Abstract
Hepadnaviruses (hepatitis B viruses) cause transient and chronic infections of the liver. Transient infections run a course of several months, and chronic infections are often lifelong. Chronic infections can lead to liver failure with cirrhosis and hepatocellular carcinoma. The replication strategy of these viruses has been described in great detail, but virus-host interactions leading to acute and chronic disease are still poorly understood. Studies on how the virus evades the immune response to cause prolonged transient infections with high-titer viremia and lifelong infections with an ongoing inflammation of the liver are still at an early stage, and the role of the virus in liver cancer is still elusive. The state of knowledge in this very active field is therefore reviewed with an emphasis on past accomplishments as well as goals for the future.
Hepatitis B virus (HBV) causes transient and chronic infections of the liver. Transient infections may produce serious illness, and approximately 0.5% terminate with fatal, fulminant hepatitis. Chronic infections may also have serious consequences: nearly 25% terminate in untreatable liver cancer (10). Worldwide deaths from liver cancer caused by HBV infection probably exceed one million per year (52, 162).
An effective vaccine has been available for nearly 20 years, and attempts at universal vaccination are now under way in developed countries, including areas of endemic infection such as China, which has about 120 million carriers (163). However, vaccination is not a treatment for established infections, and its effectiveness in preventing blood-borne transmission from an infected mother to her newborn is about 90% (196). Moreover, the high cost of the vaccination programs has impeded introduction in many poorer countries where HBV infections are prevalent, afflicting 5 to 20% of the population.
Attempts at treatment of chronic infections have had only limited success. Alpha interferon therapy is effective in inducing virus elimination in a subset of HBV carriers, especially those with the most active disease, but is ineffective in more than 50% of carriers (87). The long-term potential of more recent approaches to eliminating the virus, involving therapy with nucleoside analogs and other inhibitors of virus replication, has yet to be determined. More sophisticated approaches to treatment, involving manipulation of the host immune system to induce the set of responses needed for virus clearance, are still in development. Thus, in the absence of “spontaneous” virus clearance, chronic and productive infections in many patients will persist for life. The number of carriers worldwide is estimated at 350 million.
One of the reasons for chronic HBV infections is that the virus causes chronic, noncytocidal infections of hepatocytes, the principal cell type of the liver. Hepatocytes continuously shed virus into the bloodstream, ensuring that 100% of the hepatocyte population is infected. Also, hepatocytes are normally long-lived, with half-lives estimated at 6 to 12 months or longer. The combination of a long-lived, usually nondividing host cell and a stable virus-host cell interaction virtually ensures the persistence of an infection in the absence of a robust host immune response. Indeed, liver disease in transient and chronic carriers is thought to be largely due to the host immune response to the infection. This response can induce a high level of hepatocyte destruction (154), leading to scarring, disruption of blood flow, and obstruction of bile drainage without necessarily eliminating the infection. It remains unknown why this response is unable to clear the virus in many who become chronic carriers or, for that matter, why elimination sometimes occurs spontaneously after many years of virus production. Therefore, a major focus of HBV research is to understand the virus-host interactions which determine whether an infection will persist or terminate. The hope is that this knowledge will lead to better treatments and a reduced incidence of hepatocellular cancer (HCC).
An important development in hepatitis B research was the discovery that human HBV virus is the prototype for a family of viruses, referred to as Hepadnaviridae. Those most closely related to HBV have been found in woodchucks (202) and ground squirrels (135, 208, 210). These viruses have about 70% sequence homology to HBV but are not known to infect humans or other primates; in contrast, HBV is infectious for the great apes. Because of similarities in DNA sequence and genome organization, the viruses infecting mammals are grouped in the genus Orthohepadnavirus. More distantly related viruses, with somewhat similar genome organization but almost no sequence homology, are found in ducks and geese (138, 193). These are grouped in the genus Avihepadnavirus. Duck hepatitis B virus (DHBV) has been used primarily as a model system to characterize how hepadnaviruses replicate, rather than as a disease model, since chronic DHBV infection has generally been studied after in ovo transmission. In ovo transmission, with exposure to DHBV from the beginning of embryonic development and infection of the developing liver (221), apparently prevents the development of an effective immune response to infected hepatocytes (78). Neither chronic liver disease nor liver cancer is normally detected, although antibodies to the viral nucleocapsid may be produced (A. Jilbert, personal communication). Woodchuck hepatitis virus (WHV) infections have been studied intensely as a model for chronic liver disease and the attendant liver cancer. Both WHV and DHBV have been used to develop antiviral agents and therapies. The HBV transgenic mouse has also been introduced for this purpose, as well as for characterizing the susceptibility of HBV, not just to alpha interferon but also to various other cytokines produced during an immune response to an infection.
The purpose of this review is to introduce the reader to various aspects of hepadnavirus replication, pathogenesis, liver cancer, and therapy that have been elucidated over the last several years. In addition, we attempt to emphasize the major problems that need to be addressed in the future to meet the ultimate challenge in this field, which is to provide the basic knowledge required to eventually cure chronic infections. As a prelude, we include a brief description of basic liver structure and function, which we deem essential for an understanding of hepadnavirus infections.
THE LIVER AS A TARGET FOR HEPADNAVIRUS INFECTION
The liver plays an essential role in energy storage and conversion, blood homeostasis, chemical detoxification, and immunity to microbial infections. Although the liver is composed of many different types of cells, much of the functional activity resides in hepatocytes (which constitute 70% of the liver), bile ductule epithelium, and Kupffer cells (macrophages) (62). Among these, hepatocytes and bile ductule epithelial cells are unique to the liver and are also closely related. In fact, they may originate during embryonic life from a common progenitor (188, 189, 211) and also may be replaced by proliferation and differentiation of a common progenitor cell in response to very acute forms of liver injury (211).
Because hepatocytes are the major cell type in the liver, it might be expected that they would also be the major target of infection by a liver-tropic virus such as HBV. Indeed, this appears to be the case. Hepatocytes are the only confirmed site of replication for all members of this virus family. Bile ductule epithelial cells may also be a target of infection, as may a subset of cells in the pancreas, kidneys, and lymphoid system (16, 74, 77, 98, 111, 122, 151, 160, 170, 187, 243). However, the evidence for replication of the orthohepadnaviruses in bile ductules and at extrahepatic sites is in some cases controversial or incomplete, and these sites are not usually considered in discussions of viral reproduction and pathogenesis. This approach to infections is at least compatible with the notion that many of the extrahepatic symptoms of infection that are not attributed to liver dysfunction are the result of deposition of antibody-antigen complexes. Thus, for the purpose of simplicity, we discuss infection only in the context of hepatic manifestations. The implications of extrahepatic infections have yet to be determined.
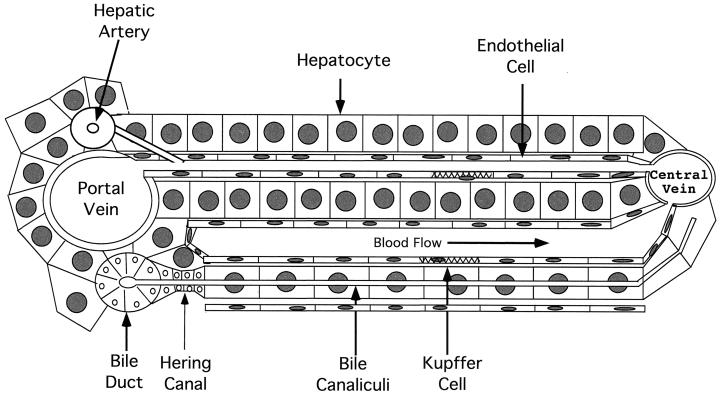
The liver itself is usually considered, for convenience, to be divided into small compartments called lobules. This subdivision emphasizes the role of the liver in relation to blood flow. While this view of liver anatomy is somewhat arbitrary, it provides a convenient way of considering liver function, development, regeneration, and pathogenesis. An essentially one-dimensional view of a section of a “classical” lobule is illustrated in Fig. 1. In this view, blood enters the lobule through portal veins and hepatic arteries and is distributed by smaller vessels to enter the sinusoidal spaces, created by plates of hepatocytes. The plates are generally one hepatocyte thick in mammals and two hepatocytes thick in birds. Blood passes through these spaces, which are lined by fenestrated endothelial cells and fixed macrophages (Kupffer cells), where the various functional interactions take place. The blood is then collected in central veins and exits the liver. (It should be noted that because of the rather homogeneous nature of liver anatomy, blood from each portal vein and artery will flow to several surrounding central veins; a more thorough discussion of liver structure and function is presented in reference 55. Generally, the distance from a portal vein to a central vein is 20 to 30 hepatocytes. During catabolism of heme produced by red blood cell breakdown in the liver, bile is formed and secreted into bile canaliculi, small channels formed at the junctions of hepatocytes. Bile flows in the opposite direction from blood, passing through a region known as the canal of Hering to enter bile ducts. From there, it flows into larger ducts and is eventually transported to the gallbladder and intestine.
FIG. 1.
Anatomy of the liver lobule. The major cell types within the liver are illustrated. Blood enters at the portal tracts (encompassing the portal vein, hepatic artery, and bile ductules), distributes laterally through smaller veins to the sinusoidal spaces, and flows toward the central vein. The sinusoidal spaces are lined with endothelial cells and fixed macrophages (Kupffer cells). The endothelial cells are fenestrated, allowing free diffusion between the blood and hepatocytes.
Although hepatocytes are “terminally” differentiated, they retain the capacity for extensive proliferation in response to liver injury. Under normal conditions, hepatocytes may have lifetimes exceeding 6 to 12 months. However, when required, virtually the entire population may enter the cell cycle and divide. Following partial hepatectomy of 70% of the liver, virtually every hepatocyte passes through the cell cycle at least once, and the liver cell mass is restored within a few days (67, 132). In an even more extreme example of liver cell proliferation, Rhim et al. (175) observed, under conditions of very acute liver injury, the replacement of virtually the entire hepatocyte population of the mouse by clonogenic outgrowth of mature hepatocytes. This required an average of at least 12 cycles of cell division.
Hepatocyte replacement may also occur via proliferation of progenitor cells, a situation that appears in the context of long-term liver injury and/or acute injury (e.g., caused by some hepatotoxic drugs) in which hepatocyte proliferation is retarded. These progenitors are thought to be a population of facultative stem cells present in the region of the portal tracts, possibly closely associated with or identical to cells in bile ducts or the canals of Hering (36, 42, 54, 56, 212). When induced to proliferate, they may first appear as the so-called oval cells (176) and may then differentiate to form hepatocytes.
Thus, while hepatocytes may represent a fairly homogeneous population of quiescent cells at the time of infection, the situation can rapidly change as the immune system attacks infected cells. This means that an understanding of transient infections and chronic infections requires an understanding of how the liver regenerates, as well as how components of the virus replication machinery, including transcriptionally active viral DNAs, behave as the infected cells begin to divide. For example, any explanation of transient infections requires some explanation of the fate of infected hepatocytes as the virus is cleared. If the entire hepatocyte population is infected, virus clearance requires either a mechanism for eliminating virus from hepatocytes or complete replacement of infected hepatocytes by proliferation of hypothetically uninfected population of progenitor cells. While these two alternatives appear clear-cut, they are not in practice that easy to distinguish. Likewise, understanding of chronic infections and how they may be treated requires some knowledge of liver cell proliferation during an infection and how cell proliferation affects the virus life cycle. Again, the answers are either unavailable or not clear-cut. Nonetheless, as we attempt to stress later in this review, without answers to these questions, our knowledge of hepatitis B is at best superficial and the possibility of effectively dealing with disease progression in patients is probably limited.
GENOME REPLICATION
Introduction
The life cycle of hepadnaviruses is characterized by the synthesis of a ∼3-kb partially double-stranded, relaxed-circular DNA (rcDNA) genome by reverse transcription of an RNA intermediate, the pregenome (Fig. 2 and 3). The mechanism of RNA-directed DNA synthesis has been well characterized through genetic as well as biochemical studies, which have been described in several reviews (61, 182). In contrast, early events of the viral life cycle, including entry, uncoating, and delivery of the viral genome into the cell nucleus, are not well understood. This is, in part, due to the absence of cell lines that are susceptible to hepadnavirus infection.
FIG. 2.
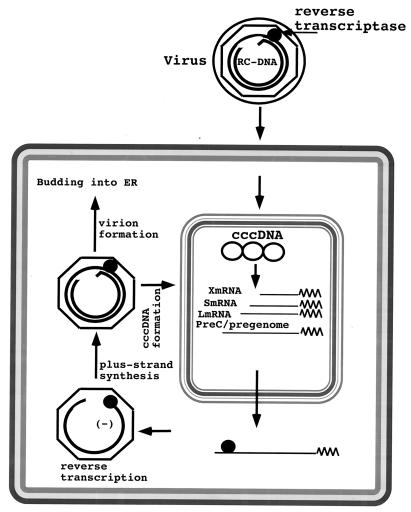
Hepadnavirus life cycle. The details of the replication cycle are discussed in the text. Briefly, during initiation of infection the viral rcDNA (or linear DNA) genome, with a protein (the viral reverse transcriptase) attached to the 5′ end of the minus strand and a short RNA attached to the 5′ end of the plus strand, is converted into cccDNA. During this process, both the protein and the RNA are removed. The cccDNA serves as the template for transcription of viral mRNAs (see Fig. 4). One of these, the pregenome, serves as the mRNA for the synthesis of core protein (nucleocapsid subunit) and the viral reverse transcriptase. The reverse transcriptase binds to the 5′ end of its own mRNA template, and the complex is then packaged into nucleocapsids, where viral DNA synthesis occurs (see Fig. 3). Once partially double-stranded DNA has been produced, nucleocapsids can undergo a maturation event that facilitates their acquisition of an outer envelope via budding into the ER. These nucleocapsids can also migrate to the nucleus to increase the copy number of cccDNA. Since cccDNA does not undergo semiconservative replication, all cccDNA is derived from viral DNA made in the cytoplasm via the reverse transcription pathway (217). Accumulation of viral envelope proteins prevents excessive cccDNA formation, which can be toxic to hepatocytes (200, 201).
FIG. 3.
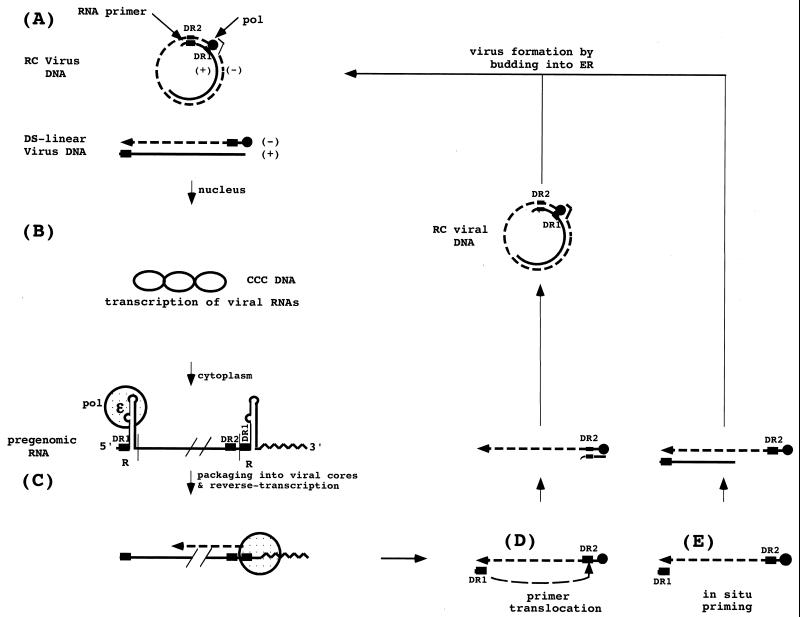
Hepadnavirus DNA replication. (A) Virus particles contain predominantly rcDNA with a complete minus strand and a partially synthesized plus strand. A small amount of linear DNA, formed as a result of aberrant plus-strand priming (in situ priming) is also found in the virus. (B) During initiation of infection, both forms of virion DNA are converted to a cccDNA; however, conversion of the linear form involves illegitimate recombination, which can lead to subsequent defects in the ability of the virus to replicate. cccDNA serves as template for the transcription of the pregenome. The reverse transcriptase binds to the epsilon stem-loop structure near the 5′ end of its own mRNA to facilitate packaging into nucleocapsids and initiation of reverse transcription by a protein-priming mechanism, utilizing a tyrosine located near the amino terminus of the reverse transcriptase itself. (C) Following the synthesis of 4 bases, the polymerase translocates to the 3′ end of the RNA template, where the 4 bases can anneal with complementary sequences. During the elongation of the minus strand to the 5′ end of the pregenome, all but the 5′ 17 to 18 bases of the pregenome, including the CAP and DR1, are degraded by the viral RNase H. (D) The remaining fragment then serves as the primer for plus-strand synthesis, usually following its translocation to DR2, with which it can hybridize because of the sequence identity between DR1 and DR2. A third translocation occurs when the plus strand reaches the 5′ end of the minus strand, to circularize the molecule and allow continued plus-strand elongation. This translocation may be facilitated by the short (∼9-base) terminal redundancy on the minus strand. The plus strand is not completed prior to virion release; a repair reaction to produce a fully double stranded DNA occurs during initiation of a subsequent round of infection. (E) A fraction of the virions have linear genomes because priming of plus-strand DNA synthesis occurs in the absence of primer translocation.
Infection
Since hepatocytes are the primary site of viral DNA replication, it is tempting to conclude that tissue specificity is controlled primarily by a hepatocyte-specific receptor. However, hepatotropism is also manifested on the level of viral gene expression. Indeed, a variety of studies indicated a selectivity of the virus for productive infection of only very specific cells. In vivo, viral DNA replication outside the liver has been most thoroughly documented in DHBV-infected ducks. It was found in a subset of exocrine cells and in the endocrine islets of the pancreas and in the proximal tubular epithelium of the kidney (75–77, 98, 204). While it has been straightforward to identify certain extrahepatic tissues as possible sites of virus replication, it has been much harder to rule out other sites.
For instance, as noted above, periodic reports suggested that lymphocytes are targets of infection by the hepadnaviruses and could provide a second reservoir for virus to persist. However, whether the detection of viral nucleic acids in lymphocytes reflects active viral replication or is the result of phagocytic activity of immune system cells is still a matter of debate. Some recent studies have suggested, for example, that HBV nucleic acids found in lymphocyte preparations are due to adsorbed virus (108), while others have asserted that the HBV nucleic acids detected in these cell preparations include viral RNA transcribed in the cells from a covalently closed circular DNA (cccDNA) template (197). Indeed, the difficulty in resolving this question may arise in large part from the need to use very sensitive techniques such as PCR and reverse transcriptase PCR to detect any virus signal. A more biological approach suggested that WHV can infect woodchuck lymphocytes in vivo. This can be deduced from the observation that virus replication is induced upon mitogen stimulation of lymphocytes cultured from WHV-infected woodchucks (111, 143).
Formation of cccDNA
The first event in virus DNA replication accessible to experimental investigation is the conversion of the viral rcDNA genome into cccDNA (Fig. 2 and 3). Since cccDNA is the template for the transcription of viral mRNAs, its formation indicates a successful initiation of infection. Conversion of rcDNA to cccDNA in the liver is detected within the first 24 h following virus inoculation (137, 203). The mechanism of DNA repair involved in this conversion is not known. The ability of inhibitors of the viral DNA polymerase to block initiation of infection by an orthohepadnavirus (2, 146) implies that this enzyme plays a role. However, we can also anticipate that general concepts of cellular DNA repair may apply to the hepadnavirus system in some aspect of the completion of plus-strand DNA and the ligation of the ends of the two DNA strands. The removal of the covalently attached reverse transcriptase (Fig. 3) from the 5′ end of minus-strand DNA is, in contrast, a unique reaction and, depending on the mechanism, has consequences for our understanding of cccDNA formation. For instance, ligation of the two ends of minus-strand DNA first requires the removal of the reverse transcriptase from the 5′ end. Is this step catalyzed by the polymerase itself, and does it require the hydrolysis of the DNA-protein bond, or does a cellular endonuclease cleave viral DNA close to the 5′ end? Also, does this step occur before or after the translocation of the DNA across the nuclear membrane?
Recent reports describing the ability of peptides to direct plasmid DNA into the nucleus via the nuclear import pathway raise questions about a similar role for the reverse transcriptase in nuclear transport of hepadnavirus DNA (249). Some experimental data reported by Kann et al. seem to support such a possibility (103), but the evidence has been derived from an in vitro transport system with a very low efficiency and thus awaits further confirmation. Interestingly, Kann et al. recently found that phosphorylated core protein can bind to the nuclear core complex in a karyopherin-dependent reaction, suggesting that the core could also act as a vector for the nuclear transport of viral DNA (104). Whatever the mechanism for the nuclear transport of virion DNA may be, formation of cccDNA, which accumulates only in the nucleus, completes the initiation of infection. cccDNA then acts as a template for the transcription of all the viral mRNAs.
Viral Gene Expression
The viral RNAs include pregenomic RNA (pgRNA), which serves as the template for reverse transcription, as well as three subgenomic mRNAs (two for the avihepadnaviruses) necessary for the translation of the envelope proteins and the mRNA for the X protein. X is encoded by all known mammalian hepadnaviruses (Fig. 4) but not by the avihepadnaviruses. Because the cccDNA template is circular and because all these RNAs have a common polyadenylation signal, use of a single template with multiple promoters raises obvious but unresolved questions about how potential problems, such as promoter occlusion, are avoided. For instance, transcription of pregenomic RNA can suppress transcription of the downstream envelope mRNAs of DHBV (95). However, as discussed below, by the time virus production begins, infected cells contain multiple copies of cccDNA in the nucleus, and these, at least for DHBV, exist as a heterogeneous population of small chromosomes (149). It is thus formally possible that part of the potential structural impediments posed by using a single template for the synthesis of all these mRNAs is relieved by template specialization.
FIG. 4.
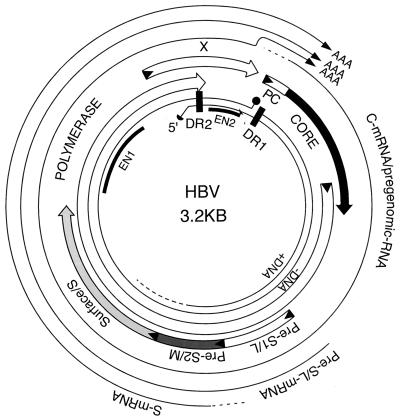
Transcriptional and translational map of HBV. The figure shows the physical map of the HBV genome. The inner circle depicts the rcDNA with the reverse transcriptase attached to the 5′ end of the complete minus-strand DNA (solid sphere) and a capped RNA oligomer attached to the 5′ end of the incomplete plus-strand DNA (solid half sphere). The positions of the direct repeats, DR1 and DR2, as well as the positions of the two enhancers, EN1 and EN2, are indicated. The outer circle depicts the three major viral RNAs, the core (C) or pgRNA, the pre-S (L) mRNA, and the S mRNA. The common 3′ ends of the three mRNAs are indicated by the letters A. Not shown in the figure is the putative X mRNA that spans the X coding region and terminates at the site indicated for the other three mRNAs. The four protein-coding regions are shown between the inner and outer circles. They include the precore (PC) and core genes, the polymerase gene, and the X gene. The envelope genes pre-S1 (L), pre-S2 (M), and surface (S) overlap with the polymerase open reading frame.
Control of gene expression has been studied most thoroughly for HBV. Basically, this research has identified elements in the viral genome that display promoter and/or enhancer activity in cell culture systems and/or liver extracts. Thus, it is now known that each gene of HBV has one or more promoters regulating its activity and that these promoters are in turn regulated by one or both of the viral enhancer elements, En1 and En2, that are located upstream of the core promoter (Fig. 4). Details about transcription factors capable of operating through the two enhancers and the four promoters of HBV have been obtained primarily by studying expression in liver-derived cell lines, which mimic some but certainly not all of the transcriptional regulation of mature hepatocytes. In brief, the results confirm the idea that HBV transcription is dependent to a large extent upon liver-enriched transcription factors. These results have been reviewed in detail elsewhere (178, 184) and will not be further considered here. Preliminary studies with DHBV (40, 126) and WHV (48, 232) have also suggested the importance of liver-specific transcription factors, although very little else is known about the specific factor interactions that take place in the hosts of these two viruses.
A problem of particular importance with a circular DNA template is termination of transcription, especially for the pregenome and precore mRNAs. For these, transcription must pass through the polyadenylation signal on the first pass in order to make a full-size, terminally redundant RNA. Studies with DHBV indicated that this is facilitated by a sequence on cccDNA, named PET (positive effector of transcription), located between the transcription initiation site and the poly(A) signal (Fig. 4) (95). PET somehow facilitates passage of transcription complexes through the termination site on the first but not the second passage. If PET is deleted, transcription of the smaller, envelope mRNAs is increased. A second signal sequence, NET (negative effector of transcription), which is needed for termination, is suppressed by PET. If NET is deleted, longer transcripts accumulate due to additional transits around the cccDNA template (13).
Spliced transcripts have sometimes been detected, but there is so far no evidence that RNA splicing plays an important role in HBV gene expression. On the other hand, a spliced form of the pregenome has been detected in DHBV-infected cells and could serve as a second mRNA for the synthesis of the large envelope protein of DHBV. It was reported that mutation of the splice donor site to block production of the spliced RNA did not inhibit virus release from a transfected cell line whereas virus release from primary hepatocyte cultures infected with the mutant virus was blocked (156). There is no a priori explanation for the mutant phenotype. The authors did not indicate if the mutation affected early viral DNA synthesis and/or cccDNA amplification in the primary hepatocyte cultures. An effect on DNA synthesis might have occurred because of the location of the splice donor site in a region critical for initiation of DNA synthesis, and an effect on early cccDNA amplification might have been anticipated if the new mRNA had primarily a regulatory function.
Regulation of viral gene expression also occurs at the level of translation. The pregenome (Fig. 4) serves as the mRNA not only for the viral core protein but also for the viral polymerase, which initiates from an AUG located in the distal portion of the core gene, although not in the same reading frame as core (30, 161). Reading of the polymerase open reading frame appears to be inefficient compared to that of the core open reading frame. However, since core particles are assembled from 240 core protein subunits (41) and only one or perhaps two polymerase proteins (7), pgRNA may serve as an mRNA for the translation, on average, of approximately 200 to 300 core polypeptides before allowing the translation of a polymerase polypeptide. Since the polymerase preferentially binds to the 5′ end of its own mRNA to initiate reverse transcription and packaging (6, 224), synthesis of the polymerase is apparently sufficient to stop further translation of the pregenome.
Viral Proteins
With a genome that is only 3 kbp in length, hepadnaviruses express a very limited repertoire of proteins. The core and polymerase genes are essential for viral DNA replication, and the envelope proteins, which consists of two or three subspecies, depending on the hepadnavirus, are essential for envelopment of nucleocapsids. Two additional gene products expressed during natural infections are of unknown function, hepatitis X protein (HBx) and hepatitis e-antigen (HBeAg). X is required for the establishment of an infection in vivo (32, 254) but is dispensable for virus replication in transfected cells (17, 237). HBeAg is dispensable for in vivo infections (28, 33). Curiously, avian hepadnaviruses do not possess an X gene, although DNA sequence comparisons have suggested that this gene may have been present at some time in the evolution of these viruses (148). Experiments in mice suggest that HBeAg may cause depletion of Th1 helpers cells, thereby suppressing the ability to mount a strong cytotoxic T-lymphocyte (CTL) response to the infected hepatocytes (144, 145). This idea has not yet been tested in a natural host for a hepadnavirus.
The function of the structural genes is more apparent. The core gene encodes the viral capsid protein, known as hepatitis B virus core antigen (HBcAg). Assembly of cores into the icosahedral subviral core particles requires the formation of dimers of the core subunit (29), stabilized through two disulfide bonds, followed by the assembly of 120 dimers into a shell with a diameter of 36 nm and a triangulation number (T) of 4 (41). Expression of cores with small truncations at their C termini can induce the formation of smaller shells consisting of 90 dimer subunits and a triangulation number of 3 (252). The structure of the cores was resolved first to 7.4 Å resolution by cryoelectron microscopy (19, 39) and then to 3.3 Å resolution of X-ray crystallography (235). The folding of the protein is characterized by four α-helices and the absence of β-sheets. In agreement with previous biochemical analyses, the structural data revealed two regions required for the dimerization of core monomers and for the subsequent assembly of the dimers into core particles (110, 253). Two separate regions of the core polypeptide are exposed on the surface of core particles and thus provide targets for interaction with envelope components (see “Assembly and reverse transcription” below). One of the two sites forms spikes that extend from the interface of the dimerization sites; the other is located just downstream of the α-helix believed to mediate the multimerization of dimers (38).
The polymerase polypeptide, translated from an internal initiator codon on pgRNA (Fig. 4), consists of two major domains that are tethered by an intervening spacer region. The amino-terminal domain (also called the terminal protein [TP] domain) plays a critical role in the packaging of pgRNA and in the priming of minus strand DNA, as discussed below. The carboxy-terminal domain is a reverse transcriptase that, like the retroviral polymerases, is endowed with an RNase H activity. Structural and biochemical investigations of the hepadnavirus polymerases have been complicated by the apparent requirement of cellular factors for enzymatic activity. For example, the protein-priming reaction of DHBV depends on the molecular chaperone hsp90 and some of its cofactors including p23 (93). For these and perhaps other reasons, purification of the polymerase from cellular extracts has so far almost always resulted in the loss of the protein-priming and DNA polymerase activities (however, see reference 155).
Orthohepadnaviruses express three envelope components called S, M, and L. S (226 amino acids long), the smallest, defines the S domain. The two larger proteins contain S plus amino-terminal extensions of S created by initiation at upstream start codons. For HBV, these codons are located approximately 165 (M) and 489 (L) nucleotides upstream of the initiation codon for S. The extra domain of M is known as pre-S2, while the domain unique to L is called pre-S1. All three envelope components are glycosylated, type II transmembrane proteins that can form multimers stabilized by disulfide bridges formed by cysteine residues present in the S domain. S, L, and M are all found as components of the 42-nm-diameter infectious viral particles, also known as Dane particles after the author of a paper describing their appearance in the electron microscope (45). L and M are present in roughly equal amounts in Dane particles and together constitute approximately 30% of the envelope protein content (82). S by itself, and together with the larger envelope proteins, also forms filamentous and spherical “surface antigen” particles that are secreted from infected cells in at least 100-fold excess over virions. These spheres and filaments can accumulate to concentrations of several hundred micrograms per milliliter in the blood of HBV-infected patients. Complexes of these particles with their cognate antibodies are probably responsible for the immune complex syndromes that sometimes occur during transient infections.
Antibodies to surface antigen particles composed of S protein alone are sufficient to provide protection against HBV infection. However, there is good reason to believe that the pre-S1 domain is, at least in part, the substrate for the still elusive viral receptor. Epitopes in pre-S1 displayed on the outside of surface antigen can also elicit virus-neutralizing antibodies (115, 116, 247) and alter the host range of the virus upon genetic recombination (97).
Besides its role in infection, the pre-S1 domain provides the ligand for core particles during the assembly of the viral envelope (see below), which requires display of pre-S1 epitopes on the cytosolic side of the viral membrane. As a consequence of this dual function, a fraction of L polypeptides must maintain the pre-S1 domain in the virus interior while the remaining L polypeptides undergo a conformational shift to transfer the receptor binding pre-S1 region across the lipid bilayer to the outside of the virus particle (24, 72). This transfer appears to occur as viruses mature during their passage along the secretory pathway. The function of the M polypeptide is not well understood, and the pre-S2 domain is apparently not essential for virus infection of hepatocyte cultures (58).
In addition to glycosylation, L protein of HBV is modified by N-terminal myristylation (114). Myristylation is not required for efficient virion assembly but is required for infectivity (23, 66).
The avihepadnaviruses encode only two envelope proteins, analogous to S and L. Glycosylation of these proteins has not been detected (174). The pre-S domain is phosphorylated and is also myristylated at its N terminus. Phosphorylation has not been shown to play a role in envelope assembly and infectivity (18). As with HBV, myristylation is not required for DHBV assembly but is required for infectivity (133).
Unique to orthohepadnaviruses is the presence of a fourth gene, termed X. The function of X during a natural viral infection is not known. As mentioned above, X is essential for virus replication in animals but dispensable for viral DNA synthesis in transfected tissue culture cells (32, 254). Evidence that the gene is expressed in vivo stems from the detection of antibodies present in sera of HBV-infected humans and naturally infected animals (166, 223) and from protein analysis of liver samples obtained from WHV-infected woodchucks (44). In vitro, X exhibits a plethora of activities. From cell culture studies, it is believed that X can activate the transcription of host genes, including the major histocompatibility complex (94, 219, 251) and c-myc (4), as well as viral genes (37, 192), and one investigator has even reported that X stabilizes viral RNAs (186). Most investigators agree that X is not a DNA binding protein and that it is therefore not a typical transactivator. Thus, its effects on transcription are thought to be indirect. Suggestions have included interacting with and altering the DNA binding of cyclic AMP-responsive element binding protein and ATF-2 (134), activating NF-κ B (218), and contacting basal transcription factors (80).
Aside from the transactivation of many promoters, activities linked to X include stimulation of signal transduction (14) and binding, to various degrees, to well-known protein targets such as p53 (57, 206, 229), proteasome subunits (190, 205), and UV-damaged DNA binding protein (119). Many different explanations for these and other effects of the X protein have been proposed. Whether all or any point to the role of X in the virus life cycle is unknown. It is clear that X-defective virus is unable to initiate infection in vivo (32, 254). However, the physiological role of X during the course of an infection remains a major unresolved issue in hepadnavirus biology.
Assembly and Reverse Transcription
Inferring from genetic experiments, it has been suggested that translation of the polymerase and pgRNA packaging into viral nucleocapsids are tightly coupled events (6, 84, 101). These experiments revealed not only that the reverse transcriptase is required for RNA packaging but also that packaging occurs in cis. It is now well established that the polymerase binds to an RNA stem-loop structure at the 5′ end of pgRNA, termed epsilon, and that this event triggers the sequestration of viral RNA and polymerase into core particles (167, 226) (Fig. 2 and 3).
RNA packaging also depends on host factors, in particular polypeptides belonging to the molecular chaperone complex of hsp90 (92). hsp90 appears to stabilize a transient conformation of the polymerase that facilitates the binding to epsilon on pgRNA. Thus, drugs that interfere with hsp90 function, such as the ansamycin geldanamycin, inhibit RNA packaging and consequently the replication of both DHBV and HBV (93).
In addition to its role in RNA packaging, epsilon plays a major role in the initiation of viral DNA synthesis. This reaction is unique to hepadnaviruses, since it uses the reverse transcriptase itself as a protein primer for DNA synthesis (118, 207, 225). A hydroxyl residue of a tyrosine located near the amino terminus of the polymerase polypeptide serves as a substrate for the formation of a phosphodiester linkage with dGMP (230, 255). The template for this reaction is located in the proposed bulge region of epsilon RNA that is believed to separate the lower and upper stem regions (168, 224). The priming reaction terminates following the synthesis of only 4 nucleotides, but it somehow triggers the transfer of the nascent DNA strand to the 3′ end of the pgRNA, where the 4 nucleotides base pair with complementary sequences (Fig. 3). Exactly how this transfer occurs is not known, since information about the spatial organization of nucleic acids and polymerase inside the core particles is not available.
Plus-strand priming, like minus-strand priming, also involves a transfer reaction. In this case, an RNA oligomer derived from the 5′ end of pgRNA recombines with an internal acceptor site on minus-strand DNA (123, 179). Annealing of the primer with complementary sequences at the acceptor is possible due to an 11- to 12-nucleotide sequence homology referred to as direct repeats 1 and 2 (DR1 and DR2) (Fig. 3). The 3′ end of the RNA primer most probably represents the final RNase H cleavage site on pgRNA (194). Plus-strand DNA synthesis involves a template switch from the 5′ end to the 3′ end of minus-strand DNA, which is facilitated by short sequence repetitions at the ends of the DNA template (79). The final result of the reverse transcription reaction is an rcDNA genome with two modified 5′ ends and, in mammalian viruses, with a less-than-genome-length plus-strand DNA, leaving up to 50% of the rcDNA single stranded (89, 117, 131, 199). With DHBV, the plus strand is more nearly complete in viral rcDNA, with completion apparently blocked at DR2 because of the RNA primer of plus-strand synthesis that is annealed to the minus strand (124).
In approximately 10% of cases, the transfer of RNA primer from DR1 to DR2 does not occur, leading to an in situ priming reaction (194). The result is a double-stranded linear DNA (dslDNA) genome rather than rcDNA (Fig. 3). Although the significance of dslDNA in a natural infection is not known, genetic experiments showed that variants of DHBV producing only dslDNA can infect primary hepatocyte cultures and ducks. cccDNA formation seems to occur through illegitimate recombination of the ends of dslDNA (242).
Amplification of cccDNA and Production of Infectious Virus
The most intriguing aspect of the hepadnavirus replication cycle is the regulation of cccDNA synthesis. It is the basis for the establishment and persistence of hepadnaviruses in infected hepatocytes and consequently can be expected to play a major role in recovery from an infection. The concept that cccDNA synthesis is regulated is consistent with the observation that the nuclei of DHBV-infected hepatocytes contain on average 10 to 50 copies of cccDNA while 10 times as much of the rcDNA precursor to cccDNA may reside in the cytoplasm. In addition, amplification of cccDNA levels takes place in the first few days following infection of primary duck hepatocyte cultures with DHBV (217). By using DHBV variants that fail to express the large envelope polypeptide, Summers et al. showed that cccDNA accumulation was indeed a regulated process (200, 201). In the absence of envelope proteins, accumulation continued to levels toxic to primary hepatocyte cultures as well as hepatocytes in vivo (121, 201). These experiments pointed to a role for the envelope in the regulation of cccDNA. A direct implication of such a model is that cccDNA amplification preferentially occurs during the early phase of an infection, when the concentration of the envelope is still low. Once higher levels are reached, subviral core particles are segregated into the secretory pathway through their interaction with envelope polypeptides residing in membranes of the endoplasmic reticulum (ER). Of note is the general observation that virion formation depends on viral DNA synthesis, i.e., that cores have to “mature” in order to interact with viral envelope to form virions. Thus, virus is enriched for the partially double-stranded rcDNA while infected cells contain approximately equal amounts of partially double-stranded and single-stranded DNAs (198, 231). The molecular signals that emerge on the surface of cores and presumably regulate the interaction with envelope components leading to virion formation are unknown. Changes in the phosphorylation of the DHBV core protein are believed to play a role, since core protein in viral particles has a different phosphorylation pattern than core protein within cells (173). However, mutation from serine to alanine at one of the phosphorylation sites (Ser-259) that is apparently essential for the intracellular pattern (173) did not block virus assembly, although the resultant virus was unable to infect primary duck hepatocyte cultures (245, 246). In brief, there is still no evidence that the interaction between nucleocapsids and envelope proteins is actually regulated by changes in core protein phosphorylation.
Binding of cores to the N-terminal domain of the large envelope polypeptides results in the translocation (budding) of the core particles across the ER membrane. Enveloped particles containing all three envelope proteins are thought to be transported through the ER into the Golgi complex (96). Glycosylation at an asparagine residue located in the S domain of the envelope proteins occurs during this phase of the assembly process, which is completed with the secretion of mature virions into the bloodstream.
TRANSIENT INFECTIONS
The majority of hepadnavirus infections (90 to 95%) in adults are transient, while ∼90% of perinatal infections are chronic (195). One view of the immunoregulation of viral infections, generally applied to infections by cytocidal viruses, is that CTL responses quickly recognize epitopes of early viral proteins that are displayed by major histocompatibility complex type I on the surface of infected cells. Many cells are then killed before virus production can initiate, thereby preventing the spread of viruses to other susceptible cells. Further control of virus spread is provided by a rapid production of virus-neutralizing antibodies, which serve to protect susceptible cells. Local protection of susceptible cells is also provided by interferons and other cytokines, which may induce resistance to infection in nearby cells. In brief, the infection is prevented from spreading to all potential targets.
This model is not easily adapted to transient hepadnavirus infections, which generally run a course of 1 to 6 months, including an asymptomatic incubation period, often accompanied by a high-titer viremia (up to 1010 per ml), which may itself last several weeks. During this period, the immune system is either unable or not activated to gain control of the infection, and the entire hepatocyte population of the liver may be infected. The lack of symptoms in HBV-infected patients for the first few weeks suggests that activation of the immune response is delayed, although there are very few objective data on which to base this conclusion. Thus, the virus may spread from only a few hepatocytes to involve, in at least some cases, the entire hepatocyte population of the liver (5, 15, 71, 88, 100, 102, 171). The virus is then rapidly cleared from the serum, often in a few weeks, with either coincident or delayed clearance of infected hepatocytes from the liver.
Hepadnavirus infections, and transient infections in particular, therefore raise difficult issues. First, why is there not a more effective control of virus spread during the early period? Second, how is the immune system later able to clear an infection, after it has spread to essentially the entire hepatocyte population? A possible immunosuppressive role of the e-antigen has been suggested (144, 145) but has not yet been proven in an experimental model of infection. The concept that e-antigen is immunosuppressive in a natural infection is consistent with the observation that ducks infected with DHBV that is unable to produce e-antigen develop and sustain higher antibody titers to viral core antigen than do ducks infected with wild-type DHBV (250; J. Summers, personal communication). This may happen because of suppression of the antibody response by e-antigen, which is cross-reactive with the core, or because infections with e-antigen-deficient strains produce more liver disease than do infections with wild-type strains, thereby increasing the amount of antigen stimulation. Both mechanisms would be consistent with the proposal that e-antigen functions to suppress the antiviral immune response.
The second issue is equally difficult. How is the immune system finally activated, and virus eliminated, after infection of the entire hepatocyte population? In approaching this issue, it is necessary to at least take account of various beliefs and observations about the virus and the host. (i) Hepatocytes have, as mentioned above, a very long half-life (6 to 12 months or more). Indirect evidence suggests that the hepatocyte half-life may be as little as 10 days in some chronically infected patients with active liver disease (154). However, even this high a turnover would not by itself be sufficient to explain the clearance of infected cells during resolution of transient infections (see below). (ii) Viral cccDNA is thought to be stable in infected and nonproliferating hepatocytes (130, 146; however, see reference 35), although virus expression can be suppressed and degradation of viral proteins and nucleic acids in the cytoplasm can be induced by certain cytokines (68). (iii) Clearance of virus from the liver during recovery from a transient infection does not appear to involve the replacement of the infected cell population by proliferation and differentiation into hepatocytes of a hypothetical population of uninfected progenitor cells. In particular, periportal hepatocytes, located near the putative origin of progenitor cells at or adjacent to bile ductules, can be the last in the lobule to clear an infection (99, 102). (iv) Massive cell destruction has not so far been linked to virus clearance even after the entire hepatocyte population appears to have been infected, although it has also not been definitively ruled out. (v) Hepatocytes can actively proliferate to maintain liver cell mass.
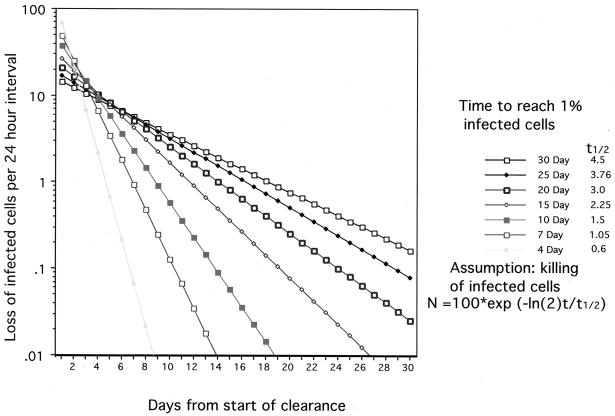
If we assume that clearance involves replacement of infected hepatocytes via proliferation of initially rare uninfected hepatocytes and that infected cells do not divide, it is possible to generate a reasonably simple model for clearance. In particular, if elimination of infected cells occurs by CTL killing via a first-order reaction, a series of curves can be drawn relating the kinetics of clearance to the rate of hepatocyte replacement (Fig. 5). Thus, a single time point determination of the fraction of infected cells and the rate of liver cell regeneration during the clearance phase will yield an estimate of the half-life for infected hepatocytes. The first measurement is fairly straightforward. While there is no simple way to precisely measure rates of cell turnover, it is possible to estimate this by counting mitotic figures, apoptotic cells, or proliferating-cell nuclear antigen-positive hepatocytes or by in vivo pulse-labeling with bromodeoxyuridine (BUdR).
FIG. 5.
CTL killing and hepatocyte replacement. The kinetics of clearance of infected hepatocytes have been calculated by assuming that CTL killing is a first-order reaction. Each point indicates the fraction of the initial number of infected hepatocytes that were killed in the preceding 24-h period. The different curves were determined using t1/2 values for killing that would produce 99% loss of infected hepatocytes in the indicated times.
If this model were correct and if complete clearance (e.g., to less than 1% infected hepatocytes) occurred in less than 15 to 30 days (Fig. 5), as suggested by studies with the woodchuck and duck models (100, 102, 171), the half-life of infected hepatocytes could be only 2.25 to 4.5 days or less during the clearance phase. Thus, extremely high levels of hepatocyte loss and replacement would take place only for a short period, perhaps a week or less, at the times when significant numbers (e.g., >10%) of the hepatocytes were still infected. To our knowledge, such measurements have not been made in patients recovering from transient HBV infections. In our own experiments with woodchucks and ducks, we have obtained liver biopsy specimens during the partial clearance phase of an infection only three times (73, 100, 102). In one example, a liver biopsy specimen taken during the recovery phase of the infection revealed ∼20% infected cells and a rate of hepatocyte replacement (and, by inference, cell death), estimated from BUdR and proliferating-cell nuclear antigen labeling indices, of ∼3 to 6% per day. Such a rate would be compatible with half-lives of ∼2.25 to 4.5 days for infected hepatocytes. These rates would be sufficient to produce a 99% decline in the number of infected hepatocytes in 2 to 4 weeks.
Unfortunately, the actual rate of virus clearance is unknown and methods for accurately determining rates of cell replacement are not available. In addition, this simple model does not take account of recent observations made with an HBV transgenic mouse. These studies revealed that cytokines, particularly alpha and gamma interferons and tumor necrosis factor alpha (TNF-α) play key roles in pathways that induce the apparently noncytocidal loss of virus replication intermediates, including proteins, replicating DNA, and RNAs, from nonproliferating hepatocytes (26, 68, 216). Analogous effects on cccDNA, if any, were not identified, since cccDNA is not synthesized in this transgenic mouse. Thus, although hepatocyte death appears to be a characteristic feature of transient infections, there is at least a possibility that virus is completely eliminated without this killing. Recent data on HBV infections of chimpanzees appear consistent with this possibility (71).
The idea that virus can be lost from hepatocytes is also consistent with studies of WHV infections suggesting that infected hepatocytes, which were BUdR pulse-labeled at the peak of a transient infection, were still present after the virus had been eliminated (102). However, another explanation of this result is that the BUdR-labeled hepatocytes lost the virus due to a combination of cytokine-induced destruction of replication intermediates, which form the precursors to cccDNA, and the loss of cccDNA itself during cell division. One of the unknown factors is whether cccDNA is passed to daughter cells when hepatocytes divide.
If cccDNA is lost during mitosis or passes inefficiently to daughter cells, it is possible to generate models for virus clearance in which infected hepatocytes are lost with less cumulative cell death than in the model described above, in which all infected hepatocytes (up to 100% of hepatocytes) are destroyed. Thus, if both uninfected and infected hepatocytes divide to replace the infected hepatocytes that are killed by CTLs and if infected hepatocytes are no longer infected as a result of cell division, a 99% loss of infected hepatocytes will occur after only 70% are killed by CTLs (139). However, 70% cell replacement in only a few weeks is still a major change compared to the quiescent nature of the healthy liver. Thus, unless there is a novel mechanism for virus elimination from quiescent cells, cell death plays an obligatory role. This conclusion, if correct, has major implications not just for transient infections but also for the design of antiviral treatments to terminate chronic infections.
Irrespective of the mechanism of virus elimination, an obvious question remains. What triggers an effective antiviral response after a delay of many weeks or even months (158)? Most probably the delay is due, in part, to interactions between the immune system and the liver. In particular, similarly prolonged time courses and extensive infection of the hepatocyte population are observed with two other liver-specific viruses, hepatitis A virus (a picornavirus) and hepatitis E virus (a calicivirus), neither of which produces a chronic infection. Hepatitis delta virus follows a similar time course as well, although its life cycle is more complicated, since it depends upon HBV for its envelope proteins. It has been proposed that the liver may suppress CTL responses in favor of antibody production, since the liver plays a major role in antibody-mediated protection against prokaryotic and eukaryotic pathogens. However, whether this explains the delay in mounting a successful antiviral response is not yet known.
CHRONIC INFECTIONS
Chronic infection is defined as the persistent presence of HBsAg in the serum of an individual for 6 months or longer (52). Commercial assays can detect as little as a few nanograms of HBsAg per milliliter of serum, and most chronically infected individuals have titers exceeding 1 μg per ml (53). Thus, the assay has the advantage of great sensitivity. One disadvantage of defining chronic infection on the basis of this assay alone is that HBsAg may be produced at high levels even if virus replication in the liver has virtually ceased. The probable reason for this continued HBsAg production in the absence of detectable levels of virus replication is that viral DNA may integrate into host chromosomes during the course of infection and provide a template for transcription of HBsAg mRNAs. The predominant precursor to integrated DNA is likely to be the linear double-stranded DNA produced as a result of in situ priming (63, 64, 241, 242). The result of integration is that viral genes, particularly for the core and polymerase, may be disrupted (Fig. 4) while the coding regions and promoters for the envelope proteins remain intact. (Integrated DNA may also be highly rearranged. It is unclear if this rearrangement occurs before or after integration or if both are possible [159, 177].) Thus, chronic infection as defined by HBsAg production may encompass a broad spectrum of virologic and pathogenic states.
It is generally the case that HBsAg carriers who no longer make virus that is detectable by conventional hybridization assays (e.g., >105 to 106 per ml of serum) also no longer have active liver disease, defined by ongoing infiltration of leukocytes and active destruction of hepatocytes. Viremic individuals with the highest titers (109 to 1010 per ml) usually are thought to have a low immunologic response to the viral antigens and a low disease activity, while those with somewhat lower titers (107 to 108 per ml) appear to have the highest disease activity.
Chronic infection begins when the immune response that normally clears the infection fails to take place or is too weak to be effective. Thus, infections are almost always chronic following exposure of children younger than 1 year or of immunocompromised individuals. Chronic infections also occur in about 5% of otherwise healthy adults. Patients infected as adults often have the most severe and progressive liver disease (142). These patients may also experience recurrent bouts of acute liver disease, possibly because their immune system attempts to produce the response that normally leads to virus clearance (86, 127).
The prognosis for many chronic carriers of HBV who were infected with HBV as adults is poor. About 10 to 25% will die of either liver cancer or cirrhosis (tissue scarring). Smaller numbers, particularly those infected as adults, will die or require liver transplantation due to rapidly progressing liver disease leading to hepatic failure. Attempts at antiviral therapy to eliminate the virus and reduce disease activity have had the highest success rate in individuals with active, immune system-mediated liver disease (22, 87). This is probably because their immune system is already close to producing a response leading to viral clearance. Antiviral therapy to eliminate the virus has not been as successful for the vast majority of viremic carriers, probably because their immune system mounts a weaker response to the infection. Immune system-mediated liver disease may be less active in these carriers because infection occurred during birth or in the first year or two of life, producing a degree of tolerance to the viral antigens. Unfortunately, most carriers fall into this group, which therefore represents the largest target for antiviral intervention. In these carriers, the interval between initiation of the HBV infection and the peak incidence of HCC is ∼30 to 50 years (11, 12).
Interestingly, the majority of untreated carriers, who were infected early in life, do not die of liver disease or liver cancer, and, indeed, cancer incidence may decline as carriers age (11, 12). One explanation is that some carriers differ in their risk due to their genetic makeup. Indeed, a case-control study carried out in China indicated that a family history of HCC was a risk factor for HCC among HBV carriers (129). One study even suggested that carriers lacking the gene for glutathione S-transferase M1 might be at increased risk for HCC, possibly because of increased sensitivity to DNA adduct formation by aflatoxin B1 (141).
Another factor that has been considered to increase the risk of liver disease and HCC is persistent high-titer virus replication. The incidence of high-titer virus production (where the virus is readily detectable by nucleic acid hybridization techniques [e.g., >106 per ml of serum]) declines with increasing age, at least among individuals infected in childhood (53). For instance, about 25% of male carriers in China maintain high-titer virus production past the age of 30 years whereas virtually none do in Senegal. Moreover, the HCC risk in the Chinese population was about 10-fold higher than in the Senegalese cohort. Thus, a correlation between HCC and prolonged high-titer virus replication might be deduced. Unfortunately, although the correlation may seem correct when populations are examined, it was not observed among individuals who were first investigated near the time of the HCC diagnosis, even when the sensitivity of the virus assay was increased through PCR. The hypothesis remains attractive because antiviral therapy would also prevent liver cancer if the risk of HCC were related to the duration of high-titer virus replication.
There are currently two approved therapeutic agents for chronic HBV, alpha interferon and lamivudine (a cytosine analog). Alpha interferon has been used for more than a decade. It probably acts directly to suppress virus replication within hepatocytes by inducing the degradation of viral mRNAs (69) and indirectly to stimulate the immune system. Indeed, as noted above, patients with an active liver disease due to their immune response to HBV are the best candidates for successful therapy with alpha interferon (22, 165, 234). A probable reason for the failure of alpha interferon therapy in most other carriers is that their ongoing immune response to the infection is quite low (128).
Recently, lamivudine has been approved for treatment of HBV carriers. In theory, suppression of virus replication for a sufficiently long time should lead to clearance as infected hepatocytes and other reservoirs for the virus die off. In practice, lamivudine-resistant variants may replace wild-type virus after a year or more of therapy (3, 8, 9, 31, 125, 152, 154, 213). Lamivudine-resistant variants of the HBV polymerase are sensitive to adefovir, another nucleoside analog inhibitor of viral DNA synthesis that is now undergoing clinical evaluation, and the two may prove useful in combination (236).
Interestingly, lamivudine alone can induce a rapid reduction in symptoms of liver disease even though the virus is still present in the host (154). It is not known whether this reduction in disease activity is due to a decline in the number of productively infected hepatocytes as targets for the immune system or whether it occurs because viral DNA replication per se increases the susceptibility of hepatocytes to injury. Another hypothetical explanation is that lamivudine alters the immune response to HBV, as suggested for the therapeutic effects of the nucleoside ribavirin (153) in the treatment of chronic hepatitis C. Reports that several months can elapse between the appearance of low levels of lamivudine-resistant HBV in the serum and a major elevation in HBV titers suggest that the spread of the mutants in the liver may be blocked by wild-type HBV through a superinfection resistance mechanism (31). In other words, the therapeutic effect is not dependent on clearance of HBV from hepatocytes.
LIVER CANCER
Epidemiological studies established a link between HBV infection and liver cancer. In humans, the lifetime risk for a chronically infected person is approximately 10 to 25%. In woodchucks chronically infected with WHV, the lifetime risk is essentially 100%. An obvious question raised by these observations concerns the role of the virus in liver cancer. Although this question has been addressed experimentally in a variety of settings over the past two decades, the amount of solid information is still disappointingly sparse. One exception is the work by Buendia and colleagues on the genetic analysis of tumors from woodchucks, which implicated the myc family of genes as contributors to liver cancer (for a review, see reference 181).
The quest to unravel the mechanism of HBV induced liver cancer was originally spurred by reports from a number of laboratories (20, 27, 51, 136) that HBV DNA was present in high-molecular-weight DNA extracted from HCC or from cell lines derived from HCC. Since the pattern of DNA integration was random with regard to the cellular DNA flanking the viral sequences, it was concluded that integration occurs prior to or at the beginning of the expansion of the tumor. By analogy to the model of oncogene activation by insertional mutagenesis, coined by Hayward et al. (81) and Payne et al. (164), it was hypothesized that HBV could act as an insertional mutagen, causing the activation of a proto-oncogene. Results obtained with the woodchuck model supported this hypothesis. They revealed that WHV sequences are present at or near N-myc2, a pseudogene of N-myc, in almost every liver tumor arising in WHV-infected animals (59, 60). In almost all cases examined so far, integration of WHV results in activation of transcription of the N-myc2 locus. In the few cases where N-myc2 is not transcribed, WHV integration is found at the c-myc locus (91) or the N-myc1 locus (60). In contrast to the observations with woodchucks, similar efforts with tumors obtained from HBV-infected patients did not reveal a common cellular target for HBV integration. Over the past 20 years, only a handful of “interesting” HBV insertions have been reported. They were found adjacent to or within the genes encoding cyclin A and the retinoic acid receptor beta (46, 227, 228). However, these findings have not been generalized beyond the original tumor samples and thus did not represent the hoped-for bonanza concealing a general mechanism for HBV-induced oncogenesis.
There are other obvious differences between WHV and HBV associated cancers. First, liver cancer in humans is almost always associated with cirrhosis whereas in woodchucks fibrosis of the liver is uncommon. Second, the time for the development of liver neoplasia in humans is on the average 20 to 40 years whereas in woodchucks it is only 1 to 3 years.
An important factor to be considered in models for cancer is the frequency of cell regeneration. Regeneration of hepatocytes is a direct consequence of cell killing caused by the immune response of the host to the infection. Cell killing occurs by apoptosis of hepatocytes or by necrosis due to liver inflammation. As mentioned above, HBV-infected patients can exhibit severalfold-increased rates of hepatocyte turnover compared to uninfected healthy people.
What, then, are other factors that could play a role in hepatocarcinogenesis? As the search for common cellular targets for HBV DNA integration remained unproductive, two alternative models that emerged from research on retroviruses and colon cancer were adapted to explain HBV-induced oncogenesis. The first model predicted that HBV, like certain tumor viruses, contains an oncogene. The second model invoked a role for tumor suppressor genes, in particular p53, in HCC formation.
The major prediction of the oncogene model is that expression of one or several HBV gene products induces malignant transformation in tissue culture cells or in transgenic mice. Transformation of cells expressing intact hepadnavirus genomes has generally not been observed. An exception is a report by Hohne et al. (85), who transfected a nonmalignant immortalized mouse hepatocyte line harboring metallothionein promoter-driven simian virus 40 large tumor antigen with HBV DNA. They observed that all transfected clones which replicated HBV displayed malignant growth characteristics in soft agar and were tumorigenic upon inoculation in nude mice. A notable difference between these cell lines and infected liver tissue was that the cell lines expressed a transcript corresponding to the viral X gene at much higher levels than were observed under physiological conditions in vivo. Indeed, subsequent studies by the same laboratory with the same cell line showed that overexpression of the viral X gene was required for cell transformation (183).
Expression of the viral X gene can also disturb normal cell growth in transgenic mice. For example, overexpression of HBx in transgenic CD-1 mice, which are susceptible to the development of spontaneous hepatomas, increased the frequency of liver tumors at least 10-fold compared to that in normal mice of the same strain (105, 109). In contrast, transgenic mice expressing lower, most probably more physiological, levels of X did not develop hepatomas at an increased rate compared to normal CD-1 mice (109). Also, another strain of transgenic mice (ICR × B6C3F1) expressing X did not develop hepatic tumors, and, most importantly, hepatomas have not yet been observed in transgenic mice (C57BL/6 and B10.D2) expressing replication-competent HBV genomes (120). Several laboratories have reported that transgenic mice expressing X exhibit an increased rate of liver cancer formation when exposed to known carcinogens such as diethylnitrosamine or p-dimethylaminoazobenzene (50, 191). Similarly, X has been shown to reduce the latency period for HCC development in transgenic mice expressing a woodchuck c-myc gene (209).
Thus, most of the experimental evidence that has accumulated so far suggests that X is not, by itself, the cause of HBV-associated carcinogenesis. Indeed, it should be kept in mind that the effects of X may be strongly dependent on the experimental system, which in no case was the natural host for HBV infection. For example, Kim et al. showed that in another experimental system, X induced apoptosis rather than cell growth (106). It therefore remains possible that X, particularly if overexpressed, is a contributing factor in tumor promotion. It is also quite possible that other viral gene products can exhibit a tumor-promoting activity in hepatocytes that are exposed to an altered environment, as occurs in a chronically infected liver.
Different from the observations reported for HBx, overexpression of the large envelope protein of HBV in transgenic mice causes hepatocyte injury and death and induces liver regeneration. These mice ultimately develop HCC (34). Injury is believed to be due to the accumulation of filamentous aggregates formed by L and HBsAg in the ER of hepatocytes, resulting in a storage-like disease, such as observed in patients with α1-antitrypsin deficiency or in mice expressing mutant α1-antitrypsin. Since L protein in natural infections does not accumulate to levels observed in transgenic mice, it is unlikely that this model is relevant for liver cancer in chronic infections. Surface antigen filaments can overaccumulate in a fraction of infected hepatocytes, sometimes known as ground-glass hepatocytes, but this contribution to liver injury is probably small compared with the injury caused by the antiviral immune response.
The second model predicting that HCC development proceeds in multiple steps is based on the identification of specific lesions that are visible in livers of patients with HCC as well as in animal models of liver cancer. Such lesions include altered hepatic foci, dysplastic (neoplastic) nodules, and low- and high-grade HCCs exhibiting different levels of cell differentiation. It is generally believed that the development of carcinomas progresses through these individual stages, as described for the progression of colon cancer. While the progression of neoplasia in colon cancer has been characterized on the molecular level, i.e., through the correlation of phenotypic alterations with mutations or deletions in specific genes, a molecular characterization of liver cancer is still at an early stage (107).
The major focus, so far, has been on the genetic analysis of the tumor suppressor p53. Interest in this gene has been spurred by two reports demonstrating that p53 mutations were present in approximately half of HCC samples obtained from HBV-infected patients in southern Africa and Qidong in China (21, 90). Most importantly, in 80 to 90% of the cases with p53 mutations, the G residue of codon 249 was changed to T. Such G-to-T transversions are known to occur by guanosine adduct formation caused by certain chemical carcinogens such as aflatoxin B1. Patients from both areas could have been exposed to aflatoxins through contaminated foods. Consistent with a role of environmental factors in p53 mutagenesis, several other reports indicated that the incidence of p53 mutations in HCC is generally much lower, ranging from 12 to 30% (25, 150). These numbers are below the rate observed in other human cancers, where p53 mutations are estimated to occur in roughly 50% of all cases (65). Whether p53 mutations are generally lower in HCC than in other malignancies or whether the lower incidence is characteristic of HBV infection is an important unanswered question. Observations made with HCC samples from HCV-infected individuals indicated that the relatively low rate of p53 mutations in HCC is not dependent on the etiology of the disease. For example, Pontisso et al. found p53 mutations in less than 10% of HCC samples obtained from HCV-infected patients of Caucasian background (169). Similarly, Sheu et al. found p53 mutations in approximately 25% of HCC samples obtained from Taiwanese patients that were negative for HBsAg but positive for HCV markers (185).
In agreement with observations made in colon and other cancers, p53 mutations in HCC are considered late events associated with a dedifferentiated phenotype of the tumor (107). For example, a report by Oda et al. showed that tumor nodules growing out of a parent nodule (nodules-in-nodule) exhibited distinct p53 genotypes from each other (157).
Information about early markers for HCC development is still scanty, mainly because liver cancers are generally diagnosed during the late phase of the disease. However, in humans as well as in the woodchuck model, insulin-like growth factor II (IGFII) is activated in altered hepatic or dysplastic foci as well as in HCCs (47, 240). Consistent with its role as a target for WHV integration, N-myc-2 can also be activated in altered hepatic foci in WHV-infected woodchucks (238). Although it is not known whether IGFII plays a role in HCC development, it is conceivable that it promotes cell proliferation in an autocrine fashion (238). On the other hand, IGFII expression may be required to suppress apoptosis induced by activated myc genes, including N-myc-2 (220, 239).
MAJOR UNRESOLVED ISSUES
During the past two decades, the major principles of hepadnavirus infections have been resolved. Infectious viral genomes have been cloned and sequenced, the major viral gene products have been characterized, and the mechanism of viral DNA replication has been uncovered. In addition, the course of viral infections in vivo has been characterized with the help of animal models and, to some extent, with samples obtained from HBV-infected patients. In spite of these accomplishments, many gaps in our knowledge of the viral life cycle still exist.
On the level of viral replication, two major unresolved issues concern early events of the replication cycle and the function of the X gene. As alluded to above (see “Infection”), the viral receptor is not known. It is generally believed that the viral receptor is a major determinant for the observed tissue tropism of hepadnaviruses. Similarly, it is believed that the resistance of tissue culture cell lines to viral infection is due to the absence of the receptor on the surface of these cells. A major impediment to the isolation of the receptor lies in the difficulty of constructing a replication-competent hepadnavirus vector with a gene that can be used for positive selection in mammalian cells. This is mainly due to the small genome size and the compactness of cis-acting sequences dispersed over the entire genome. Such recombinant viral vectors have been essential for an unambiguous identification of retroviral receptors including those for Moloney murine leukemia virus, avian leukosis virus, and human immunodeficiency virus (1, 43, 140, 244). Recently, however, the production of HBV and DHBV variants expressing green fluorescent protein or gamma interferon has been reported (172). An alternative approach could be the production of pseudotypes with viruses such as vesicular stomatitis virus or certain retroviruses carrying hepadnavirus envelope components that could be used for the identification or selection of cells expressing the viral receptor(s), as shown for the identification of CD4 as the receptor for human immunodeficiency virus.
Using a biochemical approach, Kuroki et al. (112, 113) identified a novel carboxypeptidase as a potential DHBV receptor. The polypeptide was shown to interact with both virions and recombinant envelope polypeptides and could be blocked with neutralizing antibodies. While these findings have been confirmed by other investigators (214, 222), the observation that carboxypeptidase is expressed in tissues that are resistant to DHBV infection has been interpreted to mean that additional factors control viral infection. This view may be correct, but the possibility that events subsequent to viral attachment and uptake control the host range should also be considered. For example, transgenic mice expressing HBV do not accumulate cccDNA, suggesting that the pathway responsible for the conversion of rcDNA into cccDNA is defective in these animals (70). Moreover, duck primary hepatocyte cultures maintained in the presence of glucagon develop an intracellular block to cccDNA formation (83).
As discussed above, the role of the viral X protein for viral replication in vivo remains as a major unsolved problem. Why is this gene not necessary for the replication of the avihepadnaviruses? At what stage of the replication is X required? Since cloned hepadnavirus DNA is infectious when inoculated into the livers of susceptible animals, X is not required for the initial transcription of viral mRNAs (180, 233). Consistent with this view, we have not been able to demonstrate the presence of X protein in extracts from purified virions obtained from sera of WHV infected woodchucks (E. Leber, G. Chen, and C. Seeger, unpublished observations). This problem could be addressed with the help of primary hepatocyte cultures that are permissive for infection. Unfortunately, with WHV, for which primary hepatocyte cultures are readily available, the execution of this important experiment has been stymied by the inability to produce sufficient amounts of infectious virus in transfected cell lines. This is not a problem with HBV, but the difficulty of obtaining susceptible primary human hepatocyte cultures is a major impediment.
On the level of host-virus interactions, the major unresolved issues concern the mechanism by which the virus is eliminated from the liver. Evidence for the existence of cellular factors that can suppress virus replication has been obtained from the analyses of human as well as chimpanzee HBV carriers during superinfection by other hepatotropic viruses, including HAV, HCV, and HDV (49, 215, 248). In several of the cases investigated, HBV replication was suppressed during recovery from HAV infection. Similarly, a reduction in viral titers was found in chronic WHV carrier woodchucks infected with HDV (147). These results led to the speculation that certain cytokines could activate antiviral pathways. This view has subsequently been supported by experiments with HBV transgenic mice. They have shown that TNF-α can reduce the half-life of viral mRNAs and that gamma interferon and TNF-α can induce the degradation of viral DNA replication intermediates (68, 69). However, the mechanism responsible for the apparent depletion of intracellular virus particles is not yet known. One possibility is that certain cytokines activate the cellular pathway(s) used for the disintegration of core particles following virus penetration. Unfortunately, very little is known about the biochemical steps that are responsible for virus disassembly. Research aimed at resolving this interesting problem could provide the basis for the future design of effective antiviral therapies required to cure the enormous number of patients with chronic HBV infections.
ACKNOWLEDGMENTS
We are grateful to W. T. London for critical reading of the manuscript and to Jesse Summers (University of New Mexico, Albuquerque) for helpful discussions.
Work in our laboratories is supported by grants from the National Institutes of Health and by an appropriation from the Commonwealth of Pennsylvania.
REFERENCES
- 1.Albritton L M, Tseng L, Scadden D, Cunningham J M. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell. 1989;57:659–666. doi: 10.1016/0092-8674(89)90134-7. [DOI] [PubMed] [Google Scholar]
- 2.Aldrich C E, Coates L, Wu T T, Newbold J, Tennant B C, Summers J, Seeger C, Mason W S. In vitro infection of woodchuck hepatocytes with woodchuck hepatitis virus and ground squirrel hepatitis virus. Virology. 1989;172:247–252. doi: 10.1016/0042-6822(89)90126-8. [DOI] [PubMed] [Google Scholar]
- 3.Allen M I, Deslauriers M, Andrews C W, Tipples G A, Walters K A, Tyrrell D L, Brown N, Condreay L D. Identification and characterization of mutations in hepatitis B virus resistant to lamivudine. Lamivudine Clinical Investigation Group. Hepatology. 1998;27:1670–1677. doi: 10.1002/hep.510270628. [DOI] [PubMed] [Google Scholar]
- 4.Balsano C, Avantaggiati M L, Natoli G, De M E, Will H, Perricaudet M, Levrero M. Full-length and truncated versions of the hepatitis B virus (HBV) X protein (pX) transactivate the cmyc protooncogene at the transcriptional level. Biochem Biophys Res Commun. 1991;176:985–992. doi: 10.1016/0006-291x(91)90379-l. [DOI] [PubMed] [Google Scholar]
- 5.Barker L F, Chisari F V, McGrath P P, Dalgard D W, Kirschstein R L, Almeida J D, Edgington T S, Sharp D G, Peterson M R. Transmission of type B viral hepatitis to chimpanzees. J Infect Dis. 1973;127:648–652. doi: 10.1093/infdis/127.6.648. [DOI] [PubMed] [Google Scholar]
- 6.Bartenschlager R, Junker-Niepmann M, Schaller H. The P gene product of hepatitis B virus is required as a structural component for genomic RNA encapsidation. J Virol. 1990;64:5324–5332. doi: 10.1128/jvi.64.11.5324-5332.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartenschlager R, Schaller H. Hepadnaviral assembly is initiated by polymerase binding to the encapsidation signal in the viral RNA genome. EMBO J. 1992;11:3413–3420. doi: 10.1002/j.1460-2075.1992.tb05420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartholomeusz A, Groenen L C, Locarnini S A. Clinical experience with famciclovir against hepatitis B virus. Intervirology. 1997;40:337–342. doi: 10.1159/000150566. [DOI] [PubMed] [Google Scholar]
- 9.Bartholomew M M, Jansen R W, Jeffers L J, Reddy K R, Johnson L C, Bunzendahl H, Condreay L D, Tzakis A G, Schiff E R, Brown N A. Hepatitis-B-virus resistance to lamivudine given for recurrent infection after orthotopic liver transplantation. Lancet. 1997;349:20–22. doi: 10.1016/S0140-6736(96)02266-0. [DOI] [PubMed] [Google Scholar]
- 10.Beasley R. Hepatitis B virus: the major etiology of hepatocellular carcinoma. Cancer. 1988;61:1942–1956. doi: 10.1002/1097-0142(19880515)61:10<1942::aid-cncr2820611003>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 11.Beasley R P. Hepatitis B virus as the etiologic agent in hepatocellular carcinoma: epidemiologic considerations. Hepatology. 1982;2:21S–26S. [Google Scholar]
- 12.Beasley R P, Hwang L-Y. Overview on the epidemiology of hepatocellular carcinoma. In: Hollinger F B, Lemon S M, Margolis H S, editors. Viral hepatitis and liver disease. Baltimore, Md: The Williams & Wilkins Co.; 1991. pp. 532–535. [Google Scholar]
- 13.Beckel-Mitchener A, Summers J. A novel transcriptional element in circular DNA monomers of the duck hepatitis B virus. J Virol. 1997;71:7917–7922. doi: 10.1128/jvi.71.10.7917-7922.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benn J, Schneider R J. Hepatitis B virus HBx protein activates Ras-GTP complex formation and establishes a Ras, Raf, MAP kinase signaling cascade. Proc Natl Acad Sci USA. 1994;91:10350–10354. doi: 10.1073/pnas.91.22.10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berquist K R, Peterson J M, Murphy B L, Ebert J W, Maynard J E, Purcell R H. Hepatitis B antigens in serum and liver of chimpanzees acutely infected with hepatitis B virus. Infect Immun. 1975;12:602–605. doi: 10.1128/iai.12.3.602-605.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blum H E, Geballe A P, Stowring L, Figus A, Haase A, Vyas G. Hepatitis B virus in nonhepatocytes: demonstration of viral DNA in spleen, bile duct epithelium, and vascular elements by in situ hybridization. In: Vyas G N, Dienstag J L, Hoofnagle J H, editors. Viral hepatitis and liver disease. New York, N.Y: Grune & Stratton; 1984. p. 634. [Google Scholar]
- 17.Blum H E, Zhang Z S, Galun E, von W F, Garner B, Liang T J, Wands J R. Hepatitis B virus X protein is not central to the viral life cycle in vitro. J Virol. 1992;66:1223–1227. doi: 10.1128/jvi.66.2.1223-1227.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borel C, Sunyach C, Hantz O, Trepo C, Kay A. Phosphorylation of DHBV pre-S: identification of the major site of phosphorylation and effects of mutations on the virus life cycle. Virology. 1998;242:90–98. doi: 10.1006/viro.1997.9004. [DOI] [PubMed] [Google Scholar]
- 19.Bottcher B, Wynne S A, Crowther R A. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature. 1997;386:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- 20.Brechot C, Pourcel C, Louise A, Rain B, Tiollais P. Presence of integrated hepatitis B virus DNA sequences in cellular DNA of human hepatocellular carcinoma. Nature. 1980;286:533–535. doi: 10.1038/286533a0. [DOI] [PubMed] [Google Scholar]
- 21.Bressac B, Kew M, Wands J, Ozturk M. Selective G to T mutations of p53 gene in hepatocellular carcinoma from southern Africa. Nature. 1991;350:429–431. doi: 10.1038/350429a0. [DOI] [PubMed] [Google Scholar]
- 22.Brook M G, Karayiannis P, Thomas H C. Which patients with chronic hepatitis B virus infection will respond to a-interferon therapy? A statistical analysis of predictive factors. Hepatology. 1989;10:761–763. doi: 10.1002/hep.1840100502. [DOI] [PubMed] [Google Scholar]
- 23.Bruss V, Hagelstein J, Gerhardt E, Galle P R. Myristylation of the large surface protein is required for hepatitis B virus in vitro infectivity. Virology. 1996;218:396–409. doi: 10.1006/viro.1996.0209. [DOI] [PubMed] [Google Scholar]
- 24.Bruss V, Lu X, Thomssen R, Gerlich W H. Post-translational alterations in transmembrane topology of the hepatitis B virus large envelope protein. EMBO J. 1994;13:2273–2279. doi: 10.1002/j.1460-2075.1994.tb06509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buetow K H, Sheffield V C, Zhu M, Zhou T, Shen F M, Hino O, Smith M, McMahon B J, Lanier A P, London W T, et al. Low frequency of p53 mutations observed in a diverse collection of primary hepatocellular carcinomas. Proc Natl Acad Sci USA. 1992;89:9622–9626. doi: 10.1073/pnas.89.20.9622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cavanaugh V J, Guidotti L, Chisari F V. Inhibition of Hepatitis B virus replication during adenovirus and cytomegalovirus infections with transgenic mice. J Virol. 1998;72:2630–2637. doi: 10.1128/jvi.72.4.2630-2637.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chakraborty P R, Ruiz-Opazo N, Shouval D, Shafritz D A. Identification of integrated hepatitis B virus DNA and expression of viral RNA in HBsAg-producing human hepatocellular carcinoma cell line. Nature. 1980;286:531–533. doi: 10.1038/286531a0. [DOI] [PubMed] [Google Scholar]
- 28.Chang C, Enders G, Sprengel R, Peters N, Varmus H E, Ganem D. Expression of the precore region of an avian hepatitis B virus is not required for viral replication. J Virol. 1987;61:3322–3325. doi: 10.1128/jvi.61.10.3322-3325.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang C, Zhou S, Ganem D, Standring D N. Phenotypic mixing between different hepadnavirus nucleocapsid proteins reveals C protein dimerization to be cis preferential. J Virol. 1994;68:5225–5231. doi: 10.1128/jvi.68.8.5225-5231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang L J, Pryciak P, Ganem D, Varmus H E. Biosynthesis of the reverse transcriptase of hepatitis B viruses involves de novo translational initiation not ribosomal frameshifting. Nature. 1989;337:364–368. doi: 10.1038/337364a0. [DOI] [PubMed] [Google Scholar]
- 31.Chayama K, Suzuki Y, Kobayashi M, Tsubota A, Hashimoto M, Miyano Y, Koike H, Koida I, Arase Y, Saitoh S, Murashima N, Ikeda K, Kumada H. Emergence and takeover of YMDD motif mutant hepatitis B virus during long-term lamivudine therapy and re-takeover by wild type after cessation of therapy. Hepatology. 1998;27:1711–1716. doi: 10.1002/hep.510270634. [DOI] [PubMed] [Google Scholar]
- 32.Chen H S, Kaneko S, Girones R, Anderson R W, Hornbuckle W E, Tennant B C, Cote P J, Gerin J L, Purcell R H, Miller R H. The woodchuck hepatitis virus X gene is important for establishment of virus infection in woodchucks. J Virol. 1993;67:1218–1226. doi: 10.1128/jvi.67.3.1218-1226.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen H S, Kew M C, Hornbuckle W E, Tennant B C, Cote P J, Gerin J L, Purcell R H, Miller R H. The precore gene of the woodchuck hepatitis virus genome is not essential for viral replication in the natural host. J Virol. 1992;66:5682–5684. doi: 10.1128/jvi.66.9.5682-5684.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chisari F V, Klopchin K, Moriyama T, Pasquinelli C, Dunsford H A, Sell S, Pinkert C A, Brinster R L, Palmiter R D. Molecular pathogenesis of hepatocellular carcinoma in hepatitis B virus transgenic mice. Cell. 1989;59:1145–1156. doi: 10.1016/0092-8674(89)90770-8. [DOI] [PubMed] [Google Scholar]
- 35.Civitico G M, Locarnini S A. The half-life of duck hepatitis B virus supercoiled DNA in congenitally infected primary hepatocyte cultures. Virology. 1994;203:81–89. doi: 10.1006/viro.1994.1457. [DOI] [PubMed] [Google Scholar]
- 36.Coleman W B, Wennerberg A E, Smith G J, Grisham J W. Regulation of the differentiation of diploid and some aneuploid rat liver epithelial (stemlike) cells by the hepatic microenvironment. Am J Pathol. 1993;142:1373–1382. [PMC free article] [PubMed] [Google Scholar]
- 37.Colgrove R, Simon G, Ganem D. Transcriptional activation of homologous and heterologous genes by the hepatitis B virus X gene product in cells permissive for viral replication. J Virol. 1989;63:4019–4026. doi: 10.1128/jvi.63.9.4019-4026.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Conway J F, Cheng N, Zlotnick A, Stahl S J, Wingfield P T, Belnap D M, Kanngiesser U, Noah M, Steven A C. Hepatitis B virus capsid: localization of the putative immunodominant loop (residues 78 to 83) on the capsid surface, and implications for the distinction between c- and e-antigens. J Mol Biol. 1998;279:1111–1121. doi: 10.1006/jmbi.1998.1845. [DOI] [PubMed] [Google Scholar]
- 39.Conway J F, Cheng N, Zlotnick A, Wingfield P T, Stahl S J, Steven A C. Visualization of a 4-helix bundle in the hepatitis B virus capsid by cryo-electron microscopy. Nature. 1997;386:91–94. doi: 10.1038/386091a0. [DOI] [PubMed] [Google Scholar]
- 40.Crescenzo-Chaigne B, Pillot J, Lilienbaum A. Interplay between a new HNF3 and the HNF1 transcriptional factors in the duck hepatitis B virus enhancer. Virology. 1995;213:231–240. doi: 10.1006/viro.1995.1563. [DOI] [PubMed] [Google Scholar]
- 41.Crowther R A, Kiselev N A, Bottcher B, Berriman J A, Borisova G P, Ose V, Pumpens P. Three-dimensional structure of hepatitis B virus core particles determined by electron cryomicroscopy. Cell. 1994;77:943–950. doi: 10.1016/0092-8674(94)90142-2. [DOI] [PubMed] [Google Scholar]
- 42.Dabeva M D, Alpini G, Hurston E, Shafritz D A. Models for hepatic progenitor cell activation. Proc Soc Exp Biol Med. 1993;204:242–252. doi: 10.3181/00379727-204-43660. [DOI] [PubMed] [Google Scholar]
- 43.Dalgleish A G, Beverley P C, Clapham P R, Crawford D H, Greaves M F, Weiss R A. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 44.Dandri M, Schirmacher P, Rogler C E. Woodchuck hepatitis virus X protein is present in chronically infected woodchuck liver and woodchuck hepatocellular carcinomas which are permissive for viral replication. J Virol. 1996;70:5246–5254. doi: 10.1128/jvi.70.8.5246-5254.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dane D S, Cameron C H, Briggs M. Virus-like particles in serum of patients with Australia-antigen-associated hepatitis. Lancet. 1970;i:695–698. doi: 10.1016/s0140-6736(70)90926-8. [DOI] [PubMed] [Google Scholar]
- 46.Dejean A, Bougueleret L, Grzeschik K H, Tiollais P. Hepatitis B virus DNA integration in a sequence homologous to v-erb-A and steroid receptor genes in a hepatocellular carcinoma. Nature. 1986;322:70–72. doi: 10.1038/322070a0. [DOI] [PubMed] [Google Scholar]
- 47.D'Errico A, Grigioni W F, Fiorentino M, Baccarini P, Lamas E, De M S, Gozzetti G, Mancini A M, Brechot C. Expression of insulin-like growth factor II (IGF-II) in human hepatocellular carcinomas: an immunohistochemical study. Pathol Int. 1994;44:131–137. doi: 10.1111/j.1440-1827.1994.tb01697.x. [DOI] [PubMed] [Google Scholar]
- 48.Di Q, Summers J, Burch J B, Mason W S. Major differences between WHV and HBV in the regulation of transcription. Virology. 1997;229:25–35. doi: 10.1006/viro.1996.8422. [DOI] [PubMed] [Google Scholar]
- 49.Dienes H P, Purcell R H, Popper H, Ponzetto A. The significance of infections with two types of viral hepatitis demonstrated by histologic features in chimpanzees. J Hepatol. 1990;10:77–84. doi: 10.1016/0168-8278(90)90076-4. [DOI] [PubMed] [Google Scholar]
- 50.Dragani T A, Manenti G, Farza H, Della Porta G, Tiollais P, Pourcel C. Transgenic mice containing hepatitis B virus sequences are more susceptible to carcinogen-induced hepatocarcinogenesis. Carcinogenesis. 1990;11:953–956. doi: 10.1093/carcin/11.6.953. [DOI] [PubMed] [Google Scholar]
- 51.Edman J C, Gray P, Valenzuela P, Rall L B, Rutter W J. Integration of hepatitis B virus sequences and their expression in a human hepatoma cell line. Nature. 1980;286:535–538. doi: 10.1038/286535a0. [DOI] [PubMed] [Google Scholar]
- 52.Evans A A, London W T. Epidemiology of hepatitis B. In: Zuckerman A J, Thomas H C, editors. Viral hepatitis. London, United Kingdom: Harcourt Brace & Co., Ltd.; 1998. pp. 107–114. [Google Scholar]
- 53.Evans A A, O'Connell A P, Pugh J C, Mason W S, Shen F M, Chen G C, Lin W Y, Dia A, M'Boup S, Drame B, London W T. Geographic variation in viral load among hepatitis B carriers with differing risks of hepatocellular carcinoma. Cancer Epidemiol Biomarkers Prev. 1998;7:559–565. [PubMed] [Google Scholar]
- 54.Evarts R P, Nagy P, Marsden E, Thorgeirsson S S. A precursor product relationship between oval cells and hepatocytes in rat liver. Carcinogenesis. 1987;8:1737–1740. doi: 10.1093/carcin/8.11.1737. [DOI] [PubMed] [Google Scholar]
- 55.Fausto N. Hepatocyte differentiation and liver progenitor cells. Curr Opin Cell Biol. 1990;2:1036–1042. doi: 10.1016/0955-0674(90)90153-6. [DOI] [PubMed] [Google Scholar]
- 56.Fausto N, Lemire J M, Shiojiri N. Cell lineages in hepatic development and the identification of progenitor cells in normal and injured liver. Proc Soc Exp Biol Med. 1993;214:237–241. doi: 10.3181/00379727-204-43659. [DOI] [PubMed] [Google Scholar]
- 57.Feitelson M A, Zhu M, Duan L X, London W T. Hepatitis B × antigen and p53 are associated in vitro and in liver tissues from patients with primary hepatocellular carcinoma. Oncogene. 1993;8:1109–1117. [PubMed] [Google Scholar]
- 58.Fernholz D, Galle P R, Stemler M, Brunetto M, Bonino F, Will H. Infectious hepatitis B virus variant defective in pre-S2 protein expression in a chronic carrier. Virology. 1993;194:137–148. doi: 10.1006/viro.1993.1243. [DOI] [PubMed] [Google Scholar]
- 59.Fourel G, Couturier J, Wei Y, Apiou F, Tiollais P, Buendia M A. Evidence for long-range oncogene activation by hepadnavirus insertion. EMBO J. 1994;13:2526–2534. doi: 10.1002/j.1460-2075.1994.tb06542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fourel G, Trepo C, Bougueleret L, Henglein B, Ponzetto A, Tiollais P, Buendia M A. Frequent activation of N-myc genes by hepadnavirus insertion in woodchuck liver tumours. Nature. 1990;347:294–298. doi: 10.1038/347294a0. [DOI] [PubMed] [Google Scholar]
- 61.Ganem D, Pollack J R, Tavis J. Hepatitis B virus reverse transcriptase and its many roles in hepadnaviral genomic replication. Infect Agents Dis. 1994;3:85–93. [PubMed] [Google Scholar]
- 62.Gartner L P, Hiatt J L. Color textbook of histology. Philadelphia, Pa: The W. B. Saunders Co.; 1997. [Google Scholar]
- 63.Gong S S, Jensen A D, Rogler C E. Loss and acquisition of duck hepatitis B virus integrations in lineages of LMH-D2 chicken hepatoma cells. J Virol. 1996;70:2000–2007. doi: 10.1128/jvi.70.3.2000-2007.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gong S S, Jensen A D, Wang H, Rogler C E. Duck hepatitis B virus integrations in LMH chicken hepatoma cells: identification and characterization of new episomally derived integrations. J Virol. 1995;69:8102–8108. doi: 10.1128/jvi.69.12.8102-8108.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gottlieb T M, Oren M. p53 in growth control and neoplasia. Biochim Biophys Acta. 1996;1287:77–102. doi: 10.1016/0304-419x(95)00019-c. [DOI] [PubMed] [Google Scholar]
- 66.Gripon P, Le S J, Rumin S, Guguen G C. Myristylation of the hepatitis B virus large surface protein is essential for viral infectivity. Virology. 1995;213:292–299. doi: 10.1006/viro.1995.0002. [DOI] [PubMed] [Google Scholar]
- 67.Grisham J W. A morphologic study of deoxyribonucleic acid synthesis and cell proliferation in regenerating rat liver: autoradiography with thymidine-H3. Cancer Res. 1962;22:842–849. [PubMed] [Google Scholar]
- 68.Guidotti L G, Borrow P, Hobbs M V, Matzke B, Gresser I, Oldstone M B, Chisari F V. Viral cross talk: intracellular inactivation of the hepatitis B virus during an unrelated viral infection of the liver. Proc Natl Acad Sci USA. 1996;93:4589–4594. doi: 10.1073/pnas.93.10.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guidotti L G, Guilhot S, Chisari F V. Interleukin-2 and alpha/beta interferon down-regulate hepatitis B virus gene expression in vivo by tumor necrosis factor-dependent and -independent pathways. J Virol. 1994;68:1265–1270. doi: 10.1128/jvi.68.3.1265-1270.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guidotti L G, Matzke B, Schaller H, Chisari F V. High-level hepatitis B virus replication in transgenic mice. J Virol. 1995;69:6158–6169. doi: 10.1128/jvi.69.10.6158-6169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guidotti L G, Rochford R, Chung J, Shapiro M, Purcell R, Chisari F V. Viral clearance without destruction of infected cells during acute HBV infection. Science. 1999;284:825–829. doi: 10.1126/science.284.5415.825. [DOI] [PubMed] [Google Scholar]
- 72.Guo J-T, Pugh J C. The topology of the large envelope protein of duck hepatitis B virus suggests a mechanism for membrane translocation during particle morphogenesis. J Virol. 1997;71:1107–1114. doi: 10.1128/jvi.71.2.1107-1114.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guo J-T, Zhou H, Liu C, Aldrich C, Saputelli J, Whitaker T, Barrasa M I, Mason W S, Seeger C. Apoptosis and regeneration of hepatocytes during recovery from transient hepadnavirus infection. J Virol. 2000;74:1495–1505. doi: 10.1128/jvi.74.3.1495-1505.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hadchouel M, Pasquinelli C, Fournier J G, Hugon R N, Scotto J, Bernard O, Brechot C. Detection of mononuclear cells expressing hepatitis B virus in peripheral blood from HBsAg positive and negative patients by in situ hybridisation. J Med Virol. 1988;24:27–32. doi: 10.1002/jmv.1890240105. [DOI] [PubMed] [Google Scholar]
- 75.Halpern M S, Egan J, Mason W S, England J M. Viral antigen in endocrine cells of the pancreatic islets and adrenal cortex of Pekin ducks infected with duck hepatitis B virus. Virus Res. 1984;1:213–223. doi: 10.1016/0168-1702(84)90040-6. [DOI] [PubMed] [Google Scholar]
- 76.Halpern M S, Egan J, McMahon S B, Ewert D L. Duck hepatitis B virus is tropic for exocrine cells of the pancreas. Virology. 1985;146:157–161. doi: 10.1016/0042-6822(85)90064-9. [DOI] [PubMed] [Google Scholar]
- 77.Halpern M S, England J M, Deery D T, Petcu D J, Mason W S, Molnar-Kimber K L. Viral nucleic acid synthesis and antigen accumulation in pancreas and kidney of Pekin ducks infected with duck hepatitis B virus. Proc Natl Acad Sci USA. 1983;80:4865–4869. doi: 10.1073/pnas.80.15.4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Halpern M S, Mason W S, Coates L, O'Connell A P, England J M. Humoral immune responsiveness in duck hepatitis B virus-infected ducks. J Virol. 1987;61:916–920. doi: 10.1128/jvi.61.3.916-920.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Havert M B, Loeb D D. cis-acting sequences in addition to donor and acceptor sites are required for template switching during synthesis of plus-strand DNA for duck hepatitis B virus. J Virol. 1997;71:5336–5344. doi: 10.1128/jvi.71.7.5336-5344.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Haviv I, Shamay M, Doitsh G, Shaul Y. Hepatitis B virus pX targets TFIIB in transcription coactivation. Mol Cell Biol. 1998;18:1562–1569. doi: 10.1128/mcb.18.3.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hayward W S, Neel B G, Astrin S M. Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature. 1981;290:475–480. doi: 10.1038/290475a0. [DOI] [PubMed] [Google Scholar]
- 82.Heermann K H, Kruse F, Seifer M, Gerlich W H. Immunogenicity of the gene S and Pre-S domains in hepatitis B virions and HBsAg filaments. Intervirology. 1987;28:14–25. doi: 10.1159/000149993. [DOI] [PubMed] [Google Scholar]
- 83.Hild M, Weber O, Schaller H. Glucagon treatment interferes with an early step of duck hepatitis B virus infection. J Virol. 1998;72:2600–2606. doi: 10.1128/jvi.72.4.2600-2606.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hirsch R C, Lavine J E, Chang L J, Varmus H E, Ganem D. Polymerase gene products of hepatitis B viruses are required for genomic RNA packaging as well as for reverse transcription. Nature. 1990;344:552–555. doi: 10.1038/344552a0. [DOI] [PubMed] [Google Scholar]
- 85.Hohne M, Schaefer S, Seifer M, Feitelson M A, Paul D, Gerlich W H. Malignant transformation of immortalized transgenic hepatocytes after transfection with hepatitis B virus DNA. EMBO J. 1990;9:1137–1145. doi: 10.1002/j.1460-2075.1990.tb08220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hoofnagle J, Shafritz D, Popper H. Chronic type B hepatitis and the “healthy” HBsAg carrier state. Hepatology. 1987;7:758–763. doi: 10.1002/hep.1840070424. [DOI] [PubMed] [Google Scholar]
- 87.Hoofnagle J H, DiBisceglie A M. The treatment of chronic viral hepatitis. N Engl J Med. 1997;336:347–356. doi: 10.1056/NEJM199701303360507. [DOI] [PubMed] [Google Scholar]
- 88.Hoofnagle J H, Michalak T, Nowoslawski A, Gerety R J, Barker L F. Immunofluorescence microscopy in experimentally induced, type B hepatitis in the chimpanzee. Gastroenterology. 1978;74:182–187. [PubMed] [Google Scholar]
- 89.Hruska J F, Clayton D A, Rubenstein J L, Robinson W S. Structure of hepatitis B Dane particle DNA before and after the Dane particle DNA polymerase reaction. J Virol. 1977;21:666–672. doi: 10.1128/jvi.21.2.666-672.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hsu I C, Metcalf R A, Sun T, Welsh J A, Wang N J, Harris C C. Mutational hotspot in the p53 gene in human hepatocellular carcinomas. Nature. 1991;350:427–428. doi: 10.1038/350427a0. [DOI] [PubMed] [Google Scholar]
- 91.Hsu T, Moroy T, Etiemble J, Louise A, Trepo C, Tiollais P, Buendia M. Activation of c-myc by woodchuck hepatitis virus insertion in hepatocellular carcinoma. Cell. 1988;55:627–635. doi: 10.1016/0092-8674(88)90221-8. [DOI] [PubMed] [Google Scholar]
- 92.Hu J, Seeger C. Hsp90 is required for the activity of a hepatitis B virus reverse transcriptase. Proc Natl Acad Sci USA. 1996;93:1060–1064. doi: 10.1073/pnas.93.3.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hu J, Toft D O, Seeger C. Hepadnavirus assembly and reverse transcription require a multi-component chaperone complex which is incorporated into nucleocapsids. EMBO J. 1997;16:59–68. doi: 10.1093/emboj/16.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hu K Q, Vierling J M, Siddiqui A. Trans-activation of HLA-DR gene by hepatitis B virus X gene product. Proc Natl Acad Sci USA. 1990;87:7140–7144. doi: 10.1073/pnas.87.18.7140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huang M, Summers J. pet, a small sequence distal to the pregenome cap site, is required for expression of the duck hepatitis B virus pregenome. J Virol. 1994;68:1564–1572. doi: 10.1128/jvi.68.3.1564-1572.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Huovila A P, Eder A M, Fuller S D. Hepatitis B surface antigen assembles in a post-ER, pre-Golgi compartment. J Cell Biol. 1992;118:1305–1320. doi: 10.1083/jcb.118.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ishikawa T, Ganem D. The pre-S domain of the large viral envelope protein determines host range in avian hepatitis B viruses. Proc Natl Acad Sci USA. 1995;92:6259–6263. doi: 10.1073/pnas.92.14.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jilbert A R, Freiman J S, Gowans E J, Holmes M, Cossart Y E, Burrell C J. Duck hepatitis B virus DNA in liver, spleen, and pancreas: analysis by in situ and Southern blot hybridization. Virology. 1987;158:330–338. doi: 10.1016/0042-6822(87)90205-4. [DOI] [PubMed] [Google Scholar]
- 99.Jilbert A R, Miller D S, Scougall C A, Turnbull H, Burrell C J. Kinetics of duck hepatitis B virus infection following low dose virus inoculation: one virus DNA genome is infectious in neonatal ducks. Virology. 1996;226:338–345. doi: 10.1006/viro.1996.0661. [DOI] [PubMed] [Google Scholar]
- 100.Jilbert A R, Wu T-T, England J M, de la M. Hall P, Carp N Z, O'Connell A P, Mason W S. Rapid resolution of duck hepatitis B virus infections occurs after massive hepatocellular involvement. J Virol. 1992;66:1377–1388. doi: 10.1128/jvi.66.3.1377-1388.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Junker-Niepmann M, Bartenschlager R, Schaller H. A short cis-acting sequence is required for hepatitis B virus pregenome encapsidation and sufficient for packaging of foreign RNA. EMBO J. 1990;9:3389–3396. doi: 10.1002/j.1460-2075.1990.tb07540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kajino K, Jilbert A R, Saputelli J, Aldrich C E, Cullen J, Mason W S. Woodchuck hepatitis virus infections: very rapid recovery after a prolonged viremia and infection of virtually every hepatocyte. J Virol. 1994;68:5792–5803. doi: 10.1128/jvi.68.9.5792-5803.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kann M, Bischof A, Gerlich W H. In vitro model for the nuclear transport of the hepadnavirus genome. J Virol. 1997;71:1310–1316. doi: 10.1128/jvi.71.2.1310-1316.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kann M, Sodeik B, Vlachou A, Gerlich W H, Helenius A. Phosphorylation-dependent binding of hepatitis B virus core particles to the nuclear pore complex. J Cell Biol. 1999;145:45–55. doi: 10.1083/jcb.145.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kim C M, Koike K, Saito I, Miyamura T, Jay G. HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature. 1991;351:317–320. doi: 10.1038/351317a0. [DOI] [PubMed] [Google Scholar]
- 106.Kim H, Lee H, Yun Y. X-gene product of hepatitis B virus induces apoptosis in liver cells. J Biol Chem. 1998;273:381–385. doi: 10.1074/jbc.273.1.381. [DOI] [PubMed] [Google Scholar]
- 107.Kinzler K W, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 108.Kock J, Theilmann L, Galle P, Schlicht H J. Hepatitis B virus nucleic acids associated with human peripheral blood mononuclear cells do not originate from replicating virus. Hepatology. 1996;23:405–413. doi: 10.1002/hep.510230303. [DOI] [PubMed] [Google Scholar]
- 109.Koike K, Moriya K, Iino S, Yotsuyanagi H, Endo Y, Miyamura T, Kurokawa K. High-level expression of hepatitis B virus HBx gene and hepatocarcinogenesis in transgenic mice. Hepatology. 1994;19:810–819. [PubMed] [Google Scholar]
- 110.Konig S, Beterams G, Nassal M. Mapping of homologous interaction sites in the hepatitis B virus core protein. J Virol. 1998;72:4997–5005. doi: 10.1128/jvi.72.6.4997-5005.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Korba B E, Cote P J, Shapiro M, Purcell R H, Gerin J L. In vitro production of infectious woodchuck hepatitis virus by lipopolysaccharide-stimulated peripheral blood lymphocytes. J Infect Dis. 1989;160:572–576. doi: 10.1093/infdis/160.4.572. [DOI] [PubMed] [Google Scholar]
- 112.Kuroki K, Cheung R, Marion P L, Ganem D. A cell surface protein that binds avian hepatitis B virus particles. J Virol. 1994;68:2091–2096. doi: 10.1128/jvi.68.4.2091-2096.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kuroki K, Eng F, Ishikawa T, Turck C, Harada F, Ganem D. gp180, a host cell glycoprotein that binds duck hepatitis B virus particles, is encoded by a member of the carboxypeptidase gene family. J Biol Chem. 1995;270:15022–15028. doi: 10.1074/jbc.270.25.15022. [DOI] [PubMed] [Google Scholar]
- 114.Kuroki K, Russnak R, Ganem D. Novel N-terminal amino acid sequence required for retention of a hepatitis B virus glycoprotein in the endoplasmic reticulum. Mol Cell Biol. 1989;9:4459–4466. doi: 10.1128/mcb.9.10.4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lambert V, Chassot S, Kay A, Trepo C, Cova L. In vivo neutralization of duck hepatitis B virus by antibodies specific to the N-terminal portion of pre-S protein. Virology. 1991;185:446–450. doi: 10.1016/0042-6822(91)90796-e. [DOI] [PubMed] [Google Scholar]
- 116.Lambert V, Fernholz D, Sprengel R, Fourel I, Deleage G, Wildner G, Peyret C, Trepo C, Cova L, Will H. Virus-neutralizing monoclonal antibody to a conserved epitope on the duck hepatitis B virus pre-S protein. J Virol. 1990;64:1290–1297. doi: 10.1128/jvi.64.3.1290-1297.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Landers T A, Greenberg H B, Robinson W S. Structure of hepatitis B Dane particle DNA and nature of the endogenous DNA polymerase reaction. J Virol. 1977;23:368–376. doi: 10.1128/jvi.23.2.368-376.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lanford R E, Notvall L, Beames B. Nucleotide priming and reverse transcriptase activity of hepatitis B virus polymerase expressed in insect cells. J Virol. 1995;69:4431–4439. doi: 10.1128/jvi.69.7.4431-4439.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lee T H, Elledge S J, Butel J S. Hepatitis B virus X protein interacts with a probable cellular DNA repair protein. J Virol. 1995;69:1107–1114. doi: 10.1128/jvi.69.2.1107-1114.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lee T H, Finegold M J, Shen R F, DeMayo J L, Woo S L, Butel J S. Hepatitis B virus transactivator X protein is not tumorigenic in transgenic mice. J Virol. 1990;64:5939–5947. doi: 10.1128/jvi.64.12.5939-5947.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lenhoff R J, Luscombe C A, Summers J. Competition in vivo between a cytopathic variant and a wild-type duck hepatitis B virus. Virology. 1998;251:85–95. doi: 10.1006/viro.1998.9394. [DOI] [PubMed] [Google Scholar]
- 122.Lieberman H M, Tung W W, Shafritz D A. Splenic replication of hepatitis B virus in the chimpanzee chronic carrier. J Med Virol. 1987;21:347–359. doi: 10.1002/jmv.1890210407. [DOI] [PubMed] [Google Scholar]
- 123.Lien J M, Aldrich C E, Mason W S. Evidence that a capped oligoribonucleotide is the primer for duck hepatitis B virus plus-strand DNA synthesis. J Virol. 1986;57:229–236. doi: 10.1128/jvi.57.1.229-236.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lien J M, Petcu D J, Aldrich C E, Mason W S. Initiation and termination of duck hepatitis B virus DNA synthesis during virus maturation. J Virol. 1987;61:3832–3840. doi: 10.1128/jvi.61.12.3832-3840.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ling R, Mutimer D, Ahmed M, Boxall E H, Elias E, Dusheiko G M, Harrison T J. Selection of mutations in the hepatitis B virus polymerase during therapy of transplant recipients with lamivudine. Hepatology. 1996;24:711–713. doi: 10.1002/hep.510240339. [DOI] [PubMed] [Google Scholar]
- 126.Liu C, Mason W S, Burch J B. Identification of factor-binding sites in the duck hepatitis B virus enhancer and in vivo effects of enhancer mutations. J Virol. 1994;68:2286–2296. doi: 10.1128/jvi.68.4.2286-2296.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lok A S, Lai C L, Chung H T, Lau J Y, Leung E K, Wong L S. Morbidity and mortality from chronic hepatitis B virus infection in family members of patients with malignant and nonmalignant hepatitis B virus-related chronic liver diseases. Hepatology. 1991;13:834–837. doi: 10.1002/hep.1840130506. [DOI] [PubMed] [Google Scholar]
- 128.Lok A S, Wu P C, Lai C L, Lau J Y, Leung E K, Wong L S, Ma O C, Lauder I J, Ng C P, Chung H T. A controlled trial of interferon with or without prednisone priming for chronic hepatitis B. Gastroenterology. 1992;102:2091–2097. doi: 10.1016/0016-5085(92)90337-x. [DOI] [PubMed] [Google Scholar]
- 129.London W T, Evans A A, McGlynn K, Buetow K, An P, Gao L, Lustbader E, Ross E, Chen G, Shen F. Viral, host and environmental risk factors for hepatocellular carcinoma: a prospective study in Haimen City, China. Intervirology. 1995;38:155–161. doi: 10.1159/000150426. [DOI] [PubMed] [Google Scholar]
- 130.Luscombe C, Pedersen J, Uren E, Locarnini S. Long-term ganciclovir chemotherapy for congenital duck hepatitis B virus infection in vivo: effect on intrahepatic-viral DNA, RNA, and protein expression. Hepatology. 1996;24:766–773. doi: 10.1053/jhep.1996.v24.pm0008855174. [DOI] [PubMed] [Google Scholar]
- 131.Lutwick L I, Robinson W S. DNA synthesized in the hepatitis B Dane particle DNA polymerase reaction. J Virol. 1977;21:96–104. doi: 10.1128/jvi.21.1.96-104.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.MacDonald R A. “Lifespan” of liver cells. Arch Intern Med. 1960;107:335–343. doi: 10.1001/archinte.1961.03620030023003. [DOI] [PubMed] [Google Scholar]
- 133.Macrae D R, Bruss V, Ganem D. Myristylation of a duck hepatitis B virus envelope protein is essential for infectivity but not for virus assembly. Virology. 1991;181:359–363. doi: 10.1016/0042-6822(91)90503-4. [DOI] [PubMed] [Google Scholar]
- 134.Maguire H F, Hoeffler J P, Siddiqui A. HBV X protein alters the DNA binding specificity of CREB and ATF-2 by protein-protein interactions. Science. 1991;252:842–844. doi: 10.1126/science.1827531. [DOI] [PubMed] [Google Scholar]
- 135.Marion P L, Oshiro L S, Regnery D C, Scullard G H, Robinson W S. A virus in Beechey ground squirrels that is related to hepatitis B virus of humans. Proc Natl Acad Sci USA. 1980;77:2941–2945. doi: 10.1073/pnas.77.5.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Marion P L, Salazar F H, Alexander J J, Robinson W S. State of hepatitis B viral DNA in a human hepatoma cell line. J Virol. 1980;33:795–806. doi: 10.1128/jvi.33.2.795-806.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mason W S, Halpern M S, England J M, Seal G, Egan J, Coates L, Aldrich C, Summers J. Experimental transmission of duck hepatitis B virus. Virology. 1983;131:375–384. doi: 10.1016/0042-6822(83)90505-6. [DOI] [PubMed] [Google Scholar]
- 138.Mason W S, Seal G, Summers J. Virus of Pekin ducks with structural and biological relatedness to human hepatitis B virus. J Virol. 1980;36:829–836. doi: 10.1128/jvi.36.3.829-836.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Mason W S, Zhou T, Nunes F, Condreay L D, Litwin S, Summers J. Antiviral therapy for chronic hepadnavirus infections. In: Schinazi R F, Sommadossi J-P, Thomas H C, editors. Therapies for viral hepatitis. London, United Kingdom: International Medical Press; 1998. pp. 177–184. [Google Scholar]
- 140.McDougal J S, Maddon P J, Dalgleish A G, Clapham P R, Littman D R, Godfrey M, Maddon D E, Chess L, Weiss R A, Axel R. The T4 glycoprotein is a cell-surface receptor for the AIDS virus. Cold Spring Harbor Symp Quant Biol. 1986;51:703–711. doi: 10.1101/sqb.1986.051.01.083. [DOI] [PubMed] [Google Scholar]
- 141.McGlynn K A, Rosvold E A, Lustbader E D, Hu Y, Clapper M L, Zhou T, Wild C P, Xia X L, Baffoe B A, Ofori A D, et al. Susceptibility to hepatocellular carcinoma is associated with genetic variation in the enzymatic detoxification of aflatoxin B1. Proc Natl Acad Sci USA. 1995;92:2384–2387. doi: 10.1073/pnas.92.6.2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.McMahon B J, Alward W L, Hall D B, Heyward W L, Bender T R, Francis D P, Maynard J E. Acute hepatitis B virus infection: relation of age to the clinical expression of disease and subsequent development of the carrier state. J Infect Dis. 1985;151:599–603. doi: 10.1093/infdis/151.4.599. [DOI] [PubMed] [Google Scholar]
- 143.Michalak T I, Pardoe I U, Coffin C S, Churchill N D, Freake D S, Smith P, Trelegan C L. Occult lifelong persistence of infectious hepadnavirus and residual liver inflammation in woodchucks convalescent from acute viral hepatitis. Hepatology. 1999;29:928–938. doi: 10.1002/hep.510290329. [DOI] [PubMed] [Google Scholar]
- 144.Milich D R, Chen M K, Hughes J L, Jones J E. The secreted hepatitis B precore antigen can modulate the immune response to the nucleocapsid: a mechanism for persistence. J Immunol. 1998;160:2013–2021. [PubMed] [Google Scholar]
- 145.Milich D R, Jones J E, Hughes J L, Price J, Raney A K, McLachlan A. Is a function of the secreted hepatitis B e antigen to induce immunologic tolerance in utero? Proc Natl Acad Sci USA. 1990;87:6599–6603. doi: 10.1073/pnas.87.17.6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Moraleda G, Saputelli J, Aldrich C E, Averett D, Condreay L, Mason W S. Lack of effect of antiviral therapy in nondividing hepatocyte cultures on the closed circular DNA of woodchuck hepatitis virus. J Virol. 1997;71:9392–9399. doi: 10.1128/jvi.71.12.9392-9399.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Negro F, Korba B E, Forzani B, Baroudy B M, Brown T L, Gerin J L, Ponzetto A. Hepatitis delta virus (HDV) and woodchuck hepatitis virus (WHV) nucleic acids in tissues of HDV-infected chronic WHV carrier woodchucks. J Virol. 1989;63:1612–1618. doi: 10.1128/jvi.63.4.1612-1618.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Netter H J, Chassot S, Chang S F, Cova L, Will H. Sequence heterogeneity of heron hepatitis B virus genomes determined by full-length DNA amplification and direct sequencing reveals novel and unique features. J Gen Virol. 1997;78:1707–1718. doi: 10.1099/0022-1317-78-7-1707. [DOI] [PubMed] [Google Scholar]
- 149.Newbold J E, Xin H, Tencza M, Sherman G, Dean J, Bowden S, Locarnini S. The covalently closed duplex form of the hepadnavirus genome exists in situ as a heterogeneous population of viral minichromosomes. J Virol. 1995;69:3350–3357. doi: 10.1128/jvi.69.6.3350-3357.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ng I O, Chung L P, Tsang S W, Lam C L, Lai E C, Fan S T, Ng M. p53 gene mutation spectrum in hepatocellular carcinomas in Hong Kong Chinese. Oncogene. 1994;9:985–990. [PubMed] [Google Scholar]
- 151.Nicoll A J, Angus P W, Chou S T, Luscombe C A, Smallwood R A, Locarnini S A. Demonstration of duck hepatitis B virus in bile duct epithelial cells: implications for pathogenesis and persistent infection. Hepatology. 1997;25:463–469. doi: 10.1002/hep.510250235. [DOI] [PubMed] [Google Scholar]
- 152.Niesters H G, Honkoop P, Haagsma E B, de Man R A, Schalm S W, Osterhaus A D. Identification of more than one mutation in the hepatitis B virus polymerase gene arising during prolonged lamivudine treatment. J Infect Dis. 1998;177:1382–1385. doi: 10.1086/517819. [DOI] [PubMed] [Google Scholar]
- 153.Ning Q, Brown D, Parodo J, Cattral M, Gorczynski R, Cole E, Fung L, Ding J W, Liu M F, Rotstein O, Phillips M J, Levy G. Ribavirin inhibits viral-induced macrophage production of TNF, IL-1, the procoagulant fgl2 prothrombinase and preserves Th1 cytokine production but inhibits Th2 cytokine response. J Immunol. 1998;160:3487–3493. [PubMed] [Google Scholar]
- 154.Nowak M A, Bonhoeffer S, Hill A M, Boehme R, Thomas H C, McDade H. Viral dynamics in hepatitis B virus infection. Proc Natl Acad Sci USA. 1996;93:4398–4402. doi: 10.1073/pnas.93.9.4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Oberhaus S M, Newbold J E. Detection of DNA polymerase activities associated with purified duck hepatitis B virus core particles by using an activity gel assay. J Virol. 1993;67:6558–6566. doi: 10.1128/jvi.67.11.6558-6566.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Obert S, Zachmann-Brand B, Deindl E, Tucker W, Bartenschlager R, Schaller H. A splice hepadnavirus RNA that is essential for virus replication. EMBO J. 1996;15:2565–2574. [PMC free article] [PubMed] [Google Scholar]
- 157.Oda T, Tsuda H, Sakamoto M, Hirohashi S. Different mutations of the p53 gene in nodule-in-nodule hepatocellular carcinoma as a evidence for multistage progression. Cancer Lett. 1994;83:197–200. doi: 10.1016/0304-3835(94)90319-0. [DOI] [PubMed] [Google Scholar]
- 158.Ogata N, Ostberg L, Ehrlich P H, Wong D C, Miller R H, Purcell R H. Markedly prolonged incubation period of hepatitis B in a chimpanzee passively immunized with a human monoclonal antibody to the a determinant of hepatitis B surface antigen. Proc Natl Acad Sci USA. 1993;90:3014–3018. doi: 10.1073/pnas.90.7.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ogston C W, Jonak G J, Rogler C E, Astrin S M, Summers J. Cloning and structural analysis of integrated woodchuck hepatitis virus sequences from hepatocellular carcinomas of woodchucks. Cell. 1982;29:385–394. doi: 10.1016/0092-8674(82)90155-6. [DOI] [PubMed] [Google Scholar]
- 160.Ogston C W, Schechter E M, Humes C A, Pranikoff M B. Extrahepatic replication of woodchuck hepatitis virus in chronic infection. Virology. 1989;169:9–14. doi: 10.1016/0042-6822(89)90035-4. [DOI] [PubMed] [Google Scholar]
- 161.Ou J H, Bao H, Shih C, Tahara S M. Preferred translation of human hepatitis B virus polymerase from core protein- but not from precore protein-specific transcript. J Virol. 1990;64:4578–4581. doi: 10.1128/jvi.64.9.4578-4581.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Parkin D M, Pisani P, Ferlay J. Estimates of the worldwide incidence of 25 major cancers in 1990. Int J Cancer. 1999;80:827–841. doi: 10.1002/(sici)1097-0215(19990315)80:6<827::aid-ijc6>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 163.Parkin D M, Pisani P, Munoz N, Ferlay J. The global health burden of infection associated cancers. Cancer Surv. 1999;33:5–33. [Google Scholar]
- 164.Payne G S, Bishop J M, Varmus H E. Multiple arrangements of viral DNA and an activated host oncogene in bursal lymphomas. Nature. 1982;295:209–214. doi: 10.1038/295209a0. [DOI] [PubMed] [Google Scholar]
- 165.Perrillo R P. Interferon in the management of chronic hepatitis B. Dig Dis Sci. 1993;38:577–593. doi: 10.1007/BF01316785. [DOI] [PubMed] [Google Scholar]
- 166.Persing D H, Varmus H E, Ganem D. Antibodies to pre-S and X determinants arise during natural infection with ground squirrel hepatitis virus. J Virol. 1986;60:177–184. doi: 10.1128/jvi.60.1.177-184.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Pollack J R, Ganem D. An RNA stem-loop structure directs hepatitis B virus genomic RNA encapsidation. J Virol. 1993;67:3254–3263. doi: 10.1128/jvi.67.6.3254-3263.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Pollack J R, Ganem D. Site-specific RNA binding by a hepatitis B virus reverse transcriptase initiates two distinct reactions: RNA packaging and DNA synthesis. J Virol. 1994;68:5579–5587. doi: 10.1128/jvi.68.9.5579-5587.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Pontisso P, Belluco C, Bertorelle R, De Moliner L, Chieco-Bianchi L, Nitti D, Lise M, Alberti A. Hepatitis C virus infection associated with human hepatocellular carcinoma: lack of correlation with p53 abnormalities in Caucasian patients. Cancer. 1998;83:1489–1494. [PubMed] [Google Scholar]
- 170.Pontisso P, Poon M C, Tiollais P, Brechot C. Detection of hepatitis B virus DNA in mononuclear blood cells. Br Med J Clinical Res. 1984;288:1563–1566. doi: 10.1136/bmj.288.6430.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ponzetto A, Cote P J, Ford E C, Purcell R H, Gerin J L. Core antigen and antibody in woodchucks after infection with woodchuck hepatitis virus. J Virol. 1984;52:70–76. doi: 10.1128/jvi.52.1.70-76.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Protzer U, Nassal M, Chiang P-W, Kirschfink M, Schaller H. Interferon gene transfer by a hepatitis B virus vector efficiently suppresses wild-type virus infection. Proc Natl Acad Sci USA. 1999;96:10818–10823. doi: 10.1073/pnas.96.19.10818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Pugh J, Zweidler A, Summers J. Characterization of the major duck hepatitis B virus core particle protein. J Virol. 1989;63:1371–1376. doi: 10.1128/jvi.63.3.1371-1376.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Pugh J C, Sninsky J J, Summers J W, Schaeffer E. Characterization of a pre-S polypeptide on the surfaces of infectious avian hepadnavirus particles. J Virol. 1987;61:1384–1390. doi: 10.1128/jvi.61.5.1384-1390.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Rhim J A, Sandgren E P, Degen J L, Palmiter R D, Brinster R L. Replacement of diseased mouse liver by hepatic cell regeneration. Science. 1994;263:1149–1152. doi: 10.1126/science.8108734. [DOI] [PubMed] [Google Scholar]
- 176.Rogler C E. Cellular and molecular mechanisms of hepatocarcinogenesis associated with hepadnavirus infection. Curr Top Microbiol Immunol. 1991;168:103–140. doi: 10.1007/978-3-642-76015-0_6. [DOI] [PubMed] [Google Scholar]
- 177.Rogler C E, Summers J. Novel forms of woodchuck hepatitis virus DNA isolated from chronically infected woodchuck liver nuclei. J Virol. 1982;44:852–863. doi: 10.1128/jvi.44.3.852-863.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Schaller H, Fischer M. Transcriptional control of hepadnavirus gene expression. Curr Top Microbiol Immunol. 1991;168:21–39. doi: 10.1007/978-3-642-76015-0_2. [DOI] [PubMed] [Google Scholar]
- 179.Seeger C, Ganem D, Varmus H E. Biochemical and genetic evidence for the hepatitis B virus replication strategy. Science. 1986;232:477–484. doi: 10.1126/science.3961490. [DOI] [PubMed] [Google Scholar]
- 180.Seeger C, Ganem D, Varmus H E. The cloned genome of ground squirrel hepatitis virus is infectious in the animal. Proc Natl Acad Sci USA. 1984;81:5849–5852. doi: 10.1073/pnas.81.18.5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Seeger C, Mason W S. Chronic hepadnavirus infections of the woodchuck and duck. In: Ahmed R, Chen I, editors. Persistent viral infection. New York, N.Y: John Wiley & Sons, Inc.; 1999. pp. 607–621. [Google Scholar]
- 182.Seeger C, Mason W S. Replication of the hepatitis virus genome. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 815–831. [Google Scholar]
- 183.Seifer M, Hohne M, Schaefer S, Gerlich W H. In vitro tumorigenicity of hepatitis B virus DNA and HBx protein. J Hepatol. 1991;13:S61–S65. doi: 10.1016/0168-8278(91)90026-8. [DOI] [PubMed] [Google Scholar]
- 184.Shaul Y. Regulation of hepadnavirus transcription. In: McLachlan A, editor. Molecular biology of the hepatitis B virus. Boca Raton, Fla: CRC Press, Inc.; 1992. pp. 193–211. [Google Scholar]
- 185.Sheu J C, Huang G T, Lee P H, Chung J C, Chou H C, Lai M Y, Wang J T, Lee H S, Shih L N, Yang P M, et al. Mutation of p53 gene in hepatocellular carcinoma in Taiwan. Cancer Res. 1992;52:6098–6100. [PubMed] [Google Scholar]
- 186.Shimazu T, Takada S, Ueno Y, Hayashi Y, Koike K. Post-transcriptional control of the level of mRNA by hepatitis B virus X gene in the transient expression system using human hepatic cells. Genes Cells. 1998;3:477–484. doi: 10.1046/j.1365-2443.1998.00203.x. [DOI] [PubMed] [Google Scholar]
- 187.Shimoda T, Shikata T, Karasawa T, Tsukagoshi S, Yoshimura M, Sakurai I. Light microscopic localization of hepatitis B virus antigens in human pancreas. Gastroenterology. 1981;81:998–1005. [PubMed] [Google Scholar]
- 188.Shiojiri N. Enzymo- and immunocytochemical analyses of the differentiation of liver cells in the prenatal mouse. J Embryol Exp Morphol. 1981;62:139–152. [PubMed] [Google Scholar]
- 189.Shiojiri N. The origin of intrahepatic bile duct cells in the mouse. J Embryol Exp Morphol. 1984;79:25–39. [PubMed] [Google Scholar]
- 190.Sirma H, Weil R, Rosmorduc O, Urban S, Israel A, Kremsdorf D, Brechot C. Cytosol is the prime compartment of hepatitis B virus X protein where it colocalizes with the proteasome. Oncogene. 1998;16:2051–2063. doi: 10.1038/sj.onc.1201737. [DOI] [PubMed] [Google Scholar]
- 191.Slagle B L, Lee T H, Medina D, Finegold M J, Butel J S. Increased sensitivity to the hepatocarcinogen diethylnitrosamine in transgenic mice carrying the hepatitis B virus X gene. Mol Carcinog. 1996;15:261–269. doi: 10.1002/(SICI)1098-2744(199604)15:4<261::AID-MC3>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 192.Spandau D F, Lee C H. trans-activation of viral enhancers by the hepatitis B virus X protein. J Virol. 1988;62:427–434. doi: 10.1128/jvi.62.2.427-434.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Sprengel R, Kaleta E F, Will H. Isolation and characterization of a hepatitis B virus endemic in herons. J Virol. 1988;62:932–937. doi: 10.1128/jvi.62.10.3832-3839.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Staprans S, Loeb D D, Ganem D. Mutations affecting hepadnavirus plus-strand DNA synthesis dissociate primer cleavage from translocation and reveal the origin of linear viral DNA. J Virol. 1991;65:1255–1262. doi: 10.1128/jvi.65.3.1255-1262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Stevens C E, Beasley R P, Tsui J, Lee W C. Vertical transmission of hepatitis B antigen in Taiwan. N Engl J Med. 1975;292:771–774. doi: 10.1056/NEJM197504102921503. [DOI] [PubMed] [Google Scholar]
- 196.Stevens C E, Toy P T, Tong M J, Taylor P E, Vyas G N, Nair P V, Gudavalli M, Krugman S. Perinatal hepatitis B virus transmission in the United States: prevention by passive-active immunization. JAMA. 1985;253:1740–1745. [PubMed] [Google Scholar]
- 197.Stoll-Becker S, Repp R, Glebe D, Schaefer S, Kreuder J, Kann M, Lampert F, Gerlich W H. Transcription of hepatitis B virus in peripheral blood mononuclear cells from persistently infected patients. J Virol. 1997;71:5399–5407. doi: 10.1128/jvi.71.7.5399-5407.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Summers J, Mason W S. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell. 1982;29:403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- 199.Summers J, O'Connell A, Millman I. Genome of hepatitis B virus: restriction enzyme cleavage and structure of DNA extracted from Dane particles. Proc Natl Acad Sci USA. 1975;72:4597–4601. doi: 10.1073/pnas.72.11.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Summers J, Smith P M, Horwich A L. Hepadnavirus envelope proteins regulate covalently closed circular DNA amplification. J Virol. 1990;64:2819–2824. doi: 10.1128/jvi.64.6.2819-2824.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Summers J, Smith P M, Huang M J, Yu M S. Morphogenetic and regulatory effects of mutations in the envelope proteins of an avian hepadnavirus. J Virol. 1991;65:1310–1317. doi: 10.1128/jvi.65.3.1310-1317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Summers J, Smolec J, Snyder R. A virus similar to human hepatitis B virus associated with hepatitis and hepatoma in woodchucks. Proc Natl Acad Sci USA. 1978;75:4533–4537. doi: 10.1073/pnas.75.9.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Tagawa M, Omata M, Okuda K. Appearance of viral RNA transcripts in the early stage of duck hepatitis B virus infection. Virology. 1986;152:477–482. doi: 10.1016/0042-6822(86)90151-0. [DOI] [PubMed] [Google Scholar]
- 204.Tagawa M, Robinson W S, Marion P L. Duck hepatitis B virus replicates in the yolk sac of developing embryos. J Virol. 1987;61:2273–2279. doi: 10.1128/jvi.61.7.2273-2279.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Takada S, Koike K. X protein of hepatitis B virus resembles a serine protease inhibitor. Jpn J Cancer Res. 1990;81:1191–1194. doi: 10.1111/j.1349-7006.1990.tb02675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Takada S, Tsuchida N, Kobayashi M, Koike K. Disruption of the function of tumor-suppressor gene p53 by the hepatitis B virus X protein and hepatocarcinogenesis. J Cancer Res Clin Oncol. 1995;121:593–601. doi: 10.1007/BF01197776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Tavis J E, Ganem D. Expression of functional hepatitis B virus polymerase in yeast reveals it to be the sole viral protein required for correct initiation of reverse transcription. Proc Natl Acad Sci USA. 1993;90:4107–4111. doi: 10.1073/pnas.90.9.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Tennant B C, Mrosovsky N, McLean K, Cote P J, Korba B E, Engle R E, Gerin J L, Wright J, Michener G R, Uhl E, et al. Hepatocellular carcinoma in Richardson's ground squirrels (Spermophilus richardsonii): evidence for association with hepatitis B-like virus infection. Hepatology. 1991;13:1215–1221. [PubMed] [Google Scholar]
- 209.Terradillos O, Billet O, Renard C A, Levy R, Molina T, Briand P, Buendia M A. The hepatitis B virus X gene potentiates c-myc-induced liver oncogenesis in transgenic mice. Oncogene. 1997;14:395–404. doi: 10.1038/sj.onc.1200850. [DOI] [PubMed] [Google Scholar]
- 210.Testut P, Renard C A, Terradillos O, Vitvitski T L, Tekaia F, Degott C, Blake J, Boyer B, Buendia M A. A new hepadnavirus endemic in arctic ground squirrels in Alaska. J Virol. 1996;70:4210–4219. doi: 10.1128/jvi.70.7.4210-4219.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Thorgeirsson S S. Hepatic stem cells in liver regeneration. FASEB J. 1996;10:1249–1256. [PubMed] [Google Scholar]
- 212.Thorgeirsson S S, Evarts R P, Bisgaard H C, Fujio K, Hu Z. Hepatic stem cell compartment: activation and lineage commitment. Proc Soc Exp Biol Med. 1993;204:253–260. doi: 10.3181/00379727-204-43661. [DOI] [PubMed] [Google Scholar]
- 213.Tipples G A, Ma M M, Fischer K P, Bain V G, Kneteman N M, Tyrrell D L J. Mutation in HBV DNA-dependent DNA polymerase confers resistance to lamivudine in vivo. Hepatology. 1996;24:714–717. doi: 10.1002/hep.510240340. [DOI] [PubMed] [Google Scholar]
- 214.Tong S, Li J, Wands J R. Interaction between duck hepatitis B virus and a 170-kilodalton cellular protein is mediated through a neutralizing epitope of the pre-S region and occurs during viral infection. J Virol. 1995;69:7106–7112. doi: 10.1128/jvi.69.11.7106-7112.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Tsiquaye K N, Harrison T J, Portmann B, Hu S, Zuckerman A J. Acute hepatitis A infection in hepatitis B chimpanzee carriers. Hepatology. 1984;4:504–509. doi: 10.1002/hep.1840040325. [DOI] [PubMed] [Google Scholar]
- 216.Tsui L V, Guidotti L G, Ishikawa T, Chisari F V. Posttranscriptional clearance of hepatitis B virus RNA by cytotoxic T lymphocyte-activated hepatocytes. Proc Natl Acad Sci USA. 1995;92:12398–12402. doi: 10.1073/pnas.92.26.12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Tuttleman J S, Pourcel C, Summers J. Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell. 1986;47:451–460. doi: 10.1016/0092-8674(86)90602-1. [DOI] [PubMed] [Google Scholar]
- 218.Twu J S, Chu K, Robinson W S. Hepatitis B virus X gene activates kappa B-like enhancer sequences in the long terminal repeat of human immunodeficiency virus 1. Proc Natl Acad Sci USA. 1989;86:5168–5172. doi: 10.1073/pnas.86.13.5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Twu J S, Schloemer R H. Transcriptional trans-activating function of hepatitis B virus. J Virol. 1987;61:3448–3453. doi: 10.1128/jvi.61.11.3448-3453.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Ueda K, Ganem D. Apoptosis is induced by N-myc expression in hepatocytes, a frequent event in hepadnavirus oncogenesis, and is blocked by insulin-like growth factor II. J Virol. 1996;70:1375–1383. doi: 10.1128/jvi.70.3.1375-1383.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Urban M K, O'Connell A P, London W T. Sequence of events in natural infection of Pekin duck embryos with duck hepatitis B virus. J Virol. 1985;55:16–22. doi: 10.1128/jvi.55.1.16-22.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Urban S, Breiner K M, Fehler F, Klingmuller U, Schaller H. Avian hepatitis B virus infection is initiated by the interaction of a distinct pre-S subdomain with the cellular receptor gp180. J Virol. 1998;72:8089–8097. doi: 10.1128/jvi.72.10.8089-8097.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Vitvitski T L, Kay A, Pichoud C, Chevallier P, de D S, Shamoon B M, Mandart E, Trepo C, Galibert F. Early and frequent detection of HBxAg and/or anti-HBx in hepatitis B virus infection. Hepatology. 1990;12:1278–1283. doi: 10.1002/hep.1840120605. [DOI] [PubMed] [Google Scholar]
- 224.Wang G H, Seeger C. Novel mechanism for reverse transcription in hepatitis B viruses. J Virol. 1993;67:6507–6512. doi: 10.1128/jvi.67.11.6507-6512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Wang G H, Seeger C. The reverse transcriptase of hepatitis B virus acts as a protein primer for viral DNA synthesis. Cell. 1992;71:663–670. doi: 10.1016/0092-8674(92)90599-8. [DOI] [PubMed] [Google Scholar]
- 226.Wang G H, Zoulim F, Leber E H, Kitson J, Seeger C. Role of RNA in enzymatic activity of the reverse transcriptase of hepatitis B viruses. J Virol. 1994;68:8437–8442. doi: 10.1128/jvi.68.12.8437-8442.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Wang J, Chenivesse X, Henglein B, Brechot C. Hepatitis B virus integration in a cyclin A gene in a hepatocellular carcinoma. Nature. 1990;343:555–557. doi: 10.1038/343555a0. [DOI] [PubMed] [Google Scholar]
- 228.Wang J, Zindy F, Chenivesse X, Lamas E, Henglein B, Brechot C. Modification of cyclin A expression by hepatitis B virus DNA integration in a hepatocellular carcinoma. Oncogene. 1992;7:1653–1656. [PubMed] [Google Scholar]
- 229.Wang X W, Forrester K, Yeh H, Feitelson M A, Gu J R, Harris C C. Hepatitis B virus X protein inhibits p53 sequence-specific DNA binding, transcriptional activity, and association with transcription factor ERCC3. Proc Natl Acad Sci USA. 1994;91:2230–2234. doi: 10.1073/pnas.91.6.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Weber M, Bronsema V, Bartos H, Bosserhoff A, Bartenschlager R, Schaller H. Hepadnavirus P protein utilizes a tyrosine residue in the TP domain to prime reverse transcription. J Virol. 1994;68:2994–2999. doi: 10.1128/jvi.68.5.2994-2999.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Wei Y, Tavis J E, Ganem D. Relationship between viral DNA synthesis and virion envelopment in hepatitis B viruses. J Virol. 1996;70:6455–6458. doi: 10.1128/jvi.70.9.6455-6458.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Wei Y, Tennant B, Ganem D. In vivo effects of mutations in woodchuck hepatitis virus enhancer II. J Virol. 1998;72:6608–6613. doi: 10.1128/jvi.72.8.6608-6613.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Will H, Cattaneo R, Koch H, Darai G, Schaller H, Schellekens H, van Eerd P, Deinhardt F. Cloned HBV DNA causes hepatitis in chimpanzees. Nature. 1982;299:740–742. doi: 10.1038/299740a0. [DOI] [PubMed] [Google Scholar]
- 234.Wong D K, Cheung A M, O'Rourke K, Naylor C D, Detsky A S, Heathcote J. Effect of alpha-interferon treatment in patients with hepatitis B e antigen-positive chronic hepatitis B. A meta-analysis. Ann Intern Med. 1993;119:312–323. doi: 10.7326/0003-4819-119-4-199308150-00011. [DOI] [PubMed] [Google Scholar]
- 235.Wynne S A, Crowther R A, Leslie A G. The crystal structure of the human hepatitis B virus capsid. Mol Cell. 1999;3:771–780. doi: 10.1016/s1097-2765(01)80009-5. [DOI] [PubMed] [Google Scholar]
- 236.Xiong X, Flores C, Yang H, Toole J J, Gibbs C S. Mutations in hepatitis B virus DNA polymerase associated with resistance to lamivudine do not confer resistance to Adefovir in vitro. Hepatology. 1998;28:1669–1673. doi: 10.1002/hep.510280629. [DOI] [PubMed] [Google Scholar]
- 237.Yaginuma K, Shirakata Y, Kobayashi M, Koike K. Hepatitis B virus (HBV) particles are produced in a cell culture system by transient expression of transfected HBV DNA. Proc Natl Acad Sci USA. 1987;84:2678–2682. doi: 10.1073/pnas.84.9.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Yang D, Alt E, Rogler C E. Coordinate expression of N-myc 2 and insulin-like growth factor II in precancerous altered hepatic foci in woodchuck hepatitis virus carriers. Cancer Res. 1993;53:2020–2027. [PubMed] [Google Scholar]
- 239.Yang D, Faris R, Hixson D, Affigne S, Rogler C E. Insulin-like growth factor II blocks apoptosis of N-myc2-expressing woodchuck liver epithelial cells. J Virol. 1996;70:6260–6268. doi: 10.1128/jvi.70.9.6260-6268.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Yang D Y, Rogler C E. Analysis of insulin-like growth factor II (IGF-II) expression in neoplastic nodules and hepatocellular carcinomas of woodchucks utilizing in situ hybridization and immunocytochemistry. Carcinogenesis. 1991;12:1893–1901. doi: 10.1093/carcin/12.10.1893. [DOI] [PubMed] [Google Scholar]
- 241.Yang W, Mason W S, Summers J. Covalently closed circular viral DNA formed from two types of linear DNA in woodchuck hepatitis virus-infected liver. J Virol. 1996;70:4567–4575. doi: 10.1128/jvi.70.7.4567-4575.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Yang W, Summers J. Illegitimate replication of linear hepadnavirus DNA through nonhomologous recombination. J Virol. 1995;69:4029–4036. doi: 10.1128/jvi.69.7.4029-4036.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Yoffe B, Noonan C A, Melnick J L, Hollinger F B. Hepatitis B virus DNA in mononuclear cells and analysis of cell subsets for the presence of replicative intermediates of viral DNA. J Infect Dis. 1986;153:471–477. doi: 10.1093/infdis/153.3.471. [DOI] [PubMed] [Google Scholar]
- 244.Young J A, Bates P, Varmus H E. Isolation of a chicken gene that confers susceptibility to infection by subgroup A avian leukosis and sarcoma viruses. J Virol. 1993;67:1811–1816. doi: 10.1128/jvi.67.4.1811-1816.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Yu M, Summers J. Multiple functions of capsid protein phosphorylation in duck hepatitis B virus replication. J Virol. 1994;68:4341–4348. doi: 10.1128/jvi.68.7.4341-4348.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Yu M, Summers J. Phosphorylation of the duck hepatitis B virus capsid protein associated with conformational changes in the C terminus. J Virol. 1994;68:2965–2969. doi: 10.1128/jvi.68.5.2965-2969.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Yuasa S, Cheung R C, Pham Q, Robinson W S, Marion P L. Peptide mapping of neutralizing and nonneutralizing epitopes of duck hepatitis B virus pre-S polypeptide. Virology. 1991;181:14–21. doi: 10.1016/0042-6822(91)90465-n. [DOI] [PubMed] [Google Scholar]
- 248.Zachoval R, Roggendorf M, Deinhardt F. Hepatitis A infection in chronic carriers of hepatitis B virus. Hepatology. 1983;3:528–531. doi: 10.1002/hep.1840030409. [DOI] [PubMed] [Google Scholar]
- 249.Zanta M A, Belguise-Valladier P, Behr J P. Gene delivery: a single nuclear localization signal peptide is sufficient to carry DNA to the cell nucleus. Proc Natl Acad Sci USA. 1999;96:91–96. doi: 10.1073/pnas.96.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Zhang Y-Y, Summers J. Selective advantage of a precore-minus mutant in mixed infections with duck hepatitis B virus. J Virol. 1999;73:3616–3622. doi: 10.1128/jvi.73.5.3616-3622.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Zhou D X, Taraboulos A, Ou J H, Yen T S. Activation of class I major histocompatibility complex gene expression by hepatitis B virus. J Virol. 1990;64:4025–4028. doi: 10.1128/jvi.64.8.4025-4028.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Zlotnick A, Cheng N, Conway J F, Booy F P, Steven A C, Stahl S J, Wingfield P T. Dimorphism of hepatitis B virus capsids is strongly influenced by the C-terminus of the capsid protein. Biochemistry. 1996;35:7412–7421. doi: 10.1021/bi9604800. [DOI] [PubMed] [Google Scholar]
- 253.Zlotnick A, Stahl S J, Wingfield P T, Conway J F, Cheng N, Steven A C. Shared motifs of the capsid proteins of hepadnaviruses and retroviruses suggest a common evolutionary origin. FEBS Lett. 1998;431:301–304. doi: 10.1016/s0014-5793(98)00755-8. [DOI] [PubMed] [Google Scholar]
- 254.Zoulim F, Saputelli J, Seeger C. Woodchuck hepatitis virus X protein is required for viral infection in vivo. J Virol. 1994;68:2026–2030. doi: 10.1128/jvi.68.3.2026-2030.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Zoulim F, Seeger C. Reverse transcription in hepatitis B viruses is primed by a tyrosine residue of the polymerase. J Virol. 1994;68:6–13. doi: 10.1128/jvi.68.1.6-13.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]