Abstract
A defining characteristic of eukaryotic cells is the possession of a nuclear envelope. Transport of macromolecules between the nuclear and cytoplasmic compartments occurs through nuclear pore complexes that span the double membrane of this envelope. The molecular basis for transport has been revealed only within the last few years. The transport mechanism lacks motors and pumps and instead operates by a process of facilitated diffusion of soluble carrier proteins, in which vectoriality is provided by compartment-specific assembly and disassembly of cargo-carrier complexes. The carriers recognize localization signals on the cargo and can bind to pore proteins. They also bind a small GTPase, Ran, whose GTP-bound form is predominantly nuclear. Ran-GTP dissociates import carriers from their cargo and promotes the assembly of export carriers with cargo. The ongoing discovery of numerous carriers, Ran-independent transport mechanisms, and cofactors highlights the complexity of the nuclear transport process. Multiple regulatory mechanisms are also being identified that control cargo-carrier interactions. Circadian rhythms, cell cycle, transcription, RNA processing, and signal transduction are all regulated at the level of nucleocytoplasmic transport. This review focuses on recent discoveries in the field, with an emphasis on the carriers and cofactors involved in transport and on possible mechanisms for movement through the nuclear pores.
The defining characteristic of eukaryotic cells is the segregation of nucleic acid synthesis and processing into a membrane-bound compartment, the nucleus. The nuclear envelope is a double membrane contiguous with, and presumably evolved from, the endoplasmic reticulum (ER). One might speculate that the binding of ER vesicles to chromatin during mitosis perhaps helped ensure a balanced division of the ER to the two daughter cells and that during early evolution the vesicles began to fuse on and therefore enclose the surface of the chromatin. However, complete enclosure would be lethal, and proteins were therefore needed to maintain holes in the envelope, holes of sufficient size to permit the passage of ribosomes and mRNA molecules. Although these holes may originally have been no more than open passageways to allow diffusion, they ultimately became the nuclear pore complexes (NPCs)—arguably the largest protein structures in the eukaryotic cell (200).
Compartmentalization provides opportunity for regulation, and the import and export of proteins and nucleic acids through the pores of modern eukaryotes is tightly controlled. This nucleocytoplasmic traffic has also become functionally and mechanistically diversified, serving not only to permit operation of the basal replication, transcription, and processing machinery but also to regulate the cell cycle, transcriptional activation and repression, circadian rhythms, and a host of other processes.
Diversification of nucleocytoplasmic transport has evolved by a different route from that taken for the translocation of ions and small molecules across membranes. For these species, cells have evolved numerous channels and carriers that span the membrane bilayer. However, the NPCs appear on the basis of electron microscopy, yeast genetics, and biochemistry to be identical and to permit passage of all of the various types of cargo that need to cross between the nuclear and cytoplasmic compartments (55, 215, 280).
The selectivity of nucleocytoplasmic transport rests in part on the structure of the NPCs but additionally depends on a variety of soluble carriers. These carrier proteins are designed to recognize cargo destined for translocation and to shuttle back and forth efficiently through the nuclear pores. Rapid progress has been made within the past few years in the identification and analysis of these soluble carriers and of accessory factors that aid their work, and this review will focus mainly on this aspect of nucleocytoplasmic transport. Our understanding of the structure and function of the NPCs has been hindered by their great complexity, but is beginning to yield to new imaging methods and yeast genetics (47, 60, 215, 248, 280). These advances, together with the development of technologies to observe the movement of single protein molecules through NPCs (118), will ultimately allow a detailed molecular understanding of the process of nucleocytoplasmic transport.
GATEWAY TO THE NUCLEUS: THE NUCLEAR PORE COMPLEX
During interphase, the only means of access to the nucleoplasm is through the NPCs. These are octagonally symmetric structures composed of a cylindrical channel that is attached to an outer rim by eight spokes (for reviews, see references 48, 58, 183, 215, 247, and 270). Viewed in cross section, the central channel, spokes, and rim are sandwiched between two rings. From the ring that faces the cytoplasm there extend eight fibrils, while from the nuclear ring extend eight long filaments that are linked at their extremities to another ring, so as to form a structure like a basket or fishing net. The basket ring appears to be able to open and close, like an iris, in response to changes in calcium ion concentration, but the function of this conformational change is not known (248).
The yeast NPC is rather smaller than that found in higher eukaryotes, but, weighing in at 30 to 60 MDa, it is still about 15 times the size of a ribosome. It was surprising, therefore, when exhaustive analysis of purified yeast NPCs revealed an upper limit of only about 30 distinct protein components (60, 215). Ribosomes, by comparison, contain ∼75 different proteins. Localization of the NPC components by electron microscopy also unexpectedly revealed that most are symmetrically localized on the nucleoplasmic and cytoplasmic faces of the NPC. Only the peripheral, filamentous structures appear to have distinct compositions. The small number of components may be related to the high degree of symmetry of the NPC and to the fact that the average polypeptide size (∼100 kDa) is much larger than that of ribosomal proteins. NPC symmetry may be fundamental to the transport mechanism, a topic discussed below.
The proteins of the NPC are called nucleoporins. Their synthesis, association into subcomplexes and assembly into NPCs, structural features, and functions are all the focus of intensive studies but will not be reviewed exhaustively here. Briefly, the nucleoporins can be divided into three groups: those that span the nuclear membrane, nonmembrane proteins that contain multiple repeats of a Phe-Gly motif, and nonmembrane proteins that do not. Associated with the NPC there are also shared-function, peripheral, or other proteins that may be transport cofactors or assembly factors rather than bona fide structural components (60, 215, 249, 280).
The number of NPCs per nucleus varies widely with the organism, cell type, and growth conditions, but mammalian cells typically contain ∼3,000 to 5,000 NPCs (153, 216). The structure is consistent with there being a single channel per NPC through which all transport proceeds, and passive diffusion of small molecules is consistent with a channel of about 9 nm in diameter and 45 nm long (117). Only proteins with a diameter of less than ∼5 nm, however, can diffuse across the nuclear boundary within a period of a few minutes. However, ribosomal subunits that are 25 nm in diameter, giant ribonuclear protein particles such as the Balbiani ring, and 26-nm gold particles can all be accommodated by the pores (61). The fundamental question, then, in understanding NPC function is how such a remarkable degree of flexibility and discrimination is provided. The answer must account for the lack of any motor proteins, ATPases, or GTPases among the components of the NPC.
SHIPPING DOCUMENTS: NLSS AND NESS
The concept of specific nuclear transport signals arose from the observation that certain proteins, larger than the passive diffusion limit (∼50 kDa) for the NPC, can accumulate within the nucleus. A classic series of experiments on nucleoplasmin provided the first proof of the existence of such signals, and the first signal sequence for nuclear import was identified in the simian virus 40 (SV40) large-T antigen (53, 112). This sequence, PKKKRK, and that found in nucleoplasmin, KRPAATKKAGQAKKKKLD (210), are the prototypes for the monopartite and bipartite nuclear-localization signals (NLS) now known to be present in many—probably thousands—of different proteins (Table 1). A useful Web server (PSORTII) for identifying potential NLS sequences in protein sequences is available at http://psort.nibb.ac.jp:8800/.
TABLE 1.
Nucleocytoplasmic signal sequences
| Signal | Proposed consensus sequencesa | Source or reference |
|---|---|---|
| Classical monopartite NLS | B4, P(B3x), Pxx(B3x), B3(H/P) | PSORT II server |
| Classical bipartite NLS | BBx10(B3x2) | PSORT II server |
| M9 NLS | (Y/F/W)x2JxSxZG(P/K)(M/L/V)(K/R) | 24 |
| Viral NLS | RxxRRx1,2RBR | 182, 258 |
| Ribosomal L23a NLS | VHSHKKKKIRTSPTFRRPKTLRLRRQPKYRRKSAPRRNK | 105 |
| Leucine-rich NES | Lx2,3(F/I/L/V/M)x1,2,3Lx(I/V/L) | 25 |
| Unusual NESs | IxxxIxxLxT, WxKIxLxP | 54, 123 |
B, basic residue (K or R); J, hydrophilic residue; Z, hydrophobic residue; x, any residue; subscript numbers show number of residues; letters in parentheses can be in any order; letters separated by a slash are alternate permitted residues.
One could imagine two likely mechanisms by which such signals might operate. Either they would bind directly to components of the NPC and be translocated through the pores like passengers on a moving walkway, or they could be recognized by soluble receptors that would carry them through the pores, like cargo carried on a truck. The development of an in vitro assay for nuclear import, using digitonin-permeabilized cells, provided the critical advance that demonstrated the second mechanism to be correct (3). Accumulation of a fluorescently tagged protein containing an NLS into the nuclei of permeabilized cells did not occur in the absence of cytosol. Fractionation of the cytosol led to the discovery of four soluble transport factors that could, together with ATP, reconstitute nuclear import (2, 160). One of these factors, importinα (also called karyopherinα, Kapα, and PTAC58), was shown to bind directly to an NLS, confirming the soluble receptor-carrier hypothesis (1, 2, 81, 164).
The other transport factors that were isolated from cytosol using the in vitro assay were Ran, a small GTPase (also called TC4) (154, 160), NTF2 (also called p10 or pp15) (161, 189), and importinβ (also called karyopherinβ, p97, and PTAC97) (35, 78, 198). The functions of each of these factors will be discussed in more detail below. For the present, it is sufficient to note that importinα itself does not interact with the NPC but instead functions as an adapter that binds to importinβ and that importinβ is the carrier that allows translocation through the pore.
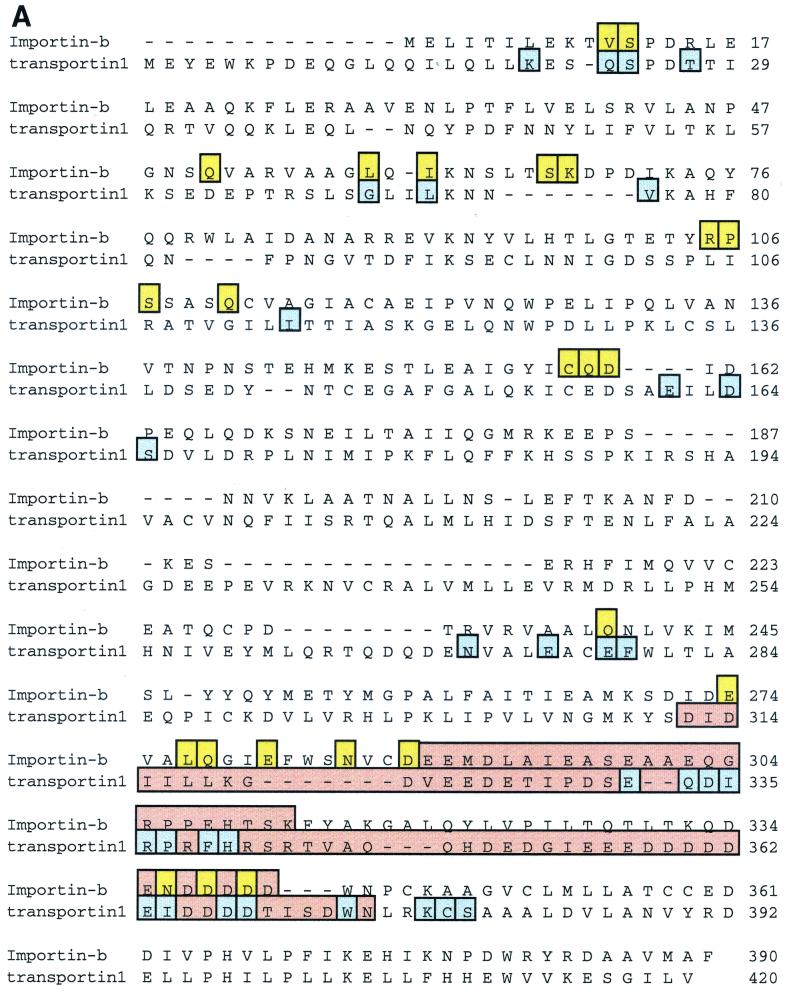
These studies revolutionized the field, but it became apparent that many nuclear proteins do not contain classical mono- or bipartite NLSs and must either use alternate entry mechanisms or piggyback on cargo that does contain a classical NLS. One example is hnRNPA1. This abundant protein shuttles efficiently between the nuclear and cytoplasmic compartments, but the sequence responsible for shuttling, called M9, is glycine and asparagine rich and does not bind to importinα (Table 1) (155). Rather, M9 was shown to be recognized by a novel protein, named transportin (karyopherinβ2), that is related to importinβ (196). A similar protein, Kap104, was also found in budding yeast (5) (Table 1). Each protein from this family that has been studied to date functions as a carrier in nucleocytoplasmic transport.
A priori, one might have expected that the carriers would be capable of transporting cargo in both directions through the pores, but to date this property has been demonstrated for only two family members, the yeast Kap142/Msn5 and mammalian importin13(157, 285). All of the other carriers appear to function exclusively either as importins or as exportins. Kap142 can import the yeast trimeric replication protein A (RPA) to the nucleus. As an exportin, it carries various proteins—Pho4, Mig1, Far1, and Ste5—to the cytoplasm (52, 110, 150, 236). It is unusual in that it exports only phosphorylated cargoes, but the consensus sequence context that allows recognition of specific phosphoserines by this carrier has not yet been defined.
Crm1 (exportin1, or Xpo1 in budding yeast) was the first export carrier to be identified (69, 72, 179, 241). It recognizes a short motif rich in leucine or related hydrophobic residues, which is found in the protein kinase A inhibitor PKI, in the human immunodeficiency virus (HIV) protein Rev, in RanBP1, and in dozens of other proteins (65, 73, 166, 208, 269). LxxxLxxLxL is the prototypical nuclear export signal (NES) sequence (Table 1), but other hydrophobic residues can substitute for several of the Leu residues, the number of intervening residues is somewhat variable, and prolines situated between the hydrophobic residues disrupt function (25). There are also efficient NESs that do not conform even to this rather vague consensus. The NES in the NFAT transcription factor, IVAAINALTT, is one example (123). Additionally, motifs that exactly match the PKI/Rev pattern sometimes have no export function, for instance in Ste5 (unpublished observation), perhaps because they are not exposed on the protein surface. The unambiguous definition of an NES is confounded by the high frequency of hydrophobic residues in protein sequences.
Many other import or export signal sequences must exist, but so far they have not been carefully dissected and are often much larger than the classical NLS and NES motifs. The import signal for uridine-rich small nuclear ribonucleoproteins (U snRNPs) comprises both the m3G cap on the RNA of the U snRNP and sequences within the Sm core protein of the RNP, which are recognized by an adapter protein called snurportin that, like importinα, binds to importinβ (100, 181). Similarly, the export signal for snurportin, which is recognized by Crm1, is a large domain that encompasses most of the protein rather than a short hydrophobic sequence (185).
Most known import signals, except for M9, do have a basic character, however. For example, the NLSs present in ribosomal proteins such as L25 and L23a are highly basic (105, 224). Some viral cargoes bind directly to importinβ rather than through an importinα adapter, and they also possess highly basic, arginine-rich NLSs (88, 182, 258). Additionally, certain cargoes with short basic sequences are able to bind to a distinct site on importinα and be transported simultaneously with proteins possessing a monopartite NLS (K. Plafker and I. G. Macara, unpublished data). This common theme may reflect the evolutionary relationship between the importin family and the signals they recognize. On the other hand, it is just as likely to be a quirk that reflects the biased nature of the very small group of NLSs that have been characterized to date.
To add to this complexity, there are transport factors that are unrelated to the importin family and proteins that can translocate through the NPCs in the absence of other soluble factors. In the first category are the yeast protein Mex67 and its mammalian homologue TAP, which most probably bind directly to mRNA sequences (84, 101, 114); cytoplasmic calreticulin is an export carrier for steroid receptors (96) (see next section). In the second category there are proteins such as hnRNPK and β-catenin, both of which most probably interact with the NPCs directly (59, 156, 282).
Clearly, we have not yet exhausted the repertoire of nuclear import and export signals. Nor have we identified all of the diverse mechanisms by which these signals can be regulated. Numerous proteins possess classical NLSs that are exposed or hidden or show altered import rates when nearby serine or threonine residues are phosphorylated (for a review, see reference 109). For example, phosphorylation of Pho4 not only permits recognition by Msn5 but also inhibits binding to the importin for this protein, Pse1 (Kap121) (110, 127). Phosphorylation by the PKB/Akt protein kinase within an NLS in the forkhead transcription factor, AFX, blocks nuclear import and results in a rapid shift of the protein to the cytoplasm (29, 30, 253). Masking of the NLS in NF-κB by IκB maintains the NF-κB in the cytoplasm until IκB is phosphorylated and degraded. This story is complicated, however, by the discovery that IκB itself shuttles in and out of the nucleus (219, 261). Another example of masking involves the 14-3-3 family of proteins, which recognize phosphorylated serine residues within the sequence context RSxS∗xP (where the asterisk indicates the phosphorylated residue). Cdc25, a protein tyrosine phosphatase that regulates the cell cycle, becomes associated with 14-3-3 when phosphorylated by a checkpoint kinase. Cdc25 possesses both an NES and a dominant NLS, but when bound to 14-3-3 the NLS is masked and Cdc25 accumulates in the cytoplasm (167). A similar mechanism may be involved in the cytoplasmic retention of forkhead transcription factors.
NESs can also be masked. One example is provided by telomerase reverse transcriptase, to which 14-3-3 can bind in a non-phosphoserine-dependent manner and block an NES (231). Another case is the transcription factor NFAT, which, in the presence of calcium ions, interacts with calcineurin. NFAT contains both an NES and one or two NLSs (depending on the isoform) and shuttles constitutively, so that masking of the NES by calcineurin permits nuclear accumulation (288). A twist on this story is that NFAT can also be negatively regulated by protein kinase A, which creates 14-3-3 binding sites on NFAT that may mask the NLS (167).
A reverse of this type of situation occurs when NLS or NES function is not masked but requires the formation of a complex. As an illustration, the import of a fission yeast protein called Mei2, which is necessary for meiosis, has to associate with a small RNA (mei-RNA) before it can accumulate in the nucleus (278). A different mechanism controls the localization of yAP1, a transcription factor that responds to oxidative stress. Cysteine residues within the NES of yAP1 are sensitive to the redox state of the cell, and their oxidation blocks interaction with the exportin, Crm1 (279).
Only during the last few years has the extraordinary range of processes that are controlled at the level of nucleocytoplasmic transport been recognized. Everything from apoptosis to circadian rhythms and from signal transduction to the cell cycle is regulated, at least in part, by switching NLSs and NESs on or off. Nuclear transport is the one common link between these diverse processes.
CARGO CARRIERS: KARYOPHERINS
Most of the proteins that carry cargo through the NPCs are members of the importinβ/karyopherinβ family (Table 2). For consistency, we will refer to this family as the karyopherins, defining those that are involved in nuclear import as importins and those involved in export as exportins. Other, structurally unrelated carriers include TAP/Mex67 and calreticulin.
TABLE 2.
Ran binding proteins
| Name (mammalian) | Name (S. cerevisiae) | Affinity for Ran | GTP or GDP | Function | Reference(s) |
|---|---|---|---|---|---|
| Regulators | |||||
| RanGEF, RCC1 | Prp20 | 900 nM | Both | Guanine nucleotide exchange factor | 121, 122 |
| RanGAP | RNA1 | 430 nM | GTP | GTPase-activating protein | 120 |
| RanBP1 | Yrb1 | 1 nM | GTP | Coactivator of RanGAP | 132 |
| RanBP2 | 1 nM | GTP | Coactivator of RanGAP, binds SUMO-modified RanGAP | 277, 283 | |
| RanBP3 | Yrb2 | >5 μM | GTP | Cofactor for Crm1/Xpo1: activates RanGEF | 142, 256 |
| Nup50/NPAP60 | Nup2 | Low | GTP | Binds importinα and other karyopherins; cofactor? | 27, 85, 98, 240 |
| NTF2 | Ntf2 | 25 nM | GDP | Import carrier for Ran | 187, 206, 238 |
| NXT1, p15 | 12 nM | GTP | Cofactor for TAP (NXF1) and Crm1 | 23, 114, 252 | |
| G3BP1, G3BP2a, G3BP2b | Low | ? | ? | 114, 186 | |
| Mog1 | Mog1 | 200 nM | (GTP) | ? | 177, 244 |
| Cargo carriers | |||||
| Importinβ | Kap95 | 0/8 nM | GTP | Imports importinα; importinβ also imports XRIPα and snurportin adapters; ribosomal proteins, HIV Rev, HIV Tat, and HIV Rex proteins; Binds importin7, and RanBP8 | 40, 104, 105, 107, 182, 258, 263 |
| Transportin1, Kapβ2 | Kap104 | 1 nM | GTP | Imports mRNA binding proteins, ribosomal proteins | 5, 39, 105, 196 |
| Transportin SR | Mtr10 | 200 nM | GTP | Imports mRNA binding proteins (phospho-RS domains): Mtr10 imports Np13 | 115, 192, 233 |
| Hmtr10 | Mtr10 | ? | GTP | ? | 139, 192, 233 |
| Kap114 | ? | GTP | Imports TBP, histones H2A and H2B | 163, 165, 193 | |
| Yrb4, Kap123 | 2–3 nM | GTP | Imports ribosomal proteins | 227 | |
| Importin7 | 25 nM | GTP | Imports ribosomal proteins | 105 | |
| Importin11 | Lph2, Kap121 | >100 nM | GTP | Imports UbcM2 | 195 |
| Importin5, Kapβ3 | Pse1, Kap121 | ? | GTP | Imports ribosomal proteins, Pho4, and Spo12 | 49, 105, 281 |
| Nmd5, Kap119 | ? | GTP | Imports Hog1 and TFIIS | 6, 64 | |
| Sxm1, Kap108 | ? | GTP | Imports Lhp1 and ribosomal proteins | 213, 229 | |
| Pdr6, Kap122 | ? | GTP | TFIIA subunits | 257 | |
| RanBP16 | 200 nM | GTP | ? | 136 | |
| RanBP8, etc. | ? | GTP | ??? | 76 | |
| Exportin5 | Msn5, Kap142 | 40 nM | GTP | Kap142 exports phosphorylated proteins; imports RPA | 52, 110, 285 |
| Crm1 | Xpo1, exportin1 | 5–100 nM (cargo specific) | GTP | Export carrier for leucine-rich NES proteins, snurportin 1 | 69, 72, 129, 179, 241 |
| Cas | Cse1 | 1 nM | GTP | Exports importinα | 134, 135, 239 |
| Exportin t | Los1 | ? | GTP | Exports tRNAs | 8, 87, 138 |
| Exportin4 | 1.5–40 nM (± cargo) | GTP | Exports eIF5A | 144 |
One feature common to the karyopherins is their ability to bind to nucleoporins. Where tested, most appear able to interact with regions of these proteins that are rich in FxFG repeats, but there are also distinct specificities (47, 70, 102, 199, 203, 228, 276). Another common feature is that karyopherins can form a complex with the GTP-bound state of the Ran GTPase. In some cases this interaction is of low affinity and is hard to detect; in others the Kd for Ran is in the low nanomolar range (76, 195, 233) (Table 2). The purpose of the interaction is to regulate the binding of cargo. Ran is the most abundant member of the Ras superfamily of GTPases, constituting about 0.4% of the total cell protein (21). Like other members of the superfamily, Ran functions as a molecular switch and undergoes a conformational change between the GDP- and GTP-bound states. Conversion between these forms is regulated by a guanine nucleotide exchange factor (RanGEF) and a GTPase-activating protein (RanGAP) (14, 18, 20). Another feature, common to most members of the superfamily but lacking in Ran, is a posttranslational modification at the C terminus by a prenyl group. Instead, Ran possesses an acidic C-terminal domain that is integral to its switch function (209).
A key to understanding the role of Ran in nucleocytoplasmic transport was the discovery that RanGEF is restricted to the nucleus and RanGAP is localized to the cytoplasmic compartment(99, 176). This asymmetry creates a steep Ran-GTP gradient across the NPC, and it is this gradient that provides the vectorial information for nuclear import and export (80). As will be discussed in detail below, Ran-GTP triggers the disassembly of import carriers from their cargoes but promotes the assembly of exportin-cargo complexes.
We will discuss transportin1 (Kapβ2) first, as a relatively simple example of an importin; then importinβ, which can use various adapters; and then Crm1, as an example of an exportin.
Importins
Transportin1 (Kapβ2).
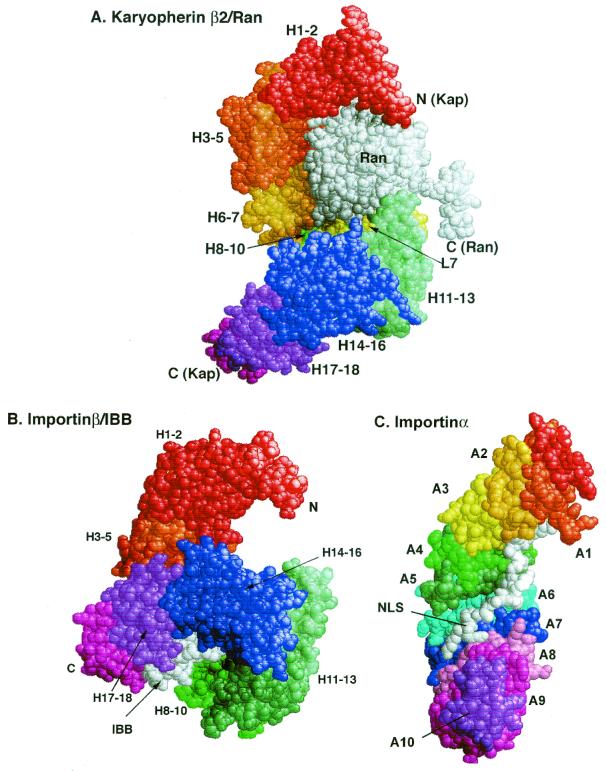
The crystal structure of a complex between transportin1 and Ran:GppNHp has been solved. (39). The GppNHp is a slowly hydrolyzing analog of GTP. Transportin1 has a mass of about 101 kDa and is entirely α-helical. It has a remarkable structure, consisting of 18 HEAT/Armadillo (Arm) domains that are twisted in a superhelix like an S in which the bottom arch has been rotated forward, out of the plane of the page, by 90° (Fig. 1A). (HEAT domains are named after the proteins in which similar sequences were first identified: huntingtin, elongation factor 3, the A subunit of protein phosphatase 2A, and the Tor1 kinase. They also resemble the HEAT/Arm repeats of β-catenin and importinα.) Each HEAT domain is made up of two α-helices (A and B) that are joined by a sharp turn to form a hairpin structure. Individual HEAT domains are linked either by loops or by additional short helices. The B helices form the concave surfaces of the arches. Ran is nestled in the N-terminal arch. The binding site for the M9 NLS sequence has been roughly mapped to the C-terminal arch.
FIG. 1.
Structures of carrier complexes with Ran (A), the N-terminal domain of importinα (IBB domain) (B), and a bipartite NLS (C). H1 to H18 identify the HEAT motifs of transportin1 and importinβ. L7 is the loop within HEAT motif7. Ran (bound to GppNHp, a nonhydrolyzable analogue of GTP), the IBB domain, and the NLS are shown in white. Note how the HEAT motifs H11 to H18 are twisted up and around the IBB domain, compared to their positions in panel A (39, 263). A1 to A10 identify the ARM motifs, which are closely related to HEAT motifs. Note that the superhelical twist is in the same direction in all three structures and that the ligands all interact with the concave inner surface of the carriers.
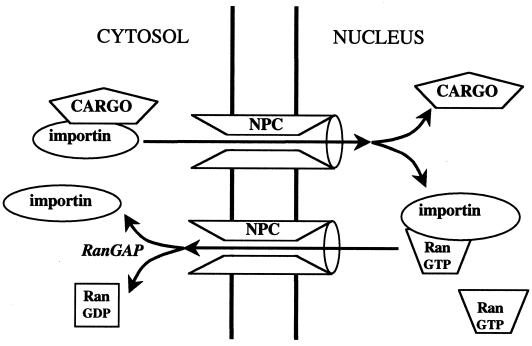
A key feature of the importins is that binding of Ran-GTP and binding of cargo are mutually exclusive, so that cargo can load onto the importin in the cytosol, where Ran-GTP concentrations are likely to be very low, and is released inside the nucleus, where they are high (Fig. 2). The affinity of transportin1 for Ran-GTP (about 1 nM) is 10,000-fold higher than that for Ran-GDP, so the switch is very efficient and must entail a conformational change in the transportin1 structure. Although the mechanism is not yet understood, it may involve a long, acidic loop in the middle of HEAT repeat 7 (Loop 7), which makes contacts with Ran and curls down over the C-terminal arch. This loop may block access to the cargo binding site. Transportin1 can also carry ribosomal proteins, such as L23a, into the nucleus, through recognition of a highly basic region of the proteins called the BIB domain, but the region of transportin1 with which it interacts has not yet been precisely mapped (105).
FIG. 2.
Mechanism of cargo import by direct interaction with an importin carrier protein. RanGTP is present at high concentrations only in the nucleus, where it disassembles the cargo-importin complex. The importin-RanGTP complex returns to the cytoplasm, where the GTP is hydrolyzed, releasing the RanGDP from the importin.
Importinβ.
Importinβ is a more complex type of carrier, because it can carry cargo either directly (at distinct sites) or via an adapter protein (Fig. 2 and 3). The best known adapter is importinα, a 55-kDa protein that binds to classical, basic NLS sequences. There is only one gene encoding an importinα in budding yeast, but six isoforms, which differ from one another mainly at their C termini, are known in mammals (151). Although each recognizes the monopartite SV40 NLS with similar affinity, they differ in their affinities for other NLSs (125, 126, 159, 168, 260, 268). The structural basis for these distinctions is not yet known. In addition, the C termini of the importinα isoforms can form binding sites for distinct cargo proteins, such as Stat1 and Nup50, which may be carried into the nucleus simultaneously with other cargo containing monopartite NLSs (232; K. Plafker and I. G. Macara, submitted for publication). Other adapters for importinβ include snurportin, which binds to m7G-capped Usn RNAs; the frog protein XRIPα, which binds to RPA; and importin-7, which facilitates histone H1 import (81, 104, 107, 182, 258, 259, 266). XRIPα is of particular interest as an example of divergent evolution, because no homologue of this protein exists in budding yeast and because its cargo, RPA, is instead imported by the karyopherin Kap142/Msn5 (285).
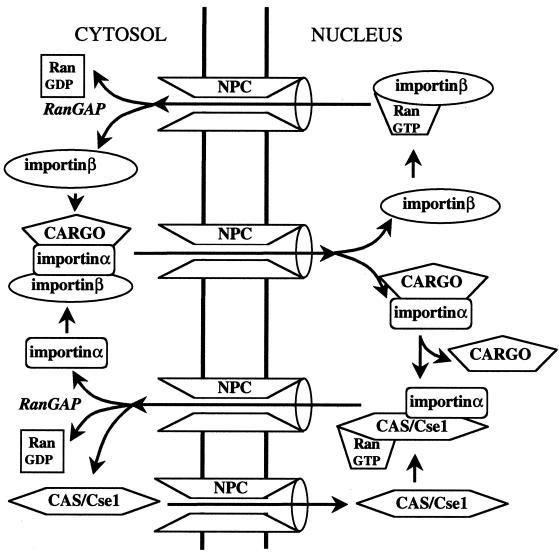
FIG. 3.
Mechanism of cargo import by the importinα-importinβ pathway. In this mechanism, the cargo binds to an adapter, importinα, rather than directly to the carrier. The cargo-importinα-importinβ complex is disassembled in the nucleus by RanGTP. Importinβ returns to the cytosol, as shown in Fig. 2. The importinα requires a carrier for export, called CAS (CseI in budding yeast). Assembly of the export complex requires RanGTP. The ternary importinα-CAS-RanGTP complex is disassembled in the cytoplasm by hydrolysis of the GTP.
The N termini of the importinα proteins are highly basic and comprise a 40-amino-acid residue domain (named IBB) that is both necessary and sufficient for interaction with importinβ (77, 267). The isolated IBB domain can function as cargo for nuclear transport by importinβ, and its sequence is distantly related to the NLS motifs of other cargo proteins that can bind directly to importinβ, such as Tat and Rev.
The structures of the IBB-importinβ complex (Fig. 1B), and of a fragment of importinβ with Ran (40, 263) have been solved. We can therefore just begin to glimpse the dynamics of the switch mechanism that controls the carrier-cargo interaction. Despite the low degree of sequence identity between transportin1 and importinβ (∼14%), their overall structures are quite similar. Both are composed almost entirely of HEAT/Arm repeats twisted into right-handed superhelices, so that the B helices of the repeats face inwards (Fig. 1). Both bind Ran primarily through the B helices of the first three HEAT repeats and through an acidic loop within HEAT repeat 7 (transportin1) or 8 (importinβ). Also, both bury similar, extensive regions of the Ran surface in the interface of the complexes, consistent with the 0.5 to 1.0 nM Kd values for the interactions. Yet, remarkably, the amino acid contacts between Ran and these carrier proteins are not conserved. Of the 31 contacts in transportin1 made to the Ran polypeptide, only 7 are similar in location and type to residues within importinβ (Fig. 4A). Even more remarkable is the fact that the contact residues on Ran itself differ between the two carriers (Fig. 4B), although similar regions of the Ran surface are involved. Of the 21 contacts on Ran made by importinβ, only half are shared by transportin1 — primarily those in the Switch 2 and basic patch areas of Ran.
FIG. 4.
Comparison of the Ran-GTP interacting surfaces between importin-β and transportin1. (A) Alignment of the Ran binding domains of importinβ and transportin1. Residues in importinβ that bind Ran are shown in yellow. Residues in transportin1 that bind Ran are shown in blue. Loop7 is indicated in pink. (B) Amino acid sequence of Ran showing the Switch 1 and Switch 2 regions that undergo a conformational switch between the GTP and GDP binding states, and the basic patch. Residues that bind importinβ are shown in yellow; residues that bind transportin1 are shown in blue (39, 263).
This heterogeneity underscores the difficulty in identifying new members of the karyopherin superfamily from the databases. The degree of sequence similarity between the putative Ran binding domains of the entire superfamily is only about 10%, and there are no invariant residues that can be identified as a core Ran binding motif (76, 276). This situation contrasts dramatically with the highly conserved Ran binding domains of proteins such as RanBP1 and RanBP2/Nup358 or with other domains such as the SH2, SH3, and DBL domains. One interpretation is that the differences are functional. The importins may have diverged so as to provide different Ran binding affinities (which would change the ability of the importin to drive accumulation of cargo against a concentration gradient) (Table 2). Alternatively, the extensive areas of surface that are buried at the Ran/importin interface may have permitted some jiggling or rotation of the proteins with respect to one another during evolution, without a catastrophic loss of binding. Clearly, it will be instructive to see other structures from the karyopherin family, and particularly those of the exportins, which bind cargo more tightly, rather than more weakly, in the presence of Ran-GTP.
Importinα.
The structure of the adapter protein, importinα, has also been solved, and it bears a striking resemblance to that of importinβ and transportin1 (45, 124). The NLS binding domain consists of 10 Armadillo motifs stacked into a right-handed superhelix, and repeats 2 to 5 can be superimposed well onto HEAT repeats 9 to 12 of importinβ (Fig. 1C). This similarity supports the proposal that all HEAT and Arm repeat proteins are related evolutionarily. The NLS sits in the concave inner groove of importinα, which is lined with conserved tryptophan and asparagine residues. There are two binding pockets, a major one formed by Arm 2 to 4, occupied by monopartite NLSs, and a second, closer to the C terminus (Arm 7 to 8), that can be occupied by the N-terminal portion of bipartite NLSs (44).
The N terminus of importinα, which contains the IBB domain, is not structured in the isolated protein, although a short stretch of basic residues contacts the major NLS binding pocket and may function as an autoinhibitor of cargo loading (45, 124). The crystal structure of the IBB domain in complex with importinβ highlights several interesting points (Fig. 1B). (i) The IBB domain becomes structured in the complex, with residues 24 to 51 folding into an α-helix. (ii) The major contacts on importinβ are, as predicted from deletion mapping, in the C-terminal half of the protein. (iii) The acidic loop that makes contacts with the basic patch on Ran also forms direct contacts with the IBB domain, consistent with the fact that the binding of these two proteins to importinβ is mutually exclusive and with the idea that interactions with this loop may trigger a conformational change in importinβ. (iv) The way in which the importinβ is twisted snail-like around the IBB domain suggests that it undergoes a substantial conformational change on binding importinα. (v) Finally, the interactions of importinβ with the IBB domain are remarkably similar to those between importinα and its NLS cargo — in both cases, acidic residues on the “receptor” form ionic bonds with basic residues on the “ligand,” whose side chains are constrained by other, hydrophobic residues. This last point supports the proposal that the entire superfamily of “αs” and “βs” be referred to by a single name, such as “karyopherins.”
Importinβ can bind other adapters, as discussed above, and also binds directly to specific cargoes such as the HIV-1 proteins, Rev and Tat. Several of these proteins contain Arg-rich motifs that may form an IBB-like α-helix, and the isolated IBB domain is itself efficiently imported into nuclei by importinβ (77, 267). Why, then, did NLSs evolve that bind to importinα rather than directly to importinβ? One factor may be that the monopartite NLS is substantially shorter than the IBB domain and is unstructured, which may have broadened the utility of the import pathway. Additionally, the use of an adapter can in principle increase the concentration gradient against which cargo can be imported. As will be discussed below, the fact that importinα is exported by another carrier, CAS, in a Ran-dependent fashion, means that the overall transport cycle for importinα-mediated NLS-cargo import consumes two GTP molecules rather than one.
Other importins.
There are 10 known members of the yeast karyopherin family that function as importins (276). An 11th, Lph2, remains to be characterized but probably also functions in nuclear import, because the mammalian ortholog is importin-11, which mediates the nuclear accumulation of a ubiquitin ligase E2 subunit, UbcM2 (195). The number of mammalian importins is likely to be at least double that of yeast importins (79). Remarkably, many of the yeast importins are not essential for survival, although their deletion sometimes leads to growth defects. Why is this? First, there are many types of import cargo whose size is below the free diffusion limit for the NPC. Examples are ribosomal proteins and histones. Free diffusion of such proteins will be slow but may be sufficient to maintain life. Second, there is redundancy of function between importins, which reduces the reliance on any single carrier. For example, Kap123 (Yrb4) and Kap121 (Pse1) both import ribosomal proteins and Kap121 and Kap123 can import histones H2A/H2B (165, 191, 217, 227). In mammalian cells, at least one ribosomal protein, rpL23a, can also be imported by multiple carriers, including importinβ and transportin (105). The purpose underlying such redundancy is unclear but may be related to the sheer volume of proteins that need to be imported in order to build ribosomes and maintain protein synthesis at optimal levels.
Many unresolved issues relating to nuclear import will be solved by further structure determinations and by identification of more cargoes and NLSs. There are probably over 2,000 proteins transported into and/or out of the yeast nucleus, for example, and the carriers and NLSs are known for only a small minority of cases. We need to know the molecular bases for the selectivity of these cargoes, for redundancy among the importins, and for the multiplicity of distinct cargoes that can bind to individual importins. There may also be cofactors for specific importins that have yet to be identified, and additional pathways may exist that function independently of Ran.
Exportins
To date, no structures of an exportin have been solved. The best-studied exportins are Crm1/Xpo1, which recognizes leucine-rich NESs, and Tap-1/Mex67, which is required for mRNA export and which will be discussed later. A third, unrelated factor is calreticulin, which can facilitate the export of steroid receptors, in addition to functioning as a chaperone within the ER.
Crm1/Xpo1.
Isolated Crm1 has only a very low affinity for Ran-GTP and a similarly low affinity for most NES cargoes. Together, however, they can form a relatively stable ternary complex (9, 10, 69). This complex can exit the nucleus through the NPCs and is dissociated by hydrolysis of the Ran-GTP (Fig. 5). The empty Crm1 then returns to the nucleus. Studies of Crm1-mediated export were helped enormously by the discovery that an antifungal agent, leptomycinB, is a highly specific and potent inhibitor of Crm1 function (69, 72, 131, 179, 274). LeptomycinB reacts irreversibly with a Cys residue (Cys 529) near or within the cargo binding domain of the protein (130). Interestingly, this Cys is lacking in the homologous gene product from budding yeast (Xpo1), which, consequently, is resistant to the agent.
FIG. 5.
Mechanism of cargo export. Cargo binds weakly to the export carrier in the absence of RanGTP but tightly in its presence. The ternary cargo-exportin-RanGTP complex is disassembled in the cytoplasm by GTP hydrolysis on Ran. The exportin is presumed to return empty to the nucleus.
Crm1 exports a wide variety of cargoes, and most possess their own NES. However, at least one major cargo—the 60S ribosomal subunit—does not bind directly to Crm1 but (at least in budding yeast) uses an adapter protein, Nmd3 (74, 94). Nmd3 possesses a classical hydrophobic NES and associates with ribosomal protein Rpl10. Why Rpl10 is not exported before incorporation into the 60S subunit, what prevents the export of immature 60S subunits, and whether other cargoes use adapters are questions that remain to be answered. Another yeast karyopherin, Lph2/Kap120, has also been implicated indirectly in 60S ribosomal subunit export (242), but it may be required for the import of specific ribosomal proteins that are required for maturation and export of the 60S subunit, rather than for export. This idea is consistent with our recent finding that the related mammalian karyopherin, importin-11, specifically imports ribosomal protein L12 (Rpl12) (Plafker and Macara, submitted). Rpl12 is associated with the ribosomal substructure referred to as the ribosomal stalk and is probably added late in 60S subunit assembly.
Cofactors.
There are several problems with the simple model that has been proposed for Ran-dependent NES cargo loading and disassembly. First, the concentration of Ran-GTP in the nucleus is calculated to be in the 10 to 15 μM range, so that a significant amount of Crm1 may be bound to Ran-GTP even in the absence of cargo. This Crm1/Ran-GTP complex could move through the NPCs to the cytosol, where the Ran-GTP would be hydrolyzed. Such a futile cycling of Crm1 would deplete the Ran gradient. Second, the affinities of NESs for Crm1 are often in the 100 nM to 1.0 μM range even in the presence of Ran-GTP (with the exception of snurportin, which has a much higher affinity). This can present a problem if the nuclear concentrations of these cargoes are low. Futile cycling of empty carrier is reduced by the low affinity of Crm1 for nucleoporins in the absence of Ran-GTP (67, 116). In addition, Crm1 recently has been found to use cofactors, namely, RanBP3, Nxt1, RanBP1, and possibly eIF-5A, that together potentiate NES binding, help Crm1 that is loaded with cargo to bind to nucleoporins in the NPC, and stimulate cargo unloading in the cytoplasm (22, 95, 142). RanBP3 can associate directly with Crm1, and the complex possesses an enhanced affinity for Ran-GTP and NES-cargo (142). In an in vitro assay using a limiting Crm1 concentration, RanBP3 stimulates export. Additionally, it inhibits the binding of unloaded Crm1 to the NPC. One potential mechanism for the effect on NPC binding is through a Ran-mediated conformational change in the “F domain” of RanBP3, which contains two nucleoporin-like FxFG motifs and which can bind constitutively to Crm1. We have proposed that in the absence of Ran, the F domain blocks the nucleoporin interaction site on Crm1, but that on binding Ran, the F domain is released, thus permitting the Crm1 to associate with the NPC. RanBP3 also stimulates the Ran exchange factor, RanGEF, and permits the formation of a Crm1-Ran-RanBP3-RCC1 complex (M. Nemergut and M. Lindsay, unpublished data). Therefore, RanBP3 may also facilitate export by directly stimulating the production of Crm1/Ran-GTP.
Nxt1 is also called p15 and was originally described as a cofactor for viral RNA export (see below) (114). It is related to NTF2, which functions in Ran-GDP import (see below). Nxt1 has been reported to bind Ran-GTP, although this is disputed (23, 114). It behaves differently from RanBP3 in Crm1-mediated export, since it has no effect on the affinity of Crm1 for NES cargo but appears to potentiate the release of Crm1-Ran-cargo complexes from the NPC. Black et al. (22) propose that Nxt1 binds directly to Crm1 and functions to deliver the export complex to a site on the cytoplasmic side of the NPC. Another cofactor, RanBP1, can then facilitate the dissociation of the complex and its release from the NPC into the cytosol (116).
So far, the function of eIF-5A appears limited to the NESs present in the retroviral Rev and Rex proteins (56, 214), and eIF-5A may not be a physiologically relevant cofactor in normal cells.
All of these cofactors can shuttle, and the movements of at least RanBP1 and RanBP3 are inhibitable by leptomycinB, suggesting that they accompany Crm1 from the nucleoplasm to the cytoplasm (95, 142). However, many questions remain about their functions. Do they all bind simultaneously to Crm1, or is there an ordered assembly and disassembly as the complex associates and translocates through the pores? Are their effects on export additive? Are there similar cofactors for other exportins or importins? Are the cofactors regulated in some way?
Other exportins.
Budding yeast possesses four known exportins in the karyopherin family, including Crm1 and Kap142/Msn5. These proteins all have mammalian homologs. At least one additional mammalian carrier, exportin4, has no yeast equivalent (144). Exportin4 can carry eIF-5A out of the nucleus. Interestingly, binding by exportin4 depends on a hypusine modification of eIF-5A. Crm1 has been reported to bind and export unmodified eIF-5A, although this is disputed (95, 144). Cse1 (equivalent to the mammalian Cas) is responsible for recycling importinα back to the cytosol (Fig. 3A) (134, 135, 239). It recognizes conserved sequences within Arm repeats 8 to 10 and must displace NLS cargo in order to bind (90). This latter property ensures that the cargo remains in the nucleus when the importinα returns to the cytosol.
As described above, Msn5 is unique among the yeast karyopherins in being an importin for at least one cargo, RPA, and being an exportin for other cargoes, which it recognizes only when they are phosphorylated. A closely related karyopherin (exportin-5) is expressed in human tissues and exports a double-stranded RNA binding protein, but cargo recognition by this carrier does not seem to depend on phosphorylation (A. M. Brownawell and I. G. Macara, unpublished data). Finally, Los1 (exportin-t) is responsible for the export of tRNAs and is the only karyopherin known to bind directly to RNA (8, 87, 138, 222). Surprisingly, Los1 is not required for viability, but no backup tRNA exporters have been identified to date, and yeast lacking LOS1 may survive by depending on passive diffusion of tRNAs through the NPCs. Los1 and exportin-t may have evolved not only to facilitate tRNA export, however, but also to provide quality control of their cargo. tRNAs undergo extensive processing, base modification, and splicing in the nucleus prior to export, and exportin-t recognizes only mature tRNA (8, 138, 143, 148). This selectivity will reduce contamination of the translation machinery in the cytoplasm by immature or nonfunctional tRNAs. Surprisingly, charging of the tRNAs with their cognate amino acids also occurs in the nucleus and is required for efficient export (83, 148, 237). However, Los1/exportin-t can bind the unloaded tRNAs, so the basis for this export specificity remains to be discovered. Possibly aminoacyl-tRNA synthetases in the nucleus, together with accessory factors such as Arc1, facilitate the loading of exportin-t (51). The translation elongation factor eEF-1α binds aminoacyl-tRNAs and is also required for efficient tRNA export (83), perhaps via an exportin-t-independent mechanism or by helping drive passive export through sequestration of the tRNAs in the cytosol. Clearly, there are issues of fundamental importance to tRNA processing and export that remain to be resolved.
OTHER CARRIERS
Tap/Mex67
Other classes of RNA, such as rRNA and mRNA, do not compete with tRNA for export when microinjected into Xenopus oocyte nuclei, indicating that they use a mechanism distinct from that used by exportin-t (106). Nor do they compete with each other. Most mRNAs are produced as large precursors that must be spliced, modified, and assembled into RNP complexes before they can be exported. Different types of mRNA recruit different RNP proteins, some of which may need to be removed prior to export while others remain bound during translocation and must be reimported from the nucleus after disassembly from the mRNA in the cytoplasm. These complications make analysis of mRNA export more difficult than that of single proteins.
One approach to simplify such analysis is to study individual retroviral genomic RNAs that are exported as unspliced or incompletely spliced species. Simian type D retroviruses contain a unique RNA sequence that functions as a constitutive transport element (CTE) (190). The CTE recruits a cellular factor called TAP that overcomes nuclear retention by the splicing machinery and drives export of the viral RNA (28, 84, 113). TAP (also named NXF1) is therefore a nuclear export factor. But is it specific for viral RNAs, or does it normally function in the export of endogenous RNAs? High concentrations of the CTE RNA can cause nuclear accumulation of mammalian endogenous mRNA, suggesting that the RNAs are competing for a common factor (190, 218). Moreover, TAP is weakly related to a yeast protein, Mex67, that is required for the exit of multiple classes of yeast mRNAs, and TAP can complement a MEX67 disruption (101, 114, 221, 230). Taken together, these data imply that TAP may play an important role in the cellular pathway for mRNA export. However, it remains to be proven that TAP is the major export carrier for endogenous mRNAs, and the recent identification of a family of TAP/NXF genes suggests that there may be complications to this story (89).
Exactly how TAP works in export remains unclear. It bears no structural resemblance to the karyopherins. Rather, it contains an unusual RNP binding domain and a leucine-rich repeat domain in its N-terminal half, unexpectedly similar to a heterodimer involved in RNA splicing, although the significance of this relationship remains unclear (141). Both domains are required for specific CTE binding. However, most cellular mRNAs do not contain the stem-loop structure found in the CTE, and CTE export is not promoted by the yeast homologue, Mex67, or by other members of the TAP family; therefore, the issue of direct mRNA binding to TAP remains to be addressed.
Mex67 binds with high affinity to a second essential mRNA export factor, called Mtr2, which appears to help dock the complex at the NPC (221, 250). Remarkably, TAP binds a functionally related factor, but one with no sequence similarity to the yeast Mtr2 (114). This small protein, p15 or NXT1, is closely allied to NTF2 but has been reported to bind Ran-GTP rather than Ran-GDP, although this property has been disputed (23, 114). RNA export assays suggest that NXT1 is an essential cofactor for TAP function, and TAP/NXT1 can partially rescue the viability of a Mex67 Mtr2 double deletion (114, 178). Surprisingly, TAP itself possesses an NTF2-like domain in the C-terminal half of the protein, which is the region involved in binding NXT1 (252). We know that NTF2 forms homodimers, so NXT1 perhaps heterodimerizes with the NTF2-like domain of TAP. NXT1 has been reported to facilitate the export of multiple classes of RNA in an in vitro export assay (178), and mutations that disrupt Ran-GTP binding are inactive. However, other studies indicate that the export of spliced mRNAs is completely independent of Ran (42).
Another factor that binds Mex67 is Yra1, a nuclear protein that can associate with RNA and possesses RNA-RNA annealing activity (251). A mouse homologue, ALY, can bind either Mex67 or TAP and partially complement a YRA1 deletion. Temperature-sensitive mutants of YRA1 are defective in mRNA export. Yra1 may recruit and facilitate the interaction of Mex67 or TAP/NXF proteins with mRNA, but it is not yet clear whether the RNA-RNA annealing function has physiological relevance.
Many other factors are involved in one way or another with RNA export. After pre-mRNA splicing, the molecules must be packaged into an exportable configuration and then unpackaged in the cytoplasm prior to translation. In yeast, Rip1, Pab1, Np13, Nab2, Hrp1, Dbp5, Gle1, and Gle2 are all required for efficient mRNA export (43). Some of these proteins (Np13, Nab2, and Hrp1) are mRNA binding proteins, others (such as Rip1) are nucleoporins, others (Gle1, Gle2, and Dbp5) bind the NPC, and one (Dbp5) is a DEAD-box protein that may be involved in unpackaging the RNA on the cytoplasmic side of the NPC. However, the mechanistic role of most of these proteins remains unresolved. To complicate matters further, recent studies suggest an unexpected linkage between phosphoinositide metabolism and mRNA export, through inositol hexakisphosphate (284). Finally, there are indications that nuclear actin may somehow influence RNA export, although the mechanism remains entirely obscure (95). Much of the confusion in the field is generated by the plethora of RNA species; the multiple transcription, processing, and splicing steps that precede and may be tightly coupled to RNA export; the large number of factors involved; and the problems associated with reconstituting in vitro export assays. It is encouraging that in the face of such complexities, so much progress has been made.
Calreticulin
Functional assays can often turn up new and quite unexpected discoveries. The development of an in vitro nuclear protein export assay (97) led recently to the isolation of a new factor that can promote the export both of NES cargo and of steroid receptors (96). This factor was identified as calreticulin, a protein commonly thought to reside exclusively in the ER. Clearly, however, preconceptions can be misleading, and there is evidence of a distinct, soluble pool of calreticulin in cells. This protein can bind directly to the glucocorticoid receptor and to other members of the nuclear receptor superfamily. The glucocorticoid receptor is cytoplasmic in the absence of its ligand (cortisol), but binding of the ligand exposes at least two classical NLSs, which permit its accumulation in the nucleus (208, 223). There it activates gene expression. On withdrawal of ligand, the receptor slowly returns to the cytoplasm by a mechanism that most investigators find to be independent of Crm1 (145). Knockout mice that lack calreticulin accumulate the glucocorticoid receptors in their nuclei constitutively, and the reintroduction of calreticulin into cells derived from these mice rescues the ability of the receptor to recycle back to the cytoplasm. Most interestingly, the binding site for calreticulin on these receptors lies within the DNA binding domain, and calreticulin inhibits association with the promoter elements of receptor-responsive genes (50). Therefore, calreticulin may help switch off gene expression after ligand dissociation both by blocking the rebinding to DNA and by promoting export from the nucleus. However, we do not know if calreticulin is a true carrier or is instead a chaperone that promotes the loading of cargoes onto a karyopherin. Critically, it will be essential to determine whether it can bind directly to nucleoporins—a defining characteristic of bona fide nuclear transport carriers. Important questions also remain concerning the mechanisms for loading and unloading of calreticulin binding to steroid receptors.
PAYING FOR THE TRIP: THE RAN GTP/GDP CYCLE
There are no motors in the nuclear pores. Rather, carriers appear to translocate from one side to the other by a process of facilitated diffusion. This process does not require energy and is nondirectional (57, 205). How, then, are macromolecules moved against a concentration gradient into or out of the nucleus?
First, a binding protein resident in only one compartment could sequester the transport cargo. The free cargo concentration would remain equal on both sides of the nuclear envelope, of course, but the total concentration would be higher in the compartment in which it was sequestered. One example of this mechanism may be the 14-3-3 proteins, which may function as cytoplasmic retention factors for certain phosphorylated proteins (167). Another example is β-catenin, which can bind and be retained by specific transcription factors in the nucleus (59, 282).
Second, the cargo could be covalently modified in one compartment, for example by phosphorylation, such that it no longer interacts with the carrier. This modification would reduce the free concentration of cargo available to the carrier in that compartment and drive accumulation by mass action. An example of this type of mechanism may be the nuclear accumulation of cyclin B (254).
Third, the binding of cargo to the carrier may be regulated by a second factor, such that assembly occurs in only one compartment and disassembly occurs in the other. As discussed above, Ran-GTP fulfills this regulatory function by promoting the assembly of export complexes or the disassembly of import complexes. It has the advantage of flexibility, because chemical modifications to the cargo are avoided, and it probably accounts for the bulk of all nucleocytoplasmic transport through the NPCs. Any such system involves work, however, because, directly or indirectly, the negative free energy of loading the cargo onto the carrier must be paid for during unloading, to complete the thermodynamic cycle.
The Ran GTPase system depends on the maintenance of a high nuclear concentration of Ran-GTP, and the cost of the transport cycle is paid for by the hydrolysis of the Ran-GTP to Ran-GDP in the cytoplasm. The requirement for a Ran-GTP gradient across the nuclear envelope has been demonstrated experimentally, for both nuclear import and export (103, 207). A simple example of the role of Ran in nuclear transport is provided by transportin, which imports the mRNA binding protein hnRNPA1. Binding of cargo in the cytosol is spontaneous. Release in the nucleus is driven by the free energy of binding Ran-GTP (ΔG○ ≈ −51 kJ mol−1). On returning to the cytoplasm, the transportin/Ran-GTP complex must be disassembled. This step requires hydrolysis of the Ran-GTP (ΔG○ ≈ −33 kJ mol−1) and is the only functionally irreversible reaction in the transport cycle. One Ran-GTP molecule is consumed per cycle. The hydrolysis of the GTP throws a conformational switch in Ran that prevents transportin binding.
A mechanism of this type has two important consequences. First, Ran is required only for accumulation of the cargo against a concentration gradient and not for the actual translocation of the carrier or cargo through the NPC. Second, each complete transport cycle involves the transport of one Ran-GTP molecule out of the nucleus. The first consequence has been demonstrated experimentally, using permeabilized cells and stoichiometric amounts of transportin with an M9 fusion protein as cargo (57, 128, 170, 171, 205). The second consequence has been demonstrated for importin-β, for which case the export of a carrier/Ran-GTP complex has been observed.
How is the Ran gradient maintained? This question has two parts. The first concerns the way in which Ran-GTP is produced in the nucleus and hydrolyzed in the cytoplasm, while the second concerns the mechanism by which Ran is returned to the nucleus after it has been released from transportin or any of the other karyopherins. The endogenous GTPase activity and guanine nucleotide off-rates for Ran are extremely low (kcat ≈ 1.5 × 10−5 s−1; koff ≈ 1.8 × 10−5 s−1), and, as with other small GTPases, there are regulatory factors that catalyze these processes (120). GTP hydrolysis on Ran is catalyzed by a cytoplasmic GTPase-activating protein called RanGAP, and guanine nucleotide exchange is catalyzed by a nuclear RanGEF that in mammals is called RCC1 (Prp20 in budding yeast). Both are potent factors that can accelerate the basal rates by as much as 500,000-fold. The low intrinsic GTPase and exchange rates for Ran are essential to prevent dissipation of the gradient between the nucleus and cytoplasm; the high catalytic rate constants of the regulatory factors ensure efficient gradient generation.
RanGEF
The Ran exchange factor was initially identified in mammalian cells from a temperature-sensitive allele that caused premature chromosome condensation at the nonpermissive temperature (hence the name RCC1, for “regulator of chromosome condensation”) (176, 262). In yeast it is an essential gene, temperature-sensitive alleles of which have pleiotropic effects related to RNA transport and cell cycle defects (7, 71, 108). Like the exchange factors for other small GTPases, RanGEF stimulates the release of guanine nucleotides from Ran by stabilizing the nucleotide-free, or apo-Ran, state (120, 122).
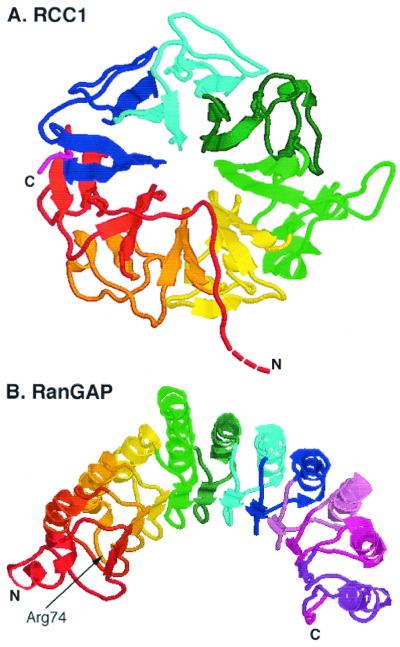
RanGEF is a constitutively nuclear protein, associated with chromatin (176). The crystal structure reveals a donut shape, created from seven “propeller blades” equally distributed around a central hole (Fig. 6A) (202). Several other proteins appear to contain RCC1-like motifs, but there is no evidence to date that they catalyze nucleotide exchange on Ran (86, 158, 212). Extensive mutagenesis and docking experiments have suggested that Ran binds to one face of the donut while the other face is responsible for binding to chromatin (11). The structure of the Ran-RanGEF complex has recently been solved, confirming the predicted binding surfaces and suggesting that an acidic residue in RanGEF plays a critical role in the exchange reaction by interacting with a lysine in the phosphate binding loop of Ran (201).
FIG. 6.
Structures of the RanGEF RCC1 (A) and of RanGAP (B). (A) The first 20 amino acids, containing the NLS, are absent. (B) The arginine finger residue is indicated. This residue is required for catalytic activity (93, 202).
The N terminus forms an extended chain, hanging off the donut core, and contains a monopartite NLS. The mammalian RanGEF, RCC1, requires this NLS for nuclear import via the importinα-importinβ system, with a preference for the importinα3 isoform. However, deletion of this NLS does not prevent nuclear import, suggesting that a second, Ran-independent mechanism exists, perhaps to guard against accumulation in the cytoplasm, which would probably be toxic to the cell (126, 172, 255).
RanGEF binds directly to nucleosomes, an interaction that is mediated preferentially by histones H2A and H2B, to which RanGEF binds with high affinity (173). The N-terminal tails of the histones appear to be dispensable for binding, and an interesting speculation is that the RanGEF binds to the exposed internucleosome faces, like hubcaps on a car wheel. Intriguingly, the histones can activate exchange activity by about twofold, presumably by inducing a conformational change in the RanGEF. RanBP3 also binds to and activates RCC1 (M. E. Nemergut, M. Lindsay, and I. G. Macara unpublished data). The effect is additive with the histone stimulation, and these data, together with computational modeling of Ran import, suggest that the modulation of RCC1 exchange activity may provide the cell with a mechanism for the global control of nuclear transport.
Frog sperm chromatin is very highly compacted and contains little or no RanGEF. It also lacks histones H2A and H2B. Decondensation of sperm chromatin can be induced in vitro, either by addition of an egg cytosolic extract or by addition of the acidic nuclear protein, nucleoplasmin, plus exogenous histones H2A and H2B. During this process, the exogenous histones become incorporated into the decondensed chromatin and RanGEF associates with the chromatin. We think that this process is key to the regulated formation of a nuclear envelope around the chromatin. Nuclear envelope formation occurs by the fusion of vesicles on or near the chromatin surface (26, 75, 271), and the trigger for this event was found recently to be the formation and/or turnover of Ran-GTP (91, 286). Remarkably, nuclear envelopes can even be formed in oocyte extracts around Sepharose beads to which glutathione S-transferase–Ran–GTP has been attached, demonstrating that no other chromatin components are required for this process. The staged association of RanGEF with sperm chromatin after its initial decondensation will produce a Ran-GTP “atmosphere” close to the chromatin surface, which may then induce vesicle fusion and nuclear envelope formation (91, 286).
RanGAP
The fission yeast RanGAP (rna1) has remarkably symmetric structure, composed of 11 leucine-rich repeats that together form a crescent (93). Each repeat forms a β-α hairpin, with the α-helices on the convex face of the crescent (Fig. 6B). Many GAPs appear to use a common “arginine finger” mechanism for the stimulation of small GTPases, in which a key Arg residue inserts into the nucleotide binding pocket of the GTPase and stabilizes the transition state (4, 226). An Arg residue in the third leucine-rich repeat may fulfill this function for RanGAP. The structures of several small GTPase-GAP complexes have been solved, and in each case the GAPs bind to the so-called Switch 1 and Switch 2 regions of the GTPases. These are the key regions that undergo a conformational rearrangement triggered by the change in state of the guanine nucleotide, and it is likely that the corresponding sequences in Ran are involved in binding RanGAP.
RanGAP is cytoplasmic. Mammalian RanGAP behaves in solution as a dimer and is larger than the diffusion limit for passive movement through the NPCs (18). However, the fission yeast rna1 is smaller and does not appear to behave as a dimer. In budding yeast, RanGAP contains both an NLS and several NES sequences, indicating that it may shuttle into and out of the nucleus, and its cytoplasmic localization is dependent on the exportin activity of Crm1 (62). Why RanGAP should shuttle is unclear. Moreover, in the fission yeast RanGAP, the hydrophobic residues that correspond to the NES sequences are not exposed but are mostly buried beneath the concave inner surface of the crescent. Given the close relatedness between RanGAP orthologs, the budding yeast protein most probably has a very similar structure to that of the fission yeast protein and the putative NES is unlikely to be functional in the context of the full-length protein.
In mammalian cells a substantial fraction of RanGAP is posttranslationally modified. Antibodies detect a form of the protein with lower mobility in sodium dodecyl sulfate-polyacrylamide gel electropheresis than would be predicted from its molecular mass, and analysis by mass spectrometry revealed that this form comprised an adduct of RanGAP with a novel, 8-kDa, ubiquitin-related polypeptide that was named GMP1, or SUMO-1 (149, 152). A growing number of other proteins have since been found to be SUMO modified, including Mdm2 and p53, and in several cases SUMOylation plays a regulatory role, inhibiting ubiquitination and protein degradation (32, 211). In RanGAP, it appears that the SUMO modification targets the protein to Nup358/RanBP2 in the cytoplasmic fibrils of the NPC (149, 152, 220). This localization may increase the efficiency with which the RanGAP can hydrolyze Ran-GTP that is exported with karyopherins from the nucleus. However, Nup358/RanBP2, and a closely related gene product, RanBP2L1, are not expressed ubiquitously, so it remains to be established that targeting is the only function of the SUMO modification (63, 174). Clearly it is not essential in budding yeast, which does not SUMO modify its RanGAP. Neither yeast nor Drosophila contains a homologue of Nup358/RanBP2.
An important limitation to RanGAP function is that its activity is blocked by karyopherins (66, 80). The Switch 2 region of Ran-GTP is buried in complexes with karyopherin-β2 or importinβ and is inaccessible to RanGAP (39, 263). Therefore, a second factor must interact with the complex and permit RanGAP function. This factor is a Ran binding protein called RanBP1.
RanBP1
RanBP1 consists principally of a Ran binding domain (RanBD) (15, 46, 180). The mammalian version possesses a nuclear export signal in a C-terminal extension and may dimerize through a short N-terminal extension (208, 289). In budding yeast the export signal appears to be contained within the RanBD (133). The export signals are not merely a safety mechanism in case of leakage of RanBP1 into the nucleus. RanBP1 shuttles rapidly in and out through the nuclear pores. The import mechanism is not fully understood but appears to be carrier independent (133, 194). The RanBD is conserved in all eukaryotes, although, oddly, the nematode Caenorhabditis elegans possesses a Ran binding protein that more closely resembles RanBP2 than RanBP1, and contains two RanBDs (15).
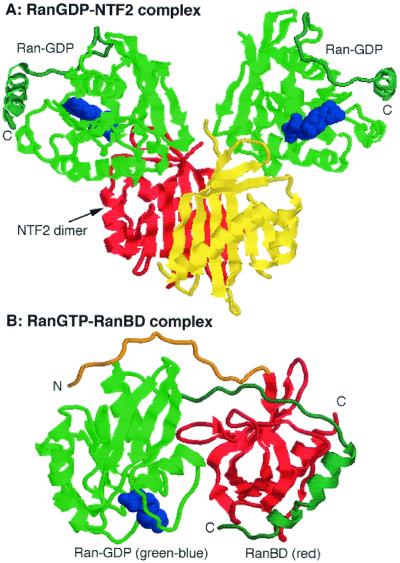
The affinity of the RanBD for Ran-GTP is very high (132). The crystal structure of Ran in a complex with a RanBD reveals an unusually close embrace between the two polypeptides, in which an acidic “hand” at the C terminus of Ran wraps around to a basic patch on the RanBD while an acidic “hand” at the N terminus of the RanBD is stretched across to bind a basic patch on Ran (Fig. 7B) (264). The body of the RanBD, which forms a so-called β-barrel, is held closely against the Switch 1 region and against other residues in the C-terminal half of the Ran protein. Comparison of the conformation of Ran-GDP with that of Ran in complex with the RanBD explains why the interaction is specific for the GTP-bound state. Like other small GTPases, the orientation of the Switch 1 loop is very different between the two states. In addition, and uniquely to Ran, the C terminus possesses a highly acidic sequence that may be attached, when Ran is in the GDP-bound state, to a basic patch between residues 139 to 142 (Fig. 7A). This conformation stabilizes the association with the nucleotide. Interaction of Ran with GTP triggers reorientation of Switch 1 and displacement of the C-terminal arm, which, when Ran binds the RanBD, then swings out through an angle of nearly 180° (Fig. 7B).
FIG. 7.
Structures of RanGDP-NTF2 (A) and RanGTP-RanBD (B). NTF2 forms a dimer, and each subunit associates with one molecule of RanGDP. Ran is shown in green, the nucleotide is shown in blue, and the NTF2 subunits are shown in red and yellow. Note the location of the Ran C-terminal arm (blue-green) in close juxtaposition to the basic patch on the RanGDP. When bound to a RanBD, the C-terminal arm (blue-green) of RanGTP is extended away from the body of the Ran (green) and embraces the RanBD (red). The N terminus of the RanBD (orange) loops around the Ran and contacts the basic patch (246, 264).
A RanBD can stimulate RanGAP activity by about 10-fold, although it has no effect on GTP hydrolysis on its own (15, 19). How does it work? One possibility is that in the absence of a RanBD, the acidic -DEDDDL sequence remains tethered to Ran-GTP, although the upstream part of the C-terminal arm must have moved to accommodate the reorientation of the Switch 1 region that occurs on binding GTP. This conformation may reduce the affinity for RanGAP. Binding of a RanBD then pulls the C terminus of Ran aside, to facilitate RanGAP binding. Consistent with this hypothesis, deletion of the -DEDDDL sequence potentiates Ran-GTP hydrolysis by RanGAP but destroys responsiveness to RanBP1 (209).
In the Ran-importin complexes, the C-terminal arm is also released from its basic patch on Ran but remains exposed (92). The Switch 2 region is almost entirely buried, and RanGAP is ineffective in catalyzing GTP hydrolysis How does the RanBD overcome the blockade of RanGAP activity? The interfaces between Ran-GTP and the RanBD do not overlap substantially with those of Ran and the importins, and the three proteins can form a stable ternary complex (37, 147). Whether RanGAP can interact with RanGTP in a quarternary complex with RanBD and importins, however, or whether the importins must first be released remains unclear. Competition between the RanBD and importinβ for interaction with Ran residues N154 and E158 could increase the off-rate of Ran-GTP from the importin. Additionally, the attachment of the C-terminal tail of Ran to the RanBD may play an important role. Deletion of the -DEDDDL sequence significantly increases the affinity of Ran for importinβ, most probably because the C terminus no longer has to be displaced from its basic pocket. One would expect that either deletion of the -DEDDDL or displacement by a RanBD would increase the on-rate for importin binding to a similar extent. We therefore suggest that in the ternary complex of Ran-GTP–importin–RanBD, both the on- and off-rates are substantially increased compared to those in the Ran-GTP–importin binary complex. This increase enables RanGAP to access the Ran-GTP and terminate the transport cycle by hydrolyzing the nucleotide to GDP.
The termination step for importinβ is rather more complicated, however, because the off-rate of Ran-GTP is too slow, even in the presence of a RanBD, to be useful. Addition of importinα can facilitate the process, probably by competing with Ran-GTP and blocking its rebinding to importinβ (17, 146). The association of importinβ with nucleoporins may also increase the off-rate of Ran-GTP from importinβ (68)
A further complication is presented by the ability of RanBDs, together with importins, to form stable ternary complexes with Ran-GDP, even though the affinity of either protein alone for Ran-GDP is extremely low (∼107-fold lower than that for Ran-GTP) (36). The displacement of the C terminus again probably plays a critical role in the formation of this ternary complex. The physiological role of the complex remains, however, completely obscure.
Adding Up the Costs
Classical NLS cargo is imported by the importinα-importinβ dimer. Importinβ returns to the cytoplasm bound to Ran-GTP, but importinα also needs to be exported. Another member of the karyopherin family, Cas (Cse1 in budding yeast), performs this task (134, 135, 239). Like Crm1, it forms a ternary complex with Ran-GTP and its cargo, importinα, translocates through the NPC, and is dissociated in the cytoplasm by hydrolysis of the GTP on Ran to GDP. Therefore the complete transport cycle for a classical NLS cargo molecule requires the hydrolysis of two GTP molecules. Why use this system? One obvious answer is that it can drive cargo import against a higher concentration gradient than would be possible using a single importin. Whether there are other advantages remains to be discovered.
A variation on this theme is used for the nuclear export of U snRNA precursors, which in metazoa are processed in the cytoplasm before being returned to the nucleus. These RNAs associate with a heterodimer called the cap binding complex (CBC), which in turn binds to an adapter called PHAX (175). Nuclear PHAX is phosphorylated and can interact with the exportin, Crm1, and Ran-GTP. All of these interactions are cooperative. After translocation through the NPC into the cytoplasm, the PHAX is dephosphorylated, Ran-GTP is hydrolyzed, and the export complex disassembles. The CBC is then reimported by the classical importinα-importinβ pathway. It is not yet clear how dephosphorylated PHAX is reimported. In terms of the energetics of transport, two nucleotides are hydrolyzed per U snRNA export step (one ATP and one GTP), two GTP molecules are hydrolyzed to return the CBC to the nucleus (one for release of the cargo and one for recycling of the importinα), and at least one GTP molecule is probably hydrolyzed to return PHAX to the nucleus. In this case, therefore, the free energy from hydrolysis of a minimum of five high-energy phosphate bonds is used to push U snRNA export. This unusual system may be required by the high degree of cooperativity in assembly of the export complex and may be designed to ensure absolute directionality to the movement of the U snRNA precursors, which could be toxic if they accumulated in the nucleus.
Recycling Ran to the Nucleus
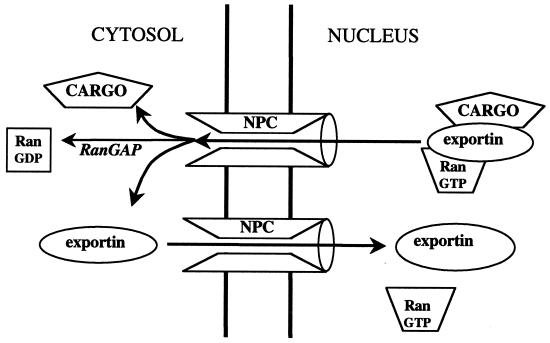
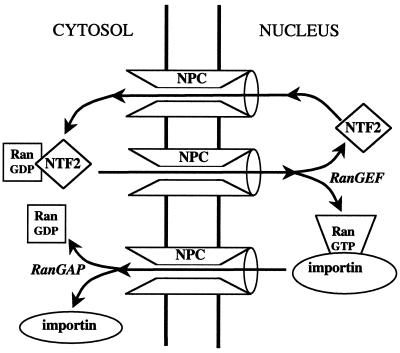
Each Ran-dependent transport cycle delivers at least one Ran molecule to the cytoplasm, which must be returned to the nucleus and converted to the GTP-bound state. Although Ran is in principle small enough to diffuse through the nuclear pores, the rate of passive diffusion is far too low to sustain the required nucleocytoplasmic traffic. The cell therefore provides a transport factor, NTF2 (also called p10), that facilititates Ran transport (31, 189, 206, 238). NTF2 recognizes only the GDP-bound form of Ran (Fig. 7A), and can bind simultaneously to the FXFG motifs present in nucleoporins (41, 187). Abundant evidence supports the idea that the NTF2-Ran-GDP complex associates with the NPC and translocates by a facilitated diffusive process to the nuclear compartment, where RanGEF triggers the disassembly of the complex by converting Ran to the GTP-bound state (Fig. 8) (34, 140, 197, 206, 238, 243). NTF2 appears to be optimized for rapid transport: it is small (15 kDa) and so can diffuse rapidly, and it forms homodimers so that two Ran-GDP molecules can be translocated per cycle (Fig. 7A) (246). Microinjection assays and computer modeling of Ran transport in intact mammalian fibroblasts suggest that the steady-state unidirectional flux of endogenous Ran is ∼100 molecules/nuclear pore/s (A. Smith et al., unpublished data). This rate is ∼40-fold greater than that for the passive diffusion of the T24N Ran mutant (which does not bind NTF2) and is also substantially higher than the rate of import of a classical NLS under similar conditions.
FIG. 8.
Mechanism of Ran import. The carrier NTF2 binds specifically to RanGDP, present in the cytoplasm. In the nucleus, RanGEF catalyzes the exchange of GDP for GTP on Ran, which releases the NTF2. NTF2 then returns empty to the cytoplasm to pick up another molecule of RanGDP. The RanGTP associates with carriers (importins or exportins) which move to the cytosol, where the GTP is hydrolyzed, releasing RanGDP.
In vitro assays suggest that NTF2 itself can transit the pores much faster than this (∼620 dimers/pore/s at 2.5 μM NTF2 dimer, which is a concentration similar to that present in cells, and ∼2,500 dimers/pore/s at 100 μM NTF2 dimer) (204). The difference between the in vitro and in vivo data probably reflects the fact that in the intact cell the NTF2 must compete for pore access with many other translocating carriers.
The fact that passive diffusion can occur suggests that a sufficiently high concentration of Ran might at least partially overcome the requirement for NTF2. Indeed, Paschal et al. (188) have found that the requirement in yeast for NTF2 can be precluded by the overexpression of Ran, and the overexpression of NTF2 can suppress the effects of several types of Ran mutant allele (275). A high concentration of Ran can also replace the need for NTF2 in permeabilized-cell assays of nuclear protein import (82, 161, 188).
The rate of Ran import provides a lower estimate for the net traffic flow through the nuclear pores. In the simplest case, where a cargo protein is imported by direct binding to a karyopherin, four passages through the pores are required per import cycle (cargo enters the nucleus with the karyopherin, which leaves with Ran-GTP, which enters with NTF2, which then leaves). This value suggests that at least 400 molecules translocate per pore per second. It is important to realize that this number represents net flow. Because movement through the pore itself is not vectorial (no work is expended), it must be random, and transport carriers will flicker back and forth across the pore until they encounter a docking site at one end or the other and/or the transport is terminated by complex disassembly.
How fast could a protein the size of, say, a Ran2-NTF2 complex diffuse from one end of the NPC to the other? Using the Einstein-Smoluchowski equation and assuming a distance of 45 nm gives a jump time of about 10 μs. Therefore, if the traffic through the NPC is of the order of 1 protein per 2.5 ms, each could jump back and forth randomly through the pore several hundred times before escaping or becoming trapped at one end or the other. Even for the translocation of NTF2 in vitro, at 2,500 dimers/pore/s, there is time for at least 40 random jumps.
Are there other mechanisms for Ran import? Another Ran binding protein, called Mog1, was recently discovered that can also rescue the growth defects caused by certain mutant alleles of yeast Ran (177, 245). While Mog1 is not essential for survival, its deletion causes a defect in nuclear protein import that can be suppressed by the overexpression of NTF2. Mog1 is also a nucleocytoplasmic shuttling protein. Nonetheless, it is unlikely to be a carrier for Ran import, because it recognizes Ran-GTP rather than Ran-GDP (244). It also has the odd property of stabilizing Ran in the nucleotide-free state. Thus it can stoichiometrically release GTP from Ran but does not act as an exchange factor. RanBP1 can form a ternary complex with Mog1 and Ran-GTP and stabilizes the GTP-bound state. Mog1 may therefore be involved in some unknown function of RanBP1, which can also cycle into and out of the nucleus (133, 194), rather than in Ran transport.
Another possible mechanism for Ran import is via the formation of a ternary complex of Ran-GDP with RanBP1 and karyopherins such as importinβ. This complex is reportedly competent to bind importinα and NLS-cargo (36). RanBP1 does not inhibit import in the permeabilized-cell assay, even at high concentrations, from which one can infer that the ternary complex is able to transit the nuclear pores (37). But how would this work? Complexes of Ran with RanBP1 or with importins are resistant to RanGEF-mediated nucleotide exchange (19). Within the nucleus, Ran-GTP would displace the bound Ran-GDP and be exported with importinβ. No net change in nuclear Ran concentration could therefore occur.
CROSSING THE CHANNEL
How do carrier-cargo complexes move through the NPC? Do they all interact with the same nucleoporins? Can multiple complexes transit the same pore simultaneously? What controls the selectivity of the pores? Several new experimental approaches are being used to address these questions, but fundamental insights into the translocation process will require a high resolution structure of the intact NPC, a goal that is beyond current capabilities.
Any mechanism must account for the diversity of macromolecules that can transit the pores, the nature of the facilitated diffusion of cargo, and the lack of any obvious pore binding domain in the carriers and must explain the observations that carriers and cargo can “dock” at the NPC (1, 82, 160, 162, 184). Docking is of particular interest because high-affinity binding sites are likely to have low off-rates, which are incompatible with rapid translocation through the pores. Some carriers, such as importinβ, require Ran-GTP to trigger release from the NPC.
The NPC binding region of importinβ has been mapped to residues around 152 to 352 (38, 137). The structure of an N-terminal fragment, importinβ(1–442), has been recently determined in a complex with five FxFG motifs from the yeast nucleoporin Nsp1 (12). Interestingly, whereas both Ran-GTP and the IBB domain of importinα bind to the concave inner face of the importinβ superhelix, the FxFG motifs bind to the convex surface at two sites, one between HEAT repeats 5–6 and one between repeats 6–7. The phenylalanines are buried in hydrophobic pockets. Comparison with the structure of importinβ bound to Ran-GTP suggests that Ran induces a twist in the importinβ, particularly between HEAT repeats 5 and 6, that would displace the first phenylalanine of the FxFG motif.
Fragments such as importinβ(45–462) bind sufficiently tightly that they act as potent inhibitors of many types of carrier-mediated transport (36, 38, 137). This observation implies that there exists a common step in translocation or docking for all of these carriers. This may be any or all of the FxFG motif-rich nucleoporins, but in Xenopus oocytes, at least, the principal binding partner on the NPC for importinβ is the nucleoporin Nup153, which is located at the basket structure on the nuclear face of the NPC rather than in the interior of the pore (234, 235). Nup153 most probably constitutes a terminal docking site for importins, therefore, rather than a component of the facilitated diffusive translocation mechanism. Studies on Crm1-mediated export suggest that another nucleoporin, Nup214, on the cytoplasmic fibrils, functions in a similar way as a terminal docking site for export complexes (70, 116).
An elegant approach to nucleoporin-carrier specificity has been to use fluorescence resonance energy transfer between karyopherin fusions to cyan fluorescent protein and nucleoporin fusions to yellow fluorescent protein in budding yeast (47). The advantages of using yeast are that each fusion can be proven by gene replacement to be functional and that the complete set of karyopherins and nucleoporins is known. These studies support the idea that karyopherins bind preferentially to distinct nucleoporins. But do these interactions indicate different transit routes through the pores, or are they docking sites? And why do docking sites exist? Although these sites provide a degree of asymmetry to the structure of the NPC, in that importin docking sites are situated on the nuclear side of the pore and exportin docking sites are situated on the cytoplasmic fibrils, it is thermodynamically impossible for them to drive asymmetric transport (i.e., transport against a concentration gradient). Indeed, by inverting the normal Ran gradient, it is possible to reverse the usual direction of movement of transport carriers through the pores (169).
Several models that address these issues have been described recently. One, called a Brownian affinity-gating model, was proposed by Rout et al. (215) and argues for a random (Brownian motion) movement of cargo-carrier complexes through the pores, with docking sites at each end. Transient interactions with nucleoporins along the pore surface provide selectivity. A similar concept, based on detailed kinetics of nuclear transport in vitro, is termed the selective-phase model (204). Again, the carrier-cargo complex is assumed to move randomly through the pore, but the phenylalanine-rich nucleoporins are proposed to form a semiliquid phase into which the carriers can partition. The nucleoporins may form a sieve-like structure within the pores, and transient interactions of the carrier with the FG repeats would allow the carrier to “dissolve” into this structure.
Along the same lines, we speculate that the NPC is an unusually open structure in which nucleoporin FG repeats form extended chains that fill the pore like loose, oily spaghetti (Fig. 9). The mean diameter of the central open tube through the pore is about 10 nm (117). The nucleoporin “spaghetti” could therefore form a layer around this tube that would be about 7 nm thick. The ΔG○ for conformational transitions in the nucleoporin chains would be sufficiently low that they would move freely at physiological temperatures. Thus, carrier-cargo complexes could push them aside easily. How would carriers move through the pore? Assuming that transient associations occur with the FG repeats (or other sequences) within the nucleoporin spaghetti, each time the carrier unbinds it can move randomly within the pore for a short distance before rebinding, and this process of transient association and random motion would produces a facilitated diffusive movement through the pore. If 200 carriers move through each pore per second, then the mean transit time for each is ∼5 ms. The jump time for a freely diffusing karyopherin from one end of the pore to the other is, as discussed above, about 10 μs, and so each could bind and unbind several hundred times during transit. There are probably >1,000 FG repeats per NPC (13); therefore, the number of binding sites is not limiting. It is instructive to consider the dissociation rate that a carrier might possess from a single FG repeat. Assuming a diffusion-limited on-rate (∼7 × 109 M−1 s−1 at 37°C) and a Kd for the interaction of a carrier with an FG repeat of about 25 μM (based on the value for NTF2/Nsp1 binding), the dissociation constant would be about 2 × 105 s−1 (34). This value is similar to the calculated jump time and supports the idea that multiple transient interactions with nucleoporins could occur during diffusion through the pore.
FIG. 9.
The oily-spaghetti model for translocation through the nuclear pores. The approximate dimensions of the pore are shown. Nucleoporins containing FxFG repeats are shown as randomly oriented lines. Molecules with a diameter of <10 nm can diffuse freely through the pore. Larger molecules are hindered by the nucleoporins. Carrier proteins bind weakly and transiently to the nucleoporins. It is assumed that the free energy changes associated with conformational motions of the nucleoporins are very low, so that the carrier can diffuse from one binding site to another relatively unhindered. The time between binding events is on the order of a few microseconds, and the carrier is assumed to diffuse randomly back and forth across the pore. Vectoriality is imparted by the assembly and disassembly reactions driven by RanGTP and is not a property of the pore. Docking sites, which are situated at some distance outside of the pore, are not shown.
The estimated Km for transportin translocation through the pores in vitro is ∼4 μM, which also supports the idea that interactions of a carrier with the nucleoporins are very weak and transient (204). Dissociation constants of >1 μM are difficult to measure directly and are much higher than those for importinβ binding to Nup153 or those for binding of Crm1 to Nup214 (70, 116). Therefore, these and several other nucleoporin-karyopherins interactions that have been studied probably represent docking sites rather than sites involved directly in translocation. One obvious reason for providing a docking site at one end of the pore is to increase the probability that the carrier-cargo complex will encounter soluble, compartment-specific factors required for complex assembly or disassembly. For example, a high-affinity site for importinβ on Nup153 will accumulate importinβ-importinα-cargo complexes, raising the probability that each will encounter a Ran-GTP molecule that will trigger cargo release. By also triggering release from the docking site, the Ran frees up the site for another incoming carrier. We speculate that importin-Ran-GTP may itself dock at a nucleoporin site on the cytoplasmic face of the NPC, where it can await disassembly by RanGAP and RanBP1. Note that for both importinβ and Crm1, the known docking sites are at a considerable distance (40 to 120 nm) from the pore itself, perhaps to prevent clogging at the exit areas.
It has been suggested that the nucleoporins may be arranged in a sequence of increasing affinity for importins, from the cytoplasmic side to the nuclear side of the pore, and that this arrangement may help confer vectoriality to the transport process (16). Binding to the nucleoporins is an equilibrium process that does not involve thermodynamic work and cannot therefore drive an accumulation of cargo against a concentration gradient. Such an arrangement will lead, however, to a gradient of importin within the pore, because the off-rates will be lower near the nuclear side. It is not clear how this would help the transport process. High affinities (low off-rates) will slow the facilitated diffusion through the pore. If Ran-GTP enters the pore to trigger cargo release from complexes on these higher-affinity sites, the cargo would become trapped within the pore (being too large to move freely by passive diffusion). Therefore, the optimum solution is to use low-affinity sites within the pore, for rapid translocation of the carrier-cargo complex, and high affinity sites outside them, for cargo unloading.
Even extremely large cargoes such as the Balbiani ring mRNA could in principle be translocated by this sort of diffusive mechanism, if they contain multiple carrier binding sites. The Balbiani ring RNP particle unwinds and enters the pore in a specific orientation (119, 265). Once the first carrier reaches the cytoplasmic side of the pore and is disengaged, the RNP will be unable to diffuse back through the pore. Thus, a ratchet mechanism, coupled to vectorial release from the carrier, can drive the particle in the required direction.
This model may be testable if single molecules can be observed with sufficient resolution while they are traversing the pore. At 37°C, cargo would be expected to arrest only at the external docking sites, but at lower temperatures, where the conformational motion of the nucleoporin “spaghetti” is reduced, cargo may become trapped within the pore lumen. Careful measurements at different temperatures could provide an estimate of nucleoporin flexibility, and fluorescence resonance energy transfer measurements may provide information on the mean dwell time within the pore lumen.
CONCLUSION AND FUTURE DIRECTIONS
A consistent framework has been constructed over the last few years that explains in a satisfactory way the process of Ran-dependent nucleocytoplasmic transport. It seems unlikely that this basic model will be changed in any profound way in the near future. However, there remain numerous and substantial gaps in our knowledge. At a fundamental level, while we can speculate on translocation by facilitated diffusion through the NPC, we still lack any experimental data that directly address this mechanism at a molecular level. Many aspects of Ran-independent transport are unclear. For example, how does RanBP1 transit the NPC? Is it a property of all HEAT/Arm motif proteins to be able to interact with the NPC? Are there vectorial transport systems that are independent of Ran? Are there other proteins like calreticulin that have unrelated functions but double as transport factors? In addition, many cofactors may exist that modulate the interactions of transport carriers with their cargoes, with the NPC, and with other transport factors. RanBP1/Yrb1, RanBP3/Yrb2, Nup50/NPAP60, Nup2, and Nxt1/p15 fall into this category, but several unrecognized cofactors probably remain to be discovered, and much work needs to be done to understand how they operate. What is the function of nuclear actin in transport? New adapter proteins such as PHAX may also represent the tip of an iceberg. Clear, mechanistic pictures of RNA transport remain elusive, and the discovery of a family of NXF/TAP proteins suggests that there are unexpected layers of complexity to mRNA export. What underlies the connection between inositol polyphosphate metabolism and RNA export? Are there distinct classes of mRNA that exit the nucleus by different mechanisms? There are also important questions about how viruses dock at the NPC and insert their genomes through the pores and about the global regulation of nuclear transport, competition between karyopherins, and the energetics of different transport pathways, an understanding of which may require computational biology approaches using systems such as the Virtual Cell (225). Finally, exciting links are being identified between Ran and other fundamental aspects of eukaryotic cell biology, including the control of spindle formation during mitosis and the formation of nuclear envelopes around chromatin during telophase (33, 111, 272, 273, 286, 287). The complex interdependency of these processes will undoubtedly keep everyone busy for years to come.
ACKNOWLEDGMENTS
I thank members of the Macara laboratory, Lucy Pemberton, and the reviewers for reading, correcting, and improving the manuscript.
This work was supported by grant GM50526 from NIH.
REFERENCES
- 1.Adam E J, Adam S A. Identification of cytosolic factors required for nuclear location sequence-mediated binding to the nuclear envelope. J Cell Biol. 1994;125:547–555. doi: 10.1083/jcb.125.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adam S A, Gerace L. Cytosolic proteins that specifically bind nuclear location signals are receptors for nuclear import. Cell. 1991;66:837–847. doi: 10.1016/0092-8674(91)90431-w. [DOI] [PubMed] [Google Scholar]
- 3.Adam S A, Marr R S, Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmadian M R, Stege P, Scheffzek K, Wittinghofer A. Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nat Struct Biol. 1997;4:686–689. doi: 10.1038/nsb0997-686. [DOI] [PubMed] [Google Scholar]
- 5.Aitchison J D, Blobel G, Rout M P. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–627. doi: 10.1126/science.274.5287.624. [DOI] [PubMed] [Google Scholar]
- 6.Albertini M, Pemberton L F, Rosenblum J S, Blobel G. A novel nuclear import pathway for the transcription factor TFIIS. J Cell Biol. 1998;143:1447–1455. doi: 10.1083/jcb.143.6.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amberg D C, Fleischmann M, Stagljar I, Cole C N, Aebi M. Nuclear PRP20 protein is required for mRNA export. EMBO J. 1993;12:233–241. doi: 10.1002/j.1460-2075.1993.tb05649.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arts G J, Fornerod M, Mattaj I W. Identification of a nuclear export receptor for tRNA. Curr Biol. 1998;8:305–314. doi: 10.1016/s0960-9822(98)70130-7. [DOI] [PubMed] [Google Scholar]
- 9.Askjaer P, Jensen T H, Nilsson J, Englmeier L, Kjems J. The specificity of the CRM1-Rev nuclear export signal interaction is mediated by RanGTP. J Biol Chem. 1998;273:33414–33422. doi: 10.1074/jbc.273.50.33414. [DOI] [PubMed] [Google Scholar]
- 10.Askjaer P, Rosendahl R, Kjems J. Nuclear export of the DEAD box An3 protein by CRM1 is coupled to An3 helicase activity. J Biol Chem. 2000;275:11561–11568. doi: 10.1074/jbc.275.16.11561. [DOI] [PubMed] [Google Scholar]
- 11.Azuma Y, Renault L, García-Ranea J A, Valencia A, Nishimoto T, Wittinghofer A. Model of the Ran-RCC1 interaction using biochemical and docking experiments. J Mol Biol. 1999;289:1119–1130. doi: 10.1006/jmbi.1999.2820. [DOI] [PubMed] [Google Scholar]
- 12.Bayliss R, Littlewood T, Stewart M. Structural basis for the interaction between FxFG nucleoporin repeats and importin-beta in nuclear trafficking. Cell. 2000;102:99–108. doi: 10.1016/s0092-8674(00)00014-3. [DOI] [PubMed] [Google Scholar]
- 13.Bayliss R, Ribbeck K, Akin D, Kent H M, Feldherr C M, Gorlich D, Stewart M. Interaction between NTF2 and xFxFG-containing nucleoporins is required to mediate nuclear import of RanGDP. J Mol Biol. 1999;293:579–593. doi: 10.1006/jmbi.1999.3166. [DOI] [PubMed] [Google Scholar]
- 14.Becker J, Melchior F, Gerke V, Bischoff F R, Ponstingl H, Wittinghofer A. RNA1 encodes a GTPase-activating protein specific for Gsp1p, the Ran/TC4 homologue of Saccharomyces cerevisiae. J Biol Chem. 1995;270:11860–11865. doi: 10.1074/jbc.270.20.11860. [DOI] [PubMed] [Google Scholar]
- 15.Beddow A L, Richards S A, Orem N R, Macara I G. The Ran/TC4 GTPase-binding domain: identification by expression cloning and characterization of a conserved sequence motif. Proc Natl Acad Sci USA. 1995;92:3328–3332. doi: 10.1073/pnas.92.8.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ben-Efraim I, Gerace L. Gradient of increasing affinity of importin β for nucleoporins along the pathway of nuclear import. J Cell Biol. 2001;152:411–418. doi: 10.1083/jcb.152.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bischoff F R, Gorlich D. RanBP1 is crucial for the release of RanGTP from importin β-related nuclear transport factors. FEBS Lett. 1997;419:249–254. doi: 10.1016/s0014-5793(97)01467-1. [DOI] [PubMed] [Google Scholar]
- 18.Bischoff F R, Klebe C, Kretschmer J, Wittinghofer A, Ponstingl H. RanGAP1 induces GTPase activity of nuclear Ras-related Ran. Proc Natl Acad Sci USA. 1994;91:2587–2591. doi: 10.1073/pnas.91.7.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bischoff F R, Krebber H, Smirnova E, Dong W, Ponstingl H. Co-activation of RanGTPase and inhibition of GTP dissociation by Ran-GTP binding protein RanBP1. EMBO J. 1995;14:705–715. doi: 10.1002/j.1460-2075.1995.tb07049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bischoff F R, Ponstingl H. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature. 1991;354:80–82. doi: 10.1038/354080a0. [DOI] [PubMed] [Google Scholar]
- 21.Bischoff F R, Ponstingl H. Mitotic regulator protein RCC1 is complexed with a nuclear ras-related polypeptide. Proc Natl Acad Sci USA. 1991;88:10830–10834. doi: 10.1073/pnas.88.23.10830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Black B E, Holaska J M, Levesque L, Ossareh-Nazari B, Gwizdek C, Dargemont C, Paschal B M. NXT1 is necessary for the terminal step of Crm1-mediated nuclear export. J Cell Biol. 2001;152:141–156. doi: 10.1083/jcb.152.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Black B E, Levesque L, Holaska J M, Wood T C, Paschal B M. Identification of an NTF2-related factor that binds Ran-GTP and regulates nuclear protein export. Mol Cell Biol. 1999;19:8616–8624. doi: 10.1128/mcb.19.12.8616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bogerd H P, Benson R E, Truant R, Herold A, Phingbodhipakkiya M, Cullen B R. Definition of a consensus transportin-specific nucleocytoplasmic transport signal. J Biol Chem. 1999;274:9771–9777. doi: 10.1074/jbc.274.14.9771. [DOI] [PubMed] [Google Scholar]
- 25.Bogerd H P, Fridell R A, Benson R E, Hua J, Cullen B R. Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel In vivo randomization-selection assay. Mol Cell Biol. 1996;16:4207–4214. doi: 10.1128/mcb.16.8.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boman A L, Delannoy M R, Wilson K L. GTP hydrolysis is required for vesicle fusion during nuclear envelope assembly in vitro. J Cell Biol. 1992;116:281–294. doi: 10.1083/jcb.116.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Booth J W, Belanger K D, Sannella M I, Davis L I. The yeast nucleoporin Nup2p is involved in nuclear export of importin alpha/Srp1p. J Biol Chem. 1999;274:32360–32367. doi: 10.1074/jbc.274.45.32360. [DOI] [PubMed] [Google Scholar]
- 28.Braun I C, Rohrbach E, Schmitt C, Izaurralde E. TAP binds to the constitutive transport element (CTE) through a novel RNA-binding motif that is sufficient to promote CTE-dependent RNA export from the nucleus. EMBO J. 1999;18:1953–1965. doi: 10.1093/emboj/18.7.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brownawell A M, Kops G J, Macara I G, Burgering B M. Inhibition of nuclear import by protein kinase B/Akt regulates the subcellular distribution and activity of the forkhead transcription factor, AFX. Mol Biol Cell. 2001;21:3534–3546. doi: 10.1128/MCB.21.10.3534-3546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brunet A, Bonni A, Zigmond M J, Lin M Z, Juo P, Hu L S, Anderson M J, Arden K C, Blenis J, Greenberg M E. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 31.Bullock T L, Clarkson W D, Kent H M, Stewart M. The 1.6 angstroms resolution crystal structure of nuclear transport factor 2 (NTF2) J Mol Biol. 1996;260:422–431. doi: 10.1006/jmbi.1996.0411. [DOI] [PubMed] [Google Scholar]
- 32.Buschmann T, Fuchs S Y, Lee C G, Pan Z Q, Ronai Z. SUMO-1 modification of Mdm2 prevents its self-ubiquitination and increases Mdm2 ability to ubiquitinate p53. Cell. 2000;101:753–762. doi: 10.1016/s0092-8674(00)80887-9. [DOI] [PubMed] [Google Scholar]
- 33.Carazo-Salas R E, Guarguaglini G, Gruss O J, Segref A, Karsenti E, Mattaj I W. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature. 1999;400:178–181. doi: 10.1038/22133. [DOI] [PubMed] [Google Scholar]
- 34.Chaillan-Huntington C, Braslavsky C V, Kuhlmann J, Stewart M. Dissecting the interactions between NTF2, RanGDP, and the nucleoporin XFXFG repeats. J Biol Chem. 2000;275:5874–5879. doi: 10.1074/jbc.275.8.5874. [DOI] [PubMed] [Google Scholar]
- 35.Chi N C, Adam E J, Adam S A. Sequence and characterization of cytoplasmic nuclear protein import factor p97. J Cell Biol. 1995;130:265–274. doi: 10.1083/jcb.130.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chi N C, Adam E J H, Adam S A. Different binding domains for Ran-GTP and Ran-GDP/RanBP1 on nuclear import factor p97. J Biol Chem. 1997;272:6818–6822. doi: 10.1074/jbc.272.10.6818. [DOI] [PubMed] [Google Scholar]
- 37.Chi N C, Adam E J H, Visser G D, Adam S A. RanBP1 stabilizes the interaction of Ran with p97 In nuclear protein import. J Cell Biol. 1996;135:559–569. doi: 10.1083/jcb.135.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chi N C, Adam S A. Functional domains in nuclear import factor p97 for binding the nuclear localization sequence receptor and the nuclear pore. Mol Biol Cell. 1997;8:945–956. doi: 10.1091/mbc.8.6.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chook Y M, Blobel G. Structure of the nuclear transport complex karyopherin-2-Ran.GppNHp. Nature. 1999;399:230–237. doi: 10.1038/20375. [DOI] [PubMed] [Google Scholar]
- 40.Cingolani G, Petosa C, Weis K, Muller C W. Structure of importin-beta bound to the IBB domain of importin-alpha. Nature. 1999;399:221–229. doi: 10.1038/20367. [DOI] [PubMed] [Google Scholar]
- 41.Clarkson W D, Kent H M, Stewart M. Separate binding sites on nuclear transport factor 2 (NTF2) for GDP-Ran and the phenylalanine-rich repeat regions of nucleoporins p62 and Nsp1p. J Mol Biol. 1996;263:517–524. doi: 10.1006/jmbi.1996.0594. [DOI] [PubMed] [Google Scholar]
- 42.Clouse K N, Luo M, Zhou Z, Reed R. A Ran-independent pathway for export of spliced mRNA. Nat Cell Biol. 2001;3:97–99. doi: 10.1038/35050625. [DOI] [PubMed] [Google Scholar]
- 43.Cole C N. mRNA export: the long and winding road. Nat Cell Biol. 2000;2:E55–E58. doi: 10.1038/35008681. [DOI] [PubMed] [Google Scholar]
- 44.Conti E, Kuriyan J. Crystallographic analysis of the specific yet versatile recognition of distinct nuclear localization signals by karyopherin alpha. Struct Fold Des. 2000;8:329–338. doi: 10.1016/s0969-2126(00)00107-6. [DOI] [PubMed] [Google Scholar]
- 45.Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell. 1998;94:193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- 46.Coutavas E, Ren M, Oppenheim J D, D'Eustachio P, Rush M G. Characterization of proteins that interact with the cell-cycle regulatory protein Ran/TC4. Nature. 1993;366:585–587. doi: 10.1038/366585a0. [DOI] [PubMed] [Google Scholar]
- 47.Damelin M, Silver P A. Mapping interactions between nuclear transport factors in living cells reveals pathways through the nuclear pore complex. Mol Cell. 2000;5:133–140. doi: 10.1016/s1097-2765(00)80409-8. [DOI] [PubMed] [Google Scholar]
- 48.Davis L I. The nuclear pore complex. Annu Rev Biochem. 1995;64:865–896. doi: 10.1146/annurev.bi.64.070195.004245. [DOI] [PubMed] [Google Scholar]
- 49.Deane R, Schafer W, Zimmermann H P, Mueller L, Gorlich D, Prehn S, Ponstingl H, Bischoff F R. Ran-binding protein 5 (RanBP5) is related to the nuclear transport factor importin-beta but interacts differently with RanBP1. Mol Cell Biol. 1997;17:5087–5096. doi: 10.1128/mcb.17.9.5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dedhar S, Rennie P S, Shago M, Hagesteijn C Y, Yang H, Filmus J, Hawley R G, Bruchovsky N, Cheng H, Matusik R J, et al. Inhibition of nuclear hormone receptor activity by calreticulin. Nature. 1994;367:480–483. doi: 10.1038/367480a0. [DOI] [PubMed] [Google Scholar]
- 51.Deinert K, Fasiolo F, Hurt E C, Simos G. Arc1p organizes the yeast aminoacyl-tRNA synthetase complex and stabilizes its interaction with the cognate tRNAs. J Biol Chem. 2001;276:6000–6008. doi: 10.1074/jbc.M008682200. [DOI] [PubMed] [Google Scholar]
- 52.DeVit M J, Johnston M. The nuclear exportin Msn5 is required for nuclear export of the Mig1 glucose repressor of Saccharomyces cerevisiae. Curr Biol. 1999;9:1231–1241. doi: 10.1016/s0960-9822(99)80503-x. [DOI] [PubMed] [Google Scholar]
- 53.Dingwall C, Sharnick S V, Laskey R A. A polypeptide domain that specifies migration of nucleoplasmin into the nucleus. Cell. 1982;30:449–458. doi: 10.1016/0092-8674(82)90242-2. [DOI] [PubMed] [Google Scholar]
- 54.Dupont S, Sharova N, DeHoratius C, Virbasius C M, Zhu X, Bukrinskaya A G, Stevenson M, Green M R. A novel nuclear export activity in HIV-1 matrix protein required for viral replication. Nature. 1999;402:681–685. doi: 10.1038/45272. [DOI] [PubMed] [Google Scholar]
- 55.Dworetzky S I, Feldherr C M. Translocation of RNA-coated gold particles through the nuclear pores of oocytes. J Cell Biol. 1988;106:575–584. doi: 10.1083/jcb.106.3.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Elfgang C, Rosorius O, Hofer L, Jaksche H, Hauber J, Bevec D. Evidence for specific nucleocytoplasmic transport pathways used by leucine-rich nuclear export signals. Proc Natl Acad Sci USA. 1999;96:6229–6234. doi: 10.1073/pnas.96.11.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Englmeier L, Olivo J C, Mattaj I W. Receptor-mediated substrate translocation through the nuclear pore complex without nucleotide triphosphate hydrolysis. Curr Biol. 1999;9:30–41. doi: 10.1016/s0960-9822(99)80044-x. [DOI] [PubMed] [Google Scholar]
- 58.Fabre E, Hurt E. Yeast genetics to dissect the nuclear pore complex and nucleocytoplasmic trafficking. Annu Rev Genet. 1997;31:277–313. doi: 10.1146/annurev.genet.31.1.277. [DOI] [PubMed] [Google Scholar]
- 59.Fagotto F, Gluck U, Gumbiner B M. Nuclear localization signal-independent and importin/karyopherin- independent nuclear import of beta-catenin. Curr Biol. 1998;8:181–190. doi: 10.1016/s0960-9822(98)70082-x. [DOI] [PubMed] [Google Scholar]
- 60.Fahrenkrog B, Aris J P, Hurt E C, Pante N, Aebi U. Comparative spatial localization of protein-A-tagged and suthentic yeast nuclear pore complex proteins by immunogold electron microscopy. J Struct Biol. 2000;129:295–305. doi: 10.1006/jsbi.2000.4223. [DOI] [PubMed] [Google Scholar]
- 61.Feldherr C M, Akin D. The location of the transport gate In the nuclear pore complex. J Cell Sci. 1997;110:3065–3070. doi: 10.1242/jcs.110.24.3065. [DOI] [PubMed] [Google Scholar]
- 62.Feng W, Benko A L, Lee J H, Stanford D R, Hopper A K. Antagonistic effects of NES and NLS motifs determine S. cerevisiae Rna1p subcellular distribution. J Cell Sci. 1999;112:339–347. doi: 10.1242/jcs.112.3.339. [DOI] [PubMed] [Google Scholar]
- 63.Ferreira P A, Hom J T, Pak W L. Retina-specifically expressed novel subtypes of bovine cyclophilin. J Biol Chem. 1995;270:23179–23188. doi: 10.1074/jbc.270.39.23179. [DOI] [PubMed] [Google Scholar]
- 64.Ferrigno P, Posas F, Koepp D, Saito H, Silver P A. Regulated nucleo/cytoplasmic exchange of HOG1 MAPK requires the importin beta homologs NMD5 and XPO1. EMBO J. 1998;17:5606–5614. doi: 10.1093/emboj/17.19.5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fischer U, Huber J, Boelens W C, Mattaj I W, Luhrmann R. The HIV-1 Rev activation domain Is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 66.Floer M, Blobel G. The nuclear transport factor karyopherin beta binds stoichiometrically to Ran-GTP and inhibits the Ran GTPase activating protein. J Biol Chem. 1996;271:5313–5316. doi: 10.1074/jbc.271.10.5313. [DOI] [PubMed] [Google Scholar]
- 67.Floer M, Blobel G. Putative reaction intermediates in Crm1-mediated nuclear protein export. J Biol Chem. 1999;274:16279–16286. doi: 10.1074/jbc.274.23.16279. [DOI] [PubMed] [Google Scholar]
- 68.Floer M, Blobel G, Rexach M. Disassembly of RanGTP karyopherinβ complex, an intermediate in nuclear protein import. J Biol Chem. 1997;272:19538–19546. doi: 10.1074/jbc.272.31.19538. [DOI] [PubMed] [Google Scholar]
- 69.Fornerod M, Ohno M, Yoshida M, Mattaj I W. Crm1 Is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 70.Fornerod M, Vandeursen J, Vanbaal S, Reynolds A, Davis D, Murti K G, Fransen J, Grosveld G. The human homologue of yeast Crm1 Is In a dynamic subcomplex with Can/Nup214 and a novel nuclear pore component Nup88. EMBO J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Forrester W, Stutz F, Rosbash M, Wickens M. Defects in mRNA 3′-end formation, transcription initiation, and mRNA transport associated with the yeast mutation prp20: possible coupling of mRNA processing and chromatin structure. Genes Dev. 1992;6:1914–1926. doi: 10.1101/gad.6.10.1914. [DOI] [PubMed] [Google Scholar]
- 72.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. Crm1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 73.Fukuda M, Gotoh I, Gotoh Y, Nishida E. Cytoplasmic localization of mitogen-activated protein kinase kinase directed by its NH2-terminal, leucine-rich short amino acid sequence, which acts as a nuclear export signal. J Biol Chem. 1996;271:20024–20028. doi: 10.1074/jbc.271.33.20024. [DOI] [PubMed] [Google Scholar]
- 74.Gadal O, Strauss D, Kessl J, Trumpower B, Tollervey D, Hurt E. Nuclear export of 60S ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol Cell Biol. 2001;21:3405–3415. doi: 10.1128/MCB.21.10.3405-3415.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gant T M, Wilson K L. Nuclear assembly. Annu Rev Cell Dev Biol. 1997;13:669–695. doi: 10.1146/annurev.cellbio.13.1.669. [DOI] [PubMed] [Google Scholar]
- 76.Gorlich D, Dabrowski M, Bischoff F R, Kutay U, Bork P, Hartmann E, Prehn S, Izaurralde E. A novel class of RanGTP binding proteins. J Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gorlich D, Henklein P, Laskey R A, Hartmann E. A 41 amino acid motif in importin-alpha confers binding to importin- beta and hence transit into the nucleus. EMBO J. 1996;15:1810–1817. [PMC free article] [PubMed] [Google Scholar]
- 78.Gorlich D, Kostka S, Kraft R, Dingwall C, Laskey R A, Hartmann E, Prehn S. Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr Biol. 1995;5:383–392. doi: 10.1016/s0960-9822(95)00079-0. [DOI] [PubMed] [Google Scholar]
- 79.Gorlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 80.Gorlich D, Pante N, Kutay U, Aebi U, Bischoff F R. Identification of different roles for RanGTP and RanGTP In nuclear protein import. EMBO J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- 81.Gorlich D, Prehn S, Laskey R A, Hartmann E. Isolation of a protein that is essential for the first step of nuclear protein import. Cell. 1994;79:767–778. doi: 10.1016/0092-8674(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 82.Gorlich D, Vogel F, Mills A D, Hartmann E, Laskey R A. Distinct functions for the two importin subunits in nuclear protein import. Nature. 1995;377:246–248. doi: 10.1038/377246a0. [DOI] [PubMed] [Google Scholar]
- 83.Grosshans H, Hurt E, Simos G. An aminoacylation-dependent nuclear tRNA export pathway in yeast. Genes Dev. 2000;14:830–840. [PMC free article] [PubMed] [Google Scholar]
- 84.Gruter P, Tabernero C, von Kobbe C, Schmitt C, Saavedra C, Bachi A, Wilm M, Felber B K, Izaurralde E. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- 85.Guan T, Kehlenbach R H, Schirmer E C, Kehlenbach A, Fan F, Clurman B E, Arnheim N, Gerace L. Nup50, a nucleoplasmically oriented nucleoporin with a role in nuclear protein export. Mol Cell Biol. 2000;20:5619–5630. doi: 10.1128/mcb.20.15.5619-5630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Guo Q, Xie J, Dang C V, Liu E T, Bishop J M. Identification of a large Myc-binding protein that contains RCC1-like repeats. Proc Natl Acad Sci USA. 1998;95:9172–9177. doi: 10.1073/pnas.95.16.9172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hellmuth K, Lau D M, Bischoff F R, Kunzler M, Hurt E, Simos G. Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol Cell Biol. 1998;18:6374–6386. doi: 10.1128/mcb.18.11.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Henderson B R, Percipalle P. Interactions between HIV Rev and nuclear import and export factors—the Rev nuclearl localisation signal mediates specific binding to human importin-β. J Mol Biol. 1997;274:693–707. doi: 10.1006/jmbi.1997.1420. [DOI] [PubMed] [Google Scholar]
- 89.Herold A, Suyama M, Rodrigues J P, Braun I C, Kutay U, Carmo-Fonseca M, Bork P, Izaurralde E. TAP (NXF1) belongs to a multigene family of putative RNA export factors with a conserved modular architecture. Mol Cell Biol. 2000;20:8996–9008. doi: 10.1128/mcb.20.23.8996-9008.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Herold A, Truant R, Wiegand H, Cullen B R. Determination of the functional domain organization of the importin alpha nuclear import factor. J Cell Biol. 1998;143:309–318. doi: 10.1083/jcb.143.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hetzer M, Bilbao-Cortes D, Walther T C, Gruss O J, Mattaj I W. GTP hydrolysis by Ran is required for nuclear envelope assembly. Mol Cell. 2000;5:1013–1024. doi: 10.1016/s1097-2765(00)80266-x. [DOI] [PubMed] [Google Scholar]
- 92.Hieda M, Tachibana T, Yokoya F, Kose S, Imamoto N, Yoneda Y. A monoclonal antibody to the COOH-terminal acidic portion of Ran inhibits both the recycling of Ran and nuclear protein import in living cells. J Cell Biol. 1999;144:645–655. doi: 10.1083/jcb.144.4.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hillig R C, Renault L, Vetter I R, Drell T, Wittinghofer A, Becker J. The crystal structure of rna1p: a new fold for a GTPase-activating protein. Mol Cell. 1999;3:781–791. doi: 10.1016/s1097-2765(01)80010-1. [DOI] [PubMed] [Google Scholar]
- 94.Ho J H, Kallstrom G, Johnson A W. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J Cell Biol. 2000;151:1057–1066. doi: 10.1083/jcb.151.5.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hofmann W, Reichart B, Ewald A, Muller E, Schmitt I, Stauber R H, Lottspeich F, Jockusch B M, Scheer U, Hauber J, Dabauvalle M C. Cofactor requirements for nuclear export of Rev response element (RRE)- and constitutive transport element (CTE)-containing retroviral RNAs. An unexpected role for actin. J Cell Biol. 2001;152:895–910. doi: 10.1083/jcb.152.5.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Holaska J M, Black B E, Love D C, Hanover J A, Leszyk J, Paschal B M. Calreticulin is a receptor for nuclear export. J Cell Biol. 2001;152:127–140. doi: 10.1083/jcb.152.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Holaska J M, Paschal B M. A cytosolic activity distinct from crm1 mediates nuclear export of protein kinase inhibitor in permeabilized cells. Proc Natl Acad Sci USA. 1998;95:14739–14744. doi: 10.1073/pnas.95.25.14739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hood J K, Casolari J M, Silver P A. Nup2p is located on the nuclear side of the nuclear pore complex and coordinates Srp1p/importin-α export. J Cell Sci. 2000;113:1471–1480. doi: 10.1242/jcs.113.8.1471. [DOI] [PubMed] [Google Scholar]
- 99.Hopper A K, Traglia H M, Dunst R W. The yeast RNA1 gene product necessary for RNA processing is located in the cytosol and apparently excluded from the nucleus. J Cell Biol. 1990;111:309–321. doi: 10.1083/jcb.111.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Huber J, Cronshagen U, Kadokura M, Marshallsay C, Wada T, Sekine M, Luhrmann R. Snurportin1, an m3G-cap-specific nuclear import receptor with a novel domain structure. EMBO J. 1998;17:4114–4126. doi: 10.1093/emboj/17.14.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hurt E, Strasser K, Segref A, Bailer S, Schlaich N, Presutti C, Tollervey D, Jansen R. Mex67p mediates nuclear export of a variety of RNA polymerase II transcripts. J Biol Chem. 2000;275:8361–8368. doi: 10.1074/jbc.275.12.8361. [DOI] [PubMed] [Google Scholar]
- 102.Iovine M K, Wente S R. Nuclear export signal in Kap95p is required for both recycling the import factor and interaction with the nucleoporin GLFG repeat regions of Nup116p and Nup100p. J Cell Biol. 1997;137:797–811. doi: 10.1083/jcb.137.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Izaurralde E, Kutay U, Vonkobbe C, Mattaj I W, Gorlich D. The asymmetric distribution of the constituents of the ran system is essential for transport into and out of the nucleus. EMBO J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jakel S, Albig W, Kutay U, Bischoff F R, Schwamborn K, Doenecke D, Gorlich D. The importin beta/importin 7 heterodimer is a functional nuclear import receptor for histone H1. EMBO J. 1999;18:2411–2423. doi: 10.1093/emboj/18.9.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jakel S, Gorlich D. Importin β, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 1998;17:4491–4502. doi: 10.1093/emboj/17.15.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jarmolowski A, Boelens W C, Izaurralde E, Mattaj I W. Nuclear export of different classes of RNA is mediated by specific factors. J Cell Biol. 1994;124:627–635. doi: 10.1083/jcb.124.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jullien D, Gorlich D, Laemmli U K, Adachi Y. Nuclear import of RPA in Xenopus egg extracts requires a novel protein XRIPalpha but not importin alpha. EMBO J. 1999;18:4348–4358. doi: 10.1093/emboj/18.15.4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kadowaki T, Goldfarb D, Spitz L M, Tartakoff A M, Ohno M. Regulation of RNA processing and transport by a nuclear guanine nucleotide release protein and members of the Ras superfamily. EMBO J. 1993;12:2929–2937. doi: 10.1002/j.1460-2075.1993.tb05955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kaffman A, O'Shea E K. Regulation of nuclear localization: a key to a door. Annu Rev Cell Dev Biol. 1999;15:291–339. doi: 10.1146/annurev.cellbio.15.1.291. [DOI] [PubMed] [Google Scholar]
- 110.Kaffman A, Rank N M, O'Neill E M, Huang L S, O'Shea E K. The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature. 1998;396:482–486. doi: 10.1038/24898. [DOI] [PubMed] [Google Scholar]
- 111.Kalab P, Pu R T, Dasso M. The Ran GTPase regulates mitotic spindle assembly. Curr Biol. 1999;9:481–484. doi: 10.1016/s0960-9822(99)80213-9. [DOI] [PubMed] [Google Scholar]
- 112.Kalderon D, Roberts B L, Richardson W D, Smith A E. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 113.Kang Y, Cullen B R. The human Tap protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleocytoplasmic transport sequences. Genes Dev. 1999;13:1126–39. doi: 10.1101/gad.13.9.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Katahira J, Strass K, Podtelejnikov A, Mann M, Jung J U, Hurt E. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 1999;18:2593–2609. doi: 10.1093/emboj/18.9.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kataoka N, Bachorik J L, Dreyfuss G. Transportin-SR, a nuclear import receptor for SR proteins. J Cell Biol. 1999;145:1145–1152. doi: 10.1083/jcb.145.6.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kehlenbach R H, Dickmans A, Kehlenbach A, Guan T, Gerace L. A role for RanBP1 in the release of Crm1 from the nuclear pore complex in a terminal step of nuclear export. J Cell Biol. 1999;145:645–657. doi: 10.1083/jcb.145.4.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Keminer O, Peters R. Permeability of single nuclear pores. Biophys J. 1999;77:217–228. doi: 10.1016/S0006-3495(99)76883-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Keminer O, Siebrasse J P, Zerf K, Peters R. Optical recording of signal-mediated protein transport through single nuclear pore complexes. Proc Natl Acad Acad Sci USA. 1999;96:11842–11847. doi: 10.1073/pnas.96.21.11842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kiseleva E, Goldberg M W, Allen T D, Akey C W. Active nuclear pore complexes in Chironomus: visualization of transporter configurations related to mRNP export. J Cell Sci. 1998;111:223–236. doi: 10.1242/jcs.111.2.223. [DOI] [PubMed] [Google Scholar]
- 120.Klebe C, Bischoff F R, Ponstingl H, Wittinghofer A. Interaction of the nuclear GTP-binding protein Ran with its regulatory proteins RCC1 and RanGAP1. Biochemistry. 1995;34:639–647. doi: 10.1021/bi00002a031. [DOI] [PubMed] [Google Scholar]
- 121.Klebe C, Nishimoto T, Wittinghofer F. Functional expression in Escherichia coli of the mitotic regulator proteins p24ran and p45rcc1 and fluorescence measurements of their interaction. Biochemistry. 1993;32:11923–11928. doi: 10.1021/bi00095a023. [DOI] [PubMed] [Google Scholar]
- 122.Klebe C, Prinz H, Wittinghofer A, Goody R S. The kinetic mechanism of Ran-nucleotide exchange catalyzed by RCC1. Biochemistry. 1995;34:12543–12552. doi: 10.1021/bi00039a008. [DOI] [PubMed] [Google Scholar]
- 123.Klemm J D, Beals C R, Crabtree G R. Rapid targeting of nuclear proteins to the cytoplasm. Curr Biol. 1997;7:638–644. doi: 10.1016/s0960-9822(06)00290-9. [DOI] [PubMed] [Google Scholar]
- 124.Kobe B. Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin alpha. Nat Struct Biol. 1999;6:388–397. doi: 10.1038/7625. [DOI] [PubMed] [Google Scholar]
- 125.Kohler M, Ansieau S, Prehn S, Leutz A, Haller H, Hartmann E. Cloning of two novel human importin-alpha subunits and analysis of the expression pattern of the importin-alpha protein family. FEBS Lett. 1997;417:104–108. doi: 10.1016/s0014-5793(97)01265-9. [DOI] [PubMed] [Google Scholar]
- 126.Kohler M, Speck C, Christiansen M, Bischoff F R, Prehn S, Haller H, Gorlich D, Hartmann E. Evidence for distinct substrate specificities of importin alpha family members in nuclear protein import. Mol Cell Biol. 1999;19:7782–7791. doi: 10.1128/mcb.19.11.7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Komeili A, O'Shea E K. Roles of phosphorylation sites in regulating activity of the transcription factor Pho4. Science. 1999;284:977–980. doi: 10.1126/science.284.5416.977. [DOI] [PubMed] [Google Scholar]
- 128.Kose S, Imamoto N, Tachibana T, Shimamoto T, Yoneda Y. Ran-unassisted nuclear migration of a 97-Kd component of nuclear pore-targeting complex. J Cell Biol. 1997;139:841–849. doi: 10.1083/jcb.139.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kudo N, Khochbin S, Nishi K, Kitano K, Yanagida M, Yoshida M, Horinouchi S. Molecular cloning and cell cycle-dependent expression of mammalian Crm1, a protein involved in nuclear export of proteins. J Biol Chem. 1997;272:29742–29751. doi: 10.1074/jbc.272.47.29742. [DOI] [PubMed] [Google Scholar]
- 130.Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner E P, Wolff B, Yoshida M, Horinouchi S. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci USA. 1999;96:9112–9117. doi: 10.1073/pnas.96.16.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kudo N, Wolff B, Sekimoto T, Schreiner E P, Yoneda Y, Yanagida M, Horinouchi S, Yoshida M. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp Cell Res. 1998;242:540–547. doi: 10.1006/excr.1998.4136. [DOI] [PubMed] [Google Scholar]
- 132.Kuhlmann J, Macara I, Wittinghofer A. Dynamic and equilibrium studies on the interaction of ran with its effector, RanBP1. Biochemistry. 1997;36:12027–12035. doi: 10.1021/bi970524k. [DOI] [PubMed] [Google Scholar]
- 133.Kunzler M, Gerstberger T, Stutz F, Bischoff F R, Hurt E. Yeast Ran-binding protein 1 (Yrb1) shuttles between the nucleus and cytoplasm and is exported from the nucleus via a Crm1 (XPO1)-dependent pathway. Mol Cell Biol. 2000;20:4295–4308. doi: 10.1128/mcb.20.12.4295-4308.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kunzler M, Hurt E C. Cse1p functions as the nuclear export receptor for importin alpha in yeast. FEBS Lett. 1998;433:185–190. doi: 10.1016/s0014-5793(98)00892-8. [DOI] [PubMed] [Google Scholar]
- 135.Kutay U, Bischoff F R, Kostka S, Kraft R, Gorlich D. Export of importin alpha from the nucleus is mediated by a specific nuclear transport factor. Cell. 1997;90:1061–1071. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]
- 136.Kutay U, Hartmann E, Treichel N, Calado A, Carmo-Fonseca M, Prehn S, Kraft R, Gorlich D, Bischoff F R. Identification of two novel RanGTP-binding proteins belonging to the importin beta superfamily. J Biol Chem. 2000;275:40163–40168. doi: 10.1074/jbc.M006242200. [DOI] [PubMed] [Google Scholar]
- 137.Kutay U, Izaurralde E, Bischoff F R, Mattaj I W, Gorlich D. Dominant-negative mutants of importin-beta block multiple pathways of import and export through the nuclear pore complex. EMBO J. 1997;16:1153–1163. doi: 10.1093/emboj/16.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kutay U, Lipowsky G, Izaurralde E, Bischoff F R, Schwarzmaier P, Hartmann E, Gorlich D. Identification of a tRNA-specific nuclear export receptor. Mol Cell. 1998;1:359–369. doi: 10.1016/s1097-2765(00)80036-2. [DOI] [PubMed] [Google Scholar]
- 139.Lai M C, Lin R I, Huang S Y, Tsai C W, Tarn W Y. A human importin-beta family protein, transportin-SR2, interacts with the phosphorylated RS domain of SR proteins. J Biol Chem. 2000;275:7950–7957. doi: 10.1074/jbc.275.11.7950. [DOI] [PubMed] [Google Scholar]
- 140.Lane C M, Cushman I, Moore M S. Selective disruption of nuclear import by a functional mutant nuclear transport carrier. J Cell Biol. 2000;151:321–332. doi: 10.1083/jcb.151.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Liker E, Fernandez E, Izaurralde E, Conti E. The structure of the mRNA export factor TAP reveals a cis arrangement of a non-canonical RNP domain and an LRR domain. EMBO J. 2000;19:5587–5598. doi: 10.1093/emboj/19.21.5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lindsay M. RanBP3 is a cofactor for Crm1-mediated nuclear protein export. J Cell Biol. 2001;153:1391–1402. doi: 10.1083/jcb.153.7.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lipowsky G, Bischoff F R, Izaurralde E, Kutay U, Schafer S, Gross H J, Beier H, Gorlich D. Coordination of tRNA nuclear export with processing of tRNA. RNA. 1999;5:539–549. doi: 10.1017/s1355838299982134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lipowsky G, Bischoff F R, Schwarzmaier P, Kraft R, Kostka S, Hartmann E, Kutay U, Gorlich D. Exportin 4: a mediator of a novel nuclear export pathway in higher eukaryotes. EMBO J. 2000;19:4362–4371. doi: 10.1093/emboj/19.16.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Liu J, DeFranco D B. Protracted nuclear export of glucocorticoid receptor limits its turnover and does not require the exportin 1/CRM1-directed nuclear export pathway. Mol Endocrinol. 2000;14:40–51. doi: 10.1210/mend.14.1.0398. [DOI] [PubMed] [Google Scholar]
- 146.Lounsbury K M, Macara I G. Ran-binding protein 1 (RanBP1) forms a ternary complex with Ran and karyopherin beta and reduces Ran GTPase-activating protein (RanGAP) inhibition by karyopherin beta. J Biol Chem. 1997;272:551–555. doi: 10.1074/jbc.272.1.551. [DOI] [PubMed] [Google Scholar]
- 147.Lounsbury K M, Richards S A, Perlungher R R, Macara I G. Ran binding domains promote the interaction of Ran with p97/beta-karyopherin, linking the docking and translocation steps of nuclear import. J Biol Chem. 1996;271:2357–2360. doi: 10.1074/jbc.271.5.2357. [DOI] [PubMed] [Google Scholar]
- 148.Lund E, Dahlberg J E. Proofreading and aminoacylation of tRNAs before export from the nucleus. Science. 1998;282:2082–2085. doi: 10.1126/science.282.5396.2082. [DOI] [PubMed] [Google Scholar]
- 149.Mahajan R, Delphin C, Guan T L, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting Rangap1 to nuclear pore complex protein Ranbp2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 150.Mahanty S K, Wang Y, Farley F W, Elion E A. Nuclear shuttling of yeast scaffold Ste5 is required for its recruitment to the plasma membrane and activation of the mating MAPK cascade. Cell. 1999;98:501–512. doi: 10.1016/s0092-8674(00)81978-9. [DOI] [PubMed] [Google Scholar]
- 151.Malik H S, Eickbush T H, Goldfarb D S. Evolutionary Specialization Of the Nuclear Targeting Apparatus. Proc Natl Acad Sci USA. 1997;94:13738–13742. doi: 10.1073/pnas.94.25.13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Matunis M J, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Maul G G. The nuclear and the cytoplasmic pore complex: structure, dynamics, distribution, and evolution. Int Rev Cytol Suppl. 1977;6:75–186. [PubMed] [Google Scholar]
- 154.Melchior F, Paschal B, Evans J, Gerace L. Inhibition of nuclear protein import by nonhydrolyzable analogues of GTP and identification of the small GTPase Ran/TC4 as an essential transport factor. J Cell Biol. 1993;123:1649–1659. doi: 10.1083/jcb.123.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Michael W M, Choi M Y, Dreyfuss G. A nuclear export signal in hnRNP A1—a signal-mediated, temperature-dependent nuclear protein export pathway. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- 156.Michael W M, Eder P S, Dreyfuss G. The K nuclear shuttling domain—a novel signal for nuclear import and nuclear export In the hnRNP K protein. EMBO J. 1997;16:3587–3598. doi: 10.1093/emboj/16.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Mingot J-M, Kostka S, Kraft R, Hartmann E, Gorlich D. Importin 13: a novel mediator of nuclear import and export. EMBO J. 2001;20:3685–3694. doi: 10.1093/emboj/20.14.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mitsui K, Nakanishi M, Ohtsuka S, Norwood T H, Okabayashi K, Miyamoto C, Tanaka K, Yoshimura A, Ohtsubo M. A novel human gene encoding HECT domain and RCC1-like repeats interacts with cyclins and is potentially regulated by the tumor suppressor proteins. Biochem Biophys Res Commun. 1999;266:115–122. doi: 10.1006/bbrc.1999.1777. [DOI] [PubMed] [Google Scholar]
- 159.Miyamoto Y, Imamoto N, Sekimoto T, Tachibana T, Seki T, Tada S, Enomoto T, Yoneda Y. Differential modes of nuclear localization signal (NLS) recognition by three distinct classes of NLS receptors. J Biol Chem. 1997;272:26375–26381. doi: 10.1074/jbc.272.42.26375. [DOI] [PubMed] [Google Scholar]
- 160.Moore M S, Blobel G. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature. 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- 161.Moore M S, Blobel G. Purification of a Ran-interacting protein that is required for protein import into the nucleus. Proc Natl Acad Sci USA. 1994;91:10212–10216. doi: 10.1073/pnas.91.21.10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Moore M S, Blobel G. The two steps of nuclear import, targeting to the nuclear envelope and translocation through the nuclear pore, require different cytosolic factors. Cell. 1992;69:939–950. doi: 10.1016/0092-8674(92)90613-h. [DOI] [PubMed] [Google Scholar]
- 163.Morehouse H, Buratowski R M, Silver P A, Buratowski S. The importin/karyopherin Kap114 mediates the nuclear import of TATA-binding protein. Proc Natl Acad Sci USA. 1999;96:12542–12547. doi: 10.1073/pnas.96.22.12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Moroianu J, Blobel G, Radu A. Previously identified protein of uncertain function is karyopherin alpha and together with karyopherin beta docks import substrate at nuclear pore complexes. Proc Natl Acad Sci USA. 1995;92:2008–2011. doi: 10.1073/pnas.92.6.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mosammaparast N, Jackson K R, Guo Y, Brame C J, Shabanowitz J, Hunt D F, Pemberton L F. Nuclear import of histone H2A and H2B is mediated by a network of karyopherins. J Cell Biol. 2001;153:251–262. doi: 10.1083/jcb.153.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Murphy R, Wente S R. An RNA-export mediator with an essential nuclear export signal. Nature. 1996;383:357–360. doi: 10.1038/383357a0. [DOI] [PubMed] [Google Scholar]
- 167.Muslin A J, Xing H. 14-3-3 proteins: regulation of subcellular localization by molecular interference. Cell Signalling. 2000;12:703–709. doi: 10.1016/s0898-6568(00)00131-5. [DOI] [PubMed] [Google Scholar]
- 168.Nachury M V, Ryder U W, Lamond A I, Weis K. Cloning and characterization of hSRP1 gamma, a tissue-specific nuclear transport factor. Proc Natl Acad Sci USA. 1998;95:582–587. doi: 10.1073/pnas.95.2.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Nachury M V, Weis K. The direction of transport through the nuclear pore can be inverted. Proc Natl Acad Sci USA. 1999;96:9622–9627. doi: 10.1073/pnas.96.17.9622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Nakielny S, Dreyfuss G. Import and export of the nuclear protein import receptor transportin by a mechanism independent of GTP hydrolysis. Curr Biol. 1998;8:89–95. doi: 10.1016/s0960-9822(98)70039-9. [DOI] [PubMed] [Google Scholar]
- 171.Nakielny S, Dreyfuss G. Nuclear export of proteins and RNAs. Curr Opin Cell Biol. 1997;9:420–429. doi: 10.1016/s0955-0674(97)80016-6. [DOI] [PubMed] [Google Scholar]
- 172.Nemergut M E, Macara I G. Nuclear import of the Ran exchange factor, RCC1, is mediated by at least two distinct mechanisms. J Cell Biol. 2000;149:835–850. doi: 10.1083/jcb.149.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Nemergut M E, Mizzen C A, Stukenberg T, Allis C D, Macara I G. Chromatin docking and exchange activity enhancement of RCC1 by histones H2A/2B. Science. 2001;292:1540–1543. doi: 10.1126/science.292.5521.1540. [DOI] [PubMed] [Google Scholar]
- 174.Nothwang H G, Rensing C, Kubler M, Denich D, Brandl B, Stubanus M, Haaf T, Kurnit D, Hildebrandt F. Identification of a novel Ran binding protein 2 related gene (RANBP2L1) and detection of a gene cluster on human chromosome 2q11–q12. Genomics. 1998;47:383–392. doi: 10.1006/geno.1997.5119. [DOI] [PubMed] [Google Scholar]
- 175.Ohno M, Segref A, Bachi A, Wilm M, Mattaj I W. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell. 2000;101:187–198. doi: 10.1016/S0092-8674(00)80829-6. [DOI] [PubMed] [Google Scholar]
- 176.Ohtsubo M, Okazaki H, Nishimoto T. The RCC1 protein, a regulator for the onset of chromosome condensation locates in the nucleus and binds to DNA. J Cell Biol. 1989;109:1389–1397. doi: 10.1083/jcb.109.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Oki M, Nishimoto T. A protein required for nuclear-protein import, Mog1p, directly interacts with GTP-Gsp1p, the Saccharomyces cerevisiae Ran homologue. Proc Natl Acad Sci USA. 1998;95:15388–15393. doi: 10.1073/pnas.95.26.15388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ossareh-Nazari B, Maison C, Black B E, Levesque L, Paschal B M, Dargemont C. RanGTP-binding protein NXT1 facilitates nuclear export of different classes of RNA in vitro. Mol Cell Biol. 2000;20:4562–4571. doi: 10.1128/mcb.20.13.4562-4571.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ossarehnazari B, Bachelerie F, Dargemont C. Evidence for a role of Crm1 In signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 180.Ouspenski II, Mueller U W, Matynia A, Sazer S, Elledge S J, Brinkley B R. Ran-binding protein-1 is an essential component of the Ran rcc1 molecular switch system in budding yeast. J Biol Chem. 1995;270:1975–1978. doi: 10.1074/jbc.270.5.1975. [DOI] [PubMed] [Google Scholar]
- 181.Palacios I, Hetzer M, Adam S A, Mattaj I W. Nuclear import of U SnRNPs requires importin beta. EMBO J. 1997;16:6783–6792. doi: 10.1093/emboj/16.22.6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Palmeri D, Malim M H. Importin beta can mediate the nuclear import of an arginine-rich nuclear localization signal in the absence of importin alpha. Mol Cell Biol. 1999;19:1218–1225. doi: 10.1128/mcb.19.2.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Pante N, Aebi U. Molecular dissection of the nuclear pore complex. Crit Rev Biochem Mol Biol. 1996;31:153–199. doi: 10.3109/10409239609106583. [DOI] [PubMed] [Google Scholar]
- 184.Pante N, Aebi U. Sequential binding of import ligands to distinct nucleopore regions during their nuclear import. Science. 1996;273:1729–1732. doi: 10.1126/science.273.5282.1729. [DOI] [PubMed] [Google Scholar]
- 185.Paraskeva E, Izaurralde E, Bischoff F R, Huber J, Kutay U, Hartmann E, Luhrmann R, Gorlich D. CRM1-mediated recycling of snurportin 1 to the cytoplasm. J Cell Biol. 1999;145:255–264. doi: 10.1083/jcb.145.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Parker F, Maurier F, Delumeau I, Duchesne M, Faucher D, Debussche L, Dugue A, Schweighoffer F, Tocque B. A Ras-GTPase-activating protein SH3-domain-binding protein. Mol Cell Biol. 1996;16:2561–2569. doi: 10.1128/mcb.16.6.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Paschal B M, Delphin C, Gerace L. Nucleotide-specific interaction of Ran/TC4 with nuclear transport factors NTF2 and p97. Proc Natl Acad Sci USA. 1996;93:7679–7683. doi: 10.1073/pnas.93.15.7679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Paschal B M, Fritze C, Guan T, Gerace L. High levels of the GTPase Ran/Tc4 relieve the requirement for nuclear protein transport factor 2. J Biol Chem. 1997;272:21534–21539. doi: 10.1074/jbc.272.34.21534. [DOI] [PubMed] [Google Scholar]
- 189.Paschal B M, Gerace L. Identification of NTF2, a cytosolic factor for nuclear import that interacts with nuclear pore complex protein p62. J Cell Biol. 1995;129:925–937. doi: 10.1083/jcb.129.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Pasquinelli A E, Ernst R K, Lund E, Grimm C, Zapp M L, Rekosh D, Hammarskjold M L, Dahlberg J E. The constitutive transport element (CTE) of Mason-Pfizer monkey virus (MPMV) accesses a cellular mRNA export pathway. EMBO J. 1997;16:7500–7510. doi: 10.1093/emboj/16.24.7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Pemberton L F, Blobel G, Rosenblum J S. Transport routes through the nuclear pore complex. Curr Opin Cell Biol. 1998;10:392–399. doi: 10.1016/s0955-0674(98)80016-1. [DOI] [PubMed] [Google Scholar]
- 192.Pemberton L F, Rosenblum J S, Blobel G. A distinct and parallel pathway for the nuclear import of an mRNA-binding protein. J Cell Biol. 1997;139:1645–1653. doi: 10.1083/jcb.139.7.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Pemberton L F, Rosenblum J S, Blobel G. Nuclear import of the TATA-binding protein: mediation by the karyopherin Kap114p and a possible mechanism for intranuclear targeting. J Cell Biol. 1999;145:1407–1417. doi: 10.1083/jcb.145.7.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Plafker K, Macara I G. Facilitated nucleocytoplasmic shuttling of the Ran binding protein RanBP1. Mol Cell Biol. 2000;20:3510–3521. doi: 10.1128/mcb.20.10.3510-3521.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Plafker S M, Macara I G. Importin-11, a nuclear import receptor for the ubiquitin-conjugating enzyme, UbcM2. EMBO J. 2000;19:5502–5513. doi: 10.1093/emboj/19.20.5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Pollard V W, Michael W M, Nakielny S, Siomi M C, Wang F, Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- 197.Quimby B B, Lamitina T, L'Hernault S W, Corbett A H. The mechanism of Ran import into the nucleus by nuclear transport factor 2. J Biol Chem. 2000;275:28575–28582. doi: 10.1074/jbc.M005055200. [DOI] [PubMed] [Google Scholar]
- 198.Radu A, Blobel G, Moore M S. Identification of a protein complex that is required for nuclear protein import and mediates docking of import substrate to distinct nucleoporins. Proc Natl Acad Sci USA. 1995;92:1769–1773. doi: 10.1073/pnas.92.5.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Radu A, Moore M S, Blobel G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 1995;81:215–222. doi: 10.1016/0092-8674(95)90331-3. [DOI] [PubMed] [Google Scholar]
- 200.Reichelt R, Holzenburg A, Buhle E L, Jr, Jarnik M, Engel A, Aebi U. Correlation between structure and mass distribution of the nuclear pore complex and of distinct pore complex components. J Cell Biol. 1990;110:883–894. doi: 10.1083/jcb.110.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Renault L, Kuhlmann J, Henkel A, Wittinghofer A. Structural basis for guanine nucleotide exchange on Ran by the regulator of chromosome condensation (RCC1) Cell. 2001;105:245–255. doi: 10.1016/s0092-8674(01)00315-4. [DOI] [PubMed] [Google Scholar]
- 202.Renault L, Nassar N, Vetter I, Becker J, Klebe C, Roth M, Wittinghofer A. The 1.7 A crystal structure of the regulator of chromosome condensation (RCC1) reveals a seven-bladed propeller. Nature. 1998;392:97–101. doi: 10.1038/32204. [DOI] [PubMed] [Google Scholar]
- 203.Rexach M, Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- 204.Ribbeck K, Gorlich D. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 2001;20:1320–1330. doi: 10.1093/emboj/20.6.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Ribbeck K, Kutay U, Paraskeva E, Gorlich D. The translocation of transportin-cargo complexes through nuclear pores is independent of both Ran and energy. Curr Biol. 1999;9:47–50. doi: 10.1016/s0960-9822(99)80046-3. [DOI] [PubMed] [Google Scholar]
- 206.Ribbeck K, Lipowsky G, Kent H M, Stewart M, Gorlich D. NTF2 mediates nuclear import of Ran. EMBO J. 1998;17:6587–6598. doi: 10.1093/emboj/17.22.6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Richards S A, Carey K L, Macara I G. Requirement of guanosine triphosphate-bound Ran for signal-mediated nuclear protein export. Science. 1997;276:1842–1844. doi: 10.1126/science.276.5320.1842. [DOI] [PubMed] [Google Scholar]
- 208.Richards S A, Lounsbury K M, Carey K L, Macara I G. A nuclear export signal is essential for the cytosolic localization of the Ran binding protein, RanBP1. J Cell Biol. 1996;134:1157–1168. doi: 10.1083/jcb.134.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Richards S A, Lounsbury K M, Macara I G. The C terminus of the nuclear RAN/TC4 GTPase stabilizes the GDP-bound state and mediates interactions with RCC1, RAN-GAP, and HTF9A/RANBP1. J Biol Chem. 1995;270:14405–14411. doi: 10.1074/jbc.270.24.14405. [DOI] [PubMed] [Google Scholar]
- 210.Robbins J, Dilworth S M, Laskey R A, Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- 211.Rodriguez M S, Desterro J M, Lain S, Midgley C A, Lane D P, Hay R T. SUMO-1 modification activates the transcriptional response of p53. EMBO J. 1999;18:6455–6461. doi: 10.1093/emboj/18.22.6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Rosa J L, Casaroli-Marano R P, Buckler A J, Vilaro S, Barbacid M. p619, a giant protein related to the chromosome condensation regulator RCC1, stimulates guanine nucleotide exchange on ARF1 and Rab proteins. EMBO J. 1996;15:4262–4273. [PMC free article] [PubMed] [Google Scholar]
- 213.Rosenblum J S, Pemberton L F, Blobel G. A nuclear import pathway for a protein involved in tRNA maturation. J Cell Biol. 1997;139:1655–1661. doi: 10.1083/jcb.139.7.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Rosorius O, Reichart B, Kratzer F, Heger P, Dabauvalle M C, Hauber J. Nuclear pore localization and nucleocytoplasmic transport of eIF-5A: evidence for direct interaction with the export receptor CRM1. J Cell Sci. 1999;112:2369–2380. doi: 10.1242/jcs.112.14.2369. [DOI] [PubMed] [Google Scholar]
- 215.Rout M P, Aitchison J D, Suprapto A, Hjertaas K, Zhao Y, Chait B T. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Rout M P, Blobel G. Isolation of the yeast nuclear pore complex. J Cell Biol. 1993;123:771–783. doi: 10.1083/jcb.123.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Rout M P, Blobel G, Aitchison J D. A distinct nuclear import pathway used by ribosomal proteins. Cell. 1997;89:715–725. doi: 10.1016/s0092-8674(00)80254-8. [DOI] [PubMed] [Google Scholar]
- 218.Saavedra C, Felber B, Izaurralde E. The simian retrovirus-1 constitutive transport element, unlike the HIV-1 RRE, uses factors required for cellular mRNA export. Curr Biol. 1997;7:619–628. doi: 10.1016/s0960-9822(06)00288-0. [DOI] [PubMed] [Google Scholar]
- 219.Sachdev S, Bagchi S, Zhang D D, Mings A C, Hannink M. Nuclear import of IkappaBalpha is accomplished by a Ran-independent transport pathway. Mol Cell Biol. 2000;20:1571–1582. doi: 10.1128/mcb.20.5.1571-1582.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Saitoh H, Sparrow D B, Shiomi T, Pu R T, Nishimoto T, Mohun T J, Dasso M. Ubc9p and the conjugation of SUMO-1 to RanGAP1 and RanBP2. Curr Biol. 1998;8:121–124. doi: 10.1016/s0960-9822(98)70044-2. [DOI] [PubMed] [Google Scholar]
- 221.Santos-Rosa H, Moreno H, Simos G, Segref A, Fahrenkrog B, Pante N, Hurt E. Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol Cell Biol. 1998;18:6826–6838. doi: 10.1128/mcb.18.11.6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Sarkar S, Hopper A K. tRNA nuclear export in saccharomyces cerevisiae: in situ hybridization analysis. Mol Biol Cell. 1998;9:3041–3055. doi: 10.1091/mbc.9.11.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Savory J G, Hsu B, Laquian I R, Giffin W, Reich T, Hache R J, Lefebvre Y A. Discrimination between NL1- and NL2-mediated nuclear localization of the glucocorticoid receptor. Mol Cell Biol. 1999;19:1025–1037. doi: 10.1128/mcb.19.2.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Schaap P J, van't Riet J, Woldringh C L, Raue H A. Identification and functional analysis of the nuclear localization signals of ribosomal protein L25 from Saccharomyces cerevisiae. J Mol Biol. 1991;221:225–237. doi: 10.1016/0022-2836(91)80216-h. [DOI] [PubMed] [Google Scholar]
- 225.Schaff J C, Slepchenko B M, Loew L M. Physiological modeling with virtual cell framework. Methods Enzymol. 2000;321:1–23. doi: 10.1016/s0076-6879(00)21184-1. [DOI] [PubMed] [Google Scholar]
- 226.Scheffzek K, Ahmadian M R, Wittinghofer A. GTPase-activating proteins: helping hands to complement an active site. Trends Biochem Sci. 1998;23:257–262. doi: 10.1016/s0968-0004(98)01224-9. [DOI] [PubMed] [Google Scholar]
- 227.Schlenstedt G, Smirnova E, Deane R, Solsbacher J, Kutay U, Gorlich D, Ponstingl H, Bischoff F R. Yrb4p, a Yeast Ran-GTP-binding protein involved in import of ribosomal protein L25 into the nucleus. EMBO J. 1997;16:6237–6249. doi: 10.1093/emboj/16.20.6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Seedorf M, Damelin M, Kahana J, Taura T, Silver P A. Interactions between a nuclear transporter and a subset of nuclear pore complex proteins depend on Ran GTPase. Mol Cell Biol. 1999;19:1547–1557. doi: 10.1128/mcb.19.2.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Seedorf M, Silver P A. Importin/karyopherin protein family members required for mRNA export from the nucleus. Proc Natl Acad Sci USA. 1997;94:8590–8595. doi: 10.1073/pnas.94.16.8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Segref A, Sharma K, Doye V, Hellwig A, Huber J, Luhrmann R, Hurt E. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 1997;16:3256–3271. doi: 10.1093/emboj/16.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Seimiya H, Sawada H, Muramatsu Y, Shimizu M, Ohko K, Yamane K, Tsuruo T. Involvement of 14-3-3 proteins in nuclear localization of telomerase. EMBO J. 2000;19:2652–2661. doi: 10.1093/emboj/19.11.2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Sekimoto T, Imamoto N, Nakajima K, Hirano T, Yoneda Y. Extracellular signal-dependent nuclear import of Stat1 is mediated by nuclear pore-targeting complex formation with Npi-1, but not Rch1. EMBO J. 1997;16:7067–7077. doi: 10.1093/emboj/16.23.7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Senger B, Simos G, Bischoff F R, Podtelejnikov A, Mann M, Hurt E. Mtr10p functions as a nuclear import receptor for the mRNA-binding protein Npl3p. EMBO J. 1998;17:2196–2207. doi: 10.1093/emboj/17.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Shah S, Forbes D J. Separate nuclear import pathways converge on the nucleoporin nup153 and can be dissected with dominant-negative inhibitors. Curr Biol. 1998;8:1376–1386. doi: 10.1016/s0960-9822(98)00018-9. [DOI] [PubMed] [Google Scholar]
- 235.Shah S, Tugendreich S, Forbes D. Major binding sites for the nuclear import receptor are the internal nucleoporin Nup153 and the adjacent nuclear filament protein Tpr. J Cell Biol. 1998;141:31–49. doi: 10.1083/jcb.141.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Shimada Y, Gulli M P, Peter M. Nuclear sequestration of the exchange factor Cdc24 by Far1 regulates cell polarity during yeast mating. Nat Cell Biol. 2000;2:117–124. doi: 10.1038/35000073. [DOI] [PubMed] [Google Scholar]
- 237.Simos G, Hurt E. Transfer RNA biogenesis: a visa to leave the nucleus. Curr Biol. 1999;9:R238–R241. doi: 10.1016/s0960-9822(99)80152-3. [DOI] [PubMed] [Google Scholar]
- 238.Smith A, Brownawell A, Macara I G. Nuclear import of Ran:GDP is mediated by NTF2. Curr Biol. 1998;8:1403–1406. doi: 10.1016/s0960-9822(98)00023-2. [DOI] [PubMed] [Google Scholar]
- 239.Solsbacher J, Maurer P, Bischoff F R, Schlenstedt G. Cse1p is involved in export of yeast importin alpha from the nucleus. Mol Cell Biol. 1998;18:6805–6815. doi: 10.1128/mcb.18.11.6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Solsbacher J, Maurer P, Vogel F, Schlenstedt G. Nup2p, a yeast nucleoporin, functions in bidirectional transport of importin alpha. Mol Cell Biol. 2000;20:8468–8479. doi: 10.1128/mcb.20.22.8468-8479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Stade K, Ford C S, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 242.Stage-Zimmermann T, Schmidt U, Silver P A. Factors affecting nuclear export of the 60S ribosomal subunit in vivo. Mol Biol Cell. 2000;11:3777–3789. doi: 10.1091/mbc.11.11.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Steggerda S M, Black B E, Paschal B M. Monoclonal antibodies to NTF2 inhibit nuclear protein import by preventing nuclear translocation of the GTPase Ran. Mol Biol Cell. 2000;11:703–719. doi: 10.1091/mbc.11.2.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Steggerda S M, Paschal B M. The mammalian Mog1 protein is a guanine nucleotide release factor for Ran. J Biol Chem. 2000;275:23175–23180. doi: 10.1074/jbc.C000252200. [DOI] [PubMed] [Google Scholar]
- 245.Stewart M, Baker R P. 1.9 A resolution crystal structure of the Saccharomyces cerevisiae Ran- binding protein Mog1p. J Mol Biol. 2000;299:213–223. doi: 10.1006/jmbi.2000.3733. [DOI] [PubMed] [Google Scholar]
- 246.Stewart M, Kent H M, McCoy A J. Structural basis for molecular recognition between nuclear transport factor 2 (NTF2) and the GDP-bound form of the Ras-family GTPase Ran. J Mol Biol. 1998;277:635–646. doi: 10.1006/jmbi.1997.1602. [DOI] [PubMed] [Google Scholar]
- 247.Stoffler D, Fahrenkrog B, Aebi U. The nuclear pore complex: from molecular architecture to functional dynamics. Curr Opin Cell Biol. 1999;11:391–401. doi: 10.1016/S0955-0674(99)80055-6. [DOI] [PubMed] [Google Scholar]
- 248.Stoffler D, Goldie K N, Feja B, Aebi U. Calcium-mediated structural changes of native nuclear pore complexes monitored by time-lapse atomic force microscopy. J Mol Biol. 1999;287:741–752. doi: 10.1006/jmbi.1999.2637. [DOI] [PubMed] [Google Scholar]
- 249.Strambio-de-Castillia C, Blobel G, Rout M P. Proteins connecting the nuclear pore complex with the nuclear interior. J Cell Biol. 1999;144:839–855. doi: 10.1083/jcb.144.5.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Strasser K, Bassler J, Hurt E. Binding of the Mex67p/Mtr2p heterodimer to FXFG, GLFG, and FG repeat nucleoporins is essential for nuclear mRNA export. J Cell Biol. 2000;150:695–706. doi: 10.1083/jcb.150.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Strasser K, Hurt E. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 2000;19:410–420. doi: 10.1093/emboj/19.3.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Suyama M, Doerks T, Braun I C, Sattler M, Izaurralde E, Bork P. Prediction of structural domains of TAP reveals details of its interaction with p15 and nucleoporins. EMBO Rep. 2000;1:53–58. doi: 10.1093/embo-reports/kvd009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Takaishi H, Konishi H, Matsuzaki H, Ono Y, Shirai Y, Saito N, Kitamura T, Ogawa W, Kasuga M, Kikkawa U, Nishizuka Y. Regulation of nuclear translocation of forkhead transcription factor AFX by protein kinase B. Proc Natl Acad Sci USA. 1999;96:11836–11841. doi: 10.1073/pnas.96.21.11836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Takizawa C G, Weis K, Morgan D O. Ran-independent nuclear import of cyclin B1-Cdc2 by importin beta. Proc Natl Acad Sci USA. 1999;96:7938–7943. doi: 10.1073/pnas.96.14.7938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Talcott B, Moore M S. The nuclear import of RCC1 requires a specific nuclear localization sequence receptor, karyopherin alpha3/Qip. J Biol Chem. 2000;275:10099–10104. doi: 10.1074/jbc.275.14.10099. [DOI] [PubMed] [Google Scholar]
- 256.Taura T, Krebber H, Silver P A. A member of the Ran-binding protein family, Yrb2p, is involved in nuclear protein export. Proc Natl Acad Sci USA. 1998;95:7427–7432. doi: 10.1073/pnas.95.13.7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Titov A A, Blobel G. The karyopherin Kap122p/Pdr6p imports both subunits of the transcription factor IIA into the nucleus. J Cell Biol. 1999;147:235–246. doi: 10.1083/jcb.147.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Truant R, Cullen B R. The arginine-rich domains present in human immunodeficiency virus type 1 Tat and Rev function as direct importin beta-dependent nuclear localization signals. Mol Cell Biol. 1999;19:1210–1217. doi: 10.1128/mcb.19.2.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Truant R, Kang Y, Cullen B R. The human tap nuclear RNA export factor contains a novel transportin-dependent nuclear localization signal that lacks nuclear export signal function. J Biol Chem. 1999;274:32167–32171. doi: 10.1074/jbc.274.45.32167. [DOI] [PubMed] [Google Scholar]
- 260.Tsuji L, Takumi T, Imamoto N, Yoneda Y. Identification of novel homologues of mouse importin alpha, the alpha aubunit of the nuclear pore-targeting complex, and their tissue-specific expression. FEBS Lett. 1997;416:30–34. doi: 10.1016/s0014-5793(97)01092-2. [DOI] [PubMed] [Google Scholar]
- 261.Turpin P, Hay R T, Dargemont C. Characterization of IkBα nuclear import pathway. J Biol Chem. 1999;274:6804–6812. doi: 10.1074/jbc.274.10.6804. [DOI] [PubMed] [Google Scholar]
- 262.Uchida S, Sekiguchi T, Nishitani H, Miyauchi K, Ohtsubo M, Nishimoto T. Premature chromosome condensation is induced by a point mutation in the hamster RCC1 gene. Mol Cell Biol. 1990;10:577–584. doi: 10.1128/mcb.10.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Vetter I R, Arndt A, Kutay U, Gorlich D, Wittinghofer A. Structural view of the Ran-Importin beta interaction at 2.3 A resolution. Cell. 1999;97:635–646. doi: 10.1016/s0092-8674(00)80774-6. [DOI] [PubMed] [Google Scholar]
- 264.Vetter I R, Nowak C, Nishimoto T, Kuhlmann J, Wittinghofer A. Structure of a Ran-binding domain complexed with Ran bound to a GTP analogue: implications for nuclear transport. Nature. 1999;398:39–46. doi: 10.1038/17969. [DOI] [PubMed] [Google Scholar]
- 265.Visa N, Alzhanova-Ericsson A T, Sun X, Kiseleva E, Bjorkroth B, Wurtz T, Daneholt B. A pre-mRNA-binding protein accompanies the RNA from the gene through the nuclear pores and into polysomes. Cell. 1996;84:253–264. doi: 10.1016/s0092-8674(00)80980-0. [DOI] [PubMed] [Google Scholar]
- 266.Weis K, Mattaj I W, Lamond A I. Identification of hSRP1 alpha as a functional receptor for nuclear localization sequences. Science. 1995;268:1049–1053. doi: 10.1126/science.7754385. [DOI] [PubMed] [Google Scholar]
- 267.Weis K, Ryder U, Lamond A I. The conserved amino-terminal domain of hSRP1 alpha is essential for nuclear protein import. EMBO J. 1996;15:1818–1825. [PMC free article] [PubMed] [Google Scholar]
- 268.Welch K, Franke J, Kohler M, Macara I G. RanBP3 contains an unusual nuclear localization signal that is imported preferentially by importin-alpha3. Mol Cell Biol. 1999;19:8400–8411. doi: 10.1128/mcb.19.12.8400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Wen W, Meinkoth J L, Tsien R Y, Taylor S S. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 270.Wente S R. Gatekeepers of the nucleus. Science. 2000;288:1374–1377. doi: 10.1126/science.288.5470.1374. [DOI] [PubMed] [Google Scholar]
- 271.Wiese C, Goldberg M W, Allen T D, Wilson K L. Nuclear envelope assembly in Xenopus extracts visualized by scanning EM reveals a transport-dependent ‘envelope smoothing’ event. J Cell Sci. 1997;110:1489–1502. doi: 10.1242/jcs.110.13.1489. [DOI] [PubMed] [Google Scholar]
- 272.Wiese C, Wilde A, Moore M S, Adam S A, Merdes A, Zheng Y. Role of importin-β in coupling Ran to downstream targets in microtubule assembly. Science. 2001;291:653–656. doi: 10.1126/science.1057661. [DOI] [PubMed] [Google Scholar]
- 273.Wilde A, Zheng Y. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science. 1999;284:1359–1362. doi: 10.1126/science.284.5418.1359. [DOI] [PubMed] [Google Scholar]
- 274.Wolff B, Sanglier J J, Wang Y. Leptomycin B Is an inhibitor of nuclear export — inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem Biol. 1997;4:139–147. doi: 10.1016/s1074-5521(97)90257-x. [DOI] [PubMed] [Google Scholar]
- 275.Wong D H, Corbett A H, Kent H M, Stewart M, Silver P A. Interaction between the small GTPase Ran/Gsp1p and NTF2p Is required for nuclear transport. Mol Cell Biol. 1997;17:3755–3767. doi: 10.1128/mcb.17.7.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Wozniak R W, Rout M P, Aitchison J D. Karyopherins and kissing cousins. Trends Cell Biol. 1998;8:184–188. doi: 10.1016/s0962-8924(98)01248-3. [DOI] [PubMed] [Google Scholar]
- 277.Wu J, Matunis M J, Kraemer D, Blobel G, Coutavas E. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran- GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J Biol Chem. 1995;270:14209–14213. doi: 10.1074/jbc.270.23.14209. [DOI] [PubMed] [Google Scholar]
- 278.Yamashita A, Watanabe Y, Nukina N, Yamamoto M. RNA-assisted nuclear transport of the meiotic regulator Mei2p in fission yeast. Cell. 1998;95:115–123. doi: 10.1016/s0092-8674(00)81787-0. [DOI] [PubMed] [Google Scholar]
- 279.Yan C, Lee L H, Davis L I. Crm1p mediates regulated nuclear export of a yeast AP-1-like transcription factor. EMBO J. 1998;17:7416–7429. doi: 10.1093/emboj/17.24.7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Yang Q, Rout M P, Akey C W. Three-dimensional architecture of the isolated yeast nuclear pore complex — functional and evolutionary implications. Mol Cell. 1998;1:223–234. doi: 10.1016/s1097-2765(00)80023-4. [DOI] [PubMed] [Google Scholar]
- 281.Yaseen N R, Blobel G. Cloning and characterization of human karyopherin beta-3. Proc Natl Acad Sci USA. 1997;94:4451–4456. doi: 10.1073/pnas.94.9.4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Yokoya F, Imamoto N, Tachibana T, Yoneda Y. β-catenin can be transported into the nucleus in a Ran-unassisted manner. Mol Biol Cell. 1999;10:1119–1131. doi: 10.1091/mbc.10.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Yokoyama N, Hayashi N, Seki T, Pante N, Ohba T, Nishii K, Kuma K, Hayashida T, Miyata T, Aebi U, et al. A giant nucleopore protein that binds Ran/TC4. Nature. 1995;376:184–188. doi: 10.1038/376184a0. [DOI] [PubMed] [Google Scholar]
- 284.York J D, Odom A R, Murphy R, Ives E B, Wente S R. A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science. 1999;285:96–100. doi: 10.1126/science.285.5424.96. [DOI] [PubMed] [Google Scholar]
- 285.Yoshida K, Blobel G. The karyopherin Kap142p/Msn5p mediates nuclear import and nuclear export of different cargo proteins. J Cell Biol. 2001;152:729–740. doi: 10.1083/jcb.152.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Zhang C, Clarke P R. Chromatin-independent nuclear envelope assembly induced by Ran GTPase in Xenopus egg extracts. Science. 2000;288:1429–1432. doi: 10.1126/science.288.5470.1429. [DOI] [PubMed] [Google Scholar]
- 287.Zhang C, Hughes M, Clarke P R. Ran-GTP stabilises microtubule asters and inhibits nuclear assembly in Xenopus egg extracts. J Cell Sci. 1999;112:2453–2461. doi: 10.1242/jcs.112.14.2453. [DOI] [PubMed] [Google Scholar]
- 288.Zhu J, McKeon F. NF-AT activation requires suppression of Crm1-dependent export by calcineurin. Nature. 1999;398:256–260. doi: 10.1038/18473. [DOI] [PubMed] [Google Scholar]
- 289.Zolotukhin A S, Felber B K. Mutations In the nuclear export signal of human Ran-binding protein RanBP1 block the Rev-mediated posttranscriptional regulation of human immunodeficiency virus type 1. J Biol Chem. 1997;272:11356–11360. doi: 10.1074/jbc.272.17.11356. [DOI] [PubMed] [Google Scholar]