Abstract
In the past few years, many retrovirus receptors, coreceptors, and cofactors have been identified. These molecules are important for some aspects of viral entry, although in some cases it remains to be determined whether they are required for binding or postbinding stages in entry, such as fusion. There are certain common features to the molecules that many retroviruses use to gain entry into the cell. For example, the receptors for most mammalian oncoretroviruses are multiple membrane-spanning transport proteins. However, avian retroviruses use single-pass membrane proteins, and a sheep retrovirus uses a glycosylphosphatidylinositol-anchored molecule as its receptor. For some retroviruses, particularly the lentiviruses, two cell surface molecules are required for efficient entry. More recently, a soluble protein that is required for viral entry has been identified for a feline oncoretrovirus. In this review, we will focus on the various strategies used by mammalian retroviruses to gain entry into the cell. The choice of receptors will also be discussed in light of pressures that drive viral evolution and persistence.
There has been intensive study of retrovirus-host cell interactions because retroviral infections can lead to diseases such as cancer and immunodeficiency in many species, including humans. In addition, retroviruses are used as vehicles for gene transfer because the DNA form of the viral genome becomes an integrated part of the host cell genome. Thus, there has been a considerable body of research focused on defining the functional interactions between retroviral envelope proteins and their receptors that are required to initiate a productive infection. As a result of these efforts, many receptors for retroviruses have been identified (Table 1).
TABLE 1.
Cloned retrovirus receptors
| Retrovirus | Receptor | Type | Function |
|---|---|---|---|
| HIV, SIV | CD4 and CXCR4, CCR5, others | TM1 TM7 | Immune recognition, G protein-coupled chemokine receptors |
| E-MLV | CAT-1 (SLC7A1) | TM14 | Basic amino acid transport |
| GALV, 10A1, MLV, FeLV-B, woolly monkey virus | Pit1 (SLC20A1) | TM10–13 | Phosphate transport |
| A-MLV, 10A1, MLV, FeLV-B | Pit2 (SLC20A2) | TM10-–13 | Phosphate transport |
| BLV | Blvr | TM1 | ? |
| ASLV-A | Tva | TM1 | LDL receptor-like protein |
| ASLV-B, ASLV-D, ASLV-E | Tvb | TM1 | Fas/NFR-like receptor |
| MMTV | Mtvr | TM1 | ? |
| RD-114, type D SRV, BaEV, HERV-W | RDR (SLC1A5) or RDR2 (SLC1A4) | TM9–10 TM9–10 | Neutral amino acid transport Glutamate, neutral amino acid transport |
| Xenotropic and polytropic MLVs | XPR1 | TM8 | G protein-cuopled signaling?, transport? |
| FeLV-C | Flvcr | TM12 | Organic anion transporter? |
| FeLV-T | FeLIX and Pit1 (SLC20A1) | Soluble TM10–13 | Env-like protein Phosphate transport |
| JSRV | HYAL2 | GPI anchored | Hyaluronidase (weak) |
In the past few years, it has become clear that there are many different types of cell surface molecules that function as retroviral receptors, although members of the same genera of retroviruses tend to use cell molecules with some similarities in structure and function. The current recommended taxonomy for retroviruses divides them into seven genera (Table 2 and information found at the ICN website, ICTV [www.ncbi.nih.gov/ICTV]), although previously, they have been grouped largely based on virion morphology (type B, C, and D). Historically, the type C retroviruses have been the most extensively studied, in part because many of these viruses are oncogenic. Under the current designation, the type C avian viruses are grouped as alpharetroviruses whereas the type C mammalian oncogenic viruses are members of the gammaretroviruses. Many of the members of these two genera also have several common features: they are horizontally and vertically transmitted; at some point they infected the germ line of the host, leaving endogenous copies of their sequences in the host genome; and they may capture either these endogenous viral sequences or cellular proto-oncogene sequences in their genome during reverse transcription. Several of the receptors for type C oncoretroviruses have been identified and will be discussed in some detail here. The Lentivirus genus has also been particularly well studied because it includes human immunodeficiency virus (HIV) and because lentiviruses are generally pathogenic. Lentiviruses have a more complex genome than the type C oncoretroviruses, and they tend to cause degenerative rather than proliferative diseases. In addition, the lentiviruses differ from the oncoretroviruses by the fact that there are not related endogenous sequences in the host genome. However, many aspects of viral entry are shared by oncoretroviruses and lentiviruses. The receptors for several lentiviruses are also known, and these receptors will be represented in this review. Less is known about the receptors for the other retroviral groups, although receptors for several betaretroviruses and one deltaretrovirus have been described. The receptors for epsilonretroviruses and spumaviruses remain largely uncharacterized.
TABLE 2.
Retrovirus genera
| Genus | Morphology | Examplesa |
|---|---|---|
| Alpharetrovirus | C type | RSV, ASLV |
| Betaretrovirus | B and D type | MMTV, SRV-1 to SRV-5, BaEV, JSRV, ENTV |
| Gammaretrovirus | C type | MoMLV, A-MLV, 10A1 MLV, X-MLV, P-MLV, AKV, GALV, MDEV, FeLV, PERV, RD-114, SNV, REV |
| Deltaretrovirus | HTLV-1, HTLV-2, STLV-1 to STLV-3, BLV | |
| Epsilonretrovirus | WDSV | |
| Lentivirus | HIV-1, HIV-2, SIV | |
| Spumavirus | HFV, SFV |
RSV, Rous sarcoma virus; MMTV, mouse mammary tumor virus; ENTV, enzootic nasal tumor virus; MoMLV, Moloney MLV; X;MLV, xenotropic MLV; P-MLV, polytropic MLV; AKV, AKV MLV; MDEV, M. dunni endogenous virus; REV, reticuloendotheliosis virus; STVL, simian T-lymphotropic virus; BLV, bovine leukemia virus; WDSV, walleye dermal sarcoma virus; HFV, human foamy virus; SFV, simian foamy virus.
The envelope protein of the virus interacts with at least one specific host cell receptor to initiate infection. In some cases, more than one cell surface molecule is required to permit viral entry. This may reflect the requirement that a cell surface molecule(s) must specifically bind the viral envelope protein, and virus binding must then lead to fusion of the viral and cell membranes for infection to occur. Thus, in some cases, one molecule may be both the binding and fusion receptor, as appears to be the case for oncoretroviruses, whereas in other cases, different cell surface proteins may carry out these distinct functions, as occurs for HIV. The pattern of expression of the binding and fusion receptor(s) helps define the host cell specificity of retroviruses, although there may be subsequent restrictions to replication in cells that express the appropriate receptor(s).
The envelope protein of retroviruses is encoded as a polyprotein precursor, which is then cleaved into a surface unit (SU), which is on the outside of the virion, and a transmembrane (TM) protein, which anchors the SU to the viral membrane. The envelope precursor enters the secretory pathway during translation, the signal sequence is cleaved, and the precursor is glycosylated in the endoplasmic reticulum. After glycosylation, the envelope precursor protein is transported to the Golgi, where the oligosaccharides are modified and the glycoprotein precursor is cleaved by a host cell protease to generate SU and TM. The mature envelope proteins are then transported to the cell surface as an oligomeric complex, where they are captured when the virus buds from the cell membrane (for more details, see reference 180).
The SU protein initiates entry by binding to a specific cell surface protein; SU is therefore the primary determinant of the range of cells susceptible to infection by a retrovirus. It is generally acknowledged that the interaction of the SU portion of the retroviral envelope to its receptor induces a conformational change that exposes a viral fusion peptide, present in the ectodomain of TM, allowing the viral membrane to fuse with the cell membrane. For most retroviruses, fusion occurs at neutral pH. The fusion process is not energetically favorable; it is subject to strong repulsive hydration, electrostatic, and steric barriers (133). It is likely that the mechanism(s) that the receptor-bound virus uses to surmount these barriers determines why only certain cell surface proteins can function as receptors for enveloped viruses. After fusion of the viral and cell membranes, the nucleocapsid, which contains the diploid viral RNA genomes and the viral reverse transcriptase, is transferred to the cytoplasmic side of the cell membrane. Because reverse transcription of the retroviral RNA genome is presumed to occur within the nucleocapsid, it is necessary for this relatively large viral structure to pass through the cytoskeletal cortex prior to being transferred to the cytosol. This final stage of viral entry, the internalization of the nucleocapsid, remains a relatively poorly defined stage of viral infection (88).
Once a retrovirus productively infects the cell, the cell typically becomes resistant to reinfection by a virus that uses the same receptor. Superinfection interference was described as the blocking of the viral receptor by envelope or virus produced in an infected cell (163). This interference can occur by one of several mechanisms, including internalization of the receptor from the cell surface, disruption of transport of the receptor to the membrane, and/or competitive inhibition for the receptor binding domain (for more details, see reference 75). Interference has been studied in cell culture systems where there is a high level of infection and/or viral SU expression, but less is known about the role played by superinfection interference among competing viruses in the host. However, there is evidence that endogenously expressed envelope proteins may interfere with exogenous viruses that bind the same receptor (61, 114). Before receptors were identified, retroviruses were often categorized into interference groups as a means of classifying viruses that used the same receptors. In the past decade, receptors for more than half of the different interference groups have been defined. The notable cases where the receptors have not been defined are the primate leukemia viruses (e.g., human T-cell leukemia virus [HTLV]), which are linked to human cancers and neurological diseases, and the various endogenous viruses of pigs (PERVs), which are of high interest due to their potential spread during xenotransplantation.
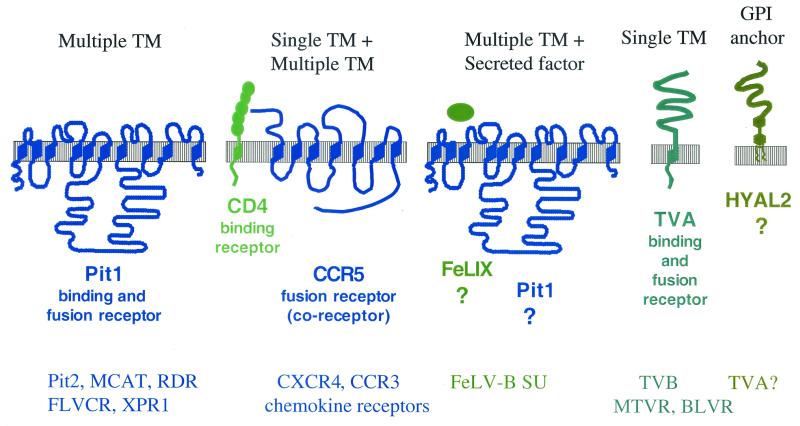
Both multiple transmembrane-spanning (CAT-1 and Pit1) and single transmembrane-spanning (CD4) receptors were among the first receptors to be identified. More recently, it has become clear that some viruses require two molecules for entry. In the past year, a secreted and a glycosylphosphatidylinositol (GPI)-anchored cellular protein have been shown to be required for entry by some retroviruses. These various classes of receptors are shown in Fig. 1. In this review, we will discuss each of these receptor classes.
FIG. 1.
Schematic depiction of the different classes of retroviral receptors. The general class of receptor is described above the cartoon, which depicts the predicted topology of a specific receptor of that class, as indicated below the cartoon. For the indicated receptor, its commonly assumed role in binding and/or fusion is indicated. A question mark indicates that there are no published data on whether the molecule is a binding or fusion receptor. At the bottom of the figure, other members of the general receptor class are shown, using colors to refer to the specific molecule in the cartoon above. These receptors are assigned to a specific class on the basis of whether they are single- or multiple-membrane proteins, not because they have an identical structure as the receptor shown. For example, the members of the multiple TM class may vary in the predicted number of TM domains. Only two coreceptors for lentiviruses are mentioned, and the reader is referred to references 20 and 71 for a more complete list. Also, as discussed in the text, there are some cases where lentiviruses use only the coreceptor and not CD4 to infect cells. This is not specifically indicated, although such cases would be best described as falling in the class of multiple TM receptors.
MANY ONCORETROVIRUSES USE MULTIPLE MEMBRANE-SPANNING CELLULAR PROTEINS AS RECEPTORS
More protein receptors have been identified for oncoretroviruses, particularly the family Gammaretroviridae, than for any other, and all are multiple membrane-spanning proteins. The first multiple membrane-spanning receptor to be identified was a receptor for murine leukemia virus (MLV). There are several isolates of MLV that differ in their host range (a feature that generally corresponds to differences in receptor usage) and their associated pathology. Ecotropic MLVs infect primarily murine and rat cells, whereas 10A1 MLV and amphotropic MLVs infect cells from a wide range of diverse species in addition to murine cells. Xenotropic MLVs have a similar host range to amphotropic MLVs, but they fail to infect most types of murine cells. Polytropic MLVs have a host range similar to amphotropic MLVs, but they are alternatively designated mink cell focus-forming viruses because of the unique cytopathic effect they induce in mink cell cultures.
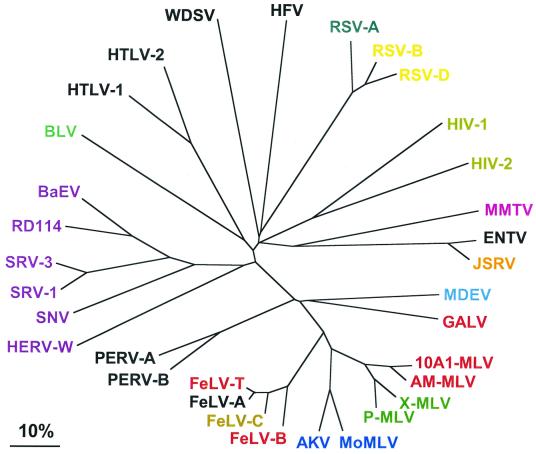
Feline leukemia virus (FeLV) is related to MLV, and its members include isolates that have distinct host range properties; these correspond to the four subgroups (A, B, C, and T [122, 155, 156]). Like MLVs, some FeLVs are restricted to replication in cells from their natural feline host (FeLV-A and FeLV-T) whereas others can replicate in a variety of nonfeline cells (FeLV-B and FeLV-C). Besides the murine and feline retroviruses, a number of other replication-competent mammalian oncoretroviruses use multiple transmembrane proteins as receptors; these include the related viruses, gibbon ape leukemia virus (GALV) and woolly monkey virus, that have a common receptor with some MLVs and FeLVs. In addition, a group of viruses (RD-114, simion retrovirus [SRV], baboon endogenous retrovirus [BaEV], reticuloendotheliosis virus A [REV-A], and human endogenous retrovirus W [HERV-W]), which are highly divergent from MLVs, FeLVs, and GALV and form a distinct evolutionary cluster (Fig. 2), all use a common multiple membrane-spanning receptor.
FIG. 2.
Amino acid sequence similarity among retrovirus envelope proteins. Envelope proteins, including the endoplasmic reticulum signal sequences, were compared using CLUSTAL W. The scale bar indicates 10% sequence difference. Viruses that are shown in the same color use a similar receptor. The receptors for viruses shown in black remain to be identified. Abbreviations: MoMLV, Moloney MLV; AKV, AKR MLV; MCF, mink cell focus-forming virus; NZB, New Zealand black mouse MLV; AM-MLV, amphotropic MLV; 10A1, 10A1 MLV; MDEV, M. dunni endogenous virus; ENTV, enzootic nasal tumor virus; MMTV, mouse mammary tumor virus; RSV-A, RSV-B, and RSV-D, Rous sarcoma virus types A, B, and D; HFV, “human” foamy virus (actually of simian origin); WDSV, walleye dermal sarcoma virus; BLV, bovine leukemia virus; RD114, cat endogenous virus RD-114; MPMV, Mason-Pfizer monkey virus, also called SRV-3; SNV, avian spleen necrosis virus;
Ecotropic MLVs Use a Cationic Amino Acid Transporter, CAT-1, as a Receptor
The receptor for ecotropic MLVs (E-MLVs) is predicted to contain 14 transmembrane domains. This receptor, designated CAT-1 (for “cationic amino acid transporter”), is a membrane-spanning glycoprotein (4) that functions as a sodium-dependent transporter of the basic amino acids, lysine, ornithine, and arginine (85, 182). Chronic infection of murine cells by E-MLVs results in a complete loss of E-MLV receptor function. However, the transporter remains functional, although a reduction in amino acid uptake activity is observed (183). Furthermore, murine CAT-1 proteins that have lost the ability to mediate viral entry retain transporter function (35, 81).
CAT-1 receptor orthologs expressed on the surfaces of cells from species other than mice and rats demonstrate the ability to transport cationic amino acids but lack the ability to mediate E-MLV entry. For example, human CAT-1 has 86% amino acid identity to murine CAT-1, but it does not function as an E-MLV receptor (196). Therefore, human/mouse CAT-1 chimeric proteins have been useful tools for mapping regions within the CAT-1 protein that are involved in E-MLV entry. Using such an approach, two specific residues have been found in the proposed third extracellular loop of murine CAT-1 that can render human CAT-1 functional as an E-MLV receptor (3, 196). CAT-1 receptor function can also be modulated by glycosylation (119, 181).
The CAT-1 ortholog expressed on cells derived from the Asian mouse, Mus dunni, functions as an efficient receptor for all E-MLV isolates with the sole exception of Moloney MLV (55). The block to efficient Moloney MLV infection of M. dunni-derived cells can be overcome by preventing N-linked glycosylation of CAT-1 in these cells. This can be achieved by either treating M. dunni target cells with tunicamycin prior to exposing the cells to Moloney MLV or ablating the site of N-linked glycosylation in the third extracellular loop of M. dunni CAT-1 (56).
As mentioned above, replication-competent E-MLV isolates have similar host range and pathogenic features. Most E-MLVs cause leukemias and lymphomas in mice; however the E-MLV PVC-211 induces a neurodegenerative disease following inoculation of neonatal mice and rats. The ability to infect brain capillary endothelial cells is a determinant of PVC-211 neuropathogenicity and appears to correlate with the ability of PVC-211 to infect CHO K1 Chinese hamster cells (111). CHO K1 cells are resistant to all other E-MLVs. The ability of PVC-211 to infect CHO K1 cells is due to the unique properties of the PVC-211 SU protein that allow it to interact with the glycosylated form of the CAT-1 ortholog present on these cells. The unique host range properties of PVC-211 can be conferred to other E-MLVs by substituting two PVC-211-derived amino acid residues (a glycine at position 116 and a lysine at position 129) for those present in other E-MLVs. (112). Further studies of the interaction between PVC-211 and hamster CAT-1 may provide insights into PVC-211 BCEC tropism and neuropathogenicity.
Xenotropic and Polytropic MLVs Have a Common Receptor, XPR1
A human receptor that facilitates entry for both xenotropic and polytropic MLVs has been recently cloned by three groups using expression cloning methodology (17, 167, 194). This receptor, XPR1, is a glycoprotein predicted to contain either eight or nine transmembrane domains and is related to the Syg1p and pho81 proteins of yeast. The function of Syglp is unknown; however, pho81 has been implicated in regulating inorganic phosphate (Pi) transport. The SU envelope region of the polytropic MLV has been demonstrated to bind specifically to the murine XPR1 protein expressed in Xenopus laevis oocytes, suggesting that it functions as a receptor that mediates virus binding (194). Orthologs of the XPR1 receptor expressed in specific cell types regulate the differences in polytropic and xenotropic virus host range. For example, the murine ortholog of XPR1 expressed on cells derived from the mouse Mus casteneus renders them resistant to polytropic but not xenotropic MLVs. Similarly, cells derived from laboratory mice are resistant to xenotropic but not polytropic MLVs due to orthomorphic variation in the form of XPR1 present on these mouse cells.
The Gammaretroviruses RD-114, BaEV, SNV, REV-A, and HERV and the Betaretroviruses SRV-1, SRV-2, SRV-3 (MPMV), SRV-4, and SRV-5 All Use RDR as a Common Receptor
SRV-1, SRV-2, SRV-3 (MPMV), -4, and -5 have been demonstrated to cross-interfere not only among themselves but with a member of a related endogenous human retrovirus family called HERV-W and several gammaretroviruses; the feline endogenous retrovirus RD-114, BaEV, and members of the REV group of avian retroviruses, spleen necrosis virus (SNV) and REV-A (24, 84, 91, 161). The envelope proteins of these diverse retroviruses are relatively similar and cluster apart from other retriviral envelope proteins (Fig. 2). The receptor for these retroviruses, denoted RDR (for “RD-114 and D-type retrovirus receptor”), has been shown to be the previously identified sodium-dependent neutral amino acid transporter SLC1A5 (also called ATB0), which is predicted to contain 9 or 10 transmembrane domains (141, 168). RDR is widely expressed in human tissues with the exception of the liver and brain (82, 168). Interestingly, transport of neutral amino acids is reduced in cells infected with retroviruses that use RDR for entry, and this might provide an explanation for the immunodeficiency induced by SNV and the SRVs in animals, which could involve selective toxicity to rapidly dividing T and B lymphocytes (141).
In the case of the HERV-W family, all of the integrated proviruses appear to be replication defective (23), but the Env protein of one member can induce fusion among cells expressing RDR, indicating that this Env can mediate fusion required for virus entry and that RDR is required for this process (24). The HERV-W Env protein is not incorporated into retrovirus vectors based on Moloney MLV (6, 24) but can be incorporated into and promote infection by vectors based on HIV (6); however, the interference properties of this pseudotyped virus have not yet been reported.
While all of the viruses in this interference group can infect human cells, only BaEV can infect mouse cells, revealing a difference in receptor use by viruses in this group. Other members of the group can infect mouse cells pretreated with tunicamycin, indicating that infection by the other viruses is blocked by glycosylation of the receptor. Surprisingly, however, while the mouse ortholog of human RDR shows 81% identity to human RDR at the amino acid level and displays similar amino acid transport properties, the mouse protein does not serve as a receptor for any of the viruses in this interference group (110). Furthermore, mutation of the glycosylation sites in the mouse RDR to prevent glycosylation did not convert this protein to a functional receptor (110). Analysis of human and mouse proteins that showed similarity to RDR at the amino acid level revealed that a transporter for neutral amino acids and glutamate from humans and mice could serve as relatively good receptors for BaEV and as weak receptors for RD-114 and SRV-2 (110). We have denoted this related receptor RDR2, and its conventional designation is SLC1A4. Mutation of the glycosylation sites in mouse RDR2 to prevent glycosylation converted this protein to a relatively efficient receptor for RD-114 and SRV-2, mimicking the results observed in mouse cells that express RDR2. Thus, members of this interference group can variably use RDR and RDR2 as receptors for entry, although RDR is the primary receptor in human cells. This situation is similar to that observed for viruses discussed above that use the related phosphate transporters Pit1 and Pit2. Use of a receptor in a family of related proteins is advantageous for a virus in that small alterations in the viral envelope allow recognition of additional receptors and facile expansion of tissue and species host range.
Amphotropic MLVs, GALV, Woolly Monkey Virus, and FeLV-B All Use Sodium-Dependent Phosphate Transporters as Receptors
In 1990 O'Hara and coworkers cloned, sequenced, and characterized the human cDNA encoding the receptor for GALV (125). This receptor was subsequently determined to function as a receptor for FeLV-B (172), woolly monkey virus (172), and MLV 10A1 (118, 189). More recently it has been determined that this viral receptor also function for FeLV-T (7) (see below). The normal function of this multiple membrane-spanning receptor, designated Pit1 (80, 127), is that of a type III sodium-dependent phosphate transporter. A second, closely related type III phosphate transporter, Pit2 (80, 188), has more recently been determined to function as the receptor for amphotropic MLV (A-MLV); (117, 178) and as a second receptor for 10A1 (118, 128) and some FeLV-B variants (25, 165).
Several features distinguish type III Pi transporters from their more widely studied type I and II counterparts. In contrast to the type I and II kidney-specific Pi transporters, Pit1 and Pit2 are ubiquitously expressed in mammalian tissues (78, 80). Second, even though Pit1 and Pit2 exhibit Pi uptake affinities similar to the kidney-specific transporters, they have no obvious sequence similarity to them (124). In addition, Pit1 and Pit2 play a fundamental housekeeping role in Pi transport, such as absorbing Pi from interstitial fluid for normal cellular functions such as cellular metabolism, signal transduction, and nucleic acid and lipid synthesis, whereas the type I and II transporters are responsible for reabsorbing Pi in the kidney (124). Another distinguishing feature of type III Pi transporters is that type III transport function is specifically blocked following infection with the appropriate virus. The loss of Pi transport mediated by the type III transporter does not compromise the viability of the infected cell, suggesting that other housekeeping Pi transporters exist that are capable of compensating for the loss of Pit1 and Pit2 Pi uptake. In contrast, the loss of either type I or II Pi transport in the kidneys has severe metabolic consequences (80, 127, 188). Finally type I and II and Pit1 and Pit2 are differentially regulated. Increases in alkaline pH stimulate increased Pi uptake by the kidney-specific Pi transporters, whereas type III Pi uptake is decreased under these conditions (174). Pit2- but not Pit1-mediated Pi uptake is specifically regulated by protein kinase C epsilon (77), and Pit1 is specifically regulated by insulin-like growth factor I in osteoblast-like cells (131).
Despite their similar structures and transporter and virus receptor functions, Pit1 and Pit2 have distinct virus recognition properties. Pit2 can be utilized by A-MLV but not GALV while Pit1 can be utilized by GALV but not A-MLV for entry into cells. Interest has therefore focused on amino acid differences between Pit1 and Pit2 that might account for their distinct virus receptor properties. Region A, comprising residues 550 to 558 of Pit1, has been proposed to be important in GALV entry (79). This proposition is based primarily on the observation that substitution of Pit1 region A residues for the corresponding residues of Pit2, the Neurospora crassa phosphate permease Pho4, or the murine ortholog of Pit1 (all proteins that fail to facilitate GALV entry) renders these proteins functional as GALV receptors. Furthermore, mutations in Pit1 region A can abolish virus receptor function (107, 169, 185).
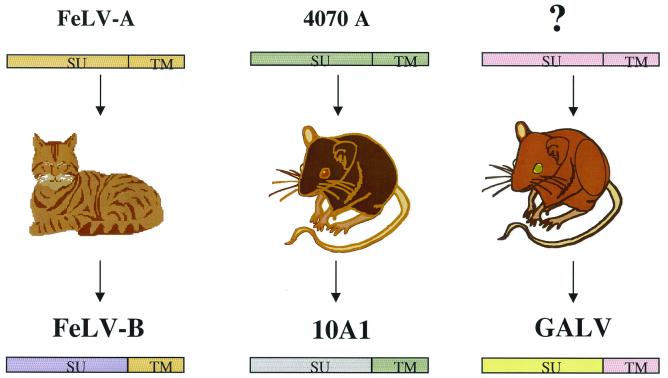
Gammaretroviruses that employ Pit1 as a receptor without the requirement for accessory factors are recombinant retroviruses. These recombinant retroviruses most commonly result from recombination between the infecting exogenous viruses and endogenous viral sequences but can also occur between exogenous viruses and host cell genomic sequences, as is the case for retroviruses that transduce oncogenes. FeLV-B resulted from recombination between exogenous FeLV-A and endogenous FeLV-related sequences (Fig. 3). Individual FeLV-B isolates contain heterogeneous envelope genes containing various regions contributed by either FeLV-A or endogenous FeLV genes. The FeLV-B subgroup is defined by the expanded host range of these various FeLV-A recombinant viruses. FeLV-A efficiently infect primarily feline cells, whereas FeLV-B infect both feline and several nonfeline cells. FeLV-B do not recognize FeLV-A receptors but, instead, display the expanded host range of viruses that utilize Pit1 and in some cases Pit2 as a receptor. Another example of a recombinant retrovirus that uses Pit1 as a a receptor is 10A1 MLV, which is a unique type of MLV isolated from a mouse infected with A-MLV. Its envelope protein appears to have been derived by recombination between A-MLV and an endogenous murine retroviral sequence (144). It retains close sequence identity to the A-MLV envelope in the N terminus of its SU, differing by only six residues encoded in the first 200 codons derived from A-MLV (128). However, the hypervariable region immediately downstream of the N terminus resembles that present in polytropic MLV. 10A1 MLV retains the ability to interact with the A-MLV receptor, Pit2, but can also enter cells through Pit1 (118). Finally, GALV recombinant viruses contain envelope sequences derived from endogenous retroviral elements present in a certain Southeast Asian species of feral mouse harboring class I endogenous retroviral sequences (29, 104). There are two types of endogenous retroviral sequences in the genome of Southeast Asian mice, designated class I and class II. Class I sequences contain genomic retroviral sequences related to those from woolly monkey virus and GALV. Class II endogenous retroviral sequences are more closely related to endogenous viral sequences commonly isolated from inbred and feral population of Mus musculus (30).
FIG. 3.
Evolution of FeLV-B, GALV, and MLV. Viruses that use only Pit1 as a receptor are those that developed from recombination between exogenous retroviruses and endogenous retroviral sequences. FeLV-B is a recombinant virus derived from the exchange of genetic material between exogenous FeLV-A and endogenous FeLV-related envelope sequences. 10A1 MLV arose as a consequence of recombination between A-MLV and an endogenous murine retroviral sequence, and GALVs contain envelope sequences derived from a certain Southeast Asian species of feral mice harboring MLV class I endogenous retroviral sequences.
There have been numerous studies of the envelope domains of gammaretroviruses that determine host cell or receptor specificity. These studies suggest that the major determinant for receptor specificity resides in the N-terminal half of SU, and specifically within variable region A (VRA), although additional domains of SU, including variable region B (VRB) and a downstream proline-rich region, have been implicated as secondary determinants for some SU-receptor interactions (14–16, 18, 25, 26, 41, 42, 66, 99, 100, 117, 125, 134, 166). In addition, truncated forms of MLV SU that lack C-terminal sequences can specifically bind to their cognate receptor, which further supports a key role for VRA and VRB in determining receptor specificity at the level of binding (14, 15, 18, 42, 66, 100). Thus, sequences encompassing VRA and VRB are often collectively referred to as the receptor binding domain. However, recent studies suggest that C-terminal sequences may also determine receptor specificity for FeLV (65, 165). For MLV, C-terminal sequences have been implicated in postbinding stages in viral entry, such as fusion activation (101).
Given the high degree of divergence among the envelope proteins of viruses that use Pit1 as a receptor, it was of considerable interest to determine if a shared Pit1 receptor recognition determinant is conserved among 10A1, FeLV-B, and GALV SU proteins that have been demonstrated to be critical for Pit1-virus interactions. Regions within FeLV-B, GALV, and 10A1 that are involved in Pit1 receptor utilization have been identified. Protein alignment of the regions involved in Pit1-mediated entry have determined that various combinations of receptor recognition determinants exist among viruses that use Pit1 and that no apparent linear receptor recognition determinant exists among viruses that use Pit1 as a primary receptor (25, 66, 175),
As stated above, the human ortholog of Pit1 functions as a receptor for the recombinant retroviruses GALV, FeLV-B, and 10A1 MLV. Despite their common ability to use human Pit1 as a receptor, these viruses do not exhibit similar infectivity patterns on cells from other species. For example, 10A1 MLV can utilize murine Pit1 as a receptor whereas GALV, simian sarcoma-associated virus (SSAV), and FeLV-B cannot. Similarly, certain FeLV-B isolates are restricted in their ability to utilize rat NRK orthologs of Pit1 as receptors whereas both 10A1 and GALV efficiently infect these cells (172). Thus, human Pit1 functions for GALV infection but the murine ortholog does not. In addition, viruses that use exclusively human Pit1 or human Pit2 do not cross-interfere in human cells but may show cross-interference properties in cells derived from nonhuman species. For example, 10A1 MLV is capable of employing both human Pit1 and Pit2 whereas GALV is restricted to infecting human cells exclusively through Pit1. Human cells chronically infected with 10A1 are therefore resistant to GALV superinfection, whereas human cells infected by GALV remain susceptible to further infection by 10A1 MLV. In E36 hamster cells, 10A1 is still capable of using both Pit1 and Pit2, and, in contrast to human cells, GALV can also use both receptors. Consequently, hamster E36 cells infected by either GALV or 10A1 are resistant to superinfection by both viruses. Thus, while the human Pit2 receptor is restrictive for GALV infection, its E36 hamster ortholog is not.
The Receptor for FeLV-C Also Appears To Be a Transport Protein
The human and feline orthologs of the receptor for the anemia-inducing FeLV-C has been identified. This receptor designated Flvcr, is a glycoprotein and is predicted to contain 12 regions that traverse the cell membrane. Flvcr shows sequence similiarity to the bacterial glycerol-3-phosphate and d-glucarate transporter members of the major facilitator superfamily of transporters. It is currently speculated that Flvcr may function as an organic anion transporter (138, 170).
RETROVIRUSES THAT REQUIRE TWO CELLULAR PROTEINS FOR ENTRY
Receptors for Primate Lentiviruses
There have been numerous recent reviews describing the HIV receptor complex (21, 47, 121, 193), and for this reason we have not provided an exhaustive review on this topic here (where appropriate, readers are referred to the relevant review for a more detailed overview). Rather, the summary of the receptors for primate lentiviruses is provided as a comparison, to illustrate common themes in retroviral receptors, as well as some of the complexities that appear to be unique to HIV-receptor interactions. HIV HIV-1 and HIV-2 and the related simian immunodeficiency viruses (SIV) typically require two receptor molecules: CD4 (40, 89, 108, 113, 157), which normally plays a role in MHC class II recognition, and one of a variety of coreceptors that are G-protein-coupled chemokine receptors (Fig. 1). HIV-1 typically uses one of two chemokine receptors, CCR5 (R5 viruses) or CXCR4 (X4 viruses), and viruses with dual coreceptor specificity (X4R5 viruses) are also observed (5, 34, 44, 49, 50, 59).
CD4 serves as the binding receptor for HIV SU and is required for efficient entry into cells by almost all natural isolates of HIV-1, HIV-2, and SIV. Binding to CD4 is believed to lead to a conformational change in the viral surface unit that exposes a binding site for the chemokine receptor, which acts as a fusion receptor (97, 153, 158, 176, 190). This second binding event then leads to fusion due to a series of subsequent conformational changes that result in the fusion peptide being inserted into the target cell membrane. This initiates pH-independent fusion between the virus and the cell. The series of events starting from the stage of coreceptor binding are very similar to the events in the entry of most oncoretroviruses. The major difference for the lentiviruses HIV-1, HIV-2, and SIV is the requirement for a binding receptor, CD4, prior to interaction with the second receptor. For most simple retroviruses, one molecule appears to be sufficient for both binding and the steps leading to fusion. However, as discussed below, there are examples for lentiviruses that can enter cells by using only one receptor molecule and there is an example of an oncoretrovirus that requires two molecules for entry.
The domains of the HIV surface unit that interact with CD4 and the chemokine receptor are complex. CD4 binds to a recessed pocket in the SU, but it also appears to contact a large portion of SU, including residues that surround the pocket (193). Residues in a disulfide-bonded loop of the HIV-1 SU that has been defined as the third variable region (V3) largely determine chemokine receptor specificity (34, 36). It remains unclear whether a direct interaction between V3 and the coreceptor is critical for binding; in any case, it is thought that other domains of SU are important for this interaction. The overall charge of this V3 loop sequence and well as the identities of specific amino acids have been shown to influence viral tropism and coreceptor specificity (reviewed in reference 71). Typically, viruses with V3 domains that are basic use CXCR4, and it has been noted that the predicted extracellular loops of CXCR4 are acidic (71). Other domains of SU, particularly variable domains 1 and 2, also determine cell tropism and, by implication, coreceptor specificity (115). The fact that HIV-1, HIV-2, and SIV all can use CCR5 as a coreceptor suggests that there must also be a conserved sequence or structure in the SU that determines interactions with the coreceptor (193).
Because CD4 is required for virus binding, the host range of HIV and SIV is largely restricted to CD4+ cells, which include a subset of T lymphocytes (T-helper cells), cells of the monocyte lineage (microglia and macrophages), and dendritic cells. Both CCR5 and CXCR4 are expressed in macrophages (102, 195, 198). CCR5 and another less commonly used HIV-1 coreceptor, CCR3, are expressed on the surface of microglia (68). Studies of peripheral blood mononuclear cells suggest that CCR5 is expressed in memory T lymphocytes whereas CXCR4 is expressed in naive T cells (22, 102, 191). In the mucosal compartment, which is presumed to harbor the cells that are early targets for HIV transmission, CCR5 and CXCR4 are also both expressed on T lymphocytes and macrophages. Interestingly, only CXCR4 and not CCR5 was detected on dendritic cells from the genital compartment, even though these cells had been postulated to be early target cells for transmission of R5 viruses (69). As discussed below, this apparent paradox may be explained by the discovery of a lectin molecule on dendritic cells that binds and sequesters HIV-1 particles, perhaps permitting dendritic cells to serve as a nonproductively infected reservoir for virus transfer to lymph nodes.
Before the discovery of the coreceptors, HIV-1 isolates were discriminated on the basis of their ability to infect T-cell lines or macrophages (60, 116). We now know that the T-cell lines expressed CXCR4 and that T-cell-line-tropic viruses use CXCR4 as a coreceptor (20). Macrophage-tropic viruses almost invariably use CCR5, although both CCR5 and CXCR4 are expressed in macrophages. It is unclear why CXCR4 is not functional for macrophage infections in most cases, although there are examples of CXCR4-mediated entry into macrophages (179, 195). It may be that levels of expression of either CXCR4 or CD4 are not adequate to permit infection because X4 primary viruses appear to require higher CD4 levels than R5 viruses do (92). In that regard, the conditions under which the cells are cultured and the state of activation may affect the levels of receptor-chemokine receptor expression and thus the susceptibility to HIV (98, 102). There have been suggestions that the oligomeric form of the CXCR4 receptor expressed in macrophages differs from that expressed in lymphocytes and that this may affect its ability to interact with CD4 (98). Posttranslational changes in CXCR4, such as processing and/or N-linked glycosylation, have been suggested to affect CXCR4 function (31, 195). There are also data to suggest that the interaction of HIV-1 SU with CCR5 and CXCR4 in macrophages leads to different signaling responses, which could restrict postentry stages in replication (105). It remains unclear which, if any, of these mechanisms limit infection of macrophages by X4 HIV-1.
Other chemokine receptors serve as coreceptors for infection by some HIV-1 strains (reviewed in reference 21), but this has largely been in transformed cell lines engineered to express CD4 and these molecules. Thus, the relevance of these coreceptors in HIV-1 replication in the host is unclear. At issue is the fact that HIV-1 enters these engineered cell lines with reduced efficiency compared to cells similarly engineered to express CD4 and CCR5 or CXCR4. In addition, it is unclear if the level of expression of these proteins in the transfected cell lines approximates what is present on the surface of primary cells. Moreover, some of the chemokine receptors may not be coexpressed with CD4 in the same cell in vivo, making it unclear how these molecules could function for HIV-1 entry.
It has been difficult to clearly define the role of specific chemokine receptors in HIV-1 replication in vivo. The exception is CCR5, where an inactivating genetic mutation (A32) present in a small fraction of Caucasians has been associated with reduced susceptibility to HIV-1 infection in high-risk individuals with the homozygous Δ32 CCR5 allele (43, 74, 106, 154). Lymphocytes and macrophages from these individuals are not permissive to replication of R5-HIV-1 strains (140), demonstrating a requirement for CCR5 for HIV-1 replication in these primary cells. For CCR5, CXCR4, and CCR3, ligands and/or antibodies that bind to these receptors block HIV-1 infection in primary cells in culture, which provides indirect evidence that they are functional coreceptors in vivo. For example, the ligand for CCR3 can block the entry of CCR3-using strains in microglial cells (68), suggesting that CCR3 may be used as a coreceptor for some neurotropic strains of HIV-1.
HIV-2 and SIV also use CD4 and CCR5 to gain entry into cells (32, 33, 109, 151). CCR5 is thought to play a major role in SIV replication and pathogenesis in macaques. There is no genetic parallel to Δ32 CCR5 in the macaques, so the basis for this hypothesis is primarily the observation that almost all SIVs use CCR5 as a coreceptor with high efficiency in vitro. GPR15 is also commonly used by SIV strains in vitro (45, 86), but Pohlmann et al. (137) have shown that when a pathogenic SIV strain that uses both CCR5 and GPR15 is mutated to abolish GPR15 recognition, the virus can still replicate to the same levels as the parental virus. This provides direct evidence that entry via GPR15 is dispensable for replication of SIV in monkeys. There are also additional coreceptors for SIV (reviewed in reference 71); as with HIV, the biological relevance of these molecules as receptors in vivo is unclear. It is of interest that SIV can replicate efficiently in cells from individuals with the deletion in CCR5 (Δ32), suggesting at least one other coreceptor may be important for SIV replication in primary target cells (33, 199; S. E. Forte and J. Overbaugh, unpublished data).
The chemokine receptors resemble the oncoretroviruses in the fact that they are multiple membrane spanning receptors that are predicted to have several extracellular loops. As with the multiple membrane-spanning receptors used by oncoretroviruses, there are interactions between the HIV-1 SU and more than one domain in the chemokine receptors. The N-terminal domain of CCR5 is important for entry by R5 HIV-1, but the N-terminus of CXCR4 does not appear to be important for entry by X4 viruses (reviewed in refrence121). Analyses of chimeric chemokine receptors that combine nonfunctional with functional receptors indicate that the domains of these coreceptors that participate in entry are complex. For example, each of the three extracellular loops of CCR5 has been shown to be critical for HIV infection in at least one study (121). Different groups have identified different domains as determinants for fusion or entry, which may reflect the viral strain tested. It may also reflect the sequence of the nonfunctional receptor used in the chimera because some of these molecules may encode sequences in one or more loop that are functional when paired with the loops from a functional receptor. Finally, some studies have only assessed fusion, and it is possible that there are additional domains that are required for subsequent steps in entry after fusion. To complicate matters, SIV and HIV appear to interact with different domains of CCR5 (52). In this regard, the coreceptors resemble the Pit receptors for simple retroviruses discussed above, where multiple domains are important for viral entry, and these domains may differ among different viruses competent to use these receptors, as exemplified by interactions between Pit1 and FeLV-B versus Pit1 and GALV (see above). Similarly, studies of chimeric Pit receptors have also illustrated that the nonpermissive receptor that is used to make the chimeric protein may also determine which domains from a functional receptor can confer the ability to permit viral infection.
The Kabat laboratory has reported an interdependence between the levels of CD4 and the coreceptor that are required to permit HIV-1 infection. Their studies of engineered cell lines suggest that when either CD4 or CCR5 are present at very low concentrations on the cell, there must be higher levels of the other receptor component for HIV-1 to gain entry into the cell (135). Recent studies from the same group suggest that interactions with the coreceptor leading to productive infection may require virus binding to a cluster of four to six coreceptors (93). This model of receptor clustering is similar to one they proposed previously for MLV infection via the mouse CAT receptor (160), suggesting that this could be a common feature of fusion receptors. It is unclear if multiple envelope trimers participate in binding, although this may be predicted for retroviruses based on studies of influenza virus HA-receptor interactions, which have served as a model for retroviral envelope-receptor interactions. If binding between multiple envelope and coreceptor molecules is required for HIV-1 entry, then a higher envelope affinity for the coreceptor and/or a high density of both proteins may be predicted to lead to more efficient infection (93).
Some Primate and Feline Lentiviruses May Enter Cells by Using Only a G-Protein-Coupled Chemokine Receptor
Several strains of HIV and SIV are CD4 independent and require only the chemokine receptor for binding, fusion, and entry (51, 53, 57, 90, 96, 142, 143). Typically, these viruses infect more efficiently in the presence of CD4 than in its absence. CD4-independent viruses appear to be most common in HIV-2, although there are several examples in SIV and a few in HIV-1. The frequency of these variants in natural infection is unclear, especially for HIV-1, where all the CD4-independent viruses reported to date were adapted in the laboratory (51, 90, 96). However, for HIV-2, which is a relatively less pathogenic virus than HIV-1, it is possible that CD4-independent viruses are replicating without the use of CD4 in HIV-2-infected individuals, because such viruses can be isolated from infected subjects (142). Because CD4-independent variants do not require interactions with CD4, it is thought that their SUs are already in a conformation that exposes the chemokine receptor binding sites. Interestingly, some of these viruses are particularly neutralization sensitive, suggesting that a conformation that results in exposure of the chemokine receptor binding site also leads to exposure of an antibody epitope (72). If this is the case, it may explain why CD4-independent viruses are rare among primary isolates from patients, because such viruses would be strongly selected against by the host immune response. In contrast, such viruses may be viable in cell culture systems, where there is no immune selection. Thus, it is tempting to speculate that the adaptation of primate lentiviruses to use CD4 binding to expose a chemokine receptor binding site may reflect immune pressures rather than structural or functional requirements for CD4 in viral entry.
Some strains of the feline lentivirus feline immunodeficiency virus (FIV) also appear to infect cells by using the chemokine receptor, CXCR4, in the absence of CD4 (54, 73, 146, 186). While it is clear that CD4 does not function as a receptor for FIV, it is less clear whether other receptors are involved in FIV entry. Studies to date have shown CXCR4 functions for infection of several FIV isolates (146, 186). However, not all isolates can infect cells expressing CXCR4, suggesting that there may be an as yet identified receptor and/or another coreceptor for FIV. It will be interesting to see if most FIVs will require only a chemokine receptor for entry. This may suggest that this feline lentivirus represents a transition virus between the oncoretroviruses and the primate lentiviruses that uses a chemokine receptor for all aspects of entry but does not require a separate binding receptor. This may suggest that the conformation of the FIV SU that leads to exposure of the receptor binding domain does not simultaneously expose a key neutralization epitope.
Are CD4 and Coreceptor Necessary and Sufficient for Efficient HIV-1 Entry?
The requirements for HIV and SIV infection appear more complex than a simple need to have CD4 and the coreceptor present on the cell surface. Studies of HIV-1 infection in nonhuman primate cells have shown that the coreceptors participate in postentry stages of replication (32). These studies showed that such restrictions to replication occur at the level of translocation of the preintegation complex to the nucleus. This block in replication can be overcome by expression of the appropriate coreceptor, suggesting that the chemokine receptors may in some manner affect steps in replication after entry but before integration of the viral DNA. The cellular processes and/or the viral factors that participate in these stages of viral replication have yet to be identified.
There are also additional molecules, other than CD4 and the chemokine receptors, that have been implicated in the earliest stages of HIV infection. Recently, a protein that is expressed on dendritic cells, DC-SIGN, has been shown to bind HIV-1 SU (63). DC SIGN and a related C-type lectin (136) do not function as receptors per se, because binding of HIV to these surface molecules does not permit entry. Rather, they are thought to bind virus particles and transfer them to cell targets expressing CD4 and chemokine coreceptor. It is not yet known what role this trans infection may play in the host, but it has been suggested that such a mechanism could permit the transfer of HIV-1 from mucosal sites that are rich in dendritic cells to lymphoid tissues that have lymphocyte and macrophage cell targets (63, 164).
Other molecules have also been implicated as binding partners for HIV-1. In some cases, such binding partners appear to facilitate entry, which contrasts with DC-SIGN, which is perhaps best described as a potential trans-infection cofactor. For example, galactosylceramide can bind HIV-1 SU, and antibodies to it inhibit HIV-1 entry (67). Therefore, it has been suggested that this glycolipid could permit entry into CD4-negative cells, particularly cells in the nervous system where it is found. However, because the efficiency of infection with galactosylceramide appears to be very low, the physiological relevance of this molecule as a receptor is unclear. Multiple studies have also shown that heparin sulfate proteoglycans (HSPGs) can bind HIV and HSPGs may play a role in virus attachment and entry into some cell targets (13, 28, 120, 126, 132, 147, 148). HSPGs also appear to play a role in FIV binding to some cells (46), although for both FIV and HIV, the role for HSPGs in attachment and binding appears to be strain specific (46, 123).
FeLV-T: the First Example of an Oncoretrovirus That Uses a Two-Component Receptor Complex
As discussed above, the receptors for most variants of FeLV and MLV are multiple membrane-spanning transport proteins. These molecules were identified as viral receptors because when they were introduced into nonsusceptible cells, they rendered them permissive for infection by specific viruses. It has largely been assumed that FeLV and related mammalian oncoretroviruses require just a single receptor for viral binding and entry, although there are examples of MLVs and FeLVs that can bind to a specific receptor but cannot infect cells that express this molecule (101, 165). Thus, it remains to be seen whether there are secondary molecules involved in entry for some oncoretroviruses or whether the inability to execute postbinding events such as fusion reflects an incompatibility between different domains of the SU that must interact to activate fusion, as has been proposed (99).
Recently, it has been demonstrated that T-tropic variants of FeLV require a second molecule, in addition to the phosphate transport receptor, Pit1, for entry (7) (Fig. 1). Analyses of postentry stages in replication, such as reverse transcription, indicate that this second protein, termed FeLIX (for “feline leukemia virus infection X-cessory protein”), acts at a stage prior to DNA synthesis, suggesting that it is an entry cofactor (A. S. Lauring and J. Overbaugh, unpublished data). In the absence of either Pit1 or FeLIX, there is no detectable reverse transcription or other evidence of infection of cells by FeLV-T, whereas when both are present, cells become highly permissive to productive infection. Surprisingly, FeLIX is secreted from the cell, and this molecule is encoded by endogenous FeLV-like cellular sequences (7). Endogenous FeLV-like sequences are present at multiple copies in the genome of domestic cats, but they do not encode infectious virus and many of the open reading frames (ORFs) are truncated relative to exogenous FeLV (19, 94). The FeLIX ORF is predicted to encode a 273-amino-acid protein that includes an endoplasmic reticulum signal sequence and is similar in sequence to the N terminus of FeLV SU (7). However, FeLIX lacks the corresponding C-terminal SU sequences and all of the TM domain. Studies using conditioned media from cells expressing FeLIX have shown that FeLIX can act in trans to facilitate FeLV-T infection in cells expressing Pit1. Thus, although FeLIX expression is restricted largely to lymphoid cells (98a, 114), other cell types may be targets for FeLV-T infection in the cat if they are exposed to FeLIX. Moreover, cells infected with FeLV-B are also targets for FeLV-T infection, because FeLV-B SU can substitute for FeLIX in FeLV-T infection (7). This is because FeLV-B evolves from FeLV-A by transduction of endogenous FeLV-like sequences (130), and as a result the N terminus of FeLV-B SU is nearly identical (96% sequence identity) to FeLIX. Because Pit1 is widely expressed, and secreted FeLIX or FeLV-B SU can act as an entry cofactor for FeLV-T, FeLV-T is predicted to have a broad host cell specificity in the cat.
There are as yet no other examples of naturally arising mammalian oncoretroviruses that use a two-component receptor. However, some MLVs that have been engineered to be defective in postbinding stages of virus replication can be rescued by exogenous SU proteins that resemble FeLIX (101). Interestingly, a variety of soluble SUs and their cognate receptors can rescue these MLVs, which encode a single amino acid deletion in the N terminus of SU. In addition, MLVs where the viral receptor binding domain has been replaced by erythropotetin sequences can be rescued in a similar manner when both the erythropoietin receptor and the receptor that binds the soluble SU are present on the cell (11). This suggest that the engineered forms of MLV, which are unable to carry out postbinding stages in entry, can pair with nearby SU-receptors to carry out the steps in entry after binding, such as fusion. Presumably, this requires some direct interaction between the virus and its receptor and the soluble SU and its receptor.
These defective MLVs are not found in natural infections and in that way are quite different from FeLV-T, which evolves in the infected cat (149). Moreover, FeLV-T is competent for replication in feline cells and in cats (130, 149) whereas these MLVs are not replication competent in murine cells (11, 101). Nonetheless, the ability of soluble SU proteins to rescue infection by these defective MLVs may provide some parallels with natural infection by FeLV-T. In particular, the changes that make these MLVs defective are in an N-terminal domain of SU, where FeLV-T encodes similar novel sequence differences relative to other FeLVs (48, 65, 101). However, the receptor interactions that lead to infection appear to be quite different for FeLV-T and defective MLVs. For example, these defective MLV SUs bind like their wild-type counterparts to the cognate receptor (101) whereas FeLV-T binding to Pit1 is weak to undetectable (A. S. Lauring, H. H. Cheng, and J. Overbaugh, unpublished data). In addition, these defective MLVs can be rescued by a variety of receptor-SU combinations (101) whereas the receptor complex for FeLV-T is quite specific: only Pit1 and an endogenous FeLV SU, either FeLIX or FeLV-B, can function as an FeLV-T receptor (98a). Even FeLV-B SUs that can bind and recognize Pit2 as receptor cannot render cells permissive to FeLV-T infection in the presence of Pit2 (98a). This suggests that there may be specific interactions between FeLV-T and the Pit1 receptor, even though binding between the two proteins cannot readily be detected using standard methods that measure equilibrium binding. It is perhaps of interest that direct binding of HIV-1 SU to CXCR4 has also been difficult to detect using standard equilibrium binding methods (47). Thus, methods that can detect lower-affinity interactions may be needed to detect FeLV-T binding to Pit1. One model to explain the present data is that as a result of FeLIX binding to Pit1, there is a conformational change in the receptor that permits or augments virus binding. However, given the novel nature of this receptor complex, more experiments are needed to define the interactions between FeLV-T, FeLIX, and Pit1 that lead to entry.
One of the interesting features of FeLV-T is it inability to establish superinfection interference, and this has been linked to its cytopathic properties (48, 145). The lack of interference could reflect the very weak or transient interaction between the virus and the Pit1 receptor, which is sufficient to permit binding and fusion but insufficient to cause downregulation or otherwise block subsequent interactions with the Pit1 receptor. It is tempting to speculate that other oncoretroviruses that also are impaired in establishing interference, such as SNV (83), will also require a two-component receptor and/or have a low-affinity interaction with their primary receptor. As proposed by Temin, failure to establish interference may cause the cytopathic effects due to such viruses (173).
RECEPTORS WITH A SINGLE MEMBRANE-SPANNING REGION
Single Transmembrane-Spanning Receptors Are Used by the Avian Oncoretroviruses
The Alpharetrovirus genus consists of the avian sarcoma and leukosis viruses (ASLV). The sarcoma viruses are distinguished from the leukosis viruses by the presence of an oncogene, for example, the src oncogene of Rous sarcoma virus. Otherwise, these viruses are simple retroviruses with only gag, pro, pol, and env genes, exhibit a C-type particle morphology, and bud at the cytoplasmic membrane. The ASLV have been divided into groups A through J based on interference properties, and the receptors for ASLV-A, ASLV-B, ASLV-D, and ASLV-E have been identified. All are single- transmembrane-spanning proteins, although the receptor for ASLV-A may exist in a GPI-anchored form as well.
Initially the gene encoding the receptor for ASLV-A viruses was cloned from chicken genomic DNA, but no transcripts were detected from this DNA and so the nature of the protein product could not be determined (197). The quail ortholog of this gene was later cloned and shown to produce two RNA species that encode two distinct proteins, Tva isoforms 800 and 950, with identical 83-amino-acid extracellular domains but different membrane anchors (12). Both of these proteins serve as receptors for ASLV-A viruses and are related in sequence to the low-density lipoprotein (LDL) receptor. One of the receptors (Tva 950) spans the membrane once with the amino terminus outside of the cell, while the other (Tva 800) is predicted to be anchored to the outer side of the cytoplasmic membrane by a GPI anchor. A soluble form of the extracellular portion of Tva blocks infection and directly binds to the Env from ASLV-A (38). Direct binding of the ASLV-A env to Tva, a protein with similarity to the LDL receptor, suggests that the Env protein could have some sequence relationship to LDL, the natural ligand for the receptor, but no sequence similarity has been identified. It is unclear whether Tva is both a binding and a fusion receptor for ASLV-A, but no other requirement for virus entry following transfer of Tva to nonsusceptible cells has been identified.
Although viruses in the ASLV-B, ASLV-D, and ASLV-E groups were initially thought to use different receptors based on their interference and host range properties, it is now clear that they all use closely related orthologs of a member of the tumor necrosis factor (TNF) receptor family denoted Tvb (also called CAR1 [for “cytopathic ASLV receptor”] for ASLV-B and ASLV-D and SEAR [for “subgroup E ASLV receptor”] for ASLV-E) (2, 27). Tvb is most closely related to the human TNF receptor family members TRAIL-R1 and TRAIL-R2 and is likely to mediate the cell killing induced by ASLV-B and ASLV-D. As with ASLV-A and LDL proteins, it is interesting to hypothesize that the ASLV-B, ASLV-D, and ASLV-E Env proteins might show some amino acid similarity to TNF family proteins, the natural ligands for TNF receptor family proteins, and this might explain the Env specificity for Tvb; however, no such similarity is apparent.
Interestingly, infection with ASLV-B or ASLV-D can block entry by ASLV-E, but infection by ASLV-E does not block infection by ASLV-B or ASLV-D in cells expressing an ortholog of Tvb (Tvbs1) that confers susceptibility to all of these ASLV subgroup viruses. This pattern of interference is called nonreciprocal interference and is not expected if all of these viruses use the same receptor for entry. This paradox was recently resolved by the finding that the tvbs1 gene encodes two types of Tvbs1 protein, type 1, serving as a receptor for all of these ASLV groups, and type 2, specific for ASLV-B and ASLV-D (1). These conclusions were reached by showing that ASLV-E Env could be used to immunoprecipitate radiolabeled receptor in cell lysates but that ASLV-B Env could immunoprecipitate additional receptor after lysate clearing with ASLV-E Env while ASLV-E Env could not precipitate additional receptor after lysate clearing with ASLV-B. Gel analysis of the receptor proteins revealed two species of the receptor proteins with different molecular weights, but no differences in size or relative abundance were observed to correlate with receptor phenotype. The nature of the differences in the Tvb proteins that account for their differential receptor activity is not known, although heterogeneous N-linked glycosylation of the receptor proteins does not appear to account for these differences (1). As for ASLV-A entry mediated by Tva, it is unclear whether Tvb is both a binding and fusion receptor for ASLV-B, ASLV-D, and ASLV-E, but no other requirement for virus entry has been identified following transfer of Tvb into nonsusceptible cells.
Examples of Oncoretroviruses That Use a Single Transmembrane Receptor
The murine cDNA encoding the receptor for mouse mammary tumor virus was identified using a technique based on cDNA expression cloning (64). This receptor, designated Mtvr, is different from gammaretrovirus receptors in that it contains a single membrane-spanning domain. However, like many other retroviral receptors described in this review, it is transcribed in a wide variety of tissues. The normal function of Mtvr is unkown.
A putative receptor for bovine leukemia virus has been identified (9, 10). Blvr shows sequence similarity to the delta subunit of the adapter-related AP-3 protein complex (8), a cytoplasmic heterotetrameric protein complex involved in intracellular protein sorting, and further study is required to resolve the potential role of Blvr as a receptor for BLV.
RECEPTORS LINKED TO THE MEMBRANE BY A GLYCOSYLPHOSPHATIDYLINOSITOL ANCHOR
Jaagsiekta sheep retrovirus (JSRV) represents the first documented example of a retrovirus that uses a GPI-anchored protein, HYAL2, as a receptor (139). GPI-anchored proteins have an amino-terminal endoplasmic reticulum signal sequence and a hydrophobic carboxy-termial end that is removed during GPI anchor addition; the hydrophobic acyl tails of the GPI moiety anchor the protein to the exterior of the cytoplasmic membrane. HYAL2 was initially thought to be a lysosomal hyaluronidase, but this appears to be an artifact of the way the localization was assessed by using a green fluorescent protein tag linked to the carboxy end of the protein, which would probably be removed and degraded during GPI anchor addition. Several lines of evidence support the existence of the GPI anchor; in particular, an amino-terminal FLAG-tagged HYAL2 protein can be cleaved from the cell surface using bacterial phosphatidylinositol-specific phospholipase C, and cells expressing this protein or the endogenous human HYAL2 can be rendered resistant to JSRV vector infection by treatment with phosphatidylinositol-specific phospholipase C while infection by an otherwise identical vector bearing an amphotropic virus Env, which binds to the non-GPI-anchored Pit2 receptor, is unaffected. There is some controversy about the hyaluronidase activity of HYAL2, which, if anything, is quite low compared to that of other hyaluronidases (103, 139). It is unclear whether HYAL2 is both a binding and fusion receptor for JSRV, but transfer of HYAL2 into nonsusceptible mouse, hamster, and several human cell lines renders these cells fully susceptible to JSRV vector infection, indicating that if there is a coreceptor, it is broadly expressed in mammalian cells.
UNIDENTIFIED RECEPTORS
As depicted in Fig. 2, receptors for several of the important retroviruses have not yet been identified. HTLV-1 and HTLV-2 are human pathogens, and knowledge of receptor use will be important for understanding disease caused by HTLV and possibly for devising therapeutic strategies. HTLV-1 and HTLV-2 use the same receptor, which localizes to human chromosome 17q (62, 162, 171). Suitable mouse, rat, and bovine cell lines that are resistant to HTLV infection are available for pursuing standard techniques for receptor cloning involving complementation of the receptor defect in these cells, for example, using cDNA expression libraries.
Transplantation of porcine tissues into humans is being considered for treatment of human disease, but the existence of several PERVs that can infect human cells poses a potential disease threat. The receptors for these viruses have not been identified, although suitable nonsusceptible cell lines exist for receptor-cloning approaches.
Foamy virus has a very broad tissue and species host range, and vectors based on foamy virus are being developed for gene therapy purposes. Identification of the receptor(s) for foamy viruses will be more difficult than for the viruses mentioned above because of the lack of nonsusceptible cells required for receptor screening.
Identification of receptors for other retroviruses, both known and yet to be discovered, will continue to provide insight into the critical properties of proteins required for retrovirus binding and entry into cells. Furthermore, retrovirus vectors can be pseudotyped with surface glycoproteins from other virus families, for example, those from vesicular stomatitis virus, filoviruses, alphaviruses, influenza viruses, and Sendai virus. Thus replacement of the env genes in the genomes of existing retroviruses with glycoprotein genes from these other viruses is likely to result in the generation of replication-competent retroviruses, and it is interesting to speculate on the fitness of such viruses, whether they could have evolved in the wild, and why they are apparently not represented among the naturally occurring retroviruses. Study of such hybrid viruses and pseudotyped vectors will improve our understanding of entry requirements and the evolution of retroviruses to use diverse receptors for cell entry.
CONSEQUENCES OF RECEPTOR SPECIFICITY ON SELECTION FOR VIRUS VARIANTS IN THE HOST
To understand the role that viral selection plays in determining the receptor specificity of related retroviruses, we must review the processes that lead to retroviral variation. Retroviruses, like many RNA viruses, are highly genetically variable. Genetic variation occurs because the viral polymerase that transcribes the viral RNA genome into DNA, reverse transcriptase, is error prone; moreover, reverse transcription is not subjected to proofreading, which is a way in which DNA polymerases limit errors during cell division (reviewed in reference 187). Typically, a retrovirus will acquire approximately one mutation in every few replication cycles. Reverse transcriptase also permits a large amount of recombination between the diploid retroviral genomes that are packaged into viral particles, because strand exchange is required for retroviral DNA synthesis. For retroviruses such as HIV, which have a high turnover rate in the infected host, a viral genome with almost every possible mutation is predicted to be represented in the viral quasispecies (37, 70, 184). Thus, the generation of the precise mutation that is required for adaptation is not likely to be rate limiting for retroviruses. More likely, the selective pressure on the virus population determines the makeup of the variants that will successfully compete and persist from among the complex pool of genetic variants that arise continually in the infected host (129).
Clearly, retroviruses must bind host cell receptor proteins, because this is required for their propagation. Therefore, genetic variation that limits their ability to recognize a cell receptor will be deleterious whereas changes that allow the virus to find new target cells may provide a fitness advantage. Retroviruses establish superinfection interference as a means of preventing reinfection of the same cell by a virus that uses the same receptor. Once a cell becomes infected, the receptor is in some manner downregulated so that it is unavailable for use by another viral particle. This sets up a situation in which viruses that evolve to use new receptors may be favored in a persistently infected host. Thus, the selection pressures related to cell tropism may differ at different stages. For example, the viruses that are selected for their fitness for transmission from host to host may be determined by the available target cells at the site of viral exposure. These cells may differ from those that are targets for the virus during persistent infection, when there may be viral interference in the cells that were targets during primary infection.
For lentiviruses such as HIV-1, there is evidence that different target cells may be important for transmission versus persistence. The identity of the earliest target cells for HIV-1 infection is not clear, and this information may remain elusive because it is very difficult to identify individuals within the very early window after infection and it is obviously difficult to sample a broad number of cell types or tissues even if one does identify them. However, there is evidence for a bottleneck to transmission of viruses that use a particular coreceptor, CCR5. The evidence to support this is that the strains found when HIV-1 infection is first detected almost always require CCR5 for entry and very few can infect cells using CXCR4 (reviewed in references 21 and 71). In addition, as mentioned previously, individuals who lack functional CCR5 are less susceptible to infection (43, 74, 106, 154). For about half of the individuals, there is evidence that either the virus expands it coreceptor specificity to use CXCR4 and perhaps other coreceptors in addition to CCR5 or the virus evolves to use CXCR4 and not CCR5 (39). Because CCR5 and CXCR4 are expressed on different T-cell subsets, this may provide some advantage for X4 viruses in a host that has been infected for long periods with R5 virus. However, the advantage may also be due in part to the increased replicative capacity of X4 viruses. In some individuals, the later-stage R5 viruses may also have increased replication fitness compared to the infecting R5 strain (159), even though there is no apparent switch in coreceptor preference. This may be similar to the situation for SIV, where the strains present at all stages of infection use CCR5 yet the late-stage variants replicate to much higher levels in an infected host (87, 152). For SIV, there is as yet no evidence that the virus changes coreceptors over the course of infection (86). Thus, it will be interesting to define the ways that these late-stage, highly replicative viruses find new target cells in the host, particularly given that they also deplete the lymphocyte targets as disease progresses. One possibility is that late-stage viruses may be forced to infect cells that have a lower level of the receptor or coreceptor, because the early-stage viruses have infected and killed many of the cells expressing larger amounts of these proteins.
For oncorctroviruses, there is evidence for selection for new variants that use a common receptor within an infected host as well as between hosts infected by different oncoretroviruses. In particular, multiple mammalian C-type retroviruses converge to use the phosphate transport proteins, Pit1 and Pit2. For example, during FeLV infection of cats, there is evolution of new variants that use Pit receptors. For FeLV, one subgroup, FeLV-A, appears to be preferentially transmitted (76), suggesting that cells expressing the as yet unidentified FeLV-A receptor are important target cells at the site of initial infection or in early virus amplification in the host. As the host becomes systemically infected, new subgroups of FeLV evolve from this transmitted genome either through a series of point mutations or through recombination during reverse transcription (25, 130, 150). FeLV-B emerges in most infected cats, and this virus uses either of two Pit receptors (Pit1 or Pit2) for infection (M. M. Anderson, et al., submitted for publication). (25, 172). Because the Pit receptors are widely expressed, FeLV-B would be expected to have a large number of possible cell targets, even if the cat has been infected for a long period with FeLV-A. Cells infected with FeLV-B are in turn targets for FeLV-T replication because FeLV-T requires Pit1 and either FeLV-B SU or FeLIX for entry. It is unclear how frequently FeLV-T variants emerge in the infected cat, because this subgroup of FeLV was only recently identified (122). Because these viruses can infect cells that are already infected with FeLV-B, as well as cells expressing FeLIX, they have a broad range of possible cell targets. Perhaps more importantly, FeLV-T variants are unable to establish superinfection interference, which permits many rounds of reinfection of the same cell (48, 145).
It is interesting that the receptor for two of the FeLV subgroups that evolve in infected cats are phosphate transporter molecules. It is perhaps more striking that these are also the receptors used by some MLVs and by GALV, all of which have evolved by recombination with endogenous host sequences, as discussed above (see Fig. 3). The domain of FcLV-B, GALV, and MLV that is the primary determinant of Pit1 and/or Pit2 receptor utilization is a region that is highly variable both within and between these virus groups (the VRA region, as discussed above). An alignment of VRA sequences of FeLV, MLV, and GALV suggests that there is no apparent linear sequence that defines the receptor recognition determinant among viruses that use Pit1 as a primary receptor (25, 66, 175). The observation that various retroviruses employ the same receptor without any marked conservation of the viral ligand implies that they have evolved different means of interacting with the desired receptor. Surprisingly, all of these interactions lead to the same functional consequences: viral fusion and entry. This type of general association between the SU and receptor contrasts, for example, with ligand-receptor interactions, in which there is a cascade of events that result from a very specific protein-protein interaction. The observation that the Pit receptors are a common target for viral entry but that details of the binding are not conserved among viruses that use Pit receptors implies that the mode of binding is immaterial to Pit-mediated viral entry (134). Similar findings have been obtained using chimeric ecotropic enveloped retroviral vectors, and it was shown that the binding of chimeric enveloped particles to cells does not lead to successful membrane fusion (200) and that most cell surface proteins fail to facilitate targeted retroviral entry due to postbinding blocks (177). These findings imply that receptor conservation may be maintained at a second, postbinding step in viral entry and that finding an effective envelope-receptor fit that permits binding is less difficult than is finding a cellular receptor competent to permit the subsequent stages in viral entry.
There may be other reasons why many different viruses converged to use similar receptors. Pit1 and Pit2 are coexpressed on almost all cell types and apparently represent only two of a possible large number of Pi transporters. This redundancy in transport receptors provides a critical safeguard for the cell because virus binding has been demonstrated to block Pit1 and Pit2-mediated Pi uptake (80, 127, 188). In addition, there are advantages to the virus in using a receptor that is found on almost all cell types in the host.
CONCLUSIONS AND FUTURE DIRECTIONS
Many retroviral receptors and coreceptors have been identified in the past decade by using gene transfer methods. The success of this approach relies on the identification of nonsusceptible cell targets that are resistant by virtue of the absence of a functional receptor and on the availability of a high-titer virus bearing the envelope of interest. Although it is not trivial to generate such reagents, once this is accomplished it is generally feasible to clone receptors using gene transfer if the virus requires only a single receptor molecule. The situation is clearly much more challenging for cases in which more than one molecule is required for viral entry. Nonetheless, both receptor molecules have been identified for HIV and SIV and for FeLV-T.
The challenge for the next decade is to better define the virus-receptor interactions that permit binding, fusion, entry and perhaps postentry stages in virus replication. There is evidence that these determinants may be structural rather than sequence based. For example, HIV-1, HIV-2, and SIV both bind and use a common coreceptor for fusion yet have less than 50% identity in the envelope SU. Similarly, FeLV has overlapping receptor specificity with both GALV and MLV yet there is less than 40% identity among these SUs (16). Thus, if we are to understand the functional interactions that are required for steps in entry and fusion, it will be important to determine the structure of retroviral envelope proteins, ideally in conjunction with their receptor. Only two such structures have been resolved, the N-terminal half of the E-MLV SU (58) and the core portion of HIV-1 SU in complex with portions of CD4 and antibody (95, 192). These structures have proven enormously useful in advancing the study of envelope-receptor interactions. It is worthwhile nothing that although 10 years ago only three retroviral receptors were known, perseverance in cloning receptors, combined with advances in technology, has led to many successes since then, as highlighted in this review. Advances in our understanding of envelope-receptor interactions in the next decade will rely on having similar rewards from our current efforts in structural analyses.
REFERENCES
- 1.Adkins H B, Blacklow S C, Young J A. Two functionally distinct forms of a retroviral receptor explain the nonreciprocal receptor interference among subgroups B, D, and E avian leukosis viruses. J Virol. 2001;75:3520–3526. doi: 10.1128/JVI.75.8.3520-3526.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adkins H B, Brojatsch J, Naughton J, Rolls M M, Pesola J M, Young J A. Identification of a cellular receptor for subgroup E avian leukosis virus. Proc Natl Acad Sci USA. 1997;94:11617–11622. doi: 10.1073/pnas.94.21.11617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albritton L M, Kim J W, Tseng L, Cunningham J M. Envelope-binding domain in the cationic amino acid transporter determines the host range of ecotropic murine retroviruses. J Virol. 1993;67:2091–2096. doi: 10.1128/jvi.67.4.2091-2096.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albritton L M, Tseng L, Scadden D, Cunningham J M. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell. 1989;57:659–666. doi: 10.1016/0092-8674(89)90134-7. [DOI] [PubMed] [Google Scholar]
- 5.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1a, MIP-1B receptor as a fusion cofactor for macrophage-tropic HIV 1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 6.An D S, Xie Y, Chen I S. Envelope gene of the human endogenous retrovirus HERV-W encodes a functional retrovirus envelope. J Virol. 2001;75:3488–3489. doi: 10.1128/JVI.75.7.3488-3489.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson M M, Lauring A S, Burns C C, Overbaugh J. Identification of a cellular cofactor required for infection by feline leukemia virus. Science. 2000;287:1828–1830. doi: 10.1126/science.287.5459.1828. [DOI] [PubMed] [Google Scholar]
- 8.Ban J, Hlavaty J, Orlik O, Splitter G A, Altaner C. The human homologue of the bovine leukemia virus receptor BLVRcp1 is the delta-subunit of adaptor-related AP-3 protein that does not bind the BVLgp51. Arch Virol. 1999;144:2013–2022. doi: 10.1007/s007050050722. [DOI] [PubMed] [Google Scholar]
- 9.Ban J, Portetelle D, Altaner C, Horion B, Milan D, Krchnak V, Burny A, Kettmann R. Isolation and characterization of a 2.3-kilobase-pair cDNA fragment encoding the binding domain of the bovine leukemia virus cell receptor. J Virol. 1993;67:1050–1057. doi: 10.1128/jvi.67.2.1050-1057.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ban J, Truong A T, Horion B, Altaner C, Burny A, Portetelle D, Kettmann R. Isolation of the missing 5′-end of the encoding region of the bovine leukemia virus cell receptor gene. Arch Virol. 1994;138:379–383. doi: 10.1007/BF01379141. [DOI] [PubMed] [Google Scholar]
- 11.Barnett A L, Davey R A, Cunningham J M. Modular organization of the Friend murine leukemia virus envelope protein underlies the mechanism of infection. Proc Natl Acad Sci USA. 2001;98:4113–4118. doi: 10.1073/pnas.071432398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bates P, Young J A T, Varmus H E. A receptor for subgroup A Rous sarcoma virus is related to the low density lipoprotein receptor. Cell. 1993;74:1043–1051. doi: 10.1016/0092-8674(93)90726-7. [DOI] [PubMed] [Google Scholar]
- 13.Batinic D, Robey F A. The V3 region of the envelope glycoprotein of human immunodeficiency virus type 1 binds sulfated polysaccharides and CD4-derived synthetic peptides. J Biol Chem. 1992;267:6664–6671. [PubMed] [Google Scholar]
- 14.Battini J L, Danos O, Heard J M. Definition of a 14-amino-acid peptide essential for the interaction between the murine leukemia virus amphotropic envelope glycoprotein and its receptor. J Virol. 1998;72:428–435. doi: 10.1128/jvi.72.1.428-435.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Battini J L, Danos O, Heard J M. Receptor-binding domain of murine leukemia virus envelope glycoproteins. J Virol. 1995;69:713–719. doi: 10.1128/jvi.69.2.713-719.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Battini J L, Heard J M, Danos O. Receptor choice determinants in the envelope glycoproteins of amphotropic, xenotropic, and polytropic murine leukemia viruses. J Virol. 1992;66:1468–1475. doi: 10.1128/jvi.66.3.1468-1475.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Battini J L, Rasko J E, Miller A D. A human cell-surface receptor for xenotropic and polytropic murine leukemia viruses: possible role in G protein-coupled signal transduction. Proc Natl Acad Sci USA. 1999;96:1385–1390. doi: 10.1073/pnas.96.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Battini J L, Rodrigues P, Muller R, Danos O, Heard J M. Receptor-binding properties of a purified fragment of the 4070A amphotropic murine leukemia virus envelope glycoprotein. J Virol. 1996;70:4387–4393. doi: 10.1128/jvi.70.7.4387-4393.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benveniste R E, Sherr C J, Todaro G J. Evolution of type C viral genes: origin of feline leukemia virus. Science. 1975;190:886–888. doi: 10.1126/science.52892. [DOI] [PubMed] [Google Scholar]
- 20.Berger E A. HIV entry and tropism: the chemokine receptor connection. AIDS. 1997;11:S3–S16. [PubMed] [Google Scholar]
- 21.Berger E A, Murphy P M, Farber J M. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 22.Bleul C C, Wu L, Hoxie J A, Springer T A, Mackay C R. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc Natl Acad Sci USA. 1997;94:1925–1930. doi: 10.1073/pnas.94.5.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blond J L, Beseme F, Duret L, Bouton O, Bedin F, Perron H, Mandrand B, Mallet F. Molecular characterization and placental expression of HERV W, a new human endogenous retrovirus family. J Virol. 1999;73:1175–1185. doi: 10.1128/jvi.73.2.1175-1185.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blond J L, Lavillette D, Cheynet V, Bouton O, Oriol G, Chapel-Fernandes S, Mandrand B, Mallet F, Cosset F L. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J Virol. 2000;74:3321–3329. doi: 10.1128/jvi.74.7.3321-3329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boomer S, Eiden M, Burns C C, Overbaugh J. Three distinct envelope domains, variably present in subgroup B feline leukemia virus recombinants, mediate Pit1 and Pit2 receptor recognition. J Virol. 1997;71:8116–8123. doi: 10.1128/jvi.71.11.8116-8123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brojatsch J, Kristal B S, Viglianti G A, Khiroya R, Hoover E A, Mullins J I. Feline leukemia virus subgroup C phenotype evolves through distinct alterations near the N terminus of the envelope surface glycoprotein. Proc Natl Acad Sci USA. 1992;89:8457–8461. doi: 10.1073/pnas.89.18.8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brojatsch J, Naughton J, Rolls M M, Zingler K, Young J A T. CAR1, a TNFR-related protein, is a cellular receptor for cytopathic avian leukosis-sarcoma viruses and mediates apoptosis. Cell. 1996;87:845–855. doi: 10.1016/s0092-8674(00)81992-3. [DOI] [PubMed] [Google Scholar]
- 28.Callahan L N, Phelan M, Mallinson M, Norcross M A. Dextran sulfare blocks antibody binding to the principal neutralizing domain of human immunodeficiency virus type 1 without interfering with gp120-CD4 interactions. J Virol. 1991;65:1543–1550. doi: 10.1128/jvi.65.3.1543-1550.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Callahan R, Meade C, Todaro G J. Isolation of an endogenous type C virus related to the infectious primate type C viruses from the Asian rodent Vandeleuria oleracea. J Virol. 1979;30:124–131. doi: 10.1128/jvi.30.1.124-131.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Callahan R, Todaro G. Origin of inbred mice. 1978. Four major endogenous retrovirus classes each genetically transmitted in various species of mus; pp. 689–712. [Google Scholar]
- 31.Chabot D J, Chen H, Dimitrov D S, Broder C C. N-linked glycosylation of CXCR4 masks coreceptor function for CCR5-dependent human immunodeficiency virus type 1 isolates. J Virol. 2000;74:4404–4413. doi: 10.1128/jvi.74.9.4404-4413.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chackerian B, Long E M, Luciw P A, Overbaugh J. Human immunodeficiency virus type 1 coreceptors participate in postentry stages in the virus replication cycle and function in simian immunodeficiency virus infection. J Virol. 1997;71:3932–3939. doi: 10.1128/jvi.71.5.3932-3939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Z, Zhou P, Ho D D, Landau N T, Marx P A. Genetically divergent strains of simian immunodeficiency virus use CCR5 as a coreceptor for entry. J Virol. 1997;71:2705–2714. doi: 10.1128/jvi.71.4.2705-2714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 35.Closs E I, Lyons C R, Kelly C, Cunningham J M. Characterization of the third member of the MCAT family of cationic amino acid transporters. Identification of a domain that determines the transport properties of the MCAT proteins. J Biol Chem. 1993;268:20796–20800. [PubMed] [Google Scholar]
- 36.Cocchi F, DeVico A L, Garzino-Demo A, Cara A, Gallo R C, Lusso P. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat Med. 1996;2:1244–1247. doi: 10.1038/nm1196-1244. [DOI] [PubMed] [Google Scholar]
- 37.Coffin J M. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 38.Connolly L, Zingler K, Young J A. A soluble form of a receptor for subgroup A avian leukosis and sarcoma viruses (ALSV-A) blocks infection and binds directly to ALSV-A. J Virol. 1994;68:2760–2764. doi: 10.1128/jvi.68.4.2760-2764.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. Change in coreceptor use correlates with disease progression in HIV-1 infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dalgleish A G, Beverley P C, Clapham P R, Crawford D H, Greaves M F, Weiss R A. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 41.Davey R A, Hamson C A, Healey J J, Cunningham J M. In vitro binding of purified murine ecotropic retrovirus envelope surface protein to its receptor, MCAT-1. J Virol. 1997;71:8096–8102. doi: 10.1128/jvi.71.11.8096-8102.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davey R A, Zuo Y, Cunningham J M. Identification of a receptor-binding pocket on the envelope protein of friend murine leukemia virus. J Virol. 1999;73:3758–3763. doi: 10.1128/jvi.73.5.3758-3763.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dean M, Carrington M, Winkler C, Huttley G A, Smith M W, Allikmets R, Goedert J J, Buchbinder S P, Vittinghoff E, Gomperts E, Donfield S, Vlahov D, Kaslow R, Saah A, Rinaldo C, Detels R. Genetic restriction of HIV-1 infection and progression to AIDS by deletion allele of the CKR5 structural gene. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 44.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 45.Deng H K, Unutmaz D, KewalRamani V N, Littman D R. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature. 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 46.de Parseval A, Elder J H. Binding of recombinant feline immunodeficiency virus surface glycoprotein to feline cells: role of CXCR4, cell surface heparans, and an unidentified non-CXCR4 receptor. J Virol. 2001;75:4528–4539. doi: 10.1128/JVI.75.10.4528-4539.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Doms R W. Beyond receptor expression: the influence of receptor conformation, density, and affinity in HIV-1 infection. Virology. 2000;276:229–237. doi: 10.1006/viro.2000.0612. [DOI] [PubMed] [Google Scholar]
- 48.Donahue P R, Quackenbush S L, Gallo M V, deNoronha C M C, Overbaugh J, Hoover E A, Mullins J I. Viral genetic determinants of T-cell killing and immunodeficiency disease induction by the feline leukemia virus FeLV-FAIDS. J Virol. 1991;65:4461–4469. doi: 10.1128/jvi.65.8.4461-4469.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 50.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 51.Dumonceaux J, Nisole S, Chanel C, Quivet L, Amara A, Baleux F, Briand P, Hazan U. Spontaneous mutations in the env gene of the human immunodeficiency virus type 1 NDK isolate are associated with a CD4-independent entry phenotype. J Virol. 1998;72:512–519. doi: 10.1128/jvi.72.1.512-519.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Edinger A L, Amedee A, Miller K, Doranz B J, Endres M, Sharron M, Samson M, Lu Z H, Clements J E, Murphey-Corb M, Peiper S C, Parmentier M, Broder C C, Doms R W. Differential utilization of CCR5 by macrophage and T cell tropic simian immunodeficiency virus strains. Proc Natl Acad Sci USA. 1997;94:4005–4010. doi: 10.1073/pnas.94.8.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Edinger A L, Mankowski J L, Doranz B J, Margulies B J, Lee B, Rucker J, Sharron M, Hoffman T L, Berson J F, Zink M C, Hirsch V M, Clements J E, Doms R W. CD4-independent, CCR5-dependent infection of brain capillary endothelial cells by a neurovirulent simian immunodeficiency virus strain. Proc Natl Acad Sci USA. 1997;94:14742–14747. doi: 10.1073/pnas.94.26.14742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Egberink H F, De Clercq E, Van Vliet A L, Balzarini J, Bridger G J, Henson G, Horzinek M C, Schols D. Bicyclams, selective antagonists of the human chemokine receptor CXCR4, potently inhibit feline immunodeficiency virus replication. J Virol. 1999;73:6346–6352. doi: 10.1128/jvi.73.8.6346-6352.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eiden M V, Farrell K, Warsowe J, Mahan L C, Wilson C A. Characterization of a naturally occurring ecotropic receptor that does not facilitate entry of all ecotropic murine retroviruses. J Virol. 1993;67:4056–4061. doi: 10.1128/jvi.67.7.4056-4061.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eiden M V, Farrell K, Wilson C A. Glycosylation-dependent inactivation of the ecotropic murine leukemia virus receptor. J Virol. 1994;68:626–631. doi: 10.1128/jvi.68.2.626-631.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Endres M J, Clapham P R, Marsh M, Ahuja M, Davis-Turner J, McKnight A, Thomas J F, Stoebenau-Haggarty B, Choe S, Vance P E, Well T N C, Power C A, Sutterwala S S, Doms R W, Landau N R, Hoxie J A. CD4-Independent infection by HIV-2 is mediated by fusin/CXCR4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 58.Fass D, Davey R A, Hamson C A, Kim P S, Cunningham J M, Berger J M. Structure of a murine leukemia virus receptor-binding glycoprotein at 2.0 angstrom resolution. Science. 1997;277:1662–1666. doi: 10.1126/science.277.5332.1662. [DOI] [PubMed] [Google Scholar]
- 59.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 60.Fenyö E M, Albert J, Asjö B. Replicative capacity, cytopathic effect and cell tropism of HIV. AIDS. 1989;3:S5–S12. doi: 10.1097/00002030-198901001-00002. [DOI] [PubMed] [Google Scholar]
- 61.Gardner M B, Kozak C A, O'Brien S J. The Lake Casitas wild mouse: evolving genetic resistance to retroviral disease. Trends Genet. 1991;7:22–27. doi: 10.1016/0168-9525(91)90017-k. [DOI] [PubMed] [Google Scholar]
- 62.Gavalchin J, Fan N, Waterbury P G, Corbett E, Faldasz B D, Peshick S M, Poiesz B J, Papsidero L, Lane M J. Regional localization of the putative cell surface receptor for HTLV-I to human chromosome 17q23.2–17q25.3. Virology. 1995;212:196–203. doi: 10.1006/viro.1995.1468. [DOI] [PubMed] [Google Scholar]
- 63.Geijtenbeek T B, Kwon D S, Torensma R, van Vliet S J, van Duijnhoven G C, Middel J, Cornelissen I L, Nottet H S, KewalRamani V N, Littman D R, Figdor C G, van Kooyk Y. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell. 2000;100:587–97. doi: 10.1016/s0092-8674(00)80694-7. [DOI] [PubMed] [Google Scholar]
- 64.Golovkina T V, Dzuris J, van den Hoogen B, Jaffe A B, Wright P C, Cofer S M, Ross S R. A novel membrane protein is a mouse mammary tumor virus receptor. J Virol. 1998;72:3066–3071. doi: 10.1128/jvi.72.4.3066-3071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gwynn S R, Hankenson F C, Lauring A S, Rohn J L, Overbaugh J. Feline leukemia virus envelope sequences that affect T-cell tropism and syncytium formation are not part of known receptor-binding domains. J Virol. 2000;74:5754–5761. doi: 10.1128/jvi.74.13.5754-5761.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Han J Y, Cannon P M, Lai K M, Zhao Y, Eiden M V, Anderson W F. Identification of envelope protein residues required for the expanded host range of 10A1 murine leukemia virus. J Virol. 1997;71:8103–8108. doi: 10.1128/jvi.71.11.8103-8108.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Harouse J M, Bhat S, Spitalnik S L, Laughlin M, Stefano K, Silberberg D H, Gonzalez-Scarano F. Inhibition of entry of HIV-1 in neural cell lines by antibodies against galactosyl ceramide. Science. 1991;253:320–323. doi: 10.1126/science.1857969. [DOI] [PubMed] [Google Scholar]
- 68.He J, Chen Y, Farzan M, Choe H, Ohagen A, Gartner S, Busciglio J, Yang X, Hofmann W, Newman W, Mackay C R, Sodroski J, Gabuzda D. CCR3 and CCR5 are co-receptors for HIV-1 infection of microglia. Nature. 1997;385:645–649. doi: 10.1038/385645a0. [DOI] [PubMed] [Google Scholar]
- 69.Hladik F, Lentz G, Akridge R E, Peterson G, Kelley H, McElroy A, McElrath M J. Dendritic cell-T-cell interactions support coreceptor-independent human immunodeficiency virus type 1 transmission in the human genital tract. J Virol. 1999;73:5833–5842. doi: 10.1128/jvi.73.7.5833-5842.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ho D D, Neumann A U, Perelson A S, Chen W, Leonard J M, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 71.Hoffman T L, Doms R W. Chemokines and coreceptors in HIV/SIV-host interactions. AIDS. 1998;12:S17–S26. [PubMed] [Google Scholar]
- 72.Hoffman T L, LaBranche C C, Zhang W, Canziani G, Robinson J, Chaiken I, Hoxie J A, Doms R W. Stable exposure of the coreceptor-binding site in a CD4-independent HIV-1 envelope protein. Proc Natl Acad Sci USA. 1999;96:6359–6364. doi: 10.1073/pnas.96.11.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hosie M J, Broere N, Hesselgesser J, Turner J D, Hoxie J A, Neil J C, Willett B J. Modulation of feline immunodeficiency virus infection by stromal cell-derived factor. J Virol. 1998;72:2097–2104. doi: 10.1128/jvi.72.3.2097-2104.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang Y, Paxton W A, Wolinsky S M, Neumann A U, Zhang L, He T, Kang S, Ceradini D, Jin Z, Yazdanbakhsh K, Kunstman K, Erickson D, Dragon E, Landau N R, Phair J, Ho D D, Koup R A. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat Med. 1996;2:1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- 75.Hunter E. Viral entry and receptor. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Old Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1997. p. 843. [PubMed] [Google Scholar]
- 76.Jarrett O, Russell P H. Differential growth and transmission in cats of feline leukemia viruses of subgroups A and B. Int J Cancer. 1978;21:466–472. doi: 10.1002/ijc.2910210411. [DOI] [PubMed] [Google Scholar]
- 77.Jobbagy Z, Olah Z, Petrovics G, Eiden M V, Leverett B D, Dean N M, Anderson W B. Up-regulation of the Pit-2 phosphate transporter/retrovirus receptor by protein kinase C epsilon. J Biol Chem. 1999;274:7067–7071. doi: 10.1074/jbc.274.11.7067. [DOI] [PubMed] [Google Scholar]
- 78.Johann S V, Gibbons J J, O'Hara B. GLVR1, a receptor for gibbon ape leukemia virus, is homologous to a phosphate permease of Neurospora crassa and is expressed at high levels in the brain and thymus. J Virol. 1992;66:1635–1640. doi: 10.1128/jvi.66.3.1635-1640.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Johann S V, van-Zeijl M, Cekleniak J, O'Hara B. Definition of a domain of GLVR1 which is necessary for infection by gibbon ape leukemia virus and which is highly polymorphic between species. J Virol. 1993;67:6733–6736. doi: 10.1128/jvi.67.11.6733-6736.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kavanaugh M P, Miller D G, Zhang W, Law W, Kozak S L, Kabat D, Miller A D. Cell-surface receptors for gibbon ape leukemia virus and amphotropic murine retrovirus are inducible sodium-dependent phosphate symporters. Proc Natl Acad Sci USA. 1994;91:7071–7075. doi: 10.1073/pnas.91.15.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kavanaugh M P, Wang H, Zhang Z, Zhang W, Wu Y N, Dechant E, North R A, Kabat D. Control of cationic amino acid transport and retroviral receptor functions in a membrane protein family. J Biol Chem. 1994;269:15445–15450. [PubMed] [Google Scholar]
- 82.Kekuda R, Prasad P D, Fei Y J, Torres-Zamorano V, Sinha S, Yang-Feng T L, Leibach F H, Ganapathy V. Cloning of the sodium-dependent, broad-scope, neutral amino acid transporter Bo from a human placental choriocarcinoma cell line. J Biol Chem. 1996;271:18657–18661. doi: 10.1074/jbc.271.31.18657. [DOI] [PubMed] [Google Scholar]
- 83.Keshet E, Temin H M. Cell killing by spleen necrosis virus is correlated with a transient accumulation of spleen necrosis virus DNA. J Virol. 1979;31:376–388. doi: 10.1128/jvi.31.2.376-388.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kewalramani V N, Panganiban A T, Emerman M. Spleen necrosis virus, an avian immunosuppressive retrovirus, shares a receptor with the type D simian retroviruses. J Virol. 1992;66:3026–3031. doi: 10.1128/jvi.66.5.3026-3031.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim J W, Closs E I, Albritton L M, Cunningham J M. Transport of cationic amino acids by the mouse ecotropic retrovirus receptor. Nature. 1991;352:725–728. doi: 10.1038/352725a0. [DOI] [PubMed] [Google Scholar]
- 86.Kimata J T, Gosink J J, KewalRamani V N, Rudensey L M, Littman D R, Overbaugh J. Coreceptor specificity of temporal variants of simian immunodeficiency virus Mne. J Virol. 1999;73:1655–1660. doi: 10.1128/jvi.73.2.1655-1660.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kimata J T, Kuller L, Anderson D B, Dailey P, Overbaugh J. Emerging cytopathic and antigenic simian immunodeficiency virus variants influence AIDS progression. Nat Med. 1999;5:535–541. doi: 10.1038/8414. [DOI] [PubMed] [Google Scholar]
- 88.Klasse P J, Bron R, Marsh M. Mechanisms of enveloped virus entry into animal cells. Adv Drug Deliv Rev. 1998;34:65–91. doi: 10.1016/S0169-409X(98)00002-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Klatzmann D, Champagne E, Chamaret S, Gruest J, Guetard D, Hercend T, Gluckman J C, Montagnier L. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature. 1984;312:767–768. doi: 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- 90.Kolchinsky P, Mirzabekov T, Farzan M, Kiprilov E, Cayabyab M, Mooney L J, Choe H, Sodroski J. Adaptation of a CCR5-using, primary human immunodeficiency virus type 1 isolate for CD4-independent replication. J Virol. 1999;73:8120–8126. doi: 10.1128/jvi.73.10.8120-8126.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Koo H M, Gu J, Varela-Echavarria A, Ron Y, Dougherty J P. Reticuloendotheliosis type C and primate type D oncoretroviruses are members of the same receptor interference group. J Virol. 1992;66:3448–3454. doi: 10.1128/jvi.66.6.3448-3454.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kozak S L, Platt E J, Madani N, Ferro F E, Jr, Peden K, Kabat D. CD4, CXCR-4, and CCR-5 dependencies for infections by primary patient and laboratory-adapted isolates of human immunodeficiency virus type 1. J Virol. 1997;71:873–882. doi: 10.1128/jvi.71.2.873-882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kuhmann S E, Platt E J, Kozak S L, Kabat D. Cooperation of multiple CCR5 coreceptors is required for infections by human immunodeficiency virus type 1. J Virol. 2000;74:7005–7015. doi: 10.1128/jvi.74.15.7005-7015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kumar D V, Berry B T, Roy-Burman P. Nucleotide sequence and distinctive characteristics of the env gene of endogenous feline leukemia provirus. J Virol. 1989;63:2379–2384. doi: 10.1128/jvi.63.5.2379-2384.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.LaBranche C C, Hoffman T L, Romano J, Haggarty B S, Edwards T G, Matthews T J, Doms R W, Hoxie J A. Determinants of CD4 independence for a human immunodeficiency virus type 1 variant map outside regions required for coreceptor specificity. J Virol. 1999;73:10310–10319. doi: 10.1128/jvi.73.12.10310-10319.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lapham C K, Ouyang J, Chandrasekhar B, Nguyen N Y, Dimitrov D S, Golding H. Evidence for cell-surface association between fusin and the CD4-gp120 complex in human cell lines. Science. 1996;274:602–605. doi: 10.1126/science.274.5287.602. [DOI] [PubMed] [Google Scholar]
- 98.Lapham C K, Zaitseva M B, Lee S, Romanstseva T, Golding H. Fusion of monocytes and macrophages with HIV-1 correlates with biochemical properties of CXCR4 and CCR5. Nat Med. 1999;5:303–308. doi: 10.1038/6523. [DOI] [PubMed] [Google Scholar]
- 98a.Lauring A S, Anderson M M, Overbaugh J. Specificity in receptor usage by T-cell-tropic feline leukemia viruses: implications for in vivo tropism of immunodeficiency-inducing variants. J Virol. 2001;19:8888–8898. doi: 10.1128/JVI.75.19.8888-8898.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lavillette D, Boson B, Russell S J, Cosset F L. Activation of membrane fusion by murine leukemia viruses is controlled in cis or in trans by interactions between the receptor-binding domain and a conserved disulfide loop of the carboxy terminus of the surface glycoprotein. J Virol. 2001;75:3685–3695. doi: 10.1128/JVI.75.8.3685-3695.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lavillette D, Maurice M, Roche C, Russell S J, Sitbon M, Cosset F L. A proline-rich motif downstream of the receptor binding domain modulates conformation and fusogenicity of murine retroviral envelopes. J Virol. 1998;72:9955–9965. doi: 10.1128/jvi.72.12.9955-9965.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lavillette D, Ruggieri A, Russell S J, Cosset F L. Activation of a cell entry pathway common to type C mammalian retroviruses by soluble envelope fragments. J Virol. 2000;74:295–304. doi: 10.1128/jvi.74.1.295-304.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee B, Sharron M, Montaner L J, Weissman D, Doms R W. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc Natl Acad Sci USA. 1999;96:5215–5220. doi: 10.1073/pnas.96.9.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lepperdinger G, Strobl B, Kreil G. HYAL2, a human gene expressed in many cells, encodes a lysosomal hyaluronidase with a novel type of specificity. J Biol Chem. 1998;273:22466–22470. doi: 10.1074/jbc.273.35.22466. [DOI] [PubMed] [Google Scholar]
- 104.Lieber M M, Sherr C J, Todaro G J, Benveniste R E, Callahan R, Coon H G. Isolation from the asian mouse Mus caroli of an endogenous type C virus related to infectious primate type C viruses. Proc Natl Acad Sci USA. 1975;72:2315–2319. doi: 10.1073/pnas.72.6.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu Q H, Williams D A, McManus C, Baribaud F, Doms R W, Schols D, De Clercq E, Kotlikoff M I, Collman R G, Freedman B D. HIV-1 gp120 and chemokines activate ion channels in primary macrophages through CCR5 and CXCR4 stimulation. Proc Natl Acad Sci USA. 2000;97:4832–4837. doi: 10.1073/pnas.090521697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu R, Paxton W A, Choe S, Ceradini D, Martin S R, Horuk R, MacDonald M E, Stuhlmann H, Koup R E, Landau N R. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 107.Lundorf M D, Pedersen F S, O'Hara B, Pedersen L. Single amino acid insertion in loop 4 confers amphotropic murine leukemia virus receptor function upon murine Pit1. J Virol. 1998;72:4524–4527. doi: 10.1128/jvi.72.5.4524-4527.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Maddon P J, Dalgleish A G, McDougal J S, Clapham P R, Weiss R A, Axel R. The CD4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell. 1986;47:333–348. doi: 10.1016/0092-8674(86)90590-8. [DOI] [PubMed] [Google Scholar]
- 109.Marcon L, Choe H, Martin K A, Farzan M, Ponath P D, Wu L, Newman W, Gerard N, Gerar C, Sodroski J. Utilization of C-C Chemokine receptor 5 by the envelope glycoproteins of a pathogenic simian immunodificiency virus, SIVmac239. J Virol. 1997;71:2522–2527. doi: 10.1128/jvi.71.3.2522-2527.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Marin M, Tailor C S, Nouri A, Kabat D. Sodium-dependent neutral amino acid transporter type 1 is an auxiliary receptor for baboon endogenous retrovirus. J Virol. 2000;74:8085–8093. doi: 10.1128/jvi.74.17.8085-8093.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Masuda M, Hanson C A, Alvord W G, Hoffman P M, Ruscetti S K. Effects of subtle changes in the SU protein of ecotropic murine leukemia virus on its brain capillary endothelial cell tropism and interference properties. Virology. 1996;215:142–151. doi: 10.1006/viro.1996.0017. [DOI] [PubMed] [Google Scholar]
- 112.Masuda M, Hanson C A, Hoffman P M, Ruscetti S K. Analysis of the unique hamster cell tropism of ecotropic murine leukemia virus PVC-211. J Virol. 1996;70:8534–8539. doi: 10.1128/jvi.70.12.8534-8539.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.McDougal J S, Kennedy M S, Sligh J M, Cort S P, Mawle A, Nicholson J K. Binding of HTLV-III/LAV to T4+ T cells by a complex of the 110K viral protein and the T4 molecule. Science. 1986;231:382–385. doi: 10.1126/science.3001934. [DOI] [PubMed] [Google Scholar]
- 114.McDougall A S, Terry A, Tzavaras T, Cheney C, Rojko J, Neil J C. Defective endogenous proviruses are expressed in feline lymphoid cells: evidence for a role in natural resistance to subgroup B feline leukemia viruses. J Virol. 1994;68:2151–2160. doi: 10.1128/jvi.68.4.2151-2160.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.McKnight A, Weiss R A, Shotton C, Takeuchi Y, Hoshino H, Clapham P R. Change in tropism upon immune escape by human immunodeficiency virus. J Virol. 1995;69:3167–3170. doi: 10.1128/jvi.69.5.3167-3170.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Miedema F, Meyaard L, Koot M, Klein M R, Roos M L, Groenink M, Fouchier R A M, Van't-Wout A B, Tersmette M, Sciellekens P T A, Schuitemaker H. Changing virus-host interactions in the course of HIV-1 infection. Immunol Rev. 1994;140:35–72. doi: 10.1111/j.1600-065x.1994.tb00864.x. [DOI] [PubMed] [Google Scholar]
- 117.Miller D G, Edwards R H, Miller A D. Cloning of the cellular receptor for amphotropic murine retroviruses reveals homology to that for gibbon ape leukemia virus. Proc Natl Acad Sci USA. 1994;91:78–82. doi: 10.1073/pnas.91.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Miller D G, Miller A D. A family of retroviruses that utilize related phosphate transporters for cell entry. J Virol. 1994;68:8270–8276. doi: 10.1128/jvi.68.12.8270-8276.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Miller D G, Miller A D. Tunicamycin treatment of CHO cells abrogates multiple blocks to retrovirus infection, one of which is due to a secreted inhibitor. J Virol. 1992;66:78–84. doi: 10.1128/jvi.66.1.78-84.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mondor I, Ugolini S, Sattentau Q J. Human immunodeficiency virus type 1 attachment to HeLa CD4 cells is CD4 independent and gp120 dependent and requires cell surface heparans. J Virol. 1998;72:3623–3634. doi: 10.1128/jvi.72.5.3623-3634.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Moore J P, Trkola A, Dragic T. Co-receptors for HIV-1 entry. Curr Opin Immunol. 1997;9:551–562. doi: 10.1016/s0952-7915(97)80110-0. [DOI] [PubMed] [Google Scholar]
- 122.Moser M, Burns C, Boomer S, Overbaugh J. The host range and interfence properties of two closely related feline leukemia virus variants suggest that they use distinct receptors. Virology. 1998;242:366–377. doi: 10.1006/viro.1997.9008. [DOI] [PubMed] [Google Scholar]
- 123.Moulard M, Lortat-Jacob H, Mondor I, Roca G, Wyatt R, Sodroski J, Zhao L, Olson W, Kwong P D, Sattentau Q J. Selective interactions of polyanions with basic surfaces on human immunodeficiency virus type 1 gp120. J Virol. 2000;74:1948–1960. doi: 10.1128/jvi.74.4.1948-1960.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Murer H, Biber J. Molecular mechanisms of renal apical Na/phosphate cotransport. Annu Rev Physiol. 1996;58:607–618. doi: 10.1146/annurev.ph.58.030196.003135. [DOI] [PubMed] [Google Scholar]
- 125.O'Hara B, Johann S V, Klinger H P, Blair D G, Rubinson H, Dunn K J, Saas P, Vitek S M, Robins T. Characterization of a human gene conferring sensitivity to infection by gibbon ape leukemia virus. Cell Growth Differ. 1990;1:119–127. [PubMed] [Google Scholar]
- 126.Ohshiro Y, Murakami T, Matsuda K, Nishioka K, Yoshida K, Yamamoto N. Role of cell surface glycosaminoglycans of human T cells in human immunodeficiency virus type-1 (HIV-1) infection. Microbiol Immunol. 1996;40:827–835. doi: 10.1111/j.1348-0421.1996.tb01148.x. [DOI] [PubMed] [Google Scholar]
- 127.Olah Z, Lehel C, Anderson W B, Eiden M V, Wilson C A. The cellular receptor for gibbon ape leukemia virus is a novel high affinity sodium-dependent phosphate transporter. J Biol Chem. 1994;269:25426–25431. [PubMed] [Google Scholar]
- 128.Ott D, Friedrich R, Rein A. Sequence analysis of amphotropic and 10A1 murine leukemia viruses: close relationship to mink cell focus-inducing viruses. J Virol. 1990;64:757–766. doi: 10.1128/jvi.64.2.757-766.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Overbaugh J, Bangham C R. Selection forces and constraints on retroviral sequence variation. Science. 2001;292:1106–1109. doi: 10.1126/science.1059128. [DOI] [PubMed] [Google Scholar]
- 130.Overbaugh J, Riedel N, Hoover E A, Mullins J I. Transduction of endogenous envelope genes by feline leukaemia virus in vitro. Nature. 1988;332:731–734. doi: 10.1038/332731a0. [DOI] [PubMed] [Google Scholar]
- 131.Palmer G, Bonjour J P, Caverzasio J. Expression of a newly identified phosphate transporter/retrovirus receptor in human SaOS-2 osteoblast-like cells and its regulation by insulin-like growth factor I. Endocrinology. 1997;138:5202–5209. doi: 10.1210/endo.138.12.5561. [DOI] [PubMed] [Google Scholar]
- 132.Patel M, Yanagishita M, Roderiquez G, Bou-Habib D C, Oravecz T, Hascall V C, Norcross M A. Cell-surface heparan sulfate proteoglycan mediates HIV-1 infection of T-cell lines. AIDS Res Hum Retroviruses. 1993;9:167–174. doi: 10.1089/aid.1993.9.167. [DOI] [PubMed] [Google Scholar]
- 133.Pecheur E I, Sainte-Marie J, Bienvenue A, Hoekstra D. Peptides and membrane fusion: towards an understanding of the molecular mechanism of protein-induced fusion. J Membr Biol. 1999;167:1–17. doi: 10.1007/s002329900466. [DOI] [PubMed] [Google Scholar]
- 134.Pedersen L, Johann S V, van-Zeijl M, Pedersen F S, O'Hara B. Chimeras of receptors for gibbon ape leukemia virus/feline leukemia virus B and amphotropic murine leukemia virus reveal different modes of receptor recognition by retrovirus. J Virol. 1995;69:2401–2405. doi: 10.1128/jvi.69.4.2401-2405.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Platt E J, Wehrly K, Kuhmann S E, Chesebro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Pohlmann S, Soilleux E J, Baribaud F, Leslie G J, Morris L S, Trowsdale J, Lee B, Coleman N, Doms R W. DC-SIGNR, a DC-SIGN homologue expressed in endothelial cells, binds to human and simian immunodeficiency viruses and activates infection in trans. Proc Natl Acad Sci USA. 2001;98:2670–2675. doi: 10.1073/pnas.051631398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pohlmann S, Stolte N, Munch J, Ten Haaft P, Heeney J L, Stahl-Hennig C, Kirchhoff F. Co-receptor usage of BOB/GPR15 in addition to CCR5 has no significant effect on replication of simian immunodeficiency virus in vivo. J Infect Dis. 1999;180:1494–1502. doi: 10.1086/315097. [DOI] [PubMed] [Google Scholar]
- 138.Quigley J G, Burns C C, Anderson M M, Sabo K M, Lynch E D, Overbaugh J, Abkowitz J L. Cloning of the cellular receptor for feline leukemia virus subgroup C (FeLV-C), a retrovirus that induces red cell aplasia. Blood. 2000;95:1093–1099. [PubMed] [Google Scholar]
- 139.Rai S K, Duh F M, Vigdorovich V, Danilkovitch-Miagkova A, Lerman M I, Miller A D. Candidate tumor suppressor HYAL2 is a glycosylphosphatidylinositol (GPI)-anchored cell-surface receptor for jaagsiekte sheep retrovirus, the envelope protein of which mediates oncogenic transformation. Proc Natl Acad Sci USA. 2001;98:4443–4448. doi: 10.1073/pnas.071572898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rana S, Besson G, Cook D G, Rucker J, Smyth R J, Yi Y, Turner J D, Hai-Hong G, Jian-Guo D, Peiper S C, Lavi E, Samson M, Libert F, Liesnard C, Vassart G, Doms R W, Parmentier M, Collman R. Role of CCR5 in infection of primary macrophages and lymphocytes by macrophage-tropic strains of human immunodeficiency virus: resistance to patient-derived and prototype isolates resulting from the Δccr5 mutation. J Virol. 1997;71:3219–3227. doi: 10.1128/jvi.71.4.3219-3227.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rasko J E J, Battini J-L, Gottschalk R J, Mazo I, Miller A D. The RD114/simian type D retrovirus receptor is a neutral amino acid transporter. Proc Natl Acad Sci USA. 1999;96:2129–2134. doi: 10.1073/pnas.96.5.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Reeves J D, Hibbitts S, Simmons G, McKnight A, Azevedo-Pereira J M, Moniz-Pereira J, Clapham P R. Primary human immunodeficiency virus type 2 (HIV-2) isolates infect CD4- negative cells via CCR5 and CXCR4: comparison with HIV-1 and simian immunodeficiency virus and relevance to cell tropism in vivo. J Virol. 1999;73:7795–7804. doi: 10.1128/jvi.73.9.7795-7804.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Reeves J D, Schulz T F. The CD4-independent tropism of human immunodeficiency virus type 2 involves several regions of the envelope protein and correlates with a reduced activation threshold for envelope-mediated fusion. J Virol. 1997;71:1453–1465. doi: 10.1128/jvi.71.2.1453-1465.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Rein A, Schultz A. Different recombinant murine leukemia viruses use different cell surface receptors. Virology. 1984;136:144–152. doi: 10.1016/0042-6822(84)90255-1. [DOI] [PubMed] [Google Scholar]
- 145.Reinhart T A, Ghosh A K, Hoover E A, Mullins J I. Distinct superinfection interference properties yet similar receptor utilization by cytopathic and noncytopathic feline leukemia viruses. J Virol. 1993;67:5153–5162. doi: 10.1128/jvi.67.9.5153-5162.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Richardson J, Pancino G, Merat R, Leste-Lasserre T, Moraillon A, Schneider-Mergener J, Alizon M, Sonigo P, Heveker N. Shared usage of the chemokine receptor CXCR4 by primary and laboratory-adapted strains of feline immunodeficiency virus. J Virol. 1999;73:3661–3671. doi: 10.1128/jvi.73.5.3661-3671.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rider C C, Coombe D R, Harrop H A, Hounsell E F, Bauer C, Feeney J, Mulloy B, Mahmood N, Hay A, Parish C R. Anti-HIV-1 activity of chemically modified heparins: correlation between binding to the V3 loop of gp120 and inhibition of cellular HIV-1 infection in vitro. Biochemistry. 1994;33:6974–6980. doi: 10.1021/bi00188a029. [DOI] [PubMed] [Google Scholar]
- 148.Roderiquez G, Oravecz T, Yanagishita M, Bou-Habib D C, Mostowski H, Norcross M A. Mediation of human immunodeficiency virus type 1 binding by interaction of cell surface heparan sulfate proteoglycans with the V3 region of envelope gp120-gp41. J Virol. 1995;69:2233–2239. doi: 10.1128/jvi.69.4.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Rohn J L, Moser M S, Gwynn S R, Baldwin D N, Overbaugh J. In vivo evolution of a novel, syncytium-inducing and cytopathic feline leukemia virus variant. J Virol. 1998;72:2686–2696. doi: 10.1128/jvi.72.4.2686-2696.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rohn J L, Moser M S, Gwynn S R, Baldwin D N, Overbaugh J. In vivo evolution of a novel, syncytium-inducing and cytopathic feline leukemia virus variant. J Virol. 1998;72:2686–2696. doi: 10.1128/jvi.72.4.2686-2696.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Rucker J, Edinger A, Sharron M, Samson M, Lee B, Berson J, Yi Y, Margulies B, Collman R, Doranz B, Parmentier M, Doms R. Utilization of chemokine receptors, orphan receptors, and herpervirus-encoded receptors by diverse human and simian immunodeficiency viruses. J Virol. 1997;71:8999–9007. doi: 10.1128/jvi.71.12.8999-9007.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Rudensey L M, Kimata J T, Long E M, Chackerian B, Overbaugh J. Changes in the extracellular envelope glycoprotein of variants that evolve during the course of SIVMne infection affect neutralizing antibody recognition, syncytia formation and macrophage tropism, but not replication, cytopathicity or CCR-5 coreceptor recognition. J Virol. 1997;72:209–217. doi: 10.1128/jvi.72.1.209-217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Salzwedel K, Smith E D, Dey B, Berger E A. Sequential CD4-coreceptor interactions in human immunodeficiency virus type 1 Env function: soluble CD4 activates Env for coreceptor-dependent fusion and reveals blocking activities of antibodies against cryptic conserved epitopes on gp120. J Virol. 2000;74:326–333. doi: 10.1128/jvi.74.1.326-333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Samson M, Libert F, Doranz B J, Rucker J, Liesnard C, Farber C M, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, Muyldermans G, Verhofstede C, Burtonboy G, Georges M, Imai T, Rana S, Yi Y, Smyth R J, Collman R G, Doms R W, Vasart G, Parmentier M. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 155.Sarma P S, Log T. Subgroup classification of feline leukemia and sarcoma viruses by viral interference and neutralization tests. Virology. 1973;54:160–169. doi: 10.1016/0042-6822(73)90125-6. [DOI] [PubMed] [Google Scholar]
- 156.Sarma P S, Log T, Jain D, Hill P R, Huebner R J. Differential host range of viruses of feline leukemia-sarcoma complex. Virology. 1975;64:438–446. doi: 10.1016/0042-6822(75)90121-x. [DOI] [PubMed] [Google Scholar]
- 157.Sattentau Q J, Clapham P R, Weiss R A, Beverley P C, Montagnier L, Alhalabi M F, Gluckmann J C, Klatzmann D. The human and simian immunodeficiency viruses HIV-1, HIV-2 and SIV interact with similar epitopes on their cellular receptor, the CD4 molecule. AIDS. 1988;2:101–105. doi: 10.1097/00002030-198804000-00005. [DOI] [PubMed] [Google Scholar]
- 158.Sattentau Q J, Moore J P. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J Exp Med. 1991;174:407–415. doi: 10.1084/jem.174.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Scoggins R M, Taylor J R, Jr, Patrie J, van't Wout A B, Schuitemaker H, Camerini D. Pathogenesis of primary R5 human immunodeficiency virus type 1 clones in SCID-hu mice. J Virol. 2000;74:3205–3216. doi: 10.1128/jvi.74.7.3205-3216.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Siess D C, Kozak S L, Kabat D. Exceptional fusogenicity of Chinese hamster ovary cells with murine retroviruses suggests roles for cellular factor(s) and receptor clusters in the membrane fusion process. J Virol. 1996;70:3432–3439. doi: 10.1128/jvi.70.6.3432-3439.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sommerfelt M A, Weiss R A. Receptor interference groups of 20 retroviruses plating on human cells. Virology. 1990;176:58–69. doi: 10.1016/0042-6822(90)90230-o. [DOI] [PubMed] [Google Scholar]
- 162.Sommerfelt M A, Williams B P, Clapham P R, Solomon E, Goodfellow P N, Weiss R A. Human T cell leukemia viruses use a receptor determined by human chromosome 17. Science. 1988;242:1557–1559. doi: 10.1126/science.3201246. [DOI] [PubMed] [Google Scholar]
- 163.Steck F T, Rubin H. The mechanism of interference between an avian leukosis virus and Rous sarcoma virus. II. Early steps of infection by RSV of cells under conditions of interference. Virology. 1966;29:642–653. doi: 10.1016/0042-6822(66)90288-1. [DOI] [PubMed] [Google Scholar]
- 164.Steinman R M. DC-SIGN: a guide to some mysteries of dendritic cells. Cell. 2000;100:491–494. doi: 10.1016/s0092-8674(00)80684-4. [DOI] [PubMed] [Google Scholar]
- 165.Sugai J, Eiden M, Anderson M, Van Hoeven N, Meiering C D, Overbaugh J. Identification of envelope determinants of feline leukemia virus subgroup B that permit infection and gene transfer to cells expressing human Pit1 or human Pit2. J Virol. 2001;75:6841–6849. doi: 10.1128/JVI.75.15.6841-6849.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Tailor C S, Kabat D. Variable regions A and B in the envelope glycoproteins of feline leukemia virus subgroup B and amphotropic murine leukemia virus interact with discrete receptor domains. J Virol. 1997;71:9383–9391. doi: 10.1128/jvi.71.12.9383-9391.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Tailor C S, Nouri A, Lee C G, Kozak C, Kabat D. Cloning and characterization of a cell surface receptor for xenotropic and polytropic murine leukemia viruses. Proc Natl Acad Sci USA. 1999;96:927–932. doi: 10.1073/pnas.96.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Tailor C S, Nouri A, Zhao Y, Takeuchi Y, Kabat D. A sodium-dependent neutral-amino-acid transporter mediates infections of feline and baboon endogenous retroviruses and simian type D retroviruses. J Virol. 1999;73:4470–4474. doi: 10.1128/jvi.73.5.4470-4474.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Tailor C S, Takeuchi Y, O'Hara B, Johann S V, Weiss R A, Collins M K. Mutation of amino acids within the gibbon ape leukemia virus (GALV) receptor differentially affects feline leukemia virus subgroup B, simian sarcoma-associated virus, and GALV infections. J Virol. 1993;67:6737–6741. doi: 10.1128/jvi.67.11.6737-6741.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Tailor C S, Willet B J, Kabat D. A putative cell surface receptor for anemia-inducing feline leukemia virus subgroup C is a member of a transporter family. J Virol. 1999;73:6500–6505. doi: 10.1128/jvi.73.8.6500-6505.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Tajima Y, Tashiro K, Camerini D. Assignment of the possible HTLV receptor gene to chromosome 17q21–q23. Somatic Cell Mol Genet. 1997;23:225–227. doi: 10.1007/BF02721374. [DOI] [PubMed] [Google Scholar]
- 172.Takeuchi Y, Vile R G, Simpson G, O'Hara B, Collins M K, Weiss R A. Feline leukemia virus subgroup B uses the same cell surface receptor as gibbon ape leukemia virus. J Virol. 1992;66:1219–1222. doi: 10.1128/jvi.66.2.1219-1222.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Temin H M. Mechanisms of cell killing/cytopathic effects by nonhuman retroviruses. Rev Infect Dis. 1988;10:399–405. doi: 10.1093/clinids/10.2.399. [DOI] [PubMed] [Google Scholar]
- 174.Tenenhouse H S, Gauthier C, Martel J, Gesek F A, Coutermarsh B A, Friedman P A. Na+-phosphate cotransport in mouse distal convoluted tubule cells: evidence for Glvr-1 and Ram-1 gene expression. J Bone Miner Res. 1998;13:590–597. doi: 10.1359/jbmr.1998.13.4.590. [DOI] [PubMed] [Google Scholar]
- 175.Ting Y T, Wilson C A, Farrell K B, Chaudry G J, Eiden M V. Simian sarcoma-associated virus fails to infect Chinese hamster cells despite the presence of functional gibbon ape leukemia virus receptors. J Virol. 1998;72:9453–9458. doi: 10.1128/jvi.72.12.9453-9458.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Trkola A, Drajic T, Arthos J, Binley L, Olsen W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. CD-4 dependent, antibody sensitive interactions between HIV-1 and its coreceptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 177.Valsesia-Wittmann S, Morling F J, Hatziioannou T, Russell S J, Cosset F L. Receptor co-operation in retrovirus entry: recruitment of an auxiliary entry mechanism after retargeted binding. EMBO J. 1997;16:1214–1223. doi: 10.1093/emboj/16.6.1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.van Zeijl M, Johann S V, Closs E, Cunningham J, Eddy R, Shows T B, O'Hara B. A human amphotropic retrovirus receptor is a second member of the gibbon ape leukemia virus receptor family. Proc Natl Acad Sci USA. 1994;91:1168–1172. doi: 10.1073/pnas.91.3.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Verani A, Pesenti E, Polo S, Tresoldi E, Scarlatti G, Lusso P, Siccardi A G, Vercelli D. CXCR4 is a functional coreceptor for infection of human macrophages by CXCR4-dependent primary HIV-1 isolates. J Immunol. 1998;161:2084–2088. [PubMed] [Google Scholar]
- 180.Vogt V M. Retroviral virions and genomes. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Old Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1997. p. 843. [PubMed] [Google Scholar]
- 181.Wang H, Klamo E, Kuhmann S E, Kozak S L, Kavanaugh M P, Kabat D. Modulation of ecotropic murine retroviruses by N-linked glycosylation of the cell surface receptor/amino acid transporter. J Virol. 1996;70:6884–6891. doi: 10.1128/jvi.70.10.6884-6891.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wang H, Paul R, Burgeson R E, Keene D R, Kabat D. Plasma membrane receptors for ecotropic murine retroviruses require a limiting accessory factor. J Virol. 1991;65:6468–6477. doi: 10.1128/jvi.65.12.6468-6477.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Wang K S, Kuhn R J, Strauss E G, Ou S, Strauss J H. High-affinity laminin receptor is a receptor for Sindbis virus in mammalian cells. J Virol. 1992;66:4992–5001. doi: 10.1128/jvi.66.8.4992-5001.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Wei X, Ghosh S K, Taylor M E, Johnson V A, Emini E A, Deutsch P, Lifson J D, Bonhoeffer S, Nowak M A, Hahn B, Saag M S, Shaw G M. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–126. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 185.Weiss R A, Tailor C S. Retrovirus receptors. Cell. 1995;82:531–533. doi: 10.1016/0092-8674(95)90024-1. [DOI] [PubMed] [Google Scholar]
- 186.Willett B J, Picard L, Hosie M J, Turner J D, Adema K, Clapham P R. Shared usage of the chemokine receptor CXCR4 by the feline and human immunodeficiency viruses. J Virol. 1997;71:6407–6415. doi: 10.1128/jvi.71.9.6407-6415.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Williams K J, Loeb L A. Retroviral reverse transcriptases: error frequencies and mutagenesis. Curr Top Microbiol Immunol. 1992;176:165–180. doi: 10.1007/978-3-642-77011-1_11. [DOI] [PubMed] [Google Scholar]
- 188.Wilson C A, Eiden M V, Anderson W B, Lehel C, Olah Z. The dual-function hamster receptor for amphotropic murine leukemia virus (MuLV), 10A1 MuLV, and gibbon ape leukemia virus is a phosphate symporter. J Virol. 1995;69:534–537. doi: 10.1128/jvi.69.1.534-537.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Wilson C A, Farrell K B, Eiden M V. Properties of a unique form of the murine amphotropic leukemia virus receptor expressed on hamster cells. J Virol. 1994;68:7697–7703. doi: 10.1128/jvi.68.12.7697-7703.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 191.Wu L, Paxton W A, Kassam N, Ruffing N, Rottman J B, Sullivan N, Choe H, Sodroski J, Newman W, Koup R A, Mackay C R. CCR5 levels and expression pattern correlate with infectability by macrophage-tropic HIV-1, in vitro. J Exp Med. 1997;185:1681–1691. doi: 10.1084/jem.185.9.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wyatt R, Kwong P D, Desjardins E, Sweet R W, Robinson J, Hendrickson W A, Sodroski J G. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 193.Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 194.Yang Y L, Guo L, Xu S, Holland C A, Kitamura T, Hunter K, Cunningham J M. Receptors for polytropic and xenotropic mouse leukaemia viruses encoded by a single gene at Rmc1. Nat Genet. 1999;21:216–219. doi: 10.1038/6005. [DOI] [PubMed] [Google Scholar]
- 195.Yi Y, Rana S, Turner J D, Gaddis N, Collman R G. CXCR-4 is expressed by primary macrophages and supports CCR5-independent infection by dual-tropic but not T-tropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:772–777. doi: 10.1128/jvi.72.1.772-777.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Yoshimoto T, Yoshimoto E, Meruelo D. Identification of amino acid residues critical for infection with ecotropic murine leukemia retrovirus. J Virol. 1993;67:1310–1314. doi: 10.1128/jvi.67.3.1310-1314.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Young J A, Bates P, Varmus H E. Isolation of a chicken gene that confers susceptibility to infection by subgroup A avian leukosis and sarcoma viruses. J Virol. 1993;67:1811–1816. doi: 10.1128/jvi.67.4.1811-1816.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Zaitseva M, Blauvelt A, Lee S, Laphan C, Klaus-Kovtun V, Mostowski H, Manischewitz J, Golding H. Expression and function of CCR5 and CXCR4 on human Langerhans cells and macrophages: implications for HIV primary infection. Nat Med. 1997;3:1369–1375. doi: 10.1038/nm1297-1369. [DOI] [PubMed] [Google Scholar]
- 199.Zhang Y, Lou B, Lal R B, Gettie A, Marx P A, Moore J P. Use of inhibitors to evaluate coreceptor usage by simian and simian/human immunodeficiency viruses and human immunodeficiency virus type 2 in primary cells. J Virol. 2000;74:6893–6910. doi: 10.1128/jvi.74.15.6893-6910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Zhao Y, Zhu L, Lee S, Li L, Chang E, Soong N W, Douer D, Anderson W F. Identification of the block in targeted retroviral-mediated gene transfer. Proc Natl Acad Sci USA. 1999;96:4005–4010. doi: 10.1073/pnas.96.7.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]