Significance
Predictability of open-chromatin regions, using their DNA sequences, is a new means for evaluating epigenomic data. While most existing methods are based on the enrichment of mapped reads in known elements, the DNA sequence–based assessment solves a bottleneck in analyzing single-cell genomics data: comprehensive identification of peaks from rare cell types. Our method, gkmQC, can identify more peaks from rare cell types with low read depths by finding optimal peak-calling thresholds, and these additional peaks explained a significant proportion of the heritability of traits relevant to the cell types. gkmQC will accelerate the genomic discovery of human diseases from the viewpoint of transcriptional regulation by enabling us to focus on high-quality epigenomic data and rare cell types.
Keywords: quality control, chromatin accessibility, sequence-based model, gkmQC
Abstract
Chromatin accessibility assays are central to the genome-wide identification of gene regulatory elements associated with transcriptional regulation. However, the data have highly variable quality arising from several biological and technical factors. To surmount this problem, we developed a sequence-based machine learning method to evaluate and refine chromatin accessibility data. Our framework, gapped k-mer SVM quality check (gkmQC), provides the quality metrics for a sample based on the prediction accuracy of the trained models. We tested 886 DNase-seq samples from the ENCODE/Roadmap projects to demonstrate that gkmQC can effectively identify “high-quality” (HQ) samples with low conventional quality scores owing to marginal read depths. Peaks identified in HQ samples are more accurately aligned at functional regulatory elements, show greater enrichment of regulatory elements harboring functional variants, and explain greater heritability of phenotypes from their relevant tissues. Moreover, gkmQC can optimize the peak-calling threshold to identify additional peaks, especially for rare cell types in single-cell chromatin accessibility data.
Open chromatin at specific genomic sites is the hallmark of cis-regulatory element (CRE) activity that modulates transcription of a target gene (1). Thus, identifying open-chromatin regions of the genome is a fundamental step toward defining the gene regulatory program encoded in the genome. Significant advances in experimental techniques to detect these regions have been made over the last decade. Sequencing-based assays, such as ATAC-seq (2) and DNase-seq (3, 4), detecting transposase-accessible and DNase-hypersensitive regions as open-chromatin regions, are now widely used to enable genome-wide mapping of regulatory elements (5). These assays have shown that epigenetic landscapes are dynamic and actively regulated across different biological states, cell types (6), developmental stages (7), aging (8), and species (9). Moreover, multiomic analyses integrating these data with genome-wide association studies (GWAS) are now significantly accelerating mechanistic understanding of how noncoding variation drives transcriptional regulation (10–12), the largest contributor to complex traits.
Defining the regulatory landscape requires rigorous assessment of the quality of chromatin accessibility data, and this remains a challenge due to several reasons, such as the lack of gold standard datasets for benchmarking and difficulties in functional validation. Several methods proposed to rectify this include quantifying quality using statistics like the fraction of reads in peaks (FRiP) (13), Signal Portion of Tags 2 (SPOT2) (14), and promoter (TSS) enrichment scores (15), which measure the degree to which reads are enriched in functional elements (i.e., identified open-chromatin peaks and promoters). Irreproducible discovery rate (IDR) is yet another quality assessment statistic measuring the reproducibility of peaks between replicates (16). However, these metrics, such as FRiP and SPOT2, may not be optimal for samples with low sequencing depth where a smaller number of peaks are detected (17, 18). This is problematic, especially for single-cell analysis or rare cell types. IDR is also limited when robust replicates are unavailable. Consequently, chromatin accessibility data with suboptimal quality currently can mislead downstream analyses.
As an improvement, we developed a complementary and biologically motivated quality metric for chromatin accessibility data based solely on their underlying DNA sequences. This metric is based on the concept that CREs, such as promoters, enhancers, and insulators, typically have multiple transcription factor binding sites (TFBSs) (19). Thus, open-chromatin peaks in high-quality (HQ) samples (those containing HQ open-chromatin peaks) are likely to harbor such TFBSs, which can be accurately captured by sequence-based predictive models (20–24). This leads to our main hypothesis that the accuracy of a sequence-based model directly correlates with the quality of the peaks derived from chromatin accessibility data. Our method, called gapped k-mer SVM quality check (gkmQC), is based on a sequence-based machine learning technique, gkm-SVM (21, 25), which can predict CREs using their primary DNA sequence only. We demonstrate that “HQ” samples defined by gkmQC, compared with low-quality samples 1), have more CREs better aligned at functional regulatory elements 2), harbor more putatively functional variants from GWAS, and 3) explain greater heritability of traits from their relevant tissues. We also show that gkmQC can identify additional peaks by optimizing a peak-calling process, especially for rare cell types in single-cell chromatin accessibility data.
Results
Open-Chromatin Peak Signals Correlate with Sequence-Based Prediction Performance.
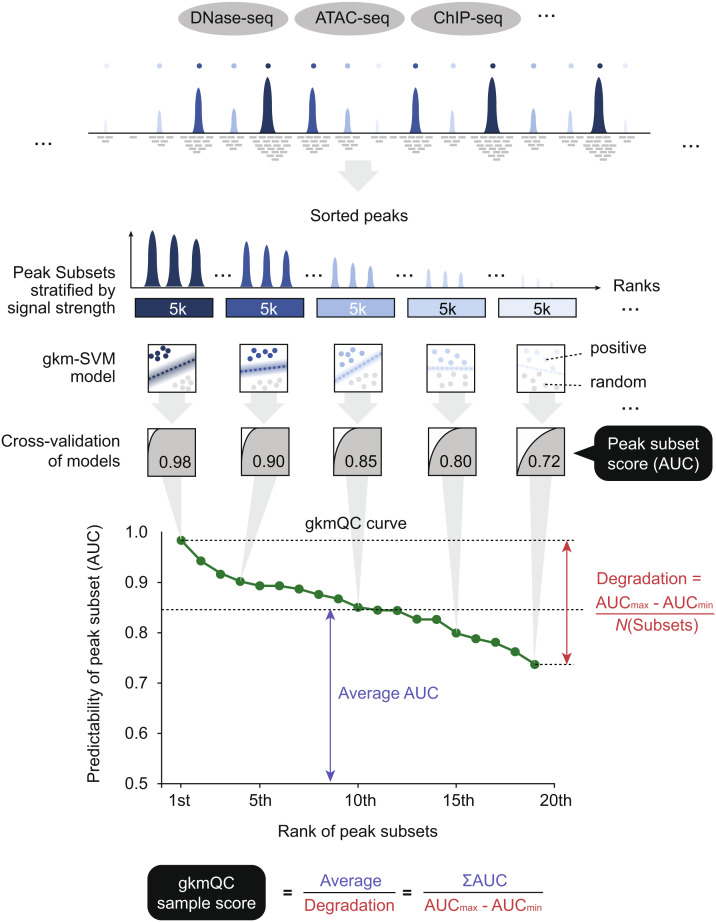
Typical chromatin accessibility data exhibit a wide range of peak signals (i.e., an abundance of mapped reads or peak heights). We hypothesized that a peak with a stronger signal, or a higher peak, is more likely to be a true CRE, harboring clearer predictive sequence features and leading to the higher classification accuracy of sequence-based predictive models. To test this, we analyzed 886 samples of ENCODE DNase-seq (26). For each sample, we first divided the entire set of peaks, stratified by peak signal strength, into subsets comprising an equal number of peaks (5,000). We call these “peak subsets.” We then trained gkm-SVM (25) for each peak subset against an equal number of random genomic regions and performed fivefold cross-validation to calculate the area under the ROC curve (AUC; i.e., peak predictability). Consistent with our hypotheses, we found a strong correlation between the AUC and peak signals for almost all datasets (SI Appendix, Fig. S1). Based on this, we defined an overall quality score for a sample as the average AUC score over its degradation rate across peak subsets, dubbed “gkmQC (sample) score” (Fig. 1 and Materials and Methods).
Fig. 1.
A schematic of peak quality assessment in gkmQC. gkmQC sorts peaks by their signal strengths, groups them into subsets of an equal number of peaks, and calculates the area under the ROC curves (AUCs) using sequence-based predictive models (gkm-SVM) with cross-validation. The AUC represents the overall predictability of peaks in a subset. For a typical dataset, AUCs strongly correlate with their peak signal strengths. The curve of AUC scores across subsets reflects its overall sample quality, and the extent of the degradation and the average of AUC across bins are represented by the gkmQC sample score.
Peak Predictability Complements Conventional Methods of Quality Assessment.
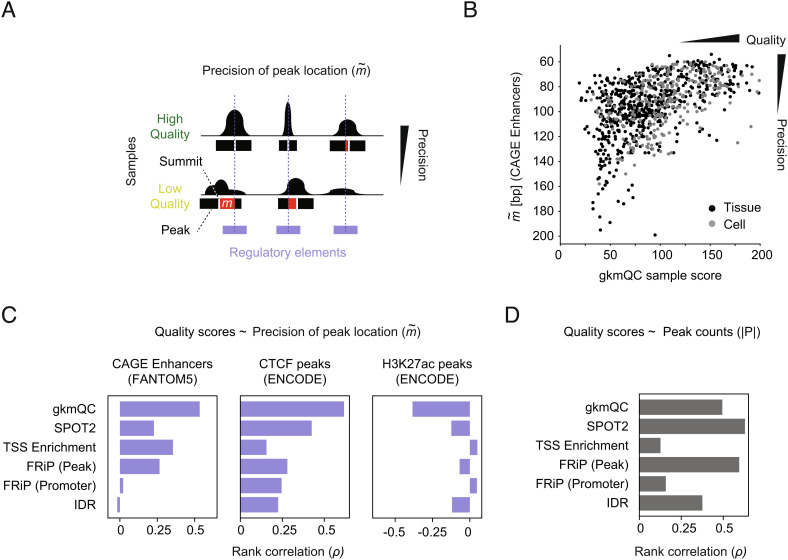
We next evaluated whether the gkmQC score could be used as an alternative sample quality metric. We reasoned that HQ samples would have greater gkmQC scores as more HQ peaks in the samples lead to high AUCs and slower degradation of AUC across peak subsets. Using the ENCODE DNase-seq dataset again, we systematically compared their gkmQC scores with the other five conventional quality metrics widely used for DNase- and ATAC-seq analyses (Materials and Methods). To evaluate QC methods, we analyzed how well peaks are aligned with known regulatory elements (i.e., precision of peak location; Fig. 2A). Specifically, we quantified genomic distance () between summits of DNase-seq peaks and the center of overlapping CAGE enhancers, CTCF-binding peaks, or H3K27ac histone modification ChIP-seq peaks. We found that the gkmQC score strongly correlated with (ρ = 0.53 for CAGE; Fig. 2 B and C) for CAGE enhancers and CTCF-binding peaks, while other quality metrics showed less correlation (ρ = 0.23 for CAGE; Fig. 2C and SI Appendix, Fig. S2A), suggesting that gkmQC can prioritize HQ samples defined by those peaks with a precise location for their functional regulatory elements. Interestingly, gkmQC scores are most negatively correlated with when using H3K27ac histone modification ChIP-seq peaks, consistent with the fact that CREs are typically flanked by histones (27).
Fig. 2.
Evaluating QC methods with the precision of peak locations. (A) A concept diagram depicts precisions of peak locations in high- and low-quality samples. The red bar shows m that represents the positional mismatch of a peak and the overlapping annotations for potential CREs. (B) gkmQC and the precision of peak locations () for CAGE enhancers were compared in the scatterplot. The Y axis for the parameter is reversed so that samples in right-top corners are HQ samples. Spearman’s rank correlation coefficients (ρ) are calculated for these comparisons. Black and gray dots are tissues and cells, respectively. (C) The rank correlation of quality metrics and was compared for six QC methods using three independent annotations. (D) The rank correlation of quality metrics and peak counts was compared across QC methods.
Compared with other metrics, gkmQC scores are less correlated with peak counts, a hallmark of peak-calling sensitivity (Fig. 2D and SI Appendix, Fig. S2B). However, the peak count is known to be significantly dependent on read depth or sample types (15). We thus hypothesized that gkmQC is better at identifying HQ samples with low peak counts than other methods. Correlations among quality metrics revealed samples with highly discordant quality scores between gkmQC and the other methods (SI Appendix, Fig. S2C). For a systematic and unbiased comparison, we determined sample quality (high vs. low) using the median score (the 50th percentile) as a cutoff (SI Appendix, Fig. S3A). About 13 to 18% of samples are classified as high quality by gkmQC but as low quality by SPOT2 (111; 13%), TSS enrichment (156, 18%), FRiP [Peak] (129, 15%), FRiP [promoters] (206, 23%), or IDR (116, 13%; Datasets S1 and S2). These samples contain fewer peaks than the remaining gkmQC HQ samples (SI Appendix, Fig. S3B) but have similar precision of peak locations, which is significantly higher than that of low-quality samples (SI Appendix, Fig. S3C). We also assessed whether other technical conditions, such as sample types, treatments, and sequencing steps, affected these quality metrics and confirmed that no other confounding factors significantly affected the gkmQC score and other metrics (SI Appendix, Fig. S4). In sum, our results suggest that gkmQC can reclassify some low-quality samples as determined by the other QC methods into HQ samples.
HQ Samples Reproduce Precise Locations of Peaks Relevant to GWAS Variants.
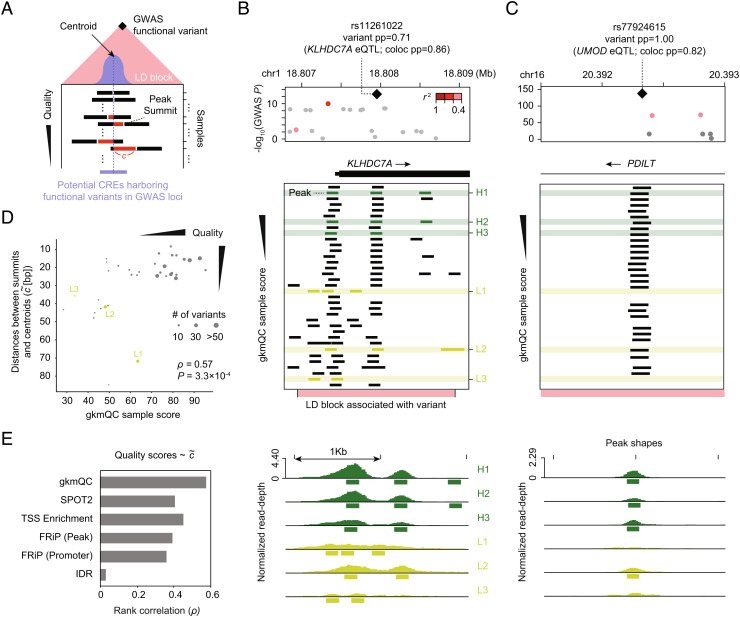
Accurate identification of regulatory elements is crucial to interpreting changes in the regulatory functions of relevant GWAS variants (28). We hypothesized that open-chromatin peaks that contain functional regulatory variants are more precisely identified in HQ than in low-quality samples (Fig. 3A). We assumed that the consensus peak summit (i.e., centroid) of open-chromatin peaks across multiple replicates represents the core functional regulatory element as previously shown by Meuleman et al. (29). We then tested whether peaks in HQ samples are in greater proximity to these centroids. We first focused on a few loci with putative causal GWAS variants for a major kidney functional trait, estimated glomerular filtration rate (eGFR) (30), using developing kidney DNase-seq samples (26). For example, rs11261022 is a likely causal regulatory variant affecting eGFR via change of KLHDC7A expression (30). This variant is in an open-chromatin peak in kidney samples (Fig. 3B). However, most lower-quality samples either do not have an overlapping peak or have a peak less optimally aligned with the variant. Similarly, rs77924615 is another putative functional regulatory variant identified by fine-mapping and colocalization analysis. Again, while HQ samples have an open-chromatin peak well aligned with this variant, low-quality samples failed to detect open-chromatin peaks in this locus (Fig. 3C). Moreover, peaks in HQ samples exhibit stronger signals than those in low-quality ones, demonstrating their greater utility in identifying regulatory elements containing GWAS variants (Fig. 3 B and C).
Fig. 3.
Peaks in HQ samples are better aligned with the centroid of peaks across biological replicates near GWAS functional variants. (A) The centroid of peaks across replicates represents the hypothetical core of a functional CRE (light purple bar). Peak summits in HQ samples are better aligned with their centroids across replicates than those in the low-quality ones. Each bar shows a peak from the corresponding sample. c (red-colored bar) is the genomic distance between a peak summit and its centroid. (B and C) Representative examples of peaks associated with functional GWAS variants in the (B) KLHDC7A and (C) PDILT loci. Rows are samples sorted by gkmQC scores, so top samples are high quality. The Bottom panels are the read pileup visualizations for the peak shapes shown in Top panels. (D) gkmQC scores are compared with for all kidney samples. H1-3 and L1-3 samples are representative examples of high- and low-quality samples classified by gkmQC, respectively. Each dot represents a developing kidney sample. The dot size corresponds to the number of GWAS loci harboring functional variants within <1 kb from the peaks in relevant samples. (E) The rank correlation of quality metrics and was compared for six QC methods.
To generalize this finding, we calculated genomic distances between the peak summits from each sample and the centroids of the consensus peaks near each putative causal GWAS variant (; Materials and Methods). We found that HQ samples determined by gkmQC have smaller average distances than low-quality samples. Overall, the gkmQC score strongly correlated with (ρ = 0.57 and P = 3.3 × 10−4; quality–rank correlation; Fig. 3D), while other conventional metrics had lower correlations (Fig. 3E). Thus, peaks in HQ samples determined by gkmQC more precisely delineate functional regulatory elements with functional GWAS variants.
HQ Samples Systematically Improve the Discovery of the Genetic Architecture of Complex Traits.
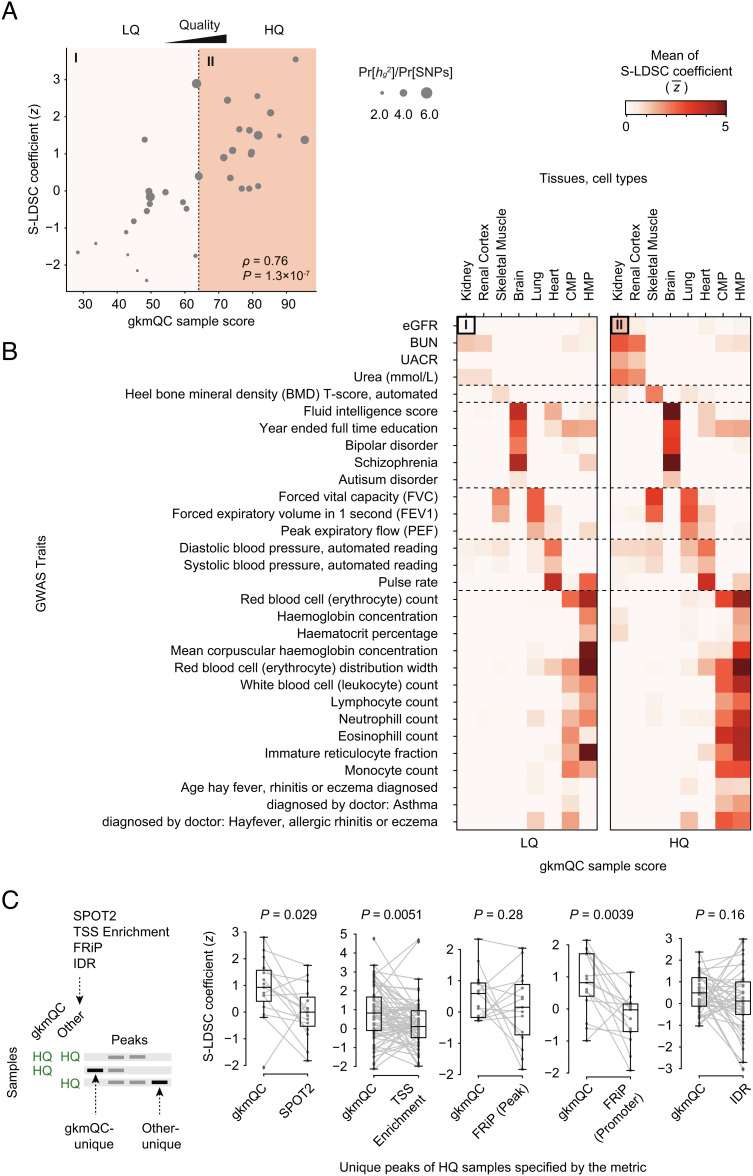
The above examples suggested that HQ samples could improve the systematic identification of genome-wide polygenic signals. To test this, we adapted the stratified LD score regression (S-LDSC) and performed partitioning heritability analyses of many human traits. Specifically, we calculated a normalized S-LDSC coefficient, which corresponds to a statistical significance of the per-SNP heritability (Materials and Methods and Dataset S3) (31, 32). Using replicate kidney samples during development, we found that the gkmQC score significantly correlated with S-LDSC coefficients for the eGFR (ρ = 0.76 and P = 1.3 × 10−7; Pearson correlation coefficient; Fig. 4A) (30). This suggests that the gkmQC scores for sample quality significantly explain the variance in heritability across biological replicates. Next, we extended our analysis to multiple tissues and traits (Materials and Methods). Using 200 samples from eight tissues/cells from the ENCODE project and 30 different traits from the UK Biobank (33), we discovered that HQ samples stratified by gkmQC consistently gave higher heritability signals, especially for relevant trait–tissue combinations (Fig. 4B and SI Appendix, Figs. S5 and S6 A and B). Most samples with gkmQC scores >70 achieved S-LDSC z >1.0 for their relevant traits (SI Appendix, Fig. S5), suggesting that this is a reasonable threshold for identifying good quality samples. With this threshold, peaks from HQ samples explained significantly greater per-SNP heritability than those from low-quality samples, as measured by median z scores of S-LDSC coefficients (z= 2.31 vs. 1.18).
Fig. 4.
Peaks in HQ samples exhibit greater heritability for relevant phenotypes. (A) A scatterplot comparing the gkmQC score and normalized S-LDSC coefficient (z score) for eGFR is shown for 35 developing kidney samples. The S-LDSC coefficient directly correlates with the enrichment score of heritability for eGFR. (B) Two heatmaps comparing the average S-LDSC coefficients between high- and low-quality samples. We used the top 50% gkmQC scores as a threshold for sample quality classification. The top-left cells of the two heatmaps summarize the scatterplot of (A). CMP and HMP are the abbreviations of common myeloid progenitor and hematopoietic multipotent myeloid progenitor cells, respectively. (C) The comparison of S-LDSC coefficients for peaks uniquely identified in gkmQC-HQ samples and HQ samples specific for another QC method. A schematic diagram shows how the unique peaks were defined for the joint S-LDSC analysis. P values were calculated from the Mann–Whitney U test.
As previously noted, several samples are differentially classified by gkmQC compared with other conventional quality metrics. We thus asked whether peaks in HQ samples determined by gkmQC are more informative than those identified by the other QC methods (Fig. 4C). Specifically, we performed the heritability analysis again using S-LDSC by further stratifying regions based on overlaps across HQ samples determined by gkmQC and other QC methods (Materials and Methods). When using phenotypes relevant to the samples, we found that gkmQC-HQ unique peaks achieved a higher per-SNP heritability than peaks in HQ samples uniquely identified by another QC method (Fig. 4C). Taken together, our results suggest that functionally important tissue-specific regulatory elements are better detected in HQ samples and that gkmQC is a better method in identifying such HQ samples.
gkmQC Optimizes the Sensitivity on Peak Calling.
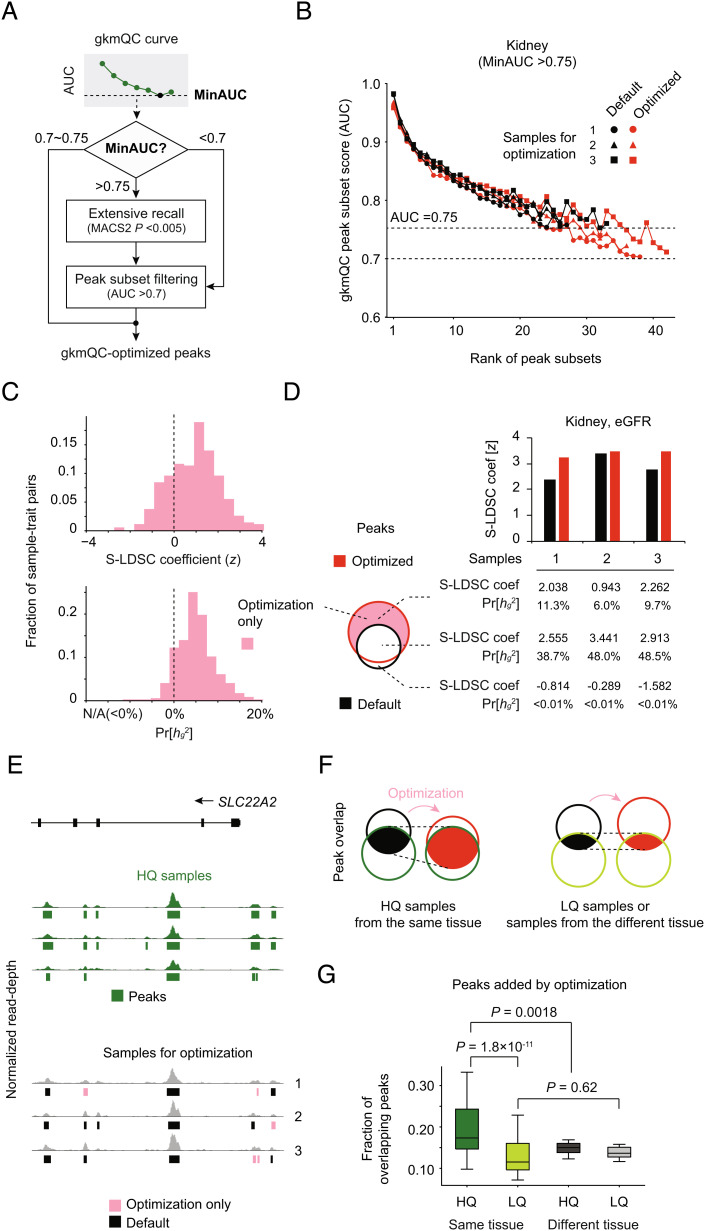
A benefit of gkmQC is that its peak predictability can be used for recovering peaks that do not pass the statistical threshold based on read enrichment but have strong sequence signatures nonetheless. While analyzing the full set of ENCODE data, we noticed that 58 samples with lower read depth exhibited high AUC for all subsets in the sample (i.e., minimum AUC [MinAUC] > 0.75; SI Appendix, Fig. S7A). This result suggested that additional CREs with strong sequence signatures were undetected. We reasoned that this might be due to an overly stringent peak-calling threshold given low read depths. Our approach can, however, naturally alleviate this issue by identifying a more accurate threshold based on peak predictability. To test this, we recalled peaks using a conventional peak caller but with a relaxed threshold and then applied gkmQC to find a new peak-calling threshold that maximizes the number of peaks with MinAUC > 0.7 (Fig. 5 A and B and Materials and Methods). We note that other quality metrics for peak subsets with moderate peak intensity were not as correlated with the rank of the peak subsets as gkmQC (SI Appendix, Fig. S8). Therefore, gkmQC is more reliable for optimizing peak-calling thresholds than conventional quality metrics.
Fig. 5.
Optimization of peak calling in bulk chromatin accessibility data. (A) Workflow diagram of peak-calling optimization process using gkmQC. (B) gkmQC curves for three representative developing kidney samples before (black) and after optimization (red). (C) Distribution of S-LDSC coefficient scores and the proportion of the heritability of newly identified peaks after optimization (i.e., optimization only; pink in the Venn diagram). Heritability was calculated for 58 samples in eight tissues and 31 relevant phenotypes requiring optimization. (D) Heritability analysis for three representative developing kidney samples. The graphs of black and red bars show the S-LDSC coefficients calculated with whole peaks before and after optimization. The table presents heritability for three subsets of peaks; optimization only (Top), commonly found regardless of the optimization (Middle), and with default parameters only (Bottom). (E) A specific example of open-chromatin peaks in the SLC22A2 locus. Green bars represent peaks from HQ samples. Dark gray and pink bars are the commonly found and optimization-only peaks in the samples targeted for the optimization step. (F) A conceptual diagram shows that gkmQC optimization increases the reproducibility of peaks in HQ samples from the same tissue. (G) Comparisons of overlaps between peaks added by optimization and those in other samples. Box plots show the fraction of peaks recovered by gkmQC optimization overlapping those in HQ replicates (green), low-quality (LQ) replicates (light green), HQ samples from different tissues (black), and LQ samples from different tissues (gray). The paired t test is used to test the significance of the differences between these groups.
Among the DNase-seq data of bulk tissues and cells, 58 of 200 samples with low read depths had MinAUC > 0.75. gkmQC optimization of these samples recovered an additional ~27.2% of total peaks per sample on average (SI Appendix, Fig. S7B and Dataset S4). To check the functional relevance of recovered peaks, we repeated the S-LDSC analyses again for traits relevant to the samples. To account for potentially inflated heritability caused by SNPs in strong LD, we analyzed the newly identified peaks in S-LDSC while controlling for default peaks (Materials and Methods). We achieved positive S-LDSC coefficients in 76.4% of all datasets and an increased proportion of the heritability for 89.2% of the datasets, suggesting that the newly identified peaks by gkmQC were trait relevant (Fig. 5C). We also observed that the optimized vs. default peaks achieved consistently higher heritability (SI Appendix, Fig. S7C). In Fig. 5E, we provide a specific example of newly identified peaks at the kidney-specific gene, SLC22A2 locus. Here, we focused on three kidney samples with MinAUC > 0.75, in which additionally discovered peaks with AUC >0.7 produced positive S-LDSC coefficients (Fig. 5 B and C). Significantly, recovered peaks at this locus (pink bars) overlapped those identified in HQ samples of the kidney (Fig. 5E and SI Appendix, Fig. S7D; KLHDC7A).
Inspired by the cases of KLHDC7A and SLC22A2, we next hypothesized that gkmQC optimization could improve the reproducibility of peaks given the quality variation across replicates (Fig. 5F). To validate this systematically, we investigated whether the newly found peaks were replicated in other HQ samples with the same tissue entity. Specifically, we calculated the fraction of the newly found peaks by optimization that overlaps with peaks in high-quality samples from the same tissue (Materials and Methods). We used low-quality samples from the same tissue and HQ samples from different tissues as negative controls for comparison. We found that the peak overlaps with high-quality replicates significantly more than low-quality replicates (Fig. 5G; P = 1.8 × 10−11; paired t test) and HQ samples from different tissues (P = 0.0018). Thus, our gkmQC optimization can considerably recover tissue-relevant peaks that are present in HQ samples.
Extensive Peak Calling by gkmQC Improves Single-Nucleus ATAC-Seq Inference.
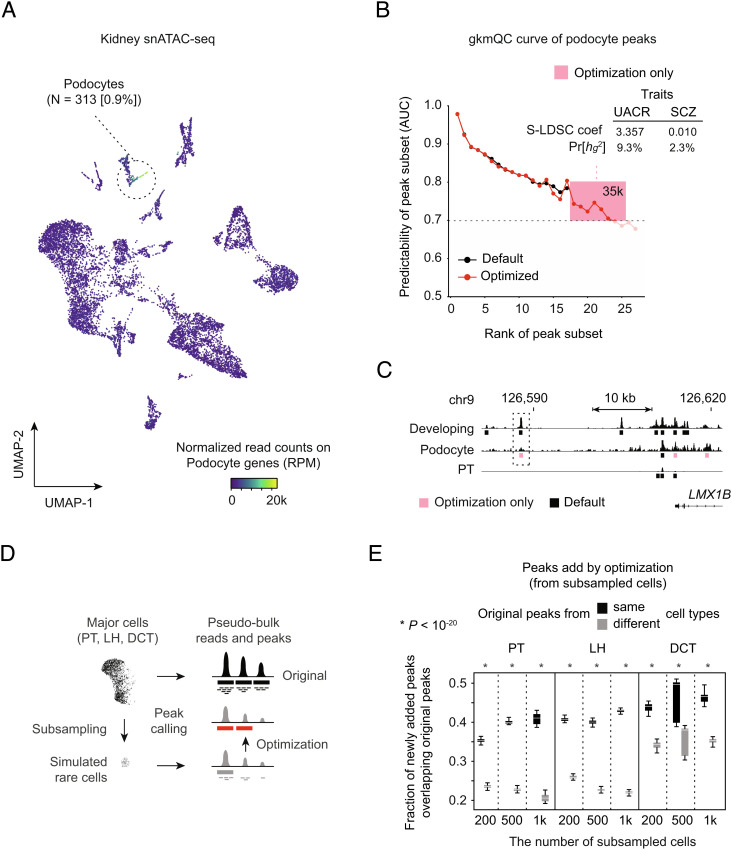
Because gkmQC successfully optimized bulk data with marginal read depth (Fig. 5), we surmised that gkmQC could also be helpful in improving peak identification in single-cell chromatin accessibility data, especially for rare cell types, which by nature have lower numbers of reads (34). To test this, we applied the peak optimization process to kidney single-nucleus ATAC-seq data (snATAC-seq) (35) (Dataset S5). We first analyzed a rare but important kidney cell type of the glomerular filtration barrier, the podocyte (<1% of the total kidney cells) (Fig. 6A and SI Appendix, Fig. S9 A–D and Materials and Methods) (35). Our gkmQC peak-calling optimization recovered ~35,000 additional peaks (~27%) in podocytes compared with peaks called with a default threshold. Also, the variants in these recovered peaks significantly contributed to the heritability of the urine albumin-to-creatinine ratio (UACR; z = 3.357 and S-LDSC coefficient; Fig. 6B), a kidney trait affected by podocytes. In contrast, no significant contribution to the heritability of unrelated traits such as schizophrenia (36) was observed (SCZ; z = 0.010). Fig. 6C shows a specific example of peaks recovered at the podocyte-relevant LMX1B locus, a well-known podocyte-specific transcription factor for kidney development and the maintenance of differentiated podocytes (37, 38). We note that a recovered peak upstream of LMX1B was also detected in bulk developing kidney tissues (Fig. 6C). Since this peak was not detected in any other kidney cell types (SI Appendix, Fig. S9E), it demonstrates that optimization using gkmQC can uncover new peaks even in rare cell types, which are more likely to have peaks missed because of their lower numbers.
Fig. 6.
Improvement of peak calling in snATAC-seq data for rare cell types. (A) A UMAP plot of kidney snATAC-seq analysis. The heatmap colors represent open-chromatin activities of podocyte-specific genes. (B) gkmQC curves for pseudobulk reads of podocyte cells are shown before and after optimization. Peak subsets within the pink box are the ones newly discovered by optimization. The Inset table shows a heritability analysis result for the podocyte-relevant (UACR) trait and one nonrelevant (schizophrenia, SCZ) trait for these newly identified peaks only. (C) A specific example of podocyte open-chromatin peaks in the LMX1B locus. The pink bar is a newly called peak by optimization. PT is the proximal tubule, the most abundant cell type in the kidney, and is used as a control to show cell-type specificity of the new peaks. (D) A schematic describing how gkmQC optimization is assessed using random subsampling of major cell types to simulate rare cell types. (E) Box plots represent fractions of optimized peaks in subsampled cells overlapping the original peaks. LH and DCT are the abbreviations of loop of Henle and distal convolute tubule cells, respectively (SI Appendix, Fig. S9A).
To systematically test if the optimization process is generalizable to other cell types and tissues, we analyzed the heritability contributed by variants in recovered peaks from the other 11 kidney cell types and peripheral blood mononuclear cells (PBMCs) (SI Appendix, Fig. S10 and Dataset S5). We discovered that, in general, cell types with lower cell counts had higher MinAUCs (SI Appendix, Figs. S9D and S10D), showing that the optimization process is more effective in rarer cell types. Expectedly, we confirmed that the newly identified peaks in rare cell types with counts <1,000 explained a significant proportion of heritability (S-LDSC coefficient > 2.0) for relevant traits (SI Appendix, Figs. S9F and S10E).
Last, we subsampled 200, 500, and 1,000 cells from five abundant cell types from the kidney and PBMCs (i.e., proximal tubule (PT), loop of Henle (LH), and distal convoluted tubule (DCT) for the kidney and CD4+ memory T cells (CD4M) and CD14+ monocytes (CD14M) for PBMCs) to simulate rare cell populations and to test whether gkmQC peak-calling optimization for subsampled cells can recapitulate peaks identified in the original populations of the cells (Materials and Methods). We found that 30 to 40% of peaks added by gkmQC optimization overlapped the original peaks from the corresponding cell types (Fig. 6 D and E and SI Appendix, Fig. S10F). The fraction of the overlapped peaks was significantly higher than that of original peaks from different cell types (P < 10−20; paired t test). Taken together, these results show that the peak-calling optimization using gkmQC helps us to identify additional cell-type-specific regulatory elements, particularly for rare cell types.
Discussion
Comprehensive quality control (QC) analysis of chromatin accessibility data is a critical need for proper analysis of genome regulatory functions because of the lack of ground-truth datasets for benchmarking. By utilizing machine learning techniques which learn sequence features underlying open-chromatin peaks, we have established a computational framework for quality assessment and refinement of chromatin accessibility data. HQ samples determined by gkmQC yield more accurate data for more robust downstream genomic analyses, such as GWAS fine-mapping of complex traits and partitioning its heritability. Our method for optimizing peak-calling thresholds also improves single-cell chromatin accessibility datasets by identifying more peaks in rare cell types.
Precise mapping of functional regulatory elements is now possible by applying several genomic technologies (39–41). A recent study demonstrated that the centroid of overlapping consensus DNase I–hypersensitive site summits can be used to robustly and accurately identify core regulatory regions where variants that disrupt TF bindings strongly perturb their regulatory activities (29). We show that peaks in HQ samples identified by gkmQC can also accurately locate these core regulatory regions (Fig. 3). Highly predictive sequence features presented in peaks in HQ samples enable us to precisely locate these elements, while quality control using these sequence features can be used to test the precision of peak location without many replicates.
In our gkmQC analyses, we found that peak subsets with lower peak intensity tend to achieve lower predictability (SI Appendix, Fig. S1). These peak subsets with medium-level AUCs (0.85 to 0.95) are enriched for distal enhancers and tissue-specific regulatory elements (SI Appendix, Figs. S11 and S12). In contrast, peak subsets with high AUCs (>0.95) mostly contain promoters and ubiquitously open regions with homogeneous sequence features. These results imply that tissue-specific CREs and distal enhancers may have consistently lower peak predictability than housekeeping ones and promoters regardless of their sample qualities, leading to an alternative hypothesis: TFs that bind to distal and tissue/cell-specific enhancers are generally less learnable than those that bind to promoters and ubiquitously open regions. It is also concordant with the known biology that regulatory activity of these tissue-specific and distal enhancers could be modulated by changes in TF expression or chromatin looping without strong sequence-specific binding of TFs (42). Thus, our strategy of evaluating multiple peak subsets stratified by signal strength independently, rather than building a single model on the whole peak set, makes it possible to assess sample qualities more accurately by allowing multiple models to capture different classes of CREs active in a sample.
Comprehensive identification of peaks for rare cell types is a big challenge in single-cell ATAC-seq analysis (34). We showed that our gkmQC optimization could find ~27% more peaks in rare cell types (Fig. 6 and SI Appendix, Figs. S9 and S10). It also enables us to estimate the minimum number of cells required for adequate peak identification, which is currently unknown. We have found that, in general, cell types with cell counts >1,000 can yield >100,000 peaks and do not need gkmQC optimization in general (SI Appendix, Figs. S9B and S10B). Considering that >100,000 peaks are typically identified in bulk DNase-seq and ATAC-seq from cell lines and primary cells, we speculate that at least 1,000 cells are needed for a comprehensive peak discovery.
We found a significant heritability enrichment for peaks added by gkmQC optimization (Figs. 5 and 6), illustrating the increased peak-calling sensitivity. However, increased sensitivity may also come with a potential loss of precision. Indeed, the per-SNP heritability of the optimization-only peak set is consistently somewhat lower than those from the default setting as shown in Fig. 5D, suggesting this is the case. However, increased sensitivity outweighs the potentially reduced precision for the low-coverage data as the additional peaks determined by gkmQC optimization can explain additional heritability for the phenotypes relevant to the samples.
We also measured the computing speed, memory, and CPU requirements of gkmQC using randomly selected samples with different peak counts from snATAC- and DNase-seq datasets. The running time of gkmQC scales linearly with respect to the peak count with the ability to process 80 k peaks per hour using eight cores of Intel Xeon Platinum 8268 2.90 GHz and ~16 GB of memory (2 GB/core) (Dataset S6). Thus, a typical sample with >100,000 peaks can be processed within ~1.2 h in a standard workstation equipped with similar resources (eight cores and 16 GB of memory).
Chromatin accessibility data have empowered us to functionally interpret disease-associated genetic variation. Over the last decade, a significant amount of chromatin accessibility data has been accumulated to create an atlas of functional regulatory elements across diverse tissues or cells. We anticipate that our quality assessment framework will further accelerate this process by prioritizing HQ samples, finding more CREs from rare cell types in snATAC-seq, and implicating sequence variants that disrupt these functions.
Materials and Methods
Sequence-Based Predictive Model for Quality Evaluation.
We constructed gkm-SVM models following our previously established framework (25, 43). Briefly, we first defined open-chromatin regions derived from a DNase-seq dataset as the positive training set using a precalculated open-chromatin peak set available in ENCODE (26). A negative set for training was then generated by random sampling of an equal number of genomic regions that match the length, GC content, and repeat fraction of the positive set. We excluded regions with >1% N-bases and >70% repeats from the training datasets. To prevent potential bias caused by variable peak lengths, we fixed the size of peaks by extending 300 bp from the summit.
We split the genomic peaks into multiple subsets, each comprising 5,000 peaks sorted by decreasing signal intensity scores from a peak caller. If a group of peaks with the same score were separated into two subsets, we randomized the order of these peaks to make sure that all neighboring peaks (i.e., peaks sorted by genomic position) were not grouped into the same subset. Then, we trained a gkm-SVM model for each peak subset using default parameters (word length = 10, informative columns = 6, truncated filter = 3, and weighted gkm kernel (wgkm) t = 4). Model performance was measured by the area under the ROC curves with five-fold cross-validation (i.e., an AUC score of the peak subset). We present AUC scores and ranks of peak subsets as a gkmQC curve on the Y and X axes, respectively.
To quantify overall sample quality, we derived a gkmQC score from a gkmQC curve using the following equation:
where is the rank numbers of peak subsets, and AUCmax and AUCmin are the maximum and minimum values of AUC scores in the curve, respectively. We limit analysis to the top 100,000 peaks (=20 subsets of 5,000 peaks per subset) to generate a gkmQC curve. We did so to reduce the computation cost based on the observation that most ENCODE samples have ~100,000 peaks.
To analyze sequence features of the models, we scored all possible 10-mers ( = 10) using the trained SVM model and then performed a principal component analysis using their score vectors. We evaluated the first two principal components to compare models (SI Appendix, Fig. S11 D and E). To analyze tissue specificity of peak subsets, we calculated pairwise peak overlaps between the same rank subsets from the same tissues. To ensure these peak overlaps were comparable across different rank peak subsets, we further normalized them by calculating an overlap fold change for each rank. As a denominator, we used the average overlap between the same rank peak subsets from random tissue pairs.
Benchmarking Datasets and Evaluation Methods.
ENCODE DNase-seq datasets.
We revisited the ENCODE DNase-seq datasets accessed on March 10, 2020. We excluded samples with <10,000 peaks, archived, or revoked status in the database. We ultimately obtained 886 DNase-seq datasets across diverse samples, including in vitro differentiated cells, primary cells, and tissues. Metadata of the full datasets are in Dataset S2.
Validating peaks with orthogonal datasets.
To obtain the precision of peak location (), we calculated the average genomic distance between peak summits and the center of peaks using three independent epigenomic datasets: overlapping CAGE (cap analysis gene expression) (44) enhancers, CTCF-binding peaks, and histone mark ChIP-seq peaks. CAGE enhancers from FANTOM (https://fantom.gsc.riken.jp/5/datafiles/reprocessed/hg38_latest/extra/enhancer/F5.hg38.enhancers.bed.gz) were called based on CAGE peaks with bidirectional balanced RNA signatures distal to known exons (+/−100-bp region from boundaries) and transcription start sites (+/−300 bp) (45). We considered an overlapping enhancer the most proximal enhancer that also overlaps with the 1-kb extended peak from the summit. The peak count, , is the count of nonoverlapping peaks. For the CTCF-binding and H3K27ac histone modification sites, we downloaded the bed files of (pseudo) replicated peaks with significant IDR derived from ENCODE CTCF and H3K27ac ChIP-seq experiments with matched tissues. For the matching, we compared anatomy/cell ontology and sample types (in vitro, primary cells and tissues).
We used FANTOM5 enhancers again to analyze the enrichment of open-chromatin peaks for known enhancers. For the enrichment analysis with promoters, we used FANTOM5 promoters (https://fantom.gsc.riken.jp/5/datafiles/reprocessed/hg38_latest/extra/CAGE_peaks/hg38_fair+new_CAGE_peaks_phase1and2.bed.gz). The FANTOM5 promoters were called based on CAGE peaks of which signal is comparable to CAGE peaks near 5'-ends of known transcripts (within 500 bp) (46).
Conventional metrics for quality evaluation.
To compare our findings with the default metrics provided by ENCODE, we used Signal Portion of Tags (SPOT2), the number of cleavages observed within HOTSPOT2 peaks divided by the total number of cleavages in a sample (14). We obtained precalculated SPOT2 scores from the metadata of the ENCODE database. To calculate TSS enrichment, we used the Python package, tssenrich (https://pypi.org/project/tssenrich) (47), following the ENCODE standards (https://www.encodeproject.org/atac-seq/). We used featureCounts (48) to calculate the fraction of reads in called peaks (FRiP), known promoters, and enhancers. IDR of peak calling across biological duplicates was calculated by the IDR package (16). To quantify a representative value of IDR values for a sample, we averaged −log10 of IDR P values of all peaks in a sample. To measure the quality affected by sample treatment, especially for autopsied tissues, we curated the duration of postmortem time from cardiac cessation to freezing of the sample from the case report in the ENTEx dataset (https://www.encodeproject.org/entex-matrix/?type=Experiment&status=released&internal_tags=ENTEx) (49).
Validation of HQ Samples using GWAS Functional Variants.
GWAS datasets.
For integrative analyses of open-chromatin peaks with relevant GWAS variants, we focused on GWAS for eGFR (30), a quantitative kidney functional trait. We chose eGFR GWAS due to the significant heritability and the availability of fine-mapping datasets with eQTL colocalization. We specifically used a precalculated dataset of putatively functional SNPs from the European ancestry meta-analysis of the eGFR trait (30) based on posterior probability >0.5 from approximate Bayes factor analysis (50).
Associating core regulatory elements near functional GWAS variants.
We measured the average genomic distance () between peak summits in a sample and the centroids of overlapping peaks across biological replicates from the same tissue. We only considered peaks near putatively functional GWAS variants, defined as genomic regions harboring fine-mapped SNPs and their neighboring SNPs in high LD (r2 > 0.8) with a 1,000-base padding. To measure the centroid, we calculated average genomic positions of overlapping peak summits across 35 replicates of the developing kidney. Similar to the peaks, we only used the centroids near fine-mapped GWAS variants. We used LocusZoom to plot association P values of GWAS variants with linkage disequilibrium information from a reference population (51). IGV was used to plot read pileup signals from open-chromatin data (52).
Validation of HQ Samples Using Partitioned Heritability Analysis.
GWAS datasets.
We obtained GWAS summary statistics data for various phenotypes from the UK Biobank project (53), as processed by Neale lab (https://nealelab.github.io/UKBB_ldsc/index.html), and three quantitative kidney traits: eGFR, UACR, and blood urea nitrogen (30, 54). To analyze the relevant GWASs with a significant genetic association, we limited GWASs with heritability z scores >4 (z4 and z7) and medium/high confidence ratings (available in https://nealelab.github.io/UKBB_ldsc/h2_browser.html). We also selected tissues with ≥5 replicates, for which relevant GWASs are available. Consequently, we derived six developing tissues, two primary cells, and 30 relevant GWAS traits (Dataset S3).
S-LDSC.
To estimate a proportion of heritability () from GWAS summary statistics, we used LD score regression (LDSC) (31, 32). We employed stratified LDSC to calculate a proportion of heritability contributed to an SNP set () in open-chromatin peaks from a sample as follows:
Enrichment of the proportional heritability is presented by / , where is the proportion of SNPs in among the total SNP set. It represents a relative polygenic contribution of SNPs within open-chromatin regions to a given trait. To compute a representative parameter reflecting both an effect size and a statistical significance of the enrichment, we used a normalized S-LDSC coefficient driven by
where is a normalized coefficient of the proportional heritability to enable z-based scoring, and is the SD of calculated from a jack-knife estimation.
To take into account potential regulatory variants in the flanking regions of open-chromatin peaks, we defined genomic annotations of CREs as a 1-kb padding from the peak summits. To accurately test the contribution of open-chromatin peaks only, we calculated the partitioned heritability along with the baseline annotations of 97 functional regions, such as protein coding, evolutionary conserved, promoter, enhancer, or UTR, as recommended by the original S-LDSC study (32). For reference LD scores, European ancestry population and the corresponding allele frequencies in 1,000 genomes phase 3 data were used. All data, including the baseline LD annotation set (v2.2), were obtained from https://data.broadinstitute.org/alkesgroup/LDSCORE/. When we compare multiple functional annotations (i.e., optimized vs. default open-chromatin peaks), we conducted S-LDSC regression jointly with multiple annotations, along with the full set of the baseline annotations in one model.
gkmQC-Based Optimization of Peak-Calling Threshold.
The default pipeline for peak calling.
As a default pipeline for peak calling, we adapted the previously established framework for DNase-seq and ATAC-seq analyses (43, 55). Specifically, we ran MACS2 (56) with no restricted model (--nomodel) using paired-end read pairs with MAPQ >30. We used 100 bp of window size (–extsize 100), −50 bp of shifts toward lagging strand (--shift −50), keeping duplicate reads (--keep-dup), and q value cutoff <0.01 (57). For ATAC-seq samples, we additionally trimmed +4 bp of the forward strand and −5 bp of the reverse strand to account for 9-bp duplicated regions by Tn5 (2).
gkmQC peak-calling optimization.
We first determine whether a sample needs a peak-calling optimization based on the minimum AUC of all peak subsets (MinAUC). If the MinAUC is >0.75, we perform the following peak-calling optimization. To recover marginal peaks with suboptimal q values, we called peaks again using a relaxed threshold with nominal P < 0.005. We then calculated AUCs for all peak subsets using our gkmQC framework. Last, we recover all peaks that are more significant than the least significant peak in the minimum rank peak subset with AUC >0.7.
Analysis of the overlap of peaks added by optimization.
To measure peak overlaps between samples, we calculated the fraction of the newly added peaks that overlap with peaks in other samples from the same tissue. We defined HQ replicates as samples with the top 50% percentile of gkmQC scores among all samples from the corresponding tissue. The rest were used as low-quality replicates. Because the variation of peak counts across samples can be a potential confounding factor for this analysis, we randomly chose 100,000 peaks from a sample. We repeated this process ten times to obtain average overlaps across the ten different realizations of random peak sets.
Analysis of snATAC-seq Data.
Single-cell ATAC-seq datasets.
Human kidney snATAC-seq data are from nontumor kidney cortex samples from five patients undergoing partial or radical nephrectomy (35). We specifically downloaded sequencing data from GEO under accession number GSE151302. For the dataset of PBMCs, we used public snATAC-seq data with total ~15,000 cells from a healthy donor (Next GEM v1.1). We downloaded position-sorted BAM files derived from Cell Ranger ATAC 1.1.0 from the 10X Genomics support page (https://support.10xgenomics.com/single-cell-atac/datasets/1.1.0/atac_pbmc_10k_nextgem; access date: 01/2021).
Single-cell ATAC-seq data processing.
For the single-cell ATAC-seq data processing, we employed both Cell Ranger ATAC 1.1.0 (https://support.10xgenomics.com/single-cell-atac/software/downloads/latest) and snapATAC pipelines (58). Specifically, we first used count operation of the Cell Ranger ATAC pipeline to perform a quality assessment, preprocessing, and read alignment, yielding position-sorted, barcoded, and read-filtered BAM files. We then selected cells with 0.15 < FRiP < 0.5 and the number of reads with uniquely mapped identifiers >10,000. Cells across all datasets were harmonized by Harmony (59) and clustered based on a K-nearest neighbor algorithm with a Louvain community detection (# of eigen dimensions = 47). Consequently, we derived unsupervised snATAC-seq cell clusters enriched to known 16 kidney cell types (SI Appendix, Fig. S9A) and 12 PBMC types (SI Appendix, Fig. S10A). Specifically, we annotated cell type of the unsupervised snATAC-seq clusters by label transfer from the cluster of snRNA-seq data that have differential gene expression of known cell markers. Label transfer is based on correlating open-chromatin activities of gene bodies and mRNA expressions transcribed from the corresponding genes. To visualize the cell clusters, we used Uniform Manifold Approximation and Projection for Dimension Reduction (UMAP) (60).
Subsampling of cells and cross-validation of the optimized peaks.
To simulate rare cell types, we subsampled cells with n = 200, 500, and 1,000 from each of the five abundant cell types in the kidney and PBMC snATAC-seq datasets: PT (n ≈ 8,000), LH (n ≈ 3,800), DCT (n ≈ 1,800), CD4M (n ≈ 1,800), and CD14M (n ≈ 4,500). We called peaks using reads aggregated across the same cell type (i.e., pseudobulk) from the subsampled cells and conducted the gkmQC peak-calling optimization. We then compared the peaks recovered by the optimization to the peaks called from all cells. To quantify the degree to which the optimization process recovers true peaks, we calculated the fraction of the added peaks overlapping original peaks from all cells.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S06 (XLSX)
Acknowledgments
We thank all the members of the Sampson Laboratory for their helpful discussion. We would like to acknowledge Boston Children’s Hospital’s High-Performance Computing (BCH HPC) Resources Clusters Enkefalos 2 (E2) made available for conducting the research. Software used in the project was installed and configured by BioGrids. Funding: D.L. and A.C. are supported by the NIH grants HL086694 and HL141980. D.L. is also supported by Boston Children’s Hospital OFD/BTREC/CTREC Faculty Development Fellowship Award and a Manton Center Endowed Scholar Award. M.G.S. is supported by NIH RO1DK119380 and RC2DK122397.
Author contributions
S.K.H., A.C., and D.L. designed research; S.K.H. and D.L. performed research; S.K.H., Y.M., P.C.W., B.D.H., M.G.S., A.C., and D.L. contributed new reagents/analytic tools; S.K.H. and D.L. analyzed data; and S.K.H., Y.M., P.C.W., B.D.H., M.G.S., A.C., and D.L. wrote the paper.
Competing interest
The authors have organizational affiliations to disclose, M.G.S. is on the Scientific Advisory Board of Natera and a consultant for Maze.
Footnotes
Reviewers: B.R., University of California San Diego; and S.S., University of Illinois at Urbana Champaign.
Contributor Information
Aravinda Chakravarti, Email: aravinda.chakravarti@nyulangone.org.
Dongwon Lee, Email: dongwon.lee@childrens.harvard.edu.
Data, Materials, and Software Availability
gkmQC is available in https://github.com/Dongwon-Lee/gkmQC. IPython notebooks to reproduce the results are available in https://github.com/Dongwon-Lee/gkmQC-manuscript. All other data needed to evaluate the conclusions in the paper are present in the paper and/or the SI Appendix. [BED files (Datasets S4 and S5)] data have been deposited in [https://osf.io/egbqv/] (TBD).
Supporting Information
References
- 1.Klemm S. L., Shipony Z., Greenleaf W. J., Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 20, 207–220 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y., Greenleaf W. J., Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyle A. P., et al. , High-resolution mapping and characterization of open chromatin across the genome. Cell 132, 311–322 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hesselberth J. R., et al. , Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat. Methods 6, 283–289 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The ENCODE Project Consortium et al. , Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583, 699–710 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Domcke S., et al. , A human cell atlas of fetal chromatin accessibility. Science 370, eaba7612 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao L., et al. , Chromatin accessibility landscape in human early embryos and its association with evolution. Cell 173, 248–259.e15 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Jänes J., et al. , Chromatin accessibility dynamics across C. elegans development and ageing. eLife 7, e37344 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han S. K., Kim D., Lee H., Kim I., Kim S., Divergence of noncoding regulatory elements explains gene-phenotype differences between human and mouse orthologous genes. Mol. Biol. Evol. 35, 1653–1667 (2018). [DOI] [PubMed] [Google Scholar]
- 10.GTEx Consortium et al. , Using an atlas of gene regulation across 44 human tissues to inform complex disease- and trait-associated variation. Nat. Genet. 50, 956–967 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeggini E., Gloyn A. L., Barton A. C., Wain L. V., Translational genomics and precision medicine: Moving from the lab to the clinic. Science 365, 1409–1413 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Nasser J., et al. , Genome-wide enhancer maps link risk variants to disease genes. Nature 593, 238–243 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ji H., et al. , An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat. Biotechnol. 26, 1293–1300 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.John S., et al. , Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 43, 264–268 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Landt S. G., et al. , ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 22, 1813–1831 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Q., Brown J. B., Huang H., Bickel P. J., Measuring reproducibility of high-throughput experiments. Ann. Appl. Stat. 5, 1752–1779 (2011). [Google Scholar]
- 17.Sims D., Sudbery I., Ilott N. E., Heger A., Ponting C. P., Sequencing depth and coverage: Key considerations in genomic analyses. Nat. Rev. Genet. 15, 121–132 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Nakato R., Shirahige K., Sensitive and robust assessment of ChIP-seq read distribution using a strand-shift profile. Bioinformatics 34, 2356–2363 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Djordjevic M., A biophysical approach to transcription factor binding site discovery. Genome Res. 13, 2381–2390 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee D., Karchin R., Beer M. A., Discriminative prediction of mammalian enhancers from DNA sequence. Genome Res. 21, 2167–2180 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghandi M., Lee D., Mohammad-Noori M., Beer M. A., Enhanced regulatory sequence prediction using gapped k-mer features. PLoS Comput. Biol. 10, e1003711 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alipanahi B., Delong A., Weirauch M. T., Frey B. J., Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat. Biotechnol. 33, 831–838 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Zhou J., Troyanskaya O. G., Predicting effects of noncoding variants with deep learning–based sequence model. Nat. Methods. 12, 931–934 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Avsec Ž, et al. , Base-resolution models of transcription-factor binding reveal soft motif syntax. Nat. Genet. 53, 354–366 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee D., LS-GKM: A new gkm-SVM for large-scale datasets. Bioinformatics 32, 2196–2198 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis C. A., et al. , The Encyclopedia of DNA elements (ENCODE): Data portal update. Nucleic Acids Res. 46, D794–D801 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rada-Iglesias A., et al. , A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470, 279–283 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lichou F., Trynka G., Functional studies of GWAS variants are gaining momentum. Nat. Commun. 11, 6283 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meuleman W., et al. , Index and biological spectrum of human DNase I hypersensitive sites. Nature 584, 244–251 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wuttke M., et al. , A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat. Genet. 51, 957–972 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bulik-Sullivan B. K., et al. , LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finucane H. K., et al. , Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 47, 1228–1235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neale B. M., Heritability of >4,000 traits & disorders in UK Biobank. UKB Heritability (2020) https://nealelab.github.io/UKBB_ldsc/index.html.
- 34.Stuart T., Satija R., Integrative single-cell analysis. Nat. Rev. Genet. 20, 257–272 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Muto Y., et al. , Single cell transcriptional and chromatin accessibility profiling redefine cellular heterogeneity in the adult human kidney. Nat. Commun. 12, 2190 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schizophrenia Working Group of the Psychiatric Genomics Consortium et al. , Comparative genetic architectures of schizophrenia in East Asian and European populations. Nat. Genet. 51, 1670–1678 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miner J. H., et al. , Transcriptional induction of slit diaphragm genes by Lmx1b is required in podocyte differentiation. J. Clin. Invest. 109, 1065–1072 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burghardt T., et al. , LMX1b is essential for the maintenance of differentiated podocytes in adult kidneys. JASN 24, 1830–1848 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tewhey R., et al. , Direct identification of hundreds of expression-modulating variants using a multiplexed reporter assay. Cell 165, 1519–1529 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vierstra J., et al. , Global reference mapping of human transcription factor footprints. Nature 583, 729–736 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andersson R., Sandelin A., Determinants of enhancer and promoter activities of regulatory elements. Nat. Rev. Genet. 21, 71–87 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Kribelbauer J. F., Rastogi C., Bussemaker H. J., Mann R. S., Low-affinity binding sites and the transcription factor specificity paradox in eukaryotes. Annu. Rev. Cell Dev. Biol. 35, 357–379 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee D., et al. , Human cardiac cis -regulatory elements, their cognate transcription factors, and regulatory DNA sequence variants. Genome. Res. 28, 1577–1588 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andersson R., et al. , An atlas of active enhancers across human cell types and tissues. Nature 507, 455–61 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Forrest A. R. R., et al. , A promoter-level mammalian expression atlas. Nature 507, 462–470 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu J., et al. , Landscape of monoallelic DNA accessibility in mouse embryonic stem cells and neural progenitor cells. Nat. Genet. 49, 377–386 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 48.eGTEx Project, Enhancing GTEx by bridging the gaps between genotype, gene expression, and disease. Nat. Genet. 49, 1664–1670 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wakefield J., Bayes factors for genome-wide association studies: Comparison with P-values. Genet. Epidemiol. 33, 79–86 (2009). [DOI] [PubMed] [Google Scholar]
- 50.Pruim R. J., et al. , LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics 26, 2336–2337 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thorvaldsdottir H., Robinson J. T., Mesirov J. P., Integrative genomics viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bycroft C., et al. , The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Teumer A., et al. , Genome-wide association meta-analyses and fine-mapping elucidate pathways influencing albuminuria. Nat. Commun. 10, 4130 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nandakumar P., et al. , Analysis of putative cis-regulatory elements regulating blood pressure variation. Hum. Mol. Genet. 29, 1922–1932 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Y., et al. , Model-based analysis of ChIP-Seq (MACS). Genome. Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gaspar J. M., Improved peak-calling with MACS2. bioXriv [Preprint] (December 9, 2020). 10.1101/496521 (Accessed 17 December 2018). [DOI]
- 57.Fang R., et al. , Comprehensive analysis of single cell ATAC-seq data with SnapATAC. Nat. Commun. 12, 1337 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Korsunsky I., et al. , Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McInnes L., Healy J., Melville J., Umap: Uniform manifold approximation and projection for dimension reduction. arXiv:1802.03426 (2020) (July 22, 2021). 10.48550/arXiv.1802.03426. [DOI]
- 60.Kodzius R., et al. , CAGE: Cap analysis of gene expression. Nat. Methods 3, 211–222 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S06 (XLSX)
Data Availability Statement
gkmQC is available in https://github.com/Dongwon-Lee/gkmQC. IPython notebooks to reproduce the results are available in https://github.com/Dongwon-Lee/gkmQC-manuscript. All other data needed to evaluate the conclusions in the paper are present in the paper and/or the SI Appendix. [BED files (Datasets S4 and S5)] data have been deposited in [https://osf.io/egbqv/] (TBD).