Significance
Molecular cell phenotype identification is important for the investigation of the role of cells and for their medical applications. For this identification, single-cell whole-transcriptome analysis, which is an unbiased method, is a powerful tool. However, this method kills the target cell, which prevents further investigation of the cell. Here, we showed a successful noninvasive approach; label-free live-cell imaging with deep learning–based prediction can distinguish cell phenotypes defined by whole-transcriptome sequencing. This approach was enabled by linking cell images and the whole transcriptome for the same cell using our newly developed robot, the automated live imaging and cell picking system (ALPS). This noninvasive and unbiased determination of live-cell molecular phenotypes will be useful for cell dynamics studies.
Keywords: robotics, microscopy, deep learning, single-cell RNA sequencing, cell picking
Abstract
Single-cell whole-transcriptome analysis is the gold standard approach to identifying molecularly defined cell phenotypes. However, this approach cannot be used for dynamics measurements such as live-cell imaging. Here, we developed a multifunctional robot, the automated live imaging and cell picking system (ALPS) and used it to perform single-cell RNA sequencing for microscopically observed cells with multiple imaging modes. Using robotically obtained data that linked cell images and the whole transcriptome, we successfully predicted transcriptome-defined cell phenotypes in a noninvasive manner using cell image–based deep learning. This noninvasive approach opens a window to determine the live-cell whole transcriptome in real time. Moreover, this work, which is based on a data-driven approach, is a proof of concept for determining the transcriptome-defined phenotypes (i.e., not relying on specific genes) of any cell from cell images using a model trained on linked datasets.
Omics analyses are powerful tools to measure biological samples in an unbiased manner, and single-cell whole-transcriptome analysis by RNA sequencing (RNA-seq) has been widely used to identify cell phenotypes at the molecular level (1, 2). Furthermore, an unbiased understanding of cell dynamics using RNA-seq is important, which may be efficiently achieved through the continuous measurement of the same cell. However, the unbiased analysis for cell dynamics is difficult because the cells are lysed or fixed for RNA-seq. As a noninvasive approach, the image-based prediction of biological phenotypes, including disease stages, at the molecular level using deep learning has become a powerful tool. Generally, each image is labeled, e.g., as cancer or normal cells; the correlation between image features and labels is modeled using a large number of labeled images, and the label of a new image is predicted based on the modeled correlation (3–6). For further applications using this type of image-based deep learning, determining labels based on parameters measured without bias, e.g., by using RNA-seq, is essential because the capability to identify biological phenotypes based on a small number of molecular markers or by experienced individuals is limited (6, 7). However, to the best of our knowledge, image-based prediction of phenotypes of untreated single cells defined by RNA-seq has not been performed thus far. To perform this prediction using deep learning with modeling, a large dataset of single cells, in which each cell has high-quality label-free live-cell images, and the whole transcriptome is needed. Recently, single-cell RNA-seq (scRNA-seq) was performed on many cells after cell isolation in microwells and live-cell imaging (SI Appendix, Table S1) (8–10). However, this approach is unsuitable for cells that cannot be cultured in isolation, which is critical for live-cell imaging. Therefore, an alternative approach that allows a cell to be imaged in a dish (a common vessel for cell culture) in the presence of other cells, and subsequently isolated from the dish to a sequencing platform, is desired (10). Indeed, in a recent study, single live cells (yeast) in a dish were isolated after imaging, and scRNA-seq was performed on the isolated cells (SI Appendix, Table S1) (11). However, in this approach, each cell was imaged and isolated sequentially without automation, which prevents rapid data acquisition and ease of use.
Here, for efficient high-quality data acquisition for deep learning, we developed a robot, the automated live imaging and cell picking system (ALPS), which enabled scRNA-seq, with programmable functions, for microscopically observed, targeted, and freely moving cells in a normal dish. Using the datasets robotically obtained by the ALPS for cell lines and primary murine peripheral blood mononuclear cells (PBMCs), we showed that accurate deep learning predictions of transcriptome-defined cell phenotypes can be achieved from bright-field live-cell images.
Results
Isolation of Biological Samples by the ALPS.
Eventually, our developed ALPS enabled the automated isolation of single cells observed by optical microscopy in each well of a 96-well plate by synchronizing an optical microscope (Eclipse Ti2-E, Nikon Co.) and a cell picker (TOPickV, YODAKA Co., Ltd.) via real-time interactive communication (Fig. 1A, SI Appendix, Fig. S1, and Movie S1, details later). First, we programmed cell picking and deposition for rapid and automated cell isolation. Picking up a cell at the center of the field of view (from the start of step-(auto-7-2) to the end of step-(auto-7-3), see Materials and Methods) took 0.52 ± 0.05 s (mean ± SD, n = 20), and picking and depositing the cell into a well (from the start of step-(auto-7-2) to the end of step-(auto-7-5), see Materials and Methods) took 3.58 ± 0.11 s (mean ± SD, n = 20). We set up an additional imaging system that recorded cell deposition to confirm that a single cell was actually deposited for downstream analyses (Materials and Methods, Movie S2). To obtain a clear cell image during deposition, we flattened the bottom of a 96-well plate by vertically grinding the tips of the wells by approximately 0.3 mm (Materials and Methods). We showed that the ALPS successfully picked up and deposited objects with a wide range of sizes, such as 1-, 3-, 45-, and 200- to 300-μm-diameter beads, and biological samples, such as cells (~10 μm), organoids (~200 μm), and crypts (~30 μm, a piece of tissue from the small intestine) (Movies S2–S8 and SI Appendix, Texts 1 and 2). We confirmed that mouse leukemia cells and human Jurkat cells proliferated after isolation by the ALPS (SI Appendix, Fig. S2 and Text 3). Thus, the ALPS may be used for the quick and efficient isolation of microscopically targeted samples with a range of more than two orders of magnitude in size and for the isolation of live biological samples for subsequent culturing and analyses.
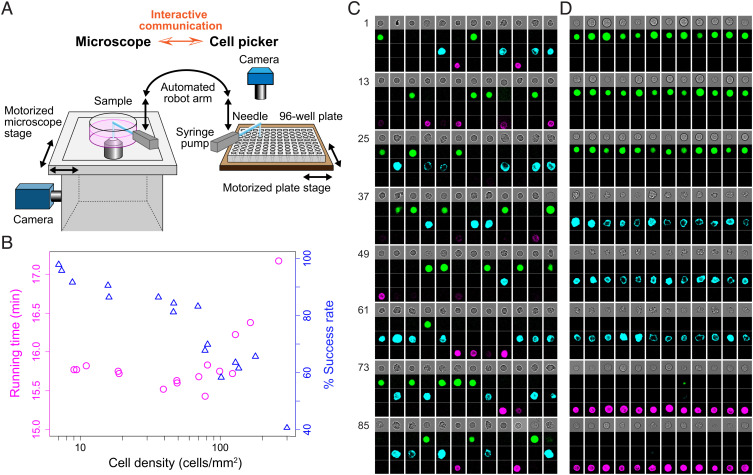
Fig. 1.
ALPS. (A) Schematic of the ALPS (details in Materials and Methods). (B) The running time and success rate for the automated isolation of 96 T cells in a dish based on the cell density. Running time, total time from the start of step-(auto-1) of the first isolation to the end of step-(auto-7) of the 96th isolation. Success rate, the number of isolations, where one cell was picked and deposited, in each of the 96 automated isolations divided by 96. Cell density, average number of cells per mm2 of all scanned squares for each of the 96 automated isolations (see section “Automated Single-Cell Isolation Programs” in Materials and Methods). (C and D) Four images (as column), bright-field image (Top) and fluorescent images of GFP (green), PE (cyan), and Alexa647 (magenta) of each cell from 3mix-ALPS-random-multimode (C) or 3mix-ALPS-target-multimode (D). The numbers indicate the order of isolation. Image size, 26 μm × 26 μm. The contrast of the images shown in (C) and (D) changed linearly.
Automated Cell Isolation by the ALPS.
Next, we further automated the ALPS by synchronizing cell picking and deposition with an optical microscope (Eclipse Ti2-E) via real-time electronic communication, which enabled the successive rapid isolation of many observed cells using programmable functions, including multimodal imaging, real-time cell targeting, and time-lapse imaging (Fig. 1A and Movies S1 and S2). The essential automated steps were as follows (Materials and Methods): i) the positions of individual cells in a dish were determined by NIS-Elements (Nikon, Co.); ii) cells that were at least a particular distance from any other cells were labeled; iii) a labeled cell was moved to the center of the field of view by moving the microscope stage, and its position was redetermined; iv) one or more images of the cell were captured; v) the signal was sent from NIS-Elements to the cell picker (TOPickV); vi) the cell at the center was picked with 9 nL or 11 nL of solution by a glass needle (40 or 60 μm in diameter); vii) the needle was moved to a 96-well plate; viii) the picked cell was deposited with 15 nL or 19 nL of solution from the needle into prepared medium (2 μL) in a well of a 96-well plate; and ix) the signal was sent from the cell picker to NIS-Elements. Between (vi) and (vii), the 96-well plate was translated so that the picked cell was deposited in the next well. The procedure from (iii) to (ix) was repeated to collect 96 cells, one cell in each well of the plate. After all of the above procedures were carried out, the plate was sealed, quickly frozen in liquid nitrogen within 1 min, and stored at −80 °C in the cases where RNA-seq for transcriptome analysis was subsequently performed (see below).
We performed five applications of the ALPS with different functions and samples. First, we successively isolated 96 cultured murine T cells (Materials and Methods) by capturing a bright-field image for each cell and measured the total time from steps (i) to (ix) with 96 repetitions and the success rate for single-cell picking and deposition depending on the cell density (9 to 262 cells/mm2) (Fig. 1B). The total time ranged from 15 min 26 s to 15 min 49 s, and the success rate was 81 to 98% when the cell density was 9 to 71 cells/mm2 (Fig. 1B). The high success rate may have been due to the interactive communication function, which was used to determine the cell position immediately before every cell picking. When the cell density was higher, failures were mainly due to the picking of multiple cells (Dataset S1). Second, to obtain more information by imaging, we investigated the protein expression levels of randomly selected cells by multimodal imaging before cell isolation. We prepared a mixture of three types of fluorescently labeled cultured cells (mouse T cells labeled with the PE-CD8a antibody, mouse leukemia cells expressing the EGFP fusion protein, and mouse hematopoietic progenitor cells (HPCs) labeled with the Alexa647-B220, referred to hereafter as three-type–mixed cells, see Materials and Methods), captured images with the bright field and three fluorescent colors, and picked and deposited cells for four 96-well plates (3mix-ALPS-random-multimode) (Fig. 1C, SI Appendix, Fig. S3, Materials and Methods, and Dataset S2). Third, we showed that the ALPS enabled targeted cell isolation by real-time feature identification. We prepared the three-type–mixed cells above and isolated 36 leukemia cells, 36 T cells, and 24 HPCs in this order for each 96-well plate (four plates in total) by the real-time calculation of fluorescent intensities using NIS-Elements (3mix-ALPS-target-multimode) (Fig. 1D, SI Appendix, Fig. S3, Materials and Methods, and Dataset S2). Fourth, to exploit the advantages of the ALPS, which uses an optical microscope, we performed time-lapse imaging followed by cell isolation. We used the three-type–mixed cells without labeling, captured bright-field cell images every 1 min for 29 min (30 frames), and scanned an area of 4.36 mm × 3.68 mm, which contained ~200 cells. We picked 96 cells sequentially and deposited them into different wells of 96-well plates, scanning the area after the isolation of every 12 cells to track the cells during cell isolation (Materials and Methods, Dataset S2). In total, we obtained sixteen 96-well plates with the cells (3mix-ALPS-timelapse). Finally, we performed isolation after the time-lapse imaging of primary PBMCs in eight 96-well plates (Materials and Methods, Dataset S2). For all plates in the 2nd to 5th applications, the success rate for single-cell picking and deposition was 86 ± 7% (mean ± SD, n = 32) (Dataset S2). Thus, the ALPS enabled the rapid, high-throughput isolation of cell lines and primary cells through its flexible programmable operations, which allowed the introduction of multimodal imaging, the identification of image-based cell features in real time, and time-lapse–based cell dynamics observation.
Combination of the ALPS and RNA-seq.
To perform high-throughput scRNA-seq for cells isolated with the ALPS (ALPS&RNA-seq), we set up an automated library preparation system for scRNA-seq using the Bravo NGS workstation (Agilent Technologies) (SI Appendix, Text 4). We first performed scRNA-seq on the cells from 3mix-ALPS-random-multimode (4 plates) and 3mix-ALPS-target-multimode (4 plates). For the eight plates (3mix-ALPS-random&target-multimode), after filtering to remove wells that contained multiple cells, no cells, apparent dust, or an unusually small amount of detected RNA, a total of 649 cells were analyzed further (Datasets S3 and S4 and SI Appendix, Texts 5 and 6). We found that the quality of the sequencing results was not affected by the cell isolation time because the number of total RNA molecules and genes detected did not depend on the order of wells, i.e., earlier picked cells and later picked cells (SI Appendix, Fig. S4). We confirmed that there were three clusters obtained from whole-transcriptome–based clustering, which was the expected result, and we determined the cell types in the clusters using marker genes (Fig. 2A and SI Appendix, Fig. S5). Importantly, we showed that the ALPS and cell barcodes (a combination of well-position barcodes and plate barcodes in SI Appendix, Text 4) were used to correctly link image-captured individual cells and their sequencing results for the 649 cells. This was confirmed through cell types identified by fluorescence imaging (SI Appendix, Text 7) and by whole-transcriptome sequencing, where completely consistent results were obtained (Figs. 1 C and D and 2A). To evaluate the effects of the ALPS on scRNA-seq, we compared the sequencing results of single cells from 3mix-ALPS-random&target-multimode and those of the three-type–mixed cells isolated by a sorter (SI Appendix, Text 8). We found that the total numbers of detected RNA molecules and genes and the copy numbers of detected RNA molecules for each gene were consistent between cells obtained by the ALPS and the cell sorter (SI Appendix, Fig. S6). These results showed that the acquisition of a linked dataset of cell images and the whole transcriptome was successful by the ALPS&RNA-seq.
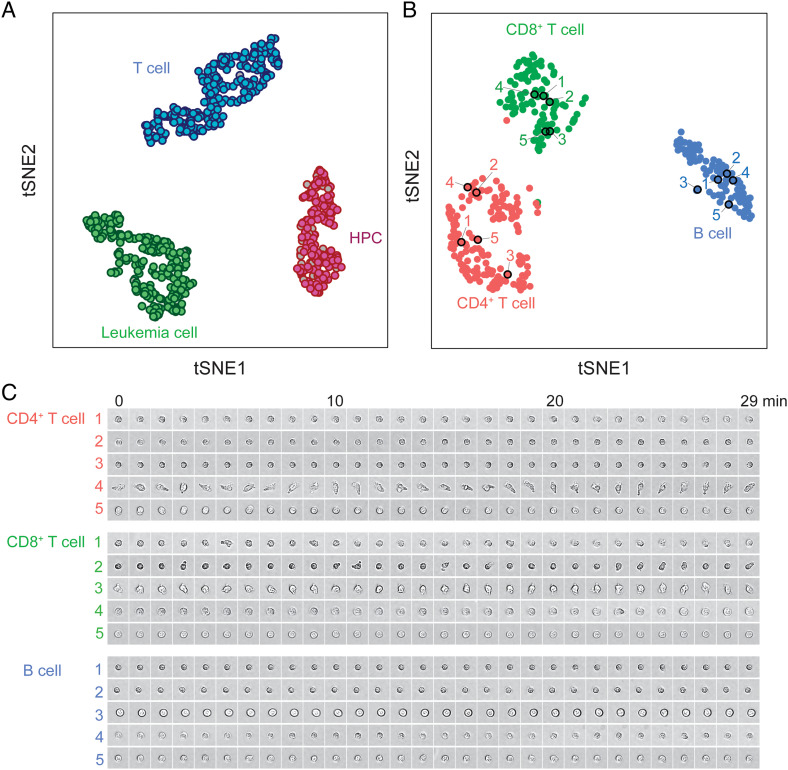
Fig. 2.
ALPS-linked cell images and whole transcriptome for the same cell. (A) t-SNE visualization of single cells from 3mix-ALPS-random&target-multimode clustered based on whole transcriptomes. Filled colors represent the cell types determined by fluorescent images; the gray color indicates that the cell type was undetermined due to low fluorescent intensities. The edge colors represent the cell types determined by the transcriptome. (B) t-SNE visualization of all PBMCs clustered based on whole transcriptomes. The colors represent the cell types determined by the transcriptome. The black circles and numbers correspond to the cells shown in (C). (C) Examples of bright-field time-lapse cell images of PBMCs (SI Appendix, Text 10). Image size, 20 μm × 20 μm. The contrast of the images shown in (C) changed linearly.
We then performed scRNA-seq for cells from 3mix-ALPS-timelapse (Materials and Methods, Datasets S3 and S4). In total, 1,008 cells with both images and high-quality sequencing results were further analyzed (SI Appendix, Fig. S7). The sequencing results from 3mix-ALPS-timelapse and 3mix-ALPS-random&target-multimode were consistent (SI Appendix, Figs. S6 A and B and S7). We also performed scRNA-seq for PBMCs obtained by the ALPS, as described above; 346 cells with images and sequencing results were further analyzed (Fig. 2 B and C, Materials and Methods, Datasets S5 and S6). We clustered the scRNA-seq results of PBMCs into three subpopulations, which were identified by marker genes as CD4+ T cells, CD8+ T cells, and B cells, the main cell types in PBMCs (12) (Fig. 2B and SI Appendix, Fig. S8 and Text 6). Thus, we linked the cell dynamics observed by time-lapse imaging and the whole transcriptome by the ALPS&RNA-seq for both cell lines and primary cells.
Prediction of Transcriptome-Defined Cell Phenotypes (Types and States).
Next, we predicted scRNA-seq–determined cell phenotypes (types or states) from the time-lapse label-free cell images using deep learning classification methods with the datasets obtained by the ALPS&RNA-seq. To efficiently extract features from the cell images and dynamics, we used two deep learning architectures: ResNet–LSTM is a combination of a deep residual network (ResNet) (13) [deep convolutional neural network (CNN)] and a long short-term memory (LSTM) (14) [recurrent neural network (RNN)], and LeNet–LSTM is a combination of a LeNet (15) (simple CNN) and an LSTM (Materials and Methods, SI Appendix, Fig. S9 and Text 9). First, we classified the cells from 3mix-ALPS-timelapse using the linked dataset (1,008 single cells) of time-lapse images and scRNA-seq–determined cell types (318 T cells, 423 leukemia cells, and 267 HPCs) between which 29 to 40% of detected genes were significantly different (padj < 0.05 and |log2foldchange| > 1) (SI Appendix, Fig. S10). To assess the generalization power of our classification in independent experiments, we trained ResNet–LSTM and LeNet–LSTM to generate models, tested those models, and repeated this process 16 times using 16 plates in a leave-one-out cross-validation strategy (16) (Materials and Methods, SI Appendix Text 9). We found that the classification of three cell types achieved a high accuracy of 0.81 ± 0.07 for ResNet–LSTM (“All cells” in Fig. 3A) and 0.75 ± 0.12 for LeNet–LSTM (SI Appendix, Fig. S11B) and high weighted F1 scores of 0.81 ± 0.06 for ResNet–LSTM and 0.75 ± 0.12 for LeNet–LSTM (mean ± SD, n = 16) (“All cells” in SI Appendix, Fig. S11 A and B). As a comparison, we also classified these cells into three clusters based on their morphological and dynamical features (Datasets S7–S9), which were extracted from the cell images by three representative and well-used conventional image analysis software programs (17): NIS-Elements (18), CellProfiler 4 (17), and TrackMate 7 (19) in Fiji (ImageJ) (20) (SI Appendix, Fig. S12 A, C, and E and Text 7). For each analysis, we assigned the feature-determined clusters to the three scRNA-seq–determined cell types by any possible one-to-one correspondence, and for each correspondence, we calculated the accuracy (that is, the number of cells matched with their scRNA-seq–determined cell types divided by the total number of cells) (SI Appendix, Fig. S12 B, D, and F). We found that the accuracies of all correspondences were in the range of 0.15 to 0.47 for the NIS-Elements analysis, 0.11 to 0.70 for CellProfiler 4, and 0.16 to 0.48 for TrackMate 7, which were all lower than the deep learning classification accuracy shown above (0.81). These results indicated that the deep learning model can be used to predict the cell types defined by RNA-seq from the cell images and dynamics, and the accuracy obtained was higher than that of conventional image analysis methods.
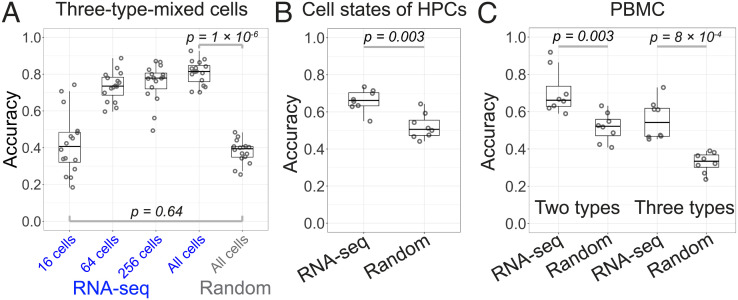
Fig. 3.
Deep learning–based classification for time-lapse imaged samples. (A) Accuracy of predicting the scRNA-seq–determined cell types of the cells from the 3mix-ALPS-timelapse using ResNet–LSTM. RNA-seq, cell types determined by scRNA-seq; Random, cell types labeled randomly; 16, 64, 256, and All cells, 16, 64, 256, and all cells were randomly selected from 15 plates for training (cells in one plate were used for testing). The P values were calculated using the Kruskal–Wallis rank sum test [in (B) and (C) as well]. (B) Accuracy of predicting scRNA-seq–determined cell states of HPCs (HPC-1 and HPC-2 in SI Appendix, Fig. S13A) using ResNet–LSTM. (C) Accuracy of predicting scRNA-seq–determined cell types of PBMCs using ResNet–LSTM. Two types, PBMCs were clustered into T cells and B cells. Three types, PBMCs were clustered into CD4+ T cells, CD8+ T cells, and B cells.
We then investigated the effects of the number of cells and number of frames on the prediction by ResNet–LSTM. The accuracy and F1 score decreased when a smaller number of cells and frames were used (Fig. 3A and SI Appendix, Fig. S11 A and C), which suggested that the high-throughput data acquisition and time-lapse imaging achieved by the development of the ALPS&RNA-seq were essential for accurate cell classification using deep learning.
Second, to test the capability of deep learning for the image-based classification of cells from the same cell type but different biological states, we classified the cells from 3mix-ALPS-timelapse into six clusters using their whole transcriptomes and found two subclusters in each cell type, i.e., T cells, leukemia cells, and HPCs (SI Appendix, Fig. S13A). We found that 4.7% of the detected genes were differentially expressed (padj < 0.05 and |log2foldchange| > 1) between the two subclusters of HPCs (HPC-1: 143 cells and HPC-2: 124 cells), while only 0.3% were differentially expressed between the two subclusters of T cells (T cell-1: 188 cells and T cell-2: 130 cells) and leukemia cells (leukemia cell-1: 220 cells and leukemia cell-2: 203 cells) (SI Appendix, Fig. S13 B–D). We found that these differentially expressed genes were predominantly linked to several biological processes, such as immune-related processes, which were identified by gene ontology analysis (21) (SI Appendix, Fig. S13 B–D), suggesting that the two subclusters generated from each cell type were in different biological states. We then performed image-based cell classification for the two subclusters of each cell type using ResNet–LSTM; we trained ResNet–LSTM to generate models, tested those models, and repeated this process eight times using the 16 plates in a leave-one-out cross-validation strategy (16) (every two plates were merged in order) (Materials and Methods, SI Appendix, Text 9 and Dataset S3). For the HPCs, the accuracy and F1 score were 0.66 ± 0.06 and 0.64 ± 0.07 (mean ± SD, n = 8), respectively, which were significantly higher than the results obtained using the same learning strategy with artificial random labeling of subclusters (Fig. 3B and SI Appendix, Fig. S14B). For the T cells and leukemia cells, the accuracies and F1 scores were not significantly different from those obtained using the same learning strategy with artificial random labeling of subclusters (SI Appendix, Fig. S14). These results indicated that the ResNet–LSTM deep learning model was generalized for predicting the biological cell states of HPCs that were defined by 4.7% differentially expressed detected genes from cell images and dynamics but not for predicting the biological cell states of T cells and leukemia cells defined by only 0.3% differentially expressed detected genes.
Third, we performed cell classification for the PBMCs using the linked dataset (346 single cells) of time-lapse label-free cell images and scRNA-seq–determined cell types, each of which was labeled by one of three types (152 CD4+ T cells, 103 CD8+ T cells, and 91 B cells) or two types (255 (= 152+103) T cells and 91 B cells) based on the whole transcriptome between cell types [12 to 15% detected genes were differentially expressed (padj < 0.05 and |log2foldchange| > 1) (SI Appendix, Fig. S15)]. For the PBMCs labeled by both two and three cell types, we trained ResNet–LSTM and LeNet–LSTM, respectively, to generate models, tested those models, and repeated this process eight times using the eight plates in a leave-one-out cross-validation strategy (16) (Materials and Methods, SI Appendix, Text 9). Although the accuracies and F1 scores for both labels were not very high (e.g., accuracy of 0.71 ± 0.12 for two types and 0.56 ± 0.10 for three types (mean ± SD, n = 8) when ResNet–LSTM was used), they were significantly higher than the results obtained using the same learning strategy with artificial random labeling of cell types (Fig. 3C and SI Appendix, Fig. S16). For comparison, we then performed conventional image analyses for PBMCs using the same strategy as for the cells from the 3mix-ALPS-timelapse above. We classified the PBMCs into three clusters based on extracted morphological and dynamical features (Datasets S10–S12) from the cell images, assigned them as CD4+ T cells, CD8+ T cells, and B cells by any possible one-to-one correspondence, and calculated the accuracy for each correspondence (SI Appendix, Fig. S17). We found that the accuracies of all correspondences were in the range of 0.14 to 0.40 for the NIS-Elements analysis, 0.21 to 0.53 for CellProfiler 4, and 0.24 to 0.46 for TrackMate 7, which were lower than the deep learning classification accuracy shown above (0.56). These results indicated that the deep learning model for predicting the cell types of murine primary PBMCs defined by RNA-seq from cell images and dynamics was generalized, and the accuracy of the determination of cell types using deep learning was higher than that using the conventional image analysis methods.
Individual Cell States Determined by Predicted RNA Expression Levels from Cell Images.
First, to test the capability of predicting the RNA expression level of each gene from the cell images and dynamics, we performed regression analysis for each of the top 300 variant genes for the cells from 3mix-ALPS-timelapse using ResNet–LSTM. Again, we trained, tested, and repeated the process 16 times using the different plates (Materials and Methods, SI Appendix, Text 9). Although the expression levels of most genes could not be accurately predicted (SI Appendix, Fig. S18 A and B), we found 14 genes (one example in Fig. 4A and others in SI Appendix, Fig. S18 C and D), which showed that > 40% (median out-of-sample R2 = 0.41 to 0.58, median Pearson’s correlation coefficient r = 0.66 to 0.77 for predicted expression levels versus measured expression levels) of the variance in expression levels among cells was explained by the cell images and dynamics, suggesting the possibility of predicting the RNA expression levels of individual genes from cell images and dynamics.
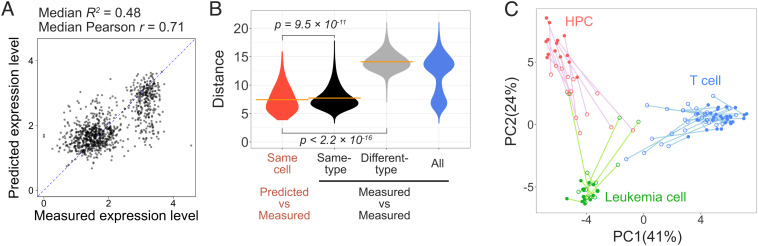
Fig. 4.
Deep learning–based regression for time-lapse imaged three-type–mixed cells (3mix-ALPS-timelapse). (A) Scatterplots of the measured and predicted RNA expression levels of the cells for the Gm1821 gene (log-transformed; SI Appendix, Text 6), including 16 predictions using different plates. Median R2, median of the out-of-sample R2 value for each prediction (SI Appendix, Fig. S18C); median Pearson’s r, median of the Pearson’s correlation coefficient r for each prediction (SI Appendix, Fig. S18D). Dashed line, diagonal line. (B) Distribution of the Euclidean distances between the predicted and measured expression levels of 300 genes for the same cell and between the measured expression levels of 300 genes of all possible pairs of all cells, same-type cells, and different-type cells. The orange lines indicate the means. The P values were calculated using the Kruskal–Wallis rank sum test. (C) PCA of the cells from 3mix-timelapse-plate5 with predicted (open circle) and measured (closed circle) expression levels of 300 genes (each cell has two dots) (data from the other 15 plates are shown in SI Appendix, Fig. S19). Colors correspond to the three different cell types determined by the measured whole transcriptome. Lines link the predicted and measured expression levels of the same cell.
Then, instead of classifying cells into subpopulation-based cell phenotypes, we determined the state for each individual cell using the predicted RNA expression levels of the 300 variant genes together (Fig. 4 B and C). To quantitatively evaluate the accuracy of the prediction of RNA expression–defined cell states, we investigated the difference between the predicted and measured cell states for each cell by calculating the Euclidean distance between the predicted and measured cell states defined by 300 gene expression levels and compared the distances of all cells with the distances between all possible pairs of measured cell states; the former was distributed at the short end of the latter range (Fig. 4B, red and blue). Furthermore, we found that although the distances between the predicted and measured cell states were nonnegligible (Fig. 4B, red), their average [7.429 ± 0.067 (mean ± SEM, n = 1,008)] was significantly smaller than both the average distance between the measured cell states of all possible pairs of same-type cells [Fig. 4B, black; 7.722 ± 0.004 (mean ± SEM, n = 175,167)] and that of different-type cells [Fig. 4B, gray; 14.103 ± 0.003 (mean ± SEM, n = 332,361)]. To visualize the prediction relative to the measurement for individual cells, we performed PCA for all cells in each plate with the predicted and measured 300 gene expression levels together (one plate in Fig. 4C and others in SI Appendix, Fig. S19). As expected, the predicted RNA expression–defined cell states were shown to be globally closer to the measured RNA expression–defined cell states of same-type cells than those of different-type cells (Fig. 4C and SI Appendix, Fig. S19). These results suggested that the states of individual cells can be directly determined by predicting the RNA expression levels of individual genes without subpopulation-based cell phenotype information.
Discussion
Both our deep learning–based classification and regression results showed that transcriptome-defined cell phenotypes (types and states) (i.e., labels without bias) could be predicted from time-lapse live-cell images. These results were enabled by our developed ALPS&RNA-seq, by which a linked dataset of live images and whole transcriptomes for the same single cells was acquired. This prediction provides a way to develop machine learning–refined images as omics-based live-cell markers that do not rely on one or more specific genes or surface proteins or on antibody labeling. Furthermore, label-free imaging is tremendously scalable at very low cost. Importantly, our successful prediction opens a window for decoding phenotypes of any cell defined by the whole transcriptome from only cell images using a trained model, once the linked datasets of images and transcriptome analyses for many phenotypes of cells are accumulated. This deep learning–based determination of live-cell molecular phenotypes will provide a noninvasive and an unbiased approach for cell dynamics studies on transcriptomes. An open challenge is to predict similar transcriptomic states of cells with high accuracy.
The ALPS, which is a key part of our developed system, has the following advantages. First, the ALPS is sufficiently fast (0.52 s) and can pick up most biological samples, unless they are rapidly moved, e.g., by an artificial flow. Second, the ALPS can be easily equipped with different types of microscopes, such as a differential interference contrast (DIC) (22), total internal reflection fluorescence (TIRF) microscope (23), confocal microscope (24), or superresolution microscope (25), to maximize the advantages of microscopy to acquire different types of informative cell images. Third, cells isolated by the ALPS can be further cultured and/or applied to different kinds of downstream analyses, such as whole-genome sequencing (26, 27), epigenetic sequencing (28), and proteomic analysis (29). Fourth, the ALPS has a flexibly programmable interactive communication function; one may target individual cells by real-time position determination and feature detection, e.g., molecular localization, cell movement, or cell shape. Finally, the ALPS may pick freely interacting cells in a cell population, which can be used to study, e.g., the molecular mechanism of cell–cell interactions. These advantages will easily expand the usage of ALPS for data acquisition, obtaining multiple features by imaging, measuring different types of molecules, and applying for different samples, which may advance not only deep learning–based prediction but also single-cell characterization in fundamental research and medical applications. A subsequent technical challenge is the automatic isolation of adherent cells such as neurons and fibroblasts from a substrate after live-cell imaging. In a recent study (30), the automatic isolation of single adherent cells by laser microdissection was shown but without live-cell imaging. When using the ALPS, an additional process to detach the adherent cells will be required.
Materials and Methods
Preparation of Cells, Organoids, and Tissue Samples.
Mouse T cells (a cell line) were established from BW-1100.129.237 (provided by Leszek Ignatowicz, Georgia State University, USA); mouse leukemia cells were established from the C1498 cell line (ATCC), and mouse hematopoietic progenitor cells (HPCs) were prepared as previously described (31) (details in SI Appendix, Text 1).
Mouse (C57BL6/J) peripheral blood mononuclear cells (PBMCs) were prepared from whole blood by a general protocol (SI Appendix, Text 1) using density centrifugation and a cell sorter [FACS Aria III (BD Biosciences)]. Mouse small intestinal crypts were isolated and cultured as described with slight modification (32) (SI Appendix, Text 1). Mouse small intestinal organoids were grown from the crypts as described (33) with slight modifications (SI Appendix, Text 1).
C57BL6/J mice were purchased from CLEA Japan and maintained in the RIKEN facility by being fed a CE-2 diet (CLEA Japan). All animal experiments were approved by the RIKEN Institutional Animal Care and Use Committee and were performed in accordance with institutional guidelines.
Hardware and Software of the ALPS.
The ALPS is composed of an inverted optical microscope (Eclipse Ti2-E, Nikon), a cell picker including a motorized stage for the displacement of a 96-well plate (TOPickV, YODAKA Co., Ltd.), an interactive communication system between the microscope and the picker, and an imaging system for monitoring a single cell in a well of a 96-well plate. For cell culture, a customized stage-top incubator and a surrounding temperature control system were used.
On the Nikon microscope, an objective lens (CFI Plan Apochromat Lambda 20X, Nikon), LEDs for bright-field imaging (TI2-D-LHLED, Nikon) with a green filter (MBN11200, Nikon) and for fluorescence imaging (460 nm for GFP and 550 nm for PE and Alexa647; pE-300, CoolLED), filter units (GFP-B C-FLL-C for GFP, mCherry C-FLL-C for PE, and Cy5 C-FLL-C for Alexa647, Nikon), a motorized stage (TI2-S-SE-E, Nikon), a perfect-focus system (TI2-N-ND-P, Nikon), and a CMOS camera (Zyla5.5, Andor) were equipped, and all of them were controlled by NIS-Elements (Nikon).
The picker consisted of an S-shaped glass needle (diameter: 40, 60, 75, and 400 μm; YODAKA) connected to an automated syringe pump (TOPick pump type B; YODAKA), an automated robot arm for the movement of the needle, and a motorized stage for the movement of a 96-well plate (plate stage). The minimum handling volume (one unit) of the pump [0.57 ± 0.05 nL (mean ± SE)] was measured (SI Appendix, Fig. S20). The arm had rotational and z axis degrees of freedom, which were used to move the needle between a dish on the microscope and a well of the 96-well plate on the plate stage (Fig. 1A). A motorized manipulator was installed between the pump and the arm for minor movement of the needle along the x, y, and z axes and was controlled by a 3-axis operation box (QT-AK3, CHUO PRECISION INDUSTRIAL CO., LTD.). The plate stage had both x and y axis degrees of freedom for the movement of the 96-well plate and could move each well of the plate to a fixed position for cell deposition (Fig. 1A). The pumping, arm movement, and plate stage movement were automated by software TOPickV.
To synchronize the picker and the microscope by interactive communication, two cables with DAQ devices (PCIe-6353 and USB-6003, National Instruments) were equipped between the picker and the microscope to transfer transistor–transistor logic (TTL) signals from the microscope to the picker (TTL 1) and from the picker to the microscope (TTL 2). The signal transfer was controlled by NIS-Elements and software TOPickV.
To monitor the process of cell deposition from the needle to the well, another CMOS camera (DMK33UX264, The Imaging Source) with a zoom lens (TS-93005; Sugitoh Co., Ltd.), controlled by IC capture (v2.4, The Imaging Source), was equipped above the plate. An LED light source (TSPA22X8-57W-4, AI Tec System Co., Ltd.) used with a condenser (ML-70, MORITEX Corporation) and a green filter (IF550, Olympus) was equipped under the plate.
To record the processes of both picking and deposition simultaneously, screen capture software, Shadowplay (NVIDIA) or Flashback express recorder (Blueberry), was used to capture the screen on a computer monitor, where the images captured by both the camera on the microscope and the camera above the 96-well plate were shown in real time (Movie S2).
For temperature control, a stage-top incubator (INUBG2TF-WSKM, Tokai Hit) was equipped on the microscope stage, which was kept at 37 °C and supplied with 5% CO2; we opened a 20-mm-diameter hole at the center of the incubator lid for cell picking. To maintain the surrounding temperature of the ALPS (35 to 37 °C), the ALPS was covered by a cage, and the temperature inside the cage was controlled using a heater (OHT-1566, Hidamari).
Automated Single-Cell Isolation Using the ALPS.
Preparation of plates for cell deposition.
For clear imaging, the bottom tip of a 96-well PCR plate (4ti-LB0960RIG, 4titude) was planed (approximately 0.3 mm from the bottom) using a PROXXON mini router (MM100, Proxxon) with a grinding bit (SMP1515, Yanase). To stabilize the RNA of the deposited cells, 2 μL of a solution of 1× buffer for Maxima H Minus Reverse Transcriptase (Thermo Fisher Scientific) with 8 U RNAsin® Plus RNase inhibitor (Promega) was added in advance to each well of the plate using the Bravo NGS workstation (Agilent Technologies). Subsequently, to position the solution at the bottom of the wells, the plate was sealed using the PlateLoc Thermal Microplate Sealer (Agilent Technologies), centrifuged for 30 s at 4,000 rpm (Plate Centrifuge PlateSpin II, KUBOTA), vortexed for 15 s at 3,000 rpm using MixMate® (Eppendorf), and centrifuged again for 30 s at 4,000 rpm. The plate was kept at room temperature (within 1 h) or at 4 °C until use. When it was kept at 4 °C, the plate was warmed at room temperature for 30 min before use.
Preparation of samples.
T cells were filtered using a 35-μm cell strainer (Falcon), counted by a Brand™ Bürker-Türk Counting Chambers (Thermo Fisher Scientific), and diluted to final concentrations of 0.05, 0.10, 0.25, 0.50, and 1.0 × 105 cells/mL using medium A [1× RPMI 1640 medium without phenol red (Thermo Fisher Scientific) supplemented with final concentrations of 10% (v/v) fetal bovine serum (Corning), 1× monothioglycerol solution (FUJIFILM Wako Pure Chemical Industries), 1× GlutaMAX™ supplement (Gibco), 1× Penicillin/Streptomycin Mixed Solution (Nacalai Tesque Inc.)], which was filtered using a 0.22-µm Steriflip filter unit (Merck). The diluted cell solutions were filtered again using 35-μm cell strainers. For each concentration, 3 mL of the filtered cell solution was added to a glass bottom dish (D35-27-1.5-U, Matsunami), which was coated in advance with 1 mL of 0.1% w/v BSA (Sigma-Aldrich) in PBS for 10 min at room temperature and washed with PBS three times. For cell sedimentation, the dish was placed on the microscope and incubated at room temperature for 15 min.
For both random and targeted single-cell isolation using multimode imaging, three types of cells, T cells, leukemia cells, and HPCs, were mixed at equal concentrations and diluted to a final concentration of 4,000 to 8,000 cells/mL in medium A. Before mixing, approximately 106 T cells were filtered using a 35-μm cell strainer, stained at a final concentration of 2 μg/mL PE anti-mouse CD8a antibody (Lot. B243038, BioLegend) and washed with medium A, and approximately 106 HPCs were filtered using a 35-μm cell strainer, stained at a final concentration of 5 μg/mL Alexa647-B220 (Lot. B243963, BioLegend) and washed with medium A. Then, 3 mL of the mixed cell solution was added to a coated glass bottom dish (see above), and the cells were sedimented on the microscope stage at room temperature for 15 min.
For automated single-cell isolation of three-type–mixed cells after time-lapse imaging, the T cells, leukemia cells, and HPCs were filtered using 35-μm cell strainers, mixed at equal concentrations, and diluted to a final concentration of 2,000 to 4,000 cells/mL in medium B [1× RPMI 1640 medium without phenol red (Sigma) supplemented with final concentrations of 10% (v/v) fetal bovine serum (Corning), 1× monothioglycerol solution (FUJIFILM Wako Pure Chemical Industries), 1× GlutaMAX™ supplement (Gibco), 1× Penicillin/Streptomycin Mixed Solution (Nacalai Tesque Inc.), and 0.26% (w/v) methyl cellulose 400 (FUJIFILM Wako Pure Chemical Industries)], which was filtered using a 0.22-µm Steriflip filter unit. Then, 1 mL of diluted cells was placed in an Attofluor Cell Chamber (Thermo Fisher Scientific) equipped with a 25-mm-diameter coverslip (no. S1, Matsunami): the coverslip was sonicated in 1 M KOH (Nacalai Tesque Inc.) for 15 min at room temperature and rinsed with Milli-Q water 3 times. The cells were sedimented in the chamber for 2 h at 37 °C and 5% CO2 in the stage-top incubator on the microscope.
For automated single-cell isolation of PBMCs, the purified PBMCs (see section “Preparation of Cells, Organoids, and Tissue Samples”) were diluted to a final concentration of 2,000 to 4,000 cells/mL in medium C [1× RPMI 1640 medium without phenol red (Sigma) supplemented with final concentrations of 10% (v/v) fetal bovine serum (Corning), 1× monothioglycerol solution (FUJIFILM Wako Pure Chemical Industries), 1× GlutaMAX™ supplement (Gibco), 1× Penicillin/Streptomycin Mixed Solution (Nacalai Tesque Inc.), 5 mM HEPES (Gibco), 1× MEM nonessential AA (Gibco), 1 mM sodium pyruvate (Gibco), and 0.26% (w/v) methyl cellulose 400 (FUJIFILM Wako Pure Chemical Industries)], which was filtered using a 0.22-µm Steriflip filter unit. Then, 1 mL of diluted PBMCs was added to an Attofluor Cell Chamber as described above but sedimented for 3 to 3.5 h.
Automated single-cell isolation programs.
Programs using JOBS in NIS-Elements were developed for four automated types of experiments: isolation of T cells, three-type–mixed cells with multimode imaging and random selection, three-type–mixed cells with multimode imaging and targeted selection, and three-type–mixed cells and PBMCs with time-lapse imaging. The four programs are described in detail below in this order.
First, for T cell isolation, the initial settings were as follows: (initial-1) a circle area, the positions of 70 squares (819 μm × 691 μm each) that covered the whole circle, and the scanning order of these squares were determined using the “point generation” job with the parameters “radius = 4 mm and scan direction = meander,” and the positions and scanning order of those squares were used for the following steps. (initial-2) Two focal positions, 1 and 2, were determined; focal position 1 was determined where most cells in the field of view (FOV) at the center of the dish were on focus determined by the human eyes. Focal position 1 was used for other squares using the Nikon perfect focus system. Focal position 2 was determined to be 20 µm higher than focal position 1. (initial-3) A threshold for cell segmentation (described in the step-(auto-1) below) was determined. (initial-4) The needle (40 μm) and pump were washed and filled with Milli-Q water using a connected syringe. (initial-5) Three needle positions with the arm were set by TOPickV: needle position 1, where the needle was touched to the surface of the dish at the center of the FOV; needle position 2, which was 3 mm higher than needle position 1; and needle position 3, where the needle was touched to the center of the surface of the first well (well A1) of the 96-well plate. The needle was placed at needle position 2 when automated cell isolation was started. (initial-6) The acceleration and deceleration times of the arm were set (rotation: 500 ms and z axis: 10 ms) in TOPickV. (initial-7) The prepared 96-well plate was unsealed and placed on the motorized stage immediately before the next step.
After the initial setting, automated cell isolation was performed by following the program below: (auto-1) The microscope stage was moved to observe the first square determined above [the next square when this step was repeated, see step-(auto-9)], and an image of the full FOV (819 μm × 691 μm) at focal position 2 was captured using imaging mode 1: bright-field illumination (30% of the LED light source) with 10 ms exposure time and a 20× objective. Then, the cells in the acquired image were segmented using the “general analysis 3” job with a threshold of intensity and size by which all cells in at least five randomly selected squares imaged using imaging method 1 were segmented (this threshold was determined before running the program). The positions and automatically determined IDs of the segmented cells that were more than 70 µm from any other segmented cells were recorded. (auto-2) The microscope stage was moved to place the first recorded cell [the next recorded cell when this step was repeated, see step-(auto-8)] at the center of the FOV. (auto-3) An image of an area (139 μm × 99 μm) at the center of the FOV was captured using imaging mode 1 at focal position 2. Subsequently, the cell(s) in the acquired image were segmented using the “general analysis 1” job with the same threshold above. Then, if the number of cells was one, the following steps (auto-4 to auto-7) were performed using its position; if not, the following steps (auto-4 to auto-7) were skipped. (auto-4) The microscope stage was moved to locate the cell at the center of the FOV. (auto-5) An image of the area (139 μm × 99 μm) at the center of the FOV was captured using imaging mode 1 at focal position 1. (auto-6) A signal was sent from NIS-Elements to the picker (TOPickV) through TTL1 (see section “Hardware and Software of the ALPS”). (auto-7) The following procedures were performed by the picker: (auto-7-1) the needle was moved from needle position 2 to needle position 1; (auto-7-2) 9 nL of solution containing the cell at the center of the FOV was aspirated from the dish by the needle-connected pump; (auto-7-3) for the first round of 96-cell isolation, well A1 of the plate was moved to needle position 3, and for the 2nd to 96th round, the next well in the following order, A1-A12-B12-B1-C1-C12-D12-D1-E1-E12-F12-F1-G1-G12-H12-H1, was moved to needle position 3; (auto-7-4) the needle was moved from needle position 1 to needle position 3; (auto-7-5) 15 nL of the solution with the cell was deposited from the needle into the prepared buffer in one well of the 96-well plate (see section “Preparation of Plates for Cell Deposition”); (auto-7-6) the needle was moved from needle position 3 to needle position 2; (auto-7-7) a signal was sent from the picker to NIS-Elements through TTL2. (auto-8) Steps (auto-2 to 7) were repeated for the next segmented cells until all segmented cells in step-(auto-1) were picked or skipped. (auto-9) Steps (auto-1 to auto-8) were repeated for the next squares until 96 cells were picked or all the determined squares were used.
We note that steps (auto-2), (auto-4), (auto-7-1), (auto-7-2), and (auto-7-4) were imaged in real time by the microscope using imaging mode 1 with the full FOV at focal position 1, and steps (auto-7-3) and (auto-7-5) were imaged in real time by the camera at the top of a 96-well plate. These real-time images by two cameras were shown on the same screen and were recorded by screen capture software (see the section “Hardware and Software of the ALPS”).
Second, for three-type–mixed cells with multimode imaging and random selection, a program that was the same as that for T cell isolation was performed except for the following modifications: in step (initial-6), the acceleration and deceleration time of the arm were 1,000 ms for rotation and 500 ms for the z axis; in step (auto-5), additionally, three fluorescent images of the same area at the same focus position as the bright-field imaging were captured using imaging mode 2 [GFP (10% of the CoolLED), PE (100%), and Alexa647 (100%) fluorescent illuminations, respectively, with 30 ms exposure time and 20 × objective].
Third, for three-type–mixed cells with multimode imaging and targeted selection, a program that was the same as that for multimode imaging and random selection was performed except for the following modifications: in step-(auto-1), additionally, three fluorescent images of the same area at the same focus position as the bright-field imaging were captured using imaging mode 2. After the cells were segmented in this step, for the first 36 rounds of the 96-cell isolations, GFP-positive cells [mean gray value (0-4095) of the pixels of a cell in the acquired GFP fluorescent image was higher than 950] were selected as targeted cells for the next steps; for the 37 to 72 rounds, PE-positive cells [mean gray value (0-4095) of the pixels of a cell in the acquired PE fluorescent image was higher than 650] were selected as targeted cells for the next steps; for the last 24 rounds, Alexa647-positive cells [mean gray value (0-4095) of the pixels of a cell in the acquired Alexa647 fluorescent image was higher than 350] were selected as targeted cells for the next steps.
Fourth, for three-type–mixed cells and PBMCs with time-lapse imaging, a program that was the same as that for T cell isolation was performed except for the following modifications: in step-(initial-1), the conditions were replaced by the following: the positions and scanning order of 5 × 5 squares (819 μm × 691 μm each) were determined using the “point generation” job with the parameters “radius = 5 mm, scan direction = meander, and area restriction = rectangle.” Immediately before step-(initial-7), time-lapse images of 6 × 6 squares (defined by the “point generation” job with the parameters radius = 5 mm, scan direction = meander, area restriction = rectangle, and overlap = 15%) that covered the 5 × 5 squares in a total of 29 min (1-min interval and 30 frames) were captured using imaging mode 1 at focal positions 1 and 2 (during the acquisition, the needle was set in a dish with Milli-Q water). Between step-(initial-7) and step-(auto-1), if the needle position was adjusted to needle position 1, the needle was washed using Milli-Q water in the connected syringe, and one scanning image of the 6 × 6 squares was captured by the method used for time-lapse imaging. After step-(auto-8), for the 12th, 24th, 36th, 48th, 60th, 72nd, 84th, and 96th rounds of 96-cell isolation, one scanning image of the 6 × 6 squares was captured by the method used for time-lapse imaging (during this acquisition, the needle was placed at needle position 2). We note that the time gap between the time-lapse imaging and the step-(auto-1) was less than 5 min. For temperature control of all steps, the customized stage-top incubator and the cage with the heater were used (see section “Hardware and Software of the ALPS”). In addition, for the last 8 plates of the three-type–mixed cells and the last 3 plates of the PBMCs, a 60-μm-diameter needle was used. When the 60-μm needle was used, the parameter “70 µm” in step-(auto-1) was changed to “100 µm,” the aspirating and depositing volumes in step-(auto-7) were changed to 11 and 19 nL, respectively, and needle position 2 was 10 mm higher than needle position 1.
Storage of isolated cells.
In experiments on three-type–mixed cells with multimode imaging and random selection, three-type–mixed cells with multimode imaging and targeted selection, and three-type–mixed cells and PBMCs with time-lapse imaging, after 96 cells were isolated, the plate was sealed, quickly frozen in liquid nitrogen within one minute, and stored at −80 °C until RNA-seq was subsequently performed.
Single-Cell RNA-seq.
For single cells isolated by both the ALPS and the cell sorter (SI Appendix, Text 8), library preparation was performed using the Bravo NGS workstation, and the libraries were sequenced on an Illumina platform (SI Appendix, Text 4). After the sequencing results were processed, the expression levels of genes and the type of each cell were determined (SI Appendix, Texts 5 and 6).
Machine Learning.
Cell types were classified using a deep residual network (ResNet) (13) or a LeNet (15) with three convolutional layers as a convolutional neural network (CNN) (34) that extracts image-based features of the cells, combined with a long short-term memory (LSTM) architecture (14) as a recurrent neural network (RNN) that models the temporal information of the cells, for both three-type–mixed cells and PBMCs. Regression of each of the top 300 variant genes using the ResNet architecture and the LSTM architecture was performed for the three-type–mixed cells. For both classification and regression, the leave-one-out cross-validation strategy (16), where cells in one plate were used for testing and cells in all other plates used for training, was applied. Details are described in SI Appendix, Text 9.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (CSV)
Dataset S05 (XLSX)
Dataset S06 (CSV
Dataset S07 (XLSX)
Dataset S08 (XLSX)
Dataset S09 (XLSX)
Dataset S10 (XLSX)
Dataset S11 (XLSX)
Dataset S12 (XLSX)
Dataset S13 (XLSX)
A demonstration movie of automated single cell isolation using ALPS from top view. Left side, the Nikon microscope; right side, the motorized stage for the displacement of a 96-well plate (Materials and Methods). Weak green light of the microscope: bright field illumination; blue light: GFP fluorescent illumination; strong green light of the microscope: PE fluorescent illumination; red light: Alx648 fluorescent illumination; green light on the stage, bright field illumination by LED. The movie was recorded from the step-(auto-2) to the step-(auto-7) with two rounds of isolation using the program used for multi-mode imaging and random selection (Materials and Methods) (acceleration and deceleration time of the arm was from the time-lapse experiment).
A movie of the picking and depositing by ALPS. Left side, live imaging under the microscope; middle, current state (e.g., preview of captured image, current stage position) of the running program in NIS-Elements; right side, live imaging of a well of the 96-well plate. The movie was a clip from the step-(auto-2) of the 7th to the step-(auto-7) of the 8th of the T cell isolation (T-cellplate2, Dataset S1). Red square, 139 μm × 99 μm.
A movie of picking and depositing a 1 μm bead using ALPS. Left side, live imaging under the microscope; right side, live imaging of a well of the 96-well plate. Needle size, 40 μm. The deposited bead was around “2 o’clock”.
A movie of picking and depositing a 3 μm bead using ALPS. Left side, live imaging under the microscope; right side, live imaging of a well of the 96-well plate. Needle size, 40 μm.
A movie of picking and depositing a 45 μm bead using ALPS. Left side, live imaging under the microscope; right side, live imaging of a well of the 96-well plate. Needle size, 75 μm.
A movie of picking and depositing a 350 μm bead using ALPS. Left side, live imaging under the microscope; right side, live imaging of a well of the 96-well plate. Needle size, ~400 μm.
A movie of picking and depositing an intestinal organoid using ALPS. Left side, live imaging under the microscope; right side, live imaging of a well of the 96-well plate. Needle size, ~400 μm.
A movie of picking and depositing a crypt using ALPS. Left side, live imaging under the microscope; right side, live imaging of a well of the 96-well plate. Needle size, 40 μm.
Acknowledgments
We thank Ms. Kaori Fukuhara for technical assistance in cell preparation, sequencing, and image analysis; Dr. Reiko Yamamoto for technical assistance in image analysis; Ms. Wakako Oda for encouragement and laboratory management; Mr. Mamoru Hirafuji for building the cell isolation system; Dr. Yichao Xu for helpful discussion on deep learning; the technical support staffs at the Laboratory for Phyloinformatics (RIKEN BDR) for a HiSeq run; and the DECODE project members for general discussion and support. This work was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI grant 18H05411, the Mochida Memorial Foundation for Medical and Pharmaceutical Research, The Sumitomo Foundation (to K. Shiroguchi), and the Special Postdoctoral Researchers Program of RIKEN (T. Ogawa and N.H.).
Author contributions
J.J., T.Ogawa, and K.Shiroguchi designed research; J.J., T.Ogawa, and N.H. performed research; J.J., T.Ogawa, K.K., K.Shimizu, T. Ikawa, T. Imanishi, T.Okazaki, and K.Shiroguchi contributed new reagents/analytic tools; J.J. and T.Ogawa analyzed data; K.Shiroguchi supervised the entire study; and J.J. and K.Shiroguchi wrote the paper.
Competing interest
The authors have patent filings to disclose, RIKEN has co-filed a patent application based on this work.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All sequencing data are available from the NCBI Sequence Read Archive under accession GSE179943. The cell images of 3mix-ALPS-timelapse cells and PBMCs are available via SSBD:repository (35) (https://doi.org/10.24631/ssbd.repos.2022.06.237).
Supporting Information
References
- 1.Tammela T., Sage J., Investigating tumor heterogeneity in mouse models. Annu. Rev. Cancer Biol. 4, 99–119 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tanay A., Regev A., Scaling single-cell genomics from phenomenology to mechanism. Nature 541, 331–338 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones W., Alasoo K., Fishman D., Parts L., Computational biology: Deep learning. Emerg. Top. Life Sci. 1, 257–274 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang B., Pan Z., Yin K., Khateeb A., Recent advances of deep learning in bioinformatics and computational biology. Front. Genet. 10, 1–10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Angermueller C., Pärnamaa T., Parts L., Stegle O., Deep learning for computational biology. Mol. Syst. Biol. 12, 878 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moen E., et al. , Deep learning for cellular image analysis. Nat. Methods 16, 1233–1246 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang W., et al. , Live-cell imaging and analysis reveal cell phenotypic transition dynamics inherently missing in snapshot data. Sci. Adv. 6, eaba9319 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Z., et al. , Integrating single-cell RNA-seq and imaging with SCOPE-seq2. Sci. Rep. 10, 1–15 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan J., Sheng J., Sims P. A., SCOPE-Seq: A scalable technology for linking live cell imaging and single-cell RNA sequencing. Genome Biol. 19, 227 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lane K., et al. , Measuring signaling and RNA-Seq in the same cell links gene expression to dynamic patterns of NF-κB activation. Cell Syst. 4, 458–469.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saint M., et al. , Single-cell imaging and RNA sequencing reveal patterns of gene expression heterogeneity during fission yeast growth and adaptation. Nat. Microbiol. 4, 480–491 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Kleiveland C. R., “Peripheral blood mononuclear cells BT” in The Impact of Food Bioactives on Health: In Vitro and Ex Vivo Models, Verhoeckx K., et al., Eds. (Springer International Publishing, 2015), pp. 161–167. [PubMed] [Google Scholar]
- 13.He K., Zhang X., Ren S., Sun J., “Deep residual learning for image recognition” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (IEEE, Las Vegas, NV, USA, 2016), pp. 770–778. [Google Scholar]
- 14.Hochreiter S., Schmidhuber J., Long short-term memory. Neural Comput. 9, 1735–1780 (1997). [DOI] [PubMed] [Google Scholar]
- 15.LeCun Y., Bottou L., Bengio Y., Haffner P., Gradient-based learning applied to document recognition. Proc. IEEE 86, 2278–2324 (1998). [Google Scholar]
- 16.Webb G. I., et al. , “Leave-one-out cross-validation” in Encyclopedia of Machine Learning (Springer, US, 2011), pp. 600–601. [Google Scholar]
- 17.Stirling D. R., et al. , Cell Profiler 4: Improvements in speed, utility and usability. BMC Bioinf. 22, 1–11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kitahara Y., et al. , A real-time monitoring system for automatic morphology analysis of yeast cultivation in a jar fermenter. Appl. Microbiol. Biotechnol. 106, 4683–4693 (2022). [DOI] [PubMed] [Google Scholar]
- 19.Ershov D., et al. , TrackMate 7: Integrating state-of-the-art segmentation algorithms into tracking pipelines. Nat. Methods 19, 829–832 (2022). [DOI] [PubMed] [Google Scholar]
- 20.Schindelin J., et al. , Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Y., et al. , Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pluta M., “Nomarski’s DIC microscopy: A review” in Phase Contrast and Differential Interference Contrast Imaging Techniques and Applications, Pluta M., Szyjer M., Eds. (SPIE, 1994), pp. 10–25. [Google Scholar]
- 23.Axelrod D., Cell-substrate contacts illuminated by total internal reflection fluorescence. J. Cell Biol. 89, 141–145 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pawley J. B., Ed., Handbook of Biological Confocal Microscopy (Springer US, 1995), 10.1007/978-1-4757-5348-6. [DOI] [Google Scholar]
- 25.Stockert J. C., Blazquez-Castro A., “Super-resolution microscopy” in Fluorescence Microscopy in Life Sciences (Bentham Science Publishers, 2017), pp. 687–714. [Google Scholar]
- 26.Huang L., Ma F., Chapman A., Lu S., Xie X. S., Single-cell whole-genome amplification and sequencing: Methodology and applications. Annu. Rev. Genomics Hum. Genet. 16, 79–102 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Gawad C., Koh W., Quake S. R., Single-cell genome sequencing: Current state of the science. Nat. Rev. Genet. 17, 175–188 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Wen L., Tang F., Single cell epigenome sequencing technologies. Mol. Aspects Med. 59, 62–69 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Pham T., Tyagi A., Wang Y., Guo J., Single-cell proteomic analysis. WIREs Mech. Dis. 13, e1503 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mund A., et al. , Deep visual proteomics defines single-cell identity and heterogeneity. Nat. Biotechnol. 40, 1231–1240 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ikawa T., Kawamoto H., Wright L. Y. T., Murre C., Long-term cultured E2A-deficient hematopoietic progenitor cells are pluripotent. Immunity 20, 349–360 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Sugimoto S., Sato T., Establishment of 3D intestinal organoid cultures from intestinal stem cells. Methods Mol. Biol. 1612, 97–105 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Li Y., et al. , A growth factor-free culture system underscores the coordination between Wnt and BMP signaling in Lgr5+ intestinal stem cell maintenance. Cell Discov. 4, 49 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buggenthin F., et al. , Prospective identification of hematopoietic lineage choice by deep learning. Nat. Methods 14, 403–406 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tohsato Y., Ho K. H. L., Kyoda K., Onami S., SSBD: A database of quantitative data of spatiotemporal dynamics of biological phenomena. Bioinformatics 32, btw417 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (CSV)
Dataset S05 (XLSX)
Dataset S06 (CSV
Dataset S07 (XLSX)
Dataset S08 (XLSX)
Dataset S09 (XLSX)
Dataset S10 (XLSX)
Dataset S11 (XLSX)
Dataset S12 (XLSX)
Dataset S13 (XLSX)
A demonstration movie of automated single cell isolation using ALPS from top view. Left side, the Nikon microscope; right side, the motorized stage for the displacement of a 96-well plate (Materials and Methods). Weak green light of the microscope: bright field illumination; blue light: GFP fluorescent illumination; strong green light of the microscope: PE fluorescent illumination; red light: Alx648 fluorescent illumination; green light on the stage, bright field illumination by LED. The movie was recorded from the step-(auto-2) to the step-(auto-7) with two rounds of isolation using the program used for multi-mode imaging and random selection (Materials and Methods) (acceleration and deceleration time of the arm was from the time-lapse experiment).
A movie of the picking and depositing by ALPS. Left side, live imaging under the microscope; middle, current state (e.g., preview of captured image, current stage position) of the running program in NIS-Elements; right side, live imaging of a well of the 96-well plate. The movie was a clip from the step-(auto-2) of the 7th to the step-(auto-7) of the 8th of the T cell isolation (T-cellplate2, Dataset S1). Red square, 139 μm × 99 μm.
A movie of picking and depositing a 1 μm bead using ALPS. Left side, live imaging under the microscope; right side, live imaging of a well of the 96-well plate. Needle size, 40 μm. The deposited bead was around “2 o’clock”.
A movie of picking and depositing a 3 μm bead using ALPS. Left side, live imaging under the microscope; right side, live imaging of a well of the 96-well plate. Needle size, 40 μm.
A movie of picking and depositing a 45 μm bead using ALPS. Left side, live imaging under the microscope; right side, live imaging of a well of the 96-well plate. Needle size, 75 μm.
A movie of picking and depositing a 350 μm bead using ALPS. Left side, live imaging under the microscope; right side, live imaging of a well of the 96-well plate. Needle size, ~400 μm.
A movie of picking and depositing an intestinal organoid using ALPS. Left side, live imaging under the microscope; right side, live imaging of a well of the 96-well plate. Needle size, ~400 μm.
A movie of picking and depositing a crypt using ALPS. Left side, live imaging under the microscope; right side, live imaging of a well of the 96-well plate. Needle size, 40 μm.
Data Availability Statement
All sequencing data are available from the NCBI Sequence Read Archive under accession GSE179943. The cell images of 3mix-ALPS-timelapse cells and PBMCs are available via SSBD:repository (35) (https://doi.org/10.24631/ssbd.repos.2022.06.237).