Abstract
Dysregulation in lipid metabolism is the leading cause of chronic kidney disease (CKD) and also the important risk factors for high morbidity and mortality. Although lipid abnormalities were identified in CKD, integral metabolic pathways for specific individual lipid species remain to be clarified. We conducted ultra-high-performance liquid chromatography-high-definition mass spectrometry-based lipidomics and identified plasma lipid species and therapeutic effects of Rheum officinale in CKD rats. Adenine-induced CKD rats were administered Rheum officinale. Urine, blood and kidney tissues were collected for analyses. We showed that exogenous adenine consumption led to declining kidney function in rats. Compared with control rats, a panel of differential plasma lipid species in CKD rats was identified in both positive and negative ion modes. Among the 50 lipid species, phosphatidylcholine (PC), lysophosphatidylcholine (LysoPC) and lysophosphatidic acid (LysoPA) accounted for the largest number of identified metabolites. We revealed that six PCs had integral metabolic pathways, in which PC was hydrolysed into LysoPC, and then converted to LysoPA, which was associated with increased cytosolic phospholipase A2 protein expression in CKD rats. The lower levels of six PCs and their corresponding metabolites could discriminate CKD rats from control rats. Receiver operating characteristic curves showed that each individual lipid species had high values of area under curve, sensitivity and specificity. Administration of Rheum officinale significantly improved impaired kidney function and aberrant PC metabolism in CKD rats. Taken together, this study demonstrates that CKD leads to PC metabolism disorders and that the dysregulation of PC metabolism is involved in CKD pathology.
Keywords: chronic kidney disease, lipidomics, phosphatidylcholine metabolism, phospholipase A2, Rheum officinale
Introduction
Chronic kidney injury contributes to renal dysfunction and high patient morbidity in a myriad of clinical settings ranging from chronic kidney insufficiency to end-stage renal disease (ESRD) or kidney transplantation [1–3]. Renal injury leads to various extracellular and intracellular disorders, including alterations in kidney haemodynamics and tubular epithelial cell injury as well as the activation of signalling molecules such as the renin-angiotensin system, transforming growth factor-β1 (TGF-β1), Wnt, aryl hydrocarbon receptor, and lipid metabolism [4–8].
Recent studies have implicated lipid metabolism dysregulation in chronic kidney disease (CKD) [9]. Lipids, which account for over 70% of endogenous metabolites, are a major component in plasma. Lipids are diverse and abundant metabolites that play important cellular and biological roles, including composing membrane bilayers, mediating signal transduction, and generating functional implementations of membrane proteins and their potential interactions [10]. Circulating lipids mainly consist of glycerolipids, glycerophospholipids, phosphatidylcholines (PCs), lysophosphatidylcholines (LysoPCs) and sterol lipids transported by lipoproteins or albumin. Previously, lipid analysis in CKD hindered the determination of the alterations of a limited number of lipids, including triglycerides, total cholesterol, high-density lipoprotein cholesterol and low-density lipoprotein cholesterol, with contradictory findings in the relationship between CKD and dyslipidaemia [11, 12]. Owing to the limited methods, the effect of CKD on various intraclass alterations of these lipid classes and the changes in other classes of lipids on CKD remains enigmatic. Recently, advanced lipidomics has provided characteristic lipid signatures and revealed biochemical mechanisms in a variety of diseases, including CKD [13]. Several studies have demonstrated that changes in lipid levels can classify patients in the early and advanced stages of disease, such as chronic kidney insufficiency and ESRD [8, 14, 15]. Our previous work and that of others suggested that PC and LysoPC were among the most abundant circulating phospholipids in patients with CKD in addition to polyunsaturated fatty acids [14–16]. These findings provide proof-of-principle of the clinical applicability of these candidate biomarkers for risk prediction.
Despite convincing evidence of their biological importance, to our knowledge, there has been no systematic study aimed at identifying the metabolic pathways of individual lipid species in CKD. Extensive studies have demonstrated that phospholipase A2 (PLA2) catalyses the hydrolysis of acyl ester bonds at the sn-2 position of phospholipids and produces active metabolites, including lysophospholipids and arachidonic acid (AA), that alter various cell functions [17, 18]. This activity releases mainly cis-unsaturated fatty acids and the corresponding lysophospholipids in mammalian cells. Increasing enzyme activity leads to membrane phospholipid turnover and alters membrane fluidity and has been demonstrated to be the rate-limiting step for AA mobilization to trigger the eicosanoid cascade [19]. Two major classes of PLA2 have an important effect on human cells and tissues [20]. Cytosolic PLA2 (cPLA2) is cardinal candidate for the regulation of PC metabolism [21], and cPLA2 is activated in renal injury [22, 23]. Interestingly, adenine provokes an imbalance between lipid synthesis and decomposition, and adenine-induced CKD in rats causes metabolic abnormalities similar to human kidney disease in clinical settings [24]. Using a rat model of CKD induced by adenine, this study comprehensively identified metabolic pathways of individual plasma lipid species to reveal CKD-associated biochemical mechanisms using ultra-high-performance liquid chromatography-high-definition mass spectrometry (UPLC-HDMS), which is considered to be suitable for lipid profiling, especially large-scale unbiased lipid profiling in terms of its selectivity, reproducibility and sensitivity [25]. In addition, we investigated the intervening effects of Rheum officinale on individual lipid species within the identified metabolic pathways.
Materials and methods
Reagents and chemicals
Adenine was obtained from Sigma Chemical Company (St. Louis, Missouri, USA). Acetonitrile was obtained from Baker Company (Phillipsburg, New Jersey, USA). All other reagents were of analytical grade, and their purity was above 99.5%. Primary antibodies, including those for alpha-smooth muscle actin (α-SMA), collagen I, fibronectin and cPLA2, were obtained from Abcam Company (Cambridge, United Kingdom). The goat anti-rabbit secondary antibody was obtained from Abbkine Scientific Company (California, USA). The method of ethyl acetate extraction (EAA) for R. officinale is presented in a previous publication [26].
Animal experiment and sample collection
Male Sprague–Dawley rats were obtained from the Central Animal Breeding House of Xi’an Jiaotong University (Xi’an, China). The rats (initially weighing 190–210 g) were maintained with free access to water under standard conditions with a standard rodent diet and housed in a humidity-controlled room (approximately 60%) and temperature (approximately 23 °C) with a 12 h light/dark cycle. The rat CKD model was induced by adenine as described previously [27]. Rats were randomly divided into control, CKD, EAA-treated control, EAA-treated CKD and uraemic clearance granule-treated CKD groups (n = 8/group). The CKD rats were given a dose of 200 mg/kg body weight adenine dissolved in 1% (w/v) gum acacia solution by oral gavage once a day for 3 weeks. The EAA (100, 200, 400 mg/kg) or uraemic clearance granules were given to CKD rats by oral gavage once a day for 6 weeks. The control rats were given an equal volume of gum acacia solution. In addition, EAA (200 mg/kg) was administered to control rats by oral gavage once a day for 6 weeks. In week 6, the rats were placed in metabolic cages (1 rat/cage) to collect urine for 24 h. The rats were anaesthetized with 60 mg/kg sodium pentobarbital by intraperitoneal injection. Blood and kidney tissue samples were obtained and then stored at −80 °C for further analysis. All experiments involving animals were conducted according to the ethical policies and procedures approved by the ethics committee of Northwest University, China.
Clinical biochemistry parameters
Clinical features, including serum creatinine, urea, uric acid, total cholesterol and triglyceride as well as urine creatinine, were determined according to manufacturer guidelines by using an Olympus AU640 automatic analyser. The creatinine clearance rate (CCr) was calculated.
Immunohistochemical analysis
For histological analysis, paraffin-embedded rat kidney tissues (5 μm) were prepared according to a standard approach. Immunohistochemical staining was carried out by using a previous method [28]. The tissue sections were then incubated overnight at 4 °C with primary antibodies against α-SMA, collagen I and fibronectin. All pathological analyses were evaluated in ten randomly chosen nonoverlapping fields at ×400 magnification.
Western blot analysis
Total protein was extracted from kidney tissues by using RIPA buffer, and the protein concentration was determined with a PierceTM BCA Protein Assay Kit (23227, Thermo Scientific, USA). Total proteins (20–30 µg) were isolated via Tris-glycine gel and then transferred to PVDF membranes (0.45 μm). After incubation for 2 h in 5% nonfat milk blocking buffer, the membranes were incubated with anti-cPLA2 overnight at 4 °C. The membranes were further incubated with anti-rabbit secondary antibodies (1:8000, A21120, Abbkine, USA) at room temperature for 2 h. Bands were visualized with enhanced chemiluminescence reagent for Western blot analysis. The intensities of the immunoblots were quantified by ImageJ software. Band densities were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
Lipid extraction
Total lipids were extracted by using an Ostro 96-well plate via a single-step in-well extraction. Plasma (100 µL) was added to each well in a 2 mL Ostro plate fitted onto a vacuum manifold. Then, 300 µL of chloroform/methanol (1/1, v/v) was added to each well and mixed using aspiration mixing with a micropipette. The abovementioned extraction procedure was repeated two times. The extracts were dried under nitrogen and reconstituted in 200 µL of chloroform/methanol (1:1, v/v) for UPLC-HDMS analysis.
Lipid species detected by UPLC-HDMS
The plasma extracts were analysed on a Waters AcquityTM Ultra Performance LC system (Waters Corporation, Milford, MA, USA) equipped with a Waters XevoTM G2 QTof mass spectrometer (Waters MS Technologies, Manchester, UK). Chromatographic separation was performed at 45 °C with an ACQUITY UPLC HSS T3 column (2.1 mm × 100 mm, 1.8 µm). A gradient procedure was used with 10 mM ammonium formate in 2-propanol/acetonitrile (90/10) in 0.1% formic acid (A) and 10 mM ammonium formate in ACN/H2O (60/40) in 0.1% formic acid (B) as follows: a linear gradient of 0–10 min, 40.0%–99.0% A and 10.0–12.0 min, 99.0%–40.0% A. The flow rate was 0.5 mL/min. The temperatures of the autosampler and chromatographic column were maintained at 4 °C and 55 °C, respectively. Two microlitres of solution was injected into the UPLC-HDMS system.
The raw mass spectrometry data were collected by using a quadrupole time-of-flight synapt high-definition mass spectrometer. The scan range was from 50 to 1200 m/z in both positive and negative ion modes. The cone and capillary voltages were set at 45 V and 2.5 kV, respectively. The flow rate of the desolvation gas was set at 900 L/h at a temperature of 550 °C. The flow rate of the cone gas was set at 50 L/h, and the source temperature was 120 °C. The data acquisition rate was set to 0.1 s with a 0.1 s interscan delay. All analyses were obtained by using lockspray to ensure accuracy and reproducibility. All acquisition data were analysed by Prognosis QI and MarkerLynx XS (Waters Corporation, MA, USA).
Lipid identification
Lipid species were identified by combining accurate MS, isotopic pattern and MSE fragment information in both positive and negative ion modes obtained from UPLC-HDMS analysis with several databases (HMDB (http://www.hmdb.ca/), MASSBANK (http://www.massbank.jp), Chemspider (http://www.chemspider.com/) and KEGG (http://www.kegg.com/)) following previous identification approaches applied by our group and others [27–31].
Pattern recognition analysis and data processing
Fold change (FC) was calculated according to the mean ratios for the CKD/control groups. Chemometrics analysis, including principal component analysis (PCA) and orthogonal partial least square-discriminant analysis (OPLS-DA), and hierarchical clustering analysis, including heatmap and dendrogram, were performed by using MetaboAnalyst software (version 5.0). Pathway was performed on metabolomics pathway analysis. The metabolic pathways in CKD were analysed with a quantitative enrichment analysis algorithm from Lipid Maps.
Receiver operating characteristic (ROC) curve
A partial least square-discriminant analysis (PLS-DA)-based ROC curve was performed to analyse the predictive performance of the selected lipid species. The values of area under the curve (AUC), sensitivity, specificity and 95% confidence interval (CI) were calculated by using SPSS software version 25 (SPSS Inc. IBM, USA).
Statistical analysis
All statistical analyses were performed by using GraphPad Prism v 6.0 and SPSS software. The number of replicates was eight groups for each dataset, and the results are expressed as the mean ± SEM. Comparisons between two groups were analysed by either unpaired two-tailed Student’s t test or two-tailed Mann–Whitney U test. The results in the different experimental groups with more than two groups were compared by using one-way analysis of variance (ANOVA) followed by Dunnett’s post-hoc test. P < 0.05 was considered to indicate a significant difference.
Results
Adenine leads to renal function decline
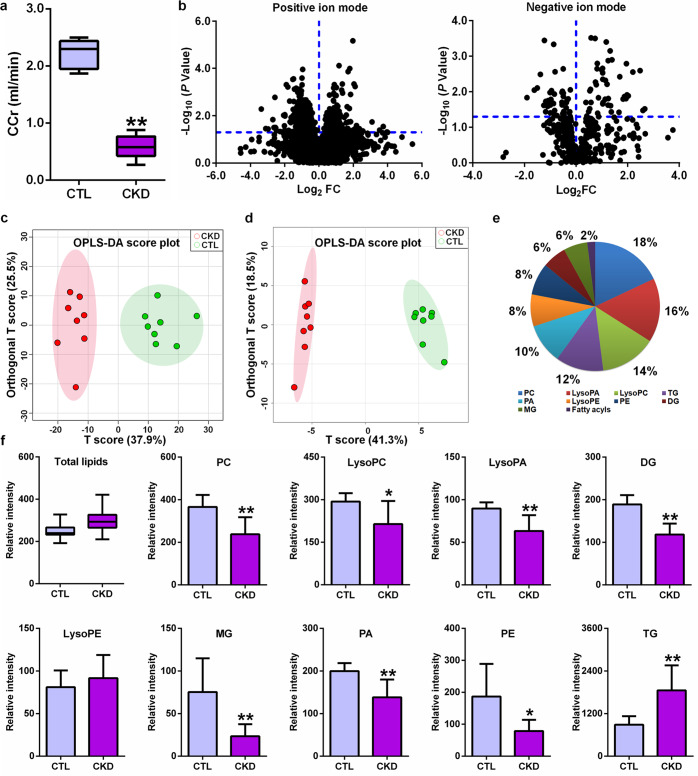
The final metabolite of adenine is uric acid. Under normal physiological conditions, the levels of adenine are very low in the blood and urine. When adenine is present in excess, adenine can be oxidized to 2,8-dihydroxyadenine via the intermediate 8-hydroxyadenine through the action of xanthine dehydrogenase [32, 33]. These metabolites are excreted to urine. However, 2,8-dihydroxyadenine has very low solubility and precipitates in renal tubules [32]. Figure 1a shows a lowered CCr in rats induced by adenine, indicating that adenine leads to renal damage and a decline in renal function.
Fig. 1. Adenine leads to renal function decline and altered lipid profiles in CKD rats.
a Creatinine clearance rate in control and CKD rats induced by adenine. b The geometric mean ratio of each variable in CKD rats versus control rats is presented in positive and negative ion modes. c OPLS-DA of 710 variables with P < 0.05 by using two-tailed unpaired Student’s t test between two groups in positive electrospray ionization mode. d OPLS-DA of 89 variables with P < 0.05 by using two-tailed unpaired Student’s t test between two groups in negative electrospray ionization mode. e Dendrogram analysis based on 50 lipid species identified in positive and negative ion modes. f The levels of total lipids and intraclass comparison of the distribution of 50 lipid species. *P < 0.05, **P < 0.01 compared with CKD rats.
Multivariate analysis and identification of important differential lipid species
UPLC-HDMS was used to obtain lipid profiles in both positive and negative ion modes. Initially, features were selected according to a value of P < 0.05. Fig. 2b shows the distribution of statistical significance by the log2 mean fold change of the features in CKD versus control rats. A total of 710 and 89 features from plasma samples had P values less than 0.05 in positive ion mode and negative ion mode, respectively (Fig. 1b). The OPLS-DA score plots showed that 710 and 89 features could both discriminate CKD rats from control rats (Fig. 1c, d). After excluding xenobiotics and different fragment ions with the same identified lipid species, 44 and 6 lipid species were identified based on our previously published approaches in positive ion mode and negative ion mode, respectively (Table 1) [28]. The lipids were mainly classified as PC, lysophosphatidic acid (LysoPA), LysoPC, triacylglycerol (TG), phosphatidic acid (PA), lysophosphatidylethanolamine (LysoPE), phosphatidylethanolamine (PE), diacylglycerol (DG) and monoacylglycerol (MG) (Fig. 1e). The levels of the total lipids remained essentially unaltered in CKD rats compared with control rats (Fig. 1f). The levels of total PC, LysoPC, LysoPA, DG, MG, PA and PE were markedly decreased in CKD rats, while the levels of total TGs and LysoPE were markedly increased in CKD rats compared with control rats (Fig. 1f).
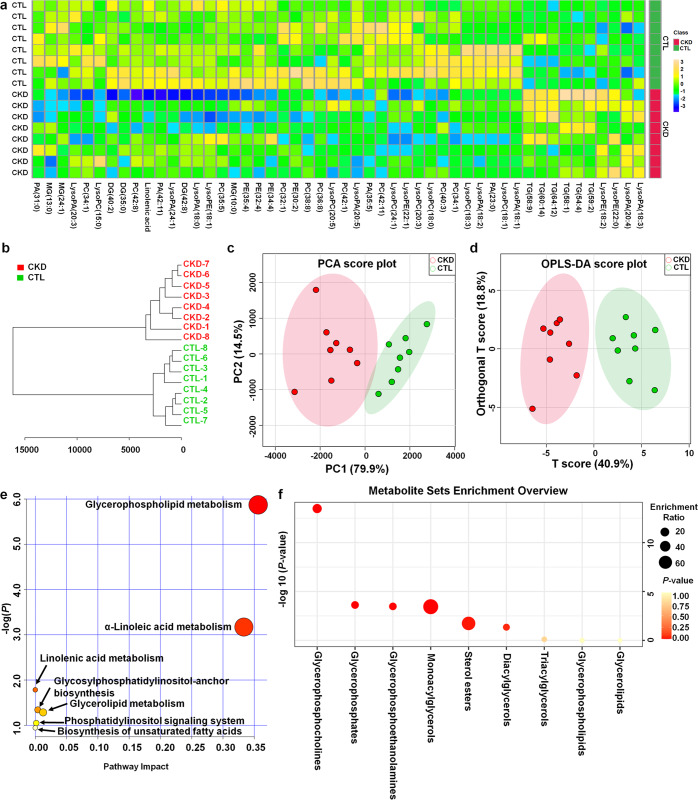
Fig. 2. Aberrant levels of lipid species are involved in CKD.
a Heatmap clustering analysis based on 50 lipid species between the two groups. b Dendrogram of hierarchical clustering analysis of 50 lipid species between the two groups. c PCA of 50 lipid species between the two groups. d OPLS-DA of 50 lipid species between the two groups. e Summary of ingenuity pathway analysis with lipidomics pathway analysis. f Lipid species set enrichment overview. The size and colour of each circle are based on the enrichment ratio and P values, respectively. The enrichment ratio was calculated by hits/expected, where hits = observed hits and expected = expected hits.
Table 1.
Identification of differential lipid species in serum of control and adenine-induced CKD rats.
| Metabolite | FCa | Pb | Pc | Class | Ion mode |
|---|---|---|---|---|---|
| LysoPA(24:1) | 0.285 | 3.60 × 10−4 | 2.00 × 10−3 | Glycerophospholipids | NEG |
| PA(31:0) | 0.214 | 4.89 × 10−4 | 2.00 × 10−3 | Glycerophospholipids | POS |
| MG(13:0) | 0.200 | 2.41 × 10−3 | 5.00 × 10−3 | Glycerolipids | POS |
| PC(42:8) | 0.641 | 2.95 × 10−3 | 3.00 × 10−3 | Glycerophospholipids | POS |
| DG(35:0) | 0.641 | 2.95 × 10−3 | 3.00 × 10−3 | Glycerolipids | POS |
| TG(58:1) | 2.018 | 3.62 × 10−3 | 2.00 × 10−3 | Glycerolipids | POS |
| TG(54:4) | 1.897 | 4.26 × 10−3 | 3.00 × 10−3 | Glycerolipids | POS |
| TG(58:9) | 3.552 | 4.98 × 10−3 | 1.20 × 10−2 | Glycerolipids | POS |
| LysoPC(20:5) | 0.422 | 5.69 × 10−3 | 1.60 × 10−2 | Glycerophospholipids | POS |
| PC(38:8) | 0.558 | 6.59 × 10−3 | 6.00 × 10−3 | Glycerophospholipids | POS |
| MG(10:0) | 0.455 | 6.59 × 10−3 | 1.20 × 10−2 | Glycerolipids | POS |
| PC(40:3) | 0.555 | 7.23 × 10−3 | 1.60 × 10−2 | Glycerophospholipids | POS |
| PA(33:2) | 0.430 | 7.46 × 10−3 | 1.20 × 10−2 | Glycerophospholipids | POS |
| LysoPC(18:1) | 0.611 | 7.59 × 10−3 | 6.00 × 10−3 | Glycerophospholipids | POS |
| LysoPA(18:0) | 0.728 | 7.61 × 10−3 | 2.10 × 10−2 | Glycerophospholipids | POS |
| LysoPE(18:2) | 1.522 | 8.58 × 10−3 | 1.60 × 10−2 | Glycerophospholipids | NEG |
| LysoPA(20:4) | 2.013 | 9.40 × 10−3 | 1.20 × 10−2 | Glycerophospholipids | NEG |
| LysoPA(18:2) | 0.639 | 1.05 × 10−2 | 1.20 × 10−2 | Glycerophospholipids | POS |
| PE(32:4) | 0.352 | 1.15 × 10−2 | 2.10 × 10−2 | Glycerophospholipids | POS |
| LysoPE(18:1) | 0.390 | 1.19 × 10−2 | 3.60 × 10−2 | Glycerophospholipids | POS |
| PE(34:4) | 0.443 | 1.46 × 10−2 | 1.60 × 10−2 | Glycerophospholipids | POS |
| PC(32:1) | 0.471 | 1.54 × 10−2 | 2.10 × 10−2 | Glycerophospholipids | POS |
| LysoPE(22:0) | 2.586 | 1.55 × 10−2 | 3.60 × 10−2 | Glycerophospholipids | NEG |
| LysoPC(20:3) | 0.677 | 1.55 × 10−2 | 2.70 × 10−2 | Glycerophospholipids | POS |
| LysoPA(20:5) | 0.556 | 1.60 × 10−2 | 3.60 × 10−2 | Glycerophospholipids | POS |
| LysoPE(22:1) | 0.589 | 1.62 × 10−2 | 2.70 × 10−2 | Glycerophospholipids | POS |
| PE(35:4) | 0.345 | 1.68 × 10−2 | 2.70 × 10−2 | Glycerophospholipids | POS |
| PA(42:11) | 0.460 | 1.78 × 10−2 | 1.80 × 10−2 | Glycerophospholipids | NEG |
| LysoPA(20:3) | 0.595 | 1.81 × 10−2 | 1.20 × 10−2 | Glycerophospholipids | POS |
| MG(24:1) | 0.269 | 1.82 × 10−2 | 3.60 × 10−2 | Glycerolipids | POS |
| LysoPC(15:0) | 0.399 | 1.85 × 10−2 | 9.00 × 10−3 | Glycerophospholipids | POS |
| PC(36:8) | 0.586 | 1.85 × 10−2 | 2.70 × 10−2 | Glycerophospholipids | POS |
| PE(30:2) | 0.273 | 1.94 × 10−2 | 1.20 × 10−2 | Glycerophospholipids | POS |
| LysoPA(18:3) | 3.025 | 2.37 × 10−2 | 4.50 × 10−2 | Glycerophospholipids | POS |
| Linoleic acid | 0.321 | 2.39 × 10−2 | 5.90 × 10−2 | Fatty acyls | NEG |
| TG(60:14) | 2.282 | 2.42 × 10−2 | 1.60 × 10−2 | Glycerolipids | POS |
| PC(35:5) | 0.565 | 2.49 × 10−2 | 2.70 × 10−2 | Glycerophospholipids | POS |
| TG(64:12) | 2.445 | 2.70 × 10−2 | 2.10 × 10−2 | Glycerolipids | POS |
| DG(40:2) | 0.502 | 2.77 × 10−2 | 3.60 × 10−2 | Glycerolipids | POS |
| TG(59:2) | 2.057 | 3.03 × 10−2 | 3.60 × 10−2 | Glycerolipids | POS |
| PC(34:1) | 0.618 | 3.16 × 10−2 | 2.70 × 10−2 | Glycerophospholipids | POS |
| PC(42:1) | 0.535 | 3.55 × 10−2 | 1.60 × 10−2 | Glycerophospholipids | POS |
| LysoPC(18:0) | 0.415 | 3.92 × 10−2 | 5.20 × 10−2 | Glycerophospholipids | POS |
| DG(42:8) | 0.635 | 3.94 × 10−2 | 2.70 × 10−2 | Glycerolipids | POS |
| PA(23:0) | 0.714 | 4.14 × 10−2 | 4.60 × 10−2 | Glycerophospholipids | POS |
| PC(42:11) | 0.768 | 4.63 × 10−2 | 4.60 × 10−2 | Glycerophospholipids | POS |
| PA(35:5) | 0.762 | 4.78 × 10−2 | 2.70 × 10−2 | Glycerophospholipids | POS |
| LysoPC(18:3) | 0.805 | 4.86 × 10−2 | 3.60 × 10−2 | Glycerophospholipids | POS |
| LysoPC(24:1) | 0.786 | 5.38 × 10−2 | 7.40 × 10−2 | Glycerophospholipids | POS |
| LysoPA(18:1) | 0.737 | 1.83 × 10−1 | 3.60 × 10−2 | Glycerophospholipids | POS |
aFC was obtained by comparing those lipid species in CRF rats with control rats; FC with a value >1 indicated a relatively higher intensity present in CRF rats, whereas a value <1 indicated a relatively lower intensity compared with control rats.
bP values from one-way ANOVA.
cP values from Mann–Whitney U-test.
Hierarchical clustering analyses, including heatmap and dendrogram, showed that 50 lipid species exhibited very high discrimination between the CKD and control rats, with a combination of the identified positively and negatively charged lipid species providing the best discrimination (Fig. 2a, b), which was further demonstrated by chemometrics analyses such as PCA and OPLS-DA (Fig. 2c, d). To understand the functional role of these plasma lipid species, the KEGG metabolic library was analysed. We tested for both the overrepresentation of changed lipid species in a pathway based on the hypergeometric tests and the impact of altered lipid species on the function of the pathway via changes in key junction points of the pathway based on relative betweenness centrality. The 82 rat pathways from KEGG are presented together to show the most significant metabolic pathways by p value from the hypergeometric test (indicated by the vertical axis in shades of red) and the impact (indicated by the horizontal axis as circles with different diameters) (Fig. 2e). The top three pathways by impact (top one) or p value (top two) are presented, including the following: (1) glycerophospholipid metabolism, (2) α-linolenic acid, and (3) linoleic acid metabolism (Table 2). Alterations in these pathways during CKD suggest that perturbation of certain central lipid species could have an impact on multiple metabolic pathways that are interconnected. The results revealed that glycerophospholipid metabolism was the most altered pathway in CKD induced by adenine (Fig. 2e). In addition, alterations in α-linolenic acid and linoleic acid metabolism suggest that CKD also leads to the dysregulation of biosynthesis of unsaturated fatty acids. Among glycerophospholipid metabolism pathways, enrichment analysis further demonstrated the dysregulation of glycerophosphocholines as the most important changes in CKD (Fig. 2f). This result demonstrated that adenine-induced renal injury might have a critical effect on PC metabolism, which is consistent with the identified lipid species (Table 1). Collectively, these findings indicated that exogenous adenine-induced CKD leads to significant alterations in metabolic lipid profiles and that aberrant PC metabolism is involved in CKD.
Table 2.
Pathway analysis of altered plasma lipid species from CKD rats compared with control rats.
| Pathway Name | Match status | p | -log(p) | Holm p | FDR | Impact |
|---|---|---|---|---|---|---|
| Glycerophospholipid metabolism | 4/36 | 1.3 × 10−6 | 5.87 | 1.1 × 10−4 | 1.1 × 10−4 | 3.6 × 10−1 |
| α-Linolenic acid metabolism | 2/13 | 6.8 × 10−4 | 3.17 | 5.6 × 10−2 | 2.8 × 10−2 | 3.3 × 10−1 |
| Linoleic acid metabolism | 1/5 | 1.6 × 10−2 | 1.78 | 1.0 | 4.6 × 10−1 | 0.0 |
| Glycosylphosphatidylinositol-anchor biosynthesis | 1/14 | 4.6 × 10−2 | 1.34 | 1.0 | 8.7 × 10−1 | 4.0 × 10−3 |
| Glycerolipid metabolism | 1/16 | 5.2 × 10−2 | 1.28 | 1.0 | 8.7 × 10−1 | 1.2 × 10−2 |
| Phosphatidylinositol signalling system | 1/28 | 9.0 × 10−2 | 1.05 | 1.0 | 1.0 | 1.5 × 10−3 |
| Arachidonic acid metabolism | 1/36 | 1.1 × 10−1 | 0.94 | 1.0 | 1.0 | 0.0 |
| Biosynthesis of unsaturated fatty acids | 1/36 | 1.1 × 10−1 | 0.94 | 1.0 | 1.0 | 0.0 |
Adenine induces an impaired PC metabolism pathway in CKD
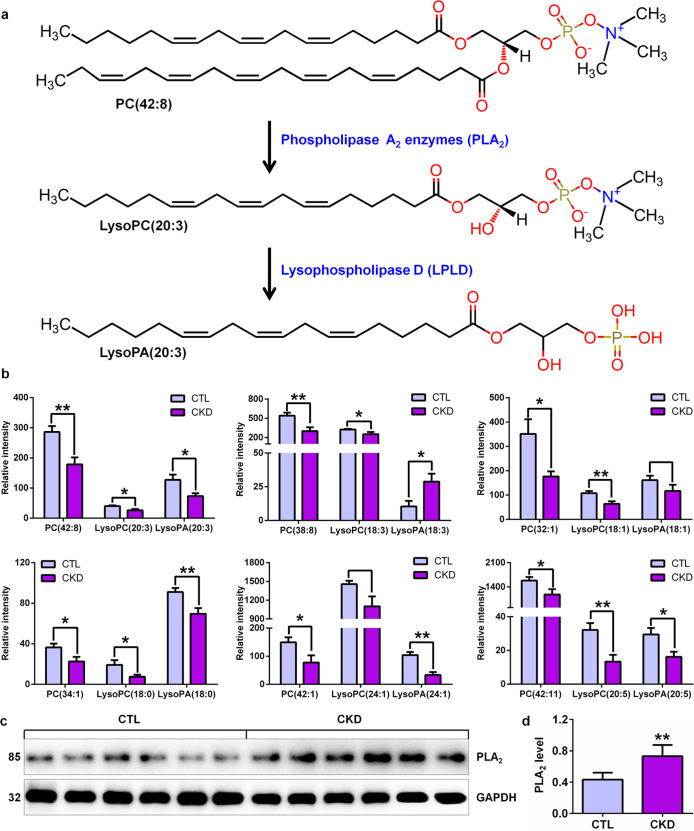
In 50 lipid species, PC, LysoPC and LysoPA were among the 3 most important classes of lipids that could be used for separating CKD rats from control rats, as they had the largest number of metabolites (Table 1 and Fig. 1e). Therefore, these lipid species are the major altered components in the plasma of adenine-induced CKD rats. PLA2 enzymes hydrolyse the sn-2 acyl bond of PC to release fatty acids and LysoPC. LysoPC is converted into LysoPA by lysophospholipase D (LPLD). Here, we take PC(42:8) as an example to illustrate the PC metabolism process (Fig. 3a). Based on PC metabolism, a total of six PCs, including PC(42:8), PC(38:8), PC(32:1), PC(34:1), PC(42:1) and PC(42:11), were found in the identified lipid species (Fig. 3b). Except for significantly increased LysoPA(18:3) levels and unaltered LysoPC(24:1) and LysoPA(18:1) levels in CKD rats, significantly decreased levels of individual PC and their metabolites, including LysoPC and LysoPA, were found in CKD rats compared with control rats (Fig. 3b). To further demonstrate whether PLA2 expression is associated with PC metabolism, we detected PLA2 protein expression levels by Western blotting. The results showed significantly increased PLA2 protein expression levels in adenine-induced CKD rats compared with control rats (Fig. 3c, d). These results demonstrate that adenine-induced CKD is accompanied by disordered PC metabolism associated with PLA2 expression.
Fig. 3. Aberrant PC metabolism is involved in CKD.
a PC metabolic pathway with phospholipase A2 enzymes and LPLD. Phospholipase A2 enzymes hydrolyse the sn-2 acyl bond of glycerophospholipids [PC(42:8)] to release fatty acids and lysophospholipids [LysoPC(20:3)]. Lysophospholipids [LysoPC(20:3)] are converted into lysophosphatidic acids [LysoPA(20:3)] by LPLD. b The levels of PC and its metabolites, including LysoPC and LysoPA, between the two groups by PC metabolism analysis. c Intrarenal PLA2 protein in adenine-induced CKD and control rats. d Quantitative analysis of intrarenal PLA2 protein expression levels in adenine-induced CKD and control rats. **P < 0.01 compared with control rats.
Impaired PC metabolism contributes to CKD
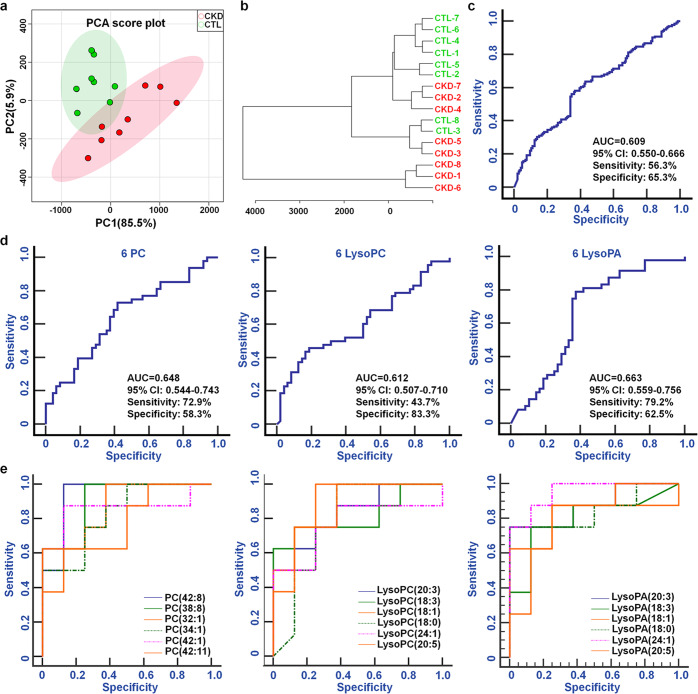
Based on 18 lipid species, including six PCs and their metabolites, both PCA score plots and dendrogram displays showed that CKD rats were separated from control rats (Fig. 4a, b). The ROC curves showed that the combination of the 18 lipid species had AUC (95% CI), sensitivity and specificity values of 0.609, 56.3% and 65.3%, respectively (Fig. 4c). Similarly, the combination of the 6 PCs, 6 LysoPCs or 6 LysoPAs had AUC values >0.600 (Fig. 4d). The ROC curves show that each individual lipid species has high AUC (95% CI), sensitivity and specificity values (Fig. 4e and Table 3). The AUC value of each individual lipid species was higher than that of the combination of the lipid species. These results demonstrate that individual lipid species are robust in distinguishing CKD rats from control rats. CKD might have an important effect on the dysregulation of these lipid species.
Fig. 4. The aberrant levels of PC metabolites could distinguish CKD rats from CTL rats.
a PCA of 18 lipid species, including 6 PCs, 6 LysoPCs and 6 LysoPAs, between the two groups. b Dendrogram of hierarchical clustering analysis of 18 lipid species, including 6 PCs, 6 LysoPCs and 6 LysoPAs, between the two groups. c Analysis of PLS-DA-based ROC curves for distinguishing CKD rats from control rats with the combination of the 18 lipid species. d Analysis of PLS-DA-based ROC curves for distinguishing CKD rats from control rats with the combination of the 6 PCs, 6 LysoPCs or 6 LysoPAs. e Analysis of PLS-DA-based ROC curves for distinguishing CKD rats from control rats with PCs, LysoPCs or LysoPAs individually.
Table 3.
Analysis of PLS-DA-based ROC curves of each individual PC, LysoPC or LysoPA.
| Metabolites | AUC | 95%CI | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|
| LysoPA(24:1) | 0.953 | 0.720–1.000 | 75.0 | 100.0 |
| PC(42:8) | 0.938 | 0.698–0.998 | 100.0 | 87.5 |
| LysoPC(18:1) | 0.906 | 0.656–0.993 | 100.0 | 75.0 |
| PC(38:8) | 0.906 | 0.656–0.993 | 100.0 | 75.0 |
| LysoPA(20:3) | 0.875 | 0.617–0.984 | 75.0 | 100.0 |
| LysoPC(20:5) | 0.859 | 0.598–0.979 | 75.0 | 87.5 |
| PC(42:1) | 0.859 | 0.598–0.979 | 87.5 | 87.5 |
| LysoPA(18:0) | 0.844 | 0.579–0.973 | 75.0 | 100.0 |
| PC(32:1) | 0.844 | 0.579–0.973 | 100.0 | 62.5 |
| LysoPC(20:3) | 0.828 | 0.561–0.967 | 50.0 | 100.0 |
| PC(34:1) | 0.828 | 0.561–0.967 | 50.0 | 100.0 |
| LysoPA(18:1) | 0.813 | 0.544–0.960 | 62.5 | 100.0 |
| LysoPA(20:5) | 0.813 | 0.544–0.960 | 87.5 | 75.0 |
| LysoPC(18:3) | 0.813 | 0.544–0.960 | 62.5 | 100.0 |
| LysoPA(18:3) | 0.797 | 0.526–0.952 | 75.0 | 87.5 |
| PC(42:11) | 0.797 | 0.526–0.952 | 62.5 | 100.0 |
| LysoPC(18:0) | 0.789 | 0.518–0.948 | 100.0 | 62.5 |
| LysoPC(24:1) | 0.766 | 0.493–0.936 | 50.0 | 100.0 |
PC metabolism might be a therapeutic target for CKD
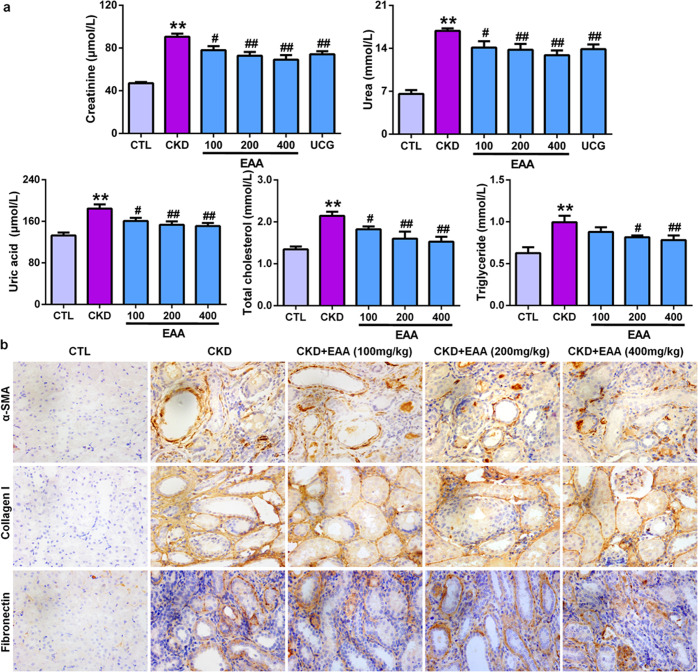
The understanding of disease molecular mechanisms aims to discover novel therapeutic agents and strategies. To further demonstrate whether the restoration of the levels of 18 lipid species from PC metabolism is related to improving kidney function, we chose R. officinale, a well-known natural product with a renoprotective effect, to assess its effects on 18 lipid species in adenine-induced CKD rats. As shown in Fig. 5a, the levels of creatinine, urea, uric acid, total cholesterol and triglycerides in serum were significantly increased in adenine-induced CKD rats compared with control rats. EAA treatment significantly improved the levels of these biochemical indexes, resulting in a significant reduction in their levels in a dose-dependent manner. Therefore, the 200 mg/kg EAA showed a strong intervention effect compared with the 100 mg/kg dose of EAA, whereas the intervention effect of the 400 mg/kg dose was similar to the intervention effect of the 200 mg/kg dose. Immunohistochemical analysis showed that treatment with the three doses of EAA significantly ameliorated the protein expression of collagen I, α-SMA and fibronectin and further alleviated renal fibrosis (Fig. 5b). Therefore, the 200 mg/kg dose of EAA was chosen as the optimal dose for further experiments.
Fig. 5. Treatment with EAA improves impaired kidney function in CKD rats induced by adenine.
a The levels of serum creatinine, urea, uric acid, total cholesterol, and triglycerides in control, CKD and CKD+EAA rats. b Immunohistochemical analyses with anti-α-SMA, collagen I and fibronectin antibodies in rat kidney tissues in control, CKD and CKD+EAA (200 mg/kg) rats. Magnification, ×400. **P < 0.01 compared with control rats; #P < 0.05, ##P < 0.01 compared with CKD rats.
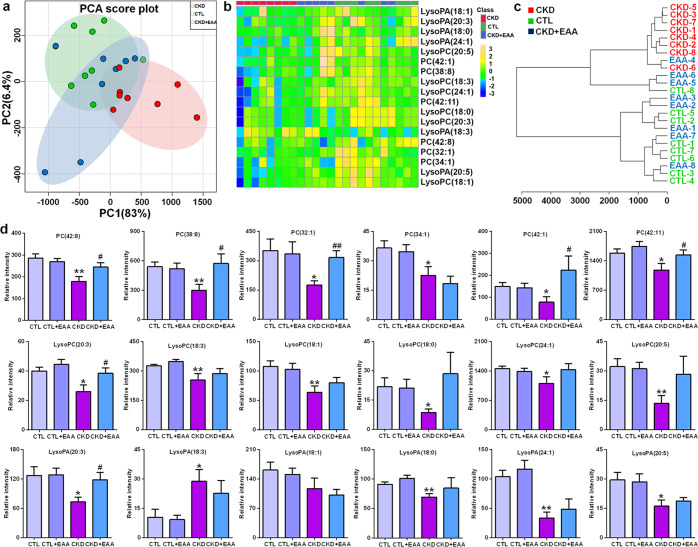
To further determine whether EAA has an effect on PC metabolism, we next examined the effects of EAA on the levels of 18 lipid species. As shown in Fig. 6a, the PCA score plot could distinguish among the control, CKD and EAA-treated CKD groups. It can also be seen that the EAA-treated CKD group was located between the CKD group and the control group but much closer to the control group, which was demonstrated by using a heatmap and dendrogram (Fig. 6b, c). Although EAA treatment restored the levels of 18 lipid species, the abnormal levels of five PCs, PC(42:8), PC(38:8), PC(32:1), PC(42:1) and PC(42:11), as well as LysoPC(20:3) and LysoPA(20:3), were also significantly reversed by EAA treatment (Fig. 6d). These results demonstrated that EAA treatment can improve aberrant PC metabolism, indicating that EAA treatment has an important effect on the PC metabolic pathway.
Fig. 6. Treatment with EAA improves PC metabolism in CKD rats induced by adenine.
a PCA of 18 lipid species, including 6 PCs, 6 LysoPCs and 6 LysoPAs, in control, CKD and CKD+EAA (200 mg/kg) rats. b Heatmap clustering analysis based on 18 lipid species, including 6 PCs, 6 LysoPCs and 6 LysoPAs, in control, CKD and CKD+EAA (200 mg/kg) rats. c Dendrogram of hierarchical clustering analysis based on 18 lipid species, including 6 PCs, 6 LysoPCs and 6 LysoPAs, in control, CKD and CKD+EAA (200 mg/kg) rats. d The levels of 18 lipid species, including 6 PCs, 6 LysoPCs and 6 LysoPAs, in the different rats. *P < 0.05, **P < 0.01 compared with control rats; #P < 0.05, ##P < 0.01 compared with CKD rats
Discussion
CKD, with increasing rates of morbidity and mortality, is associated with a variety of risk factors, such as hypertension, hypercholesterolemia, obesity and smoking [34–36]. Although patients with CKD are treated by many regimens, patients inevitably develop ESRD and require renal replacement therapies such as dialysis and transplantation [37–39]. Dyslipidaemia contributes to the increased risk of morbidity and mortality of renal diseases and its complications, but until recently, most differentially regulated lipid studies in CKD have mainly focused on the balance of several lipoproteins and determined the total levels of major classes. To test the hypothesis that molecular information of specific lipid species would highlight novel details of lipid metabolism disorder in CKD, we used an unbiased lipidomics search strategy that allowed the identification of the differentially regulated lipid species that can distinguish CKD rats from healthy controls. In this study, the levels of total PC, LysoPC, LysoPA, DG, MG, PA and PE were significantly decreased in CKD rats. Earlier studies have found that compared with healthy controls, plasma levels of total PC and/or LysoPC were significantly lower [40, 41], whereas the levels of total PE and LysoPA were significantly increased in haemodialysis patients [40]. The latest study reported that in CKD patients, the levels of total PC and LysoPC tended to decrease from CKD stages 2–3 to CKD stages 4–5 to finally ESRD maintained on haemodialysis [15]. These findings are consistent with the results of the current study, which showed lower plasma levels of total PC and LysoPC in CKD rats. Associations with clinical features have been previously demonstrated: the levels of total PC and LysoPC were reduced with the rise in serum creatinine levels, whereas these levels were increased with the levels of serum albumin, calcium, phosphate and protein. More importantly, the levels of total LysoPC presented the strongest correlations with many clinical data [15]. Michalczyk et al. demonstrated that plasma LysoPA levels were significantly increased in patients treated conservatively with peritoneal dialysis and in renal transplant recipients compared with healthy controls, whereas LysoPA levels were not different in haemodialysis patients [41].
In an animal model study, Tsutsumi et al. reported lower levels of total LysoPC and LysoPA in rats with unilateral ureteral obstruction [42]. Mirzoyan et al. reported that LysoPA levels were significantly increased in the urine but not the plasma of mice with subtotal nephrectomy and were remarkably correlated with albuminuria and tubulointerstitial fibrosis [43]. Collectively, our current findings are consistent with previous studies reporting significantly decreased levels of lipid classes, such as PC, LysoPC and LysoPA, that are involved in the PC metabolic pathway in both patients and animal models.
In the present study, we identified a total of 50 lipid species that could distinguish between CKD and control rats. There was a decrease in 6 PCs and their corresponding metabolites, including 6 LysoPCs and 5 LysoPAs, in CKD rats compared with control rats. Natural products have been considered an important source for new drugs in treating kidney diseases by targeting various signalling pathways [44–50]. Our recent study shows that R. officinale improves the dysregulation of these lipid species in adenine-induced CKD rats. PC, LysoPC and LysoPA are important bioactive components that have a variety of biological functions, such as cell survival, proliferation, apoptosis and cytoskeletal construction, in various diseases, including CKD [51–53]. An increasing number of publications suggested that the dysregulation of PC, LysoPC and LysoPA is implicated in CKD [14, 15, 41, 52, 54]. Structurally, PCs are composed of a wide range of fatty acyl chains that change in length and double bond position. PCs are typically composed of a saturated fatty acyl chain at the sn-1 position and a polyunsaturated fatty acid at the sn-2 position [51]. PC is the most abundant phospholipid and accounts for 40%–50% of total cellular phospholipids in all mammals [51]. In mammalian cells, two pathways synthesize PCs de novo. The major pathway is the cytidine diphosphate-choline pathway in all nucleated cells [53]. This pathway requires choline and includes 3 enzyme catalytic steps for PC synthesis [53, 55]. PC can also be synthesized in a second pathway through three sequential methylations of PE via phosphatidylethanolamine N-methyltransferase [55]. Therefore, pathways with impaired PC synthesis or metabolism might be associated with lower PC levels in CKD [56].
Several previous studies have demonstrated lower PC and/or LysoPC levels in patients at different stages of CKD and/or haemodialysis [14–16, 57, 58]. Our previous study showed lower levels of PC(36:0), PC(44:3), PC(48:0) and LysoPC(14:0) in CKD patients than in healthy controls, whereas the levels of PC(40:2), PC(46:0), LysoPC(18:2), LysoPC(20:4) and LysoPC(24:1) were increased [59]. Our previous study further demonstrated significantly increased LysoPA(18:2) levels in CKD patients and two animal models, including adenine- and 5/6 nephrectomized-induced CKD rats, while LysoPA(16:0) levels were significantly increased in adenine-induced CKD rats [60]. Several previous studies have demonstrated the dysregulation of PC metabolism in diabetic kidney disease [52]. Grove et al. reported increased LysoPC(18:0) and LysoPA(18:0) levels in the kidney tissues of eNOS-/- db/db mice [61]. In addition, another study demonstrated no change in PC and LysoPC levels in patients with prehaemodialysis CKD compared with healthy controls [62]. Duranton’s study further found that the LysoPC(20:3) and LysoPC(20:4) levels progressively decreased in patients from CKD 2–3 to ESRD maintained on haemodialysis, whereas altered LysoPC(20:3) and LysoPC(18:2) levels showed strong correlations with many clinical parameters and were related to the occurrence of transition to haemodialysis or even death [15]. Rhee et al. showed that low LysoPC(18:2) levels were related to CKD progression [56]. Similarly, low LysoPC(20:3) and LysoPC(18:2) levels were also observed in metabolic syndrome, heart failure and hepatic cancer [63–65], which is in line with the significantly decreased levels of 6 LysoPCs, including LysoPC(20:3), in our current study. Therefore, our current and previous work and that of others suggested impaired PC metabolism in CKD. It has been reported that the LysoPC/albumin ratio does not show a significant change in patients at different stages of CKD and haemodialysis although it remains related to a better outcome [15], indicating that plasma LysoPC levels are related to the risk of mortality independent of clinical biochemical parameters such as albumin levels, which might be an alternative predictor of mortality related to LysoPC levels. These findings elucidate that although the clinical biochemistry data may not appear abnormal [66–68], individual lipid species are significantly altered in CKD. The current work and that of others suggested that low levels of LysoPCs, such as LysoPC(20:3), are found in rats and patients with CKD. Therefore, our current animal experiments confirm previous clinical findings [15, 59]. Our study further shows that EAA treatment partly improves the levels of individual PCs and their metabolites. In particular, EAA treatment significantly increased the levels of PC(42:8), LysoPC(20:3) and LysoPA(20:3), which are involved in the PC(42:8) metabolic pathway. Taken together, these findings showed that PC metabolism, demonstrated by the lower levels of PCs and their metabolites, is the major pathway in aberrant lipid metabolism.
The molecular mechanisms of PC metabolism may be associated with altered enzymatic activities. Several earlier studies have demonstrated that PLA2 expression in PC metabolism was activated in animal models of kidney injury, such as polycystic kidney disease, remnant kidney and diabetic kidney disease [20, 69, 70]. PC can also breakdown LysoPC by the enzymatic activities of endothelial lipase and lecithin:cholesterol acyl transferase (LCAT) [71, 72]. Both increased PLA2 and deficient LCAT activity have been demonstrated in CKD patients [73, 74]. PLA2 and LCAT have major roles in LysoPC production, or alternatively, increased LysoPC decomposition by LPLD led to decreased LysoPC levels in patients with renal disease [15]. Duranton’s study found a lower molar ratio of plasma LysoPC/PC in haemodialysis patients, indicating that when PC levels were lower, LysoPC production was further reduced [15]. Although Sasagawa et al. demonstrated lower levels of plasma PC and LysoPC in haemodialysis patients than in healthy controls, a higher molar ratio of plasma LysoPC/PC was observed. In addition, a strong correlation was found between plasma LysoPA and LysoPC levels [40]. However, another study demonstrated no significant correlation between plasma LysoPA and LysoPC levels in conservatively treated haemodialysis and peritoneal dialysis patients and renal transplant recipients [41]. Our current study shows that PLA2 protein expression in the kidney tissues of adenine-induced CKD rats is increased, which may be associated with PC metabolism. Taking these findings together, we conclude that impaired PC metabolism is involved in CKD and that PC metabolism is one of the pathways influenced by R. officinale.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (Nos. 81703691 and 82074002), Shaanxi Key Science and Technology Plan Project (No. 2019ZDLSF04-04-02) and ShaanXi Science and Technology Innovation Team (No. 2019TD-016).
Author contributions
YYZ designed the study. YNW, ZHZ, HJL, and ZYG performed the experiments. YYZ performed the statistical analysis. YYZ prepared the manuscript. LZ and YMZ revised the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
References
- 1.Kalantar-Zadeh K, Jafar TH, Nitsch D, Neuen BL, Perkovic V. Chronic kidney disease. Lancet. 2021;398:786–802. doi: 10.1016/S0140-6736(21)00519-5. [DOI] [PubMed] [Google Scholar]
- 2.Hansrivijit P, Chen YJ, Lnu K, Trongtorsak A, Puthenpura MM, Thongprayoon C, et al. Prediction of mortality among patients with chronic kidney disease: a systematic review. World J Nephrol. 2021;10:59–75. doi: 10.5527/wjn.v10.i4.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mantovani A, Chiara Z. PNPLA3 gene and kidney disease. Explor Med. 2020;1:42–50. doi: 10.37349/emed.2020.00004. [DOI] [Google Scholar]
- 4.Lu CC, Hu ZB, Wang R, Hong ZH, Lu J, Chen PP, et al. Gut microbiota dysbiosis-induced activation of the intrarenal renin-angiotensin system is involved in kidney injuries in rat diabetic nephropathy. Acta Pharmacol Sin. 2020;41:1111–8. doi: 10.1038/s41401-019-0326-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geng XQ, Ma A, He JZ, Wang L, Jia YL, Shao GY, et al. Ganoderic acid hinders renal fibrosis via suppressing the TGF-β/Smad and MAPK signaling pathways. Acta Pharmacol Sin. 2020;41:670–7. doi: 10.1038/s41401-019-0324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li SS, Sun Q, Hua MR, Suo P, Chen JR, Yu XY, et al. Targeting the Wnt/β-catenin signaling pathway as a potential therapeutic strategy in renal tubulointerstitial fibrosis. Front Pharmacol. 2021;12:719880. doi: 10.3389/fphar.2021.719880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miao H, Cao G, Wu XQ, Chen YY, Chen DQ, Chen L, et al. Identification of endogenous 1-aminopyrene as a novel mediator of progressive chronic kidney disease via aryl hydrocarbon receptor activation. Br J Pharmacol. 2020;177:3415–35. doi: 10.1111/bph.15062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng YL, Chen H, Chen DQ, Vaziri ND, Su W, Ma SX, et al. Activated NF-κB/Nrf2 and Wnt/β-catenin pathways are associated with lipid metabolism in CKD patients with microalbuminuria and macroalbuminuria. Biochim Biophys Acta Mol Basis Dis. 2019;1865:2317–32. doi: 10.1016/j.bbadis.2019.05.010. [DOI] [PubMed] [Google Scholar]
- 9.Baek J, He C, Afshinnia F, Michailidis G, Pennathur S. Lipidomic approaches to dissect dysregulated lipid metabolism in kidney disease. Nat Rev Nephrol. 2022;18:38–55. doi: 10.1038/s41581-021-00488-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao YY, Cheng XL, Lin RC. Lipidomics applications for discovering biomarkers of diseases in clinical chemistry. Int Rev Cell Mol Biol. 2014;313:1–26. doi: 10.1016/B978-0-12-800177-6.00001-3. [DOI] [PubMed] [Google Scholar]
- 11.Rahman M, Yang W, Akkina S, Alper A, Anderson AH, Appel LJ, et al. Relation of serum lipids and lipoproteins with progression of CKD: The CRIC study. Clin J Am Soc Nephrol. 2014;9:1190–8. doi: 10.2215/CJN.09320913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaysen GA. Lipid and lipoprotein metabolism in chronic kidney disease. J Ren Nutr. 2009;19:73–7. doi: 10.1053/j.jrn.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 13.Zhao YY, Miao H, Cheng XL, Wei F. Lipidomics: novel insight into the biochemical mechanism of lipid metabolism and dysregulation-associated disease. Chem Biol Interact. 2015;240:220–38. doi: 10.1016/j.cbi.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 14.Afshinnia F, Rajendiran TM, Karnovsky A, Soni T, Wang X, Xie D, et al. Lipidomic signature of progression of chronic kidney disease in the chronic renal insufficiency cohort. Kidney Int Rep. 2016;1:256–68. doi: 10.1016/j.ekir.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duranton F, Laget J, Gayrard N, Saulnier-Blache JS, Lundin U, Schanstra JP, et al. The CKD plasma lipidome varies with disease severity and outcome. J Clin Lipidol. 2019;13:176–85. doi: 10.1016/j.jacl.2018.07.010. [DOI] [PubMed] [Google Scholar]
- 16.Reis A, Rudnitskaya A, Chariyavilaskul P, Dhaun N, Melville V, Goddard J, et al. Top-down lipidomics of low density lipoprotein reveal altered lipid profiles in advanced chronic kidney disease. J Lipid Res. 2015;56:413–22. doi: 10.1194/jlr.M055624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang F, Wang K, Shen J. Lipoprotein-associated phospholipase A2: The story continues. Med Res Rev. 2020;40:79–134. doi: 10.1002/med.21597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bermúdez MA, Balboa L, Balsinde J. Lipid droplets, phospholipase A2, arachidonic acid, and atherosclerosis. Biomedicines. 2021;9:1891. doi: 10.3390/biomedicines9121891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dorsam G, Harris L, Payne M, Fry M, Franson R. Development and use of ELISA to quantify type II phospholipase A2 in normal and uremic serum. Clin Chem. 1995;41:862–6. doi: 10.1093/clinchem/41.6.862. [DOI] [PubMed] [Google Scholar]
- 20.Aukema HM, Adolphe J, Mishra S, Jiang J, Cuozzo FP, Ogborn MR. Alterations in renal cytosolic phospholipase A2 and cyclooxygenases in polycystic kidney disease. FASEB J. 2003;17:298–300. doi: 10.1096/fj.02-0460fje. [DOI] [PubMed] [Google Scholar]
- 21.Murakami M, Nakatani Y, Atsumi GI, Inoue K, Kudo I. Regulatory functions of phospholipase A2. Crit Rev Immunol. 2017;37:127–95. doi: 10.1615/CritRevImmunol.v37.i2-6.20. [DOI] [PubMed] [Google Scholar]
- 22.Kohjimoto Y, Honeyman TW, Jonassen J, Gravel K, Kennington L, Scheid CR. Phospholipase A2 mediates immediate early genes in cultured renal epithelial cells: possible role of lysophospholipid. Kidney Int. 2000;58:638–46.. doi: 10.1046/j.1523-1755.2000.00210.x. [DOI] [PubMed] [Google Scholar]
- 23.Wang R, Chen J, Ding F, Zhang L, Wu X, Wan Y, et al. Renal tubular injury induced by glyphosate combined with hard water: the role of cytosolic phospholipase A2. Ann Transl Med. 2021;9:130. doi: 10.21037/atm-20-7739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao YY, Cheng XL, Wei F, Xiao XY, Sun WJ, Zhang Y, et al. Serum metabonomics study of adenine-induced chronic renal failure in rats by ultra performance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry. Biomarkers. 2012;17:48–55. doi: 10.3109/1354750X.2011.637180. [DOI] [PubMed] [Google Scholar]
- 25.Zhao YY, Wu SP, Liu S, Zhang Y, Lin RC. Ultra-performance liquid chromatography-mass spectrometry as a sensitive and powerful technology in lipidomic applications. Chem Biol Interact. 2014;220:181–92. doi: 10.1016/j.cbi.2014.06.029. [DOI] [PubMed] [Google Scholar]
- 26.Zhang ZH, Wei F, Vaziri ND, Cheng XL, Bai X, Lin RC, et al. Metabolomics insights into chronic kidney disease and modulatory effect of rhubarb against tubulointerstitial fibrosis. Sci Rep. 2015;5:14472. doi: 10.1038/srep14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao YY, Cheng XL, Wei F, Bai X, Tan XJ, Lin RC, et al. Intrarenal metabolomic investigation of chronic kidney disease and its TGF-β1 mechanism in induced-adenine rats using UPLC Q-TOF/HSMS/MSE. J Proteome Res. 2013;12:692–703. doi: 10.1021/pr3007792. [DOI] [PubMed] [Google Scholar]
- 28.Chen DQ, Cao G, Chen H, Argyopoulos CP, Yu H, Su W, et al. Identification of serum metabolites associating with chronic kidney disease progression and anti-fibrotic effect of 5-methoxytryptophan. Nat Commun. 2019;10:1476. doi: 10.1038/s41467-019-09329-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balgoma D, Guitton Y, Evans JJ, Le Bizec B, Dervilly-Pinel G, Meynier A. Modeling the fragmentation patterns of triacylglycerides in mass spectrometry allows the quantification of the regioisomers with a minimal number of standards. Anal Chim Acta. 2019;1057:60–9. doi: 10.1016/j.aca.2019.01.017. [DOI] [PubMed] [Google Scholar]
- 30.Cao W, Cheng S, Yang J, Feng J, Zhang W, Li Z, et al. Large-scale lipid analysis with C=C location and sn-position isomer resolving power. Nat Commun. 2020;11:375. doi: 10.1038/s41467-019-14180-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kind T, Liu KH, Lee DY, DeFelice B, Meissen JK, Fiehn O. LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat Methods. 2013;10:755–8. doi: 10.1038/nmeth.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Engle SJ, Stockelman MG, Chen J, Boivin G, Yum MN, Davies PM, et al. Adenine phosphoribosyltransferase-deficient mice develop 2,8-dihydroxyadenine nephrolithiasis. Proc Natl Acad Sci USA. 1996;93:5307–12. doi: 10.1073/pnas.93.11.5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Redhead NJ, Selfridge J, Wu CL, Melton DW. Mice with adenine phosphoribosyltransferase deficiency develop fatal 2,8-dihydroxyadenine lithiasis. Hum Gene Ther. 1996;7:1491–502. doi: 10.1089/hum.1996.7.13-1491. [DOI] [PubMed] [Google Scholar]
- 34.Medina Rangel PX, Priyadarshini A, Tian X. New insights into the immunity and podocyte in glomerular health and disease: From pathogenesis to therapy in proteinuric kidney disease. Integr Med Nephrol Androl. 2021;8:5. doi: 10.4103/imna.imna_26_21. [DOI] [Google Scholar]
- 35.Bhargava V, Singh K, Meena P, Sanyal R. Nephrogenic systemic fibrosis: A frivolous entity. World J Nephrol. 2021;10:29–36. doi: 10.5527/wjn.v10.i3.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Majid DSA, Castillo A, Prieto MC, Navar LG. High salt induced augmentation of angiotensin II mediated hypertension is associated with differential expression of tumor necrosis factor-alpha receptors in the kidney. Explor Med. 2022;3:205–18. doi: 10.37349/emed.2022.00086. [DOI] [Google Scholar]
- 37.Chuengsaman P, Narenpitak S, Sritippayawan S. Efficacy and safety of recombinant human erythropoietin (Hema-Plus(®)) for management of anemia in Thai patients on peritoneal dialysis. World J Nephrol. 2021;10:109–21. doi: 10.5527/wjn.v10.i6.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chauveau P. Nutrition in chronic kidney disease: Nephrology Dialysis Transplantation notable advances in 2018. Nephrol Dial Transpl. 2019;34:893–96. doi: 10.1093/ndt/gfz077. [DOI] [PubMed] [Google Scholar]
- 39.Canaud B, Kooman JP, Selby NM, Taal M, Maierhofer A, Kopperschmidt P, et al. Hidden risks associated with conventional short intermittent hemodialysis: A call for action to mitigate cardiovascular risk and morbidity. World J Nephrol. 2022;11:39–57. doi: 10.5527/wjn.v11.i2.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sasagawa T, Suzuki K, Shiota T, Kondo T, Okita M. The significance of plasma lysophospholipids in patients with renal failure on hemodialysis. J Nutr Sci Vitaminol. 1998;44:809–18. doi: 10.3177/jnsv.44.809. [DOI] [PubMed] [Google Scholar]
- 41.Michalczyk A, Dołęgowska B, Heryć R, Chlubek D, Safranow K. Associations between plasma lysophospholipids concentrations, chronic kidney disease and the type of renal replacement therapy. Lipids Health Dis. 2019;18:85. doi: 10.1186/s12944-019-1040-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsutsumi T, Adachi M, Nikawadori M, Morishige J, Tokumura A. Presence of bioactive lysophosphatidic acid in renal effluent of rats with unilateral ureteral obstruction. Life Sci. 2011;89:195–203. doi: 10.1016/j.lfs.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 43.Mirzoyan K, Baïotto A, Dupuy A, Marsal D, Denis C, Vinel C, et al. Increased urinary lysophosphatidic acid in mouse with subtotal nephrectomy: potential involvement in chronic kidney disease. J Physiol Biochem. 2016;72:803–12. doi: 10.1007/s13105-016-0518-0. [DOI] [PubMed] [Google Scholar]
- 44.Izzo AA, Teixeira M, Alexander SPH, Cirino G, Docherty JR, George CH, et al. A practical guide for transparent reporting of research on natural products in the British Journal of Pharmacology: Reproducibility of natural product research. Br J Pharmacol. 2020;177:2169–78. doi: 10.1111/bph.15054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luo LP, Suo P, Ren LL, Liu HJ, Zhang Y, Zhao YY. Shenkang injection and its three anthraquinones ameliorates renal fibrosis by simultaneous targeting IκB/NF-κB and Keap1/Nrf2 signaling pathways. Front Pharmacol. 2021;12:800522. doi: 10.3389/fphar.2021.800522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fang CY, Lou DY, Zhou LQ, Wang JC, Yang B, He QJ, et al. Natural products: potential treatments for cisplatin-induced nephrotoxicity. Acta Pharmacol Sin. 2021;42:1951–69. doi: 10.1038/s41401-021-00620-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang YN, Feng HY, Nie X, Zhang Y, Zou L, Li X, et al. Recent advances in clinical diagnosis and pharmacotherapy options of membranous nephropathy. Front Pharmacol. 2022;13:907108. doi: 10.3389/fphar.2022.907108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cao G, Miao H, Wang YN, Chen DQ, Wu XQ, Chen L, et al. Intrarenal 1-methoxypyrene, an aryl hydrocarbon receptor agonist, mediates progressive tubulointerstitial fibrosis in mice. Acta Pharmacol Sin. 2022. 10.1038/s41401-022-00914-6. [DOI] [PMC free article] [PubMed]
- 49.Yang Y, Wu C. Traditional chinese medicine in ameliorating diabetic kidney disease via modulating gut microbiota. Integr Med Nephrol Androl. 2021;8:8. doi: 10.4103/imna.imna_28_21. [DOI] [Google Scholar]
- 50.Yu XY, Sun Q, Zhang YM, Zou L, Zhao YY. TGF-β/Smad signaling pathway in tubulointerstitial fibrosis. Front Pharmacol. 2022;13:860588. doi: 10.3389/fphar.2022.860588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang B, Tontonoz P. Phospholipid remodeling in physiology and disease. Annu Rev Physiol. 2019;81:165–88. doi: 10.1146/annurev-physiol-020518-114444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee JH, Kim D, Oh YS, Jun HS. Lysophosphatidic acid signaling in diabetic nephropathy. Int J Mol Sci. 2019;20:2850. doi: 10.3390/ijms20112850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heden TD, Neufer PD, Funai K. Looking beyond structure: membrane phospholipids of skeletal muscle mitochondria. Trends Endocrinol Metab. 2016;27:553–62. doi: 10.1016/j.tem.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim D, Li HY, Lee JH, Oh YS, Jun HS. Lysophosphatidic acid increases mesangial cell proliferation in models of diabetic nephropathy via Rac1/MAPK/KLF5 signaling. Exp Mol Med. 2019;51:1–10. doi: 10.1038/s12276-019-0217-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tavasoli M, Lahire S, Reid T, Brodovsky M, McMaster CR. Genetic diseases of the Kennedy pathways for membrane synthesis. J Biol Chem. 2020;295:17877–86.. doi: 10.1074/jbc.REV120.013529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rhee EP, Clish CB, Ghorbani A, Larson MG, Elmariah S, McCabe E, et al. A combined epidemiologic and metabolomic approach improves CKD prediction. J Am Soc Nephrol. 2013;24:1330–8. doi: 10.1681/ASN.2012101006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rhee EP, Souza A, Farrell L, Pollak MR, Lewis GD, Steele DJ, et al. Metabolite profiling identifies markers of uremia. J Am Soc Nephrol. 2010;21:1041–51.. doi: 10.1681/ASN.2009111132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Piperi C, Kalofoutis C, Tzivras M, Troupis T, Skenderis A, Kalofoutis A. Effects of hemodialysis on serum lipids and phospholipids of end-stage renal failure patients. Mol Cell Biochem. 2004;265:57–61. doi: 10.1023/B:MCBI.0000044315.74038.78. [DOI] [PubMed] [Google Scholar]
- 59.Chen H, Chen L, Liu D, Chen DQ, Vaziri ND, Yu XY, et al. Combined clinical phenotype and lipidomic analysis reveals the impact of chronic kidney disease on lipid metabolism. J Proteome Res. 2017;16:1566–78. doi: 10.1021/acs.jproteome.6b00956. [DOI] [PubMed] [Google Scholar]
- 60.Zhang ZH, Chen H, Vaziri ND, Mao JR, Zhang L, Bai X, et al. Metabolomic signatures of chronic kidney disease of diverse etiologies in the rats and humans. J Proteome Res. 2016;15:3802–12. doi: 10.1021/acs.jproteome.6b00583. [DOI] [PubMed] [Google Scholar]
- 61.Grove KJ, Voziyan PA, Spraggins JM, Wang S, Paueksakon P, Harris RC, et al. Diabetic nephropathy induces alterations in the glomerular and tubule lipid profiles. J Lipid Res. 2014;55:1375–85. doi: 10.1194/jlr.M049189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang L, Hu C, Liu S, Chang M, Gao P, Wang L, et al. Plasma lipidomics investigation of hemodialysis effects by using liquid chromatography-mass spectrometry. J Proteome Res. 2016;15:1986–94. doi: 10.1021/acs.jproteome.6b00170. [DOI] [PubMed] [Google Scholar]
- 63.El-Najjar N, Orsó E, Wallner S, Liebisch G, Schmitz G. Increased levels of sphingosylphosphorylcholine (SPC) in plasma of metabolic syndrome patients. PLoS One. 2015;10:e0140683. doi: 10.1371/journal.pone.0140683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zordoky BN, Sung MM, Ezekowitz J, Mandal R, Han B, Bjorndahl TC, et al. Metabolomic fingerprint of heart failure with preserved ejection fraction. PLoS One. 2015;10:e0124844. doi: 10.1371/journal.pone.0124844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Patterson AD, Maurhofer O, Beyoglu D, Lanz C, Krausz KW, Pabst T, et al. Aberrant lipid metabolism in hepatocellular carcinoma revealed by plasma metabolomics and lipid profiling. Cancer Res. 2011;71:6590–600. doi: 10.1158/0008-5472.CAN-11-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao YY. Metabolomics in chronic kidney disease. Clin Chim Acta. 2013;422:59–69. doi: 10.1016/j.cca.2013.03.033. [DOI] [PubMed] [Google Scholar]
- 67.Wang YN, Ma SX, Chen YY, Chen L, Liu BL, Liu QQ, et al. Chronic kidney disease: Biomarker diagnosis to therapeutic targets. Clin Chim Acta. 2019;499:54–63. doi: 10.1016/j.cca.2019.08.030. [DOI] [PubMed] [Google Scholar]
- 68.Johnson CH, Ivanisevic J, Siuzdak G. Metabolomics: beyond biomarkers and towards mechanisms. Nat Rev Mol Cell Biol. 2016;17:451–9. doi: 10.1038/nrm.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dorsam G, Krieg RJ, Jr, Chan W, Ereso G, Lin KC, Chan JC. Phospholipase A2 activity, heat shock protein, and superoxide dismutase in rat remnant kidney. Pediatr Nephrol. 2000;14:128–31. doi: 10.1007/s004670050027. [DOI] [PubMed] [Google Scholar]
- 70.Lu YL, Ye L, Wu H, Xia FZ, Yu J, Yang LZ. The up-regulated expression of both phospholipase A2 and cyclooxygenase-2 is involved in renal injury in streptozotocin-induced diabetic rats. Acta Endocrinologica-Buchar. 2013;9:23–32. doi: 10.4183/aeb.2013.23. [DOI] [Google Scholar]
- 71.Schmitz G, Ruebsaamen K. Metabolism and atherogenic disease association of lysophosphatidylcholine. Atherosclerosis. 2010;208:10–8. doi: 10.1016/j.atherosclerosis.2009.05.029. [DOI] [PubMed] [Google Scholar]
- 72.Astudillo AM, Balgoma D, Balboa MA, Balsinde J. Dynamics of arachidonic acid mobilization by inflammatory cells. Biochim Biophys Acta Mol Cell Biol Lipids. 2012;1821:249–56. doi: 10.1016/j.bbalip.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 73.Gillett MP, Obineche EN, Lakhani MS, Abdulle AM, Amirlak I, Al Rukhaimi M, et al. Levels of cholesteryl esters and other lipids in the plasma of patients with end-stage renal failure. Ann Saudi Med. 2001;21:283–6. doi: 10.5144/0256-4947.2001.283. [DOI] [PubMed] [Google Scholar]
- 74.Papavasiliou EC, Gouva C, Siamopoulos KC, Tselepis AD. Erythrocyte PAF-acetylhydrolase activity in various stages of chronic kidney disease: effect of long-term therapy with erythropoietin. Kidney Int. 2005;68:246–55. doi: 10.1111/j.1523-1755.2005.00399.x. [DOI] [PubMed] [Google Scholar]