Abstract
Cellular membranes function as permeability barriers that separate cells from the external environment or partition cells into distinct compartments. These membranes are lipid bilayers composed of glycerophospholipids, sphingolipids and cholesterol, in which proteins are embedded. Glycerophospholipids and sphingolipids freely move laterally, whereas transverse movement between lipid bilayers is limited. Phospholipids are asymmetrically distributed between membrane leaflets but change their location in biological processes, serving as signalling molecules or enzyme activators. Designated proteins — flippases and scramblases — mediate this lipid movement between the bilayers. Flippases mediate the confined localization of specific phospholipids (phosphatidylserine (PtdSer) and phosphatidylethanolamine) to the cytoplasmic leaflet. Scramblases randomly scramble phospholipids between leaflets and facilitate the exposure of PtdSer on the cell surface, which serves as an important signalling molecule and as an ‘eat me’ signal for phagocytes. Defects in flippases and scramblases cause various human diseases. We herein review the recent research on the structure of flippases and scramblases and their physiological roles. Although still poorly understood, we address the mechanisms by which they translocate phospholipids between lipid bilayers and how defects cause human diseases.
Subject terms: Phospholipids, Lipid signalling
Phospholipids are asymmetrically distributed between membrane leaflets but change their location in various biological processes, which requires designated proteins — flippases and scramblases. Recent insights into the functional mechanisms of these proteins pave the way for better understanding of the roles of membrane asymmetry and the (patho)physiological consequences of its disruption.
Introduction
Plasma membranes separate cells from the external environment, whereas the membranes at the endoplasmic reticulum (ER), mitochondria, the Golgi, lysosomes and nuclei separate organelles from the cytosol, ensuring cellular compartmentalization. Cellular membranes comprise lipid bilayers that consist of glycerophospholipids, sphingolipids and cholesterol. The ratio of various lipids differs among the membranes of different cellular organelles, with abundant sphingolipids at plasma membranes, the Golgi and lysosomes but fewer at mitochondria and the ER1,2. Phosphatidylcholine (PtdCho), phosphatidylethanolamine (PtdEtn), phosphatidylinositol (PtdIns) and phosphatidylserine (PtdSer) are major glycerophospholipids in cellular membranes, whereas sphingomyelin (SM) is the main sphingolipid.
Early studies utilizing the treatment of intact erythrocytes or their membrane fraction with chemical reagents that specifically label amino groups indicated that PtdCho and SM were mainly present in the extracellular leaflet, whereas PtdSer and PtdEtn were exclusively in the cytoplasmic leaflet3. Freeze-fracture electron microscopy subsequently confirmed the predominant localization of PtdSer in their cytoplasmic leaflets4,5. This asymmetrical distribution of phospholipids6–9 is essential for maintaining the integrity of plasma membranes and cellular organelles and also for signal transduction10,11. In various biological processes, PtdSer is exposed on the cell surface and serves as a signalling molecule or activates enzymes8,12. Phospholipids have a charged head group and two hydrophobic long acyl chains (Fig. 1a). As phospholipids with the charged head group hardly move spontaneously through the lipid bilayers of membranes, maintenance of their asymmetrical distribution and reshuffling rely on the presence of designated membrane proteins6.
Fig. 1. Flippases and scramblases that regulate the phospholipid distribution in the lipid bilayer.
a, Phospholipids. Glycerophospholipids are phosphatic acid (PA) derivatives composed of glycerol attached to two fatty acyl chains (diacylglycerol; DAG) and a phosphate. Serine, choline, or ethanolamine is conjugated with the phosphate of PA to produce phosphatidylserine (PtdSer), phosphatidylcholine (PtdCho) or phosphatidylethanolamine (PtdEtn). Sphingomyelin (SM) is a ceramide to which phosphocholine is attached. b, Flippases and scramblase. Under normal conditions, ATP-driven flippases translocate PtdSer and PtdEtn from the outer or lumen side to the inner leaflets of the lipid bilayer to maintain their confined localization to the inner leaflet. Scramblases non-specifically translocate phospholipids to disrupt the asymmetric distribution of phospholipids and to expose PtdSer on the outer or lumen side of cell membranes. c, Transfer of PtdSer from the endoplasmic reticulum (ER) to plasma membranes. PtdSer synthesized by PtdSer synthase at the ER is transferred to plasma membranes via a reciprocal exchange with PtdIns(4)P by a lipid transfer protein of the Oxysterol-binding protein (OSBP)-related protein (ORP) family265. PSS, phosphatidyl serine synthase.
At least three protein families regulate the distribution of phospholipids in cellular membranes. To confine specific phospholipids to the cytoplasmic leaflet of membranes, most of the type IV P-type ATPases (P4-ATPases; also referred to as flippases) translocate or flip anionic phospholipids (PtdSer and PtdEtn) from the outer to inner leaflets, coupled with their ATPase activity13,14. To disrupt the asymmetrical distribution of phospholipids, transmembrane protein 16 (TMEM16) and XK-related (XKR) family members (also referred to as scramblases) non-specifically scramble phospholipids along their concentration gradient between lipid bilayers in a manner dependent on Ca2+ (a ubiquitous second messenger, typically associated with cell activation) or caspases (mediators of cell dismantling during apoptosis), respectively12. Some of these proteins are ubiquitously expressed, whereas others show specific expression limited to only particular cells or subcellular localizations.
The importance of these proteins is underlined by the fact that, in humans, various recessive and dominant mutations have been identified in proteins belonging to all three families. Yet the physiological roles of these proteins and the molecular mechanisms by which they translocate phospholipids in lipid bilayers have been challenging to address owing to their large family size and the difficulties in determining the tertiary structure of membrane proteins. The recent development of CRISPR–Cas9 technology for knockout genes15, single-cell RNA sequence technology to examine the gene expression16, cryogenic electron microscopy technology17 and the AlphaFold artificial intelligence program18 to elucidate or predict the tertiary structures of membrane proteins has resulted in notable advances in the field. Also, the mechanisms by which each member of the P4-ATPases, TMEM16 and XKR families regulate the distribution of phospholipids in cellular membranes are gradually being clarified.
PtdSer is a phospholipid with the most extreme asymmetrical distribution — almost exclusively localizing to the inner membrane leaflet — and its exposition on the cell surface performs various biological functions. PtdSer is thus a prominent substrate for flippases and scramblases, and the mechanisms involved in PtdSer movement across the bilayer are the best characterized. We herein review how P4-ATPase flippases maintain PtdSer in the inner leaflet of plasma membranes and discuss when this asymmetrical distribution is disturbed (in response to both physiological signals and disease) by exposition of PtdSer on the ectoplasmic leaflet of the plasma membrane. We describe the mechanisms by which PtdSer is exposed on the cell surface by scramblases, how PtdSer supports biological processes and how abnormal distribution of PtdSer caused by mutations in flippases and scramblases contributes to pathology.
Families of flippases and scramblases
As all phospholipids are synthesized intracellularly, and phospholipids cannot easily transverse the lipid bilayer, early on in the field of membrane biology it has been postulated that proteins that are able to ‘flip’ (inward moving), ‘flop’ (outward moving) and ‘scramble’ (bidirectional moving) phospholipids between the lipid bilayer must exist19 (Fig. 1b). Flippases and floppases specifically translocate PtdSer and PtdEtn from the outer to the inner leaflet of the membranes and PtdCho in the opposite direction, respectively. Scramblases non-specifically and bidirectionally translocate phospholipids between the lipid bilayer and collapse the membrane asymmetry20. Flippases and floppases require the energy from ATP to perform the ‘uphill’ reaction or transport phospholipids against the concentration to establish the asymmetrical distribution of phospholipids. By contrast, phospholipid scrambling is a ‘downhill’ reaction and does not require ATP. The identity of flippases, floppases and scramblases had been elusive for a long time. The flippase and scramblase activities can be assayed by the incorporation of the fluorescently labelled phospholipids or with staining agents for PtdSer and PtdEtn (that is, Annexin V and cinnamycin, respectively), and are well characterized. We now know that P4-ATPases serve as the flippases at various cellular compartments, whereas two different families (TMEM16 and XKR) work as Ca2+-dependent or caspase-dependent scramblase, respectively12. Although the ABC transporters appear to work as floppases6, their characterization was hampered because there is no convenient assay system for the floppase activity. Here, we describe flippases and scramblases, which are relatively well characterized.
Flippases
The flippase activity that specifically incorporates PtdSer and PtdEtn in an ATP-dependent manner was first detected in human erythrocytes21. The ATPase was purified from human erythrocyte membranes, and flippase activity was reconstituted in proteoliposomes22. A protein (ATP8A1) with similar biochemical characteristics was purified from bovine chromaffin granules, and its molecular cloning identified it as a member of the P4-ATPase family23.
The role of P4-type ATPases in lipid ‘flipping’ between the bilayer
P-type ATPases constitute a large protein family that transport cations and lipids across the membrane24. They are named ‘P-type’ because a conserved aspartic acid is transiently modified by phosphorylation during the reaction cycle. A phylogenic analysis divided the family into five subfamilies (P1–P5 types). Similar to other P-type ATPases, P4-ATPases are membrane proteins with cytoplasmic amino and carboxy termini. ATPase activity is executed by a large domain comprising two cytoplasmic loops.
We performed forward genetic screening of a human cell line to identify the flippase that functions at the plasma membrane and identified ATP11C and CDC50A (ref. 25). By expressing 12 members of the human P4-ATPase family in ATP11C–/– cells, ATP11A was also shown to function as a flippase at the plasma membrane as a complex with CDC50A, the latter functioning as a chaperone to localize ATP11A and ATP11C to the plasma membrane26 (Fig. 2a). ATP11A and ATP11C are ubiquitously expressed, suggesting that they redundantly work in various cells. Several other members of the family are present in the plasma membrane (ATP8B1, ATP8B2, ATP8B4 and ATP10D) with ATP8B1 at the canalicular (apical) membrane of hepatocytes27; however, their intact forms did not exhibit flippase activity against PtdSer at the cellular level26. Other members have been detected in endosomes (ATP8A1, ATP8A2, ATP9A, ATP11B), the Golgi (ATP9B) and lysosomes (ATP10B)28,29. All members, except for ATP9A and ATP9B, require CDC50A or its paralogue for their localization to the specific cellular compartment. Most members are expressed in a tissue-specific manner: ATP8A1 and ATP8A2 in neuronal cells in the brain; ATP8B1 and ATP10B in epithelial cells in the intestine; ATP8B4 in myeloid cells; and ATP8B3 in the testis (Table 1).
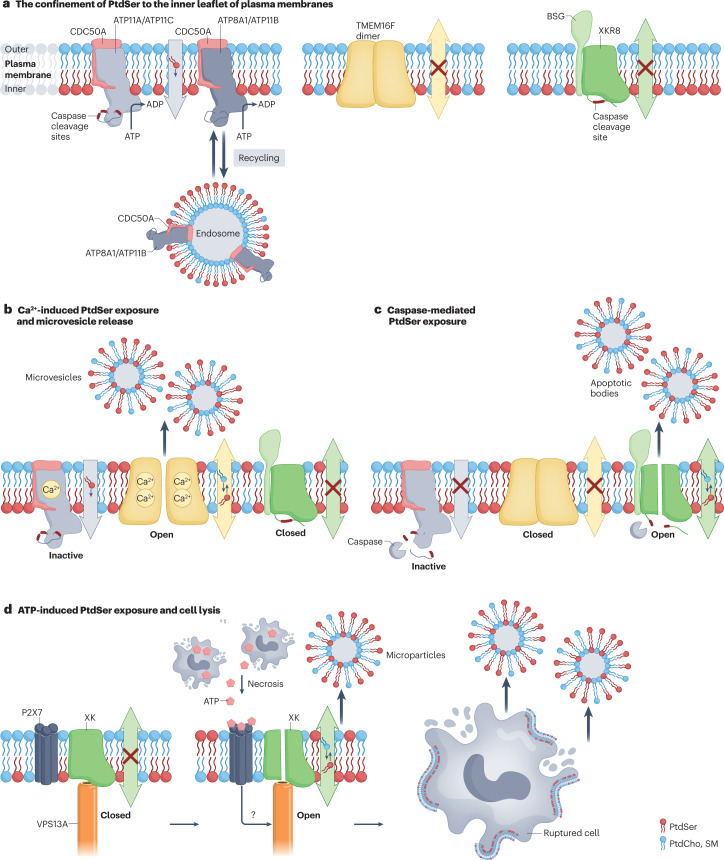
Fig. 2. Asymmetrical distribution of PtdSer and its breakdown.
a, Type IV P-type ATPase (P4-ATPase)-mediated confinement of phosphatidylserine (PtdSer) to the inner leaflet of the plasma membrane. ATP11A and ATP11C are complexed with CDC50A at the plasma membrane and serve as flippases to specifically translocate PtdSer from the outer leaflet to the inner leaflet in growing cells. ATP8A1 and ATP11B seem to recycle between the endosomes and plasma membranes and maintain the asymmetrical distribution of PtdSer at the plasma membrane. b, Ca2+-induced PtdSer exposure and microvesicle release. When platelets, osteoblasts and other cells are activated, the intracellular Ca2+ concentration increases. Binding of Ca2+ to ATP11A and ATP11C inhibits their flippase activity while activating TMEM16F, which causes temporal PtdSer exposure on the cell surface and the release of microvesicles. These microvesicles also expose PtdSer, which conveys further functions (see also Fig. 4c). c, When cells undergo apoptosis, caspase 3 is activated and cleaves the carboxy-terminal part of XKR8 to trigger its scrambling activity. At the same time, caspase 3 cleaves ATP11A and ATP11C in the middle of the molecule to inactivate them, allowing the cells to expose PtdSer irreversibly. This process is accompanied by the release of apoptotic bodies, which also expose PtdSer and are engulfed by phagocytes. d, ATP-induced PtdSer exposure and cell lysis. XK, a member of the XK-related (XKR) family, is complexed with VPS13A lipid transporter via its β-hairpin in the centre of the molecule. ATP released from necrotic cells binds the P2X7 homotrimer, and an unidentified signal (question mark) from the ATP-engaged P2X7 receptor activates the XK–VPS13A complex to scramble phospholipids in the plasma membrane. This causes the PtdSer exposure on the cell surface, followed by cell rupture. PtdCho, phosphatidylcholine; SM, sphingomyelin; TMEM16, transmembrane protein 16.
Table 1.
Mammalian flippases and scramblases
| Protein | Tissues and cells | Subcellular localization | Substrates or scramblase activity | Global and cellular phenotypes associated with mutation/misexpression | Refs. |
|---|---|---|---|---|---|
| P4-ATPases | |||||
| ATP8A1 | Ubiquitous | Endosomes | PtdSer | Recessive biallelic mutations in humans: inefficient memory; PtdSer exposure on hippocampal cells | 248 |
| ATP8A2 |
Retina, cerebral cortex, cerebellum, hypothalamus Pituitary gland |
Endosomes Outer segment of photoreceptors |
PtdSer | Recessive biallelic mutations in humans: chorea, cerebellar ataxia and mental retardation; chromatolysis, axonal degradation and optic atrophy; mice with recessive mutation (wabbler-lethal) die within 130 days | 249–251 |
| ATP8B1 | Intestines, colon, stomach | Plasma membrane | PtdSer, PtdCho | Recessive biallelic mutations in humans: cholestasis and hearing loss; increased sensitivity of canalicular membranes to bile acid, degeneration of cochlear hair cells | 252,253 |
| ATP8B2 | Ubiquitous | Plasma membrane | PtdCho | NR | |
| ATP8B3 | Testis | NR | NR | NR | |
| ATP8B4 | Bone marrow (myeloid) | Plasma membrane | NR | NR | |
| ATP9A | Ubiquitous | Endosomes | NR | Recessive biallelic mutations in humans: intellectual disability and microcephaly | 254,255 |
| ATP9B | Ubiquitous | Golgi | NR | NR | |
|
ATP10A (also known as ATP10C) |
Ubiquitous | Plasma membrane | PtdCho |
Recessive biallelic mutations in humans: neurobehavioural abnormality Obesity, insulin resistance |
256,257 |
| ATP10B | Intestines and colon | Endosomes, lysosomes | PtdCho, GluCer | NR | 32,258 |
| ATP10D | Kidney and placenta | Plasma membrane | GluCer | C57BL/6 mouse strain carries the nonsense mutation: susceptible to diabetes exposed to high-fat diet | 259,260 |
| ATP11A | Ubiquitous except for hepatocytes, B cells and red blood cells | Plasma membrane | PtdSer, PtdEtn |
Dominant heterozygous mutation in humans: developmental delays, neurological deterioration and hearing loss Knockout mice: embryonic lethal due to poor development of syncytiotrophoblasts |
102,180,215 |
| ATP11B | Ubiquitous | Endosomes | PtdSer, PtdEtn | NR | |
| ATP11C | Ubiquitous except for trophoblast in placenta | Plasma membrane | PtdSer, PtdEtn |
Recessive biallelic mutations in humans: anaemia Knockout mice: B cell lymphopenia, cholestasis |
37,205,208,209 |
| TMEM16 scramblases | |||||
| TMEM16A (also known as anoctamin 1) | Ubiquitous | Plasma membrane | Inactive | NR | |
| TMEM16B | Retina, nasopharynx, bronchus and fallopian tube | Plasma membrane | Inactive | NR | |
| TMEM16C |
Cerebral cortex Basal ganglia |
Plasma membrane | Active (Ca2+) |
Dominant heterozygous mutation in humans: dystonia Recessive biallelic mutations in humans: febrile seizure Knockout rat: dysregulated Ca2+ homeostasis |
217,218,261 |
| TMEM16D | Cerebral cortex, basal ganglia, adrenal gland, seminal vessel, ovary | Plasma membrane | Active (Ca2+) | NR | |
| TMEM16E |
Cerebral cortex, cerebellum Heart and skeletal muscle |
ER | Active (Ca2+) |
Dominant heterozygous mutation in humans: gnathodiaphyseal dysplasia Recessive biallelic mutations in humans: muscle dystrophy caused by dysregulated Ca2+ homeostasis in myotubes |
71 |
| TMEM16F | Ubiquitous | Plasma membrane | Active (Ca2+) | Recessive biallelic mutations in humans and mice: bleeding disorder caused by inefficient PtdSer exposure in platelets | 44,226–228 |
| TMEM16G | Stomach, intestines, prostate, skin | Plasma membrane: cell contact site | Active (Ca2+) | Low expression associated with human prostate cancer (function as a tumour suppressor?) | 232 |
| TMEM16H | Ubiquitous | ER | NR | NR | |
| TMEM16J | Skin and intestines: enterocytes | Plasma membrane | Active (Ca2+) | High expression associated with human colorectal and pancreatic cancer (function as a tumour promoter?) | 262,263 |
| TMEM16K | Ubiquitous | Contact site between ER and endosomes | Active (Ca2+) |
Recessive biallelic mutations in humans: spinocerebellar ataxia Recessive mutations in mice: defective endosomal sorting |
73,264 |
| XKR scramblases | |||||
| XK | Ubiquitous | Plasma membrane | Active, with VPS13A (ATP) | Recessive biallelic mutations of XK or VPS13A in humans: neuroacanthocytosis; Xk or Vps13a-knockout mice: thinner myelin sheet | 124,238,239 |
| XKR2 | Skin | ER | NR | NR | |
| XKR3 | Testis | Plasma membrane | NR | NR | |
| XKR4 | Cerebral cortex, basal ganglia, thalamus, amygdala | Plasma membrane | Active (caspase) | NR | |
| XKR5 | Neutrophils | Plasma membrane | NR | NR | |
| XKR6 | Cerebral cortex, cerebellum | Plasma membrane | NR | NR | |
| XKR7 | Cerebral cortex, cerebellum, basal ganglia | Plasma membrane | NR | NR | |
| XKR8 | Ubiquitous | Plasma membrane | Active, with Basigin (caspase) | Knockout mice: SLE-type autoimmune disease, male infertility; splenomegaly and glomerulonephritis | 78,240 |
| XKR9 | Intestines, stomach, testis | Plasma membrane | Active (caspase) | NR | |
The tissue distribution (The Human Protein Atlas) for each P4-ATPase, TMEM16 and XKR family member is listed with their subcellular localization. The known substrate for P4-ATPase is shown. For TMEM16 and XKR family members, the proteins with documented scramblase activity shown at the cellular level are indicated as ‘Active’. The triggers (Ca2+, caspase or ATP) that activate the scramblase are shown in parentheses. ER, endoplasmic reticulum; GluCer, glucosylceramide; NR, not reported; P4-ATPase, type IV P-type ATPase; PtdCho, phosphatidylcholine; PtdEtn, phosphatidylethanolamine; PtdSer, phosphatidylserine; SLE, systemic lupus erythematosus; TMEM16, transmembrane protein 16; XKR, XK-related.
In addition to ATP11A and ATP11C, four members (ATP8A1, ATP8A2, ATP8B1 and ATP11B) exhibit PtdSer/PtdEtn flippase activity in reconstituted proteoliposomes or can be activated by PtdSer for their ATPase activity27,29–31. By contrast, ATP10B localized at late endosomes and lysosomes flips glucosylceramide (GluCer) and PtdCho towards the cytoplasmic leaflets32. Surprisingly, two yeast P4-ATPases (Dnf1 and Dnf2)33 and three mammalian members (ATP8B1, ATP8B2 and ATP10A)34,35 were reported to flip PtdCho at the plasma membrane.
Flippase-mediated establishment of the asymmetrical distribution of phospholipids
A mouse T cell line (WR19L) that lacks CDC50A constitutively exposes PtdSer on the surface25, indicating that a CDC50A-dependent enzyme is involved in establishing the asymmetrical distribution of phospholipids in plasma membranes. Among the six P4-ATPases expected to flip PtdSer, WR19L cells express ATP11A and ATP11C at the plasma membrane and ATP8A1 and ATP11B at endosomes (Fig. 2a). Cells deficient in these four P4-ATPases did not expose PtdSer36. However, when the cells were transiently treated with a Ca2+ ionophore to increase the intracellular Ca2+ to activate the scramblase, PtdSer was exposed to the surface for several hours36. As constitutively exposed PtdSer serves as an ‘eat me’ signal, even in living cells25,37, cells are equipped with a device in the form of P4-ATPases that counteracts this exposure (Fig. 2a). ATP11A and ATP11C quickly flip PtdSer or internalize it to the inner leaflet; this is further supported by ATP8A1 and ATP11B, which recycle between the plasma membrane and endosomes. Nevertheless, PtdSer is not constitutively exposed on cells lacking these flippases36. It may be that the long-term asymmetry in PtdSer distribution is supported by the transport of PtdSer from the site of its synthesis in the ER — a process confined to the cytoplasmic leaflet (Fig. 1c).
Scramblases
Once an asymmetrical distribution of phospholipids is established, the charged head group hardly moves through the hydrophobic lipid bilayer without lipid transporters38. Yet platelets treated with collagen and thrombin expose PtdSer within minutes39, and cells undergoing apoptosis expose it within hours of pro-apoptotic signal reception40,41. A scramblase that provides a non-specific path for phospholipids to access the ectoplasmic leaflet was postulated to explain this swift exposure of PtdSer. A protein called PLSCR1 (phospholipid scramblase) was purified based on a cell-free assay with proteoliposomes42. However, its molecular and physiological characteristics were inconsistent with the scramblase activity43.
Scramblases of the TMEM16 family
To identify the scramblase, we repeatedly sorted mouse Ba/F3 haematopoietic cells to obtain the population that strongly responded to a Ca2+ ionophore for PtdSer exposure. A cDNA library was prepared from these cells. The transformation of Ba/F3 cells with the cDNA library identified two different cDNAs: TMEM16F, also called anoctamin 6 or ano6, and XKR8 (refs. 44,45) (Table 1 and Fig. 2a–c).
TMEM16F belongs to the TMEM16 family. As its founding member (TMEM16A) is a Ca2+-dependent Cl– channel, several groups reported that TMEM16F functions as an ion channel46–49. However, their results are conflicting. It was claimed to be a volume-regulated anion channel48, a Ca2+-activated chloride channel46, an outwardly rectifying chloride channel49 or a non-selective cation channel47. The Ca2+-dependent Cl− channel activity was not detected under the conditions where TMEM16A and TMEM16B show strong ion channel activity50–52. We reason that the main function of TMEM16F is likely lipid scrambling, whereas ion transport is the secondary activity triggered under extreme, less physiological conditions53.
The TMEM16 family has ten members. Seven members localize to plasma membranes, five of which (TMEM16C, TMEM16D, TMEM16F, TMEM16G and TMEM16J) respond to a Ca2+ ionophore for PtdSer exposure50. An analysis of the scramblase activity of the chimaera between the TMEM16A Cl− channel and TMEM16F scramblase predicted that a 35 amino acid domain between transmembrane 4 (TM4) and TM5 was responsible for scrambling activity52. Replacing the corresponding region of TMEM16A with that of other TMEM16 family members showed that TMEM16E and TMEM16K localized at the ER can scramble phospholipids54 (Table 1). The scramblase activities of TMEM16F and TMEM16K were confirmed by reconstituting the purified proteins in proteoliposomes55–57. Thermodynamic analysis indicated that mouse TMEM16F transported 4.5 × 104 lipids per second at 25 °C58. This value is much higher than that observed with ATP-dependent transporter (<102 molecules per second)59, which is consistent with the model whereby TMEM16F provides a cleft or conducting pathway for phospholipids upon Ca2+ binding (see below).
Among the five TMEM16 members that function as Ca2+ scramblases at the plasma membrane, TMEM16C, TMEM16D, TMEM16G and TMEM16J are expressed in a limited number of tissues, such as the brain, skin and intestines50. TMEM16F is ubiquitously expressed, and is solely responsible for the Ca2+-dependent scrambling of phospholipids at the plasma membranes of most cells, including hepatocytes, lymphocytes and haematopoietic cells. Of the ER-specific Ca2+-dependent scramblases, TMEM16K is ubiquitously expressed, whereas TMEM16E is mainly expressed in bone, muscle and the testis60.
TMEM16F plays an indispensable role in the exposure of PtdSer in various physiological contexts. One notable example is activated platelets. When endothelial cells are damaged, collagen is released leading to platelet activation. The activated state is associated with transient increases of the intracellular Ca2+ concentrations to several hundred micromoles per litre beneath the plasma membrane61. This increase in Ca2+ activates TMEM16F scramblase and inactivates the flippases of ATP11A and ATP11C (ref. 26), thereby rapidly exposing PtdSer (Fig. 2b). In response to the increased Ca2+ concentration62, differentiating osteoblasts also expose PtdSer in a TMEM16F-dependent manner63. In addition, TMEM16F supports the release of microparticles/microvesicles from platelets, neutrophils and osteoblasts63–65. Furthermore, TMEM16F-mediated scrambling of phospholipids has been implicated in the repair of pores in the plasma membrane introduced by stress or toxins, or after triggering cell death via pyroptosis, which is followed by the expansion of plasma membranes and the release of damaged membranes as vesicles66–68.
The physiological roles of other TMEM16 family members remain unknown, with a few hints for TMEM16E and TMEM16K. For example, when the sarcolemma, the plasma membrane of skeletal muscle cells, is damaged, TMEM16E moves from the ER to the damaged sites. Then, it undertakes the repair of the membranes by arranging annexins69 on the patches with a not well-understood mechanism70–72. The scrambling activity of TMEM16K contributes to the distribution of phospholipids between intracellular organelles4, regulating endosomal sorting at the ER–endosome contact sites73. Various human diseases are caused by mutations in the TMEM16 family members (Table 1), necessitating further research in this field.
Scramblases of the XKR family
The TMEM16F-null mouse T cell line cannot respond to Ca2+ but responds to apoptotic stimuli for the exposure of PtdSer50. This PtdSer-exposing activity in the apoptotic cells was lost when the XKR8 gene was knocked out74. XKR8 carries a caspase 3-recognition site at its C-terminal region. When cells undergo apoptosis, caspase 3 cleaves XKR8 to activate it. These findings established the presence of two independent systems for the PtdSer exposure: Ca2+-mediated or caspase-mediated mechanisms75,76 (Fig. 2b,c). In apoptotic cells, caspase 3 cleaves ATP11A and ATP11C to inactivate them, indicating the apoptotic PtdSer exposure is irreversible.
The nine XKR family genes in human chromosomes appear to be generated by co-duplicating neighbouring genes77. Apart from XKR2, all XKR members localize to the plasma membrane (Table 1). XKR4, XKR8 and XKR9 carry a caspase recognition site in the C-terminal region and can support the exposure of PtdSer during apoptosis74. The expression of XKR8 is ubiquitous, whereas XKR4 and XKR9 are confined to the brain and intestines, respectively. Therefore, XKR8 is responsible for the apoptotic exposure of PtdSer in most cell types78. XKR8 associates with Basigin or Neuroplastin of the immunoglobulin superfamily for its localization to plasma membranes79. It remains unclear whether Basigin and Neuroplastin, and any other protein(s), contribute to the scrambling activity of XKR8. XKR8 can function as a constitutively active scramblase in an interleukin (IL)-3-dependent mouse Ba/F3 cell line because it is phosphorylated at three serine/threonine residues downstream of the caspase recognition site80. As discussed below, the C-terminal domain of human XKR8 downstream of the caspase recognition site seems to bind to the cytoplasmic region of the transmembrane helices to inhibit the scrambling activity of XKR8. It is likely that the phosphorylation of the residues in this region blocks this interaction. In some biological processes such as the capacitation of sperm that occurs in the female reproductive tract, PtdSer is exposed on the surface of sperm heads in a kinase-dependent manner81. How and when XKR8 is activated by phosphorylation remains to be determined.
Many cells undergo necrosis in inflamed tissues or the tumour microenvironment, and a large amount of ATP released from the necrotic cells accumulates locally, reaching several hundred micromoles per litre82,83. ATP at this high concentration binds to P2X7 ATP receptors in various cells, particularly CD25+CD4+ regulatory T cells and macrophages, to induce PtdSer exposure, followed by cell lysis. Screening for molecules responsible for the ATP-induced exposure of PtdSer on T cells identified XK — a paralogue of XKR8 —as a scramblase responsible for this ATP-induced PtdSer exposure. Interestingly, XK partners with lipid transporter VPS13A at the plasma membrane to function as a scramblase84. An unidentified signal(s) seems to be transferred from ATP-engaged P2X7 receptors to the XK–VPS13A complex to scramble phospholipids at plasma membranes84 (Fig. 2d). This process is followed by necrosis in T cells and pyroptosis in macrophages. VPS13A transports phospholipids between membranes of intracellular organelles and the ER85. It is tempting to speculate that the XK–VPS13A complex directly transfers phospholipids from the outer leaflet of the plasma membrane to the cytoplasmic leaflets of the ER, and vice versa. To understand the physiological role of this system, we have to know how the extracellular ATP, or P2X7-mediated signal, activates the system.
Models for lipid transport
The mechanisms underlying phospholipid translocation by flippases and scramblases have been a topic of much debate in recent years. Based on the amino acid sequence, the structure of P4-ATPases was thought to be similar to other P-type ATPases. However, the canonical substrate-binding pocket is too small for lipid substrates, which are considerably larger than the inorganic ions transported by other P-type ATPases, posing a problem referred to as the ‘giant substrate’ problem86. To overcome this issue, a ‘credit card’ model has been proposed in which only the hydrophilic head groups of phospholipids penetrate and traverse the hydrophilic groove87 (Fig. 3a). At the same time, their acyl chains remain within the membrane environment. The structure of the P4-ATPases, TMEM16 and XKR essentially supports the credit card model and refines the model with the concept of a ‘central cavity model’88, ‘alternating pore/cavity mechanism’89, ‘clamshell model’ and ‘thinning the membrane model’90.
Fig. 3. Models for lipid transport.
a, Credit card model for lipid transport. The hydrophilic head group of the phospholipid (corresponding to the magnetic strip on the credit card) is protected by the hydrophilic groove of the transporter (track of the card reader) during lipid transport. Image reproduced from Pomorski et al., ref. 87. b, Structure of ATP11C–CDC50A and a model for lipid flipping. Tertiary structure of human ATP11C and CDC50A complex in a phosphatidylserine (PtdSer) occluded E2-Pi state [PDB:7BSV] shown on the left. P4-ATPases are coloured grey with selected helices (α1, α2, α4 and α6) in blue, whereas CDC50A is red. Actuator domain (A) which dephosphorylates the phosphorylated P site shown in yellow. PtdSer is coloured green–red–white. Right panel shows the proposed lipid transport cycle. In an outward open state (E2P), in which the phosphorylation (P) site is phosphorylated (P in yellow circle), a phospholipid binds to the external open cavity. In the phospholipid occluded state (E2-Pi), the phosphate is detached from the P site (Pi in a yellow circle), and helices α1 and α2 move to cover the ectoplasmic side of the cavity (red arrow). In the inward open state (E2), the phosphate (Pi in yellow circle) is released from the molecule (blue arrow), which is coupled by the movement of the A domain to the periphery (black arrow). This changes the arrangement of helices α1 and α2 (green arrow), and the cavity is now accessible to the cytosolic leaflet releasing the phospholipid. c, Structure of transmembrane protein 16 (TMEM16) and model for lipid scrambling. The Ca2+-bound open state structure of Nectria haematococca (nh)TMEM16 [PDB:4WIS] is viewed from the front and side (left). Each protomer of TMEM16 is coloured grey or dark grey. Selected helices (α3, α4, α5 and α6) are coloured blue and labelled. On the right, the activation and scrambling, viewed from the side, are schematically depicted. In a Ca2+-free closed state, the cavity is shielded from the membrane with helices α4 and α6 tightly interacting. The binding of Ca2+ triggers the conformational change to the open state; helices α4 and α6 are separated, and the cavity is accessible to the phospholipid head group. d, Structure of XK-related 8 (XKR8) and a model for lipid scrambling. The tertiary structure of the human XKR8–Basigin complex [PDB:7DCE] is shown with phosphatidylcholine (PtdCho) bound to the hydrophobic cleft. Selected helices (α1, α2, α4 and α11) are coloured blue and labelled. A model for the caspase-activated scrambling mechanism is shown on the right. A phospholipid is recruited from the outer leaflet to the hydrophobic cleft, even in a closed state. When cells undergo apoptosis, caspase 3 cleaves helix 11, which likely induces the movement of helix 2 (red arrows) to expose the path for phospholipid transport.
The central cavity model proposes that the central membrane cavity of the P4-ATPase accommodates the lipid head groups during lipid transportation across the membrane. The alternating pore/cavity mechanism suggests that the TMEM16 family adopts two alternating conformations with varying degrees of groove opening when activated, one with an ion-conductive pore and the other with a lipid-conductive cavity. The clamshell model proposes that the interface of two helices composing the groove for phospholipid permeation can open and close similar to a clamshell, changing the accessibility of phospholipid head groups to the hydrophilic interior of the track. In the thinning the membrane model, TMEM16 thins the membrane around the groove, which lowers the energy barrier for lipid scrambling.
Tertiary structures of P4-ATPases and a model for lipid transport
The tertiary structures of several P4-ATPases (human ATP8A1, ATP8B1, ATP11C, yeast Drs2, Dnf1 and fungus Dnf1) complexed with their accessory subunits, CDC50 protein family members, were elucidated by X-ray crystallography and cryogenic electron microscopy, and revealed the phospholipid flipping mechanism30,31,88,91–97 (Fig. 3b). The overall structures of P4-ATPases represent the characteristic P-type ATPase architecture, which contains ten TM helices as well as three cytoplasmic domains for ATP catalysis: the actuator (A) domain, which dephosphorylates the phosphorylated P domain, the nucleotide-binding (N) domain and the phosphorylation (P) domain. The N-terminal and C-terminal regions (ATP8B1) or the C-terminal regions (ATP8A1 and Drs2) can regulate their ATPase activities. The CDC50 subunit has a large extracellular domain, two TM helices and cytoplasmic N-terminal and C-terminal regions. The two TMs of CDC50A are located close to the TM10 of P4-ATPases and form hydrophobic interactions with the residues. The heavily glycosylated extracellular region of CDC50A covers all of the extracellular loops between the TM helices of the ATPase, except for that between α1 and α2. The N-terminal cytoplasmic tail runs along the membrane and interacts with the cytoplasmic side of the TMs of P4-ATPases.
The structures of several catalytic states of P4-ATPases, stabilized with ATP analogues and ATPase inhibitors, were elucidated. Although the scale of conformational changes during the reaction cycle differed among various P4-ATPases88,91–93,95–97, the flipping mechanism essentially follows the Post–Albers cycle described for cation-transporting P-type ATPases98,99. In this model, substrate-binding sites alternate between the cytosolic site-open state (E1, E1-ATP and E1P-ADP), temporary occlusion (E1P and E2) and the ectoplasmic or lumen site-open state (E2P and E2-Pi), in which E1P and E2P represent the phosphorylated form.
A notable feature of P4-ATPases is the significant movement of the A domain during the E1P to E2P transition, which separates TM1 and TM2 from TM4 and TM6, forming a cavity comprising TM2, TM4 and TM6 (refs. 88,91,93,96,97) (Fig. 3b). The cavity spans from the ectoplasmic side to the middle of the membrane. It is wide enough to accommodate phospholipid head groups, providing an outward pathway for phospholipids. At the bottom of the cavity, an uncoiled conserved PISL/PVSM (Pro-Ile-Ser-Leu/Pro-Val-Ser-Met) motif is present at the centre of TM4 and appears to act as a gate to control lipid access100. In the outward open state, this gate blocks the access of the lipid to the cytoplasmic side, whereas in the inward open state, this gate is predicted to be open allowing the lipid to diffuse into the cytoplasmic leaflet. In the E2P state, the cavity has an outward-facing conformation in which PtdSer is captured in the cavity with a head group coordinated by several polar residues at the bottom of the cavity and acyl chains orienting to the hydrophobic core of the membrane (Fig. 3b). In the following E2-Pi state, PtdSer is occluded in the cavity because conformational changes in TM1 and TM2 close the entrance gate from the ectoplasmic side. In the following form (E2), the cavity adopts the inward open conformation, with hydrophilic residues exposed to the cytoplasmic side to release PtdSer96,97. In this flipping mechanism, the lipid head group moves in the groove between TM2 and TM4 containing charged residues, whereas the lipid tail moves along the hydrophobic surface of TM2 and TM4. This mechanism is essentially consistent with the credit card model87.
The structure of human ATP8A1, ATP11C and yeast Drs2p and Neo1p which flip PtdSer and PtdEtn, and yeast Dnf1p and fungi CtDnf1p which flip PtdCho, revealed a phospholipid entry site at the end of TM1 facing the ectoplasmic side88,93,96,97,101 (see Supplementary Fig. S1). The replacement of Gln84 in this region of ATP11A by glutamic acid triggers the flipping of not only PtdSer but also PtdCho102. The structure of ATP11C and a molecular dynamic simulation indicate that the serine moiety of PtdSer is coordinated with Gln79 (corresponding to Gln84 in ATP11A), Thr86, Thr90, Ser91 and Asn352 (refs. 93,102), and confirmed that glutamic acid at position 79, but not glutamine, accommodates PtdCho. The amino acid sequences critical for coordination with the serine residues in ATP11A and ATP11C differ from those in ATP8B1 and ATP10A, which flip PtdCho34,35, supporting the notion that the region surrounded by these amino acids in TM1, TM2 and TM4 serves as a substrate-specific entry site. In addition, a comparison of the structures between the PtdSer and PtdCho flippases identified another region (exit site) of P4-ATPases critical for the substrate specificity96,97,101, which is consistent with previous analysis with yeast P4-ATPase mutants100,103. How the exit site extending into the cytosolic domain determines the substrate specificity remains elusive.
Tertiary structure of TMEM16 scramblase and models for lipid scrambling
The X-ray structure of Nectria haematococca (nh)TMEM16, a fungus homologue of TMEM16F, provided the first insight into the scrambling mechanism for TMEM16 (ref. 104). The Ca2+-bound or unbound structures of Aspergillus fumigatus (af)TMEM16, mouse TMEM16F, and human TMEM16K were then elucidated by X-ray and cryogenic electron microscopy analyses55–57,105,106. These analyses and molecular dynamic simulation led to the proposal of a ‘clamshell’ model or a ‘stepwise gating’ mechanism, which describes how the groove conformation changes from a closed to an open state, thereby enabling lipids to undergo scrambling by TMEM16 (refs. 107–109) (Fig. 3c).
TMEM16 scramblases share a rhomboidal architecture (130 × 40 Å) with a ‘butterfly fold’, a homodimer with each protomer comprising ten TM helices. At the periphery of each protomer, they have a trans-bilayer hydrophilic groove. The two Ca2+-binding sites composed of five negatively charged residues are located within the membrane in TM6–TM8 (refs. 104,110) (see Supplementary Fig. S2). Without Ca2+, phospholipids cannot access the groove (closed state), because the cavity is shielded from the membrane owing to tight contact between ΤΜ4 and ΤΜ6 (Fig. 3c). The binding of Ca2+ generates an open conformation, in which ΤΜ6 rearranges into a straight conformation through interactions between its acidic residues and Ca2+. ΤΜ4 and ΤΜ6 lose contact (Fig. 3c), and the groove becomes accessible to the phospholipid head group, as predicted in the credit card model.
The pathway in the groove includes the scrambling domain, predicted as the region required for the scrambling of phospholipids based on comparisons of the amino acid sequences of TMEM16 family members52,54 (see Supplementary Fig. S2). Atomic simulations and mutagenesis assays supported the interaction of lipids with amino acid residues in the interior surface of the groove111 or the ‘alternative pore/cavity’ model89, in which charged residues were predicted to serve as ‘stepping stones’ for the hydrophilic head group of phospholipids. The molecular dynamic simulation suggests that the conformation of the groove is modified by the passing phospholipids, which results in the complete opening of the groove allowing lipid scrambling108,109. In addition, the lipid bilayer, particularly around the groove, deforms and thins, which shortens the path that lipid head groups need to traverse, lowering the energy barrier for lipid scrambling56,90,105,106,111,112. TMEM16 family proteins induce membrane distortion in the presence or absence of Ca2+, suggesting that this is an intrinsic property of TMEM16.
Using a cell-free, proteo-liposome assay, it has been reported that afTMEM16 and nhTMEM16 scrambled phospholipid derivatives conjugated with polyethylene glycol (PEG) in the head region113. As the PEG-conjugated head group was too large to pass through the groove, an ‘out-of-the-groove’ model was proposed in which membranes deform to allow phospholipids to translocate outside the groove113. As the open structure was not captured for the Ca2+-bound TMEM16F, this ‘out-of-the-groove’ model was applied to the TMEM16F-mediated scrambling55,56. Although this is an interesting model, it is inconsistent with the findings that charged amino acids in the track or the scrambling domain are essential for TMEM16F-mediated phospholipid transport52,54,55,107. Recent molecular dynamics simulation analysis of mouse TMEM16F also supported the Ca2+-induced conformational change in which the groove opening correlates to the rearrangement of the Ca2+ binding site located distant from the groove114.
The tertiary structures of TMEM16 family members are similar to those of OSCA mechanically activated ion channels115–117 and TMC mechanotransduction channels118–120. The lipid scramblase activity was recently demonstrated in TMC1 in the ear’s inner and outer hair cells to regulate the phospholipid distribution in the membrane121. The defect of TMC1 causes deafness. It is interesting to note that, contrary to the TMEM16 family, the scramblase activity of TMC1 was inhibited by Ca2+ under physiological conditions and was activated by Ca2+ buffering.
Tertiary structure of XKR scramblase and a model for lipid scrambling
We and others elucidated the tertiary structures of the human XKR8–BSG complex and rat XKR9 in their resting states122,123. The amino acid sequence of XKR8 and XKR9 is well conserved (see Supplementary Fig. S3), and they have a similar protein fold, featuring eight transmembrane helices and two helices that partially enter the lipid bilayer (Fig. 3d). Their structure is divided into two similar domains (N-terminal, TM1–TM5; and C-terminal, TM6–TM10) (see Supplementary Fig. S4). A hydrophobic crevice was identified in the surface exposed to the outer leaflet between the two domains, which carried a phospholipid molecule122.
The structures of human XKR family members, predicted by AlphaFold, are similar to those of XKR8 and XKR9. The XK’s residues Arg222 and Glu327, which mutated in McLeod syndrome — a human disorder that affects erythrocytes, and the central and peripheral nervous system124 — are conserved among the members. The tertiary structure of human XKR8 indicates that a salt bridge between these residues stabilizes the protein structure122. Caenorhabditis elegans CED8 exhibits phospholipid scrambling activity45, and its structure is similar to human XKR8. Six charged residues are arranged in a stepwise manner in transmembrane helices (ΤΜ1, TM4 and TM5) in the N-terminal, but not C-terminal, domains of XKR8, XKR9 and CED8 (see Supplementary Fig. S4). These residues are essential for XKR8 to scramble phospholipids in the inward and outward directions122. An alanine mutation of Trp45 in XKR8 at the extracellular end of the hydrophilic stairway resulted in a constitutively active form of the protein122, suggesting that Trp45 functions as a ‘gate-keeper’ and shields hydrophilic residues in the stairway from lipid access. A segment in the N terminus of CED8 or the C terminus of XKR8 and XKR9 carries a caspase 3-recognition site and appears to stabilize its closed structure. The cleavage by caspase 3 at the C terminus seems to cause a conformational change in the molecule, exposing hydrophilic residues to the membrane to provide the ‘stepping stones’ for phospholipids (Fig. 3d). The cleavage of human XKR8 with caspase 3 caused its aggregation, indicating that the caspase-cleaved form of XKR8 is more unstable than the uncleaved form, supporting the notion that the caspase cleavage induces a conformational change in the protein. Of note, in one study, caspase 3-cleaved or non-cleaved XKR9 proteins were reported to have very similar structures123. Yet in this study, an anti-XKR9 sybody was used, which may have prevented the conformational change associated with caspase cleavage.
In contrast to caspase-activatable XKR members (XKR4, XKR8 and XKR9), charged residues are not present in the corresponding intramolecular region of XK (see Supplementary Fig. S3). XK carries a β-hairpin structure that interacts with VPS13A for its scrambling activity84,125,126, suggesting that the scrambling mechanism of XK may differ from that of XKR8 activated by caspase cleavage. The exact mechanism of XK-mediated scrambling and the contribution of VPS13A to this process remain elusive.
Consequences of PtdSer exposition at plasma membranes
Phospholipids are asymmetrically distributed at plasma membranes and this distribution is tightly coupled to various functions (Box 1). Thus, changes in this asymmetry will have an impact on numerous biological processes. We focus here on the best-studied example of the loss of phospholipid asymmetry — the exposition of PtdSer on the cell surface. Exposed PtdSer functions as an ‘eat me’, ‘fuse me’ or ‘repair me’ signal. The PtdSer is recognized and bound by different receptors to promote cell–cell interactions and/or uptake of PtdSer-exposing cells or vesicles67,127,128
Box 1 Roles of the intracellular phospholipids in cell death and cell signalling.
The primary function of phospholipids is to compartmentalize the cellular cytoplasm or organelles to separate them from the extracellular environment or cytosol. The size and charge of the head group and the number of double bonds in acyl chains affect membrane curvature or propensity for deformation266. The negatively charged head group and its phosphorylated derivatives serve as a platform for various enzymes in the cytoplasm and contribute to signal transduction (see the figure). Phosphatidylserine (PtdSer) activates (star symbol in the figure) protein kinase C (PKC) family members for cell proliferation267. Rac1, Rac2 and K-ras are recruited to plasma membranes via the interaction of their positively charged domains with PtdSer and PtdIns(4,5)P2 (refs. 268–271), and are activated. Gasdermin D and gasdermin A, cleaved by caspases or Streptococcal exotoxin B, and phosphorylated MLKL (mixed lineage kinase domain-like protein) bind to the phospholipids of PtdSer, PtdIns(4)P and PtdIns(4,5)P2 in the plasma membrane and form pores to induce pyroptosis or necroptosis150,272,273. PtdCho, phosphatidylcholine; SM, sphingomyelin.
PtdSer as an ‘eat me’ signal
Every day, 1010–1011 cells undergo apoptosis and are engulfed by phagocytes, mainly macrophages129–131. This process is called efferocytosis132, and inefficient efferocytosis results in the accumulation of cell debris in the extracellular milieu, which triggers autoimmunity leading to systemic lupus erythematosus (SLE)133. It is difficult to find unengulfed apoptotic cells in vivo130,134, indicating that macrophages recognize and engulf them as soon as they undergo apoptosis; this is mediated by the ‘eat me’ signals delivered specifically by apoptotic cells135. This signal was attributed to PtdSer exposition on the ectoplasmic leaflet41 (Fig. 4a). As discussed above, there are at least two mechanisms for the PtdSer exposure — Ca2+-induced or caspase-induced. The Ca2+-induced PtdSer exposure is transient: when the intracellular Ca2+ concentration decreases, the flippase quickly internalizes or flips PtdSer to the inner leaflet of the plasma membranes. The caspase-induced PtdSer exposure is irreversible: the flippases at the plasma membranes are destroyed by caspase and XKR8 scramblase is activated irreversibly by caspase cleavage. This irreversible PtdSer exposure is essential for the PtdSer to serve as the ‘eat me’ signal37,136.
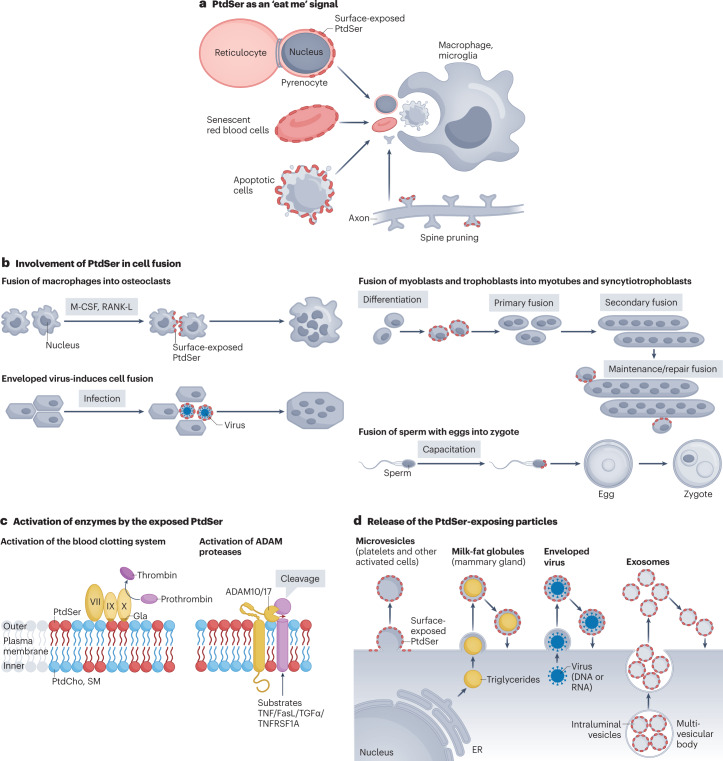
Fig. 4. Biological role of PtdSer.
a, Phosphatidylserine (PtdSer) as an ‘eat me’ signal. When cells undergo apoptosis, they expose PtdSer (red ellipse) on the surface. They are then dismantled into apoptotic bodies with PtdSer on their surface. These bodies are recognized by phagocytes via TAM receptors and swiftly eliminated to ensure the removal of cellular debris and immunogenic reactions. At the final stage of definitive erythropoiesis, erythroblasts divide into reticulocytes and pyrenocytes. Soon after the separation from reticulocytes, the pyrenocytes expose PtdSer and are engulfed by macrophages. Human red blood cells have a lifetime of about 120 days. The senescent red blood cells expose PtdSer for engulfment by macrophages. Similar mechanisms operate in neurons, whose membranes undergo regular, experience-driven pruning to remove unconnected or damaged dendritic spines (the primary location for excitatory synapses). These dendritic spines expose PtdSer, which is recognized by microglia for their removal. b, Involvement of PtdSer in cell fusion. Fusion of macrophages into osteoclasts: when macrophages are treated with macrophage colony-stimulating factor (M-CSF) or receptor activator of nuclear factor-κB ligand (RANKL), they fuse into giant osteoclasts, during which PtdSer (red ellipse) is transiently exposed to the cell surface promoting fusion. Enveloped virus-mediated cell fusion: when an enveloped virus infects cells, they fuse in a PtdSer-dependent manner. Fusion of myoblasts and trophoblasts: myoblasts and trophoblasts differentiate and fuse to form myotubes and syncytiotrophoblasts, respectively, which involves exposed PtdSer. The fusion process is also important for further growth and membrane repair in myotubes and syncytiotrophoblasts. Fusion of sperm with eggs into zygotes: before fertilization, sperm are capacitated in the female reproductive tract, which is accompanied by the PtdSer exposure on the head region. The blocking PtdSer inhibits the fusion process. Many molecules have been proposed for this process. However, very little is known about how cell fusion proceeds. c, Activation of enzymes by the exposed PtdSer. Activation of the blood clotting system: during blood clot formation, platelets are activated to expose PtdSer. This allows the binding and activation of factors VII, IX and X, which recognizes PtdSer via its ‘Gla’ domain. Factor X forms a complex with factor VII and factor IX and mediates the cleavage of prothrombin (factor II) into thrombin for blood clotting. Activation of a disintegrin and metalloproteinase (ADAM) proteases: membrane proteases, ADAM10 and ADAM17, are activated by the exposed PtdSer and work to cleave TNF, Fas ligand, TGFα, secretin, TNF receptor superfamily member 1A (TNFRSF1A) and others, releasing them into the microenvironment to allow paracrine signalling. d, Release of the PtdSer-exposing particles. When platelets and osteoblasts are activated, they expose PtdSer and produce microvesicles, which also expose PtdSer. The platelet-derived microvesicles are involved in propagating the blood clot cascade (see part c), whereas microvesicles released by osteoblasts (called matrix vesicles) carry concentrated phosphate and Ca2+ that form hydroxyapatite crystal — a process required for bone mineralization and supported by PtdSer, which has been proposed to act as a nucleator for the crystallization process. Mammary glands secrete milk fat globules (MFGs) — triglycerides covered by plasma membranes exposing PtdSer. Enveloped viruses are nucleic acids surrounded by the PtdSer-exposing plasma membrane. Exosomes derived from the multivesicular bodies are secreted from various cells and expose PtdSer. MFGs, enveloped viruses and exosomes may rebind to the cell surface in a PtdSer-dependent manner, supporting their uptake. ER, endoplasmic reticulum; PtdCho, phosphatidylcholine; SM, sphingomyelin.
PtdSer-dependent efferocytosis
Macrophages are heterogeneous: microglia, osteoclasts, Kupffer cells, Hofbauer cells, and tingible-body macrophages are tissue-resident macrophages in the brain, bones, liver, placenta and germinal centres, respectively137, and are all involved in efferocytosis. The peritoneal cavity carries two types of macrophages (resident and exudate), with exudate macrophages referring to those recruited by the peritoneal cavity during infection. Efferocytosis by peritoneal macrophages requires one of the three TAM receptor tyrosine kinases (Tyro3, Axl and MerTK)138–140 together with PtdSer-binding proteins (Protein S or GAS6)138. Milk fat globule–EGF factor 8 (MFGE8) and its paralogue DEL1 expressed in exudate and resident macrophages, respectively, bind PtdSer on apoptotic cells and promote efferocytosis by bridging dead cells to the integrin complex (αvβ3) on macrophages141–143. TIM4 (T cell immunoglobulin domain, mucin domain 4) expressed in resident macrophages has a high affinity to PtdSer and functions as a PtdSer receptor40,138; however, its short cytoplasmic region cannot transduce the signal for efferocytosis. Resident peritoneal macrophages, Kupffer cells and skin macrophages138 have at least three PtdSer-recognition systems (TIM4, DEL1 and GAS6/Protein S). By contrast, exudate macrophages use two systems (MFGE8 and GAS6/Protein S). This may explain why efferocytosis by resident macrophages is more efficient than that by exudate macrophages144. Other membrane proteins (BAI1 and stabilins) have been proposed to function as PtdSer receptors145,146. However, different from the molecules described above (TIM4, MFGE8, Protein S and Gas6), BAI1 and stabilins recognize lipopolysaccharides, lipoproteins and proteoglycans to serve as scavenger receptors147,148.
PtdSer recognition in other pathways of cellular clearance
Cells die via apoptosis, necroptosis, pyroptosis, ferroptosis and other methods of programmed cell death149. In necroptosis, an RIP kinase (receptor-interacting (serine/threonine) protein kinase) cascade is activated downstream of the TNF receptor. Phosphorylated MLKL (mixed lineage kinase domain-like protein) oligomerizes and permeabilizes the plasma membrane by binding to PtdIns phosphates150. In this process, damaged plasma membranes with exposed PtdSer are released together with other DAMPs (damage-associated molecular patterns) in an ESCRT (endosomal sorting complexes required for transport)-dependent manner151. A similar mechanism appears to apply to the release of PtdSer-exposing vesicles in pyroptosis or gasdermin-induced pore formation152.
Another mechanism by which PtdSer is exposed is ATP-triggered cell death. The extracellular ATP binds P2X7 ATP receptor153 to expose PtdSer, followed by cell lysis84. The extracellular concentration of ATP reaches a high level in the inflamed tissues and tumour environment82,83, and the P2X7 receptor is highly expressed in regulatory T cells but not CD8 effector T cells84,154. We thus propose that ATP-induced, XK-mediated cell death in regulatory T cells (which are implicated in immunosuppression) and release of cytokines from the pyroptotic macrophages contribute to the eradication of tumour cells or infected cells by effector T cells155.
In addition to the recognition of dying cells, PtdSer was shown to be important in the clearance of senescent red blood cells. When red blood cells age, they behave similarly to apoptotic cells (Fig. 4a). This process, called eryptosis, is accompanied by cell shrinkage, an increased intracellular Ca2+ concentration and the exposure of PtdSer156. Macrophages clear these senescent red blood cells in the liver and spleen. PtdSer is also involved in the recognition and clearance of pyrenocytes (nuclei surrounded by plasma membranes), which are generated in the last step of definitive erythropoiesis (occurring in the fetal liver and bone marrow). Pyrenocytes expose PtdSer on their surface (owing to the lack of an ATP-generating system; see next section) soon after separation from reticulocytes (immature red blood cells)157 (Fig. 4a). Macrophages then engulf PtdSer-exposing pyrenocytes in PtdSer-dependent and MerTK-dependent manners158. Another process requiring PtdSer exposition is neuronal pruning. During development, many neuronal networks are rebuilt by eliminating and reconnecting synapses. This pruning process also occurs in adulthood in response to external stimuli159. Several groups demonstrated that the degenerating dendrites and dendritic spines locally expose PtdSer on their surface, and microglia engulf them in a PtdSer-dependent manner160–164 (Fig. 4a).
PtdSer in cell fusion
In the development of various tissues, cells need to fuse, giving rise to multinucleated cells (Fig. 4b). For example, the formation of osteoclasts in the bones relies on the fusion of monocytes/macrophages165. Myoblasts in the muscle fuse into myotubes to incorporate actin fibres166 and trophoblasts in the placenta fuse to form syncytiotrophoblasts at the maternal–fetal interface167,168. During fertilization, sperm fuse with eggs to generate zygotes169. PtdSer is exposed at the initial stage of the cell fusion, and masking PtdSer blocks the fusion of osteoclasts, myoblasts, trophoblasts and fertilization168,170–173. In addition, enveloped viruses and liposomes exposing PtdSer promote syncytium formation174, supporting that extracellular PtdSer promotes cell fusion. The involvement of various PtdSer-recognition systems has been proposed for the fusion process, but how they work remains elusive128,175,176.
When muscle-derived C2C12 cells are induced to differentiate into myotubes, they transiently expose PtdSer on their surface177. To examine the involvement of flippases in the cell fusion process, two groups established C2C12 derivatives that constitutively expose PtdSer by knocking out the flippase activity (ATP11A or CDC50A) and reported controversial results (negligible or uncontrolled cell fusion)178,179, which suggests that the transient or regulated exposure of PtdSer, rather than its constant exposure, might be important for the efficient fusion of myoblasts. Similarly, a deficiency in plasma membrane flippase (ATP11A) results in the poor development of syncytiotrophoblasts in the mouse placenta, probably due to the constitutive exposure of PtdSer180.
PtdSer in membrane repair
Plasma membranes are damaged by internal (gasdermin and MLKL) or external (toxins, complement, and perforin) factors, leading to cell death. Furthermore, muscle cells are under daily mechanical stress and are prone to membrane lesions. To resist cell death, damaged membranes are repaired by covering the injured site with the patches formed by aggregates of PtdSer-exposing vesicles and by removing the excess patches by macrophages181,182. Two groups showed in C. elegans that severed axons exposed PtdSer before being repaired183,184. An involvement of TMEM16 scramblases in membrane repair has been proposed60,66–68,70–72, suggesting that the PtdSer-exposing vesicles mentioned above were produced by the TMEM16F-dependent mechanism.
The role of PtdSer in activating enzymes
PtdSer is indispensable for the propagation of the blood coagulation cascade. Activated platelets expose PtdSer to the surface185, which is driven by the activation of TMEM16F scramblase and concomitant inactivation of ATP11A and ATP11C flippases26. The blood clotting factors, factors II (prothrombin), VII, IX and X, carry a ‘Gla’ domain of 9–12 amino acids, which contains a carboxylated glutamic acid generated by a vitamin K-dependent reaction. These clotting factors, as the enzyme or substrate, bind to PtdSer on the activated platelets in Ca2+-dependent and ‘Gla’-dependent manners and convert prothrombin to thrombin, which then converts fibrinogen into fibrin — a key and final step in the coagulation cascade186 (Fig. 4c).
A disintegrin and metalloproteinase 17 (ADAM17), called TACE or TNF-converting enzyme, is a metalloproteinase anchored to plasma membranes187 (Fig. 4c). It is responsible for the shedding of TNF from the membrane by cleavage at the extracellular site proximal to the membranes. It also generates the soluble form of TGFα and cleaves TNF receptor superfamily member 1A (TNFRSF1A) and L-selectin. Its deficiency causes neonatal inflammatory skin and bowel diseases in humans and embryonic lethality in mice187. ADAM17 was shown to be activated by the PtdSer exposed on the cell surface188. The treatment of T cells with ATP causes the exposure of PtdSer, which is accompanied by L-selectin shedding189. Similarly, activating keratinocytes with Ca2+-ionomycin causes the PtdSer-dependent release of TGFα188. The shedding activity of ADAM10, another member of the ADAM family that sheds cadherins and Fas ligand190,191, is also dependent on externalized PtdSer192.
PtdSer exposure in extracellular particles and enveloped viruses
Various membrane-bound structures (microvesicles and exosomes) secreted from cells are characterized by the exposition of PtdSer (Fig. 4d), which can be linked to their functions. For example, to nurse infants, mammary glands produce milk fat globules (MFGs), which are triglycerols covered by plasma membranes193. Similar to pyrenocytes157, MFGs carry ATPases in the plasma membranes but lack the ATP-generating system (mitochondria and glycolysis system), which will cause the swift depletion of internal ATP, leading to inactivation of flippase and the Ca2+ pump (ATPase). This will activate Ca2+-dependent scramblase such as TMEM16F to expose PtdSer. MFGE8 protein, also known as lactadherin, is secreted from mammary epithelial cells and binds to PtdSer on MFGs194. When infants cease sucking milk, MFGs remain in the mammary glands and are phagocytosed by mammary epithelial cells and macrophages in an MFGE8-dependent and PtdSer-dependent manner194.
Nearly all cells are capable of secreting extracellular vesicles, which are understood to serve as means of cell–cell communication195. A subtype of these vesicles, exosomes, are generated in the endosomal pathway, via the inward budding of endosomal membranes to form multivesicular bodies. As a result of the inward budding, the intraluminal vesicles inside the multivesicular bodies and the exosomes released upon multivesicular body fusion with the plasma membrane expose PtdSer195,196 due to the lack of an ATP-regeneration system as described for MFGs. PtdSer, as an ‘eat me’ signal, seems to help the uptake of the exosomes by target cells195,197. Furthermore, as described earlier, activated platelets expose PtdSer to the surface185,198. At the same time, they produce many microparticles (extracellular vesicles with a diameter of 50–1,000 nm), exposing PtdSer to robustly activate blood clotting factors199 (see previous subsection).
Enveloped viruses, such as retroviruses, coronaviruses, herpesviruses and influenza viruses, expose PtdSer200, most likely due to the lack of an ATP-generating system, as discussed for pyrenocytes. PtdSer, on the surface, has been proposed to enhance the infectivity of an enveloped virus by supporting its binding to TIM or TAM family members of the PtdSer-receptor system on host cells201. However, another possibility is that PtdSer receptors, such as TIM family proteins, trap the virus to inhibit its release from virus-producing cells202. It remains unclear whether the PtdSer-receptor system affects the enveloped virus infection in vivo.
Diseases caused by malfunction of flippases and scramblases
Loss-of-function or gain-of-function mutations in P4-ATPases, TMEM16 and XKR family members cause various human diseases, including cerebellar ataxia, chorea-acanthocytosis, mental retardation, optical atrophy, lymphopenia, muscular dystrophy, bleeding disorder and tumorigenesis (Table 1). Although the molecular mechanisms leading to these diseases are elusive in most cases, we have obtained some novel insights into how perturbed lipid asymmetry contributes to pathology.
Diseases caused by malfunction of flippases at plasma membranes
Several members of the P4-ATPase are expressed in specific tissues (ATP8A2 in the retina, ATP8B1 and ATP10B in the intestines, ATP8B3 in the testis, ATP8B4 in haematopoietic cells and ATP10D in the kidney), and their subcellular localization also differs among the members (Table 1). The other members are apparently detected ubiquitously at the tissue level, but the Human Cell Atlas established by single-cell RNA sequence analysis203 indicated that most of them are specifically expressed in a limited type of cells, implying that these proteins have specific, non-redundant functions in selected cell types. In line with this, mutations in the P4-ATPase genes have been identified in various human diseases.
ATP11A and ATP11C are widely expressed and work redundantly in most cell types at plasma membranes. Yet a deficiency in either gene causes diseases because some cells only express one of them. For example, ATP11A is not expressed in hepatocytes, B cell progenitors or red blood cells204. Therefore, a loss-of-function mutation in Atp11c causes haemolytic anaemia, B cell lymphopenia and cholestasis in mice and humans205–209. When red blood cells pass through narrow vessels, the intracellular Ca2+ concentration transiently increases due to shear stress210, which may activate Ca2+-dependent scramblases to expose PtdSer. The exposure of PtdSer will be sustained without flippases at the plasma membrane of Atp11c-null cells, and cells will be removed by macrophages from the circulation by entosis in a mechanism equivalent to that proposed for senescent red blood cells211. During development in the bone marrow, B cell progenitors are activated, which induces the exposure of PtdSer. In normal B cell progenitors, PtdSer quickly withdraws from the surface through the effects of ATP11C, but it remains exposed on the surface of Atp11c–/– B cells. PtdSer-exposing B cell progenitors are then engulfed alive by macrophages in the bone marrow resulting in entosis37. CDC50A, a common subunit for P4-ATPases, is highly localized at synapses212. When the gene encoding CDC50A is knocked down in the nerve, the synapses constitutively expose PtdSer due to the lack of the flippases and are eliminated by microglia. These results suggest an involvement of the flippase system to regulate the PtdSer exposure in the synapse-pruning process160–164.
Trophoblasts in mouse placentas express ATP11A but not ATP11C (ref. 180). Therefore, Atp11a–/– trophoblasts lack flippases at the plasma membrane and cannot establish syncytiotrophoblasts, resulting in embryonic lethality. By contrast, the epiblast-specific deletion of Atp11a does not affect mouse development180, indicating that ATP11C is sufficient to maintain homeostasis in normal mouse tissues. A point mutation (Gln84Glu) in the substrate entry site of ATP11A was identified in a patient with developmental delays and neurological deterioration102. Knock-in mice carrying the Gln84Glu point mutation in ATP11A fully re-capitulated the patient’s phenotype, displayed neurological deficit phenotypes and died within 5 weeks of birth. This mutation allows ATP11A to translocate PtdSer and PtdCho. Internalized PtdCho strongly upregulates the expression of SM synthase (SMGS1) and replaces PtdCho with SM at the outer leaflet of plasma membranes102, which renders cells susceptible to cell lysis by the serum sphingomyelinase, the concentration of which increases during inflammation213,214. Sphingomyelinase, secreted into serum, is transported through the body and damages specific tissues, neurons, lungs and kidneys that expose a high level of SM because they strongly express ATP11A (ref. 102). These findings indicate that the substrate specificities of plasma membrane flippases, at least ATP11A, need to be strictly regulated. Another autosomal dominant mutation in ATP11A that removes 82 amino acids in the C terminus was recently detected in human families with hearing loss215. This mutant ATP11A has not yet been biochemically characterized, and it is unclear whether haploinsufficiency or dominant-negative characteristics cause the disease. Because the C-terminal region of ATP11A is responsible for localizing it to plasma membranes216, it is tempting to speculate that this mutant ATP11A is mislocalized from the plasma membrane, causing flipping of phospholipids at an intracellular site, thereby interfering with lipid distribution in the membranes.
Many other mutations have been identified in other P4-ATPases in various human diseases, including chorea, ataxia, cholestasis, hearing loss, obesity and others (Table 1). Currently, we know little about how these mutations lead to pathology.
Diseases caused by malfunction of TMEM16 scramblases
Some members of the TMEM family are ubiquitously expressed whereas others are specifically expressed in the brains, intestines, bones, muscles and testis. The loss-of-function mutations in TMEM16 family members as well as their gain-of-function mutations have been identified in various human diseases (Table 1). The involvement of specific TMEM16 genes with human tumours has also been reported.
A genome-wide association scanning identified Tmem16c as a risk factor for febrile seizure in humans217 (Table 1). Accordingly, Tmem16c-null rats show greater sensitivity to hyperthermia-induced seizures218. TMEM16C is localized at the plasma membrane of neurons in the cerebral cortex and basal ganglia (The Human Protein Atlas). It will be interesting to study how the phospholipid scrambling at the plasma membrane regulates thermo-sensitivity. In many patients with dystonia, autosomal dominant mutations were found throughout the TMEM16C molecule (see Supplementary Fig. S2) (The Human Gene Mutation Database). Although it must be confirmed, these mutations may result in a constitutive active form of the scramblase as found with the dominant mutations of TMEM16E (ref. 219). The constitutive active scrambling by TMEM16F causes the PtdSer exposure136. Thus, it is tempting to think that the dominant mutations of TMEM16C cause the neurons to expose PtdSer resulting in thrombosis as reported with the PtdSer-exposing blood cells220.
Loss-of-function mutations in the Tmem16e gene underlie muscle with limb-girdle muscular dystrophy 2L, Miyoshi muscular dystrophy 3 or non-specific myopathy221,222, whereas autosomal dominant gain-of-function mutations cause the bone disorder gnathodiaphyseal dysplasia223. Myoblasts from a patient with a loss-of-function mutation in Tmem16e efficiently fused to myotubes but were defective in repairing the injured sarcolemma71. Tmem16e–/– animals have conflicting phenotypes. We and others found no apparent phenotype in bones and muscles, although the fertility of male mice was reduced owing to a defect in sperm mobility60,224. By contrast, other studies reported that the Tmem16e–/– mutation affected the repair system of muscle fibres, leading to muscle dystrophy in mice and rabbits72,225. What caused this difference among different laboratories is not clear.
A loss-of-function mutation of Tmef16f is responsible for Scott syndrome, a mild bleeding disorder44,226–228. The platelets of a patient with Scott syndrome are defective in exposing PtdSer upon treatment with thrombin and collagen229. Canine hereditary bleeding disorder with similar characteristics to Scott syndrome was also shown to be driven by a mutation in the Tmem16f gene230. Platelet-specific or conventional Tmem16f–/– mice confirmed the role of TMEM16F in the exposure of PtdSer on tissue factor-activated platelets and in the release of PtdSer-exposing microparticles from platelets47,64.
TMEM16G is highly expressed in the prostate, localizes to the plasma membrane, in particular the cell-to-cell contact site231, and has Ca2+-dependent scramblase activity50. Recently, one of the single-nucleotide polymorphisms strongly associated with prostate cancer was shown to cause exon skipping in the Tmem16g gene to reduce its protein expression232. How the loss-of-function mutation of Tmem16g, or the lack of scramblase in the prostate, promotes tumorigenesis remains to be studied.
TMEM16K is ubiquitously expressed and localized at the ER–endosomes contact sites. Several patients suffering from cerebral ataxia carry the loss-of-function mutation caused by nonsense and missense mutations in the Tmem16k gene233–235. The mutation in its Drosophila melanogaster homologue (Axs) causes the defect in the mitotic spindle assembly and chromosome seggregation236. Little is known about how the malfunction of the TMEM16K scramblase leads to these phenotypes.
Diseases caused by malfunction of XKR scramblase
XK is highly expressed in the plasma membranes of red blood cells, and is complexed with a blood antigen, Kell237. XK is also expressed in various tissues, including skeletal muscle, the spinal cord and the cerebral cortex, in which Kell is not associated with XK. XK is the gene responsible for McLeod syndrome, an X-linked recessive multisystem disorder with central nervous system, neuromuscular, cardiovascular and haematological manifestations in males124. VPS13A, the partner of XK for its scramblase activity, is also responsible for chorea-acanthocytosis238, a multisystem disorder similar to McLeod syndrome. Many missense and nonsense mutations were identified in the XK and VPS13A genes of human patients124,238,239. However, whether these mutations affect the scrambling activity of the XK–VPS13A complex has not been addressed.
The Xkr8 null mutation in mice blocks the exposure of PtdSer on apoptotic cells and causes male infertility due to the accumulation of uncleared dead cells in the testis240. Its deficiency also accelerates SLE-type autoimmune diseases in autoimmune-prone female mice78, similar to the mice deficient in the PtdSer-recognition system for clearance of apoptotic cells241,242.
Conclusion and perspectives
Our understanding of the distribution of phospholipids in the plasma membrane has greatly increased since the initial demonstration of membrane asymmetry in 1972 (ref. 3), particularly in the last 10 years. However, many open questions remain. For example, we now know that flippases, P4-ATPases, are required to recover an asymmetrical distribution of PtdSer and PtdEtn at the plasma membrane36 when various stimuli disrupt the asymmetry. But we have yet to learn the mechanisms contributing to establishing an asymmetrical distribution of phospholipids in response to larger-scale membrane remodelling events, such as cell division.
We also now understand how the activated platelets expose PtdSer on their surface to trigger blood clotting factors44. The molecular mechanisms by which PtdSer is exposed on the surface of apoptotic cells and how phagocytes recognize it have been elucidated12. These findings explain why PtdSer on apoptotic cells, but not on the activated platelets, serves as an ‘eat me’ signal. PtdSer is also exposed on the cell surface in various biological processes, such as cell fusion, lymphocyte activation, axon pruning, cell senescence, membrane repair and formation of extracellular particles, such as pyrenocytes, exosomes and enveloped viruses. However, limited information is currently available on how PtdSer is exposed in these different contexts. The role of PtdSer in the respective biological processes also remains poorly understood.
The identification of P4-ATPase as a flippase and TMEM16 and XKR as Ca2+-dependent and caspase-dependent scramblases revealed that the individual molecules belong to a family that includes 9–14 members12. Some members are ubiquitously expressed, whereas the expression of others is confined to cells in the brain or intestines. In addition, some members localize to the ER, endosomes and/or lysosomes. Although the involvement of the lipid transporters in the various events, such as endocytic recycling, endocytosis, exocytosis and fission of organelles, has been proposed29,243–245, it is not well understood whether the proteins in the scramblase and flippase families have an instructive role in these processes and how exactly they impact these membrane dynamics events. This is because, compared with the proteins in plasma membranes, more difficulties are associated with examining the biological activity of these proteins in specific organelles using a cell-based assay system. Our understanding of the organelle fusion process was advanced with the cell-free system with proteoliposomes246. To study the biological role of the intracellular scramblases and flippases, we may need a cell-free system, ideally with liposomes of the lipid bilayer in which phospholipids are asymmetrically distributed between the two leaflets.
Defects in members of the P4-ATPases, TMEM16 and XK families cause many different diseases; apart from a few exceptions, their aetiologies remain unclear. In particular, we have evidence that if ATP11A flippase flips PtdCho in addition to PtdSer, it will cause harmful effects in humans102. Yet ATP8B1, ATP8B2 and ATP10A were reported to flip PtdCho at the plasma membrane34,35,247, suggesting that the expression of these flippases is restricted to a specific type of cells. In addition, the substrate is unknown for some of the P4-ATPases (ATP8B3, ATP8B4, ATP9A, ATP9B and ATP10D), but knowing their substrate is essential to understand their physiological function. The tertiary structure of nine members of the XKR family, predicted by AlphaFold, is similar. So far, four of them (XK, XKR4, XKR8 and XKR9) were shown to have scramblase activity with or without their partner (VPS13A or Basigin). But how they are activated to function as a scramblase is elusive, except for XKR8 which is activated by caspase 3 during apoptosis. In addition, whether or not other members (XKR2, XKR3, XKR5, XKR6 and XKR7) have the scramblase activity remains to be explored.
Supplementary information
Acknowledgements
This work was partly supported by grants-in-aid from the Japan Society for the Promotion of Science (T22K061020 to T.S. and A21H04770 to S.N.). The authors thank their laboratory’s current and past members who have been involved in this project.
Glossary
- AlphaFold artificial intelligence program
An artificial intelligence program that predicts the tertiary structure of proteins from their amino acid sequence.
- Annexins
A family of intracellular Ca2+-regulated phospholipid-binding proteins that regulate membrane trafficking and membrane repair. Annexin V specifically binds phosphatidylserine (PtdSer) and is widely used to detect PtdSer exposed on the cell surface.
- Basigin
A transmembrane protein belonging to the immunoglobulin superfamily. It is closely related to neuroplastin and works as a chaperone for XK-related 8 (XKR8) to localize at plasma membranes.
- Chromaffin granules
Large intracellular vesicles carrying dopamine, adrenaline and serotonin present in chromaffin cells in the adrenal gland.
- DAMPs
(Damage-associated molecular patterns). Molecules released from damaged or dying cells that activate the immune reaction.
- Dendritic spines
Small membranous protrusions present along the neuron’s dendrites. They receive synaptic input from other neurons.
- ‘Eat me’ signal
Molecules on the cell surface which are recognized by phagocytes for engulfment. Phosphatidylserine (PtdSer) is the best characterized ‘eat me’ signal.
- Entosis
A process in which a living cell invades another cell’s cytoplasm or a phagocyte engulfs living cells.
- ESCRT
(Endosomal sorting complexes required for transport). A set of five protein complexes comprising an ancient system for membrane remodelling and scission.
- Ferroptosis
A type of cell death, whereby free cytosolic iron promotes peroxidation of the membranes, producing reactive intermediates that damage the cell.
- Gasdermin
A protein cleaved by caspases, and the cleaved part forms pores in the plasma membrane to cause pyroptosis (a type of cell death).
- L-selectin
(CD62L). A cell adhesion molecule expressed on the cell surface of leukocytes. It is required for leukocytes to bind and roll on the endothelium.
- Milk fat globule–EGF factor 8
(MFGE8; also known as lactadherin). Binds phosphatidylserine (PtdSer) on apoptotic cells and integrins in macrophages, thus bridging apoptotic cells to macrophages.
- MLKL
(Mixed lineage kinase domain-like protein). A protein that is phosphorylated by RIPK3 and permeabilizes plasma membranes to cause necroptosis (a type of cell death).
- Necroptosis
A type of programmed cell death induced by TNF. RIPK1, RIPK3 and MLKL (mixed lineage kinase domain-like protein) mediate this process.
- Neuroplastin
A type 1 transmembrane protein belonging to the immunoglobulin superfamily. It is closely related to Basigin and can work as a chaperone to transport XKR8 from the endoplasmic reticulum (ER) to plasma membranes.
- OSCA mechanically activated ion channels
A hyperosmolarity-activated ion channel identified in Arabidopsis. Its mammalian orthologues are transmembrane protein 63 (TMEM63) family members.
- Oxysterol-binding protein (OSBP)-related protein (ORP) family
A family of proteins that are ubiquitously expressed in eukaryotes. They work as intracellular lipid sensors or transporters.
- P2X7
A P2X ATP receptor composed of homotrimeric membrane proteins with two transmembrane regions. It is reported to work as a cation transporter.
- Pyroptosis
A type of cell death accompanied by the formation of pores in the plasma membrane and cell lysis. This process is triggered by intracellular pathogens, and mediated by inflammasomes and inflammatory caspases.
- RIP kinase
(Receptor-interacting (serine/threonine) protein kinase). Among five human members, RIPK1 and RIPK3 are involved in necroptosis.
- Sybody
A synthetic single-domain antibody that can often recognize a specific protein conformation.
- Systemic lupus erythematosus
(SLE). An autoimmune disease in which the immune system attacks healthy tissues such as the skin, bones, kidneys and lungs.
- TAM
A family of proto-oncogene tyrosine kinase receptors (Tyro3, Axl and MerTK). Macrophages require one of them for the engulfment of apoptotic cells.
- TIM4
(T cell immunoglobulin domain, mucin domain 4). A transmembrane protein carrying the phosphatidylserine (PtdSer)-binding site in the extracellular region, serving as a PtdSer receptor.
- TMC mechanotransduction channels
One of the four mechanosensitive ion channels (TREK, Piezo, TMEM63 and TMC). TMC1 is expressed in the hair cells of the ear, and Beethoven’s deafness is caused by its mutation.
- VPS13A
A member of the evolutionally well-conserved family of large cytoplasmic proteins (human VPS13A, 3,174 amino acids). It has a hydrophobic cylinder-like cavity that may be a path for transferring lipids between organelles.
Author contributions
Both authors contributed equally to all aspects of the article (researching data for the report, discussing the content, writing, reviewing and editing).
Peer review
Peer review information
Nature Reviews Molecular Cell Biology thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
The Human Gene Mutation Database: https://www.hgmd.cf.ac.uk/
The Human Protein Atlas: https://www.proteinatlas.org/
Change history
5/22/2023
A Correction to this paper has been published: 10.1038/s41580-023-00621-y
Supplementary information
The online version contains supplementary material available at 10.1038/s41580-023-00604-z.
References
- 1.Vance JE. Phospholipid synthesis and transport in mammalian cells. Traffic. 2015;16:1–18. doi: 10.1111/tra.12230. [DOI] [PubMed] [Google Scholar]
- 2.Yang Y, Lee M, Fairn GD. Phospholipid subcellular localization and dynamics. J. Biol. Chem. 2018;293:6230–6240. doi: 10.1074/jbc.R117.000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bretscher MS. Asymmetrical lipid bilayer structure for biological membranes. Nat. N. Biol. 1972;236:11–12. doi: 10.1038/newbio236011a0. [DOI] [PubMed] [Google Scholar]
- 4.Tsuji T, et al. Predominant localization of phosphatidylserine at the cytoplasmic leaflet of the ER, and its TMEM16K-dependent redistribution. Proc. Natl Acad. Sci. USA. 2019;116:13368–13373. doi: 10.1073/pnas.1822025116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murate M, et al. Transbilayer distribution of lipids at nano scale. J. Cell Sci. 2015;128:1627–1638. doi: 10.1242/jcs.163105. [DOI] [PubMed] [Google Scholar]
- 6.Clarke RJ, Hossain KR, Cao K. Physiological roles of transverse lipid asymmetry of animal membranes. Biochim. Biophy. Acta, Biomembr. 2020;1862:183382. doi: 10.1016/j.bbamem.2020.183382. [DOI] [PubMed] [Google Scholar]
- 7.Lorent JH, et al. Plasma membranes are asymmetric in lipid unsaturation, packing and protein shape. Nat. Chem. Biol. 2020;16:644–652. doi: 10.1038/s41589-020-0529-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bevers EM, Williamson PL. Getting to the outer leaflet: physiology of phosphatidylserine exposure at the plasma membrane. Physiol. Rev. 2016;96:605–645. doi: 10.1152/physrev.00020.2015. [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi T, Menon AK. Transbilayer lipid asymmetry. Curr. Biol. 2018;28:R386–R391. doi: 10.1016/j.cub.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 10.Doktorova M, Symons JL, Levental I. Structural and functional consequences of reversible lipid asymmetry in living membranes. Nat. Chem. Biol. 2020;16:1321–1330. doi: 10.1038/s41589-020-00688-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meca J, et al. Avidity-driven polarity establishment via multivalent lipid–GTPase module interactions. EMBO J. 2019;38:e99652. doi: 10.15252/embj.201899652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagata S, Suzuki J, Segawa K, Fujii T. Exposure of phosphatidylserine on the cell surface. Cell Death Differ. 2016;23:952–961. doi: 10.1038/cdd.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coleman JA, Quazi F, Molday RS. Mammalian P4-ATPases and ABC transporters and their role in phospholipid transport. Biochim. Biophys. Acta. 2013;1831:555–574. doi: 10.1016/j.bbalip.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palmgren M, Østerberg JT, Nintemann SJ, Poulsen LR, López-Marqués RL. Evolution and a revised nomenclature of P4 ATPases, a eukaryotic family of lipid flippases. Biochim. Biophys. Acta, Biomembr. 2019;1861:1135–1151. doi: 10.1016/j.bbamem.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 15.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanay A, Regev A. Scaling single-cell genomics from phenomenology to mechanism. Nature. 2017;541:331–338. doi: 10.1038/nature21350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng Y, Grigorieff N, Penczek, Pawel A, Walz T. A primer to single-particle cryo-electron microscopy. Cell. 2015;161:438–449. doi: 10.1016/j.cell.2015.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jumper J, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bretscher MS. Membrane structure: some general principles. Science. 1973;181:622–629. doi: 10.1126/science.181.4100.622. [DOI] [PubMed] [Google Scholar]
- 20.Leventis PA, Grinstein S. The distribution and function of phosphatidylserine in cellular membranes. Annu. Rev. Biophys. 2010;39:407–427. doi: 10.1146/annurev.biophys.093008.131234. [DOI] [PubMed] [Google Scholar]
- 21.Seigneuret M, Devaux P. ATP-dependent asymmetric distribution of spin-labeled phospholipids in the erythrocyte membrane: relation to shape changes. Proc. Natl Acad. Sci. USA. 1984;81:3751–3755. doi: 10.1073/pnas.81.12.3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Auland M, Roufogalis B, Devaux P, Zachowski A. Reconstitution of ATP-dependent aminophospholipid translocation in proteoliposomes. Proc. Natl Acad. Sci. USA. 1994;91:10938–10942. doi: 10.1073/pnas.91.23.10938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang X, Halleck MS, Schlegel RA, Williamson P. A subfamily of P-type ATPases with aminophospholipid transporting activity. Science. 1996;272:1495–1497. doi: 10.1126/science.272.5267.1495. [DOI] [PubMed] [Google Scholar]
- 24.Palmgren MG, Nissen P. P-type ATPases. Annu. Rev. Biophys. 2011;40:243–266. doi: 10.1146/annurev.biophys.093008.131331. [DOI] [PubMed] [Google Scholar]
- 25.Segawa K, et al. Caspase-mediated cleavage of phospholipid flippase for apoptotic phosphatidylserine exposure. Science. 2014;344:1164–1168. doi: 10.1126/science.1252809. [DOI] [PubMed] [Google Scholar]
- 26.Segawa K, Kurata S, Nagata S. Human type IV P-type ATPases that work as plasma membrane phospholipid flippases, and their regulation by caspase and calcium. J. Biol. Chem. 2016;291:762–772. doi: 10.1074/jbc.M115.690727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J, et al. Proteomic analysis and functional characterization of P4-ATPase phospholipid flippases from murine tissues. Sci. Rep. 2018;8:10795. doi: 10.1038/s41598-018-29108-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coleman JA, Kwok MC, Molday RS. Localization, purification, and functional reconstitution of the P4-ATPase Atp8a2, a phosphatidylserine flippase in photoreceptor disc membranes. J. Biol. Chem. 2009;284:32670–32679. doi: 10.1074/jbc.M109.047415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee S, et al. Transport through recycling endosomes requires EHD1 recruitment by a phosphatidylserine translocase. EMBO J. 2015;34:669–688. doi: 10.15252/embj.201489703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng M-T, et al. Structural insights into the activation of autoinhibited human lipid flippase ATP8B1 upon substrate binding. Proc. Natl Acad. Sci. USA. 2022;119:e2118656119. doi: 10.1073/pnas.2118656119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dieudonné T, et al. Autoinhibition and regulation by phosphoinositides of ATP8B1, a human lipid flippase associated with intrahepatic cholestatic disorders. eLife. 2022;11:e75272. doi: 10.7554/eLife.75272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin S, et al. Mutated ATP10B increases Parkinson’s disease risk by compromising lysosomal glucosylceramide export. Acta Neuropathol. 2020;139:1001–1024. doi: 10.1007/s00401-020-02145-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Best JT, Xu P, Graham TR. Phospholipid flippases in membrane remodeling and transport carrier biogenesis. Curr. Opin. Cell Biol. 2019;59:8–15. doi: 10.1016/j.ceb.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takatsu H, et al. Phospholipid flippase activities and substrate specificities of human type IV P-type ATPases localized to the plasma membrane. J. Biol. Chem. 2014;289:33543–33556. doi: 10.1074/jbc.M114.593012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naito T, et al. Phospholipid flippase ATP10A translocates phosphatidylcholine and is involved in plasma membrane dynamics. J. Biol. Chem. 2015;290:15004–15017. doi: 10.1074/jbc.M115.655191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyata Y, Yamada K, Nagata S, Segawa K. Two types of type IV P-type ATPases independently re-establish the asymmetrical distribution of phosphatidylserine in plasma membranes. J. Biol. Chem. 2022;298:102527. doi: 10.1016/j.jbc.2022.102527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Segawa K, et al. Phospholipid flippases enable precursor B cells to flee engulfment by macrophages. Proc. Natl Acad. Sci. USA. 2018;115:12212–12217. doi: 10.1073/pnas.1814323115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kornberg RD, McConnell HM. Inside–outside transitions of phospholipids in vesicle membranes. Biochemistry. 1971;10:1111–1120. doi: 10.1021/bi00783a003. [DOI] [PubMed] [Google Scholar]
- 39.Bevers EM, Comfurius P, Zwaal RF. Changes in membrane phospholipid distribution during platelet activation. Biochim. Biophys. Acta. 1983;736:57–66. doi: 10.1016/0005-2736(83)90169-4. [DOI] [PubMed] [Google Scholar]
- 40.Miyanishi M, et al. Identification of Tim4 as a phosphatidylserine receptor. Nature. 2007;450:435–439. doi: 10.1038/nature06307. [DOI] [PubMed] [Google Scholar]
- 41.Fadok VA, et al. Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. J. Immunol. 1992;148:2207–2216. doi: 10.4049/jimmunol.148.7.2207. [DOI] [PubMed] [Google Scholar]
- 42.Basse F, Stout JG, Sims PJ, Wiedmer T. Isolation of an erythrocyte membrane protein that mediates Ca2+-dependent transbilayer movement of phospholipid. J. Biol. Chem. 1996;271:17205–17210. doi: 10.1074/jbc.271.29.17205. [DOI] [PubMed] [Google Scholar]
- 43.Bevers EM, Williamson PL. Phospholipid scramblase: an update. FEBS Lett. 2010;584:2724–2730. doi: 10.1016/j.febslet.2010.03.020. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki J, Umeda M, Sims PJ, Nagata S. Calcium-dependent phospholipid scrambling by TMEM16F. Nature. 2010;468:834–838. doi: 10.1038/nature09583. [DOI] [PubMed] [Google Scholar]
- 45.Suzuki J, Denning DP, Imanishi E, Horvitz HR, Nagata S. Xk-related protein 8 and CED-8 promote phosphatidylserine exposure in apoptotic cells. Science. 2013;341:403–406. doi: 10.1126/science.1236758. [DOI] [PubMed] [Google Scholar]
- 46.Schreiber R, et al. Expression and function of epithelial anoctamins. J. Biol. Chem. 2010;285:7838–7845. doi: 10.1074/jbc.M109.065367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang H, et al. TMEM16F forms a Ca2+-activated cation channel required for lipid scrambling in platelets during blood coagulation. Cell. 2012;151:111–122. doi: 10.1016/j.cell.2012.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Almaça J, et al. TMEM16 proteins produce volume-regulated chloride currents that are reduced in mice lacking TMEM16A. J. Biol. Chem. 2009;284:28571–28578. doi: 10.1074/jbc.M109.010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martins JR, et al. Anoctamin 6 is an essential component of the outwardly rectifying chloride channel. Proc. Natl Acad. Sci. USA. 2011;108:18168–18172. doi: 10.1073/pnas.1108094108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suzuki J, et al. Calcium-dependent phospholipid scramblase activity of TMEM16 protein family members. J. Biol. Chem. 2013;288:13305–13316. doi: 10.1074/jbc.M113.457937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Duran C, Qu Z, Osunkoya AO, Cui Y, Hartzell HC. ANOs 3–7 in the anoctamin/Tmem16 Cl– channel family are intracellular proteins. Am. J. Physiol. Cell Physiol. 2012;302:C482–C493. doi: 10.1152/ajpcell.00140.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu K, et al. Identification of a lipid scrambling domain in ANO6/TMEM16F. eLife. 2015;4:e06901. doi: 10.7554/eLife.06901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scudieri P, et al. Ion channel and lipid scramblase activity associated with expression of TMEM16F/ANO6 isoforms. J. Physiol. 2015;593:3829–3848. doi: 10.1113/JP270691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gyobu S, Ishihara K, Suzuki J, Segawa K, Nagata S. Characterization of the scrambling domain of the TMEM16 family. Proc. Natl Acad. Sci. USA. 2017;114:6274–6279. doi: 10.1073/pnas.1703391114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alvadia C, et al. Cryo-EM structures and functional characterization of the murine lipid scramblase TMEM16F. eLife. 2019;8:e44365. doi: 10.7554/eLife.44365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feng S, et al. Cryo-EM studies of TMEM16F calcium-activated ion channel suggest features important for lipid scrambling. Cell Rep. 2019;28:567–579. doi: 10.1016/j.celrep.2019.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bushell SR, et al. The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K. Nat. Commun. 2019;10:3956. doi: 10.1038/s41467-019-11753-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Watanabe R, Sakuragi T, Noji H, Nagata S. Single-molecule analysis of phospholipid scrambling by TMEM16F. Proc. Natl Acad. Sci. USA. 2018;115:3066–3071. doi: 10.1073/pnas.1717956115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Veshaguri S, et al. Direct observation of proton pumping by a eukaryotic P-type ATPase. Science. 2016;351:1469–1473. doi: 10.1126/science.aad6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gyobu S, et al. A role of TMEM16E carrying a scrambling domain in sperm motility. Mol. Cell Biol. 2016;36:645–659. doi: 10.1128/MCB.00919-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marsault R, Murgia M, Pozzan T, Rizzuto R. Domains of high Ca2+ beneath the plasma membrane of living A7r5 cells. EMBO J. 1997;16:1575–1581. doi: 10.1093/emboj/16.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zayzafoon M. Calcium/calmodulin signaling controls osteoblast growth and differentiation. J. Cell. Biochem. 2006;97:56–70. doi: 10.1002/jcb.20675. [DOI] [PubMed] [Google Scholar]
- 63.Ehlen HW, et al. Inactivation of Anoctamin-6/Tmem16f, a regulator of phosphatidylserine scrambling in osteoblasts, leads to decreased mineral deposition in skeletal tissues. J. Bone Miner. Res. 2013;28:246–259. doi: 10.1002/jbmr.1751. [DOI] [PubMed] [Google Scholar]
- 64.Fujii T, Sakata A, Nishimura S, Eto K, Nagata S. TMEM16F is required for phosphatidylserine exposure and microvesicle release in activated mouse platelets. Proc. Natl Acad. Sci. USA. 2015;112:12800–12805. doi: 10.1073/pnas.1516594112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Headland SE, et al. Neutrophil-derived microvesicles enter cartilage and protect the joint in inflammatory arthritis. Sci. Transl. Med. 2015;7:315ra190. doi: 10.1126/scitranslmed.aac5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang X, et al. Bacterial endotoxin activates the coagulation cascade through Gasdermin D-dependent phosphatidylserine exposure. Immunity. 2019;51:983–996. doi: 10.1016/j.immuni.2019.11.005. [DOI] [PubMed] [Google Scholar]
- 67.Wu N, et al. Critical role of lipid scramblase TMEM16F in phosphatidylserine exposure and repair of plasma membrane after pore formation. Cell Rep. 2020;30:1129–1140.e5. doi: 10.1016/j.celrep.2019.12.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Deisl C, Hilgemann DW, Syeda R, Fine M. TMEM16F and dynamins control expansive plasma membrane reservoirs. Nat. Commun. 2021;12:4990. doi: 10.1038/s41467-021-25286-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gerke V, Creutz CE, Moss SE. Annexins: linking Ca2+ signalling to membrane dynamics. Nat. Rev. Mol. Cell Biol. 2005;6:449–461. doi: 10.1038/nrm1661. [DOI] [PubMed] [Google Scholar]
- 70.Foltz SJ, Cui YY, Choo HJ, Hartzell HC. ANO5 ensures trafficking of annexins in wounded myofibers. J. Cell Biol. 2021;220:e202007059. doi: 10.1083/jcb.202007059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chandra G, et al. Dysregulated calcium homeostasis prevents plasma membrane repair in Anoctamin 5/TMEM16E-deficient patient muscle cells. Cell Death Discov. 2019;5:118. doi: 10.1038/s41420-019-0197-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Griffin DA, et al. Defective membrane fusion and repair in Anoctamin5-deficient muscular dystrophy. Hum. Mol. Genet. 2016;25:1900–1911. doi: 10.1093/hmg/ddw063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Petkovic M, Oses-Prieto J, Burlingame A, Jan LY, Jan YN. TMEM16K is an interorganelle regulator of endosomal sorting. Nat. Commun. 2020;11:3298. doi: 10.1038/s41467-020-17016-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Suzuki J, Imanishi E, Nagata S. Exposure of phosphatidylserine by Xk-related protein family members during apoptosis. J. Biol. Chem. 2014;289:30257–30267. doi: 10.1074/jbc.M114.583419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Williamson P, et al. Phospholipid scramblase activation pathways in lymphocytes. Biochemistry. 2001;40:8065–8072. doi: 10.1021/bi001929z. [DOI] [PubMed] [Google Scholar]
- 76.Schoenwaelder SM, et al. Two distinct pathways regulate platelet phosphatidylserine exposure and procoagulant function. Blood. 2009;114:663–666. doi: 10.1182/blood-2009-01-200345. [DOI] [PubMed] [Google Scholar]
- 77.Pervaiz N, et al. Evolutionary history of the human multigene families reveals widespread gene duplications throughout the history of animals. BMC Evol. Biol. 2019;19:128. doi: 10.1186/s12862-019-1441-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kawano M, Nagata S. Lupus-like autoimmune disease caused by a lack of Xkr8, a caspase-dependent phospholipid scramblase. Proc. Natl Acad. Sci. USA. 2018;280:2132–2137. doi: 10.1073/pnas.1720732115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Suzuki J, Imanishi E, Nagata S. Xkr8 phospholipid scrambling complex in apoptotic phosphatidylserine exposure. Proc. Natl Acad. Sci. USA. 2016;113:9509–9514. doi: 10.1073/pnas.1610403113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sakuragi T, Kosako H, Nagata S. Phosphorylation-mediated activation of mouse Xkr8 scramblase for phosphatidylserine exposure. Proc. Natl Acad. Sci. USA. 2019;33:2907–2912. doi: 10.1073/pnas.1820499116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gadella BM, Harrison RA. The capacitating agent bicarbonate induces protein kinase A-dependent changes in phospholipid transbilayer behavior in the sperm plasma membrane. Development. 2000;127:2407–2420. doi: 10.1242/dev.127.11.2407. [DOI] [PubMed] [Google Scholar]
- 82.Di Virgilio F, Sarti AC, Falzoni S, Marchi ED, Adinolfi E. Extracellular ATP and P2 purinergic signalling in the tumour microenvironment. Nat. Rev. Cancer. 2018;18:601–618. doi: 10.1038/s41568-018-0037-0. [DOI] [PubMed] [Google Scholar]
- 83.Kamata-Sakurai M, et al. Antibody to CD137 activated by extracellular adenosine triphosphate is tumor selective and broadly effective in vivo without systemic immune activation. Cancer Discov. 2021;11:158–175. doi: 10.1158/2159-8290.CD-20-0328. [DOI] [PubMed] [Google Scholar]
- 84.Ryoden Y, Segawa K, Nagata S. Requirement of Xk and Vps13a for the P2X7-mediated phospholipid scrambling and cell lysis in mouse T cells. Proc. Natl Acad. Sci. USA. 2022;119:e2119286119. doi: 10.1073/pnas.2119286119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kumar N, et al. VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J. Cell Biol. 2018;217:3625–3639. doi: 10.1083/jcb.201807019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Puts CF, Holthuis JC. Mechanism and significance of P4 ATPase-catalyzed lipid transport: lessons from a Na+/K+-pump. Biochim. Biophys. Acta. 2009;1791:603–611. doi: 10.1016/j.bbalip.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 87.Pomorski T, Menon AK. Lipid flippases and their biological functions. Cell. Mol. Life Sci. 2006;63:2908–2921. doi: 10.1007/s00018-006-6167-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hiraizumi M, Yamashita K, Nishizawa T, Nureki O. Cryo-EM structures capture the transport cycle of the P4-ATPase flippase. Science. 2019;365:1149–1155. doi: 10.1126/science.aay3353. [DOI] [PubMed] [Google Scholar]
- 89.Kalienkova V, Mosina VC, Paulino C. The groovy TMEM16 family: molecular mechanisms of lipid scrambling and ion conduction. J. Mol. Biol. 2021;433:166941. doi: 10.1016/j.jmb.2021.166941. [DOI] [PubMed] [Google Scholar]
- 90.Falzone ME, et al. TMEM16 scramblases thin the membrane to enable lipid scrambling. Nat. Commun. 2022;13:2604. doi: 10.1038/s41467-022-30300-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Timcenko M, et al. Structure and autoregulation of a P4-ATPase lipid flippase. Nature. 2019;571:366–370. doi: 10.1038/s41586-019-1344-7. [DOI] [PubMed] [Google Scholar]
- 92.Nakanishi H, et al. Crystal structure of a human plasma membrane phospholipid flippase. J. Biol. Chem. 2020;295:10180–10194. doi: 10.1074/jbc.RA120.014144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nakanishi H, et al. Transport cycle of plasma membrane flippase ATP11C by cryo-EM. Cell Rep. 2020;32:108208. doi: 10.1016/j.celrep.2020.108208. [DOI] [PubMed] [Google Scholar]
- 94.Bai L, et al. Autoinhibition and activation mechanisms of the eukaryotic lipid flippase Drs2p–Cdc50p. Nat. Commun. 2019;10:4142. doi: 10.1038/s41467-019-12191-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Timcenko M, et al. Structural basis of substrate-independent phosphorylation in a P4-ATPase lipid flippase. J. Mol. Biol. 2021;433:167062. doi: 10.1016/j.jmb.2021.167062. [DOI] [PubMed] [Google Scholar]
- 96.He Y, Xu J, Wu X, Li L. Structures of a P4-ATPase lipid flippase in lipid bilayers. Protein Cell. 2020;11:458–463. doi: 10.1007/s13238-020-00712-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bai L, et al. Transport mechanism of P4 ATPase phosphatidylcholine flippases. eLife. 2020;9:e62163. doi: 10.7554/eLife.62163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Post RL, Hegyvary C, Kume S. Activation by adenosine triphosphate in the phosphorylation kinetics of sodium and potassium ion transport adenosine triphosphatase. J. Biol. Chem. 1972;247:6530–6540. doi: 10.1016/S0021-9258(19)44725-X. [DOI] [PubMed] [Google Scholar]
- 99.Albers RW. Biochemical aspects of active transport. Annu. Rev. Biochem. 1967;36:727–756. doi: 10.1146/annurev.bi.36.070167.003455. [DOI] [PubMed] [Google Scholar]
- 100.Vestergaard AL, et al. Critical roles of isoleucine-364 and adjacent residues in a hydrophobic gate control of phospholipid transport by the mammalian P4-ATPase ATP8A2. Proc. Natl Acad. Sci. USA. 2014;111:E1334–E1343. doi: 10.1073/pnas.1321165111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bai L, et al. Structural basis of the P4B ATPase lipid flippase activity. Nat. Commun. 2021;12:5963. doi: 10.1038/s41467-021-26273-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Segawa K, et al. A sublethal ATP11A mutation associated with neurological deterioration causes aberrant phosphatidylcholine flipping in plasma membranes. J. Clin. Invest. 2021;131:e148005. doi: 10.1172/JCI148005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Baldridge RD, Graham TR. Two-gate mechanism for phospholipid selection and transport by type IV P-type ATPases. Proc. Natl Acad. Sci. USA. 2013;110:E358–E367. doi: 10.1073/pnas.1216948110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Brunner JD, Lim NK, Schenck S, Duerst A, Dutzler R. X-ray structure of a calcium-activated TMEM16 lipid scramblase. Nature. 2014;516:207–212. doi: 10.1038/nature13984. [DOI] [PubMed] [Google Scholar]
- 105.Kalienkova V, et al. Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM. eLife. 2019;8:e44364. doi: 10.7554/eLife.44364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Falzone ME, et al. Structural basis of Ca2+-dependent activation and lipid transport by a TMEM16 scramblase. eLife. 2019;8:e43229. doi: 10.7554/eLife.43229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Le T, et al. An inner activation gate controls TMEM16F phospholipid scrambling. Nat. Commun. 2019;10:1846. doi: 10.1038/s41467-019-09778-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lee B-C, et al. Gating mechanism of the extracellular entry to the lipid pathway in a TMEM16 scramblase. Nat. Commun. 2018;9:3251. doi: 10.1038/s41467-018-05724-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Khelashvili G, et al. Membrane lipids are both the substrates and a mechanistically responsive environment of TMEM16 scramblase proteins. J. Comput. Chem. 2020;41:538–551. doi: 10.1002/jcc.26105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ishihara K, Suzuki J, Nagata S. Role of Ca2+ in the stability and function of TMEM16F and 16K. Biochemistry. 2016;55:3180–3188. doi: 10.1021/acs.biochem.6b00176. [DOI] [PubMed] [Google Scholar]
- 111.Bethel NP, Grabe M. Atomistic insight into lipid translocation by a TMEM16 scramblase. Proc. Natl Acad. Sci. USA. 2016;113:14049–14054. doi: 10.1073/pnas.1607574113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jiang T, Yu K, Hartzell HC, Tajkhorshid E. Lipids and ions traverse the membrane by the same physical pathway in the nhTMEM16 scramblase. eLife. 2017;6:e28671. doi: 10.7554/eLife.28671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Malvezzi M, et al. Out-of-the-groove transport of lipids by TMEM16 and GPCR scramblases. Proc. Natl Acad. Sci. USA. 2018;115:E7033–E7042. doi: 10.1073/pnas.1806721115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Khelashvili G, Kots E, Cheng X, Levine MV, Weinstein H. The allosteric mechanism leading to an open-groove lipid conductive state of the TMEM16F scramblase. Commun. Biol. 2022;5:990. doi: 10.1038/s42003-022-03930-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jojoa-Cruz S, et al. Cryo-EM structure of the mechanically activated ion channel OSCA1.2. eLife. 2018;7:e41845. doi: 10.7554/eLife.41845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liu X, Wang J, Sun L. Structure of the hyperosmolality-gated calcium-permeable channel OSCA1.2. Nat. Commun. 2018;9:5060. doi: 10.1038/s41467-018-07564-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Maity K, et al. Cryo-EM structure of OSCA1.2 from Oryza sativa elucidates the mechanical basis of potential membrane hyperosmolality gating. Proc. Natl Acad. Sci. USA. 2019;116:14309–14318. doi: 10.1073/pnas.1900774116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ballesteros A, Fenollar-Ferrer C, Swartz KJ. Structural relationship between the putative hair cell mechanotransduction channel TMC1 and TMEM16 proteins. eLife. 2018;7:e38433. doi: 10.7554/eLife.38433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pan B, et al. TMC1 forms the pore of mechanosensory transduction channels in vertebrate inner ear hair cells. Neuron. 2018;99:736–753. doi: 10.1016/j.neuron.2018.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jeong H, et al. Structures of the TMC-1 complex illuminate mechanosensory transduction. Nature. 2022;610:796–803. doi: 10.1038/s41586-022-05314-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ballesteros A, Swartz KJ. Regulation of membrane homeostasis by TMC1 mechanoelectrical transduction channels is essential for hearing. Sci. Adv. 2022;8:eabm5550. doi: 10.1126/sciadv.abm5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sakuragi T, et al. The tertiary structure of the human Xkr8–Basigin complex that scrambles phospholipids at plasma membranes. Nat. Struct. Mol. Biol. 2021;28:825–834. doi: 10.1038/s41594-021-00665-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Straub MS, Alvadia C, Sawicka M, Dutzler R. Cryo-EM structures of the caspase activated protein XKR9 involved in apoptotic lipid scrambling. eLife. 2021;10:e69800. doi: 10.7554/eLife.69800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jung, H. H., et al. McLeod neuroacanthocytosis syndrome. National Library of Medicinehttps://www.ncbi.nlm.nih.gov/books/NBK1354/ (2021).
- 125.Park J-S, Hu Y, Hollingsworth NM, Miltenberger-Miltenyi G, Neiman AM. Interaction between VPS13A and the XK scramblase is important for VPS13A function in humans. J. Cell Sci. 2022;135:jcs260227. doi: 10.1242/jcs.260227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Guillén-Samander A, et al. A partnership between the lipid scramblase XK and the lipid transfer protein VPS13A at the plasma membrane. Proc. Natl Acad. Sci. USA. 2022;119:e2205425119. doi: 10.1073/pnas.2205425119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Segawa K, Nagata S. An apoptotic ‘eat me’ signal: phosphatidylserine exposure. Trends Cell Biol. 2015;25:649–650. doi: 10.1016/j.tcb.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 128.Whitlock JM, Chernomordik LV. Flagging fusion: phosphatidylserine signaling in cell–cell fusion. J. Biol. Chem. 2021;296:100411. doi: 10.1016/j.jbc.2021.100411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wood W, et al. Mesenchymal cells engulf and clear apoptotic footplate cells in macrophageless PU.1 null mouse embryos. Development. 2000;127:5245–5252. doi: 10.1242/dev.127.24.5245. [DOI] [PubMed] [Google Scholar]
- 130.Nagasaka A, Kawane K, Yoshida H, Nagata S. Apaf-1-independent programmed cell death in mouse development. Cell Death Differ. 2010;17:931–941. doi: 10.1038/cdd.2009.186. [DOI] [PubMed] [Google Scholar]
- 131.Nagata S, Hanayama R, Kawane K. Autoimmunity and the clearance of dead cells. Cell. 2010;140:619–630. doi: 10.1016/j.cell.2010.02.014. [DOI] [PubMed] [Google Scholar]
- 132.deCathelineau AM, Henson PM. The final step in programmed cell death: phagocytes carry apoptotic cells to the grave. Essays Biochem. 2003;39:105–117. doi: 10.1042/bse0390105. [DOI] [PubMed] [Google Scholar]
- 133.Muñoz LE, Lauber K, Schiller M, Manfredi AA, Herrmann M. The role of defective clearance of apoptotic cells in systemic autoimmunity. Nat. Rev. Rheumatol. 2010;6:280–289. doi: 10.1038/nrrheum.2010.46. [DOI] [PubMed] [Google Scholar]
- 134.Surh CD, Sprent J. T-cell apoptosis detected in situ during positive and negative selection in the thymus. Nature. 1994;372:100–103. doi: 10.1038/372100a0. [DOI] [PubMed] [Google Scholar]
- 135.Ren Y, Savill J. Apoptosis: the importance of being eaten. Cell Death Differ. 1998;5:563–568. doi: 10.1038/sj.cdd.4400407. [DOI] [PubMed] [Google Scholar]
- 136.Segawa K, Suzuki J, Nagata S. Constitutive exposure of phosphatidylserine on viable cells. Proc. Natl Acad. Sci. USA. 2011;108:19246–19251. doi: 10.1073/pnas.1114799108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Davies LC, Jenkins SJ, Allen JE, Taylor PR. Tissue-resident macrophages. Nat. Immunol. 2013;14:986–995. doi: 10.1038/ni.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yanagihashi Y, Segawa K, Maeda R, Nabeshima Y-I, Nagata S. Mouse macrophages show different requirements for phosphatidylserine receptor Tim4 in efferocytosis. Proc. Natl Acad. Sci. USA. 2017;114:8800–8805. doi: 10.1073/pnas.1705365114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lemke G. How macrophages deal with death. Nat. Rev. Immunol. 2019;36:1–11. doi: 10.1038/s41577-019-0167-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Nishi C, Toda S, Segawa K, Nagata S. Tim4- and MerTK-mediated engulfment of apoptotic cells by mouse resident peritoneal macrophages. Mol. Cell. Biol. 2014;34:1512–1520. doi: 10.1128/MCB.01394-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hanayama R, et al. Identification of a factor that links apoptotic cells to phagocytes. Nature. 2002;417:182–187. doi: 10.1038/417182a. [DOI] [PubMed] [Google Scholar]
- 142.Hanayama R, Tanaka M, Miwa K, Nagata S. Expression of developmental endothelial locus-1 in a subset of macrophages for engulfment of apoptotic cells. J. Immunol. 2004;172:3876–3882. doi: 10.4049/jimmunol.172.6.3876. [DOI] [PubMed] [Google Scholar]
- 143.Kourtzelis I, et al. DEL-1 promotes macrophage efferocytosis and clearance of inflammation. Nat. Immunol. 2019;20:40–49. doi: 10.1038/s41590-018-0249-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Proto JD, et al. Regulatory T cells promote macrophage efferocytosis during inflammation resolution. Immunity. 2018;49:666–677. doi: 10.1016/j.immuni.2018.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Park D, et al. BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature. 2007;450:430–434. doi: 10.1038/nature06329. [DOI] [PubMed] [Google Scholar]
- 146.Park SY, et al. Rapid cell corpse clearance by stabilin-2, a membrane phosphatidylserine receptor. Cell Death Differ. 2008;15:192–201. doi: 10.1038/sj.cdd.4402242. [DOI] [PubMed] [Google Scholar]
- 147.Das S, et al. Brain angiogenesis inhibitor 1 (BAI1) is a pattern recognition receptor that mediates macrophage binding and engulfment of Gram-negative bacteria. Proc. Natl Acad. Sci. USA. 2011;108:2136–2141. doi: 10.1073/pnas.1014775108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Harris EN, Cabral F. Ligand binding and signaling of HARE/Stabilin-2. Biomolecules. 2019;9:273. doi: 10.3390/biom9070273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Galluzzi L, et al. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018;25:486–541. doi: 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Shan B, Pan H, Najafov A, Yuan J. Necroptosis in development and diseases. Genes Dev. 2018;32:327–340. doi: 10.1101/gad.312561.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Gong Y-N, et al. ESCRT-III acts downstream of MLKL to regulate necroptotic cell death and its consequences. Cell. 2017;169:286–300. doi: 10.1016/j.cell.2017.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Rühl S, et al. ESCRT-dependent membrane repair negatively regulates pyroptosis downstream of GSDMD activation. Science. 2018;362:956–960. doi: 10.1126/science.aar7607. [DOI] [PubMed] [Google Scholar]
- 153.Di Virgilio F, Ben DD, Sarti AC, Giuliani AL, Falzoni S. The P2X7 receptor in infection and inflammation. Immunity. 2017;47:15–31. doi: 10.1016/j.immuni.2017.06.020. [DOI] [PubMed] [Google Scholar]
- 154.Aswad F, Kawamura H, Dennert G. High sensitivity of CD4+CD25+ regulatory T cells to extracellular metabolites nicotinamide adenine dinucleotide and ATP: a role for P2X7 receptors. J. Immunol. 2005;175:3075–3083. doi: 10.4049/jimmunol.175.5.3075. [DOI] [PubMed] [Google Scholar]
- 155.Ryoden Y, Nagata S. The XK plasma membrane scramblase and the VPS13A cytosolic lipid transporter for ATP-induced cell death. BioEssays. 2022;44:e2200106. doi: 10.1002/bies.202200106. [DOI] [PubMed] [Google Scholar]
- 156.Thiagarajan P, Parker CJ, Prchal JT. How do red blood cells die? Front. Physiol. 2021;12:655393. doi: 10.3389/fphys.2021.655393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Yoshida H, et al. Phosphatidylserine-dependent engulfment by macrophages of nuclei from erythroid precursor cells. Nature. 2005;437:754–758. doi: 10.1038/nature03964. [DOI] [PubMed] [Google Scholar]
- 158.Toda S, Segawa K, Nagata S. MerTK-mediated engulfment of pyrenocytes by central macrophages in erythroblastic islands. Blood. 2014;123:3963–3971. doi: 10.1182/blood-2014-01-547976. [DOI] [PubMed] [Google Scholar]
- 159.Ball JB, Green-Fulgham SM, Watkins LR. Mechanisms of microglia-mediated synapse turnover and synaptogenesis. Prog. Neurobiol. 2022;218:102336. doi: 10.1016/j.pneurobio.2022.102336. [DOI] [PubMed] [Google Scholar]
- 160.Frost JL, Schafer DP. Microglia: architects of the developing nervous system. Trends Cell Biol. 2016;26:587–597. doi: 10.1016/j.tcb.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Li T, et al. A splicing isoform of GPR56 mediates microglial synaptic refinement via phosphatidylserine binding. EMBO J. 2020;39:e104136. doi: 10.15252/embj.2019104136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Scott-Hewitt N, et al. Local externalization of phosphatidylserine mediates developmental synaptic pruning by microglia. EMBO J. 2020;39:e105380. doi: 10.15252/embj.2020105380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kurematsu C, et al. Synaptic pruning of murine adult-born neurons by microglia depends on phosphatidylserine. J. Exp. Med. 2022;219:e20202304. doi: 10.1084/jem.20202304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sapar ML, et al. Phosphatidylserine externalization results from and causes neurite degeneration in Drosophila. Cell Rep. 2018;24:2273–2286. doi: 10.1016/j.celrep.2018.07.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Pereira M, et al. Common signalling pathways in macrophage and osteoclast multinucleation. J. Cell. Sci. 2018;131:jcs216267. doi: 10.1242/jcs.216267. [DOI] [PubMed] [Google Scholar]
- 166.Petrany MJ, Millay DP. Cell fusion: merging membranes and making muscle. Trends Cell Biol. 2019;29:964–973. doi: 10.1016/j.tcb.2019.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Gerbaud P, Pidoux G. An overview of molecular events occurring in human trophoblast fusion. Placenta. 2015;36:S35–S42. doi: 10.1016/j.placenta.2014.12.015. [DOI] [PubMed] [Google Scholar]
- 168.Das M, et al. Phosphatidylserine efflux and intercellular fusion in a BeWo model of human villous cytotrophoblast. Placenta. 2004;25:396–407. doi: 10.1016/j.placenta.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 169.Satouh Y, Ikawa M. New insights into the molecular events of mammalian fertilization. Trends Biochem. Sci. 2018;43:818–828. doi: 10.1016/j.tibs.2018.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Helming L, Winter J, Gordon S. The scavenger receptor CD36 plays a role in cytokine-induced macrophage fusion. J. Cell Sci. 2009;122:453–459. doi: 10.1242/jcs.037200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Verma SK, et al. Cell-surface phosphatidylserine regulates osteoclast precursor fusion. J. Biol. Chem. 2018;293:254–270. doi: 10.1074/jbc.M117.809681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Jeong J, Conboy IM. Phosphatidylserine directly and positively regulates fusion of myoblasts into myotubes. Biochem. Biophys. Res. Commun. 2011;414:9–13. doi: 10.1016/j.bbrc.2011.08.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Rival CM, et al. Phosphatidylserine on viable sperm and phagocytic machinery in oocytes regulate mammalian fertilization. Nat. Commun. 2019;10:4456. doi: 10.1038/s41467-019-12406-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Martens S, McMahon HT. Mechanisms of membrane fusion: disparate players and common principles. Nat. Rev. Mol. Cell Biol. 2008;9:543–556. doi: 10.1038/nrm2417. [DOI] [PubMed] [Google Scholar]
- 175.Deneke VE, Pauli A. The fertilization enigma: how sperm and egg fuse. Annu. Rev. Cell Dev. Biol. 2021;37:391–414. doi: 10.1146/annurev-cellbio-120219-021751. [DOI] [PubMed] [Google Scholar]
- 176.Szondy Z, et al. Involvement of phosphatidylserine receptors in the skeletal muscle regeneration: therapeutic implications. J. Cachexia Sarcopenia Muscle. 2022;13:1961–1973. doi: 10.1002/jcsm.13024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.van den Eijnde SM, et al. Transient expression of phosphatidylserine at cell–cell contact areas is required for myotube formation. J. Cell Sci. 2001;114:3631–3642. doi: 10.1242/jcs.114.20.3631. [DOI] [PubMed] [Google Scholar]
- 178.Tsuchiya M, et al. Cell surface flip-flop of phosphatidylserine is critical for PIEZO1-mediated myotube formation. Nat. Commun. 2018;9:2049. doi: 10.1038/s41467-018-04436-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Grifell-Junyent M, et al. CDC50A is required for aminophospholipid transport and cell fusion in mouse C2C12 myoblasts. J. Cell Sci. 2022;135:jcs258649. doi: 10.1242/jcs.258649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ochiai Y, Suzuki C, Segawa K, Uchiyama Y, Nagata S. Inefficient development of syncytiotrophoblasts in the Atp11a-deficient mouse placenta. Proc. Natl Acad. Sci. USA. 2022;119:e2200582119. doi: 10.1073/pnas.2200582119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Middel V, et al. Dysferlin-mediated phosphatidylserine sorting engages macrophages in sarcolemma repair. Nat. Commun. 2016;7:12875. doi: 10.1038/ncomms12875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Croissant C, Carmeille R, Brévart C, Bouter A. Annexins and membrane repair dysfunctions in muscular dystrophies. Int. J. Mol. Sci. 2021;22:5276. doi: 10.3390/ijms22105276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Neumann B, et al. EFF-1-mediated regenerative axonal fusion requires components of the apoptotic pathway. Nature. 2015;517:219–222. doi: 10.1038/nature14102. [DOI] [PubMed] [Google Scholar]
- 184.Hisamoto N, et al. Phosphatidylserine exposure mediated by ABC transporter activates the integrin signaling pathway promoting axon regeneration. Nat. Commun. 2018;9:3099. doi: 10.1038/s41467-018-05478-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Bevers EM, Comfurius P, van Rijn JL, Hemker HC, Zwaal RF. Generation of prothrombin-converting activity and the exposure of phosphatidylserine at the outer surface of platelets. Eur. J. Biochem. 1982;122:429–436. doi: 10.1111/j.1432-1033.1982.tb05898.x. [DOI] [PubMed] [Google Scholar]
- 186.Zwaal RF, Comfurius P, Bevers EM. Lipid–protein interactions in blood coagulation. Biochim. Biophys. Acta. 1998;1376:433–453. doi: 10.1016/S0304-4157(98)00018-5. [DOI] [PubMed] [Google Scholar]
- 187.Peschon JJ, et al. An essential role for ectodomain shedding in mammalian development. Science. 1998;282:1281–1284. doi: 10.1126/science.282.5392.1281. [DOI] [PubMed] [Google Scholar]
- 188.Sommer A, et al. Phosphatidylserine exposure is required for ADAM17 sheddase function. Nat. Commun. 2016;7:11523. doi: 10.1038/ncomms11523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Elliott JI, et al. Membrane phosphatidylserine distribution as a non-apoptotic signalling mechanism in lymphocytes. Nat. Cell Biol. 2005;7:808–816. doi: 10.1038/ncb1279. [DOI] [PubMed] [Google Scholar]
- 190.Kirkin V, et al. The Fas ligand intracellular domain is released by ADAM10 and SPPL2a cleavage in T-cells. Cell Death Differ. 2007;14:1678–1687. doi: 10.1038/sj.cdd.4402175. [DOI] [PubMed] [Google Scholar]
- 191.Schulte M, et al. ADAM10 regulates FasL cell surface expression and modulates FasL-induced cytotoxicity and activation-induced cell death. Cell Death Differ. 2007;14:1040–1049. doi: 10.1038/sj.cdd.4402101. [DOI] [PubMed] [Google Scholar]
- 192.Bleibaum F, et al. ADAM10 sheddase activation is controlled by cell membrane asymmetry. J. Mol. Cell Biol. 2019;11:979–993. doi: 10.1093/jmcb/mjz008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Patton S, Keenan TW. The milk fat globule membrane. Biochim. Biophys. Acta. 1975;415:273–309. doi: 10.1016/0304-4157(75)90011-8. [DOI] [PubMed] [Google Scholar]
- 194.Hanayama R, Nagata S. Impaired involution of mammary glands in the absence of milk fat globule EGF factor 8. Proc. Natl Acad. Sci. USA. 2005;102:16886–16891. doi: 10.1073/pnas.0508599102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 196.Nakai W, et al. A novel affinity-based method for the isolation of highly purified extracellular vesicles. Sci. Rep. 2016;6:33935. doi: 10.1038/srep33935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Wei X, et al. Surface phosphatidylserine is responsible for the internalization on microvesicles derived from hypoxia-induced human bone marrow mesenchymal stem cells into human endothelial cells. PLoS ONE. 2016;11:e0147360. doi: 10.1371/journal.pone.0147360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Puhm F, Boilard E, Machlus KR. Platelet extracellular vesicles. Arterioscler. Thromb. Vasc. Biol. 2020;41:87–96. doi: 10.1161/ATVBAHA.120.314644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Sims P, Wiedmer T, Esmon C, Weiss H, Shattil S. Assembly of the platelet prothrombinase complex is linked to vesiculation of the platelet plasma membrane. Studies in Scott syndrome: an isolated defect in platelet procoagulant activity. J. Biol. Chem. 1989;264:17049–17057. doi: 10.1016/S0021-9258(18)71457-9. [DOI] [PubMed] [Google Scholar]
- 200.Mercer J, Helenius A. Vaccinia virus uses macropinocytosis and apoptotic mimicry to enter host cells. Science. 2008;320:531–535. doi: 10.1126/science.1155164. [DOI] [PubMed] [Google Scholar]
- 201.Morizono K, Chen ISY. Role of phosphatidylserine receptors in enveloped virus infection. J. Virol. 2014;88:4275–4290. doi: 10.1128/JVI.03287-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Li M, et al. TIM-family proteins inhibit HIV-1 release. Proc. Natl Acad. Sci. USA. 2014;111:E3699–E3707. doi: 10.1073/pnas.1404851111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Rood JE, Maartens A, Hupalowska A, Teichmann SA, Regev A. Impact of the Human Cell Atlas on medicine. Nat. Med. 2022;28:2486–2496. doi: 10.1038/s41591-022-02104-7. [DOI] [PubMed] [Google Scholar]
- 204.Liou AY, Molday LL, Wang J, Andersen JP, Molday RS. Identification and functional analyses of disease-associated P4-ATPase phospholipid flippase variants in red blood cells. J. Biol. Chem. 2019;294:6809–6821. doi: 10.1074/jbc.RA118.007270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Siggs OM, et al. The P4-type ATPase ATP11C is essential for B lymphopoiesis in adult bone marrow. Nat. Immunol. 2011;12:434–440. doi: 10.1038/ni.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Siggs OM, Schnabl B, Webb B, Beutler B. X-linked cholestasis in mouse due to mutations of the P4-ATPase ATP11C. Proc. Natl Acad. Sci. USA. 2011;108:7890–7895. doi: 10.1073/pnas.1104631108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Yabas M, et al. Mice deficient in the putative phospholipid flippase ATP11C exhibit altered erythrocyte shape, anemia, and reduced erythrocyte life span. J. Biol. Chem. 2014;289:19531–19537. doi: 10.1074/jbc.C114.570267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Yabas M, et al. ATP11C is critical for the internalization of phosphatidylserine and differentiation of B lymphocytes. Nat. Immunol. 2011;12:441–449. doi: 10.1038/ni.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Arashiki N, et al. ATP11C is a major flippase in human erythrocytes and its defect causes congenital hemolytic anemia. Haematologica. 2016;101:559–565. doi: 10.3324/haematol.2016.142273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Brain MC, Pihl C, Robertson L, Brown CB. Evidence for a mechanosensitive calcium influx into red cells. Blood Cell. Mol. Dis. 2004;32:349–352. doi: 10.1016/j.bcmd.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 211.Boas FE, Forman L, Beutler E. Phosphatidylserine exposure and red cell viability in red cell aging and in hemolytic anemia. Proc. Natl Acad. Sci. USA. 1998;95:3077–3081. doi: 10.1073/pnas.95.6.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Li T, et al. Phospholipid-flippase chaperone CDC50A is required for synapse maintenance by regulating phosphatidylserine exposure. EMBO J. 2021;40:e107915. doi: 10.15252/embj.2021107915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Mühle C, et al. Characterization of acid sphingomyelinase activity in human cerebrospinal fluid. PLoS ONE. 2013;8:e62912. doi: 10.1371/journal.pone.0062912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Kornhuber J, Rhein C, Müller CP, Mühle C. Secretory sphingomyelinase in health and disease. Biol. Chem. 2015;396:707–736. doi: 10.1515/hsz-2015-0109. [DOI] [PubMed] [Google Scholar]
- 215.Pater JA, et al. Autosomal dominant non-syndromic hearing loss maps to DFNA33 (13q34) and co-segregates with splice and frameshift variants in ATP11A, a phospholipid flippase gene. Hum. Genet. 2022;141:431–444. doi: 10.1007/s00439-022-02444-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Okamoto S, et al. The N- or C-terminal cytoplasmic regions of P4-ATPases determine their cellular localization. Mol. Biol. Cell. 2020;31:2115–2124. doi: 10.1091/mbc.E20-04-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Feenstra B, et al. Common variants associated with general and MMR vaccine-related febrile seizures. Nat. Genet. 2014;46:1274–1282. doi: 10.1038/ng.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Wang TA, et al. TMEM16C is involved in thermoregulation and protects rodent pups from febrile seizures. Proc. Natl Acad. Sci. USA. 2021;118:e202334211. doi: 10.1073/pnas.2023342118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Zanni ED, Gradogna A, Picco C, Scholz-Starke J, Boccaccio A. TMEM16E/ANO5 mutations related to bone dysplasia or muscular dystrophy cause opposite effects on lipid scrambling. Hum. Mutat. 2020;41:1157–1170. doi: 10.1002/humu.24006. [DOI] [PubMed] [Google Scholar]
- 220.Zwaal RF, Schroit AJ. Pathophysiologic implications of membrane phospholipid asymmetry in blood cells. Blood. 1997;89:1121–1132. doi: 10.1182/blood.V89.4.1121. [DOI] [PubMed] [Google Scholar]
- 221.Marconi C, et al. A novel missense mutation in ANO5/TMEM16E is causative for gnathodiaphyseal dyplasia in a large Italian pedigree. Eur. J. Hum. Genet. 2013;21:613–619. doi: 10.1038/ejhg.2012.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Savarese M, et al. Next generation sequencing on patients with LGMD and nonspecific myopathies: findings associated with ANO5 mutations. Neuromuscul. Disord. 2015;25:533–541. doi: 10.1016/j.nmd.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Tsutsumi S, et al. The novel gene encoding a putative transmembrane protein is mutated in gnathodiaphyseal dysplasia (GDD) Am. J. Hum. Genet. 2004;74:1255–1261. doi: 10.1086/421527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Xu J, et al. Genetic disruption of Ano5 in mice does not recapitulate human ANO5-deficient muscular dystrophy. Skelet. Muscle. 2015;5:43. doi: 10.1186/s13395-015-0069-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Sui T, et al. Development of muscular dystrophy in a CRISPR-engineered mutant rabbit model with frame-disrupting ANO5 mutations. Cell Death Dis. 2018;9:609. doi: 10.1038/s41419-018-0674-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Millington-Burgess SL, Harper MT. Gene of the issue: ANO6 and Scott syndrome. Platelets. 2020;31:964–967. doi: 10.1080/09537104.2019.1693039. [DOI] [PubMed] [Google Scholar]
- 227.Castoldi E, Collins PW, Williamson PL, Bevers EM. Compound heterozygosity for 2 novel TMEM16F mutations in a patient with Scott syndrome. Blood. 2011;117:4399–4400. doi: 10.1182/blood-2011-01-332502. [DOI] [PubMed] [Google Scholar]
- 228.Boisseau P, et al. A new mutation of ANO6 in two familial cases of Scott syndrome. Br. J. Haematol. 2016;180:750–752. doi: 10.1111/bjh.14439. [DOI] [PubMed] [Google Scholar]
- 229.Rosing J, et al. Impaired factor X and prothrombin activation associated with decreased phospholipid exposure in platelets from a patient with a bleeding disorder. Blood. 1985;65:1557–1561. doi: 10.1182/blood.V65.6.1557.bloodjournal6561557. [DOI] [PubMed] [Google Scholar]
- 230.Brooks MB, et al. A TMEM16F point mutation causes an absence of canine platelet TMEM16F and ineffective activation and death-induced phospholipid scrambling. J. Thromb. Haemost. 2015;13:2240–2252. doi: 10.1111/jth.13157. [DOI] [PubMed] [Google Scholar]
- 231.Das S, et al. NGEP, a prostate-specific plasma membrane protein that promotes the association of LNCaP cells. Cancer Res. 2007;67:1594–1601. doi: 10.1158/0008-5472.CAN-06-2673. [DOI] [PubMed] [Google Scholar]
- 232.Wahlström G, et al. The variant rs77559646 associated with aggressive prostate cancer disrupts ANO7 mRNA splicing and protein expression. Hum. Mol. Genet. 2022;31:2063–2077. doi: 10.1093/hmg/ddac012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Renaud M, et al. Autosomal recessive cerebellar ataxia type 3 due to ANO10 mutations: delineation and genotype–phenotype correlation study. JAMA Neurol. 2014;71:1305–1310. doi: 10.1001/jamaneurol.2014.193. [DOI] [PubMed] [Google Scholar]
- 234.Balreira A, et al. ANO10 mutations cause ataxia and coenzyme Q10 deficiency. J. Neurol. 2014;261:2192–2198. doi: 10.1007/s00415-014-7476-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Vermeer S, et al. Targeted next-generation sequencing of a 12.5 Mb homozygous region reveals ANO10 mutations in patients with autosomal-recessive cerebellar ataxia. Am. J. Hum. Genet. 2010;87:813–819. doi: 10.1016/j.ajhg.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Kramer J, Hawley RS. The spindle-associated transmembrane protein Axs identifies a membranous structure ensheathing the meiotic spindle. Nat. Cell Biol. 2003;5:261–263. doi: 10.1038/ncb944. [DOI] [PubMed] [Google Scholar]
- 237.Lee S, Russo D, Redman CM. The Kell blood group system: Kell and XK membrane proteins. Semin. Hematol. 2000;37:113–121. doi: 10.1016/S0037-1963(00)90036-2. [DOI] [PubMed] [Google Scholar]
- 238.Peikert, K., et al. VPS13A disease. National Library of Medicine. https://www.ncbi.nlm.nih.gov/books/NBK1387/ (2023).
- 239.Zhu X, et al. Giant axon formation in mice lacking Kell, XK, or Kell and XK animal models of McLeod neuroacanthocytosis syndrome. Ame. J. Pathol. 2014;184:800–807. doi: 10.1016/j.ajpath.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Yamashita Y, Suzuki C, Uchiyama Y, Nagata S. Infertility caused by inefficient apoptotic germ cell clearance in Xkr8-deficient male mice. Mol. Cell. Biol. 2020;40:e00402–e00419. doi: 10.1128/MCB.00402-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Hanayama R, et al. Autoimmune disease and impaired uptake of apoptotic cells in MFG-E8-deficient mice. Science. 2004;304:1147–1150. doi: 10.1126/science.1094359. [DOI] [PubMed] [Google Scholar]
- 242.Miyanishi M, Segawa K, Nagata S. Synergistic effect of Tim4 and MFG-E8 null mutations on the development of autoimmunity. Int. Immunol. 2012;24:551–559. doi: 10.1093/intimm/dxs064. [DOI] [PubMed] [Google Scholar]
- 243.Kaneshiro N, et al. Lipid flippase dysfunction as a therapeutic target for endosomal anomalies in Alzheimer’s disease. iScience. 2022;25:103869. doi: 10.1016/j.isci.2022.103869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Devaux PF. Is lipid translocation involved during endo- and exocytosis? Biochimie. 2000;82:497–509. doi: 10.1016/S0300-9084(00)00209-1. [DOI] [PubMed] [Google Scholar]
- 245.Hu Y, et al. Scramblase TMEM16F terminates T cell receptor signaling to restrict T cell exhaustion. J. Exp. Med. 2016;213:2759–2772. doi: 10.1084/jem.20160612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Baker RW, Hughson FM. Chaperoning SNARE assembly and disassembly. Nat. Rev. Mol. Cell. Biol. 2016;17:465–479. doi: 10.1038/nrm.2016.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Shin H-W, Takatsu H. Substrates of P4-ATPases: beyond aminophospholipids (phosphatidylserine and phosphatidylethanolamine) FASEB J. 2019;33:3087–3096. doi: 10.1096/fj.201801873R. [DOI] [PubMed] [Google Scholar]
- 248.Levano K, et al. Atp8a1 deficiency is associated with phosphatidylserine externalization in hippocampus and delayed hippocampus-dependent learning. J. Neurochem. 2012;120:302–313. doi: 10.1111/j.1471-4159.2011.07543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.McMillan HJ, et al. Recessive mutations in ATP8A2 cause severe hypotonia, cognitive impairment, hyperkinetic movement disorders and progressive optic atrophy. Orphanet J. Rare Dis. 2018 doi: 10.1186/s13023-018-0825-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Guissart C, et al. ATP8A2-related disorders as recessive cerebellar ataxia. J. Neurol. 2020;267:203–213. doi: 10.1007/s00415-019-09579-4. [DOI] [PubMed] [Google Scholar]
- 251.Zhu X, et al. Mutations in a P-type ATPase gene cause axonal degeneration. PLoS Genet. 2012;8:e1002853. doi: 10.1371/journal.pgen.1002853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Klomp LW, et al. Characterization of mutations in ATP8B1 associated with hereditary cholestasis. Hepatology. 2004;40:27–38. doi: 10.1002/hep.20285. [DOI] [PubMed] [Google Scholar]
- 253.Stapelbroek JM, et al. ATP8B1 is essential for maintaining normal hearing. Proc. Natl Acad. Sci. USA. 2009;106:9709–9714. doi: 10.1073/pnas.0807919106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Vogt G, et al. Biallelic truncating variants in ATP9A cause a novel neurodevelopmental disorder involving postnatal microcephaly and failure to thrive. J. Med. Genet. 2022;59:662–668. doi: 10.1136/jmedgenet-2021-107843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Mattioli F, et al. Biallelic truncation variants in ATP9A are associated with a novel autosomal recessive neurodevelopmental disorder. NPJ Genom. Med. 2021;6:94. doi: 10.1038/s41525-021-00255-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Meguro M, et al. A novel maternally expressed gene, ATP10C, encodes a putative aminophospholipid translocase associated with Angelman syndrome. Nat. Genet. 2001;28:19–20. doi: 10.1038/ng0501-19. [DOI] [PubMed] [Google Scholar]
- 257.Dhar MS, Yuan JS, Elliott SB, Sommardahl C. A type IV P-type ATPase affects insulin-mediated glucose uptake in adipose tissue and skeletal muscle in mice. J. Nutr. Biochem. 2006;17:811–820. doi: 10.1016/j.jnutbio.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 258.Real R, et al. ATP10B and the risk for Parkinson’s disease. Acta Neuropathol. 2020;140:401–402. doi: 10.1007/s00401-020-02172-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Roland BP, et al. Yeast and human P4-ATPases transport glycosphingolipids using conserved structural motifs. J. Biol. Chem. 2019;294:1794–1806. doi: 10.1074/jbc.RA118.005876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Sigruener A, et al. Lipidomic and metabolic changes in the P4-type ATPase ATP10D deficient C57BL/6J wild type mice upon rescue of ATP10D function. PLoS ONE. 2017;12:e0178368. doi: 10.1371/journal.pone.0178368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Charlesworth G, et al. Mutations in ANO3 cause dominant craniocervical dystonia: ion channel implicated in pathogenesis. Am. J. Hum. Genet. 2012;91:1041–1050. doi: 10.1016/j.ajhg.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Jun I, et al. ANO9/TMEM16J promotes tumourigenesis via EGFR and is a novel therapeutic target for pancreatic cancer. Br. J. Cancer. 2017;117:1798–1809. doi: 10.1038/bjc.2017.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Li C, Cai S, Wang X, Jiang Z. Identification and characterization of ANO9 in stage II and III colorectal carcinoma. Oncotarget. 2015;6:29324–29334. doi: 10.18632/oncotarget.4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Chrysanthou A, Ververis A, Christodoulou K. ANO10 function in health and disease. Cerebellum. 2022 doi: 10.1007/s12311-022-01395-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Chung J, et al. PI4P/phosphatidylserine countertransport at ORP5- and ORP8-mediated ER–plasma membrane contacts. Science. 2015;349:428–432. doi: 10.1126/science.aab1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Pinot M, et al. Polyunsaturated phospholipids facilitate membrane deformation and fission by endocytic proteins. Science. 2014;345:693–697. doi: 10.1126/science.1255288. [DOI] [PubMed] [Google Scholar]
- 267.Corbalán-García S, Gómez-Fernández JC. Classical protein kinases C are regulated by concerted interaction with lipids: the importance of phosphatidylinositol-4,5-bisphosphate. Biophys. Rev. 2013;6:3–14. doi: 10.1007/s12551-013-0125-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Zhou Y, et al. Membrane potential modulates plasma membrane phospholipid dynamics and K-Ras signaling. Science. 2015;349:873–876. doi: 10.1126/science.aaa5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Yeung T, et al. Membrane phosphatidylserine regulates surface charge and protein localization. Science. 2008;319:210–213. doi: 10.1126/science.1152066. [DOI] [PubMed] [Google Scholar]
- 270.Yeung T, et al. Receptor activation alters inner surface potential during phagocytosis. Science. 2006;313:347–351. doi: 10.1126/science.1129551. [DOI] [PubMed] [Google Scholar]
- 271.Fairn GD, Hermansson M, Somerharju P, Grinstein S. Phosphatidylserine is polarized and required for proper Cdc42 localization and for development of cell polarity. Nat. Cell Biol. 2011;13:1424–1430. doi: 10.1038/ncb2351. [DOI] [PubMed] [Google Scholar]
- 272.Liu X, et al. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature. 2016;535:153–158. doi: 10.1038/nature18629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Deng W, et al. Streptococcal pyrogenic exotoxin B cleaves GSDMA and triggers pyroptosis. Nature. 2022;602:496–502. doi: 10.1038/s41586-021-04384-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.