Abstract
Adult mammalian skin wounds heal by forming fibrotic scars. We report that full-thickness injuries of reindeer antler skin (velvet) regenerate, whereas back skin forms fibrotic scar. Single-cell multi-omics reveal that uninjured velvet fibroblasts resemble human fetal fibroblasts, whereas back-skin fibroblasts express inflammatory mediators mimicking pro-fibrotic adult human and rodent fibroblasts. Consequently, injury elicits site-specific immune responses: back skin fibroblasts amplify myeloid infiltration and maturation during repair, whereas velvet fibroblasts adopt an immunosuppressive phenotype that restricts leukocyte recruitment and hastens immune resolution. Ectopic transplantation of velvet to scar-forming back skin is initially regenerative, but progressively transitions to a fibrotic phenotype akin to the scarless fetal-to-scar-forming transition reported in humans. Skin regeneration is diminished by intensifying, or enhanced by neutralizing, these pathologic fibroblast-immune interactions. Reindeer represent a powerful comparative model for interrogating divergent wound healing outcomes and our results nominate decoupling of fibroblast-immune interactions as a promising approach to mitigate scar.
Keywords: reindeer, wound healing, skin regeneration, fibroblast, stromal-immune crosstalk, immune modulation, inflammation, scar, fetal human fibroblast, myeloid cell maturation, single cell genomics
In Brief
The particular properties of reindeer antler velvet are leveraged here to identify which factors lead to scarring versus regenerative healing in mammalian skin, with relevance not only to reindeer, but also mouse and human systems.
Graphical Abstract
Introduction
In adult mammals, deep skin injuries heal by rapid repopulation with reactive fibroblasts that deposit extracellular matrix and form fibrotic scar tissue that impairs tissue function and diminishes quality of life1. Effective treatments to restore skin structure and function are lacking due to our limited understanding of mechanisms that prevent, or promote, skin regeneration. Although fetal mammals, including humans, exhibit scarless healing2–5, few adult mammals exhibit reparative regeneration6. Spiny mice (Acomys spp.) can regenerate skin and complex tissues7, while other rodents exhibit modest regeneration in certain injury contexts8–10. Unfortunately, rodent models fail to fully recapitulate adult human skin repair11 because of key differences in skin anatomy (e.g. absence of a subdermal panniculus carnosus muscle that enables wound contraction in rodents)12, immunologic composition11, genetics, and microbial/environmental13 influences. These shortcomings limit our ability to effectively translate therapeutic discoveries into human application.
Discovery of a tight-skinned, free-living mammal that exhibits both skin regeneration and fibrotic repair would enable comparative insights that better contextualizes human healing. Although complete tissue regeneration in adult mammals is rare, one exception is in reindeer (Rangifer tarandus), where antlers are regenerated annually in both sexes and grow at an explosive rate exceeding 1cm in length each day14,15. Growing antlers are covered by specialized skin called velvet, which is pigmented, highly innervated and vascularized, and richly embedded with hair follicles (HFs) and sebaceous glands16 (Figure S1A,B). Moreover, individual antler tines can regenerate following amputation17. Here, we hypothesized that antler velvet itself may harbor innate regenerative capacity and could represent a unique model to study the molecular events enabling adult skin regeneration.
Our results demonstrate that velvet undergoes remarkable regenerative repair following full-thickness injury, whereas identical injury in back skin heals by formation of scar. Comparative multi-omic analysis revealed that divergent fibroblast states act as key determinants of wound healing outcomes, via induction of developmental programming and distinct patterns of stromal immune signaling. The insights gained from this unique comparative model, made accessible through our interactive website (biernaskielab.ca/reindeer_atlas), provide exciting avenues for targeted regenerative medicine approaches toward improving outcomes after severe skin injury.
Results
Antler velvet exhibits regeneration after wounding
Identical 12-mm-diameter full-thickness excisional skin wounds were created in both dorsal back skin and antler velvet of adult reindeer (Figure 1A–C, S1C, E, F). At 30 days after excision injury, back skin wounds generated an irregular, often hypertrophic scar devoid of HFs and lacking pigmentation (Figure 1B, D, and S1E), reminiscent of human scar. Conversely, identical wounds in velvet lacked hypertrophic scarring and exhibited nearly full surface re-pigmentation with neogenic (newly-formed) HFs decorating the entire surface of the lesion (Figure 1C, E, F-H, and S1D–F). By 60 days post-wound (dpw), wounds were nearly indistinguishable from uninjured velvet (Figure 1C). Histological analysis confirmed the presence of neogenic HFs, embedded within a fully pigmented epidermis that followed an outside-in pattern of regeneration with the most immature HFs within the centre. Despite the appearance of epithelial invaginations within the back skin wounds, these structures failed to evolve into mature HFs (Figure 1D; Figure S1C, E). In contrast, neogenic HFs within the velvet all contained proliferating epithelial cells (Figure 1F; Figure S1C–F), engulfed versican-expressing dermal papillae (Figure 1G) and were associated with lipid-laden sebaceous glands (Figure 1H). Quantification of HFs at 30 dpw confirmed that HF density was restored to 73% of pre-injury levels in velvet, but limited to 12% in back skin (Figure 1I). Back skin wounds had significantly greater contraction than velvet wounds (Figure 1J) suggesting that velvet regeneration resulted from the production of new tissue rather than contraction of wound edges. The density and architectural arrangement of collagen fibers was evaluated in back skin and velvet at baseline and 30dpw (n=3 animal-matched back and velvet) (Figure 1K–M)18. There was deviation in ECM ultrastructure between uninjured versus healed back skin dermis, while healed velvet exhibited strong overlap with uninjured velvet (Figure 1L–M). Histological staining at 60dpw confirmed near-scarless healing in velvet wounds, but visible fibrosis and few appendages in back skin wounds (Figure 1N–Q). Taken together, HF neogenesis, re-pigmentation and matrix architecture restoration in reindeer antler velvet demonstrates bona fide skin regeneration.
Figure 1 -. Antler velvet regeneration following full thickness injury.
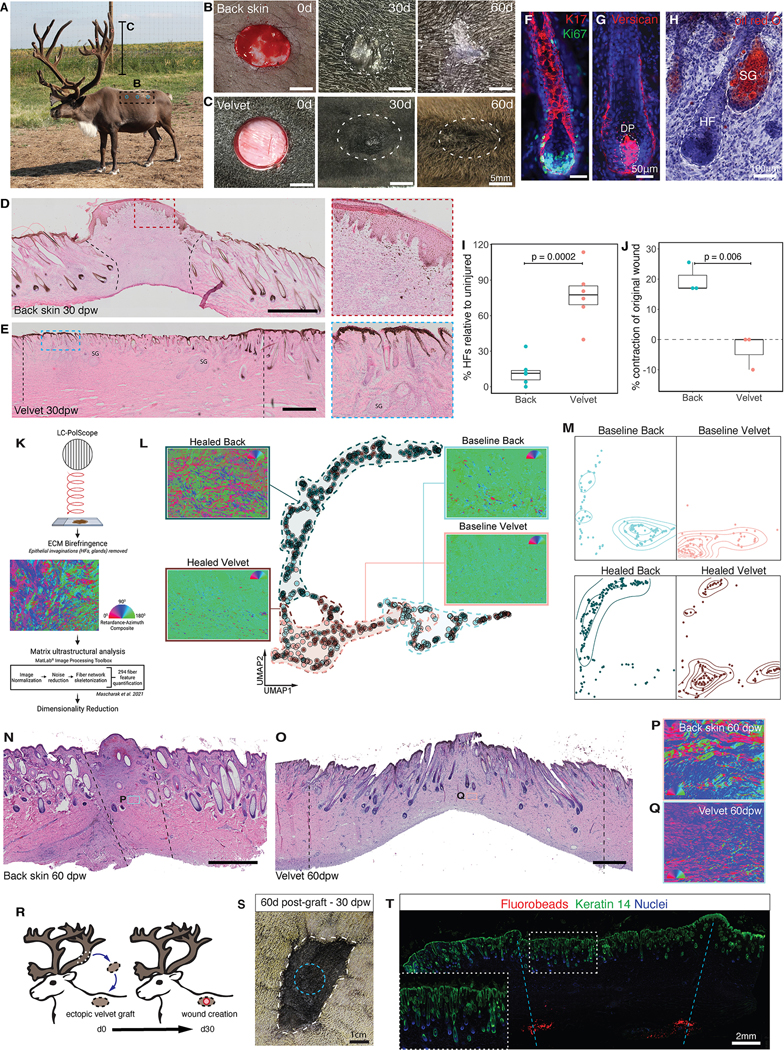
(A) Image of adult male reindeer during antlerogenesis. (B, C) Full-thickness excision wounds on back and antler at day 0-, 30- or 60-days post-wounding (dpw). (D and E) H&E-stained skin sections from 30dpw back (D) and antler (E) excision wounds. SG=secretory gland. (F-H) Neogenic HFs within velvet wounds stained for (F) Ki67 (green) and keratin-17 (red), (G) versican (red) and (H) Oil red O (red). DP=dermal papilla; SG=sebaceous gland. (I) HF density quantification within wounds relative to surrounding uninjured tissue (n=6 back, n=6 velvet). (J) Percentage of wound contraction relative to original wound size (n=3 back, n=3 velvet). Data are mean±SEM and compared with unpaired t-tests. (K) Schematic summarizing ECM structural analysis. (L-M) Combined (L) and sample-separated (M) UMAPs based on ECM ultrastructural properties of back skin and velvet at baseline and 35dpw (n=3 animal-matched back and velvet). Dashed lines (L) approximate distribution for conditions and contour lines (M) show matrix architecture density on sample-separated UMAPs. (N-O) Replicate of (D-E) for back skin (N) and velvet (O) at 60dpw. (P-Q) Light retardation and azimuthal polarization composites for back skin (P) and velvet (Q) at 60dpw. (R) Schematic of autologous velvet-to-back grafting and wounding. (S) Ectopic velvet graft (white dashed line) at 60 days post-transplant, and 30dpw (blue dashed line). (T) Healed wound (30dpw) on ectopic velvet graft stained with Keratin-14 (green). Fluorobeads (red) demarcate wound margins. Inset shows high magnification of regenerated appendages. Dashed lines represent the original wound margins (B-E). Nuclei are stained with Hoechst (blue).
We also created a 12mm diameter full-thickness burn injury using hot contact which was full depth (confirmed at 3dpw; Figure S1K). At 30dpw, back skin burns exhibited hypertrophic scarring with a near-absence of skin appendages within the wounds (Figure S1G, I). Any HFs present within wounds were limited to the margins and often appeared abnormal (Figure S1I). Despite the additional burden of removing necrotic cellular material, velvet burn wounds still showed robust regeneration by 30dpw, including restoration of appendages and pigment (Figure S1H, J, L). Finally, by 60dpw back skin burns were disorganized, and fibrotic, but velvet burns showed near-complete regeneration (Figure S1M,N).
Regenerative competence is intrinsic to velvet cells
Previous work demonstrated the preservation of intrinsic fibrotic propensity in adult dermal fibroblasts following transplantation to regenerative fetal19 and oral environments20. Here, we asked whether regenerative capacity was intrinsic to velvet fibroblasts. To test this, full-thickness velvet was ectopically transplanted to the back skin on the same animal (Figure 1R, S1O–P). After 30 days, an excisional wound was created and healing assessed 30dpw. There was excellent graft take of 6/8 grafts at 30 days (Figure S1O,P). Remarkably, after 30dpw, ectopic velvet showed near-equivalent regenerative capacity to that of native velvet (on antler), with little visible scar (Figure 1S and S1Q). Indeed, neodermal tissue between fluorobead-marked boundaries contained a full complement of neogenic HFs that reached a similar density to pre-injury ectopic velvet (Figure 1T). Hence, regenerative competence is an autonomous feature of velvet cells.
Velvet dermis is comprised of fetal-like fibroblasts
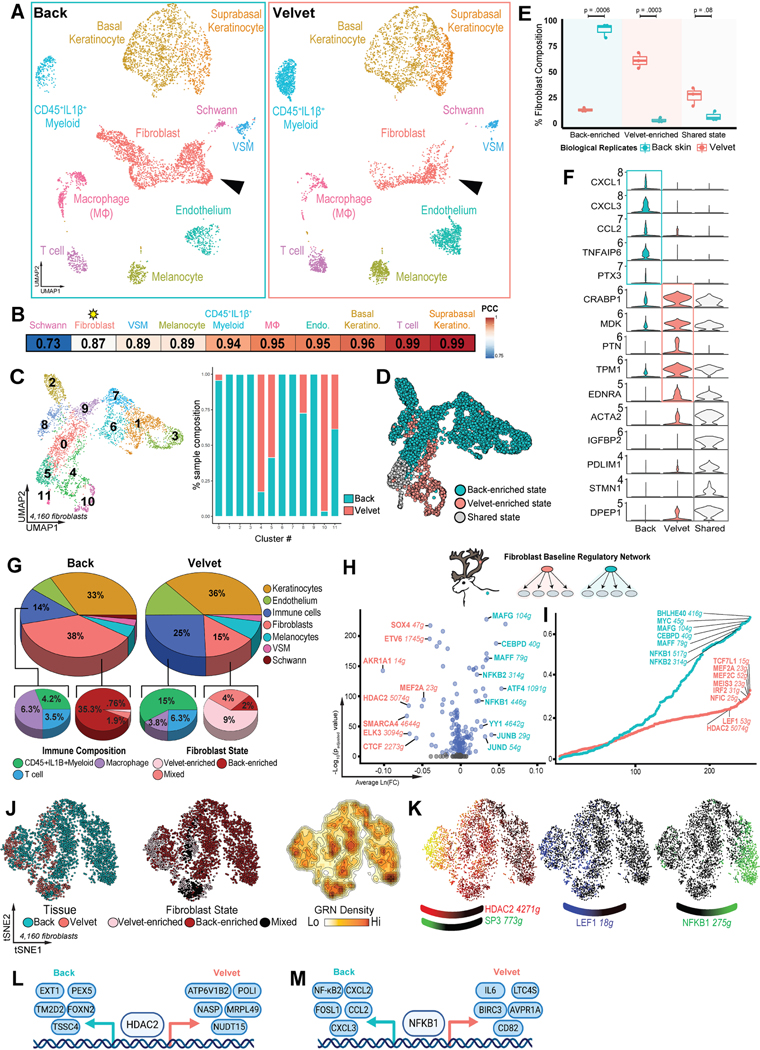
To understand the molecular features underlying this unique regenerative capacity, we performed analyses of back skin and velvet using single-cell RNA-sequencing (scRNA-seq). Interrogation of cell composition within each unwounded tissue revealed ten distinct resident cell populations (Figure 2A, S2C). While shared subpopulations of all cell types were identified21 across velvet and back skin, Schwann cells and fibroblasts were identified as the most highly discordant (Pearson correlation r =0.73 and 0.87; Figure 2B). Given 1) that 10/12 fibroblast subclusters exhibited tissue-specific enrichment (Figure 2C–D), 2) their established role in determining healing outcomes22, and 3) the limited recovery of Schwann cells, we focused our primary analysis on fibroblasts. Genetic variants were used to demultiplex reindeer genotypes23, confirming that tissue-specific fibroblast states were highly reproducible across n=3 biological replicates (Figure 2E). Back-enriched fibroblast states were 7-fold higher in back skin (p=0.0006), whereas velvet-enriched states were 22-fold higher in velvet (p=0.0003). Genes enriched in velvet fibroblasts were associated with regenerative competence (CRABP1, MDK, PTN; Figure 2F)8,24,25. In contrast, back skin fibroblasts uniquely expressed pro-inflammatory genes (CXCL1, CXCL3, CCL2). Of note, 98% of baseline velvet-enriched and 99% of back skin-enriched fibroblast differentially expressed genes (DEGs) have conserved orthologs in the human genome. While velvet harbored more resident immune cells (25% of total cells) than back skin (14%, Figure 2G), CSF1R+ macrophages were enriched in back skin (3.6% in velvet vs 6.1% in back skin, p=0.0472, Figure S2D). Parallel bulk RNA-seq on 3–4 additional biological replicates (Figure S2E–F) validated the transcriptional differences observed with scRNA-seq and confirmed upregulation of regeneration-associated genes in velvet (CRABP1, RUNX1, PRSS35) and pro-fibrotic genes (PTGDS, SCARA5) in back skin (Figure S2G–I).
Figure 2 – Resting fibroblast states in velvet and back skin.
(A) UMAP projection of 11,158 single-cell transcriptomes comparing uninjured back skin (left) and antler velvet (right) colored by manual cell type annotation. (B) Pearson correlation coefficients comparing cell types across velvet and back skin. (C) Unsupervised assignment of fibroblast clusters by contributions of velvet and backskin fibroblasts to each cluster. (D) UMAP projection of 4,160 fibroblasts colored by states enriched in velvet and back skin. (E) Boxplot showing percentage of each fibroblast state (n=3) colored by tissue. All P-values are Bonferroni adjusted. (F) Violin plots showing signature genes unique to back skin, velvet or shared across both sites. (G) Relative proportions of immune subsets and fibroblast states in resting back skin and antler velvet. (H) Volcano plot of differentially activated regulons in uninjured velvet (red) and back skin (cyan). (I) Rank plot of baseline fibroblast regulons ordered by Regulon Specificity Score (RSS), including top regulons for velvet (red) or back skin (cyan). (J) tSNE projection of gene regulatory network-based clustering of uninjured fibroblasts colored by anatomical site, fibroblast state, and regulon density (surrogate for stable fibroblast regulatory states). (K) Signature regulons for pro-regenerative (HDAC2, SP3 and LEF1) or pro-fibrotic (NFKB1) fibroblast states. (L and M) Top downstream targets of transcription factors HDAC2 (L) and NFKB1 (M) in resting back and velvet fibroblasts.
We next examined gene regulatory network (GRN) activity by re-clustering fibroblasts based on SCENIC-predicted transcription factor (TF) activation scores26. Indeed, back and velvet fibroblasts occupied distinct regulatory states with core TF networks unique to each (Figure 2H–M). While pro-inflammatory networks like NF-κB1/2 were robustly active in back skin, velvet fibroblasts activated GRNs implicated in cell plasticity (HDAC2-SP3 co-activation)8,27, HF induction (LEF1)28 and attenuation of inflammation (NFI-C)29 (Figure 2J–M). Despite shared activation of HDAC2 and NFKB1, these TFs exhibited distinct targetomes (Figure 2L–M) suggesting regionalized transcriptional circuitries govern fibroblast ground states. Together, these data suggest that velvet fibroblasts are readied for regeneration.
Cross-species comparisons reveal molecular hallmarks of divergent healing outcomes
Scarless wound healing in fetal human skin3–5 and in spiny mice (Acomys)7,30 has been well documented. To understand whether reindeer fibroblast programs (Figure 2) represent universally shared mammalian priming mechanisms that predetermine healing outcomes, we compared fibroblast DEGs from uninjured velvet and back skin to uninjured fibroblasts from rodent (Acomys versus Mus) (Simkin et al. Under review) and human (fetal versus adult) skin31. Fibrosis-primed fibroblasts exhibited striking molecular overlap (602 genes, 20%) (Figure 3A). Gene ontology of 602 conserved fibrosis-priming genes revealed enrichment of cytokine- and chemokine-mediated signaling (GO:0019221, padj=6×10−21) driven by NFκB (GO:0043122, padj=9×10−7) and prostaglandin (GO:0006693, padj=2×10−5) pathways (Figure 3B). To explore if the fibroblast immune dialogue was specific to reindeer healing, GO analysis examined 273 (9%) features conserved between reindeer and human fibrosis-primed fibroblasts, revealing a state readied to incite innate immune responses, indicated by defense signaling typically triggered by microorganisms (GO:0140546, padj=7×10−5) via pattern recognition receptors like TLR3 (GO:0034138, padj=0.03) (Figure S3A).
Figure 3 – Cross-species comparisons of repair- versus regeneration-primed skin reveals conserved and divergent fibroblast ground states.
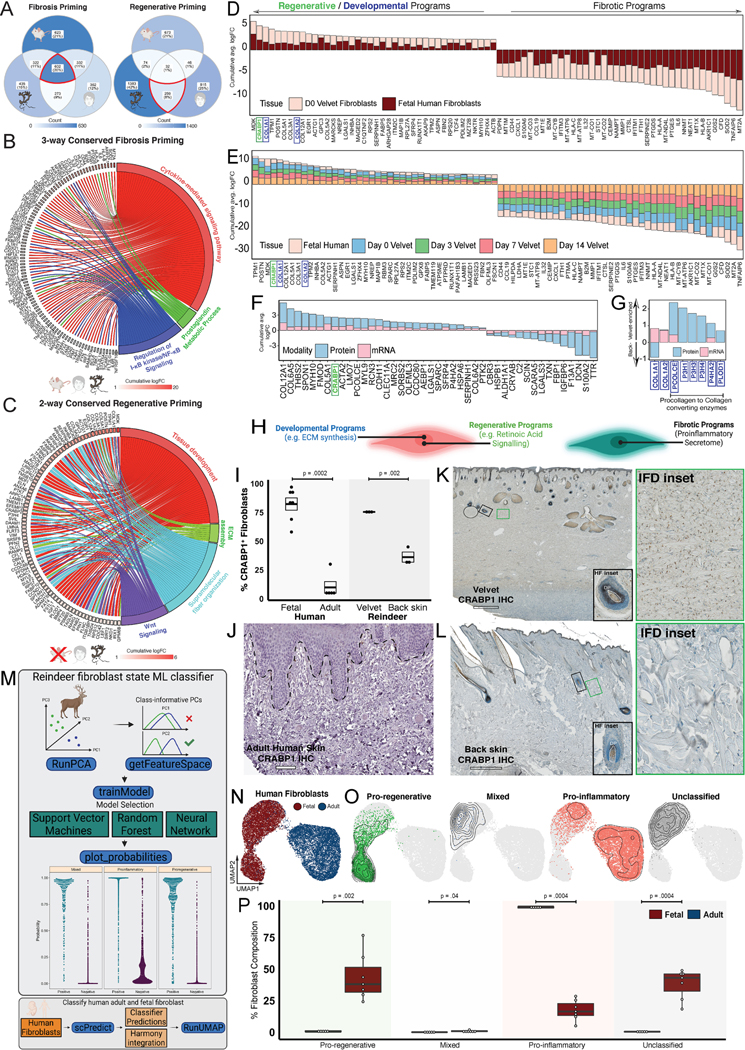
(A) Venn diagram showing conserved and species-specific gene signatures of repair- versus regeneration-primed fibroblasts in rodent (Mus versus Acomys), reindeer (velvet versus back), and human (fetal versus adult). (B-C) Repair-primed fibroblasts signaling across rodent, reindeer, and humans (B) or shared regenerative signaling across reindeer velvet and fetal human (C). (D-E) Consensus plot depicting DEGs enriched in regenerative (velvet and fetal human) versus scar-forming (backskin and adult human) fibroblasts at baseline (D) and during healing (E). Bars represent cumulative log-fold-change colored by velvet and fetal fibroblast contributions. (F-G) Global (F) and matrisome-specific (G) features distinguishing resting velvet and back skin fibroblasts identified in both mRNA and protein enrichments. (H) Schematic highlighting developmental and regenerative programs enriched in velvet fibroblasts and inflammatory signaling in pro-fibrotic back skin fibroblasts. (I) Boxplot showing enrichment of CRABP1+ fibroblasts in regeneration-primed reindeer and human skin. n=7, n=5, n=3, and n=3 biological replicates of human fetal, human adult, reindeer velvet and reindeer back, respectively. Bonferroni-adjusted P-values are shown. (J-L) Immunohistochemistry for CRABP1 (brown) across adult human (J), reindeer velvet (K) and back skin (L). Insets show HF and interfollicular dermal fibroblasts. (M) Schematic of scPred-based machine learning classifier trained to distinguish velvet-enriched ‘Pro-regenerative’, back skin-enriched ‘Pro-inflammatory’, and shared ‘Mixed’ fibroblast states, subsequently applied to fetal and adult human skin fibroblasts. (N and O) Harmony-integrated human fibroblasts, colored by tissue of origin (N) and scPred-inferred fibroblast states (O). (P) Boxplot showing percentage of scPred-inferred fibroblast states in adult and fetal human skin. Bonferroni-adjusted P-values are shown. Human IHC staining obtained from the Human Protein Atlas. Scale bars are 100 μm (J), and 500μm (K-L).
Only 32 (1%) homologs were shared in regenerative fibroblasts across species, likely because velvet and fetal human fibroblasts are engaged in active tissue morphogenesis, while adult mouse fibroblasts are not. Conserved regenerative signaling across human and reindeer (259 genes, 8%) revealed signaling related to tissue development (GO:0009888, padj=9×10−8) via extracellular matrix assembly (GO:0032502, padj=3×10−4) and Wnt signaling (GO:0016055, padj=8×10−3) (Figure 3C). We next assessed DEGs that distinguished regenerative (reindeer velvet and fetal human31) from scar-forming (reindeer back skin and adult human skin) fibroblasts (Figure 3D). This highlighted a remarkable conservation of regenerative (MDK, CRABP1, TPM1) and inflammatory/fibrotic (IL6, PTGES, PTGDS) signatures that were maintained over the entirety of healing, suggesting ground-state fibroblast programming has a durable impact on reparative response (Figure 3D–E, Table S1). We also found conserved enrichment of YAP1, a downstream effector of mechanotransduction in adult human and reindeer back skin that was maintained over the entirety of healing (Table S5), suggesting heightened mechano-sensation in resting and wound-responsive myofibroblasts is a feature of fibrotic healing. Proteomics corroborated site-specific enrichment of pro-regenerative (CRABP1) and pro-inflammatory (coagulation cascade C3 and F13A1) fibroblast programs at baseline (Figure 3F) and revealed additional ECM programs that were either discrepant across modalities or missed by transcriptomics (Figure 3G). Interestingly, while COL1A1 and COL1A2 transcripts were detected in velvet fibroblasts, higher or equivalent amounts of type I collagen were observed in back skin. On the protein level, resting velvet was enriched for hydroxylases (PCOLCE, P3H1/3/4, P4HA2) that mature collagen fibrils, paralleling observations from fetal skin32. Therefore, while velvet and human fetal fibroblasts generate large quantities of ECM (Figure 3D–E), elevated matrisome programs likely reflect ongoing skin morphogenesis as opposed to those that prime regeneration (Figure 3H).
Immunohistochemical examination of CRABP18,24,25,28 across unwounded adult human skin, reindeer velvet, and back skin revealed stark differences in magnitude and expression pattern (Figure 3I–L). While CRABP1 protein was robustly expressed throughout the interfollicular dermis and HF mesenchyme in velvet (Figure 3I, K), in back skin it was found almost exclusively in HF mesenchyme (Figure 3I, L) and was not detected in human interfollicular dermis (Figure 3I, J). Comparisons to another adult human fibroblast dataset33 and Human Protein Atlas34 confirmed enrichment of SOD2, PTGDS, and EFEMP1 in scar-forming (reindeer back skin and adult human) fibroblasts (Figure S3B–F). Conversely, regeneration-associated features (POSTN, LMO7) were either absent or spatially restricted to papillary dermis in adult humans (Figure S3G–H).
We then compared global CRABP1+ fibroblast frequency in fetal versus adult human skin31. This revealed a disproportionate enrichment in fetal (82%) versus adult (10%) skin (Figure S3I–K), suggesting that CRABP1 marks a highly conserved regenerative fibroblast state in mammalian skin8,24,28. To further elucidate fibroblast signatures of pro-regenerative versus pro-fibrotic states across reindeer and humans, we first employed an automatic machine learning classifier35,36 to annotated fibroblast states in a cluster-agnostic manner (Figure S3L). As proof-of-principle, the machine exhibited high discriminatory capacity categorizing ‘pro-regenerative’, ‘pro-fibrotic’, and ‘mixed’ reindeer fibroblast states (Figure S3M). When applied to human fibroblasts, 93% of adult human fibroblasts were classified as ‘pro-fibrotic’ whereas 68% of fetal human fibroblasts were classified as ‘pro-regenerative’ (Figure S3N–O).
We next performed an unbiased feature selection using scPred37 and trained three prediction models (Figure 3M). Classification with svmRadial uncovered a transcriptional continuum that spanned pro-regenerative fibroblasts predicted to reside exclusively in human fetal skin (p=0.002) to pro-inflammatory fibroblasts found predominately within the human adult skin (p=0.0004) (Figure 3N–P). We also found 38% of fibroblasts in the human fetal skin lie between pro-regenerative and proinflammatory states and are labeled ‘Unclassified’ (Figure 3O–P). Together, these analyses suggest a remarkable cross-species conservation of fibroblast programming and define state-specific fibroblast marker genes which elicit regenerative or fibrotic healing responses.
Fibroblast fate reversion and immunosuppression drives velvet regeneration
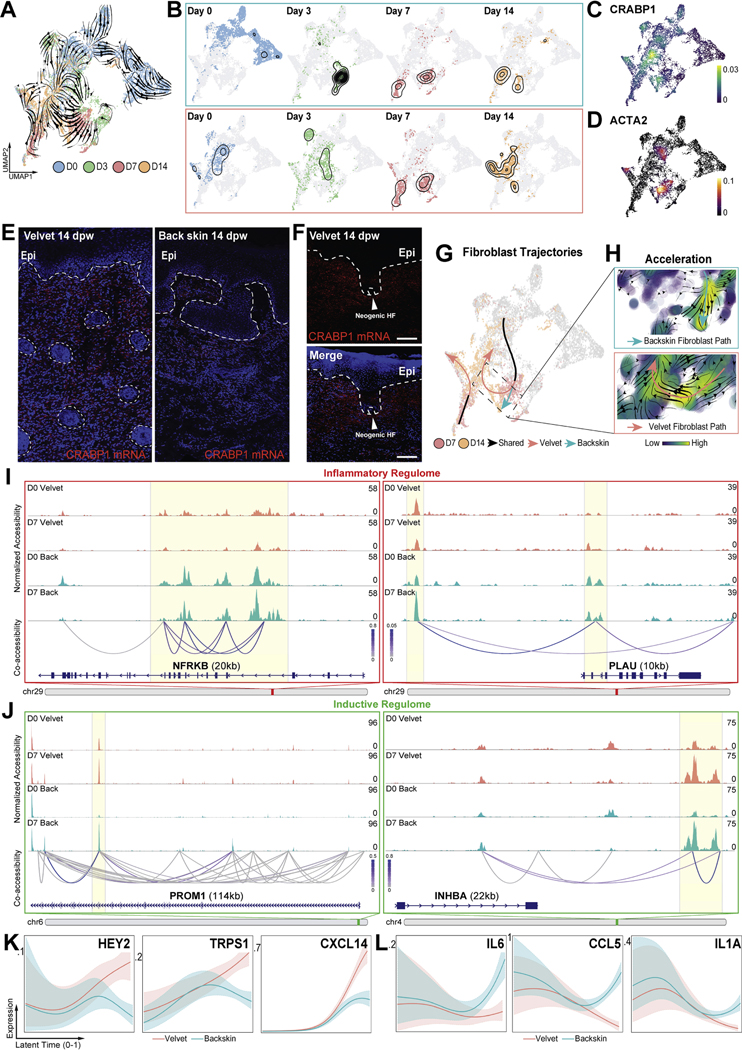
To reconstruct fibroblast dynamics during divergent healing contexts, we constructed in silico trajectories from spliced and unspliced mRNA ratios (using scVelo38,39). Despite maintaining distinct transcriptional programs at baseline and acutely after injury (3dpw), back skin and velvet fibroblasts exhibited a surprising convergence at 7dpw (Figure 4A–B) as both tissues contributed to Clusters 3 and 5 (Figure S4A). This convergence was corroborated by GRN-based hierarchical clustering (Figure S4I). Interestingly, in transition from 7 to 14dpw there was a re-divergence, accompanied by high rates of state transition and reacquisition of ground state in velvet fibroblasts (Figure 4A–C, 4G–H, and S4B–E). Robust reactivation of CRABP1 (Figure 3A–C) within velvet neodermis and in fibroblasts surrounding neogenic HFs was confirmed by RNAscope (Figure 4E–F). In contrast, back skin fibroblasts sustained an activated myofibroblast-like state, markedly different from its pre-injury ground state (Figure 4A–B, 4D, 4G–H). Velvet fibroblasts upregulated morphogenic dermal condensate/papilla markers (DAAM2, ADAM12, SFRP4)40–42 whereas back skin fibroblasts engaged pro-inflammatory programs (NF-κB1, PTGS2, IL6, CXCL2, CSF1) (Figure S4F–G, Table S2). Notably, these fibroblast features included tissue-specific splicing kinetics over the entirety of the healing time course, suggesting velvet and back skin fibroblasts maintained distinct molecular profiles even as they occupied a shared ACTA2+ myofibroblast state (Figure 4A–D, Table S2).
Figure 4 – Inaccessible inflammatory regulome and re-engagement of morphogenic programs in fibroblasts enables regeneration.
(A) RNA velocities calculated from the dynamical model. (B) Temporal progression of velvet and back skin fibroblasts along velocity-inferred topologies. (C, D) Density plots showing expression of CRABP1, (C) and ACTA2 (D). (E, F) In situ hybridization (RNAscope®) for CRABP1 mRNA (red) within velvet and back skin neodermis (E) and in dermal fibroblasts surrounding neogenic HFs in velvet (F) at 14dpw. (G) scVelo-inferred connectivity between 7 and 14dpw showing reversion to ground state in velvet wounds. (H) Acceleration vector fields driving divergent terminal fates in velvet versus back skin. (I, J) Normalized fibroblast pseudobulk chromatin accessibility tracks with cis-regulatory DNA interactions identifying differential accessibility of inflammatory (I) and regeneration-associated (J) loci at baseline and 7dpw. (K, L) Smoothed expression trends in latent time for genes predicted to drive divergent terminal fibroblast fates in velvet (K) and back skin (L).
Trajectory analysis of velvet and back skin fibroblasts identified 7dpw as a defining juncture where their terminal fates were imprinted (Figure 4G–H). To understand epigenetic regulation of fibroblast fate choices, we profiled chromatin accessibility with single-cell Assay for Transposase-Accessible Chromatin using sequencing (scATAC-Seq) at 0- and 7dpw43,44. Unbiased interrogation of fibroblast epigenetic landscapes45 identified 3,882 differentially accessible peaks (DAPs); 1,752 velvet-enriched, 2,130 back skin-enriched) at baseline. These differences became more pronounced during healing as 8,671 DAPs (4,585 velvet-enriched, 4,086 back skin-enriched) distinguished myofibroblasts across the two sites at 7dpw. Of these DAPs, only 36% overlapped protein-coding segments and the remainder were in non-protein-coding regions, suggestive of cis- or trans-acting regulatory functions. We observed significant epigenomic variability within the inflammatory and fibroblast regenerative competence-associated coding and regulatory regions. Consistent with parallel single-cell transcriptomics we observed significantly elevated resting accessibility and cis-regulatory connectivity within the inflammatory epigenome (NFRKB p<1.22×10−11) in back skin fibroblasts and elevated accessibility for regeneration-associated features such as PROM1 (p<1.75×10−9) and INHBA (p<4.30×10−6) in velvet fibroblasts (Figure 4I–J). Surprisingly, wound-activated fibroblasts from both back and velvet at 7dpw revealed profound epigenetic remodeling of regeneration-associated regions through elevated accessibility relative to their respective ground states (Figure 4J). Interestingly, regeneration-associated loci which were enriched in velvet at baseline (PROM1, INHBA, GREB1L) were no longer enriched in velvet and back skin by 7dpw, largely due to their robust induction in back skin fibroblasts (Figure 4J, Online Atlas). Thus, despite their propensity for fibrotic repair, back skin fibroblasts also initiate a latent pro-regenerative epigenetic reconfiguration.
Transition probabilistic modeling on fibroblast temporal dynamics46 revealed that velvet fate reversion during 7 to 14dpw is enabled by both morphogenic (HEY2, TRPS1) and immunosuppressive (CCL2647, CXCL1448) factors that may orchestrate velvet fibroblasts re-activation of baseline regenerative programs. In contrast, immunostimulatory genes (IL6, CCL5, IL1A) propel back skin fibroblast terminal fates (Figure 4K–L). Paradoxically, back skin fibroblasts exhibited greater activation of regenerative programs, in both magnitude and frequency of fibroblasts expressing regeneration-associated markers (Table S3), during the 7–14dpw transition. Given that they still succumbed to fibrotic fates despite transient acquisition of regeneration-permissive epigenomic and transcriptional architecture, it is possible that adoption of regenerative competence depends on either exceeding, rather than simply initiating, a critical threshold of network activity, or timing of this initiation to drive regeneration.
Reconstruction of regulatory networks underlying fibroblasts fate commitments revealed that GRN signatures of pre-injury fibroblasts were either potentiated (NF-κB1 in back skin, HDAC2 in velvet) or remained differentially activated (LEF1 in velvet) suggesting homeostatic programs bias the fibroblast reparative response (Figure S4I–L). A core set of velvet and back skin fibroblast DEGs at ground state were differentially maintained across all stages of healing (Figure S4H, Table S4). Core back skin fibroblast programs included complement and coagulation cascade members (C3, C1R, PLAU) and immunomodulatory cytokines (CCL2, CXCL12) whereas velvet fibroblasts maintained their core development/regeneration (CRABP1, MDK, LGALS1) and ECM (COL11A1, COL12A1, COL27A1) signatures. Interestingly, velvet fibroblasts exhibited stable enrichment of negative regulators of TGFβ signaling (LMO7, PMEPA1) and actin-depolymerizers (DSTN) potentially indicating a resolution of fibroblast reactivity. In contrast, back skin fibroblasts adopted and sustained TGFβ-driven myofibroblast states (TGFBR3, CD44, CRYAB). Conservation of ground state signatures over the entirety of reindeer healing and with divergently primed human fibroblasts suggests pre-injury fibroblast priming preordains regenerative or fibrotic responses (Figure 3E, Table S4,5).
Fibroblast priming exacerbates inflammation and myeloid maturation to promote scar
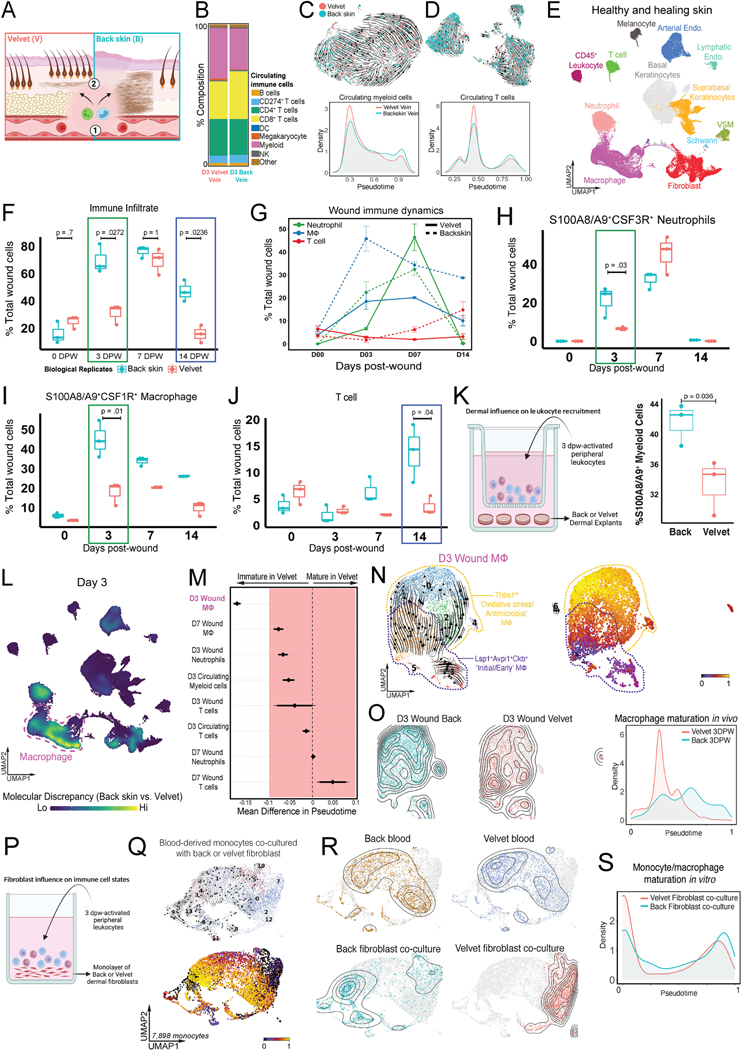
To determine whether resting fibroblast inflammatory priming influences local immune responses within these divergent healing scenarios, we asked whether there was a comparable pool of circulating immune cells supplied to both tissues (Figure 5A–D). scRNA-seq at 3dpw demonstrated no significant difference in cell type composition (Chi square, p=0.97, Figure 5B) of peripheral blood isolated from systemic or antler-specific locations. Moreover, both exhibited near-identical myeloid (Figure 5C) and lymphoid (Figure 5D) pseudotemporal distribution suggesting comparable circulating immune cell maturity.
Figure 5 – Back skin wound signals promote rapid myeloid cell infiltration and maturation to drive fibrotic repair.
(A) Immune cell composition in site-specific circulation (1) or within back and velvet wounds (2). (B) Relative composition of circulating immune cells in saphenous (back skin) versus distal antler velvet vein at 3dpw. (C, D) UMAP projections plotting velocyto vector fields and pseudotime distributions of circulating myeloid cells (C) and T lymphocytes (D) across velvet-specific and systemic venous blood. (E) UMAP projection of all cells in uninjured and healing skin wounds from back and velvet. (F) Boxplot showing percentage of total immune cells in n=3 biological replicates grouped by dpw and colored by wound site. Bonferroni adjusted p-values shown. (G) Temporal distribution of immune cell types within velvet versus back skin wounds at 0, 3, 7, and 14dpw. Error bars represent±SEM (n=3). (H-J) Boxplots (as in F) showing percentage of neutrophils (H), macrophages (I) and T lymphocytes (J). (K). Quantification of leukocyte migration when co-cultured with back skin or velvet dermal explants. n=3 biological replicates. (L) Nebulosa plot depicting cell types exhibiting greatest transcriptional divergence between back and velvet wounds at 3dpw. M. Forrest plot showing quantified pseudotime analysis of immune cell maturation states. N-O. UMAP projections plotting velocyto vector fields and pseudotime distributions of macrophages at 3dpw (N) and distribution of macrophage maturation states between wound sites (O). (P) Schematic showing circulating leukocyte-dermal fibroblast co-culture assay. (Q) UMAP showing velocyto vector fields and pseudotime distributions of monocytes following exposure to back or velvet fibroblasts. (R) UMAP showing transcriptional overlap in monocytes derived from back versus velvet blood, but a marked divergence following exposure to site-specific fibroblasts. S. Pseudotime analysis showing distribution of monocyte maturation. n=3 biological replicates.
Despite drawing from a similar pool of circulating cells, we observed dramatic differences in magnitude, distribution, and maturity states of immune cells recruited into back skin or velvet wounds (Figure 5E). Notably, there was a marked increase in immune cells in back skin wounds relative to velvet wounds at 3dpw (71% vs 31% of total cells were immune cells, p=0.0272, Figure 5F). Although velvet immune infiltration matched that of back skin by 7dpw (69.5% velvet vs 76.3% back skin, p=1), they diverged again at 14dpw (15.8% velvet versus 47.1% back skin, p=0.0236) such that the immune infiltrate persisted in back skin wounds but was largely resolved in velvet.
Most intriguing was the marked heterogeneity in immune cell composition and state within each wound location (Figure 5G–J; Figure SA–D). Back skin wounds harbored a >2-fold enrichment in S100A8/A9+CSF3R+ neutrophils at 3dpw (Figure 5G, H), a persistent elevation in S100A8/A9+CSF1R+ macrophages at 3, 7 and 14dpw (Figure 5G, I) and then a doubling of CD3E+ T cells at 14dpw (Figure 5G, J). Given the differential recruitment, we asked whether intrinsic differences in dermal signaling could modulate immune recruitment directly. Dermal explants from back skin or velvet were co-cultured with circulating immune cells from the same reindeer. Quantification of migrating S100A8/A9+ leukocytes revealed a marked increase following exposure to back skin explant (Figure 5K) suggesting tissue-specific modulation of immune behavior.
In addition to differential recruitment in vitro, our serial in vivo transcriptomic analysis also revealed notable alterations in myeloid differentiation states (Figure 5L–O; S5A–D). Perturbation scores identified robust transcriptomic discrepancy in macrophages and neutrophils at 3 and 7dpw compared to uninjured tissue (Figure 5 L; Figure S5A, B). Pseudotime analysis of macrophages at 3dpw showed enrichment of immature states in velvet wounds highly reminiscent of Lsp1+Avpi1+Ckb+ “initial/early” macrophages49 (Figure 5 M,N,O). Conversely, back wounds were dominated by Thbs1+ macrophages typically associated with oxidative stress and antimicrobial activity.
Three lines of evidence suggested that fibroblasts may be a principal driver of macrophage function during wound healing: 1) fibroblasts were the most discrepant cell type between tissues, 2) immune cell fate diversification happened only after wound recruitment (Figure 5B,C,R), and 3) temporal fibroblast connectome revealed a distinct set of immunomodulatory signals both at rest and during repair (Figure S5C). Therefore, we surmised that fibroblast-derived signals may be sufficient to recapitulate the divergent macrophage maturation states observed during healing. To test this, primary dermal fibroblasts isolated from back skin or velvet were co-cultured with freshly isolated circulating immune cells from the same reindeer that had been wounded 3 days prior (Figure 5P). After 24 hours of exposure, the cells were subjected to scRNA-Seq (Figure 5Q–S). Drastic divergence in monocyte/macrophage transcriptomics was observed after exposure to fibroblasts, with velvet reinstalling transcriptional state dynamics observed in vivo, characterized by promotion/maintenance of immature macrophage states (Figure 5S).
Similarly, pseudotemporal ordering of neutrophils (Figure S5D) from 3dpw revealed a striking enrichment of GRO1/CXCL1hiCXCL2hiIL1Bhi cluster 0 neutrophils (66% vs. 8%; back vs velvet), predicted to be the most terminally differentiated subset (Figure S5E). Biological replicate-separated quantification confirmed a 3.5-fold increase in stage G0-G3 immature neutrophils at 3dpw (9.6% vs 2.7%)50 within velvet wounds (Figure S5F–G). Of note, despite intensified T cell recruitment in back skin by 14dpw (15% vs 3%, p=0.04, Figure 5K), there was minimal discrepancy in T cell maturation states at any time during healing (Figure S5H,I) or following in vitro exposure to back and velvet fibroblast cultures directly (Figure S5J,K), suggesting that T cell recruitment may be a consequence of modified myeloid composition rather than direct fibroblast interactions.
Further, given velvet regenerative capacity is maintained after 30-day engraftment to back skin, it is likely that the grafted velvet experienced similar immune events to native velvet supporting that local immune states are not due to selective endothelial admission or differences in circulating immune cells. Together, this suggests that dermal fibroblasts impart distinct, tissue-specific modulation of incoming immune cells during the acute phase of wound healing that consequently biases downstream healing outcomes.
Pro-inflammatory stromal-immune crosstalk creates a pro-fibrotic wound environment
Both bulk and sc-RNA sequencing experiments showed pronounced differences in transcriptional programs between velvet and back skin, particularly amongst immune and fibroblast-derived ligands (Figure S2H–I; Table S6). To explore the interplay between pro-fibrotic or pro-regenerative fibroblasts and the local wound immune environment, we first reconstructed the fibroblast–immune interface51. This revealed that back skin fibroblasts were uniquely primed for leukocyte interaction compared to velvet fibroblasts (Figure 6A–B), which spanned healing (Table S6). At baseline, velvet fibroblasts secrete pro-regenerative and inductive mesenchymal signals (MDK, PTN, BMP3, RSPO3), whereas back skin fibroblasts secreted immunostimulatory factors (GAS6, CSF1, CXCL12, PLAU), suggesting differential ground-state priming. At 3dpw, fibroblast secretomes were further polarized; velvet sustained production of inductive/morphogenic factors (MDK, RSPO3), while back skin fibroblasts continued to produce potent pro-inflammatory cytokines (IL1A, IL6, IL1B, CXCL12). We then quantified fibroblast-immune cell connectivity52, which revealed CXC ligands as conserved mediators of fibro-immune cell dialogue in both reindeer back skin and adult human fibroblasts (Figure S6A). By 7dpw, there was a 1.6-fold increase in pro-inflammatory (CD68+iNOS+) macrophages in back skin compared to velvet in response to differential fibroblast signaling. In essence, pro-inflammatory cytokines dominated the immune-fibroblast axis in back skin at a seemingly critical time for terminal fate imprinting. At equivalent timepoints in velvet, pro-inflammatory cues were absent (Figure 6A–B), possibly fostering re-engagement of pro-regenerative ground state programs. Tissue staining for MAC387 (S100A8/A9), a pan-granulocyte and macrophage marker, confirmed their sustained presence in back skin relative to velvet at 14dpw (Figure S6C–D) coinciding with either maintenance of myofibroblastic programs in back skin or a near-return to ground state regeneration-competency in velvet fibroblasts (Figure 4E). At this time the neovascularity of the wound bed in back skin was extensively arborized—typical of granulation tissue that is subsequently remodeled into fibrotic scar; the wound bed of velvet wounds revealed vascular patterning restored to baseline (Figure S6B–D). These data implicate resident fibroblasts as orchestrators of the reparative inflammatory response and suggest that inflammatory fibroblast priming predisposes scar-formation and suppression of priming enables regeneration.
Figure 6 – Pro-inflammatory fibroblast signaling attenuates regenerative potential and promotes scar-formation.
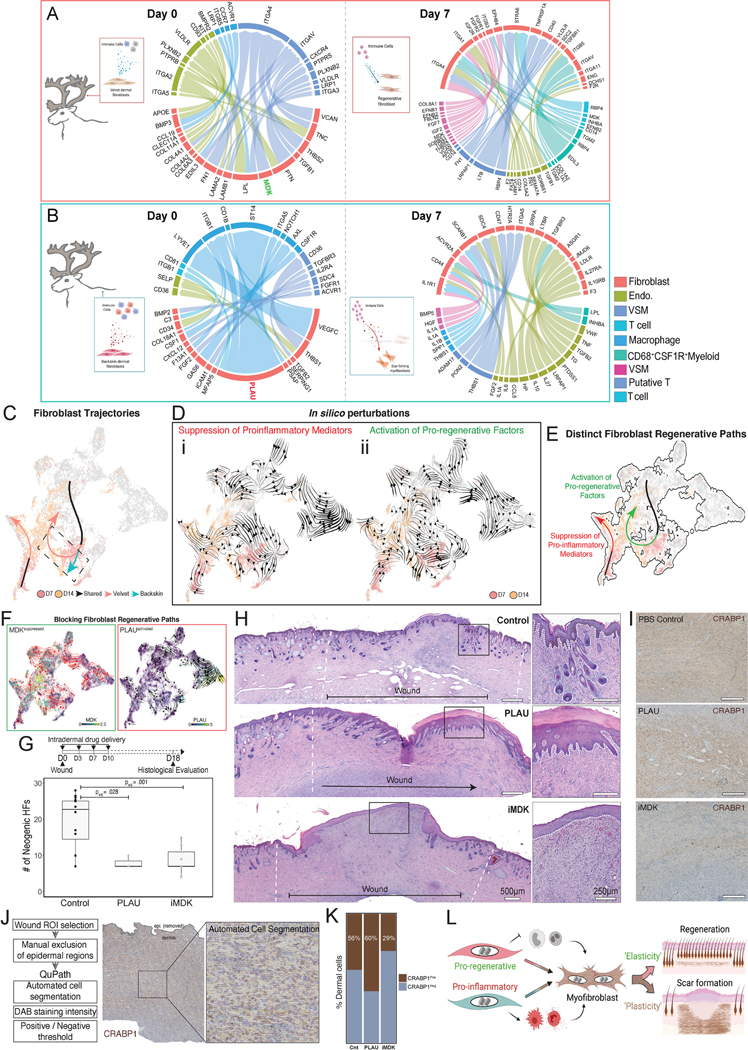
(A, B) Circos plots showing differential fibroblast and immune cell secretome-driven interactions in unwounded (Day 0) and 7dpw in velvet (A) and back skin (B). (C) Schematic summarizing velvet and back skin fibroblast fate trajectories (shown in Figure 4A–H). (D) Fibroblast fate diversions in response to in silico suppression of (i) proinflammatory mediators (CSF1, PLAU, CXCL12) and (ii) hyperactivation of pro-regenerative factors (CRABP1, MDK, TPM1). (E) Replicate of (C) showing fibroblast reversion back to baseline along distinct paths regulated either by suppression of pro-inflammatory (red) or hyperactivation of pro-regenerative (green) factors. (F) Predicted outcomes of pro-regenerative MDK suppression, or hyperactivating pro-inflammatory factor PLAU on fibroblast velocity vector fields. (G) Quantification of neogenic HFs within the wound bed at 18dpw following intradermal vehicle, PLAU or iMDK application. (H) H&E-stained wound sections from vehicle-, PLAU-, and iMDK-treated (I) wounds examined 18dpw. Scale bars represent 500μm (tile scan) and 250μm (inset). Wound histology is representative of n=16 control (n=3 PBS, n=4 IgG, n=6 empty, n=3 DMSO), n=3 PLAU, and n=8 iMDK. (I-K) Representative IHC staining for CRABP1 in wound neodermis (I), image processing and QuPath-based automated analysis (J), and stacked bar plot showing percentage CRABP1+ dermal cells across treatments (K). (L) Model of fibroblast inflammatory priming as a driver of maladaptive immune cell recruitment and maturation promoting pathologic wound repair.
Pharmacologic recapitulation of back skin milieu ameliorates velvet regeneration
Although velvet and back skin reindeer fibroblasts activated similar transcriptional responses to injury at 7dpw, back skin fibroblasts maintained a hyperinflammatory state persisting throughout wound healing. To ask which factors might drive velvet myofibroblasts towards regenerative states acquired by 14dpw (Figure 4B, 6C), we performed in silico genetic perturbations to predict fibroblast fate diversions53. Both suppression of pro-inflammatory mediators (CSF1, PLAU, CXCL12) and hyperactivation of pro-regenerative programs (CRABP1, MDK, TPM1) were sufficient to terminate myofibroblastic states and reacquire pre-injury ground states. Interestingly, these two programs drove distinct regenerative trajectories enabling near-complete restoration of pre-injury fibroblast diversity (Figure 6D–E).
Based on this in silico prediction, we hypothesized that hyperactivation of fibroblast immunomodulatory secretome (through exogenous addition of PLAU) and suppression of pro-regenerative programs (by blocking MDK signaling) promotes scar (Figure 6F). Indeed, intradermal application of PLAU to full-thickness velvet wounds at 0, 3, 7, 10dpw reduced HFs across the wound (2.4-fold suppression, padj=0.028), while increasing epidermal thickness and invaginations reminiscent of back skin wounds (Figure 6G–H). Of note, when treated wounds were allowed to heal for 42dpw (Figure S6E), the early exposure to PLAU delayed, but did not prevent, eventual regeneration (Figure S6F–H). We treated a parallel set of wounds with a small-molecule inhibitor of midkine (iMDK), which led to a robust 2.2-fold suppression of appendage regeneration (padj=0.001) (Figure 6G–H), an effect that persisted even at 42dpw (Figure S6I).
We next probed whether provision of PLAU or iMDK compromised velvet fibroblasts’ regenerative competence by quantifying CRABP1+ cells (Figure 6I–K) or altered ECM matrix architecture (Figure S6J–K) at 18dpw. Consistent with PLAU-induced delay in regeneration, we found more CRABP1+ cells in PLAU-treated wounds relative to vehicle controls (60% vs 44%) and ECM architecture identical to that of control (Figure S6J–K) suggesting that exacerbated inflammation masks fibroblast regenerative potential. In contrast, iMDK-treated wounds had fewer CRABP1+ cells (29% vs 44% in controls) and a distinct ECM (Figure S6J–K) at 18dpw suggesting pro-regenerative suppression is sufficient to drive regenerative failure (Figure 6L).
Acquisition of fibroblast inflammatory priming predetermines fibrotic repair
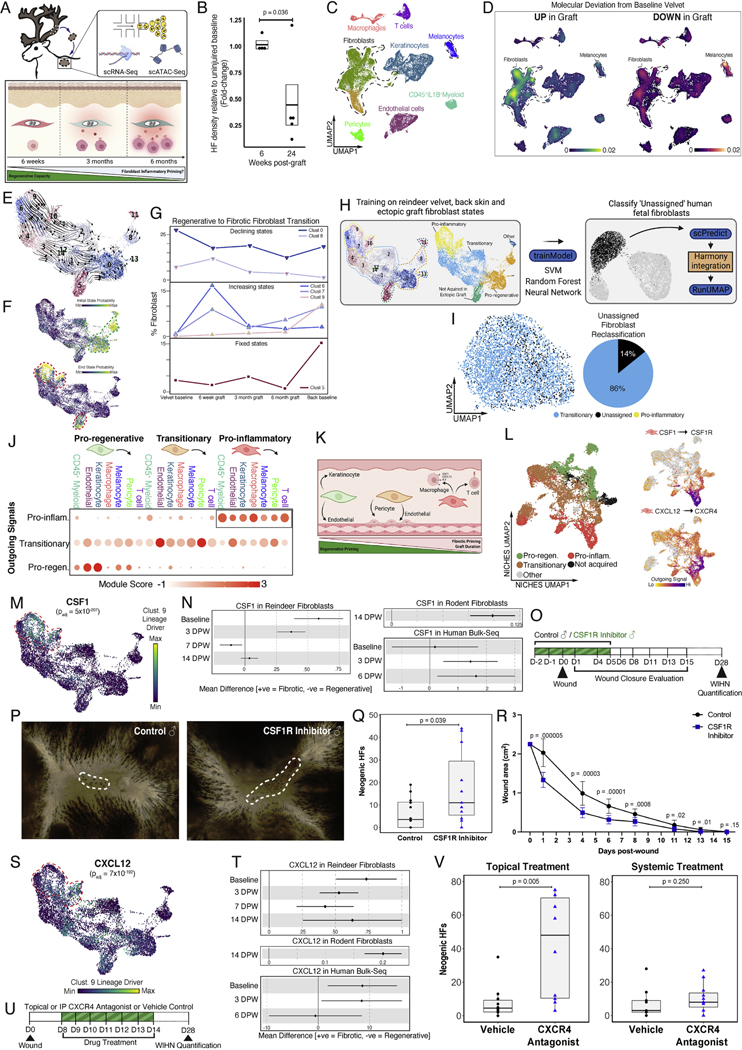
While regenerative capacity of fetal skin is well documented2, the mechanisms by which regenerative competence is lost postnatally are unknown. Short-term follow-up of velvet-to-back ectopic grafts revealed a remarkable preservation of regenerative capacity (Figure 1M–O), however we observed a progressive decline in regenerative priming when grafts were allowed to “age”. That is, by 6 months post-engraftment, baseline HF density was only partially restored post-injury (Figure 7A–B, S7A–C).
Figure 7 – CSF1- and CXCL12-driven fibroblast-immune interactions prime fibrotic repair.
(A) Schematic depicting the hypothesis that fibroblasts in ectopic velvet grafts progressively acquire an inflammatory phenotype that correlates with declining regenerative potential. (B) Boxplot showing HF density relative to uninjured baseline following wounds at 6 (n=4 technical replicates) or 24 weeks (n=5 technical replicates) post-graft. (C) UMAP of 28,164 cells from back skin, baseline velvet, or at 6, 18, or 24 weeks post-grafting. (D) Kernel density estimates depicting magnitude of molecular deviation in ectopic grafts relative to baseline velvet, calculated by summing DEG FCs for all cell types. (E) RNA velocity analysis of 11,100 subclustered fibroblasts undergoing state transitions, colored by Louvain cluster. (F) CellRank-calculated initial and terminal state probabilities. (G) Relative contribution of fibroblast clusters at different graft timepoints, anchored by baseline velvet and back skin and grouped as ‘declining’, ‘increasing’, or ‘fixed’ compared to baseline velvet. (H, I) scPred-based ML classifier (from Figure 3M) used to reclassify previously ‘Unassigned’ fetal human fibroblasts. (J-K) Dot plot depicting fibroblast state-specific outgoing signal intensity as module scores (J) and a schematic summarizing the evolution in fibroblasts’ interactions during regenerative-to-inflammatory transition (K). (L) NICHES ‘cell-to-system’ signaling UMAP generated using fibroblasts as principal sender, colored by scVelo-based annotations (Figure S7F). CSF1-CSF1R and CXCL12-CXCR4 axes are signatures of inflammatory fibroblasts. (M) CellRank-identified regenerative-to-inflammatory transition driver CSF1. (N) Cross-species meta-analysis of CSF1 querying regenerative oral mucosa versus fibrotic skin healing in humans56, fibroblasts in regenerative versus fibrotic domains during WIHN8, and velvet versus back skin fibroblasts in reindeer (this study). Enrichment reaches statistical significance if confidence intervals do not cross the vertical line of no effect. (O-R) Pharmacologic inhibition of CSF1 (O) enhances WIHN (P, Q) and accelerates wound closure (R) (n=12 control, n=11 CSF1R inhibitor). (S-T) Replicate of (M-N) for CXCL12. (U-V) Topical (n=10 control, n=10 topical CXCR4 inhibitor) but not systemic (n=9 control, n=11 systemic CXCR4 inhibitor) inhibition of CXCL12 receptor CXCR4 enhances WIHN. Data are mean±SEM. *p<0.05; **p<0.01; ***p<0.001.
Despite minor changes in ECM architecture across time, ectopic graft ECM did not resemble baseline antler or by 6 months post-grafting (Figure S7D–E). To profile changes accompanying graft maturation, we performed scRNA-Seq of unwounded grafts at 6 weeks, 3 months, and 6 months post-engraftment. The resulting datasets were integrated with baseline velvet and back skin for context (Figure 7C). Interestingly, of all cells, fibroblasts exhibited the most pronounced transcriptional alterations in the graft relative to baseline velvet (Figure 7D). To interrogate fibroblast dynamics within the graft, we: 1) subclustered 10,997 fibroblasts, 2) reconstructed fibroblast state transitions46, and 3) categorized subclusters as ‘Increasing states’ if they showed enrichment in graft relative to velvet, ‘Declining states’ if they were depleted in grafts, or ‘Fixed states’ if their proportions remained unchanged (Figure 7E–G). Intriguingly, these analyses revealed a unidirectional drift of fibroblast states within the graft whereby back skin-enriched pro-inflammatory end-point clusters 6, 7 and 9 were progressively acquired to replace velvet-enriched root clusters 0 and 8 (Figure S7F). We also observed a distinct back skin-enriched fibroblast state (cluster 5) that was not acquired within the graft at any of the surveyed time points (Figure 7G). Based on this, four main macrostates were considered: ‘pro-regenerative’, ‘pro-inflammatory’, ‘transitionary’ connecting pro-regenerative to pro-inflammatory states, and ‘not acquired in ectopic graft’ (Figure 7G, H). Due to the surprising emergence of ‘transitionary’ fibroblasts within ectopic grafts, we revisited our previously ‘Unclassified’ fetal human fibroblasts that were at the nexus between ‘pro-regenerative’ fetal and ‘pro-inflammatory’ adult human fibroblast states (Figure 3M–N). We asked whether including the full fibroblast spectrum (as opposed to only using velvet and back skin fibroblasts) for training would remap the previously ‘Unclassified’ subset (Figure 7H). Indeed, our revised training reclassified 86% of previously ‘Unclassified’ fetal fibroblasts as ‘transitionary’ (Figure 7I) implying similar fibroblast state transitions underly the switch to fibrotic priming by late gestation in humans. Interestingly, transitionary fibroblast signatures included mechanosensitive YAP1 (padj=5×10−4), TEAD1 (padj=1×10−5), EN1 (padj=5×10−5), as well as FAK/PTK2 in transitionary subcluster 2 (padj=3×10−5) (Figure S7G) suggesting that activation of fibroblast force sensors triggered inflammatory cytokine production which in turn primed fibrotic repair.
Next, we examined how the emergence of immunomodulatory programs in fibroblasts repatterned cell-cell communication. First, we asked whether resident immune cell composition co-evolved with fibroblast states within the graft. Because macrophage phenotypes were discrepant between baseline velvet and back skin (Figure 2B), we reclustered macrophages and re-annotated as ‘Phagocytic’, ‘Oxidative’, or ‘Remodeling’49. While overall macrophage abundance does not correlate with graft evolution, from 3 to 6 months we found a dramatic enrichment in IL6R+ remodeling macrophages, and by 6 months their abundance mirrored baseline back skin (Figure S7H). We then generated UMAPs based on ligand-receptor patterns54, and asked: 1) how did the fibroblast interactome evolve during regenerative-to-inflammatory transition, and 2) which signaling families drove this evolution? Our data predicts that the pro-regenerative fibroblast secretome was dominated by inductive mesenchymal ligands such as BMP3/4, NDP, INHBA, FGF10 and LAMC3 targeting overlying keratinocytes as well as immune repellents such as SLIT2/3 directed towards endothelial cells expressing ROBO1/2/455 (Figure 7J–K, Table S7). Transition to an inflammatory fate was characterized by weaning of epithelial-mesenchymal crosstalk and a switch from immunosuppressive to fibrosis (ECM-based signaling) using collagens (COL5A1/1A1/1A2/3A1) and collagen synthesis signals (CTHRC1) to endothelial cells and vessel-ensheathing pericytes (Figure 7I–J, Table S7). In contrast, pro-inflammatory fibroblasts were predicted to dialogue with innate and adaptive leukocytes using lineage-restricted growth factors like CSF1 that specifically targeted macrophages as well as cytokines (CCL2, CXCL12, PTGS2) and members of complement (C3, C4A, CFH) and coagulation (PLAU, PLAT, PROS1) cascades that activate multiple immune and vascular surface receptors (Figure 7J–K, Table S7).
Together, our findings suggest that inflammatory programs (driven in part by mechanotransduction signaling) within inductive fibroblasts rewire cell circuit topologies to drive an intensified immune response during healing. These interactions promote perpetual contractile myofibroblastic states which tip the balance from regeneration to fibrotic repair.
Intercepting mediators of fibroblast inflammatory priming enhances skin regeneration
Several converging lines of evidence nominate CSF1 and CXCL12 as fibroblast-derived cytokines that function as master mediators of inflammatory priming. First, CSF1/CSF1R and CXCL12/CXCR4 axes were differentially expressed in pro-inflammatory fibroblasts (Figure 7L). While CSF1/CSF1R axis is macrophage-specific, CXCL12 (SDF-1) cognate receptor CXCR4 was expressed by both innate and adaptive leukocytes (Figure S7I). Second, CSF1 (padj=5×10−207, Figure 7M) and CXCL12 (padj=7×10−192, Figure 7S) had tight covariation between their activation and regenerative-to-inflammatory transition. Third, our cross-species meta-analysis of regenerative and fibrotic programs that distinguish oral mucosa versus skin healing in humans56, fibroblasts in regenerative versus fibrotic domains of large wounds in rodents8, and velvet versus back skin fibroblasts in reindeer revealed conserved enrichment of CSF1 (Figure 7N) and CXCL12 (Figure 7T) during fibrotic repair.
Given the potential clinical use of these targets for scarless healing, we assessed whether pharmacologic inhibition of CSF1 and CXCL12 improved outcomes using a rodent full-thickness excisional wound model where new HFs regenerate within central domain of large wounds, while periphery forms scar9,10,57. Local inhibition of CSF1R (PLX5622 inhibitor) or CXCR4 (small-molecule antagonist) enhanced skin regeneration by 2.8- and 5.1-fold (in neogenic HFs) respectively (6.4±2.0 control vs 17.8±5.0 CSF1R inhibitor; 8.1±3.2 vs 41.6±9.7 CXCR4 antagonist) (Figure 7N–V). CSF1R inhibitor also accelerated the rate of wound closure (Figure 7R). Interestingly, systemic administration of CXCR4 antagonist by intraperitoneal (ip) injection had no effect on wound-induced HF neogenesis (7.3±3.0 controls vs 10.3±2.6 CXCR4 antagonist ip) reinforcing that CXCL12/CXCR4 interactions act locally to intensify reparative inflammation (Figure 7V). These findings support a functional role for fibroblast-secreted immunomodulators in driving fibrotic repair.
Discussion
Adult reindeer represent a powerful mammalian model to directly compare skin regeneration and fibrosis. Our comparative wounding and transplantation studies revealed that antler velvet harbours a remarkable regenerative capacity, including restoration of key functional units (e.g., glands, HFs, pigmentation). Comparative single-cell transcriptomics and epigenomics revealed highly discordant fibroblast states and distinct stromal-immune linkages that underpin these divergent healing outcomes.
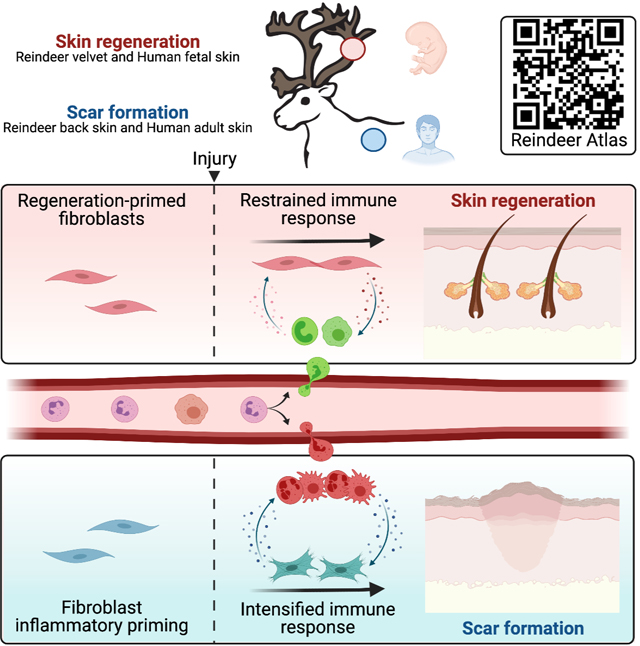
Our results suggest that skin regeneration depends on two requisite mesenchymal processes. First, engagement of a core transcriptional network is required to establish regenerative competence. We identified a set of conserved regulatory networks distinguishing fibroblasts capable of scarless healing versus those biased to form scar. As a consequence of this ground-state regenerative programming, our data suggests wound-responsive velvet fibroblasts undergo a reversion to their pre-injury ground state, enabling restoration of homeostatic skin function. Second, fibroblasts harbor site-specific inflammatory priming, which coordinates local immune cell function during wound healing. Velvet fibroblasts exhibited transcriptomic profiles indicative of an immunosuppressive secretome, whereas back skin fibroblasts maintained a heightened inflammatory state prior to fibrotic repair. Regenerative competence has been associated with an attenuated immune response during fetal healing3,58,59 or following genetic depletion of macrophages and neutrophils60. Our work documenting scarless velvet healing demonstrates that mammalian tissue regeneration can occur in the presence of a fully competent immune system and is at least partly enabled by local fibroblast modulation of incoming immune cells. Indeed, velvet wounds exhibited both diminished myeloid recruitment, arrested maturation of neutrophils and macrophages and accelerated clearance of immune infiltrate relative to identical wounds in back skin. Our co-culture experiments show that fibroblasts harboring site-specific ground states exhibit distinct effects on immune cell recruitment and functional specification. The absence of fibroblast inflammatory priming within velvet fibroblasts may serve to restrain deleterious inflammatory signals and safeguard activation of transcriptional networks necessary for regenerative healing. These observations suggest that fibroblasts direct healing outcomes by engaging core developmental programs and restraining leukocyte communication to promote regeneration, or alternatively by sustaining myofibroblast programs and potentiating leukocyte dialogue to drive scar formation.
This work imparts new insight into how regenerative competence is lost during fetal to postnatal mammalian development. When grafting velvet to a non-regenerative site (back skin) and allowing it to naturally “age”, fibroblasts undergo gradual loss of regeneration-associated networks and acquire an inflammatory primed ground state resembling that of surrounding back skin. Interestingly, the transition from regeneration-competent to scar-forming fibroblast states was associated with the activation of mechanotransduction-related signaling networks (eg. YAP–TEAD), an established driver of fibrosis18,20,61,62. We posit that mechanosensory signaling drives inflammatory signals in reparative fibroblasts, including factors like CSF1, a fibroblast-specific YAP target that promotes macrophage recruitment63. Establishment of fibroblast immune priming, as seen during graft evolution, paired with fibroblast mechano-inflammatory programs during healing drives an intensified immune response leading to fibrotic repair. Purposeful re-activation of developmental programs underlying regenerative competence paired with suppression of fibroblast-immune interactions represent important avenues for mitigating scar. Our Reindeer Wound Atlas (biernaskielab.ca/reindeer_atlas) provides an accessible reference for exploring molecular programs underlying skin regeneration versus fibrosis.
Limitations of the Study
In the absence of a thoroughly annotated reindeer genome, bulk- and scRNA, and scATAC-Seq reads were mapped to a bovine (Bos taurus) reference genome. Although it provides the greatest degree of homology to reindeer, comparisons using a comprehensively annotated reindeer reference may yield an additional ~20% read mapping. Such re-analysis may reveal processes that uniquely evolved in reindeer. Paucity of reindeer-compatible antibodies limits verification of protein activity, including those required to annotate non-coding regulatory regions (e.g., TF motifs) within the reindeer genome. Comparing genomic differences between ectopic grafts that maintained regenerative capacity versus those that transitioned to fibrotic repair could further elucidate mechanisms maintaining durable regenerative competence. However, this was not feasible because wounding (Figures 7B, S7A–C) and single-cell (Figure 7C–G) experiments were performed with different biologic replicates across two different cohorts. Finally, while long-term grafting experiments imparted mechanistic insights into decline in regenerative competence, future studies are needed to understand what triggers onset of fibroblast inflammatory priming to bias fibrotic repair.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Jeff Biernaskie (jeff.biernaskie@ucalgary.ca).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
Sequencing data reported in this work are available at NCBI GEO data repository (GSE168748). Mass spectrometry datasets are available at the ProteomeXchange Consortium in the PRIDE partner repository (PXD035749).
Analysis code generated in this work are available at: http://doi.org/10.6084/m9.figshare.14196344 and github.com/BiernaskieLab.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
Reindeer - Adult reindeer (Rangifer tarandus) were housed in an outdoor enclosure at the University of Calgary Veterinary Sciences Research Station for the duration of experimental procedures. Experiments were performed during the period of intense early antler growth unique to each sex (early spring for males, late spring for females). Sex was equally distributed across experimental groups.
Mice - Male C57Bl/6 mice were between 28–35 days of age at the time of wounding. Mice were group housed (4–5 mice of same sex per cage) in a controlled environment (12h light/dark cycle at 21oC) with unrestricted access to water and standard chow diet. Mice were randomly designated to each experimental group.
All experiments were done in accordance with guidelines set out by the University Animal Welfare Committee, the Veterinary Sciences Animal Care Committee and Health Sciences Animal Care Committee at the University of Calgary, the Canadian Council on Animal Care and the Province of Alberta Animal Protection Act and Regulation.
METHOD DETAILS
Skin wound creation and tissue collection
All animal handling was limiting to early morning where ambient temperature was cooler to reduce heat stress. Animals were restrained in a hydraulic squeeze designed for safe wildlife handling and then anesthetized (Medetomidine, 0.07–0.15mg/kg I.M. with Azaperone, 0.2mg/kg I.M., if additional duration of anesthesia required). Anesthetized animals were blindfolded, their eyes lubricated (Optixcare eye lube), and nasal insufflation of pure oxygen was administered throughout the procedure. Sternal recumbency was maintained to reduce the risk of aspiration of regurgitated rumen contents. Anesthetic depth, heart and respiratory rate and temperature were monitored continuously. Local anesthesia was provided (2% Lidocaine – Wyeth®) with a ring block and a tourniquet applied around the base of the antler; an inverted “L” block was done along the dorsal back. Analgesia was provided by administration of a non-steroidal anti-inflammatory (Meloxicam, 0.5mg/kg S.Q.) and fly strike was prevented with an extended release parasite control product (Eprinomectin, 1mg/kg topical). Back skin and antler velvet were clipped to remove hair, and asepetically prepared using betadine surgical scrub and isopropyl alcohol. Four full-thickness excisional wounds were created on each antler, and three identical full-thickness wounds were made on the dorsal back skin using a 12 mm biopsy punch. The margins of each wound were marked by placement of skin staples. In a subset of animals, creation of full-thickness burn wounds was achieved by heating a 12mm diameter brass rod in a heating block for 4 minutes at 145°C until the metal rod reached 100°C. The brass rod was immediately applied to either back skin or velvet for 30 seconds. In all cases, full thickness injuries were made on velvet that ensheaths the pedicle bone; a site that is distant from the growth zone located at the antler tip. Verification of a full thickness thermal injury was performed by gross and histologic assessment (see also Supplementary Figure 2). Reversal of the sedation/anesthesia was provided (Atipamezole, IM at 5X the dose of medetomidine used). All wounds were left to heal for 30 or 60 days and the healed tissue was then harvested by excisional biopsy utilizing the same anesthetic protocol. Collected tissue was fixed in 4% paraformaldehyde for 48 hours and then snap frozen in clear Frozen Section Compound (VWR) on dry ice and stored followed by storage at ȡ80°C until further use.
Ectopic velvet graft and wounding
In four animals, a total of 8 full-thickness velvet grafts were transplanted onto the dorsal back. General anesthesia was administered as described above. Local anesthesia was provided with a ring block (2% Lidocaine – Wyeth®) and a tourniquet applied around the base of the antler. The antler was then removed just distal to the shovel using a hand saw and hemostasis ensured with application of a temporary pressure bandage. The excised portion of antler was clipped and prepared aseptically and 2, 6×4cm elliptical full-thickness velvet grafts were harvested from the distal aspect of the antler (nearest the regenerating tip). Sharp dissection was used to remove the underlying periosteal-like layer of tissue from the graft before temporary storage in sterile saline for immediate grafting. Simultaneously, an area along the dorsal aspect of the mid-lumbar region of the back on both the right and left sides were clipped and aseptically prepared to accept grafting. The graft bed was created using a sterilized template orientated in a position along the lines of tension to facilitate later excision and wound closure and positioned dorsally enough to prevent the animal scratching the area. Following excision of the back skin, the graft bed was filled with the antler velvet graft, which was secured in place with a combination of simple interrupted skin sutures of 0 polypropylene (0 Prolene, Ethicon) and skin staples. A sterile stent bandage was sutured over top the wound and fly repellent ointment (Swat Original Fly Repellant Ointment, Farnam) was applied around the grafted area to help prevent fly strike. At four weeks post-graft, the animals were again anesthetized, and staples/sutures removed.
Successful graft take was observed in 7/8 grafts. These 7 grafts were subsequently injured using an 8mm diameter biopsy punch to remove a full thickness piece of tissue. Fluorescent polystyrene beads (FluoroSpheres®, 0.2μm, dark red 660/680, ThermoFisher) were injected intradermally at 4 points around the perimeter of the wound to identify original wound margins. Animals were recovered and returned to their enclosure for an additional 4 weeks. At this timepoint, the animals were again anesthetized, the wounded area photographed, and the entire ectopic graft was excised. The remaining back skin was closed with a combination of tension relieving (vertical mattress) sutures of 1 polydioxananone (PDS® II, Ethicon) and simple interrupted sutures of 2–0 polydioxananone (PDS® II, Ethicon). Four grafts retained flourobeads demarcating the original wound margins and these were quantified for HF density.
In vivo velvet drug delivery
Following anesthesia and analgesia described above, a single 12mm full-thickness wound was made on the medial surface of each antler branch approximately 10 cm from the distal tip. Immediately following injury, 200ul of drug (and its corresponding vehicle control) was injected intradermally within the surrounding uninjured tissue at the margin of each wound. Intradermal drug application was repeated every 3 days for 10 days. Control wounds received either PBS, DMSO, or no treatment. Experimental wounds received either midkine inhibitor (iMDK; imidazothiazolyl-chromenone compound; 2mg/mL, 200μg per wound) or recombinant human urokinase-type plasminogen activator (PLAU; 2ng/uL, 200ng per wound). The concentrations for iMDK and PLAU were determined based on manufacturer specifications and previously published work for in vivo use. For example, iMDK concentrations were adjusted based on previous work showing effective in vivo blockade of lung tumor cell growth following i.p. administration of iMdk (9 mg/kg i.p.) in mice 65. PLAU (Urokinase) has been used clinically for pulmonary embolism and cerebral infarction. Clinically, the initial loading dose is 4,400 international units per kilogram of Kinlytic™ (urokinase injection) is given at a rate of 90 mL per hour over a period of 10 minutes. This is followed with a continuous infusion of 4,400 international units per kilogram per hour at a rate of 15 mL for 12 hours. Reindeer velvet wounds were collected either 18- or 37-days post-injury and stained with H&E.
Tissue processing for single-cell genomics
To liberate cells from healthy and healing reindeer skin, 12mm circular biopsy punches collected from n=3 reindeers per sample were dissociated to single cells by mincing the skin into 1–2 mm pieces using a pair of scalpels. Minced tissue was submerged in Dispase II (2 mg/ml at 37°C for 10 minutes, Stem Cell Technologies) to remove epidermis and then in collagenase type IV (2 mg/ml at 37 for 1–3 hours with mechanical trituration at set intervals, Sigma) to digest the dermal extracellular matrix. To further liberate single cells, gentleMACS™ Octo Dissociator (Miltenyi Biotec) was used for 6 minutes on homogenization program and DNase (500 μl of DNase per 10 ml of medium) was added to each sample to maintain the single-cell suspension. Dissociated cell suspensions were passed through 100μm and then 70μm cell strainers, centrifuged at 1200rpm for 6 minutes to pellet the liberated cells, and resuspended in FACS buffer (1% bovine serum album in PBS) on ice. Wound samples were FACS‐enriched for cell viability into 1% BSA-PBS. Of note, absolute neutrophil abundance is likely underestimated because we did not employ specific procedures to optimize neutrophil yield (e.g., room temperature, RNase inhibitor).
PBMCs were collected from venous blood supply from saphenous vein or from distal antler velvet into EDTA tubes and briefly stored on ice. Blood was spun at 300g for 10 min and plasma removed and stored at −80oC. Buffy coat was collected, and red blood cells were lysed with ACK buffer (1:10 ACK to blood cells) for 5 minutes and then spun at 300g for 5min and then supernatant. Cells were washed twice with 5mL PBS, then spun at 300g 5 min. Cells were resuspended in 2% BSA in PBS at a final concentration of 30,000 cells/100uL and run through a 40μm cell strainer, before loading into the 10X controller (~12,000 cells).
Single cell RNA- and ATAC-Seq library construction and sequencing
Healthy and healing skin samples were split for joint processing using 10X Genomics Chromium™ Single Cell 3′ v3 Chemistry for scRNA-Seq and Chromium™ Single Cell ATAC Library and Gel Bead Kit Chip E (v1) for scATAC-Seq as per manufacturer’s protocol. PMBCs were processed for scRNA-Seq using Chromium Next GEM Single Cell 3’ v3.1 as per manufacturer’s protocol. Single cells were partitioned into Gel Bead in Emulsions (GEMs) which resulted in the generation of full-length cDNA from poly-adenylated mRNA. DynaBeads® MyOneTM Silane magnetic beads were used to remove biochemical reagents/primers from the GEM reaction mix and cDNA was amplified using PCR for 12 cycles. During GEM incubation, the read 1 primer sequences were added to the cDNA, subsequently followed by the addition of P5 primers, P7 primers, i7 sample index and read 2 primer sequences during library construction. For scATAC-Seq libraries, sample index PCR step was completed with 12 cycles, as per v1 recommendation for 2,001 to 6,000 target nuclei recovery. For scRNA-Seq libraries, sample index PCR step was completed with 12–14 cycles, as recommended for 25–150ng of starting cDNA. Quality and quantity of RNA samples were analyzed using Tapestation. First pass (shallow) sequencing for scRNA-Seq libraries was performed on Illumina NovaSeq SP to determine number of cells recovered and additional reads were added on Illumina NovaSeq S2 at the Centre for Health Informatics (CHGI) at University of Calgary (UofC) to achieve a minimum sequencing depth of 25,000 read pairs per cell post-aggregation. Shallow sequencing for scATAC-Seq libraries were performed using HiSeq 4000 PE100 followed by deeper sequencing on Illumina NovaSeq S1(100 cycles) at CHGI at UofC to achieve a sequencing depth of 8,398 median fragments per cell for 50,794 cells post-aggregation.
Co-culture of reindeer skin explants or fibroblasts with peripheral leukocytes
To determine dermal- or fibroblast-autonomous influence on leukocyte recruitment and function, we performed in vitro leukocyte migration assays using either whole dermis or primary fibroblast co-cultures. Full thickness 6 mm diameter skin biopsies from back skin or velvet were cut into small cubes (1–2 mm2) and placed in 24-well plates containing 600μl of serum-free DMEM (low glucose, with L-Glutamine; Gibco 11885084). Tissue was allowed to acclimate in medium for 30 minutes and then exposed to PBMCs. Circulating PBMCs were collected from whole blood from the same reindeer in heparin-containing vacutubes. Blood was spun at 300g for 5min to allow collection of serum and buffy coat separately. Isolated buffy coat was suspended in 5ml of red blood cell lysis buffer for 5 minutes at room temperature. The cell suspension was diluted in media and then centrifuged at 300g for 5 minutes. Cells were resuspended in HBSS (Gibco, 14175–095) and 50,000 PBMCs in serum-free DMEM were added on top of a Transwell® membrane (3um pore size, 6.5mm diameter, polycarbonate, Corning CLS3415). Cells were co-cultured for 3 hours and then fixed with 4% PFA. Cells were stained with a mouse anti-S100A8/A9 (Abcam, ab22506), counterstained with Hoechst (Invitrogen, H3570) and visualized with an anti-mouse Alexa 488 secondary antibody (Invitrogen, A32723) and then number of cells migrating through the membrane (total cell number and S100A8/A9) were counted.
Similar co-culture experiments were done with primary dermal fibroblasts derived from back skin or velvet. To isolate fibroblasts, skin was minced into small pieces and digested in Dispase II (StemCell Technologies; 07913) for 20 minutes at 37oC. Epidermis was removed and then underlying dermis was minced and incubated in Collagenase IV (Sigma, C5138) for 2 hours at 37oC. Cells were dissociated via trituration, passed through a 70μm cell strainer and then centrifuged at 300g for 5 min. Cells were suspended in complete media (DMEM containing 1% L-glutamine (Gibco, 25030081), 10% FBS (Gibco, 12483020), 1% penicillin-streptomycin (Gibco, 15070063) and 1% fungizone (Gibco, 15290-018)). Cells were plated in T25 flasks and grown in complete media for 5 days (~70% confluence). Fibroblasts were then replated into 24 well plates at 50,000 cells/well and allowed to acclimate for 8 hours at which time 50,000 PBMCs were either i) added to Transwell®membranes and incubated for 3 hours (migration assay as above) or ii) grown directly on top of dermal fibroblasts for 24 hours and then collected for scRNAseq to assess transcriptional state changes.
CSF1R and CXCR4 inhibition assessed using rodent WIHN model
To assess whether CSF1R inhibition impacts wound closure and HF neogenesis, mice were given large (1.5cm diameter) full-thickness excisional wounds on mid-dorsal skin and were fed control (AIN-76A, D10001i) or CSF1R inhibitor (PLX5622, D11100404i) chow from −2 to 5 DPW. All mice received meloxicam analgesia (Metacam, 25mg/kg) prior to surgery and once daily for 48 hours post-injury. To assess whether topical or systemic CXCR4 inhibition impacts HF neogenesis, mice were treated with CXCR4 antagonist (Plerixafor, Tocris Bioscience, 50 mg/kg) either intraperitoneally or using a topical DMSO-based gel from 8 till 14 DPW. All mice used were of C57BL/6 background and were between P28–35 on the day of wounding. Animals were group-housed (4–5 mice of same sex per cage) in a controlled environment (12h light/dark cycle at 21oC) with unrestricted access to water and chow diet. Photos were taken every morning at 9:00 AM for wound closure evaluation from D1 to D15 post-wounding. Wound area was quantified using ImageJ. Mice were euthanized via CO2 induction followed by cervical dislocation and wound tissue was collected at 28 DPW for wound-induced HF neogenesis (WIHN) analysis as previously described 8. Briefly, collected samples were fixed with 4% PFA in PBS for 48h. Neogenic HFs were quantified by whole-mount imaging using a V5 Slide Scanner (Olympus Life Science).
Sample preparation for shotgun proteomics
Tissue biopsies of antler velvet (n = 3) or back skin (n = 3) were subjected to shotgun proteomics analyses. Protein lysates from tissue were prepared using lysis buffer (1% SDS, 0.1 M DTT in 200 mM HEPES (pH 8) and protease inhibitor tablets [Roche]), followed by dilution to a final concentration of 3 M guanidine HCl (pH 8.0), 100 mM HEPES (pH 8.0), and 10 mM DTT. Alkylation was performed with 15 mM iodoacetamide for 25 min in the dark at room temperature. Proteins were trypsinized overnight. Next, the pH was adjusted to 6.0 with HCl. Samples were incubated for 18 h at 37 °C with isotopically heavy [40 mM 13CD2O +20 mM NaBH3CN (sodium cyanoborohydride)] or light labels [40 mM light formaldehyde (CH2O) + 20 mM NaBH3CN], to label peptide α- and ε-amines. Proteins were precipitated and acidified with 100% acetic acid to pH < 3.0 and cleaned using C18 Sep-Pak columns (Mississauga, Ontario).
QUANTIFICATION AND STATISTICAL ANALYSIS
Immunohistochemistry and quantitative image analysis
Frozen tissue was cryosectioned at 30μm thickness, placed on positively charged slides and subsequently stored at −80°C. Tissue sections were thawed, washed with Phosphate Buffered Saline (PBS). Non-specific binding sites were blocked by incubating sections for 45 minutes in 10% goat serum containing 0.5% Triton-X in PBS to allow permeabolization. Primary antibodies were diluted in PBS and incubated overnight at 4°C. The following day, the samples were washed three times with PBS for 3 minutes each and incubated with the secondary antibodies for one hour at room temperature. Samples were then washed with PBS for 3 minutes and incubated with Hoechst dye (1:500) for 15 minutes. The samples were washed with PBS three times for 3 minutes each and cover slipped using Permafluor mounting media (Thermo Scientific).
For CRABP1 immunohistochemistry, paraffin embedded sections (10μm thickness) were rehydrated with xylene and ethanol (100%, 95%, 70%, 50%). Non-specific binding sites were blocked by incubating sections for 30 minutes in 10% goat serum in PBS and 15 minutes each with Avidin and Biotin solution. Primary and secondary antibodies, diluted in 5% goat serum in PBS, were incubated for above-mentioned durations. Sections were washed with PBS and incubated with ABC solution for 30 minutes at room temperature. Following washing steps, sections were stained with DAB for 15 seconds and counterstained with haematoxylin for 10 seconds. The coverslips were mounted using Permaflour mounting media. Primary antibodies included Keratin-5 (1:500, Covance PRB-160P), Ki67 (1:500, Dako MIB-1), MAC387 (Anti-S100A9/Calprotectin antibody, 1:200, Abcam ab22506), and von Willebrand factor (1:500, Dako, A0082), CRABP1 (1:500, Cell signaling 13163).
Immunohistochemistry whole slide imaging analysis was performed using HistomicsTK python toolkit66 and QuPath67 using count_image, count_slide, and PositivePixelCount functions as part of the positive pixel count module. Nuclear localization was performed using the ‘Nuclei Segmentation’ module in HistomicsTK. Image analysis was restricted to cells in the dermis by manually defining and masking epithelial regions of interest prior to morphological reconstruction. To survey the entirety of the wound, smaller ROIs sampling the entire wound were batch-processed with optimized HSI color parameters (specified in template_params).
Imaging and HF quantification
Fluorescence images were photographed using a Zeiss Axio Observer Fluorescent Microscope and full tissue section fluorescent and brightfield images were photographed at 20X magnification using an Olympus BX61 virtual slide scanning system. For the HF quantification, data was collected from six animals and for each tissue sample the counts were repeated on a minimum of three sections. Follicular density was calculated as the number of follicles per mm of tissue and was compared between the normal skin, the burn scar and the excision scar of the distal antler and back using the Kruskal-Wallis test with the Dunn’s post-test.
Gross wound measurements
To assess wound contraction, surgical staples were applied to the skin on either side of each created wound and digital images of the wound and a ruler were captured of the wound at day 0 and day 35. Morphometric software (Image J) was utilized to measure the distance between the surgical staples at Day 0 compared with Day 35 for each wound. The percentage of wound contraction was then calculated as the difference between these 2 staples at day 30 compared with day 0 for each wound, then dividing by the original wound size (12mm).
Reference genome generation and RNA-/ATAC-Seq read alignment
Cell Ranger V.3.0.1 pipeline was used to process scRNA-Seq datasets generated using the 10x Chromium platform (support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest). Cell Ranger ATAC V.1.2.0 pipeline was used to process scATAC-Seq datasets. Since a high-quality reindeer genome was not available, sequencing reads were mapped to the bovine (Bos taurus) genome. To understand the extent of information loss that either reduce power for assessing transcriptional changes (unmapped reads) or limit generalizability of identified programs (no corresponding orthologs between bovine and human genomes), we quantified confidently mapped reads to the bovine genome, transcriptome, and percentage of DEG signatures that have bovine–human orthologs. While ~33% of reads did not map contiguously to the bovine reference, the bigger bottleneck was that ~39% of genome-aligned reads did not map to annotated protein-coding genes (Figure S2A–B). Briefly, STAR-compatible FASTA and Gene transfer format (GTF) files for all 30 chromosomes (ncbi.nlm.nih.gov/assembly/GCF_002263795.1/) were pre-filtered with cellranger and cellranger-atac mkgtf and then indexed with cellranger and cellranger-atac mkref. Following this step, FASTQs were aligned to this custom reference using cellranger-atac and cellranger count and multi-sample aggregation was performed using cellranger and cellranger-atac aggr to normalize mapped reads across libraries. Genome generation and alignment logs are available from GitHub (github.com/BiernaskieLab).
Bioinformatics analysis of single-cell RNA-Seq datasets
Gene-barcode matrix was imported into Seurat v321 for quality control, dimensionality reduction, mutual nearest neighbors-based integration, cell clustering, and differential expression analysis. Unsupervised clustering was performed using the original Louvain algorithm implemented in FindNeighbors and FindClusters functions in Seurat. Preprocessed back and velvet datasets were integrated using FindIntegrationAnchors and functional fibroblast states were annotated based on sample composition of unsupervised subclusters; classified as ‘pro-regenerative’ or ‘pro-fibrotic’ if >70% of fibroblasts in that cluster originated from Day 0 velvet or Day 0 back (Figure S3c–e), respectively. Overlap between reindeer and human fibroblast subsets was calculated by running the cor function on average expression of all genes (taken from “RNA” slot of the Seurat object) calculated using the AverageExpression function. The evolving immune cell composition was plotted using ggplot2 v.3.3.2 and compared across back and velvet using the chisq.test (chi square) function. Tests of equivalence were employed to assess differences in pseudotime distributions (Figure 5c, d, h, j, l) using the ‘two-one-sided t-tests’ (TOST) as implemented in TOSTtwo.raw function in TOSTER R package 68 (Supplementary Figure 5d). The degree of overlap across back and velvet pseudotime distributions were estimated using the overlap coefficient estimator overlapEst in overlap R package 69.
Systematic pooling of evidence across rodent, reindeer, and human models of regenerative and fibrotic wound healing was performed to identify transcriptional networks conserved across all three species and those that exhibited specie-specific activation. For rodent healing, we used our previously published scRNA-Seq datasets (GEO: GSE108677) capturing fibroblasts from central and peripheral domains of large excision wounds 14 DPW 8. For human healing, we used Iglesias-Bartolome et al.’s bulk transcriptomics analysis of human oral and cutaneous wound healing (GEO: GSE97615) profiled at 0 (baseline), 3, and 6 DPW. For each comparison, difference between normalized means approximated effect size and standard error approximated variance. Forest plots were generated using Nightingale’s R package “ggforestplot” (v0.1.0). Fibroblast inflammatory features that exhibit consensus enrichment in fibrotic healing across all three species were nominated for functional validation using rodent WIHN model.
Inferring cell–cell communication
Differential fibroblast to immune cell signaling between back and velvet across homeostatic and healing time course was reconstructed using the Connectome R toolkit v0.2.2 developed by Niklason Lab52. Briefly, DifferentialConnectome function was used to query four separate Seurat v3 objects, each housing time-matched back-velvet integrated datasets, to define nodes and edges for network analysis. A ‘perturbation score’ was calculated for each edge by taking the product of absolute ligand log FC and receptor log FC and only edges with a minimum perturbation score of 0.25 were retained. Differential edge list was passed through CircosDiff (a wrapper around the R package circlize) to filter receptor ligand pairs and generate Circos plots. Longitudinal connectomes on Reindeer Atlas were generated by defining the Longitudinal function which first subsetted the ligand and receptor of interest and then scaled its expression across timepoints to reveal changes in ligand and receptor trends over time. To interrogate community influence of fibroblasts at baseline and across the healing time course, outgoing centrality metric was quantified using the Kleinberg hub score (derived from cumulative outgoing edge weight for each cell) for ligands annotated as CXCL and Vasoactive. Baseline hub scores of human skin 33 and ectopic velvet to back skin graft fibroblasts were calculated separately and co-plotted as dashed lines alongside velvet and back skin fibroblasts. The full list of context-dependent crosstalk, as well as Kleinberg hub and authority profiles of all cell types, can be visualized on Reindeer Atlas (http://biernaskielab.ca/reindeer_atlas).
Differences in intra-cluster fibroblast signaling during regenerative-to-inflammatory fibroblast transitions in ectopic grafts were characterized using NICHES 54. Briefly, ALRA-imputed gene expression data was used as input for RunNICHES (CellToSystem argument set to TRUE) to calculate summed output of fibroblast state-specific signaling edges to every other cell type treated as a single sink 70. CellToSystem object was isolated, embedded using UMAP, and annotated by scVelo-based assignments to visualize evolution in fibroblasts outgoing signals. To ask which cell types received fibroblast signals, ligand-receptor signatures for all fibroblast states were calculated using FindAllMarkers (using ROC test) and their average expression levels scored using AddModuleScore.
Gene Regulatory Network inference
Single cell regulatory network inference and clustering (SCENIC) was used to infer transcription factor (TF) networks active in reindeer fibroblasts26. Analysis was performed using recommended parameters (https://github.com/aertslab/SCENIC) using the hg19 RcisTarget database. Violin plot showing differential AUC score distribution across conditions were plotted with ggplot2 v.3.1.1 using the regulon activity matrix (‘3.4_regulonAUC.Rds’, an output of the SCENIC workflow) in which columns represent cells and rows the AUC regulon activity. The complete set of regulons (across all comparisons described) can be queried on Reindeer Atlas (http://biernaskielab.ca/reindeer_atlas). To interrogate the regulatory architecture of differentially activated TFs, SCENIC pipeline was run separately on datasets being compared to maximize detection of discrepant GENIE3 modules. The top five most correlated targets from each gene set is displayed (Figure 4d–f). Magnitude of regulon activity change (natural log FC of average regulon AUC) and statistical significance (adjusted p-value, Bonferroni corrected) were plotted as volcano plots using EnhancedVolcano v1.4.0 (Supplemental Figure 5a,c,e,g). Regulon density, a surrogate indicator of SCENIC-inferred regulatory fibroblast state stability, was calculated using 2D binned kernel density estimate (using the bkde2D function in KernSmooth R package) and by overlaying a contour plot on SCENIC-calculated tSNE projections. Since kernel density estimates approximate the underlying probability density function, the values to which the colour legend is scaled to are arbitrary and are best interpreted as likelihood of detecting stable fibroblast states ranging on a spectrum from low (yellow), medium (orange) to high (brown). To determine essential regulators of back and velvet fibroblasts, Regulon Specificity Score (RSS) was calculated (by reimplementing pySCENIC’s regulon_specificity_scores function in R71 and plotted as a rank ordered scatter plot using ggplot2 v.3.3.2 (Supplemental Figure 5b,d,f,h).
Reconstructing fibroblast dynamics during healing
To analyze fibroblast transitions during healing, a Seurat object containing only the fibroblasts was exported as an H5AD file using the SaveH5Seurat function. Velocity estimations were derived from scVelo’s likelihood-based dynamical model which resolves directional differentiation trajectories using ratios of spliced mRNA to un-spliced pre-mRNAs39 https://scvelo.org/). First and second order vector moments were calculated by preprocessing the matrix using pp.filter_and_normalize (min_shared_counts=20, n_top_genes=2000) and running pp.moments (n_pcs=30, n_neighbors=30) and tl.velocity (mode=‘dynamical’). Differential velocity expression was calculated using scv.tl.rank_velocity_genes which employed differential velocity t-tests to generate tissue- and cluster-specific gene ranking list (Table S2). Vector field topology, including the acceleration field landscape, was learned using Dynamo with default and recommended settings53. Fibroblast fate commitments were animated using Dynamo-reconstructed vector fields and accompanying attractor points were overlaid to highlight stable nodes driving terminal fibroblast fates. Unsupervised fate map of wound-activated fibroblasts was plotted after identifying initial and terminal states using CellRank46. Smoothened expression of lineage drivers predicted to co-vary with back skin and velvet fibroblasts latent time were plotted using the cr.pl.gene_trends function in CellRank. Anchor genes distinguishing regenerative (CRABP1) versus myofibroblastic (ACTA2) states were visualized using the Nebulosa R package72.
Cross-species fibroblast state classification
To assess whether fibroblast states identified in reindeer velvet and backskin are conserved in human skin, we first employed the feature selection approach implemented in scPred 37 and used top 50 class-informative principal component (PCs) that each explain at least 0.01% of the variance to train three different prediction models. Briefly, discriminant feature space was generated using getFeatureSpace() where class-informative PCs explaining variance between velvet-enriched (‘Pro-regenerative’), backskin-enriched (‘Pro-inflammatory’), and shared (‘Mixed’) fibroblast states were shortlisted. Classifiers for the three fibroblast states were trained using trainModel() using Support Vector Machines with Radial Basis Function Kernel (model = svmRadial), Model Averaged Neural Network (model = avNNet) and Random Forest (model = ranger) and are available via our Figshare repository. Training probabilities for fibroblast states in the reindeer dataset were evaluated using get_scpred() and plotted using plot_probabilities(). Human fetal and adult fibroblast classification were classified and Harmony-integrated using scPredict() with default probability threshold.
To generate an interpretable (geneset-based) definitation of fibroblast states that generalize across reindeer and human fibroblasts, Garnett (https://cole-trapnell-lab.github.io/garnett/), an automated cell type/state classification tool was employed 35. First, the classifier was trained to distinguish ‘pro-regenerative’, ‘pro-fibrotic’, and ‘mixed’ reindeer fibroblast states (Figure S3K–L) based on Seurat- and Garnett-calculated gene signatures provided in a hierarchical markup file. Second, the classifier was applied to fetal and adult human dermal fibroblast datasets. Garnett’s check_markers() and plot_markers() were used to calculate and visualize ambiguity statistics for all candidate markers, respectively. Garnett’s train_cell_classifier() was used to train a multinomial fibroblast classifier. The trained classifier was first validated by classifying cells in the same reindeer dataset. Once the classifier exhibited high discriminatory capacity after iterative training on reindeer dataset, it was applied to classify fibroblast states (using classify_cells()) in fetal and adult human dermis. Garnett-predicted fibroblast states (stored in cluster_ext_type column) were plotted using a stacked bar plot. Fibroblast state definitions (or ‘marker_file’) is available via GitHub (github.com/BiernaskieLab).
Bioinformatics analysis of bulk RNA-Seq datasets
For bulk RNA-sequencing analysis, a bowtie-2 reference was constructed, based on the same .gtf file noted above. The file was reduced to only protein coding transcripts, and where a gene_name field was present, the gene_id was replaced with the gene_name. Genomic sequences were downloaded from NCBI (GCF_002263795.1_ARS-UCD1.2), and the 30 primary chromosomes were extracted (i.e., scaffolds and the mitochondrial genome were omitted). The reference was built using the rsem-prepare-reference tool from RSEM v1.3.073, and bowtie2 v2.3.4.174. Fastq files were trimmed and filtered using an in-house algorithm. RNA-Seq expression estimation was performed by RSEM v1.3.0 (parameters: seed-length=20, no-qualities, bowtie2-k=200, bowtie2-sensitivity-level=sensitive)73. with bowtie2 v2.3.4.1 for the alignment step74. Six of the 34 samples were rejected for having too few aligned reads. Differential Expression analysis was performed using the EBSeq package v1.26.075 in R v3.6.2. Principal component analysis (PCA) was performed on day-matched back and velvet normalized counts using pca and biplot functions in PCAtools R package. Bulk-seq measurements were integrated with scRNA-Seq by hierarchically clustering cell type-specific DEGs and displaying scaled expression values using a heatmap generated with pheatmap R package.
Bioinformatics analysis of single-cell ATAC-Seq datasets
Cell Ranger ATAC generated filtered peak-barcode matrix and fragment files were imported into Signac v1.3.021. bosTau9 genome annotations (distributed as a biostrings object in the BSgenome.Btaurus.UCSC.bosTau9 package) was used. Quality control was performed using nucleosome banding pattern (nucleosome_signal < 4 retained), ratio of fragments centered around transcriptional start site TSS (>2 retained), percentage of reads in peaks (> 15 retained), and fraction of fragments in peaks. Since ENCODE project-generated blacklist regions are not provided for the Bos taurus genome, this QC metric was not computed. Post-QC peak-barcode matrix was normalized and then dimensionality reduced using latent semantic indexing. Gene activity (inferred by counting fragments that fall within gene coordinates plus 2 kb upstream) for known fibroblast signatures and label classification scores following integration with scRNA-seq datasets housing all cells across all 4 healing time points were jointly used to identify bona fide fibroblasts. Differentially accessible fibroblast peaks across samples and timepoints were identified using logistic regression as implemented in Seurat’s FindMarkers function. Genome-wide co-accessible (cis-regulatory) networks were calculated using run_cicero, a Cicero wrapper that generates co-accessibility scores 76. DNA accessibility and cis-regulome were jointly visualized with Signac’s CoveragePlot and genome-wise accessibility can be queried on Reindeer Atlas (http://biernaskielab.ca/reindeer_atlas).
Human Protein Atlas corroboration
Immunoperoxidase stainings of differentially expressed genes between ‘pro-fibrotic’ reindeer plus human fibroblasts versus ‘pro-regenerative’ reindeer fibroblasts were mined from the Human Protein Atlas v.19.3 34 (Supplementary Figure 4).
Reindeer Atlas
A web portal housing reindeer dataset (http://www.biernaskielab.ca/reindeer_atlas) was created using RShiny (v1.1.0), shinyLP (v.1.1.2), shinyWidgets (v.0.5.3), and shinythemes (v.1.1.2) packages.
Genetic demultiplexing of pooled single-cell RNA-seq datasets
Single-cell RNA-seq libraries were generated by multiplexing n = 3 biological replicates to lower sample-specific batch variation and library preparation costs. Sample identify of each cell was computational demultiplexed based on natural genetic variation using souporcell (v2) 23. Briefly, souporcell analyses were performed with the number of clusters set to k = 3 and without the use of ‘common_variants’ or ‘skip_remap’ options as species-specific variants are not available for Rangifer tarandus or Bos taurus genome. Genotype assignments from step 4 ‘Clustering cells by genotype’ were imported into Seurat object’s metadata matrix and are made accessible via authors’ figshare archive. Demultiplexed data was visualized with box plots that include a line across the box, upper hinge, and lower hinge which represent median, 75th percentile (Q3), and 25th percentile (Q1), respectively. The upper whisker extends from the hinge to the largest value no further than Q3 + 1.5× interquartile range (IQR). The lower whisker extends from the hinge to the smallest value at most Q1 – 1.5 * IQR.
Quantitative analysis of matrisome structure
Collagen birefringence was assessed using liquid crystal-based polarization microscopy. Formalin-fixed, paraffin-embedded and picrosirius red–stained skin sections of velvet and back skin were imaged on a Zeiss Axioplan 2 (Cambridge Research Instruments LC-PolScope) using a 5x objective. Microscope settings and calibration were optimized against an empty background that did not contain a birefringent specimen in the viewing field as per recommended protocol77. The density and architectural arrangement of collagen fibers was evaluated using Picrosirius red-stained sections of back skin and velvet at baseline and 30–35 dpw (n=3 animal-matched back and velvet samples). Briefly, sections were imaged using LC-PolScope and pseudocolored by combining retardance (brightness) and azimuth orientation (primary RGB colors). Composite images were processed to isolate ECM fibers, which were noise-reduced and then binarized to digitally resolve thousands of fibers and branchpoints as done previously18,61. Dimensionality reduction of 294 quantified fiber network properties was performed using the umap function in the M3C (v1.12.0) R library.
High Performance Liquid Chromatography (HPLC) and Mass Spectrometry (MS)
Liquid chromatography and mass spectrometry experiments were carried out by the Southern Alberta Mass Spectrometry (SAMS) core facility at the University of Calgary, Canada. Samples were analyzed by the Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Scientific) operated with Xcalibur (version 4.0.21.10) and coupled to a Thermo Scientific Easy-nLC (nanoflow Liquid Chromatography) 1200 system. A total of 2 μg of tryptic peptides were loaded onto a C18 trap (75 um × 2 cm; Acclaim PepMap 100, P/N 164946; ThermoScientific) at a flow rate of 2 μl/min of solvent A (0.1% formic acid and 3% acetonitrile in LC-MS grade water). Peptides were eluted using a 120 min gradient from 5 to 40% (5% to 28% in 105 min followed by an increase to 40% B in 15 min) of solvent B (0.1% formic acid in 80% LC-MS grade acetonitrile) at a flow rate of 0.3 μL/min and separated on a C18 analytical column (75 um × 50 cm; PepMap RSLC C18; P/N ES803; Thermo Scientific). Peptides were then electrosprayed using 2.3 kV voltage into the ion transfer tube (300°C) of the Orbitrap Lumos operating in positive mode. The Orbitrap first performed a full MS scan at a resolution of 120,000 FWHM to detect the precursor ion having a m/z between 375 and 1575 and a +2 to +7 charge. The Orbitrap AGC (Auto Gain Control) and the maximum injection time were set at 4e5 and 50 ms, respectively. The Orbitrap was operated using the top speed mode with a 3 sec cycle time for precursor selection. The most intense precursor ions presenting a peptidic isotopic profile and having an intensity threshold of at least 5000 were isolated using the quadrupole and fragmented with HCD (30% collision energy) in the ion routing multipole. The fragment ions (MS2) were analyzed in the ion trap at a rapid scan rate. The AGC and the maximum injection time were set at 1 × 104 and 35 ms, respectively, for the ion trap. Dynamic exclusion was enabled for 45 sec to avoid of the acquisition of same precursor ion having a similar m/z (plus or minus 10 ppm).
Relative quantification and bioinformatics for proteomic analysis
Spectral data were matched to peptide sequences in the bovine UniProt protein database (reviewed, unreviewed, canonical and isoforms) using the Andromeda algorithm 78 as implemented in the MaxQuant 79 software package v.1.6.0.1. For the peptide-spectrum match, a false discovery rate (FDR) of 1% was applied. Search parameters included a mass tolerance of 10 p.p.m. for the parent ion, 0.5 Da for the fragment ion, carbamidomethylation of cysteine residues (+57.021464 Da), variable N-terminal modification by acetylation (+42.010565 Da), and variable methionine oxidation (+15.994915 Da). N-terminal and lysine heavy (+34.063116 Da) and light (+28.031300 Da) dimethylation were defined as labels for relative quantification. The cleavage site specificity was set to Trypsin/P (search for free N-terminus and for only lysines), with up to two missed cleavages allowed. Quantified proteins were analyzed using the DEP package80 and R language (v4.0.3). Proteins identified as “reverse” and “potential contaminant” were removed. Proteins with at least 2 quantification values in one of the groups were selected. Normalization was also performed with the DEP package, which utilizes the variance stabilizing normalization (vsn)81. Imputation was performed following minimal value centered around a Gaussian distribution (q = 0.01). In parallel, significant outlier cutoff values were determined by boxplot-and-whiskers analysis using the BoxPlotR approach82.
ADDITIONAL RESOURCES
Single-cell datasets are accessible through an interactive portal at: www.biernaskielab.ca/reindeer_atlas.
Supplementary Material
Figure S1 – Velvet regeneration following full-thickness excision or burn injury and maintenance of velvet regenerative capacity following short-term transplant to non-regenerative back skin, Related to Figure 1. (A-B) Unwounded H&E-stained back skin (A) and velvet (B). Scale bars are 500μm. (C-D) Skin sections from 30dpw on back (C) and velvet (D) stained for Keratin-5 (red) and Ki67 (green). (E-F) Representative staining (E) and quantification (F) of Ki67+ cells in interfollicular dermis (IFD) and epidermis (IFE) 30dpw on back and velvet. (G-H) Gross images of full thickness thermal injury in back (G) and velvet (H) on Day 0, 30, and 60 days after injury. White dashed lines indicate original wound area. (I-J) Brightfield image of skin sections from back (I) and velvet (J) at 30d after thermal injury. Dashed lines (black) indicate wound margin and colored dashed lines indicate high magnification insets. (K) Quantification of HF density restoration within burn wounds. Data are mean±SEM. (L) Ventral view of velvet at 3dpw confirming full-thickness thermal damage. (M-N) H&E-stained sections of back skin (M) and velvet (N) wounds at 60dpw. Black dashed lines indicate wound margins. (O) Example of an unwounded ectopic velvet graft at 60d post-grafting. (P) H&E-stained section showing velvet-to-back skin graft. Dashed lines indicate margin between velvet and back skin. Note distinct differences in HF morphology between the two skin types. (Q) Ectopic velvet grafts from two additional animals 30dpw. Original wound sites are marked by blue dashed lines. Velvet graft margins are marked by white dashed lines. Scale bars are 100μm or as indicated.
Figure S2 – Alignment metrics, cell type annotations, and integrating bulk- and single-cell measurements to elucidate reproducible site-specific gene programs, Related to Figure 2. (A) Alignment metrics of scRNA-Seq reads mapped to a custom single-cell Bos taurus reference. (B) Bar plots showing the distribution of sequencing reads that confidently map to the bovine genome and transcriptome. Horizontal dotted lines represent average mapping of rodent reads to GRCm38/mm10 assembly. Box plots represent median, Q3, Q1. (C) Violin plots showing normalized expression levels of top three marker genes across the ten annotated cell types in Figure 2A. (D) Boxplots showing relative composition of macrophage and CD45+IL1B+ myeloid cells at baseline in n=3 biological replicates colored by harvest site. P-values are shown for both comparisons. (E) Schematic for dissecting reproducible transcriptional differences across velvet and back skin by combining bulk- and sc-RNA sequencing. (F) Principal component analysis of bulk back skin (cyan circle) and velvet (red circle) gene expression profiles at 0, 3, 7, 14dpw. (G) Hierarchical clustering of differentially expressed fibroblast genes at pre-injury baseline, scaled by row to display relative changes across samples. (H-I) Hierarchical clustering of fibroblast-derived ligands pre-injury (H) and immune cellderived ligands at 7dpw (I), scaled by row to display relative changes across samples. Cellular source predicted by sc-RNA-Seq, gene expression changes shown on each heatmap corroborated by bulk-RNA-Seq (G, H, I).
Figure S3 – Cross-species validation of pro-fibrotic and pro-regenerative fibroblast transcriptional programs, Related to Figure 3. (A) Shared fibrotic signaling across reindeer and human fibroblasts. (B) UMAP projection of 8,320 fibroblasts integrated across species (4,160 human cells; 4,160 reindeer), colored by fibroblast states. (C) Comparison of human 33 and reindeer fibroblast states using Pearson correlation coefficient. (D-H) Immunoperoxidase staining of human skin and violin plots showing normalized expression for genes enriched in scar-forming (human and reindeer back skin) fibroblasts (D-F) and genes enriched in regenerative (reindeer velvet) fibroblasts (F-G). Adjusted p-values calculated using two-sided nonparametric Wilcoxon rank-sum test with Bonferroni correction. Stainings obtained from the Human Protein Atlas. Scale bars represent 100 μm (D-H). (I) Schematic illustrating cross-species comparison between reindeer and human skin. (J and K) Stacked bar plots depicting frequency of CRABP1+ fibroblasts in seven fetal (J) and five adult (K) human skin samples. (L) Schematic of Garnett-based machine learning classifier trained to distinguish reindeer velvet-enriched (‘Pro-regenerative’), backskin-enriched (‘Pro-inflammatory’), or shared (‘Mixed’) fibroblast states and then tested on fetal and adult human skin fibroblasts. (M, N) UMAPs of subclustered reindeer (M) and human (N) resting fibroblasts used for training and testing the Garnett classifier, respectively. UMAPs coloured by tissue origin and Garnett-predicted fibroblast states. (O) Stacked bar plot depicting Garnett-predicted fibroblast state composition in adult and fetal human skin. Scale bars represent 100 μm (D-H).
Figure S4– Transcriptional and regulatory drivers of regenerative and fibrotic fibroblast fates, Related to Figure 4. (A) Streamlines showing RNA velocity field on a UMAP projection of fibroblasts colored by Louvain cluster IDs. (B) Absorption probability of velvet and back skin fibroblasts from 7 to 14dpw. Color intensity represents degree of fate priming. (C-D) Fate maps leading to inferred terminal states in velvet (C) and back skin (D). (E) Inferred differentiation potential. Terminal commitment is shown in dark purple. (F, G) Phase portrait, RNA velocity and gene expression plots of differentially spliced genes driving velvet (F) and back skin (G) healing. (H) Venn diagrams displaying overlap between ground state fibroblast DEGs with 3, 7, and 14dpw velvet and back skin fibroblasts. (I) Hierarchical clustering of tissue and time-point separated fibroblasts based on gene regulatory network activity. (J) Gene regulatory network activity of HDAC2, LEF1, NFKB1 in velvet (red) and back (turquoise) fibroblasts quantified as area under the recovery curve (AUC) over time. (K) Volcano plot of Wilcoxon rank-sum determined differentially activated regulons in velvet (red) and back (cyan) at 3, 7 and 14dpw. (L) Rank plot of fibroblast regulons at 3, 7 and 14dpw ordered by Regulon Specificity Score (RSS) where higher RSS indicates greater regional (back or velvet) specificity. Top regulons are labelled in either velvet (red) or back skin (cyan).
Figure S5 – Discrepant immune response and altered fibroblast-immune signaling during regenerative versus fibrotic healing, Related to Figure 5. (A) UMAP projection of all cells from uninjured and healing back and velvet wounds. (B) Global perturbation scores from 7 and 14dpw. (C) Circos plots showing differential fibroblast secretome-driven interactions with immune cells in velvet and back skin wounds at 0, 3, 7, and 14dpw. (D) Schematic depicting morphologic 64 and molecular 50 stages during neutrophil maturation used for reference mapping reindeer neutrophils during healing. (E) Subclustered neutrophil UMAP overlaid with velocyto vector fields and stacked bar plot depicting neutrophil cluster composition in velvet and back skin wounds at 3 and 7dpw. Neutrophil cluster enriched in 3dpw back skin is outlined with dash line. (F) UMAP and density plot depicting pseudotemporal ordering of neutrophils from velvet and back skin wounds at 3 and 7dpw. (G) Boxplot showing percentage of ‘immature’ neutrophils (Stages G0-G350) recruited to back or velvet wounds at 3 and 7dpw. n=3 biological replicates at each time point. Bonferroni adjusted P-value is shown. (H) Subclustered T cell UMAP overlaid with velocyto vector fields and stacked bar plot depicting T cell cluster composition in velvet and back skin wounds at 3 and 7dpw. (I) UMAP and kernel density distribution depicting pseudotemporal ordering of T cells from velvet and back skin wound beds at 3 and 7dpw. (J-K) Analysis of T cell dynamics following co-culture with back or velvet fibroblasts. (J) Subclustered T cell UMAP overlaid with velocyto vector fields colored by Louvain cluster ID and pseudotemporal ordering of T cell. (K) Sample separated UMAPs overlaid with density contour plots, demonstrating minimal difference in T cell diversification following exposure to back skin versus velvet fibroblasts.
Figure S6 – Differential revascularization and long-term histological evaluation following pharmacological perturbation of fibroblast programming, Related to Figure 6. (A) Kleinberg hub score, an indicator of community influence via a specific set of signaling molecules, of back and velvet fibroblasts for CXCL ligands at 0, 3, 7, 14dpw. Dashed lines indicate baseline hub scores of human fibroblasts profiled in Solé-Boldo et al. 2020. (B) Percentage of endothelial and smooth muscle cells within wounds over time. (C-D) Immunostaining of von Willebrand factor (green; endothelial cells) and MAC384 (red; macrophages/neutrophils) in back skin (C) and velvet (D) at 14dpw. (E) Experimental timeline for assessing long-term impact of recapitulating the back skin fibroblast signaling milieu within velvet wounds. (F-I) Images of H&E-stained sections of velvet wounds at 42dpw after receiving intradermal application of PBS (F), DMSO (G), PLAU (H), or iMDK (I). Dashed white lines indicate wound margins. Scale bars are 500μm. (J) UMAP plot visualizing dermal ECM ultrastructural properties and representative pseudocolored images of wound sections. (K) Treatment-separated density contour plots of ECM ultrastructure from each condition.
Figure S7 – Remodeling of ectopic velvet grafts and pro-regenerative impact of CSF1 neutralization during rodent healing, Related to Figure 7. (A) Schematic of autologous velvet-to-back grafting and subsequent wounding. (B) H&E-stained excision wound made on an ectopic velvet graft 6 months post-grafting (representative of n=5 grafts quantified in Figure 7B). (C) Insets showing HF density and dermal architecture of ectopic velvet graft pre- and post-wounding. (D-E) UMAPs visualizing dermal ECM ultrastructural properties of images of wound sections from baseline (uninjured) back skin, 3-month-old ectopic velvet and 6-month-old ectopic velvet. (E) Sample-separated density contour plots of ECM ultrastructural properties. (F) A summary of fibroblast evolution in ectopic grafts based on velocity vector fields and changes in fibroblast cluster composition. (G) Kernel density estimates depicting activation of mechanosensitive genes EN1, YAP1 and its binding partner TEAD1 within transitionary fibroblasts. (H) Relative contribution of macrophage clusters at different graft timepoints. (I) Inflammatory fibroblasts’ interacting partners using CSF1/CSF1R- and CXCL12/CXCR4-signaling axis. (J) Top five positively regulated signaling pathways enriched in fibroblast states that were not acquired in ectopic grafts at any of the surveyed time points. (K-L) Distribution of body weights (K) and vehicle and PLX5622 CSF1R inhibitor chow consumption per cage in male mice (L) from 2 days pre-wound until 6dpw. (M) Gross examination of wound closure between 2 to 13dpw in male mice.
Data S1 – Consensus DEGs differentially expressed in reindeer velvet and fetal human fibroblasts relative to reindeer back skin and adult human fibroblasts, related to Figures 2 and 3. Genes with log-fold-change less than 0.25 or Wilcoxon rank-sum-determined two-sided p-value > 0.05 were filtered out.
Data S2 – Differentially spliced genes identified using RNA velocity grouped by tissue (velvet versus back skin) and fibroblasts subsets (Clusters 0–14), related to Figure 2.
Data S3 – Magnitude of gene expression change and frequency of fibroblasts expressing regeneration-associated markers CRABP1, MDK and SPARC, related to Figure 4.
Data S4 – A set of core DEGs distinguishing velvet and back skin fibroblasts at ground state that were differentially maintained in wound-activated fibroblasts, related to Figures 3 and 4.
Data S5 – Transcriptional signature shared between uninjured velvet and fetal human fibroblasts that are differentially maintained over the healing trajectory relative to reindeer backskin, related to Figure 3. Genes with log-fold-change less than 0.25 or Wilcoxon rank-sum-determined two-sided p-value > 0.05 were filtered out.
Data S6 – List of cell-cell signaling edges originating either from immune and vasculature-associated cells (tabs 1–4) or fibroblasts (tabs 5-8), related to Figures 5 and 6.
Data S7 – Fibroblast signaling mechanisms associated with regenerative-to-inflammatory transition in ectopic grafts, related to Figures 7 and S7. Differential ligand-receptor mechanisms were identified using ROC analysis and the table lists putative mechanisms and their classification power ranging from 0 (random) to 1 (perfect classification).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-Keratin5 | Covance or Biolegend | Cat#: 905501 or PRB- 160P; RRID:AB_2565050 |
| Mouse anti-Ki67 | Dako | Cat# M7240, RRID:AB_2142367 |
| Mouse Anti-S100A9/Calprotectin | Abcam | Cat# ab22506, RRID:AB_447111 |
| Rabbit Anti-Von Willebrand Factor | Dako | Cat# A0082, RRID:AB_2315602 |
| Rabbit Anti-CRABP1 | Cell signaling | Cat#13163 RRID: AB_2750569 |
| Anti-mouse Alexa 488 secondary antibody | Invitrogen | Cat# A32723 |
| Chemicals, peptides, and recombinant proteins | ||
| Serum-free DMEM (low glucose, with LGlutamine) | Gibco™ | 11885084 |
| HBSS | Gibco™ | 14175–095 |
| L-Glutamine | Gibco™ | 25030081 |
| Fetal Bovine Serum | Gibco™ | 12483020 |
| Penicillin-streptomycin | Gibco™ | 15070063 |
| Fungizone | Gibco™ | 15290018 |
| Collagenase IV | Sigma-Aldrich | C5138 |
| Dispase 5 | StemCell Technologies | 07913 |
| Medetomidine 10mg/ml | Alberta Veterinary Laboratories (Avetlabs) |
N/A |
| Azaperone | Chiron | 13849.19 |
| Optixcare eye lube | Aventix Animal Health | OPX-4242 |
| Lidocaine HCI Injection with Preservative (2%) | Teligent Canada | DIN: 02422026, 0127AJ01 |
| Meloxicam | Boehringer Ingelheim | DIN: 02330059 |
| Eprinomectin | Merial (Boehringer Ingelheim) | DIN: 02450798 |
| Atipamezole 10mg/ml | Alberta Veterinary Laboratories (Avetlabs) |
N/A |
| Paraformaldehyde (PFA) | Sigma-Aldrich | 441244 |
| Clear Frozen Section Compound | VWR | 95057–838 |
| Swat Original Fly Repellent Ointment | Farnam | 100532426 |
| FluoSpheres™ Carboxylate-Modified Microspheres, 0.2 μm, dark red fluorescent (660/680), 2% solids | ThermoFisher | F8807 |
| TGFbeta MAB (InVivoMab antimouse/human/rat/monkey/hamster/cani ne/bovine TGF-β) | Bio X Cell | BE0057–5MG-A |
| TGFbeta Isotype Control (InVivoMab mouse IgG1 isotype control) | Bio X Cell | BE0083–5MG-A |
| Bovine M-CSF (CSF-1) (Yeast-derived Recombinant Protein) | Kingfisher Biotech, Inc. | RP1353B-025 |
| Human CXCL3 (GRO gamma) (Yeastderived Recombinant Protein) | Kingfisher Biotech, Inc. | RP1673H-025 |
| Bovine CCL2 (MCP-1) (Yeast-derived Recombinant Protein) | Kingfisher Biotech, Inc. | RP0027B-025 |
| Prostaglandin D2 (PGD2) | Cayman Chemical | 12010–25 |
| Midkine Inhibitor, iMDK | MilliporeSigma | 5.08052.0001 |
| Recombinant Human u-Plasminogen Activator (Urokinase) |
BioLegend | 755304 |
| PBS | Thermo Fisher Scientific | 10010–049 |
| Control chow | Plexxikon Inc | AIN-76A, D10001i |
| CSF1R inhibitor | Plexxikon Inc | PLX5622, D11100404i |
| CXCR4 antagonist (Plerixafor) | Tocris Bioscience | 3299 |
| Dimethyl sulfoxide (DMSO) anhydrous, ≥99.9% |
Sigma-Aldrich | 276855 |
| Normal Goat Serum | Jackson ImmunoResearch | 005–000-121 |
| Triton X-100 | Sigma-Aldrich | X100 |
| Hoechst 33258 | Sigma-Aldrich, Invitrogen | 14530, H3570 |
| Permafluor mounting media | Thermo Scientific | ta030FM |
| Bovine Albumin Serum(BSA) Solution | Sigma-Aldrich | 19576 |
| ACK Buffer | Stemcell Technologies | 07800 |
| RNAlater | Thermofisher | AM7024 |
| TRIzol™ | Life Technologies | 15596026 |
| RNeasy | Qiagen | 74104 |
| Xylene | VWR | 89370–088 |
| 3,3′-Diaminobenzidine (DAB) | Vector Laboratories | SK-4105 |
| Elite ABC system | Vector Laboratories | PK-6100 |
| Avidin/Biotin Blocking Kit | Vector Laboratories | SP-2001 |
| Hematoxylin QS Counterstain | Vector Laboratories | H-3404–100 |
| Critical commercial assays | ||
| BD Facs Aria III | BD Biosciences | N/A |
| RNAScope 2.0 HD Detection Kit | ACDBio | N/A |
| 10X Chromium Controller | 10X Genomics | N/A |
| gentleMACS™ Octo Dissociator | Miltenyi Biotec | N/A |
| Chromium Single Cell Chip A kit, 48 rnxs | 10X Genomics | 120236 |
| Chromium Single cell 3′ Library & Gel beaded kit V3 and 3.1 Next GEM, 16 rnxs | 10X Genomics | 1000075; 1000128 |
| Chromium i7 multiplex Kit 96 rnxs | 10X Genomics | 120262 |
| Chromium Chip E Single Cell ATAC kit | 10X Genomics | 1000086 |
| Chromium Single Cell ATAC library & Gel Bead Kit V1 | 10X Genomics | 1000111 |
| Chromium i7 multiplex Kit N, Set A 96 rnxs | 10X Genomics | 1000084 |
| Illumina NovaSeq SP, S1, S2 Flowcells | Illumina, Centre for Health Genomics and Informatics, University of Calgary |
N/A |
| Qubit Fluorometer | Life Technologies | N/A |
| Corning® Transwell® polycarbonate membrane cell culture inserts | Corning® | CLS3415 |
| Deposited data | ||
| Reindeer scRNA-Seq Wound datasets | This paper | GEO: GSE142854 (SuperSeries: GSE168748) |
| Reindeer scRNA-Seq PBMC datasets | This paper | GEO: GSE180653 (SuperSeries: GSE168748) |
| Reindeer Bulk RNA-Seq datasets | This paper | GEO: GSE168746 (SuperSeries: GSE168748) |
| Reindeer scATAC-Seq datasets | This paper | GEO: GSE176360 (SuperSeries: GSE168748) |
| Velocyto-generated LOOM files from scRNA-Seq datasets | This paper | http://doi.org/10.6084/m9.figshare.14196344 |
| Fetal and Adult Human Skin scRNA-Seq datasets | Reynolds, Vegh, Fletcher, Poyner et al. Science 202131 | https://zenodo.org/record/4536165 https://developmentcellatlas.ncl.ac.uk/datasets/hca_skin_portal/ |
| Acomys (spiny mice) versus Mus (lab mice) D0 fibroblast comparisons | GEO: GSE216723 | |
| Human skin scRNA-Seq datasets | Solé-Boldo et al. 202033 | GEO: GSE130973 |
| Human wound healing bulk-seq datasets | Iglesias-Bartolome et al., 201856 | GEO: GSE97615 |
| Experimental models: Organisms/strains | ||
| Rangifer tarandus | University of Calgary | N/A |
| Mus Musculus C57BL/6 strain | University of Calgary | N/A |
| Software and algorithms | ||
| R Studio | https://www.rstudio.com/products/rstudio/ | Version 3.6.2–4.1.0 |
| R | https://www.r-project.org/ | Version 3.6.2–4.0.5 |
| Python | https://www.python.org/ | Version 3.8.5 |
| Cell Ranger | 10X Genomics | Version 3.0.1 |
| Cell Ranger ATAC | 10X Genomics | Version 1.2.0 |
| Seurat | https://github.com/satijalab/seurat | Version 3.2.3 – 4.0.3 |
| SCENIC | https://github.com/aertslab/SCENIC | Version 1.2.4 |
| Connectome | https://github.com/msraredon/Connectome | Version 0.2.2 |
| EnhancedVolcano | https://bioconductor.org/packages/release/bioc/html/EnhancedVolcano.html | Version 1.4.0 |
| scVelo | https://github.com/theislab/scvelo | Version 0.2.3 |
| pySCENIC | https://github.com/aertslab/pySCENIC | Version 0.10.4 |
| CellRank | https://github.com/theislab/cellrank | Version 1.2.0 |
| Nebulosa | https://github.com/powellgenomicslab/Nebulosa | Version 1.0.2 |
| ggplot2 | https://cran.r-project.org/web/packages/ggplot2/index.html | Version 3.3.2; 3.1.1 |
| RSEM | https://github.com/deweylab/RSEM | Version 1.3.0, 1.2.29 |
| bowtie2 | https://github.com/BenLangmead/bowtie2 | Version 2.3.4.1 |
| Bowtie | https://bioconductor.org/packages/release/bioc/html/Rbowtie.html | Version 1 |
| EBSeq | https://www.bioconductor.org/packages/release/bioc/html/EBSeq.html | Version 1.26.0 |
| pheatmap | https://github.com/raivokolde/pheatmap | Version 1.0.12 |
| RShiny | https://github.com/rstudio/shiny | Version 1.1.0 |
| shinyLP | https://github.com/jasdumas/shinyLP | Version 1.1.2 |
| shinyWidgets | https://github.com/dreamRs/shinyWidgets | Version 0.5.3 |
| shinythemes | https://github.com/rstudio/shinythemes | Version 1.1.2 |
| CICERO | https://github.com/stjude/CICERO | Version 1.3.4.11 |
| Signac | https://github.com/timoast/signac | Version 1.2.1 |
| ggforestplot | https://nightingalehealth.github.io/ggforestplot/index.html | Version 0.1.0 |
| overlap | https://www.rdocumentation.org/packages/overlap/versions/0.3.4 | Version 0.3.4 |
| TOSTER | https://github.com/Lakens/TOSTER | Version 0.3.4 |
| PCAtools | https://github.com/kevinblighe/PCAtools | Version 2.2.0 |
| souporcell | https://github.com/wheaton5/souporcell | Version 2 |
| scPred | https://powellgenomicslab.github.io/scPred/ | Version 1.9.2 |
| NICHES | https://github.com/msraredon/NICHES | Version 0.0.2 |
| Nebulosa | https://github.com/powellgenomicslab/Nebulosa | Version 1.0.2 |
| GOplot | https://wencke.github.io/ | Version 1.0.2 |
| ggVennDiagram | https://cran.r-project.org/web/packages/ggVennDiagram/ | Version 1.0.2 |
| Image J | ImageJ | N/A |
| QuPath | https://qupath.readthedocs.io/ | Version 0.3 |
| Olympus software | Olympus | N/A |
| BSgenome.Btaurus.UCSC.bosTau9 | Bioconductor | Version 1.4.2 |
| Leica software | Leica | N/A |
| LC-Polscope software | PerkinElmer | openpolscope.org |
| BioRender | BioRender.com | |
| Other | ||
| Human Protein Atlas | https://www.proteinatlas.org/ | Version 19.3 |
| Reindeer Web Atlas | This paper | http://www.biernaskielab.ca/reindeer_atlas/ |
| GitHub links | This paper | https://github.com/BiernaskieLab |
| PDS II (polydioxanone) Suture | Ethicon Inc | Z451G |
| Sutures - 0 polypropylene (0 Prolene) | Ethicon Inc | Z467H |
Highlights.
Reindeer antler velvet regenerates after wounding whereas back skin forms a scar
Fibroblasts direct site-specific immune cell recruitment and differentiation
Fibroblast fate reversion and absence of inflammatory signals enables regeneration
Obstructing fibroblast inflammatory signals enhances skin regeneration
Acknowledgements:
We wish to thank Barbara Smith, Cynddae McGowan, Victoria Dunkley, Dr. Dean Brown, and Lauren Carvell for their expert assistance with animal handling and care. We also thank the following for their assistance: Min Cheng for bulk RNA extraction, Jessica Yoon for FACS, John Steill and Dillon Herbst for data archival, Dragana Ponjevic for histology, and the Centre for Health Genomics and Informatics (University of Calgary) for next-generation sequencing and high-performance computing. This work was funded by National Science and Engineering Research Council (RGPIN/04825-2017 to JB and RGPIN/04992-2014 to JRM), Calgary Firefighters Burn Treatment Society (JB), NIAMS-R01AR070313 (AWS), National Center for Advancing Translational Sciences, National Institutes of Health (UL1TR001998; AWS/SP) and a grant from Marv Conney (RS).
Footnotes
Declaration of Interests
The authors declare no competing interests.
Inclusion and Diversity
We support inclusive, diverse and equitable conduct of research.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Gurtner GC, Werner S, Barrandon Y, and Longaker MT (2008). Wound repair and regeneration. Nature 453, 314–321. 10.1038/nature07039. [DOI] [PubMed] [Google Scholar]
- 2.Larson BJ, Longaker MT, and Lorenz HP (2010). Scarless fetal wound healing: a basic science review. Plastic and reconstructive surgery 126, 1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore AL, Marshall CD, Barnes LA, Murphy MP, Ransom RC, and Longaker MT (2018). Scarless wound healing: Transitioning from fetal research to regenerative healing. Wiley Interdiscip Rev Dev Biol 7. 10.1002/wdev.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lo DD, Zimmermann AS, Nauta A, Longaker MT, and Lorenz HP (2012). Scarless fetal skin wound healing update. Birth Defects Res C Embryo Today 96, 237–247. 10.1002/bdrc.21018. [DOI] [PubMed] [Google Scholar]
- 5.Rowlatt U (1979). Intrauterine wound healing in a 20 week human fetus. Virchows Arch A Pathol Anat Histol 381, 353–361. 10.1007/bf00432477. [DOI] [PubMed] [Google Scholar]
- 6.Iismaa SE., Kaidonis X., Nicks AM., Bogush N., Kikuchi K., Naqvi N., Harvey RP., Husain A., and Graham RM. (2018). Comparative regenerative mechanisms across different mammalian tissues. NPJ Regenerative medicine 3, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seifert AW, Kiama SG, Seifert MG, Goheen JR, Palmer TM, and Maden M (2012). Skin shedding and tissue regeneration in African spiny mice (Acomys). Nature 489, 561–565. 10.1038/nature11499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abbasi S, Sinha S, Labit E, Rosin NL, Yoon G, Rahmani W, Jaffer A, Sharma N, Hagner A, Shah P, et al. (2020). Distinct Regulatory Programs Control the Latent Regenerative Potential of Dermal Fibroblasts during Wound Healing. Cell Stem Cell 27, 396412.e396. 10.1016/j.stem.2020.07.008. [DOI] [PubMed] [Google Scholar]
- 9.Plikus MV, Guerrero-Juarez CF, Ito M, Li YR, Dedhia PH, Zheng Y, Shao M, Gay DL, Ramos R, and Hsi T-C (2017). Regeneration of fat cells from myofibroblasts during wound healing. Science 355, 748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ito M, Yang Z, Andl T, Cui C, Kim N, Millar SE, and Cotsarelis G (2007). Wnt-dependent de novo hair follicle regeneration in adult mouse skin after wounding. Nature 447, 316–320. 10.1038/nature05766. [DOI] [PubMed] [Google Scholar]
- 11.Zomer HD, and Trentin AG (2018). Skin wound healing in humans and mice: challenges in translational research. Journal of dermatological science 90, 3–12. [DOI] [PubMed] [Google Scholar]
- 12.Foster DS, Januszyk M, Yost KE, Chinta MS, Gulati GS, Nguyen AT, Burcham AR, Salhotra A, Ransom RC, and Henn D (2021). Integrated spatial multiomics reveals fibroblast fate during tissue repair. Proceedings of the National Academy of Sciences 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosshart SP, Herz J, Vassallo BG, Hunter A, Wall MK, Badger JH, McCulloch JA, Anastasakis DG, Sarshad AA, and Leonardi I (2019). Laboratory mice born to wild mice have natural microbiota and model human immune responses. Science 365, eaaw4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Price J, Faucheux C, and Allen S (2005). Deer antlers as a model of Mammalian regeneration. Current topics in developmental biology 67, 1–48. 10.1016/S00702153(05)67001-9. [DOI] [PubMed] [Google Scholar]
- 15.Li C, Zhao H, Liu Z, and McMahon C (2014). Deer antler--a novel model for studying organ regeneration in mammals. The international journal of biochemistry & cell biology 56, 111–122. 10.1016/j.biocel.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 16.Goss RJ (1983). Deer Antlers: Regeneration, Function and Evolution. . [Google Scholar]
- 17.Suttie J, and Fennessy P (1985). Regrowth of amputated velvet antlers with and without innervation. Journal of Experimental Zoology 234, 359–366. [DOI] [PubMed] [Google Scholar]
- 18.Mascharak S, desJardins-Park HE, Davitt MF, Griffin M, Borrelli MR, Moore AL, Chen K, Duoto B, Chinta M, and Foster DS (2021). Preventing Engrailed-1 activation in fibroblasts yields wound regeneration without scarring. Science 372, eaba2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Longaker MT, Whitby DJ, Ferguson M, Lorenz HP, Harrison MR, and Adzick NS (1994). Adult skin wounds in the fetal environment heal with scar formation. Annals of surgery 219, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rinkevich Y., Walmsley G., Hu M., Maan Z., Newman A., Drukker M., Januszyk M., Krampitz G., Gurtner G., and Lorenz H. (2015). Skin fibrosis. Identification and isolation of a dermal lineage with intrinsic fibrogenic potential. Science 348, aaa2151. This study identifies a dermal lineage with high fibrogenic potential. Ablation of these CD26+ cells attenuates cutaneous fibrosis.
- 21.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e1821. 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Plikus MV, Wang X, Sinha S, Forte E, Thompson SM, Herzog EL, Driskell RR, Rosenthal N, Biernaskie J, and Horsley V (2021). Fibroblasts: Origins, definitions, and functions in health and disease. Cell 184, 3852–3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heaton H, Talman AM, Knights A, Imaz M, Gaffney DJ, Durbin R, Hemberg M, and Lawniczak MK (2020). Souporcell: robust clustering of single-cell RNA-seq data by genotype without reference genotypes. Nature methods 17, 615–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guerrero-Juarez CF, Dedhia PH, Jin S, Ruiz-Vega R, Ma D, Liu Y, Yamaga K, Shestova O, Gay DL, Yang Z, et al. (2019). Single-cell analysis reveals fibroblast heterogeneity and myeloid-derived adipocyte progenitors in murine skin wounds. Nat Commun 10, 650. 10.1038/s41467-018-08247-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collins CA, and Watt FM (2008). Dynamic regulation of retinoic acid-binding proteins in developing, adult and neoplastic skin reveals roles for beta-catenin and Notch signalling. Dev Biol 324, 55–67. 10.1016/j.ydbio.2008.08.034. [DOI] [PubMed] [Google Scholar]
- 26.Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, Rambow F, Marine JC, Geurts P, Aerts J, et al. (2017). SCENIC: single-cell regulatory network inference and clustering. Nat Methods 14, 1083–1086. 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamakawa H, Cheng J, Penney J, Gao F, Rueda R, Wang J, Yamakawa S, Kritskiy O, Gjoneska E, and Tsai L-H (2017). The transcription factor Sp3 cooperates with HDAC2 to regulate synaptic function and plasticity in neurons. Cell reports 20, 1319–1334. [DOI] [PubMed] [Google Scholar]
- 28.Phan QM, Fine GM, Salz L, Herrera GG, Wildman B, Driskell IM, and Driskell RR (2020). Lef1 expression in fibroblasts maintains developmental potential in adult skin to regenerate wounds. Elife 9. 10.7554/eLife.60066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J, Zhang Y, Lv H, Yu Q, Zhou Z, Zhu Q, Wang Z, Cooper PR, Smith AJ, Niu Z, and He W (2013). Human stem cells from the apical papilla response to bacterial lipopolysaccharide exposure and anti-inflammatory effects of nuclear factor I C. J Endod 39, 1416–1422. 10.1016/j.joen.2013.07.018. [DOI] [PubMed] [Google Scholar]
- 30.Simkin J, Gawriluk TR, Gensel JC, and Seifert AW (2017). Macrophages are necessary for epimorphic regeneration in African spiny mice. Elife 6, e24623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reynolds G, Vegh P, Fletcher J, Poyner EFM, Stephenson E, Goh I, Botting RA, Huang N, Olabi B, Dubois A, et al. (2021). Developmental cell programs are co-opted in inflammatory skin disease. Science 371. 10.1126/science.aba6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buchanan EP, Longaker MT, and Lorenz HP (2009). Fetal skin wound healing. Advances in clinical chemistry 48, 137–161. [DOI] [PubMed] [Google Scholar]
- 33.Solé-Boldo L, Raddatz G, Schütz S, Mallm JP, Rippe K, Lonsdorf AS, Rodríguez-Paredes M, and Lyko F (2020). Single-cell transcriptomes of the human skin reveal age-related loss of fibroblast priming. Commun Biol 3, 188. 10.1038/s42003-020-0922-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, et al. (2015). Proteomics. Tissue-based map of the human proteome. Science 347, 1260419. 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 35.Pliner HA, Shendure J, and Trapnell C (2019). Supervised classification enables rapid annotation of cell atlases. Nature methods 16, 983–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharma A., Seow JJW., Dutertre C-A., Pai R., Blériot C., Mishra A., Wong RMM., Singh GSN., Sudhagar S., and Khalilnezhad S. (2020). Onco-fetal reprogramming of endothelial cells drives immunosuppressive macrophages in hepatocellular carcinoma. Cell 183, 377–394. e321. [DOI] [PubMed] [Google Scholar]
- 37.Alquicira-Hernandez J, Sathe A, Ji HP, Nguyen Q, and Powell JE (2019). scPred: accurate supervised method for cell-type classification from single-cell RNA-seq data. Genome biology 20, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lönnerberg P, Furlan A, et al. (2018). RNA velocity of single cells. Nature 560, 494–498. 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bergen V, Lange M, Peidli S, Wolf FA, and Theis FJ (2020). Generalizing RNA velocity to transient cell states through dynamical modeling. Nat Biotechnol. 10.1038/s41587-020-0591-3. [DOI] [PubMed] [Google Scholar]
- 40.Shin W, Rosin NL, Sparks H, Sinha S, Rahmani W, Sharma N, Workentine M, Abbasi S, Labit E, Stratton JA, and Biernaskie J (2020). Dysfunction of Hair Follicle Mesenchymal Progenitors Contributes to Age-Associated Hair Loss. Dev Cell 53, 185–198.e187. 10.1016/j.devcel.2020.03.019. [DOI] [PubMed] [Google Scholar]
- 41.Hagner A, Shin W, Sinha S, Alpaugh W, Workentine M, Abbasi S, Rahmani W, Agabalyan N, Sharma N, Sparks H, et al. (2020). Transcriptional Profiling of the Adult Hair Follicle Mesenchyme Reveals R-spondin as a Novel Regulator of Dermal Progenitor Function. iScience 23, 101019. 10.1016/j.isci.2020.101019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sennett R, Wang Z, Rezza A, Grisanti L, Roitershtein N, Sicchio C, Mok KW, Heitman NJ, Clavel C, Ma’ayan A, and Rendl M (2015). An Integrated Transcriptome Atlas of Embryonic Hair Follicle Progenitors, Their Niche, and the Developing Skin. Dev Cell 34, 577–591. 10.1016/j.devcel.2015.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Satpathy AT, Granja JM, Yost KE, Qi Y, Meschi F, McDermott GP, Olsen BN, Mumbach MR, Pierce SE, and Corces MR (2019). Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nature biotechnology 37, 925–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sinha S, Satpathy AT, Zhou W, Ji H, Stratton JA, Jaffer A, Bahlis N, Morrissy S, and Biernaskie JA (2021). Profiling Chromatin Accessibility at Single-cell Resolution. Genomics, Proteomics & Bioinformatics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stuart T, Srivastava A, Madad S, Lareau CA, and Satija R (2021). Single-cell chromatin state analysis with Signac. Nature methods 18, 1333–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lange M, Bergen V, Klein M, Setty M, Reuter B, Bakhti M, Lickert H, Ansari M, Schniering J, Schiller HB, et al. (2020). CellRank for directed single-cell fate mapping. bioRxiv, 2020.2010.2019.345983. 10.1101/2020.10.19.345983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ogilvie P, Bardi G, Clark-Lewis I, Baggiolini M, and Uguccioni M (2001). Eotaxin is a natural antagonist for CCR2 and an agonist for CCR5. Blood 97, 1920–1924. 10.1182/blood.v97.7.1920. [DOI] [PubMed] [Google Scholar]
- 48.Costa A, Kieffer Y, Scholer-Dahirel A, Pelon F, Bourachot B, Cardon M, Sirven P, Magagna I, Fuhrmann L, Bernard C, et al. (2018). Fibroblast Heterogeneity and Immunosuppressive Environment in Human Breast Cancer. Cancer Cell 33, 463–479.e410. 10.1016/j.ccell.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 49.Sanin DE., Ge Y., Marinkovic E., Kabat AM., Castoldi A., Caputa G., Grzes KM., Curtis JD., Thompson EA., and Willenborg S. (2022). A common framework of monocyte-derived macrophage activation. Science Immunology 7, eabl7482. [DOI] [PubMed] [Google Scholar]
- 50.Xie X, Shi Q, Wu P, Zhang X, Kambara H, Su J, Yu H, Park S-Y, Guo R, and Ren Q (2020). Single-cell transcriptome profiling reveals neutrophil heterogeneity in homeostasis and infection. Nature immunology 21, 1119–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raredon MSB, Yang J, Garritano J, Wang M, Kushnir D, Schupp JC, Adams TS, Greaney AM, Leiby KL, and Kaminski N (2021). Connectome: computation and visualization of cell-cell signaling topologies in single-cell systems data. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raredon MSB, Adams TS, Suhail Y, Schupp JC, Poli S, Neumark N, Leiby KL, Greaney AM, Yuan Y, Horien C, et al. (2019). Single-cell connectomic analysis of adult mammalian lungs. Sci Adv 5, eaaw3851. 10.1126/sciadv.aaw3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qiu X, Zhang Y, Hosseinzadeh S, Yang D, Pogson AN, Wang L, Shurtleff M, Yuan R, Xu S, Ma Y, et al. (2021). Mapping Transcriptomic Vector Fields of Single Cells. bioRxiv, 696724. 10.1101/696724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raredon MSB, Yang J, Kothapalli N, Kaminski N, Niklason LE, and Kluger Y (2022). Comprehensive visualization of cell-cell interactions in single-cell and spatial transcriptomics with NICHES. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu JY, Feng L, Park H-T, Havlioglu N, Wen L, Tang H, Bacon KB, Jiang Z. h., Zhang X. c., and Rao Y (2001). The neuronal repellent Slit inhibits leukocyte chemotaxis induced by chemotactic factors. Nature 410, 948–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iglesias-Bartolome R, Uchiyama A, Molinolo AA, Abusleme L, Brooks SR, Callejas-Valera JL, Edwards D, Doci C, Asselin-Labat M-L, and Onaitis MW (2018). Transcriptional signature primes human oral mucosa for rapid wound healing. Science translational medicine 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nelson AM, Reddy SK, Ratliff TS, Hossain MZ, Katseff AS, Zhu AS, Chang E, Resnik SR, Page C, and Kim D (2015). dsRNA released by tissue damage activates TLR3 to drive skin regeneration. Cell stem cell 17, 139–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Armstrong J, and Ferguson M (1995). Ontogeny of the skin and the transition from scar-free to scarring phenotype during wound healing in the pouch young of a marsupial, Monodelphis domestica. Developmental biology 169, 242–260. [DOI] [PubMed] [Google Scholar]
- 59.Jennings RW, Adzick NS, Longaker MT, Duncan BW, Scheuenstuhl H, and Hunt TK (1991). Ontogeny of fetal sheep polymorphonuclear leukocyte phagocytosis. Journal of pediatric surgery 26, 853–855. [DOI] [PubMed] [Google Scholar]
- 60.Martin P, D’Souza D, Martin J, Grose R, Cooper L, Maki R, and McKercher SR (2003). Wound healing in the PU. 1 null mouse—tissue repair is not dependent on inflammatory cells. Current biology 13, 1122–1128. [DOI] [PubMed] [Google Scholar]
- 61.Mascharak S, Talbott HE, Januszyk M, Griffin M, Chen K, Davitt MF, Demeter J, Henn D, Bonham CA, and Foster DS (2022). Multi-omic analysis reveals divergent molecular events in scarring and regenerative wound healing. Cell Stem Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen K, Kwon SH, Henn D, Kuehlmann BA, Tevlin R, Bonham CA, Griffin M, Trotsyuk AA, Borrelli MR, and Noishiki C (2021). Disrupting biological sensors of force promotes tissue regeneration in large organisms. Nature Communications 12, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou X, Franklin RA, Adler M, Carter TS, Condiff E, Adams TS, Pope SD, Philip NH, Meizlish ML, and Kaminski N (2022). Microenvironmental Sensing by Fibroblasts Controls Macrophage Population Size. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ng LG, Ostuni R, and Hidalgo A (2019). Heterogeneity of neutrophils. Nature Reviews Immunology 19, 255–265. [DOI] [PubMed] [Google Scholar]
- 65.Hao H, Maeda Y, Fukazawa T, Yamatsuji T, Takaoka M, Bao X-H, Matsuoka J, Okui T, Shimo T, and Takigawa N (2013). Inhibition of the growth factor MDK/midkine by a novel small molecule compound to treat non-small cell lung cancer. PloS one 8, e71093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gutman DA., Khalilia M., Lee S., Nalisnik M., Mullen Z., Beezley J., Chittajallu DR., Manthey D., and Cooper LA. (2017). The digital slide archive: A software platform for management, integration, and analysis of histology for cancer research. Cancer research 77, e75–e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, McQuaid S, Gray RT, Murray LJ, and Coleman HG (2017). QuPath: Open source software for digital pathology image analysis. Scientific reports 7, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lakens D, Scheel AM, and Isager PM (2018). Equivalence testing for psychological research: A tutorial. Advances in Methods and Practices in Psychological Science 1, 259–269. [Google Scholar]
- 69.Ridout MS, and Linkie M (2009). Estimating overlap of daily activity patterns from camera trap data. Journal of Agricultural, Biological, and Environmental Statistics 14, 322–337. [Google Scholar]
- 70.Linderman GC, Zhao J, Roulis M, Bielecki P, Flavell RA, Nadler B, and Kluger Y (2022). Zero-preserving imputation of single-cell RNA-seq data. Nature Communications 13, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Suo S, Zhu Q, Saadatpour A, Fei L, Guo G, and Yuan GC (2018). Revealing the Critical Regulators of Cell Identity in the Mouse Cell Atlas. Cell Rep 25, 1436–1445.e1433. 10.1016/j.celrep.2018.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alquicira-Hernandez J, and Powell JE (2021). Nebulosa recovers single cell gene expression signals by kernel density estimation. Bioinformatics. 10.1093/bioinformatics/btab003. [DOI] [PubMed] [Google Scholar]
- 73.Li B, and Dewey CN (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323. 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Leng N, Dawson JA, Thomson JA, Ruotti V, Rissman AI, Smits BM, Haag JD, Gould MN, Stewart RM, and Kendziorski C (2013). EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 29, 1035–1043. 10.1093/bioinformatics/btt087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pliner HA, Packer JS, McFaline-Figueroa JL, Cusanovich DA, Daza RM, Aghamirzaie D, Srivatsan S, Qiu X, Jackson D, and Minkina A (2018). Cicero predicts cis-regulatory DNA interactions from single-cell chromatin accessibility data. Molecular cell 71, 858–871. e858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oldenbourg R (2007). Analysis of microtubule dynamics by polarized light. In Microtubule Protocols, (Springer; ), pp. 111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, and Mann M (2011). Andromeda: a peptide search engine integrated into the MaxQuant environment. Journal of proteome research 10, 1794–1805. [DOI] [PubMed] [Google Scholar]
- 79.Cox J, and Mann M (2008). MaxQuant enables high peptide identification rates, individualized ppb-range mass accuracies and proteome-wide protein quantification. Nature biotechnology 26, 1367–1372. [DOI] [PubMed] [Google Scholar]
- 80.Zhang X., Smits AH., van Tilburg G., Ovaa H., Huber W., and Vermeule M. (2018). Proteome-wide identification of ubiquitin interactions using UbIA-MS. Nature protocols 13, 530–550. [DOI] [PubMed] [Google Scholar]
- 81.Huber W, Von Heydebreck A, Sültmann H, Poustka A, and Vingron M (2002). Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18, S96–S104. [DOI] [PubMed] [Google Scholar]
- 82.Spitzer M, Wildenhain J, Rappsilber J, and Tyers M (2014). BoxPlotR: a web tool for generation of box plots. Nature methods 11, 121–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 – Velvet regeneration following full-thickness excision or burn injury and maintenance of velvet regenerative capacity following short-term transplant to non-regenerative back skin, Related to Figure 1. (A-B) Unwounded H&E-stained back skin (A) and velvet (B). Scale bars are 500μm. (C-D) Skin sections from 30dpw on back (C) and velvet (D) stained for Keratin-5 (red) and Ki67 (green). (E-F) Representative staining (E) and quantification (F) of Ki67+ cells in interfollicular dermis (IFD) and epidermis (IFE) 30dpw on back and velvet. (G-H) Gross images of full thickness thermal injury in back (G) and velvet (H) on Day 0, 30, and 60 days after injury. White dashed lines indicate original wound area. (I-J) Brightfield image of skin sections from back (I) and velvet (J) at 30d after thermal injury. Dashed lines (black) indicate wound margin and colored dashed lines indicate high magnification insets. (K) Quantification of HF density restoration within burn wounds. Data are mean±SEM. (L) Ventral view of velvet at 3dpw confirming full-thickness thermal damage. (M-N) H&E-stained sections of back skin (M) and velvet (N) wounds at 60dpw. Black dashed lines indicate wound margins. (O) Example of an unwounded ectopic velvet graft at 60d post-grafting. (P) H&E-stained section showing velvet-to-back skin graft. Dashed lines indicate margin between velvet and back skin. Note distinct differences in HF morphology between the two skin types. (Q) Ectopic velvet grafts from two additional animals 30dpw. Original wound sites are marked by blue dashed lines. Velvet graft margins are marked by white dashed lines. Scale bars are 100μm or as indicated.
Figure S2 – Alignment metrics, cell type annotations, and integrating bulk- and single-cell measurements to elucidate reproducible site-specific gene programs, Related to Figure 2. (A) Alignment metrics of scRNA-Seq reads mapped to a custom single-cell Bos taurus reference. (B) Bar plots showing the distribution of sequencing reads that confidently map to the bovine genome and transcriptome. Horizontal dotted lines represent average mapping of rodent reads to GRCm38/mm10 assembly. Box plots represent median, Q3, Q1. (C) Violin plots showing normalized expression levels of top three marker genes across the ten annotated cell types in Figure 2A. (D) Boxplots showing relative composition of macrophage and CD45+IL1B+ myeloid cells at baseline in n=3 biological replicates colored by harvest site. P-values are shown for both comparisons. (E) Schematic for dissecting reproducible transcriptional differences across velvet and back skin by combining bulk- and sc-RNA sequencing. (F) Principal component analysis of bulk back skin (cyan circle) and velvet (red circle) gene expression profiles at 0, 3, 7, 14dpw. (G) Hierarchical clustering of differentially expressed fibroblast genes at pre-injury baseline, scaled by row to display relative changes across samples. (H-I) Hierarchical clustering of fibroblast-derived ligands pre-injury (H) and immune cellderived ligands at 7dpw (I), scaled by row to display relative changes across samples. Cellular source predicted by sc-RNA-Seq, gene expression changes shown on each heatmap corroborated by bulk-RNA-Seq (G, H, I).
Figure S3 – Cross-species validation of pro-fibrotic and pro-regenerative fibroblast transcriptional programs, Related to Figure 3. (A) Shared fibrotic signaling across reindeer and human fibroblasts. (B) UMAP projection of 8,320 fibroblasts integrated across species (4,160 human cells; 4,160 reindeer), colored by fibroblast states. (C) Comparison of human 33 and reindeer fibroblast states using Pearson correlation coefficient. (D-H) Immunoperoxidase staining of human skin and violin plots showing normalized expression for genes enriched in scar-forming (human and reindeer back skin) fibroblasts (D-F) and genes enriched in regenerative (reindeer velvet) fibroblasts (F-G). Adjusted p-values calculated using two-sided nonparametric Wilcoxon rank-sum test with Bonferroni correction. Stainings obtained from the Human Protein Atlas. Scale bars represent 100 μm (D-H). (I) Schematic illustrating cross-species comparison between reindeer and human skin. (J and K) Stacked bar plots depicting frequency of CRABP1+ fibroblasts in seven fetal (J) and five adult (K) human skin samples. (L) Schematic of Garnett-based machine learning classifier trained to distinguish reindeer velvet-enriched (‘Pro-regenerative’), backskin-enriched (‘Pro-inflammatory’), or shared (‘Mixed’) fibroblast states and then tested on fetal and adult human skin fibroblasts. (M, N) UMAPs of subclustered reindeer (M) and human (N) resting fibroblasts used for training and testing the Garnett classifier, respectively. UMAPs coloured by tissue origin and Garnett-predicted fibroblast states. (O) Stacked bar plot depicting Garnett-predicted fibroblast state composition in adult and fetal human skin. Scale bars represent 100 μm (D-H).
Figure S4– Transcriptional and regulatory drivers of regenerative and fibrotic fibroblast fates, Related to Figure 4. (A) Streamlines showing RNA velocity field on a UMAP projection of fibroblasts colored by Louvain cluster IDs. (B) Absorption probability of velvet and back skin fibroblasts from 7 to 14dpw. Color intensity represents degree of fate priming. (C-D) Fate maps leading to inferred terminal states in velvet (C) and back skin (D). (E) Inferred differentiation potential. Terminal commitment is shown in dark purple. (F, G) Phase portrait, RNA velocity and gene expression plots of differentially spliced genes driving velvet (F) and back skin (G) healing. (H) Venn diagrams displaying overlap between ground state fibroblast DEGs with 3, 7, and 14dpw velvet and back skin fibroblasts. (I) Hierarchical clustering of tissue and time-point separated fibroblasts based on gene regulatory network activity. (J) Gene regulatory network activity of HDAC2, LEF1, NFKB1 in velvet (red) and back (turquoise) fibroblasts quantified as area under the recovery curve (AUC) over time. (K) Volcano plot of Wilcoxon rank-sum determined differentially activated regulons in velvet (red) and back (cyan) at 3, 7 and 14dpw. (L) Rank plot of fibroblast regulons at 3, 7 and 14dpw ordered by Regulon Specificity Score (RSS) where higher RSS indicates greater regional (back or velvet) specificity. Top regulons are labelled in either velvet (red) or back skin (cyan).
Figure S5 – Discrepant immune response and altered fibroblast-immune signaling during regenerative versus fibrotic healing, Related to Figure 5. (A) UMAP projection of all cells from uninjured and healing back and velvet wounds. (B) Global perturbation scores from 7 and 14dpw. (C) Circos plots showing differential fibroblast secretome-driven interactions with immune cells in velvet and back skin wounds at 0, 3, 7, and 14dpw. (D) Schematic depicting morphologic 64 and molecular 50 stages during neutrophil maturation used for reference mapping reindeer neutrophils during healing. (E) Subclustered neutrophil UMAP overlaid with velocyto vector fields and stacked bar plot depicting neutrophil cluster composition in velvet and back skin wounds at 3 and 7dpw. Neutrophil cluster enriched in 3dpw back skin is outlined with dash line. (F) UMAP and density plot depicting pseudotemporal ordering of neutrophils from velvet and back skin wounds at 3 and 7dpw. (G) Boxplot showing percentage of ‘immature’ neutrophils (Stages G0-G350) recruited to back or velvet wounds at 3 and 7dpw. n=3 biological replicates at each time point. Bonferroni adjusted P-value is shown. (H) Subclustered T cell UMAP overlaid with velocyto vector fields and stacked bar plot depicting T cell cluster composition in velvet and back skin wounds at 3 and 7dpw. (I) UMAP and kernel density distribution depicting pseudotemporal ordering of T cells from velvet and back skin wound beds at 3 and 7dpw. (J-K) Analysis of T cell dynamics following co-culture with back or velvet fibroblasts. (J) Subclustered T cell UMAP overlaid with velocyto vector fields colored by Louvain cluster ID and pseudotemporal ordering of T cell. (K) Sample separated UMAPs overlaid with density contour plots, demonstrating minimal difference in T cell diversification following exposure to back skin versus velvet fibroblasts.
Figure S6 – Differential revascularization and long-term histological evaluation following pharmacological perturbation of fibroblast programming, Related to Figure 6. (A) Kleinberg hub score, an indicator of community influence via a specific set of signaling molecules, of back and velvet fibroblasts for CXCL ligands at 0, 3, 7, 14dpw. Dashed lines indicate baseline hub scores of human fibroblasts profiled in Solé-Boldo et al. 2020. (B) Percentage of endothelial and smooth muscle cells within wounds over time. (C-D) Immunostaining of von Willebrand factor (green; endothelial cells) and MAC384 (red; macrophages/neutrophils) in back skin (C) and velvet (D) at 14dpw. (E) Experimental timeline for assessing long-term impact of recapitulating the back skin fibroblast signaling milieu within velvet wounds. (F-I) Images of H&E-stained sections of velvet wounds at 42dpw after receiving intradermal application of PBS (F), DMSO (G), PLAU (H), or iMDK (I). Dashed white lines indicate wound margins. Scale bars are 500μm. (J) UMAP plot visualizing dermal ECM ultrastructural properties and representative pseudocolored images of wound sections. (K) Treatment-separated density contour plots of ECM ultrastructure from each condition.
Figure S7 – Remodeling of ectopic velvet grafts and pro-regenerative impact of CSF1 neutralization during rodent healing, Related to Figure 7. (A) Schematic of autologous velvet-to-back grafting and subsequent wounding. (B) H&E-stained excision wound made on an ectopic velvet graft 6 months post-grafting (representative of n=5 grafts quantified in Figure 7B). (C) Insets showing HF density and dermal architecture of ectopic velvet graft pre- and post-wounding. (D-E) UMAPs visualizing dermal ECM ultrastructural properties of images of wound sections from baseline (uninjured) back skin, 3-month-old ectopic velvet and 6-month-old ectopic velvet. (E) Sample-separated density contour plots of ECM ultrastructural properties. (F) A summary of fibroblast evolution in ectopic grafts based on velocity vector fields and changes in fibroblast cluster composition. (G) Kernel density estimates depicting activation of mechanosensitive genes EN1, YAP1 and its binding partner TEAD1 within transitionary fibroblasts. (H) Relative contribution of macrophage clusters at different graft timepoints. (I) Inflammatory fibroblasts’ interacting partners using CSF1/CSF1R- and CXCL12/CXCR4-signaling axis. (J) Top five positively regulated signaling pathways enriched in fibroblast states that were not acquired in ectopic grafts at any of the surveyed time points. (K-L) Distribution of body weights (K) and vehicle and PLX5622 CSF1R inhibitor chow consumption per cage in male mice (L) from 2 days pre-wound until 6dpw. (M) Gross examination of wound closure between 2 to 13dpw in male mice.
Data S1 – Consensus DEGs differentially expressed in reindeer velvet and fetal human fibroblasts relative to reindeer back skin and adult human fibroblasts, related to Figures 2 and 3. Genes with log-fold-change less than 0.25 or Wilcoxon rank-sum-determined two-sided p-value > 0.05 were filtered out.
Data S2 – Differentially spliced genes identified using RNA velocity grouped by tissue (velvet versus back skin) and fibroblasts subsets (Clusters 0–14), related to Figure 2.
Data S3 – Magnitude of gene expression change and frequency of fibroblasts expressing regeneration-associated markers CRABP1, MDK and SPARC, related to Figure 4.
Data S4 – A set of core DEGs distinguishing velvet and back skin fibroblasts at ground state that were differentially maintained in wound-activated fibroblasts, related to Figures 3 and 4.
Data S5 – Transcriptional signature shared between uninjured velvet and fetal human fibroblasts that are differentially maintained over the healing trajectory relative to reindeer backskin, related to Figure 3. Genes with log-fold-change less than 0.25 or Wilcoxon rank-sum-determined two-sided p-value > 0.05 were filtered out.
Data S6 – List of cell-cell signaling edges originating either from immune and vasculature-associated cells (tabs 1–4) or fibroblasts (tabs 5-8), related to Figures 5 and 6.
Data S7 – Fibroblast signaling mechanisms associated with regenerative-to-inflammatory transition in ectopic grafts, related to Figures 7 and S7. Differential ligand-receptor mechanisms were identified using ROC analysis and the table lists putative mechanisms and their classification power ranging from 0 (random) to 1 (perfect classification).
Data Availability Statement
Sequencing data reported in this work are available at NCBI GEO data repository (GSE168748). Mass spectrometry datasets are available at the ProteomeXchange Consortium in the PRIDE partner repository (PXD035749).
Analysis code generated in this work are available at: http://doi.org/10.6084/m9.figshare.14196344 and github.com/BiernaskieLab.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.