Summary
Background
Intratumor heterogeneity (ITH) has been associated with poor prognosis in advanced non-small cell cancer (NSCLC) patients receiving immune checkpoint blockade (ICB) therapies. However, there is currently no evidence supporting an ITH metric as a predictor of clinical benefit from ICB. The unique advantages of blood make it a promising material for ITH estimation and relevant applications. This study aims to develop and validate a blood-based ITH index for predicting ICB response.
Methods
NSCLC patients from the OAK and POPLAR clinical trials were used as the training cohorts for algorithm development. Survival analyses with overall survival (OS) and progression-free survival (PFS) as endpoints were performed to assess clinical response. The predictive value of bITH was subsequently validated with an independent cohort of 42 NSCLC patients treated with PD-1 blockade.
Findings
bITH was significantly associated with the differential OS and PFS elicited by atezolizumab vs. docetaxel in both univariable and multivariable analyses in the OAK patients, suggesting bITH as an independent predictor for response to ICB. Moreover, compared with blood tumor mutation burden (bTMB), bITH enabled greater OS segregation and comparable PFS segregation, and obtained a predictive role regardless of bTMB status. Moreover, the association between bITH and PFS was validated with an independent cohort.
Interpretation
Patients with low blood-based ITH metric manifest significant OS and PFS benefit from immunotherapy versus chemotherapy. Future research is awaited to corroborate our findings and to enrich the clinical utility of ITH.
Funding
This study was supported by the National Natural Science Foundation of China (Nos. 81972718 and 81572321), the Natural Scientific Foundation of Zhejiang Province, China (No. LY19H160007), the Science and Technology Program for Health and Medicine in Zhejiang Province, China (No. 2021KY541), the Scientific Research Project, Science and Technology Department of Sichuan Province (No. 21YYJC1616), the Scientific Research Project, Sichuan Medical Association (No. S20002), Wu Jieping Medical Foundation (No. 320.6750), and 2018 Entrepreneurial Leading Talent of Guangzhou Huangpu District and Guangzhou Development District (No. 2022-L023).
Keywords: Intratumor heterogeneity, Immunotherapy, Immune checkpoint blockade, Predictive biomarker, Blood biopsy, NSCLC
Research in context.
Evidence before this study
Immunotherapy had become the recommended first-line treatment for advanced NSCLC without EGFR and ALK alterations. PD-L1 expression level and TMB had been extensively studied as biomarkers for predicting response to immunotherapy, but the evidence was conflicting. Tumor ITH, which reflects the complexity of tumor clones, was also associated with immunotherapy efficacy. In this study, we aimed to evaluate ITH level using liquid biopsy and explore the potential of bITH as a predictor to immunotherapy benefits.
Added value of this study
The study provided the proof of concept in the POPLAR cohort that bITH-low patients showed survival benefits from atezolizumab compared to docetaxel. Using the OAK cohort, we further confirmed a significant interaction between bITH and treatment group. Moreover, the analysis demonstrated that bITH-low is independent of bTMB status and had predictive power in bTMB-low patients. We also retrospectively collected an independent NSCLC cohort treated with anti-PD-1 inhibitors and validated the association between bITH and progression-free survival.
Implications of all the available evidence
The genomic biomarker bITH derived from a convenient and non-invasive blood biopsy was associated with immunotherapy benefits, which was consistent with previous studies of ITH estimated with tissue samples. Current study demonstrated that bITH-low patients may be useful in selecting patients who would benefit from immunotherapy, even among the bTMB-low group.
Introduction
Immune checkpoint blockade (ICB) with antibodies inhibiting programmed cell death 1 (PD-1) or its ligand PD-L1 has been transforming the treatment landscape in non-small cell lung cancer (NSCLC). Despite the success, however, the clinical utility of ICB is obscured by the lack of established biomarkers for efficacy prediction. The search so far has yielded a handful of candidates, of which PD-L1 expression1, 2, 3, 4 and tumor mutation burden (TMB)5, 6, 7 have received the most extensive characterization. Meanwhile, as an emerging candidate, intratumor heterogeneity (ITH) has recently attracted interest due to its role in fueling the evolution of therapeutically resistant neoplastic clones.8 Several clinical investigations have proposed different tissue-based ITH metrics and shown their negative effects on survival in multiple cancer types.8, 9, 10, 11, 12 Wolf et al. associated higher ITH with dampened T cell reactivity and tumor rejection in mice. Using an index based on genomic alterations detected from tumor tissues, the authors found a significant correlation between high ITH and poor survival in melanoma patients treated with ICB.13 In NSCLC, Fang and colleagues developed an ITH index based on genomic abnormalities mostly from tumor biopsies. However, when applied to blood samples, the investigators reported a lack of significant segregation of ICB-associated survival benefits.12
As the term suggests, intratumor heterogeneity poses challenges to its comprehensive characterization using individual tumor biopsy samples.8 One strategy is multi-region sampling, although this approach requires larger tumor bulks and may incur prohibitive risks for the patient. The task is even more daunting after factoring in the potentially vast heterogeneity among the nodal and/or distant metastasis and between primary and disseminated tumors.14,15 These challenges make blood biopsies an attractive choice for ITH estimation. In addition to non-invasiveness and freedom from the potential sampling biases inherent to tissue biopsy, blood biopsies theoretically provide genetic material from multiple lesions and may therefore render a more representative snapshot of ITH across disease sites. Indeed, circulating tumor DNA (ctDNA) has been used for phylogenetic profiling to track the subclonal architecture in the lung cancer.16 Moreover, blood samples could enable the detection of additional clinically relevant alterations compared with tissues.14,17
Comparable to ITH, blood TMB (bTMB), as an alternative to tissue TMB (tTMB), has been extensively investigated in clinical trials recently as a biomarker for immunotherapy.6,18,19 Although bTMB was found to be a predictive biomarker for PFS in NSCLC patients treated with immunotherapy in the OAK cohort,6 the recently published phase 2 B-F1RST trial revealed no statistically significant difference in PFS between bTMB-low and bTMB-high groups.19 A stronger predictive surrogate biomarker of immunotherapy is desired. Together, these advantages prompted our investigation into the clinical applicability of blood-based ITH metrics for immunotherapy.
This study aimed to examine the relevance of ITH, when derived from genomic alterations detected from blood biopsies, in predicting clinical response to ICB. We developed blood-based metric bITH, and provided evidence supporting its independence from blood-based tumor mutation burden (bTMB) and superiority in ICB responder enrichment, and validated its association with survival in an independent cohort.
Methods
Study design
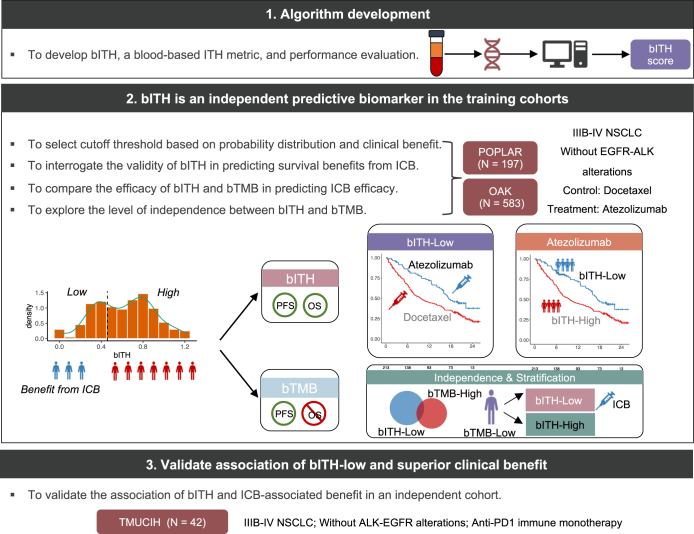
We developed bITH, a blood-based ITH estimation score, to infer ITH from non-invasive and readily available blood biopsies (Fig. 1). We first leveraged an exploratory training cohort and a larger training cohort to explore the ability of bITH as an independent predictive biomarker. The exploratory training cohort consisted of patients from the biomarker-evaluated populations (BEP) of the POPLAR (NCT01903993; intention-to-treat: n = 287; BEP: n = 211) clinical trial. The cohort from the biomarker-evaluated populations of the OAK (NCT02008227; intention-to-treat: n = 850; BEP: n = 583) clinical trial was used as the training cohort. Both studies compared the therapeutic efficacy of the PD-L1 inhibitor atezolizumab and the standard-of-care docetaxel for patients with previously treated, advanced, or metastatic NSCLC.2,20 All relevant data regarding clinicopathologic characteristics, survival, and response evaluation were obtained from Gandara et al.’s paper.6 Patients harboring either epidermal growth factor receptor (EGFR) mutations or anaplastic lymphoma kinase (ALK) rearrangement were excluded from subsequent analyses, leading to 583 patients from OAK and 197 from POPLAR (Table S1).
Fig. 1.
Study design and characteristics of bITH, a blood-based biomarker for immunotherapy.
To independently validate the association of bITH-low and the ICB benefit, we retrospectively collected blood biopsies from 42 patients with stage IIIB-IV NSCLC treated with PD-1 inhibitor at the Tianjin Medical University Cancer Institute & Hospital (TMUCIH) between January 2017 and May 2020. We initially included 54 advanced NSCLC patients without ALK and EGFR alterations. All of them received anti-PD-1 monotherapy, including nivolumab, sintilimab, and pembrolizumab. Twelve patients with no somatic alterations due to low ctDNA volume were subsequently excluded. The clinical characteristics of the cohort can be found in Tables S2 and S3. Besides, sex was self-reported by study participants and collected before treatment. This study was approved by the Institutional Review Board of TMUCIH. All patients provided written informed consent. A more detailed description of the training and the validation cohorts is provided in the Supplementary Materials.
Panel-based next-generation sequencing analysis of blood biopsies
Blood biopsies from patients in the TMUCIH cohort were subjected to sequencing with a targeted panel of 520 cancer-related genes (OncoScreen Plus, covering a 1.86 MB region of the human genome, Burning Rock Biotech, Guangzhou, China). Blood samples were stored at −80 °C after collection till further handling. DNA extraction, quantification, library construction, sequencing, and data processing were performed as previously described.21 A more detailed description is provided in the Supplementary Materials.
Computation of bITH and bTMB using blood biopsy sequencing data
Shannon Diversity Index (SDI) is a widely used approach in ITH estimation.13,22 SDI divided variant allele frequencies (VAFs) into several equal bins and measure their entropy equally, which exhibits prognostic potential with insignificant treatment selection power (Supplementary Materials, Figure S1a and b). To calculate ITH in blood samples, we developed a bITH score by modifying SDI in two ways (Figure S2a): Firstly, VAFs were scaled with MSAF, a measure of the fraction of ctDNA in blood samples. The resulting MASF-corrected VAFs (MCVs) were equivalent to cancer cell fractions (CCFs). Secondly, bITH was subsequently calculated by introducing a weight function to the SDI formula to treat alterations with distinct clonality differently:
where n is the number of bins (default 10), is the probability of MCVs located in respective bins, and is the average of the corresponding MCV bin endpoints. Detailed description of correlations among these features is provided in Supplementary Materials (Figures S1 and S2).
bTMB scores for the OAK and POPLAR cohorts were obtained from a previous study.6 bTMB score for the TMUCIH cohort was calculated using the following equation:
Briefly, after the removal of known and probably oncogenic driver events and germline single nucleotide polymorphisms, bTMB was determined as the number of somatic single-nucleotide variations and indels detected at an allele frequency of ≥0.2% within the coding regions of the genes targeted by the panel used.
Assessment of clinical outcomes
In the two training cohorts, definitions of overall survival (OS), progression-free survival (PFS), objective response rate (ORR), and disease control rate (DCR) rate were adopted from the OAK and POPLAR trials. Specifically, OS was defined as the time interval from the date of randomization to the date of death from any cause. PFS was defined as the time interval from the date of randomization to the date of first documented disease progression, as assessed by the revised Response Evaluation Criteria in Solid Tumors guidelines (RECIST v1.1), or death from any cause, whichever occurred earlier. All relevant data regarding survival status and response evaluation were downloaded from Gandara et al.’s paper.6
In the validation cohort, OS was defined as the time from the start of anti-PD-1 treatment until death from any cause; PFS was defined as the time from the start of anti-PD-1 treatment until disease progression according to RECIST v1.1 or death from any cause.
Statistical analysis
Differences in objective response rate and disease control rate between two groups were tested with Fisher's exact test. Survival curves were estimated with the Kaplan–Meier approach, and survival analysis was performed with the log-rank test and Cox proportional hazards regression (survival (3.2-7) and survminer (0.4.8) extension packages). Left censoring was adopted in all survival analyses. The proportional hazards assumption was assessed with the scaled Schoenfeld residuals. For multivariable Cox regression, all publicly available variables from the OAK and POPLAR datasets were used in the initial screening, including age, sex, race, smoker status, ECOG score, previous lines of treatment, histology, PD-L1 status, bTMB, and bITH. Variables with p < 0.1 in the univariable regression were then used for multivariable Cox analysis. All reported p-values were two-tailed, and statistical significance was defined as p < 0.05. All analyses were implemented with the R software (version 4.0.3, https://www.r-project.org/).
Statement of ethics
This study was approved by the Institutional Review Board of Tianjin Medical University Cancer Institute & Hospital (No. E20210666). All patients had provided written informed consent for participating in the study.
Role of funders
The funders had no role in study design, data collection, data analysis, interpretation or writing of manuscript.
Results
Development of bITH and determination of cutoff of bITH score
We developed bITH based on SDI, a popular metric to evaluate diversity (e.g. ITH). After correcting for MSAF and balancing genomic aberrations of various MCVs (see Materials and Methods and Supplementary Materials and Methods for details), we observed a bimodal distribution pattern in both POPLAR and OAK cohorts (Figure S3) indicating that tumors can be naturally separated according to the bITH score. Respective percentile at 1/3 and 1/2 lies between the two peaks of the POPLAR and OAK bITH distributions, suggesting an optimal range for subsequent cutoff point selection.
To evaluate the association between clinical outcomes and the bITH score, we selected several cutoff points distributed from 1/4 to 1/2 and calculate treatment benefits in the BEP (197 patients) of the POPLAR study. Based on the greater PFS and OS treatment effect in cutoff point around 1/3 percentile than other values (Figure S4a and b), BEP patients were grouped as bITH-low and -high status. Improved OS and PFS benefits were observed for bITH-low relative to bITH-high (Figure S4c and d).
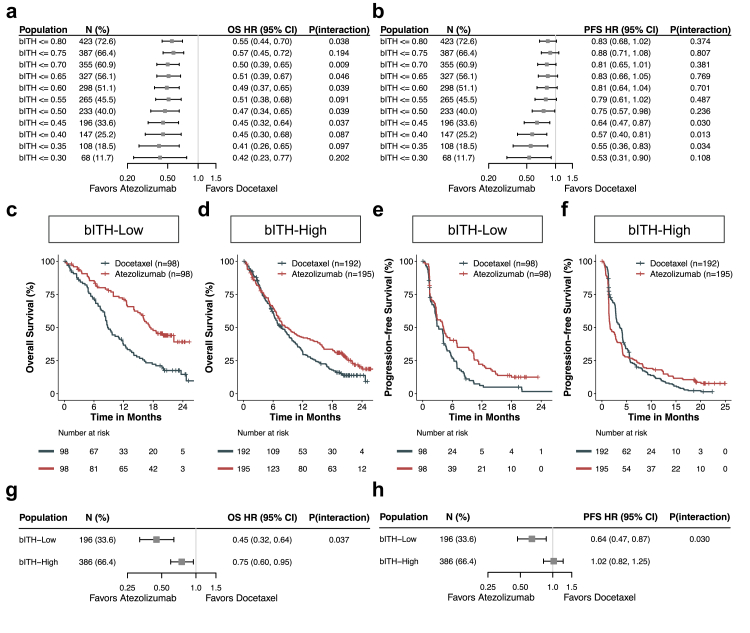
The same cutoff of 1/3 percentile was selected in the OAK cohort (Fig. 2a and b). Monotonic trends toward treatment differences in PFS were observed at various cutoff points. A consistent monotonic relationship was observed in OS with less divergence. The interaction terms in several bITH cutoff points were predictive of atezolizumab-associated OS or PFS, attesting to the amenability of this metric for predicting response to ICB.
Fig. 2.
bITH status predicted survival benefits from atezolizumab in the OAK cohort. (a & b) HRs for OS (g) and PFS (h) in cut-points from bITH ≤0.30 to bITH ≤0.80. (c & d) Kaplan–Meier curves of overall survival comparing patients treated with atezolizumab versus docetaxel in bITH-low and -high patients respectively. (e & f) Kaplan–Meier curves of progression-free survival comparing patients treated with atezolizumab versus docetaxel in bITH-low and -high patients respectively. (g & h) Forest plot of atezolizumab-over-docetaxel benefit in bITH-low than in bITH-high patients. P (interaction), a p-value of the interaction term in the corresponding Cox survival analysis. N (%) refers to the absolute number of patients and prevalence (%) of the OAK BEP cohorts. The error bars indicate 95% CIs of HRs.
bITH acts as a predictive biomarker of immunotherapy
Having determined the cutoff of the bITH score, we first stratified the POPLAR cohort into bITH-low and bITH-high groups and observed that bITH may be a potential predictor of ICB (Supplementary Materials and Methods). To further explore the predictive power of bITH in a larger cohort, we divided the OAK patients into bITH-low and bITH-high groups and evaluated the performance of bITH in predicting clinical outcomes of immunotherapy. Patients with bITH-low status manifested significant OS (HR: 0.45 (95% CI: 0.32–0.64), log-rank test p < 0.001) and PFS (HR: 0.64 (95% CI: 0.47–0.87), log-rank test p = 0.001) benefit from atezolizumab versus docetaxel (Fig. 2c and e). Nevertheless, bITH-high patients obtained significantly reduced OS (HR: 0.75 (95% CI: 0.60–0.95), log-rank test p = 0.015) and PFS (HR: 1.02 (95% CI: 0.82–1.25), log-rank test p = 0.910) difference between treatment arms (Fig. 2d and f). Although both groups derived OS benefit from atezolizumab, the inter-treatment differences were significant for both OS and PFS (cox interaction term p = 0.037 for OS and 0.030 for PFS; Fig. 2g and h). The above results demonstrated that at the cutoff of 1/3 percentile, bITH is a predictive biomarker of immunotherapy.
bITH-low status was significantly associated with survival benefits with atezolizumab
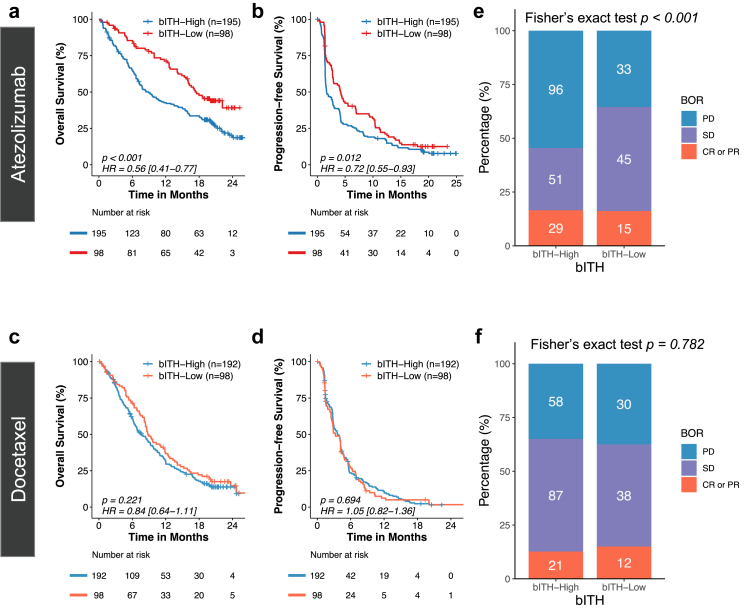
Having demonstrated bITH as a predictor of atezolizumab, we further examined the association of bITH status and clinical benefits within each treatment arm as confirmatory evidence. Among patients receiving atezolizumab in the OAK cohort, bITH-low patients achieved significantly longer OS (HR: 0.56 (95% CI: 0.41–0.77), log-rank test p < 0.001) and PFS (HR: 0.72 (95% CI: 0.55–0.93), log-rank test p = 0.012) compared with bITH-high patients (Fig. 3a and b). Median OS was 17.3 months for bITH-low patients and 8.9 months for bITH-high patients; median PFS (mPFS) was 4.2 months for bITH-low patients and 1.7 months for bITH-high patients. In contrast, comparable OS and PFS were observed for the docetaxel arm irrespective of bITH status (Fig. 3c and d). Consistent with the survival analysis, a significantly lower percentage of bITH-low patients showed progressive disease as the best response to atezolizumab (Fisher's exact test p < 0.001) compared to bITH-high cases, whereas in the docetaxel arm the proportions were highly comparable (Fisher's exact test p = 0.782; Fig. 3e and f).
Fig. 3.
A bITH-low status was associated with significantly greater survival benefit only in patients treated with atezolizumab. (a & b) Within the atezolizumab-treated arm, the bITH-low patients achieved significantly more favorable overall survival (OS) and progression-free survival (PFS) compared with bITH-high patients. (c & d) Docetaxel-treated patients manifested comparable OS and PFS irrespective of bITH status. (e & f) A significantly lower percentage of bITH-low patients showed progressive disease as the best overall response (BOR) to atezolizumab compared to bITH-high cases, whereas in the docetaxel arm the proportions were highly comparable irrespective of bITH status.
bITH demonstrates greater OS prediction efficacy than bTMB
A pioneering study by Gandara and colleagues proposed bTMB as a predictor of clinical response to atezolizumab in the OAK and POPLAR cohorts.6 Since both bITH and bTMB are blood-based metrics, comparison of their performance is naturally desired. We evaluated the prediction efficacy of bITH and bTMB in the OAK cohort. Both bTMB scores and the cut-point (≥16 as bTMB-high) were adopted from the study by Gandara et al.6
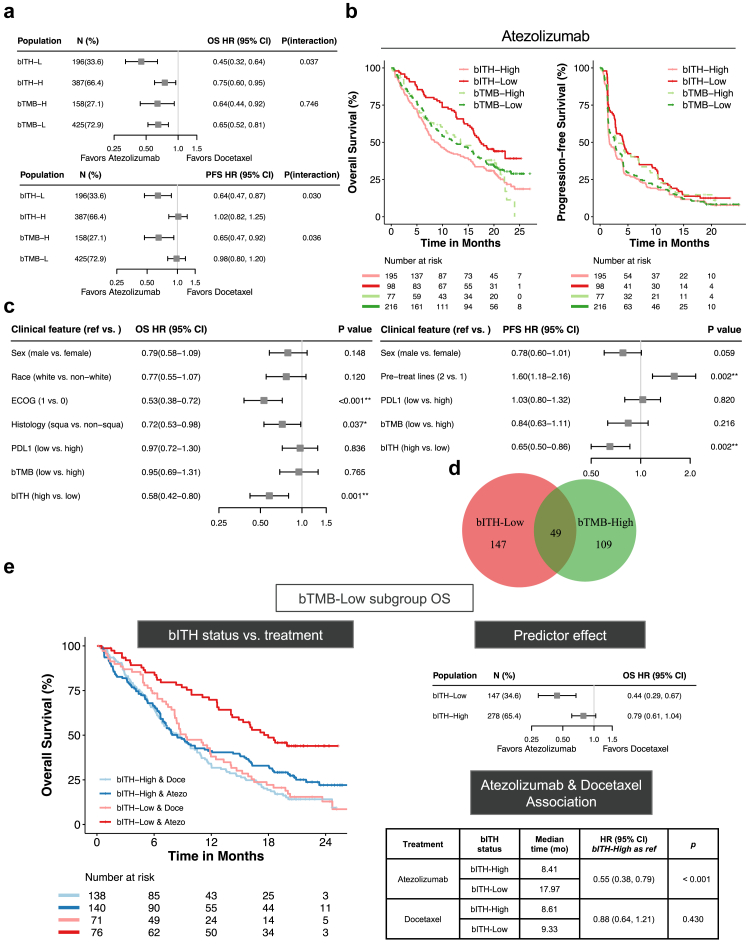
Upon comparing efficacy in enriching for potential responders to ICB, the inter-treatment difference in the risk of death was more remarkable for bITH-low than bTMB-high patients (Fig. 4a). Also, the prevalence of bITH-low was higher than bTMB-high indicating the ability to identify more potential responders. More importantly, analysis of interaction effects further showed that bTMB lacked predictive significance for atezolizumab-associated OS (cox interaction term p = 0.746). Regarding PFS, similar inter-treatment differences and prediction efficacy were observed between bITH-low versus bTMB-high patients.
Fig. 4.
bITH is independent of blood tumor mutation burden (bTMB) and superior in predicting overall survival to immune checkpoint blockade therapy in the OAK cohort. (a) Forest plots compare the atezolizumab-over-docetaxel associated overall survival (OS) and progression-free survival (PFS) with bITH and bTMB strata. P (interaction), a p-value of the interaction term indicates the statistical power of predictive efficacy. (b) Merge of Kaplan–Meier curves comparing OS and PFS segregation of bITH and bTMB status among OAK patients receiving atezolizumab. (c) Forest plots illustrating multivariable cox regression model of atezolizumab-associated OS and PFS that include factors with p ≤ 0.1 in univariable Cox regression and immunotherapy-related characteristics like bTMB and PD-L1 positivity. (d) Venn diagram illustrating the overlap between bITH-low and bTMB-high patients. (e) Kaplan–Meier curves comparing OS of four groups stratified with bITH status and treatment among bTMB-low patients. Forest plots of HRs indicating treatment difference of bITH status. Table illustrating median OS, HRs, and corresponding log-rank test p-values.
As shown in Fig. 4b, further analysis of association to atezolizumab-arm outcomes demonstrated that bITH showed greater segregation than bTMB in both OS (HR: 0.56 (95% CI: 0.41–0.77), log-rank test p < 0.001 vs. HR: 0.94 (95% CI: 0.68–1.29), log-rank test p = 0.687) and PFS (HR: 0.72 (95% CI: 0.55–0.93), log-rank test p = 0.013 vs. HR: 1.18 (95% CI: 0.89–1.56), log-rank test p = 0.238). Longer median OS were observed between bITH-low (17.31 months) vs. -high patients (8.90 months). In contrast, median OS were similar for bTMB-high and -low patients (13.50 months vs. 12.62 months). Consistently, median PFS showed greater difference for patients separated by bITH (4.21 months vs. 1.68 months) than bTMB (2.86 months vs. 2.69 months) status. Taken together, these lines of evidence suggested that bITH outperformed bTMB in atezolizumab-associated OS prediction and enriching those more likely to manifest favorable response to ICB.
bITH was an independent biomarker and predictive of ICB irrespective of bTMB status
Having confirmed bITH obtained a superior predictive effect to ICB-associated OS than bTMB, we conducted the subsequent analysis to investigate the dependency relationship between bITH and other characteristics, including age, sex, race, smoking history, ECOG performance status, previous line(s) of treatment, histologic subtype, PD-L1 positivity, and bTMB. Among those receiving atezolizumab treatment in the OAK cohort, clinical features associated with survival outcomes in the log-rank test (p ≤ 0.1) and previously proposed immunotherapy-related biomarkers like PD-L1 and bTMB were included in the multivariable cox regression model (Table S4 and Fig. 4c). We found that bITH remained significantly associated with OS and PFS benefit from atezolizumab vs. docetaxel after correcting for other potential confounders. In contrast, the association of clinical outcomes with bTMB was statistically insignificant in both univariable and multivariable analyses. In terms of patient makeup, there was but a small overlap between the bITH-low and bTMB-high populations in the ICB arm, accounting for 25.0% and 31.0% of the respective populations (Fig. 4d). Also, bITH-low patients were present in similar proportions in the bTMB-high vs. -low subgroups (31.0% vs. 34.6%, Fisher's exact test p = 0.432).
To further confirm feature dependency, we estimated the inter-treatment effect of bITH status in both bTMB stratification. Among those bTMB-low patients, which were considered non-responders to ICB, bITH-low obtained significant OS segregation between atezolizumab and docetaxel (log rank test p < 0.001). Also, the interaction effect between bITH status and treatment was significant (Fig. 4e, cox interaction term p = 0.036). Similarly improved atezolizumab-associated PFS benefit was observed in bITH-low than -high patients (Figure S5a, cox interaction term p = 0.104). Among atezolizumab-treated patients, bITH-low obtained significantly superior OS and PFS than bITH-high irrespective of bTMB status. However, the clinical outcomes were similar in the docetaxel arm (Fig. 4e and Figure S5a). Moreover, bITH-low also remained significantly associated with OS (HR: 0.48 (95% CI: 0.23–0.99), log-rank test p = 0.041; Figure S5b) and PFS (HR: 0.42 (95% CI: 0.23–0.79), log-rank test p = 0.005; Figure S5c) benefit from atezolizumab versus docetaxel among those proposed immunotherapy-benefit bTMB-high patients, which further demonstrated bITH could allow for a more specific selection of likely responders to ICB. Altogether, these evidences suggested that bITH-low was an independent biomarker of ICB and obtained a predictive role regardless of bTMB-status.
The association of bITH and PFS was validated in an independent cohort
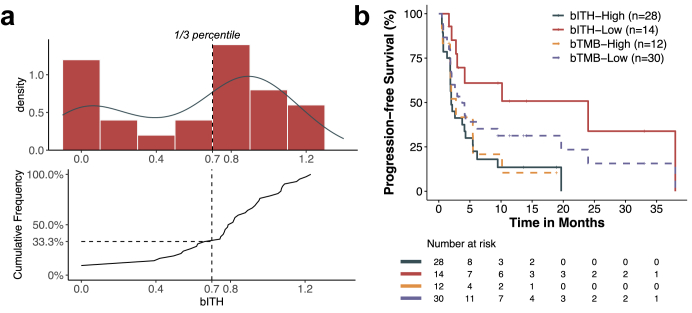
The validation cohort consisted of 42 Chinese NSCLC patients treated at TMUCIH with PD-L1 inhibitors such as nivolumab, which a recent study showed to elicit comparable OS benefit compared with atezolizumab (Table S2).23 Like the training cohort, no patient in the TMUCIH cohort carried EGFR mutations or ALK rearrangements. As with the training cohort, the bITH cut-point was set at the 1/3 percentile, defining 14 (33.4%) patients as bITH-low (Fig. 5a). Interestingly, as the cumulative frequency curve shows, the 1/3 percentile was around the increment “elbow” point, further demonstrating the rationale of cutoff selection. The bITH-low group showed significantly more favorable PFS compared with bITH-high (mPFS: 24.0 (95% CI: 2.97-not reached) vs. 2.0 (95% CI: 1.87–5.47) months, log-rank test p = 0.005; Fig. 5b). However, no significant PFS segregation between bTMB-high and bTMB-low was observed (mPFS: 2.77 (95% CI: 1.90-not reached) vs. 3.77 (95% CI: 2.07–24.0) months, log-rank test p = 0.342; Fig. 5b). Univariable analysis of bTMB-high (cut-points: ≥10 & ≥16) showed an insignificant correlation with favorable PFS (Figure S6). Moreover, other clinical features were not associated with either bITH status or immunotherapy-related PFS (Table S2). bITH-low status was further validated as enriching ICB responders and independent from other clinical factors.
Fig. 5.
The predictive value of bITH was validated in an independent cohort of patients treated with anti-PD-1 monotherapy. (a) The distribution of bITH scores in the cohort and the cutoff of 1/3 percentile was selected. (b) Merge of Kaplan–Meier curves comparing PFS segregation of bITH and bTMB status in the cohort.
Discussion
In this study, we proposed that bITH, a blood-based intratumor heterogeneity metric, could serve as a predictive biomarker for clinical benefit from those receiving immune checkpoint monotherapy. We testified treatment differences in the POPLAR study and select the 1/3 percentile as the optimal cutoff point. Subsequent confirmatory analysis in the OAK cohort demonstrated significant prediction efficacy of bITH status to PFS and OS from immunotherapy. The Independent validation cohort further certificated a strong correlation between bITH status and immunotherapy-associated PFS. This study demonstrated that bITH could serve as a predictor of both OS and PFS after receiving an anti-PD-1 inhibitor.
Intratumor heterogeneity refers to the diversity among the tumor cells within a patient and has been associated with increased therapeutic resistance to multiple therapeutic modalities.24,25 At the genetic level, ITH represents the diverseness of mutation abundance. High ITH melanoma tumors had been associated with low immune infiltration levels and poor prognosis for immunotherapy, which had also been confirmed in non-small cell lung cancer.12,13 Considering non-invasive ctDNA detection had become a widespread clinical strategy especially in guiding treatment for advanced NSCLC, we developed bITH based on the integration of tissue-based ITH metrics and blood-based mutation characteristics. Mutation abundance from ctDNA which correspond to cancer cell fraction from tissue-based genomic data was obtained by scaling VAF with MSAF. This adjustment could also eliminate the prognostic effect introduced by MSAF.26 In addition, introducing weight function distinguished the clonality of mutations with different abundance.27,28 With this blood-based method, we observed similar bimodal distribution of bITH scores in both OAK and POPLAR cohorts, which indicates the naturally separated bITH-low and bITH-high status among NSCLC patients. Moreover, the 1/3 percentile lies between the two peaks in both studies suggesting the stability of the algorithm in enriching patients benefiting from immunotherapy. Utilizing plasma ctDNA source makes bITH a particularly attractive alternative for advanced NSCLC patients whose tumor tissue is otherwise unavailable.
TMB can reflect mutational burden and has been associated with efficacy outcomes for pembrolizumab (KEYNOTE-10 and -04229) or nivolumab (CheckMate-02630). However, the clinical value of TMB as a predictive biomarker remains under debate due to evidence to the contrary, such as the latest update on the BFAST trial, which did not meet its primary PFS endpoint in the bTMB ≥16 population. As Wolf et al. suggested, among those tumors with a high mutation burden, only ITH-Low tumors exhibited a better prognosis, indicating the segregation role of ITH within bTMB stratification.13 In our study, bITH-low only overlapped with bTMB-high patients in a small proportion, indicating a selection of non-overlapped ICB benefit patients. In addition, the observation that bITH-low distinguished ICB responders irrespective of bTMB status confirmed the independence of bITH status. Most importantly, bITH-low could still obtain predictive value among bTMB-low patients, who were traditionally proposed unsuitable for receiving ICI.
This study raises several questions yet to be answered, including the determination of absolute cut-off thresholds in different populations. Although identical 1/3 percentile was validated in segregating ICB-associated response, the absolute cutoff values vary in training and validation cohorts, which could be due to different gene panels and sequencing platforms. The prediction power across different cut-points should be further explored to determine the predive values of bITH in clinical practices. Also, the retrospective independent validation cohort with limited sample size (n = 42) was single-arm and treated with different anti-PD-1 monotherapy, which could bring potential bias. Still, the significant association of bITH-low and immunotherapy-related efficacy demonstrated its reliability, which awaits prospective study to further validate. Additionally, a handful of chemo-immunotherapies have been established as frontline treatment for advanced NSCLC.31 There is currently a lack of satisfactory predictive biomarker for chemoimmunotherapy. Available evidence suggests lack of predictive power for bTMB.32 Development of biomarkers is challenged by the complex mechanisms of tumor biology, and our innovative study may be biologically valuable since it exploits intratumoral heterogeneity and blood-based assays. On the other hand, a recent retrospective study of 37 patients has shown strong association between bITH dynamics and efficacy of chemoimmunotherapy.33 This study illustrates the advantage of blood-based ITH scores, which are amenable to follow-up surveillance to reflect the changes in the tumor and predict the efficacy of chemoimmunotherapy. However, due to lacking available data, these questions warrant further investigations.
ITH is widely regarded as the fuel for therapeutic resistance and tumor progression. Two ITH metrics have been proposed and associated with survival outcomes in NSCLC patients receiving ICB therapies. The challenges of using tumor tissues for gauging ITH and the unique advantages of blood biopsies make the latter a promising strategy. In this study, we developed bITH, a blood-based ITH index, and present evidence of its validity as a predictive factor for clinical response to ICB, superiority over bTMB in identifying patients with greater OS benefit, efficient PFS segregation enabled by the bITH/bTMB composite marker, and significant association with PFS in a validation cohort. This study proposed and validated a blood-based metric as a predictor for clinical response to ICB. More research is awaited to further characterize the predictive significance of bITH and to enrich its clinical applications.
Contributors
DH, YF, YL, LW, and YC conceived of and designed the study. YF, YL, LW, YC, and WC collected, analyzed, and interpreted the data. YF and DH accessed and verified the underlying data. YF, YL, LW, YC, WS, XP, and DH drafted the manuscript. XZ, BL, ZZ, S Cai, S Chuai, and YH participated in manuscript revision. XP and DH provided administrative support and supervised the project. All authors read and approved the final version of the manuscript and are accountable for all aspects of the work.
Data sharing statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declaration of interests
YC, WS, XZ, BL, ZZ, S Cai, S Chuai, and YH were employees of Burning Rock Biotech during the conduct of the study. The other authors declare no potential conflicts of interest.
Acknowledgements
We would like to thank the patients and their families for their support. We are also grateful to the staff at Burning Rock Biotech, including Guoqiang Wang, Yu Xu, Yuchen Liao, Chengcheng Li, Yan Zheng, Jinlei Song, Jianxing Xiang, and Lu Zhang for their technical assistance and valuable intellectual input.
We would like to thank the National Natural Science Foundation of China (Nos. 81972718 and 81572321), the Natural Scientific Foundation of Zhejiang Province, China (No. LY19H160007), the Science and Technology Program for Health and Medicine in Zhejiang Province, China (No. 2021KY541), the Scientific Research Project, Science and Technology Department of Sichuan Province (No. 21YYJC1616), the Scientific Research Project, Sichuan Medical Association (No. S20002), Wu Jieping Medical Foundation (No. 320.6750), and 2018 Entrepreneurial Leading Talent of Guangzhou Huangpu District and Guangzhou Development District (No. 2022-L023).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104564.
Contributor Information
Xiaojie Pan, Email: pxj1028@hotmail.com.
Dingzhi Huang, Email: dingzhih72@163.com.
Appendix A. Supplementary data
References
- 1.Reck M., Rodríguez-Abreu D., Robinson A.G., et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med. 2016;375:1823–1833. doi: 10.1056/NEJMoa1606774. [DOI] [PubMed] [Google Scholar]
- 2.Fehrenbacher L., Spira A., Ballinger M., et al. Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet. 2016;387:1837–1846. doi: 10.1016/S0140-6736(16)00587-0. [DOI] [PubMed] [Google Scholar]
- 3.Taube J.M., Klein A., Brahmer J.R., et al. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res. 2014;20:5064–5074. doi: 10.1158/1078-0432.CCR-13-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herbst R.S., Soria J.C., Kowanetz M., et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rizvi N.A., Hellmann M.D., Snyder A., et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gandara D.R., Paul S.M., Kowanetz M., et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat Med. 2018;24:1441–1448. doi: 10.1038/s41591-018-0134-3. [DOI] [PubMed] [Google Scholar]
- 7.Wang Z., Duan J., Wang G., et al. Allele frequency-adjusted blood-based tumor mutational burden as a predictor of overall survival for patients with NSCLC treated with PD-(L)1 inhibitors. J Thorac Oncol. 2020;15:556–567. doi: 10.1016/j.jtho.2019.12.001. [DOI] [PubMed] [Google Scholar]
- 8.Jamal-Hanjani M., Wilson G.A., McGranahan N., et al. Tracking the evolution of non-small-cell lung cancer. N Engl J Med. 2017;376:2109–2121. doi: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- 9.Zhang J., Fujimoto J., Zhang J., et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014;346:256–259. doi: 10.1126/science.1256930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mroz E.A., Rocco J.W. MATH, a novel measure of intratumor genetic heterogeneity, is high in poor-outcome classes of head and neck squamous cell carcinoma. Oral Oncol. 2013;49:211–215. doi: 10.1016/j.oraloncology.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Landau D.A., Carter S.L., Stojanov P., et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang W., Jin H., Zhou H., et al. Intratumoral heterogeneity as a predictive biomarker in anti-PD-(L)1 therapies for non-small cell lung cancer. Mol Cancer. 2021;20:37. doi: 10.1186/s12943-021-01331-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolf Y., Bartok O., Patkar S., et al. UVB-induced tumor heterogeneity diminishes immune response in melanoma. Cell. 2019;179:219–235.e221. doi: 10.1016/j.cell.2019.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Savas P., Teo Z.L., Lefevre C., et al. The subclonal architecture of metastatic breast cancer: results from a prospective community-based rapid autopsy program "CASCADE". PLoS Med. 2016;13 doi: 10.1371/journal.pmed.1002204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crystal A.S., Shaw A.T., Sequist L.V., et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science. 2014;346:1480–1486. doi: 10.1126/science.1254721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abbosh C., Birkbak N.J., Wilson G.A., et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature. 2017;545:446–451. doi: 10.1038/nature22364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thierry A.R., El Messaoudi S., Mollevi C., et al. Clinical utility of circulating DNA analysis for rapid detection of actionable mutations to select metastatic colorectal patients for anti-EGFR treatment. Ann Oncol. 2017;28:2149–2159. doi: 10.1093/annonc/mdx330. [DOI] [PubMed] [Google Scholar]
- 18.Herbst R.S., Giaccone G., de Marinis F., et al. Atezolizumab for first-line treatment of PD-L1-selected patients with NSCLC. N Engl J Med. 2020;383:1328–1339. doi: 10.1056/NEJMoa1917346. [DOI] [PubMed] [Google Scholar]
- 19.Kim E.S., Velcheti V., Mekhail T., et al. Blood-based tumor mutational burden as a biomarker for atezolizumab in non-small cell lung cancer: the phase 2 B-F1RST trial. Nat Med. 2022;28:939–945. doi: 10.1038/s41591-022-01754-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rittmeyer A., Barlesi F., Waterkamp D., et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet. 2017;389:255–265. doi: 10.1016/S0140-6736(16)32517-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X., Fang L., Zhu Y., et al. Blood tumor mutation burden can predict the clinical response to immune checkpoint inhibitors in advanced non-small cell lung cancer patients. Cancer Immunol Immunother. 2021;70:3513. doi: 10.1007/s00262-021-02943-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oh B.Y., Shin H.-T., Yun J.W., et al. Intratumor heterogeneity inferred from targeted deep sequencing as a prognostic indicator. Sci Rep. 2019;9:4542. doi: 10.1038/s41598-019-41098-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramagopalan S., Gupta A., Arora P., Thorlund K., Ray J., Subbiah V. Comparative effectiveness of atezolizumab, nivolumab, and docetaxel in patients with previously treated non-small cell lung cancer. JAMA Netw Open. 2021;4 doi: 10.1001/jamanetworkopen.2021.34299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalbasi A., Ribas A. Tumour-intrinsic resistance to immune checkpoint blockade. Nat Rev Immunol. 2020;20:25–39. doi: 10.1038/s41577-019-0218-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dagogo-Jack I., Shaw A.T. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol. 2018;15:81–94. doi: 10.1038/nrclinonc.2017.166. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Q., Luo J., Wu S., et al. Prognostic and predictive impact of circulating tumor DNA in patients with advanced cancers treated with immune checkpoint blockade. Cancer Discov. 2020;10:1842–1853. doi: 10.1158/2159-8290.CD-20-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guiaşu S. Weighted entropy. Rep Math Phys. 1971;2:165–179. [Google Scholar]
- 28.Gerstung M., Jolly C., Leshchiner I., et al. The evolutionary history of 2658 cancers. Nature. 2020;578:122–128. doi: 10.1038/s41586-019-1907-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mok T.S.K., Wu Y.-L., Kudaba I., et al. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): a randomised, open-label, controlled, phase 3 trial. Lancet. 2019;393:1819–1830. doi: 10.1016/S0140-6736(18)32409-7. [DOI] [PubMed] [Google Scholar]
- 30.Carbone D.P., Reck M., Paz-Ares L., et al. First-line nivolumab in stage IV or recurrent non-small-cell lung cancer. N Engl J Med. 2017;376:2415–2426. doi: 10.1056/NEJMoa1613493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thai A.A., Solomon B.J., Sequist L.V., Gainor J.F., Heist R.S. Lung cancer. Lancet. 2021;398:535–554. doi: 10.1016/S0140-6736(21)00312-3. [DOI] [PubMed] [Google Scholar]
- 32.Jiang T., Chen J., Xu X., et al. On-treatment blood TMB as predictors for camrelizumab plus chemotherapy in advanced lung squamous cell carcinoma: biomarker analysis of a phase III trial. Mol Cancer. 2022;21:4. doi: 10.1186/s12943-021-01479-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou J., Bao M., Gao G., et al. Increased blood-based intratumor heterogeneity (bITH) is associated with unfavorable outcomes of immune checkpoint inhibitors plus chemotherapy in non-small cell lung cancer. BMC Med. 2022;20:256. doi: 10.1186/s12916-022-02444-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.