Significance
Bacterial cell wall peptidoglycan is a major gut microbial signal that crosses the intestinal barrier to act directly on the host, influencing behavior, physiological development, and inflammation. Our study defines fundamental parameters governing the translocation of peptidoglycan between the gut lumen and host system. First, we describe the peptidoglycan composition of a gut bacterial community. This peptidoglycan arriving naturally via the gut has a specific tropism for host organs, when compared with routes that bypass the gut. Most significantly, we show that the peptidoglycan trafficking pathway is activated by the microbiota itself. Our data afford a valuable insight into host-gut microbiota interaction mechanisms mediated by peptidoglycan as a driver of health and disease.
Keywords: gut microbiota, peptidoglycan, systemic trafficking
Abstract
Peptidoglycan, the major structural polymer forming the cell wall of bacteria, is an important mediator of physiological and behavioral effects in mammalian hosts. These effects are frequently linked to its translocation from the intestinal lumen to host tissues. However, the modality and regulation of this translocation across the gut barrier has not been precisely addressed. In this study, we characterized the absorption of peptidoglycan across the intestine and its systemic dissemination. We report that peptidoglycan has a distinct tropism for host organs when absorbed via the gut, most notably by favoring access to the brain. We demonstrate that intestinal translocation of peptidoglycan occurs through a microbiota-induced active process. This process is regulated by the parasympathetic pathway via the muscarinic acetylcholine receptors. Together, this study reveals fundamental parameters concerning the uptake of a major microbiota molecular signal from the steady-state gut.
Within the intestinal tract, the gut microbiota generates signals that are recognized by different classes of host immune and nonimmune cells (1). These signals can come in the form of microbe-associated molecular patterns (MAMPs), a diverse group of microorganism-specific molecules that are recognized by pattern recognition receptors (PRRs) expressed mainly by innate immune cells (2). One class of MAMP, the bacterial cell wall component peptidoglycan, is well-characterized as an inflammatory molecule in acute and chronic conditions, and increasingly recognized as an important mediator of steady-state phenomena in the host. These effects are not limited to the gut where the microbiota resides (3), but include the broader host physiology and behaviors, facilitated by the continuous dissemination of peptidoglycan fragments across the intestinal epithelial barrier (4–10), for which a variety of mechanisms have been proposed (11). Nevertheless, a clear understanding of the basic principles governing the absorption and systemic dissemination of microbiota peptidoglycan is lacking.
In this study, we characterize the absorption of peptidoglycan across the gut epithelial barrier and its systemic dissemination in the steady-state. In a mouse model, we show that translocation across the intestinal barrier favors the accumulation of peptidoglycan in specific host organs, most notably the brain. We further demonstrate that the steady-state absorption of peptidoglycan is dependent on the microbial colonization status of the host. Focusing on the small intestinal epithelial barrier, we observed peptidoglycan absorption by enteric epithelial cells, among which goblet cells displayed a prominent affinity, and found that trafficking of peptidoglycan from the gut is regulated via muscarinic acetylcholine receptors. Together, our data provide insights into parameters governing the systemic presence of peptidoglycan.
Results
Gut Peptidoglycan Has a Distinct Accumulation Profile in Major Organs.
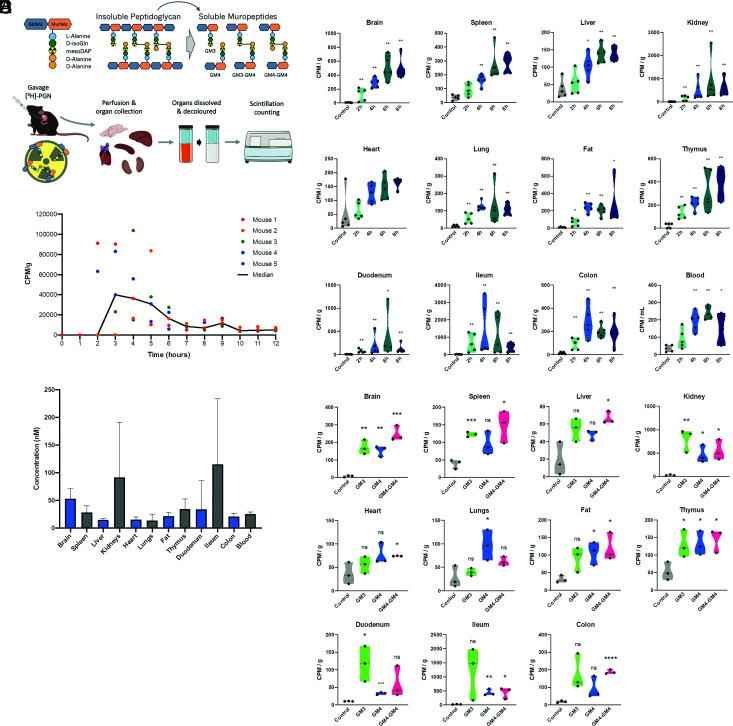
We first established the dynamics of peptidoglycan dissemination from the gut to the host system, by tracking the fate of an orally delivered, single dose of peptidoglycan in conventional adult mice. Escherichia coli peptidoglycan was radiolabeled by incorporation of 3H-mesoDAP, and solubilized enzymatically by glycoside hydrolysis of the glycan chain to generate soluble peptidoglycan moieties (termed muropeptides). We selected this strategy because 1) labeling with 3H-mesoDAP does not alter the native composition of peptidoglycan, allowing us to track peptidoglycan undergoing natural processing within the host, and 2) scintillation counting is highly sensitive. E. coli peptidoglycan is a relevant proxy for microbiota peptidoglycan, since the most abundant muropeptides are well represented among the structures naturally present in the intestinal microbiota peptidoglycome (Fig. 1 and SI Appendix, Fig. S1 and Table S1). 3H-mesoDAP-labeled peptidoglycan ([3H]-PGN; 400,000 counts per minute [cpm]) was administered per os to specific-pathogen-free (SPF) C57BL/6J mice. The blood and PBS-perfused organs were collected at time-points between 2 h and 8 h, and the [3H]-PGN content assessed by scintillation counting (Fig. 2A).
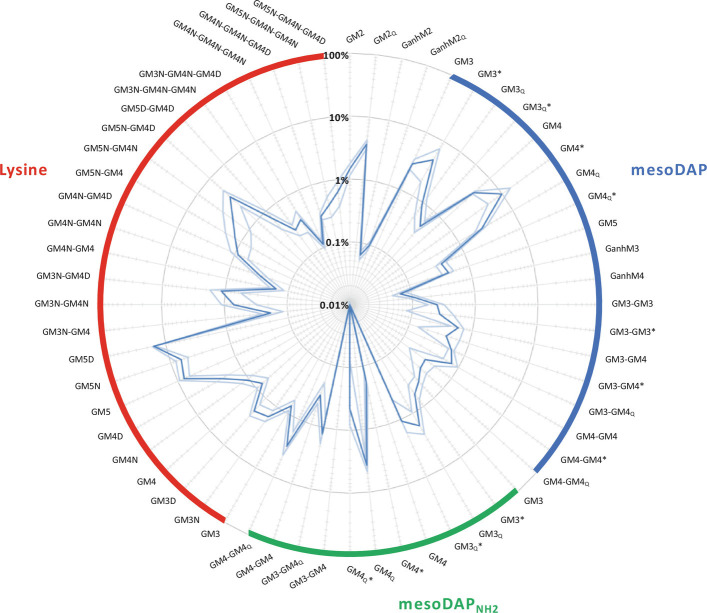
Fig. 1.
The relative abundance of selected peptidoglycan fragments from mouse gut microbiota. Muropeptide analysis was performed on gut microbiota peptidoglycan extracted from four C57BL/6J female mice, and the relative abundance of sixty muropeptides calculated. The targeted muropeptides are expected to be common in the gut microbiota. Dark blue plot: average relative abundance. Light blue plot: SD. Muropeptides are grouped according to the characteristic diamino acid at position 3 of the peptide stem: mesoDAP (blue), amidated mesoDAP (green) or lysine (red). G, N-acetyl-glucosamine; M, N-acetyl-muramitol (reduced form of N-acetyl-muramic acid), 1,6-anhydro-N-acetylmuramic acid; 2 to 5, peptide stem length; N, Asn; D, Asp; *, N-deacetylated glucosamine. mesoDAP and amidated mesoDAP types have isoglutamate at position 2 of the peptide stem unless annotated with subscript “Q”, indicating the presence of isoglutamine. Lysine types all have isoglutamine at position 2. See SI Appendix, Table S1 for more information.
Fig. 2.
The kinetics of [3H]-PGN biodistribution following administration by gavage in mice. (A) Schematics summary of radiolabeling methodology, indicating the position of the 3H-labeled mesoDAP, and radiotracking strategy. Mice (female C57BL/6j, 8 to 12 wk) were gavaged with [3H]-PGN. At the desired timepoint, mice were perfused to clear transiently circulating [3H]-PGN from the organs. The tissues to be analyzed were dissolved, then decolored by peroxide bleaching to reduce quenching effects, prior to scintillation counting. (B) [3H]-PGN measured by scintillation counting of dissolved, decolored organs and blood between 2 h and 8 h postgavage. Measured CPM values are normalized per g tissue weight, or per mL blood. Pairwise comparison with control performed using the Mann–Whitney U test. *P ≤ 0.05; **P ≤ 0.0050. Welch’s ANOVA comparing time-point groups, excluding control: Brain P = 0.0045; Spleen P = 0.0050; Liver P = 0.0068; Kidney P = 0.087; Heart P = 0.0029; Lung P = 0.0331; Fat P = 0.0043; Thymus P = 0.0165; Duodenum P = 0.4681; Ileum P = 0.3505; Colon P = 0.0375; Blood P = 0.0082. (C) Intestinal transit time of gavaged [3H]-PGN. 3H was measured by scintillation counting of fecal pellets deposited in the cage every hour. (D) Estimated concentration of [3H]-PGN in each organ. Concentration of [3H]-PGN per CPM was determined by HPLC analysis of a predetermined CPM value of [3H]-PGN. The total peak area was calculated and the value in mg determined relative to an MDP standard curve. (E) The biodistribution of [3H]-GM3, [3H]-GM4 and [3H]-GM4-GM4, administered per os in SPF mice. Scintillation counting was performed on dissolved, decolored organs. Results are normalized to CPM per g of tissue. Pairwise comparison with control performed using unpaired t test. *P ≤ 0.05; **P ≤ 0.0050; ***P ≤ 0.0005; P < 0.0001. Welch’s ANOVA comparing muropeptides groups, excluding control: Brain P = 0.0582; Spleen P = 0.2711; Liver P = 0.1508; Kidney P = 0.2478; Heart P = 0.3525; Lung P = 0.1022; Fat P = 0.6536; Thymus P = 0.8542; Duodenum P = 0.1234; Ileum P = 0.3295; Colon P = 0.2227.
At 2 h postgavage with [3H]-PGN, radioactivity was detected differentially across the host organs (Fig. 2B). Normalized by tissue weight, the highest levels of exogenously administered peptidoglycan were observed in the brain, spleen, kidneys, thymus and fat, whereas the lowest levels were observed in the liver, heart, and lungs (for absolute counts per organ, see SI Appendix, Fig. S2A). This relationship did not change over time. The presence of radioactivity in the organs accumulated, reaching maximal values at 6 to 8 h postgavage. Similarly, radioactivity in the blood peaked at 6 h, declining slightly at 8 h. Values in the intestinal tract showed the greatest variability over time, likely reflecting the local presence of peptidoglycan undergoing peristaltic transit through the intestinal lumen. To assess the intestinal transit time, mice were gavaged with 60,000 cpm of [3H]-PGN, and fecal pellets deposited on the cage bed were collected every hour for scintillation counting (Fig. 2C). First appearance of 3H in the stools occurred between 2 h and 4 h postgavage, with maximal values detected in stools collected between 2 h and 5 h postgavage. Fecal [3H]-PGN dropped to near-baseline levels in all mice by 7 h postgavage, indicating that the vast majority of [3H]-PGN had exited the intestinal tract within the study period.
To quantify exogenously administered peptidoglycan in the tissues, the ratio of 3H-mesoDAP per unit weight of [3H]-PGN was determined by scintillation counting, combined with reversed-phase HPLC quantification of [3H]-PGN muropeptides against a standard curve of muramyldipeptide (MDP). Based on the values measured in the organs at 6 h, we determined that peptidoglycan accumulated in the order of 10 to 100 ng/g of tissue, and 20 ng/mL in the blood (Fig. 2D). Our data are consistent with reported levels of serum peptidoglycan in SPF mice, measured by indirect, competitive enzyme-linked immunosorbent assay, which also fall in the ng/mL range (180 to 300 ng/mL), demonstrating the physiological relevance of our experimental approach (8).
The selectivity of the gut epithelial barrier toward peptidoglycan fragments could determine the nature of the peptidoglycan moieties found systemically. A variety of mechanisms have been proposed for epithelial transit of peptidoglycan fragments. These range from nonspecific mechanisms such as paracellular transport and carrier-mediated mechanisms, to highly specific mechanisms targeting a restricted set of monomeric muropeptides, such as the SLC15A family peptide transporters, suggested to transport muramyl dipeptides and tripeptides, reviewed in refs. 11 and 12. To gain insight into the selectivity of the intestinal epithelial barrier toward different muropeptides, we gavaged mice with 50,000 cpm of individual, 3H-labeled muropeptides possessing a peptide stem length of three, four or eight amino acids; the monomers [3H]-GM3, [3H]-GM4, or the dimer [3H]-GM4-GM4. The systemic presence of the 3H-labeled muropeptides was assessed 4 h postgavage (Fig. 2E and SI Appendix, Fig. S2B). In all cases, 3H was detected in approximately equal proportions across the distinct organs, demonstrating that muropeptide size is not a limiting factor for intestinal absorption and dissemination processes.
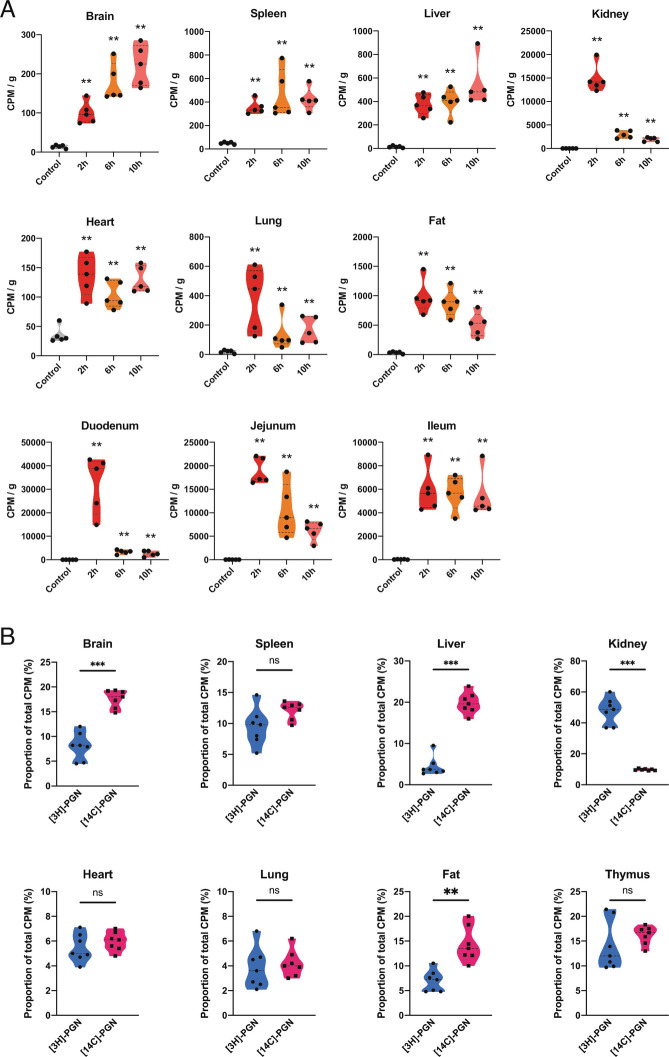
To assess the possibility that [3H]-PGN fragments are degraded in the stomach and gut, resulting in the liberation and absorption of mainly free [3H]-mesoDAP, we assessed whether [3H]-mesoDAP and [3H]-PGN displayed distinct kinetics of absorption and dissemination (Fig. 3A and SI Appendix, Fig. S3A). Gavage with [3H]-mesoDAP did not recapitulate the kinetics or distribution profile observed for [3H]-PGN. Notably, [3H]-mesoDAP reached maximal values in the kidney at 2 h postgavage, followed by rapid clearance (Fig. 3A and SI Appendix, Fig. S3A). Abundant accumulation in the fat peaked between 2 h and 6 h, and then declined. Uptake in the spleen and liver reached their maximum by 2 h and remained steady for the duration of the experiment. Only comparably low levels of [3H]-mesoDAP accumulated in the brain, differing from the case of [3H]-PGN where relative to the other organs, the brain was a major reservoir. The striking differences between the dynamics of [3H]-mesoDAP and [3H]-PGN indicate that peptidoglycan does not undergo acute catabolism in the gut leading to absorption of mainly free mesoDAP.
Fig. 3.
Effect of labeling strategy parameters on biodistribution of peptidoglycan. (A) Biodistribution of [3H]-mesoDAP amino acid administered to mice per os. [3H]-mesoDAP biodistribution was measured by scintillation counting of dissolved, decolored organs, 2 h, 6 h, and 10 h postgavage. The measured CPM values are normalized per g tissue weight. (B) Biodistribution of [3H]-PGN versus [14C]-PGN administered to mice by gavage. The relative abundance of [3H]-mesoDAP or [14C]-GlcNAc-labeled peptidoglycan detected for each organ or tissue is shown. Data are presented as the proportion of the sum of all [3H] or [14C] CPM measurements in the organs per mouse. Pairwise comparison with control performed using the Mann–Whitney U test. **P ≤ 0.005; ***P ≤ 0.0005. Welch’s ANOVA comparing time-point groups, excluding control, Brain P = 0.0052; Spleen P = 0.3334; Liver P = 0.3307; Kidney P = 0.0002; Heart P = 0.1921; Lung P = 0.1624; Fat P = 0.0323; Duodenum P = 0.0040; Jejunum P = 0.0003; Ileum P = 0.9333.
PGLYRP-2 serum amidase cleaves muropeptides larger than MDP, generating peptides that are preferentially cleared via urinary excretion (13). To gain insight into the integrity of disseminated peptidoglycan fragments, we compared the distribution profile of orally administered peptidoglycan when the glycan chain was radiolabeled versus the peptide stem. Mice were gavaged with 400,000 cpm of Escherichia coli peptidoglycan labeled with [14C]-N-acetyl-D-glucosamine ([14C]-PGN) or [3H]-PGN. Since the two radiolabels have different activities and levels of incorporation, their dissemination profiles cannot be compared directly. We therefore plotted the proportion of 3H or 14C in each organ, relative to the total amount of 3H or 14C radioactivity measured systemically, excluding the intestinal tract (Fig. 3B and SI Appendix, Fig. S3 B–D). For most of the organs, the ratio of [3H]-PGN to [14C]-PGN was approximately 1:1, suggesting that these are intact peptidoglycan fragments that escaped serum amidase activity. The brain and fat showed small deviations, with the proportion of [14C]-PGN approximately double that of [3H]-PGN, suggesting separate accumulation of glycan versus intact muropeptide or peptide fractions. The kidneys and liver demonstrated notable deviations, with [3H]-PGN to [14C]-PGN ratios of 5:1 and 1:4, respectively. Our data are consistent with a report that peptides are preferentially cleared via the kidney (urinary excretion), whereas free disaccharide is preferentially directed to the liver (14). Finally, to assess whether peptidoglycan subtype alters the dissemination profile to organs, we gavaged mice with [14C]-N-acetyl-D-glucosamine-labeled peptidoglycan of Lactobacillus rhamnosus Lr32 ([14C]-Lr32), which has the A4α peptidoglycan subtype (SI Appendix, Fig. S3E) (15). Note that data for the brain were published previously (10). We observed no major differences in the dissemination profile compared with Escherichia coli [14C]-PGN.
Bypassing the Gut Restricts Peptidoglycan Access to the Brain.
We next asked whether the systemic distribution of [3H]-PGN is altered when the intestinal barrier is bypassed completely. We administered 40,000 cpm of [3H]-PGN intravenously, an amount that exceeds the maximal values detected systemically in gavage experiments, and measured accumulation in organs at 1 h, 4 h and 8 h postinjection (p.i.). Surprisingly, with the exception of the kidney, [3H]-PGN was detected at low levels across the organs (Fig. 4A and SI Appendix, Fig. S4A). Peak accumulation of [3H]-PGN in the kidney occurred 1 h p.i., returning to near-background levels by 4 h p.i., suggesting rapid clearance of this dose of [3H]-PGN via urinary excretion. Only when the system was forced by intravenous administration of 400,000 cpm of [3H]-PGN, the same dose administered in gavage experiments, did we observe uptake in the organs at 1 h p.i. (Fig. 4B and SI Appendix, Fig. S4B). When compared with [3H]-PGN biodistribution at 4 h postgavage, intravenously administered [3H]-PGN was detected at similar or slightly elevated levels in the spleen, liver, kidney, lungs, fat and thymus, and at a reduced level in the heart. Strikingly, intravenously administered [3H]-PGN was barely detected in the brain.
Fig. 4.
Dose-dependent biodistribution of [3H]-PGN administered intravenously or intraperitoneally. (A) Mice were administered 40,000 CPM of [3H]-PGN intravenously and biodistribution to organs and tissues measured at 1 h, 4 h, 8 h, and 24 h postinjection. Welch’s ANOVA comparing time-point groups, excluding control: Brain P = 0.9236; Spleen P = 0.9972; Liver P = 0.4364; Kidney P = 0.0842; Heart P = 0.6337; Lung P = 0.1320; Fat P = 0.2897; Duodenum P = 0.1497; Ileum P = 0.8240; Colon P = 0.9038. (B) Mice were administered 400,000 CPM of [3H]-PGN intravenously (two injections of 200,000 cpm) 30 min apart, or 400,000 CPM of [3H]-PGN by gavage. Organs were harvested at 1 h after the first injection, or 4 h after gavage. Scintillation counting was performed on the dissolved, decolored organs. (C) Biodistribution of [3H]-PGN administered intraperitoneally. Mice were injected intraperitoneally with 400,000 CPM of [3H]-PGN and scintillation counting performed on the dissolved, decolored organs harvested at 2 h and 6 h postgavage. Data are normalized as CPM values per g tissue weight. Pairwise comparison with control performed using the Mann–Whitney U test. *P ≤ 0.05; **P ≤ 0.005. Pairwise comparison with between time-points performed using the Mann–Whitney U test, Brain P = 0.0159; Spleen P = 0.8413; Liver P = 0.8413; Kidney P = 0.0079; Heart P = 0.7222; Lung P = 0.5952; Fat P = 0.0317; Duodenum P = 0.0159; Ileum P = 0.6905; Colon P = 0.8413. (D) [3H]-PGN detected in brain regions following administration by gavage. Schematic is a sagittal view of the mouse brain showing the average relative proportion of [3H]-PGN detected in each brain region. Right panel shows the [3H]-PGN measured in each region in CPM/g. Each data point represents the indicated brain region pooled from 5 mice. “Central brain areas” include the thalamus, hypothalamus and midbrain. Pairwise comparison of different regions to olfactory bulb performed using the Mann–Whitney U test *P ≤ 0.05.
We next administered 400,000 cpm of [3H]-PGN intraperitoneally. Maximal uptake to the kidneys, spleen, liver, fat and thymus was observed at the earliest time point (2 h p.i.), similar to the kinetic profile obtained by intravenous administration of [3H]-PGN (Fig. 4C and SI Appendix, Fig. S4C). Radioactivity in kidneys and fat was lower at 6 h p.i., indicating clearance, whereas [3H]-PGN levels were sustained in the spleen and liver at 6 h p.i. [3H]-PGN was not detected in the heart, lungs, or intestinal tract. As with intravenously administered [3H]-PGN, intraperitoneally administered [3H]-PGN was not detected in the brain, indicating that entry to the brain is restricted when [3H]-PGN is administered directly into the host system, bypassing the gut, while absorption across the intestinal barrier facilitates trafficking to the brain. We assessed whether the uptake of orally administered [3H]-PGN was localized to any particular region of the brain, by comparing levels of [3H]-PGN in six major brain regions at 6 h postgavage (Fig. 4E). We observed enrichment of [3H]-PGN in the olfactory bulb, where approximately 33% of total brain radioactivity was detected when normalized by tissue weight. The relative proportion of [3H]-PGN was approximately equal throughout other brain regions.
Goblet Cells Have an Affinity for Peptidoglycan.
Having identified that the intestinal tract functions as a gateway favoring the accumulation of peptidoglycan through the host system, we investigated the initial step of peptidoglycan translocation across the small intestinal epithelial barrier using confocal microscopy. Fluorescent conjugates of MDP (MDP-rho), or Escherichia coli peptidoglycan (Fluo-PGN) were injected into ileal ligatures of anesthetized mice. After 20-min incubation, the tissue was washed, fixed, and stained for imaging by confocal microscopy. We observed transcellular uptake of MDP-rho and Fluo-PGN in the ileal villus epithelium (Fig. 5 A and B and SI Appendix, Fig. S5 A and B). Goblet cells (identified by peripheral cell mucus-staining with wheat germ agglutinin, cellular morphology and basal positioning of the nucleus) displayed a prominent affinity for MDP-rho and Fluo-PGN, with fluorescent peptidoglycan observed as a ring around the apical portion of the cell, excluded from the regions containing mucus, and more diffuse in the basal cytoplasm. The intensity of fluorescence in these cells was variable, perhaps reflecting the local concentration of peptidoglycan or dynamics of uptake by these cells. We did not observe peptidoglycan translocation in enteroendocrine cells, tuft cells, or in microfold- (M-) cells (SI Appendix, Fig. S5C). We next assessed uptake of Fluo-PGN in the small intestine (duodenal and ileal ligatures) and large intestine (colonic ligatures). While Fluo-PGN was detected in the majority of goblet cells in duodenum and ileum, uptake was not observed in colonic goblet cells, and only rarely observed in any part of the colonic epithelium or lamina propria (SI Appendix, Fig. S6). Thus, the small intestine and large intestine goblet cells appear to be functionally distinct with regards to absorption of peptidoglycan.
Fig. 5.
Peptidoglycan uptake and dissemination by intestinal epithelial cells. (A) Upper panel: MDP-rho internalization was observed in a subset of epithelial cells in the ileal villi. Lower panel; internalization of MDPrho in goblet cells (WGA+). No fluorescence was observed in the villus epithelium including the goblet cells, in the rhodamine channel of vehicle-treated controls. (B) Internalization of PGN-AF647 in the ileum of SPF mice by a subset of epithelial cells, which includes goblet cells (WGA+; panel I) and other cell types (WGA−, panel II). (C) The biodistribution of [3H]-PGN administered per os in SPF mice, is suppressed by parasympathetic inhibition. SPF mice were administered tropicamide, atropine or vehicle control prior to gavage with [3H]-PGN. Scintillation counting was performed on dissolved, decolored organs. (D) [3H]-PGN biodistribution from the gut is suppressed by GW4869 treatment. Results are normalized to CPM per g of tissue. Pairwise comparison with vehicle control performed using the Mann–Whitney U test. *P ≤ 0.05; **P ≤ 0.005.
Gut Microbiota and Muscarinic Receptors Regulate Peptidoglycan Absorption.
Microscopy analysis indicated that peptidoglycan transits the epithelial barrier via active transepithelial processes, and prominent uptake by goblet cells. Goblet cell-associated antigen passages (GAPs) translocate luminal antigens to underlying antigen presenting cells, facilitating small intestinal tolerogenesis (16), a process regulated by muscarinic acetylcholine receptor (mAchR)-4 activation (17). Expression of all five mAchR subtypes is reported in the mouse small intestine epithelia (18–20). We asked whether mAchR-mediated pathways regulate the translocation and dissemination of peptidoglycan. We performed per os [3H]-PGN biodistribution analysis in mice treated with the parasympatholytic agents tropicamide (selective antagonist of mAchR-4) or atropine (pan-mAchR antagonist). Pan-mAchR antagonism with atropine led to significant inhibition of [3H]-PGN biodistribution (Fig. 5C and SI Appendix, Fig. S7A). Atropine treatment significantly reduced the levels of [3H]-PGN circulating in the blood, suggesting that mAchR inhibition restricts peptidoglycan absorption at the intestinal barrier level. In the case of tropicamide treatment targeting peptidoglycan uptake via goblet cells, we observed a trend of reduced distribution to the organs and blood, although for the majority of organs this reduction did not meet the significance cutoff of P ≤ 0.05. Our data suggest a partial role for GAPs in the systemic dissemination of peptidoglycan.
Migrating cells and exosomes have both been proposed as potential vehicles for systemic dissemination of peptidoglycan, either of which could facilitate entry of peptidoglycan into the brain (21, 22). To assess these possibilities, we administered neutral-sphingomyelinase 2 (N-SMase2) inhibitor GW4869 to mice, followed by gavage with [3H]-PGN. Inhibition of N-SMase2 was shown to disrupt directional orientation of motile macrophages, and to block budding of exosomes (23, 24). Radioactivity presence was diminished in almost all organs tested following GW4869 treatment, but not in the gut, suggesting that inhibition of N-SMase2 activity confines peptidoglycan in the gut tissue (Fig. 5D and SI Appendix, Fig. S7B). Although we cannot rule out indirect effects, our data support a hypothesis that gut peptidoglycan dissemination involves a cellular or vesicular vehicle.
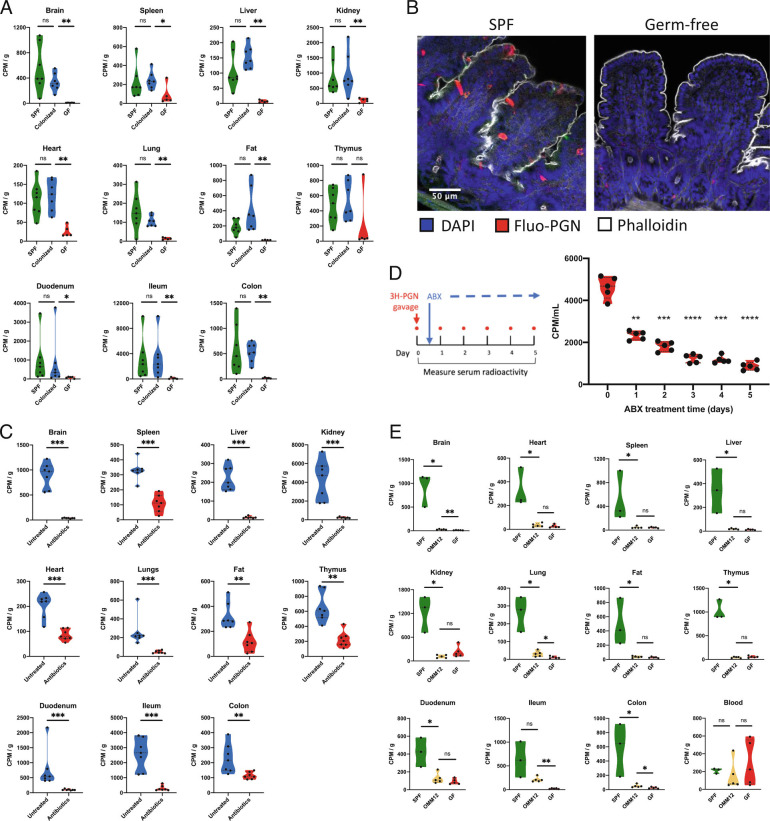
In mice lacking a microbiota [germfree (GF)], the gut is considered to be leaky, as microbial colonization triggers tightening of the intraepithelial tight junctions, augmenting epithelial paraselectivity. To explore the connection between the gut microbiota, intestinal permeability and systemic biodistribution of peptidoglycan, we compared the dissemination of [3H]-PGN administered by gavage to SPF mice, GF mice, and formerly GF mice, conventionalized with SPF microbiota. Remarkably, peptidoglycan biodistribution was suppressed in GF mice, with low levels of [3H]-PGN detected across the distinct organs. By comparison, peptidoglycan biodistribution was restored to SPF levels upon conventionalization (Fig. 6A and SI Appendix, Fig. S8 A and B). The observed effect was not due to impaired intestinal motility in GF mice, since tracking of [3H]-PGN administered by gavage showed that gut peptidoglycan reached the jejunum and cecum at 2 h and 8 h respectively with low presence in organs (SI Appendix, Fig. S8C). Epithelial cell uptake of MDP-rho was observed in SPF, but not GF mice, suggesting that the passage of peptidoglycan was restricted at the level of the intestinal epithelial barrier (Fig. 6B). To investigate whether the uptake of intestinal peptidoglycan observed in SPF mice requires the sustained presence of the microbiota or is due to an irreversible physiological alteration triggered by microbial colonization, we depleted the microbiota of adult SPF mice by administering a cocktail of antibiotics in the drinking water for 7 d prior to gavage with [3H]-PGN. Antibiotic depletion of the microbiota suppressed the systemic biodistribution of gut peptidoglycan (Fig. 6C and SI Appendix, Fig. S8D), demonstrating that absorption of gut peptidoglycan is malleable to changes in microbial colonization status. When peptidoglycan systemic dissemination was measured over time during oral antibiotic treatment, we observed an immediate suppressory effect on peptidoglycan transit to the blood (within 24 h of antibiotic initiation), followed by a steady decline in [3H]-PGN uptake over the study period (Fig. 6D).
Fig. 6.
Microbiota colonization is required for efficient peptidoglycan biodistribution. (A) The biodistribution of [3H]-PGN in germfree (GF) mice, specific pathogen free (SPF) mice and colonized mice (previously GF mice co-housed with SPF for 3 wk). (B) Epithelial cell uptake of MDP-rho observed in the ileal villi of SPF mice is absent in germfree mice. (C) Biodistribution of [3H]-PGN in SPF mice treated with a broad-spectrum antibiotic cocktail. (D) Time-series showing absorption of orally administered [3H]-PGN before and during broad-spectrum antibiotic treatment. Radioactivity in the serum was measured daily, 6 h postgavage with [3H]-PGN. (E) The biodistribution of [3H]-PGN in OMM12 mice, compared with SPF and germfree mice. Scintillation counting was performed on dissolved, decolored organs. Results are normalized to CPM per g of tissue. Pairwise comparisons performed using the Mann–Whitney U test. *P ≤ 0.05; **P ≤ 0.005; ***P ≤ 0.0005. In panel D, results are normalized to CPM per mL of serum. Pairwise comparisons are performed using the Mann–Whitney U test to Day 0 (pre-antibiotic treatment). *P ≤ 0.05; **P ≤ 0.005; ***P ≤ 0.0005; ****P < 0.0001.
Finally, we assessed [3H]-PGN absorption and dissemination in Oligo-Mouse-Microbiota (OMM12) mice, which are colonized by a functional synthetic microbiota of twelve bacterial species representing the five major phyla naturally abundant in the mouse gut (25). We observed that OMM12 microbiota colonization was not sufficient to restore peptidoglycan absorption and dissemination pathways, having a profile similar to that of GF mice (Fig. 6D and SI Appendix, Fig. S8E). Taking these data together, we conclude that the presence and composition of the gut microbiota are critical determinants to activate and sustain the systemic biodistribution of peptidoglycan.
Discussion
In this study, we explored fundamental parameters governing the biodistribution of peptidoglycan originating in the mammalian host gut. Our study highlights that the kinetics and tropism of systemically distributed peptidoglycan depend on the route of administration. Orally administered peptidoglycan gradually disseminated to major organs over 6 h. Direct intravenous delivery of peptidoglycan was detected primarily in the kidneys where the peptidoglycan presence was high at 1 h, then declined rapidly, indicating that intravenously administered peptidoglycan is mainly excreted via the urine. Consistent with our data, previous studies tracking intravenously administered peptidoglycan moieties showed that up to 80% of 14C-labeled muropeptide monomers were excreted within the first hour (14, 26). Fewer studies have explored the fate of peptidoglycan entering the host system via the gut. Valinger et al. (27) showed that 14C-labeled peptidoglycan disaccharide pentapeptide monomer, administered by oral gavage, gradually accumulated in organs over several hours. Only 15% of radioactivity was excreted in the urine, while up to 30% of radioactivity was exhaled as CO2 within 48 h. Host metabolic pathways that degrade peptidoglycan to CO2 or other products have not been identified. Serum amidase PGLYRP-2, lysozyme, and macrophage N-acetyl-D-glucosaminidase remain the only host enzymes known to directly degrade peptidoglycan in mammals (13, 28, 29). Targeted mass spectrometry experiments performed on plasma or serum of different mammalian hosts have detected only a small number of anticipated masses associated with classical NOD1 and NOD2 ligands (8, 9). In this study, we demonstrated that when mice were gavaged with radiolabeled muropeptides with a peptide stem length of three, four, or eight amino acids, radioactivity was detected in approximately equal proportions in the organs, and in blood. Thus, muropeptides of different sizes are equally capable of acting as a source of the peptidoglycan moieties disseminated systemically. Our untargeted approach followed radioactivity from the mesoDAP moiety of peptidoglycan, and thus potentially detects uncharacterized products of host peptidoglycan metabolism derived from the mesoDAP component. The host is increasingly found to respond to nonclassical peptidoglycan derivates, independent of NOD1 and NOD2 receptors (30, 31), and even subtle structural differences between naturally occurring MDPs and muramyltripeptides are reported to induce different innate immune response profiles upon PRR activation (32). The importance of enzymatic processing of peptidoglycan in the intestine is yet to be fully explored, and may influence tropism by enriching the pool of moieties that are substrates for receptors or transporters at epithelial and endothelial barriers. Lysozyme activity was already shown to be critical to the systemic availability of NOD1 ligands (33). Thus, our data further highlight the need to better understand the true scope of peptidoglycan metabolism by the host.
Growing evidence suggests a gut–brain axis connecting microbiota peptidoglycan translocation to effects on the brain (10, 22, 34). Our data indicate that for studies of peptidoglycan effects on the brain, the route of delivery is a critical factor, as peptidoglycan was detected in the brain only when administered by gavage. Intravenously administered peptidoglycan was not detected in the brain, and absence from the brain was even more striking when administered intraperitoneally, since a higher relative abundance of peptidoglycan was observed in the spleen, liver, kidney, and fat. We also showed that peptidoglycan administered by gavage was enriched in the olfactory bulb. The brain shows regional differences for the uptake of blood-borne proteins and peptides. Like peptidoglycan, serum albumin and insulin are enriched in the olfactory bulb of rats and mice, whereas IL-1α, amylin, leptin accumulate differentially in other brain regions (35–38). However, all these molecules are transported into the brain from the blood when administered intravenously via saturable transport systems, whereas we were unable to detect intravenously administered peptidoglycan in the brain. The SLC15A family proton-coupled oligopeptide transporter proteins are proposed to transport muropeptides. In the brain, PepT2/SLC15A2 predominates, while expression of PHT1/SLC15A4 and PHT2/SLC15A3 has been reported in the brain or brain/brain barrier-derived cells in humans and rodents (39–41). PepT2 in rat choroid plexus endothelial cells localizes to the apical membrane of Blood–CSF barrier choroid plexus epithelial cells, where it performs efflux of peptide substrates from the CSF into the endothelia (42, 43). Notably, SLC15A transporter expression has not been detected at the Blood–Brain Barrier. Since our data showed that systemic peptidoglycan has poor access to the brain, one hypothesis is that brain barrier SLC15A transporters are not positioned to facilitate uptake into the brain directly from the blood, but function to clear peptidoglycan from the CNS. Conversely, intestinal transit facilitates peptidoglycan entry to the brain, with the same kinetic profile as for other organs, suggesting that they share a pathway of translocation. How might intestinal uptake facilitate dissemination and transit of CNS barriers? When absorbed by the gut, the peptidoglycan may undergo structural modifications that favor absorption and retention, which could explain why classical muropeptides are poorly detected from host biomaterials using mass spectrometry approaches. Intestinal transit could also direct peptidoglycan to carrier vehicles such as proteins, membrane vesicles, or migrating cell populations. It has been proposed that macrophages, dendritic cells (DCs), and neutrophils could perform this function (22), and exosomes were also proposed as a vehicle for peptidoglycan (21). Neutral-sphingomyelinase 2 (N-SMase2) activity, a key enzyme for ceramide production, is critical for exosome budding. Here, we show that inhibition of N-SMase2 suppresses peptidoglycan absorption from the gut. However, N-SMase2 is involved in a broad range of physiological processes, including orientation of migrating cells, inflammatory signaling, and cell differentiation, growth, and death (23, 44). Thus, the direct or indirect role of N-SMase2 activity in the peptidoglycan absorption and dissemination pathway requires further elucidation.
In this study, we demonstrated that the pathway facilitating intestinal translocation of peptidoglycan is induced by the gut microbiota. Peptidoglycan absorption and dissemination in GF mice occurred at extremely low levels compared with SPF mice. This basal level of absorption in GF mice is nevertheless sufficient to facilitate peptidoglycan-mediated effects in the gut and systemically (3, 6). The GF gut is considered leaky due to the observation that microbiota colonization, or microbial metabolites induce fortification of the intestinal barrier junction integrity (45–48). Our data suggest that in the absence of epithelial junction tightening, GF epithelial barrier integrity is sufficient to restrict the passive permeation of peptidoglycan fragments. This finding is consistent with a previous report that the GF mouse colon is less permeable to MDP and lipopolysaccharide than that of conventional mice (49). In GF mice, the peptidoglycan uptake system was induced by colonization with conventional microbiota, but not by OMM12 synthetic microbiota representing five major phyla of the conventional mouse gut. Conversely, peptidoglycan absorption was suppressed in adult SPF mice by antibiotic depletion of the microbiota. Using confocal microscopy, we observed that absorption of muropeptides is largely restricted to the small intestine (SI Appendix, Fig. S7), where the mucus barrier is thinner and the microbiota is less abundant, compared with the colon. Functional compartmentalization of peptidoglycan absorption to the small intestine may favor diffusion of peptidoglycan through the lumen to absorptive epithelial cells, while imposing an intrinsic limit on the abundance of peptidoglycan fragments entering the host system, due to the reduced number of gut bacteria. Abundant absorption of short dietary peptides occurs in the small intestine via PepT1(SLC15A1), which is also a proposed muropeptide transporter. However, microbial colonization status does not seem to effect intestinal PepT1 expression (50). Therefore, the peptidoglycan uptake pathway that is modulated by microbiota composition must either be independent of PepT1 or occurs downstream of PepT1 transport. It should also be noted that our study does not characterize dissemination of peptidoglycan via the lymphatic system. The small intestinal microbiota has been shown to sustain mature lacteals in the small intestinal villi, by induction of vascular endothelial growth factor C (VEGF-C) in macrophages, dependent on (TLR signaling adaptor protein) MyD88 activation (51). Microbiota and mature lacteal vessels are necessary for absorption of triglyceride, cholesterol, and free fatty acids (51, 52). A similar phenomenon could contribute to the suppression of peptidoglycan absorption in GF or microbiota-depleted mice. However, our data suggest that the microbiota-dependent absorption of peptidoglycan is not a simple consequence of lacteal integrity; we observed a sharp decline in peptidoglycan fragments reaching the blood within 24 h of antibiotic treatment (Fig. 6), whereas significant alteration of lacteal integrity required over two weeks of antibiotic treatment in postnatal mice. Acute effects of microbiota depletion were not tested in adult mice (52). On the other hand, while we showed that muropeptides injected intravenously are rapidly cleared from the host system and do not reach the brain, we do not know the fate of peptidoglycan in the lymphatic system, and this remains an avenue for further study.
Fluorescent muropeptide uptake was observed in gut epithelia of SPF mice, but not GF mice, suggesting that in the absence of microbiota, peptidoglycan absorption is restricted at the level of the epithelial interface with the gut lumen. Our study found that uptake of Fluo-PGN was most prominent in goblet cells. However, Fluo-PGN was observed in an irregular manner in nongoblet cells of the epithelial layer, and we hypothesize that a subgroup of absorptive enterocytes that escaped characterization may also absorb peptidoglycan. Consistent with this hypothesis, targeted inhibition of GAPs suggested a partial contribution of goblet cells in peptidoglycan systemic dissemination (Fig. 5C). GAPs facilitate delivery of luminal antigen to innate immune regulatory cell populations such as CD103+ DCs, to promote lamina propria immune cell tolerance to dietary antigens (16, 18). Lactobacillus salivarius Ls33 peptidoglycan was found to protect mice against experimental colitis via a CD103+ DC-dependent mechanism (53), supporting a hypothesis that goblet cell absorption of peptidoglycan is primarily an immune-sampling function. We also cannot exclude GAP-independent trafficking of peptidoglycan by goblet cells. M-cells, another specialist cell class involved in immune surveillance, are proposed to absorb nanomineral encapsulated peptidoglycan for presentation to antigen-presenting cells (54). We did not observe peptidoglycan absorption by M-cells, however our methodology (injection of fluorescent muropeptide conjugate into ileal ligatures over a short time frame) may not favor detection of nanomineral encapsulated peptidoglycan. Together, our data support a hypothesis of functional separation between small intestine epithelial cells involved in luminal peptidoglycan sampling for immune surveillance, and those that coordinate the steady-state dissemination of peptidoglycan.
This study highlights the importance of the gut for the systemic tropism of peptidoglycan and reveals major processes regulating the dissemination of peptidoglycan from the gut. By exploring such parameters, we will better understand the peptidoglycan-driven mechanisms through which gut microbiota mediate steady-state processes in health, and how their dysbiosis drives pathological effects.
Materials and Methods
Unless otherwise stated, radiolabeling was performed in the Escherichia coli FB8-LysA strain which is unable to convert meso-diaminopimelic acid (mesoDAP) to lysine by decarboxylation (55). Therefore, mesoDAP added to the growth media is incorporated specifically into the peptidoglycan layer. E. coli FB8-LysA was cultured on LB Miller agar (BD Difco) containing 25 mg/mL kanamycin (Merck). Several colonies were inoculated into 50 mL LB medium plus 25 mg/mL kanamycin and incubated at 37 °C. At approximately OD600 1.0, the preculture was used to inoculate (1:100) 1L prewarmed M9 minimal media supplemented with 100 µg/mL threonine, methionine, and lysine (Merck). For 3H-mesoDAP labeling, 50 µCi/L 3H-meso diaminopimelic acid (3H-mesoDAP; Moraveck Inc.) was added. For 14C-GlcNAc labeling of Escherichia coli, culture was performed in M9 media using 100 μM GlcNAc (Merck) as the carbon source, and spiked with 10 μCi/L 14C-N-acetylglucosamine (14C-GlcNAc; ARC). Cultures were incubated overnight at 37 °C with aeration. Final OD600 was approximately 2.0. Bacteria were harvested by centrifugation at 4,000 × g, resuspension in a small volume of cold H2O, and dropped into 20 mL of 4% SDS in a boiling bain-marie. After 1 h boiling with vigorous agitation, the suspension was cooled to room temperature, and centrifugated at 13,000 × g. The supernatant was discarded, and the pellet resuspended with 20 mL H2O. The radiolabeled cell wall material was washed by centrifugation and resuspension of the pellet with H2O, until the presence of SDS in the supernatant could no longer be detected using the method of Hayashi (56). The pellet was then resuspended in 4 mL of 50 mM Tris pH 7.5 (Merck), and incubated at 37 °C with 100 µg/mL alpha-amylase (Merck) for 2 h, followed by 2 h incubation with RNase, DNase, and MgCl2, then incubated overnight with 100 µg/mL 3× crystallized trypsin (Worthing Biochemical Corporation) and CaCl2. The pellet was incubated for 15 min at 100 °C, then washed once with H20. Radiolabeled peptidoglycan was stored at −20 °C. For gavage of mice, peptidoglycan was digested overnight with 100 U/mg mutanolysin (from Streptomyces globisporus ATCC 21553, Merck) in 12.5 mM sodium phosphate pH 5.6 (Merck) at 37 °C. Radiolabeling of L. rhamnosus Lr32 was described previously (10).
Animals.
C57BL/6J female mice were purchased from Charlies River Laboratories at the ages of 6 to 7 wk and housed at the Institut Pasteur animal facilities under specific pathogen-free conditions. NOD1/2 KO mice, GF mice, and OMM12 mice were bred and maintained at Institut Pasteur facilities. GF mice and OMM12 mice were housed in the Centre for Gnotobiology Platform of the Institut Pasteur. Experiments were performed using female mice at 8 to 12 wk of age.
Ethics Statement.
Animal experiments were performed in accordance with Directive 2010/63/EU of the European Parliament and the French regulation for the protection of laboratory animals decree of February 1, 2013. The project number APAFIS #8551 was approved by the Institut Pasteur ethical committee for animal experimentation (Comité d’Ethique en Expérimentation Animale CETEA registry number #89) and authorized by the Ministère de l'Enseignement Supérieur, de la Recherche et de l'Innovation.
Radioactive Peptidoglycan Tracking.
Unless otherwise stated, mice were gavaged with a 200 μL volume containing approximately 400,000 cpm of labeled peptidoglycan. Approximately 30 min before the specified time point, profound anesthesia was induced in mice by intraperitoneal administration of 100 mg/kg Imalgene1000 (Boehringer-Ingelheim) and 8 mg/kg of Rompun 2% (Bayer). To avoid the detection of radioactivity transiently present in the circulation, blood was cleared from the organs by transcardial perfusion with 40 mL PBS (Lonza Pharma & Biotech) by a syringe with a 26-G needle, with section of the inferior vena cava. Blood collection was performed immediately upon section of the vena cava. Organs were removed into 2 mL microfuge tubes. For small intestine samples, approximately 8 cm of tissue adjacent to the stomach (“Duodeum”), and 8 cm of tissue adjacent to the cecum (“Ileum”) were collected. Approximately 6 cm of colon was collected. The gut pieces were opened longitudinally, then the gut contents removed by vigorous washing three times in PBS. The gut tissue was dried on tissue paper then transferred to 2 mL microfuge tubes. For blood, collection tubes contained 10 µL of 0.5 M EDTA pH 8.0 (Lonza Pharma & Biotech). The weights of all organs and tissues were recorded. If further processing was not performed immediately, the samples were stored at −20 °C.
Drug Treatments.
Parasympatholytic reagents: 100× stocks of tropicamide (Merck) in ethanol (5.5 mg/mL) or atropine sulfate (Merck) in ethanol (6 mg/mL) were prepared and stored at 4 °C. On the day of injection, reagents were diluted 1:100 in PBS. Tropicamide (550 μg/kg) or atropine sulfate (600 μg/kg) were injected intraperitoneally in a 200 μL volume, 20 min prior to administration of [3H]-PGN. Control mice received vehicle only.
GW4869 treatment: Five mg/mL GW4869 (N,N′-Bis[4-(4,5-dihydro-1H-imidazol-2-yl)phenyl]-3,3′-p-phenylene-bis-acrylamide dihydrochloride; Merck) solution was diluted in sterile PBS to a final concentration of 0.25 mg/mL. Two-hundred microliters of GW4869 solution was administered intraperitoneally 16 h and 2 h prior to gavage with [3H]-PGN. Control mice received vehicle only.
Antibiotic treatment: intestinal microbiota of SPF mice was depleted by administration of antibiotic cocktail in the drinking water of mice for 21 d. Ampicillin (1 mg/mL, Merck), streptomycin (5 mg/mL, Euromedex), colistin sulfate (1 mg/mL, Merck), and vancomycin (0.25 mg/mL, Merck) were prepared in water and filtered through 0.22-µm sterile filters (Corning) and protected from light with aluminum foil. Fresh solution was changed every 3 d. Microbiota depletion was regularly checked by spotting 10-µL serial dilutions of fecal homogenate in PBS onto trypticase soy agar (BD) enriched with 5% defibrinated horse blood (Thermo Fisher) and incubation at 37 °C under aerobic and anaerobic atmosphere. Colony forming units were counted after 24 h and 48 h incubation.
Scintillation Counting.
Organs, tissues, or blood (200 μL) were transferred into glass vials, then dissolved by addition of 2 mL (3 mL for the liver samples) of Solvable (PerkinElmer) to each vial. Samples were incubated overnight at 60 °C. After cooling to room temperature, the solutions were rendered transparent by addition of 100 μL of 0.5 M EDTA pH 8, followed by 2× 200 μL of 30% hydrogen peroxide (Millipore). Samples were incubated for 30 min at 60 °C. After cooling again to room temperature, the solutions were poured into 20 mL HDPE scintillation vials with a urea cap containing a polyethylene cone (Duran Wheaton Kimble), followed by addition of 10 mL Ultima Gold LLT scintillation cocktail (Perkin Elmer). For stools, 1 mL PBS was added to 1.5-mL centrifuge tubes containing fecal pellets and the material homogenized with a disposable pestle. The samples were centrifugated at 300 × g for 5 min. Then supernatant was collected into a 5 mL HDPE scintillation vial then 20 μL of 0.5 M EDTA pH 8, and 100 μL of 30% hydrogen peroxide added, then incubated at room temperature for 60 min, followed by 30 min at 60 °C. After cooling to room temperature, 5 mL Ultima Gold LLT scintillation cocktail was added. The samples were equilibrated in the dark at room temperature for 4 h. Scintillation counting was performed using a Tri-Carb 3110 TR Liquid Scintillation Analyzer with QuantaSmart TriCarb LCS 3.00 software. 3H was measured in the range 2.0 to 18.6 keV for 5 min on the high sensitivity setting. 14C was measure in the range 0.0 to 156 keV for 2 min on the high-sensitivity setting.
Intestinal Ligatures.
For intestinal ligature surgery in mice, profound anesthesia was induced by intraperitoneal administration of 100 mg/kg Imalgene1000 (Boehringer-Ingelheim) and 8 mg/kg of Rompun 2% (Bayer). Mice were placed on a heated mat to prevent hypothermia. A horizontal laparotomy was performed on the abdomen and a 3 to 4-cm region of proximal intestine (duodenum), distal small intestine (ileum) or colon clamped on the extremities using Schwartz vessel clips (World Precision Instruments). Care was taken to avoid compromising the blood supply to the ligated region. Two hundred microliters of MDP-rhodamine (InvivoGen) or mutanolysin (100 U/mg) digested peptidoglycan-AF647 conjugate (1 mg/mL) or PBS control solution was injected into the ligature using a 29-G needle. Ligatures were covered gently with gauze soaked with warm DMEM solution (Gibco). After 20 min, the ligature was collected and the mice sacrificed by cervical dislocation. The excised tissue was either opened longitudinally, washed 3 times in PBS, and pinned flat onto an agarose pad, or washed directly without opening. Tissues were fixed overnight using 4% v/v paraformaldehyde in PBS at 4 °C, then washed 5 times in PBS. Fixed tissues were embedded in 4% LMP agarose (Merck) blocks, then cut into 100-μm-thick sections using a Microm HM 650 V Vibration microtome (Thermo Fisher). Tissue sections were carefully removed from agarose and stored short term in PBS at 4 °C in the dark.
Immunofluorescence Imaging.
Tissue sections were blocked and permeabilized by incubation in blocking solution, which comprised PBS containing 3% w/v bovine serum albumin (Merck) and 0.4% v/v Triton X-100 (Merck), at 4 °C. The following primary antibodies were used: anti-mouse E-cadherin (rat mAb Eccd2, Takara Bio #M108 1:250), anti-mouse Microfold (M) cell (rat mAb NKM 16-2-4, Miltenyi Biotec #130-096-150 1:250), anti-mouse Siglec-F (rat mAb E50-2440, BD Pharmingen #552125, 1:100), Anti-mouse Chromaganin-A (goat pAb sc-1488, Santa Cruz Biotechnology, 1:200). For secondary staining the following antibodies were used: Alexa Fluor 488 goat anti-rat (Invitrogen, 1:500), Alexa-Fluor 488 donkey, anti-goat (Invitrogen, 1:500). For antibody staining, tissues were incubated in blocking solution for 3 h, then transferred to blocking solution containing primary antibody at the appropriate dilution and incubated overnight at 4 °C with gentle agitation. Primary antibody was removed by passage of tissue sections five times in 4 mL PBS for 5 min. Tissue sections were then incubated for 1 h at room temperature with secondary antibody and/or other markers as appropriate: Wheat Germ Agglutinin-Alexa Fluor 488 (1:200, Invitrogen), DAPI (1:1,000, BD Biosciences), Phalloidin-iFluor 647 (1:200, Abcam). Tissue sections were washed by passage in PBS as before, then mounted on Superfrost Plus microscope slides (Thermo Fisher) using ProLong Gold Antifade reagent (Thermo Fisher) and #1.5 coverslips (VWR). Confocal acquisitions were performed using a Leica TCS SP8 and Leica HyD SP5 confocal microscopes.
Peptidoglycome Purification.
Mice were sacrificed and the entire intestinal tract removed. The lumen content of the intestinal tract was pressed into 5 mL PBS and frozen at −80 °C. To extract the peptidoglycan, 4 mL of 20% w/v SDS (Interchim) and 11 mL of H2O were added directly to the pellet, which was thawed and then incubated in a boiling bain-marie for 1 h with vigorous agitation. After cooling to room temperature, the suspension was filtered through a 30-µm cell strainer; then the standard protocol for purification of peptidoglycan from Gram-positive bacteria was performed, which includes all the steps necessary to purify peptidoglycan from Gram-negative peptidoglycan (57). The final extract contains purified insoluble peptidoglycan of the gut microbiota. Soluble muropeptides were generated by overnight incubation of the purified extract in 160 µL of 12.5 mM NaH2PO4 pH 5.6 and 100 U mutanolysin from S. globisporus ATCC 21553 (Merck) at 37 °C. The reaction was stopped by incubation at 100 °C for 10 min, then centrifugated for 5 min at 16,000 × g. The supernatant was collected and stored at −20 °C.
Mass Spectrometry Analysis.
Muropeptides were reduced by addition of 150 µL of 500 mM borate buffer pH 9, and 50 µL of 20 mg/mL sodium borohydride (NaBH4) solution (Merck) prepared immediately before use. After 30-min incubation at room temperature, the reaction was stopped by adjusting the pH to 4 by addition of 85% orthophosphoric acid (Prolabo). The reaction was centrifugated for 5 min at 16,000 g and the supernatant containing reduced muropeptides collected. Ten microliters of muropeptide solution was diluted fivefold in mobile phase (formic acid 0.1% in water) before analysis.
Muropeptides analysis was performed by UHPLC-HRMS, using a Dionex Ultimate 3,000 UHPLC coupled to a qExactive Focus (Thermo Fisher). Analysis was performed by injecting 10 µL muropeptide solution onto a Hypersil GOLD C18 aQ C18 (175 Å, 1.9 μm, 2.1 × 150 mm) column, at a temperature of 50 °C at a flow rate of 0.2 mL/min. Mobile phase A: 0.1% formic acid (Optima, Fisher chemical) in water (Optima LC-MS, Fisher chemical); Mobile phase B = 0.1% formic acid in acetonitrile (Optima LC-MS, Fisher chemical). Analytes were separated using a gradient of 0 to 15% mobile phase B over 30 min. For Escherichia coli peptidoglycan analysis: The Q Exactive Focus was operated under electrospray ionization (H-ESI II) positive mode. Full scan (m/z 200 to 2,000) used resolution 70,000 (FWHM) at m/z 200, with an automatic gain control (AGC) target of 1 × 106 ions and an automated maximum ion injection time (IT). Data-dependent MS/MS were acquired on a “Top 3” data-dependent mode using the following parameters: resolution 17,500; AGC 1×105 ions, maximum IT 50 ms, NCE 25%, and a dynamic exclusion time 5 s. For mouse gut microbiota peptidoglycan analysis: Mass spectrometry was set to positive electrospray ionization mode with a scan range from 150 to 2,000, with full scan data-dependent acquisition of the three most abundant precursor ions for tandem MS by HCD fragmentation. Trace Finder (3.3; Thermo Scientific) software was used for the peptidoglycome analysis. The screening method was based on our homemade muropeptides database.
HPLC.
HPLC analysis of 3H-mesoDAP-labeled peptidoglycan was performed using an LC20 Shimadzu HPLC system, equipped with a Hypersil GOLD aQ C18 column (4.6 × 250 mm; Thermo Fisher) at 52 °C, using a flow rate of 0.5 mL/min. Muropeptides were detected at a 206-nm wavelength using a Shimadzu SPD-20A-UV-Vis detector. For collection of muropeptides, peaks were collected from the outlet of the UV detector into 2-mL microfuge tubes. For radioactive measurements of HPLC purified muropeptides, a 10 μL aliquot of muropeptide solution was transferred to a 6 mL scintillation vial and combined with 5 mL Ultima Gold LLT scintillation cocktail (Perkin Elmer). After equilibration in the dark at room temperature for 4 h, samples were analyzed using a Tri-Carb 3110 TR Liquid Scintillation Analyzer with QuantaSmart TriCarb LCS 3.00 software, with measurement performed in the range 2.0 to 18.6 keV for 5 min on the high sensitivity setting.
Data analysis and Figure Preparation.
Scintillation counting data were plotted using Prism GraphPad. Details of statistical analyses performed are indicated on the relevant figure legend. Schematics were created using BioRender.com and Servier Medical Art, provided by Servier, licensed under a Creative Commons Attribution 3.0 unported license.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank the UTechS PBI, a member of the France–BioImaging infrastructure network supported by the French National Research Agency (ANR-10–INSB–04, Investments for the future) for microscope usage and assistance. We thank the members of the Centre for Gnotobiology Platform of the Institut Pasteur (especially Thierry Angélique, Eddie Maranghi, Martine Jacob, and Marisa Gabriela Lopez Dieguez), and of the Institut Pasteur Central Animal Facility for all their assistance with animal studies. P.A.D.B. was part of the Pasteur-Paris University International PhD Program. This project has received funding from the Institut Carnot Pasteur Microbes & Santé, and the European Union's Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement no 665807. A.R. and I.G.B. laboratory were supported by Investissement d’Avenir program, Laboratoire d’Excellence “Integrative Biology of Emerging Infectious Diseases” (ANR-10-LABX-62-IBEID). I.G.B. laboratory was also supported by the Investissement d’Avenir program (RHU Torino Lumière ANR-16-RHUS-0008), by the French National Research Agency (ANR-16-CE15-0021) and by R&D grants from Danone and MEIJI. Additional funding was provided by DIM1Health.
Author contributions
R.W., P.A.D.B., and I.G.B. designed research; R.W., P.A.D.B., O.D., A.R., I.G., and J.S. performed research; M.B. contributed new reagents/analytic tools; R.W., P.A.D.B., A.R., M.L., and I.G.B. analyzed data; R.W., M.L, and I.G.B. acquired funding; and R.W., P.A.D.B., and I.G.B. wrote the paper.
Competing interest
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Richard Wheeler, Email: wheelerrsci@gmail.com.
Ivo Gomperts Boneca, Email: bonecai@pasteur.fr.
Data, Materials, and Software Availability
Confocal miroscopy raw data are available at the Zenodo repository, doi: 10.5281/zenodo.7509202. Mass spectrometry data for every replicate of the gut peptidoglycome analysis have been deposited in MassIVE under accession number MSV000091014. All other study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Curciarello R., Canziani K. E., Docena G. H., Muglia C. I., Contribution of non-immune cells to activation and modulation of the intestinal inflammation. Front. Immunol. 10, 647 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chu H., Mazmanian S. K., Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nat. Immunol. 14, 668–675 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouskra D., et al. , Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature 456, 507–510 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Hergott C. B., et al. , Peptidoglycan from the gut microbiota governs the lifespan of circulating phagocytes at homeostasis. Blood 127, 2460–2471 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clarke T. B., et al. , Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat. Med. 16, 228–231 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arentsen T., et al. , The bacterial peptidoglycan-sensing molecule Pglyrp2 modulates brain development and behavior. Mol. Psychiatry 22, 257–266 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schrijver I. A., et al. , Reduced systemic IgG levels against peptidoglycan in rheumatoid arthritis (RA) patients. Clin. Exp. Immunol. 123, 140–146 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang Z., et al. , Antibody neutralization of microbiota-derived circulating peptidoglycan dampens inflammation and ameliorates autoimmunity. Nat. Microbiol. 4, 766–773 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Molinaro R., Mukherjee T., Flick R., Philpott D. J., Girardin S. E., Trace levels of peptidoglycan in serum underlie the NOD-dependent cytokine response to endoplasmic reticulum stress. J. Biol. Chem. 294, 9007–9015 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gabanyi I., Bacterial sensing via neuronal Nod2 regulates appetite and body temperature. Science 376, eabj3986 (2022). [DOI] [PubMed] [Google Scholar]
- 11.Bastos P. A. D., Wheeler R., Boneca I. G., Uptake, recognition and responses to peptidoglycan in the mammalian host. FEMS Microbiol. Rev. 45, fuaa044 (2020), 10.1093/femsre/fuaa044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith D. E., Clémençon B., Hediger M. A., Proton-coupled oligopeptide transporter family SLC15: Physiological, pharmacological and pathological implications. Mol. Aspects Med. 34, 323–336 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z.-M., et al. , Human peptidoglycan recognition protein-L is an N-acetylmuramoyl-L-alanine amidase. J. Biol. Chem. 278, 49044–49052 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Tomasić J., Ladesić B., Valinger Z., Hrsak I., The metabolic fate of 14C-labeled peptidoglycan monomer in mice. I. Identification of the monomer and the corresponding pentapeptide in urine. Biochim. Biophys. Acta 629, 77–82 (1980). [DOI] [PubMed] [Google Scholar]
- 15.Schleifer K. H., Kandler O., Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36, 407–477 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kulkarni D. H., et al. , Goblet cell associated antigen passages support the induction and maintenance of oral tolerance. Mucosal Immunol. 13, 271–282 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knoop K. A., McDonald K. G., McCrate S., McDole J. R., Newberry R. D., Microbial sensing by goblet cells controls immune surveillance of luminal antigens in the colon. Mucosal Immunol. 8, 198–210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDole J. R., et al. , Goblet cells deliver luminal antigen to CD103+ dendritic cells in the small intestine. Nature 483, 345–349 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller M. J., Knoop K. A., Newberry R. D., Mind the GAPs: Insights into intestinal epithelial barrier maintenance and luminal antigen delivery. Mucosal Immunol. 7, 452–454 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Muise E. D., Gandotra N., Tackett J. J., Bamdad M. C., Cowles R. A., Distribution of muscarinic acetylcholine receptor subtypes in the murine small intestine. Life Sci. 169, 6–10 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Bu H.-F., Wang X., Tang Y., Koti V., Tan X.-D., Toll-like receptor 2-mediated peptidoglycan uptake by immature intestinal epithelial cells from apical side and exosome-associated transcellular transcytosis. J. Cell Physiol. 222, 658–668 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laman J. D., Hart B. A., Power C., Dziarski R., Bacterial peptidoglycan as a driver of chronic brain inflammation. Trends Mol. Med. 26, 670–682 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Sitrin R. G., Sassanella T. M., Petty H. R., An obligate role for membrane-associated neutral sphingomyelinase activity in orienting chemotactic migration of human neutrophils. Am. J. Respir. Cell Mol. Biol. 44, 205–212 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X., et al. , Cardiomyocytes mediate anti-angiogenesis in type 2 diabetic rats through the exosomal transfer of miR-320 into endothelial cells. J. Mol. Cell Cardiol. 74, 139–150 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brugiroux S., et al. , Genome-guided design of a defined mouse microbiota that confers colonization resistance against Salmonella enterica serovar Typhimurium. Nat. Microbiol. 2, 1–12 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Ladesić B., Tomasić J., Kveder S., Hrsak I., The metabolic fate of 14C-labeled immunoadjuvant peptidoglycan monomer. II. In vitro studies. Biochim. Biophys. Acta 678, 12–17 (1981). [DOI] [PubMed] [Google Scholar]
- 27.Valinger Z., Ladesić B., Hrsak I., Tomasić J., Relationship of metabolism and immunostimulating activity of peptidoglycan monomer in mice after three different routes of administration. Int. J. Immunopharmacol. 9, 325–332 (1987). [DOI] [PubMed] [Google Scholar]
- 28.Bourbouze R., et al. , N-acetyl-beta-D-glucosaminidase (NAG) isoenzymes release from human monocyte-derived macrophages in response to zymosan and human recombinant interferon-gamma. Clin. Chim. Acta Int. J. Clin. Chem. 199, 185–194 (1991). [DOI] [PubMed] [Google Scholar]
- 29.Irwin D. M., Biegel J. M., Stewart C.-B., Evolution of the mammalian lysozyme gene family. BMC Evol. Biol. 11, 166 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolf A. J., et al. , Hexokinase is an innate immune receptor for the detection of bacterial peptidoglycan. Cell 166, 624–636 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Read C. B., et al. , Cutting Edge: Identification of neutrophil PGLYRP1 as a ligand for TREM-1. J. Immunol. 194, 1417–1421 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bersch K. L., et al. , Bacterial peptidoglycan fragments differentially regulate innate immune signaling. ACS Cent. Sci. 7, 688–696 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Q., et al. , Intestinal lysozyme liberates Nod1 ligands from microbes to direct insulin trafficking in pancreatic beta cells. Cell Res. 29, 516–532 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tosoni G., Conti M., Diaz Heijtz R., Bacterial peptidoglycans as novel signaling molecules from microbiota to brain. Curr. Opin. Pharmacol. 48, 107–113 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Ueno M., et al. , The persistence of high uptake of serum albumin in the olfactory bulbs of mice throughout their adult lives. Arch. Gerontol. Geriatr. 13, 201–209 (1991). [DOI] [PubMed] [Google Scholar]
- 36.Maness L. M., Banks W. A., Zadina J. E., Kastin A. J., Selective transport of blood-borne interleukin-1 alpha into the posterior division of the septum of the mouse brain. Brain Res. 700, 83–88 (1995). [DOI] [PubMed] [Google Scholar]
- 37.Banks W. A., Kastin A. J., Huang W., Jaspan J. B., Maness L. M., Leptin enters the brain by a saturable system independent of insulin. Peptides 17, 305–311 (1996). [DOI] [PubMed] [Google Scholar]
- 38.Banks W. A., Kastin A. J., Differential permeability of the blood-brain barrier to two pancreatic peptides: Insulin and amylin. Peptides 19, 883–889 (1998). [DOI] [PubMed] [Google Scholar]
- 39.Yamashita T., et al. , Cloning and functional expression of a brain peptide/histidine transporter. J. Biol. Chem. 272, 10205–10211 (1997). [DOI] [PubMed] [Google Scholar]
- 40.Sakata K., et al. , Cloning of a lymphatic peptide/histidine transporter. Biochem. J. 356, 53–60 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oppermann H., Heinrich M., Birkemeyer C., Meixensberger J., Gaunitz F., The proton-coupled oligopeptide transporters PEPT2, PHT1 and PHT2 mediate the uptake of carnosine in glioblastoma cells. Amino Acids 51, 999–1008 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Shu C., et al. , Role of PEPT2 in peptide/mimetic trafficking at the blood-cerebrospinal fluid barrier: Studies in rat choroid plexus epithelial cells in primary culture. J. Pharmacol. Exp. Ther. 301, 820–829 (2002). [DOI] [PubMed] [Google Scholar]
- 43.Ocheltree S. M., Shen H., Hu Y., Keep R. F., Smith D. E., Role and relevance of peptide transporter 2 (PEPT2) in the kidney and choroid plexus: In vivo studies with glycylsarcosine in wild-type and PEPT2 knockout mice. J. Pharmacol. Exp. Ther. 315, 240–247 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Shamseddine A. A., Airola M. V., Hannun Y. A., Roles and regulation of neutral sphingomyelinase-2 in cellular and pathological processes. Adv. Biol. Regul. 57, 24–41 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hooper L. V., et al. , Molecular analysis of commensal host-microbial relationships in the intestine. Science 291, 881–884 (2001). [DOI] [PubMed] [Google Scholar]
- 46.Shimada Y., et al. , Commensal bacteria-dependent indole production enhances epithelial barrier function in the colon. PLoS One 8, e80604 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh R., et al. , Enhancement of the gut barrier integrity by a microbial metabolite through the Nrf2 pathway. Nat. Commun. 10, 89 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karczewski J., et al. , Regulation of human epithelial tight junction proteins by Lactobacillus plantarum in vivo and protective effects on the epithelial barrier. Am. J. Physiol. Gastrointest. Liver Physiol. 298, G851–G859 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Hayes C. L., et al. , Commensal microbiota induces colonic barrier structure and functions that contribute to homeostasis. Sci. Rep. 8, 14184 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wuensch T., et al. , The peptide transporter PEPT1 is expressed in distal colon in rodents and humans and contributes to water absorption. Am. J. Physiol. Gastrointest. Liver Physiol. 305, G66–73 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Martinez-Guryn K., et al. , Small intestine microbiota regulate host digestive and absorptive adaptive responses to dietary lipids. Cell Host Microbe 23, 458–469.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suh S. H., et al. , Gut microbiota regulates lacteal integrity by inducing VEGF-C in intestinal villus macrophages. EMBO Rep. 20, e46927 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Macho Fernandez E., et al. , Anti-inflammatory capacity of selected lactobacilli in experimental colitis is driven by NOD2-mediated recognition of a specific peptidoglycan-derived muropeptide. Gut 60, 1050–1059 (2011). [DOI] [PubMed] [Google Scholar]
- 54.Powell J. J., et al. , An endogenous nanomineral chaperones luminal antigen and peptidoglycan to intestinal immune cells. Nat. Nanotechnol. 10, 361–369 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mengin-Lecreulx D., Siegel E., van Heijenoort J., Variations in UDP-N-acetylglucosamine and UDP-N-acetylmuramyl-pentapeptide pools in Escherichia coli after inhibition of protein synthesis. J. Bacteriol. 171, 3282–3287 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hayashi K., A rapid determination of sodium dodecyl sulfate with methylene blue. Anal. Biochem. 67, 503–506 (1975). [DOI] [PubMed] [Google Scholar]
- 57.Wheeler R., Veyrier F., Werts C., Boneca I. G., “Peptidoglycan and Nod Receptor” in Glycoscience: Biology and Medicine, Taniguchi N., Endo T., Hart G. W., Seeberger P. H., Wong C. H., Eds. (Springer, Japan, 2015), pp. 737–747, 10.1007/978-4-431-54841-6_147. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Confocal miroscopy raw data are available at the Zenodo repository, doi: 10.5281/zenodo.7509202. Mass spectrometry data for every replicate of the gut peptidoglycome analysis have been deposited in MassIVE under accession number MSV000091014. All other study data are included in the article and/or SI Appendix.