Abstract
The extremities of eukaryotic chromosomes are called telomeres. They have a structure unlike the bulk of the chromosome, which allows the cell DNA repair machinery to distinguish them from ‘broken’ DNA ends. But these specialised structures present a problem when it comes to replicating the DNA. Indeed, telomeric DNA progressively erodes with each round of cell division in cells that do not express telomerase, a specialised reverse transcriptase necessary to fully duplicate the telomeric DNA. Telomerase is expressed in tumour cells but not in most somatic cells and thus telomeres and telomerase may be proposed as attractive targets for the discovery of new anticancer agents.
INTRODUCTION
The telomere is a DNA–protein structure found at the ends of all eukaryotic linear chromosomes. It is involved in several essential functions: (i) telomere DNA-associated proteins help to prevent telomere DNA from being recognised as DNA breaks and allow cells to distinguish between a normal end and the result of a double-strand DNA break, i.e. telomeres ‘cap’ chromosomal ends; (ii) it provides a means of complete replication of the chromosome, since many mammalian cells that are without telomerase lose telomeric DNA at each division; (iii) it contributes to the spatial and functional organisation of chromosomes within the nucleus; (iv) it participates in transcriptome regulation.
The replication and capping functions of telomeres are essential to maintain the integrity of the genome and must be present in all eukaryotic organisms. These two points, in connection with the regulation and manipulation of telomerase in normal and cancerous human cells, will be discussed in detail in this paper. The two other characteristics may be considered as acquired functions, which may play a fundamental role in the physiology of some organisms, but could, at least in theory, be supported by other nuclear or chromosomal components.
Human telomeric DNA consists of a few kilobases of a short repetitive motif which is double-stranded, except for a 3′-terminal G-rich overhang (1–3) (Table 1). Telomere maintenance is necessary for long-term cell proliferation. In the absence of a specific replication machinery at the telomere ends it was predicted (4), and later demonstrated (5), that gradual sequence loss due to incomplete replication of the lagging strand would eventually lead to critically short telomeres and trimming of essential chromosomal sequences. The mechanism whereby cells count divisions uses the gradual erosion of telomeres, which ultimately triggers replicative senescence in many cell types. In order to compensate for this loss, different mechanisms for the addition of new telomere sequences have evolved. In humans, telomere maintenance is mainly performed by a specific reverse transcriptase, telomerase, which was initially identified in ciliates (6,7). Human telomerase is a ribonucleoprotein (8) composed of a catalytic subunit, hTERT (9–11), and a 451 nt long RNA (hTR; also known as hTER or hTERC) (12), which acts as a template for the addition of a short repetitive motif d(GGGTTA)n on the 3′-end of a primer.
Table 1. Telomeric repeats.
| Species | Motifa(5′→3′) | 3′-Overhangb | T-loopc | G4d |
|---|---|---|---|---|
| Homo sapiens | GGGTTA | Yes (1–3) | Yes (270) | ? |
| Saccharomyces cerevisiae | G1–3T | Yes (383) | ? | ? |
| Trypanosoma brucei | GGGTTA | Yes (384) | Yes (384) | ? |
| Oxytricha fallax/nova | GGGGTTTT | Yes (385,386) | Yes (387) | ? |
| Tetrahymena thermophila | GGGGTT | Yes (388) | ? | ? |
| Stylonychia lemnae/pustulata | GGGGTTTT | Yes (133,385) | ? | Yes (133) |
aSequence of the telomeric repeat motif.
bEvidence for the presence of a 3′ G-rich overhang (and relevant reference).
cEvidence for T-loop formation (and relevant reference).
dEvidence for telomeric quadruplex formation in the cell (and relevant reference).
Telomerase is active in the germline, as well as some stem cells, but is inactive in most somatic cells. It is assumed, but not firmly demonstrated, that the original length of the telomeres of these cells will be sufficient to act as a proper buffer against excessive loss during cell division: the initial telomere length is probably sufficient for a normal lifetime. Telomeres are indeed shorter in fibroblasts from an old donor compared to fibroblasts from a young donor (13). Interestingly, telomerase is reactivated in a large majority of cancer cells (for a review see 14). Furthermore, recent key experiments demonstrated that: (i) telomerase is sufficient for immortalisation of many cell types (15) and sufficient to allow transformed cells to escape from crisis (16), however, telomerase alone does not induce changes associated with a transformed phenotype (17,18); (ii) inhibition of telomerase limits the growth of human cancer cells (19); (iii) ectopic expression of the telomerase catalytic subunit (hTERT; also known as hEST2 or hTRT) in combination with several oncogenes (the simian virus 40 large T and small t oncoproteins and an oncogenic allele of H-ras) results in direct tumourigenic conversion of normal human epithelial and fibroblast cells (20,21). All these results point to a key role of telomerase in the tumourigenic process. In a recent review, unlimited proliferative potential, which depends on telomere maintenance, was defined as one of the six hallmarks of cancer (22). Mutations leading to reactivation or up-regulation of the enzyme may represent a required event in the multistep development of many cancers, such as colorectal carcinomas (23). In some cases (notably neuroblastomas, gastric and breast tumours), higher levels of telomerase activity are associated with poor prognosis, showing that telomerase could be used as a predictive marker. Results obtained with mice lacking a functional telomerase enzyme and in successive generations of mice doubly inactivated for telomerase and INK4a clearly show that telomere dysfunction impairs tumour development (24,25).
Understanding telomere/telomerase regulation is expected to give major insights into the tumourigenesis process and its manipulation becomes a challenge for the design of future anti-oncogenic approaches. In this review we will first present the mechanisms that regulate telomerase activity in human cells, before presenting the different strategies that have been proposed in order to inhibit telomerase in cancer cells.
TELOMERASE REGULATION
Telomerase activity is absent in many normal human somatic cells. Repression of telomerase activity during somatic development in other mammals, like rodents, is not as tightly regulated as in humans. Interestingly, animals that grow indeterminately, such as lobsters (26) and rainbow trout, appear to express telomerase ubiquitously.
Telomerase activity is growth regulated in certain human tissues and is the target of many cellular programmes. For instance, telomerase activity is enhanced in activated lymphocytes (27–29) and in endometrial tissue during the menstrual cycle (30). In contrast, terminal maturation or differentiation of cells has been correlated with repression of telomerase activity (31–34), but these two events might be uncoupled in some cases (35). A number of extra- or intracellular signals modulate telomerase, such as UV radiation (36), calcium (37), zinc (38), interferon α (39), oestrogen (40,41) and cytokine (42).
In vitro reconstitution of human telomerase is possible in cell extracts with two partners: the template RNA component hTR and the catalytic protein subunit hTERT (43,44). The following sections will discuss the regulation of these two essential components before presenting other factors that influence telomerase activity in living cells, with special emphasis on the human case (Fig. 1).
Figure 1.
Telomerase components. Telomerase is composed of two major components: the catalytic subunit and the template RNA (hTR). Several proteins are associated with hTERT or hTR and facilitate their folding or assembly. Many different proteins interact with telomeric DNA and participate in telomerase recruitment. Mutations in two telomerase component (hTR and dyskerin, in red) have been demonstrated to be involved into dyskeratosis congenita (DKC), a progressive bone-marrow failure syndrome (103,104).
hTERT
Normal human diploid cells transiently expressing hTERT acquire telomerase activity, demonstrating that hTERT is the limiting component necessary for restoration of telomerase activity in these cells (45,46). hTERT is a relatively large protein (127 kDa), with a net basic charge (pI 11.3) and reverse transcriptase motifs in its C-terminal part.
Gene amplification. The hTERT gene is present in the human genome as a single copy sequence on chromosome 5p15.33 (47). It encompasses >37 kb and consists of 16 exons (48,49). It is actually the most distal gene on chromosome 5p. One may speculate that this proximity to the telomere influences its transcription thanks to telomeric position effects, recently described for human telomeres (50; C.Koering, A.Pollice, M.P.Zibella, L.Sabatier, C.Brun, S.Bauwens, J.Pulitzer and E.Gilson, submitted for publication).
This localisation at the tip of 5p can also explain the amplification of hTERT observed in 31% of tumour cell lines and 30% of primary tumours (47,51). This suggests that increasing the copy number of hTERT may well be a way to up-regulate telomerase levels in tumour cells.
Transcription regulation. A large number of studies have been performed on regulation of the hTERT promoter. In agreement with a key role of telomerase in cell programming, this promoter is the target for a large number of signalling pathways and integrates multiple levels of gene regulation. However, the mechanisms involved in differential hTERT transcription in normal and tumour cells are still not understood. In normal cells hTERT expression appears to be repressed: the number of hTERT mRNA molecules per cell is below the sensitivity of quantitative RT–PCR (<0.004 molecules/cell), as compared to 0.2–6 molecules of spliced hTERT RNA molecules per telomerase-positive cell (52). Up-regulation could be a consequence of epigenetic changes in the hTERT gene.
A complex network of transcription factors. A variety of transcription factors have been shown to participate in hTERT gene expression (53). They include Myc, Sp1, the estrogen receptor (40), E2F-1 (54), WT-1 (55), NF-κB (56) and MZF-2 (57). Among them, Myc has been studied in the greatest detail.
Myc induces telomerase by increasing the expression level of hTERT (58). hTERT expression correlates with myc overexpression in human prostate cancer (59) and cervical cancer (60). Although c-Myc is a direct mediator of telomerase activation (61,62), a recent report indicates that c-Myc-mediated regulation of telomerase activity is disabled in immortalised cells (63). Myc-induced activation of the hTERT promoter requires an evolutionarily conserved E-box, which is a preferred target for Myc/Max heterodimers (50,64). Overexpression of c-Myc results in a significant increase in transcriptional activity of the core promoter (65). c-Myc genes are frequently deregulated in human tumours and myc overexpression may cause telomerase reactivation. However, TERT is unable to substitute for c-Myc in the transformation of primary rodent fibroblasts, suggesting that the transforming activities of c-Myc extend beyond its ability to activate TERT gene expression and hence telomerase activity (62). Both exogenous and endogenous ceramides modulate hTERT promoter activity via a rapid proteolysis of the ubiquitin-conjugated c-Myc transcription factor (66). It has also been reported that arsenic inhibits transcription of the hTERT gene and that this effect is mediated at least in part by decreased c-Myc and Sp1 transcription factor activities (67).
Myc and Mad1 proteins have an antagonistic effect on hTERT transcription (68). The E-boxes at the hTERT proximal promoter are occupied in vivo by c-Myc in exponentially proliferating HL60 cells. In contrast, Mad1 protein is induced and bound to the hTERT promoter in differentiated HL60 cells. These data suggest that reciprocal E-box occupancy by Myc/Max and Mad/Max is responsible for activation and repression of the hTERT gene (69). Mad is induced by 12-O-tetradecanoylphorbol-13-acetate treatment, which represses hTERT promoter-driven reporter gene activity (70).
Spl cooperates with c-Myc to activate transcription of hTERT (64). The 3′-region of the hTERT promoter contains a GC-box, which binds Sp1 and is essential for transactivation. Overexpression of Sp1 leads to significant activation of transcription in a cell type-specific manner. Western blot analysis using various cell lines reveals a positive correlation between c-Myc and Sp1 expression and transcriptional activity of hTERT (64). Interestingly, an interaction with p53 could eliminate the binding of Sp1, resulting in hTERT repression (71,72).
Epigenetic modifications. Inhibition of histone deacetylases (HDAC) by trichostatin A (TSA) in telomerase-negative cells results in activation of telomerase activity and up-regulation of hTERT mRNA (69,73,74). This TSA sensitivity can be explained by the recruitment of HDAC by the Myc/Mad heterodimer (75). Indeed, TSA alleviates TERT repression by Mad (73). Interestingly, a hTERT promoter devoid of E-boxes still exhibits TSA sensitivity (73), raising the possibility that factors other than Myc/Mad are able to recruit HDAC. A likely candidate is Sp1 (73,74), which would suggest a dual role for this factor in the regulation of hTERT transcription, both as an activator and as a repressor.
Analysis of the DNA sequence surrounding the putative transcriptional start region reveals a TATA-less, CAAT-less, GC-rich promoter located in a CpG island (65,76). Demethylation of DNA with 5-azacytidine in two cell lines induces expression of hTERT, suggesting that DNA methylation can contribute to hTERT repression in some cells. However, the TERT CpG island is unmethylated in some telomerase-negative primary tissues and non-immortalised cultured cells, indicating that mechanisms independent of DNA methylation can prevent hTERT expression (77).
Alternative splicing. There is a likely role of post-transcriptional factors in the control of enzyme function (78). Multiple products of hTERT mRNA have been identified by RT–PCR. At least six splice variants of hTERT have been shown, including a ‘deletion’ variant (hTERTα) that is missing conserved residues from the catalytic core of the protein. hTERTα inhibits endogenous telomerase activity, which results in telomere shortening and senescence (79,80). Only tissues expressing hTERT containing complete reverse transcriptase motifs demonstrate telomerase activity (81). Most telomerase-positive cell lines only contain a few molecules of potentially functional hTERT mRNA (389). Various tumour cell lines and tissues show considerable differences in their splicing patterns (10,82). These results suggest a possible role for hTERT splice variants in the regulation of telomerase.
Assembly. Various purification protocols reveal that the telomerase enzyme exists as a large complex that acts as a dimer or a multimer (83,84). Two separate, catalytically inactive TERT proteins can complement each other in trans to reconstitute catalytic activity (71). This complementation requires the N-terminus of one hTERT and the reverse transcriptase and C-terminal domains of the second hTERT. Moreover, the telomerase RNA templates in the active enzyme are interdependent and function cooperatively with each other (84). These data suggest that the catalytic core of human telomerase may exist as a functionally cooperative dimer in vivo, in a manner similar to other reverse transcriptases.
Remarkably, assembly of the holoenzyme appears to be regulated by hsp90-related chaperones (85). It is noteworthy that the concentration of these proteins is markedly elevated during transformation, as well as in advanced prostate carcinomas. Addition of purified chaperone components enhances reconstitution of telomerase activity in cell extracts, which suggests a novel mechanism for increasing telomerase assembly via hsp90 chaperoning (85). The hsp90 yeast homolog (Hsc82) also affects telomere length (86). In humans, both hsp90 and p23 bind specifically to the hTERT protein and influence its proper assembly with the template RNA, hTR. Hsp90 is distinguished from other chaperones in that most of its known substrates are signal transduction proteins (for a review see 87). The hsp70 chaperone also associates with hTERT in the absence of hTR and dissociates when telomerase is folded into its active state, which is similar to what occurs with other chaperone targets. In contrast, hsp90 and p23 remain associated with functional telomerase complexes, which differs from other hsp90-folded enzymes that require only transient hsp90/p23 binding (88).
Cellular localisation. The enhanced telomerase activity in human T lymphocytes is associated with cellular relocalisation of the enzyme from the cytoplasm to the nucleus and does not require increased expression of hTERT (89). Thus, the mechanism of telomerase activation in these cells appears different from that in tumour cells. One candidate to regulate telomerase trafficking is the 14-3-3 signal protein, which has also been identified as a hTERT-binding partner (90). A dominant negative 14-3-3 redistributes the nuclear form of hTERT into the cytoplasm. 14-3-3 probably enhances nuclear localisation of hTERT by inhibiting CRM1/exportin-mediated nuclear export of hTERT (90). Down-regulation of 14-3-3 σ prevents clonal evolution and leads to immortalisation of primary human keratinocytes (91).
Phosphorylation. The hTERT subunit of telomerase appears to be a phosphoprotein whose activity is modulated by a complex set of protein kinases, providing additional connections between telomerase activity and signal transduction pathways.
Protein kinase C (PKC) is a serine/threonine kinase involved in signal transduction pathways that govern a wide range of physiological processes, including differentiation, proliferation and gene expression. PKC appears to enhance telomerase activity through phosphorylation of hTERT (92,93). PKCζ is believed to be the PKC isotype that functions in vivo in NPC cells (94). The PKC inhibitor bis-indolylmaleimide I inhibits telomerase activity in cultured nasopharyngeal cancer cells (92) and during T cell activation (45). In an inverse fashion, protein phosphatase 2A (PP2A) inhibits telomerase activity (95). Therefore, it is possible that PKC and PP2A are involved in the reciprocal control of telomerase activity, which is consistent with the notion that a balance between PKC and PP2A plays an important part in tumourigenesis.
Besides PKC, protein kinase B (PKB or Akt) is also involved in up-regulating telomerase activity (96). Treatment of human melanoma cells with the protein phosphatase inhibitor okadaic acid stimulates both hTERT peptide phosphorylation and telomerase activity, whereas treatment of the cells with the PI3 kinase inhibitor Wortmannin inhibits phosphorylation and telomerase activity. These observations suggest that the serine residue at position 824 of hTERT may be phosphorylated by Akt in human cancer cells and that this phosphorylation is involved in mediating cellular signalling produced by growth factor activation of the PI3 kinase pathway (96). Telomerase may thus be one of the molecular targets of Akt in the processes of apoptosis, cell survival and proliferation during ageing and tumourigenesis.
Finally, it has been reported that c-Abl tyrosine kinase, activated by DNA damage, associates and phosphorylates hTERT, leading to telomerase inhibition (97). Furthermore, mouse cells deficient in c-Abl show telomere lengthening. These findings suggest a direct link between DNA damage signalling and telomeres in human cells, consistent with previous studies in yeast showing that telomeres are modified in response to DNA double-strand breaks (98,99).
Other factors modulating catalytic activity. In vitro both the catalytic activity and processivity of human telomerase are modulated by temperature, substrate (dNTP and primer) concentration and potassium concentration (100). A close correlation was found between telomerase processivity and telomere length in vivo, suggesting that enzyme processivity is a key factor in telomere regulation (101). Unfortunately, little is known concerning the factors that modulate this processivity in the cell. Telomerase activity isotonically extracted from human leukemia CEM line nuclei shows low processivity, while 500 g nuclear extracts or CHAPS extracts are highly processive, suggesting that the association of telomerase with nuclear chromatin affects telomerase activity (102).
hTR
Several names have been proposed for the RNA component of human telomerase; we have chosen to use ‘hTR’ throughout the text. A mutation in hTR has recently been demonstrated to be involved in dyskeratosis congenita (DKC), a progressive bone marrow failure syndrome that is characterised by abnormal skin pigmentation, leukoplakia and nail dystrophy (103), and a mutation of another telomerase component is involved in the X-linked form of the disease (see hTR-binding proteins below; 104). DKC cells have a lower level of telomerase RNA, produce lower levels of telomerase activity and have shorter telomeres than matched normal cells (105).
Expression and maturation. In contrast to hTERT, hTR is not a rate limiting factor for telomerase (12). hTR is sometimes considered as a basal, housekeeping RNA with more or less constant expression levels. This assumption does not completely reflect reality and the human telomerase RNA component (hTR) is present in normal somatic cells at lower levels than in cancer-derived cell lines. The hTR gene is present in the human genome as a single copy sequence on chromosome 3q26. In humans and yeast, the RNA subunit is transcribed by RNA polymerase II, in contrast with ciliates, where RNA polymerase III is responsible for this RNA synthesis. The mature form of hTR has a 5′-trimethylguanosine cap, a hallmark of small nuclear ribonucleoprotein (snoRNP) particles involved in RNA splicing (106). The half-life of hTR in telomerase-negative cells is ∼5 days and is increased 1.6-fold in the presence of hTERT (107). The transcription rate of hTR is greatly increased in cells expressing endogenous hTERT and an increased steady-state hTR level also appears to be due to the increased half-life (107). The 4-week half-life of hTR in H1299 tumour cells is the longest half-life yet reported for any RNA.
Like hTERT, the hTR gene contains CpG islands (108). The elements responsible for promoter activity are contained in a 231 bp region upstream of the transcriptional start site. The hTR promoter is methylated in some ALT cell lines and this is associated with a total absence of hTR expression in these lines (109). However, there is no evidence for hTR promoter methylation in normal somatic tissues.
hTR-binding proteins. The 2–3-fold increase in size of mammalian telomerase RNAs relative to ciliate telomerase RNAs is due to the presence of a 3′ domain resembling a box H/ACA small nucleolar RNA (snoRNA) (110). The human telomerase RNA (hTR) H/ACA motif is dispensible for in vitro assemby of telomerase. However, this H/ACA domain is essential in vivo for hTR accumulation, hTR 3′-end processing and telomerase activity. A small percentage of hTR is associated with nucleoli (110). Remarkably, all four common proteins associated with H/ACA snoRNAs (hGAR1, NAP57/dyskerin, hNOP10 and hNHP2) also bind to hTR. Among these proteins, dyskerin is a nucleolar protein involved in pseudo-uridylation of specific residues of rRNA. Mutations in the gene encoding dyskerin lead to the X-linked form of the human disease DKC (104). The pathology of DKC is consistent with compromised telomerase function. Antibodies that specifically recognise the human GAR1 (hGAR1) protein can immunoprecipitate H/ACA snoRNAs and hTR from HeLa cell extracts, which demonstrates that hGAR1 is a component of H/ACA snoRNPs and telomerase in vivo (111). Cells from individuals with the rare inherited disorder DKC, which is due to mutations in the dyskerin gene, have reduced levels of telomerase and shortened telomeres, which might provide the first direct genetic test of the function of telomeres in humans (104). hNOP10 and hNHP2 specifically associate with H/ACA RNAs, including hTR (112). The accumulation in yeast of the mature RNA component of human telomerase depends on its association with three of the four core H/ACA snoRNP proteins and it is likely that this is also the case in human cells (113).
Besides the H/ACA-binding proteins, heterogeneous nuclear ribonucleoproteins (hnRNPs) C1 and C2 associate with the RNA component of human telomerase (114) and binding of hnRNPs C1 and C2 to telomerase correlates with the ability of telomerase to access the telomere. La, hStau and L22 also interact with hTR (115–117). These hTR-associated proteins may play a role in hTR processing, telomerase assembly or localisation in vivo.
Dimerisation and association with hTERT. There is evidence that yeast telomerase contains more than one active site per telomerase complex. At least two functionally interacting RNA molecules are present and both act as templates for DNA polymerisation (118). Functional human telomerase also contains two hTR molecules (and at least two hTERT subunits; 83). These two RNA templates are interdependent and functionally cooperate with each other (84,119). This suggests that one telomerase complex is able to couple the elongation of both chromatid ends in a single round of replication.
The RNA template which is located at the 5′-end of the hTR RNA is stably associated with the catalytic subunit, making telomerase a very peculiar reverse transcriptase. The protein and RNA domains responsible for this rather tight association (although no association constant may be found in the literature) have been mapped (120–122). It is not yet clear whether telomerase activity can be normally modulated by a variable association level of its essential components.
The telomere itself regulates telomerase activity
In several organisms, the telomere length of telomerase-positive cells is maintained at a constant mean value. This can be viewed as a balance between elongation and shortening (123,124). In budding yeast, this balance is determined by negative regulation of telomerase activity by the telomere itself when its length exceeds a threshold value (125–127). This negative feedback appears to be mediated by a complex set of telomeric proteins acting in cis on telomerase activity both at the G-tail and at the duplex part of telomeric DNA. Whether these two pathways of cis-acting regulation of telomerase activity are coordinated or have separate outcomes is certainly an exciting question for future investigations.
Telomerase cis-regulation by the G-tail. The active telomerase holoenzyme is not free to elongate chromosome ends because its activity requires specific G-tail functions. Therefore, it is not surprising that the conformation of this distal part of the telomere is subject to a complex set of controls.
DNA structure. G-tails can form non-canonical DNA structures such as G-quadruplexes (128–131) that are known to inhibit telomerase activity (132). The presence of telomeric antiparallel quadruplexes has recently been demonstrated in the macronucleus of a ciliate, Stylonychia lemnae (133). The intramolecular telomeric G-quadruplex is fairly stable under physiological conditions (134) and may be recognised by nuclear proteins. Other polypeptides, such as hnRNP D, destabilise intrastrand G-G pairing and disrupt these structures (135). It is also interesting to mention that the cytosine-rich complementary telomeric strand may also form an unusual structure, called i-DNA (136,137), although no data currently support its formation in vivo.
Telomerase activation and recruitment. In budding yeast, Cdc13 specifically binds a G-tail in vitro (138,139) and telomeres in vivo (140). Therefore, Cdc13 appears to be a G-tail protein in vivo. One role of Cdc13 is to activate the telomerase holoenzyme or to recruit it to chromosome ends (141–144). It is likely that the contact between Cdc13 and telomerase is transient and, presumably, restricted to the period of the cell cycle where telomerase is active (145,146).
Cdc13 has no obvious orthologues in species other than budding yeast. Nevertheless, it is highly likely that other types of telomeric DNA-binding proteins carry out the functions of Cdc13 in other than Saccharomyces species. A possible Cdc13 functional homologue is hnRNP A1, a single-stranded DNA-binding factor that modulates telomere replication in mammalian cells (147). Interestingly, the human protein Pot1 (148) shares amino acid sequence homologies with the ciliate end-binding proteins (149). Deletion of the fission yeast pot1+ gene has an immediate effect on chromosome stability, causing rapid loss of telomeric DNA (148). Whether Pot1, hnRNP A1 or other proteins are involved in the activation of telomerase at human chromosome ends remains to be determined.
Cdc13 is not the only nucleic acid-binding protein known to stimulate a reverse transcriptase. Indeed, the nucleocapsid protein (NCp7) of human immunodeficiency virus type 1 covers the genomic RNA and is able to stimulate reverse transcription (150). NCp7 interacts both with RNA and DNA and with the reverse transcriptase subunits, suggesting that NCp7 plays a role in recruitment and/or activation of the enzyme during viral DNA synthesis (150). Both NCp7 and the ciliate end-binding proteins have nucleic acid protection and annealing properties (151,152), raising the question of whether similar nucleic acid chaperoning activities are involved in Cdc13 functions. In any case, the parallels between the roles of Cdc13, NCp7 and the ciliate end-binding proteins have to be added to the growing list of resemblances existing between retrotransposition and telomere maintenance (153,154).
Other proteins are also involved in correct processing of the G-tail for telomerase. In contrast to Cdc13, these proteins are well conserved from yeast to human and play additional roles in recombinational repair. Among them, Ku is a heterodimeric protein involved in non-homologous DNA end joining. Yeast Ku mutants are defective in telomere structure: the 3′-overhang that is normally present only during the S phase of the cell cycle in yeast remains present throughout the cycle (155). yKu is bound to telomeric DNA and plays a direct role in telomeric silencing (156). Mammalian Ku86 also plays a fundamental role at the telomere by preventing telomeric fusions (157). All these results illustrate the pivotal role of Ku proteins, which bind to telomeric DNA as well as to the telomeric RNA template and essential proteins.
Mutations in the yeast gene Tel1, the closest homologue of the human ataxia telangiectasia mutated gene, result in shortened telomeres (158) and human ataxia telangiectasia fibroblasts also have a reduced telomere length (159). Tel1 appears to act through the Rad50–Mre11–Xrs2/Nbs1 complex, probably by controlling the activity of this complex at telomeres. In yeast, Tel1 is partially redundant, with Mec1 coding for a DNA damage response kinase. Mec1 also plays a role in telomere maintenance and a Tel1/Mec1 double mutant undergoes senescence.
Telomerase inhibition. Cdc13 has antagonistic roles in telomere maintenance. Loss-of-function mutations in either Stn1 or Ten1, two Cdc13-interacting factors, produce dramatic lengthening of telomeres in a telomerase-dependent manner (160,161). Stn1 does not, however, appear to be involved in the telomerase activation functions of Cdc13 (162). Altogether, Cdc13, Stn1 and Ten1 appear to form a complex within the cell that inhibits telomerase. Since Cdc13 also positively regulates telomerase (see above), it is thought that Cdc13 exists in two configurations having opposite effects on telomerase activity. The balance between the telomerase-positive and the telomerase-negative forms of Cdc13 is expected to play an important role in telomere length regulation.
Coupling between telomerase and C-strand synthesis. C-strand synthesis occurs concomitantly with, or shortly after, G-strand elongation by telomerase and is believed to be carried out by the lagging strand replication machinery (163). In budding yeast, mutations in DNA polymerase α (cdc17/pol1) that cause no obvious defect in general replication can perturb telomere replication and lead to telomere lengthening and accumulation of single-stranded DNA at telomeric regions (145,164–166). In the ciliate Euplotes, partial inhibition of C-strand synthesis by aphidicolin leads to over-elongation of the G-strand by telomerase (167). These findings have two important implications. First, a telomere-specific mechanism for lagging strand synthesis is coupled with chromosome end structure replication. Secondly, lagging strand synthesis inhibits telomere elongation.
Several mechanisms may account for the increase in G-tail length upon inhibition of C-strand synthesis. A failure to fully replicate the C-strand is expected to increase the length of the single-stranded DNA in the corresponding G-strand. Consequently, longer G-tails may bind additional Cdc13 proteins (168), which may favour telomerase recruitment and/or activation. These processes are expected to create a positive feedback loop of G-tail elongation. A more direct inhibitory effect of the C-strand replication machinery on telomerase can also be imagined. Finally, G-tail elongation may be amplified in a replication-defective context by unmasking a nuclease activity, as suggested by the alteration in chromatin structure and the rapid increase in single-stranded DNA at telomeres upon DNA polymerase α inactivation (166).
The telomeric phenotypes of DNA polymerase α mutant strains are very similar to those exhibited by strains bearing telomere-elongating alleles of CDC13 and STN1 (143,144,160,164). This raises the interesting possibility that CDC13 and STN1 are required for C-strand synthesis. A strong argument in favour of this model is the interaction between Cdc13 and DNA polymerase α, as revealed by two-hybrid analysis and by co-immunoprecipitation in vivo (164). Moreover, point mutations in either CDC13 or CDC17/POL1 that reduce this interaction result in telomere lengthening (164). Therefore, Cdc13, presumably assisted by Stn1, may load the DNA polymerase α–primase complex to initiate C-strand synthesis at chromosome ends. This proposed role of the Cdc13–Stn1 complex in the initiation of C-strand synthesis provides a simple explanation for the negative effect of this complex on telomerase activity, since lagging strand synthesis is believed to inhibit telomerase (see above).
Cdc13, Stn1 and Ten1 play additional roles in capping: their mutation leads to G2 growth arrest and an over-elongated G-tail. Taking into account the other role of Stn1 in telomere replication, one can propose that Stn1 both regulates G-tail length and ensures chromosome end stability by controlling C-strand synthesis.
The proteins binding to the duplex part of telomeric DNA repress telomere elongation in cis. The budding yeast protein Rap1p, which binds along the double-stranded telomeric DNA repeats, enacts an additional level of regulation. An excess of Rap1p molecules bound to the telomere causes repression of telomerase activity in cis (126,169–171). This creates a negative feedback loop that contributes to the setting of telomere length. Two factors interacting with Rap1p, Rif1p and Rif2p are also involved in this regulation (172). Several lines of evidence indicate that the mechanism where proteins binding to the double-stranded parts of telomeres limit telomere elongation might also apply to organisms other than budding yeast. In fission yeast, a deletion of taz1+, a gene encoding a telomeric DNA-binding protein, dramatically increases telomere length (173).
In human cells, the known telomeric DNA-binding proteins are the TTAGGG repeat factors 1 and 2 (TRF1 and TRF2) (174–177). The C-terminal sequences of TRF1 and TRF2, called teloboxes, specifically recognise a telomeric DNA fragment but require an N-terminal dimerisation domain to firmly bind DNA (175,178,179). Reminiscent of the properties of yeast Rap1p protein, an excess of TRF1 and TRF2 inhibits telomere elongation (180,181). Moreover, their targeting to a single telomere leads to telomere shortening, indicating that they act in cis to repress telomere elongation (K.Ancelin, M.Brunon, S.Bauwens, C.Koering, C.Brun, M.Ricoul, J.P.Pommier, L.Sabatier and E.Gilson, submitted for publication).
Inhibition of telomerase activity by an antisense telomerase oligonucleotide did not further increase the pace of telomere erosion caused by TRF1 targeting, suggesting that telomerase itself is the target of TRF1 regulation (K.Ancelin, M.Brunon, S.Bauwens, C.Koering, C.Brun, M.Ricoul, J.P.Pommier, L.Sabatier and E.Gilson, submitted for publication). We do not know at present whether this inhibitory effect is mediated by TRF1 itself or by associated proteins. The latter hypothesis should be seriously considered in the light of the capacity of TRF1-interacting factor TIN2 to inhibit telomerase-mediated telomere elongation (182). In any case, this effect of TRF1 is reminiscent of the cis inhibition of yeast telomerase by Rap1 (170,171). In contrast, TRF2 targeting and telomerase inhibition have additive effects (K.Ancelin, M.Brunon, S.Bauwens, C.Koering, C.Brun, M.Ricoul, J.P.Pommier, L.Sabatier and E.Gilson, submitted for publication). This raises the interesting possibility that TRF2 could control telomere length independently of telomerase. For instance, TRF2 might activate nucleases at telomeres. The expression of a trans-negative form of TRF2 is associated with a significantly decreased amount of detected G-rich single-stranded telomeric DNA, which is likely to reflect either a trimmed or a masked G-tail (183). Therefore, TRF2 may be directly involved in the processing of chromosomal DNA ends, for example by targeting the RAD50–MRE11 complex, which is known to exhibit nuclease activity (184), to telomeres (185). These studies predict that TRF2 could still contribute to telomere length regulation in telomerase-negative cells. The mechanisms which control the rate of telomere shortening in human cells are still unknown (186) and, interestingly enough, appear to be developmentally regulated (187). Modulations of TRF2 dosage and expression might be part of this regulation. Strikingly, the human orthologue of Rap1 does not bind DNA but is targeted to telomeres via its interaction with TRF2 (188). The TRF2–hRap1 complex might be another means to limit telomere proliferation, perhaps by repressing telomerase activity in cis, like yeast Rap1 protein.
Tankyrase (189,190), a telomeric poly(ADP-ribose) polymerase (PARP), was identified through its interaction with TRF1. Tankyrase-mediated ADP ribosylation inhibits binding of TRF1 to telomeric repeats in vitro and acts as a positive regulator of telomere elongation in vivo, apparently by inhibiting TRF1 (190). Long-term overexpression of tankyrase in telomerase-positive human cells results in a gradual and progressive elongation of telomeres. A second tankyrase gene product (Tank2, with 85% homology to Tank1) was recently found to interact with TRF1 (191,390). Overexpression of Tank2 causes rapid cell death. In contrast, the initial observation that PARP1 plays a major role in telomere regulation (192) was recently challenged by another study (193).
PinX1 is a nuclear protein that interacts with Pin2/TRF1 and hTERT. It inhibits telomerase and affects tumourigenicity. Overexpression of PinX1 leads to delayed growth arrest, expression of senescence markers and telomere shortening (194). PinX1 expression is reduced in many human tumours and an antisense construct against PinX1 mRNA favours tumour formation in nude mice (194)
It is unknown at the molecular level how telomeric proteins repress distally located telomerase in cis. One can imagine that gradual folding of the telomeric chromatin into a restrictive higher order configuration can impair telomerase activity. It has been proposed that folding of human telomeres into t-loops, favoured by the binding of telomeric proteins, provides a means to sequester the G-tail and thus to impair telomerase access to its substrate.
Connections between telomerase regulation and end capping. It is remarkable that factors involved in capping, like Rap1 and the Cdc13–Stn1–Ten1 complex in yeast and TRF2 in vertebrates, behave as cis-acting repressors of telomerase and telomere elongation (160,161,170,181; K.Ancelin and E.Gilson, unpublished results). In addition, several checkpoint and DNA repair factors appear to activate telomerase. This is the case for Mec3 (195), for the Rad50–Mre11–Xrs2/Nbs1 complex, for Rad1/Rad17/Mrt2 (196) and for the single strand-binding protein RPA (V.Schramke, J.Tonnelier, V.Brevet, M.P.Longhese, E.Gilson and V.Géli, submitted for publication). Accordingly, a transient relaxation of the end protection complexes at the time of telomere replication might be a crucial step for telomerase activation. In this respect, telomerase can be viewed as a double-strand break repair enzyme specifically acting on partially and/or transiently unprotected telomeres. This predicts fascinating new connections between telomerase and chromosome end stability.
INHIBITING TELOMERASE
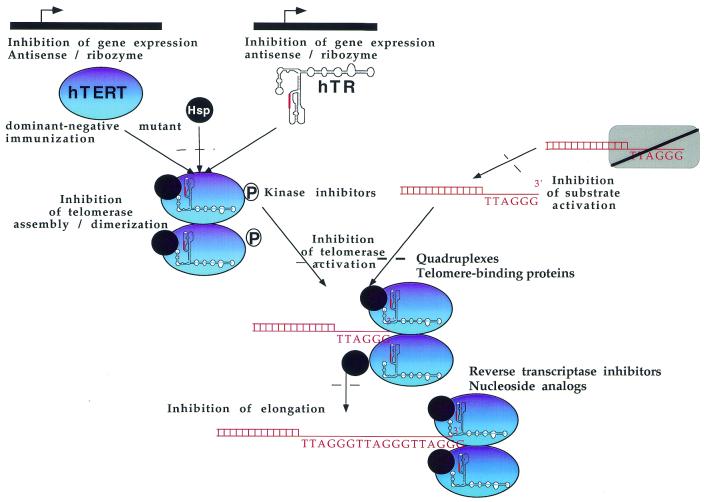
Telomerase is overexpressed in a large number of tumours whereas it is not expressed in most somatic cells, which usually have longer telomeres. This characteristic differential expression gives a rationale for further evaluation of telomerase as a target for new anticancer drugs. Several reviews concerning telomerase inhibitors have been published in the last few years (197–205). For this reason we will mainly focus on recent developments in the field. Different strategies have been developed in order to inhibit telomerase activity and interfere with tumour development (Fig. 2). Potential targets are: the catalytic subunit, the RNA component of telomerase, other proteins associated with telomerase and telomeric DNA. In the latter case, targeting telomeres may lead to other problems, since telomeres and proteins associated with telomeres are present in both normal and cancer cells. Other factors than telomerase activity (perhaps telomere length and accessibility) should therefore be proposed as a rationale for a specific effect on cancer cells.
Figure 2.
Strategies for telomerase inhibition. Possible pathways of pharmacological inhibition of telomerase: targeting of the catalytic subunit; antisense or ribozyme strategies against hTR; targeting telomeric DNA. See text for details.
The search for telomerase inhibitors was made possible by the introduction of enzymatic tests that allow semi-quantitative measurement of telomerase activity in cell extracts. The now famous telomere repeat amplification protocol (TRAP) assay uses a polymerase amplification step after telomerase extension of a primer (206). Many variations and improvements of this test have been proposed (207–212) and many laboratories use related but not identical protocols for telomerase activity measurement. For these reasons, and because the TRAP assay involves amplification of the products by PCR, direct comparisons of the concentrations that inhibit 50% of telomerase activity (IC50) should be made with caution. Recently, these enzymatic tests have been applied in a high throughput screening format, allowing the systematic evaluation of thousands of chemicals and speeding up the discovery of inhibitors (213).
Targeting the catalytic subunit (hTERT)
hTERT seems the target of choice as this protein is the rate limiting factor for telomerase activity. A strong motivation for targeting telomerase is the existence of clinically useful inhibitors of reverse transcriptases, which fall into two major classes: (i) nucleoside analogs; (ii) other inhibitors which bind to a pocket located between the palm and thumb of reverse transcriptases.
Nucleoside analogs. Nucleoside analogs were initially tested against ciliate telomerase (214). A number of nucleoside reverse transcriptase inhibitors do show anti-telomerase activity (215,216).
Azidothymidine (AZT) has the potential to target the telomeric ends of chromosomes in cancer cells, promoting cell death (217–219). AZT, dideoxyinosine (ddI) and AZT-5′ triphosphate (AZT-TP) do not cause major changes in telomerase activity or telomere length in MCAS cells. However, ddI and AZT-TP reduce telomerase activity and shorten the length of the telomere in HEC-1 cells in a dose- and time-dependent manner, showing that the effect of reverse transcriptase inhibitors can be cell type specific (220). AZT causes progressive telomere shortening in some but not all T and B cell cultures (221). AZT-treated tumour cells have reduced tumourigenicity in syngeneic BALB/c mice (222).
Other reverse transcriptase inhibitors have also been tested against telomerase, but the selectivity of these agents for telomerase versus other polymerases is a major concern. Among the l-enantiomers of NTPs, l-dTTP and l-dGTP inhibit telomerase activity while the others show slight or no inhibitory effect (223). Recently, a very potent nucleoside telomerase inhibitor, 6-thio-2′-deoxyguanosine 5′-triphosphate (TDG-TP), has been described. The chemical formula of this compound, as well as of some other inhibitors, is shown in Figure 3. Its low IC50 (0.06 µM) and relative specificity versus other reverse transcriptases suggest that a major step forward has been made (216). TDG-TP is incorporated into the DNA by telomerase in the absence of dGTP, but unlike dGTP there is little extension of the DNA chain after its incorporation (224). 2′,3′-Dideoxyguanosine 5′-triphosphate, carbovir 5′-triphosphate and d-carbocyclic-2′-deoxyguanosine 5′-triphosphate also inhibit telomerase activity (216).
Figure 3.
Chemical formulae of some inhibitors. 1, TDG-TP {R=[P(=O)O2]34–}; 2, EGCG; 3, BIBR 1532; 4, β-rubromycins; 5, isothiazolones (TMPI); 6, rhodacyanines (FJ5002); 7, bis-indoles; 8, telomestatin. The properties of these molecules are detailed in Table 2.
Other catalytic inhibitors. Screening of a chemical library of 16 000 synthetic compounds yielded six telomerase inhibitors, which included four isothiazolone derivatives (213). The most potent inhibitor, 2-[3-(trifluoromethyl)phenyl]isothiazolin-3-one (TMPI), has an IC50 of 1 µM against telomerase. Analysis using partially purified telomerase from AH7974 rat hepatoma cells demonstrates non-competitive inhibition with the telomere repeat primer and mixed inhibition with the dNTPs. Inhibition by TMPI is highly selective for telomerase and is quenched by dithiothreitol and glutathione, suggesting that TMPI inhibits telomerase by acting at a cysteine residue. A group of rubromycins and their analogues (a class of quinone antibiotics that possess benzofuran and benzodipyran rings) inhibit human telomerase as assessed with a modified telomeric repeat amplification protocol (IC50 = 3 µM) (225). A kinetic study revealed a competitive interaction with respect to the telomerase substrate primer. β-Rubromycin is a potent inhibitor of other reverse transcriptases but has virtually no effect on DNA and RNA polymerases, deoxyribonucleases and topoisomerases.
Furthermore, scientists from Boerhinger Ingelheim have recently described carboxylic amide derivatives (BIBR 1532) that interact with the catalytic subunit (IC50 = 0.093 µM). These derivatives induce progressive telomere shortening and delayed growth arrest (226).
Antisense oligonucleotides/ribozymes. To our knowledge, there have been very few reports of short antisense oligonucleotides targeted to the catalytic subunit mRNA. Phosphorothioate anti-hTERT oligomers 20–22 bases long (5–15 µM) induce delayed inhibition of cell viability in the DU145 prostate cancer cell line (227). However, no reduction in telomere length is observed even after 45 days treatment. Among four different antisense oligonucleotides directed against the mouse TERT mRNA, a 19mer overlapping the translation intitiation codon inhibits production of the protein in developing brain neurons (228). On the other hand, ribozymes have been tested against hTERT by transient and stable transfection procedures. A ribozyme targeting 13 nt downstream from the 5′-end of hTERT mRNA exhibits the strongest telomerase inhibitory activity. A stable transfection study confirmed that this ribozyme suppresses telomerase (229). Ribozyme cleavage of telomerase mRNA also sensitises breast epithelial cells to inhibitors of topoisomerase (230).
Inhibiting hTERT phosphorylation. Two PKC inhibitors (bis-indolylmaleimide I and H-7) produce a strong inhibition of telomerase activity in treated cells. The inhibition of telomerase activity by PKC inhibitors appears to be somewhat specific, since the treated cells are mostly viable and still retain significant levels of protein synthesis capability (92). However, it is difficult to imagine how hTERT phosphorylation may be specifically affected without interfering with the phosphorylation of other physiologicaly relevant PKC targets (for a review on PKC inhibitors see ref. 391).
Inhibiting hTERT transcription. Inhibiting hTERT transcription in a specific manner is a promising approach to achieve telomerase inhibition. Only a few compounds have been tested for this transcriptional inhibition. Arsenic is effective in the treatment of some leukaemias, despite its carcinogenic properties. Interestingly, it has recently been demonstrated that arsenic inhibits transcription of the hTERT gene and this effect may in part be explained by decreased c-Myc and Sp1 transcription factor activities (67).
Targeting RNA (hTR)
The RNA component of telomerase (hTR) is absolutely required for telomerase reverse transcription and is therefore a natural target for anti-telomerase agents. In contrast to hTERT, hTR is present in many normal tissues that do not express telomerase activity. No role for hTR in telomerase-negative cells has been demonstrated so far. As a consequence, inhibition of hTR in telomerase-negative cells such as normal somatic cells is not expected to be toxic. Different strategies have been successfully tested to target hTR.
The antisense approach. The antisense approach has been widely exploited and directed to the 451 nt long human telomerase RNA. This strategy benefits from the wealth of data accumulated on antisense oligonucleotides. However, the hTR target has some original characteristics. (i) It is not mRNA and will not be translated into protein: an antisense oligomer will not have to compete with the ribosomal machinery. As a consequence, RNase H-independent inhibition of telomerase activity should be possible. (ii) hTR provides a template (nt 46–56, r-5′-CUAACCCUAAC-3′) for reverse transcription. Therefore, this region of the RNA is expected to be highly accessible. A recent comparison of vertebrate telomerase RNA genes from a variety of species indicates evolutionary conservation of the global architecture of telomerase RNA and should help in the identification of other regions that may be targeted by antisense oligomers (231).
The original antisense approach used an expression vector that allowed the synthesis of a long antisense RNA. This key experiment demonstrated that hTR was indeed the RNA component of telomerase (12). An antisense vector against human telomerase RNA has also been introduced into human malignant glioma cells exhibiting telomerase activity. After 30 doublings, some sub-populations of transfectants express a high level of interleukin lp-converting enzyme (ICE) and undergo apoptosis. In contrast, other sub-populations show enhanced ICE protein but escape from apoptotic crisis and continue to grow, although their DNA synthesis, invasive ability and tumourigenicity in nude mice are significantly reduced (232).
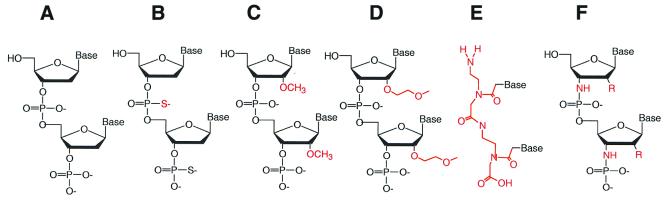
Short oligomers have also been targeted to hTR RNA. The chemical formulae of some of these agents are presented in Figure 4. Peptide nucleic acids (PNAs) in which the sugar–phosphate backbone has been replaced by N-(2-aminoethyl)glycine recognise the RNA component of human telomerase (hTR) and inhibit activity of the enzyme with IC50 values in the picomolar to nanomolar range provided that the hTR template is targeted (233). The exact identification of determinants for inhibitor binding within the RNA active site of human telomerase has been performed using PNA scanning (234). Anti-hTR PNAs are co-electroporated into immortal human cells as PNAs do not readily enter cells. Introduction of these agents effectively inhibits telomerase activity in intact cells, shortens telomeres, reduces colony size and arrests cell proliferation after a lag period of 5–30 cell generations, consistent with suppression of their ‘immortality’ (235). Other methods of PNA cellular delivery have been proposed besides electroporation (236,237).
Figure 4.
Antisense oligonucleotides. (A) regular DNA; (B) phosphorothioate; (C) 2-OMe RNA; (D) 2′-MOE RNA; (E) PNA; (F) N3′→P5′ phosphoramidates. R may be -H, -OH, -F or -OCH3 (246).
In contrast, phosphorothioate oligonucleotides (PS) inhibit telomerase in a non-sequence-selective fashion (233) and probably act by interacting with the catalytic subunit rather than the RNA. 2′-O-methyl-RNA (2′-O-MeRNA) inhibits telomerase with a potency superior to those possessed by analogous PNAs, despite a lower binding affinity for complementary RNA (238,239). 2′-O-(2-methoxyethyl)-RNAs (2′-MOE) possess favourable pharmacokinetic properties and inhibit human telomerase with IC50 values of 5–10 nM (240). After introduction into DU145 prostate cancer cells inhibition of telomerase activity persisted for up to 7 days.
2′,5′-Oligoadenylate (2-5A) antisense oligomers directed against telomerase RNA are also efficient binders (241,242). In these experiments, a 2′,5′-oligoadenylate was attached to the oligonucleotide in order to recruit and activate a specific endoribonuclease (RNase L) at the target site of the RNA sequence. A 19mer antisense oligonucleotide targeted against the RNA component of human telomerase (hTR) has been linked to a 2-5A molecule (2-5A–anti-hTR) and tested on prostate cancer cells. After continuous treatment for 1–2 weeks, these cells underwent massive apoptosis (242). This delay is probably too short to be the result of gradual telomere shortening. This treatment reduces cell viability and induces apoptosis by activated caspase family members. Furthermore, treatment of subcutaneous tumours in nude mice with 2-5A–anti-hTR significantly suppresses tumour growth through apoptosis induction (243). This antisense agent also exhibits an antitumour effect on an intracranial malignant glioma model in nude mice (244,245).
Various phosphoramidate derivatives, including 2′-deoxy-, hydroxy-, methoxy- and fluoro-N3′→P5′ phosphoramidates have recently been tested against telomerase in vitro. These compounds demonstrate sequence-specific and dose-dependent activities with IC50 values <1 nM (246). Again, oligonucleotides with the highest anti-telomerase activity are complementary to a significant portion of the template region, especially the rCCC segment. Interestingly, two other sites in the hTR primary sequence are susceptible to phosphoramidate inhibition (IC50 values down to 0.4 nM). These compounds are also efficient in various cell lines, provided they are vectored with a lipid carrier (IC50 = 0.1–0.5 µM in seven different cell lines).
Ribozymes. A hammerhead ribozyme directed against the RNA component of human telomerase shows a specific cleavage activity for the telomerase RNA component and inhibits telomerase activity in cell extracts (247). When ribozyme RNAs are introduced into endometrial carcinoma cells, only those targeting the RNA template region are able to inhibit telomerase activity (248). An anti-hTR ribozyme delivered to growing melanoma cells inhibited telomerase activity (249). The ribozyme sequence was then cloned into an expression vector and transfected into JR8 cells. The cell clones obtained showed reduced telomerase activity and significantly longer doubling times, but no telomere shortening.
The RNA/DNA duplex. Telomeric DNA synthesis by telomerase reverse transcription involves the formation of a transient DNA/RNA duplex of up to 11 bp. Molecules that bind to this duplex could inhibit the enzyme by either preventing strand dissociation or by sufficiently distorting the substrate, thereby causing a misalignment of key catalytic residues. These agents do not strictly target hTR, but its interaction with the substrate. Four intercalators show promising anti-telomerase activity in the low micromolar range (250). Equilibrium dialysis (251) should help to discover ligands that preferentially bind to this heteroduplex. A comparison of the binding of 84 different compounds to a poly(rA)·poly(dT) hybrid led to the discovery of five compounds with higher than average affinity (252). However, other key cellular processes involve RNA/DNA duplexes, such as Okasaki fragments occurring during replication of the lagging strand, and these processes would also be affected. In addition, RNase H activity is inhibited by these ligands (252).
Modifying the hTR RNA template. Telomerase hTR gene constructs containing various mutant templates decrease cellular viability and increase apoptosis even when expressed at low levels in prostate (LNCaP) and breast (MCF-7) cancer cell lines (253). This occurs despite retention of normal levels of endogenous wild-type telomerase RNA and endogenous wild-type telomerase activity and unaltered stable telomere lengths. Mutant template telomerase RNAs exert a strongly dominant negative effect on cell proliferation and tumour growth. Even very limited synthesis of mutated sequences can affect telomere structure in human cells and the toxicity of mutant telomerases is due to telomere dysfunction. These results support the potential use of mutant template telomerase RNA expression as an anti-neoplastic strategy. In another study, cells expressing mutant telomerases had a slower growth rate, abnormal cell cycle, aberrant nuclei, fused chromosomes and reduced viability (254). All phenotypes were apparent within the first few cell divisions after expression of mutant hTR. However, mutant telomerase activity is progressively selected against with cell culturing and this correlates with the disappearance of cells with aberrant phenotypes, illustrating a type of resistance mechanism.
The use of a small region of telomerase RNA as a template (11 nt) implies that telomerase establishes boundaries that prevent the reverse transcription of surrounding regions. There may be a physical barrier consisting of a protein–RNA interaction or an RNA structure that efficiently prevents utilisation of the next RNA nucleotide. If this barrier could be partially released, copying of these extra nucleotides would lead to mutated telomeric DNA repeats, which could in turn lead to the same apoptotic phenomenon described by Blackburn and co-workers (253). In that case, it would not be necessary to introduce an extra mutated hTR gene. However, this requires a drug that would disrupt such putative RNA–protein or RNA–RNA interactions. However, no molecule which could act in a similar fashion to some antibiotics that target rRNA has yet been described, although we believe that this strategy is promising.
Targeting other telomerase components
TEP1. TEP1 has been identified as a telomerase component, although its seems dispensable for telomerase activity. A mTep1–/– mouse showed no apparent change in telomere length or telomerase activity over six generations (255). TEP1 acts in vivo rather as an integral vault protein (256). It is therefore unlikely that targeting of TEP1 would lead to telomerase inhibition and no attempt has been reported.
Chaperones. Inhibition of hsp90 blocks assembly of active telomerase (257). Geldanamycin, which targets hsp90, partially reduces hsp90-stimulated telomerase activity (258). MKT-077 binds to an hsp70 family member and inhibits telomerase (259). However, all drugs targeting chaperones will interfere with many cellular processes besides telomerase activity.
Targeting telomere counting proteins. Several human proteins participate in the telomere counting mechanism. The higher the number bound to the telomere, the less likely it is that telomerase is recruited to that telomere. Therefore, modulating the number of DNA-bound proteins such as TRF1 or TRF2 could in turn inhibit or enhance telomerase activity. However, achieving such modulation by pharmacological drugs does not seem a simple task. It has been proposed that PARP inhibitors (provided they are also active on TANK1 and TANK2) may indirectly inhibit telomerase activity. Tankyrase promotes telomere elongation in human cells (190): an anti-tankyrase molecule should therefore induce telomere shortening. However, tankyrase inhibitors that do not affect other PARPs remain to be discovered.
Targeting telomeric DNA rather than telomerase
Targeting the substrate of an enzyme is an original way to inhibit its activity. There are fundamental differences between the targeting of telomeres and the targeting of telomerase subunits (hTR, hTERT or associated factors). Telomeres exist in the absence of telomerase activity and play at least one fundamental role in telomerase-negative cells, the protection (capping) of chromosome ends. Telomere-interacting molecules might then have an effect on immortal cells that regulate telomere length via the ALT pathway (a potential benefit) but also on normal/mortal cells that do not maintain telomere length, leading to undesired toxicity.
Cisplatin and other antitumour agents. Yeast telomeres are susceptible to cisplatin-induced intrastrand crosslinks and SsI2 (Rad25) and the nucleotide excision repair pathway may play a critical role in repair of this damage (260). Ciliate telomeric tandem repeats are guanosine rich and susceptible to crosslinking by cisplatin (261). Cisplatin reduces telomerase activity in a specific and concentration-dependent manner in human testicular tumour cells (262) but not in nasopharyngeal cancer cells (92). Sub-lethal doses of vincristine, an antineoplastic alkaloid, induce a decline in telomerase activity in three different human lymphoma cell lines, without a concomitant decrease in hTERT mRNA levels (263). Etoposide, a well known topoisomerase II poison, is able to stimulate cleavable complex formation at telomeric DNA sequences. This is due to the sequence selectivity of epipodophyllotoxin derivatives, which preferentially induces topoisomerase II-mediated DNA cleavage at (C/G)T sites. By itself, etoposide does not inhibit telomerase, but topoisomerase lesions at telomeres may interfere with telomere replication by a mechanism analogous to the fork collision model (264,265). Other DNA topoisomerase I and II inhibitors induce senescence-associated β-galactosidase expression and cell division arrest in normal human fibroblast. The cells resume growth upon removal of the inhibitors and accelerated telomere shortening is not observed in the arrested cells, in contrast to telomerase inhibitors (266).
Ionising radiation also modulates telomerase activity. A dose-dependent decrease in telomerase activity is usually observed in proportion to tumour regression after radiotherapy (267). However, low doses of radiation may have an opposite effect in some lymphoma or myeloma cell lines (268).
G4 DNA. Chromosomal DNA of ciliates, yeasts and vertebrates ends in a single-stranded 3′-overhang (Table 1). The overhang length and the identity of the 3′-terminal nucleotide are tightly regulated in ciliates (269). The human overhang is longer and may be involved in different DNA conformations, such as T-loops (270), triplexes (271) or G-quadruplexes (128–131). G-quadruplexes are highly polymorphic; there are three classes of G-quadruplex structures: (i) G4 DNA, a parallel quadruplex formed by four independent strands (272,273); (ii) G′2 DNA, an antiparallel bimolecular quadruplex (274,275); (iii) G4′ DNA, resulting from intramolecular folding of a single strand (276,277; for a review see 278). A DNA strand carrying at least four blocks of consecutive guanines may fold into an intramolecular G4′ structure, schematically presented in Figure 5, upper left. In the case of the human telomeric overhang, this motif is compatible with the formation of three adjacent G-quartets. Each G-quartet involves four co-planar guanines (Fig. 5, upper right). The 3-dimensional structure of the telomeric quadruplex has been solved (277). In the presence of sodium, this G-tetraplex is stabilised by three stacked G-tetrads which are connected by two lateral loops and a central diagonal loop. Of the four grooves that are formed, one is wide, two are of medium width and one is narrow. Three of the four adenines stack on top of adjacent G-tetrads, while the majority of the thymines show multiple conformations. There are a number of proteins that either bind to preformed quadruplex DNA (279–281), induce its formation (282), unwind (283–285) or cleave it (for a review see 278). Some of these proteins play a role in telomere maintenance (286–289) or other key processes, such as meiosis or immunoglobulin switch recombination.
Figure 5.
G-quadruplexes. (Upper) The G-rich telomeric strand may fold into an intramolecular G-quadruplex (right) leading to the formation of three adjacent G-quartets. A G-quartet involving four guanines is shown on the left. (Lower) Formulae of some G4 ligands: 1, amidoanthraquinone; 2, PIPER; 3, porphyrin (TMPyP4); 4, acridine; 5, dibenzophenanthroline; 6, triazine; 7, ethidium; 8, bis-acridine; 9, RHPS4 (pentacyclic acridine). The properties of these molecules are detailed in Table 2.
In vitro folding of telomeric G-rich single strand quadruplex DNA has been found to inhibit telomerase activity (132). It was deduced from this observation that a molecule that favours quadruplex formation locks the telomeric substrate into an inactive conformation which is no longer recognised nor extended by the enzyme. Stabilisation of G-quadruplexes can then be considered an original strategy to achieve antitumour activity (290,291). G4 ligands require structural selectivity, i.e. preferential binding to quadruplexes over duplexes and single strands. DNA structure-specific (rather than sequence-specific) ligands have been identified previously (292). The quadruplex itself, which is very different from classical double-stranded B-DNA, provides a good structural basis for selective recognition and such an assumption has been shown to be correct (293,294). These pioneering studies have opened up a new field in the area of ligand–DNA interactions. In recent years a large number of quadruplex ligands have been found, such as porphyrins (295–297), perylenes (298), amidoanthracene-9,10-diones (299), 2,7-disubstituted amidofluorenones (300) and indoloquinolines (301) (for a review see 278). Nevertheless, most of these molecules have a relatively modest potency, with IC50 values above (and sometimes greatly above) 1 µM. An important improvement has been achieved in the last few months, as several laboratories, including ours, have identified much better G4-based telomerase inhibitors, with IC50 values between 0.02 and 0.2 µM (Table 2 and Fig. 5, lower part) (302–306). We have used a fluorescence resonance energy transfer-based method to discover new G4-based telomerase inhibitors (303,307). Thousands of molecules have been screened and at least four independent families of ligands found [ethidium derivatives (304), dibenzophenanthrolines (303), triazines (308) and bis-acridines (309)]. Some of these molecules may act as quadruplex probes, as their fluorescence is strongly enhanced in the presence of quadruplex DNA (304). Their affinities for quadruplex DNA range from 106 to 108 M–1.
Table 2. New telomerase inhibitors.
| Familya | IC50b (µM) | Targetc | Cell effectd | Reference |
|---|---|---|---|---|
| BIBR 1532 | 0.093 | hTERT | D | (226) |
| β-Rubromycin | 3 | ? | ? | (225) |
| Isothiazolones (TMPI) | 1 | hTERT? | ? | (213) |
| Rhodacyanines (FJ5002) | 2 | hTERT? | ? | (342) |
| Bis-indoles | 2 | ? | ? | (338) |
| Catechins (EGCG) | 1 | ? | ? | (339) |
| Telomestatin | 0.005 | ? | ? | (343) |
| TDG-TP | 0.06 | Nucleoside | ? | (216) |
| Ribozymes | ? | hTR | I | (249) |
| PNA | <0.001 | hTR | D | (234,235,239) |
| 2′-OMe (oligonucleotide) | hTR | D | (238,239) | |
| 2′-MOE (oligonucleotide) | 0.005 | hTR | D | (240) |
| 2′-5′A-oligonucleotide | ? | hTR | I/D | (242) |
| N3′→P5′ phosphoramidates | <0.001 | hTR | ? | (246) |
| Dibenzophenanthrolines | 0.03 | G4 | ? | (303) |
| Acridines | 0.06 | G4 | ? | (302) |
| RHPS4 (pentacyclic acridine) | 0.3 | G4 | D | (306) |
| Ethidium | 0.03 | G4 | ? | (304) |
| Triazines | 0.04 | G4 | D | (305) |
| Bis-acridine | 0.75 | G4 | ? | (309) |
aOnly the most active compound of each family is presented. Chemical formulae of some of these agents are shown in Figures 3 and 4. 2′-OME and 2′-MOE are oligoribonucleotides with a modified sugar in the 2′ position (see Fig. 4).
bIC50 of the most active compound belonging to that family.
cMechanism of action/target: G4, quadruplex ligands; hTR, the RNA component of telomerase is targeted; hTERT, the catalytic subunit is targeted; Nucleoside, nucleoside analog.
dCellular effect: ?, not determined; I, immediate inhibition of cell growth or viability; D, delayed inhibition.
Most quadruplex ligands are polyaromatic molecules bearing one or more positive charges. A notable exception to that rule is N-methyl mesoporphyrin IX (NMM) (296), an anionic porphyrin which binds almost exclusively to quadruplexes. Equilibrium dialysis assays allow one to conclude that this derivative, perhaps thanks to its negative charge and despite a relatively low affinity, is the most selective quadruplex ligand studied so far (251). Very little structural data is available on the mode of interaction of these molecules with quadruplex DNA (298). The geometry of these compounds suggests that they interact by stacking on a terminal quartet. The surface of a quartet is much larger than the surface offered by a base pair, explaining in part how a large aromatic molecule may have a preference for quadruplex DNA, thanks to favourable stacking interactions. True intercalation between adjacent quartets should be disfavoured as a result of the energetic penalty required to unstack two quartets and eject a monocation (310). Nevertheless, some results suggest that this mode of binding might be observed in specific cases (311). Experimental observations, by either NMR or site-specific cleavage studies (312), tend to favour more plausible modes of binding of ligands to G-quadruplexes, i.e. external stacking on a terminal quartet. However, interactions such as groove binding have to be considered as well, since G-quartets are likely to form four different G grooves and/or expose adenine/thymine loops that may be specifically recognised by the ligands.
Motifs for the formation of G-quadruplex-prone DNA sequences other than telomeres are present in different regions of eukaryotic genomes: they are especially abundant in regions of biological significance, such as promoters of many important genes (the c-myc oncogene), rDNA repeats (313) and recombination hotspots. The nuclease-hypersensitive element of the c-myc promoter region, which forms a stable Watson–Crick double helix under physiological conditions, can be effectively converted to G-quadruplex DNA by a quadruplex ligand (313). This could in turn modulate the expression of that gene. Finally, telomere-like repeats have been identified at several internal sites on human chromosomes using a long synthetic (T2AG3)n probe (314). Three classes of interstitial telomeric sequences have been characterised: (i) short repeats, composed of a few essentially exact T2AG3 units; (ii) sub-telomeric repeats, composed of larger arrays (several hundred base pairs) including many degenerate units within sub-telomeric domains; (iii) fusion repeats, in which two extended stretches of telomeric repeats are oriented head-to-head (315). Finally, guanine-rich RNAs may also form quadruplexes (316,317). Recognition of these RNA motifs by specific proteins such as FMR1 (318,319) could also be altered by quadruplex ligands. As a result, quadruplex ligands could then have cellular effects mediated by quadruplex induction independently of telomeres. Other DNA-interacting enzymes also appear to be inhibited by quadruplex ligands, such as RecQ family helicases (320,321). Only a few of these ligands have been tested in cells. Porphyrins are readily absorbed into tumour cell nuclei in culture (322) and their cytotoxicity has been assessed in normal human cells (fibroblasts and breast cells) and human tumour cells representing models selected for high telomerase activity and short telomeres (breast and prostate carcinoma and lymphoma). The observed inhibition of telomerase activity is paralleled by cell growth arrest in G2/M.
On the other hand, guanine substitutes that can impair the formation of G-quartets could modify the cellular equilibrium between G-quadruplexes and B-DNA. 6-Mercaptopurine is a clinically active anticancer agent and is the pro-drug for 6-thioguanine, which blocks the formation of G-quadruplexes in vitro when incorporated into DNA. So, one part of its mechanism of action may be due to disruption of the G-quadruplex structures formed at the telomere or in other parts of the genome (323).
Telomere mimics. Studies have been carried out on inhibiting telomerase activity using a series of PS oligonucleotides with telomere sequence motifs of various lengths and sequences. The role of the 3′ end and secondary structure of telomere mimics has also been analysed, showing that telomerase inhibition requires guanine nucleotides at the 3′ end (324). The best telomere mimic is a 9 base long phosphorothioate oligodeoxynucleotide (GGGTTAGGG) with an IC50 value of 0.3 µM.
Agents specific for double-stranded telomeric DNA. DNA minor groove-binding compounds (polyamides) that specifically target vertebrate telomeric repeats have recently been synthesised (325). Epifluorescence microscopy studies show that fluorescent derivatives of these polyamides stain the telomeres of insect and vertebrate chromosomes and nuclei sharply, allowing the rapid estimation of relative telomere length. A possible interference of these compounds with the binding of telomeric proteins and/or regulation of telomere length remains to be tested.
Indirect, miscellaneous or unknown
There are a growing number of inhibitors that are active against telomerase in the standard TRAP assay. Some elegant ways to achieve telomerase inhibition do not fall into the above mentioned classes and are presented below. Unfortunately, in many cases their precise target is not known as they may act on the catalytic subunit, the RNA or the primer recognition site of the holoenzyme. These agents are presented in the last paragraph.
Suicide therapy. As telomerase is mainly expressed in tumour cells, the use of the transcription regulatory sequences of telomerase (hTR and hTERT) to drive the synthesis of a suicide gene might lead to the selective killing of cancer cells. Expression vectors containing the diphtheria toxin A-chain (DT-A) gene have been linked to hTR and hTERT transcription regulatory sequences. Inhibition of protein synthesis occurs in bladder and hepatocellular carcinoma cells transfected with these plasmids (326). Induction of Bax gene expression via the hTERT promoter elicits tumour-specific apoptosis in vitro and suppresses tumour growth in nude mice and prevents the toxicity of the Bax gene in vitro and in vivo (327). Apoptosis is restricted to telomerase-positive tumour cells using a hTERT promoter-driven caspase-8 expression vector (hTERT/caspase-8) and is not seen in normal fibroblast cells without telomerase activity. Furthermore, treatment of subcutaneous tumours in nude mice with the hTERT/caspase-8 construct inhibits tumour growth and induces apoptosis (328). Another expression vector consisting of the constitutively active caspase-6 under control of the hTERT promoter (hTERT/rev-caspase-6) has been tested in malignant glioma cells (329). The rev-caspase-6 gene is an attractive candidate as it induces apoptosis independent of the initiator caspases. The hTERT/rev-caspase-6 construct induces apoptosis in hTERT-positive malignant glioma cells but not in hTERT-negative astrocytes, fibroblasts and ALT cells. In addition, the growth of tumours in nude mice is significantly suppressed by treatment with the hTERT/rev-caspase-6 construct (329). The selective killing of cancer cells can be achieved by gene-directed enzyme pro-drug therapy (392,393). Another study used the thymidine kinase gene fused to the hTERT promoter, which sensitises the cells to ganciclovir (330). Thus, the hTERT promoter is apparently a strong and tumour-selective promoter with potential applications in targeted cancer gene therapy.
Immunisation. hTERT is an attractive target for cancer immunotherapy because it is preferentially expressed in malignant cells. The telomerase catalytic subunit is capable of triggering antitumour cytotoxic T lymphocyte (CTL) responses (331,332). Interestingly, no CTL effect was found in normal telomerase-positive CD34+ haematopoietic cells. Immunisation against TERT stimulates cytotoxic T lymphocytes, which lyse melanoma and thymoma tumour cells and inhibit the growth of unrelated tumours in mice of distinct genetic backgrounds (333). Although immunisation with TERT RNA represents an important part of the response obtained with tumour RNA, the latter is generally more effective, suggesting that an optimal immunisation protocol may have to include TERT as well as additional tumour antigens (333). Fortunately, preliminary studies do not reveal autoimmune manifestations resulting from vaccination with TERT (333). hTERT might be described as a candidate ‘universal cancer vaccine’ (334). Two hTERT-derived peptides carrying motifs for HLA-A24 (HLA-A*2402) are able to elicit anti-leukaemic CTLs. The CD8+ CTL clones specific for these hTERT peptides exert cytotoxicity against leukaemia cells in a HLA-A24-restricted manner (335). However, a lack of tumour recognition by hTERT peptide 540–548-specific CD8+ T cells from melanoma patients has recently been reported, raising questions as to the use of this peptide for cancer-specific immunotherapy. This reveals inefficient antigen processing by the proteasome (336).
Inhibition of proliferation. Telomerase activity appears to be correlated with the proliferation state of cells. Therefore, inhibition of cell division by anti-proliferative agents is expected to lead to telomerase inhibition, and this assumption has been verified for a number of drugs. This inhibition should be seen as a consequence rather than a cause of the anti-proliferative properties and is beyond the scope of this article.
Unknown target. Reports on several telomerase inhibitors have been published in the last few years without a description of their precise mechanism of action.
Two quinolones, ofloxacin and levofloxacin, inhibit telomerase activity in transitional cell carcinoma cell lines, but only at relatively high concentrations and probably through an indirect mechanism (337). Anionic bis-indole derivatives have also been shown to inhibit telomerase (IC50 = 2.5 µM) (338). Tea cathechins such as epigallocatechin gallate (EGCG) (IC50 = 1 µM) also interact with telomerase and induce telomere shortening and senescence of cancer cells (339). Alterperynol, a fungus metabolite, inhibits human telomerase activity (IC50 = 30 µM), but not viral reverse transcriptases (340). Diazaphilonic acid also exhibits telomerase inhibitory activity (341).
MKT077, a rhodacyanine derivative currently under clinical trials, shows an inhibitory effect with 50% inhibition at ∼5 µM (see Chaperones, above, for a possible mechanism of action). FJ5002 (Fig. 3), a close derivative of MKT077, is more potent (IC50 = 2 µM). Long-term cultivation of a human leukaemia cell line (U937) with sub-acute concentrations of FJ5002 results in population doubling-dependent changes characterised by progressive telomere erosion (from ∼10 to ∼4.0 kb), increased chromosome abnormalities and senescence/crisis-like features (342). Telomestatin is another potent telomerase inhibitor which has been isolated from Streptomyces anulatus (343). Its IC50 of 0.005 µM makes it the most potent small molecule telomerase inhibitor described so far.
It should be mentionned that DMSO itself, which is often used as a solvent for many poorly water soluble molecules, may affect telomerase activity. Contradictory effects have been reported. DMSO (a few percent) may act as a differentiating agent and differentiation usually leads to telomerase repression (33). However, lower doses (down to 0.07%) may induce a 2–3-fold increase in telomerase activity in ovarian epithelial cell lines (344). This is an important concern for many pharmacological inhibitors of telomerase whose dilutions may contain comparable amounts of this solvent.
Hypothetical: stimulating telomere degradation. Telomere length is tightly regulated in yeast by coordinated mechanisms of telomere lengthening and shortening. Rather than inhibiting telomere lengthening, it might be interesting to stimulate telomere degradation mechanisms. A rapid truncation of overelongated telomeres, called telomeric rapid deletion (TRD), has been previously identified in the yeast Saccharomyces cerevisiae (345). The mechanism of TRD is only partially understood: TRD is an intrachromatid deletion process in which sequences near the extreme terminus invade end-distal sequences and excise the intervening sequences (346). It will be important to determine whether human cells are also prone to TRD and which mechanisms control its action. Such studies would lead to the search for TRD-stimulating agents.
SHORTCOMINGS
There are several potential (or verified) problems with the anti-telomerase approach against cancer. We will not discuss molecules that inhibit telomerase in an indirect or non-specific way but focus on ‘ideal’ drugs that only affect telomerase and/or telomeres. Besides the specific problems discussed below, telomerase inhibitors may encounter the usual or classical obstacles found for all types of pharmacological agents, such as cellular uptake and localisation, binding to other intra- or extracellular components, biodistribution, metabolism, haematological, renal or hepatic toxicity, in vivo half-life and pharmacokinetics, etc. (a discussion of these points is beyond the scope of this review). Even if we assume that all these telomere-independent problems are solved, some new telomere/telomerase-specific problems have been identified.
Telomerase-independent maintenance of telomeres
Several organisms use telomerase-independent mechanisms to maintain their telomeres. These alternative processes may only occur when telomerase is inactivated, as a back-up solution, or may be the primary mechanism(s) of maintenance. In humans, some cancer cells (up to 15%), as well as some immortalised cells, do not express telomerase. This observation suggests that tumourigenesis does not necessarily require telomerase reactivation. On the other hand, one may argue that probably all tumour cells require a solution to the end replication problem, telomerase-dependent or otherwise. Finally, if telomerase is detected in vitro in 85% of tumours, it does not imply that telomerase is necessary for the long-term survival of all tumours. It is possible, but unverified, that some cancer cells could use telomerase-independent ways to maintain telomeres even if telomerase activity can be detected in vitro (remember the discrepancy between activity in vitro and in vivo). For all these tumour cells, an approach targeting telomerase activity is unlikely to succeed.
Even for cells that use telomerase as the sole mechanism of telomere maintenance, one cannot exclude the possibility that long-term exposure to anti-telomerase agents could lead to the selection of mutants that resist the drug through a variety of ways. Besides multidrug resistance phenotypes, which are not specific to telomerase inhibitors, the tumour cell could adapt by overexpressing telomerase or producing a mutated protein that is no longer sensitive to the drug, in a manner similar to the resistance to HIV reverse transcriptase catalytic subunit inhibitors. Fortunately, the human catalytic subunit is not likely to evolve at the same rate as its retroviral homologue, even in cancer cells, where many repair pathways have been altered. Long-term treatment of HL60 cells with sub-toxic concentrations of the telomerase/reverse transcriptase inhibitor AZT leads rather to the selection of AZT-resistant cells than to senescence induction (J.F.Riou and F.Chevallier, unpublished results). On the other hand, resistance to long-term treatment with cisplatin and 5-fluorouracil is associated with a higher proliferation rate, telomerase activity and telomere length (347).
The main cause of worry is the prediction that telomerase-dependent cells could spontaneously become telomerase-independent if a strong selection pressure is exerted by an anti-telomerase agent. The likehood of this phenotypic transformation will be one of the key factors that will govern the success of this approach: if evolution to a telomerase-independent maintenance pathway is probable during the course of cancer treatment with anti-telomerase agents, then these drugs will probably be of little therapeutic use, offering temporary remission at best. There is little data that deals with this field, however, we will nevertheless present some observations that indicate that such a resistance mechanism is possible, but probably not very likely. Firstly, the main cause of resistance in human cells would be the so-called ALT (alternative lengthening of telomeres) pathway (348). Cells utilising ALT have telomeres of heterogeneous length, ranging from very short to very long (349). ALT occurs by means of homologous recombination and copy switching (350). Other telomerase maintenance solutions observed in model organisms, such as chromosome circularisation and retrotransposition, are unlikely to happen in human cancer cells. Interestingly, the ALT pathway is repressed in telomerase-positive cells (351) and this repression is not mediated by the catalytic subunit itself, but by another unknown factor (352). Fusion of ALT cells with telomerase-positive immortal cells usually results in hybrids that appear immortal and telomerase positive but repress the ALT telomere phenotype (353). Recent experiments have demonstrated that ALT cells retain factors associated with telomerase to maintain telomere length. When telomerase is de novo expressed in these telomerase-negative cells, recombination-based elongation of telomeres can be functionally inhibited and the cells switch to a telomerase-based pathway of telomere maintainance (351). Secondly, this ALT pathway does not occur with the same probability in cell lines and tumours of various origins. As cancers in humans are of epithelial origin in >90% of cases, it is interesting to note that the ALT mechanism is unlikely in this type of cell. Up to 46% of in vitro immortalised human cell lines use ATL for telomere maintenance, whereas <10% (probably ~5%) (348) of human tumours are estimated to rely on ALT. This suggests that ALT is difficult to mobilise or selected against during the tumourigenic process in vivo. One may then argue that such resistance will be unlikely. Furthermore, telomerase-negative immortal cells contain one or several genes functioning as telomerase repressors and maintain telomere length by a dominant mechanism other than telomerase reactivation, showing that ALT and telomerase are usually mutually exclusive (354). The second line of observation that indicates that alternative lengthening of telomeres might not be a widespread cause of resistance comes from experiments in which telomerase is repressed. For example, in some experiments where a dominant negative hTERT subunit was introduced into immortalised cells, resistance mechanisms were sometimes observed, but never through a telomerase-independent pathway. Resistance was rather the result of overexpression of the wild-type subunit (355). In the case of antisense oligonucleotides, no resistance was ever reported, despite very long treatment (4 months) (239). Finally, the occurrence of ALT resistance will depend on the mechanism of action of the agent. For agents that directly target telomeric DNA, such as G-quadruplex ligands, resistance through ALT remains unlikely.
Delayed effect
According to the initial paradigm for telomerase inhibitors, these agents should not initially affect growth rate but induce progressive telomere shortening. Decreased proliferation should only be observed when the telomeres reach a critically short length. Therefore, the delay necessary for the anti-proliferative effect should depend on the initial telomere length. This paradigm has been verified by several key experiments used for target validation purposes. Unfortunately, among the dozens of telomerase inhibitors described so far, only a minority have been tested in cultured cells. Many do inhibit telomerase activity in cells as well as in cell extracts, but only a handful induce telomere shortening and delayed cell growth. In the case of the antisense approach, using an expression vector against hTR, cell crisis is observed after 23–26 population doublings in HeLa cells (12). The antisense vector against human telomerase RNA has also been introduced into human malignant glioma cells exhibiting telomerase activity (232). After 30 doublings, some sub-populations of transfectants express a high level of ICE protein and undergo apoptosis. In contrast, other sub-populations show enhanced ICE protein but escape apoptotic crisis and continue to grow, although their DNA synthesis, invasive ability and tumourigenicity in nude mice are significantly reduced. These data show that treatment with antisense telomerase inhibits telomerase activity and subsequently induces either apoptosis or differentiation. In the same line, the onset of cell division arrest by a dominant negative mutant of hTERT was related to the initial telomere length (19,355): fast in LoVo cells (TRF = 2–3 kb) but slower in 36M ovarian cells (TRF = 5–7 kb); arrest was only obtained after 30–40 days in the latter case. In a model cell line with very short telomeres, the effect of such dominant negative mutants is obtained after a few days (355). Concerning telomerase inhibitors, this delayed effect has been verified in a few cases. PNA antisense oligomers act according to this paradigm (239). However, even longer delays are required: full inhibition in DU145 cells takes several months, even if these cells harbour relatively short telomeres (TRF = 3.6 kb). Such further delay might be the result of incomplete inhibition (5–30% telomerase activity remained), arguing for the need for a potent inhibitor that would clear the cell of any residual activity. Even with that limitation in mind, telomerase inhibitors should not be discarded. First, many tumour cells harbour short telomeres and in some cases extremely short telomeres. These compounds should therefore be active at least in this telomere context. Measuring telomere length of the tumour cells before starting treatment might help to determine whether telomerase inhibition could be helpful or not. However, average telomere length may not be the most relevant factor. It might be more interesting to analyse the length of the shorter telomeres, which are expected to be the first to reach a critically short length after telomerase elongation. Another important parameter that should be taken into account is the rate of telomere loss at each division. This rate may vary greatly between telomerase-inactive cells and is directly proportional to the length of the 3′ G-overhang (356). Regulation of the overhang length, ranging from 50 to 150 bases, is not well understood. Bearing in mind the extreme variations in telomere lengths of various tumours and the discrepancies between the rates of telomere shortening, one may then easily imagine that the delay in growth arrest, expressed in cell divisions, induced by telomerase inhibitors may vary a lot, perhaps by a factor of 10 or more. Nevertheless, recent experiments support the idea that an immediate response to telomere interference may be observed.
First, the telomerase complex itself could participate in the capping of short telomeres (357). Several observations support this hypothesis: (i) the telomeres of some immortal telomerase-positive cells are actually shorter than in control cells that have gone into crisis; (ii) in yeast, telomeres of normally growing cells with hypomorphic telomerase are shorter than telomeres in control cells lacking telomerase that are dividing (118,358); (iii) human cells may be rescued from crisis by ectopic expression of telomerase without lengthening of telomeres (359). Enzymatically competent telomerase has a protective effect on short telomeres, such as those observed in some cancer cells, and this capping function could be absent or superfluous for normal telomeres (360). In that case, targeting telomerase within these cells should lead to an immediate effect rather than delayed growth inhibition.
Secondly, the mutation of some key telomeric proteins such as TRF2 has immediate and profound effects on cell physiology. One may then imagine that, by targeting the telomere rather than telomerase, the other essential function of chromosomal ends, capping, could be disrupted. In that case, chromosomal aberrations such as telomere fusions should be expected, leading to immediate cell cycle arrest or mitotic disaster if too many aberrations accumulate. Agents targeting telomeres rather than telomerase should also be active on telomerase-inactive cells: a mixed blessing, as this would allow the targeting of ALT cancer cells but also of normal dividing cells. In that case, one is not taking advantage of the crucial difference between normal and cancer cells: the necessity of a telomere maintenance mechanism in the latter case. Nevertheless, such an approach would benefit from two differences between cancer and normal cells: a faster division rate and shorter, and probably more exposed, telomeres.
One approach seems to overcome many of the above mentioned objections. The introduction of a mutant hTR may lead to the incorporation of abnormal telomeric repeats, which would in turn lead to immediate and massive apoptosis (253). In that case, the effect is telomerase dependent but immediate rather than delayed and normal telomerase-inactive cells should be refractory.
Another solution to accelerate the phenotype of telomerase inhibition could be to associate this treatment with other anti-proliferative drugs or with radiotherapy. Some data argue for a synergistic effect between telomerase inhibitors and other anticancer compounds. Cells with short telomeres are more susceptible to drugs (361) and ionising radiation (362–364). The impact of telomere dysfunction on chemotherapeutic responses has been assessed in normal and neoplastic cells derived from telomerase RNA null (mTERC–/–) mice. Telomere dysfunction, rather than telomerase per se, has been found to be the principal determinant governing chemosensitivity, specifically to agents that induce DNA double-strand breaks (DSB). These results point to the combined use of DSB-inducing agents and telomere maintenance inhibition as an effective anticancer therapeutic approach, particularly in p53-positive cells (361). Moreover, telomerase-expressing cells have elongated telomeres and are resistant to apoptosis induced by hydroxyl radicals (365). Telomerase also has an anti-apoptotic role in pheochromocytoma cells (366) and neurons (367). Exposure of cells to etoposide results in a 2–3-fold increase in telomerase activity and the percentage of dead cells after etoposide treatment is inversely correlated with the level of telomerase activation (368). These findings suggest the existence of an anti-apoptotic pathway through which telomerase is up-regulated in response to DNA damage. In murine lymphoma cells, radiosensitive cells have shorter telomeres in comparison with radioresistant cells. A significant inverse correlation between telomere length and chromosomal radiosensitivity was also observed in lymphocytes from breast cancer patients (364). For all these reasons it appears reasonable to expect a positive synergy between telomerase inhibitors and other agents that target cancer cells.
Toxicity
The naive idea that only tumour cells are telomerase positive is of course oversimplified. Germ cells and some important somatic cells do possess an active telomerase and telomerase inhibitors are expected to be effective on these cells as well. Telomerase activity has been detected in haematopoeitic stem cells, in lymphocytes and basal layer cells in skin, hair follicles and small intestine. This raises several crucial questions: is this telomerase activity necessary for these cells; what would be the effect of a transitory inhibition? A few observations indicate that such toxicity could be tolerated: (i) these normal cells (especially germ cells) generally have a longer initial telomere length than cancer cells; (ii) normal cells usually divide less frequently than tumour cells. Therefore, critical telomere shortening could be achieved in cancer cells before normal cells. Interestingly, when a telomerase inhibitor is removed from the cell culture medium, the telomeres rapidly regain their length, suggesting that normal cells may rapidly recover once treatment is completed (239).
Lessons from the telomerase-negative mouse
Fundamental differences between telomere regulation in the mouse and human suggest that the mouse is not a straightforward model to critically test the role of telomere loss and telomerase in human malignancy (369). In contrast to human primary fibroblasts, mouse embryonic fibroblasts have telomerase activity, immortalise spontaneously in culture and can be transformed by oncogenic stress. Shay and Wright suggest that humans have to be >104 times more resistant to developing cancer than mice based on body weight and life expectancy (370). What has been called cellular senescence in cultured rodent cells is not dependent on a telomere clock but reflects inappropriate culture conditions (371,372). Mouse telomeres are also much longer than human telomeres, explaining why it takes six generations to observe a pronounced phenotype in knock-out mice (373,374). Telomerase inhibition in immortal or transformed human cells using dominant negative hTERT mutants leads to telomere shortening and cell death. The expression of a dominant negative mutant of the catalytic subunit of mouse telomerase, mTERT-DN, in a murine kidney tumour cell line, RenCa, initially leads to telomerase activity inhibition and telomere shortening. However, all clones expressing mTERT-DN eventually reactivate telomerase and show normal viability, in contrast to that described for human cells (375). Having stated these differences, useful lessons may still be extracted from mice without telomerase (376) and we can analyse the relationships between telomeres and cancer in murine cells.
Studies agree that telomerase-deficient mice develop at least as many tumours as wild-type animals (373), showing that telomerase is not a necessary condition for tumour formation in rodents. Telomere dysfunction may even promote non-reciprocal translocations and epithelial cancers in mice (377) by a process of fusion bridge breakage that leads to the formation of complex non-reciprocal translocations usually found in human carcinomas. In the same line, a functional genomics approach identified mTERT as a suppressor of fibroblast transformation (378). The rampant chromosomal instability caused by telomere dysfunction may facilitate the generation of a pro-cancer genotype. Deletion of p53 significantly attenuates the adverse cellular and organismal effects of telomere dysfunction, but only during the earliest stages of genetic crisis, thus establishing a key role for p53 in the cellular response to telomere dysfunction in both normal and neoplastic cells (379).
Even if the relationship between telomerase and tumourigenesis is complex in rodents, some reports illustrate the importance of telomeres in the mouse: telomerase-deficient mice with short telomeres are resistant to skin tumourigenesis (24) and increased epidermal tumours are found in transgenic mice overexpressing mTERT in basal keratinocytes (380). Furthermore, short dysfunctional telomeres impair tumourigenesis in the INK4a(Δ2/3) cancer-prone mouse (25). In APC (min) TERC–/– mice, progressive telomere dysfunction leads to an increase in initiated lesions (microscopic adenomas), yet a significant decline in the multiplicity and size of macroscopic adenomas (381). These data are consistent with a model in which telomere dysfunction promotes the chromosomal instability that drives early carcinogenesis, while telomerase activation restores genomic stability to a level permissive for tumour progression. As antitumour therapy is likely to commence with the latter cells, telomerase inhibition should have an anti-proliferative action on them. This link between telomere dysfunction and genomic instability has recently been confirmed in yeast (382). Based on these observations, telomerase may be envisioned both as an oncogene and an anti-oncogene, depending on the state of the cell.
CONCLUSION
Telomere-driven senescence is believed to be a by-product of a system devised to provide a tumour suppression function. This preventive system may be bypassed by cancer cells through reactivation of a telomere maintenance mechanism: telomerase inhibition therefore aims to restore mortality to tumour cells. While telomerase may not be a universal target for cancer treatment, targeting the telomere maintenance mechanisms will be important in future cancer research. Fundamental advances have been achieved in the comprehension of telomere physiology in recent years. Nevertheless, important points remain to be elucidated, illustrating the undiminished need for strong basic science in that field.
Major results have been obtained in the applied field of pharmalogical inhibition of telomerase and various potent inhibitors have recently been isolated. They can tentatively be classified into two major classes: telomerase- or telomere-interacting molecules. Fundamental differences in terms of cell specificify and delayed versus immediate effects may be foreseen and various advantages or pitfalls may result from these differences. It is not yet possible to support the claim that the telomere is a better target than telomerase or vice versa. Many experiments presented in this review have been performed on a number of different cellular systems, using various markers to measure the effects of the compounds, which makes a comparison of these molecules difficult. Perhaps the choice of a few ‘standard’ models should be made in order to rationalise this area of research. Nevertheless, the recent discovery of potent and specific agents of both classes should permit further target validation experiments in tumour-bearing mice and ultimately in cancer patients.
Acknowledgments
ACKNOWLEDGEMENTS
We thank F. Hamy (Aventis, Vitry), L. Guittat, P. Alberti, L. Lacroix, P. B. Arimondo, T. Garestier and C. Hélène (MNHN, Paris) for helpful discussions and Martin Mills for careful reading of the manuscript. This work was supported by a CNRS-PCV grant (J.L.M. and E.G.), a Ligue Nationale contre le Cancer grant (E.G.), an ARC grant (no. 4321) and an Aventis research grant (J.L.M.).
REFERENCES
- 1.Wright W.E., Tesmer,V.M., Huffman,K.E., Levene,S.D. and Shay,J.W. (1997) Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev., 11, 2801–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Makarov V.L., Hirose,Y. and Langmore,J.P. (1997) Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell, 88, 657–666. [DOI] [PubMed] [Google Scholar]
- 3.McElligott R. and Wellinger,R.J. (1997) The terminal DNA structure of mammalian chromosomes. EMBO J., 16, 3705–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olovnikov A.M. (1973) A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J. Theor. Biol., 41, 181–190. [DOI] [PubMed] [Google Scholar]
- 5.Allsopp R.C., Chang,E., Kashefiaazam,M., Rogaev,E.I., Piatyszek,M.A., Shay,J.W. and Harley,C.B. (1995) Telomere shortening is associated with cell division in vitro and in vivo. Exp. Cell Res., 220, 194–200. [DOI] [PubMed] [Google Scholar]
- 6.Greider C.W. and Blackburn,E.H. (1985) Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell, 43, 405–413. [DOI] [PubMed] [Google Scholar]
- 7.Lingner J., Hughes,T.R., Shevchenko,A., Mann,M., Lundblad,V. and Cech,T.R. (1997) Reverse transcriptase motifs in the catalytic subunit of telomerase. Science, 276, 561–567. [DOI] [PubMed] [Google Scholar]
- 8.Morin G.B. (1989) The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell, 59, 521–529. [DOI] [PubMed] [Google Scholar]
- 9.Meyerson M., Counter,C.M., Eaton,E.N., Ellisen,L.W., Steiner,P., Caddle,S.D., Ziaugra,L., Beijersbergen,R.L., Davidoff,M.J., Liu,Q.Y., Bacchetti,S., Haber,D.A. and Weinberg,R.A. (1997) hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell, 90, 785–795. [DOI] [PubMed] [Google Scholar]
- 10.Kilian A., Bowtell,D.D.L., Abud,H.E., Hime,G.R., Venter,D.J., Keese,P.K., Duncan,E.L., Reddel,R.R. and Jefferson,R.A. (1997) Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cell types. Hum. Mol. Genet., 6, 2011–2019. [DOI] [PubMed] [Google Scholar]
- 11.Nakamura T.M., Morin,G.B., Chapman,K.B., Weinrich,S.L., Andrews,W.H., Lingner,J., Harley,C.B. and Cech,T.R. (1997) Telomerase catalytic subunit homologs from fission yeast and human. Science, 277, 955–959. [DOI] [PubMed] [Google Scholar]
- 12.Feng J.L., Funk,W.D., Wang,S.S., Weinrich,S.L., Avilion,A.A., Chiu,C.P., Adams,R.R., Chang,E., Allsopp,R.C., Yu,J.H., Le,S.Y., West,M.D., Harley,C.B., Andrews,W.H., Greider,C.W. and Villeponteau,B. (1995) The RNA component of human telomerase. Science, 269, 1236–1241. [DOI] [PubMed] [Google Scholar]
- 13.Kruk P.A., Rampino,N.J. and Bohr,V.A. (1995) DNA damage and repair in telomeres: relation to aging. Proc. Natl Acad. Sci. USA, 92, 258–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dhaene K., Van Marck,E. and Parwaresch,R. (2000) Telomeres, telomerase and cancer: an up-date. Virchows Arch., 437, 1–16. [DOI] [PubMed] [Google Scholar]
- 15.Bodnar A.G., Ouellette,M., Frolkis,M., Holt,S.E., Chiu,C.P., Morin,G.B., Harley,C.B., Shay,J.W., Lichtsteiner,S. and Wright,W.E. (1998) Extension of life-span by introduction of telomerase into normal human cells. Science, 279, 349–352. [DOI] [PubMed] [Google Scholar]
- 16.Halvorsen T.L., Leibowitz,G. and Levine,F. (1999) Telomerase activity is sufficient to allow transformed cells to escape from crisis. Mol. Cell. Biol., 19, 1864–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang X.R., Jimenez,G., Chang,E., Frolkis,M., Kusler,B., Sage,M., Beeche,M., Bodnar,A.G., Wahl,G.M., Tlsty,T.D. and Chiu,C.P. (1999) Telomerase expression in human somatic cells does not induce changes associated with a transformed phenotype. Nature Genet., 21, 111–114. [DOI] [PubMed] [Google Scholar]
- 18.Morales C.P., Holt,S.E., Ouellette,M., Kaur,K.J., Yan,Y., Wilson,K.S., White,M.A., Wright,W.E. and Shay,J.W. (1999) Absence of cancer-associated changes in human fibroblasts immortalized with telomerase. Nature Genet., 21, 115–118. [DOI] [PubMed] [Google Scholar]
- 19.Hahn W.C., Stewart,S.A., Brooks,M.W., York,S.G., Eaton,E., Kurachi,A., Beijersbergen,R.L., Knoll,J.H.M., Meyerson,M. and Weinberg,R.A. (1999) Inhibition of telomerase limits the growth of human cancer cells. Nature Med., 5, 1164–1170. [DOI] [PubMed] [Google Scholar]
- 20.Hahn W.C., Counter,C.M., Lundberg,A.S., Beijersbergen,R.L., Brooks,M.W. and Weinberg,R.A. (1999) Creation of human tumour cells with defined genetic elements. Nature, 400, 464–468. [DOI] [PubMed] [Google Scholar]
- 21.Elenbaas B., Spirio,L., Koerner,F., Fleming,M.D., Zimonjic,D.B., Donaher,J.L., Popescu,N.C., Hahn,W.C. and Weinberg,R.A. (2001) Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev., 15, 50–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanahan D. and Weinberg,R.A. (2000) The hallmarks of cancer. Cell, 100, 57–70. [DOI] [PubMed] [Google Scholar]
- 23.Chadeneau C., Hay,K., Hirte,H.W., Gallinger,S. and Bacchetti,S. (1995) Telomerase activity associated with acquisition of malignancy in human colorectal cancer. Cancer Res., 55, 2533–2536. [PubMed] [Google Scholar]
- 24.Gonzalez-Suarez E., Samper,E., Flores,J.M. and Blasco,M.A. (2000) Telomerase-deficient mice with short telomeres are resistant to skin tumorigenesis. Nature Genet., 26, 114–117. [DOI] [PubMed] [Google Scholar]
- 25.Greenberg R.A., Chin,L., Femino,A., Lee,K.H., Gottlieb,G.J., Singer,R.H., Greider,C.W. and DePinho,R.A. (1999) Short dysfunctional telomeres impair tumorigenesis in the INK4a(Delta 2/3) cancer-prone mouse. Cell, 97, 515–525. [DOI] [PubMed] [Google Scholar]
- 26.Klapper W., Kuhne,K., Singh,K.K., Heidorn,K., Parwaresch,R. and Krupp,G. (1998) Longevity of lobsters is linked to ubiquitous telomerase expression. FEBS Lett., 439, 143–146. [DOI] [PubMed] [Google Scholar]
- 27.Buchkovich K.J. and Greider,C.W. (1996) Telomerase regulation during entry into the cell cycle in normal human T cells. Mol. Biol. Cell, 7, 1443–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weng N.P., Granger,L. and Hodes,R.J. (1997) Telomere lengthening and telomerase activation during human B cell differentiation. Proc. Natl Acad. Sci. USA, 94, 10827–10832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu K., Schoonmaker,M.M., Levine,B.L., June,C.H., Hodes,R.J. and Weng,N.P. (1999) Constitutive and regulated expression of telomerase reverse transcriptase (hTERT) in human lymphocytes. Proc. Natl Acad. Sci. USA, 96, 5147–5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kyo S., Takakura,M., Kohama,T. and Inoue,M. (1997) Telomerase activity in human endometrium. Cancer Res., 57, 610–614. [PubMed] [Google Scholar]
- 31.Xu D., Gruber,A., Peterson,C. and Pisa,P. (1996) Suppression of telomerase activity in HL60 cells after treatment with differentiating agents. Leukemia, 10, 1354–1357. [PubMed] [Google Scholar]
- 32.Sharma H.W., Sokoloski,J.A., Perez,J.R., Maltese,J.Y., Sartorelli,A.C., Stein,C.A., Nichols,G., Khaled,Z., Telang,N.T. and Narayanan,R. (1995) Differentiation of immortal cells inhibits telomerase activity. Proc. Natl Acad. Sci. USA, 92, 12343–12346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bestilny L.J., Brown,C.B., Miura,Y., Robertson,L.D. and Riabowol,K.T. (1996) Selective inhibition of telomerase activity during terminal differentiation of immortal cell lines. Cancer Res., 56, 3796–3802. [PubMed] [Google Scholar]
- 34.Koyanagi Y., Kobayashi,D., Yajima,T., Asanuma,K., Kimura,T., Sato,T., Kida,T., Yagihashi,A., Kameshima,H. and Watanabe,N. (2000) Telomerase activity is down regulated via decreases in hTERT mRNA but not TEP1 mRNA or hTERC during the differentiation of leukemic cells. Anticancer Res., 20, 773–778. [PubMed] [Google Scholar]
- 35.Pendino F., Flexor,M., Delhommeau,F., Buet,D., Lanotte,M. and Segal-Bendirdjian,E. (2001) Retinoids down-regulate telomerase and telomere length in a pathway distinct from leukemia cell differentiation. Proc. Natl Acad. Sci. USA, 98, 6662–6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ueda M., Ouhtit,A., Bito,T., Nakazawa,K., Lubbe,J., Ichihashi,M., Yamasaki,H. and Nakazawa,H. (1997) Evidence for UV-associated activation of telomerase in human skin. Cancer Res., 57, 370–374. [PubMed] [Google Scholar]
- 37.Bickenbach J.R., Vornwald-Dogan,V., Bachor,C., Bleuel,K., Schnapp,G. and Boukamp,P. (1998) Telomerase is not an epidermal stem cell marker and is downregulated by calcium. J. Invest. Dermatol., 111, 1045–1052. [DOI] [PubMed] [Google Scholar]
- 38.Nemoto K., Kondo,Y., Himeno,S., Suzuki,Y., Hara,S., Akimoto,M. and Imura,N. (2000) Modulation of telomerase activity by zinc in human prostatic and renal cancer cells. Biochem. Pharmacol., 59, 401–405. [DOI] [PubMed] [Google Scholar]
- 39.Xu D.W., Erickson,S., Szeps,M., Gruber,A., Sangfelt,O., Einhorn,S., Pisa,P. and Grander,D. (2000) Interferon down-regulates telomerase reverse transcriptase and telomerase activity in human malignant and nonmalignant hematopoietic cells. Blood, 96, 4313–4318. [PubMed] [Google Scholar]
- 40.Misiti S., Nanni,S., Fontemaggi,G., Cong,Y.S., Wen,J.P., Hirte,H.W., Piaggio,G., Sacchi,A., Pontecorvi,A., Bacchetti,S. and Farsetti,A. (2000) Induction of hTERT expression and telomerase activity by estrogens in human ovary epithelium cells. Mol. Cell. Biol., 20, 3764–3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kyo S., Takakura,M., Kanaya,T., Zhuo,W., Fujimoto,K., Nishio,Y., Orimo,A. and Inoue,M. (1999) Estrogen activates telomerase. Cancer Res., 59, 5917–5921. [PubMed] [Google Scholar]
- 42.Yang H., Kyo,S., Takatura,M. and Sun,L.Z. (2001) Autocrine transforming growth factor beta suppresses telomerase activity and transcription of human telomerase reverse transcriptase in human cancer cells. Cell Growth Differ., 12, 119–127. [PubMed] [Google Scholar]
- 43.Weinrich S.L., Pruzan,R., Ma,L., Ouellette,M., Tesmer,V.M., Holt,S.E., Bodnar,A.G., Lichtsteiner,S., Kim,N.W., Trager,J.B., Taylor,R.D., Carlos,R., Andrews,W.H., Wright,W.E., Shay,J.W., Harley,C.B. and Morin,G.B. (1997) Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nature Genet., 17, 498–502. [DOI] [PubMed] [Google Scholar]
- 44.Beattie T.L., Zhou,W., Robinson,M.O. and Harrington,L. (1998) Reconstitution of human telomerase activity in vitro. Curr. Biol., 8, 177–180. [DOI] [PubMed] [Google Scholar]
- 45.Bodnar A.G., Kim,N.W., Effros,R.B. and Chiu,C.P. (1996) Mechanism of telomerase induction during T cell activation. Exp. Cell Res., 228, 58–64. [DOI] [PubMed] [Google Scholar]
- 46.Vaziri H. and Benchimol,S. (1999) Alternative pathways for the extension of cellular life span: inactivation of p53/pRb and expression of telomerase. Oncogene, 18, 7676–7680. [DOI] [PubMed] [Google Scholar]
- 47.Bryce L.A., Morrison,N., Hoare,S.F., Muir,S. and Keith,W.N. (2000) Mapping of the gene for the human telomerase reverse transcriptase, hTERT, to chromosome 5p15.33 by fluorescence in situ hybridization. Neoplasia, 2, 197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wick M., Zubov,D. and Hagen,G. (1999) Genomic organization and promoter characterization of the gene encoding the human telomerase reverse transcriptase (HTERT). Gene, 232, 97–106. [DOI] [PubMed] [Google Scholar]
- 49.Cong Y.S., Wen,J.P. and Bacchetti,S. (1999) The human telomerase catalytic subunit hTERT: organization of the gene and characterization of the promoter. Hum. Mol. Genet., 8, 137–142. [DOI] [PubMed] [Google Scholar]
- 50.Baur J.A., Zou,Y., Shay,J.W. and Wright,W.E. (2001) Telomere position effect in human cells. Science, 292, 2075–2077. [DOI] [PubMed] [Google Scholar]
- 51.Zhang A., Zheng,C., Lindvall,C., Hou,M., Ekedahl,J., Lewensohn,R., Yan,Z., Yang,X., Henriksson,M., Blennow,E., Nordenskjold,M., Zetterberg,A., Bjorkholm,M., Gruber,A. and Xu,D. (2000) Frequent amplification of the telomerase reverse transcriptase gene in human tumors. Cancer Res., 60, 6230–6235. [PubMed] [Google Scholar]
- 52.Ducrest A.L., Amacker,M., Mathieu,Y.D., Cuthbert,A.P., Trott,D.A., Newbold,R.F., Nabholz,M. and Lingner,J. (2001) Regulation of human telomerase activity: repression by normal chromosome 3 abolishes nuclear telomerase reverse transcriptase transcripts but does not affect c-Myc activity. Cancer Res., 61, 7594–7602. [PubMed] [Google Scholar]
- 53.Poole J.C., Andrews,L.G. and Tollefsbol,T.O. (2001) Activity, function and gene regulation of the catalytic subunit of telomerase (HTERT). Gene, 269, 1–12. [DOI] [PubMed] [Google Scholar]
- 54.Crowe D.L. and Nguyen,D.C. (2001) Rb and E2F-1 regulate telomerase activity in human cancer cells. Biochim. Biophys. Acta, 1518, 1–6. [DOI] [PubMed] [Google Scholar]
- 55.Oh S., Song,Y., Yim,J. and Kim,T.K. (1999) The Wilms’ tumor 1 tumor suppressor gene represses transcription of the human telomerase reverse transcriptase gene. J. Biol. Chem., 274, 37473–37478. [DOI] [PubMed] [Google Scholar]
- 56.Yin L., Hubbard,A.K. and Giardina,C. (2000) NF-kappa B regulates transcription of the mouse telomerase catalytic subunit. J. Biol. Chem., 275, 36671–36675. [DOI] [PubMed] [Google Scholar]
- 57.Fujimoto K., Kyo,S., Takakura,M., Kanaya,T., Kitagawa,Y., Itoh,H., Takahashi,M. and Inoue,M. (2000) Identification and characterization of negative regulatory elements of the human telomerase catalytic subunit (HTERT) gene promoter: possible role of MZF-2 in transcriptional repression of hTERT. Nucleic Acids Res., 28, 2557–2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang J., Xie,L.Y., Allan,S., Beach,D. and Hannon,G.J. (1998) Myc activates telomerase. Genes Dev., 12, 1769–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Latil A., Vidaud,D., Valeri,A., Fournier,G., Vidaud,M., Lidereau,R., Cussenot,O. and Bieche,I. (2000) hTERT expression correlates with myc over-expression in human prostate cancer. Int. J. Cancer, 89, 172–176. [PubMed] [Google Scholar]
- 60.Sagawa Y., Nishi,H., Isaka,K., Fujito,A. and Takayama,M. (2001) The correlation of TERT expression with c-myc expression in cervical cancer. Cancer Lett., 168, 45–50. [DOI] [PubMed] [Google Scholar]
- 61.Wu K.J., Grandori,C., Amacker,M., SimonVermot,N., Polack,A., Lingner,J. and Dalla Favera,R. (1999) Direct activation of TERT transcription by c-MYC. Nature Genet., 21, 220–224. [DOI] [PubMed] [Google Scholar]
- 62.Greenberg R.A., O’Hagan,R.C., Deng,H.Y., Xiao,Q.R., Hann,S.R., Adams,R.R., Lichtsteiner,S., Chin,L., Morin,G.B. and De Pinho,R.A. (1999) Telomerase reverse transcriptase gene is a direct target of c-Myc but is not functionally equivalent in cellular transformation. Oncogene, 18, 1219–1226. [DOI] [PubMed] [Google Scholar]
- 63.Drissi R., Zindy,F., Roussel,M.F. and Cleveland,J.L. (2001) c-Myc-mediated regulation of telomerase activity is disabled in immortalized cells. J. Biol. Chem., 276, 29994–30001. [DOI] [PubMed] [Google Scholar]
- 64.Kyo S., Takakura,M., Taira,T., Kanaya,T., Itoh,H., Yutsudo,M., Ariga,H. and Inoue,M. (2000) Spl cooperates with c-Myc to activate transcription of the human telomerase reverse transcriptase gene (HTERT). Nucleic Acids Res., 28, 669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Takakura M., Kyo,S., Kanaya,T., Hirano,H., Takeda,J., Yutsudo,M. and Inoue,M. (1999) Cloning of human telomerase catalytic subunit (HTERT) gene promoter and identification of proximal core promoter sequences essential for transcriptional activation in immortalized and cancer cells. Cancer Res., 59, 551–557. [PubMed] [Google Scholar]
- 66.Ogretmen B., Kraveka,J.M., Schady,D., Usta,J., Hannun,Y.A. and Obeid,L.M. (2001) Molecular mechanisms of ceramide-mediated telomerase inhibition in the A549 human lung adenocarcinoma cell line. J. Biol. Chem., 276, 32506–32514. [DOI] [PubMed] [Google Scholar]
- 67.Chou W.C., Hawkins,A.L., Barret,J.F., Griffin,C.A. and Dang,C.V. (2001) Arsenic inhibition of telomerase transcription leads to genetic instability. J. Clin. Invest., 108, 1541–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Oh S., Song,Y.H., Yim,J. and Kim,T.K. (2000) Identification of Mad as a repressor of the human telomerase (HTERT) gene. Oncogene, 19, 1485–1490. [DOI] [PubMed] [Google Scholar]
- 69.Xu D.W., Popov,N., Hou,M., Wang,Q., Bjorkholm,M., Gruber,A., Menkel,A.R. and Henriksson,M. (2001) Switch from Myc/Max to Mad1/Max binding and decrease in histone acetylation at the telomerase reverse transcriptase promoter during differentiation of HL60 cells. Proc. Natl Acad. Sci. USA, 98, 3826–3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gunes C., Lichtsteiner,S., Vasserot,A.P. and Englert,C. (2000) Expression of the hTERT gene is regulated at the level of transcriptional initiation and repressed by Mad1. Cancer Res., 60, 2116–2121. [PubMed] [Google Scholar]
- 71.Kanaya T., Kyo,S., Hamada,K., Takakura,M., Kitagawa,Y., Harada,H. and Inoue,M. (2000) Adenoviral expression of p53 represses telomerase activity through down-regulation of human telomerase reverse transcriptase transcription. Clin. Cancer Res., 6, 1239–1247. [PubMed] [Google Scholar]
- 72.Xu D.W., Wang,Q., Gruber,A., Bjorkholm,M., Chen,Z.G., Zaid,A., Selivanova,G., Peterson,C., Wiman,K.G. and Pisa,P. (2000) Downregulation of telomerase reverse transcriptase mRNA expression by wild type p53 in human tumor cells. Oncogene, 19, 5123–5133. [DOI] [PubMed] [Google Scholar]
- 73.Cong Y.S. and Bacchetti,S. (2000) Histone deacetylation is involved in the transcriptional repression of hTERT in normal human cells. J. Biol. Chem., 275, 35665–35668. [DOI] [PubMed] [Google Scholar]
- 74.Takakura M., Kyo,S., Sowa,Y., Wang,Z., Yatabe,N., Maida,Y., Tanaka,M. and Inoue,M. (2001) Telomerase activation by histone deacetylase inhibitor in normal cells. Nucleic Acids Res., 29, 3006–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Laherty C.D., Yang,W.M., Sun,J.M., Davie,J.R., Seto,E. and Eisenman,R.N. (1997) Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell, 89, 349–356. [DOI] [PubMed] [Google Scholar]
- 76.Horikawa I., Cable,P.L., Afshari,C. and Barrett,J.J. (1999) Cloning and characterization of the promoter region of human telomerase reverse transcriptase gene. Cancer Res., 59, 826–830. [PubMed] [Google Scholar]
- 77.Dessain S.K., Yu,H.Y., Reddel,R.R., Beijersbergen,R.L. and Weinberg,R.A. (2000) Methylation of the human telomerase gene CpG island. Cancer Res., 60, 537–541. [PubMed] [Google Scholar]
- 78.Liu K.B., Schoonmaker,M.M., Levine,B.L., June,C.H., Hodes,R.J. and Weng,N.P. (1999) Constitutive and regulated expression of telomerase reverse transcriptase (HTERT) in human lymphocytes. Proc. Natl Acad. Sci. USA, 96, 5147–5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Colgin L.M., Wilkinson,C., Englezou,A., Kilian,A., Robinson,M.O. and Reddel,R.R. (2000) The hTERT alpha splice variant is a dominant negative inhibitor of telomerase activity. Neoplasia, 2, 426–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yi X.M., White,D.M., Aisner,D.L., Baur,J.A., Wright,W.E. and Shay,J.W. (2000) An alternate splicing variant of the human telomerase catalytic subunit inhibits telomerase activity. Neoplasia, 2, 433–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ulaner G.A., Hu,J.F., Vu,T.H., Oruganti,H., Giudice,L.C. and Hoffman,A.R. (2000) Regulation of telomerase by alternate splicing of human telomerase reverse transcriptase (HTERT) in normal and neoplastic ovary, endometrium and myometrium. Int. J. Cancer, 85, 330–335. [PubMed] [Google Scholar]
- 82.Villa R., Della-Porta,C., Folini,M., Daidone,M.G. and Zaffaroni,N. (2001) Possible regulation of telomerase activity by transcription and alternative splicing of telomerase reverse transcriptase in human melanoma. J. Invest. Dermatol., 116, 867–873. [DOI] [PubMed] [Google Scholar]
- 83.Beattie T.L., Zhou,W., Robinson,M.O. and Harrington,L. (2001) Functional multimerization of the human telomerase reverse transcriptase. Mol. Cell. Biol., 21, 6151–6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wenz C., Enenkel,B., Amacker,M., Kelleher,C., Damm,K. and Lingner,J. (2001) Human telomerase contains two cooperating telomerase RNA molecules. EMBO J., 20, 3526–3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Akalin A., Elmore,L.W., Forsythe,H.L., Amaker,B.A., McCollum,E.D., Nelson,P.S., Ware,J.L. and Holt,S.E. (2001) A novel mechanism for chaperone-mediated telomerase regulation during prostate cancer progression. Cancer Res., 61, 4791–4796. [PubMed] [Google Scholar]
- 86.Grandin N. and Charbonneau,M. (2001) Hsp90 levels affect telomere length in yeast. Mol. Genet. Genomics, 265, 126–134. [DOI] [PubMed] [Google Scholar]
- 87.Young J.C., Moarefi,I. and Hartl,F.U. (2001) Hsp90: a specialized but essential protein folding tool. J. Cell Biol., 154, 267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Forsythe H.L., Jarvis,J.L., Turner,J.W., Elmore,L.W. and Holt,S.E. (2001) Stable association of hsp90 and p23, but not hsp70, with active human telomerase. J. Biol. Chem., 276, 15571–15574. [DOI] [PubMed] [Google Scholar]
- 89.Liu K., Hodes,R.J. and Weng,N. (2001) Cutting edge: telomerase activation in human T lymphocytes does not require increase in telomerase reverse transcriptase (hTERT) protein but is associated with hTERT phosphorylation and nuclear translocation. J. Immunol., 166, 4826–4830. [DOI] [PubMed] [Google Scholar]
- 90.Seimiya H., Sawada,H., Muramatsu,Y., Shimizu,M., Ohko,K., Yamane,K. and Tsuruo,T. (2000) Involvement of 14-3-3 proteins in nuclear localization of telomerase. EMBO J., 19, 2652–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dellambra E., Golisano,O., Bondanza,S., Siviero,E., Lacal,P., Molinari,M., DAtri,S. and DeLuca,M. (2000) Downregulation of 14-3-3 sigma prevents clonal evolution and leads to immortalization of primary human keratinocytes. J. Cell Biol., 149, 1117–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ku W.C., Cheng,A.J. and Wang,T.C.V. (1997) Inhibition of telomerase activity by PKC inhibitors in human nasopharyngeal cancer cells in culture. Biochem. Biophys. Res. Commun., 241, 730–736. [DOI] [PubMed] [Google Scholar]
- 93.Li H., Zhao,L.L., Yang,Z.Y., Funder,J.W. and Liu,J.P. (1998) Telomerase is controlled by protein kinase C alpha in human breast cancer cells. J. Biol. Chem., 273, 33436–33442. [DOI] [PubMed] [Google Scholar]
- 94.Yu C.C., Lo,S.C. and Wang,T.C.V. (2001) Telomerase is regulated by protein kinase C-zeta in human nasopharyngeal cancer cells. Biochem. J., 355, 459–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li H., Zhao,L.L., Funder,J.W. and Liu,J.P. (1997) Protein phosphatase 2A inhibits nuclear telomerase activity in human breast cancer cells. J. Biol. Chem., 272, 16729–16732. [DOI] [PubMed] [Google Scholar]
- 96.Kang S.S., Kwon,T., Kwon,D.Y. and Do,S.I. (1999) Akt protein kinase enhances human telomerase activity through phosphorylation of telomerase reverse transcriptase subunit. J. Biol. Chem., 274, 13085–13090. [DOI] [PubMed] [Google Scholar]
- 97.Kharbanda S., Kumar,V., Dhar,S., Pandey,P., Chen,C., Majumder,P., Yuan,Z.M., Whang,Y., Strauss,W., Pandita,T.K., Weaver,D. and Kufe,D. (2000) Regulation of the hTERT telomerase catalytic subunit by the c-Abl tyrosine kinase. Curr. Biol., 10, 568–575. [DOI] [PubMed] [Google Scholar]
- 98.Mills K.D., Sinclair,D.A. and Guarente,L. (1999) MEC1-dependent redistribution of the Sir3 silencing protein from telomeres to DNA double-strand breaks. Cell, 97, 609–620. [DOI] [PubMed] [Google Scholar]
- 99.Martin S.G., Laroche,T., Suka,N., Grunstein,M. and Gasser,S.M. (1999) Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell, 97, 621–633. [DOI] [PubMed] [Google Scholar]
- 100.Sun D., Lopez-Guajardo,C.C., Quada,J., Hurley,L.H. and Von Hoff,D.D. (1999) Regulation of catalytic activity and processivity of human telomerase. Biochemistry, 38, 4037–4044. [DOI] [PubMed] [Google Scholar]
- 101.Peng Y., Mian,S. and Lue,N.F. (2001) Analysis of telomerase processivity: mechanistic similarity to HIV-1 reverse transcriptase and role in telomere maintenance. Mol. Cell, 7, 1201–1211. [DOI] [PubMed] [Google Scholar]
- 102.Fletcher T.M., Trevino,A. and Woynarowski,J.M. (1999) Enzymatic activity of endogenous telomerase associated with intact nuclei from human leukemia CEM cells. Biochem. Biophys. Res. Commun., 265, 51–56. [DOI] [PubMed] [Google Scholar]
- 103.Vulliamy T., Marrone,A., Goldman,F., Dearlove,A., Bessler,M., Mason,P.J. and Dokal,I. (2001) The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature, 413, 432–435. [DOI] [PubMed] [Google Scholar]
- 104.Mitchell J.R., Wood,E. and Collins,K. (1999) A telomerase component is defective in the human disease dyskeratosis congenita. Nature, 402, 551–555. [DOI] [PubMed] [Google Scholar]
- 105.Vulliamy T.J., Knight,S.W., Mason,P.J. and Dokal,I. (2001) Very short telomeres in the peripheral blood of patients with X-linked and autosomal dyskeratosis congenita. Blood Cells Mol. Dis., 27, 353–357. [DOI] [PubMed] [Google Scholar]
- 106.Seto A.G., Zaug,A.J., Sobel,S.G., Wolin,S.L. and Cech,T.R. (1999) Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature, 401, 177–180. [DOI] [PubMed] [Google Scholar]
- 107.Yi X.M., Tesmer,V.M., Savre-Train,I., Shay,J.W. and Wright,W.E. (1999) Both transcriptional and posttranscriptional mechanisms regulate human telomerase template RNA levels. Mol. Cell. Biol., 19, 3989–3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhao J.Q., Hoare,S.F., McFarlane,R., Muir,S., Parkinson,E.K., Black,D.M. and Keith,W.N. (1998) Cloning and characterization of human and mouse telomerase RNA gene promoter sequences. Oncogene, 16, 1345–1350. [DOI] [PubMed] [Google Scholar]
- 109.Hoare S.F., Bryce,L.A., Wisman,G.B.A., Burns,S., Going,J.J., van der Zee,A.G.J. and Keith,W.N. (2001) Lack of telomerase RNA gene hTERC expression in alternative lengthening of telomeres cells is associated with methylation of the hTERC promoter. Cancer Res., 61, 27–32. [PubMed] [Google Scholar]
- 110.Mitchell J.R., Cheng,J. and Collins,K. (1999) A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol. Cell. Biol., 19, 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dragon F., Pogacic,V. and Filipowicz,W. (2000) In vitro assembly of human H/ACA small nucleolar RNPs reveals unique features of U17 and telomerase RNAs. Mol. Cell. Biol., 20, 3037–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pogacic V., Dragon,F. and Filipowicz,W. (2000) Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10. Mol. Cell. Biol., 20, 9028–9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dez C., Henras,A., Faucon,B., Lafontaine,D.L.J., Caizergues-Ferrer,M. and Henry,Y. (2001) Stable expression in yeast of the mature form of human telomerase RNA depends on its association with the box H/ACA small nucleolar RNP proteins Cbf5p, Nhp2p and Nop10p. Nucleic Acids Res., 29, 598–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ford L.P., Suh,J.M., Wright,W.E. and Shay,J.W. (2000) Heterogeneous nuclear ribonucleoproteins C1 and C2 associate with the RNA component of human telomerase. Mol. Cell. Biol., 20, 9084–9091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ford L.P., Shay,J.W. and Wright,W.E. (2001) The La antigen associates with the human telomerase ribonucleoprotein and influences telomere length in vivo. RNA, 7, 1068–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Le S.Y., Sternglanz,R. and Greider,C.W. (2000) Identification of two RNA-binding proteins associated with human telomerase RNA. Mol. Biol. Cell, 11, 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bachand F., Triki,I. and Autexier,C. (2001) Human telomerase RNA–protein interactions. Nucleic Acids Res., 29, 3385–3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Prescott J. and Blackburn,E.H. (1997) Functionally interacting telomerase RNAs in the yeast telomerase complex. Genes Dev., 11, 2790–2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tesmer V.M., Ford,L.P., Holt,S.E., Frank,B.C., Yi,X., Aisner,D.L., Ouellette,M., Shay,J.W. and Wright,W.E. (1999) Two inactive fragments of the integral RNA cooperate to assemble active telomerase with the human protein catalytic subunit (hTERT) in vitro. Mol. Cell. Biol., 19, 6207–6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bachand F. and Autexier,C. (2001) Functional regions of human telomerase reverse transcriptase and human telomerase RNA required for telomerase activity and RNA–protein interactions. Mol. Cell. Biol., 21, 1888–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lai C.K., Mitchell,J.R. and Collins,K. (2001) RNA binding domain of telomerase reverse transcriptase. Mol. Cell. Biol., 21, 990–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Martin-Rivera L. and Blasco,M.A. (2001) Identification of functional domains and dominant negative mutations in vertebrate telomerase RNA using an in vivo reconstitution system. J. Biol. Chem., 276, 5856–5865. [DOI] [PubMed] [Google Scholar]
- 123.Larson D.D., Spangler,E.A. and Blackburn,E.H. (1987) Dynamics of telomere length variation in Tetrahymena thermophila. Cell, 50, 477–483. [DOI] [PubMed] [Google Scholar]
- 124.Greider C.W. (1996) Telomere length regulation. Annu. Rev. Biochem., 65, 337–365. [DOI] [PubMed] [Google Scholar]
- 125.Murray A.W., Claus,T.E. and Szostak,J.W. (1988) Characterization of two telomeric DNA processing reactions in Saccharomyces cerevisiae. Mol. Cell. Biol., 8, 4642–4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kyrion G., Boakye,K.A. and Lustig,A.J. (1992) C-terminal truncation of RAP1 results in the deregulation of telomere size, stability and function in Saccharomyces cerevisiae. Mol. Cell. Biol., 12, 5159–5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.McEachern M.J. and Blackburn,E.H. (1995) Runaway telomere elongation caused by telomerase RNA gene mutations. Nature, 376, 403–409. [DOI] [PubMed] [Google Scholar]
- 128.Oka Y. and Thomas,C.A.Jr (1987) The cohering telomeres of Oxytricha. Nucleic Acids Res., 15, 8877–8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sen D. and Gilbert,W. (1988) Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its applications for meiosis. Nature, 334, 364–366. [DOI] [PubMed] [Google Scholar]
- 130.Sundquist W.I. and Klug,A. (1989) Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature, 342, 825–829. [DOI] [PubMed] [Google Scholar]
- 131.Williamson J.R. (1994) G-quartet structures in telomeric DNA. Annu. Rev. Biophys. Biomol. Struct., 23, 703–730. [DOI] [PubMed] [Google Scholar]
- 132.Zahler A.M., Williamson,J.R., Cech,T.R. and Prescott,D.M. (1991) Inhibition of telomerase by G-quartet DNA structures. Nature, 350, 718–720. [DOI] [PubMed] [Google Scholar]
- 133.Schaffitzel C., Berger,I., Postberg,J., Hanes,J., Lipps,H.J. and Plückthun,A. (2001) In vitro generated antibodies specific for telomeric guanine quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl Acad. Sci. USA, 98, 8572–8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mergny J.L., Phan,A.T. and Lacroix,L. (1998) Following G-quartet formation by UV-spectroscopy. FEBS Lett., 435, 74–78. [DOI] [PubMed] [Google Scholar]
- 135.Eversole A. and Maizels,N. (2000) In vitro properties of the conserved mammalian protein hnRNP D suggest a role in telomere maintenance. Mol. Cell. Biol., 20, 5425–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Leroy J.L., Guéron,M., Mergny,J.L. and Hélène,C. (1994) Intramolecular folding of a fragment of the cytosine-rich strand of telomeric DNA into an i-motif. Nucleic Acids Res., 22, 1600–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ahmed S., Kintanar,A. and Henderson,E. (1994) Human telomeric C-strand tetraplexes. Nature Struct. Biol., 1, 83–88. [DOI] [PubMed] [Google Scholar]
- 138.Lin J.J. and Zakian,V.A. (1996) The Saccharomyces CDC13 protein is a single-strand TG(1–3) telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc. Natl Acad. Sci. USA, 93, 13760–13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang M.J., Lin,Y.C., Pang,T.L., Lee,J.M., Chou,C.C. and Lin,J.J. (2000) Telomere-binding and Stn1p-interacting activities are required for the essential function of Saccharomyces cerevisiae Cdc13p. Nucleic Acids Res., 28, 4733–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bourns B.D., Alexander,M.K., Smith,A.M. and Zakian,V.A. (1998) Sir proteins, Rif proteins and Cdc13p bind Saccharomyces telomeres in vivo. Mol. Cell. Biol., 18, 5600–5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nugent C.I., Hughes,T.R., Lue,N.F. and Lundblad,V. (1996) Cdc13p: a single-strand telomeric DNA binding protein with a dual role in yeast telomere maintenance. Science, 274, 249–252. [DOI] [PubMed] [Google Scholar]
- 142.Evans S.K. and Lundblad,V. (1999) Est1 and Cdc13 as comediators of telomerase access. Science, 286, 117–120. [DOI] [PubMed] [Google Scholar]
- 143.Grandin N., Damon,C. and Charbonneau,M. (2000) Cdc13 cooperates with the yeast Ku proteins and Stn1 to regulate telomerase recruitment. Mol. Cell. Biol., 20, 8397–8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chandra A., Hughes,T.R., Nugent,C.I. and Lundblad,V. (2001) Cdc13 both positively and negatively regulates telomere replication. Genes Dev., 15, 404–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Diede S.J. and Gottschling,D.E. (1999) Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell, 99, 723–733. [DOI] [PubMed] [Google Scholar]
- 146.Marcand S., Brevet,V., Mann,C. and Gilson,E. (2000) Cell cycle restriction of telomere elongation. Curr. Biol., 10, 487–490. [DOI] [PubMed] [Google Scholar]
- 147.La Branche H., Dupuis,S., Ben-David,Y., Bani,M.R., Wellinger,R.J. and Chabot,B. (1998) Telomere elongation by hnRNP A1 and a derivative that interacts with telomeric repeats and telomerase. Nature Genet., 19, 199–202. [DOI] [PubMed] [Google Scholar]
- 148.Baumann P. and Cech,T.R. (2001) Pot1, the putative telomere end-binding protein in fission yeast and humans. Science, 292, 1171–1175. [DOI] [PubMed] [Google Scholar]
- 149.Gottschling D.E. and Zakian,V.A. (1986) Telomere proteins: specific recognition and protection of the natural termini of Oxytricha macronuclear DNA. Cell, 47, 195–205. [DOI] [PubMed] [Google Scholar]
- 150.Lener D., Tanchou,V., Roques,B.P., Le Grice,S.F. and Darlix,J.L. (1998) Involvement of HIV-I nucleocapsid protein in the recruitment of reverse transcriptase into nucleoprotein complexes formed in vitro. J. Biol. Chem., 273, 33781–33786. [DOI] [PubMed] [Google Scholar]
- 151.Fang G.W. and Cech,T.R. (1993) The beta-subunit of Oxytricha telomere-binding protein promotes G-quartet formation by telomeric DNA. Cell, 74, 875–885. [DOI] [PubMed] [Google Scholar]
- 152.Darlix J.L., Cristofari,G., Rau,M., Pechoux,C., Berthoux,L. and Roques,B. (2000) Nucleocapsid protein of human immunodeficiency virus as a model protein with chaperoning functions and as a target for antiviral drugs. Adv. Pharmacol., 48, 345–372. [DOI] [PubMed] [Google Scholar]
- 153.Biessmann H., Mason,J.M., Ferry,K., d’Hulst,M., Valgeirsdottir,K., Traverse,K.L. and Pardue,M.L. (1990) Addition of telomere-associated HeT DNA sequences “heals” broken chromosome ends in Drosophila. Cell, 61, 663–673. [DOI] [PubMed] [Google Scholar]
- 154.Morris D.K. and Lundblad,V. (1997) Programmed translational frameshifting in a gene required for yeast telomere replication. Curr. Biol., 7, 969–976. [DOI] [PubMed] [Google Scholar]
- 155.Gravel S., Larrivee,M., Labrecque,P. and Wellinger,R.J. (1998) Yeast Ku as a regulator of chromosomal DNA end structure. Science, 280, 741–744. [DOI] [PubMed] [Google Scholar]
- 156.Mishra K. and Shore,D. (1999) Yeast Ku protein plays a direct role in telomeric silencing and counteracts inhibition by Rif proteins. Curr. Biol., 9, 1123–1126. [DOI] [PubMed] [Google Scholar]
- 157.Samper E., Goytisolo,F.A., Slijepcevic,P., van Buul,P.P.W. and Blasco,M.A. (2000) Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO Rep., 1, 244–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Greenwell P.W., Kronmal,S.L., Porter,S.E., Gassenhuber,J., Obermaier,B. and Petes,T.D. (1995) TEL1, a gene involved in controlling telomere length in S. cerevisiae, is homologous to the human ataxia telangiectasia gene. Cell, 82, 823–829. [DOI] [PubMed] [Google Scholar]
- 159.Xia S.J.J., Shammas,M.A. and Reis,R.J.S. (1996) Reduced telomere length in ataxia-telangiectasia fibroblasts. Mutat. Res. DNA Repair, 364, 1–11. [DOI] [PubMed] [Google Scholar]
- 160.Grandin N., Reed,S.I. and Charbonneau,M. (1997) Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev., 11, 512–527. [DOI] [PubMed] [Google Scholar]
- 161.Grandin N., Damon,C. and Charbonneau,M. (2001) Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J., 20, 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Pennock E., Buckley,K. and Lundblad,V. (2001) Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell, 104, 387–396. [DOI] [PubMed] [Google Scholar]
- 163.Wellinger R.J., Ethier,K., Labrecque,P. and Zakian,V.A. (1996) Evidence for a new step in telomere maintenance. Cell, 85, 423–433. [DOI] [PubMed] [Google Scholar]
- 164.Qi H.Y. and Zakian,V.A. (2000) The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated Est1 protein. Genes Dev., 14, 1777–1788. [PMC free article] [PubMed] [Google Scholar]
- 165.Carson M.J. and Hartwell,L. (1985) CDC17: an essential gene that prevents telomere elongation in yeast. Cell, 42, 249–257. [DOI] [PubMed] [Google Scholar]
- 166.Martin A.D., Dionne,I., Wellinger,R.J. and Holm,C. (2000) The function of DNA polymerase alpha at telomeric G-tails is important for telomere homeostasis. Mol. Cell. Biol., 20, 786–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Fan X.Q. and Price,C.M. (1997) Coordinate regulation of G- and C-strand length during new telomere synthesis. Mol. Biol. Cell, 8, 2145–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hughes T.R., Weilbaecher,R.G., Walterscheid,M. and Lundblad,V. (2000) Identification of the single-strand telomeric DNA binding domain of the Saccharomyces cerevisiae Cdc13 protein. Proc. Natl Acad. Sci. USA, 97, 6457–6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Krauskopf A. and Blackburn,E.H. (1996) Control of telomere growth by interactions of RAP1 with the most distal telomeric repeats. Nature, 383, 354–357. [DOI] [PubMed] [Google Scholar]
- 170.Marcand S., Gilson,E. and Shore,D. (1997) A protein-counting mechanism for telomere length regulation in yeast. Science, 275, 986–990. [DOI] [PubMed] [Google Scholar]
- 171.Marcand S., Brevet,V. and Gilson,E. (1999) Progressive cis-inhibition of telomerase upon telomere elongation. EMBO J., 18, 3509–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wotton D. and Shore,D. (1997) A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev., 11, 748–760. [DOI] [PubMed] [Google Scholar]
- 173.Cooper J.P., Nimmo,E.R., Allshire,R.C. and Cech,T.R. (1997) Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature, 385, 744–747. [DOI] [PubMed] [Google Scholar]
- 174.Chong L., van Steensel,B., Broccoli,D., Erdjument-Bromage,H., Hanish,J., Tempst,P. and de Lange,T. (1995) A human telomeric protein. Science, 270, 1663–1667. [DOI] [PubMed] [Google Scholar]
- 175.Bilaud T., Koering,C.E., Binetbrasselet,E., Ancelin,K., Pollice,A., Gasser,S.M. and Gilson,E. (1996) The telobox, a Myb-related telomeric DNA binding motif found in proteins from yeast, plants and human. Nucleic Acids Res., 24, 1294–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Broccoli D., Smogorzewska,A., Chong,L. and de Lange,T. (1997) Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nature Genet., 17, 231–235. [DOI] [PubMed] [Google Scholar]
- 177.Bilaud T., Brun,C., Ancelin,K., Koering,C.E., Laroche,T. and Gilson,E. (1997) Telomeric localization of TRF2, a novel human telobox protein. Nature Genet., 17, 236–239. [DOI] [PubMed] [Google Scholar]
- 178.Bianchi A., Stansel,R.M., Fairall,L., Griffith,J.D., Rhodes,D. and de Lange,T. (1999) TRF1 binds a bipartite telomeric site with extreme spatial flexibility. EMBO J., 18, 5735–5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Vassetzky N.S., Gaden,F., Brun,C., Gasser,S.M. and Gilson,E. (1999) Taz1p and Teb1p, two telobox proteins in Schizosaccharomyces pombe, recognize different telomere-related DNA sequences. Nucleic Acids Res., 27, 4687–4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.van Steensel B. and de Lange,T. (1997) Control of telomere length by the human telomeric protein TRF1. Nature, 385, 740–743. [DOI] [PubMed] [Google Scholar]
- 181.Smogorzewska A., Van Steensel,B., Bianchi,A., Oelmann,S., Schaefer,M.R., Schnapp,G. and De Lange,T. (2000) Control of human telomere length by TRF1 and TRF2. Mol. Cell. Biol., 20, 1659–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kim S.H., Kaminker,P. and Campisi,J. (1999) TIN2, a new regulator of telomere length in human cells. Nature Genet., 23, 405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.van Steensel B., Smogorzewska,A. and de Lange,T. (1998) TRF2 protects human telomeres from end-to-end fusions. Cell, 92, 401–413. [DOI] [PubMed] [Google Scholar]
- 184.Trujillo K.M. and Sung,P. (2001) DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50·Mre11 complex. J. Biol. Chem., 276, 35458–35464. [DOI] [PubMed] [Google Scholar]
- 185.Zhu X.D., Kuster,B., Mann,M., Petrini,J.H.J. and de Lange,T. (2000) Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nature Genet., 25, 347–352. [DOI] [PubMed] [Google Scholar]
- 186.McEachern M.J., Krauskopf,A. and Blackburn,E.H. (2000) Telomeres and their control. Annu. Rev. Genet., 34, 331–358. [DOI] [PubMed] [Google Scholar]
- 187.Frenck R.W., Blackburn,E.H. and Shannon,K.M. (1998) The rate of telomere sequence loss in human leukocytes varies with age. Proc. Natl Acad. Sci. USA, 95, 5607–5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Li B.B., Oestreich,S. and de Lange,T. (2000) Identification of human Rap1: implications for telomere evolution. Cell, 101, 471–483. [DOI] [PubMed] [Google Scholar]
- 189.Smith S., Giriat,I., Schmitt,A. and de Lange,T. (1998) Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science, 282, 1484–1487. [DOI] [PubMed] [Google Scholar]
- 190.Smith S. and de Lange,T. (2000) Tankyrase promotes telomere elongation in human cells. Curr. Biol., 10, 1299–1302. [DOI] [PubMed] [Google Scholar]
- 191.Kaminker P.G., Kim,S.H., Taylor,R.D., Zebarjadian,Y., Funk,W.D., Morin,G.B., Yaswen,P. and Campisi,J. (2001) TANK2, a new TRF1-associated poly(ADP-ribose) polymerase, causes rapid induction of cell death upon overexpression. J. Biol. Chem., 276, 35891–35899. [DOI] [PubMed] [Google Scholar]
- 192.d’Adda di Fagagna F., Hande,M.P., Tong,W.M., Lansdorp,P.M., Wang,Z.Q. and Jackson,S.P. (1999) Functions of poly(ADP-ribose) polymerase in controling telomere length and chromosomal stability. Nature Genet., 23, 76–80. [DOI] [PubMed] [Google Scholar]
- 193.Samper E., Goytisolo,F.A., Ménissier-de Murcia,J., Gonzalez-Suarez,E., Cigudosa,J.C., de Murcia,G. and Blasco,M.A. (2001) Normal telomere length and chromosomal end capping in poly(ADP-ribose) polymerase deficient mice and primary cells despite increased chromosomal instability. J. Cell Biol., 154, 49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Zhou X.Z. and Lu,K.P. (2001) The Pin2/TRF1-interacting protein PinX1 is a potent telomerase inhibitor. Cell, 107, 347–359. [DOI] [PubMed] [Google Scholar]
- 195.Corda Y., Schramke,V., Longhese,M.P., Smokvina,T., Paciotti,V., Brevet,V., Gilson,E. and Geli,V. (1999) Interaction between Set1p and checkpoint protein Mec3p in DNA repair and telomere functions. Nature Genet., 21, 204–208. [DOI] [PubMed] [Google Scholar]
- 196.Ahmed S. and Hodgkin,J. (2000) MRT-2 checkpoint protein is required for germline immortality and telomere replication in C. elegans. Nature, 403, 159–164. [DOI] [PubMed] [Google Scholar]
- 197.Raymond E., Faivre,S., Dieras,V. and Von Hoff,D. (1997) Inhibition of telomeres and telomerase. Seeking for new anticancer drugs. Bull. Cancer, 84, 1123–1133. [PubMed] [Google Scholar]
- 198.Lloyd A.W. (1998) Human telomerase inhibitors. Drug Discov. Today, 3, 523. [Google Scholar]
- 199.Perry P.J. and Kelland,L.R. (1998) Telomeres and telomerase: targets for cancer chemotherapy? Expert Opin. Ther. Patents, 8, 1567–1586. [Google Scholar]
- 200.Pitts A.E. and Corey,D.R. (1999) The telomerase challenge—an unusual problem in drug discovery. Drug Discov. Today, 4, 155–161. [DOI] [PubMed] [Google Scholar]
- 201.Helder M.N., de Jong,S., de Vries,E.G.E. and van der Zee,A.G.J. (1999) Telomerase targeting in cancer treatment: new developments. Drug Resist. Update, 2, 104–115. [DOI] [PubMed] [Google Scholar]
- 202.Perry P.J. and Jenkins,T.C. (1999) Recent advances in the development of telomerase inhibitors for the treatment of cancer. Expert Opin. Investig. Drugs, 8, 1981–2008. [DOI] [PubMed] [Google Scholar]
- 203.Rowley P.T. and Tabler,M. (2000) Telomerase inhibitors. Anticancer Res., 20, 4419–4429. [PubMed] [Google Scholar]
- 204.Kelland L.R. (2000) Telomerase inhibitors: targeting the vulnerable end of cancer. Anticancer Drugs, 11, 503–513. [DOI] [PubMed] [Google Scholar]
- 205.White L.K., Wright,W.E. and Shay,J.W. (2001) Telomerase inhibitors. Trends Biotechnol., 19, 114–120. [DOI] [PubMed] [Google Scholar]
- 206.Kim N.W., Piatyszek,M.A., Prowse,K.R., Harley,C.B., West,M.D., Ho,P.L.C., Coviello,G.M., Wright,W.E., Weinrich,S.L. and Shay,J.W. (1994) Specific association of human telomerase activity with immortal cells and cancer. Science, 266, 2011–2015. [DOI] [PubMed] [Google Scholar]
- 207.Falchetti M.L., Levi,A., Molinari,P., Verna,R. and D’Ambrosio,E. (1998) Increased sensitivity and reproducibility of TRAP assay by avoiding direct primers interaction. Nucleic Acids Res., 26, 862–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Gonzalez-Quevedo R., DeJuan,C., Massa,M.J., Sanchez Pernaute,A., Torres,A., Balibrea,J.L., Benito,M. and Iniesta,P. (2000) Detection of telomerase activity in human carcinomas using a TRAP-ELISA method: correlation with hTR and hTERT expression. Int. J. Oncol., 16, 623–628. [DOI] [PubMed] [Google Scholar]
- 209.Hisatomi H., Kawaoto,Y., Ohmura,Y., Kuwahara,M., Yamauchi,T. and Hikiji,K. (1996) Detection of telomerase activity in hepatocellular carcinoma by fluorescence-based TRAP method. Int. Hepatol. Commun., 6, 29–35. [Google Scholar]
- 210.Hou M., Xu,D.W., Bjorkholm,M. and Gruber,A. (2001) Real-time quantitative telomeric repeat amplification protocol assay for the detection of telomerase activity. Clin. Chem., 47, 519–524. [PubMed] [Google Scholar]
- 211.Krupp G., Kuhne,K., Tamm,S., Klapper,W., Heidorn,K., Rott,A. and Parwaresch,R. (1997) Molecular basis of artifacts in the detection of telomerase activity and a modified primer for a more robust ‘TRAP’ assay. Nucleic Acids Res., 25, 919–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Savoysky E., Akamatsu,K., Tsuchiya,M. and Yamazaki,T. (1996) Detection of telomerase activity by combination of TRAP method and scintillation proximity assay (SPA). Nucleic Acids Res., 24, 1175–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Hayakawa N., Nozawa,K., Ogawa,A., Kato,N., Yoshida,K., Akamatsu,K., Tsuchiya,M., Nagasaka,A. and Yoshida,S. (1999) Isothiazolone derivatives selectively inhibit telomerase from human and rat cancer cells in vitro. Biochemistry, 38, 11501–11507. [DOI] [PubMed] [Google Scholar]
- 214.Strahl C. and Blackburn,E.H. (1994) The effects of nucleoside analogs on telomerase and telomeres in Tetrahymena. Nucleic Acids Res., 22, 893–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Fletcher T.M., Salazar,M. and Chen,S.F. (1996) Human telomerase inhibition by 7-deaza-2′-deoxypurine nucleoside triphosphates. Biochemistry, 35, 15611–15617. [DOI] [PubMed] [Google Scholar]
- 216.Fletcher T.M., Cathers,B.E., Ravikumar,K.S., Mamiya,B.M. and Kerwin,S.M. (2001) Inhibition of human telomerase by 7-deaza-2′-deoxyguanosine nucleoside triphosphate analogs: potent inhibition by 6-thio-7-deaza-2′-deoxyguanosine 5′-triphosphate. Bioorg. Chem., 29, 36–55. [DOI] [PubMed] [Google Scholar]
- 217.Gomez D.E., Kassim,A. and Olivero,O.A. (1995) Preferential incorporation of 3′-azido-2′,3′-dideoxythymidine (AZT) in telomeric sequences of CHO cells. Int. J. Oncol., 7, 1057–1060. [DOI] [PubMed] [Google Scholar]
- 218.Gomez D.E., Tejera,A.M. and Olivero,O.A. (1998) Irreversible telomere shortening by 3′-azido-2′,3′-dideoxythymidine (AZT) treatment. Biochem. Biophys. Res. Commun., 246, 107–110. [DOI] [PubMed] [Google Scholar]
- 219.Multani A.S., Furlong,C. and Pathak,S. (1998) Reduction of telomeric signals in murine melanoma and human breast cancer cell lines treated with 3′-azido-2′-3′-dideoxythymidine. Int. J. Oncol., 13, 923–925. [DOI] [PubMed] [Google Scholar]
- 220.Murakami J., Nagai,N., Shigemasa,K. and Ohama,K. (1999) Inhibition of telomerase activity and cell proliferation by a reverse transcriptase inhibitor in gynaecological cancer cell lines. Eur. J. Cancer., 35, 1027–1034. [DOI] [PubMed] [Google Scholar]
- 221.Strahl C. and Blackburn,E.H. (1996) Effects of reverse transcriptase inhibitors on telomere length and telomerase activity in two immortalized human cell lines. Mol. Cell. Biol., 16, 53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Tejera A.M., Alonso,D.F., Gomez,D.E. and Olivero,O.A. (2001) Chronic in vitro exposure to 3′-azido-2′,3′-dideoxythymidine induces senescence and apoptosis and reduces tumorigenicity of metastatic mouse mammary tumor cells. Breast Cancer Res. Treat., 65, 93–99. [DOI] [PubMed] [Google Scholar]
- 223.Yamaguchi T., Yamada,R., Tomikawa,A., Shudo,K., Saito,M., Ishikawa,F. and Saneyoshi,M. (2000) Recognition of 2′-deoxy-L-ribonucleoside 5′-triphosphates by human telomerase. Biochem. Biophys. Res. Commun., 279, 475–481. [DOI] [PubMed] [Google Scholar]
- 224.Tendian S.W. and Parker,W.B. (2000) Interaction of deoxyguanosine nucleotide analogs with human telomerase. Mol. Pharmacol., 57, 695–699. [DOI] [PubMed] [Google Scholar]
- 225.Ueno T., Takahashi,H., Oda,M., Mizunuma,M., Yokoyama,A., Goto,Y., Mizushina,Y., Sakaguchi,K. and Hayashi,H. (2000) Inhibition of human telomerase by rubromycins: implication of spiroketal system of the compounds as an active moiety. Biochemistry, 39, 5995–6002. [DOI] [PubMed] [Google Scholar]
- 226.Damm K., Hemmann,U., Garin-Chesa,P., Hauel,N., Kauffmann,I., Priepke,H., Niestroj,C., Daiber,C., Enenkel,B., Guilliard,B., Lauritsch,I., Müller,E., Pascolo,E., Sauter,G., Pantic,M., Martens,U.M., Wenz,C., Lingner,J., Kraut,N., Rettig,W.J. and Schnapp,A. (2001) A highly selective telomerase inhibitor limiting human cancer cell proliferation. EMBO J., 20, 6958–6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Schindler A., Fiedler,U., Meye,A., Schmidt,U., Fussel,S., Pilarsky,C., Herrmann,J. and Wirth,M.P. (2001) Human telomerase reverse transcriptase antisense treatment downregulates the viability of prostate cancer cells in vitro. Int. J. Oncol., 19, 25–30. [PubMed] [Google Scholar]
- 228.Fu W.M., Killen,M., Culmsee,C., Dhar,S., Pandita,T.K. and Mattson,M.P. (2000) The catalytic subunit of telomerase is expressed in developing brain neurons and serves a cell survival-promoting function. J. Mol. Neurosci., 14, 3–15. [DOI] [PubMed] [Google Scholar]
- 229.Yokoyama Y., Takahashi,Y., Shinohara,A., Wan,X.Y., Takahashi,S., Niwa,K. and Tamaya,T. (2000) The 5′-end of hTERT mRNA is a good target for hammerhead ribozyme to suppress telomerase activity. Biochem. Biophys. Res. Commun., 273, 316–321. [DOI] [PubMed] [Google Scholar]
- 230.Ludwig A., Saretzki,G., Holm,P.S., Tiemann,F., Lorenz,M., Emrich,T., Harley,C.B. and von Zglinicki,T. (2001) Ribozyme cleavage of telomerase mRNA sensitizes breast epithelial cells to inhibitors of topoisomerase. Cancer Res., 61, 3053–3061. [PubMed] [Google Scholar]
- 231.Chen J.L., Blasco,M.A. and Greider,C.W. (2000) Secondary structure of vertebrate telomerase RNA. Cell, 100, 503–514. [DOI] [PubMed] [Google Scholar]
- 232.Kondo S., Tanaka,Y., Kondo,Y., Hitomi,M., Barnett,G.H., Ishizaka,Y., Liu,J., Haqqi,T., Nishiyama,A., Villeponteau,B., Cowell,J.K. and Barna,B.P. (1998) Antisense telomerase treatment: induction of two distinct pathways, apoptosis and differentiation. FASEB J., 12, 801–811. [DOI] [PubMed] [Google Scholar]
- 233.Norton J.C., Piatyszek,M.A., Wright,W.E., Shay,J.W. and Corey,D.R. (1996) Inhibition of human telomerase activity by peptide nucleic acids. Nature Biotechnol., 14, 615–619. [DOI] [PubMed] [Google Scholar]
- 234.Hamilton S.E., Pitts,A.E., Katipally,R.R., Jia,X.Y., Rutter,J.P., Davies,B.A., Shay,J.W., Wright,W.E. and Corey,D.R. (1997) Identification of determinants for inhibitor binding within the RNA active site of human telomerase using PNA scanning. Biochemistry, 36, 11873–11880. [DOI] [PubMed] [Google Scholar]
- 235.Shammas M.A., Simmons,C.G., Corey,D.R. and Reis,R.J.S. (1999) Telomerase inhibition by peptide nucleic acids reverses ‘immortality’ of transformed human cells. Oncogene, 18, 6191–6200. [DOI] [PubMed] [Google Scholar]
- 236.Hamilton S.E., Simmons,C.G., Kathiriya,I.S. and Corey,D.R. (1999) Cellular delivery of peptide nucleic acids and inhibition of human telomerase. Chem. Biol., 6, 343–351. [DOI] [PubMed] [Google Scholar]
- 237.Harrison J.G., Frier,C., Laurant,R., Dennis,R., Raney,K.D. and Balasubramanian,S. (1999) Inhibition of human telomerase by PNA-cationic peptide conjugates. Bioorg. Med. Chem. Lett., 9, 1273–1278. [DOI] [PubMed] [Google Scholar]
- 238.Pitts A.E. and Corey,D.R. (1998) Inhibition of human telomerase by 2′-O-methyl-RNA. Proc. Natl Acad. Sci. USA, 95, 11549–11554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Herbert B.S., Pitts,A.E., Baker,S.I., Hamilton,S.E., Wright,W.E., Shay,J.W. and Corey,D.R. (1999) Inhibition of human telomerase in immortal human cells leads to progressive telomere shortening and cell death. Proc. Natl Acad. Sci. USA, 96, 14276–14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Elayadi A.N., Demieville,A., Wancewicz,E.V., Monia,B.P. and Corey,D.R. (2001) Inhibition of telomerase by 2′-O-(2-methoxyethyl) RNA oligomers: effect of length, phosphorothioate substitution and time inside cells. Nucleic Acids Res., 29, 1683–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Kondo S., Kondo,Y., Li,G., Silverman,R.H. and Cowell,J.K. (1998) Targeted therapy of human malignant glioma in a mouse model by 2-5A antisense directed against telomerase RNA. Oncogene, 16, 3323–3330. [DOI] [PubMed] [Google Scholar]
- 242.Kushner D.M., Jayashree,J.M., Bandyopadhyay,B., Cramer,H., Leaman,D.W., Kennedy,A.W., Silverman,R.H. and Cowell,J.K. (2000) 2-5A antisense directed against telomerase RNA produces apoptosis in ovarian cancer cells. Gynecol. Oncol., 76, 183–192. [DOI] [PubMed] [Google Scholar]
- 243.Kondo Y., Koga,S., Komata,T. and Kondo,S. (2000) Treatment of prostate cancer in vitro and in vivo with 2-5A-anti-telomerase RNA component. Oncogene, 19, 2205–2211. [DOI] [PubMed] [Google Scholar]
- 244.Mukai S., Kondo,Y., Koga,S., Komata,T., Barna,B.P. and Kondo,S. (2000) 2-5A antisense telomerase RNA therapy for intracranial malignant gliomas. Cancer Res., 60, 4461–4467. [PubMed] [Google Scholar]
- 245.Komata T., Kondo,Y., Koga,S., Ko,S.C., Chung,L.W.K. and Kondo,S. (2000) Combination therapy of malignant glioma cells with 2-5A-antisense telomerase RNA and recombinant adenovirus p53. Gene Ther., 7, 2071–2079. [DOI] [PubMed] [Google Scholar]
- 246.Gryaznov S.M., Pongracz,K., Matray,T., Schultz,R., Pruzan,R., Aimi,J., Chin,A., Harley,C., Shea-Herbert,B., Shay,J., Oshima,Y., Asai,A. and Yamashita,Y. (2001) Telomerase inhibitors—oligonucleotide phosphoramidates as potential therapeutic agents. Nucl. Nucl., 20, 401–410. [DOI] [PubMed] [Google Scholar]
- 247.Kanazawa Y., Ohkawa,K., Ueda,K., Mita,E., Takehara,T., Sasaki,Y., Kasahara,A. and Hayashi,N. (1996) Hammerhead ribozyme-mediated inhibition of telomerase activity in extracts of human hepatocellular carcinoma cells. Biochem. Biophys. Res. Commun., 225, 570–576. [DOI] [PubMed] [Google Scholar]
- 248.Yokoyama Y., Takahashi,Y., Shinohara,A., Lian,Z.L., Wan,X.Y., Niwa,K. and Tamaya,T. (1998) Attenuation of telomerase activity by a hammerhead ribozyme targeting the template region of telomerase RNA in endometrial carcinoma cells. Cancer Res., 58, 5406–5410. [PubMed] [Google Scholar]
- 249.Folini M., Colella,G., Villa,R., Lualdi,S., Daidone,M.G. and Zaffaroni,N. (2000) Inhibition of telomerase activity by a hammerhead ribozyme targeting the RNA component of telomerase in human melanoma cells. J. Invest. Dermatol., 114, 259–267. [DOI] [PubMed] [Google Scholar]
- 250.Francis R., West,C. and Friedman,S.H. (2001) Targeting telomerase via its key RNA/DNA heteroduplex. Bioorg. Chem., 29, 107–117. [DOI] [PubMed] [Google Scholar]
- 251.Ren J.S. and Chaires,J.B. (1999) Sequence and structural selectivity of nucleic acid binding ligands. Biochemistry, 38, 16067–16075. [DOI] [PubMed] [Google Scholar]
- 252.Ren J., Qu,X., Dattagupta,N. and Chaires,J.B. (2001) Molecular recognition of a RNA:DNA hybrid structure. J. Am. Chem. Soc., 123, 6742–6743. [DOI] [PubMed] [Google Scholar]
- 253.Kim M.M., Rivera,M.A., Botchkina,I.L., Shalaby,R., Thor,A.D. and Blackburn,E.H. (2001) A low threshold level of expression of mutant-template telomerase RNA inhibits human tumor cell proliferation. Proc. Natl Acad. Sci. USA, 98, 7982–7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Guiducci C., Cerone,M.A. and Bacchetti,S. (2001) Expression of mutant telomerase in immortal telomerase-negative human cells results in cell cycle deregulation, nuclear and chromosomal abnormalities and rapid loss of viability. Oncogene, 20, 714–725. [DOI] [PubMed] [Google Scholar]
- 255.Liu Y., Snow,B.E., Hande,M.P., Baerlocher,G., Kickhoefer,V.A., Yeung,D., Wakeham,A., Itie,A., Siderovski,D.P., Lansdorp,P.M., Robinson,M.O. and Harrington,L. (2000) Telomerase-associated protein TEP1 is not essential for telomerase activity or telomere length maintenance in vivo. Mol. Cell. Biol., 20, 8178–8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Kickhoefer V.A., Liu,Y., Kong,L.B., Snow,B.E., Stewart,P.L., Harrington,L. and Rome,L.H. (2001) The telomerase/vault-associated protein TEP1 is required for vault RNA stability and its association with the vault particle. J. Cell Biol., 152, 157–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Holt S.E., Aisner,D.L., Baur,J., Tesmer,V.M., Dy,M., Ouellette,M., Trager,J.B., Morin,G.B., Toft,D.O., Shay,J.W., Wright,W.E. and White,M.A. (1999) Functional requirement of p23 and Hsp90 in telomerase complexes. Genes Dev., 13, 817–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Masutomi K., Kaneko,S., Hayashi,N., Yamashita,T., Shirota,Y., Kobayashi,K. and Murakami,S. (2000) Telomerase activity reconstituted in vitro with purified human telomerase reverse transcriptase and human telomerase RNA component. J. Biol. Chem., 275, 22568–22573. [DOI] [PubMed] [Google Scholar]
- 259.Wadhwa R., Sugihara,T., Yoshida,A., Nomura,H., Reddel,R.R., Simpson,R., Maruta,H. and Kaul,S.C. (2000) Selective toxicity of MKT-077 to cancer cells is mediated by its binding to the hsp70 family protein mot-2 and reactivation of p53 function. Cancer Res., 60, 6818–6821. [PubMed] [Google Scholar]
- 260.Ishii K., Yang,W.L., Cvijic,M.E., Kikuchi,Y., Nagata,I. and Chin,K.V. (2000) Telomere shortening by cisplatin in yeast nucleotide excision repair mutant. Exp. Cell Res., 255, 95–101. [DOI] [PubMed] [Google Scholar]
- 261.Redon S., Bombard,S., Elizondo-Riojas,M.A. and Chottard,J.C. (2001) Platination of the (T2G4)4 telomeric sequence: a structural and crosslinking story. Biochemistry, 40, 8463–8470. [DOI] [PubMed] [Google Scholar]
- 262.Burger A.M., Double,J.A. and Newell,D.R. (1997) Inhibition of telomerase activity by cisplatin in human testicular cancer cells. Eur. J. Cancer, 33, 638–644. [DOI] [PubMed] [Google Scholar]
- 263.Lin Z.S., Lim,S., Viani,M.A., Sapp,M. and Lim,M.S. (2001) Down-regulation of telomerase activity in malignant lymphomas by radiation and chemotherapeutic agents. Am. J. Pathol., 159, 711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Yoon H.J., Choi,I.Y., Kang,M.R., Kim,S.S., Muller,M.T., Spitzner,J.R. and Chung,I.K. (1998) DNA topoisomerase II cleavage of telomeres in vitro and in vivo. Biochim. Biophys. Acta, 1395, 110–120. [DOI] [PubMed] [Google Scholar]
- 265.Lee C.C. and Huang,T.S. (2001) A novel topoisomerase II poison GL331 preferentially induces DNA cleavage at (C/G)T sites and can cause telomere DNA damage. Pharm. Res., 18, 846–851. [DOI] [PubMed] [Google Scholar]
- 266.Michishita E., Nakabayashi,K., Ogino,H., Suzuki,T., Fujii,M. and Ayusawa,D. (1998) DNA topoisomerase inhibitors induce reversible senescence in normal human fibroblasts. Biochem. Biophys. Res. Commun., 253, 667–671. [DOI] [PubMed] [Google Scholar]
- 267.Sawant S.G., Gregoire,V., Dhar,S., Umbricht,C.B., Cvilic,S., Sukumar,S. and Pandita,T.K. (1999) Telomerase activity as a measure for monitoring radiocurability of tumor cells. FASEB J., 13, 1047–1054. [DOI] [PubMed] [Google Scholar]
- 268.Terashima M., Ogawa,Y., Toda,K., Nishioka,A., Inomata,T., Kubonishi,I., Taguchi,H., Yoshida,S. and Shizuta,Y. (1998) Effects of irradiation on telomerase activity in human lymphoma and myeloma cell lines. Int. J. Mol. Med., 2, 567–571. [DOI] [PubMed] [Google Scholar]
- 269.Jacob N.K., Skopp,R. and Price,C.M. (2001) G-overhang dynamics at Tetrahymena telomeres. EMBO J., 20, 4299–4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Griffith J.D., Comeau,L., Rosenfield,S., Stansel,R.M., Bianchi,A., Moss,H. and de Lange,T. (1999) Mammalian telomeres end in a large duplex loop. Cell, 97, 503–514. [DOI] [PubMed] [Google Scholar]
- 271.Voloshin O.N., Veselkov,A.G., Belotserkovskii,B.P., Danilevskaya,O.N., Pavlova,M.N., Dobrynin,V.N. and Frank-Kamenetskii,M.D. (1992) An eclectic DNA structure adopted by human telomeric sequence under superhelical stress and low pH. J. Biomol. Struct. Dyn., 9, 643–652. [DOI] [PubMed] [Google Scholar]
- 272.Aboul-ela F., Murchie,A.I.H., Norman,D.G. and Lilley,D.M.J. (1994) Solution structure of a parallel-stranded tetraplex formed by d(TG(4)T) in the presence of sodium ions by nuclear magnetic resonance spectroscopy. J. Mol. Biol., 243, 458–471. [DOI] [PubMed] [Google Scholar]
- 273.Wang Y. and Patel,D.J. (1993) Solution structure of a parallel-stranded G-quadruplex DNA. J. Mol. Biol., 234, 1171–1183. [DOI] [PubMed] [Google Scholar]
- 274.Smith F.W., Lau,F.W. and Feigon,J. (1994) d(G(3)T(4)G(3)) forms an asymmetric diagonally looped dimeric quadruplex with guanosine 5′-syn-syn-anti and 5′-syn-anti-anti N-glycosidic conformations. Proc. Natl Acad. Sci. USA, 91, 10546–10550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Schultze P., Hud,N.V., Smith,F.W. and Feigon,J. (1999) The effect of sodium, potassium and ammonium ions on the conformation of the dimeric quadruplex formed by the Oxytricha nova telomere repeat oligonucleotide d(G(4)T(4)G(4)). Nucleic Acids Res., 27, 3018–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Smith F.W. and Feigon,J. (1992) Quadruplex structure of oxytricha telomeric DNA oligonucleotides. Nature, 356, 164–168. [DOI] [PubMed] [Google Scholar]
- 277.Wang Y. and Patel,D.J. (1993) Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure, 1, 263–282. [DOI] [PubMed] [Google Scholar]
- 278.Kerwin S.M. (2000) G-quadruplex DNA as a target for drug design. Curr. Pharm. Des., 6, 441–471. [DOI] [PubMed] [Google Scholar]
- 279.Frantz J.D. and Gilbert,W. (1995) A novel yeast gene product, G4p1, with a specific affinity for quadruplex nucleic acids. J. Biol. Chem., 270, 20692–20697. [DOI] [PubMed] [Google Scholar]
- 280.Schierer T. and Henderson,E. (1994) A protein from Tetrahymena thermophila that specifically binds parallel-stranded G4-DNA. Biochemistry, 33, 2240–2246. [DOI] [PubMed] [Google Scholar]
- 281.Muniyappa K., Anuradha,S. and Byers,B. (2000) Yeast meiosis-specific protein Hop1 binds to G4 DNA and promotes its formation. Mol. Cell. Biol., 20, 1361–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Arimondo P., Riou,J.F., Mergny,J.L., Tazi,J., Sun,J.S., Garestier,T. and Hélène,C. (2000) Interaction of human DNA topoisomerase I with intermolecular G-quartet structures. Nucleic Acids Res., 28, 4832–4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Harrington C., Lan,Y. and Akman,S.A. (1997) The identification and characterization of a G4-DNA resolvase activity. J. Biol. Chem., 272, 24631–24636. [DOI] [PubMed] [Google Scholar]
- 284.Baran N., Pucshansky,L., Marco,Y., Benjamin,S. and Manor,H. (1997) The SV40 large T-antigen helicase can unwind four stranded DNA structures linked by G-quartets. Nucleic Acids Res., 25, 297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Sun H., Bennett,R.J. and Maizels,N. (1999) The Saccharomyces cerevisiae Sgs1 helicase efficiently unwinds G-G paired DNAs. Nucleic Acids Res., 27, 1978–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Giraldo R. and Rhodes,D. (1994) The yeast telomere-binding protein RAP1 binds to and promotes the formation of DNA quadruplexes in telomeric DNA. EMBO J., 13, 2411–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Johnson F.B., Marciniak,R.A., McVey,M., Stewart,S.A., Hahn,W.C. and Guarente,L. (2001) The Saccharomyces cerevisiae WRN homolog Sgs1p participates in telomere maintenance in cells lacking telomerase. EMBO J., 20, 905–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Cohen H. and Sinclair,D.A. (2001) Recombination-mediated lengthening of terminal telomeric repeats requires the Sgs1 DNA helicase. Proc. Natl Acad. Sci. USA, 98, 3174–3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Huang P.H., Pryde,F.E., Lester,D., Maddison,R.L., Borts,R.H., Hickson,I.D. and Louis,E.J. (2001) SGS1 is required for telomere elongation in the absence of telomerase. Curr. Biol., 11, 125–129. [DOI] [PubMed] [Google Scholar]
- 290.Mergny J.L. and Hélène,C. (1998) G-quadruplex DNA: a target for drug design. Nature Med., 4, 1366–1367. [DOI] [PubMed] [Google Scholar]
- 291.Mergny J.L., Mailliet,P., Lavelle,F., Riou,J.F., Laoui,A. and Hélène,C. (1999) The development of telomerase inhibitors: the G-quartet approach. Anticancer Drug Des., 14, 327–339. [PubMed] [Google Scholar]
- 292.Mergny J.L., Duval-Valentin,G., Nguyen,C.H., Perrouault,L., Faucon,B., Rougée,M., Montenay-Garestier,T., Bisagni,E. and Hélène,C. (1992) Triple helix specific ligands. Science, 256, 1691–1694. [DOI] [PubMed] [Google Scholar]
- 293.Sun D., Thompson,B., Cathers,B.E., Salazar,M., Kerwin,S.M., Trent,J.O., Jenkins,T.C., Neidle,S. and Hurley,L.H. (1997) Inhibition of human telomerase by a G-quadruplex-interactive compound. J. Med. Chem., 40, 2113–2116. [DOI] [PubMed] [Google Scholar]
- 294.Wheelhouse R.T., Sun,D., Han,H., Han,F.X. and Hurley,L.H. (1998) Cationic porphyrins as telomerase inhibitors: the interaction of tetra (N-methyl-4-pyridyl) porphyrin with quadruplex DNA. J. Am. Chem. Soc., 120, 3261–3262. [Google Scholar]
- 295.Anantha N.V., Azam,M. and Sheardy,R.D. (1998) Porphyrin binding to quadrupled T4G4. Biochemistry, 37, 2709–2714. [DOI] [PubMed] [Google Scholar]
- 296.Arthanari H., Basu,S., Kawano,T.L. and Bolton,P.H. (1998) Fluorescent dyes specific for quadruplex DNA. Nucleic Acids Res., 26, 3724–3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Han F.X.G., Wheelhouse,R.T. and Hurley,L.H. (1999) Interactions of TMPyP4 and TMPyP2 with quadruplex DNA. Structural basis for the differential effects on telomerase inhibition. J. Am. Chem. Soc., 121, 3561–3570. [Google Scholar]
- 298.Fedoroff O.Y., Salazar,M., Han,H., Chemeris,V.V., Kerwin,S.M. and Hurley,L.H. (1998) NMR-based model of a telomerase inhibiting compound bound to G-quadruplex DNA. Biochemistry, 37, 12367–12374. [DOI] [PubMed] [Google Scholar]
- 299.Perry P.J., Reszka,A.P., Wood,A.A., Read,M.A., Gowan,S.M., Dosanjh,H.S., Trent,J.O., Jenkins,T.C., Kelland,L.R. and Neidle,S. (1998) Human telomerase inhibition by regioisomeric disubstituted amidoanthracene-9,10-diones. J. Med. Chem., 41, 4873–4884. [DOI] [PubMed] [Google Scholar]
- 300.Perry P.J., Read,M.A., Davies,R.T., Gowan,S.M., Reszka,A.P., Wood,A.A., Kelland,L.R. and Neidle,S. (1999) 2,7-Disubstituted amidofluorenone derivatives as inhibitors of human telomerase. J. Med. Chem., 42, 2679–2684. [DOI] [PubMed] [Google Scholar]
- 301.Caprio V., Guyen,B., OpokuBoahen,Y., Mann,J., Gowan,S.M., Kelland,L.M., Read,M.A. and Neidle,S. (2000) A novel inhibitor of human telomerase derived from 10H-indolo[3,2-b]quinoline. Bioorg. Med. Chem. Lett., 10, 2063–2066. [DOI] [PubMed] [Google Scholar]
- 302.Read M., Harrison,R.J., Romagnoli,B., Tanious,F.A., Gowan,S.H., Reszka,A.P., Wilson,W.D., Kelland,L.R. and Neidle,S. (2001) Structure-based design of selective and potent G quadruplex-mediated telomerase inhibitors. Proc. Natl Acad. Sci. USA, 98, 4844–4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Mergny J.L., Lacroix,L., Teulade-Fichou,M.P., Hounsou,C., Guittat,L., Hoarau,M., Arimondo,P.B., Vigneron,J.P., Lehn,J.M., Riou,J.F., Garestier,T. and Hélène,C. (2001) Telomerase inhibitors based on quadruplex ligands selected by a fluorescent assay. Proc. Natl Acad. Sci. USA, 98, 3062–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Koeppel F., Riou,J.F., Laoui,A., Mailliet,P., Arimondo,P.B., Labit,D., Petigenet,O., Hélène,C. and Mergny,J.L. (2001) Ethidium derivatives bind to G-quartets, inhibit telomerase and act as fluorescent probes for quadruplexes. Nucleic Acids Res., 29, 1087–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Mailliet P., Riou,J.F., Mergny,J.L., Laoui,A., Lavelle,F. and Petitgenet,O. (2001) Arylamine derivatives and their use as antitelomerase agents. Patent WO 0140218, Aventis-Pharma SA.
- 306.Gowan S., Heald,R., Stevens,M. and Kelland,L. (2001) Potent inhibition of telomerase by small-molecule pentacyclic acridines capable of interacting with g-quadruplexes. Mol. Pharmacol., 60, 981–988. [DOI] [PubMed] [Google Scholar]
- 307.Mergny J.L. and Maurizot,J.C. (2001) Fluorescence resonance energy transfer as a probe for G-quartet formation by a telomeric repeat. Chem. Biochem., 2, 124–132. [DOI] [PubMed] [Google Scholar]
- 308.Riou J.F., Guittat,L., Renou,E., Mailliet,P., Laoui,A., Petigenet,O., Hélène,C. and Mergny,J.L. (2002) Cell senescence and telomere shortening induced by a new series of specific quadruplex DNA ligands. Proc. Natl Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Alberti P., Ren,J., Teulade-Fichou,M.P., Guittat,L., Riou,J.F., Chaires,J.B., Hélène,C., Vigneron,J.P., Lehn,J.M. and Mergny,J.L. (2001) Interaction of an acridine dimer with DNA quadruplex structures. J. Biomol. Struct. Dyn., 19, 505–513. [DOI] [PubMed] [Google Scholar]
- 310.Han H., Langley,D.R., Rangan,A. and Hurley,L.H. (2001) Selective interactions of cationic porphyrins with G-quadruplex structures. J. Am. Chem. Soc., 123, 8902–8913. [DOI] [PubMed] [Google Scholar]
- 311.Haq I., Trent,J.O., Chowdhry,B.Z. and Jenkins,T.C. (1999) Intercalative G-tetraplex stabilization of telomeric DNA by a cationic porphyrin. J. Am. Chem. Soc., 121, 1768–1779. [Google Scholar]
- 312.Tuntiwechapikul W., Jeong,T.L. and Salazar,M. (2001) Design and synthesis of the G-quadruplex-specific cleaving reagent perylene-EDTA.iron(II). J. Am. Chem. Soc., 123, 5606–5607. [DOI] [PubMed] [Google Scholar]
- 313.Rangan A., Fedoroff,O.Y. and Hurley,L.H. (2001) Induction of duplex to G-quadruplex transition in the c-myc promoter region by a small molecule. J. Biol. Chem., 276, 4640–4646. [DOI] [PubMed] [Google Scholar]
- 314.Azzalin C.M., Mucciolo,E., Bertoni,L. and Giulotto,E. (1997) Fluorescence in situ hybridization with a synthetic (T(2)AG(3))(n) polynucleotide detects several intrachromosomal telomere-like repeats on human chromosomes. Cytogenet. Cell Genet., 78, 112–115. [DOI] [PubMed] [Google Scholar]
- 315.Azzalin C.M., Nergadze,S.G. and Giulotto,E. (2001) Human intrachromosomal telomeric-like repeats: sequence organization and mechanisms of origin. Chromosoma, 110, 75–82. [DOI] [PubMed] [Google Scholar]
- 316.Kim J., Cheong,C. and Moore,P.B. (1991) Tetramerization of an RNA oligonucleotide containing a GGGG sequence. Nature, 351, 331–332. [DOI] [PubMed] [Google Scholar]
- 317.Sugimoto N., Ohmichi,T. and Sasaki,M. (1996) The stability of DNA and RNA G-quartets. Nucleosides Nucleotides, 15, 559–567. [Google Scholar]
- 318.Schaeffer C., Bardoni,B., Mandel,J.L., Ehresmann,B., Ehresmann,C. and Moine,H. (2001) The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J., 20, 4803–4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Darnell J.C., Jensen,K.B., Jin,P., Brown,V., Warren,S.T. and Darnell,R.B. (2001) Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell, 107, 489–499. [DOI] [PubMed] [Google Scholar]
- 320.Han H.Y., Bennett,R.J. and Hurley,L.H. (2000) Inhibition of unwinding of G-quadruplex structures by Sgs1 helicase in the presence of N,N′-bis[2-(1-piperidino)ethyl]-3,4,9,10-perylenetetracarboxylic diimide, a G-quadruplex-interactive ligand. Biochemistry, 39, 9311–9316. [DOI] [PubMed] [Google Scholar]
- 321.Wu X. and Maizels,N. (2001) Substrate-specific inhibition of RecQ helicase. Nucleic Acids Res., 29, 1765–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Izbicka E., Wheelhouse,R.T., Raymond,E., Davidson,K.K., Lawrence,R.A., Sun,D.Y., Windle,B.E., Hurley,L.H. and Von Hoff,D.D. (1999) Effects of cationic porphyrins as G-quadruplex interactive agents in human tumor cells. Cancer Res., 59, 639–644. [PubMed] [Google Scholar]
- 323.Marathias V.M., Sawicki,M.J. and Bolton,P.H. (1999) 6-Thioguanine alters the structure and stability of duplex DNA and inhibits quadruplex DNA formation. Nucleic Acids Res., 27, 2860–2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Page T.J., Mata,J.E., Bridge,J.A., Siebler,J.C., Neff,J.R. and Iversen,P.L. (1999) The cytotoxic effects of single-stranded telomere mimics on OMA-BL1 cells. Exp. Cell Res., 252, 41–49. [DOI] [PubMed] [Google Scholar]
- 325.Maeshima K., Janssen,S. and Laemmli,U.K. (2001) Specific targeting of insect and vertebrate telomeres with pyrrole and imidazole polyamides. EMBO J., 20, 3218–3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Abdul-Ghani R., Ohana,P., Matouk,I., Ayesh,S., Ayesh,B., Laster,M., Bibi,O., Giladi,H., Molnar Kimber,K., Sughayer,M.A., de Groot,N. and Hochberg,A. (2000) Use of transcriptional regulatory sequences of telomerase (HTER and hTERT) for selective killing of cancer cells. Mol. Ther., 2, 539–544. [DOI] [PubMed] [Google Scholar]
- 327.Gu J., Kagawa,S., Takakura,M., Kyo,S., Inoue,M., Roth,J.A. and Fang,B.L. (2000) Tumor-specific transgene expression from the human telomerase reverse transcriptase promoter enables targeting of the therapeutic effects of the Bax gene to cancers. Cancer Res., 60, 5359–5364. [PubMed] [Google Scholar]
- 328.Koga S., Hirohata,S., Kondo,Y., Komata,T., Takakura,M., Inoue,M., Kyo,S. and Kondo,S. (2000) A novel telomerase-specific gene therapy: gene transfer of caspase-8 utilizing the human telomerase catalytic subunit gene promoter. Hum. Gene Ther., 11, 1397–1406. [DOI] [PubMed] [Google Scholar]
- 329.Komata T., Kondo,Y., Kanzawa,T., Hirohata,S., Koga,S., Sumiyoshi,H., Srinivasula,S.M., Barna,B.P., Germano,I.M., Takakura,M., Inoue,M., Alnemri,E.S., Shay,J.W., Kyo,S. and Kondo,S. (2001) Treatment of malignant glioma cells with the transfer of constitutively active caspase-6 using the human telomerase catalytic subunit (human telomerase reverse transcriptase) gene promoter. Cancer Res., 61, 5796–5802. [PubMed] [Google Scholar]
- 330.Majumdar A.S., Hughes,D.E., Lichtsteiner,S.P., Wang,Z., Lebkowski,J.S. and Vasserot,A.P. (2001) The telomerase reverse transcriptase promoter drives efficacious tumor suicide gene therapy while preventing hepatotoxicity encountered with constitutive promoters. Gene Ther., 8, 568–578. [DOI] [PubMed] [Google Scholar]
- 331.Vonderheide R.H., Hahn,W.C., Schultze,J.L. and Nadler,L.M. (1999) The telomerase catalytic subunit is a widely expressed tumor-associated antigen recognized by cytotoxic T lymphocytes. Immunity, 10, 673–679. [DOI] [PubMed] [Google Scholar]
- 332.Minev B., Hipp,J., Firat,H., Schmidt,J.D., Langlade Demoyen,P. and Zanetti,M. (2000) Cytotoxic T cell immunity against telomerase reverse transcriptase in humans. Proc. Natl Acad. Sci. USA, 97, 4796–4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Nair S.K., Heiser,A., Boczkowski,D., Majumdar,A., Naoe,M., Lebkowski,J.S., Vieweg,J. and Gilboa,E. (2000) Induction of cytotoxic T cell responses and tumor immunity against unrelated tumors using telomerase reverse transcriptase RNA transfected dendritic cells. Nature Med., 6, 1011–1017. [DOI] [PubMed] [Google Scholar]
- 334.Gozlan M. (2000) Telomerase reverse transcriptase: candidate for “universal cancer vaccine”? Lancet, 355, 1337.10776754 [Google Scholar]
- 335.Arai J., Yasukawa,M., Ohminami,H., Kakimoto,M., Hasegawa,A. and Fujita,S. (2001) Identification of human telomerase reverse transcriptase-derived peptides that induce HLA-A24-restricted antileukemia cytotoxic T lymphocytes. Blood, 97, 2903–2907. [DOI] [PubMed] [Google Scholar]
- 336.Ayyoub M., Migliaccio,M., Guillaume,P., Lienard,D., Cerottini,J.C., Romero,P., Levy,F., Speiser,D.E. and Valmori,D. (2001) Lack of tumor recognition by hTERT peptide 540–548-specific CD8(+) T cells from melanoma patients reveals inefficient antigen processing. Eur. J. Immunol., 31, 2642–2651. [DOI] [PubMed] [Google Scholar]
- 337.Yamakuchi M., Nakata,M., Kawahara,K., Kitajima,I. and Maruyama,I. (1997) New quinolones, ofloxacin and levofloxacin, inhibit telomerase activity in transitional cell carcinoma cell lines. Cancer Lett., 119, 213–219. [DOI] [PubMed] [Google Scholar]
- 338.Sasaki S., Ehara,T., Sakata,I., Fujino,Y., Harada,N., Kimura,J., Nakamura,H. and Maeda,M. (2001) Development of novel telomerase inhibitors based on a bisindole unit. Bioorg. Med. Chem. Lett., 11, 583–585. [DOI] [PubMed] [Google Scholar]
- 339.Naasani I., Seimiya,H. and Tsuruo,T. (1998) Telomerase inhibition, telomere shortening and senescence of cancer cells by tea catechins. Biochem. Biophys. Res. Commun., 249, 391–396. [DOI] [PubMed] [Google Scholar]
- 340.Togashi K., Kakeya,H., Morishita,M., Song,Y.X. and Osada,H. (1998) Inhibition of human telomerase activity by alterperylenol. Oncol. Res., 10, 449–453. [PubMed] [Google Scholar]
- 341.Tabata Y., Ikegami,S., Yaguchi,T., Sasaki,T., Hoshiko,S., Sakuma,S., ShinYa,K. and Seto,H. (1999) Diazaphilonic acid, a new azaphilone with telomerase inhibitory activity. J. Antibiot., 52, 412–414. [DOI] [PubMed] [Google Scholar]
- 342.Naasani I., Seimiya,H., Yamori,T. and Tsuruo,T. (1999) FJ5002: a potent telomerase inhibitor identified by exploiting the disease-oriented screening program with COMPARE analysis. Cancer Res., 59, 4004–4011. [PubMed] [Google Scholar]
- 343.Shin-ya K., Wierzba,K., Matsuo,K., Ohtani,T., Yamada,Y., Furihata,K., Hayakawa,Y. and Seto,H. (2001) Telomestatin, a novel telomerase inhibitor from Streptomyces anulatus. J. Am. Chem. Soc., 123, 1262–1263. [DOI] [PubMed] [Google Scholar]
- 344.Alfonso De Matte M.Y., Cheng,J.Q. and Kruk,P.A. (2001) Ultraviolet irradiation- and dimethyl sulfoxide-induced telomerase activity in ovarian epithelial cell lines. Exp. Cell Res., 267, 13–27. [DOI] [PubMed] [Google Scholar]
- 345.Li B.B. and Lustig,A.J. (1996) A novel mechanism for telomere size control in Saccharomyces cerevisiae. Genes Dev., 10, 1310–1326. [DOI] [PubMed] [Google Scholar]
- 346.Bucholc M., Park,Y. and Lustig,A.J. (2001) Intrachromatid excision of telomeric DNA as a mechanism for telomere size control in Saccharomyces cerevisiae. Mol. Cell. Biol., 21, 6559–6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Kurunaga N., Shinomiya,N. and Mochizuki,H. (2001) Long term cultivation of colorectal carcinoma cells with anti-cancer drugs induces drug resistance and telomere elongation: an in vitro study. BMC Cancer, 1, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Reddel R.R., Bryan,T.M., Colgin,L.M., Perrem,K.T. and Yeager,T.R. (2001) Alternative lengthening of telomeres in human cells. Radiat. Res., 155, 194–200. [DOI] [PubMed] [Google Scholar]
- 349.Bryan T.M., Englezou,A., Dalla Pozza,L., Dunham,M.A. and Reddel,R.R. (1997) Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nature Med., 3, 1271–1274. [DOI] [PubMed] [Google Scholar]
- 350.Dunham M.A., Neumann,A.A., Fasching,C.L. and Reddel,R.R. (2000) Telomere maintenance by recombination in human cells. Nature Genet., 26, 447–450. [DOI] [PubMed] [Google Scholar]
- 351.Ford L.P., Zou,Y., Pongracz,K., Gryaznov,S.M., Shay,J.W. and Wright,W.E. (2001) Telomerase can inhibit the recombination based pathway of telomere maintenance in human cells. J. Biol. Chem., 276, 32198–32203. [DOI] [PubMed] [Google Scholar]
- 352.Perrem K., Colgin,L.M., Neumann,A.A., Yeager,T.R. and Reddel,R.R. (2001) Coexistence of alternative lengthening of telomeres and telomerase in hTERT-transfected GM847 cells. Mol. Cell. Biol., 21, 3862–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Perrem K., Bryan,T.M., Englezou,A., Hackl,T., Moy,E.L. and Reddel,R.R. (1999) Repression of an alternative mechanism for lengthening of telomeres in somatic cell hybrids. Oncogene, 18, 3383–3390. [DOI] [PubMed] [Google Scholar]
- 354.Katoh M., Katoh,M., Kameyama,M., Kugoh,H., Shimizu,M. and Oshimura,M. (1998) A repressor function for telomerase activity in telomerase-negative immortal cells. Mol. Carcinog., 21, 17–25. [DOI] [PubMed]
- 355.Zhang X.L., Mar,V., Zhou,W., Harrington,L. and Robinson,M.O. (1999) Telomere shortening and apoptosis in telomerase-inhibited human tumor cells. Genes Dev., 13, 2388–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Huffman K.E., Levene,S.D., Tesmer,V.M., Shay,J.W. and Wright,W.E. (2000) Telomere shortening is proportional to the size of the G-rich telomeric 3′-overhang. J. Biol. Chem., 275, 19719–19722. [DOI] [PubMed] [Google Scholar]
- 357.Blackburn E.H., Chan,S., Chang,J., Fulton,T.B., Krauskopf,A., McEachern,M., Prescott,J., Roy,J., Smith,C. and Wang,H. (2000) Molecular manifestations and molecular determinants of telomere capping. Cold Spring Harb. Symp. Quant. Biol., 65, 253–263. [DOI] [PubMed] [Google Scholar]
- 358.Prescott J. and Blackburn,E.H. (1997) Telomerase RNA mutations in Saccharomyces cerevisiae alter telomerase action and reveal nonprocessivity in vivo and in vitro. Genes Dev., 11, 528–540. [DOI] [PubMed] [Google Scholar]
- 359.Zhu J.Y., Wang,H., Bishop,J.M. and Blackburn,E.H. (1999) Telomerase extends the lifespan of virus-transformed human cells without net telomere lengthening. Proc. Natl Acad. Sci. USA, 96, 3723–3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Blackburn E.H. (2000) Telomere states and cell fates. Nature, 408, 53–56. [DOI] [PubMed] [Google Scholar]
- 361.Lee K.H., Rudolph,K.L., Ju,Y.J., Greenberg,R.A., Cannizzaro,L., Chin,L., Weiler,S.R. and DePinho,R.A. (2001) Telomere dysfunction alters the chemotherapeutic profile of transformed cells. Proc. Natl Acad. Sci. USA, 98, 3381–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Goytisolo F.A., Samper,E., Martin-Caballero,J., Finnon,P., Herrera,E., Flores,J.M., Bouffler,S.D. and Blasco,M.A. (2000) Short telomeres result in organismal hypersensitivity to ionizing radiation in mammals. J. Exp. Med., 192, 1625–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Wong K.K., Chang,S., Weiler,S.R., Ganesan,S., Chaudhuri,J., Zhu,C.M., Artandi,S.E., Rudolph,K.L., Gottlieb,G.J., Chin,L., Alt,F.W. and de Pinho,R.A. (2000) Telomere dysfunction impairs DNA repair and enhances sensitivity to ionizing radiation. Nature Genet., 26, 85–88. [DOI] [PubMed] [Google Scholar]
- 364.McIlrath J., Bouffler,S.D., Samper,E., Cuthbert,A., Wojcik,A., Szumiel,I., Bryant,P.E., Riches,A.C., Thompson,A., Blasco,M.A., Newbold,R.F. and Slijepcevic,P. (2001) Telomere length abnormalities in mammalian radiosensitive cells. Cancer Res., 61, 912–915. [PubMed] [Google Scholar]
- 365.Ren J.G., Xia,H.L., Tian,Y.M., Just,T., Cai,C.P. and Dai,Y.R. (2001) Expression of telomerase inhibits hydroxyl radical-induced apoptosis in normal telomerase negative human lung fibroblasts. FEBS Lett., 488, 133–138. [DOI] [PubMed] [Google Scholar]
- 366.Fu W.M., Begley,J.G., Killen,M.W. and Mattson,M.P. (1999) Anti-apoptotic role of telomerase in pheochromocytoma cells. J. Biol. Chem., 274, 7264–7271. [DOI] [PubMed] [Google Scholar]
- 367.Zhu H.Y., Fu,W.M. and Mattson,M.P. (2000) The catalytic subunit of telomerase protects neurons against amyloid beta-peptide-induced apoptosis. J. Neurochem., 75, 117–124. [DOI] [PubMed] [Google Scholar]
- 368.Sato N., Mizumoto,K., Kusumoto,M., Nishio,S., Maehara,N., Urashima,T., Ogawa,T. and Tanaka,M. (2000) Up-regulation of telomerase activity in human pancreatic cancer cells after exposure to etoposide. Br. J. Cancer., 82, 1819–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Wright W.E. and Shay,J.W. (2000) Telomere dynamics in cancer progression and prevention: fundamental differences in human and mouse biology. Nature Med., 6, 849–851. [DOI] [PubMed] [Google Scholar]
- 370.Shay J.W. and Wright,W.E. (2001) Perspectives: Aging—when do telomeres matter? Science, 291, 839–840. [DOI] [PubMed] [Google Scholar]
- 371.Mathon N.F., Malcolm,D.S., Harrisingh,M.C., Cheng,L.L. and Lloyd,A.C. (2001) Lack of replicative senescence in normal rodent glia. Science, 291, 872–875. [DOI] [PubMed] [Google Scholar]
- 372.Wei W., Hemmer,R.M. and Sedivy,J.M. (2001) Role of p14ARF in replicative and induced senescence of human fibroblasts. Mol. Cell. Biol., 21, 6748–6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Blasco M.A., Lee,H.W., Hande,M.P., Samper,E., Lansdorp,P.M., DePinho,R.A. and Greider,C.W. (1997) Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell, 91, 25–34. [DOI] [PubMed] [Google Scholar]
- 374.Lee H.W., Blasco,M.A., Gottlieb,G.J., Horner,J.W., Greider,C.W. and DePinho,R.A. (1998) Essential role of mouse telomerase in highly proliferative organs. Nature, 392, 569–74. [DOI] [PubMed] [Google Scholar]
- 375.Sachsinger J., Gonzalez-Suarez,E., Samper,E., Heicappell,R., Muller,M. and Blasco,M.A. (2001) Telomerase inhibition in RenCa, a murine tumor cell line with short telomeres, by overexpression of a dominant negative mTERT mutant, reveals fundamental differences in telomerase regulation between human and murine cells. Cancer Res., 61, 5580–5586. [PubMed] [Google Scholar]
- 376.Artandi S.E. and de Pinho,R.A. (2000) Mice without telomerase: what can they teach us about human cancer. Nature Med., 6, 852–855. [DOI] [PubMed] [Google Scholar]
- 377.Artandi S.E., Chang,S., Lee,S.L., Alson,S., Gottlieb,G.J., Chin,L. and de Pinho,R.A. (2000) Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature, 406, 641–645. [DOI] [PubMed] [Google Scholar]
- 378.Li Q.X., Robbins,J.M., Welch,P.J., Wong Staal,F. and Barber,J.R. (2000) A novel functional genomics approach identifies mTERT as a suppressor of fibroblast transformation. Nucleic Acids Res., 28, 2605–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Chin L., Artandi,S.E., Shen,Q., Tam,A., Lee,S.L., Gottlieb,G.J., Greider,C.W. and de Pinho,R.A. (1999) p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell, 97, 527–538. [DOI] [PubMed] [Google Scholar]
- 380.Gonzalez-Suarez E., Samper,E., Ramirez,A., Flores,J.M., Martin-Caballero,J., Jorcano,J.L. and Blasco,M.A. (2001) Increased epidermal tumors and increased skin wound healing in transgenic mice overexpressing the catalytic subunit of telomerase, mTERT, in basal keratinocytes. EMBO J., 20, 2619–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Rudolph K.L., Millard,M., Bosenberg,M.W. and De Pinho,R.A. (2001) Telomere dysfunction and evolution of intestinal carcinoma in mice and humans. Nature Genet., 28, 155–159. [DOI] [PubMed] [Google Scholar]
- 382.Hacket J.A., Feldser,D.M. and Greider,C.W. (2001) Telomere dysfunction increases mutation rate and genomic instability. Cell, 106, 275–286. [DOI] [PubMed] [Google Scholar]
- 383.Wellinger R.J., Wolf,A.J. and Zakian,V.A. (1993) Saccharomyces telomeres acquire single-strand TG(1–3) tails late in S-phase. Cell, 72, 51–60. [DOI] [PubMed] [Google Scholar]
- 384.Munoz-Jordan J.L., Cross,G.A.M., de Lange,T. and Griffith,J.D. (2001) T-loops at trypanosome telomeres. EMBO J., 20, 579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Klobutcher L.A., Swanton,M.T., Donini,P. and Prescott,D.M. (1981) All gene-sized DNA molecules in four species of hypotrichs have the same terminal sequence and an unusual 3′ terminus. Proc. Natl Acad. Sci. USA, 78, 3015–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Pluta A.F., Kaine,B.P. and Spear,B.B. (1982) The terminal organization of macronuclear DNA in Oxytricha fallax. Nucleic Acids Res., 10, 8145–8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Murti K.G. and Prescott,D.M. (1999) Telomeres of polytene chromosomes in a ciliated protozoan terminate in duplex DNA loops. Proc. Natl Acad. Sci. USA, 96, 14436–14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Henderson E.R. and Blackburn,E.H. (1989) An overhanging 3′ terminus is a conserved feature of telomeres. Mol. Cell. Biol., 9, 345–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Yi X.M., Shay,J.W. and Wright,W.E. (2001) Quantitation of telomerase components and hTERT mRNA splicing patterns in immortal human cells. Nucleic Acids Res., 29, 4818–4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Cook B.D., Dynek,J.N., Chang,W., Shostak,G. and Smith,S. (2002) Role for the related poly(ADP-ribose) polymerase tankyrase 1 and 2 at human telomeres. Mol. Cell. Biol., 22, 332–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Goekjian P.G. and Jirousek,M.R. (2001) Protein kinase C inhibitors as novel anticancer drugs. Expert Opin. Investig. Drugs, 10, 2117–2140. [DOI] [PubMed] [Google Scholar]
- 392.Boyd M., Mairs,R.J., Mairs,S.C., Wilson,L., Livingstone,A., Cunningham,S.H., Brown,M., Quigg,M. and Keith,W.N. (2001) Expression in UVW glioma cells of the noradrenaline transporter gene, driven by the telomerase RNA promoter, induces active uptake of [I-131]MIBG and clonogenic cell kill. Oncogene, 20, 7804–7808. [DOI] [PubMed] [Google Scholar]
- 393.Plumb J.A., Bilsland,A., Kakani,R., Zhao,J.Q., Glasspool,R.M., Knox,R.J., Evans,T.R.J. and Keith,W.N. (2001) Telomerase-specific suicide gene therapy vectors expressing bacterial nitroreductase sensitize human cancer cells to the pro-drug CB1954. Oncogene, 20, 7797–7803. [DOI] [PubMed] [Google Scholar]