Abstract
Studies of the budding yeast Saccharomyces cerevisiae have greatly advanced our understanding of the posttranscriptional steps of eukaryotic gene expression. Given the wide range of experimental tools applicable to S. cerevisiae and the recent determination of its complete genomic sequence, many of the key challenges of the posttranscriptional control field can be tackled particularly effectively by using this organism. This article reviews the current knowledge of the cellular components and mechanisms related to translation and mRNA decay, with the emphasis on the molecular basis for rate control and gene regulation. Recent progress in characterizing translation factors and their protein-protein and RNA-protein interactions has been rapid. Against the background of a growing body of structural information, the review discusses the thermodynamic and kinetic principles that govern the translation process. As in prokaryotic systems, translational initiation is a key point of control. Modulation of the activities of translational initiation factors imposes global regulation in the cell, while structural features of particular 5′ untranslated regions, such as upstream open reading frames and effector binding sites, allow for gene-specific regulation. Recent data have revealed many new details of the molecular mechanisms involved while providing insight into the functional overlaps and molecular networking that are apparently a key feature of evolving cellular systems. An overall picture of the mechanisms governing mRNA decay has only very recently begun to develop. The latest work has revealed new information about the mRNA decay pathways, the components of the mRNA degradation machinery, and the way in which these might relate to the translation apparatus. Overall, major challenges still to be addressed include the task of relating principles of posttranscriptional control to cellular compartmentalization and polysome structure and the role of molecular channelling in these highly complex expression systems.
Much of the excitement in research on eukaryotic gene expression in recent years has been generated by work on the steps of this process that follow transcription. Taken literally, posttranscriptional gene expression includes all of the steps downstream of transcription that are involved in the realization of the coding potential of the genome, encompassing processes from mRNA modification and processing through to protein folding, sorting, transport, and turnover. However, this review focuses on the fate of pre-mRNA and mRNA during its path through the nucleus into the cytoplasm and its subsequent translation and degradation. In particular, research on the interactions between the translational apparatus and mRNA has uncovered many forms of posttranscriptional control. Moreover, it has become increasingly apparent that many of these different types of control are, in a number of ways, interdependent or coupled to each other.
The yeast Saccharomyces cerevisiae has played a key role as subject and/or host of increasing numbers of investigations in this area and remains a popular organism because of the ease with which it lends itself to genetic manipulation and analysis, in vivo phenotypic analysis, and biochemical experimentation. Moreover, the extensive nature of current knowledge of this relatively simple eukaryote, combined with the attention it is receiving from programs of intensive analysis at the genome, “transcriptome,” and “proteome” levels (175, 176, 501, 571), places it first in line for achieving the status of being at least close to comprehensively characterized at some future date. All these points underline the importance of baker’s yeast as an organism for study in an area of research like posttranscriptional control, offering, as it does, an increasingly complete picture of how the investigated mechanisms contribute to the physiology and growth of a whole organism. At the same time, it should be remembered that there are aspects of posttranscriptional gene expression that were exclusive discoveries of the yeast research community, including mRNA-specific translational stress responses, mRNA destabilization mediated by upstream open reading frames (uORFs), positive modulation of mitochondrial mRNAs via nucleus-encoded activator proteins, and autocatalytic protein splicing (all of which are discussed in this review).
This review explores the diversity of posttranscriptional control pathways in S. cerevisiae, focusing primarily on those currently known to be mediated or influenced by ribosome-mRNA interactions. Its content and structure reflect the philosophy that the processes of posttranscriptional gene expression should be considered components of a whole rather than being isolated systems. It has therefore been a general aim to examine the interrelationships between the mechanisms underlying posttranscriptional control and their thermodynamic and kinetic consequences at the molecular level. Given that control can be understood only in quantitative terms, the first section sets the stage by considering appropriate theoretical tools for handling control data. In the body of the review, comparisons with analogous prokaryotic and higher eukaryotic systems are made where these highlight key mechanistic principles. Since this review focuses on the issue of control, it does not attempt to serve as a comprehensive compendium of the literature on the cellular components involved. This has inevitably led to the omission of direct citations of many interesting papers, but the reader is encouraged to seek access to these via the cited reviews by other authors.
CONCEPTS OF CONTROL IN GENE EXPRESSION
The rapid development of techniques of molecular biology, biochemistry, genetics, and structural biology over the last few decades has resulted in an explosive increase in the rate of generation of descriptive information relating to cellular components. However, one of the major challenges of contemporary biology is the formulation of physiologically relevant models that describe how these components function in cellular processes. This depends on a successful transition from qualitative to more quantitative representation of living systems which, in turn, requires that biological mechanisms are increasingly described in terms of their thermodynamic and kinetic properties. However, this remains an uneasy interface between the disciplines of the physical and biological sciences, a problem that is exacerbated by the lack of conventions in the use of appropriate terminology. A prime example is the concept of kinetic control as applied to gene expression. As argued previously (361), there already exists a clear definition of control within the framework of metabolic control theory (276, 277), and this provides a suitable basis for unambiguous terminology that can be used to describe posttranscriptional events. This also allows the term regulation to be used in a consistent and unambiguous manner.
Two types of approach to the description and analysis of gene expression pathways will be considered briefly in this review. They offer complementary views that can both be helpful in planning and evaluating quantitative experiments. The systemic approach to metabolic control analysis described by Kacser and Burns (276, 277) was conceived as a means of analyzing complex multienzyme systems which are generally too complex to be amenable to accurate analysis by standard kinetic descriptions of the component reactions. This approach can be usefully applied to the partially processive reactions of pathways such as translation, and some of the concepts are introduced here so that the corresponding terminology can be used later in this review without causing confusion.
The key conceptual tools of the analysis are the coefficients used to define “control” in such a complex system. The most relevant of these in the present context is the control coefficient. This describes the relationship between the activity of each catalytic component (E) and the resulting effect on the flux. The activity variation of each E could be caused by a change in concentration, modulation of its kinetic properties, or the binding of an effector. Each change in a given E component (enzyme) can be expressed as a fractional change, δEi/Ei, and this is reflected in a shift to a new steady-state flux, expressed as δF/F. The ratio of the latter to the former represents a measure of the effectiveness of the imposed change in altering the flux, and under the condition δEi → 0 it represents a definitive property of the system, called the control coefficient: Zi = d ln F/d ln Ei. Zi can theoretically assume any value between 1 and 0: a value near 1 means that Ei has a very strong controlling influence on the overall flux of the system, whereas a value near 0 corresponds to a comparatively minimal contribution to control and probably applies frequently to components of gene expression pathways. An important constraint defined by the Kacser and Burns analysis is the summation principle, which states that the sum of the Zi values must equal 1. Most importantly, this type of analysis emphasizes that control is distributed among the respective Eis, with the relative individual contributions being reflected in their respective Zi values. Accordingly, the use of the term “rate-limiting” for any chosen step or Ei in the vast majority of living systems is likely to be misleading and can be meaningless. The Zi values are determined by a range of factors; in a multienzyme pathway, for example, these factors include the distance of each enzyme from substrate saturation, enzyme concentration, the relationship between the mass-action ratio and the equilibrium constant for each step, and the role of effectors. Analogous properties of the gene expression pathway also contribute to an equivalent set of Zi values. Unless exceptional forces are at play, there is likely to be selection pressure on a cellular pathway to evolve toward a system in which the Zi values are not excessively unequal. For example, the provision of certain catalytic components in great excess of their required operational capacities generally makes little sense in terms of cellular energetic housekeeping. Whatever the pathway, this treatment tells us that estimates of Zi for the respective components are required so that we can model the balance of control.
The above summary of key points intrinsic to the systemic analysis of multienzyme pathways can be seen as a stepping stone to a more consistent theoretical approach to the even more complex pathways involved in gene expression. As will become apparent, although the transition is not entirely straightforward because of the processive nature of at least some of the reactions, the concepts and terminology are useful for even relatively qualitative descriptions of the pathways under examination. For convenience, this review continues to use the term “control” in its generally accepted sense to describe the factors determining the (constitutive) rates of biological processes. However, when applied in discussions of the kinetic details of specific pathways, “control” is applied in the sense explained above and is not intended to imply exclusive rate-limitation by any given step or entity.
The systemic type of model contrasts with the more conventional approximations of pathway kinetics based on several assumptions regarding sites of strong controlling influence (see, for example, references 173, 339, and 340). This second approach is discussed in connection with the specific posttranscriptional pathways as these are addressed in the review. As will become apparent, the latter type of model for a partially processive pathway can be regarded as a special case of the more general approach, but the assumptions on which it is based require careful scrutiny.
DELIVERING A FUNCTIONAL MRNA TO THE SITE OF TRANSLATION
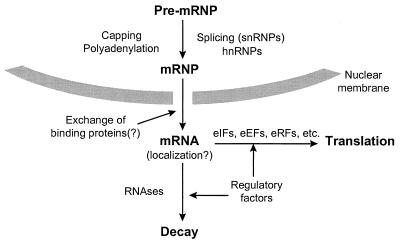
One of the least well understood areas of gene expression is how polymerase II (PolII) transcripts are transported from the sites of their synthesis in the nucleus to the sites where they are translated (Fig. 1) (see, for example, reference 404). The significance of this problem in terms of posttranscriptional control can be seen in a number of ways. First, the rate of export to the cytoplasm influences the steady-state availability of translatable mRNA. Second, pre-mRNA and mRNA interact with a range of splicing components and/or heterogeneous nuclear ribonucleoproteins (hnRNPs) (141, 581), most of which have to be effectively replaced at some stage by ribosomes and translation factors in the cytoplasm if translation is to occur. Some hnRNPs shuttle between the nucleus and the cytoplasm (439), and these might be of particular importance to the architecture and translation of the cytoplasmic mRNPs (596). Third, mRNA export and translation may occur simultaneously, possibly with vectorial and energetic consequences for the export process.
FIG. 1.
Scheme outlining the pathway of eukaryotic transcripts from the nucleus to the sites of translation and decay in the cytoplasm. This review focuses primarily on the posttranscriptional steps of gene expression after nuclear transport. Reproduced from reference 364 with permission of the publisher.
Of these three points, the first two remain at an early stage of characterization and provide us with only a few hints about potential sites of posttranscriptional control on mRNA transport. It is generally agreed that, with very few exceptions, only mature mRNAs (bound by RNA-binding proteins [mRNPs]) leave the nucleus (254, 255). A key feature in this respect is the 5′ cap structure. PolII transcripts are capped with methylated terminal structures (516), comprising in S. cerevisiae either m7G(5′)pppAp (relative frequency, 75%) or m7G(5′)pppGp (25%). Cap methylation is essential for cell viability (351). The cap promotes mRNA export (198), although experiments with an S. cerevisiae strain containing a temperature-sensitive capping enzyme (Ceg1p, which transfers GMP from GTP to the 5′ end of the mRNA) indicate that the cap is not essential for splicing, polyadenylation, or transport (158, 493). In another study, a hammerhead ribozyme was used to catalyze in cis cleavage of a fusion mRNA in S. cerevisiae (144). The capless downstream product was undetectable by standard blotting techniques, as would be expected if the cap is important for nuclear export and/or stability. From the above-mentioned work on CEG1 mutants (158, 493) and further studies described in the section on mRNA stability in this review, it would now seem that the cap influences stability more than transport. At least for histone mRNA, 3′ end formation also stimulates the transport process (143).
A heterodimeric nuclear m7G cap-binding complex (CBC), comprising two cap-binding proteins (CBP20 and CBP80), has been identified (256), but its role is unclear and it is not essential in yeast (404). In a wider context, discussions about whether nuclear mRNA moves through a “track” (461, 601) or via “channelled diffusion” (612) and discussions about the interactions between mRNA and various nuclear components, including the nucleoskeleton (47), spliceosome components (237, 324, 511, 614, 615), nucleolus (537) and nuclear membrane and/or pore complexes (238), all of which could theoretically influence the transport process, are still under way. To what degree the gene expression processes in the nucleoplasm are structurally or functionally compartmentalized is controversial (505).
The third point raises the issue of how mRNAs find their way to translationally competent ribosomes. Perhaps the most relevant data have come from studies of the Balbiani ring granule, a large RNP particle, in the dipteran Chironomus tentans (367). These results indicate that the particle generally exits the nucleus 5′-end first and is bound by ribosomes before the 3′ end passes through the nuclear pore. However, there is no evidence that mRNA export per se requires translation (41). Studies of S. cerevisiae continue to yield new clues about the process of mRNA export. For example, recent evidence indicates that the ATP-dependent RNA helicase Dbp5p, which is a DEAD-box protein closely related to eIF4A, is involved in mRNA export through the nuclear pore complexes (507a, 552a).
While translatable mRNAs are undoubtedly generated primarily by PolII promoters, there is evidence that the pathway outlined above is not the only possible route from nuclear gene to cytoplasmic protein in the eukaryotic cell. Notable in this context is the demonstration that the PolIII promoter of the adenovirus type 2 VA RNAI gene generates uncapped and nonpolyadenylated RNA, which is nevertheless translated, albeit poorly, in HeLa cells (191). This indicates that capping and polyadenylation are not essential for nuclear transport or translation, although it has yet to be determined whether the route taken by such PolIII transcripts may allow them to escape restrictions otherwise imposed on their PolII counterparts. Similarly, it has since been shown that in S. cerevisiae, HIS4 can be transcribed from a PolI promoter to generate primarily uncapped but polyadenylated mRNAs that are poorly translated and rapidly degraded (338a). Overall, these data are in accord with a theme that threads its way through a number of the processes of gene expression: redundancy of function and/or parallel routes provide alternatives for many key steps. What therefore appears to be a major pathway may not be dictated by fixed mechanistic limitations but, rather, may be guided by kinetic or thermodynamic principles of control.
It is clear from the above that there remains considerable uncertainty about the role of any of the nuclear events in posttranscriptional control. Looking beyond the nuclear membrane, there are various lines of evidence for selective distribution of exported mRNA to specific sites within a cell (129, 504, 594) or within whole organisms, for example in Drosophila embryos (504, 589). The fact that mRNA is observed associated with microtubules and actin filaments suggests that these components of the cytoskeleton may be responsible for (selective) mRNA transport (35a). In higher eukaryotes, mRNA partitioning is involved in developmental processes including the establishment of cell polarity and morphogenesis (118, 196, 288) and in the response to signals from the cell surface (89a). Recent evidence also indicates that at least one yeast mRNA (ASH1) becomes localized within the cell by virtue of its association with the cytoskeleton (535). The 3′ untranslated region (3′UTR) of this mRNA is necessary for its transport to the distal tip of daughter buds in postanaphase cells. To what extent mRNA sorting plays a role in the yeast cell cycle remains to be established.
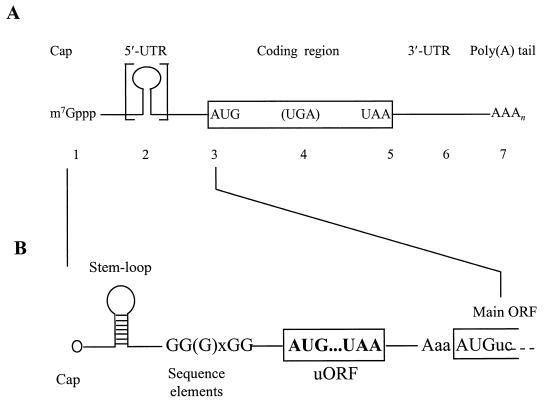
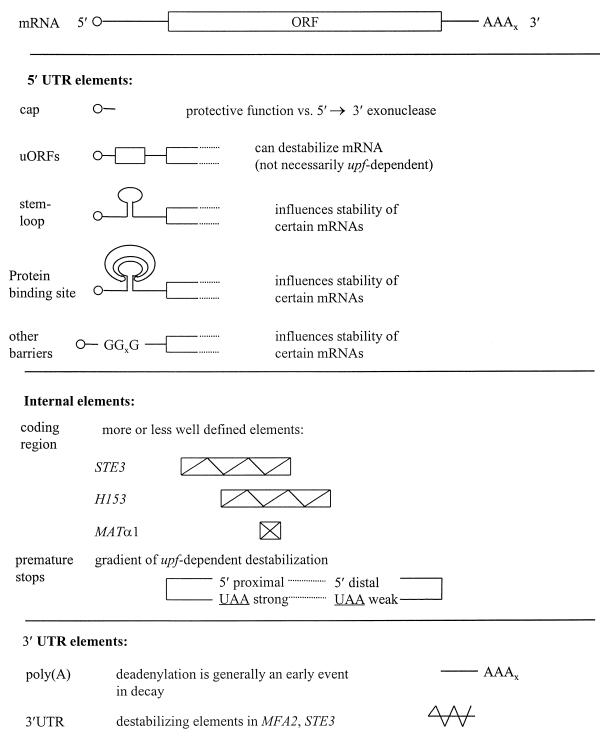
Posttranscriptional gene expression begins with a nascent transcript, which goes through a highly complex series of nuclear interactions before emerging into the cytoplasm. However, each transcript is much more than simply an intermediate carrier of genomic coding sequences. A single mRNA can contain several different types of signal element that contribute to one or more forms of posttranscriptional control (Fig. 2). The 5′- and 3′-terminal modifications have a number of general functions that affect the whole mRNA pool. Apart from its role in nuclear export (198), the cap is required for efficient translation (471, 484) and also influences mRNA stability (162), although the extent to which each function overlaps with the others remains unclear. At the 3′ end, the mRNA carries a poly(A) tail (initially 60 to 90 adenylate residues in yeast [186, 472]), which also influences the cytoplasmic expression and fate of yeast mRNAs (264, 476). The functions of the untranslated, flanking regions of yeast mRNAs have come under increasing scrutiny in recent years since they, unlike the mRNA modifications, can contain a number of signals that modulate the expression of specific genes in individual ways. Finally, the main reading frame of the mRNA not only constitutes a decodable codon sequence, but also can contain further information in the form of linear signals or conformational blueprints that influence posttranscriptional gene expression, although little is known about them at present. Overall, mRNA carries much more information than merely its coding sequence. The central challenge is understanding how this additional information is able to exert its influence via interactions with the cellular machineries responsible for translation and mRNA turnover.
FIG. 2.
Features of yeast mRNAs involved in the translation pathway relevant to control. (A) The 5′UTR stretches from the cap to the AUG start codon (positions 1 to 3). (B) Structural features in the 5′UTR that can influence translational efficiency (and mRNA stability) include secondary structures such as stem-loops and poly(G) sequences and short uORFs. uORFs can have a number of important properties, depending on their structure and sequence environment. The main coding region (positions 3 to 5) can sometimes include an in-frame stop codon that either is avoided by frameshifting or, in aberrant mRNAs, leads to premature termination (and mRNA destabilization). The 3′UTR and poly(A) tail (positions 5 to 7) influence the behavior of posttermination ribosomes at the end of the transcript, and at least the poly(A) tail has been implicated in the control of initiation. All of the numbered sites in panel A can be involved in key events of translation or mRNA turnover or act as targets for control mechanisms. The schemes shown are composites of the features of yeast mRNAs that can be involved in posttranscriptional control. Individual mRNAs differ with respect to the combination of the respective sites present. Panel A reproduced from reference 364 with permission of the publisher.
YEAST TRANSLATION APPARATUS
Review articles dealing with the cellular translation apparatus have appeared very recently for mammalian cells (369, 418, 508) and plants (69, 166) and somewhat less recently for yeast (228, 332, 553), and the reader is directed to these reviews for more detailed information about the individual components of the respective systems. In this section I focus on the properties of yeast translation that have most relevance to the known posttranscriptional control mechanisms and how the latest research has shaped the current view of them. The primary pathway of translation in S. cerevisiae is initiated via a cap-dependent mechanism that seems to follow broadly the main pathway that has been delineated on the basis of biochemical studies with mammalian cell extracts. Initiation is not only the most complex step of translation but also a major site for regulation of individual and global gene expression at this level. The subsequent elongation and termination of the polypeptide chain also follow the same general pattern seen in mammalian cells. The overall similarities between the translation machineries of the higher and lower eukaryotes offer the advantage that results gained with both types of system contribute to a general eukaryotic picture of translation. Nevertheless, yeast translation is by no means a carbon copy of its higher eukaryote counterpart, and the increasingly apparent greater or smaller differences between the respective systems can provide additional insight into the structural basis for specific functions in this process. In the following, “yeast translation” refers to protein synthesis in the cytoplasm. However, it should not be forgotten that a very small proportion of yeast proteins are synthesized in the mitochondria.
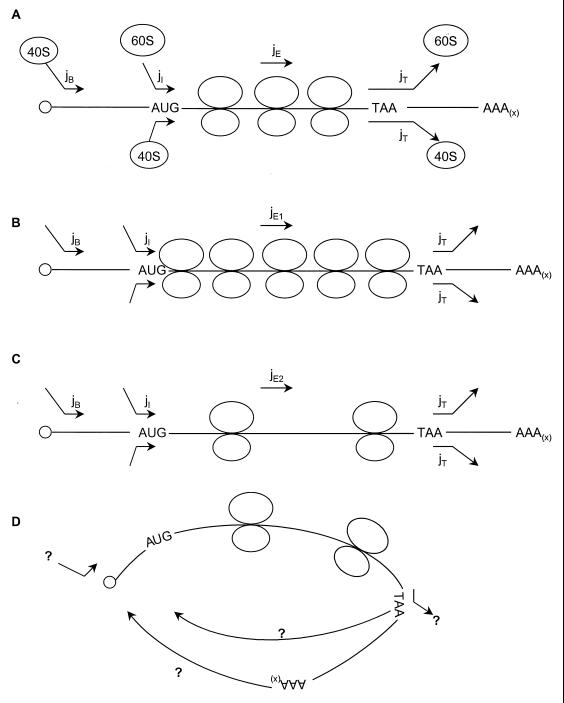
Before moving on to describe the yeast translational apparatus in more detail, it is useful to consider how the three stages of translation are kinetically related to each other (Fig. 3). Their respective kinetic characteristics are relevant to the functions of the individual components of the translation apparatus; the potential contributions of initiation, elongation, and termination to the control of flux through the whole system; and the interdependency of these three phases. A striking aspect of eukaryotic translation is the processive nature of the events occurring after initiation and the at least partly processive nature of the initiation phase. This has consequences for the ways in which rate control can be exercised on translation (Fig. 3).
FIG. 3.
Rate control exercised at different steps of translation. (A) The general scheme indicates the flow rates (j values, in events per unit time for binding and release and in nucleotides per unit time for elongation) assigned to the respective steps of 40S ribosomal subunit binding (jB), 60S junction (jI), elongation (jE), and 40S/60S release (jT). For the purpose of illustrating certain general points, the release rates for 40S and 60S are assumed here to be identical, although this is unlikely to apply to at least some mRNAs. (B) At a low relative rate of elongation (jE1), ribosome packing on the mRNA is high. (C) A reduced packing density occurs at a higher elongation rate (jE2). However, variation in jE need not have a strong effect on overall ribosomal throughput on a given mRNA if the jB and jI rates are not too high. (D) On the other hand, if termination and initiation are coupled, jT may exercise an important control function on translation as a whole.
For example, there are kinetic arguments why initiation can be expected to figure so prominently in terms of posttranscriptional control. Given certain apparently reasonable assumptions, it is relatively easy to arrive at a simplified (nonrigorous) model of the translation pathway (Fig. 3). This scheme summarizes some basic principles concerning potential control points in the protein synthesis pathway. For the sake of simplicity, the steps of translation are represented by flow rates (j values, as defined in Fig. 3). It is assumed that the rate of release of 40S ribosomal subunits from the mRNA during the scanning phase is negligible and that the scanning rate itself is relatively high. The maximal attainable rate of initiation of protein synthesis is dictated by the time taken for the 80S ribosome to clear the AUG region, making space for the next approaching 40S subunit. The region blocked by one ribosome is approximately 30 nucleotides (597); therefore, jI≤jE/30. The jE term used here may be adequately described as an average elongation term, but at least in certain mRNAs it may have to be qualified as referring to only part of the open reading frame (ORF). There is evidence that pausing occurs within eukaryotic reading frames, causing ribosomes to “stack,” at least in certain regions of the mRNA (597). If termination of protein synthesis (jT) were to proceed at a much lower rate than initiation (jI), there would be a blockage that would feed back from the 3′ end of the ORF, distorting the structure of polysomes. This is not known to happen (49), but there is evidence for a pause near the termination codon (597), and so it would seem reasonable to assume that for an mRNA with an unstructured leader, jT ≈ jI. The scanning component of jB is unlikely to be slow compared to jI on unstructured leaders, since this would result in a high loading density of 40S subunits on the 5′UTRs of all mRNAs, for which there is no evidence. Given that pausing seems to occur at the start codon (232, 260, 296, 597), we can assume that jB ≥ jI for an unstructured leader. However, the jB term will be greatly reduced in the presence of structure, thus changing this relationship to jB < jI. The above set of principles and assumptions yields a model in which the individual steps of protein synthesis are well matched, allowing the most efficient throughput of ribosomes on an mRNA molecule.
It should be pointed out, however, that while restrictions on jB, jI, or jT can seriously disturb this balance, changes in jE (at least over a certain range) can be more readily accommodated. In other words, applying the terminology of the Kacser and Burns (276, 277) approach to this processive series of reactions, the control coefficient for elongation is considerably smaller than for the other reactions. A low average rate of elongation (jE1 [Fig. 3B]) may not greatly affect the relative rate of production of complete polypeptide chains on a specific mRNA unless it results in restricted access to initiating ribosomes (jI). Increasing the rate of elongation (jE2 [Fig. 3C]) will also have no effect on the number of polypeptide chains completed in unit time under steady-state conditions unless jI (and/or jT) changes. The major difference between the cases in Fig. 3B and C in the steady state will therefore be the density of ribosomal binding. This, in turn, will influence primarily the number of ribosomes bound up in the process of protein synthesis at any one time and hence the availability of free ribosomal subunits in the cellular pool. Where the rate of elongation on individual mRNAs is modulated, for example via the internal nucleotide sequence (codon usage), the impact on the cellular ribosome pool will be negligible, so that the overall steady-state rate of polypeptide production will be relatively unresponsive. Finally, one way of maintaining tight control over the efficiency of the overall process would be to couple termination and initiation, as might occur in the closed-loop model of polysome structure (Fig. 3D).
These are the chief theoretical constraints required to explain how initiation can feature as a step with strong controlling influence (a large control coefficient). The processive nature of elongation means that translation can be represented by a simplified model in which the elongation process is viewed as a single step (i.e., elongation is represented by simply jE [Fig. 3]). This and other assumptions result in a greatly simplified treatment, which has formed the basis for previous analyses of eukaryotic translation (see, for example, references 173, 339, and 340). One of the questions raised by these analyses is whether the modus of rate control leads to differential attenuation of the translation of individual mRNAs upon shifting a cell from good growth conditions to more restrictive ones. If it is assumed that the poor translation of an mRNA carrying stable secondary structure or a uORF in its leader is based on poor selection of that mRNA by the ribosome, it might be expected that the shift from saturating or near-saturating activity of the translational apparatus to a reduced capacity will affect the poorly translatable mRNAs more severely. However, as discussed below, it is not clear whether selection of an mRNA (i.e., the initial step[s] of initiation) is affected by the structural elements located in the leader. Thus, while the semirigorous analyses reviewed so far bring the issue of control into focus, they do not yet enable us to model translation adequately. This point will be revisited once further information on the translation pathway has been considered.
Initiation Components
As discussed later in this review, eukaryotic translational initiation can occur in at least three ways on cellular mRNAs. By far the most common route is the 5′-end-dependent pathway, in which ribosomes apparently select the initiation site via processive scanning along the 5′ region of the mRNA. The most striking aspect of the cellular system involved in this pathway is the number and complexity of its components. Apart from the subunits of the eukaryotic ribosome, there are at least 11 eukaryotic initiation factors (eIFs) comprising more than 25 polypeptides (369). These are presented in Table 1. Unfortunately, the disparities between the genetic nomenclatures for the respective organisms makes this subject area highly confusing for the non-specialist. One potential solution would be to adopt a new systematic nomenclature that is related in a readily deducible fashion to the biochemical designations. A proposal of this nature has been made for Schizosaccharomyces pombe which could easily be applied generally (Table 1) (334). Of the yeast initiation factors identified so far, eIF4A, eIF4G, and eIF5A have been found to be encoded by duplicated genes. Like many other duplicated genes in S. cerevisiae, they are at least partially phenotypically redundant (180, 333). Indeed, the eIF4A products are identical. It has been suggested that the maintenance of at least some duplicated genes may reflect past adaptation of the organism to a changing environment (595), but to what extent this type of selective pressure is directly applicable to the initiation factor genes is unknown.
TABLE 1.
Translation initiation factors in yeast
| Initiation factor with subunits (reference) | Proposed functiona | S. cerevisiae gene name(s) | Proposed new gene designation(s)b |
|---|---|---|---|
| eIF1d (106, 607) | Met-tRNAi and mRNA binding to 40S | SUI1 | tif1 |
| eif1ad (580) | 40s-60s dissociation; met-trnai binding | tif11 | tif1 |
| eif2c, α, β, γ (94, 135, 200) | aug selection | sui2, sui3, gcd11 | tif211, tif212, tif213 |
| eif2b, α, β, γ, δ, ɛ (226) | guanine nucleotide exchange on eif2 | gcn3, gcd7, gcd1, gcd2, gcd6 | tif221, tif222, tif223, tif224, tif225 |
| eif3, α, β, χ, δ, ɛ, ξ, η, θ (169, 199, 399) | met-trnai and mrna binding to 40s; 40s-60s dissociation | prti, gcd10 | tif31, tif32, tif33, tif34, tif35 |
| eif4a (333) | rna-binding helicase, atpase | tif1, tif2 | tif41a, tif41b |
| eif4b (13, 98) | rna binding promotes helicase | tif3 (stmi) | tif42 |
| eif4e (8, 63) | cap binding | cdc33 | tif45 |
| eif4g1 (p150), eif4g2 (p130) (180) | interactions with eif3, eif4e, and pab1p | tif4631 and tif4632, respectively | tif47a and tif47b, respectively |
| eif4h (458) | stimulation of activities of eif4f components and eif4b | tif48 | |
| eif5 (85) | ejection of eifs | tif5 | tif5 |
| eif5a (490, 620) | function unclear; mutation affects mrna stability | tif51a, tif51b | tif51a, tif51b |
| eif6 (369) | 40s:60s dissociation | tif6 |
these are functions proposed primarily on the basis of in vitro investigations of (partially) purified components.
these are the designations proposed for the s. pombe initiation factors (334).
note the recently discovered existence of yif2 (89b).
mammalian eif1 and eif1a have now been implicated in start codon selection (435a).
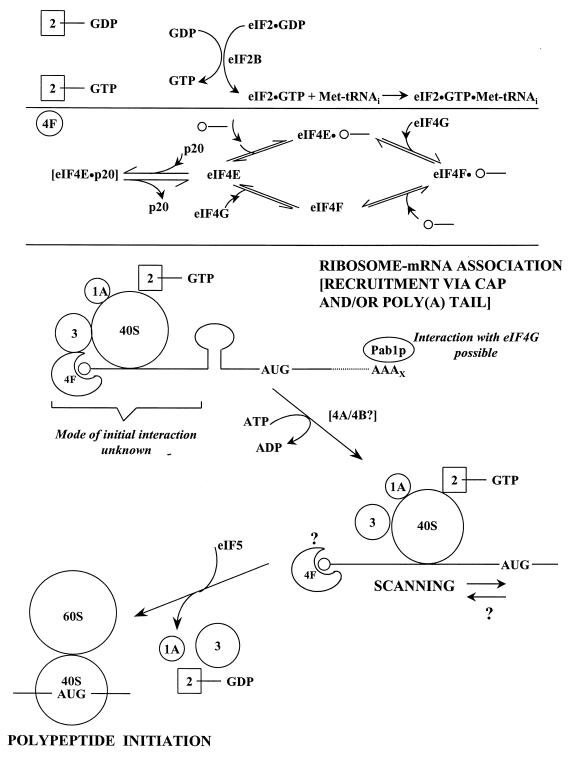
Given the complexity of the initiation process, it has inevitably been easier to characterize partial reactions, each involving a subset of the total pool of eIFs, than to piece together the whole puzzle. The current picture of the pathway derives primarily from experimental work performed on mammalian proteins. The pathway depicted in Fig. 4 has been adapted to take into account the differences so far identified in the yeast system. This undoubtedly simplified version of the real process considers four steps: (i) binding of an active ternary complex to the ribosome; (ii) association of mRNA with the cap-binding complex; (iii) selection of the translational start site; and (iv) initiation of polypeptide synthesis. These steps are discussed in detail below.
FIG. 4.
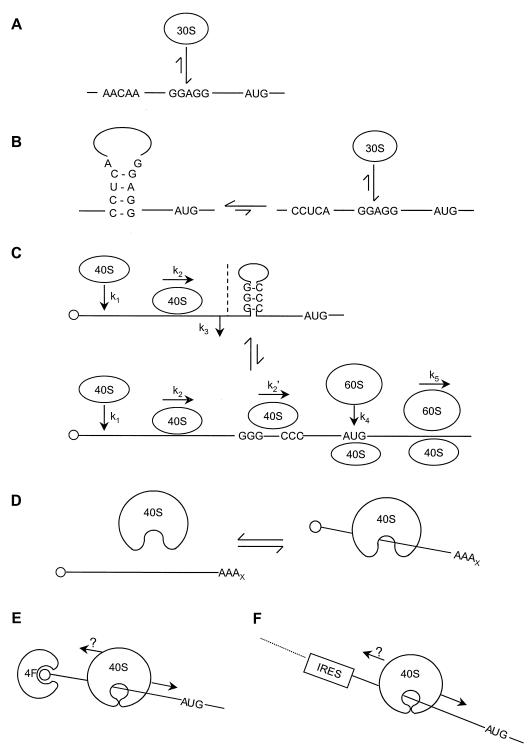
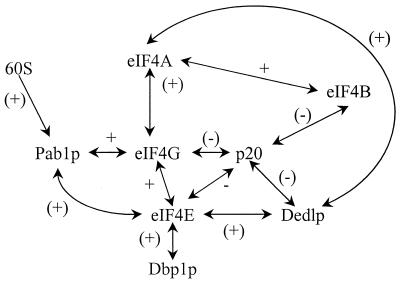
Steps of translational initiation in S. cerevisiae. The recycling of eIF-GDP and the suspected dynamics of eIF4F complex formation are indicated schematically. p20 competes with eIF4G for binding to eIF4E, but the means by which this competitive interaction is regulated has yet to be determined. It is now thought to be primarily eIF4F that binds the cap (see next section). The interaction of Pab1p with eIF4G may provide an alternative route to mRNA-ribosome joining, but the significance of such a process in vivo is unknown. In yeast, the relationship between eIF4A-eIF4B helicase/annealing activity and scanning is still controversial. By analogy to mammalian models, other initiation factors have been included in the 43S preinitiation complex that is thought to perform scanning. Many questions remain the subject of further investigation, such as what happens to eIF4F during each cycle of initiation (see Fig. 8) and how eIF3 promotes initiation. It is not known whether scanning is strictly unidirectional. The release of most of the eIFs and the joining of the ribosomal 60S subunit lead to polypeptide initiation. The recent reports on eIF4H (458) and yIF2 (89b), which are not included in this figure, emphasize that we do not yet know the full complement of factors involved in initiation.
Binding of an active ternary complex to the ribosome.
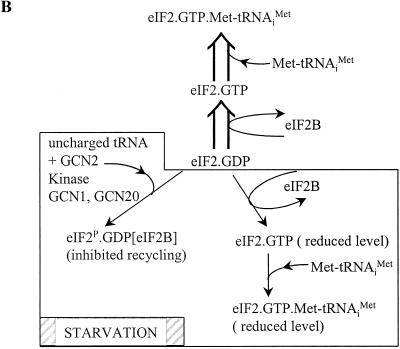
eIF2 is a heterotrimeric complex that is required for the binding of Met-tRNAi and mRNA to ribosomes in vitro. The GTP-charged form of eIF2, which binds Met-tRNAi in vitro to form the 5S ternary complex, is generated in an exchange reaction catalyzed by eIF2B (45, 480). There is also evidence suggesting that eIF2B promotes the cycling of eIF2-GDP off the ribosome during the initiation process (423a). The component subunits of eIF2 (α, β, and γ) are all essential for cell viability, and mutations in them affect start codon selection by the ribosome (93, 185, 369). Moreover, Cigan et al. (93) also used mutations in the anticodon of one of the four tRNAi genes in S. cerevisiae to establish the key role of tRNAi-start codon interactions in initiation site selection. Appropriate compensatory mutations in the anticodon allowed initiation on the HIS4 reading frame by using non-AUG codons that would otherwise not be recognized by the preinitiation complex. The order of the interactions between eIF2–GTP–Met-tRNAi, the ribosome, and the mRNA is not fully resolved. The bulk of the available in vitro data points to 40S–ternary-complex binding preceding mRNA binding, but Trachsel discusses conditions where this might not apply (552). Certainly, the phenomenon of reinitiation constitutes evidence that close association of 40S subunits and mRNA can occur in vivo before the binding of the ternary complex, at least downstream of a termination event (see below). Finally, earlier this year there was an exciting development in this area. S. cerevisiae was found to possess a homologue of E. coli IF2, called yIF2 (89b). The encoding gene, FUN12, is not essential, but its deletion imposes a severe slow-growth phenotype and a marked translation initiation defect. Biochemical experiments indicate that yIF2, like eIF2, functions to promote Met-tRNAi binding to the 40S ribosomal subunit (89b). The tantalizing question yet to be resolved is why S. cerevisiae uses both yIF2 and eIF2; does this represent an intermediate stage in ongoing evolution of the translation system in this organism, or are there particular reasons for the maintenance of these parallel functions?
Association of mRNA with the cap-binding complex.
eIF4E is the cap-binding component of the initiation factor complex eIF4F, anchoring this complex to the 5′ end of capped mRNAs. It is the least variable of the eIF4F components (in terms of presence in the complex and/or protein sequence) and one of the less abundant eIFs (estimated to be present at levels greatly substoichiometric with respect to those of the ribosome [see below]). eIF4E is required for efficient translational initiation in vitro (11), and it is thought that it fulfills the same functional role within the assembled eIF4F complex in vivo. In mammalian eIF4F, eIF4E is associated with two other factors, eIF4G and eIF4A, whereby eIF4G holds the respective factors together in the complex (Fig. 5). Recent reports (121, 168) describe a second human eIF4G gene (encoding eIF4GII), which is 46% identical to the initially cloned gene (eIF4GI [605]) (Fig. 5), and also a second human eIF4E gene, which encodes a protein differing at only two positions. eIF4GII is likely to be a functional homologue of eIF4GI, and there is speculation that the different forms of this protein may be required for a differentiated response to developmental signals (181). The second eIF4E species is likely to be fully functional, and the reason for the duplication has yet to be ascertained. On the other hand, a human protein (4E homologous protein [4EHP]), with 30% identity to eIF4E, has also been described, but as yet it has no apparent function (460a).
FIG. 5.
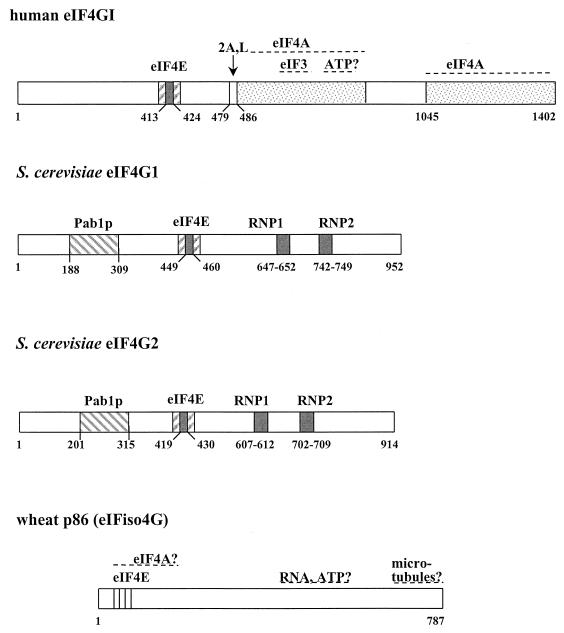
This scheme compares the overall structures and known or predicted binding sites of mammalian eIF4GI (242, 312, 346, 605), its two counterparts in S. cerevisiae (180), and wheat p86 (6, 373). The sites of cleavage by the proteases 2A and L and of the RNA-binding motifs (RNP) are also indicated. The potential eIF3 binding and RNA-binding motifs in yeast proteins have been deduced from sequence comparisons. There is no evidence for the existence of an eIF4A-binding site in the yeast eIF4Gs, whereas there are two such domains in mammalian eIF4G (242). The protein structures are approximately arranged in order to line up the homologous regions. Wheat p86 is thought to have a binding site for microtubules at the C terminus (58). There has been uncertainty about the N-terminal sequence of eIF4GI (181), which is now thought to include a Pabp-binding site (509a). Further examples of eIF4G or of eIF4G-like proteins are discussed in the text.
Mammalian eIF4A and eIF4F exhibit ATP-dependent bidirectional RNA helicase activity that is enhanced by eIF4B (467). There has also been a very recent addition to the eIF4 group of factors, called eIF4H (458). This factor stimulates the activities of eIF4F and eIF4B in a rabbit reticulocyte lysate, although its role in in vivo translation remains unclear. In S. cerevisiae, the isolatable eIF4F complex contains eIF4E and only one other type of initiation factor, eIF4G. The binding domain of the latter has a high affinity for eIF4E (the Kd is estimated by surface plasmon resonance analysis to be approximately 10−9 [449]). However, two other entities, a protein called p20 and the poly(A)-binding protein Pab1p, are also found associated with the eIF4F components of S. cerevisiae; p20 competes with eIF4G for binding to eIF4E, and Pab1p binds to eIF4F via a site on eIF4G (10, 15, 539). Moreover, other interactions are also suspected to occur at least transiently (see the sections on eIF4A and eIF4B below). eIF4G also occurs in two forms in S. cerevisiae, eIF4G1 and eIF4G2, and the former type (p150) is larger than the latter (p130) (180) (Fig. 5).
Mammalian eIF4G is much larger than its yeast homologues and has binding sites for eIF4E, eIF3, and eIF4A (312, 346). It is therefore appealing to regard eIF4G as a sort of docking protein or adapter (214, 312, 383, 479, 509) for the assembly of the complex between mRNA and the preinitiation complex. There is also a further family of eIF4G-like proteins in mammalian systems. For example, one of them, p97, shows 28% identity to the C-terminal two-thirds of eIF4G and thus apparently lacks an eIF4E-binding site (243). Other versions of this 97-kDa protein have been isolated from various sources (see the summary in reference 383). p97 may act as a translational repressor by forming translationally inactive complexes with eIF4A and eIF3, although other properties of this protein may be involved (383). In this context, it should be noted that mammalian eIF4G can be cleaved by viral proteases (e.g., protease L of poliovirus or protease 2A of foot-and-mouth disease virus) to leave a C-terminal product lacking the eIF4E-binding domain (312, 409). The resulting C-domain can support cap-independent or internal ribosome entry site (IRES)-dependent translation in vitro (409). One of the known plant eIF4G factors, p86, is considerably smaller than the other known eIF4G proteins (6, 68, 373). This protein, also known as eIFiso-4G, seems to be lacking much of the N-terminal domain present in its yeast and mammalian counterparts (Fig. 5). It is found associated with eIFiso-4E in the alternative plant complex referred to as eIFiso-4F (68, 373). Despite the inclusion of the much smaller version of eIF4G, the latter complex is functionally equivalent to eIF4F in a range of in vitro assays (320, 373). The other plant eIF4G protein, p220, has yet to be fully characterized (69).
It is not known in which order the eIF4F-mRNA complex is assembled in vivo, nor is the potential functional significance of any given order fully clear. However, the occurrence of cooperativity effects (see later in this section) is likely to dictate a preferred route for the formation of a cap-associated complex. Moreover, the association of eIF4E with eIF4G can be at least partially blocked by the binding of eIF4E-binding proteins (4E-BPs) (421). In S. cerevisiae, there is currently only one candidate for this role, p20 (10, 315), which in fact has a molecular mass of approximately 18 kDa (613). Whether p20 constitutes the full yeast equivalent of a mammalian 4E-BP is discussed in the section on translational regulation (see below).
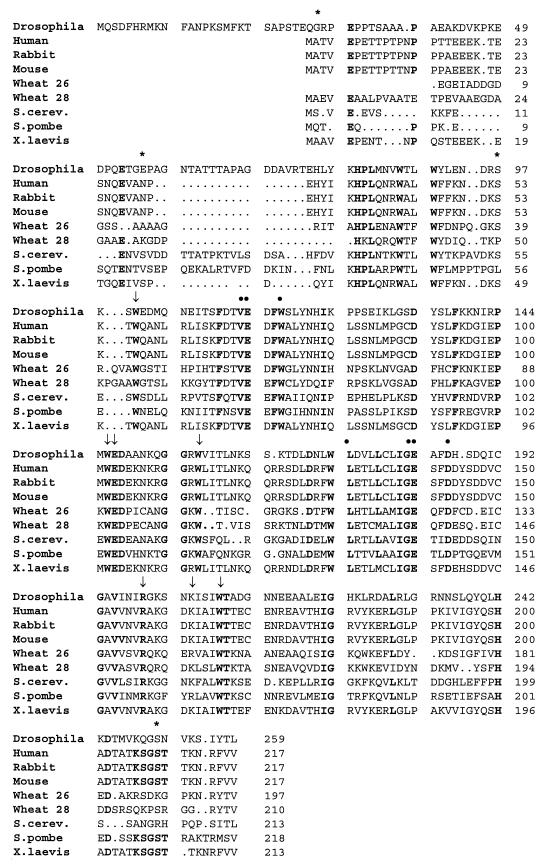
The X-ray structure of an N-terminal truncated form (amino acids 28 to 217) of mouse eIF4E has recently been determined at 2.2-Å resolution (354). The structure correlates well with previous genetic and biochemical data and also provides the basis for a number of important predictions. It is therefore worthwhile considering certain of its details at this point. Since the truncated mouse protein still binds the cap analogue 7-methyl-GDP and since an equivalent N-terminally truncated form of S. cerevisiae eIF4E (amino acids 30 to 213) has been shown to support growth in an otherwise eIF4E-deficient strain of yeast (569), it is clear that the N-terminal region is not essential for the maintenance of (at least partial) structure and function by the eIF4E protein. The determined structure comprises one domain with an overall shape resembling a cupped hand. Within this domain there is an eight-stranded antiparallel β-sheet, three long α-helices, and one short α-helix. The short α-helix and the concave surface of the β-sheet form the cap-binding slot. Of particular interest are the locations of a number of the residues that are conserved between the various eIF4E sequences that have been sequenced so far (Fig. 6). The methylated base of the cap analogue fits between the tryptophan residues at positions 56 and 102, and the interaction is most likely driven by π-π stacking enthalpy as predicted previously by Ishida et al. (246–248; see also reference 9). Apart from these stacking interactions, there are hydrogen bonds or van der Waals contacts between the methylated G of the cap and Trp102, Glu103, and Trp166; direct interactions between the ribose and diphosphate groups of the cap structure and Trp56, Arg157, and Lys162; and water-mediated contacts with Trp166 and Arg112. All of these residues are either fully conserved or subject to only conservative changes between the respective eIF4E species (Fig. 6). The results of mutational studies on a number of these residues in the yeast and human eIF4E proteins, in particular the tryptophans, have generally been consistent with their playing a role in cap binding (9, 382).
FIG. 6.
Conserved sequence and structural motifs in eIF4E. Comparison of eIF4E sequences from a range of different organisms reveals the presence of many strictly conserved or conservatively maintained features (boldface type). A number of these are involved in binding the mRNA cap structure (arrows), while others are surface residues (dots), some of which have the potential to be involved in interactions with other proteins (such as eIF4G, 4E-BPs, or p20) (354, 358, 449). Asterisks mark the positions of serine residues in the respective eIF4E sequences that have either been shown or are suspected to be sites of phosphorylation. The sequences shown belong to eIF4E proteins that bind preferentially to the m7GpppX type of mRNA cap. Caenorhabditis elegans has multiple forms of the cap-binding protein (not shown here), at least two of which also recognize m32,2,7GpppX caps (269a). Relatively few differences in the primary sequences apparently suffice to confer this broader specificity on the C. elegans cap-binding proteins.
Equally interesting are the absolutely conserved surface residues: a nonpolar group of Val69, Trp73, Leu131, and Gly139 and an acidic group of Glu70, Glu140, and Asp143 (354). These surface areas are potential candidates for the binding sites for other factors, including eIF4G and 4E-BP1 (see below). On the other hand, the crystal structure also reveals that Ser209 is located near the cap-binding slot. Moreover, on the basis of model building, Marcotrigiano et al. (354) propose that Lys159, which lies on the other side of the slot, could form a salt bridge with the phosphorylated form of Ser209, perhaps stabilizing the binding of mRNA in the slot. This hypothesis has been tested in vitro using the human protein. It was reported that the addition of anionic charge at the position Ser209 by means of mutation reduced the off-rate of eIF4E from the m7GpppG cap (501a). Moreover, examination of this question in S. pombe may also prove to be highly informative, especially with regard to analysis of the physiological significance of phosphorylation at this site (see below). Overall, a key challenge generated by progress in the X-ray crystallography of initiation factors will be to establish the functional significance of the molecular features that have been identified.
The structure of the S. cerevisiae eIF4E-cap analogue complex was analysed by NMR using a protein-CHAPS micelle (358). While largely similar to the mouse eIF4E X-ray structure, the proposed yeast nuclear magnetic resonance spectroscopy (NMR) structure shows some small differences. Five of the β-strands and one α-helix are shorter in the yeast protein, and Trp58 in the cap-binding site has a different orientation from that of its mouse counterpart. Matsuo et al. (358) also described a complex between yeast eIF4E and mammalian 4E-BP2 and showed that the NMR resonances were particularly perturbed in yeast eIF4E at amino acid positions 32 to 50 and 62 to 79, although the resonances of 13 further amino acids in the region 85 to 169 were also affected. At least some of these are surface residues, and it will now be necessary to establish whether any of them participate directly in 4E-BP binding.
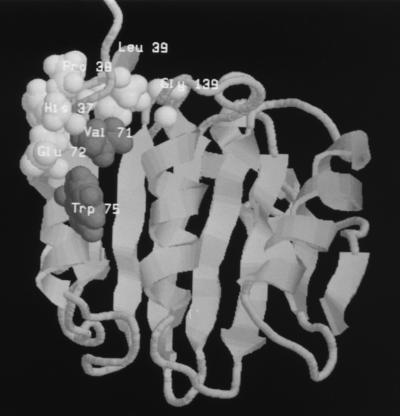
More recent work has combined classical genetic, immunological, and biochemical methods with surface plasmon resonance analysis to obtain information about the residues in yeast eIF4E that contribute to or influence the binding sites for eIF4G and p20 (449). Mutations at HPL37–39, W75, E72, V71, and G139 were found to decrease the affinity of eIF4E for a recombinant protein bearing the eIF4E-binding domain of eIF4G. These residues are all highly conserved among the eIF4E proteins sequenced so far and map to a cluster on eIF4E that is located on the dorsal side of the structure relative to the cap-binding slot (Fig. 7). It is, however, striking that the binding site for p20 overlaps but is not identical to that of eIF4G (Fig. 7).
FIG. 7.
Amino acids involved in binding to eIF4G and p20 map to a predicted surface-accessible cluster on the dorsal surface of S. cerevisiae eIF4E (449). Based on the crystal structure of the mouse (Δ27) eIF4E protein (354) and the NMR structure of yeast eIF4E (358), these groups of residues are predicted to lie together on the opposite face of yeast eIF4E from the cap-binding slot. They belong to α-helices 1 and 2, respectively, or are associated with a β-strand (β1) that follows the variable N-terminal region of the eIF4E sequence. The ribbon model shown here is based on the coordinates of the published NMR structure (358). The view is of the dorsal face angled to show the site clearly. The structure of the N-terminal region of the protein is unclear and has been cut off at the top of this representation. The amino acids that affect the binding of the eIF4E-binding domains of eIF4G and of p20 (V71 and W75) are dark grey. The other amino acids seem to influence only binding to the eIF4E-binding domain of eIF4G (E72, H37, P38, L39, and G139).
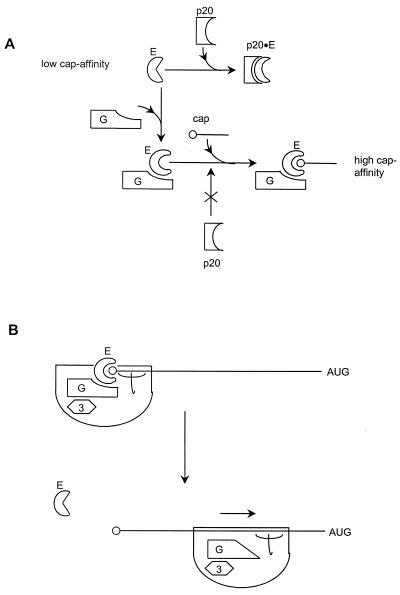
The characterization of binding sites for both eIF4E and eIF3 on eIF4G (Fig. 5) has led to the suggestion that eIF4G mediates mRNA-ribosome association by linking eIF3 and eIF4E (312, 369). Mammalian eIF4G also has two binding sites for eIF4A near its C terminus (Fig. 5) (242), whereas an equivalent site has yet to be identified in S. cerevisiae eIF4G, and eIF4A has not been found to copurify with the yeast complex. It is accordingly not yet clear to what degree the mammalian and yeast eIF4F complexes should be regarded as functionally equivalent. One potentially critical feature of the formation of these various complexes may be that the interactions cause changes in the binding characteristics of the respective components. For example, cross-linking experiments have indicated that the binding of mammalian eIF4G to eIF4E increases the latter factor’s affinity for the 5′ mRNA cap (194), and the cap-binding affinity of S. cerevisiae eIF4E has been estimated to increase at least 10-fold upon binding of the eIF4E-binding domain of eIF4G (449). This means that a tightly bound eIF4E-eIF4G complex is likely to be the primary species interacting with capped mRNA. It is also possible that the binding of eIF4E or other factors to eIF4G modulates the interactions of eIF4G with mRNA (see, for example, references 540 and 541). One attractive possibility is that the influence of interactions within eIF4F on the mRNA affinities of the component proteins underlies the cycling (or regulation) of the cap-binding complex. For example, a high-affinity form of eIF4E (bound to eIF4G) may promote initial 40S interactions with capped mRNA but subsequently convert to the low-affinity form after a rearrangement of the preinitiation complex (Fig. 8) (449).
FIG. 8.
Heterotropic cooperativity in eIF4E and the translational initiation cycle. A recent study has suggested testable models that can explain how p20 regulation (A) and cyclical eIF4F function (B) might be achieved (449). Binding of eIF4G to eIF4E induces a high-affinity cap-binding state in eIF4E (A). This promotes 40S-mRNA interactions and ultimately translational initiation. p20 can bind to part of the eIF4G-binding site on eIF4E (Fig. 7), potentially generating a dead-end complex unable to participate in the eIF4G-mediated initiation pathway. Since p20 binds with a lower affinity to eIF4E, it does not block translation but, rather, exerts fine regulation via competition with eIF4G for a shared site on eIF4E. Measurements of the relative binding affinities between these proteins (449) have provided the basis for understanding how a cyclical cap-eIF4E-binding pathway might function (B). The binding of eIF4G mediates both enhanced cap binding and association of the 40S ribosomal subunit. The relatively high affinity of eIF4G binding to eIF4E ensures that the latter binds to the 5′ cap almost exclusively as part of the eIF4F complex. Subsequently, and perhaps during scanning or as a result of 60S junction, a rearrangement of the preinitiation complex induces dissociation of eIF4E from eIF4G, which results in the loss of the high-affinity cap-binding state in eIF4E. As a result, eIF4E can be released relatively easily from the mRNA, thus becoming free to rebind eIF4G and thus restart another cycle. Reproduced from reference 449 with permission of the publisher.
Finally, the recent advances in our understanding of the structures and interactions of the eIF4F components have raised many new questions, especially about the role of eIF4G. Moreover, beyond the functional complexity of eIF4F itself, there may be other translation components that can perform parallel functions. For example, at least the mammalian eIF4B protein may also be capable of mediating complex formation between mRNA and ribosomes via eIF3 and/or interactions with rRNA (371, 372). It remains to be seen whether this capability acts in concert with the equivalent functions of eIF4G or defines a parallel (alternative) means to the same end.
Recruitment via the poly(A) tail—an alternative route?
Recent work has shown that S. cerevisiae eIF4G1 and eIF4G2 also have N-terminal binding sites for the poly(A)-binding protein (539, 541) (Fig. 5). This and other observations (165, 167, 320, 392) have revitalized the debate about the roles of the poly(A) tail and the poly(A)-binding protein in translation (see, for example, the recent review of this theme by Jacobson [264]). Speculation about possible “long-range” interactions between Pab1p and the ribosome had already been stimulated by the isolation of pab1 suppressor strains that harbored mutations in genes encoding 60S subunit proteins (474). Moreover, as discussed below, there is in vitro evidence that the poly(A) tail participates in the recruitment of ribosomes onto mRNAs via a pathway that can function independently of the cap. Investigations in vivo (445, 446, 541), however, paint a more complex picture (see below). The poly(A)-binding protein of higher and lower eukaryotes has four RNA recognition motifs. These contribute to differing degrees to poly(A)-specific and non-poly(A)-specific RNA binding (72, 114, 306, 403, 478). The second RNA recognition motif of Pab1p is required for binding to eIF4G in S. cerevisiae (284), but the actual site has yet to be defined. Most remarkable is the conclusion from mutagenesis studies that Pab1p does not require its specificity for poly(A) to perform functions necessary for cell viability (114). Moreover, while Pab1p binding is shared by wheat eIF-iso4G (320), it has not been clear whether it is also evident in human eIF4GI/II or S. pombe eIF4G (181, 383). There is, however, now agreement that in the former case the site was originally overlooked because of uncertainties about the N-terminal sequence of human eIF4G (509a). Continuing work on S. pombe eIF4G should also resolve uncertainty about Pab1p binding for this fission yeast (449a). A further study has now complicated the story somewhat, since a 480-amino-acid human poly(A)-binding protein (PABP) which shows similarity to the central region of eIF4G has been described (102). The results of continuing investigations of these various Pab1p-related proteins and interactions are awaited with interest.
Selection of the translational start site.
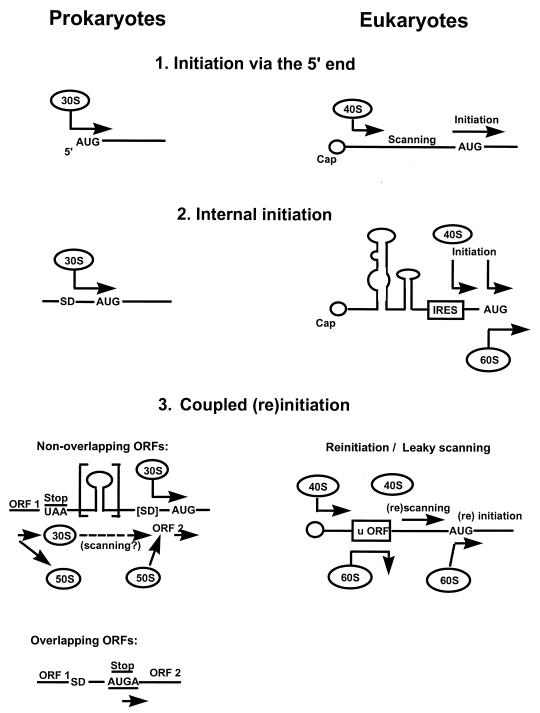
The prokaryotic ribosome can locate a start codon via direct interactions with sequence elements such as the Shine-Dalgarno region located within a translational initiation region (TIR) that has evolved to guide and modulate the initiation process (177, 178, 362, 363). In contrast, despite the theoretical proposition that “translation-initiation promotion sites” may enhance the expression of certain genes (545), there is no experimental evidence for such rRNA-mRNA interactions mediating start site selection in the eukaryotic cell. Instead, initiation on the vast majority of cellular mRNAs involves a process currently modelled by the “scanning hypothesis” (to be considered in more detail below). Investigations of mammalian in vitro systems have indicated that the process of scanning through structural leader regions to the start codon may be driven by the helicase activity of eIF4A (which is enhanced in association with eIF4B), possibly associated with the small ribosomal subunit (509). eIF4A belongs to the DEAD-box family of proteins, which possess ATPase and ATP-dependent RNA helicase activities (161, 487). It is required for translation in a yeast cell extract (57). However, it is not included in the eIF4F complex isolated from S. cerevisiae, and it is therefore questionable whether it cycles through the yeast eIF4F complex, as has been suggested for the mammalian protein (420). On the other hand, it seems unlikely that the function of eIF4A is linked solely to its potential role in destabilizing secondary structure in the 5′UTR, since in the experiments of Blum et al. (57) it was required for the translation of an mRNA that had a relatively short, unstructured leader. Overall, the role of the RNA helicase activity of eIF4A and eIF4B in scanning remains poorly defined.
Scanning continues until the preinitiation complex has selected an AUG codon. In the apparent absence of an equivalent to the prokaryotic rRNA–Shine-Dalgarno (SD) region interaction, AUG selection by the eukaryotic ribosome is directed by the anticodon-codon specificity of Met-tRNAi (93). It is known that the selection process involves participation of eIF2, since mutations in this factor can alter the specificity of the codon selection process (135, 236). Moreover, very recent work by the Donahue group suggests an unexpected role for eIF5 in determining the stringency of AUG selection (236). It seems that eIF5 acts to control the fidelity of the AUG selection process (there is a functional analogy here to the role of prokaryotic IF3). A mutant form of eIF5 was found to allow recognition of UUG as a start codon in vivo and to enhance GTP hydrolysis on the 43S preinitiation complex in vitro (236). It was proposed that an abnormal eIF5 can promote GTP hydrolysis, thus triggering release of eIF2-GDP (plus other initiation factors [Fig. 4]) at a non-AUG codon. This is then thought to allow the ribosome to recognize the non-AUG codon as a start site, effectively switching it into the polypeptide initiation mode. The β subunit of eIF2 carries a binding site for eIF5, suggesting that the above functions involve direct interactions between the two factors (112a).
Initiation of polypeptide synthesis.
Once the 40S subunit has located a start codon, the 60S subunit joins it to form the 80S initiation complex, which can then begin with peptide bond formation between the initial methionine and the second encoded amino acid. This ribosome-joining step is promoted by eIF5, which, as we have seen, acts to ensure the fidelity of the Met-tRNAi–start codon interaction (85, 236, 369). A number of other factors are likely to be released at this point. Of particular interest is the dissociation of eIF2, which is now complexed with GDP after hydrolysis of the GTP that was originally bound and which will have to be recycled back to the GTP form by eIF2B in preparation for a further round of initiation. The mechanism of this GDP-GTP exchange reaction is still the subject of lively discussion (552).
Additional factors involved in translation.
There is quite a collection of proteins that, like Pab1p, seem to be involved in, or influence, the initiation process but are not formally classified as eIFs. This is something of a grey area in terms of formal classification, but it may also be indicative of important but as yet uncharacterized functional interactions between the translational apparatus and other cellular components. For example, Donahue and colleagues (190, 609) isolated four unlinked genes (SSL1 to SSL4) which, when mutated, can suppress the inhibitory effect of a stem-loop structure in the 5′UTR of HIS4. SSL1 and SSL2 were more recently determined to encode components of the transcription factor TFIIH (578). SSL2 was also found to be a yeast homologue of the human ERCC-3 gene, thought to be involved in DNA repair (190). It is still unclear to what extent the observed effects of the SSL mutants on translation reflect the normal roles of these genes in wild-type cells. SIS1, which encodes a yeast homologue of Escherichia coli DnaJ, has also been linked to translation (617). The temperature-sensitive phenotype of a SIS1 mutant was suppressed by one of the ribosomal gene deletion mutants that was previously shown to suppress a pab1 mutant (474). Again, while links between translation and other cellular processes are suggested, the mechanism underlying this effect remains unknown. It is interesting that the theme of chaperones apparently influencing translation is also evident in the phenotypes of mutations in two genes encoding 70-kDa heat shock proteins (hsp70s), i.e., SSB1 and SSB2. In this case however, the translation defect was suppressed by overexpression of the HBS1 gene, which encodes a protein resembling eIF1A and eRF3 (401). The Ssb proteins may be core components of the translating ribosome, perhaps preventing misfolding of nascent polypeptide chains (436a). Finally, one additional factor has been found that seems to be required for translation in wild-type yeast cells (91, 119). This is Ded1p, which is a DEAD-box protein originally identified as a potential suppressor of defects in pre-mRNA splicing (269) and PolIII (549). Ded1p is required for translational initiation in vitro (91), and ded1 mutants are defective in translational initiation (91, 119).
The above examples of gene products that can apparently influence translation might be telling us that the cellular translational apparatus is not adequately defined in terms of the formally classified translation factors alone. They can also be interpreted in terms of the view that functional overlaps are an essential feature of molecular evolution, so that it would be inadvisable to apply excessively rigid definitions of what might constitute “true” translational components. In this context, it is worthwhile to consider how a family of proteins, such as that sharing the DEAD-box motifs, can be involved in a range of processes associated with RNA (161, 487). Indeed, the protein encoded by FAL1 in S. cerevisiae has 55% amino acid sequence identity and 73% similarity to eIF4A but is active in pre-rRNA processing rather than translation (303). These observations raise the question whether eIF4A itself may be inadequately defined as “merely” a translation factor and whether, in a wider context, an unknown number of the other proteins currently classified as translation factors are fully “dedicated” components of the translational apparatus. Further research should reveal how the DEAD-box-type motifs can be combined with a variety of other protein domain structures to confer different functional roles on the members of this family.
Reaction pathways and kinetic control.
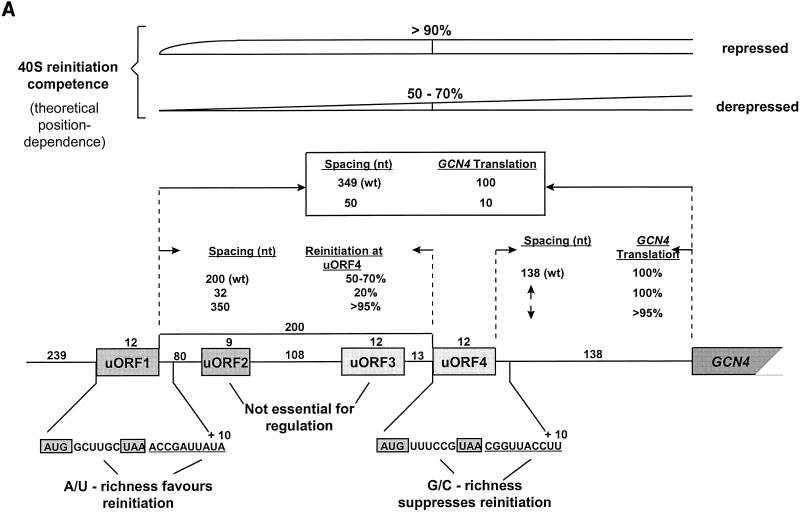
While it is convenient to break down the initiation process into distinct steps, there is in fact little information on the spatial and temporal relationships between the respective partial reactions within the living cell. For example, the translational apparatus can hardly be envisaged as a farrago of randomly acting components, but is there a highly ordered multifactor supercomplex (a “translatosome”), or does the reality lie somewhere between these two possible extremes? It is impossible to resolve issues such as this on the basis of in vitro experiments alone, since disruption of the complex and delicate pathways in the cell may leave only partially or completely uncoupled component reactions. Effectively torn out of its natural cellular environment, translation in vitro is likely to reflect, but unlikely to reproduce, the bona fide process in vivo. This problem may be particularly applicable to the initiation step, since this encompasses not only the transition from the nuclear phase of the life of an mRNA to its recruitment by the translational machinery but also the orchestration of the most complex phase in protein synthesis. Also linked to this problem is the fact that initiation is not an autonomous process occurring independently of the other phases of translation. Statistically, it would be expected that most ribosomes initiating protein synthesis on an mRNA had recycled after previously terminating at least one other polypeptide chain. In general, this means that the pool of ribosomes available for initiation is subject to control by the termination process and posttermination events. Moreover, there are theoretically two extreme cases where this control might be exercised within the confines of a single polysome: as a consequence of reinitiation on a multicistronic mRNA (see below), and in the context of a “closed-loop” (265) type of polysome structure which might allow a ribosome terminating at the 3′ end of the mRNA to be recycled back to the 5′ end (see, for example, reference 233) or to influence de novo initiation at the 5′ end. Reinitiation is known to occur (see below), while the latter type of mechanism is feasible but has not been confirmed as a potential pathway. These considerations emphasize the importance of the cyclical nature of the operations performed by the translational machinery and thus of the relationship between initiation and the other phases of translation.
The kinetic (and thus the temporal) control of the various steps of a process as complex as translational initiation is an even more difficult problem. The apparent order of the current schemes for a major pathway is seductive but may also be misleading. There are at least three potentially serious issues. The first is that in vitro analyses of partial reactions may use conditions that distort the behavior of the translation components under study. For example, the use of an RNA-binding translation factor at excessively high ratios over the mRNA template may lead to the attribution of significant reaction rates to a process that is kinetically insignificant in vivo. Alternatively, an excess of mRNA template in an in vitro cell-free system may titrate out RNA-binding proteins that normally influence the selectivity of the translation apparatus (see, for example, reference 534). Second, reactions that may be largely temporally compartmentalized in vivo may be literally thrown together in an in vitro system, thus generating an apparently viable process that nevertheless does not accurately reflect the course of events in vivo. A hypothetical example, used here to illustrate this point, would be the role of eIF4E. Although this factor can be shown to be required for translation in a cell-free system, its major role in vivo might be restricted to the earliest stages of interaction between ribosomes and mRNA. It is not yet certain that initiation cycles on polyribosomes follow the same type of eIF4E-dependent pathway as applies to the earliest initiation events on mRNA molecules that have just left the nucleus (see also the next section). In other words, is every initiation cycle on a given mRNA mechanistically equivalent? Finally, the route followed by the translational machinery under any given conditions in the cell may be dictated by kinetic control rather than the absence of mechanistic alternatives. Thus, for example, 40S binding to the mRNA might theoretically be primarily cap mediated in vivo because of the relatively rapid kinetics of at least the initial recruitment of capped mRNA into polysomes. However, uncapped mRNA may be an acceptable alternative that is normally discriminated against on competitive kinetic grounds. These and other complications with in vitro experimental work generate uncertainties with respect to the interpretation of the resulting data.
Translational Elongation and Termination
Elongation factors.
The process of elongation in eukaryotic translation has generally received comparatively little attention and is assumed to function in an analogous fashion to that of its counterpart in E. coli (459). While this assumption is certainly likely to apply to the basic biochemical principles, the eukaryotic systems have their own, more complex, set of elongation factors. A highly abundant homologue of the bacterial factor EF1A (formerly called EF-Tu) is present (eEF1A). This forms a ternary complex with GTP and aminoacyl-tRNA and promotes binding of the latter to the ribosomal A site. The other eukaryotic factors (eEF1B and eEF2) do not show readily identifiable sequence homology to their prokaryotic counterparts (EF1B and EF2, formerly known as EF-Ts and EF-G, respectively). The heterotrimeric factor eEF1B catalyzes GDP-GTP exchange on eEF1A yet is dissimilar to the prokaryotic factor EF1B (EF-Ts), which performs an analogous function for EF1A. However, in yeast, eEF1B is rendered redundant if eIF1A is overexpressed (286). The other G-protein, eEF2, is thought to be required for translocation of the peptidyl-tRNA to the P site and, by analogy to the prokaryotic system, of deacylated tRNA to the E site. eEF2 is potentially a major site of regulation mediated by phosphorylation in higher cells (395, 418). An intriguing property of the eukaryotic factors eEF1A and eEF2 is their ability to bind to cytoskeletal components (36, 500), since this may provide a mechanism for the intracellular transport of mRNA, perhaps within polysomes. eIF1A and eEF2 are encoded by duplicated genes. In both cases, the encoded proteins are identical whereas there are only minimal differences in the respective reading frames (99, 394, 433).
A remarkable feature of yeasts and fungi is that they have an additional elongation factor, eEF3 (506), that is apparently absent in mammalian systems, although it does have an apparent homologue in the Chlorella virus CVU2 (604). eEF3 has a curious combination of structural features, with domains similar to ATP-binding/catalytic domains of the ATP-binding cassette (ABC) superfamily of proteins, the E. coli S5 ribosomal protein, and regions of predicted interaction with rRNA, tRNA, and mRNA (40). There is considerable uncertainty about the function of eEF3, with suggestions that it is involved in stimulating either the binding of cognate aminoacyl-tRNA to the A site (381) or the release of uncharged tRNA from the ribosomal E site (402). It remains to be seen whether the function of this factor is fulfilled by an integral component of the mammalian ribosome. For obvious reasons, eEF3 constitutes a favored target for the development of specific antifungal agents by pharmaceutical companies (201, 555).
Release factors.
There have been considerable advances in the study of eukaryotic translational termination in recent years. It is now thought that in-frame stop codons are recognized following the binding of the heterodimeric release factor (RF) complex, comprising eRF1 and eRF3, to the ribosome (517, 618). eRF1 is involved in recognition of all three stop codons (UAA, UAG, or UGA) and mediates peptidyl release (159). eRF3, a GTPase showing homology to eEF1A, stimulates this reaction in a GTP-dependent, codon-independent way. The GTPase activity of eRF3 is triggered by the formation of a ternary complex with eRF1 and the ribosome (160). The current model of termination therefore envisages that eRF3, with its three GTP-binding consensus elements, binds the ribosome in a similar way to eEF1A (517). During elongation, eEF1A would bind aminoacyl-tRNA and GTP in preparation for the formation of the next peptide bond. eRF3, in contrast, can bind only eRF1 and GTP, and through this interaction on the ribosome it promotes peptidyl release. In S. cerevisiae, eRF1 and eRF3 are essential, low-abundance proteins present at a molar ratio of less than 1:20 with respect to the ribosome. However, other reports indicate that the eRF1-eRF3 interaction is not essential for eRF1 function in human (174) or S. pombe (250) cells.
It has emerged recently that eRF3 can assume a prion-like conformation, thus providing the molecular basis for the yeast cryptic hereditable phenotype referred to as [PSI] (335, 554, 591, 592). [PSI] was originally identified as a cytoplasmically inherited determinant that enhances the ochre suppressor activity of a mutated tRNASer (SUQ5) (100a). In [PSI+] cells, eRF3 aggregates as a result of interactions involving a specific domain in the N-terminal region (amino acids 1 to 254) of the protein. Moreover, by virtue of its ability to bind eRF3, eRF1 apparently coaggregates with eRF3 in [PSI+] cells (422). Interestingly, there is evidence that the [PSI] activity can be modulated by the chaperone functions of heat shock proteins (88). There is at least one other yeast prion ([URE3], encoded by URE2) (592).
Sequence contexts and termination efficiency.
Current evidence points to at least two factors influencing the rate of translational termination: the context of the termination codon, and the structure of the C terminus of the peptide chain. In S. cerevisiae, the overall use of the three possible stop codons is biased: UAA (53.1%) > UGA (26.8%) > UAG (20.2%). The bias toward UAA is even stronger (87.2%) if the analysis is restricted to highly expressed genes. However, there is evidence that the triplet sequence alone is not solely responsible for determining termination efficiency and stop codon usage. There is clearly discernible nonrandomness in the bases observed at positions close to the stop codon in E. coli, S. cerevisiae, and mammalian cells (64–67, 442, 543). In particular, there are biases in the identity of the base immediately downstream of the stop codon. Analysis of 784 S. cerevisiae genes (66) revealed overall preferences in terms of the frequency of use of the following tetranucleotide sequences: UAAA (18.2%), UAAG (13.5%), UAAU (16.6%), UGAA (9.9%), UGAU (9.6%), and UAGA (8.4%). Moreover, the bias toward a number of these was greatly increased if only highly expressed genes were included in the analysis: UAAA (33.3%), UAAG (35.9%), and UAAU (16.7%). This type of data has been interpreted to mean that the fourth-base context influences the recognition of the triplet stop codons, perhaps via modulation of the interactions of the RFs in various organisms (543). Bonetti et al. (59) tested the influence of stop codon (fourth-base) context in S. cerevisiae by examining its ability to modulate the suppression of stop codons inserted early in the lacZ gene. They found the following apparent relative termination efficiencies for the respective stop codons: G > U > A > C (UGA), G > A > U > C (UAA), and A > U > C > G (UAG). Comparison with the previous data sets reveals a correlation with the frequency of use of tetranucleotide signals in more highly expressed genes, whereas there is evidently a wider spread of tetranucleotides used in the yeast genes as a whole. A relatively small effect on termination efficiency was also observed when the third base downstream of the stop codon was altered (59). Overall, these data suggest that a significant level of posttranscriptional control is exercised at this step of translation. At least in E. coli, the influence of the stop codon context is thought to be related to evolutionary and adaptive principles affecting the use of the genetic code (543). The role of the termination codon context in yeast has yet to be intensively studied, but there are a number of ways in which it could be significant. One aspect worthy of attention is the influence of context effects on reinitiation, a phenomenon that is of particular significance for mRNAs that bear uORFs.
The potential influence of sequences upstream of the stop codon is more complex to analyze, since the upstream context also affects the encoded C-terminal amino acid sequence. Statistical and experimental analysis of termination in E. coli has indicated that the identity of at least the last two codons in the nascent peptide chain can influence termination efficiency (21, 53, 384, 542). For example, there seems to be selection for a serine codon (UCC) and for both phenylalanine codons (UUC and UUU) immediately 5′ of UGA and for lysine (AAG) immediately 5′ of UAA. Possible explanations for this type of bias include the potential influence of the peptidyl-tRNA in the P site on the function of RFs in the A site and the influence of the physical properties of C-terminal amino acids in the nascent polypeptide chain on termination. Evidence for the latter has come from measurements of termination efficiency associated with UGAA preceded by codons encoding different amino acids at positions −1 and −2 (53). The relationship between amino acid identity and termination seems to be quite complex, but there are apparent correlations with properties such as the propensity to form α-helices and β-strands. As with S. cerevisiae, there are indications that the C-terminal codon choices also influence termination (59), and more recent investigations have revealed that the relationship between C-terminal peptide structure and termination efficiency in S. cerevisiae differs from that observed in E. coli (385).
Analogous functions in prokaryotes and eukaryotes.
Comparisons between the respective prokaryotic and eukaryotic translation termination factors are informative (71, 398). eRF1 (Sup45p in yeast) seems to perform the same function as the E. coli factors RF1 and RF2. There is no extensive homology between eRF1 and RF1/2, but certain conserved sequence elements have been identified (249, 398), suggesting that there is common ancestry and/or convergence. This has provided one argument supporting the idea that RF1/RF2 and eRF1 may bind to the ribosomal A site by mimicking at least the tRNA component of the tRNA-elongation factor complexes involved in elongation (398). On the other hand, S. cerevisiae, Xenopus laevis, and Homo sapiens each have an eRF3 homologue (in yeast encoded by SUP35) with a C-terminal domain that contains a GTP-binding motif and shows significant similarity to apparently equivalent domains in the prokaryotic factors RF3, EF-Tu, EF-G, and the eukaryotic factor eEF1A. It is important to note that RF3 (which seems to be the prokaryotic counterpart of eRF3 [Sup35p]) resembles EF-Tu and the N-terminal domain of EF-G. It has been suggested that either the complex RF1/2-RF3, or possibly RF3 alone, can mimic the complex between EF-Tu and tRNA in a manner analogous to that proposed for EF-G, thus binding to the ribosomal A site (249, 397, 398, 405). However, unlike Sup35p, RF3 is not essential for growth and is therefore thought to act to promote the action of the other RFs. Overall, there is reason to believe that despite the original expectations of functional homology raised by the identification of regions of sequence similarity, Sup35p and RF3 do not fulfill (fully) equivalent functions.
One clue to the difference between the prokaryotic and eukaryotic termination processes may lie in the observed prokaryotic requirement for a fourth factor, the essential ribosome-recycling factor (RRF, or RF4) (270). RRF may promote the translocation event that moves deacylated tRNA out of the P site of the prokaryotic ribosome (into the E site) and/or the removal of whichever RF is in the A site, thus promoting ribosome release and recycling (398), although alternative pathways can be envisaged (71). The latest in vitro experiments have revealed that RRF, together with EF2 (EF-G), promote posttermination mobility of the ribosome, which can then lead to either release from the mRNA or reinitiation (424, 425). Ouzounis and colleagues have described a yeast gene encoding an RRF-like factor, but this protein is probably mitochondrial (417, 518). Perhaps, therefore, eRF1 and eRF3 are sufficient to bring about all of the prokaryotic RF functions (stop codon recognition in the A site, peptidyl-tRNA hydrolysis, termination complex disassembly, and ribosome release) without the involvement of further factors. Further parallel investigations of the eukaryotic and prokaryotic factors will shed light on this and other questions concerning the respective termination processes. This will also be significant to our understanding of the relationship between termination and posttermination events on the mRNA, including reinitiation and mRNA degradation processes. One area of interest will be the nature of the interactions between the various termination factors and the ribosome. Mutational analysis of the E. coli ribosome has revealed that rRNAs from both subunits are involved in peptidyl-tRNA hydrolysis during termination (22). Analysis of the role of a nucleotide in S. cerevisiae 18S rRNA that is equivalent to one of the sites mutated in the E. coli study has indicated that the yeast rRNAs also participate in an analogous fashion in the termination process.
Termination need not be followed by ribosome release.
Given that translation is performed primarily by recycling ribosomes, the question arises how the transition from termination of one polypeptide chain to initiation of another is achieved. As will become evident later, this turns out to be an issue of fundamental importance to certain types of posttranscriptional control, but here it is appropriate to focus on the roles of initiation factors in preparing the ribosome for the different operations it has to perform. Work on GCN4 expression in S. cerevisiae has indicated that termination does not necessarily lead to rapid release of ribosomal subunits from the mRNA (see reference 227 for a review). The fate of the ribosome after termination is influenced by the sequence context of the stop codon. One option is for at least the 40S subunit to remain on the mRNA, where it seems to be capable of resuming scanning. That this is possible demonstrates that Met-tRNAi binding can occur on a 40S subunit that is already associated with the mRNA. It also shows that the eukaryotic posttermination ribosome can remain much more firmly associated with the mRNA than can its prokaryotic counterpart. These observations leave a number of questions open, including whether the 60S subunit is necessarily released by a posttermination 40S subunit that is associated with the mRNA (and, if so, how rapidly) and whether, at the 3′ end of an mRNA, posttermination scanning of the ribosomal subunit(s) continues until the poly(A) tail is reached before being released. Unfortunately, the sequence of events associated with termination is still poorly understood, and much remains to be learned about the cycling of ribosomes. For example, eIF1A, eIF3, and eIF6 are all thought to promote separation of the ribosomal subunits, yet it is not known at which point they bind in the posttermination phase. Moreover, the kinetics of binding of such factors is expected to influence the potential of posttermination ribosomes to participate in (re)initiation events both before and after release from the mRNA.
Mitochondrial Translation
Yeast is dependent on mitochondrial translation only under conditions in which oxidative phosphorylation is required for the metabolism of respiratory substrates. There are only eight major mitochondrial mRNAs, and more than 100 nucleus-encoded proteins have to be transported into this organelle (see, for example, references 32, 208, 492, and 530 for reviews of mitochondrial protein transport; see also reference 221 for references relevant to other types of protein transport) to allow mitochondrial translation to take place. Mitochondrial translation in yeast has been reviewed recently by Fox (157), and only a few points are summarized here. Translation in the S. cerevisiae mitochondrion shows some striking differences from cytoplasmic translation. The mRNAs are generally uncapped, and most of them have 5′UTRs longer than 300 nucleotides. It is thought that initiation involves internal ribosome binding, but there is no clear evidence for the existence of SD-like or alternative motifs in the mRNAs that might promote the prokaryotic (E. coli) type of initiation pathway, although A residues are conserved at eight positions within the region from −25 to +18 flanking the start codon (155, 380). Many of the imported translation factors are homologous to prokaryotic factors, but at the same time, the mitochondrial ribosomes contain subunits that have no identifiable homology to any prokaryotic counterparts. Overall, it seems that mitochondrial translation is more prokaryotic than eukaryotic in type, but the pathway is uncharacterized. The mitochondrial translation apparatus also interprets certain parts of the genetic code differently from its cytoplasmic counterpart, as discussed below in the section on alternative readings of open reading frames.
Of further note is that at least five, and possibly all, of the S. cerevisiae mitochondrial mRNAs require the presence of activator proteins for translation (157, 350, 426, 528, 557). Moreover, at least one nucleus-encoded protein (Cbp1p) stabilizes a mitochondrial mRNA (COB) (87, 128). Remarkably, the translational activators seem to be specific for each mRNA. The current working model is that the activator proteins tether each target mRNA to the inner mitochondrial membrane, perhaps thereby facilitating cotranslational insertion of the encoded proteins (157). The fact that translational activation is generally a comparatively rare phenomenon (363, 364), makes this unusual arrangement all the more puzzling. The explanation presumably lies somewhere in the selective forces and mechanisms that have maintained the shared coding potential of mitochondria and nuclei.
PATHWAYS OF TRANSLATIONAL INITIATION
Eukaryotic ribosomes enter into the translation process by at least three different routes, which can usefully be compared to the pathways of initiation in E. coli (Fig. 9). Where eukaryotic and prokaryotic initiation were compared in the past, it was usual to stress the immediately obvious differences between the two systems. However, there are also some striking similarities, and a balanced analysis highlights important elements of kinetic and thermodynamic control that play decisive roles in mRNA-ribosome interactions.
FIG. 9.
Pathways of translational initiation in prokaryotic and eukaryotic cells. These are formalized comparisons of the main options available to prokaryotic and eukaryotic ribosomes encountering mRNA molecules. 5′-end or cap-dependent initiation is typical of eukaryotic mRNAs, but there is only an apparent counterpart process on certain prokaryotic mRNAs (pathway 1). Internal initiation, by contrast, is common on prokaryotic mRNAs (pathway 2) but is efficient only on the small percentage of eukaryotic mRNAs that possess an IRES. Finally, initiation can be coupled to a previous termination event on the same mRNA (pathway 3). Reinitiation is apparently complex in both prokaryotic and eukaryotic systems. Both prokaryotic and eukaryotic ribosomes can be involved in reinitiation. However, the stability of binding, the effective off-rates, and the (re)start site selection process differ significantly. Moreover, de novo internal initiation on a prokaryotic mRNA can also be tied in to termination on the upstream ORF via a “facilitated-binding” mechanism (see the text). Many details of eukaryotic reinitiation are unknown.
In the eukaryotic cell, the 5′ and 3′ mRNA modifications (Fig. 2 to 4) play important roles in translation. Recent work has made it important to examine the roles of protein interactions with these terminal structures in some detail before considering the alternative pathways of initiation. It has been known for some time that capping and polyadenylation are positive modulators of higher eukaryotic translation (264). Analogous effects were observed on the translation of in vitro-synthesized mRNA electroporated into spheroplasts from S. cerevisiae (165) or added to S. cerevisiae translation cell extracts (172). The poly(A)-binding protein was originally found to promote poly(A)-dependent translation in rabbit reticulocyte lysates (187). Later work then showed that polyadenylation stimulates translation even in a yeast cell extract lacking active wild-type eIF4E and that cap-dependent stimulation also functions in the absence of active Pab1p (538). Since both eIF4E and Pab1p can bind eIF4G (539, 541) (Fig. 5), it is tempting to conclude that eIF4G is the key player in both stimulatory effects. However, in the absence of further information on the physical and kinetic causality of ribosome-mRNA association, it is not yet possible to pinpoint the mechanisms involved, and this remains a challenging area of investigation.
An aspect of eIF4E-eIF4G interactions not yet understood is how these relate to the efficiency of selection of capped as opposed to uncapped mRNAs. Very recent studies with mutants of eIF4G compromised in their ability to bind eIF4E have revealed reduced selectivity for the translation of capped as opposed to uncapped mRNAs in vitro (540). In the same series of experiments, the Sachs group also found that the product of the mutant eIF4E gene cdc33-1 (defined in reference 11), which shows reduced binding to both the cap and to eIF4G, also supported lower cap selectivity. The experiments with the cdc33-1 mutant are consistent with the effects of deletion mutations in eIF4E on the selectivity of the translational apparatus toward capped and uncapped mRNAs electroporated into yeast spheroplasts (569): the cap-binding affinity of eIF4E was found to play a central role in controlling this selectivity. However, the effects of mutations in the eIF4G-binding site for eIF4E on in vitro selectivity have been interpreted to mean that eIF4E acts as a negative regulator of the capacity of eIF4G to promote the translation of uncapped mRNAs (539). An alternative possibility is that the changes in eIF4E-eIF4G interaction skew the competitive selection of capped and uncapped mRNAs. The latter effect would be expected of a system in which eIF4E-eIF4G guides the initial ribosome-mRNA interactions. The significance of this behavior in terms of in vivo translation will therefore become evident only once the effects of eIF4E-binding (in an appropriate environment) on the interactions of eIF4G with mRNA, and perhaps other protein factors, have been established. Of further relevance at this point are recent experiments with wheat germ extracts that indicate that the binding of PABP to eIF4F enhances the affinity of eIF4F for a cap analogue (580a). This suggests that at least plant PABP, like eIF4G, may act directly or indirectly (partially via eIF4G?) as a positive modulator of the cap-binding affinity of eIF4E.
As stressed earlier in this review, the posttermination pathway followed by ribosomes should not be overlooked as a potentially critical point for the control of translation. It is conceivable that the poly(A) tail (and thus the Pab1p bound to it) may play an important role in the recycling of posttermination (40S) ribosomes through to their renewed initiation on either the same or another mRNA molecule. Another question regarding poly(A)-mediated initiation is whether it could define a cap-independent pathway in vivo that places the 40S subunit downstream of the cap structure, thus effectively mediating internal initiation. Perhaps this type of pathway contributes to the poly(A) stimulation seen in yeast cell-free systems (172, 538). The question remains, however, whether this route is kinetically favored in vivo. Interestingly, at least one plant virus (PAV barley yellow dwarf virus) uses an element in the 3′UTR to promote cap-independent translation (577).
There is a need for further characterization of the role of Pab1p in living cells. Recent studies of S. cerevisiae strains bearing a mutant pap1 gene that encodes a temperature-sensitive form of poly(A) polymerase have been informative. Using this mutation in different host backgrounds, Proweller and Butler (445, 446) have been able to explore the relationship between poly(A) length and recruitment of mRNAs into polyribosomes. At the nonpermissive temperature, pap1-1 cells contained approximately half as much total mRNA and their translational apparatus showed apparently only minimal discrimination between poly(A)+ and poly(A)− mRNA. However, discrimination was increased in a strain in which the abundance of ribosomal subunits was reduced, creating a nearly wild-type ratio of mRNA to ribosomes. In this strain, poly(A)− mRNAs were generally associated with smaller polysomes. These results emphasize the significance of the relative activities of the components of the translational machinery in terms of rate control. They also exemplify the problem of at least quantitative disparity between the results obtained with in vivo and in vitro systems (538). This problem also arises in the interpretation of a further report of poly(A) tail-mediated stimulation of translation in vitro (442a). This study essentially confirmed earlier demonstrations of poly(A) stimulation of translation in cell extracts from S. cerevisiae (172, 538) while also finding that the translation of a noncapped, polyadenylated mRNA was resistant to inhibition by antisense 2′-O-allyl-oligoribonucleotides targeted to sites in the 5′UTR more than a minimal distance upstream from the reporter gene start codon. This result was taken to mean that the poly(A) tail supports cap-independent, internal initiation. However, as indicated in the earlier discussion of the distorted conditions for translation created by the preparation of cell extracts, it is difficult to interpret such data in terms of the in vivo functional role of the poly(A) tail. Perhaps the PolI transcription system described by the Donahue laboratory (338a) could provide more direct information about the in vivo role of the poly(A) tail (on uncapped mRNAs).
The issue of in vivo versus in vitro behavior is also raised by the respective studies of the role of eIF4G-Pab1p interactions in vitro and in vivo (538, 541). While cap-independent, poly(A)-dependent translation is clearly evident in a yeast cell extract (172, 538) and eIF4G and Pab1p are capable of interacting with each other (539), more recent work has shown that the Pab1p-binding site of eIF4G is not essential for yeast viability (541). Moreover, a point mutation in eIF4G1 (Tif4631-213p) which eliminated Pab1p binding reduced the growth rate by only 20% at 30°C and still allowed the synergistic stimulation of mRNA translation by the cap and a poly(A) tail in vitro. This contrasts with the lethality of point mutations in eIF4G (540) or eIF4E (449) that eliminate eIF4G-eIF4E binding. At the same time, mutational analysis of the eIF4E- and Pab1p-binding sites in eIF4G and the demonstration of synthetically lethal interactions between the respective genes suggest that the in vivo functional roles of eIF4E and Pab1p might at least partially overlap (541). In this respect, it is relevant that in vitro experiments with wheat germ extracts indicate the occurrence of mutual cooperativity in this plant system between the binding of PABP to eIF4F (eIF-iso4F) and the binding of eIF4F (eIF-iso4F) to a cap analogue (580a). However, the significance of the observed redundancy of function in vitro in terms of a wild-type cellular environment is unknown. There could be a parallel here to the observation that the contribution of an alternative prokaryotic translation-promoting element is of little significance if a correctly positioned, strong SD region dominates E. coli TIR function (see below). Further investigations of the roles of the respective components in vivo should resolve issues of mechanistic and kinetic control that cannot be resolved conclusively by in vitro experiments. This may turn out to be challenging because of multiple (functional) interactions between proteins like Pab1p and eIF4G with various components of the translational apparatus (see the consideration of “networking” in the section on translational regulation).
Another aspect to be considered is how the function of either terminal modification of mRNA is accommodated within the polysome structure. The closed-loop type of model suggests that they are held close together (264), perhaps via an eIF4G-linked RNP complex (312, 479, 539). For example, if either of them acts to promote the initial mRNA-ribosome interactions following (or during) mRNA nuclear export, the subsequent rounds of translation on the polysome-incorporated mRNA might become less dependent on a cap- or poly(A)-mediated pathway. Indeed, it cannot be ruled out that cap- and/or poly(A)-binding proteins may be able to diffuse in or out of polysomes without disrupting repeated rounds of translation within each particle. This matter takes on particular significance when considered in the context of reinitiation (covered in a later section) and initiation events on circular mRNA (86).
The following sections review the three identifiable types of eukaryotic (cytoplasmic) initiation pathway, which are ordered according to the chronological sequence of their original description in the literature. Convenient though this classification may be, it is not a definitive characterization; there may be pathways intermediate between the three major classes, and a given mRNA may be translated (or translatable) via more than one pathway. Moreover, it should be emphasized that the options available to the translation apparatus in cell-free translation systems are at least quantitatively different from those encountered in vivo. This is of central importance to our interpretation of in vitro experimental data.
5′-End-Dependent Initiation
The major pathway of eukaryotic initiation is currently best described by the “scanning model” (291, 302). This 5′-end-dependent, apparently processive mechanism, has no direct counterpart in E. coli, and the nearest prokaryotic case where the ribosome may actually bind at the 5′ end immediately prior to initiation is where the mRNA sequence begins at or just before the initiation codon (as in the bacteriophage λ cI mRNA as transcribed from the maintenance promoter and in a number of other mRNAs in various organisms [515]). However, in the latter case, there is no reason to believe that the interaction with the AUG start codon is mediated by a 5′-end-specific mechanism. Instead, 30S subunit binding, poor though it is, may be driven at least partly by an interaction between the 16SrRNA and a sequence downstream of the AUG (497), although the role of secondary structure in this TIR is undoubtedly a key factor in determining the level of start codon recognition (362, 363). Interestingly, it has been shown that yeast mRNAs either lacking a leader (TCM1) (347) or with leaders of 7 nucleotides or less (145, 565) are still translated, showing that yeast ribosomes can also initiate at a cap-adjacent start codon. Before considering the distinctions between the prokaryotic and eukaryotic pathways further, it is appropriate to define the characteristics of the scanning process as they seem to apply to S. cerevisiae.
Principles of the scanning model.
The scanning model, and the evidence from mammalian systems consistent with it, have been extensively discussed by Kozak (see references 296, 300, and 301 for recent reviews of the relevant data). Since in vitro translational initiation is dependent on ATP hydrolysis (258, 292), it is assumed that the scanning process itself is either directly or indirectly ATP driven. The same basic principles of the scanning model all seem to apply to the translation of cellular mRNAs in S. cerevisiae, but there is some divergence in terms of detailed sequence and structure requirements for initiation and the magnitude of their respective influences on translational efficiency. These are summarized in a comparative tabulation of related data from S. cerevisiae, vertebrate cells, and E. coli (Table 2) and are also considered below.
TABLE 2.
Characteristics of translational initiation sites
| Organism | AUG context and initiation site signals | AUG priority | Sensitivity to mRNA secondary structure (90% inhibition at ca.): | Position effect guiding inhibition via structure |
|---|---|---|---|---|
| E. coli | . . SD . . AUG . .a AU rich or other elementsb | AUG selection efficiency determined by local TIRd sequence and structure; coupling between AUGs in cis can be very influential | 8 kcal mol−1 in TIR | Structure restricting access to SD/AUG |
| S. cerevisiae | Preferred: AA/UAAUG . .c (context effects) | Scanning dictates a 5′-proximity selection gradient for multiple AUGs which is subject to modulation by context effects | 15 kcal mol−1e 5′ of AUG | Degree of inhibition largely independent of position of secondary structure in 5′UTR |
| Vertebrates | Preferred: C .A/GCCAUGc | Same as in yeast, but different context effects | 50 kcal mol−1 5′ of AUG | Secondary structure more inhibitory in 5′-end-proximal position |
SD-to-AUG distances are generally 5 to 13 nucleotides. Initiation at GUG (relative frequency, ∼8%), UUG (relative frequency, ∼1%), or AUU (one case) possible.
Other sequence elements may play a role in initiation, especially in the absence of a strong SD region (see summaries in references 362 and 363).
The significance of downstream nucleotide contexts is likely to be complex, given that these will affect the N-terminal sequence of the encoded protein. These nucleotides are therefore omitted here.
TIR, translational initiation region (see reference 363 for the definition used).
This value depends on the G+C content of the stem-loop structure (570).
(i) AUG context.
Essentially, initiation in S. cerevisiae is restricted to the AUG codon and is reduced in efficiency if the sequence surrounding it deviates significantly from certain preferred nucleotides (134). However, the effects of changes in the context sequence are considerably smaller in S. cerevisiae than in vertebrate systems (299). Even at the most influential position, −3 with respect to the AUG, the best choice (A) is maximally twofold better than other nucleotides (31, 93). Overall, analyses of large numbers of S. cerevisiae genes indicate a general preference for A upstream of the start codon (82, 92, 608) whereas the typical vertebrate upstream context is considerably more CG rich (82, 295). Analysis of the downstream context yields no apparent preference for A at positions +4, +5 and +6 but, rather, shows a predominance of pyrimidines (92, 228, 608). This may be linked to selective forces affecting the N-terminal sequence of the encoded protein (92, 93) (also compare the upstream biases for termination codons discussed in the earlier section on termination). It seems likely that the bias toward A in the S. cerevisiae upstream start codon context, which is especially pronounced in highly expressed mRNAs, is at least partly a reflection of the relatively high sensitivity of the yeast translational apparatus towards secondary structure.
(ii) AUG priority rule.
A further factor influencing the selection of an AUG as a site of initiation is its position within the 5′UTR. In general, the first AUG downstream of the 5′ end acts as the major initiation site (134, 136, 293, 299, 338, 440, 499, 608). This can be explained most readily by an overall 5′→3′ movement of ribosomes during scanning, which effectively imposes a “priority rule” on start site selection. As explained in the section on initiation components, the selectivity of the translational apparatus is tightly controlled by structural characteristics of the initiator-tRNA and of eIF2. There is a sizeable group of natural mRNAs whose leaders contain upstream AUGs (uAUGs) or short uORFs (92, 299, 572). As expected on the basis of the scanning model, these upstream elements generally inhibit translation, albeit to greatly differing degrees. Moreover, analysis of the GCN4 leader, which has four uAUGs (see below), has provided some of the most convincing support for the scanning model of initiation. Equally consistent with the scanning model is the observation that the introduction of a uAUG into a natural yeast leader sequence, or into a completely artificial yeast leader, also leads to inhibition of translation of the main ORF (92, 411, 498). This latter type of result supports the contention that there are unlikely to be any special yeast context sequences required for the priority rule to be observed. Additionally, this type of experiment, as well as other work with leader deletions (31, 565), indicates that there is no eukaryotic equivalent to a prokaryotic SD region. However, if the first AUG is located less than 15 to 20 nucleotides from the 5′ end, depending on the organism, recognition by the ribosome will be impaired, thus attenuating the initiation rate (299, 565). One explanation offered for this is that close proximity of the AUG to the 5′ end may lead to steric hindrance of 40S subunits entering the scanning pathway by 80S complexes paused at the start site (565). Alternatively, start site recognition may be compromised because the shortness of the leader prevents optimal interactions between the 40S subunit and the mRNA to be coincident with appropriate AUG positioning. Since the former explanation assumes that 80S complexes exert rate limitation at the start site, “queuing” of 40S subunits on leaders longer than the apparent minimum length might also be expected to result in a similar inhibitory effect. Although a clear-cut distinction may not be possible here, these considerations would argue for the model in which optimal start site recognition is dependent on a longer region of interaction 5′ of the AUG. At the other extreme, there is no indication that extending yeast 5′UTR lengths beyond the average length of approximately 50 nucleotides has any detrimental effects on translation.
The factors of AUG context, AUG priority, and leader length also come into play when the 5′UTR has more than one potential initiation site. For example, a combination of these influences allows MOD5 mRNAs whose 5′ ends are at −10 or −11 with respect to the first AUG to support translation at both the first AUG and at a second AUG at position +34 (396, 507). Indeed, all three factors are thought to be balanced in such a way as to allow some ribosomes to generate an N-terminally extended form of the tRNA-modifying enzyme Δ2-isopentenyl pyrophosphate:tRNA isopentenyl transferase, which is targeted to the mitochondria. However, other ribosomes scan through the first AUG and initiate a shorter form of the enzyme that is localized to the cytoplasm and nucleus. Although the details of the mechanisms determining this distribution of initiation events have yet to be worked out, it is clear that leader design can be used to ensure the generation of two different proteins from one and the same mRNA template. A number of examples of the use of alternative initiation codons are known in mammalian cells (562).
(iii) Secondary structure blocks initiation.
The ability of stem-loop structures in the 5′UTR to block the progress of 40S subunits along the mRNA and thereby to inhibit translation constitutes a further indicator of the operation of a scanning mechanism in mammalian initiation (294, 296). Moreover, differential sensitivity to the presence of a stem-loop as a function of its position in the leader was taken to reflect interference by this structure in two distinct stages of the mammalian initiation pathway (297). In a rabbit reticulocyte translation system, a cap-proximal stem-loop with a stability of −30 kcal mol−1 was found to be more inhibitory than an equivalent structure placed 52 nucleotides further downstream. It was argued that the cap-proximal structure interferes with initial 40S binding at the 5′ end of the mRNA whereas a downstream stem-loop is encountered by a scanning ribosome, which is evidently driven by a thermodynamic force that is sufficiently large to unwind the structure (297). On the other hand, a cap-distal stem-loop can inhibit translation by more than 85% if its stability is increased to over −50 kcal mol−1 (297). These effects are not only readily explained in terms of the scanning model but also provide useful clues about the energetics of component steps of the initiation pathway. This becomes particularly evident in a comparison between the respective responses of the mammalian and yeast translational machineries to mRNA structure (Table 2). Not only is translational initiation more sensitive to leader structure in S. cerevisiae (31, 52, 93, 412, 570, 608), but also there is no cap-proximal potentiation effect in this organism (412, 570), so that variations in the position of a stem-loop structure in the 5′UTR hardly modulate its inhibitory effect. As discussed below, this difference is also reflected in the mechanisms of action available for translational regulation by RNA-binding proteins in higher and lower eukaryotic cells. Finally, it should be added that a stem-loop structure positioned behind an AUG can enhance the efficiency of start site recognition; the optimum gap was found to be 14 nucleotides (298). A plausible hypothesis has been offered to explain this. It is proposed that the stem-loop causes scanning ribosomes to pause over the AUG, increasing the probability that the start codon will be recognized (298).
In conclusion, the observations of AUG context effects, the AUG priority rule, and the blocking effects of secondary structure are all consistent with the basic scanning model (299, 608), although they do not tell us much about the mechanism of ribosome movement along the mRNA. While most yeast mRNAs tend to have 5′UTRs that are relatively free of secondary structure (82, 92), approximately 10% are predicted to have sufficient structure to cause significant translational inhibition. Many of the leaders in this group have the potential to form localized hairpin structures with stabilities of up to −10 kcal mol−1, which can reduce translational efficiency by approximately 50% (Table 2; the level of inhibition depends on the G+C content of the stem-loop [412, 481, 570]). The corollary of the above is that, as in vertebrate cells (299), translational restriction of this kind affects the expression of a sizeable pool of natural mRNAs in the cell. Finally, there is an interesting aside to this story of the thermodynamic control of translation. The mRNA-folding energies that have been found capable of restricting initiation on selected eukaryotic mRNAs are large in biomolecular terms. For example, the free energy of folding of an entire monomeric protein may be no more than approximately −15 kcal mol−1 (although the component enthalpy and entropy terms are much bigger [103]). Viewing this from another angle, proteins involved in the formation of mRNPs may be able to destabilize structures below a certain stability threshold, so that further unwinding may not be required for translational initiation to take place. However, the free energies of protein-mRNA binding, including perhaps those of protein conformational changes, will have to be found in the reactions driving scanning that have to displace the bound proteins.
Mechanisms relating translation rates to mRNA structure.
Returning to the comparison of prokaryotic and eukaryotic initiation pathways (Fig. 9 and 10; Table 2), it is useful to examine how the respective translation systems react to structured regions of initiation in the mRNA.
FIG. 10.
Principles of kinetic control affecting prokaryotic and eukaryotic initiation. (A) A prokaryotic 30S ribosomal subunit has direct access to the SD sequence of an mRNA with an unstructured TIR. (B) Inhibition via structure in the TIR can be adequately modelled by assuming a thermodynamic control mechanism in which the steady-state distribution of folded and unfolded TIR dictates the amount of mRNA accessible to ribosome binding. (C) In the eukaryotic case, 40S ribosomal subunit binding can occur unhindered on a leader bearing localized structure, but the structured region is thought then to inhibit the scanning process. The off-rate (k3) for ribosomal subunits in this situation is unknown. Disruption of the secondary structure can be driven by an apparently ATP-dependent process, allowing resumption of scanning through the structured region. (D) Random internal binding of 40S ribosomal subunits to mRNA seems possible but is most probably not favored kinetically, and there may be kinetic or mechanistic restrictions on the ability of the subunits to become tightly associated with the mRNA. Initiation factors will influence the type of interaction entered into (compare Fig. 4). (E and F) 5′-end-dependent (E) or IRES-directed (F) initiation results in a tight association between the 40S subunit and the mRNA (the clasp is closed around the mRNA). It is not known whether scanning is normally unidirectional (E and F).
(i) Structure in the prokaryotic TIR.
Prokaryotic 30S subunits are able to recognize internal TIRs within polycistronic mRNAs on the basis of 16S rRNA-mRNA interactions, most importantly involving Watson-Crick base pairing with the SD region. However, as in the eukaryotes, the rate at which the ribosomes can effectively access the start codon and initiate polypeptide synthesis varies over at least a 1,000-fold range (123, 363), and a key contributory factor to this variation in rate is the mRNA structure. Even in complex polycistronic mRNAs, the efficiency of translational initiation is determined primarily by local properties of the mRNA (121, 140, 268, 313, 365). Investigations of the E. coli atp operon provided insight into the mechanism by which this local control can be achieved (313). Small in vitro-synthesized fragments of the atp operon containing the TIRs of individual genes were allowed to interact with purified 30S subunits and fMet-tRNAfMet, and the resulting complexes were analyzed by toeprinting (203), sucrose gradients (189), and runoff polypeptide incorporation assays. The results revealed that structure in the atpG TIR fragment, which supported very poor initiation, interfered with 30S binding whereas the efficient atpE TIR supported both efficient initiation and strong 30S binding. These data support the idea that prokaryotic TIRs exercise control at least partly by regulating the affinity of 30S subunit binding. Consistent with this, detailed analysis of the inhibitory influence of various mutant forms of the hairpin in the RNA bacteriophage MS2 coat protein TIR revealed a correlation between the predicted free energy of folding of secondary structure in this TIR and the efficiency of coat protein synthesis (122). The function of a prokaryotic TIR can be approximately modelled on the basis of competition between folding in the TIR and 30S subunit binding to the SD region (123; compare reference 527). At 37°C, translation efficiency in a TIR controlled by structure is reduced by a factor of 10 with every increase in folding stability of 1.4 kcal mol−1 (above a threshold of −5 to −6 kcal mol−1 [124, 305]). This form of thermodynamic control of ribosome accessibility to the initiation site is apparently achieved without any requirement for additional factors or sources of free energy.
A whole series of alternative TIR recognition sequences has been proposed since the first description of a potential cellular “enhancer” upstream of the E. coli atpE gene (365). It is possible, but generally not proven, that such sequences engage in additional interactions with the 16S rRNA of the 30S subunit, thereby promoting the formation of the initiation-competent 30S-mRNA complex (152, 362). However, these additional interactions are at least as likely as the SD–anti-SD (ASD) interaction to be modulated by secondary structure in the TIR. This obvious principle, which can easily influence many of the weak interactions that are proposed and can seriously hamper attempts to delineate precisely apparent enhancer elements (486), has often been ignored, leading to misinterpretation of much of the data on putative alternative “enhancer” sequences (363). Apart from reassessing the claims made for various sequence elements, there is a serious need for rigorous analysis of their structural and functional properties.
(ii) Structure in the eukaryotic 5′UTR.
An appreciation of the molecular basis of translational inhibition via secondary structure within the 5′UTR is essential to our understanding of the scanning process. The driving force for initiation site location and stable initiation complex formation in E. coli is primarily the free energy of formation of Watson-Crick base pairing between the 30S subunit 16S rRNA and sequences in the mRNA. This explains the relatively great sensitivity of prokaryotic initiation to secondary structure involving the TIR (Table 2), although this relationship is a function of the extent and stability of the mRNA-rRNA interactions that can be achieved (see, for example, reference 123). In the absence of this direct mechanism of AUG localization, start site selection on eukaryotic cellular mRNAs is the result of scanning that depends on a relatively tight association of 40S subunits with the mRNA and apparently on coupling to ATP hydrolysis. A processive, ATP-driven scanning mechanism would be ineffective and energetically wasteful if the off-rate of ribosomal subunits on the 5′UTR were comparable to the on-rate at the 5′ end. Unlike prokaryotic ribosomal subunits, therefore, eukaryotic start site selection must be preceded by a mechanism that effectively promotes tight association of the 40S subunits to the mRNA (Fig. 10). This explains why 40S subunits have to negotiate secondary structure within the 5′UTR, irrespective of its position relative to the cap or the start codon. Such a model predicts that the sensitivity of eukaryotic ribosomes to cap-distal structure in the 5′UTR is therefore determined by the specific rate of free energy input channelled into the scanning process. In S. cerevisiae, the translation rate decreases approximately 10-fold for an increase of 15 kcal mol−1 in the stability of a discrete stem-loop structure in the 5′UTR (at 30°C) (482, 570). This value is influenced by the G+C content of the folded region. Moreover, at least mammalian 40S subunits react differently to structure that is cap proximal.
There are some apparent extremes of structure in natural yeast 5′UTRs, whose existence at first sight seems inconsistent with quantitative assessments of the range of stem-loop stabilities that can be tolerated in the leader (412, 570). For example, the PMA1 mRNA leader is predicted to be able to form secondary structure with a total stability exceeding −50 kcal mol−1 (75). According to the relationship between stem-loop stability and translational inhibition established by Vega Laso et al. (570), this might lead to the prediction that translation should be inhibited by more than 99%. However, the expected structure would be of a very extended nature, involving primarily A · U base pairs, and the PMA1 leader was in fact found to support significant translation in vivo (481). It therefore seems likely that a minimum density of C · G base pairs is required for secondary structure to block scanning ribosomes effectively. An extensive AU-rich structure may be relatively easily opened up, analogous to a long zip fastener, possibly because the free energy per unit distance of mRNA required for disruption of hydrogen bonds is comparatively low. This would fit with a model in which the scanning ribosome acts like a molecular machine subject to a driving force whose maximum value is related by an as yet undefined “coupling factor” to the nonequilibrium free energy available from ATP hydrolysis (Fig. 10). Accordingly, internal initiation (see below) or “ribosome shunting” (133, 163, 260, 610) need not necessarily constitute the explanation for the translatability of the PMA1 leader. In a 40S-driven unwinding model, the unknown (numerical) coupling factor would be a function of the mechanism of the device coupling ATP hydrolysis to ribosome translocation, including a term reflecting the number of ATP molecules hydrolyzed per unit length of mRNA (which might be variable), and is expected to differ between yeast and mammalian cells. Alternatively, as we shall see, the free energy available for driving scanning might be channelled, at least partly, via translation factor complexes. It follows from the above that reliance on predicted mRNA folding energies alone, especially when these have been calculated over a large stretch of nucleotides, can be misleading.
Mechanism of scanning.
Having briefly considered the evidence which has justified adoption of the scanning model as the working hypothesis of eukaryotic 5′-end-dependent translation, it comes as something of a surprise to realize that the mechanism underlying the scanning process is still obscure. This is not for the lack of potentially feasible models of the scanning process. One alternative model used to explain how at least mammalian ribosomal subunits negotiate stable secondary structure in the 5′UTR has been referred to as the mRNA helicase hypothesis (509). This proposes that eIF4A and eIF4B pave the way for the scanning ribosome by virtue of their unwinding activity, thus deviating from the order of binding intrinsic to the schemes proposed by most other authors (see, for example, references 300 and 369). It is envisaged that these two factors initially become assembled at the 5′ end of the mRNA as part of a complex with eIF4F, which in higher eukaryotes comprises eIF4E, eIF4G, and eIF4A. The ribosome is then thought to bind the 5′ region of the mRNA which has been freed from inhibitory structure. Accordingly, this model foresees that mRNA lacking stable secondary structure in its 5′UTR will not require the helicase-catalyzed unwinding activity in order to be bound by ribosomes. In a more extreme version of this model, multiple copies of eIF4A and eIF4B extend from the 5′ end along the mRNA and allow internal binding of 40S subunits (534). A testable prediction of this proposal is that in the presence of excess eIF4A and eIF4B, it should be possible to detect mRNA heavily loaded with both of these initiation factors.
As noted already, S. cerevisiae eIF4F is structured differently from its mammalian counterpart, raising the question whether it could follow the above type of model or whether eIF4A and eIF4B in yeast might function independently of eIF4F to unwind leader structure. Yeast eIF4B, like its mammalian equivalent, is an RNA-binding protein and promotes 40S and 80S binding to mRNA in vitro when present in excess over added mRNA (14). However, disruption of the S. cerevisiae gene encoding eIF4B is not lethal but, rather, generates a slow-growth and temperature-sensitive phenotype (13, 98). There seems to be only a single copy of the eIF4B gene in the S. cerevisiae genome. This would argue that unless there is a relatively distantly related protein with homologous function, eIF4B fulfills only a helper or modulator function in yeast translation. Its mammalian counterpart enhances the helicase activity of eIF4A in vitro (370, 467). In vitro investigations of purified recombinant yeast eIF4B, on the other hand, revealed that it catalyzes a slow RNA-annealing reaction at high molar protein-to-RNA ratios (at least 50:1) (14). This annealing reaction was inhibited in the presence of eIF4A, and, indeed, the complex between the two factors catalyzed the unwinding of duplex RNA. On the basis of these data, Altmann et al. (14) speculate that during translation, eIF4B switches between its annealing and duplex-melting modes as a function of (presumably controlled) interactions with eIF4A. Whether this idea reflects the true functions of these two proteins in vivo remains to be determined. Neither the kinetics observed nor the protein-to-RNA ratios required in these experiments to achieve the effects reported are reassuring in this respect. Perhaps other factors are necessary for the proposed model to work, but these have yet to be identified.
In summary, the known characteristics of the yeast translation system are not readily reconciled with the mRNA helicase hypothesis that has been proposed to describe mammalian scanning (509). If proteins such as eIF4A and eIF4B are used to free mRNA of potentially inhibitory structure, how are their activities restrained to that part of the mRNA, i.e., the 5′UTR, where they might be required? This lack of containment, so to speak, would also provide no obvious means of effectively maintaining the unwound state of a leader region up to the time of arrival of the ribosome. As discussed above, eIF4F in S. cerevisiae seems not to provide the anchorage that is intrinsic to the specificity required by the mRNA helicase model. Moreover, quite elaborate and unlikely schemes have to be proposed to explain start site selection in complex leaders like that of GCN4 in terms of this model (see below). At present, it is therefore easier to envisage a ribosome-associated unwinding activity, where this might be necessary, as providing the required specificity and kinetic properties to function effectively, at least in the yeast cell. However, the Sonenberg type of model does focus our attention on the fact that there may be alternative explanations for at least some of the properties of eukaryotic (yeast) mRNAs.
Overall, there is still much to learn about the mechanism(s) of the scanning process. One of the most important questions is whether scanning is exclusively processive and unidirectional or whether eukaryotic 40S subunits can passively shuffle back and forth as do their prokaryotic counterparts. A form of discontinuous scanning or “shunting” has been described in cells of higher eukaryotes (133, 163, 610), but there is no evidence for this phenomenon in yeast. Beyond this, it will be essential to establish whether yeast eIF4A and eIF4B are obligatory components of the scanning ribosomal complex or are “targeted” to the 5′ region of the mRNA by some other means. Finally, reliable estimates of the on- and off-rates of eukaryotic ribosomes at different stages of translational initiation are required. Both the latest methods of molecular biology and new biophysical techniques (such as surface plasmon resonance analysis [153] and, once certain technical problems have been overcome, time-resolved fluorescence analysis of movements on ordered mRNA molecules [using, for example, the techniques described in reference 275]) could be of help here.
Does all of the above mean that it is premature to attempt to model translational control? Only partly. A number of principles have been established that can contribute to the interpretation of experimental data. Moreover, the response of the eukaryotic translational apparatus to variables such as structure in different 5′UTRs provides information that can be used to refine the modelling process. At least in S. cerevisiae, the absence of a significant position effect on inhibition caused by structure in the 5′UTR indicates that the (quantitative) modelling of ribosome binding and scanning as part of the same process (Fig. 3) is justifiable. It is therefore reasonable to model the quantitative control (but not necessarily the mechanism) of initiation according to the principle of thermodynamic control of access to the start codon, much as has been done for E. coli. The essential mechanistic difference is that eukaryotic 40S ribosomal subunits can begin the initiation pathway on a structured leader whereas structure in a prokaryotic TIR displaces the initial binding “equilibrium” away from the complexed form of the mRNA. It should also be noted that a stem-loop structure, once disrupted, may be held unfolded by the traffic of scanning ribosomal subunits (481) and may therefore not participate in the type of mass-action control envisaged to operate on the prokaryotic TIR.
Reinitiation
Reinitiation is a widespread phenomenon among E. coli mRNAs and plays an important role in the overall control of gene expression in this organism (50, 171, 252, 337, 365, 436, 491, 606). On the other hand, while recognized as playing a key role in the control of a few eukaryotic mRNAs (170, 226), reinitiation has tended not to be regarded as a significant factor in terms of the overall eukaryotic transcriptome. However, as discussed in this review, reinitiation is a manifestation of the posttermination behavior of eukaryotic ribosomes, which not only is of broad significance in terms of the diversity of mRNAs that it affects but also reflects important mechanistic properties of the eukaryotic translation apparatus.
Prokaryotic reinitiation.
Reinitiation provides a means of coupling distinct reading frames on a prokaryotic polycistronic mRNA, but it is not the only mechanism available or required for achieving coupled expression. The terminating E. coli ribosome seems to be able to shuffle back and forth in the vicinity of the stop codon of the gene on which it has just terminated (2), and in doing so, it can bind to a fresh initiation site without leaving the mRNA, providing that this site is located within a region that is short in terms of the ribosomal off-rate (on the order of one ribosome-equivalent’s length). The start codon can either lie upstream or downstream of the previous gene’s stop codon or directly overlap with it (AUGA [see, for example, references 331 and 484). Reinitiation at this site reaches full efficiency if there is an adequate SD region and the stop and start codons of the respective genes are close to each other (certainly less than the equivalent of one ribosome length [510]). The latter characteristic presumably reflects the relatively fast release kinetics of the ribosomal subunits. However, at the same time, the presence of an SD in an internal TIR also opens the door to de novo initiation. The extent to which alternative recognition sequences, such as the so-called downstream box (514), can participate in reinitiation seems not to have been explored. An extremely poor TIR may effectively be “activated” by means of reinitiation (253) but will not support very efficient translation. One striking consequence of prokaryotic coupling is that whole series of genes in polycistronic operons can be more or less tightly linked to the translation of the leading cistron on a polycistronic mRNA (212, 331, 455). It should, however, be noted that translational coupling can also be of a negative kind. Interference by ribosomes on an upstream cistron can be expected to become increasingly likely where termination is close to the start of an efficiently translated gene but there is no coupling pathway to ensure that ribosomes have free access to the downstream TIR (147).
In general, reinitiation itself is not necessarily directly responsible for rendering translation of the downstream gene strongly dependent on the reading frame preceding it (124, 456). This is especially true where the gap between the respective stop and start codons allows ribosomes from the cellular pool to gain access to the downstream TIR. Coupling can be imposed by the presence of structure in the TIR of the downstream ORF that inhibits initiation unless it is disrupted by ribosomes as they translate the preceding ORF. In this case, reinitiation need not be the exclusive or major mechanism for translation of the downstream ORF. Indeed, a coupled region can evolve in such a way that translation of the second gene in a coupled pair is performed almost exclusively by ribosomes entering directly from the cellular pool (the facilitated-binding mechanism [456]). Translation of the upstream ORF seems to eliminate restricted accessibility for ribosomes from the cellular pool to the second TIR. Therefore, whereas reinitiation can support maximally a 1:1 ratio of translation of two coupled genes, the facilitated-binding mechanism can allow the downstream gene in a coupled gene pair to be much more efficiently translated than its upstream partner. The ratio of translation rates in the latter case will be linked to both the stability and the (re)folding kinetics of structure in the downstream TIR (124, 456).
Whereas the driving force for the facilitated-binding mechanism is generally provided by translating ribosomes, reinitiation on prokaryotic mRNAs in which the stop and start codons are not overlapping requires nondissociative ribosome movements that resemble eukaryotic scanning. Very little is understood about this type of posttermination ribosome-mRNA interaction, but the known properties can be usefully compared with eukaryotic scanning and reinitiation. In anticipation of the discussion to come, we should consider briefly the characteristics of reinitiation as opposed to de novo initiation on the UUG codon in E. coli. This codon is a very poor de novo start site but functions quite well as a reinitiation site (2, 519). It has been suggested that IF3 proofreading may be responsible for this difference in selection of UUG as a start codon (2). Thus, if IF3 is unlikely to rebind to a posttermination 30S ribosomal subunit (or 70S ribosome) that has not dissociated from the mRNA, its absence may prevent the proofreading function from discriminating against UUG. Therefore, the principle underlying UUG selection by a reinitiating prokaryotic ribosome may be the modulation of (30S) ribosomal subunit behaviour via a translation factor.
Finally, the relationship between termination of the polypeptide chain and the posttermination behaviour of ribosomes on the mRNA will be influenced by the RFs. Of particular note are the observations of Ryoji et al. (471) on the reinitiation capacity of ribosomes in the absence of RF4. The system studied was a mutant form of bacteriophage R17, which has an amber codon at position 7 of the coat cistron. Readthrough to produce a full-length coat protein required a suppressor tRNA. In the absence of RF4, reinitiation occurred at the codon immediately 3′ of the amber codon, producing a coat protein lacking the first 7 amino acids. Thus, RF4 presumably acts to prevent recognition of a further codon (presumably at least initially in the A site) once a stop codon has been reached. The intriguing question is how rapidly RF4 promotes dissociation of the ribosome from the mRNA subsequent to release of peptidyl-tRNA from the P site. The relative kinetics of these component steps of the termination pathway are clearly of direct significance to the ability of ribosomes to reinitiate on adjacent reading frames.
Eukaryotic reinitiation.
Cellular eukaryotic mRNAs are generally monocistronic, in the sense that each carries only a single major ORF. However, according to the most recent estimates, there are likely to be at least a few hundred genes in S. cerevisiae with short uORFs (572) (see examples in Table 3), as well as many more uORF-containing mammalian mRNAs (299). Yeast mRNAs can contain one to at least six uORFs, and they can appear as distinct entities or overlapped with each other on the main reading frame. There seem to be two different modes of action of eukaryotic uORFs. In the first, the function of the uORF is independent of the coding potential of the uORF. In the second, the influence of the uORF on gene expression depends on the uORF-encoded peptide (343).
TABLE 3.
Examples of uORF-containing leaders in S. cerevisiae
| Gene | Length of major 5′UTR (nt)a | No. and size of uORFs (no. of codons) | Product (reference) |
|---|---|---|---|
| CBS1 | 101 | uORF (4) | PET gene involved in the 5′ end processing of the cytochrome b (57) |
| CLN3 | 864 | uORF (1) | G1 cyclin (441) |
| CPA1 | 244 | uORF (26) | Small subunit of cytosolic carbamoyl phosphate synthetase (406) |
| DCD1 | 33 | uORF (4) | dCMP deaminase (366) |
| GCN4 | 591 | uORF1 (4), uORF2 (3), uORF3 (4), uORF4 (4) | Transcriptional activator of amino acid biosynthetic pathway (391) |
| HAP4 | ∼280 | uORF1 (10), uORF2 (4) | Subunit of transcriptional activator complex binding CCAAT (156) |
| HOL1 | ∼385 | uORF (6) | Major facilitator family (drug resistance subfamily) of putative transport proteins (598) |
| LEU4 | 85 | uORF1 (13) | α-Isopropylmalate synthase (cytoplasmic) (43) |
| PET111 | 459 | uORF1 (6), uORF2 (31), uORF3 (11), uORF4 (30) | Mitochondrial translational activator (528) |
| PPR1 | 50 | uORF (6) | Regulatory protein controlling transcription of two genes in pyrimidine biosynthesis pathway (278) |
| SCHO9 | ∼600 | uORF (55) | Protein kinase that positively regulates the progression of yeast through G1 phase (56) |
| SCO1 | ∼150 | uORF (3) | PET gene involved in the accumulation of cytochrome c oxidase subunits I and II (304) |
| TIF4631 | 295 | uORF1 (12), uORF2 (20), uORF3 (16), uORF4 (8), uORF5 (12), uORF6 (22) | Translation initiator factor p150 (180) |
| YAP1 | 164 | uORF (7) | Stress-related transcription factor (386) |
| YAP2 | 157 | uORF1 (6), uORF2 (23) | Stress-related transcription factor (62) |
nt, nucleotides.
The most intensively studied eukaryotic uORF-containing mRNA is that of GCN4, which has an exceptionally long 5′UTR with special properties (222, 223, 391, 548, 559). Work on this system has provided unequivocal evidence of reinitiation both as an alternative pathway to the standard scanning mechanism and as a basis for translational regulation in response to environmental stress (226) (Fig. 11). On the other hand, quite different aspects of uORF function have been revealed by studies on other mRNAs. Some of these are also covered in this section, while others are discussed below in the section on mechanisms controlling mRNA stability.
FIG. 11.
Features of GCN4 regulation involving modulation of eIF2 activity and the roles of short uORFs (222–227). (A) Diagram indicating the lengths (in nucleotides) of the respective uORFs and noncoding regions of the GCN4 leader. Since uORF2 and uORF3 are not essential for regulation, the scheme focuses on the roles of uORF1 and uORF4. The effects of alterations in the intercistronic distances on the estimated level of reinitiation on uORF4 and the measured rate of translation of GCN4 (as a GCN4::lacZ fusion) are indicated in columns above the illustration of the GCN4 leader. Also shown is a sketch representing simple theoretical gradients of reinitiation competence as a function of time and/or nucleotide sequence travelled in the repressed and derepressed states. These gradients have been used as the basis for a model of regulation of GCN4 repression mediated by changes in the status of eIF2 phosphorylation. It is, however, still uncertain how accurately they reflect the true dynamics of change in the status of ribosomal subunits on the GCN4 leader. These will undoubtedly be a complex function of both distance and various sequence effects. (B) Proposed cycle of events regulating the level of activity of eIF2 in the yeast cell. Under starvation conditions, part of the eIF2-GDP population is “sidelined” into a form restricting GDP-GTP exchange via phosphorylation of eIF2 by GCN2 kinase in response to increased levels of uncharged tRNA. See the text for further details and references.
GCN4.
A remarkable feature of the response of S. cerevisiae to the deprivation of amino acids or purine is that while overall protein synthesis is partially inhibited, GCN4 expression is up regulated. This induction of synthesis of Gcn4p, which is a transcriptional activator, leads to the activation of at least 40 genes involved in amino acid biosynthesis (224). The currently available evidence is consistent with a model in which the kinetic control of reinitiation underlies the posttranscriptional regulation of this gene. The two key components of this model are the special structure of the GCN4 leader (Fig. 11A) and the ability of the cell to regulate the phosphorylation status of eIF2 in response to nutrient limitation (Fig. 11B). The GCN4 leader has evidently evolved in such a way as to ensure that initiation on the main reading frame is performed almost exclusively by posttermination ribosomal subunits which have already gone through at least one cycle of initiation and termination on an uORF. The recovery of the posttermination ribosomal subunits to resume initiation competence is modulated by the availability of the active ternary complex eIF2–GTP–Met-tRNAi. Thus, uORF-containing 5′UTRs can act as sites of termination and repriming of ribosomal subunits with the ternary complex. It is the relocalization of this repriming reaction from the cellular ribosome pool(s) to the GCN4 5′UTR which is thought to couple the reinitiation process to the phosphorylation state of eIF2 (Fig. 11B).
The historical development of studies of the mechanisms regulating eIF2 activity is detailed in a number of reviews (224–226, 228), and only the essential details are summarized here. The ability of the yeast cell to respond to starvation conditions by triggering GCN4 induction was found to be affected by mutations in a large group of unlinked genes. These fell into two groups, with the phenotypes general control nonderepressible (Gcn) and general control derepressed (Gcd). The former mutations affect genes encoding positive regulators of GCN4 expression and prevent GCN4 induction, whereas the latter mutations cause constitutive derepression. The action of all of these mutations on GCN4 regulation was found to be dependent on the presence of the uORFs. The gcn mutations typically affect either the activity of the GCN2 kinase or that of eIF2B, while the gcd mutations were generally found to affect eIF2B or eIF2 (Fig. 11B). Consistent with the proposed role of eIF2 in GCN4 regulation, other mutations in genes encoding constituent subunits of this heterotrimeric factor (SUI2 and SUI3) were also found to confer the Gcd− phenotype. An additional bonus of analyzing many of the gcd and gcn mutations is that they were found to encode component subunits of eIF2, eIF2B, and eIF3 which had not previously been characterized. For the sake of clarity, the principles of both kinetic control and regulation of GCN4 expression are presented together in this one section.
The work of the Hinnebusch group on GCN4 has provided many important details of the reinitiation process in S. cerevisiae, and the reader is referred to a series of earlier reviews for comprehensive and incisive discussions of the large body of experimental data on this system (225–227). The analysis of the GCN4 leader has made use of a large number of substitutions, deletions, and insertions as well as lacZ fusions with both the main reading frame and the respective uORFs (Fig. 11A). The following key features, which are most easily interpreted in terms of the scanning model of initiation, emerged from this work.
(i) There are essentially two types of uORF.
Each of the four uORFs is initiated upon with a similar efficiency while individually inhibiting downstream translation (of GCN4) to a greater or lesser degree under nonstarvation conditions. uORF1 and, to a lesser extent, uORF2 are significantly less inhibitory than uORF3 and uORF4. Two lines of evidence indicate that uORF1 is efficiently translated and promotes downstream reinitiation (183). First, moving uORF1 to within 50 nucleotides of GCN4 inhibited GCN4 translation by approximately 90%, indicating that leaky scanning past uORF1 does not provide the mechanism for ribosomes to reach the main ORF (Fig. 11A). Second, extending uORF1 via fusion so that the resulting uORF overlaps GCN4 by 130 nucleotides abolished GCN4 expression. Ribosomes scanning past the uORF1 AUG would be expected to have remained unaffected by this change. Moreover, under derepressing (starvation) conditions, uORF1 and uORF2 act as positive control elements that relieve the strong negative effects of uORF3 and uORF4. It became clear during the early experiments that much of the regulatory character of the leader was maintained if only one of each of the two classes of uORF (uORF1 and uORF4) was left intact after elimination of the start codons of uORF2 and uORF3 by point mutations. Thus, with only uORF1 and uORF4 in the leader, amino acid starvation still induces GCN4 expression by a factor of 7 (228).
Given the strong indications that uORF1 promotes downstream reinitiation whereas uORF4 does not, how are these two types of function generated? The environment of the stop codon plays a decisive role (Fig. 11A). To a first approximation, AU-richness in the penultimate uORF codon and in the downstream region is correlated with efficient reinitiation whereas GC-richness confers an inhibitory phenotype (182). The leader sequence upstream of uORF1 was also found to influence the ability of this uORF to promote reinitiation (184), but the mechanistic basis of this result is not yet clear. It has been suggested that a nucleotide sequence environment rich in Gs and Cs might cause the ribosomes terminating on uORF4 to pause long enough to allow a factor equivalent to RF4 to bind and cause ribosomal release (226). On the other hand, by analogy to the influence of terminal amino acids in the nascent polypeptide in E. coli on termination efficiency (53), the C-terminal codon(s) of an uORF might exert an influence on the ability of a ribosome to reinitiate via interactions with the encoded peptide product. Progress toward an understanding of the molecular mechanism underlying the choice between ribosomal release and reinitiation will be dependent on the provision of kinetic data on the component steps of termination, including peptidyl-tRNA hydrolysis, eRF binding, and deacylated tRNA release, as a function of the context of the stop codon. A further unanswered question is to what extent the shortness of uORFs such as those in the GCN4 leader might modify the normal course of termination, as occurs at the end of a main reading frame near the 3′ end of the mRNA, thus creating a special sensitivity to the stop codon environment.
(ii) The spacing between sites of termination and initiation is critical.
The spacing between the respective uORFs and between them and GCN4 is a crucial factor in GCN4 regulation. This is because the spacing has to be matched to the rate of change of the reinitiation competence of ribosomes as they move through the leader (Fig. 11A). The positive influence of uORF1 (and uORF2) on GCN4 expression is attributable to the ability of posttermination ribosomes to ignore the inhibitory uORFs further downstream when they have not yet reattained initiation competence through the binding of eIF2–GTP–Met-tRNAi. Figure 11A includes a theoretical gradient for the change in reinitiation competence as a function of distance travelled after termination on uORF1. Under repressing (nonstarvation) conditions, reinitiation competence is regained more rapidly in the derepressed state. This explains the marked sensitivity of the system to changes in uORF1-uORF4 spacing under derepressed conditions. Reinitiation at uORF4 is strongly compromised when this spacing is reduced, whereas extending the spacing has the opposite effect (1, 182). In the latter case, the increase in uORF recognition is paralleled by a decrease in GCN4 expression. The spacing between uORF4 and GCN4 is also critical: reducing it restricts the space (and time) available for scanning ribosomes to regain initiation competence, thus attenuating GCN4 expression.
(iii) Regulation is dependent on a fine balance between uORF types and intersite spacing.
The GCN4 posttranscriptional regulatory system can work effectively only when uORF types 1 and 4 are placed in their natural order and their spacing is kept within defined limits. The mechanism of this sensitively poised system seems generally well accounted for by the basic model proposed by Hinnebusch and colleagues (226). Derepression of GCN4 is explained by proposing that the phosphorylation of eIF2α slows the kinetics of active ternary-complex binding to the ribosomes scanning on after termination on uORF1, thus causing reinitiation competence to peak, or reach a plateau, beyond uORF4 (Fig. 11B). However, there may be some additional forces at play which cause certain observed small deviations from the behavior that is predicted on the basis of the model. For example, even uORF1 or uORF4 alone in a GCN4 leader otherwise devoid of uORFs was found to support a GCN4 induction ratio of up to 2 (see, for example, references 226 and 228). Although this induction ratio is small compared to that of the intact GCN4 system, the underlying cause may be generally relevant to the function of uORF-containing 5′UTRs in yeast.
A more major issue was raised by a comparative study of reinitiation on uORF4 and the GCN4 main reading frame when these are positioned downstream of uORF1 in a series of test constructs (183). Analysis of the effects of varying the distance between uORF1 and the downstream ORF revealed that ribosomes become competent to reinitiate on uORF4 over much shorter stretches of intervening sequence than they do when uORF1 is followed by GCN4. Moreover, the efficiency of reinitiation on uORF4 at shorter intercistronic distances seems to be much more sensitive to the availability of active eIF2. Grant et al. (183) interpret these data in terms of two types of model. The first postulates that a slow step in initiation or elongation on uORF4 restricts the rate of access of scanning ribosomes to the AUG under nonstarvation conditions, thus causing a queuing of ribosomal subunits that have a correspondingly increased period subsequent to termination on uORF1 during which they can rebind the ternary complex. The second possibility is that the sequence of the uORF4 region promotes the binding of an additional, as yet uncharacterized, factor necessary for reinitiation. This could be a factor which, like eIF3, is needed to promote the binding of eIF2. Whatever the potential role of additional factors, the former model touches upon a kinetic principle that may be of key importance to the role of uORFs. The very translation of a short uORF may be defined by a different set of kinetic parameters from those applicable to a full-sized gene. The most obvious consequence of the short length of a three- or four-codon uORF is that it cannot allow simultaneous initiation, elongation, and termination and can be translated only by a single ribosome at any one time. The potential for direct feedback from elongation and/or termination on the initiation process is therefore obvious and may represent a factor that contributed to the evolution of such small uORFs in the GCN4 system. It would be of particular interest to know what role the termination process on the uORFs plays in controlling the reacquisition of initiation competence by scanning ribosomes. Finally, the above discussion emphasizes the importance of detailed quantitative experimentation to the analysis of such a finally balanced regulatory system.
(iv) Regulation of GCN2 kinase activity.
Since the discovery that an intact GCN2 kinase (Gcn2p) is essential for GCN4 derepression (224, 466), much evidence has accumulated which indicates that this kinase plays a key role in the regulation of eIF2 activity via phosphorylation. GCN2 kinase has a multidomain structure, including a protein kinase domain and a histidyl-tRNA synthetase (HisRS)-like domain (583). It also possesses a C-terminal region responsible for binding to the ribosome (452). The current model of the stimulation of GCN2 kinase-mediated phosphorylation of eIF2α (226), (Fig. 11B) involves the binding of an uncharged tRNA to the HisRS domain, which in turn allosterically activates the kinase domain. The resulting stimulation of GCN2 kinase activity leads to increased phosphorylation of Ser51 on eIF2α. There is a correlation between the GCN2 kinase-dependent phosphorylation of Ser51 and the level of induction of GCN4 (125). Moreover, phosphorylation of this site by the mammalian kinases PKR (double-stranded RNA dependent kinase) and HRI (hemin-controlled repressor) in yeast led to constitutive induction of GCN4 (126). Significantly, the level of phosphorylation catalyzed by these two enzymes was unphysiologically high, causing strong growth inhibition. This emphasizes the fine balance maintained by the fully homologous GCN4 regulatory system. Mutation of eIF2α Ser51 to Ala51 prevents phosphorylation by any of these kinases and abolishes GCN4 derepression (125, 126). Although eIF2α has other sites of phosphorylation (150, 566), none of these are involved in GCN4 induction. By analogy to the known effects of Ser51 phosphorylation in mammalian systems (218, 418), it was proposed that phosphorylation of this site in eIF2α impairs eIF2B-dependent GDP-GTP recycling (see, for example, reference 125). Several lines of experimental evidence have subsequently supported this view (227). For example, mutations in the eIF2B subunits can either mimic or reverse the derepression effects of eIF2α phosphorylation (227, 423). Alternatively, overexpressing the eIF2B subunits renders eIF2α phosphorylation less inhibitory (127). Ongoing work is revealing more details of the functions of the component subunits of eIF2B. For example, a subcomplex comprising the subunits γ (GCD1) and ɛ (GCD6) catalyzes GDP-GTP exchange on eIF2 while a regulatory subcomplex comprising α (GCN3), β (GCD7) and δ (GCD2) subunits mediates the inhibition of nucleotide exchange by the phosphorylated form of eIF2 (423a).
Other results are providing additional pieces of the GCN4 puzzle, allowing the proposal of a more detailed model (227). A significant part of this model is based on recent studies of GCN1 and GCN20, both of which are required for activation of GCN2 kinase in starved cells (356) (Fig. 11B). Gcn20p contains nucleotide-binding domains that place it in the ABC superfamily, while these domains are also similar to those present in eEF3. Gcn1p has no ABC domains, but part of its C-terminal half shows similarity to the N-terminal region of eEF3. It is this very part that is required for interactions with Gcn20p (356). Consideration of this and other data has led to the proposal that Gcn1p and Gcn20p bind as a complex to the ribosome in an analogous fashion to eEF3 (see the section on elongation factors). This interaction, possibly near the A site, might stimulate either the binding of uncharged tRNA to the A site or its channeling to the HisRS-like domain of GCN2 kinase (227). Either of these Gcn1p-Gcn20p-mediated events would then promote eIF2α phosphorylation and thus GCN4 induction.
While the GCN2 kinase-mediated phosphorylation pathway is most likely to represent the causal link between starvation and lowered ternary-complex activity, this is by no means the end of the story. The extent to which phosphatases are involved in determining the level and perhaps the regulation of eIF2α phosphorylation remains unclear. A type I protein phosphatase (encoded by GLC7) has already been shown genetically to be able to act antagonistically to GCN2 kinase (226). Moreover, GCN4 can be derepressed, at least transiently, via a GCN2 kinase-independent pathway (560). We can conclude that further novel aspects of the relationship between ternary-complex activity and GCN4 regulation are likely to be revealed as these studies continue.
A final quantitative aspect of the relationship between ternary-complex activity and uORF function in the GCN4 system deserves further consideration. It has been pointed out (226) that the yeast starvation response induces GCN4 translation at least 10-fold while leaving general cellular translation rates little changed. The uORF-dependent mechanism of derepression effectively amplifies small changes in eIF2 activity. This is evidently achieved by virtue of the distinct mechanistic properties and kinetics of reinitiation, which are both subject to coarse and fine regulation by the very same mRNA leader on which the reinitiation process is played out. It has recently been suggested that the sensitivity of the GCN4 system to the status of eIF2α phosphorylation is linked to specific binding of GCN2 kinase to the GCN4 mRNA (544), but this model seems incapable of explaining all of the relevant data (215), and the proposed molecular interaction has yet to be demonstrated. The localization of reinitiation events to a specific leader provides the basis for gene-specific translational regulation that is subject to modulation by forces that only marginally affect translational initiation on the majority of other leaders. We shall also see that this is not the only type of control that can be achieved in this way.
CPA1.
Expression of the gene encoding the glutaminase subunit of the carbamoyl-phosphate synthetase A (arginine pathway) is repressed approximately fivefold by arginine. CPA1 was one of the earliest yeast mRNAs to be found to have an uORF (406, 587). However, in contrast to GCN4 and the YAP1 and YAP2 mRNAs, the 250-nucleotide CPA1 leader has a single, relatively long uORF comprising 26 codons (including the stop codon). Moreover, mutational analyses performed on it have revealed the operation of a different type of regulatory principle, indicating that it acts according to the second type of mechanism outlined at the beginning of this section (Fig. 12). Mutation of the uORF AUG to UUG eliminates repressibility by arginine while having no significant effect on the absolute level of nonrepressed expression (588). This apparent lack of any stimulation of CPA1 expression was originally taken to mean that the uORF is not efficiently translated by ribosomes (588). However, further reflection by this research group on subsequent experimental findings (110) led them to suggest that the CPA1 uORF might in fact simply promote efficient reinitiation, analogously to uORF1 in the GCN4 leader. This is fully consistent with the fact that the CPA1 leader has an AU-rich downstream sequence, which also promotes reinitiation in the GCN4, YAP1, and YAP2 mRNAs (225, 572), but does not rule out the possibility that leaky scanning through the uORF contributes to downstream translation.
FIG. 12.
The model of CPA1 regulation by arginine, originally proposed by Werner et al. (588). The key feature of this proposal is that the peptide product of the uORF is involved in blocking the passage of ribosomes beyond the end of the uORF in the presence of arginine. The induced pausing of the ribosome may involve a regulatory protein called CPAR, but no details are currently known.
The most striking outcome of the analysis of CPA1 was that missense and nonsense mutations in the CPA1 uORF eliminate arginine repressibility whereas silent nucleotide substitutions have no effect (120, 588). It therefore seems that the peptide encoded by the uORF plays an important role in arginine regulation of CPA1 (Fig. 12). The failure of mRNAs carrying a wild-type uORF to complement mutations affecting the composition of the encoded peptide shows that the coding sequence can act only in cis. Remarkably, arginine-dependent repression is to a large extent retained when the CPA1 uORF is fused to the lacZ gene (120). This means that the encoded peptide sequence acts to repress gene expression even when fused to a large protein that is unrelated to the system.
A limited deletion analysis of the 5′ and 3′ regions surrounding the uORF indicated that at least some of the CPA1 uORF-flanking sequences are not essential for arginine repressibility (120), although this study paid scant attention to the nucleotides closest to the uORF. In an alternative type of approach, the CPA1 uORF was inserted at the site of either uORF1 or of uORF4 in a derivative GCN4 leader from which the original uORFs had been eliminated. This experiment indicated that the GCN4 uORF1 type of leader environment allows the CPA1 uORF to promote efficient reinitiation at the GCN4::lacZ fusion ORF and to mediate arginine repressibility. In contrast, at the GCN4 uORF4 position, the CPA1 uORF is strongly inhibitory and does not confer arginine repressibility on the mRNA. It should be added that the fact that the later experiments (120) evidently did not include controls for potential differences in mRNA abundance and/or stability introduces an element of uncertainty into the interpretation of the data, which remains to be addressed. It is therefore clear that resolution of the effects of position and sequence environment on the function of the CPA1 uORF will require further investigation.
While the most likely explanation of the data on the CPA1 system is that the peptide product of the uORF mediates the repressive effect of arginine, the mechanism remains undefined. The fact that the uORF only functions in cis might be explained by a requirement for interactions between the peptide product and the translational machinery during or immediately after synthesis. To what extent the stability or potential rate of diffusion of the peptide product might influence its effective range of action is unknown. Why should the uORF-encoded peptide block expression in an arginine-dependent manner? It has been suggested that the peptide is retained on the translating 80S ribosome in the presence of arginine and possibly also of the regulatory gene product CPAR, forming a complex that somehow blocks the initiation of further ribosomal subunits on CPA1 (Fig. 12). The role of CPAR is, however, unclear, because the effects of cpaR mutants on CPA1 expression may be attributable to indirect effects on ribosomal reinitiation downstream of uORFs (120a). Since the uORF-encoded peptide also functions when fused to β-galactosidase, the blocking effect is presumably imposed on translating 80S ribosomes. It would be of interest to know if this sequence also functions when inserted further 3′ of the lacZ start codon. Biochemical analyses of the proposed “pausing” behavior, as well as genetic suppression analysis starting from mutations in the uORF (or cpaR), should prove illuminating. Investigations of the human cytomegalovirus gpUL4 (gp48) mRNA provide an interesting parallel (74). Current evidence is consistent with a peptide-dependent mechanism leading to ribosomal arrest at termination on uORF2 of this transcript. Cao and Geballe could detect a species representing the uORF2 peptide still covalently bound to the tRNAPro that decodes the penultimate codon of uORF2. Translation is therefore thought to arrest at the stop codon with the peptidyl-tRNA still bound to the ribosome (74).
How does CPA1 regulation relate to that of the GCN4 system? Apart from the distinctive role of the CPA1 uORF-encoded peptide, there may be a common principle of regulating reinitiation downstream of the uORF. Both the nonrepressed inhibitory influence and the arginine repressibility of the CPA1 uORF respond to the sequence environment of GCN4 uORF1 and uORF4 in ways that reinforce this idea. Perhaps, therefore, at least one function of the CPA1 uORF-encoded peptide sequence is to regulate the fate of terminating ribosomes in an arginine-dependent fashion. The peptide-based regulatory mechanism may effectively switch the posttermination events between the two extremes otherwise typical of uORF1 and uORF4 in the GCN4 leader. This would provide an alternative model for CPA1 regulation to the “blocking” mechanism proposed originally (Fig. 12). Again, it should be possible to distinguish between such alternative models experimentally. It may, however, be necessary to consider less obvious mechanisms. For example, the CPA1 uORF-encoded peptide might have a quite different significance, perhaps targeting the mRNA to a particular compartment or binding an as yet unidentified additional factor. Finally, since the majority of the work on this system has focused on translational regulation, it would be easy to forget that other results have identified differential mRNA stability as a potential explanation of arginine-induced CPA1 repression (101). It would therefore be premature at this stage to assume that even the broad outline of CPA1 posttranscriptional control is settled.
An analogous uORF-ORF constellation in the arg-2 mRNA encodes the carbamoyl phosphate synthetase small subunit in two other fungi (345, 579), and comparative analysis promises to provide further insight into at least some of the questions concerning this type of regulation. The arg-2 uORF encodes a peptide with a sequence closely related to that of CPA1. Toeprinting analysis data were consistent with ribosomal stalling occurring at the stop codon (and perhaps one other site) of the uORF at high arginine concentrations (579). Since modification of the start codon context of the uORF to allow enhanced recognition inhibited the translation of the arg-2 ORF, it seems likely that the uORF does not support efficient reinitiation. Thus, leaky scanning and regulatable ribosomal stalling on the uORF seem to be the primary events dictating the translational behavior of the arg-2 mRNA. The mechanism by which the stalling is able to respond to changes in the arginine concentration remains to be determined.
YAP1 and YAP2.
Two further uORF-containing mRNAs have recently been investigated (572). The YAP1 and YAP2 genes encode transcription factors manifesting strong homology to AP1-like factors in higher eukaryotes and to yeast Gcn4p (62, 202, 386, 599). These regulatory genes are involved in the mechanisms used by the yeast cell to protect itself in various situations of stress. For example, overexpression of YAP1 and YAP2 confers general stress resistance to a variety of unrelated compounds from metal ions to different inhibitors and drugs (62, 192, 219, 230, 326, 489, 556, 599). The YAP1 leader has one 7-codon uORF, whereas the YAP2 leader has one 6-codon uORF (uORF1) and an overlapping short reading frame (uORF2) of 23 codons which is positioned −1 with respect to the main reading frame (572). The uORFs of the respective mRNAs influence expression in quite different ways. This was initially indicated by examining the effects of eliminating them from the leaders (Fig. 13A). Thus, by using centromeric plasmid constructs, it could be shown that while removal of the YAP1 uORF had little effect on the resistance of yeast to H2O2, elimination of the YAP2 uORF start codons greatly enhanced cellular resistance to heavy metals.
FIG. 13.
The S. cerevisiae mRNAs YAP1 and YAP2 have different types of uORF, which affect translation and mRNA stability in distinct ways. (A) Elimination of the respective uORFs via mutation of the start codons (AUG→AAG; crosses in the construct maps) reveals striking differences which are most readily detected by using a reporter gene (in this case LUC, encoding luciferase. Removal of the uORF from the natural YAP1 leader (puY1) has little effect on translation or mRNA stability (pΔuY1). The YAP2 uORFs, in contrast, have been found to influence translation and mRNA stability (here reflected in changed steady-state mRNA levels). Both translation and relative mRNA levels were increased upon removing either YAP2 uORF1 (pΔu1Y2) or YAP2 uORF2 (pΔu2Y2) or both uORFs [pΔu(1+2)Y2] from the 5′UTR. The wild-type and mutant sequences of the YAP1 uORF and YAP2 uORF1 are indicated. (B) Mechanism by which the YAP2 uORFs affect mRNA decay. The Northern blots show the disappearance of the YAP2 mRNA signal as a consequence of inactivating the PolII promoter in a heat-sensitive rpb1 mutant. The destabilization effect is largely UPF1 independent. Reproduced from reference 572 with permission of the publisher.
Further examination of the effects of these changes on the expression of reporter genes indicated the nature of the underlying control mechanisms. First of all, the apparent lack of influence of the YAP1 uORF was shown not to be due to the absence of recognition of this uORF by ribosomes. Instead, the YAP1 uORF shows similar functional characteristics to those of GCN4 uORF1; it is recognized by approximately half of the ribosomal subunits scanning along the YAP1 leader, and these show efficient reinitiation subsequent to termination on the uORF. Translation of the downstream main reading frame (YAP1 or a reporter gene) is therefore performed by both ribosomes that have scanned through the uORF (leaky scanning) and by reinitiating ribosomes. In contrast, the YAP2 uORFs do not allow efficient reinitiation, and act to block the expression of downstream reading frames. The YAP2 type of uORF therefore seems to fall into the same class as GCN4 uORF4. Indeed, comparison of the flanking sequences of the YAP and GCN4 uORFs suggests that there is a correlation between the sequence environment of these uORFs and their functional influence on gene expression (572). This proposal was borne out by experiments in which the flanking sequences and the penultimate codon of the YAP uORFs were manipulated. It was found to be possible to interconvert the characteristics of the respective uORF types so that the YAP1 uORF assumed the characteristics of the YAP2 type of uORF, and vice versa (572, 573).
Consideration of the effects of the YAP uORFs on translation suggests that we have found additional examples of principles of translational control originally described for the GCN4 system. However, this applies only in the sense that the individual uORFs manifest similar properties in the respective systems; the YAP1 and YAP2 uORFs are not configured in such a way as to allow GCN4-type regulation. Thus, neither of the YAP mRNAs is subject to translational regulation coupled to eIF2 activity (573). However, the intrinsic properties of the respective YAP uORFs are very similar to those of the GCN4 uORF1 and uORF4 types, so that, organized appropriately, they can form the basis of a regulatable system of the GCN4 type. Thus, combining the YAP1 uORF1 with YAP2 uORF1 in a newly created leader was found to support the inducibility of the downstream reporter gene in response to amino acid starvation (546). As with the GCN4 system, the degree of inducibility was found to be a function of the distances between the uORFs.
In conclusion, while the YAP uORFs have the intrinsic potential to perform adequately as building blocks in a GCN4-type regulatory system, they are not used to this effect in the wild-type YAP mRNAs, although some form of stress-related modulation of at least YAP2 has not been ruled out. However, further investigation has revealed a further property of the YAP2 type of uORF: it acts to destabilize mRNA. This new function for a natural uORF will be revisited in the section on mRNA stability.
CLN3 and other examples of uORF-related control.
Since there are many further examples of uORF-containing mRNAs (441, 572), we can expect to hear more about the roles of uORFs in modulating the synthesis of a whole range of proteins. CLN3 has recently been identified as an mRNA whose translation is restricted by a four-codon uORF (441). The cyclin encoded by CLN3 is required for the normal passage of the cell cycle through the G1→S transition. By attenuating the translation of CLN3, the uORF contributes to the control of the cell cycle at this step. Elimination of the uORF start codon, for example, increased budding in late log phase, most probably due to accelerated progress through Start. The wild-type CLN3 leader is therefore capable of inhibiting cell division as growth becomes limited. An important question is whether the attenuation of CLN3 translation associated with inhibition of the overall capacity of the protein synthesis machinery is a specific effect. Polysomal fractionation experiments with a mutant strain (cdc63-1) with a defective η subunit of eIF3 revealed different degrees of shifting of the distribution of the CLN3, ACT1, and SSA2 mRNAs between monosomes and polysomes in response to the mutation (441), indicating that there are different degrees of inhibition. The extent to which translation of the CLN3 mRNA is differentially attenuated relative to other cellular mRNAs is therefore an issue worthy of more intensive investigation. The theoretical possibility that differential control of the translation of an mRNA such as CLN3 can be exercised at the selection (initial binding) step is considered in the section on prokaryotic and eukaryotic translation.
Internal Initiation in Yeast?
The third type of initiation described so far in eukaryotic systems occurs without any identifiable mediation by the 5′ end of the mRNA. This is mediated by IRESs. It is striking that eukaryotic internal initiation, unlike the standard prokaryotic form of TIR-guided initiation, generally does not occur on mRNAs that are polycistronic in the sense that they contain more than one structural gene (260, 261, 407, 562). In other words, despite the demonstrable capability of the internal initiation pathway to promote the translation of both genes on a dicistronic mRNA, its primary physiological role is apparently to render the translation of certain major ORFs partially independent of the main host initiation pathway. Despite the high level of interest in the mechanism of internal initiation on viral and mammalian mRNAs, its relevance to yeast remains unclear. Attempts to promote internal initiation in S. cerevisiae cells by using the IRESs of poliovirus (100) and encephalomyocarditis virus (146) have failed. On the other hand, there have been reports that cap-independent initiation can be supported in yeast cell-free systems by mammalian and plant viral IRES sequences as well as by natural yeast leaders (12, 241). Moreover, the in vitro study of Iizuka et al. (241) also describes experiments with dicistronic constructs that apparently support internal initiation. A more recent study has indicated that the capacity of the yeast translational machinery to initiate on uncapped mRNAs in vivo can be increased by mutations in the cap-binding affinity of eIF4E (569). The implication of this result is that a latent potential for cap-independent translation is present in vivo but that it becomes measurably active only after modification of the cap specificity of the translation apparatus. However, this in itself does not constitute evidence for the operation of an internal initiation mechanism.
One of the unexpected aspects of the work of Iizuka et al. (241) is the fact that the yeast 5′UTRs reported to contain IRES elements show no readily identifiable characteristics that might distinguish them markedly from the majority of other leaders in this organism (156, 488). One of these leaders (HAP4) contains uORFs (Table 3), which may impose properties affecting posttranscriptional gene expression that are unrelated to IRES function. Overall, it remains uncertain whether bona fide internal initiation, as opposed to simply weakly cap-dependent initiation, can occur in vivo in S. cerevisiae. There has been some debate whether S. pombe might be better equipped for the internal pathway, but we await further news on the ongoing work in this area. A further twist to this tale is provided by the observation that S. cerevisiae contains a 60-nucleotide RNA species (called I-RNA) which has been shown to inhibit preferentially the translation mediated by the poliovirus IRES in mammalian systems (111, 112). Cross-linking studies with HeLa cell extracts revealed that I-RNA interacts with the human La autoantigen, which is suspected to be a trans-acting factor that somehow promotes IRES-dependent translation. S. cerevisiae seems to possess an La protein homologue, but this is nonessential (90), and thus, overall, the significance of I-RNA is still a mystery. Another study has implicated the yeast La homologue in tRNA maturation (567).
Despite the uncertainty about the potential significance of internal initiation in yeast, there are aspects of this phenomenon that are relevant to the discussion of yeast translation. One of these is the fact that 5′-end-dependent initiation seems to require the 40S ribosomal subunit to form a “tight” association with the mRNA that allows scanning to occur in the absence of a high off-rate (Fig. 10). The analogy can be drawn to the operation of a type of clasp, which in molecular terms would be closed around the mRNA. Potential mechanisms of encirclement of the mRNA by the 40S ribosomal subunit have been discussed previously (260, 300, 301). Whatever the exact mechanism involved, there is currently no reason to assume that this close association can be achieved only via the 5′ end. Mediation of this clasping event can be envisaged to occur via internal sequences, and this may effectively be the function of IRES elements. The extent to which the localization of the early steps of initiation to the 5′ end of most mRNAs is a reflection of structural characteristics of the eukaryotic ribosome, as opposed to kinetic control exercised by other components of the translational apparatus, is not known (Fig. 10). However, the latter explanation raises the possibility that there is a background level of internal initiation at non-IRESs that is normally insignificant but that could theoretically be enhanced by changes in the availability or state of components of the translational machinery. In this context, it should be noted that even if 40S ribosomal subunits in the cellular pool have a finite affinity for internal regions of mRNA, this is normally likely to be relatively low. Overall, these considerations reveal that, rather than being restricted to a choice between (fixed) mechanistically distinct pathways (see, for example, reference 479), the route taken can be explained in terms of kinetic control.
A model in which kinetic control determines the dominance of 5′-end- or IRES-dependent initiation can provide a basis for explaining how the balance can be shifted to normally less favored initiation pathways. Underlying the preference for the 5′-end- and IRES-dependent initiation events could be the reduced off-rates of ribosomal subunits that become bound (by a clasp mechanism) to the mRNA via these two types of site (Fig. 10). In contrast, the combination of both a high off-rate and the minimal probability of binding to an AUG-containing sequence so as to form an initiation-competent complex may normally discriminate strongly against non-5′-end- and non-IRES-dependent initiation (Fig. 10). Changes in the activities of components of the translation machinery or in the loading of mRNA with RNA-binding proteins may be sufficient to cause a shift in emphasis to less specific initiation. One particular case where such distortions may be particularly significant is translation in cell-free systems. For example, it is known that translation in reticulocyte lysates manifests poor fidelity and an increased incidence of initiations at normally insignificant internal start sites (113). Moreover, the ratio of mRNA to general RNA-binding proteins in this system has been observed to influence the level of priority given to cap-dependent initiation (534). Again, such considerations warn against the danger of misinterpreting in vitro expression data, since these may reflect a distorted emphasis on translation pathways that are less significant in vivo.
Another relevant property of internal initiation in mammalian cells is that it involves essentially the same set of canonical factors as cap-dependent initiation (260, 435). However, work on picornavirus IRESs has revealed that the interactive properties of eIF4G (Fig. 5) influence the specificity of the translation apparatus. Cleavage of eIF4G by picornavirus proteases generates an N-terminal fragment that binds eIF4E and a C-terminal fragment that contains binding sites for eIF3 and eIF4A (312) (Fig. 5). In vitro studies have indicated that cleavage of eIF4G allows efficient initiation on IRES-containing (327, 619) and uncapped (408, 619) mRNAs but disrupts cap-dependent initiation. It has been proposed that this cleavage uncouples the eIF4E-dependent cap binding of the N-terminal domain from the C-terminal functions of ribosome binding (via eIF3) and interaction with helicase activity (312). The shift away from cap dependence is not as pronounced in vivo, since eIF4G cleavage alone does not suffice for complete inhibition of host protein synthesis (60, 434). Nevertheless, it achieves a similar effect to that seen when eIF4E cap specificity is reduced in S. cerevisiae (569): prioritization for cap-mediated initiation is relaxed. This can be explained in terms of elimination of the kinetic bias toward cap-mediated initiation, which, in turn, allows the translation apparatus to focus more on cap-independent or internal initiation. The molecular basis and kinetic control of the selection of capped mRNAs are discussed in the earlier sections on the initiation components and on pathways of translational initiation.
This is consistent with the general argument that internal initiation and 5′-end-dependent initiation differ primarily with respect to the route by which the 40S ribosomal subunits are guided into a “committed” association with the mRNA. 5′ end (cap) recognition and IRES recognition by ribosomes may therefore constitute two alternative initial steps leading to what is fundamentally the same overall initiation process. In neither case is the mechanism that clasps the 40S ribosomal subunit tightly onto the mRNA understood, but the molecular machinery involved seems to be more or less the same. However, the existence of these two types of initial pathway seems to confirm the principle that specific recognition events secure rapid access to bona fide start codons. As yet, there is no convincing evidence that S. cerevisiae has the capacity to recognize the IRES type of binding element in vivo. Moreover, it remains to be determined whether the yeast eIF4G domains equivalent to those generated by protease cleavage in mammalian cells (Fig. 5) can be used to shift the specificity of translational initiation. Finally, random access via alternative routes dependent on relatively nonspecific interactions is normally restricted by mechanistic or structural barriers (the conformation of the 40S subunit or masking of the mRNA by proteins) and/or kinetic control (Fig. 10).
Alternative Coding Potential of Open Reading Frames
There is an alternative dimension to the posttranscriptional control of gene expression in yeast. As in other eukaryotic and prokaryotic organisms, specific signals can induce “programmed” shifts away from the reading frame established by initiation at the major start codon (582). Both +1 and −1 frameshifting have been described in S. cerevisiae, and each type involves a distinct mechanism (148). The mechanisms of programmed frameshifts stand out against the background of an estimated frameshift error frequency of approximately 5 × 10−5 per codon (307). Moreover, as discussed in this section, there is at least potential scope for regulation of programmed frameshifting events.
+1 frameshifting.
Yeast transposable (Ty) elements are retrotransposons whose life cycle requires the expression of two overlapping genes in different reading frames. TYA and TYB are analogues of the retroviral gag and pol genes, respectively, whereby TYB is produced as a fusion to the TYA protein after the ribosomes have shifted +1 in the overlap region (95) (Fig. 14A). Not only is the effective shifting direction in the Ty elements different from that of retroviruses, but also the mechanisms are distinct. Frameshifting in Ty1 requires two key components: a core “slippage” sequence comprising 7 nucleotides (CUU AGG C), and a low-abundance Arg-tRNACCU that is thought to manifest slow decoding kinetics (39). The working model for this system is that pausing occurs with the Leu-tRNAUAG in the ribosome P site pairing with the CUU codon in the 0 frame and the A site poised empty over the AGG (Fig. 14A). Repairing occurs between the Leu-tRNAUAG and the UUA, which leads to decoding of the following GGC (glycine) in the A site, thus establishing the +1 shift. Two further aspects of this system attract particular attention in terms of control. The first is that the cellular abundance of the Arg-tRNACCU strongly influences the rate of frameshifting. Increasing the copy number of the corresponding gene fivefold reduced the efficiency from more than 40% to 1% (39, 603). This result is consistent with kinetic control of the frameshifting event which depends on the relative rates of binding of Arg-tRNACCU to the A site and the rate of slippage of pairing by Leu-tRNAUAG from CUU to UUA. The second point is that frameshifting is inhibited by proximity to the start codon (39, 95). Frameshifting was completely inhibited by reducing the distance between the essential 7-nucleotide element and the start codon to two codons and became unaffected only once the gap was extended to four codons. It is unclear why the competence to undergo a frameshift is suppressed in this early region of the reading frame. Since the translational apparatus very decisively sets the 0 reading frame at the start codon, it will be of interest to determine how this maintenance of frame is imposed on the first three or four codons.
FIG. 14.
Different forms of frameshifting in S. cerevisiae. Two types of +1 frameshifting have been described. (A) The first type, observed in the yeast transposable element Ty1, involves re-pairing from the CUU codon to the +1 UUA codon. (B) The second type, described for Ty3, does not seem to involve re-pairing by the decoding tRNA interacting with the first codon of the “slippage site.” (C) Finally, the retroviral type of −1 frameshifting occurs in the dsRNA viruses of S. cerevisiae.
Another mechanism of +1 frameshifting operates in Ty3 between GAG3 and POL3 (149). Analogously to Ty1, there is a 7-nucleotide core slippage site (GCG AGU U), but the shift mechanism does not involve re-pairing from GCG to CGA, and the 14-nucleotide stretch downstream of the shift site promotes frameshifting. It is proposed that the A base between GCG and GUU is skipped (Fig. 14B) by an as yet unidentified mechanism. A further feature shared with the Ty1 system is that overexpression of Ser-tRNAGCU, which is the cognate tRNA for the AGU codon, reduces the estimated frameshift activity approximately 10-fold. Thus, the principle of inducing a pause at the second codon in the 7-nucleotide core sequence seems to be shared by the Ty1 and Ty3 types of frameshifting mechanism. Moreover, it seems unlikely to be an accident that two such similar codons (AGG and AGU) are used (149), indicating that the codon-tRNA interaction at this site must have specific properties. However, there is a striking difference in the codon-tRNA interaction immediately upstream of the shift site. The GCG codon in the Ty3 site is decoded by Ala-tRNACGC, which cannot slip onto the +1 frame codon, CGA. Overproduction of a synthetic Ala-tRNACGC was found to reduce frameshifting, indicating that the codon choice is important for the mechanism (419). The Farabaugh group also investigated whether codons other than GCG can assume the same role and found that in total only eight tRNAs can stimulate the +1 frameshift (575). It has been proposed that frameshifting is most probably mediated either by direct out-of-frame binding of Val-tRNAIAC to the +1 codon GUU (perhaps promoted by tRNA “neighbor” effects associated with the upstream Ala-tRNACGC) or by means of 4-base decoding involving a tRNA capable of participating in an extended codon-anticodon interaction.
−1 frameshifting.
Double-stranded RNA (dsRNA) viruses are widespread among strains of S. cerevisiae. They show many similarities to the dsRNA viruses found in cells of higher eukaryotes and are responsible for the yeast killer phenotype (590). The dsRNA viruses have the overall gag-pol structure typical of the retroviruses. For example, in the L-A viral dsRNA, ORF1 of the (+) strand comprises 681 codons and encodes the viral particle major coat protein (80 kDa) whereas ORF2 comprises 869 codons and encodes RNA-binding and RNA Pol domains. ORF1 and ORF2 overlap in the −1 frame by 130 nucleotides. Frameshifting at the by now familiar retroviral type of slippery site (257) located in the overlap region yields a 180-kDa protein which incorporates the domains encoded by both ORF1 and ORF2. The latter is equivalent to the reverse transcriptase domain encoded in an analogous position in retroviruses. The L-A virus frameshifting site possesses the standard features of a retroviral type of −1 shift site (Fig. 14C). The shift sequence is followed by a structural element (possibly a pseudoknot [561]) that is thought to cause the translating ribosome to pause at the appropriate position. The most likely pathway is that tRNAGly shifts −1 back from the GGU codon, thus reestablishing its nonwobble base pairs while changing the wobble interaction from a U to a G (130, 240). The efficiency of this process is equivalent to approximately 1.8% of the total number of ribosomes passing through the site. The mechanistic details of the various types of frameshifting have not been determined, but models are discussed by Farabaugh (148). One area of special interest is the potential role of trans-acting factors. These will certainly include proteins that act as general modulators of translational accuracy (such as those discussed in references 106 and 584 to 586), but it is uncertain whether there are factors that specifically promote frameshifting at −1 (or for that matter, +1) sites.
Could the efficiency of frameshifting at this site be subject to regulation? There is certainly no evidence for this in yeast. However, an intriguing proposition is that variation in the state of modification of bases in the tRNA anticodon loop (4) in cells of higher eukaryotes could be brought about by retroviral infection (205, 206), thus modulating the ability of at least certain tRNAs to re-pair on the −1 shift site. This is, however, a controversial proposition (63a). It remains to be determined whether there is any form of modulation of the capacity of tRNAs to re-pair in yeast.
Alternative decoding.
Recoding is not evident in S. cerevisiae cytoplasmic translation but does occur in the mitochondrion, in which UGA encodes Trp (instead of stop), AUA encodes Met (instead of Ile), and CUN encodes Thr (instead of Leu). The only known occurrence of cytoplasmic recoding in a yeast is the reassignment of CUG from Leu to Ser in several species of Candida (415, 485). Finally, there has been keen interest in the possibility that yeast species incorporate the 21st amino acid, selenocysteine, into certain proteins. This is known to occur in representatives of the prokaryotes and archaea, as well as in other eukaryotic organisms. In all cases, it involves recoding of the UGA codon. Unfortunately, no unequivocal evidence for this phenomenon has been reported for either S. cerevisiae or S. pombe.
Protein splicing.
It was recently discovered that there is a very different way in which the encoded information of a reading frame can be interpreted to generate a noncolinear product (97). Protein splicing was first observed in S. cerevisiae. The TFP1 gene theoretically encodes a protein of 119 kDa, but the protein product generated is the 69-kDa catalytic subunit of the vacuolar H+-ATPase (V-ATPase) (231, 279). The remarkable solution to this puzzle was found to be the processing of the initial protein precursor. A 454-amino-acid segment (the intein) is excised via a process in which the N- and C-terminal exeins are joined to yield the 69-kDa mature product. Protein splicing has been observed in the prokaryotes, archaea and eukaryotes, and much current work focuses on determining its mechanism (97).
Prokaryotic and Eukaryotic Translation
At this point, it is informative to return to the comparative assessment of the initiation pathways available to prokaryotic and eukaryotic ribosomes. Overall, there seem to be two different strategies for start codon localization. Interactions between rRNA and mRNA sequences, of which the ASD-SD interaction is the paradigm, constitute the primary driving force in the prokaryotic system. The prokaryotic 30S ribosomal subunit can theoretically obtain access to all internal sites on the mRNA, but its binding is guided by the thermodynamics of interaction with the mRNA (and possibly by the kinetics of transition from the preinitiation complex to the initiation complex proper [460]) to those TIRs that are relatively structure free and contain SDs that manifest the appropriate range of ASD-binding energies (Fig. 10). The energetics of this type of interaction evidently allow broad flexibility in the control of translational initiation in cells where transcription and translation are tightly coupled and nascent transcripts are accessible to ribosomes. It is likely that these early ribosome-mRNA interactions influence the folding pathways and functions of nascent mRNA and also have significant effects on the temporal control of gene expression on polycistronic mRNAs (185, 456). Moreover, the fact that ribosomes can directly access internal TIRs means that a polycistronic mRNA can support greatly varying translation rates from its respective genes, whereby the coupling ratio between neighboring genes can deviate significantly in either a positive or a negative sense from the obligatory ratio of 1:1 obtained under conditions of tight reinitiation (363, 456).
Overall, the eukaryotic system has not evolved to provide this form of flexibility. It is, however, equipped to deal with particular properties of mRNAs that emerge from the nucleus having undergone modification, possibly splicing, and interactions with a host of nuclear proteins and RNAs. The process of start site selection in eukaryotic cells has shifted away from the prokaryotic type of rRNA-mRNA interaction mediated process to principles of protein-mRNA mediation. The translation apparatus (or factors physically or functionally associated with it) is energetically competent to displace at least the majority of mRNA-binding proteins that accompany the mRNA out of the nucleus, apparently irrespective of the rate at which individual mRNAs are translated. The exceptions are the proteins that serve to “mask” or “silence” mRNAs in vertebrates and amphibia at early stages of development (457, 512, 589). The adaptation of the eukaryotic translation apparatus to form a tightly binding complex during the preinitiation stage may even be a consequence of the need for it to function on “prepackaged” mRNPs emerging from the nucleus. According to the general model outlined in Fig. 10, nonspecific initiations at internal AUGs can be avoided by kinetic prioritization of the 5′-end (or IRES)-dependent pathway, which leads to tight association of the 40S ribosomal subunits with the mRNA and allows them to “scan” over long distances. At the same time, the alternative route to internal initiation is discouraged by high off-rates (and/or low on-rates) and possibly by relatively low resistance to blockage by certain RNA-binding proteins (534). The IRES-mediated channelling of 40S ribosomal subunits into the initiation pathway may involve ribosome-mRNA interactions analogous to those of the prokaryotic system, but this has yet to be experimentally proven. However, as with the prokaryotic TIR, the efficiency of IRES-mediated internal initiation does vary over a wide range as a function of IRES structure (61).
While prokaryotic and eukaryotic ribosomal subunits may share a low-affinity “scanning” facility (Fig. 10), the tightly associated eukaryotic 40S ribosomal preinitiation complex formed during the major initiation pathway is capable of scanning through structured regions which would be impenetrable to its prokaryotic counterpart (Table 2). As discussed above, although eukaryotic ATP-dependent helicase activities are thought to provide the driving force for this capability, the mechanism is unknown. The prokaryotic ribosome has a comparatively poor capacity to unwind secondary structure, and the quantitative relationship between the free energy of localized mRNA folding in the TIR and the initiation rate is most easily explained by assuming that only the fraction of the mRNA molecules in the unfolded state can be bound to form a preinitiation complex (123). In contrast, a eukaryotic 40S ribosomal subunit can bind an mRNA that bears strongly inhibitory secondary structure (297, 481) and may have to negotiate that structure only during the scanning phase. This may mean that the selection step (i.e., initial binding) cannot distinguish between mRNAs that can be efficiently translated and those whose translation is restricted by structural elements in the 5′UTR. The idea that this type of selection does occur forms the basis of some of the analyses mentioned at the beginning of this review and is frequently used to explain the behavior of specific mRNAs under various translation conditions. There is, however, no unequivocal evidence for mRNA selection of this kind, and this principle of control requires further consideration.
There is indirect evidence that scanning 40S subunits pause in front of a stem-loop structure (297), and there is little doubt that unwinding of stem-loops up to a given stability (Table 2) can occur. The inhibition curve of eukaryotic translational initiation in response to stem-loops of various stabilities in the 5′UTR indicates that this is the eukaryotic form of thermodynamic control via secondary structure (481, 570). The extent to which blocked 40S subunits are forced to queue up on a structured leader, as opposed to abandoning it by virtue of a significant k3 (Fig. 10C), is undetermined. The release rate that pertains in this situation may be different from that applicable to unstructured leaders but may have to be comparable to the scanning rate (k2) in order to prevent the formation of a queue of 40S subunits back to the 5′ end. It is also highly relevant to the question of mRNA selection and rate control exerted during initiation addressed above.
There is currently considerable excitement about the mechanisms and functional roles of termination and reinitiation in eukaryotic cells. How is the release of ribosomes and/or the capacity to reinitiate controlled? By analogy to E. coli, we might expect eRF1 and eRF3 to be involved in the response of the translational apparatus to signals in the vicinity of the stop codon. For example, a recycling-factor type of activity analogous to RF4 might be activated upon termination in certain sequence environments, such as those provided by GCN4 uORF4 or YAP2 uORF1. Alternatively, rapid progression from termination to resumption of the scanning mode might be possible in at least some AU-rich stop codon environments (as in the GCN4 uORF1 or the YAP1 uORF), whereupon the capacity to reinitiate may be governed by the kinetics of rebinding of specific factors. The arguments for the involvement of eIF2 and perhaps of other factors were considered earlier in this review. At first sight, the apparent involvement of IF3 in influencing the ability of prokaryotic ribosomes to reinitiate on a UUG start codon seems to suggest that the modulation of mRNA-bound ribosomes via initiation factor binding represents a conserved theme in evolution. However, for the prokaryotes, it is thought that the inability of the factor to bind to 30S ribosomal subunits in the scanning mode is required for reinitiation. For yeast GCN4, it is not yet known why reinitiation of ribosomes on the leader is so much more sensitive to ternary-complex activity than is general initiation, but it seems likely that kinetic control and/or involvement of distinct combinations of initiation factors in the respective processes is responsible (226). Further research into these factor-ribosome interactions will probably uncover novel mechanisms of control.
Unresolved Issues of Quantitative Control
The systemic type of analysis of multistep pathways considered earlier in this review formalizes the view that control is rarely concentrated in just one component step of a pathway. The definition of control used by Kacser and Burns (276, 277) is particularly appropriate for the binding and catalytic steps of the translational pathway that are nonprocessive. It provides a framework to represent the distribution of control among a number of steps. On the other hand, the Kacser and Burns type of treatment is less useful for processive reactions, especially elongation and possibly initiation. These steps can be approximately modelled by more standard procedures, provided that a number of assumptions are made (173, 340). Given the quantitation of translation in terms of the rate of production of complete polypeptide chains, protein synthesis can be considered a process whose rate is determined by the rate of initiation and/or termination.
Although insufficient data are available for translation to be accurately modelled, the use of these theoretical treatments focuses attention on important questions. For example, it has been assumed that eIF4E functions can be attributed to large “sensitivity” coefficients, meaning that changes in eIF4E activity exert strong control on the rate of translation, but the experimental data available do not justify this. Another issue is the extent to which the limited availability of ribosomal subunits restricts the translation process, thus influencing rate control and/or selectivity (see, for example, the discussion of the pap1-1 mutant). A further open question is the degree to which termination, either directly or indirectly, controls the initiation rate in individual polysomes. This aspect was not considered in earlier work. Finally, one key factor in earlier models was the so-called discriminatory factor (173, 339, 340), which was presumed to be responsible for determining relative rates of translation on individual mRNAs but was not clearly defined. There is no evidence that eIF4E binding to different capped mRNAs in S. cerevisiae is generally controlled via features of the 5′UTR (314), so that eIF4F seems unlikely to function as a discriminatory factor. It presently seems more likely that “discrimination” in terms of initiation rate is determined via the modulation of ribosome progress along the mRNA leader. These and other questions make it clear that the relationship between mechanistic and kinetic models requires further clarification.
More information is required about the “control coefficients” of the components of the translational apparatus in terms of overall rate before quantitative data can be reliably interpreted in terms of mechanistic models. There are a number of ways in which this information can be obtained. Mutational analysis and biochemical experiments can provide information about the effective control coefficients of translation factors, and, for example, rRNA-processing mutants might help with controlling ribosome concentration. In performing this type of analysis, it is essential to consider appropriate procedures for relating quantitative control effects to mechanistic function in vivo. Useful information on the control of ribosome throughput on individual mRNAs can be obtained by studying the response of translation rate to structure or protein binding in the leader. Since the yeast system shows no pronounced position dependence and an apparently straightforward, partially linear dependence of inhibitory effect on structural stability, it is tempting to apply a thermodynamic model of rate control equivalent to that used for initiation in a prokaryotic TIR (123, 527). However, even when approximations like this seem to fit the data, more information is required for the scanning mechanism to be characterized. For example, in vitro investigations of ribosome on- and off-rates will be necessary to fill in the missing numbers in the schemes shown in Fig. 3 and 10.
Finally, genetic and biochemical studies with S. cerevisiae show that the interactions between components of the translation apparatus are suggestive of functional networks. Consider, for example, the complex interactions involving eIF4F proteins (Fig. 15). They illustrate the extensive functional interactions and overlaps that can be identified for many of the proteins involved in posttranscriptional control and warn us that a process such as initiation cannot be adequately described solely in terms of a linear mechanistic pathway.
FIG. 15.
“Networked” interactions involving components of S. cerevisiae eIF4F. The diagram shows interactions defined in the following ways: physical interactions demonstrated biochemically in vitro (binding, +; competition or negative regulation, −) and genetic (functional) interactions that are synthetically lethal or potentiate a negative effect (−) or are of a positive, phenotype-suppressing nature (+). The various interactions are discussed in the text. These are not the only interactions detected for the respective proteins, but they suffice to illustrate the functional complexity of the yeast translation system. These and other results are consistent with the existence of multiple interactions, functional overlaps, and alternative routes to initiation in vivo. Translation, therefore, like many other cellular processes, is not definable in terms of an independent linear pathway involving dedicated components.
MECHANISMS OF TRANSLATIONAL REGULATION
The interactions between the translational machinery and the more than 6,000 different mRNA species in S. cerevisiae are governed by a number of different structural attributes of these mRNAs. Returning to the scheme of a “typical” mRNA (Fig. 2), the numbered regions indicate the diversity of sites at which even small structural changes can make a significant impression on the translation of a given mRNA. These domains of the mRNA can be involved in two types of translational regulation. General regulation is exercised via modulation of the activities of components of the translational machinery which, in turn, interact with more or less precisely defined regions of each mRNA. In contrast, variations in individual mRNA sequences are linked to gene-specific posttranscriptional regulatory events, which may involve site-specific protein binding or, as with GCN4, mRNA-specific responses to changes in translation factor activity.
Modulation of Translation Factor Activities
The main site of general translational regulation of the pool of eukaryotic cellular mRNAs is thought to be the initiation process (262, 330, 364). While this view may potentially do injustice to the role of other steps in translation, there is a considerable body of data implicating initiation factors in global regulation. The majority of these data indicate that eIF4E and eIF2 constitute key regulatory targets. However, the balance of evidence pertaining to these factors is different for yeast and mammalian systems.
eIF4E.
Regulation of eIF4E activity is expected to allow the cell to modulate the 5′-end-dependent binding of 40S ribosomal subunits to mRNA. Two types of regulatory mechanism have been proposed: modulation of eIF4E cap-binding activity mediated by changes in the phosphorylation state of this protein, and regulation mediated by interactions between eIF4E and the so-called 4E-binding proteins. Before considering either of these, it is important to address the general perception that eIF4E must be a “limiting factor” in initiation because it is present in substoichiometric amounts relative to the cellular content of ribosomes. The assumption that low relative abundance is a reliable indicator of a special regulatory status is unjustified without knowledge of the mechanism of action of this factor. Given that eIF4E exchanges between mRNAs, there is certainly no pressing theoretical argument for this proposition. Moreover, the earlier data on eIF4E abundance in mammalian systems, which were originally interpreted as evidence that this factor is limiting, could not be confirmed in more recent work (453). As for S. cerevisiae, studies of the influence of variations in cellular levels of eIF4E on translation have revealed no evidence of the proposed “limiting” role (314, 569).
Another type of result taken to indicate a special limiting status for eIF4E is the observation that its overproduction in various cultured cells leads to effects such as cellular transformation or changes in cell morphology (115, 319). However, such transformation effects are also seen after the overproduction of other proteins in higher cells, including a nonphosphorylatable mutant form of human eIF2 (138) and hepatitis C virus proteins including the nonstructural protein NS3 (483). In none of these cases is the causal link to transformation characterized, and even if it were, it would not necessarily confer a special “limiting status” on the overproduced molecule. Again, studies with S. cerevisiae have revealed no analogous effects of overproduction of eIF4E: no measurable change in growth is seen unless eIF4E abundance is increased by a factor of approximately 100-fold compared to the wild-type, when a slight reduction in growth rate is observed (314). As in mammalian cells, the cause of the anomalous behavior of yeast cells associated with eIF4E overproduction is not certain. A favored explanation for higher cells is that the excess eIF4E potentiates the capacity of eIF4F-supported unwinding activity that releases translational attenuation of mRNAs with structured leaders which encode proteins involved in growth regulation (483, 484). It has also been reported that eIF4E overproduction can influence the nucleocytoplasmic transport of cyclin D1 mRNA in NIH 3T3 cells (465). The principle of selective translational enhancement does not apply to S. cerevisiae (569) and has in fact not been found to apply to all highly structured eukaryotic mRNAs (51). Overall, while translation-related effects may be involved, other mechanisms have not been ruled out. Since eIF4E is both nuclear and cytoplasmic (314, 325), these could potentially be linked to a wide range of processes (447). In conclusion, it seems likely that eIF4E is present in quantities that are well matched to its role in translation (314, 569). Indeed, the yeast cell shows remarkably low sensitivity to variations in eIF4E function, and even large reductions in cap-binding affinity caused by deletion mutations can be partially or fully compensated for by overproduction of the resulting truncated versions of this protein (542). On the other hand, in vitro (172) and in vivo (35) data suggest that heat shock mRNAs may be subject to an eIF4E-related selective response under stress conditions, although the magnitude and mechanism (see also the section on prokaryotic and eukaryotic translation) of such an effect are uncertain.
The proposal that the level of eIF4E phosphorylation is related to its cellular activity is derived primarily from reported correlations between its phosphorylation status and translation rates under different cellular growth conditions (218). Direct experimental evidence is thin on the ground. One study has indicated that the affinity of mammalian eIF4E for the cap structure is enhanced approximately threefold by phosphorylation (375), whereas other work has thrown doubt on the concept that phosphorylation controls the incorporation of eIF4E into the cap-bound eIF4F complex (418). A further controversial issue has been the actual site of phosphorylation on eIF4E. This was originally thought to be Ser53 in the mammalian factor, because mutation of this residue to alanine nullified various properties of wild-type eIF4E (115, 274, 319, 470). However, there have been conflicting data on the effects of the Ser-to-Ala mutation at position 53 (280). Moreover, subsequent biochemical analyses of the mammalian protein have demonstrated that the true major site of phosphorylation is Ser209 (154, 273). Parallel studies of the S. cerevisiae factor, which lacks the C-terminal serine, revealed that phosphorylation is generally weak, apparently not regulated in response to changes in cellular growth conditions, and located primarily at the N-terminal sites Ser2 and Ser15 (613). Both of these sites can be mutated to alanine with little effect on cellular growth or protein synthesis (613). However, in S. pombe, eIF4E does possess the Ser209 site (448), although current evidence indicates that S. pombe eIF4E is poorly phosphorylated (151). It must therefore be concluded that the question whether eIF4E phosphorylation provides the basis for a physiologically significant regulatory mechanism remains unresolved.
4E-BPs.
A set of mammalian 4E-BPs which can act as regulators of eIF4E function have been identified. For example, treatment of adipose cells with insulin leads to enhanced phosphorylation of rat PHAS-I (phosphorylated heat- and acid-stable protein regulated by insulin [one example of a 4E-BP] [42, 235]), which correlates with enhanced translation (328, 329, 421). Since it could be shown that 4E-BPs compete with eIF4G for binding to eIF4E (195), a model was proposed in which eIF4G-eIF4E binding is regulated by the eIF4E–4E-BP interaction, which in turn is subject to modulation by phosphorylation of the 4E-BP. Serum, growth factors, hormones, and stress can all modulate 4E-BP binding via a number of signal transduction pathways (172a, 508, 576a). There are sequence similarities between the eIF4E-binding domain of eIF4G (Fig. 4) and a region in PHAS-I and other 4E-BPs (346, 508).
The comparison with yeast is again an interesting exercise. Analogously to the 4E-BPs, the so-called p20 protein of S. cerevisiae is subject to levels of phosphorylation that vary in response to changes in growth conditions (613). The N terminus of p20 also carries a sequence resembling the motif found in both forms of S. cerevisiae eIF4G that is thought to comprise an essential part of the eIF4E binding site (15). These results are suggestive of a mechanism of regulation analogous to that of the 4E-BPs. It follows that the absence of p20 should allow the cap complex to achieve higher activities. However, under standard laboratory growth conditions, the observed effects have been small. One group reported a small increase in the growth rate of S. cerevisiae in rich medium associated with disruption of the gene encoding p20 (CAF20) (15, 315), while another group saw no significant change (119). On the other hand, the latter group found that a CAF20 disruption partially suppresses growth defects associated with mutations in other initiation factor genes. There is agreement that overexpression of this gene from a 2 μm plasmid slows growth by between 10 and 20% (15, 119). Further biochemical evidence for specific binding between eIF4E and p20 was obtained with a GST::p20 fusion protein (15). The GST::p20 fusion protein competed with eIF4G for binding to eIF4E, a result that is consistent with the idea that p20 acts like a yeast 4E-BP.
A more recent study has provided information relevant to the relatively limited capacity of p20 to compete with eIF4G for binding to eIF4E in vivo (449). Surface plasmon resonance analysis was used to compare the eIF4E-binding affinities of p20 and the eIF4E-binding domain of eIF4G. p20 was estimated to bind to eIF4E with an approximately 10-fold-lower affinity (Kd = 10−8 liter mol−1) than it bound to the eIF4E-binding domain of eIF4G (Kd = 10−9 liter mol−1). A further factor in the p20 regulatory equation is the fact that the control coefficient (Zi [see above]) for eIF4E activity in terms of the translation rate is likely to be considerably smaller than 1 (569), which therefore constrains the impact on translation that can be expected to be achieved by regulating the cap-binding interaction in this way. At the same time, p20 is present at relatively low levels in the yeast cell, partly as the result of rapid degradation of this unstable protein. Having rationalized the limited inhibitory capacity of p20, we still have to explain the function of a regulatory protein which, at least under laboratory conditions, seems not to exert a strong effect. It is possible that it normally acts as a fine regulator and/or that it is more potent under limiting growth conditions that are more similar to those of the native environment of natural yeast strains. There are clearly interesting issues of quantitative control here that remain to be addressed in future work. Moreover, alternative leads may have to be followed up. For example, apart from the N-terminal motif resembling part of eIF4G mentioned above, p20 also has a site of homology (amino acids 26 to 31) to eIF4E (amino acids 73 to 78). It is not yet known whether this is significant.
Are there other modulators of eIF4E function in S. cerevisiae? A genetic screen for extragenic suppressors of a temperature-sensitive mutant allele of cdc33 yielded two genes, DED1 and DBP1, both encoding putative ATP-dependent RNA helicases belonging to the DEAD-box family (119). The latter gene suppressed the mutant phenotype only when expressed from a multicopy plasmid. However, neither of the encoded proteins is known to be a direct modulator of eIF4E. The mutant Ded1p was found to cause distortion of cellular polysomal gradient profiles typical for mutant cells bearing a mutation in the translational initiation pathway (91, 119). It can be speculated that the identified helicase activities somehow compensate for the partial loss of eIF4A and eIF4B helicase activity that would otherwise be functionally linked to translational initiation via eIF4E. The possible existence of such a functional linkage now needs to be investigated. The complexity of helicase function is, however, underlined by the fact that DED1 has previously been identified as a suppressor of a gene defect in nuclear pre-mRNA splicing (269). Moreover, overexpression of the p20 gene (CAF20) strongly inhibited the growth of a ded1 mutant strain (119). These and other data are possibly telling us that we have to look at the pathway leading up to translational initiation on cytoplasmic mRNA to find the functional links between eIF4E, helicases, and p20. We are also reminded that the networks of genetic and biochemical interactions seen in such studies are indicative of the existence of finely balanced functional overlaps in yeast (as shown, for example, in Fig. 15).
Finally, while possible mechanisms of regulation of yeast eIF4E are suggested by analogy to mammalian cells, it is still not clear whether this route is used to mediate a cellular response to environmental signals. It has been proposed that the TOR (target of rapamycin)-containing signal transduction pathway (308) regulates eIF4E activity via modulation of this factor’s phosphorylation status (33). However, the authors present no evidence of any such change in eIF4E activity. In cells of higher eukaryotes, rapamycin-induced inhibition of translation seems to be mediated via 4E-BP1 (46). While rapamycin causes large reductions in the general rate of protein synthesis in S. cerevisiae, nitrogen deprivation has been found to repress expression of the cyclin-encoding CLN3 mRNA more specifically (164). The resulting reduction in Cln3 production is possibly one of a number of routes leading to arrest of the cell cycle in G1 phase. Both decreased CLN3 mRNA translation and increased turnover of Cln3 protein are involved (164). However, Gallego et al. (164) found no evidence of specific translational repression mediated by the CLN3 5′UTR, while concluding that the effect is independent of the TOR pathway. This contrasts with previous results relating to uORF-mediated inhibition of CLN3 translation (441). It is clear that there is still much to be learned about the role of posttranscriptional control mechanisms in the cell cycle. Indeed, the Trachsel and Altmann group has reported that the addition of rapamycin to S. cerevisiae causes degradation of eIF4G without affecting the abundance (or presumably the activity) of eIF4E (51a).
eIF2.
While a convincing case for physiologically meaningful regulation of eIF4E via phosphorylation has yet to be presented, there is little doubt about the role of eIF2 phosphorylation in regulating protein synthesis. The key players in this form of regulation, apart from eIF2 itself, are the kinases (and phosphatases) that act upon eIF2 and the guanine nucleotide exchange factor eIF2B. eIF2 is a heterotrimeric complex whose α subunit is phosphorylated by three known eukaryotic kinases: HRI and PKR in mammals and GCN2 kinase in S. cerevisiae (see references 96, 226, 227, 359, and 552 for recent reviews). The phosphorylation event prevents eIF2 from undergoing GDP-GTP exchange on eIF2B. Moreover, since the phosphorylated form has a greatly increased affinity for eIF2B, it acts as an effective competitive inhibitor of the cellular regeneration of active eIF2–GTP–Met-tRNAi. The essential details of eIF2 phosphorylation and their relationship to GCN4 regulation are summarized above in the section on reinitiation, while comprehensive analyses of the literature in this area are available elsewhere (96, 142, 226). Two points remain to be mentioned here. First, given the considerable number of uORF-bearing mRNAs in S. cerevisiae, there is a significant probability that GCN4 is not the only case of posttranscriptional control mediated via the status of eIF2 phosphorylation, whereby the regulatory effects in other systems may turn out to be less marked. Second, the extent to which eIF2-mediated regulation of overall translation rates features as a response by yeast to stress conditions is unclear. This situation contrasts with the range of eIF2-mediated regulatory phenomena being studied in higher eukaryotes (96).
Further gene-specific regulatory systems linked to Gcn4p activity.
The regulation of GCN4, mediated via eIF2 activity, is also coupled to other regulatory circuits by virtue of the transcriptional activation function of Gcn4p. One example of this was provided by a study of the expression of GCD5 (otherwise known as KRS1), which encodes lysyl-tRNA synthetase (316). The GCD5 gene has a consensus Gcn4p-binding site (TGACTC box) (378), and it was shown that GCD5 transcription is Gcn4p dependent (316). Analysis of the effects of a point mutation located in the lysine-binding domain of this synthetase led to the proposal of an autoregulatory model. According to this model, a reduced level of tRNALys charging by the synthetase activates GCN2 kinase and thereby induces GCN4, the consequence of which is activation of GCD5 transcription. The resulting increased levels of synthetase stimulate tRNALys charging, thus providing negative feedback within the regulatory loop (316). This coupling between the translational regulation of GCN4 and the transcriptional regulation of GCD5 may also apply to other genes encoding aminoacyl-tRNA synthetases.
Gene-Specific Regulation via trans-Acting Factors
Regulation via RNA-binding proteins.
Studies of gene expression in E. coli and its bacteriophages provided the first examples of translational regulation mediated by RNA-binding proteins targeted to specific motifs in the mRNA (363). The majority of the prokaryotic regulatory circuits are of a negative type, whereby binding of the effector interferes with the 30S subunit-TIR interaction. An alternative to this competitive binding mechanism is exemplified by E. coli S15, whose binding to its own TIR stabilizes a pseudoknot structure. The latter structure does not prevent binding of the 30S ribosomal subunit but, rather, acts to “trap” it in an SD region-associated complex which cannot proceed to polypeptide initiation (437). Since RNA-binding proteins can alter local mRNA conformations by binding to adjacent regions of the mRNA sequence, this means that access of the 30S ribosomal subunit to binding sites in an mRNA can also be positively influenced by a regulatory protein (16, 363, 600). Functional equivalents to the negative types of prokaryotic regulation are more easily imagined for eukaryotic translation than are positive-type mechanisms, and the characterized systems have so far been consistent with the expected pattern (364). Some of the principles of RNA-protein recognition in these prokaryotic and eukaryotic systems are becoming evident (see, for example, references 27, 48, 73, 139, 282, 360, 393, 495, 563, 564, and 568).
Although this review focuses on posttranscriptional control in yeast, it is appropriate to begin a consideration of gene-specific regulation via RNA-binding proteins with the higher eukaryotic system that has been most extensively characterized to date. The iron-dependent regulation of mRNAs containing iron-responsive elements (IREs) in vertebrates and insects involves the binding of the iron-regulatory proteins (IRPs) (289, 464). There are IREs located in the 5′UTRs of the mRNAs encoding the vertebrate ferritins, the erythroid form of δ-aminolevulinic acid synthase, and succinate dehydrogenase subunit b. The 5′ location of an IRE allows translational regulation to be achieved via binding of an IRP, since this protein has a sufficiently high affinity for IREs (Kd = 10−10 to 10−11 at low iron concentrations) to form a tight complex capable of blocking initiation by vertebrate 40S ribosomal subunits (464). The IRP-IRE affinity is reduced 50- to 100-fold in the presence of iron levels in excess of the requirements of the vertebrate cell. This range of iron-related affinity change is sufficient to mediate the required iron-sensitive regulation. The other location of IREs is the 3′UTR of the mRNA encoding the vertebrate transferrin receptor, whereby IRP binding at this site regulates the stability, rather than the translation, of the target mRNA. Translational regulation of vertebrate mRNAs mediated by an IRP is subject to a strong position effect. Analogously to the position dependence of inhibition via a stem-loop structure, cap proximity is a requirement for effective translational inhibition via IRP. Increasing the distance of the IRE from the cap greatly reduces the repressive effect of IRP binding (179).
The above work demonstrated that a translational repressor can function by targeting the 5′UTR of the mRNA of a higher eukaryote. However, the situation in yeast is less clear. Evidence has been presented that is consistent with translational regulation of the mRNAs encoding catalase (197) and the ribosomal protein L32 (109) in S. cerevisiae. However, in neither case has the regulatory mechanism been defined, and the results reported were not indicative of the operation of a high degree of control. Moreover, the regulatory role of L32 seems to be multifunctional, with recent evidence indicating that it also inhibits pre-35S rRNA processing and the splicing of its own pre-mRNA (574). Analysis of the expression patterns of other yeast ribosomal protein genes has also provided no unequivocal evidence for translational regulation (263). On the other hand, despite the relative paucity of information about endogenous translational regulation systems in yeast, recent analyses of regulatory components “imported” from other organisms into S. cerevisiae have been informative. Coupling an IRE-containing 5′UTR to a reporter gene in S. cerevisiae renders the encoded mRNA subject to translational regulation by IRP (413). Translational repression of the reporter mRNA can be achieved by adding recombinant IRP to a yeast cell extract or by expressing the IRP gene on a second plasmid in vivo. This work established that an IRP and an IRE are sufficient components to create a regulatory circuit and that this two-component system is fully active in a foreign cellular environment. Further studies were performed to investigate the regulatory properties of quite different RNA-binding proteins in yeast and higher cells. It turns out that the RNA-binding protein targeted to the 5′UTR need not normally fulfill the function of a eukaryotic translational repressor to be able to block eukaryotic translational initiation. Thus, the insertion of the respective binding motifs of the bacteriophage MS2 coat protein and the spliceosomal protein U1A into the 5′UTR of a reporter gene allowed both of these proteins to act effectively as repressors when expressed in yeast and HeLa cells (529). This indicates that in this context, it is the ability of the chosen protein to bind specifically to its corresponding target motif rather than its evolved cellular function that determines whether it is capable of repressing translation successfully. Clearly, in their original cellular environments, these RNA-binding proteins have evolved to provide specific regulatory responses under defined conditions.
A further study in S. cerevisiae has also contributed to our understanding of the mechanism of translational repression (290). Unlike IRP-mediated repression in cells of higher eukaryotes, the distance of the target IRE from the cap could be extended to 59 nucleotides without affecting repression in yeast. However, other experiments indicated that the relationship between IRP binding and the degree of translational repression in yeast are comparable to those seen in the vertebrate cellular environment. In particular, a range of IRE mutations that reduced IRP binding affinity by up to 160-fold manifested a progressive attenuation in the degree of translational repression, reaching almost complete abolition of the inhibitory effect (290). This mirrors the quantitative range of iron-induced adjustment of IRP-IRE affinity in mammalian cells. It was not possible to mimic the regulation of IRP binding affinity by changing the iron concentration in yeast, most probably because the internal iron levels in this organism do not respond in the same way as vertebrate cells to changes in external iron concentrations. Overall, the distinctions in the position dependence of repression between yeast and higher-eukaryote cells are indications of differences between the translational machineries of the respective organisms. One testable model explaining this is that the binding and scanning phases (Fig. 10, k1 and k2) of the respective higher- and lower-eukaryote 40S ribosomal subunits are driven by different thermodynamic forces. Most importantly, the scanning of a vertebrate 40S subunit may be coupled to a greater thermodynamic force than is its yeast counterpart, thus explaining the ability of the former to overcome a cap-distal IRP-IRE complex. We can conclude that translational regulation can be imposed on yeast mRNAs by the binding of RNA-binding proteins to specific motifs located in the 5′UTR. However, the parameters governing this regulation are different in yeast, reflecting key properties of its translational apparatus. It will be important to explore the general validity of these principles in whatever endogenous translational repression systems are found to function in this organism.
Regulating the expression of dsRNA genomes.
The genome of the cytoplasmic dsRNA viral particles frequently found in S. cerevisiae is transcribed to generate a single-stranded (plus-strand) mRNA that serves as the template for translation of the gag and pol genes (see the section on frameshifting). This mRNA is neither capped nor polyadenylated (70, 547). The lack of the 5′ and 3′ mRNA modifications means that the viral mRNA is potentially disadvantaged in terms of translation and also is at risk of being rapidly degraded by cellular 5′→3′ exoribonuclease activities (see the section on mRNA decay). Indeed, exoribonuclease-1 is encoded by SKI1 (XRN1), one of the yeast chromosomal genes originally identified as being involved in suppressing the dsRNA copy number (271). Mutations in SKI1, as well as in seven other SKI genes, were isolated on the basis of their ability to enhance the expression of killer toxin encoded by the M-type dsRNA satellite of dsRNA viruses (590, 592), thus creating a “superkiller” phenotype. For this reason, the SKI genes have generally been regarded as constituting a cellular antiviral system.
The functional interactions between the SKI gene products and viral replication and expression are only beginning to be understood (357, 593), but recent work has generated some remarkable results. Ski2p, Ski3p, and Ski8p, which are essential to the cell only in their capacity to suppress dsRNA viruses, may act by repressing the translation of nonpolyadenylated mRNA (357, 593). Clues to the molecular basis of this effect have come from studies of the relationship between the availability of active 60S ribosomal subunits and the propagation of dsRNA virus in the cell. Mutations reducing the size of the active 60S population inhibit virus propagation (81, 410), while this effect can be relieved by ski2, ski3, or ski8 mutations (364, 551). The model proposed by the Wickner group to explain this assumes that the Ski proteins influence the proposed recruitment of 60S subunits to the initiation process that is mediated via the poly(A) tail by virtue of their roles in 60S biogenesis (44, 364, 410). Recent work on ski6-2 has shown that this mutation causes defective 60S biogenesis and suppresses the poor expression of the poly(A)− viral mRNA seen in a strain whose 60S activity is compromised by a mutation in the L4 protein (44). It is argued that the ski6 mutation eliminates the specificity for polyadenylated mRNA normally imposed by Ski6p by virtue of its role in 60S assembly. This would mean that the specificity of ribosomes for different mRNA types can be readily modified, a principle that might have very significant implications for translational control.
Another notable aspect of the dsRNA viral system is the apparent relationship between Ski1p and the major coat protein (Gag) of the L-A virus. Gag (and its equivalent in the L-BC virus) becomes covalently attached to and removes the 5′ cap of a proportion of the cellular mRNAs (54, 55, 357). This activity is essential for synthesis of the M1 killer toxin in a SKI1+ host but becomes dispensable in a ski1 mutant (357). It has accordingly been postulated that Gag effectively generates decapped mRNAs that seem to decoy Xrn1p away from at least some of the viral uncapped mRNAs (357), allowing them to survive longer in the yeast cytoplasm. Given that the viral mRNAs are not delivered to the cytoplasm through the nuclear membrane, it would be interesting to know whether this affects their “presentation” to either the degradation machinery or the translational apparatus. Perhaps differences in the compartmentalization and/or kinetic control of the decay or translation of viral mRNAs contributes to the viral survival strategy. Other genes involved in controlling or regulating dsRNA viruses (such as the MAK genes [239]) are discussed by Wickner (592).
Gene-specific regulation via antisense RNA in yeast?
Studies of plasmid replication in E. coli demonstrated the principle of antisense regulation more than 17 years ago (251, 310). Since then, a whole range of cases of specific antisense RNA-mediated regulation has been uncovered in prokaryotic systems (244, 502, 576b). In general, prokaryotic antisense RNAs are between 70 and 110 nucleotides long and are capable of forming stem-loop structures with apical loops of 6 to 8 nucleotides. The stem regions are thought to confer both a clearly defined structure and stability on the RNAs, while the loop nucleotides are believed to be important for the initial interactions with the target molecules. The binding of prokaryotic antisense RNAs can modulate a number of different target functions, including RNA processing, translational initiation, transcriptional termination, and mRNA stability (502). The prokaryotic work therefore demonstrates that antisense RNA can act as a versatile modulator of gene expression.
In contrast, the role of antisense RNA in eukaryotic cells is much less clear. There are numerous examples of eukaryotic genes that are transcribed on both strands, meaning that potential antisense RNAs are undoubtedly generated in the cell. However, unequivocal evidence for natural antisense regulation is scarce. Despite this, artificial antisense RNA constructions have been used successfully in both plant and animal cells, although the mechanisms underlying the inhibitory effects observed have received little attention. The approach used has generally been based on simple empirical principles or trial and error.
S. cerevisiae is a particularly poor host for experiments of this type (23, 26), yielding only the occasional success (400). The apparent ineffectiveness of antisense RNA strategies in S. cerevisiae has led to the suggestion that at least this yeast has characteristics that somehow foil the standard procedures (26). S. pombe might be more amenable to this type of imposed gene suppression (23). However, it should be emphasized that there has been little systematic investigation of the possible mechanistic pathways that might lead to antisense RNA suppression in eukaryotic cells in general and in yeast in particular. The standard approach involves the generation of RNAs that are complementary to smaller or larger regions of the target mRNA via transcription of the second strand. However, comparison of the natural prokaryotic systems suggests that this strategy is unlikely to provide optimal conditions for sense-antisense regulation because it ignores the influence of antisense RNA structure on stability and the kinetics of antisense-sense interactions. Consistent with this view, recent studies with translation cell extracts derived from S. cerevisiae have shown that at least in vitro, a modified form of the E. coli IS10 RNAOUT/RNAIN regulatory system can be used to regulate translational initiation on a eukaryotic mRNA in a yeast environment (28). By targeting the RNAIN sequence inserted upstream of a reporter gene, it could be shown that an antisense RNA can block yeast translation via interactions with a 5′UTR target site. This suggests that the prokaryotic type of specifically targeted antisense regulation may be possible in yeast, given the appropriate set of conditions. It follows, therefore, that it may be possible to establish a set of rules that allow the design of reliable antisense RNA constructs capable of imposing regulation on selected eukaryotic (yeast) genes. However, this will depend on further characterization of the mechanistic principles required for antisense RNA regulation. At the same time, we may learn the extent to which antisense-RNA-mediated regulation could, at least theoretically, offer an alternative route to protein-mediated regulation.
CONTROL OF MRNA DECAY
Much of this review has been concerned with the functional lives of mRNA molecules, but these are all eventually terminated by enzyme-catalyzed degradation reactions that lead to functional inactivation and then complete hydrolysis. This turnover process would be of limited interest if it were not for the fact that mRNA decay is subject to a number of controlling influences. As a result, the estimated physical half-lives of mRNA in S. cerevisiae vary from less than 1 min to over 60 min (215, 267). This means that mRNA decay contributes to the differential control of gene expression. Moreover, there are strong indications that at least some mRNA turnover rates are regulated in response to environmental changes. Specialized reviews on eukaryotic mRNA degradation have been published at regular intervals (the more recent ones include references 78, 266, 352, 353, 454, 462, 463, 468, and 546), and the reader is directed to these for comprehensive coverage of the relevant literature. There is increasing interest in the role of mRNA degradation in the yeast mitochondrion (355). Information about the various methods used to study mRNA decay in yeast is available elsewhere (29, 78, 215).
The main objectives of this section are to summarize the basic principles that have emerged from studies of nonaberrant mRNA decay in the cytoplasm of S. cerevisiae and to consider how these fit into the overall picture of posttranscriptional control in this organism, especially in relation to translation. mRNA decay can be conveniently considered in terms of cis-acting mRNA determinants, trans-acting factors, the order and causality of decay events, and the potential mechanisms of regulation, and these themes will provide the framework for this brief overview.
cis-Acting mRNA Determinants
Stability determinants in the main ORF and 3′UTR.
One of the major unanswered questions in the study of mRNA decay is how the specific decay rates of individual mRNAs are determined. Experimental analysis of a number of decaying mRNAs extracted from cells in which transcription has been blocked reveals that, in general, whereas full-length or near-full-length mRNAs are readily detectable, smaller intermediate products generated by endo- and exonucleolytic cleavages of natural mRNAs are comparatively short-lived. This indicates that degradation, once triggered by an early event that does not greatly change the overall structural integrity of the mRNA, is rapidly completed. Control over the observed stabilities of different mRNAs is therefore exercised early in the overall pathway. It is also important to note that the stabilities of eukaryotic mRNAs are generally defined in terms of the half-lives of physically intact mRNAs comprising their full DNA-encoded sequences, rather than in terms of functional inactivation. By analogy to the protective effects of ribosomes and RNA-binding proteins on prokaryotic mRNA (80, 245), it might be expected that the stability of eukaryotic mRNA should be related to the accessibility of potential cleavage sites in each mRNA molecule. In an extreme model, these sites would be highly redundant and would therefore be frequently represented in all mRNAs. Such a model would predict that the number of potentially accessible sites, and the relative periods of their exposure, would be influenced by the density of ribosomal loading. However, general correlations between mRNA stability and features likely to affect the number and proportion of mRNA sites occupied by ribosomes at any one time, including transcript length, codon usage, and the kinetics of translational initiation and elongation, are not identifiable (78, 215, 438, 482, 570). This in itself does not rule out any participation of such determinants in the turnover of various mRNAs, but it does suggest that other features are responsible for controlling the rates of decay. As will become apparent, much less is known about key rate-controlling features in mRNA decay than in translation. The acid test for this is to ask whether it is possible to make an educated guess at the likely stability of an mRNA on the basis of knowledge of its sequence. This will generally not be possible, whereas a number of clues to the translation efficiency of an mRNA are readily identifiable.
The poly(A) tail is a general feature of PolII mRNAs and is thought to play a general role in controlling the onset of decay in many mRNAs (see the section on decay pathways). Other stability determinants exist in various forms and act at a number of different locations within the mRNA (Fig. 16). In most cases, their influence is position dependent. The analysis of a small number of relatively unstable mRNAs (HIS3, MATα1, MFA2, and STE3) has revealed the existence of segments of the coding region or the 3′UTR that are capable of inducing rapid decay. “Cut-and-paste” experiments and deletion analyses have been used to achieve approximate delineation of the HIS3 and STE3 destabilizing regions (211, 213, 267). The relatively compact MATα1 element has been characterized to a considerably greater degree than any of the other putative internal stability elements. It has been defined as an entity comprising 65 nucleotides, of which the first 33 are rich in rare codons and the C-terminal region is AU rich. Destabilization by this element seems to require translation through it, since the introduction of a stop codon 5′ of it stabilizes the MATα1 mRNA two- to threefold (213). It has been proposed that the rare-codon region helps decelerate translating ribosomes, which then interact somehow with the AU-rich region, perhaps via an unknown factor. It is not known how this proposed mode of action then leads to destabilization. The 3′UTR can also contain destabilizing elements. The 3′UTR of STE3 was found to be capable of destabilizing a truncated PGK1 mRNA (211), while mutations in the MFA2 3′UTR resulted in stabilization of the MFA2 mRNA (388).
FIG. 16.
Features of yeast mRNAs influencing stability. The major types of mRNA stability determinant reported so far for S. cerevisiae are shown. The majority of these elements act to destabilize the mRNAs in which they have been studied. Internal stop codons (here indicated as UAA) can cause strong destabilization in 5′-proximal positions (strong) but have less or no effect at more distal positions (weak). Apart from the cap or poly(A) tail, it is not clear whether discrete (and transferable) stabilizing elements exist.
It is not clear whether there are any common features shared by these destabilizing elements. No mechanisms of action have been characterized, but certain consequences in terms of the decay process have been reported. Deadenylation and decapping were found to be accelerated in the presence of the MFA2 and MATα1 destabilizing elements (79, 389). Both events feature prominently in the proposed pathways of yeast mRNA decay (see below). Also unclear is the extent to which the potential decay behavior of a given mRNA can be defined in terms of the identified determinants alone. For example, the inactivation or removal of the MATα1 or MFA2 elements generates mRNAs with only intermediate stabilities (213). Such results suggest that other, as yet undefined, elements also contribute to the stability of each mRNA. Indeed, taking this further, could it be that a combination of structural attributes, perhaps even spread over the whole mRNA, determines stability? If so, the determination of mRNA stability may generally be a complex function of relatively large domains of molecular structure. This model would explain why at least some elements are only fully functional in certain mRNA environments (211). An alternative explanation of such context effects could be the existence of specific stabilizing sequences which, when combined with destabilizing elements, might neutralize the effects of the latter elements. However, until it can be proved that a range of appropriately placed, discrete stabilizing and/or destabilizing elements exist, there is little reason to accept the latter model. Relevant to this point is the observation that the PGK1 mRNA requires translation to maintain its high stability. It remains to be seen whether this behavior is attributable to the combined action of a number of discrete stability elements or to the overall structure of much of the mRNA.
Modulation of decay via the 5′UTR.
The 5′UTR can be home to a number of structural elements that influence stability (Fig. 16). The effects of many of these elements depend on the nature of the mRNA downstream of their location in the sequence. The most generally relevant feature is the cap structure, which is recognized to fulfill a protective function in terms of 5′→3′ exonuclease activity (78, 162). Other features, such as a stem-loop, a poly(G) stretch, or the formation of a protein-target complex in the 5′UTR, affect the stabilities of only certain types of mRNA (37, 336, 390, 482, 570, 576). They are thought to exert their influence in two ways.
First, by inhibiting translation, they may change the activity of (as yet undefined) stability determinants within the body of the mRNA (see, for example, references 76, 213, 266, and 336). Consistent with this is the observation that the degree of destabilization of the PGK1 mRNA caused by a stem-loop or IRP-IRE interaction in the 5′UTR correlates with the degree of translational inhibition imposed on the mRNA by these elements (336). However, in mutant strains whose translational apparatus is dependent on partially inactivated eIF4E proteins, the reduction in translation capacity did not correlate with destabilisation of PGK1 mRNA. This suggests that it is the disruption of the initiation process on the mRNA, rather than the reduction in the frequency of translational initiation per se, that controls the destabilization potential of the PGK1 stability determinants. Blocking the scanning process may disrupt intra- or intermolecular interactions that disturb the formation of translation-competent polysomes in a way that is distinct from the effects of reducing the frequency of ribosome-mRNA interactions via eIF4E. Alternatively, it has not been ruled out that such mutations in eIF4E affect events prior to translation, such as mRNA transport or compartmentalisation. It should, however, be pointed out that the PGK1 mRNA may be atypical in its response to elements in the 5′UTR. Stem-loop structures have no effect on the stabilities of MFA2, YAP1, or cat mRNAs in yeast (37, 336, 482, 570, 572), and they stabilize the LUC mRNA (336). These differences are likely to reflect the distinct parameters dictating the decay behavior of the respective mRNAs.
The second effect of structural elements in the 5′UTR is to inhibit directly the progress of exonuclease activities. Poly(G) stretches and, to a lesser extent, hairpin loops of stabilities sufficient to cause strong translational inhibition (−10 to −20 kcal mol−1), act to impede 5′→3′ exonucleolytic degradation (390, 576). The balance between the contributions of the two types of effect discussed above will be a function of the presence or absence of stability elements and the decay pathways of individual mRNAs. This complicates the interpretation of the observed influence of structure in the 5′UTR on stability.
Further experimental data that are consistent with there being, in broad terms, a relationship between translation and mRNA decay have been obtained by imposing general blocks on translation. Cycloheximide (215, 429) or a defective tRNA nucleotidyltransferase (429) inhibits translational elongation, thereby increasing the average size of cellular polysomes. This has been found to stabilize a number of mRNAs. However, it is difficult to interpret these results reliably in terms of protective interactions between the translational apparatus and the degradation machinery on individual mRNAs because of the indiscriminate nature of the inhibition caused by these measures. Such general attenuation of protein synthesis may lead to rapid distortion of the balance of cellular components that directly or indirectly affect mRNA stability. It should also be pointed out that verrucarin A has also been found to stabilize certain mRNAs even though it greatly reduces polysome sizes (532). As discussed above, another means of inhibiting translation is to inactivate one of the translation factors. Work with a prt1 mutant strain (defective in eIF3 [Table 1]) revealed preferential destabilization of two mRNAs (SSA1 and SSA2) encoding proteins belonging to the Hsp70 heat shock protein family (34). Cereghino et al. (84) found that inactivation of prt1 led to destabilization of SDH2 mRNA but not of ACT1 or CUP1 mRNA. Such differential effects are presumably due to the influence of specific elements or structural characteristics in the respective mRNAs. However, the responses mediated by such elements may differ according to the step of translation that is disrupted. For example, it is not known whether defects in eIF4E (compare reference 336) are capable of generating comparable differential destabilization to that seen with the prt1 mutant. Mutants with mutations in GCD2 or SUI2 were found not to destabilize SDH2 mRNA (84). In contrast, a single amino acid substitution in eIF5A has been found to stabilize a number of mRNAs, causing the host cell to accumulate uncapped mRNAs (620).
An uORF constitutes a quite different type of 5′UTR element. The presence of an uORF can, but need not, lead to accelerated degradation of an mRNA (411, 469, 572). The experiments of Oliveira and McCarthy (411) showed that the already relatively unstable cat mRNA can be destabilized by approximately a factor of 4 as a result of a single nucleotide change (AAG to AUG) in the 5′UTR that generates a seven-codon uORF. Given the foreign nature of the cat mRNA, this and other results suggested that natural yeast stability elements may not be required for the destabilization process. Investigations of natural uORFs have revealed a more complex picture. The PPR1 5′UTR has been identified as having transferable destabilizing properties (438). It contains a six-codon uORF that overlaps +1 at its 3′ end (AUA UGA) with the start codon of the main ORF. Fusion of this leader in the same configuration with the PGK1 ORF generates a highly unstable mRNA, while the effect is nullified if the AUG codons at positions 1 and 2 of the uORF are mutated to AAG (336). This suggests that the stability determinant of the PPR1 5′UTR that is capable of destabilizing the PGK1 mRNA includes this natural overlapping uORF. This contrasts with the earlier results of Pierrat et al. (438), who observed no change in the stability of the unfused PPR1 mRNA when the two AUGs in the overlapping uORF were mutated to AGGs (438). Resolution of this apparent discrepancy will require further analysis of the role of the PPR1 coding region in controlling decay.
Two natural 5′UTRs containing nonoverlapping uORFs that were considered in the earlier section on reinitiation have also been subject to recent investigations: the GCN4 leader, which normally has no detectable effect on the stability of its mRNA, and the YAP2 leader, which naturally destabilizes its own mRNA. It has been proposed that the GCN4 leader does not destabilize mRNA because it lacks a downstream element of the type previously reported to be required for nonsense-dependent destabilization in mRNAs carrying nonsense codons (469). To be able to assess this potential relationship between premature termination and the role of uORFs, it is necessary to briefly consider the current models that attempt to explain how aberrant mRNAs containing premature stop codons are dealt with in the cell.
The destabilizing effect of internal nonsense codons in mRNA was observed first in E. coli (229) and then somewhat later in yeast (342). Nonsense codon-dependent accelerated decay has since been observed in a number of yeast mRNAs (266, 322, 323, 427, 611, 616), and unspliced pre-mRNAs (209), but most investigations of this phenomenon have been performed with the PGK1 mRNA. Destabilization of the PGK1 mRNA was initially reported to be dependent on the presence of a downstream element comprising some 80 nucleotides 3′ of a premature termination codon (430). Two AUGs in this element were originally found to be required for destabilization. However, a later report concluded that the critical element is the sequence motif UGYYGAUGYYYYY (616), and the most recent analysis concludes that the AUG in this motif is not required for activity (469).
This leaves a highly degenerate 13-nucleotide pyrimidine-rich region as the proposed motif. Moreover, this motif was found to be nonfunctional in the absence of flanking sequences (616), and therefore the extent to which these additional regions dictate the stability of the mRNA is not clear. The motif was also found not to be necessary for nonsense codon-dependent destabilization in at least one PGK1 construct (430). Sections of more than 100 nucleotides containing regions resembling the motif were taken from the ADE3 and HIS4 genes and tested for their ability to promote nonsense codon-dependent destabilization of a “mini-PGK1” gene that carries an N-terminal amber codon but lacks most of the normal PGK1 coding region (616). These regions also supported accelerated decay. Models proposed for the mode of action of this motif have predicted either that it interacts with the 40S subunit or an undefined factor to promote translational reinitiation or that this interaction causes ribosomal pausing (432). The interaction with the ribosome was suggested to be mediated via the partial complementarity of the motif to a region of 18S rRNA (430), but this idea will require particularly careful testing, given the high degree of redundancy apparently permissible in the motif, the fact that the corresponding region in the 18S rRNA is predicted to be involved in intramolecular basepairing (110), and the observation that the AUG is nonessential (469). The lack of a requirement for an AUG also means that reinitiation within the motif plays no essential role. Moreover, the role of secondary structure in the mRNA downstream of nonsense codons remains uncharacterized. Whatever signals are involved, premature termination in PGK1 or a truncated version of it leads to accelerated decay via a mechanism involving the UPF gene products (see the discussion of trans-acting factors). This dependence on the UPF genes is a feature of nonsense codon-mediated decay in the mRNAs studied so far (266). However, UPF-dependence is apparently not exclusive to the decay of nonsense codon-containing mRNAs; at least two non-aberrant mRNAs that lack early nonsense codons are degraded via upf-dependent pathways (78; compare reference 33a).
There is a nonlinear gradient of sensitivity to the presence of premature stop codons in PGK1 (Fig. 16), so that stop codons in the last quarter of the reading frame have no effect, stop codons in the first 55% accelerate decay approximately 12-fold, and there is a transition in effect over the intervening region (431). This polarity effect in the destabilization potential of nonsense codons has been explained in terms of the existence of localized stabilizing sequences in the PGK1 reading frame that are proposed to modulate the function of the destabilizing motifs (431). The proposed stabilizing elements have yet to be characterized. The extent to which the secondary structure of regions containing the putative elements and the potential interactions between such regions play a role is a matter for discussion. A further matter worthy of consideration is that even upf-independent accelerated decay of the PGK1 mRNA, caused by inhibition of translational initiation, reacts very sensitively to changes in the translation rate (see above). As suggested in the previous section, this may mean that an unusually complex set of factors modulates PGK1 decay behavior. On the other hand, since the HIS4 and CYC1 mRNAs show similar position gradients of nonsense codon-dependent destabilization (193, 611), it is likely that certain generally acting principles are involved. Further research is needed to determine the extent to which the polarity effect is attributable to the specific action of localized stability determinants as opposed to more general principles of control related to the overall structure of mRNP and polysomes (see Fig. 18).
FIG. 18.
Polysome disruption as a key process in mRNA degradation. An integrated scheme of mRNA decay events in yeast is shown. Triggering events are generally likely to disrupt the mRNP and polysome structure, which may therefore constitute the common intermediate step leading to further decay events. Since mRNA interacts with both nuclear and cytoplasmic proteins on its pathway through the nucleus into the cytoplasm, the interactions with both groups of proteins may be relevant to the control of mRNA degradation. Further research is expected to reveal whether this mechanism or more specific coupling mechanisms underlie decay pathways.
Returning to the GCN4 mRNA, it has been proposed that termination on one of the uORFs of its leader can be coupled to upf-dependent destabilization via the same means as that which applies in the case of premature termination within a reading frame, provided that a downstream motif of the type described above is introduced 3′ of the uORF (469). It has been argued that the wild-type GCN4 mRNA lacks the elements required for destabilization (469). (For the sake of completeness, it should be noted that another group has described transient destabilization of the wild-type GCN4 mRNA following translational derepression [309]). A segment of more than 200 nucleotides derived from PGK1 was inserted 3′ of uORF1 or uORF4 to achieve destabilization. However, upon inspection, this region was found to have the potential to form stable secondary structure (including individual stem-loops with predicted stabilities of up to −24 kcal mol−1) (573). It is possible that these structured sections in the inserted RNA or other, as yet undefined properties play a role in the destabilization caused by the PGK1 segment. In a later paper, the Peltz group reported that the action of the PGK1 downstream element can be inhibited by inserting a “stabilizing element” between the termination codon of the uORF and the destabilizing element (469a). The stabilizing element they describe is in fact an AU-rich segment derived from the GCN4 leader (nucleotides +498 to +565). It is of interest to consider these results in the light of the following discussion of studies with the YAP2 system.
Termination in the 5′UTR.
Since only modified forms of the GCN4 leader can destabilise mRNA, it has been unclear for some time to what extent termination-linked accelerated decay might play a role in the turnover of natural, non-aberrant mRNAs. This situation was changed by the observation that the YAP2 mRNA is subject to uORF-dependent destabilisation (572). Not only do the YAP2 uORFs inhibit translation, they also accelerate mRNA decay (Fig. 13B). This mRNA is therefore under further investigation because it throws light on the use of termination-linked mRNA destabilization as a means of imposing posttranscriptional control on non-aberrant cellular mRNAs (572, 573). A number of features of the YAP2 mRNA are of interest. First, destabilization by the uORFs of either the YAP2 mRNA or reporter mRNAs is to a great extent upf independent. This distinguishes the YAP2 system from nonsense codon-dependent destabilization in aberrant mRNAs. Second, in this natural mRNA, there is no evidence that uORF-dependent destabilization is dependent on the presence of the CU-rich type of sequence element. This may reflect the lack of dependence on the UPF genes for decay but might also be attributable to a high level of redundancy in the types of mRNA element and structure capable of supporting destabilization. It has also been observed that nonsense codon-dependent decay of PGK1 mRNA can be triggered in the absence of the UC-rich motif (430). Third, there is a strong correlation between a combination of efficient recognition of the uORF start codon plus inhibition of downstream reinitiation and the destabilizing effect of the uORF. This was established by manipulating uORF structure and examining the consequences in terms of translational inhibition and decay rate. One of the structural features promoting accelerated decay in the YAP2 system is the presence of a CG-rich sequence downstream of the uORF termination codon. This is a feature shared by GCN4 uORF4, which was also found to be capable of destabilizing both YAP mRNAs (573). A proposed explanation for this behavior is that the decay rate is influenced by ribosomal termination and release. A further piece of evidence in favor of this model is that the insertion of a stem-loop structure 3′ of the uORF leads to destabilization of the mRNA even if the uORF structure alone is not capable of causing destabilization (573). This effect is generally diminished if the stem-loop structure is located at a greater distance from the uORF. The influence of structure on uORF function seen here is also relevant to the experiments performed on the GCN4 leader (469), since the downstream elements inserted 3′ of the GCN4 uORFs are potentially structured (see above). Fourth, reinitiation (or suppression of the frequency of termination [and ribosome release] on the uORF) acts to suppress destabilization. This may be linked to the ability of the ribosomal subunits to remain on the mRNA after termination, or to scan through the potentially destabilizing uORF, or could be coupled to the reinitiation process.
Given the existence of a considerable number of different uORF-containing mRNAs in S. cerevisiae (Table 3), the apparent role of uORFs in mRNA decay may be of more general relevance. The mechanism underlying uORF function in YAP2 has not yet been fully characterized, but certain properties of this leader have become apparent. Experiments with gcd and gcn mutations have indicated that the activity of eIF2 in the cell influences both the reinitiation capacity of ribosomes terminating on YAP2 uORF1 and the destabilizing potential of posttermination events (573). This becomes evident when using one of the constructs mentioned above in which a YAP uORF was followed by a stem-loop structure. For example, in a gcd2 strain with reduced eIF2 activity, the destabilizing effect was enhanced, much as would be expected if fewer ribosomes had acquired reinitiation competence by the time they reached the structural impediment in the leader. This supports the notion that the effects of the YAP uORFs on translation and mRNA decay are tied into the same set of functional principles as those that govern GCN4 translation. Indeed, there are probably some further similarities between the modes of action of the GCN4 and YAP2 uORFs.
Two examples taken from the available experimental data illustrate this. First, it was found that the destabilizing effect of the PGK1 downstream element was reduced upon extending its distance from the uORF stop codon (469a). If the secondary structure of the large downstream element is important, this result is consistent with the effects of increasing distance between a stem-loop and the YAP uORFs (573). In both cases, the increased distance between the uORF and the element inserted downstream gives the ribosomes more time to reacquire (re)initiation competence, thus becoming resistant to release caused by the downstream structure. Second, the stabilizer element described by the Peltz group is an AU-rich sequence. When placed downstream of an uORF, this type of sequence promotes reinitiation and does not support uORF-related destabilisation (226, 572, 573). These considerations raise the question whether the segments tested in the GCN4 system really represent specific mRNA stability elements or, rather, are further examples of general structural features that influence the posttermination behavior of ribosomes. The latter concept would provide a potentially unifying model to explain the behavior of the respective uORF-containing systems.
It therefore remains to ask why, if the YAP2 uORF and GCN4 uORF4 are really of the same type, does GCN4 uORF4 not succeed in destabilizing its own mRNA? One possibility is that the GCN4 main ORF is relatively resistant to attack by the degradation machinery in an mRNP particle or polysome with a low ribosomal loading density and that more than termination on the uORF is required to destabilize this “protected” status. There is good reason to believe that there is significant variation in the susceptibility of different mRNAs to accelerated decay in disrupted or poorly translated polysomes (as exemplified, for example, by a comparison with PGK1). Further experimentation should be directed to testing this and the localized-stability-element model.
Various structural features can influence mRNA stability.
Summarizing the current understanding of stability elements in yeast mRNAs is not a simple matter. It is not yet clear how numerous or diverse the proposed stability elements found in the main coding region or 3′UTR of nonaberrant mRNAs really are. Of the stability elements studied so far, the MATα1 element has been most intensively studied. At least one type of downstream element associated with nonsense codon-dependent decay contains highly degenerate UC-rich tracts, but the requirement for other sequence components remains undefined. Overall, the current data do little to convince us that stability is related to the action of a small group of destabilizing elements. The challenge, therefore, is to establish whether discrete internal elements of the MATα1 type are a common feature of yeast mRNAs. Moreover, future work will also have to determine whether discrete stabilizing elements are present in yeast mRNAs. If so, it will be important to determine whether they are transferable, independently acting stabilizers, or (more degenerate) sequences and/or structures that act to suppress the function of destabilizing elements, perhaps by obscuring or affecting the structures in which the latter are involved.
Should the apparent lack of a readily discernible pattern in the roles of stability elements in yeast mRNAs give us cause for concern? Comparison with the situation in E. coli reassures us that this is nothing unusual. It seems likely that, as in the prokaryotic case, alternative structural options are available to achieve a given rate and/or course of decay and that clearly identifiable motifs or structural elements may be difficult to identify, especially within the body of the mRNA. A useful comparative example is the cleavage pattern of RNase E in E. coli. This enzyme cleaves at a large number of what appear to be only loosely related cleavage sites. Thus, recognition of sites that play a key role in controlling decay rates does not depend on the existence of highly conserved sequence motifs. While yeast mRNAs may not be subject to this type of decay, the elements controlling their decay may be at least as diverse in sequence and structure. This generally flexible strategy also seems likely to be advantageous, since it enables the cell to determine individual mRNA half-lives by using combinations of different, most probably varied and possibly delocalized structural features within the body of the mRNA (modelled in reference 80). Additionally, other layers of control can be imposed by more discrete elements, including the poly(A) tail, the 5′ cap, certain uORFs, and what so far seems likely to be a small number of internal discrete stability elements such as that in MATα1. Each of these elements exerts a relatively powerful effect as an independent unit. The flexibility of the overall system also allows mRNA turnover rates to be made either dependent or independent of translation rates. Interestingly, discrete consensus stability elements are more a feature of mammalian and plant mRNAs than of S. cerevisiae mRNAs (462, 463). It remains to be seen whether S. pombe is different in this respect.
The 5′UTR elements can be considered in relation to their effects on the translational initiation process, and on the decay process itself. Inhibitory elements like stem-loops or protein-target complexes affect the stability only of an mRNA which, like PGK1, has internal elements or overall structural features which allow it to react to translational inhibition at this step. As discussed below, the effects of the latter type of activity are dependent on the pathway of decay of each mRNA. Certain uORFs seem to be able to act generally to destabilize mRNAs and apparently do not necessarily require (translation-independent) internal elements to do this. The YAP2 type of uORF can apparently link termination to mRNA decay, and there is evidence that it acts (in combination with short flanking sequences) as a discrete, transferable agent. At the same time, it acts as a very strong inhibitor of translational initiation. Work on derivatives of the GCN4 mRNA, on the other hand, suggests that other elements can also be involved. Studies of uORF-dependent destabilization may therefore provide an alternative route to characterizing at least certain forms of decay in terms of clearly defined mRNA elements. Moreover, as we have seen, further detailed study of these systems may help us identify a unifying model for 5′UTR-mediated decay that links disruption of the translation process to destabilization (see Fig. 18).
trans-Acting Factors
A number of yeast mRNA degradation activities have been partially characterized (Table 4). The Stevens laboratory described a 5′→3′ exonuclease activity (175 kDa) that attacks mRNA (234, 520, 524) and an mRNA-decapping enzyme (522). The gene encoding the exonuclease was subsequently cloned and given the designation XRN1 (5′-exoribonuclease 1 [317, 523]). Later work described further genes, DCP1 (38, 311) and DCP2 (142a), which encode decapping activities. Gene disruption experiments have revealed that none of these genes is essential individually, but the resulting mutants are growth impaired and contain mRNAs with longer half-lives. There is, however, a second 5′→3′ exonuclease activity (5′-exoribonuclease 2; 116 kDa) in S. cerevisiae, which is encoded by the essential gene HKE1 (283, 521, 522, 525). The amino acid sequences of Xrn1p and Hke1p show significant similarities, and both proteins are multifunctional (5, 17, 281). Xrn1p is a cytoplasmic protein involved in a number of activities other than mRNA degradation, including DNA strand exchange and 5′→3′ exonucleolytic degradation of DNA (220, 272). Indeed, XRN1 has also been identified in different roles as DST2, KEM1, RAR5, SEP1, and SKI1 (234, 285, 287, 318, 520, 550). The functions of Hke1p, which is nuclear (272, 283), are less clear, but mutations that mislocalize it to the cytoplasm allow it to suppress the phenotype of an xrn1 deletion (272). HKE1 is identical to RAT1 (17) and TAP1 (132). The functional interchangeability is also reciprocal; targeting Xrn1p to the nucleus complements an hke1 mutant (272).
TABLE 4.
trans-acting factors known or thought to be involved in mRNA decay in S. cerevisiae
| Name | Function | Reference(s) |
|---|---|---|
| Enzymes involved in general decay | ||
| Xrn1p | 5′→3′ exonuclease (plus other functions) | 234, 520, 524 |
| Hke1p (Xrn2p) | 5′→3′ exonuclease (plus other functions) | 283, 521, 522, 525 |
| Dcp1p and Dcp2p | Decapping enzymes | 38, 311, 522 |
| Rrp41p/Ski6p | 3′→5′ exonuclease (5.8S rRNA processing) | 20, 379 |
| Other activities influencing decay | ||
| PAN | Pab1p-dependent poly(A) nuclease | 344, 475 |
| Ski2p, Ski3p, Ski8p | 20, 75 | |
| Mrt1p, Mrt3p | 204 | |
| Nonsense codon-dependent decay | ||
| Upf1p | Polysome-associated ATPase/helicase (interacts with eRF1 and eRF3) | 25, 105, 108, 108a, 131, 266, 322, 323, 584, 585 |
| Upf2p | Can bind other Upf proteins relatively strongly | 104, 107, 210, 266 |
| Upf3p | Predominantly nuclear (influences nuclear transport) | 25, 266, 321, 323 |
A number of proteins are now thought to influence mRNA decay via the 3′ end. An RNase activity which might catalyze deadenylation in vivo has been detected in vitro (PAN). This activity is at least partially Pab1p dependent (344) but has yet to be characterized in any detail (475). Pab1p may be involved in controlling poly(A) tail lengths via interactions with Rna15p, which is one of the components of the yeast 3′-end RNA-processing complex CF1 (18, 377). Early work on Rna14p and Rna15p suggested that these proteins might be involved in the regulation of mRNA deadenylation (348, 376). However, other evidence points to a role in polyadenylation (18, 78, 349, 377), and therefore these two proteins can be given only very tentative positions among the cast of players controlling mRNA decay.
The most striking recent findings in this area have concerned a number of proteins that either catalyze or modulate 3′→5′ exonucleolytic degradation of mRNA. The Tollervey group described a heteropentameric complex in S. cerevisiae, named the exosome, which is required for 3′ processing of the 5.8S rRNA (379). Three of the exosome proteins, Rrp4p, Rrp41p, and Rrp44p, were shown to exhibit intrinsic 3′→5′ exonuclease activity. Rrp41p is identical to one of the Ski proteins (Ski6p) (see the section on dsRNA viruses, above). Moreover, sequence similarities were identified between Rrp44p and bacterial RNase II and between three other exosome components (Rrp41p, Rrp42p, and Rrp43p) and bacterial RNase PH. These data suggest that the exosome is a multi-RNase complex. A subsequent analysis revealed that mutations in SKI6/RRP41 and RRP4, as well as in three further SKI mutants (ski2, ski3, and ski8), attenuated 3′→5′ degradation of a poly(G)-protected mRNA fragment derived from MFA2 or PGK1 (20). These and other data suggest that exosome components are involved in 3′→5′ mRNA decay and that Ski2p, Ski3p, and Ski8p modulate this activity. Since Ski2p is a DEVH box protein, it is tempting to speculate that its involvement is somehow related to an intrinsic RNA helicase activity (20). This would be reminiscent of the bacterial “degradosome” (19, 374, 450, 451), which comprises a helicase (RhlB), an endonuclease (RNase E), an exonuclease (polynucleotide phosphorylase, PNPase), and an enolase. In vitro data indicate that the helicase activity in this complex facilitates 3′→5′ degradation through structured RNA. Whether the association with enolase in some way links (localized) energy metabolism to the ATP-dependent functions of the degradosome remains to be seen. Future work should tell us whether the exosome constitutes a eukaryotic equivalent to the degradosome and whether its specificity is subject to modulation via other proteins, including those of the Ski type (20).
The relevance of Ski-modulated 3′→5′ decay to cellular mRNA metabolism seems to be underlined by the previous observation that mutations in SKI2 and SKI3 are synthetically lethal with deletions of XRN1 (271). One possible concern here might be that, given the multifunctionality of Xrn1p, we cannot assume that this interaction is attributable purely to the combined effects of the mutations on mRNA decay alone. However, reassuringly, the Parker group found that the combination of xrn1Δ and ski8Δ and that of dcp1Δ with any of the ski2, ski3, or ski8 deletions were also lethal. They also showed that at a growth-permissive temperature the double mutation dcp1 ski8Δ resulted in greatly lengthened mRNA half-lives (20).
The accelerated decay associated with premature translational termination depends on the activities of three nonessential genes (UPF1, UPF2, and UPF3 [up-frameshift]). Although the encoded proteins are generally involved only in the decay of aberrant mRNAs, they are of interest because they seem to mediate one type of interaction between translational events and mRNA stability. Inactivation of any one of these genes can give rise to stabilization (in a nonadditive fashion) of mRNAs containing premature stop codons (327). The Upf proteins are not thought to have nuclease activities, and they are generally not required for the decay of wild-type mRNAs (78, 266), although there may be exceptions (such as PPR1 and CTF13 [78, 322; see also reference 33a]). UPF genes were initially identified via a screen for suppressors of the His− phenotype caused by a +1 frameshift in the HIS4 gene (his4-38) (107, 322, 323). This 5′-proximal frameshift leads to translational termination at an adjacent stop codon, which in turn destabilizes the mRNA. UPF1 was also identified as a high-copy suppressor of mitochondrial RNA splicing reactions (7).
Upf1p interacts with both of the other Upf proteins, but the binding to Upf2p is significantly stronger (25, 266). It is a cytoplasmic, multifunctional protein bearing a cysteine-rich region as well as ATPase-helicase and RNA-binding domains (584, 585). A mutation in the cysteine-rich domain of Upf1p causes enhanced −1 frameshifting as well as suppression of nonsense codon-dependent accelerated decay (105). Upf2p is thought to have a cytoplasmic function (104), while Upf3p is primarily nuclear (321, 323). Upf3p mutations that affect nuclear transport also suppress nonsense codon-dependent decay (25). How might these proteins be involved in the coupling of (premature) translational termination to mRNA destabilization? It has been proposed that they participate in a posttermination “surveillance complex” (469a) that scans the mRNA downstream of (premature) stop codons for a downstream element before triggering accelerated decay (586). A further clue to the functions of the Upf proteins is the very recent demonstration that Upf1p can interact with eRF1 and eRF3 (108a). At least Upf1p may act to modulate eRF1- and eRF3-mediated termination reactions (108a, 584–586). The binding of Upf1p to the eRF proteins also inhibits Upf1p binding to RNA and inhibits the ATPase activity of this factor (108a). This has led to the proposal that after peptide hydrolysis and the release of the eRFs, Upf1p forges a path for the remaining ribosome complex along the mRNA downstream of the termination event and mediates the recognition of a downstream element that triggers nonsense codon-dependent decay (108a). As we have seen, it is unclear whether a discrete consensus sequence motif is required for this triggering step (see also the next section). Alternatively, it might be envisaged that the Upf proteins are involved in the cellular response to the disruptive effects on polysome structure and/or nuclear transport caused by premature termination. Ongoing work by the respective research groups can be expected to elucidate the means by which the Upf proteins are involved in the coupling of translational termination and mRNA decay and also to clarify the full range of functions of these proteins in the cell.
A number of other proteins are known, or suspected, to be involved in mRNA degradation (Table 4). The screening of temperature-sensitive S. cerevisiae strains for the presence of mutations that affect mRNA decay has led to the identification of a number of mrt (mRNA turnover) alleles. Three of the mutations were in the XRN1 and DCP1 genes (see above), while four were localized to two further, as yet uncharacterized genes (MRT1 and MRT3) (204). The MRT1 and MRT3 mutations do not stabilize nonsense codon-destabilised mRNAs, but they do extend the half-lives of at least some nonaberrant mRNAs. However, stabilisation was found to be selective: mrt1-3 stabilized five different mRNAs between 1.9- and 4.6-fold but did not stabilize PAB1 mRNA, while mrt3-1 stabilized four of the tested mRNAs but had only a minimal effect on the half-life of GAL10 mRNA and slightly destabilized PAB1 mRNA (204). Both of these mrt mutations also inhibited decapping of MFA2 and PGK1 mRNAs containing poly(G) inserts in their respective 3′UTRs but did not affect the decapping of the PGK1 mRNA when this contained an early nonsense codon. The same authors found that PAB1 mRNA was also unaffected by inactivation of Dcp1p. Since extracts prepared from the mrt1-3 and mrt3-1 strains contained normal levels of decapping activity, it was suggested that the Mrt proteins modulate the decapping reaction of nonaberrant mRNAs (204). It remains to be determined why the mrt alleles affect mRNAs differentially and whether they really are part of a generally acting decapping pathway.
It is not yet clear whether the list of exo- and/or endonucleolytic activities involved in yeast mRNA turnover is complete (379, 523), and further activities may well be discovered. Finally, there may be factors that influence the stability of only certain mRNAs. For example, Ume2p and Ume5p are suspected to mediate the decay rates of mRNAs involved in yeast meiosis (see the section on regulation of the mRNA decay rate).
Pathways of mRNA Decay
Role of deadenylation.
A picture of the pathways responsible for mRNA decay in yeast only began to take form over the last 5 years. There are a number of reasons to believe that deadenylation plays an important role in the degradation of at least some yeast mRNAs (78). Jacobson (264) has discussed how earlier studies in higher eukaryotic systems suggested a role for poly(A) shortening in triggering decay. A study of S. cerevisiae cells with a deletion in XRN1 revealed that they accumulated uncapped mRNAs bearing shortened poly(A) tails (234). This suggested that deadenylation might normally precede decapping, which in turn exposes mRNA to 5′→3′ exonucleolytic degradation. Further experiments used poly(G) as a physical block to trap decay intermediates from specific mRNAs (576). After insertion of poly(G) into the 5′UTR of MFA2, deadenylated and decapped decay products whose 5′ ends had been trimmed to the 5′ side of the poly(G) were detected (117, 118). Moreover, in an xrn1 deletion strain, some of the MFA2 mRNA accumulated as a decapped and deadenylated species. In a dcp1 strain, a number of mRNAs were stabilized (36). The importance of the cap in stabilizing mRNA is underlined by the observation that conditional mutants of the capping enzyme Ceg1p manifest accelerated decay rates (493). These and other detailed studies (78) of two short-lived mRNAs (MFA2 and MATα1) and one long-lived mRNA (PGK1) are consistent with the existence of a causal link between deadenylation and decapping in different types of yeast mRNA.
The concept of a major pathway comprising the causally linked steps deadenylation→decapping→5′→3′ exonucleolytic decay has been formulated by the Parker group (Fig. 17) (78). This incorporates elements of direct signalling that are believed to trigger and/or modulate key steps. The nature of the causal relationship between deadenylation and further processes in decay remains uncharacterized. The respective kinetics of deadenylation and of the remaining decay process suggest that deadenylation triggers overall degradation but does not determine the rate at which it occurs (78). Other factors, including perhaps the MRT genes (207), are likely to control decapping and the further steps of degradation. Moreover, deadenylation-dependent decapping is apparently not the only route that has been observed in detailed studies of PGK1 and MFA2 decay (389, 390), meaning that there is at least a minor contribution from other pathways. For example, in PGK1, 3′→5′ exonucleolytic decay also followed deadenylation (390). The mechanisms controlling or regulating the rate of deadenylation by poly(A) nuclease activities are not known, but current models implicate the mediation of signals in the 3′UTR or main body of the mRNA via Pab1p and/or additional factors (79, 344, 476). The triggering of further degradation (decapping) occurs once the poly(A) tail has reached an oligo(A) length and is also thought to involve Pab1p. Such a view is compatible with the suspected role of Pab1p in mediating 5′-3′ interactions in mRNPs (see the sections on translation) and provides an explanation for the observations that mutations in PAB1 result in the accumulation of decapped mRNAs with long poly(A) tails (78). It is argued that the loss of the coupling between deadenylation and decapping allows the latter to occur independently of the former, although this is not necessarily the only explanation of the observed effect.
FIG. 17.
A major route of mRNA decay in yeast. Specific elements in the body of the mRNA are proposed to dictate (possibly via interactions with Pab1p) the rate of deadenylation by a PAN-type enzyme (complex). Once a shortened tail has been generated, the major pathway involves the triggering of decapping by Dcp1p (or Dcp2p), which is then followed by 5′→3′ exonucleolytic decay (catalyzed by Xrn1p). The Mrt proteins may modulate decapping. Alternatively, exonucleolytic degradation from the 3′ end has been observed. It is now suspected that the multienzyme “exosome,” or something like it, is involved in this process.
However, attractive as this model may seem, these links between 3′ and 5′ events could be indirect. One way of representing this is to consider a minimum model of coupling between triggering events and decay processes in which a number of possible processes that disturb the structure and function of the mRNP and polysome initiate degradation (polysome disruption model [Fig. 18] [see, for example, references 266 and 336). It is a major objective of current work in this area to establish the extent to which more direct and/or differentiated mechanisms are responsible for the respective decay patterns of the mRNAs studied so far (78). Future efforts will have to distinguish between models of the two types illustrated here (Fig. 17 and 18).
Other triggers of decay.
The accelerated decay induced by the presence of a premature stop codon can trigger rapid decapping that is apparently not dependent on deadenylation of the tail to the oligo(A) state (387). Analysis of the effects of xrn1 and dcp1 mutants indicates that the decay events observed here involve the same components of the degradation machinery as normal decay does (78). Might this reflect the fact that premature termination also triggers decay via destabilization of mRNP/polysome structure (see, for example, reference 266)? The disruptive effect seems to occur via a parallel route to that linked to deadenylation (Fig. 18).
This area has received renewed impetus from the most recent investigations of the Upf proteins, which have provided indications of how these factors might influence both translation and mRNA decay (108a). There are, however, still many questions to be answered, not least of which is how interactions between the mRNA and the posttermination ribosome complex (also called the surveillance complex [469a]) trigger accelerated decay. As discussed above, it is still not clear whether a specific type of discrete downstream element is required; certainly the pyrimidine-rich motif alone is insufficient. Moreover, the field is missing a unifying model explaining the responses of the degradation machinery both to termination on uORFs and/or the main ORFs of nonaberrant mRNAs and to aberrant termination on premature nonsense codons. Perhaps this model might be found if more was known of the kinetics governing normal and aberrant termination events, in particular the kinetics of binding and release of the eRFs and of other factors, like the Upfps, which can modulate termination. One possibility, for example, would be that peptide hydrolysis and eRF release at a subset of stop codons can proceed efficiently and be coupled to accelerated decay without positive modulation by the Upfps. This would, in turn, explain why termination at such stop codons can trigger accelerated decay at least partially independently of at least one of the UPF genes. In other words, the UPF dependence of termination-triggered accelerated decay may be kinetically controlled rather than constituting mechanistic necessity. Other characteristics of nonsense codon-dependent decay, such as the strong position dependence of the degradation kinetics, might also be related to the respective contributions of the eRFs and Upfps to the component steps of termination. The kinetics of the interactions between these respective factors and the ribosome might be controlled by polysome localization and/or structure. Much further work is needed to determine whether any of these speculations approximate to reality.
The other known means of triggering decay is endonucleolytic cleavage. So far, it has been suggested that endonucleolytic cleavage triggers the decay of the PGK1 (576; but see reference 390) and L2 (444) mRNAs. For PGK1, there is a considerable body of evidence indicating that 5′→3′ exonuclease-driven decay from the decapped 5′ end is the dominant pathway. More cases of endonucleolytic cleavage have been found in higher eukaryotes (117, 266, 462, 463). It remains to be seen whether endonucleases participate in rate-controlling steps in the decay pathways of a significant number of yeast mRNAs.
Regulation of mRNA Decay Rate
An area of growing interest is the extent to which regulation of gene expression can be achieved by modulating the steps of mRNA decay. One potential means of achieving this is via coupling between mRNA turnover and translation. As discussed above, changes in the decay rate of an mRNA can be coupled to modulation of translational rate via the 5′UTR, provided that the mRNA has appropriate stability determinants. At present it is not clear how many yeast mRNAs show the sensitivity toward translation manifested by PGK1. On the basis of current evidence, 5′UTR-mediated translational regulation via trans-acting (site-specific) factors seems likely to be rare in yeast. However, a potential role for translation in observed regulatory changes in mRNA stability should always be considered possible until experimentally disproven. The role of uORFs has yet to be fully clarified, but stress- or even cell cycle-related modulation of decay mediated by uORFs can certainly be envisaged as a feasible, if not necessarily widely used, regulatory mechanism. Despite these considerations, it is noteworthy that the cases of stability regulation reported so far are evidently not directly mediated by translational modulation.
Five different meiotic mRNAs (early transcripts SPO13, SPO11, and IME1, the middle transcript SPO12, and the middle-late transcript DIT2) are stabilized up to approximately twofold in response to the shift from vegetative growth of S. cerevisiae in glucose medium to sporulation conditions in acetate medium (532, 533). This stabilization effect is dependent on proteins encoded by UME2 and UME5, whereby the product of the latter gene shows similarity to members of the CDC28 serine/threonine-specific protein kinases. The regulation of meiotic mRNA stability could be shown to be triggered by the removal of glucose from the medium rather than by the shift to meiosis. Surosky et al. (532, 533) also showed that UME5-dependent stabilization of SPO13 does not involve any change in the rate of deadenylation. A more striking example of destabilization associated with glucose repression is the 12-fold decrease in the half-life of the SDH2 mRNA observed upon a glycerol-to-glucose shift (84, 341). The 5′UTR of the SDH2 mRNA is thought to play a major role in the glucose-induced destabilization effect (84). Other work has shown that glucose induces slightly accelerated decay of the PCK1 and FBP1 mRNAs (368) and a larger decrease in the stability of the SUC2 mRNA (83). It remains to be seen how general this glucose-dependent destabilization of yeast mRNAs is and to what extent mRNA-specific factors are involved. So far, it is known that neither Ume5p nor most of the other known factors involved in the glucose repression pathway are required for the regulation of SDH2 mRNA stability (83). The REG1 gene product, on the other hand, was found to be required for regulation of the decay of this mRNA, although the mode of influence of this factor on mRNA turnover was not characterized.
There is also evidence that the stability of a number of ribosomal protein mRNAs is subject to regulation. Mild heat shock results in a transient destabilization of the S10 and L25 mRNAs (217). In a more intensively investigated case, the decay rate of the L2 mRNA was found to be regulated autogenously by the L2 subunit (443, 444). The destabilization caused by L2 requires the region of the L2 mRNA from −21 to +339 and has been shown to function in both the nucleus and the cytoplasm (444). A strategy involving the insertion of poly(G) to stabilize mRNA 3′ of the region responsible for L2-dependent destabilization was used to examine the pathway of decay. The results were consistent with a model in which an endonucleolytic cleavage on the 3′ side of this region constitutes the initial cleavage step (444).
Back to Rate Control
Explaining how the trans-acting factors interact with the various structural features of mRNAs (mRNPs) to determine the pathways and rates of degradation remains a major goal of this field of research. The effective degrees of control (control coefficients) exercised by the steps of deadenylation, decapping, exonucleolytic decay, and endonucleolytic cleavage events can apparently vary from mRNA species to mRNA species. This matter is complicated by the processivity of at least some of the reaction steps (see for example, reference 379) and the fact that the causality of apparently sequential events is not understood. Moreover, one mRNA species may be degraded by more than one pathway at any one time (390). Despite this apparent complexity, mRNA turnover is not simply a free-for-all for RNases, and there is persuasive evidence that a considerable number of yeast mRNAs are degraded via a deadenylation-dependent 5′→3′ pathway (Fig. 17) (78). However, this still leaves us without a general model that could explain the turnover behaviour of all mRNAs, whether aberrant or nonaberrant, and how this is influenced by the translational apparatus.
Although the polysome-disruption type of model (Fig. 18) is undoubtedly an oversimplification, the concept of a critical mRNP disruption event (or series of sequential or alternative disruption events) being initiated by a number of possible triggers allows some flexibility in explaining the relationships between mRNA structure and the kinetics of decay. Structural and/or functional interactions between the 5′ and 3′ ends of the mRNA may, in as yet unknown ways, contribute to the control of the decay pathway. Moreover, the roles of the respective translation factors remain unclear. Analogously to the parallel pathways of translational initiation, there are likely to be several potential routes to full hydrolysis of an mRNA. Obviously, the primary pathway followed by any given mRNA could theoretically be affected by a large number of factors. However, kinetic control and/or molecular channeling (in the context of guiding processes with the help of large molecular assemblies) can be expected to play decisive roles in mRNA degradation. For example, if normal mRNP structure inhibits decapping until an event such as deadenylation leads to exposure of the cap to attack by Dcp1p, this may explain the molecular basis of causality between these two steps (Fig. 17 and 18). It remains to be seen whether one relevant form of “disruption” caused by deadenylation is associated with its negative effect on translation. The rate of subsequent events, including decapping, may then be subject to control via mRNA structural elements that have little or nothing to do with the deadenylation phase (78). It should be possible to distinguish between the direct action of specific mRNA signals on individual steps (Fig. 17) and the mediation of polysome structure in controlling mRNA decay (Fig. 18), although both are likely to be relevant.
Finally, in this context it is of interest that prokaryotic polyadenylation may influence decay via mechanisms that, at least at first sight, appear to be different from those acting in eukaryotes. It has been demonstrated that 3′ polyadenylation of ColE1 RNAI in E. coli destabilizes this RNA (602). Moreover, polyadenylation by a poly(A) polymerase(s) of mRNA decay intermediates generated by endonucleolytic cleavage (by RNase E) in E. coli accelerates the decay of these fragments (208a). The poly(A) tails on mRNA decay intermediates may recruit the 3′→5′ exonucleases PNPase and RNase II, perhaps as components of the prokaryotic degradosome. Interestingly, however, polyadenylation cannot be essential for the initial cleavage(s) of full-length de novo transcripts, since rapid degradation of parts of at least some polycistronic mRNAs occurs before transcription is completed (see, for example, references 365a and 490a). Thus, while the poly(A) tail is likely to be a general modulator of the translation and decay of full-length eukaryotic mRNAs, its role in E. coli seems to be limited to that of a tagging function that promotes the rapid degradation of decay intermediates. There may, however, be some common ground between the two domains; it remains to be determined whether the eukaryotic poly(A) tail acts to promote the recruitment of RNases located in “exosome” particles. Perhaps at least this recruitment property of the poly(A) tail is a feature that has been conserved by means of convergent or divergent evolution.
CONCLUSIONS AND PERSPECTIVES
The posttranscriptional control field is in a phase of rapid development, and the key role of yeast in much of the research performed so far is clearly evident. However, while focusing on yeast, this review has endeavored to emphasize the value of comparative reference to the wealth of information generated by work with bacterial, mammalian, and plant systems. Analysis of the similarities and the differences between the respective cell types will continue to provide additional insight into central principles of mechanism and control.
Recent years have seen the cloning and at least preliminary characterization of many genes and proteins involved in eukaryotic translation and mRNA decay. The three-dimensional structures of only very few of these proteins have been solved so far, but new ones will undoubtedly appear with increasing frequency, and we can also look forward to structural information on multicomponent complexes such as eIF4F. However, progress in understanding the complex process of posttranscriptional gene expression will require that structural studies be complemented by detailed quantitative investigations of intermolecular interactions and rate control. Work on the thermodynamics and kinetics of function in these systems will therefore be as least as important as the analysis of their structures, and it will continue to be essential to examine carefully the relevance of in vitro data to the conditions that exist in the cellular environment.
Any reviewer of this field cannot help but be struck by the overlaps and networks between functions of the cellular components that are involved in cytoplasmic gene expression. This principle is certainly not unexpected for evolving cellular systems, and awareness of it warns us not to be unduly eager to categorize too narrowly the roles of individual factors. Indeed, the redundancy of function becoming so evident in yeast is a significant feature worthy of analysis in its own right. In proceeding with work in this area, it is important to take note of the recent developments in large-scale yeast gene functional analysis (414). Of the approaching 1,000 genes analyzed by disruption to date, only approximately 12% have been found to be essential. Other data have indicated that up to 40% of the S. cerevisiae ORFs can be disrupted without resulting in detectable phenotypes in a range of assays. One of the reasons for this is likely to derive from the large gap between the conditions under which this organism has naturally evolved and the conditions that apply in standard laboratory experiments and assays. It is therefore essential that we begin to bridge that gap by examining regulatory phenomena in yeast under precisely specified growth-limiting conditions. Such work could potentially redefine the meaning of significant parts of the current dataset on posttranscriptional control and could also lead to an understanding of the functions of many new genes whose roles are apparent only transiently or under tightly defined conditions. This is certainly a major challenge for the future.
Multifunctionality can take the form of cellular functions (or capabilities) shared by two or more proteins (e.g., RNA helicases) or could constitute the basis for coupling between different processes. Coupling has been a common theme in this review and is also seen in other forms, for example between protein synthesis and transport (30). Beyond this versatility of the components of the cellular machineries, future work will have to address their supramolecular organization. In particular, and reminiscent of multifunctional enzyme complexes in metabolism (416), the posttranscriptional pathways are subject to levels of control and channeling that are only just becoming discernible. In considering these and other aspects in this review, it is evident that many revelations about these exquisitely balanced systems are still to come.
ACKNOWLEDGMENTS
While taking full responsibility for the views expressed, and any errors, in this review, I am grateful to Patrick Linder (Geneva, Switzerland) and Mick Tuite (Canterbury, United Kingdom) for their comments on early versions of it and to one of the reviewers, who provided valuable constructive criticism. I also thank colleagues who sent me manuscripts prior to publication. Barbara Skoyles provided assistance with the figures and references.
Some of the work from my laboratory mentioned in this review was supported by the BBSRC (UK).
REFERENCES
- 1.Abastado J-P, Miller P F, Jackson B M, Hinnebusch A G. Suppression of ribosomal reinitiation at upstream open reading frames in amino acid-starved cells forms the basis for GCN4 translational control. Mol Cell Biol. 1991;11:486–496. doi: 10.1128/mcb.11.1.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adhin M R, Van Duin J. Translational regulation of the lysis gene in RNA bacteriophage fr requires a UUG initiation codon. Mol Gen Genet. 1989;218:137–142. doi: 10.1007/BF00330576. [DOI] [PubMed] [Google Scholar]
- 3.Adhin M R, Van Duin J. Scanning model for translational reinitiation in eubacteria. J Mol Biol. 1990;213:811–818. doi: 10.1016/S0022-2836(05)80265-7. [DOI] [PubMed] [Google Scholar]
- 4.Agris P F. The importance of being modified: roles of modified nucleosides and Mg2+ in RNA structure and function. Prog Nucleic Acid Res Mol Biol. 1996;53:79–129. doi: 10.1016/s0079-6603(08)60143-9. [DOI] [PubMed] [Google Scholar]
- 5.Aldrich T L, Di Segni G, McConaughy B L, Keen N J, Whelen S, Hall B D. Structure of the yeast TAP1 protein: dependence of transcription activation on the DNA context of the target gene. Mol Cell Biol. 1993;13:3434–3444. doi: 10.1128/mcb.13.6.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allen M L, Metz A M, Timmer R T, Rhoads R E, Browning K S. Isolation and sequence of the cDNAs encoding the subunits of the isozyme form of wheat protein synthesis initiation factor 4F. J Biol Chem. 1992;267:23232–23236. [PubMed] [Google Scholar]
- 7.Altamura N, Groudinsky O, Dujardin G, Slonimski P P. NAM7 n1-6n-ligand motif and is involved in mitochondrial functions in Saccharomyces cerevisiae. J Mol Biol. 1992;224:575–587. doi: 10.1016/0022-2836(92)90545-u. [DOI] [PubMed] [Google Scholar]
- 8.Altmann M, Handschin C, Trachsel H. mRNA cap-binding protein: cloning of the gene encoding protein synthesis initiation factor eIF-4E from Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:998–1003. doi: 10.1128/mcb.7.3.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altmann M, Edery I, Trachsel H, Sonenberg N. Site-directed mutagenesis of the tryptophan residues in yeast eukaryotic initiation factor 4E. J Biol Chem. 1988;263:17229–17323. [PubMed] [Google Scholar]
- 10.Altmann M, Krieger M, Trachsel H. Nucleotide sequence of the gene encoding a 20 kDa protein associated with the cap binding protein eIF-4E from Saccharomyces cerevisiae. Nucleic Acids Res. 1989;17:7520. doi: 10.1093/nar/17.18.7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altmann M, Sonenberg N, Trachsel H. Translation in Saccharomyces cerevisiae: initiation factor 4E-dependent cell-free system. Mol Cell Biol. 1989;9:4467–4472. doi: 10.1128/mcb.9.10.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altmann M, Blum S, Wilson T M A, Trachsel H. The 5′-leader sequence of tobacco mosaic-virus RNA mediates initiation-factor-4E-independent, but still initiation-factor-4A-dependent translation in yeast extracts. Gene. 1990;91:127–129. doi: 10.1016/0378-1119(90)90173-o. [DOI] [PubMed] [Google Scholar]
- 13.Altmann M, Müller P P, Wittmer B, Ruchti F, Laner S, Trachsel H. A Saccharomyces cerevisiae homologue of mammalian translation initiation factor 4B contributes to RNA helicase activity. EMBO J. 1993;12:3887–4003. doi: 10.1002/j.1460-2075.1993.tb06077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altmann M, Wittmer B, Méthot N, Sonenberg N, Trachsel H. The Saccharomyces cerevisiae translation initiation factor Tif3 and its mammalian homologue, eIF-4B, have RNA annealing activity. EMBO J. 1995;14:3820–3827. doi: 10.1002/j.1460-2075.1995.tb00051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altmann M, Schmitz N, Berset C, Trachsel H. A novel inhibitor of cap-dependent translation initiation in yeast: p20 competes with eIF4G for binding to eIF4E. EMBO J. 1997;16:1114–1121. doi: 10.1093/emboj/16.5.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altuvia S, Kornitzer D, Kobi S, Oppenheim A B. Functional and structural elements of the mRNA of the cIII gene of bacteriophage lamda. J Mol Biol. 1991;218:723–733. doi: 10.1016/0022-2836(91)90261-4. [DOI] [PubMed] [Google Scholar]
- 17.Amberg D C, Goldberg A L, Cole C N. Isolation and characterization of rat1—an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of messenger-RNA. Genes Dev. 1992;6:1173–1189. doi: 10.1101/gad.6.7.1173. [DOI] [PubMed] [Google Scholar]
- 18.Amrani N, Minet M, Le Gouar M, Lacroute F, Wyers F. Yeast Pab1 interacts with Rna15 and participates in the control of the poly(A) tail length in vitro. Mol Cell Biol. 1997;17:3694–3701. doi: 10.1128/mcb.17.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson J S J, Parker R. RNA turnover: the helicase story unwinds. Curr Biol. 1996;6:780–782. doi: 10.1016/s0960-9822(02)00593-6. [DOI] [PubMed] [Google Scholar]
- 20.Anderson J S J, Parker R. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SK12 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arkov A L, Korolev S V, Kisselev L L. Termination of translation in bacteria may be modulated via specific interaction between peptide chain release factor-2 and the last peptidyl-transfer RNA (Ser/Phe) Nucleic Acids Res. 1993;21:2891–2897. doi: 10.1093/nar/21.12.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arkov A L, Freistroffer D V, Ehrenberg M, Murgola E J. Mutations in RNAs of both ribosomal subunits cause defects in translation termination. EMBO J. 1998;17:1507–1514. doi: 10.1093/emboj/17.5.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arndt G M, Xiao W, Rank G H. Antisense RNA regulation of the ILV2 gene in yeast—a correction. Curr Genet. 1994;25:289. doi: 10.1007/BF00357175. [DOI] [PubMed] [Google Scholar]
- 24.Arndt G M, Atkins D, Patrikakis M. Gene regulation by antisense RNA in the fission yeast Saccharomyces cerevisiae. Mol Gen Genet. 1995;248:293–300. doi: 10.1007/BF02191596. [DOI] [PubMed] [Google Scholar]
- 25.Atkin A L, Schenkman L R, Eastham M, Dahlseid J F, Lelivelt M J, Culbertson M R. Relationship between yeast polyribosomes and Upf proteins required for nonsense mRNA decay. J Biol Chem. 1997;272:22163–22172. doi: 10.1074/jbc.272.35.22163. [DOI] [PubMed] [Google Scholar]
- 26.Atkins D, Arndt G M, Izant J G. Antisense gene-expression in yeast. Biol Chem Hoppe-Seyler. 1994;375:721–729. doi: 10.1515/bchm3.1994.375.11.721. [DOI] [PubMed] [Google Scholar]
- 27.Avis J M, Allain F H T, Howe P W A, Varani G, Nagai K, Neuhaus D. Solution structure of the N-terminal Rnp domain of U1A protein—the role of C-terminal residues in structure stability and RNA-binding. J Mol Biol. 1996;257:389–411. doi: 10.1006/jmbi.1996.0171. [DOI] [PubMed] [Google Scholar]
- 28.Aygün-Yücsel, B., B. Zeiler, R. Simons, and J. E. G. McCarthy. Unpublished data.
- 29.Bach M L, Lacroute F, Botstein D. Evidence for transcriptional regulation of orotidine-5′-phosphate decarboxylase in yeast by hybridization of mRNA to the yeast structural gene cloned in Escherichia coli. Proc Natl Acad Sci USA. 1979;76:386–390. doi: 10.1073/pnas.76.1.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bacher G, Lütcke H, Jungnickel B, Rapoport T A, Dobberstein B. Regulation by the ribosome of the GTPase of the signal-recognition particle during protein targeting. Nature. 1996;381:248–251. doi: 10.1038/381248a0. [DOI] [PubMed] [Google Scholar]
- 30a.Bailey-Serres J, Gallie D R. A look beyond transcription: mechanisms determining mRNA stability and translation in plants. Rockville, Md: American Society of Plant Physiologists; 1998. [Google Scholar]
- 31.Baim S B, Sherman F. mRNA structures influencing translation in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:1591–1601. doi: 10.1128/mcb.8.4.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baker K P, Schatz G. Mitochondrial proteins essential for viability mediate protein import into yeast mitochondria. Nature. 1991;349:205–208. doi: 10.1038/349205a0. [DOI] [PubMed] [Google Scholar]
- 33.Barbet N C, Schneider U, Helliwell S P, Stansfield I, Tuite M F, Hall M N. TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33a.Barnes C A. Upf1 and Upf2 proteins mediate normal yeast mRNA degradation when translation initiation is limited. Nucleic Acids Res. 1998;26:2433–2441. doi: 10.1093/nar/26.10.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barnes C A, Singer R A, Johnston G C. Yeast prt1 mutations alter heat-shock gene expression through transcript fragmentation. EMBO J. 1993;12:3323–3332. doi: 10.1002/j.1460-2075.1993.tb06002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barnes C A, MacKenzie M M, Johnston G C, Singer R H. Efficient translation of an SAA1-derived heat-shock mRNA in yeast cells limited for cap-binding protein and eIF-4F. Mol Gen Genet. 1995;246:619–627. doi: 10.1007/BF00298969. [DOI] [PubMed] [Google Scholar]
- 35a.Bassell G, Singer R H. MRNA and cytoskeletal filaments. Curr Opin Cell Biol. 1997;9:109–115. doi: 10.1016/s0955-0674(97)80159-7. [DOI] [PubMed] [Google Scholar]
- 36.Bassell G, Powers C M, Taneja K L, Singer R H. Single mRNAs visualized by ultrastructural in situ hybridization are principally localized at actin filament intersections in fibroblasts. J Cell Biol. 1994;126:863–876. doi: 10.1083/jcb.126.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beelman C A, Parker R. Differential effects of translational inhibition in cis and in trans on the decay of the unstable yeast MFA2 mRNA. J Biol Chem. 1994;269:9687–9692. [PubMed] [Google Scholar]
- 38.Beelman C A, Stevens A, Caponigro G, LaGrandeur T E, Hatfield L, Fortner D M, Parker R. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature. 1996;382:642–646. doi: 10.1038/382642a0. [DOI] [PubMed] [Google Scholar]
- 39.Belcourt M F, Farabaugh P J. Ribosomal frameshifting in the yeast retrotransponson Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell. 1990;62:339–352. doi: 10.1016/0092-8674(90)90371-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Belfield G P, Ross-Smith N J, Tuite M F. Translation elongation factor-3 (EF-3): an evolving eukaryotic ribosomal protein? J Mol Evol. 1995;41:376–387. [PubMed] [Google Scholar]
- 41.Belgrader P, Cheng J, Maquat L E. Evidence to implicate translation by ribosomes in the mechanism by which nonsense codons reduce the nuclear level of human triosephosphate isomerase mRNA. Proc Natl Acad Sci USA. 1993;90:482–486. doi: 10.1073/pnas.90.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Belsham G J, Denton R M. The effect of insulin and adrenaline on the phosphorylation of a 22 000-molecular weight protein within isolated fat cells; possible identification as the inhibitor-1 of the “general phosphatase.”. Biochem Soc Trans. 1980;8:382–383. doi: 10.1042/bst0080382. [DOI] [PubMed] [Google Scholar]
- 43.Beltzer J P, Chang L-F L, Hinkkanen A E, Kohlhaw G B. Structure of yeast LEU4 - the 5′ flanking regions contain features that predict 2 modes of control and 2 productive translation starts. J Biol Chem. 1986;261:5160–5167. [PubMed] [Google Scholar]
- 44.Benard L, Carroll K, Valle R C P, Wickner R B. Ski6p is a homolog of RNA-processing enzymes that affects translation of non-poly(A) mRNAs and 60S ribosomal subunit biogenesis. Mol Cell Biol. 1998;18:2688–2696. doi: 10.1128/mcb.18.5.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benne R, Wong C, Luedi M, Hershey J W B. Purification and characterization of initiation factor IF-E2 from rabbit reticulocytes. J Biol Chem. 1976;251:7675–7681. [PubMed] [Google Scholar]
- 46.Beretta L, Gingras A-C, Svitkin Y V, Hall M N, Sonenberg N. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits ca-dependent initiation of translation. EMBO J. 1996;15:658–664. [PMC free article] [PubMed] [Google Scholar]
- 47.Berezney R. The nuclear matrix: a heuristic model for investigating genome organization and function in the cell nucleus. J Cell Biochem. 1991;47:109–123. doi: 10.1002/jcb.240470204. [DOI] [PubMed] [Google Scholar]
- 48.Berglund H, Rak A, Serganov A, Garber M, Hard T. Solution structure of the ribosomal RNA binding protein S15 from Thermus thermophilus. Nat Struct Biol. 1997;4:20–23. doi: 10.1038/nsb0197-20. [DOI] [PubMed] [Google Scholar]
- 49.Bergmann J E, Lodish H. A kinetic model of protein synthesis. J Biol Chem. 1979;254:11927–11937. [PubMed] [Google Scholar]
- 50.Berkhout B, Schmidt B F, van Strien A, van Boom J, van Westrenen J, van Duin J. Lysis gene of bacterial MS2 is activated by translation termination at the overlapping coat gene. J Mol Biol. 1987;195:517–524. doi: 10.1016/0022-2836(87)90180-x. [DOI] [PubMed] [Google Scholar]
- 51.Bernstein J, Shefler I, Elroy-Stein O. The translational repression mediated by the platelet-derived growth factor 2/c-sis mRNA leader is relieved during megakaryocytic differentiation. J Biol Chem. 1995;270:10559–10565. doi: 10.1074/jbc.270.18.10559. [DOI] [PubMed] [Google Scholar]
- 51a.Berset C, Trachsel H, Altmann M. The TOR (target of rapamycin) signal transduction pathway regulates the stability of translation initiation factor eIF4G in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1998;95:4264–4269. doi: 10.1073/pnas.95.8.4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bettany A J E, Moore P A, Cafferkey R, Bell L D, Goodey A R, Carter B L A, Brown A J P. 5′-secondary structure formation, in contrast to a short string of non-preferred codons, inhibits the translation of the pyruvate-kinase messenger-RNA in yeast. Yeast. 1989;5:187–198. doi: 10.1002/yea.320050308. [DOI] [PubMed] [Google Scholar]
- 53.Björnsson A, Mottagui-Tabar S, Isaksson L A. Structure of the C-terminal end of the nascent peptide influences translation termination. EMBO J. 1996;15:1696–1704. [PMC free article] [PubMed] [Google Scholar]
- 54.Blanc A, Goyer C, Sonenberg N. The coat protein of the yeast double-stranded RNA virus L-A attaches covalently to the cap structure of eukaryotic mRNA. Mol Cell Biol. 1992;12:3390–3398. doi: 10.1128/mcb.12.8.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blanc A, Ribas J C, Wickner R B, Sonenberg N. His-154 is involved in the linkage of the Saccharomyces cerevisiae L-A double stranded RNA virus Gag protein to the cap structure of mRNAs and is essential for M1 satellite virus expression. Mol Cell Biol. 1994;14:2664–2674. doi: 10.1128/mcb.14.4.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blasi F, Carra E, Vendittis E, Masturzo P, Burderi E, Lambrinoudaki M, Mirisola M G, Seidita G, Fasano O. The SCH9 protein kinase mRNA contains a long 5′ leader with a small open reading frame. Yeast. 1993;9:21–32. doi: 10.1002/yea.320090104. [DOI] [PubMed] [Google Scholar]
- 57.Blum S, Mueller M, Schmid S R, Linder P, Trachsel H. Translation in Saccharomyces cerevisiae: initiation factor 4A-dependent cell-free system. Proc Natl Acad Sci USA. 1989;86:6043–6046. doi: 10.1073/pnas.86.16.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bokros C L, Hugdahl J D, Kim H-H, Haneworth V R, Van Heerden A, Browning K S, Morejohn L C. Function of the p86 subunit of eukaryotic initiation factor (iso) 4F as a microtubule-associated protein in plant cells. Proc Natl Acad Sci USA. 1995;92:7120–7124. doi: 10.1073/pnas.92.15.7120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bonetti R, Fu L, Moon J, Bedwell D M. The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae. J Mol Biol. 1995;251:334–345. doi: 10.1006/jmbi.1995.0438. [DOI] [PubMed] [Google Scholar]
- 60.Bonneau A-M, Sonenberg N. Proteolysis of the p220 component of the cap-binding protein complex is not sufficient for complete inhibition of host cell protein synthesis after poliovirus infection. J Virol. 1987;61:986–991. doi: 10.1128/jvi.61.4.986-991.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Borman A M, Bailly J-L, Girar M, Kean K M. Picornavirus internal ribosome entry segments: comparison of translation efficiency and the requirements for optimal internal initiation of translation in vitro. Nucleic Acids Res. 1995;23:3656–3663. doi: 10.1093/nar/23.18.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bossier P, Fernandes L, Rocha D, Rodrigues-Pousada C. Overexpression of YAP2, coding for a new yap protein, and YAP1 in Saccharomyces cerevisiae alleviates growth inhibition caused by 1,10-phenanthroline. J Biol Chem. 1993;268:23640–23645. [PubMed] [Google Scholar]
- 63.Brenner C, Nakayama N, Goebl M, Tanaka K, Toh-e A, Masumoto K. CDC33 encodes mRNA cap-binding protein eIF-4E of Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:3556–3559. doi: 10.1128/mcb.8.8.3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63a.Brierley I, Meredith M R, Boys A J, Hagervall T G. Expression of a coronavirus ribosomal frameshift signal in Escherichia coli: influence of tRNA anticodon modification on frameshifting. J Mol Biol. 1997;270:360–373. doi: 10.1006/jmbi.1997.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brown C M, Stockwell P A, Trotman C N A, Tate W P. The signal for the termination of protein synthesis in prokaryotes. Nucleic Acids Res. 1990;18:2079–2086. doi: 10.1093/nar/18.8.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brown C M, McCaughan K K, Tate W P. Two regions of the Escherichia coli 16S ribosomal RNA are important for decoding stop signals in polypeptide chain termination. Nucleic Acids Res. 1993;21:2109–2115. doi: 10.1093/nar/21.9.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brown C M, Dalphin M E, Stockwell P A, Tate W P. The translational termination signal database. Nucleic Acids Res. 1993;21:3119–3123. doi: 10.1093/nar/21.13.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brown C M, Stockwell P A, Dalphin M E, Tate W P. The translational termination signal database (TransTerm) now also includes initiation contexts. Nucleic Acids Res. 1994;22:3620–3624. doi: 10.1093/nar/22.17.3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Browning K S, Webster C, Roberts J K M, Ravel J M. Identification of an isozyme form of protein synthesis initiation factor 4F in plants. J Biol Chem. 1992;267:10096–10100. [PubMed] [Google Scholar]
- 69.Browning K S. The plant translational apparatus. Plant Mol Biol. 1996;32:107–144. doi: 10.1007/BF00039380. [DOI] [PubMed] [Google Scholar]
- 70.Bruenn J, Keitz B. The 5′ end of yeast killer factor RNAs are pppGp. Nucleic Acids Res. 1976;3:2427–2436. doi: 10.1093/nar/3.10.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Buckingham R H, Grentzmann G, Kisselev L. Polypeptide chain release factors. Mol Microbiol. 1997;24:449–456. doi: 10.1046/j.1365-2958.1997.3711734.x. [DOI] [PubMed] [Google Scholar]
- 72.Burd C G, Matunis E L, Dreyfuss G. The multiple RNA-binding domains of the mRNA poly(A) binding protein have different RNA-binding activities. Mol Cell Biol. 1991;11:3419–3424. doi: 10.1128/mcb.11.7.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bycroft M, Hubbard T J P, Proctor M, Freund S M V, Murzin A G. The solution structure of the S1 RNA binding domain: a member of an ancient nucleic acid-binding fold. Cell. 1997;88:235–242. doi: 10.1016/s0092-8674(00)81844-9. [DOI] [PubMed] [Google Scholar]
- 74.Cao J, Geballe A P. Coding sequence-dependent ribosomal arrest at termination of translation. Mol Cell Biol. 1996;16:603–608. doi: 10.1128/mcb.16.2.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Capieaux E, Vignais M-L, Sentenac A, Goffeau A. The yeast H+-ATPase gene is controlled by the promoter binding factor TUF. J Biol Chem. 1989;264:7437–7446. [PubMed] [Google Scholar]
- 76.Caponigro G, Muhlrad D, Parker R. A small segment of the MATα1 transcript promotes mRNA decay in Saccharomyces cerevisiae: a stimulatory role for rare codons. Mol Cell Biol. 1993;13:5141–5148. doi: 10.1128/mcb.13.9.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Caponigro G, Parker R. Multiple functions for the poly(A)-binding protein in mRNA decapping and deadenylation in yeast. Genes Dev. 1995;9:2421–2432. doi: 10.1101/gad.9.19.2421. [DOI] [PubMed] [Google Scholar]
- 78.Caponigro G, Parker R. Mechanisms and control of mRNA turnover in Saccharomyces cerevisiae. Microbiol Rev. 1996;60:233–249. doi: 10.1128/mr.60.1.233-249.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Caponigro G, Parker R. mRNA turnover in yeast promoted by MATα1 instability element. Nucleic Acids Res. 1996;24:4304–4312. doi: 10.1093/nar/24.21.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Carrier T A, Keasling J D. Mechanistic modeling of prokaryotic mRNA decay. J Theor Biol. 1997;189:195–209. doi: 10.1006/jtbi.1997.0509. [DOI] [PubMed] [Google Scholar]
- 81.Carroll K, Wickner R B. Translation and M1 double-stranded RNA propagation: MAK18 = RPL41B and cycloheximide curing. J Bacteriol. 1995;177:2887–2891. doi: 10.1128/jb.177.10.2887-2891.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cavener D R, Ray S C. Eukaryotic start and stop translation sites. Nucleic Acids Res. 1991;19:3185–3192. doi: 10.1093/nar/19.12.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cereghino G P, Scheffler I E. Genetic analysis of glucose regulation in Saccharomyces cerevisiae: control of transcription versus mRNA turnover. EMBO J. 1996;15:363–374. [PMC free article] [PubMed] [Google Scholar]
- 84.Cereghino G P, Atencio D P, Saghbini M, Beiner J, Scheffler I E. Glucose-dependent turnover of the mRNAs encoding succinate dehydrogenase peptides in Saccharomyces cerevisiae: sequence elements in the 5′ untranslated region of the Ip mRNA play a dominant role. Mol Biol Cell. 1995;6:1125–1143. doi: 10.1091/mbc.6.9.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chakravarti D, Maitra U. Eukaryotic translation initiation factor 5 from Saccharomyces cerevisiae: cloning, characterization and expression of the gene encoding the 45,346 dalton protein. J Biol Chem. 1993;268:10524–10533. [PubMed] [Google Scholar]
- 86.Chen C-Y, Sarnow R. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 1995;268:415–417. doi: 10.1126/science.7536344. [DOI] [PubMed] [Google Scholar]
- 87.Chen W, Dieckmann C L. Genetic evidence for interaction between Cpb1 and specific nucleotides in the 5′ untranslated region of mitochondrial cytochrome b mRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:6203–6211. doi: 10.1128/mcb.17.11.6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chernoff Y O, et al. Role of the chaperone HSP104 in propagation of the yeast prion-like factor [PSI(+)] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 89.Chernoff Y O, Newman G P, Liebman S W. The translational function of nucleotide C1054 in the small subunit rRNA is conserved throughout evolution: genetic evidence in yeast. Proc Natl Acad Sci USA. 1996;93:2517–2522. doi: 10.1073/pnas.93.6.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89a.Chicurel M E, Singer R H, Meyer C J, Ingber D E. Integrin binding and mechanical tension induce movement of mRNA and ribosomes to focal adhesions. Nature. 1998;392:730–733. doi: 10.1038/33719. [DOI] [PubMed] [Google Scholar]
- 89b.Choi S K, Lee J H, Zoll W L, Meerick W C, Dever T E. Promotion of Met-tRNAIMet binding to ribosomes by yIF2, a bacterial IF2 homolog in yeast. Science. 1998;280:1757–1760. doi: 10.1126/science.280.5370.1757. [DOI] [PubMed] [Google Scholar]
- 90.Christopher Y J, Wolin S L. La proteins from Drosophila melanogaster and Saccharomyces cerevisiae: a yeast homolog of the La autoantigen is dispensable for growth. Mol Cell Biol. 1994;14:5412–5424. doi: 10.1128/mcb.14.8.5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chuang R-Y, Weaver P L, Liu Z, Chang T-H. Requirement of the DEAD-box protein Ded1p for messenger RNA translation. Science. 1997;275:1469–1471. doi: 10.1126/science.275.5305.1468. [DOI] [PubMed] [Google Scholar]
- 92.Cigan A M, Donahue T F. Sequence and structural features associated with translational initiator regions in yeast—a review. Gene. 1987;59:1–18. doi: 10.1016/0378-1119(87)90261-7. [DOI] [PubMed] [Google Scholar]
- 93.Cigan A M, Pabisch E K, Donahue T F. Mutational analysis of the HIS4 translational initiation region in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:2964–2975. doi: 10.1128/mcb.8.7.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cigan A M, Pabich E K, Feng L, Donahue T F. Yeast translation initiation suppressor sui2 encodes the α subunit of eukaryotic initiation factor 2 and shares sequence identity with the human α subunit. Proc Natl Acad Sci USA. 1989;86:2784–2788. doi: 10.1073/pnas.86.8.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Clare J J, Belcourt M, Farabaugh P J. Efficient translational frameshifting occurs within a conserved sequence of the overlap between the two genes of a yeast Ty1 transposon. Proc Natl Acad Sci USA. 1988;85:6816–6820. doi: 10.1073/pnas.85.18.6816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Clemens M J. Protein kinases that phosphorylate eIF2 and eIF2B, and their role in eukaryotic cell translational control. In: Hershey J W B, Matthews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 139–172. [Google Scholar]
- 97.Cooper A A, Stevens T H. Protein splicing: self-splicing of genetically mobile elements at the protein level. Trends Biochem Sci. 1995;20:351–357. doi: 10.1016/s0968-0004(00)89075-1. [DOI] [PubMed] [Google Scholar]
- 98.Coppolecchia R, Buser P, Stotz A, Linder P. A new yeast translation initiation factor suppresses a mutation in the eIF-4A RNA helicase. EMBO J. 1993;12:4005–4011. doi: 10.1002/j.1460-2075.1993.tb06078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cottrelle P, Thiele D, Price V L, Memet S, Micouin J Y, Marck C, Buhler J M, Sentenac A, Fromageot P. Cloning, nucleotide-sequence, and expression of one of two genes coding for yeast elongation-factor 1-alpha. J Biol Chem. 1985;260:3090–3096. [PubMed] [Google Scholar]
- 100.Coward P, Dasgupta A. Yeast cells are incapable of translating RNAs containing the poliovirus 5′-untranslated region: evidence for a translational inhibitor. J Virol. 1992;66:286–295. doi: 10.1128/jvi.66.1.286-295.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100a.Cox B S. Psi, a cytoplasmic suppressor of super-suppressor in yeast. Heredity. 1965;20:505–521. [Google Scholar]
- 101.Crabeel M, Lavalle R, Glansdorff N. Arginine-specific repression in Saccharomyces cerevisiae: kinetic data on ARG1 and ARG3 mRNA transcription and stability support a transcriptional control mechanism. Mol Cell Biol. 1990;10:1226–1233. doi: 10.1128/mcb.10.3.1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Craig A W B, Haghighat A, Yu A T K, Sonenberg N. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature. 1998;392:520–523. doi: 10.1038/33198. [DOI] [PubMed] [Google Scholar]
- 103.Creighton T E, editor. Proteins, structures and molecular properties. 2nd ed. New York, N.Y: W. H. Freeman & Co.; 1993. [Google Scholar]
- 104.Cui Y, Hagan K W, Zhang S, Peltz S W. Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translation termination codon. Genes Dev. 1995;9:423–436. doi: 10.1101/gad.9.4.423. [DOI] [PubMed] [Google Scholar]
- 105.Cui Y, Dinman J D, Peltz S W. mof4-1 is an allele of the UPF1/IFS2 gene which affects both mRNA turnover and −1 ribosomal frameshifting efficiency. EMBO J. 1996;15:5726–5736. doi: 10.1002/j.1460-2075.1996.tb00956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cui Y, Dinman J D, Kinzy T G, Peltz S W. The Mof2/Sui1 protein is a general monitor of translational accuracy. Mol Cell Biol. 1998;18:1506–1516. doi: 10.1128/mcb.18.3.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Culbertson M R, Underbrink K M, Fink G R. Frameshift suppression in Saccharomyces cerevisiae. Genetic properties of group II suppressors. Genetics. 1980;95:833–853. doi: 10.1093/genetics/95.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Czaplinski K, Weng Y, Hagan K W, Peltz S W. Purification and characterization of the Upf1 protein: A factor involved in translation and mRNA degradation. RNA. 1995;1:610–623. [PMC free article] [PubMed] [Google Scholar]
- 108a.Czaplinski K, Ruiz-Echevarria M J, Paushkin S V, Han X, Weng Y, Perlick H A, Dietz H C, Ter-Avanesyan M D, Peltz S W. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 1998;12:1665–1677. doi: 10.1101/gad.12.11.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dabeva M D, Warner J R. Ribosomal protein L32 of Saccharomyces cerevisiae regulates both splicing and translation of its own transcript. J Biol Chem. 1993;268:19669–19674. [PubMed] [Google Scholar]
- 110.Dams, E., L. Hendricks, Y. V. de Peer, J.-M. Neefs, G. Smits, I. Vandenbempt, and R. D. Wachter. 1988. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 16(Suppl.):r87–r173. [DOI] [PMC free article] [PubMed]
- 111.Das S, Coward P, Dasgupta A. A small yeast RNA selectively blocks internal initiation of translation programmed by poliovirus RNA: specific interaction with cellular proteins that bind the viral 5′-untranslated region. J Virol. 1994;68:7200–7211. doi: 10.1128/jvi.68.11.7200-7211.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Das S, Kenan D J, Bocskai D, Keene J D, Dasgupta A. Sequences within a small yeast RNA required for inhibition of internal initiation of translation—interaction with La and other cellular proteins influences its inhibitory activity. J Virol. 1996;70:1624–1632. doi: 10.1128/jvi.70.3.1624-1632.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112a.Das S, Maiti T, Das K, Maitra U. Specific interaction of eukaryotic translation initiation factor 5 (eIF5) with the beta-subunit of eIF2. J Biol Chem. 1997;272:31712–31718. doi: 10.1074/jbc.272.50.31712. [DOI] [PubMed] [Google Scholar]
- 113.Dasso M C, Jackson R J. On the fidelity of mRNA translation in the nuclease-treated rabbit reticulocyte lysate system. Nucleic Acids Res. 1989;17:3129–3144. doi: 10.1093/nar/17.8.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Deardorff J A, Sachs A B. Differential effects of aromatic and charge residue substitutions in the RNA binding domains of the yeast poly(A)-binding protein. J Mol Biol. 1997;269:67–81. doi: 10.1006/jmbi.1997.1013. [DOI] [PubMed] [Google Scholar]
- 115.De Benedetti A, Rhoads R E. Overexpression of eukaryotic protein synthesis initiation factor 4E in HeLa cell results in aberrant growth and morphology. Proc Natl Acad Sci USA. 1990;87:8212–8216. doi: 10.1073/pnas.87.21.8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Decker C J, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 117.Decker C J, Parker R. Mechanisms of mRNA degradation in eukaryotes. Trends Biochem Sci. 1994;19:336–349. doi: 10.1016/0968-0004(94)90073-6. [DOI] [PubMed] [Google Scholar]
- 118.Decker C J, Parker R. Diversity of cytoplasmic functions for the 3′ untranslated region of eukaryotic transcripts. Curr Opin Cell Biol. 1995;7:386–392. doi: 10.1016/0955-0674(95)80094-8. [DOI] [PubMed] [Google Scholar]
- 119.De la Cruz J, Iost I, Kressler D, Linder P. The p20 and DED1 proteins have antagonistic roles in eIF4E-dependent translation in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:5201–5206. doi: 10.1073/pnas.94.10.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Delbecq P, Werner M, Feller A, Filipkowski R K, Messenguy F, Piérard A. A segment of mRNA encoding the leader peptide of the CPA1 gene confers repression by arginine on a heterologous yeast gene transcript. Mol Cell Biol. 1994;14:2378–2390. doi: 10.1128/mcb.14.4.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120a.Delbecq P, Flipkowski R, Messenguy F, Pièrard A. Program and Abstracts of the 1993 Yeast Genetics and Molecular Biology Meeting. 1998. Pleiotropic effect of cpaR mutation on regulation of amino acids metabolism: effect on GCN4 expression; p. 219. [Google Scholar]
- 121.De Smit M H, Van Duin J. Control of prokaryotic translational initiation by mRNA secondary structure. Prog Nucleic Acid Res Mol Biol. 1990;38:1–35. doi: 10.1016/s0079-6603(08)60707-2. [DOI] [PubMed] [Google Scholar]
- 122.De Smit M H, Van Duin J. Secondary structure of the ribosome binding site determines translational efficiency: a quantitative analysis. Proc Natl Acad Sci USA. 1990;87:7668–7672. doi: 10.1073/pnas.87.19.7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.De Smit M H, Van Duin J. Control of translation by mRNA secondary structure in Escherichia coli: a quantitative analysis of literature data. J Mol Biol. 1994;244:144–150. doi: 10.1006/jmbi.1994.1714. [DOI] [PubMed] [Google Scholar]
- 124.De Smit M H. Translational control by mRNA structure in eubacteria: molecular biology and physical chemistry. In: Simons R W, Grunberg-Manago M, editors. RNA structure and function. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 495–540. [Google Scholar]
- 125.Dever T E, Feng L, Wek R C, Cigan A M, Donahue T D, Hinnebusch A G. Phosphorylation of initiation factor 2α by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell. 1992;68:585–596. doi: 10.1016/0092-8674(92)90193-g. [DOI] [PubMed] [Google Scholar]
- 126.Dever T E, Cheng J-J, Barber G N, Cigan A M, Feng L, Donahue T F, London I M, Katze M G, Hinnebusch A G. Mammalian eukaryotic initiation factor 2α kinases functionally substitute for GCN2 in the GCN4 translational control mechanism of yeast. Proc Natl Acad Sci USA. 1993;90:4616–4620. doi: 10.1073/pnas.90.10.4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dever T E, Yang W M, Astrom S, Bystrom A S, Hinnebusch A G. Modulation of tRNA(i), eIF-2, and eIF-2B expression shows that GCN4 translation is inversely coupled to the level of eIF-2-GTP-Met-tRNA(i) ternary complexes. Mol Cell Biol. 1995;15:6351–6363. doi: 10.1128/mcb.15.11.6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dieckmann C L, Homison G, Tzagoloff A. Assembly of the mitochondrial membrane system. J Biol Chem. 1984;259:4732–4738. [PubMed] [Google Scholar]
- 129.Ding D, Lipshitz H D. Localized RNAs and their functions. Bioessays. 1993;15:651–658. doi: 10.1002/bies.950151004. [DOI] [PubMed] [Google Scholar]
- 130.Dinman J D, Icho T, Wickner R B. A-1 ribosomal frameshift in a double-stranded RNA virus of yeast forms a gag-pol fusion protein. Proc Natl Acad Sci USA. 1991;88:174–178. doi: 10.1073/pnas.88.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Dinman J D. Ribosomal frameshifting in yeast viruses. Yeast. 1995;11:1115–1127. doi: 10.1002/yea.320111202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Di Segni G, McConaughy B L, Shapiro R A, Alrich T L, Hall B D. TAP1, a yeast gene that activates the expression of a tRNA gene with a defective internal promoter. Mol Cell Biol. 1993;13:3424–3433. doi: 10.1128/mcb.13.6.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dolph P J, Huang J, Schneider R J. Translation by the adenovirus tripartite leader: elements which determine independence from cap-binding protein complex. J Virol. 1990;64:2669–2677. doi: 10.1128/jvi.64.6.2669-2677.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Donahue T F, Cigan A M. Genetic selection for mutations that reduce or abolish ribosomal recognition of the HIS4 translational initiator region. Mol Cell Biol. 1988;8:2955–2963. doi: 10.1128/mcb.8.7.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Donahue T F, Cigan A M, Pabich E K, Valavicius B C. Mutations at a Zn(II) finger motif in the yeast eIF-2β gene alter ribosomal start-site selection during the scanning process. Cell. 1988;54:621–632. doi: 10.1016/s0092-8674(88)80006-0. [DOI] [PubMed] [Google Scholar]
- 136.Donahue T F, Cigan A M. Sequence and structural requirements for efficient translation in yeast. Methods Enzymol. 1990;185:366–372. doi: 10.1016/0076-6879(90)85032-j. [DOI] [PubMed] [Google Scholar]
- 137.Donovan W P, Kushner S R. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc Natl Acad Sci USA. 1986;83:120–124. doi: 10.1073/pnas.83.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Donzé O, Jagus R, Koromilas A E, Hershey J W B, Sonenberg N. Abrogation of translation initiation factor eIF-2 phosphorylation causes malignant transformation of NIH 3T3 cells. EMBO J. 1995;14:3828–3834. doi: 10.1002/j.1460-2075.1995.tb00052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Draper D E. Protein-RNA recognition. Annu Rev Biochem. 1995;64:593–620. doi: 10.1146/annurev.bi.64.070195.003113. [DOI] [PubMed] [Google Scholar]
- 140.Dreyfus M. What constitutes the signal for the initiation of protein synthesis on Escherichia coli mRNAs? J Mol Biol. 1988;204:79–94. doi: 10.1016/0022-2836(88)90601-8. [DOI] [PubMed] [Google Scholar]
- 141.Dreyfuss G, Matunis M J, Piñol-Roma S, Burd C G. HnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 142.Duncan R F. Translational control during heat shock. In: Hershey J W B, Matthews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 271–293. [Google Scholar]
- 142a.Dunckley, T., and R. Parker. Personal communication.
- 143.Eckner R, Ellmeier W, Birnstiel M L. Mature mRNA 3′ end formation stimulates RNA export from the nucleus. EMBO J. 1991;10:3513–3522. doi: 10.1002/j.1460-2075.1991.tb04915.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Egli C M, Braus G H. Uncoupling of mRNA 3′ cleavage and polyadenylation by expression of a hammerhead ribozyme in yeast. J Biol Chem. 1994;269:27378–27383. [PubMed] [Google Scholar]
- 145.Ellis S R, Hopper A K, Martin N C. Amino-terminal extension generated from an upstream AUG codon is not required for mitochondrial import of yeast N2,N2-dimethylguanosine-specific tRNA methyltransferase. Proc Natl Acad Sci USA. 1987;84:5172–5176. doi: 10.1073/pnas.84.15.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Evstafieva A G, Beletsky A V, Borovjagin A V, Bogdanov A A. Internal ribosome entry site of encephalomyocarditis virus RNA is unable to direct translation in Saccharomyces cerevisiae. FEBS Lett. 1993;335:273–276. doi: 10.1016/0014-5793(93)80745-g. [DOI] [PubMed] [Google Scholar]
- 147.Eyre-Walker A. The distance between Escherichia coli genes is related to gene expression levels. J Bacteriol. 1995;177:5368–5369. doi: 10.1128/jb.177.18.5368-5369.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Farabaugh P J. Programmed translational frameshifting. Microbiol Rev. 1996;60:103–134. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Farabaugh P J, Zhao H, Vimaladithan A. A novel programed frameshift expresses the POL3 gene of retrotransposon Ty3 of yeast: frameshifting without tRNA slippage. Cell. 1993;74:93–103. doi: 10.1016/0092-8674(93)90297-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Feng L, Yoon H, Donahue T F. Casein kinase II mediates multiple phosphorylation of Saccharomyces cerevisiae eIF-2α (encoded by SUI2), which is required for optimal eIF-2 function in S. cerevisiae. Mol Cell Biol. 1994;14:5139–5153. doi: 10.1128/mcb.14.8.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Fierro-Monti, I., and J. E. G. McCarthy. Unpublished data.
- 152.Firpo M A, Dahlberg A E. The role of ribosomal RNA in the control of gene expression. NATO ASI Ser Ser H. 1990;49:185–195. [Google Scholar]
- 153.Fisher R J, Fivash M. Surface plasmon resonance based methods for measuring the kinetics and binding affinities of biomolecular interactions. Curr Opin Biotechnol. 1994;5:389–395. doi: 10.1016/0958-1669(94)90047-7. [DOI] [PubMed] [Google Scholar]
- 154.Flynn A, Proud C G. Serine 209, not serine 53, is the major site of phosphorylation in initiation factor eIF-4E in serum-treated Chinese hamster ovary cells. J Biol Chem. 1995;270:21684–21688. doi: 10.1074/jbc.270.37.21684. [DOI] [PubMed] [Google Scholar]
- 155.Folley L S, Fox T D. Site-directed mutagenesis of a Saccharomyces cerevisiae mitochondrial translation initiation codon. Genetics. 1991;129:659–668. doi: 10.1093/genetics/129.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Forsburg S L, Guarente L. Identification and characterization of HAP4: a third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 1989;3:1166–1178. doi: 10.1101/gad.3.8.1166. [DOI] [PubMed] [Google Scholar]
- 157.Fox T D. Genetics of mitochondrial translation. In: Hershey J W B, Matthews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 733–758. [Google Scholar]
- 158.Fresco L D, Buratowski S. Conditional mutants of the yeast messenger-RNA capping enzyme show that the cap enhances, but is not required for, messenger-RNA splicing. RNA. 1996;2:584–596. [PMC free article] [PubMed] [Google Scholar]
- 159.Frolova L, Le Goff X, Rasmussen H H, Cheperegin S, Drugeon G, Kress M, Arman I, Haenni A-L, Celis J E, Philippe M, Justesen J, Kisselev L. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature. 1994;372:701–703. doi: 10.1038/372701a0. [DOI] [PubMed] [Google Scholar]
- 160.Frolova L, Le Goff X, Zhouravleva G, Davydova E, Philippe M, Kisselev L. Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA. 1996;2:334–341. [PMC free article] [PubMed] [Google Scholar]
- 161.Fuller-Pace F V. RNA helicases: modulators of RNA structure. Trends Cell Biol. 1994;4:271–274. doi: 10.1016/0962-8924(94)90210-0. [DOI] [PubMed] [Google Scholar]
- 162.Furuichi Y, LaFiandra A, Shatkin A J. 5′-terminal structure and mRNA stability. Nature. 1977;266:235–239. doi: 10.1038/266235a0. [DOI] [PubMed] [Google Scholar]
- 163.Fütterer J, Potrykus I, Bao Y, Li L, Burns T M, Hull R, Hohn T. Position-dependent ATT initiation during plant pararetrovirus rice tungro bacilliform virus translation. J Virol. 1996;70:2999–3010. doi: 10.1128/jvi.70.5.2999-3010.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Gallego C, Gari E, Colomina N, Herrero E, Aldea M. The Cln3 cyclin is down-regulated by translational repression and degradation during the G1 arrest caused by nitrogen deprivation in budding yeast. EMBO J. 1997;16:7196–7206. doi: 10.1093/emboj/16.23.7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Gallie D R. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 166.Gallie D R. Translational control of cellular and viral mRNAs. Plant Mol Biol. 1996;32:145–158. doi: 10.1007/BF00039381. [DOI] [PubMed] [Google Scholar]
- 167.Gallie D R, Tanguay R. Poly(A) binds to initiation factors and increases cap-dependent translation in vitro. J Biol Chem. 1994;269:17166–17173. [PubMed] [Google Scholar]
- 168.Gao M, Rychlik W, Rhoads R E. Cloning and characterization of human eIF4E genes. J Biol Chem. 1998;273:4622–4628. doi: 10.1074/jbc.273.8.4622. [DOI] [PubMed] [Google Scholar]
- 169.Garcia-Barrio M T, Narada T, Vazques de Aldana C R, Cuesta R, Hinnebusch A G, Hershey J B W, Tamame M. GCD10, a translational repressor of GCN4, is the RNA-binding subunit of eukaryotic translation initiation factor-3. Genes Dev. 1995;9:1781–1796. doi: 10.1101/gad.9.14.1781. [DOI] [PubMed] [Google Scholar]
- 170.Geballe A P. Translational control mediated by upstream open reading frames. In: Hershey J W B, Matthews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 173–197. [Google Scholar]
- 171.Gerstel B, McCarthy J E G. Independent and coupled translational initiation of atp genes in Escherichia coli: experiments using chromosomal and plasmid-borne lacZ fusions. Mol Microbiol. 1989;3:851–859. doi: 10.1111/j.1365-2958.1989.tb00234.x. [DOI] [PubMed] [Google Scholar]
- 172.Gerstel B, Tuite M F, McCarthy J E G. The effects of 5′-capping, 3′-polyadenylation and leader composition upon the translation and stability of mRNA in a cell-free extract derived from the yeast Saccharomyces cerevisiae. Mol Microbiol. 1992;6:2339–2348. doi: 10.1111/j.1365-2958.1992.tb01409.x. [DOI] [PubMed] [Google Scholar]
- 172a.Gingras A-C, Kennedy S G, O’Leary M A, Sonenberg N, Hay N. 4E-BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the Akt(PKB) signaling pathway. Genes Dev. 1998;12:502–513. doi: 10.1101/gad.12.4.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Godefroy-Colburn T, Thach R E. The role of mRNA competition in regulating translation. J Biol Chem. 1981;256:11762–11773. [PubMed] [Google Scholar]
- 174.Goff X L, Philipe M, Jean-Jean O. Overexpression of human release factor 1 alone has an antisuppressor effect in human cells. Mol Cell Biol. 1997;17:3164–3172. doi: 10.1128/mcb.17.6.3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Goffeau A, Barrell B H, Bussey H, Davis R W, Dujon B, Feldmann H, Galibert F, Hoheisel J D, Jacq O, Johnston M, Louis E J, Mewes H W, Murakami Y, Philippsen P, Tettlin H, Oliver S G. Life with 6000 genes. Science. 1996;274:546–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 176.Goffeau, A., A. Brown, M. E. Gent, K. J. Indge, C. M. James, S. G. Oliver, L. I. Stateva, et al. 1997. The yeast genome directory. Nature 387(Suppl.):5–105.
- 177.Gold L, Pribnow D, Schnieder T, Shinedling S, Stromo B S, Stromo G. Translational initiation in prokaryotes. Annu Rev Microbiol. 1981;35:365–403. doi: 10.1146/annurev.mi.35.100181.002053. [DOI] [PubMed] [Google Scholar]
- 178.Gold L. Posttranscriptional regulatory mechanisms in Escherichia coli. Annu Rev Biochem. 1988;57:199–233. doi: 10.1146/annurev.bi.57.070188.001215. [DOI] [PubMed] [Google Scholar]
- 179.Goossen B, Hentze M W. Position is the critical determinant for function of iron-responsive elements as translational regulators. Mol Cell Biol. 1992;12:1959–1966. doi: 10.1128/mcb.12.5.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Goyer C, Altmann M, Lee H S, Blanc A, Deshmukh M, Woolford J L, Trachsel H, Sonenberg N. TIF4631 and TIF4632: two yeast genes encoding the high-molecular-weight subunits of the cap-binding protein complex (eukaryotic initiation factor 4F) contain an RNA recognition motif-like sequence and carry out an essential function. Mol Cell Biol. 1993;13:4860–4874. doi: 10.1128/mcb.13.8.4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Gradi A, Imataka H, Svitkin Y V, Rom E, Raught B, Morino S, Sonenberg N. A novel functional human eukaryotic translation initiation factor 4G. Mol Cell Biol. 1998;18:334–342. doi: 10.1128/mcb.18.1.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Grant C M, Hinnebusch A G. Effect of sequence context at stop codons on efficiency of reinitiation in GCN4 translational control. Mol Cell Biol. 1994;14:606–618. doi: 10.1128/mcb.14.1.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Grant C M, Miller P F, Hinnebusch A G. Requirements for intercistronic distance and level of eukaryotic initiation factor 2 activity in reinitiation on GCN4 vary with the downstream cistron. Mol Cell Biol. 1994;14:2616–2628. doi: 10.1128/mcb.14.4.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Grant C M, Miller P F, Hinnebusch A G. Sequences 5′ of the first upstream open reading frame in GCN4 mRNA are required for efficient translational reinitiation. Nucleic Acids Res. 1995;23:3980–3988. doi: 10.1093/nar/23.19.3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Groeneveld H, Thimon K, van Duin J. Translational control of maturation-protein synthesis in phage HS2: a role for the kinetics of RNA folding? RNA. 1995;1:79–88. [PMC free article] [PubMed] [Google Scholar]
- 186.Groner B, Hynes N, Phillips S. Length heterogeneity in the poly(adenylic acid) region of yeast messenger ribonucleic acid. Biochemistry. 1974;13:5378–5383. doi: 10.1021/bi00723a020. [DOI] [PubMed] [Google Scholar]
- 187.Grossi de Sa M-F, Standart N, Martins C, Akhayat O, Huesca M, Scherrer K. The poly(A)-binding protein facilitates in vitro translation of poly(A)-rich mRNA. Eur J Biochem. 1988;176:521–526. doi: 10.1111/j.1432-1033.1988.tb14309.x. [DOI] [PubMed] [Google Scholar]
- 188.Grunberg-Manago M, et al. Prokaryotic and eukaryotic translation factors. Biochimie. 1996;78:1119–1120. [PubMed] [Google Scholar]
- 189.Gualerzi C O, Calogero R A, Canonaco M A, Brombach M, Pon C L. Selection of mRNA by ribosomes during prokaryotic translational initiation. NATO ASI Ser Ser H. 1988;14:317–330. [Google Scholar]
- 190.Gulyas K D, Donahue T F. SSL2, a suppressor of a stem-loop mutation in the HIS4 leader encodes the yeast homolog of human ERCC-3. Cell. 1992;69:1031–1042. doi: 10.1016/0092-8674(92)90621-i. [DOI] [PubMed] [Google Scholar]
- 191.Gunnery S, Mathews M B. Functional mRNA can be generated by RNA polymerase III. Mol Cell Biol. 1995;15:3597–3607. doi: 10.1128/mcb.15.7.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Haase E, Servos J, Brendel M. Isolation and characterization of additional genes influencing resistance to various mutagens in the yeast Saccharomyces cerevisiae. Curr Genet. 1992;21:319–324. doi: 10.1007/BF00351689. [DOI] [PubMed] [Google Scholar]
- 193.Hagan K W, Ruiz-Echevarria M J, Quan Y, Peltz S W. Characterization of cis-acting sequences and decay intermediates involved in nonsense-mediated mRNA turnover. Mol Cell Biol. 1995;15:809–823. doi: 10.1128/mcb.15.2.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Haghighat A, Sonenberg N. eIF4G dramatically enhances the binding of eIF4E to the mRNA 5′-cap structure. J Biol Chem. 1997;272:21677–21680. doi: 10.1074/jbc.272.35.21677. [DOI] [PubMed] [Google Scholar]
- 195.Haghighat A, Mader S, Pause A, Sonenberg N. Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 1995;14:7501. doi: 10.1002/j.1460-2075.1995.tb00257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Hamill D, Davis J, Drawbridge J, Suprenant K A. Polyribosome targeting to microtubules—enrichment of specific messenger-RNAs in a reconstituted microtubule preparation from sea- urchin embryos. J Cell Biol. 1994;127:973–984. doi: 10.1083/jcb.127.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Hamilton B, Hofbauer R, Ruis H. Translational control of catalase synthesis by hemin in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1982;79:7609–7613. doi: 10.1073/pnas.79.24.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Hamm J, Mattaj I W. Monomethylated cap structures facilitate RNA export from the nucleus. Cell. 1990;63:109–118. doi: 10.1016/0092-8674(90)90292-m. [DOI] [PubMed] [Google Scholar]
- 199.Hannig E M. Protein synthesis in eukaryotic organisms: new insights into the function of translation initiation factor eIF-3. Bioessays. 1995;17:915–919. doi: 10.1002/bies.950171103. [DOI] [PubMed] [Google Scholar]
- 200.Hannig E M, Cigan A M, Freeman B A, Kinzy T G. GCD11, a negative regulator of GCN4 expression, encodes the gamma subunit of eIF-2 in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:506–520. doi: 10.1128/mcb.13.1.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Harford J B. The regulated turnover of the transferrin receptor mRNA. In: Belasco J, Brawermann G, editors. Control of mRNA stability. New York, N.Y: Academic Press, Inc.; 1993. pp. 239–266. [Google Scholar]
- 202.Harshman K D, Moye-Rowley W S, Parker C S. Transcriptional activation by the SV40 AP-1 recognition element in yeast is mediated by a factor similar to AP-1 that is distinct from GCN4. Cell. 1988;53:231–330. doi: 10.1016/0092-8674(88)90393-5. [DOI] [PubMed] [Google Scholar]
- 203.Hartz D, McPheeters D S, Traut R, Gold L. Extension inhibition analysis of translation initiation-complexes. Methods Enzymol. 1987;164:419–425. doi: 10.1016/s0076-6879(88)64058-4. [DOI] [PubMed] [Google Scholar]
- 204.Hatfield D, Feng Y-X, Lee B J, Rein A, Levin J G, Oroszlan S. Chromatographic analysis of the aminoacyl-transfer RNAs which are required for translation of codons at and around the ribosomal frameshift sites of HIV, HTLV-1, and BLV. Virology. 1989;173:736–742. doi: 10.1016/0042-6822(89)90589-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Hatfield D, Oroszlan S. The where, what and how of ribosomal frameshifting in retroviral protein-synthesis. Trends Biochem Sci. 1990;15:186–190. doi: 10.1016/0968-0004(90)90159-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Hatfield D, Feng Y-X, Lee B J, Rein A, Levin J D, Oroszlan S. Chromatographic analysis of the aminoacyl-tRNAs which are required for translation of codons at and around the ribosomal frameshift sites of HIV, HTLV-1, and BLV. Virology. 1993;173:736–742. doi: 10.1016/0042-6822(89)90589-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Hatfield L, Beelman C A, Stevens A, Parker R. Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5830–5838. doi: 10.1128/mcb.16.10.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Haucke V, Rospert S, Tokatlidis K, Horst M, Azem A, Matouschek A, Koehler C, Schatz G. The mitochondrial protein import machine. Mol Biol Cell. 1996;7:1942. [Google Scholar]
- 208a.Haugel-Nielsen J, Hajndorf E, Régnier P. The rpsO mRNA of Escherichia coli is polyadenylated at multiple sites resulting from endonucleolytic processing and exonucleolytic degradation. EMBO J. 1996;15:3144–3152. [PMC free article] [PubMed] [Google Scholar]
- 209.He F, Peltz S W, Donohue J L, Rosbash M, Jacobson A. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1− mutant. Proc Natl Acad Sci USA. 1993;90:7034–7038. doi: 10.1073/pnas.90.15.7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.He F, Brown A H, Jacobson A. Upf1, Nmd2p, and Upf3 are interacting components of the Upf1, Nmd2p, and Upf3 are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol Cell Biol. 1997;17:1580–1594. doi: 10.1128/mcb.17.3.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Heaton B, Decker C, Muhlrad D, Donahue J, Jacobson A, Parker R. Analysis of chimeric mRNAs identifies two regions within the STE3 mRNA which promote rapid mRNA decay. Nucleic Acids Res. 1992;20:5365–5373. doi: 10.1093/nar/20.20.5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Hellmuth K, Rex G, Surin B, Zinck R, McCarthy J E G. Translational coupling varying in efficiency between different pairs of genes in the central region of the atp operon of Escherichia coli. Mol Microbiol. 1992;5:813–824. doi: 10.1111/j.1365-2958.1991.tb00754.x. [DOI] [PubMed] [Google Scholar]
- 213.Hennigan A N, Jacobson A J. Functional mapping of the translation-dependent instability element of yeast MATα1 mRNA. Mol Cell Biol. 1996;16:3833–3843. doi: 10.1128/mcb.16.7.3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Hentze M. A multipurpose ribosome adapter? Science. 1997;275:500–501. doi: 10.1126/science.275.5299.500. [DOI] [PubMed] [Google Scholar]
- 215.Herrick D, Parker R, Jacobson A. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:2269–2284. doi: 10.1128/mcb.10.5.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Herrick D, Jacobson A. A coding region segment is necessary, but not sufficient for rapid decay of the HIS3 mRNA in yeast. Gene. 1992;114:35–41. doi: 10.1016/0378-1119(92)90704-s. [DOI] [PubMed] [Google Scholar]
- 217.Herruer M H, Mager W H, Raué H E, Vreken P, Wilms E, Planta R J. Mild temperature shock affects transcription of yeast ribosomal protein genes as well as the stability of their mRNAs. Nucleic Acids Res. 1988;16:7917–7929. doi: 10.1093/nar/16.16.7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Hershey J W B. Translational control in mammalian cells. Annu Rev Biochem. 1991;60:717–757. doi: 10.1146/annurev.bi.60.070191.003441. [DOI] [PubMed] [Google Scholar]
- 219.Hertle K, Haase E, Brendel M. The SNQ3 gene of Saccharomyces cerevisiae confers hyper-resistance to several functionally unrelated chemicals. Curr Genet. 1991;19:429–433. doi: 10.1007/BF00312733. [DOI] [PubMed] [Google Scholar]
- 220.Heyer W-D, Johnson A W, Reinhart U, Kolodner R D. Regulation and intracellular localization of the Saccharomyces cerevisiae strand exchange protein 1 (Sep1/Xrn1/Kem1), a multifunctional exonuclease. Mol Cell Biol. 1995;15:2728–2736. doi: 10.1128/mcb.15.5.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Hicks G R, Raikhel N V. Protein import into the nucleus. Annu Rev Cell Dev Biol. 1995;11:155–188. doi: 10.1146/annurev.cb.11.110195.001103. [DOI] [PubMed] [Google Scholar]
- 222.Hinnebusch A G. Evidence for translational regulation of the activator of general amino acid control in yeast. Proc Natl Acad Sci USA. 1984;81:6442–6446. doi: 10.1073/pnas.81.20.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Hinnebusch A G. A hierarchy of trans-acting factors modulates translation of an activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol Cell Biol. 1985;5:2349–2360. doi: 10.1128/mcb.5.9.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Hinnebusch A G. Mechanisms of gene regulation in the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Microbiol Rev. 1988;52:248–273. doi: 10.1128/mr.52.2.248-273.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Hinnebusch A G. Translational control of GCN4: an in vivo barometer of initiation-factor activity. Trends Biochem Sci. 1994;19:409–414. doi: 10.1016/0968-0004(94)90089-2. [DOI] [PubMed] [Google Scholar]
- 226.Hinnebusch A G. Translational control of GCN4: gene-specific regulation by phosphorylation of eIF2. In: Hershey J W B, Matthews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 199–204. [Google Scholar]
- 227.Hinnebusch A G. Translational regulation of yeast GCN4. J Biol Chem. 1997;272:21661–21664. doi: 10.1074/jbc.272.35.21661. [DOI] [PubMed] [Google Scholar]
- 228.Hinnebusch A G, Liebman S W. Protein synthesis and translational control in Saccharomyces cerevisiae. In: Broach J R, Pringle J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces. Genome dynamics, protein synthesis, and engergetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1991. pp. 627–735. [Google Scholar]
- 229.Hiraga S, Yanofsky C. Hyper-labile messenger RNA in polar mutants of the tryptophan operon of Escherichia coli. J Mol Biol. 1972;72:103–110. doi: 10.1016/0022-2836(72)90072-1. [DOI] [PubMed] [Google Scholar]
- 230.Hirata D, Yano K, Miyakawa T. Stress-induced transcriptional activation mediated by YAP1 and YAP2 genes that encode the Jun family of transcriptional activators in Saccharomyces cerevisiae. Mol Gen Genet. 1994;242:250–256. doi: 10.1007/BF00280413. [DOI] [PubMed] [Google Scholar]
- 231.Hirata R, Ohsumi Y, Nakano A, Kawasaki H, Suzuki K, Anraku Y. Molecular structure of a gene, VMA1, encoding the catalytic subunit of H+-translocating adenosine triphosphatase from vacuolar membranes of Saccharomyces cerevisiae. J Biol Chem. 1990;265:6726–6733. [PubMed] [Google Scholar]
- 232.Hoerz W, McCarty K S. Evidence for a proposed initiation complex for protein synthesis in reticulocyte polyribosome profiles. Proc Natl Acad Sci USA. 1969;63:1206–1213. doi: 10.1073/pnas.63.4.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Howard G A, Adamson S D, Herbert E. Subunit recycling during translation in a reticulocyte cell-free system. J Biol Chem. 1970;245:6237–6239. [PubMed] [Google Scholar]
- 234.Hsu C L, Stevens A. Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol Cell Biol. 1993;13:4826–4835. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Hu C, Pang S, Kong X, Velleca M, Lawrence J., Jr Molecular cloning and tissue distribution of PHAS-1, and intracellular target for insulin and growth factors. Proc Natl Acad Sci USA. 1994;91:3730–3734. doi: 10.1073/pnas.91.9.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Huang H-K, Yoon H, Hannig E M, Donahue T F. GTP hydrolysis controls stringent selection of the AUG start codon during translation initiation in Saccharomyces cerevisiae. Genes Dev. 1997;11:2396–2413. doi: 10.1101/gad.11.18.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Huang S, Spector D L. Nascent pre-mRNA transcripts are associated with nuclear regions enriched in splicing factors. Genes Dev. 1991;5:2288–2302. doi: 10.1101/gad.5.12a.2288. [DOI] [PubMed] [Google Scholar]
- 238.Huang S, Deerinck T J, Ellisman M H, Spector D L. In vivo analysis of the stability and transport of nuclear poly(A)+ RNA. J Cell Biol. 1994;126:877–900. doi: 10.1083/jcb.126.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Icho T, Wickner R B. The MAK11 gene is essential for cell growth and replication of M double-stranded RNA and is apparently a membrane-associated protein. J Biol Chem. 1988;263:1467. [PubMed] [Google Scholar]
- 240.Icho T, Wickner R B. The double-stranded RNA genome of yeast virus L-A encodes its own putative RNA polymerase by fusing two open reading frames. J Biol Chem. 1989;264:6716. [PubMed] [Google Scholar]
- 241.Iizuka N, Najita L, Franzusoff A, Sarnow P. Cap-dependent and cap-independent translation by internal initiation of mRNAs in cell extracts prepared from Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:7322–7330. doi: 10.1128/mcb.14.11.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Imataka H, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol Cell Biol. 1997;17:6940–6947. doi: 10.1128/mcb.17.12.6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Imataka H, Olsen H S, Sonenberg N. A new translational regulator with homology to eukaryotic translation initiation factor 4G. EMBO J. 1997;16:817–825. doi: 10.1093/emboj/16.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Inouye M, Delihas N. Small RNAs in the prokaryotes: a growing list of diverse roles. Cell. 1988;53:5–7. doi: 10.1016/0092-8674(88)90480-1. [DOI] [PubMed] [Google Scholar]
- 245.Iost I, Dreyfus M. mRNAs can be stabilized by DEAD-box proteins. Nature. 1994;372:193–196. doi: 10.1038/372193a0. [DOI] [PubMed] [Google Scholar]
- 246.Ishida T, Shibata M, Fujii K, Inoue M. Inter- and intramolecular stacking interaction between indole and adenium rings. Biochemistry. 1983;22:3571–3581. doi: 10.1021/bi00284a006. [DOI] [PubMed] [Google Scholar]
- 247.Ishida T, Doi M, Ueda H, Inoue M, Sheldrick G M. Specific ring stacking interaction on the trytophan-7-methylgaunine system: comparative crystallographic studies of indole derivatives-7-methylguanine base, nucleoside complexes. J Am Chem Soc. 1988;110:2286–2294. [Google Scholar]
- 248.Ishida T, Iyo H, Ueda H, Doi M, Inoue M, Nishimura S, Kitamura K. Interaction of indole derivatives with biologically important aromatic compounds, part 22. J Chem Soc Perkin Trans. 1991;1991:1847–1853. [Google Scholar]
- 249.Ito K, Ebihara K, Uno M, Nakamura Y. Conserved motifs in prokaryotic and eukaryotic polypeptide release factors: tRNA-protein mimicry hypothesis. Proc Natl Acad Sci USA. 1996;93:5443–5448. doi: 10.1073/pnas.93.11.5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Ito, K., K. Ebihara, and Y. Nakamura. Dissection of tRNA mimicry element of protein release factor eRF1 of fission yeast: eRF3 binding is not necessary for eRF1 function and cell viability. Submitted for publication.
- 251.Itoh T, Tomizawa J. Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proc Natl Acad Sci USA. 1980;77:2450–2454. doi: 10.1073/pnas.77.5.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Ivey-Hoyle M, Steege D A. Translation of phage f1 gene VII occurs from an inherently defective initiation site made functional by coupling. J Mol Biol. 1989;208:233–244. doi: 10.1016/0022-2836(89)90385-9. [DOI] [PubMed] [Google Scholar]
- 253.Ivey-Hoyle M, Steege D A. Mutational analysis of an inherently defective translation initiation site. J Mol Biol. 1992;224:1039–1054. doi: 10.1016/0022-2836(92)90468-y. [DOI] [PubMed] [Google Scholar]
- 254.Izaurralde E, Mattaj I W. Transport of RNA between nucleus and cytoplasm. Semin Cell Biol. 1992;3:279–288. doi: 10.1016/1043-4682(92)90029-u. [DOI] [PubMed] [Google Scholar]
- 255.Izaurralde E, Mattaj I W. RNA export. Cell. 1995;81:153–159. doi: 10.1016/0092-8674(95)90323-2. [DOI] [PubMed] [Google Scholar]
- 256.Izaurralde E, Lewis J, Gamberi C, Jarmolowski A, McGuigan C, Mattaj I. A cap-binding protein complex mediating U snRNA export. Nature. 1995;376:709–712. doi: 10.1038/376709a0. [DOI] [PubMed] [Google Scholar]
- 257.Jacks T, Varmus H E. Expression of the rous sarcoma virus pol gene by ribosomal frameshifting. Science. 1985;230:1237–1242. doi: 10.1126/science.2416054. [DOI] [PubMed] [Google Scholar]
- 258.Jackson R J. The ATP requirement for initiation of eukaryotic translation varies according to the mRNA species. Eur J Biochem. 1991;200:285–294. doi: 10.1111/j.1432-1033.1991.tb16184.x. [DOI] [PubMed] [Google Scholar]
- 259.Jackson R J. Cytoplasmic regulation of mRNA function: the importance of the 3′ untranslated region. Cell. 1993;74:9–14. doi: 10.1016/0092-8674(93)90290-7. [DOI] [PubMed] [Google Scholar]
- 260.Jackson R J. A comparative view of initiation site selection mechanisms. In: Hershey J W B, Matthews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 71–112. [Google Scholar]
- 261.Jackson R J, Kaminski A. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- 262.Jackson R J, Wickens M. Translational controls impinging on the 5′-untranslated region and initiation factor proteins. Curr Opin Genet Dev. 1997;7:233–241. doi: 10.1016/s0959-437x(97)80133-5. [DOI] [PubMed] [Google Scholar]
- 263.Jacobs-Lorena M, Fried H M. Translational regulation of ribosomal protein gene expression in eukaryotes. In: Ilan J, editor. Translational regulation in eukaryotes. New York, N.Y: Plenum Press; 1993. pp. 63–85. [Google Scholar]
- 264.Jacobson A. Poly(A) metabolism and translation: the closed-loop model. In: Hershey J W B, Matthews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 451–480. [Google Scholar]
- 265.Jacobson A, Favreau M. Possible involvement of poly(A) in protein synthesis. Nucleic Acids Res. 1983;11:6353–6368. doi: 10.1093/nar/11.18.6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Jacobson A, Peltz S W. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- 267.Jacobson A, Brown A H, Donahue J L, Herrick D, Parker R, Peltz S W. Regulation of mRNA stability in yeast. NATO ASI Ser Ser H. 1990;49:45–54. [Google Scholar]
- 268.Jacques N, Dreyfus M. Translation initiation in Escherichia coli: old and new questions. Mol Microbiol. 1990;4:1063–1067. doi: 10.1111/j.1365-2958.1990.tb00679.x. [DOI] [PubMed] [Google Scholar]
- 269.Jamieson D J, Rahe B, Beggs J D. A suppressor of a yeast splicing mutation (prp8-1) encodes a putative ATP-dependent RNA helicase. Nature. 1991;349:715–717. doi: 10.1038/349715a0. [DOI] [PubMed] [Google Scholar]
- 269a.Jankowska-Anyszka M, Lamphear B J, Aamodt E J, Harrington T, Darzynkiewicz E, Stolarski R, Rhoads R E. Multiple isoforms of eukaryotic protein synthesis initiation factor 4E in Caenorhabditis elegans can distinguish between mono- and trimethylated mRNA cap structures. J Biol Chem. 1998;17:10538–10542. doi: 10.1074/jbc.273.17.10538. [DOI] [PubMed] [Google Scholar]
- 270.Janosi L, Shimizu I, Kaji A. Ribosome recycling factor (ribosome releasing factor) is essential for bacterial growth. Proc Natl Acad Sci USA. 1994;91:4249–4253. doi: 10.1073/pnas.91.10.4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Johnson A W, Kolodner R D. Synthetic lethality of sep1 (xrn1) ski2 and sep1 (xrn1) ski3 mutants of Saccharomyces cerevisiae is independent of killer virus and suggests a general role of these genes in translation control. Mol Cell Biol. 1995;15:2719–2727. doi: 10.1128/mcb.15.5.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Johnson A W. Rat1p and Xrn1p are functionally interchangeable exoribonucleases that are restricted to and required in the nucleus and cytoplasm, respectively. Mol Cell Biol. 1997;17:6122–6130. doi: 10.1128/mcb.17.10.6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Joshi B, Cai A-L, Keiper B D, Minich W B, Mendez R, Beach C M, Stepinski J, Stolarski R, Darzynkiewicz E, Rhoads R E. Phosphorylation of eukaryotic protein-synthesis initiation-factor 4E at Ser-209. J Biol Chem. 1995;270:14597–14603. doi: 10.1074/jbc.270.24.14597. [DOI] [PubMed] [Google Scholar]
- 274.Joshi-Barve S, Rychlik W, Rhoads R E. Alteration of the major phosphorylation site of eukaryotic protein synthesis initiation factor 4E prevents its association with the 48 S initiation complex. J Biol Chem. 1990;265:2979–2983. [PubMed] [Google Scholar]
- 275.Kabata H, Kurosawa O, Arai I, Washizu M, Margarson S A, Glass R E, Shimamoto N. Visualization of single molecules or RNA polymerase sliding along DNA. Science. 1993;262:1561–1563. doi: 10.1126/science.8248804. [DOI] [PubMed] [Google Scholar]
- 276.Kacser H, Burns J A. The control of flux. Symp Soc Exp Biol. 1973;32:65–104. [PubMed] [Google Scholar]
- 277.Kacser H, Burns J A. Molecular democracy: who shares the controls? Biochem Rev. 1979;7:1149–1160. doi: 10.1042/bst0071149. [DOI] [PubMed] [Google Scholar]
- 278.Kammerer B, Guyonvarch A, Hubert J C. Yeast regulatory gene PPR1, nucleotide sequence, restriction map and codon usage. J Mol Biol. 1984;180:239–250. doi: 10.1016/s0022-2836(84)80002-9. [DOI] [PubMed] [Google Scholar]
- 279.Kane P M, Yamashiro C T, Wolczyk D F, Neff N, Goebl M, Stevens T H. Protein splicing converts the yeast TFP1 gene-product to the 69-kD subunit of the vacuolar H+-adenosine triphosphatase. Science. 1990;250:651–657. doi: 10.1126/science.2146742. [DOI] [PubMed] [Google Scholar]
- 280.Kaufman R J, Murtha-Riel P, Pittman D D, Davies M V. Characterization of wild-type and Ser53 mutant eukaryotic initiation factor 4E overexpression in mammalian cells. J Biol Chem. 1993;268:11902–11909. [PubMed] [Google Scholar]
- 281.Kearsey S, Kipling D. Recombination and RNA processing: a common strand? Trends Cell Biol. 1991;1:110–112. doi: 10.1016/0962-8924(91)90101-e. [DOI] [PubMed] [Google Scholar]
- 282.Kenan D J, Query C C, Keene J D. RNA recognition: towards identifying determinants of specificity. Trends Biochem Sci. 1991;16:214–220. doi: 10.1016/0968-0004(91)90088-d. [DOI] [PubMed] [Google Scholar]
- 283.Kenna M, Stevens A, McCammon M, Douglas M G. An essential yeast gene with homology to the exonuclease-encoding XRN1/KEM1 gene also encodes a protein with exoribonuclease activity. Mol Cell Biol. 1993;13:341–350. doi: 10.1128/mcb.13.1.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Kessler S H, Sachs A B. RNA recognition motif 2 of yeast Pab1p is required for its functional interaction with eIF4G. Mol Cell Biol. 1997;18:51–57. doi: 10.1128/mcb.18.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Kim J, Ljungdahl P O, Fink G R. Kem mutations affect nuclear fusion in Saccharomyces cerevisiae. Genetics. 1990;126:799–812. doi: 10.1093/genetics/126.4.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Kinzy T G, Woolford J L J. Increased expression of Saccharomyces cerevisiae translation elongation factor 1α bypasses the lethality of a TEF5 null allele encoding EF-1β. Genetics. 1995;140:481–489. doi: 10.1093/genetics/141.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Kipling D, Tambini C, Kearsey S E. rar mutations which increase artificial chromosome stability in Saccharomyces cerevisiae identify transcription and recombination proteins. Nucleic Acids Res. 1991;19:1385–1391. doi: 10.1093/nar/19.7.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Kislaukis E H, Li Z, Singer R H, Taneja K L. Isoform-specific 3′-untranslated sequences sort α-cardiac and β-cytoplasmic actin messenger RNAs to different cytoplasmic compartments. J Cell Biol. 1993;123:165–172. doi: 10.1083/jcb.123.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Kohler S A, Henderson B R, Kühn L C. Succinate dehydrogenase b mRNA of Drosophila melanogaster has a functional iron-responsive element in its 5′-untranslated region. J Biol Chem. 1995;270:30781–30786. doi: 10.1074/jbc.270.51.30781. [DOI] [PubMed] [Google Scholar]
- 290.Koloteva N, Müller P P, McCarthy J E G. The position dependence of translational regulation via RNA-RNA and RNA-protein interactions in the 5′-untranslated regions of eukaryotic mRNA is a function of the thermodynamic competence of 40S ribosomes in translational initiation. J Biol Chem. 1997;272:16531–16539. doi: 10.1074/jbc.272.26.16531. [DOI] [PubMed] [Google Scholar]
- 291.Kozak M. How do eukaryotic ribosomes select initiation regions in messenger RNA? Cell. 1978;15:1109–1123. doi: 10.1016/0092-8674(78)90039-9. [DOI] [PubMed] [Google Scholar]
- 292.Kozak M. Role of ATP in binding and migration of 40S ribosomal subunits. Cell. 1980;22:459–467. doi: 10.1016/0092-8674(80)90356-6. [DOI] [PubMed] [Google Scholar]
- 293.Kozak M. Selection of initiation sites by eukaryotic ribosomes: effect if inserting AUG triplets upstream from the coding sequence for preproinsulin. Nucleic Acids Res. 1984;12:3873–3893. doi: 10.1093/nar/12.9.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Kozak M. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc Natl Acad Sci USA. 1986;83:2850–2854. doi: 10.1073/pnas.83.9.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Kozak M. Circumstances and mechanisms of inhibition of translation by secondary structure in eukaryotic mRNAs. Mol Cell Biol. 1989;9:5134–5142. doi: 10.1128/mcb.9.11.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Kozak M. Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc Natl Acad Sci USA. 1990;87:8301–8305. doi: 10.1073/pnas.87.21.8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Kozak M. An analysis of vertebrate mRNA sequences: intimations of translational control. J Cell Biol. 1991;115:887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Kozak M. A consideration of alternative models for the initiation of translation in eukaryotes. Crit Rev Biochem Mol Biol. 1992;27:385–402. doi: 10.3109/10409239209082567. [DOI] [PubMed] [Google Scholar]
- 301.Kozak M. Regulation of translation in eukaryotic systems. Annu Rev Cell Biol. 1992;8:197–225. doi: 10.1146/annurev.cb.08.110192.001213. [DOI] [PubMed] [Google Scholar]
- 302.Kozak M, Shatkin A J. Migration of 40S ribosomal subunits in the presence of edeine. J Biol Chem. 1978;253:6568–6577. [PubMed] [Google Scholar]
- 303.Kressler D, de la Cruz J, Rojo M, Linder P. Fal1p is an essential DEAD-box protein involved in 40S-ribosomal-subunit biogenesis in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:7283–7294. doi: 10.1128/mcb.17.12.7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Krummeck G, Gottenöf T, Rödel G. AUG codons in the RNA leader sequences of the yeast PET genes CBS1 and SCO1 have no influence on translation efficiency. Curr Genet. 1991;20:465–469. doi: 10.1007/BF00334773. [DOI] [PubMed] [Google Scholar]
- 305.Kubo M, Higo Y, Imanaka T. Biological threshold values of prokaryotic gene expression which is controlled by the DNA inverted repeat sequence and the mRNA secondary structure. J Ferment Bioeng. 1990;69:305–307. [Google Scholar]
- 306.Kuhn U, Pieler T. Xenopus poly(A) binding protein: functional domains in RNA binding and protein-protein interactions. J Mol Biol. 1996;256:20–30. doi: 10.1006/jmbi.1996.0065. [DOI] [PubMed] [Google Scholar]
- 307.Kurland C G. Translational accuracy and the fitness of bacteria. Annu Rev Genet. 1992;26:29–50. doi: 10.1146/annurev.ge.26.120192.000333. [DOI] [PubMed] [Google Scholar]
- 308.Kunz J, Herniquez R, Schnieder U, Deuter-Reinhard M, Movva N R, Hall M N. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell. 1993;73:585–596. doi: 10.1016/0092-8674(93)90144-f. [DOI] [PubMed] [Google Scholar]
- 309.Kyrpides N, Tavernarakis N, Papamatheakis J, Thireos G. A transient GCN4 mRNA destabilization follows GCN4 translational depression. J Biol Chem. 1995;270:17317–17320. doi: 10.1074/jbc.270.29.17317. [DOI] [PubMed] [Google Scholar]
- 310.Lacatena R M, Cesareni G. Base pairing of RNA I with its complementary sequence in the primer precursor inhibits ColE1 replication. Nature. 1981;294:623–626. doi: 10.1038/294623a0. [DOI] [PubMed] [Google Scholar]
- 311.LaGrandeur T L, Parker R. Isolation and characterization of Dcp1p, the yeast mRNA decapping enzyme. EMBO J. 1998;17:1487–1496. doi: 10.1093/emboj/17.5.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Lamphear B J, Kirchweger R, Skern T, Rhoads R E. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. J Biol Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- 313.Lang V, Gualerzi C, McCarthy J E G. Ribosomal affinity and translational initiation in Escherichia coli. J Mol Biol. 1989;210:659–663. doi: 10.1016/0022-2836(89)90140-x. [DOI] [PubMed] [Google Scholar]
- 314.Lang V, Zanchin N, Lünsdorf H, Tuite M F, McCarthy J E G. Initiation factor eIF-4E of Saccharomyces cerevisiae: distribution within the cell, binding to mRNA and consequences of its overproduction. J Biol Chem. 1994;269:6117–6123. [PubMed] [Google Scholar]
- 315.Lanker S, Müller P P, Altmann M, Goyer C, Sonenberg N, Trachsel H. Interactions of the eIF-4F subunits in the yeast Saccharomyces cerevisiae. J Biol Chem. 1992;267:21167–21171. [PubMed] [Google Scholar]
- 316.Lanker S, Bushman J L, Hinnebusch A G, Trachsel H, Mueller P P. Autoregulation of the yeast lysyl-tRNA synthetase gene GCD5/KRS1 by translational and transcriptional control mechanisms. Cell. 1992;70:647–657. doi: 10.1016/0092-8674(92)90433-d. [DOI] [PubMed] [Google Scholar]
- 317.Larimer F W, Stevens A. Disruption of the gene XRN1, encoding for a 5′-3′ exoribonuclease, restricts yeast cell growth. Gene. 1990;59:85–90. doi: 10.1016/0378-1119(90)90417-p. [DOI] [PubMed] [Google Scholar]
- 318.Larimer F W, Hsu C L, Maupin M K, Stevens A. Characterization of the XRN1 gene encoding 5′→3′ exoribonuclease: sequence data and analysis of disparate protein and mRNA levels of gene-disrupted cells. Gene. 1992;120:51–75. doi: 10.1016/0378-1119(92)90008-d. [DOI] [PubMed] [Google Scholar]
- 319.Lazaris-Karazas A, Montine K S, Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature. 1990;345:544–547. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- 320.Le H, Tanguay R L, Balasta M L, Wei C-C, Browning K S, Metz A M, Goss D J, Gallie D R. Translation initiation factors eIF-iso4G and eIF-4B interact with the poly(A)-binding protein and increase its RNA binding activity. J Biol Chem. 1997;272:16247–16255. doi: 10.1074/jbc.272.26.16247. [DOI] [PubMed] [Google Scholar]
- 321.Lee B-S, Culbertson M R. Identification of a new gene required for eukaryotic nonsense mRNA turnover. Proc Natl Acad Sci USA. 1995;92:10354–10358. doi: 10.1073/pnas.92.22.10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Leeds P, Peltz S W, Jacobson A, Culbertson M R. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991;5:2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- 323.Leeds P, Wood J M, Lee B-S, Culbertson M R. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2165–2177. doi: 10.1128/mcb.12.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Legrain R, Rosbash M. Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell. 1989;57:573–583. doi: 10.1016/0092-8674(89)90127-x. [DOI] [PubMed] [Google Scholar]
- 325.Lejbkowicz F, Goyer C, Darveau A, Neron S, Lemieux R, Sonenberg N. A fraction of the mRNA 5′ cap-binding protein, eukaryotic initiation factor 4E, localizes to the nucleus. Proc Natl Acad Sci USA. 1992;89:9612–9616. doi: 10.1073/pnas.89.20.9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Lesuisse E, Labbe P. Effects of cadmium and of YAP1 and CAD1/YAP2 genes on iron metabolism in the yeast Saccharomyces cerevisiae. Microbiology. 1995;141:2937–2043. doi: 10.1099/13500872-141-11-2937. [DOI] [PubMed] [Google Scholar]
- 327.Liebig H D, Ziegler E, Yan R, Hartmuth K, Klump H, Kowalski H, Blaas D, Sommergruber W, Frasel L, Lamphear B, Rhoads R E, Keuchler E, Skern T. Purification of two picornaviral 2A proteinases: interaction with eIF-4gamma and influence on in vitro translation. Biochemistry. 1993;32:7581–7588. doi: 10.1021/bi00080a033. [DOI] [PubMed] [Google Scholar]
- 328.Lin T A, Kong X, Haystead T A J, Pause A, Belsham G, Sonenberg N, Lawrence J C., Jr PHAS-I as a link between mitogen-activated protein kinase and translation initiation. Science. 1994;266:653–656. doi: 10.1126/science.7939721. [DOI] [PubMed] [Google Scholar]
- 329.Lin T A, Kong X, Saltiel A R, Blackshear P J, Lawrence J C., Jr Control of PHAS-I by insulin in 3T3-L1 adipocytes. Synthesis, degradation, and phosphorylation by a rapamycin-sensitive and mitogen-activated kinase-independent pathway. J Biol Chem. 1995;270:18531–18538. doi: 10.1074/jbc.270.31.18531. [DOI] [PubMed] [Google Scholar]
- 330.Lindahl L, Hinnebusch A. Diversity of mechanisms in the regulation of translation in prokaryotes and lower eukaryotes. Curr Opin Genet Dev. 1992;2:720–726. doi: 10.1016/s0959-437x(05)80132-7. [DOI] [PubMed] [Google Scholar]
- 331.Lindahl L, Zengel J. Ribosomal genes in Escherichia coli. Annu Rev Genet. 1986;20:297–326. doi: 10.1146/annurev.ge.20.120186.001501. [DOI] [PubMed] [Google Scholar]
- 332.Linder P. Molecular biology of translation in yeast. Antonie Leeuwenhoek. 1992;62:47–62. doi: 10.1007/BF00584462. [DOI] [PubMed] [Google Scholar]
- 333.Linder P, Slominski P P. An essential yeast protein, encoded by duplicated genes TIF1 and TIF2 and homologous to the mammalian translation initiation factor eIF4A, can suppress a mitochondrial missense mutation. Proc Natl Acad Sci USA. 1989;86:2286–2290. doi: 10.1073/pnas.86.7.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Linder, P., H.-P. Vornlocher, J. W. B. Hershey, and J. E. G. McCarthy. A systematic nomenclature for new translation initiation factor genes from S. pombe and other fungi. Submitted for publication. [DOI] [PubMed]
- 335.Lindquist S. Mad cows meet psi-chotic yeast: the expansion of the prion hypothesis. Cell. 1997;89:495–498. doi: 10.1016/s0092-8674(00)80231-7. [DOI] [PubMed] [Google Scholar]
- 336.Linz B, Koloteva N, Vasilescu S, McCarthy J E G. Disruption of ribosomal scanning on the 5′-untranslated region, and not restriction of translational initiation per se, modulates the stability of nonaberrant mRNAs in the yeast Saccharomyces cerevisiae. J Biol Chem. 1997;272:9131–9140. doi: 10.1074/jbc.272.14.9131. [DOI] [PubMed] [Google Scholar]
- 337.Little S, Hyde S, Campbell C J, Lilley R J, Robinson M K. Translational coupling in the threonine operon of Escherichia coli K-12. J Bacteriol. 1989;171:3518–3522. doi: 10.1128/jb.171.6.3518-3522.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Liu C, Simonsen C C, Levinson A D. Initiation of translation at internal AUG codon in mammalian cells. Nature. 1984;309:82–85. doi: 10.1038/309082a0. [DOI] [PubMed] [Google Scholar]
- 338a.Lo H-J, Huang H-K, Donahue T F. RNA polymerase I-promoted HIS4 expression yields uncapped, polyadenylated mRNA that is unstable and inefficiently translated in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:665–675. doi: 10.1128/mcb.18.2.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Lodish H F. Model of the regulation of mRNA translation applied to haemoglobin synthesis. Nature. 1974;251:385–388. doi: 10.1038/251385a0. [DOI] [PubMed] [Google Scholar]
- 340.Lodish H F. Translational control of protein synthesis. Annu Rev Biochem. 1976;45:39–72. doi: 10.1146/annurev.bi.45.070176.000351. [DOI] [PubMed] [Google Scholar]
- 341.Lombardo A, Cereghino G P, Scheffler I E. Control of mRNA turnover as a mechanism of glucose repression in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2941–2948. doi: 10.1128/mcb.12.7.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Losson R, Lacroute F. Interference of nonsense mutations with eucaryotic messenger RNA stability. Proc Natl Acad Sci USA. 1979;76:5134–5137. doi: 10.1073/pnas.76.10.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Lovett P S, Rogers E J. Ribosome regulation by the nascent peptide. Microbiol Rev. 1996;60:366. doi: 10.1128/mr.60.2.366-385.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Lowell J, Rudner D, Sachs A. 3′UTR-dependent deadenylation by the yeast poly(A) nuclease. Genes Dev. 1992;6:2088–2099. doi: 10.1101/gad.6.11.2088. [DOI] [PubMed] [Google Scholar]
- 345.Luo Z, Sachs M S. Role of an upstream open reading frame in mediating arginine-specific translational control in Neurospora crassa. J Bacteriol. 1996;178:2172–2177. doi: 10.1128/jb.178.8.2172-2177.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Mader S, Lee H, Pause A, Sonenberg N. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4γ and the translational repressors 4E-binding proteins. Mol Cell Biol. 1995;15:4990–4997. doi: 10.1128/mcb.15.9.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Maicas E, Shago M, Friesen J D. Translation of the Saccharomyces cerevisiae TCM1 gene in the absence of a 5′-untranslated leader. Nucleic Acids Res. 1990;18:5823–5828. doi: 10.1093/nar/18.19.5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Mandart E, Dufour M-E, Lacroute F. Inactivation of SSM4, a new Saccharomyces cerevisiae gene, suppresses mRNA instability due to rna14 mutations. Mol Gen Genet. 1994;245:323–333. doi: 10.1007/BF00290112. [DOI] [PubMed] [Google Scholar]
- 349.Mandart E, Parker R. Effects of mutations in the Saccharomyces cerevisiae RNA14, RNA15, and PAP1 genes on polyadenylation in vivo. Mol Cell Biol. 1995;15:6979–6986. doi: 10.1128/mcb.15.12.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Manthy G M, McEwen J E. The product of the nuclear gene PET309 is required for translation of mature mRNA and stability or production of intron-containing RNAs derived from the mitochondrial COX1 locus of Saccharomyces cerevisiae. EMBO J. 1995;14:4031–4043. doi: 10.1002/j.1460-2075.1995.tb00074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Mao X, Schwer B, Shuman S. Mutational analysis of the Saccharomyces cerevisiae ABD1 gene: cap methyltransferase activity is essential for cell growth. Mol Cell Biol. 1996;16:475–480. doi: 10.1128/mcb.16.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Maquat L E. When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. RNA. 1995;1:453–465. [PMC free article] [PubMed] [Google Scholar]
- 353.Maquat L E. Defects in RNA splicing and the consequence of shortened translational reading frames. Am J Hum Genet. 1996;59:279–286. [PMC free article] [PubMed] [Google Scholar]
- 354.Marcotrigiano J, Gingras A-C, Sonenberg N, Burley S K. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell. 1997;89:951–961. doi: 10.1016/s0092-8674(00)80280-9. [DOI] [PubMed] [Google Scholar]
- 355.Margossian S P, Butow R A. RNA turnover and the control of mitochondrial gene expression. Trends Biochem Sci. 1996;21:392–396. [PubMed] [Google Scholar]
- 356.Marton M J, Vazquez de Aldana C R, Qui H, Chakraburtty K, Hinnebusch A G. Evidence that GCN1 and GCN20, translational regulators of GCN4, function on elongating ribosomes in activation of eIF2α kinase GCN2. Mol Cell Biol. 1997;17:4474–4489. doi: 10.1128/mcb.17.8.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Masison D C, Blanc A, Ribas J C, Carroll K, Sonenberg N, Wickner R B. Decoying the cap− mRNA degradation system by a double-stranded RNA virus and poly(A)− mRNA surveillance by a yeast antiviral system. Mol Cell Biol. 1995;15:2763–2771. doi: 10.1128/mcb.15.5.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Matsuo H, Li H, McGuire A M, Fletcher C M, Gingras A-C, Sonenberg N, Wagner G. Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein. Nat Struct Biol. 1997;4:717–724. doi: 10.1038/nsb0997-717. [DOI] [PubMed] [Google Scholar]
- 359.Mathews M B. Interactions between viruses and the cellular machinery of protein synthesis. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 505–548. [Google Scholar]
- 360.Mattaj I W. RNA recognition: a family matter? Cell. 1993;73:837–840. doi: 10.1016/0092-8674(93)90265-r. [DOI] [PubMed] [Google Scholar]
- 361.McCarthy J E G. Post-transcriptional control in the polycistronic operon environment: studies of the atp operon of Escherichia coli. Mol Microbiol. 1990;4:1233–1240. doi: 10.1111/j.1365-2958.1990.tb00702.x. [DOI] [PubMed] [Google Scholar]
- 362.McCarthy J E G, Brimacombe R. Prokaryotic translation: the interactive pathway leading to initiation. Trends Genet. 1994;10:402–407. doi: 10.1016/0168-9525(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 363.McCarthy J E G, Gualerzi C. Translational control of prokaryotic gene expression. Trends Genet. 1990;6:78–85. doi: 10.1016/0168-9525(90)90098-q. [DOI] [PubMed] [Google Scholar]
- 364.McCarthy J E G, Kollmus H. Cytoplasmic mRNA-protein interactions in eukaryotic gene expression. Trends Biochem Sci. 1995;20:191–197. doi: 10.1016/s0968-0004(00)89006-4. [DOI] [PubMed] [Google Scholar]
- 365.McCarthy J E G, Schairer H U, Sebald W. Translational initiation frequency of atp genes from Escherichia coli: identification of an intercistronic sequence that enhances translation. EMBO J. 1985;4:519–526. doi: 10.1002/j.1460-2075.1985.tb03659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365a.McCarthy J E G, Gerstel B, Surin B, Wiedemann U, Ziemke P. Differential gene expression from the Escherichia coli atp operon mediated by segmental differences in mRNA stability. Mol Microbiol. 1991;5:2447–2458. doi: 10.1111/j.1365-2958.1991.tb02090.x. [DOI] [PubMed] [Google Scholar]
- 366.McIntosh E N, Haynes R H. Sequence and expression of the dCMP deaminase gene (DCD1) of Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:1771–1721. doi: 10.1128/mcb.6.5.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Mehlin H, Daneholt B, Skoglund U. Translocation of a specific premessenger ribonucleoprotein particle through the nuclear pore studied with electron microscope tomography. Cell. 1992;69:605–613. doi: 10.1016/0092-8674(92)90224-z. [DOI] [PubMed] [Google Scholar]
- 368.Mercado J J, Smith R, Sagliocco F A, Brown A J P, Gancedo J M. The levels of yeast gluconeogenic mRNAs respond to environmental factors. Eur J Biochem. 1994;224:473–481. doi: 10.1111/j.1432-1033.1994.00473.x. [DOI] [PubMed] [Google Scholar]
- 369.Merrick W C, Hershey J W B. The pathway and mechanism of eukaryotic protein synthesis. In: Hershey J W B, Matthews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 31–70. [Google Scholar]
- 370.Méthot N, Pause A, Hershey J W B, Sonenberg N. The translation initiation factor eIF-4B contains an RNA-binding region that is distinct and independent from its ribonucleoprotein consensus sequence. Mol Cell Biol. 1994;14:2307–2316. doi: 10.1128/mcb.14.4.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Méthot N, Pickett G, Keene J D, Sonenberg N. In vitro RNA selection identifies RNA ligands that specifically bind to eukaryotic translation initiation factor eIF4B: the role of the RNA recognition motif. RNA. 1996;2:38–50. [PMC free article] [PubMed] [Google Scholar]
- 372.Méthot N, Song M S, Sonenberg N. A region rich in aspartic acid, arginine, tyrosine and glycine (DRYG) mediates eukaryotic initiation factor 4B (eIF4B) self-association and interaction with eIF3. Mol Cell Biol. 1996;16:5328–5334. doi: 10.1128/mcb.16.10.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Metz A M, Browning K S. Mutational analysis of the functional domains of the large subunit of the isozyme form of wheat initiation factor eIF4F. J Biol Chem. 1996;271:31033–31036. doi: 10.1074/jbc.271.49.31033. [DOI] [PubMed] [Google Scholar]
- 374.Miczak A, Kaberdin V R, Wei C-L, Lin-Chao S. Proteins associated with RNase E in a multicomponent ribonucleotic complex. Proc Natl Acad Sci USA. 1996;93:3865–3869. doi: 10.1073/pnas.93.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Minich W B, Balasta M L, Goss D J, Rhoads R E. Chromatographic resolution of in vivo phosphorylated and nonphosphorylated eukaryotic translation initiation factor eIF-4E: increased cap affinity of the phosphorylated form. Proc Natl Acad Sci USA. 1994;91:7668–7672. doi: 10.1073/pnas.91.16.7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Minvielle-Sebastia L, Winsor B, Bonneaud N, Lacroute F. Mutations in the yeast RNA14 and RNA15 genes results in an abnormal mRNA decay rate: sequence analysis reveals an RNA-binding domain in the RNA15 protein. Mol Cell Biol. 1991;11:3075–3087. doi: 10.1128/mcb.11.6.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Minvielle-Sebastia L, Preker P J, Keller W. RNA14 and RNA15 proteins as components of a yeast pre-mRNA 3′-end processing factor. Science. 1994;266:1702–1705. doi: 10.1126/science.7992054. [DOI] [PubMed] [Google Scholar]
- 378.Mirande M, Waller J P. The yeast lysyl-tRNA synthetase gene. J Biol Chem. 1988;263:18443–18451. [PubMed] [Google Scholar]
- 379.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 380.Mittelmeier T M, Dieckmann C L. In vivo analysis of sequences required for translation of cytochrome b transcripts in yeast mitochondria. Mol Cell Biol. 1995;15:780–789. doi: 10.1128/mcb.15.2.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Miyazaki M, Uritani M, Kitaoka Y, Ogawa K, Kagiyama H. Functional role and biochemical properties of yeast peptide elongation factor 3 (EF-3) In: McCarthy J E G, Tuite M F, editors. Posttranslational control of gene expression. Berlin, Germany: Springer-Verlag KG; 1990. pp. 557–566. [Google Scholar]
- 382.Morino S, Hazama H, Ozaki M, Teraoka Y, Shibata S, Doi M, Ueda H, Ishida T, Uesugi S. Analysis of the mRNA cap-binding ability of human eukaryotic initiation factor-4E by use of recombinant wild-type and mutant forms. Eur J Biochem. 1996;239:597–601. doi: 10.1111/j.1432-1033.1996.0597u.x. [DOI] [PubMed] [Google Scholar]
- 383.Morley S J, Curtis P S, Pain V M. eIF4G: Translation’s mystery factor begins to yield its secrets. RNA. 1997;3:1085–1104. [PMC free article] [PubMed] [Google Scholar]
- 384.Mottagui-Tabar S, Björnsson A, Isaksson L A. The second to last amino acid in the nascent peptide as a codon context determinant. EMBO J. 1994;13:249–257. doi: 10.1002/j.1460-2075.1994.tb06255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Mottagui-Tabar, S., M. F. Tuite, and L. A. Isakkson. The influence of the 5′ codon context on translation termination in Saccharomyces cerevisiae. Eur. J. Biochem., in press. [DOI] [PubMed]
- 386.Moye-Rowley W S, Harshmann K D, Parker C S. Yeast YAP1 encodes a novel form of the jun family of transcriptional activator proteins. Genes Dev. 1989;3:283–292. doi: 10.1101/gad.3.3.283. [DOI] [PubMed] [Google Scholar]
- 387.Muhlrad D, Parker R. Premature translational termination triggers mRNA decapping. Nature. 1994;370:578–581. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- 388.Muhlrad D, Decker C J, Parker R. Mutations affecting stability and deadenylation of the yeast MAF2 transcript. Genes Dev. 1992;6:2000–2111. doi: 10.1101/gad.6.11.2100. [DOI] [PubMed] [Google Scholar]
- 389.Muhlrad D, Decker C J, Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- 390.Muhlrad D, Decker C J, Parker R. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol Cell Biol. 1995;15:2145–2156. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Müller P P, Hinnebusch A. Multiple AUG codons mediate translational control of GCN4. Cell. 1986;45:201–207. doi: 10.1016/0092-8674(86)90384-3. [DOI] [PubMed] [Google Scholar]
- 392.Munroe D, Jacobson A. mRNA poly(A) tail, a 3′ enhancer of translational initiation. Mol Cell Biol. 1990;10:3441–3455. doi: 10.1128/mcb.10.7.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Nagai K, Mattaj I A, editors. RNA-protein interactions. New York, N.Y: IRL Press at Oxford University Press; 1994. [Google Scholar]
- 394.Nagashima K, Kasai M, Nagata S, Kasiro Y. Structure of the two genes coding for polypeptide-chain elongation factor-1-alpha (EF-1-Alpha) from Saccharomyces cerevisiae. Gene. 1986;45:265–273. doi: 10.1016/0378-1119(86)90024-7. [DOI] [PubMed] [Google Scholar]
- 395.Nairn A C, Palfrey H C. Regulation of protein synthesis by calcium. In: Hershey J W B, Matthews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 295–318. [Google Scholar]
- 396.Najarian D, Dihanich M E, Martin N C, Hopper A K. DNA sequence and transcript mapping of MOD5: features of the 5′ region which suggest two translational starts. Mol Cell Biol. 1987;7:185–191. doi: 10.1128/mcb.7.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Nakamura Y, Ito K, Matsumura K, Kawazu Y, Ebihara K. Regulation of translation termination: conserved structural motifs in bacterial and eukaryotic polypeptide release factors. Biochem Cell Biol. 1995;73:1113–1122. doi: 10.1139/o95-120. [DOI] [PubMed] [Google Scholar]
- 398.Nakamura Y, Ito K, Isaksson L A. Emerging understanding of translation termination. Cell. 1996;87:147–150. doi: 10.1016/s0092-8674(00)81331-8. [DOI] [PubMed] [Google Scholar]
- 399.Naranda T, MacMillan S E, Hershey J W B. Purified yeast translational initiation factor eIF-3 is an RNA-binding protein complex that contains the PRT1 protein. J Biol Chem. 1994;269:32286–32292. [PubMed] [Google Scholar]
- 400.Nasr F, Bécam A-M, Brown S C, DeNay D, Slominski S S, Herbert C J. Artificial antisense RNA regulation of YBR1012 (YBR136w), an essential gene from Saccharomyces cerevisiae which is important for progression through G1/S. Mol Gen Genet. 1995;249:51–57. doi: 10.1007/BF00290235. [DOI] [PubMed] [Google Scholar]
- 401.Nelson R J, Ziegelhoffer T, Nicolet C, Werner-Washburne M, Craig E A. The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell. 1992;71:97–105. doi: 10.1016/0092-8674(92)90269-i. [DOI] [PubMed] [Google Scholar]
- 402.Nierhaus K H. Solution of the ribosome riddle: how the ribosome selects the correct aminoacyl-tRNA out of 41 similar contestants. Mol Microbiol. 1993;9:661–669. doi: 10.1111/j.1365-2958.1993.tb01726.x. [DOI] [PubMed] [Google Scholar]
- 403.Nietfield W, Mentzel H, Pieler T. The Xenopus laevis poly(A) binding protein is composed of multiple functionally independent RNA binding domains. EMBO J. 1990;9:3699–3705. doi: 10.1002/j.1460-2075.1990.tb07582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Nigg E A. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 405.Nissen B, Kjeldgaard M, Thirup S, Polekhina G, Reshnetnikova L, Clark B F C, Nyborg J. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and GTP analog. Science. 1995;270:1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- 406.Nyunoya H, Lusty C J. Sequence of the small subunit of yeast carbamyl-phosphate synthetase and identification of its catalytic domain. J Biol Chem. 1984;259:9790–9798. [PubMed] [Google Scholar]
- 407.Oh S-K, Scott M P, Sarnow P. Homeotic gene Antennapedia mRNA contains 5′-noncoding sequences that confer translational initiation by internal ribosome binding. Genes Dev. 1992;6:1643–1653. doi: 10.1101/gad.6.9.1643. [DOI] [PubMed] [Google Scholar]
- 408.Ohlmann T, Rau M, Morley S J, Pain V M. Proteolytic cleavage of initiation factor eIF-4γ in the reticulocyte lysate inhibits translation of capped mRNAs but enhances that of uncapped mRNAs. Nucleic Acids Res. 1995;23:334–340. doi: 10.1093/nar/23.3.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Ohlmann T, Rau M, Pain V M, Morley S J. The C-terminal domain of eukaryotic protein synthesis initiation factor (eIF4) 4G is sufficient to support cap-independent translation in the absence of eIF4E. EMBO J. 1996;15:1371–1382. [PMC free article] [PubMed] [Google Scholar]
- 410.Ohtake Y, Wickner R B. Yeast virus propagation depends critically on free 60S ribosomal subunit concentration. Mol Cell Biol. 1995;15:2772–2781. doi: 10.1128/mcb.15.5.2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Oliveira C C, McCarthy J E G. The relationship between eukaryotic translation and mRNA stability: a short upstream open reading frame strongly inhibits translational initiation and greatly accelerates mRNA degradation in the yeast Saccharomyces cerevisiae. J Biol Chem. 1995;270:8936–8943. doi: 10.1074/jbc.270.15.8936. [DOI] [PubMed] [Google Scholar]
- 412.Oliveira C C, van der Heuvel J J, McCarthy J E G. Inhibition of translational initiation in Saccharomyces cerevisiae by secondary structure: the role of the stability and position of stem-loops in the mRNA leader. Mol Microbiol. 1993;9:521–532. doi: 10.1111/j.1365-2958.1993.tb01713.x. [DOI] [PubMed] [Google Scholar]
- 413.Oliveira C C, Goossen B, Zanchin N I T, McCarthy J E G, Hentze M W, Stripecke R. Translational repression by the human iron-regulatory factor (IRF) in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:5316–5322. doi: 10.1093/nar/21.23.5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Oliver S G, Winson M K, Kell D B, Baganz F. Systematic functional analysis of the yeast genome. 1998. Trends Biotechnol., in press. [DOI] [PubMed] [Google Scholar]
- 415.Osawa S, Jukes T H, Watanabe K, Muto A. Recent evidence for evolution of the genetic code. Microbiol Rev. 1992;56:229–264. doi: 10.1128/mr.56.1.229-264.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Ovadi J. Cell architecture and metabolic channeling. New York, N.Y: Springer-Verlag; 1995. [Google Scholar]
- 417.Ouzounis C, Bork P, Cesari G, Saunders C. New protein functions in yeast chromosome VIII. Protein Sci. 1995;4:2424–2428. doi: 10.1002/pro.5560041121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Pain V M. Initiation of protein synthesis in eukaryotic cells. Eur J Biochem. 1996;236:747–771. doi: 10.1111/j.1432-1033.1996.00747.x. [DOI] [PubMed] [Google Scholar]
- 419.Pande S, Vimaladithan A, Zhao H, Farabaugh P J. Pulling the ribosome out of frame +1 at a programmed frameshift site by cognate binding of aminoacyl-tRNA. Mol Cell Biol. 1995;15:298–304. doi: 10.1128/mcb.15.1.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Pause A, Méthot N, Svitkin Y, Merrick W C, Sonenberg N. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 1994;13:1205–1215. doi: 10.1002/j.1460-2075.1994.tb06370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Pause A, Belsham G J, Gingras A-C, Donzé O, Lin T-A, Lawrence J C, Jr, Sonenberg N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 422.Paushkin S V, Kushnirov V V, Smirnov V N, Ter-Avanesyan M D. Interaction between yeast Sup45p (eRF1) and Sup35p (eRF3) polypeptide chain release factors: implications for prion-dependent regulation. Mol Cell Biol. 1997;17:2798–2805. doi: 10.1128/mcb.17.5.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Pavitt G D, Yang W, Hinnebusch A G. Homologous segments in three subunits of the guanine nucleotide exchange factor eIF2B mediated translational regulation of phosphorylation of eIF2. Mol Cell Biol. 1997;17:1298–1313. doi: 10.1128/mcb.17.3.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423a.Pavitt G D, Ramaiah K V A, Kimball S R, Hinnebusch A G. eIF2 independently binds two distinct eIF2B subcomplexes that catalyze and regulate guanine-nucleotide exchange. Genes Dev. 1998;12:514–526. doi: 10.1101/gad.12.4.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Pavlov M Y, Freistroffer D V, MacDougall J, Buckingham R H, Ehrenberg M. Fast recycling of E. coli ribosomes requires both ribosome recycling factor (RRF) and release factor RF3. EMBO J. 1997;16:4134–4141. doi: 10.1093/emboj/16.13.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Pavlov M Y, Freistroffer D V, Huergué-Hamard V. Release factor RF3 abolishes competition between release factor RF1 and ribosome recycling factor (RRF) for a ribosome binding site. J Mol Biol. 1997;273:389–401. doi: 10.1006/jmbi.1997.1324. [DOI] [PubMed] [Google Scholar]
- 426.Pel H J, Grivell L A. Protein synthesis in mitochondria. Mol Biol Rep. 1994;19:183–194. doi: 10.1007/BF00986960. [DOI] [PubMed] [Google Scholar]
- 427.Pelsy F, Lacroute F. Effect of ochre nonsense mutations on yeast URA1 mRNA stability. Curr Genet. 1984;8:277–282. doi: 10.1007/BF00419725. [DOI] [PubMed] [Google Scholar]
- 428.Peltz S W, Brewer G, Bernstein P, Hart P A, Ross J. Regulation of mRNA turnover in eukaryotic cells. Crit Rev Eukaryotic Gene Expression. 1991;1:99–126. [PubMed] [Google Scholar]
- 429.Peltz S W, Donahue J L, Jacobson A. A mutation in the tRNA nucleotidyltransferase gene promotes stabilization of mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:5778–5784. doi: 10.1128/mcb.12.12.5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Peltz S W, Brown A H, Jacobson A. mRNA destabilization triggered by premature translation termination depends on three mRNA sequence elements and at least one trans-acting factor. Genes Dev. 1993;7:1737–1754. doi: 10.1101/gad.7.9.1737. [DOI] [PubMed] [Google Scholar]
- 431.Peltz S W, Trotta C, He F, Brown A, Donahue J, Welch E, Jacobson A. Identification of the cis-acting sequences and trans-acting factors involved in nonsense-mediated mRNA decay. NATO ASI Ser Ser H. 1993;71:1–10. [Google Scholar]
- 432.Peltz S W, He F, Welch E, Jacobson A. Nonsense-mediated mRNA decay in yeast. Proc Nucleic Acid Res. 1994;47:271–298. doi: 10.1016/s0079-6603(08)60254-8. [DOI] [PubMed] [Google Scholar]
- 433.Perentesis J P, Phan L D, Gleason W B, LaPorte D C, Kivingston D M, Bodley J W. Saccharomyces cerevisiae elongation factor 2. J Biol Chem. 1992;267:1190–1197. [PubMed] [Google Scholar]
- 434.Perez L, Carrasco L. Lack of direct correlation between p220 cleavage and the shutoff of host translation after poliovirus infection. Virology. 1992;189:178–186. doi: 10.1016/0042-6822(92)90693-j. [DOI] [PubMed] [Google Scholar]
- 435.Pestova T V, Shatsky I N, Hellen C U T. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol. 1996;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435a.Pestova T V, Borukhov S I, Hellen C U T. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998;394:854–859. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- 436.Petersen C. Long-range translational coupling in the rplJL-rpoBC operon of Escherichia coli. J Mol Biol. 1989;206:323–332. doi: 10.1016/0022-2836(89)90482-8. [DOI] [PubMed] [Google Scholar]
- 436a.Pfund C, Lopez-Hoyo N, Ziegelhoffer T, Schilke B A, Lopez-Buesa P, Walter W A, Wiedmann M, Craig E A. The molecular chaperone Ssb from Saccharomyces cerevisiae is a component of the ribosome-nascent chain complex. EMBO J. 1998;17:3981–3989. doi: 10.1093/emboj/17.14.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Philippe C, Eyermann F, Bénard L, Portier C, Ehresmann B, Ehresmann C. Ribosomal protein S15 from Escherichia coli modulates its own translation by trapping the ribosome on the mRNA initiation loading site. Proc Natl Acad Sci USA. 1993;90:4394–4398. doi: 10.1073/pnas.90.10.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Pierrat B, Lacroute F, Losson R. The 5′ untranslated region of the PPR1 regulatory gene dictates rapid mRNA decay in yeast. Gene. 1993;131:43–51. doi: 10.1016/0378-1119(93)90667-r. [DOI] [PubMed] [Google Scholar]
- 439.Piňol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–832. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- 440.Pinto I, Na J G, Sherman F, Hampsey M. cis- and trans-acting suppressors of a translation initiation defect at the cyc1 locus of Saccharomyces cerevisiae. Genetics. 1992;132:97–112. doi: 10.1093/genetics/132.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Polymenis M, Schmidt E V. Coupling of cell division to cell growth by translational control of the G1 cyclin CLN3 in yeast. Genes Dev. 1997;11:2522–2531. doi: 10.1101/gad.11.19.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Poole E S, Brown C M, Tate W P. The identity of the base following the stop codon determines the efficiency of in vivo translational termination in Escherichia coli. EMBO J. 1995;14:151–158. doi: 10.1002/j.1460-2075.1995.tb06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442a.Preiss T, Hentze M. Dual function of the messenger RNA cap structure in poly(A) tail-promoted translation in yeast. Nature. 1998;392:516–519. doi: 10.1038/33192. [DOI] [PubMed] [Google Scholar]
- 443.Presutti C, Ciafré S A, Bozzoni I. The ribosomal protein-L2 in Saccharomyces cerevisiae controls the level of accumulation of its own messenger-RNA. EMBO J. 1991;10:2215–2221. doi: 10.1002/j.1460-2075.1991.tb07757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Presutti C, Villa T, Hall D, Pertica C, Bozzoni I. Identification of the cis-elements mediating the autogenous control of ribosomal protein L2 mRNA stability in yeast. EMBO J. 1995;14:4022–4030. doi: 10.1002/j.1460-2075.1995.tb00073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Proweller A, Butler S. Efficient translation of poly(A)-deficient mRNAs in Saccharomyces cerevisiae. Genes Dev. 1994;8:2629–2640. doi: 10.1101/gad.8.21.2629. [DOI] [PubMed] [Google Scholar]
- 446.Proweller A, Butler S. Ribosome concentration contributes to discrimination against poly(A)− mRNA during translation initiation in Saccharomyces cerevisiae. J Biol Chem. 1997;272:6004–6010. doi: 10.1074/jbc.272.9.6004. [DOI] [PubMed] [Google Scholar]
- 447.Ptushkina M, Vasilescu S, Fierro-Monti I, Rohde M, McCarthy J E G. Intracellular targeting and mRNA interactions of the eukaryotic translation initiation factor eIF4E in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 1996;1308:142–150. doi: 10.1016/0167-4781(96)00096-6. [DOI] [PubMed] [Google Scholar]
- 448.Ptushkina M, Fierro-Monti I, van den Heuvel J, Vasilescu S, Birkenhäger R, Mita K, McCarthy J E G. Schizosaccharomyces pombe has a novel eukaryotic initiation factor 4F complex containing a cap-binding protein with the human eIF4E C-terminal motif KSGST. J Biol Chem. 1996;271:32818–32824. doi: 10.1074/jbc.271.51.32818. [DOI] [PubMed] [Google Scholar]
- 449.Ptushkina M, Von der Haar T, Vasilescu S, Frank R, Birkenhäger R, McCarthy J E G. Cooperative modulation by eIF4G of eIF4E binding to the mRNA 5′ cap in yeast involves a site partially shared by p20. EMBO J. 1998;17:4798–4808. doi: 10.1093/emboj/17.16.4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449a.Ptushkina, M., et al. Unpublished data.
- 450.Py B, Causton H, Mudd E A, Higgins C F. A protein complex mediating mRNA degradation in Escherichia coli. Mol Microbiol. 1994;14:717–729. doi: 10.1111/j.1365-2958.1994.tb01309.x. [DOI] [PubMed] [Google Scholar]
- 451.Py B, Higgins C F, Krisch H M, Carpousis A J. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature. 1996;381:169–172. doi: 10.1038/381169a0. [DOI] [PubMed] [Google Scholar]
- 452.Ramirez M, Wek R C, Hinnebusch A G. Ribosome-association of GCN2 protein kinase, a translational activator of the GCN4 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:3027–3036. doi: 10.1128/mcb.11.6.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Rau M, Ohlmann T, Morley S J, Pain V M. A reevaluation of the cap-binding protein, eIF4E, as a rate-limiting factor for initiation of translation in reticulocyte lysate. J Biol Chem. 1996;271:8983–8990. doi: 10.1074/jbc.271.15.8983. [DOI] [PubMed] [Google Scholar]
- 454.Raué H A. Metabolic stability of mRNA in yeast—a potential target for modulating productivity? Trends Biotechnol. 1994;12:444–449. doi: 10.1016/0167-7799(94)90019-1. [DOI] [PubMed] [Google Scholar]
- 455.Ray P N, Pearson M L. Evidence for post-transcriptional control of the morphogenetic genes of bacteriophage lambda. J Mol Biol. 1974;85:163–175. doi: 10.1016/0022-2836(74)90135-1. [DOI] [PubMed] [Google Scholar]
- 456.Rex G, Surin B, Besse G, Scheppe B, McCarthy J E G. The mechanism of translational coupling in Escherichia coli. J Biol Chem. 1994;269:18118–18127. [PubMed] [Google Scholar]
- 457.Richter J D. Translational control during early development. Bioessays. 1991;13:179–183. doi: 10.1002/bies.950130406. [DOI] [PubMed] [Google Scholar]
- 458.Richter-Cook N J, Dever T E, Hensold J O, Merrick W C. Purification and characterisation of a new eukaryotic protein translation factor—eukaryotic initiation factor 4H. J Biol Chem. 1998;273:7579–7587. doi: 10.1074/jbc.273.13.7579. [DOI] [PubMed] [Google Scholar]
- 459.Riis B, Rattan S I S, Clark B F C, Merrick W C. Eukaryotic protein elongation factors. Trends Biochem Sci. 1990;15:420–424. doi: 10.1016/0968-0004(90)90279-k. [DOI] [PubMed] [Google Scholar]
- 460.Ringquist S, Shinedling S, Barrick D, Green L, Binkley J, Stormo G D, Gold L. Translation initiation in Escherichia coli; sequences within the ribosome-binding site. Mol Microbiol. 1992;6:1219–1229. doi: 10.1111/j.1365-2958.1992.tb01561.x. [DOI] [PubMed] [Google Scholar]
- 460a.Rom E, Kim H C, Gingras A-C, Marcotrigiano J, Favre D, Olsen H, Burley S K, Sonenberg N. Cloning and characterization of 4EHP, a novel mammalian eIF4E-regulated cap-binding protein. J Biol Chem. 1998;273:13104–13109. doi: 10.1074/jbc.273.21.13104. [DOI] [PubMed] [Google Scholar]
- 461.Rosbash R, Singer R H. RNA travel: tracks from DNA to cytoplasm. Cell. 1993;75:399–401. doi: 10.1016/0092-8674(93)90373-x. [DOI] [PubMed] [Google Scholar]
- 462.Ross J. mRNA stability in mammalian cells. Microbiol Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Ross J. Control of messenger RNA stability in higher eukaryotes. Trends Genet. 1996;12:171–175. doi: 10.1016/0168-9525(96)10016-0. [DOI] [PubMed] [Google Scholar]
- 464.Rouault T A, Klausner R D, Harford J B. Translational control of ferritin. In: Hershey J W B, Matthews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 335–362. [Google Scholar]
- 465.Rousseau D, Kaspar R, Rosenwald I, Gehrke L, Sonenberg N. Translation initiation of ornithine decarboxylase and nucleocytoplasmic transport of cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc Natl Acad Sci USA. 1996;93:1065–1070. doi: 10.1073/pnas.93.3.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Roussou I, Thireos G, Hague B M. Transcriptional-translational regulatory circuit in Saccharomyces cerevisiae which involves the GCN4 transcriptional activator and the GCN2 protein kinase. Mol Cell Biol. 1988;8:2132–2139. doi: 10.1128/mcb.8.5.2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Rozen F, Edery I, Meerovitch K, Dever T E, Merrick W C, Sonenberg N. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol Cell Biol. 1990;10:1134–1144. doi: 10.1128/mcb.10.3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Ruiz-Echevarria M J, Czaplinski K, Peltz S W. Making sense of nonsense in yeast. Trends Biochem Sci. 1996;21:433–438. doi: 10.1016/s0968-0004(96)10055-4. [DOI] [PubMed] [Google Scholar]
- 469.Ruiz-Echevarria M J, Peltz S W. Utilizing the GCN4 leader region to investigate the role of the sequence determinants in nonsense-mediated mRNA decay. EMBO J. 1996;15:2810–2819. [PMC free article] [PubMed] [Google Scholar]
- 469a.Ruiz-Echevarria M J, Gonzalez C I, Peltz S W. Identifying the right stop: determining how the surveillance complex recognizes and degrades an aberrant mRNA. EMBO J. 1998;17:575–589. doi: 10.1093/emboj/17.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Rychlik W, Russ M S, Rhoads R E. Phosphorylation site of eukaryotic initiation factor 4E. J Biol Chem. 1987;262:10434–10437. [PubMed] [Google Scholar]
- 471.Ryoji M, Berland R, Kaji A. Reinitiation of translation from the triplet next to the amber termination codon in the absence of ribosome-releasing factor. Proc Natl Acad Sci USA. 1981;78:5973–5977. doi: 10.1073/pnas.78.10.5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Sachs A B. The role of poly(A) in the translation and stability of mRNA. Curr Opin Cell Biol. 1990;2:1092–1098. doi: 10.1016/0955-0674(90)90161-7. [DOI] [PubMed] [Google Scholar]
- 473.Sachs A B. Messenger RNA degradation in eukaryotes. Cell. 1993;74:413–421. doi: 10.1016/0092-8674(93)80043-e. [DOI] [PubMed] [Google Scholar]
- 474.Sachs A B, Davis R W. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989;58:857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- 475.Sachs A B, Deardorff J. Translation initiation requires the PAB-dependent poly(A) ribonuclease in yeast. Cell. 1992;70:961–973. doi: 10.1016/0092-8674(92)90246-9. . (Erratum, 83:48, 1995.) [DOI] [PubMed] [Google Scholar]
- 476.Sachs A B, Wahle E. Poly(A) tail metabolism and function in eukaryotes. J Biol Chem. 1993;268:22955–22958. [PubMed] [Google Scholar]
- 477.Sachs A B, Bond M W, Kornberg R D. A single gene from yeast for both nuclear and cytoplasmic polyadenylate-binding proteins: domain structure and expression. Cell. 1986;45:827–835. doi: 10.1016/0092-8674(86)90557-x. [DOI] [PubMed] [Google Scholar]
- 478.Sachs A B, Davis R W, Kornberg R D. A single domain of yeast poly(A) binding protein is necessary and sufficient for RNA binding and cell viability. Mol Cell Biol. 1987;7:3268–3276. doi: 10.1128/mcb.7.9.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 479.Sachs A B, Sarnow P, Hentze M W. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 480.Safer B, Anderson W C, Merrick W C. Purification and physical properties of homogeneous initiation factor MP from rabbit reticulocytes. J Biol Chem. 1975;250:9067–9075. [PubMed] [Google Scholar]
- 481.Sagliocco F, Vega Laso M R, Zhu D, Tuite M F, McCarthy J E G, Brown A J P. The influence of 5′ secondary structures upon ribosome binding to mRNA during translation in yeast. J Biol Chem. 1993;268:26522–26530. [PubMed] [Google Scholar]
- 482.Sagliocco F, Zhu D, Vega Laso M R, McCarthy J E G, Tuite M F, Brown A J P. Rapid mRNA degradation in yeast can proceed independently of translational elongation. J Biol Chem. 1994;269:18630–18637. [PubMed] [Google Scholar]
- 483.Sakamuro D, Furukawa T, Takegami T. Hepatitis-C virus nonstructural protein NS3 transforms NIH 3T3 cells. J Virol. 1995;69:3893–3896. doi: 10.1128/jvi.69.6.3893-3896.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Sanger F, Coulson A R, Hong G F, Hill D F, Petersen G B. Nucleotide sequence of bacteriophage λ DNA. J Mol Biol. 1982;162:729–773. doi: 10.1016/0022-2836(82)90546-0. [DOI] [PubMed] [Google Scholar]
- 485.Santos M A S, Ueda T, Watanabe K, Tuite M F. The non-standard genetic code of Candida species: an evolving genetic code or a novel mechanism for adaptation? Mol Microbiol. 1997;26:423–431. doi: 10.1046/j.1365-2958.1997.5891961.x. [DOI] [PubMed] [Google Scholar]
- 486.Schauder B, McCarthy J E G. The role of bases upstream of the Shine-Dalgarno region and in the coding sequence in the control of gene expression in Escherichia coli: translation and stability of mRNAs in vivo. Gene. 1989;78:59–72. doi: 10.1016/0378-1119(89)90314-4. [DOI] [PubMed] [Google Scholar]
- 487.Schmid S R, Linder P. D-E-A-D protein family of putative RNA helicases. Mol Microbiol. 1992;6:283–292. doi: 10.1111/j.1365-2958.1992.tb01470.x. [DOI] [PubMed] [Google Scholar]
- 488.Schmidt M C, Kao C C, Pei R, Berk A J. Yeast TATA-box transcription factor gene. Proc Natl Acad Sci USA. 1989;86:7785–7789. doi: 10.1073/pnas.86.20.7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Schnell N, Entian K-D. Identification and characterization of a Saccharomyces cerevisiae gene (PAR1) conferring resistance to iron chelators. Eur J Biochem. 1991;200:487–493. doi: 10.1111/j.1432-1033.1991.tb16209.x. [DOI] [PubMed] [Google Scholar]
- 490.Schnier J, Schwelberger H G, Smit-McBride Z, Ah Kang H, Hershey J W B. Translation initiation factor 5A and its hypusine modification are essential for cell viability in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:3105–3114. doi: 10.1128/mcb.11.6.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490a.Schramm H-C, Schneppe B, Birkenhäger R, McCarthy J E G. The promoter-proximal, unstable IB region of the atp mRNA of Escherichia coli: an independently degraded region that can act as a destabilizing element. Biochim Biophys Acta. 1995;1307:162–170. doi: 10.1016/0167-4781(96)00034-6. [DOI] [PubMed] [Google Scholar]
- 491.Schümperli D, McKenny K, Sobieski D A, Rosenberg M. Translational coupling at an intercistronic boundary of the Escherichia coli galactose operon. Cell. 1982;30:865–871. doi: 10.1016/0092-8674(82)90291-4. [DOI] [PubMed] [Google Scholar]
- 492.Schwarz E, Neupert W. Mitochondrial protein import—mechanisms, components and energetics. Biochim Biophys Acta Bioenerg. 1994;1187:270–274. doi: 10.1016/0005-2728(94)90125-2. [DOI] [PubMed] [Google Scholar]
- 493.Schwer B, Shuman S. Conditional inactivation of messenger-RNA capping enzyme affects yeast pre-messenger-RNA splicing in vivo. RNA. 1996;2:574–583. [PMC free article] [PubMed] [Google Scholar]
- 494.Serdyuk I, Baranov V, Tsalkova T, Gulamova D, Pavlov M, Spirin A, May R. Structural dynamics of translating ribosomes. Biochimie. 1992;74:299–306. doi: 10.1016/0300-9084(92)90107-p. [DOI] [PubMed] [Google Scholar]
- 495.Shamoo Y, Krueger U, Rice L M, Williams K P, Steitz T A. Crystal structure of the two RNA binding domains of human hnRNP A1 at 1.75 angstrom resolution. Nat Struct Biol. 1997;4:215–222. doi: 10.1038/nsb0397-215. [DOI] [PubMed] [Google Scholar]
- 496.Shatkin A J. Capping of eukaryotic mRNAs. Cell. 1976;9:645–653. doi: 10.1016/0092-8674(76)90128-8. [DOI] [PubMed] [Google Scholar]
- 497.Shean C A, Gottesmann M E. Translation of the prophage λ cI transcript. Cell. 1992;70:513–522. doi: 10.1016/0092-8674(92)90175-c. [DOI] [PubMed] [Google Scholar]
- 498.Sherman F, Stewart J W, Scheingruber A M. Mutants of yeast initiating translation of iso-1-cytochrome c within a region spanning 37 nucleotides. Cell. 1980;20:215–222. doi: 10.1016/0092-8674(80)90249-4. [DOI] [PubMed] [Google Scholar]
- 499.Sherman F, Stewart J W. Mutations altering the initiation of translation of yeast iso-1-cytochrome c: contrasts between the eukaryotic and prokaryotic initiation process. In: Strathern J N, Jones E W, Broach J R, editors. Molecular biology of the yeast Saccharomyces cerevisiae: metabolism and gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. pp. 301–333. [Google Scholar]
- 500.Shestakova E A, Motuz L P, Minin A A, Gelfand V I, Gavrilova L P. Some of eukaryotic elongation factor-II is colocalized with actin microfilament bundles in mouse embryo fibroblasts. Cell Biol Int Rep. 1991;15:75–84. doi: 10.1016/0309-1651(91)90084-v. [DOI] [PubMed] [Google Scholar]
- 501.Shevchenko A, Jensen O N, Podtelejnikov A V, Sagliocco F, Wilm M, Vorm O, Mortensen P, Shevchenko A, Boucherie H, Mann M. Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels. Proc Natl Acad Sci USA. 1996;93:14440–14445. doi: 10.1073/pnas.93.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501a.Shibata S, Morino S, Tomoo K, In Y, Ishida I. Effect of mRNA cap structure on eIF-4E phosphorylation and cap binding analyses using Ser209-mutated eIF-4Es. Biochem Biophys Res Commun. 1998;247:213–216. doi: 10.1006/bbrc.1998.8761. [DOI] [PubMed] [Google Scholar]
- 502.Simons R W. The control of prokaryotic and eukaryotic gene expression by naturally occurring antisense RNA. In: Crooke S T, Lebleu B, editors. Antisense research and applications. Boca Raton, Fla: CRC Press, Inc.; 1993. pp. 97–124. [Google Scholar]
- 503.Reference deleted.
- 504.Singer R H. The cytoskeleton and mRNA localization. Curr Opin Cell Biol. 1992;4:15–19. doi: 10.1016/0955-0674(92)90053-f. [DOI] [PubMed] [Google Scholar]
- 505.Singer R H, Green M R. Compartmentalization of eukaryotic gene expression: causes and effects. Cell. 1997;91:291–294. doi: 10.1016/s0092-8674(00)80411-0. [DOI] [PubMed] [Google Scholar]
- 506.Skogerson L, Wakatama E. A ribosome-dependent GTPase from yeast distinct from elongation factor 2. Proc Natl Acad Sci USA. 1976;73:73–76. doi: 10.1073/pnas.73.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Slusher L B, Gillman E C, Martin N C, Hopper A K. mRNA leader length and initiation codon context determine alternative AUG selection for the yeast gene MOD5. Proc Natl Acad Sci USA. 1991;88:9789–9793. doi: 10.1073/pnas.88.21.9789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507a.Snay-Hodge C A, Colot H V, Goldstein A L, Cole C N. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J. 1998;17:2663–2676. doi: 10.1093/emboj/17.9.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 508.Sonenberg N, Gingras A-C. The mRNA 5′ cap-binding protein eIF4E and control cell growth. Curr Opin Cell Biol. 1998;10:268–275. doi: 10.1016/s0955-0674(98)80150-6. [DOI] [PubMed] [Google Scholar]
- 509.Sonenberg N. mRNA 5′ cap-binding protein eIF4E and control of cell growth. In: Hershey J W B, Matthews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 245–269. [Google Scholar]
- 509a.Sonenberg, N. Personal communication.
- 510.Spanjaard R A, van Duin J. Translation reinitiation in the presence and absence of a Shine and Dalgarno sequence. Nucleic Acids Res. 1989;17:5501–5507. doi: 10.1093/nar/17.14.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 511.Spector D L. Macromolecular domains within the cell nucleus. Annu Rev Cell Biol. 1993;9:265–315. doi: 10.1146/annurev.cb.09.110193.001405. [DOI] [PubMed] [Google Scholar]
- 512.Spirin A S. Masked and translatable messenger ribonucleoproteins in higher eukaryotes. In: Hershey J W B, Matthews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 319–334. [Google Scholar]
- 513.Spirin A S, Baranov V I, Polubesov G S, Serdyuk I N, May R P. Translocation makes the ribosome less compact. J Mol Biol. 1987;194:119–128. doi: 10.1016/0022-2836(87)90720-0. [DOI] [PubMed] [Google Scholar]
- 514.Sprengart M L, Fuchs E, Porter A H. The downstream box: an efficient and independent translation initiation signal in Escherichia coli. EMBO J. 1996;15:665–674. [PMC free article] [PubMed] [Google Scholar]
- 515.Sprengart M L, Porter A G. Functional importance of RNA interactions in selection of translation initiation codons. Mol Microbiol. 1997;24:19–28. doi: 10.1046/j.1365-2958.1997.3161684.x. [DOI] [PubMed] [Google Scholar]
- 516.Sripati C E, Groner Y, Warner J R. Methylated, blocked 5′ termini of yeast mRNA. J Biol Chem. 1976;251:2898–2904. [PubMed] [Google Scholar]
- 517.Stansfield I, Jones K M, Kushnirov V V, Dagkesamanskaya A R, Poznyakovski I R, Paushkin S V, Nierras C R, Cox B S, Ter-Avanesyan M D, Tuite M F. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 1995;14:4365–4373. doi: 10.1002/j.1460-2075.1995.tb00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 518.Stansfield, I., S. Pitts, and M. F. Tuite. Personal communication.
- 519.Steege D. 5′-Terminal nucleotide sequence of Escherichia coli lactose repressor mRNA: features of translational initiation and reinitiation sites. Proc Natl Acad Sci USA. 1977;74:4163–4167. doi: 10.1073/pnas.74.10.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520.Stevens A. Purification and characterization of a Saccharomyces cerevisiae exoribonuclease which yields 5′-mononucleotides by a 5′→3′ mode of hydrolysis. J Biol Chem. 1980;255:3080–3085. [PubMed] [Google Scholar]
- 521.Stevens A. An mRNA decapping enzyme from ribosomes of Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1980;81:656–661. doi: 10.1016/0006-291x(80)90072-8. [DOI] [PubMed] [Google Scholar]
- 522.Stevens A. mRNA-decapping enzyme from Saccharomyces cerevisiae: purification and unique specificity for long RNA chains. Mol Cell Biol. 1988;8:2005–2010. doi: 10.1128/mcb.8.5.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 523.Stevens A. Eukaryotic nucleases and mRNA turnover. In: Belasco J, Brawerman G, editors. Control of messenger mRNA stability. San Diego, Calif: Academic Press, Inc.; 1993. pp. 449–471. [Google Scholar]
- 524.Stevens A, Maupin M K. A 5′→3′ exoribonuclease of Saccharomyces cerevisiae: size and novel substrate specificity. Arch Biochem Biophys. 1987;252:339–347. doi: 10.1016/0003-9861(87)90040-3. [DOI] [PubMed] [Google Scholar]
- 525.Stevens A, Poole T L. 5′-Exonuclease-2 of Saccharomyces cerevisiae. J Biol Chem. 1995;270:16063–16069. doi: 10.1074/jbc.270.27.16063. [DOI] [PubMed] [Google Scholar]
- 526.Stewart J W, Sherman F, Shipman N, Jackson M. Identification and mutational relocation of the AUG codon initiating translation of iso-1-cytochrome c in yeast. J Biol Chem. 1971;246:7429–7445. [PubMed] [Google Scholar]
- 527.Stormo G D. Translation initiation. In: Reznikoff W, Gold L, editors. Maximising gene expression. London, United Kingdom: Butterworths; 1986. pp. 195–224. [Google Scholar]
- 528.Strick C, Fox T D. Saccharomyces cerevisiae positive regulatory gene PET111 encodes a mitochondrial protein that is translated from an mRNA with a long 5′ leader. Mol Cell Biol. 1987;7:2728–2734. doi: 10.1128/mcb.7.8.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 529.Stripecke R, Oliveira C C, McCarthy J E G, Hentze M. Proteins binding to 5′ UTR sites: a general mechanism for translational regulation of mRNAs in human and yeast cells. Mol Cell Biol. 1994;14:5894–5909. doi: 10.1128/mcb.14.9.5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 530.Stuart R A, Neupert W. Topogenesis of inner membrane proteins of mitochondria. Trends Biochem Sci. 1996;21:261–267. [PubMed] [Google Scholar]
- 531.Suissa M, Altuvia S, Koby G, Oppenheim A B. Translational signals of a major head protein gene of bacteriophage lambda. Mol Gen Genet. 1988;214:570–573. doi: 10.1007/BF00330496. [DOI] [PubMed] [Google Scholar]
- 532.Surosky R T, Esposito R E. Early meiotic transcripts are highly unstable in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:3948–3958. doi: 10.1128/mcb.12.9.3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 533.Surosky R T, Strich R, Eposito R E. The yeast UME5 gene regulates the stability of meiotic mRNAs in response to glucose. Mol Cell Biol. 1994;14:3446–3458. doi: 10.1128/mcb.14.5.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 534.Svitkin Y V, Ovchinnikov L P, Dreyfuss G, Sonenberg N. General RNA binding proteins render translation cap dependent. EMBO J. 1996;15:7147–7155. [PMC free article] [PubMed] [Google Scholar]
- 535.Takizawa P A, Sil A, Swedlow J R, Herskowitz I, Vale R D. Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature. 1997;389:90–93. doi: 10.1038/38015. [DOI] [PubMed] [Google Scholar]
- 536.Tanguay R, Gallie D R. Translational efficiency is regulated by the length of the 3′ untranslated region. Mol Cell Biol. 1996;16:146–156. doi: 10.1128/mcb.16.1.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 537.Tani T, Derby R B, Hiraoka Y, Spector D. Nucleolar accumulation of poly (A)+ in heat-shocked yeast cells: implication of nucleolar involvement in mRNA transport. Mol Biol Cell. 1995;6:1515–1534. doi: 10.1091/mbc.6.11.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 538.Tarun S Z, Sachs A B. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev. 1995;9:2997–3007. doi: 10.1101/gad.9.23.2997. [DOI] [PubMed] [Google Scholar]
- 539.Tarun S Z, Sachs A B. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 540.Tarun S Z, Sachs A B. Binding of eukaryotic translation initiation factor 4E (eIF4E) to eIF4G represses translation of uncapped mRNA. Mol Cell Biol. 1997;17:6876–6886. doi: 10.1128/mcb.17.12.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 541.Tarun S Z, Wells S E, Deardorff J A, Sachs A B. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc Natl Acad Sci USA. 1997;94:9045–9051. doi: 10.1073/pnas.94.17.9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 542.Tate W P, Mannering S A. Three, four or more: the translational stop signal at length. Mol Microbiol. 1996;21:213–219. doi: 10.1046/j.1365-2958.1996.6391352.x. [DOI] [PubMed] [Google Scholar]
- 543.Tate W P, Poole E S, Mannering S A. Hidden infidelities of the translational stop signal. Prog Nucleic Acid Res Mol Biol. 1996;52:293–333. doi: 10.1016/s0079-6603(08)60970-8. [DOI] [PubMed] [Google Scholar]
- 544.Tavernarakis N, Thireos G. Genetic evidence for functional specificity of the yeast GCN2 kinase. Mol Gen Genet. 1996;251:613–618. doi: 10.1007/BF02173652. [DOI] [PubMed] [Google Scholar]
- 545.Thanaraj T W, Pandit M W. Translation-initiation promoting site on transcripts of highly expressed genes from Saccharomyces cerevisiae and the role of hairpin stems to position the site near the initiation codon. J Biomol Struct Dyn. 1990;7:1279–1289. doi: 10.1080/07391102.1990.10508565. [DOI] [PubMed] [Google Scholar]
- 546.Theodorakis N G, Cleveland D W. Translationally coupled degradation of mRNA in eukaryotes. In: Hershey J W B, Matthews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 631–652. [Google Scholar]
- 547.Thiele D J, Hannig E M, Leibowitz M J. Multiple L double-stranded RNA species of Saccharomyces cerevisiae: evidence for separate encapsidation. Mol Cell Biol. 1984;4:92–100. doi: 10.1128/mcb.4.1.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 548.Thireos G, Driscoll-Penn M, Greer H. 5′ untranslated sequences are required for the translational control of a yeast regulatory gene. Proc Natl Acad Sci USA. 1984;81:5096–5100. doi: 10.1073/pnas.81.16.5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 549.Thullier V, Stettler S, Sentenac A, Thuriaux P, Werner M. A mutation in the C31 subunit of Saccharomyces cerevisiae RNA polymerase III affects transcription initiation. EMBO J. 1995;14:351–359. doi: 10.1002/j.1460-2075.1995.tb07009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 550.Tishkoff D X, Johnson A W, Kolodner R D. Molecular and genetic analysis of the gene encoding the Saccharomyces cerevisiae strand exchange protein Sep1. Mol Cell Biol. 1991;11:2593–2608. doi: 10.1128/mcb.11.5.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 551.Toh-e A, Wickner R B. “Superkiller” mutations suppress chromosomal mutations affecting double-stranded RNA killer plasmid replication in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1980;77:527–530. doi: 10.1073/pnas.77.1.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 552.Trachsel H. Binding of initiator methionyl-tRNA to ribosomes. In: Hershey J W B, Matthews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 113–138. [Google Scholar]
- 552a.Tseng S S-I, Weaver P L, Liu Y, Hitomi M, Tartakoff A M, Chang T-H. Dbp5p, a cytosolic RNA helicase, is required for poly(A)+ RNA export. EMBO J. 1998;17:2651–2662. doi: 10.1093/emboj/17.9.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 553.Tuite M F. Protein synthesis. In: Rose A H, Harrison J S, editors. The yeasts. Vol. 3. San Diego, Calif: Academic Press, Inc.; 1989. pp. 161–204. [Google Scholar]
- 554.Tuite M F, Lindquist S L. Maintenance and inheritance of yeast prions. Trends Genet. 1996;12:467–471. doi: 10.1016/0168-9525(96)10045-7. [DOI] [PubMed] [Google Scholar]
- 555.Tuite M F, Belfield G P, Colthurst D R, Ross-Smith N. Defining a new molecular target in fungal protein synthesis: eukaryotic elongation factor 3. In: Dixon G K, Copping L G, Hollomon D W, editors. Antifungal agents. Oxford, United Kingdom: Bios Scientific Publishers; 1995. pp. 119–129. [Google Scholar]
- 556.Turton H E, Dawes I W, Grant C M. Saccharomyces cerevisiae exhibits a yAP-1 mediated adaptive response to malondialdehyde. J Bacteriol. 1997;179:1096–1101. doi: 10.1128/jb.179.4.1096-1101.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 557.Tzagoloff A, Dieckmann C L. Pet genes of Saccharomyces cerevisiae. Microbiol Rev. 1990;54:211–225. doi: 10.1128/mr.54.3.211-225.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 558.Tzamarias D, Thireos G. Evidence that the GCN2 protein kinase regulates reinitiation by the yeast ribosomes. EMBO J. 1988;7:3547–3551. doi: 10.1002/j.1460-2075.1988.tb03231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 559.Tzamarias D, Alexandraki D, Thireos G. Multiple cis-acting elements modulate the translational efficiency of GCN4 mRNA in yeast. Proc Natl Acad Sci USA. 1986;83:4849–4853. doi: 10.1073/pnas.83.13.4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 560.Tzamarias D, Roussou I, Thireos G. Coupling of GCN4 mRNA translational activation with decreased rates of polypeptide chain initiation. Cell. 1989;57:947–954. doi: 10.1016/0092-8674(89)90333-4. [DOI] [PubMed] [Google Scholar]
- 561.Tzeng T-H, Tu C-L, Bruenn J E. Ribosomal frameshifting requires a pseudoknot in the Saccharomyces cerevisiae double-stranded RNA virus. J Virol. 1992;66:999–1006. doi: 10.1128/jvi.66.2.999-1006.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 562.Vagner S, Gensac M-C, Maret A, Bayard F, Amalric F, Prats H, Prats A-C. Alternative translation of human fibroblast growth factor 2 mRNA occurs by internal entry of ribosomes. Mol Cell Biol. 1995;15:35–44. doi: 10.1128/mcb.15.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 563.Valegard K, Murray J B, Stockley P G, Stonehouse N J, Liljas L. Crystal-structure of a bacteriophage-RNA coat protein-operator complex. Nature. 1994;371:623–626. doi: 10.1038/371623a0. [DOI] [PubMed] [Google Scholar]
- 564.Valegard K, Murray J B, Stonehouse N J, van den Worm S, Stockley P G, Liljas L. The three-dimensional structures of two complexes between recombinant MS2 capsids and RNA operator fragments reveal sequence-specific protein-RNA interactions. J Mol Biol. 1997;270:724–738. doi: 10.1006/jmbi.1997.1144. [DOI] [PubMed] [Google Scholar]
- 565.Van den Heuvel J J, Bergkamp R J M, Planta R J, Raué H A. Effect of deletions in the 5′-noncoding region on the translational efficiency of phosphoglycerate kinase mRNA in yeast. Gene. 1989;79:83–95. doi: 10.1016/0378-1119(89)90094-2. [DOI] [PubMed] [Google Scholar]
- 566.Van den Heuvel J J, Lang V, Richeter G, Price N, Peacock L, Proud C, McCarthy J E G. The highly acidic C-terminal region of the yeast initiation factor subunit 2 (eIF-2) contains casein kinase phosphorylation sites and is essential for maintaining normal regulation of GCN4. Biochim Biophys Acta. 1995;1261:337–348. doi: 10.1016/0167-4781(95)00026-d. [DOI] [PubMed] [Google Scholar]
- 567.Van Horn D J, Yoo C J, Xue D H, Shi H, Wolin S L. The La protein in Schizosaccharomyces pombe: A conserved yet dispensable phosphoprotein that functions in tRNA maturation. RNA. 1997;3:1434–1443. [PMC free article] [PubMed] [Google Scholar]
- 568.Van Tilbeurgh H, Manival X, Aymerich S, Lhoste J M, Dumas C, Kochoyan M. Crystal structure of a new RNA-binding domain from the antiterminator protein SacY of Bacillus subtilis. EMBO J. 1997;16:5030–5036. doi: 10.1093/emboj/16.16.5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 569.Vasilescu S, Ptushkina M, Linz B, Müller P P, McCarthy J E G. Mutants of eukaryotic initiation factor eIF-4E with altered mRNA cap binding specificity reprogram mRNA selection by ribosomes in Saccharomyces cerevisiae. J Biol Chem. 1996;271:7030–7037. doi: 10.1074/jbc.271.12.7030. [DOI] [PubMed] [Google Scholar]
- 570.Vega Laso M R, Zhu D, Sagliocco F, Brown A J P, Tuite M F, McCarthy J E G. Inhibition of translational initiation in the yeast Saccharomyces cerevisiae as a function of the stability and position of hairpin structures in the mRNA leader. J Biol Chem. 1993;268:6453–6462. [PubMed] [Google Scholar]
- 571.Velculescu V E, Zhang L, Zhou W, Vogelstein J, Basrai M A, Bassett D E, Jr, Hieter P, Vogelstein B, Kinzler K W. Characterization of the yeast transcriptome. Cell. 1997;88:243–251. doi: 10.1016/s0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- 572.Vilela C, Linz B, Rodrigues-Pousada C, McCarthy J E G. The yeast transcription factor genes YAP1 and YAP2 are subject to differential control at the levels of both translation and mRNA stability. Nucleic Acids Res. 1998;26:1150–1159. doi: 10.1093/nar/26.5.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 573.Vilela, C., et al. Unpublished observations.
- 574.Vilardell J, Warner J R. Ribosomal protein L32 of Saccharomyces cerevisiae influences both the splicing of its own transcript and the processing of rRNA. Mol Cell Biol. 1997;17:1959–1965. doi: 10.1128/mcb.17.4.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 575.Vimaladithan A, Farabough P J. Special peptidyl-tRNA molecules promote translational frameshifting without slippage. Mol Cell Biol. 1994;14:8107–8116. doi: 10.1128/mcb.14.12.8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 576.Vreken P, Raué H A. The rate-limiting step in yeast PGK1 mRNA degradation is an endonucleolytic cleavage in the 3′-terminal part of the coding region. Mol Cell Biol. 1992;12:2986–2996. doi: 10.1128/mcb.12.7.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 576a.Vries R G J, Flynn A, Patel J C, Wang X, Denton R M, Proud C G. Heat shock increases the association of binding protein-1 with initiation factor 4E. J Biol Chem. 1998;272:32779–32784. doi: 10.1074/jbc.272.52.32779. [DOI] [PubMed] [Google Scholar]
- 576b.Wagner G, Simons R W. Antisense RNA control in bacteria, phages, and plasmids. Annu Rev Microbiol. 1994;48:713–742. doi: 10.1146/annurev.mi.48.100194.003433. [DOI] [PubMed] [Google Scholar]
- 577.Wang S, Browning K S, Miller W A. A viral sequence in the 3′-untranslated region mimics a 5′ cap in facilitating translation of uncapped mRNA. EMBO J. 1997;16:4107–4116. doi: 10.1093/emboj/16.13.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 578.Wang Z, Buratowski S, Svejstrup J Q, Feaver W J, Wu X, Kornberg R D, Donahue T F, Friedberg E C. The yeast TFB1 and SSL1 genes, which encode subunits of transcription factor IIH, are required for nucleotide excision repair and RNA polymerase II transcription. Mol Cell Biol. 1995;15:2288–2293. doi: 10.1128/mcb.15.4.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 579.Wang Z, Sachs M S. Ribosome stalling is responsible for arginine-specific translational attenuation in Neurospora crassa. Mol Cell Biol. 1997;17:4904–4913. doi: 10.1128/mcb.17.9.4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 580.Wei C-L, Kainuma M, Hershey J W B. Characterization of yeast translation initiation factor 1A and cloning of its essential gene. J Biol Chem. 1995;270:5764–5771. doi: 10.1074/jbc.270.39.22788. [DOI] [PubMed] [Google Scholar]
- 580a.Wei C C, Balasta M L, Ren J H, Goss D J. Wheat germ poly(A) binding protein enhances the binding affinity of eukaryotic initiation factor 4F and (iso)4F for cap analogues. Biochemistry. 1998;37:1910–1916. doi: 10.1021/bi9724570. [DOI] [PubMed] [Google Scholar]
- 581.Weighardt F, Biamonti G, Riva S. The roles of heterogeneous nuclear ribonucleoproteins (hnRNP) in RNA metabolism. Bioessays. 1996;18:747–756. doi: 10.1002/bies.950180910. [DOI] [PubMed] [Google Scholar]
- 582.Weiss R B. Ribosomal frameshift and frame-jump sites as control points during elongation. NATO ASI Ser. 1990;49:579–590. [Google Scholar]
- 583.Wek R C, Jackson B M, Hinnebusch A G. Juxtaposition of domains homologous to protein kinases and histidyl-tRNA synthases in GCN2 protein suggests a mechanism for coupling GCN4 expression to amino acid availability. Proc Natl Acad Sci USA. 1989;86:4579–4583. doi: 10.1073/pnas.86.12.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 584.Weng Y, Czaplinski K, Peltz S W. Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol Cell Biol. 1996;16:5477–5490. doi: 10.1128/mcb.16.10.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 585.Weng Y, Czaplinski K, Peltz S W. Identification and characterization of mutations in the UPF1 gene that affect nonsense suppression and the formation of the Upf protein complex but not mRNA turnover. Mol Cell Biol. 1996;16:5491–5506. doi: 10.1128/mcb.16.10.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 586.Weng Y, Czaplinski K, Peltz S W. ATP is a cofactor of the Upf1 protein that modulates its translation termination and RNA binding activities. RNA. 1998;4:205–214. [PMC free article] [PubMed] [Google Scholar]
- 587.Werner M, Feller A, Piérard A. Nucleotide sequence of yeast gene CPA1 encoding the small subunit of arginine-pathway carbamoyl-phosphate synthetase: homology of the deduced amino acid sequence to other glutamine amidotransferases. Eur J Biochem. 1985;146:371–381. doi: 10.1111/j.1432-1033.1985.tb08663.x. [DOI] [PubMed] [Google Scholar]
- 588.Werner M, Feller A, Messenguy F, Piérard A. The leader peptide of yeast gene CPA1 is essential for the translational repression of its expression. Cell. 1987;49:805–813. doi: 10.1016/0092-8674(87)90618-0. [DOI] [PubMed] [Google Scholar]
- 589.Wickens M, Kimble J, Strickland S. Translational control and developmental decisions. In: Hershey J W B, Matthews M B, Sonenberg N, editors. Cold Spring Harbor Laboratory Press. N.Y: Cold Spring Harbor; 1996. pp. 411–450. [Google Scholar]
- 590.Wickner R B. Double-stranded and single-stranded RNA viruses of Saccharomyces cerevisiae. Annu Rev Microbiol. 1992;46:347–375. doi: 10.1146/annurev.mi.46.100192.002023. [DOI] [PubMed] [Google Scholar]
- 591.Wickner R B. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 592.Wickner R B. Prions and RNA viruses of Saccharomyces cerevisiae. Annu Rev Genet. 1996;30:109–139. doi: 10.1146/annurev.genet.30.1.109. [DOI] [PubMed] [Google Scholar]
- 593.Widner W R, Wickner R B. Evidence that the SKI antiviral system of Saccharomyces cerevisiae acts by blocking expression of viral mRNA. Mol Cell Biol. 1993;13:4331–4341. doi: 10.1128/mcb.13.7.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 594.Wilhelm J E, Vale R D. RNA on the move: the mRNA localization pathway. J Cell Biol. 1993;123:269–274. doi: 10.1083/jcb.123.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 595.Wolfe K H, Shields D C. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 596.Wolffe A P, Meric F. Coupling transcription to translation: a novel site for the regulation of eukaryotic gene expression. Int J Biochem Cell Biol. 1996;28:247–257. doi: 10.1016/1357-2725(95)00141-7. [DOI] [PubMed] [Google Scholar]
- 597.Wolin S L, Walter R. Ribosome pausing and stacking during translation of a eukaryotic mRNA. EMBO J. 1988;7:3559–3569. doi: 10.1002/j.1460-2075.1988.tb03233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 598.Wright M B, Howell E A, Gabber R F. Amino-acid substitutions in membrane spanning domains of HOL1, a member of the major facilitator superfamily of transporters, confer nonselective cation uptake in Saccharomyces cerevisiae. J Bacteriol. 1996;178:7197–7205. doi: 10.1128/jb.178.24.7197-7205.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 599.Wu A, Wemmie J, Edginton N P, Goebl M, Guevara J L, Moye-Rowley W S. Yeast bZip proteins mediate pleiotropic drug and metal resistance. J Biol Chem. 1993;268:18850–18858. [PubMed] [Google Scholar]
- 600.Wulczyn F G, Kahmann R. Translational stimulation: RNA sequence and structural requirements for binding of com protein. Cell. 1991;65:259–269. doi: 10.1016/0092-8674(91)90160-z. [DOI] [PubMed] [Google Scholar]
- 601.Xing Y, Johnson C V, Dobner P R, Lawrence J B. Higher-level organization of individual gene transcription and RNA splicing. Science. 1993;259:1326–1330. doi: 10.1126/science.8446901. [DOI] [PubMed] [Google Scholar]
- 602.Xu F, Cohen S N. RNA degradation in Escherichia coli regulated by 3′ adenylation and 5′ phosphorylation. Nature. 1995;374:180–183. doi: 10.1038/374180a0. [DOI] [PubMed] [Google Scholar]
- 603.Xu H, Boeke J D. Host genes that influence transposition in yeast: the abundance of a rare tRNA regulates Ty1 transposition frequency. Proc Natl Acad Sci USA. 1990;87:8360–8364. doi: 10.1073/pnas.87.21.8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 604.Yamada T, Fukuda T, Tamura K, Furukawa S, Songsri P. Expression of the gene encoding a translational elongation factor 3 homolog of Chlorella virus CVK2. Virology. 1993;197:742–750. doi: 10.1006/viro.1993.1650. [DOI] [PubMed] [Google Scholar]
- 605.Yan R, Rychlik W, Etchinson D, Rhoads R E. Amino acid sequence of the human protein synthesis initiation factor eIF-4γ. J Biol Chem. 1992;267:23226–23231. [PubMed] [Google Scholar]
- 606.Yates J L, Dean D, Strycharz W A, Nomura M. E. coli ribosomal protein L10 inhibits translation of L10 and L7/L12 mRNAs by acting at a single site. Nature. 1981;294:190–192. doi: 10.1038/294190a0. [DOI] [PubMed] [Google Scholar]
- 607.Yoon H, Donahue T F. The sui1 suppressor locus in Saccharomyces cerevisiae encodes a translation factor that functions during tRNAiMet recognition of the start codon. Mol Cell Biol. 1992;12:248–260. doi: 10.1128/mcb.12.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 608.Yoon H, Donahue T F. Control of translation initiation in Saccharomyces cerevisiae. Mol Microbiol. 1992;6:1413–1419. doi: 10.1111/j.1365-2958.1992.tb00861.x. [DOI] [PubMed] [Google Scholar]
- 609.Yoon H, Miller S P, Pabich E K, Donahue T F. SSL1, a suppressor of a HIS4 5′-UTR stem-loop mutation, is essential for translation initiation and affects UV resistance in yeast. Genes Dev. 1992;6:2463–2477. doi: 10.1101/gad.6.12b.2463. [DOI] [PubMed] [Google Scholar]
- 610.Yueh A, Schneider R J. Selective translation initiation by ribosome jumping in adenovirus-infected and heat-shocked cells. Genes Dev. 1996;10:1557–1567. doi: 10.1101/gad.10.12.1557. [DOI] [PubMed] [Google Scholar]
- 611.Yun D-F, Sherman F. Initiation of translation can occur only in a restricted region of the CYC1 mRNA of Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:1021–1033. doi: 10.1128/mcb.15.2.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 612.Zachar Z, Kramer J, Mims I P, Bingham P M. Evidence for channelled diffusion of pre-mRNAs during nuclear RNA transport in metazoans. J Cell Biol. 1993;121:729–742. doi: 10.1083/jcb.121.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 613.Zanchin N I T, McCarthy J E G. Characterization of the in vivo phosphorylation sites of the mRNA-cap-binding complex proteins eukaryotic initiation factor-4E and p20 in Saccharomyces cerevisiae. J Biol Chem. 1995;270:26505–26510. doi: 10.1074/jbc.270.44.26505. [DOI] [PubMed] [Google Scholar]
- 614.Zeitlin S, Parent A, Silverstein S, Estratiadis A. Pre-mRNA splicing and the nuclear matrix. Mol Cell Biol. 1987;7:111–120. doi: 10.1128/mcb.7.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 615.Zeitlin S, Wilson R C, Efstratiadis A. Autonomous splicing and complementation of in vivo-assembled spliceosomes. J Cell Biol. 1989;108:765–777. doi: 10.1083/jcb.108.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 616.Zhang S, Ruiz-Echevarria M J, Quan Y, Peltz S W. Identification and characterization of a sequence motif involved in nonsense-mediated mRNA decay. Mol Cell Biol. 1995;15:2231–2244. doi: 10.1128/mcb.15.4.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 617.Zhong T, Arndt K T. The yeast SIS1 protein, a DnaJ homolog, is required for the initiation of translation. Cell. 1993;73:1175–1186. doi: 10.1016/0092-8674(93)90646-8. [DOI] [PubMed] [Google Scholar]
- 618.Zhouravleva G, Frolova L, Le Goff X, Le Guellex R, Inge-Vechtomov S G, Kisselev L, Philippe M. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 1995;14:4065–4972. doi: 10.1002/j.1460-2075.1995.tb00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 619.Ziegler E, Borman A M, Kirchweger R, Skern T, Kean K M. Foot-and-mouth disease virus Lb proteinase can stimulate rhinovirus and enterovirus IRES-driven translation and cleave several proteins of cellular and viral origin. J Virol. 1995;69:3465–3474. doi: 10.1128/jvi.69.6.3465-3474.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 620.Zuk D, Jacobson A. A single amino acid substitution in yeast eIF-5A results in mRNA stabilization. EMBO J. 1998;17:2914–2925. doi: 10.1093/emboj/17.10.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]