Summary
Cardiovascular disorders such as heart failure are leading causes of mortality. Patient stratification via identification of novel biomarkers could improve management of cardiovascular diseases of complex etiologies. Long-noncoding RNAs (lncRNAs) are highly tissue-specific in nature and have emerged as important biomarkers in human diseases. In this study, we aimed to identify cardiac-enriched lncRNAs as potential biomarkers for cardiovascular conditions.
Deep RNA sequencing and quantitative PCR identified differentially expressed lncRNAs between failing and non-failing hearts. An independent dataset was used to evaluate the enrichment of lncRNAs in normal hearts.
We identified a panel of 2906 lncRNAs, named FIMICS, that were either cardiac-enriched or differentially expressed between failing and non-failing hearts. Expression of lncRNAs in blood samples differentiated patients with myocarditis and acute myocardial infarction.
We hereby present the FIMICS panel, a readily available tool to provide insights into cardiovascular pathologies and which could be helpful for diagnosis, monitoring and prognosis purposes.
Keywords: Biomarker, Heart failure, Cardiovascular condition, Long-noncoding RNA, Deep RNA sequencing, Cardiac enriched RNA
1. Introduction
Despite significant advances in healthcare, cardiovascular disorders remain the major cause of mortality and morbidity across the world [1]. Among these, heart failure (HF) plays a predominant role. HF can occur as a consequence of various cardiac abnormalities leading to a complex clinical outcome with multiple phenotypes [2]. Different aetiologies of cardiological complications imply the need for diverse and specific biomarkers in order to assess cardiac function, establish accurate diagnostic and prognostic measure, stratify patient's outcome, and monitor treatment regimens. Such biomarkers may foster the implementation of personalized medicine and improve patient management in the clinics [3].
Natriuretic peptide testing, especially N-terminal pro-B-type natriuretic peptide (Nt-proBNP), is widely used to support diagnosis and management of patients with HF and is recommended in the guidelines of European Society of Cardiology and American Heart Association [4,5]. However, the diagnostic value of natriuretic peptides may be limited by the heterogeneity of the underlying heart-associated disease, patient's age, sex, genetic background, and lifestyle [6]. Therefore, novel, non-invasive yet highly sensitive and specific biomarkers are needed. When combined with classical disease indicators and risk factors, these biomarkers may provide a detailed and more accurate fingerprint of the patient's disease state.
Since the initial sequencing of the human genome [7,8], considerable progress has been made in the understanding of its complexity. It is widely accepted that only a minor part of the human DNA encodes proteins, while the remaining is transcribed into non-protein coding RNAs also known as noncoding RNAs [9]. Noncoding RNAs have been dichotomized as short noncoding RNAs (<200 nucleotides) – among which the well-known 20–22 nucleotide-long microRNAs (miRNAs) – and long noncoding RNAs (lncRNAs, >200 nucleotides). While miRNAs down-regulate gene expression mostly by destabilization of target messenger RNA, the regulation of gene expression by lncRNAs appears to be much more complex, involving both activation and repression of gene expression, and other epigenetics mechanisms including modulation of chromatin architecture [10]. Since their discovery, lncRNAs have emerged as attractive biomarkers for human diseases [11] and therapeutic targets of cardiac diseases [10]. This relates to their specificity in tissue distribution and function. More recently, circular RNAs also showed interesting regulation in HF and, due to their stability, can be considered as HF biomarkers [12]. Overall, our knowledge of the role of lncRNAs in cardiovascular diseases and their biomarker value is still in its infancy [13,14].
Even if only a few lncRNAs have been shown to be biologically relevant and functionally annotated, there is growing evidence that the majority of lncRNAs are likely to be functional. While the exact function of most lncRNAs still remains unknown, they have been shown to be implicated in various biological processes, mainly relating to transcriptional, post-transcriptional and epigenetic regulation [10]. To date, most of the functionally characterized lncRNAs regulate developmental processes. In addition, a deep profiling of the murine cardiac transcriptome after myocardial infarction supports the involvement of lncRNAs in controlling cardiac remodelling and regeneration [15]. Hundreds of human orthologs of these lncRNAs were regulated in human cardiac disease [15].
RNA sequencing (RNA-seq) has become a widely used technology for gene expression profiling. In publicly available RNA-seq data associated with HF, most studies used a poly-adenylation (poly-A)-based method whilst others had a sequencing depth less than 50 million reads on average. However, only ∼60% of lncRNAs are covered by poly-A-based RNA-seq [16]. In addition, lncRNAs are weakly expressed as compared to protein-coding RNAs. Thus, a reliable detection of known lncRNA transcripts and the discovery of novel transcribed elements using RNA-seq require a minimum sequencing depth of 100–200 million reads. To fill these gaps, we performed deep non-poly-A-restricted RNA-seq on biopsies from failing and non-failing human hearts. This allowed us to identify a comprehensive panel of potential RNA biomarkers predictive of cardiac health and ailments.
2. Materials and methods
2.1. Human samples
Left ventricular biopsies were obtained from 11 patients with ischemic cardiomyopathy (ICM), 10 patients with dilated cardiomyopathy (DCM) and 5 organ donors (controls) having either head injury (n = 2) or subarachnoid haemorrhage (n = 3). The aetiology of HF (ICM or DCM) was established based on patient's history, echocardiography, coronary angiography and appropriate additional tests. All patients presented chronic (over 6 months) left ventricular failure and were optimally treated accordingly to ESC guidelines. In case of coronary artery disease, ICM was defined. DCM patients were identified from non-ICM patients, and were subjected to genetic testing. Controls had no history of heart disease and detailed echocardiography and coronary arteries angiography were performed before heart explantation. A final verification was performed during tissue sampling by the researcher in charge of samples collection. The protocol was approved by the Local Ethics Committee at Cardinal Stefan Wyszynski Institute of Cardiology (approval number IK-NP-0021-48/846/13; April 09, 2013). No donors or their relatives completed the national refusal list. Biopsies were snap-frozen and stored at −80 °C until RNA isolation. For validation of selected lncRNAs, left ventricular biopsies from a validation cohort of 93 patients were used (32 ICM, 43 DCM and 18 controls). To test the FIMICS panel, whole blood and plasma samples from healthy volunteers (n = 148 and n = 99, respectively), patients with myocarditis (MYO) (n = 13) and patients with acute myocardial infarction (AMI) (n = 10) were used. Patients adjudication has been reported elsewhere [17]. Cohort demographics are described in Supplementary Table 1.
2.2. RNA isolation from cardiac biopsies
Total RNA was extracted from cardiac biopsies using the mirVana isolation kit (Life technologies, Merelbeke, Belgium). Potential contaminating genomic DNA was digested with DNase I (Qiagen, Venlo, The Netherlands). RNA concentration was measured with a Nanodrop spectrophotometer (Nanodrop products, Wilmington, USA) and RNA integrity was verified using a 2100 Bioanalyzer (Agilent technologies, Santa Clara, USA).
2.3. RNA sequencing
Sequencing libraries were prepared from 0.5 μg of total RNA using the Illumina TruSeq stranded total 5 RNA library preparation kit combined with the human/mouse/rat RiboZero ribosomal RNA removal kit (Illumina Inc. San Diego, USA). All steps were performed with the low-throughput protocol and according to the manufacturer's instructions. Briefly, ribosomal RNA-depleted RNA samples were fragmented by heat digestion with divalent cations (8 min at 94 °C) and reverse transcribed into cDNA using Superscript II reverse transcriptase (Thermo Fisher Scientific, Luxembourg), and then converted to double-stranded cDNA with the second strand marking mix that incorporates dUTP in place of dTTP. Resulting blunt ended cDNA was purified using AMPure XP magnetic beads. After a 3'end adenylation step, Illumina's adapter ligation was performed. The single indexed libraries were enriched by PCR (15 cycles). For quality control, 1 μl of each library was run on the Agilent Technologies 2100 Bioanalyzer using a DNA 1000 chip according to the manufacturer's recommendations. All libraries were sequenced with the Illumina NextSeq500 (2x75bp).
2.4. RNA sequencing data processing
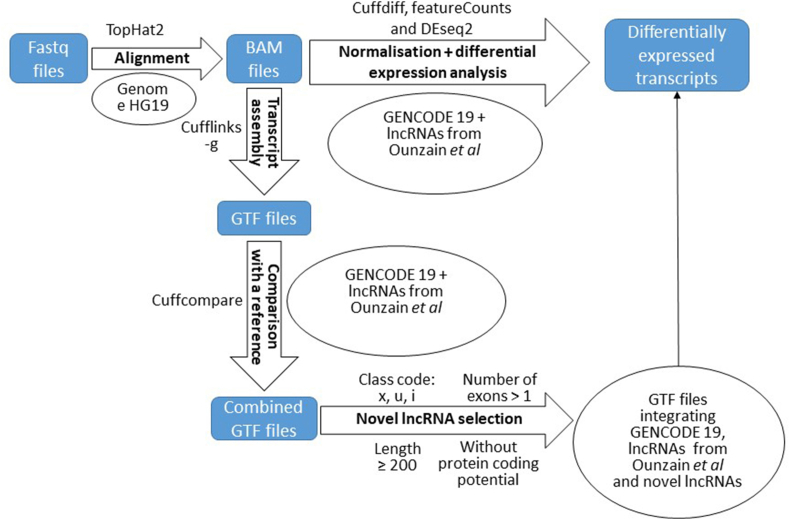
Quality control of the RNA-seq data was performed using FastQC tool (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Trimmomatic (https://doi.org/10.1093/bioinformatics/btu170) was used to trim adaptor sequenes: java -jar trimmomatic-0.35.jar PE -phred33 in_R1.fq.gz in_R2.fq.gz out_R1_paired.fq.gz out_R1_unpaired.fq.gz out_R2_paired.fq.gz out_R2_unpaired.fq.gz ILLUMINACLIP:TruSeq3-PE-2.fa:2:30:10 MINLEN:50. FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) was used to eliminate low-quality bases with quality score less than 28: fastq_quality_filter -q 28 -p 0.8 -z -i in.fq.gz -o out.fq.gz. Then the reads were aligned to the human reference genome (hg19) using Tophat 2.1.0 program with default set-up [18]: tophat --b2-sensitive hg19_index in_R1.fq.gz in_R2.fq.gz. Bowtie index of UCSC reference sequences were downloaded from Illumina iGenomes (https://support.illumina.com/sequencing/sequencing_software/igenome.html).
2.4.1. Pipeline of novel lncRNA prediction
Reference annotation based transcript assembly [19] was performed using cufflinks with -g option: cufflinks -g GENCODE19_-M hg19_rRNA_RepeatMasker.gtf Ounzain.gtf –N tophat_out.bam, which output all reference transcripts as well as any novel genes and isoforms that were assembled. The annotation was performed using the 3 samples from each group (Control, ICM, DCM) that have the highest number of reads. The known reference annotation was generated from GTF files from GENCODE v19 comprehensive gene annotation release 19 [20] and human lncRNAs identified by Ounzain et al. [15]. The Cuffcompare program [21] was used to compare the assembled transcripts and genes to the known reference annotation to generate a new GTF file with all transcripts from 9 samples for further analysis: cuffcompare -r GENCODE19_Ounzain.gtf cufflinks_1.gtf cufflinks_2.gtf … cufflinks_9.gtf.
Filtering was done on Transfrag class codes (the type of match between the assembled transcript and the reference transcript) generated by Cuffcompare [21], transcript length, number of exons and protein coding potential. Firstly, the transcripts with code ‘i’, ‘j’, ‘o’, ‘u’, ‘x’ and ‘.’ were extracted, all of which could potentially include novel lncRNAs. The ‘i’ category, for example, could contain the lncRNAs entirely within the intron of known genes. Similarly, the ‘j’ category could be long noncoding isoforms of known genes. The ‘o’ category could include novel lncRNAs having generic exonic overlap with known transcripts. The ‘u’ category could be long intergenic noncoding RNAs. The ‘x’ category could contain novel lncRNAs on the opposite strand of reference genes. The ‘.’ category may be sequences with multiple classifications. The combined GTF file was converted to a BED file using UCSC table browser (https://genome.ucsc.edu/cgi-bin/hgTables). Following this, only the transcripts with a length ≥200 nucleotides and with at least 2 exons were kept for the next step. Finally, the BED files of transcripts from the last 2 filtering steps were uploaded to the coding-potential assessment tool (CPAT) [22] to calculate the coding potential score. The transcripts with CPAT scores <0.364 were considered as noncoding [22]. A new GTF file was generated with the final list of selected novel lncRNAs.
2.4.2. Differential expression of lncRNAs
The transcript assembly was performed using cufflinks 2.2.1 21 with -G option and/or the featureCounts function of Rsubread R package [23] with the known reference annotation and the novel lncRNAs described in the previous section. Cuffdiff [21] and/or DESeq2 [24] were used for differential expression analysis. Transcripts from the known reference annotation with an adjusted p-value <0.05 and fold-change ≥2 between controls and failing hearts (either ICM or DCM), or with p-value <0.05 between ICM and DCM, were considered to be differentially expressed. The novel lncRNAs with p-value <0.05 were considered to be differentially expressed.
2.4.3. Analysis of GSE45326 dataset
FASTQ files of the publicly-available GSE45326 [25] dataset were downloaded from ArrayExpress [26]. The dataset includes total RNA-seq data of 12 normal human tissues. Reads alignment and transcript assembly using TopHat and cufflinks for known genes and novel lncRNAs were performed as described before. The transcripts with fragments per kilobase per million reads mapped (FPKM) ≥ 1 and at least twice higher than the FPKM of any other tissues were considered as cardiac-enriched.
2.5. Functional analysis of lncRNAs
Gene enrichment analysis was performed using DAVID (The Database for Annotation, Visualization and Integrated Discovery) web tool [27]. The coding neighbour of a lncRNA was defined as the protein-coding gene adjacent to the lncRNA with a distance less than 50 kb. For lncRNAs with more than 1 adjacent coding gene, the closest one was considered. Heart failure-associated genes were obtained from NCBI gene database (https://www.ncbi.nlm.nih.gov/gene) using the keywords: “heart failure” or “failing heart”. The Spearman's rank correlation between the expression of protein-coding genes and lncRNAs was calculated with WGCNA R package [28].
2.6. Quantitative RT-PCR (qRT-PCR)
Human cardiac biopsies were homogenized using a Polytron® in TriReagent® (Sigma). Total RNA was extracted using the RNeasy® Mini kit (Qiagen). After extraction, RNA was quantified with the ND-1000 spectrophotometer (NanoDrop® Technologies). One microgram of total RNA was reverse-transcribed using the Superscript II RT kit (Life technologies, Belgium). Real-time PCR was performed in a CFX96 apparatus (Biorad, Temse, Belgium) with IQ SYBR Green Supermix (Biorad) and primers were designed with the Beacon Designer software (Premier Biosoft; Supplementary Table 2). Glyceraldehyde-3-phosphate deshydrogenase (GAPDH) was chosen as a housekeeping gene for normalization. Expression levels were calculated by the relative quantification method (ΔΔCt) using the CFX Manager 2.1 software (Bio-Rad).
2.7. Development of FIMICS kit for lncRNA detection from blood
2.7.1. RNA extraction and library preparation
To optimize cardiac lncRNA quantification in peripheral blood samples, a targeted sequencing kit to specifically quantify the lncRNAs of interest was developed. For this purpose, capture probes of 120 nucleotides specific and complementary to the sequence of lncRNAs were designed. Multiple probes per lncRNA were designed for each sequence over 200 bp to cover specific regions. Maximum probe coverage was limited to 2000 nucleotides. Region to design probes were determined by blast to define unique and specific region of each lncRNA of interest. 55974 probes were designed to cover all the lncRNAs sequences. This panel uses Celemics beads-based hybridization capture technology.
Briefly, library preparation was performed as following: in order to convert total RNA into a library of template molecules of known strand origin, the KAPA Stranded RNA-Seq Kit with RiboErase (HMR) was used. 500 ng of total RNA extracted from PAXgene blood RNA was used. A ribosomal depletion step was performed in order to remove cytoplasmic rRNA, then RNA molecules were cleaned up with the AMPure XP beads, a DNAse digestion step was conducted and fragmentation parameters were defined according to RNA quality. After these initial steps, the first and the second strands cDNA were synthesized prior to perform an A-tailing reaction in order to adenylate the 3′ ends of the double-stranded cDNA molecules. This A-tailing step is crucial for the ligation of the Illumina adapters. Finally, the DNA fragments were enriched by PCR with 15 cycles.
For quality control, 1 μl of each library was run on the Agilent Technologies 2100 Bioanalyzer using a DNA 1000 chip according to the manufacturer's recommendations. Absence of adapter dimers was checked and the average library size was determined by a region table. Libraries were quantified on Qubit 3.0 using Qubit 1X dsDNA High Sensitivity assay kit (Invitrogen). Library size previously determined on the Bioanalyzer was used to calculate molar concentrations from mass concentrations.
2.7.2. FIMICS capture
The lncRNA capture panel, named FIMICS™ and manufactured by Celemics Inc. (Seoul, Korea), was used to capture cardiac lncRNAs of interest. Briefly, biotinylated target capture probes, specific for the lncRNAs of interest, were hybridized to the sequencing libraries at 65 °C for 24h. Then, captured lncRNA sequences were purified on T1 streptavidin-coated magnetic beads. Seven successive washes were performed to eliminate all libraries sequences not specific to the panel probes. Finally, the on-beads captured sequences were enriched by PCR (14 cycles). Then, PCR products were purified with Beckman Coulter™ Agencourt AMPure XP beads and the captured libraries were eluted in 30 μl of nuclease-free water.
2.7.3. Sequencing
Denaturation and dilution of the prepared libraries for sequencing on the Illumina NextSeq 500 were performed according to the manufacturer's recommendation. Briefly, libraries were denatured and diluted (prepared libraries and PhIX library) to a final loading volume of 1.3 ml at a recommended concentration of 1.8 pM for high output kits. The PhiX library provides a quality control for cluster generation, sequencing, and alignment, and a calibration control for cross-talk matrix generation, phasing, and prephasing. High Output Flow cells were used for sequencing at a read length set to 2 x 75 bp.
2.7.4. Demultiplexing step
The sample sheet was prepared using the Illumina Experiment Manager (version 1.16). The BCL files were converted into FASTQ files using the bcl2fastq software (by Illumina, version 2.18.0.12). The option –no-lane-splitting was used to not split the FASTQ files by lane. All other default parameters were used.
2.7.5. Quality checking and trimming reads
The samples were imported in project in Partek Flow software. Before the alignment, a pre-alignment QA/QC was done to check the quality of the input data. All reads were examined. The raw FASTQ files were trimmed at the 3’ end in function of their quality score (Phred score). The parameters used were an end minimum quality level of 25 and a minimum read length of 50.
2.7.6. Mapping reads and quantification
The raw reads were aligned to the Homo sapiens hg19 reference genome using the software STAR version 2.5.3 [29] (using Partek Flow software). The default parameters were used. After the mapping step, a new quality checking was done (Post-alignment QA/QC). Then, mapped reads were quantified against the annotation with all lncRNAs from FIMICS panel with the Partek Expectation/Maximization (E/M) algorithm [30]. The Ensembl IDs of known lncRNAs of FIMICS panel identified from GENCODE19 (FIMICSv1.0) were mapped to Ensembl IDs of the lncRNAs from GENCODE39 to generate FIMICSv2.0 panel. The sequences of novel lncRNAs of FIMICSv1.0 were aligned to all RNA sequences from GENCODE39 and the sequences having more than 90% similarity in identity and coverage with known RNAs were excluded from FIMICSv2.0 panel.
2.8. Statistical analysis
SigmaPlot v12.0 software was used for statistical analyses of qRT-PCR data. All tests were preceded by the Shapiro-Wilk normality test. Comparisons between 3 groups were performed using one-way analysis of variance (ANOVA) for Gaussian data and ANOVA on rank for non-Gaussian data. Correlation coefficients between two variables were determined using the Spearman test. A p-value <0.05 was considered significant.
3. Results
Characteristics of patients and controls included in this study are gathered in Supplementary Table 1.
3.1. Deep RNA-seq
To identify lncRNAs predictive of cardiac conditions and diseases we first performed a deep RNA-seq experiment in left ventricular biopsies from 11 patients with ICM, 10 patients with DCM and 5 controls. On average, 210 ± 81 M reads per sample were obtained. Raw RNA-seq data are deposited in the Gene Expression Omnibus under the reference GSE165556.
3.2. Expression of known lncRNAs in human left ventricular biopsies
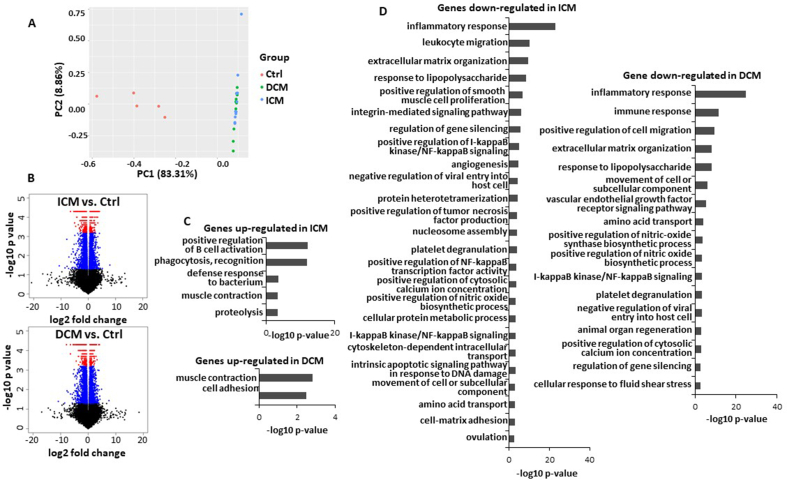
Differentially expressed lncRNAs between controls and failing heart biopsies were analyzed from the RNA-seq data. The analytical workflow for the discovery is described in Supplementary Fig. 1. To assess the expression of known lncRNAs, transcripts were assembled using the sequences from GENCODE. The transcripts having more than 2 reads in half of samples of any group were considered. This resulted in 71157 identified transcripts. A principal component analysis (PCA) showed that the study participants can be correctly separated into control and heart failure groups by using RNA-seq expression data (Fig. 1A). The expression profiles of patients in ICM and DCM groups were similar (Fig. 1A).
Fig. 1.
Differential expression of transcripts between heart failure and controls. A. Principal component analysis (PCA) showing that gene expression profiles (containing both known and unknown transcripts) can discriminate heart failure patients (DCM in green and ICM in blue) from controls (Ctrl in red). RNA-seq data from discovery cohort of 11 ICM patients, 10 DCM patients and 5 controls are included in this analysis. B. Volcano plots showing differentially expressed transcripts between ICM patients and controls (left panel), and between DCM patients and controls (right panel) (11 ICM, 10 DCM and 5 controls). Blue dots indicate transcripts with a p-value <0.05; red dots indicate transcripts with a false discovery rate (FDR) < 0.05. C. Gene enrichment analysis of transcripts upregulated in ICM or DCM compared to controls. D. Gene enrichment analysis of transcripts downregulated in ICM or DCM compared to control group. (FDR <0.05 and fold-change ≥2). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The Volcano plots in Fig. 1B show the differentially expressed known transcripts as analyzed with Cuffdiff, either with a p-value <0.05 (blue dots) or with a false discovery rate (FDR) < 0.05 (red dots). Using a FDR <0.05 and a fold change ≥2 as cut-offs, 715 transcripts were differentially expressed between ICM patients and controls, and 672 were differentially expressed between DCM patients and controls.
To understand the functional aspect of gene regulation, we performed enrichment analysis with genes regulated with a FDR <0.05 and a fold change ≥2. In both ICM and DCM groups, up-regulated genes (compared to control group) were implicated in muscle contraction while down-regulated genes were mostly enriched in inflammatory response (Fig. 1 C, D).
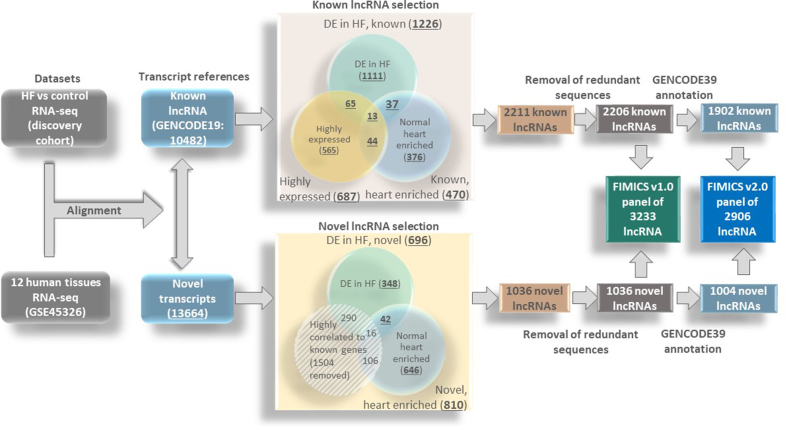
Mapping of our RNA-seq data to GENCODE19 allowed the detection of 10,482 known lncRNAs (Fig. 2, green box upper panel). Using a FDR <0.05 and a fold change ≥2 as cut-offs, we found 1226 differentially expressed lncRNAs in either ICM or DCM or control groups (Fig. 2, green circle in upper Venn diagram). 606 lncRNAs were differentially expressed in ICM vs control and 642 lncRNAs were differentially expressed in DCM vs controls. Moreover, 484 lncRNAs were differentially expressed between ICM and DCM groups (Supplementary Table 3). To further enrich the panel with highly expressed lncRNAs in heart tissue, 687 known lncRNAs were identified with at least 2 FPKM in more than half of the samples from each group (Fig. 2, yellow circle in upper Venn diagram, Supplementary Table 3). After identification of differentially expressed known lncRNAs, we extrapolated our analysis to identify differentially expressed novel lncRNAs.
Fig. 2.
Selection of lncRNAs for the FIMICS panel. The flowchart represents the workflow and displays the numbers of lncRNAs at each step of the selection process. From left to right: the grey boxes represent the RNA-seq datasets used for the analysis. The green box shows known lncRNAs detected from GENCODE19 (10,482), the light blue box shows novel transcripts identified (13,664), the Venn diagram in pink box shows the known lncRNAs selected by different strategies [differentially expressed (DE) in HF (green), highly expressed (yellow) and cardiac enriched (blue)], the Venn diagram in yellow box shows the novel lncRNAs [DE in HF (green), cardiac enriched (blue), and removed due to high correlation to known genes (grey striped circle)]. 2211 known and 1036 novel lncRNAs were identified (orange boxes), of which 3233 (dark blue box) were included in the FIMICS panel after removal of 14 lncRNAs due to sequence redundancy. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. Identification of novel lncRNAs in human left ventricular biopsies
To identify differentially expressed novel lncRNAs, we initially identified all detected novel transcripts from the RNA-seq data. Using the lncRNA annotation pipeline displayed in Supplementary Fig. 1, we identified 13,664 novel multi-exonic sequences (Fig. 2, light blue box lower panel) longer than 200 base pairs lacking protein-coding potential as predicted by CPAT (Supplementary Table 4). Among these transcripts, 10,622 were mapped as novel isoforms of known genes from GENCODE and were subsequently removed. 3042 lncRNAs were derived from novel genes (Supplementary Table 4).
Expression levels of the 13,664 novel lncRNAs in left ventricular biopsies from failing and control hearts were retrieved from RNA-seq data. After removal of the lncRNAs with less than 2 reads in more than half of the samples from all groups, we identified 696 novel differentially expressed lncRNAs between HF and controls (Fig. 2, green circle in lower Venn diagram). Of these, 665 novel lncRNAs were differentially expressed between ICM and control hearts and 658 between DCM and control hearts using Cuffdiff algorithms with a FDR <0.05 (Supplementary Table 5). In addition, 24 novel lncRNAs were differentially expressed between ICM and DCM patients.
3.4. Cardiac enriched lncRNAs and their differential expression
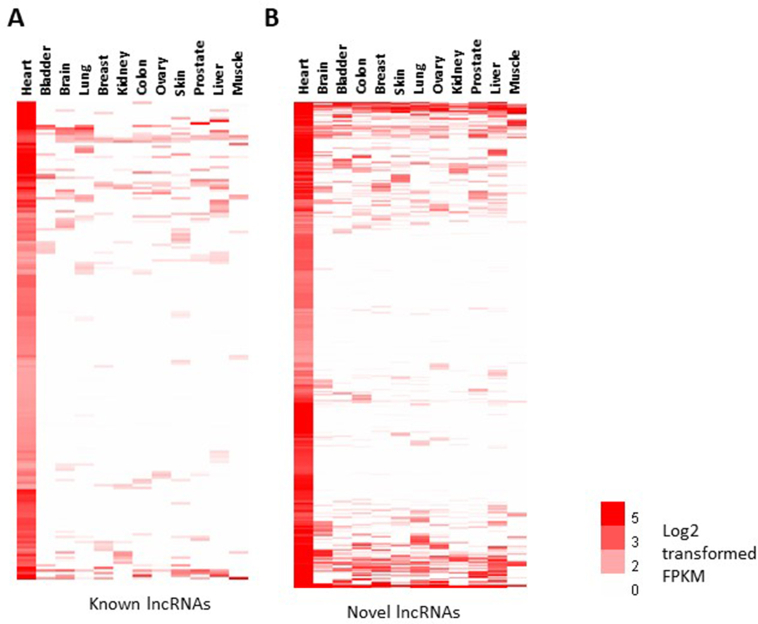
To address a potential enrichment in the heart of the known lncRNAs from GENCODE19 and novel lncRNAs, we analyzed the public RNA-seq dataset GSE45326 [25] which includes 12 different healthy human organs - heart, brain, bladder, colon, breast, skin, lung, ovary, kidney, prostate, liver and muscle, using the same transcript references as those used in our analysis of control and failing left ventricles. We observed that 470 known (Fig. 2, blue circle upper Venn diagram) and 810 novel lncRNAs (Fig. 2, blue circle lower Venn diagram) were expressed in the heart at a level higher than 1 FPKM and higher than twice the maximal expression in the other organs (Fig. 3). These lncRNAs were considered to be cardiac-enriched.
Fig. 3.
Expression levels of cardiac-enriched lncRNAs and selection of lncRNAs. Heatmaps representing the expression levels of 470 known (A) and 810 novel (B) lncRNAs cardiac-enriched and differentially expressed between failing and controls hearts in HF vs control RNA-seq data. Expression level in heart more than 1 FPKM and higher than twice the maximal expression in the other 11 organs are considered cardiac enriched. The GSE45326 RNA-seq dataset from 12 different human organs was used for this analysis.
Together, we identified 2211 known lncRNAs (Fig. 2, upper Venn diagram green, blue, yellow together) and 1448 novel lncRNAs (Fig. 2, lower Venn diagram blue and green circles together) which were either differentially expressed in failing hearts as compared to control hearts or enriched in the heart. Among the 1448 novel lncRNAs, 412 lncRNAs (Fig. 2, lower Venn diagram, grey striped circle) had overlapping and/or shared exons with known genes on the same strand. Their expression were correlated with their known genes with a Spearman correlation coefficient above 0.7. Due to this high correlation and therefore high probability of co-transcription with the known genes, they were not considered novel and were removed from the panel.
Overall, we identified 3247 human lncRNAs, including 2211 known and 1036 novel lncRNAs (Fig. 2, brown boxes), either associated with HF (i.e. differentially expressed between failing and control hearts) or enriched in the cardiac tissue (Fig. 3, Supplementary Tables 3 and 5). 14 of these lncRNAs were removed due to sequence redundancy. Thus, a total of 3233 lncRNAs (Fig. 2, dark green box) (2206 known and 1027 novel lncRNAs) constitute the FIMICSv1.0 panel (Supplementary Table 6). The lncRNAs identified in FIMICSv1.0 were mapped to the latest GENCODE39. We excluded 312 GENCODE19 known lncRNAs from FIMICSv1.0 which were not present in GENCODE39. We excluded 23 novel lncRNAs from FIMICSv1.0, which were mapped to GENCODE39 sequences with both identity and coverage matching of 90% or more. Among these, 8 were classified as lncRNAs in GENCODE39 and were added to the known lncRNA list of FIMICS panel v2.0. Finally, the FIMICSv2.0 was formulated with 2906 (1902 known and 1004 novel) lncRNAs (Fig. 2, blue box; Supplementary Table 6). Expressions of these lncRNAs with >1CPM were confirmed in left ventricle (ENCODE ID: ENCSR436QDU, ENCSR391VGU) and right atrium auricular region (ENCODE ID: ENCSR457ENP, ENCSR571RXE) in independent publicly available cardiac tissue datasets from ENCODE database (https://www.encodeproject.org/) (Supplementary Fig. 2).
3.5. Functional analysis of novel lncRNAs
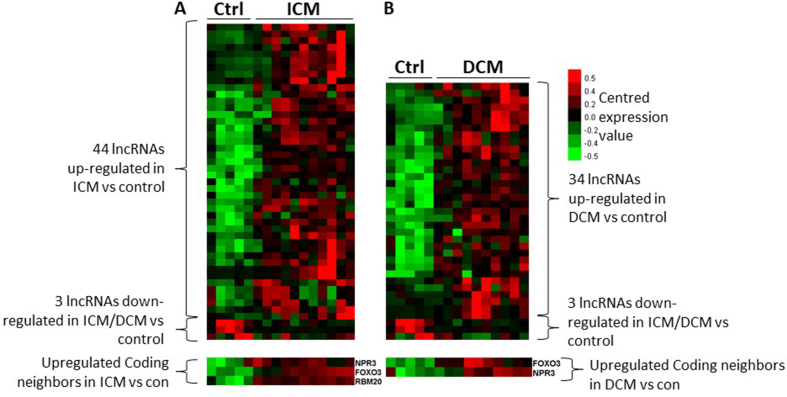
As described above and shown in Supplementary Tables 4 and 3042 lncRNAs were derived from novel genes. LncRNAs with more than 0.1 FPKM in more than half of the samples from 1 group, i.e. in at least 3 samples in control group or in at least 6 samples in heart failure group, were retained for subsequent analysis. Using a FDR <0.05 and a fold change ≥2 as cut-offs for differential expression, we identified 44 (Fig. 4A) and 34 (Fig. 4B) novel cardiac-enriched lncRNAs derived from novel genes which were significantly increased in ICM and DCM groups respectively, compared to controls. In addition, 3 such novel lncRNAs were decreased in both ICM and DCM groups as compared to the control group (Fig. 4).
Fig. 4.
Differentially expressed novel lncRNAs. Heatmaps representing the novel lncRNAs differentially expressed between failing (11 ICM, 10 DCM) and control (5) hearts. A. control vs. ICM, B. control vs. DCM group. A false discovery rate (FDR) < 0.05 and a fold change ≥2 are used as cut-offs for differential expression. Centered gene expression values corresponding to the subtraction of the row-wise median value of all lncRNAs from the expression value of each lncRNA are represented as a color scale. The red color indicates an up-regulation and the green color a down-regulation in failing vs non-failing hearts. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
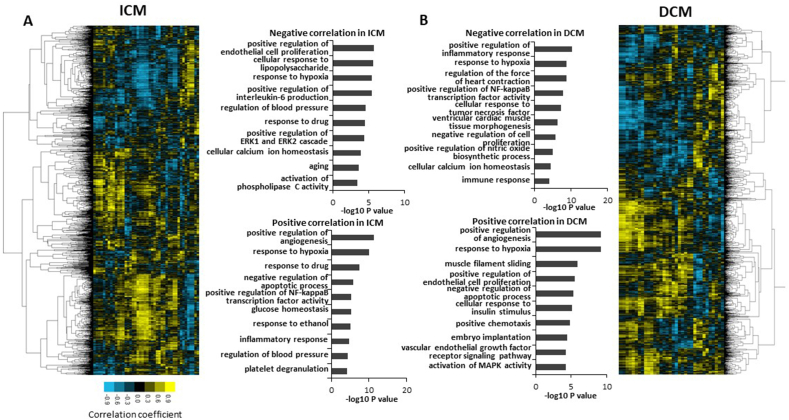
LncRNAs can regulate the expression of protein-coding genes both in cis and in trans. For cis regulation, lncRNAs should be close to protein-coding genes. Among the differentially expressed lncRNAs shown in Fig. 4, we found that 24 novel lncRNAs were close to 41 protein-coding genes with a distance of less than 50 thousand base pairs. 3 of these protein-coding neighbours − FOXO3, NPR3 and RBM20 − were up-regulated in the ICM group with a FDR <0.05 and a fold change ≥2. FOXO3 and NPR3 were also up-regulated in the DCM group (Fig. 4). For trans regulation, lncRNAs shall display a high correlation with the protein-coding targets. We identified 1168 heart failure-associated transcripts (found in the NCBI gene database using the keywords “heart failure” or “failing heart”, see Methods section), corresponding to 453 genes, which correlated to at least one differentially expressed novel lncRNA with a Spearman rank correlation coefficient above 0.7 or below −0.7 in ICM group. In the DCM group, we identified 1644 heart failure-associated transcripts, corresponding to 469 genes and correlating with at least one differentially expressed novel lncRNA with a Spearman rank correlation coefficient above 0.7 or below −0.7 (Supplementary Fig. 3).
As displayed in Supplementary Fig. 3, clustering analysis separated these transcripts in two clusters, one mainly including the transcripts negatively correlated with lncRNAs (upper part of the heatmaps), and the other one mainly including the transcripts positively correlated with lncRNAs (lower part of the heatmaps). Gene ontology analysis of the genes in the two clusters revealed that the positively correlated genes were enriched in positive regulation of angiogenesis process in both ICM and DCM group. On the other hand, the negatively correlated genes were mostly enriched in cell proliferation processes in the ICM group, and in inflammatory responses and heart contraction processes in the DCM group (Supplementary Fig. 2).
3.6. Validation of selected novel lncRNAs associated with heart failure
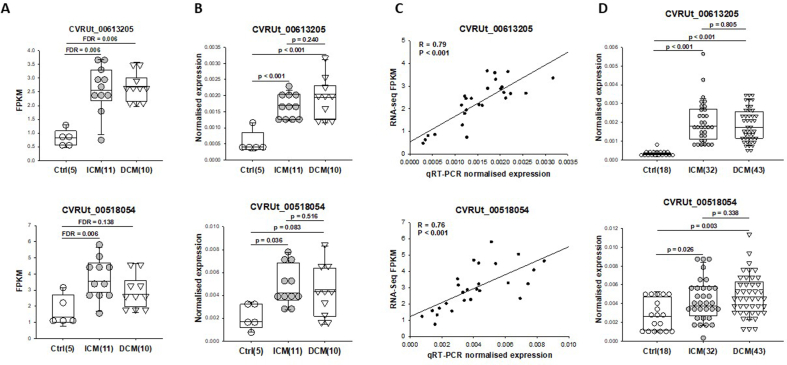
To identify candidate lncRNAs for validation experiments, we searched for novel lncRNAs with stringent criteria. We selected candidates which i. were derived from novel genes, i.e. the genes didn't match any reference gene from Cuffcompare analysis; ii. were differentially expressed either in ICM or DCM group; iii. had a median expression level more than 2 FPKM in at least one group; iv. were shorter than 10,000 bp. Two lncRNAs satisfied these criteria. They were arbitrarily named CVRUt_00613205 and CVRUt_00518054 and were selected for validation. In RNA-seq data, both lncRNAs were up-regulated in the ICM and DCM groups as compared to the control group (Fig. 5A). The two lncRNAs were measured by qRT-PCR, first in the same 26 left ventricular biopsies (11 ICM, 10 DCM and 5 controls) used for RNA-seq (“technical validation” in discovery cohort) and second in 93 other left ventricular biopsies (32 ICM, 43 DCM and 18 controls), constituting an “independent validation” group (validation cohort).
Fig. 5.
Validation of selected lncRNAs. A. The expression of two selected lncRNAs, CVRUt_00613205 and CVRUt_00518054, assessed by RNA-seq. FDR as indicated. B. lncRNAs are measured using qRT-PCR in the discovery cohort samples that are also used for RNA-seq. P-values are from one way ANOVA. C. correlation between RNA-seq and qPCR data. R correlation coefficient is calculated using Spearman rank correlation. D. measurement by qRT-PCR of the 2 lncRNAs in independent left ventricular biopsies from 32 ICM patients, 43 DCM patients and 18 controls. P-values are from one way ANOVA.
As observed with RNA-seq (Fig. 5A), CVRUt_00613205 measured by qRT-PCR in the 26 RNA-seq samples was up-regulated in both ICM and DCM groups (p < 0.001; Fig. 5B). No difference between the ICM and DCM groups was observed. CVRUt_00518054 was also up-regulated in the ICM group (p = 0.036; Fig. 5B). In the DCM group, the up-regulation of CVRUt_00518054 was not significant (p = 0.083), consistently with the RNA-seq data showing that it was increased in DCM group as compared to control group with a FDR of 0.138 (Fig. 5A). For these 2 lncRNAs, RNA-seq and qRT-PCR data were highly correlated (Fig. 5C).
When the 2 lncRNAs were measured by qRT-PCR in the 93 independent validation samples, both were significantly up-regulated in ICM and DCM groups compared with the control group (Fig. 5D). No difference was observed between ICM and DCM groups. Therefore, lncRNAs CVRUt_00613205 and CVRUt_00518054 were consistently up-regulated in left ventricular biopsies from failing hearts, independently of the ischemic or dilated aetiology.
3.7. Biomarker potential of the FIMICS panel
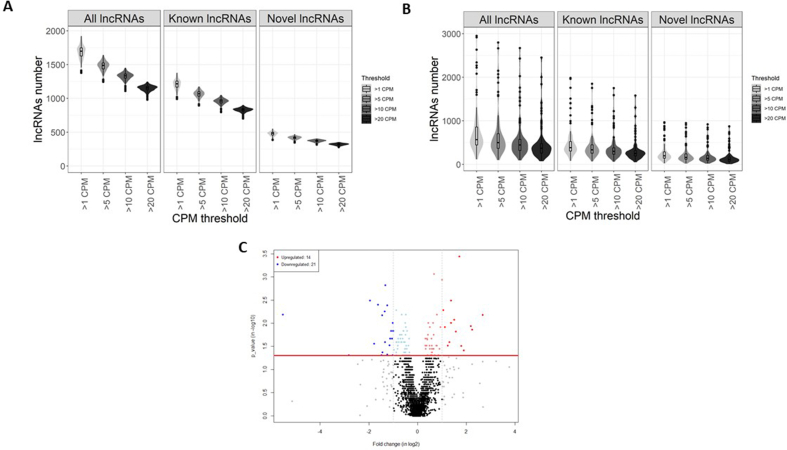
To assess the potential of the FIMICS panel to identify novel biomarkers, we first applied it to PAXgene whole blood RNA samples (n = 148) and plasma samples (n = 99) obtained from healthy donors. A majority of the lncRNAs were identified in whole blood and plasma samples with more than 1 count per million (CPM) (Fig. 6A and B, Supplementary Table 7).
Fig. 6.
Expression of lncRNAs in blood of healthy volunteers and in myocarditis vs. AMI patients. Violin plots representing the number of lncRNAs detected in total RNA extracted from PAXgene whole blood samples (n = 148) (A) or plasma samples (n = 99) (B) from healthy volunteers. Numbers of lncRNAs with detection counts more than 1, 5, 10 and 20 CPM are indicated. C. Volcano plot representing the differentially expressed lncRNAs in blood samples of myocarditis patients (n = 13) vs. patients with acute myocardial infarction (n = 10). A fold change >2 and a p-value <0.05 were considered as cut-offs for differential expression. The red color indicates an up-regulation and the blue color a down-regulation in myocarditis vs. AMI patients. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.8. Testing of the FIMICS panel in blood samples from myocarditis and acute myocardial infarction patients
We next investigated whether the FIMICS panel was able to identify relevant lncRNA biomarkers in patients with other cardiac conditions than heart failure. FIMICS panel was used to quantify lncRNAs in whole blood samples from patients with myocarditis (n = 13) or acute myocardial infarction (n = 10) (Fig. 6C). Characteristics of the two groups of patients included in this study are gathered in Supplementary Table 1. In these patients, a total of 1648 lncRNAs were detected with a threshold of at least 5 CPM in half of the patients in one group. Comparison of the 2 groups of patients revealed that 14 lncRNAs were upregulated and 21 were downregulated in myocarditis group as compared to acute myocardial infarction group (fold change >2 and P ≤ 0.05). The 16 lncRNAs with the highest capacity to discriminate patients between groups are shown in Table 1 and have an individual area under the curve (AUC) between 0.70 and 0.95. These results support the potential of the FIMICS panel to discriminate different types of cardiac disorders.
Table 1.
Biomarker potential validation Expression of 16 best candidate lncRNAs in whole blood samples of myocarditis (MYO) (n = 13) and acute myocardial infarction (AMI) patients (n = 10).
| lncRNA ID | MYO |
AMI |
p value | Fold change | AUC | ||
|---|---|---|---|---|---|---|---|
| Mean expression | SD | Mean expression | SD | ||||
| SEQ0194 | 1028.49 | 354.91 | 1457.80 | 400.79 | 1.21E-02 | 1.417 | 0.808 |
| SEQ0335 | 3.09 | 2.62 | 10.07 | 4.34 | 3.61E-04 | 3.265 | 0.946 |
| SEQ0446 | 19.48 | 9.47 | 6.32 | 7.86 | 4.00E-03 | 0.324 | 0.858 |
| SEQ0453 | 13.61 | 5.40 | 27.11 | 9.37 | 1.15E-03 | 1.991 | 0.885 |
| SEQ0476 | 12.11 | 12.23 | 0.26 | 0.57 | 6.47E-03 | 0.022 | 0.831 |
| SEQ0767 | 77.13 | 20.48 | 56.96 | 13.55 | 1.47E-02 | 0.738 | 0.800 |
| SEQ0867 | 107.64 | 74.36 | 50.50 | 58.97 | 1.13E-01 | 0.469 | 0.700 |
| SEQ1057 | 186.44 | 62.28 | 293.81 | 118.87 | 1.78E-02 | 1.576 | 0.792 |
| SEQ1280 | 617.00 | 224.57 | 223.83 | 285.14 | 6.70E-03 | 0.363 | 0.838 |
| SEQ1344 | 3.71 | 2.57 | 9.59 | 5.76 | 3.24E-03 | 2.587 | 0.854 |
| SEQ1599 | 22.06 | 7.21 | 35.15 | 10.77 | 8.62E-04 | 1.593 | 0.892 |
| SEQ2161 | 17.56 | 3.71 | 12.60 | 5.73 | 6.47E-03 | 0.718 | 0.831 |
| SEQ2164 | 136.19 | 49.14 | 53.53 | 61.81 | 5.55E-03 | 0.393 | 0.846 |
| SEQ2556 | 8.20 | 4.08 | 4.19 | 3.34 | 3.76E-02 | 0.511 | 0.762 |
| SEQ2585 | 4.20 | 2.71 | 8.01 | 3.43 | 1.21E-02 | 1.910 | 0.808 |
| SEQ2725 | 1.59 | 2.70 | 5.91 | 5.22 | 3.85E-02 | 3.714 | 0.746 |
4. Discussion
We identified a panel of 2906 lncRNAs which could be useful to provide insights into the molecular mechanisms of cardiovascular conditions and which could be used as a tool to discover novel disease biomarkers. Biomarkers indicative of cardiovascular health or dysfunction are important components of strategies for risk assessment, patient stratification and management. A single biomarker might not be the best predictor of cardiac ailments due to the complexity of associated aetiologies and molecular mechanisms. In the current study, we have first focused on heart failure to identify heart disease-associated lncRNAs and, thereafter, to obtain a comprehensive overview of cardiovascular health, we have extended our investigations to myocarditis and acute myocardial infarction. We report that the identified 2906 cardiac-enriched lncRNAs could be useful not only to monitor, predict and better understand heart failure, but also could be useful for other cardiac conditions.
In our analysis, we have adopted different strategies to identify lncRNAs that could have important functions in cardiovascular health and disease. From our RNA-seq data analysis, we have identified differentially expressed known and novel lncRNAs from cardiac biopsies of heart failure patients. Gene ontology analysis of differentially expressed transcripts between failing and non-failing control hearts highlighted interesting potential insights into the molecular mechanisms of heart failure, which remains to be thoroughly investigated. To validate the expression of the identified lncRNAs and to obtain a comprehensive overview of the cardiac lncRNA transcriptome, we analyzed lncRNAs identified from an independent study. Using the publicly available dataset GSE45326, lncRNAs enriched in normal heart tissue compared to twelve other tissues, were identified. We also identified additional lncRNAs that were highly expressed in cardiac tissue compared to other tissues, indicative of their potential role in cardiac function. Many of these lncRNAs were novel and previously uncharacterized. Through these strategies, we identified a panel of 3233 lncRNAs (2206 known and 1027 novel lncRNAs) associated with heart failure or expressed predominantly in heart tissue. We mapped these lncRNAs to the latest GENCODE39 database and updated the panel to 2906 lncRNAs in the FIMICSv2.0, which includes 1902 known lncRNAs and 1004 novel lncRNAs.
In our genetic association analysis, we found 24 novel differentially expressed lncRNAs which were overlapping or close to 41 protein-coding genes in ICM and DCM groups compared to controls. Among these, natriuretic peptide receptor 3 (NPR3) and Forkhead box O3 (FOXO3) were found to be upregulated in both ICM and DCM groups. This is of particular interest since NPR3 is a receptor for natriuretic peptides which are heart failure biomarkers [4]. NPR3 is responsible for clearance of circulating natriuretic peptides [31]. FOXO3 is a transcription factor known to activate apoptotic pathways [32]. A clustering analysis highlighted a positive correlation between angiogenesis and heart failure and a negative correlation with inflammation related processes (Supplementary Fig. 3). These clustering data showing potential associations of the identified lncRNAs with already known pathways related to heart failure [33,34] support the usefulness of the FIMICS panel as a novel research tool.
We validated the expression of two novel lncRNAs by quantitative PCR in the discovery cohort as well as in an independent validation cohort. Functional analysis of these lncRNAs could give important indications of their involvement in the cardiac pathology. The detectability of many lncRNAs in the peripheral circulation, together with their ability to discriminate different forms of cardiac diseases, validate their potential as biomarkers for multiple cardiac conditions. This supports the wide application domain of the FIMICS panel.
Clinical implications. As cardiovascular disease remains responsible for a large number of deaths worldwide, novel strategies to personalize healthcare are needed. In that respect, the FIMICS panel is a novel and unique tool to identify novel lncRNA biomarkers for diagnostic and risk stratification purposes. Being able to identify patients at high risk of developing cardiovascular problems at an early stage of the disease allows adapting healthcare strategies (e.g. imaging, medication, follow-up) to reduce disease burden. In addition, the panel can be used to deepen our understanding of the molecular mechanisms involved in cardiovascular disease development and progression, hence providing novel opportunities for drug development. The RNA field has recently emerged with the development of RNA-based vaccines and it may well be extended to RNA biomarkers and therapeutics.
Study limitations. In this study, we have presented a panel of 2906 lncRNAs either cardiac-enriched or differentially expressed between failing and non-failing hearts. We acknowledge the small number of patients included in the discovery cohort due to challenges in acquiring human cardiac biopsies and performing deep sequencing in a large number of samples. In addition, only 2 lncRNAs were validated by qRT-PCR in a larger group of patients. This study is also limited by the small number of patients with myocarditis and acute myocardial infarction included in the proof-of-concept of the biomarker potential of the FIMICS panel. To extend the coverage of lncRNAs by the FIMICS panel, we have considered the less stringent p-value instead of adjusted p-value in some statistical analyses. For the discovery of novel lncRNAs, we have used Cufflinks and bowtie softwares. There might be more advanced technologies available, however, we have found consistent and significant results with these analyses which are also widely cited and recognized by the scientific community.
In conclusion, we propose a FIMICSv2.0 panel of 2906 lncRNAs including 1902 known and 1004 novel transcripts, indicative of cardiovascular conditions. Such range of transcripts could help to elucidate the complexity of cardiac pathologies with multiple aetiologies and could facilitate the personalization of healthcare through the use of novel RNA-based biomarkers. As the epigenomics field is rapidly evolving, we expect that the FIMICS panel may become useful for research purposes and clinical application in combination with other omics approaches such as RNA profiling, ChIP-seq profiling, or epitranscriptomics.
Funding
This work is supported by funding from the EU Horizon 2020 project COVIRNA (Grant Agreement # 101016072), the National Research Fund (grants # C14/BM/8225223, C17/BM/11613033 and COVID-19/2020-1/14719577/miRCOVID), the Ministry of Higher Education and Research, and the Heart Foundation-Daniel Wagner of Luxembourg to Y.D.
Disclosures
Y.D., L.Z., P.L., E.S., S.O., T.P. and H.F. filed a patent on long noncoding RNAs for diagnosic, prognostic and therapeutic uses for pathologies and toxicities inducing heart disorders (WO2018229046). Firalis SA is commercializing the FIMICS panel of long noncoding RNAs for heart diseases.
Y.D. and L.Z. own patents related to diagnostic and therapeutic applications of RNAs.
T.F.L. has received research and educational grants outside this work from Abbott, Amgen, Boehringer-Ingelheim, Daichi-Sanky, Eli-Lilly, Metronic, Novartis, Sanofi, Servier and Vifor.
Acknowledgments
This publication is based upon work from the EU-CardioRNA COST ACTION CA17129, supported by COST (European Cooperation in Science and Technology).
Footnotes
This manuscript has been published as a preprint on the ResearchSquare server and is available at https://doi.org/10.21203/rs.3.rs-1474147/v1.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.heliyon.2023.e13087.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
figs1
figs2
figs3
References
- 1.Virani S.S., et al. Heart disease and stroke statistics-2020 update: a report from the American heart association. Circulation. 2020;141:e139–e596. doi: 10.1161/CIR.0000000000000757. [DOI] [PubMed] [Google Scholar]
- 2.Mentz R.J., O'Connor C.M. Pathophysiology and clinical evaluation of acute heart failure. Nat. Rev. Cardiol. 2016;13:28–35. doi: 10.1038/nrcardio.2015.134. [DOI] [PubMed] [Google Scholar]
- 3.Goretti E., Wagner D.R., Devaux Y. miRNAs as biomarkers of myocardial infarction: a step forward towards personalized medicine? Trends Mol. Med. 2014;20:716–725. doi: 10.1016/j.molmed.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 4.McDonagh T.A., et al. ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure. Eur. Heart J. 2021;42:3599–3726. doi: 10.1093/eurheartj/ehab368. 2021. [DOI] [PubMed] [Google Scholar]
- 5.Virani S.S., et al. Heart disease and stroke statistics-2021 update: a report from the American heart association. Circulation. 2021;143:e254–e743. doi: 10.1161/CIR.0000000000000950. [DOI] [PubMed] [Google Scholar]
- 6.Enroth S., Johansson Å., Enroth S.B., Gyllensten U. Strong effects of genetic and lifestyle factors on biomarker variation and use of personalized cutoffs. Nat. Commun. 2014;5:4684. doi: 10.1038/ncomms5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lander E.S., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 8.Venter J.C., et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 9.Consortium E.P., et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devaux Y., et al. Long noncoding RNAs in cardiac development and ageing. Nat. Rev. Cardiol. 2015;12:415–425. doi: 10.1038/nrcardio.2015.55. [DOI] [PubMed] [Google Scholar]
- 11.Bolha L., Ravnik-Glavac M., Glavac D. Long noncoding RNAs as biomarkers in Cancer. Dis. Markers. 2017;2017 doi: 10.1155/2017/7243968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devaux Y., et al. Circular RNAs in heart failure. Eur. J. Heart Fail. 2017;19:701–709. doi: 10.1002/ejhf.801. [DOI] [PubMed] [Google Scholar]
- 13.Robinson E.L., Emanueli C., Martelli F., Devaux Y. Leveraging non-coding RNAs to fight cardiovascular disease: the EU-CardioRNA network. Eur. Heart J. 2021 doi: 10.1093/eurheartj/ehab326. [DOI] [PubMed] [Google Scholar]
- 14.Vanhaverbeke M., et al. Peripheral blood RNA biomarkers for cardiovascular disease from bench to bedside: a Position Paper from the EU-CardioRNA COST Action CA17129. Cardiovasc. Res. 2021 doi: 10.1093/cvr/cvab327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ounzain S., et al. Genome-wide profiling of the cardiac transcriptome after myocardial infarction identifies novel heart-specific long non-coding RNAs. Eur. Heart J. 2015;36:353–368a. doi: 10.1093/eurheartj/ehu180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng J., et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 17.Patriki D., et al. Approximation of the incidence of myocarditis by systematic screening with cardiac magnetic resonance imaging. JACC. Heart failure. 2018;6:573–579. doi: 10.1016/j.jchf.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 18.Kim D., et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roberts A., Pimentel H., Trapnell C., Pachter L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics. 2011;27:2325–2329. doi: 10.1093/bioinformatics/btr355. [DOI] [PubMed] [Google Scholar]
- 20.Harrow J., et al. GENCODE: the reference human genome annotation for the ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trapnell C., et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L., et al. CPAT: coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucl. Acids Res. 2013;41:e74. doi: 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao Y., Smyth G.K., Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucl. Acids Res. 2013;41:e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nielsen M.M., et al. Identification of expressed and conserved human noncoding RNAs. RNA. 2014;20:236–251. doi: 10.1261/rna.038927.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolesnikov N., et al. ArrayExpress update--simplifying data submissions. Nucl. Acids Res. 2015;43:D1113–D1116. doi: 10.1093/nar/gku1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 28.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dobin A., et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xing Y., et al. An expectation-maximization algorithm for probabilistic reconstructions of full-length isoforms from splice graphs. Nucleic Acids Res. 2006;34:3150–3160. doi: 10.1093/nar/gkl396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pandey K.N. Emerging roles of natriuretic peptides and their receptors in Pathophysiology of Hypertension and cardiovascular regulation. J. Am. Soc. Hypertens. 2008;2:210–226. doi: 10.1016/j.jash.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fitzwalter B.E., Thorburn A. FOXO3 links autophagy to apoptosis. Autophagy. 2018;14:1467–1468. doi: 10.1080/15548627.2018.1475819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang C., et al. Pathological bases and clinical application of long noncoding RNAs in cardiovascular diseases. Hypertension. 2021;78:16–29. doi: 10.1161/HYPERTENSIONAHA.120.16752. [DOI] [PubMed] [Google Scholar]
- 34.Pena E., Brito J., El Alam S., Siques P. Oxidative stress, kinase activity and inflammatory Implications in right ventricular hypertrophy and heart failure under hypobaric hypoxia. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21176421. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.