Abstract
Although homologous recombination and DNA repair phenomena in bacteria were initially extensively studied without regard to any relationship between the two, it is now appreciated that DNA repair and homologous recombination are related through DNA replication. In Escherichia coli, two-strand DNA damage, generated mostly during replication on a template DNA containing one-strand damage, is repaired by recombination with a homologous intact duplex, usually the sister chromosome. The two major types of two-strand DNA lesions are channeled into two distinct pathways of recombinational repair: daughter-strand gaps are closed by the RecF pathway, while disintegrated replication forks are reestablished by the RecBCD pathway. The phage λ recombination system is simpler in that its major reaction is to link two double-stranded DNA ends by using overlapping homologous sequences. The remarkable progress in understanding the mechanisms of recombinational repair in E. coli over the last decade is due to the in vitro characterization of the activities of individual recombination proteins. Putting our knowledge about recombinational repair in the broader context of DNA replication will guide future experimentation.
TWO-STRAND DNA DAMAGE, RECOMBINATIONAL REPAIR, SOS RESPONSE, AND DNA REPLICATION
Homologous recombination was described in Escherichia coli in the mid-1940s (351), and for many years it was thought to be the result of a sexual process, analogous to that found in eukaryotes. When the sensitivity to DNA damage of the first recombination-deficient mutants was noticed, it was realized that recombination in this bacterium may serve the needs of DNA repair as well (105, 107, 266, 267). Subsequently, genetic studies delineated two recombinational pathways—the primary, RecBC pathway, serving the needs of “sexual” recombination, and the secondary, RecF pathway, kicking in when the primary pathway is inactive and moonlighting at “postreplication repair” of daughter strand gaps (102, 106, 108). Still later, biochemical characterization of recombinational activities suggested that their primary role is in DNA repair (131, 132). Finally, the realization that disintegrated replication forks are reassembled by recombination justified the “repair” purpose for the RecBC pathway (130, 333) and prompted a revision of our ideas about the relationships of DNA replication and recombination.
The goal of this review is to consolidate genetic data on homologous recombination, physical data on DNA damage and repair, and biochemical data on recombinational enzymes under a different idea in an attempt to highlight new areas for the future in vitro and in vivo experiments. The different idea is that the primary role of the homologous recombination system in E. coli is to repair lesions associated with DNA replication of damaged template DNA (130, 336). Therefore, this review differs from other recent reviews on homologous recombination in E. coli (108, 320, 377) in that its two main emphases are on (i) the evidence for recombinational repair in bacteria and (ii) the interactions of various recombinational repair proteins with each other and with the replication machinery. The recombinational repair machinery is conserved among eubacteria, and so the same two basic pathways are present in such dissimilar species as E. coli and Bacillus subtilis. Therefore, although concentrating on the E. coli recombinational repair paradigm, occasionally I use evidence from other eubacteria.
Mechanisms of DNA Damage and Repair
Damage reversal and one-strand repair.
Bacterial genomic DNA, like any macromolecule, is subject to constant chemical and physical assault. Repair of the resulting lesions is essential if DNA is to serve as the template for transcription and its own reduplication. In the course of evolution, a complex enzymatic machinery has evolved to maintain this centrally important molecule in usable form (195). Repair of some DNA modifications simply reverses the damage, returning DNA directly to its original state. For instance, photolyase, using near UV-visible light, splits UV-induced pyrimidine dimers (reviewed in reference 545). Another example is the suicidal Ada protein of E. coli, which transfers a methyl group from the modified base O6-methylguanine to itself (reviewed in reference 580).
Repair of other types of lesions requires removal of a segment of the DNA strand around the lesion. The double-strandedness of DNA provides the means for repairing the resulting single-strand gaps: the removed bases can be resynthesized by using the intact complementary strand as a template. One example of such a strategy is the repair of modified bases that do not cause DNA distortion. The so-called base excision repair system acts with precision—an enzyme called DNA glycosylase removes a modified base to produce an abasic site, the phosphodiester bond at the 5′ side of the site is broken, and the repair is completed by a single-base nick translation by DNA polymerase (151) and sealing of the nick by DNA ligase. Another repair system, nucleotide excision repair, deals with DNA-distorting lesions. An excinuclease removes a 12- to 13-nucleotide segment of a single strand centered around the lesion, and the resulting gap is filled in by repair synthesis (reviewed in reference 544). The third repair system, methyl-directed mismatch repair, can liberate up to 1,000 nucleotides from one strand in its efforts to correct a single mismatch arising during DNA replication (reviewed in reference 440). A lesion affecting a single DNA strand is referred to in this review as one-strand lesion, and repair of such DNA damage is referred to as one-strand repair.
Two-strand repair.
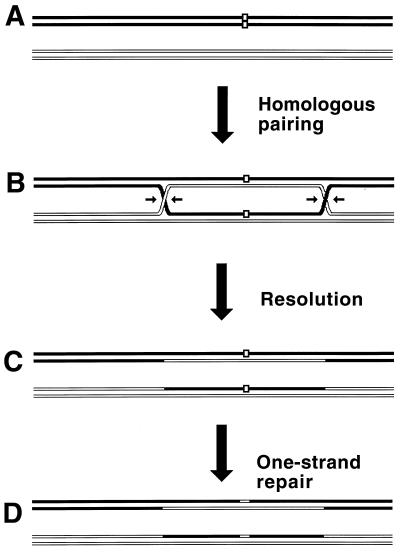
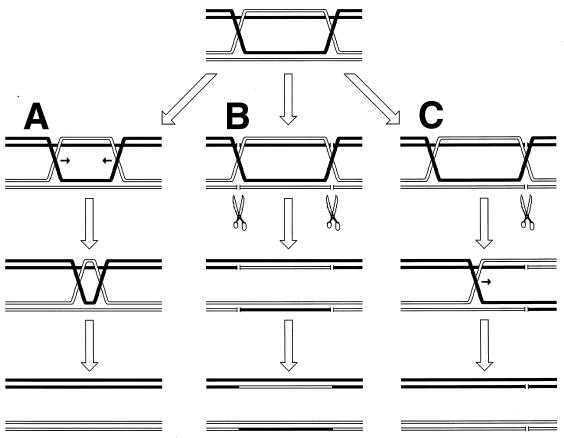
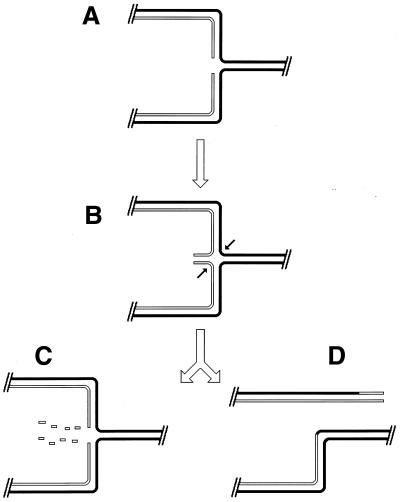
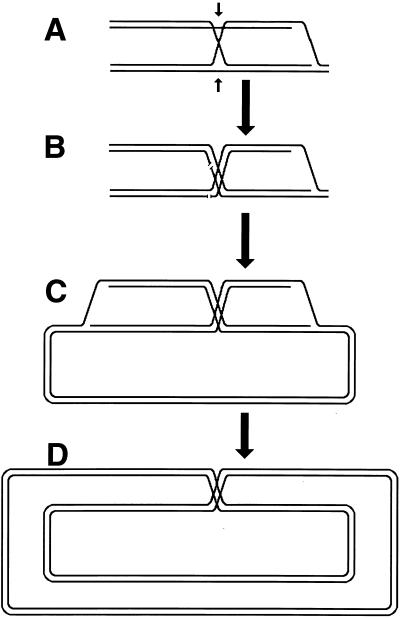
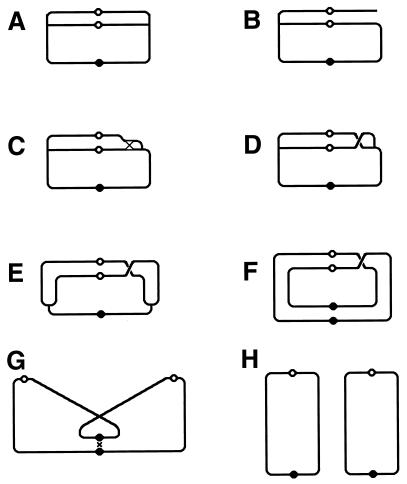
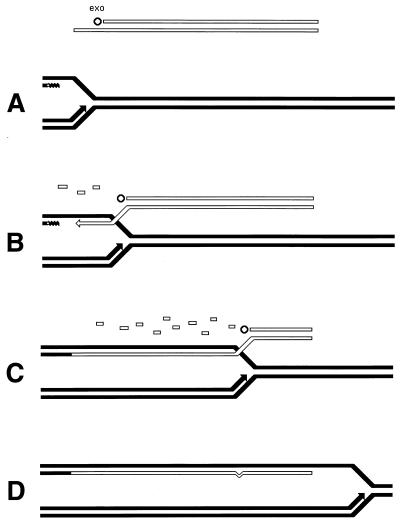
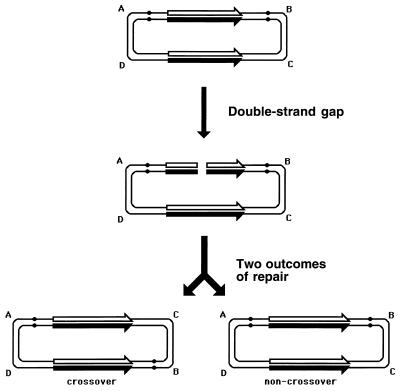
Although the bulk of DNA damage affects one strand of a duplex DNA segment, occasionally both DNA strands are damaged opposite each other, resulting in two-strand damage, a term proposed by Howard-Flanders (266). To repair two-strand damage without the loss of sequence information, a cell needs a higher level of redundancy, an extra homologous sequence whose strands could be used to fix both DNA strands of the damaged sequence. The principle of such two-strand repair is depicted in Fig. 1. An affected duplex homologously pairs and exchanges strands with an intact homologous duplex (Fig. 1B). The resulting joint molecule is “resolved” by symmetric single-strand cuts in homologous strands, yielding two new DNA molecules, each containing a single one-strand lesion (Fig. 1C). Now the damaged strands can be mended by one-strand repair with the complementary strands as templates (Fig. 1D).
FIG. 1.
The idea of two-strand repair. (A) A DNA molecule with a two-strand lesion (small open rectangles in the solid duplex) is shown side-by-side with an intact homolog (open duplex). (B) The two sequences have exchanged strands in a homologous region, converting the two-strand lesion into a pair of one-strand lesions. (C) Junction resolution (the strands to cut are shown in panel B) separates the chromosomes from each other. (D) Excision repair removes the one-strand lesions, completing the overall repair reaction. Note that if black and white “parental” DNAs are not identical, the resulting chromosomes may become “recombinant.”
Thus, the strategy of the two-strand repair is to convert a two-strand lesion into a pair of one-strand lesions by strand exchange with an intact homologous DNA sequence. Three common phases of the two-strand repair are evident from this scheme. The central phase, during which a damaged DNA sequence trades strands with an intact homologous sequence to form a joint molecule, is called synapsis. In E. coli and other eubacteria, this phase is catalyzed by RecA protein. Accordingly, the preparatory phase preceding the synapsis is called presynapsis, while the resolution of joint molecules is referred to as postsynapsis (103). The four-strand junctions holding the joint molecules together are usually referred to as Holliday junctions, after Holliday, who recognized their importance in one of the early models of homologous recombination (256).
Homologous recombination versus recombinational repair.
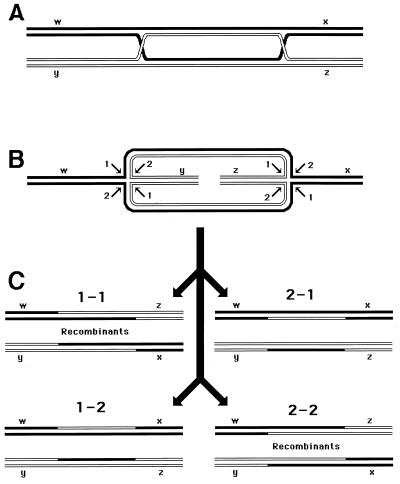
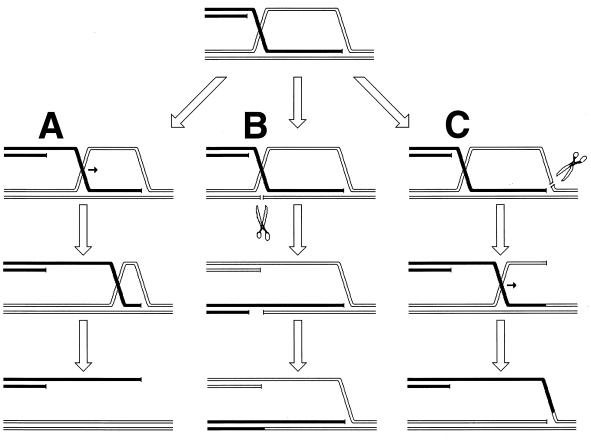
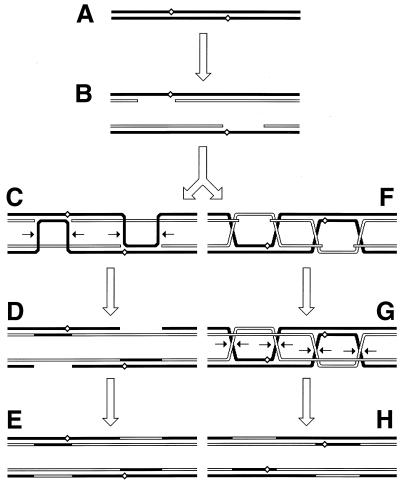
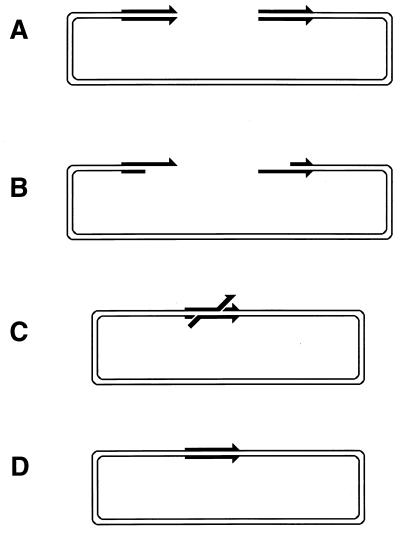
Since the machinery for the two-strand repair is complex and not copious and since the repair incidents are rather infrequent, this type of repair is more accessible to genetic than to biochemical study. The principal genetic assay for two-strand repair is to monitor the formation of new chromosomes resulting from alternative resolution of joint molecules. A joint molecule (Fig. 2A) can be redrawn to show that the DNA junctions are able to isomerize (Fig. 2B). This isomerization of the junctions creates two possible ways of resolving each junction (shown by numbers beside the arrows [Fig. 2B]). If the resolution is random, in 50% of the cases the participating chromosomes will exchange shoulders, forming two “recombinant” chromosomes (Fig. 2C). If the parental chromosomes were genetically marked, progeny carrying recombinant chromosomes would be detected genetically as having traits that initially resided on separate parental chromosomes.
FIG. 2.
The four ways to resolve a joint molecule with a double junction. Lowercase letters w, x, y, and z designate unique sites which serve as markers on the homologous chromosomes. A junction is resolved by two symmetrical single-strand cuts (small black arrows in panel B) across each other. Each such diagonal pair of cuts is numbered either 1 or 2 for each junction. If junctions freely isomerize and are resolved independently of each other, four outcomes of the resolution are expected. In two of the outcomes, the chromosome arms will be exchanged, resulting in recombinant chromosomes. (A) A joint molecule with two junctions as shown in Fig. 1B. (B) The same joint molecule isomerized to show both junctions in the open planar configuration (498). (C) The four resolution outcomes, numbered according to the resolution options realized at the left and the right junctions.
Because of this association of the two-strand repair with homologous recombination, the former is better known as recombinational repair. The availability of the homologous recombination assay was a mixed blessing for the development of recombinational-repair concept. On the one hand, most recombinational-repair mutants of E. coli were isolated because of their deficiencies in homologous recombination. On the other hand, since genetic recombination has important evolutionary consequences (181, 418, 736), the recombinational-repair system of E. coli was for a long time viewed from the perspective of its long-term evolutionary value rather than its short-term repair value.
The typical “repair” features of the recombinational-repair system in E. coli are sometimes used as an argument that it could not have evolved due to its role in genetic exchange (131, 132). However, the recombination system of E. coli might have arisen purely for repair purposes but eventually have been integrated into the evolutionary tools of the long-term survival system. Therefore, the strongest argument in favor of the repair role of homologous recombination and against its evolutionary role should come from comparison of the selective values of repair and genetic exchange for the long-term survival of bacteria. Since, due to their decreased viability (see “Frequency of two-strand lesions” below) and high sensitivity to DNA-damaging agents, recombination-deficient mutants are unlikely to survive outside the laboratory, repair must have a high selective value. In contrast, the role of homologous recombination in the E. coli evolution is obscure, since the E. coli genome in nature evolves as a collection of clonal lineages with little recombinational cross talk among the clones and little proven selective value for such horizontal transfer (419, 436). Thus, for the short-term survival, the system of homologous recombination in E. coli has a higher evolutionary value as a DNA repair mechanism than as a mechanism for creating new allelic combinations. This does not mean that the “exchange” consequences of homologous recombination are unimportant for the long-term survival of E. coli; it only means that their selection coefficient is smaller.
Formation of recombinant chromosomes must be a direct consequence of DNA damage repair, because (i) DNA damage greatly stimulates homologous recombination (111, 360, 451, 529) and (ii) the genetic requirements of this damage-stimulated recombination are the same as those of the “spontaneous” recombination. Still, it is possible that this stimulation of homologous recombination in E. coli by DNA damage occurs because DNA damage makes cells “hyper-rec” towards all DNA molecules rather than only towards the damaged ones. However, damage on one DNA stimulates its recombination only with homologous DNA, arguing against the idea of nonspecific activation of recombination by DNA damage (530). Even when damage on one DNA molecule stimulates recombination between sequences absent from the damaged molecule which are situated on other, intact molecules, such “teleactivation” is observed only when the damaged molecule carries homology to the recombining molecules (213). This strict homology requirement for the recombination activation by DNA damage also suggests that repair of certain DNA lesions requires interactions with homologous chromosomes.
The two mechanisms of two-strand damage.
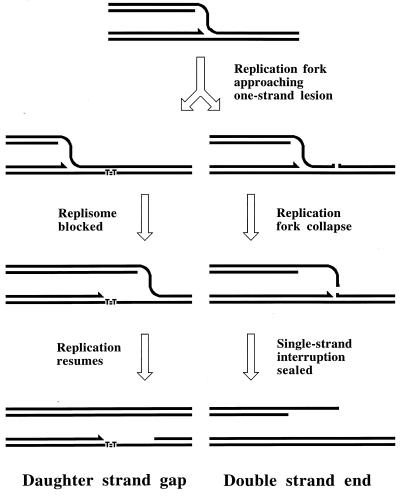
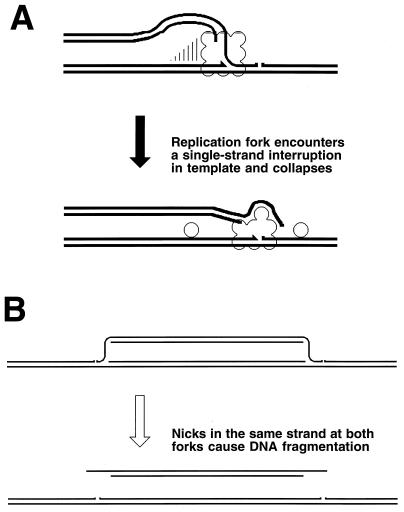
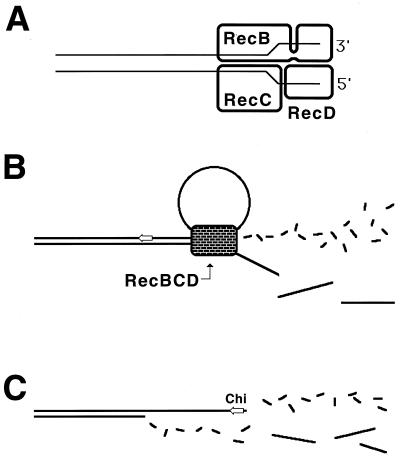
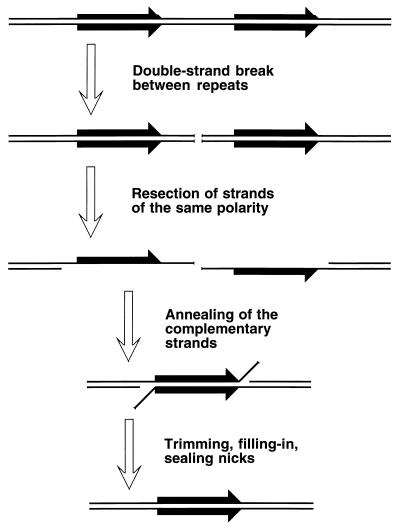
Two-strand lesions appear in DNA in two distinct ways. DNA synthesis in a region increases recombination in this region (447), suggesting that one source of two-strand lesions is DNA replication. The fact that replication of DNA containing one-strand lesions stimulates recombination between this DNA and an intact homolog (360) suggests that DNA replication causes two-strand lesions when it runs into unrepaired one-strand lesions. There are at least two mechanisms of replication-dependent conversion of one-strand damage into two-strand damage. In vivo, a noncoding lesion (for example, an abasic site) is an absolute block to DNA replication in growing cells (347); similarly, in vitro, a noncoding lesion in template DNA blocks the progress of the major E. coli DNA polymerases (59). In the chromosome, replication is likely to reinitiate downstream of a noncoding lesion (see “Elongation phase of DNA replication in E. coli” below), leaving behind an unfillable single-strand gap (Fig. 3) (see “Origin of daughter strand gaps and mechanism of their repair: early studies” below). Such an unfillable gap is called a daughter strand gap, since it appears in one of the two daughter branches after the replication fork passage (538, 734). Another type of one-strand lesion, a single-stranded interruption in template DNA, is proposed to cause a disintegration (collapse) of a replication fork (see “Evidence for replication fork disintegration” below) (234, 597). As a result, a double-strand end is detached from the full-length duplex molecule (Fig. 3). Finally, inhibited replication forks are broken, similarly releasing one of the replicating branches as a free double-stranded end (263, 334).
FIG. 3.
The two major types of replication-induced two-strand lesions. A replication fork moves from left to right along the template DNA with unrepaired one-strand lesions. The left template contains a noncoding lesion (T=T, thymine dimer), the right template has a single-strand interruption. Additional explanations are given in the figure and in the text.
The other principal source of two-strand DNA damage is direct induction. Ionizing radiation (X rays and gamma rays), when passing through a solution, generates free radicals, which damage and break molecules in their immediate vicinity. The energy deposition by gamma radiation allows the formation of clusters of several radicals, so that a big molecule near such clusters can suffer multiple instances of damage (717). Besides chemically modified bases and interruptions in one DNA strand, ionizing radiation also causes double-strand breaks (48, 219). On the average, for every 20 single-strand breaks induced by X rays in DNA, there is one double-strand break (reviewed in reference 324). Another direct two-strand lesion, a cross-link, is observed in DNA treated with psoralen plus UV-light or with mitomycin C (101, 671). In summary, two-strand damage is induced in DNA either directly or as a result of DNA replication on a template DNA containing one-strand damage.
The two recombinational repair pathways of E. coli.
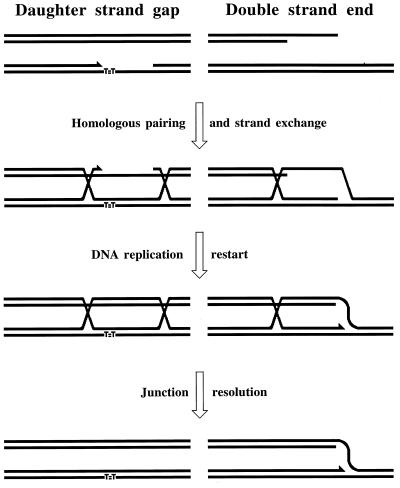
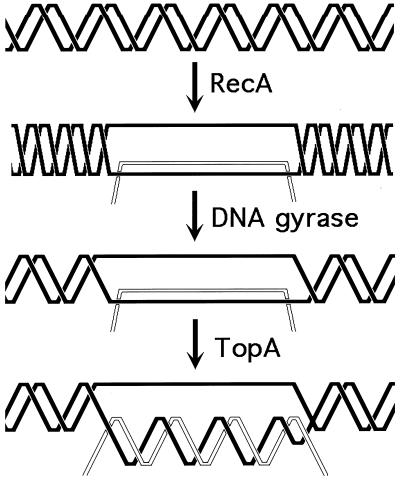
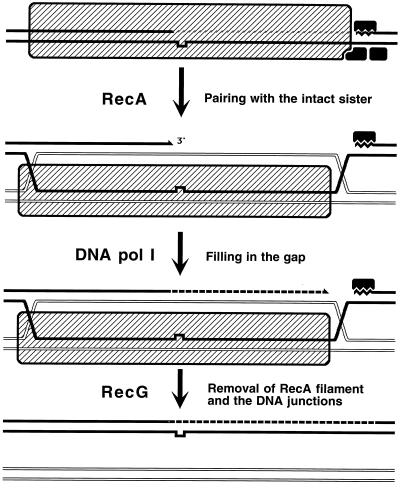
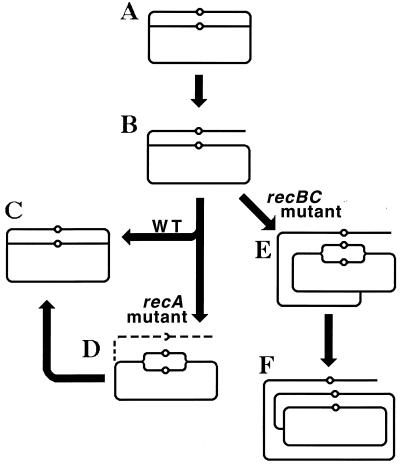
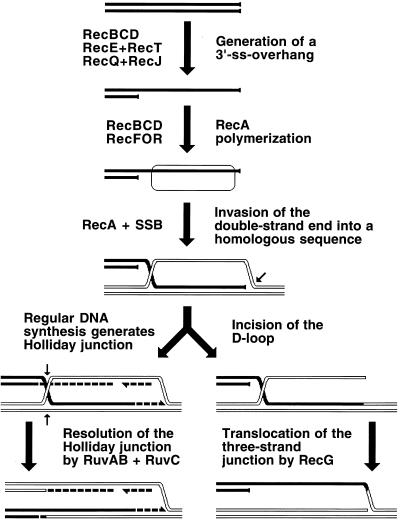
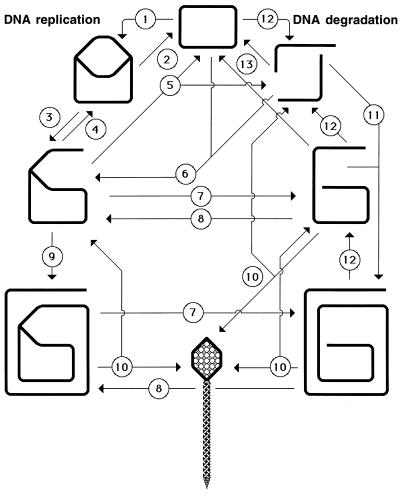
The two types of replication-induced two-strand lesions are repaired in E. coli by two separate pathways, both dependent on the recA gene but named after the critical genes that distinguish between them (Fig. 4). Daughter strand gaps are repaired by the RecF pathway (see “Repair of daughter strand gaps” below), while disintegrated replication forks are repaired by the RecBC pathway (see Double-strand and repair” below). The three common phases (see “Two-strand repair” above) of the two repair reactions are (Fig. 4) (i) presynapsis, during which the damaged DNA is prepared for homology search, followed closely by synapsis, during which homologous pairing and strand exchange with the intact sister duplex occur; (ii) DNA replication restart; and (iii) postsynapsis, during which the recombination intermediates are resolved.
FIG. 4.
The two pathways of recombinational repair in E. coli. On the left, the RecF (daughter strand gap repair) pathway is shown; on the right, the RecBC (double-strand end repair) pathway is shown. Additional explanations are given in the text.
Direct two-strand lesions are repaired by the same two pathways. Double-strand breaks are fixed by the RecBC pathway (555); most probably, they are treated as pairs of independent double-strand ends (see “Overview of double-strand end repair” below). Cross-links are repaired by the combined effort of both the RecBC and the RecF pathways (595), since a fraction of them are apparently converted to double-strand breaks while the rest are converted to unfillable single-strand gaps.
The two recombinational repair pathways are equally important for the repair of DNA damage during normal growth of enteric bacteria, since (i) both the recBC and recF null mutants reduce the viability of E. coli to approximately the same degree (85, 86, 547), and (ii) in Salmonella typhimurium, recombinational repair in the chromosome, detected as a deletion formation between long repeats, is not blocked by single recB or recF mutations but is prevented in a recB recF double mutant (198); physically detected sister chromatid exchange in the E. coli chromosome depends on both the recB and recF genes (630).
Frequency of two-strand lesions.
Under conditions of laboratory growth, two-strand lesions are too infrequent to be detectable in wild-type (WT) cells directly by physical techniques, although they are detectable in recombinational repair mutants (434). After massive DNA damage, daughter strand gaps are detected as single-stranded regions of several hundreds of nucleotides in the chromosomal DNA (278, 710) or as interruptions in the newly synthesized DNA (538, 714), double-strand breaks are detected as an immediate chromosome fragmentation (61, 323, 683), and disintegrated replication forks are detected as replication-induced chromosome fragmentation (60, 715).
A more sensitive although less precise indication of the frequency of two-strand lesions during normal growth is the viability of various recombinational repair mutants. Under laboratory conditions, mutants defective at the presynaptic and synaptic phases of recombinational repair (see “Two-strand repair” above) have 25 to 50% viability (85, 86, 547) while those blocked at the postsynaptic phase are 25% viable (370). These approximate values suggest that under laboratory conditions, E. coli experiences two-strand lesions in almost every generation. The importance of this seemingly rare occurrence is raised by the following considerations: (i) a single unrepaired two-strand lesion is a “kiss of death” for the chromosome (268, 595), and (ii) judging by the significant capacity of the E. coli cells to undergo recombinational repair, E. coli cells occasionally experience massive two-strand DNA damage in the wild (see “SOS response: reaction of E. coli to DNA damage” below).
Recombinational repair capacity of E. coli cells.
WT E. coli cells grown in a nutritionally poor medium are able to survive 53 to 71 cross-links per chromosome (595). It can be calculated on the basis of the data with excision repair-deficient strains (714) that E. coli cells are still viable after repairing 100 to 200 daughter strand gaps per chromosome. E. coli cells should also be able to tolerate multiple disintegration of replication forks, because recombinational repair should reattach the resulting double-stranded ends to the circular domains of the chromosome. The only two-strand DNA lesion that has proved to be deadly for E. coli is a double-strand break. E. coli survives only two or three double-strand breaks in its chromosome (325, 683), which suggests that whenever a double-strand break occurs in an unreplicated portion of the chromosome, it cannot be repaired. Whether E. coli is an exception among bacteria in its inability to repair multiple double-strand breaks remains to be determined. There is a eubacterium, Deinococcus radiodurans, which can repair >100 double-strand breaks per chromosome (437), but this extreme resistance to DNA damage stands out in the bacterial world.
SOS Response: Reaction of E. coli to DNA Damage
When growing in the laboratory an average E. coli cell may experience two-strand damage once or twice (see “Frequency of two-strand lesions” above). However, its capacity to repair this damage is many times this value (see “Recombinational repair capacity of E. coli cells” above), suggesting that in nature, E. coli may suffer massive DNA damage.
The two main E. coli reservoirs in nature are (i) the animal gut, where the microbe is dividing and concentrated; and (ii) the natural water of lakes and ponds, where the microbe is starving and diluted (560). In the gut, that is, in the environment rich in nutrients and protected from the elements, E. coli is likely to replicate its DNA for many generations without much need to repair it. However, when E. coli finds itself in the water, where DNA replication stops and DNA repair is anemic while the possibilities for damage of DNA are significant, the E. coli genome must accumulate a tremendous amount of DNA damage. Unfortunately, the gut is a discontinuous niche, since the animal the gut belongs to will eventually die; therefore, to survive in the long run, E. coli has to exit the old gut and recolonize a young one. When the battered E. coli from the water eventually makes it to a new gut and starts replicating, it finds its DNA riddled with unrepaired lesions.
The sporadic occurrence of massive DNA damage separated by long periods of undisturbed growth calls for a modest standby repair system, capable of rapid induction in response to increased DNA repair needs. Such an arrangement is indeed found in E. coli; the rapid increase in its DNA repair capacity is called the SOS response (506).
Repair instead of DNA damage checkpoints: the prokaryotic strategy.
The bulk of two-strand DNA lesions in enterobacteria are probably generated as a result of DNA replication on template DNA containing one-strand lesions. An easy way to prevent this aggravation would be to stop DNA synthesis altogether when one-strand lesions are sensed. This is exactly what eukaryotic cells do—they employ checkpoint mechanisms to delay chromosomal replication when their DNA is damaged (reviewed in references 295 and 401). Since prokaryotes would also benefit from such a strategy, it was argued that E. coli might have a system to delay DNA synthesis when its chromosome is damaged (73).
However, several observations contradict this attractive idea. The initial inhibition of the DNA synthesis rate in E. coli is dose dependent, so that even after irradiation with almost lethal UV doses, when one would expect a complete replication stop if a checkpoint mechanism operated, the rate of DNA replication is still 20 to 50% of the maximal rate (162, 300). Furthermore, no E. coli mutation has been isolated that would prevent the inhibition of chromosomal replication by DNA damage (as rad9 mutations in yeast or p53 mutations in mammalian cells do). Not surprisingly, preventing the initiation of DNA synthesis with chloramphenicol during irradiation and for a couple of hours thereafter significantly improves the survival of E. coli, especially of recombinational repair-deficient mutants (47, 207, 234, 496, 607). If cells restarted DNA replication only when a “safe” level of DNA damage was attained as a result of repair, there would have been no effect of this drug-mediated inhibition of DNA synthesis on cell survival. Finally, the idea that E. coli has a mechanism to inhibit replication of damaged DNA is incompatible with the observations that E. coli initiates extra rounds of DNA replication from the origin when its DNA is heavily damaged (46, 300, 501). All these phenomena seem to indicate that, in contrast to eukaryotes, E. coli lacks a mechanism to stop chromosomal replication when its DNA is damaged and instead relies on enhanced repair and damage tolerance in its attempt to faithfully replicate the damaged genome.
E. coli and other eubacteria may have evolved such a minimalistic strategy because DNA replication is often the limiting step in their cell cycle (68). Eukaryotes can easily afford a replication delay, since their S phase is only a fraction of their overall cell cycle. In contrast, rapidly dividing E. coli cells have to race against time, since their chromosomal replication may take 1.5 times as long as their cell cycle (see “Cellular processes that surround and complicate recombinational repair” below).
Organization of the SOS regulon.
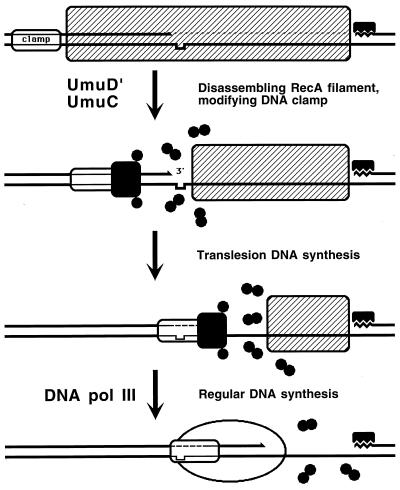
DNA lesions inhibit DNA replication. Inhibition of DNA replication in E. coli induces the SOS response: an increased expression of some 20 genes aimed at restoring the capacity of the chromosome to replicate (Table 1). The resulting enhancement of the ability of the cell to repair and tolerate DNA damage is achieved in several independent ways. The capacity of the cell for excision repair (see “Damage reversal and one-strand repair” above) is enhanced by overproduction of the UvrD helicase and the UvrA and UvrC subunits of the UvrABC excinuclease. Induced amounts of DNA polymerase II increase the capacity of the cell for DNA synthesis across abasic sites (58, 484, 660). Up to a 50-fold increase in the amount of RecA protein (292, 543) and a similar increase in the expression of RecN protein (494) enhance the recombinational repair. The SOS induction makes possible repeated disengagement of replisomes stalled at the lesions in template DNA to allow resumption of the synthesis downstream, a phenomenon known as replisome reactivation (see “Replisome reactivation and model for RecFOR catalysis of RecA polymerization at daughter strand gaps” below). When recombinational repair cannot fix certain DNA lesions, the UmuD′C complex catalyzes translesion DNA synthesis (see “Backup repair of daughter strand gaps: translesion DNA synthesis” below). Overproduction of SfiA protein inhibits cell division (41), providing extra time for completion of recombinational repair. If all these measures fail to restore DNA replication, the lingering SOS induction awakens colicinogenic plasmids and dormant prophages, whose expression lyses the cell. The lysis of doomed cells benefits the viable cells of the same clone when resources are limited, since inviable bacterial cells can multiply for several generations, wasting precious nutrients. The lysis by induction of a prophage or colicinogenic plasmid is therefore an example of “bacterial apoptosis,” which could have evolved to increase the number of viable cells in a clone.
TABLE 1.
E. coli proteins with known functions induced during the SOS response
| Gene | Gene product/function | No. of copies/cella
|
Increase in expressionb | Strength of SOS boxc/LexA affinityd | |
|---|---|---|---|---|---|
| Basal level | SOS-induced cells | ||||
| Expressed first | |||||
| lexA | LexA/SOS repressor | 1,300 | 1e | 5.8 | 6.4 and 8.3/15 |
| uvrA | UvrABC excinuclease/excision repair | 20 | 250 | 4.8 | 7.0/14.6 |
| uvrB | UvrABC excinuclease/excision repair | 250 | 1,000 | 3.7 | 6.1/8.8 |
| uvrD | Helicase II/excision repair, fidelity of recombinational repair | 5,000–8,000 | 25,000–65,000 | 5.9 | 8.8/17.9 |
| polB | DNA polymerase II/translesion DNA synthesis | 40 | 300 | 7.3 | 12.1/?f |
| ruvA | Subunit of RuvAB helicase/recombinational repair | 700 | 5,600 | 2–3 | 9.2/? |
| ruvB | Subunit of RuvAB helicase/recombinational repair | 200 | 1,600 | See ruvA | See ruvA |
| dinI | Inhibition of UmuD processing | <500 | 2,300 | ? | ? |
| Expressed second | |||||
| recA | RecA coprotease, synaptase/SOS derepressor, recombinational repair | 1,000–10,000 | 100,000 | 12.0 | 4.3/3.8 |
| recN | RecN/recombinational repair | ? | ? | 10 | 4.2 and 9.4/? |
| Expressed last | |||||
| sfiA | SfiA (SulA)/cell division inhibitor | ? | ? | 125 | 4.7/1 |
| umuD | Subunit of UmuD′C/translesion DNA synthesis | 180 | 2,400 | 22.5 | 2.8/1.1 |
| umuC | Subunit of UmuD′C/translesion DNA synthesis | 0 | 200 | ? | See umuD |
| Apoptosis | |||||
| cea | Colicin E1 | ? | ? | ? | 7.6 and 11.6/? |
| caa | Colicin A | ? | ? | ? | 9.6 and 11.5/? |
Sources for the protein copy number and its SOS increase: LexA, 558; UvrA and UvrB, 693; UvrD, 305 and 330; PolB, 58 and 503; RuvAB, 590, Benson and West (unpublished), cited in reference 721; DinI, 753; RecA, 292 and 543; RecN, 494; UmuD and UmuC, 742.
Increase in expression is given as the ratio of the gene expression without LexA repression to the gene expression with full repression, both measured as β-galactosidase activity. Except for ruvA and recN, the values are averages of two measurements done at 30°C and 42°C (563).
The strength of the SOS box in the promoter region of a gene is represented by the heterology index. Higher values reflect more deviations from the SOS box consensus and hence weaker LexA binding. The data are from reference 356.
LexA affinity is expressed as relative LexA affinity in vitro compared to the affinity to the sfiA operator. Higher values mean weaker LexA binding, while lower values mean stronger LexA binding. The data are from reference 563.
LexA protein is degraded during the SOS induction.
?, not known.
During the undisturbed growth, induced expression of the SOS genes is prevented by the LexA repressor. LexA dimer binds to a palindromic sequence, the SOS box, in the promoter regions of the SOS genes, precluding initiation of transcription. The SOS box has an inverted repeat consensus 5′-TACTGTATATATATACAGTA-3′, where the positions in bold are absolutely conserved (356). The substantial uninduced levels of certain SOS gene products (Table 1) are maintained due to imperfect SOS boxes in their operator regions or due to alternative promoters. Tight regulation of the genes with low-affinity SOS boxes is achieved by the high intracellular concentration of the LexA repressor (more than 1,000 molecules per cell) (558) and by the presence of two SOS boxes in the operators of lexA, recN and colicin genes (563).
The increased expression of the SOS genes in response to DNA synthesis inhibition is a result of inactivation of LexA repressor. The inactivation of LexA repressor is by autocleavage catalyzed by a recombinationally active form of RecA (see “Cleavage of LexA repressor by RecA filament” below). Only LexA molecules that are free in solution can be inactivated (367), which accounts for the late induction of the SOS genes with high-affinity SOS boxes. The two major types of two-strand DNA lesions (Fig. 3) induce the SOS response along the corresponding repair pathways (see “The two recombinational repair pathways of E. coli” above) (426).
Levels of SOS induction.
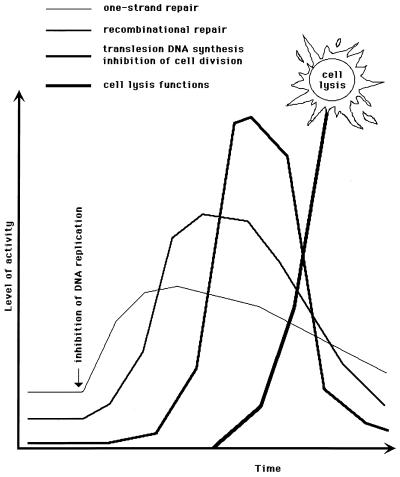
The SOS response is by no means a desperate attempt to stay alive, as its name inaccurately implies (506), but, rather, an orderly and measured reaction of the cell to DNA synthesis inhibition. General information on the E. coli genes induced during the SOS response is summarized in Table 1. The strength of SOS boxes in the operator regions of the SOS genes correlates well with the in vitro LexA repressor affinities for the corresponding promoters and is likely to determine the timing of expression of a given gene during the SOS induction. According to thus inferred order of expression during the SOS induction, the genes of the SOS regulon could be loosely grouped into three categories. The first genes to be induced are mostly those responsible for one-strand repair (uvrA, uvrB, and uvrD) or damage tolerance (polB) (Fig. 5). The LexA repressor itself is also induced immediately. The DinI gene product, which delays activation of translesion DNA synthesis (753), is likely to be synthesized at this stage, too. Increase in expression of the immediately induced genes is usually less than 10 times that of their constitutive expression. If the increased expression of the one-strand repair genes does not help to regain normal rates of DNA synthesis, the genes of recombinational repair, recA and recN, are induced (Fig. 5). The maximal induction of these genes is higher, 20- to 50-fold over their regular levels. When DNA damage is massive, so that even the enhanced recombinational repair cannot overcome the inhibition of DNA replication, the third group of genes, represented by sfiA and umuDC, is called into action. Since these genes are expressed at very low levels during regular DNA synthesis, their SOS induction could be more than 100-fold. Expression of the umuDC operon inhibits recombinational repair and makes possible translesion DNA synthesis (see “Backup repair of daughter strand gaps: translesion DNA synthesis” below), while SfiA protein delays cell division. As DNA replication rates return to normal, the three expression groups of the SOS genes are likely to become repressed in the reverse order (Fig. 5). Alternatively, if a cell cannot repair its DNA damage and is doomed to generate a dead lineage, prophages and colicin plasmids are induced to lyse it (Fig. 5).
FIG. 5.
An idealized induction kinetics of the four groups of LexA-controlled genes (also, see reference 611). The graph illustrates the well-regulated nature of the SOS response. Both the x axis (time) and y axis (the level of the SOS induction) are in arbitrary units; therefore, the heights of the three curves are not to be compared.
Cellular Processes That Surround and Complicate Recombinational Repair
The poor capacity of E. coli to repair double-strand breaks (see “Recombinational repair capacity of E. coli cells” above) suggests that this type of two-strand DNA damage is unusual in this organism. If one excludes double-strand breaks and DNA cross-links, the remaining two-strand lesions (daughter strand gaps and disintegrated replication forks) are the result of DNA replication on a damaged template DNA. In other words, recombinational repair acts to carry DNA replication through the template DNA containing unrepaired one-strand lesions. From this perspective, recombinational repair is surrounded by DNA replication: it starts when DNA replication stalls, and when it is finished, DNA replication resumes. Therefore, no discussion of recombinational repair is complete without a discussion of the DNA replication mechanisms.
The entire 4.7-Mbp circular chromosome of E. coli is traversed by a single replication bubble emanating from the unique replication origin. Both replication forks of the replication bubble are active; they meet in a chromosome region called the terminus, which is situated across from the origin. The terminus is delineated by termination sites arranged so as to form a replication fork trap—replication forks can enter the terminus, but they cannot exit it (253). To replicate the whole chromosome within a less-than-1-h bacterial cell cycle, replication forks have to proceed at about 650 bp/s (68). However, even the higher speed of almost 800 bp/s is insufficient when the cell cycle of E. coli is squeezed into 24 min in a rich medium. To prevent underreplication, E. coli starts a new round of DNA replication well before the completion of the ongoing round. Thus, in cells growing in a rich medium, there are one to three replication bubbles (two to six replication forks) (244).
Conceptually, replication of the E. coli chromosome is subdivided into three major phases: initiation, elongation, and termination (reviewed in references 25 and 406). Termination is the least understood phase (253) and is not immediately relevant to the needs of recombinational repair, although inhibition of DNA replication associated with termination sometimes causes disintegration of replication forks with their subsequent recombinational repair (263, 573). Elongation is the phase at which the two-strand lesion formation occurs and the recombinational repair machinery meets the replication machinery. Initiation of chromosomal DNA replication is helpful in defining interactions of the key replication proteins. An initiation strategy of multicopy plasmids is relevant because it utilizes the host reinitiation mechanism after the completion of recombinational repair.
Initiation of chromosomal DNA replication in E. coli.
For a replication fork to start, the DNA duplex must be open. At the origin of chromosomal DNA replication, this opening is effected by binding of the initiator protein, DnaA. DnaA recognizes and binds to a degenerate nonanucleotide (T/C)(T/C)(A/T/C)T(A/C)C(A/G)(A/C/T)(A/C) (562). At the origin of chromosomal DNA replication, four DnaA recognition sites are found in a cluster. Binding of 10 to 20 DnaA monomers to this cluster of DnaA binding sites leads to an opening of DNA duplex nearby.
In vivo, single-stranded DNA (ssDNA) is immediately complexed with ssDNA-binding protein (SSB), which precludes the binding of many other proteins to this ssDNA. To load DNA replication machinery onto SSB-complexed ssDNA, help from other proteins bound to neighboring duplex regions is needed. In the E. coli chromosomal origin, DnaA itself, still sitting on the adjacent duplex region, assists in this loading.
Since DNA polymerases cannot start DNA synthesis without primers, the 10-nucleotide riboprimers are laid by a special RNA polymerase called primase (DnaG protein). DnaG primase works in a complex with a DNA helicase (encoded by dnaB) that drives DNA unwinding at the replication fork. The complex of DnaG and DnaB proteins is called a mobile primosome; it propagates along the ssDNA in the 5′-to-3′ direction, laying primers every 1.5 to 2.0 kb. The mechanics of primosome assembly at the origin of chromosomal DNA replication is as follows. In solution, DnaB protein is always complexed with its inhibitor, DnaC protein. DnaC delivers DnaB helicase to ssDNA if DnaA protein is bound nearby. When DnaB helicase is loaded onto ssDNA and is associated with dnaG primase, the replicative primosome is formed.
The final stage of the replication fork formation is the association of a multisubunit DNA polymerase III (pol III) with the nascent replication bubble. First, a DnaN protein dimer is clamped around a primed segment of DNA to form a ring that slides along the RNA-DNA hybrid or duplex DNA. The DnaN clamp is called the processivity subunit of DNA pol III, since it ensures that DNA polymerase stays bound to DNA during polymerization. DNA synthesis begins when DNA polymerase holoenzyme is loaded onto the DnaN clamp at the primer. There are up to 300 DnaN monomers per cell (79), some 10-fold excess of DnaN dimers over DNA pol III holoenzyme, which is present at 10 to 20 copies per cell (747).
Elongation phase of DNA replication in E. coli.
In an established replication fork, DnaB helicase (maybe with the help of auxiliary helicases like Rep and UvrD) unwinds parental duplex DNA while the associated DnaG primase lays primers for both the leading and the lagging strands. DnaN clamps are formed around the primed DNA segments, while the single-stranded regions between primers are complexed with SSB. When the stretch of DNA between the two adjacent primers is duplicated (SSB is apparently displaced), DNA pol III is transferred from its current DnaN ring onto a new DnaN ring, awaiting on the next primer (this explains the requirement for the excess of DnaN subunit over the holoenzyme). The two adjacent newly synthesized DNA stretches, called Okazaki fragments, are separated by a single-strand interruption between the 3′ side of one of the fragments and the RNA primer, attached to the 5′ side of the other fragment. A one-subunit repair DNA polymerase (DNA pol I) starts DNA synthesis from the 3′ side of the interruption, simultaneously degrading the downstream RNA primer with its unique 5′-to-3′ exonuclease activity. After the complete removal of the RNA primer, the single-strand interruption is sealed by DNA ligase.
This description corresponds to the mechanism of the lagging-strand DNA synthesis elucidated in vitro. In the reconstituted in vitro systems of the E. coli DNA replication, the lagging-strand synthesis is discontinuous, requiring periodic reloading of DNA pol III, while the leading-strand synthesis is continuous, so that DNA pol III is loaded only once and then is able to replicate megabases of DNA before dissociation (406). It is said that in vitro the processivity of the leading-strand DNA synthesis is greater than that of the lagging-strand DNA synthesis. If DNA synthesis on the leading and the lagging strands has different processivity in vivo as well, the distribution of the length of daughter strand gaps (see “The two mechanisms of two-strand damage” above), produced during replication of templates with noncoding lesions, would be bimodal, with the gaps in the leading strand being longer than those in the lagging strand. However, the length distribution of daughter strand gaps is unimodal, suggesting that the processivity of DNA synthesis in vivo is comparable for the two strands (710). Indeed, the initial products of DNA synthesis in vivo, detectable in DNA ligase or DNA pol I mutants, are small fragments of the same length (the Okazaki fragments), which argues that E. coli DNA replication in vivo is discontinuous on both strands (383, 471, 483). This conclusion was questioned when it was found that the continuous leading-strand DNA synthesis in vitro may appear discontinuous if the nascent DNA misincorporates uracils instead of thymines, which are then subject to excision repair (472). However, in vivo Okazaki fragments are still generated even when excision of uracils, nucleotide excision repair, base excision repair, and mismatch repair are all inactivated (681, 709, 711), confirming that DNA replication in E. coli cells is discontinuous on both strands. Experiments of a different kind are needed to resolve this discrepancy between the in vitro and in vivo results.
Initiation of plasmid DNA replication.
Small multicopy plasmids of E. coli initiate their DNA replication in a different way—they use the priming mechanism employed in the host recombinational repair of disintegrated replication forks but substitute a transcription intermediate for the recombination intermediate. DNA at their replication origins is first transcribed, and a portion of the resulting several-hundred-nucleotide transcript forms a stable RNA-DNA hybrid with its template, displacing the complementary DNA strand into the so-called R loop. Then the RNA portion of the hybrid is cleaved at a specific site to provide a primer for a limited DNA synthesis carried out by DNA pol I.
The complementary DNA strand, displaced as a result of this DNA pol I-catalyzed synthesis, is complexed with SSB. For this reason, it would be inert for any further transaction if not for the second mechanism of primosome assembly available in E. coli. In contrast to the first mechanism, which starts with DnaA multimer binding to a cluster of its recognition sequences, the second mechanism begins with PriA binding to what looks like a replication fork framework. As determined in vitro (368, 466, 467), the sequence of events in the PriA-dependent primosome formation is as follows: (i) PriA binds to ssDNA near the branching point with the duplex DNA; (ii) PriB binds to PriA and stabilizes PriA binding to ssDNA; (iii) DnaT binds to the PriA-PriB complex, which is then further stabilized by binding of PriC; (iv) DnaB (replicative helicase) is delivered from the complex with DnaC (replicative helicase inhibitor) onto this branched DNA-PriABC-DnaT complex to form a preprimosome; and (v) DnaG (primase) associates with the preprimosome to lay primers for DNA synthesis. The key event in the primosome assembly is the competition for DnaB, with PriABC plus DnaT at the DNA substrate versus DnaC in solution. In vitro, the PriA-dependent primosome has a composition different from the DnaA-dependent primosome in that PriA and PriB proteins seem to stay associated with DnaB during the ensuing DNA synthesis (6).
Nucleoid segregation and the problem of accessibility.
Nucleoid segregation sets the “window of opportunity” for recombinational repair. In rapidly growing E. coli cells, nuclear bodies, or nucleoids, are seen in the electron microscope as dimers (688, 741), and even after being freed from their “cells,” many purified nucleoids have a doublet appearance (240, 458, 490), indicating that the separation of nascent nucleoids is concomitant with DNA replication. It has been proposed that replicated daughter branches of the parental chromosome do not stay entangled but from the very beginning form separate bodies, growing out from the replication point in opposite directions (381, 740). Spatial separation of the origin-proximal markers after origin duplication was visualized in live cells (215).
This continuous separation of daughter nucleoids should create a problem for reactions that rely on interactions between sister chromosomes. Still, it could be argued that although sister chromosomes may appear separate, they interact at the molecular level as if residing in the same space. This question was addressed by comparing site-specific recombination between plasmid molecules in vitro with the same interplasmidic reaction in vivo (250). The efficiency of the in vitro reaction depends on the plasmid concentration; the plasmid DNA concentration in vivo can be estimated from the plasmid copy number. It was expected that the effective in vivo concentration would be higher due to a variety of cellular factors. Contrary to that expectation, it was found that the effective in vivo concentration is an order of magnitude lower, suggesting that when homologous DNAs are searching for each other in vivo, they face the problem of restricted accessibility (250). In other words, if recombinational repair is to mend two-strand damage in one of the daughter DNAs, it has a certain time frame to do it before the daughter sequences are segregated into separate nucleoids.
Summary
DNA damage can be classified as affecting either one or both strands in a particular sequence. Similarly, cellular DNA repair mechanisms are categorized as either one-strand or two-strand repair. Since the two-strand repair frequently spins off recombinant chromosomes, it is generally known as recombinational repair. The bulk of the two-strand damage is generated by DNA replication, when a replisome stumbles upon an unrepaired one-strand lesion. The two major replication-induced two-strand lesions are daughter strand gaps and disintegrated replication forks. In E. coli, daughter strand gaps are repaired by the RecF pathway whereas disintegrated replication forks are repaired by the RecBCD pathway. Two-strand DNA lesions occur infrequently during regular growth in the laboratory, but in real life E. coli must occasionally experience massive DNA damage—hence the inducible DNA repair capacity, called the SOS response.
Recombinational repair acts to carry the replication apparatus through the template DNA containing unrepaired one-strand lesions and, in this respect, must collaborate with the chromosomal replication and the nucleoid segregation machinery. This puts recombinational repair reactions in a specific context, with their own idiosyncrasies, unresolved problems, and gray areas. One such controversy, bearing on the damage formation mechanisms, is whether in vivo replication is discontinuous on both DNA strands. One of the major complications for the recombinational repair, which depends on the availability of an intact sister duplex, is the accessibility of this duplex, because the sister nucleoids are continuously segregated as the cell grows. This aspect of the in vivo chromosomal metabolism is almost unstudied.
RECA: HOMOLOGOUS PAIRING ACTIVITY
For damaged DNA to be repaired with the help of an intact homologous sequence, the two DNAs need to (i) find each other among numerous unrelated sequences and (ii) trade strands to make possible one-strand repair of the damage in the affected sequence. In E. coli, these intricate and seemingly intelligent reactions are catalyzed by a single, relatively small enzyme called RecA. The 38-kDa RecA searches for homology both catalytically and stoichiometrically, since the active species is a polymer comprising hundreds of RecA monomers.
recA Gene and Mutants
recA and peculiarities of recA null mutants.
recA happened to be the first E. coli recombinational repair gene to be discovered (107). recA is not a part of any operon, is surrounded by genes unrelated to DNA metabolism, and has its own promoter and terminator (260, 546). Normally, recA expression maintains 1,000 to 10,000 RecA monomers per cell (292, 445, 543, 558). RecA production is induced by DNA-damaging treatments such as UV irradiation or nalidixic acid, resulting in up to a 50-fold increase in the amount of the protein (see “Organization of the SOS regulon” above) (292, 543).
No extragenic suppressors that would cancel the phenotypes of null recA alleles have been found, suggesting that recA is the only gene of its kind in the E. coli genome. recA mutations are unusually pleiotropic (for an early yet informative review, see reference 102). recA cells are extremely sensitive to DNA damage (107, 267, 689); nevertheless, recA null mutants are viable, although they grow slower than the WT cells. The slower growth of recA cultures is due to the continuous generation of dead cells (229) rather than because of growth defects. The fraction of dead cells in laboratory cultures of recA mutants reaches 50% (85).
WT cells stop cell division in response to inhibition of DNA synthesis, but recA mutant cells continue to divide under these conditions, producing anucleate cells (272); RecA inhibits cell division via SOS induction when there are irregularities with DNA replication (see “Organization of the SOS regulon” above). Under regular growth conditions, about 10% of recA cells lack chromosomal DNA; up to 20% of the total DNA in recA mutant cultures is degraded at any given moment (84, 105). This DNA degradation must target particular nucleoids, hence the asynchrony phenotype displayed by recA mutant cultures: whereas most cells in the WT cultures grown in a rich medium have either four or eight nucleoids, recA mutant cells have all numbers of nucleoids from zero to eight (598, 599).
Cellular processes dependent on RecA.
The induction of the SOS response, the reaction of the cell to massive DNA damage, is absolutely dependent on RecA. RecA is activated by damaged DNA, and the activated form of RecA catalyzes self-cleavage of LexA repressor (see “Cleavage of LexA repressor by RecA filament” below). The SOS response increases the capacity of the cell to repair and tolerate DNA damage and also delays cell division. The damage to bacterial DNA also causes prophage induction, i.e., the lytic development of latent bacteriophages. Similarly to LexA repressor, bacteriophage repressors cleave themselves in the presence of activated RecA, but the phage induction has nothing to do with repair of cellular DNA and is in fact lethal to the host cell.
RecA plays a pivotal role in recombinational repair of such two-strand DNA lesions as daughter strand gaps, double-strand breaks, and interstrand cross-links. For example, while WT E. coli cells survive 53 to 71 cross-links per chromosome, recA cells are killed by a single cross-link (595). RecA-dependent mechanisms of recombinational repair are discussed below (see “Resolving recombination intermediates” and “Repair of daughter strand gaps”).
A bacterial cell can acquire a linear piece of chromosome from another cell in a variety of ways (reviewed in reference 9). During conjugation, this piece is transferred from another live cell, which has a conjugative plasmid integrated into its chromosome. During transduction, this piece is delivered by a bacteriophage, whose capsid had mistakenly packaged a fragment of the host DNA instead of the phage chromosome. During transformation, a cell picks up a piece of DNA from the environment, from a dead decomposing cell. Such an exogenous piece of DNA can be inserted, in whole or in part, into the chromosome in a RecA-dependent process; recA mutants are profoundly defective in all types of chromosomal recombination (reviewed in reference 399).
Two-strand DNA damage induces RecA-dependent mutagenesis. In general, DNA modification is mutagenic in that it causes point mutations, and especially strong mutagens are those that make DNA bases change their coding interactions. For example, guanine recognizes cytosine in the opposite position, but oxidation of guanine could make it recognize adenine, causing a misincorporation and, ultimately, a point mutation (433). However, RecA-dependent mutagenesis has a completely different nature. Some DNA modifications generate the so-called noncoding lesions, i.e., bases that are missing or so distorted that they are no longer recognized by DNA polymerases. DNA replication can bypass such a noncoding lesion in a RecA-dependent reaction, during which a DNA polymerase sometimes has to incorporate a random nucleotide in the new DNA chain across the damaged position, which often results in mutations (see “Backup repair of daughter strand gaps: translesion DNA synthesis” below).
In Vitro Activities of RecA
The variety of phenotypes of recA mutant cells stems from a single deficiency, the inability to form an active RecA filament. The in vitro properties and activities of RecA filament still bewilder and fascinate those who study them. For in-depth treatment of the enchanting RecA biochemistry, see the excellent reviews by Kowalczykowski (319) and Roca and Cox (524).
RecA without DNA.
In high-concentration solutions, RecA aggregates to form oligomers, filaments, and bundles (70, 71, 246, 737). One of the major species in these aggregates consists of rings of six to eight monomers (70, 71, 246). These rings are characterized by electron microscopy for RecA from Thermus aquaticus, due to their greater stability (758). Surprisingly, they resemble in gross details both the hexameric rings of helicases like DnaB or RuvB and the F1-ATPase (168, 760).
The crystal structure of RecA, solved at 2.3-Å resolution, shows a spiral filament with six RecA monomers per turn (632). There is enough space inside the filament to accommodate two interacting DNA molecules. Although the crystals were formed either in the presence of ADP or without nucleotide cofactor, and so represent RecA species inactive in recombinational reactions, they show the overall arrangement of the structural elements within the RecA monomer as well as the way in which the monomers are arranged into filaments.
Filament formation by RecA around ssDNA.
In vitro, in the presence of physiological concentrations of Mg2+ and ATP, RecA assembles around ssDNA into a helical filament (Fig. 6), an entity proficient in all known RecA activities (in the absence of the nucleotide cofactor or in the presence of ADP, RecA forms similar filaments but with different parameters; since such filaments are inactive in RecA-promoted reactions, they are not discussed in this review). At physiological pH, RecA filament does not readily assemble on duplex DNA; however, a filament assembled on ssDNA extends into a contiguous double-stranded region (362, 572). Every RecA monomer within a filament binds a single ATP molecule (307). RecA filament assembled on ssDNA slowly hydrolyses ATP at a rate of about 30 ATP molecules per min per monomer (69); duplex DNA-bound filament has an even lower ATPase activity (362, 502).
FIG. 6.
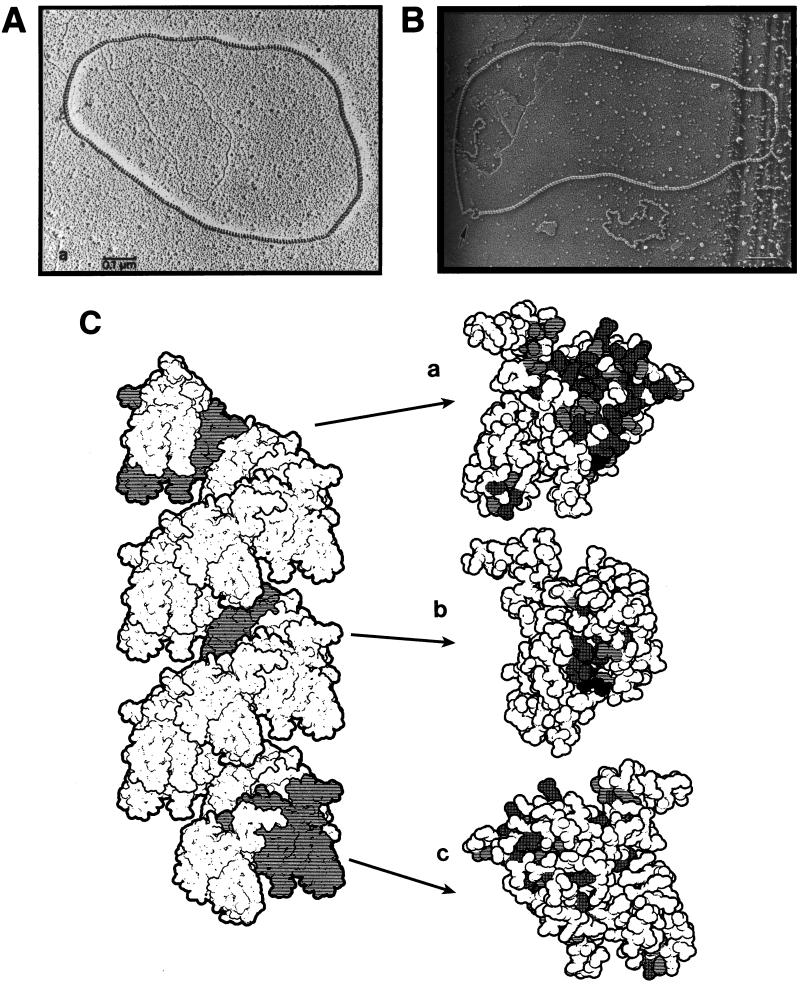
RecA filament in vitro: photographs and molecular model. (A) A relaxed circular duplex DNA completely covered by RecA filament. Naked duplex DNA of the same length, lying mostly inside the RecA filament, illustrates that DNA within RecA filament is stretched 1.5 times. Reprinted from reference 625 with permission of the publisher. (B) A RecA-covered circular ssDNA molecule; the arrow points to a segment complexed by SSB. Below, another ssDNA circle of the same length can be seen, but since it is entirely complexed with SSB, it appears very different, i.e., much smaller and kinky. Note that while RecA filament stretches ssDNA, SSB compacts it. Reprinted from reference 246 with permission of the publisher. (C) A crystal structure-based molecular model of an 18-monomer segment of RecA filament with three symmetry-related monomers in gray. Each of these monomers is enlarged on the right to show residues conserved among eubacterial RecA proteins (see reference 524 for details). In such a filament, the 5′ end of DNA1 would be at the top. Reprinted from reference 524 with permission of the publisher.
The ATP hydrolysis is not needed for the assembly of RecA filament on DNA, since the same recombination-proficient filament is formed in the presence of a nonhydrolyzable ATP analog, ATPγS (169). One role for ATP hydrolysis may be to promote filament disassembly, since RecA filaments formed in the presence of ATPγS do not disassemble on their own (reference 523 and references therein). Most of the measurements of recombination-proficient RecA filaments were done in the presence of ATPγS because of the greater filament stability in the absence of ATP hydrolysis; however, parameters of ATP-containing filaments are very similar (627).
The width of ATP-containing RecA filament is about 10 nM (100 Å) (165, 169), which is five times the width of duplex DNA. The ATPγS-containing filament has about six RecA monomers per 95-Å turn, with each RecA monomer binding about 3 nucleotides of ssDNA (311). The stoichiometry of duplex DNA binding is the same: one RecA monomer binds 3 bp, or a single 95-Å helical turn of RecA filament holds about 18 bp (the axial spacing between adjacent base pairs is 5.1 Å) (153, 161). Since the axial spacing between adjacent base pairs in native DNA duplex is 3.4 Å, it is said that duplex DNA inside RecA filament is extended 1.5 times (165, 626) (Fig. 6A). This extension, which is surprisingly close to the maximally extended DNA state of 1.7 (113, 608), is thought to facilitate homology recognition between two DNA molecules, captured by RecA filament (see “Detection by RecA filament of homology to ssDNA bound in the primary site” below).
RecA filament grows at its ends. Growth in the 5′-to-3′ direction relative to the bound ssDNA is several times more efficient than growth in the opposite direction (362, 514, 571, 572). The maximal rate of RecA filament assembly in vitro is 30 to 40 monomers per s (514). Assuming a DNA binding stoichiometry of one RecA monomer per 3 nucleotides, a growing RecA filament engulfs about 100 nucleotides of ssDNA per second. In vitro, at the same time as the 3′ end of RecA filament is growing, the 5′ end may begin slowly disassembling (362, 569). In effect, the growing RecA filament under these conditions treadmills along the DNA.
Two DNA-binding sites in RecA filament.
Soon after the beginning of biochemical characterization of RecA protein, it was realized that, to promote homologous pairing, RecA must have at least two DNA-binding sites: the primary site accommodating DNA1, around which the filament was assembled, and the secondary site for DNA2, to be compared with DNA1 (269). Since then, the idea of at least two DNA-binding sites within RecA filament has been substantiated with a variety of evidence.
In vitro, RecA promotes both three-strand exchange (between an ssDNA1 and a duplex DNA2) and four-strand exchange (between a duplex DNA1 with a single-stranded tail and a fully duplex DNA2), implying the ability of RecA filament to handle up to four DNA strands. However, the only DNA strands in these reactions fully protected by RecA filaments against DNase degradation are the ssDNA1 or the outgoing identical strand (in these experiments, SSB was absent), suggesting that either the hybrid duplex or the alternative duplex is excluded from the filament (98, 99). Moreover, RecA cannot catalyze strand exchange restricted to fully duplex DNA regions (120, 363), indicating that it cannot handle four DNA strands at the same time (reviewed in reference 129).
RecA filament has the primary site that binds ssDNA during filament assembly but can also accommodate duplex DNA. In addition, RecA filament has the secondary binding site, which can transiently bind duplex DNA if the primary site is occupied by ssDNA. If the primary site is occupied by duplex DNA, the secondary binding site can transiently bind ssDNA. If the primary site is occupied by ssDNA, the secondary site can stably bind an unrelated ssDNA (326, 421). Finally, in the presence of ATPγS and high Mg2+ concentrations, RecA filament can stably bind two DNA duplexes, but it is unclear whether they have to be homologous (762). Therefore, it seems that RecA has a primary binding site deep within the filament, accommodating up to two DNA strands, and a secondary binding site at the periphery of the filament, again accommodating up to two DNA strands.
ssDNA1 is bound by RecA filament along its sugar-phosphate backbone, so that DNA bases face inward (154, 349) and are ordered perpendicularly to the filament axis (326). Duplex DNA1 is bound by RecA filament along its minor groove (154, 161, 329). In contrast, binding by a RecA-dsDNA1 filament of the second duplex does not involve its minor groove (762).
Cleavage of LexA repressor by RecA filament.
RecA filament holding a single DNA strand promotes autocleavage of the SOS response repressor, LexA (259, 366), as well as autocleavage of phage repressors (172, 522, 559) and of the UmuD protein (77). It is said that in these reactions RecA plays a role of coprotease, because there are conditions under which LexA, phage λ repressor, and UmuD cleave themselves in the absence of RecA (77, 364). The LexA-binding site lies deep within the filament groove and overlaps with the secondary DNA-binding site (759), explaining why, when both DNA-binding sites are occupied by ssDNA, LexA cleavage is inhibited (152, 515, 642). RecA filament assembled on duplex DNA promotes LexA cleavage at 5 to 20% of the rate observed with a filament assembled on ssDNA (152, 642). This feature of the SOS repressor cleavage makes biological sense—if all the single-stranded regions associated with DNA lesions are made double stranded (supposedly by pairing with intact homologous sequences), there is no reason to boost the repair capacity of the cell any further.
The RecA-promoted LexA autocleavage is rapid and is independent of Mg2+ concentrations in the range of 1 to 10 mM (345). In contrast, RecA-promoted autocleavage of UmuD and phage repressors is slow and is observed only at Mg2+ concentrations of 10 mM or higher (77, 172, 522, 559). The supposed biological significance of these differences, in relation to the idea that the SOS response needs to be induced early while phage repressors need to be cleaved only in moribund cells, will become clear later (see “SOS-induced conditions” below).
Detection by RecA filament of homology to ssDNA bound in the primary site.
Although RecA forms a filament around ssDNA1 in a sequence-independent manner, the filament itself is “a sequence-specific DNA-binding entity, with the specificity determined by the bound DNA” (524). The amount of nonhomologous DNA in vivo is overwhelming, since even an identical sequence, shifted a single nucleotide out of register, becomes perfectly heterologous to DNA1. Heterologous DNA is not neutral in homology searches: preincubation of presynaptic filaments with heterologous DNA inhibits subsequent homologous pairing (214). The problem of the complexity of natural DNA is compounded by the enormous intracellular DNA packing densities, measured in E. coli at 20 to 100 mg/ml (57), and the restricted accessibility due to the nucleoid segregation (see “Nucleoid segregation and the problem of accessibility” above). Under these conditions, RecA has to find homology to the damaged DNA within minutes, as illustrated by the extreme DNA damage sensitivity of a partially active recA allele, proficient in homologous recombination in vivo and capable of “slow” recombinational reactions in vitro (273). If not repaired quickly, the damaged DNA could be degraded or segregated from its intact sister or could bring about even greater damage if the upcoming replication fork runs into it. On the other hand, if recombinational repair mends DNA damage mostly at replication forks, the affected and the intact homologous DNA segments should initially be in close proximity with each other.
The mechanism of homology search by RecA filament is still an enigma. An algorithm of homology search is likely to require repeated juxtaposition of short segments of DNA1 with short segments of duplex DNA. If RecA filament is able to juxtapose two potentially nonhomologous sequences, how does it then let go a duplex which proved to be nonhomologous? One possibility was that a nonhomologous duplex is expelled from the filament with the help of ATP hydrolysis. However, in vitro RecA catalyzes homologous pairing in the presence of nonhydrolyzable ATP analogs (258, 322, 428). Moreover, a mutant RecA protein with a 100-fold in vitro defect in ATP hydrolysis, which is activated by ATP as if it were ATPγS, still catalyzes homologous pairing, both in vivo and in vitro (82), so the homology search does not require ATP hydrolysis.
Kinetic experiments show that the homology search in vitro is reversible, follows second-order kinetics (i.e., it depends on the concentrations of both interacting DNAs), and is rapid compared to the next stage of pairing (31, 752). Under these conditions, a short segment of RecA-DNA1 complex is estimated to try 102 to 103 various duplex DNA segments per s, with the high iteration frequency demanding that the search be based on soft interactions only (752). However, these interactions are strong enough to cause partial unwinding of nonhomologous DNA within RecA filaments (137, 536). This unwinding is most probably caused by DNA2 extension inside the filament, as RecA puts it in register with DNA1. The duplex DNA2 is approached along its minor groove by the RecA-complexed ssDNA1 (27, 329, 495), and in the synaptic complex, all three DNA strands are underwound to the same extent of 19 nucleotides per turn (301). This underwinding may allow ssDNA1 to be accommodated in the otherwise too narrow minor groove of dsDNA2 (40).
The configuration of the three DNA strands in the synaptic complex has been a matter for debate and experimentation (129). An interesting idea was that the three strands form a DNA triplex, in which the homology recognition occurs. However, since the minor groove of the duplex DNA does not have enough determinants for homology recognition, the idea of a triplex requires ssDNA1 to interact with dsDNA2 via the major groove of the latter, which contradicts the available experimental evidence (see above). Also, no triplexes are detected in the synaptic complexes; instead, ssDNA1 is seen already paired with the complementary strand of dsDNA2 whereas the identical strand of dsDNA2 is displaced into the major groove of the nascent duplex (2, 495).
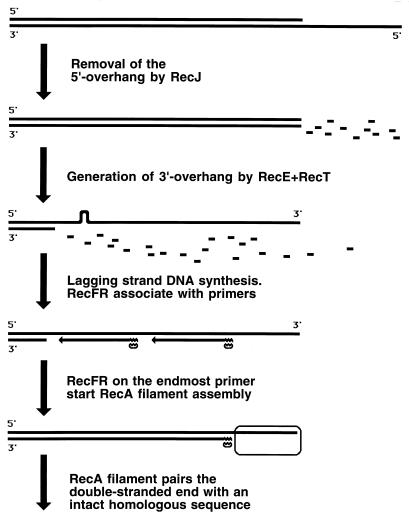
Strand exchange between DNA1 and DNA2 catalyzed by RecA filament.
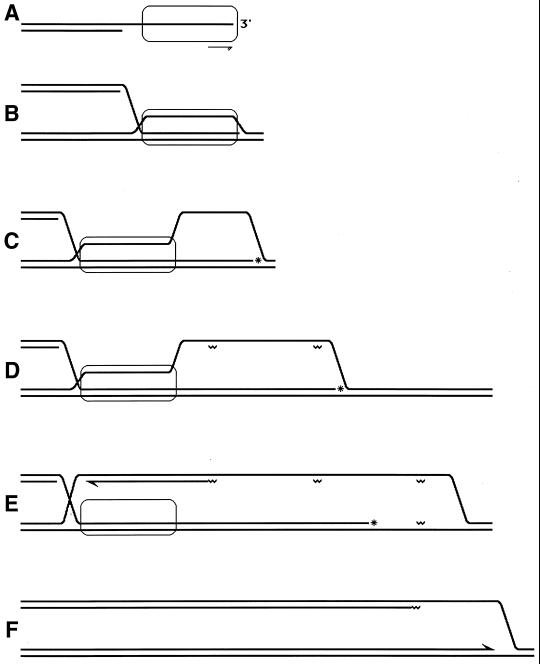
When homology is found (i.e., when a homologous duplex DNA2 is aligned with ssDNA1), RecA filament catalyzes the exchange of strands between the two DNA molecules. In this process, ssDNA1 forms hydrogen bonds with the complementary strand of DNA2 while the identical strand of the duplex is displaced (135) (Fig. 7). The outgoing strand of DNA2 is accommodated in the secondary ssDNA binding site at the periphery of the RecA filament to be extracted from there later by SSB (346, 420).
FIG. 7.
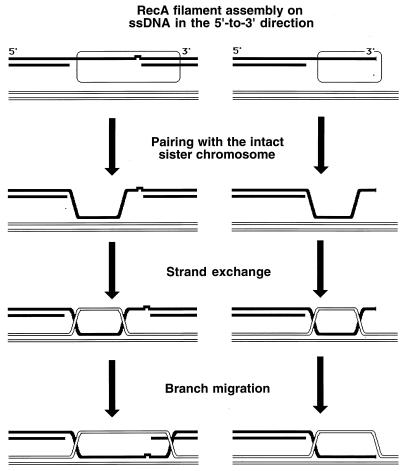
The distinguishable phases of RecA-promoted reactions, as they are thought to happen in vivo. Black lines indicate a damaged duplex; white lines indicate an intact duplex; the open rectangle with rounded corners indicates a RecA filament; the irregularity in one of the black DNA strands indicates a noncoding lesion. The left side represents daughter strand gap repair; the right side represents double-strand end repair. Explanations are given in the figure.
The RecA-catalyzed strand exchange phase can be further subdivided into (i) the “initial” strand exchange, which happens concomitantly with homologous recognition, is limited in length, and does not require ATP hydrolysis by RecA, and (ii) “hybrid duplex extension,” which requires ATP hydrolysis. Hybrid duplex extension is propagated along the RecA filament from the point of the initial homologous contact in the 5′-to-3′ direction (134, 288, 724), i.e., in the direction of RecA filament assembly. The lengths of the DNA segments that trade strands in this process can be up to several kilobases.
If DNA1 is a completely single-stranded molecule, the RecA-mediated strand exchange with a duplex DNA is called a three-strand reaction. However, since the RecA filament readily extends into the adjacent duplex regions, the initially three-strand exchange becomes four-strand exchange in the region where DNA1 becomes duplex (146, 725). In such a four-strand reaction, there are two displaced strands, which are complementary to each other; therefore, they anneal to form a second duplex, connected with the first one by crossed strands (Fig. 7). The four-strand exchange can be initiated only in the region where one of the participating duplexes is single stranded, although this initiating single-stranded region can be as short as 6 to 15 nucleotides (120, 363).
The branch or junction migration promoted by RecA is insensitive to mismatches and can overcome, although with decreased efficiency, numerous modified DNA bases and even deletions or duplications up to several hundred bases (42, 145, 369). As mentioned above, ATP hydrolysis by RecA is not needed for the limited strand exchange concomitant with homologous pairing in the three-stranded reaction (258, 322, 428). However, ATP hydrolysis increases the extent of such strand exchange above 2 kb (280) and is needed to drive strand exchange in the four-strand reaction (303). ATP hydrolysis by RecA filaments is absolutely required for strand exchange to overcome even a short heterology between two participating DNAs (302, 531).
There are two complementary explanations to account for the ATP hydrolysis requirement during strand exchange. Kowalczykowski and coworkers favor the idea that the RecA filament has to reorganize at points of filament discontinuities or where DNA1 becomes double stranded or around heterologies in DNA2. Such reorganization includes partial filament disassembly, hence the need for ATP hydrolysis (428). Cox favors the view that since DNA is a helix, strand exchange between two DNAs is driven by rotation of the participating DNA helices around their long axes (523). Thus, for every 18 bp exchanged, both participating DNA molecules rotate 360°, and ATP hydrolysis by RecA may fuel this rotation during strand exchange (133). Facilitated rotation during strand exchange is predicted to unwind the segment of heterologous DNA due to accumulation of the torsional stress; the predicted unwinding of heterologous segments is indeed detected (393).
Assistance for RecA by SSB at all stages.
In vivo, the ssDNA is promptly complexed with SSB (see “Initiation of chromosomal DNA replication in E. coli” above). Consequently, if RecA is to polymerize on ssDNA, it either has to displace SSB (Fig. 6B) or has to coexist with SSB on the same ssDNA. Peculiar patterns of SSB and RecA binding to ssDNA under different in vitro conditions are discussed below (see “Regular DNA replication” and “SOS-induced conditions”); for now it will suffice to say that under certain conditions SSB simply does not allow RecA onto ssDNA; under other conditions, SSB allows RecA polymerization on ssDNA in the presence of auxiliary proteins; while under a third set of conditions, it yields ssDNA to RecA without hesitation. Under the second and third sets of conditions, SSB also helps at all three stages of RecA-promoted in vitro reactions.
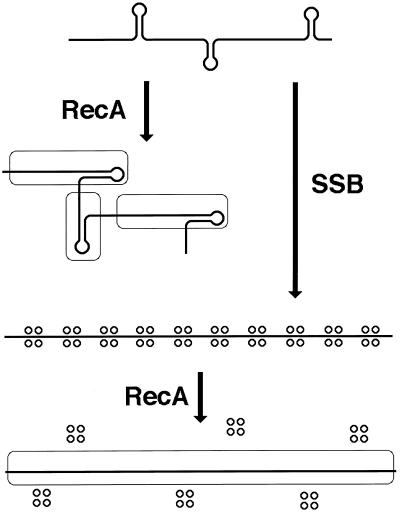
SSB helps at the presynaptic phase, assisting with RecA polymerization on ssDNA. In vitro, RecA by itself is able to form long, strand exchange-proficient filaments on naked ssDNA only under low-salt, low Mg2+ conditions, which are far from being physiological. In fact, these conditions do not allow the formed RecA filaments to carry out subsequent strand exchange! To stimulate strand exchange, the Mg2+ concentration has to be raised after RecA filaments are formed. On the other hand, if RecA filaments are preformed at these elevated Mg2+ concentrations, the subsequent strand exchange is less productive. The explanation for this paradox is that under conditions which are closer to physiological ones, ssDNA forms secondary structures, which interfere with the formation of long contiguous RecA filaments (Fig. 8). One way SSB enhances the performance of RecA is through elimination of these secondary structures, allowing the formation of long contiguous RecA filaments at high Mg2+ concentrations (456).
FIG. 8.
SSB helps in the assembly of the functional RecA filament. The thick line indicates ssDNA; stem-loop structures indicate secondary (duplex) structures in ssDNA at physiological Mg2+ concentrations; open rectangles with rounded corners indicate RecA filaments; quartets of small circles indicate SSB tetramers. Without SSB (the top and the left side), formation of long contiguous RecA filaments on ssDNA is compromised because the growth of nascent filaments is impossible past the secondary structures. SSB helps RecA to polymerize into long contiguous filaments (the right side and the bottom) by ironing out secondary structures in the ssDNA.
SSB helps with the homology search by sequestering the excess of ssDNA. The RecA filament stably binds two heterologous ssDNA, but such a filament is mute in the subsequent homology search and strand exchange (421). Therefore, it is important to guard the RecA filament from excess ssDNA, and SSB plays the role of such a guardian (420).
SSB ensures the unidirectionality of RecA-promoted strand exchange by taking the displaced strand out of the filament. Potentially, the displaced strand can reverse the reaction or can be utilized in a new round of strand exchange, serving as DNA1 for a new RecA filament. The SSB binding sequesters the displaced strand from further action, making in vitro strand exchange unidirectional (346, 420).
Supervision of RecA Activity
The efficiency of the in vitro RecA-promoted pairing is unexpectedly high. RecA can pair two sequences which share as few as 8 nucleotides of homology (270); as mentioned above, RecA can also promote extensive strand exchange between homeologous (homologous but not identical) DNA sequences. This quite indiscriminate nature of the RecA-promoted pairing poses a potential problem even for the generally nonrepetitive genomes of bacteria. For example, in the E. coli genome, there are several rRNA operons which are mostly homologous to each other (53, 251), as well as many short (20- to 30-nucleotides) perfect repeats (50). If not properly supervised, RecA could repair damage in one such repeat by using another one. Such improper pairing, if accompanied by crossing over, would lead to gross chromosomal rearrangements. The components of the major mismatch repair system in E. coli supervise the quality of RecA-promoted pairing.
Inhibition by MutS and MutL of pairing between homeologous sequences.
The supervision of the legitimacy of RecA-promoted pairing between long sequences is likely to be a secondary function of the MutS and MutL proteins. The two proteins are the components of the major mismatch repair pathway in E. coli. MutS binds to mismatches, while MutL is believed to transmit the signal of the MutS-mismatch interaction to other parts of the correction system (439). mutS and mutL mutants have increased rates of recombination between homeologous sequences (179, 488, 512, 579). Mechanistically, this phenomenon is accounted for by the in vitro ability of MutS to inhibit RecA-promoted strand exchange between homeologous sequences (744, 745). MutL enhances the efficiency of this inhibition. It is suggested that MutSL complex binds to a newly formed mismatch still within RecA filament and that this binding inhibits further RecA-promoted strand exchange (745). MutL could also recruit the UvrD helicase (231) to actively disperse RecA filaments, one known in vitro activity of UvrD (446). Thus, in the event that RecA catalyzes strand exchange between homeologous sequences, completion of such a product in vivo is likely to be aborted by MutSL.
Possible disruption of pairing of insufficient length by helicase II.
The in vitro ability of RecA to pair ssDNA and a duplex DNA which have in common fewer than 10 contiguous nucleotides (270) makes one wonder how RecA discriminates in vivo against a 10-nucleotide homology in favor of a long, genuine homologous sequence. The answer may be that it does not but that other enzymes supervise RecA-promoted pairing to disrupt recombination intermediates which are “too short” or have a specific, “banned” structure.
One such supervisor is likely to be helicase II, encoded by uvrD. Helicase II is an abundant protein, estimated at 5,000 to 8,000 monomers per cell (305); this number is elevated even further during SOS induction (Table 1). Helicase II has 3′-to-5′ polarity and unwinds DNA from nicks or double-strand ends (537). Its role in excision repair and methyl-directed mismatch repair (two major types of one-strand repair [see “Damage reversal and one-strand repair” above]) is to act after the incision step and remove segments of damage- or mismatch-containing strands which are to be resynthesized (439).
Similar to mutS and mutL mutants, uvrD mutants exhibit a “hyperrecombination” phenotype, although in a different circumstance. uvrD mutants are modestly hyperrecombinant if the exchange is between lengthy homeologous sequences (488, 512), but they are strongly hyperrecombinant when the homology is expected to be limited in length (18, 45, 179, 391, 766).
One explanation for the hyperrecombination phenotype of uvrD mutants is that they accumulate DNA lesions that cause elevated recombination. Indeed, uvrD mutants are slow to close the single-stranded interruptions introduced during excision repair (533, 596, 694). Single-strand interruptions in template DNA are proposed to cause replication fork collapse with subsequent recombinational repair (130, 333, 597), hence elevating the overall genomic recombination. This explanation predicts that uvrD mutants should be inviable if they carry additional mutations in recA or recB genes, as observed for other mutants which accumulate single-strand interruptions in their DNA (333) (see “Evidence for replication fork repair by recombination” below). However, uvrD recA and uvrD recB mutants are sick but viable (417, 601); therefore, accumulation of DNA lesions cannot be the only explanation for the hyper-rec phenotype of uvrD mutants.
Another explanation is that helicase II is an antirecombinase which disrupts RecA-assembled recombination intermediates. The poor viability of uvrD lexA (Ind−) double mutants is improved by recA mutations, suggesting recombination poisoning in the absence of helicase II and some other SOS-induced functions (371). In vitro, when added to a RecA-mediated strand exchange reaction, helicase II promotes both the completion of RecA-mediated strand exchange and disassembly of strand exchange intermediates back to the initial substrates (446). Helicase II may function to increase the fidelity of RecA-promoted pairing by disrupting homologous contacts of insufficient length. For example, helicase II could recognize such complexes as having a three-strand junction on the 3′ side of the invading DNA, loading on this single-stranded tail, and, moving in the 3′-to-5′ direction, unwinding the short recombination intermediate. Helicase II could discriminate against short homologous contacts in both the daughter strand gap repair and double-strand end repair pathways (see “The two recombinational repair pathways of E. coli” above and “Repair of daughter strand gaps” and “Double-strand end repair” below).
Summary
RecA catalyzes the central reaction of recombinational repair in E. coli; recA mutants are deficient in many aspects of DNA metabolism. recA genes are ubiquitous in eubacteria (524); they are seldom found inactive (404), but rarely are they indispensable for viability (459). The propensity of RecA to form hexameric circles in vitro betrays its structural relationship to DNA helicases and F1-ATPase. RecA forms a helical filament around ssDNA, finds a duplex DNA homologous to this ssDNA, and catalyzes strand exchange between these two DNAs. The RecA-ssDNA filament also promotes self-cleavage of the SOS repressor, LexA. SSB assists RecA in all these in vitro reactions.
One unsolved issue in RecA biochemistry is the mechanism of the homology search by the RecA filament. Studying the structure and dynamics of individual DNA strands inside the filament could shed light on the homology search mechanism as well as on some RecA-dependent in vivo phenomena. Another area of interest is the in vivo supervision of the RecA-promoted strand exchange; in vitro characterization of this important function has just begun.
RESOLVING RECOMBINATION INTERMEDIATES
The RecA filament-promoted strand exchange generates DNA junctions: the locations at which individual strands switch between the two participating DNA molecules. These junctions can involve either three DNA strands (in the region where the invading DNA is single stranded) or all four strands of the two duplexes. The four-strand junctions are usually called Holliday junctions (see “Two-strand repair” above). In the course of the daughter strand gap repair (see “Repair of daughter strand gaps” below), two DNA junctions have to be formed; during the double-strand end repair (see “Double-strand end repair” below), only one DNA junction is probably formed.
To complete recombinational repair, DNA junctions and the associated RecA filament must be removed. This section discusses what is known about the in vitro activities of the E. coli enzymes that remove DNA junctions and dissociate RecA filaments. Since the in vivo configurations of the DNA junctions during a particular repair reaction and the interaction of the removal activities with other recombinational repair proteins are still a subject of speculation, they are discussed later, in the corresponding sections (see “Repair of daughter strand gaps” and “Double-strand end repair” below).
The Three Ways To Remove a Pair of DNA Junctions
When two DNAs trade strands within their internal segments, a pair of DNA junctions is formed. Independently of whether these are three-strand or four-strand junctions, such a pair of DNA junctions can be resolved in three possible ways. One way to disengage the recombining DNAs is to simply pull them apart, reversing the RecA-catalyzed strand exchange (464, 664) (Fig. 9A). In reality, instead of pulling DNAs apart, the two junctions are probably translocated towards each other, “squeezing out” the exchanged DNA segments. The alternative way to remove the junctions is to resolve them by cutting individual strands. Three-strand junctions can be resolved by cutting a single DNA strand (190), while to resolve four-strand junctions, two DNA strands of the same polarity must be cut symmetrically (256) (Fig. 9B). Finally, there is a hybrid way of removing a pair of DNA junctions: one of the junctions is resolved by cutting, but the cuts are not sealed right away, and the second junction is eliminated by being translocated to these cuts (Fig. 9C).
FIG. 9.
The three ways to remove a double DNA junction. A pair of four-strand junctions is shown, but the same applies to a pair of three-strand junctions or to a combination of one four-strand junction and one three-strand junction. The two DNA duplexes participating in the joint molecule are shown as either solid or open double lines. Small arrows near the junctions indicate the direction of junction translocation. Scissors mark the position of strand cuts. (A) Removal by translocation only (topoisomerase model). (B) Removal by symmetrical single-strand cuts only (resolution). (C) Removal by a combination of cuts and translocation. Additional explanations are given in the text.
When a DNA end trades strands with an internal segment of a homologous DNA, a single DNA junction is formed. A single DNA junction, whether it is a three-strand or four-strand junction, can be removed by pulling DNAs apart (Fig. 10A), although this is unproductive, or it can be resolved by cutting the DNA strand(s) at the junction (Fig. 10B) or by translocating the junction to the introduced interruption in the originally intact DNA strand (Fig. 10C).
FIG. 10.
Removal of a single DNA junction. A three-strand junction is shown, but the same applies to a four-strand junction. The two DNA duplexes participating in the joint molecule are shown as either solid or open double lines. Small arrows near the junctions indicate the direction of junction translocation. Scissors mark the position of strand cuts. (A) Removal by translocation only. (B) Removal by symmetric single-strand cuts only. Note that for the three-strand junction, the black strand already has an end across the cut in the white strand. (C) Removal by a combination of a cut and translocation. Additional explanations are given in the text. The last two options create a replication fork framework, whereas the first option seems to be nonproductive. However, if the original two-strand lesion was a double-strand break and the invading end has already been extended by DNA synthesis, the expelled extended end can now anneal with the other end of the break, permitting lesion repair (517).
In E. coli, there are at least two independent enzymatic systems for DNA junction removal: the RuvABC resolvasome and the RecG helicase. Their mechanisms of interaction with DNA junctions are quite different, and yet they partially complement each other, since mutants with single mutations in one or the other system show only a moderate defect in recombinational repair. This implies that more than one way of DNA junction removal is realized in vivo.
RUV LOCUS: PHENOTYPES OF MUTANTS AND GENETIC STRUCTURE
Certain mutations conferring sensitivity to mitomycin C and UV light were mapped to a locus called ruv (372, 473). ruv mutants repair daughter strand gaps normally and are able to reinitiate DNA replication after the repair, but they fail to resume cellular division, forming long nonseptate, multinucleate filaments (372, 473). The filamentation phenotype of ruv mutants can be mutationally suppressed without improving the resistance of the cell to UV irradiation (372, 474), arguing against the idea (473) that ruv mutants are deficient in some function needed for the reinitiation of cell division after DNA damage. Staining of ruv cells for DNA after UV irradiation shows that almost all DNA is concentrated in several long filamentous cells, while most normal-size cells are anucleoid (274), suggesting a defect in the chromosome partitioning. ruv mutants are deficient for conjugative recombination in recBC sbc genetic backgrounds (see “Double-strand end repair in the absence of RecBCD” below) and for plasmid recombination in otherwise WT cells (370, 372). recA null mutations reverse the lethal effect of ruv mutations in certain circumstances (37, 474) and also suppress the chromosome partitioning defect after UV irradiation (274), indicating recombinational poisoning of ruv mutants and suggesting that the product of ruv locus acts in the postsynaptic phase, after the RecA-catalyzed formation of joint molecules.
The ruv locus was shown to be induced during the SOS response (590). Molecular characterization of the locus revealed the presence of three genes: ruvA and ruvB are organized into an SOS-inducible operon, while ruvC belongs to an adjacent, noninducible operon (38, 575, 587). Mutations in any one of the three ruv genes confer the same phenotype (370, 575).
Interaction of Ruv Proteins In Vitro with Holliday Junctions
The biochemical activities of RuvABC proteins and the ways they interact with DNA junctions, both structurally and functionally, are now well characterized. The remarkable progress in our understanding of RuvABC has been the subject of several recent reviews, to which the reader is referred for details and references (336, 585, 586, 721, 723). Here, only the major moments relevant for recombinational repair will be outlined.
RuvA (22 kDa) forms tetramers (676) that bind Holliday junctions; it is the Holliday junction-recognizing activity of E. coli (276, 480). In vitro, at physiological Mg2+ concentrations, Holliday junctions assume a folded conformation (164, 703) (Fig. 11A), which impedes spontaneous branch migration (476). In solution, when Mg2+ concentrations are lowered below a certain level, junctions assume a spread-out conformation (164) allowing rapid spontaneous branch migration (476). Binding of RuvA to a “folded” junction even in the presence of Mg2+ forces it to spread out into the square planar conformation (478) (Fig. 11B). This unfolding of the junctions by RuvA is thought to facilitate their subsequent branch migration.
FIG. 11.
Interactions of RuvA, RuvB, and RuvC with Holliday junctions. The two homologous duplexes (solid and open double lines) are connected by a single Holliday junction. RuvA tetramer is shown as the four-petal flower, RuvB hexameric rings are shown as the trapezoid washers on DNA duplexes, RuvC dimer (E) is shown as the pair of open circles. The direction of DNA movement through RuvAB complex in panels D and E is indicated by arrows. (A) A Holliday junction in a folded conformation, observed in vitro under conditions mimicking physiological ones. (B) A RuvA tetramer binds the junction to open it into a square planar conformation, while two RuvB hexamers bind two opposite arms of the junction. (C) The RuvAB translocase: the same as in panel B, but the second RuvA tetramer binds to the unoccupied side of the junction, locking it in a turtle shell configuration. (D) One of the RuvA tetramers is removed to show junction isomerization, promoted by the pulling action of RuvB (compare with panel B). (E) RuvABC resolvasome: one of the RuvA tetramers has left to allow a RuvC dimer to assume a position for the junction resolution. The resolution sites (diamonds in the opposite DNA strands) are drawn into the junction by the action of RuvB. (F) RuvC cleavage at the resolution sites separates the interacting homologs. The RuvABC resolvasome is not shown.
The crystal structure of RuvA reveals a flower-like tetramer, with a negatively charged convex surface and a positively charged concave surface (508). The crystal structure of E. coli RuvA bound to a Holliday junction shows a single RuvA tetramer holding on its concave side a Holliday junction in the open-square conformation (235), although some in vitro studies indicate that at sufficient RuvA concentrations, the junction must be sandwiched between two RuvA tetramers (481, 761). The crystal structure of RuvA from Mycobacterium leprae shows the latter configuration: two RuvA tetramers form a “turtle shell” with four sideway holes, enclosing a Holliday junction (525).
RuvB (37 kDa) looks like a helicase by sequence gazing, exhibits a weak helicase activity (678, 679), and, like several other helicases (and RecA [see “RecA without DNA” above]), forms hexameric “doughnuts”. RuvB hexamers bind duplex DNA like beads on a string (628). At high concentrations and under special conditions RuvB inefficiently branch migrates (translocates) Holliday junctions (438, 453). RuvB with a mutation in one of the helicase motifs forms hexameric doughnuts but is defective in DNA binding (432). No atomic structure is yet available for RuvB.
RuvC (19 kDa) binds a Holliday junction as a dimer, spreading the junction, almost like RuvA, in a planar conformation (it is not exactly square, perhaps because RuvC is a dimer, not a tetramer like RuvA) (36). RuvC is the long-sought Holliday junction resolvase; it nicks two strands of the same polarity, the same distance from a presumed crossover junction (122, 277). The nicking occurs most efficiently at a degenerate sequence 5′-(A/T)TT↓(G/C)-3′ (568); the minimal requirement for the cleavage is a single thymine on the 3′ side of the break (584). RuvC binds to but does not cleave junctions that lack the nicking sequence (568, 584). The resolution is most efficient when the cleavage site coincides with the position where DNA strands trade partners (34). Nicks introduced by RuvC can be directly sealed by the E. coli DNA ligase (33, 277).
The atomic structure of RuvC resolvase reveals a dimer formed by two wedge-shaped subunits with two positively charged valleys on one side of the dimer (16). A Holliday junction must unfold before it can fit into the two valleys of the RuvC dimer. Studies of RuvC mutants that are able to bind Holliday junctions but are resolution deficient suggest that catalytic domains in the RuvC dimer are situated at the bottoms of the valleys and comprise four closely spaced negatively charged residues (16, 541). The DNA strands that are proposed to lie across the active center (and so are likely to be cut during the resolution) are the “noncrossover strands”. RuvC cleavage of the noncrossover strands was demonstrated with model Holliday junctions, whose branch migration was restricted by tying together two arms at a time (35).
Pairwise Interactions of Ruv Proteins: RuvABC Resolvasome
The indistinguishable phenotypes of ruv mutants in recombination (402, 575) suggested that all three proteins work in a single complex. In vitro experiments with pairwise combinations of Ruv proteins elaborate this idea. RuvA and RuvB interact in solution (581) as well as at Holliday junctions (481). In the presence of RuvA, the concentration of RuvB required for Holliday junction translocation is lowered 20- to 40-fold (438, 453). RuvA function is not limited to RuvB loading at the junctions, since RuvA is required continuously throughout the translocation (438), perhaps to maintain the junctions in the spread-out conformation. Two RuvB doughnuts, sitting on the opposite sides of the RuvA tetramer, pull duplex DNA through their holes, causing the junction to branch migrate (255, 478) (Fig. 11B and D). The force of this pulling can translocate the junctions through extended regions of nonhomology (479). Since DNA is a helix, DNA “pumping” through RuvB is likely to be achieved by duplex rotation relative to the RuvB hexamer.
No cross-linking is detected between RuvA and RuvC in solution (171), suggesting that these two proteins do not interact with each other. Since they both bind Holliday junctions, the two proteins at least have to compete for them. RuvA binds to Holliday junctions more strongly than does RuvC, inhibits RuvC resolution, and, at high concentrations, completely displaces RuvC from the junctions, apparently forming an octameric shell around them (726). However, at subsaturating concentrations, RuvA and RuvC form a cocomplex on the junctions, with the RuvA tetramer apparently occupying a specific side of the junction and RuvC dimer binding to the unoccupied side (726). Holliday junctions in the unfolded conformation have two distinct sides, distinguished by the orientation of DNA strands around the center of the junction. On the one side, DNA strands go 5′ to 3′ clockwise, while on the other side, the 5′-to-3′ orientation is counterclockwise (Fig. 12). RuvA-Holliday junction cocrystals (235) show that RuvA tetramer binds the “counterclockwise 5′-to-3′ side” of the junction, which leaves the other side for RuvC.
FIG. 12.
The “flip” and “flop” sides of a Holliday junction in the open conformation. The same junction is shown from the opposite sides. The 3′ and 5′ ends of DNA strands are marked. In the center, the 5′-to-3′ direction around the junctions is indicated by arrows. RuvC binds to the “clockwise” side of the junction, whereas RuvA binds to the “counterclockwise” side.
RuvC and RuvB proteins form complexes in solution (171) and enhance each other’s reactions with small synthetic Holliday junctions: RuvB accelerates junction resolution by RuvC, while RuvC stimulates branch migration by RuvB (692). With longer, more natural DNA substrates, stimulation of RuvC resolution by RuvB requires the participation of RuvA (764). It is proposed that, due to the site specificity of RuvC resolution, RuvAB is needed to translocate Holliday junctions to resolution sites, where RuvC can resolve them (764). Coimmunoprecipitation allows the formation of RuvABC complexes around Holliday junctions to be detected (147), but it is unclear whether Holliday junctions stimulate interactions between RuvAB and RuvC or whether the junctions simply serve as a scaffold to hold the three proteins together.
In vitro, the presence of RuvC increases the proportion of the “productive” two-ring RuvAB complexes, formed on Holliday junctions at high RuvAB concentrations, under conditions where, in the absence of RuvC, three- or four-ring complexes predominate (691). At the same time, these two-ring RuvAB complexes impose a 20- to 40-fold specificity for the Holliday junction resolution by RuvC. Moreover, in the presence of RuvAB, RuvC resolves partially homologous Holliday junctions in the region of heterology, apparently because the resolution occurs during RuvAB-catalyzed branch migration (691). The name “resolvasome” was offered for the combined activity of RuvA, RuvB, and RuvC to reflect their functioning in a multisubunit complex capable of binding, isomerizing, translocating, and resolving Holliday junctions (337, 722).
RuvAB Translocase
The idea that RuvABC proteins work as a single complex explains why all ruv mutants have the same phenotype (370, 372), but it does not address the fact that only ruvA and ruvB expression is induced by DNA damage in E. coli (Table 1). Perhaps some SOS functions require significantly more RuvA and RuvB than RuvC. The relevant difference in cell physiology between normal and SOS-induced cells is discussed later (see “SOS expression as a compensation”); only the underlying enzymatic mechanisms are dealt with here. The in vitro observation that at saturating concentrations RuvA displaces RuvC from Holliday junctions (726) by assembling an octameric “turtle shell” around the junctions (525) (Fig. 11C) is a clue to these mechanisms, but it does not provide a rationale for them.
The rationale is suggested by the observation that recA mutants with enhanced ability to displace SSB from ssDNA (344, 396) exacerbate the UV sensitivity of ruv mutants (680), as if Ruv proteins normally counteract RecA filament assembly. In fact, one of the first discovered phenotypes of ruv mutants was their inviability in combination with the hyperactive RecA441 mutant protein (474). Therefore, it was proposed that RuvAB complex uses Holliday junctions to disperse the associated RecA filaments (337). Interactions of RuvAB translocase with RecA filaments in vitro support this notion: (i) RuvAB dissociates recombinational intermediates covered with RecA filaments (279, 677), and (ii) RuvAB disperses RecA filaments from duplex DNA (1).
Electron micrographs show that at high RuvB concentrations, four hexameric rings of RuvB surround a junction-bound RuvA tetramer (478, 691), suggesting that RuvA does not direct RuvB binding to particular arms. Although RuvC encourages the formation of the two-ring RuvAB complexes at Holliday junctions (691), the main factor in two-ring complex formation could be the availability of Holliday junction arms for the RuvB binding. In vivo, Holliday junctions are likely to be associated with RecA filaments, and RecA could preclude binding of RuvB to a particular pair of arms. By pumping through themselves the two available arms of a RecA-associated Holliday junction, RuvB hexamers will translocate the Holliday junction towards the RecA filament, which could cause filament dissociation. To complete this speculative picture, after RecA filament is dispersed, one of the RuvA tetramers could leave the junction, allowing access to RuvC resolvase (Fig. 11E).
RecG Helicase
ruvAB mutants are only moderately defective in homologous recombination or in repair of DNA damage (370, 372, 473), suggesting that other activities partially substitute for RuvAB in recombinational repair. Indeed, the moderate defect of ruv mutants in conjugational recombination or in DNA damage repair is aggravated by recG mutations (370). recG mutations by themselves cause only a moderate reduction in cell survival after UV or X-ray treatment (370, 373). recG is the last gene of the spoT operon, encoding a 76-kDa protein; there is no indication that the low-level expression of recG is enhanced during the SOS response (379).
The synergism of ruv and recG mutations is partly explained by the fact that recG encodes another DNA helicase with in vitro activities mostly overlapping those of RuvAB but with some important differences. RecG helicase binds Holliday junctions and drives their branch migration, but, in contrast to RuvAB, the RecG-promoted reaction is blocked by a heterology in excess of 30 nucleotides (380, 729). RecG can also dissociate RecA-made joint molecules containing Holliday junctions, even those still covered by RecA filaments (730). However, the DNA-unwinding activity of the RecG helicase is anemic even in comparison with the weak helicase activity of RuvAB, and the two enzymes translocate along ssDNA in the opposite directions: RuvAB in the 5′-to-3′ direction (678) and RecG in the 3′-to-5′ direction (731).
Three-Strand Junctions and the Hypothetical RecG Pathway
The in vitro activities of RecG make it a possible substitution for RuvAB in the RuvC-promoted junction resolution in vivo. However, ruvC mutants are no more UV sensitive and recombination deficient than are ruvAB mutants, while ruvC recG double mutants are as deficient as ruvA recG or ruvB recG double mutants (370). This indicates that RecG does not substitute for RuvAB in junction translocation in vivo and suggests that RecG uses a different mechanism to resolve DNA junctions.
In ruv mutants, the RecG pathway of junction resolution can be stimulated by the expression of RusA resolvase, whose gene resides on a cryptic prophage (402, 576). In contrast to RuvC, which forms a single complex with a Holliday junction, RusA, depending on its relative concentration, forms four different complexes with four-way junctions (92), suggesting that RusA monomers bind four arms of the junction independently of each other, apparently recognizing DNA branching rather than Holliday junction as a single structure. RecG also tends to bind a Holliday junction from one side and shows a comparable affinity to three-way junctions (424, 729). RecG dissociates three-way junctions by binding to a particular arm and “extruding” the extra DNA strands complementary to the strands of the original arm ahead of the branching point. Consistent with this handling of three-way junctions, RecG dissociates R loops both in vivo and in vitro (197, 257, 699), although it is unable to unwind plain RNA-DNA hybrids of the same length (699). The “extruding” activity of RecG at the three-strand junctions suggests that the RecG-dependent resolution pathway works mostly with three-strand junctions.
This hypothetical mechanism for the RecG-dependent three-strand junction removal is compatible with the observation that while both RuvAB and RecG are able to translocate deproteinized three-strand junctions, only RecG can translocate them when they are still covered with RecA filament (728). In doing so, RecG disrupts RecA-promoted pairing, dissociating the joint molecules. The direction of RecG translocation towards the RecA filament is determined by RecA itself—when the filament is absent, RecG prefers to translocate the same junction in the opposite direction (728). These observations suggest that in vivo RecG pushes three-strand junctions towards the associated RecA filament (Fig. 13). For such a resolution to be productive, nicking of DNA strands at branching points has to occur (Fig. 10B and C and 13A).
FIG. 13.
Hypothetical removal of a single three-strand junction by RecG. RecA filament is shown as an open rectangle with rounded corners; RecA monomers (in panel C) are shown as open circles; RecG is shown as a dotted oval. The small arrow in panel A indicates the position of the required single-strand incision. Explanations are given in the text.
Summary
Theoretically, there are three ways two remove DNA junctions, whether they are four-strand or three-strand junctions. E. coli has two enzymatic systems for DNA junction removal. The well-characterized RuvABC resolvasome translocates four-strand junctions and symmetrically cuts them at preferred resolution sites. The next challenge with RuvABC is to productively include it in the in vitro recombinational repair reactions. The still poorly understood resolution system centered around RecG helicase is hypothesized to remove three-strand junctions by a mechanism yet to be specified.
ruvABC genes are ubiquitous among eubacteria, but their arrangements tend to differ (721). RecG homologs are also found in other eubacteria (187, 194, 290). The hypothetical action of the two resolution systems during particular recombinational repair pathways is discussed in the appropriate subsections (see “Removal of DNA junctions and used Rec filaments” and “The two pathways for DNA junction removal in double-strand end repair”) of the next two sections.
REPAIR OF DAUGHTER STRAND GAPS
If the essence of recombinational repair is RecA polymerization with subsequent homologous pairing and strand exchange, then the essence of recombinational repair pathways is to orchestrate RecA polymerization and depolymerization around specific two-strand lesions. The section on homologous pairing activity of RecA (see above) discussed RecA-catalyzed synapsis and the enzymes that supervise this central step in recombinational repair. The section on resolving recombination intermediates (see above) discussed DNA junction resolution by two enzymatic systems whose job is also to help RecA to depolymerize after completion of repair. The following two sections deal with the complete repair reactions and will introduce more enzymes that function to assist RecA, but, in contrast to inhibition by MutSL or UvrD or dissociation by RuvAB or RecG, these new activities promote RecA polymerization on ssDNA in the presence of SSB.
Origin of Daughter Strand Gaps and Mechanism of Their Repair: Early Studies
When excision repair-deficient E. coli cells are irradiated with low doses of UV, the rate of their DNA synthesis is barely affected (538, 605). Moreover, the molecular weight of nondenatured chromosomal DNA from these cells is not decreased. Even when total DNA from the irradiated cells is denatured with alkali, it still has the same molecular weight as the denatured DNA from unirradiated control cells, confirming the absence of excision repair. However, the newly synthesized DNA in UV-irradiated cells has a lower molecular weight even in excision repair-deficient mutants, indicating single-strand interruptions.
The single-strand interruptions in the newly synthesized DNA after UV irradiation are due to replisome encounters with pyrimidine dimers. When a replisome encounters a noncoding lesion (a pyrimidine dimer or an abasic site) in template DNA, its progress is blocked, as illustrated by the inability of the major E. coli DNA polymerases to bypass such lesions in vitro (59) and by the lower than 0.5% transformation efficiency of an ssDNA carrying a single lesion of this type compared with the lesion-free ssDNA (265, 348). Replication of both strands of the E. coli chromosome in vivo is believed to be discontinuous (see “Elongation phase of DNA replication in E. coli” above), and the other replisomes are likely to restart downstream from the lesion as scheduled, but the DNA segment between the site of the lesion and the position of replication restart will be left single stranded (Fig. 14B). Since such a single-strand gap forms in only one of the two daughter branches of the replicating chromosome, it is called a daughter strand gap. Daughter strand gaps were first detected physically by Rupp and Howard-Flanders (538) and genetically by Cole (117). Their average length was reported to be 800 nucleotides (278), which is approximately half the average length of Okazaki fragments in E. coli (317). A different method produced an estimate of 100 to 200 nucleotides for the size of daughter strand gaps in both the leading and the lagging strands in specific DNA sequences (710), indicating that the gaps might be quite small.
FIG. 14.
Experimentally established principles of the daughter strand gap repair in E. coli. Solid lines indicate parental DNA strands; open lines indicate newly synthesized daughter DNA strands; diamonds indicate thymine dimers; small horizontal arrows indicate single-strand scissions required to resolve the joint molecules. (A) A DNA molecule containing unrepaired noncoding lesions (for example, thymine dimers). (B) Replication of this molecule generates two molecules with single-strand gaps in the daughter strands opposite the lesions (538). Eventually, these gaps are repaired in a RecA-dependent way (606). (C to H) Experiments by Rupp et al. (539) had revealed that after gap closure, the newly synthesized strands are connected with the parental strands (strand exchange), suggesting a model for recombinational repair of daughter-strand gaps (C to E). Later, Ley (357) found that the gaps are transferred from the daughter strands to the parental strands, while Ganesan (199) found that the thymine dimers are transferred in the opposite direction, from the parental strands to the daughter strands, suggesting the formation and resolution of Holliday junctions (F to H).
One way to fill a daughter strand gap would be to modify a stalled replisome so as to allow it to carry out translesion DNA synthesis (see “Backup repair of daughter strand gaps: translesion DNA synthesis” below). However, experiments by Rupp, Howard-Flanders and colleagues revealed a peculiar feature of the daughter strand gap repair in E. coli: filling in the gaps was accompanied by formation of hybrid DNA strands in which segments of the newly synthesized strands were linked with segments of the template strands (539). The number of such exchanges of strands between sister duplexes roughly coincided with the number of UV lesions in the template DNA, indicating that daughter strand gap repair is accompanied by strand exchange (Fig. 14C to E). This phenomenon led the authors to propose that daughter strand gaps in E. coli are filled, with the help of the intact sister duplexes, by recombinational repair (266, 539). This conclusion was in line with the findings that conjugative transfer of UV-irradiated DNA stimulates genetic recombination (734) and that recA mutants, deficient in homologous recombination, are also deficient in daughter strand gap repair (199, 268, 507, 606).
The repair-associated strand exchange was further confirmed by the demonstration that in excision repair-deficient cells, irradiated parental DNA acquires patches of daughter DNA containing the gaps, whose number is slightly smaller than the number of UV lesions (357, 533). Together with the complementary demonstration that during the daughter-strand gap repair the initial lesions in the old DNA strands have a 50% probability of being transferred to the newly synthesized DNA strands (199) these results suggested the formation of Holliday junctions (see “Two-strand repair” above) and their alternative resolution (Fig. 14F to H).
It is noteworthy that the described repair reaction mends the daughter strand gaps but does not deal with the original one-strand lesions that have caused them (Fig. 14E and H). The original noncoding lesions have to be removed by excision repair after the gap has been filled in. In excision repair-deficient mutants, used in the physical studies of daughter strand gap repair, the original lesions persist in DNA for many generations after UV irradiation, being gradually diluted as cells multiply. Because of that, it is said that recombination in excision repair-deficient mutants is a mechanism of damage tolerance rather than repair.
In E. coli, daughter strand gaps are mended by the RecF pathway of recombinational repair, which is named after the first discovered gene specific for this pathway (200, 261, 535). A tentative sequence of postulated events during this process is as follows (108) (Fig. 15): in preparation for synapsis, the RecOR complex descends on the SSB-complexed daughter strand gap, perhaps guided by the RecFR complex. The presence of RecO allows RecA polymerization on the SSB-complexed ssDNA. During the synaptic phase of the reaction, RecA filament finds an intact duplex, homologous to the single-strand gap, and pairs them (Fig. 15B). The synapsis is facilitated by two different topoisomerases: DNA gyrase relieves positive supercoils, generated in the intact duplex due to the strand invasion, while topoisomerase I (Topo I) relieves negative supercoils in the new duplex between the invading and the resident strands. Pairing of the damaged and the intact DNA molecules allows filling in of the gap by a DNA polymerase. In the postsynaptic phase of the reaction, RuvABC resolvasome or RecG helicase removes Holliday junctions and the associated RecA filaments, completing faithful repair of the daughter strand gap. If an intact homologous duplex cannot be found, UmuD′C complex modifies the RecA filament and the replisome to allow translesion DNA synthesis, restoring the duplex at the price of possible mutagenesis. The flesh of experimental observations that animate this conceptual skeleton for daughter strand gap repair is presented below.
FIG. 15.
Overview of daughter strand gap repair. (A) A DNA molecule with a daughter-strand gap. T=T, the thymine dimer that caused the gap. (B) Synapsis of the gapped molecule with the intact homologous DNA. (C) Filling in of the gap and repair of the lesion that caused the gap. (D) Disengagement of the two DNA molecules. The RuvC cleavages are indicated by small vertical arrows in panel C. Additional explanations are given in the text and in the figure.
Presynaptic Phase of Daughter Strand Gap Repair: RecF, RecO, and RecR
recF, recO, and recR: mutant phenotypes.
Operationally, the mutants deficient in the presynaptic phase of daughter strand gap repair should be as deficient in daughter strand gap closure as are recA mutants, which are blocked at the subsequent synapsis. In vitro, SSB protein helps RecA at both presynapsis and synapsis; ssb mutants are deficient in recombinational repair of UV damage (359, 732) but have not been specifically tested for the deficiency in daughter strand gap closure. There are four genes which, when mutated, create mutants deficient in daughter strand gap closure. One of them is lexA (200); LexA is the repressor of the SOS regulon, and its role in daughter strand gap closure is likely to allow increased RecA production in response to DNA damage (see “Levels of SOS induction” above). The other three genes are recF (200, 534), recO (680), and recR (680). In addition to the deficiency in gap closure, recF mutants are deficient in the reciprocal process, the transfer of lesions from the parental to the daughter strands (716), suggesting that the reason why daughter strand gaps in recF mutants cannot be repaired is because homologous exchange is blocked.
A null recF mutant has reduced viability (547). Genetic analysis of UV resistance in E. coli shows that the recF, recO, and recR genes belong to the same epistasis group; i.e., double mutants with mutations in these genes have the same survival after UV irradiation as do the single mutants (378, 400). recF, recO and recR mutants show no decrease in homologous recombination following conjugation or transduction, but they are deficient in plasmid recombination (106, 399). Accordingly, although recF, recO, or recR mutants are deficient at filling in daughter strand gaps (reference 680 and references therein), they have no effect on the double-strand end repair (see “Origin and repair of double-strand ends” below). The UV sensitivity of these mutants is partially suppressed by the same non-null mutations in recA (701, 702, 708). The SOS induction curves of the three mutants essentially overlap: after UV irradiation, the SOS response in recFOR mutants is delayed for a period corresponding roughly to a single round of DNA replication but then is induced to a greater degree than in the WT cells (241, 727) (see “Evidence for replication fork disintegration” [below] for a possible explanation).
The SOS induction defect of recFOR mutants suggested that their UV sensitivity is due to their inability to turn on the SOS response and, in particular, to overproduce the RecA protein in response to UV irradiation. Amplification of RecA, together with other SOS proteins, due to defective LexA repressor does suppress a recF deficiency slightly (669). However, overproduction of RecA alone due to a promoter-up mutation actually decreases UV survival of recF mutants (110) and does not improve the slow SOS induction in them (668). Therefore, RecA amplification is unlikely to be the function of RecFOR in vivo; rather, the proteins must assist RecA directly.
recF, recO, and recR: possible replisome connection revealed by gene structure.
recF is the third gene in a four-gene cluster which starts with dnaA (DnaA protein initiates chromosomal DNA replication at oriC) and also includes dnaN (a gene coding for the “sliding-clamp” subunit of DNA pol III) and gyrB (B subunit of DNA gyrase, the enzyme that introduces negative supercoiling into the E. coli chromosome [see “DNA gyrase” below]). Although recF has its own promoter inside the coding sequence of dnaN, it is thought that dnaA, dnaN, and recF constitute an operon under the control of dnaA promoters (485). The conserved structure of this chromosomal region in even distantly related eubacteria further argues for the biological significance of this combination of recF with replication genes (references 196 and 405 and references therein). The complex regulation of the recF gene expression (17, 485, 549) ensures that the RecF protein is maintained at a low level, less than 190 monomers per cell (394), which is, maybe coincidentally, close to the number of DnaN dimers (about 150 per cell) (79). When E. coli cells enter stationary phase, expression of dnaN and recF increases and becomes independent of dnaA expression (697). Overproduction of RecF protein adversely affects the viability and UV resistance of growing cells (221, 551).
recO is the last gene in a three-gene operon (449, 646). The first gene in the operon is rnc, encoding RNase III, which cleaves specific dsRNA structures (the enzyme participates in maturation of rRNA and modifies some mRNAs). The second gene is era, coding for an essential cytoplasmic membrane-associated GTPase suspected of participating in membrane signaling (361). recO is poorly expressed due to a weak promoter, a weak ribosome-binding site, and abundance of rare codons (449).
recR is also the last, poorly expressed gene in a three-gene operon. The first gene in this operon is dnaX, encoding the gamma and tau subunits of DNA pol III; these proteins are the main components of the “gamma complex,” which functions as the replisome frame and loads DnaN clamps onto primed template DNA (reviewed in reference 297). The function of the second gene in the operon, orf12, is unknown (188, 754). recR is expressed equally from both the dnaX promoter and from the second promoter, which it shares with orf12 and which is buried in the dnaX coding sequence (188). In B. subtilis, the organization of this chromosomal region is similar (7), arguing against fortuitous association of recR and dnaX.
The positions of both recF and recR promoters inside the coding sequences of the genes for the structural subunits of the replisome suggest that their expression is coregulated. Moreover, the colocalization of recF and recR in operons with the genes coding for the interacting components of the replicative DNA polymerase suggests the physical association of RecF or RecR proteins with DNA pol III, or interactions between these proteins and DNA pol III during the repair process, or even interactions between RecF and RecR themselves.
Properties of RecF, RecO, and RecR and their influence on RecA-promoted reactions in vitro.
The finding that UV sensitivity of recF mutants is partially suppressed by certain non-null recA mutations allowed Clark, as early as in 1980, to propose that RecF acts to stabilize RecA binding to ssDNA (104). The extension of this finding, on the one hand, to recO and recR mutants and, on the other hand, to other similar RecA mutants strengthened this idea and suggested that the RecFOR proteins function as a complex (708). Biochemical characterization of the mutant RecA proteins that do better in vivo in the absence of RecF, RecO, or RecR showed that they are more proficient than the WT RecA in displacing SSB from ssDNA in vitro (344). SSB is the main RecA competitor in vivo: SSB overexpression sensitizes cells to UV (65, 431), delays the SOS response, and inhibits recombination (445). Therefore, in vitro attempts at characterizing RecFOR activities were guided by the expectation that these proteins, working together, would help RecA to polymerize on ssDNA complexed by SSB.
Before discussing these in vitro attempts to emulate the presynaptic phase of daughter strand gap repair, we should consider the likely structure of daughter strand gaps in vivo (Fig. 16). The 5′ end of the discontinuous strand at the gap still has an RNA oligomer attached that was used to prime the downstream Okazaki fragment. The 3′ end of the gap is determined by the noncoding lesion in the template strand that blocked the completion of this Okazaki fragment; DNA pol III is probably still idling there waiting for instructions from the cell (the retention of the replisome at the noncoding lesions is suggested by the DNA synthesis shutdown after DNA damage in certain Rec− mutants [see the next section]; apparently, rec functions are needed to disengage the stalled replisomes from DNA lesion). In between these two ends, the whole length of the gap is complexed with SSB. If RecA is to polymerize on such a gap, it needs (i) to be directed to the gap and (ii) to be assisted in displacing SSB. The general biochemical properties of RecF, RecO, and RecR, especially of their pairwise combinations, make them the most suitable candidates for these two presynaptic roles.
FIG. 16.
Presynaptic phase of the daughter-strand gap repair: RecA filament assembly and replisome reactivation. The replisome is depicted as a big open oval, and its DnaN subunit (the clamp) is indicated as a smaller open rectangle with rounded corners. The 3′ end of the nascent DNA is stalled at the noncoding lesion (the irregularity in the lower strand). SSB is shown as quartets of open circles around single-stranded region; RecFR (small black rectangles) are associated with the RNA primer (wavy segment in the upper strand at the very right). The RecA filament is shown as a hatched rectangle with rounded corners spreading to the left. Explanations are given in the figure and in the text.
RecF is a 40-kDa protein which, in the absence of divalent cations and nucleotide cofactors, binds linear ssDNA with some preference for the ends (221). In the presence of ATP and magnesium, RecF binds both ssDNA and dsDNA (395). In fact, binding to DNA stimulates ATP binding by the protein, although no ATP hydrolysis is detected (395). ATP binding is probably important for the RecF function, since a mutation in the nucleotide-binding domain of the protein results in its inactivation (548). RecO is a 26-kDa protein that binds both dsDNA and ssDNA in the presence of magnesium and does not bind nucleotide cofactors (388). It can also promote the renaturation of complementary DNA strands independently of a nucleotide cofactor. RecR of E. coli has a calculated molecular mass of 22 kDa and is yet to be characterized biochemically.
When added in vitro to the SSB-containing RecA-promoted strand exchange reaction between circular ssDNA and linear duplex DNA, RecF does not show any effect at low concentrations and inhibits the reaction at high concentrations (394, 684). This inhibition is not observed if RecO and RecR are also present in the reaction. In contrast to the adverse effect of RecF, RecO and RecR stimulate the RecA-promoted reaction, with maximal stimulation being observed at a RecO-to-RecR ratio of 1:1 (684). RecF is completely dispensable for this stimulation. RecORs stimulate the rate of initiation of joint molecule formation but do not influence the rate of strand exchange itself (684), suggesting that RecOR, possibly working in a complex, help RecA to displace SSB protein from ssDNA.
Further studies have shown that RecO can physically interact with both RecR and SSB (685). Moreover, RecO and RecR bind to the SSB-covered ssDNA without displacing SSB. This RecO and RecR binding allows more efficient RecA binding to the SSB-covered DNA. Although RecA cannot bind to saturation to ssDNA covered with SSB-RecO-RecR complexes, the resulting RecA filaments are fully proficient in pairing and strand exchange (685). In a different study, RecO and RecR together (but not separately) prevented RecA dissociation from linear ssDNA as a result of competition with SSB (569).
Some recent studies generated more questions than answers. The initially promising report of the preferential RecF binding to duplex DNA with single-strand gaps in the presence of a nonhydrolyzable ATP analog (243) could not be confirmed in another laboratory (719). RecO was found to interact with RecF, but only in the absence of ATP (242), raising questions about the biological significance of this finding. In the presence of a nonhydrolyzable ATP analog, RecF was found to coat duplex DNA as a uniform filament, but the binding became sporadic in the presence of ATP (718). RecR protein showed no binding to duplex DNA under the same conditions, but RecF and RecR together completely coated duplex DNA in the presence of ATP (718). Not only did the limited quantities of RecF and RecR in vivo make one wonder about the significance of these results, but also in itself the capability for indiscriminate binding to duplex DNA in vivo would be more deleterious than helpful.
However, the most recent finding put some of the earlier results in a more agreeable perspective. On a circular DNA with a single-strand gap, RecFR proteins, binding randomly to the duplex portion of the molecule, limit RecA filament polymerization mostly to the gap region, which appears to fulfill the targeting function (719). To explain the contrast between the huge requirements for RecFR in their in vitro setup and the low intracellular copy number of these proteins, the authors proposed that RecFR complex is deposited on the replisome side of the gap by the replication machinery and functions there to limit the nonproductive spread of the RecA filament into the adjacent duplex region (719).
Replisome reactivation and model for RecFOR catalysis of RecA polymerization at daughter strand gaps.
In the WT E. coli cells irradiated with sublethal doses of UV, DNA synthesis is inhibited within 5 to 10 min (162, 298, 300, 738). It resumes shortly thereafter at the stalled replication forks but never again in ssb, recA, or lexA (Ind−) mutants (162, 300, 712, 738) and only slowly in recF and recR mutants (128). recO mutants have not been tested for resumption of DNA synthesis after UV irradiation but are expected to be defective too. It is thought that when a replisome stops at a noncoding lesion in a template DNA, it needs to be disengaged from the damaged DNA (“reactivated”) to restart downstream (300).
A plausible scenario for replisome reactivation is based on the fact that due to its directional polymerization (362, 514, 571, 572), the RecA filament assembles along a daughter strand gap towards the stalled replisome (Fig. 16). Upon reaching the replisome, the filament could nudge the replisome back into the completely duplex region. Once in the duplex region, the replisome should be able to disassemble on its own, since this ability is critical for the replisome cycling from completed Okazaki fragments to new primers. In vitro experiments indicate that the replisome will not cycle from duplex DNA unless it is provided with an excess of DnaN protein (78, 470, 577). There are 300 DnaN molecules (150 DNA clamps?) per cell under regular conditions (79), which is at least a 10-fold molar excess over the DNA pol III holoenzyme (747); dnaN expression is further increased in response to DNA damage (287, 641).
A way to accommodate both the absence of replisome reactivation in recF and recR mutants (128) and the proposed deposition of RecFR from the back of a replisome (719) is to suggest that RecFR are replisome-releasing factors that disengage it from the DnaN doughnut when the replisome is displaced by the growing RecA filament into the duplex DNA. RecFR could then be deposited to the duplex DNA in place of the replisome to limit the spread of the RecA filament. However, the general idea that RecFR inhibits RecA polymerization (719) while RecOR promotes RecA assembly on SSB-complexed ssDNA (569, 684, 685) predicts that recA mutants complementing recO defect should exaggerate the recF defect and vice versa. The prediction contradicts the established fact that all recA mutants complementing the recO defect also complement the recR and recF defects (701, 702, 708), while in vitro these mutant RecA proteins are more proficient than the WT RecA in displacing SSB from ssDNA (344).
There is a different way to place the RecFR complex at the daughter strand gap, which does not contradict the genetic data. The function of RecF and RecR might be to sense a stalled replisome and to bring RecO to the lingering SSB-covered ssDNA gap. RecFR complex could function as such a sensor if it is deposited on the 5′ side of every started Okazaki fragment (Fig. 16). If one has to speculate wildly, RecF could be associated with DnaN monomers in solution but would dissociate from them to bind the RNA primers when the DnaN monomers are dimerized by DnaX around the primers (see “recF, recO, and recR: possible replisome connection revealed by gene structure” above). For example, RecF could be taken from DnaN by RecR, which, in turn, could be initially associated with DnaX. As a result of these subunit exchanges, when a replisome starts DNA synthesis from a primer, RecFR complex could be left behind on the 5′ end of the primer. From there, RecFR should be able to target RecO to the junction of ssDNA and duplex DNA. RecOR complex would bind SSB-covered DNA in the vicinity of the transition between ssDNA and duplex DNA and promote RecA filament formation. If the template for the previous Okazaki fragment has been intact, the charging replisome would bump RecA, RecF, RecO, and RecR, together with SSB, off the DNA and complete the Okazaki fragment. However, if the replisome is stalled at a lesion, the RecA filament will reach the stalled replisome and promote its dissociation. Similar ideas have been discussed by others (395, 550). The placement of the RecFR on the 5′ ends of Okazaki fragments can also explain their role in the alternative mechanisms of double-strand end repair (see “Double-strand end repair in the absence of RecBCD” below).
The proposed role for RecF is consistent with the observation that a RecF deficiency is partially suppressed by overproduction of RecR alone (550). Significantly, although RecO is required for this suppression, it does not have to be overproduced, suggesting that of the three proteins, RecF directs the repair effort to daughter strand gaps, RecR acts as a liaison between RecF and RecO, and the RecOR complex assists RecA in polymerization on the SSB-bound ssDNA. These hypothetical schemes highlight the importance of the still missing information on how RecFOR proteins interact with the components of the replisome, especially DnaN and DnaX.
DNA Topoisomerases and Synaptic Phase of Daughter Strand Gap Repair
Since duplex DNA is a helix in which single strands are intertwined around each other every 10 bp, the RecA-catalyzed pairing of the daughter strand gap with an intact duplex DNA entails two opposite topological reactions, catalyzed in E. coli cells by two different DNA topoisomerases.
DNA gyrase.
First, the positive supercoiling, generated when the two strands of the intact duplex are pulled apart to accommodate the invading strand, must be neutralized (Fig. 17). In vitro, negative supercoiling of the duplex facilitates the RecA-catalyzed invasion of a single strand (583). Since the E. coli chromosomal DNA is negatively supercoiled (489), the invasion of the third strand decreases the local negative supercoiling, which is then enzymatically restored to its original level.
FIG. 17.
Topoisomerase requirements during synapsis. Regular DNA superhelicity is shown in the top and bottom double helices. In the two middle panels, the helices are either overwound (too much coiling) or underwound (missing coils). DNA gyrase removes extra coils from the original (black-black) duplex, while DNA Topo I (TopA) introduces the missing coils in the hybrid (black-white) duplex.
The maintenance of negative supercoiling in the E. coli chromosome is a function of DNA gyrase. DNA gyrase mutants or cells treated with DNA gyrase antagonists are significantly impaired in their ability to perform recombinational repair of UV-induced DNA damage (239, 500, 704). DNA gyrase is a heterotetramer (A2B2) of two A subunits (97 kDa) and two B subunits (90 kDa). The subunits are encoded by two separate genes, gyrA and gyrB, which in many bacteria are organized into an operon but in the E. coli chromosome are located far apart (reviewed in reference 513). GyrA is a catalytic subunit, which binds and manipulates DNA, while GyrB is an ATPase, which binds and manipulates the GyrA-DNA complex. The enzyme wraps around itself a segment of duplex DNA, makes a staggered double-strand cut in one portion of the segment, passes the uncut portion of the segment through the resulting gap, and reseals the gap (reviewed in reference 39). As a result, the number of times one strand is wound around the other in the DNA molecule is decreased by two, which amounts to introduction of two negative supercoils in the molecule.
The extent of the RecA-promoted in vitro reaction between a gapped circle and a homologous supercoiled circle is only 1/20 of the total length of the supercoiled substrate (136), apparently limited by the supercoiling density in the DNA of E. coli (489). However, if DNA gyrase is present in the reaction mixture, complete hybrid circles are formed, in which the entire strands come from different parental molecules, attesting to the stimulating effect of negative supercoiling on RecA-promoted strand exchange (89).
Topoisomerase I.
When two interacting strands of any nature run side by side without intertwining, they are said to be paranemic, in contrast to the situation when two interacting strands intertwine to form a double spiral (plectonemic interaction) (271). The first contacts of a daughter strand gap with the complementary strand of an intact duplex are necessarily paranemic (Fig. 17). In other words, the two DNA strands have so many negative supercoils that they can be freely separated from each other. The paranemic duplex is converted to a plectonemic duplex when the excess of negative supercoils is removed in a reaction catalyzed in E. coli by DNA topoisomerase I (Fig. 17).
Mutants with mutations in DNA Topo I are sensitive to UV light (475, 631), although the most strongly affected stage of the repair remains to be determined. DNA Topo I is a single polypeptide of 110 kDa and is encoded by the topA gene (675). Since the relaxation of negative supercoiling in DNA is a restoration of the energetically favored conformation, the enzyme does not need high-energy cofactors. The mechanism of its reaction is fundamentally different from that of DNA gyrase. DNA Topo I of E. coli cuts only one DNA strand, preserves the energy of the phosphodiester bond by covalently attaching itself to the 5′ phosphate of the break, rotates one side of the break around the intact strand, and reseals the break (reviewed in reference 39). One such manipulation reduces the number of negative supercoils in a DNA molecule by one.
In vitro RecA is able to pair a ssDNA circle with a homologous supercoiled duplex circle, but the product is unstable due to its paranemic nature; if Topo I is present, this RecA-promoted reaction yields stable products in which the incoming strand forms an intertwined duplex with its complement (138).
Postsynaptic Phase of Daughter Strand Gap Repair
To be completed, recombinational repair reactions require the participation of two more groups of enzymes. General DNA metabolism is carried out by a set of enzymatic activities which includes DNA topoisomerases, DNA helicases, DNA polymerases, and DNA ligases. These DNA-keeping enzymes replicate and repair DNA; they also participate in completion of recombinational repair. The other group of the postsynaptic activities is specific to recombinational repair—these enzymes handle DNA junctions (see “Resolving recombination intermediates” above).
One-strand repair: lesion removal and filling in of the gap.
As RecA catalyzes the invasion of the single-stranded region of a gapped DNA molecule into an intact homologous duplex, a pair of branch points at which the invading strand trades places with the homologous resident strand is formed. The features of RecA polymerization in vitro (see “Filament formation by RecA around ssDNA” above) suggest that in vivo RecA filament assembles on daughter strand gaps in the direction of the lesion that caused the formation of the gap and probably spreads past the lesion into the neighboring duplex DNA. RecA will eventually drive the lesion-proximal DNA junction in the same direction, converting the corresponding three-strand junction into a four-strand (Holliday) junction (Fig. 18).
FIG. 18.
Synaptic and postsynaptic phases of daughter strand gap repair. This figure is a sequel to Fig. 16. The RecA filament is shown as a hatched rectangle with rounded corners. From top to bottom, the stages are pairing with an intact homolog, repair of the gap, and removal of the DNA junctions and the associated RecA filament. Explanations are given in the figure and in the text.
DNase protection experiments show that during the initial stages of four-strand exchange, RecA filament fully protects only a single DNA strand—the one that has been single stranded in the gap (DNA1) (98, 99). Later, in these experiments that were done in the absence of SSB, the RecA protection switches to the strand that lost its partner as a result of the strand exchange. The important result of these studies is that the displaced strand, when it forms the alternative duplex, is apparently accommodated outside the filament and is likely to be available for DNA polymerases. Therefore, one can speculate that the next step in the daughter strand gap repair in vivo is filling in the gap (Fig. 18). Now is the time to mention mutants with mutations in DNA-keeping genes, partially defective in daughter strand gap closure, like uvrD (helicase II) (533), polA (DNA pol I) (30, 574), dnaB (replicative DNA helicase) (281), and polC/dnaE (the catalytic subunit of DNA pol III) (282), as well as the unsurprising deficiency of a DNA ligase mutant (755). The filling in of the daughter strand gaps is likely to be carried out by DNA pol I, although in polA mutants the gaps are apparently closed by DNA pol III (with the help of DnaB?), since the polA dnaE double mutant is deficient in gap closure (564). It was speculated that RecFR complexes are deposited by the replisomes at the 5′ ends of Okazaki fragments (see “Replisome reactivation and model for RecFOR catalysis of RecA polymerization at daughter strand gaps” above). UvrD helicase (see “Possible disruption of pairing of insufficient length by helicase II” above) might be needed to displace RecFR from the RNA primer on the 5′ end of the gap before DNA pol I can clip this primer off, so that DNA ligase could seal the last nick.
Removal of DNA junctions and associated RecA filaments.
Genetic data suggest that both the RuvABC resolvasome and the RecG helicase participate in junction removal after the daughter strand gap repair, since mutating away either activity makes cells sensitive to UV, but neither mutation shows synergistic interactions with recF mutations in relation to UV sensitivity (372, 373). The postsynaptic phase of the daughter strand gap repair is understood in its gross details (see “Resolving recombination intermediates” above), but the real mechanisms have yet to be modeled in vitro, and so the description offered is inevitably speculative. When the gap is filled, the other three-strand junction could be converted into a Holliday junction, or it may stay three stranded for a while, if the repair DNA synthesis fails to traverse it. If this right three-strand junction is converted to a Holliday junction, both junctions and the RecA filament in between could be removed by the RuvABC resolvasome (see “Pairwise interactions of Ruv proteins: RuvABC resolvasome” above) (Fig. 9B). The alternative pathway for the junction and RecA filament removal does not depend on a particular configuration of the right junction. The right junction can be either converted into a Holliday junction or resolved by an unspecified cleavage activity, as is sometimes assumed (730), or even left as is. The translocation by the RecG helicase (see “RecG helicase” above) of the left (Holliday) junction towards the second junction will remove both junctions anyway (Fig. 9A or C).
Backup Repair of Daughter Strand Gaps: Translesion DNA Synthesis
Recombinational repair of daughter strand gaps is impossible when the intact sister duplex cannot be found; the likelihood of this situation increases with increasing DNA damage. For example, both daughter chromatids could be damaged in the same sequence, or the sister chromatid could be degraded due to a downstream lesion. Many bacteria have a backup mechanism to repair lingering daughter strand gaps without recombination, by translesion DNA synthesis. When this backup mechanism is turned on, RecA filament is dispersed from the gap and the replisome is modified so as to be able to bypass the lesion. Although nonrecombinational by its nature, this process is both regulated by RecA and requires the direct participation of RecA (reviewed in references 602 and 743).
RecA regulates translesion DNA synthesis at two levels. The first level of regulation is via LexA cleavage and SOS induction. Prolonged SOS induction increases the expression of the umuDC operon (23, 24) (see “Levels of SOS induction” above). Besides RecA itself, UmuC and UmuD are the only SOS proteins required for the translesion DNA synthesis (612); both proteins participate in this process directly. However, first UmuD has to bind to RecA filament and to cleave itself in a reaction similar to the autocleavage of LexA and prophage repressors (77). RecA-promoted autocleavage of UmuD is the second level of RecA involvement in translesion DNA synthesis. In vitro, this inefficient reaction, as well as autocleavage of prophage repressors, requires higher concentrations of Mg2+ than does RecA-promoted autocleavage of LexA; processing of UmuD in vivo is also rather inefficient (565). This is likely to ensure that UmuD and phage repressors are cleaved late during SOS induction (see also “Levels of SOS induction” and “Cleavage of LexA repressor by RecA filament” above). The product of autocleavage, UmuD′, combines with UmuC to form a complex active in catalyzing translesion DNA synthesis (509, 647).
With the development of the in vitro system for translesion DNA synthesis, the molecular mechanisms by which the active UmuD′2C complex catalyzes lesion bypass are beginning to be elucidated. Two observations, that translesion DNA synthesis is inhibited by overproduction of the DnaN “clamp” subunit of DNA pol III (640) and that overexpression of UmuC is poisonous for strains with defective DNA pol III (469), suggested that UmuD′2C comes in direct contact with the DnaN clamp. UmuD′2C turned out to be a nonprocessive error-prone polymerase (DNA pol V), capable of bypassing noncoding lesions (648). UmuD′2C is hypothesized to replace the stalled DNA pol III at the lesion and to synthesize several nucleotides across the damaged template, to be promptly replaced by DNA pol III on the other side of the lesion (Fig. 19).
FIG. 19.
Model for the translesion DNA synthesis. This figure is a sequel to Fig. 16 and shows an alternative to the mechanism illustrated in Fig. 18. The RecA filament is shown as a hatched rectangle; the DnaN clamp is shown as a small open rectangle; UmuC is shown as a small black rectangle associated with the DnaN clamp; UmuD is shown as small black dimer circles, some of them associated with UmuC; the replisome is shown as an open oval. Explanations are given in the figure and in the text.
How does UmuD′2C find the unfillable single-strand gap? UmuD′2C complex binds ssDNA in the absence and the presence of RecA (75), but its way of getting to the unrepaired lesion appears to be via the RecA filament because, besides its two regulatory roles in translesion DNA synthesis, RecA is required directly, together with UmuD′, UmuC proteins, and DNA pol III (509, 647). The clue to the mechanism of direct RecA participation in translesion DNA synthesis is the observation that expression of the Umu proteins inhibits recombinational repair by interfering with RecA action (62, 610). Therefore, RecA is proposed to direct UmuD′2C towards the site of the unrepaired lesion, while UmuD′2C destabilizes the RecA filament (Fig. 19) (62, 610).
The observation that RecA filaments stabilize UmuD′2C complex against proteolysis in vivo supports the idea of a direct interaction of UmuD′C and RecA (192). Generally, both UmuD and UmuC are rapidly degraded in vivo by the Lon protease, while the majority of the newly formed UmuD′ dimerizes with unprocessed UmuD and is degraded by the ClpXP protease (191). The strategy of UmuD proteolysis apparently prolongs the delay in formation of the active and stable UmuD′ dimer, and for a good reason. Since DNA pol V has to insert random bases opposite some noncoding lesions, the translesion DNA synthesis often generates point mutations and is also known as the SOS mutagenesis. Translesion DNA synthesis is definitely not the best way to repair a lesion, but since umu mutations do confer moderate UV sensitivity (296), this error-prone DNA synthesis may be the life-saving option under conditions of massive DNA damage. Besides, translesion DNA synthesis is the only way to repair a lesion in a situation like conjugal transfer, when a single strand of a plasmid DNA is transferred from one bacterial cell to another. During the postconjugational synthesis of the complementary strand, there is no sister duplex to rely on if a noncoding lesion is encountered in the template strand. In fact, many conjugative broad-host-range plasmids carry their own genes for translesion DNA synthesis (633). This is helpful because even among the enterobacteria not all species are capable of induced mutagenesis, even though almost all of them carry genes homologous to the umuDC operon of E. coli (565).
Summary
Daughter strand gaps are formed when DNA replication encounters a noncoding lesion in a template DNA and reinitiates downstream, leaving behind a single-stranded region adjacent to the lesion. In E. coli, daughter strand gaps are closed by the RecF pathway of recombinational repair, via homologous pairing and strand exchange of the gapped chromatid with the intact sister chromatid. Other eubacteria seem to follow the same recombinational route: daughter strand gaps are repaired by recombination in Haemophilus influenzae (350, 600, 706) and in B. subtilis (159, 160). Moreover, judging by the universal presence of the recF homologs in other eubacteria (196, 405), the RecF pathway is ubiquitous. Genes for the translesion DNA synthesis (a backup repair of daughter strand gaps) are also widespread among eubacteria (633, 743).
The interactions of individual components of the daughter strand gap repair machinery are beginning to be elucidated in vitro. The prominent issue is the interaction of the RecF, RecO, and RecR proteins with the replisome, on the one hand, and RecA on the other. The replisome reactivation is another phenomenon that begs investigation, both in vivo and in vitro. The postsynaptic phase of the daughter strand gap repair is still a realm of pure speculations. The replacement of the recombinational repair machinery with the one for the translesion DNA synthesis recently became the area of exciting research.
DOUBLE-STRAND END REPAIR
Origin and Repair of Double-Strand Ends
The success in deciphering the mechanism of daughter strand gap repair (see “Origin of daughter strand gaps and mechanism of their repair: early studies” above) is, to a large extent, attributable to the strict use of excision repair-deficient mutants. Not only does the absence of excision repair make cells dependent on the daughter strand gap repair for the UV damage tolerance, but also it delays the appearance of a different type of two-strand lesions, which otherwise would have made the results of physical studies difficult to interpret. Armed with the understanding of the origin of daughter strand gaps and the mechanism of their repair, we can now face the complicated picture of recombinational repair in excision repair-proficient cells.
In contrast to cells which cannot excise pyrimidine dimers (247, 538, 605), excision repair-proficient cells stop DNA synthesis after UV irradiation (162, 605) and, if the UV dose was high, initiate a new round of DNA replication from the origin (46, 247, 397). Surprisingly, the old replication forks seem to be temporarily abandoned, since the DNA synthesized just before the irradiation becomes susceptible to degradation (128). This abandonment becomes permanent at higher UV doses (8, 162), and the total chromosomal DNA in excision repair-proficient cells becomes susceptible to a limited degradation (63, 105); in the absence of RecA, the chromosomal degradation after UV irradiation is uncontrollable (105, 128).
This bizarre behavior of the excision repair-proficient cells is rationalized by the data from neutral sucrose gradients, which allow the intact chromosomes to be separated from chromosomal fragments. Immediately after UV irradiation, the sedimentation pattern of the E. coli chromosome in neutral sucrose gradients is the same as that of an unirradiated control. However, if the irradiated cells are incubated in the growth medium, the neutral sucrose gradients indicate that the bacterial chromosome becomes fragmented. This fragmentation is suppressed in excision repair-deficient mutants (60, 667) or by inhibition of DNA replication (649), suggesting that DNA replication on template DNA that is undergoing excision repair causes chromosomal breakage.
The nature of this breakage is revealed in strains defective in the closure of ssDNA interruptions, like DNA ligase mutants (471, 483) or DNA pol I mutants (383, 635). If these mutants are allowed to replicate their DNA, their chromosome becomes similarly fragmented, as judged by its susceptibility to degradation by a nuclease specific for the double-strand ends (443, 450). Therefore, replication of a DNA template with single-stranded interruptions generates dsDNA interruptions.
Hanawalt may have been the first to propose how excision gaps interfere with DNA replication (234); later, similar schemes were advanced on several occasions (76, 142, 218, 333, 540, 567, 597). They all envision the collapse of a replication fork as it reaches the single-strand interruption in the template DNA, as a result of which the double-strand end is separated from the full-length duplex (Fig. 20A). If both forks of a replication bubble have collapsed due to nicks in the same DNA strand, in the neutral sucrose gradients this will look like chromosomal DNA fragmentation (Fig. 20B).
FIG. 20.
Replication-dependent origin of double-strand ends. (A) Replication fork collapse at a single-strand interruption in template DNA. The replisome is shown as the square group of circles, speeding from left to right along the DNA while replicating it. (B) Preexisting interruptions in the same DNA strand at both replication forks of a replication bubble cause chromosome fragmentation.
Exposure to ionizing radiation brings about chromosome fragmentation without DNA replication, indicating direct double-strand breaks (48, 219) (see “The two mechanisms of two-strand damage” above). In E. coli, double-strand breaks are sealed by recombinational repair in the replicated portion of the chromosome (323, 325, 683). In WT E. coli, the repair of double-strand breaks and reassembly of disintegrated replication forks is the function of the RecBC recombinational repair pathway (first reviewed in reference 102).
Evidence for replication fork disintegration.
Although replication fork disintegration has yet to be demonstrated, the indirect evidence supporting the ubiquity of this hypothetical event is significant. The idea of replication fork disintegration is the most economical explanation for two related in vivo phenomena: (i) DNA replication-induced fragmentation (apparent double-strand breakage) of a chromosome under conditions when no direct double-strand breaks are detected in the absence of DNA replication, and (ii) preferential degradation of the newly synthesized DNA, that is, DNA at daughter arms of the replication forks, caused by single-strand breaks in template DNA.
Actually, the term “replication fork disintegration” comprises two mechanistically unrelated events with a similar outcome. Replication fork collapse occurs when a replication fork runs into a single-strand interruption in a template DNA and comes apart as a result of this preexisting lesion (333). Replication fork “breakage” has the same result as “collapse”, that is, a detached double-strand end, but it occurs with inhibited replication forks, whose progress is somehow blocked (334).
Besides polA or lig mutants, mentioned above, dam mutants are also known to accumulate single-strand interruptions in their DNA, perhaps due to the disoriented mismatch repair system (408, 409, 639, 713). DNA replication in dam mutants with the inactivated RecBC pathway leads to chromosomal DNA fragmentation, whereas without replication the chromosomes in dam cells are sound (713). The nature of the DNA fragments, released as a result of replication fork collapse, is suggested by their susceptibility to exonuclease V (ExoV)—the main E. coli exonuclease that attacks duplex DNA only if a double-strand end is available (499, 656). If recombinational repair in polA, lig, or dam mutants is blocked by a recA mutation, a massive degradation of the replicating DNA by ExoV ensues (376, 409, 443, 444, 450).
An alternative mechanism for replication fork disintegration in dam mutants is suggested by the observations that in the absence of adenine methylation, MutH introduces double-strand breaks at GATC sites in the presence of mismatches (22). Therefore, DNA replication-generated mismatches in dam mutants could cause MutH-dependent double-strand breaks in the nascent duplexes behind replication forks. This possibility will be difficult to distinguish from replication fork collapse caused by preexisting single-strand interruptions in template DNA.
Another source of single-strand interruptions in DNA is damage processing. Short-lived single-strand interruptions appear in DNA during nucleotide excision repair of UV damage. As already mentioned, if DNA replication is allowed during an ongoing excision repair of UV damage, chromosome fragmentation is observed (60, 262, 667). In these cells, breakdown of replication forks precedes degradation of the bulk of the DNA, suggesting replication fork encounters with excision gaps (234, 262).
Recall that in recFOR mutants the SOS induction by UV irradiation is delayed for the period corresponding to a single round of DNA replication but then reaches levels higher than in the WT cells (241, 727). The delay in the SOS induction is accounted for by the deficiency of the recFOR mutants in repair of daughter strand gaps after UV irradiation (see “recF, recO, and recR: mutant phenotypes” above), but why the subsequent overshoot? It turns out that in UV-irradiated recF cells, the next round of DNA replication results in chromosome fragmentation (714, 715) and degradation of the newly synthesized DNA (128, 534); hence the belated but strong SOS response. Significantly, only half of the newly synthesized DNA is degraded (128), suggesting that only one of the two arms of damaged replication forks becomes susceptible to ExoV.
The mechanism of this damage is revealed in an elegant study, in which cells deficient in both excision repair and recombinational repair are allowed to replicate their DNA after a very low dose of UV irradiation (673, 674). In these excision repair-deficient cells, the few thymine dimers cause the formation of daughter strand gaps during the first round of DNA replication and the newly synthesized DNA becomes preferentially susceptible to degradation during the second round of DNA replication. Apparently, the replication forks of the second round collapse at the gaps left unrepaired from the first round, leading to the degradation of DNA labeled during the first replication round (673, 674).
Inhibiting DNA synthesis by thymine deprivation in B. subtilis leads to accumulation of single-strand breaks in DNA synthesized during inhibition; upon thymine addition and restoration of the normal rate of DNA replication, these single-strand interruptions induce double-strand breaks, suggesting replication fork collapse (76). The other consequence of thymine starvation is degradation of the newly synthesized DNA, both in E. coli and in B. subtilis (67, 516). In Bacillus, this DNA degradation begins at replication forks (510, 516).
The second mechanism of replication fork disintegration is breakage resulting from inhibition of replication fork progress (263, 264, 434). In contrast to collapse, during breakage the template DNA is initially intact, and it is the inability of a replication fork to proceed that somehow breaks it. The progress of replication forks in bacterial chromosomes can be halted by such means as inhibiting DNA gyrase, inactivating a temperature-sensitive component of the replisome, or blocking the path of a replication fork with a termination site.
DnaB is the helicase that drives the replication forks in E. coli (reviewed in references 25 and 406). Shifting dnaB(Ts) mutants to the nonpermissive temperature blocks the progress of replication forks and leads to a slow degradation of the chromosomal DNA (80, 176, 211, 435, 696) (for the assignment of the mutants to dnaB see reference 224). The degradation is likely to be connected with the chromosomal DNA fragmentation in dnaB(Ts) mutants at the nonpermissive temperatures (176, 434, 566); the degradation is not observed in an ExoV− mutant (80). The degradation predominantly affects the newly synthesized DNA, apparently beginning at the replication forks and proceeding towards the replication origin (211, 435, 696). If the degradation is allowed to proceed to completion, up to 80% of the nascent DNA, labeled during a 1-min pulse is degraded (211, 435); with longer pulses, roughly half of the newly incorporated label is made acid soluble (696). Although the degradation is at both replication forks, it affects a particular strand of the newly synthesized DNA (696).
A possible mechanism for the RecBCD-dependent degradation of the newly synthesized DNA at an arrested replication fork is a replication fork reversal (384), i.e., an extrusion of both newly replicated strands with their subsequent annealing (Fig. 21B). This generates a Holliday junction with one open-ended arm, which is degraded by ExoV. This model predicts that all the newly synthesized DNA will be susceptible to degradation, whereas only half of it is (696). However, if soon after the replication fork reversal the Holliday junction is resolved by RuvABC (see “Pairwise interactions of Ruv Proteins: RuvABC resolvasome” above), this would amount to replication fork breakage (Fig. 21D), explaining the upper 50% limit on degradation of the newly synthesized DNA and the required selectivity of the degradation (334). Indeed, it was recently found that mutating away ruvABC prevents replication fork breakage in dnaB(Ts) mutants (566), suggesting that inhibited replication forks are indeed extruded to form Holliday junctions, which are then resolved by RuvABC, breaking the forks.
FIG. 21.
Replication fork reversal as the mechanism of replication fork breakage. A replication fork is shown moving from left to right; solid lines indicate parental strands; open lines indicate newly synthesized strands. (A) The progress of the replication fork is blocked (replisome mulfunctioning, a protein bound to DNA). (B) This results in replication fork reversal to generate a double-strand end, composed of the newly synthesized DNA strands, and a Holliday junction. (C) ExoV-catalyzed degradation of the double-strand end eliminates the Holliday junction. (D) Alternatively, resolution of the Holliday junction by RuvABC (small arrows in panel B) breaks the replication fork.
Rep is an auxiliary helicase in the E. coli replication fork; rep mutants replicate their DNA more slowly than do WT cells (342). In combination with recBC mutations, rep mutants exhibit extensive fragmentation of the chromosomal DNA, which is prevented by blocking the initiation of chromosomal replication (434). Fragmentation in rep mutants is also prevented by ruv mutations, indicating that the RuvABC resolvasome is the enzyme that breaks inhibited replication forks (566).
Sometimes it is difficult to tell whether an event is a replication fork collapse or a breakage, as in the case of the DNA gyrase inhibitors quinolone antibiotics oxolinic acid and nalidixic acid. These drugs severely inhibit the rate of DNA replication in E. coli, as if blocking the progress of replication forks, and in vitro studies show that nalidixic acid indeed traps DNA gyrase in a cleavage complex with DNA (reviewed in reference 163). However, in vitro, the UvrD helicase (see “Possible disruption of pairing of insufficient length by helicase II” above), unwinding a DNA fragment with the bound Topo IV (a DNA gyrase relative) trapped by a quinolone derivative, causes detachment of a DNA strand from the complex (578). If nalidixic acid works by the same mechanism on DNA gyrase, it causes collapse rather than breakage of replication forks.
Whatever the molecular mechanism of the replication fork damage with quinolones, treatment with oxolinic acid instantly inhibits DNA replication but does not cause a loss of supercoiling for at least 10 min, which indicates that the treatment itself does not induce breaks in the chromosome (403, 609). However, nalidixic acid-treated cells of E. coli (124, 222, 254) and B. subtilis (123) slowly degrade their DNA. In B. subtilis, the degradation starts at the replication point and proceeds towards the replication origin, affecting the template and the newly synthesized DNA strands equally (511). In E. coli, only chromosomes with replication forks are susceptible to the nalidixic acid-induced degradation, since little degradation is observed if DNA replication is prevented for some period before the treatment (124). Specific DNA gyrase mutants of S. typhimurium mimic the action of quinolones: in combination with recA mutation, these mutations trigger DNA degradation if DNA replication was allowed at the nonpermissive temperature (203).
Evidence for replication fork repair by recombination.
Skalka was probably the first to propose that disintegrated replication forks can be reassembled by recombinational repair (597). E. coli has two recombinational repair pathways, RecF and RecBC (see “The two recombinational repair pathways of E. coli” above), corresponding to two major classes of replication-induced two-strand DNA lesions (see “The two mechanisms of two-strand damage” above). As discussed in the section on daughter strand gap repair (above), the RecF pathway mends daughter strand gaps. The notion that disintegrated replication forks are reassembled in E. coli by the RecBC pathway explains two major phenomena. The first is that strains in which replication fork disintegration is expected to be frequent are dependent on recA and recBC but independent of recFOR. This is true for the “classical” replication fork disintegration mutants, such as polA (83, 225, 443), lig (119, 217), and dam (26, 408, 486, 487), as well as for the “classical” replication fork disintegration conditions, such as exposure to nalidixic acid (423). Throughout this section, inviability of a double polA geneX mutant will signify possible dependence of double-strand end repair on geneX (of course, the other phenotype of geneX mutants, which would also result in the inviability of polA geneX double mutants, could be increased DNA damage).
The finding that the viability of a polA recB double mutant is enhanced several orders of magnitude under anaerobic conditions (448) indicates that unrepaired oxidative damage is the major cause of replication fork collapse. It can be calculated from the published data (477, 518) that E. coli growing aerobically experiences on the order of 2,000 oxidative DNA lesions per cell per generation (see also reference 130). The major intracellular oxidants, hydroxyl radicals (HO·), are thought to be produced in vivo with participation of hydrogen peroxide (H2O2), superoxide (O2−), and iron (Fe) (reviewed in reference 299). Treatment with hydrogen peroxide to stimulate oxidative damage causes single-strand breaks in DNA, both in vivo (12) and in vitro (354). Short exposure to hydrogen peroxide kills recA or recBC mutants but is inconsequential for a recF mutant (12, 87).
Superoxide dismutase (gp sodAB) protects E. coli from superoxide, which would otherwise contribute to the formation of hydroxyl radicals. sod mutants cannot grow aerobically if they also carry recA or recB mutations, but they have no problems in combination with recF mutations (299). Iron metabolism deregulation in fur mutants results in iron overload and, as a consequence, increases oxidative DNA damage. fur mutants are inviable if they also carry recA or recB mutations but are 100% viable in combination with recF mutations (672).
Besides DNA pol I (gp polA) (12), base excision repair of the oxidative DNA damage also requires one of the several AP endonucleases: ExoIII (gp xth) (149), EndoIII (gp nth), or EndoIV (gp nfo). A triple xth nth nfo mutant is inviable in combination with recA or recB mutations but is not affected by recF mutations (707).
rep mutants, in which inhibited replication forks are broken, display growth defects in combination with recA mutations and are inviable in combination with recBC mutations (687). rep mutants easily tolerate recF mutations (434).
The Terminus region of the E. coli chromosome contains a replication fork trap, a region, into which replication forks can enter but from which they cannot exit. The trap borders are guarded by Ter protein bound to asymmetric termination sites; these sites allow replication forks to pass through in one direction but not in the opposite direction (reviewed in reference 253). If termination sites are inserted in the chromosome such that they interfere with the completion of the chromosomal replication, the cells become inviable if they carry recA or recB mutations (263, 434, 573).
The second phenomenon indicative of recombinational repair of disintegrated replication forks is hyperrecombination between chromosomal repeats. In a widely used strain designed to look for hyperrecombination mutants (313), this recombination depends on recA and recBC genes and is independent of recF (765); that is, it follows the RecBC pathway. All the strains with single-strand interruptions in their DNA, like polA, lig, dam, and xth strains, are hyperrecombinant in this setup (313, 407, 766). A dnaB(Ts) mutant exhibits an increased rate of recA- and recB-dependent but recFOR-independent recombination between repeats in the chromosome and is barely viable in combination with a recB mutation (561). Hyperrecombination is also observed in the vicinity of the termination sites (264, 382) and is especially stimulated if termination sites are inserted in inappropriate positions in the chromosome (263). Other cases of hyperrecombination caused by apparent replication fork disintegration are discussed elsewhere (333, 334).
DNA replication primed by double-strand end-promoted recombination.
The idea that disintegrated replication forks are repaired by recombination requires double-strand end invasion intermediates to be resolved by DNA replication. This prediction is at variance with the current models of homologous recombination at double-strand ends, which emphasize exchange without DNA replication (108, 320, 655). The current models are based on the old observations that inactivation of the replisomes in vivo still allows the completion of some double-strand end-promoted recombinational events by the RecBC pathway of E. coli, suggesting that the intermediates can be resolved without DNA replication (615, 617).
At the same time, there are other old observations suggesting that DNA synthesis is required to complete the formation of the majority of recombinant chromosomes. In the recombinants formed after conjugation, which goes via double-strand end invasion catalyzed by the RecBC pathway (reviewed in reference 399), the invaded strands are always associated with the newly synthesized DNA (591). The formation of recombinants after conjugation is severely inhibited if replisomes are temporarily inactivated with a dnaB mutation (72, 624). Eventually, it was realized (603) that, should a double-strand end-promoted exchange be resolved without DNA replication, it becomes an endless game, since the new double-strand end would be ready to engage the new recombinant duplex in another round of recombination. Priming DNA replication by the invading end seemed to be the only way out of this “perpetual-exchange” trap (603).
More recent observations further supported the supposed connection between the RecBC-catalyzed recombination and DNA replication. After massive DNA damage or replication inhibition, E. coli is able to synthesize DNA for many hours without protein synthesis, a phenomenon known as inducible stable DNA replication (iSDR) (309). iSDR is dependent on DnaT (343) and PriA (410), the two enzymes used in initiation of plasmid DNA replication (see “Initiation of plasmid DNA replication” above). iSDR is also dependent on RecA (343) and RecBC (398), suggesting that the underlying replication potential accumulates as a result of multiple disintegration of replication forks with their subsequent recombinational repair (334). Indeed, DNA damage repair via the RecBC pathway was found to stimulate origin-independent synthesis of both plasmid and chromosomal DNA in E. coli (21, 397). Finally, priA mutants were found to be defective in recombination along the RecBC pathway, suggesting that the bulk of recombination intermediates are resolved by DNA replication (310, 552).
The first attempt at direct demonstration of DNA replication priming by the invading end in E. coli used in vivo induction of double-strand breaks in small cosmids (plasmids carrying the cos site of phage λ) by cutting them with terminase (the λ enzyme that cleaves at cos) (19). Although cosmid DNA replication was found to be dependent on the presence of the terminase-producing plasmid, in the absence of direct demonstration of double-strand breaks this result proved to be inconclusive, since terminase production would not be possible under the experimental conditions used (discussed in reference 339). In a different approach, chromosomes of phage λ were cut in vivo at unique restriction sites in the presence of uncut, differently marked repressed λ chromosomes (339). The following recombination along the RecBC pathway induced the replication of the uncut phage chromosomes, indicating that the double-strand end invasion intermediates in E. coli are resolved to generate replication forks (339).
Overview of double-strand end repair.
Double-strand end repair can be used to mend double-strand breaks as well. The current model of recombinational repair of double-strand breaks postulates repair DNA synthesis (637); in fact, the scheme calls for the installation of two converging replication forks (309, 603). Therefore, double-strand break repair can be viewed as a combination of two initially independent double-strand end repair events and is not discussed separately.
The complete mechanism of replication fork installation via recombinational repair has yet to be worked out, since interactions between the major enzymes of this repair pathway have just started to be examined in vitro. To simplify the discussion, the current model of double-strand end repair is arbitrarily subdivided into three phases: presynapsis, postsynapsis, and replication fork restart. In the presynaptic phase, the double-strand end is degraded by ExoV until a properly oriented Chi site converts RecBCD degradase into RecBCD* recombinase. After Chi, RecBCD* continues to degrade DNA but with a reduced speed and only the 5′-ending strand, generating a 3′ single-stranded overhang. This 3′ overhang is initially complexed by SSB, but RecBCD* is proficient in promoting the RecA filament assembly on SSB-complexed DNA; the RecA filament then searches for homology.
After the homologous duplex is found, the RecA filament promotes strand exchange, forming a D loop with a single three-strand junction. From this point on, there are two hypothetical scenarios for the postsynaptic phase. One suggests that the 3′ end of the invaded strand is used by DNA pol I to prime limited DNA synthesis, increasing the D loop. PriA binds to the displaced strand to catalyze primosome assembly; the assembled primosome attracts the replisome, restoring the replication fork and converting the three-strand junction into a four-strand (Holliday) junction. RuvAB translocase uses the Holliday junction to disperse the RecA filament and then attracts RuvC, which resolves the junction.
In the alternative scenario, the displaced strand is cleaved near the invaded 3′ end, and the 5′ side of the nick is ligated to the invaded end (Fig. 10C). Then the three-strand junction attracts the RecG helicase, which disperses the RecA filament and, by pushing the junction further, restores the replication fork framework (Fig. 13). Only then does the framework attract PriA to build an active replication fork. The two hypothetical scenarios differ in the junction-resolving enzymes (RuvABC or RecG) and in the timing of the replication fork restart (before or after RecA removal).
Preparation of Double-Strand Ends by RecBCD Nuclease for RecA Polymerization
In WT E. coli, rejoining of the chromosomal DNA fragmented by gamma irradiation is completely blocked by only three mutations: recA, recB, or recC (555). ssb mutants have not been tested yet, although they are also likely to be deficient in recA-dependent rejoining of fragmented chromosomes, since they are defective in recombination requiring double-strand end repair (174, 208, 504, 700). RecB and RecC combine with RecD to work in a single enzyme, whose complex behavior makes it a fascinating subject to study, surpassed only by RecA protein (reviewed in references 320 and 650). The RecBCD enzyme is a combination of a potent helicase with duplex DNA- and ssDNA-specific exonucleases. In vitro, the RecBCD enzyme degrades linear duplex DNA to oligonucleotides at an incredible speed. In vivo, it degrades bacteriophage DNA cut by the host restriction systems, and in recA mutants, it degrades the entire chromosome after it was fragmented as a result of DNA damage. “Professional” DNA degradation by the RecBCD nuclease is difficult to reconcile with the central role of this enzyme in double-strand end repair. E. coli employs a two-component system to turn this “Mr. Hyde” into “Dr. Jekyll” when double-strand breaks and disintegrated replication forks need to be repaired.
RecBCD: Genes and mutants.
The three contributing genes of the RecBCD enzyme are closely grouped on the E. coli chromosome: recB and recD form an operon, while recC, although situated nearby, has its own promoter (11, 183–185, 557). In related microbes, the structure of the region is similar (519). A combination of extremely weak promoters and suboptimal codons maintains the level of 10 RecBCD nuclease molecules per chromosome (650). None of its three genes is known to be SOS inducible.
The recombination phenotype of recB and recC mutants is just the opposite to the phenotype of recFOR mutants: recBC mutants are deficient in homologous recombination following conjugation or transduction, but they do not interfere with plasmid recombination (106, 399). recD mutations are hyperrecombination mutations in these assays. There are two major DNA repair-related phenotypes displayed by recBCD mutants. Mutants with null mutations in recB or recC genes are sensitive to DNA-damaging treatments (735) and have viability around 30% (85, 86). Although the repair of daughter strand gaps in these mutants is normal, they die after their DNA has been damaged, apparently because of their inability to repair disintegrated replication forks and double-strand breaks (715). Since recA cells, which lack recombinational repair completely, still are up to 60% viable (85, 86), there must be reasons other than recombination deficiency contributing to the poor viability of recBC cells (see “Role of ExoV in chromosomal DNA replication” below). In contrast, null recD mutants and a single recC missense mutant display normal viability as well as WT survival after DNA-damaging treatments (11, 93). Null mutants in both groups lack the powerful exonuclease activity of the RecBCD enzyme, although recD cells still exhibit a rate of DNA degradation 50% of that of the WT cells (520). In accord with the distinct recombination phenotypes of recBC and recD mutants, polA recBC mutants are inviable (83, 225, 443) but polA recD mutants are fully viable.
RecBCD: Biochemical activities.
RecBCD enzyme is a heterotrimer (652) of RecB (134 kDa), RecC (129 kDa), and RecD (67 kDa). The prominent activities of the RecBCD enzyme include DNA helicase, dsDNA exonuclease, and ssDNA exonuclease. DNA hydrolysis does not need an input of energy, and most nucleases do not hydrolyze ATP. The trademark of the RecBCD nuclease is that it requires ATP hydrolysis for the degradation of duplex DNA. In fact, the RecBCD nuclease of E. coli, also known as ExoV (746), together with analogous enzymes of other eubacteria (and the SbcCD nuclease [121]), are the only known ATP-dependent exonucleases (102, 650, 658).
ssDNA exonuclease is defined as the activity able to degrade linear ssDNA. RecBCD degrades ssDNA to pieces several nucleotides in length (212, 293, 746). This reaction needs ATP and proceeds at the same rate in the presence of a broad range of ATP concentrations; hence, ATP is thought to be used as an allosteric effector (173). dsDNA exonuclease is defined as the activity able to degrade linear duplex DNA. RecBCD degrades linear duplex DNA to the same oligonucleotide products as are formed by ssDNA, but the degradation is faster (212, 293, 746), and the rate of hydrolysis declines with increasing ATP concentrations (173). Circular duplex DNA, even containing single-strand nicks and short gaps, is refractory to RecBCD attack (293, 746), because the enzyme can enter duplex DNA only through double-strand ends (499, 656).
The degradation of duplex DNA by RecBCD absolutely requires both Mg2+ and ATP; the rate of the degradation is dependent on the ratio of these two ions. ATP and Mg2+ complex each other, and so in an equimolar solution there is little free magnesium or free ATP (202, 228). The nuclease activity of RecBCD is stimulated when the Mg2+ concentration exceeds that of ATP and is inhibited when the ATP concentration exceeds that of Mg2+ (158, 170, 655). The former conditions (Mg2+ in excess of ATP) parallel those inside the cell (see “Regular DNA replication” below), while the latter conditions (ATP in excess of Mg2+) probably reflect the fact that RecBCD requires Mg2+ for the continuous activity (see “RecBCD: mechanism of DNA hydrolysis after Chi” below).
The third major activity of the RecBCD enzyme is DNA unwinding. RecBCD is a potent and highly processive DNA helicase, which in vitro can unwind DNA at a rate close to 1,000 bp/s (526), unwinding on the average 30 kbp per binding event (527). The DNA unwinding by RecBCD absolutely requires ATP. The logical explanation for the unusual ATP-dependent dsDNA exonuclease activity of RecBCD is that the enzyme is able to hydrolyze duplex DNA only after its unwinding (528, 659).
RecBCD: Mechanism of DNA hydrolysis before Chi.
The amino acid sequences of RecB and RecD suggest that both proteins possess ATP-binding domains (183, 184); indeed, both proteins bind ATP (286). RecB is a DNA-dependent ATPase (412) and a weak helicase, which translocates 3′ to 5′ along ssDNA (56, 491). RecD is also a DNA-dependent ATPase (96); mutational inactivation of its ATP-binding domain results in an inactive reconstituted RecBCD enzyme (315, 316, 412). Experiments with hybrid RecBCD enzymes in which the ATP-binding sites were inactivated on either RecB or RecD or on both subunits established that both ATP-binding sites control the hydrolysis of ssDNA, with the RecB site controlling hydrolysis of the 3′-ending strand and the RecD site controlling hydrolysis of the 5′-ending strand (95).
The nuclease active site was always suspected to reside in the RecD subunit, since null mutations in the RecB or RecC subunits inactivate all the enzyme activities whereas null mutations in the RecD subunit abolish all the nuclease activities but leave the helicase activity functional although much reduced (10, 520). Surprisingly, the single nuclease active site of the enzyme was eventually found within the 30-kDa C-terminal domain of RecB (756, 757). In the distantly related AddAB enzyme from B. subtilis, the nuclease active site is also located in the C terminus of the AddA subunit, homologous to RecB in E. coli (230).
In vitro, RecBCD degrades DNA asymmetrically, with the 3′-ending strand receiving most of the cuts (14, 157, 655). This asymmetry of degradation reflects the asymmetry of the enzyme binding to dsDNA. In the absence of ATP but in the presence of Mg2+, RecBCD binds a double-strand end but does not proceed into the DNA. The pattern of UV cross-linking to DNA strands by such a bound enzyme suggests that the RecB subunit binds to the 3′-ending strand while the RecC and RecD subunits sit on the 5′-ending strand (201) (Fig. 22A). In such a complex, RecBCD protects both strands 15 to 20 nucleotides from the end and unwinds the terminal 5 to 6 bp (177).
FIG. 22.
Mode of RecBCD enzyme binding to a DNA end and patterns of RecBCD-promoted DNA hydrolysis. The direction of travel of the enzyme through the DNA duplex is from right to left. (A) Scheme of RecBCD binding to a double-strand end. The C-terminal domain of RecB, containing the nuclease active site, is shown as a separate subdomain. The final 5 to 6 bp of the duplex end are unwound. (B) Mode of the RecBCD action before it has seen a Chi. The top strand ends 3′ at the right. (C) Mode of RecBCD action after the enzyme has seen a Chi. The enzyme itself is not shown.
In the presence of ATP, Mg2+, and Ca2+ (to inhibit the nuclease activities of the enzyme), RecBCD translocates along the DNA, unwinding it (528) and forming a growing loop on the 3′-ending strand (64, 659) (Fig. 22B). In vitro, the size of the loop is approximately one-third of the length of the unwound region (657). The loop is postulated to exist in vivo and may be an important regulatory element for the RecBCD enzyme. The mechanism of DNA unwinding by the RecBCD heterotrimer, containing a single helicase subunit (RecB), is hypothesized to be “inchworming,” with the step from 1 to 4–6 bp per single ATP molecule hydrolyzed (177, 491). In conclusion, a DNA end being processed by RecBCD probably has a single-stranded loop on the 3′-ending strand held by the enzyme with little or no DNA strand past the enzyme, while the 5′-ending strand hangs as an unpaired tail and is occasionally clipped by the enzyme (650, 658) (Fig. 22B).
RecBCD: Mechanism of DNA hydrolysis after Chi.
The previous section described the way RecBCD treats DNA before it has seen a Chi (also written as χ). Chi sites were discovered in bacteriophage λ as mutations creating hot spots for E. coli recombination (reviewed in reference 461). red gam mutant λ (see “SSA enzymes of phage λ” below) cannot inactivate ExoV (gam) and lacks its own recombination system (red), and so its DNA is savaged by RecBCD after being linearized for packaging by a phage-encoded terminase (175). red gam λ grows poorly in WT E. coli cells but sports big-plaque variants, which all turn out to acquire point mutations creating Chi sites at one of the several locations along the λ chromosome. Linearization of Chi-containing λ DNA, instead of triggering its degradation, results in its homologous recombination with other λ DNAs catalyzed, surprisingly, by the same RecBCD enzyme. The paradox of a single enzyme (RecBCD) carrying out two mutually exclusive activities on linear DNA (complete destruction versus preservation through recombination) led to the proposal that the encounter of the RecBCD enzyme with a Chi-site disables the dsDNA exonuclease activity, converting the enzyme into a “recombinase” by virtue of its helicase activity (622). This idea was confirmed experimentally, both in vivo (139, 338, 763) and in vitro (157, 653).
The inactivation of the RecBCD “degradase” activity after the enzyme has seen a Chi site is not confined to the Chi-containing DNA molecule, since the cells with linearized Chi-containing DNA become transient ExoV− phenocopies (139, 338). These cells are still recombination proficient, suggesting that “Chi treatment” turns the cells into recD mutant phenocopies rather than into recBC mutant phenocopies (314, 460).
In E. coli, Chi is an octanucleotide, 5′-GCTGGTGG-3′ (604); Chi sites of other bacteria are also short and asymmetric but differ in sequence (49, 94, 613). Consistent with its asymmetry, Chi recognition by RecBCD occurs only when the enzyme approaches the sequence from its 3′ side (178, 604, 651, 748). In vitro, Chi (as written) is recognized in heteroduplexes and even in heteroduplex bubbles as large as 22 nucleotides, suggesting that RecBCD unwinds DNA before checking it for Chi (43). The probability of Chi site recognition by RecBCD nuclease is 15 to 50% both in vivo and in vitro (338, 622, 653, 749). In vitro, when RecBCD sees a Chi, it pauses for a few seconds (157); eventually it proceeds with DNA unwinding, but the overall rate of DNA degradation is reduced severalfold (653). Even more importantly, the strand preference of the degradation is reversed: whereas before seeing Chi RecBCD preferentially degrades the 3′-ending strand (the strand with the loop), after seeing Chi RecBCD degrades the 5′-ending strand only (14).
In vivo (in recD null mutants), RecBC acts as if permanently modified by Chi sites (662), suggesting that the RecBCD metamorphosis at Chi sites is achieved by ejection of the RecD subunit (622, 661, 662). This idea is supported by the observation that overproduction of RecD polypeptide decreases the effect of Chi in recombination (314, 460, 521) and compromises cell survival after exposure to ionizing radiation (66), as if RecD polypeptide readily dissociates from RecBC. However, in vitro experiments, although supporting the idea that RecD is the target for the Chi regulation (155, 158), indicate that the Chi-activated enzyme still retains the RecD subunit: Chi-activated RecBCD degrades the 5′-ending strand, whereas the RecBC enzyme shows no exonuclease activity in the same assay (13).
Observation of RecBCD in vitro after its interaction with Chi provided an unexpected insight into the way the three subunits of the enzyme are held together. Under conditions limited for Mg2+ (ATP in excess to Mg2+ [see “Biochemical activities” above]), the unwinding activity of RecBCD is not inhibited unless the DNA substrate contains Chi. In this case, the RecBCD-catalyzed unwinding stops at Chi but can be restarted by providing Mg2+ in excess of ATP (155). The same is true for the exonuclease activity of the enzyme (654). Interestingly, the RecBC enzyme (lacking the RecD subunit) cannot unwind DNA under these conditions until Mg2+ is added in excess of ATP (155), suggesting that (i) the three subunits of RecBCD are held together by Mg2+, which is normally inaccessible to chelation by ATP; (ii) upon RecBCD interaction with Chi, the RecD subunit is displaced, making Mg2+ accessible to chelation by ATP; and (iii) the removal of Mg2+ from the enzyme inactivates its helicase and nuclease activities. Analysis of the RecBCD composition by ultracentrifugation in glycerol gradients reveals that after interacting with Chi-containing DNA under Mg2+-limited conditions, RecBCD completely dissociates into its subunit components (654), substantiating the above interpretation. It should be noted that this artificial “explosion” of RecBCD at Chi is unlikely to play a role in vivo since (i) there is an excess of Mg2+ over ATP, not the other way around (see “Regular DNA replication” below), and (ii) after seeing Chi in vivo, RecBCD remains fully proficient in promoting recombination, which would be impossible if the enzyme has dissociated.
RecBCD: RecA filament assembly.
Inhibition of the nuclease activities of RecBCD by Chi in vitro does not require any other protein (155, 157, 653). In contrast, disabling of the nuclease activity of RecBCD in vivo requires SSB and RecA proteins (139, 338), suggesting that RecBCD and RecA interact. Another indication of functional interactions between RecA and RecBCD is the fact that the optimal recombinational repair in vivo is achieved only when both enzymes are from the same or closely related species (150).
As already discussed in relation to daughter strand gap repair, RecA polymerization on ssDNA is inhibited by SSB and so requires assistance; in the daughter strand gap repair, RecA polymerization on SSB-complexed ssDNA is facilitated by RecF, RecO, and RecR (see “Presynaptic phase of daughter strand gap repair: RecF, RecO, and RecR” above). By analogy to the RecF pathway, it was proposed that in the RecBC pathway, the RecBCD enzyme itself stimulates RecA polymerization on SSB-covered ssDNA (336). Indeed, it was found that the translocating RecBCD enzyme, after seeing Chi, promotes RecA filament polymerization in the presence of SSB on the 3′-ending DNA strand (15). As expected, the RecBC enzyme (without the RecD subunit) lacks this requirement for Chi: it loads RecA constitutively onto the 3′-ending DNA strand (100). The resulting RecA-ssDNA complexes are proficient in homologous invasion of the intact supercoiled duplexes, present in the same reaction mixture (15, 100, 156, 158). These spectacular experiments amount to modeling the presynaptic and synaptic phases of the double-strand end repair in vitro.
Postsynaptic Phase of Double-Strand End Repair
The inviability of a particular mutant in combination with a polA mutation is an indication of the possible involvement of the corresponding gene product in the double-strand end repair (see “Evidence for replication fork repair by recombination” above). priA, recG, or ruv mutants are inviable when the mutation is present in combination with a polA mutation, but the corresponding gene products are not required to reverse the chromosomal fragmentation due to double-strand breaks and disintegrated replication forks. Apparently, they must be involved with the postsynaptic phase, which in the double-strand end repair can be further subdivided into replication fork restart and junction resolution. Since the mechanisms, the relative order, and the relationship of the two subphases have yet to be worked out in vitro, my decision to put the replication fork restart first is arbitrary, and the discussion is highly speculative.
DNA-keeping enzymes.
Genetic data suggest that completion of double-strand end repair (that is, recBC-dependent recombination) requires at least three DNA-keeping activities: DNA gyrase, DNA pol I, and DNA ligase (174, 766). DNA gyrase is important for maintenance of negative supercoiling in DNA (see “DNA gyrase” above); supercoiled DNA is a better substrate for RecA-promoted strand invasion (583). DNA pol I and DNA ligase, working together, fill in and seal single-strand interruptions which are generated in any DNA repair reaction. In addition, DNA pol I may play a more specific role, as suggested by its involvement in initiation of plasmid DNA replication (see “Initiation of plasmid DNA replication” above).
The main role of DNA pol I, a 109-kDa enzyme encoded by polA gene (148, 223), is in one-strand repair (see “Damage reversal and one-strand repair” above). polA mutants are sensitive to both UV and ionizing radiation (223, 482), primarily because they cannot complete the excision repair. DNA pol I has three activities: 5′→3′ polymerization activity, 3′→5′ exonuclease (proofreading), and an unusual 5′→3′ exonuclease, which allows the enzyme to replace segments of RNA or damaged DNA with regular DNA “in one stroke” (reviewed in reference 317). polA mutants are inviable if carry additional recA or recBC mutations (see “Evidence for replication fork repair by recombination” above), supposedly because the unrepaired single-strand interruptions in template DNA cause replication fork collapse, which cannot be fixed.
In addition to its role in the one-strand repair, DNA pol I may play an auxiliary role in recombinational repair. RecBCD- and RecA-catalyzed double-strand end invasion into an intact duplex generates a D loop that can be processed into a replication fork, similar to R loops in plasmid DNA replication (see “Initiation of plasmid DNA replication” above), if a primosome and then a replisome are loaded onto it. A primosome is loaded at what looks like a replication fork framework. To generate such a framework from a RecA-catalyzed invasion intermediate (Fig. 23B), a limited DNA synthesis primed by the 3′ invading end might be needed (Fig. 23C). This DNA synthesis is likely to be catalyzed by DNA pol I. Polymerization-deficient polA mutants show a 10- to 20-fold decrease in recombination requiring double-strand end repair, but a polA mutant deficient in the 5′→3′ exonuclease is not defective in such recombination (174, 766), consistent with our scheme, according to which only the polymerization function of DNA pol I is important in double-strand end repair.
FIG. 23.
Overview of double-strand end repair. RecA filament is shown as an open rectangle with rounded corners. Small one-sided arrow indicates the direction of RecA filament assembly. The star indicates limited DNA synthesis by DNA pol I; little wavy segments indicate RNA primers. (A) RecA polymerizes on a 3′ single-stranded overhang, generated by the RecBCD action after Chi. (B) RecA-catalyzed invasion of the 3′ end allows limited DNA synthesis to be primed by DNA pol I. (C) As a result of this synthesis, a primosome assembly structure is created in the displaced strand. (D) PriA assembles a primosome at the displaced strand, which then lays down primers and unwinds DNA at the fork. (E) A replisome is loaded onto the primed replication fork; the three-strand junction is being converted into a Holliday junction. (F) Double-strand end repair is completed by resolution of the Holliday junction.
Replication fork restart.
Following the limited displacement synthesis by DNA pol I, replication fork restart is believed to begin by binding of PriA to the displaced strand (Fig. 23C) (20, 309). polA priA double mutants are inviable (352); priA mutants are sensitive to DNA-damaging agents and deficient in homologous recombination requiring double-strand end repair (310, 552). In vitro, PriA is required to attract the replisome to replication fork frameworks (283). Certain point mutations in dnaC, which encodes the inhibitor of the DnaB replicative helicase, suppress recombinational repair defects of priA mutants, suggesting that the defects are caused by the inability of priA mutants to load the primosome (310, 552). The two principal components of the primosome are DnaB helicase, which drives replication fork unwinding, and DnaG primase, which lays the primers for the DNA synthesis (see “Initiation of chromosomal DNA replication in E. coli” above) (Fig. 23D and E). The availability of primers signals that a replisome can be loaded onto the replication fork framework, finishing the regeneration of a replication fork. Now, to complete the double-strand end repair, the DNA junction and the associated RecA filament must be removed.
The two pathways for DNA junction removal in double-strand end repair.
polA recA or dam recB double mutants are inviable because the constantly required double-strand end repair is blocked at the presynaptic or synaptic phases. The postsynaptic phase turns out to be equally important, because polA ruv and dam ruv double mutants (274, 487) or polA recG double mutants (257, 275) are inviable, too. From a different perspective, although rejoining of the direct double-strand breaks resulting from treatment of WT E. coli with ionizing radiation requires the functions of only recA, recB and recC (555), the ultimate survival of E. coli cells after treatment with ionizing radiation is also compromised by mutations in the ruvA, ruvB, ruvC, and recG genes (372, 373, 473).
In contrast to the moderate effect of single ruv or recG mutations, inactivation of both the RuvABC and RecG activities results in a deficiency in DNA damage repair equaled by recA deficiency or the double recB recF deficiency (370), suggesting that there is a redundancy in the postsynaptic pathways of recombinational repair. The existence of two distinct DNA junction-removing mechanisms after the double-strand end repair also accounts for the opposite effects of ruv and recG mutations on the recombination-dependent mutagenesis (189, 238), as discussed elsewhere (337).
As a result of double-strand end invasion, a single DNA junction is formed, which, at least at the beginning, is likely to be three-stranded (Fig. 23B and C). Such a three-stranded junction can be removed by RecG if the displaced strand of the D loop is incised near the invading 3′ end and the 5′ end of the nick is ligated to the invading strand (Fig. 10C and 13A and B). A potential candidate for such an incision activity has been reported (97). Such an opening of the D loop allows removal of the three-stranded junction by translocating it towards the free single-strand end by RecG (Fig. 13B). Besides the junction removal, sliding the junction off the end disperses the associated RecA filament and simultaneously creates a replication fork framework (Fig. 13C); the PriA-dependent replisome assembly then follows as the second step.
Alternatively, the PriA-dependent replication fork assembly may occur first; the DNA synthesis converts the three-strand junction into a Holliday junction, which in the second step is resolved by RuvABC (Fig. 23F). This alternative resolution scheme explains how, in the absence of the RecG resolution pathway, the RuvABC pathway benefits from the inactivation of the helicase activity of PriA (4). In recG mutants, RuvAB could translocate the three-way junctions inefficiently, so the 3′-to-5′ helicase activity of PriA would counteract this translocation and would convert the three-way junctions into Holliday junctions. If the D loop was already incised (Fig. 13A and B), subsequent resolution of the Holliday junction by RuvABC could not be productive.
Role of ExoV in Chromosomal DNA Replication
There is an old observation suggesting that DNA degradation by ExoV plays an important role in the viability of E. coli cells. Both recA and recBC mutants of E. coli are deficient in double-strand end repair; however, the viability of recA mutants is around 60%, while the viability of recBC mutants, which inactivate both the double-strand end repair and DNA degradation, is only 30% (85). Moreover, the viability of recA recB double mutants is reduced to 20%, confirming the rule that recA mutations decrease the viability by one-third while recBC mutations decrease the viability by two-thirds. What could be the role of ExoV in the chromosomal DNA metabolism in E. coli?
ExoV and stability of replication forks.
In some replicative DNA helicase mutants, such as rep or dnaB mutants, replication forks are believed to be inhibited (434). Inhibited replication forks were suspected to be unstable (44, 263, 334) (see “Evidence for replication fork disintegration” above), which was later confirmed and shown to result in chromosomal DNA fragmentation (434). Remarkably, inhibited replication forks turned out to be less stable in recBC mutants than in the recBC+ cells (566). Moreover, the breakage of the inhibited replication forks depends on the RuvABC resolvasome, suggesting that the mechanism of the breakage is through the replication fork reversal with subsequent “resolution” of the resulting Holliday junction (Fig. 21B and D). ExoV is assumed to prevent the breakage of such a reversed replication fork by degrading the generated double-strand end and thus eliminating the Holliday junction (566) (Fig. 21C). Both the replication fork reversal with subsequent RecBCD entry (384) and the breakage of the reversed replication forks by RuvABC (334) have been considered previously.
Thus, one role of ExoV could be to prevent the RuvABC-dependent breakage of the inhibited replication forks, but it cannot be its only role. In the recBC mutant cells that are WT for the replicative helicases, some chromosomal fragmentation is still observed (566). Interestingly, this chromosomal fragmentation is not eliminated by mutating away RuvABC resolvasome, suggesting that (i) some replication forks in the WT cells disintegrate for a different reason (collapse?); and (ii) ExoV plays an additional role in the WT cells.
Excessive DNA degradation affects survival after ionizing radiation more than after UV.
Generally, recBC mutants are equally sensitive to both ionizing radiation and UV irradiation, which is thought to reflect their inability to repair both double-strand breaks and disintegrated replication forks. However, one allele of recB called rorA is more sensitive to ionizing radiation than to UV irradiation (209).
In vitro, rorA-RecBCD enzyme consumes more ATP during DNA degradation than the WT enzyme does (690), which may be the reason for the two- to threefold increase in chromosomal degradation after ionizing irradiation observed in rorA mutants (210). The increased DNA degradation is likely to compromise the repair of double-strand breaks in the replicated portion of the chromosome because it results in complete chromosome degradation if both daughter branches are broken close to each other (Fig. 24). The “super-ExoV” nature of the rorA mutants not only accounts for their sensitivity to ionizing radiation but also is compatible with their close-to-WT UV resistance. Although the rorA-RecBCD enzyme is likely to be defective in repair of disintegrated replication forks (probably the major lesion repaired by the RecBCD pathway after UV irradiation), complete degradation of the resulting linear tail should not kill the cell (see the next section).
FIG. 24.
Lethality caused by double-strand breaks in the replicated portion of the genome in rorA mutants. The chromosome is depicted as a theta-replicating structure, with the single line representing duplex DNA. The origin of DNA replication is denoted by small open circles at the top of the chromosome; the terminus is shown as a solid circle at the bottom. If both daughter branches of a replicating chromosome are broken at positions close to each other, the double-strand breaks have more chances of being repaired in the WT cells than in rorA mutant cells, due to the increased DNA degradation in the latter.
A hint to the possible nature of rorA mutation could have been the fact that a 60-fold overproduction of the RecD subunit causes a similar phenotype (almost WT UV resistance but sensitivity to gamma rays) and is accompanied by elevated DNA degradation (66). This RecD+++ phenotype is in accord with the common belief that the displacement of RecD subunit turns the RecBCD enzyme into a recombinase while RecD repositioning returns the enzyme to the degradase mode (see “Mechanism of DNA hydrolysis after Chi” above). However, the properties of the purified rorA-RecBCD enzyme cannot be simply explained by the RecD polypeptide overproduction (209, 690); it might be that the rorA mutation compromises Chi recognition.
DNA degradation as a possible backup strategy.
Of the Chi sites in the E. coli chromosome, 75% are oriented so as to stop the RecBCD degradation coming towards the replication origin (54). To a certain degree, this preferential orientation reflects the facts that (i) the Chi sequence contains the primase-binding site, and primase binding sites are more frequent in the template for the lagging-strand DNA synthesis (54); and (ii) the trinucleotide composition is different for the transcribed and untranscribed strands, while the majority of the genes in the E. coli chromosome are cooriented with the DNA replication (116). However, these two factors cannot account for all the skew, especially around the replication origin, where it is 10:1. This preferential orientation of Chi sites suggests that the RecBCD repair pathway primarily deals with disintegrated replication forks (333), probably because this type of lesions is more frequent in the E. coli chromosome than are direct double-strand breaks.
While an unrepaired double-strand break in the chromosome is an obvious threat, it might be unclear why a disintegrated replication fork could not be simply ignored. However, if one of the two replication forks in a replication bubble has disintegrated while the other fork continues to function, chromosome overreplication will occur. In circular bacterial chromosomes, this overreplication will be due to the switch of the chromosomal replication from theta to sigma mode (Fig. 25B). In a circular chromosome with a single replication fork, initiation of a new round of theta replication cannot provide an exit from sigma replication (the sigma replication trap) (Fig. 25B, E, and F).
FIG. 25.
Outline of the maintenance of circular chromosome. The chromosome is shown as a rectangle with rounded corners (the single line represents duplex DNA). The origin of DNA replication is denoted by small open circles at the top of the chromosome. (A and B) A theta-replicating chromosome (A) may suffer a collapse of one of the replication forks, becoming a sigma-replicating chromosome (B). (C) In a wild-type strain, the double-strand end is then degraded by RecBCD and reattached to the circular domain with the help of RecA. (D) In the absence of RecA, the linear replicating branch is completely degraded by RecBCD, while a new round of theta replication is initiated from the origin. (E and F) In the absence of RecBCD, the chromosome is trapped in the rolling-circle replication; initiation of a new bubble from the origin can only lengthen the linear tail.
One way out of the sigma replication trap is through recombinational repair of disintegrated replication forks (Fig. 25C). Another way involves degrading the linear branch all the way to the circular domain, eliminating the tail of the rolling-circle and returning the chromosome to theta-replication (340, 687) (Fig. 25D). In vitro, when presented with circular DNA molecules having linear tails, RecBCD degrades the tails but does not harm the circular domains of such DNA substrates (452), indicating the feasibility of a similar reaction in vivo. Thus, ExoV is not only suspected in stabilization of inhibited replication forks but could also be involved in degradation of the unrepairable DNA tails. The hypothetical involvement of DNA degradation in controlling chromosomal replication may account for the ubiquitous presence of ExoV nucleases in eubacteria (102, 650, 658).
Double-Strand End Repair in the Absence of RecBCD
The defect of recBC mutants in conjugational recombination (which requires double-strand end repair) is compensated by two categories of suppressor mutations (called sbc for “suppressors of recB and recC”). One category acquires a new exonuclease activity, while the other category lacks, in addition to ExoV, a couple of less prominent nucleases. These apparently opposite changes restore almost WT capacity for double-strand end repair, demonstrating that such repair can be realized in several ways. The knowledge about the alternative pathways for double-strand end repair in E. coli is illuminating for studies of recombinational repair in eukaryotic cells. Eukaryotes lack strong bacterium-type nucleases, and so their pathways of recombinational repair of double-strand ends have more in common with the alternative pathways in E. coli than with the primary, RecBCD pathway.
RecE pathway.
sbcA mutations activate the expression of a part of the cryptic Rac prophage (reviewed in reference 81), as a result of which at least two phage proteins relevant for recombinational repair are produced. The RecE is a duplex DNA-specific exonuclease which selectively degrades the 5′-ending strand, producing a DNA duplex with a 3′ overhang (285). RecT promotes annealing of complementary single DNA strands and can catalyze three-strand branch migration (233). Possible roles of these two activities in bacteriophage recombination are discussed below (see “SSA enzymes of the Rac prophage”). A likely role of the RecE exonuclease in the E. coli repair is to resect double-strand ends so that they terminate with long 3′ overhangs. These 3′ single-stranded tails would be complexed by SSB and then degraded by ExoI, an ssDNA-specific nuclease with the 3′-to-5′ polarity of degradation, whose activity is stimulated by SSB binding to ssDNA (441). In the RecBC pathway, the 3′-ending strand is held as a loop by RecBCD (Fig. 22), which apparently protects the 3′ end from other nucleases. In the RecE pathway, the role of RecT could be to compete with SSB for binding to the RecE-generated 3′- single-strand overhangs, thus protecting them from degradation by ExoI.
Since recE was the first gene in which mutations specifically eliminated double-strand end repair in recBC sbcA cells, the pathway was named after it (103). Recombinational repair of the chromosomal DNA along the RecE pathway is dependent on recA (206), recE (109, 206), recF (206), recJ (385), recO (312), recR (400), recT (109), and ruvC (375). The mechanism of the RecE pathway of double-strand end repair has yet to be explored in vitro; understanding of how some of these activities operate during the daughter strand gap repair (see “Presynaptic phase of daughter strand gap repair: RecF, RecO, and RecR” above) is helpful in drafting their hypothetical interaction during the mechanistically different task of double-strand end repair.
The RecE exonuclease acts on blunt or nearly blunt ends or on the ends with long 3′ overhangs. It can also degrade the ends with short 5′ overhangs but is inhibited by long 5′ overhangs (285), which could be present on half of the double-strand ends generated as a result of replication fork disintegration (Fig. 26). The removal of long 5′ overhangs could be a function of RecJ, an ssDNA-specific exonuclease with the 5′-to-3′ polarity of degradation (386). After an end with a long 5′ overhang has been blunted by RecJ, RecE could degrade it to generate a 3′ overhang, subsequently complexed with SSB and RecT. When the resection of the 5′-ending strand has unraveled a sequence able to form a hairpin in the 3′-ending strand, PriA would start primosome assembly, RNA primers could be laid, and DNA synthesis would be initiated on the single-strand overhang (Fig. 26). Since the overhang has a 3′ polarity, it cannot be duplicated to the very end, and so the endmost primer cannot be removed (Fig. 26). As was hypothesized for the daughter strand gap repair, RecFR proteins bind to the 5′ side of every RNA primer (see “Replisome reactivation and model for RecFOR catalysis of RecA polymerization at daughter strand gaps” above); since the RecFR complex at the endmost primer cannot be displaced, it would catalyze the binding of RecOR and, eventually, RecA, to the single-strand overhang (Fig. 26). The rest of the reaction is likely to be standard (Fig. 23). The weak effect of ruvB (375) and recG (373) mutations on recombinational repair along the RecE pathway is understood in terms of redundant functions (370) (see “Resolving recombination intermediates” above).
FIG. 26.
Scheme for the presynaptic phase of double-strand end repair along the RecE pathway. Short wavy segments with the associated little caps indicate RNA primers with RecFR proteins; an open rectangle with rounded corners indicates the RecA filament. Explanations are given in the figure and in the text.
RecF pathway.
The RecF pathway of recombinational repair of double-strand ends is turned on in recBC mutants by inactivation of two additional nucleases. One nuclease to be inactivated is ExoI (see the previous section), which degrades ssDNA starting from 3′ ends (353) and is thought to participate in methyl-directed mismatch repair (439). The gene coding for ExoI is called sbcB/xonA (332). Another nuclease to be inactivated is SbcCD (204, 374) which is an ATP-dependent dsDNA exonuclease (121) distantly related to RecBCD (463). It is not known how the ExoI and SbcCD inactivation enables the RecF pathway to promote double-strand end repair. One idea is that in the absence of ExoV the nuclease activity for generating 3′ overhangs is so weak that other nucleases that might degrade 3′ overhangs will interfere (103). A complementary idea is that the RecF pathway has no protein like RecT in the RecE pathway or RecBCD in the RecBC pathway to protect the 3′ single-strand overhangs from degradation.
The genetic requirements of the RecF recombinational repair pathway operating on the chromosome are similar to those of the RecE pathway: RecA (261), RecF (261), RecJ (385), RecO (312), RecR (400), and RuvC (402) are needed. In addition, RecN (312, 493), RecQ (427, 462), RuvA and RuvB (372), UvrD (427), and HelD (427) participate. RecQ, UvrD, and HelD (all three are DNA helicases with similar behavior in vitro [reviewed in reference 416]) are most probably needed to generate 3′ overhangs. It is proposed that in the RecF pathway, 3′ overhangs are produced by the combined action of the RecQ helicase (or UvrD and HelD helicases) and the RecJ ssDNA exonuclease (108, 386). The assumed role of the RecQ helicase in producing the ssDNA substrate for the RecA-catalyzed joint molecule formation has been modeled in vitro (236). Everything else could be as in the RecE pathway, explaining the overlapping requirements of the two pathways (Fig. 26). In this scheme, the only function that remains unaccounted for is RecN (see “SOS expression as a compensation” below).
Unified mechanism of double-strand end repair.
The above mechanisms for the double-strand end repair along the RecBC pathway and the alternative RecE or RecF pathways have a lot in common. In fact, their distinction is only in the presynaptic reactions, which, although mechanistically similar, are catalyzed by different enzymes. Therefore, one can develop a unified mechanism for double-strand end repair (Fig. 27) (108).
FIG. 27.
Unified scheme for double-strand end repair. RecA filament is shown as an open rectangle with rounded corners; the positions of single-strand scissions are indicated by small arrows. Explanations are given in the figure and in the text.
The first stage is the processing of a double-strand end to generate a 3′ single-strand overhang. The next step is the RecA filament polymerization, catalyzed by either RecBCD or RecFOR. The RecA filament searches for homology and, after finding it, promotes the invasion of the end into a homologous intact duplex. A three-strand junction is generated, and a limited DNA synthesis is primed by DNA pol I at the invading 3′ end. From this point on, two hypothetical ways of completing the double-strand end repair ensure the removal of the junction and displacement of the associated RecA filament. One pathway is controlled by the RecG helicase, which may translocate the three-way junction towards a double-strand end, transforming it into a replication fork framework. The alternative RuvABC-dependent pathway could operate when the replication fork has already restarted and the three-strand junction has been converted into a Holliday junction.
Summary
Disintegration of replication forks occurs in response to replication fork inhibition or as a result of the presence of single-strand interruptions in the template DNA. In E. coli, disintegrated replication forks are reassembled by the RecBC pathway of recombinational repair, via the homology-directed invasion of the double-strand end into the intact sister duplex. Double-strand break repair in E. coli is possible in the replicated portion of the chromosome and is probably the sum of two independent double-strand end repair events. Repair of disintegrated replication forks and double-strand breaks in other bacteria is likely to follow the E. coli scheme. ATP-dependent nucleases (analogous to the RecBCD enzyme) are detected in many eubacteria (425, 658); Chi sites are characterized in three other microbes (49, 94, 613).
Among the prominent issues in the RecBC pathway is the formation of the two-strand DNA lesion itself: the possibility of replication fork disintegration has yet to be demonstrated in vivo. The presynaptic and synaptic phases of the RecBC-dependent double-strand end repair have been reconstituted in vitro. The replication fork restart is the area of active current research; its progress should allow us to bridge the RecABCD in vitro reactions with those of RuvABC and RecG. In contrast, the RecF and RecE pathways of the double-strand end repair have yet to be reconstituted in vitro. The role of ExoV in the eubacterial chromosome metabolism begs in vivo experimentation.
SITE-SPECIFIC MONOMERIZATION OF THE CHROMOSOME AFTER RECOMBINATIONAL REPAIR
The inviability of the double polA dif mutants (328) could have suggested yet another gene of double-strand end repair, but dif turned out to be a short sequence related to the demultimerization sites of multicopy plasmids (51, 112, 328)! dif mutants experience difficulties with chromosome partitioning, but the difficulties disappear if recombinational repair is also inactivated (328). Recombinational repair is expected to result in crossing over in 50% of the cases (see “Homologous recombination versus recombinational repair” above); a single crossover between circular chromosomes will combine them into a dimeric chromosome (Fig. 28). The unique susceptibility of circular chromosomes to odd numbers of exchanges was first recognized by McClintock in her studies with maize ring chromosomes (422). Long considered to be an oddity, this problem turned out to be the real one for prokaryotes, with their almost invariably circular chromosomes.
FIG. 28.
Removal of DNA junctions after recombinational repair may result in a crossover which translates into chromosome dimerization. (A) A Holliday junction behind a replication fork. Positions of single-strand scissions due to the preferred mode of RuvC resolution are indicated by small arrows. (B) A crossover is formed behind a restored replication fork due to the RuvC resolution. (C and D) A single crossover in a replicated portion of a circular chromosome (C) translates into a dimer chromosome when replication is completed (D).
Genetics of the XerCD-dif System
The two replication forks in the E. coli chromosome start at the unique origin and converge on the replication terminus situated across from the origin (see “Cellular processes that surround and complicate recombinational repair” above). In the middle of the terminus, there is a 30-bp dif site responsible for the chromosome monomerization (51, 112, 328). A pair of resolvase-like enzymes encoded by unlinked genes xerC and xerD work on this site to demultimerize the chromosomes (51, 52) (Fig. 29).
FIG. 29.
XerCD-dif site-specific recombination ensures chromosome monomerization in E. coli. The chromosome is shown as a rectangle with rounded corners, with the thick single line representing duplex DNA; replication origins are shown as small open circles at the top of the chromosome, and the replication terminus is represented by a small solid circle at the bottom of the chromosome. (A) A chromosome replicating in theta mode. (B) One of the replication forks has disintegrated. (C) The replication fork is reassembled by recombinational repair, resulting in a Holliday junction. (D) The Holliday junction is resolved, generating a crossover. (E and F) Completion of the chromosomal replication results in a dimer chromosome. (G and H) Segregation of the daughter nucleoids translocates the crossover to the terminus region, where it is resolved, permitting separation of the daughter chromosomes from each other.
Mutating away xerC or deleting the dif site causes cell filamentation due to problems with nucleoid partitioning. Mutating away recA or recB alleviates these problems of xerC or dif mutants (51, 328), suggesting that it is mostly the RecBC pathway of recombinational repair that generates sister chromatid exchanges. However, direct physical detection of the in vivo site-specific recombination at dif shows that it is decreased equally by either recB or recF mutations and is eliminated only in a recB recF double mutant (630).
It is calculated that some 20% of E. coli cells in an exponentially growing culture experience an inducing event (crossing over between sister chromosomes) which subsequently leads to partitioning problems in dif mutants (328, 629), putting the number of repair events at twice that value, around 40%. In good agreement with this assessment, available data (599) allow us to calculate that about 45% of recA cells (which probably degrade linear tails, since they cannot repair collapsed replication forks [see “DNA degradation as a possible backup strategy” above]) have at least one nucleoid missing.
In Vivo Biochemistry of the XerCD-dif System
The XerCD-mediated reaction can be studied in vivo with plasmid substrates bearing the 30-bp dif sequence (51, 52, 112, 328). In this setting, the reaction shows no resolution selectivity: two dif sites are recombined independently of whether they are on a single DNA molecule or on two separate DNA molecules. If this lack of selectivity in the plasmid system reflects a similar situation in the chromosome, it is unclear how the XerCD-dif system is able to faithfully resolve chromosomal dimers.
It was postulated that the XerCD-dif system is able to monomerize chromosomes as a result of frequent exchanges between the two dif sites on the sister chromosomes and the active separation of the sister chromosomes by a partitioning apparatus of the cell (51, 328). However, it was found that the site-specific recombination at dif does not occur at all if the cell division is blocked, suggesting that the chromosome monomerization is a highly regulated process (629). Indeed, no recombination between dif sites is catalyzed by XerCD in vitro (118), demonstrating that the chromosomal monomerization system is more complex than initially appeared.
A Supramolecular Chromosomal Structure around dif?
Another idea of how the XerCD-dif system might function envisions a supramolecular structure at the chromosome terminus that renders the site-specific reaction unidirectional. Studies with reintroduction of dif at various locations in the chromosome of dif-deleted strains show that dif is active only in an interval 10 kb to each side of the natural dif location (125, 327). Contrasting with this positional specificity, both the psi demultimerization site of pSC101 plasmid and the loxP/Cre site-specific recombinational system of bacteriophage P1 can complement a dif deletion if inserted at the natural position of the dif site (126, 355). Although the location of dif coincides with the zone of meeting of the two replication forks, completion of DNA replication per se does not trigger the monomerization, since dif, in a strain with an inversion of almost half the chromosome, is situated at least 1 Mb away from the zone of replication fork meeting and still functions in the chromosome monomerization (125)!
The behavior of dif invites parallels with the terminal recombination zone, the several-hundred-kilobase chromosomal segment covering the terminus, characterized by the high orientation-dependent recBC-catalyzed homologous recombination, whose center coincides with the position of dif (127). If a supramolecular structure in the terminus region indeed exists, its dimensions must be on the order of several hundred kilobases, since deletions of up to 233 kb around dif are without consequences as long as dif is reinserted at the site of the deletion (125, 327).
Summary
Recombinational repair is frequently accompanied by crossing over, which, in dimeric eubacterial chromosomes, leads to the formation of chromosome dimers—hence the need for a chromosome monomerization system. XerCD is a site-specific recombinase that, acting at the dif site in the middle of the terminus region of the E. coli chromosome, monomerizes the latter in preparation for cell division. Homologs of the XerCD recombinases have been characterized from several eubacteria (reference 698 and references therein).
The in vitro biochemistry of the XerCD-dif system is challenging, probably because of the requirements for yet-to-be-characterized factors. The regional regulation of site-specific as well as homologous recombination within the terminal recombination zone is the subject of exciting in vivo research.
GLOBAL REGULATION OF RECOMBINATIONAL REPAIR
The major aspects of the two pathways of recombinational repair in E. coli are summarized for comparison in Table 2. The table underscores the fact that apart from the enzymes of DNA synthesis required for the replication restart, all “recombination” enzymes act in one way or the other to control RecA polymerization or depolymerization. However, regulating RecA activity during repair is only one aspect of the in vivo control over recombinational repair. The other aspect, to be discussed below, is suppression of RecA activity under conditions when no repair is needed (regular DNA replication) or stimulation of RecA activity when massive repair effort is required (SOS induction).
TABLE 2.
Major aspects of the two recombinational repair pathways in E. colia
| Aspect | RecF pathway | RecBCD pathway |
|---|---|---|
| Causative one-strand lesion | Noncoding lesion | Single-strand interruption |
| Replisome interaction with one-strand lesion | Stalled replisome | Collapsed replication fork |
| Resulting two-strand lesion | Daughter strand gap | Double-strand end |
| Generation of ssDNA associated with lesion | Unwinding without DNA synthesis | DNA degradation after Chi |
| RecA polymerizationb | RecFOR | RecBCD after Chi |
| Replisome reactivation | RecA filament (?) | Not required (?) |
| Replication restart | Gap filling by PolA (or by DNA pol III) | Fork reassembly by PolA + PriABC + DnaT |
| DNA junctions | One four-way plus one three-way (?) | One three-way or one four-way (?) |
| Backup strategy | Translesion DNA synthesis | Degradation of detached branch |
| Backup activity | UmuD′2C | RecBCD (ExoV) |
The omitted common features are as follows: SSB, the main accessory protein; RecA, homologous pairing and strand exchange; RecG and RuvABC, RecA filament dispersal and DNA junction resolution; and XerCD-dif, chromosome monomerization.
In the presence of SSB.
Regular DNA Replication
Since there is apparently no activity in either recombinational repair pathway of E. coli that would recognize two-strand DNA damage per se, recombinational repair is likely to be triggered by the availability of ssDNA in association with RecA polymerization-promoting functions (RecFOR or RecBCD). RecBCD cannot access DNA if the latter lacks double-strand ends, but RecFOR proteins are likely to stand by replication forks, which always contain regions of ssDNA. How is the RecFOR-promoted RecA polymerization at the replication forks suppressed when recombinational repair is not needed?
Some other proteins could discourage RecA polymerization during regular DNA replication. For example, LexA, the SOS regulon repressor (see “Organization of the SOS regulon” above), suppresses RecA polymerization in vitro under suboptimal conditions (237, 515). A recently characterized DinI protein is another candidate for such a negative regulation (753). Of course, the main RecA inhibitor is SSB (see “Assistance for RecA by SSB at all stages” above), the protein that controls access of other proteins to ssDNA (reviewed in reference 430). In vitro, in the presence of 1 mM Mg2+ and 1 mM ATP, RecA cannot displace SSB from ssDNA; however, if the concentration of Mg2+ is increased to 10 mM, RecA quickly displaces SSB from ssDNA (reviewed in reference 335). There are intermediate Mg2+ concentrations at which RecA will displace SSB slowly, due to a kinetic barrier for a nucleation event; the in vivo conditions are assumed to create such barrier. The reason why normal DNA replication does not elicit RecA polymerization could therefore be a kinetic one: although ssDNA is present at replication forks continuously, any particular DNA sequence stays single stranded only transiently, giving RecA insufficient time to overcome the nucleation barrier (558).
The information on the optimal Mg2+ and ATP concentrations for the various in vitro DNA replication and recombination reactions could reflect the active concentrations of these ions in vivo; such data, admittedly more illustrative than representative, are collected in Table 3. They give the impression that the optimal conditions for DNA replication, DNA degradation by ExoV, and some recombination-related reactions are Mg2+ in 10:1 excess over ATP and no dATP. In contrast, optimal conditions for multienzyme recombination reactions tend to require a higher ATP/Mg2+ ratio. RecG is active only when the ATP/Mg2+ ratio is reversed, while RecBCD under the “reversed” conditions retains only its helicase activity (see “RecBCD: biochemical activities” and “RecBCD: mechanism of DNA hydrolysis after Chi” above). In addition, at least two recombinational enzymes, RecA and RuvAB, prefer dATP over ATP.
TABLE 3.
Optimal or reported Mg2+ and ATP concentrations for some in vitro replication and recombination reactions
| Protein or enzyme | In vitro optimum concn (mM) for DNA replication and recombination
|
Best NTP (second best) | Reference(s) | ||
|---|---|---|---|---|---|
| ATP | Mg2+ | ATP/Mg2+ | |||
| RecBCD (ExoV) | 0.03 | 10 | 0.003 | GTP (ATP) | 170, 746 |
| RuvAB | 0.2 | 10 | 0.02 | dATP (ATP) | 453, 481 |
| SSBa (replication) | 1 | 10 | 0.1 | 6, 411, 467 | |
| RecA (SSB) | 1 | 10 | 0.1 | dATP (ATP) | 429, 582 |
| PriA | 1 | 5–10 | 0.1–0.2 | ATP (dATP) | 6, 467, 588 |
| RuvCa | 2 | 15 | 0.13 | 764 | |
| RecFORa (RecAb) | 1.3–3 | 10 | 0.13–0.3 | dATP (ATP) | 684, 719 |
| RecBCD (RecAb) | 5 | 8 | 0.6 | ? | 15 |
| SSB (RecAb) | 8 | 12c | 0.7 | ? | 670 |
| RecA K72Rd | 3–12 | 2–10 | 1.2–1.5 | dATP onlyd | 570 |
| RecG | 2–8 | 1–4 | 2.0 | ? | 729 |
| RecBCD (Hele) | 5 | 1 | 5.0 | ? | 13 |
The enzyme does not require ATP, but other enzymes in the reaction do.
Measured as the efficiency of the RecA-catalyzed reaction.
The only concentration used.
The mutant RecA cannot utilize ATP.
Minimal nuclease activity, maximal DNA unwinding.
Since the concentration of Mg2+ in almost all of these in vitro reactions can be lowered to 5 mM without affecting the outcome, the significant variable in these data is the concentration of ATP, which changes from as little as 0.03 mM for the maximal ExoV activity to around 10 mM for certain RecA- and RecG-catalyzed reactions. dATP adds further variation to the nucleotide theme of the equilibrium. The concentration of ATP in vivo is around 3 mM (55, 387, 413); that of dATP is only 0.2 to 0.3 mM (55, 413), but since DNA precursors are thought to be concentrated at the replication sites, the dATP concentration around the replication forks could be higher (415). In equimolar solutions with ATP, Mg2+ is tightly complexed by the anion (202, 228); therefore, since the in vivo free Mg2+ concentration in E. coli is 1 to 4 mM (3, 227, 392), it is present in a small excess over triphosphates. This gives the combined MgATP2− + Mg2+ pool around 5 mM and the in vivo ATP/Mg2+ ratio of 0.5 to 0.8, the one that is preferred by the multienzyme recombination reactions in vitro. Therefore, “regular” intracellular conditions could be permissive for RecA polymerization if long-lived ssDNA is present. Indeed, introduction into E. coli cells of ssDNA which is unable to replicate induces the SOS response (205, 248, 249).
SOS-Induced Conditions
The ATP/Mg2+ ratio during favorable growth conditions is optimal for the balance between DNA replication and occasional recombinational repair; however, this ratio may change when rapidly growing cells experience massive DNA damage. Massive DNA damage triggers a rapid severalfold increase in the intracellular dATP concentration (144, 465, 634) which is likely to stimulate RecA-promoted reactions. Maybe even more importantly, the intracellular ATP concentration follows suit and rises two- to threefold, preceding SOS induction (28, 29, 140, 226, 465). The resulting increase in ATP concentration to 6 to 9 mM could temporarily increase the ATP/free-Mg2+ ratio, although it is unlikely that all the free Mg2+ will be exhausted, since the total intracellular pool of this cation stands at about 100 mM (442). The cause of this rise in the concentration of DNA and RNA precursors is proposed to be direct inhibition of DNA synthesis by the DNA damage and replisome idling at the lesions (167, 245, 335, 492). The demonstration that when DNA replication is blocked, the rate of a nucleotide pool expansion equals the rate of incorporation of this nucleotide immediately before the block, while when DNA replication is allowed, the rate of the nucleotide pool contraction is commensurate with the new rate of incorporation (414), substantiates this idea. The important change could be the combined MgATP2− + Mg2+ pool, which raises over 10 mM.
It was suggested on several occasions that, through inhibiting replication, DNA damage induces the SOS conditions that favor recombinational repair over DNA replication and that the mechanism of SOS induction works by affecting the competition between SSB and RecA for ssDNA (167, 335, 492, 506) (Table 4). Thus, while recombinational repair is initiated by persistent ssDNA in situations when DNA replication is not generally inhibited, the SOS response could be induced when the first condition is followed by the nucleotide imbalance caused by replisome inhibition at frequent DNA lesions. From this perspective, recombinational repair and the following SOS response are the two stages in the same process of dealing with DNA synthesis irregularities. These ideas underscore the importance of measuring the intracellular Mg2+ and nucleoside triphosphate concentrations under conditions of normal and inhibited DNA replication.
TABLE 4.
In vitro properties of the SSB and RecA proteins of E. coli and their suggested interplay in the SOS inductiona
| Property | Regular conditions | SOS-induced conditions |
|---|---|---|
| In vivo | ||
| DNA synthesis | Regular | Inhibited |
| ATP and dATP concn | Low | High |
| SOS induction | No | Yes |
| Hypothesized | ||
| MgATP2− concn | Low | High |
| In vitro | ||
| Mode of SSB binding to ssDNA | SSB35 | SSB65 |
| Binding of RecA to ssDNAb | Slow | Fast |
| ATPase activity of RecA on ssDNAb | Low | High |
| RecA polymerization on ssDNA in presence of SSB | No | Yes |
| RecA-promoted strand invasion | No | Yes |
| Cleavage of phage repressors by activated RecA | No | Yes |
Data from reference 335.
Etheno-DNA.
SOS Expression as a Compensation
The hypothesized increase in the MgATP2− concentration in response to DNA damage should adversely affect cellular processes, which are dependent on the “regular” MgATP2− concentrations. From this perspective, the increase in RuvAB production during the SOS response could be such a compensation for the decreased activity of the enzyme due to the suboptimal conditions. Furthermore, the very different in vitro ATP/Mg2+ optimum ratios for RuvAB and RecG Holliday junction translocases (Table 3) suggest that in vivo RuvAB is more active under “regular” conditions whereas RecG is more active under the SOS-induced conditions.
The possibility that the SOS-induced conditions are inhibitory for some required functions suggests that some of the SOS-induced proteins specifically counteract this SOS inhibition of other important proteins. For example, because of the precursor imbalance, DNA replisomes could be inhibited directly during SOS induction. The recently characterized gene dinB is responsible for mutagenesis of undamaged DNA in cells with an induced SOS response (304). The product of dinB is an error-prone DNA polymerase (pol IV), which could function to increase the replication speed or processivity under SOS conditions at the price of elevated mutagenesis.
Another possible example of such an SOS-compensatory activity is RecN protein, whose function is still unknown. Viability of E. coli after exposure to ionizing radiation or mitomycin C (which cross-links DNA) is severely compromised in radB/recN mutants (493, 553, 554, 556). recN is not required for chromosomal rejoining after low doses of irradiation but becomes increasingly important at higher doses (555). recN expression, which is normally undetectable, is greatly induced during the SOS response (182). The recN promoter has two binding sites for the LexA repressor, making recN one of the most tightly regulated genes of the SOS regulon (532). Sequence analysis of recN reveals an ATP-binding domain but no other peculiarities that would hint to a possible function of its product (532). The function of RecN is unlikely to be RecBC pathway specific, since recN mutants are also defective in double-strand end repair along the RecF pathway (see “RecF pathway” above). The degradation of damaged DNA is unaffected in recN mutants (553). Since the initial rejoining of double-strand ends in recN mutants seems to be normal, while the massive rejoining efforts are blocked, RecN might be needed for slowing the nucleoid segregation (see “Nucleoid segregation and the problem of accessibility” above) in response to DNA damage, since double-strand break repair depends critically on the availability of sister duplex.
Summary
During regular growth, intracellular ionic conditions must be optimal for DNA replication but could be suboptimal for recombinational repair. When DNA replication is inhibited due to DNA lesions or replisome malfunction, the intracellular conditions, notably nucleotide disbalance, are hypothesized to become optimal for recombinational repair and to facilitate the SOS induction. Some of the SOS-induced proteins could function better under the nucleotide imbalance conditions. The intracellular concentrations of the important components of the nucleic acid metabolism need to be studied under both the regular and the inhibited growth conditions.
SINGLE-STRAND ANNEALING: THE PHAGE WAY TO LINK HOMOLOGOUS DOUBLE-STRAND ENDS
Single-Strand Annealing in DNA Metabolism of Lambdoid Phages
Overview of SSA repair.
In contrast to the enzymatically complex and cumbersome mechanisms of double-strand end repair in the E. coli chromosome, some bacteriophages connect homologous double-strand ends by a simple yet effective trick. However, while saving on enzymes, phages are wasteful with their DNA. This strategy would be unacceptable for the E. coli chromosome but is quite affordable for phage genomes, which are only 1 to 2% of the length of the E. coli genome and, by the end of infection, are present in cells in multiple copies.
The two recombinational repair pathways in bacterial cells depend on the RecA protein. Indeed, the essence of recombinational repair is to synapse a damaged DNA with an intact homologous sequence, and RecA is the only enzyme in bacterial cells that can catalyze such synapsis. Another possible way of repairing damaged DNA is to synapse it with another damaged homologous DNA. Two-strand DNA lesions are always associated with lengthy tracts of ssDNA. Daughter strand gaps are single stranded themselves; double-strand breaks are processed so as to have single-strand overhangs at the double-strand ends. If these tracts of ssDNA on the homologous molecules are complementary to each other, they can anneal to form a duplex. Annealing of complementary DNA strands does not require the participation of RecA and is promoted by a considerable number of other proteins (321). Annealing of complementary single strands associated with nonoverlapping lesions converts a pair of two-strand lesions into a pair of one-strand lesions and serves as a foundation for the whole class of simple recombinational repair reactions.
A particular example of such single-strand annealing (SSA) reaction involves linking two DNA ends carrying directly repeated sequences in the overlapping configuration, as in the repair of a double-strand break between direct repeats (638, 665) (Fig. 30). If both ends are processed so that strands of a particular polarity are degraded, the opposite strands bearing complements of the direct repeat can anneal to form duplex DNA. Filling in the gaps and clipping off single-stranded regions excluded from the duplex completes the linking process (Fig. 30). Although this reaction involves homologous recognition, it cannot be faithful: as a result of it, one copy of the repeat and whatever sequences happened to be on the wrong side of the repeat are lost. Hence, recombination associated with SSA repair is said to be of the nonconservative type.
FIG. 30.
SSA repair of a double-strand break between direct repeats. Direct repeats are shown as black arrows. Explanations are given in the figure and in the text.
Because of the nonrepetitive nature of the genome and because it does not keep multiple genomes in one cell, E. coli cannot exercise SSA repair in its chromosome. However, SSA repair is the mechanism of choice for concatemeric phage and plasmid genomes. Both phage λ and the lambdoid Rac prophage (see “RecE pathway” above) encode two recombinational repair enzymes; the repair reactions promoted by these purified enzymes in vitro are of the SSA type (90, 233). D. radiodurans, which has 4 to 10 genome equivalents of DNA in each cell (437), may use SSA at the early stages of recovery after massive DNA damage (141).
SSA enzymes of phage λ.
Soon after the discovery of the recA gene in E. coli (107), it was found that λ recombination is not affected by recA mutations (74, 645, 689). The λ genes redα and redβ were found to be required for this RecA-independent λ recombination (166, 193, 589, 592); the corresponding proteins are an exonuclease (88, 365, 589) and an ssDNA-annealing protein (306, 455). Later, yet another gene, gam, was found to be required for the maximal levels of the Red-promoted recombination (175, 767). If unchecked, the host RecBCD nuclease (see “Preparation of double-strand ends by RecBCD nuclease for RecA polymerization” above) degrades the linear concatemeric products of λ rolling-circle DNA replication (175, 220). Gam binds to RecBCD and inhibits all the known activities of this enzyme (294, 457, 686). Gam analogs are produced by many bacteriophages of E. coli replicating their genomes as linear DNA (542).
λ exonuclease, the gp redα, degrades the 5′-ending strand of linear duplex DNA, generating long 3′ overhangs (88, 365), and also slowly degrades short ssDNA (614). The exonuclease-dependent several-thousand-nucleotide 3′ overhangs are detected at the duplex DNA ends during λ infection (252). The enzyme is unable to begin DNA degradation at nicks and is inhibited by completion of strand assimilation (90, 91) (Fig. 31A). λ exonuclease has a high processivity on duplex DNA, degrading on average 3,000 nucleotides per binding event (88). The reason for the high processivity is revealed by the exonuclease atomic structure: the enzyme is a trimeric doughnut with a hole that is proposed to allow it to slide along ssDNA, hydrolyzing the complementary strand (318). During isolation from infected cells, half of the λ exonuclease activity is purified in a complex with another phage protein, called Beta (505).
FIG. 31.
Some reactions promoted by the SSA repair proteins. (A) Strand assimilation catalyzed by λ exonuclease. The exonuclease is the washer marked exo; the small arrow below shows the direction of the exonucleolytic degradation. The exonuclease will degrade the 5′-ending strand of the duplex until the branching strand is completely assimilated into the duplex (91). The exonuclease stops its progress when strand assimilation generates a single-strand interruption, accounting for the inability of the enzyme to start DNA degradation from single-strand interruptions. (B) Three-stranded branch migration catalyzed by RecT or Beta. If the RecT- or Beta-promoted annealing of complementary strands encounters a duplex region overlapping the protein filament on the other DNA, the protein catalyzes strand exchange, displacing the resident strand by the incoming strand. The reaction resembles the three-strand branch migration promoted by RecA protein. However, in contrast to the RecA-promoted strand exchange, RecT and λ Beta are able to start this reaction only if both participating molecules have complementary single-stranded regions (233, 358).
Beta protein, the gp redβ, is a 30-kDa ssDNA-binding protein which promotes the annealing of complementary DNA strands (306, 455). If the annealing reaction runs into a duplex region, Beta catalyzes strand displacement, also known as branch migration or strand exchange (358) (Fig. 31B). The strand exchange reaction reveals that Beta promotes annealing with 5′-to-3′ polarity relative to the ssDNA to which it is bound. Beta does not bind duplex DNA but stays bound to the supposedly duplex product of the annealing reaction (291), suggesting that the mechanism of strand exchange is through the polarized association of the protein with the products of strand exchange (233). Another possible function of Beta is protection of single-stranded overhangs, generated by λ Exo, from the host ssDNA-specific exonucleases, mostly ExoI (see “RecE pathway” above). This idea is substantiated by the in vitro findings that (i) 3′ ends stimulate Beta binding to ssDNA and (ii) double-strand ends with 3′ overhangs are protected by Beta from nuclease degradation four times better than are double-strand ends with 5′ overhangs (291, 358).
Yet another possible function of Beta is suggested by the fact that this protein is purified from infected cells in a complex not only with λ exo but also with the host S1 ribosomal protein and with the NusA subunit of RNA polymerase (454, 695). S1 is the largest protein of the small ribosome subunit and is responsible for mRNA binding (682). NusA is a regulatory subunit of RNA polymerase—it serves as a transcription elongation factor interacting with other proteins (143). By binding to these regulatory factors, Beta could function to clear the DNA of transcription-translation complexes ahead of the exonuclease.
SSA enzymes of the Rac prophage.
Rac is a cryptic lambdoid prophage residing in the E. coli chromosome (see “RecE pathway” above). It has at least two SSA repair genes, which are usually silent but can be activated by sbcA mutations. These two recombinational genes complement λ red mutants; in fact, the two genes are picked up from the chromosome by λ with its own recombinational genes deleted, resulting in a recombination-proficient revertant (216, 289, 767). The product of recE is ExoVIII, which degrades the 5′-ending strand of linear duplex DNA and, hence, is analogous to λ exo (285). However, the two genes do not have significant sequence similarity, and the proteins are very different: while λ Exo is a 26-kDa dwarf, ExoVIII is a 140-kDa monster (284).
Beta protein of phage λ also has its counterpart in the Rac prophage, called RecT. The 33-kDa RecT promotes the annealing of complementary single strands and also catalyzes a three-strand branch migration when the annealing reaction runs into duplex DNA (232, 233) (Fig. 31B). The biological significance of the latter reaction, also observed with λ Beta, is not obvious, although it is conceptually similar to the strand assimilation reaction catalyzed by λ exonuclease (Fig. 31A). In vitro, in the absence of Mg2+, RecT promotes detectable strand invasion of ssDNA into a homologous supercoiled duplex; the mechanism of this pairing could be through RecT binding to duplex DNA under these conditions (468).
Mechanisms of double-strand end repair in λ infection: invasion versus annealing.
Biochemical characterization of the recombination functions of λ showed that in vivo this phage was likely to recombine by SSA mechanism (90, 358). In the proposed scheme, λ exonuclease would degrade the 5′-ending strands of the two ends with overlapping homology while λ Beta would anneal the unraveled complementary 3′-ending strands. The final processing of recombination intermediates requires strand assimilation and sealing of the single-strand interruptions by DNA ligase (90), which is provided by the host.
Later it was found that in WT cells λ often recombines via RecA-catalyzed strand invasion (616, 623), analogous to the RecE pathway of double-strand end repair (see “RecE pathway” above). However, the frequency of λ recombination is not influenced by recA mutations of the host, making it unclear why the phage has to switch from the invasion type of recombination to the annealing type in the absence of RecA. Therefore, λ Beta was repeatedly proposed to substitute for RecA to catalyze strand invasion during the phage RecA-independent recombination (616, 620, 643, 663), although these proposals were ignoring the failure to demonstrate the supposed strand invasion activity of Beta in vitro (455).
In fact, these proposals were also disregarding the earlier in vivo studies in which inhibition of DNA replication revealed qualitative differences in recombination of red+ λ in the presence and absence of RecA. Whereas this recombination is largely RecA independent in standard crosses (74, 193), it becomes largely RecA dependent when the phage DNA replication is blocked (617–619). This RecA dependence is not due to the hypothetical destruction of the Red recombination intermediates in recA mutant cells by RecBCD nuclease (621), in contrast to the suggestion based on studies of the intracellular pool of λ DNA (733). Furthermore, in a different in vivo setup, when the Red system of λ has to pair a double-strand end with an intact duplex, RecA is also required (114, 497).
The question about the nature of the mechanism of λ recombination in the absence of RecA was addressed in a study which explored the opposing predictions generated by the two competing mechanisms (623) (Fig. 32). The invasion scheme predicted that, in the absence of DNA replication, (i) λ recombination would be dependent on RecA, (ii) a solitary double-strand break would be sufficient to stimulate recombination, and (iii) a track of hybrid DNA formed at the site of the invasion would be relatively short. In contrast, the SSA scheme predicted that in the absence of DNA replication, (i) λ recombination would not require RecA, (ii) for recombination in the absence of RecA, nonallelic double-strand ends would be required; and (iii) in such RecA-independent recombinants, hybrid DNA could span the entire length between the two double-strand ends. These two sets of predictions were tested in vivo by using both physical and genetic techniques; the conclusions were that (i) in the absence of RecA, recombination of λ promoted by the Red pathway follows the SSA mechanism, and (ii) when RecA is available, the Red-mediated recombination follows the more robust invasion mechanism (623).
FIG. 32.
Comparison of the double-strand end invasion with single-strand annealing reactions. Duplex DNA is shown as double lines (open or solid); the cos (packaging) site of λ is shown as a chevron. (A) Circular λ chromosomes. (B) The chromosomes in panel A are cut at different and unique sites in vivo (marked with scissors in panel A. (C) The ends are processed by λ exo to generate 3′ overhangs. (D and E) The RecA-catalyzed invasion of the 3′ overhang into an intact λ chromosome with the following resolution of the joint molecule (190). (F) Alternatively, the 3′ overhang is annealed by Beta with a complementary 3′ overhang of another linear λ chromosome, cut at a different location. (G) Completion of the SSA recombination. Note that the hybrid region in panel E is expected to be shorter than in panel G.
Possible role of SSA repair in the life cycle of λ.
If the Lambda Red system could indeed catalyze strand invasion in the absence of RecA, it should be able to promote recombination and repair in the E. coli chromosome. Generalized phage transduction requires “double-strand end repair” of the linear transducing DNA with a circular chromosome. Consistent with the notion that any strand invasion into a circular duplex requires RecA, the frequency of generalized transduction is reduced 2 to 3 orders of magnitude in recA mutants (245, 765). Remarkably, the Red recombination system of phage λ is able to catalyze a low level of transduction in the absence of RecA (705, 720). This Red-promoted recombination could occur at replication forks, where single-stranded pieces of DNA could be gradually assimilated via annealing with the template for the lagging-strand DNA synthesis (Fig. 33). The scheme predicts that the RecA-independent Red-mediated transduction will be influenced by the position of replication forks and will be inhibited if DNA replication is temporarily blocked.
FIG. 33.
Mechanism for the Red-mediated transduction in the absence of RecA. Open lines indicate the transducing duplex DNA; solid lines indicate the host chromosome; arrows indicate DNA synthesis at the 3′ ends; the circle indicates λ exo. (A) The transducing DNA is degraded by λ exo, while a replication fork is passing through the homologous region in the chromosome. (B) λ Beta catalyzes annealing of the transducing strand with the template for the lagging-strand DNA synthesis. (C) Assimilation of the incoming strand into the host DNA catalyzed by the combined action of λ exo and Beta. (D) The strand is completely assimilated. There is a heterology (shown as a bump) between the transducing DNA strand and the host DNA strand. This heterology is probably resolved by the next round of DNA replication, since the incoming DNA is likely to be fully methylated (a poor substrate for the methyl-directed mismatch repair [see “Damage reversal and one-strand repair”]).
The low level of RecA-independent transduction is the only recombinational repair transaction the Red or the RecE pathways can perform in the chromosome in the absence of RecA, confirming the SSA nature of recombinational repair they catalyze. SSA repair cannot possibly restore a disintegrated replication fork and, in general, cannot mend the chromosomal DNA lesions in recA mutants (206), underscoring the fact that its usefulness is restricted to highly repetitive linear genomes. In which circumstances would SSA repair be preferable to double-strand end repair?
An apparent but still poorly understood effect of the Red system on phage physiology is the stimulation of λ DNA replication, since red infections produce only one-third of the phage DNA produced by the WT infections (175, 767). Early in infection, phage λ replicates as a big plasmid, generating circular monomeric copies of its chromosome. Later, the phage replication switches from theta to sigma mode, spooling linear arrays that are several genomes long, which are the preferred substrate for packaging (Fig. 34). The clue to understanding the stimulatory role of the Red recombination may lie in the λ packaging enzyme, terminase, which should interfere with phage DNA replication by cutting the circular domains of the replicating “sigmas.” Due to this terminase action, λ DNA late in infection is represented mostly by linear pieces of different lengths. Initiation of λ DNA replication in vitro requires supercoiling (5, 739); since initiation in vivo is likely to require supercoiling as well, only circles could be initiation competent. Therefore, the Red-catalyzed SSA could boost λ DNA replication by restoring circular phage chromosomes (Fig. 34, step 13; also see Fig. 36) and allowing new initiations.
FIG. 34.
Role of invasion-type and annealing-type recombination in phage λ DNA metabolism. Duplex λ chromosome is shown as a thick single line or a circle; a mature phage particle is shown at the bottom. The two basic processes, DNA replication (steps 1, 2, and 9) and DNA degradation due to the acting of packaging enzyme terminase (step 12) and spurious strand breaks (steps 3, 5, and 7), are shown in stages on the left and on the right, eventually leading to packaging of the phage DNA into the capsid (step 10). Homologous recombination takes the degradation products and returns them into the DNA replication metabolism (steps 4, 6, 8, 11, and 13). Steps are as follows: 1, theta replication of the circular λ chromosome; 2, completion of the theta replication; 3, collapse of one of the replication forks, converting the theta-replicating chromosome into a sigma-replicating one; 4, RecA- and Red-dependent recombinational repair of the collapsed replication fork, returning the chromosome to theta replication; 5, collapse of the replication fork due to the nick in the linear DNA strand of the sigma structure, yielding a circular chromosome and a short linear fragment; 6, RecA- and Red-dependent invasion-type recombination, generating a sigma-replicating chromosome from a circular chromosome and a short linear chromosomal fragment; 7, collapse of the replication fork due to the nick in the circular DNA strand of the sigma structure, generating a long linear chromosomal fragment; 8, RecA- and Red-dependent invasion-type intramolecular recombination, generating a sigma-replicating chromosome from a long fragment; 9, DNA replication lengthening the linear tail of the sigma structure; 10, packaging of a genome segment from linear concatemers; 11, Red-dependent annealing-type recombination, combining two short fragments into a longer fragment; 12, unproductive terminase cutting to reduce the size of the linear chromosomal pieces; 13, Red-dependent annealing-type recombination, combining two short fragments or a single long fragment into a circular chromosome, able to initiate DNA replication from the origin.
FIG. 36.
SSA recircularizes linear DNA with terminal redundancies. This scheme is an illustration of step 13 in Fig. 34. Direct repeats are shown as arrows. (A) A linear piece of λ concatemeric DNA with terminal redundancies (direct repeats), unable to initiate DNA replication. (B) Strand-specific resection of double-strand ends. (C) Annealing of the unraveled complements of the direct repeat. (D) Removal of the DNA excluded from the synapsis, sealing the interruptions. The resulting recircularized λ chromosome is competent for initiation of DNA replication.
In addition to stimulation of phage DNA synthesis, SSA repair could stimulate the phage yield in a more direct way. Packaging of the λ genome starts and ends at a site on the phage chromosome called cos. The sequences important for cos recognition by the packaging machinery are mapped to both sides of the actual double-strand break site (32). At face value, this means that only genomes bracketed by two intact cos sites can be packaged in vivo, although packaging of monomers could be inefficient for other reasons (666). λ packaging is processive (180), and so longer arrays yield more packageable genomes. SSA repair could act on the products of sigma replication and leftovers from packaging to combine them into longer pieces and therefore to increase the yield of the phage (Fig. 35).
FIG. 35.
Possible role of SSA repair in a better utilization of λ DNA during packaging. This scheme is an illustration of step 11 in Fig. 34. Products of the late DNA replication (linear concatemeric pieces) are represented by double lines; small open circles indicate λ exo degrading the 5′-ending strands. Genome equivalents in the λ DNA concatemers are indicated by the short vertical lines. A packageable genome would be bracketed by two intact vertical lines. Thus, from the two initial DNA pieces (top), two phage genomes could be packaged, while the product of SSA repair (bottom) contains three packageable genome equivalents.
Single-Strand Annealing Recombination in Plasmids in the Absence of RecA
Although Rac prophage has several functional genes and is capable of excising its chromosome from the E. coli chromosome, it does not develop past this stage and therefore cannot be used as a substrate to study its own recombination. In studies of the Rac prophage-catalyzed recombinational repair, plasmids proved to be the substrates of choice. Similar to the λ SSA repair, the Rac prophage-catalyzed plasmid recombination in the absence of RecA has sometimes been viewed as an invasion-type repair.
RecA-promoted double-strand end repair in E. coli proved difficult to demonstrate in vivo with model substrates. In contrast, SSA repair, efficient because of its simplicity, is relatively easy to show in vivo with engineered constructs. This stems partly from the fact that smaller DNA substrates give higher yields with SSA repair while the opposite is observed for RecA-promoted double-strand end repair. Furthermore, in some experimental setups, the recombinational products do not allow us to distinguish between the two mechanisms. The difficulties in discrimination between the two mechanisms led to a series of observations that were interpreted in terms of the invasion-type repair, although the results are in fact more compatible with SSA repair.
Double-strand break repair in plasmids with direct repeats.
In the recBC sbcA genetic background, where the recombinational system of the Rac prophage is expressed (see “RecE pathway” and “SSA enzymes of the Rac prophage” above), formation of deletions between direct repeats in plasmids does not require RecA (186, 341). It is thought that in this background, plasmids form linear multimers similar to concatemeric DNA of phage λ (115) and then recircularize in a RecA-independent manner, deleting the DNA between the repeats (593) (Fig. 36). If transformed with a linear dimer plasmid, a recBC sbcA strain recircularizes the plasmid with an efficiency approaching unity (636). This efficient double-strand break repair depends only on the RecE exonuclease and is independent of any other tested Rec proteins (RecT has not been tested), including RecA (390, 636). Long heterologies at one end of such linear plasmids do not preclude efficient recircularization (389, 594). The last finding was interpreted to signify a mechanism based on double-strand end invasion, with the role of synaptase being played by the RecT protein (389, 594). However, since RecT alone does not promote RecA-like synapsis (233), SSA repair remains the more economical way to explain these data. SSA repair should be able to tolerate substantial end heterologies, with the single-strand “whiskers” being removed by ssDNA-specific nucleases.
Double-strand break repair in plasmids with inverted repeats.
In a plasmid with an inverted repeat, each inverted segment containing a drug resistance gene with nonallelic mutations, recombination between the segments can restore the functional drug resistance gene in one of them (751). When WT or recBC sbcA cells are transformed with such a plasmid and plated for determination of the drug resistance, recombinants are infrequent (308, 644). However, if recBC sbcA cells are transformed with the plasmid that was cut in the middle of one of the repeats, the recombination frequency increases 100-fold (WT cells show no increase) (308, 331, 644). Two recombinational products are formed: 30% of the recombinants have the original configuration of the outside markers, while the other 70% of the recombinants have the markers flipped (crossover) (Fig. 37) (308). This double-strand break-stimulated recombination absolutely requires the Rac prophage-encoded RecE and RecT proteins but is independent of any host recombinational gene, including recA (331, 644). The ratio of the two recombinational products is more or less constant across the “recombinational alphabet,” with the exception of recJ mutants, in which the ratio is reversed (331). Cloned recombinational genes of phage λ also promote this double-strand break repair reaction (643). RecA independence notwithstanding, the authors explain these results in terms of the invasion-type double-strand break repair, assigning the role of RecA to RecT of Rac or to λ Beta (308, 331, 643, 644). However, the formation of the two types of recombinants in this peculiar system is explained by SSA repair as well (Fig. 38); this explanation is more attractive because it does not require a biochemically undemonstrated RecA-like activity. SSA repair also explains the influence of the RecJ exonuclease, which could normally degrade the 5′ end of the intermediate in Fig. 38F, preventing the formation of the noncrossover product. In the absence of the RecJ nuclease, there is no discrimination against the noncrossover products, resulting in the flipped ratio of crossovers to noncrossovers. Finally, it is quite illustrative that the RecF pathway, which is so genetically similar to the RecE pathway in the RecA-dependent double-strand end repair (see “Double-strand end repair in the absence of RecBCD” above), is quite incapable of promoting this kind of recombination in the linearized substrates (750).
FIG. 37.
The two products of double-strand gap repair in a plasmid with inverted repeats. Inverted repeats are shown as two black-and-white arrows on the opposite sides of the plasmid circle. One side of the circle is marked by knots on DNA strands to facilitate the detection of the inversion (crossover). The letters A, B, C, and D serve the same purpose.
FIG. 38.
SSA repair of a double-strand break in a plasmid with inverted repeats. Inverted repeats are shown as black-and-white arrows on the opposite sides of the plasmid circle. The letters A, B, C, and D help to reveal the inversion. (A) A plasmid carrying two inverted repeats with a double-strand gap in the middle of one of the repeats. (B) Degradation of the 5′-ending strand from one of the ends up to the middle of the intact inverted repeat. (C) The unraveled strand anneals on itself at the inverted repeat. The degradation stops (Fig. 31A), and the nick is sealed, resulting in a linear molecule with a loop at one end. (D) Degradation of the same polarity from the other end to the middle of the intact repeat. (E) The unraveled strand anneals on itself again, using the inverted repeat. The resulting circular ssDNA is then converted to a double-stranded form (not shown). It has markers on the opposite sides of the inverted repeats flipped, as if there was a conservative repair with associated crossing over (Fig. 37, left). (F) Alternatively, the degradation in panel B goes all the way to the other end of the molecule, rendering it completely single stranded. (G) The broken repeat anneals with the intact repeat and is repaired off it. Resynthesis of the degraded strand (not shown) generates a product identical to the one of the conservative double-strand break repair without crossing over (Fig. 37, right).
Summary
The SSA recombination is a simple alternative to the RecA-catalyzed double-strand end repair and is optimally suited for the repair needs of the concatemeric precursors of the phage λ chromosome. SSA repair requires only two phage-encoded enzymes, of which the function of the exonuclease is more or less understood, but the importance of the annealing protein needs investigation. Also, our understanding of the in vivo role of SSA recombination in the phage λ DNA metabolism will benefit from further experiments.
The phage-encoded recombinases can also catalyze double-strand end repair in the E. coli plasmids in the absence of RecA. The resulting recombination products do not allow us to distinguish between the SSA mechanism and the invasion mechanism (catalyzed in E. coli exclusively by RecA); therefore, caution should be exercised in the interpretation of these results.
CONCLUSION AND FUTURE DIRECTIONS
Initially, recombinational repair was known only because of its genetic consequences—the formation of recombinant chromosomes. Over the years, genetic and then also biochemical studies have generated a wealth of information about homologous recombination and recombinational enzymes in E. coli. However, only recently has homologous recombination started to be widely perceived as a repair system for special DNA damage, brought about mostly by DNA replication. The contemporary schemes of this process attempt to connect DNA replication and recombinational repair into a single metabolic network.
Investigation of this interconnected metabolic pathways will benefit from progress in three major areas. The first is classical biochemistry, which has already developed separate in vitro systems for DNA replication and homologous recombination. Now is the turn of the more complex systems, in which DNA replication of a damaged template would trigger its recombinational repair, which, in its turn, would lead to a resumption of DNA replication.
This challenging task should be aided by experiments in the second area, namely, the study of the in vivo DNA metabolism and general cell physiology during regular DNA replication as well as under conditions of DNA damage. The goal of this research is to find metabolic cues (changes in ion concentrations?) and accessory activities that regulate the complex interplay of DNA replication and repair in vivo. Global regulation of gene expression during transition from DNA replication to recombinational repair and back to normal DNA replication, monitored with DNA chips, could be informative.
Finally, the third area of future studies, the so-called in vivo biochemistry, has recently become available for E. coli, as ways to control the powerful nuclease activities of bacterial cells have been developed. The approach is to introduce substantial amounts of repair substrates in vivo, to let the cells proceed with the repair reactions, and then to retrieve the recombining DNA from cells and analyze it in vitro. Thus, in vivo reactions can be subdivided into phases, and the genetic requirements of these phases can be studied by using cells as test tubes. This approach promises to bridge the concepts, developed both within classical biochemistry and within cell physiology, into a unifying theory of recombinational repair in E. coli.
ACKNOWLEDGMENTS
I am grateful to Michael Cox, Steve Kowalczykowski, Bénédicte Michel, Gerry Smith, Frank Stahl, and Steve West for enlightening criticism and encouragement.
I am supported by the NSF grant MCB-9402695.
REFERENCES
- 1.Adams D E, Tsaneva I R, West S C. Dissociation of RecA filament from duplex DNA by the RuvA and RuvB DNA repair proteins. Proc Natl Acad Sci USA. 1994;91:9901–9905. doi: 10.1073/pnas.91.21.9901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adzuma K. Stable synapsis of homologous DNA molecules mediated by the Escherichia coli RecA protein involves local exchange of DNA strands. Genes Dev. 1992;6:1679–1694. doi: 10.1101/gad.6.9.1679. [DOI] [PubMed] [Google Scholar]
- 3.Alatossava T, Jütte H, Kuhn A, Kellenberger E. Manipulation of intracellular magnesium content in polymyxin B nonapeptide-sensitized Escherichia coli by ionophore A23187. J Bacteriol. 1985;162:413–419. doi: 10.1128/jb.162.1.413-419.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Deib A A, Mahdi A A, Lloyd R G. Modulation of recombination and DNA repair by the RecG and PriA helicases of Escherichia coli K-12. J Bacteriol. 1996;178:6782–6789. doi: 10.1128/jb.178.23.6782-6789.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alfano C, McMacken R. The role of template superhelicity in the initiation of bacteriophage λ DNA replication. Nucleic Acids Res. 1988;16:9611–9630. doi: 10.1093/nar/16.20.9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allen G C, Jr, Kornberg A. Assembly of the primosome of DNA replication in Escherichia coli. J Biol Chem. 1993;268:19204–19209. [PubMed] [Google Scholar]
- 7.Alonso J C, Shirahige K, Ogasawara N. Molecular cloning, genetic characterization and DNA sequence analysis of the recM region of Bacillus subtilis. Nucleic Acids Res. 1990;18:6771–6777. doi: 10.1093/nar/18.23.6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altshuler M. Recovery of DNA replication in UV-damaged Escherichia coli. Mutat Res. 1993;294:91–100. doi: 10.1016/0921-8777(93)90017-b. [DOI] [PubMed] [Google Scholar]
- 9.Amabile-Cuevas C F. Origin, evolution and spread of antibiotic resistance genes. R. G. Austin, Tex: Landes Co.; 1993. [Google Scholar]
- 10.Amundsen S K, Neiman A M, Thibodeaux S M, Smith G R. Genetic dissection of the biochemical activities of RecBCD enzyme. Genetics. 1990;126:25–40. doi: 10.1093/genetics/126.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amundsen S K, Taylor A F, Chaudhury A M, Smith G R. recD: the gene for an essential third subunit of exonuclease V. Proc Natl Acad Sci USA. 1986;83:5558–5562. doi: 10.1073/pnas.83.15.5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ananthaswamy H N, Eisenstark A. Repair of hydrogen peroxide-induced single-strand breaks in Escherichia coli deoxyribonucleic acid. J Bacteriol. 1977;130:187–191. doi: 10.1128/jb.130.1.187-191.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson D G, Churchill J J, Kowalczykowski S C. Chi-activated RecBCD enzyme possesses 5′-3′ nucleolytic activity, but RecBC enzyme does not: evidence suggesting that the alteration induced by Chi is not simply ejection of the RecD subunit. Genes Cells. 1997;2:117–128. doi: 10.1046/j.1365-2443.1997.1130311.x. [DOI] [PubMed] [Google Scholar]
- 14.Anderson D G, Kowalczykowski S C. The recombination hot spot χ is a regulatory element that switches the polarity of DNA degradation by the RecBCD enzyme. Genes Dev. 1997;11:571–581. doi: 10.1101/gad.11.5.571. [DOI] [PubMed] [Google Scholar]
- 15.Anderson D G, Kowalczykowski S C. The translocation RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a χ-regulated manner. Cell. 1997;90:77–86. doi: 10.1016/s0092-8674(00)80315-3. [DOI] [PubMed] [Google Scholar]
- 16.Ariyoshi M, Vassylyev D G, Iwasaki H, Nakamura H, Shinagawa H, Morikawa K. Atomic structure of the RuvC resolvase: a Holliday junction-specific endonuclease from E. coli. Cell. 1994;78:1063–1072. doi: 10.1016/0092-8674(94)90280-1. [DOI] [PubMed] [Google Scholar]
- 17.Armengod M E, Garcia-Sogo M, Perez-Roger I, Macian F, Navarro-Avino J P. Tandem transcription termination sites in the dnaN gene of Escherichia coli. J Biol Chem. 1991;266:19725–19730. [PubMed] [Google Scholar]
- 18.Arthur H M, Lloyd R G. Hyper-recombination in uvrD mutants of Escherichia coli K-12. Mol Gen Genet. 1980;180:185–191. doi: 10.1007/BF00267368. [DOI] [PubMed] [Google Scholar]
- 19.Asai T, Bates D B, Kogoma T. DNA replication triggered by double-strand breaks in E. coli: dependence on homologous recombination functions. Cell. 1994;78:1051–1061. doi: 10.1016/0092-8674(94)90279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Asai T, Kogoma T. D-loops and R-loops: alternative mechanisms for the initiation of chromosome replication in Escherichia coli. J Bacteriol. 1994;176:1807–1812. doi: 10.1128/jb.176.7.1807-1812.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Asai T, Sommer S, Bailone A, Kogoma T. Homologous recombination-dependent initiation of DNA replication from damage-inducible origins in Escherichia coli. EMBO J. 1993;12:3287–3295. doi: 10.1002/j.1460-2075.1993.tb05998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Au K G, Welsh K, Modrich P. Initiation of methyl-directed mismatch repair. J Biol Chem. 1992;267:12142–12148. [PubMed] [Google Scholar]
- 23.Bagg A, Kenyon C J, Walker G C. Inducibility of a gene product required for UV and chemical mutagenesis in Escherichia coli. Proc Natl Acad Sci USA. 1981;78:5749–5753. doi: 10.1073/pnas.78.9.5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bailone A, Sommer S, Knezevic J, Dutreix M, Devoret R. A RecA protein mutant deficient in its interaction with the UmuDC complex. Biochimie. 1991;73:479–484. doi: 10.1016/0300-9084(91)90115-h. [DOI] [PubMed] [Google Scholar]
- 25.Baker T A, Wickner S H. Genetics and enzymology of DNA replication in Escherichia coli. Annu Rev Genet. 1992;26:447–477. doi: 10.1146/annurev.ge.26.120192.002311. [DOI] [PubMed] [Google Scholar]
- 26.Bale A, d’Alarcao M, Marinus M G. Characterization of DNA adenine methylation mutants of Escherichia coli K12. Mutat Res. 1979;59:157–165. doi: 10.1016/0027-5107(79)90153-2. [DOI] [PubMed] [Google Scholar]
- 27.Baliga R, Singleton J W, Dervan P B. RecA-oligonucleotide filaments bind in the minor groove of double-stranded DNA. Proc Natl Acad Sci USA. 1995;92:10393–10397. doi: 10.1073/pnas.92.22.10393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barbé J, Vericat J A, Cairó J, Guerrero R. Further characterization of SOS system induction in recBC mutants of Escherichia coli. Mutat Res. 1985;146:23–32. doi: 10.1016/0167-8817(85)90051-3. [DOI] [PubMed] [Google Scholar]
- 29.Barbé J, Villaverde A, Guerrero R. Evolution of cellular ATP concentration after UV-mediated induction of SOS system in Escherichia coli. Biochem Biophys Res Commun. 1983;117:556–561. doi: 10.1016/0006-291x(83)91236-6. [DOI] [PubMed] [Google Scholar]
- 30.Barfknecht T R, Smith K C. The involvement of DNA polymerase I in the postreplication repair of ultraviolet radiation-induced damage in Escherichia coli K-12. Mol Gen Genet. 1978;167:37–41. doi: 10.1007/BF00270319. [DOI] [PubMed] [Google Scholar]
- 31.Bazemore L R, Takahashi M, Radding C M. Kinetic analysis of pairing and strand exchange catalyzed by RecA. Detection by fluorescence energy transfer. J Biol Chem. 1997;272:14672–14682. doi: 10.1074/jbc.272.23.14672. [DOI] [PubMed] [Google Scholar]
- 32.Becker A, Murialdo H. Bacteriophage λ DNA: the beginning of the end. J Bacteriol. 1990;172:2819–2824. doi: 10.1128/jb.172.6.2819-2824.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bennett R J, Dunderdale H J, West S C. Resolution of Holliday junctions by RuvC resolvase: cleavage specificity and DNA distortion. Cell. 1993;74:1021–1031. doi: 10.1016/0092-8674(93)90724-5. [DOI] [PubMed] [Google Scholar]
- 34.Bennett R J, West S C. Resolution of Holliday junctions in genetic recombination: RuvC protein nicks DNA at the point of strand exchange. Proc Natl Acad Sci USA. 1996;93:12217–12222. doi: 10.1073/pnas.93.22.12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bennett R J, West S C. RuvC protein resolves Holliday junctions via cleavage of the continuous (noncrossover) strands. Proc Natl Acad Sci USA. 1995;92:5635–5639. doi: 10.1073/pnas.92.12.5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bennett R J, West S C. Structural analysis of the RuvC-Holliday junction complex reveals an unfolded junction. J Mol Biol. 1995;252:213–226. doi: 10.1006/jmbi.1995.0489. [DOI] [PubMed] [Google Scholar]
- 37.Benson F, Collier S, Lloyd R G. Evidence of abortive recombination in ruv mutants of Escherichia coli K12. Mol Gen Genet. 1991;225:266–272. doi: 10.1007/BF00269858. [DOI] [PubMed] [Google Scholar]
- 38.Benson F E, Illing G T, Sharples G J, Lloyd R G. Nucleotide sequencing of the ruv region of Escherichia coli K12 reveals a LexA regulated operon encoding two genes. Nucleic Acids Res. 1988;16:1541–1549. doi: 10.1093/nar/16.4.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berger J M. Structure of DNA topoisomerases. Biochim Biophys Acta. 1998;1400:3–18. doi: 10.1016/s0167-4781(98)00124-9. [DOI] [PubMed] [Google Scholar]
- 40.Bertucat G, Lavery R, Prévost C. A model for parallel triple helix formation by RecA: single-strand association with a homologous duplex via the minor groove. J Biomol Struct Dyn. 1998;16:535–546. doi: 10.1080/07391102.1998.10508268. [DOI] [PubMed] [Google Scholar]
- 41.Bi E, Lutkenhaus J. Cell division inhibitors SulA and MinCD prevent formation of the FtsZ ring. J Bacteriol. 1993;175:1118–1125. doi: 10.1128/jb.175.4.1118-1125.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bianchi M E, Radding C M. Insertions, deletions and mismatches in heteroduplex DNA made by RecA protein. Cell. 1983;35:511–520. doi: 10.1016/0092-8674(83)90185-x. [DOI] [PubMed] [Google Scholar]
- 43.Bianco P R, Kowalczykowski S C. The recombination hotspot Chi is recognized by the translocating RecBCD enzyme as the single strand of DNA containing the sequence 5′-GCTGGTGG-3′. Proc Natl Acad Sci USA. 1997;94:6706–6711. doi: 10.1073/pnas.94.13.6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bierne H, Michel B. When replication forks stop. Mol Microbiol. 1994;13:17–23. doi: 10.1111/j.1365-2958.1994.tb00398.x. [DOI] [PubMed] [Google Scholar]
- 45.Bierne H, Seigneur M, Ehrlich S D, Michel B. uvrD mutations enhance tandem repeat deletion in the Escherichia coli chromosome via SOS induction of the RecF recombination pathway. Mol Microbiol. 1997;26:557–567. doi: 10.1046/j.1365-2958.1997.6011973.x. [DOI] [PubMed] [Google Scholar]
- 46.Billen D. Replication of the bacterial chromosome: location of new initiation sites after irradiation. J Bacteriol. 1969;97:1169–1175. doi: 10.1128/jb.97.3.1169-1175.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Billen D. Unbalanced deoxyribonucleic acid synthesis: its role in X-ray-induced bacterial death. Biochim Biophys Acta. 1963;72:608–618. [PubMed] [Google Scholar]
- 48.Billen D, Hewitt R. Concerning the dynamics of chromosome replication and degradation in a bacterial population exposed to X-rays. Biochim Biophys Acta. 1967;138:587–595. doi: 10.1016/0005-2787(67)90554-0. [DOI] [PubMed] [Google Scholar]
- 49.Biswas I, Maguin E, Ehrlich S D, Gruss A. A 7-base-pair sequence protects DNA from exonucleolytic degradation in Lactococcus lactis. Proc Natl Acad Sci USA. 1995;92:2244–2248. doi: 10.1073/pnas.92.6.2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blaisdell B E, Rudd K E, Matin A, Karlin S. Significant dispersed recurrent DNA sequences in the Escherichia coli genome. J Mol Biol. 1993;229:833–848. doi: 10.1006/jmbi.1993.1090. [DOI] [PubMed] [Google Scholar]
- 51.Blakely G, Colloms S, May G, Burke M, Sherratt D. Escherichia coli XerC recombinase is required for chromosomal segregation at cell division. New Biol. 1991;3:789–798. [PubMed] [Google Scholar]
- 52.Blakely G, May G, McCulloch R, Arciszewska L K, Burke M, Lovett S T, Sherratt D J. Two related recombinases are required for site-specific recombination at dif and cer in E. coli K12. Cell. 1993;75:351–361. doi: 10.1016/0092-8674(93)80076-q. [DOI] [PubMed] [Google Scholar]
- 53.Blattner F R, Burland V, Plunkett III G, Sofia H J, Daniels D L. Analysis of the Escherichia coli genome. IV. DNA sequence of the region from 89.2 to 92.8. Nucleic Acids Res. 1993;21:5408–5417. doi: 10.1093/nar/21.23.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 55.Bochner B R, Ames B N. Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J Biol Chem. 1982;257:9759–9769. [PubMed] [Google Scholar]
- 56.Boehmer P E, Emmerson P T. The RecB subunit of the Escherichia coli RecBCD enzyme couples ATP hydrolysis to DNA unwinding. J Biol Chem. 1992;267:4981–4987. [PubMed] [Google Scholar]
- 57.Bohrmann B, Haider M, Kellenberger E. Concentration evaluation of chromatin in unstained resin-embedded sections by means of low-dose ratio-contrast imaging in STEM. Ultramicroscopy. 1993;49:235–251. doi: 10.1016/0304-3991(93)90230-u. [DOI] [PubMed] [Google Scholar]
- 58.Bonner C A, Randall S K, Rayssiguier C, Radman M, Eritja R, Kaplan B E, McEntee K, Goodman M F. Purification and characterization of an inducible Escherichia coli DNA polymerase capable of insertion and bypass at abasic lesions in DNA. J Biol Chem. 1988;263:18946–18952. [PubMed] [Google Scholar]
- 59.Bonner C A, Stukenberg P T, Rajagopalan M, Eritja R, O’Donnel M, McEntee K, Echols H, Goodman M F. Processive DNA synthesis by DNA polymerase II mediated by DNA polymerase III accessory proteins. J Biol Chem. 1992;267:11431–11438. [PubMed] [Google Scholar]
- 60.Bonura T, Smith K C. Enzymatic production of deoxyribonucleic acid double-strand breaks after ultraviolet irradiation of Escherichia coli K-12. J Bacteriol. 1975;121:511–517. doi: 10.1128/jb.121.2.511-517.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bonura T, Smith K C, Kaplan H S. Enzymatic induction of DNA double-strand breaks in γ-irradiated Escherichia coli K-12. Proc Natl Acad Sci USA. 1975;72:4265–4269. doi: 10.1073/pnas.72.11.4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boudsocq F, Campbell M, Devoret R, Bailone A. Quantitation of the inhibition of Hfr × F− recombination by the mutagenesis complex UmuD′C. J Mol Biol. 1997;270:201–211. doi: 10.1006/jmbi.1997.1098. [DOI] [PubMed] [Google Scholar]
- 63.Boyce R P, Howard-Flanders P. Genetic control of DNA breakdown and repair in E. coli K-12 treated with mitomycin C or ultraviolet light. Z Vererbungsl. 1964;95:345–350. doi: 10.1007/BF01268667. [DOI] [PubMed] [Google Scholar]
- 64.Braedt G, Smith G R. Strand specificity of DNA unwinding by RecBCD enzyme. Proc Natl Acad Sci USA. 1989;86:871–875. doi: 10.1073/pnas.86.3.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brandsma J A, Stoorvogel J, van Sluis C A, van de Putte P. Effect of lexA and ssb genes, present on a uvrA recombinant plasmid, on the UV survival of Escherichia coli K-12. Gene. 1982;18:77–85. doi: 10.1016/0378-1119(82)90058-0. [DOI] [PubMed] [Google Scholar]
- 66.Brcic-Kostic K, Stojiljkovic I, Salaj-Smic E, Trgovcevic Z. Overproduction of the RecD polypeptide sensitizes Escherichia coli cells to γ-radiation. Mutat Res. 1992;281:123–127. doi: 10.1016/0165-7992(92)90046-k. [DOI] [PubMed] [Google Scholar]
- 67.Breitman T R, Maury P B, Toal J N. Loss of deoxyribonucleic acid-thymine during thymine starvation of Escherichia coli. J Bacteriol. 1972;112:646–648. doi: 10.1128/jb.112.1.646-648.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bremer H, Dennis P P. Modulation of chemical composition and other parameters of the cell by growth rate. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1987. pp. 1527–1542. [Google Scholar]
- 69.Brenner S L, Mitchell R S, Morrical S W, Neuendorf S K, Schutte B C, Cox M M. RecA protein-promoted ATP hydrolysis occurs throughout RecA nucleoprotein filaments. J Biol Chem. 1987;262:4011–4016. [PubMed] [Google Scholar]
- 70.Brenner S L, Zlotnick A, Griffith J D. RecA protein self-assembly. Multiple discrete aggregation states. J Mol Biol. 1988;204:959–972. doi: 10.1016/0022-2836(88)90055-1. [DOI] [PubMed] [Google Scholar]
- 71.Brenner S L, Zlotnick A, Stafford W F., III RecA protein self-assembly. II. Analytical equilibrium ultracentrifugation studies of the entropy-driven self-association of RecA. J Mol Biol. 1990;216:949–964. doi: 10.1016/S0022-2836(99)80013-8. [DOI] [PubMed] [Google Scholar]
- 72.Bresler S E, Lanzov V A, Lukjaniec-Blinkova A A. On the mechanism of conjugation in Escherichia coli K12. Mol Gen Genet. 1968;102:269–284. doi: 10.1007/BF00433718. [DOI] [PubMed] [Google Scholar]
- 73.Bridges B A. Are there DNA damage checkpoints in E. coli? Bioessays. 1995;17:63–70. doi: 10.1002/bies.950170112. [DOI] [PubMed] [Google Scholar]
- 74.Brooks K, Clark A J. Behavior of λ bacteriophage in a recombination deficient strain of Escherichia coli. J Virol. 1967;1:283–293. doi: 10.1128/jvi.1.2.283-293.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bruck I, Woodgate R, McEntee K, Goodman M F. Purification of a soluble UmuD′C complex from Escherichia coli. Cooperative binding of UmuD′C to single-stranded DNA. J Biol Chem. 1996;271:10767–10774. doi: 10.1074/jbc.271.18.10767. [DOI] [PubMed] [Google Scholar]
- 76.Buick R N, Harris W J. Thymineless death in Bacillus subtilis. J Gen Microbiol. 1975;88:115–122. doi: 10.1099/00221287-88-1-115. [DOI] [PubMed] [Google Scholar]
- 77.Burckhardt S E, Woodgate R, Scheuermann R H, Echols H. UmuD mutagenesis protein of Escherichia coli: overproduction, purification, and cleavage by RecA. Proc Natl Acad Sci USA. 1988;85:1811–1815. doi: 10.1073/pnas.85.6.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Burgers P M J, Kornberg A. The cycling of Escherichia coli DNA polymerase III holoenzyme in replication. J Biol Chem. 1983;258:7669–7675. [PubMed] [Google Scholar]
- 79.Burgers P M J, Kornberg A, Sakakibara Y. The dnaN gene codes for the β subunit of DNA polymerase III holoenzyme of Escherichia coli. Proc Natl Acad Sci USA. 1981;78:5391–5395. doi: 10.1073/pnas.78.9.5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Buttin G, Wright M. Enzymatic DNA degradation in E. coli: its relationship to synthetic processes at the chromosome level. Cold Spring Harbor Symp Quant Biol. 1968;33:259–269. doi: 10.1101/sqb.1968.033.01.030. [DOI] [PubMed] [Google Scholar]
- 81.Campbell A. Comparative molecular biology of lamboid phages. Annu Rev Microbiol. 1994;48:193–222. doi: 10.1146/annurev.mi.48.100194.001205. [DOI] [PubMed] [Google Scholar]
- 82.Campbell M J, Davis R W. On the in vivo function of the RecA ATPase. J Mol Biol. 1999;286:437–445. doi: 10.1006/jmbi.1998.2457. [DOI] [PubMed] [Google Scholar]
- 83.Cao Y, Kogoma T. The mechanism of recA polA lethality: suppression by RecA-independent recombination repair activated by the lexA(Def) mutation in Escherichia coli. Genetics. 1995;139:1483–1494. doi: 10.1093/genetics/139.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Capaldo F N, Barbour S D. DNA content, synthesis and integrity in dividing and non-dividing cells of Rec− strains of Escherichia coli K12. J Mol Biol. 1975;91:53–66. doi: 10.1016/0022-2836(75)90371-x. [DOI] [PubMed] [Google Scholar]
- 85.Capaldo F N, Ramsey G, Barbour S D. Analysis of the growth of recombination-deficient strains of Escherichia coli K-12. J Bacteriol. 1974;118:242–249. doi: 10.1128/jb.118.1.242-249.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Capaldo-Kimball F, Barbour S D. Involvement of recombination genes in growth and viability of Escherichia coli K-12. J Bacteriol. 1971;106:204–212. doi: 10.1128/jb.106.1.204-212.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Carlsson J, Carpenter V S. The recA+ gene product is more important than catalase and superoxide dismutase in protecting Escherichia coli against hydrogen peroxide toxicity. J Bacteriol. 1980;142:319–321. doi: 10.1128/jb.142.1.319-321.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Carter D M, Radding C M. The role of exonuclease and β protein of phage λ in genetic recombination. II. Substrate specificity and the mode of action of λ exonuclease. J Biol Chem. 1971;246:2502–2510. [PubMed] [Google Scholar]
- 89.Cassuto E. Formation of covalently closed heteroduplex DNA by the combined action of gyrase and RecA protein. EMBO J. 1984;3:2159–2164. doi: 10.1002/j.1460-2075.1984.tb02106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cassuto E, Lash T, Sriprakash K C, Radding C M. Role of exonuclease and β protein of phage λ in genetic recombination. V. Recombination of λ DNA in vitro. Proc Natl Acad Sci USA. 1971;68:1639–1643. doi: 10.1073/pnas.68.7.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cassuto E, Radding C M. Mechanism for the action of λ exonuclease in genetic recombination. Nat New Biol. 1971;229:13–16. doi: 10.1038/newbio229013a0. [DOI] [PubMed] [Google Scholar]
- 92.Chan S N, Vincent S D, Lloyd R G. Recognition and manipulation of branched DNA by the RusA Holliday junction resolvase of Escherichia coli. Nucleic Acids Res. 1998;26:1560–1566. doi: 10.1093/nar/26.7.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chaudhury A M, Smith G R. A new class of Escherichia coli recBC mutants: implications for the role of RecBC enzyme in homologous recombination. Proc Natl Acad Sci USA. 1984;81:7850–7854. doi: 10.1073/pnas.81.24.7850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chédin F, Noirot P, Biaudet V, Ehrlich S D. A five-nucleotide sequence protects DNA from exonucleolytic degradation by AddAB, the RecBCD analogue of Bacillus subtilis. Mol Microbiol. 1998;29:1369–1377. doi: 10.1046/j.1365-2958.1998.01018.x. [DOI] [PubMed] [Google Scholar]
- 95.Chen H-W, Randle D E, Gabbidon M, Julin D A. Functions of the ATP hydrolysis subunits (RecB and RecD) in the nuclease reactions catalyzed by the RecBCD enzyme from Escherichia coli. J Mol Biol. 1998;278:89–104. doi: 10.1006/jmbi.1998.1694. [DOI] [PubMed] [Google Scholar]
- 96.Chen H W, Ruan B, Yu M, Wang J D, Julin D A. The RecD subunit of the RecBCD enzyme from Escherichia coli is a single-stranded DNA-dependent ATPase. J Biol Chem. 1997;272:10072–10079. doi: 10.1074/jbc.272.15.10072. [DOI] [PubMed] [Google Scholar]
- 97.Chiu S K, Low K B, Yuan A, Radding C M. Resolution of an early RecA-recombination intermediate by a junction-specific endonuclease. Proc Natl Acad Sci USA. 1997;94:6079–6083. doi: 10.1073/pnas.94.12.6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chow S A, Honigberg S M, Bainton R J, Radding C M. Patterns of nuclease protection during strand exchange. RecA protein forms heteroduplex DNA by binding to strands of the same polarity. J Biol Chem. 1986;261:6961–6971. [PubMed] [Google Scholar]
- 99.Chow S A, Honigberg S M, Radding C M. DNase protection by RecA protein during strand exchange. Asymmetric protection of the Holliday structure. J Biol Chem. 1988;263:3335–3347. [PubMed] [Google Scholar]
- 100.Churchill J J, Anderson D G, Kowalczykowski S C. The RecBC enzyme loads RecA protein onto ssDNA asymmetrically and independently of χ, resulting in constitutive recombination activation. Genes Dev. 1999;13:901–911. doi: 10.1101/gad.13.7.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cimino G D, Gamper H B, Isaacs S T, Hearst J E. Psoralens as photoactive probes of nucleic acid structure and function: organic chemistry, photochemistry, and biochemistry. Annu Rev Biochem. 1985;54:1151–1193. doi: 10.1146/annurev.bi.54.070185.005443. [DOI] [PubMed] [Google Scholar]
- 102.Clark A J. Recombination deficient mutants of E. coli and other bacteria. Annu Rev Genet. 1973;7:67–86. doi: 10.1146/annurev.ge.07.120173.000435. [DOI] [PubMed] [Google Scholar]
- 103.Clark A J. Toward a metabolic interpretation of genetic recombination of E. coli and its phages. Annu Rev Microbiol. 1971;25:437–464. doi: 10.1146/annurev.mi.25.100171.002253. [DOI] [PubMed] [Google Scholar]
- 104.Clark A J. A view of the RecBC and RecF pathways of E. coli recombination. In: Alberts B, editor. Mechanistic studies of DNA replication and genetic recombination. New York, N.Y: Academic Press, Inc.; 1980. pp. 891–899. [Google Scholar]
- 105.Clark A J, Chamberlin M, Boyce R P, Howard-Flanders P. Abnormal metabolic response to ultraviolet light of a recombination deficient mutant of Escherichia coli K12. J Mol Biol. 1966;19:442–454. doi: 10.1016/s0022-2836(66)80015-3. [DOI] [PubMed] [Google Scholar]
- 106.Clark A J, Low K B. Pathways and systems of homologous recombination in Escherichia coli. In: Low K B, editor. The recombination of genetic material. San Diego, Calif: Academic Press, Inc.; 1988. pp. 155–215. [Google Scholar]
- 107.Clark A J, Margulies A D. Isolation and characterization of recombination-deficient mutants of Escherichia coli K-12. Proc Natl Acad Sci USA. 1965;53:451–459. doi: 10.1073/pnas.53.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Clark A J, Sandler S J. Homologous recombination: the pieces begin to fall into place. Crit Rev Microbiol. 1994;20:125–142. doi: 10.3109/10408419409113552. [DOI] [PubMed] [Google Scholar]
- 109.Clark A J, Sharma V, Brenowitz S, Chu C C, Sandler S, Satin L, Templin A, Berger I, Cohen A. Genetic and molecular analyses of the C-terminal region of the recE gene from the Rac prophage of Escherichia coli K-12 reveal the recT gene. J Bacteriol. 1993;175:7673–7682. doi: 10.1128/jb.175.23.7673-7682.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Clark A J, Volkert M R, Margossian L J, Nagaishi H. Effects of a recA operator mutation on mutant phenotypes conferred by lexA and recF mutations. Mutat Res. 1982;106:11–26. doi: 10.1016/0027-5107(82)90187-7. [DOI] [PubMed] [Google Scholar]
- 111.Clark J B, Haas F, Stone W S, Wyss O. The stimulation of gene recombination in Escherichia coli. J Bacteriol. 1950;59:375–379. doi: 10.1128/jb.59.3.375-379.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Clerget M. Site-specific recombination promoted by a short DNA segment of plasmid R1 and by a homologous segment in the terminus region of the Escherichia coli chromosome. New Biol. 1991;3:780–788. [PubMed] [Google Scholar]
- 113.Cluzel P, Lebrun A, Heller C, Lavery R, Viovy J-L, Chatenay D, Caron F. DNA: an extensible molecule. Science. 1996;271:792–794. doi: 10.1126/science.271.5250.792. [DOI] [PubMed] [Google Scholar]
- 114.Clyman J, Belfort M. trans and cis requirements for intron mobility in a prokaryotic system. Genes Dev. 1992;6:1269–1279. doi: 10.1101/gad.6.7.1269. [DOI] [PubMed] [Google Scholar]
- 115.Cohen A, Clark A J. Synthesis of linear plasmid multimers in Escherichia coli K-12. J Bacteriol. 1986;167:327–335. doi: 10.1128/jb.167.1.327-335.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Colbert T, Taylor A F, Smith G R. Genomics, Chi sites and codons: “islands of preferred DNA pairing” are oceans of ORFs. Trends Genet. 1998;14:485–488. doi: 10.1016/s0168-9525(98)01606-0. [DOI] [PubMed] [Google Scholar]
- 117.Cole R S. Properties of F′ factor deoxyribonucleic acid transferred from ultraviolet-irradiated donors: photoreactivation in the recipient and the influence of recA, recB, recC, and uvr genes. J Bacteriol. 1971;106:143–149. doi: 10.1128/jb.106.1.143-149.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Colloms S D, McCulloch R, Grant K, Neilson L, Sherratt D J. Xer-mediated site-specific recombination in vitro. EMBO J. 1996;15:1172–1181. [PMC free article] [PubMed] [Google Scholar]
- 119.Condra J H, Pauling C. Induction of the SOS system by DNA ligase-deficient growth of Escherichia coli. J Gen Microbiol. 1982;128:613–622. doi: 10.1099/00221287-128-3-613. [DOI] [PubMed] [Google Scholar]
- 120.Conley E C, West S C. Underwinding of DNA associated with duplex-duplex pairing by RecA protein. J Biol Chem. 1990;265:10156–10163. [PubMed] [Google Scholar]
- 121.Connelly J C, de Leau E S, Okely E A, Leach D R F. Overexpression, purification, and characterization of the SbcCD protein from Escherichia coli. J Biol Chem. 1997;272:19819–19826. doi: 10.1074/jbc.272.32.19819. [DOI] [PubMed] [Google Scholar]
- 122.Connolly B, Parsons C A, Benson F E, Dunderdale H J, Sharples G J, Lloyd R G, West S C. Resolution of Holliday junctions in vitro requires the Escherichia coli ruvC gene product. Proc Natl Acad Sci USA. 1991;88:6063–6067. doi: 10.1073/pnas.88.14.6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cook T M, Brown K G, Boyle J V, Goss W A. Bactericidal action of nalidixic acid on Bacillus subtilis. J Bacteriol. 1966;92:1510–1514. doi: 10.1128/jb.92.5.1510-1514.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cook T M, Deitz W H, Goss W A. Mechanism of action of nalidixic acid on Escherichia coli. IV. Effects on the stability of cellular constituents. J Bacteriol. 1966;91:774–779. doi: 10.1128/jb.91.2.774-779.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cornet F, Louarn J, Patte J, Louarn J-M. Restriction of the activity of the recombination site dif to a small zone of the Escherichia coli chromosome. Genes Dev. 1996;10:1152–1161. doi: 10.1101/gad.10.9.1152. [DOI] [PubMed] [Google Scholar]
- 126.Cornet F, Mortier I, Patte J, Louarn J-M. Plasmid pSC101 harbors a recombination site, psi, which is able to resolve plasmid multimers and to substitute for the analogous chromosomal E. coli site, dif. J Bacteriol. 1994;176:3188–3195. doi: 10.1128/jb.176.11.3188-3195.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Corre J, Cornet F, Patte J, Louarn J-M. Unraveling a region-specific hyper-recombination phenomenon: genetic control and modalities of terminal recombination in Escherichia coli. Genetics. 1997;147:979–989. doi: 10.1093/genetics/147.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Courcelle J, Carswell-Crumpton C, Hanawalt P C. recF and recR are required for the resumption of replication at DNA replication forks in Escherichia coli. Proc Natl Acad Sci USA. 1997;94:3714–3719. doi: 10.1073/pnas.94.8.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cox M M. Alignment of 3 (but not 4) DNA strands within a RecA protein filament. J Biol Chem. 1995;270:26021–26024. doi: 10.1074/jbc.270.44.26021. [DOI] [PubMed] [Google Scholar]
- 130.Cox M M. A broadening view of recombinational DNA repair in bacteria. Genes Cells. 1998;3:65–78. doi: 10.1046/j.1365-2443.1998.00175.x. [DOI] [PubMed] [Google Scholar]
- 131.Cox M M. The RecA protein as a recombinational repair system. Mol Microbiol. 1991;5:1295–1299. doi: 10.1111/j.1365-2958.1991.tb00775.x. [DOI] [PubMed] [Google Scholar]
- 132.Cox M M. Relating biochemistry to biology: how the recombinational repair function of RecA protein is manifested in its molecular properties. Bioessays. 1993;15:617–623. doi: 10.1002/bies.950150908. [DOI] [PubMed] [Google Scholar]
- 133.Cox M M. Why does RecA protein hydrolyze ATP? Trends Biochem Sci. 1994;19:217–222. doi: 10.1016/0968-0004(94)90025-6. [DOI] [PubMed] [Google Scholar]
- 134.Cox M M, Lehman I R. Directionality and polarity in RecA protein-promoted branch migration. Proc Natl Acad Sci USA. 1981;78:6018–6022. doi: 10.1073/pnas.78.10.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cox M M, Lehman I R. RecA protein-promoted DNA strand exchange. J Biol Chem. 1982;257:8523–8532. [PubMed] [Google Scholar]
- 136.Cunningham R P, DasGupta C, Shibata T, Radding C M. Homologous pairing in genetic recombination: RecA protein makes joint molecules of gapped circular DNA and closed circular DNA. Cell. 1980;20:223–235. doi: 10.1016/0092-8674(80)90250-0. [DOI] [PubMed] [Google Scholar]
- 137.Cunningham R P, Shibata T, DasGupta C, Radding C M. Single strands induce RecA protein to unwind duplex DNA for homologous pairing. Nature. 1979;281:191–195. doi: 10.1038/281191a0. [DOI] [PubMed] [Google Scholar]
- 138.Cunningham R P, Wu A M, Shibata T, DasGupta C, Radding C M. Homologous pairing and topological linkage of DNA molecules by combined action of E. coli RecA protein and topoisomerase I. Cell. 1981;24:213–223. doi: 10.1016/0092-8674(81)90517-1. [DOI] [PubMed] [Google Scholar]
- 139.Dabert P, Ehrlich S D, Gruss A. χ sequence protects against RecBCD degradation of DNA in vivo. Proc Natl Acad Sci USA. 1992;89:12073–12077. doi: 10.1073/pnas.89.24.12073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Dahan-Grobgeld E, Livneh Z, Maretzek A F, Polak-Charcon S, Eichenbaum Z, Degani H. Reversible induction of ATP synthesis by DNA damage and repair in Escherichia coli. In vivo NMR studies. J Biol Chem. 1998;273:30232–30238. doi: 10.1074/jbc.273.46.30232. [DOI] [PubMed] [Google Scholar]
- 141.Daly M J, Minton K W. An alternative pathway of recombination of chromosomal fragments precedes recA-dependent recombination in the radioresistant bacterium Deinococcus radiodurans. J Bacteriol. 1996;178:4461–4471. doi: 10.1128/jb.178.15.4461-4471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.D’Arpa P, Liu L F. Topoisomerase-targeting antitumor drugs. Biochim Biophys Acta. 1989;989:163–177. doi: 10.1016/0304-419x(89)90041-3. [DOI] [PubMed] [Google Scholar]
- 143.Das A. Control of transcription termination by DNA-binding proteins. Annu Rev Biochem. 1993;62:893–930. doi: 10.1146/annurev.bi.62.070193.004333. [DOI] [PubMed] [Google Scholar]
- 144.Das S K, Loeb L A. UV irradiation alters deoxynucleoside triphosphate pools in Escherichia coli. Mutat Res. 1984;131:97–100. doi: 10.1016/0167-8817(84)90047-6. [DOI] [PubMed] [Google Scholar]
- 145.DasGupta C, Radding C M. Polar branch migration promoted by RecA protein: effect of mismatched base pairs. Proc Natl Acad Sci USA. 1982;79:762–766. doi: 10.1073/pnas.79.3.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.DasGupta C, Wu A M, Kahn R, Cunningham R P, Radding C M. Concerted strand exchange and formation of Holliday structures by E. coli RecA protein. Cell. 1981;25:507–516. doi: 10.1016/0092-8674(81)90069-6. [DOI] [PubMed] [Google Scholar]
- 147.Davies A A, West S C. Formation of RuvABC-Holliday junction complexes in vitro. Curr Biol. 1998;8:725–727. doi: 10.1016/s0960-9822(98)70282-9. [DOI] [PubMed] [Google Scholar]
- 148.De Lucia P, Cairns J. Isolation of an E. coli strain with a mutation affecting DNA polymerase. Nature. 1969;224:1164–1166. doi: 10.1038/2241164a0. [DOI] [PubMed] [Google Scholar]
- 149.Demple B, Halbrook J, Linn S. Escherichia coli xth mutants are hypersensitive to hydrogen peroxide. J Bacteriol. 1983;153:1079–1082. doi: 10.1128/jb.153.2.1079-1082.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.de Vries J, Wackernagel W. Recombination and UV resistance of Escherichia coli with the cloned recA and recBCD genes of Serratia marcescens and Proteus mirabilis: evidence for an advantage of interspecies combination of P. mirabilis RecA protein and RecBCD enzyme. J Gen Microbiol. 1992;138:31–38. doi: 10.1099/00221287-138-1-31. [DOI] [PubMed] [Google Scholar]
- 151.Dianov G, Price A, Lindahl T. Generation of single-nucleotide repair patches following excision of uracil residues from DNA. Mol Cell Biol. 1992;12:1605–1612. doi: 10.1128/mcb.12.4.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Di Capua E, Cuillel M, Hewat E, Schnarr M, Timmins P A, Ruigrok R W H. Activation of RecA protein. The open helix model for LexA cleavage. J Mol Biol. 1992;226:707–719. doi: 10.1016/0022-2836(92)90627-v. [DOI] [PubMed] [Google Scholar]
- 153.Di Capua E, Engel A, Stasiak A, Koller T. Characterization of complexes between RecA protein and duplex DNA by electron microscopy. J Mol Biol. 1982;157:87–103. doi: 10.1016/0022-2836(82)90514-9. [DOI] [PubMed] [Google Scholar]
- 154.Di Capua E, Müller B. The accessibility of DNA to dimethylsulfate in complexes with RecA protein. EMBO J. 1987;6:2493–2498. doi: 10.1002/j.1460-2075.1987.tb02531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Dixon D A, Churchill J J, Kowalczykowski S C. Reversible inactivation of the Escherichia coli RecBCD enzyme by the recombination hotspot χ in vitro: evidence for functional inactivation or loss of the RecD subunit. Proc Natl Acad Sci USA. 1994;91:2980–2984. doi: 10.1073/pnas.91.8.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Dixon D A, Kowalczykowski S C. Homologous pairing in vitro stimulated by the recombination hotspot, Chi. Cell. 1991;66:361–371. doi: 10.1016/0092-8674(91)90625-9. [DOI] [PubMed] [Google Scholar]
- 157.Dixon D A, Kowalczykowski S C. The recombination hotspot χ is a regulatory sequence that acts by attenuating the nuclease activity of the E. coli RecBCD enzyme. Cell. 1993;73:87–96. doi: 10.1016/0092-8674(93)90162-j. [DOI] [PubMed] [Google Scholar]
- 158.Dixon D A, Kowalczykowski S C. Role of the Escherichia coli recombination hotspot, χ, in RecABCD-dependent homologous pairing. J Biol Chem. 1995;270:16360–16370. doi: 10.1074/jbc.270.27.16360. [DOI] [PubMed] [Google Scholar]
- 159.Dodson L A, Hadden C T. Capacity for postreplication repair correlated with transducibility in Rec− mutants of Bacillus subtilis. J Bacteriol. 1980;144:608–615. doi: 10.1128/jb.144.2.608-615.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Dodson L A, Hadden C T. Postreplication repair of deoxyribonucleic acid and daughter strand exchange in Uvr− mutants of Bacillus subtilis. J Bacteriol. 1980;144:840–843. doi: 10.1128/jb.144.2.840-843.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Dombroski D F, Scraba D G, Bradley R D, Morgan A R. Studies of the interaction of RecA protein with DNA. Nucleic Acids Res. 1983;11:7487–7504. doi: 10.1093/nar/11.21.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Doudney C O. DNA-replication recovery inhibition and subsequent reinitiation in UV-radiation-damaged E. coli: a strategy for survival. Mutat Res. 1990;243:179–186. doi: 10.1016/0165-7992(90)90088-2. [DOI] [PubMed] [Google Scholar]
- 163.Drlica K, Zhao X. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol Mol Biol Rev. 1997;61:377–392. doi: 10.1128/mmbr.61.3.377-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Duckett D R, Murchie A I H, Diekmann S, von Kitzing E, Kemper B, Lilley D M J. The structure of the Holliday junction, and its resolution. Cell. 1988;55:79–89. doi: 10.1016/0092-8674(88)90011-6. [DOI] [PubMed] [Google Scholar]
- 165.Dunn K, Chrysogelos S, Griffith J. Electron microscopic visualization of RecA-DNA filaments: evidence for a cyclic extension of duplex DNA. Cell. 1982;28:757–765. doi: 10.1016/0092-8674(82)90055-1. [DOI] [PubMed] [Google Scholar]
- 166.Echols H, Gingery R. Mutants of bacteriophage λ defective in vegetative genetic recombination. J Mol Biol. 1968;34:239–249. doi: 10.1016/0022-2836(68)90249-0. [DOI] [PubMed] [Google Scholar]
- 167.Echols H, Goodman M F. Mutation induced by DNA damage: a many protein affair. Mutat Res. 1990;236:301–311. doi: 10.1016/0921-8777(90)90013-u. [DOI] [PubMed] [Google Scholar]
- 168.Egelman E H. Bacterial helicases. J Struct Biol. 1998;124:123–128. doi: 10.1006/jsbi.1998.4050. [DOI] [PubMed] [Google Scholar]
- 169.Egelman E H, Stasiak A. The structure of helical RecA-DNA complexes. I. Complexes formed in the presence of ATP-γ-S or ATP. J Mol Biol. 1986;191:677–697. doi: 10.1016/0022-2836(86)90453-5. [DOI] [PubMed] [Google Scholar]
- 170.Eggleston A K, Kowalczykowski S C. Biochemical characterization of a mutant RecBCD enzyme, the RecB2109CD enzyme, which lacks χ-specific, but not non-specific, nuclease activity. J Mol Biol. 1993;231:605–620. doi: 10.1006/jmbi.1993.1313. [DOI] [PubMed] [Google Scholar]
- 171.Eggleston A K, Mitchell A H, West S C. In vitro reconstitution of the late steps of genetic recombination in E. coli. Cell. 1997;89:607–617. doi: 10.1016/s0092-8674(00)80242-1. [DOI] [PubMed] [Google Scholar]
- 172.Eguchi Y, Ogawa T, Ogawa H. Cleavage of bacteriophage φ80 cI repressor by RecA protein. J Mol Biol. 1988;202:565–573. doi: 10.1016/0022-2836(88)90286-0. [DOI] [PubMed] [Google Scholar]
- 173.Eichler D C, Lehman I R. On the role of ATP in phosphodiester bond hydrolysis catalyzed by the recBC deoxyribonuclease of Escherichia coli. J Biol Chem. 1977;252:499–503. [PubMed] [Google Scholar]
- 174.Ennis D G, Amundsen S K, Smith G R. Genetic functions promoting homologous recombination in Escherichia coli: a study of inversion in phage λ. Genetics. 1987;115:11–24. doi: 10.1093/genetics/115.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Enquist L W, Skalka A. Replication of bacteriophage λ DNA dependent on the function of host and viral genes. I. Interaction of red, gam, and rec. J Mol Biol. 1973;75:185–212. doi: 10.1016/0022-2836(73)90016-8. [DOI] [PubMed] [Google Scholar]
- 176.Fangman W L, Russel M. X-irradiation sensitivity in Escherichia coli defective in DNA replication. Mol Gen Genet. 1971;110:332–347. doi: 10.1007/BF00438275. [DOI] [PubMed] [Google Scholar]
- 177.Farah J A, Smith G R. The RecBCD enzyme initiation complex for DNA unwinding: enzyme positioning and DNA opening. J Mol Biol. 1997;272:699–715. doi: 10.1006/jmbi.1997.1259. [DOI] [PubMed] [Google Scholar]
- 178.Faulds D, Dower N, Stahl M M, Stahl F W. Orientation-dependent recombination hotspot activity in bacteriophage λ. J Mol Biol. 1979;131:681–695. doi: 10.1016/0022-2836(79)90197-9. [DOI] [PubMed] [Google Scholar]
- 179.Feinstein S I, Low K B. Hyper-recombining recipient strains in bacterial conjugation. Genetics. 1986;113:13–33. doi: 10.1093/genetics/113.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Feiss M, Sippy J, Miller G. Processive action of terminase during sequential packaging of bacteriophage λ chromosome. J Mol Biol. 1985;186:759–771. doi: 10.1016/0022-2836(85)90395-x. [DOI] [PubMed] [Google Scholar]
- 181.Felsenstein J. The evolutionary advantage of recombination. Genetics. 1974;78:737–756. doi: 10.1093/genetics/78.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Finch P W, Chambers P, Emmerson P T. Identification of the Escherichia coli recN gene product as a major SOS protein. J Bacteriol. 1985;164:653–658. doi: 10.1128/jb.164.2.653-658.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Finch P W, Storey A, Brown K, Hickson I D, Emmerson P T. Complete nucleotide sequence of recD, the structural gene for the alpha subunit of exonuclease V of Escherichia coli. Nucleic Acids Res. 1986;14:8583–8594. doi: 10.1093/nar/14.21.8583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Finch P W, Storey A, Chapman K E, Brown K, Hickson I D, Emmerson P T. Complete nucleotide sequence of the Escherichia coli recB gene. Nucleic Acids Res. 1986;14:8573–8582. doi: 10.1093/nar/14.21.8573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Finch P W, Wilson R E, Brown K, Hickson I D, Tomkinson A E, Emmerson P T. Complete nucleotide sequence of the Escherichia coli recC gene and of the thyA-recC intergenic region. Nucleic Acids Res. 1986;14:4437–4451. doi: 10.1093/nar/14.11.4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Fishel R A, James A A, Kolodner R. RecA-independent general recombination of plasmids. Nature. 1981;294:184–186. doi: 10.1038/294184a0. [DOI] [PubMed] [Google Scholar]
- 187.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J-F, Dougherty B A, Merrick J M, McKenney K, Sutton G, Fitzhugh W, Fields C, Gocayne J D, Scott J, Shirley R, Liu L-I, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback T R, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Fuhrmann J L, Geoghagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 188.Flower A M, McHenry C S. Transcriptional organization of the Escherichia coli dnaX gene. J Mol Biol. 1991;220:649–658. doi: 10.1016/0022-2836(91)90107-h. [DOI] [PubMed] [Google Scholar]
- 189.Foster P L, Trimarchi J M, Maurer R A. Two enzymes, both of which process recombination intermediates, have opposite effects on adaptive mutation in Escherichia coli. Genetics. 1996;142:25–37. doi: 10.1093/genetics/142.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Fox M S. On the mechanism of integration of transforming deoxyribonucleate. J Gen Physiol. 1966;49:183–196. doi: 10.1085/jgp.49.6.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Frank E G, Ennis D G, Gonzalez M, Levine A S, Woodgate R. Regulation of SOS mutagenesis by proteolysis. Proc Natl Acad Sci USA. 1996;93:10291–10296. doi: 10.1073/pnas.93.19.10291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Frank E G, Gonzalez M, Ennis D G, Levine A S, Woodgate R. In vivo stability of the Umu mutagenesis proteins: a major role for RecA. J Bacteriol. 1996;178:3550–3556. doi: 10.1128/jb.178.12.3550-3556.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Franklin N C. Deletions and functions of the center of the φ80-λ phage genome. Evidence for a phage function promoting genetic recombination. Genetics. 1967;57:301–318. doi: 10.1093/genetics/57.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R, Lathigra R, White O, Ketchum K A, Dodson R, Hickey E K, Gwinn M, Dougherty B, Tomb J-F, Fleischmann R D, Richardson D, Peterson J, Kerlavage A R, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams M D, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fujii C, Cotton M D, Horst K, Roberts K, Hatch B, Smith H O, Venter J C. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 195.Friedberg E C, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C.: ASM Press; 1995. [Google Scholar]
- 196.Fujita M Q, Yoshikawa H, Ogasawara N. Structure of dnaA region of Pseudomonas putida: conservation among three bacteria, Bacillus subtilis, Escherichia coli and P. putida. Mol Gen Genet. 1989;215:381–387. doi: 10.1007/BF00427033. [DOI] [PubMed] [Google Scholar]
- 197.Fukuoh A, Iwasaki H, Ishioka K, Shinagawa H. ATP-dependent resolution of R-loops at the ColE1 replication origin by Escherichia coli RecG protein, a Holliday junction-specific helicase. EMBO J. 1997;16:203–209. doi: 10.1093/emboj/16.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Galitski T, Roth J R. Pathways for homologous recombination between chromosomal direct repeats in Salmonella typhimurium. Genetics. 1997;146:751–767. doi: 10.1093/genetics/146.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Ganesan A K. Persistence of pyrimidine dimers during post-replication repair in ultraviolet light-irradiated Escherichia coli K12. J Mol Biol. 1974;87:103–119. doi: 10.1016/0022-2836(74)90563-4. [DOI] [PubMed] [Google Scholar]
- 200.Ganesan A K, Seawell P C. The effect of lexA and recF mutations on post-replication repair and DNA synthesis in Escherichia coli K-12. Mol Gen Genet. 1975;141:189–205. doi: 10.1007/BF00341799. [DOI] [PubMed] [Google Scholar]
- 201.Ganesan S, Smith G R. Strand-specific binding to duplex DNA ends by the subunits of the Escherichia coli RecBCD enzyme. J Mol Biol. 1993;229:67–78. doi: 10.1006/jmbi.1993.1008. [DOI] [PubMed] [Google Scholar]
- 202.Garfinkel L, Garfinkel D. Calculation of free-Mg2+ concentration in adenosine 5′-triphosphate containing solutions in vitro and in vivo. Biochemistry. 1984;23:3547–3552. doi: 10.1021/bi00310a025. [DOI] [PubMed] [Google Scholar]
- 203.Gari E, Figueroa-Bossi N, Blanc-Potard A-B, Spirito F, Schmid M B, Bossi L. A class of gyrase mutants of Salmonella typhimurium show quinolone-like lethality and require Rec functions for viability. Mol Microbiol. 1996;21:111–122. doi: 10.1046/j.1365-2958.1996.6221338.x. [DOI] [PubMed] [Google Scholar]
- 204.Gibson F P, Leach D R F, Lloyd R G. Identification of sbcD mutations as cosuppressors of recBC that allow propagation of DNA palindromes in Escherichia coli K-12. J Bacteriol. 1992;174:1222–1228. doi: 10.1128/jb.174.4.1222-1228.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Gigliani F, Ciotta C, Del Grosso M F, Battaglia P A. pR plasmid replication provides evidence that single-stranded DNA induces the SOS system in vivo. Mol Gen Genet. 1993;238:333–338. doi: 10.1007/BF00291991. [DOI] [PubMed] [Google Scholar]
- 206.Gillen J R, Willis D K, Clark A J. Genetic analysis of the RecE pathway of genetic recombination in Escherichia coli K-12. J Bacteriol. 1981;145:521–532. doi: 10.1128/jb.145.1.521-532.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Ginsberg D M, Jagger J. Evidence that initial ultraviolet lethal damage in Escherichia coli strain 15 T−A−U− is independent of growth phase. J Gen Microbiol. 1965;40:171–184. doi: 10.1099/00221287-40-2-171. [DOI] [PubMed] [Google Scholar]
- 208.Glassberg J, Meyer R R, Kornberg A. Mutant single-strand binding protein of Escherichia coli: genetic and physiological characterization. J Bacteriol. 1979;140:14–19. doi: 10.1128/jb.140.1.14-19.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Glickman B W. rorA mutation of Escherichia coli K-12 affects the recB subunit of exonuclease V. J Bacteriol. 1979;137:658–660. doi: 10.1128/jb.137.1.658-660.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Glickman B W, Zwenk H, Van Sluis C A, Rorsch A. The isolation and characterization of an X-ray-sensitive ultraviolet-resistant mutant of Escherichia coli. Biochim Biophys Acta. 1971;254:144–154. doi: 10.1016/0005-2787(71)90121-3. [DOI] [PubMed] [Google Scholar]
- 211.Goebel W. Degradation of DNA in a temperature-sensitive mutant of Escherichia coli defective in DNA synthesis, harboring the colicinogenic factor E1. Biochim Biophys Acta. 1970;224:353–360. doi: 10.1016/0005-2787(70)90568-x. [DOI] [PubMed] [Google Scholar]
- 212.Goldmark P J, Linn S. Purification and properties of RecBC DNase of Escherichia coli K-12. J Biol Chem. 1972;247:1849–1860. [PubMed] [Google Scholar]
- 213.Golub E I, Low K B. Indirect stimulation of genetic recombination. Proc Natl Acad Sci USA. 1983;80:1401–1405. doi: 10.1073/pnas.80.5.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Gonda D K, Radding C M. The mechanism of the search for homology promoted by RecA protein. Facilitated diffusion within nucleoprotein networks. J Biol Chem. 1986;261:13087–13096. [PubMed] [Google Scholar]
- 215.Gordon G S, Sitnikov D, Webb C D, Teleman A, Straight A, Losick R, Murray A W, Wright A. Chromosome and low copy plasmid segregation in E. coli: visual evidence for distinct mechanisms. Cell. 1997;90:1113–1121. doi: 10.1016/s0092-8674(00)80377-3. [DOI] [PubMed] [Google Scholar]
- 216.Gottesman M M, Gottesman M E, Gottesman S, Gellert M. Characterization of bacteriophage λ reverse as an Escherichia coli phage carrying a unique set of host-derived recombination functions. J Mol Biol. 1974;88:471–487. doi: 10.1016/0022-2836(74)90496-3. [DOI] [PubMed] [Google Scholar]
- 217.Gottesman M M, Hicks M L, Gellert M. Genetics and physiology of DNA ligase mutants of Escherichia coli. In: Wells R D, Inman R B, editors. DNA synthesis in vitro. Baltimore, Md: University Park Press; 1973. pp. 107–122. [Google Scholar]
- 218.Gourlie B B, Pigiet V P. Polyoma virus minichromosomes: characterization of the products of in vitro DNA synthesis. J Virol. 1983;45:585–593. doi: 10.1128/jvi.45.2.585-593.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Gray W J H, Green M H L, Bridges B A. DNA synthesis in gamma-irradiated recombination deficient strains of Escherichia coli. J Gen Microb. 1972;71:359–366. doi: 10.1099/00221287-71-2-359. [DOI] [PubMed] [Google Scholar]
- 220.Greenstein M, Skalka A. Replication of bacteriophage lambda DNA: in vivo studies of the interaction between the viral Gamma protein and the host RecBC DNAase. J Mol Biol. 1975;97:543–559. doi: 10.1016/s0022-2836(75)80058-1. [DOI] [PubMed] [Google Scholar]
- 221.Griffin T J, IV, Kolodner R D. Purification and preliminary characterization of the Escherichia coli K-12 RecF protein. J Bacteriol. 1990;172:6291–6299. doi: 10.1128/jb.172.11.6291-6299.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Grigg G W. Stability of DNA in dark-repair mutants of Escherichia coli B treated with nalidixic acid. J Gen Microbiol. 1970;61:21–25. doi: 10.1099/00221287-61-1-21. [DOI] [PubMed] [Google Scholar]
- 223.Gross J, Gross M. Genetic analysis of an E. coli strain with a mutation affecting DNA polymerase. Nature. 1969;224:1166–1168. doi: 10.1038/2241166a0. [DOI] [PubMed] [Google Scholar]
- 224.Gross J D. DNA replication in bacteria. Curr Top Microbiol Immunol. 1972;57:39–74. doi: 10.1007/978-3-642-65297-4_2. [DOI] [PubMed] [Google Scholar]
- 225.Gross J D, Grunstein J, Witkin E M. Inviability of recA-derivatives of the DNA polymerase mutant of De Lucia and Cairns. J Mol Biol. 1971;58:631–634. doi: 10.1016/0022-2836(71)90377-9. [DOI] [PubMed] [Google Scholar]
- 226.Guerrero R, Llagostera M, Villaverde A, Barbé J. Changes in ATP concentration in Escherichia coli during induction of the SOS system by mitomycin C and bleomycin. J Gen Microbiol. 1984;130:2247–2251. doi: 10.1099/00221287-130-9-2247. [DOI] [PubMed] [Google Scholar]
- 227.Günther T, Dorn F. Uber die intrazellulare Mg-ionenaktivitat von E. coli-zellen. Z Naturforsch Teil B. 1969;24:713–717. [PubMed] [Google Scholar]
- 228.Gupta R K, Gupta P, Yushok W D, Rose Z B. Measurement of the dissociation constant of MgATP at physiological nucleotide levels by a combination of 31P NMR and optical absorbance spectroscopy. Biochem Biophys Res Commun. 1983;117:210–216. doi: 10.1016/0006-291x(83)91562-0. [DOI] [PubMed] [Google Scholar]
- 229.Haefner K. Spontaneous lethal sectoring, a further feature of Escherichia coli strains deficient in the function of rec and uvr genes. J Bacteriol. 1968;96:652–659. doi: 10.1128/jb.96.3.652-659.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Haijema B J, Venema G, Kooistra J. The C terminus of the AddA subunit of the Bacillus subtilis ATP-dependent DNase is required for the ATP-dependent exonuclease activity but not for the helicase activity. J Bacteriol. 1996;178:5086–5091. doi: 10.1128/jb.178.17.5086-5091.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Hall M C, Jordan J R, Matson S W. Evidence for a physical interaction between the Escherichia coli methyl-directed mismatch repair proteins MutL and UvrD. EMBO J. 1998;17:1535–1541. doi: 10.1093/emboj/17.5.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Hall S D, Kane M F, Kolodner R D. Identification and characterization of the Escherichia coli RecT protein, a protein encoded by the recE region that promotes renaturation of homologous single-stranded DNA. J Bacteriol. 1993;175:277–287. doi: 10.1128/jb.175.1.277-287.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Hall S D, Kolodner R D. Homologous pairing and strand exchange promoted by the Escherichia coli RecT protein. Proc Natl Acad Sci USA. 1994;91:3205–3209. doi: 10.1073/pnas.91.8.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Hanawalt P C. The U.V. sensitivity of bacteria: its relation to the DNA replication cycle. Photochem Photobiol. 1966;5:1–12. [PubMed] [Google Scholar]
- 235.Hargreaves D, Rice D W, Sedelnikova S E, Artymiuk P J, Lloyd R G, Rafferty J B. Crystal structure of E. coli RuvA with bound DNA Holliday junction at 6 Å resolution. Nat Struct Biol. 1998;6:441–446. doi: 10.1038/nsb0698-441. [DOI] [PubMed] [Google Scholar]
- 236.Harmon F G, Kowalczykowski S C. RecQ helicase, in concert with RecA and SSB proteins, initiates and disrupts DNA recombination. Genes Dev. 1998;12:1134–1144. doi: 10.1101/gad.12.8.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Harmon F G, Rehrauer W M, Kowalczykowski S C. Interaction of Escherichia coli RecA protein with LexA repressor. II. Inhibition of DNA strand exchange by the uncleavable LexA S119A repressor argues that recombination and SOS induction are competitive processes. J Biol Chem. 1996;271:23874–23883. [PubMed] [Google Scholar]
- 238.Harris R S, Ross K J, Rosenberg S M. Opposing roles of the Holliday junction processing systems of Escherichia coli in recombination-dependent adaptive mutation. Genetics. 1996;142:681–691. doi: 10.1093/genetics/142.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Hays J B, Boehmer S. Antagonists of DNA gyrase inhibit repair and recombination of UV-irradiated phage λ. Proc Natl Acad Sci USA. 1978;75:4125–4129. doi: 10.1073/pnas.75.9.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Hecht R M, Taggart R T, Pettijohn D E. Size and DNA content of purified E. coli nucleoids observed by fluorescence microscopy. Nature. 1975;253:60–62. doi: 10.1038/253060a0. [DOI] [PubMed] [Google Scholar]
- 241.Hegde S, Sandler S J, Clark A J, Madiraju M V V S. recO and recR mutations delay induction of the SOS response in Escherichia coli. Mol Gen Genet. 1995;246:254–258. doi: 10.1007/BF00294689. [DOI] [PubMed] [Google Scholar]
- 242.Hegde S P, Qin M, Li X, Atkinson M A L, Clark A J, Rajagopalan M, Madiraju M V V S. Interactions of RecF protein with RecO, RecR, and single-stranded DNA binding proteins reveal roles for the RecF-RecO-RecR complex in DNA repair and recombination. Proc Natl Acad Sci USA. 1996;93:14468–14473. doi: 10.1073/pnas.93.25.14468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Hegde S P, Rajagopalan M, Madiraju M V V S. Preferential binding of Escherichia coli RecF protein to gapped DNA in the presence of adenosine (γ-thio) triphosphate. J Bacteriol. 1996;178:184–190. doi: 10.1128/jb.178.1.184-190.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Helmstetter C E. Timing of synthetic activities in the cell cycle. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1987. pp. 1594–1605. [Google Scholar]
- 245.Hertman I, Luria S E. Transduction studies on the role of a rec+ gene in the ultraviolet induction of prophage lambda. J Mol Biol. 1967;23:117–133. doi: 10.1016/s0022-2836(67)80021-4. [DOI] [PubMed] [Google Scholar]
- 246.Heuser J, Griffith J. Visualization of RecA protein and its complexes with DNA by quick-freeze/deep-etch electron microscopy. J Mol Biol. 1989;210:473–484. doi: 10.1016/0022-2836(89)90124-1. [DOI] [PubMed] [Google Scholar]
- 247.Hewitt R, Gaskins P. Influence of ultraviolet irradiation on chromosome replication in ultraviolet-sensitive bacteria. J Mol Biol. 1971;62:215–221. doi: 10.1016/0022-2836(71)90140-9. [DOI] [PubMed] [Google Scholar]
- 248.Higashitani N, Higashitani A, Horiuchi K. SOS induction in Escherichia coli by single-stranded DNA of mutant filamentous phage: monitoring by cleavage of LexA repressor. J Bacteriol. 1995;177:3610–3612. doi: 10.1128/jb.177.12.3610-3612.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Higashitani N, Higashitani A, Roth A, Horiuchi K. SOS induction in Escherichia coli by infection with mutant filamentous phage that are defective in initiation of complementary-strand DNA synthesis. J Bacteriol. 1992;174:1612–1618. doi: 10.1128/jb.174.5.1612-1618.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Hildebrandt E R, Cozzarelli N R. Comparison of recombination in vitro and in E. coli cells: measure of the effective concentration of DNA in vivo. Cell. 1995;81:331–340. doi: 10.1016/0092-8674(95)90386-0. [DOI] [PubMed] [Google Scholar]
- 251.Hill C W, Harvey S, Gray J A. Recombination between rRNA genes in Escherichia coli and Salmonella typhimurium. In: Drlica K, Riley M, editors. The bacterial chromosome. Washington, D.C.: American Society for Microbiology; 1990. pp. 335–340. [Google Scholar]
- 252.Hill S A, Stahl M M, Stahl F W. Single-strand DNA intermediates in phage λ’s Red recombination pathway. Proc Natl Acad Sci USA. 1997;94:2951–2956. doi: 10.1073/pnas.94.7.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Hill T M. Arrest of bacterial DNA replication. Annu Rev Microbiol. 1992;46:603–633. doi: 10.1146/annurev.mi.46.100192.003131. [DOI] [PubMed] [Google Scholar]
- 254.Hill W E, Fangman W L. Single-strand breaks in deoxyribonucleic acid and viability loss during deoxyribonucleic acid synthesis inhibition in Escherichia coli. J Bacteriol. 1973;116:1329–1335. doi: 10.1128/jb.116.3.1329-1335.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Hiom K, West S C. Branch migration during homologous recombination: assembly of a RuvAB-Holliday junction complex in vitro. Cell. 1995;80:787–793. doi: 10.1016/0092-8674(95)90357-7. [DOI] [PubMed] [Google Scholar]
- 256.Holliday R. A mechanism for gene conversion in fungi. Genet Res. 1964;5:282–304. doi: 10.1017/S0016672308009476. [DOI] [PubMed] [Google Scholar]
- 257.Hong X, Cadwell G W, Kogoma T. Escherichia coli RecG and RecA proteins in R-loop formation. EMBO J. 1995;14:2385–2392. doi: 10.1002/j.1460-2075.1995.tb07233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Honigberg S M, Gonda D K, Flory J, Radding C M. The pairing activity of stable nucleoprotein filaments made from RecA protein, single-stranded DNA, and adenosine 5′-(γ-thio)triphosphate. J Biol Chem. 1985;260:11845–11851. [PubMed] [Google Scholar]
- 259.Horii T, Ogawa T, Nakatani T, Hase T, Matsubara H, Ogawa H. Regulation of SOS functions: purification of E. coli LexA protein and determination of its specific site cleavage by the RecA protein. Cell. 1981;27:515–522. doi: 10.1016/0092-8674(81)90393-7. [DOI] [PubMed] [Google Scholar]
- 260.Horii T, Ogawa T, Ogawa H. Organization of the recA gene of Escherichia coli. Proc Natl Acad Sci USA. 1980;77:313–317. doi: 10.1073/pnas.77.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Horii Z-I, Clark A J. Genetic analysis of the RecF pathway of genetic recombination in Escherichia coli K-12: isolation and characterization of mutants. J Mol Biol. 1973;80:327–344. doi: 10.1016/0022-2836(73)90176-9. [DOI] [PubMed] [Google Scholar]
- 262.Horii Z I, Suzuki K. Degradation of the DNA of Escherichia coli K12 rec− (JC1569b) after irradiation with ultraviolet light. Photochem Photobiol. 1968;8:93–105. doi: 10.1111/j.1751-1097.1970.tb05976.x. [DOI] [PubMed] [Google Scholar]
- 263.Horiuchi T, Fujimura Y. Recombinational rescue of the stalled DNA replication fork: a model based on analysis of an Escherichia coli strain with a chromosomal region difficult to replicate. J Bacteriol. 1995;177:783–791. doi: 10.1128/jb.177.3.783-791.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Horiuchi T, Fujimura Y, Nishitani H, Kobayashi T, Hidaka M. The DNA replication fork blocked at the Ter site may be an entrance for the RecBCD enzyme into duplex DNA. J Bacteriol. 1994;176:4656–4663. doi: 10.1128/jb.176.15.4656-4663.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Horsfall M J, Lawrence C W. Accuracy of replication past the T-C (6-4) adduct. J Mol Biol. 1994;235:465–471. doi: 10.1006/jmbi.1994.1006. [DOI] [PubMed] [Google Scholar]
- 266.Howard-Flanders P. DNA repair and recombination. Br Med Bull. 1973;29:226–235. doi: 10.1093/oxfordjournals.bmb.a071012. [DOI] [PubMed] [Google Scholar]
- 267.Howard-Flanders P, Theriot L. Mutants of Escherichia coli K-12 defective in DNA repair and in genetic recombination. Genetics. 1966;53:1137–1150. doi: 10.1093/genetics/53.6.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Howard-Flanders P, Theriot L, Stedeford J B. Some properties of excision-defective recombination-deficient mutants of Escherichia coli K-12. J Bacteriol. 1969;97:1134–1141. doi: 10.1128/jb.97.3.1134-1141.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Howard-Flanders P, West S C, Stasiak A. Role of RecA protein spiral filaments in genetic recombination. Nature. 1984;309:215–220. doi: 10.1038/309215a0. [DOI] [PubMed] [Google Scholar]
- 270.Hsieh P, Camerini-Otero C S, Camerini-Otero R D. The synapsis event in the homologous pairing of DNAs: RecA recognizes and pairs less than one helical repeat of DNA. Proc Natl Acad Sci USA. 1992;89:6492–6496. doi: 10.1073/pnas.89.14.6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Huskins C L. The coiling of chromonemata. Cold Spring Harbor Symp Quant Biol. 1941;9:13–18. [Google Scholar]
- 272.Inouye M. Pleiotripoc effect of the recA gene of Escherichia coli: uncoupling of cell division from deoxyribonucleic acid replication. J Bacteriol. 1971;106:539–542. doi: 10.1128/jb.106.2.539-542.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Ishimori K, Sommer S, Bailone A, Takahashi M, Cox M M, Devoret R. Characterization of a mutant RecA protein that facilitates homologous genetic recombination but not recombinational DNA repair: RecA423. J Mol Biol. 1996;264:696–712. doi: 10.1006/jmbi.1996.0670. [DOI] [PubMed] [Google Scholar]
- 274.Ishioka K, Fukuoh A, Iwasaki H, Nakata A, Shinagawa H. Abortive recombination in Escherichia coli ruv mutants blocks chromosome partitioning. Genes Cells. 1998;3:209–220. doi: 10.1046/j.1365-2443.1998.00185.x. [DOI] [PubMed] [Google Scholar]
- 275.Ishioka K, Iwasaki H, Shinagawa H. Roles of the recG gene product of Escherichia coli in recombinational repair: effects of the ΔrecG mutation on cell division and chromosome partition. Genes Genet Syst. 1997;72:91–99. doi: 10.1266/ggs.72.91. [DOI] [PubMed] [Google Scholar]
- 276.Iwasaki H, Takahaghi M, Nakata A, Shinagawa H. Escherichia coli RuvA and RuvB proteins specifically interact with Holliday junctions and promote branch migration. Genes Dev. 1992;6:2214–2220. doi: 10.1101/gad.6.11.2214. [DOI] [PubMed] [Google Scholar]
- 277.Iwasaki H, Takahaghi M, Shiba T, Nakata A, Shinagawa H. Escherichia coli RuvC protein is an endonuclease that resolves the Holliday structure. EMBO J. 1991;10:4381–4389. doi: 10.1002/j.1460-2075.1991.tb05016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Iyer V N, Rupp W D. Usefulness of benzoylated naphthoylated DEAE-cellulose to distinguish and fractionate double-stranded DNA bearing different extents of single-stranded regions. Biochim Biophys Acta. 1971;228:117–126. doi: 10.1016/0005-2787(71)90551-x. [DOI] [PubMed] [Google Scholar]
- 279.Iype L E, Inman R B, Cox M M. Blocked RecA protein-mediated DNA strand exchange reactions are reversed by the RuvA and RuvB proteins. J Biol Chem. 1995;270:19473–19480. doi: 10.1074/jbc.270.33.19473. [DOI] [PubMed] [Google Scholar]
- 280.Jain S K, Cox M M, Inman R B. On the role of ATP hydrolysis in RecA protein-mediated DNA strand exchange. III. Unidirectional branch migration and extensive hybrid DNA formation. J Biol Chem. 1994;269:20653–20661. [PubMed] [Google Scholar]
- 281.Johnson R C. Postreplication repair gap filling in an Escherichia coli strain deficient in dnaB gene product. In: Hanawalt P C, Setlow R B, editors. Molecular mechanisms for repair of DNA. 5A. New York, N.Y: Plenum Press; 1975. pp. 325–329. [DOI] [PubMed] [Google Scholar]
- 282.Johnson R C. Reduction of postreplication DNA repair in two Escherichia coli mutants with temperature-sensitive polymerase III activity: implications for the postreplication repair pathway. J Bacteriol. 1978;136:125–130. doi: 10.1128/jb.136.1.125-130.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Jones J M, Nakai H. The ϕX174-type primosome promotes replisome assembly at the site of recombination in bacteriophage Mu transposition. EMBO J. 1997;16:6886–6895. doi: 10.1093/emboj/16.22.6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Joseph J W, Kolodner R. Exonuclease VIII of Escherichia coli. I. Purification and physical properties. J Biol Chem. 1983;258:10411–10417. [PubMed] [Google Scholar]
- 285.Joseph J W, Kolodner R. Exonuclease VIII of Escherichia coli. II. Mechanism of action. J Biol Chem. 1983;258:10418–10424. [PubMed] [Google Scholar]
- 286.Julin D A, Lehman I R. Photoaffinity labeling of the RecBCD enzyme of Escherichia coli with 8-azidoadenosine 5′-triphosphate. J Biol Chem. 1987;262:9044–9051. [PubMed] [Google Scholar]
- 287.Kaasch M, Kaasch J, Quinones A. Expression of dnaN and dnaQ genes of Escherichia coli is inducible by mitomycin C. Mol Gen Genet. 1989;219:187–192. doi: 10.1007/BF00261175. [DOI] [PubMed] [Google Scholar]
- 288.Kahn R, Cunningham R P, DasGupta C, Radding C M. Polarity of heteroduplex formation promoted by Escherichia coli RecA protein. Proc Natl Acad Sci USA. 1981;78:4786–4790. doi: 10.1073/pnas.78.8.4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Kaiser K, Murray N E. Physical characterization of the “Rac prophage” in E. coli K-12. Mol Gen Genet. 1979;175:159–174. doi: 10.1007/BF00425532. [DOI] [PubMed] [Google Scholar]
- 290.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 291.Karakousis G, Ye N, Li Z, Chiu S K, Reddy G, Radding C M. The Beta protein of phage λ binds preferentially to an intermediate in DNA renaturation. J Mol Biol. 1998;276:721–731. doi: 10.1006/jmbi.1997.1573. [DOI] [PubMed] [Google Scholar]
- 292.Karu A E, Belk E D. Induction of E. coli recA protein via recBC and alternative pathways: quantitation by enzyme-linked immunosorbent assay (ELISA) Mol Gen Genet. 1982;185:275–282. doi: 10.1007/BF00330798. [DOI] [PubMed] [Google Scholar]
- 293.Karu A E, MacKay V, Goldmark P J, Linn S. The RecBC deoxyribonuclease of Escherichia coli K-12. Substrate specificity and reaction intermediates. J Biol Chem. 1973;248:4874–4884. [PubMed] [Google Scholar]
- 294.Karu A E, Yoshiyuki E, Echols H, Linn S. The γ protein specified by bacteriophage λ. J Biol Chem. 1975;250:7377–7387. [PubMed] [Google Scholar]
- 295.Kastan M B, Kuerbitz S J. Control of G1 arrest after DNA damage. Environ Health Perspect. 1993;101(Suppl. 5):55–58. doi: 10.1289/ehp.93101s555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Kato T, Shinoura Y. Isolation and characterization of mutants of Escherichia coli deficient in induction of mutations by ultraviolet light. Mol Gen Genet. 1977;156:121–131. doi: 10.1007/BF00283484. [DOI] [PubMed] [Google Scholar]
- 297.Kelman Z, O’Donnell M. DNA polymerase III holoenzyme: structure and function of a chromosomal replicating machine. Annu Rev Biochem. 1995;64:171–200. doi: 10.1146/annurev.bi.64.070195.001131. [DOI] [PubMed] [Google Scholar]
- 298.Kelner A. Growth, respiration, and nucleic acid synthesis in ultraviolet-irradiated and in photoreactivated Escherichia coli. J Bacteriol. 1953;65:252–262. doi: 10.1128/jb.65.3.252-262.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Keyer K, Strohmeier Gort A, Imlay J A. Superoxide and the production of oxidative DNA damage. J Bacteriol. 1995;177:6782–6790. doi: 10.1128/jb.177.23.6782-6790.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Khidhir M A, Casaregola S, Holland I B. Mechanism of transient inhibition of DNA synthesis in ultraviolet-irradiated E. coli: inhibition is independent of recA whilst recovery requires RecA protein itself and an additional, inducible SOS function. Mol Gen Genet. 1985;199:133–140. doi: 10.1007/BF00327522. [DOI] [PubMed] [Google Scholar]
- 301.Kiianitsa K, Stasiak A. Helical repeat of DNA in the region of homologous pairing. Proc Natl Acad Sci USA. 1997;94:7837–7840. doi: 10.1073/pnas.94.15.7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Kim J-I, Cox M M, Inman R B. On the role of ATP hydrolysis in RecA protein-mediated DNA strand exchange. I. Bypassing a short heterologous insert in one DNA substrate. J Biol Chem. 1992;267:16438–16443. [PubMed] [Google Scholar]
- 303.Kim J-I, Cox M M, Inman R B. On the role of ATP hydrolysis in RecA protein-mediated DNA strand exchange. II. Four-strand exchanges. J Biol Chem. 1992;267:16444–16449. [PubMed] [Google Scholar]
- 304.Kim S-R, Maenhaut-Michel G, Yamada M, Yamamoto Y, Matsui K, Sofuni T, Nohmi T, Ohmori H. Multiple pathways for SOS-induced mutagenesis in Escherichia coli: an overexpression of dinB/dinP results in strongly enhancing mutagenesis in the absence of any exogenous treatment to damage DNA. Proc Natl Acad Sci USA. 1997;94:13792–13797. doi: 10.1073/pnas.94.25.13792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Klinkert M Q, Klein A, Abdel-Monem M. Studies on the function of DNA helicase I and DNA helicase II of Escherichia coli. J Biol Chem. 1980;255:9746–9752. [PubMed] [Google Scholar]
- 306.Kmiec E, Holloman W K. β protein of bacteriophage λ promotes renaturation of DNA. J Biol Chem. 1981;256:12636–12639. [PubMed] [Google Scholar]
- 307.Knight K L, McEntee K. Covalent modification of the RecA protein from Escherichia coli with the photoaffinity label 8-azidoadenosine 5′-triphosphate. J Biol Chem. 1985;260:867–872. [PubMed] [Google Scholar]
- 308.Kobayashi I, Takahashi N. Double-stranded gap repair of DNA by gene conversion in Escherichia coli. Genetics. 1988;119:751–757. doi: 10.1093/genetics/119.4.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Kogoma T. Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol Mol Biol Rev. 1997;61:212–238. doi: 10.1128/mmbr.61.2.212-238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Kogoma T, Cadwell G W, Barnard K G, Asai T. The DNA replication priming protein, PriA, is required for homologous recombination and double-strand break repair. J Bacteriol. 1996;178:1258–1264. doi: 10.1128/jb.178.5.1258-1264.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Koller T, Di Capua E, Stasiak A. Complexes of RecA with single-stranded DNA. In: Cozzarelli N, editor. Mechanisms of DNA replication and recombination. New York, N.Y: Alan R. Liss, Inc.; 1983. pp. 723–729. [Google Scholar]
- 312.Kolodner R, Fishel R A, Howard M. Genetic recombination of bacterial plasmid DNA: effect of RecF pathway mutations on plasmid recombination in Escherichia coli. J Bacteriol. 1985;163:1060–1066. doi: 10.1128/jb.163.3.1060-1066.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Konrad E B. Method for the isolation of Escherichia coli mutants with enhanced recombination between chromosomal duplications. J Bacteriol. 1977;130:167–172. doi: 10.1128/jb.130.1.167-172.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Köppen A, Krobitsch S, Thoms B, Wackernagel W. Interaction with the recombination hot spot χ in vivo converts the RecBCD enzyme of Escherichia coli into a χ-independent recombinase by inactivation of the RecD subunit. Proc Natl Acad Sci USA. 1995;92:6249–6253. doi: 10.1073/pnas.92.14.6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Korangy F, Julin D A. Enzymatic effects of a lysine-to-glutamine mutation in the ATP-binding consensus sequence in the RecD subunit of the RecBCD enzyme from Escherichia coli. J Biol Chem. 1992;267:1733–1740. [PubMed] [Google Scholar]
- 316.Korangy F, Julin D A. A mutation in the consensus ATP-binding sequence of the RecD subunit reduces the processivity of the RecBCD enzyme from Escherichia coli. J Biol Chem. 1992;267:3088–3095. [PubMed] [Google Scholar]
- 317.Kornberg A, Baker T A. DNA replication. W. H. New York, N.Y: Freeman & Co.; 1992. [Google Scholar]
- 318.Kovall R, Matthews B W. Toroidal structure of λ-exonuclease. Science. 1997;277:1824–1827. doi: 10.1126/science.277.5333.1824. [DOI] [PubMed] [Google Scholar]
- 319.Kowalczykowski S C. Biochemistry of genetic recombination: energetics and mechanism of DNA strand exchange. Annu Rev Biophys Biophys Chem. 1991;20:539–575. doi: 10.1146/annurev.bb.20.060191.002543. [DOI] [PubMed] [Google Scholar]
- 320.Kowalczykowski S C, Dixon D A, Eggleston A K, Lauder S D, Rehrauer W M. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Kowalczykowski S C, Eggleston A K. Homologous pairing and DNA strand-exchange proteins. Annu Rev Biochem. 1994;63:991–1043. doi: 10.1146/annurev.bi.63.070194.005015. [DOI] [PubMed] [Google Scholar]
- 322.Kowalczykowski S C, Krupp R A. DNA-strand exchange promoted by RecA protein in the absence of ATP: implications for the mechanism of energy transduction in protein-promoted nucleic acid transactions. Proc Natl Acad Sci USA. 1995;92:3478–3482. doi: 10.1073/pnas.92.8.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Krasin F, Hutchinson F. Repair of DNA double-strand breaks in Escherichia coli, which requires recA function and the presence of a duplicate genome. J Mol Biol. 1977;116:81–98. doi: 10.1016/0022-2836(77)90120-6. [DOI] [PubMed] [Google Scholar]
- 324.Krisch R E, Flick M B, Trumbore C N. Radiation chemical mechanisms of single- and double-strand break formation in irradiated SV40 DNA. Radiat Res. 1991;126:251–259. [PubMed] [Google Scholar]
- 325.Krisch R E, Krasin F, Sauri C J. DNA breakage, repair and lethality after 125I decay in rec+ and recA strains of Escherichia coli. Int J Radiat Biol. 1976;29:37–50. doi: 10.1080/09553007614551541. [DOI] [PubMed] [Google Scholar]
- 326.Kubista M, Takahashi M, Nordén B. Stoichiometry, base orientation and nuclease accessibility of RecA-DNA complexes seen by polarized light in flow-oriented solution. Implications for the mechanism of genetic recombination. J Biol Chem. 1990;265:18891–18897. [PubMed] [Google Scholar]
- 327.Kuempel P, Hogaard A, Nielsen M, Nagappan O, Tecklenburg M. Use of transposon (Tndif) to obtain suppressing and nonsuppressing insertions of the dif resolvase site of Escherichia coli. Genes Dev. 1996;10:1162–1171. doi: 10.1101/gad.10.9.1162. [DOI] [PubMed] [Google Scholar]
- 328.Kuempel P L, Henson J M, Dircks L, Tecklenburg M, Lim D F. dif, a recA-independent recombination site in the terminus region of the chromosome of Escherichia coli. New Biol. 1991;3:799–811. [PubMed] [Google Scholar]
- 329.Kumar K A, Muniyappa K. Use of structure-directed DNA ligands to probe the binding of RecA protein to narrow and wide grooves of DNA and on its ability to promote homologous pairing. J Biol Chem. 1992;267:24824–24832. [PubMed] [Google Scholar]
- 330.Kumura K, Sekiguchi M. Identification of the uvrD gene product of Escherichia coli as DNA helicase II and its induction by DNA-damaging agents. J Biol Chem. 1984;259:1560–1565. [PubMed] [Google Scholar]
- 331.Kusano K, Sunohara Y, Takahashi N, Yoshikura H, Kobayashi I. DNA double-strand break repair: genetic determinants of flanking crossing-over. Proc Natl Acad Sci USA. 1994;91:1173–1177. doi: 10.1073/pnas.91.3.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Kushner S R, Nagaishi H, Templin A, Clark A J. Genetic recombination in Escherichia coli: the role of exonuclease I. Proc Natl Acad Sci USA. 1971;68:824–827. doi: 10.1073/pnas.68.4.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Kuzminov A. Collapse and repair of replication forks in Escherichia coli. Mol Microbiol. 1995;16:373–384. doi: 10.1111/j.1365-2958.1995.tb02403.x. [DOI] [PubMed] [Google Scholar]
- 334.Kuzminov A. Instability of inhibited replication forks in E. coli. Bioessays. 1995;17:733–741. doi: 10.1002/bies.950170810. [DOI] [PubMed] [Google Scholar]
- 335.Kuzminov A. A mechanism for induction of the SOS response in E. coli: insights into the regulation of reversible protein polymerization in vivo. J Theor Biol. 1995;177:29–43. doi: 10.1006/jtbi.1995.0222. [DOI] [PubMed] [Google Scholar]
- 336.Kuzminov A. Recombinational repair of DNA damage. R. G. Austin, Tex: Landes Co.; 1996. [Google Scholar]
- 337.Kuzminov A. Unraveling the late stages of recombinational repair: metabolism of DNA junctions in Escherichia coli. Bioessays. 1996;18:757–765. doi: 10.1002/bies.950180911. [DOI] [PubMed] [Google Scholar]
- 338.Kuzminov A, Schabtach E, Stahl F W. χ-sites in combination with RecA protein increase the survival of linear DNA in E. coli by inactivating exoV activity of RecBCD nuclease. EMBO J. 1994;13:2764–2776. doi: 10.1002/j.1460-2075.1994.tb06570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Kuzminov A, Stahl F W. Double-strand end repair via the RecBC pathway in Escherichia coli primes DNA replication. Genes Dev. 1999;13:345–356. doi: 10.1101/gad.13.3.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Kuzminov A, Stahl F W. Stability of linear DNA in recA mutant Escherichia coli cells reflects ongoing chromosomal DNA degradation. J Bacteriol. 1997;179:880–888. doi: 10.1128/jb.179.3.880-888.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Laban A, Cohen A. Interplasmidic and intraplasmidic recombination in Escherichia coli K-12. Mol Gen Genet. 1981;184:200–207. doi: 10.1007/BF00272905. [DOI] [PubMed] [Google Scholar]
- 342.Lane H E D, Denhardt D T. The rep mutation. IV. Slower movement of the replication forks in Escherichia coli rep strains. J Mol Biol. 1975;97:99–112. doi: 10.1016/s0022-2836(75)80025-8. [DOI] [PubMed] [Google Scholar]
- 343.Lark K G, Lark C A. recA-dependent DNA replication in the absence of protein synthesis: characteristics of a dominant lethal replication mutation, dnaT, and requirement for recA+ function. Cold Spring Harbor Symp Quant Biol. 1979;43:537–549. doi: 10.1101/sqb.1979.043.01.059. [DOI] [PubMed] [Google Scholar]
- 344.Lavery P E, Kowalczykowski S C. Biochemical basis of the constitutive repressor cleavage activity of RecA730 protein. A comparison to RecA441 and RecA803 proteins. J Biol Chem. 1992;267:20648–20658. [PubMed] [Google Scholar]
- 345.Lavery P E, Kowalczykowski S C. Biochemical basis of the temperature-inducible constitutive protease activity of the RecA441 protein of Escherichia coli. J Mol Biol. 1988;203:861–874. doi: 10.1016/0022-2836(88)90112-x. [DOI] [PubMed] [Google Scholar]
- 346.Lavery P E, Kowalczykowski S C. A postsynaptic role for single-stranded DNA-binding protein in RecA protein-promoted DNA strand exchange. J Biol Chem. 1992;267:9315–9320. [PubMed] [Google Scholar]
- 347.Lawrence C W, Borden A, Banerjee S K, LeClerc J E. Mutation frequency and spectrum resulting from a single abasic site in a single-stranded vector. Nucleic Acids Res. 1990;18:2153–2157. doi: 10.1093/nar/18.8.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Lawrence C W, Gibbs P E, Borden A, Horsfall M J, Kilbey B J. Mutagenesis induced by single UV photoproducts in E. coli and yeast. Mutat Res. 1993;299:157–163. doi: 10.1016/0165-1218(93)90093-s. [DOI] [PubMed] [Google Scholar]
- 349.Leahy M C, Radding C M. Topography of the interaction of RecA protein with single-stranded deoxyoligonucleotides. J Biol Chem. 1986;261:6954–5960. [PubMed] [Google Scholar]
- 350.LeClerc J E, Setlow J K. Postreplication repair of ultraviolet damage in Haemophilus influenzae. J Bacteriol. 1972;110:930–934. doi: 10.1128/jb.110.3.930-934.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Lederberg J. Gene recombination and linked segregations in Escherichia coli. Genetics. 1947;32:505–525. doi: 10.1093/genetics/32.5.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Lee E H, Kornberg A. Replication deficiencies in priA mutants of Escherichia coli lacking the primosomal replication n′ protein. Proc Natl Acad Sci USA. 1991;88:3029–3032. doi: 10.1073/pnas.88.8.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Lehman I R, Nussbaum R. The deoxyribonuclease of Escherichia coli. V. On the specificity of exonuclease I (phosphodiesterase) J Biol Chem. 1964;239:2628–2636. [PubMed] [Google Scholar]
- 354.Lesko S A, Lorentzen R J, Ts’o P O P. Role of superoxide in deoxyribonucleic acid strand scission. Biochemistry. 1980;19:3023–3028. doi: 10.1021/bi00554a029. [DOI] [PubMed] [Google Scholar]
- 355.Leslie N R, Sherratt D J. Site-specific recombination in the replication terminus region of Escherichia coli: functional replacement of dif. EMBO J. 1995;14:1561–1570. doi: 10.1002/j.1460-2075.1995.tb07142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Lewis L K, Harlow G R, Gregg-Jolly L A, Mount D W. Identification of high affinity binding sites for LexA which define new DNA damage-inducible genes in Escherichia coli. J Mol Biol. 1994;241:507–523. doi: 10.1006/jmbi.1994.1528. [DOI] [PubMed] [Google Scholar]
- 357.Ley R D. Postreplication repair in an excision-defective mutant of Escherichia coli: ultraviolet light-induced incorporation of bromodeoxyuridine into parental DNA. Photochem Photobiol. 1973;18:87–95. doi: 10.1111/j.1751-1097.1973.tb06397.x. [DOI] [PubMed] [Google Scholar]
- 358.Li Z, Karakousis G, Chiu S K, Reddy G, Radding C M. The Beta protein of phage λ promotes strand exchange. J Mol Biol. 1998;276:733–744. doi: 10.1006/jmbi.1997.1572. [DOI] [PubMed] [Google Scholar]
- 359.Lieberman H B, Witkin E M. Variable expression of the ssb-1 allele in different strains of Escherichia coli K12 and B: differential suppression of its effects on DNA replication, DNA repair and ultraviolet mutagenesis. Mol Gen Genet. 1981;183:348–355. doi: 10.1007/BF00270639. [DOI] [PubMed] [Google Scholar]
- 360.Lin P-F, Howard-Flanders P. Genetic exchanges caused by ultraviolet photoproducts in phage λ DNA molecules: the role of DNA replication. Mol Gen Genet. 1976;146:107–115. doi: 10.1007/BF00268079. [DOI] [PubMed] [Google Scholar]
- 361.Lin Y-P, Sharer J D, March P E. GTPase-dependent signaling in bacteria: characterization of a membrane-binding site for Era in Escherichia coli. J Bacteriol. 1994;176:44–49. doi: 10.1128/jb.176.1.44-49.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Lindsley J E, Cox M M. Dissociation pathway for RecA nucleoprotein filaments formed on linear duplex DNA. J Mol Biol. 1989;205:695–711. doi: 10.1016/0022-2836(89)90315-x. [DOI] [PubMed] [Google Scholar]
- 363.Lindsley J E, Cox M M. On RecA protein-mediated homologous alignment of two DNA molecules. Three strands versus four strands. J Biol Chem. 1990;265:10164–10171. [PubMed] [Google Scholar]
- 364.Little J W. Autodigestion of LexA and phage lambda repressors. Biochemistry. 1984;81:1375–1379. doi: 10.1073/pnas.81.5.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Little J W. An exonuclease induced by bacteriophage λ. II. Nature of the enzymatic reaction. J Biol Chem. 1967;242:679–686. [PubMed] [Google Scholar]
- 366.Little J W, Edmiston S H, Pacelli L Z, Mount D W. Cleavage of the Escherichia coli LexA protein by the RecA protease. Proc Natl Acad Sci USA. 1980;77:3225–3229. doi: 10.1073/pnas.77.6.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Little J W, Mount D W. The SOS regulatory system of Escherichia coli. Cell. 1982;29:11–22. doi: 10.1016/0092-8674(82)90085-x. [DOI] [PubMed] [Google Scholar]
- 368.Liu J, Nurse P, Marians K J. The ordered assembly of the ϕX174-type primosome. III. PriB facilitates complex formation between PriA and DnaT. J Biol Chem. 1996;271:15656–15661. doi: 10.1074/jbc.271.26.15656. [DOI] [PubMed] [Google Scholar]
- 369.Livneh Z, Lehman I R. Recombinational bypass of pyrimidine dimers promoted by the RecA protein of Escherichia coli. Proc Natl Acad Sci USA. 1982;79:3171–3175. doi: 10.1073/pnas.79.10.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Lloyd R G. Conjugational recombination in resolvase-deficient ruvC mutants of Escherichia coli K-12 depends on recG. J Bacteriol. 1991;173:5414–5418. doi: 10.1128/jb.173.17.5414-5418.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Lloyd R G. lexA dependent recombination in uvrD strains of Escherichia coli. Mol Gen Genet. 1983;189:157–161. doi: 10.1007/BF00326069. [DOI] [PubMed] [Google Scholar]
- 372.Lloyd R G, Benson F E, Shurvinton C E. Effect of ruv mutations on recombination and DNA repair in Escherichia coli K12. Mol Gen Genet. 1984;194:303–309. doi: 10.1007/BF00383532. [DOI] [PubMed] [Google Scholar]
- 373.Lloyd R G, Buckman C. Genetic analysis of the recG locus of Escherichia coli K-12 and of its role in recombination and DNA repair. J Bacteriol. 1991;173:1004–1011. doi: 10.1128/jb.173.3.1004-1011.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Lloyd R G, Buckman C. Identification and genetic analysis of sbcC mutations in commonly used recBC sbcB strains of Escherichia coli K-12. J Bacteriol. 1985;164:836–844. doi: 10.1128/jb.164.2.836-844.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Lloyd R G, Buckman C, Benson F E. Genetic analysis of conjugational recombination in Escherichia coli K12 strains deficient in RecBCD enzyme. J Gen Microbiol. 1987;133:2531–2538. doi: 10.1099/00221287-133-9-2531. [DOI] [PubMed] [Google Scholar]
- 376.Lloyd R G, Low B, Godson G N, Birge E A. Isolation and characterization of an Escherichia coli K-12 mutant with a temperature-sensitive RecA− phenotype. J Bacteriol. 1974;120:407–415. doi: 10.1128/jb.120.1.407-415.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Lloyd R G, Low K B. Homologous recombination. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 2236–2255. [Google Scholar]
- 378.Lloyd R G, Porton M C, Buckman C. Effect of recF, recJ, recN, recO and ruv mutations on ultraviolet survival and genetic recombination in a recD strain of Escherichia coli K12. Mol Gen Genet. 1988;212:317–324. doi: 10.1007/BF00334702. [DOI] [PubMed] [Google Scholar]
- 379.Lloyd R G, Sharples G J. Molecular organization and nucleotide sequence of the recG locus of Escherichia coli K-12. J Bacteriol. 1991;173:6837–6843. doi: 10.1128/jb.173.21.6837-6843.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Lloyd R G, Sharples G J. Processing of recombination intermediates by the RecG and RuvAB proteins of Escherichia coli. Nucleic Acids Res. 1993;21:1719–1725. doi: 10.1093/nar/21.8.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Lobner-Olesen A, Kuempel P L. Chromosome partitioning in Escherichia coli. J Bacteriol. 1992;174:7883–7889. doi: 10.1128/jb.174.24.7883-7889.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Louarn J, Cornet F, François V, Patte J, Louarn J-M. Hyperrecombination in the Terminus region of the Escherichia coli chromosome: possible relation to nucleoid organization. J Bacteriol. 1994;176:7524–7531. doi: 10.1128/jb.176.24.7524-7531.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Louarn J-M, Bird R E. Size distribution and molecular polarity of newly replicated DNA in Escherichia coli. Proc Natl Acad Sci USA. 1974;71:329–333. doi: 10.1073/pnas.71.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Louarn J-M, Louarn J, François V, Patte J. Analysis and possible role of hyperrecombination in the termination region of the Escherichia coli chromosome. J Bacteriol. 1991;173:5097–5104. doi: 10.1128/jb.173.16.5097-5104.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Lovett S T, Clark A J. Genetic analysis of the recJ gene of Escherichia coli K-12. J Bacteriol. 1984;157:190–196. doi: 10.1128/jb.157.1.190-196.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Lovett S T, Kolodner R D. Identification and purification of a single-stranded DNA-specific exonuclease encoded by the recJ gene of Escherichia coli. Proc Natl Acad Sci USA. 1989;86:2627–2631. doi: 10.1073/pnas.86.8.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Lowry O H, Carter J, Ward J B, Glaser L. The effect of carbon and nitrogen sources on the level of metabolic intermediates in Escherichia coli. J Biol Chem. 1971;246:6511–6521. [PubMed] [Google Scholar]
- 388.Luisi-DeLuca C, Kolodner R. Purification and characterization of the Escherichia coli RecO protein. Renaturation of complementary single-stranded DNA molecules catalyzed by the RecO protein. J Mol Biol. 1994;236:124–138. doi: 10.1006/jmbi.1994.1123. [DOI] [PubMed] [Google Scholar]
- 389.Luisi-DeLuca C, Kolodner R D. Effect of terminal non-homology on intramolecular recombination of linear plasmid substrates in Escherichia coli. J Mol Biol. 1992;227:72–80. doi: 10.1016/0022-2836(92)90682-a. [DOI] [PubMed] [Google Scholar]
- 390.Luisi-DeLuca C, Lovett S T, Kolodner R D. Genetic and physical analysis of plasmid recombination in recB recC sbcB and recB recC sbcA Escherichia coli K-12 mutants. Genetics. 1989;122:269–278. doi: 10.1093/genetics/122.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Lundblad V, Kleckner N. Mismatch repair mutations of Escherichia coli K12 enhance transposon excision. Genetics. 1984;109:3–19. doi: 10.1093/genetics/109.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Lusk J E, Williams J P, Kennedy E P. Magnesium and the growth of Escherichia coli. J Biol Chem. 1968;243:2618–2624. [PubMed] [Google Scholar]
- 393.MacFarland K J, Shan Q, Inman R B, Cox M M. RecA as a motor protein. Testing models for the role of ATP hydrolysis in DNA strand exchange. J Biol Chem. 1997;272:17675–17685. doi: 10.1074/jbc.272.28.17675. [DOI] [PubMed] [Google Scholar]
- 394.Madiraju M V V S, Clark A J. Effect of RecF protein on reactions catalyzed by RecA protein. Nucleic Acids Res. 1991;19:6295–6300. doi: 10.1093/nar/19.22.6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Madiraju M V V S, Clark A J. Evidence for ATP binding and double-stranded DNA binding by Escherichia coli RecF protein. J Bacteriol. 1992;174:7705–7710. doi: 10.1128/jb.174.23.7705-7710.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Madiraju M V V S, Lavery P E, Kowalczykowski S C, Clark A J. Enzymatic properties of the RecA803 protein, a partial suppressor of recF mutations. Biochemistry. 1992;31:10529–10535. doi: 10.1021/bi00158a016. [DOI] [PubMed] [Google Scholar]
- 397.Magee T R, Asai T, Malka D, Kogoma T. DNA damage-inducible origins of DNA replication in Escherichia coli. EMBO J. 1992;11:4219–4225. doi: 10.1002/j.1460-2075.1992.tb05516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Magee T R, Kogoma T. Requirement of RecBC enzyme and an elevated level of activated RecA for induced stable DNA replication in Escherichia coli. J Bacteriol. 1990;172:1834–1839. doi: 10.1128/jb.172.4.1834-1839.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Mahajan S K. Pathways of homologous recombination in Escherichia coli. In: Kucherlapati R, Smith G R, editors. Genetic recombination. Washington, D.C.: American Society for Microbiology; 1988. pp. 87–140. [Google Scholar]
- 400.Mahdi A A, Lloyd R G. Identification of the recR locus of Escherichia coli K-12 and analysis of its role in recombination and DNA repair. Mol Gen Genet. 1989;216:503–510. doi: 10.1007/BF00334397. [DOI] [PubMed] [Google Scholar]
- 401.Maity A, McKenna W G, Muschel R J. The molecular basis for cell cycle delays following ionizing radiation: a review. Radiother Oncol. 1994;31:1–13. doi: 10.1016/0167-8140(94)90408-1. [DOI] [PubMed] [Google Scholar]
- 402.Mandal T N, Mahdi A A, Sharples G J, Lloyd R G. Resolution of Holliday intermediates in recombination and DNA repair: indirect suppression of ruvA, ruvB, and ruvC mutations. J Bacteriol. 1993;175:4325–4334. doi: 10.1128/jb.175.14.4325-4334.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Manes S H, Pruss G J, Drlica K. Inhibition of RNA synthesis by oxolinic acid is unrelated to average DNA supercoiling. J Bacteriol. 1983;155:420–423. doi: 10.1128/jb.155.1.420-423.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Marais A, Bové J-M, Renaudin J. Characterization of the recA gene regions of Spiroplasma citri and Spiroplasma melliferum. J Bacteriol. 1996;178:7003–7009. doi: 10.1128/jb.178.23.7003-7009.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Margerrison E E C, Hopewell R, Fisher L M. Nucleotide sequence of the Staphylococcus aureus gyrB-gyrA locus encoding the DNA gyrase A and B proteins. J Bacteriol. 1992;174:1596–1603. doi: 10.1128/jb.174.5.1596-1603.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Marians K J. Prokaryotic DNA replication. Annu Rev Biochem. 1992;61:673–719. doi: 10.1146/annurev.bi.61.070192.003325. [DOI] [PubMed] [Google Scholar]
- 407.Marinus M G, Konrad E B. Hyper-recombination in dam mutants of Escherichia coli K-12. Mol Gen Genet. 1976;149:273–277. doi: 10.1007/BF00268528. [DOI] [PubMed] [Google Scholar]
- 408.Marinus M G, Morris N R. Biological function for 6-methyladenine residues in the DNA of Escherichia coli K12. J Mol Biol. 1974;85:309–322. doi: 10.1016/0022-2836(74)90366-0. [DOI] [PubMed] [Google Scholar]
- 409.Marinus M G, Morris N R. Pleiotropic effects of a DNA adenine methylation mutation (dam-3) in Escherichia coli K12. Mutat Res. 1975;28:15–26. doi: 10.1016/0027-5107(75)90309-7. [DOI] [PubMed] [Google Scholar]
- 410.Masai H, Asai T, Kubota Y, Arai K, Kogoma T. Escherichia coli PriA protein is essential for inducible and constitutive stable DNA replication. EMBO J. 1994;13:5338–5345. doi: 10.1002/j.1460-2075.1994.tb06868.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Masai H, Nomura N, Kubota Y, Arai K. Roles of ϕX174 type primosome- and G4 type primase-dependent primings in initiation of lagging and leading strand synthesis of DNA replication. J Biol Chem. 1990;265:15124–15133. [PubMed] [Google Scholar]
- 412.Masterson C, Boehmer P E, McDonald F, Chaudhury S, Hickson I D, Emmerson P T. Reconstitution of the activities of the RecBCD holoenzyme of Escherichia coli from the purified subunits. J Biol Chem. 1992;267:13564–13572. [PubMed] [Google Scholar]
- 413.Mathews C K. Biochemistry of deoxyribonucleic acid-defective amber mutants of bacteriophage T4. J Biol Chem. 1972;247:7430–7438. [PubMed] [Google Scholar]
- 414.Mathews C K. Biochemistry of DNA-defective mutants of bacteriophage T4. Arch Biochem Biophys. 1976;172:178–187. doi: 10.1016/0003-9861(76)90064-3. [DOI] [PubMed] [Google Scholar]
- 415.Mathews C K, Sinha N K. Are DNA precursors concentrated at replication sites? Proc Natl Acad Sci USA. 1982;79:302–306. doi: 10.1073/pnas.79.2.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Matson S W, Bean D W, George J W. DNA helicases: enzymes with essential role in all aspects of DNA metabolism. BioEssays. 1994;16:13–22. doi: 10.1002/bies.950160103. [DOI] [PubMed] [Google Scholar]
- 417.Mattern I E, Houtman P C. Properties of uvrE mutants of Escherichia coli K12. II. Construction and properties of pol and rec derivatives. Mutat Res. 1974;25:281–287. doi: 10.1016/0027-5107(74)90056-6. [DOI] [PubMed] [Google Scholar]
- 418.Maynard Smith J. What use is sex? J Theor Biol. 1971;30:319–335. doi: 10.1016/0022-5193(71)90058-0. [DOI] [PubMed] [Google Scholar]
- 419.Maynard Smith J, Dowson C G, Spratt B G. Localized sex in bacteria. Nature. 1991;349:29–31. doi: 10.1038/349029a0. [DOI] [PubMed] [Google Scholar]
- 420.Mazin A V, Kowalczykowski S C. The function of the secondary DNA-binding site of RecA protein during DNA strand exchange. EMBO J. 1998;17:1161–1168. doi: 10.1093/emboj/17.4.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Mazin A V, Kowalczykowski S C. The specificity of the secondary DNA binding site of RecA protein defines its role in DNA strand exchange. Proc Natl Acad Sci USA. 1996;93:10673–10678. doi: 10.1073/pnas.93.20.10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.McClintock B. A correlation of ring-shaped chromosomes with variegation in Zea mays. Proc Natl Acad Sci USA. 1932;18:677–681. doi: 10.1073/pnas.18.12.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.McDaniel L S, Rogers L H, Hill W E. Survival of recombination-deficient mutants of Escherichia coli during incubation with nalidixic acid. J Bacteriol. 1978;134:1195–1198. doi: 10.1128/jb.134.3.1195-1198.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.McGlynn P, Al-Deib A A, Liu J, Marians K J, Lloyd R G. The DNA replication protein PriA and the recombination protein RecG bind D-loops. J Mol Biol. 1997;270:212–221. doi: 10.1006/jmbi.1997.1120. [DOI] [PubMed] [Google Scholar]
- 425.McKittrick N H, Smith G R. Activation of Chi recombinational hotspots by RecBCD-like enzymes from enteric bacteria. J Mol Biol. 1989;210:485–495. doi: 10.1016/0022-2836(89)90125-3. [DOI] [PubMed] [Google Scholar]
- 426.McPartland A, Green L, Echols H. Control of recA gene RNA in E. coli: regulatory and signal genes. Cell. 1980;20:731–737. doi: 10.1016/0092-8674(80)90319-0. [DOI] [PubMed] [Google Scholar]
- 427.Mendonca V M, Klepin H D, Matson S W. DNA helicases in recombination and repair: construction of a ΔuvrD ΔhelD ΔrecQ mutant deficient in recombination and repair. J Bacteriol. 1995;177:1326–1335. doi: 10.1128/jb.177.5.1326-1335.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Menetski J P, Bear D G, Kowalczykowski S C. Stable DNA heteroduplex formation catalyzed by the Escherichia coli RecA protein in the absence of ATP hydrolysis. Proc Natl Acad Sci USA. 1990;87:21–25. doi: 10.1073/pnas.87.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Menetski J P, Kowalczykowski S C. Enhancement of Escherichia coli RecA protein enzymatic function by dATP. Biochemistry. 1989;28:5871–5881. doi: 10.1021/bi00440a025. [DOI] [PubMed] [Google Scholar]
- 430.Meyer R R, Laine P S. The single-stranded DNA-binding protein of Escherichia coli. Microbiol Rev. 1990;54:342–380. doi: 10.1128/mr.54.4.342-380.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Meyer R R, Voegele D W, Ruben S M, Rein D C, Trela J M. Influence of single-strand binding protein on recA induction in Escherichia coli. Mutat Res. 1982;94:299–313. doi: 10.1016/0027-5107(82)90293-7. [DOI] [PubMed] [Google Scholar]
- 432.Mézard C, Davies A A, Stasiak A, West S C. Biochemical properties of RuvBD113N: a mutation in helicase motif II of the RuvB hexamer affects DNA binding and ATPase activities. J Mol Biol. 1997;271:704–717. doi: 10.1006/jmbi.1997.1225. [DOI] [PubMed] [Google Scholar]
- 433.Michaels M L, Miller J H. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) J Bacteriol. 1992;174:6321–6325. doi: 10.1128/jb.174.20.6321-6325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Michel B, Ehrlich S D, Uzest M. DNA double-strand breaks caused by replication arrest. EMBO J. 1997;16:430–438. doi: 10.1093/emboj/16.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Mikolajczyk M, Schuster H. The fate of newly made DNA in Escherichia coli mutant thermosensitive in DNA synthesis. Mol Gen Genet. 1971;110:272–288. doi: 10.1007/BF00337839. [DOI] [PubMed] [Google Scholar]
- 436.Milkman R, McKane Bridges M. Molecular evolution of the Escherichia coli chromosome. III. Clonal frames. Genetics. 1990;126:505–517. doi: 10.1093/genetics/126.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Minton K W. Repair of ionizing-radiation damage in the radiation resistant bacterium Deinococcus radiodurans. Mutat Res. 1996;363:1–7. doi: 10.1016/0921-8777(95)00014-3. [DOI] [PubMed] [Google Scholar]
- 438.Mitchell A H, West S C. Role of RuvA in branch migration reactions catalyzed by the RuvA and RuvB proteins of Escherichia coli. J Biol Chem. 1996;271:19497–19502. doi: 10.1074/jbc.271.32.19497. [DOI] [PubMed] [Google Scholar]
- 439.Modrich P. Mechanisms and biological effects of mismatch repair. Annu Rev Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- 440.Modrich P. Mismatch repair, genetic stability, and cancer. Science. 1994;266:1959–1960. doi: 10.1126/science.7801122. [DOI] [PubMed] [Google Scholar]
- 441.Molineux I J, Gefter M L. Properties of the Escherichia coli DNA-binding (unwinding) protein interaction with nucleolytic enzymes and DNA. J Mol Biol. 1975;98:811–825. doi: 10.1016/s0022-2836(75)80012-x. [DOI] [PubMed] [Google Scholar]
- 442.Moncany M L J, Kellenberger E. High magnesium content of Escherichia coli B. Experientia. 1981;37:846–847. doi: 10.1007/BF01985672. [DOI] [PubMed] [Google Scholar]
- 443.Monk M, Kinross J. Conditional lethality of recA and recB derivatives of a strain of Escherichia coli K-12 with a temperature-sensitive deoxyribonucleic acid polymerase I. J Bacteriol. 1972;109:971–978. doi: 10.1128/jb.109.3.971-978.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Monk M, Kinross J, Town C. Deoxyribonucleic acid synthesis in recA and recB derivatives of an Escherichia coli K-12 strain with a temperature-sensitive deoxyribonucleic acid polymerase I. J Bacteriol. 1973;114:1014–1017. doi: 10.1128/jb.114.3.1014-1017.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Moreau P L. Effects of overproduction of single-stranded DNA-binding protein on RecA protein-dependent processes in Escherichia coli. J Mol Biol. 1987;194:621–634. doi: 10.1016/0022-2836(87)90239-7. [DOI] [PubMed] [Google Scholar]
- 446.Morel P, Hejna J A, Ehrlich S D, Cassuto E. Antipairing and strand transferase activities of E. coli helicase II (UvrD) Nucleic Acids Res. 1993;21:3205–3209. doi: 10.1093/nar/21.14.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Morel-Deville F, Ehrlich S D. Theta-type DNA replication stimulates homologous recombination in the Bacillus subtilis chromosome. Mol Microbiol. 1996;19:587–598. doi: 10.1046/j.1365-2958.1996.398936.x. [DOI] [PubMed] [Google Scholar]
- 448.Morimyo M. Anaerobic incubation enhances the colony formation of a polA recB strain of Escherichia coli K-12. J Bacteriol. 1982;152:208–214. doi: 10.1128/jb.152.1.208-214.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Morrison P T, Lovett S T, Gilson L E, Kolodner R. Molecular analysis of the Escherichia coli recO gene. J Bacteriol. 1989;171:3641–3649. doi: 10.1128/jb.171.7.3641-3649.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Morse L S, Beck L A, Pauling C. Effect of chloramphenicol and the recB gene product on DNA metabolism in Escherichia coli K12 strains defective in DNA ligase. Mol Gen Genet. 1976;147:79–82. doi: 10.1007/BF00337939. [DOI] [PubMed] [Google Scholar]
- 451.Mudgett J S, Buckholt M, Taylor W D. Ultraviolet light-induced plasmid-chromosome recombination in Escherichia coli: the role of recB and recF. Gene. 1991;97:131–136. doi: 10.1016/0378-1119(91)90020-c. [DOI] [PubMed] [Google Scholar]
- 452.Muller B, Boehmer P E, Emmerson P T, West S C. Action of RecBCD enzyme on Holliday structures made by RecA. J Biol Chem. 1991;266:19028–19033. [PubMed] [Google Scholar]
- 453.Muller B, Tsaneva I R, West S C. Branch migration of Holliday junctions promoted by the Escherichia coli RuvA and RuvB proteins. I. Comparison of RuvAB- and RuvB-mediated reactions. J Biol Chem. 1993;268:17179–17184. [PubMed] [Google Scholar]
- 454.Muniyappa K, Mythili E. Phage lambda beta protein, a component of general recombination, is associated with host ribosomal S1 protein. Biochem Mol Biol Int. 1993;31:1–11. [PubMed] [Google Scholar]
- 455.Muniyappa K, Radding C M. The homologous recombination system of phage λ: pairing activities of β protein. J Biol Chem. 1986;261:7472–7478. [PubMed] [Google Scholar]
- 456.Muniyappa K, Shaner S L, Tsang S S, Radding C M. Mechanism of the concerted action of RecA protein and helix-destabilizing proteins in homologous recombination. Proc Natl Acad Sci USA. 1984;81:2757–2761. doi: 10.1073/pnas.81.9.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Murphy K C. λ Gam protein inhibits the helicase and χ-stimulated recombination activities of Escherichia coli RecBCD enzyme. J Bacteriol. 1991;173:5808–5821. doi: 10.1128/jb.173.18.5808-5821.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Murphy L D, Zimmerman S B. Isolation and characterization of spermidine nucleoids from Escherichia coli. J Struct Biol. 1997;119:321–335. doi: 10.1006/jsbi.1997.3883. [DOI] [PubMed] [Google Scholar]
- 459.Muth G, Frese D, Kleber A, Wohlleben W. Mutational analysis of the Streptomyces lividans recA gene suggests that only mutants with residual activity remain viable. Mol Gen Genet. 1997;255:420–428. doi: 10.1007/s004380050514. [DOI] [PubMed] [Google Scholar]
- 460.Myers R S, Kuzminov A, Stahl F W. The recombination hotspot χ activates RecBCD recombination by converting E. coli to a recD mutant phenocopy. Proc Natl Acad Sci USA. 1995;92:6244–6248. doi: 10.1073/pnas.92.14.6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Myers R S, Stahl F W. χ and the RecBCD enzyme of Escherichia coli. Annu Rev Genet. 1994;28:49–70. doi: 10.1146/annurev.ge.28.120194.000405. [DOI] [PubMed] [Google Scholar]
- 462.Nakayama H, Nakayama K, Nakayama R, Irino N, Nakayama Y, Hanawalt P C. Isolation and genetic characterization of a thymineless death-resistant mutant of Escherichia coli K12: identification of a new mutation (recQ1) that blocks the RecF recombination pathway. Mol Gen Genet. 1984;195:474–480. doi: 10.1007/BF00341449. [DOI] [PubMed] [Google Scholar]
- 463.Naom I S, Morton S J, Leach D R F, Lloyd R G. Molecular organisation of sbcC, a gene that affects genetic recombination and the viability of DNA palindromes in Escherichia coli K-12. Nucleic Acids Res. 1989;17:8033–8045. doi: 10.1093/nar/17.20.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Nasmyth K A. Molecular genetics of yeast mating type. Annu Rev Genet. 1982;16:439–500. doi: 10.1146/annurev.ge.16.120182.002255. [DOI] [PubMed] [Google Scholar]
- 465.Neuhard J. Studies on the acid-soluble nucleotide pool in thymine-requiring mutants of Escherichia coli during thymine starvation. III. On the regulation of the deoxyadenosine triphosphate and deoxycytidine triphosphate pools of Escherichia coli. Biochim Biophys Acta. 1966;129:104–115. doi: 10.1016/0005-2787(66)90012-8. [DOI] [PubMed] [Google Scholar]
- 466.Ng J Y, Marians K J. The ordered assembly of the ϕX174-type primosome. I. Isolation and identification of intermediate protein-DNA complexes. J Biol Chem. 1996;271:15642–15648. doi: 10.1074/jbc.271.26.15642. [DOI] [PubMed] [Google Scholar]
- 467.Ng J Y, Marians K J. The ordered assembly of the ϕX174-type primosome. II. Preservation of primosome composition from assembly through replication. J Biol Chem. 1996;271:15649–15655. doi: 10.1074/jbc.271.26.15649. [DOI] [PubMed] [Google Scholar]
- 468.Noirot P, Kolodner R D. DNA strand invasion promoted by Escherichia coli RecT protein. J Biol Chem. 1998;273:12274–12280. doi: 10.1074/jbc.273.20.12274. [DOI] [PubMed] [Google Scholar]
- 469.Nowicka A, Kanabus M, Sledziewska-Gojska E, Ciesla Z. Different UmuC requirements for generation of different kinds of UV-induced mutations in Escherichia coli. Mol Gen Genet. 1994;243:584–592. doi: 10.1007/BF00284207. [DOI] [PubMed] [Google Scholar]
- 470.O’Donnell M, Studwell P S. Total reconstitution of DNA polymerase III holoenzyme from Escherichia coli. J Biol Chem. 1990;265:1179–1187. [PubMed] [Google Scholar]
- 471.Okazaki R, Okazaki T, Sakabe K, Sugimoto K, Kainuma R, Sugino A, Iwatsuki N. In vivo mechanism of DNA chain growth. Cold Spring Harbor Symp Quant Biol. 1968;33:129–143. [Google Scholar]
- 472.Olivera B M. DNA intermediates at the Escherichia coli replication fork: effect of dUTP. Proc Natl Acad Sci USA. 1978;75:238–242. doi: 10.1073/pnas.75.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Otsuji N, Iyehara H, Hideshima Y. Isolation and characterization of Escherichia coli ruv mutant which forms nonseptate filaments after low doses of ultraviolet light irradiation. J Bacteriol. 1974;117:337–344. doi: 10.1128/jb.117.2.337-344.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Otsuji N, Iyehara-Ogawa H. Thermoresistant revertants of an Escherichia coli strain carrying tif-1 and ruv mutations: non-suppressibility of ruv by sfi. J Bacteriol. 1979;138:1–6. doi: 10.1128/jb.138.1.1-6.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Overbye K M, Margolin P. Role of the supX gene in ultraviolet light-induced mutagenesis in Salmonella typhimurium. J Bacteriol. 1981;146:170–178. doi: 10.1128/jb.146.1.170-178.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Panyutin I G, Biswas I, Hsieh P. A pivotal role for the structure of the Holliday junction in DNA branch migration. EMBO J. 1995;14:1819–1826. doi: 10.1002/j.1460-2075.1995.tb07170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Park E-M, Shigenaga M K, Degan P, Korn T S, Kitzler J W, Wehr C M, Kolachana P, Ames B N. Assay of excised oxidative DNA lesions: isolation of 8-oxoguanine and its nucleoside derivatives from biological fluids with a monoclonal antibody column. Proc Natl Acad Sci USA. 1992;89:3375–3379. doi: 10.1073/pnas.89.8.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Parsons C A, Stasiak A, Bennett R J, West S C. Structure of a multisubunit complex that promotes DNA branch migration. Nature. 1995;374:375–378. doi: 10.1038/374375a0. [DOI] [PubMed] [Google Scholar]
- 479.Parsons C A, Stasiak A, West S C. The E. coli RuvAB proteins branch migrate Holliday junctions through heterologous DNA sequences in a reaction facilitated by SSB. EMBO J. 1995;14:5736–5744. doi: 10.1002/j.1460-2075.1995.tb00260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Parsons C A, Tsaneva I, Lloyd R G, West S C. Interaction of Escherichia coli RuvA and RuvB proteins with synthetic Holliday junctions. Proc Natl Acad Sci USA. 1992;89:5452–5456. doi: 10.1073/pnas.89.12.5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Parsons C A, West S C. Formation of a RuvAB-Holliday junction complex in vitro. J Mol Biol. 1993;232:397–405. doi: 10.1006/jmbi.1993.1399. [DOI] [PubMed] [Google Scholar]
- 482.Paterson M C, Boyle J M, Setlow R B. Ultraviolet- and X-ray-induced responses of a deoxyribonucleic acid polymerase-deficient mutant of Escherichia coli. J Bacteriol. 1971;107:61–67. doi: 10.1128/jb.107.1.61-67.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Pauling C, Hamm L. Properties of a temperature-sensitive, radiation-sensitive mutant of Escherichia coli. II. DNA replication. Proc Natl Acad Sci USA. 1969;64:1195–1202. doi: 10.1073/pnas.64.4.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Paz-Elizur T, Takeshita M, Goodman M, O’Donnell M, Livneh Z. Mechanism of translesion DNA synthesis by DNA polymerase II. Comparison to DNA polymerase I and III core. J Biol Chem. 1996;271:24662–24669. doi: 10.1074/jbc.271.40.24662. [DOI] [PubMed] [Google Scholar]
- 485.Perez-Roger I, Garcia-Sogo M, Navarro-Avino J P, Lopez-Acedo C, Macian F, Armengod M E. Positive and negative regulatory elements in the dnaA-dnaN-recF operon of Escherichia coli. Biochimie. 1991;73:329–334. doi: 10.1016/0300-9084(91)90220-u. [DOI] [PubMed] [Google Scholar]
- 486.Peterson K R, Mount D W. Analysis of the genetic requirements for viability of Escherichia coli K-12 DNA adenine methylase (dam) mutants. J Bacteriol. 1993;175:7505–7508. doi: 10.1128/jb.175.22.7505-7508.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Peterson K R, Wertman K F, Mount D W, Marinus M G. Viability of Escherichia coli K-12 DNA adenine methylase (dam) mutants requires increased expression of specific genes in the SOS regulon. Mol Gen Genet. 1985;201:14–19. doi: 10.1007/BF00397979. [DOI] [PubMed] [Google Scholar]
- 488.Petit M-A, Dimpfl J, Radman M, Echols H. Control of large chromosomal duplications in Escherichia coli by the mismatch repair system. Genetics. 1991;129:327–332. doi: 10.1093/genetics/129.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Pettijohn D E. The nucleoid. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 158–166. [Google Scholar]
- 490.Pettijohn D E, Sinden R R. Structure of isolated nucleoid. In: Nanninga N, editor. Molecular cytology of Escherichia coli. London, United Kingdom: Academic Press, Ltd.; 1985. pp. 199–227. [Google Scholar]
- 491.Phillips R J, Hickleton D C, Boehmer P E, Emmerson P T. The RecB protein of Escherichia coli translocates along single-stranded DNA in the 3′ to 5′ direction: a proposed ratchet mechanism. Mol Gen Genet. 1997;254:319–329. doi: 10.1007/pl00008605. [DOI] [PubMed] [Google Scholar]
- 492.Phizicky E M, Roberts J W. Induction of SOS functions: regulation of proteolytic activity of E. coli RecA protein by interaction with DNA and nucleoside triphosphate. Cell. 1981;25:259–267. doi: 10.1016/0092-8674(81)90251-8. [DOI] [PubMed] [Google Scholar]
- 493.Picksley S M, Attfield P V, Lloyd R G. Repair of DNA double-strand breaks in Escherichia coli K12 requires a functional recN product. Mol Gen Genet. 1984;195:267–274. doi: 10.1007/BF00332758. [DOI] [PubMed] [Google Scholar]
- 494.Picksley S M, Lloyd R G, Buckman C. Genetic analysis and regulation of inducible recombination in Escherichia coli K-12. Cold Spring Harbor Symp Quant Biol. 1984;49:469–474. doi: 10.1101/sqb.1984.049.01.053. [DOI] [PubMed] [Google Scholar]
- 495.Podyminogin M A, Meyer R B, Gamper H B. Sequence-specific covalent modification of DNA by cross-linking oligonucleotides. Catalysis by RecA and implication for the mechanism of synaptic joint formation. Biochemistry. 1995;34:13098–13108. doi: 10.1021/bi00040a022. [DOI] [PubMed] [Google Scholar]
- 496.Pollard E C, Bronner C E, Fluke D J. The influence of repair systems on the presence of sensitive and resistant fractions in the relation of survival of colony-forming ability in Escherichia coli to UV exposure. Mutat Res. 1986;165:63–70. doi: 10.1016/0167-8817(86)90061-1. [DOI] [PubMed] [Google Scholar]
- 497.Poteete A R, Fenton A C. Efficient double-strand break-stimulated recombination promoted by the general recombination systems of phages λ and P22. Genetics. 1993;134:1013–1021. doi: 10.1093/genetics/134.4.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.Potter H, Dressler D. DNA recombination: in vivo and in vitro studies. Cold Spring Harbor Symp Quant Biol. 1979;43:969–985. doi: 10.1101/sqb.1979.043.01.106. [DOI] [PubMed] [Google Scholar]
- 499.Prell A, Wackernagel W. Degradation of linear and circular DNA with gaps by the RecBC enzyme of Escherichia coli. Effects of gap length and the presence of cell-free extracts. Eur J Biochem. 1980;105:109–116. doi: 10.1111/j.1432-1033.1980.tb04480.x. [DOI] [PubMed] [Google Scholar]
- 500.Priel E. Inhibition of postreplication recombinational repair by DNA gyrase antagonists in Escherichia coli. Cell Biol Int Rep. 1984;8:773–786. doi: 10.1016/0309-1651(84)90116-4. [DOI] [PubMed] [Google Scholar]
- 501.Pritchard R H, Lark K G. Induction of replication by thymine starvation at the chromosome origin in Escherichia coli. J Mol Biol. 1964;9:288–307. doi: 10.1016/s0022-2836(64)80208-4. [DOI] [PubMed] [Google Scholar]
- 502.Pugh B F, Cox M M. Stable binding of RecA protein to duplex DNA. Unraveling a paradox. J Biol Chem. 1987;262:1326–1336. [PubMed] [Google Scholar]
- 503.Qui Z, Goodman M F. The Escherichia coli polB locus is identical to dinA, the structural gene for DNA polymerase II. J Biol Chem. 1997;272:8611–8617. doi: 10.1074/jbc.272.13.8611. [DOI] [PubMed] [Google Scholar]
- 504.Quinones A, Piechocki R. Differential suppressor effects of the ssb-1 and ssb-113 alleles on uvrD mutator of Escherichia coli in DNA repair and mutagenesis. J Basic Microbiol. 1987;5:263–273. doi: 10.1002/jobm.3620270508. [DOI] [PubMed] [Google Scholar]
- 505.Radding C M, Rosenzweig J, Richards F, Cassuto E. Separation and characterization of exonuclease, β protein, and a complex of both. J Biol Chem. 1971;246:2510–2512. [Google Scholar]
- 506.Radman M. SOS repair hypothesis: phenomenology of an inducible DNA repair which is accompanied by mutagenesis. In: Hanawalt P C, Setlow R B, editors. Molecular mechanisms for repair of DNA. A. New York, N.Y: Plenum Press; 1975. pp. 355–367. [DOI] [PubMed] [Google Scholar]
- 507.Radman M, Cordone L, Krsmanovic-Simic D, Errera M. Complementary action of recombination and excision repair of ultraviolet irradiation damage to DNA. J Mol Biol. 1970;49:203–212. doi: 10.1016/0022-2836(70)90386-4. [DOI] [PubMed] [Google Scholar]
- 508.Rafferty J B, Sedelnikova S E, Hargreaves D, Artymiuk P J, Baker P J, Sharples G J, Mahdi A A, Lloyd R G, Rice D W. Crystal structure of DNA recombination protein RuvA and a model for its binding to the Holliday junction. Science. 1996;274:415–421. doi: 10.1126/science.274.5286.415. [DOI] [PubMed] [Google Scholar]
- 509.Rajagopalan M, Lu C, Woodgate R, O’Donnell M, Goodman M F, Echols H. Activity of the purified mutagenesis proteins UmuC, UmuD′, and RecA in replicative bypass of an abasic DNA lesion by DNA polymerase III. Proc Natl Acad Sci USA. 1992;89:10777–10781. doi: 10.1073/pnas.89.22.10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Ramareddy G, Reiter H. Sequential loss of loci in thymine-starved Bacillus subtilis 168 cells. Evidence for a circular chromosome. J Mol Biol. 1970;50:525–532. doi: 10.1016/0022-2836(70)90209-3. [DOI] [PubMed] [Google Scholar]
- 511.Ramareddy G, Reiter H. Specific loss of newly replicated deoxyribonucleic acid in nalidixic acid-treated Bacillus subtilis 168. J Bacteriol. 1969;100:724–729. doi: 10.1128/jb.100.2.724-729.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Rayssiguier C, Thaler D S, Radman M. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature. 1989;342:396–401. doi: 10.1038/342396a0. [DOI] [PubMed] [Google Scholar]
- 513.Reece R J, Maxwell A. DNA gyrase: structure and function. Crit Rev Biochem Mol Biol. 1991;26:335–375. doi: 10.3109/10409239109114072. [DOI] [PubMed] [Google Scholar]
- 514.Register J C, III, Griffith J. The direction of RecA protein assembly onto single-strand DNA is the same as the direction of strand assimilation during strand exchange. J Biol Chem. 1985;260:12308–12312. [PubMed] [Google Scholar]
- 515.Rehrauer W M, Lavery P E, Palmer E L, Singh R N, Kowalczykowski S C. Interaction of Escherichia coli RecA protein with LexA repressor. I. LexA repressor cleavage is competitive with binding of a secondary DNA molecule. J Biol Chem. 1996;271:23865–23873. [PubMed] [Google Scholar]
- 516.Reiter H, Ramareddy G. Loss of DNA behind the growing point of thymine-starved Bacillus subtilis 168. J Mol Biol. 1970;50:533–548. doi: 10.1016/0022-2836(70)90210-x. [DOI] [PubMed] [Google Scholar]
- 517.Resnick M A. The repair of double-strand breaks in DNA: a model involving recombination. J Theor Biol. 1976;59:97–106. doi: 10.1016/s0022-5193(76)80025-2. [DOI] [PubMed] [Google Scholar]
- 518.Richter C, Park J-W, Ames B N. Normal oxidative damage to mitochondrial and nuclear DNA is extensive. Proc Natl Acad Sci USA. 1988;85:6465–6467. doi: 10.1073/pnas.85.17.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 519.Rinken R, de Vries J, Weichenhan D, Wackernagel W. The recA-recBCD dependent recombination pathways of Serratia marcescens and Proteus mirabilis in Escherichia coli: functions of hybrid enzymes and hybrid pathways. Biochimie. 1991;73:375–384. doi: 10.1016/0300-9084(91)90104-9. [DOI] [PubMed] [Google Scholar]
- 520.Rinken R, Thoms B, Wackernagel W. Evidence that recBC-dependent degradation of duplex DNA in Escherichia coli recD mutants involves DNA unwinding. J Bacteriol. 1992;174:5424–5429. doi: 10.1128/jb.174.16.5424-5429.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521.Rinken R, Wackernagel W. Inhibition of the recBCD-dependent activation of Chi recombinational hot spots in SOS-induced cells of Escherichia coli. J Bacteriol. 1992;174:1172–1178. doi: 10.1128/jb.174.4.1172-1178.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 522.Roberts J W, Roberts C W, Mount D W. Inactivation and proteolytic cleavage of phage λ repressor in vitro in ATP-dependent reaction. Proc Natl Acad Sci USA. 1977;74:2283–2287. doi: 10.1073/pnas.74.6.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 523.Roca A I, Cox M M. The RecA protein: structure and function. Crit Rev Biochem Mol Biol. 1990;25:415–456. doi: 10.3109/10409239009090617. [DOI] [PubMed] [Google Scholar]
- 524.Roca A I, Cox M M. RecA protein: structure, function, and role in recombinational DNA repair. Prog Nucleic Acid Res Mol Biol. 1997;56:129–223. doi: 10.1016/s0079-6603(08)61005-3. [DOI] [PubMed] [Google Scholar]
- 525.Roe S M, Barlow T, Brown T, Oram M, Keeley A, Tsaneva I R, Pearl L H. Crystal structure of an octameric RuvA-Holliday junction complex. Mol Cell. 1998;2:361–372. doi: 10.1016/s1097-2765(00)80280-4. [DOI] [PubMed] [Google Scholar]
- 526.Roman L, Kowalczykowski S C. Characterization of the helicase activity of the Escherichia coli RecBCD enzyme using a novel helicase assay. Biochemistry. 1989;28:2863–2873. doi: 10.1021/bi00433a018. [DOI] [PubMed] [Google Scholar]
- 527.Roman L J, Eggleston A K, Kowalczykowski S C. Processivity of the DNA helicase activity of Escherichia coli RecBCD enzyme. J Biol Chem. 1992;267:4207–4214. [PubMed] [Google Scholar]
- 528.Rosamond J, Telander K M, Linn S. Modulation of the action of the recBC enzyme of Escherichia coli K-12 by Ca2+ J Biol Chem. 1979;254:8646–8652. [PubMed] [Google Scholar]
- 529.Ross P, Howard-Flanders P. Initiation of recA+-dependent recombination in Escherichia coli (λ). I. Undamaged covalent circular lambda DNA molecules in uvrA+ recA+ lysogenic host cells are cut following superinfection with psoralen-damaged lambda phages. J Mol Biol. 1977;117:137–158. doi: 10.1016/0022-2836(77)90028-6. [DOI] [PubMed] [Google Scholar]
- 530.Ross P, Howard-Flanders P. Initiation of recA+-dependent recombination in Escherichia coli (λ). II. Specificity in the induction of recombination and strand cutting in undamaged covalent circular bacteriophage 186 and lambda DNA molecules in phage-infected cells. J Mol Biol. 1977;117:159–174. doi: 10.1016/0022-2836(77)90029-8. [DOI] [PubMed] [Google Scholar]
- 531.Rosselli W, Stasiak A. The ATPase activity of RecA is needed to push the DNA strand exchange through heterologous regions. EMBO J. 1991;10:4391–4396. doi: 10.1002/j.1460-2075.1991.tb05017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 532.Rostas K, Morton S J, Picksley S M, Lloyd R G. Nucleotide sequence and LexA regulation of the Escherichia coli recN gene. Nucleic Acids Res. 1987;15:5041–5049. doi: 10.1093/nar/15.13.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 533.Rothman R H, Clark A J. Defective excision and postreplication repair of UV-damaged DNA in a recL mutant strain of E. coli K-12. Mol Gen Genet. 1977;155:267–277. doi: 10.1007/BF00272805. [DOI] [PubMed] [Google Scholar]
- 534.Rothman R H, Clark A J. The dependence of postreplication repair on uvrB in a recF mutant of Escherichia coli K-12. Mol Gen Genet. 1977;155:279–286. doi: 10.1007/BF00272806. [DOI] [PubMed] [Google Scholar]
- 535.Rothman R H, Kato T, Clark A J. The beginning of an investigation of the role of recF in the pathways of metabolism of ultraviolet-irradiated DNA in Escherichia coli. In: Hanawalt P C, Setlow R B, editors. Molecular mechanisms for repair of DNA. A. New York, N.Y: Plenum Press; 1975. pp. 283–291. [DOI] [PubMed] [Google Scholar]
- 536.Rould E, Muniyappa K, Radding C M. Unwinding of heterologous DNA by RecA protein during the search for homologous sequences. J Mol Biol. 1992;226:127–139. doi: 10.1016/0022-2836(92)90129-8. [DOI] [PubMed] [Google Scholar]
- 537.Runyon G T, Bear D G, Lohman T M. Escherichia coli helicase II (UvrD) protein initiates DNA unwinding at nicks and blunt ends. Proc Natl Acad Sci USA. 1990;87:6383–6387. doi: 10.1073/pnas.87.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 538.Rupp W D, Howard-Flanders P. Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation. J Mol Biol. 1968;31:291–304. doi: 10.1016/0022-2836(68)90445-2. [DOI] [PubMed] [Google Scholar]
- 539.Rupp W D, Wilde III C E, Reno D L, Howard-Flanders P. Exchanges between DNA strands in ultraviolet-irradiated Escherichia coli. J Mol Biol. 1971;61:25–44. doi: 10.1016/0022-2836(71)90204-x. [DOI] [PubMed] [Google Scholar]
- 540.Saffhill R, Ockey C H. Strand breaks arising from the repair of the 5-bromodeoxyuridine-substituted template and methyl methanesulfonate-induced lesions can explain the formation of sister-chromatid exchanges. Chromosoma. 1985;92:218–224. doi: 10.1007/BF00348697. [DOI] [PubMed] [Google Scholar]
- 541.Saito A, Iwasaki H, Ariyoshi M, Morikawa K, Shinagawa H. Identification of four acidic amino acids that constitute the catalytic center of the RuvC Holliday junction resolvase. Proc Natl Acad Sci USA. 1995;92:7470–7474. doi: 10.1073/pnas.92.16.7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 542.Sakaki Y. Inactivation of the ATP-dependent DNase of Escherichia coli after infection with double-stranded DNA phages. J Virol. 1974;14:1611–1612. doi: 10.1128/jvi.14.6.1611-1612.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 543.Salles B, Paoletti C. Control of UV induction of RecA protein. Proc Natl Acad Sci USA. 1983;80:65–69. doi: 10.1073/pnas.80.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 544.Sancar A. Mechanisms of DNA excision repair. Science. 1994;266:1954–1956. doi: 10.1126/science.7801120. [DOI] [PubMed] [Google Scholar]
- 545.Sancar A. Structure and function of DNA photolyase. Biochemistry. 1994;33:2–9. doi: 10.1021/bi00167a001. [DOI] [PubMed] [Google Scholar]
- 546.Sancar A, Stachelek C, Konigsberg W, Rupp W D. Sequence of the recA gene and protein. Proc Natl Acad Sci USA. 1980;77:2611–2615. doi: 10.1073/pnas.77.5.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 547.Sandler S J. dnaC809 distinguishes between overlapping functions for recF and priA in cell viability and UV-inducible SOS expression in E. coli K-12. Mol Microbiol. 1996;19:871–880. doi: 10.1046/j.1365-2958.1996.429959.x. [DOI] [PubMed] [Google Scholar]
- 548.Sandler S J, Chackerian B, Li J T, Clark A J. Sequence and complementation analysis of recF genes from Escherichia coli, Salmonella typhimurium, Pseudomonas putida and Bacillus subtilis: evidence for an essential phosphate binding loop. Nucleic Acids Res. 1992;20:839–845. doi: 10.1093/nar/20.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 549.Sandler S J, Clark A J. Factors affecting expression of the recF gene of Escherichia coli K-12. Gene. 1990;86:35–43. doi: 10.1016/0378-1119(90)90111-4. [DOI] [PubMed] [Google Scholar]
- 550.Sandler S J, Clark A J. RecOR suppression of recF mutant phenotypes in Escherichia coli K-12. J Bacteriol. 1994;176:3661–3672. doi: 10.1128/jb.176.12.3661-3672.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 551.Sandler S J, Clark A J. Use of high and low level overexpression plasmids to test mutant alleles of the recF gene of Escherichia coli K-12 for partial activity. Genetics. 1993;135:643–654. doi: 10.1093/genetics/135.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 552.Sandler S J, Samra H S, Clark A J. Differential suppression of priA2::kan phenotypes in Escherichia coli K-12 by mutations in priA, lexA, and dnaC. Genetics. 1996;143:5–13. doi: 10.1093/genetics/143.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 553.Sargentini N J, Smith K C. Characterization of an Escherichia coli mutant (radB101) sensitive to γ and UV radiation, and methyl methanesulfonate. Radiat Res. 1983;104:109–115. [PubMed] [Google Scholar]
- 554.Sargentini N J, Smith K C. Genetic and phenotypic analyses indicating occurrence of the recN262 and radB101 mutations at the same locus in Escherichia coli. J Bacteriol. 1988;170:2392–2394. doi: 10.1128/jb.170.5.2392-2394.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 555.Sargentini N J, Smith K C. Quantitation of the involvement of the recA, recB, recC, recF, recJ, recN, lexA, radA, radB, uvrD, and umuC genes in the repair of X-ray-induced DNA double-strand breaks in Escherichia coli. Radiat Res. 1986;107:58–72. [PubMed] [Google Scholar]
- 556.Sargentini N J, Smith K C. Role of the radB gene in postreplication repair in UV-irradiated Escherichia coli uvrB. Mutat Res. 1986;166:17–22. doi: 10.1016/0167-8817(86)90036-2. [DOI] [PubMed] [Google Scholar]
- 557.Sasaki M, Fujiyoshi T, Shimada K, Takagi Y. Fine structure of the recB and recC gene region of Escherichia coli. Biochem Biophys Res Commun. 1982;109:414–422. doi: 10.1016/0006-291x(82)91737-5. [DOI] [PubMed] [Google Scholar]
- 558.Sassanfar M, Roberts J W. Nature of the SOS-inducing signal in Escherichia coli. The involvement of DNA replication. J Mol Biol. 1990;212:79–96. doi: 10.1016/0022-2836(90)90306-7. [DOI] [PubMed] [Google Scholar]
- 559.Sauer R T, Ross M J, Ptashne M. Cleavage of the lambda and P22 repressors by RecA protein. J Biol Chem. 1982;257:4458–4462. [PubMed] [Google Scholar]
- 560.Savageau M A. Escherichia coli habitats, cell types, and molecular mechanisms of gene control. Am Nat. 1983;122:732–744. [Google Scholar]
- 561.Saveson C J, Lovett S T. Tandem repeat recombination induced by replication fork defects in Escherichia coli requires a novel factor, RadC. Genetics. 1999;152:5–13. doi: 10.1093/genetics/152.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 562.Schaefer C, Messer W. DnaA protein/DNA interaction. Modulation of the recognition sequence. Mol Gen Genet. 1991;226:34–40. doi: 10.1007/BF00273584. [DOI] [PubMed] [Google Scholar]
- 563.Schnarr M, Oertel-Buchheit P, Kazmaier M, Granger-Schnarr M. DNA binding properties of the LexA repressor. Biochimie. 1991;73:423–431. doi: 10.1016/0300-9084(91)90109-e. [DOI] [PubMed] [Google Scholar]
- 564.Sedgwick S G, Bridges B A. Requirement for either DNA polymerase I or DNA polymerase III in post-replication repair in excision-proficient Escherichia coli. Nature. 1974;249:348–349. doi: 10.1038/249348a0. [DOI] [PubMed] [Google Scholar]
- 565.Sedgwick S G, Ho C, Woodgate R. Mutagenic DNA repair in enterobacteria. J Bacteriol. 1991;173:5604–5611. doi: 10.1128/jb.173.18.5604-5611.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 566.Seigneur M, Bidnenko V, Ehrlich S D, Michel B. RuvAB acts at arrested replication forks. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- 567.Shafer D A. Alternate replication bypass mechanisms for sister chromatid exchange formation. In: Sandberg A A, editor. Sister chromatid exchange. New York, N.Y: Alan R. Liss, Inc.; 1982. pp. 67–98. [Google Scholar]
- 568.Shah R, Bennett R J, West S C. Genetic recombination in E. coli: RuvC protein cleaves Holliday junctions at resolution hotspots in vitro. Cell. 1994;79:853–864. doi: 10.1016/0092-8674(94)90074-4. [DOI] [PubMed] [Google Scholar]
- 569.Shan Q, Bork J M, Webb B L, Inman R B, Cox M M. RecA protein filaments: end-dependent dissociation from ssDNA and stabilization by RecO and RecR proteins. J Mol Biol. 1997;265:519–540. doi: 10.1006/jmbi.1996.0748. [DOI] [PubMed] [Google Scholar]
- 570.Shan Q, Cox M M, Inman R B. DNA strand exchange promoted by RecA K72R. Two reaction phases with different Mg2+ requirements. J Biol Chem. 1996;271:5712–5724. doi: 10.1074/jbc.271.10.5712. [DOI] [PubMed] [Google Scholar]
- 571.Shaner S L, Flory J, Radding C M. The distribution of Escherichia coli RecA protein bound to duplex DNA with single-stranded ends. J Biol Chem. 1987;262:9220–9230. [PubMed] [Google Scholar]
- 572.Shaner S L, Radding C M. Translocation of Escherichia coli RecA protein from a single-stranded tail to contiguous duplex DNA. J Biol Chem. 1987;262:9211–9219. [PubMed] [Google Scholar]
- 573.Sharma B, Hill T M. Insertion of inverted Ter sites into the terminus region of the Escherichia coli chromosome delays completion of DNA replication and disrupts the cell cycle. Mol Microbiol. 1995;18:45–61. doi: 10.1111/j.1365-2958.1995.mmi_18010045.x. [DOI] [PubMed] [Google Scholar]
- 574.Sharma R C, Smith K C. Role of DNA polymerase I in postreplication repair: a reexamination with Escherichia coli ΔpolA. J Bacteriol. 1987;169:4559–4564. doi: 10.1128/jb.169.10.4559-4564.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 575.Sharples G J, Benson F E, Illing G T, Lloyd R G. Molecular and functional analysis of the ruv region of Escherichia coli K-12 reveals three genes involved in DNA repair and recombination. Mol Gen Genet. 1990;221:219–226. doi: 10.1007/BF00261724. [DOI] [PubMed] [Google Scholar]
- 576.Sharples G J, Chan S N, Mahdi A A, Whitby M C, Lloyd R G. Processing of intermediates in recombination and DNA repair: identification of a new endonuclease that specifically cleaves Holliday junctions. EMBO J. 1994;13:6133–6142. doi: 10.1002/j.1460-2075.1994.tb06960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 577.Shavitt O, Livneh Z. The β subunit modulates bypass and termination at UV-lesions during in vitro replication with DNA polymerase III holoenzyme of Escherichia coli. J Biol Chem. 1989;264:11275–11281. [PubMed] [Google Scholar]
- 578.Shea M E, Hiasa H. Interactions between DNA helicases and frozen topoisomerase IV-quinolone-DNA ternary complexes. J Biol Chem. 1999;274:22747–22754. doi: 10.1074/jbc.274.32.22747. [DOI] [PubMed] [Google Scholar]
- 579.Shen P, Huang H V. Effect of base pair mismatches on recombination via the RecBCD pathway. Mol Gen Genet. 1989;218:358–360. doi: 10.1007/BF00331291. [DOI] [PubMed] [Google Scholar]
- 580.Shevell D E, Friedman B M, Walker G C. Resistance to alkylation damage in Escherichia coli: role of the Ada protein in induction of the adaptive response. Mutat Res. 1990;233:53–72. doi: 10.1016/0027-5107(90)90151-s. [DOI] [PubMed] [Google Scholar]
- 581.Shiba T, Iwasaki H, Nakata A, Shinagawa H. Escherichia coli RuvA and RuvB proteins involved in recombinational repair: physical properties and interaction with DNA. Mol Gen Genet. 1993;237:395–399. doi: 10.1007/BF00279443. [DOI] [PubMed] [Google Scholar]
- 582.Shibata T, DasGupta C, Cunningham R P, Williams J G K, Osber L, Radding C M. Homologous pairing in genetic recombination. The pairing reaction catalyzed by Escherichia coli RecA protein. J Biol Chem. 1981;256:7565–7572. [PubMed] [Google Scholar]
- 583.Shibata T, Ohtani T, Chang P K, Ando T. Role of superhelicity in homologous pairing of DNA molecules promoted by Escherichia coli RecA protein. J Biol Chem. 1982;257:370–376. [PubMed] [Google Scholar]
- 584.Shida T, Iwasaki H, Saito A, Kyogoku Y, Shinagawa H. Analysis of substrate specificity of the RuvC Holliday junction resolvase with synthetic Holliday junctions. J Biol Chem. 1996;271:26105–26109. doi: 10.1074/jbc.271.42.26105. [DOI] [PubMed] [Google Scholar]
- 585.Shinagawa H, Iwasaki H. Molecular mechanisms of Holliday junction processing in Escherichia coli. Adv Biophys. 1995;31:49–65. doi: 10.1016/0065-227x(95)99382-y. [DOI] [PubMed] [Google Scholar]
- 586.Shinagawa H, Iwasaki H. Processing the Holliday junction in homologous recombination. Trends Biochem Sci. 1996;21:107–111. [PubMed] [Google Scholar]
- 587.Shinagawa H, Makino K, Amemura M, Kimura S, Iwasaki H, Nakata A. Structure and regulation of the Escherichia coli ruv operon involved in DNA repair and recombination. J Bacteriol. 1988;170:4322–4329. doi: 10.1128/jb.170.9.4322-4329.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 588.Shlomai J, Kornberg A. A prepriming DNA replication enzyme of Escherichia coli. I. Purification of protein n′: a sequence-specific, DNA-dependent ATPase. J Biol Chem. 1980;255:6789–6793. [PubMed] [Google Scholar]
- 589.Shulman M J, Hallick L M, Echols H, Signer E R. Properties of recombination-deficient mutants of bacteriophage lambda. J Mol Biol. 1970;52:501–520. doi: 10.1016/0022-2836(70)90416-x. [DOI] [PubMed] [Google Scholar]
- 590.Shurvinton C E, Lloyd R G. Damage to DNA induces expression of the ruv gene of Escherichia coli. Mol Gen Genet. 1982;185:352–355. doi: 10.1007/BF00330811. [DOI] [PubMed] [Google Scholar]
- 591.Siddiqi O, Fox M S. Integration of donor DNA in bacterial conjugation. J Mol Biol. 1973;77:101–123. doi: 10.1016/0022-2836(73)90365-3. [DOI] [PubMed] [Google Scholar]
- 592.Signer E R, Weil J. Recombination in bacteriophage λ. I. Mutants deficient in general recombination. J Mol Biol. 1968;34:261–271. doi: 10.1016/0022-2836(68)90251-9. [DOI] [PubMed] [Google Scholar]
- 593.Silberstein Z, Shalit M, Cohen A. Heteroduplex strand-specificity in restriction-stimulated recombination by the RecE pathway of Escherichia coli. Genetics. 1993;133:439–448. doi: 10.1093/genetics/133.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 594.Silberstein Z, Tzfati Y, Cohen A. Primary products of break-induced recombination by Escherichia coli RecE pathway. J Bacteriol. 1995;177:1692–1698. doi: 10.1128/jb.177.7.1692-1698.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 595.Sinden R R, Cole R S. Repair of cross-linked DNA and survival of Escherichia coli treated with psoralen and light: effects of mutations influencing genetic recombination and DNA metabolism. J Bacteriol. 1978;136:538–547. doi: 10.1128/jb.136.2.538-547.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 596.Sinzinis B I, Smirnov G B, Saenko A A. Repair deficiency in Escherichia coli UV-sensitive mutator strain uvr502. Biochem Biophys Res Commun. 1973;53:309–316. doi: 10.1016/0006-291x(73)91435-6. [DOI] [PubMed] [Google Scholar]
- 597.Skalka A. A replicator’s view of recombination (and repair) In: Grell R F, editor. Mechanisms in recombination. New York, N.Y: Plenum Press; 1974. pp. 421–432. [Google Scholar]
- 598.Skarstad K, Boye E. Degradation of individual chromosomes in recA mutants of Escherichia coli. J Bacteriol. 1993;175:5505–5509. doi: 10.1128/jb.175.17.5505-5509.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 599.Skarstad K, Boye E. Perturbed chromosomal replication in recA mutants of Escherichia coli. J Bacteriol. 1988;170:2549–2554. doi: 10.1128/jb.170.6.2549-2554.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 600.Small G D. Mechanism of gap-filling during postreplication repair of ultraviolet damage in Haemophilus influenzae. J Bacteriol. 1975;124:176–181. doi: 10.1128/jb.124.1.176-181.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 601.Smirnov G B, Filkova E V, Skavronskaya A G. Ultraviolet sensitivity, spontaneous mutability and DNA degradation in Escherichia coli strains carrying mutations in uvr and rec genes. J Gen Microbiol. 1973;76:407–416. doi: 10.1099/00221287-76-2-407. [DOI] [PubMed] [Google Scholar]
- 602.Smith B T, Walker G C. Mutagenesis and more: umuDC and the Escherichia coli SOS response. Genetics. 1998;148:1599–1610. doi: 10.1093/genetics/148.4.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 603.Smith G R. Conjugational recombination in E. coli: myths and mechanisms. Cell. 1991;64:19–27. doi: 10.1016/0092-8674(91)90205-d. [DOI] [PubMed] [Google Scholar]
- 604.Smith G R, Kunes S M, Schultz D W, Taylor A, Triman K L. Structure of Chi hotspots of generalized recombination. Cell. 1981;24:429–436. doi: 10.1016/0092-8674(81)90333-0. [DOI] [PubMed] [Google Scholar]
- 605.Smith K C. DNA synthesis in sensitive and resistant mutants of Escherichia coli B after ultraviolet irradiation. Mutat Res. 1969;8:481–495. doi: 10.1016/0027-5107(69)90065-7. [DOI] [PubMed] [Google Scholar]
- 606.Smith K C, Meun D H C. Repair of radiation-induced damage in Escherichia coli. I. Effect of rec mutations on post-replication repair of damage due to ultraviolet radiation. J Mol Biol. 1970;51:459–472. doi: 10.1016/0022-2836(70)90001-x. [DOI] [PubMed] [Google Scholar]
- 607.Smith K C, Sharma R C. A model for the recA-dependent repair of excision gaps in UV-irradiated Escherichia coli. Mutat Res. 1987;183:1–9. doi: 10.1016/0167-8817(87)90039-3. [DOI] [PubMed] [Google Scholar]
- 608.Smith S B, Cui Y, Bustamante C. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996;271:795–798. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 609.Snyder M, Drlica K. DNA gyrase on the bacterial chromosome: DNA cleavage induced by oxolinic acid. J Mol Biol. 1979;131:287–302. doi: 10.1016/0022-2836(79)90077-9. [DOI] [PubMed] [Google Scholar]
- 610.Sommer S, Bailone A, Devoret R. The appearance of the UmuD′C protein complex in Escherichia coli switches repair from homologous recombination to SOS mutagenesis. Mol Microbiol. 1993;10:963–971. doi: 10.1111/j.1365-2958.1993.tb00968.x. [DOI] [PubMed] [Google Scholar]
- 611.Sommer S, Boudsocq F, Devoret R, Bailone A. Specific RecA amino acid changes affect RecA-UmuD′C interaction. Mol Microbiol. 1998;28:281–291. doi: 10.1046/j.1365-2958.1998.00803.x. [DOI] [PubMed] [Google Scholar]
- 612.Sommer S, Knezevic J, Bailone A, Devoret R. Induction of only one SOS operon, umuDC, is required for SOS mutagenesis in Escherichia coli. Mol Gen Genet. 1993;239:137–144. doi: 10.1007/BF00281612. [DOI] [PubMed] [Google Scholar]
- 613.Sourice S, Biaudet V, El Karoui M, Ehrlich S D, Gruss A. Identification of the Chi site of Haemophilus influenzae as several sequences related to the Escherichia coli Chi site. Mol Microbiol. 1998;27:1021–1029. doi: 10.1046/j.1365-2958.1998.00749.x. [DOI] [PubMed] [Google Scholar]
- 614.Sriprakash K S, Lundh N, Huh M O, Radding C M. The specificity of lambda exonuclease: interactions with single-stranded DNA. J Biol Chem. 1975;250:5438–5445. [PubMed] [Google Scholar]
- 615.Stahl F W, Chung S, Crasemann J, Faulds D, Haemer J, Lam S, Malone R E, McMillin K D, Nozu Y, Siegel J, Strathern J, Stahl M. Recombination, replication, and maturation in phage lambda. In: Fox C F, Robinson W S, editors. Virus research. New York, N.Y: Academic Press, Inc.; 1973. pp. 487–503. [Google Scholar]
- 616.Stahl F W, Kobayashi I, Stahl M M. In phage λ, cos is a recombinator in the Red pathway. J Mol Biol. 1985;181:199–209. doi: 10.1016/0022-2836(85)90085-3. [DOI] [PubMed] [Google Scholar]
- 617.Stahl F W, McMilin K D, Stahl M M, Crasemann J M, Lam S. The distribution of crossovers along unreplicated lambda bacteriophage chromosomes. Genetics. 1974;77:395–408. doi: 10.1093/genetics/77.3.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 618.Stahl F W, McMilin K D, Stahl M M, Malone R E, Nozu Y, Russo V E A. A role for recombination in the production of “free-loader” lambda bacteriophage particles. J Mol Biol. 1972;68:57–67. doi: 10.1016/0022-2836(72)90262-8. [DOI] [PubMed] [Google Scholar]
- 619.Stahl F W, McMilin K D, Stahl M M, Nozu Y. An enhancing role for DNA synthesis in formation of bacteriophage lambda recombinants. Proc Natl Acad Sci USA. 1972;69:3598–3601. doi: 10.1073/pnas.69.12.3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 620.Stahl F W, Stahl M M. Non-reciprocal crossing over in phage λ. J Genet. 1985;64:31–39. [Google Scholar]
- 621.Stahl F W, Stahl M M, Malone R E. Red-mediated recombination of phage lambda in a recA− recB− host. Mol Gen Genet. 1978;159:207–211. [Google Scholar]
- 622.Stahl F W, Thomason L C, Siddiqi I, Stahl M M. Further test of a recombination model in which χ removes the RecD subunit from the RecBCD enzyme of Escherichia coli. Genetics. 1990;126:519–533. doi: 10.1093/genetics/126.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 623.Stahl M M, Thomason L, Poteete A R, Tarkowski T, Kuzminov A, Stahl F W. Annealing vs. invasion in phage λ recombination. Genetics. 1997;147:961–977. doi: 10.1093/genetics/147.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 624.Stallions D R, Curtiss R., III Chromosome transfer and recombinant formation with deoxyribonucleic acid temperature-sensitive strains of Escherichia coli. J Bacteriol. 1971;105:886–895. doi: 10.1128/jb.105.3.886-895.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 625.Stasiak A, Di Capua E D. The helicity of DNA in complexes with RecA protein. Nature. 1982;299:185–186. doi: 10.1038/299185a0. [DOI] [PubMed] [Google Scholar]
- 626.Stasiak A, DiCapua E, Koller T. The elongation of duplex DNA by RecA protein. J Mol Biol. 1981;151:557–564. doi: 10.1016/0022-2836(81)90010-3. [DOI] [PubMed] [Google Scholar]
- 627.Stasiak A, Egelman E H. Visualization of recombination reactions. In: Kucherlapati R, Smith G R, editors. Genetic recombination. Washington, D.C.: American Society for Microbiology; 1988. pp. 265–307. [Google Scholar]
- 628.Stasiak A, Tsaneva I R, West S C, Benson C J B, Yu X, Egelman E H. The Escherichia coli RuvB branch migration protein forms double hexameric rings around DNA. Proc Natl Acad Sci USA. 1994;91:7618–7622. doi: 10.1073/pnas.91.16.7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 629.Steiner W W, Kuempel P L. Cell division is required for resolution of dimer chromosomes at the dif locus of Escherichia coli. Mol Microbiol. 1998;27:257–268. doi: 10.1046/j.1365-2958.1998.00651.x. [DOI] [PubMed] [Google Scholar]
- 630.Steiner W W, Kuempel P L. Sister chromatid exchange frequencies in Escherichia coli analyzed by recombination at the dif resolvase site. J Bacteriol. 1998;180:6269–6275. doi: 10.1128/jb.180.23.6269-6275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 631.Sternglanz R, DiNardo S, Voelkel K A, Nishimura Y, Hirota Y, Becherer K, Zumstein L, Wang J C. Mutations in the gene coding for Escherichia coli DNA topoisomerase I affect transcription and transposition. Proc Natl Acad Sci USA. 1981;78:2747–2751. doi: 10.1073/pnas.78.5.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 632.Story R M, Weber I T, Steitz T A. The structure of the E. coli RecA protein monomer and polymer. Nature. 1992;355:318–325. doi: 10.1038/355318a0. [DOI] [PubMed] [Google Scholar]
- 633.Strike P, Lodwick D. Plasmid genes affecting DNA repair and mutation. J Cell Sci Suppl. 1987;6:303–321. doi: 10.1242/jcs.1984.supplement_6.20. [DOI] [PubMed] [Google Scholar]
- 634.Suzuki K, Miyaki M, Ono T, Mori H, Moriya H, Kato T. UV-induced imbalance of the deoxyribonucleoside triphosphate pool in E. coli. Mutat Res. 1983;122:293–298. doi: 10.1016/0165-7992(83)90009-x. [DOI] [PubMed] [Google Scholar]
- 635.Sweasy J B, Loeb L A. Mammalian DNA polymerase β can substitute for DNA polymerase I during DNA replication in Escherichia coli. J Biol Chem. 1992;267:1407–1410. [PubMed] [Google Scholar]
- 636.Symington L S, Morrison P, Kolodner R. Intramolecular recombination of linear DNA catalyzed by the Escherichia coli RecE recombination system. J Mol Biol. 1985;186:515–525. doi: 10.1016/0022-2836(85)90126-3. [DOI] [PubMed] [Google Scholar]
- 637.Szostak J W, Orr-Weaver T L, Rothstein R J, Stahl F W. The double-strand break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 638.Szybalski W. Structural modifications of DNA: crosslinking, circularization and single-strand interruptions. Abh. Dtsch. Akad. Wiss. Berlin. Kl Med. 1964;4:1–19. [Google Scholar]
- 639.Szyf M, Meisels E, Razin A. Biological role of DNA methylation: sequence-specific single-strand breaks associated with hypomethylation of GATC sites in Escherichia coli DNA. J Bacteriol. 1986;168:1487–1490. doi: 10.1128/jb.168.3.1487-1490.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 640.Tadmor Y, Ascarelli-Goell R, Skaliter R, Livneh Z. Overproduction of the β subunit of DNA polymerase III holoenzyme reduces UV mutagenesis in Escherichia coli. J Bacteriol. 1992;174:2517–2524. doi: 10.1128/jb.174.8.2517-2524.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 641.Tadmor Y, Bergstein M, Skaliter R, Shwartz H, Livneh Z. β subunit of DNA polymerase III holoenzyme is induced upon ultraviolet irradiation or nalidixic acid treatment of Escherichia coli. Mutat Res. 1994;308:53–64. doi: 10.1016/0027-5107(94)90198-8. [DOI] [PubMed] [Google Scholar]
- 642.Takahashi M, Schnarr M. Investigation of RecA-polynucleotide interactions from the measurement of LexA repressor cleavage kinetics. Eur J Biochem. 1989;183:617–622. doi: 10.1111/j.1432-1033.1989.tb21091.x. [DOI] [PubMed] [Google Scholar]
- 643.Takahashi N, Kobayashi I. Evidence for the double-strand break repair model of bacteriophage λ recombination. Proc Natl Acad Sci USA. 1990;87:2790–2794. doi: 10.1073/pnas.87.7.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 644.Takahashi N K, Kusano K, Yokochi T, Kitamura Y, Yoshikura H, Kobayashi I. Genetic analysis of double-strand break repair in Escherichia coli. J Bacteriol. 1993;175:5176–5185. doi: 10.1128/jb.175.16.5176-5185.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 645.Takano T. Behavior of some episomal elements in a recombination-deficient mutant of Escherichia coli. Jpn J Microbiol. 1966;10:201–210. [Google Scholar]
- 646.Takiff H E, Chen S-M, Court D L. Genetic analysis of the rnc operon of Escherichia coli. J Bacteriol. 1989;171:2581–2590. doi: 10.1128/jb.171.5.2581-2590.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 647.Tang M, Bruck I, Eritja R, Turner J, Frank E G, Woodgate R, O’Donnell M, Goodman M F. Biochemical basis of SOS-induced mutagenesis in Escherichia coli: reconstitution of in vitro lesion bypass dependent on the UmuD′2C mutagenic complex and RecA protein. Proc Natl Acad Sci USA. 1998;95:9755–9760. doi: 10.1073/pnas.95.17.9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 648.Tang M, Shen X, Frank E G, O’Donnell M, Woodgate R, Goodman M F. UmuD′2C is an error-prone DNA polymerase, Escherichia coli pol V. Proc Natl Acad Sci USA. 1999;96:8919–8924. doi: 10.1073/pnas.96.16.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 649.Tang M-S, Smith K C. The effects of lexA101, recB21, recF143 and uvrD3 mutations on liquid-holding recovery in ultraviolet-irradiated Escherichia coli K12 recA56. Mutat Res. 1981;80:15–25. doi: 10.1016/0027-5107(81)90174-3. [DOI] [PubMed] [Google Scholar]
- 650.Taylor A F. RecBCD enzyme of Escherichia coli. In: Kucherlapati R, Smith G R, editors. Genetic recombination. Washington, D.C.: American Society for Microbiology; 1988. pp. 231–263. [Google Scholar]
- 651.Taylor A F, Schultz D W, Ponticelli A S, Smith G R. RecBC enzyme nicking at Chi sites during DNA unwinding: location and orientation-dependence of the cutting. Cell. 1985;41:153–163. doi: 10.1016/0092-8674(85)90070-4. [DOI] [PubMed] [Google Scholar]
- 652.Taylor A F, Smith G R. Monomeric RecBCD enzyme binds and unwinds DNA. J Biol Chem. 1995;270:24451–24458. doi: 10.1074/jbc.270.41.24451. [DOI] [PubMed] [Google Scholar]
- 653.Taylor A F, Smith G R. RecBCD enzyme is altered upon cutting DNA at a Chi recombinational hotspot. Proc Natl Acad Sci USA. 1992;89:5226–5230. doi: 10.1073/pnas.89.12.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 654.Taylor A F, Smith G R. Regulation of homologous recombination: Chi inactivates RecBCD enzyme by disassembly of the three subunits. Genes Dev. 1999;13:890–900. doi: 10.1101/gad.13.7.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 655.Taylor A F, Smith G R. Strand specificity of nicking of DNA at Chi sites by RecBCD enzyme. Modulation by ATP and magnesium levels. J Biol Chem. 1995;270:24459–24467. doi: 10.1074/jbc.270.41.24459. [DOI] [PubMed] [Google Scholar]
- 656.Taylor A F, Smith G R. Substrate specificity of the DNA unwinding activity of the RecBC enzyme of Escherichia coli. J Mol Biol. 1985;185:431–443. doi: 10.1016/0022-2836(85)90414-0. [DOI] [PubMed] [Google Scholar]
- 657.Taylor A F, Smith G R. Unwinding and rewinding of DNA by the RecBC enzyme. Cell. 1980;22:447–457. doi: 10.1016/0092-8674(80)90355-4. [DOI] [PubMed] [Google Scholar]
- 658.Telander-Muskavitch K M, Linn S. RecBC-like enzymes: exonuclease V deoxyribonucleases. In: Boyer P D, editor. The enzymes. Vol. 9. New York, N.Y: Academic Press, Inc.; 1981. pp. 234–250. [Google Scholar]
- 659.Telander-Muskavitch K M, Linn S. A unified mechanism for the nuclease and unwinding activities of the recBC enzyme of Escherichia coli. J Biol Chem. 1982;257:2641–2648. [PubMed] [Google Scholar]
- 660.Tessman I, Kennedy M A. DNA polymerase II of Escherichia coli in the bypass of abasic sites in vivo. Genetics. 1994;136:439–448. doi: 10.1093/genetics/136.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 661.Thaler D S, Sampson E, Siddiqi I, Rosenberg S M, Stahl F W, Stahl M M. A hypothesis: Chi-activation of RecBCD enzyme involves removal of the RecD subunit. In: Friedberg E, Hanawalt P, editors. Mechanisms and consequences of DNA damage processing. New York, N.Y: Alan R. Liss, Inc.; 1988. pp. 413–422. [Google Scholar]
- 662.Thaler D S, Sampson E, Siddiqi I, Rosenberg S M, Thomason L C, Stahl F W, Stahl M M. Recombination of bacteriophage λ in recD mutants of Escherichia coli. Genome. 1989;31:53–67. doi: 10.1139/g89-013. [DOI] [PubMed] [Google Scholar]
- 663.Thaler D S, Stahl M M, Stahl F W. Double-chain-cut sites are recombination hotspots in the Red pathway of phage λ. J Mol Biol. 1987;195:75–87. doi: 10.1016/0022-2836(87)90328-7. [DOI] [PubMed] [Google Scholar]
- 664.Thaler D S, Stahl M M, Stahl F W. Tests of the double-strand-break repair model for red-mediated recombination of phage lambda and plasmid lambda dv. Genetics. 1987;116:501–511. doi: 10.1093/genetics/116.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 665.Thomas C A. The recombination of DNA molecules. In: Quarton G C, Melnechuk T, Schmitt F O, editors. The neurosciences. A study program. New York, N.Y: The Rockefeller University Press; 1967. pp. 162–182. [Google Scholar]
- 666.Thomason L C, Thaler D S, Stahl M M, Stahl F W. In vivo packaging of bacteriophage lambda monomeric chromosomes. J Mol Biol. 1997;267:75–87. doi: 10.1006/jmbi.1996.0870. [DOI] [PubMed] [Google Scholar]
- 667.Thoms B, Wackernagel W. Interaction of RecBCD enzyme with DNA at double-strand breaks produced in UV-irradiated Escherichia coli: requirements for DNA end processing. J Bacteriol. 1998;180:5639–5645. doi: 10.1128/jb.180.21.5639-5645.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 668.Thoms B, Wackernagel W. Regulatory role of recF in the SOS response of Escherichia coli: impaired induction of SOS genes by UV irradiation and nalidixic acid in a recF mutant. J Bacteriol. 1987;169:1731–1736. doi: 10.1128/jb.169.4.1731-1736.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 669.Thoms B, Wackernagel W. Suppression of the UV-sensitive phenotype of Escherichia coli recF mutants by recA(Srf) and recA(Tif) mutations requires recJ+ J Bacteriol. 1988;170:3675–3681. doi: 10.1128/jb.170.8.3675-3681.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 670.Thresher R J, Christiansen G, Griffith J D. Assembly of presynaptic filaments. Factors affecting the assembly of RecA protein onto single-stranded DNA. J Mol Biol. 1988;201:101–113. doi: 10.1016/0022-2836(88)90442-1. [DOI] [PubMed] [Google Scholar]
- 671.Tomasz M, Lipman R, Chowdary D, Pawlak J, Verdine G L, Nakanishi K. Isolation and structure of a covalent cross-link adduct between mitomycin C and DNA. Science. 1987;235:1204–1208. doi: 10.1126/science.3103215. [DOI] [PubMed] [Google Scholar]
- 672.Touati D, Jacques M, Tardat B, Bouchard L, Despied S. Lethal oxidative damage and mutagenesis are generated by iron in Δfur mutants of Escherichia coli: protective role of superoxide dismutase. J Bacteriol. 1995;177:2305–2314. doi: 10.1128/jb.177.9.2305-2314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 673.Trgovcevic Z, Petranovic D, Petranovic M, Salaj-Smic E. Degradation of Escherichia coli DNA synthesized after ultraviolet irradiation in the absence of repair. Int J Radiat Biol. 1984;45:193–196. doi: 10.1080/09553008414550211. [DOI] [PubMed] [Google Scholar]
- 674.Trgovcevic Z, Petranovic D, Salaj-Smic E, Petranovic M, Trinajstic N, Jericevic Z. DNA replication past pyrimidine dimers in the absence of repair. Mutat Res. 1983;112:17–22. doi: 10.1016/0167-8817(83)90020-2. [DOI] [PubMed] [Google Scholar]
- 675.Trucksis M, Depew R E. Identification and localization of a gene that specifies production of Escherichia coli DNA topoisomerase I. Proc Natl Acad Sci USA. 1981;78:2164–2168. doi: 10.1073/pnas.78.4.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 676.Tsaneva I R, Illing G, Lloyd R G, West S C. Purification and properties of the RuvA and RuvB proteins of Escherichia coli. Mol Gen Genet. 1992;235:1–10. doi: 10.1007/BF00286175. [DOI] [PubMed] [Google Scholar]
- 677.Tsaneva I R, Muller B, West S C. ATP-dependent branch migration of Holliday junctions promoted by the RuvA and RuvB proteins of E. coli. Cell. 1992;69:1171–1180. doi: 10.1016/0092-8674(92)90638-s. [DOI] [PubMed] [Google Scholar]
- 678.Tsaneva I R, Muller B, West S C. The RuvA and RuvB proteins of Escherichia coli exhibit DNA helicase activity in vitro. Proc Natl Acad Sci USA. 1993;90:1315–1319. doi: 10.1073/pnas.90.4.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 679.Tsaneva I R, West S C. Targeted versus non-targeted DNA helicase activity of the RuvA and RuvB proteins of Escherichia coli. J Biol Chem. 1994;269:26552–26558. [PubMed] [Google Scholar]
- 680.Tseng Y-C, Hung J-L, Wang T-C V. Involvement of the RecF pathway recombination genes in postreplication repair in UV-irradiated Escherichia coli cells. Mutat Res. 1994;315:1–9. doi: 10.1016/0921-8777(94)90021-3. [DOI] [PubMed] [Google Scholar]
- 681.Tye B-K, Chien J, Lehman I R, Duncan B K, Warner H R. Uracil incorporation: a source of pulse-labeled DNA fragments in the replication of the Escherichia coli chromosome. Proc Natl Acad Sci USA. 1978;75:233–237. doi: 10.1073/pnas.75.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 682.Tzareva N V, Makhno V I, Boni I V. Ribosome-messenger recognition in the absence of the Shine-Dalgarno interactions. FEBS Lett. 1994;337:189–194. doi: 10.1016/0014-5793(94)80271-8. [DOI] [PubMed] [Google Scholar]
- 683.Ulmer K M, Gomez R F, Sinskey A J. Ionizing radiation damage to the folded chromosome of Escherichia coli K-12: repair of double-strand breaks in deoxyribonucleic acid. J Bacteriol. 1979;138:486–491. doi: 10.1128/jb.138.2.486-491.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 684.Umezu K, Chi N-W, Kolodner R D. Biochemical interaction of the Escherichia coli RecF, RecO, and RecR proteins with RecA protein and single-stranded DNA binding protein. Proc Natl Acad Sci USA. 1993;90:3875–3879. doi: 10.1073/pnas.90.9.3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 685.Umezu K, Kolodner R D. Protein interactions in genetic recombination in Escherichia coli. Interactions involving RecO and RecR overcome the inhibition of RecA by single-stranded DNA-binding protein. J Biol Chem. 1994;269:30005–30013. [PubMed] [Google Scholar]
- 686.Unger R C, Clark A J. Interaction of the recombination pathways of bacteriophage λ and its host Escherichia coli K12: effects on exonuclease V activity. J Mol Biol. 1972;70:539–548. doi: 10.1016/0022-2836(72)90558-x. [DOI] [PubMed] [Google Scholar]
- 687.Uzest M, Ehrlich S D, Michel B. Lethality of rep recB and rep recC double mutants of Escherichia coli. Mol Microbiol. 1995;17:1177–1188. doi: 10.1111/j.1365-2958.1995.mmi_17061177.x. [DOI] [PubMed] [Google Scholar]
- 688.Valkenburg J A C, Woldringh C L, Brakenhoff G I, van der Voort H T M, Nanninga N. Confocal scanning light microscopy of the Escherichia coli nucleoid: comparison with phase-contrast and electron microscope images. J Bacteriol. 1985;161:478–483. doi: 10.1128/jb.161.2.478-483.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 689.van de Putte P, Zwenk H, Rörsch A. Properties of four mutants of Escherichia coli defective in genetic recombination. Mutat Res. 1966;3:381–392. doi: 10.1016/0027-5107(66)90048-0. [DOI] [PubMed] [Google Scholar]
- 690.Van Dorp B, Benne R, Palitti F. The ATP-dependent DNAase from Escherichia coli rorA: a nuclease with changed enzymatic properties. Biochim Biophys Acta. 1975;395:446–454. doi: 10.1016/0005-2787(75)90068-4. [DOI] [PubMed] [Google Scholar]
- 691.van Gool A J, Hajibagheri N M A, Stasiak A, West S C. Assembly of the Escherichia coli RuvABC resolvasome directs the orientation of Holliday junction resolution. Genes Dev. 1999;13:1861–1870. doi: 10.1101/gad.13.14.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 692.van Gool A J, Shah R, Mézard C, West S C. Functional interactions between the Holliday junction resolvase and the branch migration motor of Escherichia coli. EMBO J. 1998;17:1838–1845. doi: 10.1093/emboj/17.6.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 693.van Houten B. Nucleotide excision repair in Escherichia coli. Microbiol Rev. 1990;54:18–51. doi: 10.1128/mr.54.1.18-51.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 694.Van Sluis C A, Mattern I E, Paterson M C. Properties of uvrE mutants of Escherichia coli K12. I. Effects of UV irradiation on DNA metabolism. Mutat Res. 1974;25:273–279. doi: 10.1016/0027-5107(74)90055-4. [DOI] [PubMed] [Google Scholar]
- 695.Venkatesh T V, Radding C M. Ribosomal protein S1 and NusA protein complexed to recombination protein β of phage λ. J Bacteriol. 1993;175:1844–1846. doi: 10.1128/jb.175.6.1844-1846.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 696.Veomett G E, Kuempel P L. Strand-specific DNA degradation in a mutant of Escherichia coli. Mol Gen Genet. 1973;123:17–28. doi: 10.1007/BF00282985. [DOI] [PubMed] [Google Scholar]
- 697.Villarroya M, Pérez-Roger I, Macian F, Armengod M E. Stationary phase induction of dnaN and recF, two genes of Escherichia coli involved in DNA replication and repair. EMBO J. 1998;17:1829–1837. doi: 10.1093/emboj/17.6.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 698.Villion M, Szatmari G. Cloning and characterization of the Proteus mirabilis xerD gene. FEMS Microbiol Lett. 1998;164:83–90. doi: 10.1111/j.1574-6968.1998.tb13071.x. [DOI] [PubMed] [Google Scholar]
- 699.Vincent S D, Mahdi A A, Lloyd R G. The RecG branch migration protein of Escherichia coli dissociates R-loops. J Mol Biol. 1996;264:713–721. doi: 10.1006/jmbi.1996.0671. [DOI] [PubMed] [Google Scholar]
- 700.Vinogradskaia G R, Goryshin I I, Lanzov V A. The role of ssb gene in the inducible and the constitutive pathways of recombination in Escherichia coli K-12. Dokl Acad Nauk. 1986;290:228–231. . (In Russian.) [PubMed] [Google Scholar]
- 701.Volkert M R, Hartke M A. Suppression of Escherichia coli recF mutations by recA-linked srfA mutations. J Bacteriol. 1984;157:498–506. doi: 10.1128/jb.157.2.498-506.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 702.Volkert M R, Margossian L J, Clark A J. Two-component suppression of recF143 by recA441 in Escherichia coli K-12. J Bacteriol. 1984;160:702–705. doi: 10.1128/jb.160.2.702-705.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 703.von Kitzing E, Lilley D M J, Diekmann S. The stereochemistry of a four-way DNA junction: a theoretical study. Nucleic Acids Res. 1990;18:2671–2683. doi: 10.1093/nar/18.9.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 704.von Wright A, Bridges B A. Effect of gyrB-mediated changes in chromosome structure on killing of Escherichia coli by ultraviolet light: experiments with strains differing in deoxyribonucleic acid repair capacity. J Bacteriol. 1981;146:18–23. doi: 10.1128/jb.146.1.18-23.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 705.Wackernagel W, Radding C M. Transformation and transduction of Escherichia coli: the nature of recombinants formed by Rec, RecF, and λ Red. In: Grell R F, editor. Mechanisms in recombination. New York, N.Y: Plenum Press; 1974. pp. 111–122. [Google Scholar]
- 706.Waldstein E, Setlow J K, Santasier L. A special type of UV-stimulated recombination in Haemophilus influenzae. Cold Spring Harbor Symp Quant Biol. 1979;43:1059–1062. doi: 10.1101/sqb.1979.043.01.115. [DOI] [PubMed] [Google Scholar]
- 707.Wang T-C, Chang H-Y. Effect of rec mutations on viability and processing of DNA damaged by methylmethane sulfonate in xth nth nfo cells of Escherichia coli. Biochem Biophys Res Commun. 1991;180:774–781. doi: 10.1016/s0006-291x(05)81132-5. [DOI] [PubMed] [Google Scholar]
- 708.Wang T-C V, Chang H-Y, Hung J-L. Cosuppression of recF, recR and recO mutations by mutant recA alleles in Escherichia coli cells. Mutat Res. 1993;294:157–166. doi: 10.1016/0921-8777(93)90024-b. [DOI] [PubMed] [Google Scholar]
- 709.Wang T-C V, Chen S-H. Okazaki DNA fragments contain equal amounts of lagging-strand and leading-strand sequences. Biochem Biophys Res Commun. 1994;198:844–849. doi: 10.1006/bbrc.1994.1120. [DOI] [PubMed] [Google Scholar]
- 710.Wang T-C V, Chen S-H. Similar-sized daughter-strand gaps are produced in the leading and the lagging strands of DNA in UV-irradiated E. coli uvrA cells. Biochem Biophys Res Commun. 1992;184:1496–1503. doi: 10.1016/s0006-291x(05)80052-x. [DOI] [PubMed] [Google Scholar]
- 711.Wang T-C V, Smith K C. Discontinuous DNA replication in a lig-7 strain of Escherichia coli is not the result of mismatch repair, nucleotide-excision repair, or the base-excision repair of DNA uracil. Biochem Biophys Res Commun. 1989;165:685–688. doi: 10.1016/s0006-291x(89)80020-8. [DOI] [PubMed] [Google Scholar]
- 712.Wang T-C V, Smith K C. Effects of the ssb-1 and ssb-113 mutations on survival and DNA repair in UV-irradiated ΔuvrB strains of Escherichia coli K-12. J Bacteriol. 1982;151:186–192. doi: 10.1128/jb.151.1.186-192.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 713.Wang T-C V, Smith K C. Inviability of dam recA and dam recB cells of Escherichia coli is correlated with their inability to repair DNA double-strand breaks produced by mismatch repair. J Bacteriol. 1986;165:1023–1025. doi: 10.1128/jb.165.3.1023-1025.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 714.Wang T-C V, Smith K C. Mechanisms for recF-dependent and recB-dependent pathways of postreplication repair in UV-irradiated Escherichia coli uvrB. J Bacteriol. 1983;156:1093–1098. doi: 10.1128/jb.156.3.1093-1098.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 715.Wang T-C V, Smith K C. Postreplicational formation and repair of DNA double-strand breaks in UV-irradiated Escherichia coli uvrB cells. Mutat Res. 1986;165:39–44. doi: 10.1016/0167-8817(86)90007-6. [DOI] [PubMed] [Google Scholar]
- 716.Wang T-C V, Smith K C. recF-dependent and recF recB-independent DNA gap-filling repair processes transfer dimer-containing parental strands to daughter strands in Escherichia coli K-12 uvrB. J Bacteriol. 1984;158:727–729. doi: 10.1128/jb.158.2.727-729.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 717.Ward J F. DNA damage produced by ionizing radiation in mammalian cells: identities, mechanisms of formation, and reparability. Prog Nucleic Acid Res Mol Biol. 1988;35:95–125. doi: 10.1016/s0079-6603(08)60611-x. [DOI] [PubMed] [Google Scholar]
- 718.Webb B L, Cox M M, Inman R B. An interaction between the Escherichia coli RecF and RecR proteins dependent on ATP and double-stranded DNA. J Biol Chem. 1995;270:31397–31404. doi: 10.1074/jbc.270.52.31397. [DOI] [PubMed] [Google Scholar]
- 719.Webb B L, Cox M M, Inman R B. Recombinational DNA repair: the RecF and RecR proteins limit the extension of RecA filaments beyond single-strand DNA gaps. Cell. 1997;91:347–356. doi: 10.1016/s0092-8674(00)80418-3. [DOI] [PubMed] [Google Scholar]
- 720.Weisberg R A, Sternberg N. Transduction of recB− host is promoted by λ red+ function. In: Grell R F, editor. Mechanisms in recombination. New York, N.Y: Plenum Press; 1974. pp. 107–109. [Google Scholar]
- 721.West S C. Processing of recombination intermediates by the RuvABC proteins. Annu Rev Genet. 1997;31:213–244. doi: 10.1146/annurev.genet.31.1.213. [DOI] [PubMed] [Google Scholar]
- 722.West S C. RuvA gets X-rayed on Holliday. Cell. 1998;94:699–701. doi: 10.1016/s0092-8674(00)81729-8. [DOI] [PubMed] [Google Scholar]
- 723.West S C. The RuvABC proteins and Holliday junction processing in Escherichia coli. J Bacteriol. 1996;178:1237–1241. doi: 10.1128/jb.178.5.1237-1241.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 724.West S C, Cassuto E, Howard-Flanders P. Heteroduplex formation by RecA protein: polarity of strand exchanges. Proc Natl Acad Sci USA. 1981;78:6149–6153. doi: 10.1073/pnas.78.10.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 725.West S C, Cassuto E, Howard-Flanders P. Postreplication repair in E. coli: strand exchange reactions of gapped DNA by RecA protein. Mol Gen Genet. 1982;187:209–217. doi: 10.1007/BF00331119. [DOI] [PubMed] [Google Scholar]
- 726.Whitby M C, Bolt E L, Chan S N, Lloyd R G. Interactions between RuvA and RuvC at Holliday junctions: inhibition of junction cleavage and formation of a RuvA-RuvC-DNA complex. J Mol Biol. 1996;264:878–890. doi: 10.1006/jmbi.1996.0684. [DOI] [PubMed] [Google Scholar]
- 727.Whitby M C, Lloyd R G. Altered SOS induction associated with mutations in recF, recO and recR. Mol Gen Genet. 1995;246:174–179. doi: 10.1007/BF00294680. [DOI] [PubMed] [Google Scholar]
- 728.Whitby M C, Lloyd R G. Branch migration of three-strand recombination intermediates by RecG, a possible pathway for securing exchanges initiated by 3′-tailed duplex DNA. EMBO J. 1995;14:3302–3310. doi: 10.1002/j.1460-2075.1995.tb07337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 729.Whitby M C, Lloyd R G. Targeting Holliday junctions by the RecG branch migration protein of Escherichia coli. J Biol Chem. 1998;273:19729–19739. doi: 10.1074/jbc.273.31.19729. [DOI] [PubMed] [Google Scholar]
- 730.Whitby M C, Ryder L, Lloyd R G. Reverse branch migration of Holliday junctions by RecG protein: a new mechanism for resolution of intermediates in recombination and DNA repair. Cell. 1993;75:341–350. doi: 10.1016/0092-8674(93)80075-p. [DOI] [PubMed] [Google Scholar]
- 731.Whitby M C, Vincent S D, Lloyd R G. Branch migration of Holliday junctions: identification of RecG protein as a junction-specific DNA helicase. EMBO J. 1994;13:5220–5228. doi: 10.1002/j.1460-2075.1994.tb06853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 732.Whittier R F, Chase J W. DNA repair in E. coli strains deficient in single-strand DNA binding protein. Mol Gen Genet. 1981;183:341–347. doi: 10.1007/BF00270638. [DOI] [PubMed] [Google Scholar]
- 733.Wilkins A S, Mistry J. Phage lambda’s generalized recombination system. Study of intracellular DNA pool during lytic infection. Mol Gen Genet. 1974;129:275–293. doi: 10.1007/BF00265693. [DOI] [PubMed] [Google Scholar]
- 734.Wilkins B M, Howard-Flanders P. The genetic properties of DNA transferred from ultraviolet-irradiated Hfr cells of Escherichia coli K-12 during mating. Genetics. 1968;60:243–255. doi: 10.1093/genetics/60.2.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 735.Willetts N S, Mount D W. Genetic analysis of recombination-deficient mutants of Escherichia coli K-12 carrying rec mutations cotransducible with thyA. J Bacteriol. 1969;100:923–934. doi: 10.1128/jb.100.2.923-934.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 736.Williams G C. Sex and evolution. Princeton, N.J: Princeton University Press; 1975. [Google Scholar]
- 737.Wilson D H, Benight A S. Kinetic analysis of the pre-equilibrium steps in the self-assembly of recA protein from Escherichia coli. J Biol Chem. 1990;265:7351–7359. [PubMed] [Google Scholar]
- 738.Witkin E M, Roegner-Maniscalco V, Sweasy J B, McCall J O. Recovery from ultraviolet light-induced inhibition of DNA synthesis requires umuDC gene products in recA718 mutant strains but not in recA+ strains of Escherichia coli. Proc Natl Acad Sci USA. 1987;84:6805–6809. doi: 10.1073/pnas.84.19.6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 739.Wold M S, Mallory J B, Roberts J D, LeBowitz J H, McMacken R. Initiation of bacteriophage λ DNA replication in vitro with purified λ replication proteins. Proc Natl Acad Sci USA. 1982;79:6176–6180. doi: 10.1073/pnas.79.20.6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 740.Woldringh C L, Nanninga N. Structure of nucleoid and cytoplasm in the intact cell. In: Nanninga N, editor. Molecular cytology of Escherichia coli. London, United Kingdom: Academic Press, Ltd.; 1985. pp. 161–197. [Google Scholar]
- 741.Woldringh C L, Zaritsky A, Grover N B. Nucleoid partitioning and the division plane in Escherichia coli. J Bacteriol. 1994;176:6030–6038. doi: 10.1128/jb.176.19.6030-6038.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 742.Woodgate R, Ennis D G. Levels of chromosomally encoded Umu proteins and requirements for in vivo UmuD cleavage. Mol Gen Genet. 1991;229:10–16. doi: 10.1007/BF00264207. [DOI] [PubMed] [Google Scholar]
- 743.Woodgate R, Sedgwick S G. Mutagenesis induced by bacterial UmuDC proteins and their plasmid homologues. Mol Microbiol. 1992;6:2213–2218. doi: 10.1111/j.1365-2958.1992.tb01397.x. [DOI] [PubMed] [Google Scholar]
- 744.Worth L, Jr, Bader T, Yang J, Clark S. Role of MutS ATPase activity in MutS,L-dependent block of in vitro strand transfer. J Biol Chem. 1998;273:23176–23182. doi: 10.1074/jbc.273.36.23176. [DOI] [PubMed] [Google Scholar]
- 745.Worth L, Jr, Clark S, Radman M, Modrich P. Mismatch repair proteins MutS and MutL inhibit RecA-catalyzed strand transfer between diverged DNAs. Proc Natl Acad Sci USA. 1994;91:3238–3241. doi: 10.1073/pnas.91.8.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 746.Wright M, Buttin G, Hurwitz J. The isolation and characterization from Escherichia coli of adenosine triphosphate-dependent deoxyribonuclease directed by recB,C genes. J Biol Chem. 1971;246:6543–6555. [PubMed] [Google Scholar]
- 747.Wu Y H, Franden M A, Hawker J R, McHenry C S. Monoclonal antibodies specific for the α subunit of the Escherichia coli DNA polymerase III holoenzyme. J Biol Chem. 1984;259:12117–12122. [PubMed] [Google Scholar]
- 748.Yagil E, Dower N A, Chattoraj D, Stahl M, Pierson C, Stahl F W. Chi mutation in a transposon and the orientation-dependence of Chi phenotype. Genetics. 1980;96:43–57. doi: 10.1093/genetics/96.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 749.Yagil E, Shtromas I. Rec-mediated recombinational activity of two adjacent Chi elements in bacteriophage lambda. Genet Res. 1985;45:1–8. doi: 10.1017/s0016672300021911. [DOI] [PubMed] [Google Scholar]
- 750.Yamamoto K, Takahashi N, Fujitani Y, Yoshikura H, Kobayashi I. Orientation dependence in homologous recombination. Genetics. 1996;143:27–36. doi: 10.1093/genetics/143.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 751.Yamamoto K, Yoshikura H, Takahashi N, Kobayashi I. Apparent gene conversion in an Escherichia coli rec+ strain is explained by multiple rounds of reciprocal crossing-over. Mol Gen Genet. 1988;212:393–404. doi: 10.1007/BF00330842. [DOI] [PubMed] [Google Scholar]
- 752.Yancey-Wrona J E, Camerini-Otero R D. The search for DNA homology does not limit stable homologous pairing promoted by RecA protein. Curr Biol. 1995;5:1149–1158. doi: 10.1016/s0960-9822(95)00231-4. [DOI] [PubMed] [Google Scholar]
- 753.Yasuda T, Morimatsu K, Horii T, Nagata T, Ohmori H. Inhibition of Escherichia coli RecA coprotease activities by DinI. EMBO J. 1998;17:3207–3216. doi: 10.1093/emboj/17.11.3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 754.Yeung T, Mullin D A, Chen K-S, Craig E A, Bardwell J C A, Walker J R. Sequence and expression of the Escherichia coli recR locus. J Bacteriol. 1990;172:6042–6047. doi: 10.1128/jb.172.10.6042-6047.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 755.Youngs D A, Smith K C. The involvement of polynucleotide ligase in the repair of UV-induced DNA damage in Escherichia coli K-12 cells. Mol Gen Genet. 1977;152:37–41. doi: 10.1007/BF00264937. [DOI] [PubMed] [Google Scholar]
- 756.Yu M, Souaya J, Julin D A. The 30-kDa C-terminal domain of the RecB protein is critical for the nuclease activity, but not the helicase activity, of the RecBCD enzyme from Escherichia coli. Proc Natl Acad Sci USA. 1998;95:981–986. doi: 10.1073/pnas.95.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 757.Yu M, Souaya J, Julin D A. Identification of the nuclease active site in the multifunctional RecBCD enzyme by creation of a chimeric enzyme. J Mol Biol. 1998;283:797–808. doi: 10.1006/jmbi.1998.2127. [DOI] [PubMed] [Google Scholar]
- 758.Yu X, Angov E, Camerini-Otero R D, Egelman E H. Structural polymorphism of the RecA protein from the thermophilic bacterium Thermus aquaticus. Biophys J. 1995;69:2728–2738. doi: 10.1016/S0006-3495(95)80144-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 759.Yu X, Egelman E H. The LexA repressor binds within the deep helical groove of the activated RecA filament. J Mol Biol. 1993;231:29–40. doi: 10.1006/jmbi.1993.1254. [DOI] [PubMed] [Google Scholar]
- 760.Yu X, Egelman E H. The RecA hexamer is a structural homologue of ring helicases. Nat Struct Biol. 1997;4:101–104. doi: 10.1038/nsb0297-101. [DOI] [PubMed] [Google Scholar]
- 761.Yu X, West S C, Egelman E H. Structure and subunit composition of the RuvAB-Holliday junction complex. J Mol Biol. 1997;266:217–222. doi: 10.1006/jmbi.1996.0799. [DOI] [PubMed] [Google Scholar]
- 762.Zaitsev E N, Kowalczykowski S C. The simultaneous binding of two double-stranded DNA molecules by Escherichia coli RecA protein. J Mol Biol. 1999;287:21–31. doi: 10.1006/jmbi.1998.2580. [DOI] [PubMed] [Google Scholar]
- 763.Zaman M M, Boles T C. Chi-dependent formation of linear plasmid DNA in exonuclease-deficient recBCD+ strains of Escherichia coli. J Bacteriol. 1994;176:5093–5100. doi: 10.1128/jb.176.16.5093-5100.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 764.Zerbib D, Mézard C, George H, West S C. Coordinated actions of RuvABC in Holliday junction processing. J Mol Biol. 1998;281:621–630. doi: 10.1006/jmbi.1998.1959. [DOI] [PubMed] [Google Scholar]
- 765.Zieg J, Kushner S R. Analysis of genetic recombination between two partially deleted lactose operons of Escherichia coli K12. J Bacteriol. 1977;131:123–132. doi: 10.1128/jb.131.1.123-132.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 766.Zieg J, Maples V F, Kushner S R. Recombination levels of Escherichia coli K-12 mutants deficient in various replication, recombination, or repair genes. J Bacteriol. 1978;134:958–966. doi: 10.1128/jb.134.3.958-966.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 767.Zissler J, Signer E, Schaefer F. The role of recombination in growth of bacteriophage lambda. I. The gamma gene. In: Hershey A D, editor. The bacteriophage lambda. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1971. pp. 455–468. [Google Scholar]