Abstract
The prediction of RNA secondary structure and thermodynamics from sequence relies on free energy minimization and nearest neighbor parameters. Currently, algorithms used to make these predictions are based on parameters from optical melting studies performed in 1 M NaCl. However, many physiological and biochemical buffers containing RNA include much lower concentrations of monovalent cations and the presence of divalent cations. In order to improve these algorithms, thermodynamic data was previously collected for RNA duplexes in solutions containing 71, 121, 221, and 621 mM Na+. From this data, correction factors for free energy (ΔG°37) and melting temperature (Tm) were derived. Despite these newly derived correction factors for sodium, the stabilizing effects of magnesium have been ignored. Here, the same RNA duplexes were melted in solutions containing 0.5, 1.5, 3.0, and 10.0 mM Mg2+ in the absence of monovalent cations. Correction factors for Tm and ΔG°37 were derived to scale the current parameters to a range of magnesium concentrations. The Tm correction factor predicts the melting temperature within 1.2°C, and the ΔG°37 correction factor predicts the free energy within 0.30 . These newly derived magnesium correction factors can be incorporated into algorithms that predict RNA secondary structure and stability from sequence.
Significance
RNA secondary structure prediction guides a variety of experimental methods involving RNA. Nearest neighbor parameters derived from RNA solutions in 1 M NaCl can be used to predict RNA secondary structure; however, these conditions are far from native cellular conditions. Here, we scale melting temperature (Tm) and free energy (ΔG°37) predictive algorithms (derived from nearest neighbor parameters) to a concentration range of magnesium that is within those found in cellular environments and in many buffers designed to mimic cellular conditions. These new predictive algorithms can be used to more accurately predict RNA secondary structure from sequence in magnesium-containing solutions that mimic cell-like conditions.
Introduction
Ribonucleic acid (RNA) has the ability to participate in a variety of cellular functions in addition to protein synthesis (1,2,3). Due to its single-stranded nature, RNA can often fold back onto itself to form complex tertiary structures that allow for unique functions (4,5). These findings have deepened the desire to understand RNA tertiary structure and how these structures can impact RNA function. Because methods for solving RNA tertiary structure, such as nuclear magnetic resonance (NMR), x-ray crystallography, and cryogenic electron microscopy (cryo-EM), are difficult, time-intensive, and resource-heavy, research working toward accurate prediction of RNA 3D structure from sequence is ongoing (6,7,8). Although predicting 3D structure from sequence can be difficult, a promising intermediate step is predicting secondary structure from sequence. The development of accurate secondary structure predictive algorithms in various environmental conditions can serve as a strong scaffold to build upon for prediction of RNA 3D structures. This is especially true if environmental conditions mimic those found in a cell.
One of the most common methods used to predict secondary structure from sequence uses free energy minimization with the nearest neighbor model (9,10). The nearest neighbor model was derived from optical melting studies of short RNA duplexes in 1.0 M NaCl, 10 or 20 mM sodium cacodylate, and 0.5 mM Na2EDTA (10). However, intracellular [Na+] is much lower than the 1 M NaCl buffer used in the optical melting studies to develop the nearest neighbor model. Also, molecular biology techniques, such as PCR, often use buffers containing 20–100 mM monovalent cations (11,12). Optical melting studies to determine the thermodynamic parameters of short RNA duplexes in more biologically relevant [Na+] (71, 121, 221, and 621 mM) were conducted to derive Na+ correction factors (13). These correction factors can be incorporated into prediction algorithms to more accurately predict RNA secondary structure in varying [Na+].
Although these newly derived parameters for Na+ are more useful for modern biological techniques, they still neglect the stabilizing effects of divalent cations, such as Mg2+, which are essential for intracellular enzymatic reactions and modern physiological buffers. Like monovalent cations, divalent cations are responsible for binding to nucleic acids and affecting their physical properties as well as stabilizing nucleic acid duplexes and facilitating secondary and tertiary folding (14,15). Chelated and free divalent metal ions, such as Mg2+, also participate in more than just RNA folding interactions (16). For example, chelation of Mg2+ can assist with RNA catalysis, protection, and stabilization (16). Many studies have also shown Mg2+ to participate in an in-line attack, catalyzing RNA cleavage events (17,18,19). The free [Mg2+] in a cell varies depending on the organism. Free Mg2+ concentrations of 0.2–1.3 mM and 2.0–3.0 mM have been reported for eukaryotic and bacterial cells, respectively (16,20).
Foundational studies investigating the effects that Na+ (21) and Mg2+ (22) have on nucleic acid thermodynamics have shown similarities between the 1.0 M NaCl buffered conditions and 150 mM NaCl with 10.0 mM MgCl2 (22). Additional studies have investigated how different monovalent and divalent cations of varying concentrations influence nucleic acid stability (23,24,25,26,27,28). However, these studies either only investigated monovalent cations (24), used a conceptual model of a single nucleotide sequence as a case study (25), or the environment containing the cations contained both divalent and monovalent cations either in the same solution (23,26,28) or in the presence of crowding agents (27). Single-molecule studies have measured DNA stability in Mg2+ and Na+ (29), measured RNA stability in Na+ (30), and identified RNA binding sites for Mg2+ and Na+ ions (31). To our knowledge, no studies have been conducted that focus exclusively on the stability of short RNA duplexes in the presence of biologically relevant [Mg2+] and no monovalent cations ([Mon+]).
It has been shown that monovalent and divalent cations compete for interaction with nucleic acids (32,33,34). In order to determine how Na+ and Mg2+ compete to stabilize DNA, Owczarzy et al. first studied the contributions of Na+ alone (35) and Mg2+ alone (11) on duplex stability. Next, they used information from the studies completed with Na+ alone and Mg2+ alone to help understand the thermodynamics of DNA in mixtures of monovalent and divalent cations (11). Although this study was performed with DNA, it is likely that RNA in solutions containing both Mg2+ and Mon+ may act in a similar manner. Systematic thermodynamic data for RNA oligonucleotides with different ratios of [Mg2+] and [Mon+] are needed. As mentioned previously, the effect of Mg2+ alone (in the absence of monovalent salt cations), which is required to understand how mixtures of Mg2+ and Mon+ affect RNA stability, has not been investigated for short RNA duplexes.
To that end, the thermodynamic data from optical melting studies are reported for a set of 17 RNA duplexes. Each duplex was melted with a range of [Mg2+] (0.5, 1.5, 3.0, and 10.0 mM). Using the RNA Na+ correction factors of Chen and Znosko (13) and the DNA Mg2+ correction factors of Owczarzy et al. (11) as a guide, Tm and ΔG°37 correction factors are reported here to adjust the standard 1 M NaCl RNA nearest neighbor predictions to account for the presence of Mg2+. Similar to the previously mentioned Na+ correction factors, these can be incorporated into prediction algorithms that predict RNA secondary structure and stability from sequence. Building on the knowledge of how Na+ alone affects RNA stability, the understanding of how Mg2+ alone affects RNA stability will guide future studies to determine how mixtures of Mg2+ and Mon+ affect RNA stability.
Materials and methods
RNA oligonucleotide preparation
All sequences studied here were also used by Xia et al. (10) to derive the RNA nearest neighbor parameters in 1 M NaCl and by Chen and Znosko (13) to derive a correction factor for varying [Na+]. All oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA) and purified using standard procedures (36,37,38).
Optical melting studies
A melting scheme was developed to ensure that each duplex was melted at least nine times at different oligonucleotide concentrations. A Beckman-Coulter DU800 spectrophotometer was equipped with a high-performance temperature controller, allowing for variability in temperature during absorbance measurements. A heating rate of 1°C/min was used to obtain absorbance versus temperature melting curves between 15°C and 95°C at 280 nm for purely G-C duplexes and 8°C and 85°C at 260 nm for all other duplexes. MeltWin v3.5 (39) was used to analyze the absorbance versus temperature curves and produce Tm−1 versus ln CT plots to determine the thermodynamic parameters of each duplex.
All optical melting studies used to derive correction factors were conducted in the presence of 0.5, 1.5, 3.0, or 10.0 mM MgCl2 in buffer containing 2 mM Tris at pH 8.3. This range of [Mg2+] represents the range of free Mg2+ in various cells (20,40,41,42,43). No monovalent cations were added to the buffer to determine the effect of Mg2+ alone on RNA duplex stability.
pH effects on RNA stability
Tris buffer was used in this study to prevent competition between the divalent cations studied here and monovalent cations. The concentration of monovalent cations in phosphate, cacodylate, and citrate buffers is higher than the trace amounts found in Tris. In addition, Tris buffers are usually employed in modern biological applications, such as PCR (11,12). However, the pH of Tris buffer is temperature dependent, leading to a decrease in pH of approximately 0.029 per 1°C increase. To verify that a change in pH of the Tris buffer over the temperature range of a typical optical melting experiment would not affect duplex thermodynamics, pH effects on Tm were determined for the RNA duplexes (5′-CGCGCG-3′)2 and (5′-ACUUAAGU-3′)2 as previously described by Owczarzy et al. with DNA (11). Five different buffers with pH values of 6.5, 7.0, 7.6, 8.3, and 12.1 were prepared. The buffers at pH 6.5, 7.0, and 7.6 contained 1.5 mM MgCl2, 50 mM NaCl, and 10 mM sodium cacodylate. The buffer at pH 8.3 contained 10 mM 3-(N-morpholino)propanesulfonic acid (MOPS) instead of sodium cacodylate. The buffer at pH 12.1 contained 10 mM N-cyclohexyl-2-aminoethanesulfonic acid (CHES) instead of sodium cacodylate or MOPS.
UV monitoring of RNA cleavage and degradation
To verify that cleavage was not occurring during the optical melting experiments, forward (heating the duplex by 1°C/min) followed by reverse melting (cooling the single strands by 1°C/min) experiments were conducted with the duplexes (5′-CGCGCG-3′)2 and (5′-ACUUAAGU-3′)2 in both 10.0 mM MgCl2 in Tris and standard 1 M NaCl optical melting buffer (containing 1.0 M NaCl, 20 mM sodium cacodylate, and 0.5 mM Na2EDTA at pH 7.0). No reports of cleavage events for short RNA oligonucleotides in standard optical melting buffer during optical melting experiments exist, so the oligonucleotides in the standard optical melting buffer served as a reference. Cooling of the samples occurred directly after reaching the end temperature in the forward melting experiment. The samples remained at the end temperature for 1 min before beginning the reverse melt. These two oligonucleotides were chosen because they varied in length and fraction of G-C basepairs (fGC). In addition, the U-A step in (5′-ACUUAAGU-3′)2 is a known cleavage site when in the presence of Mg2+ (17). These oligonucleotides also represented the two extremes in terms of ΔG°37 and Tm for this study. One can assume that all oligonucleotides that fall within, or close to, these two extremes should behave in a similar manner. More specifically, if the ΔG°37 and Tm do not vary significantly for these two duplexes during the forward and reverse melting experiments while in solutions containing Mg2+ at high temperatures, all other duplexes should not vary significantly as well.
To further verify that cleavage was not occurring during the melting experiments and that using serial dilutions of previously melted duplexes was a valid approach, the (5′-CGCGCG-3′)2 and (5′-ACUUAAGU-3′)2 duplexes were melted in the 10.0 mM MgCl2 buffer using fresh, never before melted oligonucleotide for each of the concentrations in the Tm−1 versus ln CT plot. A total of nine concentrations were used in the melt scheme. The same absorbance wavelengths and heating rates were used as previously mentioned. The results of using fresh, never before melted oligonucleotide for each of the concentrations were compared with the results of using serial dilutions of previously melted samples.
HPLC monitoring of RNA cleavage and degradation
In addition to reverse melts and using fresh oligonucleotide in a melt scheme, high-performance liquid chromatography (HPLC) was also used to verify that no cleavage and minimal degradation occurred during the melting experiments. The duplexes (5′-CGCGCG-3′)2 and (5′-ACUUAAGU-3′)2 were analyzed by HPLC before and after optical melting experiments in 10.0 mM MgCl2 with 2.0 mM Tris buffer and in standard 1 M NaCl optical melting buffer. Before column injection, before and after melt samples were diluted to 200 ppm with autoclaved nanopure water. The melt buffers alone and the samples in the diluted melt buffers were analyzed on a Shimadzu LCMS-2010EV, with the MS function disconnected. UV detection wavelengths of 260 and 280 nm were used. A C18 reverse phase column was used with a flow rate of 0.25 mL/min. The pH of the sample and Mg2+-containing buffer was adjusted below 7.0 to prevent degradation of the C-18 column before injection. HPLC-grade 100% acetonitrile and 50 mM ammonium acetate were chosen as the solvents. An isocratic method was used, starting at 0% acetonitrile. From 4 to 24 min, the percent acetonitrile increased from 5% to 15%. The percent acetonitrile then increased to 30% for 2 min and 80% for an additional 8 min.
Predicting Tm
The RNA data collected here in varying [Mg2+] were tested against six DNA correction factors for [Na+] (44,45,46,47,48,49,50), two RNA correction factors for [Na+] (13), four DNA correction factors for [Mg2+] (11,51,52), and one RNA correction factor for [Mg2+] (53,54). All existing correction factors were tested using the experimental 1 M NaCl Tm values, and these values were corrected to predict Tm values at 0.5, 1.5, 3.0, and 10.0 mM Mg2+. The average deviation of the models was determined by comparing 68 experimental melting temperatures (17 duplexes at four different [Mg2+]) to the corresponding predicted melting temperatures using |ΔTm|ave:
| (1) |
To improve the average deviation, the coefficients for nine of the 13 models mentioned above were updated. This was conducted using all the data reported here and the LINEST function (linear regression) in Microsoft Excel. For the linear regression, the numerical coefficients in the previously published models were set as the variables, and the melting temperatures were set as the constants, allowing for the derivation of revised coefficients based on the data reported here. These updated models were then tested using Eq. 1.
Predicting ΔG°37
Similar to what was described above for Tm, previously published correction factors for ΔG°37, one for DNA in [Na+] (50), four for RNA in [Na+] (13), and one for DNA in [Mg2+], were tested for average deviation (11). The average deviation of the models was determined by comparing the 68 experimental ΔG°37 values to the corresponding predicted ΔG°37 values using |ΔΔG°37|ave:
| (2) |
Again, based on the RNA data reported here, the coefficients for five of the six models tested were updated in the same manner as the Tm correction factors and evaluated for average deviation using Eq. 2.
Leave-one-out analysis
To ensure the recommended correction factors and updated coefficients are predictive as well as postdictive, a comprehensive leave-one-out analysis was conducted for the recommended correction factors (see discussion). For this analysis, the data for an entire oligonucleotide sequence (experimental thermodynamic parameters for 0.5, 1.5, 3.0, and 10.0 mM Mg2+) were excluded in the linear regression analysis used to determine the coefficients, leaving 64 unique data points and generating new coefficients for the recommended correction factors. The process was repeated 17 times, each time leaving out one of the 17 oligonucleotides studied here. The coefficients resulting from each regression were used to predict the Tm and ΔG°37 of the oligonucleotide that was left out in each of the Mg2+ buffers. Both the updated coefficients and predicted parameters were compared with the original model proposed and the experimental values, respectively. The and were calculated for each of the 17 new models both for the oligonucleotides used in the fitting of the model and the one oligonucleotide that was left out. Predictions from the leave-one-out analysis were compared with predictions from the recommended correction factors. The average of each coefficient was also calculated across the 17 new models.
Results
Effects of buffer pH on melting temperature
RNA contains a phosphate backbone and nitrogenous bases that are capable of deprotonation and protonation at high and low pH, respectively. Deprotonation and protonation can influence hydrogen bonding interactions between bases, ultimately affecting duplex formation. The pKa value(s) of adenosine is 3.5, guanosine are 1.6 and 9.2, uridine is 9.2, and cytidine is 4.2 (55). Therefore, at pH 7, all bases are likely to exist as neutral species with no charged groups. Hydrolysis of the RNA backbone is possible in a pH range above 8.0 (56). The presence of divalent metal ions in solution can stabilize the transition state in general acid-base-catalyzed cleavage of the RNA backbone (19). The pH of Tris buffer is temperature dependent, exhibiting a decrease in pH with an increase in temperature. An investigation into the effects of buffer pH on melting temperature was necessary.
The melting temperatures for two RNA self-complementary duplexes, (5′-CGCGCG-3′)2 and (5′-ACUUAAGU-3′)2, in the pH range of 6.5–12.1 were measured (Fig. S1). Melting temperatures in the pH range of 6.5–8.3 varied only slightly for each oligonucleotide (0.6°C and 0.9°C, respectively). When the pH was increased to 12.1, significant changes were evident. In theory, at this pH, all bases should be deprotonated at positions involved in hydrogen bonding, decreasing the overall stability of the duplex. Degradation and cleavage of the phosphodiester backbone would also theoretically occur at a pH of 12.1. At this pH, the melting temperature of (5′CGCGCG-3′)2 decreased by 9.8°C. For (5′-ACUUAAGU-3′)2 at pH 12.1, the interactions between the bases were disrupted to the point where a duplex did not form, cleavage events likely took place, and no melting temperature was obtained due to the inability to fit the melt curves to an assumed two-state model. The results found here for RNA duplexes in the pH range of 6.5–8.3 compare to the optical melting results obtained for DNA duplex oligomers with a pH range of 6.5–8.3 (11), a DNA hairpin with a pH range of 5.0–8.0 (57), T2 phage DNA with pH ranges of 3.5–5.4 and 7.0 to 9.6 (58), and calf thymus DNA with a pH range of 3–12 (59), where melting temperatures did not change significantly between pH 6.5 and pH 8.3. Studies investigating the stability of RNA internal loops reported average differences in ΔG°37 and Tm at pH 7.5 and 5.5 of <0.4 and <3.0°C in the presence of Mg2+, respectively (28). The same study reported differences in ΔG°37 and Tm at pH 7.5 and 5.5 for an RNA helix of <0.2 and <1.1°C, respectively (28). Because these pH ranges had no significant effect on melting temperatures and represent a range where no deprotonation or protonation of the bases should occur, the Tris buffer used for further optical melting experiments was set at a pH of 8.3. Any decrease in pH with increasing temperature should remain in the tested range of 6.5–8.3 as demonstrated by Owczarzy et al. (11).
Monitoring of RNA cleavage and degradation
Forward optical melting experiments immediately followed by reverse optical melting experiments of the (5′-CGCGCG-3′)2 and (5′-ACUUAAGU-3′)2 duplexes in the 10.0 mM Mg2+ buffer and in standard 1 M NaCl buffer (1.0 M NaCl, 20 mM sodium cacodylate, and 0.5 mM Na2EDTA at pH 7.0) showed minimal variation in the thermodynamic parameters acquired. The (5′-CGCGCG-3′)2 duplex showed a ΔΔG°37 from the Tm−1 versus ln CT plot between the forward and reverse melt in 10.0 mM Mg2+ and in standard 1 M NaCl buffer of −0.01 and −0.10 respectively, with a ΔTm of 0.4°C and −0.6°C, respectively (Table S1). The ΔΔG°37 for the (5′-ACUUAAGU-3′)2 duplex in 10.0 mM Mg2+ and in standard 1 M NaCl buffer was −0.05 and 0.01 respectively, with no change in melting temperature for both environmental conditions (Table S1). For both duplexes studied, the ΔΔG°37 and the ΔTm in Mg2+-containing buffers were relatively small and comparable to Na+-containing buffers. This suggested no cleavage or significant degradation occurred in Mg2+-containing buffers after heating to a high temperature (>80°C). If cleavage or significant degradation were to occur, one would have expected a large ΔΔG°37 and ΔTm between the forward and reverse melts in the presence of Mg2+. It is important to note, a forward melting experiment immediately followed by a reverse melting experiment subjected the duplexes to high temperatures for twice the length of time compared with traditional, one-way melting experiments. Additionally, both the forward and reverse melt curves exhibited a single inflection point with a characteristic sigmoidal shape (data not shown), suggesting that there is a single species in solution, and no cleavage was taking place.
In comparison to the data collected while using serial dilutions to obtain the different concentrations of RNA duplex, using fresh oligonucleotide in Mg2+-containing buffers for each of the concentrations in the melt scheme also showed little variation in the thermodynamic parameters acquired. For the (5′-CGCGCG-3′)2 duplex, there was no change in ΔG°37 and a 1.1°C difference in Tm (Table S2). For the (5′-ACUUAAGU-3′)2 duplex, there was a −0.09 difference in ΔG°37 and a 0.2°C difference in Tm (Table S2). All values were within experimental error. These findings validated our method of serial diluting the RNA sample in Mg2+-containing buffers during the melt scheme. No significant differences between the fresh oligonucleotide and the reused oligonucleotide melt schemes were observed, again suggesting no cleavage or significant degradation occurred during our analysis.
Although there was no evidence of RNA cleavage in the UV spectrum while in the presence of Mg2+ during the optical melting experiments, HPLC was used to analyze two RNA oligonucleotides before and after melting to verify that no cleavage had occurred. The HPLC chromatograms were identical before and after melting the RNA sample in both 10.0 mM Mg2+ and standard 1 M NaCl buffer for (5′-CGCGCG-3′)2 (Fig. S2) and (5′-ACUUAAGU-3′)2 (Fig. S3). This indicated that the addition of heat and Mg2+ did not result in cleavage of the RNA duplexes. These findings were despite the presence of a U-A step in the (5′-ACUUAAGU-3′)2 sequence, a cleavage site found in the env22 twister ribozyme that is catalyzed by Mg2+ (17).
RNA thermodynamic parameters
Experimental Tm, ΔG°37, ΔH°, and ΔS° values for all 17 duplexes in four different [Mg2+] are available in Table S3. The experimental Tm and ΔG°37 values from the Tm−1 versus ln CT plots for all duplexes in all [Mg2+] are presented in Tables 1 and 2, respectively. Duplexes melted in 0.5, 1.5, 3.0, and 10.0 mM Mg2+ had average Tm values that were 5.2°C lower, 2.1°C lower, 0.4°C lower, and 1.6°C higher, respectively, than that of the same duplexes melted in 1 M NaCl. Likewise, the average ΔG°37 values for the duplexes melted in the same [Mg2+] were 0.85 less, 0.30 less, 0.03 more, and 0.41 more stable, respectively, than the same duplexes in 1 M NaCl. Based on this data, 3.0 mM Mg2+ stabilizes RNA comparably to 1 M NaCl.
Table 1.
Experimental melting temperatures for self-complementary RNA duplexes in solutions of varying magnesium concentrations and in 1 M sodium chloride with no magnesium
| RNA sequence (5′ to 3′)a | fGCb |
Tmc (°C) |
||||
|---|---|---|---|---|---|---|
| 0.5 mM Mg2+ | 1.5 mM Mg2+ | 3.0 mM Mg2+ | 10.0 mM Mg2+ | 1 M NaCld | ||
| CGCGCG | 1.00 | 53.3 | 57.1 | 58.3 | 60.3 | 57.8 |
| CGGCCG | 1.00 | 58.2 | 61.2 | 63.6 | 64.9 | 63.2 |
| GCGCGC | 1.00 | 56.9 | 59.4 | 61.0 | 62.3 | 62.5 |
| ACCGGU | 0.67 | 52.3 | 55.6 | 56.8 | 58.4 | 53.9 |
| AGCGCU | 0.67 | 43.3 | 47.0 | 49.2 | 51.7 | 52.0 |
| CACGUG | 0.67 | 38.7 | 41.7 | 43.2 | 44.9 | 42.8 |
| CAGCUG | 0.67 | 37.9 | 40.8 | 42.4 | 44.7 | 43.1 |
| CCAUGG | 0.67 | 43.9 | 46.3 | 47.9 | 50.1 | 46.4 |
| CCUAGG | 0.67 | 48.4 | 51.4 | 52.3 | 54.2 | 50.0 |
| CUGCAG | 0.67 | 40.5 | 42.8 | 44.2 | 46.9 | 45.3 |
| GACGUC | 0.67 | 41.6 | 45.2 | 45.8 | 48.4 | 46.2 |
| GAGCUC | 0.67 | 41.4 | 44.5 | 46.7 | 48.5 | 48.7 |
| GCAUGC | 0.67 | 40.1 | 43.3 | 45.0 | 47.2 | 45.7 |
| ACUUAAGU | 0.25 | 32.1 | 35.9 | 38.5 | 40.0 | 40.3 |
| AACUAGUU | 0.25 | 38.8 | 42.0 | 43.4 | 45.8 | 45.7 |
| ACUAUAGU | 0.25 | 38.1 | 41.0 | 43.1 | 44.7 | 44.0 |
| AGAUAUCU | 0.25 | 34.9 | 37.8 | 40.0 | 43.1 | 41.4 |
All oligonucleotides are forming self-complementary duplexes in solution.
fGC is the fraction of G-C basepairs in the duplex.
Calculated for 0.1 mM oligonucleotide concentration based on the Tm−1 versus ln CT plots. Errors are typically ∼1.0°C for melting temperature measurements.
Table 2.
Experimental ΔG°37 values for self-complementary RNA duplexes in solutions of varying magnesium concentrations and in 1 M sodium chloride with no magnesium
| RNA sequence (5′ to 3′)a | fGCb | ΔG°37 (kcal/mol)c |
||||
|---|---|---|---|---|---|---|
| 0.5 mM Mg2+ | 1.5 mM Mg2+ | 3.0 mM Mg2+ | 10.0 mM Mg2+ | 1 M NaCld | ||
| CGCGCG | 1.00 | −8.78 | −9.35 | −9.40 | −9.86 | −9.12 |
| CGGCCG | 1.00 | −9.37 | −9.77 | −10.07 | −10.25 | −9.90 |
| GCGCGC | 1.00 | −9.34 | −9.70 | −10.13 | −10.22 | −10.56 |
| ACCGGU | 0.67 | −8.50 | −8.83 | −9.21 | −9.74 | −8.51 |
| AGCGCU | 0.67 | −6.65 | −7.33 | −7.61 | −8.13 | −7.99 |
| CACGUG | 0.67 | −5.96 | −6.46 | −6.78 | −7.05 | −6.59 |
| CAGCUG | 0.67 | −5.83 | −6.32 | −6.61 | −6.99 | −6.68 |
| CCAUGG | 0.67 | −7.02 | −7.48 | −7.88 | −8.26 | −7.30 |
| CCUAGG | 0.67 | −7.86 | −8.31 | −8.61 | −8.84 | −7.80 |
| CUGCAG | 0.67 | −6.27 | −6.76 | −7.18 | −7.53 | −7.11 |
| GACGUC | 0.67 | −6.51 | −7.24 | −7.48 | −7.96 | −7.35 |
| GAGCUC | 0.67 | −6.45 | −7.14 | −7.53 | −7.87 | −7.98 |
| GCAUGC | 0.67 | −6.23 | −6.85 | −7.15 | −7.52 | −7.38 |
| ACUUAAGU | 0.25 | −4.85 | −5.47 | −5.97 | −6.34 | −6.16 |
| AACUAGUU | 0.25 | −6.00 | −6.69 | −6.86 | −7.36 | −7.16 |
| ACUAUAGU | 0.25 | −5.89 | −6.51 | −6.89 | −7.35 | −6.98 |
| AGAUAUCU | 0.25 | −5.27 | −5.83 | −6.29 | −6.78 | −6.58 |
All oligonucleotides are forming self-complementary duplexes in solution.
fGC is the fraction of G-C basepairs in the duplex.
Calculated based on the Tm-1 versus ln CT plots. Errors are typically <0.5 kcal/mol for ΔG°37 values.
Tm correction factors
It is evident that short RNA duplexes in buffers containing Mg2+ melt at temperatures that are different than those for RNA duplexes in varying amounts of Na+. Although there are Tm correction factors for RNA duplexes in varying amounts of Na+, Tm correction factors are needed for RNA in Mg2+-containing solutions for accurate predictions. There have been numerous previously published correction factors for DNA and RNA in Na+ and a few for DNA in Mg2+ that were tested using the RNA data reported here. These can be found in Table S4 (with Eqs. S1–S19 therein) with the corresponding average deviations of those models calculated as described in materials and methods. Not surprisingly, the correction factors for DNA in Mg2+ provided smaller deviations, |ΔTm|ave ≤5.0°C, than did those for DNA and RNA in Na+, with |ΔTm|ave ≥18.8°C. Nevertheless, a wide range of correction factors were updated based on the data reported here to improve the average deviations of the models (Table 3 and Eqs. 3–9 therein). With new coefficients, all models, with the exception of Eq. 8, improved, resulting in |ΔTm|ave values ≤ 2.7°C. Eq. 7 and Eq. 9 both had average deviations of 1.2°C. Because Eq. 7 is simpler than Eq. 9, it is the recommended Tm correction factor to be used for RNA in [Mg2+] ≤10.0 mM. Eq. 7 also works well predicting Tm across the entire magnesium concentration range studied here. Comparing the experimental Tm to the predicted Tm calculated using Eq. 7 resulted in an average |ΔTm| of 1.4°C, 1.4°C, 1.1°C, and 1.1°C in the 0.5 mM, 1.5 mM, 3.0 mM, and 10.0 mM Mg2+ solutions, respectively.
Table 3.
Newly derived RNA correction factors
| Eq no. | Name | Equation | Average deviationa |
|---|---|---|---|
| RNA Tm correction factorsb | |||
| 3 | Modified Schildkraut-Lifson and Tm and 12.5log[Na+] equation | 2.3°C | |
| 4 | Modified Wetmur equation | 2.2°C | |
| 5 | Modified Marmur-Schildkraut-Doty and Frank-Kamenetskii Tm equation | 2.7°C | |
| 6 | Modified SantaLucia equationc,d | 2.2°C | |
| 7 | Modified Chen Tm quadratic equation | 1.2°C | |
| 8 | Modified Chen 1/Tm quadratic equationc | 18.8°C | |
| 9 | Modified Owczarzy magnesium Tm correction factor equationc | 1.2°C | |
| RNA ΔG°37 Correction factorsb | |||
| 10 | Modified SantaLucia ΔG°37 equation | 0.48 kcal/mol | |
| 11 | Modified Chen ΔG°37 linear equation | 0.47 kcal/mol | |
| 12 | Modified Chen ΔG°37 quadratic equation | 0.30 kcal/mol | |
| 13 | Modified Chen 1/ΔG°37 quadratic equation | 0.36 kcal/mol | |
| 14 | Modified Owczarzy magnesium ΔG°37 correction factor equationd | 0.88 kcal/mol | |
As stated in materials and methods, |ΔTm|ave is used to evaluate the average deviation of the Tm correction factors, and |ΔΔG°37|ave is used to evaluate the average deviation of the ΔG°37 correction factors.
1 M NaCl melting temperatures and free energy values were used as the starting point for all correction factors tested. Original correction factors with their respective references can be found in Table S4.
In this equation, Tm should be in units of K.
In this equation, ΔH° is in cal/mol, and the experimental ΔH° value from the Tm−1 versus ln CT plot was used.
The leave-one-out analysis for assessing the updated coefficients for Eq. 7 showed minimal variation in each of the coefficients upon excluding each of the 17 sequences from the linear regression (Fig. S4). Taking the differences in predicted Tm using the coefficients in the leave-one-out analysis and the coefficient proposed in Eq. 7 yielded an average difference of 0.1°C (Table S5). The average coefficient from the leave-one-out analysis was equal to the proposed coefficients in Eq. 7, validating the use of the proposed coefficients in Eq. 7.
ΔG°37 correction factors
Despite the many models available for predicting Tm values, there are only a few models available for predicting changes in ΔG°37 values as a function of salt concentration. Those available were tested with the RNA data reported here and are presented in Table S4 with the corresponding average deviations of those models. Similar to the Tm correction factors, the one available for DNA in Mg2+ resulted in smaller deviations, |ΔΔG°37|ave = 0.59 , than did those for DNA and RNA in Na+. Again, a wide range of these correction factors were updated with new coefficients based on the data reported here to improve the average deviations of the models (Table 3 and Eqs. 10–14 therein). With the exception of Eq. 14, all newly derived ΔG°37 correction factors for RNA in Mg2+ had |ΔΔG°37|ave values ≤ 0.48 . However, due to its average deviation, |ΔΔG°37|ave = 0.30 , and consistency with the Tm correction factor discussed above, Eq. 12 is the recommended ΔG°37 correction factor to be used for RNA in [Mg2+] ≤10.0 mM. Eq. 12 also works well predicting ΔG°37 values across the entire magnesium concentration range studied here. Comparing the experimental ΔG°37 values to the predicted ΔG°37 calculated using Eq. 12 resulted in an average |ΔΔG°37| of 0.32 , 0.31 , 0.27 , and 0.29 in the 0.5 mM, 1.5 mM, 3.0 mM, and 10.0 mM Mg2+ solutions, respectively.
The leave-one-out analysis for assessing the updated coefficients, similar to Eq. 7, showed minimal variation in each of the coefficients upon excluding each of the 17 sequences from the linear regression (Fig. S4). Taking the differences in predicted ΔG°37 using the coefficients in the leave-one-out analysis and the coefficient proposed in Eq. 7 yielded an average difference of 0.04 (Table S6). The average coefficient from the leave-one-out analysis was equal to the proposed coefficients in Eq. 12, validating the use of the proposed coefficients in Eq. 12.
Discussion
RNA cleavage and degradation
It has been reported previously that if RNA solutions containing Mg2+ are heated, it can lead to RNA cleavage (60,61,62,63). These studies were performed on large RNA molecules, and extensive RNA cleavage was reported when [Mg2+] ≥50 mM (60,61,63). Additionally, the likelihood of RNA cleavage increased at pH levels ≥8.0 and at temperatures ≥37°C, especially when the temperature was elevated over an extended period of time (60,62,63). It has also been reported that cleavage of RNA is significantly reduced when [Mg2+] is <40 mM (60). Other thermodynamic studies indicate that the introduction of Mg2+ and heat does not always induce RNA cleavage (64). In this work, the highest [Mg2+] studied was 10.0 mM, and RNA duplexes were not incubated at elevated temperatures for extended periods of time. Nonetheless, we wanted to confirm that the RNA duplexes studied here were not cleaved during the melting experiments in the presence of low [Mg2+].
The results from four different experiments suggested that there is no cleavage of short RNA oligonucleotides during the optical melting experiments conducted here. First, most optical melting experiments were completed as a series of dilutions. The results of the melting experiments after each dilution were consistent with one another, suggesting that none of the short RNA duplexes studied here under these conditions were cleaved during melting experiments. Additionally, each melt curve collected here was a single, sharp sigmoidal curve. If cleavage did occur during the melting experiment, we would expect the samples resulting from serial dilutions to consist of multiple species in solution. This would likely have resulted in melt curves with multiple transitions or very broad transitions.
A forward and then a reverse melt was also conducted on two RNA duplexes, (5′-CGCGCG-3′)2 and (5′-ACUUAAGU-3′)2, at the highest [Mg2+] of 10.0 mM and in standard 1 M NaCl buffer. During the forward and reverse melting experiments, the RNA duplexes were at higher temperatures for twice the amount of time compared with standard optical melting experiments. The high temperature should theoretically increase the reaction rate for a Mg2+ catalyzed cleavage event of the phosphodiester backbone. If a cleavage event of the RNA backbone were to occur, the event would likely occur at high temperatures. These conditions were prolonged during the forward and reverse melting experiments. The data from the forward and reverse melts of the two duplexes were identical in both conditions (Table S1), suggesting that no cleavage and no significant degradation occurred. If cleavage did occur in the Mg2+-containing samples, we would expect to see large differences between the forward and reverse melting data. The sample in standard 1 M NaCl buffer sample served as a reference since there are no reported instances of RNA being cleaved when suspended in this buffer. Therefore, we assume that if the before and after optical melting data in the Mg2+ buffer was similar to the before and after optical melting data in the NaCl buffer, there was no significant cleavage or degradation of RNA in the Mg2+ buffer. Because no large differences were seen, and the differences in the Mg2+ data were similar to that in standard 1 M NaCl buffer, we assumed no cleavage or significant degradation of the RNA samples occurred.
Additionally, instead of using serial dilutions, fresh oligonucleotide was used for each concentration in a series of melts in the 10.0 mM MgCl2 buffer for the (5′-CGCGCG-3′)2 and (5′-ACUUAAGU-3′)2 duplexes. No significant differences in the acquired thermodynamic parameters were seen when compared with the data acquired from the standard melt scheme that dilutes previously melted samples (Table S2). Both experiments utilized the same concentration range and utilized nine different concentrations of oligonucleotide in the experiment. These similarities in values gave validity to the optical melting scheme used in this work and provided additional evidence for the lack of RNA cleavage in this study.
Finally, the same two RNA duplexes were monitored by HPLC before and after optical melting experiments in 10.0 mM Mg2+ and standard 1 M NaCl buffer. The chromatograms before and after optical melting experiments were identical (Figs. S2 and S3), further suggesting that RNA was not cleaved during optical melting experiments. As mentioned, these oligonucleotides represented the two extremes for ΔG°37 and Tm. These two oligonucleotides appeared relatively unaltered in Mg2+ solutions at high temperatures before and after melting. This was further validated when comparing the Mg2+ sample to the standard NaCl sample. We, therefore, assumed all other oligonucleotides should behave in a similar manner. The lack of evidence of cleavage during the serial dilutions, during the forward and reverse optical melting experiments, during the fresh oligonucleotide melt scheme, and by monitoring before and after melting experiments with HPLC indicates that short RNA duplexes heated to 80°C at an increment of 1°C/min are not cleaved or significantly degraded in the presence of 10.0 mM Mg2+. Due to these findings, we assumed that short RNA duplexes in the presence of [Mg2+] <10.0 mM would also not exhibit any cleavage.
Magnesium effects on RNA duplex stability and Tm
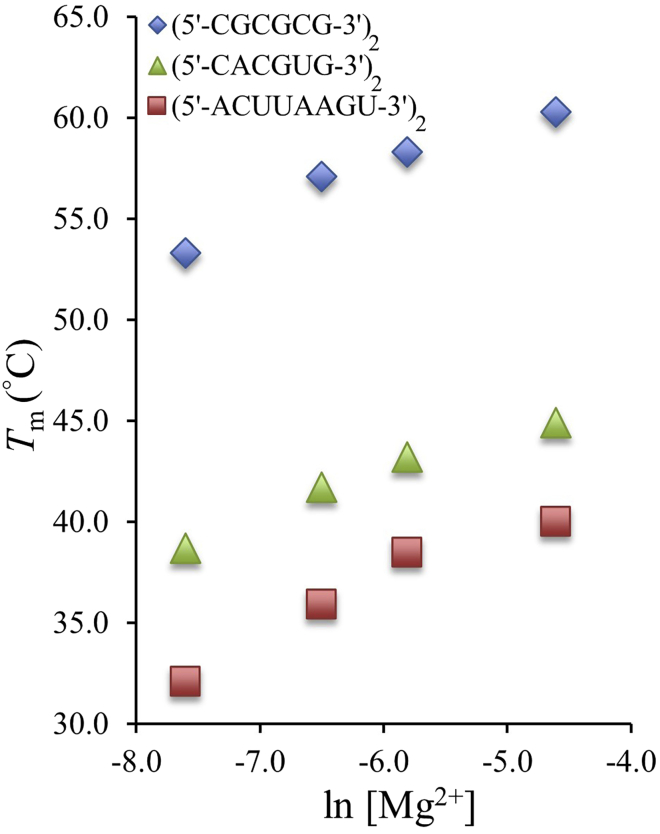
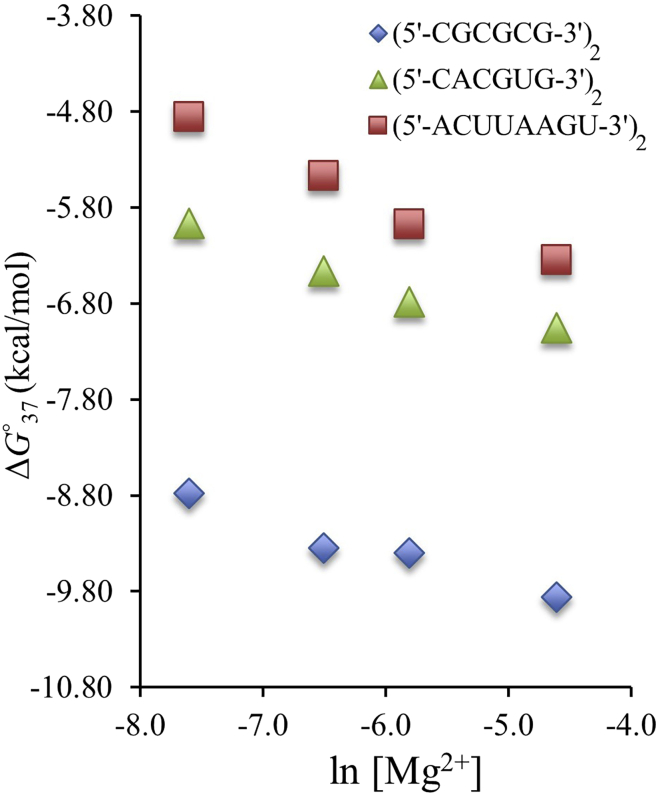
As the [Mg2+] was increased from 0.5 mM to 10.0 mM, the ΔG°37 values became more negative, and the Tm values increased as expected (Tables 1 and 2 and Figs. 1 and 2). These trends were consistent with what has been shown with DNA in Mg2+ across a larger concentration range (11). Previous studies have reported on the effects of Na+ on RNA duplex stability and have found that stability increases until a certain saturation point is reached (35,54,65). Although it appears as if the changes in ΔG°37 and Tm are in the process of leveling out as [Mg2+] approaches 10.0 mM, the range of [Mg2+] studied here does not allow for the determination of the exact saturation point, which is likely to occur above 10.0 mM. Figs. 1 and 2 demonstrate the relationship between Tm versus ln [Mg2+] and ΔG°37 versus ln [Mg2+] for a few RNA sequences that vary in fGC. The shape of these curves resembles the shape of the same curves constructed for RNA in the presence of [Na+] in the range of 71 mM to 1.02 M (13). Also, when deriving correction factors for Na+, it was determined that DNA correction factors for Na+ were a good starting point for RNA Na+ correction factors (13,35). As a result, correction factors for DNA and RNA in Na+ and DNA in Mg2+ were used as starting points to derive correction factors for RNA in Mg2+.
Figure 1.
The relationship between melting temperatures and ln [Mg2+] for RNA duplexes with varying G-C content. (5′-CGCGCG-3′)2 is 100% G-C, (5′-CACGUG-3′)2 is 67% G-C, and (5′-ACUUAAGU-3′)2 is 25% G-C. Errors in Tm are ∼ ±1°C. To see this figure in color, go online.
Figure 2.
The relationship between ΔG°37 and ln [Mg2+] for RNA duplexes with varying G-C content. (5′-CGCGCG-3′)2 is 100% G-C, (5′-CACGUG-3′)2 is 67% G-C, and (5′-ACUUAAGU-3′)2 is 25% G-C. To see this figure in color, go online.
Tm correction factors
There are several previously published Tm correction factors for DNA and RNA in Na+ and DNA and RNA in Mg2+. Generally, the correction factors fell within one of three categories: those dependent exclusively on the relationship between Tm and ln (or log) [Mg2+], those that not only depend on the relationship between Tm and ln (or log) [Mg2+] but include the additional parameters Nbp-1 and ΔH°, or those that depend on the relationship between Tm and ln (or log) [Mg2+] and fGC. The previously published Tm correction factors for DNA and RNA in Na+ and DNA and RNA in Mg2+ were compared with the experimental RNA data reported here, with deviations ranging from 2.6°C to 42.8°C (Table S4). Each of the correction factors were updated to reflect the data collected here, resulting in improved average deviations ranging from 1.2°C to 18.8°C (Table 3). Eq. S7 and Eq. S13 improved to the best average deviation of 1.2°C (Eq. 7 and Eq. 9), so it was determined that the addition of the new parameter fGC was beneficial in accurately predicting melting temperature. Fig. 1 compares fGC and the relationship between melting temperature and ln [Mg2+]. However, because of simplicity and the consistency with the ΔG°37 correction factor (discussed below), the recommended Tm correction factor is Eq. 7.
In order to test this correction factor, a sample calculation using the recommended Tm correction factor with an RNA duplex used in the derivation here is illustrated below. The sample sequence is (5′-AACUAGUU-3′)2 measured in 0.5 mM Mg2+ and 2 mM Tris. The experimental Tm in 1 M NaCl is 45.7°C and was used to predict the Tm at 0.5 mM Mg2+ in the correction calculation shown below:
| (15) |
| (16) |
| (17) |
The experimental Tm reported here for this duplex is 38.8°C, which is a difference of 0.5°C between the experimental and predicted values. Thus, this value fell within the average deviation value reported for this correction factor. If, however, the experimental Tm values are not available in 1 M NaCl, predicted values based on the nearest neighbor model are typically sufficient for prediction in Mg2+ (data not shown).
ΔG°37 correction factors
Despite the numerous Tm correction factors available in the literature for DNA and RNA in Na+, there is only one DNA ΔG°37 correction factor (Eq. S14) and four RNA ΔG°37 correction factors (Eqs. S15–S18) available. In addition to these, Eq. S19 was developed for DNA in Mg2+. These correction factors rely on the relationship between ΔG°37 and ln (or log) [Mg2+] and some include other parameters such as Nbp-1 and fGC to accurately predict ΔG°37. Like melting temperatures, Fig. 2 compares fGC and the relationship between ΔG°37 and ln [Mg2+]. The previously published ΔG°37 correction factors for DNA and RNA in Na+ and DNA and RNA in Mg2+ were compared with the experimental RNA data reported here, with deviations ranging from 0.59 to 6.78 (Table S4). These correction factors were updated using data reported here, resulting in improved average deviations ranging from 0.30 to 0.88 . Because Eq. S17 produced an updated correction factor (Eq. 12) with the best deviation and is consistent with the Tm correction factor recommended above, Eq. 12 is the correction factor recommended to predict ΔG°37 values for RNA in [Mg2+] between 0.5 and 10.0 mM.
To validate the accuracy of this model, the same oligonucleotide used in the sample Tm correction factor calculation above was used to predict ΔG°37 at 0.5 mM Mg2+. The experimental ΔG°37 value in 1 M NaCl is −7.16 and was used in the correction calculation shown below:
| (18) |
| (19) |
| (20) |
The experimental ΔG°37 reported here for this duplex is −6.00 , which is a difference of 0.06 between the experimental and predicted temperatures. Thus, this value fell within the average deviation value reported for this model. Again, if the experimental ΔG°37 values are not available in 1 M NaCl, predicted values are typically sufficient for prediction in Mg2+ (data not shown).
Oligomer length and sequence
It is apparent that different studies employed different parameters to account for duplex length and sequence composition on the stability of DNA or RNA in varying Na+ concentrations. Some correction factors utilized Nbp-1, which accounts for the effect of length on stability, whereas others included fGC, which is a sequence composition parameter. After updating the majority of the previously published correction factors of both types, it was determined that the correction factor incorporating fGC works best for the RNA duplexes studied here in Mg2+. Owczarzy et al. (35) stated that their correction factor developed for DNA in Na+ utilizing fGC could be used for duplexes ranging in length from six to at least 60 basepairs. Although our new correction factor is based upon the same model they proposed but for RNA in Mg2+, it would be necessary to test the accuracy of our model as length increased. Due to experimental constraints, melting shorter or longer duplexes was not possible for this study.
Magnesium concentrations viable for correction factors
The Mg2+ range used here to derive the correction factors to accurately predict Tm and ΔG°37 values is 0.5–10.0 mM Mg2+. Thus, the correction factors can be used for RNA duplexes within this range. Conveniently, this range of [Mg2+] is representative of the free Mg2+ in cells and the concentration employed in modern physiological buffers. However, there may exist cases that include Mg2+ concentrations higher than what was tested here or include both Mg2+ and Na+ in concentrations that are competing. Further studies need to be conducted to validate the accuracy of our model on RNA duplexes in solutions with higher [Mg2+] than studied here and solutions containing both Mg2+ and Na+.
In summary, stability in Mg2+ solutions was studied for short RNA duplexes. Previously published Tm correction factors for DNA and RNA in Na+ and DNA and RNA in Mg2+ were updated with the RNA data reported here to derive new correction factors with better accuracies. Eq. 7, which is a modified version of Eq. S7, produced the best average deviation, 1.2°C, for the data reported here and is the recommended Tm correction factor for RNA duplexes in Mg2+. Likewise, previously published ΔG°37 correction factors for DNA and RNA in Na+ and DNA in Mg2+ were updated to derive new correction factors. The modification of Eq. S17 to Eq. 12 with new coefficients had the best average deviation, 0.30 , and is the recommended ΔG°37 correction factor. These recommended correction factors can be incorporated into RNA secondary structure prediction algorithms to accurately predict the stability of RNA duplexes in [Mg2+] ranging from 0.5 to 10.0 mM.
Author contributions
B.M.Z., N.L.M., KE.R., S.H., and S.J.A. designed the studies. B.M.Z. supervised the project. N.L.M., K.E.R., S.H., and S.J.A. performed experiments and analyzed the data. N.L.M., M.S.A., S.J.A., and B.M.Z. wrote and edited the manuscript.
Acknowledgments
This work was supported by NIH Grant [R15 GM085699-04] awarded to B.M.Z.
Declaration of interests
The authors declare no competing interests.
Editor: Jason Kahn.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2022.12.025.
Supporting material
References
- 1.Eddy S.R. Non-coding RNA genes and the modern RNA world. Nat. Rev. Genet. 2001;2:919–929. doi: 10.1038/35103511. [DOI] [PubMed] [Google Scholar]
- 2.Serganov A., Patel D.J. Ribozymes, riboswitches and beyond: regulation of genen expression without proteins. Nat. Rev. Genet. 2007;8:776–790. doi: 10.1038/nrg2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He L., Hannon G.J. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 4.Pley H.W., Flaherty K.M., McKay D.B. Three-dimensional structure of a hammerhead ribozyme. Nature. 1994;372:68–74. doi: 10.1038/372068a0. [DOI] [PubMed] [Google Scholar]
- 5.Wang X.W., Liu C.X., et al. Zhang Q.C. RNA structure probing uncovers RNA structure-dependent biological functions. Nat. Chem. Biol. 2021;17:755–766. doi: 10.1038/s41589-021-00805-7. [DOI] [PubMed] [Google Scholar]
- 6.Pucci F., Schug A. Shedding light on the dark matter of the biomolecular structural universe: progress in RNA 3D structure prediction. Methods. 2019;162-163:68–73. doi: 10.1016/j.ymeth.2019.04.012. [DOI] [PubMed] [Google Scholar]
- 7.Magnus M., Kappel K., et al. Bujnicki J.M. RNA 3D structure prediction guided by independent folding of homologous sequences. BMC Bioinf. 2019;20:512. doi: 10.1186/s12859-019-3120-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y., Wang J., Xiao Y. 3dRNA: building RNA 3D structure with improved template library. Comput. Struct. Biotechnol. J. 2020;18:2416–2423. doi: 10.1016/j.csbj.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mathews D.H., Sabina J., et al. Turner D.H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 10.Xia T., SantaLucia J., et al. Turner D.H. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 11.Owczarzy R., Moreira B.G., et al. Walder J.A. Predicting stability of DNA duplexes in solutions containing magnesium and monovalent cations. Biochemistry. 2008;47:5336–5353. doi: 10.1021/bi702363u. [DOI] [PubMed] [Google Scholar]
- 12.Saiki R.K., Gelfand D.H., et al. Erlich H.A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 13.Chen Z., Znosko B.M. Effect of sodium ions on RNA duplex stability. Biochemistry. 2013;52:7477–7485. doi: 10.1021/bi4008275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sissoëff I., Grisvard J., Guillé E. Studies on metal ions-DNA interations: specific behaviour of reiterative DNA sequences. Prog. Biophys. Mol. Biol. 1976;31:165–199. doi: 10.1016/0079-6107(78)90008-1. [DOI] [PubMed] [Google Scholar]
- 15.Misra V.K., Draper D.E. On the role of magnesium ions in RNA stability. Biopolymers. 1998;48:113–135. doi: 10.1002/(SICI)1097-0282(1998)48:2%3C113::AID-BIP3%3E3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 16.Yamagami R., Sieg J.P., Bevilacqua P.C. Functional roles of chelated magnesium ions in RNA folding and function. Biochemistry. 2021;60:2374–2386. doi: 10.1021/acs.biochem.1c00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ren A., Košutić M., et al. Patel D.J. In-line alignment and Mg2+ coordination at the cleavage site of the env22 twister ribozyme. Nat. Commun. 2014;5:5534. doi: 10.1038/ncomms6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piccirilli J.A., Vyle J.S., et al. Cech T.R. Metal ion catalysis in the Tetrahymena ribozyme reaction. Nature. 1993;361:85–88. doi: 10.1038/361085a0. [DOI] [PubMed] [Google Scholar]
- 19.Frederiksen J.K., Fong R., Piccirilli J.A. Royal Society of Chemistry; 2009. Metal Ions in RNA Catalysis. [Google Scholar]
- 20.Romani A.M.P. Magnesium homeostasis in mammalian cells. Front. Biosci. 2007;12:308–331. doi: 10.2741/2066. [DOI] [PubMed] [Google Scholar]
- 21.Breslauer K.J., Frank R., et al. Marky L.A. Predicting DNA duplex stability from the base sequence. Proc. Natl. Acad. Sci. USA. 1986;83:3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams A.P., Longfellow C.E., et al. Turner D.H. Laser temperature-jump, spectroscopic, and thermodynamic study of salt effects on duplex formation by dGCATGC. Biochemistry. 1989;28:4283–4291. doi: 10.1021/bi00436a025. [DOI] [PubMed] [Google Scholar]
- 23.Nakano S., Fujimoto M., et al. Sugimoto N. Nucleic acid duplex stability: influence of base composition on cation effects. Nucleic Acids Res. 1999;27:2957–2965. doi: 10.1093/nar/27.14.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams D.J., Hall K.B. Thermodynamic comparison of the salt dependence of natural RNA hairpins and RNA hairpins with non-nucleotide spacers. Biochemistry. 1996;35:14665–14670. doi: 10.1021/bi961654g. [DOI] [PubMed] [Google Scholar]
- 25.Misra V.K., Draper D.E. A thermodynamic framework for Mg2+ binding to RNA. Proc. Natl. Acad. Sci. USA. 2001;98:12456–12461. doi: 10.1073/pnas.221234598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schroeder S.J., Turner D.H. Thermodynamic stabilities of internal loops with GU closing pairs in RNA. Biochemistry. 2001;40:11509–11517. doi: 10.1021/bi010489o. [DOI] [PubMed] [Google Scholar]
- 27.Gu X., Nguyen M.T., et al. Schroeder S.J. Effects of salt, polyethylene glycol, and locked nucleic acids on the thermodynamic stabilities of consecutive terminal adenosine mismatches in RNA duplexes. J. Phys. Chem. B. 2013;117:3531–3540. doi: 10.1021/jp312154d. [DOI] [PubMed] [Google Scholar]
- 28.O'Connell A.A., Hanson J.A., et al. Grover N. Thermodynamic examination of pH and magnesium effect on U6 RNA internal loop. RNA. 2019;25:1779–1792. doi: 10.1261/rna.070466.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huguet J.M., Ribezzi-Crivellari M., et al. Ritort F. Derivation of nearest-neighbor DNA parameters in magnesium from single molecule experiments. Nucleic Acids Res. 2017;45:12921–12931. doi: 10.1093/nar/gkx1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vieregg J., Cheng W., et al. Tinoco I. Measurement of the effect of monovalent cations on RNA hairpin stability. J. Am. Chem. Soc. 2007;129:14966–14973. doi: 10.1021/ja074809o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bizarro C.V., Alemany A., Ritort F. Non-specific binding of Na+ and Mg2+ to RNA determined by force spectroscopy methods. Nucleic Acids Res. 2012;40:6922–6935. doi: 10.1093/nar/gks289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bai Y., Greenfeld M., et al. Herschlag D. Quantitative and comprehensive decomposition of the ion atmosphere around nucleic acids. J. Am. Chem. Soc. 2007;129:14981–14988. doi: 10.1021/ja075020g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gebala M., Herschlag D. Quantitative studies of an RNA duplex electrostatics by ion counting. Biophys. J. 2019;117:1116–1124. doi: 10.1016/j.bpj.2019.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xi K., Wang F.H., et al. Tan Z.J. Competitive binding of Mg2+ and Na+ ions to nucleic acids: from helices to tertiary structures. Biophys. J. 2018;114:1776–1790. doi: 10.1016/j.bpj.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Owczarzy R., You Y., et al. Walder J.A. Effects of sodium ions of DNA duplex oligomers: improved prediction of melting temperatures. Biochemistry. 2004;43:3537–3554. doi: 10.1021/bi034621r. [DOI] [PubMed] [Google Scholar]
- 36.Wright D.J., Rice J.L., et al. Znosko B.M. Nearest neighbor parameters for inosine uridine pairs in RNA duplexes. Biochemistry. 2007;46:4625–4634. doi: 10.1021/bi0616910. [DOI] [PubMed] [Google Scholar]
- 37.Christiansen M.E., Znosko B.M. Thermodynamic characterization of tandem mismatches found in naturally occurring RNA. Nucleic Acids Res. 2009;37:4696–4706. doi: 10.1093/nar/gkp465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davis A.R., Znosko B.M. Thermodynamic characterization of single mismatches found in naturally occurring RNA. Biochemistry. 2007;46:13425–13436. doi: 10.1021/bi701311c. [DOI] [PubMed] [Google Scholar]
- 39.McDowell J.A., Turner D.H. Investigation of the structural basis for thermodynamic stabilities of tandem GU mismatches: solution structure of (rGAGGUCUC)2 by two-dimensional NMR and simulated annealing. Biochemistry. 1996;35:14077–14089. doi: 10.1021/bi9615710. [DOI] [PubMed] [Google Scholar]
- 40.Alberts B., Bray D., et al. Watson J.D. Garland Publishing; 1994. Molecular Biology of the Cell. [Google Scholar]
- 41.London R.E. Methods for measurement of intracellular magnesium: NMR and fluorescence. Annu. Rev. Physiol. 1991;53:241–258. doi: 10.1146/annurev.ph.53.030191.001325. [DOI] [PubMed] [Google Scholar]
- 42.Lusk J.E., Williams R.J., Kennedy E.P. Magnesium and the growth of Escherichia coli. J. Biol. Chem. 1968;243:2618–2624. doi: 10.1016/S0021-9258(18)93417-4. [DOI] [PubMed] [Google Scholar]
- 43.Truong D.M., Sidote D.J., et al. Lambowitz A.M. Enhanced group II intron retrohoming in magnesium-deficient Escherichia coli via selection of mutations in the ribosome core. Proc. Natl. Acad. Sci. USA. 2013;110:E3800–E3809. doi: 10.1073/pnas.1315742110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schildkraut C., Lifson S. Dependence of the melting temperature of DNA on salt concentration. Biopolymers. 1965;3:195–208. doi: 10.1002/bip.360030207. [DOI] [PubMed] [Google Scholar]
- 45.Wetmur J.G. DNA probes: applications of the principles of nucleic acid hybridization. Crit. Rev. Biochem. Mol. Biol. 1991;26:227–259. doi: 10.3109/10409239109114069. [DOI] [PubMed] [Google Scholar]
- 46.Frank-Kamenetskii M.D. Simplification of the empirical relationship between melting temperature of DNA, its GC content and concentration of sodium ions in solution. Biopolymers. 1971;10:2623–2624. doi: 10.1002/bip.360101223. [DOI] [PubMed] [Google Scholar]
- 47.Blake R.D., Delcourt S.G. Thermal stability of DNA. Nucleic Acids Res. 1998;26:3323–3332. doi: 10.1093/nar/26.14.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marmur J., Doty P. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J. Mol. Biol. 1962;5:109–118. doi: 10.1016/S0022-2836(62)80066-7. [DOI] [PubMed] [Google Scholar]
- 49.SantaLucia J., Jr. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. USA. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.SantaLucia J., Jr., Allawi H.T., Seneviratne P.A. Improved nearest-neighbor parameters for predicting DNA duplex stability. Biochemistry. 1996;35:3555–3562. doi: 10.1021/bi951907q. [DOI] [PubMed] [Google Scholar]
- 51.von Ahsen N., Wittwer C.T., Schütz E. Oligonucleotide melting temperatures under PCR conditions: nearest-neighbor corrections for Mg2+, deoxynucleotide triphosphate, and dimethyl sulfoxide concentrations with comparison to alternative empirical formulas. Clin. Chem. 2001;47:1956–1961. doi: 10.1093/clinchem/47.11.1956. [DOI] [PubMed] [Google Scholar]
- 52.Peyret N. Wayne State University; 2000. Prediction of Nucleic Acid Hybridization: Parameters and Algorithms. PhD dissertation. [Google Scholar]
- 53.Mitsuhashi M. Technial Report: part 1. Basic requirements for designing optimal oligonucleotide probe sequences. J. Clin. Lab. Anal. 1996;10:277–284. doi: 10.1002/(SICI)1098-2825(1996)10:5%3C277::AID-JCLA8%3E3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 54.Tan Z.J., Chen S.J. RNA helix stability in mixed Na+/Mg2+ solution. Biophys. J. 2007;92:3615–3632. doi: 10.1529/biophysj.106.100388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bloomfield V.A., Crothers D.M., Tinoco I., Jr. University Science Books; 2000. Nucleic Acids: Structures, Properties, and Functions. [Google Scholar]
- 56.Jarvinen P., Oivanen M., Lonnberg H. Interconversion and phosphoester hydrolysis of 2', 5'- and 3', 5'-dinucleoside monophosphates: kinetics and mechanisms. J. Org. Chem. 1991;56:5396–5401. doi: 10.1021/jo00018a037. [DOI] [Google Scholar]
- 57.Plum G.E., Breslauer K.J. Thermodynamics of an intramolecular DNA triple helix: a calorimetric and spectroscopic study of pH and salt dependence of thermally induced structural transitions. J. Mol. Biol. 1995;248:679–695. doi: 10.1006/jmbi.1995.0251. [DOI] [PubMed] [Google Scholar]
- 58.Privalov P.L., Ptitsyn O.B., Birshtein T.M. Determination of stability of the DNA double helix in an aqueous medium. Biopolymers. 1969;8:559–571. doi: 10.1002/bip.1969.360080502. [DOI] [Google Scholar]
- 59.Record M.T., Jr. Electrostatic effects on polynucleotide transitions. II. Behavior of titrated systems. Biopolymers. 1967;5:993–1008. doi: 10.1002/bip.1967.360051011. [DOI] [PubMed] [Google Scholar]
- 60.Marciniec T., Ciesiołka J., et al. Krzyzosiak W.J. Speficifity and mechanism of the cleavages induced in yeast tRNAPhe by magnesium ions. Acta Biochim. Pol. 1989;36:183–194. [PubMed] [Google Scholar]
- 61.Yamagami R., Bingaman J.L., et al. Bevilacqua P.C. Cellular conditions of weakly chelated magnesium ions strongly promote RNA stability and catalysis. Nat. Commun. 2018;9:2149. doi: 10.1038/s41467-018-04415-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Matsuo M., Yokogawa T., et al. Okada N. Highly specific and efficient cleavage of squid tRNALys catalyzed by magnesium ions. J. Biol. Chem. 1995;270:10097–10104. doi: 10.1074/jbc.270.17.10097. [DOI] [PubMed] [Google Scholar]
- 63.AbouHaidar M.G., Ivanov I.G. Non-enzymatic RNA hydrolysis promoted by the combined catalytic activity of buffers and magnesium ions. Z. Naturforsch. C J. Biosci. 1999;54:542–548. doi: 10.1515/znc-1999-7-813. [DOI] [PubMed] [Google Scholar]
- 64.Serra M.J., Baird J.D., et al. Westhof E. Effects of magnesium ions on the stabilization of RNA oligomers of defined structures. RNA. 2002;8:307–323. doi: 10.1017/s1355838202024226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tan Z.J., Chen S.J. Nucleic acid helix stability: effects of salt concentration, cation valence and size, and chain length. Biophys. J. 2006;90:1175–1190. doi: 10.1529/biophysj.105.070904. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.