Abstract
Continuous biomarkers are common for disease screening and diagnosis. To reach a dichotomous clinical decision, a threshold would be imposed to distinguish subjects with disease from nondiseased individuals. Among various performance metrics, specificity at a controlled sensitivity level (or vice versa) is often desirable because it directly targets the clinical utility of the intended clinical test. Meanwhile, covariates, such as age, race, as well as sample collection conditions, could impact the biomarker distribution and may also confound the association between biomarker and disease status. Therefore, covariate adjustment is important in such biomarker evaluation. Most existing covariate adjustment methods do not specifically target the desired sensitivity/specificity level, but rather do so for the entire biomarker distribution. As such, they might be more prone to model misspecification. In this paper, we suggest a parsimonious quantile regression model for the diseased population, only locally at the controlled sensitivity level, and assess specificity with covariate-specific control of the sensitivity. Variance estimates are obtained from a sample-based approach and bootstrap. Furthermore, our proposed local model extends readily to a global one for covariate adjustment for the receiver operating characteristic (ROC) curve over the sensitivity continuum. We demonstrate computational efficiency of this proposed method and restore the inherent monotonicity in the estimated covariate-adjusted ROC curve. The asymptotic properties of the proposed estimators are established. Simulation studies show favorable performance of the proposal. Finally, we illustrate our method in biomarker evaluation for aggressive prostate cancer.
Keywords: quantile regression, receiver operating characteristic curve, sensitivity, sensitivity at controlled specificity, specificity, specificity at controlled sensitivity
1 |. INTRODUCTION
Continuous biomarker is often utilized for disease screening and diagnosis, where a threshold is imposed to reach a dichotomous clinical decision. For its performance assessment, the receiver operating characteristic (ROC) curve provides a comprehensive evaluation across all possible thresholds. Area under the ROC curve (AUC) is a popular performance metric, but it may not be clinically sensible (Hanley and McNeil, 1982). Obviously, a continuous biomarker would not operate at all thresholds, because a diagnostic test typically needs to reach a certain sensitivity (or specificity) level to be clinically useful. Therefore, specificity at a controlled sensitivity level (or vice versa) would be a more desirable performance metric in practice. For example, for the noninvasive diagnosis of aggressive prostate cancer, the cost of a false negative is usually much higher than that of a false positive as a positive test result would be confirmed with biopsy. Thus, the clinical utility of a continuous biomarker would be best measured with specificity at a controlled high sensitivity level, for example, 95% (Sanda et al., 2017). In this work, we mainly focus on specificity at a controlled sensitivity level. The same methods proposed can be directly applied to sensitivity at a controlled specificity level by switching the roles of cases and controls.
Platt et al. (2000) and Zhou and Qin (2005), among others, studied the estimation of such a metric, in the absence of covariates. However, factors, such as age and ethnicity as well as specimen collection condition, may influence a biomarker. For example, prostate-specific antigen (PSA), as a prostate cancer biomarker, tends to be higher in older men (Partin et al., 1996). In addition, African American men have higher PSA than men of other racial backgrounds (Henderson et al., 1997; Sanda et al., 2017). While intrinsically they do not discriminate diseased from nondiseased, these covariates may impact the performance of a biomarker in a number of ways (Pepe, 2003). In fact, covariates may confound the association between the biomarker and disease status when the covariate distributions differ between diseased and nondiseased individuals. Even when the two covariate distributions are the same, ignoring the covariates may lead to biased accuracy assessment. At a minimum, when a test is intended to operate at a controlled sensitivity level, covariate adjustment for the threshold would be necessary to ensure a uniform sensitivity level across subpopulations.
There are many existing methods for covariate adjustment in the assessment of continuous biomarkers. Most of them model covariate effects on biomarker beyond the specific sensitivity/specificity level of interest. As such, they might be prone to model misspecification. For example, Tosteson and Begg (1988) and Pepe (1998) modeled the covariate effects on the diseased and nondiseased populations, and then derived covariate-specific ROC curve. Some other methods directly estimate covariate-adjusted ROC curve through generalized linear regression, for example, Pepe (1997, 2000); Cai and Pepe (2002). However, the covariate effects could be different at different sensitivity levels, as recognized by some of these authors. For example, Cai and Pepe (2002) discussed the possibility of including interactions between false-positive rates and covariates. An exception is Janes and Pepe (2009), who developed a nonparametric estimator in the case of discrete covariates. Nevertheless, when continuous covariates are involved, they used a semiparametric estimator to adjust for covariate effects over the entire biomarker distribution, not specifically targeting the sensitivity/specificity of interest. A few previous works have also adopted Bayesian modeling framework by incorporating the covariate effects in the parameters of the ROC curve associated data distributions (de Carvalho et al., 2013; de Carvalho and Rodriguez-Alvarez, 2018).
In this paper, we develop a covariate adjustment method for specificity at a controlled sensitivity level, by adopting the quantile regression model (Koenker and Bassett, 1978) at the given sensitivity for the diseased population. As such, minimal assumptions are imposed. The proposal also extends readily to the continuous spectrum of sensitivity levels so as to address covariate adjustment for the ROC curve. In the special case that the covariates have a finite number of values, the quantile regression model becomes saturated and thus does not actually impose any assumptions. Accordingly, our method then reduces to the nonparametric method as considered by Janes and Pepe (2009). However, with continuous covariates, the semiparametric model of Janes and Pepe (2009) is different as indicated before. This seemingly natural model has not been favorably considered previously due to a few technique difficulties particularly in the circumstance of covariate adjustment for the ROC curve (Pepe, 2003, p. 139). First, the computation burden may be of concern as the covariate effects are allowed to vary over quantiles. Second, the standard quantile regression of Koenker and Bassett (1978) does not respect the inherent monotonicity of the conditional quantile functions. Subsequently, the estimated ROC after covariate adjustment may not even be monotone. Both issues are resolved in our proposal.
This paper is organized as follows. Section 2 presents the proposed covariate adjustment method for specificity at a controlled sensitivity level. Section 3 extends the proposal to global covariate adjustment over all sensitivity levels, resulting in a covariate-adjusted ROC curve. Simulation studies are presented in Section 4, and a real data illustration given in Section 5. Final discussions are provided in Section 6. Technical proofs are relegated to the Web Appendix. The proposed methods are implemented and available as a user-friendly R/CRAN package caROC (https://cran.r-project.org/web/packages/caROC/index.html).
2 |. SPECIFICITY AT A CONTROLLED SENSITIVITY LEVEL
Denote the case biomarker of interest by and its associated covariate by . The case sample consists of i.i.d. replicates of , . Similarly, denote the control biomarker by and its associated covariate by , and the control sample consists of i.i.d. replicates , . These covariates may be discrete or continuous.
Write the conditional distribution function of the cases as . The corresponding conditional quantile function is . Controlling sensitivity level at , between 0 and 1, yields a test threshold to be the th quantile, . We adopt the following quantile regression model for the relationship between -level sensitivity and the covariates:
| (1) |
where is the regression coefficient; note that 1 is added to the covariate vector to incorporate an intercept. This model imposes a structure only at the controlled sensitivity level . In the case of the -sample problem, that is, with dummy indicator covariates denoting these samples, this model becomes saturated and no model structure is actually imposed. For controls, we similarly define as the conditional distribution function, that is, . Write as the true value of . The covariate-adjusted specificity at controlled sensitivity is given by
This measure gives the overall specificity with covariate-specific threshold so as to keep the same controlled sensitivity level for covariate-specific subpopulations.
Standard quantile regression of Koenker and Bassett (1978) gives a point estimator , which is a solution to the following estimating equation:
Then an estimator of can be obtained via the plug-in principle . In the special case of the -sample problem, our estimator reduces to the nonparametric estimator of Janes and Pepe (2009).
2.1 |. Asymptotic study
Now we consider the asymptotic properties of our proposed estimator. The following regularity conditions are imposed:
Condition 1. The control and case size ratio approaches a constant as .
Condition 2. Covariates and are bounded.
Condition 3. is nonsingular, where and for vector .
Condition 4a. Both and are differentiable at the threshold with derivative bounded away from 0 and uniformly in over the supports of and , respectively.
These conditions are standard and mild. In particular, the differentiability assumption in Condition 4a is only imposed at the threshold of interest, whereas and could be discontinuous elsewhere.
Theorem 1.
Suppose that the quantile regression model for the cases as given in (1) holds locally at the is consistent for converges to a normal distribution with mean zero and variance
| (2) |
where
and , respectively.
The second component of is the variance if is known and used instead of . Meanwhile, the additional variability as given by the first component arises from the estimation of .
2.2 |. Inference
Theorem 1 provides the asymptotic variance for the proposed estimator. As derivatives of the distribution functions are involved, direct estimation, however, is difficult. To overcome this difficulty, we adopt the method of Huang (2002) for variance estimation with nonsmooth estimating functions. Recast the estimator as the solution to the following set of estimating equations:
| (3) |
where . Denote the true value by . The asymptotic variance of is , where is the asymptotic variance of and is the derivative of the limit of at . Note that in (2) corresponds to the last diagonal element of . Sandwich variance estimation cannot be directly applied, because is not differentiable in . The method of Huang (2002) resolves this issue. Specifically, start with an estimator for as
| (4) |
Perform the Cholesky decomposition to give . Write with column vectors , where is the length of . Then, a sample-based variance estimator for is given by . This method overcomes the nondifferentiability issue discussed before effectively by a numerical differentiation of the inverse estimating equation using a data-adaptive bandwidth.
For the variance estimation, a computationally more intensive alternative is bootstrap with resampling for cases and controls drawn separately. This approach has been commonly adopted for related problems (e.g., Janes and Pepe 2009).
3 |. ROC CURVE
The preceding methods target a particular sensitivity level of interest. The same modeling strategy readily extends to each and every sensitivity level in a continuum, resulting in a covariate-adjusted ROC curve. As such, we impose the global model,
| (5) |
where the regression coefficient function may vary with . The covariate-adjusted specificity also varies with ,
| (6) |
Clearly, the global model is a submodel of the local one given by (1). Nevertheless, the global model is fairly general itself as being nonparametric. Just the same as the local model, this global model actually imposes no structure whatsoever in the special case of the -sample problem (Huang, 2010).
As the global model implies local models for each value in (0, 1), we apply the estimation procedure described in Section 2 in a pointwise fashion to obtain the estimators. The regression coefficient estimator is the solution of
and an estimator of is . The computation might be perceived as intensive to have a solution at each and every . Nevertheless, the estimator is a step function and its computation can be formulated as a parametric programming problem with breakpoints to examine (Koenker 2005, Section 6.3). Starting from upward, this algorithm involves alternately solving the equation at the current value and finding the next breakpoint. The computation burden does not impose a real concern for most applications.
3.1 |. Asymptotic study
For the asymptotic study with the global model, we strengthen Condition 4a.
Condition 4b. Both and have density functions and , respectively, which are continuous in for given and bounded uniformly in and over the supports of and , respectively. Meanwhile, is continuously differentiable on for any and such that .
The existence of density function is a standard condition when ROC curve is of interest (Janes and Pepe, 2009). The differentiability of is also mild and commonly imposed (Koenker, 2005). Under the condition, there is no zero-density intervals and thus no jump in quantile.
Theorem 2.
Suppose that the quantile regression model for the cases as given in (5) holds globally over converges in probability to uniformly over converges weakly to a Gaussian process over .
3.2 |. Monotonization of the estimated ROC curve
As mentioned in the Introduction, lack of monotonicity in the estimated conditional quantile functions could result in that of the estimated covariate-adjusted ROC curve. In fact, a few existing works adopted location-scale models to avoid illogical results in estimating quantiles, for example, He (1997) and Heagerty and Pepe (1999). However, their models become more restrictive. We rather restore monotonicity in the ROC estimation under the original quantile regression model and suggest two approaches below.
The root of the issue is the lack of monotonicity-respecting with the estimated quantile regression coefficient process. Huang (2017) developed a method to restore the monotonicity-respecting property by identifying and interpolating monotonicity-respecting breakpoints of the original estimated coefficient process. We apply this approach to obtain a monotonicity-respecting estimator . Plugging this estimator in (6) results in an estimated ROC curve that is monotone. As a note, the resulting monotonized ROC curve is still a step function. The second strategy is to directly apply the method of Huang (2017) to the estimated ROC curve . This method results in a piecewise-linear monotonized ROC curve. Web Appendix Section S3 provides more details about the two monotonization methods. We refer these two methods as regression- and ROC-based monotonizations thereafter.
The monotonized estimators are asymptotically equivalent to the original estimators as shown in Huang (2017). For finite sample, the monotonized estimators may have efficiency gain.
3.3 |. Inference
For a point on the estimated ROC curve, one may adopt the same inference procedure with the local model as described in (1). However, note the availability of several point estimates, depending on whether a monotonized ROC is employed. Nevertheless, any choice of these estimates does not make a difference because they are all asymptotically equivalent.
If the whole ROC curve is of interest, it is possible to construct confidence band using bootstrap. To estimate the distribution of , we can use the same bootstrap approach in local model except that the estimand now is functional. Denote the bootstrap estimator by . The distribution of conditioning on the data is asymptotically the same as . For with and satisfying , the 95% equal-precision confidence band of is given by
where is the standard error of and is the estimated 95% percentile of . is also based on bootstrap resamples. One may construct a confidence band based on a monotonized ROC curve as in Section 3.2 in the same fashion, simply with replaced by the monotonized version.
4 |. SIMULATIONS
We evaluate the finite sample properties of our proposal under practical sample sizes. Assume that the biomarker relies on two independent covariates and , both following uniform distribution between 0 and 1 among cases and controls. For cases, the biomarkers ’s are generated from formulation (1) and consists of an intercept and two slopes . For controls, the biomarkers ’s are generated from . The true specificities at controlled sensitivity levels 0.95, 0.90, 0.85, and 0.80 are 0.24, 0.36, 0.45, and 0.52, respectively.
Table 1 reports the performance of the proposed method in this setting, including bias, sample- and bootstrap-based standard errors, as well as the coverage probability of confidence intervals. We also present the logit transformation-based confidence interval, which is obtained by backtransforming the Wald-type confidence interval of the logit-transformed . As a comparison, we present the results using the seimiparametric method by Janes and Pepe (2009) (Column “JP-SP”), implemented in R/CRAN package ROCnReg (Rodríguez-Álvarez and Inacio, 2020). All results are summarized over 5000 Monte Carlo datasets. The estimation bias of the proposed method is very small and decreases with the increase of sample size. Both sample- and bootstrap-based standard errors are close to standard deviations. In addition, the coverage rate of confidence intervals is close to the nominal level under all scenarios. These demonstrate the favorable performance of the proposed method. The semiparametric method by Janes and Pepe (2009) shows larger bias and worse coverage probability. Of course, this comparison is not completely fair because the two methods impose different models and the data are simulated under our model. Web Appendix Figure S1 presents the computational time for different sample sizes using sample-based inference versus bootstrap-based inference. We observe that sample-based variance estimation has comparable performance with the bootstrap-based estimation, whereas the sample-based inference has advantages in computational efficiency.
TABLE 1.
Results of the simulation study for estimating specificity under controlled sensitivity level
| Proposed method |
JP-SP |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sample-based |
BT-based |
|||||||||
| Bias | SD | SE | Cov | LCov | SE | Cov | LCov | Bias | Cov | |
| 100 | 160 | 784 | 1111 | 94.28 | 95.62 | 818 | 94.72 | 94.22 | 424 | 85.98 |
| 200 | 85 | 570 | 739 | 93.14 | 94.42 | 597 | 94.68 | 95.06 | 409 | 80.12 |
| 500 | 43 | 358 | 444 | 94.76 | 95.24 | 382 | 95.52 | 95.18 | 396 | 68.90 |
| 1000 | 19 | 265 | 300 | 93.78 | 94.18 | 270 | 94.36 | 94.40 | 388 | 51.44 |
| 100 | 68 | 777 | 1037 | 95.54 | 96.36 | 822 | 95.62 | 96.58 | 116 | 93.48 |
| 200 | 25 | 557 | 682 | 94.64 | 95.46 | 582 | 95.00 | 95.68 | 103 | 91.36 |
| 500 | 13 | 354 | 410 | 95.00 | 95.22 | 367 | 95.08 | 95.38 | 103 | 92.04 |
| 1000 | 4 | 254 | 277 | 94.8 | 94.98 | 257 | 94.82 | 94.96 | 95 | 90.12 |
| , | ||||||||||
| 100 | 17 | 736 | 943 | 95.38 | 96.28 | 788 | 95.64 | 96.94 | −65 | 94.32 |
| 200 | 1 | 537 | 634 | 95.08 | 95.74 | 554 | 94.94 | 95.40 | −72 | 92.92 |
| 500 | 5 | 334 | 377 | 95.28 | 95.66 | 346 | 95.28 | 95.48 | −68 | 93.02 |
| 1000 | 2 | 238 | 259 | 94.82 | 94.98 | 243 | 94.76 | 94.90 | −72 | 92.82 |
| , | ||||||||||
| 100 | −10 | 706 | 874 | 95.18 | 96.24 | 750 | 95.36 | 96.64 | −172 | 94.32 |
| 200 | −6 | 510 | 588 | 95.20 | 95.78 | 525 | 95.14 | 95.70 | −176 | 92.34 |
| 500 | 2 | 315 | 352 | 94.98 | 95.12 | 326 | 94.50 | 94.70 | −172 | 90.64 |
| 1000 | 2 | 226 | 242 | 95.04 | 95.12 | 229 | 94.84 | 94.92 | −173 | 85.90 |
BT-based, bootstrap-based; JP-SP, semiparametric method using the normal linear model of Janes and Pepe (2009); Bias, ; SD, standard deviation (×104); SE, mean standard error (×104); Cov (%) and LCov (%), coverage rates of 95% confidence interval and logit transformation-based confidence interval.
As discussed in Section 3.2, the estimator may not respect the monotonicity of , leading to illogical results. We implement the two monotonization methods described in Section 3.2 and evaluate their performance. Table 2 reports the bias and coverage rate related with these two methods where and correspond to the estimators after adopting regression- and ROC-based monotonization methods, respectively. The confidence intervals of the monotonized estimators are constructed with the sample-based standard error estimates. We find both regression- and ROC-based methods show small bias and good coverage rate under different sample sizes and sensitivity levels. ROC-based approach generally results in better coverage probability than regression-based method. The coverage probability of ROC-based method is comparable or even better than that without applying monotonicity-restoration method (results presented in Table 1). Lastly, applying monotonicity-restoration method may lead to smaller variance than original estimator, as shown for the ROC-based method and largely so for the regression-based method. This observation is consistent with the finding in Huang (2017).
TABLE 2.
Comparison of two monotonization methods in the simulation study
|
|
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Bias | SD | Cov | LCov | Bias | SD | Cov | LCov | |
| , | ||||||||
| 100 | 419 | 973 | 88.82 | 88.64 | 276 | 776 | 94.68 | 94.58 |
| 200 | 84 | 616 | 92.96 | 93.48 | 154 | 562 | 94.76 | 94.94 |
| 500 | 27 | 363 | 94.52 | 95.24 | 56 | 363 | 94.58 | 94.70 |
| 1000 | 26 | 259 | 94.94 | 95.18 | 34 | 260 | 95.02 | 95.10 |
| , | ||||||||
| 100 | 52 | 810 | 94.44 | 95.44 | 131 | 768 | 95.00 | 95.96 |
| 200 | 24 | 556 | 94.90 | 95.60 | 69 | 551 | 95.30 | 95.82 |
| 500 | 20 | 349 | 95.22 | 95.34 | 27 | 350 | 95.02 | 95.18 |
| 1000 | 16 | 246 | 95.14 | 95.28 | 18 | 248 | 95.08 | 95.12 |
| , | ||||||||
| 100 | 6 | 732 | 95.66 | 96.46 | 66 | 724 | 95.86 | 96.72 |
| 200 | 24 | 525 | 95.22 | 95.78 | 43 | 526 | 95.46 | 96.00 |
| 500 | 14 | 328 | 95.32 | 95.56 | 16 | 329 | 95.24 | 95.54 |
| 1000 | 10 | 231 | 95.44 | 95.52 | 10 | 232 | 95.20 | 95.34 |
| , | ||||||||
| 100 | −14 | 685 | 95.92 | 96.68 | 22 | 685 | 96.08 | 96.96 |
| 200 | 13 | 495 | 95.96 | 96.40 | 21 | 499 | 95.74 | 96.16 |
| 500 | 5 | 311 | 95.82 | 95.98 | 5 | 312 | 95.80 | 95.94 |
| 1000 | 7 | 218 | 95.64 | 95.74 | 7 | 218 | 95.82 | 95.90 |
, the estimator with regression-based monotonization; , the estimator with ROC-based monotonization; Bias, ; SD, standard deviation ×104; Cov (%) and LCov (%), coverage rates of 95% confidence interval and logit transformation-based confidence interval.
We also consider the case with discrete covariates only. In this case, our proposed estimator coincides the nonparametric estimator in Janes and Pepe (2009) as indicated in the Introduction. The focus is on comparing the performance of our inference methods to Janes and Pepe (2009), under their simulation setup. The details of this simulation study are presented in Web Appendix Section S1 and the results in Web Tables S1 and S2. Our sample-based variance estimation shows comparable coverage rate as our bootstrap-based inference, whereas their kernel density-based inference has worse performance than the bootstrap-based approach. Thus, our sample-based method outperforms the kernel density-based variance estimation in Janes and Pepe (2009). Such advantage is especially obvious when the controlled specificity is large ( and 0.90).
5 |. ILLUSTRATION WITH A CLINICAL STUDY
Data from a clinical study for aggressive prostate cancer (Sanda et al., 2017) are used for illustration. This was a prospective, multicenter cohort of male participants for first-time prostate biopsy without preexisting prostate cancer. After excluding 14 subjects with missing values, the data consist of 150 subjects with aggressive (Gleason score ≥ 7) prostate cancer, per biopsy, and 352 controls. The biomarker under consideration herein is PSA. Figure 1a shows the density of PSA from cases and controls.
FIGURE 1.
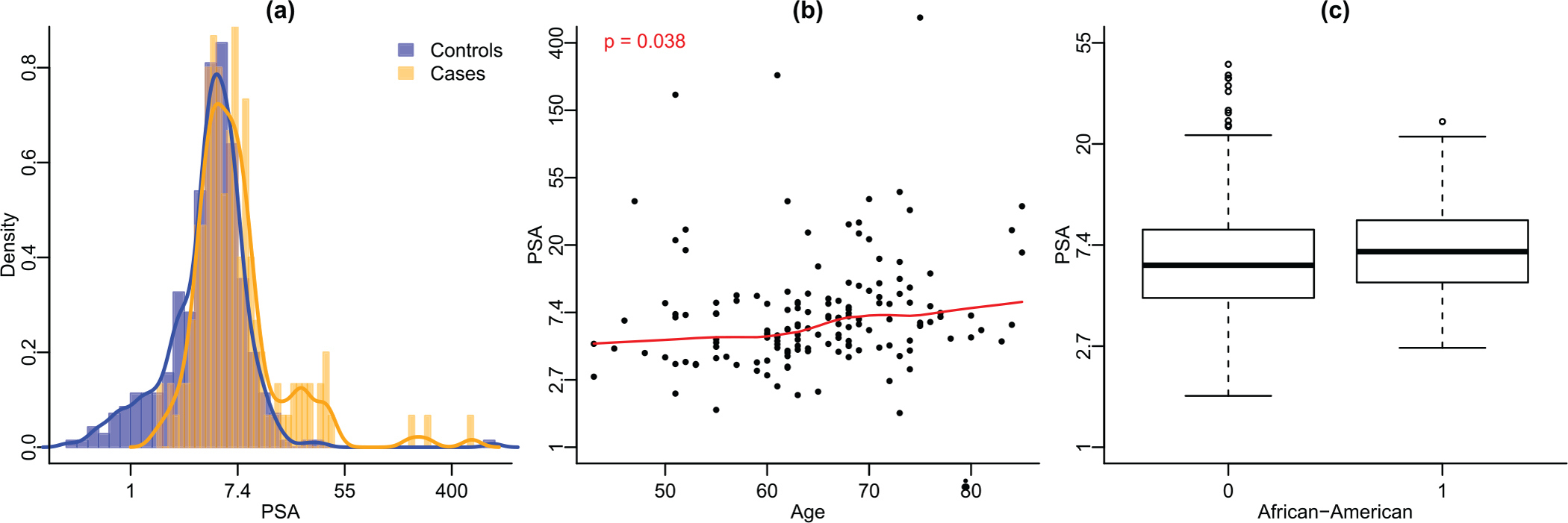
Exploratory plots for the clinical study. Panel (a): Histograms of PSA for cases and controls with density curves overlaid. Panel (b): Scatterplot of PSA versus age, cases only. The red solid line is fitted by loess and the p-value is obtained from testing zero Pearson’s correlation coefficient. Panel (c): Boxplot of PSA of African American population and non-African American group, cases only. This figure appears in color in the electronic version of this article, and any mention of color refers to that version
As mentioned in the Introduction, elder men tend to have higher PSA values than younger men (Oesterling et al., 1993; Lilja et al., 2008). Among the cases of our study, we observe significant elevation in PSA with the increase of age (Figure 1b). In addition to age, African-American men were also reported to have higher PSA than white men (Henderson et al., 1997). Figure 1c shows a small increase in PSA among African American cases compared to non-African American cases, although the increase is not statistically significant. In the following analysis, age and being African-American (AA) are included as covariates. For model checking, we have considered additional terms of squared age and the interaction of age and AA. They are not significant at the sensitivity levels of interest, , and 0.95, and thus not included in the model.
The ROC curves with and without adjusting for covariates are shown in Figure 2a (black and blue curves; this figure appears in color in the electronic version of this article, and any mention of color refers to that version). The adjusted ROC curves after regression- and ROC-based monotonization are also presented (red and purple curves). Adjusting for covariates leads to different ROC curve compared to the one without covariate adjustment. Covariate-adjusted specificity is higher than no-adjustment when sensitivity is between 0.7 and 0.9, and lower when that is between 0.2 and 0.7. Imposing the monotonicity does not make much difference. The exact specificity estimations for all the methods at controlled sensitivity levels 95%, 90%, 85%, and 80% are reported in Table 3. Consistent with the observations in Figure 2a, the covariate-adjusted specificity is lower than no-adjusted specificity for but higher for , and 80%.
FIGURE 2.
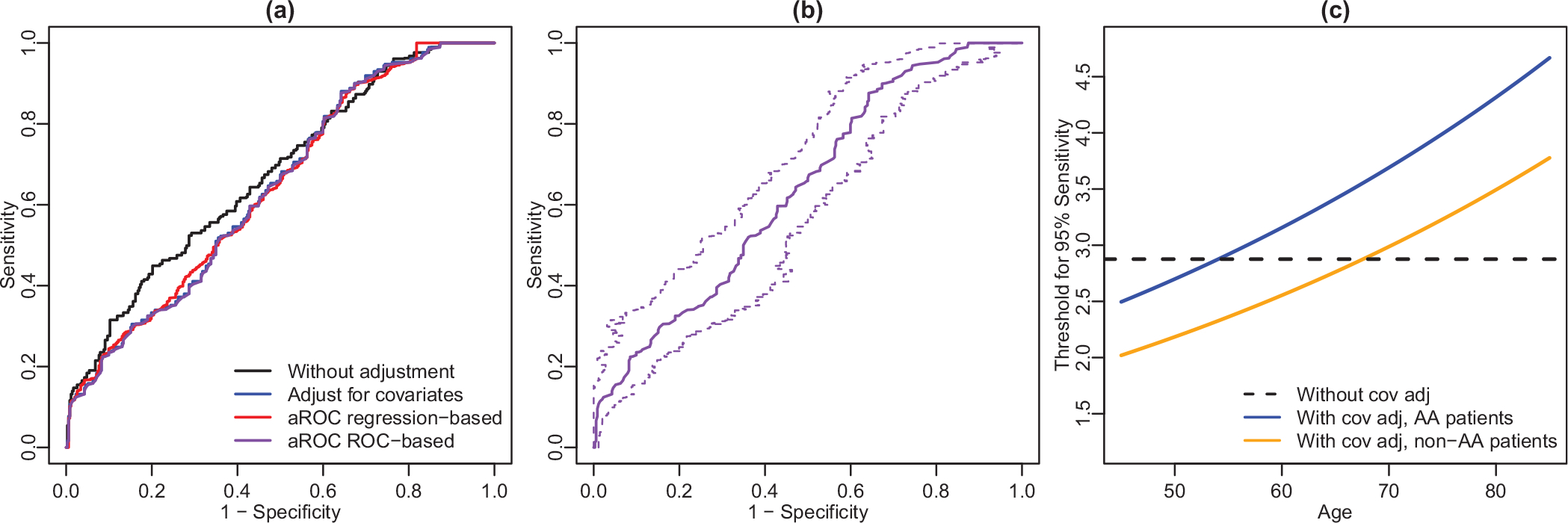
ROC curve and threshold results for the prostate study. Panel (a): ROC curve without and with adjustment for covariates (black and blue curve, respectively). Red and purple curves are adjusted ROC curves after applying regression- and ROC-based monotonization. Panel (b): Covariate-adjusted ROC curve with ROC-based monotonization is in solid purple. Dashed purple lines are the 95% confidence band. Panel (c): Estimated PSA threshold at controlled 95% sensitivity level by age based on the local model. This figure appears in color in the electronic version of this article, and any mention of color refers to that version
TABLE 3.
Estimated specificity along with 95% confidence interval at controlled sensitivity level in the aggressive prostate cancer application
| 0.95 | 0.239 | 0.196 (0.108, 0.330) | 0.208 (0.118, 0.340) | 0.218 (0.127, 0.348) |
| 0.90 | 0.284 | 0.318 (0.225, 0.429) | 0.316 (0.223, 0.427) | 0.321 (0.228, 0.431) |
| 0.85 | 0.332 | 0.361 (0.297, 0.430) | 0.358 (0.294, 0.428) | 0.361 (0.297, 0.430) |
| 0.80 | 0.392 | 0.398 (0.320, 0.481) | 0.398 (0.320, 0.480) | 0.400 (0.323, 0.483) |
, estimated specificity without covariate adjustment; , estimated specificity adjusting for age and AA without applying monotonicity restoration; and , covariate-adjusted specificity after applying regression- and ROC-based monotonization methods, respectively. The confidence interval is logit transformation-based with sample-based standard error
In contrast to pointwise confidence intervals, we also construct 95% confidence bands for the covariate-adjusted ROC curves. As the confidence bands are similar for the ROC curves with or without monotonicity restorations, we only present the confidence band for the ROC curve after applying ROC-based monotonization in Figure 2b. The confidence band works well in providing inference for the whole ROC curve.
Lastly, we present the estimated covariate-adjusted thresholds for PSA at controlled sensitivity level 95%in Figure 2c. Such thresholds could be useful for physicians to identify subjects with aggressive prostate cancer. For example, given a non-African American patient of age 50, an estimated PSA threshold of 2.2 may be used for controlled sensitivity level of 0.95. The design of our method ensures that the sensitivity is equally controlled among covariate-specific subpopulations. The covariate-adjusted threshold increases with age and is higher in African Americans, which aligns with existing understanding of these covariates.
Our implementation has excellent computational performance. With this prostate cancer data, which contain 150 diseased and 352 nondiseased samples, computing a covariate-adjusted ROC curve takes less than 1 s on a laptop computer with 4GB RAM and Intel Core i5 CPU. Computing the confidence band takes less than 3 s.
6 |. DISCUSSION
Our contributions are fourfold. First, we provide a covariate adjustment approach for a clinical utility-sensible performance metric, specificity at controlled sensitivity or vice versa, with minimal modeling assumptions. Compared with existing methods that assume uniform covariate effects across the ROC curve, our method only models the covariate effect specific to the corresponding threshold and thus is less prone to model misspecification. Second, this method extends to covariate adjustment for the whole ROC curve, where the issues of computation and monotonicity have been addressed. Third, we develop a sample-based variance estimation using a numerical differentiation with data-adaptive bandwidth. The proposed samples-based inference demonstrates comparable performance as bootstrap-based inference but is more computationally efficient. Lastly, we adopt the monotonization restoration methods of Huang (2017) and develop two strategies for restoring monotonicity in the covariate-adjusted ROC curves. These strategies are generally applicable in ROC estimation.
It is worthwhile to point out that our methods focus on the overall specificity with covariate-adjusted threshold at a controlled sensitivity level. This is different from covariate-specific performance evaluation as for subpopulations, as considered by Toledano and Gatsonis (1995), Pepe (1997), Pepe (1998), Pepe (2000), Cai and Pepe (2002), and Cai and Moskowitz (2004). Our notion of covariate adjustment is similar to that in Janes and Pepe (2009). As pointed by one of the reviewers, by analogy to Janes and Pepe (2009), the pooled specificity at the controlled sensitivity can be formulated as . denotes the covariate-specific specificity at sensitivity and is defined as , where is the distribution function of .
Our model may be extended to accommodate nonlinear covariate effects. Koenker (2005, 14, Chapter 6.6 and 7) discussed nonparametric quantile regression methods, including kernel-based approximation, additive models, penalized splines, and penalized triogram. In addition, a few recent works have also developed strategies to solve quantile regression with splines (Yoshida, 2013; Andriyana et al., 2014; Lian et al., 2015). They may be incorporated in our proposed method.
Supplementary Material
• S1 Simulation study with discrete covariates
• Table S1 Results under the simulation setting with discrete covariate values. Sensitivity under controlled specificity is estimated and presented.
• Table S1 Results under the simulation setting with discrete covariate values. Specificity under controlled sensitivity is estimated and presented.
• S2 Proof of Theorems
• S3 Details about two monotonization methods
• Figure S1 Summarization of computational time using two inference methods at different sample size selections.
ACKNOWLEDGMENT
We thank the editor, associate editor, and two anonymous reviewers who provided constructive comments to improve the manuscript. This project was partly supported by the National Institutes of Health grants R01CA230268 and U01CA113913.
Funding information
National Cancer Institute, Grant/Award Numbers: R01 CA230268, U01 CA113913
Footnotes
SUPPORTING INFORMATION
Web Appendix A with Proofs, Tables, and Figures referenced in Section 2, 3, and 4 are available with this paper at the Biometrics website on Wiley Online Library. The proposed methods are implemented and available both on Wiley Online Library and as a user-friendly R/CRAN package caROC (https://cran.r-project.org/web/packages/caROC/index.html).
DATA AVAILABILITY STATEMENT
Data subject to third-party restrictions. Restrictions apply to the availability of these data, which were used under permission for this paper. The data that support the findings of this paper are available from Dr. Martin G. Sanda (martinsanda@emory.edu) with the permission of the Emory Prostate Cancer study. The proposed methods are implemented and available as a user-friendly R/CRAN package caROC (https://cran.r-project.org/web/packages/caROC/index.html).
REFERENCES
- Andriyana Y, Gijbels I & Verhasselt A (2014) P-splines quantile regression estimation in varying coefficient models. Test, 23, 153–194. [Google Scholar]
- Cai T & Moskowitz CS (2004) Semi-parametric estimation of the binormal roc curve for a continuous diagnostic test. Biostatistics, 5, 573–586. [DOI] [PubMed] [Google Scholar]
- Cai T & Pepe MS (2002) Semiparametric receiver operating characteristic analysis to evaluate biomarkers for disease. Journal of the American statistical Association, 97, 1099–1107. [Google Scholar]
- de Carvalho VI, Jara A, Hanson TE, de Carvalho M et al. (2013) Bayesian nonparametric roc regression modeling. Bayesian Analysis, 8, 623–646. [Google Scholar]
- de Carvalho VI & Rodriguez-Alvarez MX (2018) Bayesian nonparametric inference for the covariate-adjusted roc curve. arXiv preprint arXiv:1806.00473. [Google Scholar]
- Hanley JA & McNeil BJ (1982) The meaning and use of the area under a receiver operating characteristic (roc) curve. Radiology, 143, 29–36. [DOI] [PubMed] [Google Scholar]
- He X (1997) Quantile curves without crossing. The American Statistician, 51, 186–192. [Google Scholar]
- Heagerty PJ & Pepe MS (1999) Semiparametric estimation of regression quantiles with application to standardizing weight for height and age in us children. Journal of the Royal Statistical Society: Series C (Applied Statistics), 48, 533–551. [Google Scholar]
- Henderson RJ, Eastham JA, Daniel J C, Whatley T, Mata J, Venable D, Kattan MW & Sartor O (1997) Prostate-specific antigen (psa) and psa density: racial differences in men without prostate cancer. Journal of the National Cancer Institute, 89, 134–138. [DOI] [PubMed] [Google Scholar]
- Huang Y (2002) Calibration regression of censored lifetime medical cost. Journal of the American Statistical Association, 97, 318–327. [Google Scholar]
- Huang Y (2010) Quantile calculus and censored regression. Annals of statistics, 38, 1607–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y (2017) Restoration of monotonicity respecting in dynamic regression. Journal of the American Statistical Association, 112, 613–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janes H & Pepe MS (2009) Adjusting for covariate effects on classification accuracy using the covariate-adjusted receiver operating characteristic curve. Biometrika, 96, 371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenker R (2005) Quantile regression (Econometric Society Monographs). Cambridge: Cambridge University Press. [Google Scholar]
- Koenker R & Bassett G (1978) Regression quantiles. Econometrica: journal of the Econometric Society, 1, 33–50. [Google Scholar]
- Lian H, Meng J & Fan Z (2015) Simultaneous estimation of linear conditional quantiles with penalized splines. Journal of Multivariate Analysis, 141, 1–21. [Google Scholar]
- Lilja H, Ulmert D & Vickers AJ (2008) Prostate-specific antigen and prostate cancer: prediction, detection and monitoring. Nature Reviews Cancer, 8, 268–278. [DOI] [PubMed] [Google Scholar]
- Oesterling JE, Cooner WH, Jacobsen SJ, Guess HA & Lieber MM (1993) Influence of patient age on the serum psa concentration. An important clinical observation. The Urologic clinics of North America, 20, 671–680. [PubMed] [Google Scholar]
- Partin AW, Catalona WJ, Southwick PC, Subong EN, Gasior GH & Chan DW (1996) Analysis of percent free prostate-specific antigen (psa) for prostate cancer detection: influence of total psa, prostate volume, and age. Urology, 48, 55–61. [DOI] [PubMed] [Google Scholar]
- Pepe MS (1997) A regression modelling framework for receiver operating characteristic curves in medical diagnostic testing. Biometrika, 84, 595–608. [Google Scholar]
- Pepe MS (1998) Three approaches to regression analysis of receiver operating characteristic curves for continuous test results. Biometrics, 54, 124–135. [PubMed] [Google Scholar]
- Pepe MS (2000) An interpretation for the roc curve and inference using glm procedures. Biometrics, 56, 352–359. [DOI] [PubMed] [Google Scholar]
- Pepe MS (2003) The statistical evaluation of medical tests for classification and prediction. Oxford: Oxford University Press. [Google Scholar]
- Platt RW, Hanley JA & Yang H (2000) Bootstrap confidence intervals for the sensitivity of a quantitative diagnostic test. Statistics in Medicine, 19, 313–322. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Álvarez MX & Inacio V (2020) Rocnreg: an r package for receiver operating characteristic curve inference with and without covariate information. arXiv preprint arXiv:2003.13111. [Google Scholar]
- Sanda MG, Feng Z, Howard DH, Tomlins SA, Sokoll LJ, Chan DW, Regan MM, Groskopf J, Chipman J, Patil DH et al. (2017) Association between combined tmprss2: Erg and pca3 rna urinary testing and detection of aggressive prostate cancer. JAMA Oncology, 3, 1085–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledano A & Gatsonis C (1995) Regression analysis of correlated receiver operating characteristic data. Academic Radiology, 2, S30. [PubMed] [Google Scholar]
- Tosteson ANA & Begg CB (1988) A general regression methodology for roc curve estimation. Medical Decision Making, 8, 204–215. [DOI] [PubMed] [Google Scholar]
- Yoshida T (2013) Asymptotics for penalized spline estimators in quantile regression. Communications in Statistics-Theory and Methods. 10.1080/03610926.2013.765477 [DOI] [Google Scholar]
- Zhou X-H & Qin G (2005) Improved confidence intervals for the sensitivity at a fixed level of specificity of a continuous-scale diagnostic test. Statistics in Medicine, 24, 465–477. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
• S1 Simulation study with discrete covariates
• Table S1 Results under the simulation setting with discrete covariate values. Sensitivity under controlled specificity is estimated and presented.
• Table S1 Results under the simulation setting with discrete covariate values. Specificity under controlled sensitivity is estimated and presented.
• S2 Proof of Theorems
• S3 Details about two monotonization methods
• Figure S1 Summarization of computational time using two inference methods at different sample size selections.
Data Availability Statement
Data subject to third-party restrictions. Restrictions apply to the availability of these data, which were used under permission for this paper. The data that support the findings of this paper are available from Dr. Martin G. Sanda (martinsanda@emory.edu) with the permission of the Emory Prostate Cancer study. The proposed methods are implemented and available as a user-friendly R/CRAN package caROC (https://cran.r-project.org/web/packages/caROC/index.html).


