Summary
The coefficient of determination (R2) is a well-established measure to indicate the predictive ability of polygenic scores (PGSs). However, the sampling variance of R2 is rarely considered so that 95% confidence intervals (CI) are not usually reported. Moreover, when comparisons are made between PGSs based on different discovery samples, the sampling covariance of R2 is required to test the difference between them. Here, we show how to estimate the variance and covariance of R2 values to assess the 95% CI and p value of the R2 difference. We apply this approach to real data calculating PGSs in 28,880 European participants derived from UK Biobank (UKBB) and Biobank Japan (BBJ) GWAS summary statistics for cholesterol and BMI. We quantify the significantly higher predictive ability of UKBB PGSs compared to BBJ PGSs (p value 7.6e−31 for cholesterol and 1.4e−50 for BMI). A joint model of UKBB and BBJ PGSs significantly improves the predictive ability, compared to a model of UKBB PGS only (p value 3.5e−05 for cholesterol and 1.3e−28 for BMI). We also show that the predictive ability of regulatory SNPs is significantly enriched over non-regulatory SNPs for cholesterol (p value 8.9e−26 for UKBB and 3.8e−17 for BBJ). We suggest that the proposed approach (available in R package r2redux) should be used to test the statistical significance of difference between pairs of PGSs, which may help to draw a correct conclusion about the comparative predictive ability of PGSs.
Keywords: coefficient of determination, R2, polygenic scores, PGSs, sampling variance, sampling covariance, significance test, non-nested model comparison, R2-based genomic partitioning
R2 is a well-established measure for the reliability of polygenic score models although its significance test is rarely considered in this context. We release an R package r2redux that allows formal statistical comparison of two polygenic score models, providing the 95% confidence interval and significance of R2 difference.
Introduction
Complex traits are affected by many risk factors including polygenic effects.1,2,3 Genetic profile analysis can quantify how polygenic effects are associated with future disease risk at the individual and population levels.4,5 Genetic profiling has potential benefits that can help people make informed decisions when they manage their health and medical care.6,7,8
Genome-wide association studies (GWASs) have provided an opportunity to estimate genetic profile or polygenic scores (PGSs) that represent individual risk predictions from genetic data.4,9,10,11,12,13,14 Typically, the effects of genome-wide single-nucleotide polymorphisms (SNPs) associated with complex traits are estimated in a discovery dataset, which are projected in an independent target dataset. Then, for each individual in the target samples the weighted genotypic coefficients according to the projected SNP effects (i.e., PGSs) are derived and correlated with outcome (trait including affected/unaffected for disease) to quantify the prediction accuracy. The squared correlation or coefficient of determination (R2) is a useful measure to quantify the reliability of the PGS. Note that R2 is equivalent to the squared regression coefficient if the dependent and explanatory variables are column standardized.15
Previously, we introduced a measure of R2 on the liability scale that can be comparable across different models and scales16 when using disease traits or ascertained case-control data. Choi et al.12 reported that this R2 measure on the liability scale outperforms the widely used Nagelkerke pseudo R2 in controlling for bias due to ascertained case-control samples. Nagelkerke pseudo R2 estimates depend on the proportion of affected individuals in the sample. In contrast, R2 on the liability scale does not depend on the proportion of cases in the sample but does require an estimate of the lifetime population prevalence of the disease.
Wand et al.11 suggested that any PGS study should report R2 as an indicator of the predictive ability. Choi et al.12 concluded that R2 is a useful metric to measure association and goodness of fit in the interpretation of PGS predictions. Many studies have demonstrated the predictive ability of PGSs, using R2.12,13,17,18 However, the variance of R215 has been rarely studied especially in the context of PGSs although it is the crucial parameter for estimation of confidence intervals (CI) of R2. Furthermore, estimates of the covariance between a pair of R2 values (e.g., from two sets of PGSs) are necessary to assess whether they are significantly different from each other, or if the ratio of two R2 values significantly deviates from the expectation. This significance test for the difference or ratio is important when comparing two or multiple sets of PGSs that are derived from different sets of SNPs, e.g., genomic partitioning, genome-wide association p value thresholds (pT) analysis, or PGSs based on pathway subsets.19,20
In this study, we use R2 measures and their variance-covariance matrix to assess whether the predictive abilities of PGSs based on different sources are significantly different from each other. We derive the variance and covariance of R2 values to generate estimates of its 95% CI and p value of the R2 difference, considering two sets of dependent or independent PGSs. We also derive the variance and covariance matrix (i.e., information matrix) of squared regression coefficients in a multiple regression model, testing whether the proportion of the squared regression coefficient attributable to SNPs in the regulatory region is significantly higher than expected (i.e., PGS-based genomic partitioning method). We apply this approach to real data to compare PGSs calculated in 28,880 European individuals using UK Biobank (UKBB) and Biobank Japan (BBJ) GWAS summary statistics for cholesterol and BMI.
Material and methods
We used data from the UK Biobank (https://www.ukbiobank.ac.uk), the scientific protocol of which has been reviewed and approved by the Northwest Multi-center Research Ethics Committee, National Information Governance Board for Health & Social Care, and Community Health Index Advisory Group. UK Biobank has obtained informed consent from all participants. Our access to the UK Biobank data was under the reference number 14575.
Publicly available GWAS summary statistics of Biobank Japan (BBJ)21,22 were used, following BBJ’s guidelines (http://jenger.riken.jp/en/result). The research ethics approval of this study has been obtained from the University of South Australia Human Research Ethics Committee.
PGS models
We use a linear model that regresses the observed phenotypes on a single or multiple sets of PGSs. It is assumed that the phenotypes are already adjusted for other non-genetic and environmental factors (e.g., demographic variables, ancestry principal components), and PGSs are already calculated based on GWAS summary statistics.
A PGS model can be written as
| (Equation 1) |
where y is the vector of standardized phenotypes of trait, X is a column-standardized N × M matrix including M sets of PGS, β is the vector of regression coefficients of X (i.e., PGS), and e is the vector of residuals. For example, with two sets of PGSs (M = 2), X and can be expressed as
| (Equation 2) |
| (Equation 3) |
where are correlations between y and the first PGS (x1), y and the second PGS (x2), and between the two PGSs (x1 and x2), respectively, in the sample. Using that are estimated in the multiple regression (Equation 2), the predicted phenotypes () can be obtained as
The coefficient of determination for this multiple regression model with in Equation 1 can be written as
| (Equation 4) |
With a single set of PGSs, i.e., M = 1 and or in Equation 1, the expression of R2 can be reduced as
or
It is noted that , , or is an estimate of parameter , and each estimate has a sampling variance.
Variance of R2
The distribution of R2 can be transformed to a non-central distribution with mean = M+ and variance = ) where is the non-centrality parameter. For example, the variance of the transformed value for is
Therefore,
| (Equation 5) |
where , M = 1, and is the squared correlation in the population and can be approximated as .23,24
In a similar manner, Equation 5 can be extended to multiple explanatory variables as
| (Equation 6) |
that is, Equation 6 is a generalized form of Equation 5.
Wishart25 introduced a formula to obtain the variance of R2 (also see Stuart and Ord26 and Olkin and Finn15) as
which provides an equivalent estimate as in Equation 6. Wishart25 derived his formula of the variance of R2 based on the hypergeometric series that has been used in the literature including Olkin and Finn.15 We introduce Equation 6 derived based on the transformation of a non-central distribution. Both Equation 6 and Wishart equation provide identical estimates of the variance of R2 (Figure S1). The s.e. of R2 estimate is the square root of
Variance of the difference between two R2 values
Following Olkin and Finn,15 we use the delta method to estimate the variance of the difference between R2 values based on two sets of PGS (x1 and x2). Assuming that the difference of R2 values can be formulated as a function of the correlations, i.e., , the delta method approximates the variance of the difference as
| (Equation 7) |
where
| (Equation 8) |
is the derivatives of f with respect to the correlations and
Each element of is shown in Olkin and Finn15 (also see Supplemental Note A).
From Equation 7, the following variances of differences can be estimated and used in our PGS analyses.
R2 difference when using different discovery samples to generate the PGS
The variance of R2 difference can be written as
| (Equation 9) |
which allows us to compare two PGS models that are not nested to each other (see R2 difference when using different information sources in results section), for which the conventional log likelihood ratio test cannot be applied.
In Equation 9, the values of from random samples in the population are normally distributed when the sample size is sufficient.15 Assuming that our PGS analysis is sufficiently powered (n > 25,000), the p value for the significance test of the difference can be derived from
and the 95% confidence interval is
| (Equation 10) |
When comparisons are made between R2 values based on two sets of PGSs (x1 and x2), the sampling covariance of R2 is required, which is explicitly used in Equations 7 and 9. If the sampling covariance ignored, the test statistics can be biased (Figures S2 and S3).
R2 difference when using nested models
When using nested models, the variance of R2 difference can be written as
| (Equation 11) |
where and are the estimated regression coefficients from a multiple regression (Equation 2), calculated from (see Equations 2, 3, and 4). Again, the derivative with respect to each of the correlations can be obtained for this function (Equation 8). Note that the comparison between and is equivalent to the log likelihood ratio test (i.e., vs. ).15
The values of in Equation 11 from random samples in the population follows a non-central chi-squared distribution with a non-centrality parameter . The p value for the significance test of the difference can be derived from
and the 95% confidence interval is
| (Equation 12) |
where is the value at the percentile of the inverse of non-central chi-squared cumulative distribution function with mean = and d.f. = 1.
When the sample size is large, the values of from random samples in the population are normally distributed,15 and the 95% confidence interval is
| (Equation 13) |
Note that Equations 12 and 13 are equivalent when the sample size is sufficient.15
R2 difference when using two independent sets of PGSs
In this case, there is no correlation structure between two independent sets of PGSs (, e.g., PGSs in male and female individuals), so the variance of R2 difference is simply the sum of the variances of each R2 value, which can be obtained from Equation 5. For example, assuming , the variance of R2 difference can be written as
| (Equation 14) |
where y1 and y2 are the vectors of standardized phenotypes and N1 and N2 are the sample sizes for the two independent sets of PGSs. The non-centrality parameters ( and ) for two independent PGSs can be written as
The p value for the significance test of the difference can be derived from
and the 95% confidence interval15 is
| (Equation 15) |
PGS-based genomic partitioning analysis
It is of interest to test whether a set of PGSs based on a genomic region of interest (or a pathway-based SNP subset) can better predict the phenotypes, compared to the rest of genomic regions. The proportion of the coefficient of determination explained by x1 can be estimated as from a multiple regression, , where x1 is the PGS of a genomic region of interest and x2 is the PGS of the rest of genomic regions. The expected proportion of the coefficient of determination explained by x1 can be calculated from prior information, referred to as pexp = # SNPs used for PGS1/total # SNPs. We are interested in testing whether the value of is significantly different from its expectation, pexp, which requires to estimate the sampling variance of , using Equation 7. The variance of the proportion can be written as
| (Equation 16) |
where is the estimated regression coefficient of x1, calculated from (Equation 3), and is the coefficient of determination. Therefore, it is possible to get the derivative with respect to each of the correlations, in Equation 8. This variance can be used to obtain the significance and 95 CI of the observed proportion of the coefficient of determination.
Analogous to Equation 9, the values of with random samples in the population are asymptotically normal.15 Using a Wald test, the p value for the significance test of the difference can be derived from
The 95% confidence interval of the ratio is
| (Equation 17) |
In addition, the package, r2redux, can provide , , and , i.e., the information matrix of the squared regression coefficients (see Supplemental Note B) that is useful when comparing the actual values of and .
It is noted that the delta method employed in this study is a well-established approach to derive the distribution of a function of an asymptotically normal variable.27 Following Olkin and Finn,15 we used the delta method to derive the variances of R2 and their difference as a function of regression coefficients (Equations 7, 8, 9, 11, and 16). We explicitly checked that the regression coefficients are asymptotically normal, using a realistic correlation structure among variables (Figures S4–S6).
Data
The UK Biobank is a large-scale biomedical database that comprises 0.5 million individuals who had been recruited between 2006 and 2010; their age ranged between 40 and 69 years.28,29 The data consist of health-related information for samples who are genotyped for genome-wide SNPs. A stringent quality control (QC) process was applied to UKBB data that excludes individuals with non-white British ancestries, mismatched sex between reported and inferred from genotypic information, genotype call rate < 0.95, or putative sex chromosome aneuploidy. The SNP QC criteria filtered out SNPs with an imputation reliability <0.6, missingness >0.05, minor allele frequency (MAF) < 0.01, or Hardy-Weinberg equilibrium p value <10−7. We also applied a relatedness cut-off QC (>0.05) so that there was no high pairwise relatedness among individuals. After QC, 288,792 individuals and 7,701,772 SNPs were retained.
Discovery GWAS data
Ninety percent of the individuals from the 288,792 QCed individuals were randomly selected as discovery samples (n = 259,912 to generate GWAS summary statistics (UKBB hereafter) for the 7,701,772 SNPs.. For the GWAS with the 259,912 UKBB discovery samples, we used BMI and cholesterol that were adjusted for age, sex, birth year, Townsend Deprivation Index (TDI), education, genotype measurement batch, assessment center, and the first 10 ancestry principal components using a linear regression.
We also have access to Japanese Biobank (BBJ) (http://jenger.riken.jp/en/result) GWAS summary statistics (BBJ hereafter) for BMI21 (n = 158,284) and cholesterol22 (n = 128,305) for 5,961,601 SNPs.
Target data
Ten percent of the individuals from the 288,792 QCed individuals were randomly selected as an independent target dataset (n = 28,880) that were non-overlapping and unrelated with the UKBB and BBJ discovery samples. In the PGS analyses, we used only 4,113,630 SNPs that were common between UKBB and BBJ GWAS data after excluding ambiguous SNPs and SNPs with any strand issue.
In the target dataset (n = 28,880), the phenotypes of each trait were adjusted for age, sex, birth year, TDI, education, genotype batch, assessment center, and the first 10 principal components using a linear regression. The pre-adjusted phenotypes were correlated with PGSs estimated in the following step. For each trait, we used the UKBB and BBJ GWAS summary statistics to estimate two sets of PGSs (UKBB PGSs vs. BBJ PGSs for the 28,880 target individuals ), using PLINK2 (https://www.cog-genomics.org/plink/2.0/) with the score function.30 Then, we estimated the correlation between the PGS and pre-adjusted phenotypes to obtain R2 values in the PGS analyses.
Functional annotation of the genome
We annotated the genome using pre-defined functional categories (regulatory vs. non-regulatory genomic regions).31 Regulatory region includes SNPs from coding regions, untranslated regions (UTRs), and promotors. Non-regulatory region includes all the other regions except the regulatory region. The number of SNPs belong to regulatory and non-regulatory is 158,653 and 3,954,947 (i.e., 4% of the total SNPs are located in the regulatory region).
Simulation of dependent and explanatory variables
For a quantitative trait, we simulated dependent variable (y) and PGSs (x1 and x2), varying the correlation structure of and the sample size (detailed simulation parameters are shown in Figures S7–S15). For a disease trait, the same simulation procedure was used, and the simulated quantitative phenotypes were transformed to binary responses using a liability threshold model with a population prevalence of k = 0.05. For example, case-control status was assigned to individuals according to their standardized quantitative phenotypes (i.e., liability), i.e., cases have liability greater than a threshold such that the proportion of cases is k = 0.05. The empirical variances of , , , and were obtained over 10,000 replicates, which were compared to the theoretical variances estimated using Equations 6, 9, 11, and 17, respectively.
Results
Simulation verification
The theory of the proposed method has been explicitly verified using simulations, varying sample size, and values of , , , and (Figures S7–S15). The empirical variances obtained from 10,000 simulated replicates are almost perfectly correlated with the theoretical variance for the values of , , , when varying the sample size (Figures S7–S10) and when varying R2 values (Figures S11–S14). When considering two independent PGSs, the theoretical and empirical variances are also agreed well (Figure S15).
R2 difference when using different information sources: UKBB vs. BBJ
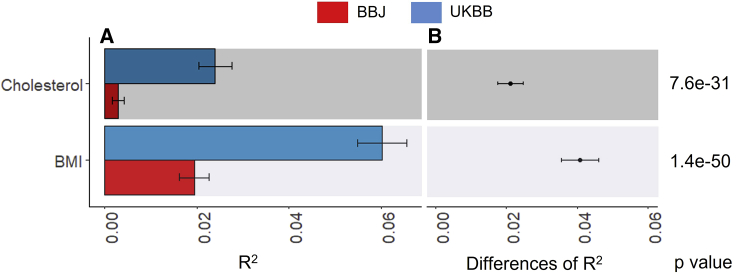
It is of interest to determine whether different information sources (e.g., ancestries) have significantly different predictive abilities in PGS analyses, which can be assessed using Equations 9 and 10. Figure 1 illustrates that when predicting the 28,880 European target samples, the coefficient of determinations (R2) with the UKBB and BBJ PGSs were 0.024 (95% CI = 0.021–0.028) and 0.003 (95% CI = 0.002–0.004), respectively, for cholesterol. However, these R2 values and CIs cannot be used to assess their difference because the two sets of PGSs are not independent. Furthermore, the two PGS models with UKBB and BBJ are not nested to each other, so the likelihood ratio test could not be used either. For this problem, we used Equations 9 and 10 to obtain the variance, 95% CI (0.0175–0.0247), and p value (7.6e−31) of the R2 difference, accounting for the dependency between UKBB and BBJ PGSs, for cholesterol (Figure 1). Similarly, the test statistics of the R2 difference was obtained for BMI, 0.035–0.046 for 95% CI and p value = 1.4e−50 (Figure 1).
Figure 1.
The predictive ability (R2) of PGSs when predicting 28,880 European individuals using UKBB or BBJ discovery GWAS dataset
(A) The main bars represent R2 values and error bars correspond 95% confidence intervals. Two sets of GWAS summary statistics were obtained from UKBB and BBJ discovery GWAS datasets to estimate two sets of PGSs.
(B) Dot points represent the differences of R2 values between UKBB and BBJ PGS models, and error bars indicate 95% confidence intervals of the difference.
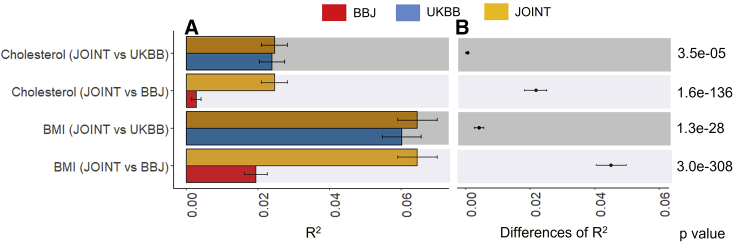
It is also interesting to whether BBJ PGSs provides a significant improvement in the predictive ability, in addition to UKBB PGSs, when predicting the 28,880 European target samples. Figure 2 compares R2 value with each UKBB or BBJ PGSs to R2 value from a joint model fitting UKBB and BBJ PGSs simultaneously. Using Equations 11 and 12, we acquired the variance, 95% CI (0.0001–0.001), and p value (3.5e−05) of R2 difference when comparing the joint model with a single model with UKBB, indicating that BBJ PGSs contributed to a significant improvement for cholesterol. Similarly, BBJ PGSs improved the predictive ability significantly (p value = 1.3e−28) for BMI. As expected, excluding UKBB PGSs from the joint model substantially decreased the prediction accuracy (p value = 1.6e−136 for cholesterol and 3.0e−308 for BMI).
Figure 2.
The predictive ability (R2) of a PGS model based on UKBB or BBJ discovery dataset, compared to the joint model of both UKBB and BBJ when predicting 28,880 European individuals
(A) The main bars represent R2 values and error bars correspond 95% confidence intervals. Two sets of GWAS summary statistics were obtained from UKBB and BBJ discovery GWAS datasets to estimate two sets of PGSs, i.e., UKBB and BBJ PGSs. In addition, a joint model fitting both UKBB and BBJ PGSs was compared.
(B) Dot points represent the differences of R2 values between the joint model and UKBB or BBJ PGS model, and error bars indicate 95% confidence intervals of the difference.
R2 difference when using two independent sets of PGSs: male vs. female
We were also interested in testing whether the PGSs could predict the adjusted phenotypes of the target individuals equally well for males and females. In this case, there is no correlation structure between male and female PGSs, so the variance of R2 difference is simply the sum of the variances of each R2 value, which can be obtained from Equation 5 or 6. Figure S16 shows that there was no significant difference between male and female PGSs in their predictive ability for cholesterol and BMI whether using UKBB or BBJ discovery GWAS dataset.
PGSs with genome-wide association p value thresholds (pT)
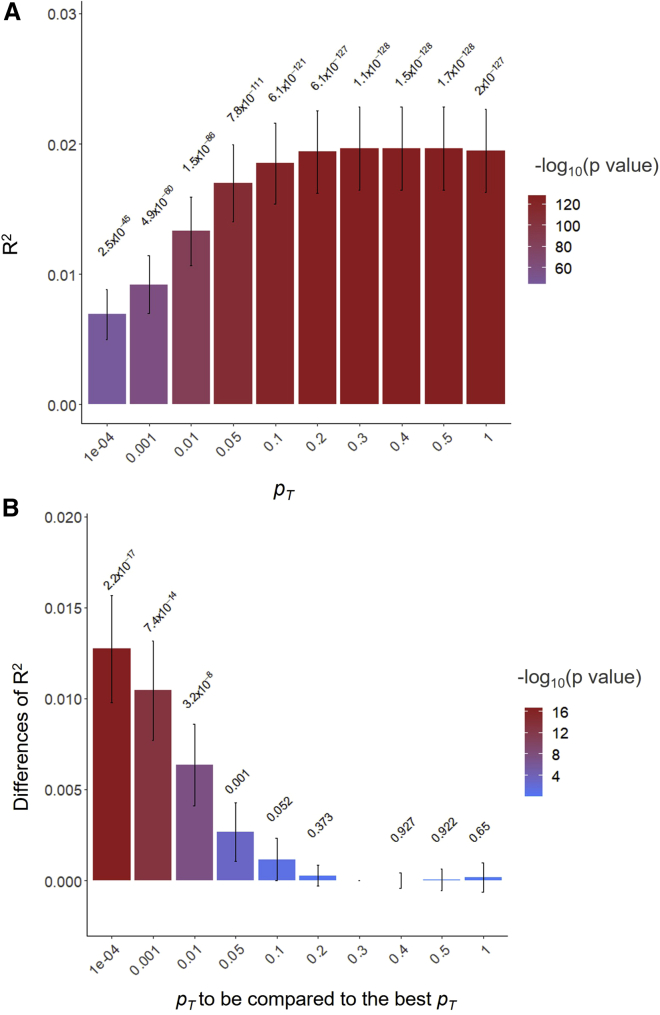
PGSs also have been widely used to determine which pT provides the highest prediction accuracy, for example, using PGS software such as PLINK.30,32 However, there is a lack of test statistics that can assess whether the predictive ability of the best-performing pT is significantly different from the other pT. Figure 3A illustrates that R2 value is the highest at pT = 0.3 when predicting 28,880 European individuals in the target dataset, using BBJ discovery GWAS dataset (BMI). However, it is not clear if the predictive ability at pT = 0.3 is significantly higher than the adjacent pT (e.g., pT = 0.2 or 0.4), and it may be important to report pT of which the predictive ability is not statistically different from the best-performing pT. Using Equations 9 and 10, we assessed the significance of difference between the best-performing pT and each of the other pT (Figure 3B). From this analysis, we found that the best-performing pT was not significantly different from pT ranging between 0.1 and 1, but significantly different from pT < 0.05 (Figure 3B). When using the UKBB discovery GWAS dataset to predict the 28,880 European individuals, the highest R2 value at the pT of 1 was significantly different from all the other pT (Figure S17B).
Figure 3.
The predictive ability (R2) of PGSs estimated based on SNPs below pT when predicting BMI in 28,880 European samples using BBJ discovery samples (GWAS summary statistics)
(A) The main bars represent R2 values and error bars correspond 95% confidence intervals. The values above 95% CIs are p values indicating that R2 values are not different from zero.
(B) The main bars represent the difference of R2 values between the corresponding pT and the best-performing pT and error bars indicate 95% confidence intervals. The values above 95% CIs are p values indicating the significance of the difference between the pairs of R2 values.
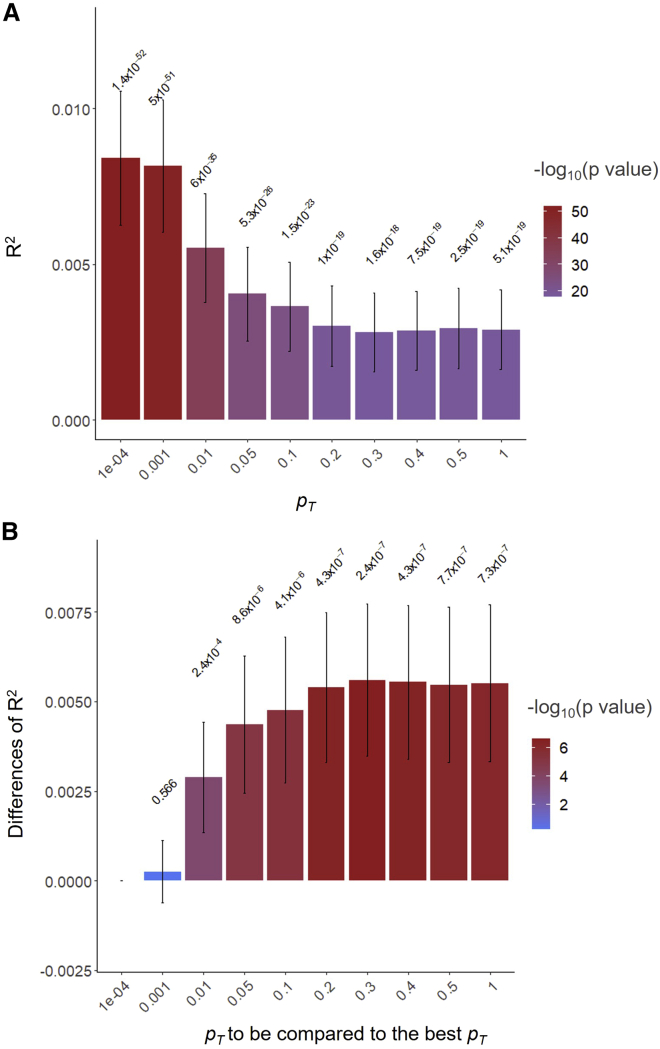
Interestingly, the highest R2 value was found at pT = 1e−04 (Figure 4A) when predicting the European target samples using BBJ discovery GWAS dataset for cholesterol, which was not statistically different from pT = 0.001 but was significantly higher than the other pT (Figure 4B). For the same target samples and trait, the best R2 value was obtained from pT = 0.01 when using the UKBB discovery GWAS dataset (Figure S18A). Except for pT = 0.01, 0.05, and 0.1, R2 values at the other pT were significantly different from the best R2 values (Figure S18B).
Figure 4.
The predictive ability (R2) of PGSs estimated based on SNPs below the pT when predicting cholesterol in 28,880 European samples using BBJ discovery samples (GWAS summary statistics)
(A) The main bars represent R2 values and error bars correspond 95% confidence intervals. The values above 95% CIs are p values indicating that R2 values are not different from zero.
(B) The main bars represent the difference of R2 values between the corresponding pT and the best-performing pT and error bars indicate 95% confidence intervals. The values above 95% CIs are p values indicating the significance of the difference between the pairs of R2 values.
PGS-based genomic partitioning analyses
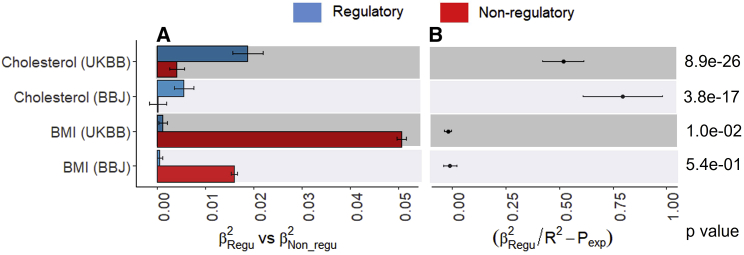
Genomic partitioning analyses have been widely applied.31,33,34,35 Such analysis could be useful in the PGS context. Using Equation 16, we can estimate the variance of the where is the estimated regression coefficient from a multiple regression (Equation 2), and assess whether the observed proportion ( ) is significant different from pexp (i.e., the coverage of the SNPs belonged to the category). For example, we partitioned the genome-wide SNPs into the regulatory (158,653) and non-regulatory (3,954,947) regions, following Gusev et al.,31 resulting pexp = 4% of SNP coverage for the regulatory region as the expectation. We simultaneously fit two sets of PGSs from regulatory and non-regulatory regions to get and , using a multiple regression, then assess whether the value of is significantly different from zero (Equation 17). Figure 5 shows that the predictive ability of regulatory SNPs was significantly higher than the expectation (p value = 8.9e−26 for UKBB and 3.8e−17 for BBJ) for cholesterol. In contrast, the predictive ability of regulatory SNPs was not better than the expectation for BMI (Figure 5).
Figure 5.
PGS-based genomic partitioning method to assess whether the predictive ability is enriched in the regulatory region for cholesterol and BMI
Here pexp = 0.04 is the expectation for the regulatory SNPs based on the proportion of SNPs allocated to this annotation.
(A) The main bars represent squared regression coefficients attributable to SNPs in the regulatory region () and non-regulatory region (), and error bars correspond to 95% confidence intervals when predicting 28,880 European samples using UKBB or BBJ GWAS summary statistics.
(B) Dot points represent the difference between the observed and expected proportions () and error bars indicate 95% confidence intervals of the difference.
Application to binary responses and ascertained case-control data
The proposed method is also explicitly verified using simulation for binary or case-control data, varying sample size and values of , , , and (Figures S19–S26). The empirical variances obtained from 10,000 simulated replicates are almost identical with the theoretical variances for the values of , , , and when varying the sample size (Figures S19–S22) and when varying R2 values (Figures S23–S26). In the case of ascertained case-control, a similar pattern is shown, i.e., the empirically observed variances obtained from 10,000 simulated replicates are agreed well with the theoretical variances for the values (Figures S27–S30). This finding shows that the proposed method can be applied to test the significance of difference between predictive abilities of PGSs for binary traits and ascertained case-control traits when R2 is not very high (<0.1). Note that the empirical and theoretical variances diverge when R2 values on the observed scale are more than 0.1 for binary responses and ascertained case control (Figures S31 and S32). Although R2 value > 0.1 is not frequently observed in the current PGS studies (Table S2), a careful interpretation is required for the variance of such high R2, and we would not recommend using the theoretical approximation.
Discussion
R2 has been widely used to measure the predictive ability of PGSs.13 However, the confidence interval of R2 has rarely been reported, and the test statistic for the difference of two R2 values has not been well documented. Here, we show how to get the variance of each estimated R2 value and covariance between two R2 estimates (from two sets of PGSs) that can be used to assess whether they are significantly different from each other.
Martin et al.18 reported that the PGS prediction accuracy is higher when discovery and target samples are from the same ancestry background, compared to when the samples are from different ancestries. However, they did not formally assess the statistical significance of the increase (no p value provided). More importantly, they did not consider the correlation structure between predictors when they compared two PGSs. We applied the proposed approach and found that the predictive ability of PGSs based on UKBB discovery GWASs is significantly higher than that of PGSs based on BBJ discovery GWASs, by formally deriving the 95% CI and p value of the R2 difference.
Many studies evaluating PGSs use the pT method12 and report the pT that maximizes performance. This provides useful information when inferring the genetic architecture of the trait of interest and when fine-tuning pT as a hyper-parameter in PGS methods.32,36,37,38 For such cases, it may be crucial to determine if the best-performing pT is genuinely better than other (adjacent) pT or if it occurs just by random chance (i.e., sampling error). For example, in Figure 3, the best-performing pT is 0.3 (the set of SNPs with pT ≤ 0.3), which is, however, not statistically different from pT ≤ 0.2 or ≤ 0.1. Note that the set of SNPs with pT ≤ 0.1 is nested within SNPs with pT ≤ 0.3, meaning that the additional SNPs in the latter would not significantly improve the prediction accuracy. Therefore, pT ≤ 0.1 should be used instead of the pT ≤ 0.3 as the former is a more parsimonious model than the latter. Our proposed approach can formally assess statistical difference among pT, providing 95% CI of the difference with a significance p value.
We also derived an information matrix of squared regression coefficients in a multiple regression model, establishing a PGS-based genomic partitioning method that could test whether the ratio of two squared regression coefficients is significantly deviated from its expectation given the proportion of SNPs allocated to each partition. This is analogous to the existing genomic partitioning approaches using GREML or LDSC31,33,34,35 that may have an overfitting issue because SNP effects and genomic partitioning are estimated in the same samples.
In conclusion, we show how to estimate the variance and covariance of R2 estimates to quantify the 95% CI and p value of the difference and ratio when comparing two PGSs, which is available in R package r2redux (see Supplemental Note B). We suggest that the proposed approach should be used to test the statistical significance of difference and ratio between pairs of PGSs, which may help to draw a correct conclusion about the predictive ability of PGSs.
Acknowledgments
This research is supported by the Australian Research Council (DP190100766). The R package development is supported by Cooperative Research Program for Agriculture Science and Technology Development (PJ01609901) from the Rural Development Administration, Republic of Korea. We thank the staffs and samples of the UK Biobank and Biobank Japan for their important contributions. Our reference number approved by UK Biobank is 14575. The UK Biobank is funded by the UK Department of Health, the Medical Research Council, the Scottish Executive, and the Wellcome Trust medical research charity. The analyses were performed using computational resources provided by the Australian Government through Gadi under the National Computational Merit Allocation Scheme (NCMAS), and HPCs (Tango and Statgen server) managed by UniSA IT. N.R.W. acknowledges funding from the NHMRC (1113400, 1173790), Australia.
Author contributions
S.H.L. and N.R.W. conceived the idea. S.H.L. derived theory and supervised the study. M.M.M. performed the analysis. M.M.M. and S.H.L. verified the theory and analytical methods, and made the R package, with support from S.L. S.H.L. and M.M.M. wrote the first draft of the manuscript. N.R.W. and S.L. provided critical feedback and suggestions. All the authors discussed the results and contributed to the final manuscript.
Declaration of interests
The authors declare that they have no competing interests.
Published: January 25, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2023.01.004.
Contributor Information
Md. Moksedul Momin, Email: cvasu.momin@gmail.com.
S. Hong Lee, Email: hong.lee@unisa.edu.au.
Supplemental information
Data and code availability
The genotype and phenotype data of the UK Biobank can be accessed through procedures described on its webpage (https://www.ukbiobank.ac.uk/) and summary statistics of BMI and cholesterol from Japanese Biobank (BBJ) can be obtained from its website (http://jenger.riken.jp/en/). r2redux can be downloaded from (https://github.com/mommy003/r2redux) or from CRAN [install.packages("r2redux") in R] (also see Supplemental Note B).
References
- 1.Plomin R., Haworth C.M.A., Davis O.S.P. Common disorders are quantitative traits. Nat. Rev. Genet. 2009;10:872–878. doi: 10.1038/nrg2670. [DOI] [PubMed] [Google Scholar]
- 2.Schork N.J. Genetics of complex disease: approaches, problems, and solutions. Am. J. Respir. Crit. Care Med. 1997;156:S103–S109. doi: 10.1164/ajrccm.156.4.12-tac-5. [DOI] [PubMed] [Google Scholar]
- 3.Gibson G. Decanalization and the origin of complex disease. Nat. Rev. Genet. 2009;10:134–140. doi: 10.1038/nrg2502. [DOI] [PubMed] [Google Scholar]
- 4.Khera A.V., Chaffin M., Aragam K.G., Haas M.E., Roselli C., Choi S.H., Natarajan P., Lander E.S., Lubitz S.A., Ellinor P.T., Kathiresan S. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 2018;50:1219–1224. doi: 10.1038/s41588-018-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding Y., Hou K., Burch K.S., Lapinska S., Privé F., Vilhjálmsson B., Sankararaman S., Pasaniuc B. Large uncertainty in individual polygenic risk score estimation impacts PRS-based risk stratification. Nat. Genet. 2022;54:30–39. doi: 10.1038/s41588-021-00961-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bilkey G.A., Burns B.L., Coles E.P., Bowman F.L., Beilby J.P., Pachter N.S., Baynam G., JS Dawkins H., Nowak K.J., Weeramanthri T.S. Genomic testing for human health and disease across the life cycle: applications and ethical, legal, and social challenges. Front. Public Health. 2019;7:40. doi: 10.3389/fpubh.2019.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allyse M.A., Robinson D.H., Ferber M.J., Sharp R.R. Vol. 1. Elsevier; 2018. pp. 113–120. (Direct-to-consumer Testing 2.0: Emerging Models of Direct-To-Consumer Genetic Testing). [DOI] [PubMed] [Google Scholar]
- 8.Frerichs F.C.P., Dingemans K.P., Brinkman K. Cardiomyopathy with mitochondrial damage associated with nucleoside reverse-transcriptase inhibitors. N. Engl. J. Med. 2002;347:1895–1896. doi: 10.1056/NEJM200212053472320. [DOI] [PubMed] [Google Scholar]
- 9.Wray N.R., Goddard M.E., Visscher P.M. Prediction of individual genetic risk to disease from genome-wide association studies. Genome Res. 2007;17:1520–1528. doi: 10.1101/gr.6665407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wray N.R., Yang J., Goddard M.E., Visscher P.M. The genetic interpretation of area under the ROC curve in genomic profiling. PLoS Genet. 2010;6:e1000864. doi: 10.1371/journal.pgen.1000864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wand H., Lambert S.A., Tamburro C., Iacocca M.A., O’Sullivan J.W., Sillari C., Kullo I.J., Rowley R., Dron J.S., Brockman D., et al. Improving reporting standards for polygenic scores in risk prediction studies. Nature. 2021;591:211–219. doi: 10.1038/s41586-021-03243-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi S.W., Mak T.S.H., O’Reilly P.F. A guide to performing Polygenic Risk Score analyses. Nat. Protoc. 2020;15:2759–2772. doi: 10.1038/s41596-020-0353-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis C.M., Vassos E. Polygenic risk scores: from research tools to clinical instruments. Genome Med. 2020;12 doi: 10.1186/s13073-020-00742-5. 44–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Purcell S.M., Wray N.R., Stone J.L., Visscher P.M., O'Donovan M.C., Sullivan P.F., Sklar P., Consortium I.S. Common polygenic variation contributes to risk of schizophrenia that overlaps with bipolar disorder. 2009;460:748. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olkin I., Finn J.D. Correlations redux. Psychol. Bull. 1995;118:155–164. [Google Scholar]
- 16.Lee S.H., Goddard M.E., Wray N.R., Visscher P.M. A better coefficient of determination for genetic profile analysis. Genet. Epidemiol. 2012;36:214–224. doi: 10.1002/gepi.21614. [DOI] [PubMed] [Google Scholar]
- 17.So H.-C., Sham P.C. Exploring the predictive power of polygenic scores derived from genome-wide association studies: a study of 10 complex traits. Bioinformatics. 2017;33:886–892. doi: 10.1093/bioinformatics/btw745. [DOI] [PubMed] [Google Scholar]
- 18.Martin A.R., Kanai M., Kamatani Y., Okada Y., Neale B.M., Daly M.J. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 2019;51:584–591. doi: 10.1038/s41588-019-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi S., Garcia-Gonzalez J., Ruan Y., Wu H., Johnson J., Hoggart C., O’Reilly P. The power of pathway-based polygenic risk scores. Research Square. 2021 doi: 10.21203/rs.3.rs-643696/v1. [DOI] [Google Scholar]
- 20.Li J., Chaudhary D.P., Khan A., Griessenauer C., Carey D.J., Zand R., Abedi V. Polygenic risk scores augment stroke subtyping. Neurol. Genet. 2021;7:e560. doi: 10.1212/NXG.0000000000000560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akiyama M., Okada Y., Kanai M., Takahashi A., Momozawa Y., Ikeda M., Iwata N., Ikegawa S., Hirata M., Matsuda K., et al. Genome-wide association study identifies 112 new loci for body mass index in the Japanese population. Nat. Genet. 2017;49:1458–1467. doi: 10.1038/ng.3951. [DOI] [PubMed] [Google Scholar]
- 22.Kanai M., Akiyama M., Takahashi A., Matoba N., Momozawa Y., Ikeda M., Iwata N., Ikegawa S., Hirata M., Matsuda K., et al. Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat. Genet. 2018;50:390–400. doi: 10.1038/s41588-018-0047-6. [DOI] [PubMed] [Google Scholar]
- 23.Olkin I., Siotani M. Asymptotic distribution of functions of a correlation matrix. Essays in probability and statistics. 1976:235–251. [Google Scholar]
- 24.Olkin I., Finn J.D. Testing correlated correlations. Psychol. Bull. 1990;108:330–333. [Google Scholar]
- 25.Wishart J. The mean and second moment coefficient of the multiple correlation coefficient, in samples from a normal population. Biometrika. 1931;22:353–361. [Google Scholar]
- 26.Stuart A., Ord J.K. 5th ed. Vol 2. 1991. Kendall's Advanced Theory of Statistics. [Google Scholar]
- 27.Ver Hoef J.M. Who invented the delta method? Am. Statistician. 2012;66:124–127. [Google Scholar]
- 28.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O’Connell J., et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fry A., Littlejohns T.J., Sudlow C., Doherty N., Adamska L., Sprosen T., Collins R., Allen N.E. Comparison of sociodemographic and health-related characteristics of UK Biobank participants with those of the general population. Am. J. Epidemiol. 2017;186:1026–1034. doi: 10.1093/aje/kwx246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gusev A., Lee S.H., Trynka G., Finucane H., Vilhjálmsson B.J., Xu H., Zang C., Ripke S., Bulik-Sullivan B., Stahl E., et al. Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am. J. Hum. Genet. 2014;95:535–552. doi: 10.1016/j.ajhg.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Euesden J., Lewis C.M., O’Reilly P.F. PRSice: polygenic risk score software. Bioinformatics. 2015;31:1466–1468. doi: 10.1093/bioinformatics/btu848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang J., Manolio T.A., Pasquale L.R., Boerwinkle E., Caporaso N., Cunningham J.M., De Andrade M., Feenstra B., Feingold E., Hayes M.G., et al. Genome partitioning of genetic variation for complex traits using common SNPs. Nat. Genet. 2011;43:519–525. doi: 10.1038/ng.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee S.H., DeCandia T.R., Ripke S., Yang J., Schizophrenia Psychiatric Genome-Wide Association Study Consortium PGC-SCZ. International Schizophrenia Consortium ISC. Molecular Genetics of Schizophrenia Collaboration MGS. Sullivan P.F., Goddard M.E., Keller M.C., et al. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat. Genet. 2012;44:247–250. doi: 10.1038/ng.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finucane H.K., Bulik-Sullivan B., Gusev A., Trynka G., Reshef Y., Loh P.-R., Anttila V., Xu H., Zang C., Farh K., et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi S.W., O'Reilly P.F. PRSice-2: Polygenic Risk Score software for biobank-scale data. GigaScience. 2019;8:giz082. doi: 10.1093/gigascience/giz082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao Z., Yi Y., Song J., Wu Y., Zhong X., Lin Y., Hohman T.J., Fletcher J., Lu Q. PUMAS: fine-tuning polygenic risk scores with GWAS summary statistics. Genome Biol. 2021;22:257–319. doi: 10.1186/s13059-021-02479-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vilhjálmsson B.J., Yang J., Finucane H.K., Gusev A., Lindström S., Ripke S., Genovese G., Loh P.-R., Bhatia G., Do R., et al. Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am. J. Hum. Genet. 2015;97:576–592. doi: 10.1016/j.ajhg.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genotype and phenotype data of the UK Biobank can be accessed through procedures described on its webpage (https://www.ukbiobank.ac.uk/) and summary statistics of BMI and cholesterol from Japanese Biobank (BBJ) can be obtained from its website (http://jenger.riken.jp/en/). r2redux can be downloaded from (https://github.com/mommy003/r2redux) or from CRAN [install.packages("r2redux") in R] (also see Supplemental Note B).