Abstract
Background:
Adenosine Deaminase Domain Containing 2 (ADAD2) is a testis-specific protein composed of a double-stranded RNA binding domain and a non-catalytic adenosine deaminase domain. A recent study showed that ADAD2 is indispensable for the male reproduction in mice. However, the detailed functions of ADAD2 remain elusive.
Objectives:
This study aimed to investigate the cause of male sterility in Adad2 mutant mice and to understand the molecular functions of ADAD2.
Materials and Methods:
Adad2 homozygous mutant mouse lines, Adad2–/– and Adad2Δ/Δ, were generated by CRISPR/Cas9. Western blot and immunohistochemistry were used to reveal the expression and subcellular localization of ADAD2. Co-immunoprecipitation tandem mass spectrometry was employed to determine the ADAD2-interacting proteins in mouse testes. RNA-seq analyses were carried out to analyze changes in the transcriptome and piRNA populations of Adad2 mutant testes compared to control testes.
Results:
Adad2–/– and Adad2Δ/Δ mice exhibit male-specific sterility due to abnormal spermiogenesis. ADAD2 interacts with multiple RNA-binding proteins involved in piRNA biogenesis, including MILI, MIWI, RNF17, and YTHDC2. ADAD2 co-localizes and forms novel granules with RNF17 in spermatocytes. Ablation of ADAD2 impairs the formation of RNF17 granules, decreases the number of cluster-derived pachytene piRNAs, and increases expression of ping-pong-derived piRNAs.
Discussion and Conclusion:
In collaboration with RNF17 and other RNA-binding proteins in spermatocytes, ADAD2 directly or indirectly functions in piRNA biogenesis.
Introduction
Spermatogenesis is a complicated, highly coordinated developmental process fundamental for accurate propagation of genetic information to the next generation1,2. Transposable elements (TEs), or transposons, are discrete segments of DNA capable of changing their genomic locations, replicating themselves, and integrating into and creating mutations in the genome3. TE-mediated mutations are of evolutionary significance, but uncontrolled proliferation of TEs has deleterious effects on host fitness. To attenuate the potential threats of TEs, the male germline recruits a sophisticated, small RNA-based defense system involving P-element-induced wimpy testis (PIWI)-like proteins and PIWI-interacting RNAs (piRNAs) to prevent the propagation of TEs across the genome3,4.
piRNAs, which are generally 25 to 31 nucleotides (nt) in length, are generated via the primary and secondary processing pathways5. Primary piRNA biogenesis involves the production of precursor piRNAs (pre-piRNAs) by endonuclease-mediated fragmentation of long, single-stranded RNAs that are transcribed from piRNA cluster loci containing retrotransposon sequences6,7. The 3’ termini of the pre-piRNAs are subsequently trimmed and 2’-O-methylated8. The resultant mature primary piRNAs are characterized by a preference for a uridine (1U) at the first position. Primary piRNAs guide the recognition and PIWI-mediated cleavage of complementary transcripts between the 10th and 11th nucleotides to generate secondary piRNAs, an amplification process known as the ping-pong cycle9. Primary and secondary piRNAs harbor a 10-nt complementary overlap, and the secondary piRNAs exhibit a bias for an adenine at the 10th position (10A) from their 5’ termini10–12. Ping-pong-derived piRNAs are abundant in fetal germ cells but are low in adult testes4.
The mouse genome encodes three testis-enriched PIWI-like proteins, MIWI (Piwi-like protein 1 or PIWIL1), MILI (Piwi-like protein 2 or PIWIL2), and MIWI2 (Piwi-like protein 4 or PIWIL4), that exhibit distinct expression timing and molecular functions during spermatogenesis13–15. Mili and Miwi2 are initially expressed in the prenatal testes; mutations in MILI or MIWI2 cause spermatogenic arrest at the zygotene/pachytene spermatocyte stage14,16. In contrast, expression of Miwi is restricted to late spermatogenesis, from pachytene spermatocytes to elongating spermatids; ablation of MIWI results in spermiogenic arrest at the round spermatid stage3,17. Based on the timing of expression, these PIWI-bound RNAs are referred to as fetal and postnatal pre-pachytene or pachytene piRNAs in spermatogenic cells18,19. Pre-pachytene piRNAs in fetal testes are primarily derived from TEs and are associated with MILI and MIWI25,20. Different from pre-pachytene piRNAs, pachytene piRNAs are predominantly transcribed from intergenic regions termed pachytene piRNA clusters, whilst 10 – 20% of them are produced from non-cluster regions such as coding RNAs, non-coding RNAs, repeats, and introns18,21. Both cluster and non-cluster-derived pachytene piRNAs bind to MILI and MIWI3. Because of their repeat-devoid origin, the detailed functions and target transcripts of pachytene piRNAs remain to be clarified. After meiosis, sperm chromatin undergoes extensive chromatin remodeling, during which histone replacement occurs10. The incorporation of histone variants is associated with open and accessible chromatin, resulting in loosened transcriptional control10,22. Pachytene piRNAs have been reported to regulate post-transcriptional silencing of mRNAs and lncRNAs, which are transcribed due to this genome-wide derepression of transcription23–26.
In addition to PIWI-like proteins, a multitude of proteins have been discovered essential for piRNA biogenesis and function, such as GASZ27,28, GPAT229,30, MAEL31,32, MVH33, RNF177,34, TDRD135−37, and TDRD938,39. MAEL, or Maelstrom, is an RNA-binding protein localized to the chromatoid bodies of the male germ cells and is required for transposon silencing and male fertility in mice31,32. MAEL binds pachytene piRNAs; depletion of MAEL results in a drastic reduction of pachytene piRNAs. Furthermore, Mael mutant mice exhibit decreased translation of various sperm acrosomal and flagellar proteins, which may contribute to arrested spermiogenesis31. MAEL interacts with ADAD2 and RNF17, which are localized to pachytene spermatocytes and implicated in RNA modification31. Similarly, mice lacking ADAD2 or RNF17 show male-specific sterility due to abnormal spermatid differentiation34,40. RNF17 is localized to granules distinct from other known nuages in late pachytene and diplotene spermatocytes34. Depletion of RNF17 unleashes the ping-pong cycle, which aberrantly produces secondary piRNAs that degrade not only TEs but also mRNAs and lncRNAs7. Similar to RNF17, ADAD2 also forms prominent granules in pachytene spermatocytes40,41. Nevertheless, it is not clear how ADAD2 governs the differentiation of spermatids. In this study employing CRISPR/Cas9-based gene editing, transcriptomic, and proteomic techniques, we have investigated the molecular functions of ADAD2 and its potential involvement in piRNA biogenesis.
Materials and Methods
Animals
Wildtype mice were purchased from Japan SLC, Inc. (Shizuoka, Japan) for experiments in the laboratories of M.I. and S.K.-M., or produced by intercrossing C57BL6 and 129S6/SvEv mice in the laboratory of M.M.M. All mice were maintained in cycles of 12 h light and 12 h darkness with ad libitum feeding. The animal experiments were approved by the Institutional Animal Care and Use Committees at Osaka University and Baylor College of Medicine.
Frozen spermatozoa from Adad1+/– and Adad2+/– male mice (B6D2-Adad1<em1Osb>, RBRC#11659, CARD#3216; B6D2-Adad2<em1Osb>, RBRC#10998, CARD#2905) are available at the Riken BioResource Center (RIKEN BRC; web.brc.riken.jp/en) and the Center for Animal Resources and Development, Kumamoto University (CARD R-BASE; cardb.cc.kumamoto-u.ac.jp/transgenic).
Generation of knockout mice
Adad2 null (Adad2+/–) and mutant (Adad2+/Δ) mouse lines and the Adad1 knockout mouse line were generated by the CRISPR/Cas9 system as previously described42. The sequences of sgRNAs and mutant alleles are enumerated in Table S1. The sequences of primers for genomic PCR are listed in Table S2.
Reverse transcription polymerase chain reaction (RT-PCR)
Mouse cDNA was prepared from various tissues of adult mice, or from 5- to 42-day-old mouse testes. Alternatively, cDNA was prepared from spermatocytes and spermatids, which were isolated and purified from mouse testes as previously described43. Briefly, testes from 24-day-old mice were dissected to remove the tunica albuginea. The decapsulated testes were treated with collagenase B (Roche, Tokyo, Japan) and DNase I (Sigma-Aldrich, St. Louis, MO), followed by treatment with trypsin (ThermoFisher, Waltham, MA) and DNase I. The isolated germ cells were then washed with phosphate-buffered saline (PBS) and stained with Hoechst 33342 in 5% bovine serum albumin (BSA) in PBS at 34°C for 1 hour. Spermatocytes and spermatids were sorted based on DNA content and light-scattering properties by BD FACSAria™ IIu (BD Biosciences, Tokyo, Japan). Reverse transcription quantitative PCR (RT-qPCR) was performed using the THUNDERBIRD™ Probe and SYBR® qPCR Mix (TOYOBO, Osaka, Japan) and the CFX384 Touch™ Real-Time PCR Detection System (Bio-Rad, Hercules, CA). The sequences of primers are listed in Table S2.
Fertility tests
Sexually mature Adad1 or Adad2 knockout males were individually caged with three B6D2F1 female mice for eight weeks. During this period, vaginal plugs were examined as an indicator of successful copulation, and the number of offspring in each litter was recorded at birth. Three knockout males were analyzed to meet the requirements for statistical validity. The fecundity of three wildtype B6D2F1 males was tested in parallel as positive controls. After eight weeks of breeding, the male mice were withdrawn from the cages, and the females were kept for another three weeks to allow the final litters to be delivered.
Histological analyses
Testes and epididymides were fixed in Bouin’s solution (Polysciences, Inc., Warrington, PA), embedded in paraffin wax, sectioned at a thickness of 5 μm on a Microm HM325 microtome (Microm, Walldorf, Germany), and stained with periodic acid (Nacalai Tesque, Kyoto, Japan) and Schiff’s reagent (Wako, Osaka, Japan), followed by counterstaining with Mayer’s hematoxylin solution (Wako, Osaka, Japan).
Antibodies
Polyclonal antibodies against the amino acids (aa) 504 – 522 of mouse ADAD1 (CCDS17315.1) and the aa 1 – 93 of mouse ADAD2 (pAb; CCDS52683.1) were raised and produced in rabbits. A monoclonal antibody (mAb#19–8) against aa 1 – 93 of mouse ADAD2 was raised and produced in rats as previously described44. Rabbit polyclonal anti-MILI (26F) antibody that recognizes both MILI and MIWI14, rabbit polyclonal anti-MILI antibody (#2071; Cell Signaling Technology, Danvers, MA), mouse monoclonal anti-MIWI antibody (#2C12; Wako, Osaka, Japan), rabbit polyclonal anti-RNF17 antibody (kindly gifted by Dr. P. Jeremy Wang34), rabbit polyclonal anti-MVH (#ab13840; Abcam, Cambridge, UK) and rabbit monoclonal anti-GAPDH antibody (#2118; Cell Signaling Technology, Danvers, MA) were used for immunoprecipitation or Western blot analyses.
Immunohistochemistry
Testes were fixed in 4% paraformaldehyde, infused with 15% and 30% sucrose in PBS, embedded in the Tissue-Tek® optimal cutting temperature (O.C.T.) compound (Sakura Finetek USA, Inc., Torrance, CA), and snap-frozen in liquid nitrogen. Testis blocks were sectioned at a thickness of 5 – 10 μm on a CryoStar NX70 cryostat (ThermoFisher, Waltham, MA). The testis sections were dried on adhesive microscope slides, permeabilized, and blocked with 0.1% Triton X-100, 3% BSA, and 10% goat serum in PBS for 1 h at room temperature. The samples were then incubated with primary antibodies for 3 h at room temperature or overnight at 4°C. After three washes in 0.1% Triton X-100 in PBS, the sections were incubated with fluorophore-conjugated secondary antibodies for 1 h at room temperature and then stained with 1 μg/mL Hoechst 33342 (ThermoFisher, Waltham, MA) for 30 min at room temperature. The sections were mounted with Epredia™ Immu-Mount (Fisher Scientific, Pittsburgh, PA) prior to imaging. Fluorescence images were captured with a Nikon Eclipse Ti microscope equipped with a Nikon C2 confocal module (Nikon, Tokyo, Japan).
Protein extraction and co-immunoprecipitation (co-IP)
Testes were homogenized in ice-cold lysis buffer [50 mM Tris (pH 7.5), 150 mM NaCl, 1% Triton X-100, and 10% glycerol (for Figure 1E, 4A, 4C, 4D, and 4E); or 20 mM Tris (pH 7.5), 200 mM NaCl, 2.5 mM MgCl2, 0.1% Triton X-100, and 0.5% NP-40 (for Figure S2C and S8B)] with a Dounce homogenizer (for Figure 1E, 4A, 4C, 4D, and 4E), or using a sterile syringe with 18G and 23G needles in succession (for Figure S2C and S8B). Co-IP was performed using Pierce™ Crosslink IP Kit (ThermoFisher, Waltham, MA; for Figure 4A) or Invitrogen™ Dynabeads™ Protein G Magnetic Beads (ThermoFisher, Waltham, MA; for Figure 4C, 4D, 4E, and S8B) in accordance with the manufacturer’s instructions. The IP eluates were subjected to mass spectrometry (MS) or sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analyses.
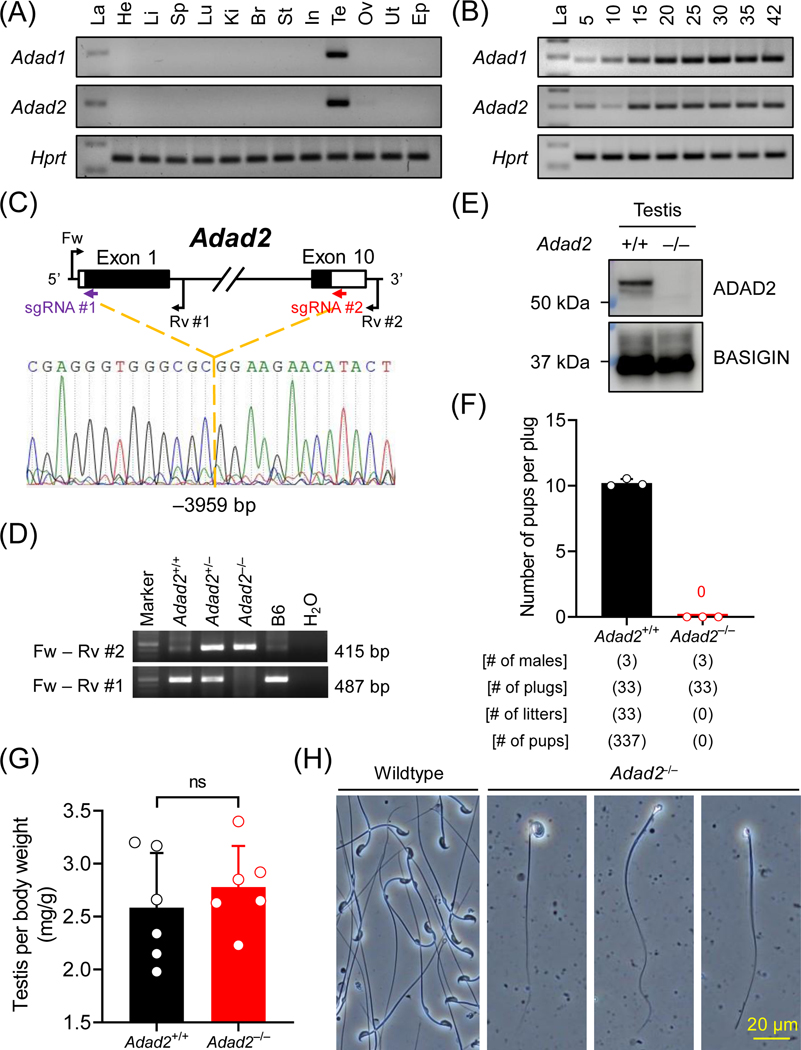
Figure 1. Adad2 is indispensable for male reproduction in mice.
(A) Multi-tissue RT-PCR analyses of mouse Adad1 and Adad2. La, ladder; He, heart; Li, liver; Sp, spleen; Lu, lung; Ki, kidney; Br, brain; St, stomach; In, intestine; Te, testis; Ov, ovary; Ut, uterus; Ep, epididymis. The expression of Hprt was analyzed in parallel as a loading control. (B) Postnatal testis RT-PCR analyses of mouse Adad1 and Adad2. The expression of Adad1 and Adad2 was analyzed in mouse testes from postnatal day 5 to 42. (C – D) Schematic representation of Adad2 null mouse generation by CRISPR/Cas9. Single guide RNAs (sgRNAs) #1 and #2, which target the first and the tenth exons, respectively, were used to delete the entire coding region of Adad2. Fw and Rv #1 primers were used to detect the wildtype allele, whereas Fw and Rv #2 primers were used to detect the null allele by genomic PCR. (E) Immunodetection of ADAD2 in wildtype and homozygous null testis lysates using mAb#19–8. BASIGIN was analyzed as a loading control. (F) Fertility tests of Adad2 null male mice. (G) Testis relative to body weight in wildtype and Adad2 null males. ns, no significant difference. (H) Observation of sperm extracted from cauda epididymis from wildtype and Adad2 null males.
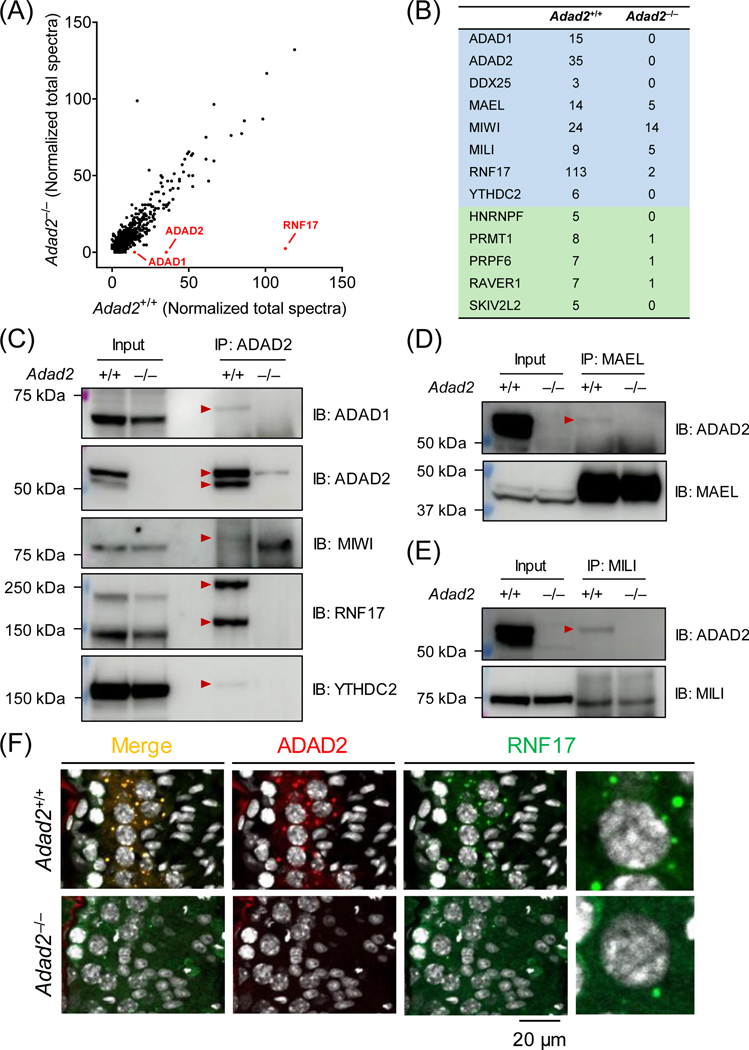
Figure 4. ADAD2 interacts with RNA-binding proteins implicated in piRNA biogenesis.
(A) Co-IP/MS analysis of ADAD2-interacting proteins. Co-IP was performed using agarose resin crosslinked with the polyclonal antibody against aa 1 – 93 of mouse ADAD2. ADAD1, ADAD2, and RNF17 are detected with high specificity in wildtype testis lysate. (B) Representative RNA-binding proteins enriched in the ADAD2-associated proteome. Blue indicates the proteins potentially involved in both piRNA biogenesis and spermatogenesis, whereas green highlights the ones implicated in spermatogenesis. (C – E) Co-IP followed by Western blot analyses. Co-IP experiments were performed using magnetic beads conjugated with the antibodies against ADAD2 (pAb), MAEL, or MILI. The immunoprecipitated proteins were then analyzed by Western blot using antibodies against ADAD1, ADAD2 (mAb#19–8), MAEL, MILI, MIWI, RNF17, and YTHDC2. Red arrowheads indicate the protein bands specifically detected in the wildtype IP samples. (F) Immunohistochemistry analyses of ADAD2 and RNF17 in wildtype and Adad2–/– testis cryosections. ADAD2 was immunodetected using mAb#19–8 and cell nuclei were visualized by Hoechst 33342.
Mass spectrometry
Proteomic analysis was performed as previously described45. Briefly, protein samples were processed, and the resultant protein peptides were subjected to nanocapillary reversed-phase liquid chromatography (LC)-MS/MS analysis using a C18 column (10 cm × 75 μm, 1.9 μm) on a nanoLC system connected to a timsTOF Pro mass spectrometer and a nano-electrospray ion source (CaptiveSpray; Bruker Daltonics, Billerica, MA). The resulting data were processed using DataAnalysis (Bruker Daltonics, Billerica, MA), and proteins were identified using the MASCOT Server (Matrix Science, Tokyo, Japan) based on the UniProtKB/Swiss-Prot database. Quantitative values were calculated by Scaffold 5 software (v5.1.2; Proteome Software, Portland, OR).
Next generation sequencing
Total RNA for whole transcriptome and small RNA analyses was purified from postnatal day 24 mouse testes using ISOGEN (Nippon Gene, Tokyo, Japan) according to the manufacturer’s instruction. For small RNA immunoprecipitation (RIP) analysis, RNAs were immunoprecipitated and purified from postnatal day 24 mouse testes using anti-MILI and MIWI antibodies. RNA-sequencing (RNA-seq) and small RNA-seq libraries were prepared using TruSeq RNA Library Prep Kit (Illumina, San Diego, SA) and TruSeq Small RNA Library Prep Kit (Illumina, San Diego, SA), respectively. The small RNA-seq library for RIP analysis was prepared using the SMARTer smRNA-Seq Kit (Takara Bio, Shiga, Japan). The RNA-seq library for whole transcriptome analysis was subjected to paired-end 75 bp sequencing on the NextSeq 500 system (Illumina, San Diego, SA), whereas the small RNA-seq libraries were subjected to single-end 51 bp sequencing on the HiSeq 2500 system (Illumina, San Diego, SA). Basecalls were performed using NextSeq 500/550 RTA software (v1.0) or HiSeq 2500 HCS (v2.2) and RTA (v1.18) software. FASTQ files were generated using bcl2fastq (v2.17.1.14 for RNA-seq and v1.8.4 for small RNA-seq). Differentially expressed genes (adjusted p value < 0.05, and > 2-fold change) were determined by DESeq2 (Table S3) and analyzed by DAVID Knowledgebase (v2022q3)46 to identify the enriched gene ontology terms. The raw sequencing data of total small RNA samples were processed using CLC Genomics Workbench software (Filgen, Nagoya, Japan) to trim the adapter sequences, to remove microRNAs (miRbase; up to 1 nt mismatches were allowed), and to exclude low-quality reads with lengths less than 15 nt and more than 35 nt. RNA-seq reads were mapped to the mouse genome assembly mm9 using piPipes47. The 24 – 35 nt long small RNAs and all reads of MILI- or MIWI-bound small RNAs were mapped to mm10 by Bowtie 2. The small RNAs were finally divided into the ones mapped to pachytene piRNA clusters [467 piRNA precursor transcripts described by Li et al.18 (Table S1)] and others that were not mapped to those regions. All RNA-seq data have been deposited in NCBI Gene Expression Omnibus (GEO) under accession number GSE209984 and Sequence Read Archive (SRA) under BioProject accession number PRJNA871197).
Statistical analyses
Data are presented as mean values and error bars indicate standard deviation (SD). Experimental groups were analyzed statistically using an unpaired two-tailed Student’s t-test. P values less than 0.05 were considered statistically significant (*, P < 0.05; ** P < 0.01; *** P < 0.001).
Results
ADAD2 is essential for male fertility in mice
As depicted by RT-PCR, mouse Adad1 and Adad2 are testis-specific genes initially expressed at postnatal day 5 (Figure 1A and 1B). In human, ADAD2 is also expressed in brain (Figure S1A). According to a previous single-cell RNA-seq (scRNA-seq) analysis of mouse spermatogenic cells48, Adad1 is highly expressed in late spermatocytes and early spermatids, whereas Adad2 shows peak expression in pachytene spermatocytes (Figure S1B), implying the two paralogous genes may have distinct functions during spermatogenesis. Both ADAD1 and ADAD2 contain an N-terminal double-stranded RNA-binding domain (dsRBD) and a C-terminal adenosine deaminase domain [or adenosine aminohydrolase (ADA)] (Figure S1C and S1D). Phyre2 homology modeling49 of ADAD1 and ADAD2 uncovered that their dsRBDs consist of three β-strands sandwiched by two α-helices, creating a typical α-β-β-β-α topology (Figure S1E). TreeFam50,51 indicated that Adad1 and Adad2 are evolutionarily conserved among species, including protists, invertebrates, and vertebrates (Figure S1F).
To study the molecular functions of ADAD2, we generated a homozygous null mouse model (Adad2–/–) lacking the entire coding region (–3959 bp) and a homozygous mutant mouse model (Adad2Δ/Δ) bearing a frameshift mutation (–27+2 bp) at the first coding exon on the genetic background of B6D2F1 and C57BL/6J, respectively (Figure 1C, 1D, S2A, and S2B). An ADAD2 protein band was absent in the homozygous null and mutant testis lysate (Figure 1E and S2C). When consecutively paired with wildtype females, the homozygous mutant males with either a complete deletion or a frameshift allele failed to sire any offspring (Figure 1F and S2D), demonstrating that Adad2 is essential for male reproduction in mice.
Ablation of ADAD2 impairs spermiogenesis
Adad2–/– males with the large deletion showed comparable testis weights as the wildtype controls [Adad2+/+; Figure 1G]. In contrast, the testis weights of Adad2Δ/Δ mice with the frameshift mutation were smaller compared with those of age-matched heterozygous littermates [Adad2+/Δ: 96.3 ± 10.5; Adad2Δ/Δ: 74.8 ± 5.4] (Figure S2E and S2F). We observed low numbers of immature or deformed sperm cells in the extracts of cauda epididymides from Adad2–/– males (Figure 1H), suggesting the male sterility phenotype originates from impaired spermiogenesis.
Histological analyses of testis and epididymis sections revealed that spermatogenesis was generally normal in Adad2–/– and Adad2Δ/Δ males until the early round spermatid stage (Figure 2A and S3A). Binucleated round spermatids were occasionally observed in the seminiferous tubules of Adad2–/– males, indicating disrupted meiotic cytokinesis (Figure S3B). Adad2–/– males showed aberrant migration of elongating spermatids in stage VI-VIII seminiferous tubules, delayed detachment of elongated spermatids from the germinal epithelium in stage IX-X tubules, abnormal head morphology in step 9–16 spermatids, and reduced numbers of elongated spermatids (Figure S3A). Adad2Δ/Δ males exhibited relatively severe anomalies in spermiogenesis compared with Adad2–/– males. In Adad2Δ/Δ seminiferous tubules, elongated spermatids were rarely observed and multinucleated cysts containing degenerated round spermatids were detected (Figure 2A and S3A). Immunohistochemistry revealed that ADAD2 was detected as granules exclusively localized to the pachytene spermatocytes (Figure 2B).
Figure 2. Adad2 deficient male mice show impaired spermiogenesis.
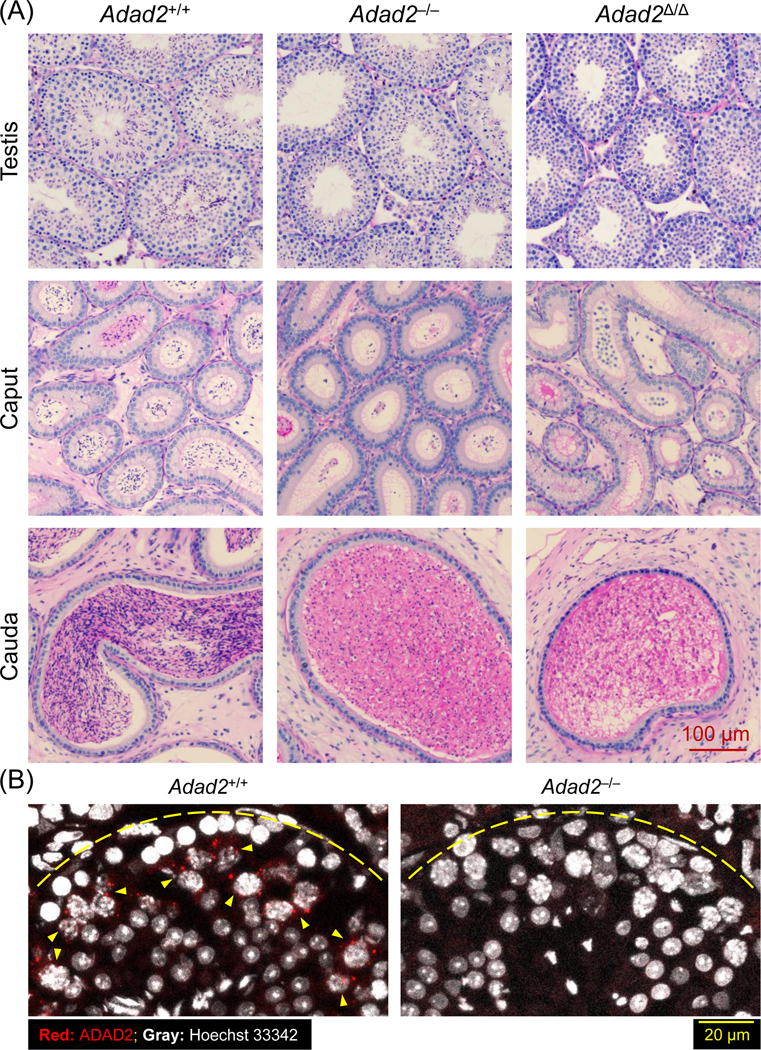
(A) Histological analyses on the testes and epididymides of Adad2+/+, Adad2–/–, and Adad2Δ/Δ males. (B) Immunostaining of ADAD2 on the testis cryosection using the antibody against aa 1 – 93 of mouse ADAD2. Yellow arrowheads indicate pachytene spermatocytes with the ADAD2 signals. Yellow dotted lines represent the basement membrane of the seminiferous tubules.
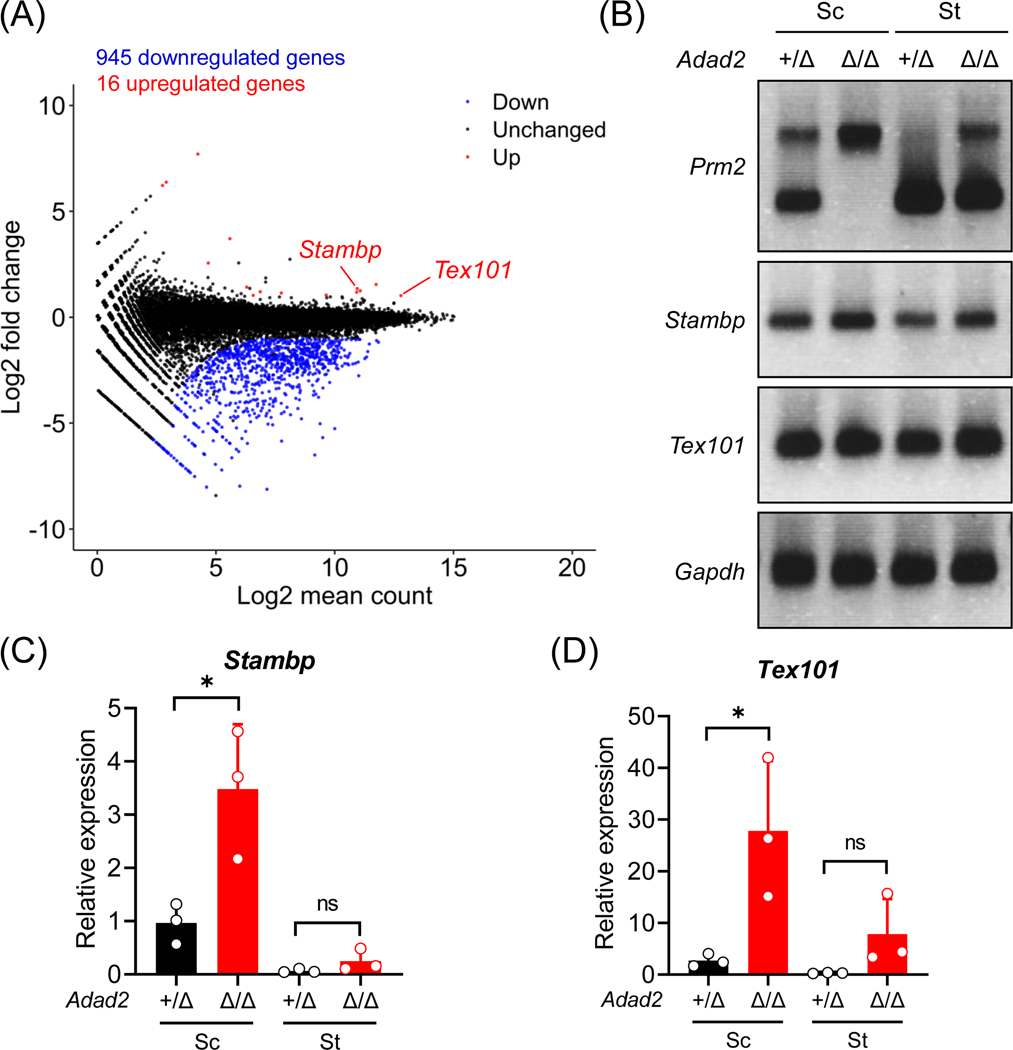
Next, we performed RNA-seq analyses using postnatal day 24 wildtype and Adad2Δ/Δ testes. The Adad2Δ/Δ males exhibit dramatic changes in the transcriptome, including 945 downregulated genes and 16 upregulated genes (Figure 3A and Table S3). GO enrichment analyses46 indicate that the downregulated genes are related to male gonad development (GO:0008584), flagellated sperm motility (GO:0030317), and spermatogenesis (GO:0007283; Figure S4A). The downregulated genes encode proteins localized to sperm head plasma membrane (GO:1990913), acrosomal vesicle (GO:0001669), and sperm principal piece (GO:0097228; Figure S4A). Multiple downregulated hits include testis-enriched genes essential for male fertility, such as Tssk1, Tssk2, Spata19, and Tbc1d21 (for mitochondrial sheath assembly52–56); Akap4, Odf1, and Odf3 (for sperm flagellar formation57); and Tnp1, Tnp2, Tssk6, Prm1, and Prm2 (for chromatin remodeling in spermatids58–63; Figure S4B). Such a massive downregulation of genes might be a consequence of reduced round or elongated spermatids in Adad2 mutant males compared to wildtype males. Moreover, the mutant spermatocytes and spermatids show intron retention in the Prm2 transcript (Figure 3B). In addition, Prelid1, Stambp, and Tex101 are upregulated in Adad2Δ/Δ male germ cells (Figure S4B, 3C and 3D).
Figure 3. Ablation of ADAD2 causes dramatic transcriptomic changes in the male germline.
(A) An MA plot depicting differentially expressed genes in the testes from wildtype and Adad2Δ/Δ male mice at 24 days postpartum. Each dot represents one gene; the x-axis and y-axis denotes the log2 value of mean expression level and fold change, respectively. Black, red, and blue indicates no significant difference, significant upregulation, and significant downregulation (more than 2-fold change), respectively. (B) RT-PCR analyses of Prm2, Stambp, and Tex101 in the spermatocytes (Sc) and spermatids (St) from Adad2+/Δ and Adad2Δ/Δ male mice. Gapdh was analyzed in parallel as a loading control. (C – D) RT-qPCR analyses of Stambp and Tex101 in the spermatocytes (Sc) and spermatids (St) from Adad2+/Δ and Adad2Δ/Δ male mice.
ADAD2 interacts with multiple RNA-binding proteins in male germ cells
To acquire a better understanding of the molecular functions of ADAD2, we analyzed its interacting proteins in wildtype testes (with Adad2–/– serving as a negative control) by co-IP/MS (Figure 3A). GO analyses unveiled that a significant portion of the interacting proteins exhibit RNA-binding ability (Figure S5A). These include DDX25, MAEL, MILI, MIWI, RNF17, and YTHDC2 (Figure 4B and S5B), which function in piRNA biogenesis during spermatogenesis7,20,31,32,64,65. Noticeably, DDX25 and RNF17, which were detected in the ADAD2 interactome, have been concurrently identified as MAEL-interacting proteins31 (Figure S5C). By co-IP tandem Western blot analyses, we confirmed that ADAD2 interacts with ADAD1, MAEL, MILI, MIWI, RNF17, and YTHDC2 (Figure 4C, 4D, and 4E). Immunohistochemistry revealed that ADAD2 co-localizes with RNF17 in large granules in the wildtype pachytene spermatocytes. In Adad2–/– spermatocytes, the numbers of RNF17 granules are markedly decreased (Figure 4F). The ADAD2 granules show minor co-localization with MILI in spermatocytes (Figure S6).
Since co-IP tandem MS and Western blot analyses revealed an interaction between ADAD1 and ADAD2 (Figure 4A and 4C), we anticipated that these two paralogous proteins are tightly associated during spermatogenesis. In line with previous reports40,66, Adad1–/– males are sterile and exhibit abnormal spermiogenesis (Figure S7A – E). However, ablation of ADAD1 does not affect the abundance of ADAD2, and vice versa (Figure S7F and S7G), indicating that ADAD1 and ADAD2 are not interdependent at the protein level.
Deletion of Adad2 alters piRNA populations in the male germline
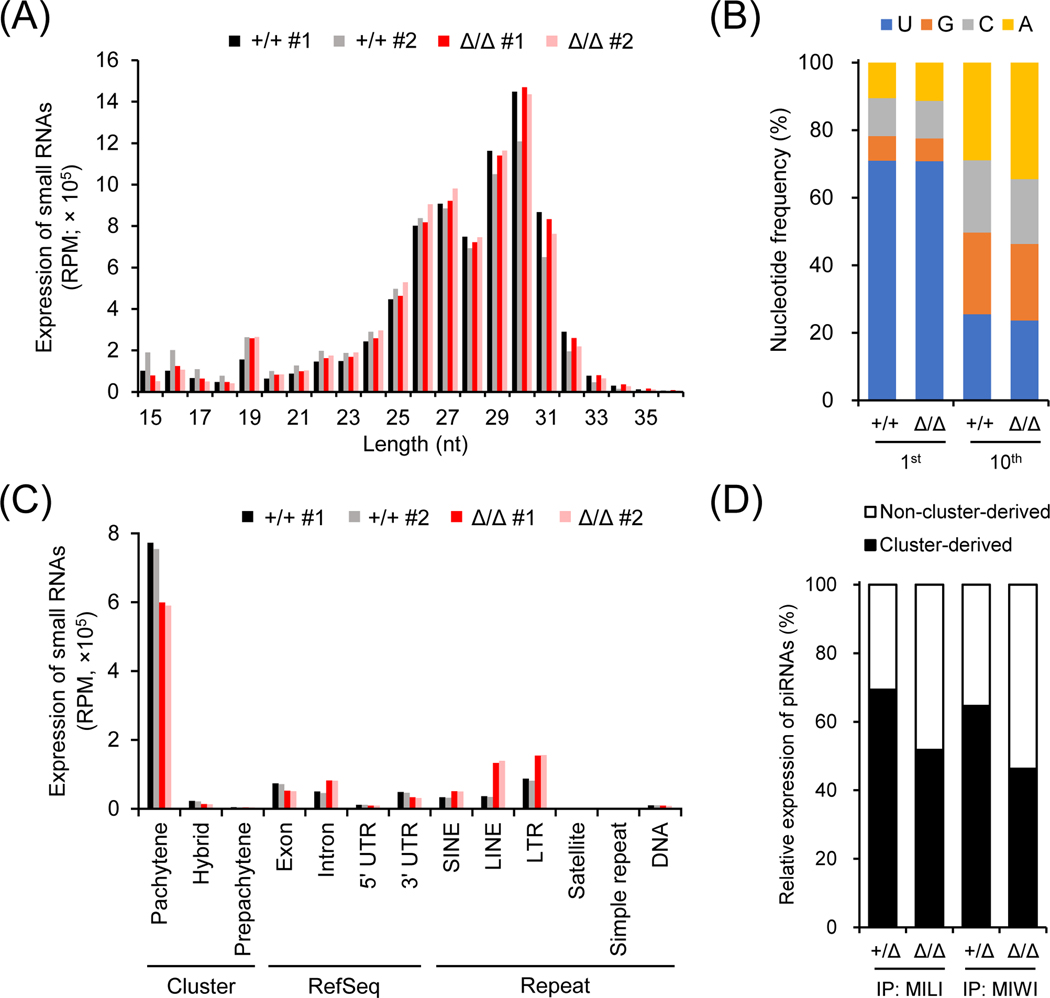
Since ADAD2 is associated with multiple RNA-binding proteins implicated in piRNA biogenesis, we carried out deep sequencing analyses of small RNAs in wildtype and Adad2Δ/Δ postnatal day 24 testes. Length distribution analysis of small RNAs indicated that the abundance of piRNAs ranging from 24 to 31 nt was comparable in wildtype and mutant testes (Figure 5A). No significant difference was observed in the nucleotide distribution of small RNAs or the ratio of 1U-biased primary piRNAs between wildtype and mutant (Figure 5A and 5B). Interestingly, the percentage of secondary piRNAs exhibiting 10th A bias was increased in Adad2Δ/Δ testes (Figure 5B). In corroboration with this observation, the mutant showed higher expression of piRNAs carrying 10-nt complementary overlaps, another signature of piRNAs derived from the ping-pong cycle (Adad2+/+ Z score = 19.1; Adad2Δ/Δ Z score = 33.1; Figure S8A). To clarify the changes in the piRNA populations, we next analyzed the expression of piRNAs transcribed from different genomic loci. While the expression of intron, SINE, LINE, and LTR-derived piRNAs was significantly increased, cluster-derived pachytene piRNAs showed decreased expression in the mutant males (Figure 5C). RNA-IP sequencing (RIP-seq) further revealed that both MILI-bound and MIWI-bound pachytene cluster-derived piRNAs were reduced in the mutant testes (MILI-bound: Adad2+/Δ, 73.9%; Adad2Δ/Δ, 57.9%; MIWI-bound: Adad2+/Δ, 71.8%; Adad2Δ/Δ, 49%; Figure 5D and S8B). LINE-1 expression is not detected in Adad2 homozygous mutant testes, indicating that the arrested spermiogenesis is not attributed to LINE-1 derepression (Figure S8C).
Figure 5. Depletion of ADAD2 leads to changes in the piRNA populations.
(A) Length distribution of small RNAs in Adad2+/+ and Adad2Δ/Δ testes. RPM, reads per million reads. (B) Distribution of the 1st and 10th nucleotides of piRNAs (24 – 35 nt in length) in Adad2+/Δ and Adad2Δ/Δ testes. (C) Expression of piRNAs in Adad2+/+ and Adad2Δ/Δ testes. LTR, long terminal repeat retrotransposons; LINE, non-LTR autonomous long interspersed element; SINE, non-LTR non-autonomous short-interspersed element. (D) Relative expression of cluster-derived and non-cluster-derived pachytene piRNAs bound to MILI or MIWI in Adad2+/+ and Adad2Δ/Δ testes.
Discussion
In this study, we demonstrate the essential role of ADAD2 in male reproduction. Male mice lacking Adad2 exhibit sterility due to compromised spermiogenesis. ADAD2 interacts with MAEL, MILI, MIWI, RNF17, and YTHDC2, which are involved in spermatogenesis and piRNA biogenesis.
As revealed by testis histology, Adad2Δ/Δ males with a frameshift mutation show more severe impairments in spermiogenesis than Adad2–/– males with a complete deletion. The milder phenotype in Adad2–/– males might be attributed to their mixed genetic backgrounds (compared to Adad2Δ/Δ in C57BL/6J inbred background). Overall, Adad2 deficient male mice exhibit multiple anomalies in sperm development, including defective meiotic cytokinesis, degeneration of round spermatids, abnormal sperm head morphogenesis, delayed adluminal migration, and premature detachment of spermatids from the geminal epithelium (Figure 1H, 2A, S3A, and S3B). Owing to the complicated knockout phenotype, it is challenging to define the primary function of ADAD2 in spermatogenesis. In the current and a recent study40, the RNA-seq analyses were carried out using mouse testes at around postnatal day 24, equivalent to the onset of spermiogenesis. Notably, in Adad2Δ/Δ males, a portion of the early round spermatids undergo abnormal cytokinesis and apoptosis, reflected by the presence of binucleated and multinucleated cells, respectively (Figure S3A and S3B). Thus, as also pointed out by Synder et al.40, such anomalies may lead to a reduced number of round spermatids in Adad2 deficient males, thereby diminishing the relevance of the RNA-seq outcomes. To overcome this potential concern, Synder et al.40 focused only on the downregulated spermatocyte-enriched genes and upregulated round spermatid-enriched genes in the mutant testes. This approach, however, neglects a number of downregulated spermatid-enriched genes with significant fold changes (e.g., Prm1, Prm2, Tnp1, and Tnp2). Instead, RT-PCR or quantitative PCR could be alternatively or additionally performed to verify the mRNA levels of genes of interest in purified spermatocytes or spermatids (e.g., Figure 3B, 3C, and 3D). In future studies, RNA-seq analyses could be further conducted using testes at 14 days postpartum to elucidate how the spermatocyte-localized ADAD2 underpins subsequent spermatid differentiation.
ADAD2 interacts and co-localizes with RNF17 in spermatocytes (Figure 4C and 4F). The partial co-localization of ADAD2 and MILI suggests that the ADAD2/RNF17 granules might be intermitochondrial cement (Figure S6). Depletion of Adad2 impairs formation of RNF17 granules (Figure 4F). Consistent with the tight association between ADAD2 and RNF17, mutant mice lacking either protein exhibit arrested spermiogenesis and massive downregulation of protein-coding transcripts (Figure 3A and S4)7,34. In both mutants, the expression of total pachytene cluster piRNAs, and MIWI-bound and MILI-bound pachytene cluster piRNAs, is decreased, whereas the abundance of transposon-derived secondary piRNAs is increased (Figure 5C, 5D, and S8A)7. In response to such alterations in the piRNA population, mRNAs encoding Prelid1, Stambp, and Tex101, which are negatively regulated by MIWI-bound pachytene cluster piRNAs67, are upregulated in Adad2Δ/Δ testes (Figure 3A and S4B). Despite the defective piRNA biogenesis, MILI and MIWI show normal levels in the testes of Adad2 homozygous mutant males (Figure 3C and 3E). Future studies are necessary to determine the direct and/or indirect roles of ADAD2 in the piRNA processing pathway.
Recently, Chukrallah et al.41 indicate that ablation of ADAD2 results in reduced translation of Mdc1, a gene involved in meiotic sex chromosome inactivation and XY-body formation68. The heterochromatin condensation is abnormal in Adad2 knockout round spermatids, manifested by increased numbers of chromocenters in the nuclei. The authors propose that such anomalies may lead to apoptosis and defective chromatin compaction of round spermatids in Adad2 knockout males. In agreement with these findings, we observed dramatic downregulation of genes involved in sperm chromatin remodeling, including Prm1, Prm2, Tnp1, and Tnp2 (Figure S4B). Nevertheless, whether the reduced abundance of MDC1 primarily contributes to the knockout phenotype remains unclear.
Overall, our work uncovers a novel role of ADAD2 in ensuring the formation of RNF17 granules in spermatocytes. Similar to Rnf17-deficient males, cluster-derived pachytene piRNAs are reduced and transposon-derived piRNAs are increased in Adad2 in the testes of homozygous mutant males. Future studies are warranted to investigate the mechanism by which ADAD2 and RNF17 coordinate piRNA biogenesis in association with other RNA-binding proteins in meiotic cells.
Supplementary Material
Table S1. Information about Adad1 and Adad2 mutant mouse lines.
Table S2. Sequences of primers used in this study.
Table S3. RNA-seq analysis of postnatal day 24 wildtype and Adad2Δ/Δ testes.
Figure S1. Characterization of ADAD1 and ADAD2. (A) Multi-tissue RT-PCR analyses of human ADAD1 and ADAD2. La, ladder; Li, liver; Ki, kidney; Br, brain; In, intestine; Te, testis; Ut, uterus; Cap, caput epididymis; Cor, corpus epididymis; Cau, cauda epididymis. The expression of GAPDH was analyzed in parallel as a loading control. (B) mRNA expression of mouse Adad1 and Adad2 based on a previously published scRNA-seq analysis. (C) Domain composition of mouse ADAD1 and ADAD2. Both proteins contain a N-terminal double-stranded RNA-binding domain (dsRBD; in red) and a C-terminal adenosine deaminase domain (ADA; in blue). (D) Alignment of mouse ADAD1 and ADAD2 protein sequences. Red and blue represents dsRBD and ADA, respectively. (E) In silico homology modeling of the dsRBDs of mouse ADAD1 and ADAD2 by Phyre2. (F) Phylogenetic analyses of Adad1 and Adad2 by TreeFam.
Figure S2. Generation of an Adad2 mutant mouse line. (A) Schematic representation of Adad2 mutant mouse generation by CRISPR/Cas9. The sgRNA #1’ targeting the first exon was used to introduce a frameshift mutation (–27+2 bp) in Adad2. Fw’ and Rv’ primers were used to detect the wildtype and mutant alleles by genomic PCR. (B) Genomic PCR distinguishing the wildtype (WT) and the mutant (Mut) alleles (blue arrowheads). (C) Immunodetection of ADAD2 in wildtype and mutant testis lysates using the polyclonal antibody against aa 1 – 93 of mouse ADAD2. MVH was analyzed as a loading control. (D) Fertility tests of Adad2 mutant male mice. (E – F) Testis weights and appearance in wildtype and mutant males.
Figure S3. Adad2 deficient males show defective spermiogenesis. (A) Detailed analyses of spermatogenesis in Adad2+/+, Adad2–/–, and Adad2Δ/Δ males. Red arrowheads indicate abnormal migration or releasing of elongated spermatids. (B) Histological analyses of Adad2–/– testes. Red arrowheads indicate binucleated early round spermatids.
Figure S4. Ablation of ADAD2 causes changes in the transcriptome. (A) GO enrichment analyses of the 945 genes downregulated in Adad2Δ/Δ males. (B) Representative downregulated genes and upregulated genes in Adad2Δ/Δ testes.
Figure S5. ADAD2 interacts with RNA-binding proteins implicated in male reproduction. (A) GO enrichment analyses of ADAD2-interacting proteins identified by co-IP/MS analysis. (B) STRING analysis of ADAD2-interacting RNA-binding proteins. Blue indicates the proteins potentially involved in both piRNA biogenesis and spermatogenesis. Green indicates the proteins implicated in spermatogenesis. (C) Venn diagram depicting the proteins concurrently detected in the co-IP/MS analyses of ADAD2 and MAEL-interacting proteins.
Figure S6. ADAD2 granules show minor co-localization with MILI in pachytene spermatocytes. Immunohistochemistry analyses of ADAD2 and MILI in wildtype and Adad2–/– testis cryosections. ADAD2 was immunodetected using mAb#19–8 and cell nuclei were visualized by Hoechst 33342.
Figure S7. Adad1 is required for male fertility and spermiogenesis. (A-B) Schematic representation of Adad1 knockout mouse generation by CRISPR/Cas9. The sgRNAs #1” and #2”, which target the second and the twelfth exons, respectively, were used to delete the entire coding region of Adad1. Fw #1 and Rv primers were used to detect the wildtype allele, whereas Fw #2 and Rv primers were used to detect the knockout allele by genomic PCR. (C) Fertility tests of Adad1 knockout male mice. (D) Microscopic observation of cauda epididymal sperm in Adad1–/– males. (E) Histological analyses on the testes and epididymides of Adad1–/– males. (F) Immunodetection of ADAD1 and ADAD2 in Adad2–/– testes. ADAD1 and ADAD2 were immunodetected with the rabbit polyclonal antibodies. GAPDH was analyzed as a loading control. (G) Immunodetection of ADAD1 and ADAD2 in Adad1–/– testes.
Figure S8. Depletion of Adad2 does not induce LINE-1 derepression. (A) The 5’−5’ overlap between the piRNAs of opposite strands. (B) Western blot detection of MILI and MIWI immunoprecipitated by anti-MILI and MIWI antibodies, respectively. Small RNAs were co-immunoprecipitated and subjected to deep sequencing analyses. (C) Immunohistochemistry analyses of ADAD2 and LINE-1 in wildtype, Adad2–/– and Mili–/– testis cryosections. ADAD2 was immunodetected using mAb#19–8 and cell nuclei were visualized by Hoechst 33342.
Acknowledgements
The authors thank Prof. Toru Nakano for critical discussion on this study. This work was supported by the Ministry of Education, Culture, Sports, Science and Technology (MEXT)/Japan Society for the Promotion of Science (JSPS) KAKENHI grants (JP18K16735 and JP22K15103 to Y.L., JP16K07199 to I.N., JP21H02382 to H.K., JP18K14715 and JP20K15804 to J.M.C., and JP19H05750, JP21H04753, and JP21H05033 to M.I.); MEXT-Supported Program for the Strategic Research Foundation at Private Universities (S1311017 to H.K.); the Takeda Science Foundation (M.I.); the Eunice Kennedy Shriver National Institute of Child Health and Human Development (R01HD088412 and P01HD087157 to M.I. and M.M.M.); and the Bill and Melinda Gates Foundation (grant INV-001902 to M.I. and M.M.M.).
References
- 1.Nishimura H, L’Hernault SW. Spermatogenesis. Curr Biol. 2017;27(18):R988–R994. [DOI] [PubMed] [Google Scholar]
- 2.Fayomi AP, Orwig KE. Spermatogonial stem cells and spermatogenesis in mice, monkeys and men. Stem Cell Research. 2018;29:207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chuma S, Nakano T. piRNA and spermatogenesis in mice. Philosophical Transactions of the Royal Society B: Biological Sciences. 2013;368(1609):20110338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pillai RS, Chuma S. piRNAs and their involvement in male germline development in mice. Development, Growth & Differentiation. 2012;54(1):78–92. [DOI] [PubMed] [Google Scholar]
- 5.Aravin A, Gaidatzis D, Pfeffer S, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442(7099):203–207. [DOI] [PubMed] [Google Scholar]
- 6.Vourekas A, Zheng K, Fu Q, et al. The RNA helicase MOV10L1 binds piRNA precursors to initiate piRNA processing. Genes Dev. 2015;29(6):617–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wasik KA, Tam OH, Knott SR, et al. RNF17 blocks promiscuous activity of PIWI proteins in mouse testes. Genes Dev. 2015;29(13):1403–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Izumi N, Shoji K, Sakaguchi Y, et al. Identification and Functional Analysis of the Pre-piRNA 3’ Trimmer in Silkworms. Cell. 2016;164(5):962–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Fazio S, Bartonicek N, Di Giacomo M, et al. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature. 2011;480(7376):259–263. [DOI] [PubMed] [Google Scholar]
- 10.Ernst C, Odom DT, Kutter C. The emergence of piRNAs against transposon invasion to preserve mammalian genome integrity. Nature Communications. 2017;8(1):1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brennecke J, Aravin AA, Stark A, et al. Discrete Small RNA-Generating Loci as Master Regulators of Transposon Activity in Drosophila. Cell. 2007;128(6):1089–1103. [DOI] [PubMed] [Google Scholar]
- 12.Gunawardane LS, Saito K, Nishida KM, et al. A Slicer-Mediated Mechanism for Repeat-Associated siRNA 5’ End Formation in Drosophila. Science. 2007;315(5818):1587–1590. [DOI] [PubMed] [Google Scholar]
- 13.Siomi MC, Kuramochi-Miyagawa S. RNA silencing in germlines—exquisite collaboration of Argonaute proteins with small RNAs for germline survival. Curr Opin Cell Biol. 2009;21(3):426–434. [DOI] [PubMed] [Google Scholar]
- 14.Kuramochi-Miyagawa S, Kimura T, Ijiri TW, et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131(4):839–849. [DOI] [PubMed] [Google Scholar]
- 15.Grivna ST, Pyhtila B, Lin H. MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc Natl Acad Sci U S A. 2006;103(36):13415–13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carmell MA, Girard A, van de Kant HJG, et al. MIWI2 Is Essential for Spermatogenesis and Repression of Transposons in the Mouse Male Germline. Dev Cell. 2007;12(4):503–514. [DOI] [PubMed] [Google Scholar]
- 17.Deng W, Lin H. miwi, a Murine Homolog of piwi, Encodes a Cytoplasmic Protein Essential for Spermatogenesis. Dev Cell. 2002;2(6):819–830. [DOI] [PubMed] [Google Scholar]
- 18.Li Xin Z, Roy Christian K, Dong X, et al. An Ancient Transcription Factor Initiates the Burst of piRNA Production during Early Meiosis in Mouse Testes. Mol Cell. 2013;50(1):67–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vourekas A, Zheng Q, Alexiou P, et al. Mili and Miwi target RNA repertoire reveals piRNA biogenesis and function of Miwi in spermiogenesis. Nat Struct Mol Biol. 2012;19(8):773–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manakov Sergei A, Pezic D, Marinov Georgi K, Pastor William A, Sachidanandam R, Aravin Alexei A. MIWI2 and MILI Have Differential Effects on piRNA Biogenesis and DNA Methylation. Cell Rep. 2015;12(8):1234–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding D, Liu J, Midic U, et al. TDRD5 binds piRNA precursors and selectively enhances pachytene piRNA processing in mice. Nature Communications. 2018;9(1):127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimmins S, Sassone-Corsi P. Chromatin remodelling and epigenetic features of germ cells. Nature. 2005;434(7033):583–589. [DOI] [PubMed] [Google Scholar]
- 23.Goh WSS, Falciatori I, Tam OH, et al. piRNA-directed cleavage of meiotic transcripts regulates spermatogenesis. Genes Dev. 2015;29(10):1032–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gou L-T, Dai P, Yang J-H, et al. Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell Res. 2014;24(6):680–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu P-H, Fu Y, Cecchini K, et al. The evolutionarily conserved piRNA-producing locus pi6 is required for male mouse fertility. Nat Genet. 2020;52(7):728–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang P, Kang J-Y, Gou L-T, et al. MIWI and piRNA-mediated cleavage of messenger RNAs in mouse testes. Cell Res. 2015;25(2):193–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma L, Buchold GM, Greenbaum MP, et al. GASZ Is Essential for Male Meiosis and Suppression of Retrotransposon Expression in the Male Germline. PLoS Genet. 2009;5(9):e1000635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ikeda S, Tanaka K, Ohtani R, et al. Disruption of piRNA machinery by deletion of ASZ1/GASZ results in the expression of aberrant chimeric transcripts in gonocytes. Journal of Reproduction and Development. 2022;68(2):125–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shiromoto Y, Kuramochi-Miyagawa S, Nagamori I, et al. GPAT2 is required for piRNA biogenesis, transposon silencing, and maintenance of spermatogonia in mice. Biol Reprod. 2019;101(1):248–256. [DOI] [PubMed] [Google Scholar]
- 30.Shiromoto Y, Kuramochi-Miyagawa S, Daiba A, et al. GPAT2, a mitochondrial outer membrane protein, in piRNA biogenesis in germline stem cells. RNA. 2013;19(6):803–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castañeda J, Genzor P, van der Heijden GW, et al. Reduced pachytene piRNAs and translation underlie spermiogenic arrest in Maelstrom mutant mice. The EMBO Journal. 2014;33(18):1999–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soper SFC, van der Heijden GW, Hardiman TC, et al. Mouse Maelstrom, a Component of Nuage, Is Essential for Spermatogenesis and Transposon Repression in Meiosis. Dev Cell. 2008;15(2):285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuramochi-Miyagawa S, Watanabe T, Gotoh K, et al. MVH in piRNA processing and gene silencing of retrotransposons. Genes Dev. 2010;24(9):887–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pan J, Goodheart M, Chuma S, Nakatsuji N, Page DC, Wang PJ. RNF17, a component of the mammalian germ cell nuage, is essential for spermiogenesis. Development. 2005;132(18):4029–4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chuma S, Hosokawa M, Kitamura K, et al. Tdrd1/Mtr-1, a tudor-related gene, is essential for male germ-cell differentiation and nuage/germinal granule formation in mice. Proc Natl Acad Sci U S A. 2006;103(43):15894–15899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hosokawa M, Shoji M, Kitamura K, et al. Tudor-related proteins TDRD1/MTR-1, TDRD6 and TDRD7/TRAP: Domain composition, intracellular localization, and function in male germ cells in mice. Dev Biol. 2007;301(1):38–52. [DOI] [PubMed] [Google Scholar]
- 37.Wang J, Saxe JP, Tanaka T, Chuma S, Lin H. Mili Interacts with Tudor Domain-Containing Protein 1 in Regulating Spermatogenesis. Curr Biol. 2009;19(8):640–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shoji M, Tanaka T, Hosokawa M, et al. The TDRD9-MIWI2 Complex Is Essential for piRNA-Mediated Retrotransposon Silencing in the Mouse Male Germline. Dev Cell. 2009;17(6):775–787. [DOI] [PubMed] [Google Scholar]
- 39.Wenda JM, Homolka D, Yang Z, et al. Distinct Roles of RNA Helicases MVH and TDRD9 in PIWI Slicing-Triggered Mammalian piRNA Biogenesis and Function. Dev Cell. 2017;41(6):623–637.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Snyder E, Chukrallah L, Seltzer K, Goodwin L, Braun RE. ADAD1 and ADAD2, testis-specific adenosine deaminase domain-containing proteins, are required for male fertility. Sci Rep. 2020;10(1):11536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chukrallah LG, Badrinath A, Vittor GG, Snyder EM. ADAD2 regulates heterochromatin in meiotic and post-meiotic male germ cells via translation of MDC1. J Cell Sci. 2022;135(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu Y, Oura S, Matsumura T, et al. CRISPR/Cas9-mediated genome editing reveals 30 testis-enriched genes dispensable for male fertility in mice. Biol Reprod. 2019;101(2):501–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kojima-Kita K, Kuramochi-Miyagawa S, Nakayama M, et al. MORC3, a novel MIWI2 association partner, as an epigenetic regulator of piRNA dependent transposon silencing in male germ cells. Sci Rep. 2021;11(1):20472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Noda T, Lu Y, Fujihara Y, et al. Sperm proteins SOF1, TMEM95, and SPACA6 are required for sperm–oocyte fusion in mice. Proc Natl Acad Sci U S A. 2020;117(21):11493–11502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X, Sun J, Lu Y, et al. LRRC23 is a conserved component of the radial spoke that is necessary for sperm motility and male fertility in mice. J Cell Sci. 2021;134(20). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sherman BT, Hao M, Qiu J, et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022;50(W1):W216–W221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nishimura T, Nagamori I, Nakatani T, et al. PNLDC1, mouse pre-piRNA Trimmer, is required for meiotic and post-meiotic male germ cell development. EMBO Rep. 2018;19(3):e44957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hermann BP, Cheng K, Singh A, et al. The Mammalian Spermatogenesis Single-Cell Transcriptome, from Spermatogonial Stem Cells to Spermatids. Cell Rep. 2018;25(6):1650–1667.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10(6):845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li H, Coghlan A, Ruan J, et al. TreeFam: a curated database of phylogenetic trees of animal gene families. Nucleic Acids Res. 2006;34:D572–D580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruan J, Li H, Chen Z, et al. TreeFam: 2008 Update. Nucleic Acids Res. 2007;36(suppl_1):D735–D740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.MacLeod G, Shang P, Booth GT, et al. PPP1CC2 can form a kinase/phosphatase complex with the testis-specific proteins TSSK1 and TSKS in the mouse testis. Reproduction. 2014;147(1):1–12. [DOI] [PubMed] [Google Scholar]
- 53.Mi Y, Shi Z, Li J. Spata19 is critical for sperm mitochondrial function and male fertility. Mol Reprod Dev. 2015;82(11):907–913. [DOI] [PubMed] [Google Scholar]
- 54.Wang Y-Y, Ke C-C, Chen Y-L, et al. Deficiency of the Tbc1d21 gene causes male infertility with morphological abnormalities of the sperm mitochondria and flagellum in mice. PLoS Genet. 2020;16(9):e1009020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shimada K, Park S, Miyata H, et al. ARMC12 regulates spatiotemporal mitochondrial dynamics during spermiogenesis and is required for male fertility. Proc Natl Acad Sci U S A. 2021;118(6):e2018355118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen Y, Chen X, Zhang H, et al. TBC1D21 is an essential factor for sperm mitochondrial sheath assembly and male fertility. Biol Reprod. 2022. [DOI] [PubMed] [Google Scholar]
- 57.Miki K, Willis WD, Brown PR, Goulding EH, Fulcher KD, Eddy EM. Targeted Disruption of the Akap4 Gene Causes Defects in Sperm Flagellum and Motility. Dev Biol. 2002;248(2):331–342. [DOI] [PubMed] [Google Scholar]
- 58.Shirley CR, Hayashi S, Mounsey S, Yanagimachi R, Meistrich ML. Abnormalities and Reduced Reproductive Potential of Sperm from Tnp1- and Tnp2-Null Double Mutant Mice. Biol Reprod. 2004;71(4):1220–1229. [DOI] [PubMed] [Google Scholar]
- 59.Adham IM, Nayernia K, Burkhardt-Göttges E, et al. Teratozoospermia in mice lacking the transition protein 2 (Tnp2). Mol Human Reprod. 2001;7(6):513–520. [DOI] [PubMed] [Google Scholar]
- 60.Zhao M, Shirley CR, Mounsey S, Meistrich ML. Nucleoprotein Transitions During Spermiogenesis in Mice with Transition Nuclear Protein Tnp1 and Tnp2 Mutations1. Biol Reprod. 2004;71(3):1016–1025. [DOI] [PubMed] [Google Scholar]
- 61.Jha KN, Tripurani SK, Johnson GR. TSSK6 is required for γH2AX formation and the histone-to-protamine transition during spermiogenesis. J Cell Sci. 2017;130(10):1835–1844. [DOI] [PubMed] [Google Scholar]
- 62.Cho C, Willis WD, Goulding EH, et al. Haploinsufficiency of protamine-1 or −2 causes infertility in mice. Nat Genet. 2001;28(1):82–86. [DOI] [PubMed] [Google Scholar]
- 63.Merges GE, Meier J, Schneider S, et al. Loss of Prm1 leads to defective chromatin protamination, impaired PRM2 processing, reduced sperm motility and subfertility in male mice. Development. 2022;149(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sato H, Tsai-Morris C-H, Dufau ML. Relevance of gonadotropin-regulated testicular RNA helicase (GRTH/DDX25) in the structural integrity of the chromatoid body during spermatogenesis. Biochim Biophys Acta. 2010;1803(5):534–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bailey AS, Batista PJ, Gold RS, et al. The conserved RNA helicase YTHDC2 regulates the transition from proliferation to differentiation in the germline. eLife. 2017;6:e26116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Connolly CM, Dearth AT, Braun RE. Disruption of murine Tenr results in teratospermia and male infertility. Dev Biol. 2005;278(1):13–21. [DOI] [PubMed] [Google Scholar]
- 67.Watanabe T, Cheng E-c, Zhong M, Lin H. Retrotransposons and pseudogenes regulate mRNAs and lncRNAs via the piRNA pathway in the germline. Genome Res. 2015;25(3):368–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ichijima Y, Ichijima M, Lou Z, et al. MDC1 directs chromosome-wide silencing of the sex chromosomes in male germ cells. Genes Dev. 2011;25(9):959–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Information about Adad1 and Adad2 mutant mouse lines.
Table S2. Sequences of primers used in this study.
Table S3. RNA-seq analysis of postnatal day 24 wildtype and Adad2Δ/Δ testes.
Figure S1. Characterization of ADAD1 and ADAD2. (A) Multi-tissue RT-PCR analyses of human ADAD1 and ADAD2. La, ladder; Li, liver; Ki, kidney; Br, brain; In, intestine; Te, testis; Ut, uterus; Cap, caput epididymis; Cor, corpus epididymis; Cau, cauda epididymis. The expression of GAPDH was analyzed in parallel as a loading control. (B) mRNA expression of mouse Adad1 and Adad2 based on a previously published scRNA-seq analysis. (C) Domain composition of mouse ADAD1 and ADAD2. Both proteins contain a N-terminal double-stranded RNA-binding domain (dsRBD; in red) and a C-terminal adenosine deaminase domain (ADA; in blue). (D) Alignment of mouse ADAD1 and ADAD2 protein sequences. Red and blue represents dsRBD and ADA, respectively. (E) In silico homology modeling of the dsRBDs of mouse ADAD1 and ADAD2 by Phyre2. (F) Phylogenetic analyses of Adad1 and Adad2 by TreeFam.
Figure S2. Generation of an Adad2 mutant mouse line. (A) Schematic representation of Adad2 mutant mouse generation by CRISPR/Cas9. The sgRNA #1’ targeting the first exon was used to introduce a frameshift mutation (–27+2 bp) in Adad2. Fw’ and Rv’ primers were used to detect the wildtype and mutant alleles by genomic PCR. (B) Genomic PCR distinguishing the wildtype (WT) and the mutant (Mut) alleles (blue arrowheads). (C) Immunodetection of ADAD2 in wildtype and mutant testis lysates using the polyclonal antibody against aa 1 – 93 of mouse ADAD2. MVH was analyzed as a loading control. (D) Fertility tests of Adad2 mutant male mice. (E – F) Testis weights and appearance in wildtype and mutant males.
Figure S3. Adad2 deficient males show defective spermiogenesis. (A) Detailed analyses of spermatogenesis in Adad2+/+, Adad2–/–, and Adad2Δ/Δ males. Red arrowheads indicate abnormal migration or releasing of elongated spermatids. (B) Histological analyses of Adad2–/– testes. Red arrowheads indicate binucleated early round spermatids.
Figure S4. Ablation of ADAD2 causes changes in the transcriptome. (A) GO enrichment analyses of the 945 genes downregulated in Adad2Δ/Δ males. (B) Representative downregulated genes and upregulated genes in Adad2Δ/Δ testes.
Figure S5. ADAD2 interacts with RNA-binding proteins implicated in male reproduction. (A) GO enrichment analyses of ADAD2-interacting proteins identified by co-IP/MS analysis. (B) STRING analysis of ADAD2-interacting RNA-binding proteins. Blue indicates the proteins potentially involved in both piRNA biogenesis and spermatogenesis. Green indicates the proteins implicated in spermatogenesis. (C) Venn diagram depicting the proteins concurrently detected in the co-IP/MS analyses of ADAD2 and MAEL-interacting proteins.
Figure S6. ADAD2 granules show minor co-localization with MILI in pachytene spermatocytes. Immunohistochemistry analyses of ADAD2 and MILI in wildtype and Adad2–/– testis cryosections. ADAD2 was immunodetected using mAb#19–8 and cell nuclei were visualized by Hoechst 33342.
Figure S7. Adad1 is required for male fertility and spermiogenesis. (A-B) Schematic representation of Adad1 knockout mouse generation by CRISPR/Cas9. The sgRNAs #1” and #2”, which target the second and the twelfth exons, respectively, were used to delete the entire coding region of Adad1. Fw #1 and Rv primers were used to detect the wildtype allele, whereas Fw #2 and Rv primers were used to detect the knockout allele by genomic PCR. (C) Fertility tests of Adad1 knockout male mice. (D) Microscopic observation of cauda epididymal sperm in Adad1–/– males. (E) Histological analyses on the testes and epididymides of Adad1–/– males. (F) Immunodetection of ADAD1 and ADAD2 in Adad2–/– testes. ADAD1 and ADAD2 were immunodetected with the rabbit polyclonal antibodies. GAPDH was analyzed as a loading control. (G) Immunodetection of ADAD1 and ADAD2 in Adad1–/– testes.
Figure S8. Depletion of Adad2 does not induce LINE-1 derepression. (A) The 5’−5’ overlap between the piRNAs of opposite strands. (B) Western blot detection of MILI and MIWI immunoprecipitated by anti-MILI and MIWI antibodies, respectively. Small RNAs were co-immunoprecipitated and subjected to deep sequencing analyses. (C) Immunohistochemistry analyses of ADAD2 and LINE-1 in wildtype, Adad2–/– and Mili–/– testis cryosections. ADAD2 was immunodetected using mAb#19–8 and cell nuclei were visualized by Hoechst 33342.