Abstract
Glucose and related sugars repress the transcription of genes encoding enzymes required for the utilization of alternative carbon sources; some of these genes are also repressed by other sugars such as galactose, and the process is known as catabolite repression. The different sugars produce signals which modify the conformation of certain proteins that, in turn, directly or through a regulatory cascade affect the expression of the genes subject to catabolite repression. These genes are not all controlled by a single set of regulatory proteins, but there are different circuits of repression for different groups of genes. However, the protein kinase Snf1/Cat1 is shared by the various circuits and is therefore a central element in the regulatory process. Snf1 is not operative in the presence of glucose, and preliminary evidence suggests that Snf1 is in a dephosphorylated state under these conditions. However, the enzymes that phosphorylate and dephosphorylate Snf1 have not been identified, and it is not known how the presence of glucose may affect their activity. What has been established is that Snf1 remains active in mutants lacking either the proteins Grr1/Cat80 or Hxk2 or the Glc7 complex, which functions as a protein phosphatase. One of the main roles of Snf1 is to relieve repression by the Mig1 complex, but it is also required for the operation of transcription factors such as Adr1 and possibly other factors that are still unidentified. Although our knowledge of catabolite repression is still very incomplete, it is possible in certain cases to propose a partial model of the way in which the different elements involved in catabolite repression may be integrated.
Saccharomyces cerevisiae and many other yeasts may thrive on a variety of carbon sources, but glucose and fructose are the preferred ones. When one of these sugars is present, the enzymes required for the utilization of alternative carbon sources are synthesized at low rates or not at all. This phenomenon is known as carbon catabolite repression, or simply catabolite repression, and since no “catabolite” derived from glucose and involved in the repression has been yet identified, the term “glucose repression” has also been proposed. In this review, I still use the term “catabolite repression” as well as glucose repression, to stress that other sugars, such as galactose or maltose, are able to affect the synthesis of enzymes repressed by glucose (Table 1).
TABLE 1.
Catabolite repression caused by different sugars
| Yeast species | Carbon source | Enzyme activity (mU/mg of protein)
|
|||||
|---|---|---|---|---|---|---|---|
| Malate synthase | Fructose bisphosphatase | Isocitrate lyase | Cytochrome oxidase | Malate dehydrogenase | Glutamate dehydrogenasec | ||
| Saccharomyces cerevisiaea | Glucose | 1 | 1 | 1 | 6 | 450 | 1 |
| Galactose | 1 | 1 | 1 | 19 | 700 | 8 | |
| Pyruvate | 180 | 40 | 80 | 38 | 11,000 | 53 | |
| Schizosaccharomyces pombeb | Glucose | 1 | 9 | 100 | 15 | ||
| Maltose | 1 | 20 | 250 | 45 | |||
| Ethanol | 80 | 90 | 3,400 | 48 | |||
A comprehensive picture of the mechanism(s) of catabolite repression is not yet available, in spite of the accumulation of information on the subject (for earlier reviews, see references 95, 96a, 96b, 124, 163, 289, and 346). Although the solution of the puzzle has progressed, important pieces are still missing and it has been found that other pieces, originally thought to belong, do not really pertain to the basic frame. The last few years have seen important advances, which are reviewed and discussed in this article. I also propose some models for catabolite repression of different genes and discuss some perspectives for future research. Although the review deals mainly with S. cerevisiae, reference to other yeast species is made, as far as information is available.
For easy reference, Table 2 provides an overview of the alternative names given to genes related to catabolite repression, since these genes have been repeatedly isolated by different groups and given different names.
TABLE 2.
Alternative names for genes discussed in this review
| Genes directly involved in catabolite repression |
| CYC8 = SSN6 |
| GLC7 = DIS2S1 = CID1 |
| GRR1 = CAT80 |
| HXK2 = HEX1 = GLR1 |
| MIG1 = CAT4 = SSN1 |
| MTH1 = DGT1 = HTR1 |
| REG1 = HEX2 |
| SNF1 = CAT1 = CCR1 |
| SNF4 = CAT3 |
| TUP1 = CYC9 = FLK1 = UMR7 = AAR1 = AER2 = AMM1 = ROX4 = SLF2 |
| Genes which play an indirect role |
| ADA3 = NGG1 |
| ADA5 = SPT20 |
| GCN5 = ADA4 |
| MSN1 = FUP1 = PHD2 = MSS10 |
| MSN3 = STD1 |
| ROX3 = SSN7 |
| SIN4 = SSN4 |
| SKO1 = ACR1 |
| SNF2 = SWI2 = GAM1 |
| SPT6 = CRE2 = SSN20 |
| SPT11 = HTA1 |
| SPT12 = HTB1 |
| SRB8 = SSN5 = GIG1 |
| SRB9 = SSN2 |
| SRB10 = UME5 = SSN3 = GIG2 |
| SRB11 = SSN8 = GIG3 |
| SWI1 = ADR6 = GAM3 |
| TFG3 = TAF30 |
LEVELS OF CONTROL
Glucose may affect enzyme levels by causing a decrease in the concentration of the corresponding mRNAs, a decrease in their translation rate, or an increase in the degradation rate of the protein. In turn, mRNA levels would depend both on the rate of transcription of the corresponding gene and on the stability of the mRNA. The main effect of glucose takes place at the transcriptional level; accordingly, this review deals mainly with this mechanism of regulation. Nevertheless, alternative mechanisms which are operative in certain cases are briefly discussed in this section.
Control of the mRNA translation rate is not common in yeast; however, in the case of the transcriptional activator Adr1, glucose appears to act at this step. While the concentration of Adr1 is at least 10-fold higher in ethanol-grown yeast than in glucose-grown yeast, there is only a 2-fold difference in the levels of the corresponding mRNAs (354). Since the half-life of Adr1 itself is not longer in ethanol-grown cells than in glucose-grown cells, it has been concluded that the observed decrease in the level of Adr1 is due mainly to a reduction in the rate of Adr1 synthesis brought about by glucose. The molecular mechanism by which glucose acts remains unclear, but it has been shown that the translational control does not depend on the long untranslated 5′ leader sequence of ADR1 mRNA (354). Removal of the gene sequence corresponding to the 681 C-terminal residues of Adr1 (more than half the length of the protein) did not disrupt the translational control, but the ADR1 coding sequence between amino acids 262 and 642 is required for the control of ADR1 translation by glucose.
While translational control by glucose is rare, glucose triggers inactivation and/or proteolysis of a number of proteins. By analogy to catabolite repression, this phenomenon has been called catabolite inactivation (151); it affects a variety of proteins, from gluconeogenic enzymes to transport molecules, but it is not yet known whether the same mechanism underlies the inactivation in the different cases (125a). Inactivation of fructose-1,6-bisphosphatase (FbPase) by glucose (121) has been most extensively studied, and it has been shown that glucose causes a very rapid phosphorylation of FbPase and a proteolytic degradation of the enzyme (119, 224, 225, 236). Two alternative mechanisms for the proteolysis have been described: (i) transfer of FbPase to the vacuole and degradation by vacuolar proteases (56), and (ii) ubiquitination of FbPase (310) followed by degradation by the proteasome (309). The relative contribution of each pathway could depend on the physiological state of the yeast (306). Proteolysis of FbPase triggered by glucose can occur even in the absence of phosphorylation (291), but it has not been established whether phosphorylation may be a requirement or a facilitator of at least one of the degradation systems. The mechanism by which glucose triggers the proteolysis is not known, but glucose appears to induce the synthesis of proteins required for the degradation process. Some of these proteins would be transported to the vacuole, via the secretory pathway, and facilitate the uptake of FbPase by the vacuole (56). Other proteins would be required for ubiquitination of FbPase and its degradation in the proteasome (310). Recently, a series of mutants have been isolated in which inactivation and degradation of FbPase are uncoupled and the inactive protein accumulates in the cytosol and in small vesicles in the cytoplasm (149). Although the proteins involved in the process have not been identified yet, there is some evidence that in wild-type cells, FbPase could be imported in cytoplasmic vesicles before being degraded in the vacuole (155).
The most general system of catabolite repression involves a parallel decrease in mRNA and protein levels. Glucose has been reported to destabilize the corresponding mRNAs in a few systems: the functional half-life of CYC1 mRNA was shown to decrease from 12 min in derepressed cells to about 2 min when glucose was present (400, 401); for MAL6S (MAL62) mRNA the decrease was from 25 to 6 min (107); glucose also had a very strong effect on the mRNAs encoding subunits of succinate dehydrogenase, decreasing the half-life from more than 60 min to less than 10 min (198); for PCK1 mRNA, glucose accelerated the degradation rate only twofold (230). In no case, however, was information available about a possible mechanism for this effect of glucose.
More recently, it has been shown that the 5′ untranslated end of the mRNA encoding the iron protein (Ip) subunit of succinate dehydrogenase is able to promote rapid degradation of a fusion mRNA upon glucose addition (48). The 5′ exonuclease Xrn1 seems to play an important role in the mRNA degradation, and it has been suggested that the rate of degradation of mRNA could be set by a competition between initiation of translation and nuclease action (48). In this case, glucose could promote mRNA degradation by blocking mRNA translation. Phosphorylation of glucose or fructose was required to trigger Ip mRNA turnover, but any of the hexose kinases would be effective. Although further metabolism of the hexose phosphate formed does not seem to be required (both glucose and fructose increased turnover in a pgi mutant), addition of 2-deoxyglucose to a derepressed yeast culture did not decrease Ip mRNA stability (49). A possible interpretation for this could be that 2-deoxyglucose would decrease ATP levels and that the triggering of mRNA degradation would be energy dependent. Most factors required for glucose repression of genes such as SUC2 or the GAL genes (Hxk2, Grr1, Tup1, and Cyc8) did not markedly affect mRNA turnover in the presence of glucose, and the protein Ume5 (Srb10), which has been reported to destabilize SPO13 mRNA in a glucose-containing medium (331), was not needed for differential turnover of Ip mRNA (49). On the other hand, the regulatory protein Reg1 was required for increased degradation of Ip mRNA upon glucose addition (49).
The mechanism(s) regulating mRNA turnover in response to the carbon source remain to be worked out, but it is clearly established that for a subset of genes regulated by glucose, control is operating on mRNA stability instead of (or in addition to) on transcription rates. However, in the rest of this review, catabolite repression is considered in the narrow sense of the repression of transcription caused by glucose, since, as mentioned above, this is the major control mechanism.
ELEMENTS OF THE SYSTEM
Glucose and other repressing sugars can affect the rate of transcription by two basic mechanisms: they interfere with activators of transcription, or they facilitate the action of proteins with a negative effect on transcription. In bacteria, one or the other of these control mechanisms regulates catabolite repression in different species (297). Our present picture is that in yeast, the different sugars do not act directly on DNA-binding proteins but produce signals that are transmitted through a series of proteins to the promoters of the corresponding genes. To unravel the mechanisms of catabolite repression, it is therefore necessary to identify the signals produced by the sugar, the proteins which respond to them, and their substrates, down to the proteins binding to the promoter of the regulated genes.
To find the different elements that participate in the cascade of reactions between glucose and the final target, mutants affected in the process have been invaluable. Two kinds of mutants were sought, mutants for which glucose was no longer repressing and mutants in which derepression did not occur even when the glucose in the medium had been used up. To obtain these mutants, a variety of strategies were used (see reference 125 for a review), and the number of different genes isolated has increased in a bewildering manner. However, in the last few years it has become apparent that a number of these genes are not specifically related to the control by glucose but play a more general role in the control of transcription.
In the following sections, we consider in detail the different elements of the system: proteins which act as specific transcriptional repressors or activators, intermediary regulatory factors, and elements involved in glucose signalling. A number of proteins which somehow affect catabolite repression but are not directly involved in the response to glucose are also discussed.
Activators
The Hap2/3/4/5 complex.
A large number of genes are regulated by a complex containing the proteins Hap2, Hap3, Hap4, and Hap5 (see reference 79 for a compilation of such genes). The complex, which activates transcription when yeast grows on a nonfermentable carbon source, binds DNA and makes contacts with a consensus ACCAA(T/C)NA sequence called the CCAAT box (255). In a search for yeasts unable to activate a CCAAT box-containing fusion gene, hap2, hap3, and hap4 mutants were isolated (114, 134, 136). Hap5 was identified by the two-hybrid assay with the core region of Hap2 as a bait (227). Hap2, Hap3, and Hap5 are absolutely required for CCAAT-binding activity, as shown in band shift experiments, and the three subunits are also sufficient for DNA binding, since a mixture of the three purified recombinant proteins allows binding to a CCAAT box in vitro (227). This is in contrast to initial reports that Hap4 was required for binding (114). It appears now that Hap4 is mainly responsible for the activation of transcription produced by the complex (257). The stoichiometry of the complex is not yet clear, but it has been reported to contain a single Hap2 molecule (384).
A 60-amino-acid core region of Hap2 is sufficient for functionality: within this region, there is a DNA binding domain and a subunit association domain (256, 257). This last domain is essential for interactions between Hap2 and other subunits of the complex and is able to form a helical structure (384). The DNA-binding domain of Hap2 is a 21-amino-acid region in which three critical histidines and three critical arginines have been identified (383). Hap3 contains also a 7-amino-acid region required for DNA binding, but its subunit association domain has not been clearly delineated (383).
It has been proposed that Hap5 interacts with both Hap2 and Hap3 and brings the proteins together, allowing the interaction between the DNA-binding domains of Hap2 and Hap3 (227). The proteins Hap3 and Hap5, but not Hap2, contain the histone fold motif, a structural feature first identified in histones and now found in a large group of proteins, involved in protein-protein and/or protein-DNA interactions (8).
It is not clear to what extent glucose affects Hap2 levels, since it was reported (272) that glucose decreased the levels of the HAP2 transcript fivefold whereas it was stated later that HAP4, “unlike HAP2 and HAP3,” was induced fivefold when yeast cells were shifted from glucose to lactate (114). The effect of glucose on HAP4 is clearer, and the presence of a strong Mig1 binding site in the promoter of HAP4 suggested that catabolite repression of HAP4 would operate through Mig1 (79). However, it has been found that HAP4 is still repressed by glucose in a mig1 mutant (200). A possible interpretation for this observation is that an analog of Mig1, Mig2 (see the section on repressors, below), is sufficient to maintain HAP4 in a repressed state. Although it has been established that genes such as CYC1 or COX6, regulated by the Hap2/3/4/5 complex, require Snf1 for derepression (382), there have been no reports on a possible role of Snf1 on HAP4 expression.
The interesting observation has been made that while the expression of CYC1-lacZ in a galactose medium decreases over 25-fold in a hap2 or a hap3 mutant, the decrease is less than 4-fold in a hap4 mutant (67). This could suggest that the Hap2/3/4/5 complex plays a double role: remodeling of the chromatin structure, which does not require Hap4, and direct activation of the RNA polymerase, in which Hap4 would be involved. In fact, a short region of the general transcription factor TFIIB, which contains an amphipathic helix unique to yeast TFIIB, is specifically required for activation of transcription by the Hap2/3/4/5 complex (315).
Homologs of the HAP genes have been identified in other yeast species. In Schizosaccharomyces pombe, the php2 gene is able to complement an S. cerevisiae hap2 mutant, but in contrast to S. cerevisiae, the capacity of cellular extracts of S. pombe to bind to a CCAAT probe was similar for glucose- and glycerol-grown cells (256). Nevertheless, since disruption of php2 made S. pombe unable to grow in glycerol, the fission yeast gene also appears to be involved in mitochondrial function. The situation is different in Kluyveromyces lactis, where disruption of the functional homologs of HAP2 or HAP3 had no significant effect on the growth of the yeast on respiratory substrates (235, 248).
Gal4.
The protein Gal4 (184) activates the transcription of a family of genes, GAL1, GAL2, GAL7, GAL10, and MEL1, involved in the catabolism of galactose and melibiose (for a review, see references 161 and 228). These genes contain one to four copies of a regulatory element, UASGAL, with the palindromic consensus binding site CGGA(G/C)GACAGTC(C/G)TCCG (129), to which Gal4 can bind.
Gal4 has a DNA binding domain at the N terminus, which is a C6 zinc cluster (162), and two acidic activation regions, one near the DNA binding domain and the other at the C terminus (206). Gal4 is found as a monomer in the absence of DNA, but it binds to DNA as a dimer, with the C6 zinc clusters making contacts with a conserved CCG triplet at each end of the site (213). A short coiled-coil dimerization element of Gal4 is responsible for the symmetrical binding (35). Gal4 forms a complex with the regulatory protein Gal80 (129), and this formation requires the carboxy-terminal 30 amino acids from Gal4 (206). In the absence of galactose in the medium, the complex binds to UASGAL but is not able to activate transcription (129); when galactose is present, the regulatory protein Gal3, a protein with strong homology to galactokinase, binds Gal80, thereby relieving its inhibitory action on Gal4 (333, 388). Since Gal4 has been shown to occur in vivo in different phosphorylation states, it has been proposed that regulated phosphorylation modulates Gal4 activity (239). Although Gal4 can be phosphorylated at multiple sites, phosphorylation at Ser-699 plays a special role, since it is required for maximal activated transcription (296). There is evidence that phosphorylation of Gal4 takes place after Gal4 has stimulated the assembly of the general transcriptional machinery (228).
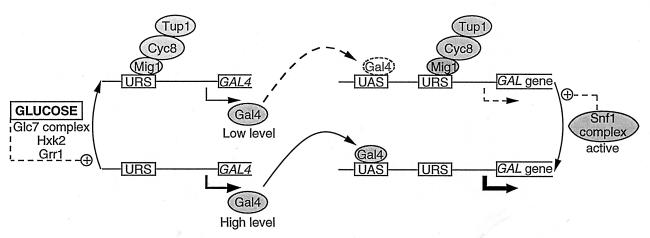
When there is glucose in the medium, transcription of the GAL genes is repressed even if galactose is also present (1). Glucose could act on Gal4 at different levels: preventing its synthesis, blocking its capacity to bind to the UASGAL site, or interfering with its activating function. Expression of GAL4 is moderately repressed by glucose, and this repression involves the binding of the regulatory protein Mig1 to the GAL4 promoter (133, 241). In addition, glucose interferes with the release of the inhibition of Gal4 by Gal80 performed by galactose (164). This could be related to the observation that in the presence of Gal80, glucose prevents the phosphorylation of Gal4 (296). In a gal80 mutant with constitutive expression of Gal4, the activity of Gal4 appears to be unaffected by glucose (328) and the replacement of Ser-699 by alanine has no effect on the capacity of Gal4 to activate transcription (296).
In K. lactis a transcriptional activator called Lac9, equivalent to Gal4, has been identified (302). Lac9 and Gal4 are operative in the heterologous yeast (286, 302), even though they have diverged considerably and homologies between them are restricted to areas of the protein implicated in nuclear localization, DNA binding, and transcriptional activation. The main regulatory features of the system have been conserved, with some interesting variations. In K. lactis, there is a KlGal80 protein that is very similar to the Gal80 protein and is able to block transcriptional activation by Lac9 in the absence of galactose (394). In contrast, there is no protein homologous to Gal3, and its function is taken by the galactokinase itself. A KlGal1-KlGal80 complex can be formed in the presence of galactose and ATP (395), and its formation is relevant for the expression of the GAL genes in K. lactis, as shown by the fact that a mutant protein KlGal1-m1 with galactokinase activity but unable to form a complex with KlGal80 has lost its regulatory function (395).
Very large differences between strains, from no repression to 100-fold repression by glucose of the GAL genes from K. lactis, have been observed (24). These differences have been shown to depend on the LAC9 allele present in the strain. A 2-base difference in the LAC9 promoter region modifies the level of expression of the gene, and the small change (two- to threefold) in the concentration of the regulatory protein produces large effects in the expression of the GAL genes (178, 392). In contrast to the situation in S. cerevisiae, in K. lactis the absence of KlGal80 is sufficient to relieve the repression by glucose (394).
Mal63.
In different S. cerevisiae strains, the genes required for maltose utilization may be found at different loci called MAL1, MAL2, MAL3, MAL4, or MAL6. The most extensively studied gene complex is MAL6; in this complex, MAL61 (MAL6T) encodes maltose permease, MAL62 (MAL6S) encodes maltase, and MAL63 (MAL6R) encodes a protein which activates the expression of the genes MAL61 and MAL62 and probably that of the MAL63 gene itself (51, 240).
The protein Mal63 belongs to the same family of C6 zinc cluster proteins as Gal4 (50, 168). In contrast to Gal4, it is found as a dimer even in the absence of DNA (323) and it lacks acidic or glutamine-rich regions which appear associated with the capacity to activate transcription in other systems. The DNA recognition motif for Mal63 has not been clearly established; it was first suggested that the sequence GAAA(A/T)TTTCGC, found twice in a 68-bp region situated between the MAL61 and MAL62 genes and necessary for their maltose-induced expression, could be important (188). However, a footprint assay revealed three protected sites in the MAL61-MAL62 promoter, none of which corresponded to this sequence (323). Among these sites, one contains the sequence CGGN9CGG, a structure similar to that bound by the zinc cluster protein Hap1 (396) and another contains the sequence CGCN9CGC; the third one looks like a half-site sequence. In addition, a sequence in the promoter of MAL63 itself which binds Mal63 (240) has a similar motif, CGGN9CGC. In all these sequences, the intermediary N9 region is very AT rich.
There is at present no information about the mechanism by which Mal63 is activated in the presence of maltose, although it has been speculated that maltose may bind to Mal63, yielding a conformation with functional activity (369). Constitutive alleles of MAL63, or the wild-type allele of MAL43 (an equivalent gene in the MAL4 locus), have multiple amino acid substitutions in the C-terminal region and may adopt the active conformation, even in the absence of maltose (369).
Glucose represses the expression of the MAL genes, even when maltose is present in the medium (107, 154), and it appears to affect both MAL63 transcription and the formation of the active conformation of Mal63 (154, 369). The effect on MAL63 transcription is mediated largely by Mig1 as the disruption of MIG1 (154) or the removal of a Mig1 binding site in the MAL63 promoter (369) increases MAL63 expression in the presence of glucose. On the other hand, glucose may also cause inducer exclusion, perhaps by inactivating the maltose permease (27, 131), thereby preventing the conformational change of Mal63. Glucose repression of the MAL genes decreases strongly in strains carrying constitutive alleles of the regulatory proteins Mal63 or Mal43 (154, 369).
In Candida albicans, an α-glucosidase encoded by the CAMAL2 gene is induced by both maltose and sucrose and repressed by glucose (127). The regulatory gene CASUC1, which is also required for the utilization of sucrose or maltose, encodes a C6 zinc cluster protein with 28% identity to the Mal63 protein from Saccharomyces and able to complement a mal63 mutation (167). An inspection of the promoter regions of CASUC1 and CAMAL2 reveals a number of sequences with some similarity to the consensus sequence able to bind Mig1 in S. cerevisiae, but none of them seems likely to have a strong affinity for this protein (200). However, a putative homolog of Mig1 has been only recently identified in C. albicans (393), and there is not yet any information on its precise requirements for DNA binding. It therefore remains possible that catabolite repression of the α-glucosidase from Candida, like that of its Saccharomyces counterpart, is partly mediated by Mig1.
Adr1.
Adr1 is a zinc finger protein belonging to the C2H2 family (138), identified as a positive effector of the expression of ADH2, a gene which is repressed by glucose and which encodes alcohol dehydrogenase II (57). Adr1 has been localized in the nucleus, and its zinc fingers are essential for DNA binding (14). Adr1 binds a 22-bp palindromic sequence in the promoter of ADH2 (94) and may also bind sequences in the promoters of genes encoding peroxisomal proteins or proteins involved in glycerol utilization (266, 320). A consensus sequence for Adr1 binding, C(T/C)CC(A/G)N6–38(T/C)GG(A/G)G, has been proposed (53).
Several regions in Adr1, named TADI to TADIII, may act as transcription activation domains (60); a fourth region (TADIV) would be required for the activation of peroxisomal genes (322). To activate transcription efficiently, Adr1 requires a coactivator complex which includes among its components Ada2 and the histone acetyltransferase Gcn5 (55). There is evidence that Ada2 interacts specifically with TADII while Gcn5 may interact with any of the four TADs. In addition, TADI and TADIV bind to the C-terminal half of the RNA polymerase II subunit TFIIB (55). It would appear, therefore, that Adr1 interacts with several components of the coactivator complex, thus facilitating the acetylation of histones and a nucleosomal rearrangement which allows transcription to proceed (362). The interaction with TFIIB could also recruit the transcriptional machinery to the promoter, contributing further to the activation of the gene regulated by Adr1.
Adr1 activity is sensitive to catabolite repression, with glucose acting at different levels; the relative importance of the different effects of glucose may vary depending on the genetic background. Glucose decreases the rate of transcription of ADR1 10- to 20-fold in some strains and has no significant effect in others (15). In a yeast strain where ADR1 was not repressed by glucose, a mutation in any of the genes SAF1, SAF2, or SAF3 made ADR1 sensitive to glucose (61). Although this may be related to the differences observed between strains, there have been no further reports on the role of the SAF genes. Since transcription of ADR1 is inhibited when the activity of the cyclic AMP (cAMP)-dependent protein kinase is unregulated (82), a possible way for glucose to affect ADR1 expression would be through an increase in the cAMP level. An effect of glucose on the stability of the ADR1 mRNA has also been observed; in the presence of glucose, the half-life of the mRNA decreases from about 2 h to 45 min (61). The main effect of glucose, however, appears to be on ADR1 mRNA translation, as discussed in the section on levels of control (above).
It had also been reported that glucose could act at a posttranslational level. Since Adr1 is a substrate for cAMP-dependent protein kinases in vitro and since increased kinase activity in vivo inhibits ADH2 expression, it was suggested that an increase in the cAMP level during growth on glucose would cause the phosphorylation of Adr1, possibly at Ser-230 (54). The phosphorylated Adr1 would still bind to the ADH2 promoter but would not interact with the transcription machinery (337). The existence of ADR1c mutations, which cause enhanced ADH2 transcription under repressed conditions and which interfere with the phosphorylation of Ser-230 in vitro, appeared to support the idea that Adr1 would be regulated by phosphorylation. However, it was concluded later that the ADR1c mutations may be acting by a different mechanism, either blocking the binding of a putative repressor to Adr1 or altering the conformation of Adr1 in such a way that it retains activity in the presence of glucose (75). It should also be noted that phosphorylation of Adr1 in vivo occurs at multiple sites and that the pattern of phosphorylation is similar in glucose- or ethanol-grown cells (354).
The existence of an Adr1 homolog in Hansenula polymorpha was suggested by the fact that the promoter of the MOX gene (encoding methanol oxidase) from H. polymorpha is able to bind Adr1 (268). Moreover, the expression of MOX-lacZ in S. cerevisiae is dependent on the endogenous Adr1 and repressed by glucose, as occurs with the ADH2 from S. cerevisiae.
Other activators.
There are a number of proteins related to catabolite repression which have features of a transcriptional activator but for which no targets have been identified. The characteristics of Sip3 and Sip4 which have been isolated by their capacity to interact with the protein kinase Snf1 in the two-hybrid assay (186, 187) are described in the section on the Snf1 complex (below).
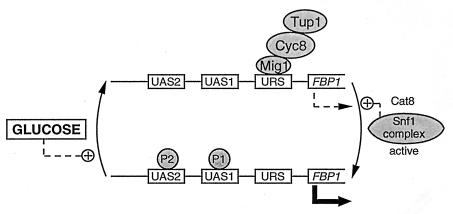
Cat8 is a protein required for the derepression of the gluconeogenic enzymes FbPase, phosphoenolpyruvate carboxykinase, and isocitrate lyase but dispensable for the derepression of invertase or maltase (141). Cat8 is a C6 zinc cluster protein, which is strongly repressed by glucose. Repression still occurs in a hxk2 mutant (278) but is reduced to twofold in the absence of the regulatory protein Mig1 (276, 278). There is a binding site for the Hap2/3/4/5 complex in the promoter of CAT8, and derepression is only partial in a hap2 mutant (278). Although derepression of Cat8, as well as that of the gluconeogenic enzymes, requires the Cat5 protein (276), Cat5 does not seem to be itself an activator of transcription. Cat5 may have only an indirect effect on catabolite repression, its role being to participate in ubiquinone biosynthesis (211).
The Cat8 protein undergoes phosphorylation under derepressing conditions yielding different modified forms (279). Two of these forms are found only in SNF1 strains, but another one can be formed in the absence of Snf1. While Snf1 is required for the activation of transcription depending on Cat8 (278), it is not yet known to what extent the Snf1-independent modification is required for Cat8 to be operative. Glucose triggers the dephosphorylation of Cat8, a process which does not appear to require protein phosphatase 1, since it occurs in a glc7 mutant (279).
Repressors
The Mig1 complex.
The MIG1 gene, an important element in glucose repression, was identified in a search for genes which would turn off the GAL1 promoter of S. cerevisiae (243). Mutations called cat4 or ssn1, which turned out to be allelic to MIG1, were also isolated as extragenic suppressors of snf1 and snf4 mutations (311, 356). Mig1 is a C2H2 zinc finger protein that is able to bind to the promoters of a variety of genes repressed by glucose. Binding requires a GC box with the consensus sequence (G/C)(C/T)GGGG, but it also requires an AT-rich region 5′ to the GC box (200). It has been suggested that finger 1 from Mig1 recognizes a G(G/A)G triplet and finger 2 recognizes a (G/C)(C/T)G triplet. The specific residues involved in the contacts would be an arginine at position 21, a histidine at position 18, and an arginine at position 15 for the first finger and an arginine, a glutamic acid, and an arginine at the same positions for the second finger. The AT-rich region would be required to stabilize the interaction, since it would allow bending of the DNA and facilitate further protein-DNA contacts (200).
A LexA-Mig1 fusion protein is able to repress a reporter gene with several Lex operators in a yeast growing on high glucose (352). The repression is much reduced at low glucose concentrations and disappears when galactose is the carbon source (344). A reporter gene containing a Mig1 binding site is also repressed in the presence of glucose; in a snf1 background, repression is maintained even when glucose is removed (365).
Deletion mapping of the MIG1 gene has been performed to delineate possible effector domains implicated in the repression by Mig1 and to delineate regulatory domains which would block the repression in the absence of glucose (258). It was found that the C-terminal 24 amino acids of Mig1 are sufficient for repression, when fused to the DNA binding domain. For repression to be relieved as the concentration of glucose in the medium decreases, two internal elements of the Mig1 protein are also required. One of these elements includes two RXXS motifs, potential substrates for some protein kinases. Another important feature of Mig1 is a basic domain 3′ to the zinc fingers, which could be involved in targeting Mig1 to the nucleus (258).
Although the operation of Mig1 appears to be controlled by the protein kinase Snf1 (258, 344, 365), as yet there is no evidence that Mig1 is actually phosphorylated by Snf1. In the Mig1 sequence, two sites which correspond to the consensus sequence for phosphorylation by Snf1 (defined in reference 68) can be identified, as well as a third site at which Snf1 would be less efficient (Fig. 1). By using a Mig1-viral protein 16 fusion protein that may function as an activator of transcription controlled by Snf1, it has been found that replacing the serines at positions 278, 310, and 311 by alanines affected the phosphorylation state of the fusion protein and strongly decreased the negative effect of Snf1 on its activity (258a). On the other hand, a Mig1 protein with both serines at position 311 and 381, which could be phosphorylated by Snf1, replaced by alanine is still subject to regulation by glucose in the same way as the wild-type protein (365).
FIG. 1.
Sequences of the Mig1 proteins from different yeasts which may be substrates of the protein kinase Snf1; the serine which may be phosphorylated is underlined. Amino acids which were found to be important in a study with artificial peptides as substrates for Snf1 (68) are shown in boldface type; amino acids in parentheses are suboptimal. Equivalent regions in the different proteins are aligned.
Genes similar to MIG1 have been cloned in the yeasts K. lactis (42), K. marxianus (43), S. pombe (336a), and C. albicans (393) and in the ascomycetes fungus Aspergillus nidulans (87). In addition, two further proteins with zinc fingers similar to those of Mig1 have been identified in S. cerevisiae (202). One of them, Mig2, binds to the promoter of SUC2 and contributes to its repression by glucose but has little affinity for the GAL1 promoter. The second protein, Yer028, has not been found to play a role in glucose repression. A comparison between the sequences of the homologous proteins from S. cerevisiae does not reveal extensive common motifs outside the zinc finger region (202). However, while potential phosphorylation sites for Snf1 are not found in these proteins, in both Mig2 and Yer028, as in Mig1, there is a stretch of basic amino acids 3′ to the zinc fingers and a 7-amino-acid sequence [LPP(I/V)(R/K)(S/N)(L/I)] in the C-terminal effector domain, which appears to be conserved. The different functional domains identified in S. cerevisiae have clear counterparts in K. lactis (42) and K. marxianus (43), and even sites which are potential substrates for Snf1 are conserved (Fig. 1). In fact, the genes from K. lactis or K. marxianus are able to complement an S. cerevisiae mig1 mutant. Although Mig1 from C. albicans is also functional in S. cerevisiae, preliminary results show that its sequence does not retain homology to that of S. cerevisiae Mig1 outside the very strongly conserved zinc fingers region and an adjacent 3′ domain with five positively charged residues (393). The equivalent protein from A. nidulans, CREA, plays a double role in the control of the expression of the ethanol regulon genes, since it acts on the promoters of both the structural genes and on a regulatory gene alcR, encoding a transcriptional activator (218). It has been further shown that CREA acts by competing directly with the binding of the ALCR activator to the promoters. In S. cerevisiae, as discussed in a later section, Mig1 also interacts with the promoters of the GAL genes, encoding enzymes required for galactose metabolism, and with the promoter of the regulatory gene GAL4. However, there is no evidence for a direct competition for DNA binding between Mig1 and the transcriptional activator Gal4. On the other hand, there could be competition between Mig1 and Mal63 at the MAL62 promoter (370), and very recent results suggest competition for GC boxes between Mig1 and a still unidentified SUC2 activator (26a).
A number of mutants with mutations which act synergistically with mig1 to relieve glucose repression of SUC2 and to suppress the effect of a snf1 mutation on invertase expression (356) or on the growth on gluconeogenic carbon sources (5) have been isolated. These mutants were affected in components of a multiprotein complex associated with RNA polymerase II (5, 176, 324) and are discussed below.
There is strong evidence that Mig1 exerts its repressive effect by recruiting a complex which contains the proteins Tup1 and Cyc8 (Ssn6) (344, 351). Mutations in TUP1 and CYC8 affect catabolite repression but are also highly pleiotropic and have been repeatedly isolated by different groups and given different names (see references 124 and 375).
Tup1 contains in its carboxyl region seven copies of a β-transducin motif (171a, 374). Although these motifs are not absolutely required for repression or for interaction with Cyc8, they appear to play an important role in Tup1 function (351, 352). Deletion analysis experiments suggest that the N-terminal 72-residue stretch of Tup1 is necessary and sufficient for the binding of Tup1 to Cyc8. In addition, Tup1 contains a domain, responsible for repression, which includes at least two separate transcriptional repression regions. These regions have little sequence similarity but are characterized by being alanine rich and almost completely uncharged (351).
Cyc8 is a large protein with a very high proportion of glutamine residues (313, 345). It contains near its amino terminus 10 copies of a 34-amino-acid motif, the tetratricopeptide repeat TPR (319). A number of TPR units are required for Cyc8 function (314), and it was proposed that they mediate protein-protein interactions (144). Later, two-hybrid assays showed that the amino-terminal region of Cyc8, comprising the first three TPR motifs interacts, with Tup1 (352). The two-hybrid system also indicates interaction between Cyc8 and Mig1 (344), and according to functional tests, TPR motifs 8 to 10 from Cyc8 would be involved in this interaction (352).
Cyc8 and Tup1 are associated in a high-molecular-weight complex (375) composed of one Cyc8 and four Tup1 subunits (359). The Cyc8-Tup1 complex is able to repress different classes of yeast genes depending on the DNA-binding protein with which it associates (166). The mechanism by which this complex exerts transcriptional repression is not yet clear, but two possibilities, not mutually exclusive, have been considered. Cyc8-Tup1 could modify the chromatin structure and control nucleosome positioning, and/or it could interfere directly with components of the basal transcription machinery. Much information on the mode of action of Cyc8-Tup1 has been obtained in studies of the α2-Mcm1-Cyc8-Tup1 complex, which is involved in sexual differentiation. In cyc8 or tup1 mutants, there is a perturbation in the placement and stability of nucleosomes around the α2 binding site (62). There is also evidence for an organized chromatin structure at the promoter of a gene such as SUC2 under repressed conditions (145, 217), and in cyc8 mutants an open chromatin structure is observed, even in the presence of glucose. Further support for the role of Tup1 on chromatin structure comes from the observation that Tup1 binds to histones H3 and H4 (90). This interaction is weakened by amino-terminal mutations in H3 and H4, which also cause a derepression of genes regulated by the Cyc8-Tup1 complex (90). Regarding possible interactions with the RNA polymerase II complex, recent experiments suggest that such interactions take place for the α2-Mcm1-Cyc8-Tup1 complex (282).
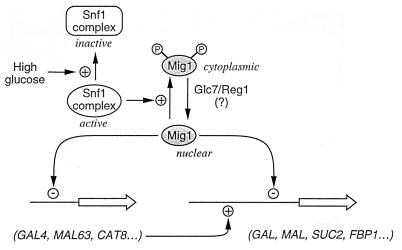
An important point to consider is how glucose regulates repression by the Mig1-Cyc8-Tup1 complex. An excess of Mig1 interferes with derepression of SUC2 and of the GAL genes and inhibits growth on maltose (243), and overproduction of Cyc8 or Tup1 reduces the transcription of SUC2 even in the absence of glucose (313, 374). Nevertheless, neither the amount of Mig1 (243) nor the amounts of Cyc8 and Tup1 (314, 375) are regulated by glucose. The facts that Mig1 is phosphorylated to different extents in repressed and derepressed cells and that relief of Mig1 repression requires the protein kinase Snf1 suggested a role for phosphorylation of Mig1 in the control of repression (344). This is strongly supported by recent observations which show that Mig1 is localized in the nucleus in repressed cells and that a few minutes after glucose removal from the medium, Mig1 is both phosphorylated and translocated to the cytosol (78). A generalized scheme for the role of Mig1 in catabolite repression is shown in Fig. 2. Although both Cyc8 and Tup1 have been reported to exist in a phosphorylated form (281, 314), there is no evidence that changes in their degree of phosphorylation have a regulatory significance.
FIG. 2.
Schematic view of the mode of action of Mig1 and its regulation. In the presence of glucose, Mig1 is found in the nucleus, where it represses the transcription of genes encoding activators such as GAL4 and MAL63 and of genes whose products are implicated in the metabolism of alternative carbon sources. Glucose removal causes both phosphorylation of Mig1, depending on the Snf1 complex, and its translocation to the cytoplasm. For details, see the text.
Other repressors.
There is not much evidence about proteins different from the Mig1 family that are able to bind to the promoters of genes subject to catabolite repression and to inhibit the transcription of these genes. The SKO1 gene, which was isolated by the same procedure as MIG1, encodes a protein that is able to bind to the promoter of SUC2, but its role in mediating repression by glucose is doubtful (242). Although transcription of SUC2 is increased up to twofold in a sko1 strain, the effect of the SKO1 disruption is more marked during growth on raffinose than in the presence of glucose. Moreover, the lack of Sko1 has no effect on the expression of GAL genes or on growth on different carbon sources (242).
Since the binding site for Sko1 (Acr1) has the characteristics of a cAMP responsive element (CRE) (242, 363), repression by Sko1 could be due to competition with a potential CRE binding activator, for which some evidence exists (252, 363). In this regard, it should be noted that it has not been yet established whether CRE motifs in S. cerevisiae are really responsive to cAMP.
A number of genes have been identified which could formally encode repressors, since mutations in them increase expression of repressible genes in the presence of glucose. However, in most cases, the encoded proteins such as Hxk2, Reg1, or Glc7 have been found not to act directly on the corresponding promoters. In other cases, the genes, such as GAL82 or SRG1 (102, 223), have not yet been cloned, and their mode of action remains obscure. Mutations in different genes (URR1, URR3, and URR4) have been isolated as relieving glucose repression conferred to a heterologous promoter by two different upstream repressor sequences (URSs) from the GAL1 promoter (111, 113). Although it has been suggested that the URR genes encode negative regulators which may be controlled by the Snf1 protein kinase, the genes have not been characterized further.
Intermediary Elements
The Snf1 complex.
The SNF1 gene (also called CAT1 or CCR1) is absolutely required for the derepression of genes repressed by glucose (40, 58, 399). SNF1 encodes a Ser/Thr protein kinase (44), the first member of a growing family of protein kinases to be identified (9). In particular, it has a mammalian homolog, which is the catalytic α subunit of the AMP-activated protein kinase (AMPK) (36, 234, 380).
In yeast cells, the Snf1 protein is found associated with other proteins: Snf4, Sip1, Sip2, and Gal 83 (45, 46, 385, 386). While snf4 mutants are unable to derepress the genes controlled by catabolite repression (98, 244), sip1, sip2, and gal83 mutants and even the sip1Δ sip2Δ gal83Δ triple mutant have no defect in the expression of a GAL gene (101) or the SUC2 gene (386). It has recently been reported that the sequence of the β subunit of the mammalian AMPK has 35% identity to the yeast Sip2 protein and that the sequence of the γ subunit is also 35% identical to that of Snf4 (126, 379). This identity is not restricted to parts of the protein but is found all along the sequence. Taking into account the homology of function and sequence of the yeast proteins Sip1, Sip2, and Gal83 (101, 102) and the data on interactions between Snf1, Snf4, and these proteins (159a), it appears possible that Snf1 participates, together with Snf4, in a family of complexes containing either Sip1, Sip2, or Gal83. These complexes may be, at least to some extent, functionally equivalent, but the observation that a sip1Δ sip2Δ gal83Δ mutant is able to maintain normal regulation of a set of glucose-repressed genes would suggest the existence of still unidentified additional regulatory proteins. It would be interesting to examine whether other genes, such as those encoding gluconeogenic enzymes, are also unaffected by the triple mutation.
In the mammalian AMPK, the β subunit interacts with both the α and γ subunits in vitro but the α and γ subunits are unable to interact under the same conditions (379). Snf1 and Snf4 do not interact in two-hybrid experiments in glucose-grown yeast (159), while Sip1 and Sip2 interact with Snf1 (386). Moreover, Sip1 and Sip2 coimmunoprecipitate with Snf1, even in the absence of Snf4 (386). However, when the glucose concentration in the medium is low, there is a direct interaction between Snf1 and Snf4 in vivo (159). Although there are no published data on interactions between the Sip proteins and Snf4, it has been suggested that the Sip1, Sip2, or Gal83 proteins would act as a bridge between Snf1 and Snf4, bringing them together in a complex (159).
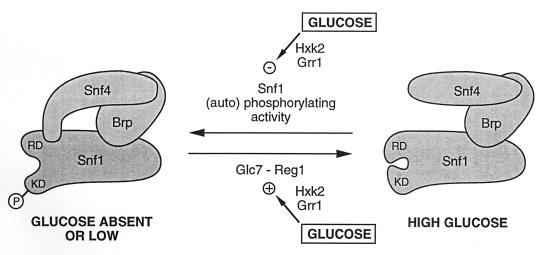
Genetic and biochemical evidence allows the construction of the model shown in Fig. 3 for the regulation of the interactions within the S. cerevisiae Snf1 complex. The protein kinase Snf1 has two domains: an amino-terminal catalytic domain (KD) and a carboxy-terminal regulatory domain (RD). Although the deletion of the regulatory domain bypasses the requirement for Snf4, it does not abolish the repression by glucose (45), and therefore Snf4 is not the (only) target of the glucose signal. At high glucose concentrations, the Snf1 regulatory domain binds to the catalytic domain and inhibits the kinase activity; at low glucose concentrations, Snf4 interacts with the regulatory domain, counteracting its inhibitory effect (159). The bridging protein (Brp), which maintains Snf1 and Snf4 together, could play a role in the equilibrium between the active and inactive forms of the Snf1 complex. Two functionally similar dominant mutations GAL82-1 and GAL83-2000, which partially relieve glucose repression of the GAL genes, have been isolated (220). The mutant protein Gal83-2000 could have an altered conformation, allowing at least a partial dissociation of KD and RD in the presence of glucose and therefore some degree of phosphorylation of at least a subset of Snf1 substrates. Regarding Gal82-1, no model can be proposed, because the corresponding gene has not been yet cloned.
FIG. 3.
Model for the regulation of the Snf1 complex by glucose. The bridging protein (Brp) between Snf1 and Snf4 can be Gal83, Sip1, Sip2, or some other, as yet unidentified, protein. Glucose affects the interaction between the catalytic domain (KD) and the regulatory domain (RD) of Snf1, presumably by inhibiting the (auto)phosphorylation of Snf1 and/or activating its dephosphorylation. Glucose may act at the level of the corresponding kinase and phosphatase but may also alter the conformation of Snf1 or even Brp, making Snf1 a worse or better substrate for the corresponding enzyme. Hxk2 and Grr1 are required for transmitting the glucose signal. Redrawn from reference 159.
How does the presence of glucose affect Snf1 activity? Earlier measurements of the protein kinase activity of Snf1 did not detect differences between extracts of cells grown on different media, and it was suggested that Snf1 would not itself be sensitive to glucose but would be required to phosphorylate a specific substrate(s), which would be the real target(s) of glucose (106). More recently, however, it has been reported that Snf1 activity increases very strongly in extracts from derepressed cells (377). The dramatic activation of Snf1 which occurs upon derepression appears to be due to phosphorylation, because it disappears when the yeast extracts are treated with a protein phosphatase. It should be noted that the difference in protein kinase activity between extracts from repressed and derepressed cells is observed only if the cells are harvested by rapid membrane filtration and frozen in liquid nitrogen. If the cells are harvested by centrifugation (the usual method), the peptide kinase activity is as high under repressing conditions as under derepressing conditions (377). This very rapid activation of Snf1 upon glucose removal would explain the earlier observations of apparently constitutive Snf1 activity. The activation of Snf1 in the absence of glucose, which depends on phosphorylation, parallels the activation of the mammalian AMPK by phosphorylation triggered by cellular stress (140).
In the case of the mammalian enzyme, the AMPK kinase has been partially purified and characterized, and it has been established that it phosphorylates a threonine residue from AMPK at position 172 (139). AMP affects the phosphorylation reaction at different levels: it activates the AMPK kinase, it makes AMPK a better substrate for the kinase, and it inhibits the dephosphorylation of the phosphorylated AMPK (71). Since ATP antagonizes these effects, the signal for phosphorylation appears to be a high AMP/ATP ratio.
For Snf1, the putative Snf1 kinase has not been identified, but a Snf1-reactivating factor has been reported which reverses the effect of in vitro dephosphorylation and is likely to be an upstream protein kinase (377). The fact that no mutants affecting the activity of this postulated protein kinase have been isolated could be explained if there are redundant protein kinases or if a defect in the kinase results in a loss of viability. Alternatively, the Snf1-reactivating factor could act by potentiating the capacity for autophosphorylation of Snf1 (45). Although Snf1, in contrast to AMPK, is not allosterically activated by AMP, it has been proposed that adenine nucleotides could also control Snf1 activity in vivo (377). This suggestion is based on the fact that under the metabolic conditions tested there was a correlation between the AMP/ATP (or ADP/ATP) ratio and the activation state of Snf1. However, there are data showing that the concentrations of adenine nucleotides do not differ markedly in yeasts growing on glucose and on a gluconeogenic carbon source (6), although Snf1 is absolutely required in the latter case and is therefore likely to be active under these conditions. The nature of the signal triggering the cascade responsible for Snf1 activation therefore remains obscure. What is clearly established is that threonine 210 from Snf1, equivalent to threonine 172 from AMPK, is absolutely required for Snf1 function in vivo. If this threonine is mutated to alanine, or even to an acidic residue such as aspartic acid, there is a complete loss of function (106). A provisional model for the mode of action of glucose (Fig. 3) would be as follows. In the absence of glucose, Snf1 is phosphorylated, by autophosphorylation or otherwise, and its catalytic and regulatory domains are dissociated. If glucose becomes available, a signal requiring Grr1/Cat80 and Hxk2 is generated (see the section on elements involved in glucose signaling, below), which facilitates the dephosphorylation of Snf1 by a protein phosphatase which could be the Glc7 complex (see the section on the Glc7 complex, below). This dephosphorylation weakens the interaction between Snf4 and the regulatory domain of Snf1, with this last domain thus remaining free to block the catalytic domain.
While snf1 mutants do not grow on gluconeogenic carbon sources, the requirement for Snf1 can be bypassed, at least partially, by a decrease in the activity of the cAMP-dependent protein kinases (340). On the other hand, derepression of alcohol dehydrogenase II, invertase, and FbPase is still totally dependent on Snf1 in a bcy1tpkw mutant, where cAMP-dependent protein kinase is attenuated and is independent of the cAMP level in the cell (74, 157, 194). It can therefore be concluded that although Snf1 may, in some cases, counteract the effects of an activated cAMP-dependent protein kinase, it is also able to operate through a cAMP-independent pathway.
The search for proteins that are able to interact in vivo with the protein kinase Snf1 has allowed the identification of additional proteins which may participate in the transcriptional regulation mediated by Snf1. One of them is Sip3, which interacts with Snf1 only transiently, since the two proteins do not coimmunoprecipitate (186). Sip3 contains a leucine zipper motif and can activate transcription when fused to a protein that is able to interact with a promoter region. This activation, however, is independent of Snf1. Although a SIP3 deletion did not affect yeast growth on different media and overexpression of SIP3 in a snf4 background caused only a slight increase in SUC2 expression, it has been suggested that Sip3 could be functionally related to Snf1 (186).
Another protein that interacts with Snf1 in the two-hybrid system is Sip4. It has the characteristics of a C6 zinc cluster protein, and a LexA-Sip4 fusion protein is able to activate transcription (187). This activation is dependent on Snf1 and is lowered about sevenfold in the presence of glucose. Glucose also represses the expression of a fusion gene, SIP4-lacZ, about 10-fold. Sip4 shows a weak interaction with Gal83, and this interaction, as well as that of Sip4 with Snf1, is independent of the presence of glucose. Nevertheless, phosphorylation of the fusion protein HA-Sip4 in vivo takes place only when there is no glucose in the medium. The fact that this phosphorylation is dependent on Snf1 would suggest that Sip4 is the long-sought substrate of Snf1. However, a sip4Δ mutant has no special phenotype, even when carrying a sip1Δ, sip2Δ, or sip3Δ mutation. This could indicate that some functional homolog of Sip4 is operative in S. cerevisiae, although no gene homologous to SIP4 was detected by Southern analysis under low stringency. The role of Sip4 remains obscure, since no promoter binding Sip4 has been yet identified and overexpression of SIP4 is not able to bypass the requirement for Snf4 to derepress invertase (187).
Very recently, a further protein, Sip5, has been identified (199). It interacts with both Snf1 and Snf4 and coimmunoprecipitates with them. It can also interact in the two-hybrid system with Reg1, a regulatory subunit of the protein phosphatase 1 complex (see the section on the Glc7 complex, below).
In K. lactis, proteins homologous to Snf1 and to the Gal83, Sip1, Sip2 group have been identified (130). The KlFOG2 gene is not only homologous to SNF1 but is also able to complement a snf1 S. cerevisiae mutant. The KlFOG1 gene encodes a protein with a sequence similar to that of Gal83, and no genes homologous to FOG1 have been detected by Southern blotting. It is likely, therefore, that in K. lactis there is a single complex equivalent to the family of Snf1 complexes in S. cerevisiae. Besides, both fog1 and fog2 mutants are unable to grow on galactose, melibiose, maltose, raffinose, glycerol, ethanol, or lactate. This can be correlated with the fact that the mutants are unable to derepress maltase, or l- and d-lactate ferricytochrome c oxidoreductase. On the other hand, the fog1 and fog2 mutants are able to partially derepress invertase and β-galactosidase and can grow on sucrose or lactose (130). It is not known whether K. lactis also contains a protein homologous to Snf4 from S. cerevisiae.
The Glc7 complex.
GLC7 (DIS2S1) is an essential gene, encoding a protein phosphatase type 1, which controls a variety of processes including glycogen accumulation (108, 254). A mutation called cid1, which partially relieved the repression of invertase by glucose (245), turned out to be a glc7-T152K mutation (348). This mutation did not impair glycogen accumulation (348), while a different mutation, glc7-1, which interfered with glycogen synthesis (34, 267), did not affect the repression of invertase by glucose (348). Since the mutant protein Glc7-1 was shown to be defective in its interaction with Gac1, a protein that appears to be a glycogen-specific regulatory subunit of the protein phosphatase type 1 (330), it was suggested that Glc7-T152K could be impaired in its interaction with a different regulatory protein, which would direct the phosphatase to some substrate(s) specifically related to the control of catabolite repression (348).
A hex2 mutation relieving glucose repression of invertase (97) and a reg1 mutation conferring resistance to catabolite repression of galactokinase synthesis (223) were later found to be allelic (249). When the REG1 gene was cloned and sequenced (250), the sequence did not show significant features, and the role of the corresponding protein remained obscure. However, since the reg1 and glc7-T152K mutations showed similar phenotypes, both suppressed by a snf1 mutation, Reg1 appeared to be a promising candidate for the putative regulatory protein modulating Glc7 activity. A strong interaction between the Reg1 and Glc7 proteins was observed in the two-hybrid system; this interaction decreased markedly when the Glc7 fusion protein carried the T152K mutation (349). Moreover, the LexA-Reg1 and HA-Glc7 fusion proteins were shown to coimmunoprecipitate, and additional evidence that Glc7 and Reg1 work together in glucose repression was provided by the observation that overexpression of REG1 restores glucose repression of invertase in a glc7-T152K mutant (349). On the other hand, it is not yet clear whether glucose can regulate the activity of the Glc7-Reg1 complex. Expression of REG1 is not regulated by glucose (250), and the observation that interaction between Reg1 and Glc7, in the two-hybrid system, is higher in repressed than in derepressed cells could be due to a variety of causes (349). Recently, further glc7 mutants, specifically affected in catabolite repression, were obtained (4). The fact that the mutated residues in the protein phosphatase are clustered on one face of the protein suggests that this is the region where Reg1 binds.
Although genetic evidence shows that the Snf1 protein kinase and the Glc7 complex play antagonistic roles, the targets of the phosphatase activity have not yet been identified. Potential candidates would be substrates of Snf1, regulators of Snf1, and the Snf1 protein itself (349). Specifically, it has been proposed that Mig1 would be phosphorylated by Snf1 and dephosphorylated by Glc7 (78). A new protein, Sip5, which is able to interact with Snf1, Snf4 (even in the absence of Snf1), and Reg1 has been identified (199). It could provide a link between the Snf1 and the Glc7 complex. There is also preliminary evidence, using the two-hybrid system, that Reg1 interacts with the catalytic domain of Snf1 (38). It has been reported (189) that Reg1 interacts with the yeast regulatory protein Grr1 (see the next section), and it has been suggested that Grr1 may recognize PEST sequences, a potential signal for protein degradation, in Reg1. The physiological significance of this finding remains unclear: if Grr1 facilitated the degradation of Reg1, reg1 and grr1 mutants would be expected to have opposite phenotypes; instead, they have similar ones (97).
A protein of 38 kDa has been identified recently and called Reg2, since it showed a strong similarity to the central region of the 114-kDa protein Reg1 (118). Although this protein interacts with Glc7, it does not appear to play a role in the catabolite repression process.
Elements Involved in Glucose Signaling
When glucose is available to yeast, a series of reactions occur whose final result is a change in the amount and/or activity of proteins that bind to gene promoters and modulate their transcription rate. The first signal triggering these reactions may be a charged receptor in the membrane, or a change in the concentration of some intracellular metabolite, including glucose itself. There could be a single signal or several signals, each of which turns on different sets of reactions.
Glucose signaling has a variety of effects. It causes the repression and the inactivation of many enzymes but also the induction and the activation of other enzymes (122). It is possible that some signals cause different kinds of effects while others are more specific for particular systems. For genes encoding glycolytic enzymes and requiring glucose for full expression, induction by glucose depends on the accumulation of intermediary metabolites (16, 18, 238). For some genes, an increase in the level of hexose-6-phosphates is required, while for others, induction is triggered by glycolytic three-carbon metabolites.
There is strong evidence for specific glucose sensors in the yeast membrane. Among the large number of hexose transporter (HXT) genes identified in S. cerevisiae (17, 175), two genes, SNF3 and RGT2, are expressed at very low levels compared to most HXT genes and play a specific regulatory role (247, 259). While SNF3 is repressed by a high concentration of glucose, RGT2 is expressed constitutively. Since Snf3 is required for the induction by low levels of glucose of some hexose transporter genes such as HXT2, it is likely to function as a sensor for low glucose levels. Rgt2, which is required for the induction of HXT1 by high glucose levels, would be a sensor for high glucose levels (259). Since both Snf3 and Rgt2, unlike other glucose transporters, have a long cytoplasmic domain at the C terminus, it has been speculated that the binding of glucose to these proteins causes a conformational change affecting a putative C-terminal signaling domain (47, 259). Since the whole yeast genome has been now sequenced, it can be ascertained that no further protein with a structure similar to that of Snf3 and Rgt2 remains to be discovered (175).
A snf3Δ mutation does not relieve the repression of SUC2, GAL10, or ADH2 by high glucose levels (192, 247), but it abolishes the ability of raffinose to repress ADH2 (192) and the induction of SUC2 by low glucose levels (264). While an rgt2Δ mutation does not affect the repression of SUC2 or GAL1 by high glucose levels (259), an snf3 rgt2 double mutant grows very poorly in glucose, and in this mutant neither GAL1 nor SUC2 are repressed by 4% glucose (258b).
It is important to note that signaling by glucose does not have the same requirements for different systems. In a yeast strain with a mutant protein Rgt2-1, a gene such as HXT2 is induced even in the absence of glucose in the medium, suggesting that the signaling domain of the mutant protein is permanently activated. However, in an RGT2-1 strain, the SUC2 gene is still normally derepressed in a medium with low glucose (214).
Could other glucose transporters be involved in the signaling pathway leading to catabolite repression? To address this question, a yeast mutant strain, unable to transport glucose, has been constructed where all the genes from HXT1 to HXT7 are disrupted and a series of derived strains expressing one or several of the genes HXT1 to HXT7 have been studied with regard to their capacity to repress the MAL2, SUC2, or GAL1 genes (283). The results indicate that glucose repression does not depend on any specific transporter but that in the different strains, the extent of repression is correlated with the glucose uptake capacity, suggesting that the rate of glucose utilization determines the strength of the relevant glucose signal. Similarly, it has been reported that in a strain which retains only the HXT6/7 gene and has a low glucose transport capacity, SUC2 is no longer repressed by glucose (368). In contrast to these results, repression of SUC2 and ADH2 was observed in a yeast strain lacking the transport genes HXT1 through HXT4 and HXT6/7, after overnight growth in a medium containing glycerol, ethanol, and 5% glucose (192). It is possible, however, that under these particular conditions other transport genes are expressed at sufficient levels to allow glucose uptake and catabolite repression.
Other reports have shown a correlation between transport capacity and catabolite repression. A yeast strain with the dominant mutation DGT1-1 expresses glucose transporters at low levels, and in this strain glucose repression of a variety of enzymes, including the gluconeogenic ones, is totally or partially relieved while repression by galactose is not affected (120). Another dominant mutation, HTR1-23, has similar but less strong effects (260). HTR1 is allelic to MTH1 (312), and it has now been found that although HTR1-23 and DGT1-1 differ in some of their effects, they are alleles of the same gene (179). MTH1 appears to be involved in the expression of the HXT genes, and although its exact role has not been yet elucidated, another protein, Std1/Msn3, structurally and functionally related to Mth1, has been reported to interact directly with the TATA binding protein (341).
Another mutation, grr1/cat80, isolated as conferring resistance to catabolite repression (3, 97), has also been found to affect the regulation of glucose transporters. Induction of the HXT1 to HXT4 genes is defective in grr1 mutants (261). There is now evidence that Grr1, a large protein with multiple leucine-rich repeats and tightly associated with a particulate fraction (112), is a component of a ubiquitin-conjugating enzyme complex which would regulate, perhaps indirectly, Rgt1 (189). Rgt1 is a DNA-binding protein that is able to repress HXT genes in the absence of glucose and to activate some genes such as HXT1 at high concentrations of glucose (263). Grr1 would be required both at low glucose concentrations to inactivate the Rgt1 repressor function and at high glucose concentrations to turn Rgt1 into an activator. The glucose signal would be transmitted to Grr1 via Snf3 or Rgt2, depending on the concentration of external glucose. In the absence of Grr1, Rgt1 permanently acts as a repressor, the glucose transport capacity of the cell is low, and glucose repression of a number of genes is relieved. In fact, the lack of Rgt1 restores both glucose-induced expression of the HXT1 gene and glucose repression in a grr1 mutant (102, 261, 356a).
In K. lactis, sensitivity to glucose repression is correlated with the hexose transporter genes present in the yeast genome. Strains containing two transporter genes, KHT1 and KHT2, in tandem are very sensitive to glucose. Natural isolates in which most of KHT2 has been lost by a recombination event between KHT1 and KHT2 which generated the gene RAG1 are moderately repressed by glucose. Mutant rag1 strains, which do not synthesize the low-affinity glucose transporter, are nearly insensitive to glucose repression (372).
Among the earliest mutants isolated as defective in catabolite repression were mutants lacking hexokinase II (398); the mutation was first called hex1 (97) and was renamed hxk2 once the affected gene had been identified. Since there are three hexose kinases in S. cerevisiae, hexokinase I, hexokinase II, and glucokinase (195), and the hxk2 mutants can still phosphorylate glucose efficiently, it was first assumed that hexokinase II had a specific regulatory domain required for glucose repression (96). However, the study of a large number of strains with different mutations in the HXK2 gene showed a parallelism between glucose repression and the residual phosphorylating capacity of the mutated hexokinase (205). Later, strains containing either Hxk1 or Hxk2 or hybrid hexokinases between Hxk1 and Hxk2 were constructed, and a strong correlation was found between the capacity of these strains to phosphorylate glucose or fructose and the repression of maltase and invertase by glucose or fructose (290). None of these results supported the hypothesis of a separate regulatory domain in hexokinase II. Hxk2 is phosphorylated in vivo and can be autophosphorylated in vitro (143, 366). Although Hxk2 is more highly phosphorylated when the glucose concentration in the medium is low (366), this phosphorylation does not seem connected with catabolite repression: while the main residue phosphorylated in vivo is Ser-15 (174), a deletion of the N-terminal 15 amino acids of hexokinase II does not affect glucose repression (204).
Kinetic studies, monitoring SUC2 mRNA levels after the addition of glucose to derepressed yeast cells, have shown that glucose has short-term and long-term effects on SUC2 expression (80, 303). Although it is difficult to compare the results of the two groups, since there are wide differences in the experimental conditions, the sugar kinase requirements appear to be different for the two processes. When glucose is added to fully derepressed cells, the early repression response occurs when any of the hexose kinases is present whereas the late response requires Hxk2 (303). On the other hand, while SUC2 expression is not repressed in an hxk2 mutant grown on glucose, growth on fructose has still a strong repressing effect in such a mutant (80).
At this stage, we can conclude that for a sugar to exert catabolite repression, it should be phosphorylated. The extent to which the structure of the phosphorylating protein plays a role in the repression process is still uncertain. However, the recent observation that Hxk2 can be found in the yeast nucleus suggests that this protein plays a regulatory role distinct from its capacity to phosphorylate sugars (279a).
The situation in other yeasts is not uniform. While it has been reported that mutants of Schwanniomyces occidentalis or Pachysolen tannophilus lacking a hexokinase isoenzyme have a defect in catabolite repression (226, 371), in Candida utilis a strong decrease in hexokinase activity does not suppress glucose repression of α-glucosidase (103), and in Aspergillus nidulans, too, hexokinase is not involved in glucose repression (295).
The next question is how extensively glucose should be metabolized to be able to repress transcription. By using strains with a range of reduced amounts of phosphoglucose isomerase, it has been observed that even when the level of phosphoglucose isomerase is less than 2% of that found in a wild-type yeast, both glucose and fructose strongly repress maltase and invertase (290). It has then been concluded that for catabolite repression, glucose signaling does not require any metabolic step in the glycolytic pathway beyond phosphorylation.
Glucose is known to trigger an immediate, transient increase in the intracellular concentration of cAMP in derepressed cells of S. cerevisiae (225, 357, 373); it is less widely appreciated that glucose also has a long-term effect on cAMP levels. The fact that in different yeasts the intracellular levels of cAMP are higher in the presence of glucose and other sugars than under derepressed conditions (100) could suggest a role for cAMP in catabolite repression.
In S. pombe, where adenylate cyclase is dispensable for growth (207), the gluconeogenic enzyme FbPase is no longer repressed by glucose in strains lacking this enzyme or other elements of the glucose-induced adenylate cyclase activating pathway (28, 147). It is not yet established, however, how general this effect of cAMP is in S. pombe. A very recent report indicates that in a strain where the cyr1 gene has been deleted, invertase is still subject to glucose repression (336a).
In S. cerevisiae, the situation is more complex: strains lacking adenylate cyclase (Cyr1) are not viable unless they also lack the regulatory subunit (Bcy1) of the cAMP-dependent protein kinases. In a cry1 bcy1 double mutant, where protein kinase A is constitutively activated, galactokinase, α-glucosidase, and invertase can be derepressed normally (222). In addition, in this genetic background, galactokinase expression is still sensitive to glucose repression. On the other hand, unregulated cAMP-dependent protein kinase in a bcy1 mutant does not allow maximal expression of POT1/FOX3, encoding the peroxisomal thiolase gene (158).
The effect of cAMP has also been tested directly, using specific strains sensitive to cAMP in the medium. It has been found that cAMP does not inhibit the synthesis of galactokinase in a GAL81 constitutive mutant whereas glucose blocks this synthesis completely (221). In fact, in the absence of glucose, cAMP increased the rate of synthesis of galactokinase. In contrast, it has recently been reported that in a cdc25 pde2 mutant, in which the internal concentration of cAMP can be modulated by external cAMP, the nucleotide prevents the synthesis of proteins which are usually derepressed upon glucose exhaustion (20). Among those proteins, isocitrate lyase, hexokinase I, and alcohol dehydrogenase II have been identified. In the same strain, cAMP is also able to interfere with the derepression of FbPase, phosphoenolpyruvate carboxykinase, and invertase (194). It has also been reported that a rapid drop in the levels of FBP1 and PCK1 mRNAs, similar to that observed after the addition of glucose to a derepressed culture, can be triggered by exogenous cAMP (390).
All these results indicate that in S. cerevisiae cAMP can facilitate catabolite repression of certain enzymes by a mechanism that remains obscure. The existence of redundant repression mechanisms, however, is shown by the fact that in a tpk1w tpk2 tpk3 bcy1 strain, where the protein kinase A level is low and insensitive to cAMP, invertase activity is high under derepressing conditions and is as sensitive to glucose repression as in a wild-type strain (157). The same observation has been made in such a strain with FbPase (194). In addition, the decrease in the levels of FBP1 and PCK1 mRNAs triggered by glucose is still observed in different mutants affecting the Ras/cAMP pathway, such as bcy1 strains (both TPK and tpkw), and cdc25-5, RAS2Val19,Ala22, and lcr1 (lack of cAMP response, nonactivable allele of adenylate cyclase) mutants (390).
It has been suggested that a high AMP/ATP ratio may be a signal for activating the protein kinase Snf1 (see the section on the Snf1 complex, above) when the glucose concentration in the medium is low or when glucose is absent (377). The idea is attractive, because it would mean that a basic regulation signal of the protein kinase Snf1 has been conserved in its mammalian counterpart, the AMP-activated protein kinase. However, the intracellular concentrations of ATP and AMP are very similar in yeasts growing on glucose or on ethanol, with an AMP/ATP ratio around 0.07 in both cases (6). In contrast, it has been reported that the AMP/ATP ratio was around 0.05 in yeast cells suspended in 2% glucose but rose to about 10 when repressed yeast cells were resuspended in 0.05% glucose (377). A possible interpretation is that the high AMP/ATP ratio recorded when yeast cells are transferred from a high- to a low-glucose medium corresponds to a transient adaptation phase and that during growth, homeostatic mechanisms tend to maintain the ratio below 0.1. In this case, an increase in the AMP level could be an early signal for activating Snf1 upon glucose depletion, but different mechanisms would operate at later stages. This can be compared with the different requirements for Hxk2 for the early and late repression responses (303).
The protein Tps1, a subunit of the trehalose synthase complex which catalyzes the synthesis of trehalose-6-phosphate, has been suggested as an element in glucose signaling (338). To examine the effect of Tps1 in glucose repression, it is not possible to compare directly a wild-type yeast with a yeast lacking Tps1, since this last one cannot grow on glucose. Although lack of growth is suppressed by a further mutation in the HXK2 gene (13, 150), it is not suitable to use the double mutant tps1 hxk2, since Hxk2 itself is involved in catabolite repression of a number of enzymes. An alternative is to compare a set of diploid strains: a wild-type diploid, a strain with one copy of the HXK2 gene interrupted, and a third one lacking Tps1 but still able to grow on glucose due to the interruption in one of the copies of the hexokinase gene. Such experiments have been carried out, and they show that repression by glucose of different genes is maintained in the absence of Tps1 (288). It appears, therefore, that Tps1 is not a requirement for glucose signaling.
A new signaling pathway has been proposed, based on the observation that addition of glucose to derepressed S. cerevisiae triggers the cleavage of a glycosyl-phosphoinositol-anchored protein, the cAMP-binding ectoprotein Gce1. It has been suggested that this reaction could generate phosphoinositolglycan peptides, which may function as soluble messenger molecules mediating nutritional signaling during glucose repression (237). Although there is not yet any evidence for this idea, it will be interesting to follow this lead.
Very recently, it has been reported that mutations in the GSF1 and GSF2 genes relieve the repression of SUC2 and GAL10 (316). GSF1 and possibly GSF2 operate upstream of Snf1, but their mode of action is still unknown. GSF2 has been cloned and shown to encode a protein which does not present obvious functional motifs, except for a hydrophobic stretch that could serve as a transmembrane domain and a C-terminal dilysine motif that could allow retention in the endoplasmic reticulum (316). Gsf1 is likely to be involved in glucose signaling, since gsf1 mutations not only relieve catabolite repression but also decrease glucose induction of HXT1 transcription, a process independent of Snf1.
We can conclude that our knowledge of glucose signaling is still very deficient. While it is clear that the rate of phosphorylation of the sugar is important for the repression process, the rate itself cannot modify downstream proteins such as Snf1. It is likely that different rates of phosphorylation are correlated with different concentrations of intracellular metabolites and that the activation or inhibition of specific regulatory proteins is carried out by these metabolites. While glucose-6-phosphate fructose-6-phosphate, and cAMP could be some of the candidates as signaling metabolites and Snf1 is likely to be one of the final targets, key intermediary elements remain to be identified.
Elements Which Play an Indirect Role
In addition to the proteins which participate in the formation, transmission, and reception of the glucose signal, a large number of proteins have been identified which affect the expression of genes regulated by catabolite repression but do not seem involved in the regulation of the genes by the carbon source. Many of these proteins belong to large complexes with a general role in transcription, while others are more disparate and are discussed below in a miscellaneous class.
The Snf/Swi complex and the SPT proteins.
In a search for genes necessary for SUC2 transcription under derepressing conditions, the SNF (sucrose nonfermenting) genes SNF2, SNF5, and SNF6 were identified (244). These genes, as well as SWI1/ADR6 and SWI3, are required for the transcription of the genes ADH2 or GAL1, sensitive to catabolite repression, but also for the expression of unrelated genes such as HO, INO1, and ADH1 (270).
The corresponding Snf and Swi proteins were later found to be components of an 11-subunit complex which includes Swi1/Adr6, Snf2/Swi2, Swi3, Snf5, Snf6 (30, 64, 269) Snf11 (343), Swp73 (31), and Tfg3/TAF30 (29). This complex may in turn be an integral component of the RNA polymerase II holoenzyme (376).
Genetic studies (see reference 378 for a review) and analyses of chromatin structure at the SUC2 promoter in wild-type and snf mutant yeast strains (145, 217) suggest that the Snf/Swi complex acts by disrupting the nucleosome structure by using the energy of the DNA-stimulated ATPase Snf2 (271). The exact mechanism by which the Snf/Swi complex affects the chromatin structure remains to be worked out, although several models have been proposed (170, 329). In any case, there is no evidence that the complex responds to the presence of glucose, and therefore it plays only an indirect role in modulating the expression of genes subject to catabolite repression.
The cre2-1 mutation, which allowed the glucose-insensitive expression of ADH2, (73) and an ssn20 mutation, which suppressed the defect in SUC2 transcription caused by snf2, snf5, or snf6 mutations (246), were later found to be allelic with spt6 mutations (378). It is now established that Spt6 interacts with the Spt4 and Spt5 proteins and that spt4 and spt5 mutations partially suppress the defect in SUC2 expression in snf2 mutant strains (336). It has been proposed that the proteins Spt4, Spt5, and Spt6 interact with histones H2A and H2B to yield a tight chromatin structure which hinders transcription (280). In fact, mutations in the SPT11/HTA1 or SPT12/HTB1 genes, which encode histones H2A and H2B, suppressed mutations in SNF2, SNF5, or SNF6 (145), strongly suggesting that the Snf/Swi complex is required to counteract the effect of the Spt proteins. The complex has a high affinity for DNA; it recognizes cruciform and four-way junction synthetic DNA without requiring any consensus sequence. It is also able to introduce supercoils in relaxed plasmid DNA, a property likely to be important in the remodeling of chromatin structure (277).
The mediator.
Looking for genes involved in catabolite repression, a series of proteins which influence repression by glucose of the SUC2 gene (5, 39, 176, 299, 324) and are components of a large multiprotein complex have been detected. This complex is associated with the C-terminal domain of the largest subunit of RNA polymerase II, is required for transcriptional activation, and has been called the mediator (169, 171, 339). The mediator includes a number of proteins called Srb, for suppressors of RNA polymerase B (Srb2, Srb4, Srb5, Srb6, and Srb7), together with TFIIF and the proteins Gal11, Rgr1, and Sin4 which form a subcomplex (190). In addition, other components of the mediator would be Srb8, Srb9, Srb10, and Srb11 (142, 193), Sug1 (169, 335), Rox3 (135), and a series of proteins which have been given the names Med1 through Med8 (191, 238a).
The mode of action of these proteins is still obscure, but the following points have been established.
The deletion of SRB2, SRB10, or SRB11 weakens the expression of a gene under the control of the GAL10 promoter (193). The genes SRB8 to SRB11 are identical to SSN5/GIG1, SSN2, UME5/SSN3/GIG2, and SSN8/GIG3, respectively (5, 49, 176, 324). Mutations in these genes, which are not essential, cause similar phenotypes: decreased growth rate, most marked on galactose and gluconeogenic carbon sources (5), and decreased induction of the GAL1 promoter (39); a slight relief of the catabolite repression of invertase, which can even be absent in a different genetic background (5, 39, 324); a weak suppression of the inability of snf1 mutants to derepress invertase (39, 324); a strong synergy with mig1 to suppress a snf1 defect, with regard both to invertase activity (39, 176, 324) and to growth on gluconeogenic carbon sources (5). While Srb8 and Srb9 have no conspicuous features, Srb10 and Srb11 encode a protein kinase and a cyclin-like protein, respectively (176, 193). It has been suggested that the Srb10/11 kinase/cyclin pair is involved in the phosphorylation of the carboxy-terminal domain of RNA polymerase II and that this phosphorylation plays a role in transcriptional regulation (193).
Although Rgr1, Gal11, and Sin4 are elements of the same subcomplex within the mediator (190), the three proteins do not appear to play exactly equivalent roles. RGR1 is an essential gene, and rgr1 mutants derepress invertase in the presence of glucose (299, 300). Disruption of the GAL11 gene is not lethal; it causes weak fermentation of galactose but not of sucrose or maltose and slow growth on nonfermentable carbon sources (332). Lack of Gal11 also strongly decreases derepressed transcription of the GAL genes but not of MEL1 (332), while it causes a very slight increase in GAL1 expression during growth on glucose (52). Disruption of SIN4, on the other hand, does not interfere with maximal expression of GAL1 and allows significant although low expression of GAL1 during growth on glucose (52). In addition, a sin4 mutation partially suppresses the effect of mutations in SNF1, SWI1 or SNF2 on the expression of invertase (39, 160, 324).
In a rox3 mutant, respiration is partially released from glucose repression, but there is only a modest increase in the level of expression of CYB2, encoding cytochrome b2, or GAL1, encoding galactokinase, during growth on glucose (25). On the other hand, maximal expression of both CYB2 and GAL1 is decreased up to threefold in this rox3 mutant. In addition, a mutation in ROX3, isolated as ssn7 (39), acts synergistically with mig1 to release glucose repression of invertase and suppress snf1 (324).
A mutation in the essential gene MED6 decreases the growth rate of the yeast on carbon sources other than glucose and strongly inhibits the transcription of GAL1 and SUC2 under derepressing conditions (185). The med6 mutation, however, does not affect catabolite repression of GAL1 and SUC2.
Sug1 was supposed to participate in the control of transcription because mutations in SUG1 corrected the defect in transcription caused by a mutated Gal4 protein (219, 334). It has been found that Sug1 is a subunit of the proteasome (172, 294) and that the association of Sug1 with the mediator reported previously (335) could be fortuitous and due to overexpression of the protein (335a). The suppressive effect of the sug1 mutation could be explained if a defect in Sug1 decreased the turnover of the mutated Gal4 protein, but a more indirect effect is also possible. Recent work has shown that Sug1 is a DNA helicase, and it has been proposed that it may be involved in different complexes, affecting different processes including mRNA metabolism (117, 210).
It has been suggested that the mediator is not a single entity but that multiple RNA polymerase II-containing complexes are involved in the expression of different genes (317). One of these complexes includes Gal11, Cdc73 and Paf1, as well as the general initiation factors TFIIB and TFIIF, but lacks TBP, TFIIH, and the Srb proteins (317). Disruption of Paf1 does not affect the expression of GAL4 but reduces the level of GAL7 and GAL10 transcripts three- to fivefold under derepression conditions (318).
A provisional conclusion could be that a defective mediator complex does not allow maximal rates of transcription and does not respond as well to transcriptional regulators. As a consequence, a mutation in a component of the mediator could result in a decrease of glucose repression for certain genes and an incomplete derepression in the absence of glucose. Most probably, the elements of the mediator are not directly sensitive to the glucose signal.
The Ada/Gcn5 complex.
The proteins Ada2 and Ada3/Ngg1, first identified as required for full transcriptional activation by some acidic activators, such as Gcn4 or Gal4-VP16 (11, 273), have also been shown to be necessary for maximal glucose repression of a GAL fusion gene in a gal80 background (22, 23). Derepressed levels of Adh2 decreased 3- to 10-fold in ada2 and gcn5 mutants, while repressed levels were not affected (55). The effect of the different mutations on the regulation of other genes subject to catabolite repression has not been examined.
Ada2 and Ada3 associate to form multicomponent complexes (301) which may contain also Ada1, Gcn5/Ada4, and Ada5/Spt20 (32, 152, 153). One of these complexes, called SAGA, contains, in addition to Ada2, Ada3, Ada5, and Gcn5, the proteins Spt3, Spt7, and Spt8 (132, 287). Gcn5 is a histone acetylase, able to acetylate free histones but not nucleosomal histones in vitro (132, 177, 387). It is likely that other elements in the Ada complex play a role in the interaction with the nucleosomes and that the acetylation of the nucleosomes facilitates transcription (33). The complex may also bring specific activation domains close to the TATA binding protein and possibly to other basal transcription factors (7, 212). The ada1 and ada5 mutants display more severe phenotypic defects than do the other ada mutants; they show slowed growth and reduced transcription of different reporter genes. It has therefore been proposed that the Ada1 and Ada5 proteins would retain activity in the absence of Ada2, Ada3, or Gcn5 while in the absence of Ada1 or Ada5 there would be no operative complex (153). What is still unresolved is how the lack of Ada2 or Ada3 relieves glucose repression in some cases.
Other elements.
Starting with mutants with weak activity of the protein kinase Snf1, a screen was carried out to identify genes that, in multicopy, would allow some expression of the SUC2 gene. This search led to the isolation of the genes MSN1 (104), MSN2 and MSN4 (105), and MSN3 and MTH1 (156). The deletion of MSN1, MSN2, and MSN4 together or MSN3 and MTH1 together caused only a moderate decrease in invertase expression; most probably the different Msn proteins do not directly control SUC2 transcription.
It appears now that Msn1 may be involved in the response to different types of nutritional deprivation. It has been isolated under different names (Fup1, Phd2, or Mss10) and shown to enhance iron-limited growth (91) and to induce pseudohyphal differentiation in response to nitrogen starvation (128) or carbon limitation (181).
Msn2 and Msn4 are homologous and belong to the family of C2H2 zinc finger proteins (105); they bind to the stress response element STRE (215). It is likely that they act as activators of the transcription of genes involved in the response to different types of stress. It has been suggested that the overexpression of MSN2 and MSN4 allows the growth of an snf1 mutant on raffinose (105) because it increases the resistance of the mutant yeast to the stress of carbon starvation (215).
Msn3 (also called Std1) interacts with Snf1, both in vitro and in vivo (156), and also with the TATA binding protein (341). However, the mechanism which connects Msn3 with the control of SUC2 expression is not clear, and there is evidence that Msn3 affects the expression of genes unrelated to catabolite repression, such as CUP1 and ACT1. Regarding the homolog of Msn3, Mth1, there is little information except that it plays a role in the expression of the hexose transporter genes (17, 179, 312).
A further series of mutants, snf7 to snf10, has also been isolated in which invertase derepression is impaired, at least in part, at the transcriptional level (355). The genes SNF7 and SNF8 have been cloned (350, 389), and it was shown that the interruption of either gene, or both together, decreases the expression of invertase to 20 to 30% of the wild-type level. The mutants grew well on galactose, and only the interruption of SNF8 caused a slight growth defect on glycerol. The snf9 and snf10 mutants behave like the SNF7 and SNF8 disruptants, respectively.
Two other proteins required for the transcription of genes necessary for growth on nonfermentable carbon sources are Ccr4 (76) and Pop2 (298). Ccr4 does not bind to DNA but is able to activate transcription in a glucose-repressible manner when fused to a DNA binding protein (88); its activity is dependent on Pop2 (89). Ccr4 and Pop2 are components of a complex which is not part of the Snf/Swi protein complex or of the mediator (88, 89); although their role in the control of transcription is not yet understood, they are not specific for catabolite repression since they are also involved in processes unrelated to carbon metabolism.
Another protein, Imp2, is required for the utilization of maltose, galactose, or raffinose only when mitochondrial inhibitors are present (84). Imp2 is also necessary for the rapid derepression of maltase, invertase, or alcohol dehydrogenase II but not for that of the NAD-dependent glutamate dehydrogenase or the l-lactate:ferricytochrome c oxidoreductase (196). Again, Imp2 is not specific for genes regulated by catabolite repression, since it seems to be involved in the repair of oxidative DNA lesions (216). Imp2 (not to be confused with an inner membrane protease also called Imp2 [253]) has been proposed to be a transcriptional activator which would interact with a protein bound to the promoter element of target genes (216).
REGULATION OF SPECIFIC GENES
The GAL Genes
The GAL genes of S. cerevisiae, coding for the enzymes required for the catabolism of galactose, are subject to dual control of expression: they are induced by galactose and repressed by glucose (for extended reviews, see references 163, 197, and 228). To dissociate the phenomena of induction and repression, I consider first the situation in a gal80 strain, lacking the regulatory protein Gal80 (Fig. 4). In such a strain, the GAL genes are still repressible by glucose but do not require galactose for induction; they are expressed, for instance, during growth on glycerol.
FIG. 4.
Control of the GAL genes by glucose in a gal80 background. In the presence of glucose, the Mig1 complex is active and decreases the rate of synthesis of GAL4 mRNA. The resulting low level of Gal4, together with the repressing effect of the Mig1 complex, leads to a very low level of expression of the GAL genes. For the Mig1 complex to exert its repressing activity, the Glc7 complex and elements in the glucose signalling pathway, such as Grr1 and Hxk2, are required. In the absence of glucose, the Snf1 complex is activated (see Fig. 3) and is able to release repression by Mig1; this allows maximal expression of GAL4, and the elevated levels of Gal4, together with the lack of activity of Mig1, turn on completely the expression of the GAL genes.
In a gal80 strain and in the absence of glucose, GAL4 is transcribed at its maximal rate and there is a high level of the activating factor Gal4. Gal4 in turn binds to the promoters of the GAL genes, which are then fully expressed. When glucose becomes available, the Mig1 complex binds to the GAL4 promoter and represses the expression of the gene, causing a fourfold decrease in the concentration of Gal4 (133, 183, 241). This decrease is sufficient to cause a large decrease in the expression of the GAL genes, 30-fold in the case of GAL1 (164). The transcription rate of the GAL genes is further reduced (fivefold for GAL1) by the binding of the Mig1 complex to the corresponding promoters (164). For the repression by glucose to take place, a series of elements, such as Grr1, Hxk2, and Reg1, involved in glucose signaling are required (3, 133, 223, 232). Mutations in other genes, such as GAL82-1 or GAL83-2000, also decrease repression by glucose (220), and the proteins Sip1 and Sip2 may also be related to catabolite repression (101). Although the role of the proteins Gal82, Gal83, Sip1, and Sip2 is still unclear, they appear to interact with the protein kinase Snf1 and may modulate its activity (see the section on the Snf1 complex, above). Once glucose has been used up, Snf1 is activated and turns off the Mig1 complex, thus allowing expression of the GAL genes. In a snf1 strain, the Mig1 complex remains permanently in its repressing state, independent of the presence of glucose and of elements involved in glucose signaling. In a mig1 (or tup1 or cyc8) background, the GAL genes are no longer repressed by glucose and Snf1 is not needed for derepression. From this, it has been concluded that Mig1 is the key factor in the control of the GAL genes and that glucose repression mechanisms acting on the Gal4 protein itself, which have been proposed (239, 328), are not likely to play a significant role (164).
In a wild-type GAL80 yeast strain, the GAL genes require induction by galactose, and glucose operates through additional mechanisms. Glucose decreases the transcription of GAL2, which encodes a galactose permease (21, 347), inactivates the permease (180), and even competes for it with galactose (283), thereby decreasing the level of the inducer galactose within the cell. Glucose also represses GAL3, which encodes a regulatory protein that participates in the galactose signal transduction pathway relieving the inhibition of Gal4 by Gal80 (333). As a result, in the presence of glucose the intracellular levels of both Gal3 and galactose are low, Gal4 is blocked by Gal80, and there is a very strong repression of the GAL genes, up to 1,000-fold compared with the ca. 150-fold repression observed in a gal80 strain (164).
In the dairy yeast K. lactis, the Lac9 transcription factor (equivalent to Gal4) activates the transcription of two families of genes, the genes GAL and LAC, required for the utilization of galactose and lactose, respectively (381). The expression of these genes can be repressed by glucose, at least in some strains (24), and some of the underlying mechanisms are similar to those operating in S. cerevisiae. For instance, a deletion of the KlSNF1 (FOG2) gene (homologous to SNF1) decreases the expression of the genes of the lactose-galactose regulon complex and a deletion in KlMIG1 relieves glucose repression of these genes to a certain extent (83). However, there are marked differences between the two systems: the LAC9 gene has a Lac9 binding site in its promoter, and this site is essential for the induction of the gene (65); the deletion of KlSNF1 does not completely block the induction of the GAL and LAC genes, while the deletion of KlMIG1 causes only a partial relief of the glucose repression of most of these genes, although LAC9 itself becomes completely glucose insensitive (83). These observations would suggest that in K. lactis there is a Snf1-independent pathway controlling induction and a Mig1-independent mechanism for glucose repression. It is not easy to understand how a deletion in KlGAL80 is sufficient to relieve catabolite repression (394), since this result would imply that in K. lactis glucose represses the transcription of the GAL/LAC genes exclusively by interfering with galactose induction.
SUC2
Although the expression of the SUC2 gene, coding for invertase, is repressed by high levels of glucose (37, 182), SUC2 expression is about eightfold lower in a yeast growing on a nonrepressing carbon source such as glycerol than in a yeast exposed to low levels (0.1% or less) of glucose or fructose (264). This induction of SUC2 transcription by low glucose levels requires the glucose-sensing protein Snf3, but a significant induction occurs even in the absence of the Rgt1 repressor. This indicates that SUC2 induction does not occur by the same mechanism as the induction of the hexose transporter genes HXT2 and HXT4 (see the section on elements involved in glucose signaling, above). While glucose is no longer required for maximal induction in a cyc8 strain, a deletion of MIG1 does not increase expression in the absence of a low concentration of glucose (264). This could suggest either that Mig2 can fully substitute for Mig1 in this particular regulatory circuit or, more probably, that Cyc8 plays a role that is not dependent on the Mig1 complex.
Different regulatory regions have been identified in the promoter of the SUC2 gene. Two of them, situated around positions −500 to −485 and positions −448 to −433, are able to bind the repressor protein Mig1 (243); the first of these is also able to bind the related protein Mig2 (202). Another less well defined URS is found in the region from −403 to −223 (264, 304). A deletion of this region increases the maximal expression of SUC2 two- to eightfold depending on the yeast strain, but as yet there is no information on the protein binding to the DNA. The deletion also suppresses the requirement for glucose for maximal induction but does not relieve repression by high levels of glucose (264). Regarding regions which may act as upstream activating sequences (UASs), it has been found that tandem copies of the sequence from −437 to −406 are able to activate the transcription of a fusion gene but that this activation is not regulated by glucose (305). Although variants of the element AAGAAAT present in the sequence from −437 to −406 are found at several places along the promoter, the evidence for their functionality is only indirect: pseudorevertants from a partially deleted promoter had acquired sequences which corresponded to the consensus for the repeated element (305). Very recently, it was found that mutations in the GC box, around position −495, that is able to bind Mig1 decrease the expression of SUC2 two- to threefold under derepressed conditions and that the decrease is stronger if the GC boxes at both −495 and −435 are mutated (26a). This provides good evidence for transcriptional activator(s) binding to these regions, but again, nothing is known about the activator protein(s).
At present, we can view the situation as follows. One element responsible for the activation of transcription could depend on the signal given by low glucose levels through Snf3 and could counteract, at least partially, the negative effect of the region from −403 to −223. Other positive elements are still poorly characterized, and it is not even clear whether they are dependent on the presence of glucose. There is some evidence that SUC2 expression is basically controlled by negative elements, mainly those binding Mig1. The activity of the Mig1 complex is controlled by the presence or absence of glucose through the Snf1 and Glc7 complexes and the different elements involved in glucose signaling. The observation that glucose repression of SUC2 is only partially relieved in a mig1 mutant appears related to the fact that the Mig2 protein is partially redundant with Mig1; in a mig1 mig2 double mutant, on the other hand, the degree of catabolite repression is only twofold (202). A result that is difficult to explain is that in a snf1 mig1 double mutant, the expression of SUC2 is still subject to a 10-fold regulation by glucose (356). A possibility is that repression by Mig2 is relieved by a mechanism independent of Snf1, and to test this it would be interesting to examine what happens in an snf1 mig1 mig2 triple mutant. The residual glucose repression in a mig1 mig2 double mutant could also occur through a mechanism which does not involve Snf1. The fact that cyc8 or tup1 mutations completely relieve catabolite repression of SUC2 implies, however, that any parallel pathway for glucose repression is also controlled by the Cyc8-Tup1 complex (26a).
FBP1
The FBP1 gene, which encodes the gluconeogenic enzyme FbPase, is repressed by glucose and other sugars such as galactose (100, 120, 123).
In the promoter of FBP1, two UASs and a URS have been found (229, 231, 251, 364). While the URS element, situated at positions −200 to −184 in the promoter, binds the Mig1 protein (200), the proteins binding to UAS1 and UAS2 have not been identified. Band shift experiments have shown that these proteins are absent or are in a conformation unable to bind the UAS sites in nuclear extracts from glucose-grown cells (251, 308, 364). It has been found that both UAS1 and UAS2 confer glucose-repressed expression to a heterologous reporter gene and that activation of expression by either element requires a functional SNF1 gene (141, 364).
Snf1 is active in a wild-type yeast growing on galactose or in strains such as hxk2 or reg1 even during growth on glucose. Since FBP1 and the UAS1-CYC1-lacZ and UAS2-CYC1-lacZ genes are still repressed under such conditions (72, 97, 141), it is apparent that an active Snf1 is a necessary but not sufficient condition for derepression of these UASs. This may be related to the fact that expression of FBP1 and of the UAS-CYC1-lacZ fusion genes is also dependent on the CAT8 gene (141, 278) and that CAT8 is still repressible by glucose in a hxk2 mutant, where Snf1 is operative in the presence of glucose (278). It should be noted that this last observation appears to contradict the release of CAT8 from catabolite repression shown to occur in mig1 mutants (141, 278).
The facts that Cat8 is a DNA binding protein and that it can function as an Snf1-dependent transcriptional activator (278) suggested that Cat8 is one of the proteins binding to UAS1 or UAS2 in FBP1. However, band shift experiments performed with a tagged Cat8 and tag-specific antibodies (278) have shown that there is no direct binding of Cat8 to the carbon source-responsive element, CSRE, a sequence motif in UAS2 which is also present in other gluconeogenic genes (141, 308, 364). On the other hand, it is unlikely that Cat8 binds to the UAS1 site, since the DNA-protein complex formed by UAS1 has a high mobility (364) while Cat8 is a large protein of 160 kDa (141).
The present model for the control of FBP1 is still very incomplete; it would run as follows (Fig. 5). In the presence of glucose, Snf1 is inactive, Cat8 is expressed at a very low level, and the proteins binding UAS1 and UAS2 are probably not synthesized; in addition, the Mig1 complex acts as a repressor, and therefore there is no significant transcription of FBP1. When glucose is removed, Snf1 is activated, repression by Mig1 is released, and Cat8 is expressed. Cat8 is also phosphorylated by Snf1 itself or by a protein kinase activated by Snf1 and also by some other protein kinase (see the section on activators, above). The phosphorylated Cat8, in turn, is required, in a direct or indirect manner, for the operation of the activator elements P1 and P2, which bind to UAS1 and UAS2, respectively, and turn on FBP1 transcription. While it has been observed that in the absence of glucose, FbPase transcription is repressed by cAMP (194), the target for the protein kinase activated by the increase in cAMP is not known.
FIG. 5.
Control of the FBP1 gene by glucose. In the presence of glucose, the Mig1 complex is active and Cat8 and the as yet unidentified activatory elements P1 and P2 are repressed; there is no expression of FBP1. When glucose is removed, the Snf1 complex is activated, and this results in release of repression by the Mig1 complex and derepression of Cat8. The expression and activation of the regulatory elements P1 and P2 depends on both Cat8 and an active Snf1 complex. Once P1 and P2 have been activated, they stimulate the transcription of FBP1.
In a galactose medium, Snf1 would be active, but there is no information on the expression of Cat8 under these conditions. The data available are therefore insufficient for interpreting the low level of transcription of FBP1 and of the UAS-CYC1-lacZ fusion genes in galactose-grown cells.
In S. pombe, fbp1 is repressed by glucose (360) and two pathways regulating the expression of fbp1 have been identified. One of them involves the cAMP-dependent protein kinase, since it has been found that fbp1 is no longer repressed by glucose in mutants affected in the protein kinase, adenylate cyclase, or genes required for the activation of adenylate cyclase by glucose (147, 148). The other pathway includes the mitogen-activated protein kinase (MAPK) sty1/spc1, the wis1 MAPK kinase (MAPKK), and a protein tyrosine phosphatase (pyp1) which is able to dephosphorylate and inactivate the MAPK (69, 327). Both the wis1 MAPKK and the sty1 MAPK are required for fbp1 expression (327), and overexpression of pyp1 inhibits the transcription of fbp1 (69). The substrates for the cAMP-dependent protein kinase and the sty1 MAPK are unknown, and it has not been established if the two pathways work in parallel, one facilitating repression and the other facilitating activation of transcription, or if the cAMP-dependent protein kinase has a negative effect on the wis1 MAPKK. Nothing is known, either, about the proteins binding the fbp1 promoter.
ADH2
The ADH2 gene from S. cerevisiae encodes alcohol dehydrogenase II, an enzyme which is repressed several hundred-fold by glucose (203). The ADH2 promoter contains two elements, UAS1 and UAS2, which act synergistically to regulate the expression of the gene (391). UAS1 includes a 22-bp inverted repeat which is able to bind the regulatory protein Adr1 (94). UAS2 confers glucose-regulated, Adr1-independent expression to a reporter gene, but the identity of the proteins binding UAS2 is not known.
In contrast to the situation in most genes subject to catabolite repression, there is no evidence for a Mig1 binding site in the ADH2 promoter. This is consistent with the observation that glucose repression of ADH2 is not relieved in a cyc8 strain (74).
Derepression of ADH2 is completely dependent on Snf1 (59), and this dependence is still observed in the case of truncated promoters which are no longer regulated by Adr1 (74). This led to the suggestion that Snf1 does not act through Adr1 (74). Although this has not been tested directly with the ADH2 promoter, a region able to bind Adr1 has also been identified in the promoter of the CTA1 gene, encoding the peroxisomal catalase A (320), and it has been found that expression of a fusion gene driven by this Adr1 binding element depends on the protein kinase Snf1 (110).
Other protein kinases have also been reported to modulate the expression of ADH2. While an increase in cAMP-dependent protein kinase activity strongly decreases the level of alcohol dehydrogenase II, the protein kinase Sch9 is required for maximal derepression (74). The mode of action of Sch9 is unknown, but there is ample evidence that the target of the cAMP-dependent protein kinases is Adr1. Adr1 may be phosphorylated at Ser-230 position and hence made less active (54, 337), but other mechanisms independent of the phosphorylation at Ser-230 can operate (75), one of them leading to a decrease in the expression of ADR1 (82).
The Reg1 protein, an element of the Glc7 complex, is also involved in the regulation of ADH2. In reg1 mutants, expression of ADH2 and of reporter genes under the control of UAS1 or UAS2 becomes partially resistant to glucose repression (81). Surprisingly, the cid1-226 mutation, a mutation in the GLC7 gene which relieves catabolite repression of SUC2 (245, 348) did not relieve the repression of ADH2 (81).
There are conflicting reports about the effect on ADH2 expression of a disruption of CAT8, a gene required for the derepression of different gluconeogenic enzymes; in one case, no effect was observed (85), while in the other, some decrease in the maximal expression achieved was reported (278). It therefore appears that except for Snf1 and Reg1, most of the elements controlling the expression of other genes subject to catabolite repression are not operating on ADH2. Although mutations in a variety of genes such as CCR4, CRE1, SPT6, or SWI1 have an effect on ADH2 expression, the corresponding proteins are not likely to be involved directly in transmitting the glucose signal (see the section on elements which play an indirect role, above).
A still very incomplete model of glucose control of ADH2 would be as follows. When glucose is present in the medium, the level of Adr1 is low; the level varies depending on the yeast strain and is set by a combination of effects of glucose on transcription and translation (see the section on Adr1 [above] for details). Such a low level of Adr1 is, however, not the only cause of the lack of ADH2 expression under these conditions: a similar low level of Adr1 allows a significant expression of ADH2 under derepressing conditions (82). Other mechanisms should therefore contribute to the repression observed. One of them could be a decrease in Adr1 activity caused by its phosphorylation by cAMP-dependent protein kinases; in addition, the protein(s) binding UAS2 most probably requires an active Snf1 to be operative. In a reg1 mutant, these proteins would be active, even in the presence of glucose, and this would explain the partial release of the repression of ADH2. Upon glucose removal, Adr1 levels increase, and this transcription factor in its active form would stimulate transcription through UAS1; in addition, the Snf1 complex is activated and UAS2 becomes operative. Under these conditions, the synergistic effect of UAS1 and UAS2 allows full expression of ADH2.
CYC1
The transcription of the CYC1 gene, encoding iso-1-cytochrome c, depends on two tandem UASs which are differentially regulated. UAS1 binds the transcription factor Cyp1/Hap1, and the expression of a UAS1-lacZ fusion gene is activated by heme, while UAS2 binds the Hap2/3/4/5 complex, and expression of a UAS2-lacZ fusion gene is repressed by glucose (134).
Glucose represses the synthesis of Hap4 (114), but no direct test has been performed to investigate whether the decrease in Hap4 levels caused by glucose accounts for the repression of CYC1. On the other hand, although different mutations affect CYC1 expression, their effect on HAP4 expression has not been examined. It has only been reported that a mig1 mutation did not affect the regulation of HAP4, although there is a Mig1 binding site in the HAP4 promoter (200). For CYC1 itself, the results observed are variable: glucose repression of a CYC1-lacZ fusion gene was partially relieved in a mig1 mutant (311) but was not affected by a MIG1 deletion in a different yeast strain (365). This could be related to the fact that the regulation by carbon source of CYC1 and other genes encoding mitochondrial proteins shows large strain-dependent variations (26).
Derepression of CYC1 requires the SNF1 gene (382), while repression is partially relieved by mutations in GRR1, HXK2, or REG1 (3, 19, 99). No repression at all is observed in cyc8 or tup1 mutants (292, 382). This could suggest that regulation of HAP4 involves a mechanism similar to that operating on GAL4 but with another element, perhaps Mig2, taking the role of Mig1. Hap4 levels in turn would control the rate of transcription of CYC1.
Other Genes
I will not discuss in detail all the genes known to be subject to catabolite repression but will show that most of them can be distributed in different families and examine them in turn.
For the genes required for the metabolism of sugars such as galactose, maltose, or sucrose, a central element for glucose control is the Mig1 protein. The regulation of the GAL genes and of SUC2 is described above. For the MAL genes, glucose plays a double role, as it did for the GAL genes: it interferes with induction of the transcriptional activator Mal63 by maltose, but even in strains where Mal63 is constitutive, glucose represses the expression of the permease Mal61 and the maltase Mal62 (see the section on Mal63, above). Binding sites for Mig1 are found upstream of the MAL61, MAL62, and MAL63 genes (154), and deletion of Mig1 in a constitutive strain relieves repression partially or totally depending on the yeast strain (154, 370). A peculiarity of the MAL genes is the competition between Mig1 and Mal63 at the MAL62 promoter (370). Repression by glucose of the genes encoding the glucose transporters Hxt2 and Hxt4 is also mediated by the binding of Mig1 to their promoters (261, 262).
Many of the genes involved in respiratory metabolism are regulated by the Hap2/3/4/5 complex (79). Mutations in the genes HAP2 and HAP3 prevent the activation of a series of genes such as COX4, COX5a, CYT1, HEM1 (115), COX6 (342), KGD1 (285), or COR2 (86), but only for COX6 is there information on additional regulatory elements involved: the expression of COX6 requires Snf1, and repression by glucose is relieved in a cyc8 mutant (382). Although the gene COX5b, encoding a subunit of cytochrome c oxidase, is repressed by glucose and contains in its promoter a potential binding site for the Hap2/3/4/5 complex, this sequence is found outside UAS15b, which mediates regulation by glucose (146). It has been reported that some alleles of cat8 caused a defective respiration and lack of cytochrome c oxidase (276); however, it is not known if Cat8 is needed for the expression of the genes regulated by Hap2/3/4/5. Repression of the CYB2 gene, encoding cytochrome b2, is partially relieved by mutations in GRR1 or HXK2, but it has been found that a cyc8 mutation does not affect glucose repression and, moreover, that Cyc8 is required for full derepression of CYB2 (25). This could explain why cyc8 mutants grow poorly on nonfermentable carbon sources (313, 345). This requirement for Cyc8 cannot be only because the Cyc8-Tup1 complex has a positive effect on the activity of Cyp1(Hap1), a transcriptional activator of genes encoding components of the respiratory chain, since Cyp1 itself is not essential for growth on nonfermentable carbon sources (397).
In relation to the genes regulated by Hap2/3/4/5, it should be noted that although overexpression of MBR1 and MBR3 improves the growth of hap mutants on glycerol (66), these genes are not likely to be involved directly in the regulatory circuit. Mbr1 appears to have a more general effect in the protection of cells against stress, perhaps by playing a role in intracellular trafficking (284). Mbr3, in turn, is identical to Isf1 (2) and could be involved in the control of Nam7/Upf1, a protein affecting mRNA turnover.
Another family of genes, which encode enzymes expressed under gluconeogenic conditions, includes in its promoters one or two copies of the CSRE (carbon source responsive element) (308). There are two variants of this element, one derived from the sequence CGG ATG AAT GGA and the other with the consensus sequence CGG CCC AAT GGA. The first one is found once in the promoters of different genes: ICL1, encoding isocitrate lyase (307); ACS1, encoding acetyl coenzyme A synthetase (173); and FBP1, as discussed above. It is found twice in the promoters of PCK1, encoding phosphoenolpyruvate carboxykinase (229, 275), and ACR1 (unrelated to the SKO1/ACR1 gene), encoding a protein essential for acetyl coenzyme A synthetase activity (109), which has turned out to be a succinate-fumarate mitochondrial transporter (265). The second variant is found once in the promoter of MDH2, encoding cytoplasmic malate dehydrogenase (EMBL database), and twice in that of MLS1, encoding malate synthase (41). One of the elements in MLS1 competed with CSREICL1 in gel retardation experiments and directed transcription in a manner strongly regulated by glucose and dependent on Cat8 (41). This element is weaker than the UASICL1, but two copies show a strong synergism. While the potential UASs in ACS1 (173) and PCK1 (275) have also be shown to be functional, those in ACR1 and MDH2 have not been tested directly. In addition to the UAS containing the CSRE sequence, a constitutive UAS has been identified in the MLS1 promoter: it may bind Abf1 and allows a partial derepression of the gene in a cat8 background. The promoter of the ACS1 gene also has a complex structure; besides the CSRE, it includes binding sites for the transcriptional activators Adr1 and Abf1 and for the negative factor Ume6 (173a).
Another feature of some of the genes encoding gluconeogenic enzymes is the existence of a Mig1 binding site in their promoters. In MLS1 (41) and MDH2 (EMBL database), the sequences found correspond to the described consensus (200); in the other genes, the sequences are less highly conserved, and their functionality has been checked only for ICL1 (41).
The isocitrate lyase in C. tropicalis is also regulated by the carbon source, and when the corresponding gene, under its own promoter, is introduced into S. cerevisiae, it is strongly repressed by glucose. In the heterologous yeast, two regions of the CtICL1 promoter are able to direct regulated expression dependent on Snf1. There is no recognizable motif in the first one, but the second one contains a CSRE sequence. In the case of a fusion gene with the first regulatory region, a mig1 mutation partially relieves glucose repression and Cat8 is not required for maximal expression. The second region behaves as a CSRE from S. cerevisiae; it does not respond to the absence of Mig1 and requires Cat8 (353). These observations show that isocitrate lyase is not regulated exactly in the same way in the two yeast species but that some regulatory pathways have been well conserved.
In S. cerevisiae, peroxisomal proteins are repressed by glucose and induced by oleate (361). It has been shown that derepression of such proteins is dependent on Adr1 and on the Snf1 complex (320–322). Induction by oleate depends on a “peroxisome box” (110), also called the oleate response element (92), which is an imperfect palindrome including the consensus sequence CGGNNNTNA. There is evidence that the transcription factor binding this sequence is a heterodimer containing the proteins Oaf1 and Oaf2/Pip2, which are both C6 zinc cluster proteins (165, 201, 293). Expression mediated by the oleate response element is also repressed by glucose, and there is evidence that it is controlled by Snf1 (110). For the FOX3 promoter, glucose repression is not relieved by a mig1 mutation but is decreased in a ume6/car80 mutant and for fusion genes, where sites of the promoter, binding Abf1 and the heterooligomer BUF, have been mutated (93).
The POX1/FOX1 gene which encodes acyl coenzyme A oxidase, a peroxisomal enzyme, constitutes a particular case among the genes encoding peroxisomal proteins. Although it is subject to catabolite repression, its expression has been reported not to depend on the transcriptional factor Adr1 or on the protein kinase Snf1, which is essential for most glucose-regulated genes (326).
Adr1 is also required for growth on glycerol (10). Glycerol utilization depends on the genes GUT1 and GUT2, encoding glycerol kinase and sn-glycerol 3-phosphate dehydrogenase, respectively, both regulated by catabolite repression (325). Derepression of GUT1 is greatly decreased in adr1 mutants (266); although there is no information on the effect of an adr1 mutation on GUT2 expression, its promoter contains a sequence potentially able to bind Adr1 (53).
A gene presenting unique regulatory characteristics is the NAD-linked glutamate dehydrogenase from S. cerevisiae, encoded by the GDH2 gene and regulated by both the nitrogen and the carbon sources used by the yeast (see reference 208 for a review). The regulatory mechanisms are interdependent to some degree, since repression by glucose is observed in a medium where the nitrogen source is glutamine or ammonium but not where the nitrogen source is glutamate (63). Although the region of the GDH2 promoter from −350 to −245 directs glucose-regulated transcription, the corresponding sequence does not show recognizable motifs (63, 233). Two mutations, rgc1 and rgc2, that blocked derepression of GDH2 in response to the carbon source were reported (63), but they have not been characterized further. A cyc8 mutation allows a partial derepression of GDH2 in the presence of glucose, while a hxk2 mutation has no effect (63). This can be related to the fact that galactose represses GDH2 as strongly as glucose does (63, 100). Mutations in HAP2, HAP3, or HAP4 do not interfere with the derepression of GDH2 (63, 70).
CONCLUSIONS AND PERSPECTIVES
Many elements of the circuits underlying catabolite repression have been identified, and in some cases, the relationships between them have been established; however, basic questions remain to be answered. A number of them pertain to the protein kinase Snf1, a central element in the different regulatory circuits. In vitro results suggest that Snf1 activity decreases strongly when glucose is present in the medium and that this decrease is correlated with a dephosphorylation of Snf1 (377), but we do not know how this dephosphorylation is triggered. Changes in the concentration of some metabolites, as a result of the presence of glucose, could affect the phosphorylation rate or the dephosphorylation rate of Snf1 or both. While it has been established that no extensive glucose metabolism is required for repression (290) and therefore presumably for the change in metabolite levels, the nature of the regulatory metabolite(s) is unknown. A point which is still controversial is if the requirement of Hxk2 for the repression of certain genes is due only to its catalytic activity or if the protein itself plays a role in glucose signaling. The use of heterologous hexokinases could shed some light on this question.
To elucidate the mechanisms which control Snf1, a prerequisite will be to establish if there is a regulated phosphorylation of Snf1 due to autophosphorylation or if there is a specific protein kinase acting on Snf1. If such an Snf1 protein kinase exists, it is likely to be essential or to present isoenzymes; both alternatives would explain why a mutant lacking this activity has not appeared in the many screens carried out to identify mutants affected in derepression. A search for conditional mutants could be rewarding. With respect to the protein phosphatase which would dephosphorylate Snf-P, genetic data point to the Glc7 complex as a potential candidate. However, as discussed in the corresponding section, the behavior of the glc7 mutants could also be explained if the Glc7 complex acted on some effector or substrate of Snf1. To understand the role of Glc7, it would be useful to compare the phosphorylation status of Snf1 in a wild-type strain and in a glc7 mutant. If these in vivo tests gave evidence for Snf1 being a substrate for Glc7, the effect of the phosphatase on phosphorylated Snf1 could be tested directly in biochemical experiments designed to demonstrate a possible influence of intermediary metabolites on the dephosphorylation.
Another subject for further research is the identification of Snf1 substrates. In spite of the thorough search for them, carried out by the two-hybrid assay (186, 187, 199, 386), the results have been disappointing. A family of proteins called Sip (for Snf1-interacting protein) has been identified, but among them only Sip4 has been shown to be a real substrate of Snf1, and even in this case the role of Sip4 remains uncertain because its deletion has no noticeable phenotypic effects. It is possible that the interaction of Snf1 with its substrates is too transient to be detected by the two-hybrid assay. An alternative approach, now that the entire genome of S. cerevisiae is known, would be to search the coding sequences for diagnostic features of a Snf1 substrate. For instance, there are several motifs in the sequence of the repressor protein Mig1 which are putative substrates for Snf1; since Mig1 is phosphorylated under derepression conditions and since relief of repression by Mig1 requires Snf1, it has been suggested that Mig1 is a substrate for Snf1 (78). However, it remains to be shown in a conclusive manner whether the regulation of Mig1 by Snf1 is direct or indirect.
Another topic which requires additional work is the identification of the transcription factors which activate the transcription of many of the genes subject to catabolite repression. We still do not know which are the activating factors interacting with the SUC2 promoter, with the CSRE domain in the promoters of the genes encoding gluconeogenic enzymes, or with other UAS sequences such as UAS1FBP1 or UAS2ADH1. Since it has been reported that different SUC genes and the MAL61 gene could have Adr1 binding sites (53), it would be worthwhile to examine in band shift assays whether Adr1 really binds to the various sites. Some general approaches to identify DNA binding proteins could include Southwestern blotting (116), affinity chromatography purification methods, and one-hybrid assays (12) as well as the screening of expression libraries with the corresponding labeled DNA probe.
The mode of regulation of the activating factors required for the different systems has been well established for Gal4 and Adr1, but information is lacking for other systems. Although the expression of the genes activated by the Hap2/3/4/5 complex appears to depend on the availability of Hap4, it is not known how the transcription of HAP4 is controlled by glucose.
A promising approach for investigating in a global way the role of proteins involved in catabolite repression is the use of DNA microarrays containing all the genes of S. cerevisiae (77). These microarrays have already been used to identify genes whose expression is affected by a deletion in the regulatory gene TUP1, and the feasibility of the approach has been clearly shown (77).
A recurrent feature of the catabolite repression regulatory systems is the existence of redundant circuits. For instance, although cAMP represses the expression of a variety of genes, lowering the activity of the cAMP-dependent protein kinases does not result in derepression of these genes (157, 194), probably because other repressing pathways, such as the one mediated by the Mig1 complex, remain operative. Another interesting case is that of Sip4, a substrate of the protein kinase Snf1. From about 150 genes encoding known or putative transcription factors, SIP4 is, together with HAP4, the only one to be strongly induced at the diauxic shift, a phase of yeast growth where catabolite repression of most genes is lifted (77). Although this points to a pivotal role for Sip4, there should be some pathway parallel to that controlled by Sip4, since an interruption of SIP4 has no marked phenotypic effects (187). Another fact which can be noted is that sugars such as galactose or maltose do not block Snf1 activity and therefore should act as repressors by some alternative pathway not involving Snf1 or at least not involving Snf1 alone. This putative pathway is probably shared with glucose itself.
It is striking that many proteins involved in yeast catabolite repression have their counterparts in mammalian organisms: the AMP-activated protein kinase corresponds to Snf1 (36, 234, 380), the Egr and Wilms’ tumor proteins correspond to Mig1 (243), and two subunits of the CCAAT box-binding factor NF-Y/CBF correspond to Hap2 and Hap3 (209, 358, 367). However, there is no evidence that the homologous proteins play the same role in the two systems. The Snf1/AMPK family of protein kinases could have diverged from an ancient stress response system, but the protein found in yeast would react to the lack of glucose, while that of the mammalian cell would respond to an increase in AMP levels (137). It seems likely that proteins which were present in some primitive organism were later recruited to perform different functions as the need for them arose.
ACKNOWLEDGMENTS
I am grateful to all the colleagues who sent me reprints or preprints pertinent to this review or who communicated to me unpublished results. I thank C. Gancedo for a thorough revision of the manuscript.
Work in my laboratory has been supported by grant PB94-0091-CO2-01 from the Dirección General de Investigación Científica y Técnica.
Footnotes
Dedicated to the memory of Helmut Holzer, who greatly contributed to the knowledge of yeast metabolism.
REFERENCES
- 1.Adams B. Induction of galactokinase in Saccharomyces cerevisiae: kinetics of induction and glucose effects. J Bacteriol. 1972;111:308–315. doi: 10.1128/jb.111.2.308-315.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altamura N, Dujardin G, Groudinsky O, Slonimski P P. Two adjacent nuclear genes, ISF1 and NAM7/UPF1, cooperatively participate in mitochondrial functions in Saccharomyces cerevisiae. Mol Gen Genet. 1994;242:49–56. doi: 10.1007/BF00277347. [DOI] [PubMed] [Google Scholar]
- 3.Bailey R B, Woodward A. Isolation and characterization of a pleiotropic glucose repression resistant mutant of Saccharomyces cerevisiae. Mol Gen Genet. 1984;193:507–512. doi: 10.1007/BF00382091. [DOI] [PubMed] [Google Scholar]
- 4.Baker S H, Frederick D L, Bloecher A, Tatchell K. Alanine-scanning mutagenesis of protein phosphatase type 1 in the yeast Saccharomyces cerevisiae. Genetics. 1997;145:615–626. doi: 10.1093/genetics/145.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balciunas D, Ronne H. Three subunits of the RNA polymerase II mediator complex are involved in glucose repression. Nucleic Acids Res. 1995;23:4421–4425. doi: 10.1093/nar/23.21.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bañuelos M, Gancedo C, Gancedo J M. Activation by phosphate of yeast phosphofructokinase. J Biol Chem. 1977;252:6394–6398. [PubMed] [Google Scholar]
- 7.Barlev N A, Candau R, Wang L, Darpino P, Silverman L, Berger S L. Characterization of physical interactions of the putative transcriptional adaptor, ADA2, with acidic activation domains and TATA-binding protein. J Biol Chem. 1995;270:19337–19344. doi: 10.1074/jbc.270.33.19337. [DOI] [PubMed] [Google Scholar]
- 8.Baxevanis A D, Arents G, Moudrianakis E N, Landsman D. A variety of DNA-binding and multimeric proteins contain the histone fold motif. Nucleic Acids Res. 1995;23:2685–2691. doi: 10.1093/nar/23.14.2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Becker W, Heukelbach J, Kentrup H, Joost H-G. Molecular cloning and characterization of a novel mammalian protein kinase harboring a homology domain that defines a subfamily of serine/threonine kinases. Eur J Biochem. 1996;235:736–743. doi: 10.1111/j.1432-1033.1996.00736.x. [DOI] [PubMed] [Google Scholar]
- 10.Bemis L T, Denis C L. Identification of functional regions in the yeast transcriptional activator ADR1. Mol Cell Biol. 1988;8:2125–2131. doi: 10.1128/mcb.8.5.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berger S L, Piña B, Silverman N, Marcus G A, Agapite J, Regier J L, Triezenberg S J, Guarente L. Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic domains. Cell. 1992;70:251–265. doi: 10.1016/0092-8674(92)90100-q. [DOI] [PubMed] [Google Scholar]
- 12.Blaiseau P-L, Isnard A-D, Surdin-Kerjan Y, Thomas D. Met31p and Met32p, two related zinc finger proteins, are involved in transcriptional regulation of yeast sulfur amino acid metabolism. Mol Cell Biol. 1997;17:3640–3648. doi: 10.1128/mcb.17.7.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blázquez M A, Gancedo C. Identification of extragenic suppressors of the cif1 mutation in Saccharomyces cerevisiae. Curr Genet. 1994;25:89–94. doi: 10.1007/BF00309531. [DOI] [PubMed] [Google Scholar]
- 14.Blumberg H, Eisen A, Sledziewski A, Bader D, Young E T. Two zinc fingers of a yeast regulatory protein shown by genetic evidence to be essential for its function. Nature (London) 1987;328:443–445. doi: 10.1038/328443a0. [DOI] [PubMed] [Google Scholar]
- 15.Blumberg H, Hartshorne T A, Young E T. Regulation of expression and activity of the yeast transcription factor ADR1. Mol Cell Biol. 1988;8:1868–1876. doi: 10.1128/mcb.8.5.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boles E, Heinisch J, Zimmermann F K. Different signals control the activation of glycolysis in the yeast Saccharomyces cerevisiae. Yeast. 1993;9:761–770. doi: 10.1002/yea.320090710. [DOI] [PubMed] [Google Scholar]
- 17.Boles E, Hollenberg C P. The molecular genetics of hexose transport in yeasts. FEMS Microbiol Rev. 1997;21:85–111. doi: 10.1111/j.1574-6976.1997.tb00346.x. [DOI] [PubMed] [Google Scholar]
- 18.Boles E, Zimmermann F K. Induction of pyruvate decarboxylase in glycolysis mutants of Saccharomyces cerevisiae correlates with the concentration of three-carbon metabolites. Arch Microbiol. 1993;160:324–328. doi: 10.1007/BF00292085. [DOI] [PubMed] [Google Scholar]
- 19.Borralho L M, Malamud D R, Panek A D, Tenan M N, Oliveira D E, Mattoon J R. Parallel changes in catabolite repression of haem biosynthesis and cytochromes in repression-resistant mutants of Saccharomyces cerevisiae. J Gen Microbiol. 1989;135:1217–1227. doi: 10.1099/00221287-135-5-1217. [DOI] [PubMed] [Google Scholar]
- 20.Boy-Marcotte E, Tadi D, Perrot M, Boucherie H, Jacquet M. High cAMP levels antagonize the reprogramming of gene expression that occurs at the diauxic shift in Saccharomyces cerevisiae. Microbiology. 1996;142:459–467. doi: 10.1099/13500872-142-3-459. [DOI] [PubMed] [Google Scholar]
- 21.Bram R J, Lue N F, Kornberg R D. A GAL family of upstream activating sequences in yeast: roles in both induction and repression of transcription. EMBO J. 1986;5:603–608. doi: 10.1002/j.1460-2075.1986.tb04253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brandl C J, Furlanetto A M, Martens J A, Hamilton K S. Characterization of NGG1, a novel yeast gene required for glucose repression of GAL4p-regulated transcription. EMBO J. 1993;12:5255–5265. doi: 10.1002/j.1460-2075.1993.tb06221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brandl C J, Martens J A, Margaliot A, Stenning D, Furlanetto A M, Saleh A, Hamilton K S, Genereaux J. Structure/functional properties of the yeast dual regulator protein NGG1 that are required for glucose repression. J Biol Chem. 1996;271:9298–9306. doi: 10.1074/jbc.271.16.9298. [DOI] [PubMed] [Google Scholar]
- 24.Breunig K D. Glucose repression of LAC gene expression in yeast is mediated by the transcriptional activator LAC9. Mol Gen Genet. 1989;216:422–427. doi: 10.1007/BF00334386. [DOI] [PubMed] [Google Scholar]
- 25.Brown T A, Evangelista C, Trumpower B L. Regulation of nuclear genes encoding mitochondrial proteins in Saccharomyces cerevisiae. J Bacteriol. 1995;177:6836–6843. doi: 10.1128/jb.177.23.6836-6843.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown T A, Trumpower B L. Strain-dependent variation in carbon source regulation of nucleus-encoded mitochondrial proteins of Saccharomyces cerevisiae. J Bacteriol. 1995;177:1380–1382. doi: 10.1128/jb.177.5.1380-1382.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26a.Bu Y, Schmidt M C. Identification of cis-acting elements in the SUC2 promoter of Saccharomyces cerevisiae required for activation of transcription. Nucleic Acids Res. 1998;26:1002–1009. doi: 10.1093/nar/26.4.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Busturia A, Lagunas R. Identification of two forms of the maltose transport system in Saccharomyces cerevisiae and their regulation by catabolite inactivation. Biochim Biophys Acta. 1985;820:324–326. [Google Scholar]
- 28.Byrne S M, Hoffman C S. Six git genes encode a glucose-induced adenylate cyclase activation pathway in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 1993;105:1095–1100. doi: 10.1242/jcs.105.4.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cairns B R, Henry N L, Kornberg R D. TFG3/TAF38/ANC1, a component of the yeast SWI/SNF complex that is similar to the leukemogenic proteins ENL and AF-9. Mol Cell Biol. 1996;16:3308–3316. doi: 10.1128/mcb.16.7.3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cairns B R, Kim Y J, Sayre M H, Laurent B C, Kornberg R D. A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5 and SNF6 gene products isolated from yeast. Proc Natl Acad Sci USA. 1994;91:1950–1954. doi: 10.1073/pnas.91.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cairns B R, Levinson R S, Yamamoto K R, Kornberg R D. Essential role of Swp73p in the formation of yeast Swi/Snf complex. Genes Dev. 1996;10:2131–2143. doi: 10.1101/gad.10.17.2131. [DOI] [PubMed] [Google Scholar]
- 32.Candau R, Berger S L. Structural and functional analysis of yeast putative adaptors. Evidence for an adaptor complex in vivo. J Biol Chem. 1996;271:5237–5245. doi: 10.1074/jbc.271.9.5237. [DOI] [PubMed] [Google Scholar]
- 33.Candau R, Zhou J, Allis C D, Berger S L. Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo. EMBO J. 1997;16:555–565. doi: 10.1093/emboj/16.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cannon J, Pringle J R, Fiechter A, Khalil M. Characterization of glycogen-deficient glc mutants of Saccharomyces cerevisiae. Genetics. 1994;136:485–503. doi: 10.1093/genetics/136.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carey M, Kakidani H, Leatherwood J, Mostashari F, Ptashne M. An amino-terminal fragment of GAL4 binds DNA as a dimer. J Mol Biol. 1989;209:423–432. doi: 10.1016/0022-2836(89)90007-7. [DOI] [PubMed] [Google Scholar]
- 36.Carling D, Aguan K, Woods A, Verhoeven A J M, Beri R K, Brennan C H, Sidebottom C, Davison M D, Scott J. Mammalian AMP-activated protein kinase is homologous to yeast and plant protein kinases involved in the regulation of carbon metabolism. J Biol Chem. 1994;289:11442–11448. [PubMed] [Google Scholar]
- 37.Carlson M, Botstein D. Two differentially regulated mRNAs with different 5′ ends encode secreted and intracellular forms of yeast invertase. Cell. 1982;28:145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- 38.Carlson M, Ludin K, Jiang R. EMBO-FEBS Workshop. 1997. Glucose-regulated interaction of a regulatory subunit of protein phosphatase 1 with the Snf1 protein kinase; p. 27. Protein phosphatases and protein dephosphorylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carlson M, Osmond B C, Botstein D. A suppressor of snf1 mutations causes constitutive high-level invertase synthesis in yeast. Genetics. 1984;107:19–32. doi: 10.1093/genetics/107.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carlson M, Osmond B C, Neigeborn L, Botstein D. Mutants of yeast defective in sucrose utilization. Genetics. 1981;98:25–40. doi: 10.1093/genetics/98.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caspary F, Hartig A, Schüller H-J. Constitutive and carbon source-responsive promoter elements involved in the regulated expression of the Saccharomyces cerevisiae malate synthase gene MLS1. Mol Gen Genet. 1997;255:619–627. doi: 10.1007/s004380050536. [DOI] [PubMed] [Google Scholar]
- 42.Cassart J-P, Georis I, Östling J, Ronne H, Vandenhaute J. The MIG1 repressor from Kluyveromyces lactis: cloning, sequencing and functional analysis in Saccharomyces cerevisiae. FEBS Lett. 1995;371:191–194. doi: 10.1016/0014-5793(95)00909-s. [DOI] [PubMed] [Google Scholar]
- 43.Cassart J-P, Östling J, Ronne H, Vandenhaute J. Comparative analysis in three fungi reveals structurally and functionally conserved regions in the Mig1 repressor. Mol Gen Genet. 1997;255:9–18. doi: 10.1007/s004380050469. [DOI] [PubMed] [Google Scholar]
- 44.Celenza J L, Carlson M. A yeast gene that is essential for release from glucose repression encodes a protein kinase. Science. 1986;233:1175–1180. doi: 10.1126/science.3526554. [DOI] [PubMed] [Google Scholar]
- 45.Celenza J L, Carlson M. Mutational analysis of the Saccharomyces cerevisiae SNF1 protein kinase and evidence for functional interaction with the SNF4 protein. Mol Cell Biol. 1989;9:5034–5044. doi: 10.1128/mcb.9.11.5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Celenza J L, Eng F J, Carlson M. Molecular analysis of the SNF4 gene of Saccharomyces cerevisiae: evidence for physical association of the SNF4 protein with the SNF1 protein kinase. Mol Cell Biol. 1989;9:5045–5054. doi: 10.1128/mcb.9.11.5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Celenza J L, Marshall-Carlson L, Carlson M. The yeast SNF3 gene encodes a glucose transporter homologous to the mammalian protein. Proc Natl Acad Sci USA. 1988;85:2130–2134. doi: 10.1073/pnas.85.7.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cereghino G P, Atencio D P, Saghbini M, Beiner J, Scheffler I E. Glucose-dependent turnover of the mRNAs encoding succinate dehydrogenase peptides in Saccharomyces cerevisiae: sequence elements in the 5′ untranslated region of the Ip mRNA play a dominant role. Mol Biol Cell. 1995;6:1125–1143. doi: 10.1091/mbc.6.9.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cereghino G P, Scheffler I E. Genetic analysis of glucose regulation in Saccharomyces cerevisiae: control of transcription versus mRNA turnover. EMBO J. 1996;15:363–374. [PMC free article] [PubMed] [Google Scholar]
- 50.Chang Y S, Dubin R A, Perkins E, Needleman R. MAL63 codes for a positive regulator of maltose fermentation in Saccharomyces cerevisiae. Curr Genet. 1988;14:201–209. doi: 10.1007/BF00376740. [DOI] [PubMed] [Google Scholar]
- 51.Charron M J, Dubin R A, Michels C A. Structural and functional analysis of the MAL1 locus of Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:3891–3899. doi: 10.1128/mcb.6.11.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen S, West R W, Jr, Johnson S L, Gans H, Kruger B, Ma J. TSF3, a global regulatory protein that silences transcription of yeast GAL genes, also mediates repression by α2 repressor and is identical to SIN4. Mol Cell Biol. 1993;13:831–840. doi: 10.1128/mcb.13.2.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheng C, Kacherovsky N, Dombek K M, Camier S, Thukral S K, Rhim E, Young E T. Identification of potential target genes for Adr1p through characterization of essential nucleotides in UAS1. Mol Cell Biol. 1994;14:3842–3852. doi: 10.1128/mcb.14.6.3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cherry J R, Johnson T R, Dollard J R, Shuster J R, Denis C L. Cyclic AMP-dependent protein kinase phosphorylates and inactivates the yeast transcriptional activator ADR1. Cell. 1989;56:409–419. doi: 10.1016/0092-8674(89)90244-4. [DOI] [PubMed] [Google Scholar]
- 55.Chiang Y-C, Komarnitsky P, Chase D, Denis C L. ADR1 activation domains contact the histone acetyltransferase GCN5 and the core transcriptional factor TFIIB. J Biol Chem. 1996;271:32359–32365. doi: 10.1074/jbc.271.50.32359. [DOI] [PubMed] [Google Scholar]
- 56.Chiang H L, Schekman R. Regulated import and degradation of a cytosolic protein in the yeast vacuole. Nature (London) 1991;350:313–318. doi: 10.1038/350313a0. [DOI] [PubMed] [Google Scholar]
- 57.Ciriacy M. Genetics of alcohol dehydrogenase in Saccharomyces cerevisiae. II. Two loci controlling synthesis of the glucose repressible ADHII. Mol Gen Genet. 1975;138:157–164. doi: 10.1007/BF02428119. [DOI] [PubMed] [Google Scholar]
- 58.Ciriacy M. Isolation and characterization of yeast mutants defective in intermediary carbon metabolism and in carbon catabolite derepression. Mol Gen Genet. 1977;154:213–220. doi: 10.1007/BF00330840. [DOI] [PubMed] [Google Scholar]
- 59.Ciriacy M. Isolation and characterization of further cis- and trans-acting regulatory elements involved in the synthesis of glucose-repressible alcohol dehydrogenase (ADHII) in Saccharomyces cerevisiae. Mol Gen Genet. 1979;176:427–431. doi: 10.1007/BF00333107. [DOI] [PubMed] [Google Scholar]
- 60.Cook W J, Chase D, Audino D C, Denis C L. Dissection of the ADR1 protein reveals multiple, functionally redundant activation domains interspersed with inhibitory regions: evidence for a repressor binding to the ADR1c region. Mol Cell Biol. 1994;14:629–640. doi: 10.1128/mcb.14.1.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cook W J, Denis C L. Identification of three genes required for the glucose-dependent transcription of the yeast transcriptional activator ADR1. Curr Genet. 1993;23:192–200. doi: 10.1007/BF00351495. [DOI] [PubMed] [Google Scholar]
- 62.Cooper J P, Roth S Y, Simpson R T. The global transcriptional regulators, SSN6 and TUP1, play distinct roles in the establishment of a repressive chromatin structure. Genes Dev. 1994;8:1400–1410. doi: 10.1101/gad.8.12.1400. [DOI] [PubMed] [Google Scholar]
- 63.Coshigano P W, Miller S M, Magasanik B. Physiological and genetical analysis of the carbon regulation of the NAD-dependent glutamate dehydrogenase of Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:4455–4465. doi: 10.1128/mcb.11.9.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Côté J, Quinn J, Workman J L, Peterson C L. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 65.Czyz M, Nagiec M M, Dickson R C. Autoregulation of GAL4 transcription is essential for rapid growth of Kluyveromyces lactis on lactose and galactose. Nucleic Acids Res. 1993;21:4378–4382. doi: 10.1093/nar/21.18.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Daignan-Fornier B, Nguyen C C, Reisdorf P, Lemeignan B, Bolotin-Fukuhara M. MBR1 and MBR3, two related yeast genes that can suppress the growth defect of hap2, hap3 and hap4 mutants. Mol Gen Genet. 1994;243:575–583. doi: 10.1007/BF00284206. [DOI] [PubMed] [Google Scholar]
- 67.Daignan-Fornier B, Valens M, Lemire B D, Bolotin-Fukuhara M. Structure and regulation of SDH3, the yeast gene encoding the cytochrome b560 subunit of respiratory complex II. J Biol Chem. 1994;269:15469–15472. [PubMed] [Google Scholar]
- 68.Dale S, Wilson W, Edelman A, Hardie D G. Similar substrate recognition motifs for mammalian AMP-activated protein kinase, higher plant HMG-CoA reductase kinase-A, yeast SNF1, and mammalian calmodulin-dependent protein kinase I. FEBS Lett. 1995;361:191–195. doi: 10.1016/0014-5793(95)00172-6. [DOI] [PubMed] [Google Scholar]
- 69.Dal Santo P, Blanchard B, Hoffman C S. The Schizosaccharomyces pombe pyp1 protein tyrosine phosphatase negatively regulates nutrient monitoring pathways. J Cell Sci. 1996;109:1919–1925. doi: 10.1242/jcs.109.7.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dang V D, Bohn C, Bolotin-Fukuhara M, Daignan-Fornier B. The CCAAT box-binding factor stimulates ammonium assimilation in Saccharomyces cerevisiae defining a new cross-pathway regulation between nitrogen and carbon metabolism. J Bacteriol. 1996;178:1842–1849. doi: 10.1128/jb.178.7.1842-1849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Davies S P, Helps N R, Cohen P T W, Hardie D G. 5′AMP inhibits dephosphorylation, as well as promoting phosphorylation, of the AMP-activated protein kinase. Studies using bacterially expressed human protein phosphatase-2Cα and native protein phosphatase-2AC. FEBS Lett. 1995;377:421–425. doi: 10.1016/0014-5793(95)01368-7. [DOI] [PubMed] [Google Scholar]
- 72.de Mesquita, J. F., O. Zaragoza, and J. M. Gancedo. Functional analysis of upstream activating elements in the promoter of the FBP1 gene from Saccharomyces cerevisiae. Curr. Genet., in press. [DOI] [PubMed]
- 73.Denis C L. Identification of new genes involved in the regulation of yeast alcohol dehydrogenase II. Genetics. 1984;108:845–858. doi: 10.1093/genetics/108.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Denis C L, Audino D C. The CCR1 (SNF1) and SCH9 protein kinases act independently of cAMP-dependent protein kinase and the transcriptional activator ADR1 in controlling yeast ADH2 expression. Mol Gen Genet. 1991;229:395–399. doi: 10.1007/BF00267461. [DOI] [PubMed] [Google Scholar]
- 75.Denis C L, Fontaine S C, Chase D, Kemp B E, Bemis L T. ADR1c mutations enhance the ability of ADR1 to activate transcription by a mechanism that is independent of effects on cyclic AMP-dependent protein kinase phosphorylation of Ser-230. Mol Cell Biol. 1992;12:1507–1514. doi: 10.1128/mcb.12.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Denis C L, Malvar T. The CCR4 gene from Saccharomyces cerevisiae is required for both non-fermentative and spt-mediated gene expression. Genetics. 1990;124:283–291. doi: 10.1093/genetics/124.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.DeRisi J L, Iyer V R, Brown P O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 78.DeVit M J, Waddle J A, Johnston M. Regulated nuclear translocation of the Mig1 glucose repressor. Mol Biol Cell. 1997;8:1603–1618. doi: 10.1091/mbc.8.8.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.de Winde J H, Grivell L A. Global regulation of mitochondrial biogenesis in Saccharomyces cerevisiae. Prog Nucleic Acid Res Mol Biol. 1993;46:51–91. doi: 10.1016/s0079-6603(08)61018-1. [DOI] [PubMed] [Google Scholar]
- 80.de Winde J H, Crauwels M, Hohmann S, Thevelein J M, Winderickx J. Differential requirements of the yeast sugar kinases for sugar sensing in establishing the catabolic-repressed state. Eur J Biochem. 1996;241:633–643. doi: 10.1111/j.1432-1033.1996.00633.x. [DOI] [PubMed] [Google Scholar]
- 81.Dombek K M, Camier S, Young E T. ADH2 expression is repressed by REG1 independently of mutations that alter the phosphorylation of the yeast transcription factor ADR1. Mol Cell Biol. 1993;13:4391–4399. doi: 10.1128/mcb.13.7.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dombek K M, Young E T. Cyclic AMP-dependent protein kinase inhibits ADH2 expression in part by decreasing expression of the transcription factor gene ADR1. Mol Cell Biol. 1997;17:1450–1458. doi: 10.1128/mcb.17.3.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dong J, Dickson R D. Glucose represses the lactose-galactose regulon in Kluyveromyces lactis through a SNF1- and MIG1-dependent pathway that modulates galactokinase (GAL1) gene expression. Nucleic Acids Res. 1997;25:3657–3664. doi: 10.1093/nar/25.18.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Donnini C, Lodi T, Ferrero I, Puglisi P P. IMP2, a nuclear gene controlling the mitochondrial dependence of galactose, maltose and raffinose utilization in Saccharomyces cerevisiae. Yeast. 1992;8:83–93. doi: 10.1002/yea.320080203. [DOI] [PubMed] [Google Scholar]
- 85.Donoviel M S, Young E T. Isolation and identification of genes activating UAS2-dependent ADH2 expression in Saccharomyces cerevisiae. Genetics. 1996;143:1137–1148. doi: 10.1093/genetics/143.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dorsman J C, Grivell L A. Expression of the gene encoding subunit II of yeast QH2: cytochrome c oxidoreductase is regulated by multiple factors. Curr Genet. 1990;17:459–464. doi: 10.1007/BF00313072. [DOI] [PubMed] [Google Scholar]
- 87.Dowzer C E A, Kelly J M. Analysis of the creA gene, a regulator of carbon catabolite repression in Aspergillus nidulans. Mol Cell Biol. 1991;11:5701–5709. doi: 10.1128/mcb.11.11.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Draper M P, Liu H, Nelsbach A H, Mosley S P, Denis C L. CCR4 is a glucose-regulated transcription factor whose leucine-rich repeat binds several proteins important for placing CCR4 in its proper promoter context. Mol Cell Biol. 1994;14:4522–4531. doi: 10.1128/mcb.14.7.4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Draper M P, Salvadore C, Denis C L. Identification of a mouse protein whose homolog in Saccharomyces cerevisiae is a component of the CCR4 transcriptional regulatory complex. Mol Cell Biol. 1995;15:3487–3495. doi: 10.1128/mcb.15.7.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Edmondson D G, Smith M M, Roth S Y. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev. 1996;10:1247–1259. doi: 10.1101/gad.10.10.1247. [DOI] [PubMed] [Google Scholar]
- 91.Eide D, Guarente L. Increased dosage of a transcriptional activator gene enhances iron-limited growth of Saccharomyces cerevisiae. J Gen Microbiol. 1992;138:347–354. doi: 10.1099/00221287-138-2-347. [DOI] [PubMed] [Google Scholar]
- 92.Einerhand A W C, Kos W T, Distel B, Tabak H F. Characterization of a transcriptional control element involved in proliferation of peroxisomes in yeast in response to oleate. Eur J Biochem. 1993;214:323–331. doi: 10.1111/j.1432-1033.1993.tb17927.x. [DOI] [PubMed] [Google Scholar]
- 93.Einerhand A W C, Kos W T, Smart W C, Kal A J, Tabak H F, Cooper T G. The upstream region of the FOX3 gene encoding peroxisomal 3-oxoacetyl-coenzyme A thiolase in Saccharomyces cerevisiae contains ABF1- and replication protein A-binding sites that participate in its regulation by glucose repression. Mol Cell Biol. 1995;15:3405–3414. doi: 10.1128/mcb.15.6.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Eisen A, Taylor W E, Blumberg H, Young E T. The yeast regulatory protein ADR1 binds in a zinc-dependent manner to the upstream activating sequence of ADH2. Mol Cell Biol. 1988;8:4552–4556. doi: 10.1128/mcb.8.10.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Entian K-D. Glucose repression: a complex regulatory system in yeast. Microbiol Sci. 1986;3:366–371. [PubMed] [Google Scholar]
- 96.Entian K-D, Fröhlich K-U. Saccharomyces cerevisiae mutants provide evidence of hexokinase PII as a bifunctional enzyme with catalytic and regulatory domains for triggering carbon catabolite repression. J Bacteriol. 1984;158:29–35. doi: 10.1128/jb.158.1.29-35.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96a.Entian K-D, Schüller H-J. Glucose repression (carbon catabolite repression) in yeast. In: Zimmermann F K, Entian K-D, editors. Yeast sugar metabolism. Lancaster, Pa: Technomic; 1997. pp. 409–434. [Google Scholar]
- 96b.Entian K-D, Schüller H-J. Genetics of di- and trisaccharide utilization. In: Zimmermann F K, Entian K-D, editors. Yeast sugar metabolism. Lancaster, Pa: Technomic; 1997. pp. 225–233. [Google Scholar]
- 97.Entian K-D, Zimmermann F K. Glycolytic enzymes and intermediates in carbon catabolite repression mutants of Saccharomyces cerevisiae. Mol Gen Genet. 1980;177:345–350. doi: 10.1007/BF00267449. [DOI] [PubMed] [Google Scholar]
- 98.Entian K-D, Zimmermann F K. New genes involved in catabolite repression and derepression in the yeast Saccharomyces cerevisiae. J Bacteriol. 1982;151:1123–1128. doi: 10.1128/jb.151.3.1123-1128.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Entian K-D, Zimmermann F K, Scheel I. A partial defect in carbon catabolite repression in mutants of Saccharomyces cerevisiae with reduced hexose phosphorylation. Mol Gen Genet. 1977;156:99–105. doi: 10.1007/BF00272258. [DOI] [PubMed] [Google Scholar]
- 100.Eraso P, Gancedo J M. Catabolite repression in yeasts is not associated with low levels of cAMP. Eur J Biochem. 1984;141:195–198. doi: 10.1111/j.1432-1033.1984.tb08174.x. [DOI] [PubMed] [Google Scholar]
- 101.Erickson J R, Johnston M. Genetic and molecular characterization of GAL83: its interaction and similarities with other genes involved in glucose repression in Saccharomyces cerevisiae. Genetics. 1993;135:655–664. doi: 10.1093/genetics/135.3.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Erickson J R, Johnston M. Suppressors reveal two classes of glucose repression genes in the yeast Saccharomyces cerevisiae. Genetics. 1994;136:1271–1278. doi: 10.1093/genetics/136.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Espinel A E, Gómez-Toribio V, Peinado J M. The inactivation of hexokinase activity does not prevent glucose repression in Candida utilis. FEMS Microbiol Lett. 1996;135:327–332. doi: 10.1111/j.1574-6968.1996.tb08009.x. [DOI] [PubMed] [Google Scholar]
- 104.Estruch F, Carlson M. Increased dosage of the MSN1 gene restores invertase expression in yeast mutants defective in the SNF1 protein kinase. Nucleic Acids Res. 1990;18:6959–6964. doi: 10.1093/nar/18.23.6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Estruch F, Carlson M. Two homologous zinc finger genes identified by multicopy suppression in a SNF1 protein kinase mutant of Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:3872–3881. doi: 10.1128/mcb.13.7.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Estruch F, Treitel M A, Yang X, Carlson M. N-terminal mutations modulate yeast SNF1 kinase function. Genetics. 1992;132:639–650. doi: 10.1093/genetics/132.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Federoff H J, Eccleshall T R, Marmur J. Carbon catabolite repression of maltase synthesis in Saccharomyces carlsbergensis. J Bacteriol. 1983;156:301–307. doi: 10.1128/jb.156.1.301-307.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Feng Z, Wilson S E, Peng Z Y, Schlender K K, Reiman E M, Trumbly R J. The yeast GLC7 gene required for glycogen accumulation encodes a type 1 protein phosphatase. J Biol Chem. 1991;266:23796–23801. [PubMed] [Google Scholar]
- 109.Fernández M, Fernández E, Rodicio R. ACR1, a gene encoding a protein related to mitochondrial carriers, is essential for acetyl-CoA synthetase activity in Saccharomyces cerevisiae. Mol Gen Genet. 1994;242:727–735. doi: 10.1007/BF00283428. [DOI] [PubMed] [Google Scholar]
- 110.Filipits M, Simon M M, Rapatz W, Hamilton B, Ruis H. A Saccharomyces cerevisiae upstream activating sequence mediates induction of peroxisome proliferation by fatty acids. Gene. 1993;132:49–55. doi: 10.1016/0378-1119(93)90513-3. [DOI] [PubMed] [Google Scholar]
- 111.Flick J S, Johnston M. Two systems of glucose repression of the GAL1 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:4757–4769. doi: 10.1128/mcb.10.9.4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Flick J S, Johnston M. GRR1 of Saccharomyces cerevisiae is required for glucose repression and encodes a protein with leucine-rich repeats. Genetics. 1991;130:295–304. doi: 10.1128/mcb.11.10.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Flick J S, Johnston M. Analysis of URSG-mediated glucose repression of the GAL1 promoter of Saccharomyces cerevisiae. Genetics. 1992;130:295–304. doi: 10.1093/genetics/130.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Forsburg S L, Guarente L. Identification and characterization of HAP4: A third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 1989;3:1166–1178. doi: 10.1101/gad.3.8.1166. [DOI] [PubMed] [Google Scholar]
- 115.Forsburg S L, Guarente L. Communication between mitochondria and the nucleus in regulation of cytochrome genes in the yeast Saccharomyces cerevisiae. Annu Rev Cell Biol. 1989;5:153–180. doi: 10.1146/annurev.cb.05.110189.001101. [DOI] [PubMed] [Google Scholar]
- 116.Francis-Lang H, Price M, Polycarpou-Schwarz M, Di Lauro R. Cell-type-specific expression of the rat thyroperoxidase promoter indicates common mechanism for thyroid-specific gene expression. Mol Cell Biol. 1992;12:576–588. doi: 10.1128/mcb.12.2.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fraser R A, Rossignol M, Heard D J, Egly J-M, Chambon P. SUG1, a putative transcriptional mediator and subunit of the PA700 proteasome regulatory complex, is a DNA helicase. J Biol Chem. 1997;272:7122–7126. doi: 10.1074/jbc.272.11.7122. [DOI] [PubMed] [Google Scholar]
- 118.Frederick D L, Tatchell K. The REG2 gene of Saccharomyces cerevisiae encodes a type 1 protein phosphatase-binding protein that functions with Reg1p and the Snf1 protein kinase to regulate growth. Mol Cell Biol. 1996;16:2922–2931. doi: 10.1128/mcb.16.6.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Funayama S, Gancedo J M, Gancedo C. Turnover of yeast fructose-1,6-bisphosphatase in different metabolic conditions. Eur J Biochem. 1980;109:61–66. doi: 10.1111/j.1432-1033.1980.tb04767.x. [DOI] [PubMed] [Google Scholar]
- 120.Gamo F-J, Lafuente M J, Gancedo C. The mutation DGT1-1 decreases glucose transport and alleviates carbon catabolite repression in Saccharomyces cerevisiae. J Bacteriol. 1994;176:7423–7429. doi: 10.1128/jb.176.24.7423-7429.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gancedo C. Inactivation of fructose-1,6-diphosphatase by glucose in yeast. J Bacteriol. 1971;107:401–405. doi: 10.1128/jb.107.2.401-405.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gancedo C, Serrano R. Energy-yielding metabolism. In: Rose A H, Harrison J S, editors. The Yeasts. Vol. 3. London, United Kingdom: Academic Press, Ltd.; 1989. pp. 205–259. [Google Scholar]
- 123.Gancedo C, Salas M L, Giner A, Sols A. Reciprocal effects of carbon sources on the levels of the AMP-sensitive fructose-1,6-diphosphatase and phosphofructokinase in yeast. Biochem Biophys Res Commun. 1965;20:15–20. doi: 10.1016/0006-291x(65)90944-7. [DOI] [PubMed] [Google Scholar]
- 124.Gancedo J M. Carbon catabolite repression in yeast. Eur J Biochem. 1992;206:297–313. doi: 10.1111/j.1432-1033.1992.tb16928.x. [DOI] [PubMed] [Google Scholar]
- 125.Gancedo J M, Gancedo C. Catabolite repression mutants in yeast. FEMS Microbiol Rev. 1986;32:179–187. [Google Scholar]
- 125a.Gancedo J M, Gancedo C. Gluconeogenesis and catabolite inactivation. In: Zimmermann F K, Entian K-D, editors. Yeast sugar metabolism. Lancaster, Pa: Technomic; 1997. pp. 359–377. [Google Scholar]
- 126.Gao G, Fernandez C S, Stapleton D, Auster A S, Widmer J, Dyck J R B, Kemp B E, Witters L E. Non-catalytic β- and γ-subunit isoforms of the 5′-AMP-activated protein kinase. J Biol Chem. 1996;271:8675–8681. doi: 10.1074/jbc.271.15.8675. [DOI] [PubMed] [Google Scholar]
- 127.Geber A, Williamson P R, Rex J H, Sweeney E C, Bennett J E. Cloning and characterization of a Candida albicans maltase gene involved in sucrose utilization. J Bacteriol. 1992;174:6992–6996. doi: 10.1128/jb.174.21.6992-6996.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gimeno C J, Fink G R. Induction of pseudohyphal growth by overexpression of PHD1, a Saccharomyces cerevisiae gene related to transcriptional regulators of fungal development. Mol Cell Biol. 1994;14:2100–2112. doi: 10.1128/mcb.14.3.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Giniger E, Varnum S M, Ptashne M. Specific DNA-binding of GAL4, a positive regulatory protein of yeast. Cell. 1985;40:767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- 130.Goffrini P, Ficarelli A, Donnini C, Lodi T, Puglisi P P, Ferrero I. FOG1 and FOG2 genes, required for the transcriptional activation of glucose-repressible genes of Kluyveromyces lactis, are homologous to GAL83 and SNF1 of Saccharomyces cerevisiae. Curr Genet. 1996;29:316–326. [PubMed] [Google Scholar]
- 131.Görts C P M. Effect of glucose on the activity and the kinetics of the maltose uptake system and α-glucosidase in Saccharomyces cerevisiae. Biochim Biophys Acta. 1969;184:299–305. doi: 10.1016/0304-4165(69)90032-4. [DOI] [PubMed] [Google Scholar]
- 132.Grant P A, Duggan L, Côté J, Roberts S M, Brownell J E, Candau R, Ohba R, Owen-Hughes T, Allis C D, Winston F, Berger S L, Workman J L. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 133.Griggs D W, Johnston M. Regulated expression of the GAL4 activator gene in yeast provides a sensitive genetic switch for glucose repression. Proc Natl Acad Sci USA. 1991;88:8597–8601. doi: 10.1073/pnas.88.19.8597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Guarente L, Lalonde B, Gifford P, Alani E. Distinctly regulated tandem upstream activation sites mediate catabolite repression of the CYC1 gene of S. cerevisiae. Cell. 1984;36:503–511. doi: 10.1016/0092-8674(84)90243-5. [DOI] [PubMed] [Google Scholar]
- 135.Gustafsson C L, Myers L C, Li Y, Redd M J, Lui M, Erdjument-Bromage H, Tempest P, Kornberg R D. Identification of Rox3 as a component of mediator and RNA polymerase II holoenzyme. J Biol Chem. 1997;272:48–50. doi: 10.1074/jbc.272.1.48. [DOI] [PubMed] [Google Scholar]
- 136.Hahn S, Pinkham J, Wei R, Miller R, Guarente L. The HAP3 regulatory locus of Saccharomyces cerevisiae encodes divergent overlapping transcripts. Mol Cell Biol. 1988;8:655–663. doi: 10.1128/mcb.8.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hardie D G, Carling D. The AMP-activated protein kinase. Fuel gauge of the mammalian cell? Eur J Biochem. 1997;246:259–273. doi: 10.1111/j.1432-1033.1997.00259.x. [DOI] [PubMed] [Google Scholar]
- 138.Hartshorne T A, Blumberg H, Young E T. Sequence homology of the yeast regulatory protein ADR1 with Xenopus transcription factor TFIIIA. Nature (London) 1986;320:283–287. doi: 10.1038/320283a0. [DOI] [PubMed] [Google Scholar]
- 139.Hawley S A, Davison M, Woods A, Davies S P, Beri R K, Carling D, Hardie D G. Characterization of the AMP-activated protein kinase kinase from rat liver, and identification of threonine-172 as the major site at which it phosphorylates and activates AMP-activated protein kinase. J Biol Chem. 1996;271:27879–27887. doi: 10.1074/jbc.271.44.27879. [DOI] [PubMed] [Google Scholar]
- 140.Hawley S A, Selbert M A, Goldstein E G, Edelman A M, Carling D, Hardie D G. 5′-AMP activates the AMP-activated protein kinase cascade, and Ca2+/calmodulin the calmodulin-dependent protein kinase I cascade, via three independent mechanisms. J Biol Chem. 1995;270:27186–27191. doi: 10.1074/jbc.270.45.27186. [DOI] [PubMed] [Google Scholar]
- 141.Hedges D, Proft M, Entian K-D. CAT8, a new zinc cluster-encoding gene necessary for derepression of gluconeogenic enzymes in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:1915–1922. doi: 10.1128/mcb.15.4.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hengartner C J, Thompson C M, Zhang J, Chao D M, Liao S M, Koleske A J, Okamura S, Young R A. Association of an activator with an RNA polymerase II holoenzyme. Genes Dev. 1995;9:897–910. doi: 10.1101/gad.9.8.897. [DOI] [PubMed] [Google Scholar]
- 143.Herrero P, Fernández R, Moreno F. The hexokinase isoenzyme PII of Saccharomyces cerevisiae is a protein kinase. J Gen Microbiol. 1989;135:1209–1216. doi: 10.1099/00221287-135-5-1209. [DOI] [PubMed] [Google Scholar]
- 144.Hirano T, Kinoshita N, Morikawa K, Kimura A. Snap helix with knob and hole: essential repeats in S. pombe nuclear protein nuc2+ Cell. 1990;60:319–328. doi: 10.1016/0092-8674(90)90746-2. [DOI] [PubMed] [Google Scholar]
- 145.Hirschhorn J N, Brown S A, Clark C D, Winston F. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 1992;6:2288–2298. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- 146.Hodge M R, Singh K, Cumsky M G. Upstream activation and repression elements control transcription of the yeast COX5b gene. Mol Cell Biol. 1990;10:5510–5520. doi: 10.1128/mcb.10.10.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hoffman C S, Winston F. Isolation and characterization of mutants constitutive for the expression of the fbp1 gene of Schizosaccharomyces pombe. Genetics. 1990;124:807–816. doi: 10.1093/genetics/124.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hoffman C S, Winston F. Glucose repression of transcription of the Schizosaccharomyces pombe fbp1 gene occurs by a cAMP signaling pathway. Genes Dev. 1991;5:561–571. doi: 10.1101/gad.5.4.561. [DOI] [PubMed] [Google Scholar]
- 149.Hoffman M, Chiang H-L. Isolation of degradation-deficient mutants defective in the targeting of fructose-1,6-bisphosphatase into the vacuole for degradation in Saccharomyces cerevisiae. Genetics. 1996;143:1555–1566. doi: 10.1093/genetics/143.4.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hohmann S, Neves M J, de Koning W, Alijo R, Ramos J, Thevelein J M. The growth and signalling defects of the ggs1(fdp1/byp1) deletion mutant on glucose are suppressed by a deletion of the gene encoding hexokinase PII. Curr Genet. 1993;23:281–289. doi: 10.1007/BF00310888. [DOI] [PubMed] [Google Scholar]
- 151.Holzer H. Catabolite inactivation in yeast. Trends Biochem Sci. 1976;1:178–181. [Google Scholar]
- 152.Horiuchi J, Silverman N, Marcus G A, Guarente L. ADA3, a putative transcriptional adaptor, consists of two separable domains and interacts with ADA2 and GCN5 in a trimeric complex. Mol Cell Biol. 1995;15:1203–1209. doi: 10.1128/mcb.15.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Horiuchi J, Silverman N, Piña B, Marcus G A, Guarente L. ADA1, a novel component of the ADA/GCN5 complex, has broader effects than GCN5, ADA2, or ADA3. Mol Cell Biol. 1997;17:3220–3228. doi: 10.1128/mcb.17.6.3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hu Z, Nehlin J O, Ronne H, Michels C A. MIG1-dependent and MIG1-independent glucose regulation of MAL gene expression in Saccharomyces cerevisiae. Curr Genet. 1995;28:258–266. doi: 10.1007/BF00309785. [DOI] [PubMed] [Google Scholar]
- 155.Huang P-H, Chiang H-L. Identification of novel vesicles in the cytosol to vacuole protein degradation pathway. J Cell Biol. 1997;136:803–810. doi: 10.1083/jcb.136.4.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hubbard E J A, Jiang R, Carlson M. Dosage-dependent modulation of glucose repression by MSN3 (STD1) in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:1972–1978. doi: 10.1128/mcb.14.3.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hubbard E J A, Yang X, Carlson M. Relationship of the cAMP-dependent protein kinase pathway to the SNF1 protein and invertase expression in Saccharomyces cerevisiae. Genetics. 1992;130:71–80. doi: 10.1093/genetics/130.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Igual J C, Navarro B. Respiration and low cAMP-dependent protein kinase activity are required for high-level expression of the peroxisomal thiolase gene in Saccharomyces cerevisiae. Mol Gen Genet. 1996;252:446–455. doi: 10.1007/BF02173010. [DOI] [PubMed] [Google Scholar]
- 159.Jiang R, Carlson M. Glucose regulates protein interactions within the yeast SNF1 protein kinase complex. Genes Dev. 1996;10:3105–3115. doi: 10.1101/gad.10.24.3105. [DOI] [PubMed] [Google Scholar]
- 159a.Jiang R, Carlson M. The Snf1 protein kinase and its activating subunit, Snf4, interact with distinct domains of the Sip1/Sip2/Gal83 component in the kinase complex. Mol Cell Biol. 1997;17:2099–2106. doi: 10.1128/mcb.17.4.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Jiang Y W, Stillman D J. Involvement of the SIN4 global transcriptional regulator in the chromatin structure of Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:4503–4514. doi: 10.1128/mcb.12.10.4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Johnston M. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol Rev. 1987;51:458–476. doi: 10.1128/mr.51.4.458-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Johnston M. Genetic evidence that zinc is an essential co-factor in the DNA-binding domain of GAL4 protein. Nature (London) 1987;328:353–355. doi: 10.1038/328353a0. [DOI] [PubMed] [Google Scholar]
- 163.Johnston M, Carlson M. Regulation of carbon and phosphate utilization. In: Jones E W, Pringle J R, Broach J R, editors. The molecular and cellular biology of the yeast Saccharomyces. 2. Gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. pp. 193–281. [Google Scholar]
- 164.Johnston M, Flick J S, Pexton T. Multiple mechanisms provide rapid and stringent glucose repression of GAL gene expression in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:3834–3841. doi: 10.1128/mcb.14.6.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Karpichev I V, Luo Y, Russell C M, Small G M. A complex containing two transcription factors regulates peroxisome proliferation and the coordinate induction of β-oxidation enzymes in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:69–80. doi: 10.1128/mcb.17.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Keleher C A, Redd M J, Schultz J, Carlson M, Johnson A D. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992;68:709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- 167.Kelly R, Kwon-Chung K J. A zinc finger protein from Candida albicans is involved in sucrose utilization. J Bacteriol. 1992;174:222–232. doi: 10.1128/jb.174.1.222-232.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kim J, Michels C A. The MAL63 gene of Saccharomyces cerevisiae encodes a cysteine-zinc finger protein. Curr Genet. 1988;14:319–323. doi: 10.1007/BF00419988. [DOI] [PubMed] [Google Scholar]
- 169.Kim Y-J, Bjorklund S, Li Y, Sayre M H, Kornberg R D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 170.Kingston R E, Bunker C A, Imbalzano A N. Repression and activation by multiprotein complexes that alter chromatin structure. Genes Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- 171.Koleske A J, Young R A. An RNA polymerase II holoenzyme responsive to activators. Nature (London) 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 171a.Komachi K, Redd M J, Johnson A D. The WD repeats of Tup1 interact with the homeo domain protein α2. Genes Dev. 1994;8:2857–2867. doi: 10.1101/gad.8.23.2857. [DOI] [PubMed] [Google Scholar]
- 172.Kominami K, DeMartino G N, Moomaw C R, Slaughter C A, Shimbara N, Fujimuro M, Yokosawa H, Hisamatsu H, Tanahashi N, Shimizu Y, Tanaka K, Toh-e A. Nin1p, a regulatory subunit of the 26S proteasome, is necessary for activation of Cdc28p kinase of Saccharomyces cerevisiae. EMBO J. 1995;14:3105–3115. doi: 10.1002/j.1460-2075.1995.tb07313.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kratzer S, Schüller H-J. Carbon source-dependent regulation of the acetyl-coenzyme A synthetase-encoding gene ACS1 from Saccharomyces cerevisiae. Gene. 1995;161:75–79. doi: 10.1016/0378-1119(95)00289-i. [DOI] [PubMed] [Google Scholar]
- 173a.Kratzer S, Schüller H-J. Transcriptional control of the yeast acetyl-coenzyme A synthetase gene, ACS1, by the positive regulators CAT8 and ADR1 and the pleiotropic repressor UME6. Mol Microbiol. 1997;26:631–641. doi: 10.1046/j.1365-2958.1997.5611937.x. [DOI] [PubMed] [Google Scholar]
- 174.Kriegel T M, Rush J, Vojtek A B, Clifton D, Fraenkel D G. In vivo phosphorylation site of hexokinase 2 in Saccharomyces cerevisiae. Biochemistry. 1994;33:148–152. doi: 10.1021/bi00167a019. [DOI] [PubMed] [Google Scholar]
- 175.Kruckeberg A L. The hexose transporter family of Saccharomyces cerevisiae. Arch Microbiol. 1996;166:283–292. doi: 10.1007/s002030050385. [DOI] [PubMed] [Google Scholar]
- 176.Kuchin S, Yeghiayan P, Carlson M. Cyclin-dependent protein kinase and cyclin homologs SSN3 and SSN8 contribute to transcriptional control in yeast. Proc Natl Acad Sci USA. 1995;92:4006–4010. doi: 10.1073/pnas.92.9.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kuo M-H, Brownell J E, Sobel R E, Ranalli T A, Cook R G, Edmondson D E, Roth S Y, Allis C D. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature (London) 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- 178.Kuzhandaivelu N, Jones W K, Martin A K, Dickson R C. The signal for glucose repression of the lactose-galactose regulon is amplified through subtle modulation of transcription of the Kluyveromyces lactis Kl-GAL4 activator gene. Mol Cell Biol. 1992;12:1924–1931. doi: 10.1128/mcb.12.5.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lafuente, M. J., and C. Gancedo. Unpublished data.
- 180.Lagunas R. Sugar transport in Saccharomyces cerevisiae. FEMS Microbiol Rev. 1993;104:229–242. doi: 10.1016/0378-1097(93)90598-v. [DOI] [PubMed] [Google Scholar]
- 181.Lambrechts M G, Bauer F F, Marmur J, Pretorius I S. Muc1, a mucin-like protein that is regulated by Mss10, is critical for pseudohyphal differentiation in yeast. Proc Natl Acad Sci USA. 1996;93:8419–8424. doi: 10.1073/pnas.93.16.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Lampen J O. External enzymes of yeast: their nature and formation. Antonie Leeuwenhoek. 1968;34:1–18. doi: 10.1007/BF02046409. [DOI] [PubMed] [Google Scholar]
- 183.Lamphier M, Ptashne M. Multiple mechanisms mediate glucose repression of the yeast GAL1 gene. Proc Natl Acad Sci USA. 1992;89:5922–5926. doi: 10.1073/pnas.89.13.5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Laughon A, Gesteland R F. Primary structure of the Saccharomyces cerevisiae GAL4 gene. Mol Cell Biol. 1984;4:260–267. doi: 10.1128/mcb.4.2.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lee Y C, Min S, Gim B S, Kim Y-J. A transcriptional mediator protein that is required for activation of many RNA polymerase II promoters and is conserved from yeast to humans. Mol Cell Biol. 1997;17:4622–4632. doi: 10.1128/mcb.17.8.4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lesage P, Yang X, Carlson M. Analysis of the SIP3 protein identified in a two-hybrid screen for interaction with the SNF1 protein kinase. Nucleic Acids Res. 1994;22:597–603. doi: 10.1093/nar/22.4.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lesage P, Yang X, Carlson M. Yeast SNF1 protein kinase interacts with SIP4, a C6 zinc cluster transcriptional activator: a new role for SNF1 in the glucose response. Mol Cell Biol. 1996;16:1921–1928. doi: 10.1128/mcb.16.5.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Levine J, Tanouye L, Michels C A. The UASMAL is a bidirectional promoter element required for the expression of both the MAL61 and MAL62 genes of the Saccharomyces MAL6 locus. Curr Genet. 1992;22:181–189. doi: 10.1007/BF00351724. [DOI] [PubMed] [Google Scholar]
- 189.Li F N, Johnston M. Grr1 of Saccharomyces cerevisiae is connected to the ubiquitin proteolysis machinery through Skp1: coupling glucose sensing to gene expression and the cell cycle. EMBO J. 1997;16:5629–5638. doi: 10.1093/emboj/16.18.5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Li Y, Bjorklund S, Jiang Y W, Lane W S, Stillman D J, Kornberg R D. Yeast global transcriptional regulators Sin4 and Rgr1 are components of mediator complex/RNA polymerase II holoenzyme. Proc Natl Acad Sci USA. 1995;92:10864–10868. doi: 10.1073/pnas.92.24.10864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Li Y, Bjorklund S, Kim Y-J, Kornberg R D. Yeast RNA polymerase II holoenzyme. Methods Enzymol. 1996;273:172–176. doi: 10.1016/s0076-6879(96)73017-3. [DOI] [PubMed] [Google Scholar]
- 192.Liang H, Gaber R F. A novel signal transduction pathway in Saccharomyces cerevisiae defined by Snf3-regulated expression of HXT6. Mol Biol Cell. 1996;7:1953–1966. doi: 10.1091/mbc.7.12.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Liao S M, Zhang J, Jeffery D A, Koleske A J, Thompson C M, Chao D M, Viljoen M, van Vuuren H J, Young R A. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature (London) 1995;374:193–196. doi: 10.1038/374193a0. [DOI] [PubMed] [Google Scholar]
- 194.Lindley, C., O. Zaragoza, and J. M. Gancedo. Unpublished data.
- 195.Lobo Z, Maitra P K. Physiological role of glucose-phosphorylating enzymes in Saccharomyces cerevisiae. Arch Biochem Biophys. 1977;182:639–645. doi: 10.1016/0003-9861(77)90544-6. [DOI] [PubMed] [Google Scholar]
- 196.Lodi T, Goffrini P, Ferrero I, Donnini C. IMP2, a gene involved in the expression of glucose-repressible genes in Saccharomyces cerevisiae. Microbiology. 1995;141:2201–2209. doi: 10.1099/13500872-141-9-2201. [DOI] [PubMed] [Google Scholar]
- 197.Lohr B, Venkov P, Zlatanova J. Transcriptional regulation in the yeast GAL gene family: a complex genetic network. FASEB J. 1995;9:777–787. doi: 10.1096/fasebj.9.9.7601342. [DOI] [PubMed] [Google Scholar]
- 198.Lombardo A, Cereghino G P, Scheffler I E. Control of mRNA turnover as a mechanism of glucose repression in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2941–2948. doi: 10.1128/mcb.12.7.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Ludin, K., and M. Carlson. 1997. Personal communication.
- 200.Lundin M, Nehlin J O, Ronne H. Importance of a flanking AT-rich region in target site recognition by the GC box-binding zinc finger protein MIG1. Mol Cell Biol. 1994;14:1979–1985. doi: 10.1128/mcb.14.3.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Luo Y, Karpichev I V, Kohanski R A, Small G M. Purification, identification and properties of a Saccharomyces cerevisiae oleate-activated upstream activating sequence-binding protein that is involved in the activation of POX1. J Biol Chem. 1996;271:12068–12075. doi: 10.1074/jbc.271.20.12068. [DOI] [PubMed] [Google Scholar]
- 202.Lutfiyya L L, Johnston M. Two zinc-finger-containing repressors are responsible for glucose repression of SUC2 expression. Mol Cell Biol. 1996;16:4790–4797. doi: 10.1128/mcb.16.9.4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Lutstorf U, Megnet R. Multiple forms of alcohol dehydrogenase in Saccharomyces cerevisiae. Arch Biochem Biophys. 1968;126:933–944. doi: 10.1016/0003-9861(68)90487-6. [DOI] [PubMed] [Google Scholar]
- 204.Ma H, Bloom L M, Dakin S E, Walsh C T, Botstein D. The 15 N-terminal amino acids of hexokinase II are not required for in vivo functions: analysis of a truncated form of hexokinase II in Saccharomyces cerevisiae. Proteins. 1989;5:218–223. doi: 10.1002/prot.340050305. [DOI] [PubMed] [Google Scholar]
- 205.Ma H, Bloom L M, Walsh C T, Botstein D. The residual enzymatic phosphorylation activity of hexokinase II mutants is correlated with glucose repression in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:5643–5649. doi: 10.1128/mcb.9.12.5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Ma J, Ptashne M. The carboxy-terminal 30 amino acids of GAL4 are recognized by GAL80. Cell. 1987;50:137–142. doi: 10.1016/0092-8674(87)90670-2. [DOI] [PubMed] [Google Scholar]
- 207.Maeda T, Mochizuki N, Yamamoto M. Adenylyl cyclase is dispensable for vegetative cell growth in fission yeast Schizosaccharomyces pombe. Proc Natl Acad Sci USA. 1990;87:7814–7818. doi: 10.1073/pnas.87.20.7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Magasanik B. Regulation of nitrogen utilization. In: Jones E W, Pringle J R, Broach J R, editors. The molecular and cellular biology of the yeast Saccharomyces. 2. Gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. pp. 283–317. [Google Scholar]
- 209.Maity S N, Vuorio T, de Crombrugghe B. The B subunit of a rat heteromeric CCAAT-binding transcription factor shows a striking sequence identity with the yeast Hap2 transcription factor. Proc Natl Acad Sci USA. 1990;87:5378–5382. doi: 10.1073/pnas.87.14.5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Makino Y, Yamano K, Kanemaki M, Morikawa K, Kishimoto T, Shimbara N, Tanaka K, Tamura T. SUG1, a component of the 26S proteasome, is an ATPase stimulated by specific mRNAs. J Biol Chem. 1997;272:23201–23205. doi: 10.1074/jbc.272.37.23201. [DOI] [PubMed] [Google Scholar]
- 211.Marbois B N, Clarke C F. The COQ7 gene encodes a protein in Saccharomyces cerevisiae necessary for ubiquinone biosynthesis. J Biol Chem. 1996;271:2995–3004. doi: 10.1074/jbc.271.6.2995. [DOI] [PubMed] [Google Scholar]
- 212.Marcus G A, Horiuchi J, Silverman N, Guarente L. ADA5/SPT20 links the ADA and SPT genes, which are involved in yeast transcription. Mol Cell Biol. 1996;16:3197–3205. doi: 10.1128/mcb.16.6.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Marmorstein R, Carey M, Ptashne M, Harrison S C. DNA recognition by GAL4: structure of a protein-DNA complex. Nature (London) 1992;356:408–414. doi: 10.1038/356408a0. [DOI] [PubMed] [Google Scholar]
- 214.Marshall-Carlson L, Neigeborn L, Coons D, Bisson L, Carlson M. Dominant and recessive suppressors that restore glucose transport in a yeast snf3 mutant. Genetics. 1991;128:505–512. doi: 10.1093/genetics/128.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Martínez-Pastor M T, Marchler G, Schüller C, Marchler-Bauer A, Ruis H, Estruch F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress-response element (STRE) EMBO J. 1996;15:2227–2235. [PMC free article] [PubMed] [Google Scholar]
- 216.Masson J-Y, Ramotar D. The Saccharomyces cerevisiae IMP2 gene encodes a transcriptional activator that mediates protection against DNA damage caused by bleomycin and other oxidants. Mol Cell Biol. 1996;16:2091–2100. doi: 10.1128/mcb.16.5.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Matallana E, Franco L, Pérez-Ortin J E. Chromatin structure of the yeast SUC2 promoter in regulatory mutants. Mol Gen Genet. 1992;231:395–400. doi: 10.1007/BF00292708. [DOI] [PubMed] [Google Scholar]
- 218.Mathieu M, Felenbok B. The Aspergillus nidulans CREA protein mediates glucose repression of the ethanol regulon at various levels through competition with the ALCR-specific transactivator. EMBO J. 1994;13:4022–4027. doi: 10.1002/j.1460-2075.1994.tb06718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Matsumoto K, Adachi Y, Toh-e A, Oshima Y. Function of positive regulatory gene gal4 in the synthesis of galactose pathway enzymes in Saccharomyces cerevisiae: evidence that the GAL81 region codes for part of the gal4 protein. J Bacteriol. 1980;141:508–527. doi: 10.1128/jb.141.2.508-527.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Matsumoto K, Toh-e A, Oshima Y. Isolation and characterization of dominant mutations resistant to catabolite repression of galactokinase synthesis in S. cerevisiae. Mol Cell Biol. 1981;1:83–93. doi: 10.1128/mcb.1.2.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Matsumoto K, Uno I, Toh-e A, Ishikawa T, Oshima Y. Cyclic AMP may not be involved in catabolite repression in Saccharomyces cerevisiae: evidence from mutants capable of utilizing it as an adenine source. J Bacteriol. 1982;150:277–285. doi: 10.1128/jb.150.1.277-285.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Matsumoto K, Uno I, Ishikawa T, Oshima Y. Cyclic AMP may not be involved in catabolite repression in Saccharomyces cerevisiae: evidence from mutants unable to synthesize it. J Bacteriol. 1983;156:898–900. doi: 10.1128/jb.156.2.898-900.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Matsumoto K, Yoshimatsu T, Oshima Y. Recessive mutations conferring resistance to carbon catabolite repression of galactokinase synthesis in Saccharomyces cerevisiae. J Bacteriol. 1983;153:1405–1414. doi: 10.1128/jb.153.3.1405-1414.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Mazón M J, Gancedo J M, Gancedo C. Inactivation of yeast fructose-1,6-bisphosphatase. In vivo phosphorylation of the enzyme. J Biol Chem. 1982;257:1128–1130. [PubMed] [Google Scholar]
- 225.Mazón M J, Gancedo J M, Gancedo C. Phosphorylation and inactivation of yeast fructose bisphosphatase in vivo by glucose and by protein ionophores. A possible role for cAMP. Eur J Biochem. 1982;127:605–608. doi: 10.1111/j.1432-1033.1982.tb06915.x. [DOI] [PubMed] [Google Scholar]
- 226.McCann A K, Hilberg F, Kenworthy P, Barnett J A. An unusual hexose-ATP-kinase with two catabolite sites and a role in carbon catabolite repression in the yeast Schwanniomyces occidentalis. J Gen Microbiol. 1987;133:381–389. [Google Scholar]
- 227.McNabb D S, Xing Y, Guarente L. Cloning of yeast HAP5: a novel subunit of a heterotrimeric complex required for CCAAT binding. Genes Dev. 1995;9:47–58. doi: 10.1101/gad.9.1.47. [DOI] [PubMed] [Google Scholar]
- 228.Mechler K. Galactose metabolism in Saccharomyces cerevisiae: a paradigm of eukaryotic gene regulation. In: Zimmermann F K, Entian K-D, editors. Yeast sugar metabolism. Lancaster, Pa: Technomic; 1997. pp. 235–269. [Google Scholar]
- 229.Mercado J J, Gancedo J M. Regulatory regions in the yeast FBP1 and PCK1 genes. FEBS Lett. 1992;311:110–114. doi: 10.1016/0014-5793(92)81379-z. [DOI] [PubMed] [Google Scholar]
- 230.Mercado J J, Smith R, Sagliocco F A, Brown A J P, Gancedo J M. The levels of yeast gluconeogenic mRNAs respond to several environmental factors. Eur J Biochem. 1994;224:473–481. doi: 10.1111/j.1432-1033.1994.00473.x. [DOI] [PubMed] [Google Scholar]
- 231.Mercado J J, Vincent O, Gancedo J M. Regions in the promoter of the yeast FBP1 gene implicated in catabolite repression may bind the product of the regulatory gene MIG1. FEBS Lett. 1991;291:97–100. doi: 10.1016/0014-5793(91)81112-l. [DOI] [PubMed] [Google Scholar]
- 232.Michels C A, Halmenberger K M, Silvestre Y. Pleiotropic mutations regulating resistance to glucose repression in Saccharomyces cerevisiae are allelic to the structural gene for hexokinase B. J Bacteriol. 1983;153:574–578. doi: 10.1128/jb.153.1.574-578.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Miller S M, Magasanik B. Role of the complex upstream region of the GDH2 gene in nitrogen regulation of the NAD-linked glutamate dehydrogenase in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:6229–6247. doi: 10.1128/mcb.11.12.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Mitchelhill K I, Stapleton D, Gao G, House C, Michell B, Katsis F, Witters L A, Kemp B E. Mammalian AMP-activated protein kinase shares structural and functional homology with the catalytic domain of yeast Snf1 protein kinase. J Biol Chem. 1994;269:2361–2364. [PubMed] [Google Scholar]
- 235.Mulder W, Sholten I H, de Boer R W, Grivell L A. Sequence of the HAP3 transcription factor of Kluyveromyces lactis predicts the presence of a novel 4-cysteine zinc-finger motif. Mol Gen Genet. 1994;245:96–106. doi: 10.1007/BF00279755. [DOI] [PubMed] [Google Scholar]
- 236.Müller D, Holzer H. Regulation of fructose-1,6-bisphosphatase in yeast by phosphorylation/dephosphorylation. Biochem Biophys Res Commun. 1981;103:926–933. doi: 10.1016/0006-291x(81)90899-8. [DOI] [PubMed] [Google Scholar]
- 237.Müller G, Gross E, Wied S, Bandlow W. Glucose-induced sequential processing of a glycosyl-phosphatidylinositol-anchored ectoprotein in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:442–456. doi: 10.1128/mcb.16.1.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Müller S, Boles E, May M, Zimmermann F K. Different internal metabolites trigger the induction of glycolytic gene expression in Saccharomyces cerevisiae. J Bacteriol. 1995;177:4517–4519. doi: 10.1128/jb.177.15.4517-4519.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238a.Myers L C, Gustafsson C M, Bushnell D A, Lui M, Erdjument-Bromage H, Tempst P, Kornberg R. The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev. 1998;12:45–54. doi: 10.1101/gad.12.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Mylin L M, Bhat J P, Hopper J E. Regulated phosphorylation and dephosphorylation of GAL4, a transcriptional activator. Genes Dev. 1989;3:1157–1165. doi: 10.1101/gad.3.8.1157. [DOI] [PubMed] [Google Scholar]
- 240.Needleman B. Control of maltase synthesis in yeast. Mol Microbiol. 1991;5:2079–2084. doi: 10.1111/j.1365-2958.1991.tb02136.x. [DOI] [PubMed] [Google Scholar]
- 241.Nehlin J O, Carlberg M, Ronne H. Control of yeast GAL genes by MIG1 repressor, a transcriptional cascade in the glucose response. EMBO J. 1991;10:3373–3377. doi: 10.1002/j.1460-2075.1991.tb04901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Nehlin J O, Carlberg M, Ronne H. Yeast SKO1 gene encodes a bZIP protein that binds to the CRE motif and acts as a repressor of transcription. Nucleic Acids Res. 1992;20:5271–5278. doi: 10.1093/nar/20.20.5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Nehlin J O, Ronne H. Yeast MIG1 repressor is related to the mammalian early growth response and Wilms’ tumour finger proteins. EMBO J. 1990;9:2891–2898. doi: 10.1002/j.1460-2075.1990.tb07479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Neigeborn L, Carlson M. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics. 1984;108:845–858. doi: 10.1093/genetics/108.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Neigeborn L, Carlson M. Mutations causing constitutive invertase synthesis in yeast: genetic interactions with snf1 mutations. Genetics. 1987;115:247–253. doi: 10.1093/genetics/115.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Neigeborn L, Rubin K, Carlson M. Suppressors of snf2 mutations restore invertase derepression and cause temperature-sensitive lethality in yeast. Genetics. 1986;112:741–753. doi: 10.1093/genetics/112.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Neigeborn L, Schwartzberg P, Reid R, Carlson M. Null mutations in the SNF3 gene of Saccharomyces cerevisiae cause a different phenotype than do previously isolated missense mutations. Mol Cell Biol. 1986;6:3569–3574. doi: 10.1128/mcb.6.11.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.NGuyen C, Bolotin-Fukuhara M, Wésolowski-Louvel M, Fukuhara H. The respiratory system of Kluyveromyces lactis escapes from HAP2 control. Gene. 1995;152:113–115. doi: 10.1016/0378-1119(94)00684-k. [DOI] [PubMed] [Google Scholar]
- 249.Niederacher D, Entian K-D. Isolation and characterization of the regulatory HEX2 gene necessary for glucose repression in yeast. Mol Gen Genet. 1987;206:505–509. doi: 10.1007/BF00428892. [DOI] [PubMed] [Google Scholar]
- 250.Niederacher D, Entian K-D. Characterization of Hex2 protein, a negative regulatory element necessary for glucose repression in yeast. Eur J Biochem. 1991;200:311–319. doi: 10.1111/j.1432-1033.1991.tb16187.x. [DOI] [PubMed] [Google Scholar]
- 251.Niederacher D, Schüller H-J, Grzesitza D, Gütlich H, Hauser H P, Wagner T, Entian K-D. Identification of UAS elements and binding proteins necessary for derepression of Saccharomyces cerevisiae fructose-1,6-bisphosphatase. Curr Genet. 1992;22:363–370. doi: 10.1007/BF00352437. [DOI] [PubMed] [Google Scholar]
- 252.Nojima H, Leem S-H, Araki H, Sakai A, Nakashima N, Kanaoka Y, Ono Y. A novel yeast bZIP protein binding to the CRE motif is a multicopy suppressor for cdc10 mutant of Schizosaccharomyces pombe. Nucleic Acids Res. 1994;22:5279–5288. doi: 10.1093/nar/22.24.5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Nunnari J, Fox T D, Walter P. A mitochondrial protease with two catalytic subunits of nonoverlapping specificities. Science. 1993;262:1997–2004. doi: 10.1126/science.8266095. [DOI] [PubMed] [Google Scholar]
- 254.Ohkura H, Kinoshita N, Minatani S, Toda S, Yanagida M. The fission yeast dis2+ gene required for chromosome disjoining encodes one of two putative type 1 protein phosphatases. Cell. 1989;57:997–1007. doi: 10.1016/0092-8674(89)90338-3. [DOI] [PubMed] [Google Scholar]
- 255.Olesen J, Hahn S, Guarente L. Yeast HAP2 and HAP3 activators both bind to the CYC1 upstream activation site, UAS2, in an interdependent manner. Cell. 1987;51:953–961. doi: 10.1016/0092-8674(87)90582-4. [DOI] [PubMed] [Google Scholar]
- 256.Olesen J T, Fikes J D, Guarente L. The Schizosaccharomyces pombe homolog of Saccharomyces cerevisiae HAP2 reveals selective and stringent conservation of the small essential core protein domain. Mol Cell Biol. 1991;11:611–619. doi: 10.1128/mcb.11.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Olesen J T, Guarente L. The HAP2 subunit of yeast CCAAT transcriptional activator contains adjacent domains for subunit association and DNA recognition: model for the HAP2/3/4 complex. Genes Dev. 1990;4:1714–1729. doi: 10.1101/gad.4.10.1714. [DOI] [PubMed] [Google Scholar]
- 258.Östling J, Carlberg M, Ronne H. Functional domains in the Mig1 repressor. Mol Cell Biol. 1996;16:753–761. doi: 10.1128/mcb.16.3.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258a.Östling J, Ronne H. Negative control of the Mig1p repressor by Snf1p-dependent phosphorylation in the absence of glucose. Eur J Biochem. 1998;252:162–168. doi: 10.1046/j.1432-1327.1998.2520162.x. [DOI] [PubMed] [Google Scholar]
- 258b.Özcan, S., J. Dover, and M. Johnston. Personal communication.
- 259.Özcan S, Dover J, Rosenwald A G, Wölfl S, Johnston M. Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc Natl Acad Sci USA. 1996;93:12428–12432. doi: 10.1073/pnas.93.22.12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Özcan S, Freidel K, Leuker A, Ciriacy M. Glucose uptake and catabolite repression in dominant HTR1 mutants of Saccharomyces cerevisiae. J Bacteriol. 1993;175:5520–5528. doi: 10.1128/jb.175.17.5520-5528.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Özcan S, Johnston M. Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Mol Cell Biol. 1995;15:1564–1572. doi: 10.1128/mcb.15.3.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Özcan S, Johnston M. Two different repressors collaborate to restrict expression of the yeast glucose transporter genes HXT2 and HXT4 to low levels of glucose. Mol Cell Biol. 1996;16:5536–5545. doi: 10.1128/mcb.16.10.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Özcan S, Leong T, Johnston M. Rgt1p of Saccharomyces cerevisiae, a key regulator of glucose-induced genes, is both an activator and a repressor of transcription. Mol Cell Biol. 1996;16:6419–6426. doi: 10.1128/mcb.16.11.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Özcan S, Vallier L G, Flick J S, Carlson M, Johnston M. Expression of the SUC2 gene of Saccharomyces cerevisiae is induced by low levels of glucose. Yeast. 1997;13:127–137. doi: 10.1002/(SICI)1097-0061(199702)13:2<127::AID-YEA68>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 265.Palmieri L, Lasorsa F M, De Palma A, Palmieri F, Runswick M J, Walker J E. Identification of the yeast ACR1 gene product as a succinate-fumarate transporter essential for growth on ethanol or acetate. FEBS Lett. 1997;417:114–118. doi: 10.1016/s0014-5793(97)01269-6. [DOI] [PubMed] [Google Scholar]
- 266.Pavlik P, Simon M, Shuster T, Ruis H. The glycerol kinase (GUT1) gene of Saccharomyces cerevisiae: cloning and characterization. Curr Genet. 1993;24:21–25. doi: 10.1007/BF00324660. [DOI] [PubMed] [Google Scholar]
- 267.Peng Z, Trumbly R J, Reimann E M. Purification and characterization of glycogen synthase from a glycogen-deficient strain of Saccharomyces cerevisiae. J Biol Chem. 1990;265:13871–13877. [PubMed] [Google Scholar]
- 268.Pereira G G, Hollenberg C P. Conserved regulation of the Hansenula polymorpha MOX promoter in Saccharomyces cerevisiae reveals insights in the transcriptional activation by Adr1p. Eur J Biochem. 1996;238:181–191. doi: 10.1111/j.1432-1033.1996.0181q.x. [DOI] [PubMed] [Google Scholar]
- 269.Peterson C L, Dingwall A, Scott M P. Five SWI/SNF gene products are components of a large multiprotein complex required for transcriptional enhancement. Proc Natl Acad Sci USA. 1994;91:2905–2908. doi: 10.1073/pnas.91.8.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Peterson C L, Herskowitz I. Characterization of the yeast SWI, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992;68:573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- 271.Peterson C L, Tamkun J W. The SWI-SNF complex: a chromatin remodeling machine? Trends Biochem Sci. 1995;20:143–146. doi: 10.1016/s0968-0004(00)88990-2. [DOI] [PubMed] [Google Scholar]
- 272.Pinkham J L, Guarente L. Cloning and molecular analysis of the HAP2 locus: a global regulator of respiratory genes in Saccharomyces cerevisiae. Mol Cell Biol. 1985;5:3410–3416. doi: 10.1128/mcb.5.12.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Piña B, Berger S, Marcus G A, Silverman N, Agapite J, Guarente L. ADA3: a gene, identified by resistance to GAL4-VP16, with properties similar to and different from those of ADA2. Mol Cell Biol. 1993;13:5981–5989. doi: 10.1128/mcb.13.10.5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Polakis E S, Bartley W. Changes in the enzyme activities of Saccharomyces cerevisiae during aerobic growth on different carbon sources. Biochem J. 1965;97:284–297. doi: 10.1042/bj0970284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Proft M, Grzesitza D, Entian K-D. Identification and characterization of regulatory elements in the phosphoenolpyruvate carboxykinase gene PCK1 of Saccharomyces cerevisiae. Mol Gen Genet. 1995;246:367–373. doi: 10.1007/BF00288610. [DOI] [PubMed] [Google Scholar]
- 276.Proft M, Kötter P, Hedges D, Bojunga N, Entian K-D. CAT5, a new gene necessary for derepression of gluconeogenic enzymes in Saccharomyces cerevisiae. EMBO J. 1995;14:6116–6126. doi: 10.1002/j.1460-2075.1995.tb00302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Quinn J, Fyrberg A M, Ganster R W, Schmidt M C, Peterson C L. DNA-binding properties of the yeast SWI/SNF complex. Nature (London) 1996;379:844–847. doi: 10.1038/379844a0. [DOI] [PubMed] [Google Scholar]
- 278.Rahner A, Schöler A, Mertens E, Gollwitzer B, Schüller H-J. Dual influence of the yeast Cat1p (Snf1p) protein kinase on carbon source-dependent transcriptional activation of gluconeogenic genes by the regulatory gene CAT8. Nucleic Acids Res. 1996;24:2331–2337. doi: 10.1093/nar/24.12.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Rández-Gil F, Bojunga N, Proft M, Entian K-D. Glucose derepression of gluconeogenic enzymes in Saccharomyces cerevisiae correlates with phosphorylation of the gene activator Cat8p. Mol Cell Biol. 1997;17:2502–2510. doi: 10.1128/mcb.17.5.2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279a.Rández-Gil F, Herrero P, Sanz P, Prieto J A, Moreno F. The bifunctional protein hexokinase 2 has a double cytosolic-nuclear localization in Saccharomyces cerevisiae cells. FEBS Lett. 1998;425:475–478. doi: 10.1016/s0014-5793(98)00289-0. [DOI] [PubMed] [Google Scholar]
- 280.Recht J, Dunn B, Raff A, Osley M A. Functional analysis of histones H2A and H2B in transcriptional repression in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2545–2553. doi: 10.1128/mcb.16.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Redd M J, Arnaud M B, Johnson A D. A complex composed of Tup1 and Ssn6 represses transcription in vitro. J Biol Chem. 1997;272:11193–11197. doi: 10.1074/jbc.272.17.11193. [DOI] [PubMed] [Google Scholar]
- 282.Redd M J, Stark M R, Johnson A D. Accessibility of α2-repressed promoters to the activator Gal4. Mol Cell Biol. 1996;16:2865–2869. doi: 10.1128/mcb.16.6.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Reifenberger E, Boles E, Ciriacy M. Kinetic characterization of individual hexose transporters of Saccharomyces cerevisiae and their relation to the triggering mechanism of glucose repression. Eur J Biochem. 1997;245:324–333. doi: 10.1111/j.1432-1033.1997.00324.x. [DOI] [PubMed] [Google Scholar]
- 284.Reisdorf P, Boy-Marcotte E, Bolotin-Fukuhara M. The MBR1 gene from Saccharomyces cerevisiae is activated by and required for growth under suboptimal conditions. Mol Gen Genet. 1997;255:400–409. doi: 10.1007/s004380050512. [DOI] [PubMed] [Google Scholar]
- 285.Repetto B, Tzagoloff A. Structure and regulation of KGD1, the structural gene for yeast α-ketoglutarate dehydrogenase. Mol Cell Biol. 1989;9:2695–2705. doi: 10.1128/mcb.9.6.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Riley M I, Hopper J E, Johnston S A, Dickson R C. GAL4 of Saccharomyces cerevisiae activates the lactose-galactose regulon of Kluyveromyces lactis and creates a new phenotype: glucose repression of the regulon. Mol Cell Biol. 1987;7:780–786. doi: 10.1128/mcb.7.2.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Roberts S M, Winston F. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada and Gcn5 proteins with the Snf/Swi and Srb/mediator complexes. Genetics. 1997;147:451–465. doi: 10.1093/genetics/147.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Rodriguez, C., and J. M. Gancedo. Unpublished data.
- 289.Ronne H. Glucose repression in fungi. Trends Genet. 1995;11:12–17. doi: 10.1016/s0168-9525(00)88980-5. [DOI] [PubMed] [Google Scholar]
- 290.Rose M, Albig W, Entian K-D. Glucose repression in Saccharomyces cerevisiae is directly associated with hexokinase phosphorylation by hexokinases PI and PII. Eur J Biochem. 1991;199:511–518. doi: 10.1111/j.1432-1033.1991.tb16149.x. [DOI] [PubMed] [Google Scholar]
- 291.Rose M, Entian K-D, Hofmann L, Vogel R D, Mecke D. Irreversible inactivation of Saccharomyces cerevisiae fructose-1,6-bisphosphatase independent of protein phosphorylation at Ser11. FEBS Lett. 1988;241:55–59. doi: 10.1016/0014-5793(88)81030-5. [DOI] [PubMed] [Google Scholar]
- 292.Rothstein R J, Sherman F. Genes affecting the expression of cytochrome c in yeast: genetic mapping and genetic interactions. Genetics. 1980;94:871–889. doi: 10.1093/genetics/94.4.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Rottensteiner H, Kal A J, Filipits M, Binder M, Hamilton B, Tabak H F, Ruis H. Pip2p: a transcriptional regulator of peroxisome proliferation in the yeast Saccharomyces cerevisiae. EMBO J. 1996;15:2924–2934. [PMC free article] [PubMed] [Google Scholar]
- 294.Rubin D M, Coux O, Wefes I, Hengartner C, Young R A, Goldberg A L, Finley D. Identification of the gal4 suppressor Sug1 as a subunit of the yeast 26S proteasome. Nature (London) 1996;379:655–657. doi: 10.1038/379655a0. [DOI] [PubMed] [Google Scholar]
- 295.Ruijter G J, Panneman H, van den Broeck H C, Bennett J M, Visser J. Characterization of the Aspergillus nidulans frA1 mutant: hexose phosphorylation and apparent lack of involvement of hexokinase in glucose repression. FEMS Microbiol Lett. 1996;139:223–228. doi: 10.1111/j.1574-6968.1996.tb08206.x. [DOI] [PubMed] [Google Scholar]
- 296.Sadowski I, Costa C, Dhanawansa R. Phosphorylation of Gal4p at a single C-terminal residue is necessary for galactose-induced transcription. Mol Cell Biol. 1996;16:4879–4887. doi: 10.1128/mcb.16.9.4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Saier M H, Jr, Chauvaux S, Deutscher J, Reizer J, Ye J-J. Protein phosphorylation and regulation of carbon metabolism in Gram-negative versus Gram-positive bacteria. Trends Biochem Sci. 1995;20:267–271. doi: 10.1016/s0968-0004(00)89041-6. [DOI] [PubMed] [Google Scholar]
- 298.Sakai A, Chibazakura T, Shimizu Y, Hishinuma F. Molecular analysis of POP2 gene, a gene required for glucose-derepression of gene expression in Saccharomyces cerevisiae. Nucleic Acids Res. 1992;20:6227–6233. doi: 10.1093/nar/20.23.6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Sakai A, Shimizu Y, Hishinuma F. Isolation and characterization of mutants which show an oversecretion phenotype in Saccharomyces cerevisiae. Genetics. 1988;119:499–506. doi: 10.1093/genetics/119.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Sakai A, Shimizu Y, Kondou S, Chibazakura T, Hishinuma F. Structure and molecular analysis of RGR1, a gene required for glucose repression of Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:4130–4138. doi: 10.1128/mcb.10.8.4130. . (Erratum, 12:4806, 1992.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Saleh A, Lang V, Cook R, Brandl C. Identification of native complexes containing the yeast coactivator/repressor proteins NGG1/ADA3 and ADA2. J Biol Chem. 1997;272:5571–5578. doi: 10.1074/jbc.272.9.5571. [DOI] [PubMed] [Google Scholar]
- 302.Salmeron J M, Jr, Johnston S A. Analysis of the Kluyveromyces lactis positive regulatory gene LAC9 reveals functional homology to, but sequence divergence from, the Saccharomyces cerevisiae GAL4 gene. Nucleic Acids Res. 1986;14:7767–7781. doi: 10.1093/nar/14.19.7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Sanz P, Nieto A, Prieto J A. Glucose repression may involve processes with different sugar kinase requirements. J Bacteriol. 1996;178:4721–4723. doi: 10.1128/jb.178.15.4721-4723.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Sarokin L, Carlson M. Upstream region required for regulated expression of the glucose-repressible SUC2 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:2750–2757. doi: 10.1128/mcb.4.12.2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Sarokin L, Carlson M. Short repeated elements in the upstream regulatory region of the SUC2 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:2324–2333. doi: 10.1128/mcb.6.7.2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Schäfer W, Kalisz H, Holzer H. Evidence for non-vacuolar proteolytic catabolite inactivation of yeast fructose-1,6-bisphosphatase. Biochim Biophys Acta. 1987;925:150–155. doi: 10.1016/0304-4165(87)90104-8. [DOI] [PubMed] [Google Scholar]
- 307.Schöler A, Schüller H-J. Structure and regulation of the isocitrate lyase gene ICL1 from the yeast Saccharomyces cerevisiae. Curr Genet. 1993;23:375–381. doi: 10.1007/BF00312621. [DOI] [PubMed] [Google Scholar]
- 308.Schöler A, Schüller H-J. A carbon source-responsive promoter element necessary for activation of the isocitrate lyase gene ICL1 is common to genes of the gluconeogenic pathway in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:3613–3622. doi: 10.1128/mcb.14.6.3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Schork S M, Bee G, Thumm M, Wolf D H. Site of catabolite inactivation. Nature (London) 1994;369:283–284. doi: 10.1038/369283a0. [DOI] [PubMed] [Google Scholar]
- 310.Schork S M, Thumm M, Wolf D H. Catabolite inactivation of fructose-1,6-bisphosphatase of Saccharomyces cerevisiae. Degradation occurs via the ubiquitin pathway. J Biol Chem. 1995;270:26446–26450. doi: 10.1074/jbc.270.44.26446. [DOI] [PubMed] [Google Scholar]
- 311.Schüller H-J, Entian K-D. Extragenic suppressors of yeast glucose derepression mutants leading to constitutive synthesis of several glucose-repressible enzymes. J Bacteriol. 1991;173:2045–2052. doi: 10.1128/jb.173.6.2045-2052.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Schulte F, Ciriacy M. HTR1/MTH1 encodes a repressor for HXT genes. Yeast. 1995;11(Spec. Issue):S239. [Google Scholar]
- 313.Schultz J, Carlson M. Molecular analysis of SSN6, a gene functionally related to SNF1 protein kinase of Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:3637–3645. doi: 10.1128/mcb.7.10.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Schultz J, Marshall-Carlson L, Carlson M. The N-terminal TPR region is the functional domain of SSN6, a nuclear phosphoprotein of Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:4744–4756. doi: 10.1128/mcb.10.9.4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Shaw S P, Wingfield J, Dorsey M J, Ma J. Identifying a species-specific region of yeast TFIIB in vivo. Mol Cell Biol. 1996;16:3651–3657. doi: 10.1128/mcb.16.7.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Sherwood P W, Carlson M. Mutations in GSF1 and GSF2 alter glucose signaling in Saccharomyces cerevisiae. Genetics. 1997;147:557–566. doi: 10.1093/genetics/147.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Shi X, Chang M, Wolf A J, Chang C-H, Frazer-Abel A A, Wade P A, Burton Z F, Jaehning J A. Cdc73p and Paf1p are found in a novel RNA polymerase II-containing complex distinct from the Srbp-containing holoenzyme. Mol Cell Biol. 1997;17:1160–1169. doi: 10.1128/mcb.17.3.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Shi X, Finkelstein A, Wolf A J, Wade P A, Burton Z F, Jaehning J A. Paf1p, an RNA polymerase II-associated factor in Saccharomyces cerevisiae, may have both positive and negative roles in transcription. Mol Cell Biol. 1996;16:669–676. doi: 10.1128/mcb.16.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Sikorski R S, Boguski M S, Goebl M, Hieter P. A repeating aminoacid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell. 1990;60:307–317. doi: 10.1016/0092-8674(90)90745-z. [DOI] [PubMed] [Google Scholar]
- 320.Simon M M, Adam G, Rapatz W, Spevak W, Ruis H. The Saccharomyces cerevisiae ADR1 gene is a positive regulator of transcription of genes encoding peroxisomal proteins. Mol Cell Biol. 1991;11:699–704. doi: 10.1128/mcb.11.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Simon M M, Binder M, Adam G, Hartig A, Ruis H. Control of peroxisome proliferation in Saccharomyces cerevisiae by ADR1, SNF1 (CAT1, CCR1) and SNF4 (CAT3) Yeast. 1992;8:303–309. doi: 10.1002/yea.320080407. [DOI] [PubMed] [Google Scholar]
- 322.Simon M M, Pavlik P, Hartig A, Binder M, Ruis H, Cook W J, Denis C L, Schanz B. A C-terminal region of the Saccharomyces cerevisiae transcription factor ADR1 plays an important role in the regulation of peroxisome proliferation by fatty acids. Mol Gen Genet. 1995;249:289–296. doi: 10.1007/BF00290529. [DOI] [PubMed] [Google Scholar]
- 323.Sirenko O I, Ni B, Needleman R B. Purification and binding properties of the Mal63p activator of Saccharomyces cerevisiae. Curr Genet. 1995;27:509–516. doi: 10.1007/BF00314440. [DOI] [PubMed] [Google Scholar]
- 324.Song W, Treich I, Qian N, Kuchin S, Carlson M. SSN genes that affect transcriptional repression in Saccharomyces cerevisiae encode SIN4, ROX3, and SRB proteins associated with RNA polymerase II. Mol Cell Biol. 1996;16:115–120. doi: 10.1128/mcb.16.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Sprague G F, Jr, Cronan J E., Jr Isolation and characterization of Saccharomyces cerevisiae mutants defective in glycerol catabolism. J Bacteriol. 1977;129:1335–1342. doi: 10.1128/jb.129.3.1335-1342.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Stanway C A, Gibbs J M, Berardi E. Expression of the FOX1 gene of Saccharomyces cerevisiae is regulated by carbon source, but not by the known glucose repression genes. Curr Genet. 1995;27:404–408. doi: 10.1007/BF00311208. [DOI] [PubMed] [Google Scholar]
- 327.Stettler S, Warbrick E, Prochnik S, Mackie S, Fantes P. The wis1 signal transduction pathway is required for expression of cAMP-repressed genes in fission yeast. J Cell Sci. 1996;109:1927–1935. doi: 10.1242/jcs.109.7.1927. [DOI] [PubMed] [Google Scholar]
- 328.Stone G, Sadowski I. GAL4 is regulated by a glucose-responsive functional domain. EMBO J. 1993;12:1375–1385. doi: 10.1002/j.1460-2075.1993.tb05782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Struhl K. Chromatin structure and RNA polymerase II connection: implications for transcription. Cell. 1996;84:179–182. doi: 10.1016/s0092-8674(00)80970-8. [DOI] [PubMed] [Google Scholar]
- 330.Stuart J S, Frederick D L, Varner C M, Tatchell K. The mutant type 1 protein phosphatase encoded by glc7-1 from Saccharomyces cerevisiae fails to interact productively with the GAC1-encoded regulatory subunit. Mol Cell Biol. 1994;14:896–905. doi: 10.1128/mcb.14.2.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Surosky R T, Strich R, Esposito R E. The yeast UME5 gene regulates the stability of meiotic mRNAs in response to glucose. Mol Cell Biol. 1994;14:3446–3458. doi: 10.1128/mcb.14.5.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Suzuki Y, Nogi Y, Abe A, Fukasawa T. GAL11 protein, an auxiliary transcription activator for genes encoding galactose-metabolizing enzymes in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:4991–4999. doi: 10.1128/mcb.8.11.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Suzuki-Fujimoto T, Fukuma M, Yano K-I, Sakurai H, Vonika A, Johnston S A, Fukasawa T. Analysis of the galactose signal transduction pathway in Saccharomyces cerevisiae: interaction between Gal3p and Gal80p. Mol Cell Biol. 1996;16:2504–2508. doi: 10.1128/mcb.16.5.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Swaffield J C, Bromberg J F, Johnston S A. Alterations in a yeast protein resembling HIV Tat-binding protein relieve requirement for an acidic activator domain in GAL4. Nature (London) 1992;357:698–700. doi: 10.1038/357698a0. [DOI] [PubMed] [Google Scholar]
- 335.Swaffield J C, Melcher K, Johnston S A. A highly conserved ATPase protein as a mediator between acidic activation domains and the TATA-binding protein. Nature (London) 1995;374:88–91. doi: 10.1038/374088a0. [DOI] [PubMed] [Google Scholar]
- 335a.Swaffield J C, Melcher K, Johnston S A. A highly conserved ATPase protein as a mediator between acidic activation domains and the TATA-binding protein (correction) Nature (London) 1996;379:658. doi: 10.1038/374088a0. [DOI] [PubMed] [Google Scholar]
- 336.Swanson M S, Winston F. SPT4, SPT5 and SPT6 interactions: effects on transcription and viability in Saccharomyces cerevisiae. Genetics. 1992;132:325–336. doi: 10.1093/genetics/132.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336a.Tanaka N, Ohuchi N, Mukai Y, Osaka Y, Ohtani Y, Tabuchi M, Bhuiyan M S A, Fukui H, Harashima S, Takegawa K. Isolation and characterization of an invertase and its repressor gene from Schizosaccharomyces pombe. Biochem Biophys Res Commun. 1998;245:246–253. doi: 10.1006/bbrc.1998.8406. [DOI] [PubMed] [Google Scholar]
- 337.Taylor W E, Young E T. cAMP dependent-phosphorylation and inactivation of yeast transcription factor ADR1 does not affect DNA binding. Proc Natl Acad Sci USA. 1990;87:4098–4102. doi: 10.1073/pnas.87.11.4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Thevelein J M, Hohmann S. Trehalose synthase: guard to the gate of glycolysis in yeast? Trends Biochem Sci. 1995;20:3–10. doi: 10.1016/s0968-0004(00)88938-0. [DOI] [PubMed] [Google Scholar]
- 339.Thompson C M, Koleske A J, Chao D M, Young R A. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell. 1993;73:1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- 340.Thompson-Jaeger S, François J, Gaughran J P, Tatchell K. Deletion of SNF1 affects the nutrient response of yeast and resembles mutations which activate the adenylate cyclase pathway. Genetics. 1991;129:697–706. doi: 10.1093/genetics/129.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Tillman T S, Ganster R W, Jiang R, Carlson M, Schmidt M C. STD1 (MSN3) interacts directly with the TATA-binding protein and modulates transcription of the SUC2 gene of Saccharomyces cerevisiae. Nucleic Acids Res. 1995;23:3174–3180. doi: 10.1093/nar/23.16.3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Trawick J D, Wright R M, Poyton R O. Transcription of yeast COX6, the gene for cytochrome c oxidase subunit VI, is dependent on heme and on the HAP2 gene. J Biol Chem. 1989;264:7005–7008. [PubMed] [Google Scholar]
- 343.Treich I, Cairns B R, de los Santos T, Brewster E, Carlson M. SNF11, a new component of the yeast SNF/SWI complex that interacts with a conserved region of SNF2. Mol Cell Biol. 1995;15:4240–4248. doi: 10.1128/mcb.15.8.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Treitel M A, Carlson M. Repression by SSN6-TUP1 is directed by MIG1, a repressor/activator protein. Proc Natl Acad Sci USA. 1995;92:3132–3136. doi: 10.1073/pnas.92.8.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Trumbly R J. Cloning and characterization of the CYC8 gene mediating glucose repression in yeast. Gene. 1988;73:97–111. doi: 10.1016/0378-1119(88)90316-2. [DOI] [PubMed] [Google Scholar]
- 346.Trumbly R J. Glucose repression in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1992;6:5–21. doi: 10.1111/j.1365-2958.1992.tb00832.x. [DOI] [PubMed] [Google Scholar]
- 347.Tschopp J, Emr S, Field C, Schekman R. GAL2 codes for a membrane-bound subunit of the galactose permease in Saccharomyces cerevisiae. J Bacteriol. 1986;166:313–318. doi: 10.1128/jb.166.1.313-318.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Tu J, Carlson M. The GLC7 type 1 protein phosphatase is required for glucose repression in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:6789–6796. doi: 10.1128/mcb.14.10.6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Tu J, Carlson M. REG1 binds to protein phosphatase type 1 and regulates glucose repression in Saccharomyces cerevisiae. EMBO J. 1995;14:5939–5946. doi: 10.1002/j.1460-2075.1995.tb00282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Tu J, Vallier L J, Carlson M. Molecular and genetic analysis of the SNF7 gene in Saccharomyces cerevisiae. Genetics. 1993;135:17–23. doi: 10.1093/genetics/135.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Tzamarias D, Struhl K. Functional dissection of the yeast Cyc8-Tup1 transcriptional co-repressor complex. Nature (London) 1994;369:758–760. doi: 10.1038/369758a0. [DOI] [PubMed] [Google Scholar]
- 352.Tzamarias D, Struhl K. Distinct TPR motifs of Cyc8 are involved in recruiting the Cyc8-Tup1 co-repressor complex to differentially regulated promoters. Genes Dev. 1995;9:821–831. doi: 10.1101/gad.9.7.821. [DOI] [PubMed] [Google Scholar]
- 353.Umemura K, Atomi H, Kanai T, Takeshita S, Kanayama N, Ueda M, Tanaka A. Derepression of gene expression mediated by the 5′ upstream region of the isocitrate lyase gene of Candida tropicalis is controlled by two distinct regulatory pathways in Saccharomyces cerevisiae. Eur J Biochem. 1997;243:748–752. doi: 10.1111/j.1432-1033.1997.00748.x. [DOI] [PubMed] [Google Scholar]
- 354.Vallari R C, Cook W J, Audino D C, Morgan M J, Jensen D E, Laudano A P, Denis C L. Glucose repression of the yeast ADH2 gene occurs through multiple mechanisms, including control of the protein synthesis of its transcriptional activator, ADR1. Mol Cell Biol. 1992;12:1663–1673. doi: 10.1128/mcb.12.4.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Vallier L G, Carlson M. New SNF genes, GAL11 and GRR1, affect SUC2 expression in Saccharomyces cerevisiae. Genetics. 1991;129:675–684. doi: 10.1093/genetics/129.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Vallier L G, Carlson M. Synergistic release from glucose repression by mig1 and ssn mutations in Saccharomyces cerevisiae. Genetics. 1994;137:49–50. doi: 10.1093/genetics/137.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356a.Vallier L G, Coons D, Bisson L F, Carlson M. Altered regulatory responses to glucose are associated with a glucose transport defect in grr1 mutants of Saccharomyces cerevisiae. Genetics. 1994;136:1279–1285. doi: 10.1093/genetics/136.4.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.van der Plaat J B. Cyclic 3′,5′-adenosine monophosphate stimulates trehalose degradation in baker’s yeast. Biochem Biophys Res Commun. 1974;56:580–587. doi: 10.1016/0006-291x(74)90643-3. [DOI] [PubMed] [Google Scholar]
- 358.van Huijsduijnen X, Li Y, Black D, Matthes H, Benoist C, Mathis D. Co-evolution from yeast to mouse: cDNA cloning of the two NF-Y (CP-1/CBF) subunits. EMBO J. 1990;9:3119–3127. doi: 10.1002/j.1460-2075.1990.tb07509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Varanasi U A, Klis M, Mikesell P B, Trumbly R J. The Cyc8 (Ssn6)-Tup1 corepressor complex is composed of one Cyc8 and four Tup1 subunits. Mol Cell Biol. 1996;16:6707–6714. doi: 10.1128/mcb.16.12.6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Vassarotti A, Friesen J D. Isolation of the fructose-1,6-bisphosphatase gene of the yeast Schizosaccharomyces pombe. J Biol Chem. 1985;260:6348–6353. [PubMed] [Google Scholar]
- 361.Veenhuis M, Mateblowski M, Kunau W-H, Harder W. Proliferation of microbodies in Saccharomyces cerevisiae. Yeast. 1987;3:77–84. doi: 10.1002/yea.320030204. [DOI] [PubMed] [Google Scholar]
- 362.Verdone L, Camilloni G, Di Mauro E, Caserta M. Chromatin remodeling during Saccharomyces cerevisiae ADH2 gene activation. Mol Cell Biol. 1996;16:1978–1988. doi: 10.1128/mcb.16.5.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Vincent A C, Struhl K. ACR1, a yeast ATF/CREB repressor. Mol Cell Biol. 1992;12:5394–5404. doi: 10.1128/mcb.12.12.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Vincent O, Gancedo J M. Analysis of positive elements sensitive to glucose in the promoter of the FBP1 gene from yeast. J Biol Chem. 1995;270:12832–12838. doi: 10.1074/jbc.270.21.12832. [DOI] [PubMed] [Google Scholar]
- 365.Vincent, O., O. Zaragoza, and J. M. Gancedo. Unpublished data.
- 366.Vojtek A B, Fraenkel D G. Phosphorylation of yeast hexokinases. Eur J Biochem. 1990;190:371–375. doi: 10.1111/j.1432-1033.1990.tb15585.x. [DOI] [PubMed] [Google Scholar]
- 367.Vuorio T, Maity S N, de Crombrugghe B. Purification and molecular cloning of the “A” chain of a rat heteromeric CCAAT-binding protein. Sequence identity with the yeast Hap3 transcription factor. J Biol Chem. 1990;265:22480–22486. [PubMed] [Google Scholar]
- 368.Walsh M C, Scholte M, Valkier J, Smits H P, van Dam K. Glucose sensing and signalling properties in Saccharomyces cerevisiae require the presence of at least two members of the glucose transporter family. J Bacteriol. 1996;178:2593–2597. doi: 10.1128/jb.178.9.2593-2597.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Wang J, Needleman R. Removal of a Mig1p binding site converts a MAL63 constitutive mutant derived by interchromosomal gene conversion to glucose insensitivity. Genetics. 1996;142:51–63. doi: 10.1093/genetics/142.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Wang J, Sirenko O, Needleman R. Genomic footprinting of Mig1p in the MAL62 promoter. Binding is dependent upon carbon source and competitive with the Mal63p activator. J Biol Chem. 1997;272:4613–4622. doi: 10.1074/jbc.272.7.4613. [DOI] [PubMed] [Google Scholar]
- 371.Wedlock D N, Thornton R J. A hexokinase associated with catabolite repression in Pachysolen tannophilus. J Gen Microbiol. 1989;135:2013–2018. [Google Scholar]
- 372.Weirich J, Goffrini P, Kuger P, Ferrero I, Breunig K D. Influence of mutations in hexose-transporter genes on glucose repression in Kluyveromyces lactis. Eur J Biochem. 1997;249:248–257. doi: 10.1111/j.1432-1033.1997.t01-1-00248.x. [DOI] [PubMed] [Google Scholar]
- 373.Wigler M, Field J, Powers S, Broek D, Toda T, Cameron S, Nikawa J, Michaeli T, Colicelli J, Ferguson K. Studies of RAS function in the yeast Saccharomyces cerevisiae. Cold Spring Harbor Symp Quant Biol. 1988;53:649–655. doi: 10.1101/sqb.1988.053.01.074. [DOI] [PubMed] [Google Scholar]
- 374.Williams F E, Trumbly R J. Characterization of TUP1, a mediator of glucose repression in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:6500–6511. doi: 10.1128/mcb.10.12.6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Williams F E, Varanasi U, Trumbly R J. The CYC8 and TUP1 proteins involved in glucose repression in Saccharomyces cerevisiae are associated in a protein complex. Mol Cell Biol. 1991;11:3307–3316. doi: 10.1128/mcb.11.6.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Wilson C J, Chao D M, Imbalzano A N, Schnitzler G R, Kingston R E, Young R A. RNA polymerase II holoenzyme contains SWI/SNF regulators involved in chromatin remodeling. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- 377.Wilson W A, Hawley S A, Hardie D G. Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions and this correlates with a high AMP/ATP ratio. Curr Biol. 1996;6:1426–1434. doi: 10.1016/s0960-9822(96)00747-6. [DOI] [PubMed] [Google Scholar]
- 378.Winston F, Carlson M. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 1992;8:387–391. doi: 10.1016/0168-9525(92)90300-s. [DOI] [PubMed] [Google Scholar]
- 379.Woods A, Cheung P C F, Smith F C, Davison M D, Scott J, Beri R K, Carling D. Characterization of AMP-activated protein kinase β and γ subunits. Assembly of the heterotrimeric complex in vitro. J Biol Chem. 1996;271:10282–10290. doi: 10.1074/jbc.271.17.10282. [DOI] [PubMed] [Google Scholar]
- 380.Woods A, Munday M R, Scott J, Yang X, Carlson M, Carling D. Yeast SNF1 is functionally related to mammalian AMP-activated protein kinase and regulates acetyl-CoA carboxylase in vivo. J Biol Chem. 1994;269:19509–19515. [PubMed] [Google Scholar]
- 381.Wray L V, Jr, Witte M M, Dickson R C, Riley M I. Characterization of a positive regulatory gene, LAC9, that controls induction of the lactose-galactose regulon of Kluyveromyces lactis: structural and functional relationships to GAL4 of Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:1111–1121. doi: 10.1128/mcb.7.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Wright R M, Poyton R O. Release of two Saccharomyces cerevisiae cytochrome genes, COX6 and CYC1, from glucose repression requires the SNF1 and SSN6 gene products. Mol Cell Biol. 1990;10:1297–1300. doi: 10.1128/mcb.10.3.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Xing Y, Fikes J D, Guarente L. Mutations in yeast HAP2/HAP3 define a hybrid CCAAT box binding domain. EMBO J. 1993;12:4647–4655. doi: 10.1002/j.1460-2075.1993.tb06153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Xing Y, Zhang S, Olesen J T, Guarente L. Subunit interaction in the CCAAT-binding heteromeric complex is mediated by a very short α-helix in HAP2. Proc Natl Acad Sci USA. 1994;91:3009–3013. doi: 10.1073/pnas.91.8.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Yang X, Hubbard J A, Carlson M. A protein kinase substrate identified by the two-hybrid system. Science. 1992;257:680–682. doi: 10.1126/science.1496382. [DOI] [PubMed] [Google Scholar]
- 386.Yang X, Jiang R, Carlson M. A family of proteins containing a conserved domain that mediates interaction with the yeast SNF1 protein kinase complex. EMBO J. 1994;13:5878–5886. doi: 10.1002/j.1460-2075.1994.tb06933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Yang X, Ogryzko V, Nishikawa J, Howard B, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral E1A oncoprotein. Nature (London) 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 388.Yano K-I, Fukusawa T. Galactose-dependent reversible interaction of Gal3p with Gal80p in the induction pathway of Gal4p-activated genes of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:1721–1726. doi: 10.1073/pnas.94.5.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Yeghiayan P, Tu J, Vallier L G, Carlson M. Molecular analysis of the SNF8 gene of Saccharomyces cerevisiae. Yeast. 1995;11:219–224. doi: 10.1002/yea.320110304. [DOI] [PubMed] [Google Scholar]
- 390.Yin Z, Smith R J, Brown A J P. Multiple signalling pathways trigger the exquisite sensitivity of yeast gluconeogenic mRNAs to glucose. Mol Microbiol. 1996;20:751–764. doi: 10.1111/j.1365-2958.1996.tb02514.x. [DOI] [PubMed] [Google Scholar]
- 391.Yu J, Donoviel M S, Young E T. Adjacent upstream activation sequence elements synergistically regulate transcription of ADH2 in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:34–42. doi: 10.1128/mcb.9.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Zachariae W, Kuger P, Breunig K D. Glucose repression of lactose/galactose metabolism in Kluyveromyces lactis is determined by the concentration of the transcriptional activator LAC9 (KlGAL4) Nucleic Acids Res. 1993;21:69–77. doi: 10.1093/nar/21.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Zaragoza, O., C. Rodriguez, and C. Gancedo. Unpublished data.
- 394.Zenke F T, Zachariae W, Lunkes A, Breunig K D. Gal80 proteins of Kluyveromyces lactis and Saccharomyces cerevisiae are highly conserved but contribute differently to glucose repression of the galactose regulon. Mol Cell Biol. 1993;13:7566–7576. doi: 10.1128/mcb.13.12.7566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Zenke F T, Engels R, Vollenbroich V, Meyer J, Hollenberg C P, Breunig K D. Activation of Gal4p by galactose-dependent interaction of galactokinase and Gal80p. Science. 1996;272:1662–1665. doi: 10.1126/science.272.5268.1662. [DOI] [PubMed] [Google Scholar]
- 396.Zhang L, Guarente L. The yeast activator HAP1—a GAL4 family member—binds DNA in a directly repeated orientation. Genes Dev. 1994;8:2110–2119. doi: 10.1101/gad.8.17.2110. [DOI] [PubMed] [Google Scholar]
- 397.Zhang L, Guarente L. Evidence that TUP1/SSN6 has a positive effect on the activity of the yeast activator HAP1. Genetics. 1994;136:813–817. doi: 10.1093/genetics/136.3.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Zimmermann F K, Scheel I. Mutants of Saccharomyces cerevisiae resistant to carbon catabolite repression. Mol Gen Genet. 1977;154:75–82. doi: 10.1007/BF00265579. [DOI] [PubMed] [Google Scholar]
- 399.Zimmermann F K, Kaufmann I, Rasenberger H, Haussmann P. Genetics of carbon catabolite repression in Saccharomyces cerevisiae: genes involved in the derepression process. Mol Gen Genet. 1977;151:95–103. doi: 10.1007/BF00446918. [DOI] [PubMed] [Google Scholar]
- 400.Zitomer R S, Nichols D L. Kinetics of glucose repression of yeast cytochrome c. J Bacteriol. 1978;135:39–44. doi: 10.1128/jb.135.1.39-44.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Zitomer R S, Montgomery D L, Nichols D L, Hall B D. Transcriptional regulation of the yeast cytochrome c gene. Proc Natl Acad Sci USA. 1979;76:3627–3631. doi: 10.1073/pnas.76.8.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]