Abstract
All cells, from bacteria and yeasts to mammalian cells, respond to cues from their environment. A variety of mechanisms exist for the transduction of these external signals to the interior of the cell, resulting in altered patterns of protein activity. Eukaryotic cells commonly transduce external cues via a conserved module composed of three protein kinases, the mitogen-activated protein kinase (MAPK) cascade. This module can then activate substrates, some of which include transcriptional activators. Multiple MAPK signalling pathways coexist in a cell. This review considers different MAPK cascade signalling pathways that govern several aspects of the life cycle of budding and fission yeasts: conjugation and meiosis by the pheromone response pathway, stress response by the high-osmolarity sensing pathway, cell wall biosynthesis in response to activation of the low-osmolarity and heat-sensing pathway, and pseudohyphal growth in response to activation of a subset of the components of the pheromone response pathway. Because the MAPK cascade components are highly conserved, a key question in studies of these pathways is the mechanism by which specificity of response is achieved. Several other issues to be addressed in this review concern the nature of the receptors used to sense the external signals and the mechanism by which the receptors communicate with other components leading to activation of the MAPK cascade. Recently, it has become apparent that MAPK cascades are important in governing the pathogenicity of filamentous fungi.
The purpose of this review is to summarize the current knowledge of signalling pathways in the yeasts for nonspecialists, particularly for fungal biologists who are beginning studies in this area. This review has been written to describe both how these pathways function and how they were figured out—how the components in these different pathways were identified, how they were ordered, and what kinds of assays are used in such studies.
I describe several pathways in both Saccharomyces cerevisiae (budding yeast) and Schizosaccharomyces pombe (fission yeast), juxtaposing pathways used for the same response in an attempt to point out similarities and differences in the use of highly conserved components in different organisms. Several reasons have motivated me as a fungal biologist to do this. I am interested in cell-cell interactions between fungal cells and between fungal and plant cells. Because these interactions are likely to involve diverse signals and response pathways, an understanding of the different pathways that operate in the yeasts may be directly relevant to understanding the interactions of filamentous fungi with their environment and their hosts. The literature in the area of signalling is vast. I have therefore tried to extract some of the most important lessons to make this review accessible to nonspecialists, although I hope that specialists will also find it useful. Because information about signal transduction pathways in filamentous fungi is fragmentary, in many cases I have interspersed nuggets of information on this topic in the relevant sections describing the yeast pathways.
Several important general issues, which provide a framework for examining the different pathways, are as follows. (i) A given cell contains multiple pathways, each of which responds to a distinct signal that is transduced to give a specific response. Because the central component of these pathways, the mitogen-activated protein kinase (MAPK) cascade (see below), is highly conserved, the cell must have mechanisms for preventing inappropriate communication between pathways. One mechanism for preventing such cross talk involves scaffold or sequestering proteins. (ii) A given signalling component can be used in more than one pathway within the same cell to respond to different signals. This observation raises the question of how the specificity of the response is regulated in such cases. Specificity may involve modulation of the activity of a transcription factor by its association with different accessory proteins. (iii) Comparisons of pathways, for example, used for response to stress will illustrate that in one organism a pathway responds to only one stress signal whereas in another organism the same components are used to respond to a multitude of stress signals. We are then confronted with the question of how multiple input signals are integrated. One possibility is the use of a different sensor for each different signal. (iv) Different organisms use the same machinery to respond to the same signal, but some of the components of the machinery may be used differently. (v) The receptors used in different pathways are of different types: G-protein-coupled (serpentine or seven-transmembrane) receptors, His-Asp phosphorelay sensors, and a novel class of integral membrane proteins. In each case, a central issue (often unsolved) is how they communicate with downstream components. (vi) The responses elicited by the different signals are numerous and in many cases are common to all eukaryotes. In some cases, the response of a pathway to a given signal may serve to coordinate different processes, for example, osmolarity and mitosis or nutritional status and meiosis. (vii) A given pathway may link two different types of phosphorylation cascades. (viii) Many examples of redundancy of components are provided by the different pathways. (ix) Lastly, filamentous fungi may use some of the machinery used in the mating response in yeasts to regulate the intricacies of hyphal growth.
I first present a general introduction to the core of the signalling pathways, the MAPK cascade. I then describe the pathways that sense osmolarity in both yeasts and follow with a description of the pheromone response pathway. As part of the latter section, I discuss regulation of filamentous growth in the Basidiomycete fungi by pheromones and receptors. I also present information on the role of various signalling components in the life cycle of filamentous fungi. Each section has been written in a self-contained manner so that the reader can read the sections separately.
MAPK CASCADE MODULE AND SIGNAL TRANSDUCTION
All organisms, from bacteria and yeasts to mammalian cells, respond to cues from the extracellular environment. These cues are then transduced from the cell surface to the interior of the cell, resulting in patterns of altered gene expression and protein activity, which result in a cellular response to the external environment.
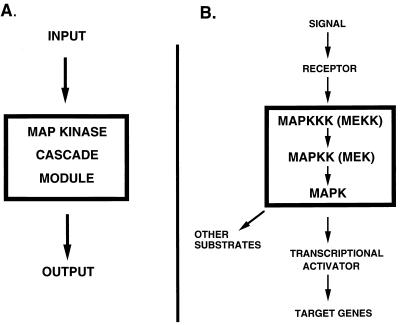
In eukaryotic cells, the MAPK cascade module is a key element in mediating the transduction of many signals generated at the cell surface to the nucleus (Fig. 1). Three protein kinases that are highly conserved in all eukaryotes make up this module: MAPK (also known as extracellular signal-regulated kinase [ERK]), MAPK kinase (MAPKK, also known as mitogen-activated, ERK-activating kinase [MEK]), and MAPK kinase kinase (MAPKKK, also known as MEK kinase [MEKK]). I will refer to them in this review as MAPK, MAPKK, and MAPKKK, respectively. Sequential activation of these kinases by phosphorylation lies at the heart of transduction of the signal through this kinase module. MAPK is activated by the dual-specificity serine/threonine tyrosine kinase MAPKK, and it is in turn activated by the serine/threonine kinase MAPKKK (reviewed in references 160, 175, and 183). The latter becomes activated in response to a signal generated by an input. Thus, input signals lead to activation of the MAPK cascade, which then generates output signals (Fig. 1).
FIG. 1.
MAPK module. (A) The MAPK module lies at the heart of many signalling pathways in eukaryotes. An input signal leads to activation of the MAPK cascade, which then generates an output response. (B) The MAPK cascade consists of MAPKKK (MEKK), MAPKK (MEK), and MAPK (ERK) (see the text for details). A receptor is activated in response to an extracellular signal, which leads to activation of the MAPK cascade (see the text for details). The activated MAPK phosphorylates substrates, some of which include transcriptional activators, resulting in altered patterns of gene expression and protein activity.
The signal that leads to activation of the MAPK cascade is perceived by a variety of types of receptors: G-protein-coupled seven-transmembrane receptors, His-Asp phosphorelay sensors, receptor-tyrosine kinases, and integral membrane proteins (reviewed in reference 73). In the yeasts, serpentine receptors, His-Asp phosphorelay sensors, and integral membrane protein sensors have been identified. The budding yeast genome does not code for any receptor-tyrosine kinases (reviewed in reference 83). The target of activation of the MAPK cascade module is often a transcription factor (reviewed in references 76 and 208), although other targets have also been identified. Phosphorylation of a transcription factor by the MAPK can in principle increase its binding affinity, activation ability, or cellular location, resulting in increased transcription of target genes. Inactivation of the MAPK cascade by dual-specificity phosphatases and by tyrosine phosphatases is one mechanism for attenuation of the signal and adaptation to the response (reviewed in references 82 and 146).
In S. cerevisiae, independently acting MAPK cascades exist that regulate response to osmotic stress, pheromones, perturbations of cell wall integrity, spore formation, and pseudohyphal growth (reviewed in reference 73). In S. pombe, fewer pathways—those for response to osmotic stress and pheromones—have been characterized. I will discuss all of the above pathways except the sporulation pathway.
The components of the MAPK cascade module are related to other kinases by virtue of their catalytic domain, which consists of approximately 250 to 300 amino acid residues. They catalyze the transfer of gamma phosphate from ATP to a hydroxyl residue on Ser and Thr or on Thr and Tyr (reviewed in reference 68).
The catalytic domain of all kinases can be subdivided into 12 conserved domains, within which 12 residues are invariant or highly conserved. These residues must thus be critical for activity of the kinase (reviewed in reference 68). Analyses of mutations of these highly conserved residues have been crucial in elucidation of the functional requirement of the kinase activity. The conserved primary structure of the catalytic domains suggests that they may fold into similar three-dimensional structures. The crystallographic structures of the catalytic domains of two mammalian MAPKs, ERK2 and p38, have demonstrated many similarities in their topology. Despite these similarities, differences were found in the ATP and substrate binding sites and in the phosphorylation lip (214, 232; see also reference 68). These variations in the topology may be important for the specificity of recognition by the activating MAPKK and for recognition of substrate.
MAPKs are activated by phosphorylation on two closely spaced Thr and Tyr residues (Thr-X-Tyr) found in catalytic subdomain VIII; X can be Pro, Gly, or Glu. Mutation of the X residue does not appear to affect activation by MAPKK (see reference 214 and references therein). The MAPKKs are activated by phosphorylation on two closely spaced serine residues or serine and threonine residues in domain VIII.
The MAPKKKs have a large N-terminal noncatalytic region and a C-terminal catalytic domain. The noncatalytic domain appears to be autoinhibitory, as inferred from the fact that deletion of this region causes constitutive activation of the kinase in the absence of a stimulus (see, for example, references 25 and 127). The mechanism by which MAPKKKs become activated remains to be elucidated and is the subject of current intensive studies. Conformational changes brought about by interaction with another protein or by self-dimerization or removal of a protein that inhibits the activation of the catalytic domain may be involved in such activation.
These introductory comments should help the reader put into perspective the MAPK cascade as we “tour the pathways.”
THE YING AND YANG OF RESPONSE TO OSMOLARITY: HOG, STY, AND PKC PATHWAYS
S. cerevisiae cells detect and respond to high and low extracellular osmolarity by activating two different MAPK pathways, HOG and PKC. High osmolarity activates the HOG pathway, and low osmolarity activates the PKC pathway. I first describe the response to high osmolarity in budding yeast and compare it with that in fission yeast. I then describe the response to low osmolarity in budding yeast; little is known of such a response in fission yeast.
Studies of the HOG pathway have provided important insights into activation of MAPK pathways. One particularly notable feature of this pathway is that it exhibits multiple redundancies. First, there is redundancy at the sensor level. Two independently acting branches activate the MAPK cascade. In one branch, the osmosensor is a member of a His-Asp phosphorelay system. In the other, the osmosensor is a membrane protein with multiple membrane-spanning domains. Second, there is redundancy at the MAPKKK level: multiple MAPKKKs can activate the MAPKK. The branch linked to the His-Asp phosphorelay system utilizes a pair of redundant MAPKKKs, whereas the other branch requires the activity of the MAPKKK of the pheromone response pathway, a very surprising finding. One important outcome of the studies with the HOG pathway is that it led to the identification of the first complete His-Asp phosphorelay system in eukaryotes. So far, no His-Asp phosphorelay systems have been identified in mammalian cells. The work carried out by Saito and colleagues provides a model of brilliant genetic detective work and biochemical analysis in the identification and characterization of many of the components of this pathway.
Response to High Osmolarity in S. cerevisiae and S. pombe
Glycerol plays an important role in the adaptation of both budding and fission yeast cells to increased external osmolarity. As both S. cerevisiae and S. pombe cells encounter hyperosmotic conditions, they increase their synthesis of glycerol, which leads to an increased internal glycerol concentration and thus to an increased internal osmolarity, which compensates for the elevated external osmolarity. S. cerevisiae and S. pombe cells unable to produce glycerol are unable to grow on hyperosmotic medium (2, 156). Thus, glycerol appears to be the major osmolyte used by the yeasts. Glycerol accumulation is in part due to increased activity of glycerol-3-phosphate dehydrogenase, encoded by GPD1 in S. cerevisiae and gpd1 in S. pombe (156). This increased activity results from activation of the MAPK cascade. Adaptation to high osmolarity is mediated by the HOG (high-osmolarity glycerol) pathway in S. cerevisiae and the Sty1 (suppressor of tyrosine phosphatase) pathway in S. pombe. In both cases, the MAPK cascade is activated in response to hyperosmolarity and leads to increased expression of a number of target genes.
Activation of the Hog1 MAPK cascade by two different sensors.
The Hog1 MAPK cascade (Fig. 2) consists of Ssk2 and Ssk22 (MAPKKK), Pbs2 (MAPKK), and Hog1 (MAPK) (16, 20, 127). Hog1 is activated by phosphorylation by Pbs2 on two residues (Thr174 and Tyr176) that reside in the catalytic domain and are conserved among all MAPKs. Pbs2 is activated by phosphorylation on two residues (Ser514 and Thr518) within the catalytic domain that are also conserved among MAPKKs (see “MAPK cascade nodule and signal transduction” above). Activation of Pbs2 can occur by two different branches (Fig. 2), both sensing hyperosmolarity and each acting independently of the other (127, 128). One branch (the Sln1 branch) involves a His-Asp phosphorelay system; the other (the Sho1 branch) employs a putative transmembrane osmosensor (Fig. 2) (128, 166). The Sln1 branch activates two redundant MAPKKKs (Ssk2 and Ssk22). These MAPKKKs, like other MAPKKKs, contain a large noncatalytic region that has been proposed to play a negative regulatory role in activation of the catalytic domain. A homolog (MTK1) of the Ssk2 and Ssk22 MAPKKKs has been recently identified and shown to mediate activation of the stress pathway in mammalian cells. Given that the similarity of these proteins extends to the noncatalytic regulatory region, it is possible that upstream regulators are also conserved (201).
FIG. 2.
Hyperosmotic stress response in S. cerevisiae and S. pombe. Response to hyperosmolarity is mediated by the HOG pathway in S. cerevisiae (left) and by the Sty pathway in S. pombe (right). Ptp2 is a phosphatase involved in down regulation of Hog1. Pyp2 is a phosphatase involved in down regulation of Sty1. Arrows indicate activation; lines with bars indicate inhibition. The dashed rectangle around Ypd1 indicates that the protein is present but not phosphorylated (see Fig. 5). See the text for details.
The other branch for activation of Pbs2 is mediated by the Sho1 osmosensor (Fig. 2). Sho1 has four transmembrane domains at its N terminus and a cytoplasmic SH3 (Src homology) domain at its C terminus (127). The SH3 domain is known to mediate interaction with other proteins by binding to proline-rich motifs (reviewed in references 36 and 179). The SH3 domain of Sho1 mediates interaction with Pbs2 and requires the Pro-rich region located in the N-terminal region of Pbs2 (127). Surprisingly, Sho1 activates Ste11, which is the MAPKKK for the pheromone response pathway (Fig. 2; see also Fig. 9). How Sho1 activates Ste11 and how Ste11 activates Pbs2 are not known. Other domains in the noncatalytic region of Pbs2 could, in principle, be important for association of Pbs2 with other proteins. Pbs2 has been shown to interact with Sho1, Hog1, and Ste11, although it is not known if the interactions with these proteins occur simultaneously. Pbs2 has been proposed to act as a scaffold that holds Ste11, Sho1, and Hog1 in a multiprotein complex (165). This proposed scaffold role is similar to the role proposed for Ste5 in the pheromone response pathway (see the section on the pheromone response pathway in S. cerevisiae, below). Because Pbs2 interacts with Sho1, which resides at the membrane, assembly of the Pbs2 multiprotein complex might occur at the membrane.
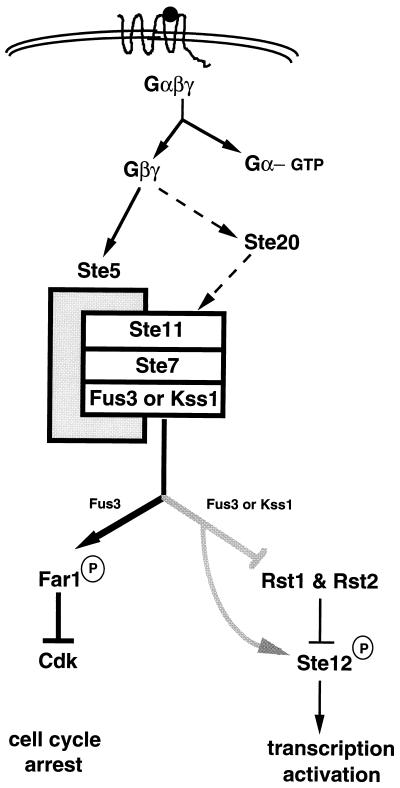
FIG. 9.
Pheromone response pathway of S. cerevisiae. The pheromone response pathway is activated upon binding of a-factor or α-factor (solid sphere) the to serpentine receptors Ste3 and Ste2, respectively. Ste5 is shown as the shaded rectangle that holds Ste11, Ste7, and Fus3 or Kss1 together. Solid arrows indicate activation; dashed arrows indicate that evidence is not conclusive; lines with bars indicate inhibition. See the text for details.
The observation that Ste11 is a component of both the pheromone response and HOG pathways raises the question whether there is cross talk between the two pathways: for example, does activation of Ste11 by pheromones lead to activation of the HOG pathway, and, conversely, does activation of the HOG pathway by hyperosmolarity lead to activation of the pheromone response pathway? Treatment of cells with pheromone induces the expression of FUS1, a target gene for the pheromone response pathway, as expected, whereas hyperosmotic shock does not. Exposure of cells to high osmolarity results in tyrosine phosphorylation of Hog1, as expected, whereas treatment with pheromone does not (165). Thus, cross talk is not observed. One explanation is that upon hyperosmotic shock, Ste11 is sequestered into a complex with Pbs2, whereas upon pheromone treatment, it is sequestered in a Ste5 complex. Pheromone treatment under hyperosmolarity conditions has not been tested. Another explanation is that upon hyperosmotic shock, Hog1 inhibits the pheromone response pathway (156a). The multiple functions of Ste11 in different pathways are summarized in Fig. 3.
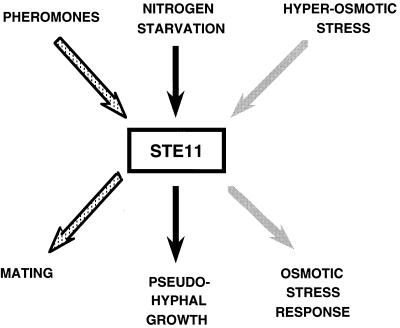
FIG. 3.
Multiple roles of the MAPKKK Ste11. The MAPKKK Ste11 can be activated by different signals resulting in different outputs. It can be activated by pheromones in the pheromone response pathway, resulting in mating (stippled arrows); a signal generated by nitrogen starvation in the pseudohyphal growth pathway, leading to morphological changes and growth properties of the cells (black arrows); and hyperosmolarity via Sho1 in the HOG pathway, resulting in the stress response (gray arrows). The mechanism by which activated Ste11 is prevented from inappropriately activating other pathways may involve scaffold proteins (see the text for details).
Osmosensing involves a multistep phosphorelay system related to the histidyl-aspartyl phosphorelay systems of bacteria.
Yeast has coupled two different types of phosphorylation cascades in the high-osmolarity signal transduction pathway. Above, I described phosphorylation through the MAPK cascade, and now I describe phosphorylation in the histidyl-aspartyl (His-Asp) phosphorelay system and how this phosphotransfer affects activation of the MAPK cascade. First, I present a brief overview of His-Asp phosphorelay systems (also known as two-component systems) in bacteria (52).
The His-Asp phosphorelay is widely used by bacteria to sense their external environment and to transduce the signal to the interior of the cell, resulting in altered patterns of gene expression. In this system, phosphate is transferred from a sensor protein to a response regulator (receiver) protein. In particular, the sensor protein contains an autophosphorylating histidine kinase domain, whose activity is modulated by an external stimulus. Activation of this kinase activity results in phosphorylation of a His residue within the kinase domain. The phosphate from this His residue is then transferred to an Asp residue in the receiver protein. The sensor protein may have an extracellular domain and a cytoplasmic domain. The receiver protein is cytoplasmic and transduces the signal. In bacteria, the receiver protein often has a DNA binding domain, so that perception of the signal results in activation of the DNA binding activity (reviewed in reference 52).
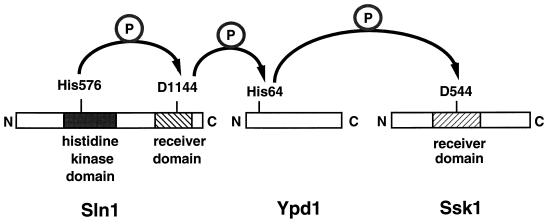
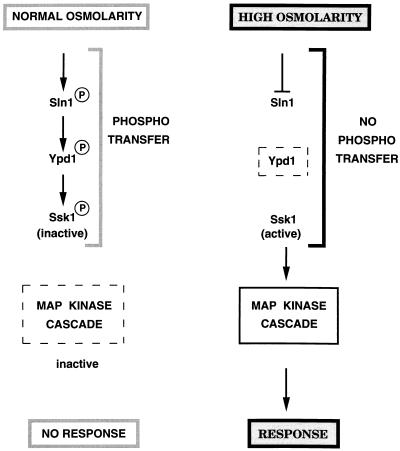
The phosphorelay system that governs the HOG pathway is complex and consists of three different proteins (Fig. 4): Sln1, Ypd1, and Ssk1 (128, 166). Sln1 contains two putative transmembrane domains that flank an extracellular domain, a cytoplasmic His kinase domain, and a receiver domain. Thus, Sln1 has both a sensor and a receiver domain. His576 in the kinase domain becomes phosphorylated, and the phosphate from His576 is then transferred to Asp1144 in the receiver domain of Sln1 (Fig. 4). The phosphate from Asp1144 is transferred to His64 in a second protein, Ypd1. Lastly, this phosphate is transferred to Asp554 within the receiver domain of the response regulator Ssk1 (Fig. 4) (166). Unlike many of the bacterial response regulators, Ssk1 does not have a DNA binding domain.
FIG. 4.
Phosphorelay system of the HOG pathway. A phosphorelay system consisting of three proteins, Sln1, Ypd1, and Ssk1, activates the HOG pathway. Sln1, the osmosensor, contains both a histidine kinase domain (black rectangle) and a receiver domain at its C terminus (hatched rectangle). Upon signal sensing, Sln1 autophosphorylates His576. Ssk1 contains a receiver domain (hatched rectangle) and is the activator of the MAPKKKs Ssk2 and Ssk22. Arrows indicate direction of phosphotransfer. Mutation of the His or Asp residues blocks phosphotransfer and results in constitutive activation of the pathway, suggesting that phosphorylation prevents activation of the HOG MAPK cascade. See the text for other details.
In bacterial His-Asp phosphorelay systems, a given signal can lead to either activation or inhibition of the His kinase activity in the sensor molecule. In the HOG pathway, hyperosmolarity inhibits the kinase activity of Sln1, the first protein in the phosphorelay system (Fig. 5) (128). Thus, no phosphate transfer occurs among the phosphorelay components, which leads to Ssk1 being unphosphorylated and able to activate the MAPKKKs (Fig. 5). Upon return to normal-osmolarity conditions, the Sln1 kinase is activated, phosphotransfer ensues, and phosphorylated Ssk1 cannot activate the MAPKKKs (Fig. 5). Several lines of evidence indicate that unphosphorylated Ssk1 activates the MAPKKKs. For example, mutations in amino acid residues involved in phosphotransfer interfere with phosphorelay and result in constitutive activation of the pathway (166). The mechanism by which Ssk1 activates the MAPKKKs is not known, but because Ssk1 interacts with the noncatalytic domain of Ssk2 and Ssk22, it has been proposed that it relieves the inhibitory activity of the N-terminal domain of these MAPKKKs (127).
FIG. 5.
High osmolarity inhibits phosphotransfer in the phosphorelay system and causes activation of the HOG MAPK cascade. Normal osmolarity conditions (left) stimulate phosphotransfer and result in phosphorylation of Ssk1. Phosphorylated Ssk1 cannot activate the MAPK cascade, and consequently there is no response. High osmolarity (right) inhibits Sln1 by an unknown mechanism. Phosphotransfer is blocked, and unphosphorylated Ssk1 is proposed to activate the MAPK cascade, resulting in the stress response. Arrows indicate activation; lines with bars indicate inhibition. The dashed rectangle around the MAPK cascade (left) indicates that it is inactive. The dashed rectangle around Ypd1 (right) indicates that the protein is present but not phosphorylated. See the text for details.
A gene, nik-1+, encoding a component of a His-Asp phosphorelay has been identified in the filamentous ascomycete Neurospora crassa. Like Sln1, Nik-1 is a hybrid kinase containing a histidine kinase domain and a response regulator domain (3; see also reference 182). nik-1+ is necessary for hyphal development. Deletion of nik-1+ results in misshapen and swollen hyphae that appear to lyse. Hyphae lacking nik-1 are osmosensitive. Thus, nik-1+ appears to regulate not only the response to hyperosmolarity but also fungal morphology. It would be interesting to determine if His-Asp phosphorelay systems also regulate hyphal development in other fungi.
Mutations that turn the pathway on or off—use of epistasis relationships to order pathway components.
In this section, I provide some examples of how components of the pathway have been identified, in some cases by genetic analysis and in others by inference from nucleotide sequence analysis. Genetic analysis relies on two key types of mutations: mutations that turn the pathway off and mutations that turn the pathway on in the absence of stimulus. There is no good reporter gene (see the next section) for measuring the activation of the HOG pathway, unlike the situation for the pheromone response pathway. Activation of the HOG pathway is assayed biochemically by determining tyrosine phosphorylation of the MAPK Hog1 with commercially available antiphosphotyrosine antibodies on proteins immobilized on a nitrocellulose membrane after a given treatment (see, for example, reference 165). Growth on plates with high osmoticum (1.5 M sorbitol or 0.9 M NaCl) is used in conjunction with the above assay. This plate assay is also used to screen for osmosensitive (Osms) mutants in the identification of pathway components.
The HOG1 and PBS2 genes were identified by isolation of mutants that were sensitive to medium of high osmolarity: both hog1 and pbs2 mutants are osmosensitive (Osms). Screening for such mutants in other fungi is likely to turn up homologs of these genes. Given that both hog1 and pbs2 mutants are Osms, the next step is to determine whether the double mutant has a similar osmosensitivity to that of the single mutants. In fact, the hog1 pbs2 mutant is as Osms as either single mutant. One interpretation of this result is that both genes act in a linear pathway affecting the same process.
Constitutive mutations are of great use in ordering components of a pathway and in identifying additional downstream components. A common theme for all the MAPKKKs is that deletion of the N-terminal noncatalytic region leads to constitutive activation of the protein and consequently of the pathway. For example, deletion of the N terminus of Ssk2 (the MAPKKK) leads to activation of the HOG pathway in the absence of stimulus (127). Because pbs2 mutations suppress the ΔN-SSK2 mutation, it was concluded that PBS2 is downstream of SSK2. Had the result been the opposite (i.e., no suppression), the conclusion would have been that PBS2 is most probably upstream. Another constitutive mutation that results in activation of the HOG pathway is ΔN-STE11 (165). This constitutive phenotype is abolished by pbs2 mutations, indicating that STE11 is upstream of PBS2. Constitutive mutations can also be used to identify unknown downstream components. For example, Δsln1 results in constitutivity of the pathway (and in lethality) (128). Downstream components were identified by screening for mutants that suppress the lethality of the constitutive mutation. Mutations in SSK1 (and in other genes of the pathway) were obtained in this manner.
Redundancy in genes or pathways can be inferred from analysis of the deletion phenotype of a given gene. For example, it was found that strains with SSK2 deleted still activated the MAPK Hog1, as measured by phosphotyrosine phosphorylation. This observation led to the suspicion that a redundant MAPKKK might exist. Low-stringency Southern blot analysis confirmed this suspicion and led to identification of SSK22, which is very similar to SSK2 (although the expression or activity of these genes or their products may be differentially regulated [see reference 127 for details]). Strikingly, the HOG pathway is still activated in a strain lacking both SSK2 and SSK22, and the mutant strain is osmoresistant (Osmr). Screening for Osms mutants of this strain led to the identification of additional components of the HOG1 pathway: SHO1 and STE11 (127, 165).
Stress response element.
Heat shock and osmotic stress induce synthesis of an overlapping set of proteins (211, 212; see also reference 77). Genes induced by osmotic stress, for example CTT1 and GPD1, contain a response element in their promoter region that appears to mediate the response to osmotic stress (129, 181). The stress response element (STRE) can also mediate the induction of transcription by other types of stresses, for example heat shock, nitrogen starvation, and oxidative stress, which are independent of the HOG pathway. Because this element mediates both HOG-dependent and HOG-independent induction, the use of STRE-reporter fusions has not proved very useful in analysis of the HOG pathway. Use of a transposon library (177) or of the more recently developed “gene microarrays” (186) should facilitate the identification of Hog1 targets and reporter genes for this pathway.
Response to Stress via the Sty1 MAPK Pathway in S. pombe—Coordination of Multiple Distinct Outputs in Response to Stress
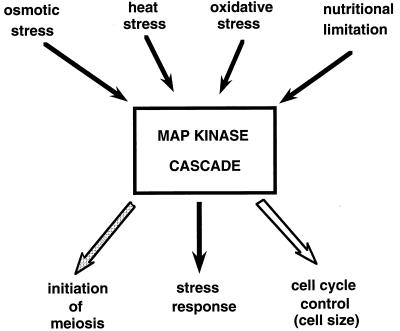
Fission yeast cells respond to osmotic stress via the Sty1 MAPK pathway (which is also called Spc1 and Phh1) (Fig. 2). Studies of this pathway have led to some important findings which clearly distinguish it from the HOG pathway of S. cerevisiae. First, in contrast to the HOG pathway, which is activated only by hyperosmotic stress, the Sty pathway is also activated by oxidative and heat shock stresses, nutrient limitation, and anisomycin (a protein synthesis inhibitor) (43, 222). Thus, in this respect the S. pombe pathway resembles the p38 and SAPK/JNK stress-activated pathways of mammalian cells (57, 67, 108). Activation of the Sty pathway by multiple stress signals raises the question whether different sensors are used for the different stress signals. Second, the Sty pathway integrates stress sensing with control of mitosis, a very important finding which may shed light on how the extracellular environment regulates mitosis (138, 190). Third, the Sty pathway utilizes a transcription factor with similarity to the transcription factor activated in mammalian cells in response to stress (189, 222). Therefore, the similarity of the pathway to that of mammalian cells extends to the transcriptional activator. Fourth, the Sty pathway links stress signalling with control of sexual differentiation (43, 97, 200). The S. pombe transcriptional activator governs the expression of target genes involved in the stress response and in the initiation of meiosis (189, 200, 222). Thus, the Sty pathway not only regulates stress responses but also integrates this response with two processes fundamental to all eukaryotes: control of mitosis and initiation of meiosis (Fig. 2 and 6).
FIG. 6.
Multiple stress conditions activate the Sty1 MAPK cascade, resulting in coordination of the stress response with mitosis and meiosis. The Sty pathway can be activated by osmotic, heat, and oxidative stress, by nutritional limitation (black arrows above rectangle), and by anisomycin (not shown). Osmotic stress not only activates the stress response (black arrow below rectangle) but also controls cell size at time of mitosis (open arrow below rectangle). In response to nutritional limitation, the stress response is activated, resulting in expression of ste11, which initiates sexual differentiation (stippled arrow below rectangle).
The components of the Sty1 MAPK pathway are homologous to those of the HOG pathway of S. cerevisiae.
The MAPK cascade module of the Sty1 pathway (Fig. 2) consists of the MAPK Sty1, which is activated by phosphorylation on the Thr171 and Tyr173 residues by the MAPKK Wis1 (138, 178a, 190). This kinase in turn is activated on Ser469 and Thr473 (178a, 187, 188) by the MAPKKK Wak1 (also known as Wik1 and Wis4). The MAPKKK in turn appears to be activated by Mcs4 (187, 188) (Fig. 2), which has amino acid sequence similarity to Ssk1. It has been proposed that a His-Asp phosphorelay system similar to that regulating the HOG pathway is involved in the response to high osmolarity in S. pombe, although a histidine kinase has yet to be identified (187, 188).
The MAPK Sty1 appears to be downregulated by Pyp2, a tyrosine phosphatase whose expression is induced by stress conditions. It thus appears that Pyp2 participates in a negative-feedback loop that allows restoration of the basal level of Sty1 activity (138, 190). Inactivation of Pyp1, another tyrosine phosphatase that acts on Sty1, has been proposed to be the mechanism by which heat shock activates the Sty1 pathway (178a).
A transcription factor with similarity to mammalian ATF2 is the target of the MAPK cascade.
A transcriptional activator has not been identified for the S. cerevisiae HOG pathway. In contrast, in S. pombe, the transcriptional activator Atf1 (also known as Gad7) is downstream of the Sty1 MAPK (189, 200, 222) (Fig. 2). Atf1 is a bZIP transcription factor (references 96 and 200 and references therein), highly homologous to mammalian ATF-2 (which is involved in the mammalian stress response). It is phosphorylated in a Sty1- and Wis1-dependent manner and appears to be a direct substrate for Sty1 (189, 200, 222). Several genes are known to be induced by different stress conditions in a Sty1-dependent manner (references 1, 43, 97, 138, 187, 189, 200, and 222 and references therein [detailing the identification and characterization of the genes]): pyp2 (encoding a tyrosine phosphatase), gpd1 (encoding glycerol-3-phosphate dehydrogenase), ctt1 (encoding catalase), fbp1 (encoding fructose-6-phosphatase), ste11 (encoding a high mobility group [HMG] transcription factor), and tps1 (encoding trehalose-6-phosphate synthase). Expression of these genes, not surprisingly, has also been shown to be Atf1 dependent (200, 222), and some evidence exists suggesting that Atf1 directly interacts with ATF-like binding sites in the promoter region of gpd1 (222).
The atf1 gene was identified independently in three different ways: (i) by searching the fission yeast genome for open reading frames with homology to mammalian ATF transcription factors (200), (ii) by screening for genes that on a high-copy-number plasmid suppress the mating defect of sty1 mutants (see below) (189), and (iii) by screening for weak sterile mutants defective in G1 arrest after nitrogen starvation (96). The atf1 mutants, in summary, are Osms, sterile, defective in G1 arrest, and defective in transcription of genes activated by stress conditions. atf1 mutants are not defective in size control at the time of mitosis, an important piece of evidence for proposing that Sty1 controls cell size at mitosis via a different target (Fig. 2) (see the next section (189, 222).
Links between the Sty pathway and cell size control at mitosis.
Understanding how eukaryotic cells coordinate the response to the extracellular environment and progress through the cell cycle is a key question, about which we have little information. Studies of the S. pombe Sty pathway may provide important insights into how this might occur. In this section, after a synopsis of fission yeast growth and mitotic cycle, I present some of the observations that have led to the proposal that the Sty pathway links the control of cell cycle progression and the response to stress (for more details see references 138 and 190).
Fission yeast cells are cylindrical rods that grow by elongation at the tips. Division occurs when the cells attain a critical cell size, which is constant from cell cycle to cell cycle under the same growth conditions (reviewed in reference 72). A medial septum is laid down, dividing the cell into two equal daughter cells (28). Mitotic initiation is controlled in all eukaryotes by activation of a cyclin-dependent kinase (Cdk), a key regulator of mitosis. At interphase, the Cdk is maintained in an inactive state by phosphorylation carried out by tyrosine kinases. Activation of the Cdk (Cdc2) in S. pombe is triggered by tyrosine dephosphorylation by the Cdc25 phosphatase. Other genes are also involved in control of the timing of mitosis (reviewed in reference 153).
Several lines of evidence establish a link between the Sty pathway and control of mitosis. First, the wis1 gene, which encodes a MAPKK, was identified as a dose-dependent initiator of mitosis (215). Overexpression of wis1 causes cells to enter mitosis at a reduced cell size, whereas loss of wis1 causes an elongated phenotype, indicating a delay in entry to mitosis. In addition, wis1 mutant cells are Osms, an important finding tying these different phenotypes to a single gene. Second, loss-of-function mutations in sty1, wak1, and mcs4 all lead to an elongated cell phenotype and to Osms cells (178a, 187, 188). The wis1 sty1 double mutant is as Osms and as elongated as each of the single mutants, suggesting that these genes most probably act in a linear pathway governing the same processes. Third, mcs4 was originally identified as a suppressor of the mitotic catastrophe phenotype of cdc2-3w wee1-50 mutants (references 187, 188, and 190 and references therein). Because mutations in mcs4, wak1, wis1, and sty1 affect not only growth on medium of high osmolarity but also size, it has been proposed that S. pombe cells are able to integrate changes in extracellular osmolarity and control of cell size at the time of mitosis (138, 190). How this is accomplished at the molecular level remains to be determined.
In contrast to mutations in the above genes, mutations in atf1 cause osmosensitivity but not an elongated cell phenotype, suggesting that Sty1 controls cell size at mitosis via an output distinct from atf1 (Fig. 2) (189, 200, 222).
Identification and order of some of the pathway components.
As in the analysis of the HOG pathway, the use of mutations that turn the pathway off or on has been extremely useful in ordering the components of the Sty pathway. One way in which the Sty1 pathway can be activated constitutively is by expressing genes under the control of the inducible nmt promoter. In the presence of thiamine, the promoter is repressed, whereas in its absence, the promoter is activated, resulting in high-level expression of the gene under its control. Overexpression of mcs4 in this manner causes lethality; this lethality can be suppressed by mutations in wak1, wis1, and sty1, indicating that these genes are probably downstream of mcs4 (188). The order of wak1 with respect to wis1 was inferred from the following analysis. As is the case with the MAPKKK of the HOG pathway, deletion of the N terminus of Wak1 causes constitutive activation of the pathway and leads to lethality. Tyrosine phosphorylation of Sty1 in the absence of stimulus was observed under these conditions and was dependent on a functional Wis1 protein. The lethality of ΔN-wak1 can be suppressed by mutations in wis1 but not in mcs4, indicating that wis1 is most probably downstream of wak1 and that mcs4 is most probably upstream of wak1 (187). These and other results support the order Mcs4-Wak1-Wis1-Sty1 (references 178a, 187, and 188 and references therein). Other observations (see references 187 and 188 for details) have led to the proposal that Wis1 may be activated by an independent pathway, by analogy to the HOG pathway of budding yeast (but see reference 178a for an alternative view).
Two independent approaches led to the identification of the Wak1 MAPKKK: (i) a search of the S. pombe genome database for genes exhibiting similarity to SSK2 and SSK22 of S. cerevisiae (187, 188) and (ii) a genetic screen (178a). Given that the genomes of other fungi are not available, another approach to identifying homologs is the candidate gene approach by using PCR with degenerate oligonucleotides that match a subset of MAPKKKs involved in a particular response, for example, stress response but not pheromone response. This strategy has been used successfully to identify a STE7/byr1 homolog in the corn smut fungus Ustilago maydis (9) and a FUS3 homolog in the rice blast fungus Magnaporthe grisea (225) (see the sections, Mate as you wish, and Tales of G proteins, pheromones, and MAPK components, respectively [below]). Another approach to the identification of desired genes is by functional complementation of mutations in the yeasts with expression cDNA libraries from other fungi.
Integration of the response to stress with sexual development.
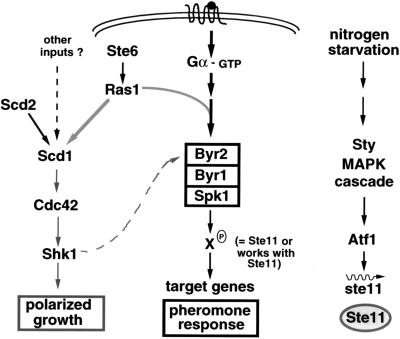
The Sty pathway has proven to be involved not only in regulation of the stress response and mitosis (see above) but also in the initial step leading to sexual differentiation (Fig. 2; also see Fig. 11). Nitrogen starvation, which is required for conjugation and sporulation (see the section on the pheromone response pathway in S. pombe, below), induces the Sty stress response. One consequence of this activation is the transcriptional induction of ste11 (97, 200) (Fig. 2). This gene encodes a transcription factor required for the expression of genes necessary for the initiation of sexual development (199). Thus, an output of the Sty1 pathway, Ste11, subsequently participates in the transcription of genes which then activate the pheromone response pathway (Fig. 2; also see Fig. 11).
FIG. 11.
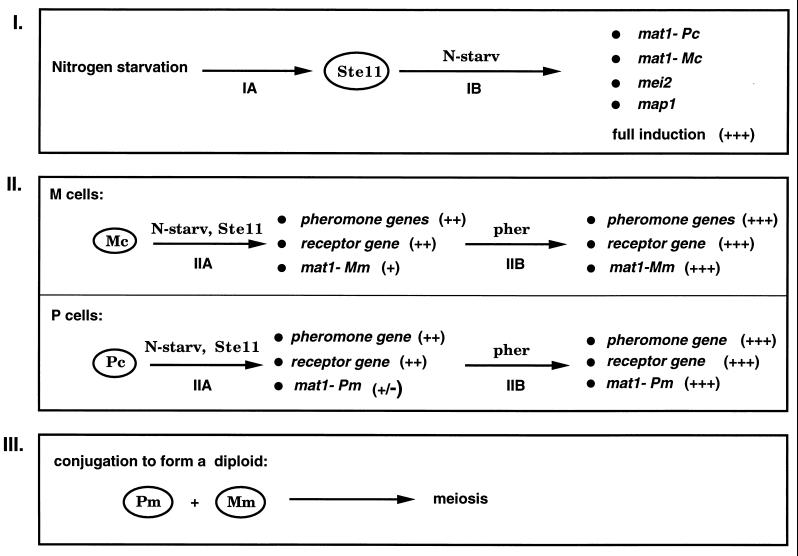
Stepwise induction of genes for mating and meiosis in S. pombe. The induction of genes necessary for mating and meiosis appears to occur in several steps (steps I to III), all of which require nitrogen-depleted conditions. Ste11 appears to be required for steps I and II. Symbols: +++, full-level induction; ++, induction but not to full level; +, low-level induction; +/−, slight induction; pher, pheromones; N-starv, nitrogen starvation. See the text for details.
The evidence linking the output of the Sty1 pathway with the initiation of sexual differentiation was first suggested by the finding that strains with mutations in components of the Sty pathway exhibited reduced mating and sporulation. On closer examination, these mutants were found to be defective in N starvation-induced G1 arrest and in induction of ste11 mRNA (96, 97, 189, 200, 222). The finding that the 5′ regulatory region of ste11 contains putative binding sites for Atf1 suggests a direct role of Atf1 in activating the transcription of ste11. Whether Atf1 acts alone or in conjunction with another ATF-like protein, Pcr1, which is also required for G1 arrest and ste11 expression, remains to be determined (for details, see references 96 and 200).
The discussion of the S. cerevisiae HOG and S. pombe Sty pathways has attempted to summarize our current knowledge of the response to high osmolarity in the yeasts. Comparison of these pathways shows that structurally related MAPK cascades can be used to respond to only one stimulus, as in the HOG pathway, or to multiple stimuli, as in the Sty pathway. More importantly, studies on the Sty pathway have shown that the cell can integrate its response to external stimuli with critical processes in the life cycle of an organism, such as mitosis and meiosis (Fig. 6). The similarities of the components of these pathways make it likely that homologs are to be found in other fungi, where they might regulate a variety of processes. The relatedness of the components of the HOG and Sty pathways should allow a rational design of degenerate primers for the candidate gene approach to identify the desired genes in the organism of choice. As the studies with nik-1 in N. crassa show, osmosensing pathways can be important in the regulation not only of hyphal growth but also of hyphal morphology (3, 182).
PKC Pathway in S. cerevisiae and Response to Low Osmolarity
In contrast to the HOG pathway, which responds to hyperosmolarity, the PKC pathway is activated by low osmolarity. It is also activated in response to a variety of stimuli: nutrient sensing, thermal stress, and pheromones (38, 40, 95, 136) (Fig. 7). It is thought that a major role of the PKC pathway in response to these stimuli is to maintain cell integrity by controlling cell wall assembly and perhaps membrane assembly.
FIG. 7.
The PKC pathway controls cell integrity. The PKC MAPK cascade is activated in response to heat and low osmotic stress and nutrient limitation. Hcs77 is a putative mechanosensor that is proposed to sense membrane stretch. Pkc1 is proposed to activate a branched pathway (one branch is the MAPK and the other is hypothetical). Rlm1 appears to be a target for Mpk1. Solid arrows indicate activation; the dashed arrow is speculative. See the text for details.
Changes in cell wall composition can occur in response to growth medium, pheromones, cell fusion, cell cycle, and, in the case of pathogenic fungi, contact and growth within their host (136, 164, 230; see also reference 34). During apical growth of a yeast bud or a hypha in the cell cycle, the cell wall is in a dynamic state of change as new material is added at the growing point and subsequently modified to produce a structure capable of sustaining mechanical and chemical stress (reviewed in references 24 and 34). These modifications of the cell wall in response to diverse conditions entail activation of cell wall-synthesizing enzymes resident in the cell membrane and vectorial transport and exocytosis of vesicles that carry other wall and membrane components (reviewed in reference 34). The inability of a cell to modify its cell wall accordingly may result in a fragile cell wall and may lead to cell lysis, misshapen cells, or altered patterns of growth (for example, altered branching patterns or septum deposition).
Because one of the functions of the PKC pathway is to control the expression of genes encoding cell wall components and possibly also to control the vectorial transport of vesicles that may contain other components essential for cell integrity, this pathway is likely to be a major player during bud formation and hyphal extension. Studies of the PKC pathway in budding yeast should provide insights into how different input signals are integrated and mediate cell integrity.
Some notable features of the PKC pathway are as follows: (i) a small GTP binding protein (Rho1) controls the activity of the Pkc1 protein and also of glucan synthase, an enzyme involved in the synthesis of a major structural component of the fungal cell wall; (ii) a homolog of a phosphatidylinositol kinase regulates the activity of the Rho1 protein, perhaps linking phospholipid metabolism with cell wall synthesis and integrity in yeast; and (iii) a recently identified presumptive integral membrane protein may act as one of the sensors for the pathway.
PKC MAPK cascade and its activation.
The MAPK cascade module that mediates the transduction of the signal generated by low osmolarity and heat stress consists of the MAPK Mpk1, two redundant MAPKKs, Mkk1 and Mkk2, and the MAPKKK Bck1 (Fig. 7) (38, 88, 89, 114, 115, 136, 206). The proposal for sequential activation by phosphorylation of the MAPK cascade is based on in vivo epistasis analysis and on structural relatedness to kinases in other pathways for which in vitro function has been established. The MAPK Mpk1 is tyrosine phosphorylated and activated in response to heat shock and hypotonic conditions in an Mkk1- Mkk2-, Bck1-, and Pkc1-dependent manner (40, 95, 230). Mpk1 tyrosine phosphorylation increases as osmolarity decreases from isotonic conditions. In contrast, as osmolarity increases, Hog1 tyrosine phosphorylation increases. Thus, these two MAPKs are activated by opposite osmolarity conditions (40).
(i) Activation by Pkc1.
The MAPK cascade is activated by Pkc1 (Fig. 7), a serine/threonine protein kinase, although the mechanism of this activation is not known. Pkc1 contains several domains: a catalytic domain, a putative Ca2+ binding domain (C2 domain), a putative diacylglycerol (DAG) binding domain (C1 domain), and a pseudosubstrate domain. Thus, budding yeast Pkc1 resembles mammalian isoforms that require phospholipid, Ca2+, and DAG as cofactors for activation (118, 152). Mammalian protein kinase C enzymes (PKCs) govern cell growth, proliferation, differentiation, and other processes (reviewed in references 44 and 150). In mammalian cells, PKCs respond to extracellular signals through receptor-mediated hydrolysis of phosphatidylinositol-4,5-bisphosphate to diacylglycerol (DAG) and inositol-1,4,5-triphosphate (IP3). DAG serves as a second messenger to activate PKC, and IP3 functions to mobilize Ca2+ from intracellular stores (reviewed in reference 46). The pseudosubstrate domain is conserved in PKCs from yeast to mammalian cells and is proposed to maintain the PKCs in an inactive state in the absence of inducing signal (reviewed in reference 44). The fact that a mutation (Pkc1R398P) resulting in constitutive activation of yeast Pkc1 maps to the pseudosubstrate region supports this contention (152). Pkc1 is activated by Rho1 (see the section on Rho1 regulation of Pkc1 and glucan synthase, below).
Deletion of any of the genes encoding components of the MAPK cascade (Δbck1, Δmkk1 Δmkk2, or Δmpk1) results in the same phenotype: cell lysis at elevated temperature, which is osmotically remedial. This phenotype is also exhibited by mutants defective in the HCS77 gene (65) (see below). Because these components are needed only at 37°C, it is likely that a partially redundant pathway operates at other temperatures; perhaps this pathway is the other branch controlled by Pkc1 (Fig. 7) (see below). In contrast to this temperature-sensitive growth defect, PKC1 and RHO1 (see below) are essential for growth at all temperatures. Δpkc1 mutants proliferate only in osmotically stabilized medium; they undergo rapid lysis after transfer to medium lacking osmotic stabilizer (117, 158). Because deletion of PKC1 causes a more severe phenotype than does deletion of MPK1, it has been proposed that Pkc1 governs a branched pathway (Fig. 7) (115), but little is known of this branch. Because Δrho1 cannot be osmotically stabilized, Rho1 has been inferred to govern targets other than Pkc1 (Fig. 7) (152) (see below).
(ii) Activation by pheromones.
Mpk1 appears to be tyrosine phosphorylated and activated in response to pheromones (Fig. 7) (230). This phosphorylation is dependent on Bck1 and, surprisingly, also on Ste20, a protein kinase that activates the MAPK cascade of the pheromone response pathway (see the section on the pheromone response pathway in S. cerevisiae, below). Phosphorylation was observed 1 h after treatment with pheromone and was not dependent on Ste12. Pheromones induce the formation of a mating projection exhibiting highly polarized growth. Since Mpk1 has been implicated in polarized growth (proper chitin deposition, organization of cortical actin patches, and vectorial transport of vesicles), it is possible that pheromone induction of Mpk1 ensures cell wall integrity during projection formation (136, 230). In this context, it is worth noting that BCK1 mutants were identified in a synthetic lethal screen with mutants defective in SPA2, a gene required for projection formation during mating (38). The PKC pathway is necessary during projection formation but not during cell fusion—an activated allele of Pkc1 blocks cell fusion (164). During cell fusion, localized degradation of cell walls occurs in the area of cell-cell contact. This degradation is essential to allow the fusion of cellular membranes to form a zygote. One possibility is that the PKC pathway is downregulated at this stage to allow cell fusion (see reference 164 for details).
Rho1 regulates Pkc1 and glucan synthase.
(i) Activation of Pkc1.
Rho1 belongs to the family of small GTP binding proteins which includes the Rho, Rac, and Cdc42 subfamilies. These proteins have GDP- and GTP-bound states and act as molecular switches regulating a variety of cellular processes (reviewed in references 172 and 203). The switch from one state to the other is controlled by two types of proteins: a guanine exchange factor (GEF), which promotes the transition from the GDP-bound form to the GTP-bound form, and a GTPase-activating protein (GAP), which promotes GTP hydrolysis and hence the transition from the GTP state to the GDP state. The GTP-bound form is usually the active form and interacts with target proteins. (Examples are known, however, in which both GTP- and GDP-bound forms have different functions [159].) Rho has been proposed to be involved in reorganization of the actin cytoskeleton in mammalian cells and, through this process, to affect cell morphology, motility, and cytokinesis (reviewed in references 172 and 203). In S. cerevisiae, Rho1 is localized at sites of active growth or secretion—the presumptive bud site, the bud tip, and the neck region—and has been proposed to play an important role in bud formation. Rho1 localization coincides with localization of actin patches during the cell cycle (229). Actin is proposed to be involved in the transport of vesicles to the actively growing regions. Whether Rho1 is involved in actin reorganization in yeast is not known at present. The similarity of phenotype of both pkc1(ts) and rho1(ts) mutants and their suppressibility by 1 M sorbitol led to the suggestion that both RHO1 and PKC1 act in the same pathway.
S. cerevisiae Rho1-GTP has been shown in biochemical experiments to activate Pkc1 (Fig. 7). This activation may occur by relieving the inhibitory action of the pseudosubstrate region of Pkc1 on its catalytic domain. Activation of Pkc1 by Rho1-GTP is stimulated by cofactors such as phosphatidylserine (94, 152). The interaction of Rho1 with Pkc1 may involve translocation of Pkc1 to the membrane, similarly to Ras-mediated translocation of Raf to the membrane (see the section on the pheromone response pathway in S. cerevisiae, below). Because the Pkc1R398P activating mutation does not suppress the lethality of Δrho1, although it suppresses the rho1(ts) mutation, it has been proposed that Rho1 has other substrates in addition to Pkc1 (152).
(ii) Rho1 activates glucan synthase.
Yeast cell walls appear to be layered, with an inner, electron-transparent layer containing mainly glucan polymers and a fibrillar, electron-dense outer layer consisting mainly of mannoproteins. 1,3-β-Glucan polymers and mannoproteins are the major structural components of the yeast cell wall (reviewed in references 24, 34, and 87). In the filamentous Ascomycetes and in the Basidiomycetes, in addition to 1,3-β-glucan and mannoproteins, chitin is another major structural component (reviewed in reference 192). Synthesis of 1,3-β-glucan polymers is mediated by 1,3-β-glucan synthase (GS), a membrane-bound enzyme whose activity is stimulated by GTP. GS catalyzes the transfer of a glucosyl residue from UDP-glucose to a growing chain of 1,3-β-linked glucosyl residues (141; reviewed in references 24 and 87). Two genes, FKS1 and FKS2, encoding components of the catalytic moiety of GS have been identified in S. cerevisiae. Deletion of both FKS1 and FKS2 is lethal, whereas deletion of either is not, although the single mutants are very sensitive to cell wall inhibitors such as echinocandin. Both Fks1 and Fks2 exhibit a high degree of similarity at the amino acid level: 16 putative membrane-spanning domains, of which 6 comprise a block separated from the other 10 by a stretch of hydrophilic amino acids (50, 135). Because their structure resembles that of ATP binding cassette (ABC) transporters of bacteria (reference 50 and references therein), it is hypothesized that the glucan chain is extruded into the periplasmic space as it is synthesized (51).
Rho1 has recently been shown to be a regulatory subunit of the GS complex (Fig. 7) (51, 168). Both Fks1 and Rho1 localize to the bud tip (168), which is an active site of cell wall remodelling. It has been proposed that Rho1 regulates the synthesis of β-glucans in two ways: (i) by activation of the PKC pathway, which regulates the synthesis of the catalytic subunit of GS; and (ii) by direct activation of GS enzymatic activity (51, 84) (see below).
Targets of the PKC pathway.
The direct target of the PKC MAPK cascade appears to be a transcriptional activator (Rlm1) that may regulate the expression of genes for cell wall biosynthesis. It is also possible that additional targets exist for this pathway.
(i) Rlm1, a transcriptional factor downstream of Mpk1.
Rlm1 is a member of the MADS box of transcription factors, of which S. cerevisiae Mcm1 and mammalian serum response factor (SRF) are also members (218). The DNA binding domain of Rlm1 is most similar to that of mammalian MEF2 and appears to bind the same consensus sequence as MEF2 (47). The yeast genome contains a homolog of Rlm1, Smp1, which recognizes an extended version of the consensus recognized by Rlm1. The two proteins can form heterodimers (47). The C-terminal region of Rlm1, which may be the transcriptional activation domain, appears to be directly phosphorylated by Mpk1 in vivo and in vitro, suggesting that Mpk1 modulates the activation domain of Rlm1 (47, 217). Deletion of RLM1 or of SMP1 does not result in the cell lysis phenotype characteristic of deletion of the components of the PKC pathway. One possibility is that Mpk1 exerts its effects via targets in addition to Rlm1 and Smp1 (for details, see references 47 and 217).
(ii) Genes for cell wall biosynthesis.
The expression of several genes involved in cell wall biosynthesis is cell cycle regulated in a SWI4- and SWI6-dependent manner and peaks at the G1/S boundary, which coincides with active apical growth and extensive wall remodelling (84). These genes are FKS1, KRE6, MNN1, VAN2, CSD2, and GAS1, which encode, respectively, a subunit of GS, a protein for 1,6-β-glucan synthesis, proteins involved in mannosylation, a protein involved in glycosylation, chitin synthase III, and a glycosylphosphatidylinositol (GPI)-anchored membrane protein (reference 84 and references therein). Not surprisingly, these genes contain cell cycle regulatory sites in their 5′ regions (84). Pkc1 is not required for this cell cycle regulation but is required for a high constitutive level of expression of FKS1, CSD2, MNN1, KRE6, and GAS1 throughout the cell cycle. It has been proposed that the PKC pathway and Swi4-Swi6 regulate the synthesis of these genes in a coordinated manner and that this might explain the synthetic lethality observed for Δswi4 or Δswi6 and components of the PKC pathway (84).
A search of the S. cerevisiae genome database revealed several genes whose 5′ regulatory regions contained MEF2 core consensus sites (the site recognized by Rlm1) (47). Among them are RLM1, SMP1, and genes whose products are involved in cell wall construction: HKR1, encoding a regulator of β-glucan synthesis; KTR2, encoding mannosyltransferase; HSP150, required for cell wall integrity; FLO1, which governs flocculation; AFR1, whose product is involved in pheromone-induced morphogenesis; and FKS1 (see above) (reference 47 and references therein). Of these genes with MEF2 consensus sites, only FKS1 is regulated by Pkc1 (84). It remains to be determined whether the others are also regulated by Pkc1. It is possible that Pkc1 regulates their expression via another unidentified transcription factor.
The work of Igual et al. (84) and others, indicating that pkc1 mutants have an altered cell wall organization due to lowered 1,3-β-glucan, 1,6-β-glucan, and mannan content (158, 176), provides key evidence for the role of the PKC pathway in cell integrity by regulation of cell wall biosynthesis. The role of Pkc1 in cell wall integrity has been argued largely on the basis of the cell lysis phenotype exhibited by strains with mutations in the pathway. Although suggestive, it has never been convincingly demonstrated whether lysis precedes or follows death. Cells can lyse for a number of reasons, some of which, of course, are due to cell wall defects.
Hcs77, a putative receptor that senses membrane stretch.
HCS77 encodes a protein of 378 amino acid residues with a single putative transmembrane domain and an N-terminal signal sequence (65, 155). A number of observations indicate that it functions in the PKC pathway. Deletion of HCS77 results in a phenotype that is similar to the phenotype observed for deletion of components of the PKC pathway—a temperature-dependent cell lysis phenotype that can be remedied by osmotic stabilizers. Δhcs77 Δbck1 and Δhcs77 Δmpk1 double mutants exhibit the same phenotype as the single mutants, consistent with the view that they act in the same linear pathway (65). Of particular importance is the observation that heat induction of Mpk1 activity was reduced in the Δhcs77 mutant strain. Taken together, these results suggest that Hcs77 may be an upstream regulator of Pkc1. Because the phenotype of Δhcs77 is not as severe as that of Δpkc1, it can be argued that Pkc1 can be activated in other ways.
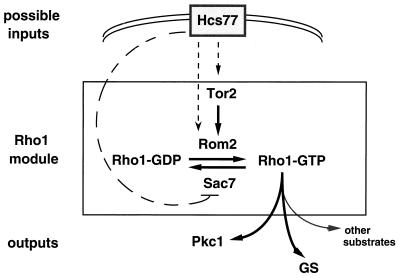
The analysis of hcs77 mutants and the likely membrane localization of Hcs77 protein make it an ideal candidate for sensing membrane perturbations in response to temperature and osmotic shock. Because Rho1 is localized to the membrane and activates Pkc1, one possibility is that Hcs77 activates Pkc1 via Rho1 (Fig. 8). As discussed above, exchange of GDP for GTP is promoted by GEFs, two of which are known for Rho1, Rom1 and Rom2 (157). The activity of Rom2 has recently been shown to be regulated by Tor2, an S. cerevisiae homolog of phosphatidylinositol kinase, which is implicated in the organization of the actin cytoskeleton (reference 180 and references therein). It has been proposed that a phosphorylated phosphoinositide might bind the PH (pleckstrin homology) domain of Rom2 and target its localization to the membrane, where it can act on Rho1 (157, 180). In one scenario, Hcs77 could affect GEF activity directly by stimulating Rom2 or indirectly by activating Tor2 (Fig. 8). The intrinsic GTP hydrolysis activity of Rho1-GTP is stimulated by GAPs, two of which, Sac7 and Bem2, have been identified (references 180 and 234 and references therein). SAC7 has GAP activity towards Rho1 (180). An alternative way in which Hcs77 could act on Rho1 is by inhibiting the GAP Sac7, thus promoting the GTP-bound state of Rho1 (Fig. 8). Future biochemical experiments designed to test interactions among these proteins should provide insights into how Hcs77 activates Pkc1.
FIG. 8.
Proposed activation of the Rho1 module by Hcs77. Hcs77, a putative mechanosensor, may activate Tor2, a phosphatidylinositol kinase homolog. Tor2 is proposed to activate Rom2, a GEF for Rho1-GDP. Alternatively, Hcs77 could activate Rom2 directly or could inhibit Sac7, a GAP for Rho1-GTP. This inhibition would promote the Rho1-GTP state. Rho1-GTP activates Pkc1, glucan synthase, and presumably other substrates. Solid arrows indicate activation; dashed arrows are highly speculative. See the text for details.
Little is known of the molecular mechanisms that govern the dynamic process of wall remodelling at the hyphal tip in filamentous fungi, but it seems likely that conserved components of the PKC pathway will play key roles.
PHEROMONE RESPONSE MAPK CASCADE FOR MATING, MEIOSIS, AND FILAMENTOUS GROWTH
Pheromone Response Pathway in S. cerevisiae
In this section I first describe the pheromone response pathway of S. cerevisiae and then that of S. pombe. The machinery used for the response—receptors, pheromones, heterotrimeric G protein, and MAPK cascade—is highly conserved, but some of the components are used differently. In particular, Gβγ transmits the signal to downstream components in budding yeast whereas Gα performs this task in fission yeast. Inputs other than the pheromones can modulate activity of the pathway in one of the yeasts. Because pheromones and receptors (and presumably a conserved response pathway) regulate filamentous growth in Basidiomycete fungi, it is likely that similar machinery is used by these fungi in their response to pheromones. Additional inputs are likely to be required to modulate the complexity of the developmental pathway leading to filamentous growth.
The pheromone response pathway of S. cerevisiae mediates cell-cell interactions during mating. This pathway has been intensively studied for many years and is one of the best-understood signalling pathways in eukaryotes. Studies with the pheromone response pathway of budding yeast have provided a framework for understanding how mitogenic factors regulate cell cycle progression in mammalian cells. An overview of this pathway follows (Fig. 9). (i) Peptide pheromones secreted by cells interact with seven-transmembrane receptors and initiate the response. (ii) The receptors interact with a heterotrimeric G protein that transmits the signal to downstream components. (iii) Partially redundant MAPKs mediate the activation of a transcription factor. (iv) One of the MAPKs phosphorylates a cyclin-dependent kinase inhibitor (CKI), leading to cell cycle arrest. (v) A component of the response pathway appears to act as a scaffold that brings together the MAPK cascade components and may prevent cross talk between this and other pathways. (vi) Many of the target genes for this pathway are known and include genes necessary for cell and nuclear fusion and for cell cycle arrest.
MAPK cascade and Ste5 scaffold.
The MAPK cascade (Fig. 9) consists of the MAPKKK Ste11, which phosphorylates and activates the MAPKK Ste7 on two residues conserved among all MAPKKs. Ste7 in turn phosphorylates and activates the redundant MAPKs Fus3 and Kss1 on conserved Thr and Tyr residues (56, 59, 122, 148, 235).
Downstream of the MAPKs is the transcriptional activator Ste12 (Fig. 9). Ste12 governs the expression of many genes in the pheromone response pathway and also genes for cell and nuclear fusion. It binds to a pheromone response element (PRE) located in the 5′ regulatory region of target genes (reviewed in references 107 and 132). Because Ste12 is phosphorylated by both MAPKs, it was proposed to be the direct target of the MAPKs (54, 81, 194). Studies of the activation of Ste12 by the MAPKs has revealed new levels of complexity: additional components are involved in the interaction of the MAPKs with Ste12. The proteins encoded by two recently discovered genes, RST1 (DIG1) and RST2 (DIG2), appear to act as inhibitors of Ste12 activation in the absence of pheromone. Both Fus3 and Kss1 interact with these proteins and phosphorylate them. Ste12 also interacts with Rst1 and Rst2 (37, 204). Rst1 and Rst2 are proposed to form a complex with Ste12 at Ste12 target sites, preventing transcriptional activation. Upon pheromone stimulation, the MAPKs phosphorylate Rst1, Rst2, and Ste12, resulting in dissociation of the complex and allowing Ste12 to activate transcription (Fig. 9) (see reference 37 for other models). It must be noted that the sites phosphorylated in Ste12 are not known, nor has the biological significance of this phosphorylation been determined, although it correlates with transcriptional activation (see references 81 and 194 for details).
Fus3 and Kss1 have been considered to be partially redundant MAPKs for transcriptional induction and mating but not for cell cycle arrest. Of the two MAPKs, only Fus3 phosphorylates Far1 (Fig. 9). This phosphorylation of Far1 regulates its association with the Cdk, causing cell cycle arrest in G1 (Fig. 9) (162). Recently, it has been proposed that Fus3, and not Kss1, is the MAPK for this pathway. A kinase-deficient point mutation of FUS3 has a severe defect in mating, indicating that Kss1 cannot substitute for Fus3 function when Fus3 protein is present, albeit in an inactive form (see reference 125 for details). Kss1 can supplant Fus3 only when Fus3 is absent due to deletion of the FUS3 gene (125).
The mechanism by which the MAPK cascade is activated is not well understood and remains an intensive area of research. It might involve Ste20 or Ste5 or both (Fig. 9). The role of Ste20 will be discussed in a separate section (see below).
The role of the Ste5 protein remained enigmatic for many years, even though it had been identified two decades ago (124). Epistasis analysis placed it after the G protein and before the MAPK cascade components (for details, see references 71, 104, and 112). STE5 codes for a protein with a LIM-like motif, which appears to mediate protein-protein interactions in other systems (112). Dimerization of Ste5 has been proposed to result in its activation, and the LIM motif, together with other domains, may promote Ste5-Ste5 interactions (227). Ste5 also has Pro-rich regions on either side of the LIM motif, which may interact with SH3 domain-containing proteins (reviewed in reference 53). The work of several groups, using the two-hybrid system and immunoprecipitation, led to several important findings on the role of Ste5 (32, 131, 167). First, Ste5 interacts with all of the components of the MAPK cascade—Ste11, Ste7, Fus3, and Kss1. Second, each of the MAPK components interacts with a different region of Ste5. Third, Ste5 interacts with the amino-terminal region of Ste11, an autoinhibitory region in all MAPKKKs. These and other observations led to the proposal that Ste5 acts as a scaffold to bring the MAPK cascade components together to promote sequential phosphorylation during activation of the pathway and also to facilitate attenuation of the response by Fus3 (reviewed in reference 53) (Fig. 9). Because the MAPK cascade components of the pheromone response pathway are structurally very similar to those of other pathways that coexist in the same cell, it has been suggested that another role of Ste5 is to prevent cross talk between pathways by confining the components of the pheromone response MAPK cascade in a complex. For example, such a complex may prevent Ste11, Ste7, and Fus3 from activating other pathways inappropriately (reviewed in reference 73). It is anticipated that other MAPK cascades will have proteins analogous or homologous to Ste5. Pbs2, the MAPKK of the HOG pathway, has been proposed to act as a scaffold (165) (see the section on response to high osmolarity in S. cerevisiae and S. pombe, above).
Attenuation of the pheromone response may involve phosphorylation of Ste7 and Ste5 by Fus3 (56, 59, 104) and also dephosphorylation of the MAPK Fus3 by the phosphatase Msg5 and other phosphatases (49, 231). In addition, the receptors and the Sst2 protein play a role in attenuation of the response (see below).
Upstream of the MAPK cascade: pheromones, receptors, and heterotrimeric G protein.
(i) Pheromones.
Two types of peptide pheromones exist in S. cerevisiae: α-factor (13 amino acid residues), which is secreted by α cells and is encoded by two genes, MFα1 and MFα2, and a-factor (12 amino acid residues), which is secreted by a cells and is encoded by two genes, MFa1 and MFa2 (reviewed in references 74, 107, and 132). Purified α-factor added to a cells induces the same changes as when a and α cells are cocultured: growth inhibition and formation of a mating projection (shmoo). Growth inhibition is due to arrest in the G1 phase of the cell cycle. Other assays have shown that upon pheromone treatment, expression of several genes is induced, including the receptor and pheromone precursor genes, genes for cell fusion, and genes for nuclear fusion (reviewed in references 107 and 132). Unlike α-factor, the a-factor precursor is modified at its C terminus by carboxymethylation and farnesylation (reviewed in references 107 and 132). The farnesyl moiety is found in many eukaryotic proteins and is proposed to target the proteins to the membrane (35). Both α and a cells contain the genes for the different pheromones and the different receptors, but expression of the genes is cell type specific, governed by regulatory proteins encoded by the mating-type locus (MAT) (reviewed in reference 74). In contrast, in the filamentous Basidiomycetes, the pheromone precursor and receptor genes are encoded by the mating-type loci themselves (reviewed in references 6, 25a, and 210).
All pheromones found so far in the Ascomycetes are structurally of the two types described in budding yeast. In contrast, the pheromones found in the Basidiomycetes are all structurally of the a-factor type; that is, all are small prenylated peptides that lack a signal sequence and are thus presumably secreted by a Ste6-like mechanism (25a, 41, 210). Whatever the reasons for this difference, one practical disadvantage is that purification of a-factor-like lipopeptides is more difficult than that of α-factor-like peptides. Thus, purified pheromone may not be as readily available for the types of experiments that have been so useful in yeast with commercially available α-factor.
(ii) Receptors and heterotrimeric G proteins.
The receptors in both a and α cells are of the seven-transmembrane type. The receptor for a-factor is encoded by STE3, and that for α-factor is encoded by STE2 (reviewed in reference 74). There is little similarity between the amino acid sequence of these receptors, although there is structural conservation. The third cytoplasmic loop of the α-factor receptor has been implicated in pheromone discrimination, receptor activation, and internalization (21, 197; see also reference 18). Phosphorylation of the C-terminal tail mediates adaptation to the pheromone response, and ubiquitination governs internalization of the receptor (31, 75, 178; reviewed in reference 110).
The receptors appear to be coupled to a heterotrimeric G protein (Fig. 9) (78, 93), which is the same in both cell types and consists of Gα, Gβ, and Gγ subunits, encoded by the GPA1 (SCG1), STE4, and STE18 genes, respectively (reviewed in references 107 and 132). Both Ste4 and Ste18 precursors have a CAAX box at their C terminus (reviewed in references 107 and 132), a hallmark for isoprenylation. Activation of the receptor results in exchange of GDP for GTP in the Gα subunit and concomitant dissociation of the heterotrimeric G protein into Gα-GTP and Gβγ subunits (reviewed in references 19, 147, and 171). Gβγ is the activator of the downstream components (reviewed in reference 73): a null mutation of GPA1 results in constitutive activation of the pathway and lethality due to cell cycle arrest, whereas Δste4 or Δste5 results in a nonmating phenotype. A key observation is that double mutants, for example Δgpa1 Δste4, behave as Δste4, indicating that Gβγ is the activator. Binding of Gα to Gβγ prevents activation by Gβγ in the absence of stimulus (reviewed in references 73, 110, and 132). The SST2 gene is required for adaptation to the response: sst2 mutations cause hypersensitivity to the pheromones (45, 48). It is hypothesized that Sst2 is a GAP for Gα-GTP, as is now known to be the case for some members of the RGS family of proteins in other eukaryotes (13; reviewed in references 101 and 191).
A very important question remaining in studies of the pheromone response pathway is the mechanism by which Gβγ activates downstream components and the molecular nature of the signal. Ste4 and Ste5 appear to interact as assayed by two-hybrid analysis and immunoprecipitation. The first 214 amino acid residues of Ste5 appear to be sufficient for this in vivo interaction, although whether the interaction is direct remains to be determined (221). Such an interaction might be important for translocation of Ste5 to the membrane, where Gβγ resides. Other work suggests that Ste5 and Ste20 interact, but again it is not known if this interaction is direct (116). One possibility is that Gβγ transmits a signal to both Ste5 and Ste20 (Fig. 9). Alternative possibilities exist, and future experiments should clarify these interactions.
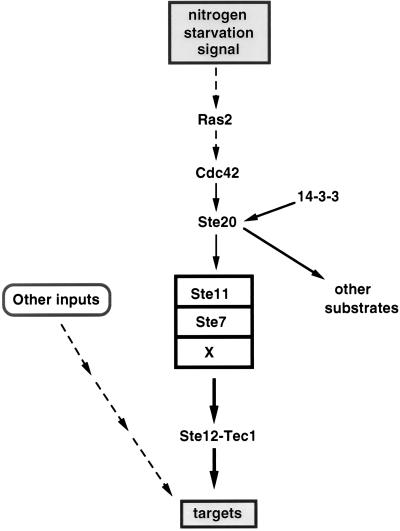
Ste20, a p65PAK protein involved in mating.
A key link of the MAPK cascade with upstream elements may be the Ste20 protein, which was placed upstream of the MAPK cascade and downstream of the G protein by epistasis analysis (reviewed in reference 110). Because it is a kinase (109, 113, 161, 169) (see below), an attractive possibility is that it activates Ste11 by phosphorylation. In vitro studies indicate that Ste20 phosphorylates Ste11 but whether this interaction is relevant in vivo remains to be determined (224).
Ste20 is a serine/threonine protein kinase of the p65PAK family found in different eukaryotic organisms and shown to be activated in vitro by GTP-bound Cdc42, a member of the Rho family of small GTP-binding proteins (reviewed in references 22 and 184). Ste20, like many members of this family, contains a C-terminal catalytic domain and an N-terminal noncatalytic region, within which is a conserved motif, the Cdc42/Rac interactive binding motif (CRIB), necessary for binding by activated Cdc42 (reviewed in reference 22). Activation of p65PAK proteins may mediate the role of Rho proteins in governing the reorganization of the actin cytoskeleton in response to diverse stimuli (184). This prompted an investigation of whether binding of Cdc42 to the CRIB site of Ste20 is responsible for activation of the pheromone response MAPK cascade (109, 161, 193, 233) and for the morphological changes that ensue, which involve reorganization of the actin cytoskeleton.
The role of Cdc42 in the pheromone response pathway has been definitively tested by examining the phenotype of STE20 mutants that lack the Cdc42 binding site (109, 161). These mutants exhibit normal transcriptional induction, mating projection formation, and G1 arrest. Thus, Cdc42 binding to Ste20 is not necessary for early steps in the pheromone response. Given that Cdc42 binds Ste20 in vitro and in vivo and that the CRIB site is necessary for such interaction, what is the role of this interaction? One possibility is that association of Cdc42 with Ste20 has a role in cell-cell contact during fusion, as inferred from the fact that mutants with deletion of the CRIB site show a reduced frequency of zygote formation (109, 161). This behavior is not due to defects in actin cytoskeleton organization, inability to orient toward a pheromone source, or inability to form mating projections, which occur normally in the mutants. Because Cdc42 binding to Ste20 is necessary for localization of Ste20 at the shmoo tip, one possible explanation for a defect in cell-cell contact is that Ste20 is necessary for polarized secretion of components necessary for adhesion.
These studies have clarified much about the role of Ste20 in the pheromone response pathway. Nonetheless, it remains to be determined how Ste20 is activated. It has been suggested that p65PAK proteins may be activated by G-protein-linked receptors in a Rho-independent manner (reviewed in reference 184). It is thus possible that Gβ γ activates Ste20 by an as yet unknown mechanism. Because the noncatalytic region of Ste20 appears to inhibit activation of the catalytic domain (109, 169), one possibility is that this region is the target of Gβ γ (reviewed in reference 110).
Identification of the components of the pathway.
Most of the components of the pathway have been identified by simple genetic screens and selections in combination with simple assays for the pathway. The “halo” assay tests growth inhibition in response to pheromones: strains that are sensitive cannot grow around the source of pheromone and form a clear zone or halo of no growth, whereas resistant strains produce no zone of inhibition (reviewed in reference 74). Transcriptional activation of the pathway is assayed by measuring the level of induction of FUS1-lacZ, a reporter gene which is highly induced by pheromone treatment (reviewed in references 107 and 132). Elucidation of the order of the components relied on the use of constitutive mutations which activate the pathway in the absence of pheromone and mutations which turn the pathway off and, more recently, on the use of two-hybrid analyses and biochemical techniques.
Because α-factor causes growth inhibition of a cells, isolation of mutants resistant to this growth inhibition led to the identification of STE2, STE4, STE5, STE7, STE11, STE12, STE18, and FUS3 (also identified as a gene required for cell fusion) (27, 55, 70). FAR1 was identified as a mutation conferring α-factor transcriptional induction, in contrast to other α-factor-resistant mutants, which block transcriptional induction (28). A subset of these components was also identified as mutations that confer a nonmater phenotype: STE2, STE3, STE4, and STE5 (124). Many of the genes involved in a-pheromone biosynthesis were also identified as mutations that result in a nonmater phenotype: STE6, STE14, and STE16 (reviewed in reference 132). The gene encoding Gα (GPA1/SCG1) was identified because its expression on a high-copy-number plasmid confers α-factor resistance. It was also identified by hybridization with a rat Gα gene as a probe (reviewed in references 107 and 132).
Constitutive mutations in STE11 allowed the ordering of STE11 with respect to STE7 and FUS3 but left its relationship with STE5 ambiguous, since the STE11 constitutive allele suppresses the FUS1 transcriptional defect of Δste5 but not the mating defect (25, 198). Constitutive mutations in STE5 allowed the ordering of STE5 with respect to Gβγ (encoded by STE4 and STE18) but left its position with respect to STE11 ambiguous (71). The use of the two-hybrid system led to a definitive explanation of these discrepancies as already described (32, 131, 167).
Pheromone Response Pathway in S. pombe
In contrast to S. cerevisiae, in S. pombe the pheromone response pathway is used not only for mating but also for initiation of meiosis, an observation that is highly relevant to Basidiomycetes, where pheromones and receptors are necessary for postfusion events. The pheromone response pathways of the two yeasts use essentially the same machinery, but as we shall see, one of the components is used differently.
Several features of the S. pombe pheromone response pathway distinguish it from the S. cerevisiae pathway: first, two inputs appear to be important for activation of the MAPK cascade, a G-protein-coupled receptor and the Ras1 protein. Second, Gα rather than Gβγ is the activator of the pathway. Third, nitrogen starvation is essential for activation of the pathway. It induces the expression of a transcriptional activator (Ste11) essential for the transcription of many genes necessary for the pheromone response, including the mating-type genes, which, unlike the MAT genes of S. cerevisiae, are not fully expressed during vegetative growth.
Before describing the pheromone response pathway in fission yeast, I will give a brief account of the different mating types and the regulators that govern cell type specificity and initiation of meiosis (see Table 1).
TABLE 1.
S. pombe genes that govern mating and meiosis
| Gene | Product | Reference(s) |
|---|---|---|
| Required by M and P cells | ||
| fus1 | Fusion protein | 163 |
| map1 | MADS box protein required for P-specific gene expression and enhancement of M-specific gene expression | 151, 226 |
| mei2 | RNA binding protein required for initiation of meiosis | 219 |
| ste6 | GEF for Ras1 | 80 |
| ste11 | HMG-box transcription activator | 199 |
| gpa1 | Gα subunit coupled to pheromone receptors | 154 |
| ras1 | Homolog of mammalian Ras | 145 |
| byr2 | MAPKKK | 213 |
| byr1 | MAPKK | 145 |
| spk1 | MAPK | 205 |
| Required by M cells | ||
| mam1 | M-factor ABC transporter | 33 |
| mam2 | P-factor receptor | 98 |
| mfm1 | M-factor precursor 1 | 42 |
| mfm2 | M-factor precursor 2 | 42 |
| mfm3 | M-factor precursor 3 | 100 |
| sxa2 | Protease that degrades P-factor | 86 |
| mat1-Mc | HMG-box regulatory protein required for M-cell-specific gene expression | 99 |
| mat1-Mm | Mating-type regulatory protein required for meiosis | 228a |
| Required by P cells | ||
| mat1-Pc | Mating-type regulatory protein required for P-cell-specific gene expression | 228a |
| mat1-Pm | Mating-type regulatory protein of the homeodomain family required for meiosis | 228a |
| map2 | P-factor precursor gene | 85 |
| map3 | Receptor for M-factor | 202 |
| sxa1 | Protease that degrades M-factor | 86 |
Haploid cells of S. pombe are of two mating types: minus (h−) and plus (h+), and they produce M and P pheromones, respectively, upon nitrogen starvation (reviewed in references 228 and 228a). The M-factor precursor is encoded by three genes: mfm1, mfm2, and mfm3. M factor, like a-factor, is farnesylated and carboxymethylated (100; reviewed in reference 41). The P-factor precursor is encoded by the map2 gene and is structurally similar to the α-factor precursor of S. cerevisiae (85). In response to mating pheromones, cells produce a short conjugation tube and fuse to form a zygote, which immediately undergoes meiosis and sporulation. Thus, meiosis is tightly coupled to mating (reviewed in references 228 and 228a).
The mat genes control mating type and also entry into meiosis. M cells have the matM allele with two genes, matMm (matMi) and matMc. P cells have the matP allele with two genes, matPm (matPi) and matPc (reviewed in references 228 and 228a). matMc and matPc are expressed constitutively at a low level and become fully induced upon nitrogen starvation (see the section on stepwise induction of mating and meiosis by nitrogen starvation and pheromones, below). They are required for expression of the pheromone and receptor genes. In contrast, matPm and matMm are expressed only under nitrogen starvation, become fully induced by pheromones, and are absolutely required for initiation of meiosis (199, 223, 228a) (see the section on stepwise induction of mating and meiosis by nitrogen starvation and pheromones, below). matPm and matMm act together to induce the expression of mei3, a positive regulator of meiosis, which functions by inhibiting an inhibitor of meiosis (Ran, also called Pat): Mei3 ———| Ran ———| Mei2 → meiosis (137; reviewed in reference 228a). The requirement for the combinatorial activity of Pm and Mm in the regulation of meiosis is similar to the requirement of a1-α2 for meiosis in S. cerevisiae.
MAPK cascade components.
The MAPK cascade consists of the Byr2 MAPKKK, the Byr1 MAPKK, and the Spk1 MAPK (64, 145, 205, 213) (Fig. 10). These kinases are presumed to act sequentially (see reference 149 for details). As in S. cerevisiae, binding of pheromones to the receptors results in dissociation of the heterotrimeric G protein into Gα-GTP and Gβγ subunits. Gα transmits the signal to downstream components. The observation that led to this conclusion is that deletion of the gene encoding Gα (gpa1) results in a nonmating phenotype (154). Both Gα and Ras1 are needed for activation of the MAPK cascade (reference 85 and references therein; reviewed in reference 79), and the signal that they transduce converges on the MAPKKK Byr2 (Fig. 10). Ras1 interacts with Byr2, but the mechanism by which this interaction leads to activation is unknown (134). The input from Gα and Ras1 may be coordinated by the pheromones, as suggested by the fact that ste6, encoding the putative GEF for Ras1, is transcriptionally induced by both nitrogen starvation and pheromones (80).
FIG. 10.
Pheromone response pathway of S. pombe. Ste11 is a key regulator of sexual differentiation in S. pombe. Its transcription is induced by the Sty pathway in response to nitrogen starvation (right) (also see Fig. 2). Its activity may be regulated by the Spk1 MAPK (middle). Ras1 and Gα-GTP are both necessary for activation of the pheromone response MAPK cascade. The target genes for this pathway include the pheromone precursor and receptor genes, the ste6 gene (encoding the GEF for Ras1), fus1 (required for cell fusion), mat1-Pm and mat1-Mm, and other genes (Table 1). Nitrogen starvation is indispensable for the pheromone response, since addition of pheromones to cells growing in rich medium does not elicit the mating response. Ras1 is proposed to regulate a morphogenetic cascade (left) by activation of Scd1 (a proposed GEF for Cdc42). Cdc42 in turn activates Shk1, a Ste20 homolog. See the text for details. Solid arrows indicate activation; dashed arrows indicate that evidence is limited or nonexistent.
The transcription factor that becomes activated by the MAPK Spk1 has not been identified. It might be Ste11 or a protein that acts with Ste11 (Fig. 10). Two MAPK cascades may thus control Ste11: one is the Sty pathway, which controls transcription of ste11 mRNA in response to nitrogen starvation (see the section on response to stress via the Sty MAPK pathway in S. pombe, above), and the other is the Spk1 pathway (Fig. 10), which may control the activity of Ste11 by phosphorylation or by controlling the activity of a protein that interacts with Ste11. The integration of the inputs of these two MAPK cascades ensures coordination of nutrient sensing and sexual development.
Stepwise induction of mating and meiosis by nitrogen starvation and pheromones.
In contrast to S. cerevisiae, S. pombe initiates sexual development only under nitrogen starvation (reviewed in references 228 and 228a). Expression of the components of the pheromone response pathway in S. pombe occurs in several steps, an initial step in response to nitrogen starvation, a second step involving cell-type-specific regulatory proteins, and a third step in response to pheromones and the pheromone response pathway (Fig. 11) (99, 223). The following description is a synthesis of work by different groups and may reflect some simplification.
In response to nitrogen starvation, ste11 is transcribed under the control of the Sty1 MAPK cascade, as described above (Fig. 3 and 11, step IA). Ste11, an HMG transcriptional activator, activates full-level transcription of mat1-Pc (in P cells), mat1-Mc (in M cells), mei2 (encoding a protein essential for meiosis), and map1 (encoding a MADS box protein, homologous to S. cerevisiae Mcm1) (Fig. 11, step IB) (151, 199, 219, 223, 226). In M cells, the Mc protein activates the transcription of pheromone and receptor genes and a low-level induction of the mat1-Mm gene (Fig. 11, step IIA) (100, 226). Induction of these genes also requires Ste11 (see reference 100 for details). In P cells, the Pc protein leads to induction of the pheromone and receptor genes and a slight increase in the induction of the mat1-Pm gene (Fig. 11, step IIA) (5, 85, 202). As in M cells, Pc-mediated induction may require Ste11. The pheromones and receptors thus synthesized activate the pheromone response pathway, leading to a boost in induction of pheromone and receptor genes and full-level induction of mat1-Mm in M cells and mat1-Pm in P cells (Fig. 11, step IIB) (5, 99, 223). Activation of the pheromone response pathway leads to induction of target genes responsible for formation of conjugation tubes and for cell fusion (Table 1). The fully differentiated M and P cells then conjugate to form a diploid zygote containing both Pm and Mm proteins, which associate to form the combinatorial activity Pm-Mm, which induces the transcription of mei3 and initiation of meiosis (Fig. 11, step III) (reviewed in reference 228a).
Several points about this stepwise induction should be mentioned. First, nitrogen starvation conditions are necessary at all stages. For example, cells growing in rich medium do not respond to pheromones. Second, Ste11 appears to be required at different stages: for the initial step in response to nitrogen starvation, for induction of cell-type-specific genes, and for pheromone induction. Recent work indicates that Mc may help recruit Ste11 to some TR sites (the Ste11 binding site; see below) (see reference 99 for details). Presumably, Pc acts similarly. Third, the Map1 protein is required for P-cell-specific gene expression and for enhancement of M-cell-specific gene expression. Little is known about the mechanism by which Map1 functions. Some evidence suggests that it may interact with the Pc protein (151, 226). Thus, cell-type-specific expression might involve a multiprotein complex of Pc or Mc with Map1 and Ste11. Future biochemical work is expected to shed light on the mechanisms governing cell-type-specific expression in S. pombe. Lastly, studies on the induction of mat1-Pm by pheromones indicate a requirement for Ste11 and the TR box (see below) (5). Whether Ste11 and the TR box are also required for pheromone induction of other genes remains to be determined.
The TR box, a 10-bp T-rich motif, is a common feature of the upstream regulatory region of all genes whose expression is Ste11 dependent upon nitrogen starvation. This motif is required for Ste11-mediated transcription (199; see also reference 99) and may also mediate the pheromone response, as indicated above. In addition to the TR box, some genes may contain a different site to which accessory proteins bind and help recruit Ste11 (99). Thus, different combinatorial activities containing Ste11 and other proteins may regulate the stepwise circuitry of activation by starvation and pheromones (99).
In contrast to S. cerevisiae, S. pombe requires the pheromones and receptors for the initiation of meiosis, which may simply reflect the requirement of pheromones for full induction of mat1-Pm and mat1-Mm. Support for this idea comes from the observation that ectopic expression of mat1-Pm and mat1-Mm triggers the induction of mei3 expression under nitrogen starvation (137, 223). In contrast to budding yeast, where pheromone and receptor genes are turned off in the a/α diploid cell (reviewed in reference 74), S. pombe h+/h− diploids transcribe these genes (85, 202).
The role of cyclic AMP (cAMP) in sexual development has been omitted from the above discussion for various reasons. Briefly, high levels of cAMP (and thus of cAMP-dependent protein kinase, PKA) inhibit sexual development whereas low levels of cAMP (and PKA) promote sexual development. Nitrogen starvation results in lowered cAMP levels. It appears that a trimeric G protein monitors nutritional conditions and may regulate the activity of adenylyl cyclase and thus cAMP and PKA levels. The mechanism by which cAMP and PKA regulate sexual development remains to be determined (reviewed in reference 228a).
In U. maydis, Prf1 is also a member of the HMG family of proteins and appears to recognize a site similar to that recognized by Ste11 of S. pombe. prf1 mRNA is apparently induced by pheromones, and Prf1 in turn regulates the expression of pheromone precursor and receptor genes and also of the genes at the b locus (69). Whether Prf1 is the direct target of a MAPK is not known.
Ras1 has a dual regulatory function: activation of the pheromone MAPK cascade and activation of a polarity and cell morphology cascade.
An important feature of Ras in mammalian cells is connecting the outputs from diverse types of receptors to MAPK cascades. This connection involves adaptor proteins such as Grb2, which recruits Sos (the exchange factor for Ras) to the receptor, leading to activation of Ras (reviewed in reference 133). Ras-GTP interacts with Raf, a serine/threonine protein kinase that is structurally similar to MAPKKKs, and brings Raf to the membrane. Activated Raf can then phosphorylate a MAPKK (reviewed in reference 142). Ras in mammalian cells is also proposed to play a role in the Rho family-mediated reorganization of the actin cytoskeleton (reviewed in reference 123).
The ras1 gene of S. pombe, like its mammalian counterpart, appears to regulate two different pathways (Fig. 10): a MAPK cascade pathway and a cell polarity and morphogenesis pathway involving a Rho family protein, Cdc42 (26, 130). The cell polarity and morphogenesis pathway includes proteins that are key regulators of polarity establishment: Cdc42, Scd1 (the proposed GEF for SpCdc42; homologous to S. cerevisiae Cdc24), and Scd2 (an SH3 domain-containing protein homologous to S. cerevisiae Bem1). Two-hybrid system analysis showed that interactions occur among these proteins and that Scd1 interacts with Ras1 (26). These observations led to the proposal that Ras1 activates Scd1, which then activates Cdc42. Activated Cdc42, in turn, activates Shk1 (Fig. 11). The shk1 gene, a homolog of S. cerevisiae STE20, is essential for growth. Δshk1 spores germinate and arrest as spherical cells, which is also the phenotype exhibited by Δcdc42 and Δscd1 mutants (130). Several arguments have been used to propose a role for Shk1 in the pheromone response pathway (Fig. 10) (130), but resolution of its participation in this pathway awaits more conclusive experiments. Because the phenotype of ras1 null mutants is weaker than that of scd1, cdc42, or shk1, null mutants it can be proposed that other inputs besides Ras1 can activate the morphogenesis pathway.
MAPK Cascade Signalling and Dimorphism
Pseudohyphal pathway of S. cerevisiae.
Pseudohyphal growth is a morphogenetic transition exhibited by a/α cells under nitrogen starvation conditions (62) and is widespread in nature (66, 207). This transition entails changes in cell morphology, budding pattern, and cell separation—cells switch from an ovoid to an elongated shape; they bud apically, and the buds remain attached, resulting in chains of attached cells; and the cells acquire the ability to invade the agar surface on which they are growing (62, 105). The switch to this type of growth is thought to allow yeast cells to scavenge better for nutrients; it has been observed that ovoid cells bud from the tips of the pseudohyphae, effectively allowing new growth away from the established colony (61). A similar but less exhuberant form of pseudohyphal growth (invasive growth) is also exhibited by a or α cells in older colonies, where nutrients have presumably been depleted (174).
(i) MAPK cascade and transcriptional activators.
Pseudohyphal growth is mediated in part by activation of a signalling pathway in response to nitrogen starvation. Remarkably, this signalling pathway involves many but not all of the components used in the pheromone response pathway: Ste20 (p65PAK), Ste11 (MAPKKK), Ste7 (MAPKK), and Ste12 (transcription factor) (121) (Fig. 12). The MAPK for this pathway could be Kss1 (125) (see below). Because Ste12 is the transcriptional activator for both the pheromone response and pseudohyphal growth pathways, a mechanism must exist for restricting the set of target genes activated. The interaction of Ste12 with another transcription factor, Tec1 (60, 126) (Fig. 12), appears to confer specificity of the response. Ste12-Tec1 recognizes a composite response element containing a Ste12 site and a Tec1 site, which is found in the TEC1 gene and in a gene activated by nutrient depletion (126). It is presumed that target genes for the pseudohyphal pathway have this composite site. The binding site for Tec1 is similar to that bound by the A. nidulans AbaA protein, a protein involved in a regulatory circuit controlling conidiophore development (4).
FIG. 12.
Signalling pathway for pseudohyphal growth. Nitrogen starvation activates the pseudohyphal pathway in a/α diploid cells, although the mechanism of activation is not known (dashed arrow from signal to Ras2). An alternative pathway may also regulate pseudohyphal growth, indicated by the dashed arrows originating from other inputs. X, unidentified MAPK, which may be Kss1. See the text for details.
Identifying the MAPK for this pathway has been elusive. Six MAPKs are known from the yeast genome sequence, Hog1, Mpk1, Fus3, Kss1, Smk1, and Mlmp1 (217; reviewed in 83), but deletion of all six MAPKs or combinations thereof does not seem to affect pseudohyphal growth (125). Several lines of evidence suggest that Kss1 may either be the MAPK of this pathway or play some other important role. The first argument begins with the observation that deletion of both RST1 and RST2 leads to constitutive pseudohyphal growth, indicating that the function of Rst1 and Rst2 is to inhibit pseudohyphal growth, presumably by inhibiting Ste12 (37, 204). One function of Kss1 and Fus3 is to inhibit Rst1 and Rst2 during the pheromone response (37, 204). Because KSS1 but not FUS3 is expressed in a/α diploid cells, Kss1 may be responsible for inhibiting Rst1 and Rst2 (39, 55). The observation that strains with KSS1 deleted exhibit normal pseudohyphal growth would argue against this role of Kss1. Recent results suggest that Kss1 plays a dual role in pseudohyphal growth: a positive role, which is Ste7 dependent, and a negative role, which is Ste7 independent (see reference 125 for details). Further analyses of Kss1 should help elucidate its role in this pathway.
(ii) Upstream of the MAPK cascade.
Neither the receptors nor the heterotrimeric G protein for the pheromone response pathway participates in signal sensing in the pseudohyphal pathway: these genes are not expressed in a/α diploids. Ste5 is also not needed for pseudohyphal growth (121). The signal generated by nitrogen starvation appears to be mediated by Ras2 and Cdc42, two small GTP binding proteins that act upstream of Ste20 (144) (Fig. 12), although it is not known if Ras2 is immediately upstream of Cdc42. The Cdc42 binding site in Ste20 has recently been demonstrated to be necessary for pseudohyphal growth. Since this site is necessary for interaction with Cdc42 in vitro and in vivo, it appears that binding of Cdc42 to Ste20 activates Ste20 for its function in pseudohyphal growth (109, 161). In addition to these components, the two yeast homologs of 14-3-3 proteins, Bmh1 and Bmh2, are implicated in the pseudohyphal pathway: they appear to regulate transcriptional induction and cell elongation independently of each other; their action may be exerted by interaction with the Ste20 protein (reference 173 and references therein).
(iii) Parallel pathways for pseudohyphal growth?
Mutations in STE20 cause a more severe defect in pseudohyphal growth than do mutations in STE7 or in STE12; thus, Ste20 may regulate other targets in addition to the MAPK cascade (144, 173). Because deletion of STE7 and STE12 genes reduces but does not completely abolish pseudohyphal growth (121), it is likely that another pathway can promote pseudohyphal growth. The concerted action of both may be important for a full response. Parallel pathways for filamentous growth have also been proposed to exist in Candida albicans and U. maydis (see below).
The ELM genes may regulate an alternative pathway of pseudohyphal growth or some aspect of the pseudohyphal transition in S. cerevisiae. The normal function of these genes appears to be the inhibition of pseudohyphal growth under optimal growth conditions, although the specific function of most of these gene products is unknown (14, 15). ELM1 encodes a protein kinase whose absence leads to a constitutive pseudohyphal morphology: elongated cells that remain attached. Because the elongated morphology of pseudohyphal cells may reflect altered levels of Cdk activity (119) and because of genetic interactions of ELM1 with other genes that impinge on Cdk, it has been proposed that ELM1 functions in a pathway involving other protein kinases that regulate Cdk activity (51a, 144a).
Other genes controlling pseudohyphal growth have also been identified and include PHD1 and SOK2 (61, 143, 216). Sok2 appears to antagonize pseudohyphal growth, whereas Phd1 appears to promote it: they may either act in a linear pathway or converge independently on the same target. Their relationship to Ste12 and Tec1 remains to be determined. Both Phd1 and Sok2 share amino acid similarity with transcriptional activators of cell cycle-regulated genes and to the StuA protein of Aspergillus nidulans (140). StuA controls a developmental program leading to conidiophore formation. A cascade of regulatory proteins similar to that governing conidiophore development in A. nidulans (reviewed in reference 139; see also reference 23) may govern pseudohyphal growth in yeast.
Signalling and dimorphism in other fungi: C. albicans and U. maydis.
Two fungi, C. albicans and U. maydis, have attracted renewed interest in fungal dimorphism because of its link to pathogenicity.
C. albicans, an imperfect fungus (i.e., no sexual stage), is an opportunistic pathogen of immunocompromised patients and has become a major pathogen in nosocomial infections. It has the ability to switch from a yeast-like to a pseudohyphal form and also to a true hyphal form. (Growth of a hypha is by apical extension, and division occurs in a fission-like manner, whereas pseudohyphae arise by budding.) The dimorphic transition in C. albicans is mediated in part by the same signalling components that regulate pseudohyphal growth in S. cerevisiae: Cst20 (a homolog of Ste20), Hst7 (a homolog of Ste7), and Cph1 (a homolog of Ste12) (102, 111, 120). Because these components are required under some conditions but are dispensable under others, it has been proposed that alternative pathways exist for activation of filamentous growth in C. albicans. The question of how the dimorphic switch and pathogenicity are coupled remains unclear. It is possible that switching to different forms allows this fungus to evade the host immune response.
In contrast to C. albicans, in the basidiomycete fungus U. maydis a distinct relationship exists between form and pathogenicity: the haploid yeast form is nonpathogenic, whereas the dikaryotic filamentous form is pathogenic on maize (reviewed in references 6, 7, and 92). Dimorphism in this fungus is governed by two mating-type loci: a controls a pheromone signalling pathway, and b encodes homeodomain regulatory proteins. The integrated input of both loci is required for filamentous growth in culture (reviewed in references 6 and 92). Formation of the filamentous pathogenic dikaryon comes about by fusion of haploid cells carrying different alleles at both loci, for example, a1 b1 and a2 b2. Cell fusion is governed by the a locus with two alleles (a1 and a2), each of which contains genes that code for a pheromone precursor and a receptor (17). Thus, as in yeast, cell fusion is controlled by the interaction of pheromones with receptors. Surprisingly, in culture, different a alleles (that is, different pheromones and receptors) are required for hyphal growth, which indicates that an autocrine-like response (i.e., self-stimulation) governs filamentous growth (11, 195). This autocrine response requires, in addition to the pheromones and receptors, a Gα (Gpa3) protein, a MAPKK (Fuz7), and a transcription factor (Prf1), all of which may be components of a pheromone response pathway (9, 69, 170). The requirement for pheromones and receptors contrasts with budding yeast, where these elements are not necessary for pseudohyphal growth of a/α diploids.
Dimorphism is also regulated by cAMP levels: high levels promote yeast-like growth, whereas low levels promote filamentous growth (63), a situation reminiscent of the regulation of sexual development by cAMP levels in S. pombe. Because nitrogen starvation is required for filamentous growth (10), one possibility is that, as in S. pombe, nitrogen starvation lowers cAMP levels. In S. pombe, the Gα protein Gpa2 regulates cAMP levels in response to nutritional conditions (90). Therefore, cAMP levels in U. maydis, in principle, could be controlled by a Gα protein that senses nutritional status.
Because different a alleles are not necessary for the development of hyphae in planta, it has been proposed that plant signals activate the pathway activated by pheromones or an alternative pathway for filamentous growth (8).
Mate as You Wish: What Matters Is What You Are after Mating
We have already seen that filamentous growth in U. maydis entails the input of pheromones and receptors and homeodomain proteins. The pheromone and receptor system is simple in that there are only two receptors (Pra1 and Pra2) and two pheromones (Mfa1 and Mfa2). Pra1 is activated by Mfa2 and Pra2 by Mfa1 (17). In the world of the mushrooms (which, like U. maydis, are Basidiomycete fungi), life is not so simple: the life cycle of Schizophyllum commune and Coprinus cinereus is regulated by two mating complexes, A and B, each containing two loci, α and β. The α and β loci are both multiallelic (for further details, see references 25a, 106, 196, 209, and 220). These fungi exist only as filaments, which are of two types, monokaryotic and dikaryotic; only the latter can produce a fruiting body (the mushroom). Mating occurs between cells of homokaryotic filaments and is not regulated (it’s a free-for-all). Strikingly, development of the dikaryotic filament after mating is exquisitely regulated by the coordinate action of the A and B mating complexes (reviewed in references 25a and 103).
Events regulated by the A and B complexes.
To understand the possible role of pheromones and receptors in the life cycle of these fungi, I first give a synopsis of the events regulated by A and B (Fig. 13A). After mating, reciprocal exchange of nuclei occurs (Fig. 13A, step 1); this is followed by migration of the invading nucleus through the hypha until it reaches the tip cell (step 2). Septa dissolve to allow passage of the migrating nucleus. In the tip cell, the invading and resident nuclei pair and establish dikaryosis (step 2). Division of this dikaryotic cell occurs through an ornate process as follows: the tip cell forms an appendage (hook cell or clamp connection) that curves towards the subapical cell, coordinate division (conjugate division) occurs, and one nucleus migrates into the appendage (step 3). Three cells are created: a dikaryotic tip cell, a subapical cell with one nucleus, and an appendage with one nucleus (step 4). The appendage fuses with the subapical cell and delivers the nucleus so that dikaryosis is established in this cell (step 5). The fusion of this appendage to the subapical cell can be viewed as mating between two yeast cells. Not surprisingly, this particular step is regulated by the B complex, which encodes pheromone precursors and receptors. This division process requires different alleles at Aα or Aβ and at Bα or Bβ (reviewed in references 25a, 103, and 210) (see the legend to Fig. 13 for the steps regulated by A).
FIG. 13.
Multiple pheromones and multiple receptors govern filamentous growth in the mushrooms (Basidiomycete fungi). (A) Plasmogamy occurs between cells of hyphae in a mating-type-independent manner (step 1). Steps 2, 3, and 4 are regulated by the A mating complex, which encodes homeodomain proteins. Steps 1 (nuclear migration but not cell fusion) and 5 are regulated by the B mating complex. Step 5 may be viewed as analogous to cell fusion during mating in the yeasts. (B) The B complex encodes multiple pheromones and receptors. Bβ1 and Bβ2 are two of the nine alleles of the Bβ locus within the B complex (see the text for details). 1-1, 1-2, 1-3, 2-2, 2-2, and 2-3 are putative pheromone precursor genes; R1 and R2 are putative pheromone receptor genes.
Multiple pheromones and receptors regulate a specialized form of cell fusion.
To simplify the following discussion, I consider only the Bβ locus of Schizophyllum commune. Two of nine different alleles have been cloned and sequenced (209). The remarkable finding is that each contains three genes encoding putative precursors for lipopeptides like the a-factor of S. cerevisiae (Fig. 13B) and also a gene encoding a Ste3-like putative seven transmembrane receptor (Fig. 13B). Although the structural organization of the receptors is conserved, the amino acid sequences of R1 and R2 are not identical (Fig. 13B). The putative pheromones within a given allele are different from each other: 1-1 is different from 1-2 and 1-3 and from the putative pheromones of the other allele (2-1, 2-2, and 2-3, and so on) (Fig. 13B). The pheromones encoded by allele 1 do not activate R1, but some can activate R2. Conversely, the pheromones encoded by allele 2 do not activate R2, but some can activate R1. It appears that a given pheromone, for example 1-1, can activate more than one receptor, for example R2, R3, etc., and that a given receptor, for example R2, can be activated by multiple pheromones, for example, 1-1, 1-2, and 1-3 (for details, see references 209 and 220). Clearly, the pheromone system of the mushrooms poses intriguing challenges.
Tales of G Proteins, Pheromones, and MAPK Components
In this section, I present information about various signalling components and their inferred roles in the life cycle of several filamentous fungi.
Heterotrimeric G proteins.
Gα subunits have been identified in a number of fungi. I discuss only those in N. crassa, Cryphonectria parasitica, and U. maydis. A common theme emerging from these studies is that multiple Gα subunits exist in a given fungus and may regulate diverse aspects of the life cycle. The gna-1 gene of N. crassa, an Ascomycete fungus, encodes a Gαi-type protein that governs various aspects of the life cycle of this organism: apical extension of hyphae, macroconidiation, female fertility, and sensitivity to hyperosmolarity (91). Gna-1 might regulate these diverse processes via a common pathway, for example by regulation of a MAPK cascade in response to different stimuli, or perhaps Gna-1 regulates different signalling pathways.
C. parasitica, another Ascomycete fungus, is the cause of chestnut blight disease. Two genes encoding Gα subunits have been identified: CPG-1 and CPG-2. Infection of this fungus by a virus results in attenuation of fungal virulence and correlates with reduced levels of Cpg-1 protein and increased cAMP levels. Drug-mediated elevation of cAMP levels mimics the attenuation of virulence caused by viral infection, suggesting that Cpg-1, like some mammalian counterparts, regulates adenylyl cyclase. Disruption of CPG-1 indicates that Cpg-1 is required for optimal hyphal growth, conidiation, female fertility, and virulence. Disruption of CPG-2 had only marginal effects on the above processes (30, 58). The processes regulated by Cpg-1 in C. parasitica are similar (except for virulence) to those regulated by Gna-1 in N. crassa, which suggests that common pathways in these fungi are regulated by these Gα subunits.
In U. maydis, four genes encoding Gα subunits, gpa1 to gpa4, have been identified (170). Gpa3 may be the pheromone receptor-coupled G protein. Because pheromones and receptors are necessary for filamentous growth (see the section on signalling and dimorphism in C. albicans and U. maydis, above) and because Gpa3 appears to be necessary for the pheromone-induced expression of pheromone genes, it was not surprising that Gpa3 is also required for filamentous growth (170). Activated alleles of gpa3 allow the mating of strains carrying identical a alleles. Because wild-type strains with identical a alleles do not mate, these results suggest that the activated gpa3 allele leads to constitutive activation of a putative pheromone response pathway, resulting in mating. Gpa3 is similar to Gpa2 of S. pombe, which is involved in nutrient sensing, and to CPG-2 of C. parasitica. It remains to be determined whether Gpa3 regulates cAMP levels or the pheromone MAPK cascade. The roles of gpa1, gpa2, and gpa4 are unknown.
MAPKK, MAPK, and pheromones.
The MAPKK Fuz7 of U. maydis is necessary for conjugation and filament formation in culture (see the section on signalling and dimorphism in C. albicans and U. maydis, above) (9). It is presumably involved in the pheromone response signalling pathway, although this remains to be conclusively demonstrated. In addition to this role, Fuz7 is necessary for pathogenicity: strains with fuz7 deleted are unable to induce tumors (9). This defect is not a consequence of an inability to produce filaments, since null mutants produce filaments in planta (8). It is thus hypothesized that Fuz7 is activated by a plant signal and that this activation is crucial for the fungus to induce tumors (9).
Pmk1 is a homolog of Fus3 and Kss1 in Magnaporthe grisea, an Ascomycete fungus that causes rice blast disease. This MAPK is required for formation of the appressorium, a specialized structure formed by many pathogenic fungi for invasion of plant cells. Deletion of PMK1 results in a defect in appressorium formation and thus in nonpathogenicity (225). The nature of the signal that activates this MAPK is not known.
A remarkable finding about M. grisea is that S. cerevisiae α-factor at high levels interferes with appressorium formation and thus inhibits pathogenicity (see reference 12 for details). The effect was mating-type specific, affecting cells of the MAT1-2 but not the MAT1-1 mating type. Culture filtrates of MAT1-1 cells contain an activity that inhibits appressorium formation of MAT1-2 cells, suggesting that pheromones and a pheromone response pathway may regulate appressorium formation (12). Pmk1 may be part of this pathway. Thus, it can be speculated that this pathway plays two roles: in response to pheromones, it activates mating and inhibits appressorium formation, and in response to another signal, it inhibits mating and activates appressorium formation.
CONCLUDING REMARKS
This tour of the different signalling pathways in the yeasts has attempted to illustrate the use of the highly conserved MAPK cascade in the transduction of diverse signals from the cell surface to the interior of the cell. Despite the high conservation of this module and the multiplicity of such modules within a single cell, the response to a particular signal is very specific, with little cross talk between pathways. The specificity of the response is due in part to sequestering proteins that facilitate complex formation of the MAPK cascade components and thus prevent inappropriate activation of other pathways. Although this is an attractive mechanism for inhibition of cross talk, only two examples of scaffold proteins have been identified—Ste5 and Pbs2. This paucity may reflect our incomplete knowledge of the other pathways or the existence of other mechanisms that ensure specificity. One possibility, mentioned briefly in the introductory comments to the MAPK cascade, is that the interaction of a given MAPK with its activator and its substrates is very specific and may be dictated by conformation of the proteins. The three-dimensional structure of two mammalian MAPKs (ERK2 and p38) shows that they have similar folding of several domains but different folding of other domains. These differences may be critical for the specificity of recognition and activation of a given MAPK by its MAPKK and for substrate recognition and activation by the MAPK. As the three-dimensional structures of other MAPKs, in both their inactive and active states, become available, our understanding of the specificity of MAPKs with their substrates and activators will be greatly illuminated.
Although cross talk is generally not observed, it might be desirable under some conditions to coordinate the output of two responses. One example is the activation of the PKC pathway by pheromones during formation of mating projections. As our knowledge of these pathways deepens, we might also uncover other instances of communication between pathways.
Some other areas where our knowledge is sparse are the following. (i) What is the mechanism by which a single pathway can be activated by multiple signals, as is the case for the osmoresponse pathway in S. pombe and the PKC pathway of S. cerevisiae? This raises several questions, one of which concerns whether each signal activates a specific sensor. If so, do targets activated in response to different input signals differ? We have seen that specificity may be conferred by different combinatorial activities of transcriptional activators. (ii) What is the nature of the receptors involved in the osmosensing pathways? These receptors may be mechanosensors, of which we know very little. Thus, the identification and characterization of these receptors is expected to shed considerable light on our understanding of how these sensors become activated and how they transduce the signal to downstream components. (iii) How do components that reside in the membrane communicate with the elements of the pathway that do not? This is likely to involve adaptor proteins, one function of which might be the translocation of cytoplasmic components to the membrane for activation. A specific question is, for example, how does Ste5 become localized to the membrane for interaction with Gβγ during activation of the pheromone response pathway in S. cerevisiae? Translocation might occur in a pheromone-dependent manner. (iv) How is the pheromone response coupled with the machinery responsible for reorganization of the actin cytoskeleton to cause morphological changes in response to pheromone treatment?
The study of signalling pathways in filamentous fungi will predictably experience an explosion of information in the near future. Current information suggests that heterotrimeric G proteins control diverse aspects of their life cycle. In addition, MAPK cascade components appear to play crucial roles in pathogenicity. It is hoped that future studies will provide a better understanding of how different phases of the life cycle of filamentous fungi are regulated by signalling pathways.
ACKNOWLEDGMENTS
I thank Linda Huang for comments on the pheromone response section, Sean O’Rourke for comments on the S. cerevisiae osmolarity response section, Ira Herskowitz for valuable comments on the entire manuscript, Hiten Madhani for sharing unpublished results and for discussion, and anonymous reviewers for comments.
REFERENCES
- 1.Aiba H, Yamada H, Ohmiya R, Mizuno T. The osmoinducible gpd1+ gene is a target of the signaling pathway involving Wis1 MAP-kinase kinase in fission yeast. FEBS Lett. 1995;376:199–201. doi: 10.1016/0014-5793(95)01277-4. [DOI] [PubMed] [Google Scholar]
- 2.Albertyn J, Hohmann S, Thevelein J M, Prior B A. GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway. Mol Cell Biol. 1994;14:4135–4144. doi: 10.1128/mcb.14.6.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alex L A, Borkovich K A, Simon M I. Hyphal development in Neurospora crassa: involvement of a two-component histidine kinase. Proc Natl Acad Sci USA. 1996;93:3416–3421. doi: 10.1073/pnas.93.8.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrianopoulos A, Timberlake W E. The Aspergillus nidulans abaA gene encodes a transcriptional activator that acts as a genetic switch to control development. Mol Cell Biol. 1994;14:2503–2515. doi: 10.1128/mcb.14.4.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aono T, Yanah H, Miki F, Davey J, Shimoda C. Mating pheromone-induced expression of the mat1-Pm gene of Schizosaccharomyces pombe: identification of signalling components and characterization of upstream controlling elements. Yeast. 1994;10:757–770. doi: 10.1002/yea.320100607. [DOI] [PubMed] [Google Scholar]
- 6.Banuett F. Genetics of Ustilago maydis, a fungal pathogen that induces tumors in maize. Annu Rev Genet. 1995;29:179–208. doi: 10.1146/annurev.ge.29.120195.001143. [DOI] [PubMed] [Google Scholar]
- 7.Banuett F. Ustilago maydis, the delightful blight. Trends Genet. 1992;8:174–180. doi: 10.1016/0168-9525(92)90220-x. [DOI] [PubMed] [Google Scholar]
- 8.Banuett F, Herskowitz I. Discrete developmental stages during teliospore formation in the corn smut fungus, Ustilago maydis. Development. 1996;122:2965–2976. doi: 10.1242/dev.122.10.2965. [DOI] [PubMed] [Google Scholar]
- 9.Banuett F, Herskowitz I. Identification of Fuz7, a U. maydis MEK/MAPKK homologue required for a-dependent and independent processes in the fungal life cycle. Genes Dev. 1994;8:1367–1378. doi: 10.1101/gad.8.12.1367. [DOI] [PubMed] [Google Scholar]
- 10.Banuett F, Herskowitz I. Morphological transitions in the life cycle of Ustilago maydis and their genetic control by the a and the b loci. Exp Mycol. 1994;18:247–266. [Google Scholar]
- 11.Banuett F, Herskowitz I. Different a alleles of Ustilago maydis are necessary for maintenance of filamentous growth but not for meiosis. Proc Natl Acad Sci USA. 1989;86:5878–5882. doi: 10.1073/pnas.86.15.5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beckerman J L, Nalder F, Ebbole D J. Inhibition of pathogenicity of the rice blast fungus by Saccharomyces cerevisiae α-factor. Science. 1997;276:1116–1119. doi: 10.1126/science.276.5315.1116. [DOI] [PubMed] [Google Scholar]
- 13.Berman D M, Wilkie T M, Gilman A G. GAIP and RGS4 are GTPase-activating proteins for the Gi subfamily of G protein α subunits. Cell. 1996;86:445–452. doi: 10.1016/s0092-8674(00)80117-8. [DOI] [PubMed] [Google Scholar]
- 14.Blacketer M J, Madaule P, Myers A M. Mutational analysis of morphologic differentiation in Saccharomyces cerevisiae. Genetics. 1995;140:1259–1275. doi: 10.1093/genetics/140.4.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blacketer M J, Koehler C M, Coats S G, Myers A M, Madaule P. Regulation of dimorphism in Saccharomyces cerevisiae: involvement of the novel protein kinase homolog Elm1p and protein phosphatase 2A. Mol Cell Biol. 1993;13:5567–5581. doi: 10.1128/mcb.13.9.5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boguslawski G. PBS2, a yeast gene encoding a putative protein kinase, interacts with the RAS2 pathway and affects osmotic sensitivity of Saccharomyces cerevisiae. J Gen Microbiol. 1992;138:2425–2432. doi: 10.1099/00221287-138-11-2425. [DOI] [PubMed] [Google Scholar]
- 17.Bölker M, Urban M, Kahmann R. The a mating type locus of U. maydis specifies cell signalling components. Cell. 1992;68:441–450. doi: 10.1016/0092-8674(92)90182-c. [DOI] [PubMed] [Google Scholar]
- 18.Boone C, Davis N G, Sprague G F. Mutations that alter the third cytoplasmic loop of the a-factor receptor lead to a constitutive and hypersensitive phenotype. Proc Natl Acad Sci USA. 1993;90:9921–9925. doi: 10.1073/pnas.90.21.9921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bourne H R. How receptors talk to trimeric G proteins. Curr Opin Cell Biol. 1997;9:134–142. doi: 10.1016/s0955-0674(97)80054-3. [DOI] [PubMed] [Google Scholar]
- 20.Brewster J L, de Valoir T, Dwyer N D, Winter E, Gustin M C. An osmosensing signal transduction pathway in yeast. Science. 1993;259:1760–1763. doi: 10.1126/science.7681220. [DOI] [PubMed] [Google Scholar]
- 21.Büküsoglu G, Jenness D D. Agonist-specific conformational changes in the yeast α-factor pheromone receptor. Mol Cell Biol. 1996;16:4818–4823. doi: 10.1128/mcb.16.9.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burbelo P D, Drechsel D, Hall A. A conserved binding motif defines numerous candidate target proteins for both Cdc42 and Rac GTPases. J Biol Chem. 1995;279:29071–29074. doi: 10.1074/jbc.270.49.29071. [DOI] [PubMed] [Google Scholar]
- 23.Busby T M, Miller K Y, Miller B L. Suppression and enhancement of the Aspergillus nidulans medusa mutation by altered dosage of the bristle and stunted genes. Genetics. 1996;143:155–163. doi: 10.1093/genetics/143.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cabib E, Drgon T, Drgonová J, Ford R A, Kollár R. The yeast cell wall, a dynamic structure engaged in growth and morphology. Biochem Soc Trans. 1997;25:200–204. doi: 10.1042/bst0250200. [DOI] [PubMed] [Google Scholar]
- 25.Cairns B R, Ramer S W, Kornberg R D. Order of action of components in the yeast pheromone response pathway revealed with a dominant allele of the Ste11 kinase and the multiple phosphorylation of the Ste7 kinase. Genes Dev. 1992;6:1305–1318. doi: 10.1101/gad.6.7.1305. [DOI] [PubMed] [Google Scholar]
- 25a.Casselton L A, Olesnicky N S. Molecular genetics of mating recognition in basidiomycete fungi. Microbiol Mol Biol Rev. 1998;62:55–70. doi: 10.1128/mmbr.62.1.55-70.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang E C, Barr M, Wang Y, Jung V, Xu H-P, Wigler M H. Cooperative interaction of S. pombe proteins required for mating and morphogenesis. Cell. 1994;79:131–141. doi: 10.1016/0092-8674(94)90406-5. [DOI] [PubMed] [Google Scholar]
- 27.Chang F. Regulation of the cell cycle by a negative growth factor in yeast. Ph.D. thesis. University of California, San Francisco; 1991. [Google Scholar]
- 28.Chang F, Nurse P. How fission yeast fission in the middle. Cell. 1996;84:191–194. doi: 10.1016/s0092-8674(00)80973-3. [DOI] [PubMed] [Google Scholar]
- 29.Chang F, Herskowitz I. Identification of a gene necessary for cell cycle arrest by a negative growth factor of yeast: FAR1 is an inhibitor of a G1 cyclin, Cln2. Cell. 1990;63:999–1011. doi: 10.1016/0092-8674(90)90503-7. [DOI] [PubMed] [Google Scholar]
- 30.Chen B, Gao S, Choi G H, Nuss D L. Extensive alteration of fungal gene transcript accumulation and elevation of G-protein-regulated cAMP levels by a virulence-attenuating hypovirus. Proc Natl Acad Sci USA. 1996;93:7996–8000. doi: 10.1073/pnas.93.15.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Q, Konopka J B. Regulation of the α-factor receptor by phosphorylation. Mol Cell Biol. 1996;16:247–257. doi: 10.1128/mcb.16.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi K-Y, Satterberg B, Lyons D M, Elion E A. Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell. 1994;78:499–512. doi: 10.1016/0092-8674(94)90427-8. [DOI] [PubMed] [Google Scholar]
- 33.Christensen P U, Davey J, Nielsen O. The Schizosaccharomyces pombe mam1 gene encodes an ABC transporter mediating secretion of M-factor. Mol Gen Genet. 1997;255:226–236. doi: 10.1007/s004380050493. [DOI] [PubMed] [Google Scholar]
- 34.Cid V J, Durán A, del Rey F, Snyder M P, Nombela C, Sánchez M. Molecular basis of cell integrity and morphogenesis in Saccharomyces cerevisiae. Microbiol Rev. 1995;59:345–386. doi: 10.1128/mr.59.3.345-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clarke S. Protein isoprenylation and methylation at carboxyl-terminal cysteine residues. Annu Rev Biochem. 1992;61:355–386. doi: 10.1146/annurev.bi.61.070192.002035. [DOI] [PubMed] [Google Scholar]
- 36.Cohen G B, Ren R, Baltimore D. Modular binding domains in signal transduction proteins. Cell. 1995;80:237–248. doi: 10.1016/0092-8674(95)90406-9. [DOI] [PubMed] [Google Scholar]
- 37.Cook J G, Bardwell L, Kron S J, Thorner J. Two novel targets of the MAP kinase Kss1 are negative regulators of invasive growth in the yeast Saccharomyces cerevisiae. Genes Dev. 1996;10:2831–2848. doi: 10.1101/gad.10.22.2831. [DOI] [PubMed] [Google Scholar]
- 38.Costigan C, Gherung S, Snyder M. A synthetic lethal screen identifies SLK1, a novel protein kinase homolog implicated in yeast cell morphogenesis and cell growth. Mol Cell Biol. 1992;12:1162–1178. doi: 10.1128/mcb.12.3.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Courchesne W E, Kunisawa R, Thorner J. A putative protein kinase overcomes pheromone-induced arrest of cell cycling in S. cerevisiae. Cell. 1989;58:1107–1119. doi: 10.1016/0092-8674(89)90509-6. [DOI] [PubMed] [Google Scholar]
- 40.Davenport K R, Sohaskey M, Kamada Y, Levin D E, Gustin M C. A second osmosensing signal transduction pathway in yeast. J Biol Chem. 1995;270:30157–30161. doi: 10.1074/jbc.270.50.30157. [DOI] [PubMed] [Google Scholar]
- 41.Davey J. M-factor, a farnesylated mating factor from the fission yeast Schizosaccharomyces pombe. Biochem Soc Trans. 1996;24:718–723. doi: 10.1042/bst0240718. [DOI] [PubMed] [Google Scholar]
- 42.Davey J. Mating pheromones of the fission yeast Schizosaccharomyces pombe: purification and structural characterization of M-factor and isolation and analysis of two genes encoding the pheromone. EMBO J. 1992;11:951–960. doi: 10.1002/j.1460-2075.1992.tb05134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Degols G, Shiozaki K, Russell P. Activation and regulation of the Spc1 stress-activated protein kinase in Schizosaccharomyces pombe. Mol Cell Biol. 1996;16:2870–2877. doi: 10.1128/mcb.16.6.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dekker L V, Parker P J. Protein kinase C—a question of specificity. Trends Biochem Sci. 1994;19:73–77. doi: 10.1016/0968-0004(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 45.Dietzel C, Kurjan J. Pheromonal regulation and sequence of the Saccharomyces cerevisiae SST2 gene: a model for desensitization to pheromone. Mol Cell Biol. 1987;7:4169–4177. doi: 10.1128/mcb.7.12.4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Divecha N, Irvine R F. Phospholipid signalling. Cell. 1995;80:269–278. doi: 10.1016/0092-8674(95)90409-3. [DOI] [PubMed] [Google Scholar]
- 47.Dodou E, Treisman R. The Saccharomyces cerevisiae MADS-box transcription factor Rlm1 is a target for the Mpk1 mitogen-activated protein kinase pathway. Mol Cell Biol. 1997;17:1848–1859. doi: 10.1128/mcb.17.4.1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dohlman H G, Song J, Ma D, Courchesne W E, Thorner J. Sst2, a negative regulator of pheromone signalling in the yeast Saccharomyces cerevisiae: expression, localization and genetic interaction and physical association with Gpa1 (the G-protein α subunit) Mol Cell Biol. 1996;16:5194–5209. doi: 10.1128/mcb.16.9.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doi K, Gartner A, Ammerer G, Errede B, Shinkawa H, Sugimoto K, Matsumoto K. Msg5, a novel protein phosphatase promotes adaptation to pheromone response in S. cerevisiae. EMBO J. 1994;13:61–70. doi: 10.1002/j.1460-2075.1994.tb06235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Douglas C M, Foor F, Marrinan J A, Morin N, Nielsen J B, Dahl A M, Mazur P, Gainsky W, Li W, El-Sherbeini M, Clemas J A, Mandal S M, Frommer B R, Kurtz M B. The Saccharomyces cerevisiae FKS1 (ETG1) gene encodes an integral membrane protein which is a subunit of 1,3-β-d-glucan synthases. Proc Natl Acad Sci USA. 1994;91:12907–12911. doi: 10.1073/pnas.91.26.12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Drgonová J, Drgon T, Tanaka K, Kollár R, Chen G-C, Ford R A, Chan C S M, Takai Y, Cabib E. Rho1p, a yeast protein at the interface between cell polarization and morphogenesis. Science. 1996;272:277–279. doi: 10.1126/science.272.5259.277. [DOI] [PubMed] [Google Scholar]
- 51a.Edgington N, Myers A. Abstracts of the Yeast Genetics and Molecular Biology Meeting. 1996. Suppressor analysis of the kinase Elm1 identifies a dominant allele of a S. pombe nim1+ homologue in S. cerevisiae; p. 108. [Google Scholar]
- 52.Egger L A, Park H, Inouye M. Signal transduction via the histidyl-aspartyl phosphorelay. Genes Cells. 1997;2:167–184. doi: 10.1046/j.1365-2443.1997.d01-311.x. [DOI] [PubMed] [Google Scholar]
- 53.Elion E A. Ste5: a meeting place for MAP kinases and their associates. Trends Cell Biol. 1995;5:322–327. doi: 10.1016/s0962-8924(00)89055-8. [DOI] [PubMed] [Google Scholar]
- 54.Elion E A, Satterberg B, Kranz J E. Fus3 phosphorylates multiple components of the mating signal transduction cascade: evidence for Ste12 and Far1. Mol Biol Cell. 1993;4:495–510. doi: 10.1091/mbc.4.5.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Elion E A, Grisafi P L, Fink G R. FUS3 encodes a Cdc2/Cdc28-related kinase required for the transition from mitosis into conjugation. Cell. 1990;60:649–664. doi: 10.1016/0092-8674(90)90668-5. [DOI] [PubMed] [Google Scholar]
- 56.Errede B, Gartner A, Zhou Z, Nasmyth K, Ammerer G. MAP kinase-related Fus3 from S. cerevisiae is activated by Ste7 in vitro. Nature. 1993;362:261–264. doi: 10.1038/362261a0. [DOI] [PubMed] [Google Scholar]
- 57.Galcheva-Gargova Z, Dérijard B, Wu I-H, Davis R J. An osmosensing signal transduction pathway in mammalian cells. Science. 1994;265:806–808. doi: 10.1126/science.8047888. [DOI] [PubMed] [Google Scholar]
- 58.Gao S, Nuss D L. Distinct roles for two G protein α subunits in fungal virulence, morphology, and reproduction revealed by targeted disruption. Proc Natl Acad Sci USA. 1996;93:14122–14227. doi: 10.1073/pnas.93.24.14122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gartner A, Nasmyth K, Ammerer G. Signal transduction in Saccharomyces cerevisiae requires tyrosine and threonine phosphorylation of Fus3 and Kss1. Genes Dev. 1992;6:1280–1292. doi: 10.1101/gad.6.7.1280. [DOI] [PubMed] [Google Scholar]
- 60.Gavrias V, Andrianopoulos A, Gimeno C J, Timberlake W E. Saccharomyces cerevisiae TEC1 is required for pseudohyphal growth. Mol Microbiol. 1996;19:1255–1263. doi: 10.1111/j.1365-2958.1996.tb02470.x. [DOI] [PubMed] [Google Scholar]
- 61.Gimeno C J, Fink G R. Induction of pseudohyphal growth by overexpression of PHD1, a Saccharomyces cerevisiae gene related to transcriptional regulators of fungal development. Mol Cell Biol. 1994;14:2100–2112. doi: 10.1128/mcb.14.3.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gimeno C J, Ljungdahl P O, Styles C A, Fink G R. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- 63.Gold S, Duncan G, Barrett K, Kronstad J W. cAMP regulates morphogenesis in the fungal pathogen Ustilago maydis. Genes Dev. 1994;8:2805–2816. doi: 10.1101/gad.8.23.2805. [DOI] [PubMed] [Google Scholar]
- 64.Gotoh Y, Nishida E, Shimanuki M, Toda T, Imai Y, Yamamoto M. Schizosaccharomyces pombe Spk1 is a tyrosine-phosphorylated protein functionally related to Xenopus mitogen-activated protein kinase. Mol Cell Biol. 1993;13:6427–6434. doi: 10.1128/mcb.13.10.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gray J V, Ogas J P, Kamada Y, Stone M, Levin D E, Herskowitz I. A role for the Pkc1-MAP kinase pathway of Saccharomyces cerevisiae in bud emergence and identification of a putative upstream regulator. EMBO J. 1997;16:4924–4937. doi: 10.1093/emboj/16.16.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guillermond A. The yeasts. New York, N.Y: John Wiley & Sons; 1920. [Google Scholar]
- 67.Han J, Lee J-D, Bibbs L, Ulevitch R J. A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science. 1994;265:808–811. doi: 10.1126/science.7914033. [DOI] [PubMed] [Google Scholar]
- 68.Hanks S K, Hunter T. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- 69.Hartmann H A, Kahmann R, Bölker M. The pheromone response factor coordinates filamentous growth and pathogenicity in Ustilago maydis. EMBO J. 1996;15:1632–1641. [PMC free article] [PubMed] [Google Scholar]
- 70.Hartwell L H. Mutants of Saccharomyces cerevisiae unresponsive to cell division control by polypeptide mating hormone. J Cell Biol. 1980;85:811–822. doi: 10.1083/jcb.85.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hasson M S, Blinder D, Thorner J, Jenness D D. Mutational activation of the STE5 gene product bypasses the requirement for G protein β and γ subunits in the yeast pheromone response pathway. Mol Cell Biol. 1994;14:1054–1065. doi: 10.1128/mcb.14.2.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hayles J, Nurse P. Genetics of the fission yeast Schizosaccharomyces pombe. Annu Rev Genet. 1992;26:373–402. doi: 10.1146/annurev.ge.26.120192.002105. [DOI] [PubMed] [Google Scholar]
- 73.Herskowitz I. MAPK kinase pathways in yeast: for mating and more. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- 74.Herskowitz I. Life cycle of the budding yeast Saccharomyces cerevisiae. Microbiol Rev. 1988;52:536–553. doi: 10.1128/mr.52.4.536-553.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hicke L, Riezman H. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell. 1996;84:277–287. doi: 10.1016/s0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- 76.Hill C S, Treisman R. Transcriptional regulation by extracellular signals: mechanisms and specificity. Cell. 1995;80:199–211. doi: 10.1016/0092-8674(95)90403-4. [DOI] [PubMed] [Google Scholar]
- 77.Hirayama T, Maeda T, Saito H, Shinozaki K. Cloning and characterization of seven cDNAs for hyperosmolarity-responsive (HOR) genes of Saccharomyces cerevisiae. Mol Gen Genet. 1995;249:127–138. doi: 10.1007/BF00290358. [DOI] [PubMed] [Google Scholar]
- 78.Hirschman J E, De Zutter G S, Simonds W F, Jenness D D. The Gβγ complex of the yeast pheromone response pathway. J Biol Chem. 1997;272:240–248. doi: 10.1074/jbc.272.1.240. [DOI] [PubMed] [Google Scholar]
- 79.Hughes D A. Control of signal transduction and morphogenesis by Ras. Semin Cell Biol. 1995;6:89–94. doi: 10.1016/1043-4682(95)90005-5. [DOI] [PubMed] [Google Scholar]
- 80.Hughes D A, Yabana N, Yamamoto M. Transcriptional regulation of a Ras nucleotide-exchange factor gene by extracellular signals in fission yeast. J Cell Sci. 1994;107:3635–3642. doi: 10.1242/jcs.107.12.3635. [DOI] [PubMed] [Google Scholar]
- 81.Hung W, Olson K A, Breitkreutz A, Sadowski I. Characterization of the basal and pheromone-stimulated phosphorylation states of Ste12p. Eur J Biochem. 1997;245:241–251. doi: 10.1111/j.1432-1033.1997.00241.x. [DOI] [PubMed] [Google Scholar]
- 82.Hunter T. Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell. 1995;80:225–236. doi: 10.1016/0092-8674(95)90405-0. [DOI] [PubMed] [Google Scholar]
- 83.Hunter T, Plowman G D. The protein kinases of budding yeast: six score and more. Trends Biochem Sci. 1997;22:18–22. doi: 10.1016/s0968-0004(96)10068-2. [DOI] [PubMed] [Google Scholar]
- 84.Igual J C, Johnson A L, Johnston L H. Coordinated regulation of gene expression by the cell cycle transcription factor Swi4 and the protein kinase C MAP kinase pathway for yeast cell integrity. EMBO J. 1996;15:5001–5013. [PMC free article] [PubMed] [Google Scholar]
- 85.Imai Y, Yamamoto M. The fission yeast mating pheromone P-factor: its molecular structure, gene structure, and ability to induce gene expression and G1 arrest in the mating partner. Genes Dev. 1994;8:328–338. doi: 10.1101/gad.8.3.328. [DOI] [PubMed] [Google Scholar]
- 86.Imai Y, Yamamoto M. Schizosaccharomyces pombe sxa1+ and sxa2+ encode putative proteases involved in the mating response. Mol Cell Biol. 1992;12:1827–1834. doi: 10.1128/mcb.12.4.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Inoue S B, Qadota H, Arisawa M, Anraku Y, Watanabe T, Ohya Y. Signalling toward yeast 1,3-β-glucan synthesis. Cell Struct Funct. 1996;21:395–402. doi: 10.1247/csf.21.395. [DOI] [PubMed] [Google Scholar]
- 88.Irie K, Takase M, Lee K S, Levin D E, Araki H, Matsumoto K, Oshima Y. MKK1 and MKK2, which encode Saccharomyces cerevisiae mitogen-activated protein kinase-kinase homologs, function in the pathway mediated by protein kinase C. Mol Cell Biol. 1993;13:3076–3083. doi: 10.1128/mcb.13.5.3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Irie K, Araki H, Oshima Y. A new protein kinase, Ssp31, modulating the SMP3 gene product involved in plasmid maintenance in Saccharomyces cerevisiae. Gene. 1991;108:139–144. doi: 10.1016/0378-1119(91)90499-2. [DOI] [PubMed] [Google Scholar]
- 90.Isshiki T, Mochizuki N, Maeda T, Yamamoto M. Characterization of a fission yeast gene, gpa2, that encodes a Gα subunit involved in the monitoring of nutrition. Genes Dev. 1992;6:2455–2462. doi: 10.1101/gad.6.12b.2455. [DOI] [PubMed] [Google Scholar]
- 91.Ivey F D, Hodge P N, Turner G E, Borkovich K A. The Gαi homologue gna-1 controls multiple differentiation pathways in Neurospora crassa. Mol Biol Cell. 1996;7:1283–1297. doi: 10.1091/mbc.7.8.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kahmann R, Romeis T, Bölker M, Kämper J. Control of mating and development in Ustilago maydis. Curr Opin Genet Dev. 1995;5:559–564. doi: 10.1016/0959-437x(95)80023-9. [DOI] [PubMed] [Google Scholar]
- 93.Kallal L, Kurjan J. Analysis of the receptor binding domain of Gpa1p, the Gα subunit involved in the yeast pheromone response pathway. Mol Cell Biol. 1997;17:2897–2907. doi: 10.1128/mcb.17.5.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kamada Y, Qadota H, Python C P, Anraku Y, Ohya Y, Levin D E. Activation of yeast protein kinase C by Rho1 GTPase. J Biol Chem. 1996;271:9193–9196. doi: 10.1074/jbc.271.16.9193. [DOI] [PubMed] [Google Scholar]
- 95.Kamada Y, Jung U S, Piotrowski J, Levin D E. The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genes Dev. 1995;9:1559–1571. doi: 10.1101/gad.9.13.1559. [DOI] [PubMed] [Google Scholar]
- 96.Kanoh J, Watanabe Y, Ohsugi M, Iino Y, Yamamoto M. Schizosaccharomyces pombe gad7+ encodes a phosphoprotein with a bZIP domain, which is required for proper G1 arrest and gene expression under nitrogen starvation. Genes Cells. 1996;1:391–408. doi: 10.1046/j.1365-2443.1996.d01-247.x. [DOI] [PubMed] [Google Scholar]
- 97.Kato T, Okazaki K, Murakami H, Stettler S, Fantes P A, Okayama H. Stress signal, mediated by a Hog1-like MAP kinase, controls sexual development in fission yeast. FEBS Lett. 1996;378:207–212. doi: 10.1016/0014-5793(95)01442-x. [DOI] [PubMed] [Google Scholar]
- 98.Kitamura K, Shimoda C. The Schizosaccharomyces pombe mam2 gene encodes a putative pheromone receptor which has a significant homology with the Saccharomyces cerevisiae Ste2 protein. EMBO J. 1991;10:3743–3751. doi: 10.1002/j.1460-2075.1991.tb04943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kjærulff S, Dooijes D, Clevers H, Nielsen O. Cell differentiation by interaction of two HMG-box proteins: Mat1-Mc activates M cell-specific genes in S. pombe by recruiting the ubiquitous transcription factor Ste11 to weak binding sites. EMBO J. 1997;16:4021–4033. doi: 10.1093/emboj/16.13.4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kjærulff S, Davey J, Nielsen O. Analysis of the structural genes encoding M-factor in the fission yeast Schizosaccharomyces pombe: identification of a third gene, mfm3. Mol Cell Biol. 1994;14:3895–3905. doi: 10.1128/mcb.14.6.3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Koelle M R. A new family of G-protein regulators—the RGS proteins. Curr Opin Cell Biol. 1997;9:143–147. doi: 10.1016/s0955-0674(97)80055-5. [DOI] [PubMed] [Google Scholar]
- 102.Köhler J R, Fink G R. Candida albicans strains heterozygous and homozygous for mutations in mitogen-activated protein kinase signaling components have defects in hyphal development. Proc Natl Acad Sci USA. 1996;93:13223–13228. doi: 10.1073/pnas.93.23.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kothe E. Tetrapolar fungal mating types: sexes by the thousands. FEMS Microbiol Rev. 1996;18:65–87. doi: 10.1111/j.1574-6976.1996.tb00227.x. [DOI] [PubMed] [Google Scholar]
- 104.Kranz J E, Satterberg B, Elion E A. The MAP kinase Fus3 associates with and phosphorylates the upstream signaling component Ste5. Genes Dev. 1994;8:313–327. doi: 10.1101/gad.8.3.313. [DOI] [PubMed] [Google Scholar]
- 105.Kron S J, Styles C A, Fink G R. Symmetric cell division in pseudohyphae of the yeast Saccharomyces cerevisiae. Mol Biol Cell. 1994;5:1003–1022. doi: 10.1091/mbc.5.9.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kües U, Asante-Owusu R N, Mutasa E S, Tymon A M, Pardo E H, O’Shea S F, Göttgens B, Casselton L A. Two classes of homeodomain proteins specify the multiple A mating types of the mushroom Coprinus cinereus. Plant Cell. 1994;6:1467–1475. doi: 10.1105/tpc.6.10.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kurjan J. The pheromone response pathway in Saccharomyces cerevisiae. Annu Rev Genet. 1993;27:147–179. doi: 10.1146/annurev.ge.27.120193.001051. [DOI] [PubMed] [Google Scholar]
- 108.Kyriakis J M, Banerjee P, Nikolakakl E, Dai T, Rubie E A, Ahmad M F, Avruch J, Woodgett J R. The stress-activated protein kinase subfamily of c-Jun kinases. Nature. 1994;369:156–160. doi: 10.1038/369156a0. [DOI] [PubMed] [Google Scholar]
- 109.Leberer E, Wu C, Leeuw T, Fourest-Lieuvin A, Segall J W, Thomas D Y. Functional characterization of the Cdc42p binding domain of yeast Ste20p protein kinase. EMBO J. 1997;16:83–97. doi: 10.1093/emboj/16.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Leberer E, Thomas D Y, Whiteway M. Pheromone signalling and polarized morphogenesis in yeast. Curr Opin Genet Dev. 1997;7:59–66. doi: 10.1016/s0959-437x(97)80110-4. [DOI] [PubMed] [Google Scholar]
- 111.Leberer E, Harcus D, Broadbent I D, Clark K L, Dignard D, Ziegelbauer K, Schmidt A, Gow N A R, Brown A J P, Thomas D Y. Signal transduction through homologs of the Ste20p and Ste7p protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans. Proc Natl Acad Sci USA. 1996;93:13217–13222. doi: 10.1073/pnas.93.23.13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Leberer E, Dignard D, Harcus D, Hoogan L, Whiteway M, Thomas D Y. Cloning of Saccharomyces cerevisiae STE5 as a suppressor of a Ste20 protein kinase mutant: structural and functional similarity of Ste5 to Far1. Mol Gen Genet. 1993;241:241–254. doi: 10.1007/BF00284675. [DOI] [PubMed] [Google Scholar]
- 113.Leberer E, Dignard D, Harcus D, Thomas D Y, Whiteway M. The protein kinase homologue Ste20p is required to link the yeast pheromone response G-protein βγ subunits to downstream signalling components. EMBO J. 1992;11:4815–4824. doi: 10.1002/j.1460-2075.1992.tb05587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lee K S, Irie K, Gotoh Y, Watanabe Y, Araki H, Nishida E, Matsumoto K, Levin D E. A yeast mitogen-activated protein kinase homolog (Mpk1p) mediates signalling by protein kinase C. Mol Cell Biol. 1993;13:3067–3075. doi: 10.1128/mcb.13.5.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lee K S, Levin D E. Dominant mutations in a gene encoding a putative protein kinase (BCK1) bypass the requirement for a Saccharomyces cerevisiae protein kinase C homolog. Mol Cell Biol. 1992;12:172–182. doi: 10.1128/mcb.12.1.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Leeuw T, Fourest-Lieuvin A, Wu C, Chenevert J, Clark K, Whiteway M, Thomas D Y, Leberer E. Pheromone response in yeast: association of Bem1p with proteins of the MAP kinase cascade and actin. Science. 1995;270:1210–1213. doi: 10.1126/science.270.5239.1210. [DOI] [PubMed] [Google Scholar]
- 117.Levin D E, Bartlett-Heubusch E. Mutants in the S. cerevisiae PKC1 gene display a cell cycle-specific osmotic stability defect. J Cell Biol. 1992;116:1221–1229. doi: 10.1083/jcb.116.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Levin D E, Fields F O, Kunisawa R, Bishop J M, Thorner J. A candidate protein kinase C gene, PKC1, is required for the S. cerevisiae cell cycle. Cell. 1990;62:213–224. doi: 10.1016/0092-8674(90)90360-q. [DOI] [PubMed] [Google Scholar]
- 119.Lew D J, Reed S I. Cell cycle control of morphogenesis in budding yeast. Curr Opin Genet Dev. 1995;5:17–23. doi: 10.1016/s0959-437x(95)90048-9. [DOI] [PubMed] [Google Scholar]
- 120.Liu H, Köhler J, Fink G R. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- 121.Liu H, Styles C A, Fink G R. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science. 1993;262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- 122.Ma D, Cook J G, Thorner J. Phosphorylation and localization of Kss1, a MAP kinase of the Saccharomyces cerevisiae pheromone response pathway. Mol Biol Cell. 1995;6:889–909. doi: 10.1091/mbc.6.7.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Machesky L M, Hall A. Rho—a connection between membrane receptor signalling and the cytoskeleton. Trends Cell Biol. 1996;6:304–310. doi: 10.1016/0962-8924(96)10026-x. [DOI] [PubMed] [Google Scholar]
- 124.MacKay V L, Manney T R. Mutations affecting sexual conjugation and related processes in Saccharomyces cerevisiae. I. Isolation and phenotypic characterization of nonmating mutants. Genetics. 1974;76:255–271. doi: 10.1093/genetics/76.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Madhani H D, Styles C A, Fink G R. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell. 1997;91:673–684. doi: 10.1016/s0092-8674(00)80454-7. [DOI] [PubMed] [Google Scholar]
- 126.Madhani H D, Fink G R. Combinatorial control required for the specificity of yeast MAPK signaling. Science. 1997;275:1314–1317. doi: 10.1126/science.275.5304.1314. [DOI] [PubMed] [Google Scholar]
- 127.Maeda T, Takekawa M, Saito H. Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science. 1995;269:554–558. doi: 10.1126/science.7624781. [DOI] [PubMed] [Google Scholar]
- 128.Maeda T, Wurgler-Murphy S M, Saito H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature. 1994;239:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- 129.Marchler G, Schüller C, Adam G, Ruis H. A Saccharomyces cerevisiae UAS element controlled by protein kinase A activates transcription in response to a variety of stress conditions. EMBO J. 1993;12:1997–2003. doi: 10.1002/j.1460-2075.1993.tb05849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Marcus S, Polverino A, Chang E, Robbins D, Cobb M H, Wigler M H. Shk1, a homolog of the Saccharomyces cerevisiae Ste20 and mammalian p65PAK protein kinases, is a component of a Ras/Cdc42 signalling module in the fission yeast Schizosaccharomyces pombe. Proc Natl Acad Sci USA. 1995;92:6180–6184. doi: 10.1073/pnas.92.13.6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Marcus S, Polverino A, Barr M, Wigler M. Complexes between Ste5 and components of the pheromone-responsive mitogen-activated protein kinase cascade module. Proc Natl Acad Sci USA. 1994;91:7762–7766. doi: 10.1073/pnas.91.16.7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Marsh L, Neiman A M, Herskowitz I. Signal transduction during pheromone response in yeast. Annu Rev Cell Biol. 1991;7:699–728. doi: 10.1146/annurev.cb.07.110191.003411. [DOI] [PubMed] [Google Scholar]
- 133.Marshall C J. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–186. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 134.Masuda T, Kariya K, Shinkai M, Okada T, Kataoka T. Protein kinase Byr2 is a target of Ras1 in the fission yeast Schizosaccharomyces pombe. J Biol Chem. 1995;270:1979–1982. doi: 10.1074/jbc.270.5.1979. [DOI] [PubMed] [Google Scholar]
- 135.Mazur P, Morin N, Baginsky W, El-Serbeini M, Clemas J A, Nielsen J B, Foor F. Differential expression and function of two homologous subunits of yeast 1,3-β-d-glucan synthase. Mol Cell Biol. 1995;15:5671–5681. doi: 10.1128/mcb.15.10.5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mazzoni C, Zarzov P, Rambourg A, Mann C. The SLT2 (MPK1) MAP kinase homolog is involved in polarized cell growth in Saccharomyces cerevisiae. J Cell Biol. 1993;123:1821–1833. doi: 10.1083/jcb.123.6.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.McLeod M, Stein M, Beach D. The product of the mei3+ gene, expressed under control of the mating-type locus, induces meiosis and sporulation in fission yeast. EMBO J. 1987;6:729–736. doi: 10.1002/j.1460-2075.1987.tb04814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Millar J B A, Buck V, Wilkinson M G. Pyp1 and Pyp2 PTPases dephosphorylate an osmosensing MAP kinase controlling cell size at division in fission yeast. Genes Dev. 1995;9:2117–2130. doi: 10.1101/gad.9.17.2117. [DOI] [PubMed] [Google Scholar]
- 139.Miller B L. Brushing up on bristles: complex genes and morphogenesis in molds. Trends Genet. 1993;9:293–295. doi: 10.1016/0168-9525(93)90236-b. [DOI] [PubMed] [Google Scholar]
- 140.Miller K Y, Wu J, Miller B L. StuA is required for cell pattern formation in Aspergillus. Genes Dev. 1992;6:1770–1782. doi: 10.1101/gad.6.9.1770. [DOI] [PubMed] [Google Scholar]
- 141.Mol P C, Parks H M, Mullins J T, Cabib E. A GTP-binding protein regulates the activity of (1-3)-β-glucan synthase, an enzyme directly involved in cell wall morphogenesis. J Biol Chem. 1994;269:31267–31274. [PubMed] [Google Scholar]
- 142.Morrison D K, Cutler R E. The complexity of Raf1 regulation. Curr Opin Cell Biol. 1997;9:174–179. doi: 10.1016/s0955-0674(97)80060-9. [DOI] [PubMed] [Google Scholar]
- 143.Mösch H-U, Fink G R. Dissection of filamentous growth by transposon mutagenesis. Genetics. 1997;145:671–684. doi: 10.1093/genetics/145.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mösch H-U, Roberts R L, Fink G R. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5352–5356. doi: 10.1073/pnas.93.11.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144a.Myers A M, Blacketer M, Bierwagen T, Edgington N. Abstracts of the Yeast Genetics and Molecular Biology Meeting. 1996. Cyclin-dependent kinase (CDK) activity regulates multiple aspects of pseudohyphal growth, abstr; p. 4. [Google Scholar]
- 145.Nadin-Davis S A, Nasim A. Schizosaccharomyces pombe ras1 and byr1 are functionally related genes of the ste family that affect starvation-induced transcription of mating-type genes. Mol Cell Biol. 1990;10:549–560. doi: 10.1128/mcb.10.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Neel B G, Tonks N K. Protein tyrosine phosphatases in signal transduction. Curr Opin Cell Biol. 1997;9:193–204. doi: 10.1016/s0955-0674(97)80063-4. [DOI] [PubMed] [Google Scholar]
- 147.Neer E J. Heterotrimeric G proteins: organizers of transmembrane signals. Cell. 1995;80:249–257. doi: 10.1016/0092-8674(95)90407-7. [DOI] [PubMed] [Google Scholar]
- 148.Neiman A M, Herskowitz I. Reconstitution of a yeast protein kinase cascade in vitro: activation of the yeast MEK homologue Ste7 by Ste11. Proc Natl Acad Sci USA. 1994;91:3398–3402. doi: 10.1073/pnas.91.8.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Neiman A M, Stevenson B J, Xu H-P, Sprague G F, Herskowitz I, Wigler M, Marcus S. Functional homology of protein kinases required for sexual differentiation in Schizosaccharomyces pombe and Saccharomyces cerevisiae suggests a conserved signal transduction module in eukaryotic organisms. Mol Biol Cell. 1993;4:107–120. doi: 10.1091/mbc.4.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Newton A C. Regulation of protein kinase C. Curr Opin Cell Biol. 1997;9:161–167. doi: 10.1016/s0955-0674(97)80058-0. [DOI] [PubMed] [Google Scholar]
- 151.Nielsen O, Friis T, Kjærulff S. The Schizosaccharomyces pombe map1 encodes an SRF/MCM1-related protein required for P-cell specific gene expression. Mol Gen Genet. 1996;253:387–392. doi: 10.1007/pl00008604. [DOI] [PubMed] [Google Scholar]
- 152.Nonaka H, Tanaka K, Hirano H, Fujiwara T, Kohno H, Umikawa M, Mino A, Takai Y. A downstream target of RHO1 small GTP-binding protein is PKC1, a homolog of protein kinase C, which leads to activation of the MAP kinase cascade in Saccharomyces cerevisiae. EMBO J. 1995;14:5931–5938. doi: 10.1002/j.1460-2075.1995.tb00281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Nurse P. The Josef Steiner Lecture. CDKs and cell-cycle control in fission yeast: relevance to other eukaryotes and cancer. Int J Cancer. 1997;71:707–708. doi: 10.1002/(sici)1097-0215(19970529)71:5<707::aid-ijc1>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 154.Obara T, Nakafuku M, Yamamoto M, Kaziro Y. Isolation and characterization of a gene encoding a G-protein α subunit from Schizosaccharomyces pombe: involvement in mating and sporulation pathways. Proc Natl Acad Sci USA. 1991;88:5877–5881. doi: 10.1073/pnas.88.13.5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ogas J. Identification and analysis of regulators of G1 progression in Saccharomyces cerevisiae. Ph.D. thesis. University of California, San Francisco; 1992. [Google Scholar]
- 156.Ohmiya R, Yamada H, Nakashima K, Aiba H, Mizuno T. Osmoregulation of fission yeast: cloning of two distinct genes encoding glycerol-3-phosphate dehydrogenase, one of which is reponsible for osmotolerance for growth. Mol Microbiol. 1995;18:963–973. doi: 10.1111/j.1365-2958.1995.18050963.x. [DOI] [PubMed] [Google Scholar]
- 156a.O’Rourke, S. Personal communication.
- 157.Ozaki K, Tanaka K, Imamura H, Hihara T, Kameyama T, Nonaka H, Hirano H, Matsuura Y, Takai Y. Rom1p and Rom2p are GDP/GTP exchange proteins for the Rho1p small GTP binding protein in Saccharomyces cerevisiae. EMBO J. 1996;15:2196–2207. [PMC free article] [PubMed] [Google Scholar]
- 158.Paravicini G, Cooper M, Friedli L, Smith D J, Carpentier J-L, Klig L S, Payton M A. The osmotic integrity of the yeast cell requires a functional PKC1 gene product. Mol Cell Biol. 1992;12:4896–4905. doi: 10.1128/mcb.12.11.4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Park H-O, Bi E, Pringle J R, Herskowitz I. Two active states of the Ras-related Bud1/Rsr1 protein bind to different effectors to determine yeast cell polarity. Proc Natl Acad Sci USA. 1997;94:4463–4468. doi: 10.1073/pnas.94.9.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Pelech S L. Signalling pathways: kinase connections on the cellular intranet. Curr Biol. 1996;6:551–554. doi: 10.1016/s0960-9822(02)00540-7. [DOI] [PubMed] [Google Scholar]
- 161.Peter M, Neiman A M, Park H-O, van Lohuizen M, Herskowitz I. Functional analysis of the interaction between the small GTP binding protein Cdc42 and the Ste20 protein kinase in yeast. EMBO J. 1996;15:7046–7059. [PMC free article] [PubMed] [Google Scholar]
- 162.Peter M, Gartner A, Horecka J, Ammerer G, Herskowitz I. FAR1 links the signal transduction pathway to the cell cycle machinery in yeast. Cell. 1993;73:747–760. doi: 10.1016/0092-8674(93)90254-n. [DOI] [PubMed] [Google Scholar]
- 163.Petersen J, Weilguny D, Egel R, Nielsen O. Characterization of fus1 of Schizosaccharomyces pombe: a developmentally controlled function needed for conjugation. Mol Cell Biol. 1995;15:3697–3707. doi: 10.1128/mcb.15.7.3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Philips J, Herskowitz I. An osmosensing signal regulates cell fusion during mating in Saccharomyces cerevisiae. J Cell Biol. 1997;138:961–974. doi: 10.1083/jcb.138.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Posas F, Saito H. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science. 1997;276:1702–1705. doi: 10.1126/science.276.5319.1702. [DOI] [PubMed] [Google Scholar]
- 166.Posas F, Wurgler-Murphy S M, Maeda T, Witten E A, Thai T C, Saito H. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell. 1996;86:885–875. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- 167.Printen J A, Sprague G F. Protein-protein interactions in the yeast pheromone response pathway: Ste5p interacts with all members of the MAP kinase cascade. Genetics. 1994;138:609–619. doi: 10.1093/genetics/138.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Qadota H, Python C P, Inoue S B, Arisawa M, Anraku Y, Zheng Y, Watanabe T, Levin D E, Ohya Y. Identification of yeast Rhop1p GTPase as a regulatory subunit of 1,3-β-glucan synthase. Science. 1996;272:278–281. doi: 10.1126/science.272.5259.279. [DOI] [PubMed] [Google Scholar]
- 169.Ramer S W, Davis R W. A dominant truncation allele identifies a gene, STE20, that encodes a putative protein kinase necessary for mating in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1993;90:452–456. doi: 10.1073/pnas.90.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Regenfelder E, Spellig T, Hartmann A, Lauenstein S, Bölker M, Kahmann R. G proteins in Ustilago maydis: transmission of multiple signals. EMBO J. 1997;16:1934–1942. doi: 10.1093/emboj/16.8.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Rens-Domiano S, Hamm H E. Structural and functional relationships of heterotrimeric G-proteins. FASEB J. 1995;9:1059–1066. doi: 10.1096/fasebj.9.11.7649405. [DOI] [PubMed] [Google Scholar]
- 172.Ridley A J. Rho—theme and variations. Curr Biol. 1996;6:1256–1264. doi: 10.1016/s0960-9822(02)70711-2. [DOI] [PubMed] [Google Scholar]
- 173.Roberts R L, Mösch H-U, Fink G R. 14-3-3 proteins bind Ste20p and are essential for Ras/MAPK cascade signaling in S. cerevisiae. Cell. 1997;89:1055–1065. doi: 10.1016/s0092-8674(00)80293-7. [DOI] [PubMed] [Google Scholar]
- 174.Roberts R L, Fink G R. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- 175.Robinson M J, Cobb M H. Mitogen-activated protein kinase pathways. Curr Opin Cell Biol. 1997;9:180–186. doi: 10.1016/s0955-0674(97)80061-0. [DOI] [PubMed] [Google Scholar]
- 176.Roemer T, Paravicini G, Payton M A, Bussey H. Characterization of the yeast (1→6)-β-glucan biosynthetic components, Kre6p and Skn1p, and genetic interactions between the PKC1 pathway and extracellular matrix assembly. J Cell Biol. 1994;127:567–579. doi: 10.1083/jcb.127.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ross-Macdonald P, Sheehan A, Roeder G S, Snyder M. A multipurpose transposon system for analyzing protein production, localization, and function in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:190–195. doi: 10.1073/pnas.94.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Roth A F, Davis N G. Ubiquitination of the yeast a-factor receptor. J Cell Biol. 1996;134:661–674. doi: 10.1083/jcb.134.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178a.Samejima I, Mackie S, Fantes P A. Multiple modes of activation of the stress-responsive MAP kinase pathway in fission yeast. EMBO J. 1997;16:6162–6170. doi: 10.1093/emboj/16.20.6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Schlessinger J. SH2/SH3 signalling proteins. Curr Opin Genet Dev. 1994;4:25–30. doi: 10.1016/0959-437x(94)90087-6. [DOI] [PubMed] [Google Scholar]
- 180.Schmidt A, Bickle M, Beck T, Hall M N. The yeast phosphatidylinositol kinase homolog Tor2 activates Rho1 and Rho2 via the exchange factor Rom2. Cell. 1997;88:531–542. doi: 10.1016/s0092-8674(00)81893-0. [DOI] [PubMed] [Google Scholar]
- 181.Schüller C, Brewster J L, Alexander M R, Gustin M C, Ruis H. The HOG pathway controls osmotic regulation of transcription via the stress response element (STRE) of the Saccharomyces cerevisiae CTT1 gene. EMBO J. 1994;13:4382–4389. doi: 10.1002/j.1460-2075.1994.tb06758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Schumacher M M, Enderlin C S, Selitrennikoff C P. The osmotic-1 locus of Neurospora crassa encodes a putative histidine kinase similar to osmosensors of bacteria and yeast. Curr Microbiol. 1997;34:340–347. doi: 10.1007/s002849900193. [DOI] [PubMed] [Google Scholar]
- 183.Seger R, Krebs W G. The MAPK signaling cascade. FASEB J. 1995;9:726–735. [PubMed] [Google Scholar]
- 184.Sells M A, Chernoff J. Emerging from the PAK: the p21-activated protein kinase family. Trends Cell Biol. 1997;7:162–167. doi: 10.1016/S0962-8924(97)01003-9. [DOI] [PubMed] [Google Scholar]
- 185.Sells M A, Knaus U G, Bagrodia S, Ambrose D M, Bokoch G M, Chernoff J. Human p21-activated kinase (Pak1) regulates actin organization in mammalian cells. Curr Biol. 1997;7:202–210. doi: 10.1016/s0960-9822(97)70091-5. [DOI] [PubMed] [Google Scholar]
- 186.Shalon D, Smith S J, Brown P D. A DNA microarray system for analyzing complex DNA samples using two-color fluorescent probe hybridization. Genome Res. 1996;6:639–645. doi: 10.1101/gr.6.7.639. [DOI] [PubMed] [Google Scholar]
- 187.Shieh J-C, Wilkinson M G, Buck V, Morgan B A, Makino K, Millar J B A. The Mcs4 response regulator coordinately controls the stress-activated Wak1-Wis1-Sty MAP kinase pathway and fission yeast cell cycle. Genes Dev. 1997;11:1008–1022. doi: 10.1101/gad.11.8.1008. [DOI] [PubMed] [Google Scholar]
- 188.Shiozaki K, Shiozaki M, Russell P. Mcs4 mitotic catastrophe suppressor regulates the fission yeast cell cycle through the Wik1-Wis1-Spc1 kinase cascade. Mol Biol Cell. 1997;8:409–419. doi: 10.1091/mbc.8.3.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Shiozaki K, Russell P. Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes Dev. 1996;10:2276–2288. doi: 10.1101/gad.10.18.2276. [DOI] [PubMed] [Google Scholar]
- 190.Shiozaki K, Russell P. Cell-cycle control linked to extracellular environment by a MAP kinase pathway in fission yeast. Nature. 1995;378:739–743. doi: 10.1038/378739a0. [DOI] [PubMed] [Google Scholar]
- 191.Siderovski D P, Hessel A, Chung S, Mak T W, Tyers M. A new family of regulators of G-protein-coupled receptors? Curr Biol. 1996;6:211–212. doi: 10.1016/s0960-9822(02)00454-2. [DOI] [PubMed] [Google Scholar]
- 192.Sietsma H H, Wessels J G H. Apical wall biogenesis. In: Wessels J G H, Meinhardt F, editors. The Mycota. Vol. 1. Heidelberg, Germany: Springer-Verlag KG; 1994. pp. 125–141. [Google Scholar]
- 193.Simon M N, de Virgilio C, Souza B, Pringle J R, Abo A, Reed S I. Role for the Rho-family GTPase Cdc42 in yeast mating-pheromone signal pathway. Nature. 1995;376:702–705. doi: 10.1038/376702a0. [DOI] [PubMed] [Google Scholar]
- 194.Song O K, Dolan J W, Yuan Y O, Fields S. Pheromone-dependent phosphorylation of the yeast STE12 protein correlates with transcriptional activation. Genes Dev. 1991;5:741–750. doi: 10.1101/gad.5.5.741. [DOI] [PubMed] [Google Scholar]
- 195.Spellig T, Bölker M, Lottspeich F, Frank R W, Kahmann R. Pheromones trigger filamentous growth in Ustilago maydis. EMBO J. 1994;13:1620–1627. doi: 10.1002/j.1460-2075.1994.tb06425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Stankis M M, Specht C A, Yang H L, Giasson L, Ullrich R C, Novotny C P. The Aα mating locus of Schizophyllum commune encodes two dissimilar multiallelic homeodomain proteins. Proc Natl Acad Sci USA. 1992;89:7169–7173. doi: 10.1073/pnas.89.15.7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Stefan C J, Blumer K J. The third cytoplasmic loop of a yeast G-protein-coupled receptor controls pathway activation, ligand discrimination, and receptor internalization. Mol Cell Biol. 1994;14:3339–3349. doi: 10.1128/mcb.14.5.3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Stevenson B J, Rhodes N, Errede B, Sprague G F. Constitutive mutants of the protein kinase Ste11 activate the yeast pheromone response pathway in the absence of the G protein. Genes Dev. 1992;6:1292–1304. doi: 10.1101/gad.6.7.1293. [DOI] [PubMed] [Google Scholar]
- 199.Sugimoto A, Iino Y, Maeda T, Watanabe Y, Yamamoto M. Schizosaccharomyces pombe ste11+ encodes a transcription factor with an HMG motif that is a critical regulator of sexual development. Genes Dev. 1991;5:1990–1999. doi: 10.1101/gad.5.11.1990. [DOI] [PubMed] [Google Scholar]
- 200.Takeda T, Toda T, Kominami K-I, Kohnosu A, Yanagida M, Jones N. Schizosaccharomyces pombe atf1+ encodes a transcription factor required for sexual development and entry into stationary phase. EMBO J. 1995;14:6193–6208. doi: 10.1002/j.1460-2075.1995.tb00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Takekawa M, Posas F, Saito H. A human homolog of the yeast Ssk2/Ssk22 MAP kinase kinase kinases, MTK1, mediates stress-induced activation of the p38 and JNK pathways. EMBO J. 1997;16:4973–4982. doi: 10.1093/emboj/16.16.4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Tanaka K, Davey J, Imai Y, Yamamoto M. Schizosaccharomyces pombe map3+ encodes the putative M-factor receptor. Mol Cell Biol. 1993;13:80–88. doi: 10.1128/mcb.13.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Tapon N, Hall A. Rho, Rac, and Cdc42 GTPases regulate the organization of the actin cytoskeleton. Curr Opin Cell Biol. 1997;9:86–92. doi: 10.1016/s0955-0674(97)80156-1. [DOI] [PubMed] [Google Scholar]
- 204.Tedford K, Kim S, Sa D, Stevens K, Tyers M. Regulation of the mating pheromone and invasive growth responses in yeast by two MAP kinase substrates. Curr Biol. 1997;7:228–238. doi: 10.1016/s0960-9822(06)00118-7. [DOI] [PubMed] [Google Scholar]
- 205.Toda T, Shimanuki M, Yanagida M. Fission yeast genes that confer resistance to staurosporine encode an AP-1-like transcription factor and a protein kinase related to the mammalian ERK1/MAP2 and budding yeast Fus3 and Kss1 kinases. Genes Dev. 1991;5:60–73. doi: 10.1101/gad.5.1.60. [DOI] [PubMed] [Google Scholar]
- 206.Torres L, Martín H, García-Saez M I, Arroyo J, Molina M, Sánchez M, Nombela C. A protein kinase gene complements the lytic phenotype of Saccharomyces cerevisiae lyt2 mutants. Mol Microbiol. 1991;5:2845–2854. doi: 10.1111/j.1365-2958.1991.tb01993.x. [DOI] [PubMed] [Google Scholar]
- 207.Towsend G F, Lindegren C C. Characteristic growth patterns of the different members of a polyploid series of Saccharomyces cerevisiae. J Bacteriol. 1954;67:480–483. doi: 10.1128/jb.67.4.480-483.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Treisman R. Regulation of transcription by MAP kinase cascades. Curr Opin Cell Biol. 1996;8:205–215. doi: 10.1016/s0955-0674(96)80067-6. [DOI] [PubMed] [Google Scholar]
- 209.Vaillancourt L J, Raudaskoski M, Specht C A, Raper C A. Multiple genes encoding pheromones and a pheromone receptor define the Bβ1 mating-type specificity in Schizophyllum commune. Genetics. 1997;146:541–554. doi: 10.1093/genetics/146.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Vaillancourt L J, Raper C A. Pheromones and pheromone receptors as mating-type determinants in Basidiomycetes. In: Setlow J K, editor. Genetic engineering. Vol. 18. New York, N.Y: Plenum Press; 1996. pp. 219–247. [DOI] [PubMed] [Google Scholar]
- 211.Varela J C S, Praekelt U M, Meacock P A, Planta R J, Mager W H. The Saccharomyces cerevisiae HSP12 gene is activated by the high-osmolarity glycerol pathway and negatively regulated by protein kinase A. Mol Cell Biol. 1995;15:6232–6245. doi: 10.1128/mcb.15.11.6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Varela J C S, van Beekvelt C A, Planta R J, Mager W H. Osmostress-induced changes in yeast gene expression. Mol Microbiol. 1992;6:2183–2190. doi: 10.1111/j.1365-2958.1992.tb01392.x. [DOI] [PubMed] [Google Scholar]
- 213.Wang Y, Xu H-P, Riggs M, Rodgers L, Wigler M. byr2, a Schizosaccharomyces pombe gene encoding a protein kinase capable of partial suppression of the ras1 mutant phenotype. Mol Cell Biol. 1991;11:3554–3563. doi: 10.1128/mcb.11.7.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Wang Z, Harkins P C, Ulevitch R J, Han J, Cobb M H, Goldsmith E J. The structure of mitogen-activated protein kinase p38 at 2.1 Å resolution. Proc Natl Acad Sci USA. 1997;94:2327–2332. doi: 10.1073/pnas.94.6.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Warbrick E, Fantes P A. The Wis1 protein kinase is a dosage-dependent regulator of mitosis in Schizosaccharomyces pombe. EMBO J. 1991;13:4291–4299. doi: 10.1002/j.1460-2075.1991.tb05007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Ward M P, Gimeno C J, Fink G R, Garrett S. SOK2 may regulate cyclic AMP-dependent protein kinase-stimulated growth and pseudohyphal development by repressing transcription. Mol Cell Biol. 1995;15:6854–6863. doi: 10.1128/mcb.15.12.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Watanabe Y, Takaesu G, Hagiwara M, Irie K, Matsumoto K. Characterization of a serum response factor-like protein in Saccharomyces cerevisiae, Rlm1, which has transcriptional activity regulated by the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol Cell Biol. 1997;17:2615–2623. doi: 10.1128/mcb.17.5.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Watanabe Y, Irie K, Matsumoto K. Yeast RLM1 encodes a serum response factor-like protein that may function downstream of the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol Cell Biol. 1995;15:5740–5749. doi: 10.1128/mcb.15.10.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Watanabe Y, Iino Y, Furuhata K, Shimoda C, Yamamoto M. The S. pombe mei2 gene encoding a crucial molecule for commitment to meiosis is under the regulation of cAMP. EMBO J. 1988;7:761–767. doi: 10.1002/j.1460-2075.1988.tb02873.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Wendland J, Vaillancourt L J, Hegner J, Lengeler K B, Laddison K J, Specht C A, Raper C A, Kothe E. The mating type locus Bα1 of Schizophyllum commune contains a pheromone receptor gene and putative pheromone genes. EMBO J. 1995;14:5271–5278. doi: 10.1002/j.1460-2075.1995.tb00211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Whiteway M S, Wu C, Leeuw T, Clark K, Fourest-Lieuvin A, Thomas D Y, Leberer E. Association of the yeast pheromone response G protein β subunits with the MAP kinase scaffold Ste5p. Science. 1995;269:1572–1575. doi: 10.1126/science.7667635. [DOI] [PubMed] [Google Scholar]
- 222.Wilkinson M G, Samuels M, Takeda T, Toone W M, Shieh J-C, Toda T, Millar J B A, Jones N. The Atf1 transcription factor is a target for the Sty1 stress-activated MAP kinase pathway in fission yeast. Genes Dev. 1996;10:2289–2301. doi: 10.1101/gad.10.18.2289. [DOI] [PubMed] [Google Scholar]
- 223.Willer M, Hoffmann L, Styrkársdóttir U, Egel R, Davey J, Nielsen O. Two-step activation of meiosis by the mat1 locus in Schizosaccharomyces pombe. Mol Cell Biol. 1995;15:4964–4970. doi: 10.1128/mcb.15.9.4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Wu C, Whiteway M, Thomas D Y, Leberer E. Molecular characterization of Ste20p, a potential mitogen-activated protein extracellular signal-regulated kinase kinase (MEK) kinase kinase from Saccharomyces cerevisiae. J Biol Chem. 1995;270:15984–15992. doi: 10.1074/jbc.270.27.15984. [DOI] [PubMed] [Google Scholar]
- 225.Xu J R, Hamer J E. MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes Dev. 1996;10:2696–2706. doi: 10.1101/gad.10.21.2696. [DOI] [PubMed] [Google Scholar]
- 226.Yabana N, Yamamoto M. Schizosaccharomyces pombe map1+ encodes a MADS-box-family protein required for cell-type specific gene expression. Mol Cell Biol. 1996;16:3420–3428. doi: 10.1128/mcb.16.7.3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Yablonski D, Marbach I, Levitzki A. Dimerization of Ste5, a mitogen-activated protein kinase cascade scaffold protein, is required for signal transduction. Proc Natl Acad Sci USA. 1996;93:13864–13869. doi: 10.1073/pnas.93.24.13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Yamamoto M. The molecular control mechanisms of meiosis in fission yeast. Trends Biochem. 1996;21:18–22. [PubMed] [Google Scholar]
- 228a.Yamamoto M, Imai Y, Watanabe Y. Mating and sporulation in Schizosaccharomyces pombe. In: Pringle J R, Broach J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces. Cell cycle and cell biology. Vol. 3. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 1037–1106. [Google Scholar]
- 229.Yamochi W, Tanaka K, Nonaka H, Maeda A, Musha T, Takai Y. Growth site localization of Rho1 small GTP-binding protein and its involvement in bud formation in Saccharomyces cerevisiae. J Cell Biol. 1994;125:1077–1093. doi: 10.1083/jcb.125.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Zarzov P, Mazzoni C, Mann C. The Slt2 (Mpk1) MAP kinase is activated during periods of polarized cell growth in yeast. EMBO J. 1996;15:83–91. [PMC free article] [PubMed] [Google Scholar]
- 231.Zhan X L, Deschenes R J, Guan K L. Differential regulation of Fus3 MAP kinase by tyrosine-specific phosphatases Ptp2/Ptp3 and dual-specificity phosphatase Msg5 in Saccharomyces cerevisiae. Genes Dev. 1997;13:1690–1702. doi: 10.1101/gad.11.13.1690. [DOI] [PubMed] [Google Scholar]
- 232.Zhang F, Strand A, Robbins D, Cobb M H, Goldsmith E J. Atomic structure of the MAP kinase ERK2 at 2.3 Å resolution. Nature. 1994;367:704–711. doi: 10.1038/367704a0. [DOI] [PubMed] [Google Scholar]
- 233.Zhao Z-S, Leung T, Manser E, Lim L. Pheromone signalling in Saccharomyces cerevisiae requires the small GTP-binding protein Cdc42p and its activator Cdc24. Mol Cell Biol. 1995;15:5246–5257. doi: 10.1128/mcb.15.10.5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Zheng Y, Hart M J, Shinip K, Evans T, Bender A, Cerione R A. Biochemical comparisons of the Saccharomyces cerevisiae Bem2 and Bem3 proteins. J Biol Chem. 1993;268:24629–24634. [PubMed] [Google Scholar]
- 235.Zhou Z, Gartner A, Cade R, Ammerer G, Errede B. Pheromone-induced signal transduction in Saccharomyces cerevisiae requires the sequential function of three protein kinases. Mol Cell Biol. 1993;13:2069–2080. doi: 10.1128/mcb.13.4.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]