Summary:
A fundamental strategy of eukaryotic anti-viral immunity involves the cGAS enzyme, which synthesizes 2’,3’-cGAMP and activates a STING effector to prevent viral replication. Diverse bacteria contain cGAS-like enzymes that produce cyclic oligonucleotides and induce antiphage activity, known as CBASS. However, this activity has only been demonstrated through heterologous expression. Whether bacteria harboring CBASS antagonize and co-evolve with phages is unknown. Here, we identified an endogenous cGAS-like enzyme in Pseudomonas aeruginosa that generates 3’,3’-cGAMP, signals to a phospholipase effector, and limits phage replication. In response, phages express an anti-CBASS protein (Acb2) that forms a hexamer with three 3’,3’-cGAMP molecules and reduces phospholipase activity. Acb2 also binds to 2’,3’-cGAMP and 3’,3’-cUU/UA/UG, suggesting broad inhibition of cGAS-based immunity. Upon Acb2 deletion, CBASS blocks lytic phage replication and lysogenic induction, but rare phages evade CBASS through major capsid gene mutations. Together, we demonstrate endogenous CBASS anti-phage function and strategies of CBASS inhibition and evasion.
Graphical Abstract
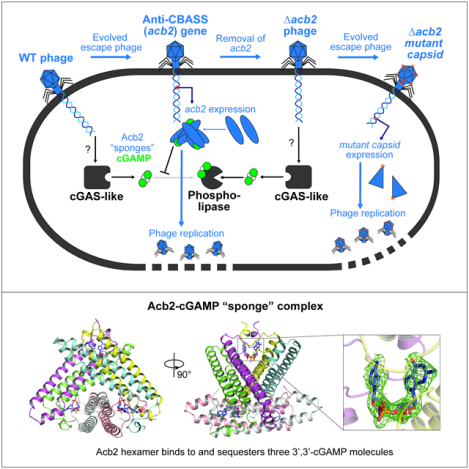
In Brief:
Bacteriophages antagonize cGAS-like bacterial immunity by sequestering immune signaling molecules and acquiring capsid gene mutations.
Introduction:
Sensing specific macromolecules produced or possessed by viruses is an conserved strategy of anti-viral immunity across all kingdoms of life1,2. In mammalian cells, viral double-stranded DNA (dsDNA) is bound by cyclic GMP-AMP synthase (cGAS) in the cytoplasm3,4. The activated cGAS enzyme produces 2’,3’-cyclic GMP-AMP (2’,3’-cGAMP) dinucleotides that bind to the STING effector protein and induces a type I interferon response5,6. Recently, thousands of cGAS-like enzymes named CD-NTases (cGAS/DncV-like nucleotidyltransferases) were identified across the entire bacterial domain and then biochemically characterized, revealing at least 8 enzymatic clades and 10 known cyclic oligonucleotides7. These enzymes are activated during phage infection through an unknown mechanism and produce cyclic oligonucleotides, like 3’,3’-cGAMP, which activate a downstream effector8–12. This strategy of bacterial immunity was coined cyclic-oligonucleotide-based anti-phage signaling system (CBASS)8. CD-NTases and effectors comprise the core CBASS genes (Type I CBASS), and additional ‘signature’ CD-NTase-associated proteins (Cap) have been identified in Type II and III CBASS that regulate CD-NTase activity10,13–16.
Phage infection introduces nucleic acids and numerous foreign proteins into the bacterial cell. However, molecules that cause a phage to be sensitive, or resistant, to a given anti-phage immune system are largely unknown. A recent study discovered a family of phage-encoded anti-CBASS phosphodiesterase enzymes (Acb1), which cleave cyclic oligonucleotides17 similarly to poxin enzymes encoded by eukaryotic viruses18. Investigating the co-evolution of phages and CBASS in a host with endogenous CBASS function will inform how cGAS-based immunity functions in nature and is the main goal of this study.
Pseudomonas aeruginosa is a human opportunistic pathogen that encodes a diversity of CBASS operons and is a generalist microbe that survives in many niches. P. aeruginosa also has a diverse phage population and is a leading candidate for phage therapy, but our limited understanding of anti-phage immunity is a barrier for basic biology and phage therapeutic development. Here, we identified a P. aeruginosa strain that harbors Type II-A CBASS (3’,3’-cGAMP producing CD-NTase; CdnA) with a phospholipase (CapV) effector that limits phage replication by ≥10,000-fold. This is a notable finding because it demonstrates that endogenous CBASS anti-phage immunity can function without overexpression. We next identified a widespread phage protein (Acb2) that forms a hexamer complex with three 3’,3-cGAMP molecules, acting as a “sponge” to reduce the available molecules to activate the phospholipase effector. In addition, Acb2 binds to multiple other cyclic dinucleotides, including 3’,3’-c-di-UMP, 3’,3’-cUA, and 3’,3’-cUG, and is necessary for optimal phage replication in the presence of Type I or II CBASS that are predicted to encode the aforementioned cyclic dinucleotides. Phages with acb2 deleted were unable to replicate in the lytic cycle or during exit from lysogeny in the presence of CBASS. However, mutations in the major capsid gene enabled phages to escape CBASS. This work provides direct evidence of phage inhibition and evasion of CBASS, demonstrating a robust arms race between the two.
Results:
Endogenous anti-phage CBASS function in Pseudomonas aeruginosa
Previous analyses13, coupled with our own bioinformatics, revealed that 252 distinct P. aeruginosa strains have >300 CBASS operons (Figure S1A–B). These systems span Type I-III and use numerous effector proteins and cyclic oligonucleotides7. The diverse CBASS types in P. aeruginosa suggest that it is important for host fitness and that it may be well suited to study phage-CBASS interactions. To identify naturally functional CBASS immunity in P. aeruginosa, CBASS loci were deleted from the genome of four strains possessing representatives of the common CBASS types (Type I-A, II-A, II-C, and III-C; Figure S1C) using a CRISPR-Cas3 tool19. Notably, these strains also encode numerous other anti-phage immune systems (Figure 1A). Therefore, due to the multitude of immune systems, we screened the CBASS mutants against a diverse panel of ~70 phages, which spanned 23 different genomic families and four different morphologies (Myoviridae, Siphoviridae, Podoviridae, and Inoviridae20). A single P. aeruginosa strain (BWHPSA011; Pa011) was identified with CBASS-dependent anti-phage activity (Figure S1D–E).
Figure 1. P. aeruginosa BWHPSA011 (Pa011) CBASS-based immunity protects against PaMx41 infection.
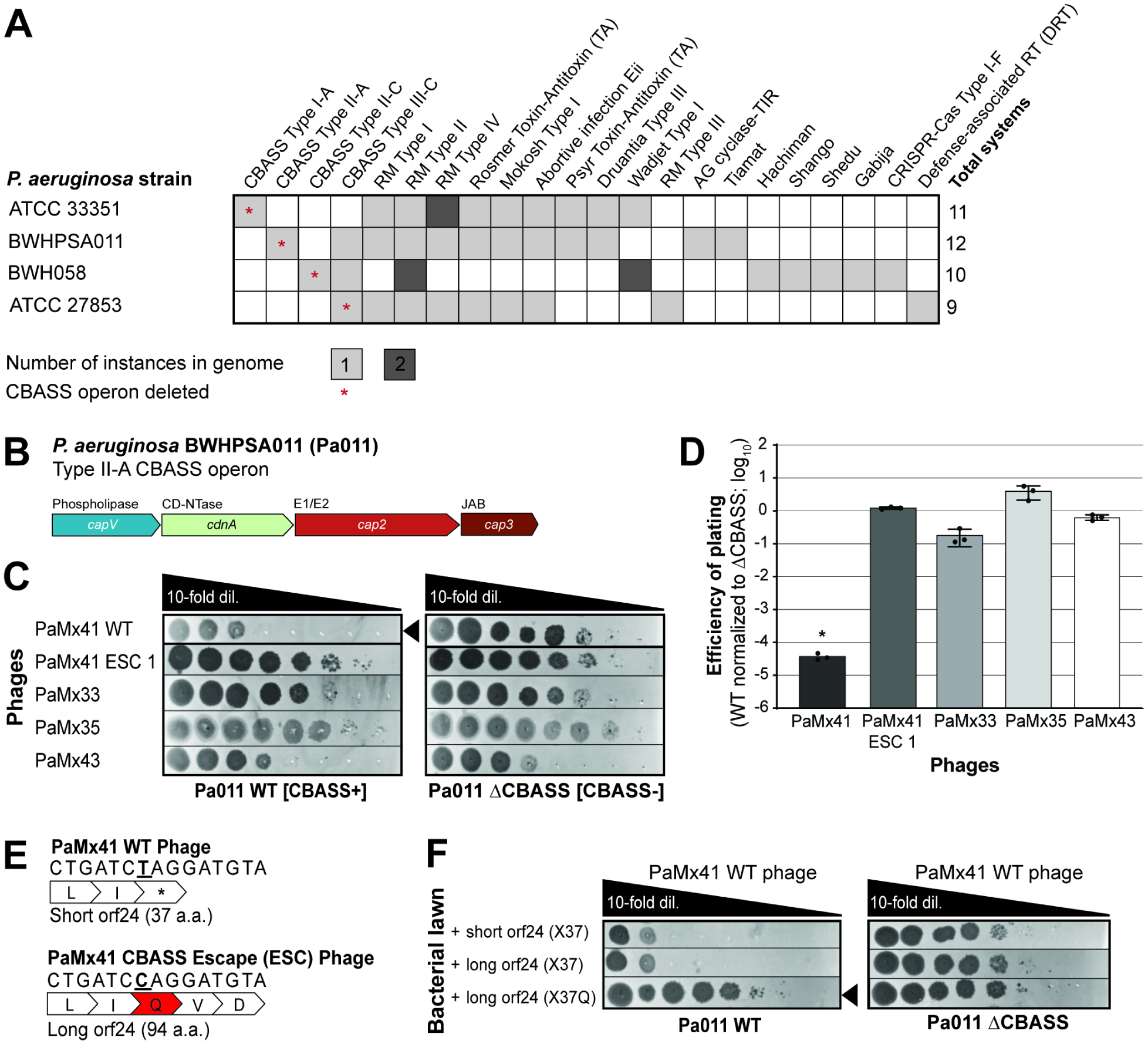
(A) The presence of different anti-phage immune systems in P. aeruginosa strains that were used in this study. Some systems are present in the genome twice as indicated by the darker shade of gray and number (2). An asterisk (*) indicates the CBASS operon was effectively deleted from the bacterial genome. (B) Pa011 CBASS operon. (C) Plaque assays with PaMx41-like phages and an evolved PaMx41 CBASS Escaper (ESC) phage spotted in 10-fold serial dilutions on a lawn of Pa011 WT [CBASS+] or ΔCBASS [CBASS-]; clearings represent phage replication. Black arrowhead highlights the reduction in PaMx41 WT phage titer. See also Figure S1. (D) Efficiency of plating was quantified as plaque-forming units (PFU) per ml on Pa011 WT divided by PFU/ml on the ΔCBASS (n=3). Data are mean ± s.d. Non-parametric ANOVA test yielded a P value of <0.0001. (E) Schematic of PaMx41 WT and CBASS escape phage genomes with the no-stop extension mutation. The bold underline indicates mutation of thymine (T) to cytosine (C), resulting in a stop codon (TAG, *) to glutamine (CAG, Q) substitution (in red). (F) Plaque assays with PaMx41 WT phage on a lawn of Pa011 WT or ΔCBASS over-expressing the indicated orf24 variants. See also Figure S2.
Deletion of the Pa011 Type II-A CBASS operon (ΔCBASS, Figure 1B) resulted in >4 orders of magnitude increase in titer of the dsDNA podophage PaMx41 (Figure 1C–D). Phage protection was restored when all four CBASS genes (capV (phospholipase effector), cdnA (CD-NTase), cap2 (E1/E2 ubiquitin-ligase-like domains), and cap3 (JAB de-ubiquitinating enzyme-like domain)) were complemented on a plasmid (Figure S1G). To determine which genes are necessary for CBASS anti-phage activity, chromosomal mutants known to disrupt catalytic activity8,15 were generated: capVS48A, cdnAD87A/D89A, cap2C450A/C453A, and cap3E38A. capV, cdnA, and cap2 mutations abolished anti-phage activity whereas the cap3 mutation did not (Figure S1H). Furthermore, using an ELISA, we observed that PaMx41 infection generated low levels of 3’,3’-cGAMP (~8 nM; Figure S2A–B) whereas the molecule was nearly undetectable in CdnA mutant strain (~0.5 nM; L.O.D. 0.24 nM). These data demonstrate that Pa011 CBASS-based immunity is naturally active, significantly limits phage replication, and requires CapV, CdnA, and Cap2 enzyme activities for phage targeting.
PaMx41-like phages encode a CBASS antagonist
How CBASS detects and targets phage is currently unknown. Therefore, to identify phage genes required for successful CBASS activity, we isolated PaMx41 mutants that resist Pa011 CBASS-based immunity. With a frequency of 3.7 × 10−5 (Figure 1D), 10 independent PaMx41 CBASS “escape” phages were isolated that replicate well on Pa011 WT (Figure 1B). Whole genome sequencing revealed one mutation in all CBASS escape phages: a no-stop extension mutation (X37Q) in orf24 (Figure 1E). X37Q lengthens gp24 from a 37 amino acid (a.a.) protein to 94 a.a. Interestingly, the naturally CBASS resistant phages, PaMx33, PaMx35, and PaMx43, share >96% nucleotide identity across the genome and naturally encode the 94 a.a. version with >98% a.a. identity.
To determine whether the short gp24 activates CBASS or the long gp24 antagonizes it, the PaMx41 gp24 variants were overexpressed in Pa011 WT and ΔCBASS cells and then plaque assays were performed. In the presence of CBASS, the long gp24 increased the titer of the PaMx41 WT phage by ≥4 orders of magnitude while the truncated gp24 versions had no effect (Figure 1F). In contrast, the PaMx41 escaper phage and PaMx33, PaMx35, and PaMx43 phages exhibited high titer on all strains (Figure S2C), demonstrating that the short gene is not a dominant CBASS activator. We next deleted orf24 from all of the resistant PaMx41-like phages using a Cas13a selection tool because these phages were surprisingly resistant to all tested DNAtargeting CRISPR-Cas systems21. In Pa011 WT cells, the titer of the Δorf24 phages was reduced 2–4 orders of magnitude; however, expression of the long gp24 in trans rescued the phages (Figure 2A–B, S2D). Taken together, these results indicate that the long gp24, or Acb2 (anti-cbass 2; PaMx33 NCBI: ANA48877) hereafter, inhibits Pa011 CBASS immunity and is necessary for phage replication in the presence of CBASS.
Figure 2. Phage-encoded acb2 is necessary for replication in the presence of CBASS.
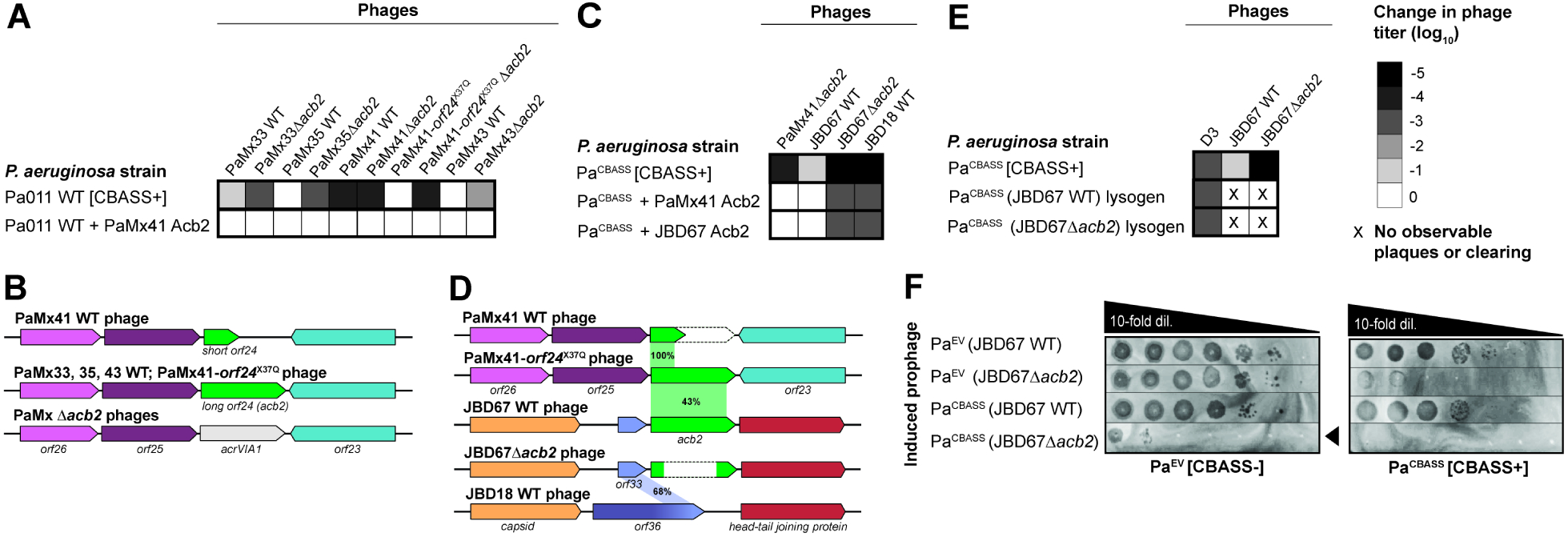
(A, C, E) Heat maps representing the order of magnitude change in phage titer, where phage titer is quantified by comparing the number of spots (with plaques, or clearing if plaques were not visible) on the CBASS+ strain divided by the CBASS- strain. Plaque assays used for these quantifications can been seen in Figure S2 (n=3). (B, D) Comparison of the acb2 locus across phage genomes. PaMx41-like Δacb2 phages have the acb2 gene substituted with the type VI-A anti-CRISPR gene (acrVIA1) as part of the knockout procedure, and JBD67Δacb2 phages have the acb2 gene removed from its genome. Genes with known protein functions are indicated with names, and genes with hypothetical proteins are indicated with “orf”. Acb2 percent amino acid identity is shown in (D). (F) Plaque assays assessing the titer induced prophages spotted on a lawn of PaEV or PaCBASS. Black arrowhead highlights reduction in JBD67Δacb2 phage titer. See also Figure S2.
Acb2 has conserved function in a broadly distributed temperate phage family
Homology searches with Acb2 revealed that it is encoded in a striking number of tailed phages, including those infecting Pseudomonas, Vibrio, Acinetobacter, Salmonella, Serratia, Erwinia, and Escherichia sp. (T2 and T4 phages; gene: vs.4), among others (Figure S3A). We observed no other examples of the truncated Acb2 variant encoded in PaMx41 WT phage. A multi-sequence alignment with diverse homologs revealed highly conserved N- and C-termini with a middle region of varied length and sequence (Figure S3B), but no molecular function could be predicted. Furthermore, acb2 is commonly encoded by P. aeruginosa B3-like temperate phages (e.g. JBD67), which are unrelated to the PaMx41-like lytic phages. Since JBD67 phage does not replicate on the Pa011 strains, we integrated the Type II-A CBASS operon with its native promoter into the chromosome of a P. aeruginosa strain (POA1) that is sensitive to this phage and naturally lacks CBASS. This engineered strain (PaCBASS) was active and reduced PaMx41 WT phage titer by 3–4 orders of magnitude (Figure S1D, S1F). By contrast, JBD67 exhibited resistance to the PaCBASS strain while a related phage that naturally lacks the acb2 gene, JBD18, was robustly inhibited (Figure 2C–D, S2E). Deletion of acb2 from JBD67 using a helicase attenuated Cascade-Cas3 system (see Methods) sensitized it to CBASS immunity (Figure 2C–D, S2E). The titer of JBD67Δacb2 and JBD18 phages were reduced by ≥5 orders of magnitude in the presence of CBASS. Expression of acb2 derived from either JBD67 or PaMx41-orf24X37Q on a plasmid fully restored the titer of the PaMx41Δacb2 phage, and only partially restored the titer of the JBD67Δacb2 and JBD18 phages (Figure 2C, S2E). We hypothesize that JBD67 and JBD18 more strongly activate CBASS, and consequently, Acb2 expression in trans becomes partially overwhelmed. Together, these results collectively demonstrate that acb2 retains its anti-CBASS function across distinct phage families.
Given that JBD67 is a temperate phage, we investigated whether acb2 is active during lysogeny as a prophage and therefore inhibits a co-encoded CBASS system. Lysogens were constructed with JBD67 in the PaCBASS strain, where CBASS targets the super-infecting phage D3. JBD67 and D3 are from hetero-immune groups, so there is no super-infection exclusion between these phages. Interestingly, the degree to which CBASS targets D3 was not impacted by the presence of acb2 in the JBD67 prophage (Figure 2E, S2F). This suggests that acb2 is not expressed during lysogeny and allows functional CBASS immunity despite a prophage-encoded inhibitor. However, upon exit from lysogeny, CBASS significantly blocks JBD67Δacb2 prophage induction, reducing the induced titer by ~5-orders of magnitude whereas JBD67 WT prophage induction was unaffected (Figure 2F). These collective findings demonstrate that acb2 is active during lysogenic induction, but not lysogenic maintenance, and importantly shows that CBASS can dramatically limit prophage induction.
Acb2 sequesters 3’,3’-cGAMP to inhibit CBASS
To determine the mechanism of Acb2, we purified Acb2 from the naturally CBASS-resistant PaMx33 phage and each CBASS protein to test direct CBASS antagonism. First, we tested binding and found that Acb2 did not bind to purified CapV, CdnA, Cap2, nor the CdnA-Cap2 complex, which was purified as described previously (Ledvina et al., 2022; Figure S4A–D). Next, we reconstituted CapV phospholipase activity in vitro and confirmed that it is only activated by 3′,3′-cGAMP in a concentration-dependent manner, but not by 2′,3′-cGAMP, c-di-AMP, or c-diGMP (Figure 3A). When Acb2 was preincubated with 3′,3′-cGAMP, CapV activity was abrogated (Figure 3A), suggesting that Acb2 may directly bind to the 3′,3′-cGAMP molecule. A native gel assay showed a significant shift of the purified Acb2 protein upon adding 3′,3′-cGAMP (Figure 3B). Isothermal calorimetry (ITC) experiments further verified that Acb2 directly binds to 3′,3′cGAMP with a KD of ~87 nM (Figure 3C, S4E). Together, these data suggest that Acb2 binding of 3′,3′-cGAMP antagonizes CBASS by reducing available signaling molecules.
Figure 3. Acb2 antagonizes CBASS activity by sequestering the 3’,3’-cGAMP signaling molecule.
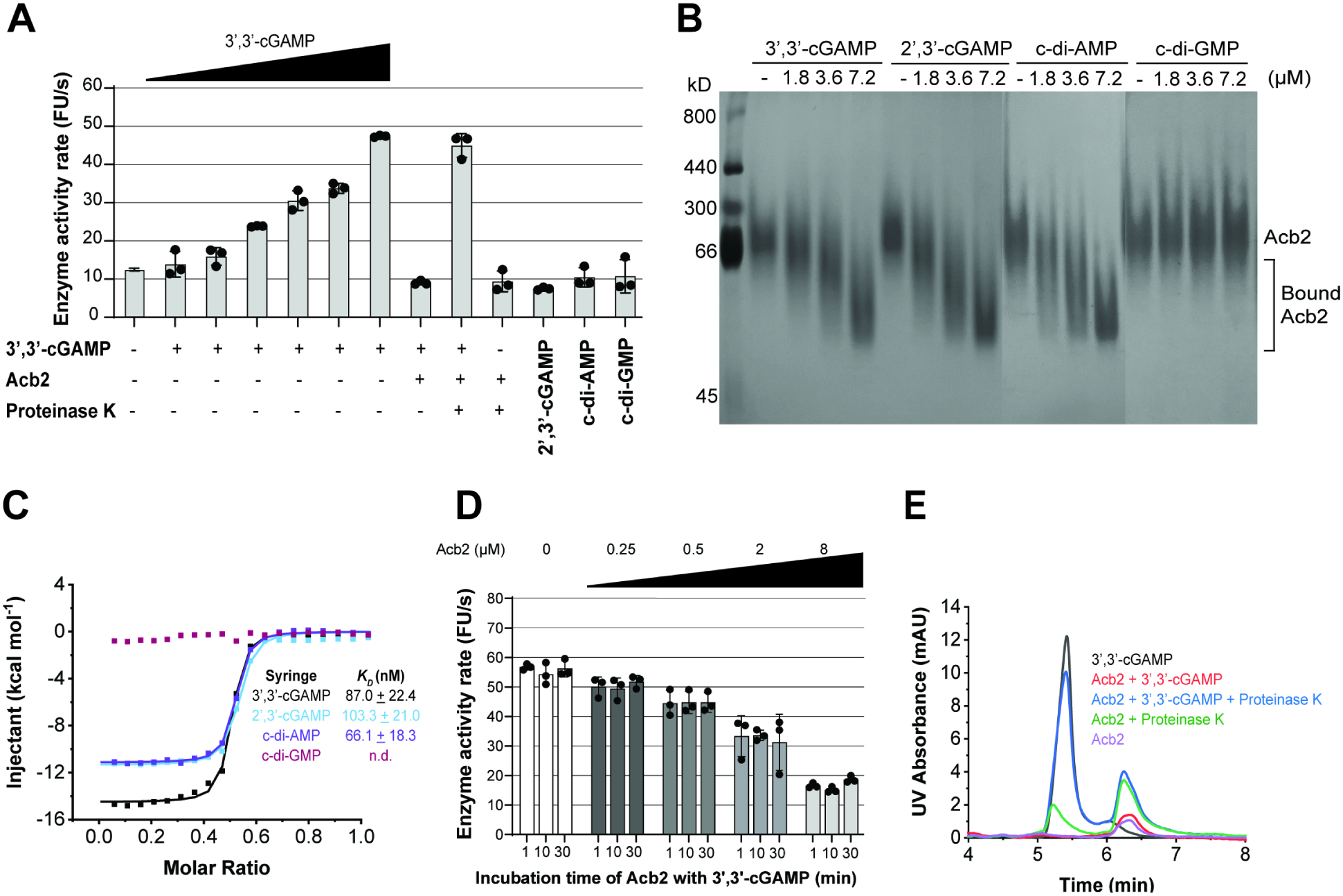
(A) CapV enzyme activity in the presence of the indicated cyclic dinucleotides and resorufin butyrate, which is a phospholipase substrate that emits fluorescence when hydrolyzed. The enzyme activity rate was measured by the accumulation rate of fluorescence units (FU) per second. The concentration of 3’,3’-cGAMP ranged from 0.025 to 0.8 μM (0.025, 0.05, 0.1, 0.2, 0.4, 0.8 μM), and the other cyclic dinucleotides were added at 0.8 μM. To test the effects of Acb2 to bind and release 3’,3’-cGAMP, Acb2 (32 μM) was incubated with 3’,3’-cGAMP (0.8 μM) for 10 minutes and then Proteinase K (0.065 mg/mL) was added to extract the nucleotides from the Acb2 protein. Filtered nucleotides products were used for the CapV activity assay. Data are mean ± s.d. (n=3). (B) Native PAGE showed the binding of Acb2 to cyclic dinucleotides. (C) Isothermal titration calorimetry (ITC) assays to test binding of cyclic dinucleotides to Acb2. Representative binding curves and binding affinities are shown. The KD values are mean ± s.d. (n=3). Raw data for these curves are shown in Figure S4. (D) CapV activity assay to test the effects of Acb2 on 3’,3’-cGAMP. The concentration of 3’,3’-cGAMP was 0.8 μM and Acb2 ranged from 0.25 to 8 μM (0.25, 0.5, 2, 8 μM). The 3’,3’-cGAMP was pre-incubated with Acb2 for 1, 10, 30 minutes, respectively. Data are mean ± s.d. (n=3). (E) The ability of Acb2 to bind and release 3’,3’-cGAMP when treated with proteinase K was analyzed by HPLC. 3’,3’-cGAMP standard was used as a control. The remaining 3’,3’-cGAMP after incubation with Acb2 were tested.
Recent studies have reported that eukaryotic18 and prokaryotic17,22 viruses express proteins that directly degrade cyclic oligonucleotides. Therefore, to determine whether Acb2 is an enzyme that cleaves 3’,3’-cGAMP molecules, we performed the CapV phospholipase activity assay using a series of Acb2 concentrations and Acb2-3′,3′-cGAMP incubation times. Inhibition of CapV activity was concentration-dependent, but not time-dependent (Figure 3D), suggesting that Acb2 is a protein that “sponges” and sequesters 3’,3’-cGAMP rather than degrade it. High-performance liquid chromatography (HPLC) clearly showed that incubation of Acb2 depletes detectable 3′,3′-cGAMP, and following proteolysis of Acb2, the molecule is released back into the buffer (Figure 3E). Filtration of the unbound 3’,3’-cGAMP enabled CapV to regain its enzymatic activity, demonstrating that the molecule is still active (Figure 3A). Consistent with these results, overexpression of Acb2 in phage-infected Pa011 WT cells reduced detectable 3’,3’-cGAMP levels, but following phenol-chloroform/chloroform nucleotide extraction, the molecule was released and levels increased ~4-fold (Figure S2A–B). Altogether, these results demonstrate that Acb2 antagonizes Type II-A CBASS immunity via binding and sequestering the 3’3’-cGAMP dinucleotide, which prevents phospholipase effector activation.
Crystal structure of Acb2 and its complex with 3′,3′-cGAMP
To further understand how Acb2 interacts with 3’,3’-cGAMP, we determined the crystal structures of apo Acb2 and its complex with the signaling molecule (Table 1). Interestingly, Acb2 folds as a homo-hexamer in its apo (Figure 4A) and 3‘,3’-cGAMP bound form (Figure 4B). Analytical ultracentrifugation (AUC) assay also indicated that Acb2 is a hexamer in its apo and 3’,3’-cGAMP-bound forms in solution (Figure S5A–E). Each protomer mainly interacts with two adjacent protomers (Figure S5B), allowing the six protomers to interlock into a compact assembly. An Acb2 protomer consists of one short N-terminal helix and two long anti-parallel helices, with a kink in the long helix at the C-terminus (Figure S5C). The ligand-bound structure showed that one Acb2 hexamer binds three 3′,3′-cGAMP molecules (Figure 4B). Each cGAMP binding pocket is formed by two Acb2 protomers that interact in a head-to-head manner, and is mainly composed of N- and C-terminal helices/loops from each protomer (Figure S5D). This is consistent with the Acb2 multiple sequence alignment, which revealed highly conserved N- and C-termini (Figure S3B). Searches using the Dali (distance-matrix alignment) server did not return entries with the same protein fold as Acb223, indicating that Acb2 adopts an overall novel cyclic dinucleotide binding fold. In support of these findings, a recent preprint16 demonstrated that the Acb2 homolog from E. coli phage T4 (gene vs.4) tightly binds to 3’,3’-cGAMP (KD of ~30 nM) and adopts a hexameric structure similar to the one reported here.
Table 1.
Data collection and refinement statistics.
| Acb2 | Acb2-3’,3’-cGAMP | Acb2-c-di-AMP | |
|---|---|---|---|
| PDB code | 8H2X | 8H2J | 8H39 |
| Data collection | |||
| Space group | P321 | P321 | P321 |
| Cell dimensions | |||
| a, b, c (Å) | 101.9, 101.9, 101.7 | 103.5, 103.5, 101.6 | 104.1, 104.1, 102.0 |
| (°) | 90.00, 90.00, 120.00 | 90.00, 90.00, 120.00 | 90.00, 90.00, 120.00 |
| Resolution (Å) | 50–2.69 (2.79–2.69)b | 12.87–2.40 (2.49–2.40) | 67.58–2.01 (2.11–2.01) |
| Rsym or Rmerge (%) | 16.9 (66.6) | 8.3 (41.6) | 9.2 (122.8) |
| I/σ(I) | 16.6 (3.0) | 31.8 (5.7) | 21.0 (2.8) |
| Completeness (%) | 99.9 (100.0) | 99.2 (100.0) | 100.0 (100.0) |
| Redundancy | 14.1 (10.2) | 22.3 (21.9) | 20.0 (20.4) |
| Refinement | |||
| Resolution (Å) | 33.37–2.69 (2.79–2.69) | 12.87–2.40 (2.49–2.40) | 67.58–2.01 (2.08–2.01) |
| Unique reflection | 17084 (1624) | 24843 (2448) | 43247 (6272) |
| Rwork / Rfreea | 0.250/0.269 | 0.244/0.279 | 0.209/0.233 |
| No. atoms | 4449 | 4704 | 4859 |
| Protein | 4302 | 4396 | 4365 |
| Ligand/ion | 62 | 180 | 208 |
| Water | 85 | 128 | 286 |
| B factors | 52.64 | 50.22 | 44.81 |
| Protein | 53.23 | 50.70 | 45.12 |
| Ligand/ion | 38.93 | 44.72 | 38.41 |
| Water | 32.44 | 41.57 | 44.80 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.010 | 0.005 | 0.004 |
| Bond angles (°) | 1.58 | 1.00 | 0.60 |
| Ramachandran plot (%) | |||
| Favored | 97.90 | 97.01 | 98.31 |
| Allowed | 2.10 | 2.99 | 1.69 |
| Outliers | 0.00 | 0.00 | 0.00 |
For each structure one crystal was used.
Values in parentheses are for highest-resolution shell.
Figure 4. Structure of Acb2 reveals hexamer bound to three molecules of 3’,3’-cGAMP.
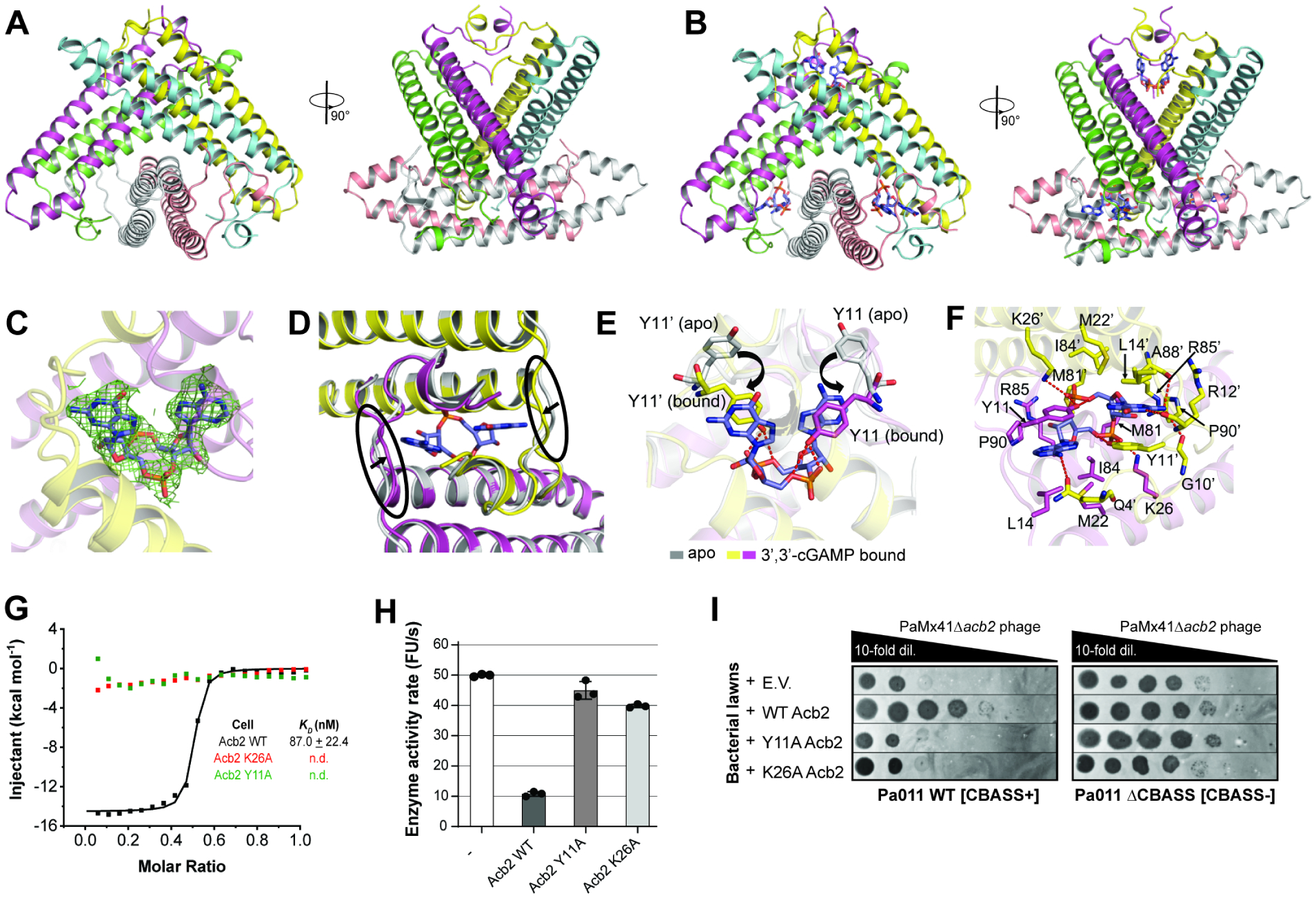
(A) Overall structure of the Acb2 hexamer. Two views are shown. (B) Overall structure of the Acb2 hexamer bound to three molecules of 3’,3’-cGAMP. 3’,3’-cGAMP molecules are colored in slate. (C) 2Fo-Fc electron density of 3’,3’-cGAMP in the binding pocket contoured at 1 σ. (D) Structural comparison between an Acb2 dimer in its apo (colored in gray) and 3’,3’-cGAMP-bound form (colored in yellow and light magenta). The loops which move upon 3’,3’-cGAMP binding are marked with black circles. (E) Structural alignment between apo and 3’,3’-cGAMP-bound Acb2, which are colored as in (B). Y11 from the two structures are highlighted as sticks. Red dashed lines represent poplar interactions. (F) Binding between Acb2 or Acb2 mutants and 3’,3’-cGAMP. Red dashed lines represent polar interactions. (G) ITC assays to test the binding of 3’,3’-cGAMP to Acb2 mutants. Representative binding curves and binding affinities are shown. The KD values are mean ± s.d. (n=3). Raw data for these curves are shown in Figure S4. (H) CapV activity assay to test the effects of Acb2 mutations. The concentration of 3’,3’-cGAMP was 0.8 μM and Acb2 or its mutants was 32 μM. The 3’,3’-cGAMP was pre-incubated with Acb2 or its mutants for 10 minutes. Bar graph represents average of three technical replicates. Data are mean ± s.d. (n=3). (I) Plaque assays on a lawn of Pa011 WT or ΔCBASS overexpressing empty vector (E.V.) or the indicated PaMx41 Acb2 mutants; clearings represent phage replication.
Acb2 binding of a cyclic dinucleotide ligand only causes a slight movement of the loop linking α1 and α2 (named loop L12) towards the ligand (Figure 4D), but several other residues display rotations and movements upon ligand binding. Among them, the most dramatic movement happens in Y11 from both protomers, which rotates and forms π-π interactions with the purine bases of cGAMP, as well as hydrogen bonds with the phosphate group of cGAMP (Figure 4E). In addition, K26 in α2 of both protomers forms a salt bridge with the phosphate group of the cGAMP, possibly stabilizing the molecule together with Y11 (Figure 4F). Additionally, cGAMP is stabilized through hydrophobic interactions by several residues from both protomers, such as L14, M22, M81, I84 and P90 (Figure 4F). Consistent with these analyses, Acb2 Y11A and K26A mutants were inactive in plaque assays (Figure 4I) and the Acb2 K26A mutant expressed in the Pa011 WT strain lost its ability to sequester 3’,3’-cGAMP in vivo (Figure S2A–B). Furthermore, Acb2 Y11A and K26A mutants abolished Acb2 binding of 3’,3’-cGAMP in vitro (Figure 4G, S4I–K) and abrogated its ability to reduce CapV activity (Figure 4H). These data collectively show that Acb2 proteins form a complex with cyclic dinucleotides and subsequently reduces downstream CBASS activation.
Acb2 sequesters variety of cyclic dinucleotides
The binding mode of 3′,3′-cGAMP within Acb2 implied that other cyclic dinucleotides could also be sequestered by this protein. Indeed, native gels and ITC experiments demonstrated that Acb2 also binds to 2′,3′-cGAMP and c-di-AMP with high affinity (KD of ~103 and 66 nM, respectively), but not c-di-GMP (Figure 3B–C, S4F–G). HPLC assays confirmed that Acb2 could also sequester c-di-AMP and 2′,3′-cGAMP, and showed a very weak sequestering effect on c-diGMP (Figure S5F–H). The Acb2-c-di-AMP structure was also solved (Table 1) and c-di-AMP showed a similar binding mode as 3′,3′-cGAMP (Figure S5I–K). Additional native gels and ITC experiments showed that Acb2 can bind to 3’,3’-c-di-UMP (cUU) and 3’,3’-c-UMP-AMP (cUA) with high affinity (KD of ~99 and 97 nM, respectively), and binds to 3’,3’-c-UMP-GMP (cUG) with lower affinity (KD of ~524 nM) (Figure S6A–E). Next, we tested whether phages expressing acb2 could inhibit CBASS subtypes in vivo that harbor a CdnE cyclase, which are predicted to generate cUU, cUA, and/or cUG (Personal Communication with A. Whiteley)7. In our strain collection, we identified that P. aeruginosa strains ATCC 33351 and JD332 encode Type I-A and Type I-B CBASS operons, respectively, with a CdnE cyclase (Figure S6F). When expressed in a P. aeruginosa strain (PAO1 WT) that naturally lacks CBASS, the titer of phages lacking acb2 was reduced 1–3 orders of magnitude – most notably by JD332 Type I-B CBASS – and the isogenic phages containing acb2 remained resistant to CBASS (Figure S6G). Together, these in vitro and in vivo results suggest that Acb2 can broadly inhibit CBASS types that utilize cyclic dinucleotides with uracil and adenine bases.
Phages escape CBASS via mutations in the capsid gene
The function and mechanism of acb2 demonstrates that CBASS places strong evolutionary pressure on phage. Therefore, we sought to determine whether any alternative mechanisms to evade CBASS exist. We again attempted to identify mutant escape phages, but this time, we used phage lacking acb2 (PaMx41Δacb2). Phages were first exposed directly to CBASS by plating on Pa011 WT, which yielded no escape plaques. However, the invisible phage population on the plate was collected, amplified in the Pa011 ΔCBASS strain, and ultimately escape phages were isolated under CBASS selection (Figure 5A). Whole genome sequencing revealed that all seven escape phages had one of four different missense point mutations in the major capsid gene (orf11, NCBI ID: YP_010088887.1; Figure 5B, Table S1). To confirm the causality of the capsid mutations for CBASS escape, we used plasmid-based recombination to introduce de novo mutations I121A, I121T, and S330P into a naive PaMx41Δacb2 phage and confirmed that these mutations induced CBASS escape (Figure 5C–D). To test whether the major capsid is sufficient to trigger CBASS, the PaMx41 WT or mutant major capsid gene with its native promoter was cloned and expressed in Pa011 WT and ΔCBASS cells. In the presence of the WT major capsid, we did not observe any CBASS-dependent cellular toxicity, indicating that capsid monomer is not sufficient to activate CBASS. We also did not observe CBASS-dependent targeting of resistant phage (Figure S7A). Likewise, expression of the mutant major capsid gene did not induce escape of CBASS sensitive phage (Figure S7B). We anticipate that the plasmidencoded major capsid gene does not express well enough to sufficiently displace the phage-expressed major capsid gene in a manner that impacts the CBASS phenotype.
Figure 5. CBASS escape phages have mutations in the major capsid gene.
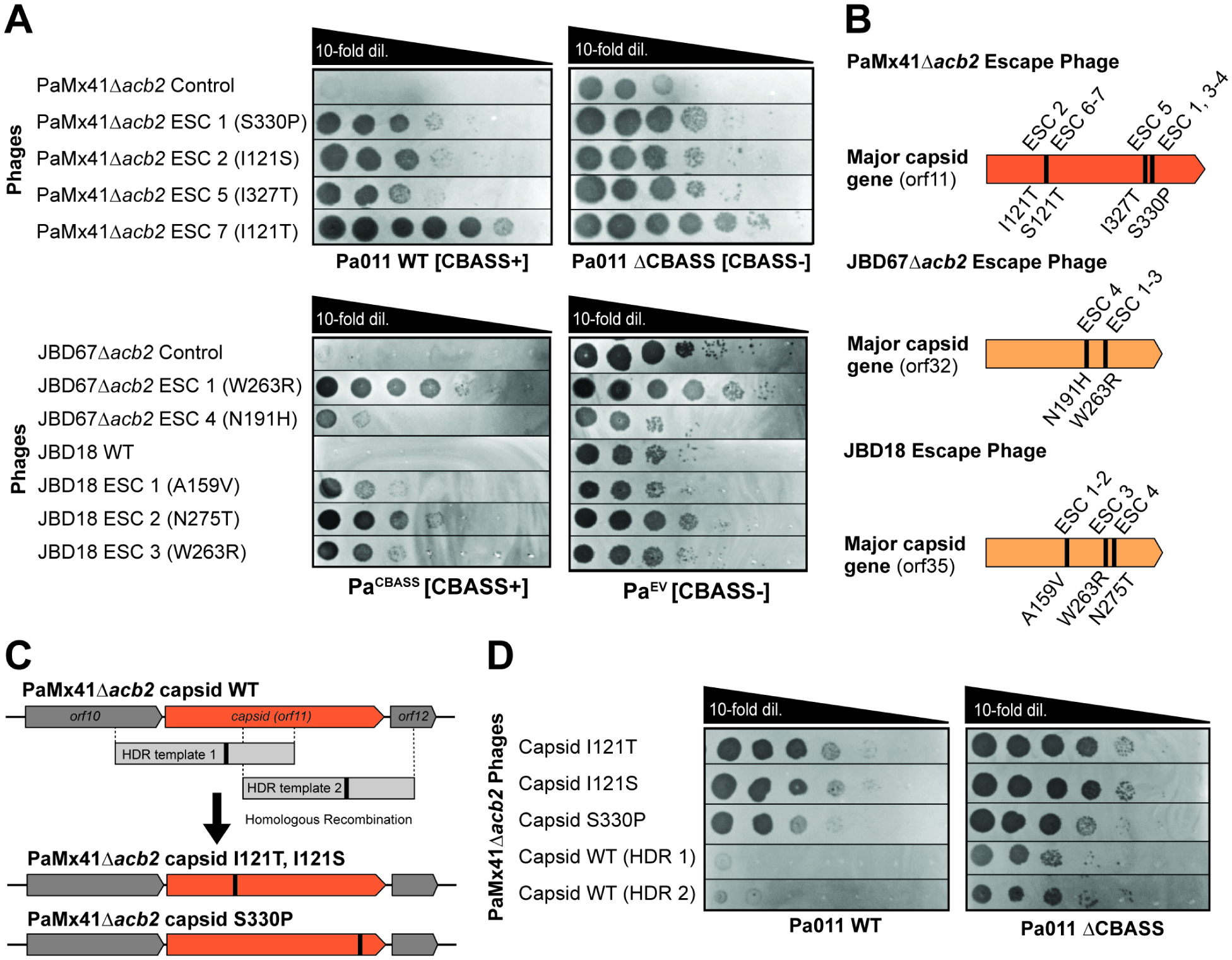
(A) Plaque assays were performed with the indicated control/WT and escape phages spotted in 10-fold serial dilutions on lawns of bacteria expressing CBASS+ (left) or lacking CBASS- (right); clearings represent phage replication. (B) Schematic of major capsid genes with corresponding missense mutations and associated CBASS Escape (ESC) phages. (C) Schematic of in vivo homologous recombination of parental phages with homology-directed repair (HDR) template 1 (encoding I121S or I121T capsid mutations) or template 2 (S330P capsid mutation) and resultant engineered/recombinant phages. (D) Plaque assays with recombinant phages possessing major capsid mutations, or WT capsid controls, spotted on lawns of Pa011 WT or ΔCBASS.
CBASS escape phages were also isolated from JBD67Δacb2 and JBD18 phages on the PaCBASS strain (Figure 5A). Strikingly, whole genome sequencing revealed missense point mutations in the genes encoding their major capsid proteins [orf32 in JBD67 (NCBI: YP_009625956) and orf35 in JBD18 (NCBI: AFR52188); Figure 5B, Table S1]. The PaMx41 major capsid protein shares no significant amino acid identity with the JBD67 and JBD18 major capsid proteins, yet the mutations all converge on the same protein. When modeling the location of the mutations on the predicted major capsid monomer structures, we did not observe overlap between the distinct phages (Figure S7C). However, modeling of the predicted major capsid hexamer structures indicated that the mutations lie on the protein-protein interface within or between hexamers (Figure S7D–E), suggesting that a higher ordered capsid structure or process may be implicated in CBASS immunity. Interestingly, some of the observed capsid mutant genotypes (i.e. I121S/T from PaMx41, N191H from JBD67, and N275D from JBD18) are common natural alleles in the capsids of other phages, suggesting that these genotypes are fit. Follow-up studies are needed to determine the mechanistic connection between the major capsid protein and CBASS.
Discussion:
The discovery and characterization of CBASS marked an exciting connection between prokaryotic and eukaryotic immunity7,8. However, many questions remain in this nascent field regarding phage-host evolution, as well as CBASS regulation and activation mechanisms, that can be best addressed through utilization of endogenous CBASS model systems. We found that the P. aeruginosa BWHPSA011 (Pa011) strain harbors a cGAS-like enzyme (CdnA) that produces 3’,3’-cGAMP dinucleotides in response to PaMx41 phage infection, which activates the phospholipase (CapV) effector. Phages related to PaMx41 (or an escape mutant derived from PaMx41) produce an anti-CBASS protein (Acb2) that is expressed as a middle gene in the phage replication cycle24 and sequesters 3’,3’-cGAMP. Acb2 likely accumulates prior to the production of the dinucleotide given that previous work demonstrated 3’,3’-cGAMP levels increase later in the E. coli phage P1 replication cycle8. Furthermore, when studying the temperate phage JBD67 with its acb2 gene removed, we found that CBASS limits prophage induction and lytic phage replication, which had not been previously noted.
The cyclic dinucleotide “sponge” protein discovered in this study, Acb2, joins an expanding repertoire of phage-encoded antagonists that directly act on signaling molecules involved in bacterial anti-phage immunity. Previous examples include the anti-CRISPR (AcrIII1) protein that cleaves c-A4 molecules22, and the anti-Pyscar (Apyc1) protein that cleaves a spectrum of cyclic nucleotides17. In parallel, a Thoeris anti-defense (Tad1) protein was found to specifically bind to and sequester gcADPR molecules25. Additionally, the anti-CBASS gene identified (Acb1) is another class of phage enzymes, similar to Apyc1, that harbors a phosphodiesterase fold that specifically binds and cleaves cyclic nucleotides17. This is a common inhibitory mechanism of the eukaryotic cGAS-STING signaling system and is utilized by poxviruses18 and a host pyrophosphatase/phosphodiesterase protein26, which enzymatically cleaves and depletes 2’,3’-cGAMP. By contrast, Acb2 binds to and sequesters bacterial 3’,3’-cGAMP, c-di-AMP, 3’,3’-c-diUMP, 3’,3’-cUA, 3’,3’-cUG, and human 2’,3’-cGAMP. The structure and mechanism of Acb2 were independently confirmed with a homolog from E. coli phage T4 in a recent preprint16. Despite the numerous different mechanisms employed by viruses to inhibit the eukaryotic cGAS-STING signaling system27, the cGAMP “sponge” mechanism is an entirely new inhibitory strategy.
The identification of capsid mutants that escape CBASS mirrors the recently identified major capsid mutations in E. coli phage T5, which enables escape from Pycsar (pyrimidine cyclase system for anti-phage resistance)28. This study also noted that expression of the T5 WT major capsid protein alone did not induce Pyscar-mediated toxicity, which we similarly observed, nor was direct binding observed between the T5 major capsid protein and any of the Pyscar proteins28, suggesting a more complex or indirect activation mechanism. However, abortive immune systems can be activated through direct binding to a capsid monomer, such as the CapRelSJ46 Toxin-Antitoxin system29 and the Lit protease30. These mechanistically diverse systems all converge on the phage capsid protein, which is one of many phage structural proteins mutated to evade targeting of anti-phage bacterial immune systems31. Additionally, binding to other phage structural proteins directly activate Avs (anti-viral STAND NTPases)32 and DSR (defense-associated sirtuins) systems33. The major capsid gene mutations identified in our study are enriched at the capsid protein interface, suggesting that a higher ordered capsid structure or process, rather than a capsid monomer, may be implicated in CBASS immunity. However, the mechanism behind the mutant phage capsid that enables CBASS evasion is unknown and remains an area of future investigation. Altogether, we present direct evidence of CBASS as an anti-phage bacterial immune system, uncover distinct paths taken by phage to inhibit or evade immunity, and expand our understanding of viral evasion strategies of cGAS-based immunity.
Limitations of the Study:
Here, we report the establishment of a native CBASS model system and used it to identify phage mutants that inhibit or evade CBASS. We were unable to establish additional native CBASS model systems because we could not identify phages targeted in a CBASS-dependent manner, which is likely because P. aeruginosa harbors several diverse anti-phage systems and a single strain alone can encode one to 17 (known) systems34,35. Furthermore, we describe a cyclic dinucleotide sponge (Acb2) as a mechanism to inhibit multiple cGAS-based immune systems. However, we have not comprehensively sampled the array of cyclic oligonucleotides that this protein family may bind to, which should be completed to further define the selectivity of the new Acb2 protein fold. An additional limitation of our study is that in vivo 3’,3’-cGAMP measurements reveal a low intracellular concentration (<10 nM), which is likely due to lower expression of CBASS genes compared to previous studies. It is also possible that uninfected cells or asynchronous infection leads to lower 3’,3’-cGAMP levels, and given that our measurement represents an average of the entire cell population, the total 3’,3’-cGAMP levels are lower. Lastly, our study identified that phage capsid mutants evade CBASS targeting. Despite this, it is unknown whether the phage capsid evades the activation of the cyclase (CdnA), regulation of the cyclase (via Cap2 or Cap3), targeting of the CapV effector, or evades another stage of CBASS that has yet to be studied.
STAR Methods:
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Joseph Bondy-Denomy (Joseph.Bondy-Denomy@ucsf.edu).
Materials Availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
The accession number for the coordinate and structure factors reported in this paper is PDB: 8H2X (Acb2), 8H2J (Acb2-3’,3’-cGAMP) and 8H39 (Acb2-c-di-AMP). This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the Lead Contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Bacterial strains and phages
The bacterial strains and phages used in this study are listed in the Key Resources Table. The P. aeruginosa strains (ATCC 33351, JD332, BWHPSA011, BWH058, ATCC 27853, PAO1) and E. coli strains (DH5ɑ and SM10) were grown in Lysogeny broth (LB) medium at 37°C both with aeration at 225 r.p.m. Plating was performed on LB solid agar with 10 mM MgSO4 when performing phage infections, and when indicated, gentamicin (50 μg ml−1 for P. aeruginosa and 15 μg ml−1 for E. coli) was used to maintain the pHERD30T plasmid. Gene expression was induced by the addition of L-arabinose (0.01% final for BWHPSA011 bacterial genes and 0.1% for phage genes, unless otherwise specified).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| P. aeruginosa ATCC 33351 WT | ATCC | NCBI: AWZD00000000.1 |
| P. aeruginosa JD332 | Jeremy R. Dettman Lab | NCBI: LJNX01000000.1 |
| P. aeruginosa BWH058 | Deborah Hung Lab | NCBI: JIES00000000.1 |
| P. aeruginosa ATCC 27853 WT | ATCC | NCBI: CP015117.1 |
| P. aeruginosa BWHPSA011 (Pa011) WT | Deborah Hung Lab | NCBI: NZ_AXQR00000000 .1 |
| Pa011 ΔCBASS (Removal of 1250436–1254723bp; CapV, CdnA, Cap2, Cap3, DUF2188) | This study | N/A |
| Pa011 CapV (phospholipase) catalytic mutant S48A | This study | N/A |
| Pa011 CdnA (cyclase) catalytic mutant D87A/D89A | This study | N/A |
| Pa011 Cap 2 (E1/E2) catalytic mutant C450A/C453A | This study | N/A |
| Pa011 Cap3 (JAB) catalytic mutant E38A | This study | N/A |
| P. aeruginosa PAO1 (Pa) WT | Joe Bondy-Denomy Lab | NCBI: NC_002516.2 |
| PaCBASS (PAO1 attTn7::Pa011 CBASS; CapV, CdnA, Cap2, Cap3, DUF2188) | This study | N/A |
| PaEV (Integration of mini-Tn7 empty vector) | This study | N/A |
| PaCBASS (JBD67 WT) lysogen | This study | N/A |
| PaCBASS (JBD67Δacb2) lysogen | This study | N/A |
| PaEV (JBD67 WT) lysogen | This study | N/A |
| PaEV (JBD67Δacb2) lysogen | This study | N/A |
| PahaCas3 (PAO1 attTn7::I-C CRISPR-Cas3 helicase attenuated (haCas3) system) | This study | N/A |
| PaCas13a (PAO1 attTn7::VI-A CRISPR-Ca13a system) | Joe Bondy-Denomy Lab21 | N/A |
| E. coli DH5α | NEB | Cat #C2987H |
| E. coli SM10 | NEB | Cat #C3019H |
| E. coli BL21 (DE3) | Weidi Biotechnology | Cat #EC1002 |
| PaMx33 | Gabriel Guarneros Peña Lab24 | NCBI: KU884561 |
| PaMx35 | Gabriel Guarneros Peña Lab24 | NCBI: KU884562 |
| PaMx41 | Gabriel Guarneros Peña Lab24 | NCBI: KU884563 |
| PaMx43 | Gabriel Guarneros Peña Lab24 | NCBI: KU884564 |
| JBD18 | Alan Davidson Lab57 | NCBI: JX495041.1 |
| JBD67 | Alan Davidson Lab57 | NCBI: NC_042135.1 |
| JBD67Δacb2 #1 (Removal of 23005–23232bp orf34) | This study | N/A |
| JBD67Δacb21 #2 (Removal of 23047–23235bp orf34) | This study | N/A |
| JBD67Δacb2 #3 (Removal of 23066–23239bp orf34) | This study | N/A |
| D3 | Alan Davison Lab58 | NCBI: AF165214.2 |
| PB-1 | Alan Davison Lab59 | NCBI: NC_011810 |
| F8 | Alan Davison Lab59 | NCBI: NC_011703 |
| 14-1 | Alan Davison Lab59 | NCBI: NC_007810 |
| Lind109 | Alan Davison Lab | N/A |
| Ab27 | Christine Pourcel Lab60 | NCBI: LN610579 |
| Ab28 | Christine Pourcel Lab60 | NCBI: LN610589 |
| PII10A | Christine Pourcel Lab61 | NCBI: LT594786 |
| PhiKZ | Alan Davison Lab62 | NCBI: AF399011 |
| PA3 | Alan Davison Lab63 | NCBI: HQ630627 |
| Ab03 | Christine Pourcel Lab60 | NCBI: LN610573 |
| Ab04 | Christine Pourcel Lab60 | NCBI: LN610581 |
| Ab06 | Christine Pourcel Lab60 | NCBI: LN610582 |
| Ab11 | Christine Pourcel Lab60 | NCBI: LN610583 |
| Ab17 | Christine Pourcel Lab60 | NCBI: LN610576 |
| M6 | Peter Weigele64 | NCBI: NC_007809 |
| YuA | Rob Lavigne Lab65 | NCBI: NC_010116 |
| Ab18 | Christine Pourcel Lab60 | NCBI: LN610577 |
| Ab19 | Christine Pourcel Lab60 | NCBI: NC_042115 |
| Ab20 | Christine Pourcel Lab60 | NCBI: LN610585 |
| Ab21 | Christine Pourcel Lab60 | NCBI: NC_042115 |
| PA5oct | Rob Lavigne Lab66 | NCBI: MK797984 |
| PA-1 | Peter Weigele64 | NCBI: MN504636.1 |
| DMS3 | Alan Davison Lab57 | NCBI: DQ631426.1 |
| JBD25 | Alan Davison Lab57 | NCBI: JX495042.1 |
| JBD30 | Alan Davison Lab67 | NCBI: NC_020198.1 |
| Ab30 | Christine Pourcel Lab60 | NCBI: LN610590 |
| JBD68 | Alan Davison Lab68 | NCBI: KY707339.1 |
| LKD16 | Rob Lavigne Lab69 | NCBI: AM265638 |
| LKD19 | Rob Lavigne Lab70 | NCBI: AM910651 |
| KMV | Rob Lavigne Lab71 | NCBI: AJ505558 |
| Ab05 | Christine Pourcel Lab60 | NCBI: LN610574 |
| Ab12 | Christine Pourcel Lab60 | NCBI: NC_047967 |
| LUZ7 | Rob Lavigne Lab72 | NCBI: NC_013691.1 |
| Lit1 | Rob Lavigne Lab72 | NCBI: NC_013692.1 |
| Ab09 | Christine Pourcel Lab60 | NCBI: HG962375 |
| P3P1 | Christine Pourcel Lab61 | NCBI: LT594787 |
| Ab22 | Christine Pourcel Lab60 | NCBI: LN610578 |
| Chemicals, peptides, and recombinant proteins | ||
| HEPES sodium salt | Sigma-Aldrich | CAS: 7365-45-9 Cat #V900477-500G |
| Tris base | Sigma-Aldrich | CAS: 77-86-1 Cat #RDD008-2.5KG |
| Sodium dihydrogen phosphate dihydrate | Sigma-Aldrich | CAS: 13472-35-0 Cat #1063420250 |
| Disodium hydrogen phosphate dodecahydrate | Sigma-Aldrich | CAS: 10039-32-4 Cat #1065790500 |
| Bis-Tris propane | Sigma-Aldrich | CAS: 64431-96-5 Cat #B6755-25G |
| Sodium bromide | Sigma-Aldrich | CAS: 7647-15-6 Cat #310506-100G |
| PEG 3350 | Biorigin | CAS: 25322-68-3 Cat #BN33640 |
| Ethylene glycol | Sigma-Aldrich | CAS: 107-21-1 Cat #102466 |
| Glycerol | Sigma-Aldrich | CAS: 56-81-5 Cat#V900122-500ML |
| Imidazole | Sigma-Aldrich | CAS: 288-32-4 Cat #V900153-500G |
| Resorufin butyrate | CHEMEGEN | CAS: 15585-42-9 Cat #CY17497 |
| Phenol:chloroform:isoamyl alcohol 25:24:1 | Sigma-Aldrich | CAS: 108-95-2; 6766-3; 123-51-3 Cat #P3803 |
| Chloroform | Sigma-Aldrich | CAS: 67-66-3 Cat #C2432 |
| Lysozyme | Sigma-Aldric | CAS: 12650-88-3 Cat #L6876 |
| 2X Phanta Max Master Mix | Vazyme | Cat #P515-03 |
| 2X Rapid Taq Master Mix | Vazyme | Cat #P222-AA |
| Gibson Assembly HiFi DNA Master Mix | NEB | Cat #E2621 |
| KOD-Plus-Neo | TOYOBO | Cat #KOD-401 |
| DpnI | NEB | Cat #R0176s |
| SacI | NEB | Cat #R3156S |
| PstI | NEB | Cat #R3140S |
| HindIII | NEB | Cat #R3104S |
| BamHI | NEB | Cat #R3136S |
| Proteinase K | NEB | Cat #P8107S |
| RNaseA | Omega Bio-TEK | Cat #AC118 |
| High Affinity Ni-NTA Resin | GenScript | Cat #L00250-100 |
| Pa011 CdnA recombinant protein | This study | N/A |
| Pa011 CapV recombinant protein | This study | N/A |
| Pa011 Cap2 (E1/E2) recombinant protein | This study | N/A |
| Pa011 Cap2C105A/C479A recombinant protein | This study | N/A |
| P1011 CdnA-Cap2C105A/C479A recombinant protein | This study | N/A |
| PaMx33 Acb2 WT recombinant protein | This study | N/A |
| PaMx33 Acb2Y11A recombinant protein | This study | N/A |
| PaMx33 Acb2K26A recombinant protein | This study | N/A |
| Critical commercial assays | ||
| DNA Clean & Concentrator Kit | Zymo Research | Cat #D4034 |
| Gel DNA Recovery Kit | Zymo Research | Cat #D4008 |
| Plasmid Miniprep Kit | Zymo Research | Cat #ZD4037 |
| Qubit 1X dsDNA High Sensitivity Assay Kit | ThermoFisher | Cat #Q33231 |
| Illumina DNA Prep Kit | Illumina | Cat #20015825 |
| Illumina Miseq v3 Reagents | Illumia | Cat #15043894 |
| Plasmid Miniprep Kit | Vazyme | Cat #DC201-01 |
| Gel DNA Extraction Mini Kit | Vazyme | Cat #DC301-01 |
| LFS Crystallization Screen Kit | Molecular Dimensions | Cat #MD1-121 |
| 3’,3’ cyclic GAMP ELISA Kit | Arbor Assays | Cat #K073-H1 |
| Deposited data | ||
| Structure of Acb2 | This study | PDB: 8H2X |
| Structure of Acb2 bound with 3’,3’-cGAMP | This study | PDB: 8H2J |
| Structure of Acb2 bound with c-di-AMP | This study | PDB: 8H39 |
| Oligonucleotides | ||
| PaMx41 orf11 crRNA guide #4: gatacgaccagtctgacgcttgac | This study | N/A |
| PaMx41 orf11 crRNA guide #5: tctgacgcttgacggtaagattga | This study | N/A |
| JBD67 orf34 crRNA guide #3: tctgacgcttgacggtaagattga | This study | N/A |
| JBD67 orf34 crRNA guide #4: ctggctgcagagccgttgcgctgggctgcgatcg | This study | N/A |
| 3’,3’-cGAMP | Sigma-Aldrich | CAS: 849214-04-6 Cat #SML1232-5UMO |
| 2’,3’-cGAMP | Sigma-Aldrich | CAS:1441190-66-4 Cat #SML1229-5UMO |
| c-di-GMP | Sigma-Aldrich | CAS: 61093-23-0 Cat #SML1228-1UMO |
| c-di-AMP | Sigma-Aldrich | CAS: 54447-84-6 Cat #SML1231-1UMO |
| 3’,3’-c-UMP-GMP | Biolog Life Science Institute | CAS: 232933-52-7 Cat #C371 |
| 3’,3’-c-di-UMP | Biolog Life Science Institute | CAS: 73120-97-5 Cat #C256 |
| 3’,3’-c-UMP-AMP | Biolog Life Science Institute | CAS: 83799-66-0 Cat #C357 |
| Recombinant DNA | ||
| pHERD30T (p30T) | 39 | N/A |
| p30T-Pa011 CBASS | This study | N/A |
| p30T-PaMx41-short orf24-v1 (37 a.a. with stop codon) | This study | N/A |
| p30T-PaMx41-short orf24-v2 (94 a.a. with stop codon) | This study | N/A |
| p30T-PaMx41-long orf24 (acb2) | This study | N/A |
| p30T-HDR-Acb2-AcrVIA1 | This study | N/A |
| p30T-crRNA-4-PaMx41-orf11 | This study | N/A |
| p30T-crRNA-5-PaMx41-orf11 | This study | N/A |
| p30T-crRNA-3-JBD67-orf34 | This study | N/A |
| p30T-crRNA-4-JBD67-orf34 | This study | N/A |
| pUC18-mini-Tn7T-LAC (pTn7) | 40 | N/A |
| pTn7-Pa011 CBASS | This study | N/A |
| pTNS3 | 41 | N/A |
| pMQ30 | 42 | N/A |
| pMQ30-HDR-Pa011-CapV-S48A | This study | N/A |
| pMQ30-HDR-Pa011-CdnA-D87A-D89A | This study | N/A |
| pMQ30-HDR-Pa011-E1-E2-C450A-C453A | This study | N/A |
| pMQ30-HDR-Pa011-JAB-E38A | This study | N/A |
| p30T-PaMx41-orf11-HDR-1-WT | This study | N/A |
| p30T-PaMx41-orf11-HDR-2-WT | This study | N/A |
| p30T-PaMx41-orf11-I121S | This study | N/A |
| p30T-PaMx41-orf11-I121T | This study | N/A |
| p30T-PaMx41-orf11-S330P | This study | N/A |
| p30T-PaMx41-Acb2-Y11A | This study | N/A |
| p30T-PaMx41-Acb2-K26A | This study | N/A |
| pET28a- His6-SUMO-Acb2 | This study | N/A |
| pET28a- His6-SUMO-Acb2-Y11A | This study | N/A |
| pET28a- His6-SUMO-Acb2-K26A | This study | N/A |
| pET28a-His6-CapV | This study | N/A |
| pET28a-His6-CdnA | This study | N/A |
| pET28a-His6-Cap2 E1/E2 | This study | N/A |
| pET28a- His6-Cap2 E1/E2-C105A/C479A | This study | N/A |
| pRSFDuet- His6-CdnA-Cap2 E1/E2-C105A/C479A | This study | N/A |
| Software and algorithms | ||
| National Center for Biotechnology Information (NCBI) database | 36 | https://blast.ncbi.nlmnih.gov/ |
| Integrated Microbial Genomes (IMG) database | 37 | https://img.jgi.doe.gov/ |
| DefenseFinder | 34,38 | https://defensefinder.mdmparislab.com/ |
| Multiple Sequence Comparison by Log-Expectation (MUSCLE) | 73 | https://www.ebi.ac.uk/Tools/msa/muscle/ |
| NCBI Multiple Sequence Alignment Viewer (MSA) | MSA Software | https://www.ncbi.nlm.nih.gov/tools/msaviewer/ |
| NCBI Constraint-based Multiple Alignment Tool (COBALT) | 50 | https://www.ncbi.nlm.nih.gov/tools/cobalt/re_cobalt.cgi |
| Interactive Tree of Life (iTOL) | 52 | https://itol.embl.de/ |
| MMSeqs2 | 74 | https://github.com/soedinglab/mmseqs2 |
| Cutadapt | 45 | https://cutadapt.readthedocs.io/en/stable/# |
| Bowtie2 | 46 | https://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Integrative Genomics Viewer (IGV) | 47 | https://software.broadinstitute.org/software/igv/ |
| SeqDiff | SeqDiff GitHub Program | https://github.com/hansenlo/SeqDiff |
| HHpred | 75 | https://toolkit.tuebingen.mpg.de/tools/hhpred |
| AlphaFold2 | 53 | https://alphafold.ebi.ac.uk/ |
| DALI | 23 | http://ekhidna2.biocenter.helsinki.fi/dali/ |
| HKL2000 | 54 | http://www.hkl-raycom/ |
| PHENIX | 56 | http://www.phenixonline.org |
| COOT | 55 | http://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot |
| PyMOL | The PyMOL Molecular Graphics System, Version 2.5.2., Schrodinger, LLC | https://pymol.org/2/ |
| OriginPro 8 | OriginPro Software | N/A |
| GraphPad Prism 9 | GraphPad Software | https://www.graphpad.com/ |
| Other | ||
| Amicon Ultra-0.5 centrifugal filter unit | Merck | Cat #UFC500396 |
| Lysing Matrix B beads | MP | Cat #6911100 |
| Amicon concentrators (3 K) | Millipore | Cat #UFC800308 |
| Amicon concentrators (10 K) | Millipore | Cat #UFC901096 |
| Amicon concentrators (30 K) | Millipore | Cat #UFC903024 |
| HisTrap FF (5 mL) | GE Healthcare | Cat #17-5255-01 |
| HiTrap Heparin HP (5 mL) | GE Healthcare | Cat #17-0407-03 |
| HiTrap Q Sepharose FF (5 mL) | GE Healthcare | Cat #17-5156-01 |
| Superdex 200 increase 10/300 GL | GE Healthcare | Cat #17517501 |
The E. coli BL21 (DE3) strain was used for recombinant protein overexpression and grown in Lysogeny broth (LB) medium. The cells were grown at 37°C until OD600nm reached 0.8 and then induced at 18°C for 12 h.
METHOD DETAILS
Identification of CBASS operons
tBLASTn was used to query the amino acid sequence of eight known CD-NTases (CdnA-H) against sequenced Pseudomonas aeruginosa genomes contained in the NCBI36 and IMG37 databases as well as our sequenced UCSF clinical isolates. Proteins with >25% amino acid sequence identity to a validated CD-NTase were accepted as “hits”7, leading to the identification of >300 CBASS operons in 252 distinct P. aeruginosa strains. The P. aeruginosa BWHPSA011 (Pa011) strain contains a Type II-A CBASS operon in contig 12 (NCBI Genome ID: NZ_AXQR000000012.1) ranging from 1250439–1254679bp, with CapV (phospholipase effector, NCBI Gene ID: Q024_30602), CdnA (cyclase, Q024_30601), Cap2 (E1/E2, Q024_30600), and Cap3 (JAB, intergenic region 1250439–1250912bp).
Identification of anti-phage immune systems
DefenseFinder was used to systematically identify all known anti-phage bacterial immune system operons in P. aeruginosa strains34,38, and the output was used to construct the table in Figure 1A.
Episomal gene expression
The shuttle vector that replicates in P. aeruginosa and E. coli, pHERD30T39 was used for cloning and episomal expression of genes in P. aeruginosa BWHPSA011 (Pa011) or PAO1 strains. This vector has an arabinose-inducible promoter and a selectable gentamicin marker. Vector was digested with SacI and PstI restriction enzymes and purified. Inserts were amplified by PCR using bacterial overnight culture or phage lysate as the DNA template, and joined into the pHERD30T vector at the SacI-PstI restriction enzyme cut sites by Hi-Fi DNA Gibson Assembly (NEB) following the manufacturer’s protocol. The resulting plasmids were transformed into E. coli DH5ɑ. All plasmid constructs were verified by sequencing using primers that annealed to sites outside the multiple cloning site. P. aeruginosa cells were electroporated with the pHERD30T constructs and selected on gentamicin.
Chromosomal CBASS integration
For chromosomal insertion of the Pa011 CBASS operon, the integrating vector pUC18-mini-Tn7TLAC40 and the transposase expressing helper plasmid pTNS341 were used to insert the BWHPSA011 CBASS operon at the Tn7 locus in P. aeruginosa PAO1 strain (PaCBASS), or an pUC18-mini-Tn7T-LAC empty vector (E.V.) control strain (PaEV). The vector was linearized using around-the-world PCR, treated with DpnI, and then purified. Two overlapping inserts encompassing the CBASS operon were amplified by PCR using Pa011 overnight culture as the DNA template, and joined into the pUC18-mini-Tn7T-LAC vector a the SacI-PstI restriction enzyme cut sites by Hi-Fi DNA Gibson Assembly (NEB) following the manufacturer’s protocol. The resulting plasmids were used to transform E. coli DH5ɑ. All plasmid constructs were verified by sequencing using primers that annealed to sites outside the multiple cloning site. P. aeruginosa PAO1 cells were electroporated with pUC18-mini-Tn7T-LAC and pTNS3 and selected for on gentamicin. Potential integrants were screened by colony PCR with primers PTn7R and PglmSdown, and then verified by sequencing using primers that anneal to sites outside the attTn7 site. Electrocompetent cell preparations, transformations, integrations, selections, plasmid curing, and FLP-recombinase-mediated marker excision with pFLP were performed as described previously40.
Chromosomal mutants of P. aeruginosa BWHPSA011
The allelic exchange vector that replicates in P. aeruginosa and E. coli, pMQ3042 was used for generating the chromosomal CBASS knockout and CBASS mutant genes in P. aeruginosa BWHPSA011 (Pa011). Vector was digested with HindIII and BamHI restriction enzymes and purified. For the CBASS knockout strain, homology arms >500bp up- and downstream of CBASS operon were amplified by PCR using Pa011 overnight culture as the template DNA. For the CBASS gene mutant strains, homology arms >500bp up- and downstream of CBASS gene catalytic residue(s), with the appropriate mutant nucleotides, were amplified by PCR using Pa011 overnight culture as the template DNA. Previously identified catalytic residues in Escherichia coli TW11681 (NZ_AELD01000000)8 were used to aid the identification of the catalytic residues in Pseudomonas aeruginosa BWHPSA011. Multiple Sequence Comparison by Log-Expectation (MUSCLE,43 and NCBI Multiple Sequence Alignment Viewer (MSA) were subsequently used to validate conserved catalytic residues between P. aeruginosa BWHPSA011, Vibrio cholerae El Tor N16961 (NC_002505.1), and E.coli TW11681. The inserts were joined into the pMQ30 vector at the HindIII-BamHI restriction enzyme cut sites by Hi-Fi DNA Gibson Assembly (NEB) following the manufacturer’s protocol. The resulting plasmids were transformed into E. coli DH5ɑ. All plasmid constructs were verified by sequencing using primers that annealed to sites outside the multiple cloning site. E. coli SM10 cells were electroporated with pMQ30 constructs and selected for on gentamicin. E. coli SM10 harboring the pMQ30 construct were mated with Pa011 to transfer the plasmid and enable allelic exchange. Potential mutant Pa011 strains were subjected to a phenotype cross streak screen with PaMx41-like phages and then verified by sequencing using primers that anneal to sites outside of the homology arms. Electrocompetent cell preparations, transformations, selections, and plasmid curing were performed as described previously44.
Phage growth
All phages were grown at 37°C with solid LB agar plates containing 20 ml of bottom agar containing 10 mM MgSO4 and any necessary inducers or antibiotics. Phages were initially grown on the permissible host P. aeruginosa PAO1 WT, which naturally lacks CBASS. 150 μl of overnight cultures of PAO1 were infected with 10 μl of low titer phage lysate (>104–7 pfu/ml) and then mixed with 3 ml of 0.7% top agar 10 mM MgSO4 for plating on the LB solid agar. After incubating at 37°C overnight, individual phage plaques were picked from top agar and resuspended in 200 μl SM phage buffer. For high titer lysates, the purified phage was further amplified on LB solid agar plates with PAO1 WT. After incubating 37°C overnight, SM phage buffer was added until the solid agar lawn was completely covered and then incubated for 5–10 minutes at room temperature. The whole cell lysate was collected and a 1% volume of chloroform was added, and then left to shake gently on an orbital shaker at room temperature for 15 min followed by centrifugation at maximum g for 3 min to remove cell debris. The supernatant phage lysate was stored at 4°C for downstream assays.
Plaque assays
Plaque assays were conducted at 37°C with solid LB agar plates. 150 μl of overnight bacterial culture was mixed with top agar and plated. Phage lysates were diluted 10-fold then 2 μl spots were applied to the top agar after it had been poured and solidified.
Lysogen construction with JDB67 phage
Lysogens were constructed by spotting serial dilutions of JDB67 WT or JBD67Δacb2 phage lysates on the engineered P. aeruginosa PAO1 strain that harbors BWHPSA011 CBASS in the chromosome (PaCBASS), or a mini-Tn7 E.V. control (PaEV) strain, and streaking out the bacteria (that is, putative lysogens) from the inside of the clearing resulting from a clutter of plaques onto a solid LB agar plate. Colonies were then screened using a cross streak test to confirm resistance to the phage used to lysogenize the strain. The putative lysogens were grown in liquid culture, and the presence of spontaneously produced phage in the supernatant that could plaque on the PAO1 wildtype strain confirmed lysogeny.
Isolation of CBASS phage escapers
For identifying PaMx41 WT phage escapers of CBASS, 150 μl of overnight cultures of the P. aeruginosa strain BWHPSA011 (Pa011) were infected with 10 μl of high titer phage lysate (>109 pfu/ml) and then plated on LB solid agar. After incubating at 37°C overnight, 10 individual phage plaques were picked from top agar and resuspended in 200 μl SM phage buffer. Phage lysates were purified for three rounds using the CBASS expressing strain. Three PaMx41 WT control phages were picked, purified, and propagated in parallel by infecting the Pa011 ΔCBASS strain. To validate the phage identity, PCR and Sanger sequencing were performed on nucleotide sequences unique to PaMx41.
To identify PaMx41 Δacb2, JBD67 Δacb2, and JBD18 WT phage escapers of CBASS, 150 μl of overnight cultures of the CBASS expressing strains (Pa011 WT or PaCBASS) were infected with 10 μl of high titer phage lysate (>109 pfu/ml) and plated on LB solid agar. After incubating at 37°C overnight, no obvious plaques were observed. SM phage buffer was added to the entire lawn and whole cell lysate collected. Next, to propagate the mutant escaper phage population, 150 μl of overnight cultures of the P. aeruginosa strains lacking CBASS (Pa011 ΔCBASS or PaEV) were infected with 10 μl of the phage lysates and plated on LB solid agar. After incubating at 37°C overnight, SM phage buffer was added to the entire lawn and whole cell lysate collected. Lastly, to isolate individual escaper plaques, 150 μl of overnight cultures of the CBASS expressing strains (Pa011 WT or PaCBASS) were infected with 10 μl of the previously collected phage lysate and plated on LB solid agar After incubating at 37°C overnight, at least four individual phage plaques were picked from top agar and resuspended in 200 μl SM phage buffer. Phage lysates were purified for three rounds using the CBASS expressing strain. At least two control or WT phages were picked, purified, and propagated in parallel by infecting the Pa011 ΔCBASS or PaEV strains. To validate the phage identity, PCR and Sanger sequencing were performed on nucleotide sequences unique to each phage.
Whole genome sequencing (WGS) and analysis
Genomic DNA from phage lysates was extracted using a modified SDS/Proteinase K method. Briefly, 200 μL high titer phage lysate (>109 pfu/ml) was mixed with an equal volume of lysis buffer (10 mM Tris, 10 mM EDTA, 100 μg/mL proteinase K, 100 μg/mL RNaseA, 0.5% SDS) and incubated at 37°C for 30 min, and then 55°C for 30 min. Preps were further purified using the DNA Clean & Concentrator Kit (Zymo Research). DNA was quantified using the Qubit 4.0 Fluorometer (Life Technologies). 20–100 ng genomic DNA was used to prepare WGS libraries using the Illumina DNA Prep Kit (formerly known as Illumina Nextera Flex Kit) using a modified protocol that utilized 5x reduced quantities of tagmentation reagents per prep, except for the bead washing step with Tagment Wash Buffer (TWB), where the recommended 100 μL of TWB was used. Subsequent on-bead PCR indexing-amplification of tagmented DNA was performed using 2x Phusion Master Mix (NEB) and custom-ordered indexing primers (IDT) matching the sequences from the Illumina Nextera Index Kit. Each 50 μL reaction was split in two tubes, amplified for 9 and 12 cycles respectively. Libraries were further purified by agarose gel electrophoresis; DNA was excised around the ~400 bp size range and purified using the Zymoclean Gel DNA Recovery Kit (Zymo Research). Libraries were quantified by Qubit and the 9-cycle reaction was used unless the yield was too low for sequencing, in which case the 12-cycle reaction was used. Libraries were pooled in equimolar ratios and sequenced with Illumina MiSeq v3 reagents (150 cycles, Read 1; 8 cycles, Index 1; 8 cycles, Index 2). WGS data were demultiplexed either on-instrument or using a custom demultiplexing Python script (written by Dr. Nimit Jain), and trimmed using cutadapt (v 3.445) to remove Nextera adapters. Trimmed reads were mapped using Bowtie 2.0 (--very-sensitive-local alignments46) and alignments were visualized using IGV (v 2.9.447). Variants were detected using the SeqDiff program (https://github.com/hansenlo/SeqDiff).
CRISPR-Cas13a phage gene editing
Construction of template plasmids for homologous recombination and selection of engineered phages via the CRISPR-Cas13a system were performed as described previously21. Specifically, homology arms of >500bp up- and downstream of PaMx41 acb2 were amplified by PCR using Pamx41 WT phage genomic DNA as the template. The acrVIA1 gene was amplified from plasmid pAM38348, a gift from Luciano Marraffini, The Rockefeller University. PCR products were purified and assembled as a recombineering substrate and then inserted into the NheI site of the pHERD30T vector. The resulting plasmids were electroporated into P. aeruginosa PAO1 cells. PAO1 strains carrying the recombination plasmid were grown in LB media supplemented with gentamicin. 150 μl of overnight cultures were infected with 10 μl of high titer phage lysate (>109 pfu/ml; PaMx33 WT, PaMx35 WT, PaMx43 WT, PaMx41 ESC or PaMx41 WT) and then plated on LB solid agar. After incubating at 37°C overnight, SM phage buffer was added to the entire lawn and whole cell lysate collected. The resulting phage lysate containing both WT and recombinant phages were tittered on PAO1 strains with a chromosomally integrated Type VI-A CRISPR-Cas13a system, and the most efficiently targeting crRNA guide (specific to orf11; guide #5) was used to screen for recombinants. PAO1 strains carrying the Cas13a system and crRNA of choice were grown overnight in LB media supplemented with gentamicin. 150 μl of overnight cultures were infected with 10 μl of low titer phage lysate (104–7 pfu/ml), and then plated onto LB solid agar containing 0.3% arabinose and 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). After incubating at 37°C overnight, individual phage plaques were picked from top agar and resuspended in 200 μl SM phage buffer. Phage lysates were purified for three rounds using the Cas13a counter-selection strain (guide #5), and further propagated on a complementary Cas13a counter-selection strain (guide #4), to select against Cas13a escaper phages. To determine whether the phages were recombinants, PCR was performed with the appropriate pairs of primers amplifying the region outside of the homology arms, an internal region of acrVIA1, and acb2.
Homologous recombination-mediated mutation of phage gene
Construction of template plasmids for homologous recombination consisted of homology arms >500bp up- and downstream of the mutation of interest encoded in PaMx41 orf11. The homology arms were amplified by PCR using PaMx41Δacb2 escapers phage genomic DNA as the template, and PaMx41 WT phage genomic DNA as the control template. Template 1 primers were designed to symmetrically flank the PaMx41 orf11 mutations I121S and I121T, and template 2 primers were designed to symmetrically flank mutation I327T and S330P. PCR products were purified and assembled as a recombineering substrate and then inserted into the SacI-PstI site of the pHERD30T vector. The resulting plasmids were electroporated into P. aeruginosa BWHPSA011 (Pa011) ΔCBASS cells. Pa011 strains carrying the recombination plasmid were grown in LB media supplemented with gentamicin. 150 μl of overnight cultures were infected with 10 μl of high titer phage lysate (>109 pfu/ml; PaMx41Δacb2) and then plated on LB solid agar. After incubating at 37°C overnight, SM phage buffer was added to the entire lawn and whole cell lysate collected. The resulting phage lysate containing both WT and recombinant phages were screened on a lawn of Pa011 WT cells harboring an active CBASS system. Specifically, 150 μl of overnight Pa011 WT cultures were infected with 10 μl of low titer phage lysate (104–7 pfu/ml), and then plated onto LB solid agar. After incubating at 37°C overnight, individual phage plaques were picked from top agar and resuspended in 200 μl SM phage buffer. Phage lysates were purified for three rounds using the Pa011 WT strain. To confirm whether the phages were recombinants, PCR was performed with the appropriate pairs of primers amplifying the region outside of the homology arms and subject to Sanger Sequencing.
Helicase attenuated Cas3 removal of phage genes
Cas3 (Type I-C)-specific guides targeting JBD67 acb2 were cloned into a pHERD30T-derived vector containing modified I-C repeats as previously described19. The guides electroporated into P. aeruginosa PAO1 strains with a chromosomally integrated Type I-C helicase attenuated Cas3 system. JBD67 WT phage lysate was tittered on the PAO1 strains and the efficiently targeting crRNA guide (specific to acb2; guide #3) was identified. PAO1 strains carrying the Type I-C CRISPR-Cas system with a helicase attenuated Cas3 enzyme and crRNA targeting phage JBD67 acb2 were grown overnight in LB media supplemented with gentamicin. 150 μl of overnight cultures were infected with 10 μl of high titer phage lysate (>109 pfu/ml; JBD67) and plated on LB agar plates containing gentamicin, 0.1% arabinose, and 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). After incubating at 37°C overnight, SM phage buffer was added to the entire lawn and whole cell lysate collected. The resulting phage lysate containing both WT and acb2 knockout phages were grown on a complementary Cas3 counter-selection strain (guide #4) to select against Cas3 escaper phages. 150 μl of overnight cultures were infected with 10 μl of low titer phage lysate (104–7 pfu/ml; JBD67 WT) and then plated on LB solid agar containing 0.1% arabinose and 1 mM IPTG. After incubating at 37°C overnight, individual phage plaques were picked from top agar and replica-plated onto LB solid agar with PAO1EV and PAO1CBASS strains. JDB67 plaque sizes that were reduced on the PAO1CBASS plate compared to the positive control (JBD18 WT phage) were identified as potential CBASS sensitive phages. Corresponding plaques on the PAO1EV plate were picked and resuspended in 200 μl SM phage buffer. To determine whether the phages harbored deletions in acb2, PCR was performed with the appropriate pairs of primers amplifying a ~1kb region outside of acb2.
Intracellular 3’,3’-cGAMP measurements
Cell lysates were prepared similarly to previous methods8, in which P. aeruginosa BWHPA011 (Pa011) cells harboring a catalytically dead capV gene (CapVS48A) were used and then transformed with a pHERD30T vector expressing acb2 WT or K26A. Cells were taken from overnight culture, diluted 1:100 in 150ml LB medium with G50 and 0.1% arabinose (flask size 500ml), and then grown at 37°C (190 r.p.m.) until reaching an OD600nm of 0.3–0.4. From the culture, 100 ml was aliquoted and 10 mM MgSO4 added. The cells were then infected with PaMx41Δacb2 to obtain an MOI of ~5 and ensure at least one or more phages were infecting each bacterial cell. After 60 minutes following infection, the culture was separated into two 50 ml samples and centrifuged at 7,500 g for 10 mins at 4°C. Following centrifugation, supernatant was removed and pellets were kept on ice until resuspended in 600 μl of phosphate buffer (50 mM sodium phosphate (pH 7.4), 300 mM NaCl, 10% (v/v) glycerol). The resuspended pellet was supplemented with 1 μl hen-lysozyme (Sigma-Aldrich), vortexed briefly, and incubated at 25°C for 10 min. The resuspended cells were then mixed with Lysing Matrix B (MP) beads and cells were disrupted mechanically using Mini-Beadbeater 16 Biospec Products (1 cycle of 2:30, 3,450 oscillations/m, at 4 °C). Cell lysates were then centrifuged at 17,500 g for 10 min at 4°C. For each condition, one 50 ml cell lysate and subsequent supernatant was (i) loaded onto a 3kDa filter (Amicon Ultra-0.5 centrifugal filter unit; Merk) and the corresponding 50 ml cell lysate and subsequent supernatant was (ii) subjected to phenol-chloroform/chloroform nucleotide extraction (Rouillon et al., 2019). For the filtration step, the unit was centrifuged at 16,000 g for 45 min at 4 °C and flow-through (containing small molecules less than 3kDa) was used as the sample for 3’,3’-cGAMP measurements. For the nucleotide extraction step, 600 μl of supernatant was added to 600 μl of phenol-chloroform, vortexed for 30 sec, and then centrifuged at 17,500 g for 45 min at 4 °C. The top aqueous layer was carefully transferred into another eppendorf tube and 600 μl of chloroform was added, vortexed for 30 sec, and then centrifuged at 17,500 g for 10 min at 4 °C. The top aqueous layer was added to the 3kD filter, centrifuged at 16,000 g for 45 min at 4 °C, and flow-through collected. Each flow-through sample was run in technical triplicate on a 3’,3’-cGAMP ELISA Kit (Arbor Assays) and standards were prepared in the same phosphate buffer. 3’,3’-cGAMP concentrations were calculated using a sigmoidal standard curve via GraphPad Prism (v 9.4.1).
Phylogenetic analysis
Phylogenetic reconstructions were conducted similar to previous work in our lab49. Homologs of Acb2 were acquired through 3 iterations of psiBLASTp search the non-redundant protein database. Hits with >70% coverage and an E value <0.0005 were included in the generation of the position specific scoring matrix (PSSM). High confidence homologs (>70% coverage, E value < 0.0005) represented in unique species of bacteria were then aligned using NCBI COBALT50 using default settings and a phylogeny was generated in Cobalt using the fastest minimum evolution method51 employing a maximum sequence difference of 0.85 and Grishin distance to calculate the tree. The resulting phylogeny was then displayed as a phylogenetic tree using iTOL: Interactive Tree of Life52.
Computational modeling of phage capsid
Models of P. aeruginosa PaMx41 (orf11) and JBD18 (orf35) phage capsid proteins were generated using AlphaFold253 and aligned using the PyMol “super” function to the different chains of the E. coli T4 phage capsid structure (PDB: 6UZC).
Protein expression and purification
The Acb2, CapV, CdnA and Cap2 genes were synthesized by GenScript. The full-length Acb2 gene was amplified by PCR and cloned into a modified pET28a vector in which the expressed Acb2 protein contains a His-SUMO tag. The Acb2 mutants were generated by two-step PCR and were subcloned, overexpressed and purified in the same way as wild-type protein. The proteins were expressed in E. coli strain BL21 (DE3) and induced by 0.2 mM isopropyl-β-Dthiogalactopyranoside (IPTG) when the cell density reached an OD600nm of 0.8. After growth at 18°C for 12 h, the cells were harvested, re-suspended in lysis buffer (50 mM Tris–HCl pH 8.0, 300 mM NaCl, 10 mM imidazole and 1 mM PMSF) and lysed by sonication. The cell lysate was centrifuged at 20,000 g for 50 min at 4°C to remove cell debris. The supernatant was applied onto a self-packaged Ni-affinity column (2 mL Ni-NTA, Genscript) and contaminant proteins were removed with wash buffer (50 mM Tris pH 8.0, 300 mM NaCl, 30 mM imidazole). The fusion protein was then digested with Ulp1 at 18°C for 2 h, and then the Acb2 protein was eluted with wash buffer. The eluant of Acb2 was concentrated and further purified using a Superdex-200 increase 10/300 GL (GE Healthcare) column equilibrated with a buffer containing 10 mM Tris-HCl pH 8.0, 200 mM NaCl and 5 mM DTT. The purified protein was analyzed by SDS-PAGE. The fractions containing the target protein were pooled and concentrated.
The CdnA, Cap2 and CdnA-Cap2 complex were purified as His-tagged proteins, which were eluted with elution buffer (50 mM Tris pH 8.0, 300 mM NaCl, 300 mM imidazole) after removing contaminant proteins with wash buffer. The cells expressing CapV were resuspended with lysis buffer containing 50 mM phosphate buffer pH 7.4, 300 mM NaCl, 10% glycerol (v/v). The CapV proteins bound to Ni-NTA beads were washed with a buffer containing 50 mM phosphate buffer pH 7.4, 300 mM NaCl, 10% glycerol (v/v), 30 mM imidazole and then eluted with the 50 mM phosphate buffer (pH 7.4), 300 mM NaCl, 10% glycerol (v/v), 300 mM imidazole. The eluant of CapV was concentrated and further purified using a Superdex-200 increase 10/300 GL (GE Healthcare) column equilibrated with a reaction buffer containing 50 mM phosphate buffer (pH 7.4), 300 mM NaCl, 10% glycerol (v/v). The purified protein was analyzed as described above.
Crystallization, data collection and structural determination
The Acb2 protein was concentrated to 24 mg/mL in 10 mM Tris-HCl pH 8.0, 200 mM NaCl and 5 mM DTT. Crystals were grown using the hanging-drop vapor diffusion method. Crystals of Acb2 were grown at 18°C by mixing an equal volume of the protein (24 mg/mL) with reservoir solution containing 0.2 M Sodium bromide, 0.1 M Bis-Tris propane pH 6.5, 10% Ethylene glycol and 20% v/v PEG 3350. Crystals of Acb2 in complex with 3’,3’-cGAMP or c-di-AMP were grown under the same reservoir solution. Prior to crystallization, 3’,3’-cGAMP or c-di-AMP were mixed with the protein at a molar ratio of 0.8:1. The crystals appeared overnight and grew to full size in about two to three days. The crystals were cryoprotected in the reservoir solution containing 20% glycerol before its transferring to liquid nitrogen.
All the data were collected at the X-ray crystallography facility at Tsinghua University (XtaLAB Synergy Custom FRX and a hybrid photon counting detector HyPix-6000, Rigaku, Japan) and SSRF beamlines BL02U1 and BL19U1, integrated and scaled using the HKL2000 package54. The initial model of Acb2 was obtained through modeling using AlphaFold253. The structures of Acb2 and its complex with ligands were solved through molecular replacement and refined manually using COOT55. The structure was further refined with PHENIX56 using non-crystallographic symmetry and stereochemistry information as restraints. The final structure was obtained through several rounds of refinement. Data collection and structure refinement statistics are summarized in Table S1.
Isothermal titration calorimetry binding assay
The dissociation constants of binding reactions of Acb2 or Acb2 mutants with the 3’,3’-cGAMP/2’,3’-cGAMP/c-di-GMP/c-di-AMP/3’,3’-c-di-UMP/3’,3’-c-UMP-AMP/3’,3’-c-UMP-GMP were determined by isothermal titration calorimetry (ITC) using a MicroCal ITC200 calorimeter. Both proteins and cyclic dinucleotides were desalted into the working buffer (20 mM HEPES pH 7.5 and 200 mM NaCl). The titration was carried out with 19 successive injections of 2 μL cyclic dinucleotides at the 0.4 mM concentration, spaced 120 s apart, into the sample cell containing the Acb2 or Acb2 mutants with a concentration of 0.1 mM by 700 rpm at 25°C. The Origin software was used for baseline correction, integration, and curve fitting to a single site binding model.
Fluorogenic biochemical assay for CapV activity
The enzymatic reaction velocity was measured as previously described8. Briefly, the esterase activity of the 6×His-tagged CapV was probed with the fluorogenic substrate resorufin butyrate. The CapV protein was diluted in 50 mM sodium phosphate pH 7.4, 300 mM NaCl, 10% (v/v) glycerol to a final concentration of 1.77 μM. To determine the enzymatic activity of CapV activated by 3’,3’-cGAMP, increasing concentrations ranging from 0.025 to 0.8 μM of 3’,3’-cGAMP was added to DMSO solubilized resorufin butyrate (stock of 20 mM mixed with 50 mM sodium phosphate pH 7.4, 300 mM NaCl, 10% v/v glycerol reaching a final concentration of 100 μM). Subsequently, the purified 6×His-tagged CapV was added to the reaction solution containing 3’,3’-cGAMP to a final assay volume of 50 μL, and fluorescence was measured in a 96-well plate (Corning 96-well half area black non-treated plate with a flat bottom). Plates were read once every 30 s for 20 min at 37°C using a EnSpire Multimode Plate Reader (PerkinElmer) with excitation and emission wavelengths of 550 and 591 nm, respectively. To determine the function of Acb2, 32 μM Acb2 and 0.8 μM 3’,3’-cGAMP were pre-incubated at 18°C, and the subsequent detection method was as described above. To examine whether the released molecule from Acb2 is able to activate CapV, 0.8 μM 3’,3’-cGAMP was incubated with 32 μM Acb2 for 10 min at 18°C. Proteinase K was subsequently added to the reaction system at a final concentration of 0.065 mg/mL and the reaction was performed at 58°C for 3 h. Reaction products were transferred to Amicon Ultra-4 Centrifugal Filter Unit 3 kDa and centrifuged at 4°C, 4,000 g. Filtered products were used for CapV activity assay.
Gel filtration assay
The Acb2, CapV, CdnA, Cap2 and the CdnA-Cap2 complex purified as described above were subjected to gel filtration analysis (Superdex-200 increase 10/300 GL, GE Healthcare). The Acb2 was incubated with CapV, CdnA, Cap2 or the CdnA-Cap2 complex at a molar ratio of 5:1 overnight on ice before the gel filtration analysis in buffer containing 10 mM Tris–HCl pH 8.0, 200 mM NaCl, and 5 mM DTT. The assays were performed with a flow rate of 0.5 mL/min and an injection volume of 1 mL for each run. Samples from relevant fractions were subjected to SDSPAGE and visualized by Coomassie blue staining.
Analytical ultracentrifugation
Proteins were extensively dialyzed against AUC buffer (10 mM Tris pH 8.0, 200 mM NaCl). Sedimentation velocity studies were performed in a Beckman XL-A analytical ultracentrifuge at 20°C and 35,000 rpm. The absorbance at 280 nm was collected every 4 min for a total of 200 scans. These values were used to fit the data to the Lamm equation in SEDFIT software using the continuous c(s) distribution model. Graphs were prepared using Origin software.
Native-PAGE assay
Acb2 was pre-incubated with cyclic dinucleotides for 10 min at 18°C, where Acb2 was 14.3 μM and the concentrations of cyclic dinucleotides ranged from 1.8 to 7.2 μM (1.8, 3.6, 7.2 μM). Products of the reaction were analyzed using 5% native polyacrylamide gels and visualized by Coomassie blue staining.
High-performance liquid chromatography (HPLC)
40 μM Acb2 was pre-incubated with 4 μM 3’,3’-cGAMP for 10 min at 18°C. Proteinase K was subsequently added to the reaction system at a final concentration of 0.25 mg/mL and the reaction was performed at 58°C for 1 h. Reaction products were transferred to Amicon Ultra-15 Centrifugal Filter Unit 3 kDa and centrifuged at 4°C, 4,000 g. The products obtained by filtration were further filtered with a 0.22 μm filter and subsequently used for HPLC experiments. The HPLC analysis was performed on an Agilent 1200 system with a ZORBAX Bonus-RP column (4.6 × 150 mm). A mixture of acetonitrile (2%) and 0.1% trifluoroacetic acid solution in water (98%) were used as mobile phase with 0.8 mL/min. The compounds were detected at 254 nm
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical details for each experiment can be found in the figure legends and outlined in the corresponding methods details section. Data are plotted with error bars representing standard deviation (s.d.).
Supplementary Material
Table S1. List of phage capsid mutations observed via Whole Genome Sequencing, Related to Figure 5.
Figure S1. Diversity of CBASS in Pseudomonas aeruginosa strains and CBASS-dependent and -independent phage targeting, Related to Figure 1. (A) Percentage of P. aeruginosa genomes that encode a CBASS operon and (B) percentage of effector genes in each CBASS type. Data are shown for P. aeruginosa in the IMG database (n = 758); some genomes may encode more than one CBASS type/operon. (C) The presence of different CBASS types in P. aeruginosa strains according to their operon composition of core and signature genes. Core genes include cGAS/DncV-like nucleotidyltransferase (CD-NTase) and effector genes. Known and predicted (*) cyclic nucleotides are denoted next to the CD-NTase gene (Whiteley et al. 2019). Signature genes are denoted as CD-NTase-associated proteins (Cap). (D) Heat map representing the order of magnitude reduction in phage titer on a CBASS-encoding (Pa011 WT Native CBASS Host or PaCBASS Heterologous CBASS Host; CBASS+) strain normalized to a strain lacking CBASS (Pa011 ΔCBASS or PaEV; CBASS-). 46 (out of 64 total) phages infected one or both strains and are represented in the heat map. (E) Plaque assays with indicated phages spotted in 10-fold serial dilutions on a lawn of Pa011 WT or ΔCBASS, or (F) on a lawn of PaCBASS or PaEV, to highlight CBASS-dependent targeting and CBASS-independent targeting (PB-1 and Ab22 phages). Plaque assays were used to quantify the order of magnitude reduction in phage titer by comparing the number of spots (with plaques, or clearing if plaques were not visible) on the CBASS+ strain divided by the CBASS- strain (n=3). (G) Plaque assays with the indicated phages spotted in 10-fold serial dilutions on a lawn of Pa011 WT or ΔCBASS, and Pa011 ΔCBASS over-expressing the CBASS operon or empty vector (E.V.). (H) Plaque assays on a lawn of Pa011 chromosomal mutants of each CBASS gene [capVS48A (phospholipase), cdnAD87A/D89A (cyclase), cap2C450A/C453A (E1/E2), and cap3E38A (JAB)]. For all plaque assays, clearings represent phage replication and black arrowheads highlight the reduction of PaMx41 WT phage titer.
Figure S2. Acb2 protects phage and reduces 3’,3’-cGAMP molecules in CBASS-containing cells, Related to Figure 1, 2, and 4. (A) Schematic of in vivo phage infection and cGAMP detection: (1) Pa011 cells with catalytically dead CapVS48A strain overexpressing a wildtype version of an anti-CBASS gene (Acb2 WT; inhibited CBASS) or mutant version (Acb2 K26A; uninhibited or active CBASS). These cells are infected with PaMx41 phage that lacks Acb2 (PaMx41Δacb2) at an MOI of ~5 for 60 minutes to provide the phage time to proceed through its replication cycle (Cruz-Plancarte et al., 2016). (2) Cell lysates are processed without (−) or with phenol-chloroform/chloroform nucleotide extraction. (3) Resultant lysates are filtered to collect cyclic oligonucleotides and then (4) 3’,3’-cGAMP is specifically measured by an ELISA. (B) 3’,3’-cGAMP detected in each respective condition performed in biological triplicate. Data are mean ± s.d. (C) Plaque assays were performed with PaMx33, 35, 41, and 43 WT phages, as well as an evolved PaMx41 CBASS escape (ESC) phage, spotted in 10-fold serial dilutions on a lawn of Pa011 WT [CBASS+] or ΔCBASS [CBASS-] over-expressing the indicated genes; black arrowhead highlights increase in PaMx41 WT phage titer. (D) Plaque assays were performed with the indicated phages spotted in 10-fold serial dilutions on a lawn of Pa011 WT, ΔCBASS, or WT over-expressing acb2. (E) Plaque assays with indicated phages on a lawn of P. aeruginosa cells (PAO1) with a chromosomally integrated Pa011 CBASS operon (PaCBASS), or empty vector (PaEV), and overexpressing acb2. Black arrowhead highlights CBASS-dependent change in phage titer. (F) Plaque assays with the indicated phages spotted in 10-fold serial dilutions on a lawn of PaEV [CBASS-], PaEV (JBD67 WT) lysogen, or PaEV (JBD67Δacb2) lysogen, or PaCBASS [CBASS+], PaCBASS (JBD67 WT) lysogen, or PaCBASS (JBD67Δacb2) lysogen. For all plaque assays, clearings represent phage replication.
Figure S3. Acb2 is found in a broad diversity of phages and bacteria, Related to Figure 2. Phylogenetic tree of acb2 across the genomes of 239 tailed phages (Caudovirales) following two iterations of PSI-BLAST. Phages families that are colored are the most frequently identified. Phages relevant to this study are bolded. (B) Multiple sequence alignment of PaMx41orf24X37Q Acb2, PaMx33, PaMx43, JDB67 Acb2, and other homologous proteins via MAFFT alignment in Jalview. All proteins except T2 and T4 phage homologs were acquired by standard BLASTp, while T2 and T4 were observed on a second round of a PSI-BLAST. Colors indicate an identical amino acid. Black boxes highlight residues Y11 and K26, which are necessary for Acb2 activity.
Figure S4. Acb2 does not bind CBASS proteins, but does bind 3’,3’-cGAMP, 2’,3’-cGAMP, and c-di-AMP, Related to Figure 3 and 4. (A)-(D) Gel filtration profile of incubated Acb2 with Cap2-CdnA complex (A), Cap2 (B), CdnA (C) or CapV (D) (Superdex-200 increase 10/300 GL, GE Healthcare). (E) ITC assays to test binding of 3’,3’-cGAMP to Acb2 WT. (F) ITC assays to test binding of 2’,3’-cGAMP to Acb2. (G) ITC assays to test binding of c-di-AMP to Acb2 WT. (H) ITC assay to test binding of c-di-GMP to Acb2. (I), (J) ITC assay to test binding of 3’,3’-cGAMP to Acb2 Y11A and K26A, respectively. (K) Acb2 and its mutants were incubated with 3’,3’′-cGAMP at indicated concentrations. Then the samples were subjected to native PAGE.
Figure S5. Structures of apo and di-nucleotide bound Acb2, Related to Figure 4. (A) Analytical Ultracentrifugation (AUC) analysis of Acb2 and its complex with 3’,3’-cGAMP. (B) Structure of the Acb2 hexamer. (C) Structure of the Acb2 monomer. (D-E) Two types of dimer of protomers as shown in (B) are shown. (F)-(H), The ability of Acb2 to bind 2’,3’-cGAMP/c-di-AMP/c-di-GMP was analyzed by HPLC. 2’,3’-cGAMP/c-di-AMP/c-di-GMP standards were used as a control. The remaining cyclic dinucleotides after incubation with Acb2 were tested. (I) Overall structure of Acb2 bound with c-di-AMP. (J)-(K) Structural comparison between an Acb2 dimer in c-di-AMP-bound form (colored in pink) and 3’,3’-cGAMP-bound form (colored in slate).
Figure S6. Acb2 binds to the predicted CdnE cyclic dinucleotide products and protects phages against Type I-A and Type I-B CBASS immunity that encode CdnE cyclase, Related to Figure 3. (A) Isothermal titration calorimetry (ITC) assays to test binding of cyclic dinucleotides to Acb2. cUU, cUA, and cUG represent 3’,3’-cyclic-di-UMP, 3’,3’-cyclic-UMP-AMP, and 3’,3’-cyclic-UMP-GMP, respectively. Representative binding curves and binding affinities are shown. The KD values are mean ± s.d. (n=3). Raw data for these curves are shown in (B-D). (E) Native PAGE showed the binding of Acb2 to cyclic dinucleotides. (F) P. aeruginosa ATCC 33351 and JD332 CBASS operons with predicted cyclic dinucleotides (Whiteley et al. 2019). (G) Plaque assays with the indicated phages spotted in 10-fold serial dilutions on a lawn of P. aeruginosa cells (PAO1), which naturally lacks CBASS, over-expressing a CBASS operon or empty vector (E.V.); clearings represent phage replication. Black arrowhead highlights change in phage titer specific to phages lacking acb2.
Figure S7. PaMx41 phage remains sensitive to CBASS immunity in the presence of major capsid escape allele expression, Related to Figure 5. (A) Plaque assays were performed with PaMx41Δacb2 phage, harboring a wildtype (WT) capsid, spotted in 10-fold serial dilutions on a lawn of Pa011 WT [CBASS+] or ΔCBASS [CBASS-] over-expressing the indicated genes; clearings represent phage replication. (B) Plaque assays were performed with PaMx41Δacb2 CBASS Escaper phage 7, harboring a mutant (I121T) capsid, spotted in 10-fold serial dilutions on a lawn of Pa011 WT or ΔCBASS. (C) Alphafold2 prediction of the PaMx41Δacb2 (blue) and JDB18 (green) major capsid protein monomer structures overlaid using PyMOL (RMSD: 4.194). Red spheres represent amino acid residues that are mutated and are labeled with the corresponding a.a. change. (D) Alphafold2 prediction of the PaMx41Δacb2 and (E) JDB18 capsid hexamer structures based on the experimentally solved E. coli T4 phage capsid structure (PDB: 6UZC). Spheres represent the a.a. residues that are mutated. Black and gray colors are indicative of mutations within a capsid monomer and highlight the inter- and intra-protein localization of the mutations.
Highlights:
Endogenous P. aeruginosa CBASS makes 3’,3’-cGAMP in response to phage infection
Phages inhibit CBASS via anti-CBASS (Acb2) proteins that sequester 3’,3’-cGAMP
Acb2 binds to various cyclic dinucleotides: 2’,3’-cGAMP and 3’,3’-cUU/UA/UG/AA
Diverse phages evade CBASS via mutations in their major capsid gene
Acknowledgments:
We thank current and past members of the Bondy-Denomy Lab for thoughtful discussions, including Drs. Bálint Csörgo, Lina León, and Yuping Li for their technical expertise and knowledge of bacterial and molecular genetics. Drs. Shweta Karambelkar and Lina León for the CRISPR-Cas3 helicase attenuated technology to delete JBD67 acb2, and Matt Johnson for bioinformatic comparison of phage genomes. We thank Dr. Deborah Hung at the Broad Institute of MIT and Harvard for the Pseudomonas aeruginosa strains BWHPSA011 and BWH058. We thank Dr. Jeremy R. Dettman at the University of Ottawa for the sample of P. aeruginosa strain JD332. We thank Dr. Gabriel Guarneros Pena at Centro de Investigacion y de Estudios Avanzados for the PaMx33, PaMx35, PaMx41, and PaMx43 phages. We thank Dr. Aaron Whiteley at the University of Colorado, Boulder for his technical expertise and knowledge of CD-NTases and cyclic oligonucleotides.
We thank members of the Yue Feng Lab, including Zhengyu Gao and Hao Wang, for their comprehensive efforts, technical expertise, and knowledge on structural and biochemical biology. We thank the staff at beamlines BL02U1 and BL19U1 of the Shanghai Synchrotron Radiation Facility for their assistance with data collection. We thank the Tsinghua University Branch of China National Center for Protein Sciences Beijing and Dr. Shilong Fan for providing facility support for X-ray diffraction of the crystal samples. We thank Drs. Yu Xiao and Teng Gao for their help with structure determination. We thank Drs. Yuanyuan Chen, Zhenwei Yang, Bingxue Zhou at the Institute of Biophysics, Chinese Academy of Sciences for technical help with ITC experiments.
Declaration of interests:
J.B.-D. is a scientific advisory board member of SNIPR Biome and Excision Biotherapeutics and a scientific advisory board member and co-founder of Acrigen Biosciences. The Bondy-Denomy lab receives research support from Felix Biotechnology.
Inclusion and Diversity:
We support inclusive, diverse, and equitable conduct of research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- 1.Barbalat R, Ewald SE, Mouchess ML, and Barton GM (2011). Nucleic acid recognition by the innate immune system. Annu. Rev. Immunol 29, 185–214. 10.1146/annurev-immunol031210-101340. [DOI] [PubMed] [Google Scholar]
- 2.Margolis SR, Wilson SC, and Vance RE (2017). Evolutionary Origins of cGAS-STING Signaling. Trends Immunol. 38, 733–743. 10.1016/j.it.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 3.Li X-D, Wu J, Gao D, Wang H, Sun L, and Chen ZJ (2013). Pivotal roles of cGAScGAMP signaling in antiviral defense and immune adjuvant effects. Science 341, 1390–1394. 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu J, Sun L, Chen X, Du F, Shi H, Chen C, and Chen ZJ (2013). Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339, 826–830. 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishikawa H, and Barber GN (2008). STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678. 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, and Vance RE (2011). STING is a direct innate immune sensor of cyclic di-GMP. Nature 478, 515–518. 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whiteley AT, Eaglesham JB, de Oliveira Mann CC, Morehouse BR, Lowey B, Nieminen EA, Danilchanka O, King DS, Lee ASY, Mekalanos JJ, et al. (2019). Bacterial cGAS-like enzymes synthesize diverse nucleotide signals. Nature 567, 194–199. 10.1038/s41586-019-0953-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen D, Melamed S, Millman A, Shulman G, Oppenheimer-Shaanan Y, Kacen A, Doron S, Amitai G, and Sorek R (2019). Cyclic GMP-AMP signalling protects bacteria against viral infection. Nature 574, 691–695. 10.1038/s41586-019-1605-5. [DOI] [PubMed] [Google Scholar]
- 9.Lau RK, Ye Q, Patel L, Berg KR, Mathews IT, Watrous JD, Whiteley AT, Lowey B, Mekalanos JJ, Kranzusch PJ, et al. (2019). Structure and mechanism of a cyclic trinucleotide-activated bacterial endonuclease mediating bacteriophage immunity. bioRxiv, 694703. 10.1101/694703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lowey B, Whiteley AT, Keszei AFA, Morehouse BR, Mathews IT, Antine SP, Cabrera VJ, Kashin D, Niemann P, Jain M, et al. (2020). CBASS Immunity Uses CARFRelated Effectors to Sense 3’−5’- and 2’−5’-Linked Cyclic Oligonucleotide Signals and Protect Bacteria from Phage Infection. Cell 182, 38–49.e17. 10.1016/j.cell.2020.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duncan-Lowey B, McNamara-Bordewick NK, Tal N, Sorek R, and Kranzusch PJ (2021). Effector-mediated membrane disruption controls cell death in CBASS antiphage defense. Mol. Cell 81, 5039–5051.e5. 10.1016/j.molcel.2021.10.020. [DOI] [PubMed] [Google Scholar]
- 12.Morehouse BR, Yip MCJ, Keszei AFA, McNamara-Bordewick NK, Shao S, and Kranzusch PJ (2022). Cryo-EM structure of an active bacterial TIR-STING filament complex. Nature 608, 803–807. 10.1038/s41586-022-04999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Millman A, Melamed S, Amitai G, and Sorek R (2020). Diversity and classification of cyclic-oligonucleotide-based anti-phage signalling systems. Nat Microbiol 5, 1608–1615. 10.1038/s41564-020-0777-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye Q, Lau RK, Mathews IT, Birkholz EA, Watrous JD, Azimi CS, Pogliano J, Jain M, and Corbett KD (2020). HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity. Mol. Cell 77, 709–722.e7. 10.1016/j.molcel.2019.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ledvina HE, Ye Q, Gu Y, Quan Y, Lau RK, Zhou H, Corbett KD, and Whiteley AT (2022). cGASylation by a bacterial E1–E2 fusion protein primes antiviral immune signaling. bioRxiv, 2022.03.31.486616. 10.1101/2022.03.31.486616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jenson JM, Li T, Du F, and Chen ZJ (2022). Ubiquitin-like conjugation by bacterial cGAS enhances anti-phage defense. Research Square. 10.21203/rs.3.rs-1578554/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hobbs SJ, Wein T, Lu A, Morehouse BR, Schnabel J, Leavitt A, Yirmiya E, Sorek R, and Kranzusch PJ (2022). Phage anti-CBASS and anti-Pycsar nucleases subvert bacterial immunity. Nature 605, 522–526. 10.1038/s41586-022-04716-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eaglesham JB, Pan Y, Kupper TS, and Kranzusch PJ (2019). Viral and metazoan poxins are cGAMP-specific nucleases that restrict cGAS-STING signalling. Nature 566, 259–263. 10.1038/s41586-019-0928-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Csörgő B, León LM, Chau-Ly IJ, Vasquez-Rifo A, Berry JD, Mahendra C, Crawford ED, Lewis JD, and Bondy-Denomy J (2020). A compact Cascade-Cas3 system for targeted genome engineering. Nat. Methods 17, 1183–1190. 10.1038/s41592-020-00980-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ha AD, and Denver DR (2018). Comparative Genomic Analysis of 130 Bacteriophages Infecting Bacteria in the Genus Pseudomonas. Front. Microbiol 9, 1456. 10.3389/fmicb.2018.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guan J, Oromí-Bosch A, Mendoza SD, Karambelkar S, Berry J, and Bondy-Denomy J (2022). RNA targeting with CRISPR-Cas13a facilitates bacteriophage genome engineering. bioRxiv, 2022.02.14.480438. 10.1101/2022.02.14.480438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Athukoralage JS, McMahon SA, Zhang C, Grüschow S, Graham S, Krupovic M, Whitaker RJ, Gloster TM, and White MF (2020). An anti-CRISPR viral ring nuclease subverts type III CRISPR immunity. Nature 577, 572–575. 10.1038/s41586-019-1909-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holm L (2022). Dali server: structural unification of protein families. Nucleic Acids Res. 50, W210–5. 10.1093/nar/gkac387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cruz-Plancarte I, Cazares A, and Guarneros G (2016). Genomic and Transcriptional Mapping of PaMx41, Archetype of a New Lineage of Bacteriophages Infecting Pseudomonas aeruginosa. Appl. Environ. Microbiol 82, 6541–6547. 10.1128/AEM.01415-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leavitt A, Yirmiya E, Amitai G, Lu A, Garb J, Herbst E, Morehouse BR, Hobbs SJ, Antine SP, Sun Z-YJ, et al. (2022). Viruses inhibit TIR gcADPR signaling to overcome bacterial defense. Nature. 10.1038/s41586-022-05375-9. [DOI] [PubMed] [Google Scholar]
- 26.Li L, Yin Q, Kuss P, Maliga Z, Millán JL, Wu H, and Mitchison TJ (2014). Hydrolysis of 2’3’-cGAMP by ENPP1 and design of nonhydrolyzable analogs. Nat. Chem. Biol 10, 1043–1048. 10.1038/nchembio.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eaglesham JB, and Kranzusch PJ (2020). Conserved strategies for pathogen evasion of cGAS-STING immunity. Curr. Opin. Immunol 66, 27–34. 10.1016/j.coi.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tal N, Morehouse BR, Millman A, Stokar-Avihail A, Avraham C, Fedorenko T, Yirmiya E, Herbst E, Brandis A, Mehlman T, et al. (2021). Cyclic CMP and cyclic UMP mediate bacterial immunity against phages. Cell 184, 5728–5739.e16. 10.1016/j.cell.2021.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang T, Tamman H, Coppieters ‘t Wallant K, Kurata T, LeRoux M, Srikant S, Brodiazhenko T, Cepauskas A, Talavera A, Martens C, et al. (2022). Direct activation of a bacterial innate immune system by a viral capsid protein. Nature, 1–9. 10.1038/s41586022-05444-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu YT, and Snyder L (1994). Translation elongation factor Tu cleaved by a phageexclusion system. Proc. Natl. Acad. Sci. U. S. A 91, 802–806. 10.1073/pnas.91.2.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stokar-Avihail A, Fedorenko T, Garb J, Leavitt A, Millman A, Shulman G, Wojtania N, Melamed S, Amitai G, and Sorek R (2022). Discovery of phage determinants that confer sensitivity to bacterial immune systems. bioRxiv, 2022.08.27.505566. 10.1101/2022.08.27.505566. [DOI] [PubMed] [Google Scholar]
- 32.Gao LA, Wilkinson ME, Strecker J, Makarova KS, Macrae RK, Koonin EV, and Zhang F (2022). Prokaryotic innate immunity through pattern recognition of conserved viral proteins. Science 377, eabm4096. 10.1126/science.abm4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garb J, Lopatina A, Bernheim A, Zaremba M, Siksnys V, Melamed S, Leavitt A, Millman A, Amitai G, and Sorek R (2022). Multiple phage resistance systems inhibit infection via SIR2-dependent NAD+ depletion. Nat Microbiol 7, 1849–1856. 10.1038/s41564022-01207-8. [DOI] [PubMed] [Google Scholar]
- 34.Tesson F, Hervé A, Mordret E, Touchon M, d’Humières C, Cury J, and Bernheim A (2022). Systematic and quantitative view of the antiviral arsenal of prokaryotes. Nat. Commun 13, 2561. 10.1038/s41467-022-30269-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson MC, Laderman E, Huiting E, Zhang C, Davidson A, and Bondy-Denomy J (2022). Core Defense Hotspots within Pseudomonas aeruginosa are a consistent and rich source of anti-phage defense systems. bioRxiv, 2022.11.11.516204. 10.1101/2022.11.11.516204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sayers EW, Bolton EE, Brister JR, Canese K, Chan J, Comeau DC, Connor R, Funk K, Kelly C, Kim S, et al. (2022). Database resources of the national center for biotechnology information. Nucleic Acids Res. 50, D20–D26. 10.1093/nar/gkab1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen I-MA, Chu K, Palaniappan K, Ratner A, Huang J, Huntemann M, Hajek P, Ritter S, Varghese N, Seshadri R, et al. (2021). The IMG/M data management and analysis system v.6.0: new tools and advanced capabilities. Nucleic Acids Res. 49, D751–D763. 10.1093/nar/gkaa939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abby SS, Néron B, Ménager H, Touchon M, and Rocha EPC (2014). MacSyFinder: a program to mine genomes for molecular systems with an application to CRISPR-Cas systems. PLoS One 9, e110726. 10.1371/journal.pone.0110726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qiu D, Damron FH, Mima T, Schweizer HP, and Yu HD (2008). PBAD-based shuttle vectors for functional analysis of toxic and highly regulated genes in Pseudomonas and Burkholderia spp. and other bacteria. Appl. Environ. Microbiol 74, 7422–7426. 10.1128/AEM.01369-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi K-H, and Schweizer HP (2006). mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat. Protoc 1, 153–161. 10.1038/nprot.2006.24. [DOI] [PubMed] [Google Scholar]
- 41.Choi K-H, Mima T, Casart Y, Rholl D, Kumar A, Beacham IR, and Schweizer HP (2008). Genetic tools for select-agent-compliant manipulation of Burkholderia pseudomallei. Appl. Environ. Microbiol 74, 1064–1075. 10.1128/AEM.02430-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shanks RMQ, Caiazza NC, Hinsa SM, Toutain CM, and O’Toole GA (2006). Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gramnegative bacteria. Appl. Environ. Microbiol 72, 5027–5036. 10.1128/AEM.00682-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edgar RC (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hmelo LR, Borlee BR, Almblad H, Love ME, Randall TE, Tseng BS, Lin C, Irie Y, Storek KM, Yang JJ, et al. (2015). Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat. Protoc 10, 1820–1841. 10.1038/nprot.2015.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin M (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12. 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 46.Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, and Mesirov JP (2011). Integrative genomics viewer. Nat. Biotechnol 29, 24–26. 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meeske AJ, Jia N, Cassel AK, Kozlova A, Liao J, Wiedmann M, Patel DJ, and Marraffini LA (2020). A phage-encoded anti-CRISPR enables complete evasion of type VIA CRISPR-Cas immunity. Science 369, 54–59. 10.1126/science.abb6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marino ND, Zhang JY, Borges AL, Sousa AA, Leon LM, Rauch BJ, Walton RT, Berry JD, Joung JK, Kleinstiver BP, et al. (2018). Discovery of widespread type I and type V CRISPR-Cas inhibitors. Science 362, 240–242. 10.1126/science.aau5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Papadopoulos JS, and Agarwala R (2007). COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics 23, 1073–1079. 10.1093/bioinformatics/btm076. [DOI] [PubMed] [Google Scholar]
- 51.Desper R, and Gascuel O (2004). Theoretical foundation of the balanced minimum evolution method of phylogenetic inference and its relationship to weighted least-squares tree fitting. Mol. Biol. Evol 21, 587–598. 10.1093/molbev/msh049. [DOI] [PubMed] [Google Scholar]
- 52.Letunic I, and Bork P (2018). 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46, D493–D496. 10.1093/nar/gkx922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. 10.1038/s41586-02103819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Otwinowski Z, and Minor W (1997). [20] Processing of X-ray diffraction data collected in oscillation mode. In Methods in Enzymology (Academic Press; ), pp. 307–326. 10.1016/S00766879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 55.Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, and Terwilliger TC (2002). PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr 58, 1948–1954. 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 57.Cady KC, Bondy-Denomy J, Heussler GE, Davidson AR, and O’Toole GA (2012). The CRISPR/Cas adaptive immune system of Pseudomonas aeruginosa mediates resistance to naturally occurring and engineered phages. J. Bacteriol 194, 5728–5738. 10.1128/JB.01184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kropinski AM (2000). Sequence of the genome of the temperate, serotype-converting, Pseudomonas aeruginosa bacteriophage D3. J. Bacteriol 182, 6066–6074. 10.1128/JB.182.21.6066-6074.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ceyssens P-J, Miroshnikov K, Mattheus W, Krylov V, Robben J, Noben J-P, Vanderschraeghe S, Sykilinda N, Kropinski AM, Volckaert G, et al. (2009). Comparative analysis of the widespread and conserved PB1-like viruses infecting Pseudomonas aeruginosa. Environ. Microbiol 11, 2874–2883. 10.1111/j.1462-2920.2009.02030.x. [DOI] [PubMed] [Google Scholar]
- 60.Essoh C, Latino L, Midoux C, Blouin Y, Loukou G, Nguetta S-PA, Lathro S, Cablanmian A, Kouassi AK, Vergnaud G, et al. (2015). Investigation of a Large Collection of Pseudomonas aeruginosa Bacteriophages Collected from a Single Environmental Source in Abidjan, Côte d’Ivoire. PLoS One 10, e0130548. 10.1371/journal.pone.0130548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pourcel C, Midoux C, Latino L, Petit M-A, and Vergnaud G (2016). Complete Genome Sequences of Pseudomonas aeruginosa Phages vB_PaeP_PcyII-10_P3P1 and vB_PaeM_PcyII-10_PII10A. Genome Announc. 4. 10.1128/genomeA.00916-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mesyanzhinov VV, Robben J, Grymonprez B, Kostyuchenko VA, Bourkaltseva MV, Sykilinda NN, Krylov VN, and Volckaert G (2002). The genome of bacteriophage phiKZ of Pseudomonas aeruginosa. J. Mol. Biol 317, 1–19. 10.1006/jmbi.2001.5396. [DOI] [PubMed] [Google Scholar]
- 63.Monson R, Foulds I, Foweraker J, Welch M, and Salmond GPC (2011). The Pseudomonas aeruginosa generalized transducing phage phiPA3 is a new member of the phiKZ-like group of “jumbo” phages, and infects model laboratory strains and clinical isolates from cystic fibrosis patients. Microbiology 157, 859–867. 10.1099/mic.0.044701-0. [DOI] [PubMed] [Google Scholar]
- 64.Lee Y-J, Dai N, Walsh SE, Müller S, Fraser ME, Kauffman KM, Guan C, Corrêa IR Jr, and Weigele PR (2018). Identification and biosynthesis of thymidine hypermodifications in the genomic DNA of widespread bacterial viruses. Proc. Natl. Acad. Sci. U. S. A 115, E3116–E3125. 10.1073/pnas.1714812115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ceyssens P-J, Mesyanzhinov V, Sykilinda N, Briers Y, Roucourt B, Lavigne R, Robben J, Domashin A, Miroshnikov K, Volckaert G, et al. (2008). The genome and structural proteome of YuA, a new Pseudomonas aeruginosa phage resembling M6. J. Bacteriol 190, 1429–1435. 10.1128/JB.01441-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lood C, Danis-Wlodarczyk K, Blasdel BG, Jang HB, Vandenheuvel D, Briers Y, Noben J-P, van Noort V, Drulis-Kawa Z, and Lavigne R (2020). Integrative omics analysis of Pseudomonas aeruginosa virus PA5oct highlights the molecular complexity of jumbo phages. Environ. Microbiol 22, 2165–2181. 10.1111/1462-2920.14979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bondy-Denomy J, Pawluk A, Maxwell KL, and Davidson AR (2013). Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 493, 429–432. 10.1038/nature11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bondy-Denomy J, Qian J, Westra ER, Buckling A, Guttman DS, Davidson AR, and Maxwell KL (2016). Prophages mediate defense against phage infection through diverse mechanisms. ISME J. 10, 2854–2866. 10.1038/ismej.2016.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ceyssens Pieter-Jan, Lavigne Rob, Mattheus Wesley, Chibeu Andrew, Hertveldt Kirsten, Mast Jan, Robben Johan, and Volckaert Guido (2006). Genomic Analysis of Pseudomonas aeruginosa Phages LKD16 and LKA1: Establishment of the φKMV Subgroup within the T7 Supergroup. J. Bacteriol 188, 6924–6931. 10.1128/JB.00831-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lammens E, Ceyssens P-J, Voet M, Hertveldt K, Lavigne R, and Volckaert G (2009). Representational Difference Analysis (RDA) of bacteriophage genomes. J. Microbiol. Methods 77, 207–213. 10.1016/j.mimet.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 71.Lavigne R, Burkal’tseva MV, Robben J, Sykilinda NN, Kurochkina LP, Grymonprez B, Jonckx B, Krylov VN, Mesyanzhinov VV, and Volckaert G (2003). The genome of bacteriophage phiKMV, a T7-like virus infecting Pseudomonas aeruginosa. Virology 312, 49–59. 10.1016/s0042-6822(03)00123-5. [DOI] [PubMed] [Google Scholar]
- 72.Ceyssens P-J, Brabban A, Rogge L, Lewis MS, Pickard D, Goulding D, Dougan G, Noben J-P, Kropinski A, Kutter E, et al. (2010). Molecular and physiological analysis of three Pseudomonas aeruginosa phages belonging to the “N4-like viruses.” Virology 405, 26–30. 10.1016/j.virol.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Madeira F, Pearce M, Tivey ARN, Basutkar P, Lee J, Edbali O, Madhusoodanan N, Kolesnikov A, and Lopez R (2022). Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 10.1093/nar/gkac240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Steinegger M, and Söding J (2017). MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol 35, 1026–1028. 10.1038/nbt.3988. [DOI] [PubMed] [Google Scholar]
- 75.Zimmermann L, Stephens A, Nam S-Z, Rau D, Kübler J, Lozajic M, Gabler F, Söding J, Lupas AN, and Alva V (2018). A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J. Mol. Biol 430, 2237–2243. 10.1016/j.jmb.2017.12.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of phage capsid mutations observed via Whole Genome Sequencing, Related to Figure 5.
Figure S1. Diversity of CBASS in Pseudomonas aeruginosa strains and CBASS-dependent and -independent phage targeting, Related to Figure 1. (A) Percentage of P. aeruginosa genomes that encode a CBASS operon and (B) percentage of effector genes in each CBASS type. Data are shown for P. aeruginosa in the IMG database (n = 758); some genomes may encode more than one CBASS type/operon. (C) The presence of different CBASS types in P. aeruginosa strains according to their operon composition of core and signature genes. Core genes include cGAS/DncV-like nucleotidyltransferase (CD-NTase) and effector genes. Known and predicted (*) cyclic nucleotides are denoted next to the CD-NTase gene (Whiteley et al. 2019). Signature genes are denoted as CD-NTase-associated proteins (Cap). (D) Heat map representing the order of magnitude reduction in phage titer on a CBASS-encoding (Pa011 WT Native CBASS Host or PaCBASS Heterologous CBASS Host; CBASS+) strain normalized to a strain lacking CBASS (Pa011 ΔCBASS or PaEV; CBASS-). 46 (out of 64 total) phages infected one or both strains and are represented in the heat map. (E) Plaque assays with indicated phages spotted in 10-fold serial dilutions on a lawn of Pa011 WT or ΔCBASS, or (F) on a lawn of PaCBASS or PaEV, to highlight CBASS-dependent targeting and CBASS-independent targeting (PB-1 and Ab22 phages). Plaque assays were used to quantify the order of magnitude reduction in phage titer by comparing the number of spots (with plaques, or clearing if plaques were not visible) on the CBASS+ strain divided by the CBASS- strain (n=3). (G) Plaque assays with the indicated phages spotted in 10-fold serial dilutions on a lawn of Pa011 WT or ΔCBASS, and Pa011 ΔCBASS over-expressing the CBASS operon or empty vector (E.V.). (H) Plaque assays on a lawn of Pa011 chromosomal mutants of each CBASS gene [capVS48A (phospholipase), cdnAD87A/D89A (cyclase), cap2C450A/C453A (E1/E2), and cap3E38A (JAB)]. For all plaque assays, clearings represent phage replication and black arrowheads highlight the reduction of PaMx41 WT phage titer.
Figure S2. Acb2 protects phage and reduces 3’,3’-cGAMP molecules in CBASS-containing cells, Related to Figure 1, 2, and 4. (A) Schematic of in vivo phage infection and cGAMP detection: (1) Pa011 cells with catalytically dead CapVS48A strain overexpressing a wildtype version of an anti-CBASS gene (Acb2 WT; inhibited CBASS) or mutant version (Acb2 K26A; uninhibited or active CBASS). These cells are infected with PaMx41 phage that lacks Acb2 (PaMx41Δacb2) at an MOI of ~5 for 60 minutes to provide the phage time to proceed through its replication cycle (Cruz-Plancarte et al., 2016). (2) Cell lysates are processed without (−) or with phenol-chloroform/chloroform nucleotide extraction. (3) Resultant lysates are filtered to collect cyclic oligonucleotides and then (4) 3’,3’-cGAMP is specifically measured by an ELISA. (B) 3’,3’-cGAMP detected in each respective condition performed in biological triplicate. Data are mean ± s.d. (C) Plaque assays were performed with PaMx33, 35, 41, and 43 WT phages, as well as an evolved PaMx41 CBASS escape (ESC) phage, spotted in 10-fold serial dilutions on a lawn of Pa011 WT [CBASS+] or ΔCBASS [CBASS-] over-expressing the indicated genes; black arrowhead highlights increase in PaMx41 WT phage titer. (D) Plaque assays were performed with the indicated phages spotted in 10-fold serial dilutions on a lawn of Pa011 WT, ΔCBASS, or WT over-expressing acb2. (E) Plaque assays with indicated phages on a lawn of P. aeruginosa cells (PAO1) with a chromosomally integrated Pa011 CBASS operon (PaCBASS), or empty vector (PaEV), and overexpressing acb2. Black arrowhead highlights CBASS-dependent change in phage titer. (F) Plaque assays with the indicated phages spotted in 10-fold serial dilutions on a lawn of PaEV [CBASS-], PaEV (JBD67 WT) lysogen, or PaEV (JBD67Δacb2) lysogen, or PaCBASS [CBASS+], PaCBASS (JBD67 WT) lysogen, or PaCBASS (JBD67Δacb2) lysogen. For all plaque assays, clearings represent phage replication.
Figure S3. Acb2 is found in a broad diversity of phages and bacteria, Related to Figure 2. Phylogenetic tree of acb2 across the genomes of 239 tailed phages (Caudovirales) following two iterations of PSI-BLAST. Phages families that are colored are the most frequently identified. Phages relevant to this study are bolded. (B) Multiple sequence alignment of PaMx41orf24X37Q Acb2, PaMx33, PaMx43, JDB67 Acb2, and other homologous proteins via MAFFT alignment in Jalview. All proteins except T2 and T4 phage homologs were acquired by standard BLASTp, while T2 and T4 were observed on a second round of a PSI-BLAST. Colors indicate an identical amino acid. Black boxes highlight residues Y11 and K26, which are necessary for Acb2 activity.
Figure S4. Acb2 does not bind CBASS proteins, but does bind 3’,3’-cGAMP, 2’,3’-cGAMP, and c-di-AMP, Related to Figure 3 and 4. (A)-(D) Gel filtration profile of incubated Acb2 with Cap2-CdnA complex (A), Cap2 (B), CdnA (C) or CapV (D) (Superdex-200 increase 10/300 GL, GE Healthcare). (E) ITC assays to test binding of 3’,3’-cGAMP to Acb2 WT. (F) ITC assays to test binding of 2’,3’-cGAMP to Acb2. (G) ITC assays to test binding of c-di-AMP to Acb2 WT. (H) ITC assay to test binding of c-di-GMP to Acb2. (I), (J) ITC assay to test binding of 3’,3’-cGAMP to Acb2 Y11A and K26A, respectively. (K) Acb2 and its mutants were incubated with 3’,3’′-cGAMP at indicated concentrations. Then the samples were subjected to native PAGE.
Figure S5. Structures of apo and di-nucleotide bound Acb2, Related to Figure 4. (A) Analytical Ultracentrifugation (AUC) analysis of Acb2 and its complex with 3’,3’-cGAMP. (B) Structure of the Acb2 hexamer. (C) Structure of the Acb2 monomer. (D-E) Two types of dimer of protomers as shown in (B) are shown. (F)-(H), The ability of Acb2 to bind 2’,3’-cGAMP/c-di-AMP/c-di-GMP was analyzed by HPLC. 2’,3’-cGAMP/c-di-AMP/c-di-GMP standards were used as a control. The remaining cyclic dinucleotides after incubation with Acb2 were tested. (I) Overall structure of Acb2 bound with c-di-AMP. (J)-(K) Structural comparison between an Acb2 dimer in c-di-AMP-bound form (colored in pink) and 3’,3’-cGAMP-bound form (colored in slate).
Figure S6. Acb2 binds to the predicted CdnE cyclic dinucleotide products and protects phages against Type I-A and Type I-B CBASS immunity that encode CdnE cyclase, Related to Figure 3. (A) Isothermal titration calorimetry (ITC) assays to test binding of cyclic dinucleotides to Acb2. cUU, cUA, and cUG represent 3’,3’-cyclic-di-UMP, 3’,3’-cyclic-UMP-AMP, and 3’,3’-cyclic-UMP-GMP, respectively. Representative binding curves and binding affinities are shown. The KD values are mean ± s.d. (n=3). Raw data for these curves are shown in (B-D). (E) Native PAGE showed the binding of Acb2 to cyclic dinucleotides. (F) P. aeruginosa ATCC 33351 and JD332 CBASS operons with predicted cyclic dinucleotides (Whiteley et al. 2019). (G) Plaque assays with the indicated phages spotted in 10-fold serial dilutions on a lawn of P. aeruginosa cells (PAO1), which naturally lacks CBASS, over-expressing a CBASS operon or empty vector (E.V.); clearings represent phage replication. Black arrowhead highlights change in phage titer specific to phages lacking acb2.
Figure S7. PaMx41 phage remains sensitive to CBASS immunity in the presence of major capsid escape allele expression, Related to Figure 5. (A) Plaque assays were performed with PaMx41Δacb2 phage, harboring a wildtype (WT) capsid, spotted in 10-fold serial dilutions on a lawn of Pa011 WT [CBASS+] or ΔCBASS [CBASS-] over-expressing the indicated genes; clearings represent phage replication. (B) Plaque assays were performed with PaMx41Δacb2 CBASS Escaper phage 7, harboring a mutant (I121T) capsid, spotted in 10-fold serial dilutions on a lawn of Pa011 WT or ΔCBASS. (C) Alphafold2 prediction of the PaMx41Δacb2 (blue) and JDB18 (green) major capsid protein monomer structures overlaid using PyMOL (RMSD: 4.194). Red spheres represent amino acid residues that are mutated and are labeled with the corresponding a.a. change. (D) Alphafold2 prediction of the PaMx41Δacb2 and (E) JDB18 capsid hexamer structures based on the experimentally solved E. coli T4 phage capsid structure (PDB: 6UZC). Spheres represent the a.a. residues that are mutated. Black and gray colors are indicative of mutations within a capsid monomer and highlight the inter- and intra-protein localization of the mutations.
Data Availability Statement
The accession number for the coordinate and structure factors reported in this paper is PDB: 8H2X (Acb2), 8H2J (Acb2-3’,3’-cGAMP) and 8H39 (Acb2-c-di-AMP). This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the Lead Contact upon request.


