Abstract
Hsp104 is a hexameric AAA+ ATPase and protein disaggregase found in yeast, which couples ATP hydrolysis to the dissolution of diverse polypeptides trapped in toxic preamyloid oligomers, phase-transitioned gels, disordered aggregates, amyloids, and prions. Hsp104 shows plasticity in disaggregating diverse substrates, but how its hexameric architecture operates as a molecular machine has remained unclear. Here, we highlight structural advances made via cryoelectron microscopy (cryo-EM) that enhance our mechanistic understanding of Hsp104 and other related AAA+ translocases. Hsp104 hexamers are dynamic and adopt open “lock–washer” spiral states and closed ring structures that envelope polypeptide substrate inside the axial channel. ATP hydrolysis-driven conformational changes at the spiral seam ratchet substrate deeper into the channel. Remarkably, this mode of polypeptide translocation is reminiscent of models for how hexameric helicases unwind DNA and RNA duplexes. Thus, Hsp104 likely adapts elements of a deeply rooted, ring-translocase mechanism to the specialized task of protein disaggregation.
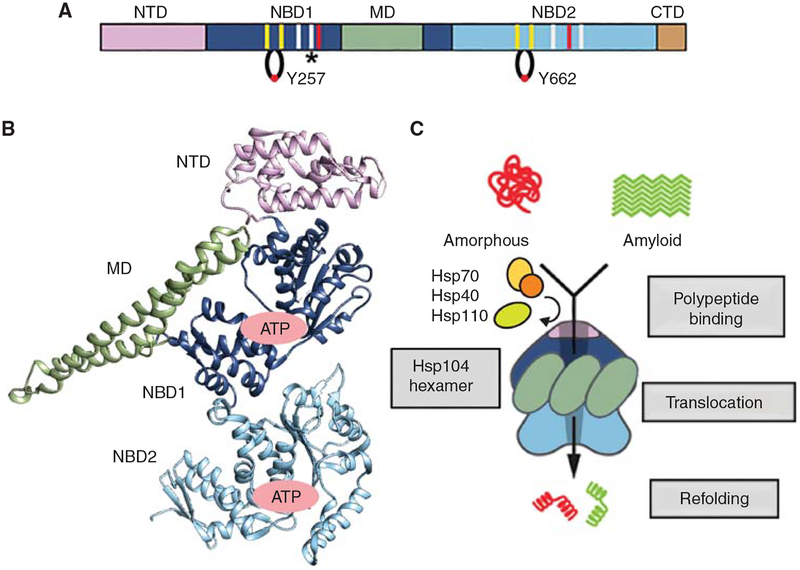
Hsp104 is a hexameric AAA+ ATPase and protein disaggregase found in yeast, which forms asymmetric ring-like structures with a prominent axial channel (Parsell et al. 1994a; Wendler et al. 2009; Sweeny and Shorter 2016; Yokom et al. 2016; Duran et al. 2017; Gates et al. 2017). It is a member of the Hsp100-class of chaperones, which include ClpB from bacteria, Hsp78 from mitochondria, and ClpA, ClpC, and ClpX, which function with the ClpP protease in bacteria (Duran et al. 2017). Hsp104 is comprised of an amino-terminal domain (NTD), nucleotide-binding domain 1 (NBD1), a middle domain (MD), NBD2, and a short carboxy-terminal domain (Fig. 1A,B; Sweeny and Shorter 2016). NBD1 and NBD2 are canonical, but distinct AAA+ domains, which originate from different clades of the AAA+ family (Erzberger and Berger 2006). Both contain conserved Walker A and B motifs, sensor residues, and an Arg finger residue that are critical for ATP hydrolysis (Sweeny and Shorter 2016). Flexible “pore loops” within the NBDs contain conserved Tyr residues that bind substrate and are required for function (Sweeny and Shorter 2016). When assembled into a hexamer, the NBDs enable Hsp104 to function as a machine that transduces energy from ATP hydrolysis to the dissolution of a myriad of proteins trapped in a variety of higher-order structures, including preamyloid oligomers (Shorter and Lindquist 2004; Lo Bianco et al. 2008; DeSantis et al. 2012), phase-transitioned gels (Kroschwald et al. 2015, 2018; Wallace et al. 2015), disordered aggregates (Parsell et al. 1994b; Glover and Lindquist 1998), amyloids (Lo Bianco et al. 2008; Liu et al. 2011b; DeSantis et al. 2012), and prions (Fig. 1C) (Shorter and Lindquist 2006, 2008; DeSantis and Shorter 2012; Klaips et al. 2014; Park et al. 2014; Zhao et al. 2017). Hsp104 (and ClpB) disaggregase activity is greatly enhanced by collaboration with the Hsp70 molecular chaperone system, which includes a J-domain chaperone (Hsp40) and a nucleotide-exchange factor (Hsp110) in addition to Hsp70 (Glover and Lindquist 1998; Cashikar et al. 2005; Haslbeck et al. 2005; Shorter and Lindquist 2008; Shorter 2011; DeSantis et al. 2012; Kaimal et al. 2017). Hsp104 couples ATP hydrolysis to the forcible extraction of polypeptides from aggregates via partial or complete translocation across its axial channel by tyrosine-bearing pore loops in NBD1 and NBD2 (Lum et al. 2004, 2008; Haslberger et al. 2008; Tessarz et al. 2008; Sweeny et al. 2015; Gates et al. 2017). Released polypeptides that are unfolded can then refold spontaneously or in collaboration with molecular chaperones.
Figure 1.
Domain structure and function of Hsp104. (A) Domain schematic for Hsp104 showing: amino-terminal domain (NTD), AAA+ nucleotide binding domains 1 (NBD1) and 2 (NBD2), middle domain (MD), and carboxy-terminal domain (CTD), and approximate location of conserved AAA+ motifs: Walker A and B (yellow), Sensor 1 and 2 (gray), Arg finger (red), and pore loops with Tyr 257 and 662. Note: in addition to the NBD1 Arg finger (R334), which contacts the γ-phosphate of ATP, another trans-interacting Arg residue (*) (R333) was identified in the Hsp104 subsrate-bound structure (Gates et al. 2017) to contact ATP and proposed to function as a noncanonical sensor 2 residue. (B) Crystal structure of the eubacterial homolog ClpB protomer colored by domain (Lee et al. 2003). (C) Schematic depicting Hsp104-mediated disaggregation of amorphous aggregates and amyloids in collaboration with the Hsp70 chaperone system. Domains are colored as in A.
Perhaps as a response to the structural heterogeneity of different aggregates, Hsp104 hexamers have been identified to show operational plasticity and can adapt different mechanisms of intersubunit collaboration to disaggregate structures of different stability (DeSantis et al. 2012). Thus, to resolve less-stable, amorphous-type aggregates, Hsp104 subunits can collaborate noncooperatively, whereas for more stable structures, such as the cross-β-sheet structures of amyloids, several subunits cooperatively engage substrate and hydrolyze ATP (DeSantis and Shorter 2012; DeSantis et al. 2012). Hsp104 can be engineered with enhanced disaggregase activity via specific mutations in its MD (Jackrel and Shorter 2014b; Jackrel et al. 2014, 2015; Tariq et al. 2018). Potentiated Hsp104 variants more effectively eliminate seminal amyloid fibrils that promote HIV infection (Castellano et al. 2015). They are also more effective in dissolving fibrils connected to neurodegenerative disease, such as those formed by TDP-43, FUS, TAF15, hnRNPA1, hnRNPA2, and α-synuclein (Jackrel et al. 2014; Jackrel and Shorter 2014a; Guo et al. 2018). Remarkably, under conditions in which wild-type Hsp104 is ineffective, these enhanced Hsp104 variants rescue dopaminergic neurodegeneration caused by α-synuclein in an intact animal nervous system (Jackrel et al. 2014) and effectively mitigate FUS aggregation and toxicity in mammalian cells (Yasuda et al. 2017).
Curiously, Hsp104 is not found in metazoa, but is present in all nonmetazoan eukaryotes, eubacteria, and some archaebacteria (Erives and Fassler 2015; Sweeny and Shorter 2016). It now appears that metazoa use several alternative machineries to directly reverse protein aggregation, including Hsp110, Hsp70, Hsp40, and sHsps (Shorter 2011; Duennwald et al. 2012; Torrente and Shorter 2013; Gao et al. 2015; Nillegoda et al. 2015; Scior et al. 2018), HtrA1 (Poepsel et al. 2015), NMNAT2 and Hsp90 (Ali et al. 2016), peptidylprolyl isomerases (Baker et al. 2017), and nuclear-import receptors (Guo et al. 2018; Yoshizawa et al. 2018). Nonetheless, these systems appear to fail or are somehow insufficient to prevent several prevalent neurodegenerative diseases. Thus, augmenting these systems with exogenous disaggregases like Hsp104 or engineered variants could find therapeutic applications for several intractable disorders (Carmichael et al. 2000; Satyal et al. 2000; Bao et al. 2002; Vacher et al. 2005; Perrin et al. 2007; Lo Bianco et al. 2008; Cushman-Nick et al. 2013; Shorter 2016, 2017). Enhanced protein disaggregases may also be extremely valuable in the purification of aggregation-prone proteins for basic, biotechnological, or pharmaceutical purposes (Sweeny and Shorter 2016).
Although not required under normal growth conditions, Hsp104 is essential for thermotolerance (Sanchez and Lindquist 1990) and enables significant selective advantages particularly under stress conditions. First, Hsp104 confers resistance to diverse chemical and thermal stresses via the recovery of properly folded proteins from disordered aggregates and by dispersing environmentally elicited protein phases harboring folded proteins (Sanchez and Lindquist 1990; Sanchez et al. 1992; Parsell et al. 1994b; Glover and Lindquist 1998; Hänninen et al. 1999; Wallace et al. 2015). Second, the ability of Hsp104 to nucleate, fragment, and dissolve prions enables yeast to exploit prions for numerous adaptive modalities (Shorter and Lindquist 2005; Halfmann and Lindquist 2010; Chakravarty and Jarosz 2018; Harvey et al. 2018). Third, Hsp104 enables proteasomal degradation of select cytosolic proteins (Lee and Goldberg 2010), extraction of insoluble proteins for endoplasmic reticulum (ER)-associated degradation (Taxis et al. 2003; Preston et al. 2018) as well as import of cytosolic misfolded proteins into mitochondria for degradation (Ruan et al. 2017). Fourth, the disaggregase activity of Hsp104 regulates the phase properties of various membraneless organelles (Wallace et al. 2015; Kroschwald et al. 2018). Thus, Hsp104 resolves cytoplasmic stress granules after stress has passed (Cherkasov et al. 2013; Kroschwald et al. 2015; Wallace et al. 2015), and maintains the liquid phase of cytoplasmic P-bodies (Kroschwald et al. 2015). Fifth, Hsp104 confers longevity by promoting the retention of damaged proteins and aggregates in the mother cell (Erjavec et al. 2007; Tessarz et al. 2009; Liu et al. 2010, 2011a; Zhou et al. 2011; Spokoini et al. 2012; Andersson et al. 2013; Coelho et al. 2013; Hill et al. 2014, 2016; Saarikangas and Barral 2015; Hanzen et al. 2016). Despite these important roles, it has remained incompletely understood how the structure of Hsp104 hexamers enable functionality.
In this review, we highlight structural advances made primarily via cryoelectron microscopy (cryo-EM) that enhance our mechanistic understanding of Hsp104 (Yokom et al. 2016; Gates et al. 2017). Hsp104 hexamers can switch between open “lock–washer” spiral states to closed ring structures that envelope polypeptide substrate inside the axial channel (Yokom et al. 2016; Gates et al. 2017). Two Hsp104 protomers undergo conformational changes to ratchet substrate deeper into the channel (Gates et al. 2017). We will discuss how these structures and proposed polypeptide translocation mechanisms align with other recent cryo-EM structures of AAA+ polypeptide translocases (Han et al. 2017; Puchades et al. 2017) and how they are akin to models for how hexameric helicases unwind DNA and RNA duplexes (Enemark and Joshua-Tor 2006; Thomsen and Berger 2009). We suggest that Hsp104 applies a conserved and adaptable ring-translocase mechanism to the specialized task of protein disaggregation.
CRYO-EM STRUCTURES REVEAL FUNDAMENTAL INSIGHT INTO SUBSTRATE TRANSLOCATION
Advancements in cryo-EM implemented in the past 4 to 5 years have led to tremendous breakthroughs in achieving near-atomic resolution structures of challenging macromolecules, resulting in what has been termed a “resolution revolution” and placing this method at the fore-front of structural biology (Kuhlbrandt 2014; Bai et al. 2015; Cheng 2015; Egelman 2016). While powerful computational analysis and 3D-reconstruction methods (Scheres 2012; Kimanius et al. 2016; Punjani et al. 2017) as well as improvements in the microscopes (Grassucci et al. 2008; Danev et al. 2017; Khoshouei et al. 2017) have been critical, the development and wide implementation of direct electron detectors (McMullan et al. 2016; Wu et al. 2016) has been arguably the primary technological advancement that has led to this recent breakthrough. This high-resolution imaging platform, including the ability to collect high frame-rate movies and correct for electron beam–induced motion of the sample (Li et al. 2013), has synergized with long-standing benefits of cryo-EM, including vitrification of samples under native-like conditions, use of small amounts of sample, potentially from endogenous sources, and single-particle approaches that do not require crystallization (Skiniotis and Southworth 2016; Takizawa et al. 2017). These benefits have been particularly evident in fields in which the macromolecules, such as membrane proteins or ribonucleoprotein complexes, have been largely refractory to X-ray crystallography methods due their size, dynamics, multimeric composition, or other complexities that impede crystallization.
For AAA+ protein complexes, including Hsp104 and related Hsp100 disaggregases, crystal structures have been achievable and have provided important high-resolution information about the structural organization, conservation, and function of the AAA+ domains (Lee et al. 2003; Carroni et al. 2014; Heuck et al. 2016). However, the crystallographic forms of the oligomer that emerge, such as a continuous spiral (Guo et al. 2002; Lee et al. 2003; Heuck et al. 2016), pose a challenge to understanding the physiologically relevant assembly. This challenge is particularly important regarding AAA+ complexes because proper intersubunit organization and communication, such as trans activation by the Arg finger, a conserved feature of AAA+ proteins, is crucial for function (Wendler et al. 2007, 2012). Indeed, initial cryo-EM studies of Hsp104 and its eubacterial homolog, ClpB, were critical for identifying the solution-state hexamer, AAA+ ring arrangement, and substrate translocation channel (Lee et al. 2003, 2007, 2010; Wendler et al. 2007, 2009; Carroni et al. 2014). However, the lower resolution achievable at the time prevented visualization of the nucleotide pockets, subunit interface, or the remarkably asymmetric and dynamic organization of the hexamer, which is now just beginning to be realized using current cryo-EM methods (Yokom et al. 2016; Deville et al. 2017; Gates et al. 2017).
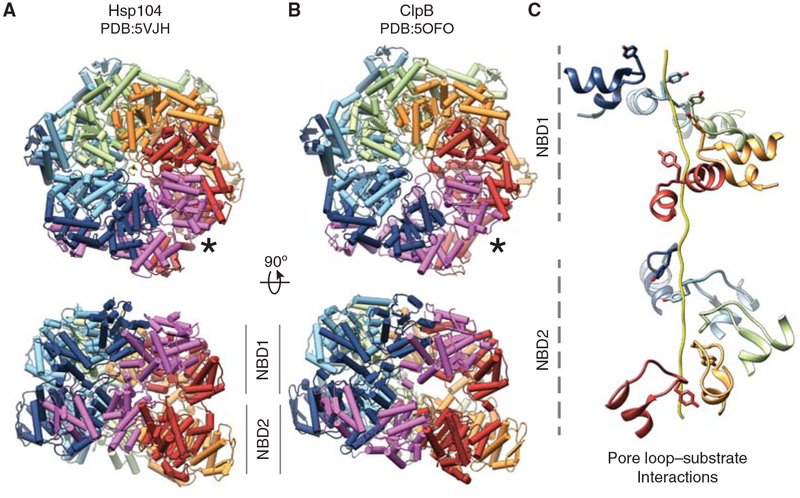
Recent cryo-EM structures of Hsp104 (Gates et al. 2017) and ClpB (Deville et al. 2017) reveal, for the first time, substrate interactions and conformational states that describe a possible translocation mechanism (Fig. 2A,B). Notably, the Hsp104 structure is of wild type, whereas ClpB is an ATPase-dead variant (BAPE279A:E678A) containing a Walker B mutation in both NBD1 and NBD2 and residues S722–N748 are replaced with V609–I635 of ClpA (Weibezahn et al. 2004; Deville et al. 2017). In the structures, the hexamer forms a right-handed spiral in which the Tyr-bearing pore loops from both AAA+ NBDs extend into the channel, forming a staggered array that contacts an ~80-Å stretch of unfolded casein (Fig. 2C; Deville et al. 2017; Gates et al. 2017). The interaction with the substrate is mediated almost entirely by the conserved Tyr residues at the ends of the loops, establishing their direct involvement in translocation (Deville et al. 2017; Gates et al. 2017), which is likely widespread for AAA+ polypeptide translocases. The Tyr residues appear to stack between the side chains of the substrate, potentially to favor nonspecific interactions with the peptide backbone to guide translocation while stabilizing the unfolded, extended arrangement (Gates et al. 2017). Five protomers contribute to these interactions and are aligned in a spiral along the polypeptide axis with an average 6–7 Å rise, which corresponds to a two-amino acid step size (Gates et al. 2017). Remarkably, this pore-loop spacing is observed in related AAA+ rings (Glynn et al. 2009; Matyskiela et al. 2013; Wehmer and Sakata 2016), and similar interdigitating Tyr-substrate interactions are identified in recent cryo-EM structures of functionally distinct translocases Vps4 (Han et al. 2017) and YME-1 (Puchades et al. 2017), indicating these are conserved features of translocases. Pore loop–substrate interactions are similarly spaced for both NBD1 and NBD2 rings in wild-type Hsp104 (Gates et al. 2017), but appear different for ClpB, with NBD1 adopting a more planar arrangement compared with NBD2 (Deville et al. 2017). Nucleotide is present in the NBD1 and NBD2 pockets, indicating an ATP-bound configuration, and the conserved Arg finger residue from the clockwise protomer is identified to directly contact the γ-phosphate for both domains while sensor residues appear to interact at the α- and β-phosphate positions (Gates et al. 2017). These residues are required for ATP hydrolysis in many AAA+ proteins; however, their precise organization around the nucleotide pocket varies in different crystal structures, owing potentially to the different crystallographic states (Lee et al. 2003; Wendler et al. 2012; Heuck et al. 2016). Indeed, from the Hsp104 structure, Arg 333 is identified to function as a trans-acting sensor 2-like residue in NBD1 (Gates et al. 2017). Thus, these structures are important in defining the active configuration of the NBDs that coincides with substrate binding (Deville et al. 2017; Gates et al. 2017).
Figure 2.
Substrate-bound architecture of ClpB and Hsp104. Cryo-electron microscope (EM) structures of (A) Hsp104 (Gates et al. 2017), and (B) ClpB (BAPE279A:E678A) (Deville et al. 2017) bound to casein and ATPγS. AAA+ domains NBD1 and NBD2 are shown and colored by protomer; substrate polypeptide bound in the channel is shown for Hsp104. The seam protomer (magenta, *) is disconnected from the substrate. (C) Spiral arrangement of Hsp104 pore loops from five protomers contacting substrate polypeptide via conserved Tyr residues 257 and 662 in the NBD1 and NBD2, respectively. The disconnected seam protomer (magenta in A and B) is not shown.
In both Hsp104 and ClpB structures (Fig. 2A,B), the sixth protomer is positioned at the spiral seam between the highest and lowest substrate contact sites and interacts with the adjacent protomers to form a closed hexameric ring (Deville et al. 2017; Gates et al. 2017). However, this protomer is disconnected from the substrate and asymmetric with respect to the hexamer spiral (Gates et al. 2017). Additionally, the cryo-EM density is at a lower resolution in this region, indicating conformational flexibility compared with the substrate-bound protomers, and the nucleotide pockets appear to be inactive with respect to the position of the adjacent Arg finger and potentially in an apo or post-hydrolysis state (Gates et al. 2017). Remarkably, this 5 bound:1 unbound arrangement of protomers is identical to recent cryo-EM structures of the single-ring AAA+ translocases YME-1 (Puchades et al. 2017) and Vps4 (Han et al. 2017), as well as the double-ring archaeal unfoldase, VAT (Ripstein et al. 2017). Similar configurations of the nucleotide pockets are also observed in these structures indicating the substrate bound protomers are primarily in an ATP state, poised for hydrolysis, while the seam protomer is likely in a posthydrolysis state. Thus, based on these recent structural efforts, this broad family of polypeptide translocases appear to adopt a remarkably universal substrate-bound spiral configuration that suggests a rotary-type translocation mechanism.
By 3D classification, we identified an additional substrate-bound conformation of Hsp104 in which all six subunits are bound to the polypeptide in an evenly spaced helical spiral (Gates et al. 2017). Comparison of these states led us to propose that the unbound subunit at the seam rearranges and binds the substrate at a position 7 Å above its clockwise neighbor, thereby advancing substrate interactions by a two-amino acid step (Fig. 3A,B; Gates et al. 2017). The two conformations were visualized in approximately equal proportion in the dataset, thus we concluded that the translocation mechanism involves a counterclockwise sequence of ratchet-like conformational changes involving these two distinct states at the spiral interface to move the polypeptide through the channel (Fig. 3C; Gates et al. 2017). Surprisingly, this six-subunit-bound arrangement has not been observed in the other substrate-bound structures, indicating potential differences in the conformational equilibrium or perhaps differences in the mechanism related to the function of Hsp104, or variations between the single- and double-ring AAA+ translocases. Notably, the YME-1 and Vps4 structures also contain the ATPase-dead Walker B mutation, similar to ClpB. Thus, additional structures of wild-type or other variants are likely necessary to identify the complete repertoire of conformational states involved in the translocation cycle for these different AAA+ complexes.
Figure 3.
Substrate-bound states of Hsp104 support a processive, rotary model for disaggregation. (A) “Closed,” and (B) “extended state” structures of Hsp104 (Gates et al. 2017) with a view of the spiral seam from inside the channel showing the ratchet-like conformational change of the seam protomer (magenta) that advanced interactions along the substrate by two amino acids. (C) Model for disaggregation involving sequential translocation steps derived from the two substrate-bound conformations of Hsp104 (molecular visualization courtesy of J. Iwasa). (For a complete movie, please see youtube.com/watch?v=VXupZku6IEg.)
The use of the slowly hydrolyzable ATP analog, ATPγS (Deville et al. 2017; Gates et al. 2017), ATP (Puchades et al. 2017), or the ADP-BeFx (Han et al. 2017) transition-state mimic were likely critical for capturing the substrate-bound states by mimicking hydrolysis or allowing partial hydrolysis to occur. Indeed, high-affinity substrate binding for Hsp104 was identified only in the presence of ATPγS compared with ATP, AMPPNP, and ADP (Gates et al. 2017; Weaver et al. 2017). Furthermore, mixtures of ATP and ATPγS can activate Hsp104 disaggregase activity in the absence of the Hsp70 chaperone system (Doyle et al. 2007; DeSantis et al. 2012). Based on structures of the single-ring AAA+ complexes YME-1 (Puchades et al. 2017) and Vps4 (Han et al. 2017; Puchades et al. 2017), the subunit at the lowest position along the substrate (down) is proposed to be in a post-hydrolysis, ADP state, while the clockwise subunit that is disengaged from the substrate is in an apo state. This situation may be similar for Hsp104. However, the nucleotide states are likely different in the NBD1 and NBD2 for these lower-positioned protomers (Gates et al. 2017). In the fully engaged structure of Hsp104, the subunit at the highest position is in the ATP state, thus the ratchet-like conformational change occurs with ATP binding and substrate engagement (Gates et al. 2017). Together, these studies support a rotary mechanism involving a counterclockwise sequence of ATP hydrolysis coupled to polypeptide translocation movement. Based on this model, ATP hydrolysis occurs in the subunits at the lower position along the substrate, possibly initiating in NBD2 (Gates et al. 2017), and thereby triggering substrate release at the spiral seam. Nucleotide exchange then enables substrate rebinding two residues above the position of the counterclockwise subunit. Higher resolution structures are needed to precisely define the nucleotide states of NBD1 and NBD2 at the spiral seam.
COMPARISON WITH AAA+ HEXAMERIC HELICASES REVEALS UNIVERSAL FEATURES
From these recent cryo-EM structures of Hsp104 and ClpB, it is emerging that the disaggregation process is remarkably similar to mechanisms of hexameric helicases, which unwind DNA and RNA duplexes. Indeed, mechanisms of stepwise separation of the nucleotide strands and steric exclusion to prevent reannealing (Trakselis 2016) parallels key requirements of disaggregation, including (1) ATPase-driven untangling of polypeptides from the larger aggregate/amyloid substrate, and (2) stabilizing the unfolded polypeptide to prevent reaggregation and promote downstream refolding or proteolysis. A number of AAA+ and RecA-like motor proteins, which are among the large family of “additional strand catalytic glutamate” (ASCE) P-loop ATPases, form hexameric ring structures that encircle and translocate substrates and contain β-hairpin pore loops that mediate substrate interactions (Enemark and Joshua-Tor 2008; Thomsen and Berger 2008). Well-characterized helicases include AAA+ superfamily 3 members SV40 large T antigen and papilloma virus E1 helicases and the MCM family involved in DNA replication in Archaea and Eukarya as well as RecA-like members: DnaB, the bacteriophage T7 gp4 helicase, and the Rho transcription termination factor (Enemark and Joshua-Tor 2008; Thomsen and Berger 2008).
The cocrystal structure of E1, a 3′−5′ helicase, identified single-stranded DNA bound to the interior of the ring by all six protomers with a helical spiral of pore loops interacting along the backbone (Enemark and Joshua-Tor 2006). Similarly, structures of Rho (Thomsen and Berger 2009) and DnaB (Itsathitphaisarn et al. 2012), which translocate 5′−3′, also identified a spiral arrangement of pore loop contacts that track along the substrate backbone. Intriguingly, the protomers in the DnaB hexamer form a spiral configuration along DNA (Itsathitphaisarn et al. 2012), while E1 and Rho hexamers are planar but with spiral arrangement of the pore loops (Enemark and Joshua-Tor 2006; Thomsen and Berger 2009), indicating translocation may be driven by either rearrangement of the spiral of protomers along the substrate or by conformational changes in the pore loops, respectively. Indeed, structures of the apo, ATP, and ADP states of the SV40 helicase identify paddle-like conformational changes, supporting that ATP hydrolysis-coupled changes in the pore loops drive translocation (Gai et al. 2004). Models based on these structures support a sequential, rotary translocation mechanism, with hydrolysis of one ATP driving a single-nucleotide step along the substrate (Enemark and Joshua-Tor 2006; Thomsen and Berger 2009). However, stochastic and concerted hydrolysis models have also been proposed based on additional biochemical and structural studies (Singleton et al. 2007; Lyubimov et al. 2011). Thus, structures of additional nucleotide-specific conformational states are needed to clarify the mechanism. Overall, the spiral configuration, substrate interactions and hydrolysis cycle are remarkably similar between helicases and polypeptide translocases, indicating the core mechanisms for the translocation of biopolymers are likely universal.
SUBSTRATE ENGAGEMENT AND RELEASE
Both Hsp104 and ClpB have an intrinsic ability to engage specific aggregated structures and disaggregate them (Shorter and Lindquist 2004, 2006; Doyle et al. 2007; DeSantis et al. 2012; Zhang et al. 2013; Jackrel et al. 2014; Krajewska et al. 2017). Typically, however, disaggregation of disordered aggregates by Hsp104 and ClpB is greatly stimulated via collaboration with the Hsp70/DnaK chaperone system (Glover and Lindquist 1998; Goloubinoff et al. 1999; Shorter 2011; Kaimal et al. 2017). Likewise, the Hsp70 system can stimulate prion and amyloid dissolution by Hsp104 and engineered variants (Lo Bianco et al. 2008; Shorter and Lindquist 2008; Shorter 2011; DeSantis et al. 2012; Jackrel et al. 2014). Downstream refolding and reactivation of substrate, if necessary, occurs following release from Hsp104 or ClpB (Glover and Lindquist 1998; Goloubinoff et al. 1999). In some cases, as with dissolution of Sup35 prions, refolding is not necessary (Sweeny et al. 2015). Here, using a partial translocation mechanism, Hsp104 simply melts the cross-β structure of the prion domain, which holds Sup35 prions together, but does not unfold the carboxy-terminal GTPase domain of Sup35, which remains correctly folded in the prion state (Sweeny et al. 2015). Thus, soluble Sup35 is liberated from the prion in an active conformation (Sweeny et al. 2015).
It is proposed that the Hsp70 chaperone system helps target Hsp104 or ClpB to aggregated structures (Weibezahn et al. 2004; Tessarz et al. 2008; Mogk et al. 2015; Kaimal et al. 2017). A direct interaction between the Hsp100 and the Hsp70 appears to be important for substrate targeting and for activation of Hsp100 ATPase activity (Seyffer et al. 2012; Lee et al. 2013; Rosenzweig et al. 2013; Doyle et al. 2015). Substrate targeting by Hsp70 involves direct interaction between its NBD and the motif 2 helices of the MD of Hsp104 or ClpB (Seyffer et al. 2012; Lee et al. 2013; Rosenzweig et al. 2013). Thus, the MD enables collaboration with Hsp70 in protein disaggregation (Haslberger et al. 2007; Oguchi et al. 2012; DeSantis et al. 2014). The MD wraps equatorially around the hexamer forming head-to-tail interactions (Carroni et al. 2014; Sweeny et al. 2015; Heuck et al. 2016; Yokom et al. 2016; Gates et al. 2017). Mutations in the MD have been identified in both ClpB (Oguchi et al. 2012) and Hsp104 (Jackrel et al. 2014, 2015; Heuck et al. 2016; Tariq et al. 2018) that alter ATP hydrolysis and potentiate function, revealing that the MD functions as an allosteric regulator during substrate engagement and disaggregation.
The Hsp70 chaperone system is proposed to act at the surface of aggregates to initiate the Hsp100 translocation process via partial solubilization and delivery of an Hsp70-bound unfolded polypeptide strand to the hexamer channel (Mogk et al. 2015). Thus, a model emerges in which polypeptides at an aggregate surface are initially made accessible to Hsp104 or ClpB by cycling of the Hsp70 chaperone system (Mogk et al. 2015; Kaimal et al. 2017). A substrate-bound Hsp70 complex might then be recognized by Hsp104 or ClpB via interactions with the MD, resulting in conformational rearrangements and ATP hydrolysis by Hsp104 or ClpB that drives substrate transfer into the hexamer channel to initiate translocation (Mogk et al. 2015).
Although appealing, this model is complicated by observations that ClpB and DnaK compete for binding to aggregates (Weibezahn et al. 2003), that DnaK causes ClpB to dissociate from substrate (Durie et al. 2018), and that Hsp104 and ClpB can show powerful disaggregase activity in the absence of Hsp70 (DeSantis et al. 2012; Duennwald et al. 2012; Zhang et al. 2013; Jackrel et al. 2014; Krajewska et al. 2017). It is also worth noting that other members of the Hsp100 family and indeed other protein-disaggregase systems show no requirement for the Hsp70 chaperone system to drive protein disaggregation (Dougan et al. 2002; Andersson et al. 2006; Haslberger et al. 2008; Park et al. 2015; Poepsel et al. 2015; Ali et al. 2016; Baker et al. 2017; LaBreck et al. 2017; Guo et al. 2018; Lee et al. 2018; Yoshizawa et al. 2018). Collectively, these observations suggest that more nuanced models are required. Indeed, the nature of chaperone-client interactions often engenders multifunctionality (Koldewey et al. 2017).
All recent substrate-bound structures of AAA+ translocases identify a narrow channel containing a single polypeptide chain that is coordinated by a spiral array of pore–loop interactions, thus it appears unlikely that more than one strand could be accommodated in the channel (Deville et al. 2017; Gates et al. 2017; Han et al. 2017; Puchades et al. 2017). However, previous studies identified that ClpB can process internal segments of substrates (Haslberger et al. 2008) in addition to translocating from the amino- or carboxy-termini (Doyle et al. 2007; Haslberger et al. 2008; Hoskins et al. 2009). Likewise, Hsp104 can drive disaggregation via partial translocation initiated at internal segments (Haslberger et al. 2008; Sweeny et al. 2015), and can also process substrates from the amino- or carboxy-termini (Doyle et al. 2007; Sweeny et al. 2015). Thus, in light of recent substrate-bound structures (Gates et al.2017), conformational rearrangements may occur during substrate engagement that open the hexamer at the spiral seam to allow accommodation of a single polypeptide strand into the channel. This plasticity in loading mechanism would allow Hsp104 or ClpB to engage substrates at multiple positions, and thus not restrict the disaggregase for hunting for free amino- or carboxy-termini to initiate translocation, which could be problematic when tackling the irregular structures of protein aggregates. Moreover, Hsp104 and ClpB hexamers are dynamic and exchange subunits on the minute timescale (Haslberger et al. 2008; Werbeck et al. 2008; DeSantis et al. 2012; Aguado et al. 2015), and recent studies by atomic force microscopy identified large conformational changes and open spiral ring conformations of ClpB (Uchihashi et al. 2018). Structures of initiation complexes, however, have not yet been determined and the Hsp100–Hsp70 interaction and substrate handover mechanisms at heterogeneous surfaces of aggregates remain a significant question.
In considering substrate binding and release mechanisms, it is worth examining recent substrate-free structures of AAA+ translocases. In the cryo-EM structure of ClpB:ATPγS that is proposed to be a substrate acceptor, ground state, the hexamer adopts an asymmetric right-handed spiral that is similar overall to the substrate-bound structure but contains a different conformation of the NBD2 pore loop at the spiral seam (Deville et al. 2017). This structure also shows nucleotide occupancy in four protomers, and a surprising NTD position that occludes the substrate channel (Deville et al. 2017). An Hsp70-mediated substrate engagement mechanism is thereby proposed that involves a large, 90° rotation of the NBD2 pore loop on substrate binding (Deville et al. 2017). However, it is unclear how this conformational change, which is at the lower portion of the channel axis and likely distal from required Hsp70-MD and NBD1–substrate interactions, would be the primary driver of substrate engagement.
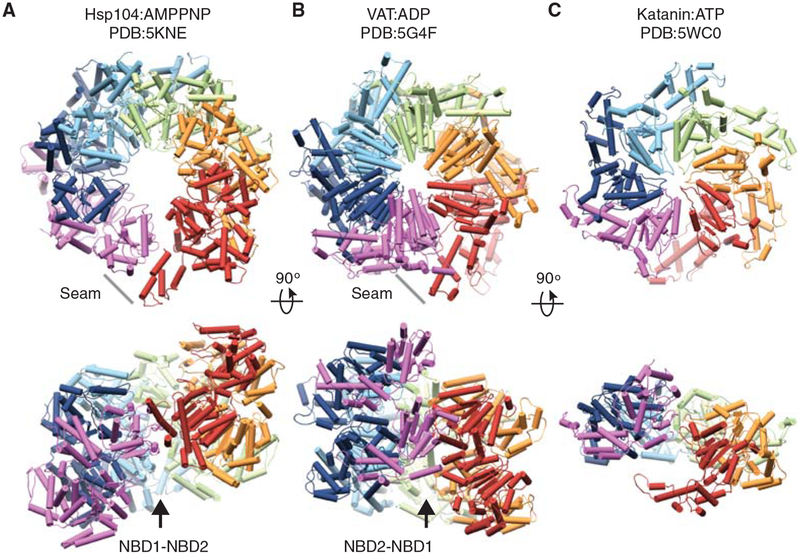
In other recent substrate-free structures (Huang et al. 2016; Yokom et al. 2016; Gates et al. 2017), the double-AAA+ ring adopts a spiral “lock–washer” conformation with a ~10 Å offset between protomers and an unusual interaction between the NBD1 and NBD2 of adjacent protomers at the spiral seam (Fig. 4). For Hsp104, identified in the presence of either AMPPNP or ADP, this conformation may serve as a key ground state for substrate binding and release (Yokom et al. 2016). The spiral is left-handed, compared with the right-handed arrangement of the substrate-bound complex (Fig. 2A), with a 30-Å-wide channel that opens toward the seam (Yokom et al. 2016; Gates et al. 2017). This cleft is proposed to be a potential access site for substrate delivery to the channel. Similar spiral lock–washer states have also been identified in cryo-EM structures of single-ring translocases, Vps4 (Su et al. 2017), and Katanin (Zehr et al. 2017) in the absence of substrate (Fig. 4C). These states are proposed to function as the power-stroke step in the translocation cycle, perhaps similar to the role of the extended state identified in Hsp104 (Fig. 3B).
Figure 4.
Substrate-free spiral “lock–washer” conformations of single and double-ring translocases. Cryo-EM structures of (A) Hsp104 bound to AMPPNP (Yokom et al. 2016), and (B) VAT, an archaeal AAA+ unfoldase, bound to ADP (Huang et al. 2016), in left-handed and right-handed spiral conformations, respectively. In both structures, the spiral seam is stabilized by a heteromeric NBD1–NBD2 interaction. (C) Cryo-EM structure of Katanin hexamer, a microtubule-severing complex, bound to ATP (Zehr et al. 2017).
The Hsp104:AMPPNP structure also identifies a MD arrangement involving salt-bridge interactions across the NBD1 of the clockwise protomer (Yokom et al.2016). Intriguingly, Hsp104: ADP adopts the same left-handed lock–washer conformation as with AMPPNP; however, the MD coiled-coil is rotated, forming the head-to-tail interactions identified for ClpB (Carroni et al. 2014; Gates et al. 2017). Remarkably, both of these nucleotide-specific MD conformations are identified in the Hsp104–substrate complex and correspond to the different nucleotides states of the protomers, supporting the notion that the MD indeed functions as allosteric ATPase control element during active translocation (Gates et al. 2017). Unlike the ClpB complex, but similar to previous crystal structures, the pore loops are unresolved in the channel of the lock–washer conformation, indicating they remain flexible in the absence of a bound substrate (Yokom et al. 2016; Gates et al. 2017).
By 3D-classification analysis, we identified that the lock–washer conformation coexists with the closed, substrate-bound states in an equilibrium that varies with different nucleotide conditions and hydrolysis mutants, thus establishing a role in the Hsp104 conformational cycle (Gates et al. 2017). Conformational changes between these states are significant, involving a 65-Å translation of the seam protomer along the channel axis and a rotation inward, narrowing the channel (Gates et al. 2017). When considering possible Hsp70:substrate interactions at the amino-terminal entrance of the Hsp104 channel, this conformational change could serve to transfer a substantial, 20-residue segment of the polypeptide into the channel, enabling engagement by both the NBD1 and NBD2 (Gates et al. 2017). Alternatively, given the flexibility and reduced interprotomer contacts at the seam, hydrolysis may trigger ring-opening, enabling transfer of the substrate into the channel thereby allowing binding to internal segments of substrates, as discussed above. Open, lock–washer conformations have also been identified for the Mcm helicases and proposed to function as a ground state that facilitates deposition on DNA during replication (Costa et al. 2011; Zhai et al. 2017). Thus, in addition to substrate-translocation steps, parallels with AAA+ helicases may also exist for the initial loading step of disaggregation.
PLASTICITY IN MECHANISMS OF DISAGGREGATION
Protein disaggregation by Hsp104 or ClpB can involve nonprocessive and processive translocase mechanisms (DeSantis et al. 2012; Li et al. 2015). Indeed, cycling between the open lock–washer and closed ring states is proposed to enable nonprocessive bind and release tugging events (Yokom et al. 2016; Gates et al. 2017). These tugging events may suffice to extract polypeptides from labile protein aggregates (DeSantis et al. 2012; Li et al. 2015; Durie et al. 2018). Interestingly, a similar mechanism has been proposed for how the AAA+ protein, Katanin, might sever microtubules (Zehr et al. 2017).
In addition, the closed and extended substrate-bound states of Hsp104 hexamers suggest a more processive translocase mechanism (Gates et al. 2017). Here, two protomers undergo ratchet-like conformational changes that enable substrate binding and release steps to occur while the hexamer remains engaged to substrate (Gates et al. 2017). This rotary-like mechanism could drive disaggregation when coupled to stepwise cycles of ATP hydrolysis around the ring (Gates et al. 2017). Such a cooperative mechanism could enable dissolution of more stable aggregates or amyloids (DeSantis et al. 2012). This mechanism differs from “stochastic” models proposed for another Hsp100 unfoldase, ClpX (Martin et al. 2005; Cordova et al. 2014; Olivares et al. 2016). However, Hsp104 can show different modes of intersubunit collaboration that are tuned to different substrates (DeSantis et al. 2012). Thus, to resolve more labile aggregates, Hsp104 subunits can collaborate non-cooperatively (DeSantis et al. 2012), which likely corresponds to probabilistic mechanisms described for ClpX (Martin et al. 2005). In contrast, for more stable structures, such as amyloids and prions, several Hsp104 subunits must cooperatively engage substrate and hydrolyze ATP (DeSantis et al. 2012; DeSantis and Shorter 2012). Although additional hexameric states are likely involved in the complex process of protein disaggregation, our recent structures reveal the remarkable structural plasticity of Hsp104 (Yokom et al. 2016; Gates et al. 2017). This plasticity likely enables adaptable mechanisms of protein disaggregation tuned to respond to the different demands of different substrates (DeSantis et al. 2012; Murray and Kelly 2012; Sweeny and Shorter 2016).
CONCLUSION
The cryo-EM revolution has enabled new insights into the mechanism of various AAA+ ATPases, including the protein disaggregase Hsp104 (Yokom et al. 2016; Gates et al. 2017). Basic mechanisms of polypeptide translocation have now been revealed (Gates et al. 2017), which indicate surprising commonalities with other translocases including hexameric helicases that unwind DNA and RNA duplexes (Enemark and Joshua-Tor 2006; Thomsen and Berger 2009). However, many questions lie ahead, which are also likely to be informed by high-resolution cryo-EM analysis of additional complexes and states. For example, not all the domains of all the protomers of the Hsp104 or ClpB hexamer have been resolved by cryo-EM (Yokom et al. 2016; Deville et al. 2017; Gates et al. 2017). Resolving these domains could yield further insights into hexamer function. How exactly might Hsp70 load polypeptide into the Hsp104 channel? Moreover, how does Hsp104 engage and dissociate various structured amyloids? Finally, how might the structures of potentiated Hsp104 variants enable their enhanced activity in comparison to wild-type Hsp104? Answers to these questions will enhance our understanding of Hsp104 disaggregases, which have a variety of important basic, therapeutic, and pharmaceutical applications (Sweeny and Shorter 2016).
ACKNOWLEDGMENTS
We thank Shorter laboratory members for comments on the manuscript and Janet Iwasa for molecular visualization artwork. J.S. is supported by National Institutes of Health (NIH) Grant R01GM099836. D.R.S. is supported by NIH Grant R01GM110001.
REFERENCES
- Aguado A, Fernández-Higuero JA, Cabrera Y, Moro F, Muga A. 2015. ClpB dynamics is driven by its ATPase cycle and regulated by the DnaK system and substrate proteins. Biochem J 466: 561–570. doi: 10.1042/BJ20141390 [DOI] [PubMed] [Google Scholar]
- Ali YO, Allen HM, Yu L, Li-Kroeger D, Bakhshizadehmahmoudi D, Hatcher A, McCabe C, Xu J, Bjorklund N, Taglialatela G, et al. 2016. NMNAT2:HSP90 complex mediates proteostasis in proteinopathies. PLoS Biol 14: e1002472. doi: 10.1371/journal.pbio.1002472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson FI, Blakytny R, Kirstein J, Turgay K, Bukau B, Mogk A, Clarke AK. 2006. Cyanobacterial ClpC/HSP100 protein displays intrinsic chaperone activity. J Biol Chem 281: 5468–5475. doi: 10.1074/jbc.M509661200 [DOI] [PubMed] [Google Scholar]
- Andersson V, Hanzen S, Liu B, Molin M, Nystrom T. 2013. Enhancing protein disaggregation restores proteasome activity in aged cells. Aging (Albany NY) 5: 802–812. doi: 10.18632/aging.100613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai XC, McMullan G, Scheres SH. 2015. How cryo-EM is revolutionizing structural biology. Trends Biochem Sci 40: 49–57. doi: 10.1016/j.tibs.2014.10.005 [DOI] [PubMed] [Google Scholar]
- Baker JD, Shelton LB, Zheng D, Favretto F, Nordhues BA, Darling A, Sullivan LE, Sun Z, Solanki PK, Martin MD, et al. 2017. Human cyclophilin 40 unravels neurotoxic amyloids. PLoS Biol 15: e2001336. doi: 10.1371/journal.pbio.2001336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao YP, Cook LJ, O’Donovan D, Uyama E, Rubinsztein DC. 2002. Mammalian, yeast, bacterial, and chemical chaperones reduce aggregate formation and death in a cell model of oculopharyngeal muscular dystrophy. J Biol Chem 277: 12263–12269. doi: 10.1074/jbc.M109633200 [DOI] [PubMed] [Google Scholar]
- Carmichael J, Chatellier J, Woolfson A, Milstein C, Fersht AR, Rubinsztein DC. 2000. Bacterial and yeast chaperones reduce both aggregate formation and cell death in mammalian cell models of Huntington’s disease. Proc Natl Acad Sci 97: 9701–9705. doi: 10.1073/pnas.170280697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroni M, Kummer E, Oguchi Y, Wendler P, Clare DK, Sinning I, Kopp J, Mogk A, Bukau B, Saibil HR. 2014. Head-to-tail interactions of the coiled-coil domains regulate ClpB activity and cooperation with Hsp70 in protein disaggregation. eLife 3: e02481. doi: 10.7554/eLife.02481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashikar AG, Duennwald M, Lindquist SL. 2005. A chaperone pathway in protein disaggregation. Hsp26 alters the nature of protein aggregates to facilitate reactivation by Hsp104. J Biol Chem 280: 23869–23875. doi: 10.1074/jbc.M502854200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano LM, Bart SM, Holmes VM, Weissman D, Shorter J. 2015. Repurposing Hsp104 to antagonize seminal amyloid and counter HIV infection. Chem Biol 22: 1074–1086. doi: 10.1016/j.chembiol.2015.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty AK, Jarosz DF. 2018. More than just a phase: Prions at the crossroads of epigenetic inheritance and evolutionary change. J Mol Biol 430: 4607–4618. doi: 10.1016/j.jmb.2018.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y 2015. Single-particle cryo-EM at crystallographic resolution. Cell 161: 450–457. doi: 10.1016/j.cell.2015.03.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkasov V, Hofmann S, Druffel-Augustin S, Mogk A, Tyedmers J, Stoecklin G, Bukau B. 2013. Coordination of translational control and protein homeostasis during severe heat stress. Curr Biol 23: 2452–2462. doi: 10.1016/j.cub.2013.09.058 [DOI] [PubMed] [Google Scholar]
- Coelho M, Dereli A, Haese A, Kuhn S, Malinovska L, De-Santis ME, Shorter J, Alberti S, Gross T, Tolic-Norrelykke IM. 2013. Fission yeast does not age under favorable conditions, but does so after stress. Curr Biol 23: 1844–1852. doi: 10.1016/j.cub.2013.07.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordova JC, Olivares AO, Shin Y, Stinson BM, Calmat S, Schmitz KR, Aubin-Tam ME, Baker TA, Lang MJ, Sauer RT. 2014. Stochastic but highly coordinated protein unfolding and translocation by the ClpXP proteolytic machine. Cell 158: 647–658. doi: 10.1016/j.cell.2014.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A, Ilves I, Tamberg N, Petojevic T, Nogales E, Botchan MR, Berger JM. 2011. The structural basis for MCM2–7 helicase activation by GINS and Cdc45. Nat Struct Mol Biol 18: 471–477. doi: 10.1038/nsmb.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushman-Nick M, Bonini NM, Shorter J. 2013. Hsp104 suppresses polyglutamine-induced degeneration post onset in a Drosophila MJD/SCA3 model. PLoS Genet 9: e1003781. doi: 10.1371/journal.pgen.1003781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danev R, Tegunov D, Baumeister W. 2017. Using the Volta phase plate with defocus for cryo-EM single particle analysis. eLife 6: e23006. doi: 10.7554/eLife.23006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis ME, Shorter J. 2012. Hsp104 drives “protein-only” positive selection of Sup35 prion strains encoding strong [PSI+]. Chem Biol 19: 1400–1410. doi: 10.1016/j.chembiol.2012.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis ME, Leung EH, Sweeny EA, Jackrel ME, Cushman-Nick M, Neuhaus-Follini A, Vashist S, Sochor MA, Knight MN, Shorter J. 2012. Operational plasticity enables Hsp104 to disaggregate diverse amyloid and nonamyloid clients. Cell 151: 778–793. doi: 10.1016/j.cell.2012.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis ME, Sweeny EA, Snead D, Leung EH, Go MS, Gupta K, Wendler P, Shorter J. 2014. Conserved distal loop residues in the Hsp104 and ClpB middle domain contact nucleotide-binding domain 2 and enable Hsp70-dependent protein disaggregation. J Biol Chem 289: 848–867. doi: 10.1074/jbc.M113.520759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deville C, Carroni M, Franke KB, Topf M, Bukau B, Mogk A, Saibil HR. 2017. Structural pathway of regulated substrate transfer and threading through an Hsp100 disaggregase. Sci Adv 3: e1701726. doi: 10.1126/sciadv.1701726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan DA, Reid BG, Horwich AL, Bukau B. 2002. ClpS, a substrate modulator of the ClpAP machine. Mol Cell 9: 673–683. doi: 10.1016/S1097-2765(02)00485-9 [DOI] [PubMed] [Google Scholar]
- Doyle SM, Shorter J, Zolkiewski M, Hoskins JR, Lindquist S, Wickner S. 2007. Asymmetric deceleration of ClpB or Hsp104 ATPase activity unleashes protein-remodeling activity. Nat Struct Mol Biol 14: 114–122. doi: 10.1038/nsmb1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle SM, Shastry S, Kravats AN, Shih YH, Miot M, Hoskins JR, Stan G, Wickner S. 2015. Interplay between E. coli DnaK, ClpB and GrpE during protein disaggregation. J Mol Biol 427: 312–327. doi: 10.1016/j.jmb.2014.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duennwald ML, Echeverria A, Shorter J. 2012. Small heat shock proteins potentiate amyloid dissolution by protein disaggregases from yeast and humans. PLoS Biol 10: e1001346. doi: 10.1371/journal.pbio.1001346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran EC, Weaver CL, Lucius AL. 2017. Comparative analysis of the structure and function of AAA+ motors ClpA, ClpB, and Hsp104: Common threads and disparate functions. Front Mol Biosci 4: 54. doi: 10.3389/fmolb.2017.00054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durie CL, Duran EC, Lucius AL. 2018. Escherichia coli DnaK allosterically modulates ClpB between high- and low-peptide affinity states. Biochemistry 57: 3665–3675. doi: 10.1021/acs.biochem.8b00045 [DOI] [PubMed] [Google Scholar]
- Egelman EH. 2016. The current revolution in cryo-EM. Biophys J 110: 1008–1012. doi: 10.1016/j.bpj.2016.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enemark EJ, Joshua-Tor L. 2006. Mechanism of DNA translocation in a replicative hexameric helicase. Nature 442: 270–275. doi: 10.1038/nature04943 [DOI] [PubMed] [Google Scholar]
- Enemark EJ, Joshua-Tor L. 2008. On helicases and other motor proteins. Curr Opin Struct Biol 18: 243–257. doi: 10.1016/j.sbi.2008.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erives AJ, Fassler JS. 2015. Metabolic and chaperone gene loss marks the origin of animals: Evidence for Hsp104 and Hsp78 chaperones sharing mitochondrial enzymes as clients. PLoS ONE 10: e0117192. doi: 10.1371/journal.pone.0117192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erjavec N, Larsson L, Grantham J, Nystrom T. 2007. Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation-remodeling factor Hsp104p. Genes Dev 21: 2410–2421. doi: 10.1101/gad.439307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erzberger JP, Berger JM. 2006. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu Rev Biophys Biomol Struct 35: 93–114. doi: 10.1146/annurev.biophys.35.040405.101933 [DOI] [PubMed] [Google Scholar]
- Gai D, Zhao R, Li D, Finkielstein CV, Chen XS. 2004. Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen. Cell 119: 47–60. doi: 10.1016/j.cell.2004.09.017 [DOI] [PubMed] [Google Scholar]
- Gao X, Carroni M, Nussbaum-Krammer C, Mogk A, Nillegoda NB, Szlachcic A, Guilbride DL, Saibil HR, Mayer MP, Bukau B. 2015. Human Hsp70 disaggregase reverses Parkinson’s-linked α-synuclein amyloid fibrils. Mol Cell 59: 781–793. doi: 10.1016/j.molcel.2015.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates SN, Yokom AL, Lin J, Jackrel ME, Rizo AN, Kendsersky NM, Buell CE, Sweeny EA, Mack KL, Chuang E, et al. 2017. Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104. Science 357: 273–279. doi: 10.1126/science.aan1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover JR, Lindquist S. 1998. Hsp104, Hsp70, and Hsp40: A novel chaperone system that rescues previously aggregated proteins. Cell 94: 73–82. doi: 10.1016/S0092-8674(00)81223-4 [DOI] [PubMed] [Google Scholar]
- Glynn SE, Martin A, Nager AR, Baker TA, Sauer RT. 2009. Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell 139: 744–756. doi: 10.1016/j.cell.2009.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goloubinoff P, Mogk A, Zvi AP, Tomoyasu T, Bukau B. 1999. Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc Natl Acad Sci 96: 13732–13737. doi: 10.1073/pnas.96.24.13732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassucci RA, Taylor D, Frank J. 2008. Visualization of macromolecular complexes using cryo-electron microscopy with FEI Tecnai transmission electron microscopes. Nat Protoc 3: 330–339. doi: 10.1038/nprot.2007.474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Maurizi MR, Esser L, Xia D. 2002. Crystal structure of ClpA, an Hsp100 chaperone and regulator of ClpAP protease. J Biol Chem 277: 46743–46752. doi: 10.1074/jbc.M207796200 [DOI] [PubMed] [Google Scholar]
- Guo L, Kim HJ, Wang H, Monaghan J, Freyermuth F, Sung JC, O’Donovan K, Fare CM, Diaz Z, Singh N, et al. 2018. Nuclear-import receptors reverse aberrant phase transitions of RNA-binding proteins with prion-like domains. Cell 173: 677–692.e20. doi: 10.1016/j.cell.2018.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann R, Lindquist S. 2010. Epigenetics in the extreme: Prions and the inheritance of environmentally acquired traits. Science 330: 629–632. doi: 10.1126/science.1191081 [DOI] [PubMed] [Google Scholar]
- Han H, Monroe N, Sundquist WI, Shen PS, Hill CP. 2017. The AAA ATPase Vps4 binds ESCRT-III substrates through a repeating array of dipeptide-binding pockets. eLife 6: e31324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänninen AL, Simola M, Saris N, Makarow M. 1999. The cytoplasmic chaperone hsp104 is required for conformational repair of heat-denatured proteins in the yeast endoplasmic reticulum. Mol Biol Cell 10: 3623–3632. doi: 10.1091/mbc.10.11.3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzen S, Vielfort K, Yang J, Roger F, Andersson V, Zamar-bide-Fores S, Andersson R, Malm L, Palais G, Biteau B, et al. 2016. Lifespan control by redox-dependent recruitment of chaperones to misfolded proteins. Cell 166: 140–151. doi: 10.1016/j.cell.2016.05.006 [DOI] [PubMed] [Google Scholar]
- Harvey ZH, Chen Y, Jarosz DF. 2018. Protein-based inheritance: Epigenetics beyond the chromosome. Mol Cell 69: 195–202. doi: 10.1016/j.molcel.2017.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslbeck M, Miess A, Stromer T, Walter S, Buchner J. 2005. Disassembling protein aggregates in the yeast cytosol. The cooperation of Hsp26 with Ssa1 and Hsp104. J Biol Chem 280: 23861–23868. doi: 10.1074/jbc.M502697200 [DOI] [PubMed] [Google Scholar]
- Haslberger T, Weibezahn J, Zahn R, Lee S, Tsai FT, Bukau B, Mogk A. 2007. M domains couple the ClpB threading motor with the DnaK chaperone activity. Mol Cell 25: 247–260. doi: 10.1016/j.molcel.2006.11.008 [DOI] [PubMed] [Google Scholar]
- Haslberger T, Zdanowicz A, Brand I, Kirstein J, Turgay K, Mogk A, Bukau B. 2008. Protein disaggregation by the AAA+ chaperone ClpB involves partial threading of looped polypeptide segments. Nat Struct Mol Biol 15: 641–650. doi: 10.1038/nsmb.1425 [DOI] [PubMed] [Google Scholar]
- Heuck A, Schitter-Sollner S, Suskiewicz MJ, Kurzbauer R, Kley J, Schleiffer A, Rombaut P, Herzog F, Clausen T. 2016. Structural basis for the disaggregase activity and regulation of Hsp104. eLife 5: e21516. doi: 10.7554/eLife.21516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill SM, Hao X, Liu B, Nystrom T. 2014. Life-span extension by a metacaspase in the yeast Saccharomyces cerevisiae. Science 344: 1389–1392. doi: 10.1126/science.1252634 [DOI] [PubMed] [Google Scholar]
- Hill SM, Hao X, Gronvall J, Spikings-Nordby S, Widlund PO, Amen T, Jorhov A, Josefson R, Kaganovich D, Liu B, et al. 2016. Asymmetric inheritance of aggregated proteins and age reset in yeast are regulated by Vac17-dependent vacuolar functions. Cell Rep 16: 826–838. doi: 10.1016/j.celrep.2016.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins JR, Doyle SM, Wickner S. 2009. Coupling ATP utilization to protein remodeling by ClpB, a hexameric AAA+ protein. Proc Natl Acad Sci 106: 22233–22238. doi: 10.1073/pnas.0911937106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R, Ripstein ZA, Augustyniak R, Lazniewski M, Ginalski K, Kay LE, Rubinstein JL. 2016. Unfolding the mechanism of the AAA+ unfoldase VAT by a combined cryo-EM, solution NMR study. Proc Natl Acad Sci 113: E4190–E4199. doi: 10.1073/pnas.1603980113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itsathitphaisarn O, Wing RA, Eliason WK, Wang J, Steitz TA. 2012. The hexameric helicase DnaB adopts a nonplanar conformation during translocation. Cell 151: 267–277. doi: 10.1016/j.cell.2012.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackrel ME, Shorter J. 2014a. Potentiated Hsp104 variants suppress toxicity of diverse neurodegenerative disease-linked proteins. Dis Model Mech 7: 1175–1184. doi: 10.1242/dmm.016113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackrel ME, Shorter J. 2014b. Reversing deleterious protein aggregation with re-engineered protein disaggregases. Cell Cycle 13: 1379–1383. doi: 10.4161/cc.28709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackrel ME, DeSantis ME, Martinez BA, Castellano LM, Stewart RM, Caldwell KA, Caldwell GA, Shorter J. 2014. Potentiated Hsp104 variants antagonize diverse proteotoxic misfolding events. Cell 156: 170–182. doi: 10.1016/j.cell.2013.11.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackrel ME, Yee K, Tariq A, Chen AI, Shorter J. 2015. Disparate mutations confer therapeutic gain of Hsp104 function. ACS Chem Biol 10: 2672–2679. doi: 10.1021/acschembio.5b00765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaimal JM, Kandasamy G, Gasser F, Andréasson C. 2017. Coordinated Hsp110 and Hsp104 activities power protein disaggregation in Saccharomyces cerevisiae. Mol Cell Biol 37: e00027–17. doi: 10.1128/MCB.00027-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoshouei M, Radjainia M, Baumeister W, Danev R. 2017. Cryo-EM structure of haemoglobin at 3.2 Å determined with the Volta phase plate. Nat Commun 8: 16099. doi: 10.1038/ncomms16099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimanius D, Forsberg BO, Scheres SH, Lindahl E. 2016. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. eLife 5: e18722. doi: 10.7554/eLife.18722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaips CL, Hochstrasser ML, Langlois CR, Serio TR. 2014. Spatial quality control bypasses cell-based limitations on proteostasis to promote prion curing. eLife 3: e04288. doi: 10.7554/eLife.04288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koldewey P, Horowitz S, Bardwell JCA. 2017. Chaperone–client interactions: Non-specificity engenders multifunctionality. J Biol Chem 292: 12010–12017. doi: 10.1074/jbc.R117.796862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajewska J, Modrak-Wojcik A, Arent ZJ, Wieckowski D, Zolkiewski M, Bzowska A, Kedzierska-Mieszkowska S. 2017. Characterization of the molecular chaperone ClpB from the pathogenic spirochaete Leptospira interrogans. PLoS ONE 12: e0181118. doi: 10.1371/journal.pone.0181118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroschwald S, Maharana S, Mateju D, Malinovska L, Nüske E, Poser I, Richter D, Alberti S. 2015. Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. eLife 4: e06807. doi: 10.7554/eLife.06807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroschwald S, Munder MC, Maharana S, Franzmann TM, Richter D, Ruer M, Hyman AA, Alberti S. 2018. Different material states of Pub1 condensates define distinct modes of stress adaptation and recovery. Cell Rep 23: 3327–3339. doi: 10.1016/j.celrep.2018.05.041 [DOI] [PubMed] [Google Scholar]
- Kuhlbrandt W 2014. Biochemistry. The resolution revolution. Science 343: 1443–1444. doi: 10.1126/science.1251652 [DOI] [PubMed] [Google Scholar]
- LaBreck CJ, May S, Viola MG, Conti J, Camberg JL. 2017. The protein chaperone ClpX targets native and non-native aggregated substrates for remodeling, disassembly, and degradation with ClpP. Front Mol Biosci 4: 26. doi: 10.3389/fmolb.2017.00026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Goldberg AL. 2010. Hsp104 is essential for the selective degradation in yeast of polyglutamine expanded ataxin-1 but not most misfolded proteins generally. Biochem Biophys Res Commun 391: 1056–1061. doi: 10.1016/j.bbrc.2009.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Sowa ME, Watanabe YH, Sigler PB, Chiu W, Yoshida M, Tsai FT. 2003. The structure of ClpB: A molecular chaperone that rescues proteins from an aggregated state. Cell 115: 229–240. doi: 10.1016/S0092-8674(03)00807-9 [DOI] [PubMed] [Google Scholar]
- Lee S, Choi JM, Tsai FT. 2007. Visualizing the ATPase cycle in a protein disaggregating machine: Structural basis for substrate binding by ClpB. Mol Cell 25: 261–271. doi: 10.1016/j.molcel.2007.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Sielaff B, Lee J, Tsai FT. 2010. CryoEM structure of Hsp104 and its mechanistic implication for protein disaggregation. Proc Natl Acad Sci 107: 8135–8140. doi: 10.1073/pnas.1003572107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kim JH, Biter AB, Sielaff B, Lee S, Tsai FT. 2013. Heat shock protein (Hsp) 70 is an activator of the Hsp104 motor. Proc Natl Acad Sci 110: 8513–8518. doi: 10.1073/pnas.1217988110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Franke KB, Kamal SM, Kim H, Lunsdorf H, Jager J, Nimtz M, Trcek J, Jansch L, Bukau B, et al. 2018. Stand-alone ClpG disaggregase confers superior heat tolerance to bacteria. Proc Natl Acad Sci 115: E273–E282. doi: 10.1073/pnas.1712051115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y. 2013. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods 10: 584–590. doi: 10.1038/nmeth.2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Weaver CL, Lin J, Duran EC, Miller JM, Lucius AL. 2015. Escherichia coli ClpB is a nonprocessive polypeptide translocase. Biochem J 470: 39–52. doi: 10.1042/BJ20141457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Larsson L, Caballero A, Hao X, Oling D, Grantham J, Nystrom T. 2010. The polarisome is required for segregation and retrograde transport of protein aggregates. Cell 140: 257–267. doi: 10.1016/j.cell.2009.12.031 [DOI] [PubMed] [Google Scholar]
- Liu B, Larsson L, Franssens V, Hao X, Hill SM, Andersson V, Hoglund D, Song J, Yang X, Oling D, et al. 2011a. Segregation of protein aggregates involves actin and the polarity machinery. Cell 147: 959–961. doi: 10.1016/j.cell.2011.11.018 [DOI] [PubMed] [Google Scholar]
- Liu YH, Han YL, Song J, Wang Y, Jing YY, Shi Q, Tian C, Wang ZY, Li CP, Han J, et al. 2011b. Heat shock protein 104 inhibited the fibrillization of prion peptide 106–126 and disassembled prion peptide 106–126 fibrils in vitro. Int J Biochem Cell Biol 43: 768–774. doi: 10.1016/j.biocel.2011.01.022 [DOI] [PubMed] [Google Scholar]
- Lo Bianco C, Shorter J, Régulier E, Lashuel H, Iwatsubo T, Lindquist S, Aebischer P. 2008. Hsp104 antagonizes α-synuclein aggregation and reduces dopaminergic degeneration in a rat model of Parkinson disease. J Clin Invest 118: 3087–3097. doi: 10.1172/JCI35781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum R, Tkach JM, Vierling E, Glover JR. 2004. Evidence for an unfolding/threading mechanism for protein disaggregation by Saccharomyces cerevisiae Hsp104. J Biol Chem 279: 29139–29146. doi: 10.1074/jbc.M403777200 [DOI] [PubMed] [Google Scholar]
- Lum R, Niggemann M, Glover JR. 2008. Peptide and protein binding in the axial channel of Hsp104. Insights into the mechanism of protein unfolding. J Biol Chem 283: 30139–30150. doi: 10.1074/jbc.M804849200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyubimov AY, Strycharska M, Berger JM. 2011. The nuts and bolts of ring-translocase structure and mechanism. Curr Opin Struct Biol 21: 240–248. doi: 10.1016/j.sbi.2011.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. 2005. Rebuilt AAA+ motors reveal operating principles for ATP-fueled machines. Nature 437: 1115–1120. doi: 10.1038/nature04031 [DOI] [PubMed] [Google Scholar]
- Matyskiela ME, Lander GC, Martin A. 2013. Conformational switching of the 26S proteasome enables substrate degradation. Nat Struct Mol Biol 20: 781–788. doi: 10.1038/nsmb.2616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullan G, Faruqi AR, Henderson R. 2016. Direct electron detectors. Methods Enzymol 579: 1–17. doi: 10.1016/bs.mie.2016.05.056 [DOI] [PubMed] [Google Scholar]
- Mogk A, Kummer E, Bukau B. 2015. Cooperation of Hsp70 and Hsp100 chaperone machines in protein disaggregation. Front Mol Biosci 2: 22. doi: 10.3389/fmolb.2015.00022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AN, Kelly JW. 2012. Hsp104 gives clients the individual attention they need. Cell 151: 695–697. doi: 10.1016/j.cell.2012.10.033 [DOI] [PubMed] [Google Scholar]
- Nillegoda NB, Kirstein J, Szlachcic A, Berynskyy M, Stank A, Stengel F, Arnsburg K, Gao X, Scior A, Aebersold R, et al. 2015. Crucial HSP70 co-chaperone complex unlocks metazoan protein disaggregation. Nature 524: 247–251. doi: 10.1038/nature14884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguchi Y, Kummer E, Seyffer F, Berynskyy M, Anstett B, Zahn R, Wade RC, Mogk A, Bukau B. 2012. A tightly regulated molecular toggle controls AAA+ disaggregase. Nat Struct Mol Biol 19: 1338–1346. doi: 10.1038/nsmb.2441 [DOI] [PubMed] [Google Scholar]
- Olivares AO, Baker TA, Sauer RT. 2016. Mechanistic insights into bacterial AAA+ proteases and protein-remodelling machines. Nat Rev Microbiol 14: 33–44. doi: 10.1038/nrmicro.2015.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YN, Zhao X, Yim YI, Todor H, Ellerbrock R, Reidy M, Eisenberg E, Masison DC, Greene LE. 2014. Hsp104 over-expression cures Saccharomyces cerevisiae [PSI+] by causing dissolution of the prion seeds. Eukaryot Cell 13: 635–647. doi: 10.1128/EC.00300-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SS, Kwon HY, Tran TD, Choi MH, Jung SH, Lee S, Briles DE, Rhee DK. 2015. ClpL is a chaperone without auxiliary factors. FEBS J 282: 1352–1367. doi: 10.1111/febs.13228 [DOI] [PubMed] [Google Scholar]
- Parsell DA, Kowal AS, Lindquist S. 1994a. Saccharomyces cerevisiae Hsp104 protein. Purification and characterization of ATP-induced structural changes. J Biol Chem 269: 4480–4487. [PubMed] [Google Scholar]
- Parsell DA, Kowal AS, Singer MA, Lindquist S. 1994b. Protein disaggregation mediated by heat-shock protein Hsp104. Nature 372: 475–478. doi: 10.1038/372475a0 [DOI] [PubMed] [Google Scholar]
- Perrin V, Regulier E, Abbas-Terki T, Hassig R, Brouillet E, Aebischer P, Luthi-Carter R, Deglon N. 2007. Neuroprotection by Hsp104 and Hsp27 in lentiviral-based rat models of Huntington’s disease. Mol Ther 15: 903–911. doi: 10.1038/mt.sj.6300141 [DOI] [PubMed] [Google Scholar]
- Poepsel S, Sprengel A, Sacca B, Kaschani F, Kaiser M, Gatsogiannis C, Raunser S, Clausen T, Ehrmann M. 2015. Determinants of amyloid fibril degradation by the PDZ protease HTRA1. Nat Chem Biol 11: 862–869. doi: 10.1038/nchembio.1931 [DOI] [PubMed] [Google Scholar]
- Preston GM, Guerriero CJ, Metzger MB, Michaelis S, Brodsky JL. 2018. Substrate insolubility dictates Hsp104-dependent endoplasmic-reticulum-associated degradation. Mol Cell 70: 242–253.e6. doi: 10.1016/j.molcel.2018.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchades C, Rampello AJ, Shin M, Giuliano CJ, Wiseman RL, Glynn SE, Lander GC. 2017. Structure of the mitochondrial inner membrane AAA+ protease YME1 gives insight into substrate processing. Science 358: eaao0464. doi: 10.1126/science.aao0464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. 2017. cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14: 290–296. doi: 10.1038/nmeth.4169 [DOI] [PubMed] [Google Scholar]
- Ripstein ZA, Huang R, Augustyniak R, Kay LE, Rubinstein JL. 2017. Structure of a AAA+ unfoldase in the process of unfolding substrate. eLife 6: e25754. doi: 10.7554/eLife.25754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig R, Moradi S, Zarrine-Afsar A, Glover JR, Kay LE. 2013. Unraveling the mechanism of protein disaggregation through a ClpB–DnaK interaction. Science 339: 1080–1083. doi: 10.1126/science.1233066 [DOI] [PubMed] [Google Scholar]
- Ruan L, Zhou C, Jin E, Kucharavy A, Zhang Y, Wen Z, Florens L, Li R. 2017. Cytosolic proteostasis through importing of misfolded proteins into mitochondria. Nature 543: 443–446. doi: 10.1038/nature21695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarikangas J, Barral Y. 2015. Protein aggregates are associated with replicative aging without compromising protein quality control. eLife 4: e06197. doi: 10.7554/eLife.06197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Y, Lindquist SL. 1990. HSP104 required for induced thermotolerance. Science 248: 1112–1115. doi: 10.1126/science.2188365 [DOI] [PubMed] [Google Scholar]
- Sanchez Y, Taulien J, Borkovich KA, Lindquist S. 1992. Hsp104 is required for tolerance to many forms of stress. EMBO J 11: 2357–2364. doi: 10.1002/j.1460-2075.1992.tb05295.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyal SH, Schmidt E, Kitagawa K, Sondheimer N, Lindquist S, Kramer JM, Morimoto RI. 2000. Polyglutamine aggregates alter protein folding homeostasis in Caenorhabditis elegans. Proc Natl Acad Sci 97: 5750–5755. doi: 10.1073/pnas.100107297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH. 2012. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180: 519–530. doi: 10.1016/j.jsb.2012.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scior A, Buntru A, Arnsburg K, Ast A, Iburg M, Juenemann K, Pigazzini ML, Mlody B, Puchkov D, Priller J, et al. 2018. Complete suppression of Htt fibrilization and disaggregation of Htt fibrils by a trimeric chaperone complex. EMBO J 37: 282–299. doi: 10.15252/embj.201797212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyffer F, Kummer E, Oguchi Y, Winkler J, Kumar M, Zahn R, Sourjik V, Bukau B, Mogk A. 2012. Hsp70 proteins bind Hsp100 regulatory M domains to activate AAA+ disaggregase at aggregate surfaces. Nat Struct Mol Biol 19: 1347–1355. doi: 10.1038/nsmb.2442 [DOI] [PubMed] [Google Scholar]
- Shorter J 2011. The mammalian disaggregase machinery: Hsp110 synergizes with Hsp70 and Hsp40 to catalyze protein disaggregation and reactivation in a cell-free system. PLoS ONE 6: e26319. doi: 10.1371/journal.pone.0026319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J 2016. Engineering therapeutic protein disaggregases. Mol Biol Cell 27: 1556–1560. doi: 10.1091/mbc.e15-10-0693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J 2017. Designer protein disaggregases to counter neurodegenerative disease. Curr Opin Genet Dev 44: 1–8. doi: 10.1016/j.gde.2017.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. 2004. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science 304: 1793–1797. doi: 10.1126/science.1098007 [DOI] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. 2005. Prions as adaptive conduits of memory and inheritance. Nat Rev Genet 6: 435–450. doi: 10.1038/nrg1616 [DOI] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. 2006. Destruction or potentiation of different prions catalyzed by similar Hsp104 remodeling activities. Mol Cell 23: 425–438. doi: 10.1016/j.molcel.2006.05.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. 2008. Hsp104, Hsp70 and Hsp40 interplay regulates formation, growth and elimination of Sup35 prions. EMBO J 27: 2712–2724. doi: 10.1038/emboj.2008.194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton MR, Dillingham MS, Wigley DB. 2007. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem 76: 23–50. doi: 10.1146/annurev.biochem.76.052305.115300 [DOI] [PubMed] [Google Scholar]
- Skiniotis G, Southworth DR. 2016. Single-particle cryo-electron microscopy of macromolecular complexes. Microscopy 65: 9–22. doi: 10.1093/jmicro/dfv366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spokoini R, Moldavski O, Nahmias Y, England JL, Schuldiner M, Kaganovich D. 2012. Confinement to organelle-associated inclusion structures mediates asymmetric inheritance of aggregated protein in budding yeast. Cell Rep 2: 738–747. doi: 10.1016/j.celrep.2012.08.024 [DOI] [PubMed] [Google Scholar]
- Su M, Guo EZ, Ding X, Li Y, Tarrasch JT, Brooks CL III, Xu Z, Skiniotis G. 2017. Mechanism of Vps4 hexamer function revealed by cryo-EM. Sci Adv 3: e1700325. doi: 10.1126/sciadv.1700325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeny EA, Shorter J. 2016. Mechanistic and structural insights into the prion-disaggregase activity of Hsp104. J Mol Biol 428: 1870–1885. doi: 10.1016/j.jmb.2015.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeny EA, Jackrel ME, Go MS, Sochor MA, Razzo BM, DeSantis ME, Gupta K, Shorter J. 2015. The Hsp104 N-terminal domain enables disaggregase plasticity and potentiation. Mol Cell 57: 836–849. doi: 10.1016/j.molcel.2014.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa Y, Binshtein E, Erwin AL, Pyburn TM, Mittendorf KF, Ohi MD. 2017. While the revolution will not be crystallized, biochemistry reigns supreme. Protein Sci 26: 69–81. doi: 10.1002/pro.3054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tariq A, Lin J, Noll MM, Torrente MP, Mack KL, Murillo OH, Jackrel ME, Shorter J. 2018. Potentiating Hsp104 activity via phosphomimetic mutations in the middle domain. FEMS Yeast Res 18: foy042. doi: 10.1093/femsyr/foy042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taxis C, Hitt R, Park SH, Deak PM, Kostova Z, Wolf DH. 2003. Use of modular substrates demonstrates mechanistic diversity and reveals differences in chaperone requirement of ERAD. J Biol Chem 278: 35903–35913. doi: 10.1074/jbc.M301080200 [DOI] [PubMed] [Google Scholar]
- Tessarz P, Mogk A, Bukau B. 2008. Substrate threading through the central pore of the Hsp104 chaperone as a common mechanism for protein disaggregation and prion propagation. Mol Microbiol 68: 87–97. doi: 10.1111/j.1365-2958.2008.06135.x [DOI] [PubMed] [Google Scholar]
- Tessarz P, Schwarz M, Mogk A, Bukau B. 2009. The yeast AAA+ chaperone Hsp104 is part of a network that links the actin cytoskeleton with the inheritance of damaged proteins. Mol Cell Biol 29: 3738–3745. doi: 10.1128/MCB.00201-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen ND, Berger JM. 2008. Structural frameworks for considering microbial protein- and nucleic acid-dependent motor ATPases. Mol Microbiol 69: 1071–1090. doi: 10.1111/j.1365-2958.2008.06364.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen ND, Berger JM. 2009. Running in reverse: The structural basis for translocation polarity in hexameric helicases. Cell 139: 523–534. doi: 10.1016/j.cell.2009.08.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrente MP, Shorter J. 2013. The metazoan protein disaggregase and amyloid depolymerase system: Hsp110, Hsp70, Hsp40, and small heat shock proteins. Prion 7: 457–463. doi: 10.4161/pri.27531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trakselis MA. 2016. Structural mechanisms of hexameric helicase loading, assembly, and unwinding. F1000Research 5: 111. doi: 10.12688/f1000research.7509.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchihashi T, Watanabe YH, Nakazaki Y, Yamasaki T, Watanabe H, Maruno T, Ishii K, Uchiyama S, Song C, Murata K, et al. 2018. Dynamic structural states of ClpB involved in its disaggregation function. Nat Commun 9: 2147. doi: 10.1038/s41467-018-04587-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacher C, Garcia-Oroz L, Rubinsztein DC. 2005. Overexpression of yeast hsp104 reduces polyglutamine aggregation and prolongs survival of a transgenic mouse model of Huntington’s disease. Hum Mol Genet 14: 3425–3433. doi: 10.1093/hmg/ddi372 [DOI] [PubMed] [Google Scholar]
- Wallace EW, Kear-Scott JL, Pilipenko EV, Schwartz MH, Laskowski PR, Rojek AE, Katanski CD, Riback JA, Dion MF, Franks AM, et al. 2015. Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell 162: 1286–1298. doi: 10.1016/j.cell.2015.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver CL, Duran EC, Mack KL, Lin J, Jackrel ME, Sweeny EA, Shorter J, Lucius AL. 2017. Avidity for polypeptide binding by nucleotide-bound Hsp104 structures. Biochemistry 56: 2071–2075. doi: 10.1021/acs.biochem.7b00225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehmer M, Sakata E. 2016. Recent advances in the structural biology of the 26S proteasome. Int J Biochem Cell Biol 79: 437–442. doi: 10.1016/j.biocel.2016.08.008 [DOI] [PubMed] [Google Scholar]
- Weibezahn J, Schlieker C, Bukau B, Mogk A. 2003. Characterization of a trap mutant of the AAA+ chaperone ClpB. J Biol Chem 278: 32608–32617. doi: 10.1074/jbc.M303653200 [DOI] [PubMed] [Google Scholar]
- Weibezahn J, Tessarz P, Schlieker C, Zahn R, Maglica Z, Lee S, Zentgraf H, Weber-Ban EU, Dougan DA, Tsai FT, et al. 2004. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell 119: 653–665. doi: 10.1016/j.cell.2004.11.027 [DOI] [PubMed] [Google Scholar]
- Wendler P, Shorter J, Plisson C, Cashikar AG, Lindquist S, Saibil HR. 2007. Atypical AAA+ subunit packing creates an expanded cavity for disaggregation by the protein-remodeling factor Hsp104. Cell 131: 1366–1377. doi: 10.1016/j.cell.2007.10.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendler P, Shorter J, Snead D, Plisson C, Clare DK, Lindquist S, Saibil HR. 2009. Motor mechanism for protein threading through Hsp104. Mol Cell 34: 81–92. doi: 10.1016/j.molcel.2009.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendler P, Ciniawsky S, Kock M, Kube S. 2012. Structure and function of the AAA+ nucleotide binding pocket. Biochim Biophys Acta 1823: 2–14. doi: 10.1016/j.bbamcr.2011.06.014 [DOI] [PubMed] [Google Scholar]
- Werbeck ND, Schlee S, Reinstein J. 2008. Coupling and dynamics of subunits in the hexameric AAA+ chaperone ClpB. J Mol Biol 378: 178–190. doi: 10.1016/j.jmb.2008.02.026 [DOI] [PubMed] [Google Scholar]
- Wu S, Armache JP, Cheng Y. 2016. Single-particle cryo-EM data acquisition by using direct electron detection camera. Microscopy 65: 35–41. doi: 10.1093/jmicro/dfv355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda K, Clatterbuck-Soper SF, Jackrel ME, Shorter J, Mili S. 2017. FUS inclusions disrupt RNA localization by sequestering kinesin-1 and inhibiting microtubule detyrosination. J Cell Biol 216: 1015–1034. doi: 10.1083/jcb.201608022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokom AL, Gates SN, Jackrel ME, Mack KL, Su M, Shorter J, Southworth DR. 2016. Spiral architecture of the Hsp104 disaggregase reveals the basis for polypeptide translocation. Nat Struct Mol Biol 23: 830–837. doi: 10.1038/nsmb.3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa T, Ali R, Jiou J, Fung HYJ, Burke KA, Kim SJ, Lin Y, Peeples WB, Saltzberg D, Soniat M, et al. 2018. Nuclear import receptor inhibits phase separation of FUS through binding to multiple sites. Cell 173: 693–705.e22. doi: 10.1016/j.cell.2018.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehr E, Szyk A, Piszczek G, Szczesna E, Zuo X, Roll-Mecak A. 2017. Katanin spiral and ring structures shed light on power stroke for microtubule severing. Nat Struct Mol Biol 24: 717–725. doi: 10.1038/nsmb.3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y, Cheng E, Wu H, Li N, Yung PY, Gao N, Tye BK. 2017. Open-ringed structure of the Cdt1–Mcm2–7 complex as a precursor of the MCM double hexamer. Nat Struct Mol Biol 24: 300–308. doi: 10.1038/nsmb.3374 [DOI] [PubMed] [Google Scholar]
- Zhang T, Kedzierska-Mieszkowska S, Liu H, Cheng C, Ganta RR, Zolkiewski M. 2013. Aggregate-reactivation activityof the molecular chaperone ClpB from Ehrlichia chaffeensis. PLoS ONE 8: e62454. doi: 10.1371/journal.pone.0062454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Rodriguez R, Silberman RE, Ahearn JM, Saidha S, Cummins KC, Eisenberg E, Greene LE. 2017. Heat shock protein 104 (Hsp104)-mediated curing of [PSI+] yeast prions depends on both [PSI+] conformation and the properties of the Hsp104 homologs. J Biol Chem 292: 8630–8641. doi: 10.1074/jbc.M116.770719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Slaughter BD, Unruh JR, Eldakak A, Rubinstein B, Li R. 2011. Motility and segregation of Hsp104-associated protein aggregates in budding yeast. Cell 147: 1186–1196. doi: 10.1016/j.cell.2011.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]