Abstract
The PII family of signal transduction proteins are among the most widely distributed signal proteins in the bacterial world. First identified in 1969 as a component of the glutamine synthetase regulatory apparatus, PII proteins have since been recognized as playing a pivotal role in control of prokaryotic nitrogen metabolism. More recently, members of the family have been found in higher plants, where they also potentially play a role in nitrogen control. The PII proteins can function in the regulation of both gene transcription, by modulating the activity of regulatory proteins, and the catalytic activity of enzymes involved in nitrogen metabolism. There is also emerging evidence that they may regulate the activity of proteins required for transport of nitrogen compounds into the cell. In this review we discuss the history of the PII proteins, their structures and biochemistry, and their distribution and functions in prokaryotes. We survey data emerging from bacterial genome sequences and consider other likely or potential targets for control by PII proteins.
Today it is widely recognized that there is probably no ecological niche on earth where bacteria have not evolved to exploit whatever nutrients are available to support life. Despite this enormous versatility of bacterial metabolism, certain fundamental mechanisms have to exist to regulate and integrate enzyme synthesis and enzyme activity. At the center of this metabolic control is the need to coordinate the catabolism and assimilation of carbon and nitrogen sources so as to maximize potential growth rates under any particular nutritional regime. There appear to be very few, if any, absolutely conserved mechanisms for achieving these ends among the major families of the eubacteria and the archaebacteria. However, the subject of this review, a small signal transduction protein commonly known as PII, is undoubtedly one of the most conserved signal proteins in the bacterial world. From its initial identification in 1969 as the second peak eluting from a gel filtration column, hence the designation PII (206), this protein family has since been found to play a significant role in the coordination of nitrogen metabolism in a wide variety of bacteria. The recent identification of members of the PII family in plants extends the biological role of these proteins even further. In this review, we have tried to bring together information, old and new, to give a broad perspective of our current knowledge of this intriguing molecule and to suggest new areas where it may also play a pivotal role in nitrogen metabolism.
HISTORICAL PERSPECTIVE
Nitrogen is one of the most important elements required for life, as it is necessary for the production of amino acids, nucleotides, amino sugars (required for the synthesis of lipopolysaccharides and peptidoglycan), NAD, and p-aminobenzoate (a precursor in folate biosynthesis). Consequently bacteria have developed a number of mechanisms by which nitrogen can be assimilated from a variety of sources, ranging from ammonium to atmospheric dinitrogen (N2). Ammonium is almost always the preferred nitrogen source, as it can be assimilated directly into glutamine and glutamate, the key donors for biosynthetic reactions, and is therefore the least energetically expensive substrate to process. By contrast, organic sources such as amino acids must first be degraded to ammonium, and inorganic sources such as NO3, NO2, and N2 must be reduced before assimilation (190).
There are two major assimilation pathways for ammonium, the glutamine synthetase/glutamate synthase (GS/GOGAT) pathway, which is ubiquitous in bacteria, and the glutamate dehydrogenase pathway, which is an alternative route of assimilation in many bacteria, including the enterobacteria. While glutamate dehydrogenase is energetically more efficient than GS/GOGAT, it has a low affinity for ammonium (Km of about 1 mM) and is consequently rather ineffective in cells growing in nitrogen-limited conditions.
Glutamine synthetase (GS) is a highly regulated enzyme at both the transcriptional and posttranslational level (for a review, see references 146, 155, 190, and 194). In enteric bacteria, GS is reversibly covalently modified by the bifunctional enzyme adenylyltransferase (ATase) in response to nitrogen availability, and the adenylylation state of GS regulates its catalytic activity. The mechanism of this regulation was determined by a series of elegant biochemical experiments initiated in the late 1960s. Initial studies showed that two protein components, PI and PII, were concerned with the adenylylation and deadenylylation process in Escherichia coli (206). The PI fraction contained an adenylytransferase whose ability to adenylylate or deadenylylate GS was specified by the PII protein and by the concentrations of PI, ATP, UTP, glutamine, and 2-ketoglutarate (5). It was subsequently determined that the PI fraction could be resolved into an ATase and a uridylyltransferase (UTase/UR) activity, the latter being capable of both uridylylation/uridylyl-removing and deuridylylation of PII (30, 148). PII stimulated adenylylation of GS by ATase, and PII-UMP stimulated the reverse reaction. PII was subsequently purified and proposed to be a tetramer with a molecular mass of about 44 kDa (1). The site of uridylylation on PII was identified as one of the two tyrosine residues in the protein, and the sequence of a tryptic peptide containing the covalently bound nucleotide was also determined (1, 193). The uridylylation and deuridylylation reactions were demonstrated to be catalyzed by a single bifunctional protein when the enzyme was finally purified to homogeneity as a single polypeptide of 95 kDa (78, 117). Hence, by 1984 the biochemical characterization of PII and the reactions that it regulated was well advanced, and a detailed model of the process of GS adenylylation had been proposed (1, 218).
In the meantime, genetic analysis of the regulation of GS and histidase in Klebsiella aerogenes had begun to shed light on the genes involved in these processes. Mutations in the gene encoding PII (glnB) were first isolated in K. aerogenes in 1973 by selection for glutamine-requiring mutants (184). Two classes of mutant were identified, those in which GS levels were undetectable, which were designated as glnA mutants, and a second class, represented by a single mutant, with very low but detectable levels of GS, which was designated the glnB mutant. It was then correctly postulated that glnA was the structural gene for GS and that glnB encoded an activator required for the synthesis of GS and for regulation of synthesis in response to nitrogen availability. Significantly, the glnB mutant was also unable to induce histidase expression when cells were nitrogen limited, giving the first indication that PII might have a role beyond the regulation of GS activity.
The product of glnB was finally identified as PII by Foor et al., who showed that in the original K. aerogenes glnB mutant, PII was altered so that it could not be uridylylated in nitrogen-limited conditions and consequently could not stimulate deadenylylation of GS by Atase (64, 65). A second class of glnB mutations resulted in loss of PII, and these glnB null mutants were still able to adenylylate and deadenylylate GS, although the rates of these reactions were reduced. However, the absence of PII also led to high levels of GS synthesis in the presence of ammonia, indicating a role of PII in regulating GS expression as well as its activity. Further evidence for PII having a role that is independent of its effect on ATase was provided by studies of glnB suppressor mutations which did not affect ATase activity but still affected levels of GS (191).
The Escherichia coli glnB gene was first sequenced by Son and Rhee (214). The deduced amino acid sequence agreed completely with that published earlier for the tryptic peptide containing the site of uridylylation (193) and showed that Tyr-51 was the modified residue. The isolation of further glutamine auxotrophs of both K. aerogenes and E. coli also led to identification of the structural genes for uridylytransferase (glnD) and adenylytransferase (glnE) that were finally cloned from E. coli and sequenced in 1993 (19, 25, 63, 64, 232).
Since the isolation of the first glnB mutant (184), there have been indications that PII has a function beyond regulating ATase activity. Subsequent genetic studies with E. coli, Salmonella enterica serovar Typhimurium, K. aerogenes, and Klebsiella pneumoniae identified a two-component nitrogen regulatory (ntr) system encoded by the ntrBC genes that was responsible for the global transcriptional control of enzymes of nitrogen assimilation and catabolism (see reference 155 for a review). The link between NtrBC and PII was demonstrated by further genetic studies in E. coli that led to the proposal that PII modulated the activity of the histidine protein kinase sensor protein NtrB (31). This established PII as the key link between changes in the intracellular nitrogen status and the activity of the transcriptional activator protein NtrC (Fig. 1).
FIG. 1.
Nitrogen regulation (Ntr) system of enteric bacteria. The activities of both GS and NtrC are regulated in response to the intracellular nitrogen status. UTase (glnD product) catalyzes the uridylylation and deuridylylation of PII (glnB product). ATase catalyzes the adenylylation and deadenylylation of GS. NtrB catalyzes the phosphorylation and dephosphorylation of NtrC.
Although by 1990 the molecular details of the enteric nitrogen regulation system seemed to be established, a number of inconsistencies were still apparent in the model. As the effects of UTase were predicted to be mediated through PII, then in strains lacking PII the glnD phenotype should be irrelevant, but this was not the case. E. coli cells lacking PII are Ntr+, as defined by their ability to utilize arginine as the sole nitrogen source, whereas both glnD mutants and glnB glnD double mutants cannot utilize arginine (13, 31). Hence, UTase has a role in Ntr regulation that is independent of PII. Similarly, cells lacking PII regulate GS adenylylation normally, whereas in vitro experiments showed that deadenylylation of GS by ATase absolutely requires PII-UMP (219, 233). A glnB glnD double mutant does not deadenylylate GS correctly in N limitation, indicating that UTase is also required for GS adenylylation in the absence of PII. Taken together, these results suggested the existence of at least one more protein that was involved in both Ntr regulation and GS adenylylation and that was also a substrate for UTase. Indications that the model was incomplete were not limited to E. coli, as studies on nitrogen control of nitrogen fixation (nif) genes in K. pneumoniae had prompted similar conclusions (55, 95).
The presence of a second PII gene in E. coli was confirmed in 1995 with the identification of a glnB homologue that was designated glnK (231, 233): ironically, this gene had been sequenced in 1993 but had gone unnoticed by researchers in nitrogen regulation (3). The E. coli GlnK protein is 67% identical to GlnB at the primary sequence level, and the protein is encoded in an operon with a second downstream gene, amtB, that is believed to encode a high-affinity ammonium transporter (231). Tyrosine 51 is conserved in GlnK, and indeed in nitrogen-limiting conditions, the protein is subject to uridylylation at this residue in a similar manner to GlnB (89, 231).
The subsequent publication of the E. coli genome sequence confirmed that the organism has just two glnB-like genes, a situation that has since been found in many bacterial species. Indeed, as we discuss below, some organisms carry up to four copies of this gene family. This mutiplicity of PII-like proteins raises complications with respect to terminology that we will discuss later, but from this point onwards we will use the term PII as a generic term to describe any member of the PII protein family.
OCCURRENCE OF PII-LIKE PROTEINS
In recent years, the explosion in bacterial genomics together with recognition of the involvement of PII proteins in a whole range of nitrogen regulation phenomena in bacteria led to the discovery that members of the PII family are ubiquitous among prokaryotes. Genes encoding PII proteins are found in the proteobacteria, the actinobacteria, the firmibacteria, the cyanobacteria, and the archaebacteria, and recently PII-like proteins have also been found in higher plants. Within these different groups, PII genes fall into a number of distinct classes with respect to both the primary amino acid sequences of their predicted products and their genetic linkage. Our current knowledge of the distribution of PII-like proteins is summarized in Table 1. Information on expression of these genes varies considerably, with very detailed studies on some organisms and nothing at all to date on others.
TABLE 1.
Organization of PII genes in prokaryotes and plants
| Organisma | Gene(s) present
|
|||||
|---|---|---|---|---|---|---|
| glnB-like | glnK-like | nif linked | ||||
| α Proteobacteria | ||||||
| Azospirillum brasilense | glnB, glnA | glnK, aat | ||||
| Azorhizobium caulinodans | glnB, glnA | glnK, amtB | ||||
| Acetobacter diazotrophicus | glnB, glnA | glnK1, amtB; glnK2, amtB2 | ||||
| Bradyrhizobium japonicum | glnB, glnA | |||||
| Rhizobium etli | glnB, glnA | glnK, amtB | ||||
| Rhodobacter capsulatus* | glnB, glnA | glnK, amtB | ||||
| Rhodobacter sphaeroides | glnB, glnA | glnK, amtB | ||||
| Rhodospirillum rubrum | glnB, glnA | glnK | ||||
| Sinorhizobium meliloti | glnB, glnA | glnK, amtB | ||||
| β Proteobacteria | ||||||
| Azoarcus sp. | nadE, glnB | glnK, amtB; gltB, glnY, amtY | ||||
| Herbaspirillum seropedicaeb | nadE, glnB | glnK? | ||||
| Neiserria meningitidis* | glnB | amtB | ||||
| δ Proteobacteria | ||||||
| Desulfovibrio gigas | nifH, nifI1 | |||||
| γ Proteobacteria | ||||||
| Azotobacter vinelandii | glnK, amtB | |||||
| Escherichia coli* | glnB | glnK, amtB | ||||
| Haemophilus influenzae* | mog, glnB, ydgD | |||||
| Klebsiella pneumoniae | glnB | glnK, amtB | ||||
| Pseudomonas aeruginosa* | glnK, amtB | |||||
| Vibrio cholerae* | glnK, amtB | |||||
| Xanthomonas citri | glnA, glnK, amtB | |||||
| Xylella fastidiosa | glnA, glnK, amtB | |||||
| Firmibacteria | ||||||
| Bacillus subtilis* | amtB, glnK | |||||
| Clostridium acetobutylicum* | amtB, glnK | nifH, nifI1, nifI2, nifD, nifK | ||||
| Clostridium cellobioparum | nifH, nifI1 | |||||
| Clostridium longisporum | glnK | |||||
| Lactococcus lactis* | amtB, glnK | |||||
| Actinobacteria | ||||||
| Corynebacterium glutamicum | amtB, glnK, glnD | |||||
| Mycobacterium tuberculosis* | amtB, glnK, glnD | |||||
| Streptomyces coelicolor* | amtB, glnK, glnD | |||||
| Archaebacteria | ||||||
| Archaeoglobus fulgidus* | amtB, glnK (3x) | |||||
| Methanobacterium ivanovii | nifH, nifI1, nifI2, nifD, nifK | |||||
| Methanobacterium thermoautotrophicum* | amtB, glnK (2x) | nifH, nifI1, nifI2, nifD, nifK | ||||
| Methanococcus jannaschii* | glnK, amtB | |||||
| amtB; glnK | ||||||
| Methanococcus maripaludis | nifH, nifI1, nifI2, nifD, nifK | |||||
| Methanococcus thermolithotrophicus | nifH, nifI1, nifI2, nifD, nifK | |||||
| Methanosarcina barkeri | nifH, nifI1, nifI2, nifD(2x) | |||||
| Deinococci | ||||||
| Deinococcus radiodurans* | glnK, amtB | |||||
| Thermatogales | ||||||
| Thermotoga maritima* | amtB, glnK | |||||
| Aquificaceae | ||||||
| Aquifex aeolicus* | glnB, glnA, amtB | glnBi, nasA, narB | ||||
| Cyanobacteria | ||||||
| Anabaena PCC7120 | glnB | |||||
| Freymella diplosiphon | glnB | |||||
| Nostoc punctiforme | glnB | |||||
| Prochlorococcus marinus | glnB | |||||
| Synechocystis PCC6803* | glnB | |||||
| Synechococcus PCC7942 | glnB | |||||
| Cyanidium caldarium | glnB | |||||
| Porphyra purpurea | glnB | |||||
| Dicotyledenous plants | ||||||
| Arabidopsis thalianad | GLB1 | |||||
| Glycine max | GLB1 | |||||
| Lycopersicon esculentum | GLB1 | |||||
| Ricinus communis | GLB1 | |||||
Complete genome sequences are available for species marked with an asterisk. In these cases, all known glnB-like genes are shown; in other cases, the list is largely derived from independently cloned genes and should not therefore be considered comprehensive.
Herbaspirillum seropedicae has two PII genes but the second has not yet been characterised (21).
In these cases the genes are located within the chloroplast genome.
A. thaliana GLB1 is a nuclear gene encoding a chloroplastic protein.
γ Proteobacteria
The E. coli glnB gene is located at 50 min on the chomosome (139), between the hmpA gene (located at the 3′ end of glnB and transcribed in the opposite direction) and yfhA (also called ORF-2 or OrfXB), located directly upstream of glnB and transcribed in the same direction (139, 232). This genetic organization is conserved in K. pneumoniae (96).
Four transcription starts involved in the transcription of E. coli glnB have been described, but none of these transcripts is regulated in response to nitrogen status (31, 87, 139, 232). Major promoters for glnB are located immediately upstream of the glnB gene (−33 and −95 from the ATG), but transcription of glnB can also be initiated at a promoter located upstream of yfhA (139). The E. coli genome sequence reveals a potential operon of three genes (yfhKGA) upstream of glnB and transcribed in the same direction. The product of yfhA has a high degree of homology to the nitrogen-regulatory protein NtrC (232), and YfhK and YfhA constitute a potential two-component regulatory pair, the function of which is unknown (161). It is therefore conceivable that under some conditions glnB is transcribed from a promoter upstream of yfhK.
A binding site for the repressor protein PurR, which is responsible for repression of enzymes required for purine nucleotide biosynthesis, is located between the two major glnB transcriptional start sites (from −68 to −83 nucleotides), and PurR downregulates the transcription of glnB twofold in conditions of purine excess (87). The physiological significance of this regulation during nitrogen excess may be to apply an additional subtle regulation of GS activity when the glutamine requirement for purine synthesis is low and little glutamine is required for protein synthesis (230).
Like E. coli, other members of the γ proteobacteria also have a second PII gene, glnK, that is located upstream of the ammonium transporter gene amtB (101, 223, 231, 233). In both E. coli and K. pneumoniae, the region upstream of the glnK gene contains a ς54 binding site and a possible NtrC binding site, and expression of the operon is nitrogen regulated in an NtrC-dependent manner (13, 101, 231). Exceptions include Azotobacter vinelandii, which appears to have only one PII gene that is linked to amtB and is expressed from a ς70-like promoter at a low constitutive level (153), and Haemophilus influenzae, which has a single glnB-like gene flanked by an upstream gene with similarity to E. coli mog and a downstream gene encoding a protein with 28% identity to E. coli YdgG and YhhT, which are both presumptive integral membrane proteins. A single PII gene is located between glnA and amtB in Xylella fastidiosa (accession no. AAF84648 to -50), and a similar linkage of glnA and glnK has been reported in Xanthomonas citri (accession no. AF182395 and -96).
α Proteobacteria
The majority of the α proteobacteria examined so far, including those in the genera Rhizobium, Bradyrhizobium, Azorhizobium, Acetobacter, Azospirillum, Rhodobacter, and Rhodospirillum, have two PII genes. One gene encodes a PII protein that is very similar to E. coli GlnB and is located upstream of and cotranscribed with glnA (the structural gene for glutamine synthetase I) (7, 27, 41, 94, 113, 127, 151, 158, 262). The second gene encodes a protein similar to GlnK and is linked to an amtB homologue (125, 157, 182, 185, 222, 254) (Y. Zhang and G. Roberts, personal communication). One exception to this pattern is Azospirillum brasilense, in which the glnK-like gene (termed glnZ) is not linked to amtB (46, 49, 228), but instead an open reading frame encoding a protein with 26% identity to the aspartate aminotransferase of Bacillus subtilis is found just 300 bp downstream of glnZ (47). The second exception is Acetobacter diazotrophicus, which has both a glnB-glnA operon and two glnK-amtB operons (D. Meletzus, personal communication).
Expression of the glnBA and glnK-amtB operons has been studied in some detail in the α proteobacteria. Transcription of the glnBA operon varies somewhat from one organism to another but shows a number of common features. Rhizobium leguminosarum and Sinorhizobium meliloti have promoters upstream of both glnB and glnA. The glnA promoter does not contain a consensus sequence characteristic of other previously described promoters, but the glnB promoter region contains a −24, −12 motif characteristic of ς54-dependent promoters. Expression of both glnB and glnA is partially NtrC dependent, but there are no clear NtrC binding sites upstream of glnB (6, 7, 36). In R. leguminosarum, the genes are cotranscribed, but neither the glnBA nor the glnA transcript is markedly nitrogen regulated (36, 195). However, expression of glnB is significantly downregulated during bacteroid differentiation (56).
Azospirillum brasilense also produces a glnBA and a glnA transcript, but in this case glnB is preceded by two promoters (48). The gene is expressed from a ς70-like promoter in nitrogen sufficiency, and transcription is elevated fivefold in nitrogen limitation as a second ς54-dependent promoter is activated. However, expression of glnB is not NtrC dependent (48, 49), and it has been proposed that glnB is activated by an NtrC homologue. The glnA promoter does not bear any similarity to the known consensus sites for ς factors, and glnA transcription is downregulated when molecular nitrogen is the sole nitrogen source (48, 49).
In Bradyrhizobium japonicum, glnB and glnA expression is almost entirely separate, but glnB is transcribed from tandem promoters. The glnA gene is constitutively expressed from a ς70-like promoter, and although glnB is expressed from the ς70-like promoter in nitrogen sufficiency and from a ς54-dependent promoter in nitrogen limitation, there is very little variation in the amount of transcript in response to the change in nitrogen status (151).
The glnB and glnA genes are also cotranscribed from tandem promoters in Rhodobacter capsulatus. The first promoter, glnBp1, is repressed by NtrC, while the second, glnBp2, is activated. In R. capsulatus, NtrC does not function with an RNA polymerase containing ς54, and hence, not surprisingly, there are no ς54 binding sites upstream of glnB (43, 73). Borghese and Wall (26) studied glnB expression using a glnB::lacZ fusion and different growth conditions from those used by Foster-Hartnett and Kranz (73). In this case, glnB expression was found to occur at higher levels under nitrogen-rich conditions than under nitrogen-poor conditions even in an ntrC mutant background. Moreover, the data suggested a posttranscriptional processing event that resulted in unequal levels of expression of glnB and glnA (26). The glnBA promoter region of Rhodobacter sphaeroides is similar to that of R. capsulatus, suggesting similar regulation, including partial nitrogen control of a glnB expression (185, 262). Interestingly, in a ribulose bisphosphate carboxylase/oxygenase (RubisCO)-deficient mutant, glnB expression was repressed and GS activity was extremely low regardless of the nitrogen source, suggesting the presence of a weak constitutive glnA promoter (185).
In Rhodospirillum rubrum, glnB and glnA are cotranscribed from a weak ς70-like promoter (glnBp1) and a strong ς54-like promoter (glnBp2) in either nitrogen-rich or nitrogen-fixing conditions (34, 113). However, glnBp2 activity is enhanced by NtrC in nitrogen-fixing conditions. Although Northern blotting analysis revealed the presence of two transcripts, no glnA promoter was detected, suggesting that the glnA mRNA is produced by processing (34, 113). As in Rhodospirillum rubrum and Rhodobacter capsulatus, Azorhizobium caulinaudans glnB and glnA appear to be cotranscribed (158). In both nitrogen excess and nitrogen limitation, the glnBA operon is transcribed from two overlapping promoters having the same start site, one of them ς54 and NtrC dependent and the other uncharacterized. Again, no promoter was detected upstream of glnA, and an mRNA processing event was proposed (158).
Regulation of glnK expression is similar in all the α proteobacteria so far studied, Azospirillum brasilense (glnZ), Azorhizobium caulinodans, and Rhizobium etli. In each case, the primary transcript is expressed from a ς54-dependent promoter that is active in nitrogen-limiting conditions and is NtrC dependent (47, 157, 222). In Rhodobacter sphaeroides, glnK is also expressed only under nitrogen-limiting conditions, and in a RubisCO-deficient mutant, glnK expression is partially derepressed in the presence of ammonium (185).
β Proteobacteria
Two representatives of the β proteobacteria, Herbasprillum seropedicae and an Azoarcus sp. have been studied in detail. H. seropedicae has two PII-like genes, one of which is linked to a homologue of E. coli nadE (a gene encoding ammonia-dependent NAD synthetase) (21). The Azoarcus sp. has three PII-like genes, one glnB homologue and two glnK homologues, each of which is linked to an amtB homologue (glnK to amtB and glnY to amtY) (119, 150, 188). Two of the Azoarcus PII genes, glnK and glnY, are preferentially transcribed under conditions of nitrogen fixation (150).
δ Proteobacteria
Within the δ proteobacteria, there is evidence for one glnB-like gene in the form of an open reading frame downstream of the nitrogenase structural gene nifH of Desulfovibrio gigas (accession no. U68183). This situation is very similar to that seen in the diazotrophic methanogens (see below).
Firmibacteria
Among the gram-positive bacteria, there are relatively few reports of PII genes to date. Bacillus subtilis has a single PII gene that was originally designated nrgB and is located downstream of an amtB homologue (nrgA) (246). The upstream region contains a B. subtilis ςA-dependent promoter, and characterization of two transcripts separated by a single nucleotide suggests a common promoter origin. The operon is markedly induced in nitrogen limitation, but regulation of an nrgA fusion is not altered in either a ΔnrgB background or a ΔnrgAB background, suggesting that these gene products are not required for their nitrogen regulation; rather, expression is activated by the Bacillus global nitrogen-regulatory protein TnrA (246–249).
Searches of the DNA sequence databases also reveal evidence for glnB-like genes in Clostridium acetobutylicum (http://www.genomecorp.com/genesequences/clostridium/clospage.html), Clostridium cellobioparum (accession no. U59414), Clostridium longisporum (29), and Listeria monocytogenes (accession no. AF104224).
Actinobacteria
PII genes have been identified in a number of actinomycetes. Mycobacterium tuberculosis has a single gene, with an amtB homologue upstream and a glnD homologue downstream, forming a potential amtB-glnK-glnD operon (38). A similar organization is found in Corynebacterium glutamicum (104) and in Streptomyces coelicolor.
Cyanobacteria
The presence of a GlnB protein in cyanobacteria was first recognized with the N-terminal sequencing of a phosphorylated 13-kDa protein from Synechococcus sp. strain PCC6301 (85). Subsequent investigations of the occurrence of glnB genes in cyanobacteria included analysis of Synechococcus sp. strain PCC7942, Calothrix sp. strain PCC7601, Pseudanabena sp. strain PCC6901, Microcystis sp. strain PCC7813, and Nostoc sp. strain PCC8009 and suggested that glnB is likely to be present in all cyanobacteria (225). Since then, the glnB genes from Synechococcus sp. strain PCC7942, Synechocystis sp. strain PCC6803, Nostoc punctiforme, and Anabaena sp. strain PCC7120 have been cloned and sequenced (76, 80, 84, 225). In all cases glnB appears to be monocistronic, and its transcription is enhanced by nitrogen limitation.
In both Synechococcus sp. strain PCC7942 and Synechocystis sp. strain PCC6803, glnB is expressed from two tandem promoters, a ς70 E. coli-type promoter, leading to constitutive expression of glnB, and a nitrogen-regulated promoter activated by the nitrogen-regulatory protein NtcA. Despite similarities, the physical organization of the promoters differs, and in Synechocystis sp. strain PCC6803 the regulated promoter is functional only in cells starved for nitrogen, whereas in Synechococcus sp. strain PCC7942 it is active both in the absence of nitrogen and in the presence of nitrate (76, 132). Moreover, in Synechocystis sp. strain PCC6803, glnB mRNA levels decrease when cells are either transferred to the dark or incubated in the presence of photosynthesis inhibitors, indicating that glnB transcription is under the control of the redox state of the cell (76).
Archaebacteria
In all the archaebacteria for which complete genome sequences are available, glnB-like genes are found linked to amtB homologues and are usually present in mutiple copies (223). However, in the diazotrophic methanogens, two additional glnB-like genes are located between nifH and nifD within the nif gene cluster (122, 123, 208, 210, 215). These genes have been anlayzed in most detail in Methanococcus maripaludis, in which they are apparently transcribed in a 7.6-kb nif mRNA transcript from a single promoter with a transcritional start site located 80 nucleotides upstream of the translational start site of nifH (122). Interestingly, a comparable gene organization is found in nitrogen-fixing microorganisms inhabiting the gut of the termite Neotermes koshunensis, in which two glnB-like genes are located between anfH and anfD (174).
Other Bacteria
The genome sequence of the extreme thermophile Aquifex aeolicus revealed two glnB-like genes (45). One of these is in a glnB-glnA operon similar to that found in the γ proteobacteria, and the other is the first gene in a cluster comprising glnBi, nasA, and narB. This latter glnB-like gene apparently encodes a totally novel PII polypeptide of 205 amino acids. The gene comprises a tandem duplication, so that the C terminus of the first “copy” is fused directly to the N terminus of the second copy. Both copies are predicted to have all the characteristic features known from structural studies of PII (see later), with the exception that the N-terminal copy is truncated prior to the last two β-sheets that comprise the C-loop. This sequence would suggest that if the gene is expressed, it could encode a trimeric protein that comprises six PII-like domains with three T-loops exposed on one face and another three similar but not identical T-loops on the opposite face. In alignments of PII amino acid sequences, the predicted product of Aguifex aeolicus glnBi is most similar to the glnB-like proteins encoded downstream of nifH in the diazotrophic archaebacteria.
Organisms in Which PII Is Absent
The PII protein is not totally ubiquitous, and the completion of a significant number of bacterial genome sequences has identified a number of organisms that do not encode a PII-like protein. These include Helicobacter pylori, Mycoplasma genitalium, Mycoplasma pneumoniae, Campylobacter jejuni, Chlamydia muridarum, Chlamydia pneumoniae, Chlamydia trachomatis, Chlamydophila pneumoniae, Treponema pallidum, Ureaplasma urealyticum, Rickettsia prowazekii, Aeropyrum pernix, Borrelia burgdorferi, Pyrococcus horikoshii, and Pyrococcus abyssi. It is notable that a large number of these organisms are pathogens whose genomes have undergone reductive evolution to compete in specific niches within their respective hosts. As such, they frequently rely on their hosts for provision of nitrogen-containing compounds, and they probably do not face the homeostatic challenges that require constant monitoring and control of their intracellular nitrogen status.
PII Genes in Plants
The first discovery of a PII gene outside the bacteria was the identification of a glnB-like gene in the chloroplast genome of the red alga Porphyra purpurea (189), and a second such sequence has since been found in the chloroplast of Cyanidium caldarium (accession no. AF022186). However, more recent studies indicate that the PII protein is probably quite widely distributed among higher plants. Genes encoding PII-like proteins have been cloned from Arabidopsis thaliana and Ricinus communis (castor bean) (97), and similar genes have been sequenced in Lycopersicon esculentum (tomato) (accession no. AI773079) and Glycine max (soybean) (accession no. AW153272). In all cases, the predicted protein sequences show significant homology to the bacterial GlnB proteins but have an additional N-terminal domain of about 70 amino acids.
The PII protein in Arabidopsis thaliana is encoded by the GLB1 gene, mapped at 10.8 centimorgans from the top of chromosome IV (97). When extracted from leaves, this nucleus-encoded chloroplast protein exhibits a molecular mass of 18 kDa. However, its predicted molecular mass of 21.4 kDa (196 amino acids) suggests that PII may be processed inside the plant cell. The transcription of GLB1 mRNA is induced by light or sucrose and is repressed by the amino acids asparagine, glutamine, and glutamate (97).
PII PROTEIN NOMENCLATURE
The recent rapid increase in the identification of genes encoding PII homologues has led to an extremely complex and confusing situation with respect to nomenclature. It is clear that a number of genes that are currently designated glnB have very different functions and in some cases are predicted to encode proteins with significantly different structures. However, the number of sequences now available allows us to assess the similarities and differences between these proteins and to begin to make some predictions about their functions. In carrying out such an assessment, we have used two criteria: conservation of gene linkage and similarity at the level of primary amino acid sequence.
With the dramatic increase in availability of prokaryotic genome sequences, conservation of genetic linkage is increasingly being recognized as offering predictions of functional relationships between gene products (44, 177). Multiple sequence alignments can also provide information on sequence similarity, which, at least in some cases, may reflect similarity of function.
Multiple sequence alignments using Clustal W and analysis of the resultant dendrograms show, perhaps not surprisingly, that PII sequences cluster into groups that reflect almost completely the taxonomic relationships of the originating organism, and furthermore, within certain groups, e.g., the archaebacteria, clear subgroups occur. We have combined this information with knowledge of gene linkage to define just three major groups of prokaryotic PII proteins (Table 1). Based on this subdivision, we consider that it is now appropriate to review the current gene nomenclature, and we suggest a revised format that we hope will aid gene description in the future. This nomenclature is not meant to imply a strict link between gene designation and function, particularly given the recognized overlapping functions that can occur, e.g., with GlnB and GlnK in some organisms (see later). However, it is hoped that these suggestions will provide a rational framework for describing members of the PII family.
glnB
The original glnB designation should be retained for genes encoding a large group of closely related proteins found predominantly in the proteobacteria and the cyanobacteria. With a few exceptions, these genes are either monocistronic operons or linked to glnA or nadE. The protein products of these genes are typified by lysine at residue 3 and glutamate or aspartate at residue 5.
glnK
The designation glnK was originally adopted for the amtB-linked gene of E. coli (231). It has since become apparent that the ammonium transporter gene amtB is almost invariably linked to a PII gene, a phenomenon that may imply a functional relationship between GlnK and AmtB (223). We suggest that the glnK nomenclature be retained for all amtB-linked PII genes. In some cases, e.g., Methanobacterium thermoautotrophicum and Archaeglobus fulgidus, organisms have multiple amtB-glnK operons, and in these cases the genes should be designated amtB1-glnK1, amtB2-glnK2, etc., to follow standard nomenclature for such situations. The GlnK proteins are often, but not invariably, distinguished from GlnB proteins in having a hydrophobic residue (leucine, isoleucine, methionine, or phenylalanine) at position 3 and isoleucine, threonine, or methionine at position 5. We suggest extending the glnK designation to include the exceptional glnZ gene of A. brasilense, which is not linked to amtB but encodes a PII protein that is very similar to the GlnK proteins of Rhizobium etli and Acetobacter diazotrophicus. Likewise, Azoarcus sp. has two glnK-like genes, one of which has been designated glnY and is linked to an amtB homologue designated amtY (150). On the basis of amino acid sequence, GlnY clusters quite closely with other GlnK proteins and could therefore perhaps more appropriately be designated GlnK2 rather than GlnY.
nifI
A quite distinct group of PII proteins are encoded by the nifH-linked genes found in the diazotrophic methanogens. These genes always occur in pairs and fall into two discrete subgroups, the nifH-proximal gene, encoding a polypeptide of about 105 amino acids, and the nifH-distal gene, encoding a polypeptide of 120 to 130 amino acids. At present these genes have been designated glnB, with a variety of subdivisions. In M. maripaludis, the products of these genes have now been shown to function in ammonia switch-off of nitrogen fixation (123, 123a). For this reason, we propose that this pair of genes should be renamed nifI1 and nifI2, respectively, to reflect both their distinction from glnB and glnK and their specific biological role. The nifI genes are not restricted to the archaebacteria, as sequences of nifI1 genes have also been identified downstream of nifH genes in Desulfovibrio gigas, Clostridium acetobutylicum, and Clostridium cellobioparum. The novel glnB-like gene in Aguifex aeolicus that is located upstream of nasA and narB and currently designated glnBi is also most similar to this group of PII proteins.
BIOCHEMISTRY OF PII PROTEINS
Initial characterization of E coli PII by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and sedimentation centrifugation indicated that the E. coli protein formed a tetramer (1). However, subsequent crystallization studies and sedimentation equilibrium experiments have shown that it is in fact a trimer (33, 46, 236). The molecular mass of the PII monomer was elucidated by matrix-assisted laser desorption mass spectrometry and shown to be 12,435 Da, which agrees very well with the prediction from the DNA sequence of 12,427 Da (235, 236).
Structure of GlnB
An X-ray crystal structure of E. coli GlnB was initially achieved at a resolution of 2.7 Å and subsequently refined to a resolution of 1.9 Å (32, 33, 46) (Fig. 2). Each monomer contains two α-helices and six β-strands arranged so that the two α-helices and β-strands 1 to 4 form a double βαβ motif connected by a large loop stretching from Gly-37 to Phe-55. At the apex of this loop is the site of uridylylation, Tyr-51, and consequently this is referred to as the T loop. A smaller loop (the B loop) stretching from Gln-82 to Asp-88 separates the second α-helix from the fourth β-sheet, and a third loop (the C loop) containing β-strands 5 and 6 is found at the C terminus. Analysis of the refined structure has revealed that hydrogen bonds are formed between Gly-27 and Val-64, stabilizing β-sheets 2 and 3, and between Glu-106 and Arg-101 and between Thr-104 and Gly-105, stabilizing the C loop.
FIG. 2.
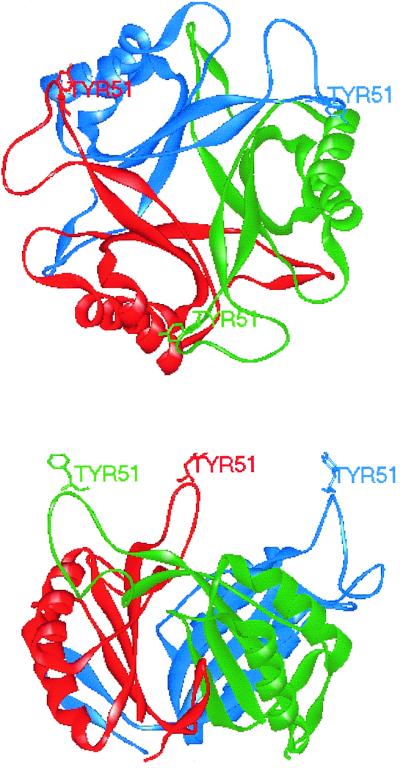
Structure of the E. coli GlnB trimer viewed (top) from above and (bottom) from the side. Individual monomers are colored red, green, and blue.
The trimer is arranged so that the four major β-strands of each monomer are flanked on one side by the T loop from the second monomer and on the other side by the C-terminal loop of the third. The arrangement of the monomers relative to one another causes a cleft, which may have functional significance, to form between the T and B loops of one monomer and the C loop of the next. Three six-stranded antiparallel β-sheets are arranged as a concave surface in the center of the trimer. The majority of the protein therefore packs into a squat barrel 30 Å high. The diameter of the cavity ranges from approximately 24 Å at the ends to 5.5 Å about 7.0 Å from either end, the diameter at the center of the cavity being about 12.0 Å. The cavity contains several ordered water molecules, and it has also been suggested that the cavity could contain metals (32). Each monomer is arranged so that an eight-residue stretch of the loop containing Tyr-51 at its apex is exposed at the same surface of the trimer about 13 Å above the surface of the barrel.
Structure of GlnK
The structure of E. coli GlnK has also been solved to a resolution of 2.0 Å (145, 252). GlnK also forms a trimer, with a core structure that is very similar to that of GlnB (Fig. 3). However, the GlnK protein was found to be present in two conformations in the crystals. They had core structures similar to each other's and to GlnB, but the structure of the T loop in each case was significantly different. It was only possible to solve the structure for one T loop which was stabilized by lattice contacts; the T loop of the other protein was disordered. The structure of the T loop differed from that of GlnB in that residues 47 to 49 of GlnK form a 310 helix and the apex of the T loop was much closer to the top of the core of the molecule than seen with GlnB. It should be noted that the T loop of GlnK was only resolvable when it was constrained, and in the GlnB crystals the T loop was also latticed to an adjacent molecule. Nevertheless, the fact that the T loops form different conformations in the crystals might suggest that they behave differently in solution and that the T loop might be flexible rather than rigid. The T loop of E. coli GlnB is known to be essential for interaction with all three of its known targets, ATase, UTase, and NtrB (103, 112). These different targets may require different T-loop conformations to facilitate specific high-affinity interactions.
FIG. 3.
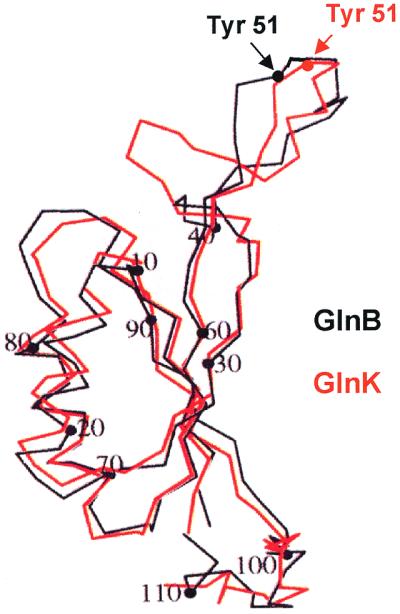
Comparison of the structures of E. coli GlnB and GlnK. Superimposition of the Cα traces for GlnB (black) and GlnK (red) with the position of the Tyr-51 residues in each molecule is indicated. Adapted from reference 252.
Both the B and C loops of GlnK also differ from those of GlnB. These loops form part of the cleft thought to be the important in the binding of effector molecules and proteins. The apex of the B loop of GlnK differs from that of GlnB by 4.3 Å, and the C loop contains a single rather than two β-strands and a 310 helix which causes the end of the peptide to turn back into the bottom of the cleft. The structure of ATP-bound GlnK was also elucidated and showed that ATP makes contacts along the length of the cleft and bonds with residues from both subunits. In particular, the nitrogen and carbonyl oxygen of Ala-64 (which is not a conserved residue and is valine in GlnB) are thought to form hydrogen bonds with the adenine group. The B loop has a consensus sequence of Thr-Gly-X-X-Gly-Asp-Gly-Lys-Ile-Phe, which is very similar to that of mononucleotide-binding proteins such as adenylate kinase. The architecture of the phosphate-binding loop is different from that of other nucleotide-binding proteins, and there is no evidence for Mg2+ binding as seen in adenylate kinase and p21ras. The motif is one of the most highly conserved regions in all the PII proteins from eubacteria, archaebacteria, and plants. This suggests that ATP binding is probably a common property of PII, and indeed it has also been demonstrated for the GlnB protein of Synechococcus sp. (67). The precise role of ATP is presently unknown, although biochemical data suggest that high-affinity binding of 2-ketoglutarate requires ATP (67, 118). The binding of ATP may also cause alterations in the structure and mobility of the T loop and in the structure of the B loop, both of which could affect molecular recognition.
Heterotrimer Formation
E. coli GlnB and GlnK can form heterotrimers in vivo when glnB and glnK are coexpressed from a plasmid or when the wild-type strain is grown in nitrogen-limiting conditions (68, 234). GS adenylylation experiments in vitro demonstrate that the fully uridylylated heterotrimer is still capable of stimulating the deadenylylation activity of ATase, albeit to a lower extent than homotrimeric PII-UMP. It has been proposed that heterotrimers could facilitate fine-tuning of the signal transduction cascade (234). While the physiological consequences of heterotrimer formation have yet to be fully established, heterotrimer formation is possibly not restricted to E. coli because it is also observed between Synechococcus GlnB and E. coli GlnB or GlnK when the cyanobacterial protein is expressed in E. coli (68). Given this observation, one might expect heterotrimer formation to occur in other organisms which express more than one form of PII concurrently. The factors controlling such events could be of considerable interest and certainly complicate the interpretation of mutant phenotypes.
Analysis of sequence comparisons revealed that residues that are either involved in the formation of the trimer (residues 30, 32, 34, 93, 95, and 98) or interact in the interior of the central cavity (residues 3, 5, 60, and 62) are not always conserved in PII proteins (252). In particular, residues 3 and 5 vary between the different subgroups of PII proteins (101). In E. coli GlnB, Lys-3 and Asp-5 form a ring of alternating charge in the central cavity of the trimer, but in GlnK these residues are replaced by neutral Leu-3 and Thr-5 residues (252). The distinction between the nature of residues 3 and 5 in GlnB and GlnK extends to other members of the PII family, e.g., NifI1 polypeptides are characterized by Met-3 and Arg-5 or Lys-5, while NifI2 is characterized by Glu-3 and Ile-5. The significance of this signature is not clear.
Structure-Function Analysis
A considerable number of mutations have been studied in E. coli glnB, and with the available crystal structure, these can help to shed some light on structure-function relationships in the protein. Uridylylation of GlnB is prevented by mutation of Tyr-51 to Phe, Asn, or Ser (14, 89, 103, 111). It is also severely affected by other alterations in the exposed part of the T loop, including E50Q, the same alteration as that produced by the K. pneumoniae glnB502 mutation (96, 111). Residue Tyr-46 is highly conserved in PII proteins, and a Y46F variant is uridylylated at a much lower rate than the wild type (103). The mutation does not alter the structure of the protein or binding of ATP or 2-ketoglutarate, suggesting that the residue may play an important role in recognition and binding by UTase (103, 111). Removal of the apex of the T loop (Δ47–53) also prevents uridylylation. Indeed, it alters interaction with all three known receptors but not the binding of small effector molecules, implying that the T loop is necessary for interaction with GlnB targets (111).
Mutations of different residues within the T loop of GlnB affect inteaction with all targets to some degree, but two of them, G41A and A49P, are notable exceptions dramatically eliminating interaction with UTase or NtrB, respectively, without having any dramatic effect on their other targets (111). A number of other alterations have also been made to residues found in the conserved region comprising the B loop and the fourth β-sheet of E. coli GlnB. The effects of these changes on the interaction of GlnB with 2-ketoglutarate, ATP, and the signal-transducing enzymes were then studied. Mutations around the B loop and the base of the T loop tend to affect the binding of the small-molecule ligands and some or all of the three protein receptors UTase/UR, ATase, and NtrB (111). It may be that the effect on enzyme interactions is a result of poor ligand binding. This second group of mutations cluster around the cleft formed by the T and B loops of one monomer and the C loop of the neighboring monomer, adding weight to the theory that this structure plays an important role in the interaction with receptor proteins and ligands.
Finally, two other alterations with marked effects on GlnB function fall outside these regions. A Thr29Met mutation was identified by the mutant's inability to activate glnA transcription, but further investigation revealed that the protein was affected in all its interactions. As this mutation destroys the hydrogen bond link with Glu-62 (found at the base of the third β-sheet), it may result in an overall change in protein conformation. A Gly89Ala mutation causes an almost total loss of function, probably linked to the inability to bind small-molecule effectors. Gly-89 is at the base of the B loop at a sharp turn, which allows the Lys-90 of β-4 to line the cleft. It is possible that this mutation causes a conformational change that prevents this turn.
Experiments investigating the functionality of heterotrimers comprising Δ47–53 GlnB and wild-type GlnB showed that the T loops can act independently of one another and a single loop is sufficient to allow interaction with the receptor proteins. Heterotrimers of Gly89Ala and wild-type monomers had different effects on interaction with different protein receptors, indicating that changes at the base of the B loop probably affect intersubunit interactions and thereby the activity of the wild-type subunit. Heterotrimers of Gly89Ala (containing a normal T loop but unable to bind small-molecule effectors) and Δ47–53 (missing the T loop) remained inactive, suggesting that there is no trans complementation between subunits (112).
Modification of PII in Response to Nitrogen
A principal characteristic of the PII proteins is their ability to be switched between two forms by covalent modification of a residue in the T loop. This modification was initially recognized as uridylylation of Tyr-51 in the T loop of E. coli GlnB. This residue is highly conserved, being present in the GlnB and GlnK proteins of the proteobacteria and the actinobacteria. Uridylylation of PII proteins has been demonstrated in Rhizobium leguminosarum (40), Klebsiella pneumoniae (55), Azospirillum brasilense (47, 50), Azotobacter vinelandii (196), Herbaspirillum seropedicae (20), and Azoarcus sp. strain BH72 (150) and suggested in Sinorhizobium meliloti (8), Rhodospirillum rubrum (114), and Corynebacterium glutamicum (104). While this process is widespread, it is not universal; PII proteins in the cyanobacteria are phosphorylated on a serine residue, Ser-49, in the T loop, and in other organisms, e.g., the archaebacteria and higher plants, modification of PII proteins has yet to be investigated.
Uridylylation of PII proteins.
Uridylylation of E. coli GlnB in vitro is a noncooperative reaction (15). The transferase reaction shows linear kinetics, with GlnB binding before ATP and pyrophosphate being released before GlnB-UMP. By contrast, the uridylyl-removing reaction proceeds with rapid equilibrium binding of substrate and random release of products. Both reactions are activated by ATP and 2-ketoglutarate, while glutamine inhibits the transferase reaction. The kinetic data are consistent with a single binding site on GlnD for GlnB and GlnB-UMP and with a single catalytic center on the enzyme, which could be located at the conserved nucleotidyl transferase site (108). In vitro, GlnK is uridylylated as effectively as GlnB, but deuridylylation of GlnK-UMP is slower than that of GlnB-UMP (14).
Under physiological conditions, the uridylylation state of PII appears to be regulated mainly by the glutamine concentration, suggesting that the UTase/UR enzyme serves as an intracellular nitrogen sensor (108). This hypothesis is consistent with the constitutive expression of glnD, so that UTase/UR is always present in the cell and its activity is modulated in response to nitrogen availability, with the result that the uridylylation level of PII reflects the intracellular nitrogen status of the cell.
The UTase/UR structural gene glnD has been cloned and sequenced from E. coli (232), K. pneumoniae (55), Azotobacter vinelandii (42), Rhizobium tropici (176), Vibrio fischeri (accession no. AF152563), Rhizobium leguminosarum biovar viciae (199), and Sinorhizobium meliloti (accession no. AF227730). Genome sequence analysis shows that glnD homologues are present in most proteobacteria and also in the actinobacteria, e.g., Streptomyces coelicolor (accession no. AL023797), Mycobacterium tuberculosis (38), and Corynebacterium glutamicum (104), but glnD is not found in the gram-positive firmibacteria (e.g., Bacillus subtilis), the cyanobacteria (e.g., Synechocystis), or the archaebacteria (Methanobacterium thermoautotrophicum and Methanococcus jannaschii). Relatively little is known about glnD expression, but the E. coli glnD gene is monocistronic and expressed at a very low level (117, 124, 232), and this is also the case for Rhizobium tropici glnD (176).
GlnD proteins have predicted molecular masses of about 100 kDa and contain a conserved nucleotidyltransferase superfamily motif (AVGGYGRXXLXPXSDIDLL) located in the amino-terminal region (93). This conserved motif is thought to be located in the active site of the enzyme, suggesting that the two catalytic activities of UTase reside at the same site unless one of the two catalytic sites does not contain the conserved nucleotidyltransferase sequence. The activities of GlnD proteins are indeed conserved as Azotobacter vinelandii glnD can complement E. coli and K. aerogenes glnD mutants, and likewise, E. coli glnD complements an Azotobacter vinelandii glnD mutant (42).
In E. coli, only one glnD mutation (glnD99::Tn10) has been physically mapped within the glnD gene (229), and the Tn10 is inserted 567 bp upstream of the glnD stop codon (T. Arcondéguy and M. Merrick, unpublished results). The leaky nature of this mutation (31, 214) led Atkinson and Ninfa (16) and Son and Rhee (214) to propose that the Tn10 does not completely inactivate UTase, which is consistent with initial data reporting a GlnB-UMP activity equivalent to 8% of the total purified GlnB from a glnD99::Tn10 mutant strain (214). The predominance of mutations at the 3′ end of the gene is a notable feature in many organisms; a Tn5 insertion in Azotobacter vinelandii glnD was located about 80 bp upstream of the stop codon (42), and in Rhizobium tropici a Tn5 insertion was located at the 3′ end of the gene (176).
In Azotobacter vinelandii, a stable glnD null mutation could only be maintained when GS adenylylation was prevented by a glnA Y407F mutation or a suppressor presumptively located in the ATase gene glnE (39, 196). An extensive glnD deletion was constructed in K. pneumoniae (55), but the possibility that this mutation was maintained as a consequence of secondary suppressor mutations cannot be excluded. A glnD null mutant of E. coli exhibits a severe growth defect in minimal medium with ammonium as a nitrogen source, but the occurrence of suppressors in this strain has also not been investigated (T. Arcondéguy and M. Merrick, unpublished results). Finally, a glnD insertion mutant of Corynebacterium glutamicum was unstable (104). It therefore appears that glnD may be essential in some (or all) organisms and that maintenance of a null mutation may require the presence of secondary mutations that can occur spontaneously at high frequencies. This may be consistent with the important role of UTase as an intracellular nitrogen sensor but could also suggest that the protein has other, as yet unidentified, roles in the cell.
Phosphorylation of PII proteins.
Although Tyr-51 is present in GlnB from the cyanobacterium Synechococcus sp. strain PCC6803, this protein is not uridylylated but is phosphorylated at the nearby Ser-49 residue (69, 71). This is consistent with the absence of a glnD homologue. Despite this difference, heterologous expression of Synechococcus sp. strain PCC7942 GlnB in E. coli results in uridylylation of this protein, albeit with low efficiency (67, 68). PII was originally identified in cyanobacteria as a 13-kDa protein in Synechococcus sp. strain PCC7942 that exhibited a posttranslational modification dependent on the nitrogen source and the spectral light quality (225). However, the spectral light quality dependency is only seen in the presence of nitrate, and hence, as in enterics, the primary role of GlnB appears to be in sensing nitrogen status (69). In the presence of ammonium, GlnB is unmodified, and in nitrate GlnB is predominantly modified. Moreover, in cells grown in ammonium but in the presence of l-methionine sulfoximine (a GS inhibitor), GlnB exhibits the modified pattern, indicating that ammonium assimilation is required to elicit dephosphorylation.
The modification state of GlnB in cyanobacteria is not controlled by a bifunctional enzyme but by a kinase and a phosphatase activity that can be biochemically separated, suggesting that these two activities reside on different proteins (100). In vivo studies of the phosphorylation state of GlnB under different growth conditions (70) and studies of the in vitro kinase activity (using a glnB null mutant extract) show that the kinase activity is stimulated by 2-ketoglutarate (71). However, unlike GlnD, the kinase does not respond to glutamine. Because dephosphorylation of GlnB is regulated by synergistic inhibition by ATP and 2-ketoglutarate and phosphorylation depends on the same metabolites, it seems that ATP and 2-ketoglutarate binding of PII (67) inversely affects its recognition by the kinase and the phosphatase (71, 100).
Modification of GlnB in cyanobacteria has also been reported in Synechococcus sp. strain 6301 (85) in Synechocystis sp. strain PCC6803 (92) and in the two filamentous nitrogen-fixing cyanobacteria Calothrix sp. strain PCC7504 (137) and Anabaena sp. strain PCC7120 (257). When Synechocystis sp. strain PCC6803 cells are exposed to conditions impairing the photosynthetic electron flow, GlnB remains unphosphorylated, suggesting that GlnB modification is also regulated by the redox state of the cells (92).
As in Synechococcus sp. strain PCC7942, Anabaena GlnB is phosphorylated when filaments are grown in the presence of nitrate and dephosphorylated in the presence of ammonium, although the kinetics of Anabaena GlnB phosphorylation-dephosphorylation is slower than in the Synechococcus sp. (80, 257). Interestingly, mutation of pknD, a eukaryote-type protein kinase, enhances phosphorylation of GlnB under nitrogen-fixing conditions. It has been proposed that PknD is probably not involved in the phosphorylation process itself but that the effect on GlnB phosphorylation is rather the consequence of impaired nitrogen metabolism or trafficking (257).
Other possible modifications.
PII modification has not yet been studied in the archaebacteria. A Tyr-51 residue (or its equivalent) is present in the PII proteins encoded by the amtB-linked genes of Methanococcus jannaschii and Methanobacterium thermoautotrophicum, but there is no glnD homologue in these organisms, suggesting that in these cases modification, if it occurs at all, is not by uridylylation (35). The archebacterial nif cluster GlnB homologues fall into two classes, NifI1, with a predicted T loop of only 15 residues, and NifI2, with a predicted T loop of 27 or more residues. Both contain conserved tyrosine residues, but they are not predicted to be at the apex of the T loop. Tyr-51 is also absent in Bacillus subtilis GlnK (246) and Clostridium longisporum GlnK (29), and consistent with this, there is no glnD homologue in the Bacillus subtilis genome. Both organisms have a serine residue in the T loop which could constitute a potential modification site.
In all the PII sequences so far identified in plants, i.e., Arabidopsis thaliana GLB1, and similar sequences in Glycine max, Lycopersicon esculentum, and Ricinus communis, the position equivalent to Tyr-51 is occupied by phenylalanine. However, all these proteins contain serine residues in the T loop at the equivalent of positions 49 and 52 that could potentially be phosphorylated. Hence, nitrogen and/or carbon status could be sensed through the activity of a serine-threonine kinase and its associated phosphatase, which are common regulatory components of signal transduction pathways in eukaryotes (221). Transgenic plants overexpressing A. thaliana GlnB exhibit a phenotype consistent with the involvement of GlnB in perceiving the status of carbon and nitrogen (97).
Role of small effector molecules.
The interaction of GlnB with three proteins, UTase/UR, GS, and NtrB, has been studied in depth, and the mechanisms that control these interactions are now beginning to be elucidated. In all cases the interactions depend on the nitrogen and carbon/energy status of the cell, which is signaled primarily by three small effector molecules, glutamine, 2-ketoglutarate, and ATP.
The ratio of glutamine to 2-ketoglutarate in the cell rises in conditions of nitrogen sufficiency and drops in conditions of nitrogen starvation (129, 205). Consequently, it was originally thought that the ratio between 2-ketoglutarate and glutamine was the critical factor influencing the uridylylation state of GlnB. However, detailed analysis in vitro showed that the concentration of 2-ketoglutarate in the cell is always sufficient for uridylylation to occur (117, 118). It has since been established that while uridylylation of GlnB requires 2-ketoglutarate and ATP and is repressed by glutamine, deuridylylation also requires ATP and 2-ketoglutarate but is stimulated by glutamine (117, 118). A low concentration of ATP or GlnB limits the uridylylation reaction, but the addition of excess GlnB does not inhibit it. This suggests that the binding of ATP to GlnB is required for GlnB to bind to UTase (108). ATP and 2-ketoglutarate are also required for the deuridylylation of GlnB-UMP, and there is evidence to suggest that this also involves their binding to GlnB (108).
Hence, glutamine is the nitrogen signal, and at physiological levels it regulates UTase/UR independently of 2-ketoglutarate (109). Glutamine inhibits uridylylation of GlnB by affecting the rate of the transfer reaction. Glutamine stimulates deuridylylation of GlnB-UMP fourfold in the presence of Mg2+, and there is evidence that glutamine interacts with UTase/UR at a single site. Both UTase and UR activities can also be stimulated by a second metal ion cofactor, Mn2+, but this is not thought to be physiologically significant for either reaction (108).
The main carbon signal appears to be 2-ketoglutarate, for which the trimeric GlnB molecule has three binding sites. The physiological significance of 2-ketoglutarate becomes clear when the interaction of GlnB with NtrB is investigated. The dephosphorylating activity of NtrB (and therefore the inactivation of NtrC) is stimulated by interaction with GlnB. This interaction is inhibited in two ways, first, by the uridylylation of GlnB, which, as discussed above, is dependent on the glutamine concentration of the cell, and second, by the binding of 2-ketoglutarate directly to GlnB. In contrast to the effect of 2-ketoglutarate on the uridylylation state of GlnB, the concentration of 2-ketoglutarate that affects the GlnB-NtrB interaction is significant within a physiologically relevant range, 0.1 to 0.9 mM (109), and very low levels of 2-ketoglutarate were found to stimulate the interaction of GlnB with NtrB in vitro (118). In vitro experiments reexamining the regulation of NtrB by GlnB in the presence of a low 2-ketoglutarate concentration showed that GlnB is an inhibitor of the kinase activity (107). Under these conditions, GlnB actively binds NtrB and activates its phosphatase activity, whereas at high 2-ketoglutarate concentrations, GlnB does not bind efficiently to NtrB, does not activate the phosphatase activity, and releases inhibition of NtrB kinase activity. As for other transmitter proteins (98, 99, 142, 197), it has been proposed that the kinase and phosphatase activities of NtrB are coordinately and reciprocally regulated (107).
GlnB also controls the adenylylation state of GS in response to glutamine and 2-ketoglutarate concentrations. Here again the glutamine concentration determines the uridylylation state of GlnB (GlnB is required for adenylylation and GlnB-UMP is required for deadenylylation of GS), but in addition to this control, 2-ketoglutarate binding prevents the stimulation of ATase activity by binding directly to GlnB and preventing the GlnB-ATase interaction (110). Moreover, glutamine alone stimulates the adenylylation of GS even at low glutamine concentrations, and GlnB is required to prevent GS adenylylation under these conditions (110).
In summary, the antagonistic effect between 2-ketoglutarate and glutamine in the regulation of either the GS adenylylation state or NtrC-dependent promoter expression is due to the effects of glutamine on the UTase and ATase enzymes and the effect of 2-ketoglutarate on the conformation and activity of GlnB. A high concentration of 2-ketoglutarate is required for the binding of GlnB to UTase/UR, for stimulation of the adenylyl-removing activity of ATase, and for inhibition of the interaction with NtrB. Conversely, lower 2-ketoglutarate levels promote interaction of GlnB-UMP with UTase/UR, activating deuridylylation, stimulation of ATase, and enhanced binding of GlnB to NtrB. This is explained by the fact that 2-ketoglutarate binding results in a strong negative cooperativity for the binding of additional 2-ketoglutarate molecules, so that the protein only becomes saturated with 2-ketoglutarate at the high end of the physiological range. Consequently, GlnB may exist in different conformations depending on the number of 2-ketoglutarate molecules bound (107). Mutagenesis experiments suggest a mutual influence of 2-ketoglutarate binding and T-loop conformation (111), but further structures with different combinations of effectors bound to the protein are likely to shed more light on how effector binding influences PII structure. The potential for a regulatory function of ATP binding to PII proteins has also been remarked upon by Xu et al. (252). The ATP-binding site is highly conserved in PII proteins, and binding of ATP and 2-ketoglutarate to GlnB is synergistic; hence, there is the potential to integrate signals that reflect both the carbon and the energy status of the cell.
Like GlnB, GlnK appears to bind one molecule of 2-ketoglutarate with a high affinity and is allosterically regulated by 2-ketoglutarate. Moreover, it seems that the binding of additional effector molecules at high 2-ketoglutarate concentrations results in another conformation of GlnK that is unable to interact with NtrB or ATase. However, unlike GlnB, low concentrations of 2-ketoglutarate do not stimulate GS adenylylation by GlnK (14).
Apart from the E. coli PII proteins, the only other in vitro studies have been carried out with Synechoccocus GlnB. This protein also binds 2-ketoglutarate and ATP, and as with E. coli GlnB, high-affinity binding of 2-ketoglutarate requires ATP (66, 67, 118). Studies of ATP-binding characteristics show that in the absence of 2-ketoglutarate, Synechococcus GlnB binds ATP with a lower affinity than E. coli GlnB. However, higher concentrations of 2-ketoglutarate reduce this difference, and by using an ATP competitor, 2-ketoglutarate can be shown to increase the specificity of the ATP-binding site. Under physiological conditions the 2-ketoglutarate concentration might determine the ligand status of GlnB, and ATP could function as a cofactor. Hence, as in E. coli, the primary function of GlnB in this Synechococcus sp. could be to sense 2-ketoglutarate (67).
When growing with CO2 as a carbon source, cyanobacteria use the reductive tricarboxylic acid (TCA) cycle to form 2-ketoglutarate directly. The only 2-ketoglutarate-consuming reaction is conversion to glutamate by GOGAT, and therefore changes in carbon and nitrogen assimilating reactions will immediately influence the pool size of 2-ketoglutarate. The central position of this metabolite may explain why cyanobacteria rely on 2-ketoglutarate as a sensor of carbon and nitrogen status (66). In contrast, in organisms using the oxidative TCA cycle, e.g., most proteobacteria, the 2-ketoglutarate pool depends on the flux through the TCA cycle, which might explain the need to use a nitrogen-containing metabolite, namely glutamine, as an indicator of nitrogen status.
ROLE OF PII PROTEINS IN GENETIC AND METABOLIC CONTROL
Although PII was originally discovered as a consequence of studies on factors controlling the activity of GS in E. coli, PII proteins are now known to have many complex roles in the regulation of microbial nitrogen metabolism. The mutiplicity of possible roles for PII proteins is presented schematically in Fig. 4. The recognition that many organisms have multiple PII genes has added a further layer of complexity, and it is extremely likely that there are still new roles for PII waiting to be discovered. In the following sections we discuss those functions of PII that have already been recognized and consider the evidence for other possible targets.
FIG. 4.
Schematic representation of the potential mutiplicity of roles for PII proteins. This cartoon summarizes information (both definitive and predicted) obtained from many different organisms.
Regulation of GS Activity
The most common form of GS in bacteria is termed GSI. It is encoded by glnA and consists of 12 identical subunits of 55 kDa, which form two superimposed hexagonal rings (256; for a review, see reference 155). In a number of species, including E. coli, GSI is posttranslationally modified by the adenylylation of a tyrosine residue (Tyr-397) on each subunit by the ATase enzyme encoded by glnE (64, 207). This allows the progressive inactivation of GSI as each subunit is independently inactivated in response to increasing levels of intracellular nitrogen. It has been proposed that the physiological significance of the posttranslational regulation of GS is to prevent a sustained decrease in the ATP concentration and to protect the cellular glutamate pool, allowing rapid growth (129, 204).
ATase is one of the best characterized of the known PII targets, but there is presently no information on the precise site of interaction of GlnB with ATase. The gene for ATase (glnE) has been cloned from E. coli and sequenced, and the protein has been shown to be a monomer of 115 kDa (232). Like UTase, ATase is a bifunctional enzyme catalyzing either the addition of AMP to Tyr-397 of GS using ATP as substrate or the removal of the adenylyl group, which results in the release of ADP. Separation of the two antagonistic activities of the protein, adenylylation and deadenylylation, has been demonstrated by the formation of two truncated proteins. The N-terminal domain of ATase deadenylylates GS, and this activity is regulated by GlnB-UMP and 2-ketoglutarate but is not affected by glutamine, whereas the C-terminal domain adenylylates GS in a glutamine-dependent manner. GlnB is required for the latter activity to occur in the complete enzyme but not when only the C-terminal region is present, giving rise to the hypothesis that the N-terminal region masks the adenylylation site and that GlnB binding causes a conformational change which exposes this site (102). glnE genes have also been found and sequenced in Streptomyces coelicolor (58), Haemophilus influenzae (62), Mycobacterium tuberculosis (38), Neisseria meningitidis (accession no. AE002380), Pseudomonas aeruginosa (accession no. U63816), and Aquifex aeolicus (accession no. AE000755).
A second form of posttranslational modification of GS in response to nitrogen status, ADP-ribosylation, has been identified in Rhodospirillum rubrum, Streptomyces griseus, and Synechocystis sp. strain. PCC6803 (181, 209, 244), although the physiological significance of this has not yet been determined. In Synechocystis sp. strain PCC6803, reversible inactivation of GSI is also controlled by interaction with two inhibitory polypeptides, GifA and GifB (77, 78). PII has not been implicated in either of these regulatory processes.
Other forms of GS in addition to GSI have been found in a number of organisms. GSII is an octamer of 36-kDa subunits encoded by glnII; GSIII, encoded by glnN, is a hexamer with a subunit molecular mass of 75 kDa; and the fourth form of the enzyme is an octamer of 47 kDa encoded by glnT. None of these enzymes has been reported to be adenylylated, and to date PII has not been implicated in regulation of their activities.
Global Nitrogen Regulation (Ntr) System
As discussed in detail above, there were early indications from the analysis of glnB mutants that PII played a role in addition to the modulation of ATase activity. A combination of genetic and biochemical experiments subsequently led to the recognition that in enteric bacteria (and indeed in most, if not all, of the proteobacteria), expression of nitrogen assimilation and catabolism genes is coordinated by a central regulatory system (147, 155, 190). This global nitrogen regulatory (Ntr) system was originally considered to consist of four proteins, UTase/UR, GlnB, and the proteins of the two-component histidine protein kinase system, NtrB and NtrC, which in enterics are cotranscribed in the glnA-ntrBC operon. The later discovery of GlnK in E. coli and many other bacteria increased the complexity of the system, although it also explained some previous anomalies. For clarity, the role of GlnB in the Ntr system (Fig. 1) will be described first, and the role of GlnK and its importance in both genetic and metabolic control will then be considered.
Enteric Ntr model.
Activation and repression of nitrogen-regulated genes is coordinated by the action of the central regulatory protein NtrC, a typical ς54-dependent transcriptional activator protein (166). The protein comprises a DNA-binding carboxy-terminal domain, a highly conserved central domain required for the activation of transcription, and an N-terminal domain which is characteristic of two-component response regulator proteins (178, 220). These proteins have a highly conserved tertiary structure in the N-terminal region, containing an aspartic acid residue, Asp-54, which, in the case of NtrC, is phosphorylated in response to low nitrogen, giving NtrC-P, the activated form of the protein (121, 239, 242). NtrC exists as a dimer in its nonphosphorylated form, but for the activation of transcription to take place, oligomerization to a tetramer or higher-order oligomer must occur. Phosphorylation induces both DNA binding and the oligomerization of NtrC, and many NtrC-dependent promoters contain more than one binding site, facilitating oligomerization (18, 156, 240, 241, 251).
Control of NtrC activity in response to nitrogen status is mediated by NtrB, which acts as a phosphate donor to NtrC. NtrB is a 36-kDa protein comprising two distinct domains, the N-terminal sensor domain and the C-terminal kinase domain, typical of histidine protein kinases (168, 178). NtrB exists as a dimer which is autophosphorylated on the conserved histidine (His-139) at the amino end of the C-terminal domain (169, 171). ATP binds to one subunit, and phosphorylation occurs on the conserved histidine residue of the second subunit (172).
NtrB is stimulated to dephosphorylate NtrC in the presence of GlnB and ATP. This is termed regulated phosphatase activity (117, 118, 120, 140, 170). Mutations in ntrB have shown that the kinase and phosphatase activities of NtrB are separate. Mutation of His-139 to arginine or mutation of the ATP-binding site prevents and significantly slows kinase activity, respectively, but does not affect the phosphatase activity of the protein. However, in these mutants GlnB is not required to stimulate phosphatase activity (17, 116). Other mutations in the N-terminal domain of NtrB affect the interaction of GlnB without affecting kinase activity (117). These data suggested that PII interacts with the N-terminal domain of NtrB, but subsequent cross-linking experiments indicated that PII regulates both the kinase and phosphatase activities of NtrB by binding to the kinase domain of the NtrB transmitter module (183). Functional dissection of the activities of NtrB confirmed this model, showing that NtrB consists of three domains: an N-terminal domain involved in signal transduction, a central phosphotransferase/phosphatase/dimerization domain, and a C-terminal kinase domain. Binding of PII to the kinase domain appears to result in an altered conformation that is transmitted to the other two domains and causes the central domain to assume a conformation with potent phosphatase activity (106).
The modification of GlnB in response to the cellular nitrogen status provides the intracellular switch that in turn regulates the phosphatase and kinase activities of NtrB and hence the transcriptional activity of NtrC. In nitrogen excess, unmodified GlnB stimulates dephosphorylation of NtrC by NtrB, and conversely, in nitrogen starvation, when GlnB is modified, GlnB-UMP no longer interacts with NtrB and the kinase activity predominates, so that NtrC is phosphorylated and transcriptionally active. In enterics, a number of genes are transcriptionally regulated by NtrBC; these include the glnK-amtB operon (13, 101, 231), the glutamine transport glnHPQ operon (37), the arginine catabolism astCADBE operon (202), the nac regulatory gene (167), S. enterica serovar Typhimurium argT (200), the nitrogen fixation regulatory genes nifLA, and the nasR and nasFEDCBA genes of K. pneumoniae (52, 136, 250).
As discussed earlier, the discovery of GlnK in E. coli helped to explain a number of anomalies in the Ntr model and rationalized certain aspects of regulation that had been observed in the absence of GlnB. Nevertheless, the problem of determining the primary physiological role of GlnK remains. Studies of single (glnB or glnK) and double (glnB glnK) mutants give some information, though by definition they do not relate to the normal physiological situation. Likewise, in vitro data can shed light on the potential activities of the proteins but do not necessarily identify their primary function. These observations are even further complicated by the recognition that GlnB and GlnK can from heterotrimers in vivo under normal physiological conditions, so that given the potential for uridylylation of both GlnB and GlnK, a large number of different PII molecules may exist in the cell in different situations.
In vivo, GlnK can complement the glnB defect in deadenylylating GS (13, 231). However, in vitro experiments show that GlnK is less efficient than GlnB in activating GS adenylylation, suggesting that in vivo, when GlnB is present, GlnK may not play a significant role in regulating ATase (14, 231). Interestingly, under some physiological conditions, neither GlnB nor GlnK is necessary for GS adenylylation (68). In vitro, GS adenylylation can be triggered by glutamine alone (110), and the adenylyl transfer process of the carboxy-terminal domain of ATase is activated by glutamine (102). Moreover, adenylylation activity of the C-terminal domain is independent of GlnB (or GlnB-UMP), whereas in the intact enzyme, GlnB is required for this activity (102). Hence, depending on the physiological conditions of the cells, the adenylylation of GS may be PII dependent or PII independent (68).
GlnK, like GlnB, can act through NtrB to regulate NtrC-P concentration, but GlnK appears to be less potent than GlnB in controlling the level of NtrC-P. Hence, GlnK may play some role in the regulation of Ntr promoters requiring a high concentration of NtrC-P (e.g., E. coli pglnK), although in K. pneumoniae pglnK expression is constitutive in the absence of GlnB (101). By contrast, GlnK has little effect on expression from the glnAp2 promoter, which is extremely sensitive to NtrC-P, which is consistent with glnA expression in the absence of GlnB (13, 14, 31).
The precise physiological role of GlnK may well not be determined until the full complement of potential PII targets is known. However, the conserved linkage of glnK to the ammonium transporter gene amtB may suggest one possible target (see later).
Regulation of the Ntr system in other organisms.
Outside the enterics, the role of the Ntr system has been studied almost exclusively in diazotrophic members of the proteobacteria. In all of these organisms except Azotobacter vinelandii, both GlnB and GlnK are present, although the phenotypes of both single and double mutants have not always been reported. Nevertheless, in many cases the roles of GlnB and GlnK are similar to those reported in E. coli.
An investigation of GlnB function in Sinorhizobium meliloti using both an in-frame glnB null mutation and a Y51F point mutation showed that in free-living conditions, GlnB is required for the expression of the NtrC-dependent GSII gene (glnII) and for regulation of the adenylylation state of GSI. As in enterics, GSI adenylylation is stimulated by native GlnB, whereas GlnB-UMP induces GSI deadenylylation. Furthermore, nonuridylylated GlnB prevents expression of NtrC-dependent genes such as glnII, presumably by stimulating NtrB phosphatase activity. However, unlike the enteric bacteria, a GlnB-deficient strain shows no glnII expression, suggesting that GlnB-UMP is required either for NtrC phosphorylation or for expression of ntrC itself (8). Unlike Sinorhizobium meliloti, a Rhizobium leguminosarum ΔglnB mutant exhibits constitutive expression of NtrC-dependent promoters, suggesting that in this organism, as in E. coli, the absence of GlnB results in NtrC activation independent of the N status (4).
In Azospirillum brasilense, neither GlnB nor GlnK is essential for GSI adenylylation or deadenylylation (47, 50). Moreover, a glnB mutation results in constitutive expression of NtrC-dependent promoters, such as that for glnK (47).
In Azorhizobium caulinodans, GlnB and GlnK are not necessary for GSI adenylylation, whereas both are required for complete GS deadenylylation. In the free-living state and under nitrogen-fixing conditions, the high level of GS adenylylation in a ΔglnBK background results in ammonium excretion. These results suggest that in this organism, GlnB and GlnK are interchangeable in regulating deadenylylation of GS (159).
In Rhodospirillum rubrum, regulation of GS adenylylation and deadenylylation was initially studied with purified GlnB and GlnB-UMP and extracts from R. rubrum grown in different conditions. Unlike the situation in Azorhizobium caulinodans, native GlnB and glutamine stimulate adenylylation of GS and 2-ketoglutarate totally inhibits the process, whereas none of the effectors tested induce the deadenylylation process (115). Mutations in glnB have no effect on glnA expression or GS modification, but the effects of glnK and glnB, glnK mutations have not yet been studied (261).
In Azotobacter vinelandii, GS is the only route for ammonium assimilation, and the organism cannot take up glutamine; therefore, glnA null mutations are lethal (224). Several attempts to construct a glnK null mutant of A. vinelandii also failed (153), and as a stable glnD-deficient strain can only be maintained in the absence of GS adenylylation, it seems that in A. vinelandii, GlnK-UMP may be absolutely required for deadenylylation of GS (39, 196).
Nitrogen Control in Other Bacteria
PII proteins are of course not restricted to the proteobacteria, but their roles in other bacteria are much less well understood. Nitrogen control systems have been characterized in members of the actinobacteria, the cyanobacteria, and the firmibacteria, but only in the actinobacteria (e.g., Corynebacterium) does it appear that PII may have a central regulatory role.
Corynebacterium glutamicum.
In Corynebacterium glutamicum, nitrogen-regulatory protein AmtR regulates transcription of the amtB-glnK-glnD operon and the amt gene. AmtR represses transcription of these genes in the presence of ammonium, and repression is relieved upon nitrogen starvation (105). AmtR does not respond to nitrogen limitation in a glnB deletion strain, indicating that GlnB acts directly or indirectly to signal the nitrogen status to AmtR (A. Burkovski, M. Jacoby, and R. Krämer, personal communication).
Cyanobacteria.
As in enteric bacteria, ammonium is a good nitrogen source for cyanobacteria, and in its presence, these organisms are unable to assimilate alternative inorganic nitrogen sources such as nitrate, nitrite, or dinitrogen. However, no homologues of the ntrBC genes are found in cyanobacteria, and nitrogen control is mediated by the NtcA protein, which belongs to the Crp family of transcriptional regulators (90b, 144, 237, 238). Moreover, GS activity is not regulated by reversible adenylylation but by protein interaction with two inhibitory peptides (77, 78). Despite the fact that no GlnB targets have been identified in cyanobacteria, the phenotype of a glnB mutant of Synechococcus sp. strain PCC7942 suggests that GlnB does not modulate the activity of NtcA (70). Indeed, the phosphorylation state of GlnB depends on NtcA (132, 198) as well as 2-ketoglutarate (66, 71, 100).
Bacillus subtilis.
There is no evidence for an NtrBC system in the firmibacteria, and despite some minor similarities with the enteric system, namely the presence of a ς54-like sigma factor (ςL) and a GlnK homologue (NrgB), B. subtilis controls nitrogen assimilation in a quite different manner (60). The GlnK homologue NrgB does not seems to regulate the activity or expression of any ςL-dependent transcriptional activators, and the only phenotype reported for a ΔnrgB mutant is one of slow growth on nitrate (79, 246).
In B. subtilis, nitrogen metabolism is not governed by a single global nitrogen-regulatory system but by three regulatory proteins, GlnR, TnrA, and CodY, which are active under different nutritional conditions. These multiple systems allow noncoordinate gene expression under different growth conditions. GlnR represses gene expression during growth in nitrogen excess, whereas TnrA activates or represses gene transcription during nitrogen-limited conditions, and finally, CodY represses transcription in cells growing rapidly with amino acids. CodY-dependent regulation appears to reflect the total nutritional state of the cell (61), whereas GlnR and TnrA respond to nitrogen availability. None of the signals regulating the activity of these three proteins has been identified, but GlnK (NrgB) has not been implicated, and the only factor so far known to be involved in the regulation of GlnR and TnrA activity is GS (249).
Regulation of Nitrogen Fixation
For those bacteria that are capable of diazotrophic growth, the fixation of atmospheric nitrogen can be a major route of nitrogen assimilation, and in nearly all cases PII proteins appear to play a significant role in the regulation of this process. Due to the sensitivity of the nitrogenase enzyme to oxygen and the high energy requirement for the process of nitrogen fixation, the nitrogen fixation activity of diazotrophs is regulated in response to both the redox and nitrogen status of the cell, so that nitrogen fixation occurs only when it is both favorable and necessary. Nitrogen regulation of nitrogen fixation has been studied most extensively in the proteobacteria, and it is becoming increasingly apparent that PII proteins play a pivotal role in this process. The primary mode of regulation is by control of the transcription of the nitrogen fixation (nif) genes, and this can be achieved by regulating both the expression and the activity of the transcriptional regulator. Some organisms also regulate the activity of the nitrogenase enzyme at a posttranslational level in response to ammonia and other fixed nitrogen sources, and here again PII proteins play their part.
nif-specific regulator NifA.
In diazotrophs of the α and γ proteobacteria, nif gene transcription is initiated at ς54-dependent promoters in conjunction with a specific transcriptional activator, NifA (51, 59, 154). Control of nif transcription is mediated by regulating either the expression or the activity of NifA in response to oxygen and fixed nitrogen. Nearly all NifA proteins have a three-domain structure typical of ς54-dependent transcriptional activators (53, 166). The three domains are functionally different, and experiments have shown that isolated domains are structurally and functionally independent (22, 23, 54). The carboxy-terminal domain contains a conserved helix-turn-helix motif which is involved in DNA binding (160, 165). The central domain contains the site for ATP binding and for interaction with ς54 (81, 166). The N-terminal domain of ς54-dependent activators is highly variable and is typically the target for regulatory signals.
In the γ proteobacteria, the product of an additional gene, nifL, is required to regulate NifA activity in response to both oxygen and fixed nitrogen. NifL proteins have some similarity to the sensor components of the two-component sensor regulator systems, such as NtrB, having N-terminal sensor and C-terminal transmitter domains tethered by a flexible Q linker. However, there is no evidence that NifL is a histidine protein kinase; indeed, the appropriate histidine residue is absent in K. pneumoniae NifL, and in Azotobacter vinelandii, NifL, where it is present, it is not required for regulation of nif gene expression (245). Both A. vinelandii and K. pneumoniae NifL are flavoproteins with flavin adenine dinucleotide as the prosthetic group (51, 91, 201). The NifL and NifA proteins form a complex in vivo and in vitro, and stoichiometric levels of the two proteins are required for effective modulation of NifA activity (82, 90, 162).
Regulation of nifA and nifLA transcription.
The factors that regulate nifA transcription reflect the lifestyle of the organism, so that in those that can fix nitrogen in the free-living state, the availability of fixed nitrogen is a major factor, whereas in symbiotic diazotrophs, the critical factor controlling nifA expression is the availability of oxygen.
Among the free-living diazotrophs, regulation of nifA expression varies considerably. In K. pneumoniae, nifLA transcription is nitrogen regulated by NtrC and is therefore dependent on GlnB (52, 95), while in Azotobacter vinelandii, nifLA transcription is independent of nitrogen sources (24, 186). In both Herbaspirillum seropedicae and Rhodobacter capsulatus, nifA expression is regulated with respect to nitrogen by NtrC, and hence GlnB may be involved (57, 72, 215). By contrast, nifA expression in Azospirillum brasilense is independent of NtrC and only partially repressed by either ammonium or oxygen, maximal repression being achieved by the synergistic effect of both effectors (57). In Rhodospirillum rubrum, nifA transcription appears to be independent of nitrogen source, and nitrogen control of nif gene expression is only exerted through NifA activity (261).
In the symbiotic diazotrophs of the genera Rhizobium, Bradyrhizobium, and Azorhizobium, nifA expression is predominantly regulated in response to oxygen availability (for a review, see reference 59). However, in Azorhizobium caulinodans, nifA expression is also regulated in response to nitrogen (187). This may be due to the fact that A. caulinodans can fix nitrogen not only symbiotically within root or stem nodules of Sesbania rostrata but also in a free-living state where the source of nitrogen is less reliable. Nitrogen control of nifA expression in A. caulinodans is mediated by two nitrogen-responsive systems, NtrB/C and NtrY/X (179, 180). In glnB or glnK mutants, nifA is expressed only during nitrogen-fixing conditions, whereas in a ΔglnBK mutant, nifA is expressed under nitrogenase-derepressing conditions and in the presence of ammonia. Consequently, it has been suggested that GlnB and GlnK are involved in the repression of NifA synthesis (159).
Nitrogen control of NifA activity.
Whereas the role of PII proteins is somewhat variable with respect to regulation of nifA expression, a much more consistent picture is emerging regarding the function of these proteins in regulating NifA activity. In this context, information comes from studies with Azospirillum brasilense, Herbaspirillum seropedicae, Rhodospirillum rubrum, and Azorhizobium caulinodans.
The first indication that a PII-like protein might be involved in the nitrogen regulation of NifA activity came from the observation that in A. brasilense, a glnB mutant was Nif− (135), while a glnK mutation had no effect on nitrogen fixation (47). As the glnB mutation did not affect nifA expression, it appeared that GlnB is required to maintain the active form of NifA. Deletions within the N-terminal domain of NifA restore nif gene expression, suggesting that GlnB is required to activate NifA by preventing the inhibitory affect of its N-terminal domain (11). Mutation of residue Tyr-18 to Phe in the N-terminal domain of NifA results in an active NifA that does not require GlnB; however, whether GlnB interacts directly with NifA or modulates the activity of another protein that in turn regulates NifA activity remains unsolved (12). A glnB Y51F mutant and a glnD mutant both exhibit a Nif− phenotype (12, 227), which is consistent with the fact that during nitrogen fixation, A. brasilense GlnB is uridylylated (47) and suggests that it is GlnB-UMP which is required for NifA activation.
A similar situation occurs in H. seropedicae: a glnB mutant is Nif−, while nifA expression, which is NtrC dependent, would be expected to be constitutive in this background (21). Studies of the H. seropedicae NifA protein in vivo show that the full-length protein expressed in A. brasilense is active only under low oxygen and in the absence of ammonium, but NifA is not active when expressed in E. coli or K. pneumoniae (217). By contrast, the amino-terminally truncated form is still active in the presence of ammonium in A. brasilense, E. coli, and K. pneumoniae, again indicating that the N-terminal domain is involved in nitrogen control. Furthermore, when expressed in trans, this domain can inhibit the activity of the truncated NifA (163, 164, 217). If GlnB were to interact with the N-terminal domain, then the inactivity of H. seropedicae NifA in E. coli could be due to the absence of the cognate PII.
In R. rubrum, a ΔglnB mutant has no nitrogenase activity, whereas a glnB Y51F mutant shows about 10% of wild-type nitrogenase activity. Expression of R. rubrum nifA from a multicopy plasmid does not restore nitrogenase activity in a ΔglnB mutant, whereas a glnB Y51F mutant is complemented. These results suggest that GlnB is essential for NifA activity, but it is not clear whether the effect of GlnB on NifA is direct or indirect (261). An attractive model is that modified GlnB leads to activation of NifA, while unmodified GlnB inhibits NifA. However, the role, if any, of GlnK in this system has still to be investigated.
The situation is somewhat different in A. caulinodans. Single glnB and glnK mutants express NifA constitutively but behave like the wild type and show ammonium repression of nif expression. However, the glnBK double mutant shows derepressed nif expression in ammonium, indicating that in this case NifA activity can be controlled by either GlnB or GlnK (159) During nitrogen fixation conditions, glnB and glnK are dispensable (even if a glnBK mutant is less able to activate nif expression), and unlike H. seropedicae or A. brasilense NifA, an amino-terminally truncated NifA is inactive (159).
The theme of PII proteins being involved in modulating the activity of NifA in response to nitrogen availability has now been extended to the γ proteobacteria. Initial studies in K. pneumoniae indicated that neither GlnB nor GlnD played a direct role in NifL-mediated regulation of NifA activity in response to fixed nitrogen (55, 95). However, in Azotobacter vinelandii, GlnD has been shown to be necessary for the sensing of nitrogen status by the A. vinelandii NifLA system, and the Nif− phenotype of a glnD mutant was suppressed by a second mutation in nifL, indicating that UTase is involved in NifL-dependent regulation of NifA activity (42).
In their studies on the role of GlnB in nif regulation, Holtel and Merrick (95) concluded that at least one other nitrogen-sensing system is present in K. pneumoniae in addition to GlnB, and Edwards and Merrick subsequently proposed that this system might involve an alternative PII-like protein (55). The discovery of a GlnK in E. coli (231) offered just such a candidate, and the demonstration by He et al. (88) that derepression of NifA activity is dependent on NtrC was then entirely consistent with a role for GlnK. Genetic studies in both E. coli and K. pneumoniae indicated that the K. pneumoniae NifL-NifA interaction is indeed modulated by the presence of GlnK in such a way that in N-limiting conditions, GlnK is required to relieve the inhibitory effect of NifL on NifA (89, 101). In an E. coli glnBK double mutant, the modulation of NifA activity by NifL is dependent on the concentration of GlnK, and when overproduced to nonphysiological levels, GlnB can substitute for GlnK (10). The effect of GlnK is independent of its uridylylation state, which raises the question of how NifL inhibition is rapidly reestablished in response to elevated N status. Studies of the NifL-NifA interaction in E. coli suggest that in normal physiological conditions, GlnB can counteract the positive action of GlnK, and it is possible that this negative effect might be mediated by GlnB-GlnK heterotrimer formation (10). Site-directed mutagenesis of GlnB indicates that residue 54 in the T loop is the single most important amino acid in determining the discrimination between GlnK and GlnB proteins in the context of the modulation of NifA activity. A single alteration of GlnB Asp-54 to Asn, found at this position in GlnK, makes GlnB quite effective in regulating the NifLA interaction, and if combined with a second T43A alteration, the resultant GlnB D54N T43A protein mimics the GlnK protein almost perfectly in relieving NifL inhibition. It has been proposed that the D54N T43A changes modify the characteristics of the T loop in such a way that the altered GlnB protein is now fully competent to interact with the target, be that NifL or NifA or the complex of both (9).
While PII is also implicated in modulating NifLA control in Azotobacter vinelandii, the situation differs from that in K. pneumoniae. A. vinelandii NifL responds to the availability of fixed nitrogen when expressed in E. coli (217), but studies in vivo and in vitro indicate that neither GlnB nor GlnK is required for derepression. However, in N excess conditions, the inhibitory function of NifL is dependent on either of the nonmodified forms of E. coli GlnB or A. vinelandii GlnK (192). In vitro analysis indicates that the inhibitory activity of A. vinelandii NifL is stimulated by interaction with the nonuridylylated form of GlnK and that the NifL-NifA system is also directly responsive to 2-ketoglutarate (138). Interestingly, sequence comparison of T-loop sequences suggests that the A. vinelandii T loop is similar to the E. coli GlnB T loop, which is consistent with the fact that it is E. coli GlnB rather than E. coli GlnK that is effective in regulating A. vinelandii NifLA function.
Hence, while a common theme is emerging of regulation of NifA activity by a PII protein, the precise details, e.g., whether GlnB or GlnK or both are involved, whether they act in concert with NifL or independently, and whether they activate or inactivate NifA, appear to differ from one organism to another. It is attractive to imagine that a common mechanism could underlie this variety, but at present it is difficult to suggest a unified model, and the challenge now is to elaborate the mechanism of regulation, preferably in more than one system.
Regulation of nitrogenase activity.
Just as GS and NifA can be regulated at the level of transcription and activity, so at least some organisms can regulate both the expression and the activity of the nitrogenase enzyme, here again PII is now recognized as a key regulator. A number of diazotrophic α proteobacteria, including Rhodospirillum rubrum, Rhodobacter capsulatus, Azospirillum brasilense, and Azospirillum lipoferum can regulate the activity of nitrogenase by ADP-ribosylation in response to ammonia, energy limitation, or anaerobiosis shifts (86, 133, 143, 152, 258, 260). This is a reversible reaction driven by the enzymes dinitrogenase reductase ADP-ribosyl transferase (DRAT), which ribosylates a specific arginine residue (Arg-101) of one subunit of the dinitrogenase reductase homodimer, inactivating the enzyme, and dinitrogenase reductase activating glucohydrolase (DRAG), which removes the ribosyl group, reactivating the enzyme (143). The R. rubrum DRAT and DRAG system has been shown to function in K. pneumoniae, in which the target residue Arg-101 of nitrogenase is conserved, and as no draG or draT genes have been identified in K. pneumoniae, it would appear that the sensing system that modulates DRAG and DRAT activity is also present in this organism (74).
In Rhodospirillum rubrum, addition of 10 mM ammonia during nitrogen fixation causes a slow decrease in nitrogenase activity, finally reaching 20 to 30% of the initial activity. In contrast, a glnB Y51F mutant shows a much more rapid loss of nitrogenase activity. Moreover, during nitrogen fixation conditions, a draG mutant exhibits normal nitogenase activity, whereas a glnB Y51F draG double mutant shows no nitrogenase activity and complete modification of the nitrogenase (261). Using a heterologous system in which DRAG and DRAT are expressed in K. pneumoniae, it has been shown that DRAT activity is unaffected in a glnK mutant, whereas regulation of DRAG activity is completely abolished, and DRAG stays active even after ammonia addition (261a). These results suggest that both GlnB and GlnK play an important role in regulating the DRAG-DRAT system. In Rhodobacter capsulatus, a glnB mutation does not affect nitrogenase switch-off, but a double glnB glnK mutant is impaired in switch-off (83, 125).
The phenomenon of switch-off has also been reported in Azorhizobium caulinodans, but it is not known whether this occurs by ADP-ribosylation (128). In Azospirillum brasilense and Rhodobacter capsulatus, a second mechanism for nitrogenase switch-off in addition to ADP-ribosylation has been shown to occur, and this could therefore also be true in other organisms (253, 255, 259). Nevertheless, in a glnBK double mutant of A. caulinodans, nitrogenase is active in the presence of 10 mM NH4+, whereas it is inactive in a wild-type strain. Hence, both GlnB and GlnK may be involved in the regulation of A. caulinaudans nitrogenase activity (159). As discussed earlier, A. caulinodans glnB and glnBA mutants induce Fix− nodules on Sesbania rostrata roots despite expressing a pnifH-lacZ fusion at a similar level to the wild type, a situation which again suggests control of nitrogenase activity by GlnB (158).
The phenomenon of ammonia switch-off is also found in the diazotrophic methanogens Methanosarcina barkeri and Methanococcus maripaludis (123, 141). However, as draG and draT homologues are not found in the genome of the related Methanobacterium thermoautotrophicum, the mechanism of this inactivation may well be novel. Despite this, PII proteins are again involved in the phenomenon. As discussed earlier, two glnB-like genes, nifI1 and nifI2, are located between the nitrogenase structural genes nifH and nifDK in a number of diazotrophic methanogens. In M. maripaludis, the involvement of these proteins in regulation of nif expression has been ruled out, but a mutant carrying an in-frame deletion of both nifI1 and nifI2 does not show the ammonia switch-off phenomenon; nitrogenase remains active after addition of ammonia, whereas in a wild-type strain nitrogenase activity is rapidly lost. Hence, at least one of the genes is involved in the switch-off process (123, 123a). A similar posttranslational control of nitrogenase may also be present in Clostridium acetobutylicum, Clostridium cellobioparum, and Desulfovibrio gigas, as glnB-like genes are also linked to nifH in these organisms.
Regulation of Transport Systems
The transport of nitrogenous compounds into the cell is another important process in nitrogen metabolism, and here again evidence is emerging for control by PII proteins.
Nitrate transport.
PII-like proteins play a role in nitrate utilization or uptake in many organisms, including Bacillus subtilis, Rhizobium leguminosarum, Azospirillum brasilense, Synechococcus sp. strain PCC7942, and Synechocystis sp. strain PCC6803. glnB mutants of B. subtilis and R. leguminosarum, fail to utilize nitrate as a sole nitrogen source, suggesting that GlnB might participate in nitrate utilization (4, 246). A glnB mutant of A. brasilense excretes ammonium when cells are grown in the presence of nitrate. It has been proposed that this effect could be linked to deregulation of the nitrate assimilation pathway (which is regulated by the ntr system), with the consequent accumulation of intracellular ammonium leading to ammonia excretion (134). This phenotype is restored by complementation with glnB or elevated expression of GS (47).
In the cyanobacterium Synechococcus sp. strain PCC7942, the nitrate/nitrite ABC transporter and nitrate reductase are rapidly inhibited upon addition of ammonium (126). In a glnB mutant, nitrate uptake activity remains high after addition of ammonium and a glnB S49A mutant (which mimics the unphosphorylated form of GlnB) does not exhibit any nitrate uptake regardless of nitrogen source. This suggests that GlnB is involved in control of the short-term inhibition of either nitrate uptake or nitrate reductase activity (131). Both glnB S49D and S49E mutants (which mimic phosphorylated GlnB) behave like the wild-type strain, indicating that both GlnB forms have a negative effect on nitrate uptake or assimilation. Because the “phosphorylated-like” forms of GlnB allow nitrate uptake only in the absence of ammonia, it has been suggested that an additional effector signaling N deficiency (such as 2-ketoglutarate) is required to establish the noninhibitory form of phosphorylated GlnB (130, 131). As in Synechococcus sp. strain PCC7942, in Synechocystis sp. strain PCC6803, nitrate uptake is correlated with the phosphorylation level of GlnB. However, when cells are grown in the presence of low concentrations of inorganic carbon, nitrate uptake is not completely abolished even though GlnB is unphosphorylated. Moreover, a glnB mutant exhibits constitutive nitrate uptake, which is consistent with a role for GlnB in regulating nitrate uptake or assimilation (92). Deletion of the C terminus of one of the ATP-binding subunits (NrtC) of the transporter allows nitrate accumulation in the presence of ammonium (126), suggesting that this domain may interact with an effector protein, perhaps GlnB.
Another organism in which PII may control nitrate metabolism is Aquifex aeolicus. One of the two glnB-like genes in this organism is located immediately upstream of nasA (encoding a nitrate transporter) and narB (encoding a nitrate reductase). The gene organization suggests that all three genes are contranscribed, and as they are all potentially translationally coupled, it is quite likely that all three gene products are functionally related.
Methylammonium and ammonium transport.
The ubiquitous linkage of genes encoding the proposed high-affinity ammonium transporter AmtB and the PII protein GlnK has led to the proposal that these proteins are functionally related (223), and some evidence to support this comes from Azospirillum brasilense and Synechococcus sp. In A. brasilense, GlnK (Glnz) negatively regulates methylammonium transport (47). In this case, although amtB expression is regulated by the ntr system, GlnK is not required for NtrC-dependent regulation (228), and therefore it has been suggested that GlnK modulates the activity of AmtB (47). A glnB mutant of Synechococcus sp. strain PCC7942 exhibits a peculiar phenotype; no methylammonium transport is observed when cells are grown in the presence of nitrate, whereas in the absence of combined nitrogen, transport activity increases rapidly to reach a higher level than in a wild-type strain (70).
Other transporters.
The glnB gene of Haemophilus influenzae is located immediately upstream (3 bp) of an open reading frame (HI0338) that encodes a protein of unknown function. However, BLAST searches reveal that this protein is a member of a large class of integral membrane proteins represented in E. coli by YdgG and YhhT. The tight linkage between glnB and HI0338 and their presumptive cotranscription is highly suggestive of a functional relationship and might imply that H. influenzae YhhT is involved in the transport of some nitrogenous compound.
Inorganic carbon uptake.
In Synechocystis sp. strain PCC6803, inorganic carbon (Ci) uptake involves a constitutively expressed low-affinity transporter and a high-affinity transporter expressed under a low concentration of Ci. In a glnB mutant, the activity of the Ci high-affinity transporter (as measured by bicarbonate uptake) is constitutive, and the regulation appears to be independent of GlnB phosphorylation. It has been proposed that in the glnB mutant, the absence of a 2-ketoglutarate–GlnB complex could activate or repress the high-affinity transporter even when the Ci concentration is high (92). Because GlnB also controls nitrate uptake in Synechocystis sp. strain PCC6803, the protein appears to be involved in the coordination of carbon and nitrogen metabolism, as was previously suggested for Synechochococcus sp. strain PCC7942 (69).
A similar link between carbon and nitrogen metabolism has been observed in Rhodobacter sphaeroides, in which a RubisCO-deficient mutant fails to express glnB under photoheterotrophic growth with either ammonium or glutamate as the nitrogen source (185). While glnK expression is strictly under nitrogen control in a wild-type strain, in the RubisCO mutant glnK is expressed in the presence of ammonium.
Nodule development.
Given the pivotal role of PII proteins in regulating nitrogen metabolism in many bacteria, the symbiotic interaction between Rhizobium spp. and legumes is a potentially important biological process in which these proteins might well be expected to play a significant part. The development of a legume nodule and the concomitant differentiation of the free-living bacterium into a nitrogen-fixing bacteroid involve a major change in the nitrogen assimilation pathways of the Rhizobium cell.
In Rhizobium leguminosarum, the glnB promoter is downregulated during bacteroid differentiation at a time coincident with the arrest of bacterial division (56). However, in Sinorhizobium meliloti, construction of both an in-frame deletion of glnB and a glnB Y51F mutant implicated GlnB in nodule development. Both mutants exhibit similar symbiotic phenotypes, albeit somewhat stronger for glnB Y51F, with defective infection on alfalfa and heterogenous nodules on 3-week-old plants. The plants are chlorotic despite being able to reduce dinitrogen to appreciable levels. On addition of ammonium, chlorosis is lost, confirming that the plants are suffering nitrogen starvation despite a high level of nitrogenase activity. This paradoxical Nif+ Fix− phenotype may be explained by the involvement of GlnB in regulation of a bacteroid ammonium transporter (8), and the critical role of the AmtB protein in nodule development has been demonstrated independently in Rhizobium etli (222). In this context, the phenotype of a glnK mutant in one or more of the Rhizobiaceae would be of considerable interest.
Regulation of Other Nitrogen-Related Systems
Mutations in the nadE genes of Salmonella enterica serovar Typhimurium (formerly nit) and Rhodobacter capsulatus (formerly adgA) prevent growth with low concentrations (<1 mM) of ammonia (2, 28, 203, 243). The nadE gene encodes an ammonia-dependent NAD synthetase with a Kmfor ammonia of 65 μM, lower than that for any known amidotransferase. It has therefore been proposed that the phenotype of the nadE mutants is due to a low residual NAD synthetase activity that is insufficient when ammonia becomes limiting. Such an ammonia-dependent enzyme is a good candidate for posttranslational regulation by PII, so that activity is coordinated with ammonia availability. It is therefore perhaps no coincidence that in Herbaspirillum seropedicae and Azoarcus, nadE is genetically linked to a glnB-like gene, and it would certainly be of interest to study the NAD synthetase activity in glnB and glnK mutants.
A further extrapolation of this observation is related to the DRAG-DRAT system that is responsible for switch-off of nitrogenase activity in Rhodospirillum rubrum and other diazotrophs (see above). The donor of the ADP-ribose moiety for the DRAT-mediated inactivation of dinitrogenase is NAD+ (213), and it has been proposed that an increase in NAD+ concentration is the signal for DRAT activation in R. rubrum (175). Given the ammonia dependence of NadE and a potential regulation of NadE activity by PII, this could suggest one step in the pathway at which PII could influence the response of the DRAG-DRAT system to ammonia shock.
Another interesting relationship is the close linkage in Azospirillum brasilense between the GlnK structural gene (termed glnZ) and a gene (aat) encoding a protein with significant similarity to an aspartate amino transferase. In E. coli this enzyme (AspC) is involved in the utilization of glutamate as a carbon source; it catalyzes the reaction between l-glutamate and oxaloacetate, producing l-aspartate and 2-ketoglutarate (149), and a similar pathway exists in K. aerogenes (226). Given that the reaction generates 2-ketoglutarate, it is clearly a candidate for regulation by a PII protein, and the gene linkage in A. brasilense may be indicative of just such an interaction.
The above considerations suggest that there could still be many other undiscovered targets for PII. Indeed, the activity of any enzyme or the transcription of any gene that is involved in nitrogen assimilation or metabolism is a potential candidate. Recruitment of any such candidate into the PII “network” would facilitate the optimal coordination of cellular nitrogen metabolism.
Role of PII Proteins in Plants
Nitrogen is largely absorbed by plants as nitrate, which in Arabidopsis thaliana is reduced primarily in the cytosol of leaf cells to nitrite by nitrate reductase. After its import into the chloroplast, nitrite is converted by nitrite reductase to ammonium, which is then assimilated into glutamine by the plant GSII. In addition, in green leaves of C3 plants, ammonium resulting from photorespiratory metabolism is imported to the chloroplast and reassimilated by GSII. Interestingly, a new member of the plant ammonium transporter (AMT) family was recently described in Arabidopsis thaliana. This protein (AtAMT2) is more highly expressed in shoots than in roots (212), and it was suggested that this protein may play a different role to the AtAMT1 proteins (173). One possibility is that AtAMT2 could be responsible for the transport of ammonium produced during photorespiration into the chloroplast. The localization of the PII-like protein GLB1 to the chloroplast (97), together with known functions of PII proteins in bacteria, suggests many possible targets for GLB1. The protein could regulate the activity of the nitrite or ammonium transporter or the metabolic enzymes nitrite reductase and GSII, all of which will affect the nitrogen status of the chloroplast. It should also be noted that regulation of the N-C balance in photosynthetic bacteria seems to be integrated by GlnB (92, 185), in which case the chloroplastic PII-like protein GLB1 could similarly be involved in coordinating nitrogen assimilation and photosynthesis.
Is PII an Essential Gene Product in Some Organisms?
An E. coli null mutant in which both PII genes are deleted is viable, albeit slow-growing. Likewise, double mutants of Azorhizobium caulinodans and Rhodobacter capsulatus, in which there are probably only two PII genes, are viable. Hence, PII is not by definition an essential protein. However, in Azotobacter vinelandii (153) and in Nostoc punctiforme (84), several attempts to create a PII mutant failed, which led the authors to suggest that in these organisms, PII may be essential. In A. vinelandii, the requirement for a certain level of GS activity has been considered the likely reason for the lethality of both glnA and glnD mutations (39). However, the apparent lethality of glnK mutations cannot be attributed to a possible effect on the adenylylation of GS, because a glnK mutation was still unstable in a mutant carrying a nonadenylylatable GS (glnA Y407F) (153).
CONCLUDING REMARKS
In summarizing the state of knowledge in the field of bacterial nitrogen control 5 years ago, we noted that one of the few apparently unifying concepts that appeared to be emerging at that time was the ubiquity of the PII proteins and their pivotal role in nitrogen metabolism (155). The very significant advances in our understanding of this protein family since then have undoubtedly served only to confirm this view. In this review, we have attempted not only to summarize the detailed information that we now have on the structures and functions of bacterial PII proteins but also to draw attention to indicators of the much wider roles that these proteins might play in coordinating a great many facets of nitrogen metabolism, including acquisition of nitrogen compounds and their catabolism and assimilation.
The explosion in genome sequence information in the last 5 years has served to reveal the ubiquitous nature of the PII protein family, and the developments in the concept of functional coupling of genes (177) are already suggesting new targets for PII proteins. However, in the near future the application of transcriptome and proteome analysis will rapidly allow us to move to a much more comprehensive analysis of the potential targets for PII, in terms of regulating both gene expression and protein activity. We would not be surprised if these analyses reveal, particularly in the proteobacteria, that we have only begun to scratch the surface.
One particularly notable feature of the PII proteins is their degree of conservation, as demonstrated by the GlnB and GlnK proteins. However, there are some interesting challenges ahead in understanding the more unusual members of the family, such as the NifI proteins, the novel GlnBI protein of Aquifex aeolicus, and the newly emerging PII proteins of higher plants with their additional N-terminal domains. While the availability of X-ray structures for both GlnB and GlnK offers insights into the mode of action of PII, further structures of other forms, e.g., with 2-ketoglutarate bound or with PII complexed to a “target” protein, would undoubtedly help to illuminate exactly how the protein functions.
There is still much to learn about this fascinating signal transduction protein that appears to have proved so versatile as a regulatory molecule. Our understanding of the biology of these proteins will undoubtedly be served by the continued endeavors of microbiologists to study as wide a spectrum of organisms as possible, and given the enormous diversity of prokaryotic organisms, there are likely many novel stories still waiting to be told.
ACKNOWLEDGMENTS
We thank all those who provided preprints and unpublished information for inclusion in this review.
We acknowledge support from EMBO (T.A.), the John Innes Foundation (R.J.), and the Biotechnology and Biological Sciences Research Council (M.M.).
REFERENCES
- 1.Adler S P, Purich D, Stadtman E R. Cascade control of Escherichia coli glutamine synthetase: properties of the PIIregulatory protein and the uridylyltransferase-uridylylremoving enzyme. J Biol Chem. 1975;250:6264–6272. [PubMed] [Google Scholar]
- 2.Allibert P, Willison J C, Vignais P M. Complementation of nitrogen-regulatory (ntr-like) mutations in Rhodobacter capsulatus by an Escherichia coligene: cloning and sequencing of the gene and characterization of the gene product. J Bacteriol. 1987;169:260–271. doi: 10.1128/jb.169.1.260-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allikmets R, Gerrard B, Court D, Dean M. Cloning and organisation of the abc and mdl genes of Escherichia coli: relationship to eukaryotic multidrug resistance. Gene. 1993;136:231–236. doi: 10.1016/0378-1119(93)90470-n. [DOI] [PubMed] [Google Scholar]
- 4.Amar M, Patriarca E J, Manco G, Bernard P, Riccio A, Lamberti A, Defez R, Iaccarino M. Regulation of nitrogen metabolism is altered in a glnB mutant strain of Rhizobium leguminosarum. Mol Microbiol. 1994;11:685–693. doi: 10.1111/j.1365-2958.1994.tb00346.x. [DOI] [PubMed] [Google Scholar]
- 5.Anderson W B, Hennig S B, Ginsburg A, Stadtman E R. Association of ATP:glutamine synthetase adenylyltransferase activity with the P1 component of the glutamine synthetase deadenylylation system. Proc Natl Acad Sci USA. 1970;67:1417–1424. doi: 10.1073/pnas.67.3.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arcondéguy T. Regulation de l'assimilation de l'azote chez le symbiote de la luzerne, R. meliloti. Ph.D. thesis. Toulouse, France: Université Paul Sabatier Toulouse III; 1996. [Google Scholar]
- 7.Arcondéguy T, Huez I, Fourment J, Kahn D. Symbiotic nitrogen fixation does not require adenylylation of glutamine synthetase I in Rhizobium meliloti. FEMS Microbiol Lett. 1996;145:33–40. doi: 10.1111/j.1574-6968.1996.tb08553.x. [DOI] [PubMed] [Google Scholar]
- 8.Arcondéguy T, Huez I, Tillard P, Gangneux C, de Billy F, Gojon A, Truchet G, Kahn D. The Rhizobium meliloti PIIprotein, which controls bacterial nitrogen metabolism, affects alfalfa nodule development. Genes Dev. 1997;11:1194–1206. doi: 10.1101/gad.11.9.1194. [DOI] [PubMed] [Google Scholar]
- 9.Arcondéguy T, Lawson D, Merrick M. Two residues in the T-loop of GlnK determine NifL-dependent nitrogen control of nifgene expression. J Biol Chem. 2000;275:38452–38456. doi: 10.1074/jbc.M001935200. [DOI] [PubMed] [Google Scholar]
- 10.Arcondéguy T, van Heeswijk W C, Merrick M. Studies on the roles of GlnK and GlnB in regulating Klebsiella pneumoniaeNifL-dependent nitrogen control. FEMS Microbiol Lett. 1999;180:263–270. doi: 10.1111/j.1574-6968.1999.tb08805.x. [DOI] [PubMed] [Google Scholar]
- 11.Arsene F, Kaminski P A, Elmerich C. Modulation of NifA activity by PII in Azospirillum brasilense: evidence for a regulatory role of the NifA N-terminal domain. J Bacteriol. 1996;178:4830–4838. doi: 10.1128/jb.178.16.4830-4838.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arsene F, Kaminski P A, Elmerich C. Control of Azospirillum brasilense NifA activity by PII: effect of replacing Tyr residues of the NifA N-terminal domain on NifA activity. FEMS Microbiol Lett. 1999;179:339–343. doi: 10.1111/j.1574-6968.1999.tb08747.x. [DOI] [PubMed] [Google Scholar]
- 13.Atkinson M, Ninfa A J. Role of the GlnK signal transduction protein in the regulation of nitrogen assimilation in Escherichia coli. Mol Microbiol. 1998;29:431–447. doi: 10.1046/j.1365-2958.1998.00932.x. [DOI] [PubMed] [Google Scholar]
- 14.Atkinson M, Ninfa A J. Characterization of the GlnK protein of Escherichia coli. Mol Microbiol. 1999;32:301–313. doi: 10.1046/j.1365-2958.1999.01349.x. [DOI] [PubMed] [Google Scholar]
- 15.Atkinson M R, Kamberov E S, Weiss R L, Ninfa A J. Reversible uridylylation of the Escherichia coli PIIsignal transduction protein regulates its ability to stimulate the dephosphorylation of the transcription factor nitrogen regulator I (NRI or NtrC) J Biol Chem. 1994;269:28288–28293. [PubMed] [Google Scholar]
- 16.Atkinson M R, Ninfa A J. Characterization of Escherichia coli glnLmutations affecting nitrogen regulation. J Bacteriol. 1992;174:4538–4548. doi: 10.1128/jb.174.14.4538-4548.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Atkinson M R, Ninfa A J. Mutational analysis of the bacterial signal-transducing protein kinase/phosphatase nitrogen regulator II (NRIIor NtrB) J Bacteriol. 1993;175:7016–7023. doi: 10.1128/jb.175.21.7016-7023.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Austin S, Dixon R. The prokaryotic enhancer binding protein NTRC has an ATPase activity which is phosphorylation and DNA dependent. EMBO J. 1992;11:2219–2228. doi: 10.1002/j.1460-2075.1992.tb05281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bancroft S, Rhee S G, Neumann C, Kustu S. Mutations that alter the covalent modification of glutamine synthetase in Salmonella typhimurium. J Bacteriol. 1978;134:1046–1055. doi: 10.1128/jb.134.3.1046-1055.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benelli, E. M., M. Buck, E. M. de Souza, M. G. Yates, and F. O. Pedrosa. Uridylylation of the PII protein from Herbaspirillum seropedicae. Can. J. Microbiol., in press. [DOI] [PubMed]
- 21.Benelli E M, Souza E M, Funayama S, Rigo I U, Pedrosa F O. Evidence for two possible glnB-type genes in Herbaspirillum seropedicae. J Bacteriol. 1997;179:4623–4626. doi: 10.1128/jb.179.14.4623-4626.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berger D K, Naberhaus F, Kustu S. The isolated catalytic domain of NifA, a bacterial enhancer-binding protein, activates transcription in vitro: activation is inhibited by NifL. Proc Natl Acad Sci USA. 1994;91:103–107. doi: 10.1073/pnas.91.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berger D K, Narberhaus F, Lee H S, Kustu S. In vitro studies of the domains of the nitrogen fixation regulatory protein NifA. J Bacteriol. 1995;177:191–199. doi: 10.1128/jb.177.1.191-199.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blanco G, Drummond M, Woodley P, Kennedy C. Sequence and molecular analysis of the nifL gene of Azotobacter vinelandii. Mol Microbiol. 1993;9:869–880. doi: 10.1111/j.1365-2958.1993.tb01745.x. [DOI] [PubMed] [Google Scholar]
- 25.Bloom F R, Levin M S, Foor F, Tyler B. Regulation of glutamine synthetase formation in Escherichia coli: characterization of mutants lacking the uridylyltransferase. J Bacteriol. 1978;134:569–577. doi: 10.1128/jb.134.2.569-577.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borghese R, Wall J D. Regulation of the glnBA operon of Rhodobacter capsulatus. J Bacteriol. 1995;177:4549–4552. doi: 10.1128/jb.177.15.4549-4552.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bozouklian H, Elmerich C. Nucleotide sequence of the Azospirillum brasilenseSp7 glutamine synthetase structural gene. Biochimie. 1986;68:1181–1187. doi: 10.1016/s0300-9084(86)80062-1. [DOI] [PubMed] [Google Scholar]
- 28.Broach J, Neuman C, Kustu S. Mutant strain (nit) of Salmonella typhimuriumwith a pleiotropic defect in nitrogen metabolism. J Bacteriol. 1976;128:86–98. doi: 10.1128/jb.128.1.86-98.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown G D, Thomson J A. Isolation and characterisation of an aryl-beta-d-glucoside uptake and utilisation system (abg) from the gram-positive ruminal Clostridium species C. longisporum. Mol Gen Genet. 1998;257:213–218. doi: 10.1007/s004380050641. [DOI] [PubMed] [Google Scholar]
- 30.Brown M S, Segal A, Stadtman E R. Modulation of glutamine synthetase adenylylation and deadenylylation is mediated by metabolic transformation of the PIIregulatory protein. Proc Natl Acad Sci USA. 1971;68:2949–2953. doi: 10.1073/pnas.68.12.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bueno R, Pahel G, Magasanik B. Role of glnB and glnD gene products in regulation of the glnALG operon of Escherichia coli. J Bacteriol. 1985;164:816–822. doi: 10.1128/jb.164.2.816-822.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carr P D, Cheah E, Suffolk P M, Vasudevan S G, Dixon N E, Ollis D L. X-ray structure of the signal transduction protein PII from Escherichia coli at 1.9Å. Acta Crystallogr Sect D. 1996;52:93–104. doi: 10.1107/S0907444995007293. [DOI] [PubMed] [Google Scholar]
- 33.Cheah E, Carr P D, Suffolk P M, Vasudevan S G, Dixon N E, Ollis D L. Structure of the Escherichia coli signal transducing protein PII. Structure (London) 1994;2:981–990. doi: 10.1016/s0969-2126(94)00100-6. [DOI] [PubMed] [Google Scholar]
- 34.Cheng J, Johansson M, Nordlund S. Expression of PII and glutamine synthetase is regulated by PII the ntrBC products, and processing of the glnBA mRNA in Rhodospirillum rubrum. J Bacteriol. 1999;181:6530–6534. doi: 10.1128/jb.181.20.6530-6534.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chien Y, Helmann J D, Zinder S H. Interactions between the promoter regions of nitrogenase structural genes (nifHDK2) and DNA-binding proteins from N2- and ammonium-grown cells of the archaeon Methanosarcina barkeri227. J Bacteriol. 1998;180:2723–2728. doi: 10.1128/jb.180.10.2723-2728.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiurazzi M, Iaccarino M. Transcriptional analysis of the glnB-glnA region of Rhizobium leguminosarum biovar viciae. Mol Microbiol. 1990;4:1727–1735. doi: 10.1111/j.1365-2958.1990.tb00550.x. [DOI] [PubMed] [Google Scholar]
- 37.Claverie-Martin F, Magasanik B. Role of integration host factor in the regulation of the glnHp2 promoter of Escherichia coli. Proc Natl Acad Sci USA. 1991;88:1631–1635. doi: 10.1073/pnas.88.5.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell B G. Deciphering the biology of Mycobacterium tuberculosisfrom the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 39.Colnaghi R, Green A, He L, Rudnick P, Kennedy C. Strategies for increasing ammonium production in free-living or plant-associated nitrogen-fixing bacteria. Plant Soil. 1997;194:145–154. [Google Scholar]
- 40.Colonna-Romano S, Patriarca E J, Amar M, Bernard P, Manco G, Lamberti A, Iaccarino M, Defez R. Uridylylation of the PII protein in Rhizobium leguminosarum. FEBS Lett. 1993;330:95–98. doi: 10.1016/0014-5793(93)80927-m. [DOI] [PubMed] [Google Scholar]
- 41.Colonna-Romano S, Riccio A, Guida M, Defez R, Lamberti A, Iaccarino M, Arnold W, Priefer U, Pühler A. Tight linkage of glnA and a putative regulatory gene in Rhizobium leguminosarum. Nucleic Acids Res. 1987;15:1951–1964. doi: 10.1093/nar/15.5.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Contreras A, Drummond M, Bali A, Blanco G, Garcia E, Bush G, Kennedy C, Merrick M. The product of the nitrogen fixation regulatory gene nfrX of Azotobacter vinelandii is functionally and structurally homologous to the uridylyltransferase encoded by glnDin enteric bacteria. J Bacteriol. 1991;173:7741–7749. doi: 10.1128/jb.173.24.7741-7749.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cullen P, Bowman W C, Foster-Hartnett D, Reilly S C, Kranz R G. Translational activation by an NtrC enhancer-binding protein. J Mol Biol. 1998;278:903–914. doi: 10.1006/jmbi.1998.1745. [DOI] [PubMed] [Google Scholar]
- 44.Dandekar T, Snel B, Huynen M, Bork P. Conservation of gene order: a fingerprint of proteins that physically interact. Trends Genet. 1998;23:324–328. doi: 10.1016/s0968-0004(98)01274-2. [DOI] [PubMed] [Google Scholar]
- 45.Deckert G, Warren P V, Gaasterland T, Young W G, Lenox A L, Graham D E, Overbeek R, Snead M A, Keller M, Aujay M, Huber R, Feldman R A, Short J M, Olsen G J, Swanson R V. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 46.de Mel V S J, Kamberov E S, Martin P D, Zhang J, Ninfa A J, Edwards B F P. Preliminary X-ray diffraction analysis of crystals of the PII protein from Escherichia coli. J Mol Biol. 1994;243:796–798. doi: 10.1016/0022-2836(94)90049-3. [DOI] [PubMed] [Google Scholar]
- 47.de Zamaroczy M. Structural homologues PII and Pz of Azospirillum brasilenseprovide intracellular signalling for selective regulation of various nitrogen-dependent functions. Mol Microbiol. 1998;29:449–463. doi: 10.1046/j.1365-2958.1998.00938.x. [DOI] [PubMed] [Google Scholar]
- 48.de Zamaroczy M, Delorme F, Elmerich C. Characterisation of three different nitrogen-regulated promoter regions for the expression of glnB and glnA in Azospirillum brasilense. Mol Gen Genet. 1990;224:421–430. doi: 10.1007/BF00262437. [DOI] [PubMed] [Google Scholar]
- 49.de Zamaroczy M, Paquelin A, Elmerich C. Functional organization of the glnB-glnA cluster of Azospirillum brasilense. J Bacteriol. 1993;175:2507–2515. doi: 10.1128/jb.175.9.2507-2515.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Zamaroczy M, Paquelin A, Peltre G, Forchhammer K, Elmerich C. Coexistence of two structurally similar but functionally different PII proteins in Azospirillum brasilense. J Bacteriol. 1996;178:4143–4149. doi: 10.1128/jb.178.14.4143-4149.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dixon R. The oxygen-responsive NifL-NifA complex: a novel two-component regulatory system controlling nitrogenase synthesis in gamma-proteobacteria. Arch Microbiol. 1998;169:371–380. doi: 10.1007/s002030050585. [DOI] [PubMed] [Google Scholar]
- 52.Drummond M, Clements J, Merrick M, Dixon R. Positive control and autogenous regulation of the nifLA promoter in Klebsiella pneumoniae. Nature. 1983;301:302–307. doi: 10.1038/301302a0. [DOI] [PubMed] [Google Scholar]
- 53.Drummond M, Whitty P, Wootton J. Sequence and domain relationships of ntrC and nifA from Klebsiella pneumoniae: homologies to other regulatory proteins. EMBO J. 1986;5:441–447. doi: 10.1002/j.1460-2075.1986.tb04230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Drummond M H, Contreras A, Mitchenall L A. The function of isolated domains and chimaeric proteins constructed from the transcriptional activators NifA and NtrC of Klebsiella pneumoniae. Mol Microbiol. 1990;4:29–37. doi: 10.1111/j.1365-2958.1990.tb02012.x. [DOI] [PubMed] [Google Scholar]
- 55.Edwards R, Merrick M. The role of uridylyltransferase in the control of Klebsiella pneumoniae nifgene regulation. Mol Gen Genet. 1995;247:189–198. doi: 10.1007/BF00705649. [DOI] [PubMed] [Google Scholar]
- 56.Ercolano E, Mirabella R, Merrick M, Chiurazzi M. The Rhizobium leguminosarum glnBgene is down-regulated during symbiosis. Mol Gen Genet. 2001;264:555–564. doi: 10.1007/s004380000333. [DOI] [PubMed] [Google Scholar]
- 57.Fadel-Picheth C M, Souza E M, Rigo L U, Funayama S, Yates M G, Pedrosa F O. Regulation of Azospirillum brasilense nifAgene expression by ammonium and oxygen. FEMS Microbiol Lett. 1999;179:281–288. doi: 10.1111/j.1574-6968.1999.tb08739.x. [DOI] [PubMed] [Google Scholar]
- 58.Fink D, Falke D, Wohlleben W, Engels A. Nitrogen metabolism in Streptomyces coelicolorA3(2): modification of glutamine synthetase I by an adenylyltransferase. Microbiology. 1999;145:2313–2322. doi: 10.1099/00221287-145-9-2313. [DOI] [PubMed] [Google Scholar]
- 59.Fischer H-M. Genetic regulation of nitrogen fixation in rhizobia. Microbiol Rev. 1994;58:352–386. doi: 10.1128/mr.58.3.352-386.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fisher S H. Regulation of nitrogen metabolism in Bacillus subtilis: vive la difference Mol. Microbiol. 1999;32:223–232. doi: 10.1046/j.1365-2958.1999.01333.x. [DOI] [PubMed] [Google Scholar]
- 61.Fisher S H, Rohrer K, Ferson A E. Role of CodY in regulation of the Bacillus subtilis hutoperon. J Bacteriol. 1996;178:3779–3784. doi: 10.1128/jb.178.13.3779-3784.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fleischmann R D. Whole-genome random sequencing and assembly of Haemophilus influenzaeRD. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 63.Foor F, Cedergren R J, Streicher S L, Rhee S G, Magasanik B. Glutamine synthetase of Klebsiella aerogenes: properties of glnDmutants lacking uridylyltransferase. J Bacteriol. 1978;134:562–568. doi: 10.1128/jb.134.2.562-568.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Foor F, Janssen K, Magasanik B. Regulation of the synthesis of glutamine synthetase by adenylylated glutamine synthetase. Proc Natl Acad Sci USA. 1975;72:4844–4848. doi: 10.1073/pnas.72.12.4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Foor F, Reuveny Z, Magasanik B. Regulation of the synthesis of glutamine synthetase by the PII protein in Klebsiella aerogenes. Proc Natl Acad Sci USA. 1980;77:2636–2640. doi: 10.1073/pnas.77.5.2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Forchhammer K. The PII protein in Synechococcus PCC7942 senses and signals 2-oxolglutarate and ATP-replete conditions. In: Peschek G, Löffelhardt W, Schmetterer G, editors. The phototrophic prokaryotes. New York, N.Y: Kluwer Academic; 1999. pp. 549–553. [Google Scholar]
- 67.Forchhammer K, Hedler A. Phosphoprotein PII from cyanobacteria: analysis of functional conservation with the PII signal protein from Escherichia coli. Eur J Biochem. 1997;244:869–875. doi: 10.1111/j.1432-1033.1997.00869.x. [DOI] [PubMed] [Google Scholar]
- 68.Forchhammer K, Hedler A, Strobel H, Weiss V. Heterotrimerization of PII-like signalling proteins: implications for PII-mediated signal transduction systems. Mol Microbiol. 1999;33:338–349. doi: 10.1046/j.1365-2958.1999.01477.x. [DOI] [PubMed] [Google Scholar]
- 69.Forchhammer K, Tandeau de Marsac N. The PII protein in the cyanobacterium Synechococcussp. strain PCC 7942 is modified by serine phosphorylation and signals the cellular N status. J Bacteriol. 1994;176:84–91. doi: 10.1128/jb.176.1.84-91.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Forchhammer K, Tandeau de Marsac N. Functional analysis of the phosphoprotein PII (glnB gene product) in the cyanobacterium Synechococcussp. strain PCC 7942. J Bacteriol. 1995;177:2033–2040. doi: 10.1128/jb.177.8.2033-2040.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Forchhammer K, Tandeau de Marsac N. Phosphorylation of the PII protein (glnB gene product) in the cyanobacterium Synechococcussp. strain PCC 7942: analysis of in vitro kinase activity. J Bacteriol. 1995;177:5812–5817. doi: 10.1128/jb.177.20.5812-5817.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Foster-Hartnett D, Kranz R G. Analysis of the promoters and upstream sequences of nifA1 and nifA2 in Rhodobacter capsulatus; activation requires ntrC but not rpoN. Mol Microbiol. 1992;6:1049–1060. doi: 10.1111/j.1365-2958.1992.tb02170.x. [DOI] [PubMed] [Google Scholar]
- 73.Foster-Hartnett D, Kranz R G. The Rhodobacter capsulatus glnB gene is regulated by NtrC at tandem rpoN-dependent promoters. J Bacteriol. 1994;176:5171–5176. doi: 10.1128/jb.176.16.5171-5176.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fu H, Burris R H, Roberts G P. Reversible ADP-ribosylation is demonstrated to be a regulatory mechanism in prokaryotes by heterologous expression. Proc Natl Acad Sci USA. 1990;87:1720–1724. doi: 10.1073/pnas.87.5.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Garcia E, Rhee S G. Cascade control of Escherichia coli glutamine synthetase: purification and properties of PIIuridylytransferase and uridylyl-removing enzyme. J Biol Chem. 1983;258:2246–2253. [PubMed] [Google Scholar]
- 76.Garcia-Dominguez M, Florencio F J. Nitrogen availability and electron transport control the expression of glnB gene (encoding PII protein) in the cyanobacterium Synechocystissp. PCC 6803. Plant Mol Biol. 1997;35:723–734. doi: 10.1023/a:1005846626187. [DOI] [PubMed] [Google Scholar]
- 77.Garcia-Dominguez M, Reyes J C, Florencio F J. Glutamine synthetase inactivation by protein-protein interaction. Proc Natl Acad Sci USA. 1999;96:7161–7166. doi: 10.1073/pnas.96.13.7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Garcia-Dominguez M, Reyes J C, Florencio F J. NtcA represses transcription of gifA and gifB, genes that encode inhibitors of glutamine synthetase type I from Synechocystissp. PCC 6803. Mol Microbiol. 2000;35:1192–1201. doi: 10.1046/j.1365-2958.2000.01789.x. [DOI] [PubMed] [Google Scholar]
- 79.Gardan R, Rapoport G, Debarbouille M. Role of the transcriptional activator RocR in the arginine-degradation pathway of Bacillus subtilis. Mol Microbiol. 1997;24:825–837. doi: 10.1046/j.1365-2958.1997.3881754.x. [DOI] [PubMed] [Google Scholar]
- 80.Gonzalez L, Phalip V, Zhang C-C. Modification of the PII protein in response to nitrogen availability in filamentous heterocystous cyanobacteria Anabaena sp. PCC 7120. In: Pedrosa F O, Hungria M, Yates M G, Newton W E, editors. Nitrogen fixation: from molecules to crop productivity. Dordrecht, The Netherlands: Kluwer Academic; 2000. p. 107. [Google Scholar]
- 81.Gonzalez V, Olvera L, Soberon X, Morett E. In vivostudies on the positive control function of NifA: a conserved hydrophobic amino acid patch at the central domain involved in transcriptional activation. Mol Microbiol. 1998;28:55–67. doi: 10.1046/j.1365-2958.1998.00772.x. [DOI] [PubMed] [Google Scholar]
- 82.Govantes F, Molina-Lopez J A, Santero E. Mechanism of coordinated synthesis of the antagonistic regulatory proteins NifL and NifA of Klebsiella pneumoniae. J Bacteriol. 1996;178:6817–6823. doi: 10.1128/jb.178.23.6817-6823.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hallenbeck P C. Mutations affecting nitrogenase switch-off in Rhodobacter capsulatus. Biochim Biophys Acta. 1992;1118:161–168. doi: 10.1016/0167-4838(92)90145-4. [DOI] [PubMed] [Google Scholar]
- 84.Hanson T E, Forchhammer K, Tandeau de Marsac N, Meeks J C. Characterisation of the glnB gene product of Nostoc punctiforme strain ATCC 29133: glnB or the PIIprotein may be essential. Microbiology. 1998;144:1537–1547. doi: 10.1099/00221287-144-6-1537. [DOI] [PubMed] [Google Scholar]
- 85.Harrison M A, Keen J N, Findlay J B, Allen J F. Modification of a glnB-like gene product by photosynthetic electron transport in the cyanobacterium Synechococcus6301. FEBS Lett. 1990;264:25–28. doi: 10.1016/0014-5793(90)80755-8. [DOI] [PubMed] [Google Scholar]
- 86.Hartmann A, Burris R H. Regulation of nitrogenase activity by oxygen in Azospirillum brasilense and Azospirillum lipoferum. J Bacteriol. 1987;169:944–948. doi: 10.1128/jb.169.3.944-948.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.He B, Choi K Y, Zalkin H. Regulation of Escherichia coli glnB, prsA, and speAby the purine repressor. J Bacteriol. 1993;175:3598–3606. doi: 10.1128/jb.175.11.3598-3606.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.He L, Soupene E, Kustu S. NtrC is required for control of Klebsiella pneumoniaeNifL activity. J Bacteriol. 1997;179:7446–7455. doi: 10.1128/jb.179.23.7446-7455.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.He L, Soupene E, Ninfa A J, Kustu S. Physiological role for the GlnK protein of enteric bacteria: relief of NifL inhibition under nitrogen-limiting conditions. J Bacteriol. 1998;180:6661–6667. doi: 10.1128/jb.180.24.6661-6667.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Henderson N, Austin S A, Dixon R A. Role of metal ions in negative regulation of nitrogen fixation by the nifL gene product from Klebsiella pneumoniae. Mol Gen Genet. 1989;216:484–491. [Google Scholar]
- 90b.Herrero A, Muro-Pastor A M, Flores E. Nitrogen control in cyanobacteria. J Bacteriol. 2001;183:411–425. doi: 10.1128/JB.183.2.411-425.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hill S, Austin S, Eydmann T, Jones T, Dixon R. Azotobacter vinelandiiNifL is a flavoprotein that modulates transcriptional activation of nitrogen- fixation genes via a redox-sensitive switch. Proc Natl Acad Sci USA. 1996;93:2143–2148. doi: 10.1073/pnas.93.5.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hisbergues M, Jeanjean R, Joset F, Tandeau de Marsac N, Bedu S. Protein PII regulates both inorganic carbon and nitrate uptake and is modified by a redox signal in SynechocystisPCC 6803. FEBS Lett. 1999;463:216–220. doi: 10.1016/s0014-5793(99)01624-5. [DOI] [PubMed] [Google Scholar]
- 93.Holm L, Sander C. DNA polymerase β belongs to an ancient nucleotidyltransferase superfamily. Trends Biochem Sci. 1995;20:345–347. doi: 10.1016/s0968-0004(00)89071-4. [DOI] [PubMed] [Google Scholar]
- 94.Holtel A, Colonna-Romano S, Guida M, Riccio A, Merrick M J, Iaccarino M. The glnB gene of Rhizobium leguminosarum biovar viciae. FEMS Microbiol Lett. 1989;58:203–208. [Google Scholar]
- 95.Holtel A, Merrick M J. The Klebsiella pneumoniae PII protein (glnB gene product) is not absolutely required for nitrogen regulation and is not involved in NifL-mediated nifgene regulation. Mol Gen Genet. 1989;217:474–480. doi: 10.1007/BF02464920. [DOI] [PubMed] [Google Scholar]
- 96.Holtel A H, Merrick M. Identification of the Klebsiella pneumoniae glnBgene: nucleotide sequence of wild-type and mutant alleles. Mol Gen Genet. 1988;215:134–138. doi: 10.1007/BF00331314. [DOI] [PubMed] [Google Scholar]
- 97.Hsieh M H, Lam H M, van de Loo F J, Coruzzi G. A PII-like protein in Arabidopsis: putative role in nitrogen sensing. Proc Natl Acad Sci USA. 1998;95:13965–13970. doi: 10.1073/pnas.95.23.13965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hsing W, Russo F D, Bernd K K, Silhavy T J. Mutations that alter the kinase and phosphatase activities of the two-component sensor EnvZ. J Bacteriol. 1998;180:4538–4546. doi: 10.1128/jb.180.17.4538-4546.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hsing W, Silhavy T J. Function of conserved histidine-243 in phosphatase activity of EnvZ, the sensor for porin osmoregulation in Escherichia coli. J Bacteriol. 1997;179:3729–3735. doi: 10.1128/jb.179.11.3729-3735.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Irmler A, Sanner S, Dierks H, Forchhammer K. Dephosphorylation of the phosphoprotein PII in SynechococcusPCC7942: identification of an ATP and 2-oxoglutarate-regulated phosphatase activity. Mol Microbiol. 1997;26:81–90. doi: 10.1046/j.1365-2958.1997.5521918.x. [DOI] [PubMed] [Google Scholar]
- 101.Jack R, de Zamaroczy M, Merrick M. The signal transduction protein GlnK is required for NifL-dependent nitrogen control of nif expression in Klebsiella pneumoniae. J Bacteriol. 1999;181:1156–1162. doi: 10.1128/jb.181.4.1156-1162.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jaggi R, van Heeswijk W C, Westerhof H V, Ollis D L, Vasudevan S G. The two opposing activities of adenylyl transferase reside in distinct homologous domains with intramolecular signal transduction. EMBO J. 1997;16:5562–5571. doi: 10.1093/emboj/16.18.5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jaggi R, Ybarlucea W, Cheah E, Carr P D, Edwards K J, Ollis D, Vasudevan S G. The role of the T-loop of the signal transducing protein PII from Escherichia coli. FEBS Lett. 1996;391:223–228. doi: 10.1016/0014-5793(96)00737-5. [DOI] [PubMed] [Google Scholar]
- 104.Jakoby M, Krämer R, Burkovski A. Nitrogen regulation in Corynebacterium glutamicum: isolation of genes involved and biochemical characterisation of corresponding proteins. FEMS Microbiol Lett. 1999;173:303–310. doi: 10.1111/j.1574-6968.1999.tb13518.x. [DOI] [PubMed] [Google Scholar]
- 105.Jakoby M, Nolden L, Meier-Wagner J, Krämer R, Burkovski A. AmtR, a global repressor in the nitrogen regulation system of Corynebacterium glutamicum. Mol Microbiol. 2000;37:964–977. doi: 10.1046/j.1365-2958.2000.02073.x. [DOI] [PubMed] [Google Scholar]
- 106.Jiang P, Atkinson M R, Srisawat C, Sun Q, Ninfa A J. Functional dissection of the dimerization and enzymatic activities of Escherichia colinitrogen regulator II and their regulation by the PII protein. Biochemistry. 2000;39:13433–13449. doi: 10.1021/bi000794u. [DOI] [PubMed] [Google Scholar]
- 107.Jiang P, Ninfa A J. Regulation of autophosphorylation of Escherichia colinitrogen regulator II by the PII signal transduction protein. J Bacteriol. 1999;181:1906–1911. doi: 10.1128/jb.181.6.1906-1911.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jiang P, Peliska J A, Ninfa A J. Enzymological characterization of the signal-transducing uridylyltransferase/uridylyl-removing enzyme (EC 2.7.7.59) of Escherichia coliand its interaction with the PII protein. Biochemistry. 1998;37:12782–12794. doi: 10.1021/bi980667m. [DOI] [PubMed] [Google Scholar]
- 109.Jiang P, Peliska J A, Ninfa A J. Reconstitution of the signal-transduction bicyclic cascade responsible for the regulation of ntr gene transcription in Escherichia coli. Biochemistry. 1998;37:12795–12801. doi: 10.1021/bi9802420. [DOI] [PubMed] [Google Scholar]
- 110.Jiang P, Peliska J A, Ninfa A J. The regulation of Escherichia coliglutamine synthetase revisited: role of 2-ketoglutarate in the regulation of glutamine synthetase adenylylation state. Biochemistry. 1998;37:12802–12810. doi: 10.1021/bi980666u. [DOI] [PubMed] [Google Scholar]
- 111.Jiang P, Zucker P, Atkinson M R, Kamberov E S, Tirasophon W, Chandran P, Schefke B R, Ninfa A J. Structure/function analysis of the PII signal transduction protein of Escherichia coli: genetic separation of interactions with protein receptors. J Bacteriol. 1997;179:4342–4353. doi: 10.1128/jb.179.13.4342-4353.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jiang P, Zucker P, Ninfa A J. Probing interactions of the homotrimeric PII signal transduction protein with its receptors by use of PII heterotrimers formed in vitro from wild-type and mutant subunits. J Bacteriol. 1997;179:4354–4360. doi: 10.1128/jb.179.13.4354-4360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Johannson M, Nordlund S. Transcription of the glnB and glnA genes in the photosynthetic bacterium Rhodospirillum rubrum. Microbiology. 1996;142:1265–1272. doi: 10.1099/13500872-142-5-1265. [DOI] [PubMed] [Google Scholar]
- 114.Johannson M, Nordlund S. Uridylylation of the PII protein of the photosynthetic bacterium Rhodospirillum rubrum. J Bacteriol. 1997;179:4190–4194. doi: 10.1128/jb.179.13.4190-4194.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Johansson M, Nordlund S. Purification of PII and PII-UMP and in vitro studies of regulation of glutamine synthetase in Rhodospirillum rubrum. J Bacteriol. 1999;181:6524–6529. doi: 10.1128/jb.181.20.6524-6529.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kamberov E S, Atkinson M R, Chandran P, Ninfa A J. Effect of mutations in Escherichia coli glnL (ntrB), encoding nitrogen regulator II (NRII or NtrB), on the phosphatase activity involved in bacterial nitrogen regulation. J Biol Chem. 1994;269:28294–28299. [PubMed] [Google Scholar]
- 117.Kamberov E S, Atkinson M R, Feng J, Chandran P, Ninfa A J. Sensory components controlling bacterial nitrogen assimilation. Cell Mol Biol Res. 1994;40:175–191. [PubMed] [Google Scholar]
- 118.Kamberov E S, Atkinson M R, Ninfa A J. The Escherichia coliPII signal transduction protein is activated upon binding 2-ketoglutarate and ATP. J Biol Chem. 1995;270:17797–17807. doi: 10.1074/jbc.270.30.17797. [DOI] [PubMed] [Google Scholar]
- 119.Karg T, Reinhold-Hurek B. Global changes in protein composition of N2-fixing Azoarcussp. strain BH72 upon diazosome formation. J Bacteriol. 1996;178:5748–5754. doi: 10.1128/jb.178.19.5748-5754.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Keener J, Kustu S. Protein kinase and phosphoprotein phosphatase activities of nitrogen regulatory proteins NtrB and NtrC of enteric bacteria: roles of the conserved amino-terminal domain of NtrC. Proc Natl Acad Sci USA. 1988;85:4976–4980. doi: 10.1073/pnas.85.14.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kern D, Volkman B F, Luginbuhl P, Nohaile M J, Kustu S, Wemmer D E. Structure of a transiently phosphorylated switch in bacterial signal transduction. Nature. 1999;402:894–898. doi: 10.1038/47273. [DOI] [PubMed] [Google Scholar]
- 122.Kessler P S, Blank C, Leigh J A. The nif gene operon of the methanogenic archaeon Methanococcus maripaludis. J Bacteriol. 1998;180:1504–1511. doi: 10.1128/jb.180.6.1504-1511.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kessler P S, Leigh J A. Genetics of nitrogen regulation in Methanococcus maripaludis. Genetics. 1999;152:1343–1351. doi: 10.1093/genetics/152.4.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123a.Kessler P C, Daniel C, Leigh J A. Ammonia switch-off of nitrogen fixation in the methanogenic archaeon Methanococcus maripaludis: mechanistic features and requirement for the novel GlnB homologues, NifI, and NifI2. J Bacteriol. 2001;183:882–889. doi: 10.1128/JB.183.3.882-889.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kim I H, Kwak S J, Kang J S, Park S C. Transcriptional control of the glnD gene is not dependent on nitrogen availability in Escherichia coli. Mol Cells. 1998;8:483–490. [PubMed] [Google Scholar]
- 125.Klipp W, Drepper T, Gross S, Masepohl B, Raabe K, Riedel K-U, Yakunin A F, Hallenbeck P C. Genetics of nitrogen fixation in Rhodobacter capsulatus: ammonium and molybdenum control of both nitrogenase systems. In: Pedrosa F O, Hungria M, Yates M G, Newton W E, editors. Nitrogen fixation: from molecules to crop productivity. Dordrecht, The Netherlands: Kluwer Academic; 2000. pp. 141–142. [Google Scholar]
- 126.Kobayashi M, Rodriguez R, Lara C, Omata T. Involvement of the C-terminal domain of an ATP-binding subunit in the regulation of the ABC-type nitrate/nitrite transporter of the cyanobacterium Synechococcussp. strain PCC 7942. J Biol Chem. 1997;272:27197–27201. doi: 10.1074/jbc.272.43.27197. [DOI] [PubMed] [Google Scholar]
- 127.Kranz R G, Pace V M, Caldicott I M. Inactivation, sequence, and lacZ fusion analysis of a regulatory locus required for repression of nitrogen fixation genes in Rhodobacter capsulatus. J Bacteriol. 1990;172:53–62. doi: 10.1128/jb.172.1.53-62.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kush A, Elmerich C, Aubert J P. Nitrogenase of Sesbania Rhizobiumstrain ORS 571: purification, properties and ‘switch-off’ by ammonia. J Gen Microbiol. 1985;131:1765–1777. [Google Scholar]
- 129.Kustu S, Hirschman J, Burton D, Jelesko J, Meeks J C. Covalent modification of bacterial glutamine synthetase: physiological significance. Mol Gen Genet. 1984;197:309–317. doi: 10.1007/BF00330979. [DOI] [PubMed] [Google Scholar]
- 130.Lee H M, Flores E, Forchhammer K, Herrero A, Tandeau de Marsac N. Phosphorylation of the signal transducer PII protein and an additional effector are required for the PII-mediated regulation of nitrate and nitrite uptake in the cyanobacterium Synechococcussp. PCC 7942. Eur J Biochem. 2000;267:591–600. doi: 10.1046/j.1432-1327.2000.01043.x. [DOI] [PubMed] [Google Scholar]
- 131.Lee H M, Flores E, Herrero A, Houmard J, Tandeau de Marsac N. A role for the signal transduction protein PIIin the control of nitrate/nitrite uptake in a cyanobacterium. FEBS Lett. 1998;427:291–295. doi: 10.1016/s0014-5793(98)00451-7. [DOI] [PubMed] [Google Scholar]
- 132.Lee H M, Vazquez-Bermudez M F, Tandeau de Marsac N. The global nitrogen regulator NtcA regulates transcription of the signal transducer PII (GlnB) and influences its phosphorylation level in response to nitrogen and carbon supplies in the cyanobacterium Synechococcussp. strain PCC 7942. J Bacteriol. 1999;181:2697–2702. doi: 10.1128/jb.181.9.2697-2702.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Liang J H, Nielsen G M, Lies D P, Burris R H, Roberts G P, Ludden P W. Mutations in the draT and draG genes of Rhodospirillum rubrumresult in loss of regulation of nitrogenase by reversible ADP-ribosylation. J Bacteriol. 1991;173:6903–6909. doi: 10.1128/jb.173.21.6903-6909.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liang Y Y, Arsene F, Elmerich C. Characterization of the ntrBC genes of Azospirillum brasilenseSp7: their involvement in the regulation of nitrogenase synthesis and activity. Mol Gen Genet. 1993;240:188–196. doi: 10.1007/BF00277056. [DOI] [PubMed] [Google Scholar]
- 135.Liang Y Y, de Zamaroczy M, Arsene F, Paquelin A, Elmerich C. Regulation of nitrogen fixation in Azospirillum brasilense Sp7: involvement of nifA, glnA and glnBgene products. FEMS Microbiol Lett. 1992;79:113–119. doi: 10.1111/j.1574-6968.1992.tb14028.x. [DOI] [PubMed] [Google Scholar]
- 136.Lin J T, Goldman B S, Stewart V. The nasFEDCBA operon for nitrate and nitrite assimilation in Klebsiella pneumonaieM5al. J Bacteriol. 1994;176:2551–2559. doi: 10.1128/jb.176.9.2551-2559.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Liotenberg S, Campbell D, Castets A-M, Houmard J, Tandeau de Marsac N. Modification of the PIIprotein in response to carbon and nitrogen availability in filamentous heterocystous cyanobacteria. FEMS Microbiol Lett. 1996;144:185–190. [Google Scholar]
- 138.Little R, Reyes-Ramirez F, Zhang Y, van Heeswijk W C, Dixon R. Signal transduction to the Azotobacter vinelandiiNifL-NifA regulatory system is influenced directly by interaction with 2-oxoglutarate and the PII regulatory protein. EMBO J. 2000;19:6041–6050. doi: 10.1093/emboj/19.22.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Liu J, Magasanik B. The glnB region of the Escherichia colichromosome. J Bacteriol. 1993;175:7441–7449. doi: 10.1128/jb.175.22.7441-7449.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Liu J, Magasanik B. Activation of the dephosphorylation of nitrogen regulator I-phosphate of Escherichia coli. J Bacteriol. 1995;177:926–931. doi: 10.1128/jb.177.4.926-931.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lobo A L, Zinder S H. Nitrogenase in the archaebacterium Methanosarcina barkeri227. J Bacteriol. 1990;172:6789–6796. doi: 10.1128/jb.172.12.6789-6796.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lois A F, Weinstein M, Ditta G S, Helinski D R. Autophosphorylation and phosphatase activities of the oxygen-sensing protein FixL of Rhizobium melilotiare coordinately regulated by oxygen. J Biol Chem. 1993;268:4370–4375. [PubMed] [Google Scholar]
- 143.Ludden P W. Reversible ADP-ribosylation as a mechanism of enzyme regulation in procaryotes. Mol Cell Biochem. 1994;138:123–129. doi: 10.1007/BF00928453. [DOI] [PubMed] [Google Scholar]
- 144.Luque I, Flores E, Herrero A. Molecular mechanism for the operation of nitrogen control in cyanobacteria. EMBO J. 1994;13:2862–2869. doi: 10.1002/j.1460-2075.1994.tb06580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.MacPherson K H, Xu Y, Cheah E, Carr P D, van Heeswijk W C, Westerhoff H V, Luque E, Vasudevan S G, Ollis D L. Crystallization and preliminary X-ray analysis of Escherichia coli GlnK. Acta Crystallogr Sect D Biol Crystallogr. 1998;54:996–998. doi: 10.1107/s0907444998001887. [DOI] [PubMed] [Google Scholar]
- 146.Magasanik B. The regulation of nitrogen utilization in enteric bacteria. J Cell Biochem. 1993;51:34–40. doi: 10.1002/jcb.240510108. [DOI] [PubMed] [Google Scholar]
- 147.Magasanik B. Regulation of nitrogen utilization. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C.: ASM Press; 1996. pp. 1344–1356. [Google Scholar]
- 148.Mangum J H, Magni G, Stadtman E R. Regulation of glutamine synthetase adenylylation and deadenylylation by the enzymatic uridylylation and deuridylylation of the PIIregulatory protein. Arch Biochem Biophys. 1973;158:514–525. doi: 10.1016/0003-9861(73)90543-2. [DOI] [PubMed] [Google Scholar]
- 149.Marcus M, Halpern Y S. The metabolic pathway of glutamate in Escherichia coliK-12. Biochim Biophys Acta. 1969;177:314–320. doi: 10.1016/0304-4165(69)90141-x. [DOI] [PubMed] [Google Scholar]
- 150.Martin D E, Hurek T, Reinhold-Hurek B. Occurrence of three PII-like signal transmitter proteins in the diazotrophic proteobacterium Azoarcussp. BH72. Mol Microbiol. 2000;38:276–288. doi: 10.1046/j.1365-2958.2000.02095.x. [DOI] [PubMed] [Google Scholar]
- 151.Martin G B, Thomashow M F, Chelm B K. Bradyrhizobium japonicum glnB, a putative nitrogen-regulatory gene, is regulated by NtrC at tandem promoters. J Bacteriol. 1989;171:5638–5645. doi: 10.1128/jb.171.10.5638-5645.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Masepohl B, Krey R, Klipp W. The draTG gene region of Rhodobacter capsulatusis required for post-translational regulation of both the molybdenum and the alternative nitrogenase. J Gen Microbiol. 1993;139:2667–2675. doi: 10.1099/00221287-139-11-2667. [DOI] [PubMed] [Google Scholar]
- 153.Meletzus D, Rudnick P, Doetsch N, Green A, Kennedy C. Characterization of the glnK-amtB operon of Azotobacter vinelandii. J Bacteriol. 1998;180:3260–3264. doi: 10.1128/jb.180.12.3260-3264.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Merrick M J. Regulation of nitrogen fixation genes in free-living and symbiotic bacteria. In: Stacey G, Burris R H, Evans H J, editors. Biological nitrogen fixation. New York, N.Y: Chapman and Hall; 1992. pp. 835–876. [Google Scholar]
- 155.Merrick M J, Edwards R A. Nitrogen control in bacteria. Microbiol Rev. 1995;59:604–622. doi: 10.1128/mr.59.4.604-622.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Mettke I, Fiedler U, Weiss V. Mechanism of activation of a response regulator: interaction of NtrC-P dimers induces ATPase activity. J Bacteriol. 1995;177:5056–5061. doi: 10.1128/jb.177.17.5056-5061.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Michel-Reydellet N, Desnoues N, de Zamaroczy M, Elmerich C, Kaminski P A. Characterisation of the glnK-amtB operon and involvement of AmtB in methylammonium uptake in Azorhizobium caulinodans. Mol Gen Genet. 1998;258:671–677. doi: 10.1007/s004380050781. [DOI] [PubMed] [Google Scholar]
- 158.Michel-Reydellet N, Desnoues N, Elmerich C, Kaminski P A. Characterization of Azorhizobium caulinodans glnB and glnA genes: involvement of the PIIprotein in symbiotic nitrogen fixation. J Bacteriol. 1997;179:3580–3587. doi: 10.1128/jb.179.11.3580-3587.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Michel-Reydellet N, Kaminski P A. Azorhizobium caulinodansPII and GlnK proteins control nitrogen fixation and ammonia assimilation. J Bacteriol. 1999;181:2655–2658. doi: 10.1128/jb.181.8.2655-2658.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Missaillidis S, Jaseja M, Ray P, Chittock R, Wharton C W, Drake A F, Buck M, Hyde E I. Secondary structure of the C-terminal DNA-binding domain of the transcriptional activator NifA from Klebsiella pneumoniae: spectroscopic analyses. Arch Biochem Biophys. 1999;361:173–182. doi: 10.1006/abbi.1998.0980. [DOI] [PubMed] [Google Scholar]
- 161.Mizuno T. Compilation of all genes encoding two-component phosphotransfer signal transducers in the genome of Escherichia coli. DNA Res. 1997;4:161–168. doi: 10.1093/dnares/4.2.161. [DOI] [PubMed] [Google Scholar]
- 162.Money T, Jones T, Dixon R, Austin S. Isolation and properties of the complex between the enhancer binding protein NifA and the sensor NifL. J Bacteriol. 1999;181:4461–4468. doi: 10.1128/jb.181.15.4461-4468.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Monteiro R A, Souza E M, Funayama S, Yates M G, Pedrosa F O, Chubatsu L S. Expression and functional analysis of an N-truncated NifA protein of Herbaspirillum seropedicae. FEBS Lett. 1999;447:283–286. doi: 10.1016/s0014-5793(99)00314-2. [DOI] [PubMed] [Google Scholar]
- 164.Monteiro R A, Souza E M, Yates M G, Pedrosa F O, Chubatsu L S. In-trans regulation of the N-truncated NifA protein of Herbaspirillum seropedicaeby the N-terminal domain. FEMS Microbiol Lett. 1999;180:157–161. doi: 10.1111/j.1574-6968.1999.tb08790.x. [DOI] [PubMed] [Google Scholar]
- 165.Morett E, Cannon W, Buck M. The DNA-binding domain of the transcriptional activator protein NifA resides in its carboxy terminus, recognises the upstream activator sequences of nifpromoters and can be separated from the positive control function of NifA. Nucleic Acids Res. 1988;16:11469–11488. doi: 10.1093/nar/16.24.11469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Morett E, Segovia L. The ς54bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J Bacteriol. 1993;175:6067–6074. doi: 10.1128/jb.175.19.6067-6074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Muse W B, Bender R A. The nac (nitrogen assimilation control) gene from Escherichia coli. J Bacteriol. 1998;180:1166–1173. doi: 10.1128/jb.180.5.1166-1173.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ninfa A J. Regulation of gene trasncription by extracellular stimuli. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C.: ASM Press; 1996. pp. 1246–1262. [Google Scholar]
- 169.Ninfa A J, Bennett R L. Identification of the site of autophosphorylation of the bacterial protein kinase/phosphatase NRII. J Biol Chem. 1991;266:6888–6893. [PubMed] [Google Scholar]
- 170.Ninfa A J, Magasanik B. Covalent modification of the glnG product, NRI, by the glnL product, NRII, regulates the transcription of the glnALG operon in Escherichia coli. Proc Natl Acad Sci USA. 1986;83:5909–5913. doi: 10.1073/pnas.83.16.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ninfa A J, Ueno-Nishio S, Hunt T P, Robustell B, Magasanik B. Purification of nitrogen regulator II, the product of the glnL (ntrB) gene of Escherichia coli. J Bacteriol. 1986;168:1002–1004. doi: 10.1128/jb.168.2.1002-1004.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ninfa E G, Atkinson M R, Kamberov E S, Ninfa A J. Mechanism of autophosphorylation of Escherichia coli nitrogen regulator II (NRII or NtrB): trans-phosphorylation between subunits. J Bacteriol. 1993;175:7024–7032. doi: 10.1128/jb.175.21.7024-7032.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ninnemann O, Jauniaux J-C, Frommer W B. Identification of a high affinity NH4+transporter from plants. EMBO J. 1994;13:3464–3471. doi: 10.1002/j.1460-2075.1994.tb06652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Noda S, Ohkuma M, Usami R, Horikoshi K, Kudo T. Culture-independent characterization of a gene responsible for nitrogen fixation in the symbiotic microbial community in the gut of the termite Neotermes koshunensis. Appl Environ Microbiol. 1999;65:4935–4942. doi: 10.1128/aem.65.11.4935-4942.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Noren A, Soliman A, Nordlund S. The role of NAD+ as a signal during nitrogenase switch-off in Rhodospirillum rubrum. Biochem J. 1997;322:829–832. doi: 10.1042/bj3220829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.O'Connell K P, Raffel S J, Saville B J, Handelsman J. Mutants of Rhizobium tropicistrain CIAT899 that do not induce chlorosis in plants. Microbiology. 1998;144:2607–2617. doi: 10.1099/00221287-144-9-2607. [DOI] [PubMed] [Google Scholar]
- 177.Overbeek R, Fonstein M, D'Souza M, Pusch G D, Maltsev N. The use of gene clusters to infer functional coupling. Proc Natl Acad Sci USA. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Parkinson J S, Kofoid E C. Communication modules in bacterial signalling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 179.Pawlowski K, Klosse U, de Bruijn F J. Characterisation of a novel Azorhizobium caulinodansORS571 two-component regulatory system, NtrY/NtrX, involved in nitrogen fixation and metabolism. Mol Gen Genet. 1991;231:124–138. doi: 10.1007/BF00293830. [DOI] [PubMed] [Google Scholar]
- 180.Pawlowski K, Ratet P, Schell J, DeBruijn F J. Cloning and characterisation of nifA and nifC genes of the stem nodulating bacterium ORS571, the nitrogen fixing symbiont of Sesbania rostrata: regulation of nitrogen fixation (nif) genes in the free-living versus symbiotic state. Mol Gen Genet. 1987;206:207–219. [Google Scholar]
- 181.Penyige A, Kalmanczhelyi A, Sipos A, Ensign J C, Barabas G. Modification of glutamine synthetase in Streptomyces griseusby ADP-ribosylation and adenylylation. Biochem Biophys Res Commun. 1994;204:598–605. doi: 10.1006/bbrc.1994.2501. [DOI] [PubMed] [Google Scholar]
- 182.Perlova O, Nawroth R, Meletzus D. Identification and characterisation of genes involved in the ammonium sensing mechanism in Acetobacter diazotrophicus. In: Elmerich C, Kondorosi A, Newton W E, editors. Biological nitrogen fixation for the 21st century. Dordrecht, The Netherlands: Kluwer Academic; 1998. p. 166. [Google Scholar]
- 183.Pioszak A A, Jiang P, Ninfa A J. The Escherichia coliPII signal transduction protein regulates the activities of the two-component system transmitter protein NRII by direct interaction with the kinase domain of the transmitter module. Biochemistry. 2000;39:13450–13461. doi: 10.1021/bi000795m. [DOI] [PubMed] [Google Scholar]
- 184.Prival M J, Brenchley J E, Magasanik B. Glutamine synthetase and the regulation of histidase formation in Klebsiella aerogenes. J Biol Chem. 1973;248:4334–4344. [PubMed] [Google Scholar]
- 185.Qian Y, Tabita F R. Expression of glnB and a glnB-like gene (glnK) in a ribulose bisphosphate carboxylase/oxygenase-deficient mutant of Rhodobacter sphaeroides. J Bacteriol. 1998;180:4644–4649. doi: 10.1128/jb.180.17.4644-4649.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Raina R, Bageshwar U K, Das H K. The Azotobacter vinelandii nifL-like gene: nucleotide sequence analysis and regulation of expression. Mol Gen Genet. 1993;237:400–406. doi: 10.1007/BF00279444. [DOI] [PubMed] [Google Scholar]
- 187.Ratet P, Pawlowski K, Schell J, de Bruijn F J. The Azorhizobium caulinodans nitrogen-fixation regulatory gene, nifA, is controlled by the cellular nitrogen and oxygen status. Mol Microbiol. 1989;3:825–838. doi: 10.1111/j.1365-2958.1989.tb00231.x. [DOI] [PubMed] [Google Scholar]
- 188.Reinhold-Hurek B, Dörr J, Egener T, Martin D, Hurek T. Interactions of diazortrophic Azoarcus spp. with rice. In: Pedrosa F O, Hungria M, Yates M G, Newton W E, editors. Nitrogen fixation: from molecules to crop productivity. Dordrecht, The Netherlands: Kluwer Academic; 2000. pp. 405–408. [Google Scholar]
- 189.Reith M E, Munholland J. Complete nucleotide sequence of the Porphyra purpureachloroplast genome. Plant Mol Biol Rep. 1995;13:333–335. [Google Scholar]
- 190.Reitzer L J. Ammonia assimilation and the biosynthesis of glutamine, glutamate, aspartate, asparagine, l-alanine and d-alanine. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C.: ASM Press; 1996. pp. 391–407. [Google Scholar]
- 191.Reuveny Z, Foor F, Magasanik B. Regulation of glutamine synthetase by regulatory protein PII in Klebsiella aerogenesmutants lacking adenylyltransferase. J Bacteriol. 1981;146:740–745. doi: 10.1128/jb.146.2.740-745.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Reyes-Ramirez F, Little R, Hill S, van Heeswijk W, Dixon R. Regulation of Azotobacter vinelandii NifA activity by NifL: role of PII-like proteins in nitrogen sensing. In: Pedrosa F O, Hungria M, Yates M G, Newton W E, editors. Nitrogen fixation: from molecules to crop productivity. Dordrecht, The Netherlands: Kluwer Academic; 2000. pp. 97–98. [Google Scholar]
- 193.Rhee S G. 5′-Nucleotidyl-O-tyrosine bond in glutamine synthetase. Methods Enzymol. 1984;107:183–201. doi: 10.1016/0076-6879(84)07012-9. [DOI] [PubMed] [Google Scholar]
- 194.Rhee S G, Chock P B, Stadtman E R. Regulation of Escherichia coliglutamine synthetase. Adv Enzymol. 1989;62:37–92. doi: 10.1002/9780470123089.ch2. [DOI] [PubMed] [Google Scholar]
- 195.Rossi M, Defez R, Chiurazzi M, Lamberti A, Fuggi A, Iaccarino M. Regulation of glutamine synthetase isoenzymes in Rhizobium legumonosarum biovar viciae. J Gen Microbiol. 1989;135:629–637. [Google Scholar]
- 196.Rudnick P, Colnaghi R, Green A, Kennedy C. Molecular analysis of the glnB, amtB, glnD and glnA genes in Azotobacter vinelandii. In: Elmerich C, Kondorosi A, Newton W E, editors. Biological nitrogen fixation for the 21st century. Dordrecht, The Netherlands: Kluwer Academic; 1998. pp. 123–124. [Google Scholar]
- 197.Russo F D, Silhavy T J. EnvZ controls the concentration of phosphorylated OmpR to mediate osmoregulation of the porin génes. J Mol Biol. 1991;222:567–580. doi: 10.1016/0022-2836(91)90497-t. [DOI] [PubMed] [Google Scholar]
- 198.Sauer J, Görl M, Forchhammer K. Nitrogen starvation in SynechococcusPCC 7942: involvement of glutamine synthetase and NctA in phycobiliprotein degradation and survival. Arch Microbiol. 1999;172:247–255. doi: 10.1007/s002030050767. [DOI] [PubMed] [Google Scholar]
- 199.Schluter A, Nohlen M, Kramer M, Defez R, Priefer U B. The Rhizobium leguminosarum bv. viciae glnDgene, encoding a uridylyltransferase/ uridylyl-removing enzyme, is expressed in the root nodule but is not essential for nitrogen fixation. Microbiology. 2000;146:2987–2996. doi: 10.1099/00221287-146-11-2987. [DOI] [PubMed] [Google Scholar]
- 200.Schmitz G, Nikaido K, Ames G F L. Regulation of a transport operon in Salmonella typhimurium: identification of sites essential for nitrogen regulation. Mol Gen Genet. 1988;215:107–117. doi: 10.1007/BF00331311. [DOI] [PubMed] [Google Scholar]
- 201.Schmitz R A. NifL of Klebsiella pneumoniaecarries an N-terminally bound FAD cofactor, which is not directly required for the inhibitory function of NifL. FEMS Microbiol Lett. 1998;157:313–318. doi: 10.1111/j.1574-6968.1997.tb12791.x. [DOI] [PubMed] [Google Scholar]
- 202.Schneider B L, Kiupakis A K, Reitzer L J. Arginine catabolism and the arginine succinyltransferase pathway in Escherichia coli. J Bacteriol. 1998;180:4278–4286. doi: 10.1128/jb.180.16.4278-4286.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Schneider B L, Reitzer L J. Salmonella typhimurium nit is nadE: defective nitrogen utilization and ammonia-dependent NAD synthetase. J Bacteriol. 1998;180:4739–4741. doi: 10.1128/jb.180.17.4739-4741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Schutt H, Holzer H. Biological function of the ammonia-induced inactivation of glutamine synthetase in Escherichia coli. Eur J Biochem. 1972;26:68–72. doi: 10.1111/j.1432-1033.1972.tb01740.x. [DOI] [PubMed] [Google Scholar]
- 205.Senior P J. Regulation of nitrogen metabolism in Escherichia coli and Klebsiella aerogenes: studies with the continuous culture technique. J Bacteriol. 1975;123:407–418. doi: 10.1128/jb.123.2.407-418.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Shapiro B M. The glutamine synthetase deadenylylating enzyme system from Escherichia coli: resolution into two components, specific nucleotide stimulation, and cofactor requirements. Biochemistry. 1969;8:659–670. doi: 10.1021/bi00830a030. [DOI] [PubMed] [Google Scholar]
- 207.Shapiro B M, Stadtman E R. Glutamine synthetase deadenylylating enzyme. Biochem Biophys Res Commun. 1968;30:32–37. doi: 10.1016/0006-291x(68)90708-0. [DOI] [PubMed] [Google Scholar]
- 208.Sibold L, Henriquet M, Possot O, Aubert J-P. Nucleotide sequence of nifH regions from Methanobacterium ivanovii and Methanosarcina barkeri 227 and characterisation of glnB-like genes. Res Microbiol. 1991;142:5–12. doi: 10.1016/0923-2508(91)90091-n. [DOI] [PubMed] [Google Scholar]
- 209.Silman N J, Carr N G, Mann N H. ADP-ribosylation of glutamine synthetase in the cyanobacterium Synechocystissp. strain PCC 6803. J Bacteriol. 1995;177:3527–3533. doi: 10.1128/jb.177.12.3527-3533.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicumdeltaH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Söderbäck E, Reyes-Ramirez F, Eydmann T, Austin S, Hill S, Dixon R. The redox- and fixed nitrogen-responsive regulatory protein NifL from Azotobacter vinelandiicomprises discrete flavin and nucleotide-binding domains. Mol Microbiol. 1998;28:179–192. doi: 10.1046/j.1365-2958.1998.00788.x. [DOI] [PubMed] [Google Scholar]
- 212.Sohlenkamp C, Shelden M, Howitt S, Udvardi M. Characterization of ArabidopsisAtAMT2, a novel ammonium transporter in plants. FEBS Lett. 2000;467:273–278. doi: 10.1016/s0014-5793(00)01153-4. [DOI] [PubMed] [Google Scholar]
- 213.Soliman A, Nordlund S. Studies on the effect of NAD(H) on nitrogenase activity in Rhodospirillum rubrum. Arch Microbiol. 1992;157:431–435. doi: 10.1007/BF00249100. [DOI] [PubMed] [Google Scholar]
- 214.Son H S, Rhee S G. Cascade control of Escherichia coli glutamine synthetase: purification and properties of PIIprotein and nucleotide sequence of its structural gene. J Biol Chem. 1987;262:8690–8695. [PubMed] [Google Scholar]
- 215.Souillard N, Sibold L. Primary structure, functional organization and expression of nitrogenase structural genes of the thermophilic archaebacterium Methanococcus thermolithotrophicus. Mol Microbiol. 1989;3:541–551. doi: 10.1111/j.1365-2958.1989.tb00200.x. [DOI] [PubMed] [Google Scholar]
- 216.Souza E M, Funayama S, Rigo L U, Yates M G, Pedrosa F O. Sequence and structural organization of a nifA-like gene and part of a nifB-like gene of Herbaspirillum seropedicaestrain Z78. J Gen Microbiol. 1991;137:1511–1522. doi: 10.1099/00221287-137-7-1511. [DOI] [PubMed] [Google Scholar]
- 217.Souza E M, Pedrosa F O, Drummond M, Rigo L U, Yates M G. Control of Herbaspirillum seropedicaeNifA activity by ammonium ions and oxygen. J Bacteriol. 1999;181:681–684. doi: 10.1128/jb.181.2.681-684.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Stadtman E R. Discovery of glutamine synthetase cascade. Methods Enzymol. 1990;182:793–809. doi: 10.1016/0076-6879(90)82062-7. [DOI] [PubMed] [Google Scholar]
- 219.Stadtman E R, Ginsburg A, Ciardi J E, Yeh J, Hennig S B, Shapiro B M. Multiple molecular forms of glutamine synthetase produced by enzyme catalyzed adenylation and deadenylylation reactions. Adv Enzyme Regul. 1970;8:99–118. doi: 10.1016/0065-2571(70)90011-7. [DOI] [PubMed] [Google Scholar]
- 220.Stock J B, Ninfa A J, Stock A M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Stone J M, Walker J C. Plant protein kinase families and signal transduction. Plant Physiol. 1995;108:451–457. doi: 10.1104/pp.108.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Tate R, Riccio A, Merrick M, Patriarca E J. The Rhizobium etli amtB gene coding for an NH+4transporter is down-regulated early during bacteroid differentiation. Mol Plant-Microbe Interact. 1998;11:188–198. doi: 10.1094/MPMI.1998.11.3.188. [DOI] [PubMed] [Google Scholar]
- 223.Thomas G, Coutts G, Merrick M. The glnKamtBoperon: a conserved gene pair in prokaryotes. Trends Genet. 2000;16:11–14. doi: 10.1016/s0168-9525(99)01887-9. [DOI] [PubMed] [Google Scholar]
- 224.Toukdarian A, Saunders G, Selman-Sosa G, Santero E, Woodley P, Kennedy C. Molecular analysis of the Azotobacter vinelandii glnAgene encoding glutamine synthetase. J Bacteriol. 1990;172:6529–6539. doi: 10.1128/jb.172.11.6529-6539.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Tsinoremas N F, Castets A M, Harrison M A, Allen J F, Tandeau de Marsac N. Photosynthetic electron transport controls nitrogen assimilation in cyanobacteria by means of posttranslational modification of the glnBgene product. Proc Natl Acad Sci USA. 1991;88:4565–4569. doi: 10.1073/pnas.88.11.4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Tyler B. Regulation of the assimilation of nitrogen compounds. Annu Rev Biochem. 1978;47:1127–1162. doi: 10.1146/annurev.bi.47.070178.005403. [DOI] [PubMed] [Google Scholar]
- 227.van Dommelen A. Ammonium transport in Azospirillum brasilense. Ph.D. thesis. Heverlee, Belgium: Katholieke Universiteit Leuven; 1998. [Google Scholar]
- 228.van Dommelen A, Keijers V, Vanderleyden J, de Zamaroczy M. (Methyl)ammonium transport in the nitrogen-fixing bacterium Azospirillum brasilense. J Bacteriol. 1998;180:2652–2659. doi: 10.1128/jb.180.10.2652-2659.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.van Heeswijk W, Kuppinger O, Merrick M, Kahn D. Precise localization of the glnD gene on the physical map of Escherichia coli. J Bacteriol. 1992;174:1702–1703. doi: 10.1128/jb.174.5.1702-1703.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.van Heeswijk W C. The glutamine synthetase adenylylation cascade Ph.D. thesis. Amsterdam, The Netherlands: Free University of Amsterdam; 1998. [Google Scholar]
- 231.van Heeswijk W C, Hoving S, Molenaar D, Stegeman B, Kahn D, Westerhoff H V. An alternative PII protein in the regulation of glutamine synthetase in Escherichia coli. Mol Microbiol. 1996;21:133–146. doi: 10.1046/j.1365-2958.1996.6281349.x. [DOI] [PubMed] [Google Scholar]
- 232.van Heeswijk W C, Rabenberg M, Westerhof H V, Kahn D. The genes of the glutamine synthetase adenylylation cascade are not regulated by nitrogen in Escherichia coli. Mol Microbiol. 1993;9:443–457. doi: 10.1111/j.1365-2958.1993.tb01706.x. [DOI] [PubMed] [Google Scholar]
- 233.van Heeswijk W C, Stegeman B, Hoving S, Molenaar D, Kahn D, Westerhoff H V. An additional PII in Escherichia coli: a new regulatory protein in the glutamine synthetase cascade. FEMS Microbiol Lett. 1995;132:153–157. doi: 10.1111/j.1574-6968.1995.tb07825.x. [DOI] [PubMed] [Google Scholar]
- 234.van Heeswijk W C, Wen D, Clancy P, Jaggi R, Ollis D L, Westerhoff H V, Vasudevan S G. The Escherichia coli signal transducers PII (GlnB) and GlnK form heterotrimers in vivo: fine tuning the nitrogen signal cascade. Proc Natl Acad Sci USA. 2000;97:3942–3947. doi: 10.1073/pnas.97.8.3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Vasudevan S G, Armarego W L, Shaw D C, Lilley P E, Dixon N E, Poole R K. Isolation and nucleotide sequence of the hmp gene that encodes a haemoglobin-like protein in Escherichia coliK-12. Mol Gen Genet. 1991;226:49–58. doi: 10.1007/BF00273586. [DOI] [PubMed] [Google Scholar]
- 236.Vasudevan S G, Gedye C, Dixon N E, Cheah E, Carr P D, Suffolk P M, Jeffrey P D, Ollis D L. Escherichia coli PIIprotein: purification, crystallisation and oligomeric structure. FEBS Lett. 1994;337:255–258. doi: 10.1016/0014-5793(94)80203-3. [DOI] [PubMed] [Google Scholar]
- 237.Vega-Palas M A, Flores E, Herrero A. NtcA, a global nitrogen regulator from the cyanobacterium Synechococcusthat belongs to the Crp family of bacterial regulators. Mol Microbiol. 1992;6:1853–1859. doi: 10.1111/j.1365-2958.1992.tb01357.x. [DOI] [PubMed] [Google Scholar]
- 238.Vega-Palas M A, Madueno F, Herrero A, Flores E. Identification and cloning of a regulatory gene for nitrogen assimilation in the cyanobacterium Synechococcussp. strain PCC 7942. J Bacteriol. 1990;172:643–647. doi: 10.1128/jb.172.2.643-647.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Volkman B F, Nohaile M J, Amy N K, Kustu S, Wemmer D E. 3-Dimensional solution stucture of the N-terminal receiver domain of NtrC. Biochemistry. 1995;34:1413–1424. doi: 10.1021/bi00004a036. [DOI] [PubMed] [Google Scholar]
- 240.Weiss D S, Batut J, Klose K E, Keener J, Kustu S. The phosphorylated form of the enhancer-binding protein NTRC has an ATPase activity that is essential for activation of transcription. Cell. 1991;67:155–167. doi: 10.1016/0092-8674(91)90579-n. [DOI] [PubMed] [Google Scholar]
- 241.Weiss V, Claverie-Martin F, Magasanik B. Phosphorylation of nitrogen regulator I of Escherichia coliinduces strong cooperative binding to DNA essential for activation of transcription. Proc Natl Acad Sci USA. 1992;89:5088–5092. doi: 10.1073/pnas.89.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Weiss V, Magasanik B. Phosphorylation of nitrogen regulator I (NR1) of Escherichia coli. Proc Natl Acad Sci USA. 1988;85:8919–8923. doi: 10.1073/pnas.85.23.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Willison J C, Tissot G. The Escherichia coli efg gene and the Rhodobacter capsulatus adg gene code for NH3-dependent NAD synthetase. J Bacteriol. 1994;176:3400–3402. doi: 10.1128/jb.176.11.3400-3402.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Woehle D L, Lueddecke B A, Ludden P W. ATP-dependent and NAD-dependent modification of glutamine synthetase from Rhodospirillum rubrum in vitro. J Biol Chem. 1990;265:13741–13749. [PubMed] [Google Scholar]
- 245.Woodley P, Drummond M. Redundancy of the conserved His residue in Azotobacter vinelandiiNifL, a histidine autokinase homologue which regulates transcription of nitrogen fixation genes. Mol Microbiol. 1994;13:619–626. doi: 10.1111/j.1365-2958.1994.tb00456.x. [DOI] [PubMed] [Google Scholar]
- 246.Wray L, Atkinson M, Fisher S. The nitrogen-regulated Bacillus subtilis nrgAB operon encodes a membrane protein and a protein highly similar to the Escherichia coli glnB-encoded PII. J Bacteriol. 1994;176:108–114. doi: 10.1128/jb.176.1.108-114.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Wray L, Zalieckas J M, Ferson A E, Fisher S H. Mutational analysis of the TnrA-binding sites in the Bacillus subtilis nrgAB and gabPpromoter regions. J Bacteriol. 1998;180:2943–2949. doi: 10.1128/jb.180.11.2943-2949.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Wray L V, Zalieckas J M, Fisher S H. Purification and in vitro activities of the Bacillus subtilisTnrA transcription factor. J Mol Biol. 2000;300:29–40. doi: 10.1006/jmbi.2000.3846. [DOI] [PubMed] [Google Scholar]
- 249.Wray L V, Jr, Ferson A E, Rohrer K, Fisher S H. TnrA, a transcription factor required for global nitrogen regulation in Bacillus subtilis. Proc Natl Acad Sci USA. 1996;93:8841–8845. doi: 10.1073/pnas.93.17.8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Wu S Q, Chai W, Lin J T, Stewart V. General nitrogen regulation of nitrate assimilation regulatory gene nasR expression in Klebsiella oxytocaM5al. J Bacteriol. 1999;181:7274–7284. doi: 10.1128/jb.181.23.7274-7284.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Wyman C, Rombel I, North A K, Bustamante C, Kustu S. Unusual oligomerization required for activity of NtrC, a bacterial enhancer-binding protein. Science. 1997;275:1658–1661. doi: 10.1126/science.275.5306.1658. [DOI] [PubMed] [Google Scholar]
- 252.Xu Y, Cheah E, Carr P D, van Heeswijk W C, Westerhof H V, Vasudevan S G, Ollis D. GlnK, a PII-homologue: structure reveals ATP binding site and indicates how the T-loops may be involved in molecular recognition. J Mol Biol. 1998;282:149–165. doi: 10.1006/jmbi.1998.1979. [DOI] [PubMed] [Google Scholar]
- 253.Yakunin A F, Hallenbeck P C. Short-term regulation of nitrogenase activity by NH4+ in Rhodobacter capsulatus: multiple in vivo nitrogenase response to NH4+addition. J Bacteriol. 1998;180:6392–6395. doi: 10.1128/jb.180.23.6392-6395.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Yakunin A F, Hallenbeck P C. AmtB is necessary for NH4+ induced nitrogenase switch-off and ADP-ribosylation in Rhodobacter capsulatus. In: Pedrosa F O, Hungria M, Yates M G, Newton W E, editors. Nitrogen fixation: from molecules to crop productivity. Dordrecht, The Netherlands: Kluwer Academic; 2000. pp. 95–96. [Google Scholar]
- 255.Yakunin A F, Hallenbeck P C. Regulation of nitrogenase activity in Rhodobacter capsulatusunder dark microoxic conditions. Arch Microbiol. 2000;173:366–372. doi: 10.1007/s002030000156. [DOI] [PubMed] [Google Scholar]
- 256.Yamashita M M, Almassy R J, Janson C A, Cascio D, Eisenberg D. Refined atomic model of glutamine synthetase at 3.5Åresolution. J Biol Chem. 1989;264:17681–17690. doi: 10.2210/pdb2gls/pdb. [DOI] [PubMed] [Google Scholar]
- 257.Zhang C C, Libs L. Cloning and characterisation of the pknD gene encoding an eukaryotic-type protein kinase in the cyanobacterium Anabaenasp. PCC7120. Mol Gen Genet. 1998;258:26–33. doi: 10.1007/s004380050703. [DOI] [PubMed] [Google Scholar]
- 258.Zhang Y, Burris R H, Ludden P W, Roberts G P. Posttranslational regulation of nitrogenase activity by anaerobiosis and ammonium in Azospirillum brasilense. J Bacteriol. 1993;175:6781–6788. doi: 10.1128/jb.175.21.6781-6788.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Zhang Y, Burris R H, Ludden P W, Roberts G P. Presence of a second mechanism for the posttranslational regulation of nitrogenase activity in Azospirillum brasilensein response to ammonium. J Bacteriol. 1996;178:2948–2953. doi: 10.1128/jb.178.10.2948-2953.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Zhang Y, Burris R H, Ludden P W, Roberts G P. Regulation of nitrogen fixation in Azospirillum brasilense. FEMS Microbiol Lett. 1997;152:195–204. doi: 10.1111/j.1574-6968.1997.tb10428.x. [DOI] [PubMed] [Google Scholar]
- 261.Zhang Y, Pohlmann E L, Ludden P W, Roberts G P. Mutagenesis and functional characterization of the glnB, glnA, and nifA genes from the photosynthetic bacterium Rhodospirillum rubrum. J Bacteriol. 2000;182:983–992. doi: 10.1128/jb.182.4.983-992.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261a.Zhang Y, Pohlmann E L, Halbleib C M, Ludden P W, Roberts G P. Effect of PII and its homolog GlnK on reversible ADP-ribosylation of dinitrogenase reductase by heterologous expression of the Rhodospirillum rubrum dinitrogenase reductase ADP-ribosyl transferase–dinitrogenase reductase-activating glycohydrolase regulatory system in Klebsiella pneumoniae. J Bacteriol. 2001;183:1610–1620. doi: 10.1128/JB.183.5.1610-1620.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Zinchenko V, Churin Y, Shestopalov V, Shestakov S. Nucleotide sequence and characterisation of Rhodobacter sphaeroides glnB and glnAgenes. Microbiology. 1994;140:2143–2151. doi: 10.1099/13500872-140-8-2143. [DOI] [PubMed] [Google Scholar]




