Abstract
Little is known about three-dimensional (3D) genome organization in skeletal muscle stem cells [also called satellite cells (SCs)]. Here, we comprehensively map the 3D genome topology reorganization during mouse SC lineage progression. Specifically, rewiring at the compartment level is most pronounced when SCs become activated. Marked loss in topologically associating domain (TAD) border insulation and chromatin looping also occurs during early activation process. Meanwhile, TADs can form TAD clusters and super-enhancer–containing TAD clusters orchestrate stage-specific gene expression. Furthermore, we uncover that transcription factor PAX7 is pivotal in enhancer-promoter (E-P) loop formation. We also identify cis-regulatory elements that are crucial for local chromatin organization at Pax7 locus and Pax7 expression. Lastly, we unveil that geriatric SC displays a prominent gain in long-range contacts and loss of TAD border insulation. Together, our results uncover that 3D chromatin extensively reorganizes at multiple architectural levels and underpins the transcriptome remodeling during SC lineage development and SC aging.
3D genome rewires extensively in adult skeletal muscle stem cells during lineage development and aging.
INTRODUCTION
Skeletal muscle tissue has a remarkable regenerative capacity, which largely depends on the activation of resident muscle stem cells, named satellite cells (SCs) (1). These cells normally reside in a niche beneath the basal lamina of the myofibers in a quiescent state and are uniquely marked by the expression of paired box 7 (Pax7) protein. Upon injury, SCs rapidly activate to enter into cell cycle, undergo proliferative expansion, differentiate, and eventually fuse to form multinucleated myotube cells; these myotubes further mature into myofibers to repair the damaged muscle. A subset of SCs undergoes self-renewal and returns to the quiescent state to replenish the adult stem cell pool (2). Deregulated SC activity is linked to the development of many muscle-associated diseases, which necessitates the comprehensive understanding of the mechanisms governing SC fate transition (3). Gene regulation is the key to understand the molecular mechanisms governing SC function. For example, it is well known that PAX7 is uniquely expressed in SCs and plays multifaceted roles in regulating SC quiescence, proliferation, and self-renewal (4). PAX7 protein is abundantly enriched in quiescent SCs but gradually diminishes as cells become activated, proliferating, and committed to terminal differentiation. However, the regulatory mechanisms underpinning the temporal expression of Pax7 are largely elusive.
Recent advents in mapping genome-wide three-dimensional (3D) chromosomal interactions have led to unprecedented understanding of hierarchical organization of the genome within the 3D nucleus and how gene expression is orchestrated at the 3D level (5–9). At intermediate scales (200 kb to 1 Mb), topologically associating domains (TADs) are identified from chromosome contact maps, which correspond to sequences that interact with each other more frequently within the same TAD rather than with sequences outside the TAD. In mammalians, TAD borders are often demarcated by CCCTC-binding factor (CTCF) and cohesin complex; perturbation of TAD borders is emerging as a driver of many human diseases (10, 11). At larger scales, interactions between TADs are thought to form two main types of compartments termed A and B compartments, which are associated with distant chromatin modifications and gene expression. In contrast to TADs, which are largely conserved among different cell types and species, compartments can display high variability. Recent studies also reveal that noncontiguous TAD clusters can act in concert to organize genome topology (12–14). Whereas long-range TAD interactions are enriched in B compartments and correlate with gene repression (12), super-enhancer (SE)–containing and highly transcribed TADs can also form clusters that span tens of megabases (13). When high-resolution Hi-C data are available, chromatin loops can be further identified in Hi-C contact matrix. In general, two types of chromatin loops are revealed: One is often invariant across cell types and demarcates TAD borders, which is often formed by CTCF and cohesin complex; the other is more cell type specific and is often associated with enhancers (15). Locus-specific and genome-wide studies have established an indispensable role of CTCF and cohesin complex in the formation of chromatin loops between CTCF sites (16–18). However, a substantial number of chromatin loops and TAD boundaries are unaffected upon CTCF removal, suggesting that additional CTCF-independent mechanisms may contribute to chromosome folding and gene regulation. For instance, oligomerization of a ubiquitously expressed transcription factor (TF), Yin Yang 1 (YY1), can instruct specific enhancer-promoter (E-P) looping (19). Similarly, myogenic differentiation 1 (MyoD1), a master TF, orchestrates muscle lineage-specific 3D genome organization (20). In addition, PAX7 is shown to shape 3D genome configuration in muscle progenitor specification and myogenic differentiation (21).
Recent findings suggest that the 3D genome is commonly reorganized during stem cell lineage progression and differentiation to ensure the spatiotemporal expression of lineage-specific genes (5, 8, 22). For example, during mouse embryonic stem cell (mESC) differentiation into cortical neurons, interactions between B compartments increase with a concomitant decrease of interactions within A compartments; E-P contacts are established concomitantly with changes in gene expression (5). Despite the rapidly gained understanding of 3D genome rewiring during stem cell differentiation, the knowledge gaps remain to be filled in SCs. In particular, our knowledge on the molecular events governing early steps of quiescence maintenance and activation of SCs is scarce because of the difficulty in preserving SCs in its native niche environment (23, 24). The lengthy dissociation steps expose cells to stress and disrupt the physiological niche, which alters SC transcriptome and histone modifications inevitably; fixation before isolation has been shown to preserve the quiescent status of SCs (23, 25), making it possible to investigate whether the 3D genome organization remodels in the early stages of SC activities and whether the 3D rewiring dictates transcriptional changes.
Alterations at 3D levels are associated with aging in many tissues and cells (26–29). Skeletal muscle suffers from sarcopenia, i.e., functional and mass decline, during human aging; this can partly be attributed to the decrease in the number and function of SCs (1, 30, 31). Elucidating the molecular causes underlying SC aging thus has become the key to better understand sarcopenia and uncover approaches for managing and treating muscle weakening conditions during physiological aging. Research from recent years demonstrates that the SC dysfunction during the aging process can be ascribed to changes in transcriptional networks and chromatin states (30, 32, 33). Yet, it is still unknown whether 3D deregulation occurs in aging SCs and drives the aging process.
Here, we map the 3D genome reorganization during SC lineage progression. We reveal that the 3D chromatin extensively reorganizes at all scales including compartment, TAD, TAD cluster, and chromatin looping levels. Our results demonstrate dynamic chromatin rewiring at different genomic scales, which orchestrates the transcriptome remodeling and SC activities. Mechanistic investigation uncovers CTCF and PAX7 regulation of chromatin loop remodeling in the transition from quiescence to early activation phase. Furthermore, in-depth analysis at Pax7 locus uncovers entangling E-P connections that establish a unique Pax7-residing TAD and coordinate its expression dynamics during SC activation. Deletion of these regulatory elements (REs) disrupts 3D genome organization at Pax7 locus and attenuates high Pax7 expression in quiescent SC (QSC). Last, we map 3D remolding in SCs isolated from physiologically aging mice and uncover that 3D genome rewiring occurs in aging SCs. Together, our findings provide a comprehensive view of 3D genome remodeling during SC lineage progression and SC aging.
RESULTS
Genome compartmentalization is globally reorganized and associated with gene expression during SC lineage development
To delineate the remodeling in genome architecture during SC lineage progression, SCs at various stages were prepared (Fig. 1A and fig. S1A). We collected freshly isolated SCs (FISCs) from Pax7-nGFP mice (34) by fluorescence-activated cell sorting (FACS) with nuclear green fluorescent protein (GFP) signal; on the basis of a prior report (23), these cells were early activating due to the niche disruption by the isolation process and were accompanied by changes in transcriptome and histone modifications. To obtain QSCs, we performed in situ fixation by paraformaldehyde (PFA) before FACS isolation and this is known to preserve the quiescence status of SCs (23). Immunofluorescence (IF) staining for PAX7 protein confirmed the purity of the FISCs (91 ± 4.7% positive for PAX7) and QSCs (94 ± 8.7% positive for PAX7) (fig. S1B). FISCs were then cultured in vitro for 1, 2, or 3 days to become activated SCs (ASC-D1), proliferating cells (ASC-D2), or early differentiating cells (DSC-D3) (Fig. 1A). We then probed 3D genome organization in the above cells by in situ Hi-C with four to seven biological replicates per sample (Fig. 1A and fig. S1C). Following the standard data analysis pipeline using HiC-Pro (35), we obtained a total of more than ~2 billion uniquely aligned contacts from all samples (table S1). The first principal component (PC1) values of the biological replicates of each time point were highly correlated (R ≥ 0.95) (fig. S1C). We observed a gradual decrease in PC1 correlation (fig. S1D) as cells progressed from QSC to DSC-D3, suggesting that changes in 3D genome organization were highly reproducible and cumulative during the SC lineage development. We thus merged the data from the above biological replicates for calculation of cis/trans interaction ratios; as a result, more than 70% of cis interactions were observed across all time points (fig. S1E and table S1), reinforcing the high quality of libraries (36). We obtained a resolution of 5 kb for QSC and FISC, and 10 kb for D1, D2, and D3 after in-depth sequencing, which rendered us to explore the 3D genome architecture at multiscales, including compartments, TADs, and chromatin loops (Fig. 1A).
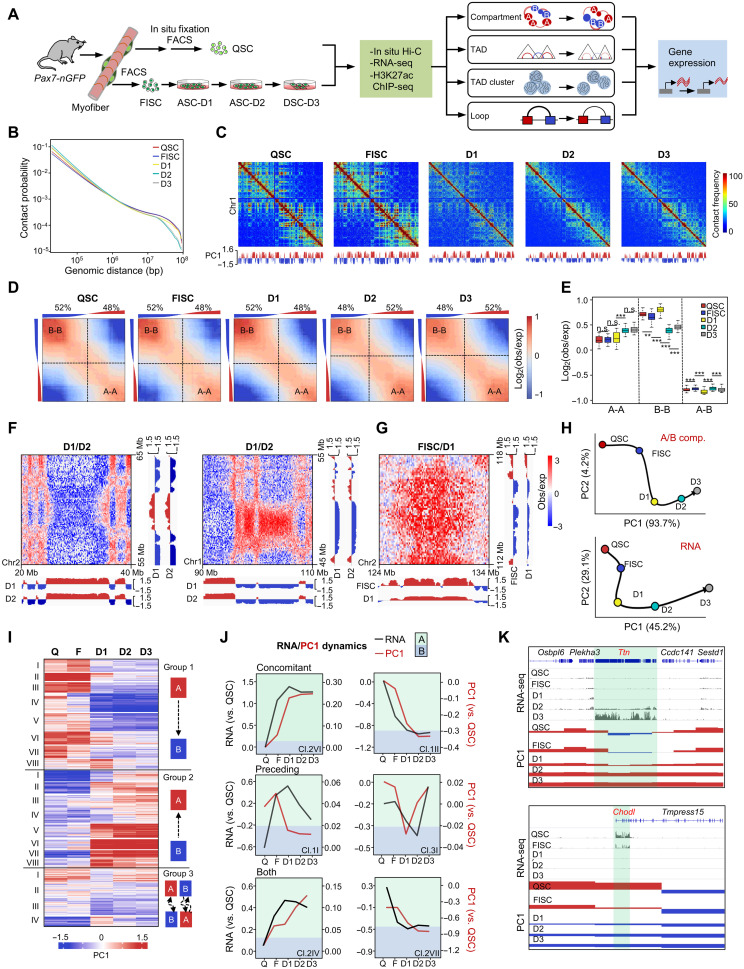
Fig. 1. Genome compartmentalization is globally reorganized and associated with gene expression during SC lineage development.
(A) Schematic overview of the study design. SCs were isolated from 2-month-old Pax7-nGFP mouse with or without in situ fixation to obtain QSCs or FISCs, respectively. FISCs were cultured in vitro for various times before collection for in situ Hi-C and RNA-seq. RNA-seq in QSC and H3K27Ac ChIP-seq data in QSC and FISC are from (23). (B) Contact probability relative to genomic distance. (C) Observed contact matrices for chromosome 1 at 500-kb resolution and the first eigenvector at 100-kb resolution. (D) Saddle plots showing the genome-wide compartmentalization. Percentage of compartment A/B and PC1 value of each 100-kb loci are shown on top. (E) Contact enrichment of 100-kb loci between compartments of the same and different types. **P < 0.001 and ***P < 0.0001; n.s., no significance, Wilcoxon signed-rank test. (F) Differential heatmaps for 10-Mb region on chromosome 2 showing increased A-A interactions (left) and decreased B-B interactions in D2 versus D1 (right). (G) Differential heatmap for 10-Mb region on chromosome 1 showing decreased A-B interactions in D1 versus FISC. (H) PCA of PC1 values and gene expression dynamics. Black arrows denote the hypothetical trajectory. (I) k-means clustering (k = 20) of PC1 values for 100-kb genomic bins that switch compartment at any time point. (J) Examples of individual switching clusters with concomitant PC1 and gene expression changes (VI/group 2 and II/group 1), PC1 changes preceding expression changes (I/group 1 and I/group 3), or clusters with both occurring (IV/group 2 and VII/group 2). (K) Genome browser view of Osbpl6-Ttn-Sestd1 (top) and Chodl-Tmpress15 (bottom) loci. PC1 and RNA-seq values at each stage are shown. Green shadings denote B-A switch (top) and A-B switch (bottom) regions.
Overall, comparing the later (D2 and D3) with the early (QSC, FISC, and D1) stages, we observed a marked decrease in the frequency of long-range interactions (>2 Mb) but an increase in the short-range interactions (<2 Mb) on all chromosomes (Fig. 1, B and C, and fig. S1F). We then determined genome segregation into A and B compartments (table S2). As expected, A-type compartments were associated with active histone marks, H3K4me3 and H3K27ac (23), which were not present in B-type compartments (fig. S1G). Although the overall proportions assigned to A (47 to 50%) and B (50 to 53%) compartments remained stable throughout the entire lineage progression (fig. S1H), compartmentalization strength [as measured by the average contact enrichment within (A-A and B-B) and between (A-B) compartments] was dynamically altered. We noted a more condensed chromatin state of compartment B at earlier stages (QSC, FISC, and D1) with a larger proportion of B-B interactions compared to later stages (D2 and D3) (52% versus 48%) (Fig. 1D) and a significantly higher interaction strength (Fig. 1E); the proportion of A-A interactions and interaction strength, on the contrary, markedly increased at later stages (Fig. 1, D and E). For instance, we observed a gain of A-A and a reduction of B-B contact frequencies in D2 versus D1 (Fig. 1F), and A-B compartment segregation was enhanced at D1 versus FISC as revealed by reduced intercompartment A-B contacts (Fig. 1G). We then examined switching of loci between A and B compartments and found that only 12% of the genome switched compartment at any two time points, with B to A and A to B each accounting for 6% (fig. S1I). Thirty-two percent of these regions were involved in multiple switching events (fig. S1I), and the switched genomic regions exhibited the most substantial PC1 changes (fig. S1J), which is consistent with previous findings (7).
Next, harnessing the existing transcriptomic profiles from QSC (23) and the in house–generated RNA sequencing (RNA-seq) from FISC and D1, D2, and D3 cells, we examined the interplay between compartmentalization and gene expression changes (table S3). Consistent with the lineage progression timing, we observed high expression of Pax7 in QSCs, but its level sharply declined in early activating FISCs; MyoD1 was absent in QSCs but highly induced in FISCs and the expression was maintained at later stages; MyoG was only induced when SCs initiated differentiation at D3; Heyl was decreased, while Egr1 was induced in FISC versus QSCs (fig. S2A). Overall, the trajectory of genome compartmentalization demonstrated that a distinct change occurred at the transition of QSC to FISC compared with transcriptomic trajectory (Fig. 1H). We then performed k-means clustering based on the PC1 values of the 20% of the genes (n = 2605) with compartment switching (table S2). We identified a total of 20 clusters with a broad range of switching dynamics (Fig. 1I and fig. S2B). Six of them (1II, 1III, 1VII, 2VI, 2VIII, and 3IV) demonstrated concomitant changes in compartmentalization and gene expression (R > 0.8; Fig. 1J), but this correlation was lost at least at one time point in other clusters (fig. S2B). Specifically, 7 of the 20 clusters displayed compartment changes preceding transcriptional changes, and these clusters involved half of the genes with compartment switching (fig. S2B, cluster 1I). Only two clusters displayed compartment changes lagging behind gene expression changes and five clusters displayed both preceding and lagging relationships. For example, the Ttn locus, which encodes a contractile and sarcomere structural protein (37), was embedded in B compartment in QSC and switched from B to A at D1 stage and remained in A compartment at D3; transcription activation of Ttn, on the other hand, was initiated at D2 and strongly enhanced at D3 (Fig. 1K). In another example, Chodl, recently identified as a QSC signature gene (23), showed a sharp expression decrease in FISC versus QSC and became completely undetectable at later stages (D1 to D3). Accordingly, we noticed a gradual transition from A to B compartment at Chodl locus from QSC to FISC, and B-compartment association was enhanced at D1 to D3 stages (Fig. 1K). Furthermore, Gene Ontology (GO) analyses demonstrated that genes in all the above 20 clusters undergoing compartment switch were associated with metabolic functions (negative regulation of DNA demethylation, regulation of the apoptotic process, protein auto phosphorylation, etc.), secretory functions (extracellular exosome, etc.), and developmental processes (cell division and cell cycle, positive regulation of fibroblast proliferation, cell development, cell differentiation, etc.) (fig. S2C and table S3). Together, the above findings demonstrate that genome compartmentalization is dynamically rewired during SC lineage progression and most pronounced when cells become activated; compartmentation remodeling proceeds expression changes in a substantial number of genes.
TADs are dynamically rewired during SC lineage development and impinge on gene expression
Next, we examined TAD organization during SC lineage development. First, we identified an average of ~3000 to 3400 borders per time point by using TopDom (38) (Fig. 2A and table S4). Partitioning of the genome into TADs was largely stable during the lineage progression: More than 60% TAD borders (n = 2175) were detected at all stages (invariant borders; Fig. 2A); a comparable number of TAD borders were stably acquired (709 of 4991, 14.2%) or lost (782 of 4991, 15.7%). As a result, we detected a steady number of borders and conserved average TAD size between 720 and 760 kb (fig. S3A). Nevertheless, we observed border reorganization at the transition of each two stages. For example, 536, 97, 114, 68, and 184 TADs rearranged, merged, split, built, or disappeared at the transition from FISC to D1 (fig. S3B). We then calculated insulation score (IS) to quantitatively measure the local chromatin insulation strength by TAD borders (table S4); a low IS is correlated with high insulation strength. In line with a previous report (7), the total number of CTCF binding motif was positively correlated with the insulation strength for all borders (fig. S3C), consistent with CTCF occupancy increasing insulation strength. Quantitative measurement of the borders that were gained (fig. S3D) or lost (fig. S3E) in each stage relative to QSC revealed that the borders gained in later stages (D1 to D3) displayed an IS decrease in the early stages (FISC and D1) (fig. S3D). Similarly, borders lost in the later stages already exhibited considerable IS increase at the beginning stages (fig. S3E). Aggregate interaction matrices also revealed a genuine loss of insulation across the TAD borders early in FISC versus QSC (Fig. 2B). IS also increased drastically in FISC versus QSC but decreased when SCs were activated, proliferated, and differentiated (D1, D2, and D3 versus FISC) (Fig. 2C). Furthermore, compared with qualitative border changes, quantitative IS changes of the invariant TADs were more evident: Hierarchical clustering identified that 52.3% (1138 of 2175) of all invariant borders showed >50% difference in between any two time points (Fig. 2D). Together, the above findings demonstrate that border reorganization occurs during the entire lineage development.
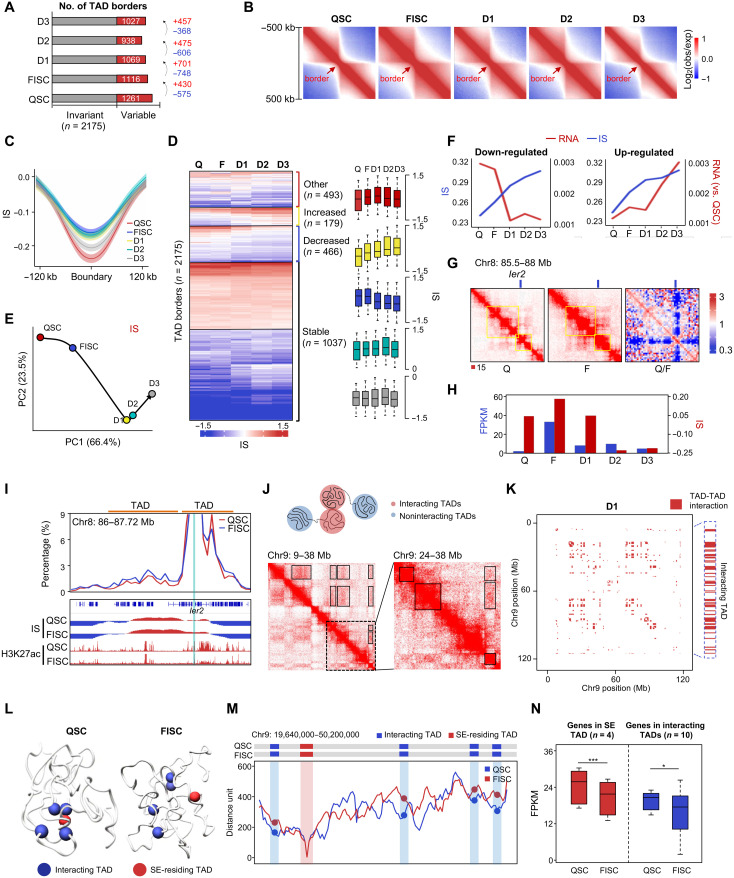
Fig. 2. Dynamic TAD organization during SC lineage development.
(A) Number of TAD borders. (B) Aggregate Hi-C maps centered on invariant TAD borders. Data are presented as the log ratio of observed and expected contacts in 40-kb bins. (C) Average IS in a 240-kb region centered on invariant TAD borders. Lines denote the mean values, while the shaded ribbons represent the 95% confidence interval. (D) k-means clustering (k = 20) of IS. Bar graphs show kinetics for groups that increased (n = 179), decreased (n = 466), transiently increased (n = 493), or did not change (n = 1037). (E) PCA of IS. Hypothetical trajectory is shown as black arrow. (F) Kinetics of expression changes at dynamic borders harboring up- or down-regulated genes with IS increased. (G) Illustration of in situ Hi-C contact maps (40-kb resolution) on the Ier2 locus. (H) IS kinetics of Ier2-residing TAD border and its expression dynamics. (I) Virtual 4C analysis on Ier2 locus. Ier2-residing and upstream TADs are indicated. Genome browser view of IS and H3K27ac ChIP-seq are shown. (J) Illustration of TAD-TAD interaction (top). A region of TAD-TAD interactions on chromosome 9 (D1) is visualized in the Hi-C data (bottom left). Boxes, TAD-TAD interactions. The dashed region is enlarged on the right. (K) Matrix of significant TAD-TAD interactions (red pixels) on chromosome 9 (D1); an example is highlighted (red pixels on the right side). (L) 3D chromatin conformation model of an SE-engaged TAD cluster. The SE-containing core TAD is marked in red, and other interacting TADs are in blue. (M) Line plot at 5-kb resolution displaying the median distance between SE-containing TAD and other interacting TADs. (N) Expression levels of genes (n = 4) in the above SE-containing TAD or in interacting TADs (n = 10). *P < 0.01 and ***P < 0.0001, Wilcoxon signed-rank test.
To further investigate whether TAD reprogramming is correlated with the local gene expression, we found that principal components analysis (PCA) of IS kinetics (Fig. 2E) showed a progressing trajectory grossly resembling the transcriptome and the PC1 trajectories determined earlier (Fig. 1H) despite that the qualitative gain (fig. S3F) or loss (fig. S3G) of TAD borders did not correlate with overall changes of local gene expression. When examining transcriptional changes at the most dynamic 545 borders that increased (Fig. 2F) or decreased (fig. S3H) in quantitative IS measurement [∣(ISD3 − ISQSC)/ISQSC ∣ ≥2], the majority (448 of 547, 81.9%) were associated with local gene down-regulation (n = 549) or up-regulation (n = 42) (log2[fold change, FC] > 1.5 between end points) (Fig. 2F and fig. S3H). Unexpectedly, IS increase did not appear to proceed the transcriptional changes; concomitant changes were observed on many loci. For example, in QSC, the TAD surrounding Ier2 locus was insulated by a strong border with a low IS; the IS considerably increased in FISC, resulting in a merge with an upstream TAD (Fig. 2G), and the transcriptional level increased concomitantly (Fig. 2H). Consistently, Hi-C–derived virtual 4C also revealed increased interaction between the Ier2 TAD with the upstream one in FISC versus QSC (23.4% versus 16.3%) (Fig. 2I). As further illustrated on Heyl locus (fig. S3I), the associated TAD border weakened with increased IS (fig. S3I), which might result in the concomitant decrease of transcription from QSC to FISC (fig. S3J).
Next, to determine the self-contacting propensity within a given TAD, we calculated a domain score (DS) to measure the portion of intra-TAD interactions in all cis interactions (table S4). As previously noted (7), DSs were positively associated with gene expressions (fig. S4A). Hierarchical clustering revealed that 72% (n = 599) of the invariant TADs displayed DS change (>20% difference between each two time points) (fig. S4B). In addition, PCA of DS kinetics revealed a trajectory (fig. S4C) resembling those of compartmentalization, transcription, and IS. Overall, The DS kinetics correlated closely with compartmentalization (PC1) changes (R > 0.84; fig. S4D) and gene expression changes (R > 0.83, fig. S4D). For example, the DS of the Egr2 TAD increased in FISC versus QSC, which corresponded to concomitant transcriptional activation of Egr2 (fig. S4E). On Abtb1 locus, on the other hand, the DS decrease was accompanied by transcriptional repression in FISC versus QSC (fig. S4E). Together, the above results suggest that the 3D genome is dynamically rewired at the TAD level and impinges on gene expression during SC lineage development.
TAD clusters are identified and dynamically reorganized in FISC versus QSC
Increasing evidence underscores the importance of large-scale inter-TAD associations that position chromatin domains in the nucleus (22, 39, 40). For example, Paulsen et al. (12) reported a level of genome organization defined as TAD cliques that involve the assemblies of linearly noncontiguous TADs through long-range TAD-TAD interactions. Therefore, to further explore the possible organization of inter-TAD interactions during SC lineage progression, we examined our Hi-C contact matrices and uncovered that TADs engaged in noncontiguous contacts (Fig. 2J, exemplified on chromosome 9 at D1 stage). Using noncentral hypergeometric (NCHG) model, we identified 3639, 3667, 2928, 1843, and 2177 notable intrachromosomal TAD-TAD interactions from QSC to D3 (table S5) and exemplified on chromosome 9 at D1 stage (Fig. 2K). The interaction distance between TADs remained constant from QSC to FISC to D1 (28.6, 28.8, and 29.2 Mb) but decreased starting D2 (19.8 Mb) (fig. S5A). Next, we defined TAD clusters, which encompassed at least three TADs that were fully connected pairwise regardless of interaction distance (fig. S5B). As a result, a large number of TADs (QSC to D3: 719, 563, 385, 261, and 285) were engaged in clusters containing 3 to 11 TADs, representing 31 to 87% of all TADs (fig. S5C). We noted a reduced number of TADs in the clusters resulting in smaller clusters as cells progressed from QSC to D3 (fig. S5C). Although 65% of TAD clusters were maintained during the lineage progression, hundreds of TADs assembled into or disassembled from clusters; waves of TAD cluster formation and dissociation were observed in FISC, D2, and D3, resulting in variation of cluster size (fig. S5D). Previous findings indicated that TAD cliques are associated with repressed chromatin domain and mostly enriched in B compartments (12). We found that the TAD clusters defined in our study were present in both compartments A and B. As illustrated in fig. S5E, a TAD cluster (C1) residing in compartment B in QSC and FISC displayed an increase of cluster size in D1 (4 to 6); a second cluster, C3, was assembled at D1 in compartment B, while C2 residing in compartment A underwent dynamic assembly/disassembly during the entire lineage progression. Furthermore, we found that the interaction distance between pairwise TAD-TAD interactions in TAD cluster significantly decreased in D1 versus FISC and D2 versus D1 (fig. S5F). TADs in short-range interactions (Q1 and Q2, QSC: <18.26 Mb; FISC: <17.98 Mb; D1: <18.04 Mb; D2: <5.1 Mb; D3: <10.24 Mb) were nearly equally associated with A and B compartments, but those in long-range interactions (Q3 and Q4, QSC: >18.26 Mb; FISC: >17.98 Mb; D1: >18.04 Mb; D2: >5.1 Mb; D3: >10.24 Mb) displayed a prominent enrichment toward B compartments (fig. S5G), suggesting that these long-range interacting TADs in Q3 and Q4 were reminiscent of the TAD cliques defined in the previous study (12). Next, we examined the gene expression in TAD clusters and found that genes in Q1 or Q2 were expressed at significantly higher levels than those in Q3 or Q4 or not in any clusters (fig. S5H); this is consistent with prior findings that TAD clusters involving long-range TAD-TAD interactions are associated with chromatin compaction and gene repression.
Last, because previous studies revealed that SEs are enriched in the highly interacting TAD triplets (13, 14), we wondered whether SE-containing TAD clusters are more active in long-range TAD-TAD interactions. Leveraging the H3K27ac chromatin immunoprecipitation sequencing (ChIP-seq) profiles in QSC and FISC (23), we defined SEs by ROSE algorithm (41, 42), and the remaining enhancers were automatically classified into typical enhancers (TEs). We found that 10 to 12% of TADs in Q3 and Q4 contained SEs and resided in compartment A (fig. S5I), and those SE-containing TAD clusters were associated with higher gene expression in both QSC and FISC compared with non-SE clusters (fig. S5J). Overall, in both QSC and FISC, we found that SE regions were involved in more TAD clusters (fig. S5K), and SE-containing TAD clusters were engaged in longer interaction distance (35 kb versus 18 kb) (fig. S5L) and a larger cluster size (fig. S5L) compared to those containing TEs or no enhancers. In addition, the presence of SE clusters was associated with more evident expression changes; transcription levels in SE-containing TAD clusters in both QSC (fig. S5M) and FISC (fig. S5N) displayed stage-specific expression patterns. As an example, a 3D model of one SE-containing TAD cluster in QSC was developed (Fig. 2L): This SE-containing TAD clustered with four other TADs to form a TAD cluster in QSC; it decommissioned in FISC due to the increased interacting distance between these TADs (Fig. 2M). The genes in this TAD cluster (both the SE residing and four interacting TADs) were expressed at significantly lower levels in FISC versus QSC (Fig. 2N). Together, our results demonstrate that TAD clusters are present and dynamically reorganized when SCs progress from QSC to FISC.
Chromatin loops are dynamically remodeled during SC lineage progression
Chromatin loops manifest as foci in high-resolution Hi-C contact maps. Considering that the most notable changes observed above at compartment and TAD levels occurred in the early stages (QSC to FISC), we increased the sequencing depth for the Hi-C libraries from these two time points to obtain >500 million uniquely mapped contacts (518 million valid pairs in QSC versus 581 million valid pairs in FISC), which enabled us to visualize chromatin looping events at a 5-kb resolution. Using FitHiC2 algorithm (43), we identified a total of 11,337 and 2412 chromatin loops in QSC and FISC, respectively (Fig. 3A and table S6), indicating a drastic decrease in the number of loops during SC early activation. In addition, we observed a reduction of interaction strength by aggregated peak analysis (APA) (4.16 versus 3.93) (Fig. 3A). Furthermore, we defined 2790 and 504 loops as QSC- and FISC-specific loops, which accounted for a minor proportion of all loops (24.6% for QSC and 20.1% for FISC) (Fig. 3B and table S6). Expectedly, QSC-specific loops showed a decreased APA score in FISC (1.23) versus QSC (4.12), while FISC-specific loops displayed a much higher APA score in FISC (3.57) versus QSC (1.91) (Fig. 3C). FISC-specific loops were larger than QSC-specific loops and contained fewer number of genes (fig. S6A). However, at both time points, the stage-specific loops localized mainly in compartment A (fig. S6B) and were positively correlated with gene expression changes (fig. S6C). Moreover, GO analysis revealed that genes involved in the stage-specific loops were enriched for distinct terms (fig. S6D and table S6). To validate the above results, we used two other algorithms, HICCUPS (6) and Mustache (44), and obtained similar conclusions (fig. S6, E to J).
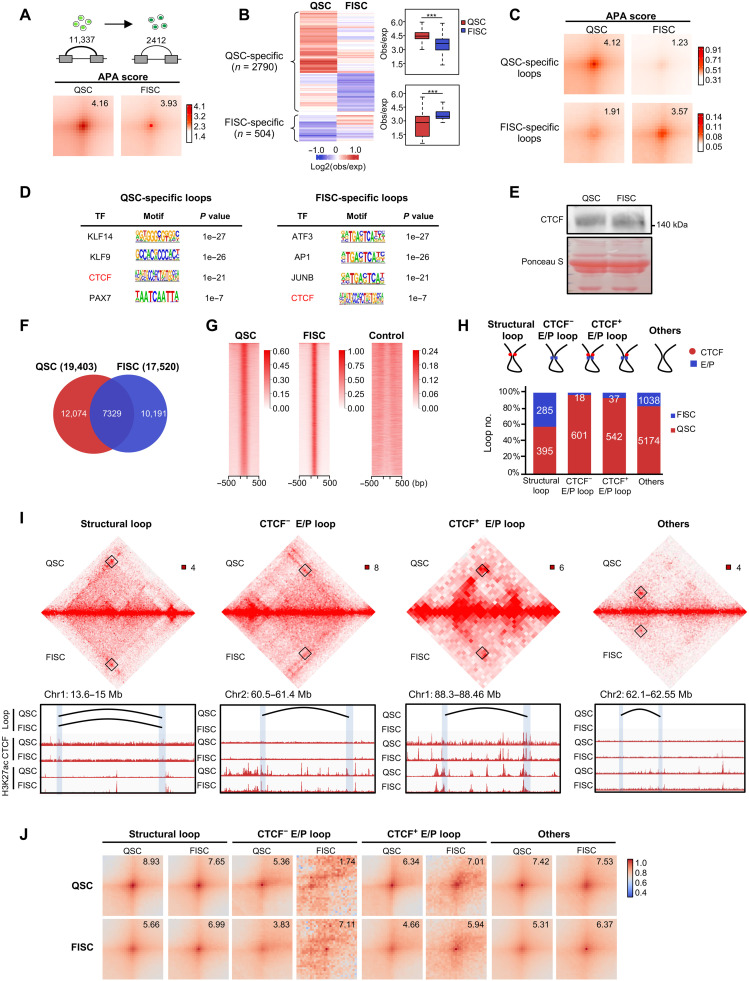
Fig. 3. Chromatin loops are dynamically remodeled during SC lineage progression.
(A) Top: Illustration of identified chromatin loops. Bottom: APA analysis of all chromatin loops detected at QSC (n = 11,337) or FISC (n = 2412). Bin size, 5 kb. (B) Heatmap (left) and boxplot (right) showing the interaction strength of stage-specific loops. ***P < 0.0001, Wilcoxon signed-rank test. (C) APA analysis of QSC- or FISC-specific loops. Bin size, 5 kb. Numbers indicate the average loop strength. (D) HOMER predicted TF motifs enriched in the anchors of QSC-specific (left) or FISC-specific (right) chromatin loops. (E) Western blot analysis of CTCF protein in QSC and FISC. Ponceau S staining is used as loading control. (F) Overlap of CTCF peaks identified in QSC (n = 19,403) and FISC (n = 17,520). A total of 7329 peaks are detected in both QSC and FISC. (G) Heatmaps showing the enrichment of CTCF binding at the identified ChIP-seq peaks in QSC and FISC. Input signals are used as a control. (H) Top: Schematic illustration of the loop classification. Loops with CTCF occupancy (red circle) at both anchors and without any cis-regulatory elements (promoters or H3K27ac for enhancers) (blue square) were defined as structural loops. Loops with cis-regulatory elements at both anchors were defined as E/P loops. Those were subdivided into CTCF− or CTCF+ E/P loops. The remaining loops were classified into “Others.” Bottom: Bar plot showing the loop number changes. (I) Hi-C contact maps showing the decrease of the above loop types in FISC versus QSC. Four representative regions are shown. Bin size, 5 kb. Genome browser tracks of chromatin loops, CTCF, and H3K27ac occupancy in QSC and FISC are shown. (J) APA analysis of each subtype of loops in QSC and FISC. Bin size, 5 kb.
To have a comprehensive view of loop remolding in the entire lineage progression, we next extended our analyses to all Hi-C datasets at a 10-kb resolution. A gain of loop number was detected at D1 (n = 10,817) after the sharp loss at FISC (n = 6487), and the number continued to increase at D2 (n = 43,502) followed by a reduction at D3 (n = 21,912) (fig. S7A). Concomitantly, the looping strength was also dynamically changed during the entire lineage development (fig. S7A). At the 10-kb resolution, we identified 2421 QSC-specific and 546 FISC-specific loops and less than 10% were present at D1 to D3 stages (fig. S7B). APA score also revealed a decrease in looping strength (fig. S7C). In addition, loops formed at later stages (D2 and D3) were larger than those from earlier stages (QSC, FISC, and D1) and contained fewer numbers of genes (fig. S7D). Stage-specific loops were positively correlated with gene expression (fig. S7E) and enriched for different sets of GO terms (fig. S7F). For instance, genes associated with D3-specific loops were enriched for “muscle cell differentiation” and “muscle tissue development” (fig. S7G). We observed that new chromatin loops formed in D2 versus D1 for Myod1 and D3 versus D2 for Myog between the gene promoters and distal regions, which may contribute to the increase of gene expression (fig. S7G). Meanwhile, on Myh1 locus, despite that an E-P loop was already established at D2, the gene expression was not detected until a stronger interaction appeared at D3 (fig. S7G). Furthermore, we performed de novo motif search within anchors of the stage-specific loops; distinct TF motifs were enriched in these loops (fig. S7H), implying their potential roles in driving the loop rewiring. For instance, YY1 motif was enriched in D2-specific loop anchors but not in D3. In line with this, YY1 ChIP-seq profiles collected in myoblasts and myotubes (45) unveiled a stronger YY1 binding at the above-predicted YY1-associated loop anchors (fig. S7I). Together, these results strengthen that chromatin loop formation is dynamically remodeled through the entire SC lineage progression.
Next, to fathom the underlying mechanism of the loop remodeling, especially the marked loss in the loop number and strength during the QSC to FISC transition, we performed de novo motif search within the QSC- and FISC-specific loop anchors, and CTCF motifs were enriched in both QSC and FISC loops (Fig. 3D). We then conducted CTCF ChIP-seq profiling to test its regulation of loop formation. Notably, CTCF protein level (Fig. 3E) remained unchanged from QSC to FISC despite mild reduction in its mRNA level (fig. S8A). Similarly, RAD21 cohesin complex component (Rad21) protein level was also comparable in QSC and FISC (fig. S8, A and B). Expectedly, we identified a comparable number of CTCF-binding peaks in QSC (n = 19,403) and FISC (n = 17,520) with a large portion (n = 7329) of overlapping (Fig. 3, F and G, and fig. S8C), despite a global increase of binding intensity (Fig. 3G and fig. S8C). In agreement with the motif analysis (Fig. 3D), CTCF binding was enriched at both QSC- and FISC-specific loop anchors (fig. S8D). Leveraging the CTCF binding profiles and the publically available H3K27ac profiles (23), we then surveyed the subtypes of 11,337 and 2412 chromatin loops identified in QSC and FISC. Among them, 2738 and 367 were demarcated with annotated promoters or H3K27ac signals at loop anchors that were defined as enahncer or promoter (E/P) loops; 1143 and 55 had enhancer or promoter mark at both anchors, while the remaining had the marks at only one anchor. We found CTCF binding at one or both anchors of 542 of 1143 loops in QSC, and the numbers sharply decreased in FISC (n = 37) (Fig. 3, H and I). In total, 601 of 1143 E/P loops in QSC were not bound by CTCF and largely disappeared in FISC (n = 18) (Fig. 3, H and I). In addition, 395 CTCF-bound loops in QSC were not marked by E/P marks, and the number also decreased to 285 in FISC; these were defined as structural loops (Fig. 3, H and I) (46). Intriguingly, our analyses also uncovered a large number of chromatin loops (n = 5174) in QSC with no CTCF binding or E/P marks, and these “others” loops also largely diminished in FISC (n = 1038) (Fig. 3, H and I). APA analyses showed that the interaction strength of structural, CTCF+ E/P, and others loops also decreased (Fig. 3J). De novo motif search at the "others" loop anchors uncovered putative binding sites for distinct TFs in QSC and FISC, indicating their potential regulatory roles in the formation of these anonymous loops independent of CTCF binding (fig. S8E).
PAX7 regulates E/P loop reorganization during SC early activation
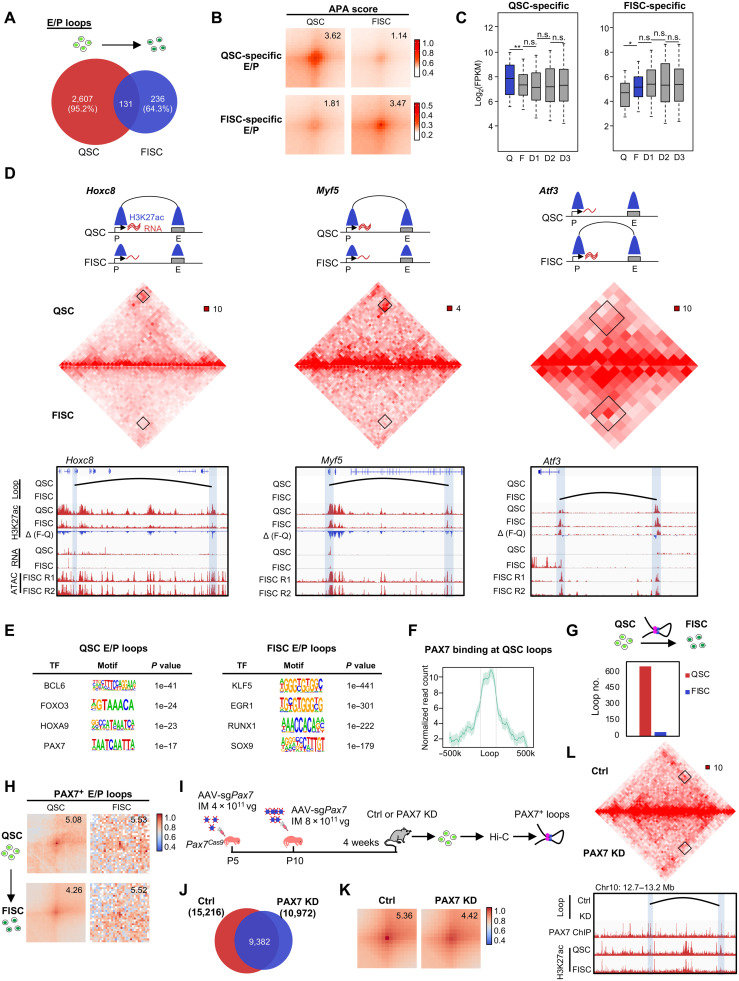
Last, we investigated the E/P loops in depth considering their importance in directing gene transcription. Of 2738 E/P loops in QSC, we found that 17% were E-P and 22% were E-E interactions; of the 367 E/P loops in FISC, 12% and 21% were E-P and E-E interactions, respectively (fig. S8F). The interaction strength of both E-P (APA: 3.73 to 3.55) and E-E loops (APA: 3.71 to 3.52) decreased in FISC versus QSC (fig. S8, G and H). Furthermore, we found that, although enhancer and promoter landscapes were largely unaltered in FISC versus QSC (10,113 shared enhancers/promoters) (fig. S8I), the loop formation was highly dynamic, with 95.2 and 64.3% specifically identified in QSC and FISC, respectively (Fig. 4A). In addition, stage-specific E-P and E-E loops also displayed more prominent difference in APA scores (1.14 versus 3.62) (Fig. 4B) and correlation with gene expression (Fig. 4, C and D).
Fig. 4. PAX7 regulates E/P loop reorganization during SC early activation.
(A) Overlapping of E/P loops between QSC and FISC. (B) APA analysis of the QSC- or FISC-specific E/P loops. (C) Box plot depicting expression dynamics of genes within QSC-specific (left, n = 1874 genes) or FISC-specific (right, n = 469 genes) loops. *P < 0.01 and **P < 0.001, Wilcoxon signed-rank test. (D) Top: Illustration of the dynamics of E-P loop formation, H3K27ac signal, and gene expression on the Hoxc8 (left), Myf5 (center), and Atf3 (right) loci. Middle: Hi-C contact maps (5-kb resolution). Bottom: E-P loops are marked in black curve. Gene promoters and interacting regions are highlighted in blue boxes. Genome browser tracks showing H3K27ac ChIP-seq and RNA-seq in QSC and FISC, and ATAC-seq of two replicates in FISC. (E) HOMER predicted TF motifs enriched in the anchors of QSC- or FISC-specific E/P loops. (F) PAX7 occupancy in the anchor regions of QSC-specific loops. (G) Number of E/P loops with PAX7 binding at the anchors in FISC versus QSC. (H) APA analysis of the PAX7+ E/P loops showing decreased interaction intensity in FISC versus QSC. (I) Schematic illustration of PAX7 in vivo knockdown (KD) in SCs by the CRISPR-Cas9/AAV9-sgRNA editing system. The mice were sacrificed for QSC collection and subjected to Hi-C analysis after 4 weeks. (J) Overlapping of chromatin loops detected in the control (n = 15,216) and PAX7 KD (n = 10,972) groups. (K) APA analysis of the PAX7+ E/P loops showing decreased interaction intensity in PAX7 KD versus control (Ctrl). (L) Hi-C contact maps of a representative region showing the diminished Pax7+ loop formation in PAX7 KD versus Ctrl. Bin size, 10 kb. Black arc denotes the loop identified in Ctrl. The genome browser tracks show PAX7 and H3K27ac occupancy.
To uncover the underlying mechanism driving E/P loop formation and reorganization, we found that TFs including B cell leukemia/lymphoma 6 (BCL6), forkhead box O3 (FOXO3), homeobox A9 (HOXA9), and PAX7 were highly enriched at the loop anchors in QSC, and Kruppel-like factor 5 (KLF5), early growth response 1 (EGR1), runt related transcription factor 1 (RUNX1), and SRY (sex determining region Y)–box 9 (SOX9) in FISC (Fig. 4E). To further investigate the possible regulatory role of PAX7 in E/P loop formation, we analyzed the publicly available PAX7 ChIP-seq profile from induced muscle progenitors (21) and found that PAX7 binding was enriched at the loop anchors of QSC-specific loops (Fig. 4F) and that the number of PAX7-bound E/P loops (PAX7+ E/P loops) sharply decreased in FISC versus QSC (Fig. 4G), which is consistent with the drastic Pax7 expression attenuation (fig. S2A). Moreover, E/P loops with PAX7 binding at either anchor showed much stronger interaction strength in QSC (Fig. 4H), suggesting that PAX7 binding may enhance the loop formation. To further elucidate the promoting role of PAX7 binding on loop formation, we deleted Pax7 in QSCs by exploiting our recently developed muscle-specific CRISPR-Cas9/adeno-associated virus (AAV)–single guide RNA (sgRNA) in vivo genome editing system (Fig. 4I) (47). Briefly, dual sgRNAs targeting the Pax7 exon 2 (fig. S8J) were applied: AAV9-sgPax7 [4 × 1011 viral genomes (vg) per mouse] was intramuscularly injected into Pax7Cas9 mouse at postnatal day 5 (P5) and followed by 8 × 1011 vg per mouse at P10. For the control group, we injected the same dose of AAV9 virus containing pAAV9-sgRNA backbone without any sgRNA insertion. We achieved a partial Pax7 knockdown (PAX7 KD) (fig. S8K), which expectedly caused decreased number of SC pool (fig. S8L) (48). We isolated QSCs from the mice 4 weeks after virus injection and performed Hi-C analysis. We found that the PAX7 KD caused a loss in the total number of chromatin loops (Fig. 4, J to L), and those E/P loops with PAX7 binding also showed notable decrease of interaction strength (Fig. 4K). Together, these results underscore that PAX7 is a key regulator of E/P loop formation and remodeling during SC early activation.
3D regulatory interactions orchestrate Pax7 expression dynamics in SC
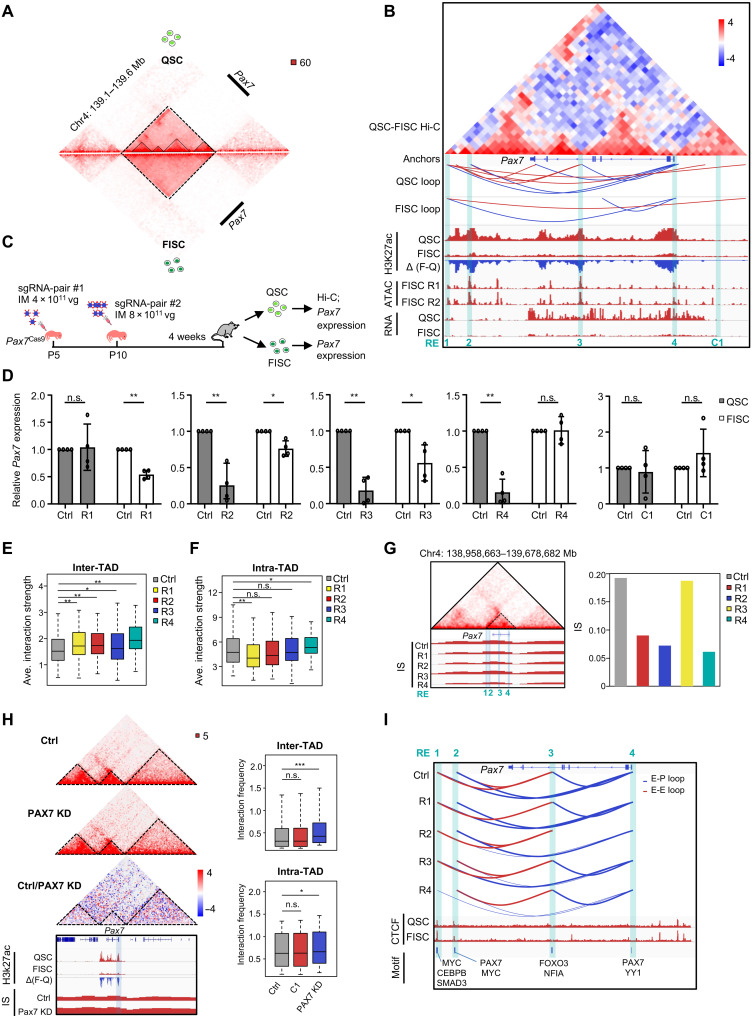
PAX7 is the key TF known to regulate the myogenic potential and function of SCs, and the mechanism dictating its high expression in QSC and attenuation in FISC (fig. S2A) remains largely unknown. We thus took a close examination at the 3D organization associated with the Pax7 locus. We identified a prominent TAD in QSC encompassing Pax7 locus only and four sub-TADs in the large Pax7 TAD (Fig. 5A). Whereas the TAD maintained constant, the four sub-TADs disappeared in FISC with a loss of interaction strength within the sub-TADs and a gain of interaction strength among them (Fig. 5, A and B). In addition, we observed a reduced number of chromatin loops in FISC (3) versus QSC (9) (Fig. 5B). The above data suggest that Pax7 locus undergoes drastic reorganization at the 3D level when SCs become activated, and this may account for its expression attenuation.
Fig. 5. 3D regulatory interactions orchestrate Pax7 expression dynamics in SC.
(A) Comparison of contact frequencies at Pax7 TAD. The dashed and solid lines indicate TAD and sub-TADs, respectively. (B) Identification of the four REs (R1 to R4) within the Pax7 TAD. Subtraction of FISC from QSC contact matrices shows changes in contact frequencies. Chromatin loops, RNA, H3K27ac, and ATAC-seq signals are shown below. (C) Schematic illustration of in vivo deletion of each RE in SCs by the CRISPR-Cas9/AAV9-sgRNA system. (D) Pax7 expression was detected by qRT-PCR. Pax7 level was normalized to 18S and presented as mean ± SD. n = 4 mice per group. *P < 0.05 and **P < 0.01, Student’s t test. (E and F) In situ Hi-C was performed in QSC from Ctrl or mutant (R1 to R4) mice. Boxplots showing inter-TAD [(E), between the Pax7 TAD and other cis-interacting TADs] or intra-TAD [(F), within Pax7 TAD] interactions. (G) Hi-C contact maps (40-kb resolution) of Pax7 TAD in control group (Ctrl, top) and IS (bottom) of Pax7 TADs in Ctrl and R1 to R4. The dashed triangle denotes Pax7 TAD. RE regions are highlighted in blue box. Bar plot showing the IS values of the above boundaries. (H) Hi-C contact maps (40-kb resolution) showing Pax7 TAD reorganization in Pax7 KD QSC in comparison with Ctrl group. Boxplots showing inter- or intra-TAD interactions. (I) E/P loops identified in QSC from Ctrl or RE mutant mice. Blue and red arcs represent E-P and E-E loops. Genome browser tracks show CTCF occupancy and predicted TF motifs enriched in each RE. Data in (E), (F), and (H) are presented in boxplots. *P < 0.01, **P < 0.001, and ***P < 0.0001, Wilcoxon signed-rank test.
To further dissect key elements governing the above-observed 3D reorganization, we identified four putative REs (R1 to R4) based on the looping interactions as well as assay for transposase-accessible chromatin using sequencing (ATAC-seq; from FISC) and H3K27ac ChIP-seq (from both QSC and FISC) signals (Fig. 5B). Among them, all four regions displayed high ATAC-seq signals. R4 resided in the Pax7 promoter region. R2 and R3 were marked with sharp H3K27ac signals in QSC, which disappeared in FISC; R1 resided in the TAD border region. We noted evident loops among R1/R4, R2/R4, and R3/R4 in QSC, but only one R1/R4 loop retained in FISC (Fig. 5B). To experimentally validate the functionality of these REs in directing the high Pax7 expression in QSC, we set to delete each in QSC by harnessing the above-described CRISPR-Cas9/AAV-sgRNA in vivo genome editing scheme (47) (Fig. 5C and fig. S9A). We also included a region located 3 kb upstream of the Pax7 promoter as a control element (C1) as it is devoid of H3K27ac and ATAC signals. The mice were sacrificed for QSC or FISC isolation and analysis 4 weeks after AAV9-sgRNA injection. We noted distinct deletion efficiency (fig. S9B): R1 and R4 manifested more efficient deletion than R2 or R3, which may be due to the inherent difference of protospacer sequences (49). Nevertheless, the deletion of R1, R2, R3, or R4 led to 0, 75, 80, and 90% decrease of Pax7 transcripts in QSC with accompanied protein reduction (fig. S9C), suggesting the individual importance of R2, R3, or R4 but not R1 in maintaining high Pax7 expression in QSC. As expected, R2, R3, or R4 deletions caused a much less pronounced Pax7 decrease in FISC (41, 25, and 0%) (Fig. 5D and fig. S9D), which is consistent with disappearance of these RE-mediated loops (Fig. 5B). As the control, C1 removal did not cause change in Pax7 expression in QSC or FISC (Fig. 5D and fig. S9, C and D).
To further determine potential effects of the RE deletion on 3D structure at the Pax7 locus, we performed in situ Hi-C in QSCs isolated from the above mice to obtain >220 million uniquely aligned contacts for downstream analyses (fig. S9E and table S7). First, comparison of the Hi-C matrices at 40- and 10-kb resolution (fig. S9F) demonstrated that the 3D conformation at the Pax7 locus from the Pax7Cas9 mice used in this experiment was comparable to that from the earlier used Pax7-nGFP mice (R = 0.85 and 0.83, respectively) (fig. S9G). Next, we found that the removal of each RE caused a substantial increase of the inter-TAD interaction strength between the Pax7 TAD and other cis-interacting TADs (Fig. 5E); the intra-Pax7 TAD interaction strength decreased significantly upon R1 loss but increased upon R4 loss and unaltered when R2 or R3 was removed (Fig. 5F). Although no obvious effect was observed on TAD organization surrounding Pax7 (fig. S9H, bottom), close examination of the IS at the boundary containing Pax7 promoter revealed a decrease upon loss of R1, R2, or R4 (Fig. 5G and table S7), indicating strengthened border insulation upon removal of these elements. Notably, R1 deletion attenuated the interaction between R1 and downstream CTCF anchor sites, but a new loop formed between the downstream and upstream CTCF anchor sites (fig. S9I). R3 deletion led to fusion of nearby sub-TADs into a new sub-TAD (fig. S9H). At the loop level, these RE-anchored intra-TAD loops were largely attenuated or even lost upon RE removal. For instance, R4 removal reduced the interaction strength among R1/R4, R2/R4, and R2/R3 (Fig. 5I, fig. S10A, and table S7). We also observed a decreased number of intrachromosomal loops anchored in Pax7 TAD upon each RE deletion (fig. S10B). Together, the above results highlight 3D rewiring at the Pax7 locus during SC activation and unveil key REs dictating the 3D remodeling and Pax7 expression.
Next, to test whether the above-observed 3D changes upon RE deletion are mediated by reduced Pax7 level, considering that PAX7 itself has 3D regulatory function (21), we reexamined the Hi-C data from Fig. 4H and found that PAX7 KD caused a significant increase of the inter-Pax7 TAD interaction strength (Fig. 5H), concordant with what was observed in RE deletion (Fig. 5E). The intra-Pax7 TAD interaction strength also increased significantly upon PAX7 KD, which resembled R4 (promoter) deletion but differed from other RE deletions (Fig. 5H). In addition, we observed a decreased insulation strength (increased IS) at the Pax7 TAD boundaries, which was also different from the RE deletions (Fig. 5H). As an alternative validation, we also collected QSCs from an inducible Pax7 knockout iKO) mouse (fig. S10D) (48), and Hi-C analysis revealed that Pax7 deletion caused a significant increase of both inter- and intra-Pax7 TAD interaction strength (fig. S10, E and F) and attenuated border insulation strength (fig. S10E). Besides the above local effect on Pax7 TAD, no evident global influence at the level of compartment, IS, and DS was detected upon deletion of each RE or Pax7 (fig. S10G).
Last, to further probe into the mechanism of RE regulation of Pax7 expression, we searched for putative TF motifs in each RE region (Fig. 5I). We identified PAX7 motifs in both R2 and R4, suggesting a possible autoregulation. Myelocytomatosis oncogene (MYC) motifs were found in both R1 and R2. Because R2 loop disappeared in FISC, it is possible that FISC-induced MYC (47) dictates Pax7 down-expression in FISC by binding to R2. In addition, we also identified motifs of FOXO3 and NFIA (nuclear factor I/A) in R3, and YY1 binding in R4. Notably, FOXO3 promotes SC to acquire quiescence during muscle regeneration by activating Notch signaling (50). YY1 is shown to repress Pax7 expression through interactions between p38α and polycomb repressive complex 2 (PRC2) on Pax7 promoter (51). More recently, NFIA is found to be highly expressed in QSC and largely diminished during QSC-FISC transition (23, 52). Thus, Pax7 regulation by these TFs can be further dissected in the future.
Multiscale analysis of 3D genome during mouse SC aging
It is well accepted that SCs decline in their regenerative capacity with aging (31), partly ascribed to the intrinsic changes at both transcriptomic and epigenomic levels. However, it is not known whether 3D genome rewiring occurs in SCs during mouse physiological aging. To fill the gap, we collected FISCs from wild-type C57BL/6 mice of 2 (young), 23 to 24 (aged), and 28 to 32 (geriatric) months of age (Fig. 6A). Consistent with prior findings (30), we detected a decrease in FISC number in the aged (3.3%) versus young (4.3%) mice and became more prominent at the geriatric stage (1.8%) (fig. S11, A and B). Transcriptional changes were profiled by RNA-seq in the above FISCs with at least two replicates from each stage. In line with previous findings (53), gene expression profiles were remarkably distinct between young and aged SCs (R = 0.74) and became more divergent in geriatric cells (R = 0.65) (fig. S11C). Notably, Pax7 expression was steadily up-regulated with SC aging, along with stress-responsive genes, Egr1 and Atf3, while Myod1 and Myc (47) were highly induced at aged followed by a decline at geriatric phase (fig. S11D). Next, results from pairwise comparison showed that 1122 genes were significantly up-regulated and 1016 were down-regulated in aged versus young SCs, whereas 910 and 1104 were in geriatric versus aged SCs (fig. S11E and table S8). Nevertheless, across the three stages, gene expression adopted distinct dynamics, with only a limited proportion of genes continuously increased (n = 120) or decreased (n = 53) (fig. S11F). Consistently, GO analysis revealed distinctly enriched terms in aged and geriatric stages (fig. S11G and table S8). For instance, the up-regulated genes in aged versus young SCs were enriched in terms such as “cell adhesion,” “inflammatory response,” and “immune response,” while those in geriatric versus aged SCs were linked with “membrane,” “cell surface,” and “extracellular space.”
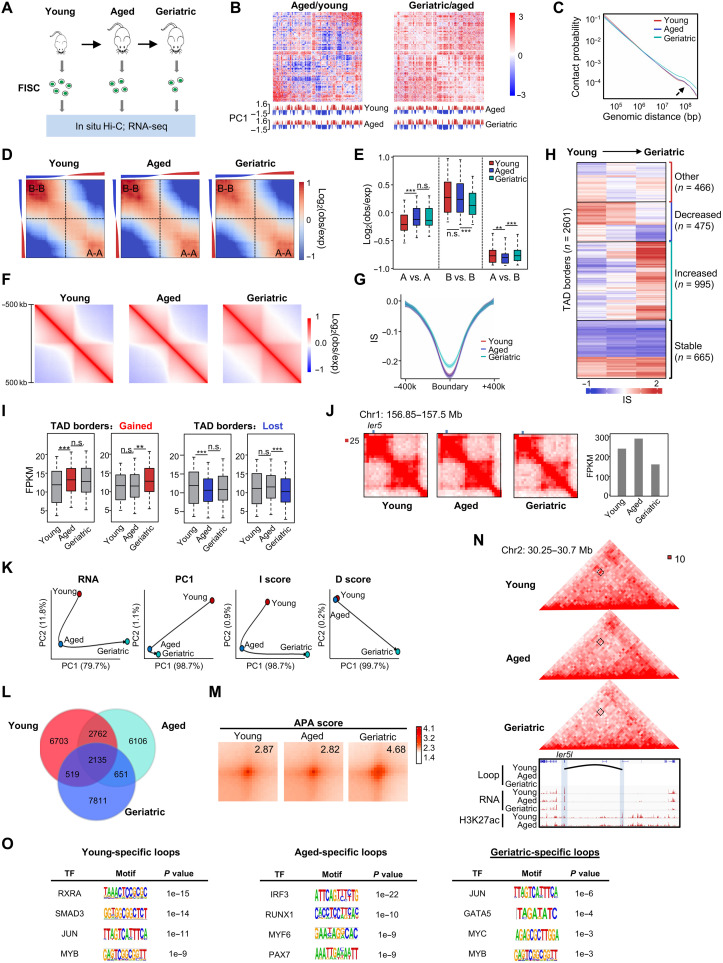
Fig. 6. Multiscale analysis of 3D genome during mouse SC aging.
(A) Schematic illustration of the aging SC study. FISCs were collected from young (2 months), aged (23 to 24 months), and geriatric (28 to 32 months) male C57BL/6 mice. (B) Differential heatmaps in aged/young (left) or geriatric/aged (right) are shown for chromosome 1. (C) Contact probability relative to genomic distance. (D) Saddle plots showing the genome-wide compartmentalization. Corners represent intracompartment (A-A/B-B) interactions. (E) Contact enrichment of 100-kb loci between compartments of the same and different types. (F) Aggregate Hi-C maps centered on invariant borders. (G) Average IS in an 800-kb region centered on invariant TAD borders. Lines denote the mean values, while the shaded ribbons represent the 95% CI. (H) k-means clustering (k = 20) of IS. Bar graphs show IS kinetics for groups that increased (n = 995), decreased (n = 475), transiently increased (n = 466), or did not change (n = 665). (I) Boxplots depicting gene expression dynamics for border regions that were gained (left) or lost (right) during SC aging. Stages that gain or lost TAD borders are indicated in blue and right, respectively. (J) Hi-C contact maps (40-kb resolution) on the Ier5 locus. The bar graphs show Ier5 expression. (K) PCA of RNA expression, PC1 values, IS, and DS. Black arrows denote the hypothetical trajectory. (L) Venn diagrams showing the overlapping of chromatin loops. (M) APA analysis of chromatin loops. Bin size, 10 kb. Numbers indicate average loop strength. (N) Hi-C contact map depicting chromatin looping between an Ier5l promoter and a putative enhancer. Dashed box and black curve denote the chromatin loop. Genome browser tracks show corresponding RNA-seq and H3K27ac signals. (O) HOMER predicted TF motifs enriched in the anchors of stage-specific loops. Data in (E) and (I) are presented in boxplots. **P < 0.001 and ***P < 0.0001, Wilcoxon signed-rank test.
Next, to delineate the changes in 3D genome organization with SC aging, we performed in situ Hi-C at the above stages with three to four replicates for each to obtain a total of >300 million uniquely aligned contacts; high correlation (R ≥ 0.95) was achieved among the replicates (fig. S12A and table S9). Overall, young SCs exhibited distinct 3D genome organization compared with aged or geriatric SCs; long-range (>3 Mb) interaction frequencies increased significantly in geriatric SCs compared with young and aged SCs (Fig. 6, B and C, and fig. S12B). The interaction strength increased between A compartments (A-A) during young to aged transition but remained constant when SCs progressed into geriatric stage; on the contrary, a significant decline in B-B interaction strength was detected during aged to geriatric progression but not young to aged transition (Fig. 6, D and E, table S9). Although the genome-wide correlation was not observed between compartmentalization and gene expression changes, we noted that the gain of A-A interactions was accompanied by an increase in the expression of several SC aging or senescence-related genes such as Il-6 and Ccnd2 (fig. S12C) (27). Overall, we found an increase in compartmentalization (decreased intercompartment contacts) at aged versus young SCs, but then reduced at geriatric stage (Fig. 6, D and E). In addition, similar to SC lineage progression (fig. S1I), we noted limited compartment switching: Only 6.5% of the genome switched compartment at any two time points with B to A and A to B, each accounting for 2.9% and 2.4%, respectively (fig. S12D). Overall, the compartmental trajectory showed drastic change at aged versus young, while transcriptomic change occurred late in geriatric stage (Fig. 6K). Whereas the lagged expression change behind the compartment switch was noticed (fig. S12E, left), concomitant changes in compartment and gene expression were also identified on some loci (fig. S12E, middle and right).
At the TAD level, we identified an average of ~3274 to 3783 TAD borders per stage (fig. S13A). Partitioning of the genome into TADs was largely stable during SC aging as >70% TAD borders (n = 2660) were detected at all stages (fig. S13A, invariant borders). The aggregate interaction matrix revealed a remarkable loss in insulation strength across the TAD borders in geriatric but not aged stage when compared with young SCs (Fig. 6F); IS calculation also showed a marked increase at geriatric stage (Fig. 6G, fig. S13B, and table S10). Furthermore, quantitative analysis of IS through hierarchical clustering also showed that a greater portion (n = 995, 38.3%) of all borders exhibited >20% increase in IS, while 18.3% (n = 475) showed a loss during SC aging (Fig. 6H). These data suggested that TAD borders largely lost insulation strength when SCs entered into geriatric state. Furthermore, we found that local gene expression underwent a significant increase or decrease when TAD borders were gained or lost, respectively (Fig. 6I), which was not observed during SC lineage progression (fig. S3, F and G). Nevertheless, in agreement with the findings in fig. S4A, DSs were positively associated with gene expression (fig. S13C) and hierarchical clustering revealed that ~47% of the TADs (n = 1754) showed DS increase or decrease (>15% difference between end points) (fig. S13D and table S10). In addition, the DS kinetics was highly correlated with changes in PC1 values (fig. S13E) or gene expression (fig. S13F) during the transition between two stages. For instance, the intra-TAD interactions surrounding Ier5 (Fig. 6J) and Hoxa10 (fig. S13G) loci were largely attenuated at geriatric stage, which might result in the concomitant decrease of transcriptional levels from aged to geriatric FISC (Fig. 6J and fig. S13G). Last, we performed PCA analysis for RNA expression, compartmentalization, IS, and DS. Transcription and IS adopted a very similar trajectory showing distinct changes from young to aged to geriatric stages; DS was largely similar between young and aged stages but underwent a drastic change from aged to geriatric stage; PC1, on the contrary, displayed the most prominent change from young to aged stage (Fig. 6K), suggesting that genome compartmentalization was altered at aged stage, while TAD connectivity changed later at geriatric stage.
Last, chromatin looping analysis at 10-kb resolution led to identification of a total of 11,180, 10,131, and 11,104 loops in young, aged, and geriatric FISCs, respectively, among which 6703, 6106, and 7811 were stage specific (Fig. 6L and table S10). We observed an increase in interaction strength of chromatin loops when SCs aged (APA score = 4.68 in geriatric versus 2.87 in young and 2.82 in aged) (Fig. 6M). Expression changes lagged behind their associated loops, as illustrated on the representative Ier5l locus (Fig. 6N). The loop between the promoter and a distal enhancer in young cells disappeared in aged cells, but a decreased gene expression did not occur until the geriatric stage. A close look at the Pax7 locus revealed the gain of a chromatin loop between R3 and R4 at geriatric stage (fig. S13I), which may account for the increased Pax7 mRNA expression. In addition, Pax7 border strength was strengthened, whereas both inter- and intra-TAD interactions were weakened during SC aging (fig. S13I). Last, to uncover putative TF regulators driving the loop remodeling during SC aging, we performed de novo motif search at stage-specific loop anchors. The distinct sets of TF binding motifs were enriched at each stage (Fig. 6O), implying their potential roles in driving loop remodeling in SC aging.
It is also interesting to note that geriatric SCs seem to favor long-range chromatin interactions more than young and aged SCs (Fig. 6C), which is similar to cultured SCs in their lineage progression (Fig. 1C). To test whether SCs are similar in other regards in the two settings, we performed PCA analyses of PC1, IS, and DS and found that their trajectories were completely distinct in the two settings (fig. S13J).
DISCUSSION
While mechanisms underlying SC lineage progression have been well studied at the transcriptional and epigenetic levels (1, 23), how genome architecture reorganization contributes to this process remains enigmatic. Here, we examined the genome conformation in SCs at distinct stages and revealed the characteristics of chromatin 3D landscape at different genomic scales and its association with altered gene expression. First, at the large scale, we identified a shift in the ratio between long- and short-range chromatin interactions and remarkable dynamic compartmentalization during SC lineage progression. The loss of long-range contacts at the late stages (D2 and D3) is probably due to the proliferative status of the cells because mitotic cells can display a rapid fall-off of chromatin contacts at ~10 Mb (9, 36); consistently, this is not observed at early stages (QSC, FISC, and D1) when SCs mostly reside in G0-G1 phases (54). In addition, SC lineage progression is accompanied with the dynamic changes in compartmentalization strength (Fig. 1, D and E), which could be resulted from the alterations in chromatin states. Previous findings have documented that the compartment strength increases concurrently with a gain of heterochromatin marks, H3K27me3 (55, 56) or H3K9me3 (57). Epigenomic profiling in SCs uncovers a loss of H3K27ac marks during SC isolation procedure (23), and a gain of H3K27me3 marks is associated with SC activation (32). Hence, it will be of great interest to further dissect the connection between the epigenomic changes with the dynamics of genome compartmentalization during SC lineage progression.
Next, at the TAD level, the most notable changes in TAD border strength occur at early activation stage (FISC). CTCF is known to play key role in chromatin insulation and TAD boundaries (16, 58), but its decrease is not observed in the process. Alternatively, CTCF-independent mechanism could contribute to the TAD border reorganization in FISC versus QSC. For example, previous observations have demonstrated that TAD boundaries are also enriched for housekeeping genes, RNA polymerase II, and active chromatin marks (5, 8); thus, active transcription could play a role in shaping the chromatin organization (59). Nevertheless, gain or loss of borders does not correlate with the overall changes in local gene expression in FISC versus QSC, arguing that transcriptional alterations per se may not be the main contributor for insulation dynamics in this process. The weakened TAD borders gain insulation strength when SCs enter differentiation, which is reminiscent of findings from neuron development (5), suggesting that TAD borders are dynamically reorganized during cellular differentiation.
Emerging studies indicate that interactions among noncontiguous TADs are prevalent (12–14). In this study, we thus defined TAD clusters as an assembly of ≥3 TADs that are fully connected pairwise using a similar approach previously used to annotate TAD cliques (12). Nevertheless, our defining approach is modified by taking into account the short-range interactions between TADs. In agreement with previous findings (12), the long-range inter-TAD interactions (Q3 and Q4) preferentially reside in B compartments and are associated with repressed gene expression, but the short-range ones (Q1 and Q2) do not display such preference. Moreover, we found that SE-containing TAD clusters are more active in long-range inter-TAD interactions. Concordantly, 3D modeling reveals that SE-containing TAD clusters reorganize in nuclear space to orchestrate the stage-specific gene expression. These findings complement recent observations that multiway interactions occur at gene-dense, active, RNA polymerase II–marked regions or regions containing SEs (13, 14).
Last, using high-resolution Hi-C datasets to dissect the chromatin looping in FISC and QSC, we revealed that the isolation procedure–induced early activation (23) is accompanied by a great loss of chromatin looping. Profiling for CTCF binding uncovers CTCF binding at the anchors of structural loops and a portion of E/P loops, but a large portion of anonymous loops contain no CTCF binding at their anchors. Hence, CTCF-independent mechanisms may regulate chromatin looping remodeling during the isolation process. For example, motif analysis uncovers the enrichment of YY1, snail family zinc finger 2 (SNAI2), zinc finger and BTB domain containing 3 (ZBTB3), and EGR1 motifs in QSC, and ISL1 transcription factor, LIM/homeodomain (ISL1), GATA binding protein 3 (GATA3), FOXA1, and SMAD family member 3 (SMAD3) motifs in FISC. YY1 is known to orchestrate specific E/P looping through its oligomerization (19). SNAI2 blocks myogenic differentiation by inhibiting MYOD-driven muscle differentiation and repressing multiple TFs necessary for terminal differentiation (60). Smad3-null SCs shows dampened propensity for SC self-renewal (61). Thus, whether these TFs partaken in the SC 3D genome organization can be further dissected in the future.
Consistent with the overall looping events, the number of E-P contacts is largely reduced in FISC versus QSC, in agreement with the prominent loss in H3K27ac levels during this process (23). Intriguingly, among the loci we examined, despite that a great portion bear enhancer marks in both QSC and FISC, the interaction is restricted to only one stage to enabling the timely gene activation. Mechanistically, our results demonstrate that PAX7 is a key regulator of E/P looping formation in QSC; sharply attenuated PAX7 protein in FISC may be one reason to drive the decommission of enhancer repertoire and rewiring of E-P contacts during SC early activation. Our findings, together with previous reports (5, 11, 39), thus highlight that the interactions provide an additional layer of fine-tuned regulation in lineage development, which cannot be simply inferred from the 1D enhancer profiling.
In addition to the normal SC lineage progression, we delineated the 3D genome remodeling during SC aging. How SCs deteriorate with aging remains largely unknown (62). Our findings reveal that despite differences between young and old SCs, geriatric cells display a more prominent gain in long-range contacts and loss of TAD insulation. This is in agreement with recent findings suggesting that SCs switch from quiescence to an irreversible pre-senescence state at geriatric age (30), and a progressive gain of far-cis contacts and loss of insulation has been shown during the onset of various types of cellular senescence (26–28). However, our results highlight a preferential gain of A-A interactions and loss of B-B interactions during SC aging, contrasting with previous findings in either oncogene-induced senescence (OIS) or replicative senescence (RS) scenario (28). In OIS, the genome organization is dominated by heterochromatin or B-B interactions resulting in a stronger genome compartmentalization, whereas in RS, dampened A-A interactions cause a decrease in compartmentalization (28). We speculate that these discrepancies may reflect the innate differences between pre-senescence state of geriatric SCs and mature or deep senescent cellular state studied in other works. In addition, SCs need to integrate the information from the deteriorated aging milieu (30, 32), which may render the state of geriatric SCs more complex and heterogeneous (33) than the in vitro systems used in other studies. Consistent with the notion, PCA analyses reveal distinct trajectories of transcriptome, compartmentalization, TAD insulation, and intra-TAD interactions (Fig. 6K). Geriatric SCs overall lose insulation at TAD borders, agreeing with the observations in OIS and RS cells (28). Recent reports suggest that HMGB1 and HMGB2 proteins act as a rheostat of topological insulation upon OIS senescence entry (27, 29). Consistently, we observed decreased expression of HMGB1 and HMGB2 with SC aging (fig. S11D); it will thus be interesting to investigate possible roles of HMGB proteins in SC aging. In summary, our study represents the first to describe the 3D genome rewiring in adult stem cells of physiologically aged mammalian organism, and we anticipate that more studies from other tissues or organs will enable us to extract the common paradigm controlling stem cell aging.
The last key contribution of the present study is the elucidation of Pax7 regulation at the 3D level. PAX7 represents the indispensable TF regulating SC maintenance and regenerative capacity (4, 63, 64). However, current knowledge regarding the upstream regulation of Pax7 expression remains largely unknown. At the transcriptional level, Pax7 RNA expression is vulnerable to diverse environmental cues and rapidly decreases upon SC activation even in response to isolation procedure (23). Recent studies have provided epigenetic clues that the Pax7 locus is embedded in an SE region in homeostatic quiescence state and that the enhancer signals diminish upon activation (23), which may account for the acute loss of Pax7 expression. Complementary to these findings, we here uncovered that Pax7 locus is self-organized into a powerful TAD with profound sub-TAD organization identified in QSCs, and the organization largely collapses upon activation. In addition, we identified chromatin loops within the Pax7 TAD involving four interconnected REs (R1, R2, R3, and R4), including promoter, enhancers, and boundary elements. Notably, during the revision of our manuscript, Dong et al. (65) reported the identification of an intronic and a distal enhancer of Pax7 in SCs using chromatin accessibility profiling by ATAC-seq and proved their regulatory functions. These two enhancers happen to be our R2 and R3. R3 was also reported by Khateb et al. (66) to control Pax7 expression and downstream myogenic and neurogenic programs in differentiating ESCs. Nevertheless, leveraging our in house–developed in vivo genome editing system, we provided in vivo evidence to show that deletion of R2, R3, and R4 not only disturbed Pax7 RNA expression in QSCs but also resulted in distinct effects on 3D genome organization at Pax7 locus. Removal of the downstream boundary region (R1) alone does not cause TAD fusion, possibly due to the enhanced contact established between the two convergent CTCF sites (5, 10). Intriguingly, it instead leads to significant reduction of intra-TAD contacts. Deletion of the downstream enhancer region (R2) largely retains the TAD structure with a marginal loss of intra-TAD contacts, especially the interactions between sub-TADs. In contrast, loss of the intronic enhancer region (R3) results in more structured sub-TAD patterns. Together, our findings unveil that Pax7 transcription is tightly controlled by the embedded REs and 3D interactions.
In conclusion, our study provides a comprehensive view of 3D genome organization in SC lineage progression and SC aging. Our findings demonstrate that chromatin rewiring at different genomic scales underpins the transcriptome remodeling and SC activities, underscoring the importance of 3D regulation in stem cell lineage development and stem cell aging. The datasets produced in this study provide a rich resource of chromatin interactions for the SC community. In addition, our work uncovers mechanistic insights driving the chromatin loop remodeling during SC early activation, pointing to PAX7 as a key regulator of E/P loop rewiring. Last, we also examined 3D regulation of Pax7 locus in-depth and defined key REs dictating the high Pax7 expression in QSCs, which underscores the importance to connect the 3D genome rewiring to the fine-tuned TF dynamics during lineage development.
MATERIALS AND METHODS
Mice
All animal experiments were performed following the guidelines for experimentation with laboratory animals set in the Chinese University of Hong Kong (CUHK) and approved by the Animal Experimentation Ethics Committee (AEEC) of the CUHK. The mice were maintained in animal room with 12-hour light/12-hour dark cycles at Animal Facility in CUHK. For SC lineage development study, 2-month-old female Pax7-nGFP mice were used to isolate SCs. For aging SC study, male wild-type C57BL/6 mice of 2 (young), 23 to 24 (aged), and 28 to 32 (geriatric) months of age were used to isolate SCs. For Pax7 locus dissection, Pax7Cas9 mice were generated by crossing homozygous Cre-dependent Rosa26Cas9-EGFP knockin mouse [B6;129-Gt (ROSA)26Sortm1(CAG-cas9*-EGFP) Fezh/J; stock number 024857, The Jackson Laboratory] with a Pax7Cre strain, as previously described (47). To generate Pax7 inducible conditional KO (Pax7 iKO) mice, Pax7fl/fl (48) mice were crossed with Pax7CreER/RosaEYFP mice. Genetic inactivation of Pax7 in SCs was induced by intraperitoneal injection of tamoxifen in adult (8- to 12-week-old) mice for seven consecutive days followed by 7 days of chase.
SC isolation by FACS
SCs were sorted by FACS based on established methods (67). Briefly, hindlimb muscles from Pax7-nGFP mice were collected and digested with collagenase II (1000 U ml−1; Worthington) for 90 min at 37°C, and then the digested muscles were triturated and washed in washing medium [Ham’s F-10 medium (Sigma-Aldrich), 10% horse serum, heat inactivated (HIHS) (Gibco), penicillin/streptomycin (1×, Gibco)] before SCs were liberated by treating with collagenase II (1000 U ml−1) and dispase (11 U ml−1) for 30 min at 37°C. Mononuclear cells were filtered with a 40-μm cell strainer, and GFP+ SCs were sorted out with a BD FACSAria Fusion cell sorter (BD Biosciences). For SC isolation from C57BL/6 mice, the filtered mononuclear cells were additionally incubated with VCAM1-biotin (105704, BioLegend), CD31-FITC (fluorescein isothiocyanate) (102506, BioLegend), CD45-FITC (103108, BioLegend), and Sca1–Alexa Fluor 647 (108118, BioLegend). The VCAM1 signal was amplified with streptavidin-PE (phycoerythrin) (554061, BD Biosciences). All antibodies were used at a dilution of 1:75. VCAM1+CD31−CD45−Sca1− cells were sorted out with a BD FACSAria Fusion cell sorter (BD Biosciences). For QSC isolation, we followed the procedures described previously (23). Briefly, the muscles were prefixed in ice-cold 0.5% PFA before cell dissociation. Collagenase II and dispase were used at 2000 and 22 U ml−1 for the digestion, respectively.
AAV9 virus production, purification, and injection
AAV9 virus was produced as described previously (47). Briefly, human embryonic kidney (HEK) 293FT cells were seeded in T75 flask and transiently transfected with AAV9-sgRNA vector (5 μg), AAV9 serotype plasmid (5 μg), and pDF6 (AAV helper plasmid) (10 μg) at a ratio of 1:1:2 using polyethyleneimine when the cell reached 80 to 90% confluency. Twenty-four hours after transfection, the cells were changed to growth medium [Dulbecco’s modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS), penicillin (100 U ml−1), streptomycin (100 μg ml−1), and 2 mM l-glutamine] and cultured for another 48 hours. The cells were harvested and washed with phosphate-buffered saline (PBS) two times. To release the AAV9 virus, the pellet was resuspended with lysis buffer [50 mM tris-HCl (pH 8.0) and 150 mM NaCl] followed by three sequential freeze-thaw cycles (liquid nitrogen/37°C). The lysates were treated with Benzonase (Sigma-Aldrich) together with MgCl2 (final concentration of 1.6 mM) at 37°C for 1 hour, followed by centrifugation at 3000 rpm for 10 min. The supernatant was filtered with 0.45-μm sterile filter and added with equal volume of 1 M NaCl and 20% (w/v) PEG8000 (polyethylene glycol, molecular weight 8000) to precipitate the virus at 4°C for overnight. After centrifugation at 12,000g for 30 min at 4°C, the pellet was resuspended with sterile PBS and then subjected to centrifugation at 3000g for 10 min. Equal volume of chloroform was then added and shaken followed by spinning down at 12,000g for 15 min at 4°C. The aqueous layer was filtered with a 0.22-μm sterile filter and passed through a 100-kDa molecular weight cutoff (Millipore). The concentrated solution was washed with sterile PBS three times. The titer of the AAV9 virus was determined by quantitative reverse transcription polymerase chain reaction (qRT-PCR) using primers targeting the cytomegalovirus (CMV) promoter. Sequences of the used primers are listed in table S11. Heterozygous Pax7Cas9 mice were intramuscularly injected with 4 × 1011 vg of AAV9-sgRNA pair #1 at P5 and followed by 8 × 1011 vg of AAV9-sgRNA pair #2 at P10. SCs were isolated 4 weeks after AAV injection. For the control group, the same dose of AAV9 virus containing pAAV9-sgRNA backbone without any sgRNA insertion was injected.
Cell culture
HEK293FT cells were obtained from the American Type Culture Collection (CRL-3216) and maintained in DMEM supplemented with 10% FBS, penicillin (100 U ml−1), and 100 μg of streptomycin in a 5% CO2 humidified incubator at 37°C. FISCs were cultured in Ham’s F10 medium (Sigma-Aldrich) with 20% FBS, penicillin/streptomycin (1×), and basic fibroblast growth factor (2.5 ng ml−1; 13256, Life Technologies).
Plasmids
Site-specific sgRNAs were selected using a web tool Crispor (http://crispor.tefor.net/) (68). Synthesized oligonucleotides encoding guide sequences were cloned into the AAV9-sgRNA transfer vector [AAV: ITR-U6-sgRNA(backbone)-CMV-DsRed-WPRE-hGHpA-ITR] using Sap I site. To generate dual AAV-sgRNA expression plasmid, the second sgRNA was constructed into the AAV9-sgRNA vector together with the gRNA cassette and U6 promoter using Xba I and Kpn I sites. Sequences for sgRNAs and primers used to detect deletion efficiency are listed in table S11. The source data underlying fig. S9B are provided as fig. S14.
RNA isolation, qRT-PCR, and RNA-seq
For FISCs and cultured SCs, total RNAs were extracted using TRIzol reagent (Invitrogen). For QSCs, total RNAs were isolated from the above-described prefixed cells using the miRNeasy FFPE Kit (Qiagen) according to the manufacturer’s instruction. For qRT-PCR, cDNAs were prepared using the HiScript III 1st Strand cDNA Synthesis Kit (Vazyme, R312-01). Real-time PCRs were performed on LightCycler 480 Instrument II (Roche Life Science) using Luna Universal qPCR Master Mix (NEB, M3003L). Sequences of all primers used are listed in table S11. For polyadenylated [poly(A)+] mRNA-seq, total RNAs were subjected to poly(A) selection (Ambion, 61006) followed by library preparation using the NEBNext Ultra II RNA Library Preparation Kit (NEB, E7770S). Libraries were paired-end sequenced with read lengths of 150 base pairs (bp) on an Illumina HiSeq X Ten instrument. For total RNA-seq, total RNAs were isolated from QSC, FISC, or cultured SCs; library preparation and paired-end sequencing were performed by Beijing Genomics Institute (Hong Kong) with a read length of 100 bp on NextSeq 500.
Immunoblotting and IF
These were performed according to our standard procedures (41, 69, 70). Briefly, total proteins were extracted using radioimmunoprecipitation assay (RIPA) lysis buffer. The following dilutions of antibodies were used for each antibody: PAX7 (1:1000; Developmental Studies Hybridoma Bank, RRID:AB_528428), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (1:2000; Santa Cruz Biotechnology, sc-137179, RRID:AB_2232048), CTCF (1:1000; ABclonal, A1133, RRID:AB_2753146), RAD21 (1:1000; Abcam, ab992, RRID:AB_2176601), and histone H3 (1:3000; Santa Cruz Biotechnology, sc-517576, RRID:AB_2848194). To compare the CTCF and RAD21 protein level between QSCs and FISCs, the FISCs were treated with 0.5% PFA before cell lysis. For IF staining, the PAX7 antibody (1:100; Developmental Studies Hybridoma Bank, RRID:AB_528428) was used. All fluorescent images were captured with a fluorescence microscope (Leica DM6000 B). The source data underlying Fig. 3E and figs. S8 (B and K), S9 (C and D), and S10D are provided in figs. S14 and S15.
ChIP followed by high-throughput sequencing
For H3K27ac ChIP-seq in FISCs, following our standard procedure (47, 69, 71), approximately 300,000 to 600,000 FISCs were cross-linked with 1% formaldehyde for 10 min at room temperature and quenched by 125 mM glycine. Cells were washed and collected by centrifugation at 700g for 5 min at 4°C, flash-frozen in liquid nitrogen, and stored at −80°C. Cells were lysed in 1 ml of LB1 [50 mM Hepes-KOH (pH 7.5), 140 mM NaCl, 1 mM EDTA, 0.25% Triton X-100, 0.5% NP-40, and 10% glycerol supplemented with cOmplete protease inhibitors (Roche)] and incubated at 4°C for 10 min. Following centrifugation at 1350g for 5 min at 4°C, the pellets were washed with 1 ml of LB2 [10 mM tris-HCl (pH 8.0), 200 mM NaCl, 1 mM EDTA (pH 8.0), and 1 mM EGTA (pH 8.0) supplemented with cOmplete proteinase inhibitors] by incubating at 4°C for 10 min. Following centrifugation at 1350g for 5 min at 4°C, nuclei were rinsed twice with 1 ml of sonication buffer [10 mM tris-HCl (pH 8.0), 0.1% SDS, and 1 mM EDTA supplemented with cOmplete proteinase inhibitors]. Then, the nuclei were resuspended in sonication buffer and sonicated with Covaris S220 (intensity: 140 W, duty cycle: 5%, cycles per burst: 200, time: 7 min). The resulting lysate was supplied with NaCl and Triton X-100 to reach a final concentration at 150 mM NaCl and 1% Triton X-100, then cleared by centrifugation for 20 min at 20,000g, and then incubated with washed Dynabeads Protein G (Invitrogen, 10004D) for 2 hours at 4°C. The cleared lysate was incubated with 0.75 μg of H3K27ac (Abcam, ab4729, RRID:AB_2118291) or immunoglobulin G (IgG) control (Santa Cruz Biotechnology, sc-2027, RRID:AB_737197) for overnight at 4°C. On the next day, 10 μl of Dynabeads Protein G was washed with IP buffer [10 mM tris-HCl (pH 8.0), 150 mM NaCl, 0.1% SDS, 1 mM EDTA, and 1% Triton X-100 supplemented with cOmplete proteinase inhibitors] and then applied to each IP reaction followed by incubation at 4°C for another 2 hours. Beads were then collected and washed twice with IP buffer, two times with high-salt wash buffer [10 mM tris-HCl (pH 8.0), 500 mM NaCl, 0.1% SDS, 1 mM EDTA, and 1% Triton X-100 supplemented with cOmplete proteinase inhibitors], two times with LiCl wash buffer [10 mM tris-HCl (pH 8), 250 mM LiCl, 1 mM EDTA, 0.5% NP-40, and 0.5% deoxycholate supplemented with cOmplete proteinase inhibitors], and one time with cold TE buffer [10 mM tris-HCl (pH 8.0), 1 mM EDTA, and 50 mM NaCl]. Immunocomplexes were eluted in ChIP elution buffer [50 mM tris-HCl (pH 8.0), 10 mM EDTA, and 1% SDS] with incubation at 65°C for 30 min, and eluents were reverse cross-linked by incubating at 65°C for 16 hours. Immunoprecipitated DNA was treated with ribonuclease A (0.2 mg ml−1) at 37°C for 2 hours, followed by proteinase K treatment (0.2 mg ml−1) at 55°C for 3 hours. Immunoprecipitated DNA was then subjected to phenol:chloroform extraction and ethanol precipitation and finally resuspended in 1× TE buffer. Libraries were prepared using the NEBNext Ultra II DNA Library Preparation Kit (NEB, E7645S) and paired-end sequenced with read lengths of 150 bp on an Illumina HiSeq X Ten instrument. For CTCF ChIP-seq in FISCs, approximately 500,000 FISCs were cross-linked with 1% formaldehyde for 10 min for CTCF at room temperature and quenched by 125 mM glycine. Cells were washed and collected by centrifugation at 700g for 5 min at 4°C, flash-frozen in liquid nitrogen, and stored at −80°C. The cell pellet was resuspended in 130 μl of shearing buffer [10 mM tris-HCl (pH 8.0), 0.1% SDS, and 1 mM EDTA supplemented with cOmplete proteinase inhibitors] and subjected to sonication with Covaris S220 (intensity: 140 W, duty cycle: 5%, cycles per burst: 200, time: 7 min). The following steps are the same as described above. For CTCF ChIP-seq in QSCs, approximately 500,000 QSCs were subjected to sonication in shearing buffer directly without additional cross-link by 1% formaldehyde. The CTCF antibody (ABclonal, A1133, RRID:AB_2753146) was used for CTCF ChIP-seq.
Assay for transposase-accessible chromatin using sequencing
ATAC-seq was performed as described previously (72). Briefly, 50,000 FISCs were washed with 50 μl of ice-cold PBS and collected by centrifugation at 700g for 5 min at 4°C. The pellet was resuspended in 50 μl of ice-cold lysis buffer [10 mM tris-Cl (pH 7.5), 10 mM NaCl, 3 mM MgCl2, 0.1% NP-40, 0.1% Tween 20, and 0.01% digitonin] and lysed on ice for 3 min. The cell lysate was added with 1 ml of wash buffer [10 mM tris-Cl (pH 7.5), 10 mM NaCl, 3 mM MgCl2, and 0.1% Tween 20] followed by centrifugation at 700g for 10 min at 4°C. The cell pellet was resuspended in transposition reaction mix [25 μl of Tagment DNA Buffer, 2.5 μl of Tagment DNA Enzyme 1 (Illumina), 0.1% Tween 20, and 0.01% digitonin] and incubated at 37°C for 30 min on thermomixer at 1000 rpm. The sample was purified by the Qiagen MinElute Reaction Cleanup Kit with 10-μl final elution volume. The transposed DNA was PCR-amplified using the NEBNext High-Fidelity 2× PCR Master Mix (NEB) with five cycles followed by cycle number determination. The resulting library was purified by VAHTS DNA Clean Beads (Vazyme, N411-01) and paired-end sequenced with read lengths of 150 bp on an Illumina HiSeq X Ten instrument.
In situ Hi-C library preparation and sequencing
Generation of Hi-C libraries with a low cell number of SCs was performed according to modified protocols (6). Approximately 100,000 SCs were cross-linked with 1% formaldehyde for 10 min at room temperature and quenched by 0.2 M glycine. The samples were lysed in 500 μl of ice-cold Hi-C lysis buffer [10 mM tris-HCl (pH 8.0), 10 mM NaCl, and 0.2% Igepal CA630 supplemented with cOmplete proteinase inhibitors] on ice for 15 min. Samples were then centrifuged at 2500g for 5 min. Pelleted nuclei were washed once with 500 μl of 1.25× NEBuffer 3.1. The supernatant was discarded, the nuclei were resuspended in 358 μl of 1.25× NEBuffer 3.1, and 11 μl of 10% SDS was added followed by incubation at 37°C for 1 hour. After incubation, 75 μl of 10% Triton X-100 was added to quench the SDS and then incubated at 37°C for 1 hour. One hundred units of Dpn II restriction enzyme (NEB, R0543) was added, and chromatins were digested at 37°C for overnight. Samples were incubated at 62°C for 20 min to inactivate Dpn II and then cooled to room temperature. Samples were then centrifuged at 2500g for 5 min. Pelleted nuclei were resuspended in 50 μl of fill-in master mix [3.75 μl of 0.4 mM biotin-14-dATP (2′-deoxyadenosine-5′-triphosphate), 1.5 μl of 1 mM 2′-deoxycytidine-5′-triphosphate (dCTP), 1.5 μl of 1 mM 2′-deoxyguanosine-5′-triphosphate (dGTP), 1.5 μl of 1 mM 3′-deoxythymidine-5′-triphosphate (dTTP) mix, 2 μl of DNA polymerase I (5 U μl−1), Large (Klenow) Fragment, and 1× NEBuffer 3.1] to fill in the restriction fragment overhangs and mark the DNA ends with biotin. Samples were mixed by pipetting and incubated at 23°C for 4 hours. Ligation master mix [398 μl of water, 50 μl of 10× NEB T4 DNA ligase buffer, 1 μl of bovine serum albumin (50 mg ml−1), 1 μl of T4 DNA ligase (400 U μl−1)] was added, and samples were incubated at 16°C for overnight in ThermoMixer C with interval shake. Nuclei were pelleted by centrifugation at 2500g for 5 min, and 380 μl of the supernatant was discarded. Pellets were then resuspended in the remaining 120 μl of ligation mix supplemented with 12 μl of 10% SDS and 5 μl of proteinase K (20 mg ml−1) and incubated at 55°C for 2 hours, with shaking at 1000 rpm. Fifteen microliters of 5 M NaCl was added, and the reaction was incubated at 65°C for 16 hours. DNA samples were subjected to phenol:chloroform extraction and ethanol precipitation and finally resuspended in 130 μl of 0.1× TE buffer. The purified DNA samples were then sheared to a length of ∼300 bp using Covaris S220 instrument (intensity: 175 W, duty cycle: 10%, cycles per burst: 200, time: 150 s). The biotinylated DNA was pulled down by 10 μl of Dynabeads MyOne Streptavidin C1 beads (10 mg ml−1; Life Technologies, 65001). The beads were then resuspended in 23 μl of 10 mM tris-Cl (pH 8.0), and libraries were prepared by on-bead reactions using the NEBNext Ultra II DNA Library Preparation Kit (NEB, E7645S). The beads were separated on a magnetic stand, and the supernatant was discarded. After washes, the beads were resuspended in 20 μl of 10 mM tris buffer and boiled at 98°C for 10 min. The elute was amplified for 10 to 13 cycles of PCR with Phanta Master Mix (Vazyme, P511-01), and the PCR products were purified using VAHTS DNA Clean Beads (Vazyme, N411-01). The Hi-C libraries were paired-end sequenced with read lengths of 150 bp on an Illumina HiSeq X Ten instrument.
Gene expression analysis of RNA-seq data
The raw reads of total RNA-seq were processed following the procedures described in our previous publication (69). Briefly, the adapter and low-quality sequences were trimmed from 3′ to 5′ ends for each read, and the reads shorter than 50 bp were discarded. The reads that passed the quality control were mapped to mouse genome (mm9) with TopHat2 (version 2.1.1). Cufflinks (version 2.2.1) were then used to estimate transcript abundance in fragments per kilobase per million (FPKM). Genes were annotated as differentially expressed if the change of expression level is greater than twofold between two stages/conditions.
ChIP-seq data analysis
Raw ChIP-seq reads were processed as previously described (71). Briefly, the adapter and low-quality sequences were trimmed from 3′ to 5′ ends by Trimmomatic (version 0.36), and the reads shorter than 36 bp were discarded. Subsequently, the preprocessed reads were aligned to the mouse genome (mm9) using Bowtie2 (v2.3.3.1). The aligned reads were then converted to bam format using samtools (version 1.5), and the duplicate reads were removed by Picard (http://broadinstitute.github.io/picard). Peaks were then identified by MACS2 (V 2.2.4) with q value equal to 0.01 by using the IgG control sample as a background. Promoters are defined as regions between −2.5 kb and +500 bp of the transcription start sites (TSSs) of genes. H3K27ac ChIP-seq is used for TE and SE identification. For TE identification, blacklisted regions from ENCODE were first excluded, and the peaks overlapping with +2-kb/−500-bp regions centered at TSS from Refseq (June 2015) were also discarded. The filtered peaks were defined as TEs and assigned to the expressed transcripts (reads per kilobase of transcript per million reads mapped (RPKM) > 0.5), whose TSSs are nearest to the center of the enhancer regions. SEs were identified as previously described with minor adjustment (42). Only the enhancers within 12.5 kb of each another and can be assigned to the same genes were stitched together and then subjected to the ROSE algorithm (https://github.com/stjude/ROSE) for SE identification.
ATAC-seq data analysis
The adaptor sequences of 150-bp paired-end ATAC-seq reads were trimmed by Trimmomatic and cut to 36-bp paired-end reads. Bowtie2 was used for alignment to the mouse reference genome (mm9) with parameters -X 2000 -no-mixed -no-discordant -local. Aligned reads were filtered for mapping quality ≥30, and redundant reads were removed by using the Picard MarkDuplicates (http://broadinstitute.github.io/picard). Peak regions of each sample were called by software MACS2 with parameters -f BAMPE -g mm -p 0.001. For visualization, the alignment bam files were converted into bedgraph files using HOMER (http://homer.ucsd.edu/homer/ngs/ucsc.html).
In situ Hi-C data processing
The in situ Hi-C data were processed with a standard pipeline HiC-Pro (v2.10.0) (35). First, adaptor sequences and poor-quality reads were removed using Trimmomatic (ILLUMINACLIP: TruSeq3-PE-2.fa:2:30:10; SLIDINGWINDOW: 4:15; MINLEN:50). The filtered reads were then aligned to reference genome (mm9) in two steps: (i) The global alignment was first conducted for all pair-end reads, and (ii) the unaligned reads were split into prospective fragments using restriction enzyme recognition sequence (GATCGATC) and aligned again. All aligned reads were then merged together and assigned to restriction fragment, while low-quality [mapping quality (MAPQ) score < 30] or multiple alignment reads were discarded. Invalid fragments including unpaired fragments (singleton), juxtaposed fragments (re-legation pairs), unligated fragments (dangling end), self-circularized fragments (self-cycle), and PCR duplicates were removed from each biological replicate. The remaining validate pairs from all replicates of each stage were then merged, followed by read depth normalization using HOMER (http://homer.ucsd.edu/homer/interactions/HiCpca.html) and matrix balancing using iterative correction and eigenvector decomposition (ICE) normalization to obtain comparable interaction matrix between different stages.
Identification and analysis of compartments and TADs
Following previous procedure (6), to separate the genome into A and B compartments, the ICE normalized intrachromosomal interaction matrices at 100-kb resolution were transformed to observe/expect contact matrices, and the background (expected) contact matrices were generated to eliminate bias caused by distance-dependent decay of interaction frequency and read depth difference. Pearson correlation was then applied to the transformed matrices, and the PC1 of these matrices was divided into two clusters. The annotation of genes and the expression profile were used to assign positive PC1 value to gene-rich component as compartment A and negative PC1 value to gene-poor component as compartment B. The compartmentalization strength was measured using cooltools (v 0.3.2; https://github.com/mirnylab/cooltools). Briefly, all 100-kb compartment bins were divided into 50 equal degrees based on the ranking of PC1 values, and the average interaction strength (observed/expected) was calculated between pairs of 100-kb loci arranged by their PC1 and normalized by genomic distance. The dynamics of interaction strength for intra/intercompartment (A versus A, B versus B, and A versus B) were measured using the top 20% of compartment bins from both compartments A and B based on absolute PC1 value. Normalized contact matrix at 40-kb resolution of each time point was used for TAD identification using TopDom (v0.0.2) (38). Briefly, for each 40-kb bin across the genome, a signal of the average interaction frequency of all pairs of genome regions within a distinct window centered on this bin was calculated; thus, TAD boundary was identified with local minimal signal within certain window. The false-detected TADs without local interaction aggregation were filtered out by statistical testing. Invariant TADs were defined using following criteria: (i) The distance of both TAD boundaries between two conditions is no more than 40 kb, and (2) the overlapping between two TADs should be larger than 80%; stage-specific TADs were defined otherwise. The insulation index for each bin was generated on the basis of a previously described method (73). Briefly, the insulation index for each bin was obtained by calculating the number of interaction across a specific bin within certain distance. The IS of the identified TAD border was also defined as previously described (73), which used the local maximum on the outside of TAD to minus the local minimum on the inside of TAD of each boundary bin. The DS was calculated for each TAD at 40-kb interaction matrix by dividing the total intra-TAD contacts by all contacts involving the TAD.
Identification of TAD-TAD interactions and TAD clusters
The significant TAD-TAD interactions and TAD clusters were identified as previously described (74) with modification. First, the significance of TAD-TAD interactions was tested by applying an NCHG model for all pairs of TADs on the same chromosome and then corrected by multiple testing with false discovery rate (FDR) < 1% using the Benjamini-Hochberg method. Significant TAD-TAD interactions were defined for those TAD pairs with adjusted P value < 0.05 and fivefold enrichment of observed interactions over expected. All significant TAD pairs regardless of interaction distance were then used to identify TAD clusters using the NetworkX Python library (http://networkx.github.io/), where each TAD was represented by a node and interactions were represented by edges. Maximal size of TAD clusters was calculated using the Bron-Kerbosch algorithm, and allvial package (https://github.com/mbojan/alluvial) was used to plot the TAD cluster dynamics.
3D modeling of TAD cluster
The normalized interaction matrix containing TAD cluster as well as their genomic 3D context were used to build 3D modeling following the previous method with modification (75): (i) to select key elements contained in the region (i.e., TADs in selected TAD cluster); (ii) to retrieve the top 1% interactors of each of these elements; (iii) to create a network between interactors where edges correspond to top 1% interactions between any of the selected interactions; (iv) to group the networks, allowing the groups in which the ratio (number of edges)/(number of nodes) smaller than 5 was filtered; and (v) to extract the connected groups that contain the most key elements. Next, the normalized interaction matrices of selected regions were modeled using TADdyn (76). A similar protocol as previously described was used (77). Briefly, a total of 1000 models were generated for each genomic region and dataset. Each ensemble of models was next clustered on the basis of structural similarity. The absence of major structural differences between clusters prompted us to use all of them for further analysis. Next, TADbit (v1.1) (78) was used to measure the following features of the models: (i) distance between particles containing genomic regions of interest in the model ensemble, (ii) distance distribution between selected pairs or particles, and (iii) notable differential distance distributions assessed by the two-sample Kolmogorov-Smirnov statistic. Last, model images were generated with Chimera (79).
Identification of chromatin loops
The loops were identified by FitHIC2 (43) (https://github.com/ay-lab/fithic) at 5-kb resolution of interaction matrix with a P value of <1 × 10−5 and required a fivefold enrichment of observed contacts over expected based on genomic distance. To obtain stage-specific loops, FC of interaction strength (observed/expected) between two stages was calculated for each identified loop. Then, the top 30% dynamics loops with absolute FC > 2 were defined as stage-specific loops.
Motif scanning
The position-specific probability matrix of motifs was obtained from the TRANSFAC database (80). Using FIMO (https://meme-suite.org/meme/doc/fimo.html), the TAD boundaries as well as enhancers and promoter regions (±2 kb) of PAX7 were scanned for motifs of all expressed TFs in both QSC and FISC. The motif was kept if the P value was smaller than 10−4.
PCA analysis
PCA was generated using the prcomp package in R (v3.6.2) with FPKM for RNA-seq, PC1 for A/B compartments, and DS/IS for conserved TADs. The eigenvalue for the first two components was used, and the explanatory ratio of each component was calculated using the eigenvalue divided by total eigenvalue.
Acknowledgments
We thank Z. Ju (Jinan University, China) for providing the aged mice and D. Leung (HKUST, Hong Kong, China) for assistance on the Hi-C sequencing. We also appreciate the support from other colleagues in H.S. and H.W. laboratories.
Funding: This work was supported by General Research Funds (GRF) from the Research Grants Council (RGC) of the Hong Kong Special Administrative Region ( 14120420, 14116918, and 14120619 to H.S. and 14115319, 14100018, 14100620, 14106117, and 14106521 to H.W.); Health and Medical Research Fund (HMRF) from Health Bureau of the Hong Kong Special Administrative Region, China (Project Code: 08190626 to H.W.); the National Natural Science Foundation of China (NSFC) (grant nos. 32100673 to Y.Z. and 31871304, 82172436 to H.W.); the research funds from Health@InnoHK program launched by Innovation Technology Commission, the Government of the Hong Kong SAR, China to H.W.; Collaborative Research Fund (CRF) from RGC to H.W. (C6018-19GF); Theme-based Research Scheme (TRS) from RGC (Project number: T13-602/21-N); Natural Science Foundation of Guangdong Province, China to Y.Z. (grant no. 2021A1515012058), NSFC/RGC Joint Research Scheme to H.S. (project code: N_CUHK 413/18); Hong Kong Epigenomics Project (EpiHK) Fund to H.W. and H.S.; Area of Excellence Scheme (AoE) from RGC (project number: AoE/M-402/20).
Author contributions: Conceptualization: H.W., Y.Z., Y.D., and H.S. Methodology: Y.Z., Y.D., and L.H. Investigation: Y.Z., L.H., Q.Z., X.C., Y.L., M.V.A., and Z.W. Bioinformatic analysis: Y.D. Visualization: Y.D. Supervision: Y.Z., H.S., and H.W. Writing—original draft: Y.Z., Y.D., L.H., and H.W. Writing—review and editing: Y.Z., Y.D., L.H., and H.W.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: In situ Hi-C and RNA-seq data reported here are deposited in the Gene Expression Omnibus database under accession no. GSE189842. All used datasets from other publications are summarized in table S12. All other data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S15
Legends for tables S1 to S12
Other Supplementary Material for this : manuscript includes the following:
Tables S1 to S12
REFERENCES AND NOTES
- 1.A. E. Almada, A. J. Wagers, Molecular circuitry of stem cell fate in skeletal muscle regeneration, ageing and disease. Nat. Rev. Mol. Cell Biol. 17, 267–279 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.H. Yin, F. Price, M. A. Rudnicki, Satellite cells and the muscle stem cell niche. Physiol. Rev. 93, 23–67 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.P. Feige, C. E. Brun, M. Ritso, M. A. Rudnicki, Orienting muscle stem cells for regeneration in homeostasis, aging, and disease. Cell Stem Cell 23, 653–664 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.P. Seale, L. A. Sabourin, A. Girgis-Gabardo, A. Mansouri, P. Gruss, M. A. Rudnicki, Pax7 is required for the specification of myogenic satellite cells. Cell 102, 777–786 (2000). [DOI] [PubMed] [Google Scholar]
- 5.B. Bonev, N. Mendelson Cohen, Q. Szabo, L. Fritsch, G. L. Papadopoulos, Y. Lubling, X. Xu, X. Lv, J. P. Hugnot, A. Tanay, G. Cavalli, Multiscale 3D genome rewiring during mouse neural development. Cell 171, 557–572.e24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.S. S. P. Rao, M. H. Huntley, N. C. Durand, E. K. Stamenova, I. D. Bochkov, J. T. Robinson, A. L. Sanborn, I. Machol, A. D. Omer, E. S. Lander, E. L. Aiden, A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.R. Stadhouders, E. Vidal, F. Serra, B. di Stefano, F. le Dily, J. Quilez, A. Gomez, S. Collombet, C. Berenguer, Y. Cuartero, J. Hecht, G. J. Filion, M. Beato, M. A. Marti-Renom, T. Graf, Transcription factors orchestrate dynamic interplay between genome topology and gene regulation during cell reprogramming. Nat. Genet. 50, 238–249 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.J. R. Dixon, S. Selvaraj, F. Yue, A. Kim, Y. Li, Y. Shen, M. Hu, J. S. Liu, B. Ren, Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.N. Naumova, M. Imakaev, G. Fudenberg, Y. Zhan, B. R. Lajoie, L. A. Mirny, J. Dekker, Organization of the mitotic chromosome. Science 342, 948–953 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Y. Guo, Q. Xu, D. Canzio, J. Shou, J. Li, D. U. Gorkin, I. Jung, H. Wu, Y. Zhai, Y. Tang, Y. Lu, Y. Wu, Z. Jia, W. Li, M. Q. Zhang, B. Ren, A. R. Krainer, T. Maniatis, Q. Wu, CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell 162, 900–910 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.B. K. Kragesteen, M. Spielmann, C. Paliou, V. Heinrich, R. Schöpflin, A. Esposito, C. Annunziatella, S. Bianco, A. M. Chiariello, I. Jerković, I. Harabula, P. Guckelberger, M. Pechstein, L. Wittler, W. L. Chan, M. Franke, D. G. Lupiáñez, K. Kraft, B. Timmermann, M. Vingron, A. Visel, M. Nicodemi, S. Mundlos, G. Andrey, Dynamic 3D chromatin architecture contributes to enhancer specificity and limb morphogenesis. Nat. Genet. 50, 1463–1473 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.J. Paulsen, T. M. Liyakat Ali, M. Nekrasov, E. Delbarre, M. O. Baudement, S. Kurscheid, D. Tremethick, P. Collas, Long-range interactions between topologically associating domains shape the four-dimensional genome during differentiation. Nat. Genet. 51, 835–843 (2019). [DOI] [PubMed] [Google Scholar]
- 13.R. A. Beagrie, A. Scialdone, M. Schueler, D. C. A. Kraemer, M. Chotalia, S. Q. Xie, M. Barbieri, I. de Santiago, L. M. Lavitas, M. R. Branco, J. Fraser, J. Dostie, L. Game, N. Dillon, P. A. W. Edwards, M. Nicodemi, A. Pombo, Complex multi-enhancer contacts captured by genome architecture mapping. Nature 543, 519–524 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.S. A. Quinodoz, N. Ollikainen, B. Tabak, A. Palla, J. M. Schmidt, E. Detmar, M. M. Lai, A. A. Shishkin, P. Bhat, Y. Takei, V. Trinh, E. Aznauryan, P. Russell, C. Cheng, M. Jovanovic, A. Chow, L. Cai, P. McDonel, M. Garber, M. Guttman, Higher-order inter-chromosomal hubs shape 3D genome organization in the nucleus. Cell 174, 744–757.e24 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.M. Yu, B. Ren, The three-dimensional organization of mammalian genomes. Annu. Rev. Cell Dev. Biol. 33, 265–289 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.E. P. Nora, A. Goloborodko, A. L. Valton, J. H. Gibcus, A. Uebersohn, N. Abdennur, J. Dekker, L. A. Mirny, B. G. Bruneau, Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169, 930–944.e22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.S. S. P. Rao, S. C. Huang, B. Glenn St Hilaire, J. M. Engreitz, E. M. Perez, K. R. Kieffer-Kwon, A. L. Sanborn, S. E. Johnstone, G. D. Bascom, I. D. Bochkov, X. Huang, M. S. Shamim, J. Shin, D. Turner, Z. Ye, A. D. Omer, J. T. Robinson, T. Schlick, B. E. Bernstein, R. Casellas, E. S. Lander, E. L. Aiden, Cohesin loss eliminates all loop domains. Cell 171, 305–320.e24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.W. Schwarzer, N. Abdennur, A. Goloborodko, A. Pekowska, G. Fudenberg, Y. Loe-Mie, N. A. Fonseca, W. Huber, C. H. Haering, L. Mirny, F. Spitz, Two independent modes of chromatin organization revealed by cohesin removal. Nature 551, 51–56 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.A. S. Weintraub, C. H. Li, A. V. Zamudio, A. A. Sigova, N. M. Hannett, D. S. Day, B. J. Abraham, M. A. Cohen, B. Nabet, D. L. Buckley, Y. E. Guo, D. Hnisz, R. Jaenisch, J. E. Bradner, N. S. Gray, R. A. Young, YY1 is a structural regulator of enhancer-promoter loops. Cell 171, 1573–1588.e28 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.R. Wang, F. Chen, Q. Chen, X. Wan, M. Shi, A. K. Chen, Z. Ma, G. Li, M. Wang, Y. Ying, Q. Liu, H. Li, X. Zhang, J. Ma, J. Zhong, M. Chen, M. Q. Zhang, Y. Zhang, Y. Chen, D. Zhu, MyoD is a 3D genome structure organizer for muscle cell identity. Nat. Commun. 13, 205 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.N. Zhang, J. Mendieta-Esteban, A. Magli, K. C. Lilja, R. C. R. Perlingeiro, M. A. Marti-Renom, A. Tsirigos, B. D. Dynlacht, Muscle progenitor specification and myogenic differentiation are associated with changes in chromatin topology. Nat. Commun. 11, 6222 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.C. B. Hug, A. G. Grimaldi, K. Kruse, J. M. Vaquerizas, Chromatin architecture emerges during zygotic genome activation independent of transcription. Cell 169, 216–228.e19 (2017). [DOI] [PubMed] [Google Scholar]
- 23.L. Machado, J. Esteves de Lima, O. Fabre, C. Proux, R. Legendre, A. Szegedi, H. Varet, L. R. Ingerslev, R. Barrès, F. Relaix, P. Mourikis, In situ fixation redefines quiescence and early activation of skeletal muscle stem cells. Cell Rep. 21, 1982–1993 (2017). [DOI] [PubMed] [Google Scholar]
- 24.C. T. J. van Velthoven, A. de Morree, I. M. Egner, J. O. Brett, T. A. Rando, Transcriptional profiling of quiescent muscle stem cells in vivo. Cell Rep. 21, 1994–2004 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.L. Yue, R. Wan, S. Luan, W. Zeng, T. H. Cheung, Dek modulates global intron retention during muscle stem cells quiescence exit. Dev. Cell 53, 661–676.e6 (2020). [DOI] [PubMed] [Google Scholar]
- 26.T. Chandra, P. A. Ewels, S. Schoenfelder, M. Furlan-Magaril, S. W. Wingett, K. Kirschner, J. Y. Thuret, S. Andrews, P. Fraser, W. Reik, Global reorganization of the nuclear landscape in senescent cells. Cell Rep. 10, 471–483 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.A. Zirkel, M. Nikolic, K. Sofiadis, J. P. Mallm, C. A. Brackley, H. Gothe, O. Drechsel, C. Becker, J. Altmüller, N. Josipovic, T. Georgomanolis, L. Brant, J. Franzen, M. Koker, E. G. Gusmao, I. G. Costa, R. T. Ullrich, W. Wagner, V. Roukos, P. Nürnberg, D. Marenduzzo, K. Rippe, A. Papantonis, HMGB2 loss upon senescence entry disrupts genomic organization and induces CTCF clustering across cell types. Mol. Cell 70, 730–744.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 28.S. Sati, B. Bonev, Q. Szabo, D. Jost, P. Bensadoun, F. Serra, V. Loubiere, G. L. Papadopoulos, J. C. Rivera-Mulia, L. Fritsch, P. Bouret, D. Castillo, J. L. Gelpi, M. Orozco, C. Vaillant, F. Pellestor, F. Bantignies, M. A. Marti-Renom, D. M. Gilbert, J. M. Lemaitre, G. Cavalli, 4D genome rewiring during oncogene-induced and replicative senescence. Mol. Cell 78, 522–538.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.K. Sofiadis, N. Josipovic, M. Nikolic, Y. Kargapolova, N. Übelmesser, V. Varamogianni-Mamatsi, A. Zirkel, I. Papadionysiou, G. Loughran, J. Keane, A. Michel, E. G. Gusmao, C. Becker, J. Altmüller, T. Georgomanolis, A. Mizi, A. Papantonis, HMGB1 coordinates SASP-related chromatin folding and RNA homeostasis on the path to senescence. Mol. Syst. Biol. 17, e9760 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.P. Sousa-Victor, S. Gutarra, L. García-Prat, J. Rodriguez-Ubreva, L. Ortet, V. Ruiz-Bonilla, M. Jardí, E. Ballestar, S. González, A. L. Serrano, E. Perdiguero, P. Muñoz-Cánoves, Geriatric muscle stem cells switch reversible quiescence into senescence. Nature 506, 316–321 (2014). [DOI] [PubMed] [Google Scholar]
- 31.P. Sousa-Victor, L. Garcia-Prat, A. L. Serrano, E. Perdiguero, P. Munoz-Canoves, Muscle stem cell aging: Regulation and rejuvenation. Trends Endocrinol. Metab. 26, 287–296 (2015). [DOI] [PubMed] [Google Scholar]
- 32.S. Schworer, F. Becker, C. Feller, A. H. Baig, U. Köber, H. Henze, J. M. Kraus, B. Xin, A. Lechel, D. B. Lipka, C. S. Varghese, M. Schmidt, R. Rohs, R. Aebersold, K. L. Medina, H. A. Kestler, F. Neri, J. von Maltzahn, S. Tümpel, K. L. Rudolph, Epigenetic stress responses induce muscle stem-cell ageing by Hoxa9 developmental signals. Nature 540, 428–432 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.I. Hernando-Herraez, B. Evano, T. Stubbs, P. H. Commere, M. Jan Bonder, S. Clark, S. Andrews, S. Tajbakhsh, W. Reik, Ageing affects DNA methylation drift and transcriptional cell-to-cell variability in mouse muscle stem cells. Nat. Commun. 10, 4361 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.P. Rocheteau, B. Gayraud-Morel, I. Siegl-Cachedenier, M. A. Blasco, S. Tajbakhsh, A subpopulation of adult skeletal muscle stem cells retains all template DNA strands after cell division. Cell 148, 112–125 (2012). [DOI] [PubMed] [Google Scholar]
- 35.N. Servant, N. Varoquaux, B. R. Lajoie, E. Viara, C. J. Chen, J. P. Vert, E. Heard, J. Dekker, E. Barillot, HiC-Pro: An optimized and flexible pipeline for Hi-C data processing. Genome Biol. 16, 259 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.T. Nagano, C. Várnai, S. Schoenfelder, B. M. Javierre, S. W. Wingett, P. Fraser, Comparison of Hi-C results using in-solution versus in-nucleus ligation. Genome Biol. 16, 175 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.J. A. Rivas-Pardo, E. C. Eckels, I. Popa, P. Kosuri, W. A. Linke, J. M. Fernández, Work done by titin protein folding assists muscle contraction. Cell Rep. 14, 1339–1347 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.H. Shin, Y. Shi, C. Dai, H. Tjong, K. Gong, F. Alber, X. J. Zhou, TopDom: An efficient and deterministic method for identifying topological domains in genomes. Nucleic Acids Res. 44, e70 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.B. M. Javierre, O. S. Burren, S. P. Wilder, R. Kreuzhuber, S. M. Hill, S. Sewitz, J. Cairns, S. W. Wingett, C. Várnai, M. J. Thiecke, F. Burden, S. Farrow, A. J. Cutler, K. Rehnström, K. Downes, L. Grassi, M. Kostadima, P. Freire-Pritchett, F. Wang, H. G. Stunnenberg, J. A. Todd, D. R. Zerbino, O. Stegle, W. H. Ouwehand, M. Frontini, C. Wallace, M. Spivakov, P. Fraser, J. H. Martens, B. Kim, N. Sharifi, E. M. Janssen-Megens, M. L. Yaspo, M. Linser, A. Kovacsovics, L. Clarke, D. Richardson, A. Datta, P. Flicek, Lineage-Specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell 167, 1369–1384.e19 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.L. Li, X. Lyu, C. Hou, N. Takenaka, H. Q. Nguyen, C. T. Ong, C. Cubeñas-Potts, M. Hu, E. P. Lei, G. Bosco, Z. S. Qin, V. G. Corces, Widespread rearrangement of 3D chromatin organization underlies polycomb-mediated stress-induced silencing. Mol. Cell 58, 216–231 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Y. Zhao, J. Zhou, L. He, Y. Li, J. Yuan, K. Sun, X. Chen, X. Bao, M. A. Esteban, H. Sun, H. Wang, MyoD induced enhancer RNA interacts with hnRNPL to activate target gene transcription during myogenic differentiation. Nat. Commun. 10, 5787 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.D. Hnisz, B. J. Abraham, T. I. Lee, A. Lau, V. Saint-André, A. A. Sigova, H. A. Hoke, R. A. Young, Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.A. Kaul, S. Bhattacharyya, F. Ay, Identifying statistically significant chromatin contacts from Hi-C data with FitHiC2. Nat. Protoc. 15, 991–1012 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.A. Roayaei Ardakany, H. T. Gezer, S. Lonardi, F. Ay, Mustache: Multi-scale detection of chromatin loops from Hi-C and Micro-C maps using scale-space representation. Genome Biol. 21, 256 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.K. Sun, L. Lu, H. Wang, H. Sun, Genome-wide profiling of YY1 binding sites during skeletal myogenesis. Genom. Data 2, 89–91 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.H. Zhang, D. J. Emerson, T. G. Gilgenast, K. R. Titus, Y. Lan, P. Huang, D. Zhang, H. Wang, C. A. Keller, B. Giardine, R. C. Hardison, J. E. Phillips-Cremins, G. A. Blobel, Chromatin structure dynamics during the mitosis-to-G1 phase transition. Nature 576, 158–162 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.L. He, Y. Ding, Y. Zhao, K. K. So, X. L. Peng, Y. Li, J. Yuan, Z. He, X. Chen, H. Sun, H. Wang, CRISPR/Cas9/AAV9-mediated in vivo editing identifies MYC regulation of 3D genome in skeletal muscle stem cell. Stem Cell Rep. 16, 2442–2458 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.C. Lepper, S. J. Conway, C. M. Fan, Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature 460, 627–631 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.M. van Overbeek, D. Capurso, M. M. Carter, M. S. Thompson, E. Frias, C. Russ, J. S. Reece-Hoyes, C. Nye, S. Gradia, B. Vidal, J. Zheng, G. R. Hoffman, C. K. Fuller, A. P. May, DNA repair profiling reveals nonrandom outcomes at Cas9-mediated breaks. Mol. Cell 63, 633–646 (2016). [DOI] [PubMed] [Google Scholar]
- 50.S. D. Gopinath, A. E. Webb, A. Brunet, T. A. Rando, FOXO3 promotes quiescence in adult muscle stem cells during the process of self-renewal. Stem Cell Rep. 2, 414–426 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.D. Palacios, C. Mozzetta, S. Consalvi, G. Caretti, V. Saccone, V. Proserpio, V. E. Marquez, S. Valente, A. Mai, S. V. Forcales, V. Sartorelli, P. L. Puri, TNF/p38α/polycomb signaling to Pax7 locus in satellite cells links inflammation to the epigenetic control of muscle regeneration. Cell Stem Cell 7, 455–469 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Z. Al Tanoury, J. Rao, O. Tassy, B. Gobert, S. Gapon, J.-M. Garnier, E. Wagner, A. Hick, A. Hall, E. Gussoni, O. Pourquié, Differentiation of the human PAX7-positive myogenic precursors/satellite cell lineage in vitro. Development 147, dev187344 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.L. Garcia-Prat, E. Perdiguero, S. Alonso-Martín, S. Dell’Orso, S. Ravichandran, S. R. Brooks, A. H. Juan, S. Campanario, K. Jiang, X. Hong, L. Ortet, V. Ruiz-Bonilla, M. Flández, V. Moiseeva, E. Rebollo, M. Jardí, H.-W. Sun, A. Musarò, M. Sandri, A. del Sol, V. Sartorelli, P. Muñoz-Cánoves, FoxO maintains a genuine muscle stem-cell quiescent state until geriatric age. Nat. Cell Biol. 22, 1307–1318 (2020). [DOI] [PubMed] [Google Scholar]
- 54.T. H. Cheung, N. L. Quach, G. W. Charville, L. Liu, L. Park, A. Edalati, B. Yoo, P. Hoang, T. A. Rando, Maintenance of muscle stem-cell quiescence by microRNA-489. Nature 482, 524–528 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Z. Du, H. Zheng, Y. K. Kawamura, K. Zhang, J. Gassler, S. Powell, Q. Xu, Z. Lin, K. Xu, Q. Zhou, E. A. Ozonov, N. Véron, B. Huang, L. Li, G. Yu, L. Liu, W. K. A. Yeung, P. Wang, L. Chang, Q. Wang, A. He, Y. Sun, J. Na, Q. Sun, H. Sasaki, K. Tachibana, A. H. F. M. Peters, W. Xie, Polycomb group proteins regulate chromatin architecture in mouse oocytes and early embryos. Mol. Cell 77, 825–839.e7 (2020). [DOI] [PubMed] [Google Scholar]
- 56.H. Zheng, B. Huang, B. Zhang, Y. Xiang, Z. du, Q. Xu, Y. Li, Q. Wang, J. Ma, X. Peng, F. Xu, W. Xie, Resetting epigenetic memory by reprogramming of histone modifications in mammals. Mol. Cell 63, 1066–1079 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Q. Bian, E. C. Anderson, Q. Yang, B. J. Meyer, Histone H3K9 methylation promotes formation of genome compartments in Caenorhabditis elegans via chromosome compaction and perinuclear anchoring. Proc. Natl. Acad. Sci. U.S.A. 117, 11459–11470 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.G. Fudenberg, M. Imakaev, C. Lu, A. Goloborodko, N. Abdennur, L. A. Mirny, Formation of chromosomal domains by loop extrusion. Cell Rep. 15, 2038–2049 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.S. Heinz, L. Texari, M. G. B. Hayes, M. Urbanowski, M. W. Chang, N. Givarkes, A. Rialdi, K. M. White, R. A. Albrecht, L. Pache, I. Marazzi, A. García-Sastre, M. L. Shaw, C. Benner, Transcription elongation can affect genome 3D structure. Cell 174, 1522–1536.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.S. Pomella, P. Sreenivas, B. E. Gryder, L. Wang, D. Milewski, M. Cassandri, K. Baxi, N. R. Hensch, E. Carcarino, Y. Song, H. C. Chou, M. E. Yohe, B. Z. Stanton, B. Amadio, I. Caruana, C. de Stefanis, R. de Vito, F. Locatelli, Y. Chen, E. Y. Chen, P. Houghton, J. Khan, R. Rota, M. S. Ignatius, Interaction between SNAI2 and MYOD enhances oncogenesis and suppresses differentiation in Fusion Negative Rhabdomyosarcoma. Nat. Commun. 12, 192 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.X. Ge, C. McFarlane, A. Vajjala, S. Lokireddy, Z. H. Ng, C. K. Tan, N. S. Tan, W. Wahli, M. Sharma, R. Kambadur, Smad3 signaling is required for satellite cell function and myogenic differentiation of myoblasts. Cell Res. 21, 1591–1604 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.P. Munoz-Canoves, J. Neves, P. Sousa-Victor, Understanding muscle regenerative decline with aging: New approaches to bring back youthfulness to aged stem cells. FEBS J. 287, 406–416 (2020). [DOI] [PubMed] [Google Scholar]
- 63.R. Sambasivan, R. Yao, A. Kissenpfennig, L. van Wittenberghe, A. Paldi, B. Gayraud-Morel, H. Guenou, B. Malissen, S. Tajbakhsh, A. Galy, Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development 138, 3647–3656 (2011). [DOI] [PubMed] [Google Scholar]
- 64.J. von Maltzahn, A. E. Jones, R. J. Parks, M. A. Rudnicki, Pax7 is critical for the normal function of satellite cells in adult skeletal muscle. Proc. Natl. Acad. Sci. U.S.A. 110, 16474–16479 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.A. Dong, J. Liu, K. Lin, W. Zeng, W. K. So, S. Hu, T. H. Cheung, Global chromatin accessibility profiling analysis reveals a chronic activation state in aged muscle stem cells. iScience 25, 104954 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.M. Khateb, J. Perovanovic, K. D. Ko, K. Jiang, X. Feng, N. Acevedo-Luna, J. Chal, V. Ciuffoli, P. Genzor, J. Simone, A. D. Haase, O. Pourquié, S. Dell’Orso, V. Sartorelli, Transcriptomics, regulatory syntax, and enhancer identification in mesoderm-induced ESCs at single-cell resolution. Cell Rep. 40, 111219 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.L. Liu, T. H. Cheung, G. W. Charville, T. A. Rando, Isolation of skeletal muscle stem cells by fluorescence-activated cell sorting. Nat. Protoc. 10, 1612–1624 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.M. Haeussler, K. Schönig, H. Eckert, A. Eschstruth, J. Mianné, J. B. Renaud, S. Schneider-Maunoury, A. Shkumatava, L. Teboul, J. Kent, J. S. Joly, J. P. Concordet, Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17, 148 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.F. Chen, J. Zhou, Y. Li, Y. Zhao, J. Yuan, Y. Cao, L. Wang, Z. Zhang, B. Zhang, C. C. Wang, T. H. Cheung, Z. Wu, C. C. L. Wong, H. Sun, H. Wang, YY1 regulates skeletal muscle regeneration through controlling metabolic reprogramming of satellite cells. EMBO J. 38, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.X. Chen, J. Yuan, G. Xue, S. Campanario, D. Wang, W. Wang, X. Mou, S. W. Liew, M. I. Umar, J. Isern, Y. Zhao, L. He, Y. Li, C. J. Mann, X. Yu, L. Wang, E. Perdiguero, W. Chen, Y. Xue, Y. Nagamine, C. K. Kwok, H. Sun, P. Muñoz-Cánoves, H. Wang, Translational control by DHX36 binding to 5′UTR G-quadruplex is essential for muscle stem-cell regenerative functions. Nat. Commun. 12, 5043 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.X. L. Peng, K. K. So, L. He, Y. Zhao, J. Zhou, Y. Li, M. Yao, B. Xu, S. Zhang, H. Yao, P. Hu, H. Sun, H. Wang, MyoD- and FoxO3-mediated hotspot interaction orchestrates super-enhancer activity during myogenic differentiation. Nucleic Acids Res. 45, 8785–8805 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.M. R. Corces, A. E. Trevino, E. G. Hamilton, P. G. Greenside, N. A. Sinnott-Armstrong, S. Vesuna, A. T. Satpathy, A. J. Rubin, K. S. Montine, B. Wu, A. Kathiria, S. W. Cho, M. R. Mumbach, A. C. Carter, M. Kasowski, L. A. Orloff, V. I. Risca, A. Kundaje, P. A. Khavari, T. J. Montine, W. J. Greenleaf, H. Y. Chang, An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.E. Crane, Q. Bian, R. P. McCord, B. R. Lajoie, B. S. Wheeler, E. J. Ralston, S. Uzawa, J. Dekker, B. J. Meyer, Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature 523, 240–244 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.J. Paulsen, M. Sekelja, A. R. Oldenburg, A. Barateau, N. Briand, E. Delbarre, A. Shah, A. L. Sørensen, C. Vigouroux, B. Buendia, P. Collas, Chrom3D: Three-dimensional genome modeling from Hi-C and nuclear lamin-genome contacts. Genome Biol. 18, 21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.G. Stik, E. Vidal, M. Barrero, S. Cuartero, M. Vila-Casadesús, J. Mendieta-Esteban, T. V. Tian, J. Choi, C. Berenguer, A. Abad, B. Borsari, F. le Dily, P. Cramer, M. A. Marti-Renom, R. Stadhouders, T. Graf, CTCF is dispensable for immune cell transdifferentiation but facilitates an acute inflammatory response. Nat. Genet. 52, 655–661 (2020). [DOI] [PubMed] [Google Scholar]
- 76.M. Di Stefano, R. Stadhouders, I. Farabella, D. Castillo, F. Serra, T. Graf, M. A. Marti-Renom, Transcriptional activation during cell reprogramming correlates with the formation of 3D open chromatin hubs. Nat. Commun. 11, 2564 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.I. Miguel-Escalada, S. Bonàs-Guarch, I. Cebola, J. Ponsa-Cobas, J. Mendieta-Esteban, G. Atla, B. M. Javierre, D. M. Y. Rolando, I. Farabella, C. C. Morgan, J. García-Hurtado, A. Beucher, I. Morán, L. Pasquali, M. Ramos-Rodríguez, E. V. R. Appel, A. Linneberg, A. P. Gjesing, D. R. Witte, O. Pedersen, N. Grarup, P. Ravassard, D. Torrents, J. M. Mercader, L. Piemonti, T. Berney, E. J. P. de Koning, J. Kerr-Conte, F. Pattou, I. O. Fedko, L. Groop, I. Prokopenko, T. Hansen, M. A. Marti-Renom, P. Fraser, J. Ferrer, Human pancreatic islet three-dimensional chromatin architecture provides insights into the genetics of type 2 diabetes. Nat. Genet. 51, 1137–1148 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.F. Serra, D. Baù, M. Goodstadt, D. Castillo, G. J. Filion, M. A. Marti-Renom, Automatic analysis and 3D-modelling of Hi-C data using TADbit reveals structural features of the fly chromatin colors. PLOS Comput. Biol. 13, e1005665 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.E. F. Pettersen, T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, T. E. Ferrin, UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 80.E. Wingender, P. Dietze, H. Karas, R. Knuppel, TRANSFAC: A database on transcription factors and their DNA binding sites. Nucleic Acids Res. 24, 238–241 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S15
Legends for tables S1 to S12
Tables S1 to S12