Abstract
Caspases are a large family of evolutionarily conserved proteases found from Caenorhabditis elegans to humans. Although the first caspase was identified as a processing enzyme for interleukin-1β, genetic and biochemical data have converged to reveal that many caspases are key mediators of apoptosis, the intrinsic cell suicide program essential for development and tissue homeostasis. Each caspase is a cysteine aspartase; it employs a nucleophilic cysteine in its active site to cleave aspartic acid peptide bonds within proteins. Caspases are synthesized as inactive precursors termed procaspases; proteolytic processing of procaspase generates the tetrameric active caspase enzyme, composed of two repeating heterotypic subunits. Based on kinetic data, substrate specificity, and procaspase structure, caspases have been conceptually divided into initiators and effectors. Initiator caspases activate effector caspases in response to specific cell death signals, and effector caspases cleave various cellular proteins to trigger apoptosis. Adapter protein-mediated oligomerization of procaspases is now recognized as a universal mechanism of initiator caspase activation and underlies the control of both cell surface death receptor and mitochondrial cytochrome c-Apaf-1 apoptosis pathways. Caspase substrates have bene identified that induce each of the classic features of apoptosis, including membrane blebbing, cell body shrinkage, and DNA fragmentation. Mice deficient for caspase genes have highlighted tissue- and signal-specific pathways for apoptosis and demonstrated an independent function for caspase-1 and -11 in cytokine processing. Dysregulation of caspases features prominently in many human diseases, including cancer, autoimmunity, and neurodegenerative disorders, and increasing evidence shows that altering caspase activity can confer therapeutic benefits.
Programmed cell death, or apoptosis, is a physiological process of cellular autodestruction. Apoptosis plays critical roles in development, maintenance of homeostasis, and host defense in multicellular organisms. Dysregulation of this process is implicated in various diseases ranging from cancer and autoimmune disorders to neurodegenerative diseases and ischemic injuries (210).
Diverse cell types can be triggered to undergo apoptosis by various signals derived from either the extracellular or intracellular milieu. Cells undergoing apoptosis exhibit a series of characteristic morphological changes, including plasma membrane blebbing, cell body shrinkage, and formation of membrane-bound apoptotic bodies, which in vivo are quickly engulfed by neighboring healthy cells (103). Thus, during apoptosis, intracellular contents are not released and potentially harmful inflammatory responses are prevented. Apoptosis is also accompanied by certain biochemical changes, notably the appearance of discrete DNA fragments on conventional gel electrophoresis (due to cleavage between nucleosomes), the flipping of phosphatidylserine from the inner leaflet to the outer leaflet of the plasma membrane, and limited cleavage of various cellular proteins.
These stereotypic changes are manifestations of an intrinsic suicide machinery that has been conserved through evolution. The core component of this machinery is a proteolytic system involving a family of proteases known as caspases. The term caspase denotes two key characteristics of these proteases: (i) they are cysteine proteases and use cysteine as the nucleophilic group for substrate cleavage and (ii) they are aspases and cleave the peptide bond C-terminal to aspartic acid residues (2). That caspases play an essential role in apoptosis is based on three observations. First, synthetic or natural inhibitors of caspases effectively abrogate apoptosis induced by diverse apoptotic stimuli. Second, animals lacking certain caspases show profound defects in apoptosis. Third, caspases are responsible for most proteolytic cleavages that lead to apoptosis. It is noteworthy that mammalian caspases have evolved additional roles in inflammatory responses.
Synthesized as latent precursors or procaspases, caspases are converted to active proteases during apoptosis through an intricately regulated proteolytic process. The proteolytic processing occurs at critical aspartic acid residues that conform to the caspase substrate recognition consensus. Consequently, caspases often function in cascades. In such a caspase cascade, an upstream caspase (initiator caspase) is activated by its interaction with a caspase adapter(s). This activation represents a key regulatory step in apoptosis and is controlled by both pro- and antiapoptotic proteins. Once activated, the initiator caspase processes and activates one or more downstream caspases (effector or executioner caspases). The activated effector caspases then cleave various cellular proteins, leading to apoptotic cell death. In this review, we will discuss various aspects of this emerging family of proteases, with particular emphasis on recent developments.
CASPASE STRUCTURE AND ENZYMATIC ACTIVITY
Structures of Procaspases
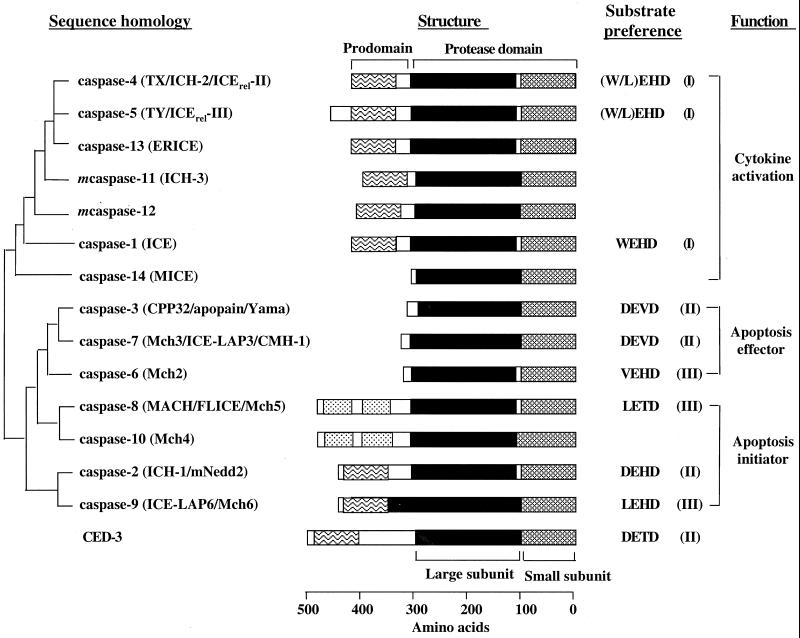
The first caspase, caspase-1, also known as ICE (interleukin-1β converting enzyme), was identified due to its ability to convert the precursor of interleukin-1β (IL-1β) to its mature form, a potent mediator of inflammation (17, 211). Subsequent cloning of ced-3 (ced, cell death abnormal), a proapoptotic gene in Caenorhabditis elegans (see below), revealed that it encodes a protein highly homologous to ICE (254). Since then, at least 14 mammalian caspases and five Drosophila caspases have been cloned. Structurally, all procaspases contain a highly homologous protease domain, the signature motif of this family of proteases (Fig. 1). This domain can be further divided into two subunits, a large subunit of approximately 20 kDa (p20) and a small subunit of approximately 10 kDa (p10). In some procaspases, there is a short linker (about 10 amino acids) between the large and small subunits. Based on the sequence similarity among the protease domains, caspases can be divided into three groups. The first group consists of inflammatory caspases: caspase-1, -4, -5, -11, -12, -13, and -14. The second group contains caspase-2 and -9. The rest of the caspases form the third group (Fig. 1).
FIG. 1.
Mammalian caspase family and C. elegans caspase CED-3. All mammalian caspases are of human origin except for murine caspase-11 and -12, for which no human counterparts have been identified yet. Phylogenetic relationships are based on sequence similarity among the protease domains. Alternative names are listed in parentheses after each caspase. Dotted box, DED domains; wavy boxe, CARD domain. Substrate preferences at the P1 to P4 positions are indicated. Based on the substrate specificity, caspases are divided into three groups (indicated in parentheses) (212).
Each procaspase also contains a prodomain or NH2-terminal peptide of variable length. Initiator apoptotic caspases and inflammatory caspases contain prodomains of over 100 amino acids, while the prodomains in effector caspases are usually less than 30 amino acids. The long prodomains contain distinct motifs, notably the death effector domain (DED) and the caspase recruitment domain (CARD). A novel motif termed the death-inducing domain (DID) was recently identified. Procaspase-8 and -10 each contain two tandem copies of DEDs (8, 54, 145), whereas the CARD domain is found in caspases-1, -2, -4, -5, and -9 as well as in CED-3 (86). DID is present in the Drosophila caspase DREDD (92). These domains mediate homophilic interaction between procaspases and their adapters and play important roles in procaspase activation (see below). In contrast, the short prodomains of executioner caspases are unlikely to mediate protein-protein interactions. Rather, they seem to inhibit caspase activation (see below).
The three-dimensional structure of a procaspase has not been determined. However, the structures of caspase prodomains, particularly those of the DED and the CARD domains, have already been solved (31, 48, 159). DED, CARD, and a related domain, DD (death domain, present in cell death adapter proteins), all consist of six alpha-helices and have similar overall folds. Nonetheless, there are some major differences among them. While charge-charge interactions govern CARD-CARD and DD-DD association, hydrophobic interactions govern DED-DED interaction.
Structures of Active Caspases
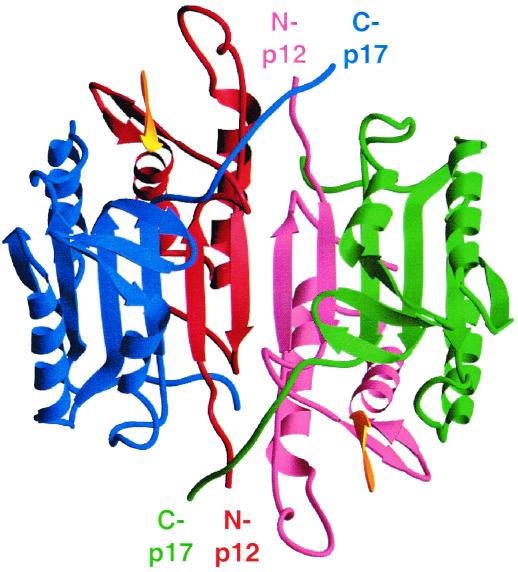
Early studies showed that active caspase-1 and -3 consist of the large and small subunits, which are released from the procaspases through proteolytic processing, and that both subunits are required for the protease activity (151, 211). Subsequently, the three-dimensional structures of these two caspases as well as that of caspase-8, each complexed with their corresponding tetrapeptide aldehyde inhibitors, have been determined (7, 169, 224, 230, 235). These structures reveal that a mature caspase is a tetramer (homodimer of the p20 and p10 heterodimers arranged in twofold rotational symmetry), with the two adjacent small subunits surrounded by two large subunits (Fig. 2). Each p20-p10 heterodimer forms a single globular domain, and the core of the globular domain is a six-stranded beta-sheet flanked on either side by alpha-helices. These two heterodimers associate with each other primarily through the interaction between the p10 subunits.
FIG. 2.
Structure of the caspase-3 tetramer in complex with Ac-DEVD-CHO. The p17 subunits are shown in blue and green, the p12 subunits in red and pink, and the bound inhibitors in yellow. The C termini of p17 and N termini of p12 are indicated. Reproduced with permission from J. Rotonda et al., Nature Structure Biology 3:619–625, 1996.
Each caspase tetramer has two cavity-shaped active sites formed by amino acids from both the p20 and p10 subunits, and these two active sites are likely to function independently. In the active site, a cysteine (Cys-285 in the p20 of caspase-1) is positioned close to the imidazole of a histidine (His-237, p20), which attracts the proton from the cysteine and enhances its nucleophilic property. The side chains of Arg-179 (p20) and Arg-341 (p10) in the S1 site participate in a direct charge-charge interaction with the aspartic acid of a substrate, contributing to the selective recognition of Asp at the P1 position of a substrate. The amino acids from the small subunit form the S2 and S3 sites. The side chains of amino acids at P2 and P3 (the second and third amino acids N-terminal to the cleavage site Asp) are mainly exposed to solvent, consistent with the less stringent requirement at these two positions in caspase substrates. One of the major differences among caspases is the S4 subsite. For example, in caspase-1, the S4 subsite is a large and shallow hydrophobic depression, and the corresponding site in caspase-3 is a narrow pocket, which accommodate a P4 tyrosyl and aspartyl side chain, respectively.
Because the small and large caspase subunits are derived from a single procaspase molecule following cleavages at the critical aspartic acid residues, the structures of active caspases are instrumental in understanding how caspases are activated (Fig. 2). The most significant features are the relative distance between the p20 C terminus and the p10 N terminus in cis (the p20-p10 pairs that form an active site) versus that in trans (the pairs that do not). The latter is only 5.3 Å, whereas the former is approximately 65 Å, too far apart for the linker region to span. Based on this critical structural feature, two alternative models have been put forth to explain the activation mechanism (224, 235). The process and association model postulates that the p20-p10 heterodimer is derived from the same procaspase molecule. This model requires an extensive reordering of the proenzyme to reconcile the fact that the p20 C terminus and the p10 N terminus in cis are distant from each other. Subsequent removal of the linker and prodomains allows the two heterodimers to form the tetrameric structure. In contrast, the association and process model predicts that the two molecules of zymogen associate and interdigitate, naturally forming a mature caspase-like intermediate. The linker and prodomain are subsequently removed to form a mature enzyme. The latter model fits well with the finding that procaspases are activated by induced proximity of two or more procaspase molecules (see below).
Enzymatic Characteristics of Caspases
Early enzyme inhibition studies revealed that caspase-1 is a cysteine protease. This caspase was potently and irreversibly inhibited by a peptide substrate carrying diazomethylketone, a highly specific modifying group for cysteine proteases (211). The caspase-1 activity was also attenuated by other cysteine-modifying reagents such as N-ethylmaleimide and iodoacetamide, but not by inhibitors of serine, aspartate, or metalloproteases. In addition, caspase-1 was readily labeled by [14C]iodoacetate, a reagent that is commonly used to identify the active-site cysteine (211). The results from these enzymatic studies were subsequently confirmed by site-directed mutagenesis studies and the three-dimensional structures of caspase-1, -3, and -8. These structures reveal a catalytic diad of Cys and His in the active site (see above). Similar to other cysteine proteases, caspases are sensitive to transition metal ions such as Zn2+ due to the interaction of these ions with the catalytic thiol. This property of caspases may account for the observation that Zn2+ can prevent apoptosis (157, 204). In contrast, Ca2+ does not appear to affect caspase activity despite its reported role in apoptosis.
Caspases are among the most specific endopeptidases. An aspartic acid residue is almost absolutely required at the P1 position. For example, any substitution in this position in the caspase-1 substrates led to a >100-fold decrease in cleavage activity (88, 187). The P2 to P4 positions also show high preference for certain amino acids. Based on their substrates, the preferred recognition sequences for caspase-1 and -3 were determined to be Tyr-Val-Ala-Asp (P4 to P1) and Asp-Glu-Val-Asp, respectively (151, 211).
The substrate specificity of 10 human caspases was determined using a systematic approach involving combinatory peptide fluorogenic substrates (212). Based on these results, caspases are divided into three groups (Fig. 1). The optimal recognition motif for the first group is WEHD. This group includes the inflammatory caspase-1 and its close homologues caspase-4 and -5. The second group prefers the sequence DEXD (where X is V, T, or H), with a high selection for Asp at the P4 position. This second group consists of apoptotic caspases with either long or short domains (caspase-2, -3, and -7). DEXD is often found in proteins that are cleaved by caspases during apoptosis, consistent with the role of caspase-3 and -7 as effector caspases. The inclusion of caspase-2, which contains a long prodomain, may imply that it can function as both an initiator and an effector caspase. For caspase-2, the amino acid at the P5 position also affects the efficiency of cleavage (205). The third group (caspase-6, -8, -9, and -10) preferentially recognize (L/V)EXD, which resembles the processing site for effector caspases and is consistent with caspase-8, -9, and -10 being initiator caspases. Categorizing caspases based on their substrate specificity gives a different result than doing so based on sequence homology among the caspases (see above). This difference is related to the fact that only small numbers of amino acids determine the substrate specificity.
In contrast to the preference for the P1 to P4 position, a methylamine group at P1′ (the position immediately C-terminal to the cleavage site) is sufficient for cleavage (211). Further analysis of protein substrates has revealed that small amino acids such as Ala, Ser, and Asn are preferred at P1′. It is noteworthy, however, that the in vitro caspase substrate specificity, determined by the tetrapeptide derivatives, may differ from that in vivo. In vivo, caspase specificity can be influenced by other factors, such as the context of the recognition sequence in the target proteins and the proximity of caspases and the target proteins. The high specificity of caspases allows them to perform limited and selective cleavages, often in the interdomain regions of the target proteins, which can lead to protein activation as well as inactivation (see below).
The enzymatic activities of the caspases within the same group vary substantially despite their similar substrate preference. For example, the peptide-based inhibitor WEHD-aldehyde has a Ki of 0.056 and 97 nM for caspase-1 and caspase-4, respectively, indicating that caspase-1 is much more active than caspase-4 (63). A similar study also revealed that caspase-8 is substantially more active than its close homologue caspase-10 (63). The significance of such differences is not clear, although they may reflect distinct roles of these caspases in apoptosis and inflammation. One possibility is that the precursor of a weak caspase may be activated before that of a more active one, leading to the amplification of death signals.
MECHANISMS OF PROCASPASE ACTIVATION
Initiator Caspases
The identification of the first caspase (caspase-1 or ICE) came with the realization that a mature caspase is generated by proteolytic processing at critical aspartic acids and hence a mature caspase may activate its precursor as well as other procaspases (211). While this observation could readily explain the activation of effector caspases (see below), how is the first initiator caspase processed from its precursor? Early studies showed that expression of caspase precursors in Escherichia coli led to their full and precise activation (154, 241, 242), supporting a self-activation model. However, the zymogen concentration in these studies was much higher than that in vivo, and the physiological relevance of this activation was unclear.
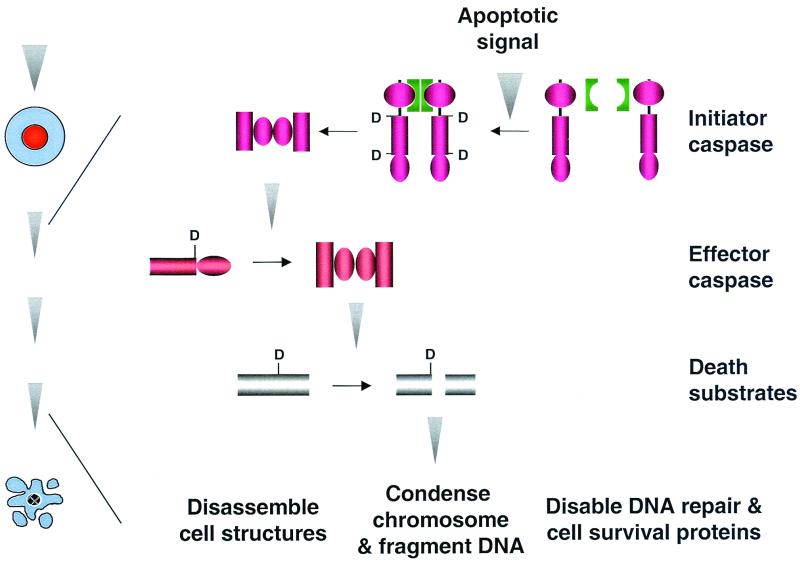
More definitive evidence for the self-activation of procaspase came from the studies on procaspase-8 (also known as FLICE, MACH, and Mch4), a caspase linked to cell surface death receptors such as Fas (see below). When Fas is aggregated by the Fas ligand, procaspase-8 is recruited to the death receptor and subsequently becomes activated (8, 145), suggesting that activation of initiator procaspases may be triggered by their oligomerization. Controlled oligomerization of procaspase-8 as well as procaspase-1 indeed leads to enhancement of their cell death activity in mammalian cells (133, 147, 244). Procaspase-8 possesses weak protease activity (147), which may allow it to cleave other procaspase-8's when in close proximity. The most convincing evidence so far for the oligomerization model is perhaps the finding that, in vitro, induced proximity of procaspase-8 triggers the otherwise stable zymogen to undergo autocleavage, generating mature caspase-8 (244) (Fig. 3). Subsequent experiments suggested that initiator procaspases linked to mitochondria (e.g., CED-3 and caspase-9) are also activated by adapter protein-mediated oligomerization (193, 245, 269) (see below). Oligomerization is now recognized as a universal mechanism of initiator caspase activation. Interestingly, this mechanism is reminiscent of the activation of receptor tyrosine kinases by ligand-mediated oligomerization, and it appears that oligomerization is a general mechanism for activating signal transduction pathways.
FIG. 3.
Activation of the caspase cascade. Apoptotic signals trigger oligomerization of death adapter proteins (e.g., CED-4, Apaf-1, and FADD). Death adapter oligomers in turn oligomerize procaspases, which leads to their autoproteolytic activation. Active initiator caspases then process and activate effector procaspases. Effector caspases cleave various death substrates to induce apoptosis.
As elegant as the oligomerization model is, exactly how oligomerization leads to caspase activation remains an outstanding question. Recent studies showed that procaspase-9 mutants lacking processing sites could still induce apoptosis (198) and that activated caspase-9 retains its prodomain and may function as a holoenzyme complexed with adapter proteins (167). These results suggest that caspase self-cleavage may not be the primary event in the oligomerization-mediated caspase activation. In addition, induced proximity of two procaspase-9 mutants, one lacking caspase activity but containing intact processing sites while the other has intact caspase activity but lacks processing sites, generated no processing (269). This result is contradictory to the simple model that procaspase molecules process each other when in close proximity.
Effector Caspases
While the initiator procaspases are activated by oligomerization, effector procaspases are often activated by other proteases, most commonly by initiator caspases but also by other proteases (trans activation) (Fig. 3). Although the extent to which this trans activation occurs in vivo is not fully understood, at least in vitro procaspase-3 and -7 can be activated by caspase-6, -8, -9, and -10 (54, 124, 146, 154, 199, 247). In vivo, the best illustrations of caspase cascades come from studies on Fas and mitochondrial apoptosis pathways (see below). Effector caspases as well as some initiator caspases can also be activated in vitro by granzyme B, an Asp-specific serine protease (47, 54, 160, 206, 247, 265). Granzyme B is an effector for cytotoxic T lymphocytes (CTL) and natural killer cells and is delivered into virally infected cells during CTL- and natural killer cell-mediated apoptosis.
The trans activation appears to be a two-step process, as illustrated in the activation of caspase-3 by granzyme B (135). The first cleavage is carried out by granzyme B and occurs in the linker region between the large and small subunits to generate a partially active intermediate that may resemble the mature caspase in structure. This linker apparently plays a critical role in preventing caspase self-activation. It is too short to allow the large and small subunits within a procaspase molecule to form a heterodimer (see above). It may also prevent the association of two procaspase molecules to spontaneously form a tetramer. Artificially reversing the order of the large and small subunits within a procaspase, which is likely to ameliorate the structural constraint imposed by the linker, results in an active enzyme (194).
In the second step, the partially active intermediate processes itself to generate the fully active, mature caspase. This self-cleavage severs the short inhibitory prodomain from the large subunit. The short prodomains present in the Drosophila effector caspases drICE and DCP-1 also play an inhibitory role in caspase activation, and deletion of these prodomains enhances the caspase apoptotic activity (57, 191). The importance of the second step is underlined by the observation that certain members of a group of apoptotic inhibitors known as inhibitors of apoptosis (IAPs) (see below) prevent apoptosis by impeding this autocleavage (40). In addition to caspase-3 autoprocessing, the cleavage that separates the prodomain from the large subunit may be carried out in trans by other caspases, as in tumor necrosis factor alpha (TNF-α)-mediated apoptosis (156). Engagement of TNF receptors by TNF-α activates both caspase-8 and caspase-9 (see below). While caspase-8 functions similarly to granzyme B and severs the linker between the large and small domains, caspase-9 removes the prodomain. Interference with caspase-9 activation by the adenovirus protein E1B prevents the completion of caspase-3 processing and apoptosis.
Interestingly, procaspase-3 can also be activated by short peptides containing the arginine-glycine-aspartate (RGD) motif (12). This motif, present in many integrin ligands, binds to integrin extracellularly. RGD peptides were found to directly induce procaspase-3 autoprocessing and subsequent apoptosis, independent of the integrin-binding ability of RGD. The mechanism of this activation is not fully understood. However, procaspase-3 contains an RGD sequence near the active site and an RGD-binding motif (aspartate-aspartate-methionine [DDM]) at the linker region. Thus, these two motifs may interact to keep procaspase-3 in a self-inhibitory conformation.
Inflammatory Caspases
Similar to apoptotic caspases, the caspases acting in inflammation, notably caspase-1, can also undergo full activation when expressed in Escherichia coli or refolded from denaturing conditions (161, 242). This activation is self-catalyzed, because mutations of the active-site cysteine abolish procaspase-1 processing. Activation of caspase-1 appears to be a quite complicated process and involves a series of processing intermediates with increasing enzymatic activity (242). Procaspase-1 also possesses a protease activity that is weaker than those of the intermediates. What initiates procaspase-1 activation in vivo is still unsettled. However, like procaspase-8, forced oligomerization of procaspase-1 leads to its activation (244), and thus there may be an adapter protein(s) facilitating procaspase-1 oligomerization in vivo.
Murine caspase-11 also plays a critical role in the activation of caspase-1. Mice deficient in caspase-11, similar to those deficient in caspase-1, fail to produce mature interleukin-1β (226). Activation of procaspase-1 is impaired in cells derived from these mice. Similarly, caspase-4 (likely a human homologue of caspase-11) enhances activation of human caspase-1. Caspase-11 and caspase-1 were found in the same complex (226). Identification of other components in this complex should help clarify the exact mechanism of procaspase-1 activation.
REGULATION OF CASPASES
Natural Inhibitors
Because apoptosis and inflammatory responses are the major host defense mechanisms against viruses, it is not surprising that viruses employ inhibitors of caspases, the central components of the apoptotic machinery, to prolong the life of host cells for maximal viral replication. These viral inhibitors may directly inhibit caspases, as exemplified by the cowpox virus protein CrmA (cytokine response modifier A) and baculovirus proteins p35 and IAPs, or inhibit caspase-adapter interactions, as exemplified by v-FLIP (viral FLICE-inhibitory protein).
CrmA is a serpin that directly targets the active site of mature caspases (163). It has an active-site loop that is easily accessible to caspases. After being cleaved by a caspase, however, CrmA stays bound to the caspase and blocks the active site (5). The early experiments using this suicide substrate of caspases provide compelling evidence that caspases play a central role in apoptosis (62, 207). CrmA is limited to the group I and group III caspases (except for caspase-6) and granzyme B (266). The dissociation constants range from 0.01 nM for caspase-1 and 0.34 nM for caspase-8 to over 1 μM for the group II caspases. Similar to CrmA, the baculovirus protein p35 also targets mature caspases and serves as a suicide substrate (13, 240). The inhibition requires a substrate-like sequence containing Asp–Gln–Met–Asp-87–Gly that fits well with the caspase active site (55). p35 is a broad-spectrum caspase-specific inhibitor; it inhibits human caspase-1, -3, -6, -7, -8, and -10 with Kis of from less than 0.1 to less than 9 nM (264). However, it does not inhibit granzyme B.
In contrast to CrmA and p35, IAPs are not active-site-specific inhibitors, and their inhibition of apoptosis does not require cleavage by caspases. The baculovirus IAPs, Op-IAP and Cp-IAP, were identified by their ability to functionally replace p35 (6, 35). While no cellular homologues of CrmA and p35 have been identified so far, the cellular homologues of IAPs constitute a major family of caspase regulators. The first human IAP, the neuronal apoptosis inhibitor protein, was cloned based on its frequent deletion in a neurodegenerative disorder, spinal muscular atrophy (125). To date, at least four other mammalian IAPs (XIAP/MIHA, c-IAP-1/MIHB, c-IAP-2/MIHC, and survivin) and two Drosophila IAPs (DIAP-1 and -2) have been identified (reviewed in reference 39). Each IAP protein contains at least one but often two to three copies of the characteristic BIR sequence (baculovirus IAP repeat), which are required for their function. In addition, several IAPs, including IAP-1 and XIAP, also have a zinc ring domain that controls the degradation of these proteins (see below).
Human XIAP and c-IAP-1 and -2 were found to inhibit mature caspase-3 and -7 (40, 41, 170). However, the inhibitory effect is substantially weaker than that of CrmA and p35. While CrmA and p35 reach maximal inhibitory effect when present at an equal molar ratio to caspases, IAPs need to be in molar excess to achieve a reasonable level of apoptosis inhibition. The exact mechanism of caspase inhibition by IAPs is not fully understood. However, IAPs may target the early steps of caspase activation, as shown by the observation that IAPs block activation of Drosophila caspases in vivo and activation of mammalian procaspase-9 in a cell-free system (40, 178). Evolutionarily ancient IAPs such as survivin may also be involved in cytokinesis and link caspase activation to cell cycle progression (120, 121). The level and activity of IAPs may determine the sensitivity of cells to apoptotic stimuli, and both can be downregulated during apoptosis to ensure effective cell killing. For example, during thymocyte death, c-IAP-1 and x-IAP are degraded in proteasomes after autoubiquitination, which is catalyzed by the ubiquitin ligase activity of their zinc ring domains (248). The pathway leading to this autoubiquitination remains to be determined. In addition, the activity of IAPs is regulated by the Drosophila cell death inducers Reaper, Grim, and HID and mammalian protein Smac/DIABLO (see below).
v-FLIPs represent another group of viral apoptotic inhibitors. v-FLIPs contain two DEDs that are similar to those in the N-terminal region of procaspase-8 (also known as FLICE and MACH). They inhibit apoptosis mediated by death receptors through competition with procaspases for recruitment to the death receptor complex. Cellular homologues of v-FLIP have been identified, and they come as both a long form and a short form, termed c-FLIPL and c-FLIPS, respectively. c-FLIPS is similar to v-FLIP and contains only the DEDs. Overexpression of this form inhibits apoptosis mediated by Fas and related death receptors. In contrast, c-FLIPL is strikingly similar to procaspase-8 and -10, comprising two NH2-terminal DEDs and a COOH-terminal caspase-like domain. The role of c-FLIPL in apoptosis still remains controversial (see below). Similar to c-FLIPS, proteins derived from alternatively spliced caspase-2 and -9 transcripts also function as apoptotic inhibitors and are named caspase-2S and caspase-9S, respectively. Both inhibitors lack the large caspase subunit and may compete with the corresponding caspases for binding to adapters.
Phosphorylation and Nitrosylation
As a major form of posttranslational modification, phosphorylation is also employed to modulate caspase activity. One example is the phosphorylation of caspase-9 by Akt (14), a serine-threonine protein kinase downstream of phosphatidylinositol 3-kinase, which is implicated in apoptosis suppression mediated by growth factor receptors. Phosphorylation of caspase-9 by Akt inhibits caspase activity in vitro and caspase activation in vivo. The phosphorylation occurs at a consesus substrate recognition sequence of Akt (RxRxxS/T), which is away from the enzymatic active site. The phosphorylation may affect assembly of the caspase tetramer. Alternatively, it may regulate caspase activity allosterically. However, this regulation is not conserved through evolution, because mouse caspase-9 does not contain the recognition site serine and is not phosphorylated by Akt (60).
Another way to modify caspases posttranslationally is by S-nitrosylation. Nitric oxide (NO) and related molecules have been found to inhibit apoptosis. A study showed that in unstimulated human cells, the active site of endogenous procaspase-3 is S-nitrosylated, but during Fas-mediated apoptosis, it becomes denitrosylated (131). The denitrosylation enhances mature caspase-3 activity, although it does not affect procaspase-3 processing. The pathway that leads to denitrosylation has not been defined, nor is it clear whether other caspases are regulated by a similar mechanism. Nevertheless, this study suggests that caspase S-nitrosylation and denitrosylation is a dynamic process during apoptosis.
Compartmentalization
Different procaspases may be present at different intracellular compartments, and their localizations may change during apoptosis. One of the best examples is that, during Fas-mediated apoptosis, procaspase-8 is recruited from the cytosol to the Fas receptor complex and becomes activated (see below). This translocation is mediated by the homotypic interaction between the DEDs in the prodomain region of procaspase-8 and that in a Fas-associated adapter protein named FADD. In addition, during TNF-induced apoptosis in HeLa cells, procaspase-1 translocates from the cytoplasm to the nucleus and becomes activated there (132). Procaspase-2 is also present in the nucleus as well as the cytoplasm (33). In both cases, the prodomains are required for nuclear localization. These findings suggest a previously unsuspected function of the nucleus in the initiation of apoptosis. Caspase-12 resides in the endoplasmic reticulum (ER) and responds specifically to ER stress (149). Furthermore, in mouse liver, procaspase-3 is present in both cytosol and mitochondria, while procaspase-7 is found only in the cytosol. During Fas-induced apoptosis, caspase-3 is confined primarily to the cytosol, whereas caspase-7 is found in microsomal fractions (20). Therefore, these two effector caspases may cleave protein substrates in different compartments. Procaspases may normally be compartmentalized away from their substrates to prevent accidental apoptosis; during apoptosis, the activation and coordinate translocation of caspases allow them to move close to their targets.
SIGNALING PATHWAYS THAT ACTIVATE CASPASES
One of the major advantages of utilizing a proteolytic system involving caspases for apoptosis is that this system is irreversible and self-amplifying, thereby ensuring rapid cell death when needed. However, inappropriate activation of this system in living cells could be just as fatal. The activation of the caspase cascade, especially that of the initiator caspases, is thus subject to intricate regulation by both pro- and antiapoptotic proteins, which often act in various signaling pathways.
Caenorhabditis elegans
During the development of the hermaphroditic nematode C. elegans, 1,090 cells are generated, and 131 of them undergo programmed cell death. Genetic studies have identified mutations that either abrogate or enhance apoptosis in C. elegans (reviewed in references 87 and 140). These and subsequent genetic analyses have helped establish the paradigm that apoptosis is an active process of cell autodestruction and is genetically programmed. Four genes are found to be at the core of this program and affect apoptosis in all cells: three of them (egl-1, ced-3, and ced-4) are required for cell death, and one (ced-9) inhibits cell death (34, 80, 253, 254). Additional genes affect apoptosis in a subset of cells, and those that control the engulfment of cell corpses have also been identified.
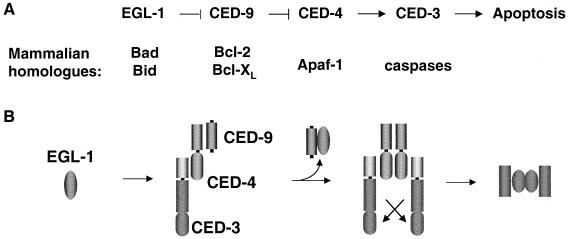
Genetic analysis revealed the functional relationship between the apoptotic genes of C. elegans (Fig. 4A). Specifically, loss of ced-9 function, which results in excessive cell death, is suppressed by mutations in either ced-3 or ced-4 but not by those in egl-1 (egg laying defective). Therefore, genetically, egl-1 functions upstream of ced-9 to inhibit its function, while ced-3 and -4 act downstream of ced-9 to promote apoptosis. The functional relationship between ced-3 and ced-4 was determined by overexpression experiments. Increased expression of ced-3 leads to cell death in ced-4 mutant cells, while increased expression of ced-4 in ced-3-deficient cells fails to induce cell death. Thus, ced-4 acts upstream of ced-3. Furthermore, overexpression of egl-1 can kill cells that normally live, and this killing requires the function of ced-3 and ced-4. The last experiment placed egl-1 in the same pathway as and upstream of ced-3 and ced-4.
FIG. 4.
(A) C. elegans apoptosis pathway and mammalian homologues of C. elegans cell death proteins. (B) CED-4 oligomerization as a unifying mechanism in the C. elegans cell death pathway.
Molecular cloning of ced-3 revealed that it encodes a homologue of mammalian ICE or caspase-1, which provided the first evidence that caspases play critical roles in apoptosis (254). Since then, additional caspases have been identified in C. elegans, and the function of these caspases remains to be determined (180). The CED-9 protein was found to be homologous to mammalian Bcl-2 family proteins, especially Bcl-2 and Bcl-xL, two predominant antiapoptotic proteins in this family (80). Subsequent experiments showed that Bcl-2 could substitute for CED-9 in C. elegans to prevent apoptosis (80). In addition, EGL-1 has been characterized as a BH3 domain-containing protein homologous to a subfamily of the mammalian Bcl-2 protein (34). A mammalian CED-4 homologue, Apaf-1, was also identified (268) (Fig. 4A). These results have enforced the idea that the apoptotic program is evolutionarily conserved.
The activation of the CED-3 caspase is regulated by the EGL-1, CED-9, and CED-4 proteins. Results from several groups suggested that CED-9, CED-4, and CED-3 coexist in a complex known as the apoptosome, with CED-4 being the crux of this complex and interacting with both CED-9 and CED-3 (28, 192, 237). EGL-1 can bind to and displace CED-9 from this complex, thereby allowing CED-4 to activate CED-3 (34). Inhibition of CED-4 by CED-9 and activation of CED-3 by CED-4 appear to occur at distinct intracellular compartments, because CED-4 and CED-9 colocalize to mitochondria in living cells, whereas CED-4 assumes a perinuclear localization in dying cells (23). How does CED-4 activate CED-3? CED-4 was found to form homooligomers both in vitro and in vivo. This oligomerization in turn aggregates the CED-4-associated CED-3 precursors, leading to CED-3 self-activation (245). Furthermore, CED-4 oligomerization is inhibited by CED-9 (245). These results suggest that CED-4 oligomerization is a unifying mechanism that connects the functions of these C. elegans apoptotic proteins (Fig. 4B) and hinted at the function of Apaf-1 (see below).
Drosophila
In recent years, Drosophila melanogaster has also become a model genetic system for the study of programmed cell death. In screening for chromosomal deletions that impair embryonic apoptosis, one such deletion in the region of 75C1,2 was identified (233). Subsequent analysis revealed that this region encompasses at least three genes: reaper, grim, and hid (head involution defective) (24, 73, 233). reaper and grim encode small proteins of only 65 and 138 amino acids, respectively, while hid encodes a protein of 410 amino acids. The proteins all have a short stretch of amino acids at their N terminus which appears to be important for their apoptotic function. Transcription of these three genes is upregulated in cells induced to die, and ectopic expression of each protein causes cell death in Drosophila as well as in mammalian cells, supporting their roles as cell death inducers. Reaper, Grim, and Hid-induced apoptosis is blocked by caspase inhibitors such as p35, indicating that these proteins engage caspases to kill cells.
At least five caspases have been identified in Drosophila. Two of them (DREDD and DRONC) contain long prodomains and are likely to be initiator caspases. DREDD is similar to mammalian procaspase-8 and contains two tandem DED domains in its N-terminal region (25). DRONC has a CARD domain (44). The other three caspases (DCP-1, drICE, and DECAY), with only short prodomains, bear strong similarity to mammalian effector caspases such as caspase-3 and -7 (45, 56, 191). Like mammalian caspase-3 and -7, DCP-1 and drICE are inhibited by IAPs, particularly DIAP-1.
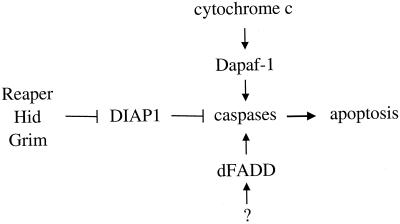
Reaper, Grim, and Hid seem to activate caspases through inhibition of DIAP-1. In vitro, recombinant DIAP-1 inhibits the protease activity of drICE, and this effect can be reversed by recombinant Hid (227). In yeast cells which lack endogenous caspases but are sensitive to caspase cleavage, expression of drICE causes cell death. The lethality of drICE is inhibited by coexpression of DIAP-1, but the expression of reaper, hid, or grim reverses this inhibitory effect (227). In another study, DIAP-1 gain-of-function mutations were identified and strongly suppressed reaper-, hid-, and grim-induced apoptosis (68). These mutations were single amino acid changes in the BIR region that decreased the interaction between DIAP-1 and Reaper, Grim, and Hid. These three proteins have homology in their first 14 amino acids, which are critical for interaction with and inhibition of DIAP-1. Together, these results support the notion that the three Drosophila apoptotic inducers activate caspases through direct inhibition of DIAP-1 (Fig. 5). This apoptotic pathway is distinct from the classic pathways mediated by death adapters such as CED-4, Apaf-1, and FADD (see the sections above and below). A similar pathway appears to exist in mammalian cells. Smac, a recently identified mammalian functional homologue of Reaper, Grim, and Hid, promotes caspase-9 and -3 activation by eliminating the inhibitory effect of mammalian IAPs (46, 71, 221). The N-terminal regions of Smac, like those in Reaper, Grim, and Hid, are indispensable for its function (18). However, unlike Reaper, Grim, and Hid, Smac resides in mitochondria in living cells and translocates to the cytosol during apoptosis to exert its effect.
FIG. 5.
Three Drosophila apoptotic pathways converge at caspase activation. Reaper, Grim, and Hid activate caspases through inhibition of DIAP-1. Caspases can also be activated by an Apaf-1-like pathway (Dapaf-1/HAC-1/Dark) and a FADD-like pathway (dFADD).
A Drosophila protein similar to CED-4 and its mammalian homologue Apaf-1 (see below) was identified and named HAC-1 (Dapaf-1, Dark) (101, 166, 263). Dapaf-1 is more closely related to Apaf-1 than CED-4 in that both Dapaf-1 and Apaf-1 have long C-terminal regions with WD-40 repeats and may require cytochrome c for activation (see below). Dapaf-1 was found to interact with DRONC in one study but with DREDD in another (101, 166), and the real target caspase remains to be determined. A death adapter, dFADD, which is homologous to the mammalian protein FADD that activates caspase-8 (see below) was also identified (92). dFADD binds to DREDD through the DID, a novel domain involved in caspase-adapter interactions, and promotes DREDD cell death activity and autoprocessing. Thus, multiple Drosophila apoptosis pathways converge on caspase activation (Fig. 5).
Mammalian I: Mitochondria/Cytochrome c
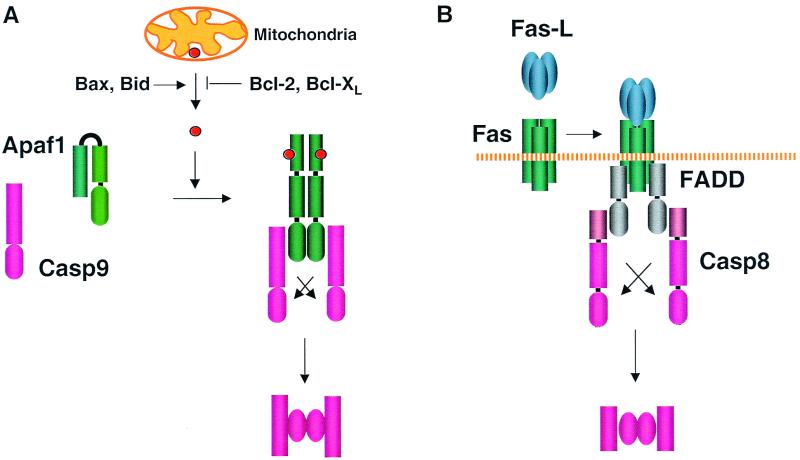
The C. elegans CED-4 pathway for caspase activation is conserved in mammalian cells. Components in this mammalian pathway were identified through biochemical purification of proteins that participate in dATP-induced caspase activation in a cell-free system (126). One such protein, Apaf-1 (apoptosis protease-activating factor-1), is homologous to CED-4 (268). Like CED-4, Apaf-1 contains an N-terminal CARD domain that can interact with the CARD domain in the N terminus of procaspase-9 (124). In addition, Apaf-1 also forms oligomers, likely octamers, in a dATP- and cytochrome c-dependent manner (193, 245, 269). The oligomerization is mediated by a CED-4-homologous region C-terminal to the CARD domain (193, 245). Unlike CED-4, however, Apaf-1 also harbors a long C-terminal region with about a dozen WD-40 repeats, a motif found in many proteins and presumably involved in protein-protein interactions. The WD-40 repeats have a self-inhibitory effect on the rest of the protein, and binding of dATP and cytochrome c to Apaf-1 antagonizes this inhibitory effect and allows Apaf-1 self-aggregation as well as Apaf-1 binding to procaspase-9 (124, 193, 269) (Fig. 6A). Apaf-1 and procaspase-9 form a complex at a 1:1 ratio (269). Thus, procaspase-9 molecules are brought into close proximity by Apaf-1 and activated by autocleavage (Fig. 6A). Active procaspase-9 then cleaves and activates procaspase-3 (124). There may be a positive feedback loop between these two caspases, as active caspase-3 cleaves partially processed caspase-9 to generate the final form of the mature caspase-9 (193). Apaf-1 was found to possess intrinsic dATPase activity that promotes oligomer formation (269). CED-4 also requires the binding of nucleotide to activate CED-3, and mutations that alter the conserved nucleotide-binding site abolish CED-4 function (27, 179). However, a dATPase activity in CED-4 is yet to be determined. CED-4 does not contain WD-40 repeats, to which cytochrome c binds, and it remains to be determined whether CED-4 requires cytochrome c for its function.
FIG. 6.
Mammalian apoptosis pathways. Procaspase-8 and -9 are activated by oligomerized FADD and Apaf-1, respectively. Upon activation, these two caspases cleave effector caspases such as caspase-3 (see Fig. 3). The red circles indicate cytochrome c.
The finding that cytochrome c, an essential component of the respiratory chain for the generation of ATP, also plays a critical role in apoptosis came as a surprise to a field in which cell life and death were generally regarded as mutually exclusive processes. The dual role of cytochrome c may be rooted in an essential logic for multicellular organisms: the purpose of individual cells' existence is the well-being of the organism. Using an essential component for cell survival as an apoptotic inducer provides an effective way to ensure that all cells that grow can undergo apoptosis when needed. Other cell death proteins, including the death adapter protein FADD (see below), may also play a dual role in cell life and death.
Cytochrome c normally resides in the space between the outer and inner membrane of mitochondria. Its release is triggered by various cellular stresses, including cytotoxic drugs, growth factor withdrawal, and DNA damage. Notably, p53-mediated apoptosis employs this mitochondrial cell death pathway (190). Cytochrome c release is also induced by development cues, and mice deficient in either Apaf-1 or caspase-9 show profound apoptosis defects during development (see below). Release of cytochrome c is regulated by the Bcl-2 family proteins (reviewed in references 1 and 72). Specifically, the antiapoptotic members of this family such as Bcl-2 and Bcl-xL prevent cytochrome c release, while the proapoptotic members such as Bax and Bid promote it (Fig. 6A) (100, 108, 122, 130, 243). Many of the Bcl-2 family proteins attach to or, during apoptosis, translocate to the outer member of mitochondria. Unlike the C. elegans Bcl-2 homologue CED-9, which prevents apoptosis through direct inhibition of CED-4, endogenous Bcl-2 family proteins are not found to physically interact with Apaf-1 (143).
The exact mechanisms by which the Bcl-2 family proteins regulate cytochrome c release are still in debate. One hypothesis states that a mitochondrial conductance channel known as the permeability transition (PT) pore opens during apoptosis. Due to the high osmolarity of the matrix, the opening of the PT pore causes swelling of the mitochondrial matrix, leading to outer membrane rupture and cytochrome c release. The dissipation of the mitochondrial inner membrane potential (Δψm) (hypopolarization), an indication of the opening of the PT pore, is observed in many apoptotic models (137, 255). Bcl-2 prevents and Bax promotes opening of the PT pore (239, 256). Another hypothesis postulates that during apoptosis, a transient hyperpolarization of the mitochondrial inner membrane causes high osmolarity in the intermembrane space and subsequent rupture of the outer membrane. Such hyperpolarization is observed in a variety of apoptosis scenarios and is suppressed by Bcl-xL (215, 216).
Both hypotheses assume that a major morphological change of mitochondria (i.e., rupture of the outer membrane) precedes cytochrome c release. However, cytochrome c release and caspase activation can occur before any detectable changes in mitochondrial morphology (72). Caspase activation can also induce cytochrome c release. It is thus possible that cytochrome c release occurs in two steps: at first only some cytochrome c molecules are released due to outer membrane rupture in a small number of mitochondria, and these cytochrome c molecules lead to caspase activation. Active caspases then cause changes in most mitochondria, and consequently almost all cytochrome c molecules are released. It is also possible that cytochrome c release occurs through a special channel instead of via outer membrane rupture. In this regard, it is intriguing that the three-dimensional structure of Bcl-xL resembles those of pore-forming bacterial toxins such as diphtheria toxin, and that Bcl-2, Bcl-xL, and Bax can each form ion channels in a synthetic membrane (141, 144). However, the channel-forming ability of these proteins alone is unlikely to account for the opposite effects of these proteins on cytochrome c release. The Bcl-2 family proteins may interact with channel-forming proteins on mitochondria to modulate cytochrome c release, and a few studies suggest that Bax and Bid promote while Bcl-xL inhibits opening of the PT channel (150, 183).
Cytochrome c is not the only harmful protein hidden in mitochondria. Other cell death effectors in mitochondria include procaspases (202) (also see above), an apoptosis-inducing factor (AIF), and the caspase coactivator Smac (also called DIABLO). AIF is a flavoprotein, and like cytochrome c, it is normally confined to mitochondria. During apoptosis, AIF translocate to the nucleus, where it induces chromatin condensation and large DNA fragmentation via a caspase-independent mechanism (203). Smac promotes activation of caspase-9 and -3 by binding to IAPs and eliminating their inhibitory effect on the caspases (18, 46, 71, 221).
Mammalian II: Death Receptors
Fas death pathway.
While the cell death pathways identified so far in C. elegans and D. melanogaster are initiated by developmental cues within individual cells, mammals have clearly evolved a mechanism by which a cell is instructed to die. This type of cell death is critical in regulating the immune response, as illustrated by the functions of a group of death receptors in the TNF receptor (TNFR) superfamily (3, 188). These receptors include Fas, TNFR1, and death receptors 3, DR-4, and -5 and contain the death domain in their intracellular region to recruit downstream apoptotic proteins.
The prototype of the death receptors, Fas, is ubiquitously expressed in various tissues, but its ligand is expressed mainly in activated T lymphocytes and natural killer cells (148). Fas-mediated apoptosis plays several important roles in regulating lymphoid homeostasis (reviewed in reference 148). First, activation-induced T-cell suicide, which is critical in deleting activated T cells at the end of an immune response, employs the Fas apoptotic pathway. Mice carrying homozygous Fas or Fas ligand mutations show lymphoproliferation and in certain genetic backgrounds autoimmunity due to accumulation of active lymphocytes. Similarly, humans heterozygous for Fas mutations can develop autoimmune and lymphoproliferative syndromes (ALPS) (see below). Second, CTL and natural killer cells use the Fas apoptosis pathway as an effector mechanism to eliminate virus-infected cells and tumor cells. In this regard, the Fas pathway acts together with the granzyme B pathway. Third, immune-privileged sites such as the eye and testis express Fas ligand, which induces apoptosis in activated T cells or neutrophils that enter such sites to prevent harmful inflammation.
The Fas apoptotic pathway has been extensively investigated as a model system for mammalian apoptosis. Fas molecules preassemble into homotrimers via the preligand-binding assembly domain in the extracellular regions (19, 186). Upon binding to the trimeric Fas ligand, Fas recruits a cytosolic adapter protein, FADD (Fas-associated death domain), through a homotypic interaction between the Fas intracellular death domain and the FADD C-terminal death domain (9, 29). FADD contains a DED in its N-terminal region. This domain interacts with the DED domains in the prodomain of procaspase-8 and recruits procaspase-8 to Fas (8, 145) (Fig. 6B). Fas, FADD, and caspase-8 do not associate with each other under normal conditions, and the biochemical basis for the sequential recruitment is not fully understood. FADD may be kept in an autoinhibitory conformation by an interaction between the death domain and the DED domain. Consistent with this hypothesis, deletion of the FADD death domain enhances the cell death activity of the FADD DED domain (29). In addition, procaspase-8 contains two DED domains, which may normally associate with each other to prevent binding to the DED domain on another procaspase-8 molecule. Therefore, it is possible that aggregation of Fas by its ligand brings the death domains in the Fas intracellular tail in close proximity to create high-affinity binding sites for the FADD death domain. Such sites may draw the FADD death domain away from the DED domain and make the DED available for binding to procaspase-8 DEDs.
The complex formed by Fas, FADD, procaspase-8, and possibly other proteins is known as DISC (death-inducing signaling complex) (106). Recruitment of procaspase-8 to the DISC leads to zymogen activation, likely occurring through autoproteolytic processing (Fig. 6B) (also see above). In one experiment, the procaspase-8 protease domain is directly linked to the Fas extracellular domain. The apoptotic ability of such chimeric molecules can still be enhanced by agonistic Fas antibody, albeit the magnitude of enhancement is substantially less than that for wild-type Fas (244). This difference suggests that intermediate motifs (such as the Fas death domain and the procaspase-8 DEDs) may have functions other than oligomerizing procaspase-8. Such functions may include recruitment of additional factors such as Daxx, a signaling protein that links Fas to the c-Jun N-terminal kinase (JNK) pathway (21, 246).
Another molecule present in the DISC is c-FLIP (also known as Casper, MRIT, CLARP, CASH, I-FLICE, FLAME, and usurpin) (67, 78, 91, 95, 96, 185, 195) (see above). c-FLIPL is a proteolytically inactive molecule that is strikingly similar to procaspase-8. It contains two DED domains and a protease-like domain that lacks amino acids essential for protease activity (including the active-site cysteine). The role of c-FLIP in apoptosis has been controversially described. It was reported to inhibit apoptosis in some studies (67, 91, 96, 195), and mouse cells defective in both FLIPL and an alternatively sliced form, FLIPS (see above), show increased sensitivity to Fas-mediated apoptosis (250). The prevailing view now holds that c-FLIP competes with procaspase-8 for binding to FADD and thereby inhibits procaspase-8 activation. However, several reports pointed to a proapoptotic activity for c-FLIPL (67, 78, 95, 185). The level of endogenous c-FLIP was found to be less than that of procaspase-8 in many cell lines and not to be correlated with sensitivity to Fas-mediated apoptosis (176). Furthermore, unlike its viral counterpart, v-FLIP, which contains only the DED domains (209), c-FLIP has retained the protease-like domain, which is not involved in FADD binding. c-FLIP may play a complex role in caspase activation.
TNFR1, DR3, DR4, and DR5 pathways.
Two other death receptors, DR4 and DR5, resemble Fas structurally and functionally (3). They mediate apoptosis upon activation by their ligand TRAIL. However, unlike FasL, which is expressed only in activated T cells, natural killer cells, immune-privileged sites, and some tumor cells, TRAIL mRNA is detected in many tissues. Similarly, DR4 and DR5 transcripts are also found in many normal tissues, suggesting that TRAIL is nontoxic to these tissues. Significantly, TRAIL triggers apoptosis in many tumor cells while sparing normal tissues, and it may prove to be an effective anticancer reagent. The different responses towards TRAIL are in part due to the expression of decoy TRAIL receptors in normal but not in tumor cells (155, 181). The signaling pathways of DR4 and DR5 are similar to that of Fas and are mediated by FADD (107).
TNFR1, together with TNFR2, mediates the pleiotropic effects of TNF-α, a primary inflammatory cytokine. TNFR1 is expressed ubiquitously, while TNF-α is produced mainly by activated macrophages and T cells in response to infection. Besides inducing apoptosis, TNF-α also activates various proinflammatory and immunomodulatory genes through transcription factors as such NF-κB and AP-1. DR3 closely resembles TNFR1 in its primary sequence, and it also activates apoptosis and NF-κB (3). However, the expression patterns of DR3 and its ligand Apo3L appear to be exactly opposite to those of TNFR1 and TNF-α: DR3 is expressed mainly in immune tissues such as spleen, thymus, and peripheral blood, while its ligand is present in many tissues constitutively. Thus, DR3 and TNFR1 may play different biological roles. Upon binding to TNF-α, TNFR1 associates with a platform adapter protein, TRADD (TNFR-associated death domain) (90), which then recruits FADD to activate caspase-8 and TRAF2 and RIP1 to activate JNK and NF-κB, respectively (89, 117, 196, 213). The signaling pathway of DR3 appears to be similar to the TNFR1 pathway.
Engagement of death receptors also leads to release of cytochrome c due to activation of Bid, a BH3 domain-containing, proapoptotic Bcl-2 family protein, by caspase-8 (122, 130). Caspase-8 cleaves Bid in the cytosol, which generates an active COOH-terminal fragment. This fragment translocates to mitochondria and induces cytochrome c release through undefined mechanisms. Cleavage of Bid thus connects the death receptor pathway to the mitochondrial apoptosis pathway and effectively amplifies apoptosis signals. The extent to which Bid contributes to death receptor-induced apoptosis varies in different tissues, as shown by analysis of Bid knockout mice (see below).
CASPASE SUBSTRATES
To date, more than 60 proteins have been shown to be substrates of one or more caspases in mammalian cells, and the list is still growing (201). These substrate proteins contain one or very few caspase cleavage sites in interdomain linker sequences. Substrate proteins are thus not degraded by caspase processing; instead, caspase cleavage may activate or inactivate the substrate protein's functions. Because of the strict requirement for an aspartate residue at P1 and distinct preferences for P2 to P4 residues, it is relatively straightforward to identify the likely caspase cleavage sites within a given substrate protein and the likely group of caspases that may recognize them. The substrates identified so far fall into two general groups: a large group of proteins thought to be involved in regulation and execution of apoptosis, and a small group of proinflammatory cytokine precursors (Table 1). For many of the identified substrates, the functional consequences of their cleavage have only been inferred from their normal functions. In other cases, the role of caspase cleavage has been experimentally assessed by expressing mutant substrate proteins that have altered caspase cleavage sites or by expressing protein fragments that represent caspase cleavage products. The evolutionary conservation of caspase substrate proteins, in particular their caspase cleavage sites, suggests that caspases were first employed for apoptosis and later coopted for cytokine processing in mammals.
TABLE 1.
Caspase substrates
| Substrate |
|---|
| Cell death proteins |
| Bcl-2 |
| Bcl-xL |
| Bid |
| CrmA |
| IAP |
| p28 Bap31 |
| p35 |
| Procaspases |
| Cell cycle regulation |
| Cdc27 |
| Cyclin A |
| MDM2 |
| p21 (Cip1/Waf1) |
| p27 (Kip1) |
| PITSLRE kinases |
| Retinoblastoma protein |
| Wee1 phosphatase |
| Cytoskeleton |
| Actin |
| β-Catenin |
| Fodrin |
| Gas2 |
| Gelsolin |
| Keratin-18 and -19 |
| Lamins |
| Plakoglobin |
| Cytokine precursors |
| Pro-IL-1β |
| Pro-IL-16 |
| Pro-IL-18 (IGIF) |
| DNA metabolism |
| Acinus |
| DNA-dependent protein kinase (DNA-PK) |
| DNA replication complex C (DSEB/RFC140) |
| ICAD |
| MCM3 DNA replication factor |
| NuMA |
| PARP |
| Topoisomerase 1 |
| Neurodegenerative disease proteins |
| APP |
| Ataxin-3 (spinocerebellar ataxia type 3) |
| Androgen receptor (Kennedy's disease) |
| Atrophin-1 (dentarorubral pallidoluysian atrophy protein |
| Huntingtin |
| Presenilins |
| RNA metabolism |
| Eukaryotic initiation factor 2α |
| Heteronuclear ribonuclear proteins C1 and C2 |
| 70-kDa U1-snRNP |
| Signal transduction |
| Adenomatous polyposis coli protein (APC) |
| Akt/PkB |
| Calmodulin-dependent kinase IV |
| c-Raf |
| D4-GDP dissociation inhibitor |
| Fyn tyrosine kinase |
| Focal adhesion kinase |
| MEKK1 |
| MST/Ksr |
| PAK-2/hPAK65 |
| Protein kinase C delta |
| Protein kinase C theta |
| Protein kinase C-related kinase 2 |
| Protein phosphatase 2A |
| Ras GTPase activating protein |
| TCR-ζ chain |
| Transcription factors |
| Heat shock factor |
| GATA-1 |
| IκB-α |
| NF-κB (p50, p65) |
| NRF-2 |
| Sp1 |
| STAT1 |
| Sterol-regulatory element-binding proteins |
| Others |
| Calpastatin |
| Hsp90 |
| Nedd4 |
| Phospholipase A2 |
| Rabaptin-5 |
| Transglutaminase |
Execution of Apoptosis
Once it was appreciated that CED-3 is a homologue of ICE (interleukin-1-converting enzyme, now known as caspase-1) (254), the search began in earnest for key caspase substrates in apoptotic execution. The involvement of proteases in apoptosis was anticipated; after all, proteolysis must occur in the irreversible disintegration of the cell. However, apoptosis could not result from the indiscriminate digestion of all cellular proteins; instead, it must involve controlled activation and inactivation of certain choice proteins to carefully dismantle the cell. The search for caspase substrates has brought several major questions into focus. What are the minimal set of proteins that must be cleaved in order to induce the phenotypic features of apoptosis? How is apoptosis coordinated with other cellular processes? How is apoptosis regulated after the activation of caspases? Although the significance of caspase-mediated cleavage is not well understood for many of the substrates, the study of caspase substrates has already shed light on these questions. Here we highlight several themes that have emerged from the accumulating information regarding caspase substrates during apoptosis.
DNA metabolism.
During apoptosis, nuclear DNA is condensed and degraded into large (50 to 300 kb) and subsequently into small oligonucleosomal fragments of several hundred base pairs. DNA degradation irreversibly dooms the cell and also destroys viruses or harmful mutations that may have found their way into the genetic blueprint. Biochemical purification by several groups led to the discovery of a caspase-regulated DNase complex termed DFF (DNA fragmentation factor) that is composed of a DNase termed CAD (caspase-activated DNase: also named DFF40 and CPAN) and its inhibitor ICAD (also named DFF45) (51, 128, 173). In healthy cells, CAD is complexed with ICAD and functionally inactive. In apoptotic cells, ICAD is cleaved by caspase-3 and -7, releasing CAD to degrade nuclear DNA (173). Two other regulators, CIDE-A and CIDE-B, have an N-terminal regulatory domain homologous to ICAD and CAD, which allows them to interact with and regulate the activity of the DFF complex (94, 129). DFF induces both chromatin condensation and DNA fragmentation in vitro (127), and cells from mice deficient for DFF activity exhibit neither of these classic apoptotic features when induced to die (259). Another caspase-3-activated factor, named acinus, induces chromatin condensation without affecting DNA fragmentation (172). Acinus contains a potential DNA/RNA-binding motif and is activated by concordant proteolytic cleavage of caspases and an unidentified serine protease activity (172). The task of DNA fragmentation appears to be separate from other phenotypic aspects of apoptosis. For example, when cells that overexpress caspase-resistant ICAD and thus lack CAD activity are induced to die, they exhibit neither large-scale nor nucleosomal DNA fragmentation, but these cells still die manifesting other features of apoptosis, such as cell body shrinkage and phosphatidylserine exposure (173). Mice deficient for DFF activity develop normally and have no obvious defects in homeostatic apoptosis (259). These results are consistent with previous genetic studies with the nuc-1 nuclease in C. elegans programmed cell death: DNA fragmentation is a downstream effect of caspase activation and is dispensable for cell killing (87). Instead, DNA fragmentation is a way of “hiding the body” and represents one of the multiple parallel execution pathways in apoptosis.
Cytoskeletal scaffold proteins.
The cytoskeleton of an apoptotic cell undergoes profound changes as the nucleus fragments, the cell body shrinks, and the cell becomes detached from surrounding cells and the basal membrane. The molecular logic behind these changes is becoming clearer with the biochemical identification of many cytoskeletal proteins that are cleaved by caspases. Lamins, the intermediate filament scaffold proteins of the nuclear envelope, are cleaved mainly by caspase-6 and inactivated, leading to nuclear fragmentation in the final phases of apoptosis (115, 204). Overexpression of lamins with mutated caspase cleavage sites delays the onset of chromatin condensation and pyknosis during apoptosis, indicating that caspase cleavage of lamins facilitates these morphologic changes specifically (162). Apoptotic execution also takes advantage of endogenous regulatory mechanisms for disassembling the cytoskeleton. Gelsolin, a cytoplasmic F actin-depolymerizing enzyme, is cleaved by caspase-3 to yield a fragment with constitutive activity (109). Gelsolin-deficient neutrophils exhibit greatly delayed membrane blebbing during apoptosis, a classic morphological feature, implying that membrane blebbing requires actin reorganization mediated by caspase-activated gelsolin (109). Caspases attack multiple targets in the cortical actin architecture: α-fodrin and focal adhesion kinase, two components of the focal adhesion complex, which links cortical actin filaments, plasma membrane, and transmembrane proteins to the extracellular matrix, are cleaved by caspases and may lead to cell body shrinkage and allow apoptotic cells to detach from neighboring cells and basement membrane (36, 136, 232). Actin itself is cleaved by effector caspases in certain cells, and the caspase-generated fragments may contribute directly to apoptotic changes in cell shape (201). In addition, PAK2, a serine/threonine kinase downstream of Rac and Cdc42, small GTPases that regulate actin dynamics, is cleaved by caspase-3 to generate a constitutively active kinase during apoptosis (171). Blocking PAK2 activity with a dominant negative kinase-dead PAK2 mutant inhibits the formation of apoptotic bodies (small sealed vesicles that contain condensed cytoplasmic material from fragmented apoptotic cells), but interestingly, nuclear features of apoptosis and phosphatidylserine externalization were not inhibited (171). From these examples, it is evident that multiple cytoskeletal proteins and their regulators are targeted by caspases to dismantle the cellular architecture, and perturbation of particular cytoskeletal targets may be responsible for specific morphological features in apoptosis. In general, preventing one substrate cleavage may delay or even abrogate a morphological feature of apoptosis but will not prevent cell death. The possibility of uncoupling morphological features in apoptosis should also caution investigators to identify clearly the microscopic or biochemical criteria used to measure apoptosis.
Cell cycle regulators.
Given some morphological similarities between apoptosis and mitotic catastrophe (in which cell division occurs without completion of S phase, leading to cell body shrinkage, DNA condensation, nuclear envelope breakdown, and eventual cell death), an early idea in the field was that apoptosis may be a kind of aborted or ectopic cell cycle. This idea was supported by the finding that cyclin-dependent kinase (Cdk) activity increases during apoptosis, and inhibition of Cdk activity attenuates apoptosis in several experimental systems (182). However, with the continued identification of caspase substrates, it is now clear that the induction of Cdk activity during apoptosis is a consequence of effector caspase activity. Cdc27, a component of the ubiquitin ligase complex that mediates the degradation of mitotic cyclins, and Weel, a kinase that provides an inhibitory phosphorylation on Cdks, are both cleaved and inactivated by caspase-3-like activities (262). Similarly, two Cdk inhibitors, p21Cipl and p27Kipl, can be cleaved by caspase-3, which decreases their association with Cdks and thus allows Cdk activity to accumulate during apoptosis (119). Because cleavage of cell cycle regulators occurs late in apoptosis by caspase-3-like activities in parallel with the dismantling of the transcription and translation machinery, caspase-activated Cdk activity cannot activate the normal mitotic program (262). For example, mitotic spindles do not form in apoptotic cells, distinguishing apoptosis from mitotic catastrophe. Instead, Cdk activity in apoptosis appears to be required for several aspects of apoptotic morphology, including DNA condensation and cell body shrinkage, but not others, such as loss of mitochondrial potential and externalization of plasma membrane phosphatidylserine (79). These results qualify caspase-mediated Cdk activation as another independent effector pathway for the execution of apoptosis. The mechanism by which activated Cdks carry out their jobs during apoptosis is unclear, but it is likely to be quite distinct from their normal targets during mitosis (262).
Repair and housekeeping enzymes.
Historically, one of the first proteins identified as being cleaved during apoptosis by diverse stimuli was poly(ADP-ribose) polymerase (PARP); PARP cleavage activity was later used to identify and purify caspase-3 as an effector caspase (151, 208). PARP is a nuclear enzyme that senses DNA nicks and catalyzes the ADP-ribosylation of histones and other nuclear proteins in order to facilitate DNA repair. PARP is cleaved at a single site by caspase-3, which separates the N-terminal DNA-binding domain from the catalytic domain and inactivates the enzymatic activity (114). With subsequent identification of other DNA repair enzymes, such as DNA-dependent protein kinases, and housekeeping enzymes, such as the 70-kDa U1-snRNP protein involved in mRNA splicing, are cleaved during apoptosis, a hypothesis was forwarded that cleavage and inactivation of repair proteins may lead to lethal DNA damage and thus contribute to apoptosis. However, this hypothesis is probably incorrect given that enucleated cells (cytoplasts) can maintain metabolic homeostasis for some time and be triggered to undergo apoptosis with all the cytoplasmic features of apoptosis (99) and cells deficient for PARP or DNA repair enzymes have no general perturbation in apoptotic execution (228). Instead, new evidence suggests that PARP inactivation by caspase-3 is important for turning off an energetically expensive repair pathway following commitment to apoptosis. Activated PARP transfers up to 100 ADP-ribose moieties to each acceptor site in target proteins, and each cycle of ADP-ribosylation is coupled with consumption of 1 NAD+ molecule, which is metabolically equivalent to 4 ATP molecules. One can imagine that during apoptotic execution, DNA fragmentation by CAD can cause significant activation of PARP and quickly deplete cellular energy stores. In the absence of an energy pool sufficient to maintain ionic homeostasis, the cell can die quickly by default necrosis (258). Indeed, when cells engineered to express caspase-resistant PARP are induced to undergo apoptosis, they undergo more extensive apoptosis and a fraction of cells also undergo necrosis (11, 81). However, because PARP contributes to DNA base excision repair, its genetic absence or inactivation by caspase cleavage can sensitize cells to apoptosis induction by DNA-damaging agents (77, 152). Consistent with the postulated requirement of maintaining cellular energy during apoptosis, cells artificially manipulated to have low ATP pools undergo necrosis instead of apoptosis in stress conditions, like cells that are unable to cleave PARP (118). The biologic roles of DNA-dependent protein kinases and U1-snRNP cleavage in apoptosis have not been examined in detail, but they may represent a concerted strategy to prevent futile repair and synthetic efforts during apoptosis. Caspase-mediated inactivation of DNA repair and housekeeping enzymes reinforces the idea that apoptosis is a carefully orchestrated program of irreversible cell suicide, and the cell's metabolic efforts may be redirected to suit that goal.
Signaling molecules.
A large number of signal transduction proteins have been found to be caspase substrates during apoptosis (Table 1). Most, if not all, of these proteins are cleaved by caspase-3-like effector caspases, in parallel with the proteolysis of housekeeping enzymes such as PARP. For several serine/threonine kinases, such as PAK2, the mitogen-activated kinase kinase kinase (MAP3Ks) MEKK1 and MST1, caspase-3 cleavage occurs between the N-terminal autoinhibitory domain and the catalytic kinase domain, resulting in kinase activation (15, 70, 171). Intriguingly, these kinases are all able to activate the JNK and p38 stress-activated protein kinase pathways, which can provide a mechanism of caspase-dependent JNK activation. As described above, activation of PAK2 is important for cytoskeletal reorganization and plasma membrane blebbing. In the case of MEKK-1, caspase-mediated kinase activation is thought to provide a positive feedback loop for signaling apoptosis, because expression of the caspase-cleaved kinase fragment induces caspase activation and apoptosis (15). Epithelial cells undergo apoptosis if they are detached from basement membrane, a process termed anoikis (Greek for homelessness)(59). MEKK1 is activated following epithelial cell detachment, and blockade of either MEKK1 or caspase activity blocks anoikis. MST1 cleavage by caspase-3 also yields a constitutive kinase and potent inducer of apoptosis (70). Apoptosis induction by activated MAP3Ks upstream of the JNK pathway may be explained in part by the ability of JNK to phosphorylate and inactivate Bcl-2 (138). Caspase-3-mediated cleavage and inactivation of the antiapoptotic kinases Akt and c-Raf provide two more examples of positive feedback loops in apoptosis (234).
Many other proteins found to be caspase substrates are cleaved by effector caspases late in apoptosis (Table 1). The kinetic data imply that this group of proteins may not have primary roles in regulating the decision to undergo cell death. A more intriguing possibility is that caspase-mediated signaling may affect subsequent events in apoptosis, such as phagocytosis of apoptotic bodies or recycling of the cellular constituents. For instance, phosphatidylserine externalization at the plasma membrane facilitates phagocytosis of apoptotic bodies and requires caspase activity, but the exact molecular pathway remains largely unknown. In this view, activation of signaling cascades by caspases can be considered a “message from the dead,” dictating to the neighboring cells what to do with this unexpected cargo. Finally, although most members of this group of substrates were discovered during studies of apoptosis, physiologic regulation of these molecules by caspases in healthy cells remains to be explored. For example, the recent discovery that caspase-mediated inactivation of GATA-1, an erythropoietic transcription factor, plays a role in the negative feedback of red blood cell development should broaden our horizons in considering the functional effects of caspase-mediated cleavage (38).
Physiologic amplification.
Given the intimate connection between caspases and the Bcl-2 family proteins, perhaps it is not surprising that Bcl-2 family members can serve as substrates for caspases. The Bcl-2 family of antiapoptotic proteins function as a threshold mechanism in preventing apoptosis, and caspase-mediated cleavage of Bcl-2 family members appears to amplify and reinforce the death signal. The antiapoptotic family members Bcl-2 and Bcl-xL can each be cleaved by effector caspases to generate fragments that have proapoptotic activity (26, 32). Specifically, the N-terminal BH4 (Bcl-2 homology) domain, present only in antiapoptotic Bcl-2 family members, is removed by caspase cleavage, resulting in C-terminal fragments that resemble the proapoptotic family members Bax and Bak (26). The conversion of antiapoptotic regulators into a proapoptotic force constitutes a positive feedback loop in the terminal phase of apoptosis, removing antiapoptotic brakes and accelerating caspase activation and apoptotic execution. The direct inactivation of Bcl-2 family members by caspases may also explain the appearance of mitochondrial damage in the terminal phase of apoptosis regardless of the initial insult.
Caspases and Bcl-2 family members can also intersect early in the initiation of apoptosis. Bid, a member of the proapoptotic BH3 domain Bcl-2 antagonists, is cleaved by the initiator caspase-8 following death receptor activation (122, 130). Bid is normally cytoplasmic and apparently inactive; following proteolytic activation, Bid translocates to the mitochondria and triggers the mitochondrial death pathway, as evidenced by the release of cytochrome c. These results suggest that Bid cleavage functions as an amplification step in caspase activation, connecting caspase-8 activity to caspase-9 activation. An important implication is that caspase cascades may not necessarily follow a domino-like sequence of one caspase cleaving the next; instead, other molecular targets may intervene and amplify the death signal via more elaborate mechanisms. A priori, it can be imagined that the physiologic role of Bid cleavage is most prominent in mitochondrion-rich tissues with weak death receptor activation of initiator caspases. Indeed, Bid knockout mice are developmentally normal and do not share the lymphoproliferative symptoms or lymphocyte apoptosis deficits of Fas-deficient mice. However, Bid−/− mice are resistant to fulminant hepatic apoptosis and death following intraperitoneal injection of agonistic anti-Fas antibody (251). How may caspase-activated Bid function to amplify the death signal? The mammalian BH3 subfamily of death inducers may function in an analogous fashion to C. elegans EGL-1, binding to and antagonizing the function of antiapoptotic Bcl-2 family members (1). Apoptosis induction by Bid can indeed be blocked by overexpression of Bcl-xL. Alternatively, activated Bid may induce mitochondrial damage and cytochrome c release directly by inserting into the mitochondrial membrane and forming a channel (177, 184).
Pathologic amplification.
The accelerating pace of apoptosis research and the molecular genetics of neurodegenerative diseases have converged on the role of caspases in the pathogenesis of a diverse group of neurodegenerative diseases thought to be caused by insoluble protein aggregates. To date, eight inherited neurodegenerative diseases, including Huntington's disease and spinocerebellar ataxias, have been found to be caused by dominantly inherited unstable CAG repeats within the coding regions of unrelated proteins (134). These diseases are characterized by neuronal apoptosis in specific, distinct regions of the brain, but their pathogenesis is unified by the correlation between the number of CAG repeats and disease severity and the apparent toxicity of the expanded polyglutamine (polyQ) sequences encoded by the CAG repeats (134). Through incompletely understood mechanisms, the expanded polyQ proteins aggregate and form nuclear inclusions in the neurons that are fated to die in each disease. Several polyQ disease proteins have been found to be substrates for caspase-3 in vitro and in vivo (Table 1). Caspase-mediated cleavage removes the polyQ sequence from the remainder of the protein, and the liberated polyQ fragments are more prone to aggregation and can induce neuronal apoptosis in vitro and in transgenic animals (more below). Furthermore, in the Huntington's disease gene product (huntingtin), increasing length of the polyQ sequence is correlated with an increased propensity for caspase-3 cleavage (66). These data suggest that the polyQ proteins encoded by the disease alleles may become preferential caspase substrates, and caspase cleavage can enhance the ability of the polyQ fragments to induce neuronal apoptosis. Indeed, mutations of the caspase recognition sites in the atrophin-1 (DRPLA disease protein) or androgen receptor (mutated in X-linked spinal and bulbar muscular atrophy, also known as Kennedy's disease) polyQ proteins abrogated their cytotoxicity in vitro (49, 50).
Alzheimer's disease is the leading cause of age-related cognitive decline and disability, and its pathoneumonic lesions are neurofibrillary tangles, composed of the microtubular protein tau, and senile plaques, which are aggregates of the β-amyloid peptide (134). Aggregates of β-amyloid peptide induce neuronal apoptosis in vitro, and increased production of β-amyloid peptide has been postulated to be the unifying pathologic mechanism underlying mutations that cause familial early-onset Alzheimer's disease (134). Effector caspases can increase β-amyloid production by several mechanisms. Loss-of-function mutations in the presenilin-1 and -2 genes are responsible for the majority of early-onset familial Alzheimer's disease and are thought to increase β-amyloid production (134). Caspase-3 can cleave and inactivate presenilins in vitro, which may mimic the effect of pathologic presenilin mutations (105). The 40 to 42-amino-acid β-amyloid peptide is derived from proteolytic processing of a large transmembrane protein termed amyloid-β precursor protein (APP) at two sites, the N-terminal β-secretase site and the C-terminal γ-secretase site. Caspase-3 cleaves APP in vitro mainly after amino acid 720, a site that is C-terminal to the γ-secretase site (64). The N-terminal caspase cleavage product, APPΔC, facilitates the production of β-amyloid peptide by approximately fivefold in vitro compared to full-length APP, and APPΔC itself appears to be a component of senile plaques found in Alzheimer patients' brains (64). Because caspase-3 activation and APPΔC formation are also induced in vitro after ischemic brain injury (a known risk factor for Alzheimer's disease), these results provide another example of a positive feedback loop between caspases and neurodegeneration. Neuronal apoptosis from ischemia or other causes activates caspase-3 and stimulates the formation of APPΔC, which increases the propensity for amyloidogenic β-amyloid peptide production. In turn, increased extracellular β-amyloid peptide production may induce neuronal apoptosis, leading to cognitive dysfunction and further deposition of senile plaques.
Proinflammatory Cytokine Processing
Three proinflammatory cytokines have been found to be processed by caspases to generate the mature, biologically active species. Interleukin (IL)-1β is a pleiotropic cytokine that mediates local and systemic effects of inflammation, including vasodilation, neutrophil migration, lymphocyte activation, and fever (43). IL-1β is synthesized as a cytosolic 33- kDa precursor that lacks a signal sequence. The first mammalian caspase, caspase-1, was originally identified as an enzyme that cleaved the IL-1β precursor to produce the mature cytokine and termed IL-1β-converting enzyme (ICE). Following cleavage by caspase-1, the 17-kDa C-terminal fragment becomes biologically active and is able to be secreted from the cell. In monocytes, caspase-1 appears to be constitutively active, but in many other cell types that can make IL-1β, caspase-1 activation appears to be inducible and occurs without apoptosis. Second, IL-18, also named gamma interferon-inducing factor (IGIF), induces gamma interferon production from T cells and natural killer cells and is thus an inducer of Th1 immune responses (42). IL-18 is structurally related to IL-1β, and like pro-IL-1β, IL-18 is synthesized as an inactive, cytosolic precursor and processed to its mature form by caspase-1 (65, 75). Finally, caspase-3, an effector caspase in apoptosis, is surprisingly also involved in cytokine processing. IL-16, another proinflammatory cytokine, is synthesized as an inactive 50-kDa precursor and can be processed by caspase-3 to generate the mature 20-kDa C-terminal protein (260). Intriguingly, proteolytic activation of caspase-3 is correlated with production of active IL-16 in T lymphocytes that appears temporally unrelated to apoptosis (238). The processing of proinflammatory cytokines by caspases appears paradoxical at first glance because apoptosis is thought to be a physiologic mechanism of cell removal that does not cause inflammation. Cells with constitutive caspase activity (such as caspase-1 in monocytes) proliferate and apparently regulate apoptosis normally; the mechanism by which caspases involved in cytokine production may be segregated from the apoptotic machinery should be addressed in future studies.
PHYSIOLOGICAL FUNCTIONS
The physiological functions of caspases have been assessed by both pharmacological inhibition and gene knockout experiments. Because of the overlapping substrate specificities of caspase family members, both pharmacological and genetic approaches have been fruitful in elucidating the redundant and specific roles of caspases. To date, the genes for caspase-1,-2, -3, -8, -9, -11, and -12 have been deleted in mice; human cell lines deficient in caspase-3, -8, and -10 have also been described. The results from these genetic experiments reveal that mammalian caspases have overlapping and tissue-specific roles in controlling apoptosis in response to specific stimuli (Table 2). In addition, caspases have been coopted for specialized functions, such as the regulatory processing of inflammatory cytokines and terminal differentiation of certain cells.
TABLE 2.
Phenotypes of mice deficient in caspase or caspase adapter proteins
| Gene | Development | Phenotype | Reference |
|---|---|---|---|
| Caspase-1 | Normal | Defective lipopolysacharide-induced secretion of IL-1α and β and γ interferon; resists endotoxic shock; thymocytes partially resistant to Fas-mediated apoptosis | 111, 123 |
| Caspase-2 | Normal | Increased number of oocytes at birth, and oocytes less susceptible to doxorubicin-induced apoptosis; B cells partially resistant to death induction by granzyme B; accelerated apoptosis of facial motor neurons | 4 |
| Caspase-3 | Perinatally lethal | Ectopic brain tissue; ES resistant to UV and osmotic shock-induced apoptosis; sensitivity to γ-irradiation unaffected; peripheral T cells partially resistant to anti-CD-3ε and Fas-induced apoptosis; transformed EF cells less susceptible to adriamycin, TNF-α, and low serum; abnormal chromatin condensation and no DNA fragmentation in dying cells | 112, 236 |
| Caspase-8 | Embryonically lethal at approx. E12.5 | Abnormal heart development and vascular hyperemia; EF resistant to Fas, TNF-α, and DR3 but exhibit normal sensitivity to UV, etoposide, low serum, and staurosporine; death receptor signaling to JNK and NF-κB intact | 219 |
| Caspase-9 | Embryonically lethal at approx. E16.5 | Ectopic brain tissue; ES cells resistant to adriamycin, UV, and γ-irradiation; EF resistant to UV and DNA-damaging agents but not to TNF-α; thymocytes resistant to etoposide, dexamethasone, and γ-irradiation, but sensitive to UV and Fas; loss of mitochondrial potential and procaspase-3 processing blocked, but cytochrome c release unaffected | 76, 110 |
| Caspase-11 | Normal | Resistant to lipopolysaccharide-induced caspase-1 processing, IL-1α and -β secretion, and endotoxic shock | 226 |
| Caspase-12 | Normal | EF partially resistant to ER stress such as tunicamycin and thapsigargin; cortical neurons resistant to β-amyloid-induced apoptosis | 149 |
| Apaf-1 | Embryonically lethal at approx. E16.5 | Ectopic brain tissue; craniofacial developmental anomalies; transient persistence of interdigital webs; ES and EF resistant to UV, DNA-damaging agents, and γ-irradiation; thymocytes and peripheral activated T cells resistant to many stimuli but not to Fas; defective processing of procaspase-2, -3, and -8 during thymocyte apoptosis induced by dexamethasone or etoposide; loss of mitochondrial potential but not cytochrome c release | 16, 252 |
| FADD | Embryonically lethal at approx. E9-E12.5 | Abnormal heart development and hemorrhage; no mature B cells and defective T-cell activation; EF and thymocytes resistant to Fas and TNF-α but normal sensitivity to E1A, myc, and adriamycin | 249, 257 |
Tissue and Signal Specificity
In C. elegans most if not all of the developmental programmed cell death requires the function of the CED-3 caspase, but knockout of mammalian caspase genes has revealed profound but selective defects in apoptosis (Table 2). Although most mammalian caspase mRNAs are ubiquitously expressed, the large number of mammalian caspase genes implied that there may be tissue-specific control or private pathways in response to specific apoptotic stimuli. A prime example is caspase-2, which appears to be critical for controlling germ cell apoptosis and thus the reproductive life span but is dispensable for normal development and death receptor killing (4). Similarly, caspase-12 responds specifically to ER stress but not to cell surface death receptor or mitochondrial apoptosis pathways (149). Careful examination of tissues from several other caspase knockout animals reveals rather specific requirements that are not easily conceptualized (261). A caspase may be essential for an apoptotic stimulus in one cell type but not another, and different apoptotic stimuli require specific caspases to different degrees. For example, Casp9−/− mouse embryonic fibroblasts but not thymocytes or splenocytes are resistant to apoptosis induction by UV irradiation, and Casp9−/− thymocytes are resistant to apoptosis induction by dexamethasone or gamma-irradiation but not by Fas (76, 110). Thus, although certain caspase activities reproducibly increase following many apoptotic stimuli, the pathway from the initiating signal to the death machinery appears to be very context dependent and becomes evident only with knockout cells. It should be noted that in caspase knockout tissues where there is evidence of increased viability in the face of apoptotic stimuli, analysis at a later time point can reveal evidence of apoptosis (110). Therefore, apoptosis is delayed but not blocked. The narrow spectrum of apoptotic defects and incomplete rescue in individual caspase gene knockout animals suggest that in mammalian cells, the large number of caspases are likely to have redundant functions or be able to compensate for one another in the case of genetic deficiency.
Caspase Cascades
How do the caspase knockouts match up with the caspase cascades predicted by biochemical experiments? Although the knockout data show that apoptosis pathways defy conceptions of linear cascades in toto, the pattern of phenotypes and apoptotic defects is broadly consistent with two known caspase cascades. Biochemical purification and reconstitution experiments by Wang and colleagues have suggested an Apaf-1/caspase-9/caspase-3 cascade (124). Indeed, mice deficient for each of these three genes have ectopic brain tissue that appears to be histologically nontransformed neurons and glia, implicating an Apaf-1 pathway in developmental neuronal apoptosis (Table 2). This phenotype is reminiscent of certain ced-3 loss-of-function alleles in C. elegans, where cells fated to die differentiated into functional neurons (87). Procaspase-3 processing induced by cytochrome c is blocked in cells lacking Apaf-1 or Casp9, confirming the role of these two proteins in initiating the mitochondrial apoptosis pathway (16, 76, 110, 252). In general, cells from of Casp9−/− or Apaf-1−1− animals are resistant to a variety of apoptotic inducers such as UV, DNA-damaging agents, γ-irradiation, and oncogene overexpression, but remain sensitive to death receptor ligands such as Fas and TNF-α. On the other hand, studies of death receptors, such as Fas and TNFR1, have suggested a FADD/caspase-8/caspase-3 cascade (3). Both FADD- and Casp8-deficient embryos die around E12.5 with abnormally thin cardiac ventricles and hyperemia. Cells from FADD−/− or Casp8−/− animals show complete resistance to at least three death receptors but have normal sensitivity to UV, DNA-damaging agents, and oncogenes (Table 2). This natural segregation of phenotypes suggests that the Apaf-1/caspase-9 mitochondrial pathway is mainly responsible for apoptosis secondary to cell damage or imbalances in homeostasis, such as following exposure to UV, chemotherapeutic drugs, or oncogene overexpression. The FADD/caspase-8 pathway is dispensable for those apoptotic stimuli but is essential for apoptosis by cell surface death receptors, such as in activation-induced cell death of lymphocytes that is critical for peripheral tolerance. Although these results at first glance contradict observations that Fas may be required for UV-, chemotherapy-, and oncogene-induced apoptosis (84, 93, 164), other Fas-mediated signal transduction pathways independent of caspases may explain such discrepancies (22, 222). FADD and caspase-8 are also essential for cardiac development, but it is unclear whether they are needed to induce apoptosis during heart development or have other signaling functions.
Finally, downstream of both pathways lie the shared effector caspases. Casp3−/− cells show defects in apoptosis in response to signals that trigger either pathway (Table 2). However, although Casp3−/− cells show reduced apoptosis in response to many stimuli, these cells still die eventually in the absence of apoptotic features such as DNA fragmentation and cellular organelle breakdown (236). Instead, the dying Casp3−/− cells exhibit plasma membrane breakdown, vacuolation, and swelling of cellular organelles, features reminiscent of necrosis (236). The absolute requirement for caspase-3 for the execution of apoptosis is surprising, because caspase-7 appears to be indistinguishable biochemically. Thus, additional control mechanisms, such as selective localization or additional cofactors, may be involved in regulating caspase-3 function (see above). The necrotic death of Casp3−/− cells may reflect a default pathway following accumulating cellular damage from the initial apoptotic stimuli, such as DNA damage or, in the case of private death pathways such as Fas, the full activation of other caspase family members. For the large list of caspase substrates (see above), it is presently unclear what fraction are cleaved in the absence of a single effector caspase. Cells from Casp3−/− and Casp7−/− mice should be instrumental in answering these questions.
Complexities in regulation.
Closer inspection of caspase knockout animals quickly reveals many details that argue against simple linear models of caspase cascades. For example, Apaf-1-deficient animals show delayed apoptosis and persistence of interdigital webs, a phenotype not present in Casp9−/− or Casp3−/− animals. These and other data argue that other caspases may substitute for caspase-9 downstream of Apaf-1 and cytochrome c, and conversely, additional CED-4-like caspase activators are likely to exist in mammalian cells. Nonlinearity is also the rule in caspase cascades downstream of death receptors. B cells fail to develop in FADD-deficient animals, and FADD−/− T cells exhibit a severe proliferation defect (257). These immunological phenotypes are not present in any of the caspase knockouts or known death receptor knockouts, suggesting that additional signal transducers exist upstream and downstream of FADD. Recently, several patients with autoimmune lymphoproliferative syndrome (ALPS), an inherited syndrome of lymphadenopathy and autoantibody production caused by defective Fas-mediated apoptosis, were found to harbor dominant mutations in Casp10 (225). Thus, both caspase-8 and caspase-10, which have most sequence similarity with one another in the caspase family, are both required for the death receptor caspase cascade. The ALPS mutations decrease the caspase activity of caspase-10 (225). The dominant effect of mutations in Casp10 contrasts with the normal phenotypes of heterozygous caspase knockout mice, which implies that caspase genes are not haploinsufficient. These differences can be understood by the oligomerization model of procaspase activation: if multiple copies of procaspases need to be brought into close proximity to activate one another (see above), half-defective procaspases in the complex (e.g., ALPS mutations) would have more deleterious effects than half the concentration of wild-type caspases (e.g., heterozygous knockout mice). Interestingly, ALPS patients with germ line Casp10 mutations have defects in lymphocyte apoptosis but no known cardiac abnormality. Thus, the FADD/caspase-8 pathway collaborates with caspase-10 in lymphocytes but does not require it in cardiac development. Although we are only beginning to define specific roles for individual caspases, it is clear that multiple caspases may function in parallel or redundantly in apoptosis, and the physiological roles shared among caspases are also controlled in a tissue-specific manner.
Inflammatory Cytokine Production
In caspase-1-deficient animals, production of IL-1β and IL-18 is concordantly blocked, emphasizing the essential role of caspase-1-mediated processing for cytokine maturation (43). Surprisingly, secretion but not processing of IL-1α, a proinflammatory cytokine that shares many functional properties with IL-1β, is also blocked in Caspl-1- cells, revealing a role for caspase-1 or a caspase-1-dependent process in IL-1α secretion (111, 123). Murine caspase-11 physically interacts with caspase-1 and has been proposed to function in the same pathway. Casp11−/− mice are developmentally normally, but procaspase-1 processing and IL-1β production are blocked in Casp11−/− cells (226). Casp1 and Casp11 knockout mice are both resistant to lethal shock induced by endotoxin (lipopolysaccharide, a component of bacterial cell wall), presumably due to reduced production of proinflammatory cytokines. These results strongly suggest that caspase-11 is an upstream activator of caspase-1, which then mediates the cleavage of cytokine precursors. Caspase-11 mRNA is strongly induced by lipopolysaccharide and therefore may be the initiator caspase in this cascade (226). The caspase-11/caspase-1 pathway appears to be dispensable for developmental and most normal homeostatic apoptosis but has evolved to handle a separate task, cytokine maturation. Interestingly, genetic deficiencies in IL-1β or IL-18 do not protect mice from endotoxic shock, suggesting that the caspase-1 pathway may control other cytokines that mediate innate immunity.
Terminal Differentiation
In the course of apoptosis, the doomed cell is fragmented and eventually taken up by neighboring cells; intriguingly, the versatile death machine can also be redirected to make useful corpses. New data suggest that caspases have been adapted to make specialized cells that lack organelles, such as lens epithelial cells and skin keratinocytes. Lens epithelial cells lose their nuclei and other organelles during terminal differentiation to maximize optical transparency; this terminal differentiation is blocked by the pan-caspase inhibitor zVAD (97). Differentiated lens cells are genetically dead but remain metabolically active, suggesting that cytosolic organization and enzymes are intact. In contrast, as keratinocytes move out from the basement membrane and differentiate, they lose their nuclei and other organelles and form a tightly compacted layer of dead cells filled with cross-linked proteinaceous material. Although the nuclei of differentiating keratinocytes do not appear pyknotic, inhibition of caspase activity by zVAD prevented nuclear destruction but not other aspects of keratinocyte differentiation (231). The role for caspases in terminal differentiation may be evolutionarily conserved. The Drosophila caspase DCP-1 is required for nurse cells to transfer cytoplasmic material into developing oocytes, a process that provides oocytes with maternally derived mRNAs and proteins and is essential for fertility (139). At present, no defects in the terminal differentiation of specialized cells have been described in caspase-deficient mice. Future studies should reveal which mammalian caspases function in terminal differentiation and how they are redirected from apoptosis. Studies in this area are likely to teach us many practical lessons for controlling caspases, organelle turnover, and differentiation.
ROLES IN HUMAN DISEASES
Caspases: The Be-All and End-All of Mammalian Apoptosis?
Alterations of cell survival feature prominently in the pathogenesis and potential treatment of many important human diseases (210); are caspases the key to the therapeutic control of apoptosis? In C. elegans, loss-of-function mutations in ced-3 or ced-4 completely rescues the lethality of ced-9 loss-of-function mutants (87). In ced-3 ced-9 double mutant animals, cells that normally die during development live, differentiate into distinct cell types, and can be shown to function normally (87). These results indicate that the only function of CED-9 is to inhibit CED-3 activity and that CED-9 has no other essential role, such as mitochondrial protection. However, it should be noted that the programmed cell death during C. elegans development is specified by lineage, and the cells doomed to die are not damaged or stressed in any way. In mammalian cells, caspase inhibitors block the morphological appearance of apoptosis in response to oxidative stress or chemotherapy drugs, but the cells still eventually die by necrosis (165). Cells overexpressing Bcl-2 or Bcl-xL are not killed by these challenges. Caspase inhibitors also do not block cytochrome c release by these stimuli, although Bcl-2 and Bcl-xL do (216). On the other hand, Bcl-2 still inhibits apoptosis after the release of cytochrome c into the cytosol (168). Collectively, these published reports suggest that the Bcl-2 family of anti-apoptotic proteins function at two levels; first, they maintain mitochondrial function, which prevents the release of mitochondrial factors (e.g., cytochrome c and reactive oxygen species) that trigger apoptosis and necrosis; second, they function downstream of mitochondrial factors to prevent caspase activation, perhaps by complexing with CED-4-like adapter proteins and preventing their oligomerization.
What may be the fate of mammalian cells under pathological duress if their caspases are inhibited? A foray into this topic quickly encounters definitional controversies regarding what exactly is meant by apoptosis (220). The utility of caspase inhibition in many disease states has been evaluated by the effects of adding peptide inhibitors of caspases, in particular broad-spectrum inhibitors such as zVAD. However, such data should be considered with the understanding that current peptide inhibitors may not block the activities of all caspases, some of which may not yet have been discovered; peptide inhibitors may not reach the necessary concentration within particular cells due to cellular factors that affect their accumulation or stability; and caspase inhibitors at high concentrations are known to inhibit other enzymes, including calpain, cathepsin, and kinases, and the reactive methylketone group in irreversible caspase inhibitors may modify various cellular proteins and lipids (10). Animals and cells deficient for caspase genes or transgenic for viral inhibitors of caspases, such as CrmA and p35, may be more specific probes of caspase functions in vitro. For therapeutic purposes, caspase inhibition needs to go beyond blocking the morphological features of apoptosis or delaying the onset of cell death; indeed, caspase inhibition has been shown in some cases to lead to clonogenic survival and normal physiological function of the surviving cells. In diseases in which the apoptotic machinery is inappropriately triggered but the cell is not otherwise damaged, caspase inhibition may prove very effective in reversing the pathophysiology. On the other hand, apoptosis may be a response to cellular damage, and blocking efficient apoptosis by caspase inhibition may lead to tissue necrosis, inflammation, and worsening disease. Here we highlight the accumulating data on beneficial effects of altering caspase activity in a diverse group of human diseases.
Therapeutic Benefits from Inhibition of Caspase Activity
Controlling cytokine production.
The phenotypes of mice deficient in Casp1 or its upstream activator Casp11 strongly suggest that this pathway is essential for the production of proinflammatory cytokines IL-1 and IL-18 (43, 226). Because mice deficient in either caspase-1 or -11 develop normally and do not exhibit any obvious perturbations of homeostatic apoptosis, caspase-1 and -11 may be ideal targets for pharmaceutical therapy. Mice deficient in Casp1 or Casp11 are resistant to endotoxic shock, which predicts that specific inhibitors of caspase-1 or -11 may be beneficial in bacterial sepsis, where excessive cytokinemia causes fever, inflammation, abscess formation, and shock. However, analogous to the clinical experience with TNF-α antagonists, clinical manifestations of septic shock in patients may already indicate advanced disease, and interventions to block caspases or cytokine production may be too late to provide protection. On the other hand, IL-1- and IL-18- dependent inflammation in chronic autoimmune diseases, such as rheumatoid arthritis and multiple sclerosis, may benefit from caspase-1 and -11 inhibition (61, 214). Indeed, mice engineered to express the pan-caspase inhibitor p35 in oligodendrocytes become resistant to experimental autoimmune encephalitis, the mouse model of multiple sclerosis (85).
Caspase-1 activation can also underlie direct tissue damage during bacterial infection. Shigella and Salmonella spp. are enteric pathogens that cause dysentery and typhoid fever, respectively. These microorganisms invade colonic epithelial cells and macrophages; bacterial invasion of macrophages causes macrophage apoptosis and massive cytokine release, leading to tissue inflammation. Shigella and Salmonella encode homologous invasion proteins that bind and activate caspase-1, and Casp1-deficient macrophages are resistant to Shigella- and Salmonella-induced apoptosis (82, 83). These results hint at the intriguing possibility that caspase-1-mediated apoptosis by bacterial pathogens may have provided the evolutionary drive to couple caspase-1 activity to IL-1β production, thus linking macrophage apoptosis with the inflammatory response that can subsequently clear the infection (83).
Inflammation is often a secondary disease process that contributes to local tissue injury and prevents restoration of tissue function. Therefore, inhibition of the inflammatory cascade after an acute insult may limit the extent of damage and speed recovery. In transgenic mice that overexpress an active-site mutant of procaspase-1 (which inhibits endogenous caspase-1 activation in a dominant negative manner), neuronal apoptosis and neurologic symptoms were ameliorated compared to control mice in models of ischemic stroke, amyotrophic lateral sclerosis (Lou Gehrig's disease), and Huntington's disease (58, 153). In the ischemic model, pathologic apoptosis may result from hypoxia or reperfusion injury, which occurs when restoration of blood flow leads to an influx of toxic oxidants and calcium ions. Whether caspase-1 directly mediates pathologic apoptosis or causes apoptosis indirectly by inducing inflammation via cytokine production is at present unclear. Nonetheless, these experiments suggest that inhibition of caspase-1 may be beneficial following acute ischemic injuries such as stroke and myocardial infarction, or during chronic degenerative processes such as amyotrophic lateral sclerosis or Alzheimer's disease.
Neurodegenerative diseases.
Neuronal loss in a number of neurodegenerative diseases is characterized by apoptosis (210), but even in single-gene hereditary diseases, the biochemical connections between the mutated gene product and caspases remain elusive. However, the prominence of apoptosis in these diseases has prompted the idea that a lowered apoptotic threshold may be a common effector pathway for cell loss. A prime example is retinitis pigmentosa, retinal degeneration caused by a group of inherited mutations in components of the phototransduction apparatus. In Drosophila, mutations of homologous genes cause retinal degeneration by apoptosis. Coexpression of the caspase inhibitor p35 inhibited photoreceptor apoptosis, and more importantly, behavioral tests and electrical recording demonstrated that the photoreceptors remained perfectly functional (37). This result argues that in retinitis pigmentosa, the primary defect may be ectopic triggering of apoptosis in the absence of cell damage; thus, inhibition of caspases can prevent photoreceptor loss and preserve vision.
In contrast, neuronal loss in the eight known hereditary neurodegenerative diseases caused by polyQ repeat expansion likely represents a combination of ectopic apoptosis and intrinsic cellular dysfunction. In Huntington's disease, spinal and bulbar muscular atrophy (Kennedy's disease), spinocerebellar ataxia types 1, 2, 3, 6, and 7, and dentatorubropallidoluysian dystrophy, unstable CAG repeats in the disease genes lead to the production of proteins containing long polyQ repeats that aggregate in the nuclei of affected neurons and lead to apoptosis (134). The disease proteins are widely expressed in the brain and other tissues, but each disease is characterized by stereotypic regions of neuronal degeneration, where the appearance of nuclear polyQ aggregates correlates with sites of neuronal death. Many of these polyQ proteins are substrates of caspase-3, which can affect the localization and enhance the aggregation of polyQ fragments (see above). Furthermore, polyQ aggregates have been reported to bind to procaspase-8, a long prodomain initiator caspase, and cause its activation by oligomerization, much like physiological cell death pathways (174). In a C. elegans model of Huntington's disease, induction of neuronal dysfunction and apoptosis by a huntingtin polyQ fragment is prevented by the ced-3 mutation, and polyQ expression and aggregation are not affected by CED-3 activity (53). The ability of the ced-3 mutation to rescue neurons in the presence of polyQ aggregates implies that polyQ aggregates do not necessarily bind conserved nuclear factors or interfere with essential nuclear functions such as transcription. In Drosophila models of polyQ neurotoxicity, p35 expression does not block neuronal apoptosis but can ameliorate the degeneration of adjacent tissues (98, 229). In cultured mammalian neurons in vitro, inhibition of caspase activation by CrmA, dominant negative procaspases, or peptide caspase inhibitors can all abrogate polyQ-induced neuronal apoptosis (104, 174, 175). These data are bolstered by the in vivo resistance of mice overexpressing dominant negative procaspase-1 to the neurodegenerative effects of a pathogenic huntingtin mutation (153). Collectively, these studies reinforce the importance of caspase activity and apoptosis in the pathogenesis of polyQ neuropathies and possibly other diseases caused by abnormal protein aggregation.
Caspase inhibition may also be beneficial in Alzheimer's disease, a far more common cause of neuron loss. Caspase-3 activity enhances the production of β-amyloid, the building block of neurotoxic senile plaques (64), and inhibition of caspase-12 activity can markedly decrease β-amyloid cytotoxicity (149). Therefore, caspase inhibition may also be beneficial in limiting neuron loss and cognitive decline in Alzheimer's disease. Finally, although broad caspase inhibition can prevent neuron loss in several experimental models of neurodegeneration, chronic caspase inhibition in human patients will block homeostatic apoptosis and risk autoimmunity and cancer. Thus, in the future it will be essential to work out the biochemical pathways that connect the disease-inciting abnormality to caspase activation. Selective interference with the initiator caspases or adapter proteins that are involved in these disease pathways may be a superior therapeutic strategy. For example, caspase-12, which is not apparently required for developmental or homeostatic apoptosis, should be an attractive pharmacological target for treating β-amyloid toxicity.
Therapeutic Benefits from Activation of Caspase Activity
Viral infection.
The critical role of caspase-mediated cell death in controlling viral infection is reflected by the number of virally encoded genes that interfere with caspase activation or inhibit caspase activity (see above). Apoptosis of infected cells, either induced autonomously or instructed by CTL, can decrease viral replication or spread by limiting the pool of host cells for productive infection, but excessive apoptosis may also lead to tissue dysfunction. The rapid decline in CD4+ T cells by apoptosis in human immunodeficiency virus (HIV) infection has been viewed as a promising target for caspase inhibition (210). Peptide inhibitors of caspases were indeed able to block T-cell apoptosis induced by HIV infection; however, they also dramatically increased the rates of viral production and host cell infection (30). These results indicate that caspases have a critical role in limiting HIV infection and suggest that anti-HIV therapy should take advantage of the unique sensitivity of HIV-infected cells to undergo apoptosis in order to root out stores of HIV provirus. One approach to this problem is an engineered procaspase-3 that has altered sequences between the mature subunits to conform to the substrate specificity of HIV-1 protease (223). Introduction of the engineered procaspase specifically induced apoptosis in HIV-infected cells and was able to decrease virus-spread in vitro. Similar strategies may be helpful in eliminating virus-reservoirs in other chronic infections.
Autoimmunity.
Apoptosis plays a critical role in the deletion of unnecessary and dangerous lymphocytes. Because the antigen receptors arise from random DNA recombination, autoimmune lymphocytes that recognize self-antigens are constantly produced. During T-cell development, autoreactive T cells are deleted in the thymus by negative selection. In the periphery, undesirable lymphocytes are further eliminated in two ways (217). Cells that are not sufficiently stimulated, such as unreactive cells or those with low-affinity antigen recognition near the end of an immune response, die by passive cell death, a process that is inhibited by Bcl-2 and mostly likely mediated by the mitochondrial caspase-9 pathway. Second, following repeated antigen stimulation, activated T lymphocytes undergo cell-autonomous Fas-mediated apoptosis, a process termed activation-induced cell death. Activated T cells also eliminate autoreactive B cells by the FasL-Fas pathway. The critical role of the Fas pathway in peripheral tolerance is demonstrated in ALPS, a disease in children characterized by massive lymphadenopathy, peripheral accumulation of doubly negative CD4− CD8− T cells, and autoantibody production leading to hepatosplenomegaly, hemolytic anemia, and thrombocytopenia (200). A majority of those related to ALPS patients harbor heterozygous mutations in the Fas gene, while a small percentage carry heterozygous mutations in the caspase-10 gene (225). The Fas pathway, mediated by initiator caspase-8 and -10 in lymphocytes, appears distinct from apoptotic pathways in thymic negative selection, because negative selection is not blocked by the lpr mutation or overexpression of CrmA and FLIP (189, 218). In the most common human autoimmune diseases, such as systemic lupus erythematosus, rheumatoid arthritis, and psoriasis, primary defects in lymphocyte apoptosis have not been demonstrated. Nonetheless, selective depletion of autoreactive lymphocytes by caspase activation and apoptosis may be a valid therapeutic strategy. Activated lymphocytes are more susceptible to Fas-mediated apoptosis, possibly in part due to downregulation of FLIP. The small molecule bisindolylmaleimide VIII enhances Fas-induced apoptosis, and systemic administration of bisindolylmaleimide VIII can ameliorate symptoms in experimental models of multiple sclerosis and arthritis (267). Better understanding of the alterations in apoptotic and survival pathways during lymphocyte development, activation, and maturation into memory cells will help refine strategies and select effective targets for treating autoimmune diseases. The link between antigen receptor activation and caspase activation during negative selection in the thymus and the molecular changes responsible for long-lived memory cells remain outstanding questions in the field that should be addressed in future studies.
Cancer.
Abnormal cell survival is a hallmark of cancer cells, and not surprisingly, both oncogenes and tumor suppressor genes have been shown to regulate apoptosis. Bcl-2 was initially discovered as the gene at a translocation breakpoint that becomes overexpressed in follicular lymphoma; its overexpression has been associated with chronic lymphocytic leukemia and confers resistance to a variety of chemotherapeutic drugs (210). p53, the most frequently mutated human tumor suppressor gene, functions in critical pathways to connect DNA damage to cell cycle arrest and apoptosis (52). Decreased apoptosis gives cancer cells a selective advantage in many steps of tumorigenesis. Initially, it allows cancer cells to survive dysregulated growth signals from oncogenes, which would normally induce apoptosis. For example, overexpression of cellular or viral oncogenes, such as c-myc and adenovirus E1A, induces apoptosis, and thus a cooperative apoptotic inhibitor such as Bcl-2 or E1B is required to transform normal cells (52). As cancer cells proliferate, decreased apoptotic sensitivity allows cancer cells to survive the hypoxic environment within tumors; indeed, experimental hypoxia selects for cells that have p53 mutations (69). Finally, Bcl-2 overexpression, p53 mutation, and certain oncogenes such as Bcr-Abl (which causes chronic myelogenous leukemia) confer resistance to chemotherapy-induced apoptosis and are likely responsible for treatment failure and disease relapse (52).
The importance of caspases in limiting oncogenic transformation has been delineated in several studies. Components of the mitochondrial pathway, Apaf-1 and caspase-9, are essential for oncogene- and chemotherapy-induced apoptosis (16, 76, 110, 252). These apoptotic pathways are known to be dependent on p53, and Apaf-1−/− or Casp9−/− cells are resistant to p53-induced apoptosis, indicating that they are essential downstream components of the p53 death pathway (190). Importantly, transformed cells deficient for Apaf-1 or caspase-9 have dramatically increased tumorigenicity in nude mice, much like p53-deficient cells (190). These results demonstrate that even following cytochrome c release from mitochondria, caspase activation is required for tumor cell death and elimination of oncogenic potential; in the absence of caspase activation, tumor cells can achieve long-term survival and growth. It should be noted that the caspase-9 knockout is more effective than zVAD in preventing chemotherapy-induced apoptosis, indicating that caspase inhibitors are not universal or gold standard reagents for determining the function of caspases. Caspase-8 and FADD, which are linked to cell surface death receptors, appear to be dispensable for oncogene- and chemotherapy-induced apoptosis in primary mouse embryonic fibroblasts (219, 249); however, they are likely involved in distinct mechanisms of limiting transformation. Fas ligand can be induced by certain types of stress, such as UV light and DNA damage and, via cognate FasL-Fas interaction, autonomously induces programmed cell death (102, 164). This may be a redundant mechanism to eliminate cells that have sustained DNA damage and thus likely to become mutated. For example, UV-induced apoptosis of keratinocytes is blocked in the absence of FasL, which allows keratinocytes with p53 mutations to accumulate abnormally in the skin (84). Somatic Fas mutations are detected in a small subset of squamous cell carcinomas, indicating that the FasL-Fas system is one of several damage-sensing mechanisms in preventing skin cancer (116). Somatic Fas mutations are also detected in approximately 10% of multiple myelomas and lymphomas, implying that the Fas pathway has an important physiologic role in limiting transformation (74, 113). Whether the Fas pathway is activated due to T-cell-mediated immune surveillance or activated autonomously by DNA damage is presently unclear.
At present, a number of outstanding questions remain to be addressed in understanding the control of caspases during oncogenic transformation. p53 appears to be a common effector pathway in oncogene-and DNA damage-induced apoptosis, but the exact molecular mechanisms connecting p53 activation to caspase activity remain incompletely understood. Bax, a Bcl-2 antagonist, is a candidate transcriptional target of p53 (142), but genetic analyses indicate that additional apoptotic effectors, perhaps the PIG genes, are involved in p53-induced apoptosis (158). Myc and E1A appear to target the mitochondrial caspase pathway; exactly how they sensitize this pathway and trigger cytochrome c release is unclear (52). Whether other oncogenes that can induce apoptosis, such E2F, also activate the caspase-9 pathway or other pathways is also an open question. Finally, apoptosis is a default response after prolonged activation of cell cycle checkpoints, but the caspase pathways responsible remain uncharacterized. Delineating the pathways from oncogenes, tumor suppressor genes, and cell cycle checkpoints to caspases will be the next challenge in considering caspase targets in cancer therapies.
Gene- and cell-based therapies.
The role of caspase-based therapies may not be limited to diseases involving apoptosis and inflammation, but may become integral parts of gene- and cell-based therapies. Current efforts at gene therapy are limited by the inefficiency of gene delivery and sustained expression level, and they are also limited by the lack of precise dosage control. Apoptosis of transgene-expressing cells has been envisioned as an ultimate failsafe mechanism to terminate gene therapy (197). For example, if a patient were to develop a life-threatening allergic reaction to a foreign gene product, its expression can be quickly shut off by inducing the producer cells to undergo programmed cell death. Caspases are the ideal tools for such application because they directly activate the apoptotic machinery, the physiologic process of cell removal without inflammation. Chimeric procaspases that can be oligomerized by small-molecule dimerizers, such as those employed experimentally to illustrate the induced-proximity principle of procaspase activation (147, 244), can be incorporated into gene therapy vectors or tissue-engineering products to allow pharmacological induction of apoptosis, which in turn allows gene therapy to be terminated at will or tissue-engineered products to be dismantled without surgery. Recalling the roles of apoptosis during development, incorporation of chimeric procaspases into tissue-engineered products, e.g., artificial arteries, cartilage, or organs, may also allow exogenously controlled changes in the implant to suit therapeutic purposes. For instance, artificial arteries may be cannulated by inducing apoptosis in the center of the graft, and they can be thrombosed by inducing apoptosis in the endothelial cells that provide antithrombotic factors. Controlled apoptosis may also be a strategy to match the size of the graft to the host or alter the shape of the graft in vivo. As gene therapy and tissue engineering progress, ingenious uses of controlled apoptosis via caspase activation will likely become a fundamental building block of these new modalities.
FUTURE DIRECTIONS
Since the initial discovery of caspase activity over a decade ago, the confluence of genetics, biochemistry, and structural biology has led to rapid progress in understanding many facets of caspase function and regulation. It is now universally recognized that caspases are the key effectors of apoptosis, but many aspects of caspase biology present avenues for important discoveries. Some of the future challenges in the field include (i) unraveling new pathways of caspase activation, such as those triggered by oncogenes, disease protein aggregates, microbial infections, and lymphocyte negative selection; (ii) identifying new caspase substrates and how they contribute to the function of caspases; (iii) finding new mechanisms of caspase regulation (are there alternative mechanisms of procaspase activation and caspase cascade propagation?); (iv) developing selective small-molecule inhibitors, especially non-peptide-based inhibitors for pharmaceutical uses; and (v) defining new functions of caspases, such as in development, differentiation, antigen presentation, and others that may be separate from apoptosis. Based on the rapid progress to date, the coming years should witness many exciting discoveries about caspases and many aspects of their fascinating biology.
REFERENCES
- 1.Adams J M, Cory S. The Bcl-2 protein family: arbiters of cell survival. Science. 1998;281:1322–1326. doi: 10.1126/science.281.5381.1322. [DOI] [PubMed] [Google Scholar]
- 2.Alnemri E S, Livingston D J, Nicholson D W, Salvesen G, Thornberry N A, Wong W W, Yuan J. Human ICE/CED-3 protease nomenclature. Cell. 1996;87:171. doi: 10.1016/s0092-8674(00)81334-3. [DOI] [PubMed] [Google Scholar]
- 3.Ashkenazi A, Dixit V M. Death receptors: signaling and modulation. Science. 1998;281:1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- 4.Bergeron L, Perez G I, Macdonald G, Shi L, Sun Y, Jurisicova A, Varmuza S, Latham K E, Flaws J A, Salter J C, Hara H, Moskowitz M A, Li E, Greenberg A, Tilly J L, Yuan J. Defects in regulation of apoptosis in caspase-2 deficient mice. Genes Dev. 1998;12:1304–1314. doi: 10.1101/gad.12.9.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertin J, Mendrysa S M, LaCount D J, Gaur S, Krebs J F, Armstrong R C, Tomaselli K J, Friesen P D. Apoptotic suppression by baculovirus P35 involves cleavage by and inhibition of a virus-induced CED-3/ICE-like protease. J Virol. 1996;70:6251–6259. doi: 10.1128/jvi.70.9.6251-6259.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Birnbaum M J, Clem R J, Miller L K. An apoptosis-inhibiting gene from a nuclear polyhedrosis virus encoding a polypeptide with Cys/His sequence motifs. J Virol. 1994;68:2521–2528. doi: 10.1128/jvi.68.4.2521-2528.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanchard H, Kodandapani L, Mittl P R, Marco S D, Krebs J F, Wu J C, Tomaselli K J, Grutter M G. The three-dimensional structure of caspase-8: an initiator enzyme in apoptosis. Struct Fold Des. 1999;7:1125–1133. doi: 10.1016/s0969-2126(99)80179-8. [DOI] [PubMed] [Google Scholar]
- 8.Boldin M P, Goncharov T M, Goltsev Y V, Wallach D. Involvement of MACH, a novel MORTI/FADD interacting protease, in Fas/APO-1 and TNF receptor-induced cell death. Cell. 1996;85:803–815. doi: 10.1016/s0092-8674(00)81265-9. [DOI] [PubMed] [Google Scholar]
- 9.Boldin M P, Varfolomev E E, Pancer Z, Mett I L, Camonis J H, Wallach D. A novel protein that interacts with the death domain of Fas/APO1 contains a sequence motif related to the death domain. J Biol Chem. 1995;270:7795–7798. doi: 10.1074/jbc.270.14.7795. [DOI] [PubMed] [Google Scholar]
- 10.Borner C, Monney L. Apoptosis without caspases: an inefficient molecular guillotine? Cell Death Differ. 1999;6:497–507. doi: 10.1038/sj.cdd.4400525. [DOI] [PubMed] [Google Scholar]
- 11.Boulares A H, Yakovlev A G, Ivanova V, Stoica B A, Wang G, Iyer S, Smulson M. Role of poly(ADP-ribose) polymerase (PARP) cleavage in apoptosis: caspase 3-resistant PARP mutant increases rates of apoptosis in transfected cells. J Biol Chem. 1999;274:22932–22940. doi: 10.1074/jbc.274.33.22932. [DOI] [PubMed] [Google Scholar]
- 12.Buckley C D, Pilling D, Henriquez N V, Parsonage G, Threlfall K, Scheel-Toellner D, Simmons D L, Akbar A N, Lord J M, Salmon M. RGD peptides induce apoptosis by direct caspase-3 activation. Nature. 1999;397:534–539. doi: 10.1038/17409. [DOI] [PubMed] [Google Scholar]
- 13.Bump N J, Hackett M, Hugunin M, Seshagiri S, Brady K, Chen P, Ferenz C, Franklin S, Ghayur T, Li P, et al. Inhibition of ICE family proteases by baculovirus antiapoptotic protein p35. Science. 1995;269:1885–1888. doi: 10.1126/science.7569933. [DOI] [PubMed] [Google Scholar]
- 14.Cardone M H, Roy N, Stennicke H R, Salvesen G S, Franke T F, Stanbridge E, Frisch S, Reed J C. Regulation of cell death protease caspase-9 by phosphorylation. Science. 1998;282:1318–1321. doi: 10.1126/science.282.5392.1318. [DOI] [PubMed] [Google Scholar]
- 15.Cardone M H, Salvesen G S, Widmann C, Johnson G, Frisch S M. The regulation of anoikis: MEKK-1 activation requires cleavage by caspases. Cell. 1997;90:315–323. doi: 10.1016/s0092-8674(00)80339-6. [DOI] [PubMed] [Google Scholar]
- 16.Cecconi F, Alvarez-Bolado G, Meyer B I, Roth K A, Gruss P. Apaf1 (CED-4 homolog) regulates programmed cell death in mammalian development. Cell. 1998;94:727–737. doi: 10.1016/s0092-8674(00)81732-8. [DOI] [PubMed] [Google Scholar]
- 17.Cerretti D P, Kozlosky C J, Mosley B, Nelson N, Van Ness K, Greenstreet T A, March C J, Kronheim S R, Druck T, Cannizzaro L A, et al. Molecular cloning of the interleukin-1 beta converting enzyme. Science. 1992;256:97–100. doi: 10.1126/science.1373520. [DOI] [PubMed] [Google Scholar]
- 18.Chai J, Du C, Wu J W, Kyin S, Wang X, Shi Y. Structural and biochemical basis of apoptotic activation by Smac/DIABLO. Nature. 2000;406:855–862. doi: 10.1038/35022514. [DOI] [PubMed] [Google Scholar]
- 19.Chan F K, Chun H J, Zheng L, Siegel R M, Bui K L, Lenardo M J. A domain in TNF receptors that mediates ligand-independent receptor assembly and signaling. Science. 2000;288:2351–2354. doi: 10.1126/science.288.5475.2351. [DOI] [PubMed] [Google Scholar]
- 20.Chandler J M, Cohen G M, MacFarlane M. Different subcellular distribution of caspase-3 and caspase-7 following Fas-induced apoptosis in mouse liver. J Biol Chem. 1998;273:10815–10818. doi: 10.1074/jbc.273.18.10815. [DOI] [PubMed] [Google Scholar]
- 21.Chang H Y, Nishitoh H, Yang X, Ichijo H, Baltimore D. Activation of apoptosis signal-regulating kinase 1 (ASK1) by the adapter protein Daxx. Science. 1998;281:1860–1863. doi: 10.1126/science.281.5384.1860. [DOI] [PubMed] [Google Scholar]
- 22.Chang H Y, Yang X, Baltimore D. Dissection of Fas signaling with an altered-specificity death-domain mutant: requirement of FADD binding for apoptosis but not Jun N-terminal kinase activation. Proc Natl Acad Sci USA. 1999;96:1252–1256. doi: 10.1073/pnas.96.4.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen F, Hersh B M, Conradt B, Zhou Z, Riemer D, Gruenbaum Y, Horvitz H R. Translocation of C. elegans CED-4 to nuclear membranes during programmed cell death. Science. 2000;287:1485–1489. doi: 10.1126/science.287.5457.1485. [DOI] [PubMed] [Google Scholar]
- 24.Chen P, Nordstrom W, Gish B, Abrams J M. grim, a novel cell death gene in Drosophila. Genes Dev. 1996;10:1773–1782. doi: 10.1101/gad.10.14.1773. [DOI] [PubMed] [Google Scholar]
- 25.Chen P, Rodriguez A, Erskine R, Thach T, Abrams J M. Dredd, a novel effector of the apoptosis activators reaper, grim, and hid in Drosophila. Dev Biol. 1998;201:202–216. doi: 10.1006/dbio.1998.9000. [DOI] [PubMed] [Google Scholar]
- 26.Cheng E H, Kirsch D G, Clem R J, Ravi R, Kastan M B, Bedi A, Ueno K, Hardwick J M. Conversion of Bcl-2 to a Bax-like death effector by caspases. Science. 1997;278:1966–1968. doi: 10.1126/science.278.5345.1966. [DOI] [PubMed] [Google Scholar]
- 27.Chinnaiyan A M, Chaudhary D, O'Rourke K, Koonin E V, Dixit V M. Role of CED-4 in the activation of CED-3. Nature. 1997;388:728–729. doi: 10.1038/41913. [DOI] [PubMed] [Google Scholar]
- 28.Chinnaiyan A M, O'Rourke K, Lane B R, Dixit V M. Interaction of CED-4 with CED-3 and CED-9: a molecular framework for cell death. Science. 1997;275:1122–1126. doi: 10.1126/science.275.5303.1122. [DOI] [PubMed] [Google Scholar]
- 29.Chinnaiyan A M, O'Rourke K, Tewari M, Dixit V M. FADD, a novel death domain-containing protein, interacts with the death domain of Fas and initiates apoptosis. Cell. 1995;81:505–512. doi: 10.1016/0092-8674(95)90071-3. [DOI] [PubMed] [Google Scholar]
- 30.Chinnaiyan A M, Woffendin C, Dixit V M, Nabel G J. The inhibition of pro-apoptotic ICE-like proteases enhances HIV replication. Nat Med. 1997;3:333–337. doi: 10.1038/nm0397-333. [DOI] [PubMed] [Google Scholar]
- 31.Chou J J, Matsuo H, Duan H, Wagner G. Solution structure of the RAIDD CARD and model for CARD/CARD interaction in caspase-2 and caspase-9 recruitment. Cell. 1998;94:171–180. doi: 10.1016/s0092-8674(00)81417-8. [DOI] [PubMed] [Google Scholar]
- 32.Clem R J, Cheng E H, Karp C L, Kirsch D G, Ueno K, Takahashi A, Kastan M B, Griffin D E, Earnshaw W C, Veliuona M A, Hardwick J M. Modulation of cell death by Bcl-XL through caspase interaction. Proc Natl Acad Sci USA. 1998;95:554–559. doi: 10.1073/pnas.95.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Colussi P A, Harvey N L, Kumar S. Prodomain-dependent nuclear localization of the caspase-2 (Nedd2) precursor: a novel function for a caspase prodomain. J Biol Chem. 1998;273:24535–24542. doi: 10.1074/jbc.273.38.24535. [DOI] [PubMed] [Google Scholar]
- 34.Conradt B, Horvitz H R. The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. Cell. 1998;93:519–529. doi: 10.1016/s0092-8674(00)81182-4. [DOI] [PubMed] [Google Scholar]
- 35.Crook N E, Clem R J, Miller L K. An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. J Virol. 1993;67:2168–2174. doi: 10.1128/jvi.67.4.2168-2174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cryns V L, Bergeron L, Zhu H, Li H, Yuan J. Specific cleavage of alpha-fodrin during Fas- and tumor necrosis factor-induced apoptosis is mediated by an interleukin-1beta-converting enzyme/Ced-3 protease distinct from the poly(ADP-ribose) polymerase protease. J Biol Chem. 1996;271:31277–31282. doi: 10.1074/jbc.271.49.31277. [DOI] [PubMed] [Google Scholar]
- 37.Davidson F F, Steller H. Blocking apoptosis prevents blindness in Drosophila retinal degeneration mutants. Nature. 1998;391:587–591. doi: 10.1038/35385. [DOI] [PubMed] [Google Scholar]
- 38.De Maria R, Zeuner A, Eramo A, Domenichelli C, Bonci D, Grignani F, Srinivasula S M, Alnemri E S, Testa U, Peschle C. Negative regulation of erythropoiesis by caspase-mediated cleavage of GATA-1. Nature. 1999;401:489–493. doi: 10.1038/46809. [DOI] [PubMed] [Google Scholar]
- 39.Deveraux Q L, Reed J C. IAP family proteins—suppressors of apoptosis. Genes Dev. 1999;13:239–252. doi: 10.1101/gad.13.3.239. [DOI] [PubMed] [Google Scholar]
- 40.Deveraux Q L, Roy N, Stennicke H R, Van Arsdale T, Zhou Q, Srinivasula S M, Alnemri E S, Salvesen G S, Reed J C. IAPs block apoptotic events induced by caspase-8 and cytochrome c by direct inhibition of distinct caspases. EMBO J. 1998;17:2215–2223. doi: 10.1093/emboj/17.8.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deveraux Q L, Takahashi R, Salvesen G S, Reed J C. X-linked IAP is a direct inhibitor of cell-death proteases. Nature. 1997;388:300–304. doi: 10.1038/40901. [DOI] [PubMed] [Google Scholar]
- 42.Dinarello C A. IL-18: A TH1-inducing, proinflammatory cytokine and new member of the IL-1 family. J Allergy Clin Immunol. 1999;103:11–24. doi: 10.1016/s0091-6749(99)70518-x. [DOI] [PubMed] [Google Scholar]
- 43.Dinarello C A. Interleukin-1 beta, interleukin-18, and the interleukin-1 beta converting enzyme. Ann NY Acad Sci. 1998;856:1–11. doi: 10.1111/j.1749-6632.1998.tb08307.x. [DOI] [PubMed] [Google Scholar]
- 44.Dorstyn L, Colussi P A, Quinn L M, Richardson H, Kumar S. DRONC, an ecdysone-inducible Drosophila caspase. Proc Natl Acad Sci USA. 1999;96:4307–4312. doi: 10.1073/pnas.96.8.4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dorstyn L, Read S H, Quinn L M, Richardson H, Kumar S. DECAY, a novel Drosophila caspase related to mammalian caspase-3 and caspase-7. J Biol Chem. 1999;274:30778–30783. doi: 10.1074/jbc.274.43.30778. [DOI] [PubMed] [Google Scholar]
- 46.Du C, Fang M, Li Y, Li L, Wang X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102:33–42. doi: 10.1016/s0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- 47.Duan H, Orth K, Chinnaiyan A M, Poirier G G, Froelich C J, He W W, Dixit V M. ICE-LAP6, a novel member of the ICE/Ced-3 gene family, is activated by the cytotoxic T cell protease granzyme B. J Biol Chem. 1996;271:16720–16724. doi: 10.1074/jbc.271.28.16720. [DOI] [PubMed] [Google Scholar]
- 48.Eberstadt M, Huang B, Chen Z, Meadows R P, Ng S C, Zheng L, Lenardo M J, Fesik S W. NMR structure and mutagenesis of the FADD (Mort1) death-effector domain. Nature. 1998;392:941–945. doi: 10.1038/31972. [DOI] [PubMed] [Google Scholar]
- 49.Ellerby L M, Andrusiak R L, Wellington C L, Hackam A S, Propp S S, Wood J D, Sharp A H, Margolis R L, Ross C A, Salvesen G S, Hayden M R, Bredesen D E. Cleavage of atrophin-1 at caspase site aspartic acid 109 modulates cytotoxicity. J Biol Chem. 1999;274:8730–8736. doi: 10.1074/jbc.274.13.8730. [DOI] [PubMed] [Google Scholar]
- 50.Ellerby L M, Hackam A S, Propp S S, Ellerby H M, Rabizadeh S, Cashman N R, Trifiro M A, Pinsky L, Wellington C L, Salvesen G S, Hayden M R, Bredesen D E. Kennedy's disease: caspase cleavage of the androgen receptor is a crucial event in cytotoxicity. J Neurochem. 1999;72:185–195. doi: 10.1046/j.1471-4159.1999.0720185.x. [DOI] [PubMed] [Google Scholar]
- 51.Enari M, Sakahira H, Yokoyama H, Okawa K, Iwamatsu A, Nagata S. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature. 1998;391:43–50. doi: 10.1038/34112. [DOI] [PubMed] [Google Scholar]
- 52.Evan G, Littlewood T. A matter of life and cell death. Science. 1998;281:1317–1322. doi: 10.1126/science.281.5381.1317. [DOI] [PubMed] [Google Scholar]
- 53.Faber P W, Alter J R, MacDonald M E, Hart A C. Polyglutamine-mediated dysfunction and apoptotic death of a Caenorhabditis elegans sensory neuron. Proc Natl Acad Sci USA. 1999;96:179–184. doi: 10.1073/pnas.96.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fernandes-Alnemri T, Armstrong R C, Krebs J, Srinivasula S M, Wang L, Bullrich F, Fritz L C, Trapani J A, Tomaselli K J, Litwack G, Alnemri E S. In vitro activation of CPP32 and Mch3 by Mch4, a novel human apoptotic cysteine protease containing two FADD-like domains. Proc Natl Acad Sci USA. 1996;93:7464–7469. doi: 10.1073/pnas.93.15.7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fisher A J, Zoog W d Cruz, S J, Schneider C L, Friesen P D. Crystal structure of baculovirus P35: role of a novel reactive site loop in apoptotic caspase inhibition. EMBO J. 1999;18:2031–2039. doi: 10.1093/emboj/18.8.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fraser A G, Evan G I. Identification of a Drosophila melanogaster ICE/CED-3-related protease, drICE. EMBO J. 1997;16:2805–2813. doi: 10.1093/emboj/16.10.2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fraser A G, McCarthy N J, Evan G I. drICE is an essential caspase required for apoptotic activity in Drosophila cells. EMBO J. 1997;16:6192–6199. doi: 10.1093/emboj/16.20.6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Friedlander R M, Gagliardini V, Wang J, Brown R H, Yuan J. Protection in an ALS model by a dominant negative ICE inhibitor. Nature. 1997;388:31. doi: 10.1038/40299. [DOI] [PubMed] [Google Scholar]
- 59.Frisch S M, Francis H. Disruption of epithelial cell-matrix interactions induces apoptosis. J Cell Biol. 1994;124:619–626. doi: 10.1083/jcb.124.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fujita E, Jinbo A, Matuzaki H, Konishi H, Kikkawa U, Momoi T. Akt phosphorylation site found in human caspase-9 is absent in mouse caspase-9. Biochem Biophys Res Commun. 1999;264:550–555. doi: 10.1006/bbrc.1999.1387. [DOI] [PubMed] [Google Scholar]
- 61.Furlan R, Martino G, Galbiati F, Poliani P L, Smiroldo S, Bergami A, Desina G, Comi G, Flavell R, Su M S, Adorini L. Caspase-1 regulates the inflammatory process leading to autoimmune demyelination. J Immunol. 1999;163:2403–2409. [PubMed] [Google Scholar]
- 62.Gagliardini V, Fernandez P A, Lee R K, Drexler H C, Rotello R J, Fishman M C, Yuan J. Prevention of vertebrate neuronal death by the crmA gene. Science. 1994;263:826–828. doi: 10.1126/science.8303301. [DOI] [PubMed] [Google Scholar]
- 63.Garcia-Calvo M, Peterson E P, Leiting B, Ruel R, Nicholson D W, Thornberry N A. Inhibition of human caspases by peptide-based and macromolecular inhibitors. J Biol Chem. 1998;273:32608–32613. doi: 10.1074/jbc.273.49.32608. [DOI] [PubMed] [Google Scholar]
- 64.Gervais F G, Xu D, Robertson G S, Vaillancourt J P, Zhu Y, Huang J Q, LeBlanc A, Smith D, Rigby M, Shearman M S, Clarke E E, Zheng H, Van Der Ploug L H T, Ruffolo S C, Thornberry N A, Xanthoudakis S, Zamboni R J, Roy S, Nicholson D W. Involvement of caspases in proteolytic cleavage of Alzheimer's amyloid-b precursor protein and amyloidogenic Ab peptide formation. Cell. 1999;97:395–406. doi: 10.1016/s0092-8674(00)80748-5. [DOI] [PubMed] [Google Scholar]
- 65.Ghayur T, Banerjee S, Hugunin M, Butler D, Herzog L, Carter A, Quintal L, Sekut L, Talanian R, Paskind M, Wong W, Kamen R, Tracey D, Allen H. Caspase-1 processes IFN-gamma-inducing factor and regulates LPS-induced IFN-gamma production. Nature. 1997;386:619–623. doi: 10.1038/386619a0. [DOI] [PubMed] [Google Scholar]
- 66.Goldberg Y P, Nicholson D W, Rasper D M, Kalchman M A, Koide H B, Graham R K, Bromm M, Kazemi-Esfarjani P, Thornberry N A, Vaillancourt J P, Hayden M R. Cleavage of huntingtin by apopain, a proapoptotic cysteine protease, is modulated by the polyglutamine tract. Nat Genet. 1996;13:442–429. doi: 10.1038/ng0896-442. [DOI] [PubMed] [Google Scholar]
- 67.Goltsev Y V, Kovalenko A V, Arnold E, Varfolomeev E E, Brodianskii V M, Wallach D. CASH, a novel caspase homologue with death effector domains. J Biol Chem. 1997;272:19641–19644. doi: 10.1074/jbc.272.32.19641. [DOI] [PubMed] [Google Scholar]
- 68.Goyal L, McCall K, Agapite J, Hartwieg E, Steller H. Induction of apoptosis by Drosophila reaper, hid and grim through inhibition of IAP function. EMBO J. 2000;19:589–597. doi: 10.1093/emboj/19.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Graeber T G, Osmanian C, Jacks T, Housman D E, Koch C J, Lowe S W, Giaccia A J. Hypoxia-mediated selection of cells with diminished apoptotic potential in solid tumours. Nature. 1996;379:88–91. doi: 10.1038/379088a0. [DOI] [PubMed] [Google Scholar]
- 70.Graves J D, Gotoh Y, Draves K E, Ambrose D, Han D K, Wright M, Chernoff J, Clark E A, Krebs E G. Caspase-mediated activation and induction of apoptosis by the mammalian Ste20-like kinase Mst1. EMBO J. 1998;17:2224–2234. doi: 10.1093/emboj/17.8.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Green D R. Apoptotic pathways: paper wraps stone blunts scissors. Cell. 2000;102:1–4. doi: 10.1016/s0092-8674(00)00003-9. [DOI] [PubMed] [Google Scholar]
- 72.Green D R, Reed J C. Mitochondria and apoptosis. Science. 1998;281:1309–12. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 73.Grether M E, Abrams J M, Agapite J, White K, Steller H. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 1995;9:1694–1708. doi: 10.1101/gad.9.14.1694. [DOI] [PubMed] [Google Scholar]
- 74.Gronbaek K, Stratern P T, Ralfkiaer E, Ahrenkiel V, Andersen M K, Hansen N E, Zeuthen J, Hou-Jensen K, Guldberg P. Somatic Fas mutation in Non-Hodgkin's lymphoma: association with extranodal disease and autoimmunity. Blood. 1998;92:3018–3024. [PubMed] [Google Scholar]
- 75.Gu Y, Kuida K, Tsutsui H, Ku G, Hsiao K, Fleming M A, Hayashi N, Higashino K, Okamura H, Nakanishi K, Kurimoto M, Tanimoto T, Flavell R A, Sato V, Harding M W, Livingston D J, Su M S. Activation of interferon-gamma inducing factor mediated by interleukin-1beta converting enzyme. Science. 1997;275:206–209. doi: 10.1126/science.275.5297.206. [DOI] [PubMed] [Google Scholar]
- 76.Hakem R, Hakem A, Duncan G S, Henderson J T, Woo M, Soengas M S, Elia A, Luis de la Pompa J, Kagi D, Khoo W, Potter J, Yoshida R, Kaufman S A, Lowe S W, Penninger J M, Mak T W. Differential requirement for caspase-9 in apoptotic pathways in vivo. Cell. 1998;94:339–352. doi: 10.1016/s0092-8674(00)81477-4. [DOI] [PubMed] [Google Scholar]
- 77.Halappanavar S S, Rhun Y L, Mounir S, Martins L M, Huot J, Earnshaw W C, Shah G M. Survival and proliferation of cells expressing caspase-uncleavable poly(ADP-ribose) polymerase in response to death-inducing DNA damage by an alkylating agent. J Biol Chem. 1999;274:37097–37104. doi: 10.1074/jbc.274.52.37097. [DOI] [PubMed] [Google Scholar]
- 78.Han D, Chaudhary P M, Wright M E, Friedman C, Trask B J, Riedel R T, Baskin D G, Schwartz S M, Hood L. MRIT, a novel death-effector domain-containing protein, interacts with caspases and BclXL and initiates cell death. Proc Natl Acad Sci USA. 1997;94:11333–11338. doi: 10.1073/pnas.94.21.11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Harvey K J, Lukovic D, Ucker D S. Caspase-dependent Cdk activity is a requisite effector of apoptotic death events. J Cell Biol. 2000;148:59–72. doi: 10.1083/jcb.148.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hengartner M O, Horvitz H R. C. elegans cell survival gene ced-9 encodes a functional homolog of the mammalian proto-oncogene bcl-2. Cell. 1994;76:665–676. doi: 10.1016/0092-8674(94)90506-1. [DOI] [PubMed] [Google Scholar]
- 81.Herceg Z, Wang Z Q. Failure of poly(ADP-ribose) polymerase cleavage by caspases leads to induction of necrosis and enhanced apoptosis. Mol Cell Biol. 1999;19:5124–5133. doi: 10.1128/mcb.19.7.5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hersh D, Monack D M, Smith M R, Ghori N, Falkow S, Zychlinsky A. The Salmonella invasin SipB induces macrophage apoptosis by binding to caspase-1. Proc Natl Acad Sci USA. 1999;96:2396–2401. doi: 10.1073/pnas.96.5.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hilbi H, Moss J E, Hersh D, Chen Y, Arondel J, Banerjee S, Flavell R A, Yuan J, Sansonetti P J, Zychlinsky A. Shigella-induced apoptosis is dependent on caspase-1 which binds to IpaB. J Biol Chem. 1998;273:32895–32900. doi: 10.1074/jbc.273.49.32895. [DOI] [PubMed] [Google Scholar]
- 84.Hill L L, Ouhtit A, Loughlin S M, Kripke M L, Ananthaswamy H N, Owen-Schaub L B. Fas ligand: a sensor for DNA damage critical in skin cancer etiology. Science. 1999;285:898–900. doi: 10.1126/science.285.5429.898. [DOI] [PubMed] [Google Scholar]
- 85.Hisahara S, Araki T, Sugiyama F, Yagami K, Suzuki M, Abe K, Yamamura K, Miyazaki J, Momoi T, Saruta T, Bernard C C, Okano H, Miura M. Targeted expression of baculovirus p35 caspase inhibitor in oligodendrocytes protects mice against autoimmune-mediated demyelination. EMBO J. 2000;19:341–348. doi: 10.1093/emboj/19.3.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hofmann K, Bucher P, Tschopp J. The CARD domain: a new apoptotic signalling motif. Trends Biochem Sci. 1997;22:155–156. doi: 10.1016/s0968-0004(97)01043-8. [DOI] [PubMed] [Google Scholar]
- 87.Horvitz H R, Shaham S, Hengartner M O. The genetics of programmed cell death in the nematode Caenorhabditis elegans. Cold Spring Harbor Symp Quant Biol. 1994;59:377–385. doi: 10.1101/sqb.1994.059.01.042. [DOI] [PubMed] [Google Scholar]
- 88.Howard A D, Kostura M J, Thornberry N, Ding G J, Limjuco G, Weidner J, Salley J P, Hogquist K A, Chaplin D D, Mumford R A, et al. IL-1-converting enzyme requires aspartic acid residues for processing of the IL-1 beta precursor at two distinct sites and does not cleave 31-kDa IL-1 alpha. J Immunol. 1991;147:2964–2969. [PubMed] [Google Scholar]
- 89.Hsu H, Shu H-B, Pan M-G, Goeddel D V. TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell. 1996;84:299–308. doi: 10.1016/s0092-8674(00)80984-8. [DOI] [PubMed] [Google Scholar]
- 90.Hsu H, Xiong J, Goeddel D V. The TNF receptor 1-associated protein TRADD signals cell death and NF-κB activation. Cell. 1995;81:495–504. doi: 10.1016/0092-8674(95)90070-5. [DOI] [PubMed] [Google Scholar]
- 91.Hu S, Vincenz C, Ni J, Gentz R, Dixi V M. I-FLICE, a novel inhibitor of tumor necrosis factor receptor-1- and CD-95-induced apoptosis. J Biol Chem. 1997;272:17255–17257. doi: 10.1074/jbc.272.28.17255. [DOI] [PubMed] [Google Scholar]
- 92.Hu, S., and X. Yang. 2000. dFADD, a novel death domain-containing adapter protein for the Drosophila caspase DREDD. J. Biol. Chem., in press. [DOI] [PubMed]
- 93.Hueber A O, Zornig M, Lyon D, Suda T, Nagata S, Evan G I. Requirement for the CD95 receptor-ligand pathway in c-Myc-induced apoptosis. Science. 1997;278:1305–1309. doi: 10.1126/science.278.5341.1305. [DOI] [PubMed] [Google Scholar]
- 94.Inohara N, Koseki T, Chen S, Wu X, Nunez G. CIDE, a novel family of cell death activators with homology to the 45 kDa subunit of the DNA fragmentation factor. EMBO J. 1998;17:2526–2533. doi: 10.1093/emboj/17.9.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Inohara N, Koseki T, Hu Y, Chen S, Nunez G. CLARP, a death effector domain-containing protein interacts with caspase-8 and regulates apoptosis. Proc Natl Acad Sci USA. 1997;94:10717–10722. doi: 10.1073/pnas.94.20.10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Irmler M, Thome M, Hahne M, Schneider P, Hofmann K, Steiner V, Bodmer J L, Schroter M, Burns K, Mattmann C, Rimoldi D, French L E, Tschopp J. Inhibition of death receptor signals by cellular FLIP. Nature. 1997;388:190–195. doi: 10.1038/40657. [DOI] [PubMed] [Google Scholar]
- 97.Ishizaki Y, Jacobson M D, Raff M C. A role for caspases in lens fiber differentiation. J Cell Biol. 1998;140:153–158. doi: 10.1083/jcb.140.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jackson G R, Salecker I, Dong X, Yao X, Arnheim N, Faber P W, MacDonald M E, Zipursky S L. Polyglutamine-expanded human huntingtin transgenes induce degeneration of Drosophila photoreceptor neurons. Neuron. 1998;21:633–642. doi: 10.1016/s0896-6273(00)80573-5. [DOI] [PubMed] [Google Scholar]
- 99.Jacobson M D, Burne J F, Raff M C. Programmed cell death and Bcl-2 protection in the absence of a nucleus. EMBO J. 1994;13:1899–1910. doi: 10.1002/j.1460-2075.1994.tb06459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jurgensmeier J M, Xie Z, Deveraux Q, Ellerby L, Bredesen D, Reed J C. Bax directly induces release of cytochrome c from isolated mitochondria. Proc Natl Acad Sci USA. 1998;95:4997–5002. doi: 10.1073/pnas.95.9.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kanuka H, Sawamoto K, Inohara N, Matsuno K, Okano H, Miura M. Control of the cell death pathway by Dapaf-1, a Drosophila Apaf-1/CED-4-related caspase activator. Mol Cell. 1999;4:757–769. doi: 10.1016/s1097-2765(00)80386-x. [DOI] [PubMed] [Google Scholar]
- 102.Kasibhatla S, Brunner T, Genestier L, Echeverri F, Mahboubi A, Green D R. DNA damaging agents induce expression of Fas ligand and subsequent apoptosis in T lymphocytes via the action of NF-kB and AP-1. Mol Cell. 1998;1:543–551. doi: 10.1016/s1097-2765(00)80054-4. [DOI] [PubMed] [Google Scholar]
- 103.Kerr J F R, Wyllie A H, Currie A R. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kim M, Lee H S, LaForet G, Mclntyre C, Martin E J, Chang P, Kim T W, Williams M, Reddy P H, Tagle D, Boyce F M, Won L, Heller A, Aronin N, DiFiglia M. Mutant huntingtin expression in clonal striatal cells: dissociation of inclusion formation and neuronal survival by caspase inhibition. J Neurosci. 1999;19:964–973. doi: 10.1523/JNEUROSCI.19-03-00964.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kim T W, Pettingell W H, Jung Y K, Kovacs D M, Tanzi R E. Alternative cleavage of Alzheimer-associated presenilins during apoptosis by a caspase-3 family protease. Science. 1997;277:373–376. doi: 10.1126/science.277.5324.373. [DOI] [PubMed] [Google Scholar]
- 106.Kischkel F C, Hellbardt S H, Behrmann I, Germer M, Pawlita M, Krammer P H, Peter M E. Cytotoxicity-dependent APO-1 (Fas/CD-95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO J. 1995;14:5579–5588. doi: 10.1002/j.1460-2075.1995.tb00245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kischkel F C, Lawrence D A, Chuntharapai A, Schow P, Kim K J, Ashkenazi A. Apo2L/TRAIL-dependent recruitment of endogenous FADD and caspase-8 to death receptors 4 and 5. Immunity. 2000;12:611–620. doi: 10.1016/s1074-7613(00)80212-5. [DOI] [PubMed] [Google Scholar]
- 108.Kluck R M, Bossy-Wetzel E, Green D R, Newmeyer D D. The release of cytochrome c from mitochondria: a primary site for Bcl-2 regulation of apoptosis. Science. 1997;275:1132–1136. doi: 10.1126/science.275.5303.1132. [DOI] [PubMed] [Google Scholar]
- 109.Kothakota S, Azuma T, Reinhard C, Klippel A, Tang J, Chu K, McGarry T J, Kirschner M W, Koths K, Kwiatkowski D J, Williams L T. Caspase-3-generated fragment of gelsolin: effector of morphological change in apoptosis. Science. 1997;278:294–298. doi: 10.1126/science.278.5336.294. [DOI] [PubMed] [Google Scholar]
- 110.Kuida K, Haydar T F, Kuan C-Y, Gu Y, Taya C, Karasuyama H, Su M S-S, Rakic P, Flavell R. Reduced apoptosis and cytochrome-c mediated caspase activation in mice lacking caspase-9. Cell. 1998;94:325–337. doi: 10.1016/s0092-8674(00)81476-2. [DOI] [PubMed] [Google Scholar]
- 111.Kuida K, Lippke J A, Ku G, Harding M W, Livingston D J, Su M S, Flavell R A. Altered cytokine export and apoptosis in mice deficient in interleukin-1 beta converting enzyme. Science. 1995;267:2000–2003. doi: 10.1126/science.7535475. [DOI] [PubMed] [Google Scholar]
- 112.Kuida K, Zheng T S, Na S, Kuan C, Yang D, Karasuyama H, Rakic P, Flavell R A. Decreased apoptosis in the brain and premature lethality in CPP32-deficient mice. Nature. 1996;384:368–372. doi: 10.1038/384368a0. [DOI] [PubMed] [Google Scholar]
- 113.Landowski T H, Qu N, Buyuksal I, Painter J S, Dalton W S. Mutations in the Fas antigen in patients with multiple myeloma. Blood. 1997;90:4266–4270. [PubMed] [Google Scholar]
- 114.Lazebnik Y A, Kaufmann S H, Desnoyers S, Poirier G G, Earnshaw W C. Cleavage of poly(ADP-ribose) polymerase by a proteinase with properties like ICE. Nature. 1994;371:346–347. doi: 10.1038/371346a0. [DOI] [PubMed] [Google Scholar]
- 115.Lazebnik Y A, Takahashi A, Moir R D, Goldman R D, Poirier G G, Kaufman S H, Earnshaw W C. Studies of the lamin proteinase reveal multiple parallel biochemical pathways during apoptotic execution. Proc Natl Acad Sci USA. 1995;92:9042–9046. doi: 10.1073/pnas.92.20.9042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lee S H, Shin M S, Kim H S, Park W S, Kim S Y, Jang J J, Rhim K J, Jang J, Lee H K, Park J Y, Oh R R, Han S Y, Lee J H, Lee J Y, Yoo N J. Somatic mutations of fas (Apo-1/CD95) gene in cutaneous squamous cell carcinoma arising from a burn scar. J Invest Dermatol. 2000;114:122–126. doi: 10.1046/j.1523-1747.2000.00819.x. [DOI] [PubMed] [Google Scholar]
- 117.Lee S Y, Reichlin A, Santana A, Sokol K A, Nussenzweig M C, Choi Y. TRAF2 is essential for JNK but not NF-kappaB activation and regulates lymphocyte proliferation and survival. Immunity. 1997;7:703–713. doi: 10.1016/s1074-7613(00)80390-8. [DOI] [PubMed] [Google Scholar]
- 118.Leist M, Single B, Castoldi A F, Kuhnle S, Nicotera P. Intracellular adenosine triphosphate (ATP) concentration: a switch in the decision between apoptosis and necrosis. J Exp Med. 1997;185:1481–1486. doi: 10.1084/jem.185.8.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Levkau B, Koyama H, Raines E W, Clurman B E, Herren B, Orth K, Roberts J M, Ross R. Cleavage of p21Cip1/Waf1 and p27Kip1 mediates apoptosis in endothelial cells through activation of Cdk2: role of a caspase cascade. Mol Cell. 1998;1:553–563. doi: 10.1016/s1097-2765(00)80055-6. [DOI] [PubMed] [Google Scholar]
- 120.Li F, Ackermann E J, Bennett C F, Rothermel A L, Plescia J, Tognin S, Villa A, Marchisio P C, Altieri D C. Pleiotropic cell-division defects and apoptosis induced by interference with survivin function. Nat Cell Biol. 1999;1:461–466. doi: 10.1038/70242. [DOI] [PubMed] [Google Scholar]
- 121.Li F, Ambrosini G, Chu E Y, Plescia J, Tognin S, Marchisio P C, Altieri D C. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 1998;396:580–584. doi: 10.1038/25141. [DOI] [PubMed] [Google Scholar]
- 122.Li H, Zhu H, Xu C, Yuan J. Cleavage of BID by caspase-8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell. 1998;94:491–501. doi: 10.1016/s0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- 123.Li P, Allen H, Banerjee S, Franklin S, Herzog L, Johnston C, McDowell J, Paskind M, Rodman L, Salfeld J, et al. Mice deficient in IL-1 beta-converting enzyme are defective in production of mature IL-1 beta and resistant to endotoxic shock. Cell. 1995;80:401–411. doi: 10.1016/0092-8674(95)90490-5. [DOI] [PubMed] [Google Scholar]
- 124.Li P, Nijhawan D, Budihardjo I, Srinivasula S M, Ahmad M, Alnemri E S, Wang X. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 125.Liston P, Roy N, Tamai K, Lefebvre C, Baird S, Cherton-Horvat G, Farahani R, McLean M, Ikeda J E, MacKenzie A, Korneluk R G. Suppression of apoptosis in mammalian cells by NIAP and a related family of IAPs. Nature. 1996;379:349–353. doi: 10.1038/379349a0. [DOI] [PubMed] [Google Scholar]
- 126.Liu X, Kim C N, Yang J, Jemmerson R, Wang X. Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell. 1996;86:147–157. doi: 10.1016/s0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- 127.Liu X, Li P, Widlak P, Zou H, Luo X, Garrard W T, Wang X. The 40-kDa subunit of DNA fragmentation factor induces DNA fragmentation and chromatin condensation during apoptosis. Proc Natl Acad Sci USA. 1998;95:8461–8466. doi: 10.1073/pnas.95.15.8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Liu X, Zou H, Slaughter C, Wang X. DFF, a heterodimeric protein that functions downstream of caspase-3 to trigger DNA fragmentation and apoptosis. Cell. 1997;89:175–184. doi: 10.1016/s0092-8674(00)80197-x. [DOI] [PubMed] [Google Scholar]
- 129.Lugovskoy A A, Zhou P, Chou J J, McCarty J S, Li P, Wagner G. Solution structure of the CIDE-N domain of CIDE-B and a model for CIDE-N/CIDE-N interactions in the DNA fragmentation pathway of apoptosis. Cell. 1999;99:747–755. doi: 10.1016/s0092-8674(00)81672-4. [DOI] [PubMed] [Google Scholar]
- 130.Luo X, Budihardjo I, Zou H, Slaughter C, Wang X. BID, a Bcl-2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell. 1998;94:481–499. doi: 10.1016/s0092-8674(00)81589-5. [DOI] [PubMed] [Google Scholar]
- 131.Mannick J B, Hausladen A, Liu L, Hess D T, Zeng M, Miao Q X, Kane L S, Gow A J, Stamler J S. Fas-induced caspase denitrosylation. Science. 1999;284:651–654. doi: 10.1126/science.284.5414.651. [DOI] [PubMed] [Google Scholar]
- 132.Mao P L, Jiang Y, Wee B Y, Porter A G. Activation of caspase-1 in the nucleus requires nuclear translocation of pro-caspase-1 mediated by its prodomain. J Biol Chem. 1998;273:23621–23624. doi: 10.1074/jbc.273.37.23621. [DOI] [PubMed] [Google Scholar]
- 133.Martin D A, Siegel R M, Zheng L, Lenardo M J. Membrane oligomerization and cleavage activates the caspase-8 (FLICE/MACHalpha1) death signal. J Biol Chem. 1998;273:4345–4349. doi: 10.1074/jbc.273.8.4345. [DOI] [PubMed] [Google Scholar]
- 134.Martin J B. Molecular basis of the neurodegenerative disorders. N Engl J Med. 1999;340:1970–1980. doi: 10.1056/NEJM199906243402507. [DOI] [PubMed] [Google Scholar]
- 135.Martin S J, Amarante-Mendes G P, Shi L, Chuang T H, Casiano C A, O'Brien G A, Fitzgerald P, Tan E M, Bokoch G M, Greenberg A H, Green D R. The cytotoxic cell protease granzyme B initiates apoptosis in a cell-free system by proteolytic processing and activation of the ICE/CED-3 family protease, CPP32, via a novel two-step mechanism. EMBO J. 1996;15:2407–2416. [PMC free article] [PubMed] [Google Scholar]
- 136.Martin S J, Finucane D M, Amarante-Mendes G P, O'Brien G A, Green D R. Phosphatidylserine externalization during CD95-induced apoptosis of cells and cytoplasts requires ICE/CED-3 protease activity. J Biol Chem. 1996;271:28753–28756. doi: 10.1074/jbc.271.46.28753. [DOI] [PubMed] [Google Scholar]
- 137.Marzo I, Brenner C, Zamzami N, Susin S A, Beutner G, Brdiczka D, Remy R, Xie Z H, Reed J C, Kroemer G. The permeability transition pore complex: a target for apoptosis regulation by caspases and bcl-2-related proteins. J Exp Med. 1998;187:1261–1271. doi: 10.1084/jem.187.8.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Maundrell K, Antonsson B, Magnenat E, Camps M, Muda M, Chabert C, Gillieron C, Boschert U, Vial-Knecht E, Martinou J C, Arkinstall S. Bcl-2 undergoes phosphorylation by c-Jun N-terminal kinase/stress-activated protein kinases in the presence of the constitutively active GTP-binding protein Racl. J Biol Chem. 1997;272:25238–25242. doi: 10.1074/jbc.272.40.25238. [DOI] [PubMed] [Google Scholar]
- 139.McCall K, Steller H. Requirement for DCP-1 caspase during Drosophila oogenesis. Science. 1998;279:230–234. doi: 10.1126/science.279.5348.230. [DOI] [PubMed] [Google Scholar]
- 140.Metzstein M M, Stanfield G M, Horvitz H R. Genetics of programmed cell death in C. elegans: past, present and future. Trends Genet. 1998;14:410–416. doi: 10.1016/s0168-9525(98)01573-x. [DOI] [PubMed] [Google Scholar]
- 141.Minn A J, Velez P, Schendel S L, Liang H, Muchmore S W, Fesik S W, Fill M, Thompson C B. Bcl-x(L) forms an ion channel in synthetic lipid membranes. Nature. 1997;385:353–357. doi: 10.1038/385353a0. [DOI] [PubMed] [Google Scholar]
- 142.Miyashita T, Reed J C. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 143.Moriishi K, Huang D C, Cory S, Adams J M. Bcl-2 family members do not inhibit apoptosis by binding the caspase activator Apaf-1. Proc Natl Acad Sci USA. 1999;96:9683–9688. doi: 10.1073/pnas.96.17.9683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Muchmore S W, Sattler M, Liang H, Meadows R P, Harlan J E, Yoon H S, Nettesheim D, Chang B S, Thompson C B, Wong S L, Ng S L, Fesik S W. X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death. Nature. 1996;381:335–341. doi: 10.1038/381335a0. [DOI] [PubMed] [Google Scholar]
- 145.Muzio M, Chinnaiyan A M, Kischkel F C, O'Rourke K, Shevchenko A, Ni J, Scaffidi C, Bretz J D, Zhang M, Gentz R, Mann M, Krammer P H, Peter M E, Dixit V M. FLICE, a novel FADD-homologous ICE/CED-3 like protease, is recruited to the CD95 (Fas/Apo1) death inducing signal complex. Cell. 1996;85:817–827. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- 146.Muzio M, Salvesen G S, Dixit V M. FLICE induced apoptosis in a cell-free system. Cleavage of caspase zymogens. J Biol Chem. 1997;272:2952–2956. doi: 10.1074/jbc.272.5.2952. [DOI] [PubMed] [Google Scholar]
- 147.Muzio M, Stockwell B R, Stennicke H R, Salvesen G S, Dixit V M. An induced proximity model of caspase-8 activation. J Biol Chem. 1998;273:2926–2930. doi: 10.1074/jbc.273.5.2926. [DOI] [PubMed] [Google Scholar]
- 148.Nagata S. Apoptosis by death factor. Cell. 1997;88:355–365. doi: 10.1016/s0092-8674(00)81874-7. [DOI] [PubMed] [Google Scholar]
- 149.Nakagawa T, Zhu H, Morishima N, Li E, Xu J, Yankner B A, Yuan J. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-beta. Nature. 2000;403:98–103. doi: 10.1038/47513. [DOI] [PubMed] [Google Scholar]
- 150.Narita M, Shimizu S, Ito T, Chittenden T, Lutz R J, Matsuda H, Tsujimoto Y. Bax interacts with the permeability transition pore to induce permeability transition and cytochrome c release in isolated mitochondria. Proc Natl Acad Sci USA. 1998;95:14681–14686. doi: 10.1073/pnas.95.25.14681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Nicholson D W, Ali A, Thornberry N A, Vaillancort J P, Ding C K, Gallant M, Gareau Y, Griffin P R, Labelle M, Lazebnik Y A, Munday N A, Raju S M, Smulson M E, Yamin T-T, Yu V, Miller D K. Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature. 1995;376:37–43. doi: 10.1038/376037a0. [DOI] [PubMed] [Google Scholar]
- 152.Oliver F J, de la Rubia G, Rolli V, Ruiz-Ruiz M C, de Murcia G, Murcia J M. Importance of poly(ADP-ribose) polymerase and its cleavage in apoptosis: lesson from an uncleavable mutant. J Biol Chem. 1998;273:33533–33539. doi: 10.1074/jbc.273.50.33533. [DOI] [PubMed] [Google Scholar]
- 153.Ona V O, Li M, Vonsattel J P, Andrews L J, Khan S Q, Chung W M, Frey A S, Menon A S, Li X J, Stieg P E, Yuan J, Penney J B, Young A B, Cha J H, M F R. Inhibition of caspase-1 slows disease progression in a mouse model of Huntington's disease. Nature. 1999;399:263–267. doi: 10.1038/20446. [DOI] [PubMed] [Google Scholar]
- 154.Orth K, O'Rourke K, Salvesen G S, Dixit V M. Molecular ordering of apoptotic mammalian CED-3/ICE-like proteases. J Biol Chem. 1996;271:20977–20980. doi: 10.1074/jbc.271.35.20977. [DOI] [PubMed] [Google Scholar]
- 155.Pan G, Ni J, Wei Y F, Yu G, Gentz R, Dixit V M. An antagonist decoy receptor and a death domain-containing receptor for TRAIL. Science. 1997;277:815–818. doi: 10.1126/science.277.5327.815. [DOI] [PubMed] [Google Scholar]
- 156.Perez D, White E. TNF-alpha signals apoptosis through a bid-dependent conformational change in Bax that is inhibited by E1B 19K. Mol Cell. 2000;6:53–63. [PubMed] [Google Scholar]
- 157.Perry D K, Smyth M J, Stennicke H R, Salvesen G S, Duriez P, Poirier G G, Hannun Y A. Zinc is a potent inhibitor of the apoptotic protease, caspase-3. A novel target for zinc in the inhibition of apoptosis. J Biol Chem. 1997;272:18530–18533. doi: 10.1074/jbc.272.30.18530. [DOI] [PubMed] [Google Scholar]
- 158.Polyak K, Xia Y, Zweier J L, Kinzler K W, Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 159.Qin H, Srinivasula S M, Wu G, Fernandes-Alnemri T, Alnemri E S, Shi Y. Structural basis of procaspase-9 recruitment by the apoptotic protease-activating factor 1. Nature. 1999;399:549–557. doi: 10.1038/21124. [DOI] [PubMed] [Google Scholar]
- 160.Quan L T, Tewari M, O'Rourke K, Dixit V, Snipas S J, Poirier G G, Ray C, Pickup D J, Salvesen G S. Proteolytic activation of the cell death protease Yama/CPP32 by granzyme B. Proc Natl Acad Sci USA. 1996;93:1972–1976. doi: 10.1073/pnas.93.5.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ramage P, Cheneval D, Chvei M, Graff P, Hemmig R, Heng R, Kocher H P, Mackenzie A, Memmert K, Revesz L, et al. Expression, refolding, and autocatalytic proteolytic processing of the interleukin-1 beta-converting enzyme precursor. J Biol Chem. 1995;270:9378–9383. doi: 10.1074/jbc.270.16.9378. [DOI] [PubMed] [Google Scholar]
- 162.Rao L, Perez D, White E. Lamin proteolysis facilitates nuclear events during apoptosis. J Cell Biol. 1996;135:1441–1455. doi: 10.1083/jcb.135.6.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ray C A, Black R A, Kronheim S R, Greenstreet T S, Sleath P R, Salvessen G S, Pickup D J. Viral inhibition of inflammation: cowpox virus encodes an inhibitor of the interleukin 1b converting enzyme. Cell. 1992;69:587–604. doi: 10.1016/0092-8674(92)90223-y. [DOI] [PubMed] [Google Scholar]
- 164.Reap E A, Roof K, Maynor K, Borrero M, Booker J, Cohen P L. Radiation and stress-induced apoptosis: a role for Fas/Fas ligand interactions. Proc Natl Acad Sci USA. 1997;94:5750–5755. doi: 10.1073/pnas.94.11.5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Reed J C. Cytochrome c: can't live with it—can't live without it. Cell. 1997;91:559–562. doi: 10.1016/s0092-8674(00)80442-0. [DOI] [PubMed] [Google Scholar]
- 166.Rodriguez A, Oliver H, Zou H, Chen P, Wang X, Abrams J M. Dark is a Drosophila homologue of Apaf-1/CED-4 and functions in an evolutionarily conserved death pathway. Nat Cell Biol. 1999;1:272–279. doi: 10.1038/12984. [DOI] [PubMed] [Google Scholar]
- 167.Rodriguez J, Lazebnik Y. Caspase-9 and APAF-1 form an active holoenzyme. Genes Dev. 1999;13:3179–3184. doi: 10.1101/gad.13.24.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Rosse T, Olivier R, Monney L, Rager M, Conus S, Fellay L, Jansen B, Borner C. Bcl-2 prolongs cell survival after Bax-induced release of cytochrome c. Nature. 1998;391:496–499. doi: 10.1038/35160. [DOI] [PubMed] [Google Scholar]
- 169.Rotonda J, Nicholson D W, Fazil K M, Gallant M, Gareau Y, Labelle M, Peterson E P, Rasper D M, Ruel R, Vaillancourt J P, Thornberry N A, Becker J W. The three-dimensional structure of apopain/CPP32, a key mediator of apoptosis. Nat Struct Biol. 1996;3:619–625. doi: 10.1038/nsb0796-619. [DOI] [PubMed] [Google Scholar]
- 170.Roy N, Deveraux Q L, Takahashi R, Salvesen G S, Reed J C. The c-IAP-1 and c-IAP-2 proteins are direct inhibitors of specific caspases. EMBO J. 1997;16:6914–6925. doi: 10.1093/emboj/16.23.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Rudel T, Bokoch G M. Membrane and morphological changes in apoptotic cells regulated by caspase-mediated activation of PAK2. Science. 1997;276:1571–1574. doi: 10.1126/science.276.5318.1571. [DOI] [PubMed] [Google Scholar]
- 172.Sahara S, Aoto M, Eguchi Y, Imamoto N, Yoneda Y, Tsujimoto Y. Acinus is a caspase-3-activated protein required for apoptotic chromatin condensation. Nature. 1999;401:168–173. doi: 10.1038/43678. [DOI] [PubMed] [Google Scholar]
- 173.Sakahira H, Enari M, Nagata S. Cleavage of CAD inhibitor in CAD activation and DNA degradation during apoptosis. Nature. 1998;391:96–99. doi: 10.1038/34214. [DOI] [PubMed] [Google Scholar]
- 174.Sanchez I, Xu C-J, Juo P, Kakizaka A, Blenis J, Yuan J. Caspase-8 is required for cell death induced by expanded polyglutamine repeats. Neuron. 1998;22:623–633. doi: 10.1016/s0896-6273(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 175.Saudou F, Finkbeiner S, Devys D, Greenberg M E. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- 176.Scaffidi C, Schmitz I, Krammer P H, Peter M E. The role of c-FLIP in modulation of CD95-induced apoptosis. J Biol Chem. 1999;274:1541–1548. doi: 10.1074/jbc.274.3.1541. [DOI] [PubMed] [Google Scholar]
- 177.Schendel S L, Azimov R, Pawlowski K, Godzik A, Kagan B L, Reed J C. Ion channel activity of the BH3 only Bcl-2 family member, BID. J Biol Chem. 1999;274:21932–21936. doi: 10.1074/jbc.274.31.21932. [DOI] [PubMed] [Google Scholar]
- 178.Seshagiri S, Miller L K. Baculovirus inhibitors of apoptosis (IAPs) block activation of Sf-caspase-1. Proc Natl Acad Sci USA. 1997;94:13606–13611. doi: 10.1073/pnas.94.25.13606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Seshagiri S, Miller L K. Caenorhabditis elegans CED-4 stimulates CED-3 processing and CED-3-induced apoptosis. Curr Biol. 1997;7:455–460. doi: 10.1016/s0960-9822(06)00216-8. [DOI] [PubMed] [Google Scholar]
- 180.Shaham S. Identification of multiple Caenorhabditis elegans caspases and their potential roles in proteolytic cascades. J Biol Chem. 1998;273:35109–35117. doi: 10.1074/jbc.273.52.35109. [DOI] [PubMed] [Google Scholar]
- 181.Sheridan J P, Marsters S A, Pitti R M, Gurney A, Skubatch M, Baldwin D, Ramakrishnan L, Gray C L, Baker K, Wood W I, Goddard A D, Godowski P, Ashkenazi A. Control of TRAIL-induced apoptosis by a family of signaling and decoy receptors. Science. 1997;277:818–821. doi: 10.1126/science.277.5327.818. [DOI] [PubMed] [Google Scholar]
- 182.Shi L, Nishioka W K, Th'ng J, Bradbury E M, Litchfield D W, Greenberg A H. Premature p34cdc2 activation required for apoptosis. Science. 1994;263:1143–1145. doi: 10.1126/science.8108732. [DOI] [PubMed] [Google Scholar]
- 183.Shimizu S, Narita M, Tsujimoto Y. Bcl-2 family proteins regulate the release of apoptogenic cytochrome c by the mitochondrial channel VDAC. Nature. 1999;399:483–487. doi: 10.1038/20959. [DOI] [PubMed] [Google Scholar]
- 184.Shimizu S, Tsujimoto Y. Proapoptotic BH3-only Bcl-2 family members induce cytochrome c release, but not mitochondrial membrane potential loss, and do not directly modulate voltage-dependent anion channel activity. Proc Natl Acad Sci USA. 2000;97:577–582. doi: 10.1073/pnas.97.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Shu H B, Halpin D R, Goeddel D V. Casper is a FADD- and caspase-related inducer of apoptosis. Immunity. 1997;6:751–763. doi: 10.1016/s1074-7613(00)80450-1. [DOI] [PubMed] [Google Scholar]
- 186.Siegel R M, Frederiksen J K, Zacharias D A, Chan F K, Johnson M, Lynch D, Tsien R Y, Lenardo M J. Fas preassociation required for apoptosis signaling and dominant inhibition by pathogenic mutations. Science. 2000;288:2354–2357. doi: 10.1126/science.288.5475.2354. [DOI] [PubMed] [Google Scholar]
- 187.Sleath P R, Hendrickson R C, Kronheim S R, March C J, Black R A. Substrate specificity of the protease that processes human interleukin-1 beta. J Biol Chem. 1990;265:14526–14528. [PubMed] [Google Scholar]
- 188.Smith C A, Farrah T, Goodwin R G. The TNF receptor superfamily of cellular and viral proteins: activation, costimulation, and death. Cell. 1994;76:959–962. doi: 10.1016/0092-8674(94)90372-7. [DOI] [PubMed] [Google Scholar]
- 189.Smith K G C, Strasser A, Vaux D L. CrmA expression in T lymphocytes of transgenic mice inhibits CD95 (Fas/APO-1) transduced apoptosis, but does not cause lymphadenopathy or autoimmune disease. EMBO J. 1996;15:51647–5176. [PMC free article] [PubMed] [Google Scholar]
- 190.Soengas M S, Alarcon R M, Yoshida H, Giaccia A J, Hakem R, Mak T W, Lowe S W. Apaf-1 and caspase-9 in p53-dependent apoptosis and tumor inhibition. Science. 1999;284:156–159. doi: 10.1126/science.284.5411.156. [DOI] [PubMed] [Google Scholar]
- 191.Song Z, McCall K, Steller H. DCP-1, a Drosophila cell death protease essential for development. Science. 1997;275:536–540. doi: 10.1126/science.275.5299.536. [DOI] [PubMed] [Google Scholar]
- 192.Spector M S, Desnoyers S, Hoeppner D J, Hengartner M O. Interactions between the C. elegans cell death regulators CED-9 and CED-4. Nature. 1997;385:653–656. doi: 10.1038/385653a0. [DOI] [PubMed] [Google Scholar]
- 193.Srinivasula S M, Ahmad M, Fernandes-Alnemri T, Alnemri E S. Autoactivation of procaspase-9 by Apaf-1-mediated oligomerization. Mol Cell. 1998;1:949–957. doi: 10.1016/s1097-2765(00)80095-7. [DOI] [PubMed] [Google Scholar]
- 194.Srinivasula S M, Ahmad M, MacFarlane M, Luo Z, Huang Z, Fernandes-Alnemri T, Alnemri E S. Generation of constitutively active recombinant caspases-3 and -6 by rearrangement of their subunits. J Biol Chem. 1998;273:10107–10111. doi: 10.1074/jbc.273.17.10107. [DOI] [PubMed] [Google Scholar]
- 195.Srinivasula S M, Ahmad M, Ottilie S, Bullrich F, Banks S, Wang Y, Fernandes-Alnemri T, Croce C M, Litwack G, Tomaselli K J, Armstrong R C, Alnemri E S. FLAME-1, a novel FADD-like anti-apoptotic molecule that regulates Fas/TNFR1-induced apoptosis. J Biol Chem. 1997;272:18542–18545. doi: 10.1074/jbc.272.30.18542. [DOI] [PubMed] [Google Scholar]
- 196.Stanger B Z, Leder P, Lee T H, Kim E, Seed B. RIP: a novel protein containing a death domain that interacts with Fas/APO-1 (CD95) in yeast and causes cell death. Cell. 1995;81:513–523. doi: 10.1016/0092-8674(95)90072-1. [DOI] [PubMed] [Google Scholar]
- 197.Steller H. Artificial death switches: induction of apoptosis by chemically induced caspase multimerization. Proc Natl Acad Sci USA. 1998;95:5421–5422. doi: 10.1073/pnas.95.10.5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Stennicke H R, Deveraux Q L, Humke E W, Reed J C, Dixit V M, Salvesen G S. Caspase-9 can be activated without proteolytic processing. J Biol Chem. 1999;274:8359–8362. doi: 10.1074/jbc.274.13.8359. [DOI] [PubMed] [Google Scholar]
- 199.Stennicke H R, Jurgensmeier J M, Shin H, Deveraux Q, Wolf B B, Yang X, Zhou Q, Ellerby H M, Ellerby L M, Bredesen D, Green D R, Reed J C, Froelich C J, Salvesen G S. Pro-caspase-3 is a major physiologic target of caspase-8. J Biol Chem. 1998;273:27084–27090. doi: 10.1074/jbc.273.42.27084. [DOI] [PubMed] [Google Scholar]
- 200.Straus S E, Sneller M, Lenardo M J, Puck J M, Strober W. An inherited disorder of lymphocyte apoptosis: the autoimmune lymphoproliferative syndrome. Ann Intern Med. 1999;130:591–601. doi: 10.7326/0003-4819-130-7-199904060-00020. [DOI] [PubMed] [Google Scholar]
- 201.Stroh C, Schulze-Osthoff K. Death by a thousand cuts: an ever increasing list of caspase substrates. Cell Death Differ. 1998;5:997–1000. doi: 10.1038/sj.cdd.4400451. [DOI] [PubMed] [Google Scholar]
- 202.Susin S A, Lorenzo H K, Zamzami N, Marzo I, Brenner C, Larochette N, Prevost M C, Alzari P M, Kroemer G. Mitochondrial release of caspase-2 and -9 during the apoptotic process. J Exp Med. 1999;189:381–394. doi: 10.1084/jem.189.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Susin S A, Lorenzo H K, Zamzami N, Marzo I, Snow B E, Brothers G M, Mangion J, Jacotot E, Costantini P, Loeffler M, Larochette N, Goodlett D R, Aebersold R, Siderovski D P, Penninger J M, Kroemer G. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature. 1999;397:441–446. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]
- 204.Takahashi A, Alnemri E S, Lazebnik Y A, Fernandes-Alnemri T, Litwack G, Moir R D, Goldman R D, Poirier G G, Kaufmann S H, Earnshaw W C. Cleavage of lamin A by Mch2 alpha but not CPP32: multiple interleukin 1 beta-converting enzyme-related proteases with distinct substrate recognition properties are active in apoptosis. Proc Natl Acad Sci USA. 1996;93:8395–8400. doi: 10.1073/pnas.93.16.8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Talanian R V, Quinlan C, Trautz S, Hackett M C, Mankovich J A, Banach D, Ghayur T, Brady K D, Wong W W. Substrate specificities of caspase family proteases. J Biol Chem. 1997;272:9677–9682. doi: 10.1074/jbc.272.15.9677. [DOI] [PubMed] [Google Scholar]
- 206.Talanian R V, Yang X, Turbov J, Seth P, Ghayur T, Casiano C A, Orth K, Froelich C J. Granule-mediated killing: pathways for granzyme B-initiated apoptosis. J Exp Med. 1997;186:1323–1331. doi: 10.1084/jem.186.8.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Tewari M, Dixit V M. Fas- and tumor necrosis factor-induced apoptosis is inhibited by the poxvirus crmA gene product. J Biol Chem. 1995;270:3255–3260. doi: 10.1074/jbc.270.7.3255. [DOI] [PubMed] [Google Scholar]
- 208.Tewari M, Quan L T, O'Rourke K, Desnoyers S, Zeng Z, Beidler D R, Dixit V M. Yama/CPP32b, a mammalian homolog of CED-3, is a crmA-inhibitable protease that cleaves the death substrate poly(ADP-ribose) polymerase. Cell. 1995;81:801–809. doi: 10.1016/0092-8674(95)90541-3. [DOI] [PubMed] [Google Scholar]
- 209.Thome M, Schneider P, Hofmann K, Fickenscher H, Meinl E, Neipel F, Mattmann C, Burns K, Bodmer J L, Schroter M, Scaffidi C, Krammer P H, Peter M E, Tschopp J. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature. 1997;386:517–521. doi: 10.1038/386517a0. [DOI] [PubMed] [Google Scholar]
- 210.Thompson C B. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267:1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- 211.Thornberry N A, Bull H G, Calaycay J R, Chapman K T, Howard A D, Kostura M J, Miller D K, Molineaux S M, Weidner J R, Aunins J, et al. A novel heterodimeric cysteine protease is required for interleukin-1 beta processing in monocytes. Nature. 1992;356:768–774. doi: 10.1038/356768a0. [DOI] [PubMed] [Google Scholar]
- 212.Thornberry N A, Rano T A, Peterson E P, Rasper D M, Timkey T, Garcia-Calvo M, Houtzager V M, Nordstrom P A, Roy S, Vaillancourt J P, Chapman K T, Nicholson D W. A combinatorial approach defines specificities of members of the caspase family and granzyme B: functional relationships established for key mediators of apoptosis. J Biol Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 213.Ting A T, Pimentel-Muinos F X, Seed B. RIP mediates tumor necrosis factor receptor 1 activation of NF-kappaB but not Fas/APO-1-initiated apoptosis. EMBO J. 1996;15:6189–6196. [PMC free article] [PubMed] [Google Scholar]
- 214.van Den Berg W B, Joosten L A, Kollias G, van De Loo F A. Role of tumour necrosis factor alpha in experimental arthritis: separate activity of interleukin 1beta in chronicity and cartilage destruction. Ann Rheum Dis. 1999;58(Suppl.):140–148. doi: 10.1136/ard.58.2008.i40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Vander Heiden M G, Chandel N S, Schumacker P T, Thompson C B. Bcl-xL prevents cell death following growth factor withdrawal by facilitating mitochondrial ATP/ADP exchange. Mol Cell. 1999;3:159–167. doi: 10.1016/s1097-2765(00)80307-x. [DOI] [PubMed] [Google Scholar]
- 216.Vander Heiden M G, Chandel N S, Williamson E K, Schumacker P T, Thompson C B. Bcl-xL regulates the membrane potential and volume homeostasis of mitochondria. Cell. 1997;91:627–637. doi: 10.1016/s0092-8674(00)80450-x. [DOI] [PubMed] [Google Scholar]
- 217.Van Parijs L, Abbas A K. Homeostasis and self-tolerance in the immune system: turning lymphocytes off. Science. 1998;280:243–248. doi: 10.1126/science.280.5361.243. [DOI] [PubMed] [Google Scholar]
- 218.Van Parijs L, Refaeli Y, Abbas A K, Baltimore D. Autoimmunity as a consequence of retrovirus-mediated expression of C-FLIP in lymphocytes. Immunity. 1999;11:763–770. doi: 10.1016/s1074-7613(00)80150-8. [DOI] [PubMed] [Google Scholar]
- 219.Varfolomeev E E, Schuchmann M, Luria V, Chiannilkulchai N, Beckmann J S, Mett I L, Rebrikov D, Brodianski V M, Kemper O C, Kollet O, Lapidot T, Soffer D, Sobe T, Avraham K B, Goncharov T, Holtmann H, Lonai P, Wallach D. Targeted disruption of the mouse caspase 8 gene ablates cell death induction by the TNF receptors, Fas/Apol, and DR3 and is lethal prenatally. Immunity. 1998;9:267–276. doi: 10.1016/s1074-7613(00)80609-3. [DOI] [PubMed] [Google Scholar]
- 220.Vaux D L. Caspases and apoptosis-biology and terminology. Cell Death Differ. 1999;6:493–494. doi: 10.1038/sj.cdd.4400523. [DOI] [PubMed] [Google Scholar]
- 221.Verhagen A M, Ekert P G, Pakusch M, Silke J, Connolly L M, Reid G E, Moritz R L, Simpson R J, Vaux D L. Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell. 2000;102:43–53. doi: 10.1016/s0092-8674(00)00009-x. [DOI] [PubMed] [Google Scholar]
- 222.Verheij M, Bose R, Lin X H, Yao B, Jarvis W D, Grant S, Birrer M J, Haimovitz-Friedman A, Fuks Z, Kolesnick R N. Requirement for ceramide-initiated SAPK/JNK signalling in stress-induced apoptosis. Nature. 1996;380:75–79. doi: 10.1038/380075a0. [DOI] [PubMed] [Google Scholar]
- 223.Vocero-Akbani A M, Heyden N V, Lissy N A, Ratner L, Dowdy S F. Killing HIV-infected cells by transduction with an HIV protease-activated caspase-3 protein. Nat Med. 1999;5:29–33. doi: 10.1038/4710. [DOI] [PubMed] [Google Scholar]
- 224.Walker N P, Talanian R V, Brady K D, Dang L C, Bump N J, Ferenz C R, Franklin S, Ghayur T, Hackett M C, Hammill L D, Herzog L, Hugunin M, Houy W, Mankovich J A, McGuiness L, Orlewicz E, Paskind M, Pratt C A, Reis P, Summani A, Terranova M, Welch J P, Xiong L, Moller A, Tracey D E, Kamen R, Wong W W. Crystal structure of the cysteine protease interleukin-1 beta-converting enzyme: a (p20/p10)2 homodimer. Cell. 1994;78:343–352. doi: 10.1016/0092-8674(94)90303-4. [DOI] [PubMed] [Google Scholar]
- 225.Wang J, Zheng L, Lobito A, Chan F K, Dale J, Sneller M, Yao X, Puck J M, Straus S E, Lenardo M J. Inherited human Caspase 10 mutations underlie defective lymphocyte and dendritic cell apoptosis in autoimmune lymphoproliferative syndrome type II. Cell. 1999;98:47–58. doi: 10.1016/S0092-8674(00)80605-4. [DOI] [PubMed] [Google Scholar]
- 226.Wang S, Miura M, Jung Y-K, Zhu H, Li E, Yuan J. Murine caspase-11, an ICE-interacting protease, is essential for the activation of ICE. Cell. 1998;92:501–509. doi: 10.1016/s0092-8674(00)80943-5. [DOI] [PubMed] [Google Scholar]
- 227.Wang S L, Hawkins C J, Yoo S J, Muller H A, Hay B A. The Drosophila caspase inhibitor DIAPI is essential for cell survival and is negatively regulated by HID. Cell. 1999;98:453–463. doi: 10.1016/s0092-8674(00)81974-1. [DOI] [PubMed] [Google Scholar]
- 228.Wang Z Q, Stingl L, Morrison C, Jantsch M, Los M, Schulze-Osthoff K, Wagner E F. PARP is important for genomic stability but mdispensable in apoptosis. Genes Dev. 1997;11:2347–2358. doi: 10.1101/gad.11.18.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Warrick J M, Paulson H L, Gray-Board G L, Bui Q T, Fischbeck K H, Pittman R N, Bonini N M. Expanded polyglutamine protein forms nuclear inclusions and causes neural degeneration in Drosophila. Cell. 1998;93:939–949. doi: 10.1016/s0092-8674(00)81200-3. [DOI] [PubMed] [Google Scholar]
- 230.Watt W, Koeplinger K A, Mildner A M, Heinrikson R L, Tomasselli A G, Watenpaugh K D. The atomic-resolution structure of human caspase-8, a key activator of apoptosis. Struct Fold Des. 1999;7:1135–1143. doi: 10.1016/s0969-2126(99)80180-4. [DOI] [PubMed] [Google Scholar]
- 231.Weil M, Raff M C, Braga V M M. Caspase activation in the terminal differentiation of human epidermal keratinocytes. Curr Biol. 1999;9:361–364. doi: 10.1016/s0960-9822(99)80162-6. [DOI] [PubMed] [Google Scholar]
- 232.Wen L P, Fahrni J A, Troie S, Guan J L, Orth K, Rosen G D. Cleavage of focal adhesion kinase by caspases during apoptosis. J Biol Chem. 1997;272:26056–26061. doi: 10.1074/jbc.272.41.26056. [DOI] [PubMed] [Google Scholar]
- 233.White K, Grether M E, J.M. A, Young L, Farrell K, Steller H. Genetic control of programmed cell death in Drosophila. Science. 1994;264:677–683. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- 234.Widmann C, Gibson S, Johnson G L. Caspase-dependent cleavage of signaling proteins during apoptosis. A turn-off mechanism for anti-apoptotic signals. J Biol Chem. 1998;273:7141–7147. doi: 10.1074/jbc.273.12.7141. [DOI] [PubMed] [Google Scholar]
- 235.Wilson K P, Black J A, Thomson J A, Kim E E, Griffith J P, Navia M A, Murcko M A, Chambers S P, Aldape R A, Raybuck S A, Livingston D J. Structure and mechanism of interleukin-1 beta converting enzyme. Nature. 1994;370:270–275. doi: 10.1038/370270a0. [DOI] [PubMed] [Google Scholar]
- 236.Woo M, Hakem R, Soengas M S, Duncan G S, Shahinian A, Kagi D, Hakem A, McCurrach M, Khoo W, Kaufman S A, Senaldi G, Howard T, Lowe S W, Mak T W. Essential contribution of caspase-3/CPP-32 to apoptosis and its associated nuclear changes. Genes Dev. 1998;12:806–819. doi: 10.1101/gad.12.6.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Wu D, Wallen H D, Nunez G. Interaction and regulation of the subcellular localization of CED-4 and CED-9. Science. 1997;275:1126–1129. doi: 10.1126/science.275.5303.1126. [DOI] [PubMed] [Google Scholar]
- 238.Wu D M, Zhang Y, Parada N A, Kornfeld H, Nicoll J, Center D M, Cruikshank W W. Processing and release of IL-16 from CD4+ but not CD8+ T cells is activation dependent. J Immunol. 1999;162:1287–1293. [PubMed] [Google Scholar]
- 239.Xiang J, Chao D T, Korsmeyer S J. BAX-induced cell death may not require interleukin 1 beta-converting enzyme-like proteases. Proc Natl Acad Sci USA. 1996;93:14559–14563. doi: 10.1073/pnas.93.25.14559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Xue D, Horvitz H R. Inhibition of the Caenorhabditis elegans cell-death protease CED-3 by a CED-3 cleavage site in baculovirus p35 protein. Nature. 1995;377:248–251. doi: 10.1038/377248a0. [DOI] [PubMed] [Google Scholar]
- 241.Xue D, Shaham S, Horvitz H R. The Caenorhabditis elegans cell-death protein CED-3 is a cysteine protease with substrate specificities similar to those of the human CPP32 protease. Genes Dev. 1996;10:1073–1083. doi: 10.1101/gad.10.9.1073. [DOI] [PubMed] [Google Scholar]
- 242.Yamin T T, Ayala J M, Miller D K. Activation of the native 45-kDa precursor form of interleukin-1- converting enzyme. J Biol Chem. 1996;271:13273–13282. doi: 10.1074/jbc.271.22.13273. [DOI] [PubMed] [Google Scholar]
- 243.Yang J, Liu X, Bhalla K, Kim C N, Ibrado A M, Cai J, Peng T I, Jones D P, Wang X. Prevention of apoptosis by Bcl-2: release of cytochrome c from mitochondria blocked. Science. 1997;275:1129–1132. doi: 10.1126/science.275.5303.1129. [DOI] [PubMed] [Google Scholar]
- 244.Yang X, Chang H Y, Baltimore D. Autoproteolytic activation of pro-caspases by oligomerization. Mol Cell. 1998;1:319–325. doi: 10.1016/s1097-2765(00)80032-5. [DOI] [PubMed] [Google Scholar]
- 245.Yang X, Chang H Y, Baltimore D. Essential role of CED-4 oligomerization in CED-3 activation and apoptosis. Science. 1998;281:1355–1357. doi: 10.1126/science.281.5381.1355. [DOI] [PubMed] [Google Scholar]
- 246.Yang X, Khosravi-Far R, Chang H Y, Baltimore D. Daxx, a novel Fas-binding protein that activates JNK and apoptosis. Cell. 1997;89:1067–1076. doi: 10.1016/s0092-8674(00)80294-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Yang X, Stennicke H R, Wang B, Green D R, Janicke R U, Srinivasan A, Seth P, Salvesen G S, Froelich C J. Granzyme B mimics apical caspases: description of a unified pathway for trans-activation of executioner caspase-3 and -7. J Biol Chem. 1998;273:34278–34283. doi: 10.1074/jbc.273.51.34278. [DOI] [PubMed] [Google Scholar]
- 248.Yang Y, Fang S, Jensen J P, Weissman A M, Ashwell J D. Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science. 2000;288:874–877. doi: 10.1126/science.288.5467.874. [DOI] [PubMed] [Google Scholar]
- 249.Yeh W-C, de la Pompa J L, McCurrach M E, Shu H-B, Elia A J, Shahinian A, Ng M, Wakeham A, Khoo W, Mitchell K, El-Deiry W S, Lowe S W, Goeddel D V, Mak T W. FADD: essential for embryo development and signaling from some, but not all, inducers of apoptosis. Science. 1998;279:1954–1958. doi: 10.1126/science.279.5358.1954. [DOI] [PubMed] [Google Scholar]
- 250.Yeh W C, Itie A, Elia A J, Ng M, Shu H B, Wakeham A, Mirtsos C, Suzuki N, Bonnard M, Goeddel D V, Mak T W. Requirement for Casper (c-FLIP) in regulation of death receptor-induced apoptosis and embryonic development. Immunity. 2000;12:633–642. doi: 10.1016/s1074-7613(00)80214-9. [DOI] [PubMed] [Google Scholar]
- 251.Yin X M, Wang K, Gross A, Zhao Y, Zinkel S, Klocke B, Roth K A, Korsmeyer S J. Bid-deficient mice are resistant to Fas-induced hepatocellular apoptosis. Nature. 1999;400:886–891. doi: 10.1038/23730. [DOI] [PubMed] [Google Scholar]
- 252.Yoshida H, Kong Y Y, Yoshida R, Elia A J, Hakem A, Hakem R, Penninger J M, Mak T W. Apaf1 is required for mitochondrial pathways of apoptosis and brain development. Cell. 1998;94:739–750. doi: 10.1016/s0092-8674(00)81733-x. [DOI] [PubMed] [Google Scholar]
- 253.Yuan J, Horvitz H R. The Caenorhabditis elegans cell death gene ced-4 encodes a novel protein and is expressed during the period of extensive programmed cell death. Development. 1992;116:309–320. doi: 10.1242/dev.116.2.309. [DOI] [PubMed] [Google Scholar]
- 254.Yuan J, Shaham S, Ledoux H M, Horvitz H R. The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-b-converting enzyme. Cell. 1993;75:641–652. doi: 10.1016/0092-8674(93)90485-9. [DOI] [PubMed] [Google Scholar]
- 255.Zamzami N, Marchetti P, Castedo M, Hirsch T, Susin S A, Masse B, Kroemer G. Inhibitors of permeability transition interfere with the disruption of the mitochondrial transmembrane potential during apoptosis. FEBS Lett. 1996;384:53–57. doi: 10.1016/0014-5793(96)00280-3. [DOI] [PubMed] [Google Scholar]
- 256.Zamzami N, Susin S A, Marchetti P, Hirsch T, Gomez-Monterrey I, Castedo M, Kroemer G. Mitochondrial control of nuclear apoptosis. J Exp Med. 1996;183:1533–1544. doi: 10.1084/jem.183.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Zhang J, Cado D, Chen A, Kabra N H, Winoto A. Fas-mediated apoptosis and activation-induced T-cell proliferation are defective in mice lacking FADD/Mort1. Nature. 1998;392:296–300. doi: 10.1038/32681. [DOI] [PubMed] [Google Scholar]
- 258.Zhang J, Dawson V L, Dawson T M, Snyder S H. Nitric oxide activation of poly(ADP-ribose) synthetase in neurotoxicity. Science. 1994;263:687–689. doi: 10.1126/science.8080500. [DOI] [PubMed] [Google Scholar]
- 259.Zhang J, Liu X, Scherer D C, van Kaer L, Wang X, Xu M. Resistance to DNA fragmentation and chromatin condensation in mice lacking the DNA fragmentation factor 45. Proc Natl Acad Sci USA. 1998;95:12480–12485. doi: 10.1073/pnas.95.21.12480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Zhang Y, Center D M, Wu D M, Cruikshank W W, Yuan J, Andrews D W, Kornfeld H. Processing and activation of pro-interleukin-16 by caspase-3. J Biol Chem. 1998;273:1144–11449. doi: 10.1074/jbc.273.2.1144. [DOI] [PubMed] [Google Scholar]
- 261.Zheng T S, Hunot S, Kuida K, Flavell R A. Caspase knockouts: matters of life and death. Cell Death Differ. 1999;6:1043–1053. doi: 10.1038/sj.cdd.4400593. [DOI] [PubMed] [Google Scholar]
- 262.Zhou B B, Li H, Yuan J, Kirschner M W. Caspase-dependent activation of cyclin-dependent kinases during Fas-induced apoptosis in Jurkat cells. Proc Natl Acad Sci USA. 1998;95:6785–6790. doi: 10.1073/pnas.95.12.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Zhou L, Song Z, Tittel J, Steller H. HAC-1, a Drosophila homolog of APAF-1 and CED-4 functions in developmental and radiation-induced apoptosis. Mol Cell. 1999;4:745–755. doi: 10.1016/s1097-2765(00)80385-8. [DOI] [PubMed] [Google Scholar]
- 264.Zhou Q, Krebs J F, Snipas S J, Price A, Alnemri E S, Tomaselli K J, Salvesen G S. Interaction of the baculovirus anti-apoptotic protein p35 with caspases: specificity, kinetics, and characterization of the caspase/p35 complex. Biochemistry. 1998;37:10757–10765. doi: 10.1021/bi980893w. [DOI] [PubMed] [Google Scholar]
- 265.Zhou Q, Salvesen G S. Activation of pro-caspase-7 by serine proteases includes a non-canonical specificity. Biochem J. 1997;324:361–364. doi: 10.1042/bj3240361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Zhou Q, Snipas S, Orth K, Muzio M, Dixit V M, Salvesen G S. Target protease specificity of the viral serpin CrmA. Analysis of five caspases. J Biol Chem. 1997;272:7797–7800. doi: 10.1074/jbc.272.12.7797. [DOI] [PubMed] [Google Scholar]
- 267.Zhou T, Song L, Yang P, Wang Z, Lui D, Jope R S. Bisindolylmaleimide VIII facilitates Fas-mediated apoptosis and inhibits T cell-mediated autoimmune diseases. Nat Med. 1999;5:42–48. doi: 10.1038/4723. [DOI] [PubMed] [Google Scholar]
- 268.Zou H, Henzel W I, Liu X, Luschg A, Wang X. Apaf-1, a human protein homologous to C. elegans CED-4, participates in the cytochrome c-dependent activation of caspase-3. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- 269.Zou H, Li Y, Liu X, Wang X. An APAF-1.cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J Biol Chem. 1999;274:11549–11556. doi: 10.1074/jbc.274.17.11549. [DOI] [PubMed] [Google Scholar]