Abstract
Comparative analyses of genomic data reveal further insights into the phylogeny and taxonomic classification of butterflies presented here. As a result, 2 new subgenera and 2 new species of Hesperiidae are described: Borna Grishin, subgen. n. (type species Godmania borincona Watson, 1937) and Lilla Grishin, subgen. n. (type species Choranthus lilliae Bell, 1931) of Choranthus Scudder, 1872, Cecropterus (Murgaria) markwalkeri Grishin, sp. n. (type locality in Mexico: Sonora), and Hedone yunga Grishin, sp. n. (type locality in Bolivia: Yungas, La Paz). The lectotype is designated for Aethilla toxeus Plötz, 1882. The type locality of Dion uza (Hewitson, 1877) is likely in southern Brazil. A number of taxonomic changes are proposed. The following taxa are subgenera, not genera: Plebulina Nabokov, 1945 of Icaricia Nabokov, 1945; Sinia Forster, 1940 of Glaucopsyche Scudder, 1872; Pseudophilotes Beuret, 1958 of Palaeophilotes Forster, 1938; and Agraulis Boisduval & Le Conte, [1835] of Dione Hübner, [1819]. Asbolis Mabille, 1904 is a subgenus of Choranthus Scudder, 1872 rather than its synonym. The following are species, not subspecies or synonyms: Glaucopsyche algirica (Heyne, 1895) (not Glaucopsyche melanops (Boisduval, 1829)), Chlosyne flavula (W. Barnes & McDunnough, 1918) (not Chlosyne palla (Boisduval, 1852)), Cercyonis hypoleuca Hawks & J. Emmel, 1998 (not Cercyonis sthenele (Boisduval, 1852)), Cecropterus coyote (Skinner, 1892) and Cecropterus nigrociliata (Mabille & Boullet, 1912) (not Aethilla toxeus Plötz, 1882), Aguna malia Evans, 1952 (not Aguna megaeles (Mabille, 1888)), Polygonus arizonensis (Skinner, 1911), Polygonus histrio Röber, 1925, Polygonus pallida Röber, 1925, and Polygonus hagar Evans, 1952 (not Polygonus leo (Gmelin, [1790])), Viola kuma (Bell, 1942), comb. nov. (not Pachyneuria helena (Hayward, 1939)), Tamela maura (Snellen, 1886) (not Tamela othonias (Hewitson, 1878)), Tamela diocles (Moore, [1866]) (not Tamela nigrita (Latreille, [1824])), Vinius phellus (Mabille, 1883) (not Vinius exilis (Plötz, 1883)), Vinius sophistes (Dyar, 1918) (not Vinius tryhana (Kaye, 1914)), and Rhinthon andricus (Mabille, 1895) and Rhinthon aqua (Evans, 1955) (not Rhinthon braesia (Hewitson, 1867)). The following are new and revised species-subspecies combinations: Cercyonis sthenele damei W. Barnes & Benjamin, 1926 (not Cercyonis meadii (W. H. Edwards, 1872)) and Chlosyne flavula blackmorei Pelham, 2008 and Chlosyne flavula calydon (W. Holland, 1931) (not Chlosyne palla). The following are valid subspecies resurrected from synonymy in new and reinstated species-subspecies combinations: Chlosyne palla pola (Boisduval, 1869) (not Chlosyne gabbii gabbii (Behr, 1863)) and Cercyonis meadii mexicana R. Chermock, 1949 (not Cercyonis sthenele damei W. Barnes & Benjamin, 1926, comb. rev.). The following are new junior subjective synonyms: Aethilla toxeus Plötz, 1882 of Cecropterus albociliatus (Mabille, 1877) and Viola dagamba Steinhauser, 1989 of Viola kuma (Bell, 1942), comb. nov., stat. rest. Leucochitonea janice Ehrmann, 1907 is a junior subjective synonym of Heliopetes alana (Reakirt, 1868) and not of Heliopetes petrus (Hübner, [1819]). The holotype of Hermeuptychia sinuosa Grishin, 2021 is illustrated after being spread.
Keywords: taxonomy, classification, genomics, phylogeny, biodiversity
This report is a direct continuation of our previous publications (Zhang et al. 2019, 2020, 2021, 2022a, 2022b, 2022c), and the general philosophy employed is best summarized in the introduction to Zhang et al. (2023). Details of experimental and computational protocols are provided in the Appendix to Li et al. (2019). We sequence specimens of any age, with collection year specified in illustrated trees, “old” means that the specimen comes from old collections, and no date is given on its labels, probably collected more than 100 years ago, around the turn of the 20th century (most specimens) or before. Phylogenetic tree construction was carried out as described in the Materials and Methods section by Zhang et al. (2022a).
For each set of specimens, we illustrate at least two trees: constructed from nuclear genomic regions (either Z chromosome or autosomes) and from the mitogenome DNA. While the two trees frequently agree, we see instances of confident (i.e., with strong statistical support and therefore unlikely caused by insufficient or internally inconsistent data) incongruence between nuclear and mitochondrial genomic trees. Comparing the two trees highlights the pitfalls of relying exclusively on mitochondrial DNA (and COI barcodes alone) in classification decisions. Even more, including a larger fraction of mitochondrial genes in a gene marker set used in the PCR amplification approach may bias the tree toward a mitogenomic signal and thus deviate from the nuclear DNA evolution scenario. In some cases, we show all three trees: autosomes, Z chromosome, and mitogenome. Their comparison may be most instructive for the analysis of evolutionary peculiarities.
It is important to note that we do not use specific gene markers. We sequence the whole genomic shotgun and assemble all protein-coding genes present in the sample by mapping them to a complete set of genes from a reference genome. This representation gives an unbiased view of the genomic content of an organism. For computing efficiency, the trees are constructed from a random sample of 100,000 codons (3 bp each) from the entire gene set, which is approximately 6 million codons. Statistical support is computed on 100 random samples of 10,000 codons each from the original pool of genes. All protein-coding genes are used in mitochondrial DNA trees, and their statistical support is computed using ultrafast bootstrap (Hoang et al. 2018).
In addition to phenotypic diagnoses of new taxa, we provide diagnostic DNA characters, both in the nuclear genome and COI barcode. DNA characters are found in nuclear protein-coding regions using our previously developed procedure (see SI Appendix to Li et al. 2019). The logic behind the character selection was detailed in Cong et al. (2019b). The character states are provided in species diagnoses as abbreviations. E.g., aly728.44.1:G672C means position 672 in exon 1 of gene 44 from scaffold 728 of the Cecropterus lyciades (Geyer, 1832) (formerly in Achalarus Scudder, 1872, thus aly) reference genome (Shen et al. 2017) is C, changed from G in the ancestor. When characters are given for the sister clade of the diagnosed taxon, the following notation is used: aly5294.20.2:A548A (not C), which means that position 548 in exon 2 of gene 20 on scaffold 5294 is occupied by the ancestral base pair A, which was changed to C in the sister clade (so it is not C in the diagnosed taxon). The same notation is used for COI barcode characters, but without a prefix ending with ‘:’. The sequences of exons from the reference genome with the positions used as character states highlighted in green are in the supplemental file deposited at <https://osf.io/nxd5y//>. Linking to these DNA sequences from this publication ensures that the numbers given in the diagnoses can be readily associated with actual sequences. Whole genome shotgun datasets we obtained and used in this work are available from the NCBI database <https://www.ncbi.nlm.nih.gov/> as BioProject PRJNA927657, and BioSample entries of the project contain the locality and other collection data of the sequenced specimens shown in the trees. COI barcode sequences have been deposited in GenBank with accessions OQ311404–OQ311413. Several photographs shown in this work are taken from iNaturalist (2022). Links to observations by observation number reported in figure legends are < https://www.inaturalist.org/observations/xxx >, where xxx is the number.
Family Lycaenidae [Leach], [1815]
Plebulina Nabokov, 1945 is a subgenus of Icaricia Nabokov, 1945
Genomic sequencing and comparison of the crown group of Polyommatina Swainson, 1827 with a focus on species found in North America, confirm that the monotypic genus Plebulina Nabokov, 1945 (type species Lycaena emigdionis F. Grinnell, 1905) is closely related to Icaricia Nabokov, 1945 (type species Lycaena icarioides Boisduval, 1852) (Fig. 1a–c). In their pioneering work, Talavera et al. (2012) proposed to treat Plebulina as a distinct genus because in their time-calibrated tree constructed from several gene markers, Plebulina was at a larger distance from Icaricia than their (somewhat flexible) cutoff for congeneric relationship.
Fig. 1.

Plebulina as a subgenus of Icaricia. a–c. Phylogenetic trees constructed from protein-coding regions in autosomes (a), Z chromosome (b), and mitochondrial genome (c): Icaricia (red, with Icaricia (Plebulina) emigdionis comb. nov. name shown in magenta), Agriades (blue), Rueckbeilia (green), and Plebejus (purple). Magenta dots mark diversification nodes of genera Icaricia, Agriades, and Plebejus. For each specimen, the name adopted in this work is given first, and a previously used name is listed in square brackets (if different), supplemented with the DNA sample number, type status (HT holotype, LT lectotype, NT neotype, ST syntype, PT paratype, and PLT paralectotype), general locality, and year. NCBI database entries in BioProject PRJNA927657 give additional data about these specimens. Synonyms are given in parentheses preceded by “=”. The type status refers to this synonym if the synonym name is provided. The same notations are used throughout this work in figures showing phylogenetic trees. d–e. Live females of Icaricia from USA: California, iNaturalist observations (data “obscured”): d. I. (Plebulina) emigdionis 26900315 Kern Co., May-2007 © Nature Ali; e. I. (Icaricia) neurona 53685349 Ventura Co., Jul-2020 © Chris. Images are rotated and cropped. CC BY-NC 4.0 https://creativecommons.org/licenses/by-nc/4.0/. f. Rueckbeilia fergana ♀ NVG-22037H11 “Tura”, B. Neumögen collection [USNM], dorsal (left) and ventral (right) views.
In our genome-level phylogeny that includes all known species of Icaricia, the clade consisting of Icaricia and Plebulina taken together is well separated from all others by a prominent branch (Fig. 1 red) in both nuclear genome trees (autosomes Fig. 1a and Z chromosome Fig. 1b). However, Icaricia is not separated from Plebulina by a prominent branch, and the branch from the last common ancestor of Icaricia + Plebulina to the last common ancestor of Icaricia is shorter compared to the previous branch. Furthermore, in the nuclear genome trees (Fig. 1a, b), the distance from the root to the last common ancestor of Icaricia + Plebulina is larger than that to the last common ancestors of genera Agriades Hübner, [1819] (type species Papilio glandon de Prunner, 1798) (Fig. 1 blue) and Plebejus Kluk, 1780 (type species Papilio argus Linnaeus, 1758) (Fig. 1 purple) (nodes marked with magenta dots in Fig 1) suggesting that the divergence between Icaricia and Plebulina might have occurred more recently than that within Agriades and Plebejus. Finally, the average distances from the leaves to the last common ancestors of Icaricia + Plebulina, Agriades, and Plebejus are approximately the same, implying that genetic differentiation in the nuclear genome is similar in the genera Agriades, Plebejus, and a group consisting of both Icaricia and Plebulina. Therefore, due to these genetic similarities and the phylogenetic tree structure, we propose to treat Plebulina Nabokov, 1945, stat. nov. as a subgenus of Icaricia Nabokov, 1945. Plebulina and Icaricia were proposed as genera in the same work issued on the same date. Being the first reviser, here we give precedence to Icaricia because more species are currently included in Icaricia than monotypic Plebulina, resulting in fewer name changes.
Despite a number of morphological differences, including a unique caterpillar foodplant of Plebulina (Nabokov 1945; Talavera et al. 2012; Ballmer 2022), phenotypic similarities between Plebulina and Icaricia are also notable. For instance, females of both species may have orange streaks along veins on the dorsal forewing wing surface (Fig. 1d, e). Furthermore, we hold an opinion that monotypic genera (such as Plebulina) should be reserved for species without apparent close relatives. In the presence of such relatives, it seems more instructive to indicate this relationship with the generic name. In contrast to Plebulina, genomic analysis supports the distinctness of (nearly monotypic) Rueckbeilia Lukhtanov, Talavera, Pierce & Vila, 2013 (type species Lycaena loewii var.? fergana Staudinger, 1881, Fig. 1f) (Fig. 1 green), because, while the statistical support for its placement in the same clade with Agriades and Icaricia + Plebulina is strong in the Z chromosome tree (1, i.e., 100%, Fig. 1b), it is not strongly associated with either of the two genera (less than 0.5). Therefore, if Icaricia and Agriades are treated as distinct genera, Rueckbeilia would also be distinct.
Finally, we offer a hypothesis about why Plebulina was more distinct from Icaricia in the phylogeny obtained by Talavera et al. (2012) than in our nuclear genome trees. In the mitochondrial genome tree (Fig. 1c), we observe this more distant relationship: the magenta dot for the red clade (Icaricia + Plebulina) is closer to the root (left) and farther from the leaves (which are at about the same level) than the magenta dots for the blue (Agriades) and purple (Plebejus) clades. Because a significant number of base pairs in the gene markers used by Talavera et al. (2012) came from mitochondrial genes, we suspect that these genes might have biased the branch lengths around Plebulina in their tree. The evolution of mitochondrial genomes experiences irregularities such as introgression and may not represent the organism as well as its nuclear genome. Therefore, we default to nuclear genome results (with an emphasis on the Z chromosome) for taxonomic classification.
Sinia Forster, 1940 is a subgenus of Glaucopsyche Scudder, 1872
Sinia Forster, 1940 (type species Glaucopsyche (Sinia) leechi Forster, 1940) was originally proposed as a subgenus of Glaucopsyche Scudder, 1872 (type species Polyommatus lygdamus E. Doubleday, 1841) that also included Lycaena lanty Oberthür, 1886 (type locality China: Sichuan Prov., Kangding) and Lycaena divina Fixsen, 1887 (type locality in North Korea). Recently, Sinia has been treated as a valid genus (Lukhtanov and Gagarina 2022). However, Lycaena divina has been segregated into a separate genus, Shijimiaeoides Beuret, 1958 (type species Lycaena barine Leech, [1893], which is a synonym or subspecies of L. divina). Based on the genomic comparison, we placed Shijimiaeoides as a junior subjective synonym of Glaucopsyche (Zhang et al. 2022b). Here, we analyze genomic data on Sinia (Fig. 2 red) and find that it originates within Glaucopsyche (Fig. 2 blue), being sister to all others except the subgenus Phaedrotes Scudder, 1876 (type species Lycaena catalina Reakirt, 1866, which is a junior subjective synonym of Lycaena piasus Boisduval, 1852) in all three trees (Fig, 2a–c). Therefore, Sinia is a subgenus unless Phaedrotes is treated as a genus. Hence, we propose to regard Sinia Forster, 1940 a subgenus of Glaucopsyche Scudder, 1872 as originally described. We also note wing pattern similarities between the type species of Sinia, G. leechi comb. rest. from China (Fig. 2d) and North American G. piasus (Fig. 2e): e.g., in the placement of white “arrowheads” on the ventral hindwing.
Fig. 2.

Trees of Scolitantidina species constructed from protein-coding regions in a. autosomes, b. Z chromosome, and c. mitochondrial genome: Glaucopsyche (blue) with subgenera Apelles (cyan) and Sinia (red), Palaeophilotes (green, nominotypical subgenus in magenta), and Euphilotes (purple) with its subgenus Philotiella (orange). Yellow highlights a case of strongly supported incongruence between nuclear and mitochondrial genome trees. d. Glaucopsyche (Sinia) leechi comb. rest. ♀ paratype ventral view, NVG-22027G01 China: Sichuan Prov., Songpan Co. [ZSMC]. e. Glaucopsyche (Phaedrotes) piasus iNaturalist observation 125272854 USA: California, Modoc Co., Modoc, 25-Jun-2022 © Paul G. Johnson. The image is rotated and cropped. CC BY-NC 4.0 https://creativecommons.org/licenses/by-nc/4.0/.
Finally, we note incongruence between the three trees (Fig. 2a–c). While Glaucopsyche is most confidently monophyletic in both nuclear genome trees (support 1, i.e., 100%, Fig. 2a, b), subgenus Phaedrotes is not in the same clade with the rest of Glaucopsyche in the mitogenome tree (Fig. 2c), cautioning about the reliance on mitochondrial DNA for organism-level phylogenies or a set of gene markers that may be dominated by mitochondrial genes. More weakly supported incongruence is in the relative position of Apelles Hemming, 1931 (type species Polyommatus melanops Boisduval, [1828]), treated as a valid subgenus by Lukhtanov and Gagarina (2022) and G. divina. Regardless of these incongruences that would be interesting to understand further, the position of Sinia is the same in all three trees. Therefore, our conclusion about its placement within Glaucopsyche is strongly supported.
Glaucopsyche algirica (Heyne, 1895), stat. nov. is a species distinct from Glaucopsyche melanops (Boisduval, 1829)
Genomic sequencing of Glaucopsyche melanops (Boisduval, 1829) (type locality in France) and Lycaena melanops var. algirica Heyne, 1895 (type locality in Algeria), currently a valid subspecies of G. melanops, reveals genetic differentiation between them more in line with that known for distinct species among its relatives (Fig. 2 cyan): e.g., their COI barcodes differ by 1.8% (12 bp), and the genetic distance between them in the mitogenome is similar to that between Glaucopsyche paphos Chapman, 1920 (type locality in Cyprus) and Glaucopsyche alexis (Poda, 1761) (type locality in Austria), or between Glaucopsyche lygdamus (Doubleday, 1841) (type locality in USA) and Glaucopsyche lycormas (Butler, 1866) (type locality in Japan) (Fig. 2c). Therefore, we propose that Glaucopsyche algirica (Heyne, 1895), stat. nov. is a species-level taxon.
Pseudophilotes Beuret, 1958 is a subgenus of Palaeophilotes Forster, 1938
Genomic sequencing of Palaeophilotes Forster, 1938 (type species Lycaena triphysina Staudinger, 1892, Fig. 3a) (Fig. 2 magenta) treated as a distinct genus by Lukhtanov and Gagarina (2022) due to the lack of its DNA sequences and pronounced phenotypic differences from other genera reveals that it is sister to the nominotypical subgenus of Pseudophilotes Beuret, 1958 (type species Papilio baton Bergsträsser, 1779, Fig. 3b) and is much closer to it genetically than the two subgenera of Pseudophilotes—nominotypical and Rubrapterus Korshunov, 1987 (type species Lycaena bavius Eversmann, 1832)—are to each other (Fig. 2). This was a surprise given the differences in the appearance of these species (Fig. 3a, b).
Fig. 3.

Ultrafast phenotypic evolution in Palaeophilotes. Dorsal view of males, all in ZSMC. a. P. (Palaeophilotes) triphysina NVG-22028C01 Central Asia, old, coll. Erhardt. b. P. (Pseudophilotes) baton NVG-22028A12 Germany: Bavaria, vic. Jura, 17-May-1975, W. Schätz leg. c. P. (Pseudophilotes) sinaicus paratype NVG-22028A10 Egypt: Sinai, Wadi Jibal, 1900 m, 26-May-1974, I. Nakamura leg. COI barcodes differ between (a) and (b) by 2.1% and between (b) and (c) by 0% (probably not a result of introgression, but the actual lack of genetic differentiation).
We observe that the tree branches in nuclear genome trees (Fig. 2a, b) leading to Palaeophilotes and the subgenus Pseudophilotes are longer (=reach farther to the right) than for all other Scolitantidina Tutt, 1907 we sequenced, indicating accelerated evolution. This elevated evolutionary rate in the group is likely the cause of the observed phenotypic disparity. Notably, the mitochondrial genome tree does not show elevated rates in this group: branches reach about the same level on the right (Fig. 2c). In accord with this mitogenome conservation, COI barcodes of Pseudophilotes (Pseudophilotes) baton (Fig. 3b) and Pseudophilotes (Pseudophilotes) sinaicus Nakamura, 1976 (Fig. 3c) are 100% identical (despite visually apparent differences in facies) and those of Palaeophilotes triphysina (Fig. 3a) and Pseudophilotes (Pseudophilotes) baton (Fig. 3b) differ by only 2.1% (14 bp), which is in the range typical for the closest congeners. To confirm these unexpected results, we sequenced four specimens of P. triphysina (NVG-22027F04, NVG-22027F05, NVG-22028B12, and NVG-22028C01), and their COI barcodes were 100% identical (Genbank OQ311404–OQ311407):
AACTTTATATTTTATTTTCGGAATTTGAGCAGGAATATTAGGAACATCTTTAAGAATTTTAATTCGTATAGAATTAGGAACACCTGGATCTTTAATTGGAGATGATCAAATTTATAACACTATTGTAACAGCTCATGCCTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGAAATTGACTAGTACCCTTAATATTAGGAGCACCTGATATAGCATTTCCACGAATAAATAATATAAGATTTTGATTATTACCTCCATCATTAATATTATTAATTTCAAGTAGAATCGTAGAAAATGGAGCAGGAACAGGATGAACAGTGTACCCCCCACTTTCATCTAATATTGCTCATAGAGGTTCATCTGTTGATTTAGCAATTTTTTCACTTCATTTAGCAGGAATTTCATCAATTTTAGGAGCAATTAATTTTATTACTACAATTATTAATATACGAGTAAATAATATATCATTTGATCAAATATCATTATTTATTTGAGCAGTAGGTATTACAGCATTACTATTATTATTATCTTTACCTGTTTTAGCAGGTGCAATTACTATATTATTAACAGATCGAAATCTTAATACCTCTTTTTTTGACCCTGCTGGAGGAGGAGATCCAATTTTATATCAACATTTATTT
Therefore, we propose to treat species of Palaeophilotes and Pseudophilotes as congeneric. However, due to the elevated rate of evolution in the nuclear genome that resulted in significant phenotypic differences, we conservatively place Pseudophilotes Beuret, 1958 as a subgenus of Palaeophilotes Forster, 1938 instead of synonymizing it. Due to the priority of names based on their dates, this action results in many name changes: we place all species of Pseudophilotes in the genus Palaeophilotes. However, if Pseudophilotes is kept as a genus, the classification of the group becomes genetically inconsistent and would require the elevation of Rubrapterus to the genus level (Fig. 2), which seems unwarranted provided its genetic and phenotypic similarities with Pseudophilotes.
In summary, the variability of evolutionary rates between taxa and rapid phenotypic evolution in some lineages pose challenges for taxonomic classification because recently diverged species can look very different from each other (Fig. 3). As a solution, we propose to keep the classification of genera largely consistent with the estimated divergence times and genetic differentiation, but to use subgenus as an indicator of phenotypic differences. This approach generally results in a smaller number of genera, frequently due to the elimination of monotypic genera, which in our opinion, should be used only to indicate the genetic uniqueness of taxa in the absence of close relatives.
Subgenus Euphilotes Mattoni, [1978] is paraphyletic in mitochondrial DNA
Zhang et al. (2019) placed Philotiella Mattoni, [1978] (type species Lycaena speciosa Hy. Edwards, 1877) as a subgenus of Euphilotes Mattoni, [1978] (type species Lycaena enoptes Boisduval, 1852) due to their genetic similarity, as recently confirmed by Lukhtanov and Gagarina (2022). Adding to these results, here we show that Philotiella and Euphilotes are not only very close to each other genetically (Fig. 2a, b yellow highlight) but also that Philotiella renders Euphilotes paraphyletic in the mitochondrial genome tree with confident statistical support (Fig. 2c yellow highlight), suggesting introgression of mitochondrial DNA between the subgenera and a possibility of future synonymization of these two names. Even phenotypically, some populations of Euphilotes pallescens (Tilden & Downey, 1955) may be superficially more similar to Philotiella than to Euphilotes species due to reduced spotting and the lack of orange spots on ventral hindwing. Finally, our current and more comprehensive trees (Fig. 2) strongly support the distinction of the genus Euphilotes (that includes Philotiella as a subgenus) from other genera in the subtribe Scolitantidina Tutt, 1907: nuclear genome trees place Euphilotes as sister to all other sequenced members of the subtribe, with higher confidence (0.92) in the Z chromosome tree (Fig. 2b). Although members of the New World genus Euphilotes have been placed within some of the Old World genera in the past, we show that unless the entire subtribe Scolitantidina is unified under a single genus (Scolitantides Hübner, 1819), Euphilotes cannot be combined with any other genus to keep it monophyletic.
Family Nymphalidae Rafinesque, 1815
Agraulis Boisduval & Le Conte, [1835] as a subgenus of Dione Hübner, [1819]
From the position of consistency and uniformity of taxonomic classification, based on the genetic closeness of Agraulis Boisduval & Le Conte, [1835] (type species Papilio vanillae Linnaeus, 1758) and Dione Hübner, [1819] (type species Papilio juno Cramer, 1779) we proposed to regard the former as a subgenus of the latter, rather than keeping the two as separate genera (Zhang et al. 2019). Treating Agraulis and Dione as congeneric was not a novel concept (Scott 1986). Recent publications are divided between the two options: some argue for keeping Agraulis as a genus (Núñez et al. 2022; Penz 2022), while others use Agraulis as a subgenus of Dione (Farfán et al. 2022a; Farfán et al. 2022b; Pelham 2022).
In our opinion, this argument reflects lumping vs. splitting viewpoints. For every monophyletic lineage, one may find a sufficient number of characters, be it morphological or molecular, to “support” the distinction of the lineage at the taxonomic rank deemed appropriate. It seems impossible to formulate criteria for which morphological character differentiates between genera and which refers to subgenera. Therefore, it is not likely that genus/subgenus disagreement will be resolved by additional studies of Agraulis morphology. It seems equally unlikely that additional genomic sequencing of Agraulis and Dione will bring us closer to resolution because the data we have at hand (Zhang et al. 2020; Núñez et al. 2022) confidently resolve the phylogeny of the group and provide reliable estimates of evolutionary distances within the group and among their relatives.
While experts in their phylogenetic groups usually find it more aesthetically appealing to split them into many smaller genera (i.e., the more you study something, the more significant seem the differences), we think that a more inclusive treatment with a smaller number of distinctive and confidently monophyletic genera is more practical, both for general biologists (who do not have to learn additional names) and newcomers. While genetic similarity may be harder to relate to, similarity in facies between North American species of Agraulis and Dione is illustrated here in Fig. 4 and needs no explanation. Provided these species are closely related (as judged by genomic analysis), it seems meaningful to treat them as congeneric. From a broader perspective, we do not see an imperative reason to place in different genera two closely related species that can sometimes be misidentified for each other. We think that genera are for broader use, and subgenera could be for specialists.
Fig. 4.

Two species of Dione, iNaturalist observations: a. Dione (Agraulis) incarnata 140735334 USA: Texas, Cameron Co. 31-Oct-2022 © Jeff Chapman; b. Dione (Dione) moneta 131933192 Mexico: Oaxaca, Tepelmeme Villa de Morelos, 6-Sep-2017 © Tepelmeme Villa de Morelos. Images are color-corrected, rotated, cropped, and/or flipped. CC BY-NC 4.0 https://creativecommons.org/licenses/by-nc/4.0/.
Genomics-based arguments for Agraulis as a subgenus of Dione from the position of internal consistency and uniformity of taxonomic classification were stated by Zhang et al. (2019). COI barcodes of Agraulis and Dione differ by less than 8%. From an even broader perspective of consistent application of ranks to clades depending on their evolutionary distance, humans (Homo sapiens Linnaeus, 1758) and chimps (Pan troglodytes (Blumenbach, 1775)), currently attributed to different genera, are 9.6% (63 bp) different in their barcodes (GenBank accessions MF479728 and HM068586), not 2% as specified by Penz (2022). Finally, subgenera Agraulis and Dione consist of a small number of species. Unifying them in one genus does not cause much inconvenience by creating a large genus that is difficult to navigate. On the contrary, the unification results in a more conveniently sized genus and emphasizes closer evolutionary connections within it. For all these reasons, we still support the treatment of Agraulis as a subgenus of Dione; however, placing them in separate genera is not obviously incorrect either.
Chlosyne flavula (W. Barnes & McDunnough, 1918), stat. rest. is a species distinct from Chlosyne palla (Boisduval, 1852)
Our recent finding that Melitaea sterope W. H. Edwards, 1870 (type locality in USA: Oregon, Wasco Co.) is not conspecific with Chlosyne acastus (W. H. Edwards, 1874) (type locality in USA: Utah, probably Utah Co.) and instead is a subspecies of Chlosyne palla (Boisduval, 1852) (type locality in USA: California, Plumas Co.) (Zhang et al. 2022b) prompted further discussions and investigations. Genomic sequencing of all C. palla subspecies partitions them into two clades in the tree constructed from protein-coding regions in autosomes (Fig. 5a blue and red). While not strongly different in mitogenomes (Fig. 5b), the two clades are genetically differentiated in the Z chromosome with Fst/Gmin of 0.2/0.016. This differentiation is approximately the same as in the following pairs of species (Fig. 5a): Chlosyne whitneyi (Behr, 1863) and Chlosyne damoetas (Skinner, 1902), Chlosyne gabbii (Behr, 1863) and C. acastus, and Chlosyne hoffmanni (Behr, 1863) and Chlosyne harrisii (Scudder, 1863). Therefore, the two clades represent two distinct species possibly coming in contact at least in Washington state as C. palla sterope (formerly C. acastus) and Chlosyne palla blackmorei Pelham, 2008 (type locality Canada: British Columbia, Lytton). The blue clade (Fig. 5) includes the lectotypes of C. palla palla and C. palla sterope. The oldest name in the red clade (Fig. 5) is Melitaea flavula W. Barnes & McDunnough, 1918 (type locality USA: Colorado, Garfield Co., Glenwood Springs, a syntype sequenced as NVG-22036E05). Therefore, we propose that Chlosyne flavula (W. Barnes & McDunnough, 1918), stat. rest. is a species distinct from Chlosyne palla (Boisduval, 1852). We place the following valid taxa (and their synonyms) as subspecies of Chlosyne flavula: Chlosyne palla blackmorei Pelham, 2008 and Melitaea calydon W. Holland, 1931 (type locality in USA: Colorado, Jefferson Co.). Other subspecies listed by Pelham (2022) remain with C. palla.
Fig. 5.

Trees of Chlosyne species constructed from protein-coding regions in a. autosomes and b. mitochondrial genome: C. palla (blue), C. flavula stat. rest. (red), C. whitneyi (green), C. damoetas (purple), C. gabbii (olive), C. acastus (orange), C. hoffmanni (cyan), and C. harrisii (brown). Primary type specimens are labeled in magenta.
Chlosyne palla pola (Boisduval, 1869), comb. nov., stat. rev. is not a junior subjective synonym of Chlosyne gabbii gabbii (Behr, 1863)
Genomic sequencing of the holotype of Melitaea pola Boisduval, 1869 (type locality “Sonora”, hypothesized to be USA: California, Los Angeles Co., La Tuna Canyon) currently regarded as a junior subjective synonym of Chlosyne gabbii gabbii (Behr, 1863) (type locality USA: California, Los Angeles Co., La Tuna Canyon) is not in the same clade with it, but instead is a specimen of Chlosyne palla (Boisduval, 1852) (type locality in USA: California, Plumas Co.) (Fig. 5). Therefore, the hypothesized type locality of M. pola, which is the same as that of C. gabbii, is most likely incorrect. Could it be that the original type locality stated as “Sonora” was accurate, but instead of Sonora, Mexico, it represents a homonymous town currently in Tuolumne Co., California? While genomic sequencing of additional specimens is necessary to solve this problem confidently, we see that the holotype of M. pola shares mitochondrial DNA with Chlosyne palla australomontana J. Emmel, T. Emmel & Mattoon, 1998 (type locality in USA: California, Tulare Co., holotype sequenced as NVG-17109C07) (Fig. 5b), and specimens of similar appearance are known from Tuolumne Co., suggesting possible synonymy. Until a confidently supported synonymization solution is found, we conservatively propose to treat M. pola as a valid subspecies Chlosyne palla pola (Boisduval, 1869), comb. nov., stat. rev.
Cercyonis hypoleuca Hawks & J. Emmel, 1998, stat. nov. is a species distinct from Cercyonis sthenele (Boisduval, 1852)
Genomic sequencing of Cercyonis Scudder, 1875 (type species Papilio alope Fabricius, 1793, which is a subspecies of Papilio pegala Fabricius, 1775) specimens reveals that Cercyonis sthenele hypoleuca Hawks & J. Emmel, 1998 (type locality USA: California, Santa Barbara Co., Santa Cruz Island) is sister to both Cercyonis sthenele (Boisduval, 1852) (type locality USA: California, San Francisco, lectotype sequenced as NVG-22036H10) and Cercyonis meadii (W. H. Edwards, 1872) (type locality USA: Colorado, Park Co. Bailey, lectotype sequenced as NVG-21011E01) in all three trees (Fig. 6). Thus, C. meadii renders C. sthenele that includes C. sthenele hypoleuca paraphyletic. Moreover, C. sthenele hypoleuca is genetically differentiated from C. sthenele at the level characteristic of distinct species: e.g., COI barcodes of C. sthenele lectotype and C. hypoleuca differ by 3.3% (22 bp). Therefore, we propose that Cercyonis hypoleuca Hawks & J. Emmel, 1998, stat. nov. is a species-level taxon. Interestingly, the ventral hindwing with a contrasting whitish pattern of C. hypoleuca (Fig. 6e) is superficially like that of nominotypical C. sthenele, which is genetically different from it. Instead, C. sthenele sthenele is genetically most similar to its geographical neighbor Cercyonis sthenele behrii (F. Grinnell, 1905) (type locality in USA: California, Marin Co.) that generally lacks the white pattern.
Fig. 6.

Trees of Cercyonis species constructed from protein-coding regions in a. autosomes, b. Z chromosome, and c. mitochondrial genome: C. sthenele (blue) with C. sthenele damei comb. rev. labeled in green, C. meadii (magenta) with C. meadii mexicana stat. rest. labeled in purple, and C. hypoleuca stat. nov. (red). The lectotype of C. sthenele sthenele and the holotype of C. sthenele damei are highlighted in yellow. Gaps in branches in (c) indicate where vertical slices of the tree were removed to reduce its horizontal dimension (to allow an increase of the font size), i.e., branches with gaps are longer than shown. d. C. sthenele damei ♂ dorsal (left) and ventral (right) views, NVG-22039B03 USA: Arizona, Coconino Co., 17-Aug-1980, J. A. Scott leg. e. C. hypoleuca stat. nov., iNaturalist observation 4540707 USA: California, Santa Barbara Co., Santa Cruz Island, Channel Islands National Park, 3-Jul-2013 © Nature Ali. The image is color-corrected and cropped. CC BY-NC 4.0 https://creativecommons.org/licenses/by-nc/4.0/.
Cercyonis sthenele damei W. Barnes & Benjamin, 1926, comb. rev. is not a subspecies of Cercyonis meadii (W. H. Edwards, 1872)
Genomic sequencing of the holotype of Cercyonis damei W. Barnes & Benjamin, 1926 (type locality USA: Arizona: Coconino Co., Grand Canyon) a taxon treated as a valid subspecies of Cercyonis meadii (W. H. Edwards, 1872) (type locality USA: Colorado, Park Co. Bailey, lectotype sequenced as NVG-21011E01) is not conspecific with it and instead is placed among specimens of Cercyonis sthenele (Boisduval, 1852) (type locality USA: California, San Francisco, lectotype sequenced as NVG-22036H10) in nuclear genome trees (Fig. 6a, b). Two more recently collected specimens from the general vicinity of the type locality of C. damei (e.g., Fig. 6d) are in the same clade with the holotype in the Z chromosome tree (Fig. 6b), which usually agrees well with speciation scenarios (Cong et al. 2019a). These specimens, including the C. damei holotype, are dark and lack reddish patches of C. meadii, being more similar phenotypically to C. sthenele. Therefore, from both its nuclear genomic sequences and superficial appearance, Cercyonis damei is not a subspecies of C. meadii but a subspecies of C. sthenele: Cercyonis sthenele damei W. Barnes & Benjamin, 1926, comb. rev. However, in the tree constructed from the protein-coding regions of autosomes (which usually harbor a larger number of introgressed genes), the three sequences specimens of C. sthenele damei do not form a clade. They are placed at the base of the C. sthenele clade (Fig. 6a), suggesting some introgression from C. meadii. These introgressed genomic regions “pull” these specimens closer to C. meadii in the tree and, thus, closer to the base of the clade. Nevertheless, the amount of introgression is insufficient to “move” any of these specimens into the C. meadii clade. Furthermore, two C. sthenele damei specimens, including the holotype, possess mitochondrial DNA of C. meadii and not C. sthenele (Fig. 6c green-labeled in the magenta clade), directly indicating introgression from C. meadii. The third specimen (Fig. 6c green-labeled in the blue clade) has mitochondrial DNA of C. sthenele. This polyphyly of C. sthenele damei in the mitochondrial DNA tree indicates a varying extent of limited hybridization and introgression with C. meadii in its various genomic regions rather than supporting this taxon’s hybrid origin.
Cercyonis meadii mexicana R. Chermock, 1949, stat. rest. is a valid subspecies and not a junior subjective synonym of Cercyonis sthenele damei W. Barnes & Benjamin, 1926, comb. rev.
Genomic sequencing of a specimen from southeastern Arizona identified as Cercyonis meadii mexicana R. Chermock, 1949 (type locality in Mexico: Chihuahua) (Fig. 6 labeled in purple), a taxon treated as a junior subjective synonym of “Cercyonis meadii damei” W. Barnes & Benjamin, 1926 (type locality USA: Arizona: Coconino Co., Grand Canyon) by Pelham (2022) reveals that it is in a clade different from the holotype of Cercyonis damei (Fig. 6), which, as we have shown above, is a subspecies of Cercyonis sthenele (Boisduval, 1852) (type locality USA: California, San Francisco, lectotype sequenced as NVG-22036H10) and instead belongs to Cercyonis meadii (W. H. Edwards, 1872) (type locality USA: Colorado, Park Co. Bailey, lectotype sequenced as NVG-21011E01), being sister to all its other subspecies in all three trees (Fig. 6 magenta) and thus is distinct from them. Therefore, we reinstate it as a valid subspecies Cercyonis meadii mexicana R. Chermock, 1949, stat. rest.
The holotype of Hermeuptychia sinuosa Grishin, 2021
The original description illustrated the holotype of Hermeuptychia sinuosa Grishin, 2021 (type locality Guatemala: El Progreso, Morazán) that was pinned through its side, unspread (Cong et al. 2021). Here, we use this opportunity and publish photographs of the holotype after it has been spread (Fig. 7). Its genitalia vial is pinned on the same pin as the specimen. The holotype is in the University of Texas Insect Collection, Austin, TX, USA.
Fig. 7.

Holotype of Hermeuptychia sinuosa Grishin, 2021 dorsal (left) and ventral (right) views, data in text.
Family Hesperiidae Latreille, 1809
Aethilla toxeus Plötz, 1882, syn. nov. is a junior subjective synonym of Cecropterus albociliatus (Mabille, 1877)
Genomic sequencing of the syntype of Aethilla toxeus Plötz, 1882 (NVG-15032A10, type locality in Mexico) in MFNB reveals that it is clustered with Cecropterus albociliatus (Mabille, 1877) (type locality in Colombia, Panama, and Guatemala) (Fig. 8 blue) and not within a species currently called Cecropterus toxeus (Fig. 8 green and purple). The sequenced specimen is a syntype (possibly the only one ever in existence) because it matches the original description and carries a label with the number 5054, as stated in the description. In the interest of stability of nomenclature, N.V.G. hereby designates this specimen in MFNB, a female, bearing the following six rectangular labels, the first is red, the third is green, and others are white: [ Typus ], [ 5054 ], [ Mexico Deppe ], [ toxeus | Pl. | type 5054. ], [ {QR code} http://coll.mfn-berlin.de/u/ | 940b65 ], and [ DNA sample ID: | NVG-15032A10 | c/o Nick V. Grishin ] as the lectotype of Aethilla toxeus Plötz, 1882. Because Ferdinand Deppe collected in Mexico in 1824–1829 (Stresemann 1954), the lectotype was likely collected during that time period. The lectotype is missing nearly all its fringes (the white portion is represented literally by a couple of remaining scales), and all its wings except the right forewing are chipped at the margins. However, paler postdiscal spots in forewing cells M3-CuA1 and CuA1-CuA2 overlap; therefore, this specimen keys out to Achalarus albociliatus in Evans (1952), and not to Evans’ “Achalarus toxeus.” Therefore, Aethilla toxeus Plötz, 1882, syn. nov. is a junior subjective synonym of Cecropterus albociliatus (Mabille, 1877).
Fig. 8.

Trees of selected Cecropterus species constructed from protein-coding regions in a. Z chromosome and b. mitochondrial genome: C. markwalkeri sp. n. (red, highlighted in yellow), C. albociliatus (blue), C. coyote stat. rest. (green), C. nigrociliata stat. rest. (purple), C. jalapus (cyan), and C. athesis (olive). Primary type specimens are labeled in magenta, and a specimen curated as a possible syntype is labeled in plum color.
Cecropterus coyote (Skinner, 1892), stat. rest. and Cecropterus nigrociliata (Mabille & Boullet, 1912), stat. nov. are species distinct from Aethilla toxeus Plötz, 1882
As shown above, a species currently known as Cecropterus toxeus (Plötz, 1882) (type locality in Mexico) loses its name to Cecropterus albociliatus (Mabille, 1877) (type locality in Colombia, Panama, and Guatemala) because the lectotype of the former is conspecific with the latter. The next oldest name for the species currently misidentified as C. toxeus is Cecropterus coyote (Skinner, 1892) (type locality in USA: Southern Texas). Genomic sequencing of a syntype of Eudamus coyote places it in the clade with specimens from the US and eastern Mexico that constitute a species distinct from others (Fig. 8 green). Moreover, we find that the holotype of Murgaria albociliata var. nigrociliata Mabille & Boullet, 1912 (type locality in Mexico), a taxon currently regarded as a junior subjective synonym of Cecropterus toxeus (Plötz, 1882) and, therefore, given our findings, possibly conspecific with C. coyote, is in the clade sister to C. coyote, together with several specimens from southwestern Mexico (Fig. 8 purple). The two clades differ genetically at the level characteristic of distinct species alike genetic differentiation between sisters Cecropterus jalapus (Plötz, 1881) (Fig. 8 cyan) and Cecropterus athesis (Hewitson, 1867) (Fig. 8 olive). COI barcodes of the primary types of E. coyote and M. albociliata var. nigrociliata differ by 1.8% (12 bp). Hence, we propose that Cecropterus coyote (Skinner, 1892), stat. rest. and Cecropterus nigrociliata (Mabille & Boullet, 1912), stat. nov. are species-level taxa.
Cecropterus (Murgaria) markwalkeri Grishin, new species
http://zoobank.org/F1805F4A-DB94-47C9-A148-82B20B46BA6E (Figs. 8 part, 9, 10a–b, 11)
Fig. 9.

Holotype of Cecropterus markwalkeri sp. n. dorsal (left) and ventral (right) views, data in text.
Fig. 10.

Genitalia of Cecropterus (Murgaria). a, b. C. markwalkeri sp. n. holotype in left lateral (a) and dorsal (b) views. c–e. C. albociliatus albociliatus from Mexico: Veracruz, BMNH(E) 1717074, Godman’s mini-slide preparation [BMNH]: genital capsule in left lateral view with left valva removed (c), left valva in right lateral view (d), and genitalia illustration from Godman & Salvin (1894: pl. 80, fig. 14), not to scale (e). Photographs c and d are © The Trustees of the Natural History Museum London and are made available under Creative Commons License 4.0 (https://creativecommons.org/licenses/by/4.0/).
Fig. 11.

Possible Cecropterus markwalkeri sp. n. from Mexico: Sonora, La Aduana near Alamos, 27-Aug-2017, dorsal (left) and ventral (right) views of the same individual, iNaturalist observation 105683875 © Ken Kertell, color-corrected. CC BY-NC 4.0 https://creativecommons.org/licenses/by-nc/4.0/.
Definition and diagnosis.
Genomic sequencing of the subgenus Murgaria E. Watson, 1893 (type species Telegonus albociliatus Mabille, 1877) reveals that two specimens from Sonora, Mexico are sister to a compact clade of Cecropterus albociliatus (Mabille, 1877) (type locality in Colombia, Panama, Guatemala) (Fig. 8). The latter clade included a specimen identified by Mabille as “Teleg. albociliatus” and curated as a type, in addition to the lectotype of Aethilla toxeus Plötz, 1882 (type locality in Mexico) and syntypes of Aethilla nocera Plötz, 1882 (type locality in Colombia) and Telegonus mithras Mabille, 1888 (type locality in Venezuela), the latter being a junior subjective synonym of the former, which is regarded as a subspecies of C. albociliatus. The two specimens from Sonora (Fig. 8 red) show prominent genetic differentiation from C. albociliatus (Fig. 8 blue) in the Z chromosome (Fst/Gmin 0.61/0.001), which is larger than that between Cecropterus jalapus (Plötz, 1881) (Fig. 8 cyan) and Cecropterus athesis (Hewitson, 1867) (Fig. 8 olive). Therefore, the Sonoran specimens represent a species distinct from C. albociliatus, and because no available name applies to this species, it is new. Curiously this new species shares COI barcodes with its sister C. albociliatus (100% identical) but differs from it in male genitalia morphology (Fig. 10). The new species keys to “Achalarus albociliatus albociliatus” C.17.3(a) in Evans (1952) sharing with it the lack of costal fold, white (with a gray tint and some scales are more translucent) hindwing and brownish forewing fringes, but differing in the shape of valva in male genitalia: the valva is broader, ampulla is less developed, and is more in line with costa, separated from it by a slight concavity (Fig. 10a), instead of ampulla strongly bulging posterodorsad, separated from costa by a large concavity in C. albociliatus (Fig. 10c–e); harpe enlarged posteriad and broader, but relatively shorter compared to a narrower and longer harpe of C. albociliatus. A diagnostic combination of nuclear genome characters is: aly1651.25.1:T210C, aly1539.8.1:C888T, aly1089.5.3:G91A, aly1089.5.3:G121A, aly1222.33.2:T630C.
Barcode sequence of the holotype: Sample NVG-18033E11, GenBank OQ311409, 658 base pairs:
AACCTTATATTTTATTTTTGGAATTTGAGCAGGATTAGTAGGAACTTCTTTAAGTTTACTTATTCGAACTGAATTAGGAACTCCAGGATCTTTAATTGGAGATGATCAAATTTATAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATGCCTATTATAATTGGAGGATTTGGAAATTGACTAGTTCCCCTTATATTAGGAGCCCCTGACATAGCTTTCCCTCGTATAAATAATATAAGATTTTGATTATTACCCCCATCTTTAACTCTTTTAATTTCAAGAAGAATTGTAGAAAATGGTGCAGGTACTGGATGAACAGTTTATCCCCCTTTATCCTCTAATATTGCCCACCAAGGAGCATCAGTAGATTTAGCAATTTTTTCTTTACATTTAGCTGGAATTTCTTCTATTCTTGGAGCTATTAACTTTATTACAACTATTATTAATATACGAATTAATAATTTATCATTTGATCAAATACCATTATTTATTTGAGCTGTCGGAATTACAGCCTTATTATTATTACTTTCTTTACCTGTTTTAGCTGGAGCTATTACTATATTATTAACTGATCGAAATTTAAATACTTCATTTTTTGATCCTGCCGGTGGAGGAGATCCTATTTTATATCAACATTTATTT
Type material.
Holotype:
♂ deposited in the National Museum of Natural History, Washington, DC, USA [USNM], illustrated in Fig. 9, bears four printed labels: three white [ Palm Canyon | 19-IX-05 Mexico | Sonora | Lush habitat on | Ruta 16 at km 196 | Mark Walker leg. ], [ DNA sample ID: | NVG-18033E11 | c/o Nick V. Grishin ], [ DNA sample ID: | NVG-22031H04 | c/o Nick V. Grishin ], and one red [ HOLOTYPE ♂ | Cecropterus | markwalkeri Grishin ]. The first NVG number corresponds to a sampled leg, and the second is for the abdomen DNA extraction followed by genitalia dissection.
Paratype:
1♂ NVG-18033E10 and NVG-22031H03, the same data as the holotype, but 17-Sep-2010.
Type locality.
Mexico: Sonora, Ruta16 at km 196, elevation 896 m, approximate GPS 28.4856, −109.3605.
Etymology.
The name honors Mark Walker, who collected the type series on expeditions with and under research permits to Paul A. Opler. Mark is a dedicated butterfly explorer who sampled specimens at many points across the globe, contributing to our knowledge of Lepidoptera by discovering new localities and range extensions. Mark passionately shares his adventures in words skillfully woven into lepisodes (essays about field adventures in Lepidoptera) that many of us eagerly await and, more recently, in pictures. Mark’s kindness in sharing his knowledge and field time with other enthusiasts is unsurpassed. The name is a noun in the genitive case.
Distribution.
Currently known only from the type locality in east-central Sonora, Mexico, but similar in appearance specimens have been photographed at other places in Sonora (Fig. 11).
Nectaring plant of Epargyreus clarus californicus MacNeill, 1975 in British Columbia, Canada
The caption to figure 38 in Zhang et al. (2022b) stated that Epargyreus clarus californicus MacNeill, 1975 was “nectaring on giant vetch in Canada: British Columbia, Cortes Island”. Instead, the plant shown is a native Fringed Bleeding Heart, Dicentra eximia (Ker-Gawl.) Torr. We thank Christian Gronau for kindly informing us about this error.
Aguna malia Evans, 1952, stat. nov. is a species distinct from Aguna megaeles (Mabille, 1888)
Genomic sequencing of Aguna megaeles (Mabille, 1888) (type locality in Brazil: Santa Catarina) specimens, including its lectotype (NVG-15029D11) reveals that Aguna megaeles malia Evans, 1952 (type locality in Venezuela) (Fig. 12 red) is genetically differentiated from the nominotypical subspecies (Fig. 12 blue) at the level characteristic of species: e.g., their COI barcodes differ by 5.3% (35 bp). Therefore, we propose that it is a species-level taxon Aguna malia Evans, 1952, stat. nov.
Fig. 12.

Trees of Aguna species constructed from protein-coding regions in a. Z chromosome and b. mitochondrial genome: A. megaeles (blue) and A. malia stat. nov. (red).
Polygonus arizonensis (Skinner, 1911), stat. nov., Polygonus histrio Röber, 1925, stat. rest., Polygonus pallida Röber, 1925, stat. nov., and Polygonus hagar Evans, 1952, stat. nov. are species distinct from Polygonus leo (Gmelin, [1790])
Genomic sequencing and analysis of Polygonus leo (Gmelin, [1790]) (type locality America, likely in Hispaniola) specimens from across the range reveal that all five subspecies of P. leo are genetically differentiated at the species level in both nuclear (Fig. 13a) and mitochondrial (Fig. 13b) DNA. E.g., COI barcodes of the closest taxa, i.e., the nominotypical P. leo (Fig. 13 purple) and Polygonus (Acolastus) histrio Röber, 1925 (type locality “vermutlich aus Panama”, but likely Cuba as suggested by DNA comparison) (Fig. 13 green) differ by 2.1% (14 bp). Therefore, we propose that Polygonus arizonensis (Skinner, 1911), stat. nov., Polygonus histrio Röber, 1925, stat. rest., Polygonus pallida Röber, 1925, stat. nov., and Polygonus hagar Evans, 1952, stat. nov. are species, not subspecies.
Fig. 13.

Trees of Polygonus leo species complex constructed from protein-coding regions in a. Z chromosome and b. mitochondrial genome: P. arizonensis stat. nov. (red), P. pallida stat. nov. (blue), P. histrio stat. rest. (green), P. leo (purple), and P. hagar stat. nov. (cyan). Primary type specimens are labeled in magenta.
Viola dagamba Steinhauser, 1989 is a new junior subjective synonym of Viola kuma (Bell, 1942), comb. nov., stat. rest.
Sequencing of the holotypes of Viola dagamba Steinhauser, 1989 (type locality in Guyana), currently a valid species, and Pellicia kuma Bell, 1942 (type locality in Venezuela), currently a junior subjective synonym of Pachyneuria helena (Hayward, 1939) (type locality in Ecuador: Rio Topo) reveals that they are conspecific (e.g., their COI barcodes are 100% identical) and belong to the genus Viola Evans, 1953 (type species Staphylus alicus Schaus, 1902), not Pachyneuria Mabille, 1888 (type species Pachyneuria obscura Mabille, 1888) (Fig. 14). Therefore, we reinstate Viola kuma (Bell, 1942), comb. nov., stat. rest. as a species and treat Viola dagamba Steinhauser, 1989, syn. nov. as its junior subjective synonym.
Fig. 14.

Trees of Viola and Pachyneuria species constructed from protein-coding regions in a. Z chromosome and b. mitochondrial genome: V. kuma comb. nov., stat. rest. (red) and Pachyneuria (blue).
Leucochitonea janice Ehrmann, 1907 is a junior subjective synonym of Heliopetes alana (Reakirt, 1868) and not of Heliopetes petrus (Hübner, [1819])
Genomic sequencing of the holotype of Leucochitonea janice Ehrmann, 1907 (NVG-15095C05, type locality Venezuela: Suapure) (Fig. 15 magenta) in CMNH, a taxon treated by Mielke (2005) as a junior subjective synonym of Heliopetes petrus (Hübner, [1819]) (type locality not given) (Fig. 15 red), reveals that it is not conspecific with it and instead is placed among specimens of Heliopetes alana (Reakirt, 1868) (type locality in Colombia) (Fig. 15 blue) in both nuclear (Fig. 15a) and mitochondrial (Fig. 15b) DNA trees. Therefore, we conclude that Leucochitonea janice Ehrmann, 1907 is a junior subjective synonym of Heliopetes alana (Reakirt, 1868) and not of Heliopetes petrus (Hübner, [1819]).
Fig. 15.
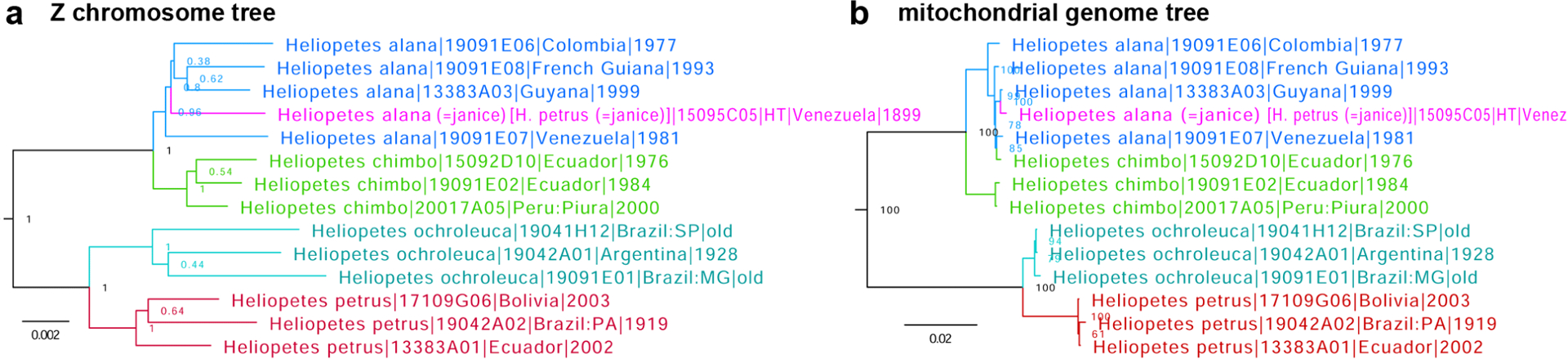
Trees of Heliopetes species constructed from protein-coding regions in a. Z chromosome and b. mitochondrial genome: H. alana (blue) with the holotype of Leucochitonea janice Ehrmann, 1907 (magenta), H. chimbo Evans, 1953 (green), H. ochroleuca J. Zikán, 1938 (cyan), and H. petrus (red).
Tamela maura (Snellen, 1886), stat. rest., and Tamela diocles (Moore, [1866]), stat. rest., are species distinct from Tamela othonias (Hewitson, 1878) and Tamela nigrita (Latreille, [1824]), respectively
Based on COI barcodes and morphological evidence, Xue et al. (2022) suggested recently that Tamela othonias (Hewitson, 1878) (type locality in Borneo) (Fig. 16 purple) and Tamela fumatus (Mabille, 1876) (type locality in the Philippines) (Fig. 16 cyan) are species distinct from Tamela nigrita (Latreille, [1824]) (type locality in Java) (Fig. 16 blue). Our genomic results confirm that but also reveal that Tamela nigrita diocles (Moore, [1866]) (type locality in Bengal) (Fig. 16 green) is not monophyletic with T. nigrita, but instead is sister to Tagiades maura Snellen, 1886 (type locality in Sumatra) (Fig. 16 red), which Xue et al. (2022) regarded as a junior subjective synonym of T. othonias. While the association of T. n. diocles with T. nigrita is clearly wrong (Fig. 16), it is conceivable to place both T. maura and T. n. diocles in T. othonias as subspecies or synonyms. However, both taxa show genetic distinction from T. othonias, so we propose treating them as distinct species: Tamela maura (Snellen, 1886), stat. rest. and Tamela diocles (Moore, [1866]), stat. rest., separate from either T. othonias or T. nigrita.
Fig. 16.

Trees of Tamela species constructed from protein-coding regions in a. Z chromosome and b. mitochondrial genome: T. fumatus (cyan), T. nigrita (blue), T. othonias (purple), T. maura stat. rest. (red), and T. diocles stat. rest. (green).
Hedone yunga Grishin, new species
http://zoobank.org/8575DF83-2451-49B1-B6C4-AA4F9C595FF1 (Figs. 17 part, 18)
Fig. 17.

Trees of nine Hedone species constructed from protein-coding regions in a. Z chromosome and b. mitochondrial genome: H. vibex (Geyer, 1832) (blue), H. praeceps Scudder, 1872 (red), H. calla (Evans, 1955) (green), H. vibicoides (de Jong, 1983) (olive), H. catilina (Plötz, 1886) (cyan), H. yunga sp. n. (highlighted in lime color), H. dictynna (Godman & Salvin, 1896) (orange), H. mira Grishin & Lamas, 2022 (magenta), and H. bittiae (Lindsey, 1925) (purple).
Fig. 18.

Holotype of Hedone yunga sp. n. dorsal (left) and ventral (right) views, data in text.
Definition and diagnosis.
Genomic sequencing of Hedone Scudder, 1872 (type species Hesperia brettus Boisduval & Le Conte, [1837]) reveals that one specimen from Bolivia was placed in the trees separately from all others, being sister to Hedone catilina (Plötz, 1886) (type locality Brazil: Santa Catarina, Blumenau; syntype NVG-18052B01 sequenced), but genetically differentiated from it at the level characteristic of species (Fig. 17). E.g., its COI barcode differs from that of the H. catilina syntype by 3.3% (22 bp). Therefore, this specimen represents a new species. This new species keys to “Polites vibex catilina” M.13.1(d) in Evans (1955) that is also known from Bolivia (Fig. 17) at a lower elevation but differs from it in females (male unknown) by the pattern of the ventral hindwing, which is whiter (instead of yellower) overscaling, with more contrasty and better defined (rather than more diffuse) brown spots and a darker area by the end of the discal cell. In the absence of known males and without probing female variation, the most reliable identification is achieved by DNA, and a combination of the following base pairs is diagnostic in the nuclear genome: aly86.14.2:A4695C, aly23605.1.46:T819C, aly23605.1.46: G3606A, aly23605.1.46:T900A, aly23605.1.46:T2070G, aly127.52.1:T621T (not C), aly671.3.4:A189A (not G), aly1139.42.1:C165C (not T), aly1060.6.1:C1050C (not T), aly1042.7.1:A2148A (not G), and COI barcode: T4C, T412C, T406C, T547T, T646C.
Barcode sequence of the holotype: Sample NVG-21127C12, GenBank OQ311411, 658 base pairs:
AACCTTATATTTTATTTTTGGTATTTGAGCAGGAATATTAGGAACTTCTTTAAGTTTATTAATTCGAACAGAATTAGGTAATCCTGGATCTTTAATTGGAGATGATCAAATTTATAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGAAATTGATTAGTTCCATTAATATTAGGAGCTCCTGATATAGCTTTCCCTCGAATAAATAATATAAGATTTTGAATATTACCCCCCTCATTAACATTATTAATTTCAAGAAGAATTGTAGAAAATGGTGCAGGAACAGGTTGAACAGTTTATCCTCCTTTATCTTCAAATATTGCTCACCAAGGATCTTCTGTTGATTTAGCAATTTTTTCTCTTCACTTAGCCGGAATTTCTTCTATTTTAGGAGCTATTAATTTTATTACAACAATTATTAATATACGAATTAAAAATTTATCTTTTGATCAAATACCTTTATTTGTATGATCTGTTGGAATTACAGCTCTTTTATTATTATTATCTTTACCTGTTTTAGCTGGAGCTATTACTATATTACTTACAGATCGAAATTTAAATACTTCATTTTTTGATCCAGCAGGTGGAGGAGATCCAATTTTATACCAACATTTATTT
Type material.
Holotype:
♀ deposited in the Museum für Naturkunde, Berlin, Germany (MFNB), illustrated in Fig. 18, bears the following five rectangular labels, four white: [ San Antonio (1800) | Bolivia (Yungas) | 1895—6. Garlepp ], [ Coll. | Staudinger ], [ ] (no text on this label), [ DNA sample ID: | NVG-21127C12 | c/o Nick V. Grishin ], and one red [ HOLOTYPE ♀ | Hedone | yunga Grishin ].
Type locality.
Bolivia: Yungas Region, La Paz Department, San Antonio.
Etymology.
The name is given for the type locality. The name is a feminine noun in apposition.
Distribution.
Currently known only from the holotype collected in the Yungas Region of Bolivia.
Comments.
Without genomic sequencing that confidently supports the distinctness of this species based on a single female specimen, it would have been a challenge to describe this species without finding other specimens, including its male. Although wing patterns of the female holotype are recognizably different from other Hedone species, it is conceivable to hypothesize that it could have beeen an individual variation or aberration. On another note, a comparison of nuclear (Fig. 17a) and mitochondrial (Fig. 17b) DNA trees reveals incongruence typical for many species complexes in Lepidoptera and non-trivial evolutionary scenarios of mitogenome evolution that is likely riddled with introgression: Hedone catilina (Plötz, 1886) is polyphyletic (Fig. 17b cyan), and H. catilina with Hedone bittiae (Lindsey, 1925) (Fig. 17b purple) show several distinct mitogenome clusters that are not evident in nuclear DNA (Fig. 17a). Similar scenarios have been documented in other species groups (Zakharov et al. 2009; Cong et al. 2017).
Vinius phellus (Mabille, 1883), stat. rest. is a species distinct from Vinius exilis (Plötz, 1883)
Genomic sequencing reveals prominent genetic differentiation between Hesperia exilis Plötz, 1883 (type locality “Californien”, in error, a possible (as hypothesized by G. Lamas) syntype in MFNB from Brazil: Santa Catarina sequenced as NVG-21116A10) (Fig. 19 blue), currently a valid species in the genus Vinius Godman, 1900 (type species Vinius arignote Godman, 1900, which is a junior subjective synonym of H. exilis) and Pamphila phellus Mabille, 1883 (type locality “Malaisie”, in error, probably French Guiana: Cayenne) (Fig. 19 red), currently a subspecies of the former species: e.g., their COI barcodes differ by 4% (26 bp). Therefore, we propose that Vinius phellus (Mabille, 1883), stat. rest. is a species-level taxon.
Fig. 19.

Trees of Vinius species constructed from protein-coding regions in a. Z chromosome and b. mitochondrial genome: V. phellus (red), V. exilis (blue), V. sophistes (green), and V. tryhana (purple). Gaps in branches indicate where vertical slices of the tree were removed to reduce its horizontal dimension (to allow an increase of the font size), i.e., branches with gaps are longer than shown.
Vinius sophistes (Dyar, 1918), stat. rest. is a species distinct from Vinius tryhana (Kaye, 1914)
Genomic sequencing reveals prominent genetic differentiation between Padraona tryhana Kaye, 1914 (type locality in Trinidad) (Fig. 19 green), currently a valid species in the genus Vinius Godman, 1900 (type species Vinius arignote Godman, 1900, which is a junior subjective synonym of Hesperia exilis Plötz, 1883) and Padraona sophistes Dyar, 1918 (type locality in Mexico: Veracruz) (Fig. 19 purple), currently a junior subjective synonym of the former species: e.g., their COI barcodes differ by 3.6% (24 bp). Therefore, we propose that Vinius sophistes (Dyar, 1918), stat. rest. is a species-level taxon.
Rhinthon andricus (Mabille, 1895), stat. rest. and Rhinthon aqua (Evans, 1955), stat. nov. are species distinct from Rhinthon braesia (Hewitson, 1867)
Genomic analysis of Rhinthon Godman, 1900 (type species Proteides chiriquensis Mabille, 1889, a junior subjective synonym of Hesperia osca Plötz, 1882) specimens reveals that Proteides andricus Mabille, 1895 (type locality in Brazil: Santa Catarina) (Fig. 20 red) currently treated as a subspecies of Rhinthon braesia (Hewitson, 1867) (type locality in Brazil: Pará) (Fig. 20 blue) is not monophyletic with it and is distinct from other species. Therefore, we reinstate it as a species-level taxon Rhinthon andricus (Mabille, 1895), stat. rest. Furthermore, Neoxeniades braesia aqua Evans, 1955 (type locality Colombia: Rio Dagua) is genetically differentiated from the nominotypical R. braesia at the level characteristic of distinct species (Fig. 20a, Fst/Gmin 0.48/0.003): e.g., compare with the pair Rhinthon molion (Godman, 1901) and Rhinthon bajula (Schaus, 1902). Therefore, we propose to treat Rhinthon aqua (Evans, 1955), stat. nov. as a species-level taxon. As a result of this analysis, R. braesia becomes monotypic. The taxon originally proposed as Neoxeniades bajula peri (Evans, 1955) (type locality in Brazil: Para) has been regarded as a valid species of Niconiades Hübner, [1821] (type species Niconiades xanthaphes Hübner, [1821]) by Zhang et al. (2022a). Finally, we note that mitochondrial DNA is largely shared among four species of Rhinthon (Fig. 20b), likely due to introgression.
Fig. 20.

Trees of Rhinthon species constructed from protein-coding regions in a. autosomes and b. mitochondrial genome: R. andricus stat. rest. (red), R. molion (green), R. bajula (olive), R. aqua stat. nov. (magenta), and R. braesia (blue).
The type locality of Dion uza (Hewitson, 1877) is likely in southern Brazil, and Dion agassus (Mabille, 1891) is confirmed as a valid species
The lectotype of Hesperia uza Hewitson, 1877 (type locality not stated) in MFNB designated by Mielke and Casagrande (2002) sequenced as NVG-18052D10 was subsequently designated as the neotype of Hesperia pruinosa Plötz, 1882 (type locality South America) in Zhang et al. (2022a), and this species was placed in the genus Dion Godman, 1901 (type species Carystus gemmatus Butler, 1872). The provenance of this specimen is unknown: no locality data were given on its labels or in the original description.
Here, we report genomic sequencing of several Dion specimens we found in MFNB that were collected at about the same time period as the D. uza lectotype. Although none of these specimens is as uniformly blue on the ventral hindwing as the lectotype, three of them cluster closely with the lectotype in both nuclear (Fig. 21a) and mitochondrial (Fig. 21b) DNA trees and therefore are conspecific with it. According to their labels, all three specimens are from southern Brazil: Espírito Santo (Southeast region of Brazil) and Santa Catarina (South region of Brazil). While it is not possible to pinpoint the type locality of D. uza with better precision using these specimens due to their genetic similarities, it is most likely that the lectotype was collected in southern Brazil, possibly in the states of Santa Catarina, Rio de Janeiro, or Espírito Santo.
Fig. 21.

Trees of Dion species constructed from protein-coding regions in a. Z chromosome and b. mitochondrial genome: D. uza (red) and D. agassus (blue). Primary type specimens are labeled in magenta.
Furthermore, Zhang et al. (2022a) treated Dion agassus (Mabille, 1891) (type locality Brazil: Amazonas, Massauary) as a species distinct from D. uza based on COI barcodes differences and phenotypic comparison of their lectotypes. Here, we confirm this treatment based on genomic sequencing of four specimens of each species (including their lectotypes) that support genetic distinction between the two species (Fig. 21) and report that COI barcodes of lectotypes of Dion uza (Hewitson, 1877) and Dion agassus (Mabille, 1891) differ by 2.9% (19 bp) (not 2.3% as stated in Zhang et al. (2022a) by mistake). The COI barcode sequence of the lectotype/neotype of D. uza/H. pruinosa, sample NVG-18052D10, GenBank accession OQ311412, 658 base pairs is:
AACTTTATATTTTATTTTTGGTATTTGAGCAGGAATATTAGGAACTTCTCTAAGTTTATTAATTCGAACAGAATTAGGTAATCCTGGCTCTTTAATTGGAGATGATCAAATTTATAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTCATAGTTATACCTATTATAATTGGAGGATTTGGTAATTGATTAGTTCCTCTAATACTAGGAGCACCTGATATAGCTTTCCCCCGAATAAATAATATAAGATTTTGAATACTGCCACCCTCCCTTATACTATTAACTTTTAGTAGAATTGTAGAAAGTGGAGCAGGTACTGGATGAACAGTTTATCCCCCTCTTTCTTCTAACATTGCTCATCAAGGTTCTTCAGTTGATTTAGCAATTTTTTCATTACATTTAGCAGGAATTTCTTCTATTTTAGGTGCTATTAATTTTATTACAACAATTATTAACATACGAATTAAAAACTTATCATTTGATCAAATACCTTTATTTGTGTGATCTGTAGGTATTACAGCCTTATTATTACTATTATCTTTACCAGTATTAGCAGGAGCTATTACAATACTTCTTACTGATCGAAATTTAAATACTTCTTTTTTTGATCCAGCAGGAGGAGGAGATCCAATTTTATATCAACATTTATTT
The COI barcode sequence of the lectotype of D. agassus, sample NVG-15036E10, GenBank accession OQ311413, 658 base pairs is:
AACTTTATATTTTATTTTTGGTATTTGAGCAGGAATATTAGGAACTTCTTTAAGTTTACTAATTCGAACAGAATTAGGTAATCCTGGCTCTTTAATTGGAGACGATCAAATTTATAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTCATAGTTATACCTATTATAATTGGAGGATTTGGTAATTGATTAGTCCCTTTAATACTAGGAGCACCTGATATAGCTTTCCCCCGAATAAATAATATAAGATTTTGAATACTACCACCCTCCCTTATACTATTAACTTTTAGTAGAATTGTAGAAAATGGAGCAGGAACTGGATGAACAGTTTACCCCCCTCTTTCTTCTAATATTGCTCATCAAGGTTCTTCAGTTGATTTAGCAATTTTTTCATTACATTTAGCAGGAATTTCTTCTATTTTAGGCGCTATTAATTTTATTACAACAATTATTAATATACGAATTAAAAACTTATCATTTGATCAAATACCTTTATTTGTGTGATCTGTAGGTATTACAGCCTTACTGTTACTACTATCTCTACCAGTATTAGCAGGAGCTATTACAATACTTCTCACTGATCGAAATTTAAATACTTCTTTTTTTGACCCAGCAGGAGGAGGAGATCCAATTTTATACCAACATTTATTT
Borna Grishin, new subgenus
http://zoobank.org/C86F2094-5A1C-417C-AAC4-A9985ED28EDA
Type species.
Godmania borincona Watson, 1937.
Definition.
This subgenus is represented in all trees by a clade sister to all other known Choranthus Scudder, 1872 (type species Hesperia radians Lucas, 1857) species (Fig. 22). Keys to M.24.6 in Evans (1955). Distinguished from its relatives by the following combination of characters: mid-tibiae smooth, ventral hindwing with dark, brownish (not bright orange) scales, in females uniformly colored, not much paler by the anal margin, dorsal hindwing in females brown with orange overscaling; in males, subapical orange spots on forewing form a nearly continuous orange band with other postdiscal spots; valva terminally deeply indented, shaped like a crab-claw. In DNA, a combination of the following base pairs is diagnostic in the nuclear genome: aly235.7.6:T37C, aly525.48.6:A129G, aly54.29.3:C201T, aly1022. 3.14:A90G, aly904.12.5:A78T, and COI barcode: G101A, T112C, T115C, A376C, C483T, T568C.
Fig. 22.

Trees of Choranthus species constructed from protein-coding regions in a. autosomes and b. mitochondrial genome colored by subgenus: Borna subgen. n. (green), Lilla subgen. n. (red), Choranthus (blue), and Asbolis (purple).
Etymology.
The name is a feminine noun in the nominative singular formed from the type species name Bor[inco]na.
Species included.
Only the type species.
Parent taxon.
Genus Choranthus Scudder, 1872.
Lilla Grishin, new subgenus
http://zoobank.org/7AAEDCAD-E796-4E4E-B3F8-37EFED5BCF15
Type species.
Choranthus lilliae Bell, 1931.
Definition.
This subgenus is represented in the autosome genes tree by a clade sister to all other known Choranthus Scudder, 1872 (type species Hesperia radians Lucas, 1857) species except those in the subgenus Borna subgen. n. (Fig. 22). Keys to M.24.5 in Evans (1955). Distinguished from its relatives by the following combination of characters: mid-tibiae smooth, ventral hindwing with dark, brownish (not bright orange) scales, in females uniformly colored, not much paler by the anal margin, dorsal hindwing in females brown with a weak orange postdiscal band; in males, subapical orange spots on forewing clearly separated from other orange spots; uncus undivided, rounded, valva nearly elliptical in shape, terminally rounded, harpe not separated from the ampulla. In DNA, a combination of the following base pairs is diagnostic in the nuclear genome: aly1449.3.1:G59C, aly1603.21.2:C96T, aly145.9.4:G1015A, aly2487. 37.1:A115T, aly274.10.13:C109T, and COI barcode: A43T, T121C, A274T, T367C, A430G, T533C.
Etymology.
The name is a feminine noun in the nominative singular formed from the type species name Lill[i]a[e].
Species included.
Only the type species.
Parent taxon.
Genus Choranthus Scudder, 1872.
Asbolis Mabille, 1904 is a subgenus of Choranthus Scudder, 1872
Both Asbolis Mabille, 1904-[IV] (type and the only species Goniloba sandarac Herrich-Schäffer, 1865, a junior subjective synonym of Eudamus capucinus Lucas, 1857) and Pyrrhocalles Mabille, 1904-[V] (type species Pamphila antiqua Herrich-Schäffer, 1863) were regarded by Zhang et al. (2022a) as junior subjective synonyms of Choranthus Scudder, 1872 (type species Hesperia radians Lucas, 1857) due to their genetic similarities. Nevertheless, while being very closely related to each other (COI barcodes differ by 5.6%, 37 bp), contrasting with their difference in appearance, Asbolis and Pyrrhocalles show larger genetic differentiation from Choranthus in the nuclear genome (Fig. 22). Treating Asbolis combined with Pyrrhocalles as a subgenus would mean that the clades of C. borincona and C. lilliae should be considered subgenera as well, because Asbolis is closer related to Choranthus than Choranthus to C. borincona and C. lilliae. Presently, because these two clades have been defined as subgenera Borna subgen. n. and Lilla subgen. n., it is meaningful to propose that Asbolis Mabille, 1904 is a subgenus of Choranthus Scudder, 1872 rather than its synonym (Fig. 22). Then, due to genetic similarities, we treat Pyrrhocalles as a junior (by about a month) subjective synonym of Asbolis. As a result, the subgenus Asbolis consists of all taxa listed by Mielke (2005) under Pyrrhocalles and Asbolis.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Ping Chen and Ming Tang for their excellent technical assistance. We are grateful to David Grimaldi and Courtney Richenbacher (AMNH: American Museum of Natural History, New York, NY, USA), Jason Weintraub (ANSP: The Academy of Natural Sciences of Drexel University, Philadelphia, PA, USA), Blanca Huertas, David Lees, and Geoff Martin (BMNH: Natural History Museum, London, UK), Jim Fetzner, Bob Androw, Vanessa Verdecia, Cat Giles, and the late John Rawlins (CMNH: Carnegie Museum of Natural History, Pittsburgh, PA, USA), Chuck Harp, and the late Boris Kondratieff (CSUC: C.P. Gillette Museum of Arthropod Diversity, Department of Agricultural Biology, Colorado State University, Fort Collins, CO, USA), Jason Dombroskie (CUIC: Cornell University Insect Collection, Ithaca, New York, USA), Crystal Maier and Rebekah Baquiran (FMNH: Field Museum of Natural History, Chicago, IL, USA), Weiping Xie (LACM: Los Angeles County Museum of Natural History, Los Angeles, CA, USA), Théo Léger, Wolfram Mey, and Viola Richter (MFNB: Museum für Naturkunde, Berlin, Germany), Andrei Sourakov, Andrew D. Warren, Debbie Matthews-Lott, Riley J. Gott, and Keith R. Willmott (MGCL: McGuire Center for Lepidoptera and Biodiversity, Gainesville, FL, USA), Rodolphe Rougerie (MNHP: Muséum National d’Histoire Naturelle, Paris, France), Matthias Nuss (MTD: Museum für Tierkunde, Dresden, Germany), Gerardo Lamas (MUSM: Museo de Historia Natural, Lima, Peru), Rob de Vos (RMNH: Naturalis Biodiversity Center, Leiden, Netherlands), Edward G. Riley, Karen Wright, and John Oswald (TAMU: Texas A&M University Insect Collection, College Station, TX, USA), Alex Wild (TMMC: University of Texas Biodiversity Center, Austin, TX, USA), Jeff Smith and Lynn Kimsey (UCDC: Bohart Museum of Entomology, University of California, Davis, CA, USA), Robert K. Robbins, John M. Burns, and Brian Harris (USNM: National Museum of Natural History, Smithsonian Institution, Washington, DC, USA), Axel Hausmann, Andreas Segerer, and Ulf Buchsbaum (ZSMC: Zoologische Staatssammlung München, Germany), for granting access to the collections under their care, sampling specimens, and stimulating discussions; to Bill R. Dempwolf, Howard Grisham, Crispin S. Guppy, Robb Hannawacker, Bernard Hermier, Steve Kohler, Kiyoshi Maruyama, and Mark Walker for specimens and leg samples, to Bernard Hermier and Jonathan Pelham for critical review of the manuscript and discussions. Evi Buckner-Opler assisted by providing emotional and logistic support and helped to collect specimens. We are indebted to the California Department of Fish and Game for collecting permit SC13645, Texas Parks and Wildlife Department (Natural Resources Program Director David H. Riskind) for the research permit 08-02Rev, to U. S. National Park Service for the research permits: Big Bend (Raymond Skiles) for BIBE-2004-SCI-0011 and Yellowstone (Erik Oberg and Annie Carlson) for YELL-2017-SCI-7076, and to the National Environment & Planning Agency of Jamaica for the permission to collect specimens. Please note that photographs from iNaturalist (2022) reproduced in this work and photographs ©The Trustees of the Natural History Museum, London are made available under Creative Commons License 4.0 (https://creativecommons.org/licenses/by/4.0/), which means in particular that when using the images you must give appropriate credit and provide a link to the license. We acknowledge the Texas Advanced Computing Center (TACC) at The University of Texas at Austin for providing HPC resources. This study was supported in part by the HHMI Investigator funds and by grants from the National Institutes of Health GM127390 and the Welch Foundation I-1505.
Footnotes
ZooBank registration: http://zoobank.org/0782F3E3-1EC3-4CB5-B548-1F9DCF6C7917
LITERATURE CITED
- Ballmer GR 2022. Life History and Ecology of the San Emigdio Blue Butterfly (Lepidoptera: Lycaenidae). The Taxonomic Report of the International Lepidoptera Survey 10(9): 1–16. [Google Scholar]
- Cong Q, Barbosa EP, Marín MA, Freitas AVL, Lamas G, and Grishin NV. 2021. Two new species of Hermeuptychia from North America and three neotype designations (Nymphalidae: Satyrinae). The Taxonomic Report of the International Lepidoptera Survey 9(7): 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Q, Shen J, Borek D, Robbins RK, Opler PA, Otwinowski Z, and Grishin NV. 2017. When COI barcodes deceive: complete genomes reveal introgression in hairstreaks. Proceedings of the Royal Society B: Biological Sciences 284(1848): 20161735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Q, Zhang J, and Grishin NV. 2019a. Genomic determinants of speciation. bioRxiv BIORXIV/2019/837666. [Google Scholar]
- Cong Q, Zhang J, Shen J, and Grishin NV. 2019b. Fifty new genera of Hesperiidae (Lepidoptera). Insecta Mundi 0731: 1–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans WH 1952. A catalogue of the American Hesperiidae indicating the classification and nomenclature adopted in the British Museum (Natural History). Part II. Pyrginae. Section I. The Trustees of the British Museum (Natural History); London. v + 178 pp., pls. 10–25. [Google Scholar]
- Evans WH 1955. A catalogue of the American Hesperiidae indicating the classification and nomenclature adopted in the British Museum (Natural History). Part IV. Hesperiinae and Megathyminae. The Trustees of the British Museum (Natural History); London. v + 499 pp., pls. 54–88. [Google Scholar]
- Farfán J, Cerdeña J, Huanca-Mamani W, Vargas HA, Gonçalves GL, and Moreira GRP. 2022a. Host plant variation and lack of genetic differentiation in populations of Dione (Agraulis) dodona Lamas & Farfan (Lepidoptera: Nymphalidae). Insects 13(819): 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farfán J, Cerdeña J, Vargas HA, Gonçalves GL, Lamas G, and Moreira GRP. 2022b. A peculiar new species of Dione (Agraulis) Boisduval & Le Conte (Lepidoptera: Nymphalidae: Heliconiinae) associated with Malesherbia Ruiz & Pavón (Passifloraceae) in xeric western slopes of the Andes. Zookeys 1113: 199–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godman FD, and Salvin O. 1894. Pl. 80 in Biologia Centrali-Americana. Insecta. Lepidoptera-Rhopalocera. Dulau & Co., Bernard Quaritch; London. v. 3 (plates). [Google Scholar]
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, and Vinh LS. 2018. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Molecular Biology and Evolution 35(2): 518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- iNaturalist. 2022. A Community for Naturalists – iNaturalist. Available from https://www.inaturalist.org. Accessed 30 January 2023.
- Li W, Cong Q, Shen J, Zhang J, Hallwachs W, Janzen DH, and Grishin NV. 2019. Genomes of skipper butterflies reveal extensive convergence of wing patterns. Proceedings of the National Academy of Sciences of the United States of America 116(13): 6232–6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukhtanov VA, and Gagarina AV. 2022. Molecular Phylogeny and Taxonomy of the Butterfly Subtribe Scolitantidina with Special Focus on the Genera Pseudophilotes, Glaucopsyche and Iolana (Lepidoptera, Lycaenidae). Insects 13(12): 1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielke OHH 2005. Catalogue of the American Hesperioidea: Hesperiidae (Lepidoptera). Sociedade Brasileira de Zoologia; Curitiba, Paraná, Brazil. xiii + 1536 pp. [Google Scholar]
- Mielke OHH, and Casagrande MM. 2002. Notas taxonômicas em Hesperiidae neotropicais, com descrições de novos taxa (Lepidoptera). Revista brasileira de Zoologia 19(Suplemento 1): 27–76. [Google Scholar]
- Nabokov V 1945. Notes on the Morphology of the Genus Lycæides (Lycænidæ, Lepidoptera). Psyche: A Journal of Entomology 51(3/4): 104–138. [Google Scholar]
- Núñez R, Willmott KR, Álvarez Y, Genaro JA, Pérez-Asso AR, Quejereta M, Turner T, Miller JY, Brévignon C, Lamas G, and Hausmann A. 2022. Integrative taxonomy clarifies species limits in the hitherto monotypic passion-vine butterfly genera Agraulis and Dryas (Lepidoptera, Nymphalidae, Heliconiinae). Systematic Entomology 47(1): 152–178. [Google Scholar]
- Pelham JP 2022. Catalogue of the Butterflies of the United States and Canada. Revised 2 February 2022. <http://www.butterfliesofamerica.com/US-Can-Cat.htm> Accessed 30 January 2023.
- Penz CM 2022. Reinstated status of the butterfly genus Agraulis (Lepidoptera, Nymphalidae, Heliconiinae). Zootaxa 5209(3): 394–398. [DOI] [PubMed] [Google Scholar]
- Scott JA 1986. The Butterflies of North America: A Natural History and Field Guide. Standford University Press; Stanford, CA. xiii + 583 pp [Google Scholar]
- Shen J, Cong Q, Borek D, Otwinowski Z, and Grishin NV. 2017. Complete genome of Achalarus lyciades, the first representative of the Eudaminae subfamily of skippers. Current Genomics 18(4): 366–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stresemann E 1954. Ferdinand Deppe’s travels in Mexico, 1824–1829. Condor 56(2): 86–92. [Google Scholar]
- Talavera G, Lukhtanov VA, Pierce NE, and Vila R. 2012. Establishing criteria for higher-level classification using molecular data: the systematics of Polyommatus blue butterflies (Lepidoptera, Lycaenidae). Cladistics 29: 166–192. [DOI] [PubMed] [Google Scholar]
- Xue G, Qu P, Li M, Chiba H, and Li W. 2022. Molecular and morphological evidences confirm the statuses of Ancistroides othonias (Hewitson, 1878) and the subspecies of A. nigrita (Latreille, [1824]), sensu Evans 1949 (Lepidoptera, Hesperiidae). Zootaxa 5205(5): 481–490. [DOI] [PubMed] [Google Scholar]
- Zakharov EV, Lobo NF, Nowak C, and Hellmann JJ. 2009. Introgression as a likely cause of mtDNA paraphyly in two allopatric skippers (Lepidoptera: Hesperiidae). Heredity (Edinb) 102(6): 590–599. [DOI] [PubMed] [Google Scholar]
- Zhang J, Cong Q, and Grishin NV. 2023. Thirteen new species of butterflies (Lepidoptera: Hesperiidae) from Texas. Insecta Mundi 0969: 1–58. [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Shen J, and Grishin NV. 2022a. Taxonomic changes suggested by the genomic analysis of Hesperiidae (Lepidoptera). Insecta Mundi 0921: 1–135. [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Shen J, Opler PA, and Grishin NV. 2019. Changes to North American butterfly names. The Taxonomic Report of the International Lepidoptera Survey 8(2): 1–11. [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Shen J, Opler PA, and Grishin NV. 2020. Genomic evidence suggests further changes of butterfly names. The Taxonomic Report of the International Lepidoptera Survey 8(7): 1–40. [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Shen J, Opler PA, and Grishin NV. 2021. Genomics-guided refinement of butterfly taxonomy. The Taxonomic Report of the International Lepidoptera Survey 9(3): 1–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Shen J, Song L, Gott, J. R, Boyer P, Guppy CS, Kohler S, Lamas G, Opler PA, and Grishin NV. 2022b. Taxonomic discoveries enabled by genomic analysis of butterflies. The Taxonomic Report of the International Lepidoptera Survey 10(7): 1–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Shen J, Song L, and Grishin NV. 2022c. Genomic DNA sequencing reveals two new North American species of Staphylus (Hesperiidae: Pyrginae: Carcharodini). The Taxonomic Report of the International Lepidoptera Survey 10(4): 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


