Abstract
Background:
Exposure to per- and polyfluoroalkyl substances (PFAS) is ubiquitous and has been associated with an increased risk of several cardiometabolic diseases. However, the metabolic pathways linking PFAS exposure and human disease are unclear.
Objective:
We examined associations of PFAS mixtures with alterations in metabolic pathways in independent cohorts of adolescents and young adults.
Methods:
Three hundred twelve overweight/obese adolescents from the Study of Latino Adolescents at Risk (SOLAR) and 137 young adults from the Southern California Children’s Health Study (CHS) were included in the analysis. Plasma PFAS and the metabolome were determined using liquid-chromatography/high-resolution mass spectrometry. A metabolome-wide association study was performed on log-transformed metabolites using Bayesian regression with a g-prior specification and g-computation for modeling exposure mixtures to estimate the impact of exposure to a mixture of six ubiquitous PFAS (PFOS, PFHxS, PFHpS, PFOA, PFNA, and PFDA). Pathway enrichment analysis was performed using Mummichog and Gene Set Enrichment Analysis. Significance across cohorts was determined using weighted -tests.
Results:
In the SOLAR and CHS cohorts, PFAS exposure was associated with alterations in tyrosine metabolism (meta-analysis ) and de novo fatty acid biosynthesis (), among others. For example, when increasing all PFAS in the mixture from low (th percentile) to high (th percentile), thyroxine (T4), a thyroid hormone related to tyrosine metabolism, increased by 0.72 standard deviations (SDs; equivalent to a standardized mean difference) in the SOLAR cohort (95% Bayesian credible interval (BCI): 0.00, 1.20) and 1.60 SD in the CHS cohort (95% BCI: 0.39, 2.80). Similarly, when going from low to high PFAS exposure, arachidonic acid increased by 0.81 SD in the SOLAR cohort (95% BCI: 0.37, 1.30) and 0.67 SD in the CHS cohort (95% BCI: 0.00, 1.50). In general, no individual PFAS appeared to drive the observed associations.
Discussion:
Exposure to PFAS is associated with alterations in amino acid metabolism and lipid metabolism in adolescents and young adults. https://doi.org/10.1289/EHP11372
Introduction
Per- and polyfluoroalkyl substances (PFAS) make up a prevalent class of persistent organic pollutants with endocrine and metabolism disrupting properties.1,2 PFAS are used in a broad range of industrial and consumer products, such as firefighting foams, nonstick pans, waterproof clothing, food packaging, and even cosmetic products, including lipstick.3,4 Because of their widespread use and resistance to chemical degradation, PFAS can be found in drinking water and food sources across the world. In the United States, an estimated people have drinking water with perfluorooctane sulfonic acid (PFOS) or perfluorooctanoic acid (PFOA) levels ,5 which is considerably higher than the U.S. Environmental Protection Agency 2022 safe drinking water health advisory levels of for PFOA and for PFOS.6,7 Because of their chemical properties, some PFAS also bioaccumulate in human tissues, with estimated half-lives of up to 25 y.8 Consequently, several legacy PFAS, including PFOS, PFOA, perfluorohexanesulfonic acid (PFHxS), perfluoroheptanesulfonic acid (PFHpS), perfluorononanoic acid (PFNA), and perfluorodecanoic acid (PFDA) are detectible in the blood of nearly all humans.9,10 Therefore, it is of utmost importance to determine how exposure to PFAS impacts human health to inform public health policy and reduce exposure levels for harmful PFAS.
Accumulating evidence suggests that PFAS exposure is associated with an increased risk of metabolic disorders.11 PFAS exposure during sensitive periods of development, such as childhood or adolescence, is of particular concern because this is an important developmental stage for cellular differentiation and development of metabolic tissues.12–15 Longitudinal studies have found that PFAS exposure during childhood is associated with the development of dysregulated glucose metabolism and insulin resistance,16,17 dyslipidemia,18 and adiposity.19,20 Similar associations have been observed in adults, suggesting that these associations persist into adulthood.21–23 However, the mechanisms linking PFAS exposure and metabolic disorders in humans remain unclear, especially in children.
Studies examining the associations of PFAS with alterations in targeted biomarkers have been used to examine the potential mechanisms linking PFAS with metabolic disorders. These targeted biomarker studies have focused primarily on metabolites known to be associated with a specific metabolic disease, with a particular focus on metabolites related to lipid metabolism. For example, cross-sectional and longitudinal human studies have consistently shown that PFAS exposure increases serum total and low-density lipoprotein cholesterol in both children and adults.11 Dyslipidemia is a hallmark of metabolic syndrome and can increase the risk for metabolic and cardiovascular disease,24 suggesting that PFAS-associated alterations in lipids may mediate the relationship between PFAS exposure and risk of metabolic disorders. However, using a targeted approach to study the impact of PFAS exposure on alterations in metabolites does not provide information about how PFAS alter metabolic networks, which is important for understanding the mechanisms linking PFAS exposure with metabolic disorders.
Untargeted metabolomics approaches can quantify thousands of metabolites, which can provide insight on the metabolic perturbations of PFAS exposure. Although previous studies have used metabolomics to examine the associations of PFAS exposure and alterations in metabolic pathways, important gaps in the literature remain.25 In vivo and in vitro models have shown that exposure to individual PFAS alters fatty acid metabolism, including in human cell lines,26 rodents,27–29 and zebrafish.30 In humans, studies using both targeted and untargeted metabolomics methods have reported similar associations between exposure to individual PFAS, including PFOS, PFHxS, and PFOA, and fatty acid metabolism.31–40 Other studies have reported associations between PFAS exposure and metabolic pathways such as amino acid31,34,37,40,41 or bile acid metabolism,39,42,43 both of which are important pathways that can contribute to the pathogenesis of diseases, such as type 2 diabetes and cancer.44–47 Together, these human studies present a myriad of sometimes conflicting results. Although differences in study populations and the use of targeted vs. untargeted metabolomic methods may account for some of the differences across studies, the majority of existing studies have examined the metabolic perturbations of PFAS congeners individually, using single exposure models. In reality, individuals are exposed to a complex mixture of potentially correlated PFAS compounds that may have synergistic effects on human metabolism.48 Previous studies examining the impact of PFAS mixtures on the metabolome have relied primarily on reducing the dimensionality of the correlated PFAS exposures prior to analysis using either principal components or by calculating the sum of PFAS congeners.25 However, it is difficult to draw conclusions about the effects of different components of the PFAS mixture from these studies owing to difficulties in interpreting principal components or summed exposure variables. Despite the analytical difficulties, examining the effects of PFAS mixtures on metabolic pathways is key to fully understand the consequences of exposure to these chemicals. Further, understanding how PFAS mixtures impact human metabolism can help to put previous studies on individual PFAS congeners in greater context.
In the present study, we aimed to examine the impact of PFAS mixtures on metabolic pathways in independent cohorts of children and young adults. By using two cohorts, we aimed to identify associations between PFAS mixtures and metabolic pathways that were consistent across cohorts despite varied background characteristics. We used high-resolution mass spectrometry (HRMS)–based untargeted metabolomics coupled with a Bayesian hierarchical regression with a g-prior specification and g-computation for modeling exposure mixtures to estimate the joint effects of PFAS mixtures on metabolic pathways.
Methods
Study Populations
Study of Latino Adolescents at Risk.
This study used data from the Study of Latino Adolescents at Risk (SOLAR). As described previously,16,49–51 the SOLAR cohort included 328 overweight/obese children recruited in two waves between 2001 and 2012. Participants completed yearly clinical visits at the General Clinical Research Center of the Clinical Trials Unit at the University of Southern California. At baseline, participants were between 8 and 13 years of age and were overweight or obese based on sex and age-specific body mass index (BMI) percentile . Participants were included in the study if they were Hispanic or Latino based on all parents and grandparents self-reporting as Hispanic or Latino and if they had a direct family history of type 2 diabetes. Participants were excluded from the study if they had type 1 or type 2 diabetes or if they were on medications known to influence glucose or insulin metabolism. For the present analysis, participants were included if they completed a 2-h oral glucose tolerance test (OGTT) during their first or second visit, resulting in a total sample size of 312 participants. The institutional review board at the University of Southern California provided ethics approval for this study, and participants and their guardians provided written informed assent/consent prior to participation.
Southern California Children’s Health Study.
To examine the generalizability of findings from the SOLAR cohort to young adults, we analyzed data from the Metabolic and Asthma Incidence Research (Meta-AIR) study.52 Meta-AIR was a study including 172 young adults between the ages of 17 and 23 years of age who were part of the Southern California Children’s Health Study (CHS).53 Meta-AIR participants were recruited for a single clinical visit between 2014 and 2018. Clinical visits occurred at the Clinical Trials Unit or the Diabetes and Obesity Research institute at the University of Southern California, during which participants performed a 2-h OGTT and completed detailed questionnaires. CHS participants were selected for inclusion in the Meta-AIR study if they had overweight or obesity between 14 and 15 years of age. Overweight and obesity were based on the U.S. Centers for Disease Control and the U.S. Preventive Services Task Force guidelines, defined as having a sex- and age-specific BMI percentile .54 Participants were excluded if they had a diagnosis of diabetes mellitus or if they took any medications that could influence glucose metabolism or insulin secretion. Meta-AIR participants were included in the present study if they provided consent for future use of biospecimens, resulting in a total sample size of 137. The University of Southern California institutional review board provided ethics approval for this study, and participants (and their guardians if participants were of age) provided written informed assent/consent prior to participation.
Covariates
Detailed information on the measurement of covariates in the SOLAR and CHS cohorts has been provided previously.16,49–53 Height (in meters) and weight (in kilograms) were measured at each visit in both cohorts and were used to calculate BMI as kilograms per meter squared. In both cohorts, participants completed questionnaires related to sociodemographics and individual and familial health history. In the SOLAR cohort, socioeconomic status (SES) was characterized using a modified version of the Hollingshead Four-Factor Index,55 as described previously.49 The Hollingshead Four-Factor Index was modified to provide a single score for children and takes into account the education, occupation, and marital status of parents or primary caregivers.49 In the CHS cohort, parental education was used to assess SES.52 For the primary analysis, SES scores were grouped into quantiles and analyzed numerically, with values ranging from 1 to 4, with 1 representing the lowest and 4 representing the highest quantile of SES. In the SOLAR cohort, SES was unavailable for 11% () of individuals. In these participants, we imputed the population median SES score; median imputation has been shown to perform similarly to more complex imputation methods when the proportion of missing values is relatively low.56 Further, as described previously, participants with missing SES data did not differ in metabolic or physical attributes, including sex (tested using a test; ), age (tested using an independent -test; ), or BMI (tested using an independent -test; ).16,51
In the SOLAR cohort, Tanner stages were determined by a physician during a physical exam to assess sexual maturity.57,58 Tanner stages range from 1 to 5 and were categorized as follows: stage 1: prepuberty, which is the developmental stage before secondary sex characteristics begin to develop; stages 2–4: puberty, which is the developmental stage when secondary sex characteristics begin developing and menarche occurs for most females; stage 5: postpuberty, the developmental stage where secondary sex characteristics, including pubic hair and secondary sex organs, reach maturity.
Covariates included in models were selected using directed acyclic graphs (DAGs; Figure S1) and included age (in years), sex (male/female, coded numerically as 0/1), BMI (in kilograms per meter squared), and SES (in quantiles, coded numerically as 1–4). In the SOLAR cohort, Tanner stage (Tanner stages 1–5, coded numerically as 1–5) and study wave (Wave 1/Wave 2, coded numerically as 0/1) were also included as covariates. No covariates for statistical models contained missing data, except SES in the SOLAR cohort (as described above).
Plasma PFAS
Six common PFAS (PFOS, PFHxS, PFHpS, PFOA, PFNA, and PFDA) were quantified in plasma samples collected at the 2-h OGTT time point. PFAS levels were quantified in batches of 70 study samples via liquid chromatography (LC) coupled to HRMS (LC-HRMS). The specific LC-HRMS protocol used to measure plasmas PFAS for this study has been described previously.59 Briefly, plasma samples were prepared by combining of plasma with of an internal standard. The internal standard contained thirty -labeled PFAS to obtain a final concentration. Proteins were then precipitated by adding of cold acetonitrile, then vortexing and centrifuging at for 15 min. LC-MS grade water was added to the resulting supernatant to achieve a dilution ratio of 2:1. PFAS analysis was completed by reverse phase (RP) chromatography with negative electrospray ionization, performed with a Thermo Scientific Vanquish Flex ultra-high-performance LC system with binary pump attached to a Thermo Scientific Q Exactive HF-X Orbitrap mass spectrometry system (Thermo Fisher Scientific).
After analysis of samples from both the SOLAR and CHS cohorts, peaks for PFAS and their corresponding -labeled PFAS were identified by matching mass m/z with a tolerance of , then extracted and integrated using TraceFinder 5.1 (Thermo Fisher Scientific). Quantification of PFAS was performed by dividing the peak for a given PFAS with its corresponding internal standard and comparing this value to a calibration curve with 6 points created using charcoal-stripped plasma. Replicate measures of National Institute of Standards and Technology (NIST) 1957 and NIST 1958 standard reference material were performed for every batch of 70 samples. Each batch also included instrumental and method blanks. The coefficient of variation for major PFAS was , whereas the analyte recovery was . The accuracy of the method was assessed by comparing with NIST standard reference materials. Method accuracy was also assessed by participating in the Center de toxicologie du Québec’s Arctic Monitoring and Assessment Program Ring Test for Persistent Organic Pollutants in Human Serum. The limits of detection (LODs) were as follows: PFOS: ; PFHxS: ; PFHpS: ; PFOA: ; PFNA: ; and PFDA: . PFDA was the only PFAS included in the present study that had values below the LOD; these values were imputed as LOD divided by the square root of 2.
To contextualize exposure levels in the SOLAR and CHS cohorts, we compared the geometric mean and 95% confidence interval of PFAS concentrations to appropriate period- and age-matched PFAS concentrations from the National Health and Nutrition Examination Survey (NHANES).60 For the SOLAR cohort, PFAS concentrations were compared with those in young persons 12–19 years of age from the NHANES survey years 2007–2008, given that this was near the middle of the study period for this cohort. For the CHS cohort, PFAS concentrations were compared with those in young persons 12–19 years of age from the NHANES survey years 2017–2018, which overlapped in time with the CHS cohort and was the only survey year in which PFHpS concentrations were reported.
Untargeted Plasma Metabolomics
Untargeted plasma metabolomics were measured using samples collected at the 2-h OGTT time point. Previous studies have shown that metabolite concentrations following a glucose challenge are potentially more informative when examining alterations in metabolic pathways because the glucose challenge is a physiological stressor that requires metabolic flexibility.61,62 Measuring untargeted metabolomics was performed using established methods,63 as described previously.59 Briefly, LC-HRMS was performed with RP and hydrophilic interaction LC (HILIC) by employing a dual polarity/dual column approach. Analyses were performed with a Thermo Fisher Vanquish Duo LC system with dual pumps and columns with independent flow paths connected to a Thermo Fisher Q Exactive HF-X Orbitrap MS system. Samples were analyzed in RP and HILIC separately to allow for the analysis of all four LC-HRMS modes.
Before analysis, samples for RP were prepared using the methods described above in the section “Plasma PFAS”.59 Samples for HILIC were prepared using the methods described above, with minor alterations in the volumes as described previously. Specifically, for HILIC mode, of acetonitrile was added to of plasma before processing was continued as described above. For HILIC mode, of the processed supernatant was added to 1:1 (vol/vol) water/acetonitrile. As described previously, samples were analyzed with both positive and negative ionization by optimizing mobile phases.59 Separation for RP was performed using (TARGA ; Higgins Analytical), whereas separation for HILIC was performed using a SeQuant ZIC-HILIC column (, 200A ; MilliporeSigma) for positive mode, and an Amide ethylene bridged hybrid (BEH) HILIC column (, 130A ; Waters) in negative mode. Measurement of mass spectral data was performed for a mass-to-charge ratio (m/z) scan range of 85 to 1,275, and the resolution was 120,000 full width at half maximum (FWHM). Additional details on this method are provided by Goodrich et al.59
Following analysis of all samples from the SOLAR and CHS cohorts, mass spectral peaks for metabolites were extracted with apLCMS and xMSanalyzer.63,64 Extraction was performed across all study samples concurrently and was performed separately for each of the four LC-HRMS modes. After extracting LC-MS features, adjustment for inter- and intra-batch variation was performed using a random forest signal correction algorithm based on quality control samples run in tandem with study samples.65 As part of the random forest signal correction algorithm, features that were not detected in of samples were removed from further analysis. Features were also removed from further analysis if the coefficient of variability in all quality control samples post-correction was . After LC-MS data processing, the total number of features included in data analysis was 23,166, including 3,711 features from the -negative mode, 5,069 features from the -positive mode, 7,442 features from the HILIC-negative mode, and 6,944 features from the HILIC-positive mode.
Statistical Analysis
Differences in PFAS levels and participant characteristics between cohorts were examined using independent -tests (for natural log-transformed PFAS concentrations and continuous participant characteristics) and chi-square tests (for categorical participant characteristics). Spearman correlation coefficients were calculated to examine associations between PFAS levels within each cohort. All analyses were performed with R (version 4.1.2; R Development Core Team).
Metabolome-Wide Association Study
To examine associations between PFAS mixtures and each metabolite, we performed a metabolome-wide association study (MWAS) using a Bayesian hierarchical regression modeling approach with g-computation (BHRM-g). We implemented a Bayesian g-computation approach66,67 to obtain both individual PFAS-specific estimates conditional on all other PFAS in the model and a single mixture effect estimate for the overall PFAS mixture. The approach is similar to quantile g-computation.68 Specifically, BHRM-g combines: a) a g-prior specification for the corresponding exposure effects to provide robust estimation of highly correlated exposures,69 b) a Bayesian stochastic selection procedure to estimate the posterior inclusion probability (PIP) of each PFAS in the PFAS mixtures,67 and c) Bayesian g-computation in a potential outcome framework for estimating the overall mixture effect based on two hypothetical exposure profiles (explained in further detail below).67 In contrast to Bayesian Kernel Machine Regression, which uses Gaussian process regression and models a nonlinear dose–response relationship, BHRM-g estimates a mixture effect from the additive terms from a regression model with each exposure and forces explicit specification of nonlinear effects.70 We modeled the dose–response with a linear function that allows for the calculation of a single monotonic effect estimate that is not dependent on the baseline exposure profile; this aids in interpretation and allows for additional downstream analysis. In this analysis, we independently fit the following model for each metabolite Y:
where is a scaled variable for PFAS exposure with corresponding estimate ; is a binary variable indicating the inclusion of PFAS exposure p in the mixture; and is a set of q covariates with corresponding effect estimates .
To obtain robust estimates in the presence of highly correlated exposures, we included a second-stage g-prior on the effect estimates of the form .71–74 Here, is the variance of the outcome; is a scalar with a specified hyper-g prior72,75,76; and is the covariance matrix.77 In this model, the scalar controls the shrinkage toward the prior mean of zero and the dispersion of the posterior covariance via a shrinkage factor of . Similar to regularized regression approaches, the resulting posterior estimate can be expressed as a function of this shrinkage , where is the maximum likelihood estimate.
To facilitate model selection, we included a binary variable in the first-stage linear predictor component of the model, indicating the inclusion of each PFAS. We included a beta-binomial prior for model selection such that .78 Within this framework, the PIP on the individual is the posterior probability that the coefficient is nonzero. Inference for the Bayesian model was achieved with Markov chain Monte Carlo techniques using JAGs and the R package rjags.79,80 The model was initialized in an adaptive mode with 4,000 iterations to increase the efficiency, and we used Markov Chain for a burn-in period with 1,000 iterations. To generate the posterior distribution of the parameters, we updated the model with 5,000 iterations.
Finally, we used g-computation to yield a single effect estimate (i.e., a mixture effect) that captures the impact of increase in levels of all exposures simultaneously.66–68 This is performed by estimating the difference in the outcome between two hypothetical exposure profiles based on the distribution of PFAS in the study samples. In this study, the low exposure profile is defined as setting each log-transformed and standardized PFAS at a -score of (i.e., the 30.8 percentile) and the high exposure profile is defined as setting each log-transformed and standardized PFAS at a -score of 0.5 (i.e., the 69.1 percentile). Specifically, we used posterior predictive distributions to estimate a single mixture risk difference (), such that , where and is the counterfactual profile for the log-transformed and standardized exposures with all exposures set to “low” (30.8% levels with an ) and to “high” (69.1% levels with an ). The PFAS concentrations for the low and high exposure profiles in the SOLAR and CHS cohorts are provided in Table S1.
Effect estimates from the BHRM-g models are reported as the posterior mean and 95% Bayesian credible intervals (BCIs). Associations between the PFAS mixture and individual metabolites were selected for further analysis based on a 95% BCI not containing zero. Inputs for the pathway enrichment analysis used the PFAS mixture effect (), dividing the posterior mean by the posterior variance to obtain a Wald test statistic and corresponding -value. False discovery rate (FDR)–adjusted -values were calculated with the Benjamini–Hochberg method to account for multiple comparisons.81 Adjusted -values were calculated within each cohort separately and are represented in the text as -values. Evidence of a significant association between the PFAS mixture and individual metabolites was defined as a 95% BCI for the PFAS mixture effect not containing zero. was considered as additional evidence of a significant association.
Prior to the MWAS, raw intensity values of LC-MS features were and normalized to a standard normal distribution to satisfy regression assumptions and obtain comparable effect estimates across all metabolites. Because metabolite intensity was and scaled prior to analysis, the effect estimate () is also equivalent to a standardized mean difference calculated between a hypothetical group of individuals with all PFAS at the th percentile vs. a hypothetical group of individuals with all PFAS at the th percentile.
Finally, for BHRM-g to provide a mixture effect estimate interpretable as the change in the outcome when increasing all PFAS in the mixture by , all exposures in the mixture must be scaled to a mean of zero and a standard deviation of one. In small sample sizes, extremely positively skewed exposures can also cause instability, and in our data, several PFAS, including PFHxS and PFNA, were highly positively skewed. Therefore, before analysis, PFAS were and scaled to a mean of zero and standard deviation of one. The R code for performing a single BHRM-g regression is provided in Supplemental Material, “Supplemental Code.” The R code for all analyses performed in this study is available on GitHub at https://github.com/chatzilab/PFAS_metabolomics_EHP_2022.
Metabolite Annotation and Pathway Enrichment
Following the BHRM-g MWAS, we used the PFAS mixture effect estimate to perform a pathway enrichment analysis using the MS peaks to pathways module from MetaboAnalyst (version 5.0).82 We used version 2 of the MS peaks to pathways module, which accounts for retention time for more accurate metabolite annotation.83 For this analysis, we included LC-MS features from both positive and negative ionization. We used a mass tolerance, a 0.05 -value threshold, and the Human reference pathways of the MetaFishNet (MFN) database.84 We used the integrated metabolic pathway enrichment analysis,85 which determines pathway enrichment using an overrepresentation analysis86 and a Gene Set Enrichment Analysis.87 Significance of metabolic pathways across cohorts was determined by combining -values using a weighted -test.88 For the pathway enrichment analysis, statistical significance was based on a -value threshold of 0.05. Metabolic pathways identified as significantly enriched were included in follow-up analysis if they included at least four significant metabolites in either the SOLAR or CHS cohorts.
Sensitivity Analysis
Due to differences in the SOLAR and CHS cohorts, including differences in developmental stages and exposure profiles, the primary method for combining results across the two cohorts in this study was to perform a meta-analysis on the effect estimates obtained from study-specific regression models with study-specific covariates. This method avoids many of the potential pitfalls of pooled analysis.89 However, to assess the impact of this analytic choice, we conducted a sensitivity analysis by performing the entire analytic workflow using pooled data from the two cohorts. Because of differences in covariates between cohorts, a minimal set of covariates was used for this analysis, including sex, developmental stage (based on Tanner stage), parental education, and study wave/cohort. Given that Tanner stage was not measured in the CHS (young adult) cohort, Tanner stage was imputed as stage 5 for all individuals from this cohort. For the pooled analysis, study wave/cohort included three levels (Wave 1 SOLAR, Wave 2 SOLAR, and CHS) and was included in the model as two numeric dummy variables. For the pooled analysis, in addition to examining metabolites associated with significantly enriched metabolic pathways, we also further examined associations of the PFAS mixtures with individual metabolites associated with any of the significantly enriched metabolic pathways from the individual cohort analysis.
Results
Characteristics of the Study Population
Participant characteristics for the SOLAR and CHS cohorts are presented in Table 1, and plasma PFAS concentrations for the SOLAR and CHS cohorts are provided in Table 2. In both cohorts, PFOS, PFHxS, PFHpS, PFOA, and PFNA were detected in all participants. PFDA was detected in 99.4% of participants from the SOLAR cohort and 98.5% of participants from the CHS cohort. PFAS levels ranged from uncorrelated to strongly positively correlated, with Spearman correlation coefficients between 0.01 and 0.93 in the SOLAR cohort and between 0.10 and 0.93 in the CHS cohort (Figure S2). Concentrations of all PFAS were significantly higher in the SOLAR cohort compared with the CHS cohort (Table 2). In both cohorts, PFAS concentrations were similar to those reported in the appropriate time period and age-matched NHANES (Table S2).60
Table 1.
Participant characteristics of adolescents from the Study of Latino Adolescents at Risk (SOLAR) cohort (recruited between 2001 and 2012) and young adults from the Southern California Children’s Health Study (CHS; recruited between 2014 and 2018).
| General characteristics | SOLAR | CHS | -Value |
|---|---|---|---|
| Sample size () | 312 | 137 | — |
| Sex [female; (%)] | 133 (40) | 61 (40) | 0.79 |
| Age [y ()] | — | ||
| BMI [ ()] | 0.0051 | ||
| Puberty status [ (%)] | — | ||
| Prepuberty (Tanner stage 1) | 99 (32) | — | |
| Puberty (Tanner stages 2–4) | 193 (62) | — | |
| Postpuberty (Tanner stage 5) | 20 (6) | — | |
| Ethnicity [ (%)] | |||
| Hispanic | 312 (100) | 79 (58) | |
| Non-Hispanic | 0 (0) | 58 (42) | |
| Study wave [ (%)] | — | ||
| Wave 1 (2001–2003) | 234 (75) | — | |
| Wave 2 (2010–2012) | 78 (25) | — | |
| Socioeconomic status | |||
| Modified Hollingshead Four-Factor Index () | — | — | |
| Household education level [ (%)] | |||
| Did not graduate high school | 146 (47) | 25 (18) | |
| High school graduate | 89 (29) | 21 (15) | |
| Partial college ( y) or specialized training | 36 (12) | 36 (26) | |
| Completed college/university | 0 (0) | 37 (27) | |
| Graduate professional training | 3 (1) | 14 (10) | |
| Missing | 34 (11) | 4 (3) | |
Note: -Values not reported for variables that were only measured in one cohort. —, not applicable; BMI, body mass index; NA, not applicable; SD, standard deviation.
Table 2.
PFAS Concentrations () in overweight and obese adolescents from the SOLAR cohort and young adults from the CHS cohort.
| PFAS subclass | PFAS name | SOLAR () | CHS () | -Value | ||||
|---|---|---|---|---|---|---|---|---|
| Geometric | Arithmetic | Median (IQR) | Geometric | Arithmetic | Median (IQR) | |||
| Perfluorosulfonic acids | PFOS | 15.1 (14.1) | 3.13 (1.90) | |||||
| PFHxS | 1.35 (1.20) | 0.95 (1.00) | ||||||
| PFHpS | 0.42 (0.31) | 0.17 (0.10) | ||||||
| Perfluorocarboxylic acids | PFOA | 3.45 (2.46) | 1.34 (0.67) | |||||
| PFNA | 0.57 (0.21) | 0.46 (0.15) | ||||||
| PFDA | 0.23 (0.11) | 0.20 (0.12) | 0.0012 | |||||
Note: -Value was calculated for the differences in PFAS concentrations between cohorts using independent -tests on log-transformed PFAS concentrations. CHS, Children’s Health Study; GSD, geometric standard deviation; IQR, interquartile range; PFAS, per- and polyfluoroalkyl substances; PFDA, perfluorodecanoic acid; PFHpS, perfluoroheptanesulfonic acid; PFHxS, perfluorohexanesulfonic acid; PFNA, perfluorononanoic acid; PFOA, perfluorooctanoic acid; PFOS, perfluorooctane sulfonic acid; SD, Arithmetic standard deviation; SOLAR, Study of Latino Adolescents at Risk.
PFAS Exposure Was Associated with Metabolic Pathways
In the SOLAR and CHS cohorts, we performed a MWAS with all 23,166 untargeted metabolite features to examine their association with a single mixture of all six PFAS compounds. In the SOLAR cohort, the MWAS identified 463 metabolite features associated with the PFAS mixture, defined as having a 95% BCI for the mixture effect not containing zero (Excel Table S1). Functional pathway analysis of the MWAS results identified significant enrichment of 11 metabolic pathways (Figure 1; Table S3). These pathways were primarily related to the metabolism of aromatic amino acids, nonaromatic amino acids, lipids, and cofactors and vitamins.
Figure 1.
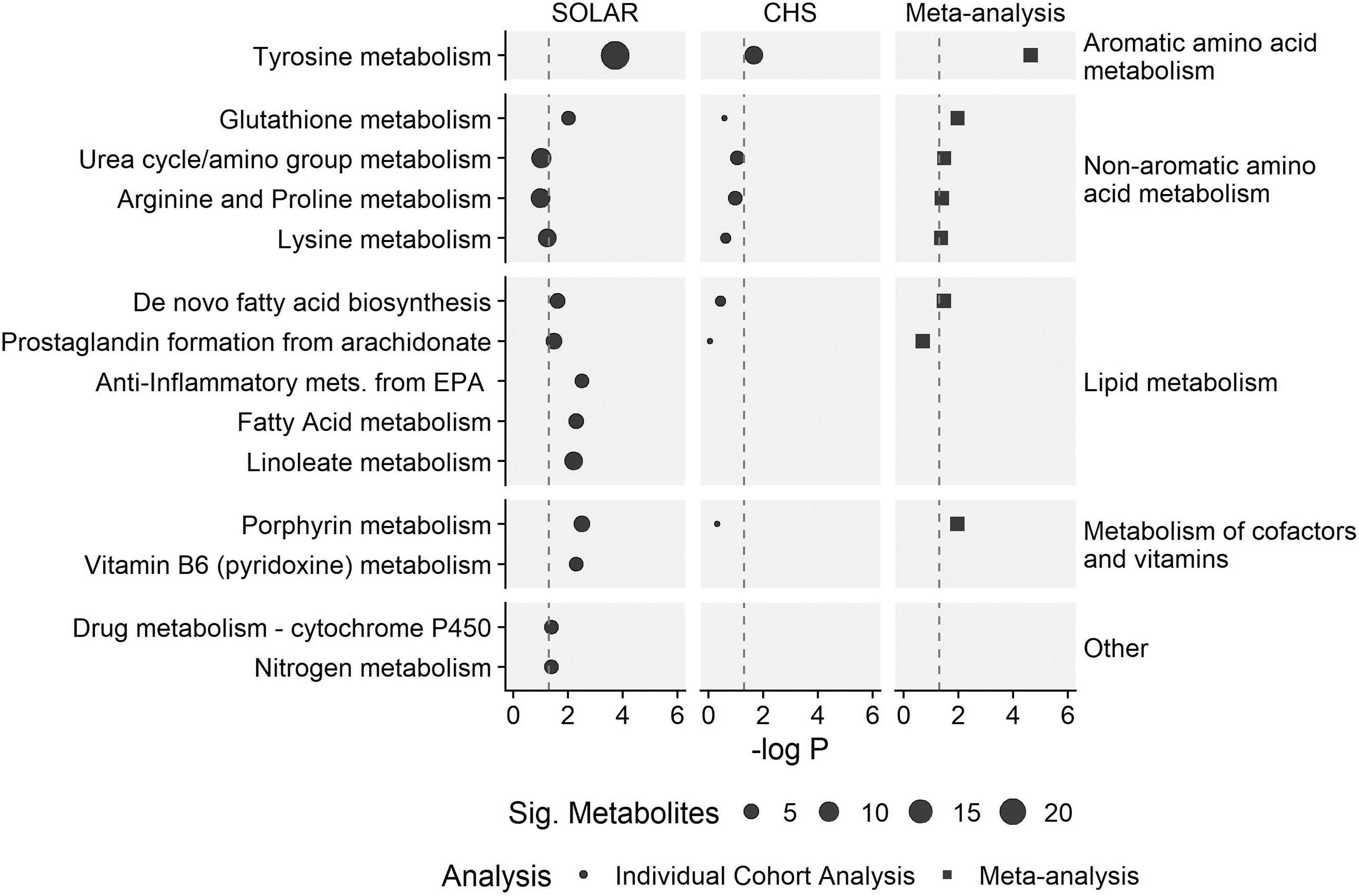
Metabolic pathways associated with exposure to a mixture of six PFAS in adolescents from the SOLAR cohort () and young adults from the CHS cohort (). Metabolic pathways are grouped into super pathways as indicated on the right of the plot. Meta-analysis values are provided for pathways identified as being associated with PFAS in both cohorts. Dot size for the SOLAR and CHS cohorts are proportional to the number of significant metabolites associated with each pathway. Only pathways that were significant in either the SOLAR cohort, the CHS cohort, or the meta-analysis are presented here; for complete results see Table S3. Note: CHS, Children’s Health Study; EPA, eicosapentaenoic acid; PFAS, per- and polyfluoroalkyl substances; Sig, Significant; SOLAR, Study of Latino Adolescents at Risk.
In the CHS cohort, 200 metabolite features were associated with the PFAS mixture (Excel Table S2). Functional pathway analysis identified the tyrosine metabolism pathway, an aromatic amino acid, as significantly enriched (; Figure 1; Table S3). In addition, four nonaromatic amino acid metabolism pathways, two lipid metabolism pathways, and one pathway related to the metabolism of cofactors were identified in the functional pathway enrichment analysis as having at least one metabolite associated with the PFAS mixture, although these pathways did not meet the threshold for statistical significance.
Meta-analysis of the -values from the pathway enrichment analysis from both cohorts identified seven statistically significant metabolic pathways, including one aromatic amino acid metabolism pathway (tyrosine metabolism), four nonaromatic amino acid metabolism pathways (glutathione, urea cycle/amino group, arginine and proline, and lysine metabolism), one lipid metabolism pathway (de novo fatty acid biosynthesis), and one pathway related to metabolism of cofactors (porphyrin metabolism). In total, PFAS exposure was associated with alterations in 14 unique metabolic pathways across four super pathways in the SOLAR cohort, the CHS cohort, or in the meta-analysis (Figure 1; Table S3).
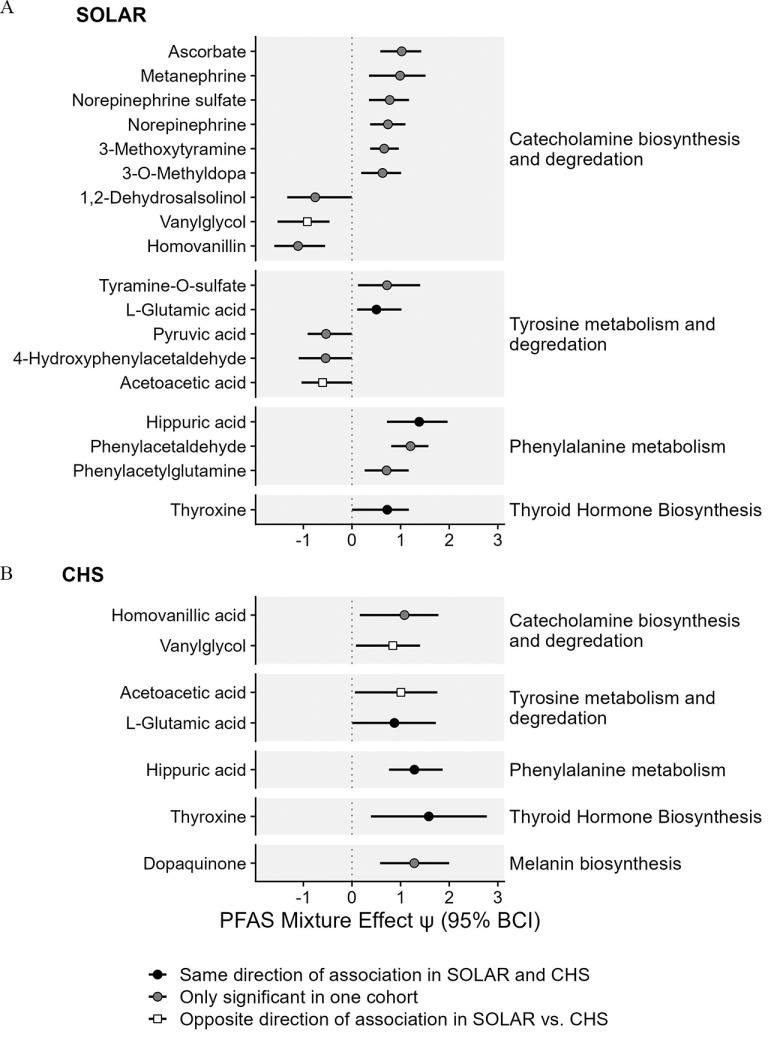
For the aromatic amino acid metabolism pathway, PFAS exposure was associated with several metabolites linked to important tyrosine metabolism subpathways across both cohorts. In the SOLAR cohort, 18 unique metabolites were linked to aromatic amino acid metabolism pathways, and seven of these associations remained significant after adjusting for multiple comparisons (Figure 2A; Table S4). In the CHS cohort, seven unique metabolites were linked to aromatic amino acid metabolism pathways, and one remained significant after adjusting for multiple comparisons (Figure 2B; Table S4). To gain additional insight about the functional implications of alterations in these metabolites, we grouped metabolites by subpathways within aromatic amino acid metabolism. These subpathways included catecholamine biosynthesis and degradation, tyrosine metabolism and degradation, thyroid hormone biosynthesis, phenylalanine metabolism, and melanin biosynthesis. Three key metabolites were positively associated with PFAS exposure in both cohorts. These included thyroxine (T4), the main thyroid hormone in circulation; l-glutamic acid, an amino acid associated with tyrosine metabolism; and hippuric acid, an acyl glycine associated with phenylalanine metabolism. Two metabolites were negatively associated with PFAS exposure in the SOLAR cohort but positively associated with PFAS exposure in the CHS cohort; these included vanylglycol, a methoxyphenol generated from the degradation of catecholamines, and acetoacetic acid, a product of the catabolism of tyrosine to fumaric acid.
Figure 2.
Associations between PFAS mixtures and metabolites associated with aromatic amino acid metabolism in (A) adolescents from the SOLAR cohort () and (B) young adults from the CHS cohort (). Metabolites are grouped by tyrosine metabolism subpathways as indicated on the right of the plot. Effect estimates for PFAS mixture () and the 95% Bayesian credible interval (BCI) estimate the change in metabolite levels (SD of the log-transformed feature intensity) when increasing all PFAS in the mixture from the 30th percentile to the 70th percentile. This estimate is also equivalent to a standardized mean difference calculated between a hypothetical group of individuals with all PFAS at the th percentile vs. a hypothetical group of individuals with all PFAS at the th percentile. Corresponding -values and -values are presented in Table S4. Note: CHS, Children’s Health Study; PFAS, per- and polyfluoroalkyl substances; SD, standard deviation; SOLAR, Study of Latino Adolescents at Risk.
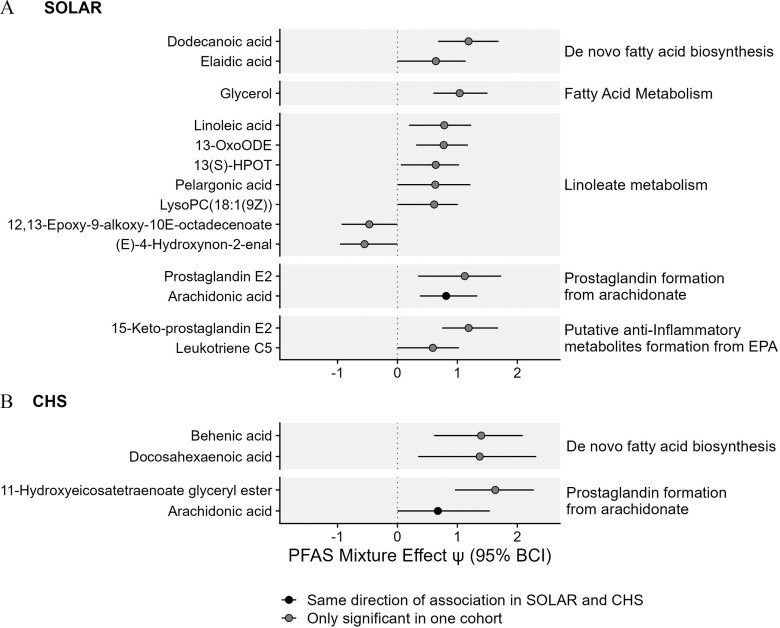
In both the SOLAR and the CHS cohorts, PFAS exposure was also associated with key metabolites associated with lipid metabolism. In the SOLAR cohort, 14 unique metabolites were linked to lipid metabolism pathways, and 3 remained significant after adjusting for multiple comparisons (Figure 3A; Table S4). In the CHS cohort, 4 unique metabolites were linked to lipid metabolism pathways, and 2 remained significant after adjusting for multiple comparisons (Figure 3B; Table S4). Across the two cohorts there were similar positive associations between PFAS exposure and metabolites associated with de novo fatty acid biosynthesis and metabolites associated with prostaglandin formation from arachidonate. Of these metabolites, the most consistent association was observed with arachidonic acid, which was positively associated with PFAS exposure in both cohorts (Figure 3; Table S4).
Figure 3.
Associations between PFAS mixtures and metabolites associated with lipid metabolism in (A) adolescents from the SOLAR cohort () and (B) young adults from the CHS cohort (). Effect estimates for PFAS mixture () and the 95% Bayesian credible interval (BCI) estimate the change in metabolite levels (SD of the log-transformed feature intensity) when increasing all PFAS in the mixture from the 30th percentile to the 70th percentile. This estimate is also equivalent to a standardized mean difference calculated between a hypothetical group of individuals with all PFAS at the th percentile vs. a hypothetical group of individuals with all PFAS at the th percentile. Corresponding -values and -values are presented in Table S4. Note: CHS, Children’s Health Study; EPA, eicosapentaenoic acid; HPOT, hydroperoxyoctadecatrienoic acid; LysoPC, lysophosphatidylcholines; OxoODE, Octadecanienoic acid; PFAS, per- and polyfluoroalkyl substances; SD, standard deviation; SOLAR, Study of Latino Adolescents at Risk.
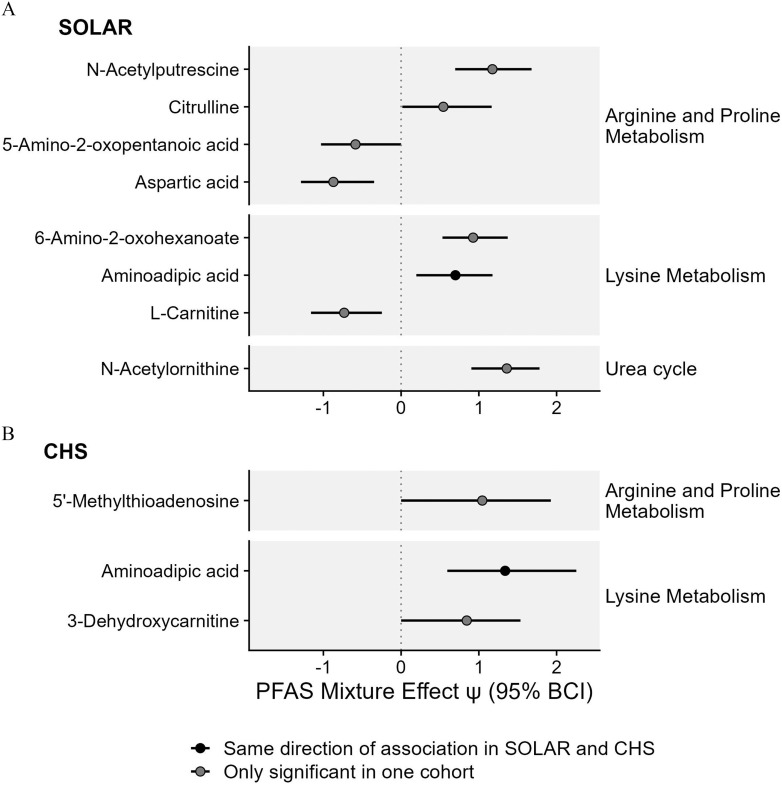
PFAS exposure was also associated with alterations in metabolites associated with the metabolism of nonaromatic amino acids (Figure 4; Table S4). In the SOLAR cohort, eight unique metabolites were linked to nonaromatic amino acid metabolism pathways, and four remained significant after adjusting for multiple comparisons (Figure 4A; Table S4). In the CHS cohort, three unique metabolites were linked to nonaromatic amino acid metabolism pathways, although none remained significant after adjusting for multiple comparisons (Figure 4B; Table S4). Across cohorts, the most consistent associations were observed with aminoadipic acid, which was positively associated with PFAS exposure in both cohorts.
Figure 4.
Associations between PFAS mixtures and metabolites associated with nonaromatic amino acid metabolism in (A) adolescents from the SOLAR cohort () and (B) young adults from the CHS cohort (). Effect estimates for PFAS mixture () and the 95% Bayesian credible interval (BCI) estimate the change in metabolite levels (SD of the log-transformed feature intensity) when increasing all PFAS in the mixture from the 30th percentile to the 70th percentile. This estimate is also equivalent to a standardized mean difference calculated between a hypothetical group of individuals with all PFAS at the th percentile vs. a hypothetical group of individuals with all PFAS at the th percentile. Corresponding -values and -values are presented in Table S4. Note: CHS, Children’s Health Study; PFAS, per- and polyfluoroalkyl substances; SD, standard deviation; SOLAR, Study of Latino Adolescents at Risk.
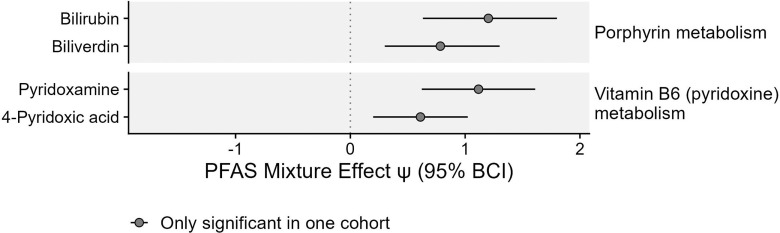
In the SOLAR cohort, PFAS exposure was also positively associated with four metabolites linked to the metabolism of cofactors, including porphyrin metabolism and pyridoxine metabolism. Two of these associations remained significant after adjusting for multiple comparisons (Figure 5; Table S4).
Figure 5.
Associations between PFAS mixtures and metabolites associated with metabolism of cofactors in adolescents from the SOLAR cohort (). No significant associations were observed in the CHS cohort. Effect estimates for PFAS mixture () and the 95% Bayesian credible interval (BCI) estimate the change in metabolite levels (SD of the log-transformed feature intensity) when increasing all PFAS in the mixture from the 30th percentile to the 70th percentile. This estimate is also equivalent to a standardized mean difference calculated between a hypothetical group of individuals with all PFAS at the th percentile vs. a hypothetical group of individuals with all PFAS at the th percentile. Corresponding -values and -values are presented in Table S4. Note: CHS, Children’s Health Study; PFAS, per- and polyfluoroalkyl substances; SD, standard deviation; SOLAR, Study of Latino Adolescents at Risk.
Contribution of Individual PFAS on Individual Metabolites
To examine the potential contribution of individual PFAS to the overall mixture effect, we examined the PIP for all PFAS with each metabolite. The PIP is the posterior probability that the coefficient for each individual PFAS in the mixture is nonzero. PFAS that do not contribute to the overall mixture effect should have a PIP reflecting the prior probability of inclusion and , where P is the total number of PFAS in the mixture. In the present analysis, this corresponds to a PIP of 0.167. PIPs indicate a greater likelihood that the individual PFAS has a nonzero effect on the overall mixture.
In the SOLAR cohort, the PIPs for individual exposures across all 44 significantly altered metabolites from enriched pathways ranged from 0.04 to 1.0 (Figures S2–S5). For 41 of the 44 metabolites (93%), two or more PFAS exhibited PIPs , indicating that for these metabolites, at least two PFAS had a nonzero effect on the overall mixture. Further examination of these PIPs revealed that no individual PFAS appeared to drive these associations across all metabolites, although some PFAS seemed more involved than others. Overall, PFDA and PFOS exhibited PIPs for 73% and 71% of the significant metabolites, indicating that these PFAS played a role in the mixture effect for the majority of metabolites. In contrast, PFNA exhibited PIPs for only 37% of metabolites, indicating that this PFAS may play less of a role in the association between PFAS exposure and alterations in metabolic pathways. Results were generally consistent within each super pathway (Figures S2–S5).
In the CHS cohort, the PIPs for individual exposures across all 14 significantly altered metabolites from enriched pathways ranged from 0.10 to 0.97. For 12 of the 14 metabolites (86%), two or more PFAS exhibited PIPs , indicating that for these metabolites, at least two PFAS had a nonzero effect on the overall mixture. Three PFAS exhibited PIPs for of metabolites, including PFHxS, PFHpS, and PFNA (Figure S3–S6).
Sensitivity Analysis
Results for the pooled analysis were similar to those of the meta-analysis. The MWAS on the pooled individual level data identified 464 metabolite features associated with the PFAS mixture, defined as having a 95% BCI for the mixture effect not containing zero (Excel Table S3). Functional pathway analysis of the MWAS results identified three metabolic pathways with four or more significant empirical compounds, two of which were significantly enriched (Table S5). These included tyrosine metabolism (; significant in both cohorts individually) and urea cycle/amino group metabolism (; significant in the meta-analysis). T4 and hippuric acid, two of the three key tyrosine metabolites associated with PFAS exposure in both cohorts, remained significantly associated with the PFAS mixture in the pooled analysis (Table S4).
Discussion
To our knowledge, this is the first study to comprehensively examine the effects of exposure to PFAS mixtures on human metabolic pathways. In two independent cohorts of children and young adults, we observed associations between PFAS exposure and alterations in aromatic amino acid metabolism, nonaromatic amino acid metabolism, and lipid metabolism pathways. These associations were present despite differences in levels of PFAS exposure between cohorts. Alterations in aromatic amino acid metabolism included changes in metabolites associated with catecholamine and thyroid hormone biosynthesis; alterations in nonaromatic amino acid metabolism included changes in metabolites related to arginine, proline, and lysine metabolism; and alterations in lipid metabolism included changes in metabolites related to de novo fatty acid biosynthesis and prostaglandin formation from arachidonate. Together, these results provide evidence that PFAS exposure is associated with alterations in several important metabolic pathways in children and young adults.
Previous studies examining metabolic perturbations of PFAS in humans have primarily relied on single exposure models. In reality, humans are exposed to a mixture of several PFAS compounds that may have synergistic effects.48 Although single exposure models provide insight into the health effects of individual PFAS compounds, these models do not account for co-exposure to multiple potentially correlated environmental exposures that may have synergistic effects. Here, we present the results of an innovative analytic strategy to examine the effects of exposure to chemical mixtures on human metabolic pathways. We used a Bayesian hierarchical regression modeling approach with a g-prior specification and Bayesian g-computation,66,67 which provided a flexible framework for obtaining robust mixture effect estimates in the presence of highly correlated exposures. In addition to providing a framework for future studies looking to examine the associations between exposure mixtures and omics scale data, our findings have important public health implications. Specifically, we did not find evidence that individual PFAS drove the associations between the PFAS mixture and metabolic pathways. Rather, alterations in metabolic pathways were primarily driven by mixtures of PFAS. Further, these effects were consistent across cohorts with different levels of exposure. As a result of changes in the regulation of PFAS in the United States starting in the early 2000s, the SOLAR cohort had higher levels of exposure to PFAS than the CHS cohort.90 These changes were especially pronounced for PFOS and PFOA, which saw decreases of between cohorts. Despite this, we found similar, although slightly attenuated, results in the CHS cohort vs. the SOLAR cohort. This trend may suggest that the toxicological effects of PFAS exposure are more related to total PFAS levels, rather than individual PFAS compounds. Given the associations of PFAS exposure and metabolic pathways related to aromatic amino acid metabolism, nonaromatic amino acid metabolism, and lipid metabolism, our findings lend support to the argument that PFAS should be regulated as a chemical class rather than being regulated on a chemical-by-chemical basis.91
It is well established that PFAS exposure impacts thyroid function, but the downstream metabolic consequences of PFAS-associated thyroid toxicity is not well characterized.11,92 In both cohorts, we observed a positive association between PFAS and T4, the main thyroid hormone in circulation, which is consistent with previous human and animal studies.93 Thyroid hormones are key regulators of metabolism, including the regulation of anabolism and catabolism of lipids, carbohydrates, and proteins.94 Thyroid hormones upregulate de novo fatty acid biosynthesis through the transcription of lipogenic genes, including Acc1, Fasn, Me1, and Thrsp.95 In the present study, we observed positive associations between PFAS exposure and increased de novo fatty acid biosynthesis. Increased de novo fatty acid biosynthesis is a hallmark of metabolic disorders and can lead to obesity, insulin resistance, nonalcoholic fatty liver disease, and cancer.96 These diseases have also been associated with PFAS exposure11 and thyroid hormones.95,97–99 Together, our findings raise the possibility that increased risk of metabolic disorders associated with PFAS exposure are caused by alterations in thyroid hormones and mediated by changes in lipid metabolism.
Several animal studies have reported associations between PFAS exposure and catecholamine biosynthesis, but human data is lacking.29,100–106 Notably, exposure to an environmentally relevant PFAS mixture has been previously shown to alter dopamine levels, one of the three main catecholamines, in mice and in wild Bank voles.102,103 In mice, PFAS exposure has also been linked to alterations in the activity of tyrosine hydroxylase,103 which is the rate-limiting enzyme in the biosynthesis of catecholamines.107 Exposure to individual PFAS, including PFOS and PFOA, has also been shown to decrease tyrosine hydroxylase activity in pheochromocytoma (PC12) cells.108 Although previous animal studies have focused on the association of PFAS exposure on dopamine levels in brain regions, tyrosine hydroxylase is also expressed in the peripheral nervous system and the adrenal medulla, where a majority of peripheral catecholamine biosynthesis occurs.109 In addition to the potential metabolic consequences of alterations in peripheral catecholamine biosynthesis,110 alterations in central catecholamine biosynthesis could play a role in the potential neurotoxic effects of PFAS exposure.111
One of the proposed mechanisms linking PFAS exposure with a variety of diseases is an increase in inflammation and oxidative stress.11,112,113 In the present study, we observed positive associations of PFAS exposure with arachidonic acid, aminoadipic acid, and hippuric acid in both cohorts, each of which is associated with inflammation or oxidative stress. Arachidonic acid is a polyunsaturated fatty acid that contributes to inflammation and plays a role in carcinogenesis and cardiovascular disease.114,115 Aminoadipic acid is an amino acid involved in lysine metabolism that is a potential biomarker of oxidative stress and has been linked to a variety of diseases including type 2 diabetes.116,117 Hippuric acid is a gut-derived amino acid that disrupts redox homeostasis and contributes to oxidative stress, and which has been shown to be a uremic toxin.118,119 Together, these results lend support to the hypothesis that PFAS exposure impact inflammation and oxidative stress.
One factor that could play a role in our findings is diet. Although each of the metabolites identified in this study can be synthesized endogenously, some can also be obtained in the diet. For example, both arachidonic acid and metabolites associated with de novo fatty acid biosynthesis can be obtained via diet.120–122 However, previous literature has linked PFAS exposure with alterations in lipid metabolism and de novo lipogenesis in a variety of experimental studies, which suggests that PFAS are impacting the regulation of these fatty acids in circulation. PFAS exposure alters the expression of genes related to de novo lipogenesis in the liver of mice, resulting in an increase in circulating serum triglycerides.29,123 Similar results have been observed in rats,124 zebrafish,125 and chicken embryos.126 Mechanistically, in addition to PFAS-associated alterations in T4 levels, PFAS also interact with several peroxisome proliferator-activated receptors (PPARs) including and ,11 both of which are key nuclear receptors associated with the regulation of de novo lipogenesis in the liver.127 also regulates the expression of ,128 the enzyme responsible for converting dietary linoleic acid to arachidonic acid. In conjunction with our findings, these studies suggest that PFAS exposure is associated with dysregulated lipid metabolism via alterations in PPAR activity. However, it is also possible that dietary intake of specific fatty acids may interact with PFAS exposure to cause further dysregulation of lipid metabolism,129 which should be examined in future studies.
Although many of the associations between PFAS mixtures and alterations in metabolic pathways were similar in the SOLAR and CHS cohorts, we also identified several differences, especially for lipid metabolism pathways. In the SOLAR cohort, three metabolic pathways (linoleate metabolism, fatty acid metabolism, and anti-inflammatory metabolite formation from eicosapentaenoic acid) were reported only in the SOLAR cohort. One possible explanation is that participants from the SOLAR cohort were either undergoing puberty or prepuberty, whereas participants in the CHS cohort were young adults postpuberty. Exposure to PFAS compounds during sensitive periods of development, such as adolescence, may be more likely to lead to deleterious health effects, including dyslipidemia, given that this is an important period of development for many metabolic tissues.12–15 These results are consistent with previous studies showing that PFAS exposure in childhood is associated with dysregulated lipid and fatty acid metabolism, which can greatly increase the risk of metabolic disorders and cardiovascular disease later in life.130
This study has some limitations. First, when using untargeted metabolomics, accurate identification of metabolites is often difficult because of the limited number of authentic standards available. Although we applied functional pathway enrichment that combines biological relationships among pathways to improve annotation confidence, in the present study the majority of metabolite annotations are limited to level 4, indicating accurate mass matching and molecular formula without informing on chemical structure.131 This is in contrast to targeted metabolomics, which have annotation levels 1 or 2, indicating a confirmed chemical structure. However, untargeted metabolomics has distinct advantages over targeted methods, including greater coverage of metabolites.132 This is ideal for hypothesis-generating studies, and allows for robust interpretation of functional activity at the pathway level.86 Second, it is difficult to assume temporality of associations because of the cross-sectional design. This also raises the possibility of exposure misclassification if plasma PFAS concentrations exhibit large changes in short periods of time. However, the half-life in plasma for PFAS included in this study is in years,133 which suggests that a single plasma measurement is likely to provide an accurate indication of a person’s long-term PFAS exposure. Third, the cohorts included in this study are relatively unique, which may limit generalizability to other populations. The SOLAR and CHS cohorts included primarily Hispanic overweight and obese children and young adults, and additional studies are required to determine whether these associations are generalizable to other populations. Fourth, there is a possibility that an unknown or unmeasured confounder could have impacted our results. For example, diet could be confounding the relationship between PFAS exposure and certain metabolites. However, dietary patterns only appeared to be a small portion of the total PFAS exposure in the general U.S. population between 2003 and 2014,134 which reduces the possibility that diet was a confounder for our study. Other potential confounders could be factors related to PFAS levels, such as plasma albumin levels,135 blood volume,136 menstruation,137 or kidney function.138 Finally, differences in covariates between cohorts may have caused some differences in the results between cohorts. For example, the measures of SES were different between cohorts. However, in the pooled analysis, which used a reduced set of covariates, we observed similar findings as those in the individual cohort analysis, especially those related to alterations in aromatic amino acid metabolism. Despite these limitations, we observed similar results across different populations with varied background characteristics and levels of PFAS exposure. Our findings are in line with previous experimental models in animals, lending additional validity to our results. The similarity of findings across different cohorts with distinct background characteristics suggests that metabolic perturbations associated with PFAS exposure are not restricted to a single cohort. In summary, this study provides evidence that PFAS exposure is associated with alterations in several key metabolic pathways, which could be an important mediating factor explaining the associations of PFAS exposure with risk of various diseases in humans.
Supplementary Material
Acknowledgments
Contributions of the authors to this article are as follows: Concept and design—J.A.G., T.L.A., K.B., Z.C., F.D.G., M.I.G., D.P.J., D.V., D.I.W., D.V.C., and L.C. Acquisition, analysis, or interpretation of data—J.A.G., J.H., X.L., B.O.B., X.H., T.L.A., Z.C., D.V., Z.C.F., S.R., H.W., K.B., F.D.G., M.I.G., D.P.J., D.I.W., D.V.C., and L.C. Drafting of the manuscript—J.A.G., J.H., B.O.B., H.W, D.I.W., D.V.C., and L.C. Critical revision of the manuscript for important intellectual content—J.A.G., J.H., X.L., B.O.B., X.H., T.L.A, Z.C., D.V., Z.C.F., S.R., H.W, K.B., F.D.G., M.I.G., D.P.J., D.I.W., D.V.C., and L.C. Statistical analysis—J.A.G., J.H., B.O.B, H.W., D.V.C., and L.C. Administrative, technical, or material support—X.L., X.H., Z.C.F., S.R., D.P.J., and D.I.W.
The results reported herein correspond to specific aims of grant R01ES029944 from the National Institutes of Health/National Institute of Environmental Health Science (NIH/NIEHS) (to L.C.). Funding for the Study of Latino Adolescents at Risk (SOLAR) came from the NIH grant R01DK59211 (to M.I.G.), and funding for the MetaAir study came from the Southern California Children’s Environmental Health Center grants funded by the NIEHS (5P01ES022845-03, 5P30ES007048, 5P01ES011627), the U.S. Environmental Protection Agency (RD83544101), and the Hastings Foundation. Additional funding from NIH supported L.C. (R01ES030691, R01ES030364, R21ES029681, R21ES028903, and P30ES007048), J.A.G. (T32ES013678, R25GM143298), Z.C. (R00ES027870), D.V. (R01ES033688, R21ES029328, K12ES033594, P30ES023515), D.I.W. (U2CES030859, R01ES032831), D.V.C. (P01CA196569, P30ES007048, R01ES030691, R01ES030364, R21ES029681, R21ES028903), T.L.A (R00ES027853, P50MD017344), and D.P.J. (U2CES030163, P30ES019776, R24ES029490, R01ES032189, R21ES031824).
J.A.G. and L.C. had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
The data that support the findings of this study are available on request from the corresponding author, J.A.G. The data are not publicly available due to them containing information that could compromise research participant privacy/consent.
References
- 1.Mimoto MS, Nadal A, Sargis RM. 2017. Polluted pathways: mechanisms of metabolic disruption by endocrine disrupting chemicals. Curr Environ Health Rep 4(2):208–222, PMID: , 10.1007/s40572-017-0137-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nadal A, Quesada I, Tudurí E, Nogueiras R, Alonso-Magdalena P. 2017. Endocrine-disrupting chemicals and the regulation of energy balance. Nat Rev Endocrinol 13(9):536–546, PMID: , 10.1038/nrendo.2017.51. [DOI] [PubMed] [Google Scholar]
- 3.Sunderland EM, Hu XC, Dassuncao C, Tokranov AK, Wagner CC, Allen JG. 2019. A review of the pathways of human exposure to poly- and perfluoroalkyl substances (PFASs) and present understanding of health effects. J Expo Sci Environ Epidemiol 29(2):131–147, PMID: , 10.1038/s41370-018-0094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whitehead HD, Venier M, Wu Y, Eastman E, Urbanik S, Diamond ML, et al. . 2021. Fluorinated compounds in North American cosmetics. Environ Sci Technol Lett 8(7):538–544, 10.1021/acs.estlett.1c00240. [DOI] [Google Scholar]
- 5.Andrews DQ, Naidenko OV. 2020. Population-wide exposure to per- and polyfluoroalkyl substances from drinking water in the United States. Environ Sci Technol Lett 7(12):931–936, 10.1021/acs.estlett.0c00713. [DOI] [Google Scholar]
- 6.U.S. EPA (U.S. Environmental Protection Agency). 2022. Interim Drinking Water Health Advisory: Perfluorooctanoic Acid (PFOA). CASRN 335-67-1. EPA/8222/R-22/003. https://www.epa.gov/system/files/documents/2022-06/interim-pfoa-2022.pdf [accessed 25 June 2022].
- 7.U.S. EPA. 2022. Interim Drinking Water Health Advisory: Perfluorooctane Sulfonic Acid (PFOS) CASRN 1763-23-1. EPA/822/R-22/004. https://www.epa.gov/system/files/documents/2022-06/interim-pfos-2022.pdf [accessed 25 June 2022].
- 8.Zhang Y, Beesoon S, Zhu L, Martin JW. 2013. Biomonitoring of perfluoroalkyl acids in human urine and estimates of biological half-life. Environ Sci Technol 47(18):10619–10627, PMID: , 10.1021/es401905e. [DOI] [PubMed] [Google Scholar]
- 9.Kannan K, Corsolini S, Falandysz J, Fillmann G, Kumar KS, Loganathan BG, et al. . 2004. Perfluorooctanesulfonate and related fluorochemicals in human blood from several countries. Environ Sci Technol 38(17):4489–4495, PMID: , 10.1021/es0493446. [DOI] [PubMed] [Google Scholar]
- 10.Kato K, Wong LY, Jia LT, Kuklenyik Z, Calafat AM. 2011. Trends in exposure to polyfluoroalkyl chemicals in the U.S. population: 1999–2008. Environ Sci Technol 45(19):8037–8045, PMID: , 10.1021/es1043613. [DOI] [PubMed] [Google Scholar]
- 11.Fenton SE, Ducatman A, Boobis A, DeWitt JC, Lau C, Ng C, et al. . 2020. Per- and polyfluoroalkyl substance toxicity and human health review: current state of knowledge and strategies for informing future research. Environ Toxicol Chem 40(3):606–630, PMID: , 10.1002/etc.4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casazza K, Goran MI, Gower BA. 2008. Associations among insulin, estrogen, and fat mass gain over the pubertal transition in African-American and European-American girls. J Clin Endocrinol Metab 93(7):2610–2615, PMID: , 10.1210/jc.2007-2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heindel JJ. 2019. The developmental basis of disease: update on environmental exposures and animal models. Basic Clin Pharmacol Toxicol 125(suppl 3):5–13, PMID: , 10.1111/bcpt.13118. [DOI] [PubMed] [Google Scholar]
- 14.Hines EP, White SS, Stanko JP, Gibbs-Flournoy EA, Lau C, Fenton SE. 2009. Phenotypic dichotomy following developmental exposure to perfluorooctanoic acid (PFOA) in female CD-1 mice: low doses induce elevated serum leptin and insulin, and overweight in mid-life. Mol Cell Endocrinol 304(1–2):97–105, PMID: , 10.1016/j.mce.2009.02.021. [DOI] [PubMed] [Google Scholar]
- 15.Kelly LA, Lane CJ, Weigensberg MJ, Toledo-Corral CM, Goran MI. 2011. Pubertal changes of insulin sensitivity, acute insulin response and β-cell function in overweight Latino youth. J Pediatr 158(3):442–446, PMID: , 10.1016/j.jpeds.2010.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodrich JA, Alderete TL, Baumert BO, Berhane K, Chen Z, Gilliland FD, et al. . 2021. Exposure to perfluoroalkyl substances and glucose homeostasis in youth. Environ Health Perspect 129(9):097002, PMID: , 10.1289/EHP9200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valvi D, Højlund K, Coull BA, Nielsen F, Weihe P, Grandjean P. 2021. Life-course exposure to perfluoroalkyl substances in relation to markers of glucose homeostasis in early adulthood. J Clin Endocrinol Metab 106(8):2495–2504, PMID: , 10.1210/clinem/dgab267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blomberg AJ, Shih YH, Messerlian C, Jørgensen LH, Weihe P, Grandjean P. 2021. Early-life associations between per- and polyfluoroalkyl substances and serum lipids in a longitudinal birth cohort. Environ Res 200:111400, PMID: , 10.1016/j.envres.2021.111400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Domazet SL, Grøntved A, Timmermann AG, Nielsen F, Jensen TK. 2016. Longitudinal associations of exposure to perfluoroalkylated substances in childhood and adolescence and indicators of adiposity and glucose metabolism 6 and 12 years later: the European Youth Heart Study. Diabetes Care 39(10):1745–1751, PMID: , 10.2337/dc16-0269. [DOI] [PubMed] [Google Scholar]
- 20.Høyer BB, Ramlau-Hansen CH, Vrijheid M, Valvi D, Pedersen HS, Zviezdai V, et al. . 2015. Anthropometry in 5- to 9-year-old Greenlandic and Ukrainian children in relation to prenatal exposure to perfluorinated alkyl substances. Environ Health Perspect 123(8):841–846, PMID: , 10.1289/ehp.1408881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cardenas A, Hivert MF, Gold DR, Hauser R, Kleinman KP, Lin PID, et al. . 2019. Associations of perfluoroalkyl and polyfluoroalkyl substances with incident diabetes and microvascular disease. Diabetes Care 42(9):1824–1832, PMID: , 10.2337/dc18-2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qi W, Clark JM, Timme-Laragy AR, Park Y. 2020. Per- and polyfluoroalkyl substances and obesity, type 2 diabetes and non-alcoholic fatty liver disease: a review of epidemiologic findings. Toxicol Environ Chem 102(1–4):1–36, PMID: , 10.1080/02772248.2020.1763997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun Q, Zong G, Valvi D, Nielsen F, Coull B, Grandjean P. 2018. Plasma concentrations of perfluoroalkyl substances and risk of type 2 diabetes: a prospective investigation among U.S. women. Environ Health Perspect 126(3):037001, PMID: , 10.1289/EHP2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balkau B, Valensi P, Eschwège E, Slama G. 2007. A review of the metabolic syndrome. Diabetes Metab 33(6):405–413, PMID: , 10.1016/j.diabet.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 25.Guo P, Furnary T, Vasiliou V, Yan Q, Nyhan K, Jones DP, et al. . 2022. Non-targeted metabolomics and associations with per- and polyfluoroalkyl substances (PFAS) exposure in humans: a scoping review. Environ Int 162:107159, PMID: , 10.1016/j.envint.2022.107159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng S, Yan L, Zhang J, Wang Z, Tian M, Shen H. 2013. An integrated metabonomics and transcriptomics approach to understanding metabolic pathway disturbance induced by perfluorooctanoic acid. J Pharm Biomed Anal 86:56–64, PMID: , 10.1016/j.jpba.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 27.Kudo N, Suzuki-Nakajima E, Mitsumoto A, Kawashima Y. 2006. Responses of the liver to perfluorinated fatty acids with different carbon chain length in male and female mice: in relation to induction of hepatomegaly, peroxisomal β-oxidation and microsomal 1-acylglycerophosphocholine acyltransferase. Biol Pharm Bull 29(9):1952–1957, PMID: , 10.1248/bpb.29.1952. [DOI] [PubMed] [Google Scholar]
- 28.Wang L, Wang Y, Liang Y, Li J, Liu Y, Zhang J, et al. . 2014. PFOS induced lipid metabolism disturbances in BALB/c mice through inhibition of low density lipoproteins excretion. Sci Rep 4(1):4582, PMID: , 10.1038/srep04582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu N, Wei S, Li M, Yang J, Li K, Jin L, et al. . 2016. Effects of perfluorooctanoic acid on metabolic profiles in brain and liver of mouse revealed by a high-throughput targeted metabolomics approach. Sci Rep 6:23963, PMID: , 10.1038/srep23963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gebreab KY, Eeza MNH, Bai T, Zuberi Z, Matysik J, O’Shea KE, et al. . 2020. Comparative toxicometabolomics of perfluorooctanoic acid (PFOA) and next-generation perfluoroalkyl substances. Environ Pollut 265(pt A):114928, PMID: , 10.1016/j.envpol.2020.114928. [DOI] [PubMed] [Google Scholar]
- 31.Alderete TL, Jin R, Walker DI, Valvi D, Chen Z, Jones DP, et al. . 2019. Perfluoroalkyl substances, metabolomic profiling, and alterations in glucose homeostasis among overweight and obese Hispanic children: a proof-of-concept analysis. Environ Int 126:445–453, PMID: , 10.1016/j.envint.2019.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Z, Yang T, Walker DI, Thomas DC, Qiu C, Chatzi L, et al. . 2020. Dysregulated lipid and fatty acid metabolism link perfluoroalkyl substances exposure and impaired glucose metabolism in young adults. Environ Int 145:106091, PMID: , 10.1016/j.envint.2020.106091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu X, Li S, Cirillo PM, Krigbaum NY, Tran V, Jones DP, et al. . 2019. Metabolome wide association study of serum poly and perfluoroalkyl substances (PFASs) in pregnancy and early postpartum. Reprod Toxicol 87:70–78, PMID: , 10.1016/j.reprotox.2019.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kingsley SL, Walker DI, Calafat AM, Chen A, Papandonatos GD, Xu Y, et al. . 2019. Metabolomics of childhood exposure to perfluoroalkyl substances: a cross-sectional study. Metabolomics 15(7):95, PMID: , 10.1007/s11306-019-1560-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y, Lu X, Yu N, Li A, Zhuang T, Du L, et al. . 2021. Exposure to legacy and novel perfluoroalkyl substance disturbs the metabolic homeostasis in pregnant women and fetuses: a metabolome-wide association study. Environ Int 156:106627, PMID: , 10.1016/j.envint.2021.106627. [DOI] [PubMed] [Google Scholar]
- 36.Lu Y, Gao K, Li X, Tang Z, Xiang L, Zhao H, et al. . 2019. Mass spectrometry-based metabolomics reveals occupational exposure to per- and polyfluoroalkyl substances relates to oxidative stress, fatty acid β-oxidation disorder, and kidney injury in a manufactory in China. Environ Sci Technol 53(16):9800–9809, PMID: , 10.1021/acs.est.9b01608. [DOI] [PubMed] [Google Scholar]
- 37.Mitro SD, Liu J, Jaacks LM, Fleisch AF, Williams PL, Knowler WC, et al. . 2021. Per- and polyfluoroalkyl substance plasma concentrations and metabolomic markers of type 2 diabetes in the Diabetes Prevention Program trial. Int J Hyg Environ Health 232:113680, PMID: , 10.1016/j.ijheh.2020.113680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salihovic S, Fall T, Ganna A, Broeckling CD, Prenni JE, Hyötyläinen T, et al. . 2019. Identification of metabolic profiles associated with human exposure to perfluoroalkyl substances. J Expo Sci Environ Epidemiol 29(2):196–205, PMID: , 10.1038/s41370-018-0060-y. [DOI] [PubMed] [Google Scholar]
- 39.Sen P, Qadri S, Luukkonen PK, Ragnarsdottir O, McGlinchey A, Jäntti S, et al. . 2022. Exposure to environmental contaminants is associated with altered hepatic lipid metabolism in non-alcoholic fatty liver disease. J Hepatol 76(2):283–293, PMID: , 10.1016/j.jhep.2021.09.039. [DOI] [PubMed] [Google Scholar]
- 40.Stratakis N, Conti DV, Jin R, Margetaki K, Valvi D, Siskos AP, et al. . 2020. Prenatal exposure to perfluoroalkyl substances associated with increased susceptibility to liver injury in children. Hepatology 72(5):1758–1770, PMID: , 10.1002/hep.31483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin R, McConnell R, Catherine C, Xu S, Walker DI, Stratakis N, et al. . 2020. Perfluoroalkyl substances and severity of nonalcoholic fatty liver in children: an untargeted metabolomics approach. Environ Int 134:105220, PMID: , 10.1016/j.envint.2019.105220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li S, Cirillo P, Hu X, Tran V, Krigbaum N, Yu S, et al. . 2020. Understanding mixed environmental exposures using metabolomics via a hierarchical community network model in a cohort of California women in 1960’s. Reprod Toxicol 92:57–65, PMID: , 10.1016/j.reprotox.2019.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matta K, Lefebvre T, Vigneau E, Cariou V, Marchand P, Guitton Y, et al. . 2022. Associations between persistent organic pollutants and endometriosis: a multiblock approach integrating metabolic and cytokine profiling. Environ Int 158:106926, PMID: , 10.1016/j.envint.2021.106926. [DOI] [PubMed] [Google Scholar]
- 44.Holeček M. 2018. Branched-chain amino acids in health and disease: metabolism, alterations in blood plasma, and as supplements. Nutr Metab (Lond) 15(1):33, PMID: , 10.1186/s12986-018-0271-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prawitt J, Caron S, Staels B. 2011. Bile acid metabolism and the pathogenesis of type 2 diabetes. Curr Diab Rep 11(3):160–166, PMID: , 10.1007/s11892-011-0187-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Di Ciaula A, Wang DQH, Molina-Molina E, Lunardi Baccetto R, Calamita G, Palmieri VO, et al. . 2017. Bile acids and cancer: direct and environmental-dependent effects. Ann Hepatol 16(suppl 1):S87–S105, PMID: , 10.5604/01.3001.0010.5501. [DOI] [PubMed] [Google Scholar]
- 47.Wei Z, Liu X, Cheng C, Yu W, Yi P. 2021. Metabolism of amino acids in cancer. Front Cell Dev Biol 8:603837, PMID: , 10.3389/fcell.2020.603837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Z, DeWitt JC, Higgins CP, Cousins IT. 2017. A never-ending story of per- and polyfluoroalkyl substances (PFASs)? Environ Sci Technol 51(5):2508–2518, PMID: , 10.1021/acs.est.6b04806. [DOI] [PubMed] [Google Scholar]
- 49.Alderete TL, Habre R, Toledo-Corral CM, Berhane K, Chen Z, Lurmann FW, et al. . 2017. Longitudinal associations between ambient air pollution with insulin sensitivity, β-cell function, and adiposity in Los Angeles Latino children. Diabetes 66(7):1789–1796, PMID: , 10.2337/db16-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goran MI, Bergman RN, Avila Q, Watkins M, Ball GDC, Shaibi GQ, et al. . 2004. Impaired glucose tolerance and reduced β-cell function in overweight Latino children with a positive family history for type 2 diabetes. J Clin Endocrinol Metab 89(1):207–212, PMID: , 10.1210/jc.2003-031402. [DOI] [PubMed] [Google Scholar]
- 51.Baumert BO, Goodrich JA, Hu X, Walker DI, Alderete TL, Chen Z, et al. . 2022. Plasma concentrations of lipophilic persistent organic pollutants and glucose homeostasis in youth populations. Environ Res 212(pt B):113296, PMID: , 10.1016/j.envres.2022.113296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim JS, Chen Z, Alderete TL, Toledo-Corral C, Lurmann F, Berhane K, et al. . 2019. Associations of air pollution, obesity and cardiometabolic health in young adults: the Meta-AIR study. Environ Int 133(pt A):105180, PMID: , 10.1016/j.envint.2019.105180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McConnell R, Shen E, Gilliland FD, Jerrett M, Wolch J, Chang CC, et al. . 2015. A longitudinal cohort study of body mass index and childhood exposure to secondhand tobacco smoke and air pollution: the Southern California Children’s Health Study. Environ Health Perspect 123(4):360–366, PMID: , 10.1289/ehp.1307031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.US Preventive Services Task Force, Grossman DC, Bibbins-Domingo K, Curry SJ, Barry MJ, Davidson KW, et al. . 2017. Screening for obesity in children and adolescents: US Preventive Services Task Force recommendation statement. JAMA 317(23):2417–2426, PMID: , 10.1001/jama.2017.6803. [DOI] [PubMed] [Google Scholar]
- 55.Hollingshead AB. 2011. Four factor index of social status. Yale J Sociol 8:21–52. [Google Scholar]
- 56.Berkelmans GFN, Read SH, Gudbjörnsdottir S, Wild SH, Franzen S, van der Graaf Y, et al. . 2022. Population median imputation was noninferior to complex approaches for imputing missing values in cardiovascular prediction models in clinical practice. J Clin Epidemiol 145:70–80, PMID: , 10.1016/j.jclinepi.2022.01.011. [DOI] [PubMed] [Google Scholar]
- 57.Marshall WA, Tanner JM. 1969. Variations in pattern of pubertal changes in girls. Arch Dis Child 44(235):291–303, PMID: , 10.1136/adc.44.235.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marshall WA, Tanner JM. 1970. Variations in the pattern of pubertal changes in boys. Arch Dis Child 45(239):13–23, PMID: , 10.1136/adc.45.239.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goodrich JA, Walker D, Lin X, Wang H, Lim T, McConnell R, et al. . 2022. Exposure to perfluoroalkyl substances and risk of hepatocellular carcinoma in a multiethnic cohort. JHEP Rep 4(10):100550, PMID: , 10.1016/j.jhepr.2022.100550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.CDC (Centers for Disease Control and Prevention). 2019. Fourth Report on Human Exposure to Environmental Chemicals, Updated Tables. Atlanta, GA: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention. [Google Scholar]
- 61.Goodpaster BH, Sparks LM. 2017. Metabolic flexibility in health and disease. Cell Metab 25(5):1027–1036, PMID: , 10.1016/j.cmet.2017.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huo S, Sun L, Zong G, Shen X, Zheng H, Jin Q, et al. . 2021. Changes in plasma metabolome profiles following oral glucose challenge among adult Chinese. Nutrients 13(5):1474, PMID: , 10.3390/nu13051474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu KH, Nellis M, Uppal K, Ma C, Tran V, Liang Y, et al. . 2020. Reference standardization for quantification and harmonization of large-scale metabolomics. Anal Chem 92(13):8836–8844, PMID: , 10.1021/acs.analchem.0c00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu Q, Walker D, Uppal K, Liu Z, Ma C, Tran V, et al. . 2020. Addressing the batch effect issue for LC/MS metabolomics data in data preprocessing. Sci Rep 10(1):13856, PMID: , 10.1038/s41598-020-70850-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Luan H, Ji F, Chen Y, Cai Z. 2018. statTarget: a streamlined tool for signal drift correction and interpretations of quantitative mass spectrometry-based omics data. Anal Chim Acta 1036:66–72, PMID: , 10.1016/j.aca.2018.08.002. [DOI] [PubMed] [Google Scholar]
- 66.Keil AP, Daza EJ, Engel SM, Buckley JP, Edwards JK. 2018. A Bayesian approach to the g-formula. Stat Methods Med Res 27(10):3183–3204, PMID: , 10.1177/0962280217694665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Keil AP, Buckley JP, Kalkbrenner AE. 2021. Bayesian g-computation for estimating impacts of interventions on exposure mixtures: demonstration with metals from coal-fired power plants and birth weight. Am J Epidemiol 190(12):2647–2657, PMID: , 10.1093/aje/kwab053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Keil AP, Buckley JP, O’Brien KM, Ferguson KK, Zhao S, White AJ. 2020. A quantile-based g-computation approach to addressing the effects of exposure mixtures. Environ Health Perspect 128(4):47004, PMID: , 10.1289/EHP5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maruyama Y, George EI. 2011. Fully Bayes factors with a generalized g-prior. Ann Statist 39(5):2740–2765, 10.1214/11-AOS917. [DOI] [Google Scholar]
- 70.Bobb JF, Valeri L, Claus Henn B, Christiani DC, Wright RO, Mazumdar M, et al. . 2015. Bayesian kernel machine regression for estimating the health effects of multi-pollutant mixtures. Biostatistics 16(3):493–508, PMID: , 10.1093/biostatistics/kxu058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bottolo L, Richardson S. 2010. Evolutionary stochastic search for Bayesian model exploration. Bayesian Anal 5(3):583–618, 10.1214/10-BA523. [DOI] [Google Scholar]
- 72.Hanson TE, Branscum AJ, Johnson WO. 2014. Informative g-priors for logistic regression. Bayesian Anal 9(3):597–612, 10.1214/14-BA868. [DOI] [Google Scholar]
- 73.Newcombe PJ, Conti DV, Richardson S. 2016. JAM: a scalable Bayesian framework for joint analysis of marginal SNP effects. Genet Epidemiol 40(3):188–201, PMID: , 10.1002/gepi.21953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zellner A. 1986. On assessing prior distributions and Bayesian regression analysis with g-prior distributions. In: Bayesian Inference and Decision Techniques: Essays in Honor of Bruno de Finetti. Goel P, Zellner A, eds. New York, NY: Elsevier Science Publishers, 233–243. [Google Scholar]
- 75.Liang F, Paulo R, Molina G, Clyde MA, Berger JO. 2008. Mixtures of g priors for Bayesian variable selection. J Am Stat Assoc 103(481):410–423, 10.1198/016214507000001337. [DOI] [Google Scholar]
- 76.Li Y, Clyde MA. 2018. Mixtures of g-priors in generalized linear models. J Am Stat Assoc 113(524):1828–1845, 10.1080/01621459.2018.1469992. [DOI] [Google Scholar]
- 77.Greenland S. 1992. A semi-Bayes approach to the analysis of correlated multiple associations, with an application to an occupational cancer-mortality study. Stat Med 11(2):219–230, PMID: , 10.1002/sim.4780110208. [DOI] [PubMed] [Google Scholar]
- 78.George E, McCulloch R. 1995. Stochastic search variable selection. In: Markov Chain Monte Carlo in Practice. Gilks WR, Richardson S, Spiegelhalter D, eds. Boca Raton, FL: Chapman and Hall/CRC, 203–214. [Google Scholar]
- 79.Plummer M, Stukalov A, Denwood M. 2021. rjags: Bayesian graphical models using MCMC. https://CRAN.R-project.org/package=rjags [accessed 18 January 2022].
- 80.Perrakis K, Ntzoufras I. 2018. Bayesian variable selection using the hyper-g prior in WinBUGS. Wiley Interdiscip Rev Comput Stat 10(6):e1442, 10.1002/wics.1442. [DOI] [Google Scholar]
- 81.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57(1):289–300, 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 82.Pang Z, Chong J, Zhou G, de Lima Morais DA, Chang L, Barrette M, et al. . 2021. MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res 49(W1):W388–W396, PMID: , 10.1093/nar/gkab382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pang Z, Chong J, Li S, Xia J. 2020. MetaboAnalystR 3.0: toward an optimized workflow for global metabolomics. Metabolites 10(5):186, PMID: , 10.3390/metabo10050186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li S, Pozhitkov A, Ryan RA, Manning CS, Brown-Peterson N, Brouwer M. 2010. Constructing a fish metabolic network model. Genome Biol 11(11):R115, PMID: , 10.1186/gb-2010-11-11-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chong J, Wishart DS, Xia J. 2019. Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr Protoc Bioinformatics 68(1):e86, PMID: , 10.1002/cpbi.86. [DOI] [PubMed] [Google Scholar]
- 86.Li S, Park Y, Duraisingham S, Strobel FH, Khan N, Soltow QA, et al. . 2013. Predicting network activity from high throughput metabolomics. PLOS Comput Biol 9(7):e1003123, PMID: , 10.1371/journal.pcbi.1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. . 2005. Gene Set Enrichment Analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102(43):15545–15550, PMID: , 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zaykin DV. 2011. Optimally weighted Z-test is a powerful method for combining probabilities in meta-analysis. J Evol Biol 24(8):1836–1841, PMID: , 10.1111/j.1420-9101.2011.02297.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bravata DM, Olkin I. 2001. Simple pooling versus combining in meta-analysis. Eval Health Prof 24(2):218–230, PMID: , 10.1177/01632780122034885. [DOI] [PubMed] [Google Scholar]
- 90.Brennan NM, Evans AT, Fritz MK, Peak SA, von Holst HE. 2021. Trends in the regulation of per- and polyfluoroalkyl substances (PFAS): a scoping review. Int J Environ es Public Health 18(20):10900, PMID: , 10.3390/ijerph182010900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kwiatkowski CF, Andrews DQ, Birnbaum LS, Bruton TA, DeWitt JC, Knappe DRU, et al. . 2020. Scientific basis for managing PFAS as a chemical class. Environ Sci Technol Lett 7(8):532–543, PMID: , 10.1021/acs.estlett.0c00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Preston EV, Webster TF, Claus Henn B, McClean MD, Gennings C, Oken E, et al. . 2020. Prenatal exposure to per- and polyfluoroalkyl substances and maternal and neonatal thyroid function in the Project Viva Cohort: a mixtures approach. Environ Int 139:105728, PMID: , 10.1016/j.envint.2020.105728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Coperchini F, Croce L, Ricci G, Magri F, Rotondi M, Imbriani M, et al. . 2021. Thyroid disrupting effects of old and new generation PFAS. Front Endocrinol (Lausanne) 11:612320, PMID: , 10.3389/fendo.2020.612320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Boelaert K, Franklyn JA. 2005. Thyroid hormone in health and disease. J Endocrinol 187(1):1–15, PMID: , 10.1677/joe.1.06131. [DOI] [PubMed] [Google Scholar]
- 95.Sinha RA, Singh BK, Yen PM. 2018. Direct effects of thyroid hormones on hepatic lipid metabolism. Nat Rev Endocrinol 14(5):259–269, PMID: , 10.1038/nrendo.2018.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ameer F, Scandiuzzi L, Hasnain S, Kalbacher H, Zaidi N. 2014. De novo lipogenesis in health and disease. Metabolism 63(7):895–902, PMID: , 10.1016/j.metabol.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 97.Wang B, Wang B, Yang Y, Xu J, Hong M, Xia M, et al. . 2021. Thyroid function and non-alcoholic fatty liver disease in hyperthyroidism patients. BMC Endocr Disord 21(1):27, PMID: , 10.1186/s12902-021-00694-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Khan SR, Chaker L, Ruiter R, Aerts JGJV, Hofman A, Dehghan A, et al. . 2016. Thyroid function and cancer risk: the Rotterdam Study. J Clin Endocrinol Metab 101(12):5030–5036, PMID: , 10.1210/jc.2016-2104. [DOI] [PubMed] [Google Scholar]
- 99.Kalra S, Aggarwal S, Khandelwal D. 2019. Thyroid dysfunction and type 2 diabetes mellitus: screening strategies and implications for management. Diabetes Ther 10(6):2035–2044, PMID: , 10.1007/s13300-019-00700-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Eggers Pedersen K, Basu N, Letcher R, Greaves AK, Sonne C, Dietz R, et al. . 2015. Brain region-specific perfluoroalkylated sulfonate (PFSA) and carboxylic acid (PFCA) accumulation and neurochemical biomarker responses in east Greenland polar bears (Ursus maritimus). Environ Res 138:22–31, PMID: , 10.1016/j.envres.2015.01.015. [DOI] [PubMed] [Google Scholar]
- 101.Foguth RM, Flynn RW, de Perre C, Iacchetta M, Lee LS, Sepúlveda MS, et al. . 2019. Developmental exposure to perfluorooctane sulfonate (PFOS) and perfluorooctanoic acid (PFOA) selectively decreases brain dopamine levels in Northern leopard frogs. Toxicol Appl Pharmacol 377:114623, PMID: , 10.1016/j.taap.2019.114623. [DOI] [PubMed] [Google Scholar]
- 102.Grønnestad R, Schlenk D, Krøkje Å, Jaspers VLB, Jenssen BM, Coffin S, et al. . 2021. Alteration of neuro-dopamine and steroid hormone homeostasis in wild Bank voles in relation to tissue concentrations of PFAS at a Nordic skiing area. Sci Total Environ 756:143745, PMID: , 10.1016/j.scitotenv.2020.143745. [DOI] [PubMed] [Google Scholar]
- 103.Grønnestad R, Johanson SM, Müller MHB, Schlenk D, Tanabe P, Krøkje Å, et al. . 2021. Effects of an environmentally relevant PFAS mixture on dopamine and steroid hormone levels in exposed mice. Toxicol Appl Pharmacol 428:115670, PMID: , 10.1016/j.taap.2021.115670. [DOI] [PubMed] [Google Scholar]
- 104.Long Y, Wang Y, Ji G, Yan L, Hu F, Gu A. 2013. Neurotoxicity of perfluorooctane sulfonate to hippocampal cells in adult mice. PLoS One 8(1):e54176, PMID: , 10.1371/journal.pone.0054176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Salgado R, López-Doval S, Pereiro N, Lafuente A. 2016. Perfluorooctane sulfonate (PFOS) exposure could modify the dopaminergic system in several limbic brain regions. Toxicol Lett 240(1):226–235, PMID: , 10.1016/j.toxlet.2015.10.023. [DOI] [PubMed] [Google Scholar]
- 106.Salgado RM, Sheard AC, Vaughan RA, Parker DL, Schneider SM, Kenefick RW, et al. . 2017. Mitochondrial efficiency and exercise economy following heat stress: a potential role of uncoupling protein 3. Physiol Rep 5(3):e13054, PMID: , 10.14814/phy2.13054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Daubner SC, Le T, Wang S. 2011. Tyrosine hydroxylase and regulation of dopamine synthesis. Arch Biochem Biophys 508(1):1–12, PMID: , 10.1016/j.abb.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Slotkin TA, MacKillop EA, Melnick RL, Thayer KA, Seidler FJ. 2008. Developmental neurotoxicity of perfluorinated chemicals modeled in vitro. Environ Health Perspect 116(6):716–722, PMID: , 10.1289/ehp.11253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Berends AMA, Eisenhofer G, Fishbein L, Horst-Schrivers ANAVD, Kema IP, Links TP, et al. . 2019. Intricacies of the molecular machinery of catecholamine biosynthesis and secretion by chromaffin cells of the normal adrenal medulla and in pheochromocytoma and paraganglioma. Cancers (Basel) 11(8):1121, PMID: , 10.3390/cancers11081121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Goldstein DS, Eisenhofer G, Kopin IJ. 2003. Sources and significance of plasma levels of catechols and their metabolites in humans. J Pharmacol Exp Ther 305(3):800–811, PMID: , 10.1124/jpet.103.049270. [DOI] [PubMed] [Google Scholar]
- 111.Patel R, Bradner JM, Stout KA, Caudle WM. 2016. Alteration to dopaminergic synapses following exposure to perfluorooctane sulfonate (PFOS), in vitro and in vivo. Med Sci (Basel) 4(3):13, PMID: , 10.3390/medsci4030013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bonato M, Corrà F, Bellio M, Guidolin L, Tallandini L, Irato P, et al. . 2020. PFAS environmental pollution and antioxidant responses: an overview of the impact on human field. Int J Environ Res Public Health 17(21):8020, PMID: , 10.3390/ijerph17218020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Omoike OE, Pack RP, Mamudu HM, Liu Y, Strasser S, Zheng S, et al. . 2021. Association between per and polyfluoroalkyl substances and markers of inflammation and oxidative stress. Environ Res 196:110361, PMID: , 10.1016/j.envres.2020.110361. [DOI] [PubMed] [Google Scholar]
- 114.Tallima H, El Ridi R. 2018. Arachidonic acid: physiological roles and potential health benefits—a review. J Adv Res 11:33–41, PMID: , 10.1016/j.jare.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang B, Wu L, Chen J, Dong L, Chen C, Wen Z, et al. . 2021. Metabolism pathways of arachidonic acids: mechanisms and potential therapeutic targets. Signal Transduct Target Ther 6(1):94, PMID: , 10.1038/s41392-020-00443-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wijekoon EP, Skinner C, Brosnan ME, Brosnan JT. 2004. Amino acid metabolism in the Zucker diabetic fatty rat: effects of insulin resistance and of type 2 diabetes. Can J Physiol Pharmacol 82(7):506–514, PMID: , 10.1139/y04-067. [DOI] [PubMed] [Google Scholar]
- 117.Zeitoun-Ghandour S, Leszczyszyn OI, Blindauer CA, Geier FM, Bundy JG, Stürzenbaum SR. 2011. C. elegans metallothioneins: response to and defence against ROS toxicity. Mol Biosyst 7(8):2397–2406, PMID: , 10.1039/c1mb05114h. [DOI] [PubMed] [Google Scholar]
- 118.Duranton F, Cohen G, De Smet R, Rodriguez M, Jankowski J, Vanholder R, et al. . 2012. Normal and pathologic concentrations of uremic toxins. J Am Soc Nephrol 23(7):1258–1270, PMID: , 10.1681/ASN.2011121175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lim YJ, Sidor NA, Tonial NC, Che A, Urquhart BL. 2021. Uremic toxins in the progression of chronic kidney disease and cardiovascular disease: mechanisms and therapeutic targets. Toxins 13(2):142, PMID: , 10.3390/toxins13020142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Calder PC. 2007. Dietary arachidonic acid: harmful, harmless or helpful? Br J Nutr 98(3):451–453, PMID: , 10.1017/S0007114507761779. [DOI] [PubMed] [Google Scholar]
- 121.Lewis RJ. 2007. Hawley’s Condensed Chemical Dictionary. 15th ed. Hoboken, NJ: Wiley. [Google Scholar]
- 122.Roe M, Pinchen H, Church S, Elahi S, Walker M, Farron-Wilson M, et al. . 2013. Trans fatty acids in a range of UK processed foods. Food Chem 140(3):427–431, PMID: , 10.1016/j.foodchem.2012.08.067. [DOI] [PubMed] [Google Scholar]
- 123.Schlezinger JJ, Hyötyläinen T, Sinioja T, Boston C, Puckett H, Oliver J, et al. . 2021. Perfluorooctanoic acid induces liver and serum dyslipidemia in humanized PPARα mice fed an American diet. Toxicol Appl Pharmacol 426:115644, PMID: , 10.1016/j.taap.2021.115644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lai KP, Li JW, Cheung A, Li R, Billah MB, Chan TF, et al. . 2017. Transcriptome sequencing reveals prenatal PFOS exposure on liver disorders. Environ Pollut 223:416–425, PMID: , 10.1016/j.envpol.2017.01.041. [DOI] [PubMed] [Google Scholar]
- 125.Cheng J, Lv S, Nie S, Liu J, Tong S, Kang N, et al. . 2016. Chronic perfluorooctane sulfonate (PFOS) exposure induces hepatic steatosis in zebrafish. Aquat Toxicol 176:45–52, PMID: , 10.1016/j.aquatox.2016.04.013. [DOI] [PubMed] [Google Scholar]
- 126.Jacobsen AV, Nordén M, Engwall M, Scherbak N. 2018. Effects of perfluorooctane sulfonate on genes controlling hepatic fatty acid metabolism in livers of chicken embryos. Environ Sci Pollut Res Int 25(23):23074–23081, PMID: , 10.1007/s11356-018-2358-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Souza-Mello V. 2015. Peroxisome proliferator-activated receptors as targets to treat non-alcoholic fatty liver disease. World J Hepatol 7(8):1012–1019, PMID: , 10.4254/wjh.v7.i8.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Saliani N, Darabi M, Yousefi B, Baradaran B, Khaniani MS, Darabi M, et al. . 2013. PPARγ agonist-induced alterations in Δ6-desaturase and stearoyl-CoA desaturase 1: role of MEK/ERK1/2 pathway. World J Hepatol 5(4):220–225, PMID: , 10.4254/wjh.v5.i4.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Roth K, Imran Z, Liu W, Petriello MC. 2020. Diet as an exposure source and mediator of per- and polyfluoroalkyl substance (PFAS) toxicity. Front Toxicol 2:601149, PMID: , 10.3389/ftox.2020.601149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rappazzo KM, Coffman E, Hines EP. 2017. Exposure to perfluorinated alkyl substances and health outcomes in children: a systematic review of the epidemiologic literature. Int J Environ Res Public Health 14(7):691, PMID: , 10.3390/ijerph14070691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Schymanski EL, Jeon J, Gulde R, Fenner K, Ruff M, Singer HP, et al. . 2014. Identifying small molecules via high resolution mass spectrometry: communicating confidence. Environ Sci Technol 48(4):2097–2098, PMID: , 10.1021/es5002105. [DOI] [PubMed] [Google Scholar]
- 132.Schrimpe-Rutledge AC, Codreanu SG, Sherrod SD, McLean JA. 2016. Untargeted metabolomics strategies—challenges and emerging directions. J Am Soc Mass Spectrom 27(12):1897–1905, PMID: , 10.1007/s13361-016-1469-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Olsen GW, Burris JM, Ehresman DJ, Froehlich JW, Seacat AM, Butenhoff JL, et al. . 2007. Half-life of serum elimination of perfluorooctanesulfonate, perfluorohexanesulfonate, and perfluorooctanoate in retired fluorochemical production workers. Environ Health Perspect 115(9):1298–1305, PMID: , 10.1289/ehp.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Susmann HP, Schaider LA, Rodgers KM, Rudel RA. 2019. Dietary habits related to food packaging and population exposure to PFASs. Environ Health Perspect 127(10):107003, PMID: , 10.1289/EHP4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Forsthuber M, Kaiser AM, Granitzer S, Hassl I, Hengstschläger M, Stangl H, et al. . 2020. Albumin is the major carrier protein for PFOS, PFOA, PFHxS, PFNA and PFDA in human plasma. Environ Int 137:105324, PMID: , 10.1016/j.envint.2019.105324. [DOI] [PubMed] [Google Scholar]
- 136.Sagiv SK, Rifas-Shiman SL, Fleisch AF, Webster TF, Calafat AM, Ye X, et al. . 2018. Early-pregnancy plasma concentrations of perfluoroalkyl substances and birth outcomes in Project Viva: confounded by pregnancy hemodynamics? Am J Epidemiol 187(4):793–802, PMID: , 10.1093/aje/kwx332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Nyström J, Benskin JP, Plassmann M, Sandblom O, Glynn A, Lampa E, et al. . 2022. Demographic, life-style and physiological determinants of serum per- and polyfluoroalkyl substance (PFAS) concentrations in a national cross-sectional survey of Swedish adolescents. Environ Res 208:112674, PMID: , 10.1016/j.envres.2022.112674. [DOI] [PubMed] [Google Scholar]
- 138.Jain RB, Ducatman A. 2019. Perfluoroalkyl substances follow inverted U-shaped distributions across various stages of glomerular function: implications for future research. Environ Res 169:476–482, PMID: , 10.1016/j.envres.2018.11.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.