Abstract
Purpose
To develop a quality assurance (QA) workflow using a robust, curated, manually-segmented anatomic ROI library as a benchmark for quantitative assessment of different image registration techniques used for head and neck radiation therapy-simulation CT (SimCT) to diagnostic CT (DxCT) co-registration.
Materials and Methods
SimCTs and DxCTs of twenty patients with head and neck squamous cell carcinoma treated with curative-intent intensity modulated radiotherapy (IMRT) between August 2011 and May 2012 were retrospectively retrieved under an institutional review board approval. 68 reference anatomic regions of interest (ROIs) in addition to gross tumor and nodal targets were then manually contoured on each scan. DxCT was registered to SimCT rigidly, and through 4 different deformable image registration (DIR) algorithms; Atlas-based, B-spline, demons, and optical flow. The resultant deformed ROIs were compared with manually contoured reference ROIs using similarity coefficient metrics (i.e. Dice similarity coefficient) and surface distance metrics (i.e. 95% maximum Hausdorff distance). Non-parametric Steel test with control was used to compare different DIR algorithms to rigid registration (RIR) with post hoc Wilcoxon rank test for stratified metric comparison.
Results
A total of 2720 anatomic and 50 tumor/nodal ROIs were delineated. All DIR algorithms showed improved performance over RIR for both anatomic and target ROIs conformance as shown for the majority of comparison metrics (Steel test, p-value <0.008 after Bonferroni correction). The performance of different algorithms varied substantially with stratification by specific anatomic structures/category, and SimCT image slice thickness.
Conclusion
Development of a formal ROI-based QA workflow for registration assessment revealed improved performance with DIR techniques over RIR. After QA, DIR implementation should be the standard for head and neck DxCT-SimCT allineation, especially for target delineation.
Introduction
Deformable image registration (DIR) is an increasingly common tool for applications in image-guided radiotherapy (IGRT) (1–3). DIR, as a tool for motion assessment/correction in tumors that move with respiration, as well as for adaptive re-contouring of target or anatomic volumes that alter over time, is becoming more widely utilized, as vendors integrate DIR solutions into commercial software packages (4–9). Additionally, emerging data suggest that, for head and neck cancers, DIR has demonstrable technical performance gain compared to rigid image registration (RIR) for adaptive radiotherapy, wherein a simulation CT dataset is co-registered with on-treatment CT or cone beam CT (10–13).
Simulation three-dimensional CT (SimCT) datasets are the initial component of radiation planning. SimCT datasets are then manually segmented to define both tumor and normal tissue volumes, with subsequent dose calculation performed using voxel-based electron density maps. Consequently, as the key imaging step in radiotherapy, all subsequent patient treatment dose delivery is entirely dependent on the quality of SimCT processes (e.g. target delineation, organ-at-risk segmentation, beam/intensity optimization, and dose calculation)(14).
The intramodality registration of pre- and post-therapy head and neck diagnostic CTs (DxCT) to SimCT data is valuable in multiple radiotherapy applications e.g. target delineation and mapping the sites of post-therapy loco-regional recurrences to the original SimCT and dose grid(15–18). Nevertheless, such intramodality fusion of DxCT and SimCT is less commonly described in the literature and presents specific obstacles to image registration. First, DxCT acquisition routinely is performed on a curved tabletop without standardized head positioning, while the SimCT is obtained in a custom thermoplastic immobilization mask on a flat toped table, resulting in positional differences of head and neck tissues (15, 16). In several head and neck cancers, institutional use of a intraoral immobilization and displacement device, such as a custom dental stent (19), results in placement of a new structure in the SimCT which was not present in the DxCT. Additionally, DxCT typically entails use of intravenous contrast for tumor assessment, while at our facility and many others, intravenous contrast is not utilized for SimCT, resulting in intensity differentials for the same structures (10). Acquisition parameters (e.g. slice thickness reconstruction (STR), field of view, kvp, etc.) may not be standardized between DxCT and SimCT. Finally, in many instances, due to either tumor progression or intervening therapy (surgical resection or induction chemotherapy) the anatomy is fundamentally altered between DxCT and SimCT. These factors, among others, make DxCT-to-SimCT registration a non-trivial task. As part of larger efforts to improve head and neck target delineation, as well as defining spatially accurate mapping of locoregional failure sites, the purpose of this study was to develop a quality assurance (QA) workflow using a robust, curated, manually-segmented anatomic ROI library as a benchmark for quantitative assessment of different image registration techniques used for head and neck radiation therapy-SimCT to DxCT co-registration.
Materials and methods
Study population
SimCT and DxCT DICOM files of twenty two head and neck cancer patients treated at our institution between August 2011 and May 2012, were retrospectively retrieved under Institutional Review Board approval. Inclusion criteria were pathologically proven diagnosis of squamous cell carcinoma of head and neck, treatment with curative-intent intensity modulated radiation therapy (IMRT), availability of non-contrast enhanced SimCT as well as contrast-enhanced DxCT for each patient within a maximum time interval of 4 weeks between both scans to minimize errors attributed to therapy or disease progression-associated anatomic changes. A total of twenty patients were eligible while two patients were excluded, one for having massive disease progression and the other for having surgical resection during the interval between DxCT and SimCT. Patients and treatment characteristics are summarized in table 1.
Table 1.
Patient characteristics.
| Characteristics | No. |
|---|---|
| Age (y) | |
| Median (range) | 63 (50–78) |
|
| |
| Sex | |
| Male | 15 |
| Female | 5 |
|
| |
| T Stage | |
| T1 | 4 |
| T2 | 5 |
| T3 | 8 |
| T4 | 2 |
| Tx | 1 |
|
| |
| N Stage | |
| N0 | 3 |
| N1 | 4 |
| N2 | 11 |
| N3 | 1 |
| Nx | 1 |
|
| |
| Primary sites | |
| Base of Tongue | 7 |
| Tonsil | 6 |
| Oral Cavity | 1 |
| Larynx | 3 |
| Nasopharyngeal/Maxillary sinus | 2 |
| Salivary gland | 1 |
|
| |
| Treatment | |
| IMRT alone | 1 |
| Concurrent chemo-IMRT | 5 |
| Induction chemotherapy + chemo- | 9 |
| IMRT | 3 |
| Induction chemotherapy + IMRT | 1 |
| Surgery + postoperative chemo-IMRT Induction chemo-IMRT + Surgery | 1 |
Imaging characteristics
Non-contrast SimCTs were acquired after immobilizing patients with thermoplastic head and neck shoulder masks with slice thickness ranging from 1–3.75 mm, X-ray tube current ranging from 100–297 mA at 120 kVp. Display field of view (DFOV) was 500 mm, axial images were acquired using a matrix of 512 × 512 pixels, and reconstructed with a pixel size of 0.98 × 0.98 mm along the x, y axes. Comparatively, contrast-enhanced DxCT were acquired with slice thickness ranging from 1.25–3.75 mm, with X-ray tube current ranging from 160–436 mA at 120 kVp. DFOV ranged from 236–300 mm, and axial images were acquired using a matrix of 512 × 512 pixels, and reconstructed with a pixel size ranged from 0.46 × 0.46 mm to 0.59 × 0.59 along the x, y axes. 120 cc contrast material was injected at a rate of 3cc/s followed by scanning after 90-second delay. (Detailed acquisition parameters are illustrated in a supplementary table 1)
Manual segmentation of reference anatomic ROIs
For each patient, a series of 68 reference anatomic ROIs (18 bony, 3 cartilaginous, 7 glandular, 30 muscular, 6 soft tissue, 4 vascular) in addition to gross primary tumor volume (GTV-P) and gross nodal target (GTV-N) were manually contoured on every patients DxCT and SimCT by both a third-year resident physician observer (MR) and a medical student (CAB). Contours were subsequently approved on a daily basis by a radiation oncologist with 7 years’ experience (ASRM), and finally, ROIs were reviewed by an expert attending head and neck radiation oncologist with 8 years’ experience (CDF). Manual segmentation was performed using a commercial treatment planning software (Pinnacle 9.0, Phillips Medical Systems, Andover, MA). (Details of the segmented ROIs are available as a supplementary figure 1 and supplementary table 2)
Image registration
For each patient DICOM dataset, image registration was performed using baseline RIR allowing automatic scalable rigid registration of the DxCT to the SimCT using a block-matching in-house GPU-based algorithm. Subsequently, each dataset was registered using a series of open-access and commercial registration algorithms. Two commercial multi-step DIR software were examined: Atlas-based (CMS ABAS 0.64, Elekta, AV, Stockholm, SE, 2013) and B-spline (VelocityAI 2.8.1, Atlanta, GA, 2012). Atlas-based DIR, presented previously(20), consisted of five registration steps to deform the original DxCT to the SimCT: linear registration, head pose correction, poly-smooth non-linear registration, dense mutual-information deformable registration, and final refinement using deformable surface model (20); and while B-spline consisted of three registration steps: manually-adjusted rigid edit, auto-rigid registration, and auto-deformable registration (21).
Likewise, two non-commercial software, Demons (ITK Demons, Kitware, Inc., Clifton Park, NY, 2013)(22), and Optical Flow (23) algorithms were investigated. Noncommercial algorithms are often dependent on numerous human-entered variable parameters that determine the efficacy of an algorithm in registration. In order to provide valid comparison of commercial to non-commercial algorithms, an optimization step was performed utilizing approximately 100 human-verified identical landmark points on both DxCT and SimCT using the methodology that our authors have previously described (24). In this methodology, parameters were iteratively varied to create varying deformation fields mapping the DxCT to the SimCT. Subsequently, these deformation fields were applied to the landmark points on the DxCT to obtain deformed landmark points on the SimCT. Euclidean distances in corresponding landmark points between the deformed landmark points and the actual landmark points on the SimCT were calculated. Parameters that minimized the total Euclidean point distances obtained from applying these non-commercial algorithms to these landmark points were used in the ROI comparison described below.
Deformation vector fields were obtained from each image registration algorithm mapping the deformation of the DxCT onto the SimCT. For the commercial algorithms, the deformation transformed definitive voxels from the original DxCT onto the SimCT, while for the non-commercial algorithms; the deformation field was from the RigidCT. Subsequently, in a custom written Matlab (MATLAB R2012a, The MathWorks Inc., Natick, MA, 2012) program, these deformation fields were applied to ROIs segmented on the DxCT to convert them into ‘deformed ROIs’ on the SimCT. Calculations (described below) were performed comparing the ‘deformed ROIs’ to the human-segmented ROIs on the SimCT.
Registration Algorithm assessment
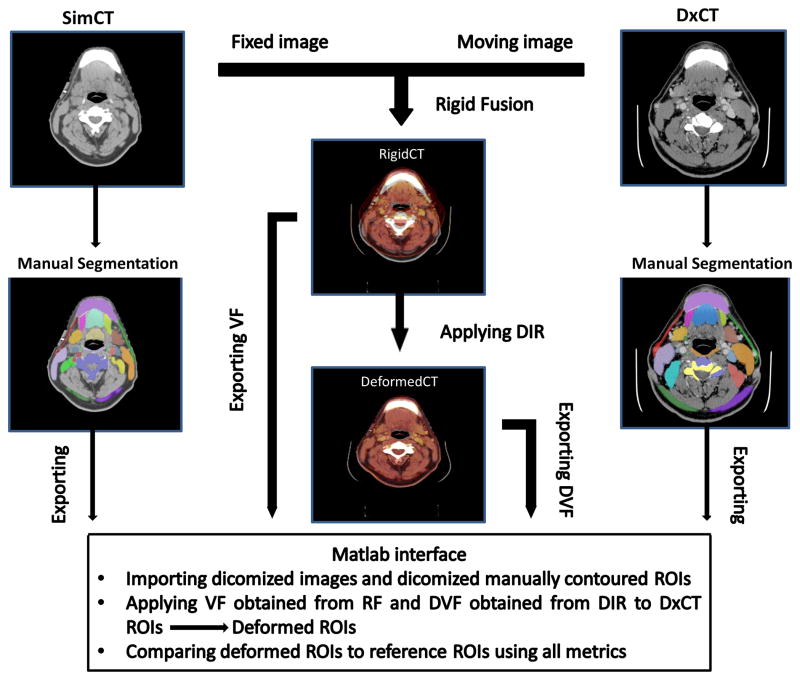
After completion of rigid and deformable registration of native DICOM image files, the resultant deformed ROIs were compared with manually contoured reference ROIs for each of the 68 anatomic structures listed plus tumor and nodal targets. Figure 1 shows a schematic illustration of the QA workflow process developed, based on a previously presented software resource (TaCTICS, Target Contour Testing/Instructional Computer Software, https://github.com/kalpathy/tacticsRT)(25–27).
Figure 1.
Schematic illustration of the quality assurance workflow process. Abbreviations: VF=vector fields; DVF=deformation vector fields; RF=rigid fusion; ROIs=regions of interest; SimCT=simulation CT; DxCT=diagnostic CT; DIR=deformable image registration.
For each registration-deformed ROI/reference ROI pair, the following ROI-based overlap metrics (28–30) was assessed:
volume overlap;
maximum and 95% maximum Hausdorff distance (95%HD);
false-positive, false-negative, and standard Dice similarity coefficients (DSC).
Individual metrics are detailed in Table 2. The different registration algorithms were then compared using different metrics for pooled ROIs overlap then compared after stratification by individual ROI, anatomical subgroup, and STR.
Table 2.
Description of employed metrics, where A= algorithm-dependent deformed ROI volume, R= reference anatomic manually segmented ROI volume.
| Metric | Symbolic Expression | Description | Full agreement | |
|---|---|---|---|---|
| Volume Overlap |
|
The portion of the reference ROI which is overlapped by the segmentation | 1 | |
|
| ||||
| Dice Similarity Coefficient |
|
The portion of overlap between the reference ROI and segmentation relative to the size of the reference ROI plus the size of the segmentation | 1 | |
|
| ||||
| False Negative Dice |
|
The volume that the segmentation missed of the reference ROI relative to the size of the reference ROI plus the size of the segmentation | 0 | |
|
| ||||
| False Positive Dice |
|
The volume of the segmentation not found within the reference ROI relative to the size of the reference ROI plus the size of the segmentation | 0 | |
|
| ||||
| 95% maximum Hausdorff Distance (HD) |
|
The maximum distance between a point in the segmentation and that of the reference ROI; for 95% maximum Hasudorrf, ±5%-ile outliers are discarded | ||
Interobserver variability
In order to interrogate the effect of interobserver variability in manual ROI delineation on the output metrics of different registration techniques post hoc, a subset of 12 ROIs (2 ROIs per each anatomic sub-group) were delineated by 3 expert observers in clinical target delineation (ASRM “7 years of experience”, SC “8 years of experience”, EKU “5 years of experience”) on paired SimCT-DxCT sets of two patients. The same workflow methodology described above was used to compare overlap and surface distance metrics obtained from expert segmentation to those of primary observer for each registration technique.
Statistical analysis
Statistical assessment was performed using JMP v 10.2 (SAS institute, Cary, NC). To assess algorithm performance for each metric listed, distributional statistics for listed metrics (Table 2) were tabulated for each anatomic ROI.
To determine the relative degree of potential difference of DIR compared to RIR, overlap metrics for anatomic and target ROIs mapped using DIR were compared to those mapped rigidly, with a p-value thresholding for multiple comparisons using Bonferroni correction through dividing the requisite α threshold of 0.05 by the number of subset comparisons, with resultant p-value specified for each comparison (vide infra). Distributional differences DIR algorithms (atlas-based, Demons, optical flow, and B-spline) compared to RIR alone were performed using the non-parametric Steel test with control (31) for between-algorithm difference, designating RIR as the standardized comparator (control). Between groups comparison was performed for all metrics for pooled ROIs, and by each ROI separately, and reported with p-value thresholding for multiple comparisons applied graphically. Additional post hoc comparison of metrics after stratification by STR and ROI anatomic subgroup (bones, cartilages, muscles, glands, vessels, soft tissues) was calculated, with comparison using Wilcoxon rank(32) (paired comparison) or Kruskal-Wallis(33) (three or more groups comparison) non-parametric tests when comparing across strata with p-value thresholding for multiple comparisons. For evaluation of interobserver dependency, Cronbach’s alpha method was performed to assess agreement between expert observers for all delineated ROIs for all tested registration methods for DSC metric.
Results
Algorithm comparative performance
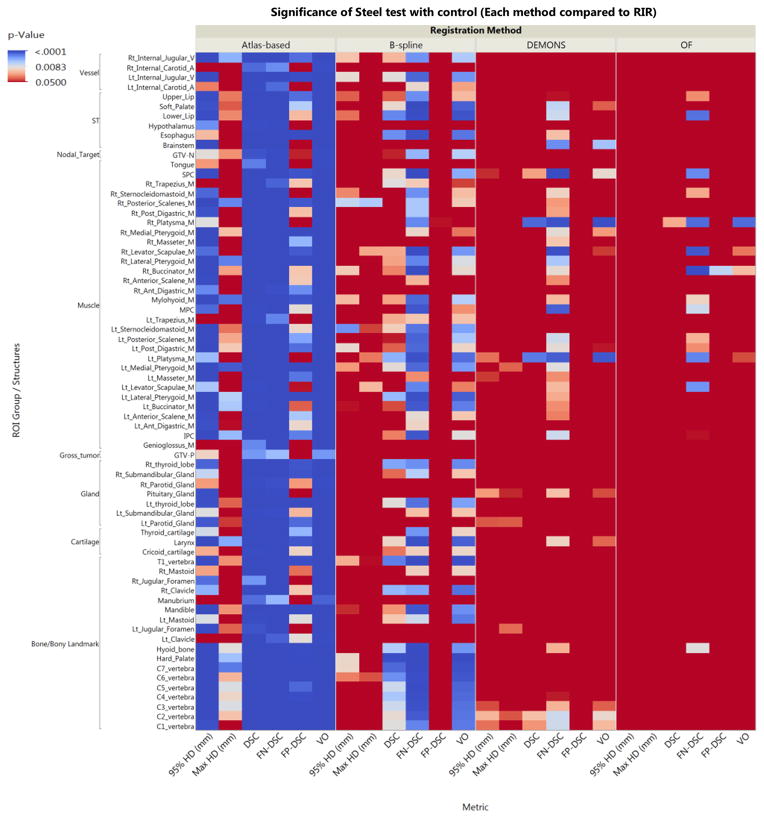
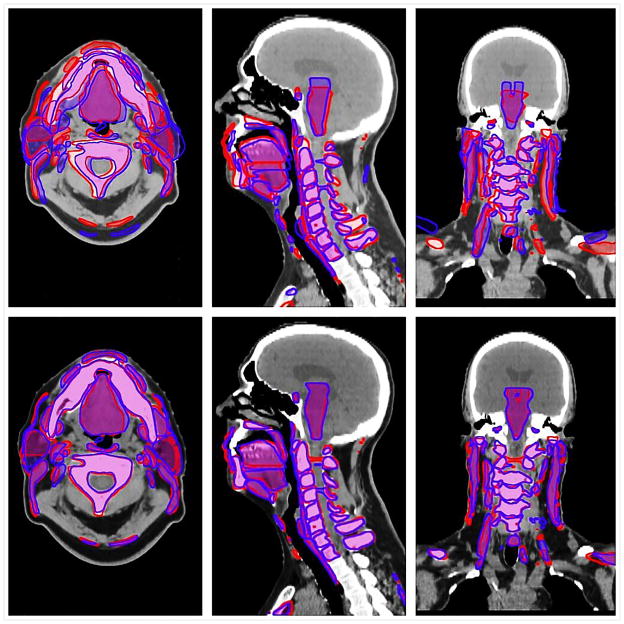
A total of 2720 anatomic ROIs were delineated (68 per each DICOM file) for all included patients’ paired SimCT-DxCT sets and a total number of 50 tumor/nodal ROIs were delineated in 15 of 20 patients paired sets (5 patients had no radiologic gross tumor or nodal targets after induction chemotherapy/surgery). Excepting optical flow, for surface distance metrics (i.e. 95% and maximum Hausdorff distance), and demons, for false negative dice, for all investigated DIR algorithms there was a detectable improvement in conformance with a manual ROI comparator over RIR for all pooled ROIs using all comparative metrics (Steel test, p < 0.008 after Bonferroni correction for multiple comparisons; p-value heat map is available as a supplementary figure 2). Estimation of differences between algorithms varied substantially with stratification by specific ROI; magnitude of effect size for the difference as compared to RIR control is illustrated as a p-value heat-map with color thresholding for multiple comparison correction (Figure 2). Figure 3 shows example of the visual comparison of the overlap between deformed ROIs and reference ROIs using the rigid versus the deformable registration.
Figure 2.
Heat map illustrates the relative performance of each of the DIR algorithms over the rigid registration for each anatomic structure using all comparison metrics. The more towards blue color means better performance. p-value thresholding for multiple comparisons was used with first threshold at p<0.008 for correction of multiple comparisons across 4 distinct registration algorithms (α = 0.05/6 “pairwise comparison of 4 DIR algorithms”), and p<0.0001 for multiple comparison across 68 distinct ROIs. Values shaded solid red are non-significantly different from rigid segmentation. Abbreviations: OF=optical flow, ST=soft tissues, DSC=dice similarity coefficient, FN-DSC=false negative dice, FP-DSC=false positive Dice, 95%HD=95% Hausdorff distance, GTV-P=gross primary tumor volume, GTV-N=gross nodal volume.
Figure 3.
Visual depiction of the deformed-reference regions of interest (ROIs) overlay: the top panel shows example of deformed ROIs in blue and reference ROIs in red using rigid registration, while the lower panel shows the same ROIs when registered using DIR.
For similarity coefficient metrics, the tested atlas based algorithm appears to have the best performance in this specific head and neck application (median DSC 0.68, median false negative Dice 0.06, and median false positive Dice 0.5). Likewise, for surface distance metrics, the atlas based algorithm had the least median distance error (4.6 mm 95%HD; 10.6 mm maximum Hausdorff distance). (Supplementary figures 3 and 4 further illustrate utilized overlap and surface distance metrics, respectively, for all ROIs for each registration method as a graphical table)
Algorithm performance by Anatomical subgroup
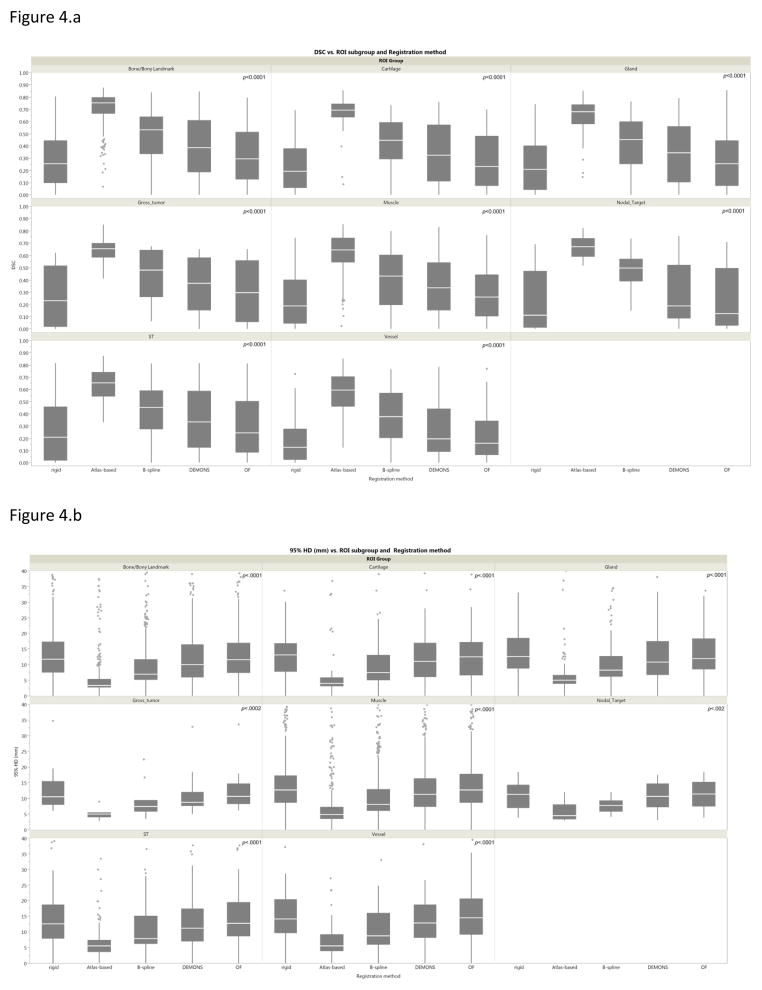
The performance of each registration method varied significantly across different ROI sub-groups (bones, cartilages, muscles, glands, vessels, soft tissues) as assessed for different metrics using Kruskal-Wallis test (p-value < 0.05) excepting, RIR (for volume overlap and 95%HD), B-spline (for volume overlap), and Demons (for 95%HD). To directly compare specific anatomical sub-group performance for each registration algorithm, further analysis using paired Wilcoxon test showed that for the vast majority of registration algorithms bony and cartilaginous ROIs were significantly more concordant than muscular and vascular ROIs (Bonferroni-corrected p-value < 0.003 for multiple comparisons across the 6 ROI subgroups). Likewise, for each anatomic category the ROIs conformance varied significantly across different registration methods for all assessed metrics (Kruskal-Wallis p-value < 0.05) with comparatively better performance of atlas based DIR followed by B-spline in all ROI sub-groups (Bonferroni-corrected p-value < 0.005 for multiple comparisons across the 5 registration methods), as illustrated visually in figure 4 for DSC and 95%HD metrics. The tumor and nodal ROIs conformance were best achieved with atlas based software with GTV-P median DSC and 95%HD of 0.65 and 4.6 mm, respectively, and GTV-N median DSC and 95% HD of 0.67 and 4.5 mm, respectively.
Figure 4.
a. Box-plot of Dice similarity coefficient analysis of each registration method by anatomic ROIs category. Pale line within the box indicates median value, while the box limits indicating the 25th and 75th percentiles. The lines represent the 10th and 90th percentiles, and the dots represent outliers.
b. Box-plot of 95% Haussdorff distance analysis of each registration method by anatomic ROIs category. Abbreviations: OF=optical flow, ST=soft tissues.
Algorithm performance by slice thickness reconstruction
STR of DxCT for all but two cases was uniform at 1.25 mm, while the SimCT STR was variable, with 10 cases reconstructed at a slice thickness of less than 3 mm (1–2.5mm) and 10 cases reconstructed at a slice thickness of 3 mm or more (3–3.75 mm). The effect of variability in SimCT STR on the performance of individual registration method was most evident for the tested B-spline algorithm as shown in table 3, a system dependency not typically considered in image-registration QA, but of practical importance in clinical image acquisition.
Table 3.
The effect of variability in SimCT slice thickness reconstruction on the performance of registration algorithms.
| Registration Method | Metric | STR < 3 mm† | STR ≥ 3 mm‡ | p-value | ||
|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | |||
| Rigid | 95%HD (mm) | 15.7 | 12.4 | 14.4 | 11.5 | 0.25 |
| DSC | 0.24 | 0.22 | 0.27 | 0.22 | 0.006* | |
| Atlas-based | 95%HD (mm) | 7.2 | 10.6 | 7.7 | 11.2 | <0.0001* § |
| DSC | 0.64 | 0.15 | 0.65 | 0.14 | 0.74 | |
| B-spline | 95%HD (mm) | 10.8 | 10.1 | 10.5 | 11.2 | 0.02* |
| DSC | 0.41 | 0.22 | 0.47 | 0.20 | <0.0001*§ | |
| Demons | 95%HD (mm) | 14.3 | 12.2 | 13 | 11.6 | 0.17 |
| DSC | 0.33 | 0.25 | 0.38 | 0.24 | 0.0019*§ | |
| Optical Flow | 95%HD (mm) | 16.1 | 13.2 | 14.3 | 11.1 | 0.14 |
| DSC | 0.28 | 0.22 | 0.31 | 0.22 | 0.0034*§ | |
n = 10 patients,
n = 10 patients,
significant p<0.05 for Wilcoxon rank test,
significant p<0.005 after Bonferroni correction (α = 0.05/10 “pairwise comparison of 5 DIR algorithms”).
Abbreviations: STR=slice thickness reconstruction, DSC=Dice similarity coefficient, 95%HD=95%Hausdorff distance.
Interobserver variability
In post hoc interobserver dependency assessment, for each registration technique there was no significant (all p > 0.05) difference between the mean values, for all metrics of all expert observers when compared to the primary observer except for the 95% HD for observer 3 in Atlas based and B-spline algorithms. Cronbach’s alpha assessment showed a minimum α of 0.6527 “acceptable interobserver agreement” for rigid registration, and maximum α of 0.88 for B-spline “Good interobserver agreement”. This confirms that the interobserver manual delineation variability was unlikely to substantively impact the outcomes of registration in this study (supplementary figure 5).
Discussion
Our results confirm that the tested DIR algorithms provided detectable performance advantages over RIR in our specific head and neck DxCT-simCT dataset, for the vast majority of ROI-based metrics. The performance of specific DIR algorithms varied across anatomical ROIs, with greater conformance of registering bony and cartilaginous ROIs to reference ROIs than for muscle and vascular ROIs. Furthermore, the difference between the registration accuracy of different structures of the anatomic class varied substantially. For example, certain bony structures (e.g. the clavicles) showed more distance error and reduced conformance as compared to other bony ROIs, likely due to the wide variation in shoulder position between both images. Another example is the relatively higher registration errors of tongue musculature ROIs and velar ROIs in the region adjacent to intraoral dental stents in the SimCT. Our QA method also revealed unanticipated algorithm dependencies, such STR, between SimCT and DxCT. STR notably impacted some algorithms disproportionately, most evident on B-spline fusions.
Several previous studies have validated the use of DIR algorithms in IGRT for head and neck cancer. Approaches used to quantify the performance of distinct DIR in these studies include landmark identification (12), ROI-based comparison (3, 10, 13, 34, 35), or computational phantom deformation (8, 9, 36, 37); each of these methods have their specific caveats and limitations of application. In the examined setting of DxCT-SimCT co-registration, the application of an evenly and densely distributed matrix of anatomic landmark points is intuitively understandable, and, with sufficient point placement, exceptionally spatially accurate and statistically robust as a validation method (24). Point placement is, nonetheless, comparatively resource intensive, requiring accurate manual identification of hundreds or thousands of points(24). Point placement in the head and neck is technically complicated secondary to substantial variation in patient position, image acquisition parameter differences (especially STR, which is a significant limit to voxel-wise point identification), and tubular internal anatomy of soft tissue ROIs in the neck, which enhances the difficulty of manually placing reproducible matched points in paired DxCT/SimCT image sets. For this effort, small-scale (~100) landmark point placement was used as an intermediary step in our quality assurance chain for optimization of non-commercial DIR settings; however, large-scale landmark point registration efforts, while underway, have yet to be completed by our group owing to listed limitations.
As a part of this effort, we sought to develop a “head and neck QA ROI library”, as a robust set of labeled anatomic ROIs, in order to properly represent the unique characteristics of the head and neck anatomy, which contains multiple structures of different shapes, sizes and Hounsfield unit (HU) intensity gradients. ROI-based assessments, as used in the present series (i.e. carefully and rigorously reviewed and curated independently by two radiation oncologists after initial manual segmentation), give insight into changes of shape, volume and location of a structure. Thus, ROI or ROI-category specific performance differences can be ascertained, a feature that may be overlooked by “anatomically agnostic” DIR QA approaches such as point registration or image intensity-matching. Likewise, the large number of ROIs/ROI-categories used in this QA workflow more nearly approximate the range of DIR algorithm performance across anatomic sub-sites in the head and neck, as opposed to “sample ROI” methods, or use of a limited number of reference ROIs, which may underrepresent inter-regional DIR variation.
Only few studies have validated the registration of DxCT-SimCT; two of which have addressed the registration of pre-therapy diagnostic PET/CT to SimCT (15, 16), while two other studies have examined the registration of SimCT to post-recurrence DxCT (17, 18). The validation methodologies for DIR assessment in those studies varied from calculating the root mean squared error from a set of observer’s marked anatomic landmarks (15) to overlap indices and center of mass comparison for sample ROIs (16–18). Ireland et al.(15) and Hwang et al.(16) showed that, matching with our results, DIR achieved superior performance to RIR. Additionally, Due et al.(18) showed that DIR has higher reproducibility than RIR in repeated registration of center of mass points used to identify the origin of locoregional recurrences mapped to original SimCT. These results should critically question the utility of RIR as an accurate tool for head and neck registration extracranially, and we advise cautious use of RIR as only “rough guide” rather than serially implemented clinical tool for DxCT-simCT head and neck workflows. However, though optical flow (12) and demons (3, 10, 36) algorithms have been studied and shown utility in adaptive radiotherapy applications, they failed to evince comparable results in the specific setting of head and neck DxCT-SimCT co-registration.
Several limitations must be noted. As a single institution study using retrospective image data, the standard caveats apply. Given this particular anatomic site (head and neck), and the fact that image sets (DxCT and simCT) were acquired using standard institutional operating procedures, excessive generalization regarding DIR algorithm performance in other anatomic sites, or differing acquisition settings is potentially specious. As the development of this QA process/ROI library was exceedingly resource-intensive, owing to the time required to manually segment and review a comparatively massive number of anatomic ROI structures, this library of paired DxCT-simCT images and ROI structure sets is provided as anonymized DICOM-RT files to any other researchers at “doi:10.6084/m9.figshare.999145–51”(38). Additionally, the effect of surgical resection or induction chemotherapy on the quality of registration was not investigated in the present study as we sought to benchmark best performance scenarios and exclude the effect of anatomical distortion caused by huge tumors.
In summary, we developed a QA framework using a robust, curated, manually-segmented anatomic ROI library to quantitatively assess different image registration strategies used for head and neck DxCT to SimCT co-registration. The presented QA framework proved that DIR algorithms for most of the tested metrics improved registration performance over RIR yet with notable variability between different algorithms, suggesting careful validation of DIR before clinical implementation (e.g. target delineation) is imperative.
Supplementary Material
Example of a 57 years old male patient with T3 N2b right tonsil tumor. The top panel of shows an example SimCT images for one patient, while the lower panel shows a DxCT images for the same patient with all the anatomic ROIs contoured. The ROIs included bony landmarks (all cervical vertebrae, T1 vertebra, right & left Jugular foramen, right & left mastoid process, hard Palate, mandible, hyoid bone, right & left clavicle), cartilage (thyroid cartilage, cricoid cartilage, larynx), glands (pituitary Gland, bilateral parotid Gland, bilateral submandibular Gland, right & left thyroid lobes) muscles (superior pharyngeal constrictor, middle pharyngeal constrictor, inferior pharyngeal constrictor, genioglossus, mylohyoid, tongue, right & left anterior digastric, right & left anterior scalene, right & left buccinator, right & left medial pterygoid, right & left lateral pterygoid, right & left levator scapulae, right & left Masseter, right & left platysma, right & left Post Digastric, right & left posterior scalene, right & left sternocleidomastoid, right & left trapezius), soft tissues (hypothalamus, brainstem, soft Palate, upper lip, lower lip, esophagus), vessels (right & left internal carotid artery, right & left internal jugular vein).
Heat map showing the relative performance of each of the DIR algorithms over the rigid registration for all pooled ROIs using all comparison metrics. p-value thresholding for multiple comparisons was used with an initial multiple comparison-corrected threshold of p<0.008 (α = 0.05/6 “pairwise comparison of 4 DIR algorithms”) and second threshold of p<0.0013 including multi-metric comparison
Graphical table shows the performance of each of the registration methods for all anatomic ROIs as measured by the volume overlap metrics. Nota bene: Color intensity scales are reversed, such that for Dice, red indicates improved performance, while for false negative and false positive Dice blue shading indicates improved overlap characteristics. Abbreviations: OF=optical flow, ST=soft tissues, VO=volume overlap, FnDICE=false negative dice, FpDICE=false positive dice.
Graphical table shows the performance of each of the registration methods for all anatomic ROIs as measured by surface distance metrics.
Example of the interobsever variability in dice similarity coefficient metric for different registration methods.
Table 1S. Details of the acquisition parameters of the SimCT and DxCT
Table 2S. Details of the anatomic ROIs contoured
Implications for Patient Care.
1- After proper quality assurance, Deformable image registration should replace rigid registration for radiotherapy applications involving head and neck Diagnostic to Therapy-Simulation Computed Tomography registration.
2- Validation of any image registration strategy is crucial before implementation for specific clinical scenarios.
Summary statement.
The presented quality assurance framework proved that deformable image registration (DIR) algorithms for most of the tested metrics improved registration performance over rigid registration yet with notable variability between different algorithms, suggesting careful validation of DIR before clinical implementation.
Acknowledgments
This work was supported in part by National Institutes of Health Cancer Center Support (Core) Grant CA016672 to The University of Texas MD Anderson Cancer Center. Dr. Fuller received/receives grant/funding support from: the SWOG/Hope Foundation Dr. Charles A. Coltman, Jr., Fellowship in Clinical Trials; the National Institutes of Health/National Cancer Institute Paul Calabresi Program in Clinical Oncology (K12 CA088084-14) and the Clinician Scientist Loan Repayment Program (L30 CA136381-02); Elekta AB MR-LinAc Development Grant; the Center for Radiation Oncology Research at MD Anderson Cancer Center; the MD Anderson Center for Advanced Biomedical Imaging/General Electric Health In-Kind funding; and the MD Anderson Institutional Research Grant Program. This work was supported in part by a UICC American Cancer Society Beginning Investigators Fellowship funded by the American Cancer Society (ACSBI) to Dr Mohamed. Dr. Kalpathy-Cramer receives support from the National Library of Medicine Pathway to Independence Ward (K99/R00 LM009889), and the National Cancer Institute Quantitative Imaging Network (U01 CA154601 and U24 CA180918). Dr. Castillo is supported by both a National Institutes of Health Clinician Scientist Loan Repayment Program Award and a Mentored Research Scientist Development Award (K01 CA181292). Dr. Guerrero is supported by the National Institutes of Health Office of the Director’s New Innovator Award (DP OD007044). Dr. Court receives support from the National Institutes of Health National Cancer Center Small Grants Program for Cancer Research (R0 CA178495). TaCTICS software was developed by Drs Kalpathy- Cramer and Fuller with previous support from the Society of Imaging Informatics in Medicine (SIIM) Product Development Grant. The listed funder(s) played no role in study design, in the collection, analysis and interpretation of data, in the writing of the manuscript, nor in the decision to submit the manuscript.
Footnotes
Conflict of interest disclosure:
The remaining authors have no conflicts of interest to declare.”
References
- 1.Cazoulat G, Lesaunier M, Simon A, et al. From image-guided radiotherapy to dose-guided radiotherapy. Cancer radiotherapie: journal de la Societe francaise de radiotherapie oncologique. 2011;15(8):691–698. doi: 10.1016/j.canrad.2011.05.011. [DOI] [PubMed] [Google Scholar]
- 2.Gregoire V, Jeraj R, Lee JA, O’Sullivan B. Radiotherapy for head and neck tumours in 2012 and beyond: conformal, tailored, and adaptive? The lancet oncology. 2012;13(7):e292–300. doi: 10.1016/S1470-2045(12)70237-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hardcastle N, Tome WA, Cannon DM, et al. A multi-institution evaluation of deformable image registration algorithms for automatic organ delineation in adaptive head and neck radiotherapy. Radiation oncology. 2012;7:90. doi: 10.1186/1748-717X-7-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ezhil M, Choi B, Starkschall G, Bucci MK, Vedam S, Balter P. Comparison of rigid and adaptive methods of propagating gross tumor volume through respiratory phases of four-dimensional computed tomography image data set. International journal of radiation oncology, biology, physics. 2008;71(1):290–296. doi: 10.1016/j.ijrobp.2008.01.025. [DOI] [PubMed] [Google Scholar]
- 5.Brock KK. Results of a multi-institution deformable registration accuracy study (MIDRAS) International journal of radiation oncology, biology, physics. 2010;76(2):583–596. doi: 10.1016/j.ijrobp.2009.06.031. [DOI] [PubMed] [Google Scholar]
- 6.Cui Y, Galvin JM, Straube WL, et al. Multi-system verification of registrations for image-guided radiotherapy in clinical trials. International journal of radiation oncology, biology, physics. 2011;81(1):305–312. doi: 10.1016/j.ijrobp.2010.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirby N, Chuang C, Ueda U, Pouliot J. The need for application-based adaptation of deformable image registration. Medical physics. 2013;40(1):011702. doi: 10.1118/1.4769114. [DOI] [PubMed] [Google Scholar]
- 8.Nie K, Chuang C, Kirby N, Braunstein S, Pouliot J. Site-specific deformable imaging registration algorithm selection using patient-based simulated deformations. Medical physics. 2013;40(4):041911. doi: 10.1118/1.4793723. [DOI] [PubMed] [Google Scholar]
- 9.Pukala J, Meeks SL, Staton RJ, Bova FJ, Mañon RR, Langen KM. A virtual phantom library for the quantification of deformable image registration uncertainties in patients with cancers of the head and neck. Medical Physics. 2013;40(11) doi: 10.1118/1.4823467. [DOI] [PubMed] [Google Scholar]
- 10.Castadot P, Lee JA, Parraga A, Geets X, Macq B, Gregoire V. Comparison of 12 deformable registration strategies in adaptive radiation therapy for the treatment of head and neck tumors. Radiotherapy and oncology: journal of the European Society for Therapeutic Radiology and Oncology. 2008;89(1):1–12. doi: 10.1016/j.radonc.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 11.Lee C, Langen KM, Lu W, et al. Evaluation of geometric changes of parotid glands during head and neck cancer radiotherapy using daily MVCT and automatic deformable registration. Radiotherapy and Oncology. 2008;89(1):81–88. doi: 10.1016/j.radonc.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 12.Ostergaard Noe K, De Senneville BD, Elstrom UV, Tanderup K, Sorensen TS. Acceleration and validation of optical flow based deformable registration for image-guided radiotherapy. Acta Oncol. 2008;47(7):1286–1293. doi: 10.1080/02841860802258760. [DOI] [PubMed] [Google Scholar]
- 13.Huger S, Graff P, Harter V, et al. Evaluation of the Block Matching deformable registration algorithm in the field of head-and-neck adaptive radiotherapy. Physica medica: PM: an international journal devoted to the applications of physics to medicine and biology: official journal of the Italian Association of Biomedical Physics. 2013 doi: 10.1016/j.ejmp.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 14.Aird EG, Conway J. CT simulation for radiotherapy treatment planning. The British journal of radiology. 2002;75(900):937–949. doi: 10.1259/bjr.75.900.750937. [DOI] [PubMed] [Google Scholar]
- 15.Ireland RH, Dyker KE, Barber DC, et al. Nonrigid image registration for head and neck cancer radiotherapy treatment planning with PET/CT. International journal of radiation oncology, biology, physics. 2007;68(3):952–957. doi: 10.1016/j.ijrobp.2007.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hwang AB, Bacharach SL, Yom SS, et al. Can Positron Emission Tomography (PET) or PET/Computed Tomography (CT) Acquired in a Nontreatment Position Be Accurately Registered to a Head-and-Neck Radiotherapy Planning CT? International Journal of Radiation Oncology*Biology*Physics. 2009;73(2):578–584. doi: 10.1016/j.ijrobp.2008.09.041. [DOI] [PubMed] [Google Scholar]
- 17.Shakam A, Scrimger R, Liu D, et al. Dose-volume analysis of locoregional recurrences in head and neck IMRT, as determined by deformable registration: a prospective multi-institutional trial. Radiotherapy and oncology: journal of the European Society for Therapeutic Radiology and Oncology. 2011;99(2):101–107. doi: 10.1016/j.radonc.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 18.Due AK, Vogelius IR, Aznar MC, et al. Methods for estimating the site of origin of locoregional recurrence in head and neck squamous cell carcinoma. Strahlentherapie und Onkologie: Organ der Deutschen Rontgengesellschaft [et al] 2012;188(8):671–676. doi: 10.1007/s00066-012-0127-y. [DOI] [PubMed] [Google Scholar]
- 19.Kaanders JH, Fleming TJ, Ang KK, Maor MH, Peters LJ. Devices valuable in head and neck radiotherapy. International journal of radiation oncology, biology, physics. 1992;23(3):639–645. doi: 10.1016/0360-3016(92)90023-b. [DOI] [PubMed] [Google Scholar]
- 20.Han X, Hoogeman MS, Levendag PC, et al. Atlas-based auto-segmentation of head and neck CT images. Medical image computing and computer-assisted intervention: MICCAI International Conference on Medical Image Computing and Computer-Assisted Intervention. 2008;11(Pt 2):434–441. doi: 10.1007/978-3-540-85990-1_52. [DOI] [PubMed] [Google Scholar]
- 21.Theodorsson-Norheim E. Kruskal-Wallis test: BASIC computer program to perform nonparametric one-way analysis of variance and multiple comparisons on ranks of several independent samples. Computer methods and programs in biomedicine. 1986;23(1):57–62. doi: 10.1016/0169-2607(86)90081-7. [DOI] [PubMed] [Google Scholar]
- 22.Yoo TS, Ackerman MJ, Lorensen WE, et al. Engineering and algorithm design for an image processing Api: a technical report on ITK--the Insight Toolkit. Studies in health technology and informatics. 2002;85:586–592. [PubMed] [Google Scholar]
- 23.Castillo E, Castillo R, Zhang Y, Guerrero T. Compressible Image Registration for Thoracic Computed Tomography Images. J Med Biol Eng. 2009;29(5):222–233. [Google Scholar]
- 24.Castillo R, Castillo E, Guerra R, et al. A framework for evaluation of deformable image registration spatial accuracy using large landmark point sets. Physics in medicine and biology. 2009;54(7):1849–1870. doi: 10.1088/0031-9155/54/7/001. [DOI] [PubMed] [Google Scholar]
- 25.Kalpathy-Cramer J, Fuller CD. Target Contour Testing/Instructional Computer Software (TaCTICS): A Novel Training and Evaluation Platform for Radiotherapy Target Delineation. AMIA Annual Symposium proceedings/AMIA Symposium AMIA Symposium. 2010;2010:361–365. [PMC free article] [PubMed] [Google Scholar]
- 26.Kalpathy-Cramer J, Awan M, Bedrick S, Rasch CR, Rosenthal DI, Fuller CD. Development of a Software for Quantitative Evaluation Radiotherapy Target and Organ-at-Risk Segmentation Comparison. Journal of digital imaging. 2013 doi: 10.1007/s10278-013-9633-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Awan M, Kalpathy-Cramer J, Gunn GB, et al. Prospective Assessment of an Atlas-Based Intervention Combined With Real-Time Software Feedback in Contouring Lymph Node Levels and Organs-at-Risk in the Head and Neck: Quantitative Assessment of Conformance to Expert Delineation. Practical radiation oncology. 2013;3(3):186193. doi: 10.1016/j.prro.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dice LR. Measures of the Amount of Ecologic Association between Species. Ecology. 1945;26(3):297–302. [Google Scholar]
- 29.Huttenlocher DP, Klanderman GA, Rucklidge WJ. Comparing Images Using the Hausdorff Distance. Ieee T Pattern Anal. 1993;15(9):850–863. [Google Scholar]
- 30.Crum WR, Camara O, Hill DL. Generalized overlap measures for evaluation and validation in medical image analysis. IEEE transactions on medical imaging. 2006;25(11):1451–1461. doi: 10.1109/TMI.2006.880587. [DOI] [PubMed] [Google Scholar]
- 31.Steel RGD. A Multiple Comparison Rank Sum Test: Treatments versus Control. Biometrics. 1959;15(4):560–572. [Google Scholar]
- 32.Wilcoxon F. Individual comparisons of grouped data by ranking methods. Journal of economic entomology. 1946;39:269. doi: 10.1093/jee/39.2.269. [DOI] [PubMed] [Google Scholar]
- 33.Kruskal WH, Wallis WA. Use of Ranks in One-Criterion Variance Analysis. J Am Stat Assoc. 1952;47(260):583–621. [Google Scholar]
- 34.Al-Mayah A, Moseley J, Hunter S, et al. Biomechanical-based image registration for head and neck radiation treatment. Physics in medicine and biology. 2010;55(21):6491–6500. doi: 10.1088/0031-9155/55/21/010. [DOI] [PubMed] [Google Scholar]
- 35.Tsuji SY, Hwang A, Weinberg V, Yom SS, Quivey JM, Xia P. Dosimetric evaluation of automatic segmentation for adaptive IMRT for head-and-neck cancer. International journal of radiation oncology, biology, physics. 2010;77(3):707–714. doi: 10.1016/j.ijrobp.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 36.Wang H, Dong L, O’Daniel J, et al. Validation of an accelerated ‘demons’ algorithm for deformable image registration in radiation therapy. Physics in medicine and biology. 2005;50(12):2887–2905. doi: 10.1088/0031-9155/50/12/011. [DOI] [PubMed] [Google Scholar]
- 37.Varadhan R, Karangelis G, Krishnan K, Hui S. A framework for deformable image registration validation in radiotherapy clinical applications. Journal of applied clinical medical physics/American College of Medical Physics. 2013;14(1):4066. doi: 10.1120/jacmp.v14i1.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mohamed ASR. DIR QA Liberary. 2014 Available from: http://figshare.com/authors/Abdallah_Mohamed/551961.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Example of a 57 years old male patient with T3 N2b right tonsil tumor. The top panel of shows an example SimCT images for one patient, while the lower panel shows a DxCT images for the same patient with all the anatomic ROIs contoured. The ROIs included bony landmarks (all cervical vertebrae, T1 vertebra, right & left Jugular foramen, right & left mastoid process, hard Palate, mandible, hyoid bone, right & left clavicle), cartilage (thyroid cartilage, cricoid cartilage, larynx), glands (pituitary Gland, bilateral parotid Gland, bilateral submandibular Gland, right & left thyroid lobes) muscles (superior pharyngeal constrictor, middle pharyngeal constrictor, inferior pharyngeal constrictor, genioglossus, mylohyoid, tongue, right & left anterior digastric, right & left anterior scalene, right & left buccinator, right & left medial pterygoid, right & left lateral pterygoid, right & left levator scapulae, right & left Masseter, right & left platysma, right & left Post Digastric, right & left posterior scalene, right & left sternocleidomastoid, right & left trapezius), soft tissues (hypothalamus, brainstem, soft Palate, upper lip, lower lip, esophagus), vessels (right & left internal carotid artery, right & left internal jugular vein).
Heat map showing the relative performance of each of the DIR algorithms over the rigid registration for all pooled ROIs using all comparison metrics. p-value thresholding for multiple comparisons was used with an initial multiple comparison-corrected threshold of p<0.008 (α = 0.05/6 “pairwise comparison of 4 DIR algorithms”) and second threshold of p<0.0013 including multi-metric comparison
Graphical table shows the performance of each of the registration methods for all anatomic ROIs as measured by the volume overlap metrics. Nota bene: Color intensity scales are reversed, such that for Dice, red indicates improved performance, while for false negative and false positive Dice blue shading indicates improved overlap characteristics. Abbreviations: OF=optical flow, ST=soft tissues, VO=volume overlap, FnDICE=false negative dice, FpDICE=false positive dice.
Graphical table shows the performance of each of the registration methods for all anatomic ROIs as measured by surface distance metrics.
Example of the interobsever variability in dice similarity coefficient metric for different registration methods.
Table 1S. Details of the acquisition parameters of the SimCT and DxCT
Table 2S. Details of the anatomic ROIs contoured