Abstract
Uracil-DNA glycosylases (UDGs) catalyse the removal of uracil by flipping it out of the double helix into their binding pockets, where the glycosidic bond is hydrolysed by a water molecule activated by a polar amino acid. Interestingly, the four known UDG families differ in their active site make-up. The activating residues in UNG and SMUG enzymes are aspartates, thermostable UDGs resemble UNG-type enzymes, but carry glutamate rather than aspartate residues in their active sites, and the less active MUG/TDG enzymes contain an active site asparagine. We now describe the first member of a fifth UDG family, Pa-UDGb from the hyperthermophilic crenarchaeon Pyrobaculum aerophilum, the active site of which lacks the polar residue that was hitherto thought to be essential for catalysis. Moreover, Pa-UDGb is the first member of the UDG family that efficiently catalyses the removal of an aberrant purine, hypoxanthine, from DNA. We postulate that this enzyme has evolved to counteract the mutagenic threat of cytosine and adenine deamination, which becomes particularly acute in organisms living at elevated temperatures.
Keywords: archaea/deamination/DNA repair/thermophiles/uracil DNA-glycosylase
Introduction
The genomes of all living organisms are constantly exposed to exogenous and endogenous DNA-damaging agents. Paradoxically, and contrary to popular belief, the greatest amount of damage is inflicted by the endogenous agents water and oxygen, which modify primarily the aromatic DNA bases. While reactive oxygen species such as hydroxyl radicals convert guanine to 8-oxoguanine and thymine to thymine glycol, water brings about the deamination of all bases carrying exocyclic amino groups. The bases that are most affected in this respect are cytosine and 5-methylcytosine, which are deaminated to uracil and thymine, respectively. It is estimated that up to 500 uracil residues are generated in the human genome each day through cytosine deamination (Lindahl, 1993; Shen et al., 1994). Adenine deaminates to hypoxanthine (Hx) ∼10-fold less frequently. Because deamination of these bases alters their base pairing properties, these reactions represent a considerable mutagenic threat. When cytosine is converted to uracil in double-stranded DNA, a U·G mispair arises. Should this pre-mutagenic lesion remain uncorrected, 50% of the progeny DNA will acquire a C→T transition mutation during the first round of replication. Similarly, adenine deamination will give rise to an Hx·T mispair, which could result in an A→G transition if unrepaired.
All organisms studied to date carry enzymes that have evolved to deal with the mutagenic threat of hydrolytic deamination of DNA bases. These enzymes, known as DNA glycosylases, recognize unnatural, damaged or mispaired bases and remove them from DNA by catalysing the cleavage of the glycosidic bond that links the base to the sugar–phosphate backbone. Interestingly, hypoxanthine residues are addressed by glycosylases that are believed to have evolved to process the methylated purines 3-methyladenine and 7-methylguanine, but which recognize and remove deaminated adenines (Hx) from DNA thanks to their relaxed substrate recognition properties (Saparbaev et al., 2000). In contrast, the removal of uracil residues is accomplished by a battery of enzymes, uracil-DNA glycosylases (UDGs), which have been classified into four distinct families (Aravind and Koonin, 2000; Pearl, 2000).
UDGs encoded by the UNG genes have been studied most extensively. These are extremely efficient enzymes, which recognize uracil in single-stranded DNA, in A·U pairs that arise when dUMP is incorporated opposite A during DNA replication, or in G·U mispairs arising through cytosine deamination. Several structures of UNG enzymes co-crystallized with different substrates have been described in the past few years (for reviews see Parikh et al., 2000a; Pearl, 2000; Scharer and Jiricny, 2001). These showed that the enzyme flips the uracil base out of the DNA helix into its active site, which is very tight and which allows the entry of uracil and 5-fluorouracil, but not of thymine or other modified pyrimidines. In order to prevent the neighbouring bases collapsing onto each other due to the loss of their stacking interactions with the uracil, the enzyme inserts a highly conserved leucine residue into the site vacated by the target base. The active site of UNG-type UDGs is composed primarily of two short sequence motifs (referred to as motifs A and B in Figure 1). The first, with a consensus sequence GQDPY, contains an aspartate residue that is thought to be responsible for the activation of the catalytic water molecule. The second motif, HPSPLSA, interacts with the minor groove once the base is flipped out into the active site and stabilizes the protein–DNA complex. Moreover, the histidine stabilizes the developing negative charge on the uracil as the glycosidic bond is being pulled apart within the enzyme’s active site.
Fig. 1. Identification of Pa-UDGb and its orthologues. (A) Complete amino acid sequence alignment of P.aerophilum uracil-DNA glycosylase b (Pa-UDGb) with homologues from Sulfolobus solfataricus (EMBL: AE006867), Thermoplasma volcanium (EMBL: AP000994), Streptomyces coelicolor (Swiss-Prot: Q9S2L3) and Mycobacterium tuberculosis (Swiss-Prot: Q11059). Identical residues are shaded and the two putative active site motifs, corresponding to motifs A and B in (B), are underlined. The conserved phenylalanine that interacts in the binding pocket of the enzyme with the flipped-out base through π–π interactions is indicated by an asterisk. The sequence alignment shown was performed using the MultAlin software (Corpet, 1988) available at www.toulouse.infra.fr. (B) Partial amino acid sequence alignment of the active site motifs A and B of representatives of the five classes of uracil-DNA glycosylases: uracil-DNA glycosylase from E.coli (udg_ecoli, EMBL: J03725), human TDG (tdg_human, EMBL: U51166), SMUG1 from Homo sapiens (smug_human, EMBL: AF125182), P.aerophilum UDGa (Sartori et al., 2001) and UDGb (this work). Highly conserved residues are shown in black boxes, residues implicated in activating the catalytic water molecule are in open boxes and the hydrophobic residues preceding motif A are shown in grey boxes. Note that motif A of the putative active site of Pa-UDGb lacks a polar amino acid residue capable of activating a water molecule towards a nucleophilic attack on the C1′ of the sugar. The two mutated sites (A68D and H196N) are indicated by arrows.
The second family, the MUG/TDG homologues, are not as efficient as UNG-type enzymes. This is not surprising, as their active site motifs are ‘detuned’ such that the optimal general base–general acid pair (D in motif A and H in motif B) are substituted for asparagines in Mug and for asparagine and methionine in TDG, respectively (Aravind and Koonin, 2000; Pearl, 2000). SMUGs constitute a hybrid between UNGs and TDG/MUGs, inasmuch as motif A carries an asparagine, but motif B, HPSPRNP, is very UNG-like. The B motif of the fourth family, which is constituted from thermostable enzymes, is again UNG-like, but motif A has lost the aspartate of UNG-type enzymes. However, the function of this residue was most probably taken over by the glutamate in the sequence GEAPG.
Despite their differences, alignment of the amino acid sequences of UDGs from all four families allows for their ready identification in databank searches (Aravind and Koonin, 2000; Pearl, 2000) and also implies a certain similarity in their mode of action. This is how we were able to identify Pa-UDG, the major uracil-processing activity in the hyperthermophilic archaeon Pyrobaculum aerophilum (Sartori et al., 2001). However, during the examination of extracts of this bacterium, we noticed that they contained more than one uracil-processing enzyme. Because the genomic DNA of P.aerophilum has now been sequenced completely (Fitz-Gibbon et al., 2002), we initiated a similarity search for a second UDG candidate among its 2587 open reading frames (ORFs). Unexpectedly, we obtained no hits when using the standard search algorithms. However, due to the conservation of motifs A and B, we decided to search the ORFs for short sequence elements that might resemble them. In this way, we were able to identify a region annotated as PAE1327, which contained the sequence HPSPLNV that resembled motif B of UNGs. Surprisingly, the likely motif A in this ORF, GLAPA, contained no polar amino acid residue. We decided to express this ORF in Escherichia coli and test its activity on uracil-containing substrates. We now report that this enzyme, annotated Pa-UDGb, is the founding member of a fifth uracil DNA glycosylase family, which has at least six members (Figure 1A). In addition to lacking what has hitherto been thought to be an essential polar residue in motif A of its active site, the P.aerophilum enzyme has unusually broad substrate specificity.
Results
Identification of a novel ORF in P.aerophilum encoding a putative UDG
Crude extracts of the hyperthermophilic archaeon P.aerophilum were shown to possess at least three distinct uracil-processing activities (Sartori et al., 2001), but analysis of its genomic DNA revealed the presence of only two ORFs that unambiguously encode uracil-processing enzymes: Pa-UDG, a member of the thermostable UDG family (Sartori et al., 2001), and Pa-MIG (Yang et al., 2000), an enzyme belonging to the EndoIII family of DNA glycosylases that removes uracil and thymine from mispairs with guanine (Horst and Fritz, 1996). As conventional sequence searches of the P.aerophilum genome failed to identify the third likely UDG candidate, we decided to search the known ORFs for short sequence motifs that are characteristic of UDGs.
Enzymes in this category possess two highly conserved amino acid sequence motifs that constitute the active site (Figure 1B). Motif A [also referred to as motif-I (Aravind and Koonin, 2000) or motif 1 (Pearl, 2000)] is generally thought to contain the polar residue that is responsible for activating a water molecule towards the nucleophilic attack at the C1′ of the sugar residue carrying the aberrant base. In the UNG-type enzymes, it is an aspartate (D) in the motif GQDPY; in the MUG and SMUG enzymes, this role has been assigned to the asparagine (N) within the motif GINPG and GMNPG, respectively, and in the thermostable UDGs to the glutamate (E) in the sequence GEAPG (Pearl, 2000). Motif B [also referred to as motif-III (Aravind and Koonin, 2000) or motif 2 (Pearl, 2000] has the consensus sequence HPSPLSA in UNG-type polypeptides and HPSPRNP in SMUGS, but is less conserved in the other enzymes. The histidine (H) within this motif is thought to make a hydrogen bond with the O2 of the uracil, while the adjacent amino acids become inserted into the duplex in place of the flipped-out base. In the case of UNG-type enzymes (Parikh et al., 2000a; Pearl, 2000), these residues take the place of the extruded base, whereas in the case of the E.coli MUG enzyme they can form specific hydrogen bond contacts with the widowed guanine in the opposite strand (Barrett et al., 1998). Although search of the P.aerophilum genome failed to identify variants of motif A, we found one ORF (Figure 1A) containing a putative motif B (HPSPLNV). Closer examination of the sequence of this ORF revealed the presence of a hydrophobic stretch of amino acid residues VMVVGLAPA (Figure 1A), which shared some similarity with the upstream region of motif A of most UDGs (Aravind and Koonin, 2000; Pearl, 2000). Moreover, a phenylalanine (F) residue was located 15 amino acids downstream from the glycine (G) of this putative motif A (Figure 1A, residue marked with an asterisk). In all UDGs characterized to date, this aromatic residue (in some enzymes the phenylalnine is substituted by tyrosine) lines the bottom of the binding pocket and helps to stabilize the flipped-out uracil by π–π interactions (Parikh et al., 2000a; Pearl, 2000). The above evidence convinced us that this ORF could encode a UDG, which we tentatively termed Pa-UDGb, in order to distinguish it form the previously described Pa-UDG (Sartori et al., 2001). We shall refer to the latter enzyme as Pa-UDGa. We amplified the Pa-UDGb sequence from the genomic plasmid PAE1327 and cloned it downstream from a His6 tag of the bacterial expression vector pET28c(+).
Homology searches with the full-length Pa-UDGb sequence revealed the existence of several putative members of this new family from Thermoplasma volcanium, Mycobacterium tuberculosis, Streptomyces coelicolor and Sulfolobus solfataricus (Figure 1A; see also Aravind and Koonin, 2000; Pearl, 2000). Thermus thermophilus also appears to encode such a polypeptide (V.Starkuviene and H.-J.Fritz, personal communication).
Expression of Pa-UDGb in E.coli
The expression construct for Pa-UDGb, pET28c(+)paudgb, was electroporated into the E.coli BL21(DE3) strain and expression of the encoded polypeptide was induced with isopropyl-β-d-thiogalactopyranoside (IPTG). The cleared lysate was adsorbed on an Ni-NTA column and the retained proteins were eluted with 250 mM imidazole. The eluted fraction (IV), which contained an abundant polypeptide with a mol. wt of ∼28 kDa (Figure 2A), catalysed the efficient removal of uracil from a 60mer oligonucleotide duplex containing a single G·U mispair (Figure 2B). In order to exclude the possibility that this activity was due to an E.coli contaminant that may have co-purified with the P.aerophilum enzyme, we transformed the host BL21(DE3) cells with the empty pET28c(+) vector and subjected their extracts to a purification protocol identical to that used for the extracts of bacteria transformed with pET28c(+)paudgb. No uracil-processing activity was detected in fraction IV from this preparation (Figure 2B). The 28 kDa protein was therefore further purified using a Mono-S FPLC ion-exchange column. It was judged to be >95% homogeneous by SDS–PAGE (Figure 2A).
Fig. 2. (A) Expression and purification of the recombinant His-tagged Pa-UDGb (see Materials and methods). I, total extract of the E.coli strain BL21(DE3)pET28c(+)-paudgb; II, total extract of the same cells, following induction with IPTG; III, cleared lysate of the same cells; IV, proteins eluted from the Ni-NTA column with 250 mM imidazole; V, Pa-UDGb eluted from a Mono-S column; M, molecular size marker. The panel shows a 12% Coomassie blue-stained denaturing polyacrylamide gel. (B) Processing of G·U mispairs by fraction IV obtained from E.coli BL21 cells transfected with the pET28c(+)-paudgb plasmid (lane 2) or with the empty pET28c(+) vector (lane 3). This experiment shows that no E.coli uracil-processing activity is present in this fraction. The 60mer oligonucleotide substrate G·U was incubated for 1 h at 37°C with 6 µl of fraction IV as described in Materials and methods. (C) Pa-UDGb is a heat-stable, monofunctional uracil-DNA glycosylase. The enzyme alone removes uracil at both indicated temperatures, but does not cleave the sugar–phosphate backbone of the mispaired DNA substrate, as witnessed by the absence of the 23mer product band in the reaction where the G·U substrate was treated with Pa-UDGb alone (lane 1). Cleavage occured only upon the addition of hot alkali (lanes 2 and 6) or of human HAP1 (lane 3). The faint product band in lane 5 is due to heat-induced spontaneous β-elimination at the labile AP sites. Incubation at 70°C significantly increased the activity of Pa-UDGb (lane 6), whereas the E.coli UDG was completely inactivated at this temperature (lane 7, cf. lane 4).
As would be expected of an enzyme encoded by a hyperthermophile with an optimal growth temperature of 100°C, Pa-UDGb is thermostable, being substantially more active at 70 than at 37°C (Figure 2C). Like Pa-UDGa, Pa-UDGb is also a monofunctional DNA glycosylase, which excises uracil without the concomitant cleavage of the sugar–phosphate backbone of the DNA: incubation of the 60mer G·U substrate with the enzyme alone produced no (Figure 2C, lane 1, 37°C) or only a small amount (lane 5, 70°C) of the cleaved 23mer product. Efficient DNA cleavage could be observed only following the subsequent treatment of the DNA with NaOH (lanes 2 and 6) or with human AP endonuclease (HAP1) (lane 3), both of which cleave DNA at abasic sites. The fact that no uracil processing was observed when the G·U substrate was incubated with the E.coli UDG at 70°C (Figure 2C, compare lanes 4 and 7) provides further evidence that the latter enzyme was not a contaminant responsible for the observed UDG activity. Interestingly, like Pa-UDGa (Sartori et al., 2001), Pa-UDGb was not inhibited by the Ugi peptide (Wang and Mosbaugh, 1989), a generic inhibitor of the UNG-type enzymes (data not shown). This implies that the Ugi-sensitive uracil-processing activity detected in crude extracts of P.aerophilum (Sartori et al., 2001) was neither Pa-UDGa nor Pa-UDGb, and thus that this organism may possess yet another uracil-processing enzyme.
Pa-UDGa and Pa-UDGb have different mismatch processing and DNA-binding properties
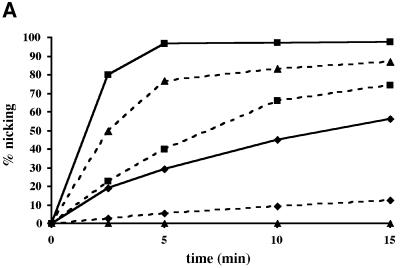
In an attempt to compare the biochemical properties of Pa-UDGa and Pa-UDGb, we studied their abilities to process different substrates. As shown in Figure 3A, Pa-UDGb catalysed the removal of uracil from all three oligonucleotide substrates tested in this experiment (dashed lines), albeit with distinctly different efficiencies. Interestingly, its preferred substrate was hydroxymethyluracil mispaired with guanine (G·hmU), followed by G·U and A·U (see also Figure 4). This contrasts with Pa-UDGa, which processed these substrates in the order of preference G·U>A·U, but possessed no detectable activity on hmU (Figure 3A, solid lines). In addition, like Pa-UDGa (Sartori et al., 2001), Pa-UDGb was able to process uracil in single-stranded DNA (ssU, see Figure 4). Although the processing of the G·U substrate by Pa-UDGb was not as efficient as that catalysed by Pa-UDGa, both enzymes displayed turnover kinetics; under optimal conditions; Pa-UDGb could process 10 mol equivalents of the G·U substrate in <30 min.
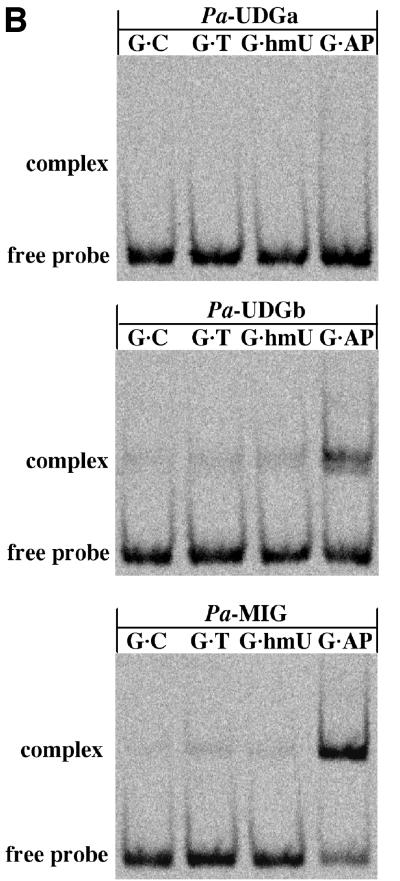
Fig. 3. Comparison of processing efficiencies and binding of different substrates by Pa-UDGa and Pa-UDGb. (A) Processing of 20 pmol of the fluorescently labelled 60mer substrates G·U (squares), A·U (diamonds) and G·hmU (triangles) with 2 pmol of Pa-UDGa (solid lines) or Pa-UDGb (dashed lines). At the indicated time points, aliquots of the reaction mixture were removed and immediately quenched with 100 mM NaOH (10 min at 90°C) to inactivate the enzyme and to cleave the resulting AP sites. The substrate and product were separated on 20% denaturing polyacrylamide gels and the band intensity was quantified using a Storm 860 PhosphorImager with ImageQuant software. The values shown represent the average of at least three independent experiments. (B) Comparison of DNA-binding specificities of Pa-UDGa, Pa-UDGb and Pa-MIG. The enzymes were incubated with the fluorescently labelled 60mer substrates under conditions (15 min at 4°C) where the base removal does not take place (data not shown). A stable protein–DNA complex was formed only between Pa-UDGb and Pa-MIG and a duplex substrate containing an AP site opposite a guanine (lane G·AP). Data were obtained from a Storm 860 PhosphorImager scan of a 6% native polyacrylamide gel.
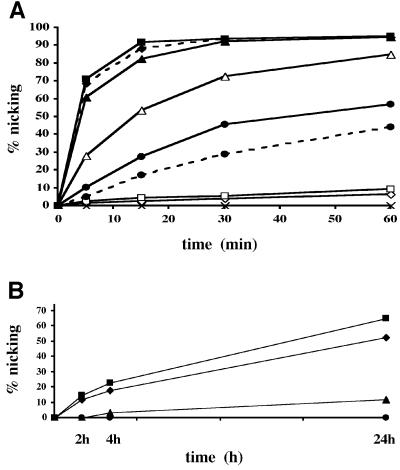
Fig. 4. Processing efficiency of various substrates by Pa-UDGb. (A) Processing of 20 pmol of the labelled substrates G·hmU (solid line, filled squares), G·εC (dashed line, filled diamonds), G·U (solid line, filled triangles), A·hmU (solid line, open triangles), A·U (solid line, filled circles), G·FU (dashed line, filled circles), T·Hx (solid line, open squares), ssU (solid line, open diamonds) and G·T (solid line, crosses) with 4 pmol of Pa-UDGb. (εC, ethenocytosine; FU, 5-fluorouracil; Hx, hypoxanthine). At the indicated time points, aliquots of the reaction mixture were removed and quenched immediately with 100 mM NaOH (10 min at 90°C) to inactivate the enzyme and to cleave the resulting AP sites. In the case of the base-labile ethenocytosine substrate, the AP sites were processed with 50 nM HAP1 (10 min at 37°C) in the presence of 2.5 mM MgCl2. The AP sites produced spontaneously under the reaction conditions in the case of the labile G·εC substrate were subtracted. The values shown represent the average of at least three independent experiments. (B) The labelled substrates (20 pmol) T·Hx (squares), C·Hx (triangles), T·G (circles) and ssU (diamonds), were incubated with 4 pmol of Pa-UDGb for 2, 4 and 24 h. The substrate and product were separated on 20% denaturing polyacrylamide gels and the band intensity was quantified using a Storm 860 PhosphorImager with ImageQuant software. The values shown represent the average of at least three independent experiments. (C) Processing of hypoxanthine-containing substrates by six different uracil-processing enzymes, Pa-UDGa (Sartori et al., 2001), Pa-UDGb, Pa-MIG (Yang et al., 2000), Ec-UDG, Hs-TDG (Neddermann and Jiricny, 1994) and Hs-MBD4 (Hendrich et al., 1999). For the G·U substrate, a 1:1 molar ratio of enzyme versus substrate was used, whereas for the T·Hx we used a 10-fold excess of enzyme over substrate. The incubations with the hyperthermophilic enzymes from P.aerophilum were carried out for 1 h at 70°C, and those with the mesophilic enzymes from E.coli and H.sapiens for 1 h at 37°C. The panel shows a 20% denaturing polyacrylamide gel scanned with a Storm 860 PhosphorImager.
The ability of Pa-UDGb to turn over sets it apart from TDG (Hardeland et al., 2000), MUG (Sung and Mosbaugh, 2000), MBD4 (Hendrich et al., 1999) and Pa-MIG (Yang et al., 2000), which fail to turn over on the G·U substrate in vitro. Similarly, under multiple-turnover conditions (excess substrate), SMUG1 exhibits only a slow turnover on double-stranded DNA (Nilsen et al., 2001). This latter phenomenon was ascribed to end product inhibition by abasic sites, which was in turn linked with the high affinity of the latter two enzymes for AP sites arising in the oligonucleotide duplexes after the removal of the bases (Hardeland et al., 2000). In agreement with this hypothesis, these enzymes can be stimulated potently by AP endonucleases, indicating that the higher affinity of an AP endonuclease for its substrate aids the displacement of the DNA glycosylase from DNA and thus facilitates its recycling. This would predict that enzymes with a relatively high turnover number should have only low affinity for AP sites in DNA. We decided to test this prediction in a series of electrophoretic mobility shift assays (EMSAs). In agreement with the prediction, Pa-UDGa was unable to bind detectably to any of the substrates tested (Figure 3B, top panel). The binding of Pa-UDGb to the G·C, G·T and G·hmU oligonucleotide probes was also weak, although small amounts of protein–DNA complexes were formed. Unexpectedly, Pa-UDGb interacted quite strongly with the G·AP DNA duplex that contains an AP site (Figure 3B, centre panel), albeit not as strongly as Pa-MIG that does not turn over on this substrate (Figure 3B, bottom panel). This result implies that turnover kinetics in this family of enzymes are controlled by factors other than simple binding to the product of the reaction. However, it should be remembered that processing of G·U and G·T substrates in mammalian cells in vivo is rapid (Brown and Jiricny, 1987; Brown and Brown-Luedi, 1989) and thus that glycosylases that fail to turn over in vitro may be induced to do so by specific interactions with other members of the base excision repair (BER) pathway (Waters et al., 1999). Indeed, as in the case of human UDG (Parikh et al., 1998), we found that a 25-fold molar excess of HAP1 in the presence of EDTA increased the uracil excision efficiency of Pa-UDGb (data not shown).
Pa-UDGb has broad substrate specificity
We tested the ability of Pa-UDGb to process a variety of different DNA substrates. As shown in Figure 4, the enzyme displayed a clear preference for double-stranded DNA substrates, especially for those containing mispairs (Figure 4A). Thus, while uracil in single-stranded DNA (ssU) was processed only sluggishly, uracil and hydroxymethyluracil opposite G (G·U and G·hmU, respectively) were processed substantially more efficiently than uracil and hydroxymethyluracil opposite adenine (A·U and A·hmU, respectively). 5-fluorouracil opposite G (G·FU) was processed with an efficiency similar to A·U, but the enzyme displayed no activity on the G·T substrate, which is processed by the mismatch-specific enzymes MIG (Horst and Fritz, 1996; Yang et al., 2000), TDG (Neddermann et al., 1996) and MBD4 (Hendrich et al., 1999). However, like TDG and Mug (Saparbaev and Laval, 1998; Lutsenko and Bhagwat, 1999), Pa-UDGb also processed ethenocytosine in a base pair with G (G·εC), albeit only with an efficiency similar to G·U.
The experiments shown in Figure 4A demonstrated that Pa-UDGb has an unusually broad substrate specificity. In order to study this phenomenon further, we decided to test whether the enzyme is also able to process purine-containing substrates. To our surprise, Pa-UDGb could excise hypoxanthine from DNA, especially from a mispair with thymine (Figure 4B, T·Hx). In order to ensure that hypoxanthine processing by UDGs was limited to Pa-UDGb, we decided to test all the representatives of the UDG family available to us. As shown in Figure 4C, equimolar amounts of all six enzymes, Pa-UDGa, Pa-UDGb and Pa-MIG from P.aerophilum, E.coli UDG and the human TDG and MBD4, processed the G·U substrate, albeit with differing efficiencies. In contrast, only Pa-UDGb and, to a very small extent, TDG, catalysed appreciable hypoxanthine removal under our assay conditions. These results place Pa-UDGb in a category of its own, as the first member of the UDG family capable of recognizing both aberrant pyrimidines and purines.
Mutagenesis of motifs A and B strongly attenuates the enzymatic activity of Pa-UDGb
As discussed above, the active sites of all members of the UDG superfamily consist of two highly conserved motifs (Figure 1B). With the exception of members of the new, fifth UDG family (Figure 1A), motif A carries a polar amino acid residue that has hitherto been thought to be essential for enzymatic activity (Parikh et al., 2000a; Pearl, 2000) by activating a water molecule towards a nucleophilic attack on the C1′ of the sugar residue carrying the uracil. Motif A of Pa-UDGb carries no polar amino acid and we wondered whether the introduction of such a residue into this motif would alter the activity of the enzyme. We chose to substitute Ala68 for aspartate, as the latter residue is found in this position in all members of family I UDGs (Figure 1B), represented by Udg from E.coli. In motif B, the residue linked to catalysis is the histidine in position 196, and we decided to change this residue for asparagine, which is found at this site in the Mug enzyme of E.coli. Both the mutant proteins could be expressed in E.coli with efficiencies comparable with the wild-type enzyme and also behaved similarly during purification. Additionally, AP site binding by both mutants was not impaired (data not shown). It therefore seems highly unlikely that the presence of the mutations altered their three-dimensional structures to any significant extent.
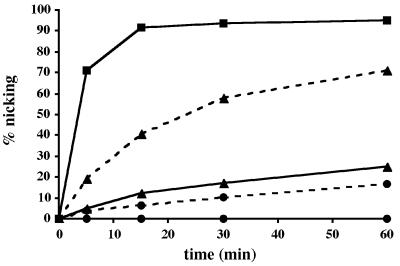
When the mutant enzymes were tested in the enzymatic assay on the best Pa-UDGb substrate, G·hmU, their processing efficiency was shown to be substantially reduced (Figure 5). The A68D mutation reduced the enzymatic activity of Pa-UDGb almost 10-fold, while substrate processing by the Pa-UDGb H196N mutant became detectable only when the enzyme:substrate ratio was raised to 1:1 (Figure 5, dashed lines).
Fig. 5. Processing of 20 pmol of the labelled G·hmU substrate with 4 pmol (solid lines, 1:5) of wild-type Pa-UDGb (squares), Pa-UDGb A68D (triangles) and Pa-UDGb H196N (circles). Processing of 10 pmol of G·hmU substrate with 10 pmol (dashed lines, 1:1) of Pa-UDGb A68D (triangles) and Pa-UDGb H196N (circles). At the indicated time points, aliquots of the reaction mixture were removed and immediately quenched with 100 mM NaOH (10 min at 90°C) to inactivate the enzyme and to cleave the resulting AP sites. The substrate and product were separated on 20% denaturing polyacrylamide gels and the band intensity was quantified using a Storm 860 PhosphorImager with ImageQuant software. The values shown represent the average of at least three independent experiments.
Discussion
Deamination of cytosines and 5-methylcytosines can lead to C→T transition mutations, and thus represents a major threat to genomic integrity, particularly at elevated temperatures. The hyperthermophilic crenarchaeon P.aerophilum appears to be particularly well equipped to counteract this threat, as it has at least three uracil-processing enzymes: Pa-UDGa (Sartori et al., 2001), Pa-UDGb (this work) and Pa-MIG (Yang et al., 2000). Given that these three enzymes have different biochemical properties, it is tempting to speculate that they are not simply redundant, but that they fulfil specific roles as antimutators.
Pa-UDGa is most probably the major uracil-processing activity of P.aerophilum. We predict that it has a role both as an antimutator in the removal of uracil residues arising through spontaneous hydrolytic deamination of cytosine, and as a general DNA surveillance enzyme that removes uracils incorporated into the newly synthesised strand in the form of dUMP during DNA replication. The latter prediction is based on our recent characterization (Yang et al., 2002) of an interaction of Pa-UDGa with the P.aerophilum orthologue of proliferating cell nuclear antigen (PCNA), which acts as a processivity factor for replicative DNA polymerases (Hubscher et al., 2000). This role has also been demonstrated for the human UDG (Otterlei et al., 1999; Krokan et al., 2001).
The second enzyme, Pa-MIG (Yang et al., 2000), removes uracil and thymine from mispairs with guanine. This enzyme is not very abundant, at least as judged by its activity in total P.aerophilum extracts (Sartori et al., 2001), and it is likely that it assumes only a minor role in uracil processing. However, should the genomic DNA of P.aerophilum contain 5-methylcytosine residues, then Pa-MIG might play an important antimutator role in the protection from the mutagenic effects of 5-methylcytosine deamination, which gives rise to G·T mispairs (Scharer and Jiricny, 2001). The presence of 5-methylcytosine in the P.aerophilum genome has so far not been documented, but our preliminary evidence indicates that the DNA of this organism may be methylated, as judged by its sensitivity to digestion with methylation-sensitive restriction enzymes. Moreover, the genome contains an ORF that is predicted to encode a DNA (cytosine-5) methyltransferase (data not shown).
What might then be the role of Pa-UDGb? This enzyme is less active than Pa-UDGa, but the difference is not so large that it could not be acting as an efficient back-up enzyme for Pa-UDGa. However, it is tempting to speculate that the enzyme has a more important role in the detoxification of P.aerophilum genomic DNA from minor products of base oxidation and hydrolysis. Its high processing efficiency of the G·hmU substrate suggests that one of the physiological roles of Pa-UDGb may lie in the removal of this base, which can arise though the oxidation of 5-methylcytosines that is followed by deamination, or through the direct oxidation of thymines.
Unexpectedly, our study revealed that Pa-UDGb might also function in reducing the number of A→G transition mutations in P.aerophilum, through removing hypoxanthine from mispairs with thymine. Hypoxanthine arises in DNA through the spontaneous hydrolytic deamination of adenine in A·T pairs and, although this reaction is an order of magnitude slower than cytosine deamination (Lindahl and Nyberg, 1974), it does represent a significant mutagenic threat. This aberrant base has so far been shown to be excised only by alkyladenine-DNA glycosylases of the AlkA and AAG type (Saparbaev et al., 2000), and it is interesting to note in this respect that P.aerophilum does not appear to encode a homologue of either enzyme (Fitz-Gibbon et al., 2002). It would therefore appear possible that Pa-UDGb is also responsible for the processing of T·Hx mispairs in vivo.
Given the broad substrate specificity of Pa-UDGb, it is remarkable that this enzyme does not remove from DNA the natural bases, thymine and guanine, such as was reported for enzymes of the AAG family, which also display broad substrate specificity. The latter proteins excise from DNA a wide range of purines, ranging from the positively charged 3-methyladenine and 7-methylguanine to the uncharged ethenoadenine and hypoxanthine. In the human AAG, guanine is thought to be excluded from the binding pocket by the steric interaction of its exocyclic amino group with an asparagine residue (N169) of the enzyme (Lau et al., 2000), and it is conceivable that Pa-UDGb distinguishes guanine from hypoxanthine by a similar mechanism. However, the exclusion of thymine on steric grounds is more difficult to understand. Unlike the binding pockets of UNG-type enzymes, which are very tight and appear actively to exclude thymine by blocking the space required to accommodate the 5-methyl group with a tyrosine residue (Pearl, 2000), Pa-UDGb must have a binding pocket large enough to accommodate hmU and εC, both of which have substituents on the pyrimidine ring that are larger than the methyl group of thymine. Could the exclusion of thymine involve electrostatic forces within the Pa-UDGb binding pocket? The flipped-out base is stabilized in the active site through π–π interactions with a phenylalanine (F) residue, and these might be expected to be stronger in the case of pyrimidines with a cloud of delocalized electrons that is slightly depleted by the electron-withdrawing effect of the substituent at the 5-position. However, although this argument might support the facile excision of εC, it does not explain the relatively sluggish removal of 5-fluorouracil (Figure 4A). The effect of electron-withdrawing substituents on the strength of the glycosidic bond is also insufficient to explain the substrate preference of Pa-UDGb; were this the case, 5-fluorouracil would be excised with high efficiency by this enzyme, as is the case with Pa-UDGa and other members of the UDG family, including the UNG-type enzymes. The elucidation of the substrate selection criteria of Pa-UDGb will have to await the results of structural studies.
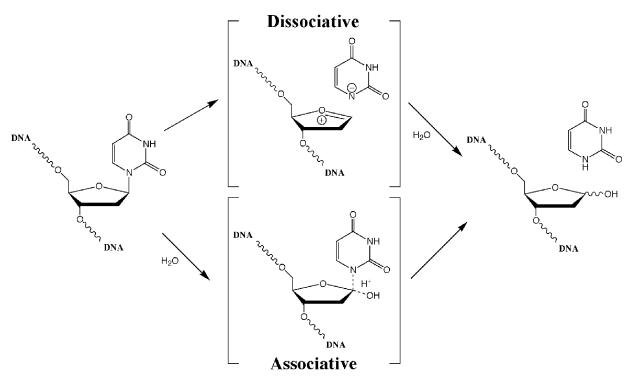
The enzyme possesses a second puzzling feature, namely its mechanism of catalysis. UDGs have been thought to act via the so-called ‘associative’ mechanism of glycosidic bond cleavage (Stivers and Drohat, 2001), which predicts that the activated water molecule attacks the C1′ of the sugar residue carrying the aberrant base, giving thus rise to a pentavalent transition state, the collapse of which results in the scission of the bond (Figure 6). This mechanism has been inferred from structural studies and from alignment of motifs A and B of the representative members of the various UDG families (Figure 1B), which shows that enzymes that excise uracil with high efficiency carry strong activating residues, typically an aspartate or a glutamate (Parikh et al., 2000a; Pearl, 2000), while those that are more sluggish, such as the MUG group proteins, carry weaker activating residues such as asparagine. However, extrapolating from this, proteins such as Pa-UDGb (Figure 4A), which carry no polar amino acid residue within motif A (Figure 1), should have no enzymatic activity. As this is clearly not the case, we must conclude either that the cleavage of the glycosidic bond might be catalysed with the help of a polar residue outside of motif A, or that the scission of the glycosidic bond can be achieved by an alternative, ‘dissociative’ mechanism (Figure 6), which does not require the help of a protein-activated water molecule. The latter possibility finds support in recent structural, kinetic and theoretical studies, which are discussed below.
Fig. 6. Putative mechanisms of ‘associative’ and ‘dissociative’ cleavage of glycosidic bonds.
The crystal structure of UDG with a flipped-out pseudouridine showed that the base is accommodated in the binding pocket of the enzyme with a geometry that forces upon the still-attached sugar residue a highly unfavourable conformation that substantially stretches and weakens the glycosidic bond (Parikh et al., 2000b). The authors suggested that the driving force of the hydrolytic reaction might be the energy gained upon relaxation of these constrictions through cleavage of the glycosidic bond. In kinetic isotope effect studies, the uracil anion could be clearly identified, but no evidence of a transition state involving a pentavalent carbon C1′ was found (Werner and Stivers, 2000). This implied that the scission of the glycosidic bond occurred directly, without the participation of a water molecule. Recent computational studies predicted this route to be favoured also by electrostatic interactions between the positive charge at the C1′ of the baseless sugar and the negatively-charged phosphate residues of the substrate DNA, buried in the vicinity of the enzyme’s active site (Dinner et al., 2001). Taken together, the above evidence points to a dissociative mechanism even in the UNG family of UDG enzymes, which carry polar amino acid residues in motif A. The considerable activity of Pa-UDGb provides more support for the dissociative mechanism of glycosidic bond cleavage. Indeed, our mutagenesis studies showed that the introduction of a polar residue into its motif A, which might have been expected to result in an increase of enzymatic activity, did not have this effect. In contrast, substitution of the histidine in motif B abolished enzymatic activity (Figure 5). Because this histidine is needed to stabilize the uracil anion in the transition state, this can be taken as further evidence in support of the dissociative pathway.
The characterisation of Pa-UDGb described in this work has opened a new chapter in our understanding of the mode of action of the UDG family of DNA glycosylases. Although it is likely that Pa-UDGb catalyses the cleavage of the glycosidic bond via the dissociative mechanism, there are still a number of questions that remain. One of these concerns the extraordinarily broad range of its substrates. We are currently trying to solve the three-dimensional structure of Pa-UDGb, in an attempt to elucidate this phenomenon.
Materials and methods
Reagents and oligonucleotides
All oligonucleotides were synthesized by Microsynth (Balgach, Switzerland), except the oligonucleotide containing 5-hydroxymethyluracil, which was obtained from Gemini Biotech Ltd. The substrate oligonucleotides were purified by PAGE. Restriction enzymes and the E.coli UDG were supplied by New England BioLabs (Beverly, MA). All other chemicals and reagents were purchased from Sigma, Roche Molecular Biochemicals, Amresco, Epicentre Technologies or Merck, and were of analytical grade purity.
The recombinant P.aerophilum mismatch-specific DNA glycosylase Pa-MIG was expressed and purified as previously described (Yang et al., 2000). The enzyme was stored at –80°C in a buffer containing 50 mM Tris–HCl pH 7.6, 1 mM EDTA, 1 mM dithiothreitol (DTT), 30 mM NaCl and 50% glycerol. Purified hsTDG and MBD4 were a kind gift of Ulrike Hardeland.
Bacterial strains
The E.coli strain DH5α was used in all cloning experiments and for plasmid amplifications, and the strain BL21 (DE3) (Sambrook et al., 1989) was used in all protein expressions.
Cloning of wild-type Pa-UDGb
The candidate protein-coding region, PAE1327, was identified by sequence analysis in the recently completed genomic sequence of P.aerophilum (Fitz-Gibbon et al., 2002). The DNA fragment encoding Pa-UDGb was amplified by PCR using P.aerophilum genomic DNA as template, and the primers (Ps) 5′-GGATCCATATGGATCTTGCTA GAGTTCACACACCCCG-3′ and (Pas) 5′-GTACGGATCCTCAT AGACAGCCGGCGTCGGC-3′ carrying NdeI and BamHI restriction sites, respectively, for subsequent cloning into the pET28c(+) vector (Novagen). The integrity of the insert was confirmed by DNA sequence analysis.
Site-directed mutagenesis
In vitro mutagenesis of P.aerophilum UDGb was performed using the QuikChange site-directed mutagenesis kit from Stratagene (San Diego, CA) according to the manufacturer’s instructions. pET28c(+)-paudgb served as template for mutagenesis, and the oligonucleotide primers used to generate the individual mutations were as follows (sense strand sequences shown only; mutation sites are underlined): Pa-UDGbA68D, 5′-GATGGTCGTGGGCCTGGATCCTGCCGCGC ACGGGG-3′; Pa-UDGbH196N, 5′-GGGTGTACGCCTCGTACAAC CCCAGTCCTCTCAACG-3′.
Expression and purification of the recombinant Pa-UDGb proteins
The plasmids expressing the His tag fusion proteins pET28c-paudgb, pET28c-paudgbA68D and pET28c-paudgbH196N were electroporated into competent E.coli BL21 (DE3) cells, and the transformed cells were used to inoculate LB medium containing 50 µg/ml kanamycin (LB-kan) supplemented with 2% d-glucose. The cells were allowed to grow overnight at 30°C. The saturated culture was diluted 1:100 in 1 l of LB-kan medium and grown with shaking at 30°C until the OD600 reached 1.6 (fraction I). The expression of UDGb protein was then induced with 0.2 mM IPTG. After 18 h incubation at 30°C, the cells were pelleted by centrifugation at 4°C (fraction II). The cell pellet was resuspended in 30 ml of ice-cold sonication buffer [50 mM sodium phosphate pH 8.0, 300 mM NaCl, 10% glycerol, 1 mM imidazole, 0.25% Tween-20, 10 mM β-mercaptoethanol and 1 mM phenylmethylsulfonyl fluoride (PMSF)] and the cells were lysed by sonication with 25× 5 s bursts on ice. The sonicate was clarified by centrifugation at 15 000 r.p.m. for 30 min at 4°C in a Sorvall SS34 rotor. The supernatant (fraction III) was incubated with gentle shaking for 1 h at 4°C with 2 ml of Ni-NTA–agarose (Qiagen), pre-equilibrated in sonication buffer. The suspension was then packed into a disposable column, and the unbound proteins were eluted with sonication buffer containing increasing concentrations of imidazole [2× 10 column volumes (cv) 5 mM imidazole, 4× 5 cv 20 mM imidazole]. The histidine-tagged Pa-UDGb protein was eluted with 4× 1 cv of sonication buffer containing 250 mM imidazole. The latter fractions were pooled (fraction IV) and dialysed overnight at 4°C against 2 l of binding buffer (50 mM sodium phosphate pH 8.0, 50 mM NaCl, 10% glycerol, 10 mM β-mercaptoethanol). Fraction IV was loaded onto a 1 ml Mono-S FPLC column (Pharmacia) and the column was washed with 10 ml of binding buffer. It was then eluted with a 30 ml linear gradient of 50 mM to 1 M NaCl at a flow rate of 0.5 ml/min. The nearly homogenous Pa-UDGb protein eluted as a major peak in fractions containing 0.35–0.45 M NaCl. These fractions were pooled (fraction V) and dialysed against storage buffer (50 mM sodium phosphate pH 8.0, 120 mM NaCl, 10% glycerol, 10 mM β-mercaptoethanol). Fraction V (2.0 ml, 200 µg/ml) containing >95% pure Pa-UDGb protein was stored in small aliquots at –80°C.
Enzymatic activity assays
The glycosylase activity of the purified enzymes was monitored using a standardized ‘nicking assay’ described previously (Sartori et al., 2001). The standard reactions were set up in 20 µl volumes containing 1× nicking buffer [50 mM Tris–HCl pH 8.0, 1 mM DTT, 1 mM EDTA, 80 mM NaCl, 0.1 mg/ml bovine serum albumin (BSA)], 1 pmol of labelled DNA and 1–5 pmol of the purified proteins. Incubation conditions varied as indicated in the text. In the time course experiments, different ratios of substrates versus enzymes were used in a total reaction volume of 50 µl in 1× nicking buffer and incubated at 70°C. At the desired time points, aliquots of the reaction mixture were removed and immediately quenched either by hot alkaline treatment (Sartori et al., 2001), or, as in the case of the labile ethenocytosine substrate, by treatment for 10 min at 37°C with 1 pmol of HAP1 in nicking buffer supplemented with 2.5 mM MgCl2.
EMSAs
In standard EMSA reactions, 5 pmol of Pa-UDGa, Pa-UDGb or Pa-MIG were incubated with 1 pmol of the labelled oligonucleotide substrates and 10 pmol of unlabelled homoduplex oligonucleotide in 50 mM Tris–HCl pH 8.0, 1 mM DTT, 1 mM EDTA and 5% glycerol at 4°C for 15 min. The protein–DNA complexes were separated by electrophoresis on 6% native polyacrylamide gels in 0.5× TBE at 4°C. The probe with an abasic site was generated by treatment of the oligonucleotide containing a G·U mismatch with E.coli UDG.
Acknowledgments
Acknowledgements
We are grateful to Primo Schär for many helpful discussions, to Ulrike Hardeland for the gift of the TDG and MBD4 enzymes, to Vytaute Starkuviene and Hans-Joachim Fritz for communicating their unpublished information, and to Orlando Schärer for critical reading of the manuscript. The generous support of UBS Stiftung to A.A.S. is also acknowledged.
References
- Aravind L. and Koonin,E.V. (2000) The α/β fold uracil DNA glycosylases: a common origin with diverse fates. Genome Biol., 1, RESEARCH0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T.E., Savva,R., Panayotou,G., Barlow,T., Brown,T., Jiricny,J. and Pearl,L.H. (1998) Crystal structure of a G:T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions. Cell, 92, 117–129. [DOI] [PubMed] [Google Scholar]
- Brown T.C. and Brown-Luedi,M.L. (1989) G/U lesions are efficiently corrected to G/C in SV40 DNA. Mutat. Res., 227, 233–236. [DOI] [PubMed] [Google Scholar]
- Brown T.C. and Jiricny,J. (1987) A specific mismatch repair event protects mammalian cells from loss of 5-methylcytosine. Cell, 50, 945–950. [DOI] [PubMed] [Google Scholar]
- Corpet F. (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res., 16, 10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinner A.R., Blackburn,G.M. and Karplus,M. (2001) Uracil-DNA glycosylase acts by substrate autocatalysis. Nature, 413, 752–755. [DOI] [PubMed] [Google Scholar]
- Fitz-Gibbon S.T., Ladner,H., Kim,U.J., Stetter,K.O., Simon,M.I. and Miller,J.H. (2002) Genome sequence of the hyperthermophilic crenarchaeon Pyrobaculum aerophilum. Proc. Natl Acad. Sci. USA, 99, 984–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardeland U., Bentele,M., Jiricny,J. and Schar,P. (2000) Separating substrate recognition from base hydrolysis in human thymine DNA glycosylase by mutational analysis. J. Biol. Chem., 275, 33449–33456. [DOI] [PubMed] [Google Scholar]
- Hendrich B., Hardeland,U., Ng,H.H., Jiricny,J. and Bird,A. (1999) The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites [published erratum appears in Nature (2000) 404, 525]. Nature, 401, 301–304. [DOI] [PubMed] [Google Scholar]
- Horst J.P. and Fritz,H.J. (1996) Counteracting the mutagenic effect of hydrolytic deamination of DNA 5-methylcytosine residues at high temperature: DNA mismatch N-glycosylase Mig.Mth of the thermophilic archaeon Methanobacterium thermoautotrophicum THF. EMBO J., 15, 5459–5469. [PMC free article] [PubMed] [Google Scholar]
- Hubscher U., Nasheuer,H.P. and Syvaoja,J.E. (2000) Eukaryotic DNA polymerases, a growing family. Trends Biochem. Sci., 25, 143–147. [DOI] [PubMed] [Google Scholar]
- Krokan H.E. et al. (2001) Properties and functions of human uracil-DNA glycosylase from the UNG gene. Prog. Nucleic Acid Res. Mol. Biol., 68, 365–386. [DOI] [PubMed] [Google Scholar]
- Lau A.Y., Wyatt,M.D., Glassner,B.J., Samson,L.D. and Ellenberger,T. (2000) Molecular basis for discriminating between normal and damaged bases by the human alkyladenine glycosylase, AAG. Proc. Natl Acad. Sci. USA, 97, 13573–13578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T. (1993) Instability and decay of the primary structure of DNA. Nature, 362, 709–715. [DOI] [PubMed] [Google Scholar]
- Lindahl T. and Nyberg,B. (1974) Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry, 13, 3405–3410. [DOI] [PubMed] [Google Scholar]
- Lutsenko E. and Bhagwat,A.S. (1999) The role of the Escherichia coli mug protein in the removal of uracil and 3,N(4)-ethenocytosine from DNA. J. Biol. Chem., 274, 31034–31038. [DOI] [PubMed] [Google Scholar]
- Neddermann P. and Jiricny,J. (1994) Efficient removal of uracil from G·U mispairs by the mismatch-specific thymine DNA glycosylase from HeLa cells. Proc. Natl Acad. Sci. USA, 91, 1642–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neddermann P., Gallinari,P., Lettieri,T., Schmid,D., Truong,O., Hsuan,J.J., Wiebauer,K. and Jiricny,J. (1996) Cloning and expression of human G/T mismatch-specific thymine-DNA glycosylase. J. Biol. Chem., 271, 12767–12774. [DOI] [PubMed] [Google Scholar]
- Nilsen H., Haushalter,K.A., Robins,P., Barnes,D.E., Verdine,G.L. and Lindahl,T. (2001) Excision of deaminated cytosine from the vertebrate genome: role of the SMUG1 uracil-DNA glycosylase. EMBO J., 20, 4278–4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otterlei M. et al. (1999) Post-replicative base excision repair in replication foci. EMBO J., 18, 3834–3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh S.S., Mol,C.D., Slupphaug,G., Bharati,S., Krokan,H.E. and Tainer,J.A. (1998) Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA. EMBO J., 17, 5214–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh S.S., Putnam,C.D. and Tainer,J.A. (2000a) Lessons learned from structural results on uracil-DNA glycosylase. Mutat. Res., 460, 183–199. [DOI] [PubMed] [Google Scholar]
- Parikh S.S., Walcher,G., Jones,G.D., Slupphaug,G., Krokan,H.E., Blackburn,G.M. and Tainer,J.A. (2000b) Uracil-DNA glycosylase–DNA substrate and product structures: conformational strain promotes catalytic efficiency by coupled stereoelectronic effects. Proc. Natl Acad. Sci. USA, 97, 5083–5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl L.H. (2000) Structure and function in the uracil-DNA glycosylase superfamily. Mutat. Res., 460, 165–181. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Saparbaev M. and Laval,J. (1998) 3,N4-ethenocytosine, a highly mutagenic adduct, is a primary substrate for Escherichia coli double-stranded uracil-DNA glycosylase and human mismatch-specific thymine-DNA glycosylase. Proc. Natl Acad. Sci. USA, 95, 8508–8513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saparbaev M., Mani,J.C. and Laval,J. (2000) Interactions of the human, rat, Saccharomyces cerevisiae and Escherichia coli 3-methyladenine-DNA glycosylases with DNA containing dIMP residues. Nucleic Acids Res., 28, 1332–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori A.A., Schar,P., Fitz-Gibbon,S., Miller,J.H. and Jiricny,J. (2001) Biochemical characterization of uracil processing activities in the hyperthermophilic archaeon Pyrobaculum aerophilum. J. Biol. Chem., 276, 29979–29986. [DOI] [PubMed] [Google Scholar]
- Scharer O.D. and Jiricny,J. (2001) Recent progress in the biology, chemistry and structural biology of DNA glycosylases. BioEssays, 23, 270–281. [DOI] [PubMed] [Google Scholar]
- Shen J.C., Rideout,W.M., 3rd and Jones,P.A. (1994) The rate of hydrolytic deamination of 5-methylcytosine in double-stranded DNA. Nucleic Acids Res., 22, 972–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stivers J.T. and Drohat,A.C. (2001) Uracil DNA glycosylase: insights from a master catalyst. Arch. Biochem. Biophys., 396, 1–9. [DOI] [PubMed] [Google Scholar]
- Sung J.S. and Mosbaugh,D.W. (2000) Escherichia coli double-strand uracil-DNA glycosylase: involvement in uracil-mediated DNA base excision repair and stimulation of activity by endonuclease IV. Biochemistry, 39, 10224–10235. [DOI] [PubMed] [Google Scholar]
- Wang Z. and Mosbaugh,D.W. (1989) Uracil-DNA glycosylase inhibitor gene of bacteriophage PBS2 encodes a binding protein specific for uracil-DNA glycosylase. J. Biol. Chem., 264, 1163–1171. [PubMed] [Google Scholar]
- Waters T.R., Gallinari,P., Jiricny,J. and Swann,P.F. (1999) Human thymine DNA glycosylase binds to apurinic sites in DNA but is displaced by human apurinic endonuclease 1. J. Biol. Chem., 274, 67–74. [DOI] [PubMed] [Google Scholar]
- Werner R.M. and Stivers,J.T. (2000) Kinetic isotope effect studies of the reaction catalyzed by uracil DNA glycosylase: evidence for an oxocarbenium ion–uracil anion intermediate. Biochemistry, 39, 14054–14064. [DOI] [PubMed] [Google Scholar]
- Yang H., Fitz-Gibbon,S., Marcotte,E.M., Tai,J.H., Hyman,E.C. and Miller,J.H. (2000) Characterization of a thermostable DNA glycosylase specific for U/G and T/G mismatches from the hyperthermophilic archaeon Pyrobaculum aerophilum. J. Bacteriol., 182, 1272–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Chiang,J.H., Fitz-Gibbon,S., Lebel,M., Sartori,A.A., Jiricny,J., Slupska,M.M. and Miller,J.H. (2002) Direct interaction between uracil-DNA glycosylase and a PCNA homolog in the crenarchaeon Pyrobaculum aerophilum. J. Biol. Chem., 277, in press. [DOI] [PubMed] [Google Scholar]