Abstract
STUDY QUESTION
Can whole exome sequencing (WES) followed by trio bioinformatics analysis identify novel pathogenic genetic causes of first trimester euploid miscarriage?
SUMMARY ANSWER
We identified genetic variants in six candidate genes that indicated plausible underlying causes of first-trimester euploid miscarriage.
WHAT IS KNOWN ALREADY
Previous studies have identified several monogenic causes of Mendelian inheritance in euploid miscarriages. However, most of these studies are without trio analyses and lack cellular and animal models to validate the functional effect of putative pathogenic variants.
STUDY DESIGN, SIZE, DURATION
Eight unexplained recurrent miscarriage (URM) couples and corresponding euploid miscarriages were included in our study for whole genome sequencing (WGS) and WES followed by trio bioinformatics analysis. Knock-in mice with Rry2 and Plxnb2 variants and immortalized human trophoblasts were utilized for functional study. Additional 113 unexplained miscarriages were included to identify the mutation prevalence of specific genes by multiplex PCR.
PARTICIPANTS/MATERIALS, SETTING, METHODS
Whole blood from URM couples and their <13 weeks gestation miscarriage products were both collected for WES, and all variants in selected genes were verified by Sanger sequencing. Different stage C57BL/6J wild-type mouse embryos were collected for immunofluorescence. Ryr2N1552S/+, Ryr2R137W/+, Plxnb2D1577E/+, and Plxnb2R465Q/+ point mutation mice were generated and backcrossed. Matrigel-coated transwell invasion assays and wound-healing assays were performed using HTR-8/SVneo cells transfected with PLXNB2 small-interfering RNA and negative control. Multiplex PCR was performed focusing on RYR2 and PLXNB2.
MAIN RESULTS AND THE ROLE OF CHANCE
Six novel candidate genes, including ATP2A2, NAP1L1, RYR2, NRK, PLXNB2, and SSPO, were identified. Immunofluorescence staining showed that ATP2A2, NAP1L1, RyR2, and PLXNB2 were widely expressed from the zygote to the blastocyst stage in mouse embryos. Although compound heterozygous mice with Rry2 and Plxnb2 variants did not show embryonic lethality, the number of pups per litter was significantly reduced when backcrossing Ryr2N1552S/+ ♂ with Ryr2R137W/+ ♀ or Plxnb2D1577E/+ ♂ with Plxnb2R465Q/+ ♀ (P < 0.05), which were in accordance with the sequencing results of Family 2 and Family 3, and the proportion of Ryr2N1552S/+ offspring was significantly lower when Ryr2N1552S/+ female mice were backcrossed with Ryr2R137W/+ male mice (P < 0.05). Moreover, siRNA-mediated PLXNB2 knockdown inhibited the migratory and invasive abilities of immortalized human trophoblasts. Besides, additional 10 variants of RYR2 and PLXNB2 were detected in 113 unexplained euploid miscarriages by multiplex PCR.
LIMITATIONS, REASONS FOR CAUTION
The relatively small number of samples is a limitation of our study which may result in the identification of variants in unique candidate genes with no definitive although plausible causal effect. Larger cohorts are needed to replicate these findings and additional functional research is needed to confirm the pathogenic effects of these variants. Moreover, the sequencing coverage restricted the detection of low-level parental mosaic variants.
WIDER IMPLICATIONS OF THE FINDINGS
For first-trimester euploid miscarriage, variants in unique genes may be underlying genetic etiologies and WES on trio could be an ideal model to identify potential genetic causes, which could facilitate individualized precise diagnostic and therapeutic regimens in the future.
STUDY FUNDING/COMPETING INTERESTS
This study was supported by grants from the National Key Research and Development Program of China (2021YFC2700604), National Natural Science Foundation of China (31900492, 82101784, 82171648), Basic Science Center Program of the National Natural Science Foundation of China (31988101), Key Research and Development Program of Shandong Province (2021LCZX02), Natural Science Foundation of Shandong Province (ZR2020QH051), Natural Science Foundation of Jiangsu Province (BK20200223), Taishan Scholars Program for Young Experts of Shandong Province (tsqn201812154) and Young Scholars Program of Shandong University. The authors declare no conflicts of interest.
TRIAL REGISTRATION NUMBER
N/A.
Keywords: candidate gene, euploid, miscarriage, single-nucleotide variant, whole exome sequencing
Introduction
Miscarriage is a common distressing pregnancy disorder that affects 15–25% of pregnant women (Practice Committee of the American Society for Reproductive Medicine, 2012). In clinical practice, ∼80% of miscarriages occur within the first trimester of pregnancy (Sarah et al., 2018). Although most miscarriages are sporadic, 1–5% of couples suffer from two or more consecutive miscarriages, a scenario that is diagnosed as recurrent miscarriage (RM) (Practice Committee of the American Society for Reproductive Medicine, 2020; Quenby et al., 2021). Multiple factors, including embryo aneuploidy, uterine anatomic abnormalities, autoimmune disorders, thrombophilia, endocrine disorders, and infection, have been causatively linked to RM. However, nearly 50% of RM lack a clear etiology; these are termed as unexplained RM (URM) (Kaandorp et al., 2010). Data supporting familial predisposition in RM pedigrees highlight the contribution of genetic factors to pregnancy loss (Christiansen et al., 1990; Kolte et al., 2011), and 60% of miscarriages are associated with chromosomal abnormalities of embryos (Kolte et al., 2015; Qiao et al., 2016). However, whether additional genetic factors, such as genetic variants, are responsible for URM still needs further investigation (Zhao et al., 2021).
Next-generation sequencing (NGS), including whole genome sequencing (WGS) and whole exome sequencing (WES), has emerged as an efficient approach for screening potential pathogenetic variants at the single-nucleotide level. For RM patients, preimplantation genetic testing for aneuploidy (PGT-A) has been utilized as a therapeutic intervention, since PGT-A can preclude the transfer of mosaic/aneuploid embryos for transfer, thereby avoiding miscarriages caused by embryos with chromosome abnormalities (Bhatt et al., 2021; Eggenhuizen et al., 2021). However, miscarriages still occur in some RM patients who underwent PGT-A treatment, highlighting the necessity of revealing unknown embryonic lethal copy number variants or single-nucleotide variants (SNVs) that may be causative factors of euploid miscarriages. A deeper understanding of the genetic causes of euploid miscarriages could help advance diagnostic and therapeutic strategies for URM clinical management.
WES is widely utilized to detect pathogenetic mutations located within exons, and its application in genetic determinants of diseases could be very helpful, especially considering that ∼85% of human genetic diseases occur in the exons of genes. Recent WES studies have identified several novel genetic causes for euploid miscarriages (Qiao et al., 2016; Quintero-Ronderos et al., 2017; Zhao et al., 2021), including compound heterozygous mutations of DYNC2H1 and ALOX15 (Qiao et al., 2016). Moreover, mutations in genes essential for development, such as KIF14, CEP55, STIL, FOXP3, GLE, RYR1, and POMT1, have been identified as novel causative genetic factors of second trimester miscarriages by sequencing several different combinations of RM specimens (Rajcan-Separovic, 2020). Although previous studies have identified candidate genetic causes of euploid miscarriages, the interpretation of these findings was primarily based on WES results in miscarriage products per se, lacking evidence from RM couples to identify the hereditary feature of SNVs. Genetic sequencing of both RM couples and their euploid miscarriages followed by trio bioinformatics analysis could effectively benefit the identification of genetic causes (Yates et al., 2017). Even though it would be ideal to sequence DNA samples from both partners in RM couples and all their miscarriages, in practice, it is more feasible to collect and sequence DNA from both partners and one of their corresponding miscarriage tissues (Rajcan-Separovic, 2020).
In the present study, we collected blood samples from eight URM couples and their euploid miscarriage products after PGT-A treatment. Using WGS and WES sequencing data of DNA prepared from these URM couples and miscarriages for trio analysis, we aimed to identify novel pathogenic genes and variants that are associated with miscarriages with euploid embryos. To verify the phenotypes of putatively pathogenic variants, we also generated knock-in mice to examine miscarriage and development-related phenotypes. Furthermore, we screened other variants in candidate genes in euploid miscarriages by multiplex PCR, which could provide further evidence supporting the causative role of mutations of these genes in euploid miscarriage. A better understanding of the genetic causes of euploid RM could promote the development of individualized, precise diagnostic and therapeutic regimens for URM couples.
Materials and methods
Subjects
URM couples with normal karyotypes, aged under 36 years old, who experienced more than two historical consecutive miscarriages and underwent PGT-A treatments followed by subsequent miscarriages at the Reproductive Hospital affiliated to Shandong University from January 2014 to December 2017 were included. All of the transferred embryos were euploid, which was verified by both PGT-A and genetic testing of miscarriage products based on Affymetrix microarrays. The exclusion criteria were as follows to eliminate interference from factors known to cause miscarriage: (i) female patients >36 years old; (ii) patients diagnosed with uterine cavity abnormalities such as congenital uterine anomalies, endometrial polyps, or adenomyosis; (iii) patients with chromosome abnormalities; (iv) females diagnosed with thrombophilia, diabetes, hypothyroidism, or autoimmune disease; (v) patients diagnosed with infectious disease (bacterial vaginosis, syphilis, human cytomegalovirus, parvovirus B19, etc.) within three months; and (vi) endometrial thickness <0.7 cm under the transplantation cycle. Finally, eight couples and their miscarriage products after PGT-A treatment were included for WGS, WES, and trio bioinformatics analysis. Whole blood from URM couples (anticoagulated by EDTA) and their <13 weeks of gestation miscarriage products previously assessed by PGT-A (based on the low-coverage WGS with a depth <1×) were both collected.
To test the carriage rate of putatively pathogenic variants in unexplained first trimester miscarriages, 1261 first trimester villi that had already undergone genetic tests based on the MiSeq or NextSeq 500 platform at the same hospital from September 2018 to September 2021 were screened. Aneuploids and mosaics, miscarriages whose parents had abnormal or unknown karyotypes, novel maternal risks (such as uterine cavity abnormalities, thrombophilia, thyroid dysfunction, and autoimmune disease), or without available clinical tests for maternal risks were excluded. Finally, 113 unexplained miscarriage first trimester villi specimens were successfully included for further analysis using multiplex PCR.
This study was approved by the Ethics Committee of the Reproductive Medicine Center of Shandong University, and informed consent was obtained from all patients.
WGS and data analysis
For WGS and WES, genomic DNA was extracted from peripheral blood and miscarriage product samples using MagPure Tissue & Blood DNA LQ Kits (Magen, Beijing, China) following the manufacturer's instructions. The ends of the DNA fragments were repaired, and Illumina Adaptor was added (Fast Library Prep Kit, iGeneTech, Beijing, China). After the sequencing library was constructed, paired-end sequencing (150 bp) was performed on the Illumina NovaSeq 6000 platform (Illumina, San Diego, CA, USA). The read depth is estimated to be 1×, and we finally obtained ∼3 GB of data per individual. Quality control and trimming of raw data were processed by FastQC software and Trimmomatic software (Bolger et al., 2014). Then, clean reads were mapped to the reference genome GRCh37 using Burrows–Wheeler Aligner (BWA) (Li and Durbin, 2009). CNV calling was performed using low-coverage massively parallel CNV sequencing (Liang et al., 2014). CNVs were selected if they were detected only in miscarriages but not in parents (i.e. overlap ratio <50%) and their population maximum allele frequencies in gnomAD database (GD_POPMAX_AF) were below 0.01. Selected CNVs were further annotated using AnnotSV (Geoffroy et al., 2018) and CNVs annotated as ‘likely pathogenic’ or ‘pathogenic’ were retained for subsequent analysis.
WES and variant prioritization
Using the sequencing library built during WGS, whole exons were captured with an AIExome Enrichment Kit V1 and sequenced on an Illumina NovaSeq6000 platform (Illumina, San Diego, CA, USA) in 150-bp paired‐end mode. We finally obtained ∼12 GB of data per individual, with over 97% of the exome covered by more than 20 reads. Quality control and trimming of raw data were processed by FastQC software and Trimmomatic software (Bolger et al., 2014). Then, clean reads were mapped to the reference genome GRCh37 by using BWA (Li and Durbin, 2009). After removing duplications, the Genome Analysis Toolkit (GATK) (DePristo et al., 2011) was used for variant calling and called variants were annotated using Annovar (Wang et al., 2010) and snpEff (Cingolani et al., 2012). Variant filtering and prioritization were performed in multiple steps based on predicted variant impact, inheritance pattern, and allele frequency; briefly, candidate pathogenic variants were prioritized based on (i) variant type, including missense, nonsense on protein, frameshift, and splice-site variants; (ii) variant impact predicted to be protein damaging or pathogenic by at least one of eight tools, including SIFT, Polyphen2, LRT, Mutation-Taster, MutationAssessor, FATHMM, fathmm-MKL, and/or Combined Annotation Dependent Depletion (CADD) (Ng and Henikoff, 2001; Chun and Fay, 2009; Schwarz et al., 2010; Adzhubei et al., 2013; Gnad et al., 2013; Kircher et al., 2014; Shihab et al., 2013, 2015); (iii) inheritance pattern including de novo, autosomal recessive, X-linked, or compound heterozygous inheritance models; (iv) minor allele frequency (MAF) in the East Asian population of gnomAD, 1000 genomes, and ExAC database <0.01 de novo mutations and MAF <0.05 for variants in other inheritance patterns; (v) number of homozygous East Asian individuals ≤3 in the gnomAD exome database; and (vi) finally, since the equivalent age of mice and humans cannot be based on the percent of elapsed pregnancy (Otis and Brent, 1954), genes harboring candidate variants were manually reviewed for their functional relevance to embryo development or embryonic lethality during whole gestation based on literature and databases, including PubMed, OMIM, ClinVar, human gene mutation data (HGMD), GeneCards, and UniProtKB. Candidate pathogenic variants in relevant genes were confirmed using Sanger sequencing of the miscarriage products and their parents. Furthermore, the remaining variants were classified according to the American College of Medical Genetics and Genomics/Association for Molecular Pathology (ACMG/AMP) guidelines (Richards et al., 2015).
Sanger sequencing
The sequenced data were aligned to the reference genome (GRCh37). Primers flanking the SNVs were created using MFEprimer-3.0 software (Wang et al., 2019) for the variants in selected genes by WES. All the primers and PCR conditions are available on request. The amplicons were purified and sequenced using an ABI 3730XL Sequencer (Applied Biosystems, Foster City, CA, United States). The primer sequences of the remaining SNVs in six genes are shown in Supplementary Table SI.
Variant mapping and evolutionary conservation analysis
Variant mapping and all structure figures were generated by PyMOL. Variant mapping was conducted utilizing the published porcine RyR2 (PDBID: 5GOA) and human SERCA2 (5ZTF) and the structures from AlphaFold for PLXNB2, NRK, and NAP1L1, whose structures were resolved. Evolutionary conservation analysis was performed with Clustal Omega and Jalview software.
Embryo immunofluorescence microscopy
The mouse embryos were washed in washing buffer, phosphate-buffered saline (PBS) containing 0.01% Triton X-100 (v/v) and 0.1% Tween 20 (v/v), and then fixed in 4% paraformaldehyde/PBS (w/v) for 30–40 min. Fixed embryos were next permeabilized with 1% Triton X-100/PBS (v/v) for 40 minutes, except embryos for PLXNB2 staining. After blocking in 1% bovine serum albumin (BSA)/PBS (w/v) for 1 h, the embryos were incubated with primary antibodies against ATP2A2 (Abcam, ab150435, 1:100), RyR2 (19765-1-AP, Proteintech, 1:100), NAP1L1 (14898-1-AP, Proteintech, 1:100), and PLXNB2 (10602-1-AP, Proteintech, 1:100) diluted in 1% BSA/PBS for 1 h. Thereafter, the embryos were stained in FITC-conjugated secondary antibody (A11008, Invitrogen, USA) at a 1:800 dilution and TRITC-phalloidin (R415, Invitrogen, USA) at a 1:400 dilution for 45 min and washed in washing buffer three times. Finally, the nuclei were stained with DAPI (p36935, Invitrogen, USA) for 10 min. All steps were performed at room temperature. Images were captured with a Dragonfly spinning disk confocal microscope (Andor Technology Ltd, North Ireland, UK) and processed using Imaris. The same experiment was independently repeated three times, and each developmental stage was stained for >30 embryos. Details of the in vitro fertilization and embryo culture protocols are reported in the Supplementary Materials and methods.
Generation of point mutant mice and validation
Ryr2N1552S/+, Ryr2R137W/+, Plxnb2D1577E/+, and Plxnb2R465Q/+ knock-in mice were constructed by the CRISPR/Cas9 technique (Cyagen, Suzhou, China). The gRNAs, donor oligos containing each mutant, to Plxnb2 (p.D1577E and p.R465Q) or Ryr2 (p.N1552S and p.R137W) gene, and Cas9 was coinjected into fertilized mouse eggs to generate targeted knock-in offspring. Genotyping was performed by PCR from genomic DNA of mouse tails followed by Sanger sequencing. The method is shown in Supplementary Fig. S1, and the primers are shown in Supplementary Table SII. All genetically altered mice had a background of C57BL/6J. WT mice for experiments were purchased from the Beijing Vital River Laboratory Animal Technology Company. Mice were housed at 22 ± 2°C controlled room temperature with a 12/12 h light/dark cycle and free access to water and food. All animal experiments were performed in accordance with the ethical guidelines approved by the Animal Care and Research Committee of Shandong University. From the F2 generation, Ryr2N1552S/+ and Ryr2R137W/+ mice were mated to each other and WT mice, and the offspring genotypes were validated by sequencing using corresponding primers. The Plxnb2D1577E/+ and Plxnb2R465Q/+ point mutant mice were mated in the same process. The detailed methods of reproductive performance and fertility testing are recorded in the Supplementary Materials and methods.
Matrigel-coated transwell invasion assay
Cell invasion was assessed using Matrigel-based transwell inserts and HTR-8/SVneo cells were transfected with small-interfering RNA (see Supplementary Materials and methods). The lower chamber (24-well plate) was filled with 750 μl of DMEM/F12 medium with 10% FBS. Approximately 8 × 104 of HTR-8/SVneo cells in 250 μl DMEM/F12 medium with 0.1% FBS were placed into the upper transwell compartment. The upper chamber membrane was precoated with 50 μl of Matrigel (Corning, 354230, USA) at 37°C overnight. The nonmigrated cells were removed from the upper chambers with a cotton-tipped swabs after 36 h of incubation. Cells that invaded across the membrane were fixed with cold methanol, and nuclei were then stained with Hoechst 33258 (Sigma-Aldrich, 94403, USA). Finally, the average number of invaded cells was counted in five randomly selected fields per insert captured by Olympus B53 fluorescence microscopy at a magnification of 100×.
Wound-healing assay
HTR-8/SVneo cells were seeded in a six-well plate and cultured under a humidified atmosphere of 5% CO2 at 37°C. The monolayers were then scratched vertically using a 200-µl sterile pipette tip, and any floating cells were washed off with PBS. Subsequently, the cells were photographed after 12, 24, 36, and 48 h at a magnification of 40×. The cell migration area was measured using ImageJ software. For further details on HTR-8/SVneo cell culture and viability assessments, please refer to Supplementary Materials and methods.
Multiplex PCR
Genomic DNA was extracted from villi of 113 unexplained first trimester miscarriages using a TIANamp Genomic DNA Kit (Tiangen, DP304, China). Then, sequencing libraries were generated using MultipSeqCustom Panel (iGeneTech, Beijing, China) following standard procedures, and amplified enriched DNA was subjected to NGS on the Illumina HiSeq2500 platform. Raw data were processed similarly to WGS. We only considered nonsynonymous variants in exonic regions that were absent or MAF < 0.01 in the East Asian population of gnomAD v2.1.1, 1000 genome, and ExAC database as well as predicted to be damaging in at least one computational tool, including SIFT, PolyPhen-2, MutationTaster2, and CADD (score >15).
Statistical analysis
Data are expressed as the mean ± SD of at least three independent experiments. A Chi-squared test or Fisher’s exact test was applied to compare qualitative variables and genotype/allele frequencies. For quantitative variables, statistical significance was determined using an unpaired t-test or one-way ANOVA. The Mann–Whitney U test or Kruskal–Wallis test was performed to analyze nonnormally distributed variables. All statistical tests were two-tailed. GraphPad Prism was employed for statistical analysis. Means were considered significantly different if P < 0.05, and columns without letters in common are significantly different.
Results
Specimen characteristics of eight URM families
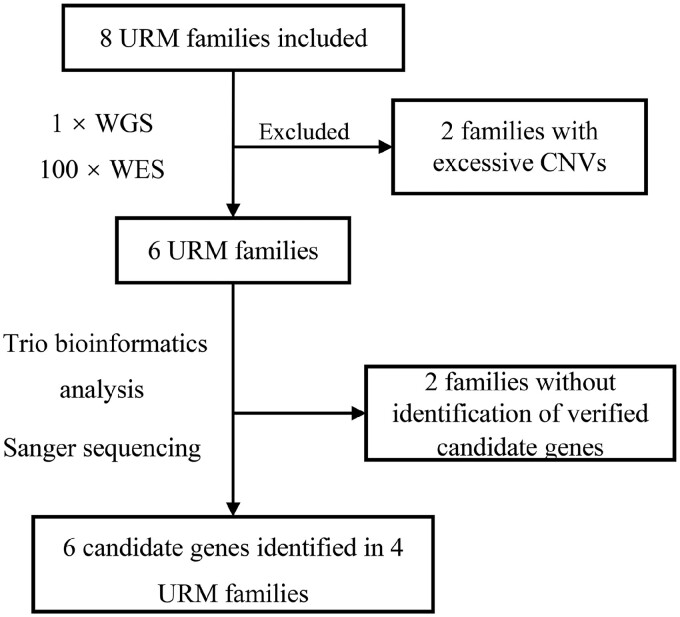
A total of eight URM couples and their euploid miscarriages were included for NGS. All female patients were under 36 years old, and miscarriages with previous PGT-A assessments that excluded aneuploid embryos were obtained between 8 and 12 weeks of gestation. WES and low-coverage WGS were performed for all the specimens. Two families were excluded for excessive CNVs in miscarriages based on WGS data. The remaining six families did not have pathogenic CNVs related to miscarriages and thus were included in the trio-WES analysis (Supplementary Table SIII). Nine variants in six candidate genes from four families were verified successfully by Sanger sequencing. The flowchart and baseline characteristics of these four families are shown in Fig. 1 and Table I.
Figure 1.
Flowchart for identifying putatively pathogenic variants and candidate genes by WGS and WES in URM families. URM, unexplained recurrent miscarriage; WES, whole exome sequencing; WGS, whole genome sequencing.
Table I.
Clinical characteristics of unexplained recurrent miscarriage couples with verified candidate genes.
| Family ID | Family 1 | Family 2 | Family 3 | Family 4 |
|---|---|---|---|---|
| Female age (years) | 27 | 32 | 28 | 26 |
| Gravidity | 5 | 6 | 4 | 4 |
| Parity | 0 | 0 | 0 | 1 |
| History of previous miscarriages | 5 | 6 | 4 | 3 |
| Gestational age (wk+days) | 9+3 | 8+3 | 9+1 | 12 |
| Female BMI | 29.69 | 29.30 | 25.12 | 21.09 |
| Cycle length | 40∼50d | 28∼30d | 30∼50d | 28∼30d |
| Female karyotype | 46, XX | 46, XX | 46, XX | 46, XX |
| Morphology of uterus | Normal | Normal | Normal | Normal |
| Endometrial thickness (cm) | 0.85 | 0.75 | 0.75 | 0.8 |
| Autoimmune disease | (–) | (–) | (–) | (–) |
| Infection | (–) | (–) | (–) | (–) |
| Male age (years) | 35 | 34 | 30 | 35 |
| Male diagnosis | Teratozoospermia | Asthenozoospermia | Oligozoospermia | Teratozoospermia |
| Male karyotype | 46, XY | 46, XY | 46, XY | 46, XY |
| Villi karyotype | Euploid | Euploid | Euploid | Euploid |
WES identified novel candidate genes for euploid miscarriages
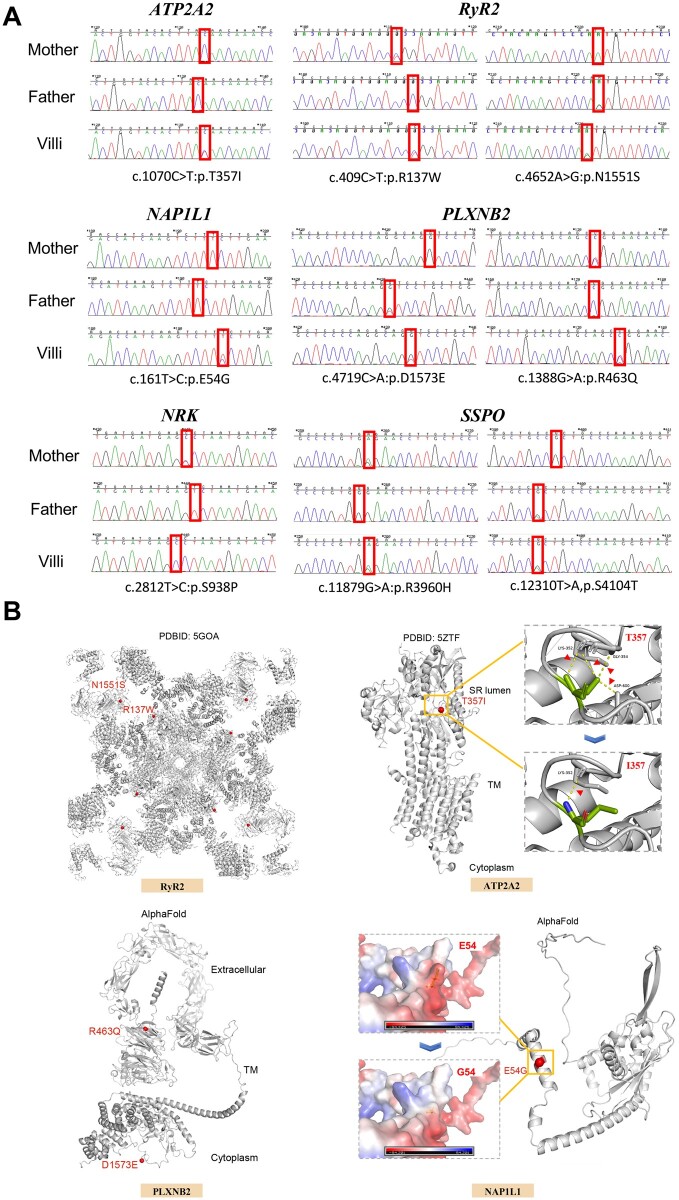
The identified variants in six novel candidate genes are shown in Table II, including two de novo heterozygous mutations in ATP2A2 (MIM 108740) from Family 1 and NAP1L1 (MIM 164060) from Family 2, three compound heterozygous mutations in RYR2 (MIM 180902), PLXNB2 (MIM 604293), and SSPO (MIM 617356) from Family 2–4, and one X-linked mutation in NRK (MIM 300791) from Family 2. These candidate genes are further described below. The results of Sanger sequencing along with the structural analysis of candidate genes are shown in Fig. 2, and the evolutionary conservation is shown in Supplementary Fig. S2. For de novo heterozygous mutations, the read depth of the mutated allele in each parent was manually checked to detect parental mosaicism. We found that the read depth of the mutated allele in each parent is ≤1, suggesting that the identified mutations in miscarriages are more likely to be de novo mutations rather than a consequence of low-level mosaicism in parents (Supplementary Table SIV) (Acuna-Hidalgo et al., 2015).
Table II.
Putative pathogenic variants in candidate genes identified in unexplained recurrent miscarriage families screened by whole exome sequencing.
| Family ID | Gene | Child | Mother | Father | Position | Reference sequences | cDNA change | AAChange | Variant ID | ACMG | Inheritance |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Family 1 | ATP2A2 | 0/1 | 0/0 | 0/0 | Chr12:110765797 | NM_001681.3 | c.1070C>T | p.T357I | – | VUS | De novo |
| Family 2 | RYR2 | 0/1 | 0/1 | 0/0 | Chr1:237538041 | NM_001035.2 | c.409C>T | p.R137W | rs761916230 | VUS | Com. het |
| 0/1 | 0/0 | 0/1 | Chr1:237765380 | c.4652A>G | p.N1551S | rs185237690 | VUS | ||||
| NRK | 1/1 | 0/1 | 0/. | ChrX:105167311 | NM_198465.2 | c.2812T>C | p.S938P | rs758879022 | VUS | X-linked | |
| NAP1L1 | 0/1 | 0/0 | 0/0 | Chr12:76461196 | NM_004537.4 | c.161A>G | p.E54G | – | VUS | De novo | |
| Family 3 | PLXNB2 | 0/1 | 0/0 | 0/1 | Chr22:50716877 | NM_012401.3 | c.4719C>A | p.D1573E | – | VUS | Com. Het |
| 0/1 | 0/1 | 0/0 | Chr22:50726459 | c.1388G>A | p.R463Q | rs192965378 | VUS | ||||
| Family 4 | SSPO | 0/1 | 0/1 | 0/0 | Chr7:149516462 | NM_198455.2 | c.11879G>A | p.R3960H | – | VUS | Com. Het |
| 0/1 | 0/0 | 0/1 | Chr7:149517589 | c.12310T>A | p.S4104T | rs753616243 | VUS |
AA, amino acid; ACMG, American College of Medical Genetics and Genomics; Com. het, compound heterozygous; Chr, Chromosome; VUS, variant of uncertain significance.
Figure 2.
Sanger sequencing of putatively pathogenic variants in six candidate genes and corresponding structural mapping. (A) Sanger sequencing validated the existence of putatively pathogenic genetic variants in six genes from four families. (B) Structures of RyR2, ATP2A2, PLXNB2, and NAP1L1 are shown. Structural mapping of the mutation T357I onto the structure of ATP2A2. T357 forms four hydrogen bonds with K352, G354, and D600, whereas only one is left after changing to I357. The variant p.N1551S (located in the SPRY3) and p.R137W (located in the NTD) are indicated on the structure of RyR2. Structural mapping of the mutation E54G onto the structure of NAP1L1, and this mutation changes a negatively charged residue to a non-charged residue. Structural mapping of the variant p.R463Q and p.D1573E onto the structure of PLXNB2. R463Q and D1573E are located on the extracellular and intracellular sides, respectively. The reference structures were downloaded from the PDB and AlphaFold protein structure databases. All the figures were prepared using PyMOL.
ATP2A2: ATP2A2 encodes sarco (endo) plasmic reticulum Ca2+ ATPase (SERCA2) with two isoforms, muscle-specific SERCA2a and ubiquitous SERCA2b (Periasamy and Kalyanasundaram, 2007), and functions to reuptake Ca2+ into the sarco/endoplasmic reticulum through hydrolysis of ATP. SERCA2 knockout zebrafish died at 6 days postfertilization, and targeted mutation is embryonic lethal (Shull et al., 2003; Ebert et al., 2005). Heterozygous mutants of ATP2A2 might also impair cardiac contractility and relaxation (Periasamy et al., 1999). We detected a heterozygous de novo SNV of ATP2A2 (p.T357I) in Family 1. This change in amino acids induced hydrogen bond reduction (Fig. 2B).
RYR2: RYR2 encodes one of the three subtypes of ryanodine receptors, which are L-type voltage-dependent Ca2+ channels in the sarcoplasmic reticulum. RyR2 is enriched in the myocardium to maintain the pivotal excitation–contraction coupling of cardiomyocytes through the calcium-induced calcium release process and is necessary for cardiac development (Takeshima et al., 1998; Priori and Napolitano, 2005). RYR2 mutations are pathogenic for catecholaminergic polymorphic ventricular tachycardia and atrial fibrillation (Kushnir et al., 2018). Ryr2−/− mice exhibit embryonic lethality (Takeshima et al., 1998). The two variants in RYR2 (p.R137W and p.N1551S) we identified are both located in the hotspot mutation regions, the N-terminal domain, which is the hotspot mutation region (Peng et al., 2016).
NAP1L1: NAP1L1, a member of the NAP1 gene family, is expressed ubiquitously in all tissues and mainly participates in nucleosome assembly and cell proliferation that are attributable to regulation (Okuwaki et al., 2010; Yan et al., 2016). Nap1l1 knockout mice showed deficiencies in embryonic neurogenesis and decreased birth rate (Qiao et al., 2018). The de novo variants c.161A>G, p.E54G from Family 2 were predicted to change the electric potential from positive to moderate in the patch (Fig. 2B).
PLXNB2: PLXNB2 belongs to a transmembrane family of plexin proteins that are widely expressed in various mammalian organs. PLXNB2 has a unique role in the development of cerebellar granule cells. In mice, it is expressed from embryo to adult (Perälä et al., 2005) and plays an essential role in both central and peripheral neural system development, involving neuronal migration, synaptogenesis, and axonal guidance regeneration (Saha et al., 2012; Van Battum et al., 2021), or as a receptor of angiogenin (Yu et al., 2017). Most Plxnb2−/− mouse embryos, especially in the C57BL/6 strain, show cephalic neural tube closure defects, leading to perinatal lethality or abnormal nervous system development (Deng et al., 2007; Van Battum et al., 2021). In Family 3, the euploid miscarriage contained a compound heterozygous mutation in PLXNB2 (p.D1573E and p.R463Q).
NRK: As our sequencing results showed, the embryo from Family 2 was male, and we detected an SNV (rs758879022) located in exon 18 of NRK, which is an X-linked gene. NRK has been suggested to participate in the developmental process of the spongiotrophoblast layer, a region derived from the fetus whose loss and expansion could cause embryonic lethality, overgrowth of the fetus, or abnormal delivery (Denda et al., 2011), potentially by regulating placental cell proliferation by modulating AKT signaling (Lestari et al., 2022).
SSPO: SSPO, a high-molecular-weight glycoprotein, is required for central nervous system development, especially in aggregation to form Reissner fiber, which impairs the body axis during embryonic development (Lu et al., 2020; Rose et al., 2020). Mutation of SSPO is associated with the etiology of human idiopathic scoliosis (Rose et al., 2020). Homozygous sspo mutant zebrafish died around 10 days postfertilization (Cantaut-Belarif et al., 2018). Our results showed that in Family 4, the euploid miscarriage carried a compound heterozygous mutation in SSPO from its parents, carrying two SNVs, p.R3960H and p.S4104T, respectively. Of note, similar to the variants in NRK, p.S4104T of SSPO was not conserved in mammals.
The expression patterns of the identified candidate genes in early mouse embryos
To elucidate the expression patterns of these proteins encoded by genes we identified during early embryo development, we performed immunofluorescence staining in mouse preimplantation embryos to examine the expression of ATP2A2, RyR2, NAP1L1, and PLXNB2, but not SSPO due to a lack of available efficient antibody. As shown in Supplementary Fig. S3, ATP2A2 and RyR2 were primarily localized to the cytoplasm, and PLXNB2 was mainly localized to the cell membrane. Our data show that the expression level of NAP1L1 was higher in the cytoplasm than in the nucleus during the early embryonic stage, which is consistent with previous findings in neural progenitor cells (Qiao et al., 2018). Each protein was continuously expressed from the zygote to the blastula stage in mouse embryos, and we did not observe expression level differences between the inner cell mass and trophectoderm.
Genotypes of offspring from crosses of heterozygous point mutant mouse parents matched Mendelian inheritance
To further evaluate the potential causative roles of variants located in RYR2 and PLXNB2 in euploid miscarriage, we generated four Ryr2 and Plxnb2 point mutant knock-in mice (Supplementary Fig. S1). The offspring of backcrossing Ryr2N1552S/+ with Ryr2R137W/+ parents and Plxnb2D1577E/+ with Plxnb2R465Q/+ parents followed Mendelian heredity (Table III) and showed normal morphology and weight at six weeks after birth, which indicates that the Plxnb2D1577E/R465Q and Ryr2N1552S/R137W mutations did not induce embryonic lethality in mice under regular conditions without an environmental burden (Supplementary Fig. S4). Interestingly, when Ryr2N1552S/+ female mice were backcrossed with Ryr2R137W/+ male mice, the birth rate of Ryr2N1552S/+ offspring was significantly lower (10% vs 25%, P = 0.02, n = 5, Table III). Although the underlying mechanisms still need to be further revealed, our findings suggest that the concurrent presence of these two variants might exert potential roles in regulating maternal endometrial condition or fetal Ryr2N1552S/+ mouse development.
Table III.
Genotype distribution of offspring from heterozygous Ryr2 and Plxnb2 knock-in mouse parents.
| Genotype distribution of total offspring from Ryr2R137W/+ mice backcross with Ryr2N1552S/+ mice | ||||
|---|---|---|---|---|
| +/+ | N1552S/+ | R137W/+ | N1552S/R137W | |
| ♂ | 16 | 15 | 21 | 21 |
| ♀ | 23 | 15 | 23 | 25 |
| Sum | 39 | 30 | 44 | 46 |
| Ratio | 24.5% | 18.9% | 27.7% | 28.9% |
|
| ||||
| Genotype of parents | Ryr2R137W/+♂ × Ryr2N1552S/+ ♀ | |||
|
| ||||
| +/+ | N1552S/+ | R137W/+ | N1552S/R137W | |
|
| ||||
| ♂ | 6 | 2 | 9 | 12 |
| ♀ | 9 | 5 | 15 | 12 |
| Sum | 15 | 7 | 24 | 24 |
| Ratio | 17.3% | 10%* | 34.3% | 34.3% |
|
| ||||
| Genotype of parents | Ryr2N1552S/+ ♂ × Ryr2R137W/+♀ | |||
|
| ||||
| +/+ | N1552S/+ | R137W/+ | N1552S/R137W | |
|
| ||||
| ♂ | 9 | 8 | 6 | 5 |
| ♀ | 7 | 7 | 5 | 9 |
| Sum | 16 | 15 | 11 | 14 |
| Ratio | 28.6% | 26.8% | 19.6% | 25% |
|
| ||||
| Genotype distribution of total offspring from Plxnb2D1577E/+ mice backcross with Plxnb2R465Q/+ mice | ||||
|
| ||||
| +/+ | D1577E/+ | R465Q/+ | D1577E/R465Q | |
|
| ||||
| ♂ | 17 | 13 | 9 | 14 |
| ♀ | 10 | 14 | 21 | 15 |
| Sum | 27 | 27 | 30 | 29 |
| Ratio | 23.8% | 23.9% | 26.5% | 25.7% |
P < 0.05.
Backcrossing Ryr2N1552S/+ with Ryr2R137W/+ parents and Plxnb2D1577E/+ with Plxnb2R465Q/+ parents showed reduced reproductive performance
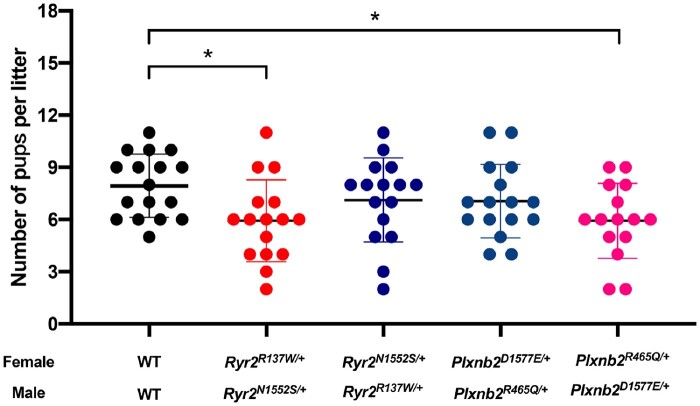
When backcrossing Ryr2N1552S/+ ♂ with Ryr2R137W/+ ♀ or Plxnb2D1577E/+ ♂ with Plxnb2R465/+ ♀, the litter sizes were significantly reduced (7.94 ± 1.82 vs 5.94 ± 2.35, P = 0.03, and 7.94 ± 1.82 vs 5.93 ± 2.15, P = 0.04, Fig. 3), findings in accordance with the sequencing results of Family 2 (p.N1551S/+ ♂ and p.R137W/+ ♀) and Family 3 (p.D1573E/+ ♂ and p.R463Q/+ ♀). Moreover, the litter sizes of backcrossing Ryr2R137W/+ ♂ with Ryr2N1552S/+ ♀ or Plxnb2R465Q/+♂ with Plxnb2D1577E/+ ♀ exhibited no difference compared with WT (7.94 ± 1.82 vs 7.13 ± 2.42, P = 0.66, and 7.94 ± 1.82 vs 7.06 ± 2.11, P = 0.61, Fig. 3). All the heterozygous female mice were sure fertile and the reproductive performance showed no differences compared with WT female mice (Supplementary Fig. S4D). These data suggest that the identified variants in Ryr2 and Plxnb2 impair murine reproductive ability under specific mating pairs. It is worth noting that the observed decreased litter size could not prove that Ryr2 and Plxnb2 point mutations are lethal, additional functional studies including the fertility ability and embryonic development examinations are needed to exclude the possibility of other plausible explanations, for example, subfertility.
Figure 3.
Reproductive performance of backcrossing Ryr2N1552S/+ with Ryr2R137W/+ parents and Plxnb2D1577E/+ with Plxnb2R465Q/+ parents. The number of pups per litter derived from pairs of Ryr2N1552S/+ with Ryr2R137W/+, Plxnb2D1577E/+ with Plxnb2R465Q/+ and WT with WT at the age of 8 weeks for 3 months. The results are shown as the mean ± SD, both female and male mice n = 8/group. *P < 0.05.
PLXNB2 knockdown impaired the migratory and invasive abilities of HTR-8/SVneo cells
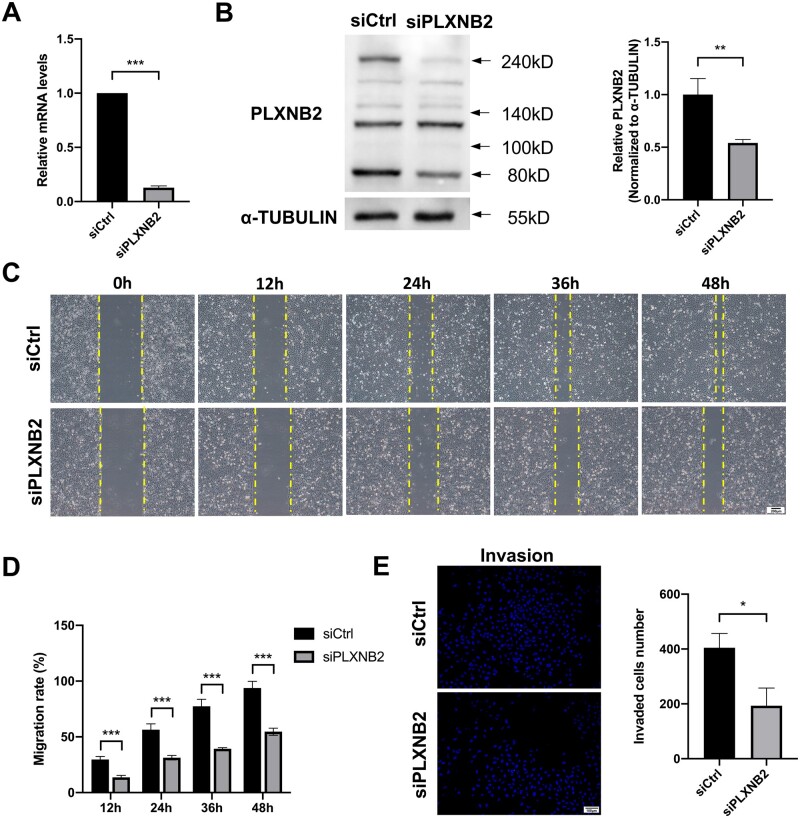
The migratory and invasive abilities of extravillous trophoblasts (EVTs) which are differentiated from the trophectoderm of blastocysts are essential for maternal–fetal interface establishment and pregnancy maintenance (Abbas et al., 2020). The revelation of trophoblast behavior-regulating molecules could better broaden our understanding of miscarriage pathogenesis. PLXNB2 has been previously reported to regulate cancer cell migration and invasion (Gurrapu et al., 2018); here we investigated the regulatory effects of PLXNB2 on the immortalized human EVT cell line HTR-8/SVneo cells. The knockdown efficiency of PLXNB2 by small-interfering RNA was confirmed by qRT-PCR analysis and western blotting (Fig. 4A and B). Wound-healing assay results indicated that the migration of HTR-8/SVneo cells was significantly inhibited after 48 h of siPLXNB2 transfection (P < 0.001, Fig. 4C and D), and the number of invaded cells was also reduced compared to the siCtrl group as measured by transwell invasion assay (P = 0.02, Fig. 4E). The EdU assay showed that PLXNB2 knockdown did not influence HTR-8/SVneo cell proliferation (50.2% vs 45.7%, P = 0.35, Supplementary Fig. S5A). However, according to flow cytometric analysis and TUNEL assay, downregulation of PLXNB2 moderately induced HTR-8/SVneo cell apoptosis (P = 0.01 and P = 0.03, respectively, Supplementary Fig. S5B and C). We also examined the effects of PLXNB2 knockdown on the endothelial-like tube formation ability of HTR-8/SVneo cells, and no difference was observed in tube branches, junctions, meshes, or mesh areas between the siCtrl and siPLXNB2 groups (Supplementary Fig. S5D).
Figure 4.
PLXNB2 knockdown inhibited the migration and invasion ability of HTR-8/SVneo cells. (A and B) Transfection efficiency was examined by RT-qPCR and western blot analysis respectively. The results showed that PLXNB2 knockdown attenuated PLXNB2 expression at both mRNA and protein levels. (C and D) Representative images of scratch wound-healing assays at 0, 12, 24, 36, and 48 h. The migration ability of HTR-8/SVneo cells was significantly inhibited 48 h after siPLXNB2 transfection. Scale bars = 200 μm. (E) Representative images of transwell invasion and migration assays. Cell invasion ability was inhibited after transfection with siPLXNB2. Scale bars = 100 μm. The results are shown as the mean ± SD of at least three independent experiments. *P < 0.05, **P < 0.01; ***P < 0.001. RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Screening potential pathogenic variants in RYR2 and PLXNB2
To screen additional pathogenic variants of RYR2 and PLXNB2 in unexplained first trimester miscarriages (Supplementary Fig. S6), we next performed multiplex PCR to sequence all exons of these genes in an additional 113 euploid miscarriages (villous tissues) from 113 families with maternal age between 22 and 42 years (mean maternal age of 31.54 ± 4.34 years). All of the variants identified by the bioinformatic pipeline were missense mutations, and we found two previously identified variants in the trio analysis and another ten new variants in RYR2 and PLXNB2 (Table IV and Supplementary Table SV). One of the previously identified variants, NM_001035.2:c.4652A>G of RYR2, was observed in two out of 113 miscarriages, and the allele frequency (MAF = 0.009) was slightly higher than the expected allele frequency in the East Asian population (MAF in gnomAD = 0.0036). Seven out of 10 addition variants were identified in only one subject with a rare allele frequency (MAF < 0.001) or absent in the 1000 Genomes, ExAC, and gnomAD databases and were predicted to be deleterious and conserved. Variant NM_012401.3:c.4612C>T in PLXNB2 showed higher allele frequency (MAF = 0.018) in miscarriages than expected (MAF in gnomAD = 0.0073) in the East Asian population. Taken together, these results support the potential pathogenic roles of variants located in RYR2 and PLXNB2 in unexplained first trimester miscarriages.
Table IV.
Variants of RYR2 and PLXNB2 identified in euploid miscarriage tissues.
| Gene name | Case ID | Position | cDNA change | AAChange | Variant ID | Nhet. | ExAC | gnomAD | SIFT | PPH2 | LRT | MutationTaster | CADD | GER | ACMG |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RYR2 | 71 | chr1:237494211 | c.G202C | p.V68L | rs752526125 | 1/113 | 0 | 0 | D | B | D | 23 | 4.35 | VUS | |
| RYR2 | 116 | chr1:237730053 | c.C3401G | p.A1134G | 1/113 | 0 | D | B | D | D | 33 | 5.29 | VUS | ||
| RYR2 | 65,120 | chr1:237777856 | c.G5428C | p.V1810L | rs754364233 | 2/113 | 0.0012 | 0.0012 | D | P | N | D | 23.1 | 5.62 | VUS |
| RYR2 | 7 | chr1:237806708 | c.G7303T | p.V2435F | rs188835713 | 1/113 | 0.0001 | 5.80E−05 | D | D | N | D | 24.2 | 3.53 | VUS |
| RYR2 | 45 | chr1:237824228 | c.G8417A | p.R2806H | rs955927781 | 1/113 | 0 | D | D | D | D | 33 | 5.46 | VUS | |
| RYR2 | 95 | chr1:237870428 | c.C9760A | p.P3254T | 1/113 | D | B | D | D | 26.6 | 5.72 | VUS | |||
| RYR2 | 114 | chr1:237947871 | c.T12859C | p.Y4287H | rs190009333 | 1/113 | 0 | 0 | D | D | D | D | 23.5 | 5.11 | VUS |
| PLXNB2 | 29,43,68,75 | chr22:50717060 | c.C4612T | p.R1538W | rs182970183 | 4/113 | 0.0076 | 0.0073 | D | D | D | D | 34 | 4.48 | VUS |
| PLXNB2 | 109 | chr22:50721219 | c.G2908A | p.V970M | rs569852454 | 1/113 | 0.0015 | 0.0015 | T | D | N | N | 23.1 | 2.65 | VUS |
| PLXNB2 | 111 | chr22:50724504 | c.G1901A | p.R634H | rs144108995 | 1/113 | 0 | 0 | T | P | N | D | 23.1 | 1.99 | VUS |
ACMG, American College of Medical Genetics and Genomics, B, Benign; D, deleterious; Nhet, number with heterozygous variant; P, possibly damaging; T, tolerated, VUS, variant of uncertain significance.
Discussion
In our study, we screened eight URM family samples by WGS and WES. After trio bioinformatics analysis and Sanger sequencing validation, we identified six candidate genes from four URM families. We observed that proteins encoded by ATP2A2, RYR2, NAP1L1, and PLXNB2 were expressed uniformly from the zygote to the blastocyst stage. Knock-in mice with Rry2 and Plxnb2 variants were generated to investigate their potential effects on early euploid miscarriages. Experiments in immortalized human trophoblasts showed the function of PLXNB2 in regulating trophoblast migratory and invasive abilities. Furthermore, we detected additional 10 variants of RYR2 and PLXNB2 in 113 unexplained euploid miscarriages.
Previous studies have supported the application of NGS in identifying genetic causes of miscarriage (Colley et al., 2019). WES has been recommended for fetuses with indications of organ system anomalies without a definitive diagnosis in other genetic examinations (Van den Veyver et al., 2022). Recently, Zhao et al. (2021) reported that 22–36% of euploid pregnancy losses could potentially be diagnosed by genetic variants through WES. Our study successfully identified nine variants in six candidate genes by utilizing WGS and WES to screen samples from eight URM couples. All these genes found in our study are proposed as causal genetic variants in unexplained first trimester miscarriage for the first time. We mainly focused on the variants in RYR2 and PLXNB2, both of which are highly conserved during evolution, especially RYR2 had been identified as a top-ranked causative gene in stillbirth (Stanley et al., 2020) and PLXNB2 has been proven to regulate multiple cell behaviors.
We found that the proportion of Plxnb2D1577E/R465Q and Ryr2N1552S/R137W offspring followed Mendelian inheritance, indicating that these compound heterozygous mutations in Plxnb2 and Ryr2 do not result in severe functional defects or embryonic lethality during the early development period. Although smaller litter sizes were observed when mating Ryr2N1552S/+ and Plxnb2D1577E/+ male mice with Ryr2R137W/+ and Plxnb2R465Q/+ female mice, respectively, it should be noted that a previous study reported Plxnb2−/− mice showed perinatal lethality from exencephaly rather than early miscarriage (Deng et al., 2007). Thus, the interpretation of the embryonic lethality of our identified SNVs requires caution. For RYR2, our two variant loci, p.N1551S and p.R137W, are both located in the N-terminal domain, which belongs to one of three hotspot mutation regions (Peng et al., 2016), but the definite role of these variants is still unclear. Further investigation of these point mutant mice and further functional studies are still needed. Considering reports that differences in the genetic background can apparently confer tolerance to some assumed loss-of-function mutations (Santesmasses et al., 2020), it is worth noting that genetic point mutations in mice may not recapitulate the phenotypes we observed in humans at the same severity.
Furthermore, when mating with Ryr2N1552S/+ and Plxnb2D1577E/+ male mice, the number of pups per litter of Ryr2R137W/+ and Plxnb2R465Q/+ female mice was decreased, which was in accordance with the clinical manifestations of URM patients, indicating that these variants in Ryr2 and Plxnb2 impaired the reproductive ability of mice under specific mating pairs. In addition, we observed that the proportion of Ryr2N1552S/+ offspring was significantly lower when Ryr2N1552S/+ female mice were backcrossed with Ryr2R137W/+ male mice. All of these results suggest that the presence of specific combinations of these variants may exert essential roles during pregnancy establishment and/or maintenance (e.g. maternal endometrial condition, placenta development). In spite of these phenotype results, it is worth noting that our current evidence cannot unequivocally establish that the examined Ryr2 and Plxnb2 point mutations are lethal, especially considering that subfertility cannot be ruled out and that developing embryos were not examined; thus, additional functional studies regarding individual variants focusing on these processes are needed.
During normal pregnancy establishment, the migration of EVT cells away from the villi and invasion into the maternal decidua are essential for remodeling the maternal spiral arteries to form a stable maternal–fetal interface during the first trimester of pregnancy (Suryawanshi et al., 2018). Dysregulation of EVT cell migration and invasion is at least partially involved in RM pathogenesis (Wang et al., 2021). It has been shown that PLXNB2 plays an essential role in regulating cellular migration (Saha et al., 2012; Van Battum et al., 2021) and tumor invasion (Gurrapu et al., 2018). Our results showed that PLXNB2 knockdown impaired HTR-8/SVneo cell migration and invasion. Simultaneously, PLXNB2 knockdown did not influence cell proliferation. Our results support that PLXNB2 plays an essential role in regulating normal placentation (Daviaud et al., 2016), and variants in PLXNB2 could potentially influence the expression of PLXNB2 and thus contribute to the pathogenesis of miscarriage.
As the increasing prevalence of WGS and WES, abundant databases of genetic variants in miscarriages are expected to be established by trio bioinformatics analysis in the near future. High sequencing coverage (i.e. at 500× or more) is still preferred to exclude the seemingly de novo mutations, which were actually inherited as a consequence of low-level mosaicism in one of the parents. Studies showed that at least 140-fold coverage is needed for detecting low-level mosaicism of ≥5% with ≥95% probability. In this study, the inheritance patterns of novelty-identified ATP2A2 and NAP1L1 variants are most likely to be de novo in Families 1 and 2, since the read coverage of SNVs in ATP2A2 and NAP1L1 was ∼140-fold and the read depth of the mutated allele in each parent is ≤1. Another challenge for sequencing research is the sample size. Since variants in candidate genes, identified from small groups of trios, are usually indefinite, findings of WES, especially rare variants, needed to be replicated in a larger group. Moreover, it is tough but indispensable to determine the function of genetic variants in different species and individuals, leading to uncertainties in their pathological evaluation. Thus, larger cohorts, multiple tissue samples, high coverage sequencing, and further functional studies are required for elucidating the genetic etiology and pathogenic mechanisms of URM, which holds promise for diagnostic and therapeutic target applications for URM clinical management. In addition, recent ultrasonography studies have been focusing on detecting structural abnormalities before 11 weeks of gestation, providing more morphological evidence for future evaluation of putative pathogenic variants identified by NGS of euploid miscarriages (Brown et al., 2021).
Conclusion
Our research identifies variants in six genes as plausible genetic contributors to euploid miscarriages and shows reduced reproductive ability in Ryr2 and Plxnb2 point mutant mice. The current findings suggested that variants in unique candidate genes may be underlying genetic etiologies for first trimester euploid miscarriage and WES on trio could be an ideal research model, which could facilitate individualized precise diagnostic and therapeutic regimens, thereby alleviating the psychological and sociopsychological burden of RM patients.
Supplementary Material
Acknowledgements
The authors are grateful to all participating families. We thank the National Research Center for Assisted Reproductive Technology and Reproductive Genetics for technical support.
Contributor Information
Xiyao Wang, Center for Reproductive Medicine, Shandong University, Jinan, China; Key Laboratory of Reproductive Endocrinology of Ministry of Education, Shandong University, Jinan, China; Medical Integration and Practice Center, Shandong University, Jinan, China.
Wenqiang Shi, State Key Laboratory of Pathogen and Biosecurity, Beijing Institute of Microbiology and Epidemiology, Beijing, China.
Shaotong Zhao, Center for Reproductive Medicine, Shandong University, Jinan, China; Key Laboratory of Reproductive Endocrinology of Ministry of Education, Shandong University, Jinan, China; Medical Integration and Practice Center, Shandong University, Jinan, China.
Deshun Gong, Key Laboratory of Growth Regulation and Transformation Research of Zhejiang Province, School of Life Sciences, Westlake University, Hangzhou, China.
Shuo Li, Center for Reproductive Medicine, Shandong University, Jinan, China; Key Laboratory of Reproductive Endocrinology of Ministry of Education, Shandong University, Jinan, China; Medical Integration and Practice Center, Shandong University, Jinan, China.
Cuiping Hu, Center for Reproductive Medicine, Shandong University, Jinan, China; Key Laboratory of Reproductive Endocrinology of Ministry of Education, Shandong University, Jinan, China; Medical Integration and Practice Center, Shandong University, Jinan, China.
Zi-Jiang Chen, Center for Reproductive Medicine, Shandong University, Jinan, China; Key Laboratory of Reproductive Endocrinology of Ministry of Education, Shandong University, Jinan, China.
Yan Li, Center for Reproductive Medicine, Shandong University, Jinan, China; Key Laboratory of Reproductive Endocrinology of Ministry of Education, Shandong University, Jinan, China; Medical Integration and Practice Center, Shandong University, Jinan, China; Suzhou Research Institute, Shandong University, Suzhou, China.
Junhao Yan, Center for Reproductive Medicine, Shandong University, Jinan, China; Key Laboratory of Reproductive Endocrinology of Ministry of Education, Shandong University, Jinan, China.
Data availability
The data underlying this article are available in the Genome Sequence Archive (GSA) in the National Genomics Data Center, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Science, at https://ngdc.cncb.ac.cn, and can be acessed with reference number HRA002683 (GSA-Human), HRA002684 (GSA-Human), and BPRJCA010337 (GSA). All these data will be shaired on reasonable request to the corresponding author Dr. Junhao Yan.
Authors’ roles
Y.L. and J.Y. conceived and designed the project. X.W., S.L., S.Z., and C.H. collected samples and performed experiments. X.W. and W.S. analyzed the data. D.G. analyzed the protein structure and mapped the location of point mutations. X.W. and W.S. wrote the article. Y.L., J.Y., and Z.-J.C. critically revised the article. All authors have been involved in interpreting the data and approved the final version.
Funding
This study was supported by grants from the National Key Research and Development Program of China (2021YFC2700604), National Natural Science Foundation of China (31900492, 82101784, 82171648), Basic Science Center Program of the National Natural Science Foundation of China (31988101), Key Research and Development Program of Shandong Province (2021LCZX02), Natural Science Foundation of Shandong Province (ZR2020QH051), Natural Science Foundation of Jiangsu Province (BK20200223), Taishan Scholars Program for Young Experts of Shandong Province (tsqn201812154), and Young Scholars Program of Shandong University.
Conflict of interest
The authors declare no conflicts of interest.
Web resources
FastQC, https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
East Asian population of gnomAD, https://gnomad.broadinstitute.org/
1000 genomes, http://www.1000genomes.org/
ExAC database, http://exac.broadinstitute.org/
PubMed (https://www.ncbi.nlm.nih.gov/pmc/)
Online Mendelian Inheritance in Man (OMIM), https://www.omim.org
ClinVar, https://www.ncbi.nlm.nih.gov/clinvar/
Human gene mutation data (HGMD), https://portal.biobase-international.com/cgi-bin/portal/login.cgi
GeneCards, http://www.genecards.org/
UniProtKB, https://www.uniprot.org
MFEprimer-3.0 software, https://mfeprimer3.igenetech.com/
PyMOL, www.pymol.org
Protein Data Bank, https://www.rcsb.org
AlphaFold, https://alphafold.ebi.ac.uk
Clustal Omega, https://www.ebi.ac.uk/Tools/msa/clustalo/
References
- Abbas Y, Turco MY, Burton GJ, Moffett A.. Investigation of human trophoblast invasion in vitro. Hum Reprod Update 2020;4:501–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acuna-Hidalgo R, Bo T, Kwint MP, van de Vorst M, Pinelli M, Veltman JA, Hoischen A, Vissers LE, Gilissen C.. Post-zygotic point mutations are an underrecognized source of de novo genomic variation. Am J Hum Genet 2015;1:67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei I, Jordan DM, Sunyaev SR.. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet 2013;Chapter 7:Unit7.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt SJ, Marchetto NM, Roy J, Morelli SS, McGovern PG.. Pregnancy outcomes following in vitro fertilization frozen embryo transfer (IVF-FET) with or without preimplantation genetic testing for aneuploidy (PGT-A) in women with recurrent pregnancy loss (RPL): a SART-CORS study. Hum Reprod 2021;8:2339–2344. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;15:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown I, Rolnik DL, Fernando S, Menezes M, Ramkrishna J, da Silva Costa F, Meagher S.. Ultrasound findings and detection of fetal abnormalities before 11 weeks of gestation. Prenat Diagn 2021;13:1675–1684. [DOI] [PubMed] [Google Scholar]
- Cantaut-Belarif Y, Sternberg JR, Thouvenin O, Wyart C, Bardet PL.. The Reissner fiber in the cerebrospinal fluid controls morphogenesis of the body axis. Curr Biol 2018;28:2479–2486.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen OB, Mathiesen O, Lauritsen JG, Grunnet N.. Idiopathic recurrent spontaneous abortion. Evidence of a familial predisposition. Acta Obstet Gynecol Scand 1990;7–8:597–601. [DOI] [PubMed] [Google Scholar]
- Chun S, Fay JC.. Identification of deleterious mutations within three human genomes. Genome Res 2009;9:1553–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang Le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM.. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colley E, Hamilton S, Smith P, Morgan NV, Coomarasamy A, Allen S.. Potential genetic causes of miscarriage in euploid pregnancies: a systematic review. Hum Reprod Update 2019;4:452–472. [DOI] [PubMed] [Google Scholar]
- Daviaud N, Chen K, Huang Y, Friedel RH, Zou H.. Impaired cortical neurogenesis in plexin-B1 and -B2 double deletion mutant. Dev Neurobiol 2016;8:882–899. [DOI] [PubMed] [Google Scholar]
- Denda K, Nakao-Wakabayashi K, Okamoto N, Kitamura N, Ryu JY, Tagawa YI, Ichisaka T, Yamanaka S, Komada M.. Nrk, an X-linked protein kinase in the germinal center kinase family, is required for placental development and fetoplacental induction of labor. J Biol Chem 2011;286:28802–28810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng S, Hirschberg A, Worzfeld T, Penachioni JY, Korostylev A, Swiercz JM, Vodrazka P, Mauti O, Stoeckli ET, Tamagnone L. et al. Plexin-B2, but not Plexin-B1, critically modulates neuronal migration and patterning of the developing nervous system in vivo. J Neurosci 2007;23:6333–6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;5:491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert AM, Hume GL, Warren KS, Cook NP, Burns CG, Mohideen MA, Siegal G, Yelon D, Fishman MC, Garrity DM.. Calcium extrusion is critical for cardiac morphogenesis and rhythm in embryonic zebrafish hearts. Proc Natl Acad Sci U S A 2005;49:17705–17710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggenhuizen GM, Go A, Koster MPH, Baart EB, Galjaard RJ.. Confined placental mosaicism and the association with pregnancy outcome and fetal growth: a review of the literature. Hum Reprod Update 2021;5:885–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoffroy V, Herenger Y, Kress A, Stoetzel C, Piton A, Dollfus H, Muller J.. AnnotSV: an integrated tool for structural variations annotation. Bioinformatics 2018;20:3572–3574. [DOI] [PubMed] [Google Scholar]
- Gnad F, Baucom A, Mukhyala K, Manning G, Zhang Z.. Assessment of computational methods for predicting the effects of missense mutations in human cancers. BMC Genomics 2013;14 Suppl 3:S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurrapu S, Pupo E, Franzolin G, Lanzetti L, Tamagnone L.. Sema4C/PlexinB2 signaling controls breast cancer cell growth, hormonal dependence and tumorigenic potential. Cell Death Differ 2018;25:1259–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaandorp SP, Goddijn M, van der Post JA, Hutten BA, Verhoeve HR, Hamulyák K, Mol BW, Folkeringa N, Nahuis M, Papatsonis DN. et al. Aspirin plus heparin or aspirin alone in women with recurrent miscarriage. N Engl J Med 2010;17:1586–1596. [DOI] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J.. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 2014;3:310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolte AM, Bernardi LA, Christiansen OB, Quenby S, Farquharson RG, Goddijn M, Stephenson MD.. Terminology for pregnancy loss prior to viability: a consensus statement from the ESHRE early pregnancy special interest group. Hum Reprod 2015;3:495–498. [DOI] [PubMed] [Google Scholar]
- Kolte AM, Nielsen HS, Moltke I, Degn B, Pedersen B, Sunde L, Nielsen FC, Christiansen OB.. A genome-wide scan in affected sibling pairs with idiopathic recurrent miscarriage suggests genetic linkage. Mol Hum Reprod 2011;6:379–385. [DOI] [PubMed] [Google Scholar]
- Kushnir A, Wajsberg B, Marks AR.. Ryanodine receptor dysfunction in human disorders. Biochim Biophys Acta Mol Cell Res 2018;1865:1687–1697. [DOI] [PubMed] [Google Scholar]
- Lestari B, Naito S, Endo A, Nishihara H, Kato A, Watanabe E, Denda K, Komada M, Fukushima T.. Placental mammals acquired functional sequences in NRK for regulating the CK2-PTEN-AKT pathway and placental cell proliferation. Mol Biol Evol 2022;2:msab371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;14:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D, Peng Y, Lv W, Deng L, Zhang Y, Li H, Yang P, Zhang J, Song Z, Xu G. et al. Copy number variation sequencing for comprehensive diagnosis of chromosome disease syndromes. J Mol Diagn 2014;5:519–526. [DOI] [PubMed] [Google Scholar]
- Lu H, Shagirova A, Goggi JL, Yeo HL, Roy S.. Reissner fibre-induced urotensin signalling from cerebrospinal fluid-contacting neurons prevents scoliosis of the vertebrate spine. Biol Open 2020;5:bio052027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S.. Predicting deleterious amino acid substitutions. Genome Res 2001;5:863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuwaki M, Kato K, Nagata K.. Functional characterization of human nucleosome assembly protein 1-like proteins as histone chaperones. Genes Cells 2010;1:13–27. [DOI] [PubMed] [Google Scholar]
- Otis EM, Brent R.. Equivalent ages in mouse and human embryos. Anat Rec 1954;1:33–63. [DOI] [PubMed] [Google Scholar]
- Peng W, Shen H, Wu J, Guo W, Pan X, Wang R, Chen SR, Yan N.. Structural basis for the gating mechanism of the type 2 ryanodine receptor RyR2. Science 2016;6310:301–310. [DOI] [PubMed] [Google Scholar]
- Perälä NM, Immonen T, Sariola H.. The expression of plexins during mouse embryogenesis. Gene Expr Patterns 2005;5:355–362. [DOI] [PubMed] [Google Scholar]
- Periasamy M, Kalyanasundaram A.. SERCA pump isoforms: their role in calcium transport and disease. Muscle Nerve 2007;4:430–442. [DOI] [PubMed] [Google Scholar]
- Periasamy M, Reed TD, Liu LH, Ji Y, Loukianov E, Paul RJ, Nieman ML, Riddle T, Duffy JJ, Doetschman T. et al. Impaired cardiac performance in heterozygous mice with a null mutation in the sarco(endo)plasmic reticulum Ca2+-ATPase isoform 2 (SERCA2) gene. J Biol Chem 1999;4:2556–2562. [DOI] [PubMed] [Google Scholar]
- Practice Committee of the American Society for Reproductive Medicine. Evaluation and treatment of recurrent pregnancy loss: a committee opinion. Fertil Steril 2012;5:1103–1111. [DOI] [PubMed] [Google Scholar]
- Practice Committee of the American Society for Reproductive Medicine. Definitions of infertility and recurrent pregnancy loss: a committee opinion. Fertil Steril 2020;3:533–535. [DOI] [PubMed] [Google Scholar]
- Priori SG, Napolitano C.. Cardiac and skeletal muscle disorders caused by mutations in the intracellular Ca2+ release channels. J Clin Invest 2005;8:2033–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H, Li Y, Feng C, Duo S, Ji F, Jiao J.. Nap1l1 controls embryonic neural progenitor cell proliferation and differentiation in the developing brain. Cell Rep 2018;22:2279–2293. [DOI] [PubMed] [Google Scholar]
- Qiao Y, Wen J, Tang F, Martell S, Shomer N, Leung PC, Stephenson MD, Rajcan-Separovic E.. Whole exome sequencing in recurrent early pregnancy loss. Mol Hum Reprod 2016;5:364–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenby S, Gallos ID, Dhillon-Smith RK, Podesek M, Stephenson MD, Fisher J, Brosens JJ, Brewin J, Ramhorst R, Lucas ES. et al. Miscarriage matters: the epidemiological, physical, psychological, and economic costs of early pregnancy loss. Lancet 2021;10285:1658–1667. [DOI] [PubMed] [Google Scholar]
- Quintero-Ronderos P, Mercier E, Fukuda M, González R, Suárez CF, Patarroyo MA, Vaiman D, Gris JC, Laissue P.. Novel genes and mutations in patients affected by recurrent pregnancy loss. PLoS One 2017;10:e0186149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajcan-Separovic E. Next generation sequencing in recurrent pregnancy loss-approaches and outcomes. Eur J Med Genet 2020;2:103644. [DOI] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E. et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;5:405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose CD, Pompili D, Henke K, Van Gennip JLM, Meyer-Miner A, Rana R, Gobron S, Harris MP, Nitz M, Ciruna B.. SCO-Spondin defects and neuroinflammation are conserved mechanisms driving spinal deformity across genetic models of idiopathic scoliosis. Curr Biol 2020;30:2363–2373.e6. [DOI] [PubMed] [Google Scholar]
- Saha B, Ypsilanti AR, Boutin C, Cremer H, Chédotal A.. Plexin-B2 regulates the proliferation and migration of neuroblasts in the postnatal and adult subventricular zone. J Neurosci 2012;47:16892–16905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santesmasses D, Mariotti M, Gladyshev VN.. Tolerance to selenoprotein loss differs between human and mouse. Mol Biol Evol 2020;2:341–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarah P, Vanessa KD, Rebecca HA.. ACOG Practice Bulletin No. 200 summary: early pregnancy loss. Obstet Gynecol 2018;5:1311–1313. [DOI] [PubMed] [Google Scholar]
- Schwarz JM, Rödelsperger C, Schuelke M, Seelow D.. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods 2010;7:575–576. [DOI] [PubMed] [Google Scholar]
- Shihab HA, Gough J, Cooper DN, Stenson PD, Barker GL, Edwards KJ, Day IN, Gaunt TR.. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum Mutat 2013;1:57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shihab HA, Rogers MF, Gough J, Mort M, Cooper DN, Day IN, Gaunt TR, Campbell C.. An integrative approach to predicting the functional effects of non-coding and coding sequence variation. Bioinformatics 2015;10:1536–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shull GE, Okunade G, Liu LH, Kozel P, Periasamy M, Lorenz JN, Prasad V.. Physiological functions of plasma membrane and intracellular Ca2+ pumps revealed by analysis of null mutants. Ann N Y Acad Sci 2003;986:453–460. [DOI] [PubMed] [Google Scholar]
- Stanley KE, Giordano J, Thorsten V, Buchovecky C, Thomas A, Ganapathi M, Liao J, Dharmadhikari AV, Revah-Politi A, Ernst M. et al. Causal genetic variants in stillbirth. N Engl J Med 2020;12:1107–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryawanshi H, Morozov P, Straus A, Sahasrabudhe N, Max KEA, Garzia A, Kustagi M, Tuschl T, Williams Z.. A single-cell survey of the human first-trimester placenta and decidua. Sci Adv 2018;10:eaau4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshima H, Komazaki S, Hirose K, Nishi M, Noda T, Iino M.. Embryonic lethality and abnormal cardiac myocytes in mice lacking ryanodine receptor type 2. EMBO j 1998;12:3309–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Battum E, Heitz-Marchaland C, Zagar Y, Fouquet S, Kuner R, Chédotal A.. Plexin-B2 controls the timing of differentiation and the motility of cerebellar granule neurons. eLife 2021;10:e60554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Veyver IB, Chandler N, Wilkins-Haug LE, Wapner RJ, Chitty LS.. International Society for Prenatal Diagnosis Updated Position Statement on the use of genome-wide sequencing for prenatal diagnosis. Prenat Diagn 2022;6:796–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H.. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;16:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li H, Xu Y, Shao Q, Yi J, Wang R, Cai W, Hang X, Zhang C, Cai H. et al. MFEprimer-3.0: quality control for PCR primers. Nucleic Acids Res 2019;W1:W610–w613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XH, Xu S, Zhou XY, Zhao R, Lin Y, Cao J, Zang WD, Tao H, Xu W, Li MQ. et al. Low chorionic villous succinate accumulation associates with recurrent spontaneous abortion risk. Nat Commun 2021;1:3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Yin P, Gong H, Xue Y, Zhang G, Fang B, Chen Z, Li Y, Yang C, Huang Z. et al. Nucleosome assembly protein 1-like 1 (Nap1l1) regulates the proliferation of murine induced pluripotent stem cells. Cell Physiol Biochem 2016;1:340–350. [DOI] [PubMed] [Google Scholar]
- Yates CL, Monaghan KG, Copenheaver D, Retterer K, Scuffins J, Kucera CR, Friedman B, Richard G, Juusola J.. Whole-exome sequencing on deceased fetuses with ultrasound anomalies: expanding our knowledge of genetic disease during fetal development. Genet Med 2017;10:1171–1178. [DOI] [PubMed] [Google Scholar]
- Yu W, Goncalves KA, Li S, Kishikawa H, Sun G, Yang H, Vanli N, Wu Y, Jiang Y, Hu MG. et al. Plexin-B2 mediates physiologic and pathologic functions of angiogenin. Cell 2017;171:849–864.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Chai H, Zhou Q, Wen J, Reddy UM, Kastury R, Jiang Y, Mak W, Bale AE, Zhang H. et al. Exome sequencing analysis on products of conception: a cohort study to evaluate clinical utility and genetic etiology for pregnancy loss. Genet Med 2021;3:435–442. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the Genome Sequence Archive (GSA) in the National Genomics Data Center, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Science, at https://ngdc.cncb.ac.cn, and can be acessed with reference number HRA002683 (GSA-Human), HRA002684 (GSA-Human), and BPRJCA010337 (GSA). All these data will be shaired on reasonable request to the corresponding author Dr. Junhao Yan.