Abstract
Immunoglobulin (Ig) superfamily proteins play diverse roles in vertebrates, including regulation of cellular responses by sensing endogenous or exogenous ligands. Siglecs are a family of glycan-recognizing proteins belonging to the Ig superfamily (i.e., I-type lectins). Siglecs are expressed on various leukocyte types and are involved in diverse aspects of immunity, including the regulation of inflammatory responses, leukocyte proliferation, host–microbe interaction, and cancer immunity. Sialoadhesin/Siglec-1, CD22/Siglec-2, and myelin-associated glycoprotein/Siglec-4 were among the first to be characterized as members of the Siglec family, and along with Siglec-15, they are relatively well-conserved among tetrapods. Conversely, CD33/Siglec-3-related Siglecs (CD33rSiglecs, so named as they show high sequence similarity with CD33/Siglec-3) are encoded in a gene cluster with many interspecies variations and even intraspecies variations within some lineages such as humans. The rapid evolution of CD33rSiglecs expressed on leukocytes involved in innate immunity likely reflects the selective pressure by pathogens that interact and possibly exploit these Siglecs. Human Siglecs have several additional unique and/or polymorphic properties as compared with closely related great apes, changes possibly related to the loss of the sialic acid Neu5Gc, another distinctly human event in sialobiology. Multiple changes in human CD33rSiglecs compared to great apes include many examples of human-specific expression in non-immune cells, coinciding with human-specific diseases involving such cell types. Some Siglec gene polymorphisms have dual consequences—beneficial in a situation but detrimental in another. The association of human Siglec gene polymorphisms with several infectious and non-infectious diseases likely reflects the ongoing competition between the host and microbial pathogens.
Keywords: Siglec, sialic acid, evolution, host-pathogen interaction, paired receptors, genetic polymorphism
1. Introduction
The immunoglobulin superfamily (IgSF) of proteins is defined by the presence of one or more domains that structurally resemble the immunoglobulin (Ig) fold (i.e., β-sandwich structure reinforced by an intersheet disulfide bond) found in immunoglobulins (i.e., antibodies) (Smith and Xue, 1997). The human genome contains ~500 genes encoding IgSF proteins (Human Genome Nomenclature Committee, https://www.genenames.org/data/genegroup/#!/group/589), accounting for ~2.5% of all protein-coding genes in the human genome (Willyard, 2018). Many IgSF proteins are expressed by various leukocytes and are involved in the recognition of endogenous or exogenous ligands that belong to various chemical classes (e.g., proteins, lipids, and sugars). Some IgSF proteins recognize glycans and are collectively called I-type lectins (Angata and Brinkman-Van der Linden, 2002; Angata et al., 2022; Powell and Varki, 1995). Thus far, the largest subgroup of such I-type lectins known is the Siglec family, which is defined by several shared features, including mutual sequence similarity, recognition of glycans containing sialic acids (Sias), presence of a conserved Arg residue that engages such Sia-bearing ligands, and a unique arrangement of Cys residues (not conserved in Siglec-15—to be discussed later).
Sias are a group of acidic sugars with a common 9-carbon backbone that are found at the outermost ends (nonreducing termini) of glycans (Angata and Varki, 2002). They are abundant in the tissues of animals of deuterostome lineage (including vertebrates and echinoderms) but generally absent in other multicellular organisms (e.g., worms, insects, and plants), and only a minority of microbes are capable of synthesizing glycoconjugates containing Sias. This restricted distribution of Sias in the living world makes them good molecular indicators for self-associated molecular patterns (SAMPs) for the deuterostome immune system to distinguish between the cells that belong to the host and to invading microbes. This may be one of the reasons why Siglecs have persisted during vertebrate evolution—to distinguish between own cells and microbes using Sia as a SAMP, preventing leukocyte attack on self cells and tissues (Varki, 2011). Conversely, certain microbial pathogens and symbionts manifest molecular mimicry of sialylated Siglec ligands, largely via convergent evolution.
Figure 1 shows a schematic representation of the Siglec family in humans, some nonhuman primates, and mice. Siglecs are type 1 transmembrane proteins, which contain extracellular regions consisting of multiple Ig-like domains, followed by a single-pass transmembrane domain and a short cytoplasmic tail. Alternative splicing or proteolytic cleavage may generate soluble form(s) (Huang et al., 2018). Alternative splicing may also yield protein isoforms with a different number of Ig-like domains (Kitzig et al., 2002; Wang and Neumann, 2010) or with a different length of cytoplasmic tail (Aizawa et al., 2002; Lai et al., 1987). Extracellular domains, in particular the amino-terminal Ig-like domain belonging to so-called V-set Ig-like domain (Smith and Xue, 1997), recognize glycans containing Sia. A conserved Arg residue in this domain is essential for Sia recognition (Supplementary Figure 1). An inter-domain disulfide bond tethering the first and second Ig-like domains of Siglecs is conserved among most Siglecs. Cytoplasmic domain of most Siglecs has one or more Tyr residues embedded in the sequence motif called immunoreceptor tyrosine-based inhibitory motifs (ITIMs), which upon phosphorylation recruits downstream effector protein(s), such as protein tyrosine phosphatase SHP-1 (gene: PTPN6) or SHP-2 (gene: PTPN11). Recruitment of SHP-1 generally results in the downregulation of cell-activating signaling elicited by other receptor proteins coupled with protein tyrosine kinases, and thus Siglecs are generally considered inhibitory receptors. An exception to this general scheme is to be discussed later. It is also a common feature that each structural element of Siglec proteins (i.e., signal peptide, each Ig-like domain, transmembrane domain) is encoded by a single exon, although there are some exceptions. Implications of this “modular design” in the evolution of Siglecs is discussed further.
Figure 1. Mammalian Siglecs.
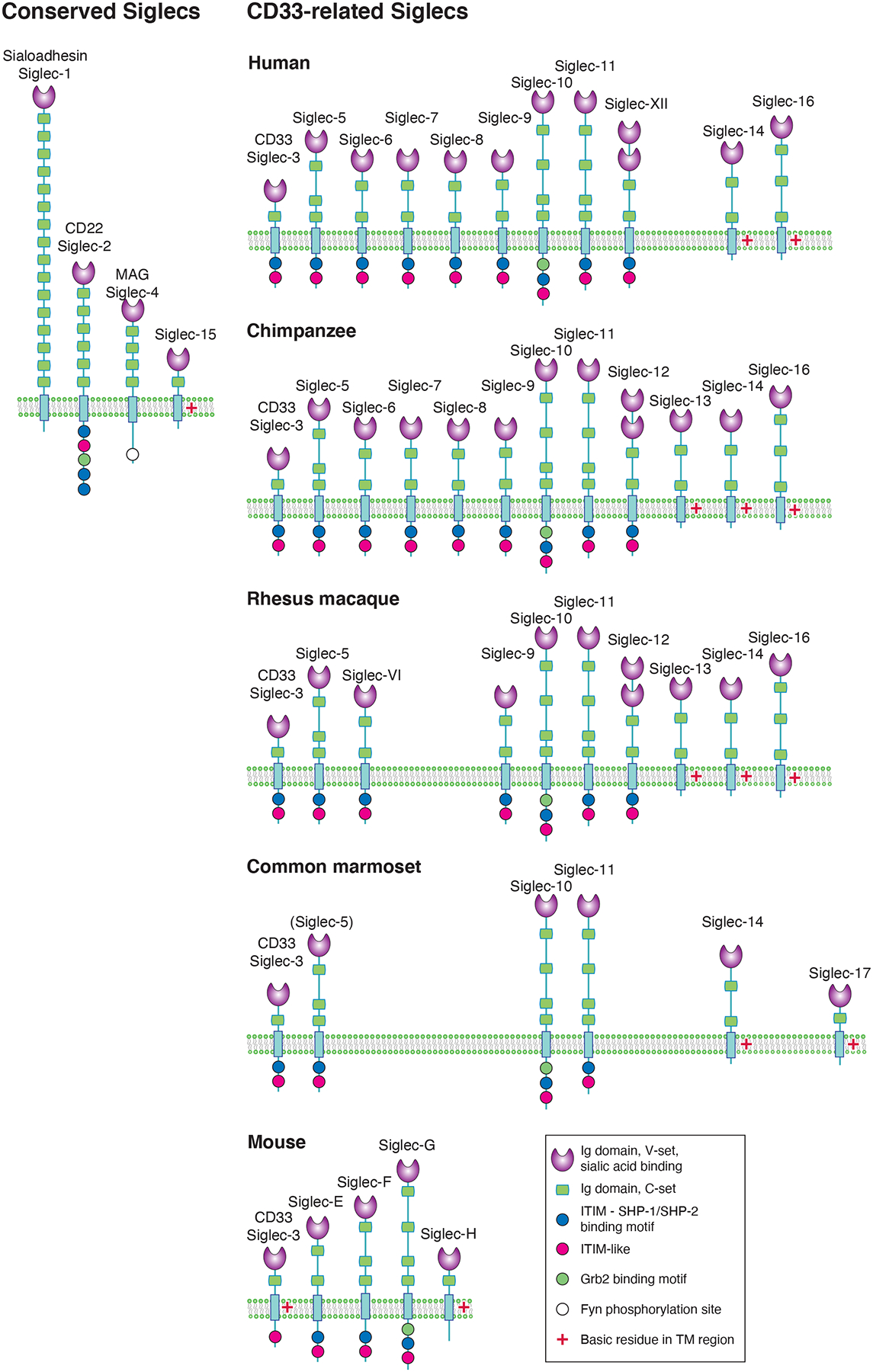
Siglecs in humans, chimpanzees, rhesus macaques, common marmosets, and mice are depicted. Mammalian Siglecs can be classified into two groups: conserved Siglecs, including Sialoadhesin/Siglec-1, CD22/Siglec-2, MAG/Siglec-4, and Siglec-15, are conserved among these species (and possibly among tetrapods); CD33/Siglec-3-related Siglecs show significant species-to-species variations. Siglec-13 was lost in humans. Siglec-17 is a functional protein in marmoset (New World monkey); the transcript of its ortholog (SIGLEC17P) is expressed in humans and great apes but does not yield functional protein due to mutations (human: single-nucleotide deletion in exon 1; chimpanzee and gorilla: five-nucleotide deletion in exon 2; orangutan: insertion of two 61-nucleotide repeats in exon 2). SIGLEC5 gene in common marmoset has a frameshift mutation in exon 2 (encoding V-set domain) but otherwise intact, and thus included.
N-terminal V-set Ig-like domain of Siglecs recognizes sialic acid (Sia) and contains several conserved residues involved in Sia recognition. Human Siglec-XII has a mutation at the essential Arg residue involved in Sia recognition, and thus lost the ability to recognize Sia, whereas great ape counterpart has the Arg and preferentially recognizes Neu5Gc.
Intracellular domain of most Siglecs contains a sequence motif called ITIM, which interacts with nonreceptor-type protein tyrosine phosphatase SHP-1 (gene: PTPN6) and negatively regulates leukocyte activities. A few Siglecs have a positively charged amino acid residue in the transmembrane domain, which interact with adapter protein DAP12 (gene: TYROBP) and transduce positive regulatory signal by way of protein tyrosine kinase SYK (gene: SYK).
Human cell types that express each Siglec are as follows: Siglec-1: macrophages; CD22/Siglec-2: B lymphocytes; CD33/Siglec-3: myeloid precursor cells, monocytes, macrophages, microglia, dendritic cells, and mast cells; MAG/Siglec-4: Schwann cells and oligodendrocytes; Siglec-5: neutrophils, monocytes, macrophages, and B lymphocytes; Siglec-6: placental trophoblasts, B lymphocytes, and mast cells; Siglec-7: natural killer cells, monocytes, macrophages, dendritic cells, and mast cells; Siglec-8: eosinophils and mast cells; Siglec-9: neutrophils, monocytes, macrophages, and dendritic cells; Siglec-10: B lymphocytes, monocytes, macrophages, and dendritic cells; Siglec-11: macrophages and microglia; Siglec-XII: macrophages and luminal epithelial cells; Siglec-14: neutrophils, monocytes, and macrophages; Siglec-15: osteoclasts and macrophage subsets; and Siglec-16: macrophages and microglia. Note that this list is not exhaustive, as tissue and systemic milieu may influence the expression of Siglecs.
Modified from Figure 35.1 (prepared by Dr. Richard Cummings (Harvard Medical School)) in Chapter 35 “I-type lectins”, Essentials of Glycobiology (4th edition), Cold Spring Harbor Press.
Various functional aspects of Siglecs in physiology and disease are described by other articles in this issue (Bochner et al.; Läubli; Macauley; Nizet; Paulson; Raïch-Regué et al.; Schnaar; Siddiqui; Siew et al.). In this review, we briefly summarize the discovery of the Siglec family, their classification, their patterns of evolution, uniquely human traits, and the relevance of polymorphisms of Siglecs to human health.
2. Discovery of Siglecs
2.1. Discovery of Sialoadhesin and CD22 as founding members of the Siglec family
Siglecs were discovered by two independent lines of studies, one leading to Sialoadhesin/Siglec-1 and the other to CD22/Siglec-2. Sialoadhesin/Siglec-1 was initially characterized as a protein on mouse macrophages that captures sheep erythrocytes in a Sia-dependent manner (Crocker and Gordon, 1986), and protein purification and characterization revealed its preferential recognition of Sia in α2–3 linkage (Crocker et al., 1991).
Meanwhile, CD22 was initially identified as a specific marker of B lymphocyte lineage, and human CD22 cDNA was obtained by expression cloning (Stamenkovic and Seed, 1990). As CD22 shows sequence similarity with myelin-associated glycoprotein (MAG) (Arquint et al., 1987; Lai et al., 1987; Salzer et al., 1987), and the involvement of MAG in intercellular interaction has been suggested, the authors tested if CD22 has a similar function, revealing its binding to erythrocytes and monocytes, which was markedly reduced by sialidase pretreatment (Stamenkovic and Seed, 1990). Further studies showed that CD22 is a lectin specifically recognizing Sia in α2–6 linkage (Powell et al., 1993; Sgroi et al., 1993; Stamenkovic et al., 1992; Stamenkovic et al., 1991).
2.2. MAG and CD33 join the family
Cloning and sequencing of Sialoadhesin cDNA revealed homology of the amino-terminal two Ig domains of Sialoadhesin, CD22, CD33, and MAG (Crocker et al., 1994), prompting the demonstration of Sia-binding activities of mouse MAG (Kelm et al., 1994) and human CD33 (Freeman et al., 1995). Based on these findings, “sialoadhesins” was proposed as a term to describe the protein family (Freeman et al., 1995; Kelm et al., 1994). Meanwhile, a broader term “I-type lectins” was proposed to describe IgSF proteins with lectin-like functions including “sialoadhesins” (Powell and Varki, 1995). The term “Siglec” (representing Sia, IgSF, and lectin) was then proposed by one of us as a better name to describe this subset of I-type lectins and was eventually accepted along with a proposal that “new members of the family should be named Siglec-5 etc, following consultation with other scientists in the field” (Crocker et al., 1998).
2.3. Discovery of other family members by transcriptome and genome sequence mining
Explosive expansion of nucleotide sequence data for transcripts (most notably in the form of “expressed sequence tag”, a single-pass read of cDNA clones mostly from 3’ end) and genomes in the 1990s allowed investigators to identify potential genes of interest based on sequence similarity with known members of the family, and the expansion of the Siglec family was greatly facilitated by this approach, as well as coordination with the Human Gene Nomenclature Committee and an email discussion group of involved investigators. Cloning of novel human Siglecs, including Siglec-5 (Cornish et al., 1998), Siglec-7 (Angata and Varki, 2000b; Nicoll et al., 1999), Siglec-8 (Floyd et al., 2000; Kikly et al., 2000), Siglec-9 (Angata and Varki, 2000a; Zhang et al., 2000), Siglec-10 (Li et al., 2001; Munday et al., 2001), Siglec-11 (Angata et al., 2002), Siglec-XII (Angata et al., 2001c), Siglec-14 (Angata et al., 2006), and Siglec-15 (Angata et al., 2007), as well as novel mouse Siglecs (Siglec-E (Ulyanova et al., 2001; Yu et al., 2001b), Siglec-F (Angata et al., 2001b), Siglec-G (Aizawa et al., 2003; Hoffmann et al., 2007), and Siglec-H (Zhang et al., 2006)) was attained by this approach. Other approaches, such as expression cloning (for human Siglec-6 (Patel et al., 1999), human Siglec-7 (Falco et al., 1999), and mouse Siglec-H (Blasius et al., 2006)) and yeast two-hybrid screening (for human Siglec-XII (Yu et al., 2001a)), also contributed to the expansion of the Siglec family. New members typically showed sialidase-sensitive rosetting of erythrocytes, which was abrogated by mutation of a critical arginine residue in the amino-terminal V-set Ig domain.
3. Classification of Siglecs
3.1. Classification of Siglecs based on sequence similarity
Human and mouse genomes and transcriptomes were the first to be extensively studied among those of vertebrates, leading to the complete identification and characterization of the Siglec family in these species. Comparison of Siglecs in human and mouse revealed that, while Sialoadhesin/Siglec-1, CD22/Siglec-2, MAG/Siglec-4, and Siglec-15 are conserved, a number of other Siglecs, now classified as CD33/Siglec-3-related Siglecs (CD33rSiglecs; based on their sequence similarity to CD33/Siglec-3), failed to show clear one-to-one orthologous correspondence (except for human Siglec-10 and mouse Siglec-G that are clearly orthologous). This is why human Siglecs are serially numbered as proposed in 1998 (Crocker et al., 1998), whereas mouse CD33rSiglecs were given alphabetical nomenclature (i.e., Siglec-E, F, G, and H). Nevertheless, comparison of CD33rSiglecs implies that the common ancestor of these two species shared “prototypes” of CD33rSiglecs in common, namely, CD33rSiglec(s) with (1) two Ig-like domains resembling CD33, (2) three Ig-like domains resembling Siglec-9/E, (3) four Ig-like domains resembling Siglec-5/F, and (4) five Ig-like domains resembling Siglec-10/G (Angata et al., 2001a). Analyses of genomic sequences of other mammalian species basically support this overall scenario (Angata, 2006; Khan et al., 2020b).
3.2. Classification of Siglecs based on signaling function
As mentioned above, the majority of Siglecs have cytosolic ITIM motifs, and recruit tyrosine phosphatase (SHP-1/2), and are considered to have suppressive signaling property. This observation fits the hypothesis that Siglecs prevent the inadvertent activation of the vertebrate innate immune system against host cells by working as the sensors of Sias as SAMPs. In contrast, a few Siglecs have a positively charged amino acid residue in the transmembrane domain, which interacts with an Asp residue in the transmembrane domain of adapter protein, such as DAP12 (gene: TYROBP), which further recruits tyrosine kinase SYK (gene: SYK) upon Tyr phosphorylation. It may be simplistic to consider tyrosine phosphatase-associated Siglecs as “inhibitory” and tyrosine kinase-associated Siglecs as “activating” (in fact, DAP12 can also mediate inhibitory signaling in myeloid cells (Takaki et al., 2006), and SHP-2 can serve as an oncogene transducing activating signal in epithelial cells (Tartaglia and Gelb, 2005; Tartaglia et al., 2001)). Furthermore, even the same Siglec may have different signaling properties depending on the cell type in which it is expressed, as is the case with Siglec-8 (Carroll et al., 2021; Carroll et al., 2018; Korver et al., 2022). Nevertheless, the association with different types of downstream signaling molecules provides a useful conceptual framework to classify Siglecs. In humans, Siglec-14–16 associate with DAP12 and considered to be “activating”; in mouse, CD33/Siglec-3, Siglec-15, and Siglec-H belong to this category.
It should be emphasized that the classification of Siglecs based on their association with SHP-1/2 versus DAP12 does not cover all Siglecs. Two Siglecs, namely, Sialoadhesin/Siglec-1 and MAG/Siglec-4, may not belong to either of these groups, although Sialoadhesin/Siglec-1 was reported to interact with DAP12 despite the lack of positively charged residue in its transmembrane domain (Wu et al., 2016; Zheng et al., 2015), and MAG/Siglec-4 was reported to associate with Src family kinase Fyn (Umemori et al., 1994). It should be also noted that CD22/Siglec-2 is known to interact with various intracellular signaling molecules (Nitschke, 2014).
4. Ligands of Siglecs
As mentioned in the Introduction section, Siglecs recognize glycans containing Sias. However, owing to the low affinity of the interaction between Siglec and monovalent sialo-oligosaccharide (with dissociation constant typically in the range of mM), for Siglec–ligand interaction to have a biological impact, a cluster of glycans (e.g., a patch of glycolipids, or a heavily glycosylated glycoprotein, etc.) is usually required. Each Siglec shows unique preference toward sialo-oligosaccharides, based on the type of Sia, linkage, and the modifications of penultimate sugars. See the article by Schnaar in this issue for further discussion on Siglec ligands (Schnaar).
One striking feature of human glycome is the absence of N-glycolylneuraminic acid (Neu5Gc), a type of Sia commonly found in the tissues of most other mammalian species. This unique feature is a consequence of the inactivating mutation of CMAH gene encoding CMP-Neu5Ac hydroxylase that generates Neu5Gc and is shared among all extant (and some extinct) humans (Chou et al., 2002; Hayakawa et al., 2001). Some primate Siglecs, such as Siglec-12 (Angata et al., 2001c), Siglec-9 (Sonnenburg et al., 2004), and Siglec-10 (Wang et al., 2021), preferentially recognize Neu5Gc. The loss of Neu5Gc in human ancestor may have had human-unique consequences, one of which may be the facilitated evolution of the Siglec family (Angata, 2018), as will be discussed later.
Although Sia is not commonly made by microbes (as mentioned in the Introduction section), there are several exceptions. Some species of bacteria produce Neu5Ac (but not Neu5Gc), whereas some others produce Sia-like nonulosonic acids (NulOs), such as pseudaminic acid, legionaminic acid, fusaminic acid, and acinetaminic acid (Angata and Varki, 2002; Lewis et al., 2022). Sia and NulOs on bacteria may serve as molecular mimicry, providing protection against the host immune system. One possible mechanism is the engagement of Siglecs on innate immune cells to dampen inflammatory responses (Carlin et al., 2007; Carlin et al., 2009b; Stephenson et al., 2014), as discussed in detail by Nizet in this issue (Nizet). Such interaction with rapidly evolving microbes that mimic host Sia and exploit Siglecs (and host response to evade the exploitation) may be a possible driving force behind the rapid evolution of Siglecs, by an evolutionary process called Red Queen Effect (Van Valen, 1974).
Some Siglecs also interact with ligands that do not contain Sia, or do not even contain glycan. Examples of endogenous mammalian ligands that does not contain Sia include leptin (ligand for Siglec-6 (Patel et al., 1999)), high-mobility group box 1 protein (ligand for Siglec-10/G (Chen et al., 2009)), heat shock protein 70 (ligand for Siglec-10/G (Chen et al., 2009) and Siglec-5 and −14 (Fong et al., 2015)), and vimentin (Tsai et al., 2020) and cardiolipin (Suematsu et al., 2019) (ligands for Siglec-14 and Siglec-5). Some microbes also express such “noncanonical” ligands, such as β protein from group B Streptococcus (GBS) (Ali et al., 2014; Carlin et al., 2009a; Chang et al., 2014) and triacylglycerol produced by fungal pathogens (Suematsu et al., 2019). See the article by Siddiqui in this issue for further discussion on noncanonical Siglec ligands that do not contain Sia (Siddiqui).
5. Evolution of Siglecs
5.1. Origin of Siglecs
The presence of orthologs of MAG/Siglec-4 (Lehmann et al., 2004) and Siglec-15 (Angata et al., 2007) in fish indicates that (1) the origin of Siglecs predates the emergence of bony vertebrates, and that (2) the common ancestor of bony vertebrates already had multiple Siglecs (Supplementary Figure 2). A recent review stated that Siglec-like sequences are found among the predicted shark genes and concluded that a common ancestor of jawed vertebrates (bony vertebrates + cartilaginous fish) already had a Siglec family (Bornhofft et al., 2018). Our genomic DNA sequence survey with TBLASTN algorithm (Gertz et al., 2006) supports this conclusion. In addition, the genome of sea lamprey (Petromyzon marinus) contains DNA segments encoding Siglec-like proteins (one resembling Siglec-1 and the other resembling Siglec-15), in which a few key amino acid residues involved in Sia recognition by mammalian Siglecs are conserved, implying Sia-binding function (Supplementary Figure 3). The absence of MAG, which is conserved among bony vertebrates, in lamprey may be related to the absence of myelin in this lineage (Bullock et al., 1984). In contrast, similar homology search in the genome sequences of sea squirt (Ciona intestinalis) and lancelet (Branchiostoma floridae), two species closely related to vertebrates, did not reveal any genomic DNA segment encoding Siglec-like V-set domain. Thus, it appears likely that Siglecs emerged in the common ancestor of all vertebrates (bony vertebrates + cartilaginous fish + jawless fish).
Which Siglec was ancestral to all others remains a question. It is tempting to speculate that Siglec-15 is ancestral, based on the facts that: (1) Siglec-15-like sequence is found in all vertebrates, including lamprey (Supplementary Figure 3); (2) V-set Ig-like domain of Siglec-15 shows significant sequence similarity with those of Tim-1 (gene HAVCR1), Tim-3 (gene HAVCR2) and Tim-4 (gene TIMD4) (Supplementary Figure 4), implying a closer evolutionary relationship to other IgSF proteins; (3) Siglec-15 lacks Cys residues that bridge the first and second Ig-like domains (Supplementary Figure 1), which is a shared property of all other mammalian Siglecs, implying its intermediate position in molecular evolution. However, some features (e.g., arrangement of Cys residues) may have emerged at a later stage of evolution in one lineage or the other (e.g., Siglec-1-like Siglec sequence in lamprey also lacks the Cys residues that bridge the first and second Ig-like domains), making this proposition still a speculation.
5.2. Diversification of Siglecs during chordate evolution
An overall pattern of Siglec evolution in vertebrates can be deduced from the genomic sequences of various vertebrates (Supplementary Figure 2), although caution is warranted in interpreting the homology search results, as it is challenging to find an ortholog of a mammalian Siglec in a distant species (particularly in fish), and the sequencing/assembly of genomes of some species discussed below may be incomplete (although these genomes were selected from those considered most complete).
As mentioned above, mammals have five Siglecs (counting CD33rSiglecs as one), namely, Sialoadhesin/Siglec-1, CD22/Siglec-2, CD33rSiglecs, MAG/Siglec-4, and Siglec-15. Orthologs of Sialoadhesin/Siglec-1, MAG/Siglec-4, and Siglec-15, as well as those similar to CD33/Siglec-3, are present in the genome of a cartilaginous fish (whale shark, Rhincodon typus), implying that the prototypes of these Siglecs were present in the common ancestor of jawed vertebrates. However, the genomes of ray-finned fish (three-spine stickleback, Gasterosteus aculeatus, and Japanese pufferfish, Takifugu rubripes) appear to lack clear orthologs of CD33rSiglecs, implying lineage-specific loss of genes. No clear ortholog of CD22/Siglec-2 was found in the genomes of cartilaginous and bony fishes. Conversely, some Siglecs appear to have undergone expansion in fish lineages; multiple Siglec-1-like genes are present in the genomes of bony fishes, and multiple Siglec-15-like genes are present in the genome of a lobe-finned fish (African coelacanth, Latimeria chalumnae). (While a previous study identified clusters of “CD33rSiglecs” and “Sialoadhesin-related Siglecs” in stickleback genome (Cao et al., 2009), the exact relationships of these and other fish Siglecs with mammalian Siglecs may benefit from further investigation, given that there appear to be many Siglecs encoded in fish genomes.)
Orthologs of all five mammalian Siglecs are found in the genome of a frog (Western clawed frog, Xenopus tropicalis), indicating that the common ancestor of tetrapods (four-limbed animals) had the prototype of these Siglecs. Among the reptiles, genomes of a lizard (green anole, Anolis carolinensis) and a turtle (green sea turtle, Chelonia mydas) maintain these five Siglecs, whereas those of a crocodilian (American alligator, Alligator mississippiensis) and a bird (Australian zebra finch, Taeniopygia guttata) lack CD22/Siglec-2 (finch genome also appears to lack CD33rSiglecs), again implying lineage-specific loss.
All mammals have five Siglecs (with differing numbers of CD33rSiglecs). Curiously, genomes of monotremes (platypus, Ornithorhynchus anatinus, and short-beaked echidna, Tachyglossus aculeatus) contain additional Siglec-15-like Siglecs in a gene cluster. Marsupials (gray short-tailed opossum, Monodelphis domestica, and koala, Phascolarctos cinereus) and placentals have one copy each of Sialoadhesin/Siglec-1, CD22/Siglec-2, MAG/Siglec-4, and Siglec-15 and varying numbers of CD33rSiglecs.
5.3. Rapid evolution of CD33rSiglecs
CD33rSiglecs are encoded in a gene cluster in most species in which they are found (including amphibians and reptiles). It is likely that CD33rSiglecs have undergone gene duplications independently in multiple lineages. For example, the genome of a frog (Western clawed frog, X.tropicalis) contains more than 30 potentially functional CD33/Siglec-3-like V-set domain in the same orientation, implying the expansion of CD33rSiglecs in this lineage. The number of CD33rSiglecs in mammals varies widely–ranging from 3 copies in monotremes, 5 or 6 in marsupials, and 5–20 among placental mammals–again implying lineage-specific expansion or contraction of the gene family. It was proposed that a chromosomal segment encoding CD33rSiglec gene cluster has undergone inverse duplication in the common ancestor of marsupials and placentals (Cao et al., 2009).
Once multiple copies of CD33rSiglecs were acquired, they underwent diversification by various mechanisms, including nucleotide substitution, nonallelic homologous recombination (resulting in hybrid genes), and gene conversion (resulting in the overwriting of a part of a gene by another gene), between related genes (Angata et al., 2004). (See reviews by others for the mechanism of nonallelic homologous recombination and gene conversion (Hastings et al., 2009; Inoue and Lupski, 2002; Sasaki et al., 2010).) The modular architecture of Siglec genes (i.e., each Ig-like domain is encoded by one exon) makes it easier for the hybrid genes to maintain functionality and also allows for frequent gene conversion events.
In contrast, some CD33rSiglec genes have acquired mutations that make them nonfunctional (e.g., frameshifts and stop codons) (Angata et al., 2004). In fact, this is not a rare event—CD33rSiglec gene cluster in many species contains multiple pseudogenes. In addition, complete deletion of Siglec-coding gene has been observed, such as primate SIGLEC13 that is lost in humans (Angata et al., 2004). Of note, some CD33rSiglecs are “semi-functional”, in that they are expressed as proteins but do not recognize Sia, due to the mutation of the essential Arg residue in the Sia-binding V-set domains. (We proposed to use Roman numerals to indicate such Siglec proteins that have the essential Arg mutated (Angata et al., 2004), whereas the gene symbols for these Siglecs remain numbered by Arabic numerals.) For example, several primate CD33rSiglecs, such as human Siglec-XII, chimpanzee Siglec-V, and baboon Siglec-VI, belong to this category (Angata et al., 2004; Angata et al., 2001c; Mitra et al., 2011). The CD33rSiglec gene cluster in many species also contains genes whose reading frame appears intact, but their protein product would not recognize Sia. This abundance of “semi-functional” Siglecs may be explained by the fact that the essential Arg in Siglecs is encoded by CGN codon, which is prone to mutate to TGN (encoding Cys/stop/Trp) or CAN (encoding His/Glu). (In vertebrates, C in CG dinucleotide is often 5-methylated by DNA cytosine 5-methyl transferase and then further spontaneously deaminated to T (Holliday and Grigg, 1993), explaining the underrepresentation of CG dinucleotide in vertebrate genomes.)
It remains a question why we need so many CD33rSiglecs—as is the case for other immune receptors that have many family members (e.g., killer Ig-like receptors of primates and Ly49 of rodents) (Akkaya and Barclay, 2013; Trowsdale et al., 2001). In primates and rodents, most CD33rSiglecs are expressed on various cells involved in innate immunity, which are the first to encounter microbial pathogens. While it remains a speculation, CD33rSiglecs may be evolving to counter rapidly evolving microbial pathogens. In the innate immune system, “a family of proteins sharing a common biological function” may have an advantage over “a single protein solely responsible for the biological function”, in that the host can afford to lose a protein that is exploited by a microbial pathogen without complete loss of the biological function (whereas the host cannot afford to do so in the latter case).
In some instances, CD33rSiglecs that are not orthologous appear to occupy the similar cell-type/functional niche in different species. For example, while human Siglec-8 and mouse Siglec-F are not orthologs, they have similar (though not identical) functions in regulating eosinophils and basophils and allergic phenomena; thus, they are functionally convergent paralogs (Tateno et al., 2005). See the article by Bochner in this issue for further details (Bochner et al.).
5.4. Paired Siglecs: a countermeasure to combat microbes that exploit inhibitory Siglecs?
As mentioned above, most CD33rSiglecs have ITIMs and mediate inhibitory function. Some pathogens have been found to interact with inhibitory CD33rSiglecs, often resulting in immune subversion (see the article by Nizet in this issue for further details (Nizet)). One way to manage the exploitation by microbial pathogens is to decommission the target Siglec (temporarily or permanently); another way may be to deploy a decoy that mimics the inhibitory Siglec. Some of the DAP12-associated Siglecs appear to be “paired” with inhibitory Siglecs; in primates, Siglec-5 (inhibitory) and Siglec-14 (activating), as well as Siglec-11 (inhibitory) and Siglec-16 (activating), show very high amino acid sequence similarity at the extracellular domain (in whole or in part), yet exhibiting counteractive signaling functions, and thus considered to be “paired Siglecs”. In the presence of bacterial pathogens that interact with paired Siglecs, cells that express inhibitory Siglecs alone show weaker inflammatory responses (e.g., the production of reactive oxygen species (ROS), cytokines, and neutrophil extracellular traps) than those that express activating counterpart, supporting the hypothesis that the activating Siglec works as a decoy (Landig et al., 2019; Schwarz et al., 2017; Yamanaka et al., 2009). The extreme sequence similarity between the “paired Siglecs” in the amino-terminal domains is likely maintained by repeated gene conversions (Angata et al., 2006; Hayakawa et al., 2017). These paired Siglecs (or equivalents) are found not only in primates but also in other mammals, and they also maintain high sequence similarity with each other (Angata, 2006; Khan et al., 2020b).
It is of note that activating-type CD33rSiglecs (i.e., SIGLEC14 and SIGLEC16) can have null allele(s) and the frequency of such null allele(s) in human populations is surprisingly high (Cao et al., 2008; Yamanaka et al., 2009). While activating-type Siglecs appear to play some positive roles, there may be some negative consequences. These polymorphisms may have relevance to human diseases, as will be discussed later.
5.5. CD33rSiglecs, ROS and mammalian lifespan
Aging is a multifactorial process that includes lifelong accumulation of molecular damage, leading to frailty, disability, disease, and eventually death. ROS derived from mitochondria and from NADPH oxidase (NOX) enzymes of innate immune cells are known to contribute to aging, with the former thought to be the dominant source. However, a highly significant correlation exists between the number of CD33rSiglec genes and maximum lifespan in mammals. Consistent with this observation, mice lacking Siglec-E, the main member of the CD33rSiglec family in the myeloid lineage in rodents, exhibit reduced survival associated with exaggerated signs of aging at the molecular, structural, and cognitive levels (Schwarz et al., 2015b). Accelerated aging was related to both an unbalanced ROS metabolism and a secondary impairment in detoxification of reactive molecules, ultimately leading to increased damage to cellular DNA, proteins, and lipids. The strong correlation of lifespan with CD33rSiglec gene number was confirmed in 26 mammalian species, independent of body weight or phylogeny (Khan et al., 2020b). This correlation is strongest when considering total CD33rSiglec gene number rather than those encoding inhibitory or activating subsets, suggesting that lifetime balancing of ROS may be important. Combining independent lines of evidence including the short half-life and spontaneous activation of neutrophils, we have calculated that even without intercurrent inflammation, a major source of lifetime ROS exposure may actually be neutrophil NOX-derived (Khan et al., 2020b). However, genomes of human supercentenarians (>110 years) were not found to harbor a significantly higher number of functional CD33rSiglec genes. This may relate to the very unusual phenomenon of prolonged post-reproductive lifespan, so far reported only in humans and certain toothed whales (Khan et al., 2020b).
6. Uniquely human evolution of Siglecs
While CD33rSiglecs are rapidly evolving in all taxa, comparative studies of Siglecs in humans and the closely related great apes have revealed unusually high number of changes unique to human Siglecs. Some of these human-specific traits may be secondary consequences of another evolutionary event unique to humans (among great apes), the loss of CMP-Neu5Ac hydroxylase that generates Neu5Gc from Neu5Ac (Chou et al., 2002; Hayakawa et al., 2001; Lewis et al., 2022). Another human-specific phenomenon that might be worth looking into is indoor air pollution, due to cooking in enclosed spaces, influencing respiratory infections (Bruce et al., 2013). Regardless of the exact cause, these changes may have consequences on the health of modern humans (Schwarz et al., 2015a; Varki, 2010).
6.1. Genetic changes in human Siglecs
Table 1 and Figure 2 summarize the major genetic changes unique to human Siglec genes. For example, an Alu-mediated recombination event resulted in the loss of human SIGLEC13, which encodes an activating-type Siglec expressed on monocytes and epithelium of great apes (Khan et al., 2020a; Wang et al., 2012b). Chimpanzee Siglec-13 protein can bind sialylated pathogenic bacteria and elicit proinflammatory responses (Wang et al., 2012b).
Table 1.
Human-unique changes of Siglec genes
| Siglec gene | Human-unique allele/state | Modern human | Archaic human* | Great apes | Ref |
|---|---|---|---|---|---|
| CD33 | rs12459419(T) rs3865444(A) | Present (minor allele) | Absent | Absent | (Schwarz et al., 2016) |
| c.61T (p.Phe21) | Fixed (100%) | Fixed (100%) | Absent | (Saha et al., 2022) | |
| SIGLEC5 | Intact essential Arg | Fixed (100%) | Fixed (100%) | Polymorphic | (Angata et al., 2006) |
| SIGLEC12 | Essential Arg mutated to Cys | Fixed (100%) | Fixed (100%) | Absent | (Angata et al., 2001c; Khan et al., 2020a; Mitra et al., 2011) |
| rs66949844(dupC) frameshift | Present (major allele) | Present (major allele) | Absent | (Khan et al., 2020a; Mitra et al., 2011). | |
| rs16982743(A) stop codon | Present (minor allele) | Present (major allele) | Absent | (Khan et al., 2020a). | |
| SIGLEC13 | Deletion | Fixed (100%) | Fixed (100%) | Absent (i.e., functional gene is present) | (Angata et al., 2004; Wang et al., 2012b) |
| SIGLEC14 | Intact essential Arg | Fixed (100%) | Fixed (100%) | Polymorphic | (Angata et al., 2006) |
| SIGLEC16 | rs12611411(T) rs12984584(C)** | Present (major allele) | Present (major allele) | Absent | (Wang et al., 2012a) |
Modified from (Khan et al., 2020a).
Neandertal (n = 6) and Denisovan (n = 2).
These SNPs are in linkage disequilibrium with an indel polymorphism causing frameshift in open reading frame of SIGLEC16.
Figure 2. Human-specific genetic changes in Siglecs.
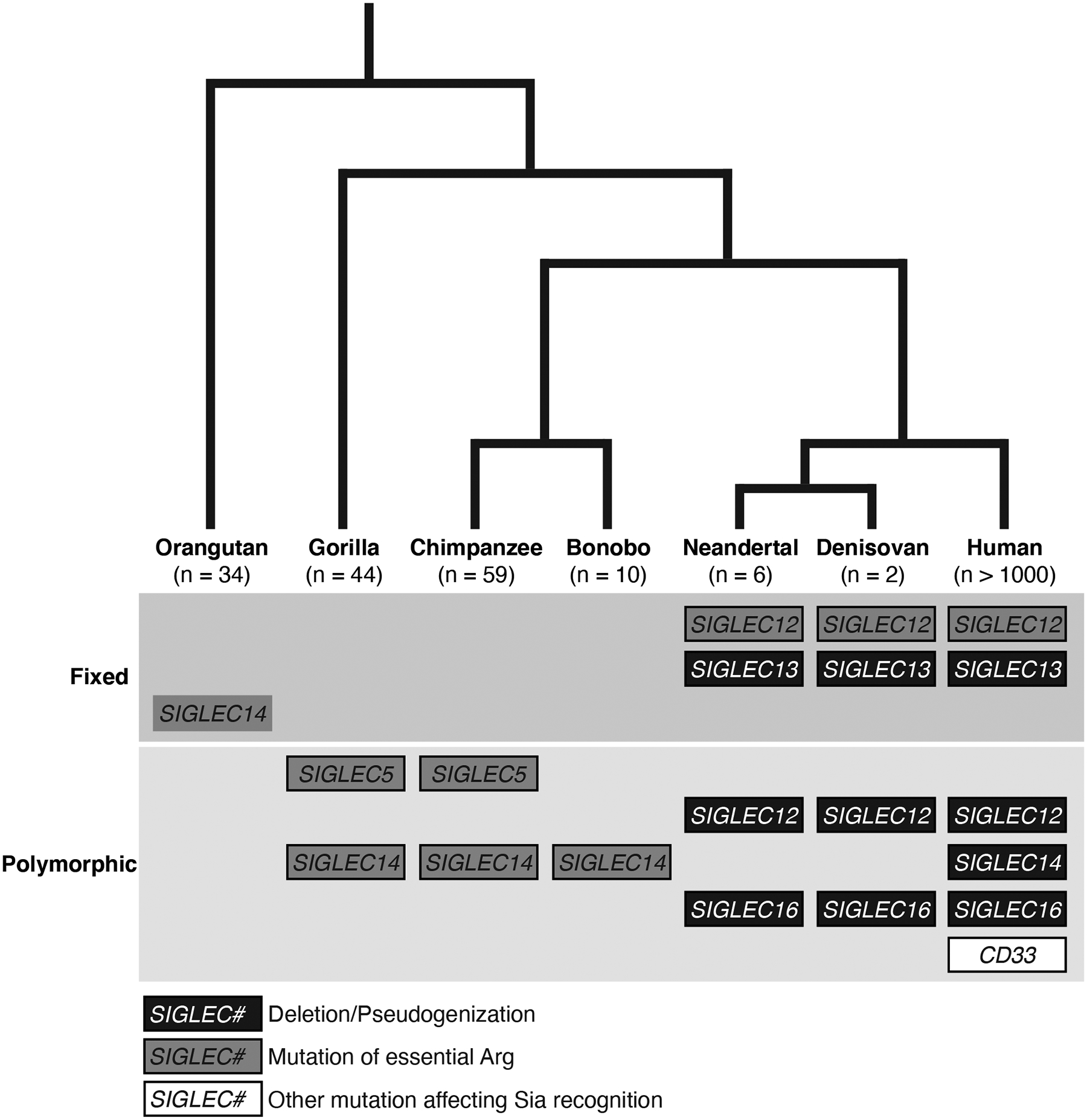
Major genetic changes in human and great ape Siglec genes are depicted. Humans appear to have accumulated a number of genetic changes in Siglec genes in a lineage-specific manner. Most of these changes are shared with extinct archaic humans (Neandertal and Denisovan).
Note that the status of orangutan SIGLEC14 mutation at the essential Arg residue is uncertain, due to low sequence coverage. In addition, the status of SIGLEC14 deletion (SIGLEC14–SIGLEC5 fusion) in archaic humans was not determined, due to the lack of SNP in linkage disequilibrium with the deletion that can be utilized as a surrogate marker. Polymorphic mutation in CD33 refers to rs12459419(T) allele, which is a human-unique allele protective against late-onset Alzheimer’s disease (Schwarz et al., 2016). This allele increases the proportion of CD33 protein without V-set domain required for Sia recognition.
Modified from (Khan et al., 2020a). See also Tables 1 and 2.
Human Siglec-12 lost Sia recognition capability, owing to a mutation in the essential Arg residue (Angata et al., 2001c), which is universal in modern and archaic humans (Khan et al., 2020a; Mitra et al., 2011). This loss-of-function mutation apparently permitted further acquisition of “null” mutations, including frameshift (Mitra et al., 2011) and in-frame stop codon (McDonough et al., 2013), which are also unique to humans. Of note, in-frame stop codon in SIGLEC12 (rs16982743(A) allele) was reported to be under positive selection (Yngvadottir et al., 2009), indicating that the complete loss of the protein may have had some selective advantage. Siglec-12 protein expression in epithelial cells and signaling through SHP-2 may facilitate tumor progression (Mitra et al., 2011), which may possibly explain the survival advantage the individuals without Siglec-12 may have had (Siddiqui et al., 2021).
CD33/Siglec-3 in great apes preferentially recognizes Neu5Gc. CD33/Siglec-3 in humans has Phe at amino acid position 21, whereas CD33/Siglec-3 in great apes has Ile at the same position. Phe21 is fixed in hominins (modern humans, Neandertals and Denisovans) and allows CD33/Siglec-3 to recognize Neu5Ac as well as Neu5Gc, implying the adaptation of human CD33/Siglec-3 to the loss of Neu5Gc with recruitment to high level microglial expression in the Neu5Ac-rich brain (Saha et al., 2022). Notably the single-nucleotide polymorphism (SNP) that yields truncated form of CD33/Siglec-3 missing the V-set domain, and protective against late-onset Alzheimer’s disease, is not found in archaic hominins (Neanderthals and Denisovans), suggesting that grandmothering emerged only in humans (Schwarz et al., 2016).
Humans also appear to have “gained” functional Siglecs. Siglec-5 and Siglec-14 in humans have essential Arg residue and are capable of recognizing Sia, and this is universal among modern humans, whereas Siglec-5 and Siglec-14 in great apes are polymorphic with regard to essential Arg; alternative allele with essential Arg mutated to His is common, if not universal, among great apes (Angata et al., 2006; Khan et al., 2020a). The physiological consequence of this change remains to be investigated.
6.2. Altered patterns of Siglec protein expression in humans
Table 2 summarizes the known human-specific changes in Siglec expression patterns. These include expression of Sialoadhesin/Siglec-1 in broader subset of splenic macrophages (Brinkman-Van der Linden et al., 2000), expression of Siglec-11 and Siglec-16 in brain microglia (Hayakawa et al., 2005; Wang et al., 2012a), Siglec-6 in placenta (Brinkman-Van der Linden et al., 2007), Siglec-5 and Siglec-14 in amniotic epithelium (Ali et al., 2014), and Siglec-7 in pancreatic islet cells (Khan et al., 2020a). These expression patterns appear to be uniquely acquired in the lineage leading to modern humans and may have consequences on human physiology (Table 2).
Table 2.
Human-unique expression patterns of Siglec proteins
| Siglec protein | Humans | Great apes | Relevant disease | Ref |
|---|---|---|---|---|
| CD33 | High expression in microglia | Low expression in microglia (chimpanzee) | Alzheimer’s disease | (Schwarz et al., 2016) |
| Siglec-5 | No/low expression in T lymphocytes | High expression in T lymphocytes | AIDS | (Nguyen et al., 2006; Soto et al., 2013; Soto et al., 2010) |
| Siglec-5, Siglec-14 | Expressed in amniotic epithelium | Not expressed in amniotic epithelium (chimpanzee) | Intrauterine infection with Group B Streptococcus | (Ali et al., 2014) |
| Siglec-6 | Expressed in placental trophoblast | Not expressed in placental trophoblast (chimpanzee, gorilla, orangutan) | Preeclampsia | (Brinkman-Van der Linden et al., 2007) |
| Siglec-7 | Expressed in pancreatic islet cells | Not expressed in pancreatic islet cells (chimpanzee) | Type I diabetes | (Khan et al., 2020a) |
| Siglec-11, Siglec-16 | Expressed in brain microglia | Not expressed in brain microglia (chimpanzee, orangutan) | E.coli K1 meningitis in neonates and infants | (Hayakawa et al., 2005; Schwarz et al., 2017; Wang et al., 2012a) |
| Siglec-11, Siglec-16 | Decreased expression in ovarian fibroblasts | Strong expression in ovarian fibroblasts (chimpanzee) | Unknown | (Wang et al., 2011) |
Modified from (Khan et al., 2020a).
Conversely, human T lymphocytes have suppressed expression of some inhibitory Siglecs, such as Siglec-5, whereas these Siglecs are highly expressed on T lymphocytes from great apes, including chimpanzee, bonobo and gorilla (Nguyen et al., 2006). The loss of inhibitory Siglecs may explain general hyperactivity of human T cells compared with those from great apes (Nguyen et al., 2006; Soto et al., 2010) and increased activation and cell death upon human immunodeficiency virus (HIV) infection (Soto et al., 2013), likely explaining the higher frequency of progression to acquired immunodeficiency syndrome (AIDS).
7. Polymorphisms of human Siglecs in association with diseases
7.1. Null polymorphisms associated with diseases
As mentioned above, the frequencies of null alleles of activating-type human Siglecs, SIGLEC14 and SIGLEC16, are surprisingly high (whereas none has been found in great apes). SIGLEC14 null allele (caused by homologous nonallelic recombination between SIGLEC14 and SIGLEC5 genes, resulting in the deletion of a gene segment of SIGLEC14 and the generation of SIGLEC14–SIGLEC5 fusion gene encoding a protein identical to Siglec-5) reaches ~0.7 in some populations (Yamanaka et al., 2009), and SIGLEC16P allele (caused by 16 base pair deletion and 12 base pair insertion in exon 2, resulting in the frameshift and premature termination) reaches as high as ~0.9 (Cao et al., 2008; Wang et al., 2012a). The frequency of SIGLEC14 null allele differs depending on the human population–generally high in Asia and low in Europe–suggesting the presence of some regional selective pressure that favors or disfavors the null allele (Yamanaka et al., 2009). We have found associations between SIGLEC14 null allele and clinical conditions induced by bacterial pathogens, such as exacerbation of chronic obstructive pulmonary disease (COPD) (Angata et al., 2013), pre-term delivery of infant in the presence of GBS infection (Ali et al., 2014), and meningitis in the presence of Mycobacterium tuberculosis infection (Graustein et al., 2017) (Table 3). It should be noted that the null allele appears to be beneficial in some situations (e.g., exacerbation of COPD) but disadvantageous in others (e.g., GBS infection), implying that the polymorphism may be maintained by balancing selection. In addition, a SIGLEC14 homozygous null mother may develop antibodies against Siglec-14 protein during pregnancy (as soluble Siglec-14 protein may be produced by a heterozygous fetus), and the blood products prepared from such donors’ blood may engage Siglec-14 on myeloid cells of transfusion recipient, possibly leading to transfusion-related adverse events (Yasui et al., 2011). Soluble Siglec-14 was also reported to be an early plasma marker for the development of bronchopulmonary dysplasia (BPD) in premature infants (Forster et al., 2018); given that soluble Siglec-14 is generated from a functional SIGLEC14 allele (Huang et al., 2018), this observation might imply that the infants with functional SIGLEC14 allele may be at a higher risk of developing BPD (although definitive proof is lacking). Thus, SIGLEC14 null polymorphism has direct and indirect implications for human health. It is notable that some polymorphisms in SIGLEC5 gene, encoding the inhibitory Siglec paired with Siglec-14, were found to be associated with bacterially induced diseases (leprosy (Liu et al., 2015) and periodontitis (Munz et al., 2019; Munz et al., 2017)), as well as with an autoimmune disorder (systemic lupus erythematosus (Sun et al., 2016)), by genome-wide association studies (GWAS). Overall, these findings imply that these Siglecs are indeed involved in the immune responses against bacterial pathogens.
Table 3.
Polymorphisms in human Siglecs associated with phenotypes
| Genome-wide or other hypothesis-free association studies | |||
|---|---|---|---|
| Gene | Polymorphism | Associated phenotype | Ref |
| SIGLEC1 | rs6037651 (nonsynonymous SNP) | Serum IgM level | (Jonsson et al., 2017) |
| CD33 | rs3865444 (5’ upstream SNP), rs12459419 (nonsynonymous SNP, also influencing splicing) | Late-onset Alzheimer’s disease | (Hollingworth et al., 2011; Malik et al., 2013; Naj et al., 2011; Raj et al., 2014) |
| rs201473304 (indel) | Neuromyelitis optica spectrum disorder | (Huang et al., 2021) | |
| SIGLEC5 | rs4284742 (intronic SNP) rs11084095 (intronic SNP) |
Periodontitis | (Munz et al., 2017) (Munz et al., 2019) |
| rs10414149 (intronic SNP) | Leprosy | (Liu et al., 2015) | |
| rs4801882 (intronic SNP) | Systemic lupus erythematosus | (Sun et al., 2016) | |
| SIGLEC6 | rs3794986 (5’ upstream SNP) | Systemic lupus erythematosus | (Flores et al., 2019; Sun et al., 2016) |
| SIGLEC11 | rs12165127 (intronic SNP) | Lung cancer in never-smokers | (McKay et al., 2017) |
| rs9304690 (synonymous SNP) | Late-onset Alzheimer’s disease | (Bellenguez et al., 2022) | |
| SIGLEC12 | rs16982743 (nonsynonymous SNP, generating stop codon) | Cardiovascular outcomes in patients with hypertension on antihypertensive therapy | (McDonough et al., 2013) |
| rs3752135 (nonsynonymous SNP) | Stress fracture predisposition | (Friedman et al., 2014) | |
| SIGLEC14 | rs10412972, rs11084102 (5’ upstream SNPs) | Blood plasminogen level | (Ma et al., 2014) |
| SIGLEC15 | rs11877682 (intronic SNP) | Esophageal or stomach cancer | (Rashkin et al., 2020) |
| rs61104666 (synonymous SNP) | Tuberculosis | (Bhattacharyya et al., 2019) | |
| Candidate gene-based association studies | |||
| Gene | Polymorphism | Associated phenotype | Ref |
| SIGLEC1 | rs656635, rs609203, rs3859664, rs4813636 (SNPs in intron or 3’UTR) | Lung function | (Bukvic et al., 2013) |
| rs3859664 (intronic SNP) | Pulmonary active tuberculosis | (Souza de Lima et al., 2017) | |
| CD22 | rs34826052 (synonymous SNP) | Limited cutaneous systemic sclerosis | (Hitomi et al., 2007) |
| rs4805119 etc. (intronic SNPs) | Pediatric B cell leukemia | (Ma et al., 2012; Uckun et al., 2010) | |
| CD33 | rs35112940, rs12459419 (nonsynonymous SNP) | Efficacy of antibody therapy for pediatric AML | (Lamba et al., 2009; Mortland et al., 2013) |
| rs12459419 (nonsynonymous SNP) | Hepatocellular carcinoma in chronic hepatitis B patients | (Tsai et al., 2021) | |
| MAG | rs720309 (intronic SNP) rs7249617 (intronic SNP) |
Schizophrenia | (Wan et al., 2005; Yang et al., 2005) (Jitoku et al., 2011) |
| SIGLEC8 | rs36498 (5’ upstream SNP) rs10409962 (nonsynonymous SNP) |
Allergic asthma | (Gao et al., 2010) (Sajay-Asbaghi et al., 2020) |
| SIGLEC9 | rs16988910 (nonsynonymous SNP) | Short-term survival of NSCLC patients; Emphysema | (Laubli et al., 2014) |
| rs2075803, rs2258983 (nonsynonymous SNPs) | Exacerbation of COPD | (Ishii et al., 2017) | |
| SIGLEC10 | rs75355870 and rs145769059 (nonsynonymous SNPs) | Guillain-Barré syndrome | (Alborzian Deh Sheikh et al., 2021) |
| SIGLEC12 | rs66949844 (frameshift)* | Advanced carcinoma | (Siddiqui et al., 2021) |
| SIGLEC14 | SIGLEC14-SIGLEC5 fusion (SIGLEC14 deletion) | Exacerbation of COPD | (Angata et al., 2013) |
| SIGLEC14-SIGLEC5 fusion (SIGLEC14 deletion) | Pre-term delivery in the presence of GBS infection | (Ali et al., 2014) | |
| SIGLEC14-SIGLEC5 fusion (SIGLEC14 deletion) | TB meningitis | (Graustein et al., 2017) | |
| SIGLEC15 | rs2919643 (nonsynonymous SNP) | Recurrent vulvovaginal candidiasis | (Jaeger et al., 2019) |
| SIGLEC16 | rs67889261 (indel)** | Gonorrhea | (Landig et al., 2019) |
Updated from previous reviews (Angata, 2014; Angata, 2017; Angata, 2018). Some of the associations discovered by GWAS listed are not mentioned in the references cited, but found in the GWAS catalog (https://www.ebi.ac.uk/gwas/).
The polymorphism causing frameshift in open reading frame and most likely explaining the absence of immunohistochemical staining of Siglec-XII protein in epithelial tissues (global allele frequency of null allele ~0.59). Another SNP (rs16982743, stop gained) is located nearby, but the frequency of null allele (~0.19) caused by this SNP is lower. Genotyping was not performed on all samples.
The polymorphism (SIGLEC16: CTGAGCAATGCGTTCT; SIGLEC16P: GTGAACAATTTG) causing frameshift in open reading frame. Actual genotyping was performed on another SNP in linkage disequilibrium with this indel polymorphism.
Siglecs are not only involved in interactions with bacterial pathogens. Lymph node macrophages use Sialoadhesin/Siglec-1 to capture enveloped viruses, including HIV, as described in the article by Izquierdo-Useros in this issue (Raïch-Regué et al.). Such interaction is suspected to enhance trans-infection of CD4+ T cells (i.e., the HIV captured by macrophages is transferred to CD4+ T cells, enhancing the infection). However, SIGLEC1 null allele (frequency ~0.01) was found not to be protective against HIV infection (Martinez-Picado et al., 2016), implying that the effect size of SIGLEC1 null on AIDS may be small (although the individuals who are SIGLEC1 null appear exceedingly rare, limiting the statistical power).
Human SIGLEC12 also has high-frequency null alleles, which are a frameshift (rs66949844; global allele frequency ~0.59) and a mutation that generates stop codon (rs16982743; global allele frequency ~0.19). Siglec-XII is unusual among CD33rSiglecs in that it is expressed on luminal epithelial cells (Angata et al., 2001c; Mitra et al., 2011). (Note that Siglec-13, also expressed on epithelium in great apes, is deleted in humans.) The presence of functional Siglec-XII does not appear to increase the risk of carriers to develop early-stage cancers (Mitra et al., 2011), but it appears to be associated with advanced cancers and poor prognosis (Siddiqui et al., 2021). Epithelial Siglec-XII interacts with SHP-2, which transduces positive regulatory signal, and its overactivation is associated with tumorigenesis (Hatakeyama, 2004; Tartaglia and Gelb, 2005; Tartaglia et al., 2001), possibly explaining the mechanism behind the association.
7.2. SNPs of Siglec genes associated with diseases
Some SNPs have been found to be associated with human diseases (Table 3). The most extensively studied among them may be the SNP in CD33 that is associated with alternative splicing generating a truncated form of CD33/Siglec-3 missing the V-set domain and protective against late-onset Alzheimer’s disease (Hollingworth et al., 2011; Malik et al., 2013; Naj et al., 2011; Raj et al., 2014). The molecular mechanism that connects the polymorphism with the disease has been the subject of intense research efforts, as discussed in the review by Macauley in this issue (Macauley). It is noteworthy that the protective allele is derived and specific to the human lineage (Schwarz et al., 2016). We have suggested that this allele was selected for protection of cognition of elderly caregivers (the grandmother hypothesis) because of the very rare presence of prolonged post-reproductive lifespan in humans.
Recent reports on the Siglec-15 polymorphism and its association with tuberculosis (TB) (Bhattacharyya et al., 2019) and recurrent vulvovaginal candidiasis (Jaeger et al., 2019) imply that Siglec-15 may be relevant to microbial defense as well, whereas the primary role of Siglec-15 was considered to be in the osteoclast differentiation (Hiruma et al., 2013; Kameda et al., 2013; Stuible et al., 2014). (Note that the two SNPs reported in these studies are different but in linkage disequilibrium.)
Many other SNPs in Siglec genes have been found to be associated with various diseases, but as always, association does not necessarily mean causation. Most of the SNPs found to be strongly associated with diseases (those identified by hypothesis-free approaches in particular) tend to be localized in the intron or intergenic region (Table 3), posing challenge in mechanistic interpretations. Further mechanistic investigation would be warranted for some of the SNPs associated with diseases.
7.3. Mendelian disorder caused by Siglec mutation
A homozygous mutation (c.399C>G; p.Ser133Arg) in MAG causes autosomal recessive spastic paraplegia-75 (SPG75) (Lossos et al., 2015). Another homozygous mutation (c.1288T>G; p.Cys430Gly) was identified in another patient with SPG75 (Novarino et al., 2014). SPG75 is the only Mendelian disorder caused by a mutation in human Siglec gene discovered so far. Overall, these findings imply that most Siglecs are dispensable for human survival, but have influence on the various aspects of the quality of life.
8. Conclusion and perspectives
We provided a brief overview of the discovery and classification of the Siglec family of Sia-recognition proteins and how the family has evolved (and is still evolving) in the timescale ranging from macroevolution (evolution of species) to microevolution (intraspecies polymorphisms and selection). Low degree of cross-species conservation of CD33rSiglecs imposes a challenge in understanding the role of human Siglecs in specific physiological and/or pathological context by reverse-genetics approach. Human-specific changes in Siglecs at genetic and protein expression levels may have human-specific consequences in physiology, even for conserved Siglecs, further complicating the extrapolation of the findings garnered in other species to humans.
Investigation of polymorphisms in human genes encoding Siglecs and their association with phenotypes (forward-genetics approach) is an alternative direction to understand the pathophysiological relevance of Siglecs in human diseases. This approach, particularly hypothesis-free GWAS, has generated plenty of data to explore, although most of the polymorphisms associated with any given phenotype are located in intergenic regions, making the interpretation of the results not straightforward. Genetic association data shall be further reinforced by fine mapping and identification of likely “causal” polymorphism, followed by mechanistic study using in vitro cellular models and in vivo animal models (e.g., transgenic animals). In addition, microarray-based genotyping platform tends to miss large insertion/deletion, such as SIGLEC14–SIGLEC5 fusion polymorphism (i.e., SIGLEC14 deletion). Technical advances in the analysis of polymorphisms allowing precise assignment of large indels is anticipated.
Supplementary Material
Acknowledgment
The authors would like to thank Enago (www.enago.com) for the English language review.
Funding:
This work was supported by the National Institutes of Health [grant number R01GM32373]; and by Academia Sinica [grant numbers AS-GC-110-MD04 and AS-GC-111-M03].
References
- Aizawa H, Plitt J, Bochner BS, 2002. Human eosinophils express two Siglec-8 splice variants. J Allergy Clin Immunol 109 (1), 176. [DOI] [PubMed] [Google Scholar]
- Aizawa H, Zimmermann N, Carrigan PE, Lee JJ, Rothenberg ME, Bochner BS, 2003. Molecular analysis of human Siglec-8 orthologs relevant to mouse eosinophils: identification of mouse orthologs of Siglec-5 (mSiglec-F) and Siglec-10 (mSiglec-G). Genomics 82 (5), 521–530. [DOI] [PubMed] [Google Scholar]
- Akkaya M, Barclay AN, 2013. How do pathogens drive the evolution of paired receptors? Eur J Immunol 43 (2), 303–313. [DOI] [PubMed] [Google Scholar]
- Alborzian Deh Sheikh A, Gomaa S, Li X, Routledge M, Saigoh K, Numoto N, Angata T, Hitomi Y, Takematsu H, Tsuiji M, Ito N, Kusunoki S, Tsubata T, 2021. A Guillain-Barre syndrome-associated SIGLEC10 rare variant impairs its recognition of gangliosides. J Autoimmun 116, 102571. [DOI] [PubMed] [Google Scholar]
- Ali S, Fong J, Carlin A, Busch T, Linden R, Angata T, Areschoug T, Parast M, Varki N, Murray J, Nizet V, Varki A, 2014. Siglec-5 and Siglec-14 are polymorphic paired receptors that modulate neutrophil and amnion signaling responses to group B Streptococcus. J Exp Med 211 (6), 1231–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angata T, 2006. Molecular diversity and evolution of the Siglec family of cell-surface lectins. Mol Divers 10 (4), 555–566. [DOI] [PubMed] [Google Scholar]
- Angata T, 2014. Associations of genetic polymorphisms of Siglecs with human diseases. Glycobiology 24 (9), 785–793. [DOI] [PubMed] [Google Scholar]
- Angata T, 2017. Polymorphisms and mutations in SIGLEC genes and their associations with diseases. Journal of Japanese Biochemical Society 89 (5), 652–659. [Google Scholar]
- Angata T, 2018. Possible Influences of Endogenous and Exogenous Ligands on the Evolution of Human Siglecs. Front Immunol 9, 2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angata T, Brinkman-Van der Linden E, 2002. I-type lectins. Biochim Biophys Acta 1572 (2–3), 294–316. [DOI] [PubMed] [Google Scholar]
- Angata T, Hayakawa T, Yamanaka M, Varki A, Nakamura M, 2006. Discovery of Siglec-14, a novel sialic acid receptor undergoing concerted evolution with Siglec-5 in primates. FASEB J 20 (12), 1964–1973. [DOI] [PubMed] [Google Scholar]
- Angata T, Hingorani R, Varki N, Varki A, 2001a. Cloning and characterization of a novel mouse Siglec, mSiglec-F: differential evolution of the mouse and human (CD33) Siglec-3-related gene clusters. J Biol Chem 276, 45128–45136. [DOI] [PubMed] [Google Scholar]
- Angata T, Hingorani R, Varki NM, Varki A, 2001b. Cloning and characterization of a novel mouse Siglec, mSiglec-F: differential evolution of the mouse and human (CD33) Siglec-3-related gene clusters. J Biol Chem 276 (48), 45128–45136. [DOI] [PubMed] [Google Scholar]
- Angata T, Ishii T, Motegi T, Oka R, Taylor R, Soto P, Chang Y, Secundino I, Gao C, Ohtsubo K, Kitazume S, Nizet V, Varki A, Gemma A, Kida K, Taniguchi N, 2013. Loss of Siglec-14 reduces the risk of chronic obstructive pulmonary disease exacerbation. Cell Mol Life Sci 70 (17), 3199–3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angata T, Kerr SC, Greaves DR, Varki NM, Crocker PR, Varki A, 2002. Cloning and characterization of human Siglec-11 - A recently evolved signaling molecule that can interact with SHP-1 and SHP-2 and is expressed by tissue macrophages, including brain microglia. Journal of Biological Chemistry 277 (27), 24466–24474. [DOI] [PubMed] [Google Scholar]
- Angata T, Margulies E, Green E, Varki A, 2004. Large-scale sequencing of the CD33-related Siglec gene cluster in five mammalian species reveals rapid evolution by multiple mechanisms. Proc Natl Acad Sci U S A 101 (36), 13251–13256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angata T, Tabuchi Y, Nakamura K, Nakamura M, 2007. Siglec-15: an immune system Siglec conserved throughout vertebrate evolution. Glycobiology 17 (8), 838–846. [DOI] [PubMed] [Google Scholar]
- Angata T, Varki A, 2000a. Cloning, characterization, and phylogenetic analysis of siglec-9, a new member of the CD33-related group of siglecs. Evidence for co-evolution with sialic acid synthesis pathways. J Biol Chem 275 (29), 22127–22135. [DOI] [PubMed] [Google Scholar]
- Angata T, Varki A, 2000b. Siglec-7: a sialic acid-binding lectin of the immunoglobulin superfamily. Glycobiology 10 (4), 431–438. [DOI] [PubMed] [Google Scholar]
- Angata T, Varki A, 2002. Chemical diversity in the sialic acids and related alpha-keto acids: An evolutionary perspective. Chem Rev 102, 439–470. [DOI] [PubMed] [Google Scholar]
- Angata T, Varki NM, Varki A, 2001c. A second uniquely human mutation affecting sialic acid biology. J Biol Chem 276 (43), 40282–40287. [DOI] [PubMed] [Google Scholar]
- Angata T, von Gunten S, Schnaar RL, Varki A, 2022. I-Type Lectins, in: th, Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Mohnen D, Kinoshita T, Packer NH, Prestegard JH, Schnaar RL, Seeberger PH (Eds.), Essentials of Glycobiology, Cold Spring Harbor (NY), pp. 475–490. [Google Scholar]
- Arquint M, Roder J, Chia LS, Down J, Wilkinson D, Bayley H, Braun P, Dunn R, 1987. Molecular cloning and primary structure of myelin-associated glycoprotein. Proc Natl Acad Sci U S A 84 (2), 600–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellenguez C, Kucukali F, Jansen IE, Kleineidam L, Moreno-Grau S, Amin N, Naj AC, Campos-Martin R, Grenier-Boley B, Andrade V, Holmans PA, Boland A, Damotte V, van der Lee SJ, Costa MR, Kuulasmaa T, Yang Q, de Rojas I, Bis JC, Yaqub A, Prokic I, Chapuis J, Ahmad S, Giedraitis V, Aarsland D, Garcia-Gonzalez P, Abdelnour C, Alarcon-Martin E, Alcolea D, Alegret M, Alvarez I, Alvarez V, Armstrong NJ, Tsolaki A, Antunez C, Appollonio I, Arcaro M, Archetti S, Pastor AA, Arosio B, Athanasiu L, Bailly H, Banaj N, Baquero M, Barral S, Beiser A, Pastor AB, Below JE, Benchek P, Benussi L, Berr C, Besse C, Bessi V, Binetti G, Bizarro A, Blesa R, Boada M, Boerwinkle E, Borroni B, Boschi S, Bossu P, Brathen G, Bressler J, Bresner C, Brodaty H, Brookes KJ, Brusco LI, Buiza-Rueda D, Burger K, Burholt V, Bush WS, Calero M, Cantwell LB, Chene G, Chung J, Cuccaro ML, Carracedo A, Cecchetti R, Cervera-Carles L, Charbonnier C, Chen HH, Chillotti C, Ciccone S, Claassen J, Clark C, Conti E, Corma-Gomez A, Costantini E, Custodero C, Daian D, Dalmasso MC, Daniele A, Dardiotis E, Dartigues JF, de Deyn PP, de Paiva Lopes K, de Witte LD, Debette S, Deckert J, Del Ser T, Denning N, DeStefano A, Dichgans M, Diehl-Schmid J, Diez-Fairen M, Rossi PD, Djurovic S, Duron E, Duzel E, Dufouil C, Eiriksdottir G, Engelborghs S, Escott-Price V, Espinosa A, Ewers M, Faber KM, Fabrizio T, Nielsen SF, Fardo DW, Farotti L, Fenoglio C, Fernandez-Fuertes M, Ferrari R, Ferreira CB, Ferri E, Fin B, Fischer P, Fladby T, Fliessbach K, Fongang B, Fornage M, Fortea J, Foroud TM, Fostinelli S, Fox NC, Franco-Macias E, Bullido MJ, Frank-Garcia A, Froelich L, Fulton-Howard B, Galimberti D, Garcia-Alberca JM, Garcia-Gonzalez P, Garcia-Madrona S, Garcia-Ribas G, Ghidoni R, Giegling I, Giorgio G, Goate AM, Goldhardt O, Gomez-Fonseca D, Gonzalez-Perez A, Graff C, Grande G, Green E, Grimmer T, Grunblatt E, Grunin M, Gudnason V, Guetta-Baranes T, Haapasalo A, Hadjigeorgiou G, Haines JL, Hamilton-Nelson KL, Hampel H, Hanon O, Hardy J, Hartmann AM, Hausner L, Harwood J, Heilmann-Heimbach S, Helisalmi S, Heneka MT, Hernandez I, Herrmann MJ, Hoffmann P, Holmes C, Holstege H, Vilas RH, Hulsman M, Humphrey J, Biessels GJ, Jian X, Johansson C, Jun GR, Kastumata Y, Kauwe J, Kehoe PG, Kilander L, Stahlbom AK, Kivipelto M, Koivisto A, Kornhuber J, Kosmidis MH, Kukull WA, Kuksa PP, Kunkle BW, Kuzma AB, Lage C, Laukka EJ, Launer L, Lauria A, Lee CY, Lehtisalo J, Lerch O, Lleo A, Longstreth W Jr., Lopez O, de Munain AL, Love S, Lowemark M, Luckcuck L, Lunetta KL, Ma Y, Macias J, MacLeod CA, Maier W, Mangialasche F, Spallazzi M, Marquie M, Marshall R, Martin ER, Montes AM, Rodriguez CM, Masullo C, Mayeux R, Mead S, Mecocci P, Medina M, Meggy A, Mehrabian S, Mendoza S, Menendez-Gonzalez M, Mir P, Moebus S, Mol M, Molina-Porcel L, Montrreal L, Morelli L, Moreno F, Morgan K, Mosley T, Nothen MM, Muchnik C, Mukherjee S, Nacmias B, Ngandu T, Nicolas G, Nordestgaard BG, Olaso R, Orellana A, Orsini M, Ortega G, Padovani A, Paolo C, Papenberg G, Parnetti L, Pasquier F, Pastor P, Peloso G, Perez-Cordon A, Perez-Tur J, Pericard P, Peters O, Pijnenburg YAL, Pineda JA, Pinol-Ripoll G, Pisanu C, Polak T, Popp J, Posthuma D, Priller J, Puerta R, Quenez O, Quintela I, Thomassen JQ, Rabano A, Rainero I, Rajabli F, Ramakers I, Real LM, Reinders MJT, Reitz C, Reyes-Dumeyer D, Ridge P, Riedel-Heller S, Riederer P, Roberto N, Rodriguez-Rodriguez E, Rongve A, Allende IR, Rosende-Roca M, Royo JL, Rubino E, Rujescu D, Saez ME, Sakka P, Saltvedt I, Sanabria A, Sanchez-Arjona MB, Sanchez-Garcia F, Juan PS, Sanchez-Valle R, Sando SB, Sarnowski C, Satizabal CL, Scamosci M, Scarmeas N, Scarpini E, Scheltens P, Scherbaum N, Scherer M, Schmid M, Schneider A, Schott JM, Selbaek G, Seripa D, Serrano M, Sha J, Shadrin AA, Skrobot O, Slifer S, Snijders GJL, Soininen H, Solfrizzi V, Solomon A, Song Y, Sorbi S, Sotolongo-Grau O, Spalletta G, Spottke A, Squassina A, Stordal E, Tartan JP, Tarraga L, Tesi N, Thalamuthu A, Thomas T, Tosto G, Traykov L, Tremolizzo L, Tybjaerg-Hansen A, Uitterlinden A, Ullgren A, Ulstein I, Valero S, Valladares O, Broeckhoven CV, Vance J, Vardarajan BN, van der Lugt A, Dongen JV, van Rooij J, van Swieten J, Vandenberghe R, Verhey F, Vidal JS, Vogelgsang J, Vyhnalek M, Wagner M, Wallon D, Wang LS, Wang R, Weinhold L, Wiltfang J, Windle G, Woods B, Yannakoulia M, Zare H, Zhao Y, Zhang X, Zhu C, Zulaica M, Eadb, Gr@Ace, Degesco, Eadi, Gerad, Demgene, FinnGen, Adgc, Charge, Farrer LA, Psaty BM, Ghanbari M, Raj T, Sachdev P, Mather K, Jessen F, Ikram MA, de Mendonca A, Hort J, Tsolaki M, Pericak-Vance MA, Amouyel P, Williams J, Frikke-Schmidt R, Clarimon J, Deleuze JF, Rossi G, Seshadri S, Andreassen OA, Ingelsson M, Hiltunen M, Sleegers K, Schellenberg GD, van Duijn CM, Sims R, van der Flier WM, Ruiz A, Ramirez A, Lambert JC, 2022. New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nat Genet 54 (4), 412–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya C, Majumder PP, Pandit B, 2019. An exome wide association study of pulmonary tuberculosis patients and their asymptomatic household contacts. Infect Genet Evol 71, 76–81. [DOI] [PubMed] [Google Scholar]
- Blasius A, Cella M, Maldonado J, Takai T, Colonna M, 2006. Siglec-H is an IPC-specific receptor that modulates type I IFN secretion through DAP12. Blood 107 (6), 2474–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner BS, O’Sullivan JA, Chang AT, Youngblood BA, “In this Issue”.
- Bornhofft KF, Goldammer T, Rebl A, Galuska SP, 2018. Siglecs: A journey through the evolution of sialic acid-binding immunoglobulin-type lectins. Dev Comp Immunol 86, 219–231. [DOI] [PubMed] [Google Scholar]
- Brinkman-Van der Linden E, Sjoberg E, Juneja L, Crocker P, Varki N, Varki A, 2000. Loss of N-glycolylneuraminic acid in human evolution - Implications for sialic acid recognition by siglecs. J Biol Chem 275, 8633–8640. [DOI] [PubMed] [Google Scholar]
- Brinkman-Van der Linden EC, Hurtado-Ziola N, Hayakawa T, Wiggleton L, Benirschke K, Varki A, Varki N, 2007. Human-specific expression of Siglec-6 in the placenta. Glycobiology 17 (9), 922–931. [DOI] [PubMed] [Google Scholar]
- Bruce NG, Dherani MK, Das JK, Balakrishnan K, Adair-Rohani H, Bhutta ZA, Pope D, 2013. Control of household air pollution for child survival: estimates for intervention impacts. BMC Public Health 13 Suppl 3, S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukvic BK, Blekic M, Simpson A, Marinho S, Curtin JA, Hankinson J, Aberle N, Custovic A, 2013. Asthma severity, polymorphisms in 20p13 and their interaction with tobacco smoke exposure. Pediatr Allergy Immunol 24 (1), 10–18. [DOI] [PubMed] [Google Scholar]
- Bullock TH, Moore JK, Fields RD, 1984. Evolution of myelin sheaths: both lamprey and hagfish lack myelin. Neurosci Lett 48 (2), 145–148. [DOI] [PubMed] [Google Scholar]
- Cao H, de Bono B, Belov K, Wong ES, Trowsdale J, Barrow AD, 2009. Comparative genomics indicates the mammalian CD33rSiglec locus evolved by an ancient large-scale inverse duplication and suggests all Siglecs share a common ancestral region. Immunogenetics 61 (5), 401–417. [DOI] [PubMed] [Google Scholar]
- Cao H, Lakner U, de Bono B, Traherne J, Trowsdale J, Barrow A, 2008. SIGLEC16 encodes a DAP12-associated receptor expressed in macrophages that evolved from its inhibitory counterpart SIGLEC11 and has functional and non-functional alleles in humans. Eur J Immunol 38 (8), 2303–2315. [DOI] [PubMed] [Google Scholar]
- Carlin A, Chang Y, Areschoug T, Lindahl G, Hurtado-Ziola N, King C, Varki A, Nizet V, 2009a. Group B Streptococcus suppression of phagocyte functions by protein-mediated engagement of human Siglec-5. J Exp Med 206 (8), 1691–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlin A, Lewis A, Varki A, Nizet V, 2007. Group B streptococcal capsular sialic acids interact with siglecs (immunoglobulin-like lectins) on human leukocytes. J Bacteriol 189 (4), 1231–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlin A, Uchiyama S, Chang Y, Lewis A, Nizet V, Varki A, 2009b. Molecular mimicry of host sialylated glycans allows a bacterial pathogen to engage neutrophil Siglec-9 and dampen the innate immune response. Blood 113 (14), 3333–3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll DJ, Cao Y, Bochner BS, O’Sullivan JA, 2021. Siglec-8 Signals Through a Non-Canonical Pathway to Cause Human Eosinophil Death In Vitro. Front Immunol 12, 737988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll DJ, O’Sullivan JA, Nix DB, Cao Y, Tiemeyer M, Bochner BS, 2018. Sialic acid-binding immunoglobulin-like lectin 8 (Siglec-8) is an activating receptor mediating beta2-integrin-dependent function in human eosinophils. J Allergy Clin Immunol 141 (6), 2196–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Olson J, Beasley F, Tung C, Zhang J, Crocker P, Varki A, Nizet V, 2014. Group B Streptococcus Engages an Inhibitory Siglec through Sialic Acid Mimicry to Blunt Innate Immune and Inflammatory Responses In Vivo. PLoS Pathog 10 (1), e1003846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GY, Tang J, Zheng P, Liu Y, 2009. CD24 and Siglec-10 selectively repress tissue damage-induced immune responses. Science 323 (5922), 1722–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou H, Hayakawa T, Diaz S, Krings M, Indriati E, Leakey M, Paabo S, Satta Y, Takahata N, Varki A, 2002. Inactivation of CMP-N-acetylneuraminic acid hydroxylase occurred prior to brain expansion during human evolution. Proc Natl Acad Sci U S A 99 (18), 11736–11741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish A, Freeman S, Forbes G, Ni J, Zhang M, Cepeda M, Gentz R, Augustus M, Carter K, Crocker P, 1998. Characterization of siglec-5, a novel glycoprotein expressed on myeloid cells related to CD33. Blood 92 (6), 2123–2132. [PubMed] [Google Scholar]
- Crocker P, Clark E, Filbin M, Gordon S, Jones Y, Kehrl J, Kelm S, Le Douarin N, Powell L, Roder J, Schnaar R, Sgroi D, Stamenkovic K, Schauer R, Schachner M, Van den Berg T, Van der Merwe P, Watt S, Varki A, 1998. Siglecs: a family of sialic-acid binding lectins [letter]. Glycobiology 8, v. [DOI] [PubMed] [Google Scholar]
- Crocker P, Mucklow S, Bouckson V, McWilliam A, Willis A, Gordon S, Milon G, Kelm S, Bradfield P, 1994. Sialoadhesin, a macrophage sialic acid binding receptor for haemopoietic cells with 17 immunoglobulin-like domains. EMBO J 13, 4490–4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker PR, Gordon S, 1986. Properties and distribution of a lectin-like hemagglutinin differentially expressed by murine stromal tissue macrophages. J Exp Med 164 (6), 1862–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker PR, Kelm S, Dubois C, Martin B, McWilliam AS, Shotton DM, Paulson JC, Gordon S, 1991. Purification and properties of sialoadhesin, a sialic acid-binding receptor of murine tissue macrophages. EMBO J 10 (7), 1661–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falco M, Biassoni R, Bottino C, Vitale M, Sivori S, Augugliaro R, Moretta L, Moretta A, 1999. Identification and molecular cloning of p75/AIRM1, a novel member of the sialoadhesin family that functions as an inhibitory receptor in human natural killer cells. J Exp Med 190, 793–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores R, Zhang P, Wu W, Wang X, Ye P, Zheng P, Liu Y, 2019. Siglec genes confer resistance to systemic lupus erythematosus in humans and mice. Cell Mol Immunol 16 (2), 154–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd H, Ni J, Cornish A, Zeng Z, Liu D, Carter K, Steel J, Crocker P, 2000. Siglec-8. A novel eosinophil-specific member of the immunoglobulin superfamily. J Biol Chem 275 (2), 861–866. [DOI] [PubMed] [Google Scholar]
- Fong JJ, Sreedhara K, Deng L, Varki NM, Angata T, Liu Q, Nizet V, Varki A, 2015. Immunomodulatory activity of extracellular Hsp70 mediated via paired receptors Siglec-5 and Siglec-14. EMBO J 34 (22), 2775–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster K, Sass S, Ehrhardt H, Mous DS, Rottier RJ, Oak P, Schulze A, Flemmer AW, Gronbach J, Hubener C, Desai T, Eickelberg O, Theis FJ, Hilgendorff A, 2018. Early Identification of Bronchopulmonary Dysplasia Using Novel Biomarkers by Proteomic Screening. Am J Respir Crit Care Med 197 (8), 1076–1080. [DOI] [PubMed] [Google Scholar]
- Freeman S, Kelm S, Barber E, Crocker P, 1995. Characterization of CD33 as a new member of the sialoadhesin family of cellular interaction molecules. Blood 85 (8), 2005–2012. [PubMed] [Google Scholar]
- Friedman E, Moran DS, Ben-Avraham D, Yanovich R, Atzmon G, 2014. Novel candidate genes putatively involved in stress fracture predisposition detected by whole-exome sequencing. Genet Res (Camb) 96, e004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Shimizu K, Grant A, Rafaels N, Zhou L, Hudson S, Konno S, Zimmermann N, Araujo M, Ponte E, Cruz A, Nishimura M, Su S, Hizawa N, Beaty T, Mathias R, Rothenberg M, Barnes K, Bochner B, 2010. Polymorphisms in the sialic acid-binding immunoglobulin-like lectin-8 (Siglec-8) gene are associated with susceptibility to asthma. Eur J Hum Genet 18 (6), 713–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertz EM, Yu YK, Agarwala R, Schaffer AA, Altschul SF, 2006. Composition-based statistics and translated nucleotide searches: improving the TBLASTN module of BLAST. BMC Biol 4, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graustein AD, Horne DJ, Fong JJ, Schwarz F, Mefford HC, Peterson GJ, Wells RD, Musvosvi M, Shey M, Hanekom WA, Hatherill M, Scriba TJ, Thuong NTT, Mai NTH, Caws M, Bang ND, Dunstan SJ, Thwaites GE, Varki A, Angata T, Hawn TR, 2017. The SIGLEC14 null allele is associated with Mycobacterium tuberculosis- and BCG-induced clinical and immunologic outcomes. Tuberculosis (Edinb) 104, 38–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Ira G, 2009. Mechanisms of change in gene copy number. Nat Rev Genet 10 (8), 551–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama M, 2004. Oncogenic mechanisms of the Helicobacter pylori CagA protein. Nat Rev Cancer 4 (9), 688–694. [DOI] [PubMed] [Google Scholar]
- Hayakawa T, Angata T, Lewis A, Mikkelsen T, Varki N, Varki A, 2005. A human-specific gene in microglia. Science 309 (5741), 1693. [DOI] [PubMed] [Google Scholar]
- Hayakawa T, Khedri Z, Schwarz F, Landig C, Liang SY, Yu H, Chen X, Fujito NT, Satta Y, Varki A, Angata T, 2017. Coevolution of Siglec-11 and Siglec-16 via gene conversion in primates. BMC Evol Biol 17 (1), 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa T, Satta Y, Gagneux P, Varki A, Takahata N, 2001. Alu-mediated inactivation of the human CMP- N-acetylneuraminic acid hydroxylase gene. Proc Natl Acad Sci U S A 98 (20), 11399–11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiruma Y, Tsuda E, Maeda N, Okada A, Kabasawa N, Miyamoto M, Hattori H, Fukuda C, 2013. Impaired osteoclast differentiation and function and mild osteopetrosis development in Siglec-15-deficient mice. Bone 53 (1), 87–93. [DOI] [PubMed] [Google Scholar]
- Hitomi Y, Tsuchiya N, Hasegawa M, Fujimoto M, Takehara K, Tokunaga K, Sato S, 2007. Association of CD22 gene polymorphism with susceptibility to limited cutaneous systemic sclerosis. Tissue Antigens 69 (3), 242–249. [DOI] [PubMed] [Google Scholar]
- Hoffmann A, Kerr S, Jellusova J, Zhang J, Weisel F, Wellmann U, Winkler T, Kneitz B, Crocker P, Nitschke L, 2007. Siglec-G is a B1 cell-inhibitory receptor that controls expansion and calcium signaling of the B1 cell population. Nat Immunol 8 (7), 695–704. [DOI] [PubMed] [Google Scholar]
- Holliday R, Grigg GW, 1993. DNA methylation and mutation. Mutat Res 285 (1), 61–67. [DOI] [PubMed] [Google Scholar]
- Hollingworth P, Harold D, Sims R, Gerrish A, Lambert J, Carrasquillo M, Abraham R, Hamshere M, Pahwa J, Moskvina V, Dowzell K, Jones N, Stretton A, Thomas C, Richards A, Ivanov D, Widdowson C, Chapman J, Lovestone S, Powell J, Proitsi P, Lupton M, Brayne C, Rubinsztein D, Gill M, Lawlor B, Lynch A, Brown K, Passmore P, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith A, Beaumont H, Warden D, Wilcock G, Love S, Kehoe P, Hooper N, Vardy E, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Ruther E, Schurmann B, Heun R, Kolsch H, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Gallacher J, Hull M, Rujescu D, Giegling I, Goate A, Kauwe J, Cruchaga C, Nowotny P, Morris J, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn P, Van Broeckhoven C, Livingston G, Bass N, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw C, Tsolaki M, Singleton A, Guerreiro R, Muhleisen T, Nothen M, Moebus S, Jockel K, Klopp N, Wichmann H, Pankratz V, Sando S, Aasly J, Barcikowska M, Wszolek Z, Dickson D, Graff-Radford N, Petersen R, van Duijn C, Breteler M, Ikram M, DeStefano A, Fitzpatrick A, Lopez O, Launer L, Seshadri S, Berr C, Campion D, Epelbaum J, Dartigues J, Tzourio C, Alperovitch A, Lathrop M, Feulner T, Friedrich P, Riehle C, Krawczak M, Schreiber S, Mayhaus M, Nicolhaus S, Wagenpfeil S, Steinberg S, Stefansson H, Stefansson K, Snaedal J, Bjornsson S, Jonsson P, Chouraki V, Genier-Boley B, Hiltunen M, Soininen H, Combarros O, Zelenika D, Delepine M, Bullido M, Pasquier F, Mateo I, Frank-Garcia A, Porcellini E, Hanon O, Coto E, Alvarez V, Bosco P, Siciliano G, Mancuso M, Panza F, Solfrizzi V, Nacmias B, Sorbi S, Bossu P, Piccardi P, Arosio B, Annoni G, Seripa D, Pilotto A, Scarpini E, Galimberti D, Brice A, Hannequin D, Licastro F, Jones L, Holmans P, Jonsson T, Riemenschneider M, Morgan K, Younkin S, Owen M, O’Donovan M, Amouyel P, Williams J, 2011. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet 43 (5), 429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang PCJ, Low PY, Wang I, Hsu STD, Angata T, 2018. Soluble Siglec-14 glycan-recognition protein is generated by alternative splicing and suppresses myeloid inflammatory responses. J Biol Chem 293 (51), 19645–19658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YJ, Lee JJ, Fan WL, Hsu CW, Tsai NW, Lu CH, Chang WN, Tsai MH, 2021. A CD33 frameshift variant is associated with neuromyelitis optica spectrum disorders. Biomed J 44 (6 Suppl 1), S93–S100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Lupski JR, 2002. Molecular mechanisms for genomic disorders. Annu Rev Genomics Hum Genet 3, 199–242. [DOI] [PubMed] [Google Scholar]
- Ishii T, Angata T, Wan ES, Cho MH, Motegi T, Gao C, Ohtsubo K, Kitazume S, Gemma A, Par EP, Lomas DA, Silverman EK, Taniguchi N, Kida K, 2017. Influence of SIGLEC9 polymorphisms on COPD phenotypes including exacerbation frequency. Respirology 22 (4), 684–690. [DOI] [PubMed] [Google Scholar]
- Jaeger M, Pinelli M, Borghi M, Constantini C, Dindo M, van Emst L, Puccetti M, Pariano M, Ricano-Ponce I, Bull C, Gresnigt MS, Wang X, Gutierrez Achury J, Jacobs CWM, Xu N, Oosting M, Arts P, Joosten LAB, van de Veerdonk FL, Veltman JA, Ten Oever J, Kullberg BJ, Feng M, Adema GJ, Wijmenga C, Kumar V, Sobel J, Gilissen C, Romani L, Netea MG, 2019. A systems genomics approach identifies SIGLEC15 as a susceptibility factor in recurrent vulvovaginal candidiasis. Sci Transl Med 11 (496), eaar3558. [DOI] [PubMed] [Google Scholar]
- Jitoku D, Hattori E, Iwayama Y, Yamada K, Toyota T, Kikuchi M, Maekawa M, Nishikawa T, Yoshikawa T, 2011. Association study of Nogo-related genes with schizophrenia in a Japanese case-control sample. Am J Med Genet B Neuropsychiatr Genet 156B (5), 581–592. [DOI] [PubMed] [Google Scholar]
- Jonsson S, Sveinbjornsson G, de Lapuente Portilla AL, Swaminathan B, Plomp R, Dekkers G, Ajore R, Ali M, Bentlage AEH, Elmer E, Eyjolfsson GI, Gudjonsson SA, Gullberg U, Gylfason A, Halldorsson BV, Hansson M, Holm H, Johansson A, Johnsson E, Jonasdottir A, Ludviksson BR, Oddsson A, Olafsson I, Olafsson S, Sigurdardottir O, Sigurdsson A, Stefansdottir L, Masson G, Sulem P, Wuhrer M, Wihlborg AK, Thorleifsson G, Gudbjartsson DF, Thorsteinsdottir U, Vidarsson G, Jonsdottir I, Nilsson B, Stefansson K, 2017. Identification of sequence variants influencing immunoglobulin levels. Nat Genet 49 (8), 1182–1191. [DOI] [PubMed] [Google Scholar]
- Kameda Y, Takahata M, Komatsu M, Mikuni S, Hatakeyama S, Shimizu T, Angata T, Kinjo M, Minami A, Iwasaki N, 2013. Siglec-15 regulates osteoclast differentiation by modulating RANKL-induced phosphatidylinositol 3-kinase/Akt and Erk pathways in association with signaling Adaptor DAP12. J Bone Miner Res 28 (12), 2463–2475. [DOI] [PubMed] [Google Scholar]
- Kelm S, Pelz A, Schauer R, Filbin M, Tang S, de Bellard M, Schnaar R, Mahoney J, Hartnell A, Bradfield P, et, a., 1994. Sialoadhesin, myelin-associated glycoprotein and CD22 define a new family of sialic acid-dependent adhesion molecules of the immunoglobulin superfamily. Curr Biol 4 (11), 965–972. [DOI] [PubMed] [Google Scholar]
- Khan N, de Manuel M, Peyregne S, Do R, Prufer K, Marques-Bonet T, Varki N, Gagneux P, Varki A, 2020a. Multiple Genomic Events Altering Hominin SIGLEC Biology and Innate Immunity Predated the Common Ancestor of Humans and Archaic Hominins. Genome Biol Evol 12 (7), 1040–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan N, Kim SK, Gagneux P, Dugan LL, Varki A, 2020b. Maximum reproductive lifespan correlates with CD33rSIGLEC gene number: Implications for NADPH oxidase-derived reactive oxygen species in aging. FASEB J 34 (2), 1928–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikly K, Bochner B, Freeman S, Tan K, Gallagher K, D’alessio K, Holmes S, Abrahamson J, Erickson-Miller C, Murdock P, Tachimoto H, Schleimer R, White J, 2000. Identification of SAF-2, a novel siglec expressed on eosinophils, mast cells, and basophils. J Allergy Clin Immunol 105 (6 Pt 1), 1093–1100. [DOI] [PubMed] [Google Scholar]
- Kitzig F, Martinez-Barriocanal A, López-Botet M, Sayós J, 2002. Cloning of two new splice variants of Siglec-10 and mapping of the interaction between Siglec-10 and SHP-1. Biochem Biophys Res Commun 296 (2), 355–362. [DOI] [PubMed] [Google Scholar]
- Korver W, Wong A, Gebremeskel S, Negri GL, Schanin J, Chang K, Leung J, Benet Z, Luu T, Brock EC, Luehrsen K, Xu A, Youngblood BA, 2022. The Inhibitory Receptor Siglec-8 Interacts With FcepsilonRI and Globally Inhibits Intracellular Signaling in Primary Mast Cells Upon Activation. Front Immunol 13, 833728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C, Brow MA, Nave KA, Noronha AB, Quarles RH, Bloom FE, Milner RJ, Sutcliffe JG, 1987. Two forms of 1B236/myelin-associated glycoprotein, a cell adhesion molecule for postnatal neural development, are produced by alternative splicing. Proc Natl Acad Sci U S A 84 (12), 4337–4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamba JK, Pounds S, Cao X, Downing JR, Campana D, Ribeiro RC, Pui CH, Rubnitz JE, 2009. Coding polymorphisms in CD33 and response to gemtuzumab ozogamicin in pediatric patients with AML: a pilot study. Leukemia 23 (2), 402–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landig CS, Hazel A, Kellman BP, Fong JJ, Schwarz F, Agarwal S, Varki N, Massari P, Lewis NE, Ram S, Varki A, 2019. Evolution of the exclusively human pathogen Neisseria gonorrhoeae: Human-specific engagement of immunoregulatory Siglecs. Evol Appl 12 (2), 337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Läubli H, “In this Issue”.
- Laubli H, Pearce O, Schwarz F, Siddiqui S, Deng L, Stanczak M, Deng L, Verhagen A, Secrest P, Lusk C, Schwartz A, Varki N, Bui J, Varki A, 2014. Engagement of myelomonocytic Siglecs by tumor-associated ligands modulates the innate immune response to cancer. Proc Natl Acad Sci U S A 111 (39), 14211–14216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann F, Gathje H, Kelm S, Dietz F, 2004. Evolution of sialic acid-binding proteins: molecular cloning and expression of fish siglec-4. Glycobiology 14 (11), 959–968. [DOI] [PubMed] [Google Scholar]
- Lewis AL, Chen X, Schnaar RL, Varki A, 2022. Sialic Acids and Other Nonulosonic Acids, in: th, Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Mohnen D, Kinoshita T, Packer NH, Prestegard JH, Schnaar RL, Seeberger PH (Eds.), Essentials of Glycobiology, Cold Spring Harbor (NY), pp. 185–204. [Google Scholar]
- Li N, Zhang W, Wan T, Zhang J, Chen T, Yu Y, Wang J, Cao X, 2001. Cloning and characterization of Siglec-10, a novel sialic acid binding member of the Ig superfamily, from human dendritic cells. J Biol Chem 276 (30), 28106–28112. [DOI] [PubMed] [Google Scholar]
- Liu H, Irwanto A, Fu X, Yu G, Yu Y, Sun Y, Wang C, Wang Z, Okada Y, Low H, Li Y, Liany H, Chen M, Bao F, Li J, You J, Zhang Q, Liu J, Chu T, Andiappan AK, Wang N, Niu G, Liu D, Yu X, Zhang L, Tian H, Zhou G, Rotzschke O, Chen S, Zhang X, Liu J, Zhang F, 2015. Discovery of six new susceptibility loci and analysis of pleiotropic effects in leprosy. Nat Genet 47 (3), 267–271. [DOI] [PubMed] [Google Scholar]
- Lossos A, Elazar N, Lerer I, Schueler-Furman O, Fellig Y, Glick B, Zimmerman BE, Azulay H, Dotan S, Goldberg S, Gomori JM, Ponger P, Newman JP, Marreed H, Steck AJ, Schaeren-Wiemers N, Mor N, Harel M, Geiger T, Eshed-Eisenbach Y, Meiner V, Peles E, 2015. Myelin-associated glycoprotein gene mutation causes Pelizaeus-Merzbacher disease-like disorder. Brain 138 (Pt 9), 2521–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Qazi S, Ozer Z, Gaynon P, Reaman G, Uckun F, 2012. CD22 Exon 12 deletion is a characteristic genetic defect of therapy-refractory clones in paediatric acute lymphoblastic leukaemia. Br J Haematol 156 (1), 89–98. [DOI] [PubMed] [Google Scholar]
- Ma Q, Ozel AB, Ramdas S, McGee B, Khoriaty R, Siemieniak D, Li HD, Guan Y, Brody LC, Mills JL, Molloy AM, Ginsburg D, Li JZ, Desch KC, 2014. Genetic variants in PLG, LPA, and SIGLEC 14 as well as smoking contribute to plasma plasminogen levels. Blood 124 (20), 3155–3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macauley MS, “In this Issue”.
- Malik M, Simpson J, Parikh I, Wilfred B, Fardo D, Nelson P, Estus S, 2013. CD33 Alzheimer’s risk-altering polymorphism, CD33 expression, and exon 2 splicing. J Neurosci 33 (33), 13320–13325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Picado J, McLaren PJ, Erkizia I, Martin MP, Benet S, Rotger M, Dalmau J, Ouchi D, Wolinsky SM, Penugonda S, Gunthard HF, Fellay J, Carrington M, Izquierdo-Useros N, Telenti A, 2016. Identification of Siglec-1 null individuals infected with HIV-1. Nat Commun 7, 12412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonough C, Gong Y, Padmanabhan S, Burkley B, Langaee T, Melander O, Pepine C, Dominiczak A, Cooper-Dehoff R, Johnson J, 2013. Pharmacogenomic association of nonsynonymous SNPs in SIGLEC12, A1BG, and the selectin region and cardiovascular outcomes. Hypertension 62 (1), 48–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay JD, Hung RJ, Han Y, Zong X, Carreras-Torres R, Christiani DC, Caporaso NE, Johansson M, Xiao X, Li Y, Byun J, Dunning A, Pooley KA, Qian DC, Ji X, Liu G, Timofeeva MN, Bojesen SE, Wu X, Le Marchand L, Albanes D, Bickeboller H, Aldrich MC, Bush WS, Tardon A, Rennert G, Teare MD, Field JK, Kiemeney LA, Lazarus P, Haugen A, Lam S, Schabath MB, Andrew AS, Shen H, Hong YC, Yuan JM, Bertazzi PA, Pesatori AC, Ye Y, Diao N, Su L, Zhang R, Brhane Y, Leighl N, Johansen JS, Mellemgaard A, Saliba W, Haiman CA, Wilkens LR, Fernandez-Somoano A, Fernandez-Tardon G, van der Heijden HFM, Kim JH, Dai J, Hu Z, Davies MPA, Marcus MW, Brunnstrom H, Manjer J, Melander O, Muller DC, Overvad K, Trichopoulou A, Tumino R, Doherty JA, Barnett MP, Chen C, Goodman GE, Cox A, Taylor F, Woll P, Bruske I, Wichmann HE, Manz J, Muley TR, Risch A, Rosenberger A, Grankvist K, Johansson M, Shepherd FA, Tsao MS, Arnold SM, Haura EB, Bolca C, Holcatova I, Janout V, Kontic M, Lissowska J, Mukeria A, Ognjanovic S, Orlowski TM, Scelo G, Swiatkowska B, Zaridze D, Bakke P, Skaug V, Zienolddiny S, Duell EJ, Butler LM, Koh WP, Gao YT, Houlston RS, McLaughlin J, Stevens VL, Joubert P, Lamontagne M, Nickle DC, Obeidat M, Timens W, Zhu B, Song L, Kachuri L, Artigas MS, Tobin MD, Wain LV, SpiroMeta C, Rafnar T, Thorgeirsson TE, Reginsson GW, Stefansson K, Hancock DB, Bierut LJ, Spitz MR, Gaddis NC, Lutz SM, Gu F, Johnson EO, Kamal A, Pikielny C, Zhu D, Lindstroem S, Jiang X, Tyndale RF, Chenevix-Trench G, Beesley J, Bosse Y, Chanock S, Brennan P, Landi MT, Amos CI, 2017. Large-scale association analysis identifies new lung cancer susceptibility loci and heterogeneity in genetic susceptibility across histological subtypes. Nat Genet 49 (7), 1126–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra N, Banda K, Altheide T, Schaffer L, Johnson-Pais T, Beuten J, Leach R, Angata T, Varki N, Varki A, 2011. SIGLEC12, a human-specific segregating (pseudo)gene, encodes a signaling molecule expressed in prostate carcinomas. J Biol Chem 286 (26), 23003–23011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortland L, Alonzo T, Walter R, Gerbing R, Mitra A, Pollard J, Loken M, Hirsch B, Raimondi S, Franklin J, Pounds S, Cao X, Rubnitz J, Ribeiro R, Gamis A, Meshinchi S, Lamba J, 2013. Clinical significance of CD33 nonsynonymous single-nucleotide polymorphisms in pediatric patients with acute myeloid leukemia treated with gemtuzumab-ozogamicin-containing chemotherapy. Clin Cancer Res 19 (6), 1620–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munday J, Kerr S, Ni J, Cornish AL, Zhang JQ, Nicoll G, Floyd H, Mattei MG, Moore P, Liu D, Crocker PR, 2001. Identification, characterization and leucocyte expression of Siglec-10, a novel human sialic acid-binding receptor. Biochem J 355 (Pt 2), 489–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munz M, Richter GM, Loos BG, Jepsen S, Divaris K, Offenbacher S, Teumer A, Holtfreter B, Kocher T, Bruckmann C, Jockel-Schneider Y, Graetz C, Ahmad I, Staufenbiel I, van der Velde N, Uitterlinden AG, de Groot L, Wellmann J, Berger K, Krone B, Hoffmann P, Laudes M, Lieb W, Franke A, Erdmann J, Dommisch H, Schaefer AS, 2019. Meta-analysis of genome-wide association studies of aggressive and chronic periodontitis identifies two novel risk loci. Eur J Hum Genet 27 (1), 102–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munz M, Willenborg C, Richter GM, Jockel-Schneider Y, Graetz C, Staufenbiel I, Wellmann J, Berger K, Krone B, Hoffmann P, van der Velde N, Uitterlinden AG, de Groot L, Sawalha AH, Direskeneli H, Saruhan-Direskeneli G, Guzeldemir-Akcakanat E, Keceli HG, Laudes M, Noack B, Teumer A, Holtfreter B, Kocher T, Eickholz P, Meyle J, Doerfer C, Bruckmann C, Lieb W, Franke A, Schreiber S, Nohutcu RM, Erdmann J, Loos BG, Jepsen S, Dommisch H, Schaefer AS, 2017. A genome-wide association study identifies nucleotide variants at SIGLEC5 and DEFA1A3 as risk loci for periodontitis. Hum Mol Genet 26 (13), 2577–2588. [DOI] [PubMed] [Google Scholar]
- Naj A, Jun G, Beecham G, Wang L, Vardarajan B, Buros J, Gallins P, Buxbaum J, Jarvik G, Crane P, Larson E, Bird T, Boeve B, Graff-Radford N, De Jager P, Evans D, Schneider J, Carrasquillo M, Ertekin-Taner N, Younkin S, Cruchaga C, Kauwe J, Nowotny P, Kramer P, Hardy J, Huentelman M, Myers A, Barmada M, Demirci F, Baldwin C, Green R, Rogaeva E, St George-Hyslop P, Arnold S, Barber R, Beach T, Bigio E, Bowen J, Boxer A, Burke J, Cairns N, Carlson C, Carney R, Carroll S, Chui H, Clark D, Corneveaux J, Cotman C, Cummings J, DeCarli C, DeKosky S, Diaz-Arrastia R, Dick M, Dickson D, Ellis W, Faber K, Fallon K, Farlow M, Ferris S, Frosch M, Galasko D, Ganguli M, Gearing M, Geschwind D, Ghetti B, Gilbert J, Gilman S, Giordani B, Glass J, Growdon J, Hamilton R, Harrell L, Head E, Honig L, Hulette C, Hyman B, Jicha G, Jin L, Johnson N, Karlawish J, Karydas A, Kaye J, Kim R, Koo E, Kowall N, Lah J, Levey A, Lieberman A, Lopez O, Mack W, Marson D, Martiniuk F, Mash D, Masliah E, McCormick W, McCurry S, McDavid A, McKee A, Mesulam M, Miller B, Miller C, Miller J, Parisi J, Perl D, Peskind E, Petersen R, Poon W, Quinn J, Rajbhandary R, Raskind M, Reisberg B, Ringman J, Roberson E, Rosenberg R, Sano M, Schneider L, Seeley W, Shelanski M, Slifer M, Smith C, Sonnen J, Spina S, Stern R, Tanzi R, Trojanowski J, Troncoso J, Van Deerlin V, Vinters H, Vonsattel J, Weintraub S, Welsh-Bohmer K, Williamson J, Woltjer R, Cantwell L, Dombroski B, Beekly D, Lunetta K, Martin E, Kamboh M, Saykin A, Reiman E, Bennett D, Morris J, Montine T, Goate A, Blacker D, Tsuang D, Hakonarson H, Kukull W, Foroud T, Haines J, Mayeux R, Pericak-Vance M, Farrer L, Schellenberg G, 2011. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet 43 (5), 436–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D, Hurtado-Ziola N, Gagneux P, Varki A, 2006. Loss of Siglec expression on T lymphocytes during human evolution. Proc Natl Acad Sci U S A 103 (20), 7765–7770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoll G, Ni J, Liu D, Klenerman P, Munday J, Dubock S, Mattei MG, Crocker PR, 1999. Identification and characterization of a novel siglec, siglec-7, expressed by human natural killer cells and monocytes. J Biol Chem 274 (48), 34089–34095. [DOI] [PubMed] [Google Scholar]
- Nitschke L, 2014. CD22 and Siglec-G regulate inhibition of B-cell signaling by sialic acid ligand binding and control B-cell tolerance. Glycobiology 24 (9), 807–817. [DOI] [PubMed] [Google Scholar]
- Nizet V, “In this Issue”.
- Novarino G, Fenstermaker AG, Zaki MS, Hofree M, Silhavy JL, Heiberg AD, Abdellateef M, Rosti B, Scott E, Mansour L, Masri A, Kayserili H, Al-Aama JY, Abdel-Salam GMH, Karminejad A, Kara M, Kara B, Bozorgmehri B, Ben-Omran T, Mojahedi F, El Din Mahmoud IG, Bouslam N, Bouhouche A, Benomar A, Hanein S, Raymond L, Forlani S, Mascaro M, Selim L, Shehata N, Al-Allawi N, Bindu PS, Azam M, Gunel M, Caglayan A, Bilguvar K, Tolun A, Issa MY, Schroth J, Spencer EG, Rosti RO, Akizu N, Vaux KK, Johansen A, Koh AA, Megahed H, Durr A, Brice A, Stevanin G, Gabriel SB, Ideker T, Gleeson JG, 2014. Exome sequencing links corticospinal motor neuron disease to common neurodegenerative disorders. Science 343 (6170), 506–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel N, Brinkman-Van der Linden E, Altmann S, Gish K, Balasubramanian S, Timans J, Peterson D, Bell M, Bazan J, Varki A, Kastelein R, 1999. OB-BP1/Siglec-6 - A leptin- and sialic acid-binding protein of the immunoglobulin superfamily. J Biol Chem 274, 22729–22738. [DOI] [PubMed] [Google Scholar]
- Paulson JC, “In this Issue”.
- Powell L, Sgroi D, Sjoberg E, Stamenkovic I, Varki A, 1993. Natural ligands of the B cell adhesion molecule CD22 beta carry N-linked oligosaccharides with alpha-2,6-linked sialic acids that are required for recognition. J Biol Chem 268 (10), 7019–7027. [PubMed] [Google Scholar]
- Powell LD, Varki A, 1995. I-type lectins. J Biol Chem 270 (24), 14243–14246. [DOI] [PubMed] [Google Scholar]
- Raïch-Regué D, Resa-Infante P, Gallemí M, Laguia F, Muñiz-Trabudua X, Muñoz-Basagoiti J, Perez-Zsolt D, Chojnaki J, Benet S, Clotet B, Martinez-Picado J, Izquierdo-Useros N, “In this Issue”. [DOI] [PMC free article] [PubMed]
- Raj T, Ryan KJ, Replogle JM, Chibnik LB, Rosenkrantz L, Tang A, Rothamel K, Stranger BE, Bennett DA, Evans DA, De Jager PL, Bradshaw EM, 2014. CD33: increased inclusion of exon 2 implicates the Ig V-set domain in Alzheimer’s disease susceptibility. Hum Mol Genet 23 (10), 2729–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashkin SR, Graff RE, Kachuri L, Thai KK, Alexeeff SE, Blatchins MA, Cavazos TB, Corley DA, Emami NC, Hoffman JD, Jorgenson E, Kushi LH, Meyers TJ, Van Den Eeden SK, Ziv E, Habel LA, Hoffmann TJ, Sakoda LC, Witte JS, 2020. Pan-cancer study detects genetic risk variants and shared genetic basis in two large cohorts. Nat Commun 11 (1), 4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Khan N, Comi T, Verhagen A, Sasmal A, Diaz S, Yu H, Chen X, Akey JM, Frank M, Gagneux P, Varki A, 2022. Evolution of Human-specific Alleles Protecting Cognitive Function of Grandmothers. Mol Biol Evol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajay-Asbaghi M, Sadeghi-Shabestrai M, Monfaredan A, Seyfizadeh N, Razavi A, Kazemi T, 2020. Promoter region single nucleotide polymorphism of siglec-8 gene associates with susceptibility to allergic asthma. Per Med 17 (3), 195–201. [DOI] [PubMed] [Google Scholar]
- Salzer JL, Holmes WP, Colman DR, 1987. The amino acid sequences of the myelin-associated glycoproteins: homology to the immunoglobulin gene superfamily. J Cell Biol 104 (4), 957–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M, Lange J, Keeney S, 2010. Genome destabilization by homologous recombination in the germ line. Nat Rev Mol Cell Biol 11 (3), 182–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaar RL, “In this Issue”.
- Schwarz F, Fong JJ, Varki A, 2015a. Human-specific evolutionary changes in the biology of siglecs. Adv Exp Med Biol 842, 1–16. [DOI] [PubMed] [Google Scholar]
- Schwarz F, Landig CS, Siddiqui S, Secundino I, Olson J, Varki N, Nizet V, Varki A, 2017. Paired Siglec receptors generate opposite inflammatory responses to a human-specific pathogen. EMBO J 36 (6), 751–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz F, Pearce OM, Wang X, Samraj AN, Laubli H, Garcia JO, Lin H, Fu X, Garcia-Bingman A, Secrest P, Romanoski CE, Heyser C, Glass CK, Hazen SL, Varki N, Varki A, Gagneux P, 2015b. Siglec receptors impact mammalian lifespan by modulating oxidative stress. Elife 4, e06184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz F, Springer SA, Altheide TK, Varki NM, Gagneux P, Varki A, 2016. Human-specific derived alleles of CD33 and other genes protect against postreproductive cognitive decline. Proc Natl Acad Sci U S A 113 (1), 74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgroi D, Varki A, Braesch-Andersen S, Stamenkovic I, 1993. CD22, a B cell-specific immunoglobulin superfamily member, is a sialic acid-binding lectin. J Biol Chem 268 (10), 7011–7018. [PubMed] [Google Scholar]
- Siddiqui SS, “In this Issue”.
- Siddiqui SS, Vaill M, Do R, Khan N, Verhagen AL, Zhang W, Lenz HJ, Johnson-Pais TL, Leach RJ, Fraser G, Wang C, Feng GS, Varki N, Varki A, 2021. Human-specific polymorphic pseudogenization of SIGLEC12 protects against advanced cancer progression. FASEB Bioadv 3 (2), 69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siew JJ, Chern Y, Khoo KH, Angata T, “In this Issue”.
- Smith DK, Xue H, 1997. Sequence profiles of immunoglobulin and immunoglobulin-like domains. J Mol Biol 274 (4), 530–545. [DOI] [PubMed] [Google Scholar]
- Sonnenburg J, Altheide T, Varki A, 2004. A uniquely human consequence of domain-specific functional adaptation in a sialic acid-binding receptor. Glycobiology 14, 339–346. [DOI] [PubMed] [Google Scholar]
- Soto P, Karris M, Spina C, Richman D, Varki A, 2013. Cell-intrinsic mechanism involving Siglec-5 associated with divergent outcomes of HIV-1 infection in human and chimpanzee CD4 T cells. J Mol Med (Berl) 91 (2), 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto PC, Stein LL, Hurtado-Ziola N, Hedrick SM, Varki A, 2010. Relative over-reactivity of human versus chimpanzee lymphocytes: implications for the human diseases associated with immune activation. J Immunol 184 (8), 4185–4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza de Lima D, Nunes VCL, Ogusku MM, Sadahiro A, Pontillo A, Alencar BC, 2017. Polymorphisms in SIGLEC1 contribute to susceptibility to pulmonary active tuberculosis possibly through the modulation of IL-1ss. Infect Genet Evol 55, 313–317. [DOI] [PubMed] [Google Scholar]
- Stamenkovic I, Seed B, 1990. The B-cell antigen CD22 mediates monocyte and erythrocyte adhesion. Nature 345 (6270), 74–77. [DOI] [PubMed] [Google Scholar]
- Stamenkovic I, Sgroi D, Aruffo A, 1992. CD22 binds to alpha-2,6-sialyltransferase-dependent epitopes on COS cells. Cell 68 (6), 1003–1004. [DOI] [PubMed] [Google Scholar]
- Stamenkovic I, Sgroi D, Aruffo A, Sy M, Anderson T, 1991. The B lymphocyte adhesion molecule CD22 interacts with leukocyte common antigen CD45RO on T cells and alpha 2–6 sialyltransferase, CD75, on B cells. Cell 66 (6), 1133–1144. [DOI] [PubMed] [Google Scholar]
- Stephenson HN, Mills DC, Jones H, Milioris E, Copland A, Dorrell N, Wren BW, Crocker PR, Escors D, Bajaj-Elliott M, 2014. Pseudaminic acid on Campylobacter jejuni flagella modulates dendritic cell IL-10 expression via Siglec-10 receptor: a novel flagellin-host interaction. J Infect Dis 210 (9), 1487–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuible M, Moraitis A, Fortin A, Saragosa S, Kalbakji A, Filion M, Tremblay G, 2014. Mechanism and function of monoclonal antibodies targeting siglec-15 for therapeutic inhibition of osteoclastic bone resorption. J Biol Chem 289 (10), 6498–6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suematsu R, Miyamoto T, Saijo S, Yamasaki S, Tada Y, Yoshida H, Miyake Y, 2019. Identification of lipophilic ligands of Siglec5 and −14 that modulate innate immune responses. J Biol Chem 294 (45), 16776–16788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Molineros JE, Looger LL, Zhou XJ, Kim K, Okada Y, Ma J, Qi YY, Kim-Howard X, Motghare P, Bhattarai K, Adler A, Bang SY, Lee HS, Kim TH, Kang YM, Suh CH, Chung WT, Park YB, Choe JY, Shim SC, Kochi Y, Suzuki A, Kubo M, Sumida T, Yamamoto K, Lee SS, Kim YJ, Han BG, Dozmorov M, Kaufman KM, Wren JD, Harley JB, Shen N, Chua KH, Zhang H, Bae SC, Nath SK, 2016. High-density genotyping of immune-related loci identifies new SLE risk variants in individuals with Asian ancestry. Nat Genet 48 (3), 323–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaki R, Watson S, Lanier L, 2006. DAP12: an adapter protein with dual functionality. Immunol Rev 214, 118–129. [DOI] [PubMed] [Google Scholar]
- Tartaglia M, Gelb BD, 2005. Noonan syndrome and related disorders: genetics and pathogenesis. Annu Rev Genomics Hum Genet 6, 45–68. [DOI] [PubMed] [Google Scholar]
- Tartaglia M, Mehler EL, Goldberg R, Zampino G, Brunner HG, Kremer H, van der Burgt I, Crosby AH, Ion A, Jeffery S, Kalidas K, Patton MA, Kucherlapati RS, Gelb BD, 2001. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat Genet 29 (4), 465–468. [DOI] [PubMed] [Google Scholar]
- Tateno H, Crocker P, Paulson J, 2005. Mouse Siglec-F and human Siglec-8 are functionally convergent paralogs that are selectively expressed on eosinophils and recognize 6’-sulfo-sialyl Lewis X as a preferred glycan ligand. Glycobiology 15 (11), 1125–1135. [DOI] [PubMed] [Google Scholar]
- Trowsdale J, Barten R, Haude A, Stewart CA, Beck S, Wilson MJ, 2001. The genomic context of natural killer receptor extended gene families. Immunol Rev 181, 20–38. [DOI] [PubMed] [Google Scholar]
- Tsai CM, Riestra AM, Ali SR, Fong JJ, Liu JZ, Hughes G, Varki A, Nizet V, 2020. Siglec-14 Enhances NLRP3-Inflammasome Activation in Macrophages. J Innate Immun 12 (4), 333–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai TY, Huang MT, Sung PS, Peng CY, Tao MH, Yang HI, Chang WC, Yang AS, Yu CM, Lin YP, Bau CY, Huang CJ, Pan MH, Wu CY, Hsiao CD, Yeh YH, Duan S, Paulson JC, Hsieh SL, 2021. SIGLEC-3 (CD33) serves as an immune checkpoint receptor for HBV infection. J Clin Invest 131 (11), e141965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uckun F, Goodman P, Ma H, Dibirdik I, Qazi S, 2010. CD22 EXON 12 deletion as a pathogenic mechanism of human B-precursor leukemia. Proc Natl Acad Sci U S A 107 (39), 16852–16857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulyanova T, Shah D, Thomas M, 2001. Molecular cloning of MIS, a myeloid inhibitory siglec, that binds protein-tyrosine phosphatases SHP-1 and SHP-2. J Biol Chem 276, 14451–14458. [DOI] [PubMed] [Google Scholar]
- Umemori H, Sato S, Yagi T, Aizawa S, Yamamoto T, 1994. Initial events of myelination involve Fyn tyrosine kinase signalling. Nature 367 (6463), 572–576. [DOI] [PubMed] [Google Scholar]
- Van Valen L, 1974. Two modes of evolution. Nature 252 (5481), 298–300. [DOI] [PubMed] [Google Scholar]
- Varki A, 2010. Colloquium paper: uniquely human evolution of sialic acid genetics and biology. Proc Natl Acad Sci U S A 107 Suppl 2, 8939–8946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, 2011. Since there are PAMPs and DAMPs, there must be SAMPs? Glycan “self-associated molecular patterns” dampen innate immunity, but pathogens can mimic them. Glycobiology 21 (9), 1121–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan C, Yang Y, Feng G, Gu N, Liu H, Zhu S, He L, Wang L, 2005. Polymorphisms of myelin-associated glycoprotein gene are associated with schizophrenia in the Chinese Han population. Neurosci Lett 388 (3), 126–131. [DOI] [PubMed] [Google Scholar]
- Wang S, Chen C, Guan M, Liu D, Wan XF, Li L, 2021. Terminal Epitope-Dependent Branch Preference of Siglecs Toward N-Glycans. Front Mol Biosci 8, 645999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chow R, Deng L, Anderson D, Weidner N, Godwin A, Bewtra C, Zlotnik A, Bui J, Varki A, Varki N, 2011. Expression of Siglec-11 by human and chimpanzee ovarian stromal cells, with uniquely human ligands: implications for human ovarian physiology and pathology. Glycobiology 21 (8), 1038–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Mitra N, Cruz P, Deng L, Varki N, Angata T, Green E, Mullikin J, Hayakawa T, Varki A, 2012a. Evolution of siglec-11 and siglec-16 genes in hominins. Mol Biol Evol 29 (8), 2073–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Mitra N, Secundino I, Banda K, Cruz P, Padler-Karavani V, Verhagen A, Reid C, Lari M, Rizzi E, Balsamo C, Corti G, De Bellis G, Longo L, Beggs W, Caramelli D, Tishkoff S, Hayakawa T, Green E, Mullikin J, Nizet V, Bui J, Varki A, 2012b. Specific inactivation of two immunomodulatory SIGLEC genes during human evolution. Proc Natl Acad Sci U S A 109 (25), 9935–9940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Neumann H, 2010. Alleviation of neurotoxicity by microglial human Siglec-11. J Neurosci 30 (9), 3482–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willyard C, 2018. New human gene tally reignites debate. Nature 558 (7710), 354–355. [DOI] [PubMed] [Google Scholar]
- Wu Y, Lan C, Ren D, Chen GY, 2016. Induction of Siglec-1 by Endotoxin Tolerance Suppresses the Innate Immune Response by Promoting TGF-beta1 Production. J Biol Chem 291 (23), 12370–12382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka M, Kato Y, Angata T, Narimatsu H, 2009. Deletion polymorphism of SIGLEC14 and its functional implications. Glycobiology 19 (8), 841–846. [DOI] [PubMed] [Google Scholar]
- Yang Y, Qin W, Shugart Y, He G, Liu X, Zhou J, Zhao X, Chen Q, La Y, Xu Y, Li X, Gu N, Feng G, Song H, Wang P, He L, 2005. Possible association of the MAG locus with schizophrenia in a Chinese Han cohort of family trios. Schizophr Res 75 (1), 11–19. [DOI] [PubMed] [Google Scholar]
- Yasui K, Angata T, Matsuyama N, Furuta R, Kimura T, Okazaki H, Tani Y, Nakano S, Narimatsu H, Hirayama F, 2011. Detection of anti-Siglec-14 alloantibodies in blood components implicated in nonhaemolytic transfusion reactions. Br J Haematol 153 (6), 794–796. [DOI] [PubMed] [Google Scholar]
- Yngvadottir B, Xue Y, Searle S, Hunt S, Delgado M, Morrison J, Whittaker P, Deloukas P, Tyler-Smith C, 2009. A genome-wide survey of the prevalence and evolutionary forces acting on human nonsense SNPs. Am J Hum Genet 84 (2), 224–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Lai C, Maoui M, Banville D, Shen S, 2001a. Identification and characterization of S2V, a novel putative siglec that contains two V set Ig-like domains and recruits protein-tyrosine phosphatases SHPs. J Biol Chem 276, 23816–23824. [DOI] [PubMed] [Google Scholar]
- Yu Z, Maoui M, Wu L, Banville D, Shen S, 2001b. mSiglec-E, a novel mouse CD33-related siglec (sialic acid-binding immunoglobulin-like lectin) that recruits Src homology 2 (SH2)-domain-containing protein tyrosine phosphatases SHP-1 and SHP-2. Biochem J 353 (Pt 3), 483–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Raper A, Sugita N, Hingorani R, Salio M, Palmowski M, Cerundolo V, Crocker P, 2006. Characterization of Siglec-H as a novel endocytic receptor expressed on murine plasmacytoid dendritic cell precursors. Blood 107 (9), 3600–3608. [DOI] [PubMed] [Google Scholar]
- Zhang JQ, Nicoll G, Jones C, Crocker PR, 2000. Siglec-9, a novel sialic acid binding member of the immunoglobulin superfamily expressed broadly on human blood leukocytes. J Biol Chem 275 (29), 22121–22126. [DOI] [PubMed] [Google Scholar]
- Zheng Q, Hou J, Zhou Y, Yang Y, Xie B, Cao X, 2015. Siglec1 suppresses antiviral innate immune response by inducing TBK1 degradation via the ubiquitin ligase TRIM27. Cell Res 25 (10), 1121–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


