, 588–595 (2017). [Google Scholar]
- 40.Chang CC et al. , Gigascience. 4, 1–16 (2015).25838885 [Google Scholar]
- 41.Danecek P et al. , Bioinformatics. 27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Narasimhan V et al. , Bioinformatics. 32, 1749–1751 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Browning SR, Genetics. 178, 2123–2132 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manichaikul A et al. , Bioinformatics. 26, 2867–2873 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng X et al. , Bioinformatics. 28, 3326–3328 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Camacho C et al. , BMC Bioinformatics. 10, 421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD, PLoS Genet. 5, 1–11 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Excoffier L, Dupanloup I, Huerta-Sánchez E, Sousa VC, Foll M, PLOS Genet. 9, e1003905 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coffman AJ, Hsieh PH, Gravel S, Gutenkunst RN, Mol. Biol. Evol 33, 591–593 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dornburg A, Brandley MC, McGowen MR, Near TJ, Mol. Biol. Evol 29, 721–736 (2012). [DOI] [PubMed] [Google Scholar]
- 51.Autenrieth M et al. , Mol. Ecol. Resour 18, 1469–1481 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Yim H-S et al. , Nat. Genet 46, 88–92 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ben Chehida Y et al. , Sci. Rep 10, 1–18 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kumar S, Stecher G, Suleski M, Hedges SB, Mol. Biol. Evol 34, 1812–1819 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Feng C et al. , Elife. 6, 1–14 (2017). [Google Scholar]
- 56.Wu FL et al. , PLoS Biol. 18, 1–38 (2020). [Google Scholar]
- 57.Vaser R, Adusumalli S, Leng SN, Sikic M, Ng PC, Nat. Protoc 11, 1–9 (2016). [DOI] [PubMed] [Google Scholar]
- 58.Cingolani P et al. , Fly (Austin). 6, 80–92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.MacArthur D, Balasubramanian S, Frankish A, Science (80-. ) 335, 1–14 (2012). [Google Scholar]
- 60.Henn BM, Botigué LR, Bustamante CD, Clark AG, Gravel S, Nat. Rev. Genet 16, 333–343 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brandvain Y, Wright SI, Trends Genet. 32, 201–210 (2016). [DOI] [PubMed] [Google Scholar]
- 62.Do R et al. , Nat. Genet 47, 126–131 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seppey M, Manni M, Zdobnov EM, BUSCO: Assessing genome assembly and annotation completeness (2019), vol. 1962. [DOI] [PubMed] [Google Scholar]
- 64.Katoh K, Standley DM, Mol. Biol. Evol 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Minh BQ et al. , Mol. Biol. Evol 37, 1530–1534 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rosen BD et al. , Gigascience. 9, 1–9 (2020). [Google Scholar]
- 67.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T, Bioinformatics. 25, 1972–1973 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Creevy C, catsequences: A tool for concatenating multiple fasta alignments for supermatrix phylogenetic analyses, (available at doi: 10.5281/zenodo.4409153). [DOI] [Google Scholar]
- 69.Chernomor O, Von Haeseler A, Minh BQ, Syst. Biol 65, 997–1008 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kalyaanamoorthy S, Minh BQ, Wong TKF, Von Haeseler A, Jermiin LS, Nat. Methods 14, 587–589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McGowen MR et al. , Syst. Biol 69, 479–501 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hoang DT, Chernomor O, Von Haeseler A, Minh BQ, Vinh LS, Mol. Biol. Evol 35, 518–522 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.R Core Team R: A language and environment for statistical computing. (2021), (available at https://www.r-project.org/). [Google Scholar]
- 74.Paradis E, Schliep K, Bioinformatics. 35, 526–528 (2019). [DOI] [PubMed] [Google Scholar]
- 75.Pennell MW et al. , Bioinformatics. 30, 2216–2218 (2014). [DOI] [PubMed] [Google Scholar]
- 76.Pinheiro J, Bates D, DebRoy S, Sarkar D, nlme: Linear and Nonlinear Mixed Effects Models. R package version 3.1–152, (available at https://cran.r-project.org/package=nlme). [Google Scholar]
- 77.Polychronopoulos D, King JWD, Nash AJ, Tan G, Lenhard B, Nucleic Acids Res. 45, 12611–12624 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Siepel A et al. , Genome Res. 15, 1034–1050 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huber CD, Kim BY, Marsden CD, Lohmueller KE, Proc. Natl. Acad. Sci 114, 4465–4470 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim BY, Huber CD, Lohmueller KE, Genetics. 206, 345–361 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Castellano D, Macià MC, Tataru P, Bataillon T, Munch K, Genetics. 213, 953–966 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Moore JE, Read AJ, Ecol. Appl 18, 1914–1931 (2008). [DOI] [PubMed] [Google Scholar]
- 83.Robinson JA et al. , Sci. Adv 5, 1–13 (2019). [Google Scholar]
- 84.Hedrick PW, Garcia-Dorado A, Trends Ecol. Evol 31, 940–952 (2016). [DOI] [PubMed] [Google Scholar]
- 85.Kardos M et al. , Proc. Natl. Acad. Sci. U. S. A 118, 1–10 (2021). [Google Scholar]
- 86.Gao Z, Waggoner D, Stephens M, Ober C, Przeworski M, Genetics. 199, 1243–1254 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Charlesworth B, Evolution in age-structured populations (Cambridge University Press, 1994). [Google Scholar]
- 88.Norris KS, Prescott J, Univ. Calif. Publ. Zool 63, 291–402 (1961). [Google Scholar]
- 89.Mitchell E, J. Fish. Res. Board Canada 32, 889–983 (1975). [Google Scholar]
- 90.Vidal O, in Biology of the Phocoenids. Reports of the International Whaling Commission (Special Issue 16). (Cambridge, UK., 1995), pp. 247–272. [Google Scholar]
- 91.D’Agrosa C, Lennert-Cody CE, Vidal O, Conserv. Biol 14, 1110–1119 (2000). [Google Scholar]
- 92.Hohn AA, Read AJ, Fernandez S, Vidal O, Findley LT, J. Zool (1996). [Google Scholar]
- 93.Gerrodette T, Rojas-Bracho L, Mar. Mammal Sci 27, 101–125 (2011). [Google Scholar]
- 94.Rojas-Bracho L, Reeves RR, Jaramillo-Legorreta A, Mamm. Rev 36, 179–216 (2006). [Google Scholar]
- 95.Lynch M, Conery J, Burger R, Evolution (N. Y) 49, 1067–1080 (1995). [DOI] [PubMed] [Google Scholar]
- 96.Lynch M, Gabriel W, Evolution (N. Y) 44, 1725–1737 (1990). [DOI] [PubMed] [Google Scholar]
- 97.Warnes GR et al. , gplots: Various R Programming Tools for Plotting Data. R package version 3.1.1 (2020), (available at https://cran.r-project.org/package=gplots). [Google Scholar]
- 98.Jones SJM et al. , Genes (Basel). 8 (2017), doi: 10.3390/genes8120378. [DOI] [Google Scholar]
- 99.Árnason Ú, Lammers F, Kumar V, Nilsson MA, Janke A, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhou X et al. , Nat. Commun 9, 1–8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Foote AD et al. , Nat. Genet 47, 272–275 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Moura AE et al. , Mol. Biol. Evol 31, 1121–1131 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Westbury MV, Petersen B, Garde E, Heide-Jørgensen MP, Lorenzen ED, iScience. 15, 592–599 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fan G et al. , Mol. Ecol. Resour 19, 944–956 (2019). [DOI] [PubMed] [Google Scholar]
- 105.Warren WC et al. , Genome Biol. Evol 9, 3260–3264 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yuan Y et al. , Genes (Basel). 9, 1–9 (2018). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.