Abstract
The Escherichia coli umuDC gene products encode DNA polymerase V, which participates in both translesion DNA synthesis (TLS) and a DNA damage checkpoint control. These two temporally distinct roles of the umuDC gene products are regulated by RecA–single-stranded DNA-facilitated self-cleavage of UmuD (which participates in the checkpoint control) to yield UmuD′ (which enables TLS). In addition, even modest overexpression of the umuDC gene products leads to a cold-sensitive growth phenotype, apparently due to the inappropriate expression of the DNA damage checkpoint control activity of UmuD2C. We have previously reported that overexpression of the ɛ proofreading subunit of DNA polymerase III suppresses umuDC-mediated cold sensitivity, suggesting that interaction of ɛ with UmuD2C is important for the DNA damage checkpoint control function of the umuDC gene products. Here, we report that overexpression of the β processivity clamp of the E. coli replicative DNA polymerase (encoded by the dnaN gene) not only exacerbates the cold sensitivity conferred by elevated levels of the umuDC gene products but, in addition, confers a severe cold-sensitive phenotype upon a strain expressing moderately elevated levels of the umuD′C gene products. Such a strain is not otherwise normally cold sensitive. To identify mutant β proteins possibly deficient for physical interactions with the umuDC gene products, we selected for novel dnaN alleles unable to confer a cold-sensitive growth phenotype upon a umuD′C-overexpressing strain. In all, we identified 75 dnaN alleles, 62 of which either reduced the expression of β or prematurely truncated its synthesis, while the remaining alleles defined eight unique missense mutations of dnaN. Each of the dnaN missense mutations retained at least a partial ability to function in chromosomal DNA replication in vivo. In addition, these eight dnaN alleles were also unable to exacerbate the cold sensitivity conferred by modestly elevated levels of the umuDC gene products, suggesting that the interactions between UmuD′ and β are a subset of those between UmuD and β. Taken together, these findings suggest that interaction of β with UmuD2C is important for the DNA damage checkpoint function of the umuDC gene products. Four possible models for how interactions of UmuD2C with the ɛ and the β subunits of DNA polymerase III might help to regulate DNA replication in response to DNA damage are discussed.
The umuDC genes encode a DNA polymerase, DNA polymerase V (Pol V), that has a remarkable ability to copy over abasic sites (43, 63), cyclobutane dimers (62), and pyrimidine-pyrimidone [6-4] photoproducts (62), a process referred to as translesion DNA synthesis (TLS). This ability, however, comes at the cost of reduced fidelity. Thus, replication by Pol V is inherently less accurate, leading to the formation of mutations, even when replicating undamaged templates (29, 62). Therefore, to limit the ability of Pol V to introduce mutations into the host genome, expression of the umuDC genes is tightly regulated as part of the Escherichia coli stress-induced SOS response (12). This highly regulated response helps the cell maintain the integrity of its genome following treatments that lead either directly or indirectly to DNA damage (12).
The E. coli SOS response consists of at least 30 unlinked genes (9, 12), collectively referred to as the SOS regulon. Expression of the various SOS-regulated genes is coordinately regulated at the level of their transcription by the LexA and RecA proteins (28). In the absence of DNA damage, LexA acts to repress the expression of the members of the SOS regulon (27). RecA protein, the main bacterial recombinase required for essentially all homologous recombination (reviewed in reference 22), binds to single-stranded DNA (ssDNA) generated by the cell's failed attempts to replicate past lesions in its genome, thus forming RecA-ssDNA nucleoprotein filaments (47). These RecA-ssDNA filaments, in addition to acting in homologous recombination, also act to facilitate the latent capacity of LexA to autodigest (26). Autodigestion of LexA serves to inactivate it as a transcriptional repressor, leading to the concomitant increase in expression of LexA-regulated genes (12).
The UmuD protein similarly undergoes a RecA-ssDNA-facilitated autodigestion that serves to remove its first 24 residues to yield UmuD′ (3, 34, 48). The UmuD′2 homodimer then interacts with UmuC (18, 59, 68), which has an ability to catalyze the formation of phosphodiester bonds, in such a way that the UmuD′2C complex is able to participate in TLS (43). In addition to participating in TLS, UmuC together with the full-length UmuD2 homodimer participates in a DNA damage checkpoint control that acts to regulate DNA replication in response to DNA damage, thereby allowing additional time for nucleotide excision repair to accurately remove lesions in the DNA prior to continued replication (37). Thus, self-cleavage of UmuD to UmuD′ can be regarded as a molecular switch that acts to temporally regulate these two distinct activities of the UmuD2C and UmuD′2C complexes (37, 57).
In addition to participating in a DNA damage checkpoint control and enabling TLS, overexpression of the umuDC gene products confers a cold sensitivity for growth (30, 38, 59). Our recent characterizations of umuDC-mediated cold sensitivity indicated that (i) moderately elevated levels of the umuDC gene products confer a cold-sensitive growth phenotype, while similarly elevated levels of the umuD′C gene products do not (59), and (ii) the catalytic DNA polymerase activity of UmuC is not required for this cold sensitivity (59). These findings, together with others, suggest that the cold sensitivity conferred by elevated levels of the umuDC gene products is a manifestation of the inappropriate expression of UmuD2C functions involved in the DNA damage checkpoint control (37, 38, 59). Therefore, in an effort to better characterize the components of the UmuD2C-dependent checkpoint control, we have embarked on an analysis of the genetic requirements of umuDC-mediated cold sensitivity.
We have previously suggested that interactions of the umuDC gene products with components of the E. coli replicative DNA polymerase, DNA polymerase III holoenzyme (Pol III), could serve as a convenient mechanism for regulating the checkpoint and TLS roles of the umuDC gene products (37, 57). On the basis of this hypothesis, we reasoned that if interactions involving the umuDC gene products and components of Pol III were important for the checkpoint role of UmuD2C, we might then be able to observe an effect on the extent of the cold sensitivity conferred by umuDC by the simultaneous overexpression of certain (i.e., relevant) components of Pol III. Using such an approach, we have recently reported that overproduction of the ɛ proofreading subunit of Pol III or deletion of its structural gene (dnaQ) suppresses umuDC-mediated cold sensitivity (56). A systematic analysis of the remaining nine Pol III subunits indicated that the homodimeric β processivity clamp was the only other Pol III subunit that when overexpressed affected the extent of umuDC-mediated cold sensitivity (56).
In this report, we describe how overexpression of the β processivity clamp (encoded by dnaN) strongly exacerbates umuDC-mediated cold sensitivity. We have exploited this ability of the β clamp to confer a cold-sensitive growth phenotype upon a umuD′C-expressing E. coli strain (a strain that is not normally cold sensitive [59]) to identify novel dnaN alleles unable to confer this phenotype. Our genetic characterizations of these novel dnaN alleles indicate that they are also unable to exacerbate the cold sensitivity conferred by elevated levels of the umuDC gene products. Our results described in this report, taken together with others (37, 38, 56, 57, 59), suggest that the UmuD2C-dependent DNA damage checkpoint control is a manifestation of protein-protein interactions involving UmuD2C and the ɛ and the β subunits of Pol III.
MATERIALS AND METHODS
Bacteriological techniques.
E. coli strains and plasmid DNAs used in this study are described in Table 1. Bacterial strains were routinely grown in Luria-Bertani (LB) medium as described previously (46), unless otherwise stated. When necessary, LB medium was supplemented with the following antibiotics at the indicated concentrations: ampicillin, 150 μg/ml; spectinomycin, 60 μg/ml; and kanamycin, 80 μg/ml. Because strains expressing elevated levels of the umuDC gene products do not grow well at 30°C (30, 38, 59), the strains bearing a umuDC-expressing plasmid used in this study were routinely grown at 42°C, unless otherwise stated. pGYD′ΔC was constructed by the same method used for construction of pGYDΔC (59). Briefly, pGY9738 was digested with MluI, followed by end filling of the linear DNA with all four deoxynucleoside triphosphates and the Klenow fragment of DNA Pol I (New England BioLabs) prior to its ligation with T4 DNA ligase (New England BioLabs). Bacterial transformation was by either calcium chloride treatment (46) or electroporation using a GenePulser (Bio-Rad) as per the manufacturer's recommendation. Plasmid DNAs were isolated using the QIA-Spin Prep kit (Qiagen) as per the manufacturer's recommendation.
TABLE 1.
E. coli strains and plasmid DNAs used in this study
| Strain or plasmid | Relevant genotype or characteristics | Source (reference) |
|---|---|---|
| E. coli strainsa | ||
| AB1157 | lexA+ recA+ dnaN+ | Laboratory stock |
| HC123 | lexA+ recA+ dnaN59(Ts) | CGSCb |
| Plasmid DNAs | ||
| pBR322 | ColE1 oriV; cloning vector | Laboratory stock |
| pJRC210 | pBR322; β overproducer | Charles McHenry |
| pGB2 | pSC101; cloning vector | Roger Woodgate (5) |
| pSE115 | pSC101; o+umuD+ umuC+ | Laboratory stock (8) |
| pGY9739 | pSC101; oc1umuD+ umuC+ | Suzanne Sommer and Adriana Bailone (52) |
| pGYDΔC | pSC101; oc1umuD+ ΔumuC | Laboratory stock (59) |
| pGY9738 | pSC101; oc1umuD′ umuC+ | Suzanne Sommer and Adriana Bailone (51) |
| pGYD′ΔC | pSC101; oc1umuD′ ΔumuC | This work |
The complete genotype for AB1157 is xyl-5 mtl-1 galK2 rpsL31 kdgK51 lacY1 tsx-33 supE44 thi-1 leuB6 hisG4(Oc) mgl-51 argE3(Oc) rfbD1 proA2 ara-14 thr-1 qsr′ qin-111. The complete genotype for HC123 is fhuA22 ompF627 relA1 metB1 dnaN59(Ts) Hfr Cavalli PO2A.
CGSC, E. coli Genetic Stock Center.
Genetic assay for the selection of novel dnaN alleles.
A derivative of AB1157 bearing the plasmid pGY9738, which constitutively expresses elevated levels of the umuD′C gene products (51), grows well at 30°C. However, if AB1157(pGY9738) also carries the compatible plasmid pJRC210, which overexpresses the β processivity clamp of Pol III from the Ptac promoter, its growth is cold sensitive (Table 2). To select for dnaN alleles unable to confer the cold-sensitive phenotype, we selected for derivatives of AB1157 bearing both the umuD′C-expressing plasmid and the β-expressing plasmid that were able to grow at 30°C. In this analysis, we used a derivative of the β-expressing plasmid (pJRC210) that had been chemically mutagenized in vitro with hydroxylamine as described previously (31) before transformation into AB1157(pGY9738). To minimize the isolation of siblings, transformation reaction mixtures were aliquoted prior to their outgrowth; automated DNA sequence analysis revealed that all of the dnaN missense alleles identified by this approach arose independently. Following a 60-min outgrowth period, a portion of each aliquot was plated at 42°C (the permissive temperature) to permit a calculation of the total number of transformants obtained. The remainder of each transformation reaction mixture was plated directly at 30°C to select for plasmid-encoded dnaN alleles that allowed growth at this otherwise nonpermissive temperature. To confirm that growth at 30°C was attributable to the plasmid-encoded dnaN allele, plasmid DNAs were purified and subsequently retransformed back into AB1157 bearing the umuD′C-expressing plasmid (pGY9738). One half of each transformation reaction mixture was plated at 30°C, and the other half was plated at 42°C. Plates were scored after incubation overnight (∼20 h).
TABLE 2.
Overexpression of β exacerbates the cold sensitivity conferred by elevated levels of the umuDC gene productsa
| Plasmid | umuD and umuC alleles | Plating efficiency (30°C/42°C) prior to transformation with pBR322 or pJRC210b | AB1157 transformants/ml (30°C/42°C)c with:
|
|
|---|---|---|---|---|
| pBR322 (control) | pJRC210 (Ptac dnaN) | |||
| pGB2 | None | 1.08 | 0.99 | 0.87 |
| pSE115 | o+umuD+ umuC+ | NDd | 1.11 | 0.81 |
| pGY9739 | oc1umuD+ umuC+ | 4.5 × 10−3 | 3.5 × 10−3 | 5.1 × 10−4 |
| pGYDΔC | oc1umuD+ ΔumuC | 0.70 | 0.92 | 0.92 |
| pGY9738 | oc1umuD′ umuC+ | 0.94 | 0.98 | 2.6 × 10−4 |
| pGYD′ΔC | oc1umuD′ ΔumuC | ND | 0.77 | 0.85 |
E. coli AB1157 and plasmid DNAs are described in Table 1.
Plating efficiency is expressed as the ratio of the CFU obtained at 30°C to those obtained at 42°C after overnight incubation. These values were reported previously (59) and are included here only for comparison to the values obtained following transformation with either pBR322 or pJRC210.
AB1157 bearing the indicated umuDC-expressing plasmid was transformed with either pBR322 or pJRC210. The ratio of the CFU obtained at 30°C to those obtained at 42°C was measured after overnight incubation at each temperature. The dnaN gene encodes the β processivity clamp of Pol III (4). The strains expressing only elevated levels of β formed colonies that were slightly smaller at 30°C than those formed by strains not expressing β. Transformation efficiencies at 42°C for all strains were on the order of ∼105 to ∼106 CFU per μg of plasmid DNA, indicating that overexpression of either the umuDC or umuD′C gene products with or without simultaneous overexpression of β is not lethal at 42°C under these conditions.
ND, not determined.
It should be stressed that high-level overproduction of β by addition of IPTG (isopropyl-β-d-thiogalactopyranoside) was not necessary for expression of the synthetic lethal phenotype. The noninduced level of β expressed from the Ptac promoter of pJRC210 was sufficient. Consequently, all of the experiments described in this report designed to measure the phenotypes of the wild-type and mutant dnaN alleles were performed in the absence of added IPTG, unless otherwise stated.
Nucleotide sequence analysis of dnaN alleles.
The promoter region and entire coding sequence for each plasmid-encoded dnaN allele were determined by automated DNA sequence analysis using at least four of the following six different oligonucleotide primers (GIBCO-BRL) corresponding to the indicated nucleotide positions (with position 1 corresponding to the first base of the coding sequence of dnaN): BETA-R1, positions 292 to 331 of the noncoding strand; BETA-F2, positions 236 to 258 of the coding strand; BETA-F3, positions 815 to 836 of the coding strand; BETA-R4, positions 879 to 901 of the noncoding strand; BETA-F5, positions 97 to 120 of the coding strand; and BETA-R6, positions 394 to 415 of the noncoding strand. Analyses of the nucleotide sequences were performed using the Lasergene software package (version 3.6.0), and mutations were identified by comparison of the sequence data files to the wild-type dnaN sequence deposited in GenBank (accession number J01602).
RESULTS
Overexpression of the β processivity clamp of Pol III exacerbates umuDC-mediated cold sensitivity.
We have previously suggested that interaction of UmuD with the β processivity clamp of Pol III is important for the checkpoint role of UmuD2C (57). We have also suggested that the cold sensitivity conferred by elevated levels of the umuDC gene products is due to the inappropriate expression of UmuD2C functions involved in this checkpoint (59). To explore this hypothesis further, we used the previously described quantitative transformation assay (38) to investigate whether or not overexpression of the β clamp would affect the extent of the cold sensitivity conferred by elevated levels of the umuDC gene products. In these experiments, elevated levels of the umuDC gene products were supplied from a pSC101 derivative named pGY9739 (Table 1) (52) that contains a single base change in the operator site near the umuDC promoter (the oc1 mutation). This mutation eliminates much of the repression normally conferred by the LexA repressor (52). As controls, we used either pGB2, the parent to pGY9739 which lacks a umuDC operon, as well as a comparable pSC101-based plasmid named pSE115, which expresses the umuDC gene products from their wild-type, LexA-regulated promoter (8). Thus, in AB1157, expression of the umuDC gene products from pSE115 is efficiently blocked by LexA protein, while expression of the umuDC gene products from the oc1umuDC allele contained in pGY9739 is not.
In this analysis, competent cells of AB1157 bearing either pGY9739, pGB2, or pSE115 were prepared following their growth at 42°C. Consistent with our previous observations that overexpression of umuDC causes cold sensitivity (59), AB1157 bearing pGY9739 does not grow well at 30°C (Table 2). We then transformed these two AB1157 derivatives with either pBR322 (as a control) or a pBR322 derivative that expresses the β processivity clamp of Pol III from the Ptac promoter (pJRC210); no IPTG was added to induce β expression in these experiments. Following transformation and a 60-min period of outgrowth at 42°C, the reaction mixtures were split and aliquots were plated at both 30 and 42°C. As expected, the extent of the growth defect at 30°C of the AB1157 derivative bearing pGY9739, which expresses elevated levels of the umuDC gene products, was unaffected by transformation with pBR322; it still plated nearly 300-fold less efficiently at 30°C than at 42°C (Table 2). The lack of a cold-sensitive growth phenotype after transformation with pBR322 for AB1157 bearing either pGB2, which does not encode a umuDC operon, or pSE115, which does not express detectable levels of the umuDC gene products in AB1157 due to the presence of the LexA repressor (data not shown), confirms that the cold sensitivity conferred by pGY9739 was due to the elevated levels of UmuD2C.
However, when the umuDC-expressing strain was transformed with pJRC210, the pBR322 derivative that overexpresses β from the Ptac promoter, it became more cold sensitive for growth (i.e., from a plating efficiency ratio at 30°C/42°C of 3.5 × 10−3 with pBR322 to 5.1 × 10−4 with pJRC210 [Table 2]), despite the fact that the Ptac promoter was not induced. This enhancement in umuDC-mediated cold sensitivity was independent of the order in which the two plasmids were introduced into AB1157; comparable plating efficiencies were observed whether the β-expressing plasmid was transformed into the umuDC-expressing strain or vice versa (data not shown). Despite the enhanced cold-sensitive growth phenotype of the strain overexpressing the umuDC gene products following its transformation with the β-expressing plasmid, its transformation efficiency with the same plasmid at 42°C was roughly 105 CFU per μg of plasmid DNA (see Table 2, footnote c). This efficiency is similar to that observed with the same strain with pBR322 in place of pJRC210, indicating that growth of the strain expressing elevated levels of both β and the umuDC gene products was essentially unaffected at 42°C. The control strain bearing pSE115, which did not express elevated levels of UmuD2C due to the presence of the LexA repressor, did not become cold sensitive when transformed with pJRC210. This indicated that the exacerbation of the cold sensitivity of the umuDC-expressing strain was due to the combination of elevated levels of UmuD2C together with elevated levels of β.
These results, indicating that the β processivity clamp of Pol III might be involved in umuDC-mediated cold sensitivity, prompted us to evaluate the effect of overexpression of β on a strain that expressed elevated levels of the umuD′C gene products. In this analysis, we used a plasmid named pGY9738 that is similar to the one described above containing the oc1 mutation except that it results in constitutive expression of the umuD′C gene products instead of the umuDC gene products. Such a strain is not normally cold sensitive (59), presumably due to the inability of UmuD′2C to function in the UmuD2C-dependent checkpoint control (37). Consistent with our previous results (59), AB1157 expressing elevated levels of the umuD′C gene products was not cold sensitive when transformed with pBR322 (Table 2). However, transformation of this umuD′C-expressing strain with the β-expressing plasmid (pJRC210) resulted in a severe cold-sensitive growth phenotype, as indicated by the nearly 4,000-fold reduction in its plating efficiency at 30°C (Table 2). This plating efficiency is similar to that which we observed for the umuDC-expressing strain following its transformation with the β-expressing plasmid (pJRC210). However, one important difference between the umuDC- and umuD′C-expressing strains is that the latter is cold sensitive only following its transformation with the β-expressing plasmid (Table 2), while the former is cold sensitive regardless of whether or not it bears the β-expressing plasmid and becomes more cold sensitive after transformation with the β-expressing plasmid.
We have previously reported that umuDC-mediated cold sensitivity requires the umuC gene product but not its catalytic proficiency as a DNA polymerase (59). Likewise, we found that overexpression of β together with elevated levels of UmuD2 or UmuD′2 alone did not confer cold sensitivity; the umuC gene product was absolutely required for cold sensitivity (Table 2). Finally, the extent of cold sensitivity conferred upon AB1157 expressing elevated levels of the umuDC or umuD′C gene products by the β-expressing plasmid (pJRC210) was unaffected by addition of IPTG, which induces high-level overproduction of β (data not shown); the noninduced level of β expressed from pJRC210 was sufficient (Table 2). Thus, it is important to stress that all the experiments described in this report designed to measure the phenotype of the pJRC210-encoded dnaN allele were performed in the absence of IPTG. Finally, although we have not yet carefully quantitated the level of expression of β from pJRC210 in the absence of IPTG, it is clear that strains bearing this plasmid and grown in rich medium, such as LB medium, express at least ∼10- to 100-fold more β from the plasmid than they do from their chromosomal allele (data not shown).
The cold sensitivity conferred by the overexpression of β together with elevated levels of the umuDC gene products is more severe than that conferred by β together with umuD′C.
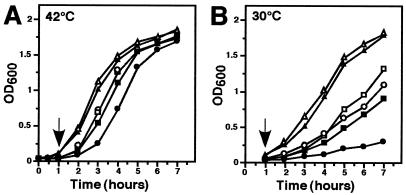
The quantitative plating assay that we typically use to gauge the extent of cold sensitivity measures the ability of a strain to form a colony on selective medium after overnight incubation (38). Consequently, it is a relatively insensitive method for characterizing small differences between strains, such as growth rates. In addition, we have previously found that depending upon the selectable antibiotic marker being used, the upper limit with respect to the extent of cold sensitivity that can be observed may vary. For example, using the same E. coli strain (GW8018) and the same umuDC-expressing plasmid (pSE117), which confers resistance to both ampicillin and kanamycin, we observed a 659-fold range in the extent of the cold sensitivity depending on whether we selected for growth using solid medium supplemented with ampicillin (32) or kanamycin (38). Therefore, to establish whether the umuDC- and umuD′C-expressing strains were similarly affected by the simultaneous overproduction of β, we further characterized the growth defects at 30°C relative to those at 42°C of some of the same strains described in Table 2 by monitoring their growth rates in liquid culture. Our focus in these experiments was on comparing the effects of elevated levels of umuDC to elevated levels of umuD′C with or without simultaneous expression of the β clamp. The AB1157 derivative bearing pSE115, which did not express detectable levels of UmuD2C (due to the presence of the LexA repressor protein), with or without the β-expressing plasmid, served as the control. Briefly, overnight cultures of each strain grown at 42°C in M9 minimal medium supplemented with 0.5% Casamino Acids, 0.2% glucose, and the appropriate antibiotics were subcultured 1:125 into the same prewarmed medium and grown at 42°C for 60 min. Cultures were then split in half, and one of each pair was shifted to 30°C while its partner was maintained at 42°C. One-milliliter aliquots of each culture were then removed at various times for determination of their optical density at 600 nm (Fig. 1).
FIG. 1.
The cold sensitivity conferred upon a umuDC-expressing strain by overexpression of β is more severe than that conferred upon a umuD′C-expressing strain. Growth curve analyses were performed as described in the text. The arrow indicates the time at which cultures were spilt into two equal parts and one of each pair was shifted to 30°C (B) while the other was maintained at 42°C (A). Symbols: □, AB1157(pGY9738)(pBR322); ▪, AB1157(pGY9738)(pJRC210); ○, AB1157(pGY9739)(pBR322); ●, AB1157(pGY9739)(pJRC210); ▵, AB1157(pSE115)(pBR322); ▴, AB1157(pSE115)(pJRC210). E. coli AB1157 and plasmid DNAs are described in Table 1. OD600, optical density at 600 nm.
The growth rates of AB1157 bearing pBR322 and expressing elevated levels of either the umuDC (pGY9739) or the umuD′C (pGY9738) gene products were slightly retarded at 42°C relative to that of the control strain bearing the umuDC genes under control of their native, LexA-regulated promoter (pSE115), which grew well at both temperatures (Fig. 1). Consistent with previous reports (38), the growth defect of the umuDC-expressing strain was more pronounced at 30°C, while the growth rate of the strain expressing elevated levels of the umuD′C gene products was somewhat less affected (Fig. 1, compare panels A and B).
In contrast to these findings, the growth rates of the strains expressing elevated levels of the umuDC or umuD′C gene products together with elevated levels of β were significantly more retarded at both 30 and 42°C compared to the same strains bearing pBR322 in place of the β-expressing plasmid. Interestingly, the growth rate of the strain expressing umuDC together with β was considerably more retarded at both 30 and 42°C than was that of the strain expressing umuD′C together with β (Fig. 1). The control strain bearing pSE115, which does not express detectable levels of the umuDC gene products because of the LexA repressor, together with the β-expressing plasmid was only slightly cold sensitive compared to the same strain bearing pBR322 in place of the β-expressing plasmid.
Taken together, these results (Fig. 1) confirm that exacerbation of the cold sensitivity requires elevated levels of both β and the umuDC gene products and indicate that the cold sensitivity conferred by the combination of elevated levels of UmuD2C together with elevated levels of β is more severe than that conferred by elevated levels of UmuD′2C together with β. Our inability to detect this difference in our plating assay is presumably due to its insensitivity with respect to small differences in growth rate (see above).
Direct selection for novel dnaN alleles deficient in conferring a cold-sensitive phenotype upon a umuD′C-expressing E. coli strain.
The ability of the β-expressing plasmid (pJRC210) to confer cold sensitivity upon AB1157 expressing elevated levels of the umuD′C gene products provided us with an extremely powerful means of identifying dnaN (which encodes β) alleles deficient in conferring this cold sensitivity. Based on our findings that (i) the umuD gene products interact with β in vitro and (ii) UmuD has a greater affinity for β than does UmuD′ (57) (an observation that correlates well with their respective degrees of cold sensitivity [Fig. 1]), we reasoned that the exacerbation of umuDC-mediated cold sensitivity conferred by elevated levels of β was in fact a manifestation of these physical interactions. Thus, we hoped that by selecting for dnaN alleles unable to exacerbate umuD′C-mediated cold sensitivity, we would be able to identify missense mutant β proteins deficient in physical interactions with the umuDC gene products. We therefore used hydroxylamine to chemically mutagenize the β-expressing plasmid in vitro and then transformed this chemically mutagenized plasmid into AB1157 bearing pGY9738, which constitutively expresses the umuD′C gene products. Transformation reaction mixtures were plated at the normally nonpermissive temperature of 30°C to select for plasmid-encoded dnaN alleles deficient in conferring cold sensitivity. To minimize selecting for siblings, transformation reaction mixtures were aliquoted prior to their outgrowth; nucleotide sequence analysis (see below) confirmed that none of the missense dnaN alleles we identified were siblings.
By this approach, we identified 76 strains whose plating efficiencies at 30 and 42°C were similar. Although we had selected these clones by directly plating the transformation reaction mixtures at 30°C, we had also plated an aliquot of each reaction mixture at the permissive temperature of 42°C to allow for a determination of the total number of viable transformants. This analysis indicated that these 76 putative mutants were selected from a total pool of ∼60,000 independent transformants. To confirm that the observed phenotypes of these 76 presumed mutant dnaN alleles were conferred by the respective pJRC210 derivatives and not by a mutation mapping to the host chromosome, we isolated the plasmid DNA from each of the 76 clones and transformed it back into AB1157 bearing the umuD′C-expressing plasmid. Of the 76 pJRC210 plasmids, 75 were unable to confer cold sensitivity upon AB1157 expressing elevated levels of the umuD′C gene products, indicating that they carried the mutation responsible for the observed phenotype.
To characterize these 75 plasmid-encoded dnaN alleles deficient in conferring the cold-sensitive phenotype further, we assessed their respective abilities to overproduce β in response to addition of IPTG; the dnaN gene in pJRC210 is under the control of the IPTG-inducible Ptac promoter. The purpose of this analysis was to differentiate between those pJRC210 derivatives unable to confer cold sensitivity because of an expression defect (i.e., a mutation affecting the expression of the dnaN gene product) or a nonsense mutation (which was not suppressed by the supE44 allele of AB1157 and would result in expression of a truncated form of β) from those expressing a full-length β protein that is expressed at a level comparable to that observed for the dnaN+ allele. The latter class represented the type of allele we were interested in and presumably corresponds to those containing one or more missense mutations that affect the ability of the β derivatives they encode to interact physically with the umuD′C gene products.
pJRC210, the parent plasmid, overexpresses β to very high levels in response to IPTG (>20% total soluble protein [data not shown]). Thus, we were able to easily distinguish those pJRC210 derivatives able to overexpress the β clamp from those unable to do so by Coomassie blue staining of sodium dodecyl sulfate-polyacrylamide gel electrophoresis-fractionated whole-cell lysates prepared from cultures of AB1157 derivatives bearing the respective β-expressing plasmids after their treatment with IPTG (data not shown). This analysis indicated that of the 75 plasmids identified by our genetic assay, 13 were capable of overproducing a full-length β protein (data not shown). Thus, only 13 of the ∼60,000 transformants carrying a hydroxylamine-treated pJRC210 derivative possessed the desired phenotypes of (i) expressing a full-length β protein and (ii) being unable to confer a cold-sensitive phenotype upon a umuD′C-expressing strain.
Nucleotide sequence determination of novel dnaN alleles.
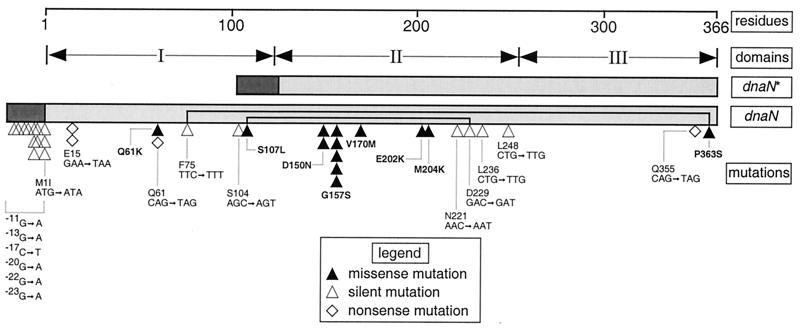
To determine the specific nucleotide alterations in each of the 13 dnaN alleles presumed to contain a missense mutation, we sequenced the promoter region and entire coding sequence of each using at least four different oligonucleotide primers (see Materials and Methods). In addition, we also sequenced 14 alleles that we suspected, based on analysis of whole-cell lysates (see above), to be either expression mutations (i.e., mutations mapping to the promoter region that affect expression) or alleles that expressed a truncated form of the β clamp (i.e., nonsense mutations). This analysis, summarized in both Table 3 and Fig. 2, confirmed our hypothesis regarding the alleles we suspected to contain either one or more mutations in their promoter region, thus affecting expression of dnaN, or a nonsense mutation resulting in expression of a truncated form of the β protein. In addition, it indicated that the 13 alleles presumed to contain a missense mutation(s) actually represent eight unique and novel missense dnaN alleles. These eight alleles identify a total of eight different amino acid residues that, when changed, affect the ability of β to confer cold sensitivity upon the umuD′C-expressing strain. Of the 35 mutations identified by nucleotide sequence analysis (see the legend to Fig. 2), only four could not be accounted for by the known propensity of in vitro hydroxylamine treatment to cause C-to-T transitions (31). Finally, this analysis indicated that, with the exceptions of pJRCHA-5.1 (S107L) and pJRCHA-6.2 (P363S), all eight alleles contained only a single missense mutation; the two exceptions contained, in addition, one silent mutation each (Table 3).
TABLE 3.
Summary of nucleotide sequence analysis of novel dnaN alleles containing missense mutationsa
| Plasmid | dnaN allele (deduced amino acid substitution)b | Nucleotide substitutionc | No. of occurrences (degree of conservation)d |
|---|---|---|---|
| pJRCHA-4.1 | Q61K | 181CAG→AAG | 1 (2/22) |
| pJRCHA-5.1 | S107L | 319TCG→TTG | 1 (3/22) |
| 685GAC→CAT (silent) | NAe | ||
| pJRCHA-8.1 | D150N | 448GAC→AAC | 2 (13/22) |
| pJRCHA-5G11 | G157S | 469GGT→AGT | 5 (16/22) |
| pJRCHA-8I11 | V170M | 508GTG→ATG | 1 (16/22) |
| pJRCHA-7.1 | E202K | 604GAA→AAA | 1 (12/22) |
| pJRCHA-6F11 | M204K | 610ATG→AAG | 1 (10/22) |
| pJRCHA-6.2 | 223TTC→TTT (silent) | NA | |
| P363S | 1087CCA→TCA | 1 (21/22) |
See Fig. 2 for a complete summary of all nucleotide alterations in dnaN identified in this study.
The name for each dnaN allele is based on its deduced amino acid substitution in one-letter code and its respective position within the primary structure of the β protein. For example, Q61K represents the dnaN allele bearing a Q-to-K substitution at position 61.
The mutation(s) resulting in each allele is indicated in the context of its respective codon(s). The numbering is relative to the start of each codon, with position 1 corresponding to the A in the ATG of the first codon of dnaN. Missense mutations that did not affect the amino acid inserted, due to the degeneracy of the genetic code, are termed silent.
The number of times that each respective dnaN allele was identified (occurrences) in the complete collection of 27 alleles subjected to automated DNA sequence analysis is indicated (see Fig. 2 for a complete summary of the DNA sequence analysis). The degree of conservation for each position among the 22 dnaN homologs whose sequence is in the National Center for Biotechnology Information database (as of 4 December 2000) is indicated in parentheses. This dnaN alignment was performed by the COGS application of the National Center for Biotechnology Information website (www.ncbi.nlm.nih.gov/COG/).
NA, not applicable.
FIG. 2.
Stick representation of the dnaN gene, summarizing the relative positions of all mutations identified in this study. The positions of all nucleotide alterations identified in the 27 dnaN alleles subjected to automated DNA sequence analysis are shown. Approximate positions of amino acid residues in β and the approximate delineation of each of the three structurally similar domains (I, II, and III) comprising each β monomer (21) are indicated. The light-shaded rectangles represent the coding sequences of the dnaN and dnaN* (see text) genes, while the dark-shaded rectangles represent their respective promoters. Approximate positions of those mutations resulting in deduced amino acid substitutions and described in Table 3 are indicated by filled triangles; approximate positions of silent mutations, including those which map to the promoter region, are indicated by open triangles; and approximate positions of nonsense mutations are indicated by open diamonds. Except for the F75 and D229 silent mutations that were identified together with the P363S and S107L mutations, respectively (indicated by the lines connecting these respective positions), the alleles containing silent or nonsense mutations are not described in Table 3. The silent mutation L236 results in formation of a rare Leu codon leading to a significantly reduced steady-state level of the expressed β protein (data not shown). The silent mutations S104, N221, and L248 were identified in combination with at least one other mutation mapping in the vicinity of the ribosome binding site (which maps to nucleotide positions −16 to −9) and appear to also express significantly lower levels of β (data not shown). The four mutations that cannot be accounted for by hydroxylamine treatment (which promotes C→T transitions when used in vitro [31]) are 181C→A (Q61K) (Table 3), 611T→A (M204K) (Table 3), 685G→C (the silent mutation in S107L) (Table 3), and 45G→T (resulting in the E15→nonsense mutation).
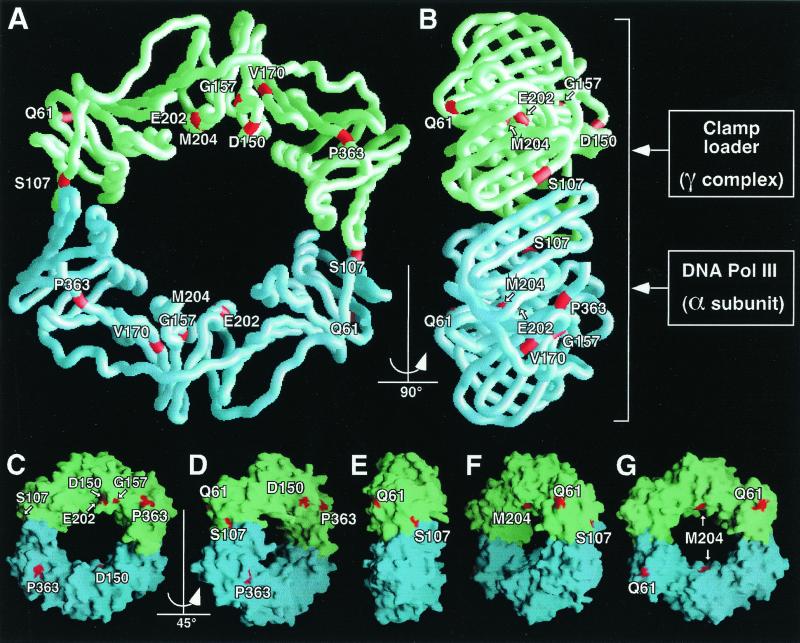
All eight amino acid substitutions identified by our genetic assay correspond to residues located at or very close to the surface of the molecule (Fig. 3) based on the crystal structure of the β clamp reported by Kong et al. (21), with the majority of them affecting the face of β that is believed to interact with DNA Pol III (Fig. 3B) (23, 24, 41, 55). Incidentally, some of the native residues that were mutated are not easily visible in the surface representations of the β clamp (Fig. 3C to G). This is because their position either is located just below the surface or is obstructed by other surface features of the β clamp in the views shown.
FIG. 3.
Relative position of each deduced amino acid substitution in the β crystal. (A and B) Worm representation of the β clamp showing the positions of missense mutations as viewed from the face bearing the extruding C-terminal tails of each protomer (A), and the side (B), corresponding to a 90° rotation of that shown in panel A. The crystal structure of the homodimeric β clamp, solved by Kong et al. (21), is shown with one β protomer in green and the other in blue. Positions of deduced amino acid substitutions are in red. The γ complex clamp loader and the α subunit of Pol III are both believed to interact with the face of β bearing the extruding C-terminal tails (B). See the text for further details. (C through G) Surface representations of the β crystal in successive rotations of 45° each (i.e., 45°, 90°, 135°, and 180°, respectively) relative to the angle shown in panel A. This view bears the extruding C-terminal tails and is arbitrarily defined as 0°. Again, one β protomer is in green and the other is in blue, and positions of deduced amino acid substitutions are in red. Note that not all of the missense mutations (for example, E204) are easily visible in the surface representation. This is because their position either is located just below the surface or is obstructed by other surface features of the β clamp in the views shown. This figure was generated with GRASP (Molecular Simulations, Inc.) using the atom coordinates of the β crystal (2POL) from the Protein Data Bank and a Silicon Graphics R10000 workstation.
The dnaN alleles described in this report likely represent a majority of the total number of dnaN alleles possible with this phenotype following treatment of the plasmid-encoded dnaN+ gene with hydroxylamine in vitro. This conclusion is based on the fact that the D150N and G157S alleles were identified two and five independent times, respectively, indicating that our selection-screen was partially saturating (Table 3). The large number of mutations mapping to the same small number of residues within the promoter region, including the ribosome binding site (a region of 8 bp [−16TAGGAGGT−9] [Fig. 2]), combined with the fact that 48 more alleles unable to overproduce dnaN to a detectable level were identified but not sequenced (see above), indicates that the target size for amino acid substitutions in β that can affect its ability to confer cold sensitivity must be very small.
Finally, it is interesting that each of the eight dnaN alleles encodes a mutant β protein with a comparatively larger residue in place of the smaller, native residue (e.g., G157S). In addition, some also radically affect the charge characteristics of the native residue (e.g., E202K), and many of the deduced amino acid substitutions identified affect residues well conserved among other prokaryotic β homologs (Table 3). It is also important to point out that mutations were identified in all three structural domains of β; each β protomer contains threefold structural similarity (21) (Fig. 2). In addition, two (Q61K and S107L) of the eight missense mutations are located in the first structural domain of β (domain I), which is lacking in an alternative form of the β clamp referred to as β* (see Fig. 2 and Discussion for further details) (39, 50).
Steady-state levels of mutant β proteins.
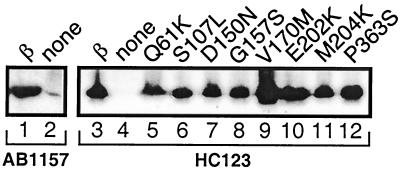
Prior to their nucleotide sequencing, we had screened each dnaN allele for its ability to overexpress a full-length β protein. Their ability to do so suggested that their promoter regions did not contain mutations that affected their respective abilities to express their gene products, an assumption corroborated by their nucleotide sequence analysis (see above). To confirm that the mutant β proteins were present at comparable levels, relative both to each other and to the wild-type control (pJRC210), in the absence of their induced expression by IPTG, we measured their respective steady-levels by qualitative Western blot analysis using a polyclonal antibody specific to β. The steady-state level of each of the eight mutant β proteins was found to be similar to that observed for the wild-type control in the absence of added IPTG (Fig. 4), indicating that their deduced amino acid substitutions do not significantly affect their overall stability in vivo. This result confirms that the respective deficiencies of these dnaN alleles to confer cold sensitivity in our genetic assay are due to their deduced amino acid substitutions and not to a large change in their abundance. Finally, it is worthwhile emphasizing that the level of β expressed from pJRC210 (as well as the levels of the mutants expressed from the pJRC210 derivatives) in the absence of IPTG is far greater than the level expressed from the chromosomally carried dnaN+ or dnaN59(Ts) allele (Fig. 4). Initial attempts to quantitate the level of expression from pJRC210 suggest that it is at least 10- to 100-fold higher than that observed from the chromosomal dnaN+ allele (data not shown).
FIG. 4.
Steady-state levels of mutant β proteins in the absence of IPTG. The steady-state level of each mutant β protein was measured in the absence of added IPTG by Western blot analysis using polyclonal antibodies specific to β and chemiluminescent detection (Tropix) as described previously (36, 57). Approximately 108 cells of each strain grown in LB medium at 30°C to mid-exponential phase (optical density at 600 nm, ∼0.5) were electrophoresed in a sodium dodecyl sulfate–15% polyacrylamide gel and then transferred to a polyvinylidene difluoride membrane as described previously (36, 57). Lane 1, AB1157(pJRC210); lane 2, AB1157(pBR322); lane 3, HC123(pJRC210); lane 4, HC123(pBR322); lane 5, HC123(pJRCHA-4.1); lane 6, HC123(pJRCHA-5.1); lane 7, HC123(pJRCHA-8.1); lane 8, HC123(pJRCHA-5G11); lane 9, HC123(pJRCHA-8I11); lane 10, HC123(pJRCHA-7.1); lane 11, HC123(pJRCHA-6F11); and lane 12, HC123(pJRCHA-6.2). E. coli AB1157 and HC123 are described in Table 1; plasmids are described in Tables 1 and 3. Note that under the conditions used in this analysis, endogenous levels of β expressed from the chromosomal dnaN+ allele (AB1157) (lane 2) were only barely detectable, while those expressed from the dnaN59(Ts) allele (HC123) (lane 4) were not detectable.
Novel dnaN alleles are deficient in exacerbating umuDC-mediated cold sensitivity.
As discussed in the report of our previous in vitro analysis (57), UmuD interacts more strongly with β than does UmuD′. Two possible explanations for this difference are (i) UmuD and UmuD′ require similar structural features of β for their interactions, but the extra N-terminal 24 amino acids in UmuD enable it to make additional contacts with β, thus accounting for its stronger interaction (57), or (ii) UmuD and UmuD′ each interact with different structural features of β, and hence their different affinities are due to the different natures of their interactions. To begin to differentiate between these two possibilities, we investigated whether or not the eight dnaN alleles identified by virtue of their inability to interact genetically with the umuD′C gene products were similarly affected with respect to their interactions with the umuDC gene products.
If UmuD and UmuD′ interact with different structural features of β, we might have expected that most, if not all, of the dnaN alleles that we isolated by virtue of their inability to confer cold sensitivity for growth upon a umuD′C-expressing strain would be proficient in causing the β-dependent exacerbation of the cold sensitivity of a umuDC-expressing strain. However, as shown in Table 4, all eight dnaN alleles that were deficient in conferring cold sensitivity upon AB1157 expressing elevated levels of umuD′C were also deficient in their ability to exacerbate the cold sensitivity conferred by elevated levels of the umuDC gene products. These observations strongly suggest that the interactions between UmuD′ and β are a subset of those between UmuD and β. Thus, we suggest that UmuD has a higher affinity for β than does UmuD′ because of additional, important interactions of β with the N-terminal 24 residues of UmuD, which are missing from UmuD′. Strikingly, some of the dnaN alleles (i.e., G157S, M204K, and, to a lesser extent, E202K) not only were deficient in exacerbation of the cold sensitivity but actually suppressed the inherent cold sensitivity conferred by elevated levels of the umuDC gene products in the absence of elevated levels of β (Table 4). This phenotype is interpreted more thoroughly in Discussion.
TABLE 4.
Novel dnaN alleles deficient in conferring cold sensitivity upon a umuD′C-expressing strain are unable to exacerbate the cold sensitivity of a umuDC-expressing strain
| Plasmid | dnaN allele | AB1157 transformants/ml (30°C/42°C)a with:
|
|
|---|---|---|---|
| pGY9738 (oc1umuD′C) | pGY9739 (oc1umuDC) | ||
| pBR322 | None | 0.98b | 3.5 × 10−3b |
| pJRC210 | dnaN+ | 2.6 × 10−4b | 5.1 × 10−4b |
| pJRCHA-4.1 | Q61K | 0.71 | 1.7 × 10−3 |
| pJRCHA-5.1 | S107L | 0.37 | 1.5 × 10−3 |
| pJRCHA-8.1 | D150N | 0.11 | 1.7 × 10−3 |
| pJRCHA-5G11 | G157S | 0.83 | 0.25 |
| pJRCHA-8I11 | V170M | 0.43 | 1.2 × 10−3 |
| pJRCHA-7.1 | E202K | 0.47 | 2.7 × 10−2 |
| pJRCHA-6F11 | M204K | 0.98 | 0.15 |
| pJRCHA-6.2 | P363S | 0.30 | 3.5 × 10−3 |
AB1157 bearing either pGY9738 or pGY9739 was transformed with either pBR322 or the indicated pJRC210 derivative. The ratio of CFU obtained at 30°C relative to those obtained at 42°C was measured as described in Table 2, footnote c. Transformation efficiencies at 42°C for all strains were on the order of ∼105 to ∼106 CFU per μg of plasmid DNA. These results indicate that overexpression of either the umuD or umuD′C gene products with or without simultaneous overexpression of β is not lethal at 42°C under these conditions.
This value is the same as that reported in Table 2 and is included here for comparison to the eight novel dnaN alleles.
Novel dnaN alleles deficient in exacerbating umuDC-mediated cold sensitivity are proficient for DNA replication in vivo when overexpressed.
To determine whether or not the novel dnaN alleles that we identified by our genetic assay were able to function in chromosomal DNA replication, we attempted to cross them onto the E. coli chromosome using the pKO3-based system devised by Link et al. (25). This plasmid was specifically designed for moving in-frame deletions and mutations onto the bacterial chromosome. Our inability to homogenetize the five dnaN alleles we tested (Q61K, D150N, G157S, E202K, and M204K) (data not shown) suggests that they are unable to function in chromosomal replication at a level adequate for viability when expressed at the normal, physiological level.
In light of this, we chose to measure the respective abilities of these dnaN alleles to function in chromosomal DNA replication when they were present at an elevated gene dosage by virtue of residing on a multicopy plasmid. For this, we used the same plasmids that we initially identified by our genetic screen and asked whether they could complement the temperature-sensitive phenotype of a dnaN59(Ts) strain. E. coli strains bearing the dnaN59(Ts) allele are temperature sensitive due to the poor stability of the mutant β protein (DnaN59) at the elevated temperature (4). Using the previously described quantitative transformation assay (38), we found that all eight dnaN alleles were similar to the plasmid-encoded dnaN+ allele with respect to their abilities to complement the temperature-sensitive phenotype of the dnaN59(Ts) allele (Table 5). In contrast, pBR322 was unable to substitute for pJRC210 in suppression of the temperature-sensitive phenotype of the dnaN59(Ts) allele. These results indicate that each of the novel dnaN alleles described in this report retains at least some biological activity with respect to DNA replication in vivo. However, their apparent inability to function in chromosomal replication at a level adequate for viability when expressed at the normal, physiological level suggests that it is difficult to genetically separate the replication function of β from its ability to interact with the umuDC gene products.
TABLE 5.
Abilities of novel dnaN alleles to complement the temperature sensitivity of the dnaN59(Ts) E. coli strain HC123
| Plasmid | dnaN allele | HC123 transformants/ml (42°C/30°C)a |
|---|---|---|
| pBR322 | None | <8.6 × 10−4 |
| pJRC210 | dnaN+ | 1.11 |
| pJRCHA-4.1 | Q61K | 0.71b |
| pJRCHA-5.1 | S107L | 1.02 |
| pJRCHA-8.1 | D150N | 1.10 |
| pJRCHA-5G11 | G157S | 0.86b |
| pJRCHA-8I11 | V170M | 0.95 |
| pJRCHA-7.1 | E202K | 1.01 |
| pJRCHA-6F11 | M204K | 0.92 |
| pJRCHA-6.2 | P363S | 0.95 |
The ratio of CFU obtained at 42°C to those obtained at 30°C was measured as described in Table 2, footnote c. Transformation efficiencies were on the order of ∼105 to ∼106 CFU per μg of plasmid DNA.
This strain formed small colonies at 42°C, suggesting that the plasmid was slightly less efficient than pJRC210 at suppressing the temperature sensitivity of E. coli HC123.
DISCUSSION
Interactions of UmuD2C with components of Pol III help to enable a DNA damage checkpoint control.
The cold sensitivity conferred by elevated levels of UmuD2C, a phenomenon that apparently is due to the inappropriate expression of their DNA damage checkpoint function (59), is exacerbated by overexpression of the β processivity clamp (Table 2). This observation, taken together with our previous findings that overexpression of the ɛ proofreading subunit of Pol III or deletion of its structural gene (dnaQ) suppresses umuDC-mediated cold sensitivity whereas overexpression of each of the other eight Pol III subunits (α, θ, τ, γ, δ, δ′, χ, and ψ) does not (56), suggests that UmuD2C associates with the replisome via direct interactions with ɛ and β and that these interactions serve to antagonize the DNA polymerase activity of Pol III, thereby arresting DNA synthesis (30, 37).
Overexpression of β also conferred a severe cold-sensitive growth phenotype upon a strain expressing moderately elevated levels of the umuD′C gene products. Such a strain is not otherwise normally cold sensitive (59), although higher-level overexpression of UmuD′2C does confer a cold sensitivity (38). We have previously suggested that the cold sensitivity conferred by higher-level overexpression of umuD′C is a result of the ability of higher levels of UmuD′2C to mimic the UmuD2C-dependent DNA damage checkpoint control (59). Thus, we now suggest that the β-expressing plasmid confers cold sensitivity upon an E. coli strain expressing moderate levels of umuD′C (a strain that is not normally cold sensitive [59]) by facilitating the ability of moderate levels of umuD′C to mimic the DNA damage checkpoint function of UmuD2C, which presumably involves, at least in part, interaction of UmuD2C with β (57, 59).
We exploited the ability of the β-expressing plasmid (pJRC210) to confer cold sensitivity upon a umuD′C-expressing strain to identify eight unique and novel missense dnaN mutations. The fact that these plasmid-borne mutant dnaN alleles could complement the temperature sensitivity of a dnaN59(Ts) mutant (Table 5) suggests that when overexpressed, these mutant β proteins retain a partial ability to participate in normal DNA replication. Some of the dnaN alleles described in this report (i.e., G157S, M204K, and, to a lesser extent, E202K) were able to suppress the inherent cold sensitivity conferred by elevated levels of the umuDC gene products in the absence of elevated levels of β (Table 4), suggesting that their presence in the replisome confers upon it an immunity to the replication block normally imposed by elevated levels of UmuD2C. However, a definitive answer to the question of whether or not these mutant β proteins are deficient in interaction with UmuD2C must await their biochemical characterization.
The eight mutant β proteins described in this report bear substitutions of amino acid residues located at or very close to the surface of the molecule (Fig. 3C to G), based on the crystal structure of the β clamp reported by Kong et al. (21). Some of the native residues that were mutated are not easily visible in the surface representations of the β clamp. This is because their position either is located just below the surface or is obstructed by other surface features of the β clamp in the views shown. With respect to the fact that all of the mutations were located at or very close to the surface of the molecule, it is worthwhile emphasizing the fact that all of the mutations correspond to substitutions of a comparatively larger residue in place of the smaller, native residue (e.g., G157S). Thus, all of the mutant β proteins described in this report are expected to have a slightly altered surface relative to that of the wild-type β protein due to the presence of their respective amino acid substitutions, consistent with the mutant β proteins being affected for interaction with the umuDC gene products.
With the exception of Q61K, all of the substitutions reside on the same face of the β clamp (Fig. 3B). Interestingly, this face is the same as that which has been previously suggested to interact with both the α catalytic subunit (23, 24, 55) and the five-subunit clamp loader, or γ complex, of Pol III (23, 24) (Fig. 3B). This finding suggests that interaction of the umuDC gene products with β may eliminate the ability of β to subsequently interact with the α catalytic subunit and/or γ complex of Pol III. Furthermore, our recent observation that the umuD gene products interact with the same structural domain of ɛ that has been previously shown to interact with α suggests that a similar situation is true for the UmuD-ɛ interaction; i.e., UmuD and α associate with ɛ in a mutually exclusive fashion (56).
If the UmuD2C complex were to interact with both the ɛ and the β subunits of Pol III in such a way as to preclude their subsequent association with α, then the umuDC-dependent checkpoint could not be a manifestation of interactions involving UmuD2C and the Pol III multiprotein complex. Rather, these observations suggest that UmuD2C exerts its checkpoint role by competing with α for binding to ɛ and β. Viewed in this way, the UmuD2C-dependent checkpoint control has certain aspects in common with the eukaryotic S-phase checkpoint control. One component of this surveillance mechanism is the p21 protein, which acts not only to regulate cyclin–cyclin-dependent kinase complexes (15, 16) but also to regulate the availability of the eukaryotic PCNA processivity clamp (66). By associating directly with the same face of PCNA as does Pol δ, p21 may preclude binding of Pol δ to PCNA, thereby helping to regulate DNA replication in response to DNA damage (66).
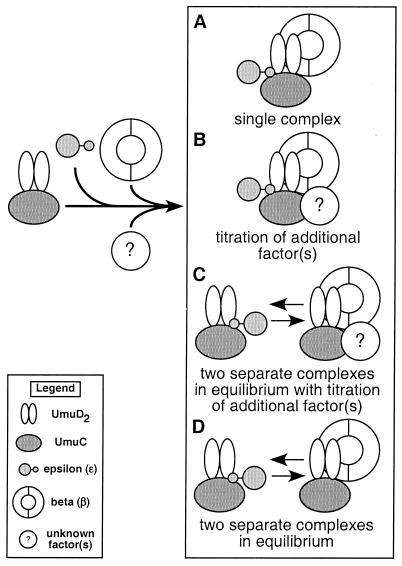
The important findings discussed above, taken together with previous observations (37, 38, 56, 57, 59), suggest at least four different models for how interactions of UmuD2C with the ɛ and the β subunits of Pol III could enable a DNA damage checkpoint control (Fig. 5). With respect to these four models, the absolute requirement of umuC for exacerbation of the cold sensitivity (30, 38, 59) suggests that UmuC may also interact directly with ɛ and β, perhaps serving to impose a directionality upon the interactions of the symmetrical UmuD2 homodimer (in the form of the UmuD2C complex) with ɛ and β.
FIG. 5.
Four possible models to describe the involvement of the ɛ and the β subunits of Pol III in UmuD2C-dependent cold sensitivity and in the UmuD2C-dependent checkpoint control. Our results discussed here, taken together with our previous findings (37, 38, 56, 57, 59), suggest at least four different models to describe the involvement of the ɛ and the β subunits of Pol III in UmuD2C-dependent cold sensitivity, which presumably is a manifestation of functions of UmuD2C involved in the DNA damage checkpoint control (37, 59). Previously described genetic interactions between umuC and ɛ (encoded by dnaQ) (10, 11, 17, 19, 35, 56), together with our findings discussed in this report regarding genetic interactions of β with both umuDC and umuD′C, collectively suggest that UmuC might interact directly with both ɛ and β. Therefore, for the purposes of these models, we have assumed that interaction of UmuC with ɛ or β imparts an asymmetry on the UmuD2 homodimer that is important for defining its subsequent interaction (when in the form of the UmuD2C complex) with ɛ or β. The two proposed structural domains of ɛ (40, 61) are represented here as circles; the N-terminal domain is represented by the large circle, while the C-terminal domain is represented by the small circle. It has previously been demonstrated that the small C-terminal domain of ɛ interacts with both the α subunit of Pol III (40, 61) and the umuD gene products (56). Interaction of the unknown factor(s) with UmuC and β (B) is arbitrary and is intended only to imply the possible involvement of additional, as-yet-unidentified components of the UmuD2C-dependent checkpoint control pathway. See Discussion for further details regarding these four different models.
In the first model (Fig. 5A), UmuD2C, ɛ, and β interact, forming a single, multiprotein complex. Since associations of both ɛ (53) and β (4, 54) with α significantly enhance its DNA polymerase activity, titration of either or both away from the replisome by UmuD2C would be expected to antagonize the catalytic DNA polymerase activity of Pol III. Overexpression of β may exacerbate umuDC-mediated cold sensitivity by stabilizing the multiprotein complex, thereby effectively removing ɛ from the replisome.
A variation of this model is suggested by our findings that deletion of the structural gene encoding ɛ (dnaQ) results in only a partial suppression of umuDC-mediated cold sensitivity (56), while overexpression of β strongly exacerbates umuDC-mediated cold sensitivity. These findings suggest that ɛ may play a comparatively less important role in umuDC-mediated cold sensitivity than does β. In light of this possibility, it could have been argued that if umuDC-mediated cold sensitivity was primarily due to UmuD2C (or possibly a UmuD2C-ɛ complex) sequestering β away from the replisome, then overexpression of β would be expected to suppress much of the cold sensitivity, the opposite to what we observed. However, if the unusually high levels of β were acting to stabilize a UmuD2C-β-ɛ complex that recruits an additional factor(s) which serves an essential role(s) (for example, the α subunit of Pol III), its sequestration into the multiprotein complex could cause the exacerbation of cold sensitivity that we have observed (Fig. 5B).
Two further models to describe how interactions of UmuD2C with the ɛ and β subunits of Pol III might serve to regulate DNA replication in response to DNA damage take into account the possibility that ɛ and β may interact with UmuD2C in mutually exclusive fashions, such that two separate complexes are formed. In such a case, one could imagine either that the UmuD2C-β complex acts to recruit an additional factor(s) (Fig. 5C), as described above for the UmuD2C-ɛ-β complex, or that the UmuD2C-ɛ and UmuD2C-β complexes simply exist in equilibrium, with neither recruiting additional factors (Fig. 5D). In both cases, one might predict that overexpression of ɛ would suppress umuDC-mediated cold sensitivity (as we have previously reported [56]) by favoring formation of the UmuD2C-ɛ complex, which may be less efficient than UmuD2C-β at conferring cold sensitivity (see above). This would have the effect of shifting the equilibrium away from formation of the UmuD2C-β complex, which may be recruiting an additional factor(s). In addition, overexpression of ɛ would replenish the levels of ɛ initially sequestered away from Pol III by elevated levels of UmuD2C. Likewise, both of these models (Fig. 5C and D) would predict that overexpression of β would act to shift the equilibrium toward formation of the UmuD2C-β (or UmuD2C-β-unknown factor[s]) complex, thus exacerbating the extent of cold sensitivity.
Although all four of the models discussed above share some common aspects, they have important differences as well. Experiments are under way to distinguish which of these four models most closely resembles the events that occur in response to DNA damage in vivo.
Does the UmuD′2C-β interaction serve an important role in the living cell?
Despite the finding that UmuD′2C can carry out efficient lesion bypass in vitro in the presence of RecA and ssDNA binding protein (SSB) but in the absence of Pol III (43, 63), various genetic and biochemical observations suggest that some form of DNA Pol III plays a role in determining when and where UmuD′2C-dependent TLS takes place (57; recently reviewed in references 2 and 58). These findings include the following: (i) some E. coli dnaE alleles (dnaE encodes the α subunit of Pol III) appeared to increase UV-induced umuDC-dependent SOS mutagenesis in vivo (45), (ii) the DNA polymerase activity of the temperature-sensitive DnaE1026 mutant α protein was stabilized by the UmuD′2C complex in vitro (63), and (iii) in vitro UmuD′2C-dependent TLS was significantly enhanced by small amounts of either the mutant DnaE1026 protein or Pol III core (63).
In contrast to the role of the α subunit of Pol III in TLS, the role, if any, of the β processivity clamp is less clear. Although Reuven et al., who have successfully reconstituted UmuD′2C-dependent lesion bypass in vitro, found it to be completely independent of the β clamp of Pol III (43), Tang et al., using a slightly different approach to reconstitute UmuD′2C-dependent TLS in vitro (63), reported a strict requirement of both the β clamp and the five-subunit clamp loader (γ complex) of Pol III for lesion bypass. This difference with respect to the requirement of the β clamp for TLS in vitro has been attributed to differences in the template DNAs and the levels of RecA protein used by these two groups (62, 67), although it could also be due to the maltose binding protein-UmuC fusion protein utilized by Reuven et al. (67).
Our results regarding the cold sensitivity conferred upon the umuD′C-expressing strain by the β-expressing plasmid could be the result of the previous proposal that the requirement for the β clamp and γ complex of Pol III for UmuD′2C-dependent TLS in vitro may not be physiologically relevant but rather may be due to the fact that UmuD2C interacts with β to enable a DNA damage checkpoint control (37, 57), and because of this, UmuD′2C retains a comparatively limited yet extraneous ability to also interact with β (13, 44). Alternatively, it is possible that interactions of β with the umuDC gene products are important for both the DNA damage checkpoint control (57) and TLS (62, 63) and that self-cleavage of UmuD to yield UmuD′ serves to modulate the interactions of UmuD2C and UmuD′2C with β to effect the release of the checkpoint control and enable TLS (57). In any event, a definitive answer regarding the role of the β clamp in TLS in the living cell will likely require a combined biochemical and genetic approach. Further characterization of the mutant β proteins described in this report will help to establish whether or not β serves an important role in TLS.
Relationship of the eight novel dnaN alleles to dnaN*.
Following exposure of E. coli to various DNA-damaging agents, the level of transcription of the dnaN gene increases (42, 60). In addition, expression of a truncated form of β corresponding almost exactly to the C-terminal two-thirds of the protein is also induced (39, 50). This protein, termed β*, is expressed, in a recA- and lexA-dependent fashion, from a promoter located within the coding region of the dnaN gene (Fig. 2) (39, 50). The gene for β*, which is contained entirely within the dnaN+ gene, has been termed dnaN* (39, 50). Biochemical characterization of β* indicates that it can form a trimer in solution (49). Furthermore, in vitro studies have demonstrated that β* can modestly enhance the processivity of Pol III*, a form of Pol III containing all subunits except the β clamp (49). However, despite these detailed genetic and biochemical analyses, the physiological role of β* is presently unknown. Interestingly, it has recently been suggested that several eukaryotic proteins originally identified because of their roles in cell cycle checkpoints act by serving as an alternative to PCNA, the normal processivity clamp (33, 64, 65), or as an alternate subunit of the five-subunit replication factor C complex (65), the normal PCNA clamp loader.
Each β protomer has threefold structural similarity (21) (Fig. 2). Because each β* protomer corresponds almost exactly to the C-terminal two-thirds of the dnaN gene product (39, 50), it retains the C-terminal two structural domains but is missing the N-terminal domain I. It is interesting that two of the mutations we identified that reduce interactions with the umuDC gene products, Q61K and S107L, both reside within the N-terminal domain (domain I), which is lacking in β*. Thus, relative to intact β, β* may also be deficient for interactions with the umuDC gene products. These results raise the interesting possibility that a clamp composed of a trimer of β* might constitute an efficient mechanism for conferring processivity upon Pol III, as well as possibly other DNA polymerases, including Pol II (1), that is insensitive to the checkpoint function(s) of UmuD2C. Such a mechanism might be particularly important during replication restart (7) (or induced replisome reactivation [20]) or following TLS, a time in which the cell is presumably involved in reestablishing a replisome wherein processive and accurate DNA synthesis is carried out primarily by Pol III (6, 14, 58). However, further genetic and biochemical characterization of β* will be required in order to establish its physiological role(s).
ACKNOWLEDGMENTS
We thank Mary Berlyn and the E. coli Genetic Stock Center for E. coli HC123; Suzanne Sommer and Adriana Bailone for plasmids pGY9738 and pGY9739; Roger Woodgate for plasmid pGB2; George Church for plasmid pKO3; Charles McHenry for plasmid pJRC210, the polyclonal anti-β antibodies, and many helpful discussions; Ann Ferentz for help in making Fig. 3; Veronica Godoy for help with cloning the dnaN alleles into pKO3; and the members of our lab for helpful discussions and advice, in particular Brad Smith for his comments on the manuscript.
This work was supported by Public Health Service grant CA21615 to G.C.W. from the National Cancer Institute. M.D.S. was supported by a fellowship (5 F32 CA79161-03) from the National Cancer Institute. B.M.B. was supported by a predoctoral training grant (5 T32 GM07287-26) from the National Institutes of Health. M.F.F. carried out her research as part of the Undergraduate Research Opportunities Program (UROP) at the Massachusetts Institute of Technology.
REFERENCES
- 1.Bonner C A, Stukenberg P T, Rajagopalan M, Eritja R, O'Donnell M, McEntee K, Echols H, Goodman M F. Processive DNA synthesis by DNA polymerase II mediated by DNA polymerase III accessory proteins. J Biol Chem. 1992;267:11431–11438. [PubMed] [Google Scholar]
- 2.Bridges B. DNA polymerases and SOS mutagenesis: can one reconcile the biochemical and genetic data? Bioessays. 2000;22:933–937. doi: 10.1002/1521-1878(200010)22:10<933::AID-BIES8>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 3.Burckhardt S E, Woodgate R, Scheuermann R H, Echols H. UmuD mutagenesis protein of Escherichia coli: overproduction, purification, and cleavage by RecA. Proc Natl Acad Sci USA. 1988;85:1811–1815. doi: 10.1073/pnas.85.6.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burgers P M, Kornberg A, Sakakibara Y. The dnaN gene codes for the beta subunit of DNA polymerase III holoenzyme of Escherichia coli. Proc Natl Acad Sci USA. 1981;78:5391–5395. doi: 10.1073/pnas.78.9.5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Churchward G, Belin D, Nagamine Y. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene. 1984;31:165–171. doi: 10.1016/0378-1119(84)90207-5. [DOI] [PubMed] [Google Scholar]
- 6.Cox M M, Goodman M F, Kreuzer K N, Sherratt D J, Sandler S J, Marians K J. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 7.Echols H, Goodman M F. Fidelity mechanisms in DNA replication. Annu Rev Biochem. 1991;60:477–511. doi: 10.1146/annurev.bi.60.070191.002401. [DOI] [PubMed] [Google Scholar]
- 8.Elledge S J, Walker G C. Proteins required for ultraviolet light and chemical mutagenesis. Identification of the products of the umuC locus of Escherichia coli. J Mol Biol. 1983;164:175–192. doi: 10.1016/0022-2836(83)90074-8. [DOI] [PubMed] [Google Scholar]
- 9.Fernandez De Henestrosa A R, Ogi T, Aoyagi S, Chafin D, Hayes J J, Ohmori H, Woodgate R. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol Microbiol. 2000;35:1560–1572. doi: 10.1046/j.1365-2958.2000.01826.x. [DOI] [PubMed] [Google Scholar]
- 10.Foster P L, Sullivan A D. Interactions between epsilon, the proofreading subunit of DNA polymerase III, and proteins involved in the SOS response of Escherichia coli. Mol Gen Genet. 1988;214:467–473. doi: 10.1007/BF00330482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foster P L, Sullivan A D, Franklin S B. Presence of the dnaQ-rnh divergent transcriptional unit on a multicopy plasmid inhibits induced mutagenesis in Escherichia coli. J Bacteriol. 1989;171:3144–3151. doi: 10.1128/jb.171.6.3144-3151.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedberg E C, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C.: ASM Press; 1995. [Google Scholar]
- 13.Goldsmith M, Sarov-Blat L, Livneh Z. Plasmid-encoded MucB protein is a DNA polymerase (pol RI) specialized for lesion bypass in the presence of MucA′, RecA, and SSB. Proc Natl Acad Sci USA. 2000;97:11227–11231. doi: 10.1073/pnas.200361997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodman M F. Coping with replication ‘train wrecks’ in Escherichia coli using Pol V, Pol II and RecA proteins. Trends Biochem Sci. 2000;25:189–195. doi: 10.1016/s0968-0004(00)01564-4. [DOI] [PubMed] [Google Scholar]
- 15.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 16.Harper J W, Elledge S J, Keyomarsi K, Dynlacht B, Tsai L H, Zhang P, Dobrowolski S, Bai C, Connell-Crowley L, Swindell E, et al. Inhibition of cyclin-dependent kinases by p21. Mol Biol Cell. 1995;6:387–400. doi: 10.1091/mbc.6.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jonczyk P, Fijalkowska I, Ciesla Z. Overproduction of the epsilon subunit of DNA polymerase III counteracts the SOS mutagenic response of Escherichia coli. Proc Natl Acad Sci USA. 1988;85:9124–9127. doi: 10.1073/pnas.85.23.9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jonczyk P, Nowicka A. Specific in vivo protein-protein interactions between Escherichia coli SOS mutagenesis proteins. J Bacteriol. 1996;178:2580–2585. doi: 10.1128/jb.178.9.2580-2585.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanabus M, Nowicka A, Sledziewska-Gojska E, Jonczyk P, Ciesla Z. The antimutagenic effect of a truncated epsilon subunit of DNA polymerase III in Escherichia coli cells irradiated with UV light. Mol Gen Genet. 1995;247:216–221. doi: 10.1007/BF00705652. [DOI] [PubMed] [Google Scholar]
- 20.Khidhir A M, Casaregola S, Holland I B. Mechanism of transient inhibition of DNA synthesis in ultraviolet-irradiated E. coli: inhibition is independent of recA whilst recovery requires RecA protein itself and an additional, inducible SOS function. Mol Gen Genet. 1985;199:133–140. doi: 10.1007/BF00327522. [DOI] [PubMed] [Google Scholar]
- 21.Kong X P, Onrust R, O'Donnell M, Kuriyan J. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 1992;69:425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- 22.Kowalczykowski S C, Dixon D A, Eggleston A K, Lauder S D, Rehrauer W M. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Latham G J, Bacheller D J, Pietroni P, von Hippel P H. Structural analyses of gp45 sliding clamp interactions during assembly of the bacteriophage T4 DNA polymerase holoenzyme. II. The Gp44/62 clamp loader interacts with a single defined face of the sliding clamp ring. J Biol Chem. 1997;272:31677–31684. doi: 10.1074/jbc.272.50.31677. [DOI] [PubMed] [Google Scholar]
- 24.Latham G J, Bacheller D J, Pietroni P, von Hippel P H. Structural analyses of gp45 sliding clamp interactions during assembly of the bacteriophage T4 DNA polymerase holoenzyme. III. The Gp43 DNA polymerase binds to the same face of the sliding clamp as the clamp loader. J Biol Chem. 1997;272:31685–31692. doi: 10.1074/jbc.272.50.31685. [DOI] [PubMed] [Google Scholar]
- 25.Link A J, Phillips D, Church G M. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J Bacteriol. 1997;179:6228–6237. doi: 10.1128/jb.179.20.6228-6237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Little J W, Edmiston S H, Pacelli L Z, Mount D W. Cleavage of the Escherichia coli lexA protein by the recA protease. Proc Natl Acad Sci USA. 1980;77:3225–3229. doi: 10.1073/pnas.77.6.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Little J W, Mount D W. The SOS regulatory system of Escherichia coli. Cell. 1982;29:11–22. doi: 10.1016/0092-8674(82)90085-x. [DOI] [PubMed] [Google Scholar]
- 28.Little J W, Mount D W, Yanisch-Perron C R. Purified lexA protein is a repressor of the recA and lexA genes. Proc Natl Acad Sci USA. 1981;78:4199–4203. doi: 10.1073/pnas.78.7.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maor-Shoshani A, Reuven N B, Tomer G, Livneh Z. Highly mutagenic replication by DNA polymerase V (UmuC) provides a mechanistic basis for SOS untargeted mutagenesis. Proc Natl Acad Sci USA. 2000;97:565–570. doi: 10.1073/pnas.97.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marsh L, Walker G C. Cold sensitivity induced by overproduction of UmuDC in Escherichia coli. J Bacteriol. 1985;162:155–161. doi: 10.1128/jb.162.1.155-161.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 32.Murli S, Opperman T, Smith B T, Walker G C. A role for the umuDC gene products of Escherichia coli in increasing resistance to DNA damage in stationary phase by inhibiting the transition to exponential growth. J Bacteriol. 2000;182:1127–1135. doi: 10.1128/jb.182.4.1127-1135.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neuwald A F, Poleksic A. PSI-BLAST searches using hidden Markov models of structural repeats: prediction of an unusual sliding DNA clamp and of beta-propellers in UV-damaged DNA-binding protein. Nucleic Acids Res. 2000;28:3570–3580. doi: 10.1093/nar/28.18.3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nohmi T, Battista J R, Dodson L A, Walker G C. RecA-mediated cleavage activates UmuD for mutagenesis: mechanistic relationship between transcriptional derepression and posttranslational activation. Proc Natl Acad Sci USA. 1988;85:1816–1820. doi: 10.1073/pnas.85.6.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nowicka A, Kanabus M, Sledziewska-Gojska E, Ciesla Z. Different UmuC requirements for generation of different kinds of UV-induced mutations in Escherichia coli. Mol Gen Genet. 1994;243:584–592. doi: 10.1007/BF00284207. [DOI] [PubMed] [Google Scholar]
- 36.Ohta T, Sutton M D, Guzzo A, Cole S, Ferentz A E, Walker G C. Mutations affecting the ability of the Escherichia coli UmuD′ protein to participate in SOS mutagenesis. J Bacteriol. 1999;181:177–185. doi: 10.1128/jb.181.1.177-185.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Opperman T, Murli S, Smith B T, Walker G C. A model for a umuDC-dependent prokaryotic DNA damage checkpoint. Proc Natl Acad Sci USA. 1999;96:9218–9223. doi: 10.1073/pnas.96.16.9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Opperman T, Murli S, Walker G C. The genetic requirements for UmuDC-mediated cold sensitivity are distinct from those for SOS mutagenesis. J Bacteriol. 1996;178:4400–4411. doi: 10.1128/jb.178.15.4400-4411.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paz-Elizur T, Skaliter R, Blumenstein S, Livneh Z. Beta*, a UV-inducible smaller form of the beta subunit sliding clamp of DNA polymerase III of Escherichia coli. I. Gene expression and regulation. J Biol Chem. 1996;271:2482–2490. [PubMed] [Google Scholar]
- 40.Perrino F W, Harvey S, McNeill S M. Two functional domains of the epsilon subunit of DNA polymerase III. Biochemistry. 1999;38:16001–16009. doi: 10.1021/bi991429+. [DOI] [PubMed] [Google Scholar]
- 41.Pietroni P, Young M C, Latham G J, von Hippel P H. Structural analyses of gp45 sliding clamp interactions during assembly of the bacteriophage T4 DNA polymerase holoenzyme. I. Conformational changes within the gp44/62-gp45-ATP complex during clamp loading. J Biol Chem. 1997;272:31666–31676. doi: 10.1074/jbc.272.50.31666. [DOI] [PubMed] [Google Scholar]
- 42.Quinones A, Kaasch J, Kaasch M, Messer W. Induction of dnaN and dnaQ gene expression in Escherichia coli by alkylation damage to DNA. EMBO J. 1989;8:587–593. doi: 10.1002/j.1460-2075.1989.tb03413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reuven N B, Arad G, Maor-Shoshani A, Livneh Z. The mutagenesis protein UmuC is a DNA polymerase activated by UmuD′, RecA, and SSB and is specialized for translesion replication. J Biol Chem. 1999;274:31763–31766. doi: 10.1074/jbc.274.45.31763. [DOI] [PubMed] [Google Scholar]
- 44.Reuven N B, Arad G, Stasiak A Z, Stasiak A, Livneh Z. Lesion bypass by the Escherichia coli DNA polymerase V requires assembly of a RecA nucleoprotein filament. J Biol Chem. 2001;276:5511–5517. doi: 10.1074/jbc.M006828200. [DOI] [PubMed] [Google Scholar]
- 45.Ruiz-Rubio M, Bridges B A. Mutagenic DNA repair in Escherichia coli. XIV. Influence of two DNA polymerase III mutator alleles on spontaneous and UV mutagenesis. Mol Gen Genet. 1987;208:542–548. doi: 10.1007/BF00328153. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 47.Sassanfar M, Roberts J W. Nature of the SOS-inducing signal in Escherichia coli. The involvement of DNA replication. J Mol Biol. 1990;212:79–96. doi: 10.1016/0022-2836(90)90306-7. [DOI] [PubMed] [Google Scholar]
- 48.Shinagawa H, Iwasaki H, Kato T, Nakata A. RecA protein-dependent cleavage of UmuD protein and SOS mutagenesis. Proc Natl Acad Sci USA. 1988;85:1806–1810. doi: 10.1073/pnas.85.6.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skaliter R, Bergstein M, Livneh Z. Beta*, a UV-inducible shorter form of the beta subunit of DNA polymerase III of Escherichia coli. II. Overproduction, purification, and activity as a polymerase processivity clamp. J Biol Chem. 1996;271:2491–2496. doi: 10.1074/jbc.271.5.2491. [DOI] [PubMed] [Google Scholar]
- 50.Skaliter R, Paz-Elizur T, Livneh Z. A smaller form of the sliding clamp subunit of DNA polymerase III is induced by UV irradiation in Escherichia coli. J Biol Chem. 1996;271:2478–2481. [PubMed] [Google Scholar]
- 51.Sommer S, Boudsocq F, Devoret R, Bailone A. Specific RecA amino acid changes affect RecA-UmuD′C interaction. Mol Microbiol. 1998;28:281–291. doi: 10.1046/j.1365-2958.1998.00803.x. [DOI] [PubMed] [Google Scholar]
- 52.Sommer S, Knezevic J, Bailone A, Devoret R. Induction of only one SOS operon, umuDC, is required for SOS mutagenesis in Escherichia coli. Mol Gen Genet. 1993;239:137–144. doi: 10.1007/BF00281612. [DOI] [PubMed] [Google Scholar]
- 53.Studwell P S, O'Donnell M. Processive replication is contingent on the exonuclease subunit of DNA polymerase III holoenzyme. J Biol Chem. 1990;265:1171–1178. [PubMed] [Google Scholar]
- 54.Stukenberg P T, Studwell-Vaughan P S, O'Donnell M. Mechanism of the sliding beta-clamp of DNA polymerase III holoenzyme. J Biol Chem. 1991;266:11328–11334. [PubMed] [Google Scholar]
- 55.Stukenberg P T, Turner J, O'Donnell M. An explanation for lagging strand replication: polymerase hopping among DNA sliding clamps. Cell. 1994;78:877–887. doi: 10.1016/s0092-8674(94)90662-9. [DOI] [PubMed] [Google Scholar]
- 56.Sutton M D, Murli S, Opperman T, Klein C, Walker G C. umuDC-dnaQ interaction and its implications for cell cycle regulation and SOS mutagenesis in Escherichia coli. J Bacteriol. 2001;183:1085–1089. doi: 10.1128/JB.183.3.1085-1089.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sutton M D, Opperman T, Walker G C. The Escherichia coli SOS mutagenesis proteins UmuD and UmuD′ interact physically with the replicative DNA polymerase. Proc Natl Acad Sci USA. 1999;96:12373–12378. doi: 10.1073/pnas.96.22.12373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sutton M D, Smith B T, Godoy V G, Walker G C. The SOS response: recent insights into umuDC-dependent DNA damage tolerance. Annu Rev Genet. 2000;34:479–497. doi: 10.1146/annurev.genet.34.1.479. [DOI] [PubMed] [Google Scholar]
- 59.Sutton M D, Walker G C. umuDC-mediated cold sensitivity is a manifestation of functions of the UmuD2C complex involved in a DNA damage checkpoint control. J Bacteriol. 2001;183:1215–1224. doi: 10.1128/JB.183.4.1215-1224.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tadmor Y, Bergstein M, Skaliter R, Shwartz H, Livneh Z. Beta subunit of DNA polymerase III holoenzyme is induced upon ultraviolet irradiation or nalidixic acid treatment of Escherichia coli. Mutat Res. 1994;308:53–64. doi: 10.1016/0027-5107(94)90198-8. [DOI] [PubMed] [Google Scholar]
- 61.Taft-Benz S A, Schaaper R M. The C-terminal domain of dnaQ contains the polymerase binding site. J Bacteriol. 1999;181:2963–2965. doi: 10.1128/jb.181.9.2963-2965.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tang M, Pham P, Shen X, Taylor J-S, O'Donnell M, Woodgate M, Goodman M F. Roles of E. coli DNA polymerase IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature. 2000;404:1014–1018. doi: 10.1038/35010020. [DOI] [PubMed] [Google Scholar]
- 63.Tang M, Shen X, Frank E G, O'Donnell M, Woodgate R, Goodman M F. UmuD′(2)C is an error-prone DNA polymerase. Escherichia coli pol V. Proc Natl Acad Sci USA. 1999;96:8919–8924. doi: 10.1073/pnas.96.16.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thelen P M, Venclovas C, Fidelis K. A sliding clamp model for the Rad1 family of cell cycle checkpoint proteins. Cell. 1999;96:769–770. doi: 10.1016/s0092-8674(00)80587-5. [DOI] [PubMed] [Google Scholar]
- 65.Venclovas C, Thelen M P. Structure-based predictions of Rad1, Rad9, Hus1 and Rad17 participation in sliding clamp and clamp-loading complexes. Nucleic Acids Res. 2000;28:2481–2493. doi: 10.1093/nar/28.13.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Waga S, Hannon G J, Beach D, Stillman B. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature. 1994;369:574–578. doi: 10.1038/369574a0. [DOI] [PubMed] [Google Scholar]
- 67.Walker G C. Skiing the black diamond slope: progress on the biochemistry of translesion DNA synthesis. Proc Natl Acad Sci USA. 1998;95:10348–10350. doi: 10.1073/pnas.95.18.10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Woodgate R, Rajagopalan M, Lu C, Echols H. UmuC mutagenesis protein of Escherichia coli: purification and interaction with UmuD and UmuD′. Proc Natl Acad Sci USA. 1989;86:7301–7305. doi: 10.1073/pnas.86.19.7301. [DOI] [PMC free article] [PubMed] [Google Scholar]