Abstract
Helicobacter pylori has a very plastic genome, reflecting its high rate of recombination and point mutation. This plasticity promotes divergence of the population by the development of subclones and presumably enhances adaptation to host niches. We have investigated the genotypic and phenotypic characteristics of two such subclones isolated from one patient as well as the genetic evolution of these isolates during experimental infection. Whole-genome genotyping of the isolates using DNA microarrays revealed that they were more similar to each other than to a panel of other genotyped strains recovered from different hosts. Nonetheless, they still showed significant differences. For example, one isolate (67:21) contained the entire Cag pathogenicity island (PAI), whereas the other (67:20) had excised the PAI. Phenotypic studies disclosed that both isolates expressed adhesins that recognized human histo-blood group Lewisb glycan receptors produced by gastric pit and surface mucus cells. In addition, both isolates were able to colonize, to equivalent density and with similar efficiency, germ-free transgenic mice genetically engineered to synthesize Lewisb glycans in their pit cells (12 to 14 mice/isolate). Remarkably, the Cag PAI-negative isolate was unable to colonize conventionally raised Lewisb transgenic mice harboring a normal gastric microflora, whereas the Cag PAI-positive isolate colonized 74% of the animals (39 to 40 mice/isolate). The genomic evolution of both isolates during the infection of conventionally raised and germ-free mice was monitored over the course of 3 months. The Cag PAI-positive isolate was also surveyed after a 10 month colonization of conventionally raised transgenic animals (n = 9 mice). Microarray analysis of the Cag PAI and sequence analysis of the cagA, recA, and 16S rRNA genes disclosed no changes in recovered isolates. Together, these results reveal that the H. pylori population infecting one individual can undergo significant divergence, creating stable subclones with substantial genotypic and phenotypic differences.
Helicobacter pylori colonizes the stomachs of about half of the world's population. The infection is lifelong unless treated. A subpopulation of infected individuals develops severe pathology, including peptic ulcers, atrophic gastritis, and gastric adenocarcinoma. Unfortunately, it is not yet possible to predict the outcome of H. pylori infection in a colonized untreated individual or to identify human populations at risk for significant pathology. All available data indicate that the outcome is determined by factors emanating from the host, the microorganism, and the environment (13, 21, 24).
H. pylori is a species with a plastic, adaptable genome that presumably helps the bacteria to persist for decades in the inhospitable niche of the human stomach. Isolates from different individuals display considerable genomic variation as defined by fingerprinting methods such as restriction fragment length polymorphism (RFLP) or arbitrary primed PCR (AP-PCR) (2, 22). Strains isolated from unrelated individuals rarely have identical fingerprints, in contrast to strains isolated from members of the same family (26). Most of this genomic variation appears to result in silent, synonymous base substitutions (40) and thus is not accompanied by great variations in the proteome. Still, it is conceivable that this variation can give rise to cohabitating organisms that differ in phenotype. This might be significant for the outcome of infection. It also poses a technical challenge in evaluating the virulence potential of a cultured bacterial population that is recovered from an infected individual at a particular point during the course of infection. If the net virulence of the population, determined by the relative ratio of more- and less-virulent subclones, shifts to a more-virulent composition, the host-microbial relationship could skew towards pathogenesis.
Recent studies have focused on the mechanisms underlying this genomic diversity (30, 31). The mutation frequency in H. pylori is unusually high (∼10−7) (3a, 43). H. pylori seems to be a panmictic species, i.e., recombination is frequent, and in addition, selective sweeps where only a subset (the most-fit variants) of the population survives are rare. Consequently, H. pylori lacks sequential bottlenecks that purify the population (1, 23, 32, 39).
One bacterial factor that has been ascribed particular significance regarding the outcome of infection is the Cag pathogenicity island (PAI) (10). The 38.5-kb Cag PAI contains 27 genes (6). Some of its genes encode a multiprotein type IV secretion system that translocates microbial compounds to host cells (6, 33, 36, 38). Inactivation of the type IV secretion machinery, by disruption of the cagE gene, produced a marked reduction in pathogenic potential in a Mongolian gerbil model (34). The cagA gene encodes a protein that, like the intimin receptor (Tir) of enteropathogenic Escherichia coli, is transferred and tyrosine phosphorylated in host cells (29, 33, 36, 38). The CagA protein has been associated with a more aggressive course of infection (6, 10).
These and other findings imply that bacteria that carry the Cag PAI are more virulent and more likely to drive the host-microbial relationship towards pathogenesis (7, 9, 10, 37). Identical 31-bp sequences located at both ends of the Cag PAI facilitate incorporation and deletion of this fragment (6). The ratio of Cag PAI-positive to -negative isolates could change by clonal expansion of the variant that best fits the requirements for survival in a given host's niche at a given time during the course of infection (19). This, in turn, could affect the overall virulence of the colonizing population in a host (7). The 31-bp repeats can also serve to introduce the Cag PAI into previously Cag PAI-deficient strains in a multistrain infection with Cag PAI-positive and -negative bacteria (30).
The emergence of divergent clones within a single individual is unlikely to be conferred solely by the PAI, but rather it is likely to reflect general plasticity of the whole genome (35). A recent case control study provided some support for this hypothesis (17). In an attempt to describe the clonal heterogeneity in the H. pylori population within an individual, antral biopsy samples were collected from patients referred for endoscopy for their upper gastrointestinal tract symptoms (16). Colonies from primary cultures were analyzed by AP-PCR (17). The results revealed the existence of subclones within individual patients. These subclones differed based on the presence or absence of the cagA gene but had identical genomic fingerprints.
In this study, we have characterized two such H. pylori subclones obtained from a single patient using DNA microarray-based whole-genome genotyping and an experimental transgenic mouse model. The results reveal substantial genotypic as well as phenotypic differences.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
H. pylori 67:20 (Cag PAI negative) and 67:21 (Cag PAI positive) were isolated as single colonies from the primary culture of two pooled antral biopsy samples obtained from a 90-year-old Swedish female with gastric ulcer. Hp1 is a previously described clinical isolate obtained from a Peruvian female with gastritis (25). The cultures were stored at −70°C until analyzed.
Bacteria were grown on Colombia II agar base BBL plates (Becton-Dickinson, Cockeysville, Md.) containing 8.5% horse blood and 10% horse serum. Cultures were incubated at 37°C under microaerophilic conditions (5% O2, 10% CO2, 85% N2).
For inoculation into mice, bacteria were grown in Brucella broth supplemented with 5% fetal calf serum and 1% IsoVitaleX (Becton-Dickinson). Bacteria from infected mice were reisolated by homogenizing half of the stomach from each animal and plating 100-μl aliquots of the homogenates on selective blood agar plates containing Skirrow's supplement (vancomycin, polymyxin B, trimethoprim; Oxoid, Basingstoke, United Kingdom).
Defining the genotypes of subclones 67:20 and 67:21. (i) Isolation of genomic DNA.
Genomic DNA from single colonies was prepared with the respiratory sample preparation kit (Roche Diagnostic Systems, Branchburg, N.J.) as described in a previous study (17). DNA for sequence and microarray analyses was prepared using the QIAamp tissue kit (Qiagen GmbH, Hilden, Germany).
(ii) Fingerprinting.
RFLP analysis of the flaA gene was performed to verify the strain identities of the two primary colonies according to the method of Enroth et al. (15). The amplified flaA fragment was digested with AluI, DpnII and HhaI, and the products were analyzed on a 1% agarose gel.
(iii) PCR.
A PCR assay was used to distinguish an intact from an absent Cag PAI. The reaction mixture contained PCR buffer (Boehringer Mannheim GmbH, Mannheim, Germany), 100 μM concentrations of deoxynucleoside triphosphates, 0.5 U of DNA polymerase (Boehringer Mannheim), 7 μM concentrations of primers UpCagF (ACT TTC ACG CCC TTT CCC TCC) and DownCagR (TTG CAT GCG TTA TTA TTT CAC), and H. pylori genomic DNA. The cycling conditions were denaturation at 94°C for 1 min, annealing at 50°C for 1 min, and elongation at 72°C for 1 min for 30 cycles followed by a final elongation step of 7 min at 72°C. This PCR assay generates a 562-bp PCR product if the PAI is absent and no product if the PAI is present.
(iv) Whole-genome genotyping using DNA microarrays.
The two primary isolates, 67:20 and 67:21, were genotyped according to the method of Salama et al. (35). Each DNA microarray contained duplicate copies of a set of 1,660 PCR products from the two fully sequenced strains, TIGR 26695 and Astra J99 (3, 42). These PCR products represent 98.9% of the open reading frames (ORFs) in the 26695 and J99 genomes. Ninety-one ORFs were unique to J99 (26695 was arbitrarily set as the reference strain). Hp1, an isolate previously genotyped using these DNA microarrays (35), was used as a reference control.
Genomic DNA (2 μg) from either the 67:20 or 67:21 isolate was labeled with Cy5. Genomic DNA (1 μg) from each of the two control strains (26695, J99) was labeled with Cy3. The Cy3-labeled control DNA was then cohybridized with the Cy5-labeled test strain DNA using previously published protocols (35). All hybridizations were performed in duplicate. Data from duplicate datasets were merged and analyzed according to the method of Salama et al. (35). Previous control experiments using Cy3-26695 or Cy3-J99 DNA alone had established that the hybridization conditions employed for the microarray analysis result in 0% false positives with the 26695 probe and 7% false positives with the J99 probe. The false-negative rates are 1 and 2% for the 26695 and J99 probes, respectively (35). After hybridization and washing, microarrays were scanned on the red and green channels, and the red/green ratio for each gene was normalized as outlined in the information available at http://genome-www4.stanford.edu/Microarray/help/results_normalization.html. If the normalized red/green ratio for a gene was less than 0.5, it was called absent in the 67:20 or 67:21 strain.
Studies using gnotobiotic transgenic mice.
FVB/N transgenic mice, expressing the human α-1,3/4-fucosyltransferase gene under the control of transcriptional regulatory elements from a Fabp gene, have been described in earlier reports (18, 25). These mice produce the human histo-blood group antigen Lewisb (Leb) (Fucα1,2Galβ1,3[Fucα1,4]GlcNAcβ) in their gastric pit and surface mucus cells. This fucosylated epitope serves as a receptor for H. pylori adhesins (27).
Mice raised under conventional conditions harbor a large and complex gastric microflora. To evaluate the capacity of 67:20 and 67:21 to compete with this flora, we compared the colonization of 5- to 8-week-old conventionally raised and germ-free Leb mice. One or the other isolate (107 CFU) was introduced into the stomachs of mice by oral administration on days 1 and 7 of an experiment. Animals were deprived of food and water for 2 h prior to each inoculation. A total of 40 conventionally raised and 14 germ-free mice were inoculated with the Cag PAI-negative isolate (67:20). Thirty-nine conventionally raised and 12 germ-free mice were treated with the Cag PAI-positive isolate (67:21). Experiments were performed on two different occasions.
Animals were sacrificed 12 weeks after inoculation. Half of the stomach from each mouse was taken for quantitation of the number of viable bacteria (in CFU). The other half of the stomach was fixed in formalin and embedded in paraffin, and serial 5-μm-thick sections were prepared. Sections were stained with hematoxylin and eosin, periodic acid-Schiff, or Alcian blue and scored for histopathologic changes by a pathologist (C.R.) in a single-blinded fashion (uninfected animals were used as reference controls).
Genotyping colonies recovered from the stomachs of colonized mice.
To assess the stability of the genome in vivo, we investigated the original primary isolates from the human host and organisms recovered from infected mice. To verify the cagA status in the reisolated H. pylori isolate 67:21 after experimental infection, DNA was prepared from 1 to 16 primary colonies recovered from each mouse. In total, 332 colonies were analyzed by PCR according to previously described methods (17).
PCR amplifications of the recA (forward primer, 5′-GAA ATT TAT GGG CAG AGT C; reverse primer, 5′-GAT AAA AAT GAG AGT GGT GTT) (41), cagA (forward primer, 5′-TTG GAA ACC ACC TTT TGT ATT AGC; reverse primer, 5′-GTG CCT GCT AGT TTG TCA GCG) (17), and 16S rRNA (forward primer, 5′-TGG CAA TCA GCG TCA GGT AAT G; reverse primer, 5′-GCT AAG AGA TCA GCC TAT GTC C) (14) genes were performed according to standard procedures. Sequence analysis of these genes was also performed on the primary isolates and on 67:21 recovered from conventionally raised mice infected for 3 or 10 months (n = 2 mice/time point and 5 colonies per animal). The same genes were also sequenced from (i) five reisolates of 67:20 (except cagA) from ex-germ-free mice, (ii) five reisolates of 67:21 from ex-germ-free animals, and (iii) the two primary subclones after 50 rounds of in vitro passage.
To identify genetic variation in the Cag PAI, we analyzed DNA using a custom microarray containing PCR products from each gene in the Cag PAI (A. Sillén, C. Nilsson, H. Enroth, P. Falk, and L. Engstrand, submitted for publication). These microarrays were used to assay 20 reisolated 67:21 colonies from a conventionally raised mouse infected for 3 months, 20 colonies from an ex-germ-free animal infected for 3 months, and 10 colonies from a conventionally raised animal infected for 10 months.
RESULTS AND DISCUSSION
Genotyping studies indicate that 67:20 and 67:21 are subclones of the same strain.
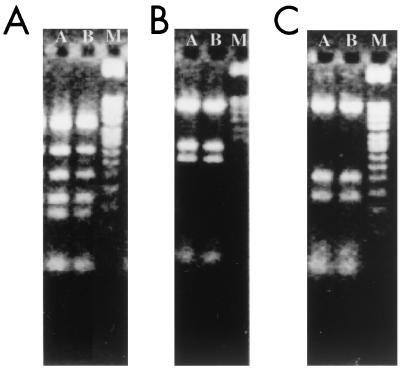
We used a stepwise approach to compare, with increasing comprehensiveness, the genomes of two isolates recovered from one H. pylori-infected patient. RFLP analysis of flaA provides one way of distinguishing distinct H. pylori strains (15, 20). RFLP analysis yielded identical patterns for the 67:20 and 67:21 isolates (Fig. 1). This finding was consistent with the results of an earlier AP-PCR analysis of these two isolates (17). In addition, analysis of the PCR fragments, representing 318 bp of the recA ORF and 396 bp of the 16S rRNA gene from the primary 67:20 and 67:21 clinical isolates, revealed that they had identical nucleotide sequences.
FIG. 1.
RFLP genotyping of the flaA gene from two H. pylori isolates, 67:20 and 67:21, obtained from a single patient. Products were obtained by digestion with AluI (A), DpnII (B), or HhaI (C). Lanes: A, isolate 67:20; B, isolate 67:21; M, 100-bp ladder.
PCR assays using primers from the two genes that flank the Cag PAI in the two fully sequenced 26695 and J99 genomes (HP0519, HP0549; see Materials and Methods) indicated that the entire PAI was deleted from 67:20 but present in 67:21 (data not shown). Genotyping studies using a custom DNA microarray containing PCR fragments from each ORF present in the Cag PAI established that all of the 27 ORFs were represented in the 67:21 genome, whereas none were detectable in the 67:20 genome.
Sequencing the 350 bp spanning the region of the excised Cag PAI in the 67:20 genome disclosed that the excision probably occurred by homologous recombination at the 31-bp direct repeats positioned at each end of the PAI. 67:20 contains both HP0519 and HP0549, and there is one copy of the 31-bp sequence at the junction between these two genes located at the 3′ end of HP0549 (glutamate racemase, glr).
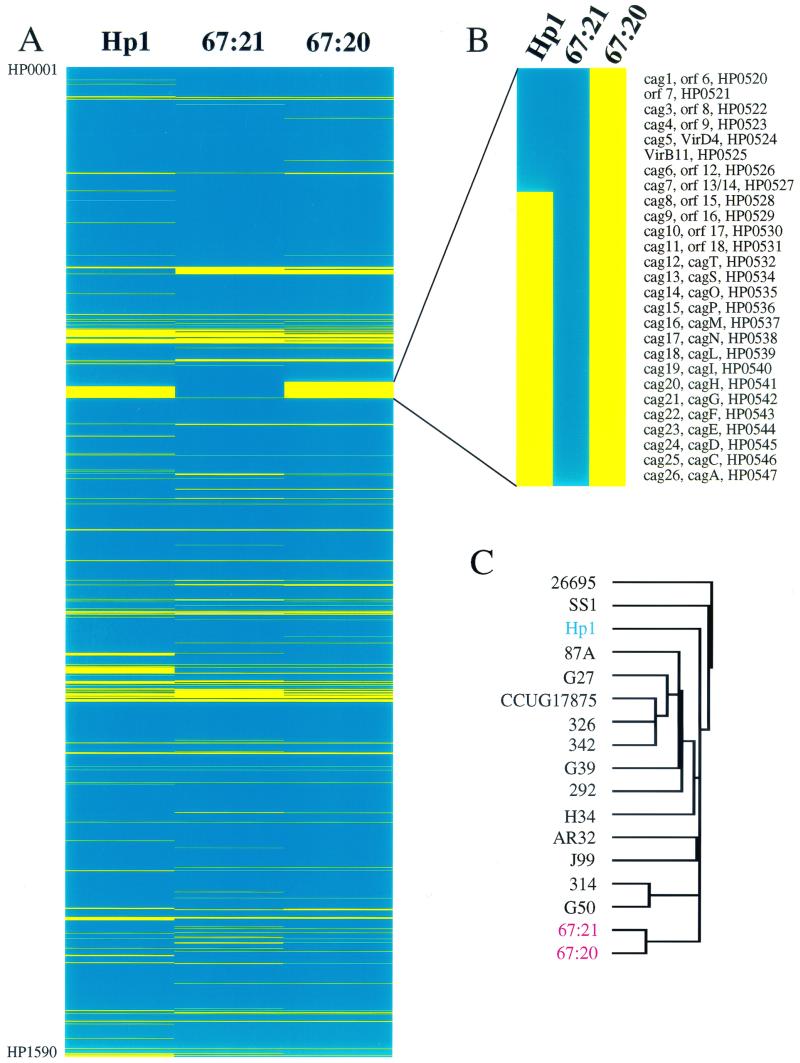
DNA microarray analysis of the whole H. pylori genome using 1,660 PCR products representing 98.9% of known bacterial ORFs indicated that a total of 133 genes from the 26695 and J99 strains were undetectable in both the 67:20 and 67:21 isolates (Fig. 2A). In addition, 67:21 lacked 29 genes that were present in 67:20, whereas 67:21 contained 37 genes missing in 67:20 (these 37 genes include 27 ORFs from the Cag PAI; Fig. 2B).
FIG. 2.
Cluster analysis of H. pylori strains subjected to whole-genome genotyping with DNA microarrays. (A) TreeView display of the results of complete genome genotyping of strain Hp1 and isolates 67:20 and 67:21. Blue color indicates the presence of a gene while yellow indicates its absence. Genes are displayed in the order of their appearance along the reference TIGR strain 26695 genome, i.e., the top of the figure presents gene HP0001 and the bottom of the figure shows gene HP1590, the last gene of strain 26695. (B) Enlargement of the TreeView display of the Cag PAI region. (C) Dendrogram of strains genotyped by Salama et al. (35) and in this study. Cluster analysis was performed using an average hierarchical clustering program (Cluster) and the strain-specific genes. The Cag PAI was excluded from the analysis to avoid skewing the results. The results, displayed using TreeView (http://www.microarrays.org/software.html), indicate that isolates 67:20 and 67:21 are more closely related to each other than to any other strain.
The identified genotypic differences between the two isolates are summarized in Table 1. Among the genes present in 67:21 and absent in 67:20 are (i) six genes without known homologies or functions, (ii) HP1352 (a methyltransferase component of one of the organism's many type II restriction-modification systems), (iii) aroK (the shikimate kinase I involved in the synthesis of aromatic amino acids), (iv) gmhA (phosphoheptose isomerase involved in synthesis of lipopolysaccharide), and (v) vapD (a gene with homology to a virulence-associated protein produced by the ovine pathogen Dichelobacter nodosus) (5, 11).
TABLE 1.
Summary of genotypic differences between the 67:20 and 67:21 subclones recovered from a 90-year-old patient with gastric ulcer
| Gene no.a | Gene nameb | Gene description | Annotation | Presence in isolatec:
|
|
|---|---|---|---|---|---|
| 67:20 | 67:21 | ||||
| HP0157 | aroK | Shikimic acid kinase I | General | − | + |
| HP0934 | NA | Conserved hypothetical protein | Hypothetical general | − | + |
| HP0315 | vapD | Virulence-associated protein D | Adaptations and atypical conditions | − | + |
| HP0520 to HP0547 | Cag PAI | Cag PAI proteins | Cag PAI | − | + |
| HP0857 | gmhA | Phosphoheptose isomerase | Cell envelope, biosynthesis of surface polysaccharides and lipopolysaccharides | − | + |
| HP1352 | HINFIM | Adenine-specific methyltransferase, type II | Restriction modification | − | + |
| HP0882, HP0085, JHP0165, JHP0956, HP0425 | Unknown | − | + | ||
| HP0491 | rpL28, rpmB | Ribosomal protein L28 | General | + | − |
| HP0962 | acpP | Acyl carrier protein | General | + | − |
| HP1192 | NA | Secreted protein involved in flagellar motility | General | + | − |
| JHP0726 | hsdS_4 | Putative type I restriction modification system, specificity subunit | Restriction modification | + | − |
| HP1382 | NA | Putative endonuclease | Restriction modification | + | − |
| HP1404 | hsdS | Type I restriction modification system | Restriction modification | + | − |
| HP0062, HP0338, JHP0959, HP0436, HP0450, HP0673, HP0681, HP0712, HP0849, HP0984, JHP0950, HP0999, HP1074, JHP0332, HP1250, HP1324, HP1396, HP1397, HP1405, HP1412, HP1439, JHP1408, HP1586 | Unknown | + | − | ||
HP numbers correspond to the numbers used by The Institute for Genomic Research for strain 26695, and JHP numbers correspond to those assigned by AstraZeneca for strain J99. For genes present in both strains, only the HP number is given.
Gene names and descriptions are according to those provided by The Institute for Genomic Research and Salama et al. (35). NA, not available.
+, gene was present; −, gene was undetectable.
Comparisons of the genotype of Hp1 with the genotypes of 67:20 and 67:21 disclosed 136 and 107 differences, respectively. Cluster analysis, carried out on 67:20, 67:21, Hp1, and 14 other strains that had been subjected to whole-genome genotyping with these microarrays in another study (35), disclosed that 67:20 and 67:21 are more closely related to each other than to any other of the genotyped strains (Fig. 2C).
Differences in gene content between these two isolates could be due to either the loss of genes or to the acquisition of new genes (30). AP-PCR studies of 10 colonies recovered from the 90-year-old patient and RFLP studies of the flaA gene in 24 additional colonies failed to reveal genomic fingerprints other than those of 67:20 and 67:21 (17; data not shown). Based on this previously reported finding and the results of the RFLP, DNA microarray genotyping, and sequence analyses described above, we concluded that 67:20 and 67:21 represent subclones rather than two distinct strains.
Phenotypic divergence manifested by differences in the ability of the subclones to colonize conventionally raised transgenic mice with an epithelial glycan receptor for H. pylori adhesin(s).
To determine if the observed differences in the genotypes of the 67:20 and 67:21 isolates confer differences in their fitness, we investigated their ability to establish a niche in the stomachs of transgenic mice that are engineered to produce a receptor to a known H. pylori adhesin.
The ability of H. pylori to attach to the gastric epithelium represents the coincidence of host and microbial factors: the capacity of a colonizing strain to produce adhesins recognized by gastric epithelial receptors and the ability of the host to express these receptors prior to or during the course of infection. A majority of clinical isolates of H. pylori appear to be able to bind to the human histo-blood group antigen Leb (27). Approximately 70% of humans produce Leb-containing glycans in their gastric epithelia, where expression is restricted to the mucus-producing pit cell lineage (4, 27). Forced expression of the human α-1,3/4 fucosyltransferase in the pit cell lineage of neonatal and adult transgenic mice belonging to the FVB/N strain allows the production of Leb-containing glycans (18).
In vitro binding assays (25) disclosed that both 67:20 and 67:21 bound equally well to Leb-expressing pit cells present in sections of transgenic mouse stomach. No binding was detectable when the same organisms were incubated with stomach sections prepared from Leb-negative nontransgenic littermates (data not shown). These latter results indicate that the observed differences in the genotypes of the subclones do not correlate with their capacity to express adhesins that recognize these Leb glycan receptors.
The efficiency of colonization of conventionally raised adult Leb transgenic mice with the Cag PAI-positive 67:21 isolate was 74% (29 of 39 animals sacrificed 3 months after inoculation). This efficiency is similar to that observed with the unrelated Hp1 isolate, containing the first 8 genes of the Cag PAI (HP0520 to HP0527) (25). Ten months after inoculation, 6 of 9 conventionally raised animals were colonized with 67:21. In contrast, the Cag PAI-negative 67:20 subclone failed to colonize any conventionally raised transgenic mice (0 of 40 animals surveyed 3 months following inoculation).
To define colonization potential in the absence of a competing indigenous microflora, 67:20 or 67:21 was introduced into the stomachs of adult germ-free Leb transgenic mice. After 3 months, 100% (12 of 12) of the gnotobiotic transgenic mice were colonized with the 67:21 isolate while 85% (12 of 14) of mice were colonized with the Cag PAI-negative 67:20 isolate. There were no statistically significant differences in the densities of colonization between mice harboring the different subclones (range = 104 to 106 CFU/stomach).
67:20 and 67:21 subclones: a model strain pair for testing the role of Cag PAI in inducing inflammation.
The 67:20 and 67:21 subclones provide an important opportunity to assess the role of the Cag PAI in inducing inflammation given the presence of the PAI in the one clone, its complete absence in the other, and the relatively high degree of genotypic similarities throughout the rest of their genomes. The ability to induce interleukin-8 (IL-8) production in cultured AGS gastric epithelial cells (8) was only seen in the 67:21 isolate (data not shown). This finding was not unexpected given that IL-8 induction in this cell line has been clearly associated with the Cag PAI (8, 9, 37). Surprisingly, a single-blind study failed to reveal any discernible differences in the histopathologic changes produced in the stomachs of ex-germ-free mice after a 3-month colonization with either the Cag PAI-negative 67:20 or Cag PAI-positive 67:21 subclone (n = 26 animals).
Other groups have reported that cagE inactivation of an already gerbil-passaged strain had a minor effect on the colonization of Mongolian gerbils, although IL-8 production and inflammation were diminished to the level obtained with Cag PAI-deficient strains (28, 34). However, when using a piglet-adapted 26695 strain in gnotobiotic piglets, or the well-established mouse-adapted strain SS1 in C57BL/6 mice, Eaton and coworkers noted that there was no effect of inactivation of the Cag PAI on the ability to colonize or to induce inflammation (12). Our findings in gnotobiotic Leb transgenic mice are consistent with these latter findings. In addition, studies in conventionally raised Leb transgenic mice support the notion that elements of the Cag PAI may help H. pylori to compete with other microbial species in patients that harbor a complex indigenous gastric flora (e.g., as in chronic atrophic gastritis).
Gnotobiotic transgenic mice reveal that the subclones exhibit a high degree of genetic stability.
The rate at which subclones emerge during the course of infection with H. pylori is not known. Studies of humans suggest that genetic drift can occur over the course of several years (31). To examine the time frame during which changes may occur in an individual subclone (67:21), we examined single colonies recovered after a 3- or 10-month infection of Leb transgenic mice (n = 140 colonies from conventionally raised animals infected for 3 months, n = 152 colonies from conventionally raised animals infected for 10 months, and n = 40 colonies from ex-germ-free animals mono-associated for 3 months). PCR studies revealed that all reisolates remained positive for cagA.
Since partial deletions in the Cag PAI would not be detected by the cagA PCR assay, we proceeded to analyze 67:21 colonies reisolated from one ex-germ-free mouse infected for 3 months, one conventionally raised mouse infected for 3 months, and one conventionally raised mouse infected for 10 months. DNA microarrays containing all of the Cag PAI genes were employed for this survey of 20 colonies from the first two experimental conditions and 10 colonies from the long-term infection. All reisolates contained all the genes of the Cag PAI, just like the primary 67:21 clinical isolate.
PCR and DNA microarray analyses can detect major genomic deletions. A large portion of the genetic diversity of H. pylori arises from single-base mutations (43). Therefore, we compared the nucleotide sequences of three genes in five reisolates per experimental condition. After passage in conventionally raised and ex-germ-free mice for 3 months or in conventionally raised mice for 10 months, no nucleotide substitutions were detected in cagA (only 67:21), recA and 16S rRNA genes in any of the reisolates. The same was true when the 67:21 isolate was passaged 50 times in vitro. (See Materials and Methods for a list of the primers used to generate the 322-bp cagA, 318-bp recA, and 396-bp 16S rRNA gene fragments used for this sequence analysis.)
It is possible that there is less selective pressure exerted on H. pylori for genetic divergence in the environment of the germ-free or conventionally raised mouse stomach than in the human stomach. Alternatively, the 67:20 and 67:21 subclones may be derived from a parent whose genotype (e.g., repertoire of restriction modification and DNA repair enzymes) favors a relatively slow rate of genetic evolution compared to other strains.
It has been postulated that in humans, there may be clonal expansion of Cag PAI-positive and -negative subclones from the same colonizing strain within the same host (7, 17). This presumably occurs in an ecosystem with few competing microbial species as long as acid production is maintained. Therefore, it is interesting to note that in conventionally raised Leb transgenic mice with dense indigenous gastric microbial populations, there were no detectable Cag PAI-negative clones after 10 months of infection with the Cag PAI-positive isolate. This might indicate selection of Cag PAI-positive clones in an environment where H. pylori benefits from a competitive edge against other microorganisms, or that the 3- to 10-month time frame of the colonization experiments is simply too short for any discernible genetic changes to occur.
Prospectus.
We have determined that two H. pylori subclones, recovered from a 90-year-old human with gastric ulcer disease, have undergone divergent genetic and phenotypic evolution since the time of their separation. Our findings lend support to the concept that clonal expansion of more-fit variants may provide a means for the bacterial population to coevolve with its host and adapt to changes in the gastric niche. These studies emphasize the importance of initiating longitudinal studies of H. pylori-infected humans over an extended time scale (decades) so that subspecies development can be examined using tools, such as DNA microarrays, that are emerging from the current revolution in genomics and proteomics.
ACKNOWLEDGMENTS
We thank Margareta Rodensjö, Gunilla Bergo, Lena Ericsson, and Ewa Österlund for technical assistance. We are grateful to Stanley Falkow and Tore Midtvedt for allowing us to conduct parts of this study in their labs.
This work was supported by grants from the Swedish Cancer Society, the Swedish Research Council, the Swedish Foundation for Strategic Research, and the National Institutes of Health.
REFERENCES
- 1.Achtman M, Azuma T, Berg D, Ito Y, Morelli G, Pan Z, Suerbaum S, Thompson S, van der Ende A, van Doorn L. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol Microbiol. 1999;32:459–470. doi: 10.1046/j.1365-2958.1999.01382.x. [DOI] [PubMed] [Google Scholar]
- 2.Akopyants N, Bukanov N, Westblom T, Kresovich S, Berg D. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 1992;20:5137–5142. doi: 10.1093/nar/20.19.5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alm R, Ling L, Moir D, King B, Brown E, Doig P, Smith D, Noonan B, Guild B, de Jonge B, Carmel G, Tummino P, Caruso A, Uria-Nickelsen M, Mills D, Ives C, Gibson R, Merberg D, Mills S, Jiang Q, Taylor D, Vovis G, Trust T. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 3a.Björkholm, B., M. Sjölund, P. G. Falk, O. G. Berg, L. Engstrand, and D. I. Andersson. Mutation frequency and biological cost of antibiotic resistance in Helicobacter pylori. Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 4.Boren T, Falk P, Roth K, Larson G, Normark S. Attachment of Helicobacter pylori to human gastric epithelium mediated by blood group antigens. Science. 1993;262:1892–1895. doi: 10.1126/science.8018146. [DOI] [PubMed] [Google Scholar]
- 5.Cao P, Cover T L. High-level genetic diversity in the vapD chromosomal region of Helicobacter pylori. J Bacteriol. 1997;179:2852–2856. doi: 10.1128/jb.179.9.2852-2856.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Censini S, Lange C, Xiang Z, Crabtree J, Ghiara P, Borodovsky M, Rappuoli R, Covacci A. Cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci USA. 1996;93:14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Covacci A, Rappuoli R. Helicobacter pylori: molecular evolution of a bacterial quasi-species. Curr Opin Microbiol. 1998;1:96–102. doi: 10.1016/s1369-5274(98)80148-3. [DOI] [PubMed] [Google Scholar]
- 8.Crabtree J, Covacci A, Farmery S, Xiang Z, Tompkins D, Perry S, Lindley I, Rappuoli R. Helicobacter pylori induced interleukin-8 expression in gastric epithelial cells is associated with CagA positive phenotype. J Clin Pathol. 1995;48:41–45. doi: 10.1136/jcp.48.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crabtree J, Kersulyte D, Li S, Lindley I, Berg D. Modulation of Helicobacter pylori induced interleukin-8 synthesis in gastric epithelial cells mediated by cag PAI encoded VirD4 homologue. J Clin Pathol. 1999;52:653–657. doi: 10.1136/jcp.52.9.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crabtree J, Taylor J, Wyatt J, Heatley R, Shallcross T, Tompkins D, Rathbone B. Mucosal IgA recognition of Helicobacter pylori 120 kDa protein, peptic ulceration, and gastric pathology. Lancet. 1991;388:332–335. doi: 10.1016/0140-6736(91)90477-7. [DOI] [PubMed] [Google Scholar]
- 11.Dundon W, Marshall D, O'Morain C, Smyth C. Population characteristics of Irish Helicobacter pylori isolates: a tRNA-associated locus. Ir J Med Sci. 2000;169:137–140. doi: 10.1007/BF03166919. [DOI] [PubMed] [Google Scholar]
- 12.Eaton K A, Kersulyte D, Mefford M, Danon S J, Krakowka S, Berg D E. Role of Helicobacter pylori cag region genes in colonization and gastritis in two animal models. Infect Immun. 2001;69:2902–2908. doi: 10.1128/IAI.69.5.2902-2908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El-Omar E, Carrington M, Chow W, McColl K, Bream J, Young H, Herrera J, Lissowska J, Yuan C, Rothman N, Lanyon G, Martin M, Fraumeni J, Rabkin C. Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature. 2000;404:398–402. doi: 10.1038/35006081. [DOI] [PubMed] [Google Scholar]
- 14.Engstrand L, Nguyen A-M H, Graham D Y, El-Zaatari F A K. Reverse transcription and polymerase chain reaction amplification of rRNA for detection of Helicobacter species. J Clin Microbiol. 1992;30:2295–2301. doi: 10.1128/jcm.30.9.2295-2301.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enroth H, Björkholm B, Engstrand L. Occurrence of resistance mutation and clonal expansion in Helicobacter pylori multiple-strain infection: a potential risk in clarithromycin-based therapy. Clin Infect Dis. 1999;28:1305–1307. doi: 10.1086/514796. [DOI] [PubMed] [Google Scholar]
- 16.Enroth H, Kraaz W, Engstrand L, Nyren O, Rohan T. Helicobacter pylori strain types and risk of gastric cancer: a case-control study. Cancer Epidemiol Biomark Prev. 2000;9:981–985. [PubMed] [Google Scholar]
- 17.Enroth H, Nyren O, Engstrand L. One stomach—one strain: does Helicobacter pylori strain variation influence disease outcome? Dig Dis Sci. 1999;44:102–107. doi: 10.1023/a:1026658301825. [DOI] [PubMed] [Google Scholar]
- 18.Falk P, Bry L, Holgersson J, Gordon J. Expression of a human α-1,3/4-fucosyltransferase in the pit cell lineage of FVB/N mouse stomach results in production of Leb-containing glycoconjugates: a potential transgenic mouse model for studying Helicobacter pylori infection. Proc Natl Acad Sci USA. 1995;92:1515–1519. doi: 10.1073/pnas.92.5.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Figura N, Vindigni C, Covacci A, Presenti L, Burroni D, Vernillo R, Banducci T, Roviello F, Marrelli D, Biscontri M, Kristodhullu S, Gennari C, Vaira D. cagA positive and negative Helicobacter pylori strains are simultaneously present in the stomach of most patients with non-ulcer dyspepsia: relevance to histological damage. Gut. 1998;42:772–778. doi: 10.1136/gut.42.6.772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forbes K, Fang Z, Pennington T. Allelic variation in the Helicobacter pylori flagellin genes flaA and flaB: its consequences for strain typing schemes and population structure. Epidemiol Infect. 1995;114:257–266. doi: 10.1017/s0950268800057927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fox J, Beck P, Dangler C, Whary M, Wang T, Shi H, Nagler-Anderson C. Concurrent enteric helminth infection modulates inflammation and gastric immune responses and reduces Helicobacter-induced gastric atrophy. Nat Med. 2000;6:536–542. doi: 10.1038/75015. [DOI] [PubMed] [Google Scholar]
- 22.Gibson J R, Slater E, Xerry J, Tompkins D S, Owen R J. Use of an amplified-fragment length polymorphism technique to fingerprint and differentiate isolates of Helicobacter pylori. J Clin Microbiol. 1998;36:2580–2585. doi: 10.1128/jcm.36.9.2580-2585.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Göttke M, Fallone C, Barkun A, Vogt K, Loo V, Trautmann M, Tong J, Nguyen T, Fainsilber T, Hahn H, Korber J, Lowe A, Beech R. Genetic variability determinants of Helicobacter pylori: influence of clinical background and geographic origin of isolates. J Infect Dis. 2000;181:1674–1681. doi: 10.1086/315425. [DOI] [PubMed] [Google Scholar]
- 24.Graham D. Helicobacter pylori infection in the pathogenesis of duodenal ulcer and gastric cancer: a model. Gastroenterology. 1997;113:1983–1991. doi: 10.1016/s0016-5085(97)70019-2. [DOI] [PubMed] [Google Scholar]
- 25.Guruge J, Falk P, Lorenz R, Dans M, Wirth H, Blaser M, Berg D, Gordon J. Epithelial attachment alters the outcome of Helicobacter pylori infection. Proc Natl Acad Sci USA. 1998;95:3925–3930. doi: 10.1073/pnas.95.7.3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han S-R, Zschausch H-C E, Meyer H G W, Schneider T, Loos M, Bhakdi S, Maeurer M J. Helicobacter pylori: clonal population structure and restricted transmission within families revealed by molecular typing. J Clin Microbiol. 2000;38:3646–3651. doi: 10.1128/jcm.38.10.3646-3651.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ilver D, Arnqvist A, Ogren J, Frick I, Kersulyte D, Incecik E, Berg D, Covacci A, Engstrand L, Boren T. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science. 1998;279:373–377. doi: 10.1126/science.279.5349.373. [DOI] [PubMed] [Google Scholar]
- 28.Israel D, Salama N, Arnold C, Moss S, Ando T, Wirth H, Tham K, Camorlinga M, Blaser M, Falkow S, Peek R J. Helicobacter pylori strain-specific differences in genetic content, identified by microarray, influence host inflammatory responses. J Clin Investig. 2001;107:611–620. doi: 10.1172/JCI11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kenny B, DeVinney R, Stein M, Reinscheid D, Frey E, Finlay B. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell. 1997;91:511–520. doi: 10.1016/s0092-8674(00)80437-7. [DOI] [PubMed] [Google Scholar]
- 30.Kersulyte D, Chalkauskas H, Berg D. Emergence of recombinant strains of Helicobacter pylori during human infection. Mol Microbiol. 1999;31:31–43. doi: 10.1046/j.1365-2958.1999.01140.x. [DOI] [PubMed] [Google Scholar]
- 31.Kuipers E, Israel D, Kusters J, Gerrits M, Weel J, van der Ende A, van der Hulst R, Wirth H, Hook-Nikanne J, Thompson S, Blaser M. Quasispecies development of Helicobacter pylori observed in paired isolates obtained years apart from the same host. J Infect Dis. 2000;181:273–282. doi: 10.1086/315173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lafay B, Atherton J, Sharp P. Absence of translationally selected synonymous codon usage bias in Helicobacter pylori. Microbiology. 2000;146:851–60. doi: 10.1099/00221287-146-4-851. [DOI] [PubMed] [Google Scholar]
- 33.Odenbreit S, Puls J, Sedlmaier B, Gerland E, Fischer W, Haas R. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science. 2000;287:1497–1500. doi: 10.1126/science.287.5457.1497. [DOI] [PubMed] [Google Scholar]
- 34.Ogura K, Maeda S, Nakao M, Watanabe T, Tada M, Kyutoku T, Yoshida H, Shiratori Y, Omata M. Virulence factors of Helicobacter pylori responsible for gastric diseases in Mongolian gerbil. J Exp Med. 2000;192:1601–1610. doi: 10.1084/jem.192.11.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salama N, Guillemin K, McDaniel T, Sherlock G, Tompkins L, Falkow S. A whole genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc Natl Acad Sci USA. 2000;97:14668–14673. doi: 10.1073/pnas.97.26.14668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Segal E, Cha J, Lo J, Falkow S, Tompkins L. Altered states: involvement of phosphorylated CagA in the induction of host cellular growth changes by Helicobacter pylori. Proc Natl Acad Sci USA. 1999;96:14559–14564. doi: 10.1073/pnas.96.25.14559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma S A, Tummuru M K, Miller G G, Blaser M J. Interleukin-8 response of gastric epithelial cell lines to Helicobacter pylori stimulation in vitro. Infect Immun. 1995;63:1681–1687. doi: 10.1128/iai.63.5.1681-1687.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stein M, Rappuoli R, Covacci A. Tyrosine phosphorylation of the Helicobacter pylori CagA antigen after cag-driven host cell translocation. Proc Natl Acad Sci USA. 2000;97:1263–1268. doi: 10.1073/pnas.97.3.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suerbaum S, Achtman M. Evolution of Helicobacter pylori: the role of recombination. Trends Microbiol. 1999;7:182–184. doi: 10.1016/s0966-842x(99)01505-x. [DOI] [PubMed] [Google Scholar]
- 40.Suerbaum S, Smith J, Bapumia K, Morelli G, Smith N, Kunstmann E, Dyrek I, Achtman M. Free recombination within Helicobacter pylori. Proc Natl Acad Sci USA. 1998;95:12619–12624. doi: 10.1073/pnas.95.21.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson S A, Blaser M J. Isolation of the Helicobacter pylori recA gene and involvement of the recA region in resistance to low pH. Infect Immun. 1995;63:2185–2193. doi: 10.1128/iai.63.6.2185-2193.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomb J, White O, Kerlavage A, Clayton R, Sutton G, Fleischmann R, Ketchum K, Klenk H, Gill S, Dougherty B, Nelson K, Quackenbush J, Zhou L, Kirkness E, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H, Glodek A, McKenney K, Fitzegerald L, Lee N, Adams M, Venter J, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 43.Wang G, Humayun M, Taylor D. Mutation as an origin of genetic variability in Helicobacter pylori. Trends Microbiol. 1999;7:488–493. doi: 10.1016/s0966-842x(99)01632-7. [DOI] [PubMed] [Google Scholar]